Abstract
B-cell acute lymphoblastic leukemia (B-ALL) remains a hard-to-treat disease with a poor prognosis in adults. Mucosa-associated lymphoid tissue lymphoma translocation protein 1 (MALT1) is a para-caspase required for B-cell receptor (BCR)-mediated NF-κB activation. Inhibition of MALT1 in preclinical models has proven efficacious in many B-cell malignancies including chronic lymphocytic leukemia, mantle cell lymphoma and diffuse large B-cell lymphoma. We sought to examine the role of MALT1 in B-ALL and determine the biological consequences of its inhibition. Targeting MALT1 with both Z-VRPR-fmk and MI-2 efficiently kills B-ALL cells independent of the cell-of-origin (pro, pre, mature) or the presence of the Philadelphia chromosome, and spares normal B cells. The mechanism of cell death was through apoptotic induction, mostly in cycling cells. The proteolytic activity of MALT1 can be studied by measuring its ability to cleave its substrates. Surprisingly, with the exception of mature B-ALL, we did not detect cleavage of MALT1 substrates at baseline, nor after proteasomal inhibition or following activation of pre-BCR. To explore the possibility of a distinct role for MALT1 in B-ALL, independent of signaling through BCR, we studied the changes in gene expression profiling following a 24-hour treatment with MI-2 in 12 B-ALL cell lines. Our transcriptome analysis revealed a strong inhibitory effect on MYC-regulated gene signatures, further confirmed by Myc protein downregulation, concomitant with an increase in the Myc degrader FBXW7. In conclusion, our evidence suggests a novel role for MALT1 in B-ALL through Myc regulation and provides support for clinical testing of MALT1 inhibitors in B-ALL.
Introduction
The dramatic improvement in B-cell acute lymphoblastic leukemia (B-ALL) outcomes with intensive therapy in children has resulted in a 5-year disease-free survival of 75-80% but drops to 25-40% in adults. Moreover, the outcome of patients following disease relapse is very poor, with an overall response rate (ORR) as low as 25-30% with classic chemotherapy.1,2 Despite the promising improvement in ORR with the novel agents blinatumomab (ORR 44%) and inotuzumab ozogamicin (ORR 80.7%), the duration of remission is short, resulting in a very short median overall survival of less than eight months for both drugs.1,2 Similarly, the chimeric antigen receptor (CAR) T-cell therapy tisagenlecleucel resulted in an ORR of 81% in children and young adults; however, 50% relapsed within a year of therapy.3 In keeping with an improvement in overall outcome, compared to classic chemotherapy, blinatumomab, inotuzumab ozogamicin, and tisagenlecleucel were granted approval by the US Food and Drug Administration (FDA) for the treatment of relapsed B-ALL. Thus, identifying alternative targets addresses an unmet need in B-ALL. Elements of the B-cell receptor (BCR) pathway have become viable targets for therapeutic applications using small molecule inhibitors. As an example, the Bruton’s tyrosine kinase (BTK) inhibitor ibrutinib, which is the most extensively studied example to date, has transformed the therapeutic approach in a myriad of B-cell malignancies, such as in chronic lymphocytic leukemia (CLL), mantle cell lymphoma (MCL), marginal zone lymphoma (MZL), and Waldenström’s macroglobulinemia (WM). The high rates of clinical response to BCR pathway inhibition in CLL match well with genetic and functional data on the role of BCR signaling in this disease, and in WM, where the hallmark MYD88 mutation activates BTK-dependent signaling.4,5 Similarly, ibrutinib’s clinical efficacy in MZL is based on antigen-mediated BCR activation caused by chronic infections (e.g., hepatitis C virus, Helicobacter pylori), as well as antigen-independent activation through acquired genetic alterations (e.g., API2-MALT1 fusion and TNFAIP3 loss of function).6,7 In contrast, the response in MCL was more surprising.8 However, our data indicate a central role for BCR activation in the development and progression of this lymphoma.9
While elements of, and signaling through, BCR are well-defined, those of precursor BCR (pre-BCR) remain ill-characterized. In B-ALL, early data suggest that signaling through pre-BCR is ongoing, and is likely altered and linked to disease biology.10 Moreover, we have previously demonstrated in B-ALL that targeting PKCβ, a critical element of the pre-BCR, with the small molecule inhibitor enzastaurin inhibits pre-BCR signaling resulting in apoptosis and cell cycle arrest through alteration of the WNT/β-catenin pathway.11 Similarly, our collaborators demonstrated that ibrutinib suppresses proliferation of B-ALL in vitro and in vivo, by targeting two components of the pre-BCR signaling pathway, BTK and BLK.12 Interestingly, by gene expression analysis, MALT1 was found to be up-regulated in primary B-ALL cells compared to the normal control, and is coupled with a stronger NF-κB activity.13 Notably, the chemical compounds MI-2 (C19H-17Cl3N4O3) and the highly selective MALT1 blocking peptide Z-VRPR-fmk bind to and suppress the protease activity of MALT1 through irreversibly inhibiting cleavage of MALT1 substrates in activated B-cell-like diffuse large B-cell lymphoma (ABC-DLBCL).14,15 We have recently characterized the efficacy and downstream effects of targeting MALT1 with MI-2 in CLL. We showed that MALT1 is constitutively active in CLL and can be effectively inhibited by MI-2, resulting in reduced BCR and NF-κB signaling. Importantly, MI-2 remained highly effective against CLL cells harboring mutations conferring ibrutinib-resistance.16
An alternative, less characterized role for MALT1, has been proposed through the activation of MYC signaling. Dai and colleagues demonstrated that MALT1 pharmacological inhibition and knockout in MCL, resulted in MYC downregulation. However, the mechanism by which MALT1 contributes to Myc stability remains ill-defined.17
The possible therapeutic role of MALT1 in ALL has not been investigated. We hypothesize that MALT1 is critical for B-ALL survival and that its inhibition could have anti-leukemic activity in ALL. Further, given the emerging role for Myc in B-ALL, we hypothesize that MALT1 inhibition could be effective in targeting Myc indirectly through effects on Myc stabilizers.18-22
Methods
Patients and samples
Following written informed consent, and in accordance with the Declaration of Helsinki and under the oversight of the Institutional Review Board at Tulane University (New Orleans, LA, USA; N M0600) peripheral blood samples were obtained from patients with treatment-naïve or relapsed ALL, as well as from patients with various blood cancers presenting with a leukemic phase. Peripheral blood mononuclear cells (PB-MC) were isolated using density gradient centrifugation with lymphocyte separation medium (ICN Biomedicals). Fresh cells were subjected to CD19 selection using magnetic beads yielding purity >96% (Miltenyi Biotec).
A total of 23 cell lines representing the wide spectrum of ALL biology (Online Supplementary Table S1) and TMD8 controls were kindly provided by Dr. Burger’s group at MD Anderson Cancer Center or purchased from ATCC and DSMZ and expanded in vitro. All cell lines were authenticated by short tandem repeat profiling and tested for mycoplasma contamination (Labcorp; Burlington, NC, USA).
Reagents
MI-2 was purchased from MedChemExpress (MCE). The irreversible polyamino-acid MALT1 inhibitor Z-VRPR-fmk (Z-Val-Arg-Pro-DL-Arg-FMK) was purchased from Tocris. Reagents were dissolved in their respective solvents (DMSO for MI-2 or diH2O for Z-VRPR-fmk) and aliquoted in microtubes to be then stored at -80°C.
Cell viability assay
CellTiter 961 AQueous One Solution Reagent (Promega) MTS dye reduction assay was used to quantify cell viability. Assay details are provided in the Online Supplementary Appendix.
Immunoblot
Immunoblot experiments were carried out as previously described.16 Primary and secondary antibodies used are detailed in the Online Supplementary Appendix.
GloSensor assay
GloSensor assay was used to quantify the enzymatic activity of MALT1 as previously described.15,23 In brief, the pGloSensor plasmid was custom designed to have 2 luciferase domains linked by the MALT1 proteolytic substrate sequence of RelB, MALT1-GloSensor reporter (Promega). Cleavage of the linker sequence by MALT1 allows conformational changes promoting a large increase in luciferase activity.
Flow cytometry
Cells were stained with anti-Annexin V or with propidium iodide (PI) as previously described,11,16 following treatment with various concentrations of MI-2 or DMSO (control). Cells were then analyzed by flow cytometry as previously described.11,16 FloJo was used for analysis of flow cytometry results.
RNA sequencing and gene expression analysis
RNA and cDNA were prepared using the High Capacity cDNA RT Kit (Applied Biosystems). Total RNA was extracted from MI-2-treated and untreated ALL cell lines using the RNeasy kit (Qiagen) then subjected to RNA sequencing as previously described.16 RNA sequencing data have been deposited in GEO under accession number GSE221273.
Statistical analysis
Student t test (paired or unpaired), Fisher’s exact test, and one-way analysis of variance were used to assess differences between groups. All P values were two-sided; P≤0.05 was considered statistically significant. Analyses were performed using GraphPad Prism (GraphPad Software Inc.) and JMP software (SAS Institute).
Results
MALT1 plays a critical role in B-cell acute lymphoblastic leukemia cell survival
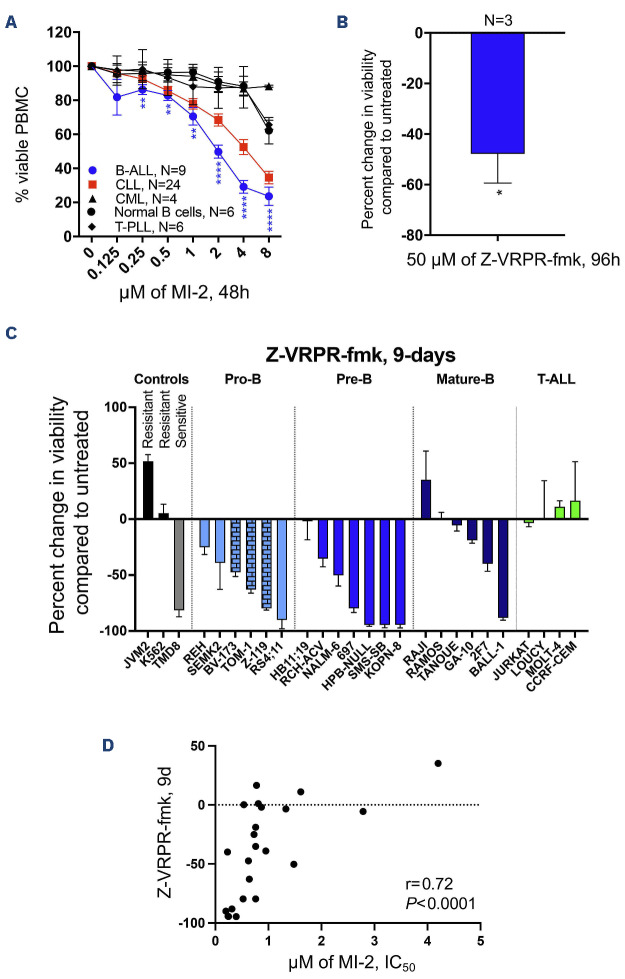
We obtained freshly collected PBMC from patients with various blood cancers presenting with a leukemic phase and determined the anti-tumor activity of MALT1 inhibition using serial concentrations of MI-2 for 48 hours (h). Samples tested were from patients with B-ALL (N=9), CLL (N=24), T-prolymphocytic leukemia (T-PLL, N=6), chronic myeloid leukemia (CML, N=4), as well as normal B cells collected from 6 volunteers. Surprisingly, B-ALL samples showed the highest sensitivity to MI-2, followed by CLL. Normal B cells, T-PLL and CML, were relatively resistant to cell killing by MI-2 (Figure 1A). To further confirm the efficacy of MALT1 inhibition against ALL, we used the highly selective MALT1 blocking peptide Z-VRPR-fmk to treat 3 CD19-selected primary ALL cells and 23 ALL cell lines representing the disease spectrum. Z-VRPR-fmk at 50 mM resulted in a significant reduction in viability of the patient-derived samples following a 96-h single treatment (Figure 1B) and in most of our B-ALL cell lines following 3 treatments over nine days (Figure 1C). Z-VRPR-fmk is a large molecule that requires a high concentration and a long incubation time to ensure cell penetration. The concentration and incubation times we used are consistent with those used across the literature.24 Notably, the cell killing in B-ALL was independent of the cell-of-origin (pro, pre, mature), or the presence or absence of the Philadelphia chromosome. The sensitivity of B-ALL contrasts with that of the T-ALL cell lines that were all resistant to MALT1 inhibition (Figure 1C). Further, we noted the same pattern of sensitivity when all cell lines were treated with MI-2 for 48 h, as demonstrated by the statistically significant correlation between the percentage of Z-VRPR-fmk-induced growth inhibition and the half maximal inhibitory concentration (IC50) of MI-2 (r=0.72, P<0.0001) (Figure 1D).
MALT1-dependent cell lines possess a protease active or inactive MALT1 in B-cell acute lymphoblastic leukemia
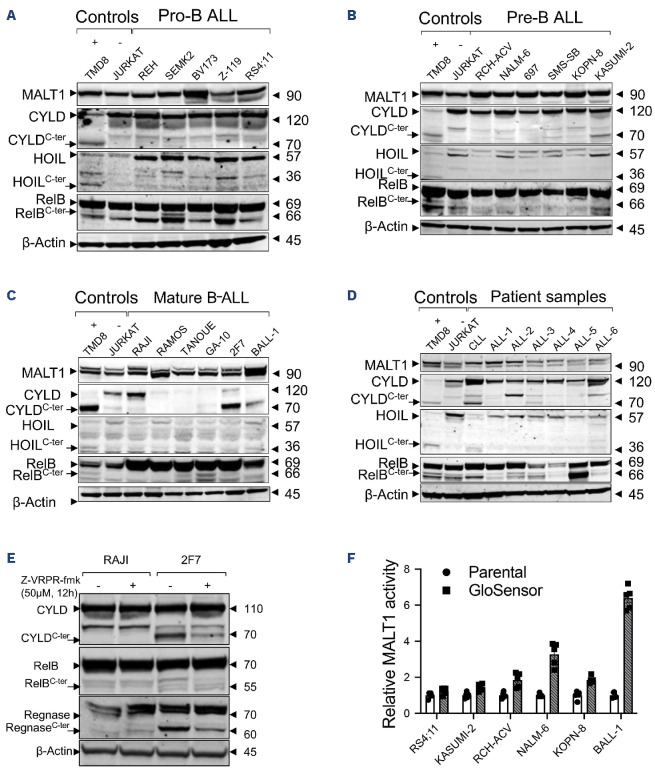
The proteolytic activity of MALT1 can be studied by measuring its ability to cleave its substrates. Known MALT1 substrates include CYLD, HOIL, RelB, A20, Bcl10, Roquin, and Regnase. In CLL, we found that MALT1 is constitutively active through its protease function, likely as part of BCR signaling, as evident by the presence of MALT1’s substrate cleavage.16 As an example, we measured cleaved CYLD as the degree of MALT1 activity.16,25 We used immunoblotting with an antibody specific for the C-terminal (C-ter) region of CYLD, to detect full length CYLD (120 kDa) and the C-ter cleaved product (CYLDC-ter ~70KDa). In B-ALL, we sought to determine whether the anti-leukemic effect observed with MALT1 inhibition correlates with the proteolytic activity of MALT1. Surprisingly, despite the expression of MALT1 in all ALL cell lines and primary cells, substrate cleavage was not detected in most samples, including the most sensitive to MALT1 inhibition (Figure 2A-D). More specifically, none of the pro or pre B-ALL cell lines showed active MALT1 protease activity. This proteolytic activity remained absent after proteasomal inhibition (to protect cleaved products from proteasomal degradation) or following crosslinking of pre-BCR with anti-IgM in pre B-ALL (Online Supplementary Figure S1). In contrast, mature B-ALL displayed a pattern similar to MCL and DLBCL, where two groups were identified: a group with active MALT1 protease and sensitive to MALT1 inhibition (BALL-1, GA-10, 2F7), and a second group with inactive MALT1 protease and resistant to MALT1 inhibition (TANOUE, RAJI, RAMOS) (Figures 1C, 2C). The effect of MALT1 inhibition on substrate cleavage was further confirmed by a reduction of the cleaved fractions of the substrates CYLD, RelB and Regnase following a 12-h incubation with Z-VRPR-fmk in the MALT1-dependent cell line 2F7, contrary to the MALT1 independent cell line RAJI where cleavage did not occur (Figure 2E). We did not observe any proteolytic activity in the tested T-ALL cell lines (data not shown), all of which were resistant to MALT1 inhibition (Figure 1C). In patient-derived samples, we detected a similar pattern in CD19 selected lymphoblasts derived from patients with B-ALL where substrate cleavage was absent in most samples (Figure 2D). The contrast between sensitivity to MALT1 inhibition and absence of proteolytic activity suggests either a protease-independent role for MALT1 or a weak enzymatic activity below the immunoblot detection level. To investigate a possible ongoing low proteolytic activity for MALT1, we used the previously described MALT1-specific GloSensor reporter assay in a set of MI-2 sensitive cell lines (RS4;11, KASUMI-2, RCH-ACV, NALM-6, KOPN-8 and BALL-1). The MALT1 GloSensor assay uses a split luciferase method, in which the luciferase activity is triggered by MALT1-induced cleavage resulting in luminescence. This luciferase activity is highly specific and a surrogate for MALT1 endogenous proteolytic function. Unsurprisingly, BALL-1 showed the highest activity with more than a 6-fold increase in bioluminescence, compared to the rest of the tested cell lines, where a low or no activity was detected (Figure 2F). These results highly suggest a novel, protease-independent role for MALT1 in B-ALL.
MYC signaling is activated by MALT1 independently of BCR signaling
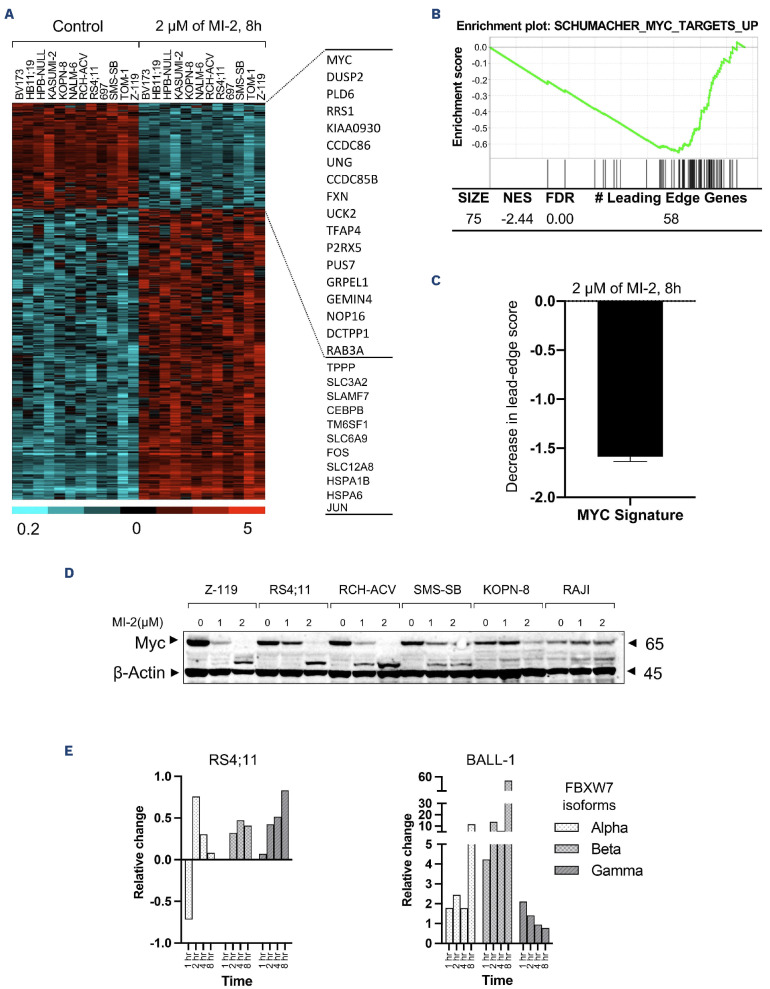
Our transcriptome analysis in CLL cells treated with MI-2 revealed the expected downregulation of NF-κB target genes, confirmed by decreased levels of nuclear p50, and select NF-κB regulated molecules, in particular Bcl-xL.16 On the other hand, the high sensitivity of most pro and pre B-ALL to MALT1 inhibition contrasts with the absence of a canonical substrate cleavage and suggests a potentially novel role for this protease in B-ALL. Similarly, the high sensitivity of 2 ibrutinib-resistant cell lines RS4;11 and 697 (as determined by Dr. Burger’s group)12 to Z-VRPR-fmk and MI-2 implicates a distinct role for MALT1 in these cell lines, arguably independent of BTK and of signaling through BCR. To explore this possibility, we used RNA sequencing to determine the changes in gene expression profiling (GEP) in 12 cell lines (pro and pre B) following an 8-h treatment with MI-2. Out of 19,428 tested genes, there were 1,036 whose expression changed ≥2-fold at false detection rate (FDR) <0.1 (275 down- and 761 up-regulated) (Figure 3A, Online Supplementary Table S2). Gene Set Enrichment Analysis (GSEA) identified 13 Oncogenic Signature gene sets at FDR <1%, and a normalized enrichment score (NES) ≥1.70, all of which were down-regulated by MI-2. Interestingly, BCR and NF-KB regulated gene signatures were not identified by GSEA as being significantly affected by MI-2. Instead, MYC-regulated gene signatures were strongly represented (5 out of 15 gene sets, including the top 2 affected signatures, strongest at NES of -2.45 and FDR of 0.00) (Figure 3B, C). Moreover, the MYC gene was found to be more than 3.5-fold down-regulated by MI-2. Most of the up-regulated genes were apoptotic/oxidative stress-related genes or pseudogenes such as JUN, CEBP-B and SLAM-7 (Figure 3A). In B-ALL, Myc is not a known target for MALT1. To investigate whether MALT1 contributes to Myc stability, we measured Myc protein expression in whole-cell lysates following treatment with increment concentration of MI-2 for 8 h. We observed a striking reduction of Myc in the tested sensitive cell lines (Z-119, RS4;11, RCH-ACV, SMS-SB, and KOPN-8), an unchanged expression in the sensitive mature cell line BALL-1 (Online Supplementary Figure S2), and an increase in the resistant cell line RAJI (Figure 3D). These findings suggest a previously unrevealed effect of MALT1 on Myc in pro and pre B-ALL, but not in mature B-ALL. Interestingly, RAJI carries a mutation in Myc at the threonine-58 residue which prohibits Myc phosphorylation at this site.26 Phosphorylation of Myc at threonine-58 and serine-62 is required for its ubiquitination by FBXW7 and subsequent proteasomal degradation.27 FBXW7, with its 3 isoforms a, |3 and y, is a component of a ubiquitin ligase complex responsible for the degradation of a number of oncoproteins such as c-Jun, Cyclin E, mTOR and Myc.28 Thus FBXW7 isoforms were examined by immunoblot in the 2 sensitive cell lines RS4;11 and BALL-1 following treatment with 1 mM of MI-2 and sampled at regular intervals for immunoblot analysis. Our results showed that the expression of all 3 isoforms of FBXW7 are enhanced following MALT1 inhibition with MI-2 (Figure 3E). While the upregulation of FBXW7 in the pro B-ALL cell line RS4;11 is concordant with a marked downregulation of Myc, we did not observe the same trend in mature B-ALL cell line BALL-1, in which Myc expression seems to be independent of MALT1 (Figure 3D, E, Online Supplementary Figure S2). These results highly suggest that FBXW7 is a new potential substrate for MALT1 and link RAJI resistance to increased Myc stability, which is shielded from MALT1 inhibition through a mutated threonine-58. Our transcriptome data showed a marked downregulation of MYC mRNA levels in most of the MI-2-treated cell lines (Figure 3A, Online Supplementary Figure S3A). Nevertheless, 3 of the cell lines with available MYC mRNA and protein expression data, that showed the least downregulation of MYC mRNA following MI-2 treatment (KOPN8, no change; SMS-SB, 3-fold change; RCH-ACV, 3.4-fold change) (Online Supplementary Figure S3A), showed a near complete depletion of Myc protein following MI-2 treatment (Figure 3D). Signaling through pre-BCR activates BCL6, which transcriptionally represses MYC.29 While our gene expression data show a marked decrease in the pre-BCR component VPREB1 and BCL6 following treatment with MI-2 (Online Supplementary Figure S3B, C), our analysis for the corresponding proteins did not yield significant changes in VPREB1 nor BCL6, as measured by flow cytometry and western blot, respectively (Online Supplementary Figure S3D, E). Collectively, these data suggest a direct effect of MALT1 on MYC/FBXW7 protein axis, rather than indirectly through effects on pre-BCR and BCL6.
Figure 1.
Targeting mucosa-associated lymphoid tissue lymphoma translocation protein 1 efficiently kills B-cell acute lymphoblastic leukemia cells. (A) Peripheral blood mononuclear cells (PBMC) from patients with B-cell acute lymphoblastic leukemia (B-ALL), T-cell acute lymphoblastic leukemia (T-ALL), chronic myeloid leukemia (CML), chronic lymphocytic leukemia (CLL), or from healthy individuals were subjected to treatment with increasing doses of MI-2 for 48 hours (h). Cell viability was quantified using an MTS assay and is shown as percentage of the untreated control. (B) CD19-selected primary B-ALL cells collected from 3 patients were treated with the highly selective mucosa-associated lymphoid tissue lymphoma translocation protein 1 (MALT1) inhibitor Z-VRPR-fmk at 50 mM for 96 h. Cell viability was quantified using an MTS assay and is shown as a percentage of untreated control. (C) Waterfall plot showing percent changes in cell viability from baseline, quantified using an MTS assay, following a 9-day (9d) treatment with 50 mM Z-VRPR-fmk. Cells were split every three days. A total of 23 ALL cell lines tested representing the disease spectrum. Positive (TMD8) and negative (K562 and JVM2) controls are shown. (D) Correlation between percent change in cell viability following treatment with Z-VRPR-fmk as shown in (C), and the half maximal inhibitory concentration (IC50) for MI-2 of B-ALL cell lines. T-PLL: T-cell prolymphocytic leukemia. *P<0.05. Error bars represent Standard Error of Mean. Columns with a brick pattern identify Philadelphia chromosome-positive cell lines.
Figure 2.
Mucosa-associated lymphoid tissue lymphoma translocation protein 1 protease activity in B-cell acute lymphoblastic leukemia cell lines and primary samples. Immunoblot analysis showing expression of mucosa-associated lymphoid tissue lymphoma translocation protein 1 (MALT1) and its substrates (CYLD, HOIL, RelB) in (A) 5 pro B-cell acute lymphoblastic leukemia (B-ALL) cell lines; (B) 6 pre-B-ALL cell lines; (C) 6 mature B-ALL cell lines; (D) CD19-selected primary samples representing 6 B-ALL and one chronic lymphocytic leukemia sample. TMD8 and JURKAT were used as positive and negative controls, respectively, for substrate cleavage. (E) Immunoblot analysis showing inhibition of CYLD, RelB, and Regnase cleavage by MALT1 following a 12-h treatment with Z-VRPR-fmk in the mature B-ALL cell line 2F7. RAJI is shown as a negative control. (F) Cell-based reporter GloSensor assay showing intrinsic MALT1 proteolytic activity in the cell lines is shown. C-ter : cleaved substrates.
MI-2 restores apoptosis and causes cell cycle arrest in B-cell acute lymphoblastic leukemia cells
Finally, we sought to determine the mechanism of cell killing with MI-2 in ALL cells.
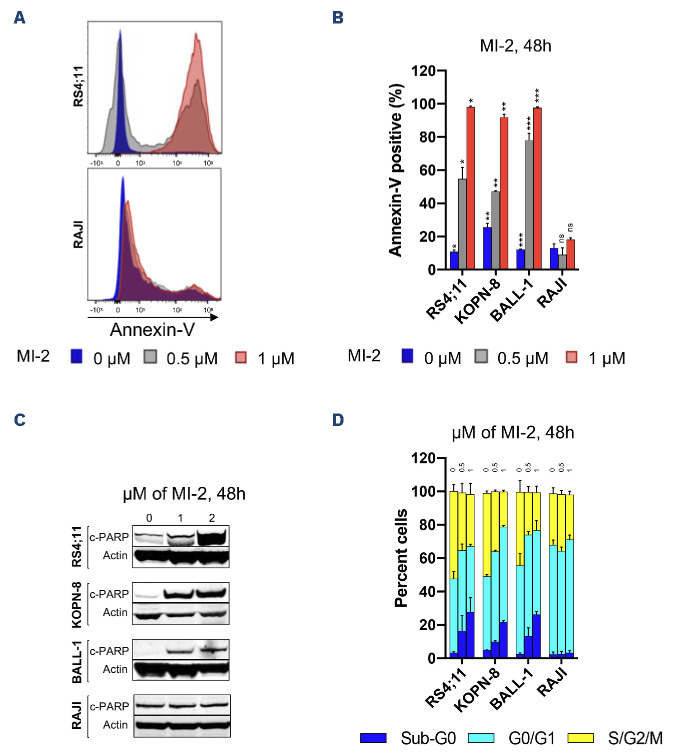
To assess cytotoxicity against B-ALL cell lines, we measured the cell viability by flow cytometry using Annexin-V following exposure to MI-2 for 48 h. We observed a dose-dependent increase in apoptosis as evident by the increase in the Annexin-V-positive fraction in the MALT1-dependent cell lines RS4;11, KOPN-8 and BALL-1, but not in the resistant RAJI cell line (Figure 4A). Apoptotic induction was further confirmed by the increase in PARP cleavage in the sensitive cell lines, but not in RAJI (Figure 4B). Next, we assessed the effect of MALT1-inhibition on cell cycle progression by PI staining and flow cytometry. The results demonstrated a significant inhibition of cell cycle progression in the sensitive cell lines RS4;11, KOPN-8 and BALL-1 after a 48-h treatment at 0.5 and 1 mM (Figure 4C). By comparison, cell cycle progression in RAJI was not affected. Notably, the reduction in the cycling population (S/ G2/M) was associated with an equivalent increase in the subG0 fraction, and a nearly unchanged G0/G1 fraction. This pattern of cell cycle changes, coupled with a strong apoptotic induction, indicate that the mechanism of MI-2 killing is through induction of apoptosis selectively in cycling cells.
Discussion
Forty percent of patients diagnosed with ALL in 2020 died.30 Death rate dramatically increases with age, with the majority of those older than 65 years succumbing to their disease. Thus, novel therapeutic strategies are needed to improve survival, particularly in those who suffer a disease relapse. The oncogenic role of BCR or its precursor, pre-BCR, in the biology of B-ALL has been the subject of intense focus in the field.31,34 We and our collaborators have shown that targeting elements of the BCR pathway such as BTK, PI3K5, and PKCb is effective in vitro on a variety of B-ALL cell lines and primary samples.11,35,36 While small molecule inhibitors of the BCR signaling pathway have transformed the way we treat B-cell malignancies, their role in B-ALL remains limited without a significant therapeutic application (clinicaltrials. gov identifiers: NCT02129062, terminated; NCT02404220, terminated; NCT01396499, terminated; NCT03267186, active; NCT02997761, active; NCT04803123, active). Hence, there is a crucial need to further understand the role of this pathway and each of its components in B-ALL biology. Here, we show for the first time that MALT1 is critical for B-ALL survival, and that its inhibition is highly efficacious in killing leukemic cells despite MALT1’s proteolytic activity being detected only in a small subset of this disease. Targeting MALT1 was also equally effective against B-ALL cells regardless of the cell of origin or the presence of the Philadelphia chromosome.
Figure 3.
Mucosa-associated lymphoid tissue lymphoma translocation protein 1 regulates MYC signaling in B-cell acute lymphoblastic leukemia. (A) RNA gene expression analysis of 12 B-cell acute lymphoblastic leukemia (B-ALL) cell lines treated with 2 mM of MI-2 for 8 hours (h) was performed by RNA sequencing. The heatmap represents 1,036 whose expression changed ≥ 2-fold at FDR < 0.1 (275 down- and 761 up-regulated). Gene expression is median centered and scaled as indicated. Each column represents a sample, and each row represents a gene. Gene symbols highlight select genes. (B) An enrichment plot representing the inhibitory effect of MI-2 on the most affected MYC gene set as identified by gene set enrichment analysis. (C) Decrease in the signature score of the MYC gene set shown in (B) computed as the average of the mRNA expression level of the leading edge genes of MI-2–treated samples minus the corresponding control. The leading edge genes represent the genes of this MYC gene set most significantly differentially expressed in the experimental data, as determined by gene set enrichment analysis. (D) Immunoblot showing Myc expression changes in whole cell lysates following 0-2 mM MI-2 treatment for 8 h. (E) Quantification of FBXW7 isoforms changes (relative change/Actin) following treatment with 1 mM MI-2 for 1-8 h. FDR: false discovery rate; NES: normalized enrichment score; MALT1: mucosa-associated lymphoid tissue lymphoma translocation protein 1.
Figure 4.
MI-2 induces apoptosis and cell cycle arrest in B-cell acute lymphoblastic leukemia cell lines. (A) Apoptotic induction quantified by flow cytometry using Annexin-V staining in the shown cell lines following treatment with 0-1 mM of MI-2 for 48 hours (h). (B) Immunoblot showing the change in expression of c-PARP in B-cell acute lymphoblastic leukemia (B-ALL) cell lines following treatment with 0-2 mM of MI-2 for 48 h. (C) Changes in cell cycle quantified by flow cytometry using propidium iodide staining in the shown cell lines following treatment with 0-1 mM of MI-2 for 48 h. *P<0.05, **P< 0.01, ***P<0.001. ns: not significant. Error bars represent Standard Error of Mean.
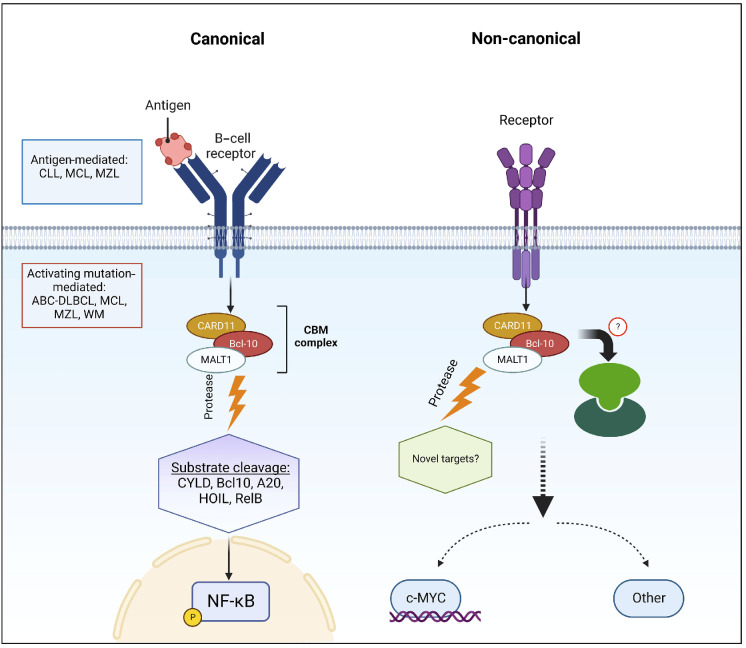
In B-cell non-Hodgkin lymphoma (NHL), triggering the BCR pathway can be achieved by either antigenic stimulation or through oncogenic mutations that confer an antigen-independent NF-κB activation.14,16,17 Within this pathway, MALT1 connects signaling through BCR to NF-κB activation. We have shown that, in CLL, MALT1 is constitutively activated, likely through antigen stimulation, and is turned off by MI-2.16 In MCL and DLBCL,14,17 two groups were identified: a group with active MALT1 protease and sensitive to MALT1 inhibition, and a second group with inactive MALT1 protease and resistance to MALT1 inhibition. We observed a similar pattern of a two-group split based on MALT1’s protease activity and sensitivity to inhibition in mature B-ALL. Whereas pro and pre B-ALL were sensitive to MALT1 inhibition despite an apparent protease-dead MALT1. Unlike other B-cell malignancies, where the canonical activity of MALT1 pivots around NF-κB, the role of MALT1 in the biology of pro and pre B-ALL is likely through a potentiation of Myc signaling. MALT1 has been shown to stabilize Myc in MCL, and that function was found to be tightly correlated with MALT1’s protease activity.17 In contrast, our data highly suggest that Myc stabilization through MALT1 is independent of its protease activity in pro and pre B-ALL, and is achieved rather through a negative impact on FBXW7. The impact on Myc irrespective of the proteolytic activity of MALT1 highly suggests a non-enzymatic, or scaffolding, non-canonical activity of this protease, or to a lesser extent, an unappreciated proteolytic activity against a distinct set of substrates compared to those identified in B-cell NHL. On the other hand, the reduction of the classic substrate cleavage (CYLD, RelB, and Regnase) in mature B-ALL following MALT1 inhibition is consistent with current knowledge, and implicates, at least in part, a canonical role for MALT1 in Myc regulation in mature B-ALL. Moreover, the discordance between pro/pre B-ALL and mature B-ALL regarding the effect of MALT1 on Myc highly suggests that a switch in MALT1 signaling from non-canonical to canonical occurs at the mature B-cell level in which a BCR pathway is fully differentiated. These findings support the view that a canonical MALT1 effect is contingent of a fully developed BCR pathway (Figure 5).
Figure 5.
Canonical and non-canonical mucosa-associated lymphoid tissue lymphoma translocation protein 1-dependent pathways in B-cell acute lymphoblastic leukemia. CLL: chronic lymphocytic leukemia; MCL: mantle cell lymphoma; MZL: marginal zone lymphoma; ABC-DLBCL: activated B-cell-like diffuse large B-cell lymphoma; WM: Waldenström’s macroglobulinemia; MALT1: mucosa-associated lymphoid tissue lymphoma translocation protein 1.
MI-2 is selectively toxic for B-ALL cells, while sparing non-malignant B cells. MI-2 is also ineffective against CML, T-PLL, and T-ALL underscoring a potential exclusive role for MALT1 in B-cell malignancies among blood cancers. The observed inefficacy of MI-2 in T-ALL, coupled with the absence of MALT1 protease activity contrast with a previous report where MI-2 resulted in apoptosis and inhibition of NF-κB activation in 2 T-ALL cell lines. No further analysis was performed on MALT1’s substrates to investigate the presence or absence of the proteolytic activity of MALT1.37 In CLL, MI-2 triggers significant apoptosis, which contrasts with the mechanism of cell growth inhibition seen with BCR inhibitors, particularly ibrutinib where little apoptosis is seen even at high concentrations.38-40 Similarly, apoptotic induction was the main mechanism of MI-2’s cell killing in B-ALL, while ibrutinib’s toxic effects were through inhibition of cell proliferation rather than apoptosis.12 Interestingly, we noted a relative killing selectivity in cycling cells further implicating Myc, a potent driver of cell cycle progression, in B-ALL biology and this underscores the MALT1-Myc interplay in this disease. In conclusion, these data identify MALT1 as a promising therapeutic candidate in B-ALL. Akin to NHL (NCT04876092, NCT03900598, NCT04859777, NCT05144347), MALT1 inhibitors should be investigated in clinical trials for patients with B-ALL.
Supplementary Material
Acknowledgments
We thank our patients for participating and donating the blood and tissue samples to make this research possible. We thank Melody Baddoo and the members of the Cancer Genetics Program - COBRE core at Tulane University for assistance with gene expression profiling and RNA sequencing analysis. We thank Ekaterina Kim for providing cell lines used in this project. We thank Prescott Deininger for his helpful advice. We also thank the Louisiana Cancer Research Center (LCRC) bio-specimen core for their help in blood sample collection and processing.
Funding Statement
Funding: This work was funded by the Ladies Leukemia League Inc., of the Gulf South Region and the Intramural Research Program of the National Heart, Lung, and Blood Institute, NIH (grant n. ZIA HL002346-13; primary recipient, A. Wiestner).
Data-sharing statement
RNA sequencing data supporting this publication are available from the GEO repository under the accession number GSE221273 . https://www.ncbi.nlm.nih.gov/geo/
References
- 1.Kantarjian H, Stein A, Gökbuget N, et al. Blinatumomab versus chemotherapy for advanced acute lymphoblastic leukemia. N Engl J Med. 2017;376(9):836-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kantarjian HM, DeAngelo DJ, Stelljes M, et al. Inotuzumab ozogamicin versus standard therapy for acute lymphoblastic leukemia. N Engl J Med. 2016;375(8):740-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maude SL, Laetsch TW, Buechner J, et al. Tisagenlecleucel in children and young adults with B-cell lymphoblastic leukemia. N Engl J Med. 2018;378(5):439-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Herishanu Y, Perez-Galan P, Liu D, et al. The lymph node microenvironment promotes B-cell receptor signaling, NF-kappaB activation, and tumor proliferation in chronic lymphocytic leukemia. Blood. 2011;117(2):563-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Treon SP, Tripsas CK, Meid K, et al. Ibrutinib in previously treated Waldenstrom’s macroglobulinemia. N Engl J Med. 2015;372(15):1430-1440. [DOI] [PubMed] [Google Scholar]
- 6.Suarez F, Lortholary O, Hermine O, Lecuit M. Infection-associated lymphomas derived from marginal zone B cells: a model of antigen-driven lymphoproliferation. Blood. 2006;107(8):3034-3044. [DOI] [PubMed] [Google Scholar]
- 7.Bertoni F, Rossi D, Zucca E. Recent advances in understanding the biology of marginal zone lymphoma. F1000Res. 2018;7:406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang ML, Rule S, Martin P, et al. Targeting BTK with ibrutinib in relapsed or refractory mantle-cell lymphoma. N Engl J Med. 2013;369(6):507-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saba NS, Liu D, Herman SE, et al. Pathogenic role of B-cell receptor signaling and canonical NF-kappaB activation in mantle cell lymphoma. Blood. 2016;128(1):82-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kohrer S, Havranek O, Seyfried F, et al. Pre-BCR signaling in precursor B-cell acute lymphoblastic leukemia regulates PI3K/ AKT, FOXO1 and MYC, and can be targeted by SYK inhibition. Leukemia. 2016;30(6):1246-1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saba NS, Angelova M, Lobelle-Rich PA, Levy LS. Disruption of pre-B-cell receptor signaling jams the WNT/beta-catenin pathway and induces cell death in B-cell acute lymphoblastic leukemia cell lines. Leuk Res. 2015;S0145-2126(15):30355-30356. [DOI] [PubMed] [Google Scholar]
- 12.Kim E, Hurtz C, Koehrer S, et al. Ibrutinib inhibits pre-BCR(+) B-cell acute lymphoblastic leukemia progression by targeting BTK and BLK. Blood. 2017;129(9):1155-1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu Y, Hu J, Wang X, et al. Overexpression of MALT1-A20-NF-kappaB in adult B-cell acute lymphoblastic leukemia. Cancer Cell Int. 2015;15:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fontan L, Yang C, Kabaleeswaran V, et al. MALT1 small molecule inhibitors specifically suppress ABC-DLBCL in vitro and in vivo. Cancer Cell. 2012;22(6):812-824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fontan L, Qiao Q, Hatcher JM, et al. Specific covalent inhibition of MALT1 paracaspase suppresses B cell lymphoma growth. J Clin Invest. 2018;128(10):4397-4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saba NS, Wong DH, Tanios G, et al. MALT1 inhibition is efficacious in both naïve and ibrutinib-resistant chronic lymphocytic leukemia. Cancer Res. 2017;77(24):7038-7048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dai B, Grau M, Juilland M, et al. B-cell receptor-driven MALT1 activity regulates MYC signaling in mantle cell lymphoma. Blood. 2017;129(3):333-346. [DOI] [PubMed] [Google Scholar]
- 18.Allen A, Gill K, Hoehn D, et al. C-myc protein expression in B-cell acute lymphoblastic leukemia, prognostic significance? Leuk Res. 2014;38(9):1061-1066. [DOI] [PubMed] [Google Scholar]
- 19.Ge Z, Guo X, Li J, et al. Clinical significance of high c-MYC and low MYCBP2 expression and their association with Ikaros dysfunction in adult acute lymphoblastic leukemia. Oncotarget. 2015;6(39):42300-42311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sheikh-Zeineddini N, Safaroghli-Azar A, Salari S, Bashash D. C-Myc inhibition sensitizes pre-B ALL cells to the anti-tumor effect of vincristine by altering apoptosis and autophagy: proposing a probable mechanism of action for 10058-F4. Eur J Pharmacol. 2020;870:172821. [DOI] [PubMed] [Google Scholar]
- 21.Folgiero V, Sorino C, Pallocca M, et al. Che-1 is targeted by c-Myc to sustain proliferation in pre-B-cell acute lymphoblastic leukemia. EMBO Rep. 2018;19(3):e44871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wossning T, Herzog S, Kohler F, et al. Deregulated Syk inhibits differentiation and induces growth factor-independent proliferation of pre-B cells. J Exp Med. 2006;203(13):2829-2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xia M, David L, Teater M, et al. BCL10 mutations define distinct dependencies guiding precision therapy for DLBCL. Cancer Discov. 2022;12(8):1922-1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rebeaud F, Hailfinger S, Posevitz-Fejfar A, et al. The proteolytic activity of the paracaspase MALT1 is key in T cell activation. Nat Immunol. 2008;9(3):272-281. [DOI] [PubMed] [Google Scholar]
- 25.Ginster S, Bardet M, Unterreiner A, et al. Two antagonistic MALT1 auto-cleavage mechanisms reveal a role for TRAF6 to unleash MALT1 activation. PLoS One. 2017;12(1):e0169026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bahram F, von der Lehr N, Cetinkaya C, Larsson LG. c-Myc hot spot mutations in lymphomas result in inefficient ubiquitination and decreased proteasome-mediated turnover. Blood. 2000;95(6):2104-2110. [PubMed] [Google Scholar]
- 27.Yada M, Hatakeyama S, Kamura T, et al. Phosphorylation-dependent degradation of c-Myc is mediated by the F-box protein Fbw7. EMBO J. 2004;23(10):2116-2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Welcker M, Clurman BE. FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nat Rev Cancer. 2008;8(2):83-93. [DOI] [PubMed] [Google Scholar]
- 29.Nahar R, Ramezani-Rad P, Mossner M, et al. Pre-B cell receptor-mediated activation of BCL6 induces pre-B cell quiescence through transcriptional repression of MYC. Blood. 2011;118(15):4174-4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.National Cancer Institute SRP. Surveillance,Epidemiology, and End Results (SEER) Program]. National Cancer Institute, Surveillance Research Program. https://seer.cancer.gov/seertools/hemelymph/51f6cf58e3e27c3994bd540e/. Accessed September 6, 2023. [Google Scholar]
- 31.Young RM, Staudt LM. Targeting pathological B cell receptor signalling in lymphoid malignancies. Nat Rev Drug Discov. 2013;12(3):229-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rickert RC. New insights into pre-BCR and BCR signalling with relevance to B cell malignancies. Nat Rev Immunol. 2013;13(8):578-591. [DOI] [PubMed] [Google Scholar]
- 33.Kohrer S, Havranek O, Seyfried F, et al. Pre-BCR signaling in precursor B-cell acute lymphoblastic leukemia regulates PI3K/ AKT, FOXO1 and MYC, and can be targeted by SYK inhibition. Leukemia. 2016;30(6):1246-1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muschen M. Rationale for targeting the pre-B-cell receptor signaling pathway in acute lymphoblastic leukemia. Blood. 2015;125(24):3688-3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim E, Koehrer S, Rosin NY, et al. Activity of Bruton’s tyrosine kinase (BTK) inhibitor ibrutinib (PCI-32765) in B-cell acute lymphoblastic leukemia (B-ALL). Blood. 2012;120(21):2569. [Google Scholar]
- 36.Kim E, Koehrer S, Wang Z, et al. The PI3K delta inhibitor idelalisib interferes with pre-B cell receptor signaling in acute lymphoblastic leukemia (ALL): a new therapeutic concept. Blood. 2013;122(21):2632. [Google Scholar]
- 37.Wang R, Zhang H, Xu J, et al. MALT1 Inhibition as a therapeutic strategy in T-cell acute lymphoblastic leukemia by blocking Notch1-induced NF-kappaB activation. Front Oncol. 2020;10:558339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Herman SE, Gordon AL, Hertlein E, et al. Bruton tyrosine kinase represents a promising therapeutic target for treatment of chronic lymphocytic leukemia and is effectively targeted by PCI-32765. Blood. 2011;117(23):6287-6296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hoellenriegel J, Coffey GP, Sinha U, et al. Selective, novel spleen tyrosine kinase (Syk) inhibitors suppress chronic lymphocytic leukemia B-cell activation and migration. Leukemia. 2012;26(7):1576-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Herman SE, Gordon AL, Wagner AJ, et al. Phosphatidylinositol 3-kinase-delta inhibitor CAL-101 shows promising preclinical activity in chronic lymphocytic leukemia by antagonizing intrinsic and extrinsic cellular survival signals. Blood. 2010;116(12):2078-2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA sequencing data supporting this publication are available from the GEO repository under the accession number GSE221273 . https://www.ncbi.nlm.nih.gov/geo/