Abstract
The activation of the PHO5 gene in Saccharomyces cerevisiae in response to phosphate starvation critically depends on two transcriptional activators, the basic helix-loop-helix protein Pho4 and the homeodomain protein Pho2. Pho4 acts through two essential binding sites corresponding to the regulatory elements UASp1 and UASp2. Mutation of either of them results in a 10-fold decrease in promoter activity, and mutation of both sites renders the promoter totally uninducible. The role of Pho4 appears relatively straightforward, but the mechanism of action of Pho2 had remained elusive. By in vitro footprinting, we have recently mapped multiple Pho2 binding sites adjacent to the Pho4 sites, and by mutating them individually or in combination, we now show that each of them contributes to PHO5 promoter activity. Their function is not only to recruit Pho2 to the promoter but to allow cooperative binding of Pho4 together with Pho2. Cooperativity requires DNA binding of Pho2 to its target sites and Pho2-Pho4 interactions. A Pho4 derivative lacking the Pho2 interaction domain is unable to activate the promoter, but testing of UASp1 and UASp2 individually in a minimal CYC1 promoter reveals a striking difference between the two UAS elements. UASp1 is fully inactive, presumably because the Pho4 derivative is not recruited to its binding site. In contrast, UASp2 activates strongly in a Pho2-independent manner. From in vivo footprinting experiments and activity measurements with a promoter variant containing two UASp2 elements, we conclude that at UASp2, Pho2 is mainly required for the ability of Pho4 to transactivate.
The expression of the PHO5 gene in Saccharomyces cerevisiae, which codes for a secreted acid phosphatase, is strongly repressed in phosphate-containing media (17). Two transcription factors, the basic helix-loop-helix protein Pho4 and the homeodomain protein Pho2, are required for transcriptional activation of the PHO5 promoter upon phosphate starvation (20). The activity of Pho4 is regulated through phosphorylation by a cyclin–cyclin-dependent kinase complex encoded by PHO80 and PHO85, respectively (15). Under repressing conditions, Pho4 is phosphorylated and located predominantly in the cytoplasm (19). In addition, Pho80 appears to repress Pho4 activity by direct interaction (13).
Deletion analysis of the PHO5 promoter revealed two regulatory elements, UASp1 and UASp2 (22), to which Pho4 has been shown to bind in vivo upon phosphate starvation, but not under high-phosphate conditions (30). The repressed PHO5 promoter is packaged in a positioned array of nucleosomes that is interrupted only by a short hypersensitive region containing UASp1 (1). Upon induction of the gene, a 600-bp region of the PHO5 promoter becomes hypersensitive to nucleases, reflecting a profound alteration in the structure of four nucleosomes (2). Binding of Pho4 to both UASp1 and UASp2 is required for this transition to occur, which appears to be a prerequisite for transcriptional activation (27).
In contrast, the role of Pho2 in PHO5 regulation is much less clear. Although Pho2 is strictly required for PHO5 promoter activation, no Pho2 target sites relevant for promoter activation have been located so far. Deletion of the one Pho2 binding site that was previously mapped in vitro (31) did not influence PHO5 promoter activity significantly (22). The finding that the activation of a heterologous promoter by a 31-bp oligonucleotide containing UASp1 was fully Pho2 dependent led to the suggestion that Pho2 acts as a trans-acting factor without binding to DNA (24).
Pho2 is a pleiotropic effector which is involved in the regulation of a diverse array of other genes. Together with Swi5, it binds cooperatively to a regulatory element in the HO promoter (7) and plays a complex role in its regulation (18). Also named Bas2, Pho2 is involved in the regulation of HIS4 (3), TRP4 (6), and certain ADE genes (8). It was recently reported that at the ADE5,7 promoter, Pho2 (Bas2) binds to a site located immediately adjacent to the Bas1 site, and indirect evidence suggests that these two proteins bind DNA cooperatively (21, 32).
In this paper, we have dissected the mechanism of Pho2 action at the PHO5 promoter and provide an answer to the long-standing question of its requirement in PHO5 activation. The strategy taken is based on our recent in vitro mapping experiments, in which we demonstrated multiple Pho2 binding sites in the PHO5 promoter. We had also found that Pho2 can bind cooperatively with Pho4 at each Pho4 binding site in vitro (5), raising the possibility that cooperativity between Pho2 and Pho4 might play a role in PHO5 activation in vivo. We now show that Pho2 acts through multiple DNA binding sites and binds to the PHO5 promoter in a cooperative manner with Pho4. Remarkably, two critical functions for Pho2 have emerged from our experiments, the first being to recruit Pho4 to the DNA and the second being to enhance its activation potential once bound to the DNA. At UASp1, recruitment of Pho4 seems to be the crucial role of Pho2, while at UASp2, it is mainly the second function of Pho2 which is required.
MATERIALS AND METHODS
Yeast strains and media.
All S. cerevisiae strains used in this study are isogenic with strain YS18 (MATα his3-11 his3-15 leu2-3 leu2-112 canr ura3Δ5). YS22 contains a disruption of the PHO4 gene, YS19 of the PHO2 gene, and YS27 of PHO4 and PHO2. Yeast strains were grown in YNB medium (Difco, Detroit, Mich.) supplemented with the required amino acids (high-phosphate conditions) or in phosphate-free synthetic medium (29).
Plasmids.
The PHO2-HIS and PHO4-HIS expression plasmids were constructed as described previously (5). The PHO4Δint-HIS plasmid was created by subcloning a PHO4Δint internal fragment into PHO4-HIS. YEp-Pho4 and Yep-Pho4Δ2 have been described by Svaren et al. (28), and Pho2-VP16 and PHO4Δint have been described by Hirst et al. (12). In that paper, PHO4Δint was referred to as PHO4Δ200.
The construction of the PHO5-lacZ reporter plasmid was described previously (26). In the PHO5-lacZ reporter containing 2× UASp2, UASp1 was replaced by UASp2 as described for YS70 (30). The PHO5-UAS CYC1-lacZ reporters were constructed as described previously (24) by using a 2μm yeast vector containing a CYC1-lacZ gene fusion (11). A 31-bp promoter fragment extending from −381 to −351 was used as the UASp1 element (24), and the UASp2 element was a 24-bp oligonucleotide ranging from position −262 to position −239, corresponding to the Pho4 footprint at UASp2 (31). PHO5 DNA restriction fragments used for DNase I footprinting were derived from PHO5-lacZ constructs containing the wild-type or mutated PHO5 promoter. PHO5 DNA fragments used in gel shift analyses were generated by PCR with the wild-type or mutated PHO5 promoter as a template. Mutations of the Pho4 and Pho2 binding sites at the PHO5 promoter were introduced by PCR by the megaprimer technique (23). Expression and purification of the Pho4-HIS, Pho4Δint-HIS, and Pho2-HIS proteins were carried out as described previously (5).
Functional assays.
β-Galactosidase activity measurements (26), DNase I footprinting and gel shift assays (5), nuclease digestion of isolated nuclei, and dimethyl sulfate (DMS) in vivo footprinting with primer 2 (5′-GCCTATTCAATTAACTC) (30) were performed as previously described (29).
RESULTS
Pho4 binding and UAS elements at the PHO5 promoter.
In a linker scanning analysis of the PHO5 upstream region, Rudolph and Hinnen had previously detected two regions which are essential for activation of the promoter by phosphate starvation (22). Each of these regions was later found to contain a Pho4 binding site, and they were termed UASp1 and UASp2 (31). Our recent in vitro binding studies revealed that Pho2 binds cooperatively with Pho4 to several sites flanking both UASp1 and UASp2. In addition, we detected a new, low-affinity Pho4 binding site located 60 bp downstream of UASp2, and there was cooperativity between Pho4 and Pho2 at this site as well (5).
Pho4 is bound to the low-affinity site in vivo under derepressed but not under repressed conditions (data not shown), as we have previously demonstrated to be true for binding of Pho4 to UASp1 and UASp2 (30). To obtain quantitative information about the contribution of each of the three sites, which we have shown to bind Pho4 in vivo, activity measurements of PHO5 promoter variants containing mutations within each Pho4 site (Fig. 1) were carried out, and the results are presented in Table 1. Mutation of either of the Pho4 sites at UASp1 or UASp2 leads to a dramatic decrease in promoter activity (approximately 10-fold), showing cooperative action of the two sites in the activation process, in agreement with the previous deletion analysis of the PHO5 upstream region (10, 22). A mutation of the third Pho4 site, in contrast, has a much smaller effect and brings down the activity to about 70% of the wild-type level. Also, the 14% residual activity of a weakened promoter variant driven by only UASp2 is not any more strongly dependent on the third Pho4 binding site than the wild-type promoter. A variant lacking a functional UASp1 as well as UASp2, but retaining the third Pho4 site, was practically inactive (1% residual activity [Table 1]).
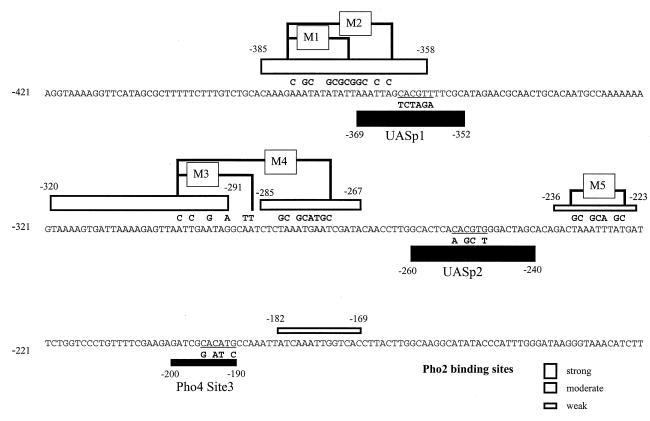
FIG. 1.
Mutation of Pho2 and Pho4 binding sites at the PHO5 promoter. The locations of the Pho4 and Pho2 binding sites as determined by in vitro footprinting (5) are indicated by solid and open bars, respectively, the width of the bars corresponding to the relative affinities of the sites for the factor. Mutated regions within the Pho2 binding sites are in boxes (M1 to M5), and the changed nucleotides are shown above the wild-type sequence. Mutations within the Pho4 consensus sequence are shown below the wild-type sequence and are referred to as UASp1-mut, UASp2-mut, and Pho4 Site3-mut.
TABLE 1.
Contribution of the individual Pho4 binding sites to PHO5 promoter activity
| Promotera | β-Galactosidase activity (%)b |
|---|---|
| Wild type | 100 |
| UASp1-mut | 14 |
| UASp2-mut | 7 |
| Pho4 Site3-mut | 71 |
| UASp1-mut + UASp2-mut | 1 |
| UASp1-mut + Pho4 Site3-mut | 10 |
Pho4 binding sites at the PHO5 promoter were mutated as shown in Fig. 1.
Activities of the mutated promoters are expressed relative to the activity of the wild-type promoter.
We have previously shown that Pho4 is absolutely required for the transition of the promoter chromatin structure to an open state as a prerequisite for transcriptional activation (9). It was therefore conceivable that the third Pho4 site, being occupied in vivo upon promoter activation, might contribute to nucleosome disruption and stabilize the open state. We therefore monitored the structure of nucleosome −2, which contains the third Pho4 binding site, in a promoter construct in which this site had been mutated by measuring the accessibility of a ClaI site to the nuclease (1). Under induced conditions, accessibility to ClaI was indistinguishable from that of the wild-type promoter (not shown). We therefore conclude that binding of Pho4 to this third Pho4 site plays no significant role in the process of chromatin transition and that this site, unlike the Pho4 sites corresponding to UASp1 and UASp2, is not essential for the activation of the PHO5 promoter.
DNA interaction of Pho2 at UASp1 is required for activity of this element.
We next turned to the newly discovered Pho2 binding sites and analyzed their functional role in vivo. To that end, the Pho2 binding sites were mutated individually or in combination (Fig. 1), and the effects on cooperative DNA binding of Pho2 and Pho4 as well as on the activity of the mutated promoter variants were determined.
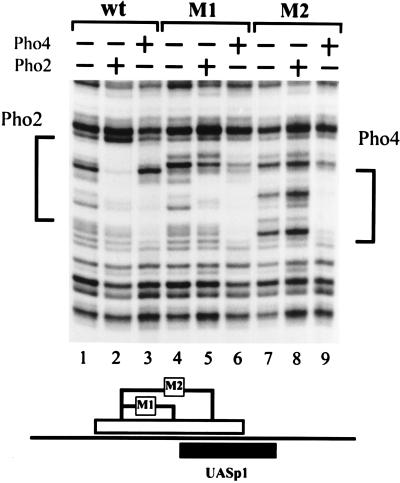
We first tested the function of the strong Pho2 binding sites mapped at UASp1 which partially overlap the Pho4 site. Two mutations were introduced in this site, as shown in Fig. 1 and the schematic of Fig. 2. In the shorter mutation (M1), 7 bases are exchanged in the Pho2-protected region upstream of the Pho4 footprint, while in the longer one (M2), an additional 5 bases extending into the Pho4 DNase I footprint are mutated. Binding of Pho2 and Pho4 to the mutated promoter fragments was then tested by DNase I footprinting in vitro. As shown in Fig. 2, M1 results in the loss of protection in the upstream half of the Pho2 binding region, while M2 brought about complete loss of Pho2 protection, indicating that the previously mapped Pho2-protected region from −385 to −358 (5) actually represents at least two adjacent Pho2 sites. Neither of these mutations affects the DNase I footprints of Pho4 (Fig. 2).
FIG. 2.
Effect of the mutations M1 and M2 on Pho2 and Pho4 binding in vitro. DNase I footprinting was performed as described in Materials and Methods. The upper strand of an SfuI (−206)-BamHI (−542) fragment, derived from the wild-type or mutated promoter, was labeled at the SfuI site. Pho4 or Pho2 was added as indicated at the top. The regions protected in the wild-type promoter are indicated on the side. The locations of the M1 and M2 mutations (Fig. 1) within the Pho2 binding site are shown schematically underneath.
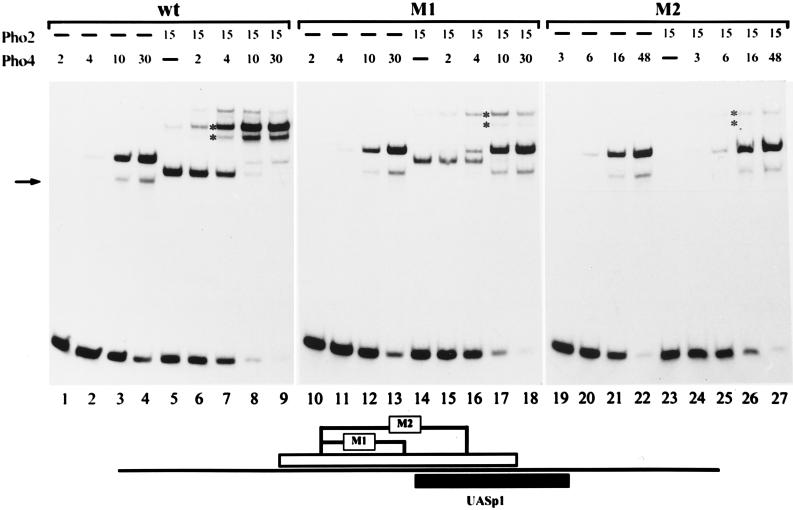
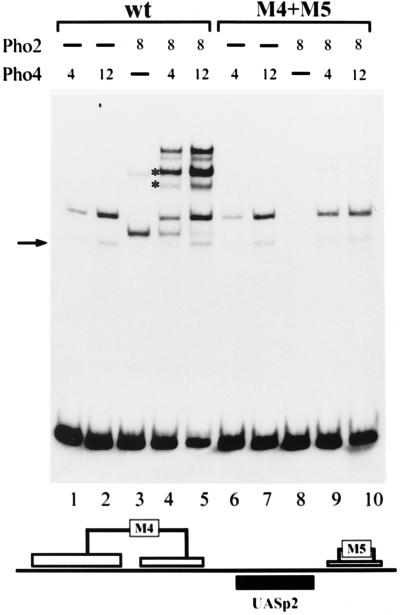
To see if partial or complete loss of Pho2 DNA binding would result in the loss of cooperative DNA binding of Pho2 and Pho4, formation of a ternary complex at the promoter variants was examined by gel shift experiments, and the results were compared to those for the wild-type promoter. As shown in Fig. 3, there was less binary Pho2-DNA complex and concomitant loss of the ternary complex with the promoter fragment containing the smaller mutation (M1). Binding of Pho4 itself was not affected by this mutation. When the promoter fragment was used with the larger mutation (M2), there was no binary Pho2-DNA complex, and the ternary complex was barely detectable. However, it should be noted that binding of Pho4 itself to the M2 fragment was slightly impaired (1.5- to 2-times-higher Pho4 concentrations were required for the same amount of binary complex).
FIG. 3.
Pho2 DNA binding is required for cooperativity between Pho2 and Pho4 at UASp1. The binding reaction and the gel shift assay were performed as described in Materials and Methods. A labeled 81-bp PCR promoter fragment (−324 to −405) was used, containing either the wild-type promoter sequence or the M1 or M2 mutation (see Fig. 1), and is schematically shown at the bottom. The amounts of protein added to an assay mixture are indicated in arbitrary units. One unit of Pho4 and Pho2 corresponds to about 5 and 6 ng of protein, respectively, as determined by sodium dodecyl sulfate gel electrophoresis. The higher-mobility protein-DNA complex observed with Pho4 added alone (marked by an arrow) represents proteolytically degraded Pho4 protein bound to DNA. The positions of the ternary complexes containing either full-length or degraded Pho4 protein are indicated by asterisks. A lower-mobility complex with only Pho2 (lane 5) migrates at approximately the same position as the ternary complex. However, the presence of a ternary complex with the proteolyzed Pho4 protein (lower band with asterisk) makes it possible to unambiguously identify the ternary complex.
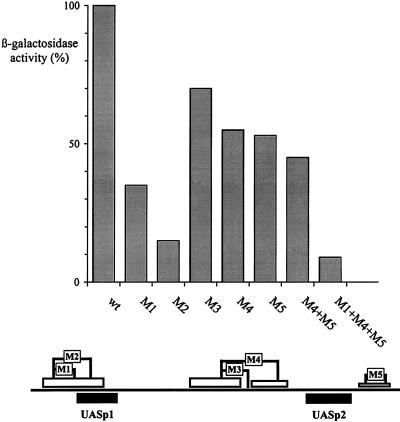
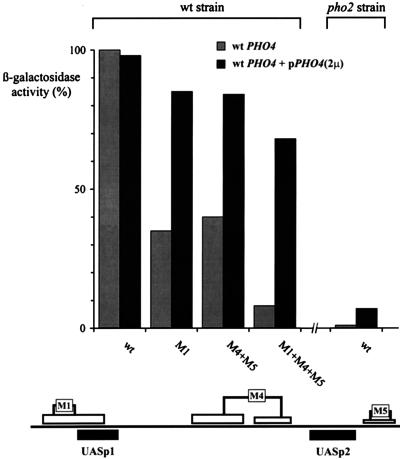
The activities of the mutated promoter variants were measured with lacZ fusion constructs. As shown in Fig. 4, the activity of the promoter containing M1 was reduced to about 35%, and that of the one containing M2 was reduced to about 15%. The residual activity of the M2 promoter corresponds to the activity of a promoter with a mutated Pho4 site at UASp1 (Table 1). This result suggests that cooperative binding of Pho2 and Pho4 at UASp1 is crucial for the activity of this element. However, since disruption of the PHO2 gene brings down promoter activity to less than 1% (Fig. 8), it is clear that Pho2 does more than just facilitate binding of Pho4 at UASp1.
FIG. 4.
Mutations in the Pho2 binding sites differentially affect PHO5 promoter activity. The activities of the wild-type PHO5 promoter fused to the lacZ gene (26) and of promoter variants containing mutations in the Pho2 binding sites were measured as described in Materials and Methods. The activities of the mutated promoter variants are expressed relative to the activity of the wild-type promoter (920 U). The mutations are schematically shown at the bottom.
FIG. 8.
Overexpression of Pho4 relieves the requirement for Pho2 cis-acting elements. Activation of PHO5 promoter variants containing mutated Pho2 cis elements, indicated schematically at the bottom, was measured in a wild-type (wt) strain (YS18) and in the same strain expressing Pho4 from a multicopy plasmid. The activation of a wild-type reporter in a pho2 strain (YS19) is shown on the right. 2μ, 2μm plasmid.
Pho2 binding sites adjacent to UASp2 are also required for full activation of the PHO5 promoter.
In vitro binding studies have revealed the existence of Pho2 binding sites closely adjacent to the Pho4 site at UASp2. Binding of Pho2 to either site alone gives rise to cooperative DNA binding of the two proteins (5). Therefore, it was important to check if mutations in these Pho2 sites would also result in loss of cooperative DNA binding and a concomitant decrease of promoter activity.
Cooperative DNA binding of Pho2 and Pho4 to the promoter fragment containing UASp2 and the mutated Pho2 sites upstream and downstream of the Pho4 site (see schematic in Fig. 5) was examined by gel shift experiments. As shown in Fig. 5, no binding of Pho2 to the fragment carrying the M4 and M5 mutations (M4+M5 variant) was observed, nor was any ternary complex detected (compare lanes 8 to 10 to lanes 3 to 5), clearly showing that Pho2-DNA binding is a prerequisite for cooperative binding of Pho2 and Pho4. Binding of Pho4 alone is unaffected by the M4 and M5 mutations.
FIG. 5.
Pho2-DNA binding is required for cooperativity between Pho2 and Pho4 also at UASp2. The binding reaction and the gel shift assay were performed as described in Materials and Methods. A labeled 109-bp PCR-generated promoter fragment (−316 to −208) containing UASp2 and either the wild-type (wt) sequence or the combined M4+M5 mutation, shown schematically at the bottom, were used. The amounts of protein added to the assay mixture are listed in arbitrary units (for details, see the legend to Fig. 3). For the explanation of the arrow and the asterisks, see the legend to Fig. 3.
When the activities of the promoter construct containing mutated Pho2 sites upstream and/or downstream of UASp2 were measured, a significant drop in activity to about 55% was observed for each of the two single-site mutations (M4 and M5) and to a level of 45% if the two mutations were combined (Fig. 4). This residual activity was still significantly higher than the activity of a promoter with a mutated Pho4 site at UASp2 (7% [Table 1]). These results therefore demonstrate that Pho2 binding adjacent to UASp2, although contributing to promoter strength, does not have the same critical importance as at UASp1.
We have further constructed a promoter variant combining mutated Pho2 sites at both UASp1 and UASp2 (M1+M4+M5). The activity of this promoter was drastically lowered to less than 10% of that of the wild-type promoter (Fig. 4) and was similar to the activity of a promoter containing mutated Pho4 sites at either UASp1 or UASp2 (Table 1). These results show definitively that binding of Pho2 to its target sites adjacent to UASp1 and UASp2 is required for activation of the PHO5 promoter. However, one of the strongest Pho2 binding sites is located between UASp1 and UASp2, and the mutations analyzed so far had not addressed the contribution of this site by itself to promoter activity in vivo. Mutation of 6 bp (M3 in Fig. 1) abolishes interaction with Pho2 within this region, except for a short Pho2-protected region that still persists immediately upstream (not shown). The activity of PHO5 promoter constructs containing this M3 mutation was reduced by about 30% (Fig. 4), showing that binding of Pho2 to this site also makes some contribution to the activation of the PHO5 promoter.
Role of the Pho2 cis elements in the chromatin transition at the PHO5 promoter.
We have previously shown that the Pho2 binding sites adjacent to UASp1 and UASp2 are required in vitro for cooperative binding of Pho2 and Pho4 to DNA (5), and we have now demonstrated that mutation of these Pho2 sites reduces the activity of the promoter in vivo significantly, especially when the sites at UASp1 are mutated. The chromatin transition at the promoter upon PHO5 activation requires binding of Pho4 to both UASp1 and UASp2 (27) and requires the presence of the Pho2 protein (9). It was therefore important to determine what consequences the mutations in the Pho2 binding sites would have on the chromatin transition of the promoter under activation conditions.
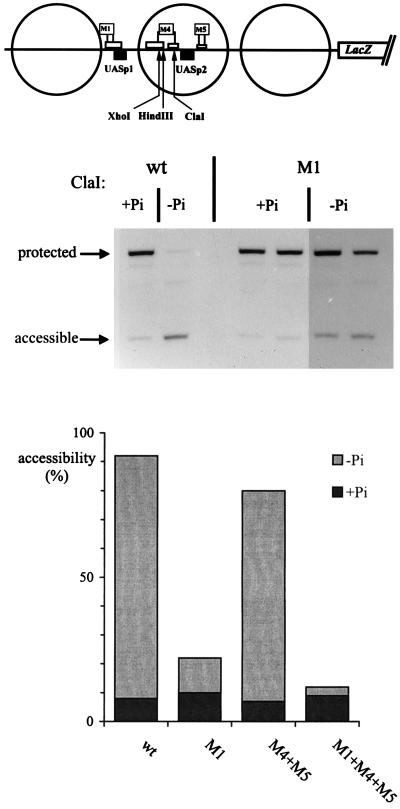
The chromatin structure of the different promoter variants with mutated Pho2 binding sites was analyzed by measuring the accessibility of restriction sites within nucleosome −2 under inducing conditions. The results presented in Fig. 6 show a clear difference in the chromatin structure between the M1 and the M4+M5 variants. By the restriction assay, nucleosome −2 was disrupted in M4+M5 to almost the same extent as in the wild-type promoter, while accessibility in the M1 variant dropped to about 20%. Furthermore, chromatin of the promoter variant containing the combined M1+M4+M5 mutations was closed and nearly indistinguishable by this assay from the structure of the repressed wild-type promoter. These results therefore indicate that binding of Pho4 to UASp1 in vivo must be strongly reduced by mutations of the adjacent Pho2 site (M1), while binding to UASp2 in the M4+M5 promoter is still productive, although impaired to a certain degree, as suggested by the closed chromatin of the M1+M4+M5 promoter compared to the partially open M1 promoter.
FIG. 6.
Chromatin opening at the PHO5 promoter depends on the Pho2 cis elements adjacent to UASp1 but not at UASp2. Strains carrying either the wild-type (wt) PHO5-lacZ plasmid or plasmids with promoter variants were grown in media containing Pi (+Pi) or not containing Pi (−Pi) as indicated, and nuclei were prepared. They were digested for 60 min at 37°C in 200 μl of buffer with 100 U of ClaI or 200 U of HindIII or XhoI (the M4 mutation introduces an XhoI site and a HindIII site, whereas the ClaI site is destroyed). In order to monitor cleavage by the restriction nuclease at the sites shown in the schematic at the top, DNA was isolated, cleaved with RsaI, analyzed in a 1% agarose gel, blotted, and hybridized with a pBR322 RsaI-BamHI fragment which hybridizes to the region immediately upstream of the BamHI site. A 1.46-kb RsaI fragment is generated if the restriction nuclease had been protected, and a fragment about half that size is generated if the site had been accessible. Analysis of the wild-type reporter and the M1 promoter is shown at the top. Accessibility values for the wild-type reporter and M1 promoter as well as M4+M5 and M1+M4+M5 are shown in the diagram below. Measurements for M4+M5 and M1+M4+M5 were derived from XhoI and HindIII digests which gave values that were within 5% of each other.
Activation of the PHO5 promoter by Pho2-VP16 requires multiple Pho2 cis elements in addition to interaction with Pho4.
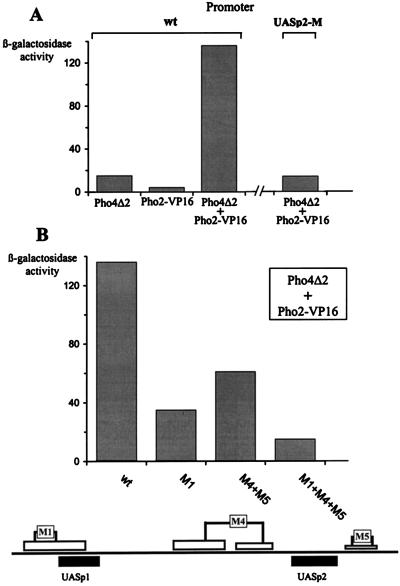
It was previously shown that Pho2-VP16 fusion could activate transcription of the endogenous acid phosphatase only when it was coexpressed with a DNA binding Pho4 derivative which was inactive by itself since it lacked an activation domain (12). This finding demonstrated that Pho2-Pho4 interactions are required for targeting Pho2-VP16 to DNA. However, it was not clear from these results whether the Pho2-VP16 fusion was directly bound to DNA or whether protein-protein interactions with Pho4 were sufficient to recruit the hybrid Pho2 molecule to the promoter. To address this question, we have measured the activity of Pho2-VP16 in the presence of Pho4Δ2, a transcriptionally inactive derivative of Pho4 (28) in a pho2 pho4 strain, by using PHO5 promoter constructs with mutated Pho2 binding sites. In agreement with previous findings, Pho2-VP16 and Pho4Δ2 together, but not individually, activated the wild-type PHO5 promoter as shown in Fig. 7A. Activation not only required the Pho4 protein derivative but also required Pho4 binding sites. The level of activation was significantly reduced after mutation of the Pho2 binding sites adjacent either to UASp1 or to UASp2 (Fig. 7B) and closely paralleled the results obtained when activation of the same mutant promoters by Pho4 was tested (see Fig. 4). The most dramatic effect was observed with multiple Pho2 binding site mutations. These results further support the conclusion that direct Pho2-DNA contacts are required in addition to Pho2-Pho4 interactions to activate the PHO5 promoter and that the multiple Pho2 binding sites mapped in vitro are functional in vivo.
FIG. 7.
Pho2 cis elements are required for activation of the PHO5 promoter by Pho2-VP16 and a transcriptionally inactive Pho4 derivative. (A) Activation of the wild-type PHO5 promoter or a promoter variant with a mutated Pho4 site at UASp2 (UASp2-M) by Pho4Δ2 and/or Pho2-VP16 as measured in YS22 (pho2) or YS27 (pho4 pho2). (B) Activation of the PHO5 promoter variants containing mutated Pho2 binding sites, shown schematically at the bottom, by Pho4Δ2 and Pho2-VP16 expressed together in YS27 (pho4 pho2).
Overexpression of Pho4 relieves the requirement for the Pho2 cis elements.
We have previously shown that overexpression of Pho4 in a pho2 strain restores its ability to disrupt the nucleosomes at the PHO5 promoter, indicative of productive binding of Pho4 to the UAS elements. However, under such conditions, acid phosphatase activity in the mutant was much lower than the level in the wild type (9). This finding raises the question to what extent overexpression of Pho4 in a PHO2 strain would relieve the requirement of the Pho2 cis elements. As shown in Fig. 8, overexpression of Pho4 increases the activity of the promoter with the mutations in the Pho2 sites at both UASp1 and UASp2 (M1+M4+M5) from 9% to 65 to 70% of the wild-type level. Furthermore, the activities of promoter variants containing mutated Pho2 sites only at UASp1 or at UASp2 reached essentially wild-type levels in the presence of overexpressed Pho4. On the other hand, overexpression of Pho4 measured in parallel with a wild-type reporter in a pho2 background gave less than 10% activity (Fig. 8). Therefore, the requirement for Pho2 binding sites in cis can be relieved by overexpression of Pho4 to a much greater extent than the requirement of Pho2 as a trans-acting factor. This finding suggests a more complex role of Pho2 in the activation process at the PHO5 promoter than merely facilitating binding of Pho4 to its target sites.
A mutant Pho4 protein lacking the Pho2 interaction domain is defective in cooperative DNA binding with Pho2.
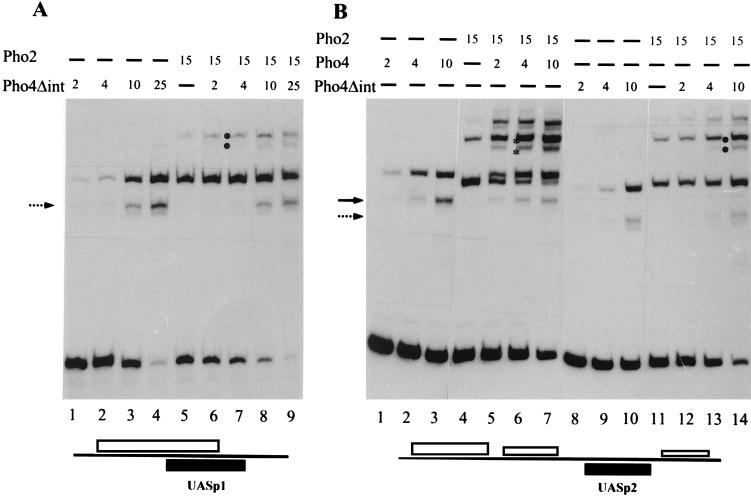
By the use of the two-hybrid system, Pho4-Pho2 interactions had been demonstrated in vivo, and the Pho4 segment essential for this interaction was mapped. A Pho4 protein lacking amino acids 200 to 247 (Pho4Δint) failed to interact with Pho2 (12). To test the importance of specific Pho4-Pho2 interactions for cooperative DNA binding of the two proteins, gel shift experiments with Pho4Δint were performed. As shown in Fig. 9A, no significant cooperativity was observed with Pho4Δint and Pho2 when binding to a promoter fragment containing UASp1 was tested. In contrast, strong cooperativity was observed with wild-type Pho4 and Pho2 in the same experiment (Fig. 3). A certain degree of cooperativity between Pho4Δint and Pho2 was retained when a promoter fragment containing UASp2 with the adjacent Pho2 binding sites was analyzed, but cooperativity was significantly reduced compared to that of wild-type Pho4 (Fig. 9B). Similar results were obtained with UASp2 promoter fragments containing only the 5′ or the 3′ Pho2 binding site (not shown). These data show that specific protein-protein interactions are important for cooperative binding of the two proteins to the PHO5 promoter.
FIG. 9.
Cooperative DNA binding with Pho2 is largely abolished with a Pho4 variant lacking the Pho2 interaction domain. The binding reaction and the gel shift assay were performed as described in Materials and Methods. Proteins were added individually or in combination in the amounts indicated (arbitrary units as in Fig. 3, with 1 U of Pho4Δint corresponding to about 5 ng of protein) to a labeled 81-bp PCR-generated promoter fragment (−324 to −405) containing UASp1 and the overlapping Pho2 sites (A), or to a labeled 109-bp PCR-generated fragment (−316 to −208) containing UASp2 and the adjacent Pho2 sites (B) (see schematics at the bottom). The arrows mark binary complexes derived from proteolytically degraded Pho4 (solid arrow) and Pho4Δint (broken arrow). Ternary complexes with full-length Pho4 and its degradation product are marked by asterisks and those for Pho4Δint are marked analogously by dots.
The Pho4 derivative lacking the Pho2 interaction domain can activate UASp2 but not UASp1.
We next wanted to test the ability of Pho4Δint to activate the PHO5 UAS elements in order to see if loss of cooperative DNA binding of Pho4 and Pho2 correlates with transcriptional activation. Our activity measurements of the PHO5 promoter in vivo suffer, however, from the fact that both UAS elements must synergize for activity, and differential effects on just one of the elements would be hard to detect. We therefore introduced each UAS element into a lacZ reporter plasmid upstream of a CYC1 minimal promoter and measured the activity of these constructs with Pho4Δint and full-length Pho4. It turned out that Pho4Δint was completely unable to activate UASp1, whereas significant activation of UASp2 was measured, which approached the level obtained with full-length Pho4 (Table 2). Activation of UASp2 by Pho4Δint is not due to the residual cooperativity with Pho2 observed in vitro (see above), since the same level of activation was found in the absence of Pho2 (not shown). These results demonstrate that there is a significant difference between the two UAS elements: Pho4Δint can bind to UASp2 and activate transcription, whereas it cannot bind to UASp1, demonstrating that interaction with Pho2 at this UAS element is stringently required for Pho4 binding. In the light of these findings, the poor activation of the PHO5 promoter with Pho4Δint (Table 2) can be explained by its inability to bind (and activate) UASp1, since, as shown before (Table 1), a UASp1 mutation cripples the promoter.
TABLE 2.
Mutant Pho4 protein lacking the Pho2 interaction domain (Pho4Δint) can activate UASp2 but not UASp1
| Reporter | β-Galactosidase activity (U)a
|
|
|---|---|---|
| Pho4Δint | Wild-type Pho4 | |
| PHO5-lacZ | 55 | 920 |
| UASp1 CYC1-lacZ | <5 | 622 |
| UASp2 CYC1-lacZ | 99 | 199 |
The activity of the PHO5 promoter and of heterologous promoter constructs containing individual PHO5 UAS elements was measured in strains expressing either wild-type Pho4 or Pho4Δint. Activity values with the parent vector CYC1-lacZ were <5U.
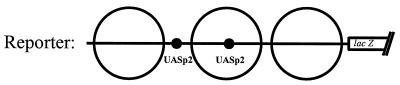
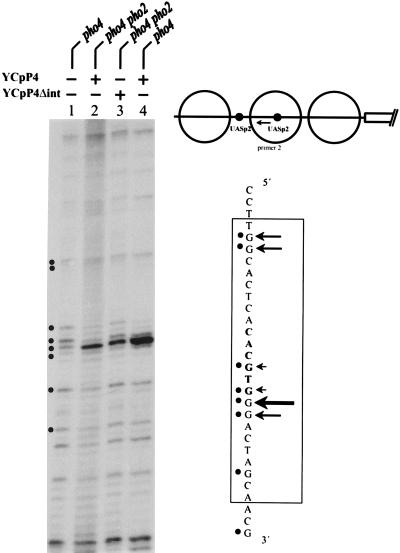
In order to examine activation of UASp2 by Pho4Δint and full-length Pho4 in the context of the PHO5 promoter, we decided to replace UASp1 in the PHO5 promoter and to construct a variant containing 2× UASp2. As expected, this promoter variant was activated significantly by Pho4Δint and again was activated in a Pho2-independent manner (Table 3). However, activation of this reporter by full-length Pho4 is still higher in a PHO2 background. This might due to an effect of Pho2 on the ability of Pho4 either to bind DNA even better, to transactivate, or both. To address this question, we used in vivo footprinting to determine the occupancy of the newly introduced UASp2 by using either full-length Pho4 or Pho4Δint. Unlike the native UASp2 element, the new one is accessible in the nucleus also under repressing conditions (28), since it resides in a constitutively hypersensitive chromatin region of the promoter (1). Binding of full-length Pho4 is indeed improved in the presence of Pho2 (compare lanes 2 and 4 in Fig. 10). However, even in the absence of Pho2, there was strong binding of Pho4 as well as Pho4Δint. As expected, binding of Pho4Δint is not increased in the presence of Pho2 (not shown), consistent with the activation results (Table 3).
TABLE 3.
Activation of a PHO5 promoter variant containing 2× UASp2
| Derivative | β-Galactosidase activity (U)a
|
|
|---|---|---|
| PHO2 | pho2 | |
| Pho4 (wild type) | 960 | 85 |
| Pho4Δint | 189 | 214 |
Activity of the PHO5 promoter variant containing 2× UASp2 (see Materials and Methods) was measured with wild-type Pho4 and Pho4Δint in either YS22 (pho4) or YS27 (pho2 pho4).
FIG. 10.
DMS footprint analysis of Pho4 binding to UASp2. Binding of wild-type Pho4 and Pho4Δint to the upstream UASp2 element in the PHO5 promoter variant in which UASp1 was replaced by UASp2 (see schematic) was examined in YS22 (pho4) or YS27 (pho2 pho4). For details of DMS footprinting and the primer used (arrow in the schematic), see Materials and Methods.
Importantly, the footprints were practically identical for Pho4 and Pho4Δint (lanes 2 and 3 in Fig. 10), indicating that both molecules bound with similar efficiency to this UAS element. Since much higher activity was measured under these conditions with Pho4Δint compared to that with full-length Pho4, this must mean that the ability of Pho4 to transactivate can be positively modified. A possible explanation is the presence of a repressive domain in Pho4 that is at least partially removed in Pho4Δint and that is counteracted by Pho2 under physiological conditions. A similar role of Pho2 in enhancing the activation potential of Pho4 was recently proposed by Shao et al. (25) using a different approach.
In conclusion, we cannot unambiguously discriminate to what extent the increased activation by wild-type Pho4 through UASp2 in the presence of Pho2 is due to improved DNA binding as opposed to enhanced transcriptional activation. However, our results with Pho4Δint show that Pho2 also has an effect on the ability of Pho4 to transactivate. Furthermore, there is a clear difference between the two UAS elements in this respect which had previously not been recognized. At UASp1, binding of Pho4 is the limiting step for which Pho2 is stringently required, while at UASp2, Pho2 plays much less of a role in affecting binding but instead exerts a novel effect by improving the activation potential of Pho4.
DISCUSSION
The strict dependence of PHO5 promoter activation on Pho2 has been known for over 25 years (20), but clues to the underlying mechanism have emerged only recently. Our recent in vitro studies revealed the existence of multiple Pho2 binding sites at the PHO5 promoter in close vicinity to the Pho4 binding sites. In addition, highly cooperative DNA binding of Pho2 and Pho4 was demonstrated at UASp1 and UASp2 (5). In this paper, we have examined the functional relevance of the newly mapped Pho2 sites and have asked whether the cooperativity between the two proteins plays a role in the activity of the PHO5 promoter in vivo. To address these questions, we have constructed a series of different promoter variants with single or multiple mutations in the Pho2 sites.
The requirement of UASp1 and UASp2 for chromatin disruption at the PHO5 promoter and subsequent transcriptional activation had previously been demonstrated by deletion analyses of the promoter (10, 22) and was confirmed here by analyzing the effects of targeted mutations. In contrast to these two elements, the recently mapped low-affinity Pho4 site (5) does not play a crucial role in the activation process. Although it is occupied in vivo under derepressed conditions, it is dispensable for chromatin disruption (data not shown), and makes only a small contribution to overall promoter activity (Table 1). We have therefore focused our investigations on the Pho2 sites adjacent to UASp1 and UASp2.
Pho2 is involved in the activation of the PHO5 promoter through cooperative DNA binding with Pho4.
Mutation of the Pho2 binding sites adjacent to UASp1 and UASp2 results in the loss of cooperative DNA binding of Pho2 and Pho4 in vitro (see Fig. 3 and 5). The finding that mutation of any individual Pho2 binding site caused a moderate to strong reduction in PHO5 promoter activity indicates that cooperative binding of the two proteins is also essential for the activity of the PHO5 promoter in vivo. Furthermore, a dramatic effect was obtained by the combined mutation of the Pho2 sites adjacent to both UASp1 and UASp2, which was similar to the effect of mutating UASp1 or UASp2 itself (Table 1 and Fig. 4). The functional importance of the Pho2 binding sites in vivo was further confirmed in experiments in which the PHO5 promoter was activated by a Pho2-VP16 hybrid in the presence of a transcriptionally inactive Pho4 derivative that could still bind DNA. Promoter variants with mutated Pho2 sites could not be efficiently activated by Pho2-VP16 (Fig. 7B). Taken together, these results provide the first direct evidence that Pho2 is involved in the activation of the PHO5 promoter as a sequence-specific DNA-binding protein. At the same time, they show that binding of Pho2 to DNA requires interactions with Pho4, because in the absence of the Pho4 derivative or a Pho4 binding site, Pho2-VP16 is transcriptionally silent.
The properties of Pho2 are typical of the family of homeodomain proteins to which Pho2 belongs. In many cases, such proteins bind DNA at multiple sites with relatively low sequence specificity in vitro and are thought to gain their selectivity through protein-protein interactions with other factors (16). The collected evidence therefore strongly indicates that Pho2 contributes to PHO5 activation as a DNA-binding factor which binds to specific sequences at the PHO5 promoter cooperatively with Pho4 (5).
UASp1 and UASp2 differ in their dependence on Pho2 cis elements.
Prevention of Pho2 interactions with its target sites results in the loss of cooperative DNA binding of the two proteins and leads to the progressive weakening of the PHO5 promoter. However, interference with Pho2 binding around UASp1 has a stronger effect than that around UASp2, as borne out in decreased promoter activity on the one hand and in its consequences on chromatin opening on the other. UASp1 is stringently required for the disruption of the four nucleosomes at the promoter (10). Similarly, the M1 mutation, in which part of the Pho2 site next to UASp1 is mutated, strongly impairs chromatin opening, and accessibility of the ClaI site in nucleosome −2 changes from around 90% to about 20% (Fig. 6). The M2 mutation that eliminates Pho2 binding entirely has an even stronger effect on activity and chromatin opening (not shown). However, this mutation extends into the region at which the Pho2 and the Pho4 sites overlap and leads to a slight decrease in Pho4 binding in vitro even in the absence of Pho2 and was therefore not further investigated.
In contrast, mutation of the two Pho2 sites adjacent to UASp2 (M4+M5) affects promoter activity in vivo less, and nucleosome −2 is still almost completely disrupted. There is a measurable effect of the M4+M5 mutation in the chromatin assay, however. When it is combined with the M1 mutation, chromatin is almost fully closed in contrast to the partial opening of the promoter with the M1 mutation. This indicates that binding of Pho4 to UASp2 in the M4+M5 promoter is not as strong as that in the wild-type promoter but is still sufficient to disrupt the nucleosome (Fig. 6). Therefore, it seems that in contrast to UASp1, binding of Pho4 to UASp2 is improved by cooperative interactions with Pho2 but is not absolutely dependent on them.
The persistence of nucleosome −2 in a pho2 strain (9) precludes directly assaying the occupancy of UASp2 by Pho4, since we have shown that the nucleosome prevents binding of Pho4 to its target site (30). However, construction of a PHO5 promoter variant with two UASp2 elements, one replacing UASp1, has made it possible to directly determine the binding of Pho2 to UASp2 now located in a nucleosome-free region. With this construct, we could demonstrate that Pho4 indeed binds strongly to UASp2, even in the absence of Pho2 (Fig. 10), in agreement with the chromatin data.
A Pho4 derivative lacking the Pho2 interaction domain clearly distinguishes between UASp1 and UASp2.
It was previously demonstrated that a Pho4 derivative lacking amino acids 200 to 247, Pho4Δint, is unable to interact with Pho2 in the two-hybrid system (12), and Pho4Δint is indeed defective in cooperative DNA binding with Pho2 in vitro (Fig. 9). Furthermore, Pho4Δint can activate PHO5 transcription only very poorly (about 6% of the level attained with full-length Pho4), supporting the importance of cooperativity between Pho2 and Pho4 in physiological activation. However, activity measurements revealed a striking difference between the two UAS elements when they were tested out of context. UASp2 was significantly activated by Pho4Δint, while UASp1 was not activated at all. Activation of UASp2 with Pho4Δint was completely Pho2 independent, showing that residual slight cooperative binding of Pho4Δint and Pho2 observed in vitro (Fig. 9B) plays no role in vivo. Consistent with this difference between UASp1 and UASp2, the PHO5 promoter variant with UASp1 replaced by UASp2 shows significant activation with Pho4Δint in the absence of Pho2. Surprisingly, under these conditions (i.e., in the absence of Pho2), wild-type Pho4 activates this construct very poorly, although in terms of binding to UASp2 it is indistinguishable from Pho4Δint. This result throws a new light on transactivation of Pho4 and makes it likely that Pho4 is negatively regulated by an internal repressive domain in its ability to transactivate. The simple deletion of the Pho4 segment interacting with Pho2 is sufficient to at least partially relieve this repression, something otherwise accomplished by interaction with Pho2.
The Pho2-independent activation of UASp2 resembles that of the recently described Pho4 mutants containing deletions in the basic region (amino acids 252 to 265), which is proposed to mediate functional interactions with Pho2 (25). These deletions result in Pho2-independent activation of the GAL1 promoter by a Gal4(DBD [for DNA binding domain])-Pho4 hybrid protein, while full-length Pho4 fused to the Gal4 DNA binding domain requires Pho2 for activation. Since a Gal4 DNA binding domain was used, a role of Pho2 in the ability of Pho4 to bind DNA was not addressed. The authors propose a model in which the Pho4 activation domain interacts with the basic region and is thereby masked. The role of Pho2 would then be to disrupt this internal interaction, expose the activation domain, and generate a transcriptionally competent molecule. In the framework of this model, the deletion of amino acids 200 to 247 in our experiments, which results in the inability of Pho4 to interact with Pho2, would destroy the internal interaction in the Pho4 protein, thus exposing the activation domain and in effect bypassing the Pho2 requirement.
A second role for Pho2 can also explain our finding that the loss of promoter activity due to mutation of Pho2 cis elements can be almost fully compensated for by overexpression of Pho4 in a PHO2 strain, while the same is not true for the loss of promoter activity due to elimination of Pho2 itself. These results show that the presence of Pho2 in trans results in higher transcriptional activity than that measured in a pho2 strain, providing further support for the additional role of Pho2 in transcriptional activation.
Pho2: a pleiotropic factor in yeast.
Pho2 is involved in the regulation of several genes, and the common principle in all cases so far seems to be that it interacts with gene-specific factors. At the HO promoter, a Pho2 binding site is located next to a Swi5 binding site, and it was shown that the two proteins bind to their sites cooperatively in vitro (7). However, recent in vivo data indicate that the role of Pho2 in HO regulation is complex (18). In the case of the HIS4 promoter, a Pho2 (Bas2)-protected region largely overlaps the Bas1 footprint. Although Bas2 and Bas1 can bind to this region simultaneously, no cooperative interactions between the two proteins were detected (3). In contrast, at the TRP4 promoter, a Pho2 binding site completely overlaps one of the two Gcn4 binding sites, and the two proteins were found to bind DNA in a mutually exclusive manner (6). The role of Pho2 (Bas2) in the activation of the ADE genes was recently investigated (21, 32). Transcriptional activation of these genes requires the concerted action of Bas1 and Bas2 and is down-regulated by adenine. From their studies, the authors conclude that Pho2 (Bas2) stimulates both DNA binding and activation by Bas1 at the ADE5,7 promoter. Interestingly, when a mutant Pho2 protein lacking the DNA binding domain was tested together with Bas1, it was still partially functional in ADE5,7 activation. This is in contrast to the mechanism of action of Pho2 at the PHO5 promoter, since our results show that DNA binding is critically required for Pho2 function. This difference is also borne out in a recent study by Justice et al. (14), who showed that mutations in the Pho2 DNA binding domain almost entirely abolish activation by PHO5 UASp1 and the UAS elements of the HIS4 as well as the HO promoter, while an ADE1-lacZ reporter retained 30 to 40% activity.
Dual role of Pho2 in the activation of the PHO5 promoter.
It is obvious that Pho2 is an exceptional protein when it comes to the diverse effects it has on cellular metabolism and the means employed to functionally complement the dedicated factor at each promoter. Cooperative binding with a specific factor appears to be the primary mechanism, but with the same partner, the requirements for cooperative binding can be different for different promoters (e.g., PHO5 versus PHO8, where Pho4 binding is largely Pho2 independent [unpublished observations; see also reference 4]), or even more remarkably, at different sites of the same promoter, as shown here for PHO5 UASp1 and UASp2. Clearly, the additional role of Pho2 in exposing the activation domain of the primary activator can be effective only in a case in which binding of the activator protein is at least to some extent Pho2 independent, as appears to be the case for UASp2 at PHO5.
ACKNOWLEDGMENTS
We thank J. Svaren and Philip Gregory for discussions and comments on the manuscript and D. Blaschke, A. Schmid, and M. Zavari for expert assistance. We are grateful to D. Stillman for the gift of PHO2-HIS.
This work was supported by the Deutsche Forschungsgemeinschaft (SFB 190) and Fonds der Chemischen Industrie.
REFERENCES
- 1.Almer A, Hörz W. Nuclease hypersensitive regions with adjacent positioned nucleosomes mark the gene boundaries of the PHO5/PHO3 locus in yeast. EMBO J. 1986;5:2681–2687. doi: 10.1002/j.1460-2075.1986.tb04551.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Almer A, Rudolph H, Hinnen A, Hörz W. Removal of positioned nucleosomes from the yeast PHO5 promoter upon PHO5 induction releases additional upstream activating DNA elements. EMBO J. 1986;5:2689–2696. doi: 10.1002/j.1460-2075.1986.tb04552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arndt K T, Styles C, Fink G R. Multiple global regulators control HIS4 transcription in yeast. Science. 1987;237:874–880. doi: 10.1126/science.3303332. [DOI] [PubMed] [Google Scholar]
- 4.Barbaric S, Fascher K D, Hörz W. Activation of the weakly regulated PHO8 promoter in S. cerevisiae: chromatin transition and binding sites for the positive regulator protein PHO4. Nucleic Acids Res. 1992;20:1031–1038. doi: 10.1093/nar/20.5.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barbaric S, Münsterkötter M, Svaren J, Hörz W. The homeodomain protein Pho2 and the basic-helix-loop-helix protein Pho4 bind DNA cooperatively at the yeast PHO5 promoter. Nucleic Acids Res. 1996;24:4479–4486. doi: 10.1093/nar/24.22.4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braus G, Mösch H U, Vogel K, Hinnen A, Hütter R. Interpathway regulation of the TRP4 gene of yeast. EMBO J. 1989;8:939–945. doi: 10.1002/j.1460-2075.1989.tb03455.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brazas R M, Bhoite L T, Murphy M D, Yu Y X, Chen Y Y, Neklason D W, Stillman D J. Determining the requirements for cooperative DNA binding by Swi5p and Pho2p (Grf10p/Bas2p) at the HO promoter. J Biol Chem. 1995;270:29151–29161. doi: 10.1074/jbc.270.49.29151. [DOI] [PubMed] [Google Scholar]
- 8.Daignan-Fornier B, Fink G R. Coregulation of purine and histidine biosynthesis by the transcriptional activators BAS1 and BAS2. Proc Natl Acad Sci USA. 1992;89:6746–6750. doi: 10.1073/pnas.89.15.6746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fascher K D, Schmitz J, Hörz W. Role of trans-activating proteins in the generation of active chromatin at the PHO5 promoter in S. cerevisiae. EMBO J. 1990;9:2523–2528. doi: 10.1002/j.1460-2075.1990.tb07432.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fascher K D, Schmitz J, Hörz W. Structural and functional requirements for the chromatin transition at the PHO5 promoter in Saccharomyces cerevisiae upon PHO5 activation. J Mol Biol. 1993;231:658–667. doi: 10.1006/jmbi.1993.1317. [DOI] [PubMed] [Google Scholar]
- 11.Guarente L, Ptashne M. Fusion of Escherichia coli lacZ to the cytochrome c gene of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1981;78:2199–2203. doi: 10.1073/pnas.78.4.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hirst K, Fisher F, McAndrew P C, Goding C R. The transcription factor, the cdk, its cyclin and their regulator: directing the transcriptional response to a nutritional signal. EMBO J. 1994;13:5410–5420. doi: 10.1002/j.1460-2075.1994.tb06876.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jayaraman P S, Hirst K, Goding C R. The activation domain of a basic helix-loop-helix protein is masked by repressor interaction with domains distinct from that required for transcription regulation. EMBO J. 1994;13:2192–2199. doi: 10.1002/j.1460-2075.1994.tb06496.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Justice M C, Hogan B P, Vershon A K. Homeodomain-DNA interactions of the Pho2 protein are promoter-dependent. Nucleic Acids Res. 1997;25:4730–4739. doi: 10.1093/nar/25.23.4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaffman A, Herskowitz I, Tjian R, O’Shea E K. Phosphorylation of the transcription factor Pho4 by a cyclin-CDK complex, Pho80-Pho85. Science. 1994;263:1153–1156. doi: 10.1126/science.8108735. [DOI] [PubMed] [Google Scholar]
- 16.Laughon A. DNA binding specificity of homeodomains. Biochemistry. 1991;30:11357–11367. doi: 10.1021/bi00112a001. [DOI] [PubMed] [Google Scholar]
- 17.Lenburg M E, Oshea E K. Signaling phosphate starvation. Trends Biochem Sci. 1996;21:383–387. [PubMed] [Google Scholar]
- 18.McBride H J, Brazas R M, Yu Y, Nasmyth K, Stillman D J. Long-range interactions at the HO promoter. Mol Cell Biol. 1997;17:2669–2678. doi: 10.1128/mcb.17.5.2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O’Neill E M, Kaffman A, Jolly E R, O’Shea E K. Regulation of Pho4 nuclear localization by the Pho80-Pho85 cyclin-CDK complex. Science. 1996;271:209–212. doi: 10.1126/science.271.5246.209. [DOI] [PubMed] [Google Scholar]
- 20.Oshima Y. Regulatory circuits for gene expression: the metabolism of galactose and phosphate. In: Strathern J N, Jones E W, Broach J R, editors. The molecular biology of the yeast Saccharomyces cerevisiae: metabolism and gene expression. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. pp. 159–180. [Google Scholar]
- 21.Rolfes R J, Zhang F, Hinnebusch A G. The transcriptional activators Bas1, Bas2, and Abf1 bind positive regulatory sites as the critical elements for adenine regulation of ADE5,7. J Biol Chem. 1997;272:13343–13354. doi: 10.1074/jbc.272.20.13343. [DOI] [PubMed] [Google Scholar]
- 22.Rudolph H, Hinnen A. The yeast PHO5 promoter: phosphate-control elements and sequences mediating mRNA start-site selection. Proc Natl Acad Sci USA. 1987;84:1340–1344. doi: 10.1073/pnas.84.5.1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sarkar G, Sommer S S. The “megaprimer” method of site-directed mutagenesis. BioTechniques. 1990;8:404–407. [PubMed] [Google Scholar]
- 24.Sengstag C, Hinnen A. A 28-bp segment of the Saccharomyces cerevisiae PHO5 upstream activator sequence confers phosphate control to the CYC1-lacZ gene fusion. Gene. 1988;67:223–228. doi: 10.1016/0378-1119(88)90399-x. [DOI] [PubMed] [Google Scholar]
- 25.Shao D, Creasy C L, Bergman L W. Interaction of Saccharomyces cerevisiae Pho2 with Pho4 increases the accessibility of the activation domain of Pho4. Mol Gen Genet. 1996;251:358–364. doi: 10.1007/BF02172527. [DOI] [PubMed] [Google Scholar]
- 26.Straka C, Hörz W. A functional role for nucleosomes in the repression of a yeast promoter. EMBO J. 1991;10:361–368. doi: 10.1002/j.1460-2075.1991.tb07957.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Svaren J, Hörz W. Transcription factors vs nucleosomes: regulation of the PHO5 promoter in yeast. Trends Biochem Sci. 1997;22:93–97. doi: 10.1016/s0968-0004(97)01001-3. [DOI] [PubMed] [Google Scholar]
- 28.Svaren J, Schmitz J, Hörz W. The transactivation domain of Pho4 is required for nucleosome disruption at the PHO5 promoter. EMBO J. 1994;13:4856–4862. doi: 10.1002/j.1460-2075.1994.tb06812.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Svaren J, Venter U, Hörz W. In vivo analysis of nucleosome structure and transcription factor binding in Saccharomyces cerevisiae. Microb Gene Techniques. 1995;6:153–167. [Google Scholar]
- 30.Venter U, Svaren J, Schmitz J, Schmid A, Hörz W. A nucleosome precludes binding of the transcription factor Pho4 in vivo to a critical target site in the PHO5 promoter. EMBO J. 1994;13:4848–4855. doi: 10.1002/j.1460-2075.1994.tb06811.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vogel K, Hörz W, Hinnen A. The two positively acting regulatory proteins PHO2 and PHO4 physically interact with PHO5 upstream activation regions. Mol Cell Biol. 1989;9:2050–2057. doi: 10.1128/mcb.9.5.2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang F, Kirouac M, Zhu N, Hinnebusch A G, Rolfes R J. Evidence that complex formation by Bas1p and Bas2p (Pho2p) unmasks the activation function of Bas1p in an adenine-repressible step of ADE gene transcription. Mol Cell Biol. 1997;17:3272–3283. doi: 10.1128/mcb.17.6.3272. [DOI] [PMC free article] [PubMed] [Google Scholar]