Abstract
The Saccharomyces cerevisiae transcription factor IIH (TFIIH) is essential both for transcription by RNA polymerase II (RNAP II) and for nucleotide excision repair (NER) of damaged DNA. We have established cell extracts which support RNAP II transcription from the yeast CYC1 promoter or NER of transcriptionally silent damaged DNA on independent plasmid templates and substrates. When plasmid templates and substrates for both processes are simultaneously incubated with these extracts, transcription is significantly inhibited. This inhibition is strictly dependent on active NER and can be complemented with purified holo-TFIIH. These results suggest that in the presence of active NER, TFIIH is preferentially mobilized from the basal transcription machinery for use in NER. Inhibition of transcription in the presence of active NER requires the RAD26 gene, the yeast homolog of the human Cockayne syndrome group B gene (CSB).
Nucleotide excision repair (NER) is a biochemically complex process by which many types of base damage are excised from the genome of living cells as oligonucleotide fragments (14). This process operates both in transcriptionally silent DNA and in DNA that is undergoing active transcription by RNA polymerase II (RNAP II) (5, 15, 21). A signature feature of the latter NER mode is that the transcribed strand is repaired significantly faster than the nontranscribed strand, a phenomenon referred to as strand-specific repair or transcription-coupled repair (5, 15, 21).
In eukaryotes, NER of both transcriptionally silent and transcriptionally active DNA requires more than 20 distinct gene products (14, 29, 51). In the yeast Saccharomyces cerevisiae, these proteins include the seven known subunits of the RNAP II basal transcription factor core TFIIH (38), encoded by the essential genes TFB1, TFB2, TFB3, TFB4, RAD3, SSL1, and SSL2 (10, 13, 17). Mutants with conditional mutations in each of these genes have been shown to be defective in NER by using a cell-free system that measures repair synthesis of damaged plasmids in vitro (13, 21a, 47, 48). While core transcription factor IIH (TFIIH) is essential for NER, this seven-subunit complex is not sufficient for RNAP II transcription in a reconstituted in vitro system (36). Such a system has an additional requirement for polypeptides encoded by the KIN28 and CCL1 genes, which comprise the transcription factor TFIIK (11, 36). The association of TFIIK with core TFIIH generates a complex designated holo-TFIIH (36, 37).
The requirement of core TFIIH for both NER and RNAP II transcription led to initial speculation that this requirement might explain the faster rate of NER observed in the transcribed strand relative to that of the nontranscribed strand of transcriptionally active genes. It was suggested that when transcription elongation complexes arrest at sites of base damage in the transcribed strand, TFIIH might promote rapid assembly of the NER machinery at such sites, thus facilitating strand-specific repair (14, 29, 51). However, several studies have shown that TFIIH dissociates from the transcription complex soon after promoter clearance (7, 18, 52) and is not normally associated with the RNAP II elongation complex. An alternative and more likely explanation for the dual roles of TFIIH in transcription and NER comes from the observation that two of the TFIIH subunits (Rad3 and Ssl2 in yeast) are DNA helicases with opposite polarity (19, 35). The concerted action of these helicases is thought to generate localized regions of denaturation (bubbles) in the DNA duplex. The margins of such bubbles comprise junctions between duplex and single-stranded DNA which, during NER, are recognized by junction-specific endonucleases with opposite single-strand polarity, thereby generating incisions (nicks) flanking sites of base damage (3, 20, 25, 26, 34). Evidence in support of TFIIH-mediated unwinding of regions of the DNA duplex during NER has recently been provided with an in vitro system reconstituted from purified human proteins (8).
The results of previous experiments from our laboratory suggest that yeast core TFIIH is a component of a large multiprotein complex designated the nucleotide excision repairosome (28a, 37). In the event that all core TFIIH is associated with either transcription initiation or NER complexes in yeast, the dual roles of TFIIH in transcription initiation and NER offer the potential of limiting transcription initiation in the presence of DNA repair. Here we report the results of experiments which directly support this notion. We have generated a cell-free system that supports either NER of damaged plasmid DNA lacking promoter sites (and hence transcriptionally inactive) or RNAP II transcription from a different undamaged plasmid carrying the yeast CYC1 promoter. We show that in the simultaneous presence of both substrates, active NER significantly limits the extent of RNAP II transcription. The inhibition of transcription can be relieved by supplementing extracts with purified holo-TFIIH, but not core TFIIH. Finally, we show that the yeast RAD26 gene, the yeast homolog of the human Cockayne syndrome group B gene (CSB), is required for NER-dependent transcription inhibition, even though extracts of rad26 mutant cells are proficient for NER of transcriptionally inactive DNA (and RNAP II transcription) in vitro. In contrast to the observation of inhibition of transcription in the presence of active NER, increased transcription had no detectable effect on NER in vitro.
MATERIALS AND METHODS
Materials and reagents.
Ultrapure ribonucleoside triphosphates, deoxynucleoside triphosphates, and sodium 3′-O-methylguanosine 5′-triphosphate (3′-O-methyl-GTP) were purchased from Pharmacia. RNase T1, RNase inhibitor, protease inhibitors (phenylmethylsulfonyl fluoride [PMSF], bestatin, pepstatin A, leupeptin, chymostatin, and antipain) and α-amanitin were obtained from Boehringer Mannheim. Acetylated bovine serum albumin (Ac-BSA), proteinase K, dithiothreitol (DTT), yeast tRNA, and ultrapure enzyme grade sucrose were purchased from Gibco BRL. Phosphocreatine (disodium salt), creatine phosphokinase, benzamidine hydrochloride, and polyethylene glycol 8000 (PEG 8000) were purchased from Sigma. Recombinant yeast lytic enzyme was obtained from ICN Biomedicals. Escherichia coli endonuclease III was kindly provided by Richard Cunningham, State University of New York at Albany. [α-32P]dCTP and [α-32P]UTP (3,000 Ci/mmol) were purchased from Amersham.
Strains and plasmids.
Yeast strains used in this study are listed in Table 1. Plasmids pUC18 (Gibco BRL) and pGEM3Zf(+) (Promega) were used for the NER assay and pCYC1/G− (a gift from Roger Kornberg, Stanford University) with a 377-bp G-less cassette driven by the yeast CYC1 promoter was used for the RNAP II transcription assay (24). Supercoiled plasmid DNA was prepared by alkaline lysis followed by purification on ethidium bromide-CsCl and neutral sucrose gradients (5 to 20%).
TABLE 1.
S. cerevisiae strains used in this study
| Strain | Genotype |
|---|---|
| W303-1B | MATa RAD can1-100 ade2-1 trp-1 leu2-3,112 his3-11,15 ura3-1 |
| MGSC104 | W303-1B rad7Δ::LEU2 |
| W303236 | W303-1B rad16Δ::URA3 |
| MGSC139 | W303-1B rad14Δ::LEU2 |
| MGSC101 | W303-1B rad23Δ::URA3 |
| MGSC102 | W303-1B rad26Δ::HIS3 |
| PB01-28 | W303-1B rad28Δ::URA3 |
| BJ2168 | MATa RAD leu2 trp1 ura3-52 pep4-3 prb1-1122 prc1-407 |
| BJ2168-RAD2::URA3 | BJ2168 rad2Δ::URA3 |
| BJ2168rad10Δ | BJ2168 rad10Δ::URA3 |
| SX46a | MATa RAD ade2 his3-532 ura3-52 trp1-289 |
| SX46a-RAD4::URA3 | SX46a rad4::URA3 |
| SX46arad2Δ | SX46a rad2Δ::TRP1 |
| SX46arad1Δ | SX46a rad1Δ::TRP1 |
| LN203-2 | MATa rad3-2 pep4-3 ade2 lys2 ura3-52 |
Cell extracts.
Yeast cells were grown in YPD (1% [wt/vol] yeast extract, 2% [wt/vol] Bacto Peptone, 2% [wt/vol] dextrose) at 30°C to a level at which the absorbance at 600 nm is 2 to 3. Cells were harvested by centrifugation in a Beckman JA-10 rotor at 3,000 rpm for 5 min at 4°C and washed once with ice-cold distilled water. Cell pellets were resuspended in 100 mM EDTA-KOH (pH 8.0)–20 mM DTT (10 ml/g of cells) and shaken slowly at 30°C for 5 min. Cells were pelleted by centrifugation at 4°C and resuspended in YPS (1% [wt/vol] yeast extract, 2% [wt/vol] Bacto Peptone, 1 M sorbitol) (1 ml/g of cells). Cells were converted to spheroplasts by digestion with yeast lytic enzyme (500 U/g of cells) for 30 min at 25°C with gentle shaking. Enzyme digestion was stopped by the addition of the prechilled YPS (20 ml/g of cells). All subsequent procedures were carried out at 0 to 4°C. Spheroplasts were collected by centrifugation in a Beckman JA-14 rotor at 4,000 rpm for 5 min, rinsed once with cold YPS, resuspended in buffer A (10 mM Tris-acetate [pH 7.8], 1 mM EDTA, 5 mM DTT) (3 ml/g of spheroplasts) and incubated on ice for 10 min. Protease inhibitors (1 mM PMSF, 2 mM benzamidine hydrochloride, leupeptin [0.5 μg/ml], chymostatin [2 μg/ml], antipain [2.5 μg/ml], bestatin [0.3 μg/ml], and pepstatin A [0.4 μg/ml]) were added followed by the addition of buffer B (50 mM Tris-acetate [pH 7.8], 10 mM MgSO4, 1 mM DTT, 25% sucrose, 50% glycerol [3 ml/g]), slowly with stirring. After 5 min, saturated (NH4)2SO4 solution (pH 7.0) was added over a period of 15 min to a concentration of 0.39 M. Cellular debris was removed by centrifugation for 5 min at 3,000 rpm in a Beckman JA-14 rotor followed by centrifugation for 90 min at 62,000 rpm in a Beckman Ti70 rotor. The supernatant was removed immediately, leaving the last 1 to 2 ml behind, adjusted to 3.05 M (NH4)2SO4 by the addition of solid ammonium sulfate, neutralized with 10 μl of 1 M KOH/g of (NH4)2SO4 and stirred slowly for 30 min at 0°C. Precipitated protein was collected by centrifugation in a Beckman JA-20 rotor at 18,000 rpm for 25 min, thoroughly resuspended in 1/30 volume of buffer C (25 mM HEPES-KOH [pH 7.6], 10 mM MgSO4, 10 mM EGTA, 5 mM DTT, 20% glycerol) and dialyzed for 6 to 8 h against four 1-liter changes of buffer C. Insoluble material was removed by centrifugation in a Beckman JA-20 rotor at 15,000 rpm for 10 min, and the supernatant was quick-frozen in small aliquots. Protein concentrations were determined by the method of Bradford, using BSA as a standard. Extracts typically contained 10 mg of protein per ml, were stable for at least 1 year at −80°C, and remained active after at least two cycles of freezing and thawing.
Purification of yeast RNAP II initiation factors.
Yeast TBP, TFIIB, and TFIIE were recombinant proteins purified from bacteria as previously described (12, 22). Histidine-tagged holo-TFIIH and core TFIIH were purified from yeast whole-cell extracts by chromatography on Bio-Rex 70, phosphocellulose, Ni2+-nitriloacetic acid agarose, phenyl HR high-performance liquid chromatography (HPLC), and Mono Q HR HPLC as described previously (36).
Damaged DNA.
To prepare acetylaminofluorene (AAF)-damaged DNA, plasmid pUC18 (50 μg/ml) was treated with 30 μM N-acetoxy-2-acetylaminofluorene (AAAF) in the dark for 3 h at 37°C in TE buffer (10 mM Tris-HCl [pH 7.6], 1 mM EDTA). Unreacted AAAF was removed by repeated extraction with diethyl ether. Modified DNA was precipitated with ethanol and purified by centrifugation on a neutral sucrose gradient (5 to 20%). To obtain UV radiation-damaged DNA, plasmid pUC18 (50 μg/ml) was irradiated in a thin layer on ice in TE buffer (pH 8.0) under a germicidal lamp (peak output at 254 nm) at a dose of 450 J/m2. UV-irradiated DNA was treated with E. coli endonuclease III (6 μg) for 2 h at 37°C in order to remove molecules containing photoproducts that are substrates for base excision repair. The remaining supercoiled DNA was purified by centrifugation through a neutral sucrose gradient (5 to 20%). Treatment with endonuclease III and purification of unnicked DNA were repeated twice, and the DNA was concentrated with a Centricon-50 (Amicon). All damaged DNA was stored at −20°C.
In vitro nucleotide excision repair.
DNA repair synthesis was performed as previously described (49) with slight modifications. Cell extract (20 to 50 μg) was mixed with 300 ng of damaged DNA as well as undamaged [pGEM3Zf(+)] DNA except when otherwise indicated. After incubation for 15 min at 20°C, NER buffer (50 mM HEPES-KOH [pH 7.6], 40 mM potassium acetate, 8 mM magnesium acetate, 1 mM DTT, 0.4 mM EDTA, 2.5 μg of Ac-BSA, 1.6 μg of creatine phosphokinase, 45 mM phosphocreatine, 6% glycerol, 4.8% PEG 8000, 4 mM ATP, 20 μM dATP, 20 μM dGTP, 20 μM TTP, 5 μM dCTP, 0.1 μl of [α-32P]dCTP) was added to a total volume of 25 μl. After 60 min of incubation at 25°C, plasmid DNA was purified and digested with HindIII as described previously (49). DNA was resolved by 1% agarose gel electrophoresis in the presence of ethidium bromide and visualized by UV illumination. Repair synthesis was detected in dried gels either by autoradiography or by PhosphorImager analysis (Molecular Dynamics).
In vitro RNA polymerase II transcription.
Reaction mixtures containing 20 to 50 μg of cell extract and 200 ng of supercoiled plasmid template were incubated at 20°C for 15 to 20 min followed by the addition of transcription buffer (NER buffer containing 400 μM CTP, 10 μM UTP, 0.5 μl of [α-32P]UTP, 10 μM 3′-O-methyl-GTP, 20 U of RNase inhibitor, but lacking deoxynucleoside triphosphates and [α-32P]dCTP) and distilled water to a final volume of 25 μl. Incubations were allowed to proceed at 25°C for 45 to 60 min. Transcription was terminated by the addition of 120 μl of RNase T1 buffer (10 mM Tris-HCl [pH 7.5], 300 mM NaCl, 5 mM EDTA) containing 15 U of RNase T1, followed by incubation at 32°C for 10 min. Then 10 μl of sodium dodecyl sulfate (10%) and 8 μl of proteinase K (10 μg/μl) were added, and reaction mixtures were incubated at 37°C for 25 min. Each reaction mixture was extracted twice with Tris-buffered phenol-chloroform. Transcripts were precipitated by the addition of 2.5 volumes of ethanol, using 50 μg of tRNA as the carrier, and collected by centrifugation. Dried pellets were resuspended in 16 μl of loading buffer (95% ultrapure formamide, 0.05% xylene cyanol, 0.05% bromphenol blue), heat denatured, and analyzed by electrophoresis through 6% (wt/vol) polyacrylamide (19:1)–7 M urea gels in 1× TBE buffer. Electrophoresis was performed with an electromotive force of 30 V/cm. Transcripts were visualized and quantified as described above. Transcription of heated nuclear extracts was performed as described previously (9).
Transcription-NER competition experiments.
All reaction mixtures contained 20 μg of cell extract and 400 ng of damaged or undamaged DNA unless otherwise noted. After preincubation at 20°C for 15 to 20 min, NER-transcription buffer (transcription buffer containing 20 μM dATP, 20 μM dGTP, and 20 μM TTP but lacking UTP) and distilled water were added to a total volume of 25 μl. To monitor NER, 5 μM dCTP, 0.1 μl of [α-32P]dCTP, and 400 μM UTP were added. To monitor transcription, 10 μM UTP, 0.5 μl of [α-32P] UTP, and 20 μM dCTP were added. Reactions were allowed to proceed for 45 to 60 min at 25°C. Further steps for the NER-transcription assays were as described above.
RESULTS
Yeast cell extracts support either RNAPII transcription or NER.
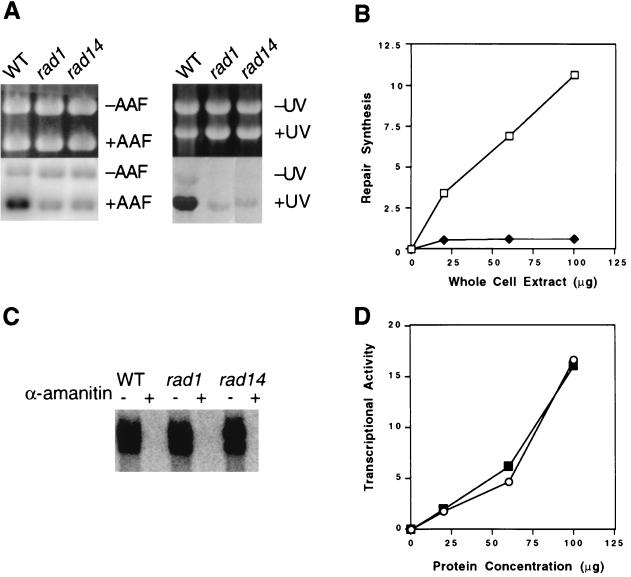
We previously reported cell-free conditions which can support either RNAP II transcription or transcriptionally independent NER, depending on the plasmid and nucleoside triphosphate substrates used (50). In this study, we have optimized these observations using extracts prepared slightly differently (see Materials and Methods). Extracts from wild-type cells supplemented with all four deoxyribonucleotide triphosphates support robust repair synthesis of supercoiled plasmid pUC18 DNA treated with either AAAF or UV radiation (Fig. 1A). The amount of repair synthesis is linearly related to protein concentration (Fig. 1B) and to the time of incubation (data not shown). Consistent with numerous previous studies (47–49), NER is defective in extracts of yeast strains carrying mutations in all RAD genes known to be indispensable for NER (Fig. 1A and B and data not shown). When wild-type or various rad mutant extracts were supplemented with ribonucleotide triphosphates instead of deoxyribonucleotide triphosphates, indistinguishable levels of transcription from a different plasmid carrying the yeast CYC1 promoter upstream of a G-less cassette were observed (Fig. 1C and D). This transcription was completely sensitive to the RNAP II inhibitor α-amanitin (Fig. 1C).
FIG. 1.
(A) NER in wild-type (strain W303-1B) and rad1 and rad14 deletion mutant extracts. NER was performed with 50 μg of extract at 25°C for 60 min. WT, wild type; +AAF, pUC18 DNA containing AAF adducts; −AAF, undamaged pGEM3Zf(+) DNA; +UV, UV-irradiated pUC18 DNA; −UV, undamaged pGEM3Zf(+) DNA. Ethidium bromide-stained gels (top) and autoradiograms of the gels (bottom) are shown. (B) Protein concentration dependence of DNA repair synthesis in cell extracts. AAF-damaged pUC18 DNA was incubated with the indicated amounts of yeast wild-type (strain W303-1B) or rad14 deletion mutant extracts for 1 h at 25°C. Quantitation was performed with a PhosphorImager. Open squares, wild type; closed diamonds, rad14 mutant. (C) RNAP II transcription of plasmid CYC1/G− was performed with 50 μg of extract at 25°C for 45 min. α-Amanitin (10 μg/ml) was included (+) or not included (−) as indicated over the gel. (D) Protein concentration dependence of transcription in cell extracts. Transcription of the template (200 ng) was performed at 25°C for 45 min with the indicated amounts of yeast wild-type (strain W303-1B) or rad14 mutant extracts. Quantitation was performed with a PhosphorImager. Open circles, wild type; closed squares, rad14 mutant.
Inhibition of RNAP II transcription in the presence of active NER.
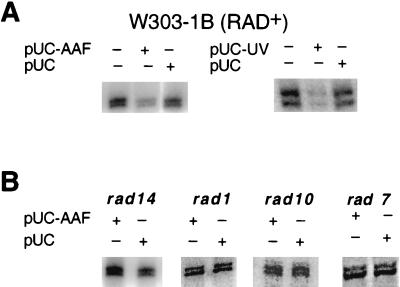
Incubation of extracts supplemented with both ribo- and deoxyribonucleoside triphosphates in the simultaneous presence of the CYC1 transcription template and plasmid pUC18 treated with either AAAF or UV radiation resulted in significant and reproducible inhibition of RNAP II transcription from the CYC1 promoter (Fig. 2A). In contrast, the presence of undamaged plasmid pUC18 had no detectable effect on the levels of transcription (Fig. 2A). To demonstrate that this inhibition is specifically related to active NER rather than the presence of damaged DNA, we examined transcription from the CYC1 promoter in extracts of rad mutants known to be defective in NER. No inhibition of transcription was observed in extracts of rad1, rad2, rad3, rad4, rad7, rad10, rad14, rad16, or rad23 mutants (Fig. 2B and data not shown).
FIG. 2.
(A) Inhibition of RNAP II transcription in the presence of NER in yeast wild-type (strain W303-1B) extracts. After 20 min of preincubation at 20°C, transcription and NER were initiated by supplying appropriate reaction buffers (see Materials and Methods). Transcription of the CYC1/G− template (200 ng) was at 25°C for 45 min with 20 μg of cell extract in the presence (+) or absence (−) of NER substrates (pUC-AAF DNA and pUC-UV DNA). NER substrates (400 ng) and control pUC18 DNA (400 ng) were included as indicated over the gels. The 350- to 375-nucleotide transcripts were resolved on a 6% polyacrylamide–7 M urea gel and detected by phosphorimaging. (B) Transcription from the CYC1/G− template (200 ng) was carried out with 20 μg of various rad mutant extracts in the presence (+) or absence (−) of 400 ng of NER substrate or control pUC18 DNA as indicated over the gels. Incubations were at 25°C for 45 min. pUC-AAF, pUC18 DNA treated with AAAF; pUC-UV, UV-irradiated pUC18 DNA; pUC, pUC18 DNA.
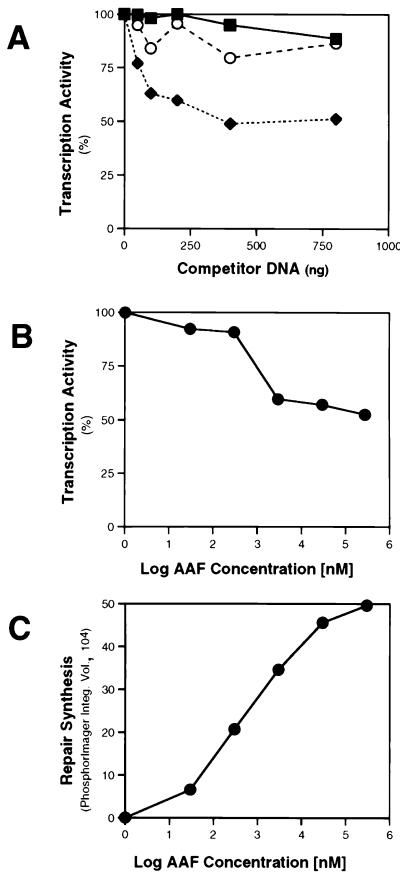
To quantitate the inhibition of RNAP II transcription in the presence of active NER, we added increasing amounts of pUC18 DNA treated with a fixed amount of AAAF to the reaction mixtures. We observed ∼50% inhibition of transcription in the presence of 400 ng or more of AAAF-treated plasmid DNA (Fig. 3A). However, the extent of transcription inhibition varied between 50 and 75% in multiple different experiments. Once again, no inhibition of transcription was observed in extracts of rad mutant cells, and the presence of undamaged DNA had little or no effect (Fig. 3A). Transcription from the CYC1 promoter was also progressively inhibited in the presence of a fixed concentration of plasmid DNA treated with increasing amounts of AAAF (Fig. 3B). Evidence that such treatment resulted in the formation of progressively more AAF adducts in the substrate DNA was derived from independent experiments showing increasing repair synthesis as a function of the amount of AAAF treatment of the plasmid substrate (Fig. 3C). At present, it is unclear why transcription is only partially inhibited in the presence of NER in vitro. Conceivably, cells contain subpopulations of RNAP II transcription complexes, some of which are sensitive to competition by active NER and some of which are not.
FIG. 3.
(A) Transcription inhibition in the presence of NER. Transcription of the CYC1/G− template (200 ng) was performed at 25°C for 45 min with 20 μg of extract in the presence of increasing amounts of pUC18 DNA (50, 100, 200, 400, or 800 ng) treated with 30 μM AAAF (pUC18-AAF), or with the same amount of undamaged pUC18 DNA. Quantitation was performed by phosphorimaging. Transcription in the absence of competitor DNA was normalized to 100%. Open circles, wild-type yeast strain with competitor pUC18 DNA; closed diamonds, wild-type strain with competitor pUC18-AAF DNA; closed squares, rad14 mutant with competitor pUC18-AAF DNA. (B) Transcription of the CYC1/G− template (200 ng) was performed with 20 μg of wild-type extract in the presence of pUC18-AAF DNA (400 ng) treated with increasing amounts of AAAF (30 nM to 3 mM) at 25°C for 45 min. Transcription in the presence of untreated pUC18 DNA was normalized to 100%. (C) NER was performed with 20 μg of wild-type extract in the presence of 400 ng of pUC18 DNA treated with increasing amounts of AAAF (30 nM to 3 mM), at 25°C for 60 min. Quantitation of repair synthesis was performed by phosphorimaging.
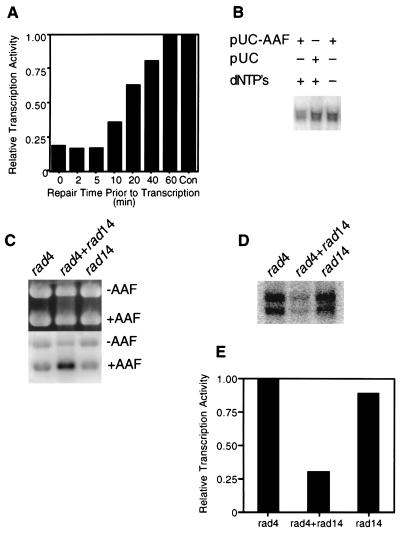
To further substantiate the observation that inhibition of transcription from the CYC1 promoter in vitro requires active NER, such repair was allowed to transpire for various periods of time prior to the initiation of transcription by the addition of ribonucleoside triphosphates. During the first 5 min of preincubation, ∼75% inhibition of transcription was observed (Fig. 4A). At later times, this inhibition was progressively alleviated, and after 60 min, no inhibition was observed relative to the level of transcription measured in the absence of NER (Fig. 4A). These results suggest that increased repair of AAAF-treated plasmid DNA progressively relieves the inhibition of RNAP II transcription. In further experiments, we demonstrated no inhibition of transcription from the CYC1 promoter in the absence of deoxyribonucleoside triphosphates required for repair synthesis during NER (Fig. 4B). Finally, we showed that whereas extracts of the NER-defective rad4 and rad14 mutants (Fig. 4C) failed to inhibit RNAPII transcription from the CYC1 promoter (Fig. 4D), mixing the extracts complemented both defective NER (Fig. 4C) and transcription inhibition (Fig. 4D and E).
FIG. 4.
Inhibition of RNAP II transcription from the CYC1/G− template requires active NER. (A) Transcription template (200 ng), pUC18 DNA treated with AAAF (pUC-AAF) DNA (400 ng), and 20 μg of wild-type extract were incubated at 20°C for 20 min. Reaction buffer (lacking CTP and UTP) was added, and NER was allowed to proceed at 25°C for the times indicated, following which CTP and UTP were added to initiate transcription for 45 min. pUC18 DNA (400 ng) was used instead of pUC-AAF DNA in the control experiment, and the amount of transcription was normalized to 1.0. (B) Transcription of CYC1/G− template (200 ng) was performed with 20 μg of wild-type extract in the presence (+) of pUC-AAF or pUC18 DNA (400 ng) at 25°C for 45 min. Deoxynucleoside triphosphates (dNTP’s) were included (+) or omitted (−) as indicated. (C) NER was performed with 20 μg of rad4, rad14, or a mixture of rad4 and rad14 mutant extracts under transcription-NER reaction conditions. +AAF, pUC18 DNA treated with AAAF; −AAF, undamaged pGEM3Zf(+) DNA. (D) Inhibition of transcription from the CYC1/G− template (200 ng) was restored to mixtures of rad4 and rad14 mutant extracts in the presence of the NER substrate (pUC18 DNA treated with UV irradiation). (E) Quantitation of the results shown in panel D. Quantitation was performed by phosphorimaging and normalized in the rad4 experiment to 1.0.
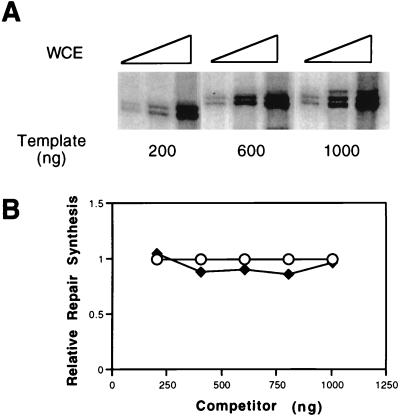
We demonstrated increased levels of transcription from the CYC1 promoter by increasing either the template concentration or the amount of cell extract (Fig. 5A). However, when extracts were incubated with a fixed amount of AAAF-treated DNA, no reduction in the level of NER was observed as a function of the amount of transcription template added (Fig. 5B) or as a function of the amount of extract between 20 and 100 μg of protein (data not shown).
FIG. 5.
(A) Increased transcription from the CYC1/G− template by increasing the amount of template or extract. Transcription was performed at 25°C for 45 min with various amounts (10, 20, and 50 μg [indicated by the height of the large open triangle over each set of three lanes]) of wild-type extract in the presence of various amounts (200, 600, or 1,000 ng) of template. (B) NER was performed at 25°C for 60 min with 20 μg of wild-type extract in the presence of various amounts (200, 400, 600, 800, or 1,000 ng) of the transcription plasmid CYC1/G− (closed diamonds) or control undamaged pGEM3Zf(+) DNA (open circles). Repair synthesis was quantitated by phosphorimaging, and the level of repair synthesis in the presence of pGEM3Zf(+) DNA was normalized to 1.0. WCE, whole-cell extract.
Transcription inhibition in the presence of NER is relieved by holo-TFIIH.
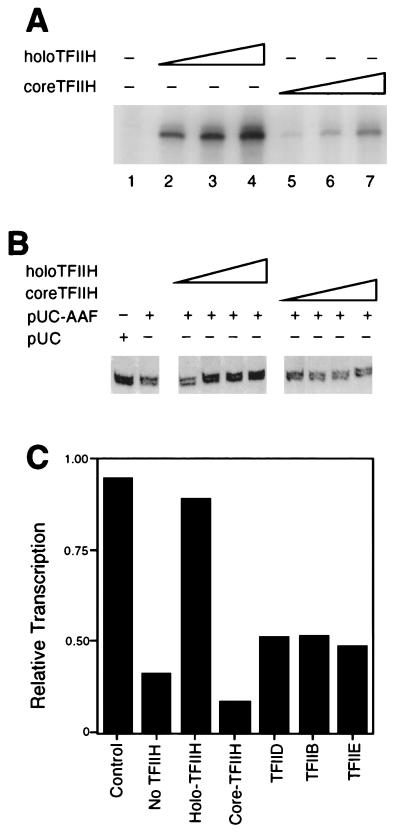
The results of the experiments described above prompted the hypothesis that the inhibition of transcription in the presence of NER is derived from the mobilization of TFIIH resident in RNAP II transcription initiation complexes for preferential use in NER. This hypothesis predicts that inhibition of transcription might be alleviated by complementing extracts with exogenous core or holo-TFIIH. Preparations of purified core or holo-TFIIH (shown to be enzymatically active for RNAP II transcription in vitro [Fig. 6A]) were added independently to reaction mixtures in which transcription from the CYC1 promoter was inhibited in the presence of NER. The addition of holo-TFIIH significantly alleviated the inhibition, but the addition of core TFIIH had little, if any, effect (Fig. 6B and C). The addition of increasing amounts of holo-TFIIH to transcription reactions in the presence or absence of nondamaged plasmid had no effect on the levels of transcription (data not shown). To demonstrate the specificity of the complementation by holo-TFIIH, we supplemented extracts with purified transcription factors IIB, IIE, and TBP. None of these transcription factors complemented inhibition of transcription in the presence of NER (Fig. 6C).
FIG. 6.
Inhibition of RNAP II transcription in the presence of NER is relieved by holo-TFIIH. (A) Both core TFIIH (lanes 2 to 4) and holo-TFIIH (lanes 5 to 7) restored transcription activity to a heat-inactivated nuclear extract (HNE). All reaction mixtures contained 6 μl of HNE and 70 ng of recombinant yeast TBP. Core and holo-TFIIH (0.5, 1.0, and 2.0 μl of Mono Q fractions [indicated by the height of the large open triangle over each set of three lanes]) were assayed. (B) Incubations contained CYC1/G− template (200 ng), pUC18 treated with AAAF (pUC-AAF) or control pUC18 DNA (400 ng), 20 μg of wild-type extract, and various amounts of holo- or core TFIIH (0.5, 1.0, 2.0, and 3.0 μl of MonoQ fraction [increasing amount of TFIIH indicated by the height of the large open triangle over each set of four lanes]). Incubations were at 20°C for 20 min, following which transcription-NER reaction buffer was added and incubation continued at 25°C for 45 min. (C) Reaction mixtures containing CYC1/G− template (200 ng), pUC-AAF or pUC plasmid (400 ng; control), and 20 μg of wild-type extract were supplemented with either distilled water, holo-TFIIH, core TFIIH, TFIIB (200 ng), TBP (200 ng), or TFIIE (200 ng). Incubations were at 20°C for 20 min, following which transcription-NER reaction buffer was added and incubation was continued at 25°C for 45 min. Quantitation of transcription was performed by phosphorimaging and normalized in the control experiment to 1.0.
Inhibition of transcription in the presence of NER requires the RAD26 gene.
The yeast RAD26 and RAD28 genes are the structural (and presumed functional) homologs of the human CSB and CSA genes, respectively (4, 43). Mutational inactivation of the human genes results in the hereditary disease Cockayne syndrome, which is characterized by postnatal growth, neurological defects, and photosensitivity (16). CS-A and CS-B cells are abnormally sensitive to UV radiation and lose the kinetic preference for repair of the transcribed strand of transcriptionally active genes displayed by cells from healthy individuals (45). Yeast rad26 mutants (but not rad28 mutants) are also defective in strand-specific NER (4, 40, 43). Additionally, rad26 mutants (but not rad28 mutants) have been shown to be deficient in the recovery of GAL10 and RNR3 mRNA synthesis following inhibition of such synthesis by exposure of yeast cells to UV radiation (28).
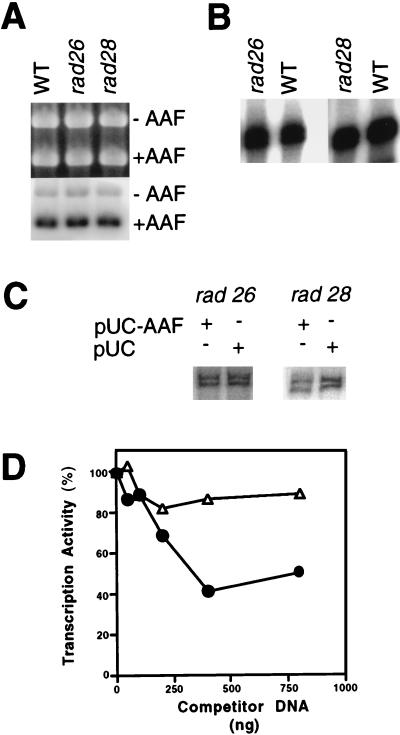
The yeast RAD26 and RAD28 and human CSB and CSA gene products are not required for NER of transcriptionally silent DNA (15). Therefore, as expected, extracts of rad26 and rad28 deletion mutants supported wild-type levels of NER of damaged pUC18 plasmid DNA (Fig. 7A). Normal levels of RNAP II transcription from the CYC1 promoter were also observed (Fig. 7B). Surprisingly, whereas inhibition of transcription at levels comparable to that with wild-type extracts was observed in rad28 mutant extracts (Fig. 7C and D), such inhibition was not observed in rad26 mutant extracts (Fig. 7C and D). Since RAD26 is required for transcription-coupled NER in yeast, we considered the possibility that the failure to observe inhibition of transcription from the CYC1 promoter in rad26 mutant extracts may reflect RAD26-dependent transcription of the damaged pUC18 plasmid. However, no measurable transcription from plasmid pUC18 could be detected in vitro, either in the presence or absence of AAAF treatment of the plasmid (data not shown). Additionally, NER of AAAF-treated pUC18 DNA was unaffected in the presence of α-amanitin (data not shown).
FIG. 7.
Inhibition of RNAP II transcription in the presence of NER requires RAD26 but not RAD28. (A) NER was performed with 50 μg of wild-type (WT), rad26, or rad28 extract in the presence of pUC18 treated with AAAF (pUC-AAF), pGEM3Zf(+) at 25°C for 60 min. −AAF, undamaged pGEM3Zf(+) DNA; +AAF, AAAF-treated pUC18 DNA. An ethidium bromide-stained gel (top) and an autoradiogram of the gel (bottom) are shown. (B) Transcription was with 50 μg of wild-type, rad26, or rad28 extract in the presence of CYC1/G− template at 25°C for 45 min. (C) Transcription from the CYC1/G− template was at 25°C for 45 min with 20 μg of rad26 or rad28 mutant extract in the presence (+) or absence (−) of pUC-AAF or control pUC plasmid (400 ng). (D) Transcription from the CYC1/G− template with 20 μg of rad26 or rad28 mutant extract was at 25°C for 45 min in the presence of various amounts of NER substrate (pUC-AAF). Transcription was quantitated as described above. Transcription in the absence of competitor DNA was normalized to 100%. Open triangles, rad26 extract; closed circles, rad28 extract.
DISCUSSION
Studies on the molecular mechanism of NER in both yeast and human cells have demonstrated that incisions flanking sites of base damage in DNA are catalyzed by endonucleases which specifically recognize junctions between duplex and single-stranded DNA (3, 20, 25, 26, 34). The elaboration of such junctions requires that the DNA strands near sites of base damage be separated over a distance of ∼28 to 30 nucleotides. Recent studies with human cell-free systems (8) have directly implicated TFIIH in this denaturation process, thereby supporting the notion that the requirement for this transcription factor in NER (and presumably RNAP II transcription) is derived from its ability to generate localized bubbles in DNA through the action of its DNA helicase subunits.
Discovery of the dual requirement for TFIIH in NER and RNAP II transcription led to the early suggestion that the preferential association of this multiprotein complex with the NER machinery might limit the rate of transcription initiation in the presence of active NER (15, 37). Recent studies with human cell-free systems are conflicting. One study using human lymphoblastoid cell extracts that support both RNAP II transcription and NER reported no competition between these processes in vitro (30). These researchers concluded that human extracts prepared as described in that study are not rate limited for TFIIH. A second study observed inhibition of RNAPII transcription in the presence of DNA damaged with either cisplatin or UV radiation but concluded that this inhibition is derived from the binding of the basal transcription factor TFIID or TBP to sites of base damage rather than the process of NER per se (46). Using yeast whole-cell extracts, we have observed inhibition of RNAP II transcription from the yeast CYC1 promoter which is strictly dependent on active NER and which is relieved by supplementing extracts with holo-TFIIH.
It has been previously reported that purified yeast holo-TFIIH supports both transcriptionally independent NER in yeast crude extracts and RNAP II transcription in a reconstituted in vitro system (36, 37). In contrast, purified core TFIIH supports transcriptionally independent NER, but not RNAP II transcription in a reconstituted system (36, 37). Our observation that inhibition of transcription from the CYC1 promoter in the presence of active NER is relieved by complementing cell extracts with purified holo-TFIIH, but not core TFIIH, is consistent with the notion that inhibition of transcription results from the redistribution of core TFIIH associated with RNAP II transcription initiation complexes to protein complexes specifically dedicated to NER. Such a redistribution has been proposed previously and is supported by the observation that core TFIIH and repairosome fractions are equally active in a core TFIIH-dependent heated nuclear extract transcription assay, suggesting that the association of core TFIIH with the repairosome in reversible (37). The utility of this process is intuitively obvious, since in living yeast cells, it may limit transcription through damaged template strands, thereby mitigating against the generation of mutant and/or truncated transcripts. Thus, yeast cells may have evolved two mechanisms for protecting against the elaboration of defective transcripts from damaged DNA. One involves inhibition of transcription initiation, while the second involves the preferential repair of template strands at sites of arrested transcription elongation.
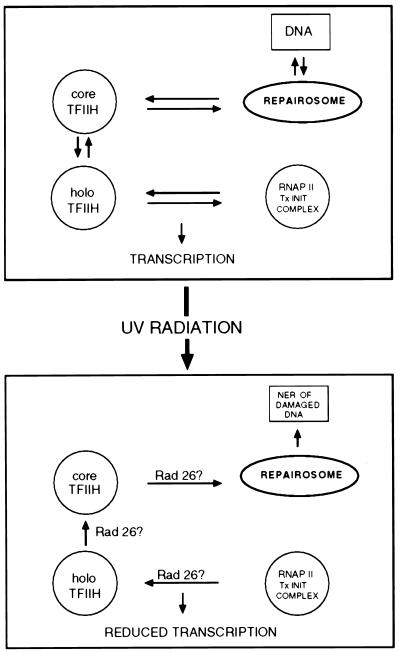
The precise mechanism by which RNAP II transcription initiation is inhibited during NER remains a challenge for the future. The observation that such inhibition is specifically associated with active NER and not simply the presence of base damage in DNA suggests that the process of active NER may alter a steady-state equilibrium which determines the distribution of TFIIH between RNAP II transcription initiation and NER complexes in cells not exposed to base damage by environmental agents such as UV radiation (Fig. 8). The further observation that inhibition of RNAP II transcription during active NER requires the product of the RAD26 gene, the yeast homolog of the human Cockayne syndrome group B gene (CSB), is provocative. Both the human CSB gene and the yeast RAD26 gene have been implicated in the preferential repair of template relative to coding strands of transcriptionally active genes (40, 41, 43). These studies have prompted the hypothesis that the CSB and Rad26 proteins are required to directly couple the NER machinery to RNAP II elongation complexes stalled at sites of base damage, thereby facilitating NER of the transcribed strand (21). However, to date, the inability to demonstrate in vitro NER which is strictly dependent on active RNAP II transcription has frustrated attempts to provide direct biochemical support of this model. Indeed, the addition of purified CSB protein to sites of arrested RNAP II transcription has no detectable effect on NER in vitro, and it does not disrupt ternary complexes made up of damaged DNA, mRNA, and RNAP II (31, 32). Hence, the molecular basis of the phenomenon of strand-specific (transcription-coupled) NER remains elusive.
FIG. 8.
Model for RNAP II transcription inhibition in the presence of active NER. The top panel portrays an equilibrium steady state which determines the distribution of holo- and core TFIIH in RNAP II transcription initiation and NER complexes, respectively, in the absence of exogenous base damage to DNA. The lower panel shows that when cells are exposed to DNA-damaging agents, such as UV radiation, this steady state is shifted such that holo-TFIIH is mobilized out of RNAP II transcription initiation complexes and core TFIIH is appropriated for active NER. This process requires Rad26 protein.
Present limitations of our understanding of the role of the CSB and Rad26 proteins in transcription-coupled NER notwithstanding, recent studies support the notion that CSB protein is involved in some aspect(s) of RNAP II transcription. Reduced levels of RNAP II transcription have been observed in CS-B cells both in vivo and in cell-free systems (2, 6). Additionally, recombinant CSB protein has been shown to promote transcription elongation on undamaged transcription templates in vitro (33). Finally, a stable association of CSB protein with RNAP II in human cell extracts has been reported in two recent studies (39, 42). There is no direct evidence for an interaction between the human CSB or yeast Rad26 protein with TFIIH subunits in vitro. However, recent studies are consistent with such an association during strand-specific repair in yeast (40). The results of this study lead us to the suggestion that the requirement for the RAD26 gene for inhibition of RNAP II transcription in the presence of active NER may reflect a role of the RAD26 gene product in the assembly and/or disassembly of the basal transcription machinery. A conceptually similar role for the human CSB protein in the turnover of NER complexes has been previously proposed (44). Hence, while the human CSB and yeast Rad26 proteins are clearly not essential for RNAP II transcription, these proteins may nonetheless participate in transcription initiation and transcription elongation when cells are exposed to various DNA-damaging agents.
An alternative model for our experimental observations is that Rad26 protein is required for a process which promotes down regulation of holo-TFIIH activity until NER is partially or fully completed. Consistent with this model, it has been reported that the kinase activity associated with human TFIIH is reduced following exposure of cells to UV radiation (1). Such a regulatory process may not require disassembly of holo-TFIIH and recruitment of core TFIIH to NER complexes. However, addition of exogenous holo-TFIIH may overwhelm the RAD26-dependent regulatory process. Further evidence suggestive of regulation of TFIIH activity is derived from the study of a gene designated MMS19 (27), which is involved in NER in yeast. Disruption of MMS19 results in lethality at elevated temperatures (23). This temperature sensitivity appears to be the result of defective RNAP II transcription. Remarkably, defective transcription is not complemented by purified MMS19 protein but is complemented by purified TFIIH, suggesting that the MMS19 gene product might regulate TFIIH activity or function and hence both RNAP II transcription and NER (23).
ACKNOWLEDGMENTS
We thank our laboratory colleagues for valuable discussions and critical review of the manuscript.
This study was supported in part by United States Public Health Service grant CA-12428. W.J.F. is a Fellow of The Jane Coffin Childs Memorial Fund for Medical Research.
REFERENCES
- 1.Adamczewski J P, Rossignol M, Tassan J P, Nigg E A, Moncollin V, Egly J-M. MAT1, cdk7 and cyclin H form a kinase complex which is UV light-sensitive upon association with TFIIH. EMBO J. 1996;15:1877–1884. [PMC free article] [PubMed] [Google Scholar]
- 2.Balajee A S, May A, Dianov G L, Friedberg E C, Bohr V A. Reduced RNA polymerase II transcription in intact and permeabilized Cockayne syndrome group B cells. Proc Natl Acad Sci USA. 1997;94:4306–4311. doi: 10.1073/pnas.94.9.4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bardwell A J, Bardwell L, Tomkinson A E, Friedberg E C. Specific cleavage of model recombination and repair intermediates by the yeast RAD1-RAD10 DNA endonuclease. Science. 1994;265:2082–2085. doi: 10.1126/science.8091230. [DOI] [PubMed] [Google Scholar]
- 4.Bhatia P K, Verhage R A, Brouwer J, Friedberg E C. Molecular cloning and characterization of Saccharomyces cerevisiae RAD28, the yeast homolog of the human Cockayne syndrome A (CSA) gene. J Bacteriol. 1996;178:5977–5988. doi: 10.1128/jb.178.20.5977-5988.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bohr V A, Smith C A, Okumoto D S, Hanawalt P C. DNA repair in an active gene: removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient than in the genome overall. Cell. 1985;40:359–369. doi: 10.1016/0092-8674(85)90150-3. [DOI] [PubMed] [Google Scholar]
- 6.Dianov G L, Houle J-F, Iyer N, Bohr V A, Friedberg E C. Reduced RNA polymerase II transcription in extracts of Cockayne syndrome and xeroderma pigmentosum/Cockayne syndrome cells. Nucleic Acids Res. 1997;25:3636–3642. doi: 10.1093/nar/25.18.3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dvir A, Conaway R C, Conaway J W. A role for TFIIH in controlling the activity of early RNA polymerase II elongation complexes. Proc Natl Acad Sci USA. 1997;94:9006–9010. doi: 10.1073/pnas.94.17.9006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Evans E, Moggs J G, Hwang J R, Egly J-M, Wood R D. Mechanism of open complex and dual incision formation by human nucleotide excision repair factors. EMBO J. 1997;16:6559–6573. doi: 10.1093/emboj/16.21.6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feaver W J, Gileadi O, Kornberg R D. Purification and characterization of yeast RNA polymerase II transcription factor b. J Biol Chem. 1991;266:19000–19005. [PubMed] [Google Scholar]
- 10.Feaver W J, Svejstrup J Q, Bardwell L, Bardwell A J, Buratowski S, Gulyas K D, Donahue T F, Friedberg E C, Kornberg R D. Dual roles of a multiprotein complex from S. cerevisiae in transcription and DNA repair. Cell. 1993;75:1379–1387. doi: 10.1016/0092-8674(93)90624-y. [DOI] [PubMed] [Google Scholar]
- 11.Feaver W J, Svejstrup J Q, Henry N L, Kornberg R D. Relationship of CDK-activating kinase and RNA polymerase II CTD kinase TFIIH/TFIIK. Cell. 1994;79:1103–1109. doi: 10.1016/0092-8674(94)90040-x. [DOI] [PubMed] [Google Scholar]
- 12.Feaver W J, Henry N L, Bushnell D A, Sayre M H, Brickner J H, Gileadi O, Kornberg R D. Yeast TFIIE: cloning, expression and homology to vertebrate proteins. J Biol Chem. 1994;269:27549–27553. [PubMed] [Google Scholar]
- 13.Feaver W J, Henry N L, Wang Z, Wu X, Svejstrup J Q, Bushnell D A, Friedberg E C, Kornberg R D. Genes for Tfb2, Tfb3, and Tfb4 subunits of yeast transcription/repair factor IIH: homology to “CAK” and human IIH subunits. J Biol Chem. 1997;272:19319–19327. doi: 10.1074/jbc.272.31.19319. [DOI] [PubMed] [Google Scholar]
- 14.Friedberg E C, Walker G C, Siede W. DNA repair and mutagenesis. Washington, D.C: ASM Press; 1995. [Google Scholar]
- 15.Friedberg E C. Relationships between DNA repair and transcription. Annu Rev Biochem. 1996;65:15–42. doi: 10.1146/annurev.bi.65.070196.000311. [DOI] [PubMed] [Google Scholar]
- 16.Friedberg E C. Cockayne syndrome—a primary defect in DNA repair, transcription, both or neither? Bioessays. 1996;18:731–738. doi: 10.1002/bies.950180908. [DOI] [PubMed] [Google Scholar]
- 17.Gileadi O, Feaver W J, Kornberg R D. Cloning of a subunit of yeast RNA polymerase II transcription factor b and CTD kinase. Science. 1991;257:1389–1392. doi: 10.1126/science.1445600. [DOI] [PubMed] [Google Scholar]
- 18.Goodrich J A, Tjian R. Transcription factors IIE and IIH and ATP hydrolysis direct promoter clearance by RNA polymerase II. Cell. 1994;77:145–156. doi: 10.1016/0092-8674(94)90242-9. [DOI] [PubMed] [Google Scholar]
- 19.Guzder S N, Sung P, Bailly V, Prakash L, Prakash S. RAD25 is a DNA helicase required for DNA repair and RNA polymerase II transcription. Nature. 1994;369:578–581. doi: 10.1038/369578a0. [DOI] [PubMed] [Google Scholar]
- 20.Habraken Y, Sung P, Prakash L, Prakash S. Yeast excision repair gene RAD2 encodes a single-stranded DNA endonuclease. Nature. 1993;366:365–368. doi: 10.1038/366365a0. [DOI] [PubMed] [Google Scholar]
- 21.Hanawalt P C. Transcription-coupled repair and human disease. Science. 1994;260:1957–1958. doi: 10.1126/science.7801121. [DOI] [PubMed] [Google Scholar]
- 21a.Huang, W., W. J. Feaver, and E. C. Friedberg. Unpublished observations.
- 22.Kelleher R J, III, Flanagan P M, Chasman D I, Ponticelli A S, Struhl K, Kornberg R D. Yeast and human TFIIDs are interchangeable for the response to acidic transcriptional activators in vitro. Genes Dev. 1992;6:296–303. doi: 10.1101/gad.6.2.296. [DOI] [PubMed] [Google Scholar]
- 23.Lauder S, Bankmann M, Guzder S N, Sung P, Prakash L, Prakash S. Dual requirement for the yeast MMS19 gene in DNA repair and RNA polymerase II transcription. Mol Cell Biol. 1996;16:6783–6793. doi: 10.1128/mcb.16.12.6783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lue N F, Flanagan P M, Sugimoto K, Kornberg R D. Initiation by yeast RNA polymerase II at the adenoviral major late promoter in vitro. Science. 1989;246:661–664. doi: 10.1126/science.2510298. [DOI] [PubMed] [Google Scholar]
- 25.Matsunaga T, Park C H, Bessho T, Mu D, Sancar A. Replication protein-A confers structure-specific endonuclease activities to the XPF-ERCC1 and XPG subunits of human DNA repair excision nucleases. J Biol Chem. 1996;271:11047–11050. doi: 10.1074/jbc.271.19.11047. [DOI] [PubMed] [Google Scholar]
- 26.O’Donovan A, Scherly D, Clarkson S G, Wood R D. Isolation of active recombinant XPG protein, a human DNA repair endonuclease. J Biol Chem. 1994;269:15965–15968. [PubMed] [Google Scholar]
- 27.Prakash L, Prakash S. Isolation and characterization of MMS-sensitive mutants of Saccharomyces cerevisiae. Genetics. 1977;86:33–55. doi: 10.1093/genetics/86.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reagan M S, Friedberg E C. Recovery of RNA polymerase II synthesis following DNA damage in mutants of Saccharomyces cerevisiae defective in nucleotide excision repair. Nucleic Acids Res. 1997;25:4257–4263. doi: 10.1093/nar/25.21.4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28a.Rodriguez K, Talamantez J, Huang W, Reed S H, Wang Z, Chen L, Friedberg E C, Tomkinson A E. Affinity purification and partial characterization of the yeast nucleotide excision repairosome using histidine-tagged Rad14 protein. 1998. Submitted for publication. [DOI] [PubMed] [Google Scholar]
- 29.Sancar A. DNA excision repair. Annu Rev Biochem. 1996;65:43–81. doi: 10.1146/annurev.bi.65.070196.000355. [DOI] [PubMed] [Google Scholar]
- 30.Satoh M S, Hanawalt P C. TFIIH-mediated nucleotide excision repair and initiation of mRNA transcription in an optimized cell-free DNA repair and RNA transcription assay. Nucleic Acids Res. 1996;24:3576–3582. doi: 10.1093/nar/24.18.3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Selby C P, Drapkin R, Reinberg D, Sancar A. RNA polymerase II stalled at a thymine dimer: footprint and effect on excision repair. Nucleic Acids Res. 1997;25:787–793. doi: 10.1093/nar/25.4.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Selby C P, Sancar A. Human transcription-repair coupling factor CSB/ERCC 6 is a DNA-stimulated ATPase but is not a helicase and does not disrupt the ternary transcription complex of stalled RNA polymerase II. J Biol Chem. 1997;272:1885–1890. doi: 10.1074/jbc.272.3.1885. [DOI] [PubMed] [Google Scholar]
- 33.Selby C P, Sancar A. Cockayne syndrome group B protein enhances elongation by RNA polymerase II. Proc Natl Acad Sci USA. 1997;94:11205–11209. doi: 10.1073/pnas.94.21.11205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sijbers A M, de Laat W L, Ariza R R, Biggerstaff M, Wei Y F, Moggs J G, Carter K C, Shell B K, Evans E, de Jong M C, Rademakers S, de Rooij J, Jaspers N G J, Hoeijmakers J H J, Wood R D. Xeroderma pigmentosum group F caused by a defect in a structure-specific DNA repair endonuclease. Cell. 1996;86:811–822. doi: 10.1016/s0092-8674(00)80155-5. [DOI] [PubMed] [Google Scholar]
- 35.Sung P, Prakash L, Matson S W, Prakash S. RAD3 protein of Saccharomyces cerevisiae is a DNA helicase. Proc Natl Acad Sci USA. 1987;84:8951–8955. doi: 10.1073/pnas.84.24.8951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Svejstrup J Q, Feaver W J, Lapoint J, Kornberg R D. RNA polymerase transcription factor IIH from yeast. J Biol Chem. 1994;269:28044–28048. [PubMed] [Google Scholar]
- 37.Svejstrup J Q, Wang Z, Feaver W J, Wu X, Bushnell D A, Donahue T F, Friedberg E C, Kornberg R D. Different forms of TFIIH for transcription and DNA repair: holo-TFIIH and a nucleotide excision repairosome. Cell. 1995;80:21–28. doi: 10.1016/0092-8674(95)90447-6. [DOI] [PubMed] [Google Scholar]
- 38.Svejstrup J Q, Vichi P, Egly J-M. The multiple role of transcription/repair factor TFIIH. Trends Biochem Sci. 1996;249:346–350. [PubMed] [Google Scholar]
- 39.Tantin D, Kansal A, Carey M. Recruitment of the putative transcription/repair coupling factor CSB/ERCC6 to RNA polymerase II elongation complexes. Mol Cell Biol. 1997;17:6803–6814. doi: 10.1128/mcb.17.12.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tijsterman M, Verhage R A, van de Putte P, Tasseron-De Jong J G, Brouwer J. Transitions in the coupling of transcription and nucleotide excision repair within RNA polymerase II-transcribed genes of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1997;94:8027–8032. doi: 10.1073/pnas.94.15.8027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Troelstra C, van Gool A, de Wit J, Vermeulen W, Bootsma D, Hoeijmakers J H J. ERCC6, a member of a subfamily of putative helicase, is involved in Cockayne’s syndrome and preferential repair of active genes. Cell. 1992;71:939–953. doi: 10.1016/0092-8674(92)90390-x. [DOI] [PubMed] [Google Scholar]
- 42.van Gool A J, Citterio E, Rademakers S, van Os R, Vermeulen W, Constantinou A, Egly J-M, Bootsma D, Hoeijmakers J H J. The Cockayne syndrome B protein, involved in transcription coupled DNA repair, resides in an RNA polymerase II-containing complex. EMBO J. 1997;16:5955–5965. doi: 10.1093/emboj/16.19.5955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Gool A J, Verhage R, Swagemakers S M A, van der Putte P, Brouwer J, Troelstra C, Bootsma D, Hoeijmakers J H J. RAD26, the functional S. cerevisiae homolog of the Cockayne syndrome B gene ERCC6. EMBO J. 1994;13:5361–5369. doi: 10.1002/j.1460-2075.1994.tb06871.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Oosterwijk M F, Versteeg A, Filon R, van Zeeland A A, Mullenders L H F. The sensitivity of Cockayne’s syndrome cells to DNA-damaging agents is not due to defective transcription-coupled repair of active genes. Mol Cell Biol. 1996;16:4436–4444. doi: 10.1128/mcb.16.8.4436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Venema J, Mullenders L H F, Natarajan A T, van Zeeland A A, Mayne L V. The genetic defect in Cockayne syndrome is associated with a defect in repair of UV-induced DNA damage in transcriptionally active DNA. Proc Natl Acad Sci USA. 1990;87:4707–4711. doi: 10.1073/pnas.87.12.4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vichi P, Coin F, Renaud J-P, Vermeulen W, Hoeijmakers J H J, Moras D, Egly J-M. Cisplatin- and UV-damaged DNA lure the basal transcription factor TFIID/TBP. EMBO J. 1997;16:7444–7456. doi: 10.1093/emboj/16.24.7444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Z, Svejstrup J Q, Feaver W J, Wu X, Kornberg R D, Friedberg E C. Requirement for RNA polymerase II transcription factor b (TFIIH) during nucleotide excision repair in the yeast Saccharomyces cerevisiae. Nature. 1994;368:74–76. doi: 10.1038/368074a0. [DOI] [PubMed] [Google Scholar]
- 48.Wang Z, Buratowski S, Svejstrup J Q, Feaver W J, Wu X, Kornberg R D, Donahue T F, Friedberg E C. Yeast TFB1 and SSL1 genes, which encode subunits of transcription factor IIH (TFIIH), are required for nucleotide excision repair and RNA polymerase II transcription. Mol Cell Biol. 1995;15:2288–2293. doi: 10.1128/mcb.15.4.2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Z, Wu X, Friedberg E C. The detection and measurement of base and nucleotide excision repair in cell free extracts of the yeast Saccharomyces cerevisiae. Methods Companion Methods Enzymol. 1995;7:177–186. [Google Scholar]
- 50.Wang Z, Wu X, Friedberg E C. A yeast whole cell extract supports nucleotide excision repair and RNA polymerase II transcription in vitro. Mutat Res. 1996;364:33–41. doi: 10.1016/0921-8777(96)00019-5. [DOI] [PubMed] [Google Scholar]
- 51.Wood R D. DNA repair in eukaryotes. Annu Rev Biochem. 1996;65:135–167. doi: 10.1146/annurev.bi.65.070196.001031. [DOI] [PubMed] [Google Scholar]
- 52.Zawel L, Kumar K P, Reinberg D. Recycling of the general transcription factors during RNA polymerase II transcription. Genes Dev. 1995;9:1479–1490. doi: 10.1101/gad.9.12.1479. [DOI] [PubMed] [Google Scholar]