Waldenström macroglobulinemia (WM) is a rare non-Hodgkin lymphoma that is often preceded by an IgM monoclonal gammopathy of undetermined significance (IgM-MGUS).1 The factors underlying the malignant progression between these IgM-monoclonal gammopathies are poorly understood, and the majority of attention has focused on protein-coding genes. One aspect that remains underexplored is the influence of the non-coding genome. Two families of non-protein-coding RNA that have been implicated in regulating an array of disease processes are microRNA (miR-NA, miR) and long non-coding RNA (lncRNA).2,3 The primary aim of this study was to determine the role of non-coding RNA, specifically miRNA and lncRNA, in IgM-gammopathies and the pathways and genes they may regulate. This study demonstrated that miRNA and lncRNA are highly differentially expressed between IgM-gammopathies and normal controls and specifically between WM and IgM-MGUS. Furthermore, pathway analysis showed miRNA-based epigenetic regulation of multiple cellular pathways including cell signaling and immune response in IgM-gammopathies. This study included prospectively collected bone marrow samples from 28 subjects (17 with WM, 6 with IgM-MGUS, 5 controls]. WM was defined as ≥10% bone marrow involvement by lymphoplasmacytic lymphoma and a serum IgM monoclonal protein of any size as per Mayo Stratification of Macroglobulinemia and Risk-Adapted Therapy (mSMART) guidelines.1 Patients were included after treatment if at least 6 months had passed between the last therapy and sample collection with symptoms indicating active disease. IgM-MGUS was defined as <10% bone marrow involvement by lymphoplasmacytic lymphoma, a serum IgM monoclonal protein <3 g/dL, and no signs/symptoms of active WM. Selection for CD19+ and/or CD138+ B cells was conducted. No clonality testing was performed. Next, total RNA was extracted, and analyses of miRNA, lncRNA and messenger RNA (mRNA) were performed.4 Differential expression between groups was evaluated and defined to be present if the fold-change in mRNA was >1 or <-1 and the fold-change in miRNA/lncRNA was >0.5 or <-0.5, with a false discovery rate <0.05. Subsequently, miRNA-mRNA target analysis was performed and differentially expressed miRNA-mRNA pairs with a correlated biological expression (i.e., either upregulated miRNA experimentally predicted to regulate downregulated mRNA or vice versa) were selected. Ingenuity Pathway Analysis was used to determine whether canonical pathways were implicated. LncRNA were further analyzed by assessing the nearest protein-coding gene or antisense protein-coding gene.
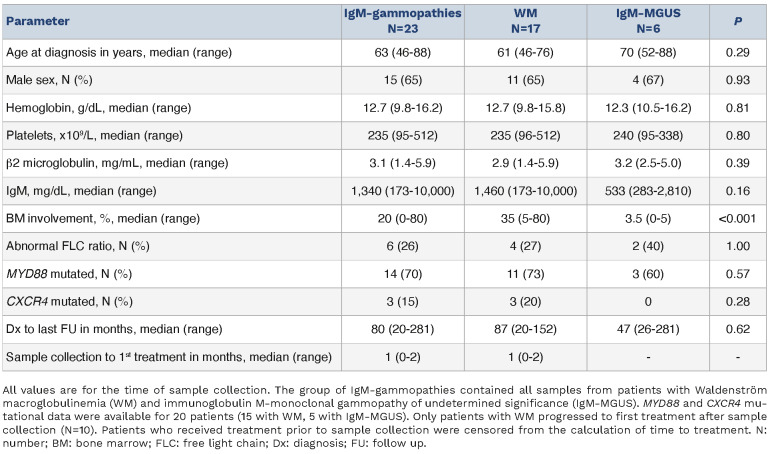
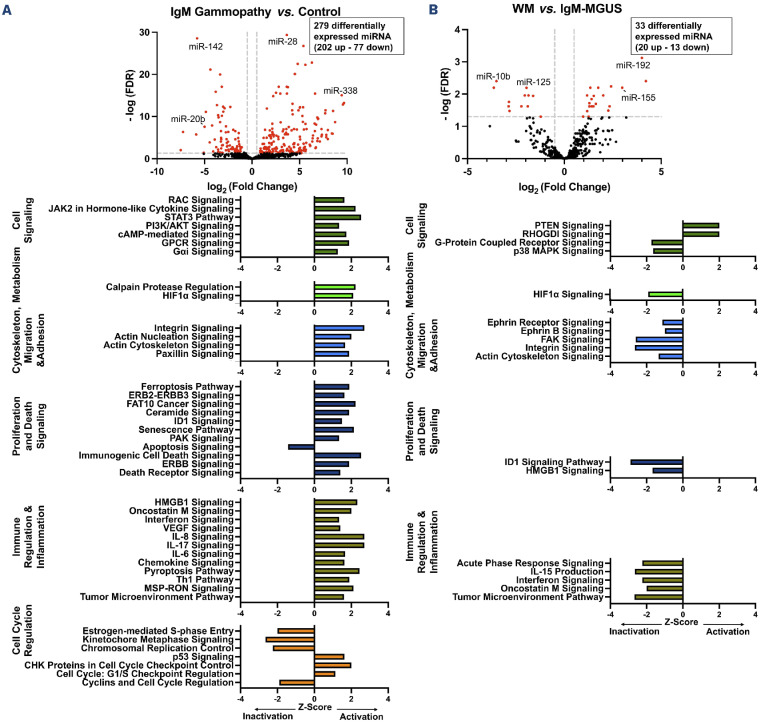
The baseline characteristics of the cohort are demonstrated in Table 1. Significant differences were seen in miRNA expression data and Figure 1 illustrates the comparisons between IgM-gammopathies and normal controls and between WM and IgM-MGUS. Online Supplementary Figure S1 shows the miRNA differential analysis between IgM-MGUS and controls and between WM and controls. In the comparisons of WM to IgM-MGUS and IgM-gammopathies to controls, there were 18 miRNA that were commonly differentially expressed. Of these miRNA, five were found to be in the same direction of expression (4 upregulated, 1 downregulated), while 13 demonstrated opposite expression patterns. Several miRNA that have been previously implicated in WM were also demonstrated to be differentially expressed in our cohort, including miR-155 and miR-192.5,6 Next, miRNA-based pathway analysis revealed several aspects of oncogenesis that were implicated when comparing both IgM-gammopathies to controls and WM to IgM-MGUS. These included cell signaling, metabolism, cytoskeleton, migration and adhesion, proliferation, immune response, and cell cycle regulation pathways (Figure 1A, B). A few pathways are discussed below, and a comprehensive analysis of implicated pathways along with underlying miRNA-mRNA pairs of interest is included in Online Supplementary Table S1.
In the assessment of miRNA-based pathways in IgM-gammopathies compared to normal controls, upregulation of signaling pathways, including STAT3, PI3K/AKT, JAK2 and RAC, was observed. Several of these pathways have been previously implicated in the pathogenesis of WM.7-9 Upregulation of the STAT 3 gene was found to be commonly dysregulated, which is a predicted target of downregulated miRNA, including miR-20B and miR-223. Additional genes implicated in these signaling pathways included BCL2 which was observed to be upregulated, and potentially epigenetically targeted by miR-16, miR-17, miR-181 and miR-34, all downregulated. Of note, BCL2 is a regulator of mitochondrial apoptotic pathways and is a critical gene in WM pathogenesis induced through signaling of the MYD88 and CXCR4 activating mutations.10
Comparing WM to IgM-MGUS, significant inactivation of multiple inflammatory and cytokine signaling pathways was observed. A pathway of interest was interferon signaling. Interferons are cytokines that have important roles in the regulation of biological processes, inflammation and tumorigenesis.11 Underlying the interferon pathway was downregulation of multiple interferon-induced genes (IFIT1, IFIT3, IFITM1) and S TAT 1 . The most notable targets of these interferon-induced genes were miR-32, miR-146, and miR-155, all of which were upregulated. Additionally, underlying multiple inflammatory signaling and cytokine pathways was upregulation of miR-146a, miR-150, and miR-194, all targets of S TAT 1, observed to be downregulated. Notably, miR-146a has previously been shown to be an inhibitor of inflammatory cytokines, including interleukin-6.12 STAT family proteins play a crucial role in the transmission of interferon-stimulated genes and cytokine signal transduction.11 Downregulation of multiple cytokine signaling pathways involved in inflammation and the tumor microenvironment, along with targeting of the STAT family, indicates a possible miRNA-based negative feedback loop to down-modulate inflammatory cytokine signaling in WM compared to IgM-MGUS.
Table 1.
Clinical characteristics of all the patients with IgM-gammopathy who provided blood samples and a comparison between patients with Waldenström macroglobulinemia and IgM-monoclonal gammopathy of undetermined significance.
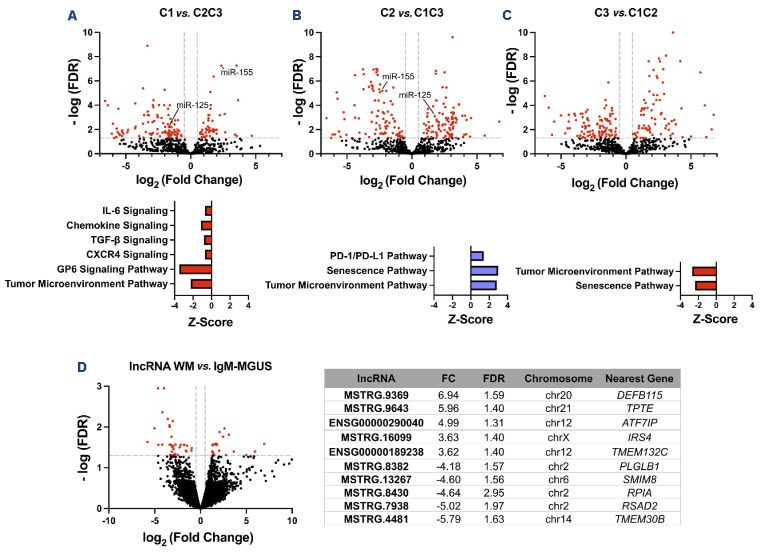
Previously, our group published a multi-omics analysis demonstrating distinct pathway activationas well as metabolomic and clinical features in each of the clinical clusters of monoclonal gammopathies analyzed (C1, C2, C3).4 MiRNA analysis performed on these clusters revealed a distinct expression profile in each group (Figure 2A-C). Assessing the expression of miRNA of interest, we found that miR-155 was upregulated in C1 and downregulated in C2. miR-155 has previously been demonstrated to regulate WM cell proliferation and growth. This observation corresponds with our findings of patients in C1 having a more aggressive phenotype. In addition, we saw downregulation of miR-125 in C1, compared to upregulation in C2. miR-125a acts via the nuclear factor-κB pathway and has been shown to be a tumor suppressor in B-cell malignancies.13 Pathway analysis additionally revealed alterations in immune response, cytokine signaling, cell cycle, senescence, and metabolism with opposite trends in C1 as compared to C2. These pathways overlapped with our previously published data based on protein expression.4
Next, assessment of lncRNA expression comparing WM and IgM-MGUS revealed 62 differentially expressed targets (24 upregulated, 38 downregulated) (Figure 2D). Several lncRNA found to be differentially expressed have been previously implicated in other malignancies (Online Supplementary Table S2). Our analysis next focused on lncRNA found to be in proximity of or antisense to coding genes. Here, we found a number of targets that regulate transcription and cell cycle regulation. Notable lncRNA included ENSG00000274987, which was found to be upregulated; this is the antisense lncRNA to KRAS, the gene that codes for the protein K-RAS, a GTPase implicated in B-cell proliferation and cell signaling, which is commonly dysregulated in lymphoid malignancies.14 Additionally, several nearest or antisense genes to lncRNA were also differentially expressed on mRNA analysis between WM and IgM-MGUS (Online Supplementary Table S2). This included ENSG00000229418, which is the antisense lncRNA to the gene DNTT, observed to be downregulated in WM (fold change: -3.6; false discovery rate <0.005) compared to IgM-MGUS. DNA nucleotidylexo-transferase (DNTT) is a DNA polymerase expressed in immature B and T lymphoid cells and has been implicated in WM in relation to V(D)J recombination.15
Figure 1.
MicroRNA expression and pathway analysis in IgM-gammopathies. (A) IgM-gammopathy compared to control samples, (B) Waldenström macroglobulinemia compared to IgM-monoclonal gammopathy of undetermined significance (B). Dotted lines on the volcano plot indicate significance thresholds for fold change and false discovery rate. Red markers indicate differentially expressed microRNA (miRNA). Black markers indicate non-differentially expressed miRNA. Figure created using GraphPad Prism. FDR: false discovery rate; miR: microRNA; IgM: immunoglobulin M; WM: Waldenström macroglobulinemia; MGUS: monoclonal gammopathy of undetermined significance.
With the increasing recognition of non-coding genomics driving progression in lymphoid malignancies, the current study offers one of the first comprehensive analyses of both miRNA and lncRNA in IgM-gammopathies. The miRNA analysis comparing IgM-gammopathies to normal controls revealed that a significant number of miRNA were differentially expressed, and diverse cellular functions were seen to be potentially epigenetically mediated through miRNA-based pathways. These included signaling pathways such as PI3K/AKT/mTOR, and the JAK/STAT pathway, which are the source of important therapeutic targets in WM.7-9 Thus, miRNA may serve as important novel targets for therapy or may play an adjunctive role with currently available therapies targeting oncogenic pathways. Extending our previous multi-omics analysis which revealed three distinct clusters in IgM-gammopathies, the miRNA analysis of these clusters revealed that each cluster has a unique non-coding expression profile, highlighting their biological differences. Further exploration of how to better classify IgM-gammopathies based on biological differences, including both miRNA and lncRNA into this framework, may allow for better molecular differentiation of IgM-gammopathies and, ultimately, a more individualized patient approach.
Figure 2.
MicroRNA analysis by cluster and long non-coding RNA analysis comparing Waldenström macroglobulinemia to IgM-monoclonal gammopathy of undetermined significance. (A-C) MicroRNA (miRNA) expression analysis by cluster with pathway analysis and (D) long non-coding RNA (lncRNA) expression data comparing Waldenström macroglobulinemia to IgM-monoclonal gammopathy of undetermined significance with the top ten dysregulated lncRNA. The cluster pathways demonstrated were based only on concordant pathway analysis as compared to previously published protein-based pathway data. Dotted lines on the volcano plot indicate thresholds for significance for fold change and false discovery rate. Red markers indicate differentially expressed miRNA/lncRNA. Black markers indicate non-differentially expressed miRNA/lncRNA. The table listing the top ten dysregulated lncRNA also presents the corresponding fold change, false discovery rate, chromosomal location and nearest gene. Figure created using GraphPad Prism. miR: microRNA; WM: Waldenström macroglobulinemia; IgM: immunoglobulin M; MGUS: monoclonal gammopathy of undetermined significance; FDR: false discovery rate.
Currently, there are no reliable biological models to predict the progression from IgM-MGUS to marginal zone lymphoma, multiple myeloma, chronic lymphocytic lymphoma or WM. In this study, given that several differentially expressed miRNA and lncRNA were identified by comparing WM to IgM-MGUS, non-coding RNA may serve a novel predictive role. Future large prospective analyses with long-term follow-up should be conducted to assess whether non-coding genomic signatures may help to indicate a higher risk of progression to malignancy. Additionally, by expanding our understanding of key non-coded RNA differences between IgM-MGUS and WM, these findings offer the potential to recognize novel therapeutic targets which may halt or slow the malignant progression between these two diseases.
Overall, we report the key miRNA and lncRNA differences underlying both IgM-MGUS and WM and explore the role of non-coding RNA in the regulation of cellular pathways in IgM-gammopathies. Further investigation of non-coding RNA of interest with functional studies will certainly be needed to help to determine their exact biological roles. As an underexplored area of pathogenesis which likely drives many of the aberrantly regulated pathways, non-coding genomics will certainly provide novel therapeutic targets, help to classify patients better based on disease biology, and improve risk stratification for purposes of prognostication.
Supplementary Material
Funding Statement
Funding: This study was supported by research funding from the BINK Foundation, and International Waldenström Macroglobulinemia Foundation (IWMF) to SMA and the Mayo Clinic.
Data-sharing statement
RNA data are available at GEO under the accession numbers GSE232994 and GSE232995. For additional data on the patients, please contact the corresponding author.
References
- 1.Kapoor P, Ansell SM, Fonseca R, et al. Diagnosis and management of Waldenström macroglobulinemia: Mayo Stratification of Macroglobulinemia and Risk-Adapted Therapy (mSMART) guidelines 2016. JAMA Oncol. 2017;3(9):1257-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O’Brien J, Hayder H, Zayed Y, Peng C. Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front Endocrinol (Lausanne). 2018;9:402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Statello L, Guo CJ, Chen LL, Huarte M. Author correction: Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol. 2021;22(2):159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mondello P, Paludo J, Novak JP, et al. Molecular clusters and tumor-immune drivers of IgM monoclonal gammopathies. Clin Cancer Res. 2023;29(5):957-970. [DOI] [PubMed] [Google Scholar]
- 5.Bouyssou JM, Liu CJ, Bustoros M, et al. Profiling of circulating exosomal miRNAs in patients with Waldenström macroglobulinemia. PLoS One. 2018;13(10):e0204589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roccaro AM, Sacco A, Chen C, et al. microRNA expression in the biology, prognosis, and therapy of Waldenström macroglobulinemia. Blood. 2009;113(18):4391-4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ghobrial IM, Gertz M, Laplant B, et al. Phase II trial of the oral mammalian target of rapamycin inhibitor everolimus in relapsed or refractory Waldenström macroglobulinemia. J Clin Oncol. 2010;28(8):1408-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ansell SM, Grote D, Elsawa SF, et al. Inhibition of the Jak/Stat pathway downregulates immunoglobulin production and induces cell death in Waldenström macroglobulinemia. Blood. 2009;114(22):1691-1691. [Google Scholar]
- 9.Tomowiak C, Poulain S, Herbaux C, et al. Obinutuzumab and idelalisib in symptomatic patients with relapsed/refractory Waldenström macroglobulinemia. Blood Adv. 2021;5(9):2438-2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gaudette BT, Dwivedi B, Chitta KS, et al. Low expression of pro-apoptotic Bcl-2 family proteins sets the apoptotic threshold in Waldenström macroglobulinemia. Oncogene. 2016;35(4):479-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schneider WM, Chevillotte MD, Rice CM. Interferon-stimulated genes: a complex web of host defenses. Annu Rev Immunol. 2014;32:513-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou C, Zhao L, Wang K, et al. MicroRNA-146a inhibits NF-kappaB activation and pro-inflammatory cytokine production by regulating IRAK1 expression in THP-1 cells. Exp Ther Med. 2019;18(4):3078-3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim SW, Ramasamy K, Bouamar H, Lin AP, Jiang D, Aguiar RC. MicroRNAs miR-125a and miR-125b constitutively activate the NF-kappaB pathway by targeting the tumor necrosis factor alpha-induced protein 3 (TNFAIP3, A20). Proc Natl Acad Sci U S A. 2012;109(20):7865-7870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vendramini E, Bomben R, Pozzo F, et al. KRAS and RAS-MAPK pathway deregulation in mature B cell lymphoproliferative disorders. Cancers (Basel). 2022;14(3):666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hunter ZR, Yang G, Xu L, Liu X, Castillo JJ, Treon SP. Genomics, signaling, and treatment of Waldenström macroglobulinemia. J Clin Oncol. 2017;35(9):994-1001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA data are available at GEO under the accession numbers GSE232994 and GSE232995. For additional data on the patients, please contact the corresponding author.