Abstract
Staf is a transcriptional activator of prime importance for enhanced transcription of small nuclear (snRNA) and snRNA-type genes transcribed by RNA polymerases II and III (Pol II and III). In addition to this activity, it also possesses the capacity to stimulate expression from an RNA polymerase II mRNA promoter. This promiscuous activator thus provides a useful model system for studying the mechanism by which one single transcription factor can activate a large variety of promoters. Here, we report the use of in vivo assays to identify the Staf activation domains involved in promoter selectivity. Analysis of Staf mutants reveals the existence of two physically and functionally distinct regions, outside of the DNA binding domain, responsible for mediating selective transcriptional activation. While a 93-amino-acid domain, with the striking presence of four repeated units, is specialized for transcriptional activation of an mRNA promoter, a segment of only 18 amino acids, with a critical Leu-213 residue, acts specifically on Pol II and Pol III snRNA and snRNA-type promoters. In addition, this study disclosed the fundamental importance of invariant leucine and aspartic acid residues located in each repeat unit of the mRNA activation domain. Staf is therefore the first transcriptional activator described so far to harbor two physically and functionally distinct activator domains. This finding suggests that the same activator can contact different, specialized transcription complexes formed on different types of basal promoters through promoter-specific transactivation pathways.
Transcriptional activation in eukaryotes is achieved by regulatory proteins recruiting the transcriptional machinery and chromatin remodeling factors to the promoter. Transcriptional activators have been shown to present a modular organization consisting of distinct domains for sequence-specific DNA binding and transcriptional activation through interaction with other proteins (for a review, see reference 34). DNA binding domains of many transactivators fall into readily identifiable classes. Instead, domains responsible for transcriptional activity may not possess such a well-defined organization. While no obvious sequence similarity has been detected among the activation domains of diverse activator proteins, they nevertheless often carry a distinctive amino acid composition. For example, activation domains particularly rich either in prolines, glutamines, acidic amino acids, or hydroxylated amino acids have been described (for reviews, see references 4, 21, 27, and 51). Despite the identification of targets for numerous activation domains and elucidation of the functional importance of particular residues, the structural basis for the capacity of an activation domain to stimulate transcription remains poorly understood. However, recent structural studies suggest that the α-helix may be a common structural motif in the binding of transactivation factors to transcriptional activation factors (22, 52).
Transcriptional activators were also described to stimulate transcription of RNA polymerases II (Pol II) and III (Pol III) small nuclear RNA (snRNA) genes. In vertebrates, these genes contain an essential proximal sequence element (PSE) located at approximately the position −59 upstream of the transcription start site. The Pol III-dependent promoters possess additionally a TATA box located at position −30, which acts as a major determinant for RNA Pol III specificity (24, 26; for a review, see reference 15). A number of other short transcription units, such as the 7SK, Y, MRP, selenocysteine tRNA (tRNASec), and H1 RNA genes, have similar basal promoter elements and can be classified as snRNA-type genes (3, 30; for a review, see reference 14). These will be here referred to as snRNA-type promoters. SNAPc, also called PTF, is a stable complex containing four protein subunits. It binds specifically to the PSE (13, 31, 37, 54, 55). Other studies have also implicated the TATA binding protein (TBP), TFIIIC1, and TFIIIBα in transcription of Pol III snRNA genes (25, 41, 48, 55).
Associated with the basal promoter, the snRNA and snRNA-type genes contain a distal sequence element (DSE) around position −220, which plays a major role in transcription efficiency. The DSE contains an octamer motif which binds the transcriptional activator Oct-1 (42; for reviews, see references 14 and 16). In addition to Oct-1, Sp1 has been shown in some instances to be involved in mediating the activation properties of the DSE (1, 20, 23, 47). Recently, we have demonstrated that the zinc finger protein Staf, originally identified as the transcriptional activator of the tRNASec gene (39), is also involved in transcriptional activation of snRNA and snRNA-type genes transcribed by RNA polymerases II and III (38). In the course of previous investigations, we observed that Staf also possesses the ability to stimulate CAT expression from a Pol II promoter (32, 39), therefore showing that it can activate a whole diversity of Pol II and Pol III promoters. At first sight, this result might be surprising in view of the fact that, although transcription of mRNA and Pol II snRNA genes is catalyzed by the same enzyme, the architectures of mRNA and snRNA transcription complexes have some dissimilarities (for reviews, see references 14 and 15). So far, the mechanism by which Staf can interact with these different Pol II and Pol III transcription complexes has remained unknown. In order to gain insight into it, the mapping and functional dissection of the Staf activation domain was undertaken. We report here that Staf contains in fact two distinct snRNA and mRNA promoter-specific activation domains, explaining why a single transcriptional activator can activate a variety of different promoters. This finding lays the foundation for further elucidation of the detailed, promoter-specific activation pathways.
MATERIALS AND METHODS
Effector constructs.
Staf-Krox-20 Δ473 to 600 (SKΔ473-600), the Staf-Krox-20 3′-end-truncated derivative, was obtained by double digestion of pBRN3–Staf-Krox-20 (39) with EcoRV (cleavage at position 1443 of the Staf cDNA sequence) and NotI (cleavage in the polylinker of pBRN3) and recircularization of the plasmid after filling-in of the NotI 5′ extremity. Staf-Krox-20 Δ6-248 (SKΔ6-248), the Staf-Krox-20 5′-end-truncated derivative, was obtained by digestion of pBRN3-Staf-Krox-20 with NcoI (cleaved at positions 138 and 771 of the Staf cDNA sequence) and recircularization of the filled-in ends of the shortened plasmid. Internal deletions Δ6-77, Δ6-102, Δ6-130, Δ6-206, Δ6-220, Δ78-220, Δ157-220, Δ6-206+Δ225-242, Δ84-176, Δ244-262, and Δ212-220 in Staf-Krox-20 Δ473-600, truncation Δ212-220 in Staf-Krox-20, and mutants PDTIS207-211AAAAA, SIEG221-224AAAA, E214A+Q215A+K219A, L213A+Y216A, S211A, L213A, E214A, Q215A, Y216A, and K219A in Staf-Krox-20 Δ473-600 were obtained by site-directed mutagenesis of the parental vector. S84-176K contains the two internal deletions, Δ6-78 and Δ177-262, obtained by site-directed mutagenesis of the parental vector.
S207-224K was obtained as follows. A polylinker created by annealing the oligonucleotides 5′GATCTTCTAGAGCCGCCACCATGGCATCGATCCCGGGAAGACACACCAAGGATCCAGATCTGC3′ and 5′CATGGCAGATCT GGATCCTTGGTGTGTCTTCCCGGGATCGATGCCATGGTGGCGGCTC TAGAA3′ was inserted into pBRN3-Krox-20–DBD cleaved at the BglII and NcoI sites (39). This creates E83. A 64-bp BbsI fragment (containing Pro 207 to Gly 224 of Staf), obtained by PCR amplification using the 5′ and 3′ primers complementary to positions 646 to 663 and 679 to 700 of the Staf cDNA sequence, was inserted into the BbsI-cleaved E83 construct. The 5′CTGAAGACACACCACCCCGACACAATCAGCGCA and 3′CTGAAGACTGTGGTCTCCTTCAATCGACACCTTTGC primers incorporated the BbsI sites. The sequence of the N-terminal part of the S207-224K protein upstream of the Krox-20-DBD is MASIPGKTHHPDTISALEQYAAKVSIEGDGGSRSAM (in bold, the Staf sequence from residues 207 to 224).
Deletion mutations in pPac-Staf (39) were obtained as follows. BamHI sites were introduced by site-directed mutagenesis of pBRN3-Staf (39) at positions 156 and 1839 of the Staf cDNA sequence to create pBRN3-Staf–BB. Deletions Δ6-248, Δ473-600, Δ84-176, Δ212-220, Δ77-102, Δ77-130, Δ77-156, Δ77-220, Δ103-220, Δ131-220, and Δ157-220 and mutants D168A+G169A+T170A, L166A, D168A, and T170A were created by site-directed mutagenesis of pBRN3-Staf–BB. The BamHI fragments encoding the Staf mutants were excised from the mutated pBRN3-Staf–BB and introduced clockwise at the BamHI site into plasmid pPac to generate the series of pPac-Staf expression constructs for transfection into Drosophila SL2 cells.
Reporter constructs.
p6E.tkCAT, AE.tkCAT, 3E.tRNASec, and 3E.U1 are described in references 53, 32, 39, and 38, respectively.
Oocyte microinjections, nuclear localization, and DNA binding assays.
Capped mRNAs were synthesized in vitro by T3 RNA polymerase as described in Schuster et al. (39) and were injected (20 nl, 1 ng) into the cytoplasm of Xenopus laevis oocytes, 20 h before nuclear injection of 20 nl containing the reporter. The concentrations of the reporters were 300 μg/ml for 6E.tkCAT and 50 μg/ml for 3E.tRNASec and 3E.U1. The 3E.tRNASec and 3E.U1 reporters were injected in the presence of [α-32P]GTP (800 Ci/mmol, 0.2 μCi/oocyte) and 5S RNA maxigene (5 μg/ml) as an internal control for nuclear microinjection and RNA recovery. The 6E.tkCAT reporter was injected in the presence of the pCH110 vector (300 μg/ml) as an internal control for nuclear injection. Incubation was for 16 h (3E.U1 and 6E.tkCAT reporters) or 3 h (3E.tRNASec reporter). Transcription of the reporter genes was analyzed as described previously (32, 39). The transcription efficiencies of the tRNASec and U1 RNA genes, relative to the 5S RNA maxigene expression, were quantitated with a Fuji Bioimage Analyzer BAS2000. For monitoring synthesis and nuclear localization of the effector proteins, oocytes microinjected with mRNAs were manually enucleated. Oocyte nuclei were homogenized in 8 μl of the extraction buffer [50 mM Tris-HCl (pH 8.0), 50 mM KCl, 0.1 mM EDTA, 5 mM MgCl2, 25% glycerol, 1 μl of bestatin per ml, 1 μl of pepstatin A per ml, 1 μl of trans-epoxysuccinyl-l-leucylamido-(4-guanidino)-butane per ml, 1 μl of 4-(2 aminoethyl)-benzenesulfonyl fluoride per ml] per nucleus. Homogenates were clarified by centrifugation at 13,000 rpm for 5 min. 32P-labeled double-stranded oligonucleotides containing the E element were used for Staf-Krox-20 and Krox-20–DBD bandshift assays as described previously (6).
Transfections, CAT assays, and Western analysis.
Schneider line 2 (SL2) Drosophila cells were transfected by the DOTAP liposomal transfection procedure (Boehringer). The cells received 500 ng of expression plasmid, 500 ng of reporter plasmid, and 200 ng of pACH110 (49). β-Galactosidase activity, normalization, and chloramphenicol acetyl-transferase (CAT) assays were performed as described previously (39). Results were quantitated with a Fuji Bioimage Analyzer BAS2000. The data represent the averages of results from three to four transfections, each performed in duplicate. For Western blot analysis, extracts from Drosophila-transfected cells were prepared essentially as described previously (2). Proteins were transferred to nitrocellulose and visualized with a polyclonal serum raised against a C-terminal Staf peptide with the use of ECL reagents (Amersham).
RESULTS
Mapping of the Staf activation domain.
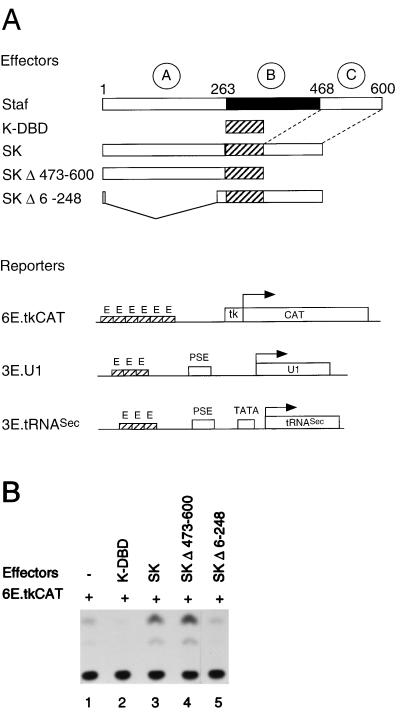
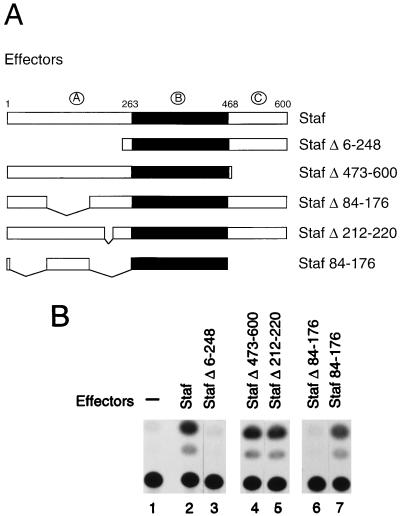
The Staf amino acid sequence can be divided into three regions (Fig. 1A): an N-terminal domain (amino acids 1 to 263), which we refer to as domain A; a central DNA binding domain B containing seven zinc fingers of the C2-H2 type (amino acids 264 to 468); and the C-terminal domain C (amino acids 469 to 600). Truncated Staf proteins were constructed, in order to define the Staf domains which are needed for the activation of snRNA, snRNA-type, and TATA box-containing mRNA promoters. The transactivation capabilities provided by the wild-type and truncated proteins (Fig. 1A) was assayed on various promoters by microinjection into X. laevis oocytes. To eliminate the background provided by the endogenous Staf in this assay, we designed chimeric Staf-Krox-20 proteins with altered DNA binding specificities. The Staf zinc finger domain was replaced by the corresponding sequences of the Krox-20 zinc finger domain (K-DBD), as illustrated in Fig. 1A (5, 39). This results in chimeric Staf-Krox-20 molecules (Fig. 1A) with different DNA binding specificities but overall structures that are likely to be relatively unchanged from that of the wild-type protein. The abilities of the different Staf-Krox-20 chimeric proteins to activate snRNA and snRNA-type promoters were assayed with Xenopus U1b1 snRNA (Pol II) and tRNASec (Pol III) promoters. They contained, in place of the residing activator element, a multimerized version of the Krox-20 binding site E element (3E.U1 and 3E.tRNASec) (Fig. 1A). The reporter 6E.tkCAT (Fig. 1A), containing six E elements upstream of the tk promoter, was used for monitoring the transcriptional activation on TATA box-containing mRNA promoters (53).
FIG. 1.
The Staf-Krox-20 chimera requires the N terminus to activate transcription from tk.CAT, U1 snRNA, and tRNASec promoters. (A) Schematic diagrams depicting the effector mRNAs synthesized in vitro and the 6E.tkCAT, 3E.U1 and 3E.tRNASec reporter genes used in the Xenopus microinjection assay. The first lane shows the structure of Staf. The A, B, and C domains are indicated with their localizations. PSE and TATA correspond to the basal promoter elements of the U1 and tRNASec genes. 3E and 6E indicate the number of multimerized E binding sites of the Krox-20 protein. SK, chimeric Staf-Krox-20 molecules. (B) CAT assays showing transcriptional activation from the 6E.tkCAT promoter by the K-DBD, SK, SKΔ473-600 and SKΔ6-248 effectors using extracts from microinjected Xenopus oocytes. Lane 1, no effector was expressed. (C) Transcriptional activation from the Pol II 3E.U1 (lanes 1 to 4) and Pol III 3E.tRNASec (lanes 5 to 8) promoters by the SK, SKΔ473-600 and SKΔ6-248 effectors using microinjected Xenopus oocytes. Lanes 1 and 5, no effector was expressed. Positions of the U1, tRNASec, and 5S maxi used as an internal standard are indicated. (D) Effector expression assayed by gel retardation with nuclear extracts from microinjected Xenopus oocytes. The identity of the expressed effector is indicated above each lane. Lanes 1 and 2, no effector was expressed. Reactions in lanes 1, 3, 5, 7, and 9 (−) contained no competitor; lanes 2, 4, 6, 8, and 10 (+) contained a 100-fold molar excess of unlabeled E competitor DNA.
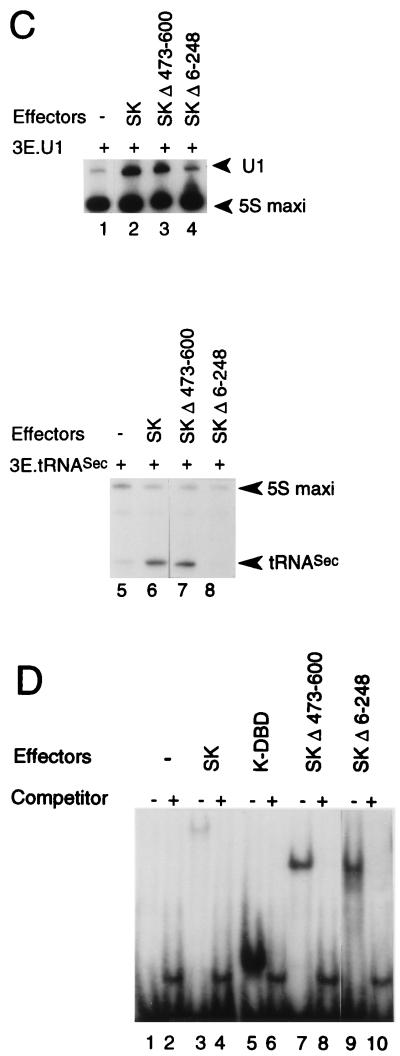
The mRNAs of the various Staf-Krox-20 effectors were transcribed in vitro, capped, and injected separately into the oocyte cytoplasm (39). Twenty hours postinjection, the 6E.tkCAT, 3E.tRNASec, and 3E.U1 reporters were injected separately into oocyte nuclei. 3E.U1 and 3E.tRNASec were injected along with [α-32P]GTP and a plasmid containing a 5S RNA maxigene as an internal standard. After a second incubation, the labeled RNAs were extracted and the transcription levels of the U1 snRNA and tRNASec genes, normalized relative to the 5S RNA maxigene expression, were used to measure the transactivation properties of the protein tested. For experiments with the mRNA promoter, a plasmid containing the β-galactosidase gene, not responding to Staf, served as an internal control and was coinjected with the 6E.tkCAT reporter into oocyte nuclei. After a second incubation, protein extracts were prepared and the normalized CAT activity was used as a measure of the transactivation properties. In the presence of Staf-Krox-20 (SK), the transcription levels of 6E.tkCAT, 3E.U1 and 3E.tRNASec were significantly enhanced (compare lanes 1 and 3 in Fig. 1B and lanes 1 and 2 and 5 and 6 in Fig. 1C). With SKΔ473-600, a Staf-Krox-20 derivative lacking the C-terminal domain C, a comparable level of activation was obtained (compare lanes 3 and 4 in Fig. 1B and lanes 2 and 3 and 6 and 7 in Fig. 1C). This effect is not mediated by Krox-20–DBD, since transcription of the 6E.tkCAT, 3E.U1, and 3E.tRNASec reporters was not enhanced by the presence of this protein containing only the DNA-binding domain (Fig. 1B, lane 2) (38, 39). In contrast, removal of the N-terminal amino acids 6 to 248 in SKΔ6-248, resulted in the complete loss of enhanced transcriptional activity for the three reporters (compare lanes 3 and 5 in Fig. 1B and lanes 2 and 4 and 6 and 8 in Fig. 1C). Note that for lane 4, the normalized value for the U1 transcription level is very similar to that obtained in lane 1, taking into account that the intensity of the 5S maxigene internal control is higher in lane 4 than in lane 1.
The quantity, quality, and nuclear localization of the effector proteins were assayed by gel retardation assays with oocyte nuclear extracts and a 32P-labeled oligonucleotide probe containing one Krox-20 E site (Fig. 1D). Competition experiments revealed the specificity of the shifted complexes since an excess of unlabeled E site competed efficiently with their formation (Fig. 1D). The relative mobilities of the different complexes indicated that full-length effector proteins were expressed in these microinjections. Quantitation of the shifted complexes showed that the relative binding efficiencies of the chimeric proteins SKΔ473-600 and SKΔ6-248 in the oocyte nuclear extracts are 10-fold higher than that of SK (Fig. 1D). However, these binding efficiencies were not used to correct the activation levels, since titration experiments showed that the levels of the effector proteins are saturating in our assays; therefore, a 10-fold increase or decrease in the mRNA concentration of the effector will not change the level of transcriptional activation (data not shown).
Taken together, these results indicate that the N terminus of Staf (domain A) is necessary and sufficient for maximal transactivation of Pol II and Pol III snRNA and snRNA-type promoters and TATA box-containing mRNA promoters.
The minimal activation domain for snRNA-type promoters is 18 amino acids long.
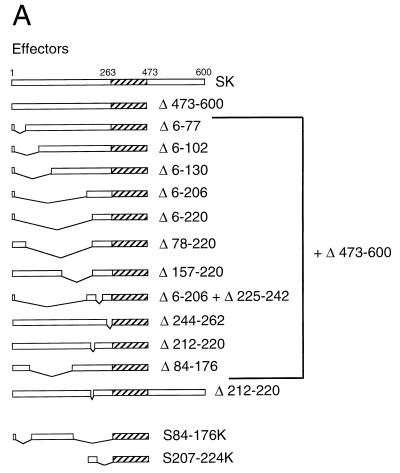
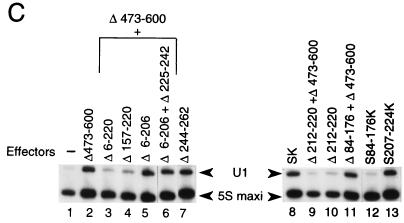
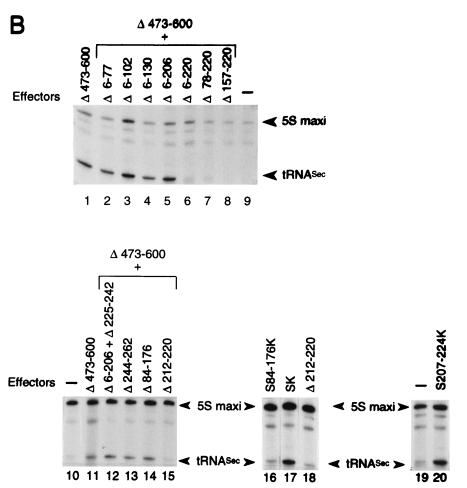
To define the minimal region of Staf sufficient for activation of snRNA and snRNA-type promoters by RNA polymerases II and III, the 263-amino-acid activation domain A was subdivided by creating a series of deletion mutants on Staf-Krox-20 Δ473-600 (Fig. 2A). These fusion proteins were again tested for their ability to activate transcription of the 3E.tRNASec and 3E.U1 reporters in a microinjection assay. Internal deletions between amino acid 5 and amino acids 78 (Δ6-77), 103 (Δ6-102), 131 (Δ6-130), and 207 (Δ6-206) did not impair activation of the Pol III tRNASec and Pol II U1 promoters (Fig. 2B, compare lane 1 and lanes 2 to 5 for the tRNASec promoter; and lanes 2 and 5 for the U1 promoter in Fig. 2C). In contrast, when the internal deletions included amino acids 207 to 220 in the Δ6-220, Δ78-220, and Δ157-220 mutants, activation of transcription was lost (Fig. 2B, lanes 6 to 8; Fig. 2C, compare lane 2 with lanes 3 and 4). The Δ6-206 truncation, associated with a deletion between amino acids 224 and 243 (Δ6-206+Δ225-242), as well as the deletion between amino acids 243 and 263 (Δ244-262), led to a protein as active as the parental Staf-Krox-20 (Fig. 2B, compare lane 11 with lanes 12 and 13; Fig. 2C, compare lane 2 with lanes 6 and 7). This truncation analysis demonstrates that the region from 207 to 224 is critical for activation of the tRNASec and U1 promoters.
FIG. 2.
Characterization of the minimal activation domain for Pol II and Pol III snRNA promoters. (A) Schematic structures of the various truncated and fused Staf-Krox-20 proteins. The Krox-20 DBD is hatched. (B) Xenopus microinjection assay of transcriptional activation from the 3E.tRNASec promoter by the effectors depicted in panel A. Identities of the effector proteins are indicated above each lane. Lanes 9, 10, and 19, assays without effector (−). Positions of the 5S maxigene and tRNASec RNAs are indicated. Autoradiographs corresponding to lanes 1 to 9, 10 to 15, 16 to 18, and 19 to 20 were from separate experiments. (C) Xenopus microinjection assay of transcriptional activation from the 3E.U1 promoter by the effectors depicted in panel A. Positions of the U1 and 5S maxi RNAs are indicated. Lane 1, assay without effector (−).
The importance of domain 207 to 224 was further established by examining the effects of deletions within this defined area. Deletions of amino acids 212 to 220 in Staf-Krox-20 (Δ212-220) and Staf-Krox-20 Δ473-600 (Δ212-220+Δ473-600) (Fig. 2A) abolished the transactivation abilities of these effector proteins (Fig. 2B, compare lanes 11 and 15 and lanes 17 and 18; Fig. 2C, compare lane 8 with lanes 9 and 10). Lastly, fusion of the 18-amino-acid domain 207 to 224 to Krox-20 DBD produced protein S207-224K (Fig. 2A) which carried the ability to activate transcription of the tRNASec (Fig. 2B, compare lanes 19 and 20) and U1 promoters (Fig. 2C, compare lanes 1, 8, and 13). The expression levels and nuclear localization of these chimera were verified by electrophoretic mobility shift assays using nuclear extracts from oocytes injected with mRNAs (data not shown).
These results show clearly that the 18-amino-acid region 207 to 224 of sequence PDTISALEQYAAKVSIEG represents the minimal Staf activation domain for Pol III tRNASec and Pol II U1 promoters.
Leu-213 is critical for transactivation activity on snRNA-type promoters.
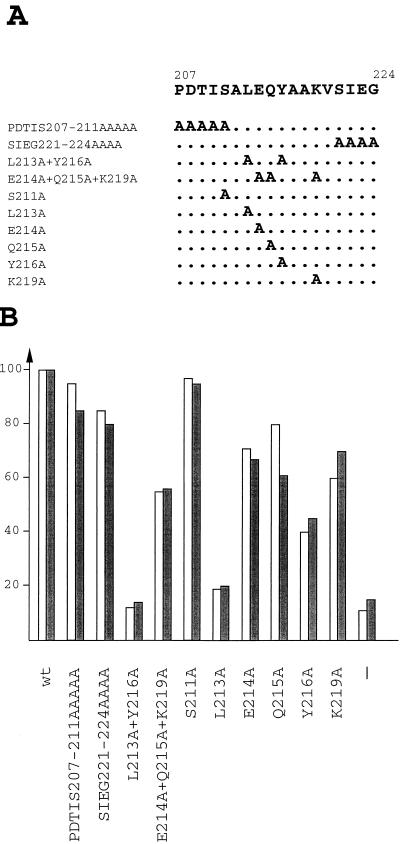
The region 207 to 224 does not have a preponderance of amino acids, such as glutamines, isoleucines, prolines, or serines-threonines that typify other well-characterized activation domains. To determine residues responsible for activation, mutants were created by alanine substitution mutagenesis in the context of Staf-Krox-20 Δ473-600. Alanine substitutions of amino acids 207 to 211, 221 to 224, and serine 211 (Fig. 3A, PDTIS207-211AAAAA, SIEG221-224AAAA, and S211A, respectively) had a very minor or no effect at all on the ability of the protein to activate the 3E.tRNASec and 3E.U1 reporters in the Xenopus microinjection assay (Fig. 3B). Replacement of the three residues at positions 214, 215, and 219 (Fig. 3A, E214A+Q215A+K219A) produced a twofold drop in the activity of the protein. In stark contrast, the simultaneous replacement of the two residues at positions 213 and 216 by an alanine L213A+Y216A (Fig. 3A) completely abolished the transactivating ability of the protein (Fig. 3B). To assess the contributions of Leu-213, Glu-214, Gln-215, Tyr-216, and Lys-219, they were individually mutated to alanines. Whereas changing Glu-214, Gln-215, Tyr-216, and Lys-219 had only a moderate effect, the L213A mutant by itself was severely defective compared to Staf-Krox-20 Δ473-600 (Fig. 3B).
FIG. 3.
Identification of amino acids essential for transactivation of snRNA and snRNA-type promoters. (A) The amino acid sequence of the Staf domain 207 to 224 is shown with the changes introduced in the mutants. Dots in the mutant sequences indicate an unchanged residue. (B) Transcriptional activation assays from the Pol II 3E.U1 and Pol III 3E.tRNASec promoters by the various effectors depicted in panel A. Activity is shown as a percentage of the activity observed with Staf-Krox-20 Δ473-600 construct containing the wild-type snRNA activation domain. Activities in microinjections assays with the 3E.U1 and 3E.tRNASec reporters are shown by solid and shaded bars, respectively. Results are the averages of two independent determinations. Identities of the effector proteins are indicated below each histogram. −, assay without effector.
These results demonstrate that Leu-213 is essential to the activity of the Staf snRNA transactivation domain.
Staf contains distinct activation domains for mRNA and snRNA-type promoters.
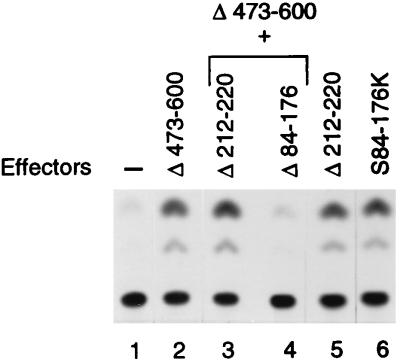
To determine if the snRNA activation domain of Staf can also activate transcription from an mRNA promoter, we tested the ability of truncated Staf-Krox-20 proteins to enhance the transcription level of the 6E.tkCAT reporter in a microinjection assay. Surprisingly, deletion of residues 212 to 220 in Staf-Krox-20 (Δ212-220) and Staf-Krox-20 Δ473-600 (Δ212-220+Δ473-600), corresponding to sequence ALEQYAAKV, had no effect on the transcription level of 6E.tkCAT, while the same mutant effectors failed to activate snRNA-type promoters (Fig. 4, compare lane 2 with lanes 3 and 5). In sharp contrast, deletion of residues 84 to 176 (Δ84-176+Δ473-600; Fig. 2A) in Staf-Krox-20 Δ473-600 completely abolished activation from the thymidine kinase (tk) promoter of 6E.tkCAT (Fig. 4, compare lanes 2 and 4) but is absolutely innocuous to activation of transcription from the tRNASec and U1 promoters (Fig. 2B, lanes 11 and 14; Fig. 2C, lanes 8 and 11). This analysis indicated that a region containing residues 84 to 176 is the major contributor to activation of transcription from an mRNA promoter. A fusion of the region containing residues 84 to 176 to the Krox-20 DBD, to yield S84-176K (Fig. 2A), led to efficient activation of transcription from 6E.tkCAT (Fig. 4, compare lanes 2 and 6). However, this same chimeric protein was unable to activate snRNA and snRNA-type promoters (Fig. 2B, lanes 16 and 17; Fig. 2C, lanes 8 and 12). Therefore, this 93-amino-acid region appears to possess an autonomous transactivation capability in the absence of the rest of Staf.
FIG. 4.
The mRNA activation domain is distinct from the snRNA counterpart. Representative CAT assays of transcriptional activation from the 6E.tkCAT promoter using extracts from microinjected Xenopus oocytes. Identities of the effectors are indicated above the lanes. The effector structures are depicted in Fig. 2A. Expression of the proteins was confirmed by gel mobility shift assays using nuclear extracts from microinjected Xenopus oocytes (data not shown).
Together, these results demonstrate unambiguously that Staf contains two physically separate and functionally distinct activation domains located in the N terminus of the molecule. The activation domain covering residues 207 to 224 functions solely in the transactivation of Pol II and Pol III snRNA-type promoters. The other activation domain, encompassing amino acids 84 to 176, is restricted to transcriptional activation from mRNA promoters.
Four repeat units for an mRNA activation domain.
So far, our investigations were carried out with the use of chimeric Stafs bearing the Krox-20 DBD. To obviate the need for this heterologous DBD, which was required to eliminate the endogenous Staf background of the Xenopus oocytes, another strategy was taken. Drosophila cells, which were shown not to possess a Staf activity, were employed (39). In these cell lines, we could assay only the mRNA activation domain since the literature provides experimental evidence that transcriptional activation of snRNA promoters in Drosophila does not proceed similarly to that in vertebrates (8). Full-length Staf or various deletion mutant versions were expressed under the control of the constitutively active actin 5C promoter (Fig. 5A). The reporter AE.tkCAT contains three Staf binding site AE upstream of the tk promoter fused to the CAT reporter gene. Cotransfection of wild-type Staf and reporter plasmid resulted in an efficient transcriptional activation of the CAT gene (Fig. 5B, compare lanes 1 and 2). Deletions of the C-terminal domain C and of amino acids 212 to 220 (Staf Δ473-600 and Staf Δ212-220, respectively) (Fig. 5A) did not reduce the ability of Staf to activate the tk promoter (Fig. 5B, compare lane 2 with lanes 4 and 5). In contrast, however, deletion of the N-terminal domain A or an internal deletion between amino acids 83 and 177 (StafΔ6-248 and StafΔ84-176, respectively) (Fig. 5A) produced molecules which were unable to stimulate the promoter of the CAT reporter gene (Fig. 5B, compare lane 2 with lanes 3 and 6). The transcriptional activity could be fully recovered with Staf 84-176 (Fig. 5A), a fusion protein consisting of only amino acids 84 to 176 fused to the Staf DBD (Fig. 5B, compare lanes 2 and 7). These results, obtained with Drosophila cells, corroborate those already obtained with the Xenopus oocyte system with chimeric Staf-Krox-20 proteins.
FIG. 5.
Transactivation abilities of Staf deletion mutants from an mRNA promoter, assayed in Drosophila cells. (A) Schematic structures of the various Staf deletion mutants. The A, B, and C domains of Staf are indicated. The Staf DNA binding domain is represented in black. (B) Representative CAT assays using extracts from Drosophila cells transfected with the effectors indicated above each lane.
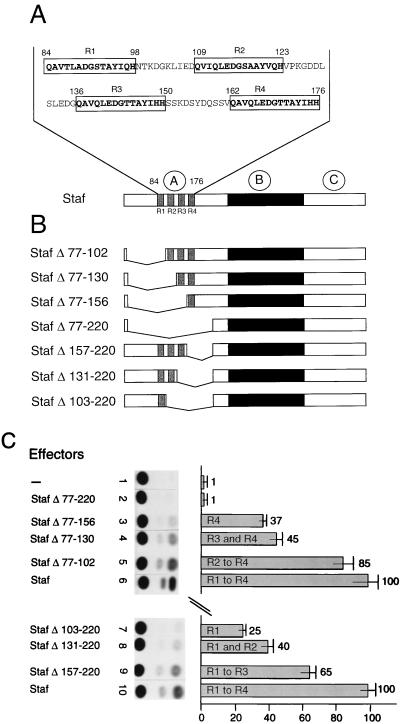
The 93-amino-acid region (84 to 176) of Staf, which can activate an mRNA promoter as an independent transactivating domain, contains four short repeats of 15 amino acids, with the consensus sequence QAVQLEDG(S/T)TAYI(Q/H)H. The distance between each repeat is 10 to 12 amino acids. The positions of the repeats, previously described (39), are the following: repeat 1 (R1), positions 84 to 98; repeat 2 (R2), positions 109 to 123; repeat 3 (R3), positions 136 to 150; repeat 4 (R4), positions 162 to 176 (Fig. 6A). The presence of four repeat units in the Staf minimal mRNA activation domain represents a novel feature for an activation domain. Therefore, to determine whether they are important for the ability of Staf to stimulate transcription, internal deletions in full-length Staf were generated and tested again in the Drosophila cotransfection assay. Fig. 6B shows the constructs that were made. Deletions from amino acid 77 to amino acids 102 (StafΔ77-102), 130 (StafΔ77-130), 156 (StafΔ77-156), and 220 (StafΔ77-220), removed the sequences containing R1, R1 and R2, R1 to R3, and R1 to R4, respectively. Truncations between amino acids 102 to 221 (StafΔ103-220), 130 to 221 (StafΔ131-220), and 156 to 221 (StafΔ157-220) eliminated the regions containing R2 to R4, R3 and R4, and R4, respectively. The expression levels and nuclear localization of these mutant proteins were verified by Western blot analysis (data not shown). The normalized values in Fig. 6C show correlation between transcription activity of the reporter and the number of repeat units. Construct StafΔ77-220, with no repeat, failed to transactivate (Fig. 6C, lane 2). Full activity was progressively restored, however, by the sequential addition of sequences containing the repeat motif (Fig. 6C, lanes 3 to 6 and 7 to 10). The presence of sequences containing repeat R1 (StafΔ103-220) or R4 (StafΔ77-156) resulted in activation levels 25- and 37-fold above the basal level, respectively (Fig. 6C, lanes 7 and 3). When Staf proteins containing the R1 and R2 or R3 and R4 repeat units were tested, transcription levels reached 40- and 45-fold of the basal level, respectively (Fig. 6C, lanes 8 and 4, respectively). More significantly, R1 to R3, but especially R2 to R4, allowed the transcription levels to attain 65- and 85-fold of the basal level (Fig. 6C, lanes 9 and 5, respectively).
FIG. 6.
Maximal transcriptional activation of an mRNA promoter by Staf requires the presence of repeats R1 to R4. (A) Sequence of the Staf mRNA activation domain (amino acids 84 to 176) with the location of repeats R1 to R4. (B) Schematic representation of the seven different effector constructs. The Staf DNA binding domain and the repeats are indicated in the black and gray areas, respectively. (C) Transcriptional stimulation of the AE.tkCAT promoter containing three AE Staf binding sites, assayed in Drosophila cells. The autoradiograph from a representative experiment is shown. The names of the effector constructs and the relative transcriptional activities are indicated above the lanes. The values represent the ratio of the CAT activity to that in the absence of effector.
Taken together, these results highlight the importance of the four repeat units of Staf to provide maximal transcription activity to mRNA promoters.
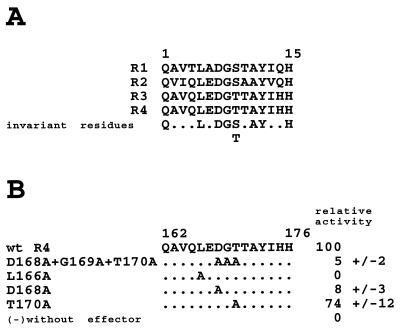
Comparison of the four repeats revealed the presence of seven invariant residues located at positions 1, 5, 7, 8, 11, 12, and 15 in each repeat (Fig. 7A). Further, position 9 of each repeat is always occupied by a hydroxylated residue. A number of alanine substitutions were designed in the context of repeat R4 in Staf Δ77-156 to test whether these residues play a role in mediating transactivation. When Asp-168, Gly-169, and Thr-170 were substituted (Fig. 7B, mutant D168A+G169A+T170A), the transactivation properties of the protein on the 3E.tkCAT promoter were dramatically reduced (5% of the R4 wild-type level) (Fig. 7B). Individual alanine replacements of Asp-168 and Thr-170 produced a significant reduction of activity, with the more pronounced effect for Asp-168 (8% of residual level) (Fig. 7B). Strikingly, the alanine substitution of Leu-166 abolished totally the transcriptional activation (Fig. 7B).
FIG. 7.
Alanine substitution mutagenesis of repeat R4 in the Staf mRNA activation domain. (A) Sequence alignments of repeats R1 to R4. (B) Staf Δ77-156 containing repeat R4 with the indicated mutations were tested in Drosophila cells for transcriptional activation of a cotransfected AE.tkCAT promoter containing three AE Staf binding sites. The activity of the Staf Δ77-156 construct containing wild-type R4 was defined as 100. Results are the averages (+/− values indicate standard deviations) of three independent determinations. wt, wild type.
From this, we conclude that residues Leu-166, Asp-168, and Thr-170 contribute actively to the activity of the Staf mRNA transactivation domain.
DISCUSSION
We have shown previously that Staf plays an important role, not only in the transcriptional activation of snRNA and snRNA-type genes transcribed by RNA polymerases II and III but also to stimulate expression from an mRNA promoter (32, 38, 39). In this study we have addressed the molecular basis of these pleiotropic effects on transcriptional activity.
Our results summarized in Fig. 8 show that domain A, corresponding to the N-terminal region of Staf, contains two physically and functionally distinct activation domains acting specifically on snRNA-type and mRNA promoters. To our knowledge, this is the first report describing such a feature in a transactivator protein. Based on our work, we propose that the two separate Staf activation domains are involved in two different transactivation mechanisms: one for the specific activation of mRNA promoters and the other for that of snRNA and snRNA-type promoters. Forsberg et al. (11) showed that Sp1 is able to stimulate transcription from mRNA and snRNA promoters. However, it must be pointed out that in this case overlapping domains are involved, while we identified two distinct, specialized domains in Staf.
FIG. 8.
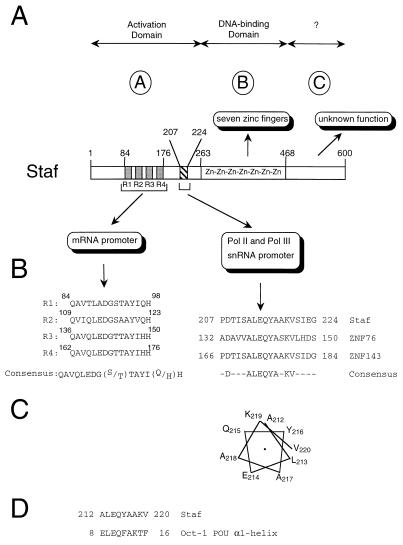
Summary of Staf subdomains. (A) Schematic representation of Staf shown with delineation of the various subdomains, based on the present and earlier studies. (B) mRNA and snRNA activation domains. Left panel, positions and sequence alignments of the four substructures R1 to R4; a consensus sequence was derived at positions for which at least three of the four repeats contain identical residues. Right panel, sequence comparisons of the snRNA activation domain with regions of ZNF76 and ZNF143 (35, 50). (C) Helical wheel representation of Staf residues 212 to 220. (D) Sequence comparison of Staf residues 212 to 220 with the α-helix-1 from the Oct-1 POU domain. The α1 helix is numbered according to Herr and Cleary (17).
A number of activators, enhancing transcription from mRNA promoters only, have been shown to contain multipartite activation domains. This is the situation found, for example, in TEF-1, Oct-2, and NGF1-A (19, 36, 46). Interestingly, we have shown in this work that Staf contains one single domain, necessary and sufficient for optimal transactivation of an mRNA promoter containing a TATA box. This 93-amino-acid activation domain (Staf residues 84 to 176) has a net negative charge and contains 11 and 17% of glutamine and hydroxylated residues, respectively, in a manner analogous to what is found in many other transactivation domains (21, 27, 51). A prominent feature of the Staf mRNA activation domain is its 15-amino-acid motif, repeated four times (Fig. 8B). A related situation was described for the Oct-2 activator, which contains a twofold reiterated 18-amino-acid motif (44). The Staf mRNA activation domain can be divided into subregions containing one, two, three, or four substructures that still have the potential to enhance transcription. This fact suggests that each substructure constitutes a functional unit, even though all four motifs are required to confer optimal transactivation potential. In line with the occurrence of repeated substructures in Staf, previous studies revealed that artificial reiteration of an activation domain can also lead to a significant increase in transcriptional activity (9, 18, 40, 43, 45).
From a mechanistic point of view, involvement of a reiterated motif in the activation process can be apprehended in two ways. In the first one, the affinity of the activator for the basal transcription complex is higher because the number of contacts with the target increases with the number of repeats. Alternatively, redundance of the interaction surfaces could raise the affinity of the activator by increasing the probability of binding to the target. Structural studies on the VP16 and p53 activation domains suggested that the FXXφφ motif (X stands for any amino acid and φ for hydrophobic amino acids) is a general recognition element of the acidic activation domain of TAFII31 (22, 52). The Staf mRNA activation domain, though, does not conform to this motif and presents no obvious sequence similarity with the other known activation domains, suggesting that it may constitute the representative of a novel category.
Oct-1 has been shown to activate snRNA promoters with multipartite snRNA activation domains (28, 29, 46). Instead, our study established that Staf performs this function with one single 18-amino-acid region (residues 207 to 224) bearing no sequence similarity to other, previously identified activation domains. The mutational analysis of this domain revealed the prime importance of the 9-amino-acid subdomain of sequence ALEQYAAKV (residues 212 to 220). This subdomain is conserved in the human proteins ZNF76 and ZNF143 (35, 50), which possess significant sequence similarities to Staf (33, 39) (Fig. 8B). This, in addition to the finding that ZNF76 is able to activate transcription of snRNA genes (33), suggests that this new short motif plays a central role in the activation process by constituting an interaction surface within the snRNA transcription complex. Based on protein modeling algorithms, the subdomain composed of residues 212 to 220 is predicted to form an α-helical region (7, 12). Displaying residues 212 to 220 as a putative α-helix reveals its amphipathic character, with hydrophilic residues E214, Q215, and K219 distributed on one face (Fig. 8C) and hydrophobic residues L213 and Y216, on the other. From this observation, we propose that hydrophobic protein-protein interactions involving Leu-213 and Tyr-216 are critical in the snRNA gene transcriptional activation by Staf.
A follow-up in the study of domain delineation in a transcriptional activator resides in the identification of the targets that it must contact to activate transcription. Our results show clearly that a short domain of Staf specifically acts as a potent activator of Pol II and Pol III snRNA and snRNA-type gene transcription. The data additionally suggest that in the activation process the molecular target of the snRNA activation domain is a component of the basal Pol II and Pol III snRNA transcription complex. The multisubunit SNAPc complex, also called the PSE transcription factor PTF, which recognizes specifically the PSE, is the only factor known to be specific and common to Pol II and Pol III snRNA transcription complexes (13, 31, 37, 55). In this respect, it is tempting to speculate that Staf interacts directly or indirectly with SNAPc. The Oct-1 POU protein has been implicated in transcriptional activation of Pol II and Pol III snRNA promoters, as well. In this case, the POU DNA binding domain of Oct-1 not only serves to target the protein to the octamer sequence on the DNA but also is intimately involved in the transcriptional activation process by stabilizing the binding of the basal transcription complex SNAPc (10, 17, 28, 29, 31). It has been proposed that the POU domain does so by recruiting SNAPc to the PSE by direct protein-protein contacts involving the N-terminal part of the POU-specific domain. It is interesting to note that the central part of the Staf snRNA activation domain bears sequence similarity to the α-helix-1 constituting the N terminus of the POU-specific domain (Fig. 8D). From this, we propose that Staf and Oct-1 recognize the same target in the SNAPc complex.
In conclusion, we have presented experimental evidence for the presence in Staf of two functionally distinct transactivation domains with specific snRNA-type and mRNA gene targets. This represents a novel feature in a transactivator protein and adds an important new dimension to structure-function studies that are being performed with transactivation domains.
ACKNOWLEDGMENTS
We are grateful to E. Myslinski for critical reading of the manuscript. We thank J. Hoffmann and C. Kappler for Drosophila cell cultures, P. Remy for microinjection facilities, A. Hoeft for oligonucleotide synthesis, and C. Loegler for excellent technical assistance.
This work was supported by grants from the Université Louis Pasteur in Strasbourg, the Association pour la Recherche sur le Cancer (ARC) and the European Union (EEC Biotech Program B102-CT92-0090).
REFERENCES
- 1.Ares M, Jr, Chung J-S, Giglio L, Weiner A M. Distinct factors with Sp1 and NF-A specificities bind to adjacent functional elements of the human U2 snRNA gene enhancer. Genes Dev. 1987;1:808–817. doi: 10.1101/gad.1.8.808. [DOI] [PubMed] [Google Scholar]
- 2.Attardi L D, Tjian R. Drosophila tissue-specific transcription factor NTF-1 contains a novel isoleucine-rich activation motif. Genes Dev. 1993;7:1341–1353. doi: 10.1101/gad.7.7b.1341. [DOI] [PubMed] [Google Scholar]
- 3.Carbon P, Krol A. Transcription of the Xenopus laevis selenocysteine tRNA(Ser)Sec gene: a system that combines an internal B box and upstream elements also found in U6 snRNA genes. EMBO J. 1991;10:599–606. doi: 10.1002/j.1460-2075.1991.tb07987.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carey M. Mechanistic advances in eukaryotic gene activation. Curr Opin Cell Biol. 1991;3:452–460. doi: 10.1016/0955-0674(91)90073-8. [DOI] [PubMed] [Google Scholar]
- 5.Chavrier P, Janssen-Timmen U, Mattei M G, Zerial M, Bravo R, Charnay P. Structure, chromosome location, and expression of the mouse, zinc finger gene Krox-20: multiple gene products and coregulation with the proto-oncogene c-fos. Mol Cell Biol. 1989;9:787–797. doi: 10.1128/mcb.9.2.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chavrier P, Vesque C, Galliot B, Vigneron M, Dollé P, Duboule D, Charnay P. The segment-specific gene Krox-20 encodes a transcription factor with binding sites in the promoter region of the Hox-1.4 gene. EMBO J. 1990;9:1209–1218. doi: 10.1002/j.1460-2075.1990.tb08228.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chou P Y, Fasman G D. Empirical predictions of protein conformation. Annu Rev Biochem. 1978;47:251–256. doi: 10.1146/annurev.bi.47.070178.001343. [DOI] [PubMed] [Google Scholar]
- 8.Dahlberg J E, Lund E. The genes and transcription of the major small nuclear RNAs. In: Birnstiel M L, editor. Structure and function of major and minor small nuclear ribonucleoprotein particles. Berlin, Germany: Springer-Verlag; 1988. pp. 38–70. [Google Scholar]
- 9.Emami K H, Carey M. A synergistic increase in potency of a multimerized VP16 transcriptional activation domain. EMBO J. 1992;11:5005–5012. doi: 10.1002/j.1460-2075.1992.tb05607.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ford E, Hernandez N. Characterization of a trimeric complex containing Oct-1, SNAPc and DNA. J Biol Chem. 1997;272:16048–16055. doi: 10.1074/jbc.272.25.16048. [DOI] [PubMed] [Google Scholar]
- 11.Forsberg M, Ström A-C, Lillhager P, Westin G. Activation functions of transcription factor Sp1 U2 snRNA and TATA box promoters. Biol Chem Hoppe-Seyler. 1995;376:661–669. doi: 10.1515/bchm3.1995.376.11.661. [DOI] [PubMed] [Google Scholar]
- 12.Garnier J, Osguthorpe D J, Robson B. Analysis of the accuracy and implications of simple methods for predicting the secondary structure of globular proteins. J Mol Biol. 1978;120:97–120. doi: 10.1016/0022-2836(78)90297-8. [DOI] [PubMed] [Google Scholar]
- 13.Henry R W, Sadowski C L, Kobayashi R, Hernandez N. A TBP-TAF complex required for transcription of human snRNA genes by RNA polymerases II and III. Nature. 1995;374:653–657. doi: 10.1038/374653a0. [DOI] [PubMed] [Google Scholar]
- 14.Hernandez N. Transcription of vertebrate snRNA genes and related genes. In: MacKnight S L, Yamamoto K R, editors. Transcriptional regulation. Vol. 1. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. pp. 281–313. [Google Scholar]
- 15.Hernandez N. TBP, a universal eukaryotic transcription factor? Genes Dev. 1993;7:1291–1308. doi: 10.1101/gad.7.7b.1291. [DOI] [PubMed] [Google Scholar]
- 16.Herr W. Oct-1 and Oct-2: differential transcriptional regulation by proteins that bind to the same DNA sequence. In: MacKnight S L, Yamamoto K R, editors. Transcriptional regulation. Vol. 1. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. pp. 1103–1135. [Google Scholar]
- 17.Herr W, Cleary M A. The POU domain: versatility in transcriptional regulation by a flexible two-in-one DNA-binding domain. Genes Dev. 1995;9:1679–1693. doi: 10.1101/gad.9.14.1679. [DOI] [PubMed] [Google Scholar]
- 18.Hollenberg S M, Evans R M. Multiple and cooperative transactivation domains of the human glucocorticoid receptor. Cell. 1988;55:899–906. doi: 10.1016/0092-8674(88)90145-6. [DOI] [PubMed] [Google Scholar]
- 19.Hwang J-J, Chambon P, Davidson I. Characterization of the transcription activation function and the DNA binding domain of the transcriptional enhancer factor-1. EMBO J. 1993;12:2337–2348. doi: 10.1002/j.1460-2075.1993.tb05888.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Janson L, Bark C, Petterson U. Identification of proteins interacting with the human U2 small nuclear RNA genes. Nucleic Acids Res. 1987;15:4997–5016. doi: 10.1093/nar/15.13.4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson P F, Sterneck E, Williams S C. Activation domains of transcriptional regulatory proteins. J Nutr Biochem. 1993;4:386–398. [Google Scholar]
- 22.Kussie P H, Gorina S, Marechal V, Elenbaas B, Moreau J, Levine A J, Pavletich N P. Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science. 1996;274:948–953. doi: 10.1126/science.274.5289.948. [DOI] [PubMed] [Google Scholar]
- 23.Lescure A, Tebb G, Mattaj I W, Krol A, Carbon P. A factor with Sp1 DNA-binding specificity stimulates Xenopus U6 snRNA in vivo transcription by RNA polymerase III. J Mol Biol. 1992;228:387–394. doi: 10.1016/0022-2836(92)90828-8. [DOI] [PubMed] [Google Scholar]
- 24.Lobo S M, Hernandez N. A 7 bp mutation converts a human RNA polymerase II snRNA promoter into an RNA polymerase III promoter. Cell. 1989;58:55–67. doi: 10.1016/0092-8674(89)90402-9. [DOI] [PubMed] [Google Scholar]
- 25.Lobo S M, Lister J M, Sullivan M L, Hernandez N. The cloned RNA polymerase II transcription factor IID selects RNA polymerase III to transcribe the human U6 gene in vitro. Genes Dev. 1991;5:1477–1489. doi: 10.1101/gad.5.8.1477. [DOI] [PubMed] [Google Scholar]
- 26.Mattaj I W, Dathan N A, Parry H D, Carbon P, Krol A. Changing the RNA polymerase specificity of U snRNA gene promoters. Cell. 1988;55:435–442. doi: 10.1016/0092-8674(88)90029-3. [DOI] [PubMed] [Google Scholar]
- 27.Mitchell P, Tjian R. Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science. 1989;245:371–378. doi: 10.1126/science.2667136. [DOI] [PubMed] [Google Scholar]
- 28.Mittal V, Cleary M A, Herr W, Hernandez N. The Oct-1 POU-specific domain can stimulate small nuclear RNA gene transcription by stabilizing the basal transcription complex SNAPc. Mol Cell Biol. 1996;16:1955–1965. doi: 10.1128/mcb.16.5.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murphy S. Differential in vivo activation of the class II and class III snRNA genes by the POU-specific domain of Oct-1. Nucleic Acids Res. 1997;25:2068–2076. doi: 10.1093/nar/25.11.2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murphy S, Di Liegro A C, Melli M. The in vitro transcription of the 7SK RNA gene by RNA polymerase III is dependent only on the presence of an upstream promoter. Cell. 1987;51:81–87. doi: 10.1016/0092-8674(87)90012-2. [DOI] [PubMed] [Google Scholar]
- 31.Murphy S, Yoon J-B, Gerster T, Roeder R G. Oct-1 and Oct-2 potentiate functional interactions of a transcription factor with the proximal sequence element of small nuclear RNA genes. Mol Cell Biol. 1992;12:3247–3261. doi: 10.1128/mcb.12.7.3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Myslinski E, Krol A, Carbon P. Optimal tRNASer/Sec gene activity requires an upstream SPH motif. Nucleic Acids Res. 1992;20:203–209. doi: 10.1093/nar/20.2.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Myslinski, E., C. Schuster, A. Krol, and P. Carbon. Unpublished data.
- 34.Ptashne M, Gann A. Transcriptional activation by recruitment. Nature. 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- 35.Ragoussis J, Senger P, Mockridge I, Sanseau P, Ruddy S, Dudley K, Sheer D, Trowsdale J. A testis-expressed Zn finger gene (ZNF76) in human 6p21.3 centromeric to the MHC is closely linked to the human homolog of the t-complex gene tcp-11. Genomics. 1992;14:673–679. doi: 10.1016/s0888-7543(05)80167-3. [DOI] [PubMed] [Google Scholar]
- 36.Russo M W, Matheny C, Milbrandt J. Transcriptional activity of the zinc finger protein NGF1-A is influenced by its interaction with a cellular factor. Mol Cell Biol. 1993;13:6858–6865. doi: 10.1128/mcb.13.11.6858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sadowski C L, Henry R W, Lobo S M, Hernandez N. Targeting TBP to a non-TATA box cis-regulatory element: a TBP-containing complex activates transcription from snRNA promoters through the PSE. Genes Dev. 1993;7:1535–1548. doi: 10.1101/gad.7.8.1535. [DOI] [PubMed] [Google Scholar]
- 38.Schaub M, Myslinski E, Schuster C, Krol A, Carbon P. Staf, a promiscuous activator for enhanced transcription by RNA polymerases II and III. EMBO J. 1997;16:173–181. doi: 10.1093/emboj/16.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schuster C, Myslinski E, Krol A, Carbon P. Staf, a novel zinc finger protein that activates the RNA polymerase III promoter of the selenocysteine tRNA gene. EMBO J. 1995;14:3777–3787. doi: 10.1002/j.1460-2075.1995.tb00047.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seipel K, Georgiev O, Schaffner W. Different activation domains stimulate transcription from remote (‘enhancer’) and proximal (‘promoter’) positions. EMBO J. 1992;11:4961–4968. doi: 10.1002/j.1460-2075.1992.tb05603.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simmen K A, Bernues J, Parry H D, Stunnenberg H G, Berkenstam A, Cavallini B, Egly J-M, Mattaj I W. TFIID is required for in vitro transcription of the human U6 gene by RNA polymerase III. EMBO J. 1991;10:1853–1862. doi: 10.1002/j.1460-2075.1991.tb07711.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sturm R A, Das G, Herr W. The ubiquitous octamer-binding protein Oct-1 contains a POU domain with a homeo box subdomain. Genes Dev. 1988;2:1582–1599. doi: 10.1101/gad.2.12a.1582. [DOI] [PubMed] [Google Scholar]
- 43.Sutherland J A, Cook A, Bannister A J, Kouzarides T. Conserved motifs in Fos and Jun define a new class of activation domain. Genes Dev. 1992;6:1810–1819. doi: 10.1101/gad.6.9.1810. [DOI] [PubMed] [Google Scholar]
- 44.Tanaka M, Clouston W M, Herr W. The Oct-2 glutamine-rich and proline-rich activation domains can synergize with each other or duplicates of themselves to activate transcription. Mol Cell Biol. 1994;14:6046–6055. doi: 10.1128/mcb.14.9.6046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tanaka M, Herr W. Reconstitution of transcriptional activation domains by reiteration of short peptide segments reveals the modular organization of a glutamine-rich activation domain. Mol Cell Biol. 1994;14:6056–6067. doi: 10.1128/mcb.14.9.6056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tanaka M, Lai J-S, Herr W. Promoter-selective activation domains in Oct-1 and Oct-2 direct differential activation of an snRNA and mRNA promoter. Cell. 1992;68:755–767. doi: 10.1016/0092-8674(92)90150-b. [DOI] [PubMed] [Google Scholar]
- 47.Tebb G, Mattaj I W. The Xenopus laevis U2 gene distal sequence element (enhancer) is composed of four subdomains that can act independently and are partly functionally redundant. Mol Cell Biol. 1989;9:1682–1690. doi: 10.1128/mcb.9.4.1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Teichmann M, Seifart K. Physical separation of two different forms of human TFIIIB active in the transcription of the U6 and the VAI gene in vitro. EMBO J. 1995;14:5974–5983. doi: 10.1002/j.1460-2075.1995.tb00286.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thisse C, Perrin-Schmitt F, Stoetzel C, Thisse B. Sequence-specific transactivation of the Drosophila twist gene by the dorsal gene product. Cell. 1991;65:1191–1201. doi: 10.1016/0092-8674(91)90014-p. [DOI] [PubMed] [Google Scholar]
- 50.Tommerup N, Vissing H. Isolation and fine mapping of 16 novel human zinc finger-encoding cDNAs identify putative candidate genes for developmental and malignant disorders. Genomics. 1995;27:259–264. doi: 10.1006/geno.1995.1040. [DOI] [PubMed] [Google Scholar]
- 51.Triezenberg S J. Structure and function of transcriptional activation domains. Curr Opin Genet Dev. 1995;5:190–196. doi: 10.1016/0959-437x(95)80007-7. [DOI] [PubMed] [Google Scholar]
- 52.Uesugi M, Nyanguile O, Lu H, Levine A J, Verdine G L. Induced α helix in the VP16 activation domain upon binding to a human TAF. Science. 1997;277:1310–1313. doi: 10.1126/science.277.5330.1310. [DOI] [PubMed] [Google Scholar]
- 53.Vesque C, Charnay P. Mapping functional regions of the segment-specific transcription factor Krox-20. Nucleic Acids Res. 1992;20:2485–2492. doi: 10.1093/nar/20.10.2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Waldschmidt R, Wanandi I, Seifart K H. Identification of transcription factors required for the expression of mammalian U6 genes in vitro. EMBO J. 1991;8:2595–2603. doi: 10.1002/j.1460-2075.1991.tb07801.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yoon J-B, Murphy S, Bai L, Wang Z, Roeder R G. Proximal sequence element-binding transcription factor (PTF) is a multisubunit complex required for transcription of both RNA polymerase II- and RNA polymerase III-dependent small nuclear RNA genes. Mol Cell Biol. 1995;15:2019–2027. doi: 10.1128/mcb.15.4.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]