Abstract
GTPases cycle between active GTP bound and inactive GDP bound forms. Exchange of GDP for GTP is catalyzed by guanine nucleotide exchange factors (GEFs). GTPase activating proteins (GAPs) accelerate GTP hydrolysis, to promote the GDP bound form. We reported that the RacGEF, PIX-1, is required for assembly of integrin adhesion complexes (IAC) in striated muscle of Caenorhabditis elegans. In C. elegans, IACs are found at the muscle cell boundaries (MCBs), and bases of sarcomeric M-lines and dense bodies (Z-disks). Screening C. elegans mutants in proteins containing RhoGAP domains revealed that loss of function of rrc-1 results in loss of IAC components at MCBs, disorganization of M-lines and dense bodies, and reduced whole animal locomotion. RRC-1 localizes to MCBs, like PIX-1. The localization of RRC-1 at MCBs requires PIX-1, and the localization of PIX-1 requires RRC-1. Loss of function of CED-10 (Rac) shows lack of PIX-1 and RRC-1 at MCBs. RRC-1 exists in a complex with PIX-1. Transgenic rescue of rrc-1 was achieved with wild type RRC-1 but not RRC-1 with a missense mutation in a highly conserved residue of the RhoGAP domain. Our results are consistent with RRC-1 being a RhoGAP for the PIX pathway in muscle.
Integrin adhesion complexes (IAC) are required to attach muscle cells to their surrounding extracellular matrix.
Previous studies reported that the guanine nucleotide exchange factors PIX-1 is required for the formation or stability of IACs in muscle of Caenorhabditis elegans.
The authors identified the GTPase activating protein for this PIX pathway, called RRC-1. Deficiency of RRC-1 resulted in loss of IAC proteins. These results reveal that RRC-1 and PIX-1 are in a common pathway that regulates IAC assembly.
INTRODUCTION
Integrin adhesion complexes (IAC), also known as focal adhesions, consist of the transmembrane heterodimeric proteins α/β integrin as well as hundreds of other proteins that are associated both from the extracellular matrix (ECM) and especially intracellularly (Anthis and Campbell, 2011; Bachir et al., 2014; Sun et al., 2014; Horton et al., 2015). IACs are important for many cell types. The adhesion of cells to a matrix is crucial for both tissue formation and for cell migration. In stationary cells like muscle, these complexes are rather stable, but in motile cells they are dynamic, with new complexes assembled at the leading edge and older complexes disassembled at the trailing edge (Anthis and Campbell, 2011). When integrins are expressed on the cell surface they are in a compact or bent or inactive state, unable to bind to their ECM targets, but can become activated to bind via several triggers (e.g., chemokines, local increase in P(4,5)IP2) that lead to binding of the cytoplasmic tail of β-integrin to talin. Binding to talin results in integrin assuming a more open conformation, able to bind to extracellular targets (Tadokoro et al., 2003). Kindlin is also involved in integrin activation by clustering of talin-activated integrins, at least in platelets (Ye et al., 2013). Although we understand the steps involved in the formation of IACs, we do not know how the composition of an IAC is determined or regulated, and we do not know what determines where an IAC forms, how many IACs form, and what their spacing will be.
In striated muscle, which includes both skeletal and cardiac muscle, myofibrils at the periphery of the cell are attached to the cell membrane and ECM via “costameres” (Ervasti, 2003; Henderson et al., 2017), muscle-specific IACs. Costameres are involved in anchorage of the muscle cell to the ECM, and transmission of force. Caenorhabditis elegans is an outstanding genetic model organism in which to learn new principles about muscle (Gieseler et al., 2017). The major striated muscle is found in the body wall and is used for locomotion (Benian and Epstein, 2011). Similar to striated muscle in other animals, the thin filaments are attached to Z-disk like structures (dense bodies), and the thick filaments are attached to M-lines. The sarcomeres are restricted to a narrow ∼1.5 μm zone adjacent to the cell membrane along the outer side of the muscle cell, and all the dense bodies and M-lines are anchored to the muscle cell membrane and ECM. The base of dense bodies and M-lines contain IACs and much is known about their protein composition (Gieseler et al., 2017). Additional IACs are located at the muscle cell boundaries (MCBs), where they form attachment plaques that anchor the muscle cell to a thin layer of ECM that lies between adjacent muscle cells (Qadota et al., 2017). Thus, in C. elegans muscle IACs are located at M-lines, dense bodies and MCBs, and although the base of each consists of integrins and a set of core proteins, they also contain proteins specific for each site (Gieseler et al., 2017). IACs at MCBs consist of a subset of proteins that are found at dense bodies (Qadota et al., 2017).
Recently, we reported that a protein in C. elegans, PIX-1 (orthologous to β-PIX in mammals), is required for the assembly or stability of IACs at MCBs (Moody et al., 2020). A PIX signaling pathway is important for the mammalian nervous (Schmalzigaug et al., 2009; Huang et al., 2011; Ramakers et al., 2012) and immune systems (Volinsky et al., 2006; Missy et al., 2008), and for the control of distal tip cell shape and migration (important for formation of the germline; Lucanic and Cheng, 2008), and for epithelial morphogenesis (Zhang et al., 2011) in C. elegans. However, no prior study had demonstrated a function for PIX in striated muscle in any organism. PIX proteins contain an SH3 domain, and a Rho GEF domain that activates the small GTPases Rac and Cdc42. In C. elegans, PIX-1 is localized to the IACs present at the muscle boundaries and the IACs at M-lines and dense bodies. As compared with wild type, a pix-1 null mutant shows reduced levels of activated, GTP-bound Rac in muscle. Interestingly, either deficiency or overexpression of PIX-1 results in disrupted MCBs and decreased nematode muscle function, suggesting that the level of PIX-1 needs to be tightly controlled. The Rho GEF domain of PIX proteins promote the exchange of GDP for GTP, thus converting inactive to active Rac or Cdc42. In the PIX pathway, the active GTP bound Rac or Cdc42 binds to and activates a PAK family protein kinase, which then phosphorylates one or more unknown substrates to somehow promote assembly of IACs.
Rho family GTPases (Rho, Rac, Cdc42) cycle between active (GTP-bound), and inactive (GDP-bound) states. Activation occurs via guanine-nucleotide exchange factors (GEFs; e.g., PIX) that promote exchange of GDP with GTP, and inactivation occurs via GTPase-activating proteins (GAPs), which promote the hydrolysis of GTP to GDP. Perhaps because of the cycling requirement, the terminal phenotypes of loss of function for a GEF and loss of function for a GAP (for a particular GTPase and cellular function), are often the same. For example, in yeast, loss of function of the GTPase Bud1p (homologous to mammalian Rap GTPases) has a similar phenotype to loss of function of its GEF, Bud5p, and its GAP, Bud2p (Michelitch and Chant, 1996). In C. elegans, the same embryonic cytokinesis defect is found for loss of function for rho-1 (RhoA), rga-3 (RhoGAP), rga-4 (RhoGAP), and ect-2 (RhoGEF; Jantsch-Plunger et al., 2000; Morita et al., 2005; Schonegg et al., 2007; Canman et al., 2008). For the PIX pathway in C. elegans, the GEF is PIX-1, and the GAP is unknown. In fact, a GAP for the PIX pathway has not been reported for any organism. We hypothesized that for the PIX pathway in nematode muscle, the loss of function for the GEF, PIX-1, and an unknown GAP, would be the same. Using an easily scorable phenotype (i.e., loss of IAC components at the MCB), we screened mutants in genes predicted to encode RhoGAP proteins, and identified one protein, RRC-1, which is required for assembly or stability of IACs at MCBs. RRC-1 contains an SH3 domain and a RhoGAP domain. We found that RRC-1 is localized to the IACs of MCBs, possibly to the muscle cell membrane. Loss of function mutants of rrc-1 show reduced accumulation of multiple IAC components at the MCB, mild disorganization of the M-lines and dense bodies, and sarcomeres, and reduced whole animal locomotion. The localization at MCBs of RRC-1 depends on pix-1, and the localization of PIX-1 depends on rrc-1. Loss of function for CED-10 (Rac) results in a lack of both PIX-1 and RRC-1 at the MCB. RRC-1 exists in a complex with PIX-1. Transgenic rescue of the rrc-1 muscle phenotype requires a putatively active RhoGAP domain. Altogether, our results suggest that RRC-1 is a RhoGAP for the PIX-1 pathway in muscle.
RESULTS
Either decreased or increased activity of PAK-1 results in disorganization of IACs at MCBs
The output of the PIX signaling pathway in mammals and nematodes is that p21-activated kinases (PAKs), which are serine/threonine protein kinases, become activated by binding to GTP-bound Rac or Cdc42. Because activation of PAK requires that a GEF first activates Rac or Cdc42, and inactivation occurs by a GAP, we hypothesized that a PAK-1 kinase dead and a PAK-1 kinase constitutively active mutant might have the same phenotype. We have an easily scorable muscle phenotype for the status of the PIX-1 pathway, that is, deficiency of any component results in disorganization of IAC at the muscle cell boundary (MCB), including a deficiency of pak-1 (Moody et al., 2020). Figure 1 is a drawing showing the locations in body wall muscle of various IAC and sarcomere components examined in this study. We used CRISPR/Cas9 to generate mutant worms carrying either a catalytically dead or a constitutively activating mutation for pak-1. For the catalytically dead mutant, we replaced K324 with A. In nearly all protein kinases, a K at this position is found in the small lobe of the kinase domain and coordinates ATP and helps transfer γ-phosphate. Mutation of this K to A or several other amino acids inactivates most known protein kinases (Iyer et al., 2005).
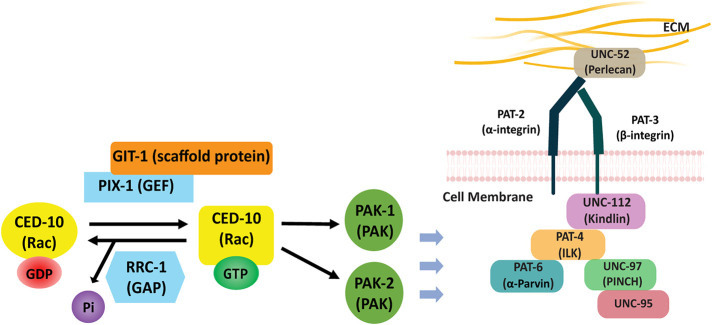
FIGURE 1:
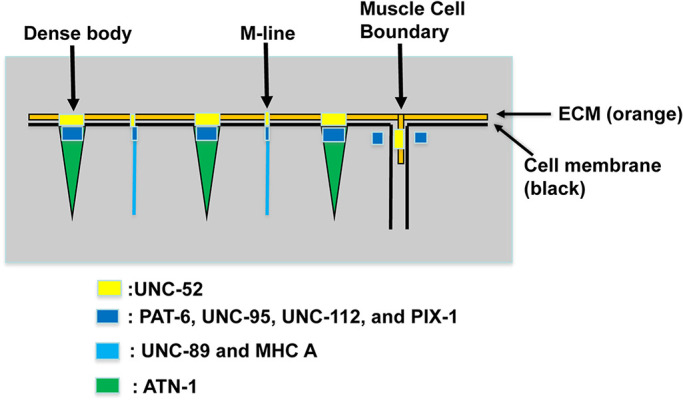
IACs in C. elegans body wall muscle and localization of proteins examined in this study. IACs are located at the bases of M-lines and dense bodies, and at the MCBs. The M-lines and dense bodies are attached to the muscle cell membrane and ECM over which lies a thin epidermal cell and thick acellular cuticle. The MCB consist of IACs at each apposing muscle cell with a thin layer of ECM in between. UNC-52 (perlecan) is in the ECM, PAT-6 (α-parvin), UNC-95, UNC-112 (kindlin), and PIX-1 (β-PIX) are located at the cytoplasmic side of the muscle cell membrane, UNC-89 (obscurin) and MHC A (myosin) are located at the M-line, and ATN-1 (α-actinin) is located at the major deeper portion of the dense body.
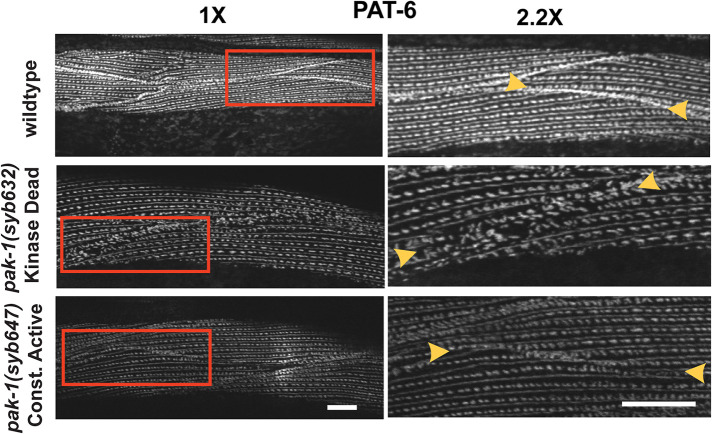
In the absence of GTP-Rac or GTP-Cdc42, PAKs exist in a closed conformation due to binding of an N-terminal (67-150) autoinhibitory domain (AID) with the more C-terminal kinase catalytic domain (Zenke et al., 1999). Binding of GTP-Rac or Cdc42 to this AID leads to an opening of the PAK structure and activation of its phosphotransferase activity. A constitutively-active human PAK has been generated by substituting the highly conserved L107 to F in this AID (Brown et al., 1996). In the nematode protein, the homologous residue is L99, and this was mutated to F as described under Materials and Methods. As shown in Figure 2, the catalytically dead mutant, pak-1(syb632) which has a K324A mutation, and the constitutively active mutant, pak-1(syb647) which has a L99F mutation, each show abnormal MCBs. Each mutant shows less accumulation of PAT-6 compared with wild type, with penetrance, or our ability to distinguish an abnormal from normal phenotype at 80–90% (Supplemental Table 2). However, the kinase dead mutant shows, in addition, some disorganization of the MCB that is not found in the constitutively active mutant. These results demonstrate that either decreased or increased activity of the PIX-1 pathway can have the same terminal phenotype. Thus, we hypothesized that increased activity of the PIX-1 pathway by inactivation of a RhoGAP would have the same phenotype as decreased activity of the PIX-1 pathway by inactivation of its Rho GEF, PIX-1.
FIGURE 2:
Either catalytically dead PAK-1 or constitutively active PAK-1 kinase results in an MCB defect. Confocal microcopy imaging of wildtype, pak-1(syb632)[K324A] kinase dead, and pak-1(syb647) [L99F] constitutively active kinase mutants immunostained with anti-PAT-6 (α-parvin), a core component of IAC. The right side of the figure shows 2.2X enlargements of regions denoted by the red boxes on the left side. Yellow arrowheads indicate MCBs. In wild type, PAT-6 is concentrated at the MCB and the structure appears “tight” whereas in the kinase-dead mutant PAT-6 appears loose or irregular at the MCB, and in the constitutively-active mutant, PAT-6 shows less accumulation at the MCB than in wild type. Scale bar, 10 μm.
Screening for a RhoGAP for the PIX pathway
A homology search revealed that there are 32 genes in C. elegans that encode proteins harboring RhoGAP domains. Of these 32, 18 of them are expressed in body wall muscle based on SAGE data (Meissner et al., 2009). We obtained mutants for all 18 genes from the Caenorhabditis Genetics Center (see Materials and Methods) and screened them for the MCB defect using anti-PAT-6 (α-parvin) immunostaining. Mutants for two genes, hum-7 and rrc-1 (Supplemental Figure 1; Supplemental Table 1) each demonstrated defects of the MCB. hum-7 encodes an 1880 residue protein containing an RA domain, a class IX myosin motor domain, 2 IQ domains, and a RhoGAP domain, and will be described elsewhere. The rrc-1 gene encodes an ∼760 residue protein that contains a RhoGAP domain and an SH3 domain (Figure 3a). Because RRC-1 is a simpler protein, we decided to focus our efforts on RRC-1. Alternative splicing of rrc-1 produces three protein isoforms containing both domains and of approximately the same size (742-759 aa; Figure 3b). A Rho GTPase effector pull-down assay of nematode RRC-1 expressed in mammalian tissue culture cells demonstrates that RRC-1 has GAP activity towards mammalian Rac and Cdc42 but not RhoA (Delawary et al., 2007).
FIGURE 3:
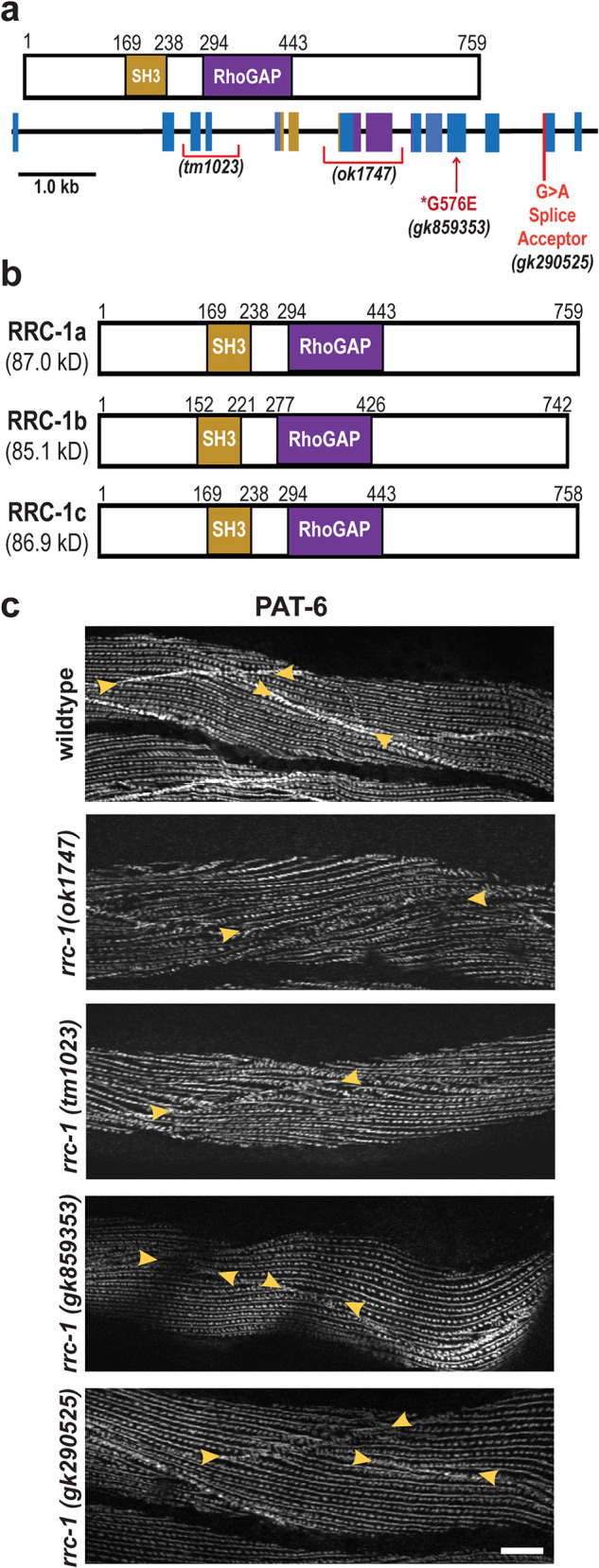
Loss of function rrc-1 mutants show a lack of or disorganization of PAT-6 at MCBs. (a) Schematic representation of domains in C. elegans RRC-1 isoform a, and the location and nature of the four rrc-1 mutants within a map of the exon-intron organization of the rrc-1 gene. (b) Schematic showing domain organization of the three predicted RRC-1 isoforms, generated by alternative splicing. (c) Confocal microscopy imaging of body wall muscle cells immunostained with antibodies to PAT-6 (α-parvin) from wildtype, two RRC-1 out-of-frame deletion allele mutants (rrc-1(ok1747) and rrc-1(tm1023)), one missense mutation, rrc-1(gk859353), and one splice site mutation, rrc-1(gk290525), each outcrossed 5x to wildtype. Arrowheads point to the boundaries between muscle cells. Note that there is also disorganization of PAT-6 localization at M-lines and dense bodies in the deletion alleles, ok1747 and tm1023. Scale bars, 10 µm.
RRC-1 is orthologous to human ARHGAP32 and ARHGAP33 proteins
Wormbase considers RRC-1 to be orthologous to three human proteins, ARHGAP31, 32, and 33, and the website notes that the best BLASTP match is to ARHGAP32. We obtained these human sequences and performed a PFAM prediction of domains and aligned and compared each of the domains to those in RRC-1a. As shown in Supplemental Figure 2, all four proteins contain a RhoGAP domain. Human ARHGAP31 only contains a RhoGAP domain, whereas RRC-1a and human ARHGAP32 and 33 also contain SH3 domains. Furthermore, human ARHGAP32 and 33 have PX domains, and ARHGAP32 has an additional RPEL repeat domain. Based on the number of shared domains and their percent identities, we consider the closest human orthologs to RRC-1 to be ARHGAP32 and ARHGAP33, and probably ARHGAP33 is closer. Inspection of The Human Protein Atlas shows that all three human proteins are expressed in multiple organs, but ARHGAP31 is not expressed in skeletal or heart muscle, whereas ARHGAP32 and 33 show “medium levels” of expression in skeletal and heart muscle. These expression patterns are consistent with ARHGAP32 and 33 being closer human orthologues for RRC-1.
rrc-1 mutants display mis-localized or missing IAC components at MCBs, and disorganized M-lines and dense bodies
The original allele that we characterized, rrc-1(ok1747), is a frame-shifting deletion that removes exons 7 and 8 (Figure 3a). We obtained 3 more rrc-1 mutant alleles from the Caenorhabditis Genetics Center, one being tm1023, which is also a frame-shifting deletion removing exons 3 and 4. Inspection of the Million Mutation Project collection (Thompson et al., 2013) revealed 16 rrc-1 mutants, one being a splicing defect, and 15 being missense mutations. We ordered the splicing defect mutant, and the four missense mutants that have nonconservative amino acid changes. Unfortunately, two of these strains were too difficult to grow, and one had background mutations in pak-1 and in unc-89, which would confound our analysis, and thus were not pursued. Therefore, we had a collection of four rrc-1 mutant alleles, including two deletions, one missense mutant, and one splicing acceptor mutant (Figure 3a). We outcrossed each mutant to wild type five times, to remove most of the background mutations. Figure 3c shows results from immunostaining of the four rrc-1 alleles with antibodies to PAT-6 (α-parvin) to visualize IACs. All four alleles show defects at the MCBs with penetrance ranging from 60–90% (Supplemental Table 2), but the deletion alleles, ok1747 and tm1023, are more severely affected, showing not only weak concentration of PAT-6 at the boundaries but also what appear to be gaps between adjacent cells, and the highest penetrance (90%).
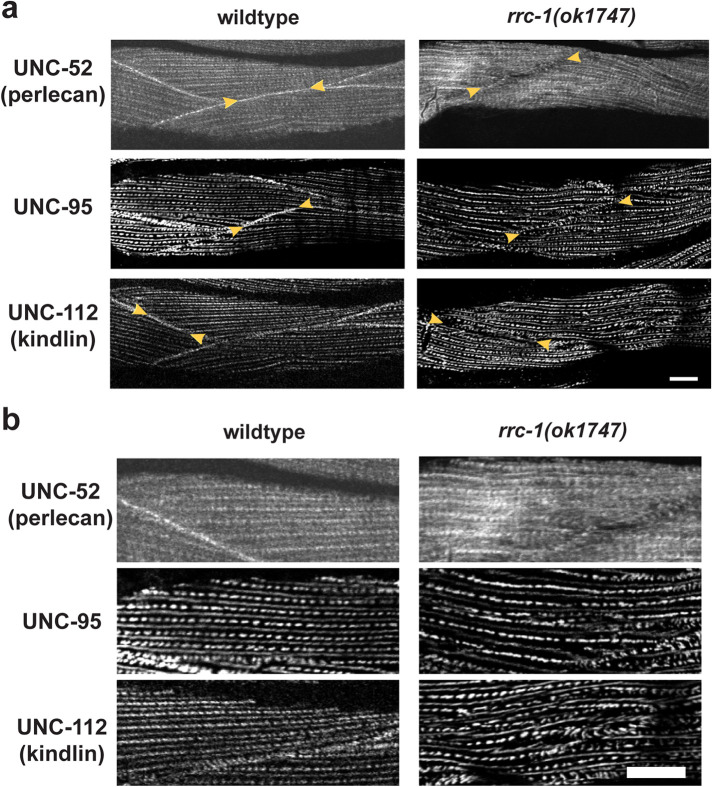
By immunostaining, we found that in the deletion mutant, rrc-1(ok1747), other IAC components are missing or mislocalized at MCBs (Figure 4), with 80–100% penetrance (Supplemental Table 2). These IAC components include UNC-52 (perlecan) in the ECM, UNC-95, and UNC-112 (kindlin; Figure 1). We conclude that the RhoGAP RRC-1 is required for the assembly or stability of IACs at the MCB.
FIGURE 4:
Mis-localization of multiple IAC components at the MCBs, M-lines and dense bodies in rrc-1 mutants. (a and b) Comparison of wild type versus rrc-1(ok1747), outcrossed 5x to wildtype, immunostained with antibodies to the indicated integrin adhesion complex (IAC) proteins and imaged by confocal microscopy. b) shows a portion of what is shown in a) enlarged 2X to emphasize the observed lack of regularity in the organization of rows of M-lines and dense bodies. In parentheses are the names of the mammalian orthologues. Arrowheads denote MCBs. Note that all three proteins are present at the MCBs in wildtype but are missing or less tightly organized at the MCBs in the rrc-1 deletion mutant. In addition, there is mislocalization of each of these IAC components at M-lines and dense bodies. Each image is a representative image obtained from at least two fixation and immunostaining experiments, and imaging of at least three different animals. Scale bars, 10 µm.
In addition, the M-lines and dense bodies are disorganized in rrc-1(ok1747) and rrc-1(tm1023), by immunostaining for PAT-6 (Figure 3c), and in rrc-1(ok1747) upon immunostaining for UNC-52, UNC-95, and UNC-112 (Figure 4). The dense bodies are not as regularly punctate, sometimes being elongated, and the distance between parallel rows of dense bodies and M-lines is irregular. Thus, RRC-1 is also required for the assembly or stability of IACs at M-lines and dense bodies.
To rule out that the defects that we observed in rrc-1 mutants by immunostaining are artefacts resulting from incomplete fixation, we localized UNC-112-GFP in live animals using a transgenic strain. As shown in Supplemental Figure 3, UNC-112-GFP is localized to the MCBs in wild type but missing at the MCBs in rrc-1(ok1747). Moreover, UNC-112-GFP is also missing at MCBs in pix-1(gk299374) (Supplemental Figure 3), consistent with our finding a defect in the MCB by immunostaining with anti-UNC-112 (Moody et al., 2020).
rrc-1 mutants also display disorganization of sarcomeres
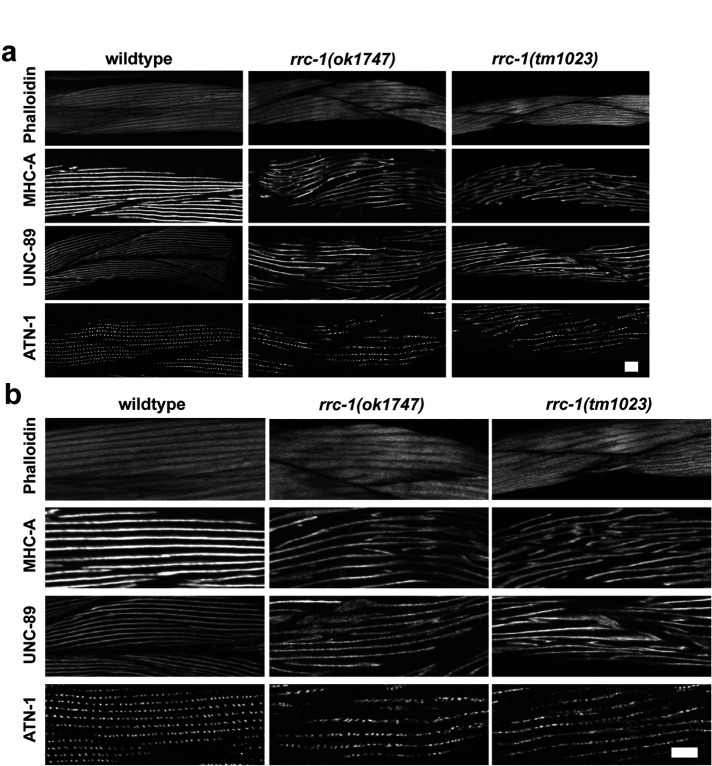
Because in rrc-1(ok1747), we found disorganization in M-lines, which normally crosslink thick filaments, and dense bodies, which normally attach to thin filaments, we examined the organization of the sarcomere further. We used phalloidin staining to determine the organization of thin filaments, and antibodies to myosin MHC A for the organization of thick filaments, to UNC-89 for the organization of M-lines (full depth), and to ATN-1 (α-actinin) for the organization of the main portion of dense bodies (Figure 1). As shown in Figure 5, all sarcomere structures are abnormal in rrc-1(ok1747) and rrc-1(tm1023), as compared with wild type, with 90–100% penetrance (Supplemental Table 2). The greatest degree of disorganization is observed in UNC-89 (M-lines), and MHC-A, which is the myosin isoform located in the middle of thick filaments where they are cross linked at the M-line. Less disorganization is observed in thin filaments (phalloidin), or with the main portions of dense bodies (ATN-1). Because similar sarcomere defects were observed in two independently generated rrc-1 mutants that were extensively outcrossed, these defects can be attributed to loss of function of rrc-1 rather than any closely linked background mutations.
FIGURE 5:
rrc-1 mutants have disorganized sarcomeres. (a and b) Confocal images of wild type, rrc-1(ok1747) and rrc-1(tm1023) reacted with phalloidin (thin filaments), and antibodies to sarcomere proteins MHC A (thick filaments), UNC-89 (M-lines), and ATN-1 (dense bodies). b) shows 2X enlargements of portions of images shown in a) to show the defects more clearly. Note the disorganization of each sarcomere component compared with wild type. Most severely affected are the thick filaments or A-bands, and the M-lines. Each image is a representative image obtained from at least two immunostaining experiments. Scale bar, 10 μm.
rrc-1 mutants are defective in whole animal locomotion
In C. elegans, the force of body wall muscle contraction that bends the worm and thus permits locomotion of the animal, is generated by the sarcomeres and transmitted through all three IAC sites, including the M-lines, the dense bodies and the adhesion plaques at the MCBs. We previously reported that pix-1 mutants have reduced whole animal locomotion, and this is likely attributable to them having poorly organized adhesion plaques at MCBs, because they have normally organized sarcomeres, M-lines and dense bodies (Moody et al., 2020). However, rrc-1 mutants show more extensive defects–in addition to the MCBs, the M-lines, dense bodies and sarcomeres are disorganized. Thus, we conducted worm motility assays. As shown in Figure 6a, both deletion alleles, ok1747 and tm1023, and the splicing mutant, gk290525, display reduced swimming when compared with wild type. However, the missense mutant, gk859353, displays swimming that is not significantly different from wild type. Although gk859353 is a nonconservative G to E change, it resides outside of a recognizable domain and for this reason or other reasons this G may not have a critical function. Crawling may be a more stringent test of worm locomotion because it is likely that the worm needs to overcome the surface tension lying between itself and the agar surface, and this would require more force than necessary for swimming. Thus, as shown in Figure 6b, all four rrc-1 mutants exhibit reduced crawling as compared with wild type. For all four alleles, the trends in both swimming and crawling are similar, with the deletion alleles and splicing mutant showing slower movement than the missense mutant. That the missense mutant, gk859353, had no defect in swimming and the least defect in crawling is consistent with it having the weakest effect on the IACs (Figure 3) – the weakest effect on MCBs with the lowest penetrance (60%; Supplemental Table 2), and no disorganization of M-lines and dense bodies.
FIGURE 6:
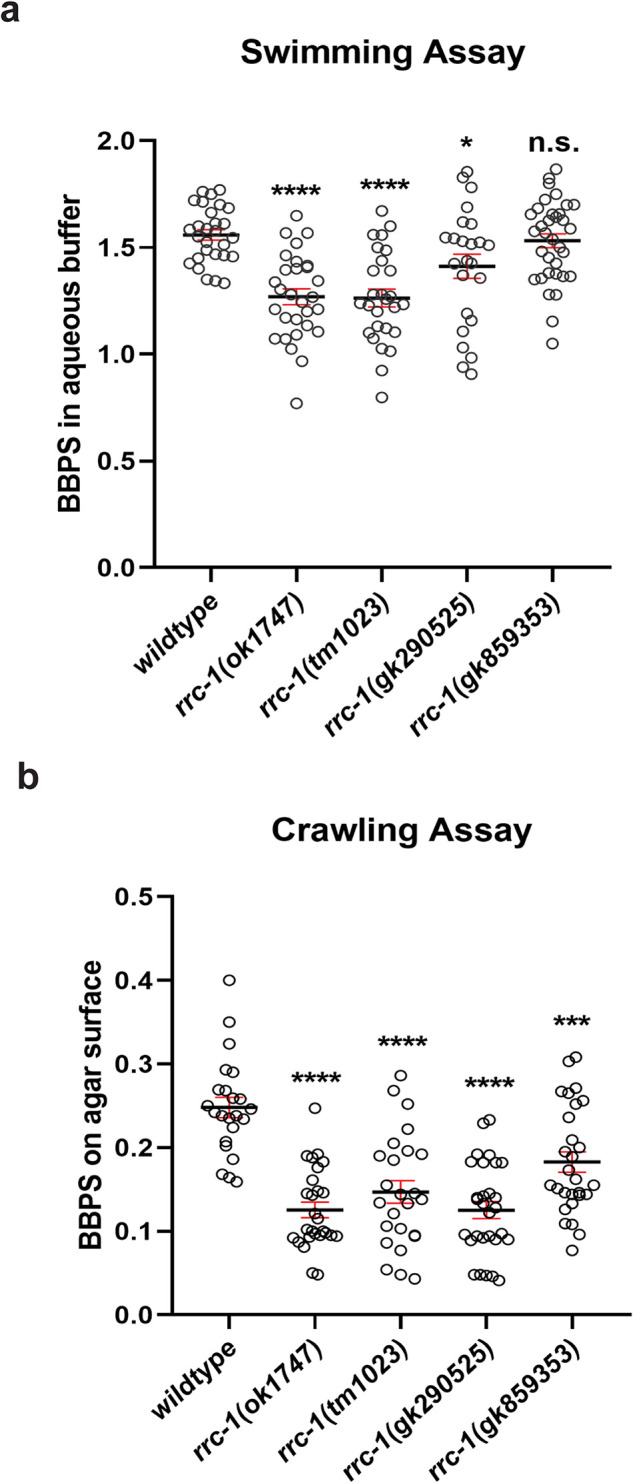
Loss of function rrc-1 mutants have reduced whole animal locomotion. a) Swimming and b) crawling assays show that RRC-1 deletion mutants rrc-1(ok1747) and rrc-1(tm1023), a splice acceptor site mutant, rrc-1(gk290525), and a missense mutant, rrc-1(gk859353), outcrossed 5x to wildtype result in reduced locomotion as compared with wildtype animals. Body bends per second (BBPS) are quantified for individual animals of each strain. In the graphs, each open circle represents the result from an independently selected animal. The exact n values vary, but n ≥ 23. Welch’s t test was used to test for significance. Error bars indicate SEM, *p ≤ 0.05, ****p ≤ 0.0001.
The localization of RRC-1 protein in muscle
We first attempted to make antibodies to RRC-1. Unfortunately, using two different immunogens, we failed to generate specific antibodies in rabbits. Therefore, to localize RRC-1 in muscle, we used CRISPR/Cas9 to create a worm strain, rrc-1(syb4499), in which the endogenous rrc-1 gene expresses RRC-1 with an HA tag fused to its C-terminus. As shown in Figure 7a, by western blot using anti-HA antibodies, we detect an RRC-1-HA fusion of expected size (∼95 kDa) from this strain but not from wild type. To test whether the HA tag might interfere with the normal function of RRC-1, we conducted locomotion assays and immunostaining of sarcomeres. As shown in Supplemental Figure 4, we observed no difference in swimming or crawling motility assays between wild type and the RRC-1-HA strain. In addition, as shown in Supplemental Figure 5, the sarcomere organization of the RRC-1-HA strain is also normal. Next, we used HA antibodies to perform immunostaining to localize RRC-1-HA in muscle. As demonstrated in Figure 7b, these antibodies localize RRC-1-HA to the MCB, colocalizing with antibodies to PAT-6 (α-parvin). We also observed weak localization of RRC-1-HA to the same focal plane as the base of the M-lines and dense bodies where PAT-6 is localized, in a generally striated pattern, but in a more diffuse, less organized manner. This weak staining is not background staining, as the same dilution of anti-HA antibodies and the same gain on the confocal microscope detected no fluorescence in wild type animals (Figure 7b, left column). The pattern of RRC-1-HA is somewhat like the pattern of UNC-52 (perlecan) immunostaining (Qadota et al., 2017). These results show that RRC-1 is localized to MCBs, and to the bases of M-lines and dense bodies, or to the muscle cell membrane where the bases of these structures are anchored, which is consistent with RRC-1 playing a role in the formation or stability of these structures and the structure of the sarcomere.
FIGURE 7:
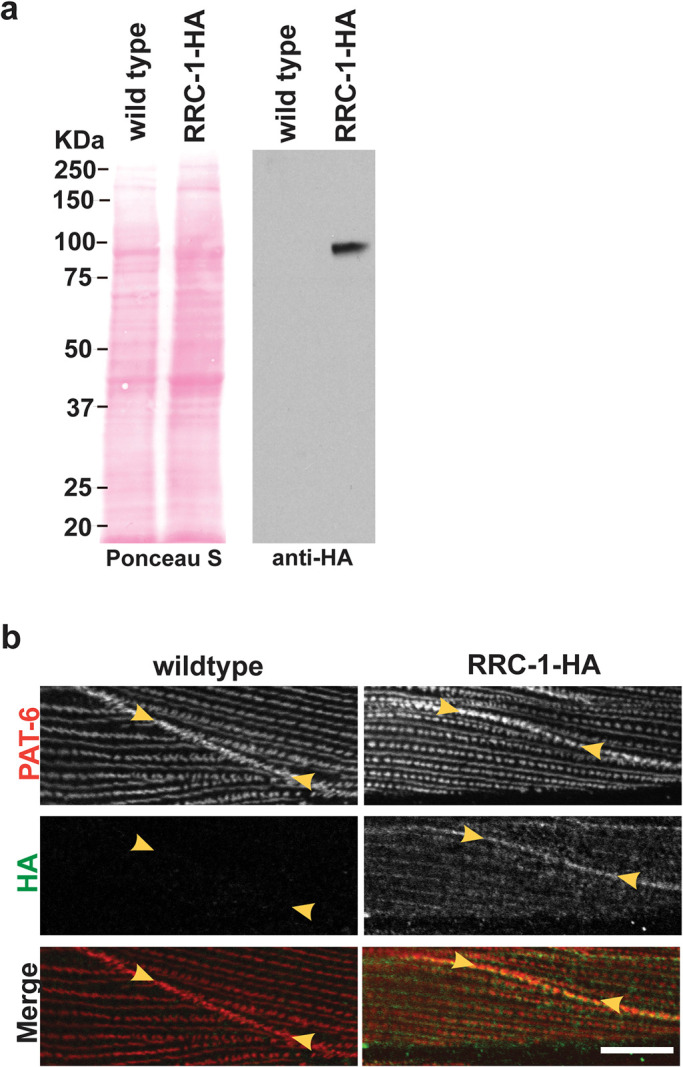
HA-tagged RRC-1 localizes to MCBs. a) Confirmation that the CRISPR/Cas9-generated strain, rrc-1(syb4499) expresses RRC-1-HA. Lysates were prepared from wildtype and rrc-1(syb4499), and portions separated by SDS–PAGE, blotted, and reacted with antibodies to HA. Anti-HA detects a protein of expected size, ∼90 kDa, from rrc-1(syb4499) and not from wild type. b) Confocal microscopy imaging of body wall muscle costained with anti-PAT-6 (α-parvin) and anti-HA antibodies in wildtype and the CRISPR generated strain that expresses RRC-1-HA. Note that RRC-1 localizes to the MCB, colocalizing with PAT-6. There is weaker localization of RRC-1-HA to the same focal plane at the bases of M-lines and dense bodies, in a striated pattern, but in a diffuse, less-organized manner. Scale bar, 10 μm.
The localization of RRC-1 depends on PIX-1, the localization of PIX-1 depends on RRC-1, and the localization of RRC-1 and PIX-1 depend on CED-10
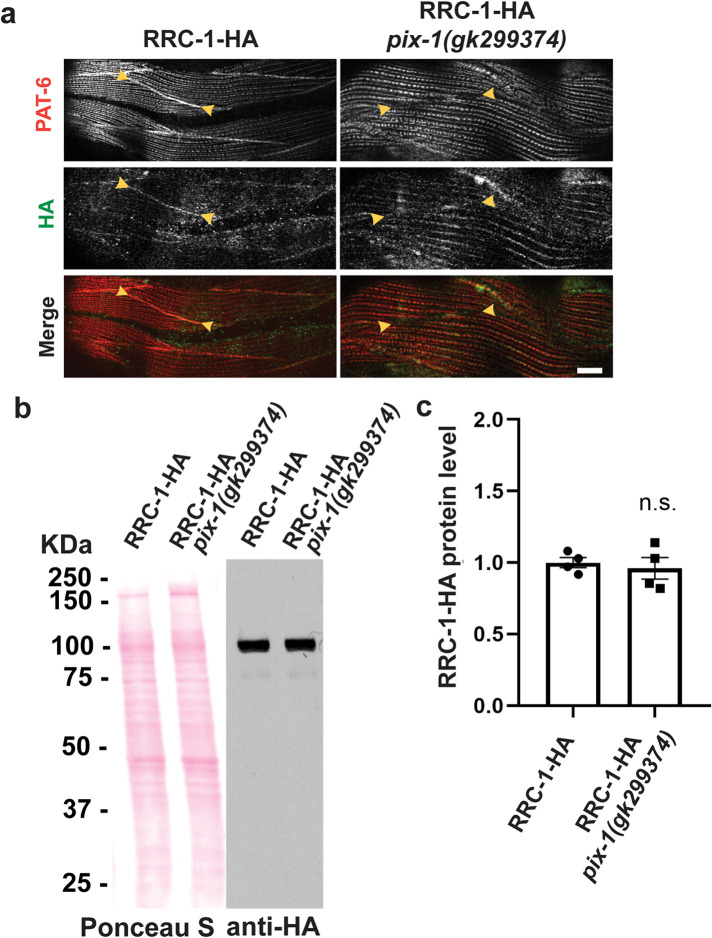
Given the similar localization of RRC-1 and PIX-1 to MCBs, and that mutants in either gene affect MCB organization, we sought to determine whether there are genetic interactions between these two genes. By genetic recombination, we created a strain that expresses the HA tagged RRC-1 in a pix-1 mutant background, which we designate, RRC-1-HA pix-1(gk299374). We compared the immunolocalization of PAT-6, and RRC-1-HA in a wild type versus the pix-1 mutant background. As shown in Figure 8a, PAT-6 is normally localized to the MCB in the strain expressing RRC-1-HA, but missing in the strain RRC-1-HA pix-1(gk299374), as was reported previously for this pix-1 mutant (Moody et al., 2020). When we immunostained with anti-HA, we found that RRC-1-HA is mostly missing from the MCB in RRC-1-HA pix-1(gk299374), with 90% penetrance (Supplemental Table 2). However, the total protein level of RRC-1-HA is not affected by pix-1 deficiency, as shown by a quantitative western blot (Figure 8, b and c). These data suggest PIX-1 is required for RRC-1 localization to the MCB.
FIGURE 8:
PIX-1 is required for the proper localization of RRC-1 but not the stability of RRC-1. a) Confocal microscopy imaging of body wall muscle costained with anti-PAT-6 (α-parvin) and anti-HA antibodies in the CRISPR generated strain that expresses RRC-1-HA, and in a strain that expresses RRC-1-HA in a pix-1 null background. Note that when PIX-1 is deficient there is less localization of RRC-1-HA to the MCB. Scale bar, 10 μm. b) Western blot showing that the level of RRC-1-HA is the same in wild type versus the pix-1 mutant. c) Quantification of HA-tagged RRC-1 protein levels in wild type versus pix-1 mutant shows no significant (n.s.) difference using a Welch’s t test for statistical analysis, N = 4.
We next asked whether rrc-1 deficiency could affect the localization of PIX-1. We compared the localization of PIX-1 in wild type versus rrc-1(ok1747) and rrc-1(tm1023). As shown in Figure 9a, there is much less accumulation of PIX-1 at MCBs in the rrc-1 mutants as compared with wild type, with ok1747 showing 60%, and tm1023 showing 80% penetrance (Supplemental Table 2). However, the total level of PIX-1 is not affected (Figure 9, b and c). Therefore, localization of PIX-1 to the MCB depends on RRC-1.
FIGURE 9:
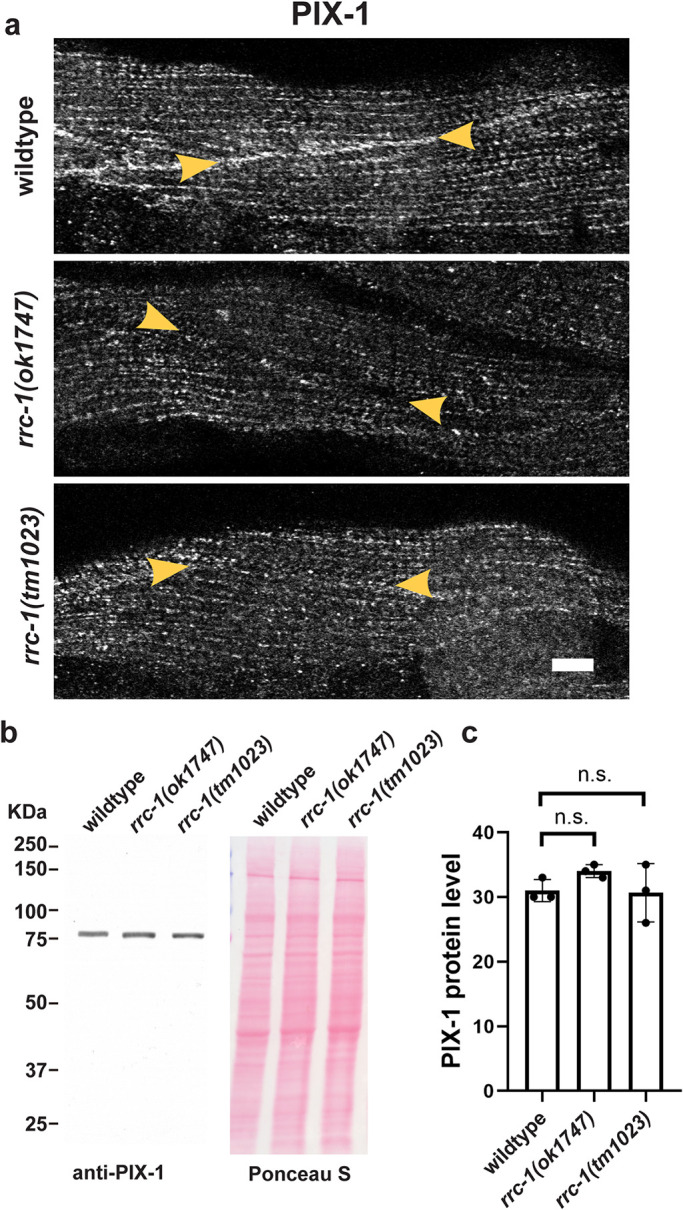
RRC-1 is required for the proper localization of PIX-1 but not the stability of PIX-1. a) Confocal microscopy imaging of body wall muscle stained with antibodies to PIX-1 in wild type and two deletion alleles of rrc-1. Note that when RRC-1 is deficient the localization of PIX-1 to the MCBs is nearly absent (indicated by yellow arrowheads). Scale bar, 10 μm. b) Western blot showing that the level of PIX-1 is the same in wild type versus the rrc-1 mutants. c) Quantification of PIX-1 protein levels in wild type versus either rrc-1 mutant showing no significant (n.s.) differences using a Welch’s t test for statistical analysis, N = 3.
The mutual requirement of PIX-1 and RRC-1 for their localization prompted us to create a rrc-1 pix-1 double mutant. Upon immunostaining with anti-PAT-6, we observed that the double mutant showed no enhancement of phenotype as compared with rrc-1 alone (Supplemental Figure 6). Therefore, rrc-1 and pix-1 belong to the same genetic pathway.
Next, we asked whether GIT-1 is required for the localization or stability of RRC-1. Previously, we reported that GIT-1, a scaffold for PIX-1, is required for the assembly or stability of IACs at the MCB, and that GIT-1 is required for the stability of PIX-1, by using a git-1 deletion allele, git-1(ok1848)(Moody et al., 2020). We now show that git-1 is also required for the localization of PIX-1 to the MCBs, as shown in Figure 10a. To examine the localization of RRC-1-HA in a git-1 mutant required a recombinant, but because the two genes are so close together on the X chromosome (<0.5 cM apart), we employed git-1 RNAi, instead. As shown in Figure 10b, knock down of git-1 results in a reduced level of PAT-6 at MCBs, as expected. However, git-1 RNAi results in RRC-1-HA still being localized at MCBs. Nevertheless, git-1 RNAi does result in a significant reduction in the level of RRC-1-HA by western blot (Figure 10, c and d). As indicated in Figure 10d, the level of RRC-1-HA is reduced by fourfold compared with empty vector.
FIGURE 10:
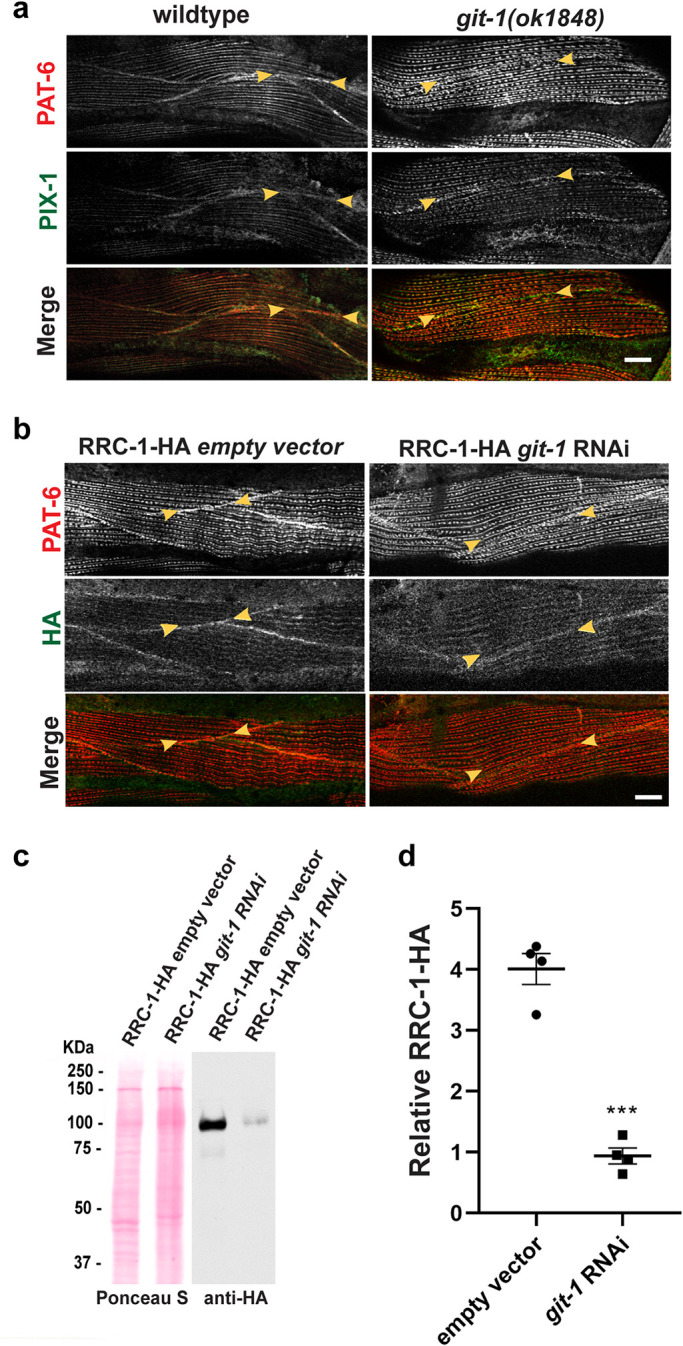
GIT-1 is not required for the localization of RRC-1, but GIT-1 is required for the stability of RRC-1. a) Confocal microscopy imaging of body wall muscle costained with anti-PAT-6 (α-parvin) and anti-PIX-1 antibodies in wildtype and in the git-1(ok1848) deletion mutant. Note that both PAT-6 and PIX-1 show reduced or absent accumulation at the MCBs. Scale bar, 10 μm. b) Confocal microscopy imaging of body wall muscle costained with anti-PAT-6 (α-parvin) and anti-HA to detect RRC-1-HA in a strain expressing RRC-1-HA with and without feeding bacteria expressing dsRNA for git-1. Note that RNAi knockdown of git-1 does not prevent the localization of RRC-1-HA to the MCBs. Scale bar, 10 μm. c) Western blot showing the level of RRC-1-HA in wild type versus git-1(RNAi). Note that git-1(RNAi) results in a reduced level of RRC-1-HA. d) Quantification of HA-tagged RRC-1 protein levels in git-1 (RNAi) versus empty vector control shows that git-1 (RNAi) reduces the level of RRC-1-HA to ∼23% of the level in wild type. A Welch’s t test for statistical analysis, N = 4, was used; Error bars indicate SEM, ***p ≤ 0.001.
Finally, we asked whether deficiency of ced-10 would result in mis-localization of RRC-1. Previously, we had reported that in a ced-10(n3246) mutant, the level of PIX-1 protein was the same as in wild type, and that ced-10(n3246) shows lack of localization of PAT-6 at the MCBs (Moody et al., 2020). As shown in Figure 11a, ced-10(n3246) also shows lack of localization of PIX-1 at the MCB, with 80% penetrance (Supplemental Table 2). In addition, ced-10(n3246) shows lack of localization of RRC-1-HA at MCBs (Figure 11b) with 50% penetrance (Supplemental Table 2), but no change in the level of RRC-1-HA protein (Figure 11, c and d). The lower penetrance of ced-10 effects is likely due to the partial loss of function of the ced-10(n3246) missense allele. An existing stronger deletion allele, ced-10(n3417) could not be used conveniently in our analysis because it is a maternal effect embryonic lethal (Lundquist et al., 2001). Overall, our results indicate that PIX-1 is required for the proper localization of RRC-1 but not its stability, that RRC-1 is required for the localization of PIX-1 but not its stability, that GIT-1 is not required for the localization of RRC-1 but is required for its stability, and that CED-10 is required for the localization but not stability of RRC-1.
FIGURE 11:
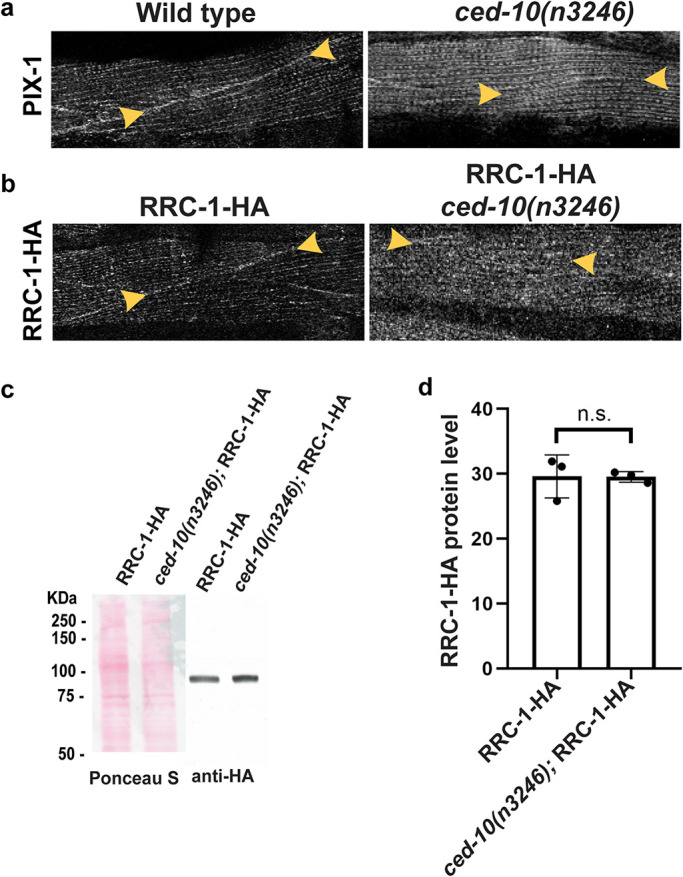
CED-10 (Rac) is required for the accumulation of PIX-1 and RRC-1 at MCBs. a) Confocal imaging of body wall muscle immunostained with anti-PIX-1 from wild type and ced-10(n3246), a loss of function missense mutation. Note the lack of accumulation of PIX-1 at the MCB in ced-10(n3246). b) Confocal imaging of body wall muscle immunostained with anti-HA to detect RRC-1-HA from wild type and ced-10(n3246). Note the lack of accumulation of RRC-1-HA at the MCB in ced-10(n3246). Scale bar, 10 μm. c) Western blot showing the level of RRC-1-HA in wild type versus ced-10(n3246). d) Quantification of HA-tagged RRC-1 protein levels in wild type versus ced-10(n3246) showing no significant (n.s.) difference using a Welch’s t test for statistical analysis, N = 3.
RRC-1 and PIX-1 form a complex
Given that the localization of RRC-1 depends on PIX-1, and that the level of RRC-1 depends on GIT-1, we asked whether RRC-1 might exist in a protein complex with PIX-1 and GIT-1. To address this question, we used CRISPR/Cas9 to add a mNeonGreen tag to the C-terminus of GIT-1 in the CRISPR/Cas 9 strain that already expresses RRC-1 with a C-terminal HA tag. The resulting strain, called PHX5908, expresses both RRC-1-HA and GIT-1-mNeonGreen. Pilot experiments showed that in order to extract RRC-1 into a soluble form suitable for immunoprecipitation required inclusion of an ionic detergent (Supplemental Figure 7). A total worm lysate of PHX5908 was made, and using antibodies to HA coupled to magnetic beads, we immunoprecipitated (IP) RRC-1-HA, running at ∼100 kDa (Figure 12). This IP was then examined by western blot for coIP of GIT-mNeoGreen using antibodies to mNeonGreen and PIX-1 using antibodies to PIX-1. Although we could not detect coIP of GIT-1-mNeonGreen, we could detect coIP of a small amount of PIX-1 (Figure 12). These results suggest that RRC-1 and PIX-1 exist in a complex.
FIGURE 12:
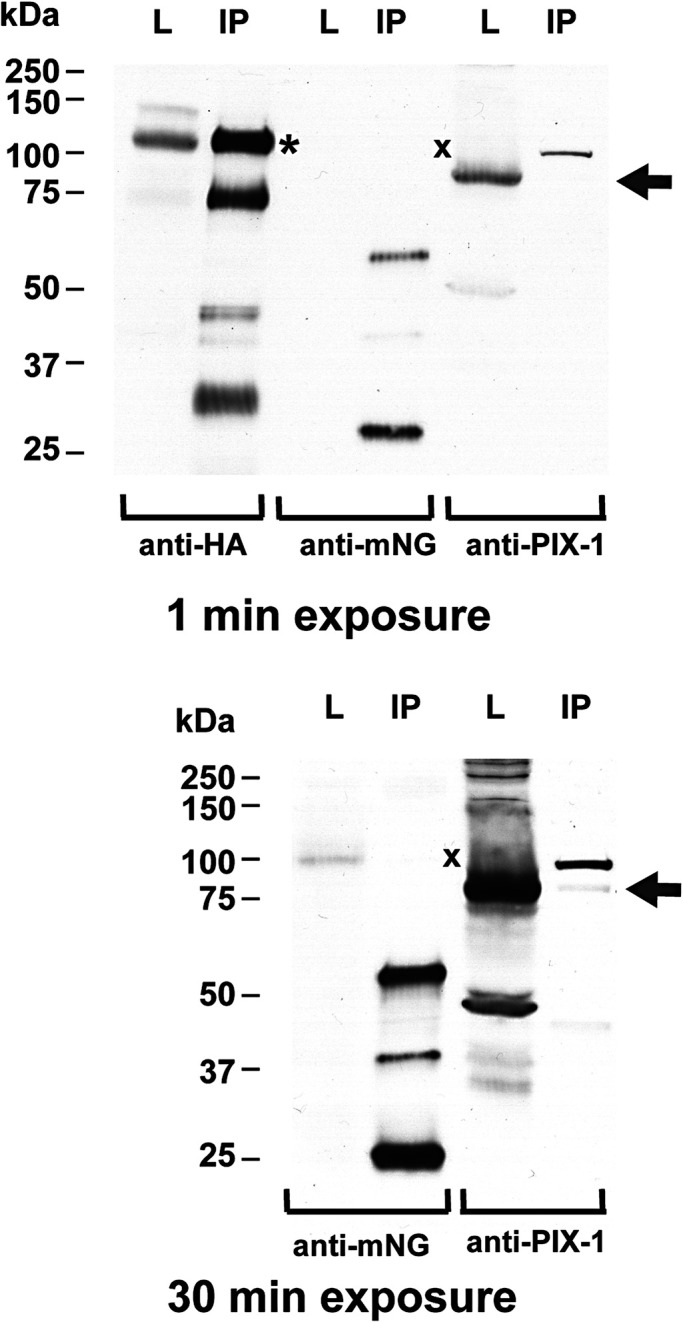
Coimmunoprecipitation (co-IP) of RRC-1 with PIX-1, but not GIT-1. RRC-1-HA was IP from a CRISPR-generated worm strain that expresses both RRC-1-HA and GIT-1-mNeonGreen. Portions of the total worm lysates (L) and IP were separated by SDS–PAGE, transferred to membrane, and reacted with the indicated antibodies. One minute and 30-min exposures are shown. As shown from left to right, the IP procedure worked well, and clearly pulled down RRC-1-HA from the lysate, a major protein of expected size (∼100 kDa, indicated by the asterisk), as well as some proteins (75, 45, 30 kDa) that are likely degradation products. Although a small amount of GIT-1-mNeonGreen is detected in the lysate after a 30-min exposure, no GIT-1-mNeonGreen is detected in the IP (position of the protein indicted by “x”). In contrast, PIX-1 was detected in the lysate even after 1-min exposure and detected in the IP after the 30-min exposure (position marked by an arrow).
Transgenic rescue of rrc-1 with wild type RRC-1 but not RRC-1 with a missense mutation in a highly conserved residue of the RhoGAP domain.
We next wondered whether the RhoGAP activity of RRC-1 was required for its muscle function. A highly conserved arginine in RhoGAP domains, called the “arginine finger”, inserts into the GTP-binding site of RhoGTPases to stabilize the transition state and catalyze the GTPase reaction (Mosaddeghzadeh and Ahmadian, 2021). When this arginine is mutated to methionine or alanine, RhoGAP in vitro activity is abolished and results in in vivo phenotypes for several genes/proteins (Muller et al., 1997; Zhang and Glotzer, 2015; Wallace et al., 2018). We made the equivalent mutation, R315M, in RRC-1, and tested its ability to rescue the rrc-1(ok1747) phenotype, in transgenic worms. We created transgenic lines in which either wild type or R315M mutant full-length RRC-1 with a C-terminal HA tag was expressed from the muscle-specific myo-3 promoter in rrc-1(ok1747). As shown in Figure 13a, a western blot using anti-HA shows that each line expresses equivalent levels of either wild type or R315M RRC-1-HA. Imaging of body wall muscle cells expressing the transgenic array detected by the expression of the transformation marker SUR-5-GFP showed complete rescue of the rrc-1 phenotype by the wild type but not the R315M mutant version of RRC-1: Expression of wild type RRC-1 resulted in PAT-6 being normally localized to the MCBs, and the dense bodies and M-lines showing normal organization (Figure 13b). Muscle cells in which the R315M version was expressed showed disorganization of PAT-6 at the MCB and disorganized M-lines and dense bodies. Therefore, these results suggest that the RhoGAP activity of RRC-1 is required for it to carry out its normal function in muscle to assemble or maintain the structure of IACs.
FIGURE 13:
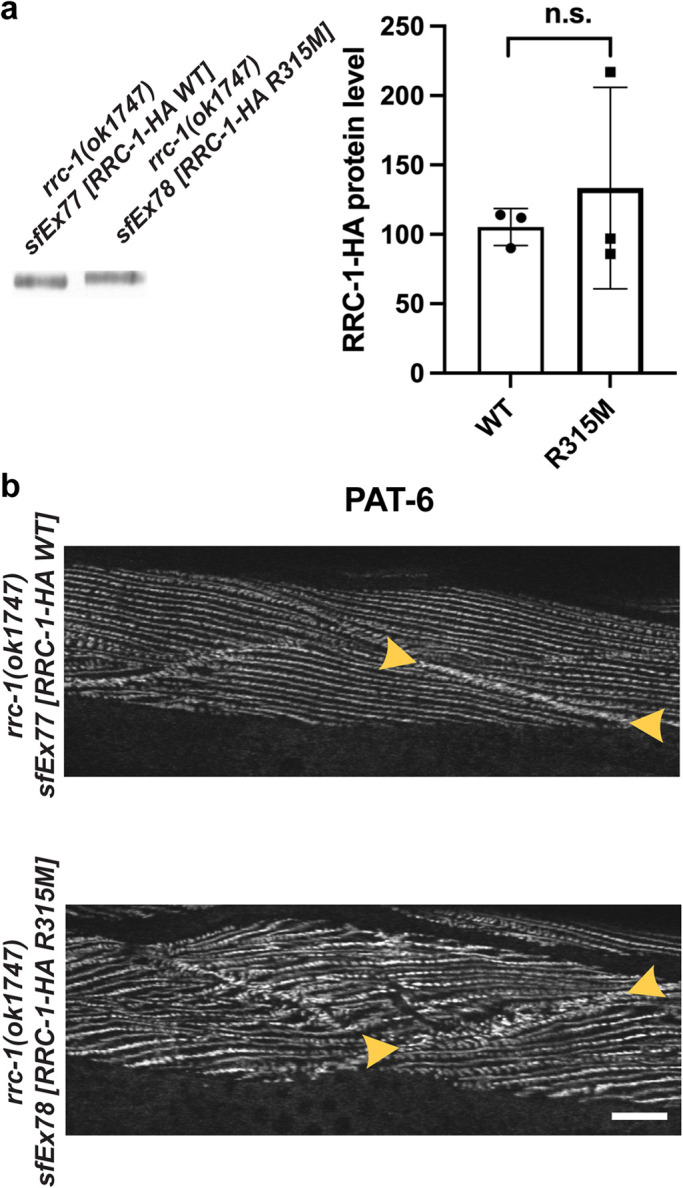
Transgenic expression of wild type but not R315M RRC-1 rescues the rrc-1 mutant phenotype. a) Similar levels of wild type and R315M versions of RRC-1-HA expressed in muscle from transgenic arrays in rrc-1(ok1747). The extrachromosomal arrays also contain the transformation marker DNA for SUR-5-GFP, which is expressed in most somatic cells. Total Laemmli-soluble proteins were prepared from 200 GFP positive adults from each strain, in triplicate, subjected to SDS–PAGE and probed with anti-HA. An unpaired t test with Welch’s correction was used to test for statistical significance. b) Rescue of the rrc-1(ok1747) muscle phenotype by transgenic expression of wild type but not R315M RRC-1. Portions of body wall muscle quadrants immunostained with anti-PAT-6, with arrowheads marking MCBs. The muscle cells shown contained the transgenic array because they express SUR-5-GFP. Note that when wild type RRC-1 is expressed in the rrc-1 mutant background the organization of the MCB and parallel alternating M-lines and rows of dense bodies is restored. Scale bar, 10 μm.
DISCUSSION
Here we identify RRC-1 as a Rho family GAP required for the assembly or stability of IACs in C. elegans muscle, and probably acting at least partially in the PIX-1 pathway. Loss of function of rrc-1 results in disorganization of the IACs at all its locations in nematode muscle–MCBs, M-lines and dense bodies (Figures 3 and 4). A likely consequence of this IAC disorganization is the disorganization of the sarcomeres and reduced muscle function (locomotion) observed in rrc-1 mutants. (Figures 5 and 6). Our conclusion that RRC-1 is a member of the PIX pathway is based upon the following: (1) Loss of function for mutations in the GEF, PIX-1, and the GAP, RRC-1, each reduce the accumulation of IAC components at the adhesion plaques of the MCB. That loss of function of a GEF, PIX-1, a positive regulator of Rac, and loss of function of a GAP, RRC-1, a negative regulator of Rac, have the same defect at MCBs, suggests that the maintenance of IACs at MCBs is a dynamic process. Consistent with these results, if we increase or eliminate the protein kinase activity of a known effector of the PIX-1 pathway, PAK-1, we also observe defects in the MCB (Figure 2). (2) PIX-1 (Moody et al., 2020; Figure 9) and RRC-1 each localize to the MCB (Figure 7). (3) RRC-1 localization to the MCB requires PIX-1 (Figure 8). (4) PIX-1 localization to the MCB requires RRC-1 (Figure 9). (5) Analysis of a rrc-1 pix-1 double mutant indicates that the two genes are members of the same genetic pathway (Supplemental Figure 6). (6) The localization of PIX-1 and RRC-1 each depend on CED-10 (Figure 11). (7) GIT-1, a scaffold for assembly of PIX-1 and PAK-1, when knocked down by RNAi results in a reduced level of RRC-1 (Figure 10). (8) RRC-1 exists in a complex with PIX-1 (Figure 12).
It is likely that the RhoGAP activity of RRC-1 is responsible for the protein’s role in assembling or maintaining the IACs: (1) Both PIX-1 and RRC-1 affect the activity of Rac: we have reported that a pix-1 null mutant has reduced levels of activated GTP-bound CED-10 (Rac) in nematode muscle (Moody et al., 2020). By expressing nematode RRC-1 in mammalian tissue culture cells, Delawary et al. (2007) reported that RRC-1 has GAP activity towards mammalian Rac and Cdc42 but not RhoA. These authors’ results are biologically relevant given that nematode CED-10 is 83% identical to human Rac1, and that CDC-42 is 86% identical to human Cdc42. (2) Rescue of the rrc-1 muscle phenotype depends on the presence of a putatively active RhoGAP domain (Figure 13). Nevertheless, we have not proven that the RhoGAP activity of RRC-1 is required for its function in IAC assembly. We have attempted, without success, in vitro RhoGAP activity assays using bacterially expressed GST-CED-10 or GST-CDC-42, with MBP-RRC-1 RhoGAP domain. We also did not detect a change (expected increase) in the level of active CED-10 in nematode muscle from a rrc-1 mutant. This last result is likely due to biochemical redundancy; in addition to RRC-1, 17 other proteins containing RhoGAP domains are expressed in body wall muscle (Supplemental Table 1; Supplemental Figure 1).
Figure 14 summarizes what we have learned about the pix-1 pathway in C. elegans body wall muscle. However, we still do not know the substrates for PAK-1 and PAK-2 protein kinases in muscle and how their phosphorylation results in the assembly or maintenance of IACs at the MCB.
FIGURE 14:
RRC-1 is RhoGAP in the PIX-1 pathway. The drawing depicts what we have learned about the PIX-1 pathway in C. elegans muscle. In Moody et al. (2020) we demonstrated that for the proper assembly or stability of integrin adhesion sites at the MCB, the Rac GEF, PIX-1, its scaffold, GIT-1, the Rac, CED-10, and the PAK effectors, PAK-1 and PAK-2 are required. The current results are most consistent with the Rac GAP, RRC-1 being a GAP for the PIX pathway in striated muscle, based on the similarity of pix-1 and rrc-1 phenotypes, and genetic interactions of rrc-1 with pix-1, ced-10, and git-1. The blue arrows indicate that, by some still unknown mechanisms, this Rac pathway is required for the assembly or stability of IACs at MCBs. Insight into these mechanisms likely will emerge by identifying the substrates for PAK-1 and PAK-2 in nematode muscle, which are currently unknown. Core components of the IAC are depicted to the right, with the names of the mammalian proteins shown in parentheses.
We wondered whether RRC-1 might exist in a complex with PIX-1 and GIT-1. This was suspected because pix-1, git-1 and rrc-1 mutants similarly affect the MCB; PIX-1 and RRC-1 colocalize to the MCB; GIT is a known scaffold for PIX and PAK in mammals; and because we showed that RNAi knock down of git-1 results in reduced levels of RRC-1 (Figure 10). IP of RRC-1-HA coIPd a small amount of PIX-1 but not GIT-1 (Figure 12), suggesting that RRC-1 exists in a complex with PIX-1. It is possible that not more PIX-1 was detected because the lysate buffer, which includes the ionic detergent sodium deoxycholate required to extract RRC-1 (Supplemental Figure 7), might have weakened the RRC-1 to PIX-1 interaction thus reducing the amount of PIX-1 that was co-IPd.
It is curious that the strongest alleles of rrc-1, in addition to having defects at MCBs, which is found in pix-1 and previously characterized PIX pathway mutants (e.g., git-1, pak-1, pak-2, ced-10), show defects in the organization of M-lines and dense bodies, and in the sarcomeres (M-lines, thick filaments, and thin filaments). It is also interesting that the pak-1 kinase-dead or constitutively active mutants only show a phenotype at the MCB. Perhaps the lack of a stronger phenotype for the pak-1 mutants is due to genetic redundancy provided by pak-2. pak-2 mutants by themselves also show the MCB defect (Moody et al., 2020). Moreover, the broader phenotype of rrc-1 mutants as compared with previously characterized pix-1 pathway mutants might be because RhoGAPs are more promiscuous than RhoGEFs (Muller et al., 2020), acting on multiple Rho GTPases (Rac, Cdc42, and RhoA). In addition, the lack of sarcomere disorganization in pix-1 mutants might be due to redundant GEFs for Rac or Cdc42 at M-lines and dense bodies. For example, UIG-1 is localized to dense bodies and is a GEF for Cdc42 (Hikita et al., 2005). Although the intracellular location in muscle for TIAM-1 and UNC-73 are unknown, each is expressed in body wall muscle (Meissner et al., 2009) and each have a RhoGEF with demonstrated GEF activity for Rac in vitro (Steven et al., 1998; Demarco et al., 2012).
Also, our results hint at the possibility that RRC-1 may be membrane-associated. This is because: (1) In addition to strong localization of RRC-1 to the MCB, we observed a diffuse or “fuzzy” localization near the outer muscle cell membrane (Figure 7b, middle row). (2) Extraction of RRC-1 into a soluble fraction required an ionic detergent (Supplemental Figure 7). Using online software prediction programs, we could not detect any transmembrane domains in RRC-1. Thus, how RRC-1 might be membrane-associated is a mystery. In the future we will test the speculation that membrane insertion might involve RRC-1 undergoing posttranslational covalent modification such as myristylation or palmitoylation.
The PIX pathway has been shown to be functionally important in multiple organisms and tissues, ranging from mammalian nervous (Ramakers et al., 2012) and immune (Missy et al., 2008) systems, to nematode germline (Lucanic and Cheng, 2008), migration of neuroblasts (Dyer et al., 2010), tension-dependent morphogenesis of epidermal cells (Zhang et al., 2011), early embryonic elongation (Martin et al., 2014), and muscle (Moody et al. 2020). Given that we have identified two possible human orthologs for RRC-1, ARHGAP32, and, ARHGAP33 (Supplemental Figure 2), and each are known to be expressed in skeletal and heart muscle, we propose that one or both of these proteins are GAPs for the PIX pathway in human muscle. As noted above, in C. elegans, pix-1 has also been shown to function in neurons. During the migration of nematode QR neuroblast descendants, noncanonical signaling through PIX-1 occurs (Dyer et al., 2010), but when migration stops, canonical Wnt signaling is engaged by activating EVA-1 in the Slt-Robo pathway, and the RhoGAP called RGA-9 (Rella et al., 2021). However, note that PIX-1 functions in activation of neuroblast migration, but RGA-9 functions to inhibit neuroblast migration. Therefore, RGA-9 might be a RhoGAP for the PIX-1 pathway in neuroblast migration, but this is difficult to conclude from the existing data. Also note that although rga-9 is expressed in muscle, we did not detect an MCB defect in a deletion allele of the gene (Supplemental Figure 1).
In addition to rrc-1, our screening of mutants in 18 RhoGAP proteins expressed in muscle revealed two other genes that when mutated result in MCB defects, hum-7 and rga-4 (Supplemental Figure 1). hum-7 mutants only affect the MCB (like pix-1 mutants), whereas the rga-4 mutant affects the MCB, M-line and dense body (like rrc-1). hum-7 encodes a 213 kDa protein with a RhoGAP domain near its C-terminus and a myosin class IX motor domain in its N-terminal half. Inspection of two independently generated mutants have the same MCB-specific defect as pix-1. However, we have only examined a single mutant allele of rga-4, the intragenic deletion rga-4(ok1935), and it had not been outcrossed to wild type. rga-4 encodes a 1126 aa protein with the only recognizable domain being the RhoGAP domain. RGA-4 has been reported to act redundantly with another RhoGAP protein, RGA-3, in the germ line and early embryo, and to inactivate RHO-1 (RhoA) of C. elegans (Schmutz et al., 2007). Interestingly, we have previously reported that RNAi knockdown of RHO-1 (RhoA) results in disorganization of the A-bands in the body wall muscle of adult nematodes (Qadota et al., 2008). We leave investigation of hum-7 and rga-4 in muscle for future studies.
MATERIALS AND METHODS
Request a protocol through Bio-protocol.
C. elegans strains
All nematode strains were grown on NGM plates using standard methods and maintained at 20°C (Brenner, 1974). Most strains were obtained from the Caenorhabditis Genetics Center. The wild type strain was N2 (Bristol). Strains containing mutations in 18 genes encoding proteins with RhoGAP domains and also expressed in muscle, are listed in Supplemental Table 1 and in Supplemental Figure 1. The following strains were generated during this study:
GB340: pak-1(syb647), which contains a L99F mutation in PAK-1, was generated by CRISPR/Cas9 by SunyBiotech (described below) as PHX647, and then outcrossed 4X to wild type.
GB341: pak-1(syb632), which contains a K324A mutation in PAK-1, was generated by CRISPR/Cas9 by SunyBiotech (described below) as PHX632, and then outcrossed 1X to wild type.
GB342: rrc-1(syb4499), which expresses RRC-1 with an HA tag fused to its C-terminus (RRC-1::HA), was generated by CRISPR/Cas9 by SunyBiotech (described below) as PHX4499.
GB343: rrc-1(ok1747) was outcrossed 5X to wild type.
GB344: rrc-1(tm1023) was outcrossed 5X to wild type.
GB345: rrc-1(gk290525) was outcrossed 5X to wild type
GB346: rrc-1(gk859353) was outcrossed 5X to wild type
GB348: rrc-1(syb4499) pix-1(gk299374) was generated by recombination starting with GB342 and GB291 (Moody et al., 2020).
DM8008: raIs8 [unc-112::GFP + rol-6(su1006)]
GB372: rrc-1(ok1747); sfEx77 [myo-3p::RRC-1::HA; sur-5::gfp]
GB373: rrc-1(ok1747); sfEx78 [myo-3p::RRC-1(R315M)::HA; sur-5::gfp]
GB374: rrc-1(ok1747) pix-1(gk299374) was generated by recombination.
MT9958: ced-10(n3246)
GB375: ced-10(n3246); rrc-1(syb4499)
CRISPR/Cas9 generation of nematode strains expressing kinase dead and kinase constitutively-active PAK-1, HA-tagged RRC-1, and mNeonGreen-tagged GIT-1.
The CRISPR/Cas9 procedures were carried out by SunyBiotech (www .sunybiotech.com). Details about the sgRNAs and repair templates used are given in Supplemental Figure 8. The resulting strains are:
PHX647, pak-1(syb647) which has an L99F mutation predicted to make the PAK-1 protein kinase constitutively active.
PHX632, pak-1(syb632) which has a K324A mutation predicted to make the PAK-1 protein kinase catalytically dead.
PHX4499, rrc-1(syb4499) which expresses RRC-1 with a C-terminal HA tag.
PHX5908, rrc-1(syb4499) git-1(syb5908) which expresses both RRC-1-HA and GIT-1-mNeonGreen.
Immunostaining and confocal microscopy of body wall muscle
Adult worms were fixed and immunostained using the method described by Nonet et al. (1993), and additional details described by Wilson et al. (2012). Antibodies were used at 1:200 dilution except as noted: anti-PAT-6 (rat polyclonal; Warner et al., 2013), anti-UNC-52 (mouse monoclonal MH2; Mullen et al., 1999; purchased from University of Iowa Hybridoma Bank), anti-UNC-95 (rabbit polyclonal Benian-13; Qadota et al., 2007), anti-UNC-112 (1:100 dilution; Hikita et al., 2005), anti-MHC A (mouse monoclonal 5–6; Miller et al., 1983; purchased from University of Iowa Hybridoma Bank), anti-UNC-89 (rabbit polyclonal EU30; Benian et al., 1996), anti-ATN-1 (mouse monoclonal MH35; Francis and Waterston, 1991; kindly provided by Pamela Hoppe, Western Michigan University), and anti-HA (rabbit monoclonal C29F4 from Cell Signaling Technology). Secondary antibodies, used at 1:200 dilution, included antirabbit Alexa 488, antirat Alexa 594, and antimouse Alexa 594, all purchased from Invitrogen. Fixation and phalloidin-rhodamine staining was conducted as described (Waterston et al., 1984). Images were captured at room temperature with a Zeiss confocal system (LSM510) equipped with an Axiovert 100M microscope and an Apochromat x63/1.4 numerical aperture oil immersion objective, in 1 × mode. For all the confocal images the color balances were adjusted by using Adobe Photoshop (Adobe, San Jose, CA).
Live imaging of UNC-112-GFP
We live imaged UNC-112-GFP using the integrated transgenic array strain DM8008 (raIs8 [unc-112::GFP + rol-6(su1006)]) which expresses full-length UNC-112 with a GFP insertion. To determine the localization of UNC-112-GFP in pix-1(gk299374) and rrc-1(ok1747), we crossed DM8008 into these pix-1 and rrc-1 mutants. The transgene was followed by using the Rol-6 marker, and the presence of the rrc-1 or pix-1 mutations was followed by PCR and/or DNA sequencing. Live imaging was performed as described in Moody et al. (2020). Essentially, animals were washed off plates using M9 buffer and immobilized with 10 μM levamisole in M9 for 10 min. Approximately 50–100 animals in 3 μl were added to 7 μl of ice-cold 25% Pluronic F127 in M9 (Hwang et al., 2014) lying on a cold glass slide, to which was added a coverslip and sealed with nail polish. After incubation at room temperature for 5–10 min to solidify, images were taken using a confocal microscope as described above.
Swimming and crawling assays
For swimming assays day 2 adults were harvested from one 6-cm NGM OP50 seeded plate with M9 buffer. Animals were subsequently washed free from bacteria using M9 buffer and then pelleted at ratio of 1:1 (worm: buffer). Two milliliters of M9 buffer followed by 5 μL of worm suspension were added to the center area of one unseeded 6-cm NGM plate. Each strain was allowed to adapt for 5 mins before recording swimming movement. The recordings were done using a dissecting stereoscopic microscope fitted with a CMOS camera (Thorlabs). For all strains a total of 15, 10-s videos were recorded from various sections of the plate with each video tracking an average of eight individual animals. Video data was analyzed by Image J WrmTracker software plug-in to obtain body bends per second (BBPS) for individual animals. Worms that moved out of frame and outliers were removed during data analysis and an average of 20 animals were analyzed for each strain. The resulting BBPS values for each mutant strain was compared with wildtype and further tested for statistically significant differences using Welch’s t test.
For crawling assays day 2 adults were harvested as described above, except for the use of 0.2 g/L gelatin in M9 buffer. Five microliters of worm suspension was added to the center of a 6 cm unseeded NGM plate and then the excess liquid was removed using a twisted KimWipe. After a 5-min adaptation time, worm crawling movement was recorded as mentioned above. BBPS values for individual animals were extracted from each video. The resulting values for each strain was compared with wild type for statistical analysis using Welch’s t test for significance.
Protein sequence analysis
Nematode RRC-1a, b, and c were obtained from Wormbase. By using a BLAST homology search, we confirmed that ARHGAP31, ARHGAP32, and ARHGAP33 are the human proteins most homologous to RRC-1. The domain organization for RRC-1 and its homologues were analyzed by the online PFAM database. PubMed pBLAST database was used to align human ARHGAP31-33 amino acid sequences with nematode RRC-1 to determine the percent identities for each domain and total protein.
Knockdown of GIT-1 via RNAi feeding
RNAi by feeding was performed as described previously (Timmons et al., 2001; Miller et al. 2009). GIT-1 cDNA was generated via PCR amplification of the 5′-most 1077 nucleotides of the GIT-1 cDNA sequence using the RB2 cDNA library as a template with the following primers–GIT-1 FWD: 5′-GCGGGATCCATGTACACAGCAGAGGCGCTT-3′ which includes a BamHI restriction enzyme (RE) site and GIT-1 REV: 5′-CGCCTCGAGTGCTGGATTGTCTCCAGTGAT-3′, which includes an XhoI RE site. The ∼1 kb amplicon was digested and ligated into the BamHI and XhoI sites of the pPD129.36 vector and used to transform competent XL1 Blue Escherichia coli cells on LB + ampicillin plates overnight at 37°C. Individual colonies from the GIT-1 cDNA pPD129.36 clones were grown overnight in liquid culture, plasmids prepared, and confirmed by restriction digestion. A resulting GIT-1 pPD129.36 clone and empty vector pPD129.36 plasmids were used to transform competent HT115 (DE3) RNAi feeding bacteria, and a resulting colony from each was grown as an overnight liquid culture. The resulting bacteria were used to seed 6 cm and 10 cm NGM plates. To conduct RNAi feeding experiments, 15 to 20 L4 stage worms were added to 25 NGM git-1 RNAi and 25 empty vector in HT115 (DE3) bacteria 6 cm plates and left overnight. Then following day, 10 worms were transferred from the 6 cm plates to the 10 cm plates under the same conditions and allowed to lay eggs for ∼8 h before being picked. The eggs on the plate were allowed to hatch for ∼48 h before being harvested for conducting fixation for immunostaining or making lysates for SDS–PAGE, followed by Western blotting analysis. (Miller et al., 2009)
Western blot analysis
The method of Hannak et al. (2002) was used to prepare total protein lysates from wild-type, rrc-1(ok1747) 5X O.C., rrc-1(tm1023) 5X O.C., RRC-1::HA, RRC-1::HA pix-1(gk299374), RRC-1::HA; RNAi empty vector, and RRC-1::HA; git-1 (RNAi) mixed-stage animals. Equal amounts of total protein were separated on 10% polyacrylamide-SDS- Laemmli gels, transferred to nitrocellulose membranes, reacted with affinity purified, E. coli-OP50-absorbed anti-PIX-1a (Moody et al., 2020), or anti-HA (rabbit monoclonal catalogue no. C29F4 from Cell Signaling Technology) at 1:1000 dilution, and then reacted with goat antirabbit immunoglobulin G conjugated to HRP (GE Healthcare) at 1:10,000 dilution, and visualized by ECL. Protein bands were quantitated by normalization to total protein in each lane detected by Ponceau S staining.
Immunoprecipitation of RRC-1-HA
A large quantity of worms (∼3.5 ml) from strain PHX5908 (rrc-1(syb4499) git-1(syb5908)) which expresses both RRC-1-HA and GIT-1-mNeonGreen, were grown on 20, 15-cm high peptone NGM plates seeded with E. coli strain OP50, and a “worm powder” was generated by grinding the worms extensively in a mortar and pestle in liquid nitrogen. A total protein lysate was prepared by adding worm powder to 1 ml of “RIPA Buffer” (∼20% vol/vol) consisting of 50 mM Tris pH 7.5, 150 mM NaCl, 1% Nonidet P-40, 0.01% sodium deoxycholate, and cOmplete Mini protease inhibitor (Roche), vortexing for 1 min, incubating on ice for 5 min, vortexing for 1 min, spinning at top speed in a microfuge at 4°C for 10 min, and saving the supernatant. A small proportion of supernatant was diluted 1:1 with 2X Laemmli sample buffer, mixed and heated at 95°C for 5 min, and the resulting material was designated as lysate (L). To the remainder of the supernatant was added 30 μl of a 1:1 suspension of anti-HA magnetic beads (ThermoFisher, Pierce, catalogue number 88836) and incubating on a rotating wheel at 4°C for 1 h, removing the beads from the solution using a rack containing neodymium magnets, and then washing the beads with 1 ml of Wash Buffer (50 mM Tris pH 7.5, 150 mM NaCl, cOmplete Mini protease inhibitors) 3X. To the washed beads was added 30 μl of 2X Laemmli sample buffer, vortexing for 5 s, heating at 95°C for 5 min, vortexing for 5 s, and then separating out the beads on the magnetic stand; the resulting liquid was designated as IP. Multiple lanes containing either 20 μl of L or 15 μl of IP, were separated on a 10% SDS–PAGE, transferred to nitrocellulose membrane, and as shown in Figure 12, portions of the blot were reacted against anti-HA (rabbit monoclonal antibody, catalogue no. 3724 from Cell Signaling, at 1:1000 dilution), anti-mNeonGreen (mouse monoclonal antibody 32F6 from Chromotek, at 1:100 dilution), and anti-PIX-1 (rabbit polyclonal antibody [Moody et al., 2020] at 1:1000 dilution), reacted with the appropriate HRP-conjugated secondary antibodies and visualized with ECL and exposure to film.
Transgenic rescue of rrc-1 attempted with wild type and R315M RRC-1-HA
To create a plasmid for transgenic expression of RRC-1 with an HA tag at its C-terminus under the control of the muscle specific myo-3 promoter, two separate cDNA fragments (N-terminal half (fragment N) and C-terminal half (fragment C)) of a full length rrc-1 cDNA were amplified by using PCR with the following primers and the RB2 cDNA library (a gift from Robert Barstead, Oklahoma Medical Research Foundation):
rrc-1-8: for fragment N:
GCGCCCGGGGCTAGCATGGAAGGCATCGAGGAATCATTTG
rrc-1-3: for fragment N of wild type:
CGCAAGCTTCTCTGAATATTCGATTGAATTC
RRC-1 R315M-R: for fragment N of R315M:
CGCAAGCTTTATTCGATTGAATTCCACACTGCATATAAATACCCGTGACAATTCC
rrc-1-4: for fragment C:
GCGCCCGGGTTTATAGGCAGTGTGGAATTC
rrc-1-9HA: for fragment C with HA tag:
CGCAAGCTTGCTAGCGGATCCTTAAGCGTAATCTGGAACATCGTATGGGTAAGATCCACCAGCGTAATCTGGAACATCGTATGGGTAAGATCCACCAGCGTAATCTGGAACATCGTATGGGTATGCAGAGAACAAATGACTGGC
rrc-1-8 and rrc-1-3 were used for amplification of fragment N (wild type). rrc-1-8 and RRC-1 R315M-R were used for amplification of fragment N (R315M). rrc-1-4 and rrc-1-9HA were used for amplification of fragment C with HA. All fragments were cloned into the SmaI and HindIII sites of pBluescript KS+ (pBS-RRC1-N-WT, pBS-RRC-1-N-R315M, and pBS-RRC-1-C-HA) and clones that were error-free were identified by Sanger sequencing. To create fragment N+C (full length), SmaI/EcoRI fragments of pBS- RRC-1-N-WT or pBS-RRC-1-N-R315M were cloned into the SmaI and EcoRI sites of pBS-RRC-1-C-HA, resulting in pBS-RRC-1-HA WT and pBS-RRC-1-HA R315M. A NheI fragment of pBS- RRC1-HA WT and pBS-RRC-1-HA R315M were cloned into the NheI site of pPD95.86 (myo-3 promoter plasmid; gift of Andrew Fire, Stanford University), resulting in plasmids pPD95.86- RRC1-HA WT and pPD95.86- RRC1-HA R315M. A mixture of pPD95.86- RRC1-HA WT or pPD95.86- RRC1-HA R315M, and pTG96 (sur-5::gfp transformation marker) at ratio of 1: 10 was injected into rrc-1(ok1747) mutant worms, and screened for GFP+ worms, resulting in GB372 (rrc-1(ok1747); sfEx77 [myo-3::rrc-1::HA(wild type); sur-5::GFP]) and GB373 (rrc-1(ok1747); sfEx78 [myo-3::rrc-1::HA(R315M); sur-5::GFP]).
Supplementary Material
Acknowledgments
We thank SunyBiotech Corporation for generation of the CRISPR/Cas9 strains PHX632, PHX647, PHX4499, and PHX5908; Sophia Figueroa and Jordan Walter for generating a full-length cDNA for rrc-1; Robert Barstead (Oklahoma Medical Research Foundation) for the cDNA library RB2; Andrew Fire (Stanford University) for the RNAi vector pPD129.36, and the myo-3 promoter plasmid pPD95.86; and Pamela Hoppe (Western Michigan University) for monoclonal MH35. We thank Richard Kahn and Dorothy Lerit for helpful comments on the manuscript. Most of the nematode strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health Office of Research Infrastructure Programs (P40 OD010440). This study was supported in-part by a National Science Foundation - Graduate Research Fellowship (DGE 1444932) to J.C.M., and from a National Institutes of Health grant (R01HL160693) to G.M.B.
Abbreviations used:
- ECM
extracellular matrix
- GAP
GTPase-activating protein
- GEF
guanine nucleotide exchange factor
- IAC
integrin adhesion complex
- MCB
muscle cell boundary
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E23-03-0095) on March 6, 2024.
REFERENCES
- Anthis NJ, Campbell ID (2011). The tail of integrin activation. Trends Biochem Sci 36, 191–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachir AI, Zareno J, Moissoglu K, Plow EF, Gratton E, Horwitz AR (2014). Integrin-associated complexes form hierarchically with variable stoichiometry in nascent adhesions. Curr Biol 24, 1845–1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benian GM, Tinley TL, Tang X, Borodovsky M (1996). The Caenorhabditis elegans gene unc-89, required for muscle M-line assembly, encodes a giant modular protein composed of Ig and signal transduction domains. J Cell Biol 132, 835–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benian GM, Epstein HF (2011). Caenorhabditis elegans muscle: a genetic and molecular model for protein interactions in the heart. Circ Res 109, 1082–1095. [DOI] [PubMed] [Google Scholar]
- Brenner S (1974). The genetics of Caenorhabditis elegans. Genetics 77, 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JL, Stowers L, Baer M, Trejo JA, Coughlin S, Chant J (1996). Human Ste20 homologue hPAK1 links GTPases to the JNK MAP kinase pathway. Curr Biol 6, 598–605. [DOI] [PubMed] [Google Scholar]
- Canman JC, Lewellyn L, Laband K, Smerdon SJ, Desai A, Bowerman B, Oegema K (2008). Inhibition of Rac by the GAP activity of centralspindlin is essential for cytokinesis. Science 322, 1543–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delawary M, Nakazawa T, Teuka T, Sawa M, Iino Y, Takenawa T, Yamamoto T (2007). Molecular characterization of a novel RhoGAP, RRC-1 of the nematode C. elegans. Biochem and Biophys Res Comm 357, 377–382. [DOI] [PubMed] [Google Scholar]
- Demarco RS, Struckhoff EC, Lundquist EA (2012). The Rac GTP exchange factor TIAM-1 acts with CDC-42 and guidance receptor UNC-40/DCC in neuronal protrusion and axon guidance. PLoS Genet 8, e1002665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer JO, Demarco RS, Lundquist EA (2010). Distinct roles of Rac GTPases and the UNC-73/Trio and PIX-1 Rac GTP exchange factors in neuroblast protrusion and migration in C. elegans. Small GTPases 1, 44–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ervasti JM (2003). Costameres: the Achilles’ heel of Herculean muscle. J Biol Chem 278, 13591–13594. [DOI] [PubMed] [Google Scholar]
- Francis R, Waterston RH (1991). Muscle cell attachment in Caenorhabditis elegans. J Cell Biol 114, 465–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gieseler K, Qadota H, Benian GM (2017). Development, structure, and maintenance of C. elegans body wall muscle. In: WormBook, ed. The C. elegans Research Community, WormBook, www.wormbook.org [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannak E, Oegema K, Kirkham M, Gonczy P, Habermann B, Hyman AA (2002). The kinetically dominant assembly pathway for centrosomal asters in Caenorhabditis elegans is gamma-tubulin dependent. J Cell Biol 157, 591–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson CA, Gomez CG, Novak SM, Mi-Mi L, Gregorio CC (2017). Overview of the muscle cytoskeleton. Compr Physiol 7, 891–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hikita T, Qadota H, Tsuboi D, Taya S, Moerman DG, Kaibuchi K (2005). Identification of a novel Cdc42 GEF that is localized to the PAT-3-mediated adhesive structure. Biochem Biophys Res Commun 335, 139–145. [DOI] [PubMed] [Google Scholar]
- Horton ER, Byron A, Askari JA, Ng DH, Millon-Frémillon A, Robertson J, Koper EJ, Paul NR, Warwood S, Knight D, et al. (2015). Definition of a consensus integrin adhesome and its dynamics during adhesion complex assembly and disassembly. Nat Cell Biol 17, 1577–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Zhou Z, Asrar S, Henkelman M, Xie W, Jia Z (2011). p21-Activated kinases 1 and 3 control brain size through coordinating neuronal complexity and synaptic properties. Mol Cell Biol 31, 388–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang H, Krajniak J, Matsunaga Y, Benian GM, Lu H (2014). On-demand optical immobilization of C. elegans for high-resolution imaging and microinjection. Lab Chip 14, 3498–3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer GH, Garrod S, Jr WVL, Taylor SS (2005). Catalytic independent functions of a protein kinase as revealed by a kinase-dead mutant: study of the Lys72His mutant of cAMP-dependent kinase. J Mol Biol 351, 1110–1122. [DOI] [PubMed] [Google Scholar]
- Jantsch-Plunger V, Gönczy P, Romano A, Schnabel H, Hamill D, Schnabel R, Hyman AA, Glotzer M (2000). CYK-4: A Rho family gtpase activating protein (GAP) required for central spindle formation and cytokinesis. J Cell Biol 149, 1391–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucanic M, Cheng HJ (2008). A RAC/CDC-42-independent GIT/PIX/PAK signaling pathway mediates cell migration in C. elegans. PLoS Genet 4, e1000269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundquist EA, Reddien PW, Hartwieg E, Horvitz HR, Bargmann CI (2001). Three C. elegans Rac proteins and several alternative Rac regulators control axon guidance, cell migration and apoptotic cell phagocytosis. Development 128, 4475–4488. [DOI] [PubMed] [Google Scholar]
- Martin E, Harel S, Nkengfac B, Hamiche K, Neault M, Jenna S (2014). pix-1 controls early elongation in parallel with mel-11 and let-502 in Caenorhabditis elegans. PLoS One 9, e94684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner B, Warner A, Wong K, Dube N, Lorch A, McKay SJ, Khattra J, Rogalski T, Somasiri A, Chaudhry I, et al. (2009). An integrated strategy to study muscle development and myofilament structure in C. elegans. PLoS Genetics 5, e1000537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelitch M, Chant J (1996). A mechanism of Bud1p GTPase action suggested by mutational analysis and immunolocalization. Curr Biol 6, 446–454. [DOI] [PubMed] [Google Scholar]
- Miller DM, Ortiz I, Berliner GC, Epstein HF (1983). Differential localization of two myosins within nematode thick filaments. Cell 34, 477–490. [DOI] [PubMed] [Google Scholar]
- Miller RK, Qadota H, Stark TJ, Mercer KB, Wortham TS, Anyanful A, Benian GM (2009). CSN-5, a component of the COP9 signalosome complex, regulates the levels of UNC-96 and UNC-98, two components of M-lines in C. elegans muscle. Mol Biol Cell 20, 3608–3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missy K, Hu B, Schilling K, Harenberg A, Sakk V, Kuchenbecker K, Kutsche K, Fischer KD (2008). AlphaPIX Rho GTPase guanine nucleotide exchange factor regulates lymphocyte functions and antigen receptor signaling. Mol Cell Biol 28, 3776–3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody JC, Qadota H, Reedy AR, Okafor CD, Shanmugan N, Matsunaga Y, Christian CJ, Ortlund EA, Benian GM (2020). The Rho-GEF PIX-1 directs assembly or stability of lateral attachment structures between muscle cells. Nature Commun 11, 5010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita K, Hirono K, Han M (2005). The Caenorhabditis elegans ect-2 Rho GEF gene regulates cytokinesis and migration of epidermal P cells. EMBO Rep 6, 1163–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosaddeghzadeh N, Ahmadian MR (2021). The RHO family GTPases: Mechanisms of regulation and signaling. Cells 10, 1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen GP, Rogalski TM, Bush JA, Gorji PR, Moerman DG (1999). Complex patterns of alternative splicing mediate the spatial and temporal distribution of perlecan/UNC-52 in Caenorhabditis elegans. Mol Biol Cell 10, 3205–3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller RT, Honnert U, Reinhard J, Bahler M (1997). The rat myosin myr5 is a GTPase-activating protein for Rho in vivo: essential role of arginine 1695. Mol Biol Cell 10, 2039–2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller PM, Rademacher J, Bagshaw RD, Wortmann C, Barth C, van Unen J, Alp KM, et al. (2020). Systems analysis of RhoGEF and RhoGAP regulatory proteins reveals spatially organized RAC1 signaling from integrin adhesions. Nat Cell Biol 22, 498–511. [DOI] [PubMed] [Google Scholar]
- Nonet ML, Grundahl K, Meyer BJ, Rand JB (1993). Synaptic function is impaired but not eliminated in C. elegans mutants lacking synaptotagmin. Cell 73, 1291–1305. [DOI] [PubMed] [Google Scholar]
- Qadota H, Mercer KB, Miller RK, Kaibuchi K, Benian GM (2007). Two LIM domain proteins and UNC-96 link UNC-97/pinch to myosin thick filaments in Caenorhabditis elegans muscle. Mol Biol Cell 18, 4317–4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qadota H, Blangy A, Xiong G, Benian GM (2008). The DH-PH region of the giant protein UNC-89 activates RHO-1 GTPase in C. elegans body wall muscle. J Mol Biol 383, 747–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qadota H, Matsunaga Y, Nguyen KCQ, Mattheyses A, Hall DH, Benian GM (2017). High resolution imaging of muscle attachment structures in C. elegans. Cytoskeleton 74, 426–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakers GJ, Wolfer D, Rosenberger G, Kuchenbecker K, Kreienkamp HJ, Prange-Kiel J, Rune G, Richter K, Langnaese K, Masneuf S, et al. (2012). Dysregulation of Rho GTPases in the αPix/Arhgef6 mouse model of X-linked intellectual disability is paralleled by impaired structural and synaptic plasticity and cognitive deficits. Hum Mol Genet 21, 268–286. [DOI] [PubMed] [Google Scholar]
- Rella L, Fernandes Póvoa EE, Mars J, Ebbing ALP, Schoppink L, Betist MC, Korswagen HC (2021). A switch from noncanonical to canonical Wnt signaling stops neuroblast migration through a Slt-Robo and RGA-9b/ARHGAP-dependent mechanism. Proc Natl Acad Sci 118, e2013239118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmalzigaug R, Rodriguiz RM, Bonner PE, Davidson CE, Wetsel WC, Premont RT (2009). Impaired fear response in mice lacking GIT1. Neurosci Lett 458, 79–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz C, Stevens J, Spang A (2007). Functions of the novel RhoGAP proteins RGA-3 and RGA-4 in the germ line and in the early embryo of C. elegans. Development 134, 3495–3505. [DOI] [PubMed] [Google Scholar]
- Schonegg S, Constantinescu AT, Hoege C, Hyman AA (2007). The Rho GTPase-activating proteins RGA-3 and RGA-4 are required to set the initial size of PAR domains in Caenorhabditis elegans one-cell embryos. Proc Natl Acad Sci USA 104, 14976–14981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steven R, Kubiseski TJ, Zheng H, Kulkarni S, Mancillas J, Ruiz Morales A, Hogue CW, Pawson T, Culotti J (1998). UNC-73 activates the Rac GTPase and is required for cell and growth cone migrations in C. elegans. Cell 92, 785–795. [DOI] [PubMed] [Google Scholar]
- Sun Z, Lambacher A, Fassler R (2014). Nascent adhesions: from fluctuations to a hierarchical organization. Curr Biol 24, R801–R803. [DOI] [PubMed] [Google Scholar]
- Tadokoro S, Shattil SJ, Eto K, Tai V, Liddington RC, de Pereda JM, Ginsberg MH, Calderwood DA (2003). Talin binding to integrin beta tails: a final common step in integrin activation. Science 302, 103–106. [DOI] [PubMed] [Google Scholar]
- Thompson O, Edgley M, Strasbourger P, Flibotte S, Ewing B, Adair R, Au V, Chaudhry I, Fernando L, Hutter H, et al. (2013). The million mutation project: a new approach to genetics in Caenorhabditis elegans. Genome Res 23, 1749–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons L, Court DL, Fire A (2001). Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in C. elegans. Gene 263, 103–112. [DOI] [PubMed] [Google Scholar]
- Volinsky N, Gantman A, Yablonski D (2006). A Pak- and Pix-dependent branch of the SDF-1alpha signaling pathway mediates T cell chemotaxis across restrictive barriers. Biochem J 397, 213–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace AG, Raduwan H, Carlet J, Soto MC (2018). The RhoGAP HUM-7/Myo9 integrates signals to modulate RHO-1/RhoA during embryonic morphogenesis in C. elegans. Development 145, dev168724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner A, Xiong G, Qadota H, Rogalski T, Vogl AW, Moerman DG, Benian GM (2013). CPNA-1, a copine domain protein, is located at integrin adhesion sites, and is required for myofilament stability in C. elegans. Mol Biol Cell 24, 601–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston RH, Hirsh D, Lane TR (1984). Dominant mutations affecting muscle structure in C. elegans that map near the actin gene cluster. J Mol Biol 180, 473–496. [DOI] [PubMed] [Google Scholar]
- Wilson KJ, Qadota H, Benian GM (2012). Immunofluorescent localization of proteins in C. elegans muscle. Meth. Mol Biol 798, 171–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye F, Petrich BG, Anekal P, Lefort CT, Kasirer-Friede A, Shattil SJ, Ruppert R, Moser M, Fassler R, Ginsberg MH (2013). The mechanism of kindlin-mediated activation of integrin α11bβ3. Curr Biol 23, 2288–2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenke FT, King CC, Bohl BP, Bokoch GM (1999). Identification of a central phosphorylation site in p21-activated kinase regulating autoinhibition and kinase activity. J Biol Chem 274, 32565–32573. [DOI] [PubMed] [Google Scholar]
- Zhang D, Glotzer M (2015). The RhoGAP activity of CYK-4/MgcRacGAP functions non-canonically by promoting RhoA activation during cytokinesis. eLife 4, e08898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Landmann F, Zahreddine H, Rodriguez D, Koch M, Labouesse M (2011). A tension-induced mechanotransduction pathway promotes epithelial morphogenesis. Nature 471, 99–103. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.