EWSR1::WT1 is essential for desmoplastic small round cell tumors and upregulates the cyclin D–CDK4/6–RB axis that can be targeted with palbociclib, providing a targeted therapeutic strategy for treating this deadly tumor type.
Abstract
Desmoplastic small round cell tumors (DSRCT) are a type of aggressive, pediatric sarcoma characterized by the EWSR1::WT1 fusion oncogene. Targeted therapies for DSRCT have not been developed, and standard multimodal therapy is insufficient, leading to a 5-year survival rate of only 15% to 25%. Here, we depleted EWSR1::WT1 in DSRCT and established its essentiality in vivo. Transcriptomic analysis revealed that EWSR1::WT1 induces unique transcriptional alterations compared with WT1 and other fusion oncoproteins and that EWSR1::WT1 binding directly mediates gene upregulation. The E-KTS isoform of EWSR1::WT1 played a dominant role in transcription, and it bound to the CCND1 promoter and stimulated DSRCT growth through the cyclin D–CDK4/6–RB axis. Treatment with the CDK4/6 inhibitor palbociclib successfully reduced growth in two DSRCT xenograft models. As palbociclib has been approved by the FDA for the treatment of breast cancer, these findings demonstrate the sensitivity of DSRCT to palbociclib and support immediate clinical investigation of palbociclib for treating this aggressive pediatric cancer.
Significance:
EWSR1::WT1 is essential for desmoplastic small round cell tumors and upregulates the cyclin D–CDK4/6–RB axis that can be targeted with palbociclib, providing a targeted therapeutic strategy for treating this deadly tumor type.
Introduction
Desmoplastic small round cell tumor (DSRCT) is an aggressive pediatric cancer characterized by the t(11;22)(p13;q12) chromosomal translocation, which leads to the establishment of the EWSR1::WT1 fusion oncogene (1). DSRCT is most commonly found in the abdominal and pelvic cavities with a high rate of metastasis at diagnosis (2). No targeted therapies have been developed and standard treatment is multimodal therapy including chemotherapy, radiotherapy, and surgery (2, 3). Most patients become refractory to treatment and prognosis is extremely poor with a 5-year survival rate of less than 30% (2–5), highlighting the urgent need to develop novel therapeutics.
The EWSR1::WT1 oncoprotein is a transcription factor consisting of the N-terminal low complexity domain (LCD) of EWSR1 and zinc fingers 2 to 4 of WT1 (1). Native EWSR1 is a ubiquitously expressed RNA binding protein associated with a myriad of functions including B lymphocyte and brown fat development (6, 7). Native WT1 is a transcription factor that functions as a tumor suppressor in Wilms tumor while counterintuitively displaying oncogenic functions in leukemia and breast cancer (8). In common with other oncogenic transcription factors, drugging EWSR1::WT1 has been elusive. With the exception of the EWSR1::WT1 translocation, DNA sequencing including whole-genome sequencing, whole-exome sequencing, and MSK-IMPACT analysis have failed to identify recurrent mutations (9, 10), posing a challenge to developing targeted therapies. An alternative strategy is to identify critical downstream effectors of the EWSR1::WT1 oncoprotein amenable to inhibition. For this strategy to succeed, (i) the EWSR1::WT1 oncoprotein must be essential to DSRCT growth and (ii) a comprehensive list of EWSR1::WT1 downstream targets must be established. Depletion of EWSR1::WT1 has been shown to reduce DSRCT proliferation in two- and three-dimension cultures in vitro (11–13). However, the essentiality of the oncogene has yet to be tested in vivo. Two studies have coupled EWSR1::WT1 knockdown with RNA sequencing (RNA-seq) to identify downstream targets of EWSR1::WT1. Surprisingly, these studies identified only 7 commonly upregulated and 11 commonly downregulated genes across three DSRCT cell lines (12, 14). Further work is critical to definitively establish the role of EWSR1::WT1 in vivo and identify sets of EWSR1::WT1 targets commonly regulated across tumors, which could serve as therapeutic candidates.
In addition, the underlying mechanisms by which EWSR1::WT1 regulates transcription and causes oncogenesis remain underinvestigated. Chromatin immunoprecipitation and sequencing (ChIP-seq) performed on the fusion protein in the JN-DSRCT-1 cell line identified enrichment of the WT1 motif at EWSR1::WT1 binding sites and demonstrated binding of EWSR1::WT1 to several genes previously shown to be regulated by the fusion (FGFR4, IGF2; ref. 15). However, a comprehensive analysis integrating EWSR1::WT1 binding and transcriptional activity has not been performed. Similar to native WT1, the WT1 domain of EWSR1::WT1 has two isoforms created by alternative splicing that includes or excludes three amino acids (Lys, Thr, and Ser, termed KTS) between WT1 zinc fingers 3 and 4 (16). In native WT1, these isoforms have different DNA binding specificity and transcriptional activity (17). In EWSR1::WT1, the resulting isoforms (herein designated E-KTS and E+KTS) lead to differences in gene expression in mouse cells but have not been investigated in human cells (11). Further, E-KTS and E+KTS isoform binding remains uninvestigated and the importance of each isoform in oncogenesis requires further evaluation.
In this work, we establish a set of four DSRCT cell lines that deplete EWSR1::WT1 in a doxycycline (dox)-inducible manner. Utilizing this novel toolkit, we demonstrate the essentiality of EWSR1::WT1 both in vitro and in vivo and perform the most comprehensive analysis of EWSR1::WT1 downstream targets. By screening druggable downstream targets, we identify palbociclib as a novel therapeutic able to reduce DSRCT growth both in vitro and in vivo by preventing retinoblastoma phosphorylation and the transition from G1 to S phase. Our novel results combined with palbociclib's previous approval by the FDA for the treatment of breast cancer establish palbociclib as an exciting DSRCT therapy, which warrants urgent clinical investigation. In this work, we further discover novel mechanistic insights into EWSR1::WT1 functionality including the uniqueness of its transcriptional alterations as compared with WT1 and other fusion oncoproteins, the direct role of EWSR1::WT1 binding in gene upregulation but not downregulation, and the dominant role of the E-KTS isoform in transcription.
Materials and Methods
Cell lines and culture conditions
DSRCT cell lines (JN-DSRCT-1, BER-DSRCT, BOD-DSRCT, and SK-DSRCT2) have been validated to contain the defining EWSR1::WT1 fusion and described previously (18–20). JN-DSRCT-1 was obtained from Jun Nishio. BER-DSRCT, BOD-DSRCT, and SK-DSRCT2 were obtained from Marc Ladanyi. Cell lines were tested for Mycoplasma by PCR and were negative as of March 2022. All cell lines were used for less than 10 passages after thawing. DSRCT cells were cultured in DMEM/F12 media supplemented with 10% FBS (Gibco), 2 mmol/L K-glutamine, 100 U/mL penicillin, and 100 μg/mL streptomycin (Thermo Fisher Scientific). The LP9 cell line is an untransformed, diploid, mesothelial cell line and was obtained from the NIGMS Human Genetic Cell Repository at the Coriell Institute (AG07086; Camden). LP9 cells were grown in DMEM/F12 media supplemented with 15% FBS, 2 mmol/L l-glutamine, 100 U/mL penicillin, 100 μg/mL streptomycin, 10 ng/mL EGF (Thermo Fisher Scientific), and 0.4 ug/mL hydrocortisone (Sigma-Aldrich).
Light and fluorescent microscopy
Light and fluorescent microscopy was performed with Nikon Eclipse 80i microscope using NIS-Elements software for image capture.
RNA isolation and real-time qPCR analysis
Total RNAs were isolated with RNA-STAT60 (Tel-Test). iScript cDNA Synthesis Kit (Bio-Rad) was used to reverse transcribe 500 ng of RNA to generate cDNA. Relative transcript levels were analyzed by real-time qPCR using SYBR Green (SsoAdvanced Universal SYBR Green Supermix; Bio-Rad) and calculated by the comparative Ct method normalized against human ACTB (β-actin). Primers are listed in Supplementary Table S1.
Protein isolation and Western blot analysis
Cell lysates were prepared in RIPA lysis buffer supplemented with complete EDTA-Free Protease Inhibitor Cocktail (Sigma-Aldrich) and phosphatase inhibitors: 1 mmol/L NaF and 2 mmol/L Na3VO4. Proteins were resolved in 10% to 15% SDS-PAGE gels and transferred onto a 0.45 μm nitrocellulose membrane (Bio-Rad). Membranes were blocked with 5% nonfat milk or 5% bovine serum albumin and incubated with primary antibodies at 4°C overnight, followed by room temperature incubation of secondary antibodies LI-COR IRDye 800CW goat anti-mouse (#926–32210, 1:15,000 dilution) or LI-COR IRDye 680RD goat anti-Rabbit (#926- 68071, 1:15,000 dilution). Images were taken on LI-COR Odyssey CLx. At least two independent immunoblots were performed for each experiment, with a representative immunoblot shown. Relative protein expression was quantified with ImageJ. Antibodies are listed in Supplementary Table S2.
Generation of dox-inducible shRNA and EWSR1::WT1 overexpression cell lines
Dox-inducible LT3-GEPIR vector (21) was used to generate stable cell lines as described previously (8). Annealed oligonucleotides for each shRNA were inserted into XhoI and EcoRI sites of the LT3-GEPIR vector. Supplementary Table S3 lists each shRNA sequence utilized. EWSR1::WT1 constructs in a pCDH-lentivirus backbone have been established and described (previously 22). Supplementary Material S1 is a map of the final shWT1 LT3-GEPIR vector. Lentivirus was generated by cotransfecting HEK293T cells with the LT3-GEPIR-shRNA lentiviral vector and ViraPower lentiviral packaging mix (Invitrogen) using Lipofectamine 3000 (Thermo Fisher Scientific). Viral supernatants were collected 48, 72, and 96 hours posttransfection, and concentrated with LentiX-Concentrator (Takara Bio). DSRCT and LP9 cells were transduced with virus in the presence of polybrene (10 μg/mL) for 16 hours with a multiplicity of infection less than 1. Cells were selected with puromycin (DSRCT: 0.5 μg/mL; LP9: 1 μg/mL) 48 hours posttransduction. Stable cell lines were validated by RT-qPCR and Western blot analyses with or without dox. In dox-inducible cell lines, treatment with 1 μg/mL of dox was used to induce shRNA expression.
Cell-cycle analysis
For dox-inducible shWT1 and shCCND1 knockdown experiments, DSRCT cells were treated for four days with or without dox treatment (n = 3). For palbociclib and abemaciclib experiments, drug treatment was performed for 2 days (n = 3). Cells were harvested with trypsin, washed with PBS, and fixed overnight with 70% ethanol. After fixation, cells were treated with RNase and stained with propidium iodide followed by flow cytometry.
Xenografts in immune-deficient mice
All animal procedures were approved by the Tulane Institutional Animal Care and Use Committee (Protocol, #1500). Male NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice (6 weeks) were purchased (RRID:IMSR_JAX:005557; Jackson Laboratory) and used for all xenograft studies. As described previously (13), DSRCT cells were counted and mixed in a 1:1 ratio of media to Matrigel (Corning). Cell mixture (200 μL) containing 1 × 106 cells was subcutaneously injected into the lower flanks of NSG mice. For dox-inducible shWT1 knockdown experiments, mice were randomized when tumors become palpable and placed with normal or dox-containing (0.2% dox/2% sucrose) drinking water (n = 5 mice per group). For palbociclib experiments, mice were randomized and treated five times per week with oral gavage of 75 mg/kg palbociclib (in water) or vehicle control (n = 4–5 mice per group). For all experiments, Tumor volume was measured weekly with calipers and calculated as: length × (width)2 × 0.5, where length is the largest diameter and width is perpendicular to the length. Mice were sacrificed when tumors became too large. Tumors were harvested and weighed. Tumor fragments were flash frozen for protein isolation and fixed in formalin for IHC analysis. Fixed tissues were embedded in paraffin, sectioned (5 μmol/L), stained with hematoxylin and eosin (H&E), and imaged (Nikon Eclipse 80i microscope; NIS-Elements software).
Cell viability assays
The CCK-8 assay (Sigma-Aldrich) was utilized to assess cell viability per the manufacturer's directions. Cells were seeded in 96-well plates at day 0 with 10,000 cells/well for DSRCT or 5,000 cells/well for LP9. After 24 hours of culture, media with either vehicle control or the appropriate drug concentration was added to cells. After 72 hours of treatment, CCK-8 assay was performed to assess viability. For all experiments, absorbance was measured using Clariostar microplate reader (BMG Labtech).
Colony formation assays
Cells were seeded at 10,000 cells per well in a 6-well plate. Media containing indicated treatment were changed every 2 days. After 14 days, plates were washed with PBS and stained with 0.5% crystal violet in 10% methanol for 20 minutes, followed by two washes with water and overnight drying. Stain was removed with 1 mL methanol and quantified by measuring absorbance at 570 nm using Clariostar microplate reader (BMG Labtech).
RNA-seq and analysis
RNAs were isolated from DSRCT shWT1 cells treated for 4 days with or without dox. RNAs were isolated from LP9 cells that were nontransduced or transduced with the following vectors: GFP only, E+KTS GFP, E-KTS mCherry, or both E+KTS GFP and E-KTS mCherry. RNAs were prepared with RNeasy Plus Kit (Qiagen). Sequencing libraries were created from 500 ng of total RNA using the Illumina TruSeq Stranded Total RNA Kit with Ribo-Zero following the manufacturer instruction. Library fragment size was verified using the Agilent 2100 Bioanalyzer (Agilent) and the concentrations were determined using Qubit (Thermo Fisher Scientific). Illumina Novaseq 6000 was utilized for 75 bp paired-end read sequencing. Gene counts were generated by alignment to human transcriptome GRCh38 using STAR in RSEM (23). Differential gene expression analysis was performed using DESeq2 (24). Overrepresentation analysis (ORA) and gene set enrichment analysis were performed using the clusterProfiler package in Bioconductor (25, 26). Publicly available RNA-seq data of EWSR1::WT1 knockdown in JN-DSRCT-1 and BER-DSRCT cell lines (GSE137561), WT1 knockdown in hematologic (17) and breast cancer cells (GSE153865, GSE93636; ref. 27), and knockdown of other fusion oncogenes in their respective fusion-driven pediatric cancers (GSE61953, GSE94277, GSE168815, GSE108028; refs. 28–31) were obtained from GEO.
ChIP-seq analysis
ChIP-seq FASTQ files of anti-WT1 in JN-DSRCT-1 (GSE156277) and anti-WT1 and anti-HA in Kasumi-1 cells (GSE153865) were obtained from GEO. Quality control was performed with FASTQC and deduplication was performed with bbmap-clumpify. Bowtie2 (32, 33) was used to align sequences to hg38 followed by narrow peak calling with MACS2 (34). HOMER was used for gene annotation and motif enrichment analysis (35). The Integrative Genomics Viewer (IGV) was used for visualization of peaks (36) and genomation was used for density heatmap creation (37).
ChIP-qPCR
ChIP assay was adapted and performed as described previously (22), using anti-WT1 (Thermo Fisher Scientific), anti-HA (Invitrogen), or rabbit IgG (Cell Signaling Technology) antibodies. Briefly, cells were crosslinked with formaldehyde, neutralized with glycine, washed twice, and lysed in Farnham Buffer. Chromatin was fragmented by sonication with Covaris M220 Focused-ultrasonicator. Samples were incubated with antibodies overnight at 4°C, pulled down with Protein G dynabeads (Invitrogen), washed, eluted, and purified with DNA spin columns (Epoch Life Science). qPCR was performed using SYBR Green (SsoAdvanced Universal SYBR Green Supermix; Bio-Rad) with ChIP-qPCR primers in Supplementary Table S4.
Statistical analysis
All experiments included two or more independent replicates. Two-way ANOVA was used to determine tumor volume differences and differences in dose response between DSRCT and LP9 cell lines. The Student t test was used as appropriate for other experiments using GraphPad Prism 7 program (GraphPad Software). A P value <0.05 was considered statistically significant.
Data availability
The data generated in this study are publicly available in Gene Expression Omnibus (GEO) at GSE252051. Publicly available data generated by others were used by the authors, including RNA-seq data from GEO (GSE137561, GSE153865, GSE93636, GSE61953, GSE94277, GSE168815, GSE108028), ChIP-seq data from GEO (GSE156277, GSE153865), and microarray data (38). All other raw data generated in this study are available upon request from the corresponding author.
Results
EWSR1::WT1 is essential for DSRCT growth
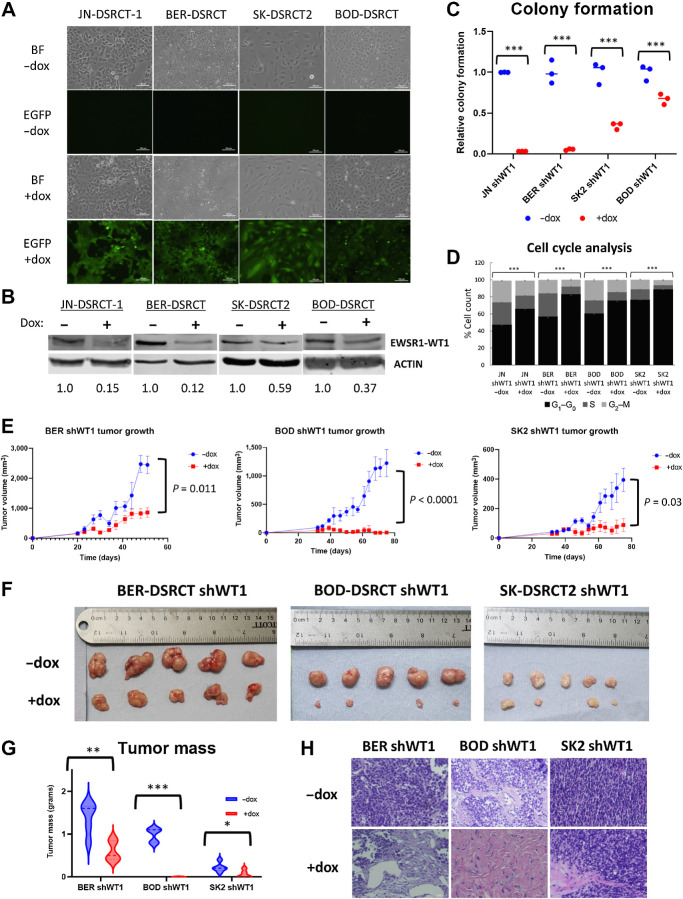
To investigate the role of EWSR1::WT1 in DSRCT, we established a tool kit of four DSRCT cell lines (JN-DSRCT-1, BER-DSRCT, BOD-DSRCT, SK-DSRCT2) with lentiviral constructs that enable dox-inducible depletion of EWSR1::WT1. Because native WT1 is not expressed in DSRCT, shRNA targeting the 3′UTR of WT1 (shWT1) is able to deplete the fusion gene without off-target effects on native WT1 (15). The establishment of stable shWT1 cell lines overcomes limitations induced by low transfection efficiency of DSRCT cells and enables evaluation of fusion gene function both in vitro and in vivo. The cell lines further express EGFP in a dox-inducible manner, enabling visual verification of system functionality (Fig. 1A). Dox-induction successfully depleted EWSR1::WT1 at the transcriptional and translational level in all four cell lines (Fig. 1B; Supplementary Fig. S1A). EWSR1::WT1 depletion completely abrogated colony formation in JN-DSRCT-1 and BER-DSRCT, and significantly reduced colony formation in SK-DSRCT2 and BOD-DSRCT (Fig. 1C). In contrast, dox-induction of scrambled shRNA did not alter EWSR1::WT1 RNA or protein expression or lead to reductions in colony formation (Supplementary Figs. S1B–S1D). EWSR1::WT1 depletion did not increase PARP cleavage, suggesting the observed colony formation reductions are not primarily due to apoptosis (Supplementary Fig. S1E). Instead, an increase in the percentage of cells in G0–G1 phase was observed (Fig. 1D).
Figure 1.
EWSR1::WT1 is essential for DSRCT growth. A, Brightfield and fluorescent images of dox-inducible shWT1 DSRCT cell lines treated with or without dox that selectively induces GFP and shRNA expression. B, Western blot analysis of EWSR1::WT1 and ACTIN protein expression in shWT1 cell lines treated with (+) or without (−) dox for 4 days (representative blot, n = 2). Average protein expression was quantified relative to ACTIN, normalized to the -dox condition, and is shown underneath each blot. C, Quantification of colony formation assays of shWT1 cell lines treated with (red) or without dox (blue; n = 3). D, Cell-cycle analysis of DSRCT shWT1 cells treated with or without dox for 4 days. E, Growth of DSRCT shWT1 tumors with or without dox treatment (n = 5). F, Images of final DSRCT shWT1 tumors treated with or without dox. G, Final tumor mass of DSRCT shWT1 tumors treated with (red) or without (blue) dox. H, Representative H&E staining of DSRCT shWT1 tumors treated with or without dox (n = 3). Scale bar, 50 μm. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
To test EWSR1::WT1 essentiality in vivo, DSRCT shWT1 cell lines were injected subcutaneously into immune-deficient NOD.SCID/IL2Rγ-null (NSG) mice (n = 10). When tumors were measurable (BER: 20 days; BOD and SK2: 32 days), randomized mice were either housed with dox-containing drinking water (n = 5) to induce EWSR1::WT1 depletion or standard water (n = 5) as control. No changes in mouse body weight were observed. Dox-treated mice showed significant reductions in tumor growth as measured by calipers twice weekly (Fig. 1E). Final tumors with dox treatment had reductions in size (Fig. 1F) and mass (Fig. 1G). The BOD-DSRCT shWT1 cell line, which showed the least pronounced reduction in colony-formation assays in vitro, unexpectedly showed the largest reductions in tumor size in vivo. Western blot analysis found reductions in fusion protein in dox-treated tumors, confirming system functionality in vivo (Supplementary Fig. S1F). H&E staining identified pockets of surviving tumor cells in dox-treated tumors derived from BER-DSRCT and SK-DSRCT2 (Fig. 1H), but dox-treated BOD-DSRCT tumors largely consisted of stroma lacking tumor cells. Kaplan–Maier analysis demonstrated significant improvement in survival for all dox-treated DSRCT tumors (with death defined as the time at which tumors reached a study endpoint; Supplementary Fig. S1G). Together, these findings demonstrate the essentiality of EWSR1::WT1 in DSRCT in vivo.
EWSR1::WT1 induces distinct transcriptional alterations
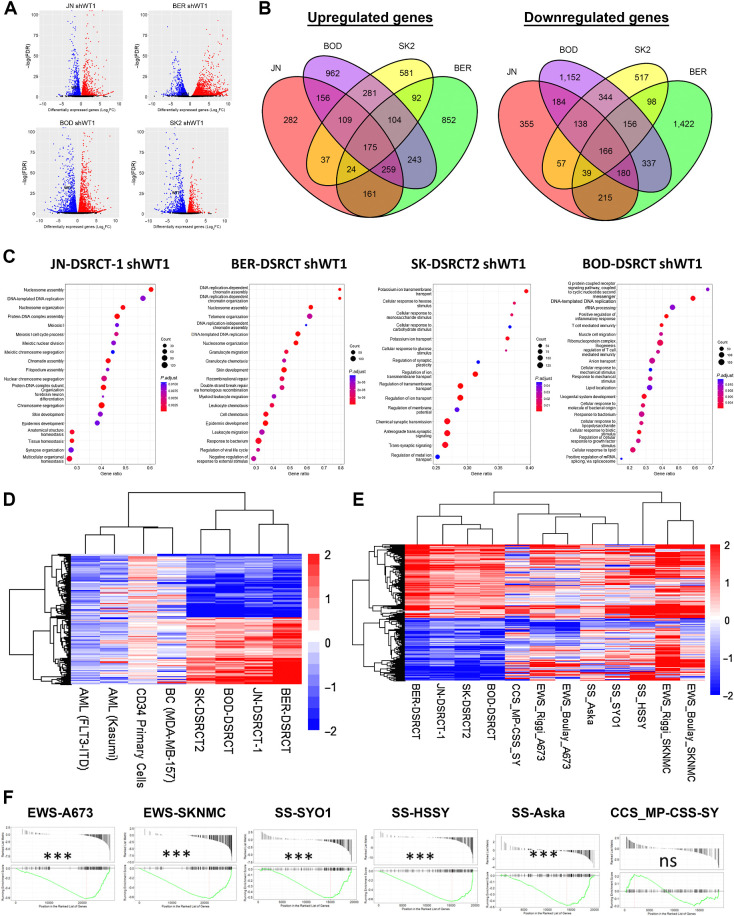
To identify downstream targets critical to EWSR1::WT1’s essentiality, RNA-seq was performed on the four DSRCT shWT1 cell lines treated with or without dox for 4 days. Differential gene expression analysis identified over 1,000 upregulated and downregulated genes in each cell line (Fig 2A, FDR < 0.05). Consistent with dox induction triggering EWSR1::WT1 depletion, WT1 expression was significantly reduced in response to dox addition in all four RNA-seq data sets (Supplementary Table S5). Comparison between cell lines demonstrated consistent transcriptomic alterations across cell lines, with a majority of EWSR1::WT1 upregulated genes in each cell line also found to be upregulated in at least one other cell line (Fig. 2B). 76.6% of EWSR1::WT1 upregulated genes in JN-DSRCT-1 were upregulated in another DSRCT cell line (BER-DSRCT: 55.3%; SK-DSRCT2: 58.6%; BOD-DSRCT: 58.0%). Although there was significant overlap in EWSR1::WT1 regulated genes between cell lines, most genes were not identified as significantly altered in all cell lines. Overall, we identified a set of 175 commonly upregulated genes and 166 commonly downregulated genes across all four DSRCT cell lines (Fig. 2B). These gene sets were consistent with RNA-seq datasets from Gedminas and colleagues performed after siRNA depletion of EWSR1::WT1 in JN-DSRCT-1 and BER-DSRCT (Supplementary Fig. S2A; ref. 12). Within each cell line there was high concordance between our results and those of Gedminas and colleagues (12). Greater than 80% of upregulated and downregulated genes identified in our differential gene expression analysis in JN-DSRCT-1 and BER-DSRCT were differentially expressed in the same direction in the Gedminas and colleagues data set (Supplementary Fig. S2B).
Figure 2.
EWSR1::WT1 induces distinct transcriptional alterations. A, Volcano plots of RNA-seq data from dox-inducible shWT1 DSRCT cell lines treated with or without dox (n = 2). Red, significantly upregulated; blue, significantly downregulated. P < 0.05. B, Venn diagrams comparing genes upregulated and downregulated by EWSR1::WT1 in JN-DSRCT-1 (red), BER-DSRCT (green), BOD-DSRCT (purple), and SK-DSRCT2 (yellow). C, Top 15 overrepresented pathways among EWSR1::WT1 upregulated genes in each DSRCT cell line. D, Heatmap of log2-fold change in gene expression caused by (i) knockdown of EWSR1::WT1 in DSRCT cell lines or (ii) knockdown of WT1 in leukemia (Kasumi, FLT3-IDT), primary CD34 cells, or breast cancer (MDA-MB-157). E, Heatmap of log2-fold change in gene expression caused by knockdown of fusion oncoproteins in DSRCT, EWS, SS, and CCS cell lines. F, GSEA of EWSR1::WT1 downregulated genes in EWS, SS, or CCS, with their respective fusion oncoproteins depleted. ***, P < 0.001; ns, nonsignficant.
ORA of our common upregulated and downregulated gene sets did not identify significantly overrepresented pathways. However, ORA on individual samples identified a variety of significantly enriched Gene Ontology Biological Processes (GO-BP) with notable heterogeneity between cell lines (Fig. 2C; Supplementary Data S1). Thirty-four pathways were positively enriched in at least three DSRCT cell lines. These pathways commonly upregulated by EWSR1::WT1 predominantly related to DNA replication (ex: cell-cycle DNA replication, DNA-templated DNA replication) and neurogenesis (ex: axonogenesis, neuron migration; Fig. 2C; Supplementary Data S1). Although there was significant heterogeneity between cell lines, JN-DSRCT-1 and BER-DSRCT shared 108 upregulated pathways including cell cycle G2–M phase transition, cell cycle G1–S phase transition, and telomere organization. ORA on downregulated genes was more concordant between cell lines with 104 pathways downregulated by EWSR1::WT1 in all four DSRCT cell lines (Supplementary Figs. S2C and S2D). Downregulated pathways predominantly related to motility (ex: taxis, wound healing), differentiation (ex: mesenchymal cell differentiation, endoderm development), and extracellular environment (ex: cell junction assembly, extracellular matrix organization; Supplementary Fig. S2C; Supplementary Data S2).
We further evaluated our EWSR1::WT1 regulated gene sets by examining the regulation of these genes by native WT1 and fusion oncoproteins from other pediatric cancers. Available RNA-seq data sets of WT1 depletion in two AML cell lines, primary CD34+ cells, and breast cancer all failed to recapitulate the EWSR1::WT1 signature (Fig. 2D). Although depletion of WT1 in the AML cell lines generally led to decreases in expression of both EWSR1::WT1 upregulated and downregulated genes, depletion of WT1 in primary CD34+ cells resulted in no change or a slight increase in expression of many of these genes. WT1 depletion in the breast cancer cell line MDA-MB-157 did not lead to a clear trend of expression change in EWSR1::WT1 regulated genes. Ewing Sarcoma (EWS), clear cell sarcoma (CCS), and synovial sarcoma (SS) are pediatric tumors driven by the expression of the fusion proteins EWSR1::FLI1, EWSR1::ATF1, and SSY::SSX, respectively. Utilizing RNA-seq data with knockdown of these fusion proteins in their respective tumor cell lines, we found that only a small portion of the EWSR1::WT1 upregulated gene set is recapitulated in these cell lines (Fig. 2E). Examining all commonly upregulated genes within each tumor type, we found no genes upregulated across all tumor types and only 13 genes that were upregulated by EWSR1::WT1 and two other fusion oncoproteins (Supplementary Fig. S2E). In contrast, a subset of EWSR1::WT1 downregulated genes were similarly downregulated by other fusion oncoproteins (Fig. 2E). Gene set enrichment analysis (GSEA) discovered significant downregulation of the EWSR1::WT1 downregulated gene set in Ewing sarcoma and synovial sarcoma cell lines, but not in the MP-CSS-SY clear cell sarcoma cell line (Fig. 2F).
Screen of druggable downstream targets identifies palbociclib as a DSRCT therapeutic
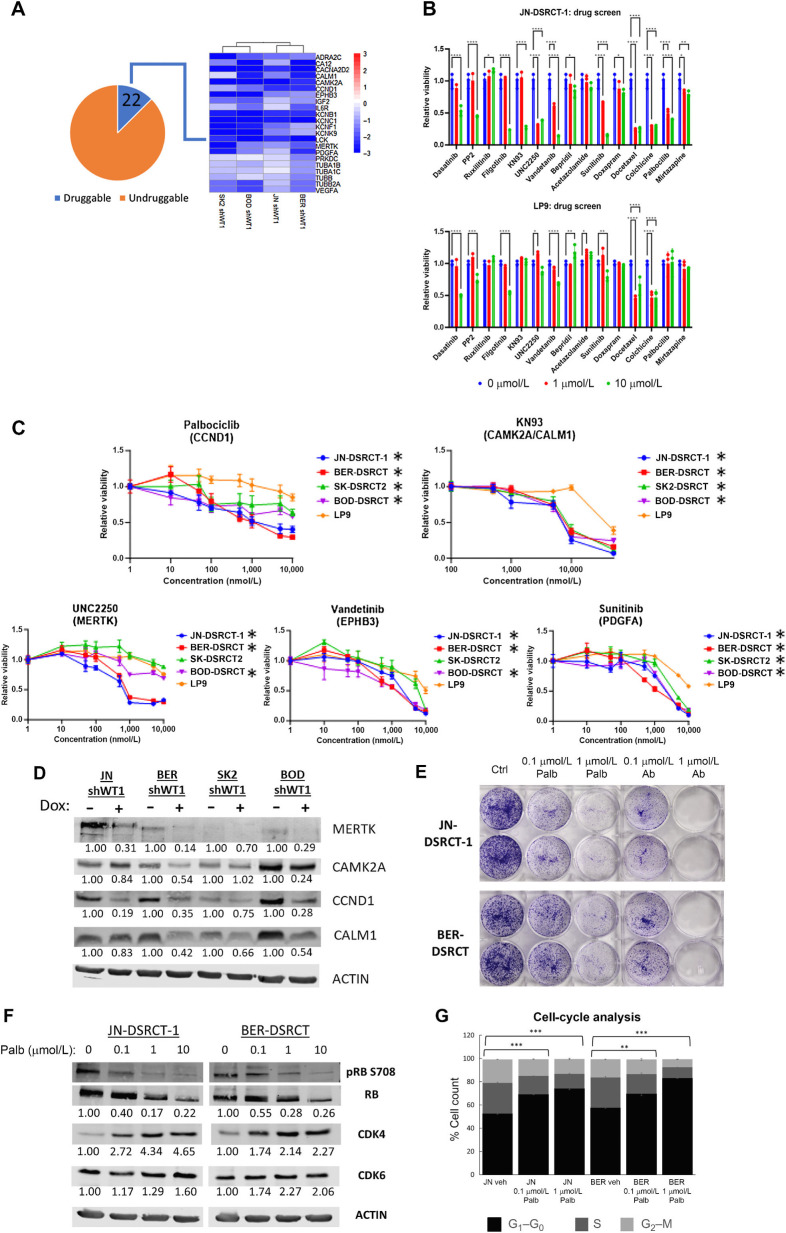
Utilizing our novel EWSR1::WT1 regulated gene sets, we sought to identify druggable dependencies that could serve as therapeutic targets. We hypothesized that genes commonly regulated across all four DSRCT cell lines are likely to be highly expressed in DSRCT tumors and critical to DSRCT proliferation. To enable faster translation to the clinic, we concentrated on targets with existing inhibitors by overlapping our list of 175 commonly upregulated EWSR1::WT1 targets with the drug–gene interaction database (39). We identified inhibitors for 22 of our 175 commonly upregulated targets (Fig. 3A) and performed a screen of these inhibitors on two DSRCT cell lines (JN-DSRCT-1, BER-DSRCT) with the normal mesothelial cell line LP9 serving as a control (Fig. 3B; Supplementary Fig. S3A). This screen identified five compounds (KN93, UNC2250, palbociclib, sunitinib, vandetinib), which caused death in DSRCT cells without substantially reducing viability in control cells at 72 hours with 1 and/or 10 μmol/L concentrations. Further examination of these compounds across four DSRCT cell lines and a broader range of doses identified KN93, UNC2250, and palbociclib as the most promising targets based on consistently lower IC50 values in DSRCT than the LP9 control cell line (Fig. 3C; Supplementary Table S6). All DSRCT cell lines responded to palbociclib at submicromolar doses (IC50s: 0.03–0.25 μmol/L) and all but SK-DSRCT2 responded to low concentrations of UNC2250 (IC50s: 0.29–0.78 μmol/L). KN93 showed higher IC50 concentrations (5.5–11.9 μmol/L) in DSRCT than the other two inhibitors, but still showed substantially greater sensitivity in DSRCT than in LP9 cells. KN93 inhibits two EWSR1::WT1 upregulated targets, CAMK2A and CALM1, whereas UNC2250 inhibits MERTK. Palbociclib is a CDK4/6i approved by the FDA for the treatment of breast cancer, and identified in our screen due to EWSR1::WT1 upregulation of CCND1, which activates CDK4 and CDK6 to phosphorylate retinoblastoma (RB) and enables cell-cycle progression for G1 to S phase (40).
Figure 3.
Drug screen identifies palbociclib as a DSRCT therapeutic. A, Heatmap of 22 EWSR-WT1 upregulated genes with available inhibitors. B, Relative viability of JN-DSRCT-1 and LP9 cells treated with 1 or 10 μmol/L of various inhibitors. C, Relative viability of four DSRCT cell lines or LP9 mesothelial cells treated for 72 hours with palbociclib, KN93, UNC2250, vandetinib, or sunitinib (n = 3). D, Western blot analysis of MERTK, CAMK2A, CALM1, and CCND1 protein expression in shWT1 DSRCT cell lines treated with (+) or without (−) dox. Average protein expression was quantified relative to ACTIN, normalized to the -dox condition, and is shown underneath each blot (n = 2). E, Colony formation of JN-DSRT-1 and BER-DSRCT cells treated with palbociclib or abemaciclib for 12 days (n = 2). F, Western blot analysis of DSRCT cells treated with palbociclib for 2 days showing decreased phosphorylation or RB (representative image of n = 2). Phosphorylated RB fraction was determined by quantifying pRB S708 relative to total RB and normalizing to the untreated condition. Average protein expression of CDK4 and CDK6 was quantified relative to ACTIN and normalized to the untreated condition. Quantifications are shown underneath each blot. G, Cell-cycle analysis of DSRCT cells treated with palbociclib for 2 days (n = 3). *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Microarray expression data of fusion-positive sarcomas DSRCT (n = 28), alveolar rhabdomyosarcoma (ARMS; n = 23), alveolar soft part sarcoma (ASPS; n = 12), Ewing sarcoma (ES; n = 28), and synovial sarcoma (SS; n = 46) showed increased expression of CALM1, CAMK2A, and MERTK in DSRCT relative to all other tumor types (Supplementary Fig. S3B; ref. 38). It also demonstrated increased expression of CCND1 in DSRCT relative to ARMS, ASPS, and SS, but not ES, where CCND1 has also been identified as critical downstream target of EWSR1::FLI1 (41). RT-qPCR in JN-DSRCT-1 and BER-DSRCT shWT1 cell lines confirmed the transcriptional control of CAMK2A, CALM1, MERTK, and CCND1 by EWSR1::WT1, with dox-induced depletion of the fusion protein leading to downregulation of these four targets (Supplementary Fig. S3C). Western blot analysis on our four DSRCT shWT1 cell lines with or without dox treatment demonstrated downregulation of MERTK, CCND1, and CALM1 protein expression in at least three out of four cell lines in response to EWSR1::WT1 depletion (Fig. 3D). In contrast, CAMK2A expression was regulated to a lesser extent and only found to be downregulated by >30% in two cell lines.
Given palbociclib's efficacy across four DSRCT cell lines and prior approval by the FDA for breast cancer treatment, we decided to further examine the potential effects of CDK4/6i in DSRCT. Treatment with the two other FDA-approved CDK4/6 inhibitors, ribociclib and abemaciclib, also led to greater reductions in viability for DSRCT cells than control cells at 72 hours (Supplementary Fig. S3D). Palbociclib and abemaciclib treatment resulted in larger growth reductions (40–80%) than ribociclib (20–40%). The effect of palbociclib and abemaciclib on DSRCT growth was even stronger in 14-day colony-formation assays where treatment with 100 nmol/L and 1 μmol/L of both drugs led to growth reductions of 75% and 95%, respectively (Fig. 3E). Treatment of JN-DSRCT-1 and BER-DSRCT with palbociclib led to a dose-dependent decrease in phosphorylation of RB at Ser708 (Fig. 3F) and an increase in the fraction of cells in G1 phase of the cell cycle (Fig. 3G). Abemaciclib treatment similarly led to decreased RB phosphorylation (Supplementary Fig. S3E) and an increase of cells in G1 phase (Supplementary Fig. S3F). These results are consistent with CDK4/6 inhibitors reducing cell growth by producing a G1 arrest.
Palbociclib acts by inhibiting the CCND/CDK4–6/RB1 axis in DSRCT
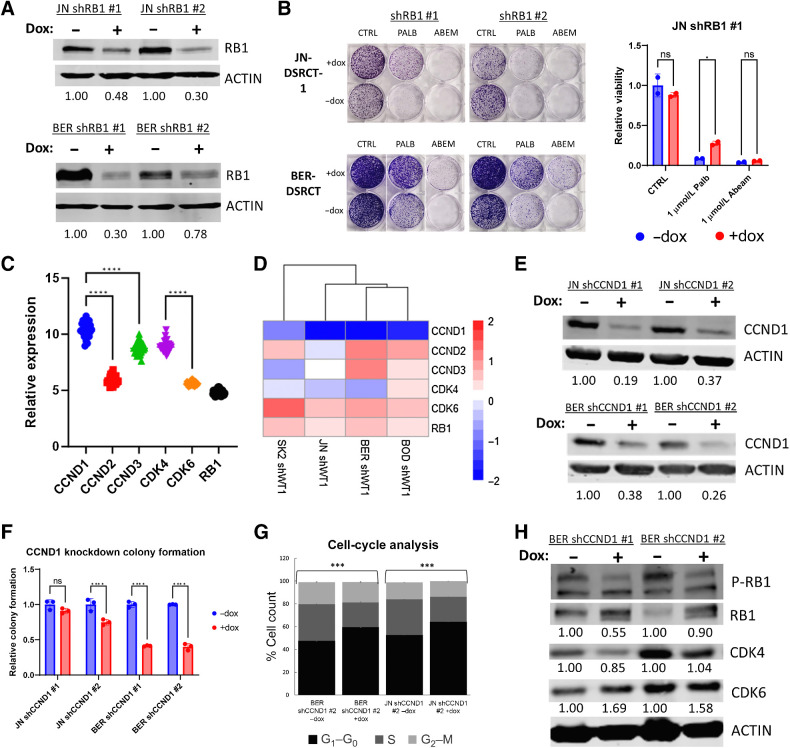
To definitively establish that palbociclib reduces DSRCT growth by dephosphorylating RB and preventing cell-cycle progression, we established dox-inducible JN-DSRCT-1 and BER-DSRCT cell lines that use two independent shRNAs to knockdown RB expression. Western blot analysis confirmed RB depletion with the addition of dox (Fig. 4A). Dox-induced depletion of RB successfully rescued palbociclib-induced growth reduction as assessed by colony-formation assay (Fig. 4B). In contrast, RB depletion led to only a minimal rescue of abemaciclib-induced growth reduction. This is consistent with previous findings that abemaciclib has off-target effects including on casein kinase (42).
Figure 4.
Palbociclib acts through the CCND-CDK4/6-RB axis. A, Western blot analysis of RB1 and ACTIN protein expression in shRB1 cell lines with (+) or without (−) dox (representative blot, n = 2). Average protein expression was quantified relative to ACTIN, normalized to the -dox condition, and is shown underneath each blot. B, Colony formation assays of shRB1 cell lines treated with or without dox in combination with vehicle control, 1 μmol/L palbociclib, or 1 μmol/L abemaciclib. C, Microarray expression analysis of 28 DSRCT tumors comparing CCND and CDK4/6 RNA levels. D, Heatmap of log2-fold change in expression of CCND–CDK4/6–RB pathway members when EWSR1::WT1 is depleted from shWT1 DSRCT cell lines. E, Western blot of CCND1 and ACTIN protein expression in shCCND1 cell lines with (+) or without (−) dox (representative blot, n = 2). Average protein expression was quantified relative to ACTIN, normalized to the -dox condition, and is shown underneath each blot. F, Colony-formation assays of shCCND1 DSRCT cells with or without dox treatment (n = 3). G, Cell-cycle analysis of shCCND1 DSRCT cells with or without dox treatment (n = 3). H, Western blot analysis of RB phosphorylation in BER-DSRCT shCCND1 cells with (+) or without (−) dox (representative blot, n = 2). Phosphorylated RB fraction was determined by quantifying pRB S708 relative to total RB and normalizing to the -dox condition. Average protein expression of CDK4 and CDK6 was quantified relative to ACTIN and normalized to the -dox condition. Quantifications are shown underneath each blot. ***, P < 0.001; ****, P < 0.0001; ns, nonsignificant.
Having established the dependence of palbociclib on RB to repress growth in DSRCT, we investigated the role of the other members of the CCND–CDK4/6-RB axis. Microarray expression analysis (n = 28) found significantly higher expression of CCND1 than CCND2 or CCND3, as well as higher expression of CDK4 than CDK6 (Fig. 4C). A heatmap of our shWT1 DSRCT RNA-seq data demonstrates strong control of CCND1 by EWSR1::WT1 but weak dependence of other pathway members on the fusion oncoprotein (Fig. 4D), suggesting CCND1 as the primary EWSR1::WT1 induced driver of G1–S phase transition. Dox-inducible cell lines with CCND1 shRNAs were generated and dox treatment successfully depleted CCND1 in JN-DSRCT-1 and BER-DSRCT cell lines (Fig. 4E). CCND1 depletion significantly reduced growth in both BER-DSRCT cell lines and one of two JN-DSRCT-1 cell lines (Fig. 4F). Consistent with the hypothesized mechanism of action, CCND1 depletion increased the percent of cells in G1 phase (Fig. 4G) and decreased phosphorylation of RB (Fig. 4H; Supplementary Fig. S4A). The magnitude of colony-formation reduction, G1 phase increase, and RB dephosphorylation with CCND1 knockdown were less than what was observed with palbociclib treatment. This may be explained by the incompleteness of CCND1 knockdown, additional effects of palbociclib, and/or by the ability of DSRCT cells to increase the expression of other pathway members in response to CCND1 depletion. In-line with the latter hypothesis, we observed an increase in CCND3 mRNA expression in BER-DSRCT cells depleted of CCND1 (Supplementary Fig. S4B) as well as alterations in CDK4 and CDK6 protein expression (Fig. 4H; Supplementary Fig. S4A).
The E-KTS isoform controls transcription alterations
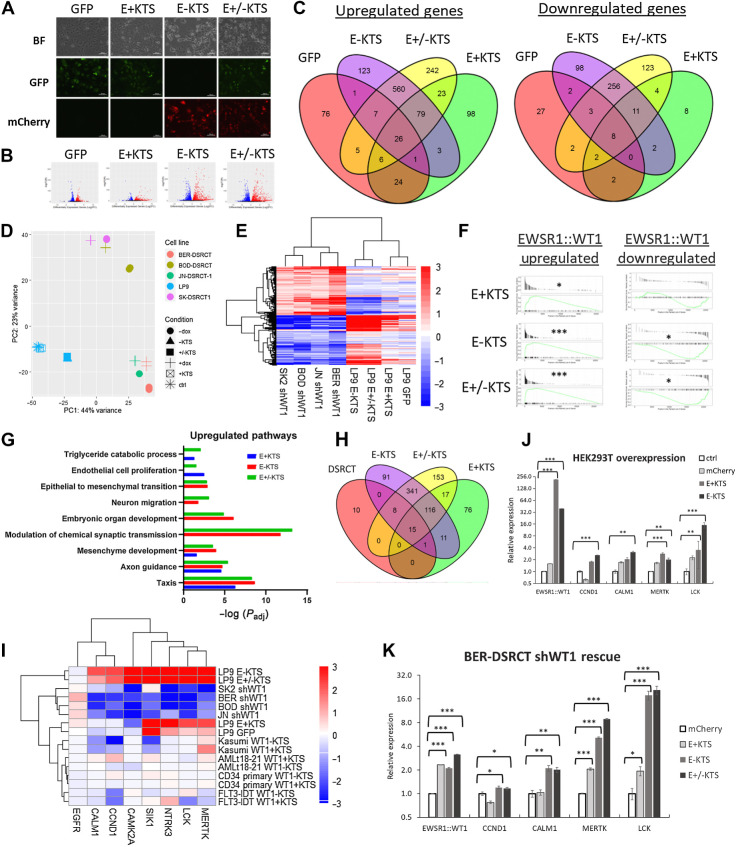
We next investigated the role of the two EWSR1::WT1 isoforms (E+KTS and E-KTS) in transcription. Using our previous RNA-seq data, we examined the relative expression of these isoforms in our four DSRCT cell lines and found slightly higher expression of the E+KTS isoform, with the E+KTS: E-KTS ratio ranging from 1.3 in BER-DSRCT to 1.7 in JN-DSRCT-1 (Supplementary Fig. S5A). This increase in the E+KTS isoform likely reflects the preferred utilization of the alternative splice site that includes the KTS as observed in native WT1 (43). We further confirmed that our dox-inducible shRNA system depletes both isoforms (Supplementary Fig. S5A), establishing that our identified EWSR1::WT1 regulated gene sets represent the net effect of gene regulation by both isoforms. To test the clinical relevance of these observations, we examined the ratio of these isoforms in a set of primary and recurrent DSRCT tumors (n = 2; ref. 44; Supplementary Fig. S5B). Consistent with our cell line data, the primary tumor (n = 1) expressed more E+KTS with an E+KTS: E-KTS ratio of 1.5. Intriguingly, the recurrent tumor (n = 1) showed greater E-KTS expression with a ratio of 0.8.
To assess isoform-specific function, we expressed one or both EWSR1::WT1 isoforms in the normal mesothelial cell line LP9, a possible DSRCT cell of origin (22). The E+KTS lentivirus co-expressed GFP and the E-KTS lentivirus co-expressed mCherry, enabling visual verifications of system functionality (Fig. 5A). EWSR1::WT1 expression was validated with RT-qPCR (Supplementary Fig. S5C) and RNA-seq, which enabled confirmation of isoform specific EWSR1::WT1 expression (Supplementary Fig. S5D). Nontransduced cells or cells transduced with lentivirus expressing only GFP served as negative controls. Differential gene expression analysis demonstrated remarkably different gene expression alterations between E-KTS and E+KTS isoforms (Fig. 5B). Although overexpression of the E-KTS isoform upregulated 765 genes and downregulated 366 genes, overexpression of the E+KTS isoform upregulated 203 genes and downregulated only 25 genes (P < 0.05, |log2FC| > 2). Comparison of altered genes identified 82 genes commonly upregulated by E-KTS and E+KTS, but only 13 commonly downregulated genes (Fig. 5C). Among genes upregulated with dual isoform expression, 560 were exclusively upregulated by E-KTS as compared with 23 uniquely upregulated by E+KTS. Principle component analysis (PCA) between LP9 cells with EWSR1::WT1 overexpression and DSRCT shWT1 cell lines demonstrated clustering of LP9 cells overexpressing E-KTS in a position shifted toward DSRCT cell lines relative to LP9 control cells (Fig. 5D). Conversely, knockdown of EWSR1::WT1 by dox-induction in DSRCT cell lines shifted their expression profile toward that of LP9. In contrast, cells overexpressing E+KTS alone clustered with nontransduced LP9 cells. A heatmap of the genes commonly upregulated and downregulated by EWSR1::WT1 across DSRCT cell lines further demonstrated the dominance of E-KTS in producing DSRCT-specific gene expression alterations (Fig. 5E). GSEA on our common EWSR1::WT1 upregulated gene set showed positive enrichment with expression of either or both isoforms, though E+KTS showed the least statistical significance (Fig. 5F). GSEA of the downregulated EWSR1::WT1 gene set showed statistically significant negative enrichment for E-KTS and E±KTS but not E+KTS alone.
Figure 5.
E-KTS isoform controls transcription alterations. A, Brightfield and fluorescent images of LP9 cells that are unaltered or transduced with GFP only, E+KTS GFP, E-KTS mCherry, or both E+KTS GFP and E-KTS mCherry. B, Volcano plots of RNA-seq data from LP9 cells with GFP, E+KTS, E-KTS, or E±KTS expression compared with nontransduced control cells (n = 2). Red, significantly upregulated; blue, significantly downregulated. FDR <0.05. C, Venn diagrams comparing genes upregulated and downregulated by expression of GFP (red), E+KTS (green), E-KTS (purple), or E±KTS (yellow) in LP9 cells. D, PCA of DSRCT or LP9 cells treated with EWSR1::WT1 knockdown by shWT1 induction or with EWSR1::WT1 isoform overexpression. E, Heatmap of log2-fold change gene expression of EWSR1::WT1 commonly upregulated and downregulated targets. F, GSEA of EWSR1::WT1 upregulated and downregulated gene sets in LP9 cells with E+KTS, E-KTS, or E±KTS overexpression (*, P < 1E−2; ***, P < 1E−8). G, Bar graph of pathways with overrepresentation of upregulated genes induced by E+KTS, E-KTS, or E±KTS expression in LP9 cells. H, Venn diagram of pathways upregulated by EWSR1::WT1 in DSRCT (red), or by E+KTS (green), E-KTS (purple), or E±KTS (yellow) in LP9 cells. I, Heatmap of log2-fold change in gene expression of druggable targets in DSRCT cells depleted of EWSR1::WT1, LP9 cells with EWSR1::WT1 isoforms overexpressed, or hematologic malignancies with WT1 isoforms overexpressed. J, RT-qPCR of EWSR1::WT1, CCND1, CALM1, MERTK, and CAMK2A in HEK293T cells with overexpression of mCherry, E+KTS, or E-KTS (n = 3). K, RT-qPCR of EWSR1::WT1, CCND1, CALM1, MERTK, and CAMK2A in BER-DSRCT shWT1 cells simultaneously treated with dox to depleted native EWSR1::WT1 and overexpressing mCherry, E+KTS, E-KTS, or both isoforms (n = 3). *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Pathway analysis identified 131 pathways commonly overrepresented by upregulated genes in LP9 cells with E+KTS, E-KTS, and E±KTS isoforms (Fig. 5G and H). This included 15 pathways also identified as upregulated by EWSR1::WT1 in three DSRCT cell lines including taxis, axon guidance, and mesenchyme development. An additional 349 pathways were upregulated in E-KTS or E±KTS cells but not E+KTS cells. Remarkably, there were no overrepresented pathways among genes downregulated by E+KTS. In contrast, 339 pathways were commonly overrepresented among genes downregulated by E-KTS and E±KTS, including 65 pathways commonly downregulated by EWSR1::WT1 in DSRCT cell lines (Supplementary Figs. S5E and S5F). These downregulated pathways, consistent with Supplementary Figs. S3C and S3D, included extracellular matrix organization, response to wounding, and muscle tissue development.
Examining our novel EWSR1::WT1 regulated druggable targets, CCND1, CALM1, and CAMK2A were specifically upregulated by the E-KTS isoform whereas MERTK was upregulated by both isoforms (Fig. 5I). Previously identified DSRCT dependencies SIK1 and NTRK3 were upregulated by both isoforms whereas EGFR was not regulated by either isoform (Fig. 5I). Using previously published WT1 isoform overexpression RNA-seq data from four cell lines of hematologic origin, we determined that the upregulation of these druggable targets is specific to EWSR1::WT1, with neither WT1-KTS or WT1+KTS expression leading to target upregulation. Intriguingly, WT1-KTS and WT1+KTS overexpression in Kasumi leukemia cells led to reduced expression of CCND1, CALM1, and SIK1, suggesting in certain circumstances EWSR1::WT1 and native WT1 may alter gene expression in the opposite direction. We further tested the isoform specificity of transcriptional changes by establishing mCherry, E+KTS, or E-KTS expression in HEK293T cells. Consistent with our observations in LP9, E-KTS expression but not E+KTS or mCherry expression led to upregulation of CCND1 and CALM1 in HEK293T cells, whereas both isoforms were able to induce MERTK and LCK expression (Fig. 5J). We also validated this isoform specific transcriptional regulation in DSRCT cells with a rescue experiment where we used our dox-inducible system to knockdown native EWSR1::WT1 while simultaneously expressing exogenous mCherry, E+KTS, or E-KTS in BER-DSRCT and SK-DSRCT2 cell lines. In BER-DSRCT, the E-KTS isoform or both isoforms together upregulated CCND1 and CALM1 expression while either isoform was able to induce MERTK and LCK (Fig. 5K). Overexpression in SK-DSRCT2 cells with EWSR1::WT1 knockdown demonstrated a similar trend with CCND1 upregulated 1.8-fold by E-KTS but not significantly increased by E+KTS (Supplementary Fig. S5G).
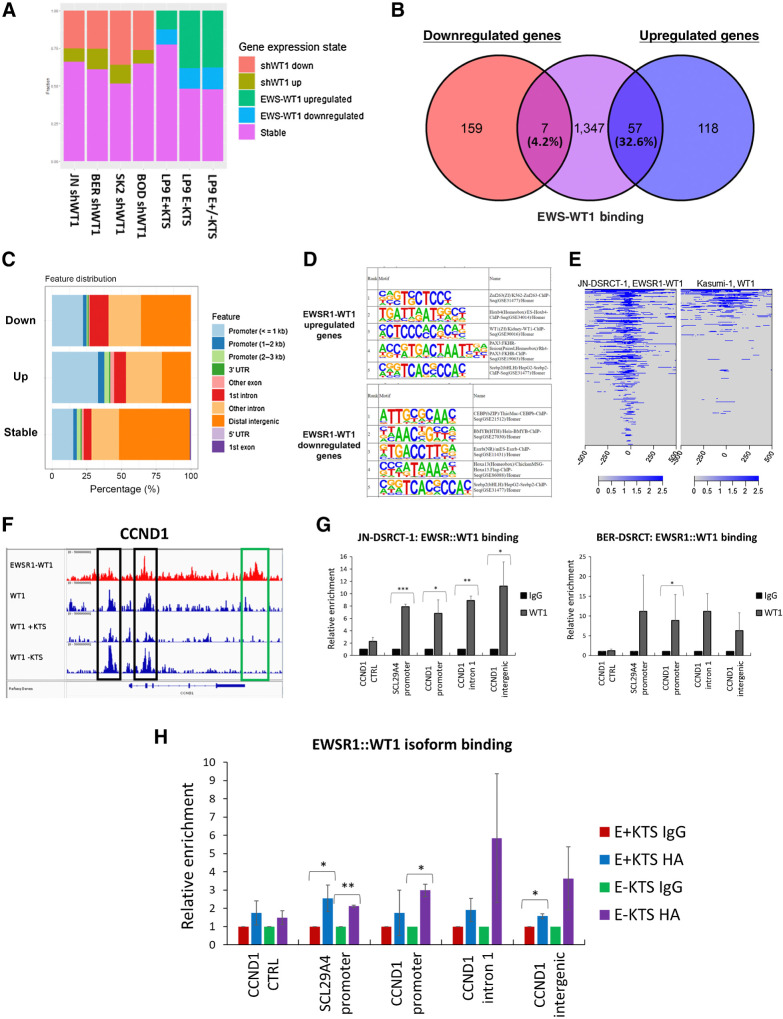
EWSR1::WT1 binding predominantly upregulates gene expression
We next integrated previously established ChIP-seq data of EWSR1::WT1 binding in JN-DSRCT-1 with our novel RNA-seq findings to understand the role of EWSR1::WT1 binding in transcription alteration. For each of the 2036 EWSR1::WT1 binding sites identified in JN-DSRCT-1 by Hingorani and colleagues (15), we identified the closest gene and examined its expression change in response to depletion of EWSR1::WT1 in our four dox-inducible DSRCT shWT1 cell lines or in response to overexpression of E+KTS, E-KTS, or E±KTS in LP9 cells (Fig. 6A). Among DSRCT cell lines, 25% to 35% of EWSR1::WT1 binding sites were associated with genes downregulated by EWSR1::WT1 depletion whereas only 8% to 12% of binding sites corresponded with genes upregulated by EWSR1::WT1 depletion (Fig. 6A). Overexpression of E-KTS or E±KTS led to upregulation of genes at 40% of binding sites and downregulation of genes at 15% of sites. E+KTS was associated with upregulation of genes at only 12% of sites and downregulation of genes at another 10% of sites. Examining our sets of genes commonly upregulated and downregulated by EWSR1::WT1 across cell lines (Fig. 3B) with EWSR1::WT1 binding, we found 57/175 upregulated genes (32.6%) were associated with EWSR1::WT1 binding compared with only 7/166 downregulated genes (4.2%; Fig. 6B). Among the set of EWSR1::WT1 binding sites, genes upregulated by EWSR1::WT1 in at least two cell lines were more likely to be bound in the promoter region whereas genes downregulated by EWSR1::WT1 in at least two cell lines were more likely to be bound in intron 1 (Fig. 6C). Genes not significantly altered in expression (stable genes) were more likely to be bound in the distal intergenic region. HOMER motif analysis identified differences in transcription factor motifs at sites associated with upregulation and downregulation, with upregulated sites associated with ZNF263, WT1, and PAX3::FOXO1 motifs, whereas downregulated sites were associated with CEBP and BMYB motifs (Fig. 6D).
Figure 6.
EWSR1::WT1 binding upregulates gene expression. A, Barplot showing the percentage of EWSR1::WT1 binding sites associated with genes that are upregulated, downregulated, or stable in response to EWSR1::WT1 knockdown in DSRCT cell lines or overexpression in LP9 cells. B, Venn diagram examining the overlap between EWSR1::WT1 bound genes (purple) and genes commonly downregulated (red) or upregulated (blue) by EWSR1::WT1. C, Bar graph showing the genomic annotation of EWSR1::WT1 binding sites associated with EWSR1::WT1-induced upregulation in at least two cell lines (up), EWSR1::WT1 induced downregulation in at least two cell lines (down), or no consistent expression alteration (stable). D, Top 5 significant HOMER motifs associated with EWSR1::WT1 bound sites that upregulate or downregulate gene expression. E, Binding profile of EWSR1::WT1 in JN-DSRCT-1 compared with WT1 in Kasumi-1 cells. F, ChIP-seq tracks of CCND1 genomic region showing binding of EWSR1::WT1 in JN-DSRCT-1 (red) or the binding of total WT1 or its isoforms in Kasumi-1 (blue). Black box indicates binding sites shared by EWSR1::WT1 and WT1, while green box indicates a EWSR1::WT1 specific binding site. G, ChIP-qPCR of IgG or anti-WT1 pulldown in JN-DSRCT-1 and BER-DSRCT (n = 3–5). H, ChIP-qPCR of IgG or anti-HA pulldown in U2OS cells overexpressing E+KTS or E-KTS (n = 2). *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Given our finding that EWSR1::WT1 alters gene expression in a pattern unique from WT1, we utilized publicly available WT1 ChIP-seq data from Kasumi-1 leukemia cells to compare binding profiles between native WT1 and the fusion oncoprotein. We found that only 20% of EWSR1::WT1 binding sites in JN-DSRCT-1 overlapped WT1 binding sites in Kasumi-1 (Fig. 6E), which could help explain the observed differences in transcription. Although this difference in binding sites could suggest EWSR1::WT1 has different binding selectivity than WT1, it could alternatively reflect differences in the chromatin landscape between DSRCT and leukemia cells.
Looking specifically at binding sites near CCND1, we find EWSR1::WT1 in DSRCT cells binds within intron 1 and in an intergenic region directly following the last exon (Fig. 6F). WT1 in leukemia cells does not bind to the intergenic site, but shares the intron 1 binding site. WT1 also binds to the CCND1 promoter in Kasumi-1 cells. Though not a peak identified by Hingorani and colleagues, EWSR1::WT1 ChIP-seq tracks suggest the fusion may also bind within the CCND1 promoter (15). Intriguingly, WT1 isoform specific ChIP-seq data from Kasumi-1 cells demonstrated stronger enrichment of the WT1 -KTS isoform at the two WT1 binding sites within CCND1. ChIP-qPCR in JN-DSRCT-1 found significant enrichment of EWSR1::WT1 at the CCND1 promoter, intron 1, and intergenic binding sites, as well as within the promoter of SLC29A4, which served as a positive control (Fig. 6G). Enrichment was not observed in intron 3 of CCND1, which served as a negative control. ChIP-qPCR in BER-DSRCT expanded these results, finding statistically significant enrichment of EWSR1::WT1 at the CCND1 intron 1 site and near statistically significant enrichment in the promoter and intergenic regions (Fig. 6G). To understand differences in binding sites between E-KTS and E+KTS isoforms, we overexpressed HA-tagged versions of each isoform in U2OS cells and performed ChIP-qPCR with anti-HA antibody or IgG control. We observed greater enrichment of the E-KTS isoform in all three regions, including a statistically significant increase in enrichment of E-KTS at the CCND1 promoter (Fig. 6H).
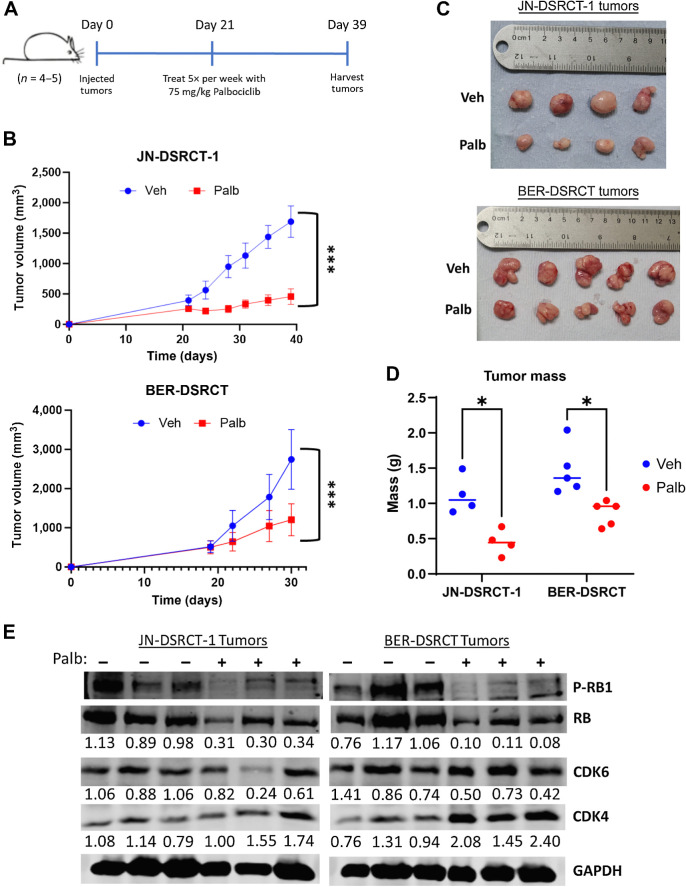
Palbociclib reduces DSRCT growth in vivo
Having identified palbociclib as a novel DSRCT therapeutic target and characterized EWSR1::WT1 regulation of CCND1, we tested the therapeutic benefit of palbociclib in vivo with xenografts derived from JN-DSRCT-1 and BER-DSRCT cell lines. DSRCT cells were injected and allowed to grow for 3 weeks, at which point, tumors were measurable (Fig. 7A). Starting at 3 weeks after injection, mice were treated five times per week via oral gavage with either 75 mg/kg palbociclib or vehicle control. Tumor growth was significantly reduced with palbociclib treatment as measured with calipers twice weekly (Fig. 7B). Correspondingly, final tumor size and mass were reduced for palbociclib-treated groups in both tumor types (Fig. 7C and D). Body weight of all mice was monitored as a measure of drug toxicity. One palbociclib-treated mouse in the JN-DSRCT-1 group experienced weight loss (Supplementary Fig. S6A). However, no mice in the BER-DSRCT group showed this effect, suggesting the observation may not be caused by palbociclib toxicity (Supplementary Fig. S6B). Western blot analysis of tumor lysates demonstrated dephosphorylation of RB in palbociclib treated tumors, consistent with its activity as a CDK4/6 inhibitor and in-line with our in vitro findings (Fig. 7E). Together, these results advance palbociclib as a novel DSRCT therapeutic and establish a need for urgent clinical investigation.
Figure 7.
Palbociclib reduces DSRCT xenograft growth. A, Timeline of xenograft injection and palbociclib treatment. B, Tumor growth monitored twice weekly with calipers in JN-DSRCT-1 (n = 4) and BER-DSRCT (n = 5) tumors. C, Image of final tumor volumes treated with vehicle or palbociclib. D, Final tumor mass of xenografts treated with vehicle or palbociclib for 2 weeks (BER-DSRCT) or 3 weeks (JN-DSRCT-1). E, Western blot analysis of RB phosphorylation, CDK4, and CDK6 in xenograft tumors treated with vehicle (−) or palbociclib (+; n = 3). Phosphorylated RB fraction was determined by quantifying pRB S708 relative to total RB and normalizing to the untreated condition. Protein expression of CDK4 and CDK6 was quantified relative to GAPDH and normalized to the untreated condition. Quantifications are shown underneath each blot. *, P < 0.05; ***, P < 0.001.
Discussion
DSRCT is an extremely deadly tumor with poor prognosis and no effective targeted therapies (2). While previous studies have shown that EWSR1::WT1 depletion can reduce DSRCT growth in vitro (11, 12), this study for the first time demonstrates that EWSR1::WT1 depletion reduces tumor growth in vivo. This finding validates therapeutic strategies targeting EWSR1::WT1 downstream effectors including NTRK3 and SIK1 (22, 45). It further establishes an urgent need to identify upstream regulators of the fusion oncoprotein that can serve as novel therapeutics.
Prior to this work, only two studies had unbiasedly characterized EWSR1::WT1 transcriptional targets and those studies arrived at quite dissimilar results with only a handful of commonly regulated targets identified (12, 14). Here we perform the most comprehensive transcriptional characterization to date and establish sets of common upregulated and downregulated targets across four DSRCT cell lines. Our findings in JN-DSRCT-1 and BER-DSRCT are consistent with the siRNA results from Gedminas and colleagues, with over 80% of our identified targets differentially expressed in the same direction within their data set (12). Similar to Gedminas and colleagues our analysis identified a variety of upregulated pathways related to cell cycle including positive regulation of G1/S phase transition and G2–M transition. Our pathway analysis also identified several upregulated neuronal pathways including synapse development and axon guidance, consistent with findings by Kang and colleagues (11). Intriguingly, neuronal proteins have also been identified as critical to Ewing sarcoma (46, 47), suggesting these tumors may be co-opting neuronal programs to support tumor growth and metastasis (48).
In addition to elucidating pathways regulated by EWSR1::WT1, our transcriptional analysis led to the identification of CCND1 as an EWSR1::WT1 regulated druggable target. We show that CCND1 is regulated by the E-KTS isoform in DSRCT, which binds to CCND1 at the promoter, within intron 1, and within an intergenic region that follows the final exon. The CCND-CDK4/6-RB axis is critical to the G1–S transition and can be inhibited by CDK4/6 inhibitors, three of which are approved by the FDA for the treatment of breast cancer (40). We found that all three inhibitors (palbociclib, ribociclib, abemaciclib) were able to reduce DSRCT cell growth across four cell lines in vitro, with palbociclib and abemaciclib having the strongest effects. RB depletion was able to rescue palbociclib-induced growth reduction but not abemaciclib-induced growth reduction, suggesting palbociclib activity in DSRCT is largely through the CCND–CDK4/6–RB axis, while abemaciclib has off-target effects. This lower level of specificity for abemaciclib is consistent with previous studies in breast cancer and has been implicated in its higher rate of gastrointestinal side effects (42, 49). Given its on-target activity, palbociclib was tested in two DSRCT xenografts where it led to a statistically significant reduction in tumor growth. These results suggest palbociclib may reduce tumor growth and improve patient survival, which warrants urgent clinical investigation. The primary method of CDK4/6 inhibitor resistance comes from RB mutation or silencing (50, 51). Studies in DSRCT including whole genome sequencing and MSK-IMPACT analysis have failed to identify RB mutations in DSRCT (9, 10), suggesting most tumors will at least initially be responsive to CDK4/6 inhibitor therapy.
In clinical trials, combination treatment with the anti-estrogen fulvestrant and palbociclib has been shown to increase both progression-free survival and overall survival in hormone receptor positive breast cancer (52, 53). By simultaneously reducing estrogen receptor stimulation of the CCND–CDK4/6–RB axis and inhibiting CDK4 and CDK6, combination therapy is able to reduce RB phosphorylation more than either monotherapy (40). If a potent inhibitor of an upstream regulator of EWSR1::WT1 is developed, a future comparable strategy in DSRCT could combine EWSR1::WT1 upstream regulator inhibition with CDK4/6 inhibitors. Combination approach could also be used with inhibitors of CDK4/6 and recently identified DSRCT vulnerabilities NTRK3, SIK1, and EGFR (20). The EGFR inhibitor cetuximab has been approved by the FDA for the treatment of colorectal cancer (54) and has been shown to reduce DSRCT growth in cell line and patient-derived xenograft models (20). Combining palbociclib and cetuximab would enable targeting of both EWSR1::WT1 dependent (CCND1) and independent (EGFR) pathways involved in DSRCT proliferation. CDK4/6 inhibitors have also shown promise clinically for the treatment of liposarcoma (55) and preclinically for the treatment of Ewing sarcoma (41) and neuroblastoma (56). In Ewing sarcoma, the EWSR1::FLI1 fusion regulates CCND1 transcription helping to establish CDK4/6 inhibitors as a vulnerability (41). A recent phase 2 trial examining combination therapy with palbociclib and ganitumab (IGF1R mAb) in patients with relapsed Ewing sarcoma found a 6-month progression free survival rate of 30% (57).
This work further establishes new insights into fundamental DSRCT biology. We compare the transcriptional alterations caused by EWSR1::WT1 depletion with those resulting from native WT1 depletion. Despite retaining three of WT1’s four zinc fingers, we show that EWSR1::WT1 in DSRCT regulates a set of genes unique from WT1 in other cell types. This may result from differences in DNA binding as WT1 in leukemia cells surprisingly binds to only 20% of the DNA regions bound by EWSR1::WT1 in DSRCT. Alternatively, this may reflect differences in chromatin accessibility or expression of coregulators in these different cell lineages. Future experiments are necessary to better elucidate the exact mechanism for these differences in transcriptional regulation. We also investigate the transcriptional role of the two EWSR1::WT1 isoforms in human cell lines and demonstrate the dominant role played by the E-KTS isoform in inducing transcriptional alteration. This finding validates previous studies in mouse-derived cells that also suggested E-KTS triggers greater transcriptional alteration than E+KTS (11, 16). Finally, we integrate our novel transcriptional data sets with EWSR1::WT1 binding profiles (15) and discover that EWSR1::WT1 binding predominantly contributes to transcriptional upregulation. Of our commonly regulated EWSR1::WT1 targets, 32.6% of upregulated genes are associated with EWSR1::WT1 binding sites but only 4.2% of downregulated genes. The mechanism by which EWSR1::WT1 induces downregulation, potentially through expression of other transcription factors or epigenetic alterations, remains unknown and an area in need of future investigation.
Supplementary Material
EWSR1::WT1 Upregulated Pathways
EWSR1::WT1 Downregulated Pathways
Supplementary Figures and Tables
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Authors' Disclosures
S.B. Lee reports grants from NCI during the conduct of the study. No disclosures were reported by the other authors.
Authors' Contributions
J.W. Magrath: Conceptualization, data curation, software, formal analysis, investigation, visualization, methodology, writing–original draft, writing–review and editing. S.S. Sampath: Investigation, writing–review and editing. D.A. Flinchum: Investigation, writing–review and editing. A.B. Hartono: Conceptualization, investigation, writing–review and editing. I.N. Goldberg: Investigation, writing–review and editing. J.R. Boehling: Investigation, writing–review and editing. S.D. Savkovic: Resources, investigation, writing–review and editing. S.B. Lee: Conceptualization, resources, data curation, supervision, funding acquisition, methodology, writing–original draft, writing–review and editing.
Acknowledgements
This work was supported by the NCI R01CA222856 (S.B. Lee) and the Tulane Carol Lavin Bernick Faculty Investment Fund (S.B. Lee).
References
- 1. Gerald WL, Haber DA. The EWS-WT1 gene fusion in desmoplastic small round cell tumor. Semin Cancer Biol 2005;15:197–205. [DOI] [PubMed] [Google Scholar]
- 2. Honoré C, Delhorme JB, Nassif E, Faron M, Ferron G, Bompas E, et al. Can we cure patients with abdominal desmoplastic small round cell tumor? results of a retrospective multicentric study on 100 patients. Surg Oncol 2019;29:107–12. [DOI] [PubMed] [Google Scholar]
- 3. Subbiah V, Lamhamedi-Cherradi SE, Cuglievan B, Menegaz BA, Camacho P, Huh W, et al. Multimodality treatment of desmoplastic small round cell tumor: chemotherapy and complete cytoreductive surgery improve patient survival. Clin Cancer Res 2018;24:4865–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kushner BH, LaQuaglia MP, Wollner N, Meyers PA, Lindsley KL, Ghavimi F, et al. Desmoplastic small round-cell tumor: prolonged progression-free survival with aggressive multimodality therapy. J Clin Oncol 1996;14:1526–31. [DOI] [PubMed] [Google Scholar]
- 5. Lal DR, Su WT, Wolden SL, Loh KC, Modak S, La Quaglia MP. Results of multimodal treatment for desmoplastic small round cell tumors. J Pediatr Surg 2005;40:251–5. [DOI] [PubMed] [Google Scholar]
- 6. Lee J, Nguyen PT, Shim HS, Hyeon SJ, Im H, Choi MH, et al. EWSR1, a multifunctional protein, regulates cellular function and aging via genetic and epigenetic pathways. Biochim Biophys Acta Mol Basis Dis 2019;1865:1938–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Li H, Watford W, Li C, Parmelee A, Bryant MA, Deng C, et al. Ewing sarcoma gene EWS is essential for meiosis and B lymphocyte development. J Clin Invest 2007;117:1314–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yang L, Han Y, Suarez Saiz F, Minden MD. A tumor suppressor and oncogene: the WT1 story. Leukemia 2007;21:868–76. [DOI] [PubMed] [Google Scholar]
- 9. Slotkin EK, Bowman AS, Levine MF, Dela Cruz F, Coutinho DF, Sanchez GI, et al. Comprehensive molecular profiling of desmoplastic small round cell tumor. Mol Cancer Res 2021;19:1146–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wu C-C, Beird HC, Lamhamedi-Cherradi S-E, Soeung M, Ingram D, Truong DD, et al. Multi-site desmoplastic small round cell tumors are genetically related and immune-cold. npj Precision Oncology 2022;6:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kang HJ, Park JH, Chen W, Kang SI, Moroz K, Ladanyi M, et al. EWS-WT1 oncoprotein activates neuronal reprogramming factor ASCL1 and promotes neural differentiation. Cancer Res 2014;74:4526–35. [DOI] [PubMed] [Google Scholar]
- 12. Gedminas JM, Chasse MH, McBrairty M, Beddows I, Kitchen-Goosen SM, Grohar PJ. Desmoplastic small round cell tumor is dependent on the EWS-WT1 transcription factor. Oncogenesis 2020;9:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Magrath JW, Kang HJ, Hartono A, Espinosa-Cotton M, Somwar R, Ladanyi M, et al. Desmoplastic small round cell tumor cancer stem cell-like cells resist chemotherapy but remain dependent on the EWSR1-WT1 oncoprotein. Front Cell Dev Biol 2022;10:1048709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bleijs M, Pleijte C, Engels S, Ringnalda F, Meyer-Wentrup F, van de Wetering M, et al. EWSR1-WT1 target genes and therapeutic options identified in a novel DSRCT In Vitro Model 2021, Cancers (Basel) 13:6072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hingorani P, Dinu V, Zhang X, Lei H, Shern JF, Park J, et al. Transcriptome analysis of desmoplastic small round cell tumors identifies actionable therapeutic targets: a report from the Children's Oncology Group. Sci Rep 2020;10:12318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kim J, Lee K, Pelletier J. The desmoplastic small round cell tumor t(11;22) translocation produces EWS/WT1 isoforms with differing oncogenic properties. Oncogene 1998;16:1973–9. [DOI] [PubMed] [Google Scholar]
- 17. Potluri S, Assi SA, Chin PS, Coleman DJL, Pickin A, Moriya S, et al. Isoform-specific and signaling-dependent propagation of acute myeloid leukemia by Wilms tumor 1. Cell Rep 2021;35:109010. [DOI] [PubMed] [Google Scholar]
- 18. Nishio J, Iwasaki H, Ishiguro M, Ohjimi Y, Fujita C, Yanai F, et al. Establishment and characterization of a novel human desmoplastic small round cell tumor cell line, JN-DSRCT-1. Lab Invest 2002;82:1175–82. [DOI] [PubMed] [Google Scholar]
- 19. Markides CS, Coil DR, Luong LH, Mendoza J, Kozielski T, Vardeman D, et al. Desmoplastic small round cell tumor (DSRCT) xenografts and tissue culture lines: Establishment and initial characterization. Oncol Lett 2013;5:1453–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Smith RS, Odintsov I, Liu Z, Lui AJ, Hayashi T, Vojnic M, et al. Novel patient-derived models of desmoplastic small round cell tumor confirm a targetable dependency on ERBB signaling. Dis Model Mech 2022;15:dmm047621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fellmann C, Hoffmann T, Sridhar V, Hopfgartner B, Muhar M, Roth M, et al. An optimized microRNA backbone for effective single-copy RNAi. Cell Rep 2013;5:1704–13. [DOI] [PubMed] [Google Scholar]
- 22. Hartono AB, Kang H-J, Shi L, Phipps W, Ungerleider N, Giardina A, et al. Salt-Inducible Kinase 1 is a potential therapeutic target in desmoplastic small round cell tumor. Oncogenesis 2022;11:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinf 2011;12:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012;16:284–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z, et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation (Camb) 2021;2:100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Artibani M, Sims AH, Slight J, Aitken S, Thornburn A, Muir M, et al. WT1 expression in breast cancer disrupts the epithelial/mesenchymal balance of tumour cells and correlates with the metabolic response to docetaxel. Sci Rep 2017;7:45255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Riggi N, Knoechel B, Gillespie SM, Rheinbay E, Boulay G, Suvà ML, et al. EWS-FLI1 utilizes divergent chromatin remodeling mechanisms to directly activate or repress enhancer elements in Ewing sarcoma. Cancer Cell 2014;26:668–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Boulay G, Sandoval GJ, Riggi N, Iyer S, Buisson R, Naigles B, et al. Cancer-specific retargeting of BAF complexes by a prion-like domain. Cell 2017;171:163–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ito K, Nagata K, Ohta S, Matsuda Y, Ukai T, Yasuda I, et al. The oncogene-dependent resistance to reprogramming unveils cancer therapeutic targets. Cell Rep 2022;39:110721. [DOI] [PubMed] [Google Scholar]
- 31. McBride MJ, Pulice JL, Beird HC, Ingram DR, D'Avino AR, Shern JF, et al. The SS18-SSX fusion oncoprotein hijacks BAF complex targeting and function to drive synovial sarcoma. Cancer Cell 2018;33:1128–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods 2012;9:357–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 2009;10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol 2008;9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 2010;38:576–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, et al. Integrative genomics viewer. Nat Biotechnol 2011;29:24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Akalin A, Franke V, Vlahoviček K, Mason CE, Schübeler D. Genomation: a toolkit to summarize, annotate and visualize genomic intervals. Bioinformatics 2015;31:1127–9. [DOI] [PubMed] [Google Scholar]
- 38. Filion C, Motoi T, Olshen AB, Laé M, Emnett RJ, Gutmann DH, et al. The EWSR1/NR4A3 fusion protein of extraskeletal myxoid chondrosarcoma activates the PPARG nuclear receptor gene. J Pathol 2009;217:83–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Griffith M, Griffith OL, Coffman AC, Weible JV, McMichael JF, Spies NC, et al. DGIdb: mining the druggable genome. Nat Methods 2013;10:1209–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. VanArsdale T, Boshoff C, Arndt KT, Abraham RT. Molecular pathways: targeting the cyclin D-CDK4/6 axis for cancer treatment. Clin Cancer Res 2015;21:2905–10. [DOI] [PubMed] [Google Scholar]
- 41. Kennedy AL, Vallurupalli M, Chen L, Crompton B, Cowley G, Vazquez F, et al. Functional, chemical genomic, and super-enhancer screening identify sensitivity to cyclin D1/CDK4 pathway inhibition in Ewing sarcoma. Oncotarget 2015;6:30178–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Knudsen ES, Hutcheson J, Vail P, Witkiewicz AK. Biological specificity of CDK4/6 inhibitors: dose response relationship, in vivo signaling, and composite response signature. Oncotarget 2017;8:43678–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Haber DA, Sohn RL, Buckler AJ, Pelletier J, Call KM, Housman DE. Alternative splicing and genomic structure of the Wilms tumor gene WT1. Proc Natl Acad Sci USA 1991;88:9618–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Magrath JW, Flinchum DA, Hartono AB, Goldberg IN, Espinosa-Cotton M, Moroz K, et al. Genomic breakpoint characterization and transcriptome analysis of metastatic, recurrent desmoplastic small round cell tumor. Sarcoma 2023;2023:6686702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ogura K, Somwar R, Hmeljak J, Magnan H, Benayed R, Momeni Boroujeni A, et al. Therapeutic potential of NTRK3 inhibition in desmoplastic small round cell tumor. Clin Cancer Res 2021;27:1184–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Suvarna K, Jayabal P, Ma X, Shiio Y. Slit2 signaling stimulates Ewing sarcoma growth. Genes Cancer 2022;13:88–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Garcia-Monclús S, López-Alemany R, Almacellas-Rabaiget O, Herrero-Martín D, Huertas-Martinez J, Lagares-Tena L, et al. EphA2 receptor is a key player in the metastatic onset of Ewing sarcoma. Int J Cancer 2018;143:1188–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mehlen P, Delloye-Bourgeois C, Chédotal A. Novel roles for Slits and netrins: axon guidance cues as anticancer targets? Nat Rev Cancer 2011;11:188–97. [DOI] [PubMed] [Google Scholar]
- 49. Thibault S, Hu W, Hirakawa B, Kalabat D, Franks T, Sung T, et al. Intestinal toxicity in rats following administration of CDK4/6 inhibitors is independent of primary pharmacology. Mol Cancer Ther 2019;18:257–66. [DOI] [PubMed] [Google Scholar]
- 50. Francis AM, Alexander A, Liu Y, Vijayaraghavan S, Low KH, Yang D, et al. CDK4/6 inhibitors sensitize rb-positive sarcoma cells to Wee1 kinase inhibition through reversible cell-cycle arrest. Mol Cancer Ther 2017;16:1751–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Asghar US, Kanani R, Roylance R, Mittnacht S. Systematic review of molecular biomarkers predictive of resistance to CDK4/6 inhibition in metastatic breast cancer. JCO Precision Oncology 2022;6:e2100002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Cristofanilli M, Turner NC, Bondarenko I, Ro J, Im SA, Masuda N, et al. Fulvestrant plus palbociclib versus fulvestrant plus placebo for treatment of hormone-receptor-positive, HER2-negative metastatic breast cancer that progressed on previous endocrine therapy (PALOMA-3): final analysis of the multicentre, double-blind, phase 3 randomised controlled trial. Lancet Oncol 2016;17:425–39. [DOI] [PubMed] [Google Scholar]
- 53. Turner NC, Slamon DJ, Ro J, Bondarenko I, Im SA, Masuda N, et al. Overall survival with palbociclib and fulvestrant in advanced breast cancer. N Engl J Med 2018;379:1926–36. [DOI] [PubMed] [Google Scholar]
- 54. Chidharla A, Parsi M, KA. Cetuximab. StatPearls. StatPearls Publishing Copyright © 2023, StatPearls Publishing LLC.: Treasure Island, FL; 2023. [Google Scholar]
- 55. Hsu JY, Seligson ND, Hays JL, Miles WO, Chen JL. Clinical utility of CDK4/6 inhibitors in sarcoma: successes and future challenges. JCO Precis Oncol 2022;6:e2100211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Rader J, Russell MR, Hart LS, Nakazawa MS, Belcastro LT, Martinez D, et al. Dual CDK4/CDK6 inhibition induces cell-cycle arrest and senescence in neuroblastoma. Clin Cancer Res 2013;19:6173–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Shulman DS, Merriam P, Choy E, Guenther LM, Cavanaugh KL, Kao PC, et al. Phase 2 trial of palbociclib and ganitumab in patients with relapsed Ewing sarcoma. Cancer Med 2023;12:15207–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
EWSR1::WT1 Upregulated Pathways
EWSR1::WT1 Downregulated Pathways
Supplementary Figures and Tables
Data Availability Statement
The data generated in this study are publicly available in Gene Expression Omnibus (GEO) at GSE252051. Publicly available data generated by others were used by the authors, including RNA-seq data from GEO (GSE137561, GSE153865, GSE93636, GSE61953, GSE94277, GSE168815, GSE108028), ChIP-seq data from GEO (GSE156277, GSE153865), and microarray data (38). All other raw data generated in this study are available upon request from the corresponding author.