Abstract

Brucea javanica oil emulsion (BJOE) is a compound Chinese medicine used for treating various cancers, such as lung cancer. However, the exact mechanism of its antilung cancer active ingredient remains unclear. This study aims to explore and validate the effective active ingredients and mechanism of action of BJOE in the treatment of lung cancer through network pharmacology, molecular docking technology, and cell experiments. The results showed that there were 13 active ingredients, 136 target genes, and 42 disease target-coexpressed genes in BJOE. The molecular docking results indicated that the main active components of the oil emulsion, YD1 (β-sitosterol), YD2 (luteolin), and YD3 (bruceitol), could stably bind to TP53 and MAPK1. Furthermore, the commercially available β-sitosterol luteolin was used as a representative compound to conduct cell experiments to verify its anticancer activity and mechanism. It was found that luteolin can inhibit the proliferation better than β-sitosterol and the activity of lung cancer cells by regulating the expression of related proteins through the P53/MAPK1 signaling pathway. This study combines network pharmacology prediction with experiments to demonstrate the “multicomponent, multitarget, multipathway” approach of B. javanica oil emulsion in treating lung cancer.
1. Introduction
Lung cancer is one of the most common malignant tumors, with increasing incidence and mortality rates year by year, posing a significant threat to human health and life safety.1,2 Early symptoms of lung cancer are often inconspicuous, and most patients are diagnosed at an advanced stage, where chemotherapy becomes their primary treatment option.3,4 The first-line chemotherapy drugs mainly include cytotoxic drugs and targeted drugs.5 Cytotoxic drugs inhibit tumor growth by directly killing tumor cells or blocking the synthesis of nucleic acids, proteins, etc. However, they also affect normal bodily functions and cause a series of adverse reactions. Targeted drugs treat specific diseases by blocking or enhancing the functions of their targets. However, due to the complexity of the pathogenesis of lung cancer, the clinical application of targeted drugs is also limited.6,7 Traditional Chinese medicine has a wide range of active ingredients that can simultaneously act on multiple anticancer sites. Moreover, most of them have low toxicity and high safety and possess a broad potential market. They play an irreplaceable role in clinical practice.8,9
Yadanzi (Brucea Fructus) is the dried mature fruit of Brucea javanica (L.) Merr, a plant belonging to the Simaroubaceae family. It possesses functions such as heat-clearing, detoxification, antimalarial, and antidiarrheal effects.10,11 It is mainly used for the treatment of diseases, such as dysentery, malaria, and warts. B. javanica oil emulsion (BJOE) is an emulsion made by adding extracts of Yadanzi to an emulsifier, containing multiple active ingredients such as bruceine, bruceantin, etc.12 As a traditional Chinese medicine, BJOE has shown good therapeutic effects on lung cancer, brain metastasis of lung cancer, and digestive tract tumors through the synergistic action of multiple components and targets, making it highly valuable in clinical practice.13 Based on the usage data from several lung cancer patients at Dongyang People’s Hospital, we have observed significant reductions in tumor markers such as carcinoembryonic antigen (CEA), carbohydrate antigen 125 (CA125), and neuron-specific enolase (NSE) after the administration of BJOE, demonstrating excellent anticancer effects (Table 1). However, the diverse active substances in BJOE and its complex mechanism of action have hindered in-depth research. Therefore, the integration of traditional Chinese medicine, targets, and diseases through network big data, coupled with systematic pharmacological research methods, to construct a systematic network of Chinese medicine–human body relationships is a new trend and approach in modern traditional Chinese medicine research. As a commonly used clinical treatment for lung cancer, further analysis of the mechanism of action of BJOE through network big data holds a great research value for the treatment of lung cancer.14
Table 1. Changes of Relevant Indexes over Time after Injection of Brucea Oil Emulsion in Three Patients with Lung Cancer.
| case | medication time | CEA | CA123 | NSE | CYFRA21-1 |
|---|---|---|---|---|---|
| one | first week | 6.7 | 39.9 | 17.3 | 12.1 |
| second week | 2.3 | 17.4 | 16.6 | 9.8 | |
| third week | 1.8 | 14.5 | 17.1 | 6.3 | |
| two | first week | 21.1 | 14.2 | 17.0 | 3.0 |
| second week | 12.5 | 13.0 | 15.2 | 2.6 | |
| third week | 8.5 | 8.0 | 12.8 | 1.0 | |
| three | first week | 9.0 | 131.0 | 24.6 | 8.5 |
| second week | 2.8 | 126.0 | 21.7 | 8.1 | |
| third week | 3.5 | 108.9 | 17.3 | 5.9 |
Network pharmacology is an emerging discipline that utilizes network integration of relevant biological information based on large-scale databases and system biology theory to comprehensively analyze the structures of small molecule drugs and targets.15 In 2008, pharmacologist Andrew L. Hopkins first proposed the concept of network pharmacology, which has shown significant development and important value in both practical and theoretical research in recent years.16 In this study, we employ network pharmacology as a means to analyze the main components and related targets of Yadanzi, aiming to identify the key anticancer substances. By integrating specific signaling nodes of lung cancer, we will elucidate the principles underlying the occurrence and development of the disease. Furthermore, molecular docking techniques will be utilized to further elucidate the binding modes between small molecules and proteins.17 The results from the in vitro experiments will provide a clear understanding of the mechanisms of action of the main components. The research process is illustrated in Figure 1.
Figure 1.
Research roadmap.
2. Results
2.1. Screening of Active Ingredients in Yadanzi Emulsion
Using the TCMSP database, a search was conducted for active ingredients of Yadanzi, and 15 compounds were obtained based on screening criteria of oral bioavailability (OB) ≥ 30% and drug-likeness (DL) ≥ 0.18. Among them, 13 compounds were found to have specific target interactions (as shown in Figure 2). The chemical structures of these compounds were drawn using ChemDraw and imported into the SwissTargetPrediction database to predict their potential targets. The target information for these compounds was collected, and after deduplication through calibration using the UniProt database, a total of 137 effective target proteins were identified.
Figure 2.
Chemical structures of active ingredients in Yadanzi.
2.2. Construction of the Protein and Compound Network
For the convenience of network convergence, we replaced the compound names with Ydn (n represents digits from 1 to 13) instead of the original names (as shown in Table 2). The codes of these compounds and their corresponding target proteins were imported into Cystoscape software to construct the “active ingredient-target” network of Yadanzi emulsion (as shown in Figure 3). This network contains 151 nodes and 231 edges, and the number of edges connected to each node is called the “degree” value, which represents the importance of the node. Therefore, by calculating the degree values of the active compounds, we can identify the core components. In Figure 3, we represent the degree values of compounds using the sizes of the graphics and characters, where YD1 and YD2 have the highest degree values. Based on this, we propose a preliminary hypothesis that the active compounds YD1 and YD2 may have a significant impact on the anticancer activity of Yadanzi.
Table 2. Compound Names and Corresponding Numbers.
| number | compound | number | compound |
|---|---|---|---|
| YD1 | β-sitosterol | YD8 | yadanzioside L |
| YD2 | luteolin | YD9 | yadanzioside M |
| YD3 | brusatol | YD10 | yadanzioside P |
| YD4 | yadanzioside B | YD11 | bruceoside B |
| YD5 | yadanzioside H | YD12 | bruceine C |
| YD6 | yadanzioside I | YD13 | brucceoside A_qt |
| YD7 | yadanzioside J |
Figure 3.
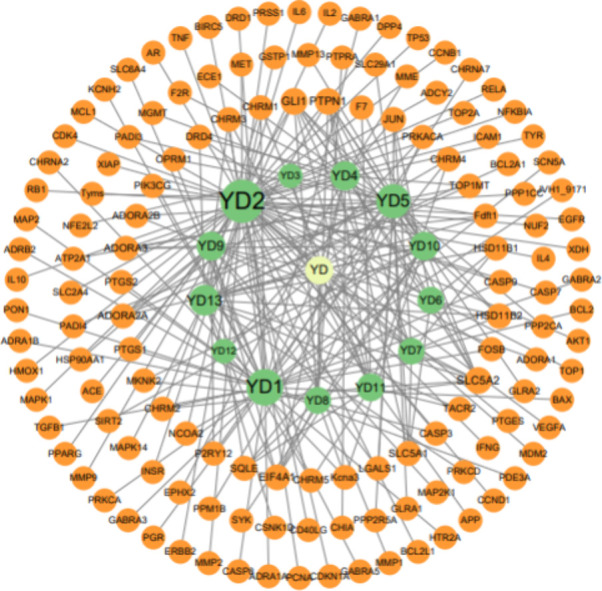
Compound–protein network.
2.3. Construction of the Venn Diagram
To further confirm the association between Yadanzi emulsion and lung cancer, we imported the target points of Yadanzi emulsion and lung cancer into the VennDetail Shiny App (http://hurlab.med.und.edu:3838/VennDetail/) and obtained 42 potential common targets, as shown in Figure 4. The results indicate that Yadanzi emulsion may inhibit the growth or metastasis of lung cancer cells by targeting the aforementioned 42 targets.
Figure 4.

Venn diagram of common targets between active components of Yadanzi and lung cancer.
2.4. Construction of the Target-Lung Cancer Protein Network Diagram
The 42 intersection targets obtained from above were imported into the STRING database, the confidence interval was adjusted to the highest confidence of 0.9 and the disconnected nodes were hidden, and the corresponding protein interaction network (PPI) was finally obtained, as shown in Figure 5. The network structure contains 42 nodes and 152 connections, and the average degree value is 6.91. The nodes represent the target of action, and the lines represent the interactions between the targets. The thicker the lines, the greater the confidence in the data support.
Figure 5.
Lung cancer protein–Yadanzi protein interaction (PPI) network.
2.5. Selection of Key Targets
The network graph obtained from STRING was imported into Cytoscape software for the analysis of PPI network topological properties. The analysis results showed that the average degree value of the network was 14.48, and the average betweenness centrality was 3.19 × 10–2. Nodes with degree and betweenness centrality values higher than the mean were selected as key targets, resulting in the identification of 9 key targets (as shown in Table 3). These key targets included JUN (Jun Proto-oncogene), TP53 (Tumor Protein p53), MAPK1 (Mitogen-activated Protein Kinase 1), IL6 (Interleukin 6), RELA (RELA Proto-oncogene), AKT1 (AKT Serine/threonine Kinase 1), TGFB1 (Transforming Growth Factor Beta 1), VEGFA (Vascular Endothelial Growth Factor A), and CCND1 (Cyclin D1). The visualization analysis in Cytoscape software is shown in Figure 6. In the visualization analysis, larger node labels and more vibrant colors indicate higher degree values of the targets, while darker border lines and thicker edges indicate stronger binding strengths. Thus, it can be observed that oncogene JUN and cancer protein P53 hold significant positions in this network.
Table 3. Degree and Betweenness Centrality of Key Targets.
| betweenness centrality | degree | target genes | betweenness centrality | degree | target genes |
|---|---|---|---|---|---|
| 0.251939 | 40 | JUN | 0.034588 | 4 | AKT1 |
| 0.21439 | 38 | TP53 | 0.041893 | 5 | VEGFA |
| 0.187476 | 34 | MAPK1 | 0.045528 | 5 | TGFB1 |
| 0.107358 | 32 | IL6 | 0.039015 | 3 | CCND1 |
| 0.047575 | 26 | RELA |
Figure 6.
Key targets of Yadanzi in lung cancer.
2.6. KEGG Enrichment Analysis of Key Targets
After the above targets were entered into the DAVID database, a total of 116 signaling pathways were enriched. Among the top 10 ranked pathways, a bar graph was plotted for analysis, as shown in Figure 7. The enrichment results revealed that the key targets of Yadanzi oil emulsion in lung cancer were involved in various diseases, including diabetes mellitus, pancreatic cancer, viral infections, leukemia, cancer pathways, cellular senescence, hepatitis B, and colorectal cancer.
Figure 7.
Analysis of KEGG enrichment results (top 10).
2.7. GO Enrichment Analysis of Key Targets
GO biological function analysis was performed using the DAVID database, and the top 10 enrichment results were imported into the ChiPlot tool Web site to generate a bubble plot. The results are shown in Figure 8. In the biological process (BP, yellow section), the main enriched terms include positive regulation of gene expression, positive regulation of transcription, aging, and positive regulation of pri-miRNA transcription from the RNA polymerase II promoter. In the cellular component (CC, blue section), the main enriched terms include transcriptional repressor complex, transcription factor complex, cytoplasm, nucleoplasm, and nucleus. In the molecular function (MF, green section), the main enriched terms include extracellular region, enzyme binding, identical protein binding, macromolecular complex binding, and protein kinase binding.
Figure 8.
GO enrichment analysis results (top 10).
2.8. Molecular Docking Analysis
Through molecular docking techniques, the main components of three Cornus officinalis oil emulsions were extracted and docked with core targets for validation. Key compounds YD1, YD2, and YD3 were selected and docked with core targets MAPK1 and TP53 (as shown in Figure 9). A smaller binding energy after docking indicates a more stable conformation between the ligand and the receptor and a higher likelihood of interaction. The binding energy of YD1 with MAPK1 was −8.3 kJ/mol, that of YD2 with MAPK1 was −7.6 kJ/mol, and that of YD3 with MAPK1 was −7.5 kJ/mol. The binding energy of YD1 with p53BP1 (a protein regulated by TP53) was −6.26 kJ/mol, that of YD2 with p53BP1 was −5.15 kJ/mol, and that of YD3 with p53BP1 was −1.06 kJ/mol. YD1 was able to dock into the cavity site of p53BP1, but whether this site is an active binding site requires further investigation.
Figure 9.
Docking analysis of the core components of C. officinalis oil emulsions with MAPK1 and P53BP1. YD1 with MAPK1 (A), YD2 with MAPK1 (B), YD3 with MAPK1 (C), YD1 with p53BP1 (D), YD2 with p53BP1 (E), and YD3 with p53BP1 (F).
2.9. Validation of the Antilung Cancer Effect of YD2
Based on the above results, YD2, which is easily available in the market, was selected as the target compound to evaluate its antilung cancer activity. As shown in Figure 10A, YD2 exhibited an IC50 of 20.95 μM against human lung squamous cells H520. Colony formation assay and scratch assay further confirmed its anticancer activity, as shown in Figure 10B,C. YD2 demonstrated concentration-dependent anticancer activity, with a good inhibitory effect on proliferation observed at a concentration of 20 μM in the colony formation assay. In the scratch assay, YD2 showed significant anticancer activity compared to the control group. Moreover, in different kinds of human lung cancer nonsmall cell H1299, compound YD2 has the same inhibitory effect on tumor, and IC50 reaches 21.05 μM (Figure S1). In addition, we also tested the anticancer activity of YD1, YD2, and BJOE in different lung cancer cells (H520, H1299, and A549 cells). The inhibitory activity of YD2 on three different cancer cells was significantly better than that of YD1. The inhibition rates of YD2 on H520, H1299, and A549 cells were 20.95, 21.02, and 26.07 μM, respectively. Surprisingly, at the cellular level, BJOE has a promoting effect on three cancer cells at different concentrations. Further colony experiments were performed on BJOE, and the number of colonies did not inhibit the effect (Figures S2 and S3).
Figure 10.
Antiproliferative activity of YD2 on H520 cells. IC50 of YD2 (A); colony formation assay (B); scratch assay (C).
2.10. Effects of YD2 on the Expression of MAPK1 and P53 Proteins
We performed western blot experiments to examine the expression of P53, P-MAPK1, and MAPK1 at different time points (0, 3, 6, 9, 12, and 24 h) after treatment with YD2 in LPS-stimulated cells. As shown in Figure 10, with increasing time, YD2 promoted the expression of P53 (Figure 11A) and inhibited the phosphorylation of MAPK1 (Figure 11B).
Figure 11.
Effects of YD2 on the expression of P53 and MAPK1. YD2 induces the expression of P53 (A); YD2 inhibits the phosphorylation of MAPK1 (B).
3. Discussion
Several studies have investigated the potential anticancer effects of the Yadanzi oil emulsion and its components in lung cancer treatment. It was found that B. javanica has significant inhibitory effects on lung cancer cell growth and induced apoptosis (programmed cell death) in lung cancer cells.18−20 Compounds in B. javanica also presented excellent antilung cancer activity. For example, β-sitosterol (YD1) has been studied for its potential antitumor and immunomodulatory effects.21 Additionally, luteolin (YD2), another component of Yadanzi oil emulsion, has been reported to possess anti-inflammatory, antioxidant, and immunostimulatory properties.22 Although the anticancer activity of B. javanica and its compounds has been extensively investigated, a precise mechanism of action is required.
In this study, we screened the key compounds of the Yadanzi emulsion using the TCMSP database and obtained 136 target proteins. Additionally, we collected 415 lung cancer-related target proteins from the GeneCards database. By using Venn diagram analysis, we found that the Yadanzi oil emulsion and lung cancer share 42 common target proteins. It is possible that the Yadanzi oil emulsion inhibits the growth or metastasis of lung cancer cells by affecting the expression of these 42 protein targets. Therefore, we imported these 42 target proteins into the STRING database for further investigation of their interactions. Visualization analysis using Cytoscape software revealed 9 proteins that play a key role in this network. KEGG and GO enrichment analyses of these key proteins revealed that Yadanzi oil emulsion mainly regulates gene expression, transcription, and other processes, affecting diseases such as diabetes, cancer, and leukemia.
The oncogene JUN regulates the nuclear transcription factor c-Jun, which can bind to the promoters of many genes and participate in the transcription process. It can be activated or influenced by various chemicals present in tissues.23 Transcription factors have smaller molecular structures, making it difficult to describe the effects of drugs on them through molecular docking techniques. Therefore, in this study, we selected TP53 and MAPK1 genes with a slightly lower degree and betweenness centrality as the main research targets. The TP53 gene is a tumor suppressor gene that can inhibit the proliferation of cancer genes or induce their apoptosis, playing an important role in maintaining normal body functions.24 In this study, the three key compounds of Yadanzi oil emulsion may have certain effects on the proteins regulated by TP53, inducing the activation of corresponding proteins and inhibiting the proliferation and migration of cancer cells. However, the binding affinity of the three key compounds to MAPK1 is lower with a higher probability of binding. MAPK1 (ERK2) is a key node in the ERK signaling pathway, and regulating the expression of MAPK1 can affect the growth, proliferation, or migration of various tumor cells.25 In molecular docking experiments, the three key compounds can bind well with MAPK1, with binding energies of less than −5.0 kJ/mol. Additionally, small drug molecules can form intermolecular hydrogen bonds with the corresponding amino acids, further enhancing the binding interaction. Therefore, the Yadanzi oil emulsion may exert therapeutic effects on lung cancer by inducing the expression of TP53 and inhibiting the activation of MAPK1.
To further validate the computational results, we conducted in vitro experiments using YD2-luteolin. The results showed that as a natural compound, luteolin exhibited significant antilung cancer activity and inhibited the proliferation of lung cancer cells in a concentration-dependent manner. Additionally, western blot experiments demonstrated that luteolin induced the expression of TP53 and inhibited the phosphorylation of MAPK1, confirming the predictions of network pharmacology. Considering the excellent anticancer effect of luteolin, we have a reason to believe that it plays a major therapeutic role in Yadanzi. Further comparison of the therapeutic effects of luteolin and Yadanzi was intended; however, Yadanzi is a complex formulation, while luteolin is a pure compound, making it difficult to achieve uniformity in terms of dosage. Therefore, it is necessary to develop a method that can compare the therapeutic effects of complex formulations and pure compounds, which can also contribute to the development of modern Chinese medicine.
4. Conclusions
In this study, we used network pharmacology to screen three major antilung cancer substances from Yadanzi, namely, YD1-β-sitosterol, YD2-luteolin, and YD3-brusatol. We validated the anticancer activity and mechanism of luteolin as a representative compound and found that it primarily inhibits the proliferation of lung cancer cells through the P53/MAPK1 signaling pathway. The anticancer effect of Yadanzi is mainly achieved through these natural compounds, and combining them in specific proportions to form a new compound formulation may have better anticancer activity than Yadanzi alone. We are also attempting to effectively combine the three compounds to develop a new compound formulation with improved efficacy and fewer side effects, providing insights and evidence for the development of traditional Chinese medicine.
5. Materials and Methods
5.1. Collection of Small Molecules and Targets in Yadanzi
Chemical components and partial targets of Yadanzi were collected through the TCMSP (Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform) database (https://old.TCMSP-e.com/TCMSP.php). According to ADME, the effective chemical components with oral bioavailability (OB) ≥ 30% and drug-like properties (DL) ≥ 0.18 were selected. The chemical structures were drawn using ChemDraw and subsequently uploaded to the SwissTargetPrediction database (http://www.swisstargetprediction.ch/) for potential target prediction. Duplicate targets were then removed, and the remaining targets were inputted into the UniProt database (https://www.uniprot.org) for standardization.
5.2. Collection and Screening of Lung Cancer-Related Targets
The keyword “lung cancer” was included in the search query of the GeneCards database to obtain relevant targets for lung cancer. Genes with relevance score ≥30.18 were selected as potential genes. The target intersection between “Yadanzi Oil Emulsion” and “Lung Cancer” was obtained using the online tool Venny 2.1.0, and the corresponding Venn diagram was generated.
5.3. Construction of the Protein–Protein Interaction (PPI) Network
The intersection targets were imported into the String database (https://string-db.org/) with the species set as Homo sapiens. The minimum required interaction score was set to >0.4 (medium confidence) to obtain the PPI network.
5.4. GO and KEGG Enrichment Analysis
The intersection targets were imported into the DAVID database (https://david.ncifcrf.gov/tools.jsp) for gene function enrichment analysis. GO and KEGG analyses were performed on the results, and visualizations were generated using Excel spreadsheets and the ChiPlot tool Web site (https://www.chiplot.online/).
5.5. Molecular Docking of Yadanzi Drug Small Molecules and Target Proteins
Core target proteins were downloaded from the RCSB PDB database (https://www.rcsb.org/), and water molecules and original ligands were removed using Pymol software. The small molecule structures were drawn using ChemDraw, converted into three-dimensional structures using ChemBio3D, and saved as mol2 format. Molecular docking of the protein and small molecule compounds was performed using AutoDock, and the structure with the highest score was selected for further structural analysis.
5.6. Examining the Inhibitory Effect of YD2 on Lung Cancer Cells
The cell growth rate was determined by the MTT assay. Cells at a density of 2 × 104 per well were added to 48-well plates in sextuplicate and cultured in an RPMI-1640 medium with 10% FBS. Following incubation for 4–5 days, 30 pL of fresh 0.5 mg/mL MTT reagent was added to each well and incubated for another 3–4 h. After incubation, 300 pL of DMSO was added to dissolve the formazan product, and absorbance at 490 nm was then measured with SpectraMax iD3 (MD, USA).
5.7. Colony Formation Assay
A thousand cells were plated into each well in a 6-well plate in triplicate with an RPMI-1640 medium containing 10% FBS. The cells were cultured in an incubator (37 °C, 5% CO2) for 7–10 days. The visible colonies were washed with phosphate-buffered saline (PBS), fixed with precooled 4% paraformaldehyde for 10 min, and then washed with PBS again. Finally, the colonies were stained with a 0.5% crystal violet solution for 10 min and washed with PBS for 3–5 times. The colony area was calculated by ImageJ software.
5.8. Wound Healing Assay
Cells at a density of 4 × 104 per well were seeded into 6-well plates and allowed to attain confluent monolayers. The cells were then cultured in an RPMI-1640 medium with 2% FBS for 12 h. A line or a shape of “#” was drawn at the center of each well using a 10 pL pipet tip to create the wound area. The cells were washed with PBS twice to remove deciduous cells. The cells were incubated for 48 h with an RPMI-1640 medium supplemented with 2% FBS. When the cell wound almost healed, pictures (×100) were taken, and the effects of NFKBIA knockdown on cell migration were evaluated.
5.9. Western Blotting Analysis
The cells were lysed with RIPA buffer (Boster, China) containing protease/phosphatase inhibitor cocktail (Boster, China). Total proteins from cultured cells were isolated, and the concentrations were measured by the Bradford protein assay kit (Bio-Rad, Hercules, CA). After 10–12% SDS-PAGE, the proteins were electrotransferred to the PVDF membrane (Bio-Rad). Then, membranes were blocked with 5% nonfat milk in Tris-buffered saline (TBS) with 1% Tween-20 (1× TBST) for 90 min at room temperature. Next, the membranes were incubated with a primary antibody in TBST overnight at 4 °C, followed by incubation with an appropriate horseradish peroxidase (HRP)-conjugated secondary antibody (×4000) for 1 h at room temperature. Specific proteins were detected with an EZ-ECL Kit (Biological Industries). Primary antibodies were obtained from Cell Signaling Technology.
Acknowledgments
This work was supported by Shaoxing Health Science and Technology Plan (2022KY011), Youth Fund of Shaoxing People’s Hospital (2022YB02), and the Zhejiang Medical Association clinical medical research special fund project (2022ZYC-D14).
Glossary
Abbreviations
- CEA
carcinoembryonic antigen
- NSE
neuron-specific enolase
- TCMSP
traditional Chinese medicine systems pharmacology database and analysis platform
- PPI
protein–protein interaction
- OB
oral bioavailability
- DL
drug-likeness
- JUN
Jun Proto-oncogene
- TP53
Tumor Protein p53
- MAPK1
mitogen-activated protein kinase 1
- IL6
interleukin 6
- AKT1
AKT serine/threonine kinase 1
- TGFB1
transforming growth factor beta 1
- VEGFA
vascular endothelial growth factor A
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.3c10105.
Antiproliferative activity of YD2 on H1299 cells; MTT assay of YD1, YD2, and BJOE on different cells (H520, H1299, and A549); colony formation assay of BJOE on H520 and H1299 cells (PDF)
Author Contributions
# Z.L. and Y.S. contributed equally to this work.
The authors declare no competing financial interest.
Supplementary Material
References
- De Giglio A.; Di Federico A.; Deiana C.; Ricciuti B.; Brambilla M.; Metro G. Advanced non-small-cell lung cancer: how to manage non-oncogene disease. Drugs Context 2022, 11. 10.7573/dic.2022-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath L.; Thienpont B.; Zhao L.; Wolf D.; Pircher A. Overcoming immunotherapy resistance in non-small cell lung cancer (NSCLC) - novel approaches and future outlook. Mol. Cancer 2020, 19 (1), 141. 10.1186/s12943-020-01260-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong J.; Barayan R.; Louie A.; Lok B. Novel therapeutic combinations with PARP inhibitors for small cell lung cancer: A bench-to-bedside review. Seminars in cancer biology 2022, 86, 521–542. 10.1016/j.semcancer.2022.07.008. [DOI] [PubMed] [Google Scholar]
- Weller D. Early diagnosis of lung cancer: diverse populations need diverse solutions. Eur. J. Cancer Care 2021, 30 (2), e13444 10.1111/ecc.13444. [DOI] [PubMed] [Google Scholar]
- Rocco G.; Pennazza G.; Santonico M.; Longo F.; Rocco R.; Crucitti P.; Antonelli Incalzi R. Breathprinting and Early Diagnosis of Lung Cancer. J. Thorac Oncol. 2018, 13 (7), 883–894. 10.1016/j.jtho.2018.02.026. [DOI] [PubMed] [Google Scholar]
- Wang S.; Zimmermann S.; Parikh K.; Mansfield A. S.; Adjei A. A. Current Diagnosis and Management of Small-Cell Lung Cancer. Mayo Clin Proc. 2019, 94 (8), 1599–1622. 10.1016/j.mayocp.2019.01.034. [DOI] [PubMed] [Google Scholar]
- Eggert J.; Palavanzadeh M.; Blanton A. Screening and Early Detection of Lung Cancer. Seminars in oncology nursing 2017, 33 (2), 129–140. 10.1016/j.soncn.2017.03.001. [DOI] [PubMed] [Google Scholar]
- He M.; Feng R.; Wang J.; Xia S.; Wang Y.; Zhang Y. Prevention and treatment of natural products from Traditional Chinese Medicine in depression: Potential targets and mechanisms of action. Frontiers in aging neuroscience 2022, 14, 950143 10.3389/fnagi.2022.950143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L.; Zhang K.; Li Y.; Chen L.; Gao H.; Chen H.; Medeiros I. Real-World Evidence of Traditional Chinese Medicine (TCM) Treatment on Cancer: A Literature-Based Review. Evidence-Based Complementary Altern. Med. 2022, 2022, 7770380. 10.1155/2022/7770380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M.; Chen R.; Wang S.; Tan W.; Hu Y.; Peng X.; Wang Y. Chemical components, pharmacological properties, and nanoparticulate delivery systems of Brucea javanica. Int. J. Nanomed. 2013, 8, 85–92. 10.2147/IJN.S31636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.; Chen S.; Yang X.; Wang S.; Wu W. Efficacy and safety of Brucea javanica oil emulsion injection as adjuvant therapy for cancer: An overview of systematic reviews and meta-analyses. Phytomedicine: international journal of phytotherapy and phytopharmacology 2022, 102, 154141 10.1016/j.phymed.2022.154141. [DOI] [PubMed] [Google Scholar]
- Fuhong D.; Xiang G.; Haiying L.; Jiangye W.; Xueming G.; Wenxiao C. Evaluation of efficacy and safety for brucea javanica oil emulsion in the control of the malignant pleural effusions via thoracic perfusion. BMC Cancer 2018, 18 (1), 411. 10.1186/s12885-018-4328-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K.; Liang Y.; Wang Q.; Li Y.; Zhou S.; Wei H.; Zhou C.; Wan X. Brucea javanica: A review on anticancer of its pharmacological properties and clinical researches. Phytomedicine: international journal of phytotherapy and phytopharmacology 2021, 86, 153560 10.1016/j.phymed.2021.153560. [DOI] [PubMed] [Google Scholar]
- Qureshi N.; Chi J.; Qian Y.; Huang Q.; Duan S. Looking for the Genes Related to Lung Cancer from Nasal Epithelial Cells by Network and Pathway Analysis. Frontiers in genetics 2022, 13, 942864 10.3389/fgene.2022.942864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H.; Liu M.; Luo P.; Yao X.; Zhou H. Network pharmacology provides a systematic approach to understanding the treatment of ischemic heart diseases with traditional Chinese medicine. Phytomedicine: international journal of phytotherapy and phytopharmacology 2022, 104, 154268 10.1016/j.phymed.2022.154268. [DOI] [PubMed] [Google Scholar]
- Hopkins A. Network pharmacology: the next paradigm in drug discovery. Nat. Chem. Biol. 2008, 4 (11), 682–90. 10.1038/nchembio.118. [DOI] [PubMed] [Google Scholar]
- Cheng Z.; Ye F.; Xu C.; Liang Y.; Zhang Z.; Chen X.; Dai X.; Ou Y.; Mou Z.; Li W.; Chen Y.; Zhou Q.; Zou L.; Mao S.; Jiang H. The potential mechanism of Longsheyangquan Decoction on the treatment of bladder cancer: Systemic network pharmacology and molecular docking. Frontiers in pharmacology 2022, 13, 932039 10.3389/fphar.2022.932039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S.; Chen F.; Ye X.; Dong Y.; Xue Y.; Xu H.; Zhang W.; Song S.; Ai L.; Zhang N.; Pan W. Intravenous microemulsion of docetaxel containing an anti-tumor synergistic ingredient (Brucea javanica oil): formulation and pharmacokinetics. Int. J. Nanomed. 2013, 8, 4045–52. 10.2147/IJN.S47956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S.; Chen F.; Ye X.; Dong Y.; Xue Y.; Xu H.; Zhang W.; Song S.; Ai L.; Zhang N.; Pan W. Intravenous microemulsion of docetaxel containing an anti-tumor synergistic ingredient (Brucea javanica oil): formulation and pharmacokinetics. Int. J. Nanomedicine. 2013, 8, 4045–52. 10.2147/IJN.S47956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen L.; Xie L.; Gong F.; Zhang S.; Xi T. Efficacy and safety of Chinese medicine injections in combination with docetaxel and cisplatin for non-small cell lung cancer: a network meta-analysis. Front Pharmacol. 2023, 11 (14), 1277284. 10.3389/fphar.2023.1277284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam P.; Al-Yousef H.; Siddiqui N.; Alhowiriny T.; Alqasoumi S.; Amina M.; Hassan W. H. B.; Abdelaziz S.; Abdalla R. H. Anticancer activity and concurrent analysis of ursolic acid, β-sitosterol and lupeol in three different Hibiscus species (aerial parts) by validated HPTLC method. Saudi Pharm. J. 2018, 26 (7), 1060–1067. 10.1016/j.jsps.2018.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H.; Zhu X.; Gong E.; Lv Y.; Li Y.; Cai X. Luteolin suppresses lung cancer progression through targeting the circ_0000190/miR-130a-3p/notch-1 signaling pathway. J. Chemother. 2023, 35 (4), 330–342. 10.1080/1120009X.2022.2102303. [DOI] [PubMed] [Google Scholar]
- Manios K.; Tsiambas E.; Stavrakis I.; Stamatelopoulos A.; Kavantzas N.; Agrogiannis G.; C Lazaris A. c-Fos/ c-Jun transcription factors in non-small cell lung carcinoma. J. BUON 2020, 25 (5), 2141–2143. [PubMed] [Google Scholar]
- Liu S.; Yu J.; Zhang H.; Liu J. TP53 Co-Mutations in Advanced EGFR-Mutated Non-Small Cell Lung Cancer: Prognosis and Therapeutic Strategy for Cancer Therapy. Frontiers in oncology 2022, 12, 860563 10.3389/fonc.2022.860563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan H.; Huan C.; Zhang W.; Hou Y.; Zhou Z.; Yao J.; Gao S. PDZK1 upregulates nitric oxide production through the PI3K/ERK2 pathway to inhibit porcine circovirus type 2 replication. Veterinary microbiology 2022, 272, 109514 10.1016/j.vetmic.2022.109514. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.











