Abstract
Genetic biocontrol aims to suppress or modify populations of species to protect public health, agriculture, and biodiversity. Advancements in genome engineering technologies have fueled a surge in research in this field, with one gene editing technology, CRISPR, leading the charge. This review focuses on the current state of CRISPR technologies for genetic biocontrol of pests and highlights the progress and ongoing challenges of using these approaches.
Keywords: CRISPR, genetic biocontrol, gene drive, homing, population suppression, population modification
1. INTRODUCTION
For millennia, humans have been plagued by arthropods and vertebrate pests, with notable examples spreading diseases that have altered the course of human history (231). However, even before they were linked to disease, the nuisance behaviors of many pests and their proclivity to damage crops made them targets for control efforts. Thus, humans have worked throughout history to reduce the detrimental effects of pests. Here, we focus on modern genetic biocontrol technologies, highlighting important themes, advances, and challenges.
1.1. The History of Genetic Biocontrol Technologies: The Pretransgenic Era
Genetic pest management strategies were conceptualized in the early twentieth century, and began to be implemented in the mid-twentieth century, with sterile insect technique (SIT) leading the charge (130, 131, 202). In SIT, sterile males are released to mate with wild females, and with frequent large-scale releases over time, this can suppress, or even eradicate, populations. Early work in this area relied on irradiation to generate sterilizing mutations (17, 131, 207). Large-scale implementations of this technique demonstrated resounding success, fully eradicating the New World screwworm from most of North America (131) and bringing about the suppression of a few other species (83, 179). However, genetic and other technical challenges prevented success in attempts to suppress certain species. In parallel to this work, a number of other control methods, based on pretransgenic era manipulations of pest genetics (e.g., translocations and inversions), were explored but generally did not meet with great success (100).
Thinking about genetic techniques for population management, particularly those meant to be self-sustaining, began >50 years ago (64, 201) and was inspired by the behavior of an ever-growing number of naturally occurring selfish genetic elements [henceforth termed gene drives (120)] found in all domains of life. Many of these gene drives were discovered early in the field of genetics, often fortuitously as a result of unexpected mutation rates, biased sex ratios, or mortality of specific genotypes. These drives advantage their transmission at the expense of other genes in the genome. This behavior can result in the spread of these drives, with respect to the corresponding chromosome counterparts, even when their presence results in a fitness cost to carriers (i.e., decreases the fitness of the population as a whole) (78, 95, 104, 178, 226). Naturally occurring gene drives are highly varied in form and mechanism and include sex ratio–distorting elements, meiotic drivers and toxin-antidote systems (3, 66, 67, 104, 117, 148), transposable elements (157, 178, 188), heritable microbes (62, 80, 225), and homing endonucleases (37, 38). The underlying mechanisms for these natural gene drives have inspired the creation of synthetic gene drive systems (120).
1.2. The History of Genetic Biocontrol Technologies: The Transgenic Era
The possibility of co-opting natural systems, or mimicking their behavior using engineered components, has been discussed for many years (37, 63, 64, 104, 112, 113, 129, 201). Researchers hypothesized that insect populations could be reduced through natural gene drives that biased sex ratios (63, 104, 112, 113), but technical limitations kept this work largely theoretical. Around this time, autosomal translocations and inversions were also proposed as a strategy to reduce populations (population suppression) or replace undesirable characteristics of populations (population modification) with more benign characteristics by linking genetic elements to the translocation (15, 64, 65, 137, 138, 192, 201, 227). Translocations and inversions achieve biased inheritance in a frequency-dependent manner associated with heterozygous disadvantage, also known as underdominance. However, generating translocations in the pretransgenic age required irradiation, which resulted in the creation of random mutations with inherently high fitness costs and unknown breakpoints. Additionally, with limited exceptions (65, 158), few researchers were able to successfully link a desired trait to the translocation, further limiting their development and use. More recent work has generated translocation drives in Drosophila melanogaster with defined breakpoints and transgenic cargo, but this requires newer genome engineering techniques and has yet to be developed in a nonmodel organism (30).
From the mid-1990s to the early 2000s, advancements in genetic engineering directed the discussion to the evaluation of other natural gene drive mechanisms to alter the composition or fate of populations. Transposable elements were considered (22, 26, 191) but met with little success due to difficulties in bringing about high-frequency mobilization in target species. Later, a gene drive element termed maternal effect dominant embryonic arrest (Medea) was characterized in the flour beetle, Tribolium castaneum (21), and in mice (115, 182). These elements (the molecular components of which remain unknown) bias inheritance by encoding a tightly linked maternal toxin and zygote antidote that induces lethality in offspring that do not inherit the Medea element. Theorized as potential pest control drives for population modification (26), Medea elements were engineered in two species of Drosophila (5, 31, 56). However, the paucity of genetic tools for engineering appropriately expressed toxins and antidotes has limited further development of these systems in other pest species. During these early years, RNA interference–based underdominance systems were also explored but have not been developed further (189).
Much of the current gene drive research is centered on homing endonuclease gene drives (HEGs), first described in yeast (59). Transposons and retrotransposons thrive by bringing about a direct increase in copy number. HEGs achieve a similar goal but through a distinct mechanism. HEGs encode a site-specific DNA nuclease that targets specific loci in the genome, creating a double-strand break (DSB) at the position on the homologous chromosome directly opposite its insertion site. When this happens in a germline heterozygote, and the host uses homologous recombination DNA repair, the HEG-bearing chromosome (which is not cleaved since the HEG is inserted into the target site) is copied, or homed, into the break created on the homologous chromosome, thereby bringing about an increase in HEG copy number (90). In short, homing converts a target loci (chromosome) lacking the gene drive to one that has it, thereby bringing about an absolute increase in HEG copy number in the population.
The use of homing endonucleases to control populations was first discussed in the early 2000s (37). The initial HEGs explored for gene drive development had modest homing rates and utilized existing naturally occurring HEGs (along with their specific target sequence), and so artificial nuclease recognition sites needed to be co-engineered into the population to direct the insertion of the drive (54, 55, 229, 230). This seminal work set the stage for all that has followed by showing that significant rates of germline homing could be achieved in an animal. By chance, one of these homing endonucleases, I-PpoI, has the useful characteristic of cleaving the ribosomal repeats, which are fortuitously located exclusively on the X chromosome in the predominant malaria vector, Anopheles gambiae. This has allowed for the development of sex-linked meiotic drives in this species (89). However, for the most part, the toolbox of endogenous homing endonuclease did not allow for the targeting of endogenous sequences of interest (e.g., genes essential for viability or femaleness/female fertility).
To facilitate the direct targeting of endogenous genes, site-specific nucleases were developed, using transcription activator–like effector nucleases (TALENs) and zinc finger nucleases (ZFNs) (205). These experiments made the critical point that cleavage and homing could be brought about at targets of choice. However, the homing rates observed were generally modest (for many, though not for all, applications, high-frequency homing >90% is desired). Additionally, the large size and repetitive sequence composition of these endonucleases often resulted in the loss of repeats (and thus function) during homing, leading to rapid breakdown of the element. To summarize, the difficulty in reprogramming stable versions of engineered HEGs to target endogenous genes in natural populations stalled their development. However, during this interval, work in prokaryotes on an adaptive immune system known as CRISPR was proceeding in parallel and would transform the field of cleavage- and homing-based genetic biocontrol.
CRISPR systems allow prokaryotes to mount a memory-based adaptive immune response that targets previously encountered foreign nucleic acids for destruction (25, 27, 212, 228). Multiple Cas endonucleases were identified and characterized, and soon the intricacies of how Cas endonucleases and their RNA guides (gRNAs) facilitated precise, site-specific RNA-guided cleavage of nucleic acids led to the development of a Nobel prize–winning genome engineering technology (121). Since then, Cas9-based genome engineering has become the leading genome engineering technology with broad application to almost any organism. Building off the theoretical work of Austin Burt (37), researchers recognized that Cas9 could be the programmable, site-specific homing endonuclease sought for gene drives (84). Cas9 expression and cleavage just needed to be adequately timed to ensure that drive conversion (homing) and inheritance would occur in the germline and at high frequency. Demonstration of engineered homing CRISPR gene drives (HCGDs) intuitively followed thereafter in several species (76, 92, 94, 105). These proof-of-concept experiments revealed complexities that have kept the field busy as it works toward implementing real-world applications. This review focuses on the most recent innovations, including technical advances, and challenges, including regulatory and ethical considerations involved in the development and deployment of such technologies.
2. HOMING CRISPR GENE DRIVES
In its simplest form, an autonomous HCGD consists of a germline-specific Cas9 endonuclease, linked to one or more programmable gRNAs expressed from a ubiquitous polymerase III promoter (Figure 1). The HGCD sits within its target site, thereby disrupting it. The gRNAs target the homologous chromosome at this same position, facilitating subsequent DNA repair using the HGCD as the repair template for homologous recombination, thereby increasing drive element frequency. While these are the standard characteristics of most HCGDs (76, 92, 94, 105), a number of imaginative variants have been conceptualized, and in some cases implemented, in a variety of organisms (10, 11, 74, 125, 144, 164, 166, 218). Initial work utilized Cas9 from Streptococcus pyogenes (121), simply because it was the first nuclease characterized. However, hundreds of new RNA-guided nucleases have been identified, and which type will ultimately prove most robust in specific genomic and physical environments remains to be determined (122, 136). As noted above, a typical autonomous HGCD encodes both Cas9 and the gRNA linked at the same loci (Figure 1). Alternatively, drive components can be split (known as a split drive), with Cas9 and the gRNA at separate, unlinked loci (44, 84) (Figure 1). These design differences, which can involve multiple versions of each part, sometimes working in cascades of dependency, result in a diversity of drive behaviors as predicted in models. HCGDs are being developed for two general purposes: population modification and population suppression. In the following sections (2.1-2.3), we discuss recent achievements in the design and engineering of HCGDs and limitations and future improvements needed for implementation of these technologies in the wild.
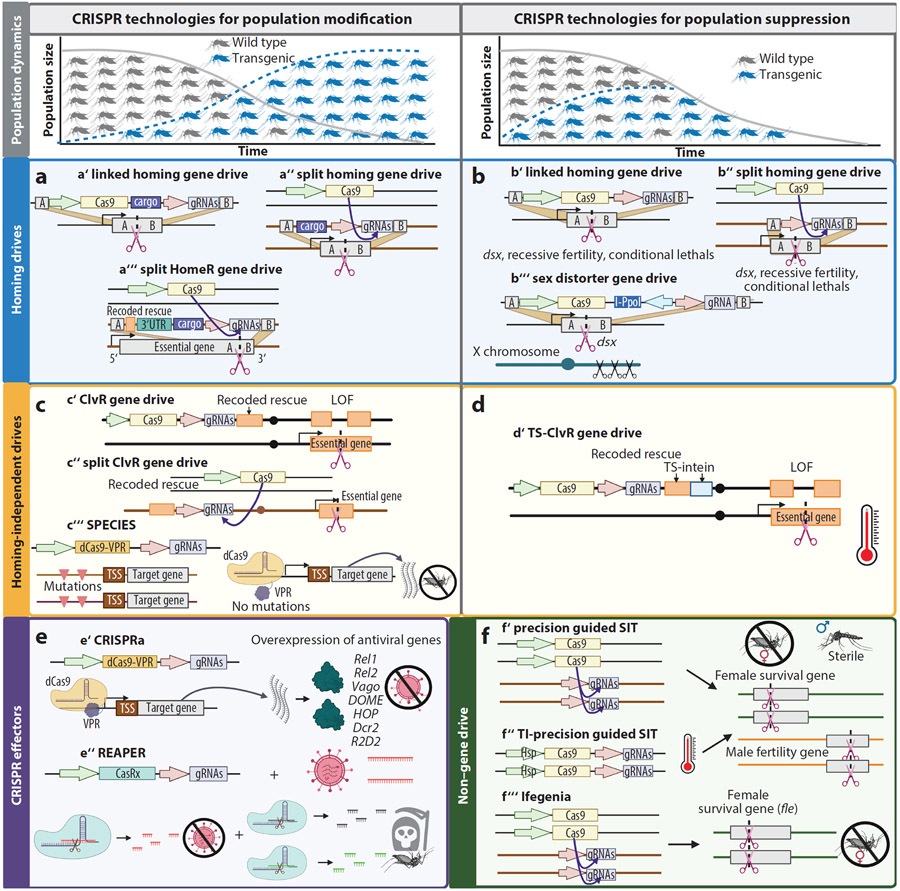
Figure 1.
Representative CRISPR technologies for the control of pest species. (Left panels) Population modification tools aim to drive desirable traits or cargo, such as disease resistance, into a population. (Right panels) Population suppression tools aim to reduce or eliminate the target population. As the transgene increases in frequency in the population, the population collapses. CRISPR technologies have been used to build (a,b) HCGDs, (c,d) homing-independent ClvR and (c) SPECIES, and (f) non–gene drive SIT technologies. (a) HCGDs for population modification typically target a unique, fitness-neutral location in the genome and then copy themselves and desirable effectors or cargo into the wild-type allele. The HomeR (a‴), for example, is a split drive designed to target an essential gene (the toxin), which is rescued by a linked cleavage-resistant version (the antidote) of the essential gene (124). HomeR was designed with a foreign 3′ UTR to prevent unintended recombination events, which can cause the generation of functional resistant alleles, and the drive was designed to target a 3′ locus in the essential gene, which minimizes the rescue recoding effort and uses the native promoter of the target gene to drive the expression of the rescue. (b) HCGDs for population suppression target either a recessive fertility gene, such as dsx, which creates sterile females when homozygous, or a conditionally lethal gene that can be driven to a high population frequency with minimal fitness load until subjected to a specific condition. HCGDs can be designed to either have both the Cas9 and gRNA gene drive elements linked at the same locus and inherited together (a′, b′) or have the Cas9 and gRNA unlinked at different loci (a″, b″) to reduce the frequency with which they are inherited together. When the Cas9 and gRNAs are unlinked, their reduced coinheritance makes the drive more spatially confinable than the linked drive, which has operational and safety considerations (see ethical considerations for low-threshold drives). Due to frequent unintended drive repair outcomes that build population resistance to a drive (see Figure 2), alternative designs are employed to reduce the formation of drive-resistant alleles in the population. For more stable and robust population suppression, the sex distorter HCGD developed by Simoni et al. (204), for example, homes into the recessive fertility gene dsx and includes a I-PpoI endonuclease to shred the X chromosome, generating infertile females when homozygous for the HCGD and biasing the population toward males (b‴). (c,d) Homing-independent methods have arisen in parallel to work on HCGDs. A TS ClvR drive (d′) contains a Cas9 and gRNA, which cleave a haplosufficient essential gene, and a linked rescue that is a recoded, cleavage-resistant version of the essential gene that also encodes a TS intein that disrupts the rescue at high temperatures (176). In this system, low temperatures allow the intein to splice out of the rescue gene for normal growth and drive fixation in the population, which is then disrupted at high temperatures when the intein is not spliced, so the essential gene is not rescued. Without this temperature specificity, ClvR can also be designed in a linked (c′) or split (c″) configuration to support population modification. The extent of component linkage can dictate the threshold, duration, and spread of the drive. The SPECIES system (c‴) uses a catalytically inactive version of Cas9 with a transactivator (dCas9-VPR) to cause untimely and lethal overexpression of an essential developmental gene. Lethality can be prevented by coinherited mutations that protect the target site from dCas9-VPR binding and lethal overexpression. (e) CRISPR-based effectors have been built to upregulate immunity genes (CRISPRa, e′) to decrease pathogen infection or to directly target and destroy RNA viruses and kill mosquitoes (REAPER, e″). When linked to a gene drive, these can replace a population that can transmit a pathogen with one that cannot sustain disease transmission. (f) pgSIT generates sterile males by simultaneously targeting genes required for male fertility and female survival early in embryogenesis (126, 128, 143). pgSIT uses the CRISPR Cas9 and gRNA components as either separate lines that generate sterile males only when crossed (f′) or a single line where the Cas9 expression is temperature inducible (TI-pgSIT, f″). In the TI-pgSIT system, Cas9 is expressed by an Hsp promoter and thus only at high temperatures (123). Ifegenia (f‴) is another binary non–gene drive CRISPR-based population control technology where separate Cas9 and gRNA lines are crossed to kill female offspring through the disruption of the fle gene (206). Here, male offspring harbor the female-killing mutation, which can lead to killing of any female offspring that result from their mating with wild females. Abbreviations: ClvR, Cleave and Rescue; CRISPRa, CRISPR activator; dsx, doublesex; fle, femaleless; gRNA, guide RNA; HCGD, homing CRISPR gene drive; HomeR, home and rescue toxin-antidote-based drive; Hsp, heat shock protein; ifegenia, inherited female elimination by genetically encoded nucleases interrupting alleles; LOF, loss of function; pgSIT, precision-guided sterile insect technique; REAPER, vRNA expression activates poisonous effector ribonuclease; SIT, sterile insect technique; SPECIES, synthetic postzygotic barriers exploiting CRISPR-based incompatibilities for engineering species; TI-pgSIT, temperature-inducible precision-guided sterile insect technique; TS, temperature sensitive; TSS, transcription start site; UTR, untranslated region; VPR, VP64-p65-Rta. Figure adapted from images created with BioRender.com.
2.1. CRISPR Homing Drives for Population Modification
Population modification strategies modify and maintain the target population. In this context, the spread of an HCGD can lead to an increase in the frequency of linked cargo genes, a form of population modification. Multiple population-modifying HCGDs have been evaluated in the laboratory in insects (2, 10, 41, 91, 94, 124, 125, 144, 184, 190, 220), fungi (203), yeast (16, 76, 98, 141, 193, 194, 234, 235), and even rodents (102, 135, 183, 224). In one application, to reduce the transmission of mosquito-borne diseases, multiple genetic elements, known as effectors, have been developed. These can, in principle, be linked to gene drives to generate disease-resistant populations.
A number of effector strategies have been developed in mosquitoes, including RNA interference (28, 85, 86, 145, 156, 195, 237), single-chain antibodies (29, 69, 238), peptides (116, 163, 239), toxic proteins (19), or upregulation of immune factors (77). Recently, CRISPR has expanded this toolbox to include antiviral effector development. For example, a CRISPR activator (CRISPRa) system engineered in the predominant dengue vector Aedes aegypti was used to bring about the over expression of a gene involved in the regulation of viral infection, Rel1, to reduce dengue viral titer by over fivefold (33) (Figure 1). With further development, the CRISPRa system could be used to harness other endogenous antipathogen effectors to reduce disease transmission. Moreover, an RNA-targeting CRISPR system was used to target chikungunya (CHIKV) viral RNA (vRNA) through a process termed vRNA expression activates poisonous effector ribonuclease (REAPER) (68) (Figure 1). The REAPER system leveraged the collateral cleavage activity of the RNA-targeting Cas13 ribonuclease, which increased mosquito mortality in the presence of the virus. Even small reductions in mosquito survival rates can have a large impact on disease transmission (149), so antipathogen effectors that reduce the survival of infected individuals could be significant. These technologies provide tools for developing antiviral effectors to support population modification, and new technologies in the pipeline should push this further.
2.2. CRISPR Homing Drives for Population Suppression
Population suppression gene drives aim to impart a fitness load on the population to reduce its size and/or eliminate it, at least locally. This can occur when the HCGD inserts itself into a haplosufficient gene required for viability or female fertility. To summarize briefly, if the rate of spread of the HGCD outpaces the fitness cost induced on the population, the population can be driven to an overall unfit state, resulting in its stochastic elimination (75). Important examples, implemented in mosquitoes, involve driving the population to a state that consists of fertile homozygous males and sterile homozygous females. Laboratory experiments have even eliminated populations when genes required for femaleness are targeted (42, 105, 133, 236). HCGDs have also been coupled with mechanisms that distort the sex ratio, which also resulted in population suppression in laboratory mosquitoes (204).
2.3. Technical Considerations for CRISPR Homing Drives
DNA repair can occur through several different pathways. Only one of these, homology-directed repair (HDR), brings about an increase in HCGD frequency when the HCGD is copied into a cut site on the homologous chromosome (Figure 2). Other end-joining (EJ) pathways, which seal the break, compete with HDR, and to the extent they are chosen, lower homing rates. They are also error prone and can result in mutation of the target site (Figure 2). Along with a failed insertion of the drive, if these alterations to the target site make it unrecognizable to the drive-encoded gRNAs (but do not disrupt target gene function), then individuals that inherit these alleles will also by definition be resistant to the drive. As these resistant alleles accumulate over time, the population will become resistant to the drive, which (if the presence of the drive results in a fitness cost to carriers) will ultimately eliminate the drive from the population (79, 81, 107, 216). This is a universal problem across HCGDs, as the generation and accumulation of drive-resistant alleles in populations have been experimentally demonstrated in nearly all HCGD studies and organisms (2, 41, 51, 92, 94, 105, 125, 133, 144, 184, 204).
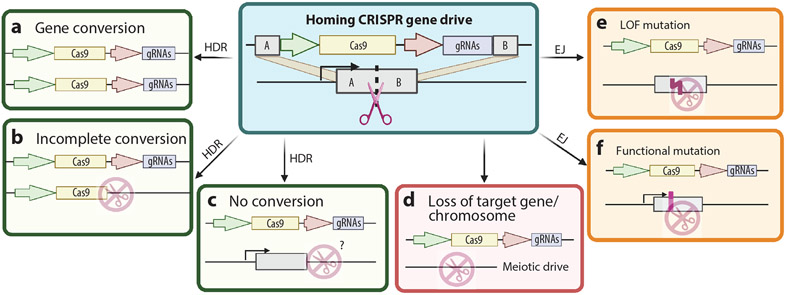
Figure 2.
HCGD outcomes. A simplified HCGD is shown using Cas9 and gRNAs to disrupt a gene via DSBs (teal box) that are repaired by HDR (green boxes) or EJ (orange boxes) but do not include cargo (e.g., resistant genes or recoded target gene). Alternatively, the repair process can fail altogether, resulting in the loss of a large chromosomal fragment (red boxes). (a) After cleavage of the target site, the HDR pathway uses the gene drive element as a template to repair the DSB, thereby copying the gene drive into the locus of the wild-type gene and biasing (increasing) the inheritance or copying of the gene drive. (b) Loss of parts of the HCGD can also occur during conversion when an alternative region of the gene drive element other than the homology arms is used as the template for the gene repair, such as part of the gRNA sequence, or as a result of internal recombination between the drive components. The chances of these events occurring increase with the size of the HCGD and multiplexing of gRNAs. Since the insertion site is still disrupted, such events would generate a drive-resistant allele that cannot be further cut by the HCGD. (c) No conversion (copying) may occur if the target site is not cleaved or the HDR process uses an alternate repair template, which could happen if the target locus has high homology to other regions in the genome; then the wild-type allele will be susceptible to cleavage and drive in subsequent generations. Otherwise, if the target site is repaired by a nonallelic template with high homology to the cut site it may be resistant to subsequent cleavage. (d) If a repair mechanism fails to repair the DSB, then the target gene, or in some cases the entire wild-type chromosome, may be lost. While this means the gRNA target site is no longer present, only gametes that inherit the drive will likely be viable since the wild-type gametes will be missing a chromosome or the target gene. This biased inheritance of gametes with the drive element is known as meiotic drive. (e,f) DSBs repaired by the EJ pathway can result in mutations at the target sites that make the next generation resistant to drive cleavage. (e) These mutations can cause a loss of gene function, which can cause the death of any offspring inheriting that mutation in a haploinsufficient gene. (f) If the mutation does not result in the loss of gene function, then the mutation inherited by the next generation will be resistant to drive cleavage. As these resistant alleles accumulate in a population over time, the drive will go extinct. Abbreviations: DSB, double-strand break; EJ, end joining; gRNA, guide RNA; HCGD, homing CRISPR gene drive; HDR, homology-directed repair; LOF, loss of function. Figure adapted from images created with BioRender.com.
Some steps are being taken to reduce the inevitable formation of resistance alleles. One focuses on bringing about germline-specific Cas9 expression, as the HDR pathway is favored in the germline. However, in experiments using promoters that were presumed to be specific for the germline, such as vasa (94, 105) and nanos (51, 125), leaky expression occurred in somatic cells. Non-germline expression of Cas9 is especially problematic when it occurs early in embryogenesis or results from maternal deposition. The early embryo, which also includes nuclei that will give rise to the next generation’s germline, has a reduced capacity for HDR as compared with EJ. Thus, Cas9 activity in the early embryo can lead to, at high frequency, heritable germline resistance alleles (42, 51, 107, 125, 172). Much effort is therefore going into the identification of promoters with stricter germline expression and limited maternal carryover. For example, recent studies using the zero population growth promoter have shown improved population suppression in an A. gambiae drive (106). Other recent work showed that the identity of the Cas9 promoter and the timing of its expression were critical for improving rates of germline homing in rodents (135). Moreover, drives developed in A. aegypti with moderately high drive transmission rates (144, 190, 220) were later improved through the use of different germline promoters driving Cas9 expression (10). In addition to testing germline-specific promoters, using multiple gRNAs (i.e., gRNA multiplexing) targeting multiple sites in the same gene can delay the production of drive-resistant versions of the target gene since multiple target sites would need to be converted to a functional but resistant state (44, 84, 152, 167). This strategy has been demonstrated to reduce or eliminate drive-resistant alleles but may come with a cost in terms of lower homing efficiency (11, 50, 172).
As more experiments are performed, the importance of other genomic variables is becoming clear. For example, the choice of the target gene into which homing is occurring matters (10). Cas9 expression from a given promoter can also be subject to what are known as position effects: Different genomic contexts, and more generally diverse genetic backgrounds, result in somewhat variable patterns of expression. These effects partially explain why rates of resistance allele formation of the same gene drive element, at the same locus, can vary by genetic background (51). Finally, it is also important to note that there are resistance allele–independent ways in which features of Cas9 expression can negatively affect drive. First, expression of Cas9 itself can be toxic to cells in some contexts (9). Second, disruption of genes other than the intended target (i.e., off-target effects) may result in unexpected fitness costs (136).
Drive performance also depends on the selection of the target gene and site (181). However, given the enormous diversity in natural populations, and the fact that new variants are continuously being created through mutation, predicting the integrity of a target site in the field is difficult. One way to address this issue involves utilization of highly conserved sites in an essential gene, where mutation to a resistant allele sequence is likely to result in a large fitness cost to carriers (50, 133). As an example, a large number of genomes (>1,000) were evaluated to identify a highly conserved intron-exon boundary within the sex determination and fertility gene, doublesex (dsx). This has been used as a target for Cas9 to build a population suppression drive in laboratory A. gambiae (133). Resistant alleles have been identified (108, 204), but these have not been observed to spread in lab cage populations undergoing suppression, suggesting that they result in large fitness costs. It remains to be determined whether high fitness-resistant alleles appear in further tests with genetically diverse, larger populations.
HDR can also cause other unintended drive outcomes (Figure 2). For example, internal recombination and partial homing during HDR can result in the imprecise copying of the drive element (219). This would be particularly problematic if the HCGD was carrying a cargo transgene to prevent disease. Incomplete transfer of intact drive elements has been identified in numerous laboratory experiments (42, 50, 51, 105, 172, 184) and would have similar deleterious consequences for population modification (Figure 2). These outcomes impact the dynamics of drive persistence and spread of the effector in the population. There is also evidence that failed DNA repair can lead to the loss of the recipient chromosome (103, 214, 233) (Figure 2). Finally, we cannot rule out that upon the release of HCGDs at larger scales in wild populations, other unintended but rare errors in drive copying may become larger issues, including mitotic recombination or meiotic drive (233) (Figure 2). That said, just because a consequence is unanticipated does not mean it is not useful if it still allows the drive to occur.
Studies have shown that females harboring the drive element can maternally transfer the Cas9 protein and gRNAs (i.e., maternal deposition) to embryos that did not inherit the drive itself. Depending on the system, this can hinder (e.g., during homing) or promote [e.g., in toxin-antidote (TA) systems] drive. In the context of homing, maternal carryover can also result in a phenomenon known as shadow drive, in which homing occurs in individuals who inherited only the gRNA drive element due to maternally deposited Cas9 biasing the inheritance of the drive (46, 103, 125, 214). This effect is unpredictable and typically occurs at the expense of overall drive performance. The outcome and effects of maternal deposition are closely linked to the timing of Cas9 expression (70, 128, 219). Shadow drive typically does not serve as a primary means of inheritance bias, but instead alludes to the challenges in tracking the mechanism of biased inheritance. In some studies that have marked chromosomes, it was discovered that HCGDs can bias inheritance by homing, and also through meiotic drive due to chromosome damage (233). In most cases, though, the mechanism of drive inheritance bias has not been tracked, which may inflate reported homing efficiencies. Due to these uncertainties, further characterization of these HCGDs is required to shed light on the mechanism of inheritance bias, which can vary by design, target gene, target locus, species, and strain.
2.4. Toxin-Antidote CRISPR Homing Gene Drives
Given the challenges associated with HCGDs, there has also been a focus on developing HCGDs that promote the removal of individuals who inherit cleavage-resistant alleles or do not inherit the drive. Proof of concept for several of these drives demonstrated their applicability for population modification (52, 124, 214). For example, the home and rescue TA-based drive (HomeR) is a potent drive designed to target an essential gene (toxin), which is rescued by a drive-linked cleavage-resistant version (antidote) of the essential gene (124) (Figure 1). HomeR is positioned in the genome opposite its target to enable homing to bias its transmission. To streamline development, HomeR was designed to target a 3′ sequence in the essential gene. This minimizes the rescue recoding effort and uses the native promoter of the target gene to drive the native expression of the rescue. This approach (to the extent that cleavage and non-HDR repair result in loss of essential gene function) suppresses the accumulation of functional resistance alleles from the population. Predictions for this drive indicate that it would have longer-term stability in large, diverse wild populations as compared to other population modification drives. In summary, HCGDs such as HomeR that utilize a TA approach to select against drive-resistant alleles may provide the durability needed for long-term, stable HCGD-based population modification. However, these remain to be developed in important pest species.
2.5. Outlook and Overview of Homing CRISPR Gene Drives
While significant progress has been made toward understanding and improving HCGDs, success will ultimately be determined by the biology of specific species, which differ dramatically in terms of frequency of HDR, conversion frequency, tract length, and mechanisms of inheritance bias. A growing body of work indicates that we have only scratched the surface in terms of understanding what happens in vivo. These caveats notwithstanding, the field is on the cusp of carrying out experiments that may determine whether current technologies are sufficient to accomplish shortterm HCGD-mediated population modification or suppression in the wild.
3. NONHOMING CRISPR GENE DRIVES
Many issues with HCGDs arise from the variable homing rates used to bias inheritance. By contrast, essentially all of the HCGD studies discussed above note that Cas9 cleavage rates are reproducibly high (128). Building from the TA drive concept commonly found throughout nature (23, 171, 200), researchers are developing next-generation gene drives that do not rely on homing but instead focus on exploiting highly efficient CRISPR cleavage. These drives focus on two simple ideas. The first is that Cas9 and other site-specific DNA-modifying enzymes can be used to create loss-of-function (LOF) alleles in an essential gene by targeting it for sequence modification (usually cleavage) at multiple positions—the basic idea being that it is generally easy to break gene function through mutation. The second is that versions of the essential gene that are functional and resistant to modification can be created through recoding. When these two activities are combined (modification and rescue), strong frequency-dependent drive can be achieved. Here, we discuss the use of cleavage for homing-independent gene drives, and in Section 5, we discuss the use of cleavage for the next generation of non–gene drive genetic biocontrol technologies.
3.1. Homing-Independent Cleave (or Otherwise Modify) and Rescue Drives for Stable Population Modification
Cleave and Rescue (ClvR) selfish genetic elements (173-177), also referred to in some implementations as TA recessive embryo (TARE) (49, 159), consist of two tightly linked components that sit at a fixed chromosomal position. The first is a DNA sequence–modifying enzyme such as Cas9 and multiple guide RNAs (gRNAs) (known as the Cleaver). These are expressed in the germline and act to disrupt—through cycles of cleavage and end joining that continue until the target site is destroyed—the endogenous version of a haplosufficient essential gene, wherever it is located. Maternal carryover of Cas9 and gRNAs also results in disruption of the paternally provided wild-type essential gene allele. The hope is that inaccurate repair at multiple positions will lead to the creation of LOF alleles (i.e., the potential toxin). gRNA-guided base or prime editors (13) could perform a similar function. The core idea is that a highly conserved essential gene can be easily broken through targeted mutation. The second component is a recoded version of the essential gene resistant to cleavage and retaining function that is expressed under the control of regulatory sequences sufficient to rescue the recessive LOF phenotype. This (i.e., the antidote/Rescue) acts to guarantee the survival of those who carry it. In such a system, LOF alleles perform their toxin function when they are present in homozygotes, and the individual lacks another source of essential gene function, such as a copy of ClvR/TARE. By contrast, those who inherit ClvR/TARE and its associated rescue always survive, provided a haplosufficient gene has been targeted for LOF allele creation. In this way, as with many other TA-based selfish genetic elements (36, 39), ClvR/TARE spreads by causing the death of those that lack it (Figure 1). Thus, ClvR/TARE drives not only circumvent the homing-associated issues of HCGDs, but they are addictive in that once they are driven to fixation in a population and completely replace the endogenous version of the haplosufficient essential gene, the presence of the recoded rescue becomes necessary for the population to persist. Individuals that lose any part of the drive or generate resistance mutations are selected against, thus solidifying the presence of ClvR/TARE in the population (173, 175).
Depending on the chromosomal details of where the ClvR/TARE element and its target gene are located, spread goes to genotype (all individuals carry at least one ClvR/TARE allele at the locus) or allele (all alleles at the locus carry ClvR/TARE) fixation. The drive is frequency dependent (very slow when rare and fast when common) and lacks an introduction threshold in the absence of fitness cost but comes with a threshold when the presence of ClvR/TARE results in a fitness cost to carriers and/or a haploinsufficient or haplolethal locus is targeted (47, 49, 159, 173-177).
While the ClvR/TARE drive has only been demonstrated in D. melanogaster, it has many features that make it amenable to transfer to other species. Other TA systems, such as Medea drives (5, 31, 56), involve more technically complicated components and rely on the strict timing of TA element activity to ensure killing only occurs under the desired conditions. By contrast, the ClvR/TARE toxin utilizes a flexible CRISPR technology that has been adapted to many organisms; the recoded rescue system is just a functional copy of the target gene, which again, is a simplified design compared to other TA systems. ClvR/TARE has also been shown to work for multiple highly conserved genes. Therefore, unlike other TA systems, ClvR/TARE is likely more versatile, and its implementation into target species more attainable. The ClvR/TARE design also enables multiple gRNAs to be utilized as toxins, thereby significantly decreasing the likelihood of manifesting resistant alleles that restore the function of the target gene.
A number of different self-sustaining ClvR or TARE elements have been implemented in Drosophila (47, 49, 159, 173-177) and shown to spread to transgene fixation. Others have fared less well due to difficulties ensuring efficient rescue of the LOF phenotype (57). ClvRs have also been created in a split drive configuration, in which the key components required for cleavage—Cas9 and gRNA—are in different places in the genome (also distinct from the target gene). This split configuration results in a strong but self-limited drive. Drive strength and duration can be further tuned by adjusting the degree of linkage of Cas9 and gRNAs, with tighter linkage resulting in a drive whose strength can approach a self-sustaining ClvR, while still ensuring that drive is always self-limiting (177). Finally, work on ClvR has provided a way to address the inevitable problem that genes needed for population modification, the cargo, and the drive mechanism, can separate, mutate to inactivity, and lose efficacy over time. In the face of these forces (relevant to any modification drive mechanism), strategies are needed for carrying out cycles of population modification, with first-generation elements being displaced in favor of new ones. A ClvR-based strategy involves placing two different ClvR elements targeting different essential genes at the same genomic position but on different homologs. The second-generation element includes a rescue for the first-generation element already established in the population. When the second-generation element is released into a population carrying a first-generation element, the former spreads to fixation, while the latter is removed (174).
Finally, we note that the core ideas of creating LOF alleles in essential genes and rescuing the lethal consequences of their presence can also be applied (so far only in models) to generate a wide diversity of related gene drive elements that differ in mechanism (e.g., killing the other or underdominance), intrinsic introduction threshold, the ability to spread in the face of migration, and tissues in which the killing that mediates the drive occurs—the zygote, gametes, or daughter cells (45, 47, 53, 161, 173). Much remains to be explored with these systems, which may also prove useful in population suppression (see Section 3.2).
3.2. Homing-Independent Cleave and Rescue Drives for Population Suppression
Population suppression drives operate by imposing a fitness load on the population. In the case of HCGD, this requires high homing rates (97). However, many (though not all) HCGDs have homing rates that are far too low to be useful for suppression (42, 72, 97, 107). ClvR-based approaches provide alternatives. For example, the ClvR drive was engineered to target a haplosufficient essential gene as described for population modification, though here the recoded rescue coding region was engineered to contain a temperature-sensitive intein (Figure 1d). At permissive (low) temperatures, the intein is spliced out, creating a functional version of the essential protein. However, at elevated temperatures, the intein is not removed, and the protein encoded by the essential gene is nonfunctional. The drive to fixation with such an element at low temperatures results in the population becoming addicted to temperature-sensitive rescue, since it provides the only source of essential gene function. Thus, when the temperature is raised beyond a threshold, a population crash occurs (176). Similar to the other ClvR designs, this approach is relatively resistance proof, since mutations that block functionality of the intein would likely still result in the loss of rescue function and therefore be eliminated from the population.
Temperature inducibility is an innovative method for bringing about conditional suppression (5), but it remains unclear if there are real-world contexts in which the temperature fluctuations in the environment match those of the engineered intein. That said, a similar logic, in which rescue activity is conditionally blocked, could bring about species-specific suppression in response to other stimuli, such as a small molecule or a virus that disrupts essential gene function at the RNA or protein level. Conditional rescue activity could also be used, in the context of simple population modification, to eliminate only cells or individuals infected with a virus (68), if the presence of viral components (such as a protease) led to inactivation of an engineered version of the essential gene (176). Other approaches to bringing about cleave and rescue population suppression have also been envisioned but not yet implemented (47).
3.3. Homing-Independent Murine Meiotic Drives
Efforts are also underway to leverage naturally occurring gene drives for population suppression. The mouse t haplotype provides an example. The t haplotype spans a large autosomal chromosomal region, with little recombination. When present in heterozygous males, sperm that lack the t haplotype are disabled, resulting in the transmission of t to progeny males and females at rates that can exceed 95%. Homozygotes for t are male sterile and female fertile. Recent modeling and experiments characterized a situation in which a male germline–expressed Cas9 and gRNAs targeting the prolactin gene (required for female fertility) are inserted into the t haplotype. In this hybrid element, tCRISPR, Cas9, and the gRNA cleave and create LOF prolactin alleles. The goal is for t-based segregation distortion in males to push the Cas9/gRNA cassette to high frequency. The latter will continuously produce LOF alleles at the Prl locus. The hope is that the combination of t-based drive and accumulation of Prl LOF alleles will drive the population to an unfit state that contains a high frequency of infertile homozygous Prl mutant females and some frequency of infertile homozygous t males (96). Proof-of-principle experiments support this approach. A Prl-targeting gRNA was incorporated into the t haplotype, with Cas9 present on another chromosome. Segregation of the gRNA was strongly biased in males and when paired with Cas9 brought about high-frequency cleavage of Prl in the male germline. These results represent a notable advancement, as mammalian gene drives based on homing have been more difficult to develop, often with very low drive conversion rates (102, 135, 183, 224).
3.4. Homing-Independent CRISPR-Based SPECIES Drive
High-threshold gene drives are interesting because they will be actively eliminated from the population unless present above some threshold frequency, providing some degree of control. There are many non-CRISPR examples of high-threshold drives (6, 44, 139). In these drives, heterozygotes (or their progeny in the case of translocations) have low fitness, while homozygotes have high fitness. CRISPR has facilitated the development of extreme underdominance systems, such as engineered genetic incompatibility (EGI) (154, 155), wherein crosses between homozygous EGI individuals and wild type result in inviable progeny. This reproductive barrier was built by coupling a dominant lethal effector and a recessive resistance allele. A related CRISPR-based extreme underdominance system, synthetic postzygotic barriers exploiting CRISPR-based incompatibilities for engineering species (SPECIES), uses a unique genetic breeding scheme to engineer the system and demonstrates threshold-dependent drive capabilities (32) (Figure 1). The SPECIES approach, however, may be easier to transition to other species and may be more evolutionarily stable than EGI since it uses a genetic process for identifying nondeleterious rescue indels and is multiplexed to target multiple genes, ensuring that if resistant mutations arise in one target gene, lethality is maintained due to overexpression of the others. Future work remains to develop EGI/SPECIES approaches in pest species.
4. TECHNICAL AND ETHICAL CONSIDERATIONS FOR IMPLEMENTING GENE DRIVES IN THE FIELD
This review has focused on the intended effect of gene drives on populations (modification or suppression) as well as their design and efficacy. As the genetic modification of entire populations of organisms comes with technical and ethical considerations, this section focuses on the broader implications of using drives to address pest species and the challenges associated with implementation.
4.1. Broader Population, Ecological, and Evolutionary Considerations for Gene Drives
There are several broader technical challenges related to the intended outcomes of gene drives. For example, since suppression gene drives impart a fitness load on the population, the homing frequency must exceed the fitness effects incurred by the drive for the drive to reach fixation, meaning that suppression drives require higher homing rates than population modification strategies (97). Unfortunately, these high population fitness costs also increase the selection for mutations that prevent suppression drives from doing their job (37, 110).
These challenges are further impacted by ecology, population genetics, and population structure (75). Inbreeding, for example, can impact the performance of suppression drives (34, 35, 37, 48, 84). The evolution of drive resistance can also be impacted by other compensatory alterations in nontargeted parts of the genome, or other mechanisms of resistance not directly linked to the activity of the drive. Susceptibility to resistance will vary greatly depending on the fitness costs incurred by the drive element and population structure (61). These challenges impact all suppression drives to some degree, and the effect of these will vary by locus, population, species, and strain, which further complicates predictions of drive fate.
Conversely, population modification gene drives may be less susceptible to the evolution of resistance than population suppression ones, but drive-associated fitness costs can still slow or halt their spread (216). Also, pathogens can evolve resistance to the antipathogen effector cargo linked to the gene drive (153, 221), though it may still be possible for these effectors to persist long enough to disrupt disease transmission and temporarily improve public health (20). Only limited laboratory studies of drive dynamics and performance have included antipathogen effector cargo (40), so knowledge about the efficacy and stability of these drive-linked effectors and their impact on drive fitness and stability is still deficient. The complexities involved are highlighted by a recent study in A. aegypti that evaluated drive integration sites previously optimized for effector efficacy and found they were not optimal for drive persistence (190). The technical approaches discussed above may mitigate the development of drive resistance in the population. However, much remains to be learned about how the local genomic environment controls the long-term efficacy of the drive and effector.
4.2. Frequency-Dependent Dynamics and the Invasiveness of Gene Drives
One critical ethical and regulatory dilemma for gene drive technologies is centered on drive confinability. One main consideration is the frequency-dependent dynamics of the drive, which reflect the threshold required for the drive to reach fixation. A gene drive with a low threshold could start at low frequencies and still be expected to reach fixation, making them relatively invasive (44, 99, 139, 186). Examples of low-threshold drives include linked HCGDs, which are frequency independent and predicted to rapidly go to population fixation and spread widely to neighboring populations. By contrast, while some TA-based drives such as ClvR and Medea can be low threshold in the absence of fitness costs, they are always frequency dependent (very weak at low frequency and only strong at higher frequencies that are system dependent) and will usually have a significant threshold if fitness costs are present. Which drive type is appropriate depends on the ecology, as well as social and regulatory concerns regarding spread outside the intended target area. In any case, low-threshold drives of either type are predicted to be challenging to remove in favor of a nontransgenic wild type, once they are established in the environment.
As high-threshold drives are (depending on migration rate) less likely to spread beyond the local target population, they are more spatially confineable and therefore controllable (44, 150, 151, 196). These drives do have higher labor and resource costs, as large numbers of drive-bearing individuals will need to be released, likely iteratively, to surpass the introduction threshold. This threshold provides an important safety feature, though, as drives could be called back by releasing large numbers of wild-type individuals to push the population frequency of the drive back below the threshold, leading to its removal from the population through natural selection. Developing gene drives with higher-frequency thresholds could be a desirable approach when, as in most cases, it is not reasonable to have global modification or elimination of all populations of a target species. That said, assuming core commitments such as ethical, efficacy, and safety standards are met and regulatory and social acceptance is established (146), an exception is—perhaps—the case of Anopheles vectors, the malaria they perpetuate, and HCGDs designed to replace or suppress their populations. These drives are low threshold and self sustaining and are thus expected to persist continually and spread beyond the initial release area, perhaps to the entire habitable range of the target species (165). These predictions are based on the fitness costs associated with the drive and imposed on the carriers observed in the lab. Thus, it is possible that the drive threshold in the wild may be higher than predicted for many gene drives typically classified as low threshold, such as Medea; linked HCGDs; and homing-independent CRISPR-based TA drives, such as ClvR (8, 73, 75, 173, 210, 222). It is also important to note that the fitness of the drive-carrying individuals can be regulated by environmental factors, so the magnitude of the threshold frequency can vary by environment and be difficult to predict (14, 75).
As alternatives to gene drive, some other non-self-sustaining technologies may be feasible for some applications. These include more traditional sterile insect releases or self-limiting drives, such as split (144, 175) or daisy (166, 218) drives, which can spread to fixation but are ultimately self-removing from the population (albeit complete loss due to natural selection may take a very long time). As an example, a recent study that directly compared linked versus split HCGDs at the same locus noted that the population dynamics were similar (213). Interestingly, though, for a wider spread and longer-term function of the drive, this study also demonstrated a strategy for converting a split HCGD to a fully linked drive (213). Perhaps a confineable split drive could be released first and later transitioned to a linked drive for a larger-scale application (185). Finally, there is evidence that many HCGDs bias inheritance through homing and meiotic drive mechanisms (220, 233). In a scenario where HCGDs were inherited solely by homing, only gRNA inheritance would be biased, but meiotic drive would bias both the Cas9 and gRNA components (220). The relative frequency of these different events needs to be further explored in target species.
4.3. Reversing and Replacing Gene Drives in the Field
Multiple call back measures have been suggested to remove gene drives from the environment (84, 217). Some of these are anti-CRISPR proteins that inhibit Cas9 function and thereby drive (18, 211). Other approaches involve the use of small molecules to modulate the activity of gene drives (70, 84). Neutralizing genetic elements have been proposed to remove a drive from the wild; however, these create unintended recombination events and leave a transgenic residue behind, creating second-order scientific and regulatory concerns (93, 233). As noted above, high-threshold drives for population modification can often be eliminated following dilution below the introduction threshold by wild types. This provides a way to return to a nontransgenic population but comes with the increased costs of implementation. Suppression systems are generally low threshold and thus not amenable to removal through simple dilution.
4.4. Modeling the Transition of Genetic Biocontrol Projects from Lab to Field
Mathematical modeling can facilitate understanding of CRISPR-based biocontrol tools by quantifying their molecular mechanisms and extrapolating them to the population scale. Malaria vectors provide a case study in which some of the most sophisticated modeling of genetic biocontrol systems has been conducted (Figure 3). Malaria incidence has plateaued at unacceptably high levels since the wide-scale distribution of interventions that began in 2000 (24, 232), and gene drive–modified mosquitoes are seen as one of the most promising tools for achieving continued reductions in transmission. As potential field trials edge closer, models with increasing levels of detail have been developed for specific gene drive systems intended for release in specific environments (140, 169, 170), and a range of logistical matters arise that stand to benefit from modeling. Regulatory approval for environmental releases will depend on demonstrations of safety and efficacy, both of which require modeling to extrapolate knowledge of molecular mechanisms to the population level. To address efficacy, modeling can inform target product profiles (TPPs) that determine parameter value ranges for which a product is likely to have its desired impact (119, 162), and, to address safety, some questions in an environmental risk assessment will be suited to modeling analyses, for example, the risk of releasing an only transiently successful genetic biocontrol system that would reduce malaria immunity in the human population and result in increased susceptibility and incidence upon technology failure (60, 114). Models can also inform the design of field trials and interventions (140, 169), requirements of monitoring efforts to assess gene drive establishment and persistence, and requirements of surveillance programs to detect either unintended spread of gene drive alleles beyond a trial site or drive-resistant alleles within a trial site (187).
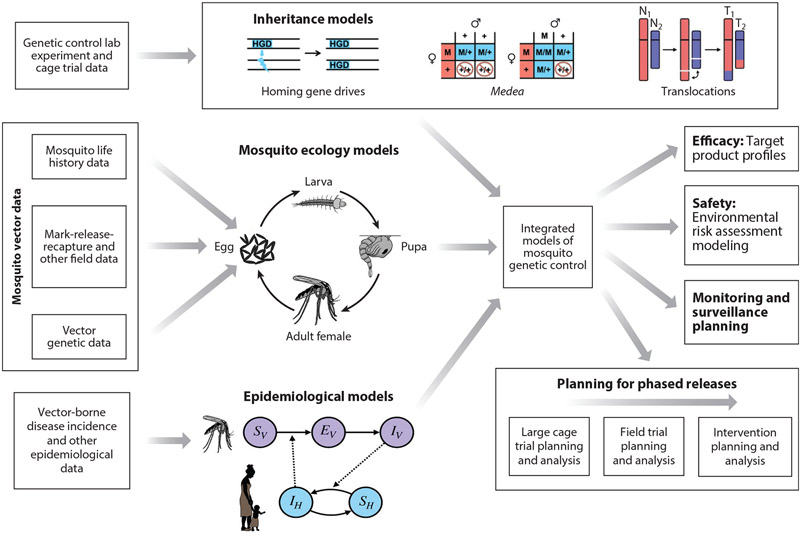
Figure 3.
Integrated models of mosquito genetic control. CRISPR-based genetic control tools are mainly applied to mosquito disease vectors, particularly malaria vectors, due to the huge global health burden of vector-borne diseases, and the potentially transformative role genetic control tools could play in their suppression. As genetic control projects progress from lab to field, models may be used to combine data from disparate sources, including lab, field, and epidemiological studies. These data inform models describing inheritance, population dynamics, and disease transmission, which may be combined into an integrated modeling framework. This integrated framework, or components thereof, may address a range of project-related modeling questions regarding product safety and efficacy and the planning and analysis of cage trials, field trials, and interventions. Pictured here are schematics for (a) homing gene drives (HGDs), which bias inheritance in their favor by converting a heterozygote into a homozygote via cleaving the wild-type allele and then serving as a template for homology-directed repair; (b) Medea, which biases inheritance in its favor through the action of a maternal toxin and zygotic antidote, rendering wild-type (+/+) offspring of heterozygous (M/+) females unviable; and (c) chromosomal translocations (T1 and T2), which result from mutual exchange between terminal segments of two nonhomologous chromosomes (N1 and N2) that provides a selective disadvantage for translocation heterozygotes and threshold-dependent dynamics. Also depicted are (a) the lumped age-class model of mosquito population dynamics, in which the egg, larval, pupal, and adult life stages are modeled, with density-dependence often incorporated at the larval stage and (b) the Ross–Macdonald model, which is the simplest model of reciprocal transmission of malaria between humans and mosquitoes. In the Ross–Macdonald model, adult female mosquitoes emerge susceptible (SV), become exposed/latently infected (EV) at a rate proportional to the number of infectious humans in the population (IH), and progress to infectiousness (IV) at a rate determined by the extrinsic incubation period. Susceptible humans (SH) become infected/infectious at a rate proportional to IV and recover to become susceptible again.
With these applications in mind, what is included in each model will depend on the question to be addressed (Figure 3). The early exploratory models of genetic biocontrol systems focused on population genetics, that is, how allele frequencies change over time as a function of inheritance processes. These models can describe the qualitative dynamics of inheritance-biasing systems in a range of species (37); however, as technologies edge closer to field applications, species-specific ecological models become paramount (87). For mosquito vectors of malaria, two key features to consider are density dependence in the mosquito life cycle and mosquito movement behavior. Density dependence is particularly relevant for population suppression systems, as it determines how a population responds to a reduction in size. Mosquito movement is relevant to both population suppression and replacement strategies, as in both cases, accurately predicting how transgenes spread spatially is essential. Finally, models of malaria transmission are becoming increasingly relevant as readiness for field trials will likely be determined by a TPP that includes a predicted epidemiological impact (119, 162), and initial field trials are expected to have a measured entomological outcome alongside a modeled epidemiological one (118). Several validated malaria transmission models are available (58, 101), and which is the most suited will depend on the data available to parameterize the model and the required specificity of the output.
4.5. Ethical and Regulatory Considerations for Low-Threshold Gene Drives
Due to the nature of gene drive technologies, the risks and benefits occur at the level of communities rather than individuals. Thus, achieving community agreement and shared values are of the utmost importance (132). Since their early development, researchers have been dedicated to the safe and responsible study of gene drive technologies. To this end, keystone leaders in the field have coauthored guidelines for the safe development of gene drives (1, 4), focusing on ensuring safe practices for their study in laboratory settings with minimal risk of release into the environment. As this field advances and discussions of future field trials of these technologies take place, developers and other stakeholders are continuing to set forth self-governance and recommendations for the safe and ethical development of these technologies (71, 82, 111, 118, 132, 146, 160, 223). These efforts have expanded to include standardizing the definition of gene drive (7) and core commitments with developers, social scientists, and other stakeholder signatories to outline the minimum requirements for fair partnership and transparency, product efficacy and safety, regulatory evaluation, risk–benefit assessment, and monitoring and mitigation (146). Even a code of ethics that includes the articulation of values for responsible science, ecological stewardship, and public engagement for gene drive research has been established (12). The most recent discussion topics include new and responsive engagement frameworks for gene drive technologies (168, 197, 199) and the value and purpose of a gene drive registry for transparency, coordination, and other benefits (209). These activities highlight the ever-growing effort to responsibly develop genetic biocontrol technologies while also weighing potential risks and benefits and reducing our reliance on harmful alternatives, such as insecticides or rodenticides. Detractors suggest that genetic and gene drive control strategies for vector and vector-borne disease control should never be considered and want a moratorium on research. However, they offer no alternatives to ease the suffering and/or economic losses of those affected. Many of the loudest voices against even studying these technologies are from areas of the world least affected by these issues. Hopefully, though, there is some public support for advancing genetic engineering technologies and gene drives even in communities not directly impacted by these issues (198).
5. NON–GENE DRIVE CRISPR TECHNOLOGIES FOR GENETIC BIOCONTROL
While gene drives are advancing in laboratory settings, to date no transgene-based gene drive technology has ever been tested in the wild. That said, multiple safe, self-limiting, non–gene drive technologies have been developed for population suppression in the wild. For example, SIT and the release of insects carrying a dominant lethal (RIDL) technology (215) have demonstrated large reductions in wild vector populations (43, 109). Female-specific flightless versions of this technology, fsRIDL, generate flightless or inviable females using a sex-specific intron with the tetracycline repressor system (88, 134, 208). CRISPR has also expedited the development of additional self-limiting genetic technologies that rely exclusively on the highly efficient cleavage of the system (and not the homing) and can be used to suppress populations.
5.1. Precision-Guided Sterile Insect Technique
Similar to gene drives that rely on the highly efficient reproducible cleavage described above, the precision-guided sterile insect technique (pgSIT) exploits the high precision and accuracy of CRISPR to disrupt genes in offspring that are crucial for female survival and male fertility (Figure 1f). This process involves a straightforward breeding method using two homozygous strains: one expressing Cas9 and another expressing gRNAs. When these two strains are mated, RNA-guided dominant biallelic knockouts are created (coined as lethal mosaicism) from two genes during development, resulting in the reproducible and efficient production of sterile males and dead or flightless females (126-128, 142, 143). pgSIT achieves this result in a single generation and can enable the deployment of eggs into the environment. Thus far, pgSIT has been developed in multiple species and shown to be incredibly robust and efficient for population suppression (126-128, 142, 143).
5.2. Temperature-Inducible Precision-Guided Sterile Insect Technique
Sex-sorting accuracy and throughput have been limiting factors in the use of many genetic biocontrol technologies (180), so new genetic sexing technologies (147) or approaches that do not require sex sorting at any stage would be beneficial and more scalable. A proof-of-concept temperature-inducible pgSIT (TI-pgSIT) system has been designed that incorporates the pgSIT system into a single strain (123) (Figure 1f). With this system, the gRNAs are continuously expressed, but the expression of Cas9 is under the inducible control of a heat shock promoter, so its expression can be regulated by temperature. At low temperatures, Cas9 expression and activity are low, and the lines can be fertile and healthy. However, at high temperatures, the induced Cas9 expression in the presence of the gRNAs results in the disruption of genes essential for female survival and male fertility, thereby generating only sterile males. If this system can be transferred to target species, it would further simplify the generation of sterile males for SIT release programs, possibly making previously unscalable SIT control strategies accessible for additional pest species.
5.3. Inherited Female Elimination by Genetically Encoded Nucleases Interrupting Alleles
In contrast to sterile male releases, nondriving female-killing strategies can also suppress populations. A recent CRISPR-based technology for female killing called inherited female elimination by genetically encoded nucleases interrupting alleles (Ifegenia) was engineered in the malaria vector, A. gambiae (206). Ifegenia is a binary system with separate Cas9 and gRNA lines that, when crossed, result in the disruption of the female essential femaleless (fle) gene, consequently killing female offspring. The males remain viable and fertile, but harbor the mutated female essential gene and editing machinery, which when inherited by subsequent generations, results in the death of future female offspring. Modeling revealed that female-killing Ifegenia alleles could reach sufficiently high frequencies in the population to achieve long-term population suppression, but similar to SIT, pgSIT, and other non–gene drive control methods, this would require iterative releases. Ifegenia also addresses long-standing difficulties with sex sorting of anopheline mosquitoes, which have less pronounced sexual dimorphisms until they are adults. Like pgSIT, Ifegenia does not require labor-intensive and costly sex-sorting strategies for the final product and therefore may be able to support the development and implementation of other genetic and gene drive control strategies for malaria vectors.
SUMMARY POINTS.
CRISPR is a powerful tool that has led to the development of diverse technologies for the control of pests and pest-associated diseases.
Homing CRISPR gene drives (HCGDs) have made enormous progress in design and capability. However, imprecise homing mechanisms make them unpredictable, so their performance in large, genetically diverse wild populations is uncertain.
Homing-independent gene drives and non–gene drive genetic biocontrol technologies provide effective and predictable alternatives to HCGDs.
FUTURE ISSUES.
Gene drives have yet to be evaluated in the field. Indeed, as with many technologies, technical, regulatory, and ethical issues need to be addressed before gene drives are used in real-world settings. Some of these issues are fairly unique to the field of genetic biocontrol due to their potential for large-scale spread and the general lack of consensus on how to weigh benefits and risks at the community level.
Due to these unknowns, confinable and predictable gene drives, such as split toxin-antidote systems, or nondrive approaches, such as precision-guided sterile insect technique (pgSIT), temperature-inducible pgSIT, and/or inherited female elimination by genetically encoded nucleases interrupting alleles (Ifegenia) should be prioritized for development, societal consideration, and use. This work will provide a template for how to evaluate other more invasive and less predictable genetic biocontrol technologies.
ACKNOWLEDGMENTS
This work was supported by funding from an NIH awards (R01AI151004, R01GM132825, DP2AI152071, RO1AI148300, RO1AI175152, RO1TR003514), EPA STAR award (RD84020401), and an Open Philanthropy award (309937-0001) awarded to O.S.A, and by funds from the Bill & Melinda Gates Foundation (INV-017683) awarded to J.M.M, and by a grant to B.A.H from the California Institute of Technology (Caltech) Resnick Sustainability Institute and the Caltech Center for Evolutionary Science. The views, opinions, and/or findings expressed are those of the authors and should not be interpreted as representing the official views or policies of the US government. This publication was developed under Assistance Agreement No. RD84020401 awarded by the US Environmental Protection Agency to O.S.A. It has not been formally reviewed by EPA. The views expressed in this document are solely those of the authors and do not necessarily reflect those of the Agency. EPA does not endorse any products or commercial services mentioned in this publication.
Footnotes
DISCLOSURE STATEMENT
O.S.A. is a founder of Agragene Inc. and Synvect Inc. with equity interest. The terms of this arrangement have been reviewed and approved by the University of California, San Diego, in accordance with its conflict of interest policies. The other authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Adelman Z, Akbari O, Bauer J, Bier E, Bloss C,et al. 2017. Rules of the road for insect gene drive research and testing. Nat. Biotechnol 35(8):716–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adolfi A, Gantz VM, Jasinskiene N, Lee H-F, Hwang K, et al. 2020. Efficient population modification gene-drive rescue system in the malaria mosquito Anopheles stephensi. Nat. Commun 11(1):5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akbari OS, Antoshechkin I, Hay BA, Ferree PM. 2013. Transcriptome profiling of Nasonia vitripennis testis reveals novel transcripts expressed from the selfish B chromosome, paternal sex ratio. G3 3(9):1597–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Akbari OS, Bellen HJ, Bier E, Bullock SL, Burt A, et al. 2015. Safeguarding gene drive experiments in the laboratory. Science 349(6251):927–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Akbari OS, Chen C-H, Marshall JM, Huang H, Antoshechkin I, Hay BA. 2014. Novel synthetic Medea selfish genetic elements drive population replacement in Drosophila; a theoretical exploration of Medea-dependent population suppression. ACS Synth. Biol 3(12):915–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Akbari OS, Matzen KD, Marshall JM, Huang H, Ward CM, Hay BA. 2013. A synthetic gene drive system for local, reversible modification and suppression of insect populations. Curr. Biol 23(8):671–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alphey LS, Crisanti A, Randazzo F, Akbari OS. 2020. Standardizing the definition of gene drive. PNAS 117(49):30864–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alphey N, Bonsall MB. 2014. Interplay of population genetics and dynamics in the genetic control of mosquitoes. J. R. Soc. Interface 11(93):20131071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Álvarez MM, Biayna J, Supek F. 2022. TP53-dependent toxicity of CRISPR/Cas9 cuts is differential across genomic loci and can confound genetic screening. Nat. Commun 13(1):4520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anderson MAE, Gonzalez E, Ang JXD, Shackleford L, Nevard K, et al. 2023. Closing the gap to effective gene drive in Aedes aegypti by exploiting germline regulatory elements. Nat. Commun 14(1):338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anderson MAE, Gonzalez E, Edgington MP, Ang JXD, Purusothaman D-K, et al. 2022. A multiplexed, confinable CRISPR/Cas9 gene drive propagates in caged Aedes aegypti populations. bioRxiv 2022.08.12.503466. 10.1101/2022.08.12.503466 [DOI] [Google Scholar]
- 12.Annas GJ, Beisel CL, Clement K, Crisanti A, Francis S, et al. 2021. A code of ethics for gene drive research. CRISPR J. 4(1):19–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anzalone AV, Koblan LW, Liu DR. 2020. Genome editing with CRISPR-Cas nucleases, base editors, transposases and prime editors. Nat. Biotechnol 38(7):824–44 [DOI] [PubMed] [Google Scholar]
- 14.Backus GA, Delborne JA. 2019. Threshold-dependent gene drives in the wild: spread, controllability, and ecological uncertainty. Bioscience 69(11):900–7 [Google Scholar]
- 15.Baker RH. 1984. Chromosome rearrangements in the control of mosquitoes. Prev. Vet. Med 2(1):529–40 [Google Scholar]
- 16.Bakerlee CW, Nguyen Ba AN, Shulgina Y, Rojas Echenique JI, Desai MM. 2022. Idiosyncratic epistasis leads to global fitness-correlated trends. Science 376(6593):630–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bakri A, Mehta K, Lance DR. 2005. Sterilizing insects with ionizing radiation. In Sterile Insect Technique: Principles and Practice in Area-Wide Integrated Pest Management, ed. Dyck VA, Hendrichs J, Robinson AS, pp. 355–98. Dordrecht, Neth.: Springer [Google Scholar]
- 18.Basgall EM, Goetting SC, Goeckel ME, Giersch RM, Roggenkamp E, et al. 2018. Gene drive inhibition by the anti-CRISPR proteins AcrIIA2 and AcrIIA4 in Saccharomyces cerevisiae. Microbiology 164(4):464–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Basu S, Reitmayer CM, Lumley S, Atkinson B, Schade-Weskott ML. 2023. A Zika virus-responsive sensor-effector system in Aedes aegypti. bioRxiv 2023.02.06.527261. 10.1101/2023.02.06.527261 [DOI] [Google Scholar]
- 20.Beaghton A, Hammond A, Nolan T, Crisanti A, Godfray HCJ, Burt A. 2017. Requirements for driving antipathogen effector genes into populations of disease vectors by homing. Genetics 205(4):1587–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beeman RW, Friesen KS, Denell RE. 1992. Maternal-effect selfish genes in flour beetles. Science 256(5053):89–92 [DOI] [PubMed] [Google Scholar]
- 22.Beerntsen BT, James AA, Christensen BM. 2000. Genetics of mosquito vector competence. Microbiol. Mol. Biol. Rev 64(1):115–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ben-David E, Burga A, Kruglyak L. 2017. A maternal-effect selfish genetic element in Caenorhabditis elegans. Science 356(6342):1051–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bhatt S, Weiss DJ, Cameron E, Bisanzio D, Mappin B, et al. 2015. The effect of malaria control on Plasmodium falciparum in Africa between 2000 and 2015. Nature 526(7572):207–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bhaya D, Davison M, Barrangou R. 2011. CRISPR-Cas systems in bacteria and archaea: versatile small RNAs for adaptive defense and regulation. Annu. Rev. Genet 45:273–97 [DOI] [PubMed] [Google Scholar]
- 26.Braig HR, Yan G. 2001. The spread of genetic constructs in natural insect populations. In Genetically Engineered Organisms: Assessing Environmental and Human Health Effects, ed. Letourneau DK, Burrows BE, pp. 251–314. Boca Raton, FL: CRC Press [Google Scholar]
- 27.Brouns SJJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJH, et al. 2008. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321(5891):960–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buchman AB, Gamez S, Li M, Antoshechkin I, Li H-H, et al. 2019. Engineered resistance to Zika virus in transgenic Aedes aegypti expressing a polycistronic cluster of synthetic small RNAs. PNAS 116(9):3656–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Buchman AB, Gamez S, Li M, Antoshechkin I, Li H-H, et al. 2020. Broad dengue neutralization in mosquitoes expressing an engineered antibody. PLOS Pathog. 16(1):e1008103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buchman AB, Ivy T, Marshall JM, Akbari OS, Hay BA. 2018. Engineered reciprocal chromosome translocations drive high threshold, reversible population replacement in Drosophila. ACS Synth. Biol 7(5):1359–70 [DOI] [PubMed] [Google Scholar]
- 31.Buchman AB, Marshall JM, Ostrovski D, Yang T, Akbari OS. 2018. Synthetically engineered Medea gene drive system in the worldwide crop pest Drosophila suzukii. PNAS 115(18):4725–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Buchman AB, Shriner I, Yang T, Liu J, Antoshechkin I, et al. 2021. Engineered reproductively isolated species drive reversible population replacement. Nat. Commun 12(1):3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bui M, Dalla Benetta E, Dong Y, Zhao Y, Yang T, et al. 2023. CRISPR mediated transactivation in the human disease vector Aedes aegypti. PLOS Pathog. 19(1):e1010842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bull JJ. 2015. Evolutionary decay and the prospects for long-term disease intervention using engineered insect vectors. Evol. Med. Public Health 2015(1):152–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bull JJ. 2016. Lethal gene drive selects inbreeding. Evol. Med. Public Health 2017(1):1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Burga A, Ben-David E, Kruglyak L. 2020. Toxin-antidote elements across the tree of life. Annu. Rev. Genet 54:387–415 [DOI] [PubMed] [Google Scholar]
- 37.Burt A 2003. Site-specific selfish genes as tools for the control and genetic engineering of natural populations. Proc. Biol. Sci 270(1518):921–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Burt A, Koufopanou V. 2004. Homing endonuclease genes: the rise and fall and rise again of a selfish element. Curr. Opin. Genet. Dev 14(6):609–15 [DOI] [PubMed] [Google Scholar]
- 39.Burt A, Trivers R. 2009. Genes in Conflict: The Biology of Selfish Genetic Elements. Cambridge, MA: Harvard Univ. Press [Google Scholar]
- 40.Carballar-Lejarazú R, Dong Y, Pham TB, Tushar T, Corder RM, et al. 2023. Dual effector population modification gene-drive strains of the African malaria mosquitoes, Anopheles gambiae and Anopheles coluzzii. PNAS 120(29):e2221118120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carballar-Lejarazú R, Ogaugwu C, Tushar T, Kelsey A, Pham TB, et al. 2020. Next-generation gene drive for population modification of the malaria vector mosquito, Anopheles gambiae. PNAS 117(37):22805–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carrami EM, Eckermann KN, Ahmed HMM, Sánchez CHM, Dippel S, et al. 2018. Consequences of resistance evolution in a Cas9-based sex conversion-suppression gene drive for insect pest management. PNAS 115(24):6189–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carvalho DO, McKemey AR, Garziera L, Lacroix R, Donnelly CA, et al. 2015. Suppression of a field population of Aedes aegypti in Brazil by sustained release of transgenic male mosquitoes. PLOS Negl. Trop. Dis 9(7):e0003864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Champer J, Buchman A, Akbari OS. 2016. Cheating evolution: engineering gene drives to manipulate the fate of wild populations. Nat. Rev. Genet 17(3):146–59 [DOI] [PubMed] [Google Scholar]
- 45.Champer J, Champer SE, Kim IK, Clark AG, Messer PW. 2021. Design and analysis of CRISPR-based underdominance toxin-antidote gene drives. Evol. Appl 14(4):1052–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Champer J, Chung J, Lee YL, Liu C, Yang E, et al. 2019. Molecular safeguarding of CRISPR gene drive experiments. eLife 8:e41439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Champer J, Kim IK, Champer SE, Clark AG, Messer PW. 2020. Performance analysis of novel toxin-antidote CRISPR gene drive systems. BMC Biol. 18(1):27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Champer J, Kim IK, Champer SE, Clark AG, Messer PW. 2021. Suppression gene drive in continuous space can result in unstable persistence of both drive and wild-type alleles. Mol. Ecol 30(4):1086–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Champer J, Lee E, Yang E, Liu C, Clark AG, Messer PW. 2020. A toxin-antidote CRISPR gene drive system for regional population modification. Nat. Commun 11:1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Champer J, Liu J, Oh SY, Reeves R, Luthra A, et al. 2018. Reducing resistance allele formation in CRISPR gene drive. PNAS 115(21):5522–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Champer J, Reeves R, Oh SY, Liu C, Liu J, et al. 2017. Novel CRISPR/Cas9 gene drive constructs reveal insights into mechanisms of resistance allele formation and drive efficiency in genetically diverse populations. PLOS Genet. 13(7):e1006796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Champer J, Yang E, Lee E, Liu J, Clark AG, Messer PW. 2020. A CRISPR homing gene drive targeting a haplolethal gene removes resistance alleles and successfully spreads through a cage population. PNAS 117(39):24377–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Champer J, Zhao J, Champer SE, Liu J, Messer PW. 2020. Population dynamics of underdominance gene drive systems in continuous space. ACS Synth. Biol 9(4):779–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chan Y-S, Huen DS, Glauert R, Whiteway E, Russell S. 2013. Optimising homing endonuclease gene drive performance in a semi-refractory species: the Drosophila melanogaster experience. PLOS ONE 8(1):e54130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chan Y-S, Naujoks DA, Huen DS, Russell S. 2011. Insect population control by homing endonuclease-based gene drive: an evaluation in Drosophila melanogaster. Genetics 188(1):33–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen C-H, Huang H, Ward CM, Su JT, Schaeffer LV, et al. 2007. A synthetic maternal-effect selfish genetic element drives population replacement in Drosophila. Science 316(5824):597–600 [DOI] [PubMed] [Google Scholar]
- 57.Chen J, Xu X, Champer J. 2023. Assessment of distant-site rescue elements for CRISPR toxin-antidote gene drives. Front. Bioeng. Biotechnol 11:1138702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chitnis N, Schapira A, Smith T, Steketee R. 2010. Comparing the effectiveness of malaria vector-control interventions through a mathematical model. Am. J. Trop. Med. Hyg 83(2):230–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Colleaux L, d’Auriol L, Betermier M, Cottarel G, Jacquier A, et al. 1986. Universal code equivalent of a yeast mitochondrial intron reading frame is expressed into E. coli as a specific double strand endonuclease. Cell 44(4):521–33 [DOI] [PubMed] [Google Scholar]
- 60.Connolly JB, Mumford JD, Glandorf DCM, Hartley S, Lewis OT, et al. 2022. Recommendations for environmental risk assessment of gene drive applications for malaria vector control. Malar. J 21(1):152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cook F, Bull JJ, Gomulkiewicz R. 2022. Gene drive escape from resistance depends on mechanism and ecology. Evol. Appl 15(5):721–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cosmides LM, Tooby J. 1981. Cytoplasmic inheritance and intragenomic conflict. J. Theor. Biol 89(1):83–129 [DOI] [PubMed] [Google Scholar]
- 63.Craig GB Jr., Hickey WA, Vandehey RC. 1960. An inherited male-producing factor in Aedes aegypti. Science 132(3443):1887–89 [DOI] [PubMed] [Google Scholar]
- 64.Curtis CF. 1968. Possible use of translocations to fix desirable genes in insect pest populations. Nature 218(5139):368–69 [DOI] [PubMed] [Google Scholar]
- 65.Curtis CF. 1975. Male-linked translocations and the control of insect pest populations. Experientia 31(10):1139–41 [DOI] [PubMed] [Google Scholar]
- 66.Dalla Benetta E, Akbari OS, Ferree PM. 2021. Mechanistically comparing reproductive manipulations caused by selfish chromosomes and bacterial symbionts. Heredity 126(5):707–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dalla Benetta E, Antoshechkin I, Yang T, Nguyen HQM, Ferree PM, Akbari OS. 2020. Genome elimination mediated by gene expression from a selfish chromosome. Sci. Adv 6(14):eaaz9808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dalla Benetta E, López-Denman A, Li H-H, Masri RA, Brogan DJ, et al. 2023. Engineered antiviral sensor targets infected mosquitoes. bioRxiv 2023.01.27.525922. 10.1101/2023.01.27.525922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.de Lara Capurro M, Coleman J, Beerntsen BT, Myles KM, Olson KE, et al. 2000. Virus-expressed, recombinant single-chain antibody blocks sporozoite infection of salivary glands in Plasmodium gallinaceum-infected Aedes aegypti. Am. J. Trop. Med. Hyg 62(4):427–33 [DOI] [PubMed] [Google Scholar]
- 70.Del Amo VL, Bishop AL, Sánchez CHM, Bennett JB, et al. 2020. A transcomplementing gene drive provides a flexible platform for laboratory investigation and potential field deployment. Nat. Commun 11:352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Delborne J, Kuzma J, Gould F, Frow E, Leitschuh C, Sudweeks J. 2018. Mapping research and governance needs for gene drives. J. Responsible Innov 5(Suppl. 1):S4–12 [Google Scholar]
- 72.Deredec A, Burt A, Godfray HCJ. 2008. The population genetics of using homing endonuclease genes in vector and pest management. Genetics 179(4):2013–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Deredec A, Godfray HCJ, Burt A. 2011. Requirements for effective malaria control with homing endonuclease genes. PNAS 108(43):E874–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dhole S, Lloyd AL, Gould F. 2019. Tethered homing gene drives: a new design for spatially restricted population replacement and suppression. Evol. Appl 12(18):1688–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dhole S, Lloyd AL, Gould F. 2020. Gene drive dynamics in natural populations: the importance of density dependence, space, and sex. Annu. Rev. Ecol. Evol. Syst 51:505–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.DiCarlo JE, Chavez A, Dietz SL, Esvelt KM, Church GM. 2015. Safeguarding CRISPR-Cas9 gene drives in yeast. Nat. Biotechnol 33(12):1250–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dong Y, Simões ML, Dimopoulos G. 2020. Versatile transgenic multistage effector-gene combinations for Plasmodium falciparum suppression in Anopheles. Sci. Adv 6(20):eaay5898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Doolittle WF, Sapienza C. 1980. Selfish genes, the phenotype paradigm and genome evolution. Nature 284(5757):601–3 [DOI] [PubMed] [Google Scholar]
- 79.Drury DW, Dapper AL, Siniard DJ, Zentner GE, Wade MJ. 2017. CRISPR/Cas9 gene drives in genetically variable and nonrandomly mating wild populations. Sci. Adv 3(5):e1601910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Eberhard WG. 1980. Evolutionary consequences of intracellular organelle competition. Q. Rev. Biol 55(3):231–49 [DOI] [PubMed] [Google Scholar]
- 81.Eckhoff PA, Wenger EA, Godfray HCJ, Burt A. 2017. Impact of mosquito gene drive on malaria elimination in a computational model with explicit spatial and temporal dynamics. PNAS 114(2):E255–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Emerson C, James S, Littler K, Randazzo FF. 2017. Principles for gene drive research. Science 358(6367):1135–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Enkerlin WR, Gutiérrez Ruelas JM, Pantaleon R, Soto Litera C, Villaseñor Cortés A, et al. 2017. The Moscamed Regional Programme: review of a success story of area-wide sterile insect technique application. Entomol. Exp. Appl 164(3):188–203 [Google Scholar]
- 84.Esvelt KM, Smidler AL, Catteruccia F, Church GM. 2014. Emerging technology: concerning RNA-guided gene drives for the alteration of wild populations. eLife 3:e03401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Franz AWE, Sanchez-Vargas I, Adelman ZN, Blair CD, Beaty BJ, et al. 2006. Engineering RNA interference-based resistance to dengue virus type 2 in genetically modified Aedes aegypti. PNAS 103(11):4198–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Franz AWE, Sanchez-Vargas I, Raban RR, Black WCIV, James AA, Olson KE. 2014. Fitness impact and stability of a transgene conferring resistance to dengue-2 virus following introgression into a genetically diverse Aedes aegypti strain. PLOS Negl. Trop. Dis 8(5):e2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Frieß JL, Lalyer CR, Giese B, Simon S, Otto M. 2023. Review of gene drive modelling and implications for risk assessment of gene drive organisms. Ecol. Model 478:110285 [Google Scholar]
- 88.Fu G, Lees RS, Nimmo D, Aw D, Jin L, et al. 2010. Female-specific flightless phenotype for mosquito control. PNAS 107(10):4550–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Galizi R, Doyle LA, Menichelli M, Bernardini F, Deredec A, et al. 2014. A synthetic sex ratio distortion system for the control of the human malaria mosquito. Nat. Commun 5:3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gallagher DN, Haber JE. 2018. Repair of a site-specific DNA cleavage: old-school lessons for Cas9-mediated gene editing. ACS Chem. Biol 13(2):397–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gamez S, Chaverra-Rodriguez D, Buchman A, Kandul NP, Mendez-Sanchez SC, et al. 2021. Exploiting a Y chromosome-linked Cas9 for sex selection and gene drive. Nat. Commun 12:7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gantz VM, Bier E. 2015. The mutagenic chain reaction: a method for converting heterozygous to homozygous mutations. Science 348(6233):442–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gantz VM, Bier E. 2016. The dawn of active genetics. Bioessays 38(1):50–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gantz VM, Jasinskiene N, Tatarenkova O, Fazekas A, Macias VM, et al. 2015. Highly efficient Cas9-mediated gene drive for population modification of the malaria vector mosquito Anopheles stephensi. PNAS 112(49):E6736–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gershenson S 1929. A new sex-ratio abnormality in Drosophila obscura. Genetics 13(6):488–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gierus L, Birand A, Bunting MD, Godahewa GI, Piltz SG, et al. 2022. Leveraging a natural murine meiotic drive to suppress invasive populations. PNAS 119(46):e2213308119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Godfray HCJ, North A, Burt A. 2017. How driving endonuclease genes can be used to combat pests and disease vectors. BMC Biol. 15(1):81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Goeckel ME, Basgall EM, Lewis IC, Goetting SC, Yan Y, et al. 2019. Modulating CRISPR gene drive activity through nucleocytoplasmic localization of Cas9 in S. cerevisiae. Fungal Biol. Biotechnol 6:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gould F 2008. Broadening the application of evolutionarily based genetic pest management. Evolution 62(2):500–10 [DOI] [PubMed] [Google Scholar]
- 100.Gould F, Schliekelman P. 2004. Population genetics of autocidal control and strain replacement. Annu. Rev. Entomol 49:193–217 [DOI] [PubMed] [Google Scholar]
- 101.Griffin JT, Hollingsworth TD, Okell LC, Churcher TS, White M, et al. 2010. Reducing Plasmodium falciparum malaria transmission in Africa: a model-based evaluation of intervention strategies. PLOS Med. 7(8):e1000324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Grunwald HA, Gantz VM, Poplawski G, Xu X-RS, Bier E, Cooper KL. 2019. Super-Mendelian inheritance mediated by CRISPR-Cas9 in the female mouse germline. Nature 566(7742):105–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Guichard A, Haque T, Bobik M, Xu X-RS, Klanseck C, et al. 2019. Efficient allelic-drive in Drosophila. Nat. Commun 10(1):1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hamilton WD. 1967. Extraordinary sex ratios: A sex-ratio theory for sex linkage and inbreeding has new implications in cytogenetics and entomology. Science 156(3774):477–88 [DOI] [PubMed] [Google Scholar]
- 105.Hammond AM, Galizi R, Kyrou K, Simoni A, Siniscalchi C, et al. 2016. A CRISPR-Cas9 gene drive system targeting female reproduction in the malaria mosquito vector Anopheles gambiae. Nat. Biotechnol 34(1):78–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hammond AM, Karlsson X, Morianou I, Kyrou K, Beaghton A, et al. 2021. Regulating the expression of gene drives is key to increasing their invasive potential and the mitigation of resistance. PLOS Genet. 17(1):e1009321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hammond AM, Kyrou K, Bruttini M, North A, Galizi R, et al. 2017. The creation and selection of mutations resistant to a gene drive over multiple generations in the malaria mosquito. PLOS Genet. 13(10):e1007039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hammond AM, Pollegioni P, Persampieri T, North A, Minuz R, et al. 2021. Gene-drive suppression of mosquito populations in large cages as a bridge between lab and field. Nat. Commun 12(1):4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Harris AF, McKemey AR, Nimmo D, Curtis Z, Black I, et al. 2012. Successful suppression of a field mosquito population by sustained release of engineered male mosquitoes. Nat. Biotechnol 30(9):828–30 [DOI] [PubMed] [Google Scholar]
- 110.Hartl DL. 1975. Modifier theory and meiotic drive. Theor. Popul. Biol 7(2):168–74 [DOI] [PubMed] [Google Scholar]
- 111.Hartley S, Taitingfong R, Fidelman P. 2022. The principles driving gene drives for conservation. Environ. Sci. Policy 135:36–45 [Google Scholar]
- 112.Hickey WA, Craig GB Jr. 1966. Distortion of sex ratio in populations of Aedes aegypti. Can. J. Genet. Cytol 8(2):260–78 [DOI] [PubMed] [Google Scholar]
- 113.Hickey WA, Craig GB Jr. 1966. Genetic distortion of sex ratio in a mosquito, Aedes aegypti. Genetics 53(6):1177–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hosack GR, Ickowicz A, Hayes KR. 2021. Quantifying the risk of vector-borne disease transmission attributable to genetically modified vectors. R. Soc. Open Sci 8(3):201525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hurst LD. 1993. scat+ is a selfish gene analogous to Medea of Tribolium castaneum. Cell 75(3):407–8 [DOI] [PubMed] [Google Scholar]
- 116.Ito J, Ghosh A, Moreira LA, Wimmer EA, Jacobs-Lorena M. 2002. Transgenic anopheline mosquitoes impaired in transmission of a malaria parasite. Nature 417(6887):452–55 [DOI] [PubMed] [Google Scholar]
- 117.Jaenike J 2001. Sex chromosome meiotic drive. Annu. Rev. Ecol. Syst 32:25–49 [Google Scholar]
- 118.James S, Collins FH, Welkhoff PA, Emerson C, Godfray HCJ, et al. 2018. Pathway to deployment of gene drive mosquitoes as a potential biocontrol tool for elimination of malaria in sub-Saharan Africa: recommendations of a scientific working group. Am. J. Trop. Med. Hyg 98(6):1–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.James SL, Marshall JM, Christophides GK, Okumu FO, Nolan T. 2020. Toward the definition of efficacy and safety criteria for advancing gene drive-modified mosquitoes to field testing. Vector Borne Zoonotic Dis. 20(4):237–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.James SL, O’Brochta DA, Randazzo F, Akbari OS. 2023. A gene drive is a gene drive: the debate over lumping or splitting definitions. Nat. Commun 14(1):1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. 2012. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337(6096):816–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Juste SS, Okamoto EM, Feng X, Del Amo VL. 2023. Next-generation CRISPR gene-drive systems using Cas12a nuclease. bioRxiv 2023.02.20.529271. 10.1101/2023.02.20.529271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kandul NP, Liu J, Akbari OS. 2021. Temperature-inducible precision-guided sterile insect technique. CRISPR J. 4(6):822–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kandul NP, Liu J, Bennett JB, Marshall JM, Akbari OS. 2021. A confinable home-and-rescue gene drive for population modification. eLife 10:e65939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kandul NP, Liu J, Buchman A, Gantz VM, Bier E, Akbari OS. 2020. Assessment of a split homing based gene drive for efficient knockout of multiple genes. G3 10(2):827–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kandul NP, Liu J, Buchman A, Shriner IC, Corder RM, et al. 2022. Precision guided sterile males suppress populations of an invasive crop pest. GEN Biotechnol. 1(4):372–85 [Google Scholar]
- 127.Kandul NP, Liu J, Sanchez CHM, Wu SL, Marshall JM, Akbari OS. 2019. Reply to “Concerns about the feasibility of using ‘precision guided sterile males’ to control insects.” Nat. Commun 10(1):3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kandul NP, Liu J, Sanchez CHM, Wu SL, Marshall JM, Akbari OS. 2019. Transforming insect population control with precision guided sterile males with demonstration in flies. Nat. Commun 10(1):84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kidwell MG, Ribeiro JM. 1992. Can transposable elements be used to drive disease refractoriness genes into vector populations? Parasitol. Today 8(10):325–29 [DOI] [PubMed] [Google Scholar]
- 130.Knipling EF. 1955. Possibilities of insect control or eradication through the use of sexually sterile males. J. Econ. Entomol 48(4):459–62 [Google Scholar]
- 131.Knipling EF. 1959. Sterile-male method of population control. Science 130(3380):902–4 [DOI] [PubMed] [Google Scholar]
- 132.Kolopack PA, Lavery JV. 2017. Informed consent in field trials of gene-drive mosquitoes. Gates Open Res. 1:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kyrou K, Hammond AM, Galizi R, Kranjc N, Burt A, et al. 2018. A CRISPR–Cas9 gene drive targeting doublesex causes complete population suppression in caged Anopheles gambiae mosquitoes. Nat. Biotechnol 36(11):1062–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Labbé GMC, Scaife S, Morgan SA, Curtis ZH, Alphey L. 2012. Female-specific flightless (fsRIDL) phenotype for control of Aedes albopictus. PLOS Negl. Trop. Dis 6(7):e1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Lai C, Alvarez O, Read K, van Fossan D, Conner CM, et al. 2022. Robust and efficient active genetics gene conversion in the rat and mouse. bioRxiv 2022.08.30.505951. 10.1101/2022.08.30.505951 [DOI] [Google Scholar]
- 136.Langmüller AM, Champer J, Lapinska S, Xie L, Metzloff M, et al. 2022. Fitness effects of CRISPR endonucleases in Drosophila melanogaster populations. eLife 11:e71809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Laven H 1969. Eradicating mosquitoes using translocations. Nature 221(5184):958–59 [DOI] [PubMed] [Google Scholar]
- 138.Laven H, Cousserans J, Guille G. 1972. Eradicating mosquitoes using translocations: a first field experiment. Nature 236:456–57 [DOI] [PubMed] [Google Scholar]
- 139.Leftwich PT, Edgington MP, Harvey-Samuel T, Carabajal Paladino LZ, Norman VC, Alphey L. 2018. Recent advances in threshold-dependent gene drives for mosquitoes. Biochem. Soc. Trans 46(5):1203–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Leung S, Windbichler N, Wenger EA, Bever CA, Selvaraj P. 2022. Population replacement gene drive characteristics for malaria elimination in a range of seasonal transmission settings: a modelling study. Malar. J 21(1):226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Lewis IC, Yan Y, Finnigan GC. 2021. Analysis of a Cas12a-based gene-drive system in budding yeast. Access Microbiol. 3(12):000301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Li M, Kandul NP, Sun R, Yang T, Benetta ED, et al. 2023. Targeting sex determination to suppress mosquito populations. bioRxiv 2023.04.18.537404. 10.1101/2023.04.18.537404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Li M, Yang T, Bui M, Gamez S, Wise T, et al. 2021. Suppressing mosquito populations with precision guided sterile males. Nat. Commun 12:5374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Li M, Yang T, Kandul NP, Bui M, Gamez S, et al. 2020. Development of a confinable gene drive system in the human disease vector Aedes aegypti. eLife 9:e51701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Liu W-L, Hsu C-W, Chan S-P, Yen P-S, Su MP, et al. 2022. Author correction: Transgenic refractory Aedes aegypti lines are resistant to multiple serotypes of dengue virus. Sci. Rep 12(1):754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Long KC, Alphey L, Annas GJ, Bloss CS, Campbell KJ, et al. 2020. Core commitments for field trials of gene drive organisms. Science 370(6523):1417–19 [DOI] [PubMed] [Google Scholar]
- 147.Lutrat C, Burckbuchler M, Olmo RP, Beugnon R, Fontaine A, et al. 2022. Combining two Genetic Sexing Strains allows sorting of non-transgenic males for Aedes genetic control. bioRxiv 2022.03.11.483912. 10.1101/2022.03.11.483912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Lyttle TW. 1993. Cheaters sometimes prosper: distortion of mendelian segregation by meiotic drive. Trends Genet. 9(6):205–10 [DOI] [PubMed] [Google Scholar]
- 149.Macdonald G 1957. The Epidemiology and Control of Malaria. London: Oxford Univ. Press [Google Scholar]
- 150.Magori K, Gould F. 2006. Genetically engineered underdominance for manipulation of pest populations: a deterministic model. Genetics 172(4):2613–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Marshall JM, Akbari OS. 2018. Can CRISPR-based gene drive be confined in the wild? A question for molecular and population biology. ACS Chem. Biol 13(2):424–30 [DOI] [PubMed] [Google Scholar]
- 152.Marshall JM, Buchman A, Sánchez CHM, Akbari OS. 2017. Overcoming evolved resistance to population-suppressing homing-based gene drives. Sci. Rep 7(1):3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Marshall JM, Raban RR, Kandul NP, Edula JR, León TM, Akbari OS. 2019. Winning the tug-of-war between effector gene design and pathogen evolution in vector population replacement strategies. Front. Genet 10:1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Maselko M, Feltman N, Upadhyay A, Hayward A, Das S, et al. 2020. Engineering multiple species-like genetic incompatibilities in insects. Nat. Commun 11(1):4468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Maselko M, Heinsch SC, Chacón JM, Harcombe WR, Smanski MJ. 2017. Engineering species-like barriers to sexual reproduction. Nat. Commun 8(1):883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Mathur G, Sanchez-Vargas I, Alvarez D, Olson KE, Marinotti O, James AA. 2010. Transgene-mediated suppression of dengue viruses in the salivary glands of the yellow fever mosquito, Aedes aegypti. Insect Mol. Biol 19(6):753–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.McClintock B 1950. The origin and behavior of mutable loci in maize. 36(6):344–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.McDonald IC, Overland DE. 1973. House fly genetics: II. Isolation of a heat-sensitive translocation homozygote. J. Hered 64(5):253–56 [DOI] [PubMed] [Google Scholar]
- 159.Metzloff M, Yang E, Dhole S, Clark AG, Messer PW, Champer J. 2022. Experimental demonstration of tethered gene drive systems for confined population modification or suppression. BMC Biol. 20(1):119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Millett P, Alexanian T, Palmer MJ, Evans SW, Kuiken T, Oye K. 2022. iGEM and gene drives: a case study for governance. Health Secur. 20(1):26–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Mingzuyu Pan AJC, Champer J. 2022. Making waves: comparative analysis of gene drive spread characteristics in a continuous space model. bioRxiv 2022.11.01.514650 10.1101/2022.11.01.514650 [DOI] [PubMed] [Google Scholar]
- 162.Mondal A, Vásquez VN, Marshall JM. 2022. Target product profiles for mosquito gene drives: incorporating insights from mathematical models. Front. Trop. Dis 3:828876 [Google Scholar]
- 163.Moreira LA, Ito J, Ghosh A, Devenport M, Zieler H, et al. 2002. Bee venom phospholipase inhibits malaria parasite development in transgenic mosquitoes. J. Biol. Chem 277(43):40839–43 [DOI] [PubMed] [Google Scholar]
- 164.Nash A, Urdaneta GM, Beaghton AK, Hoermann A, Papathanos PA, et al. 2019. Integral gene drives for population replacement. Biol. Open 8(1):bio037762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Noble C, Adlam B, Church GM, Esvelt KM, Nowak MA. 2018. Current CRISPR gene drive systems are likely to be highly invasive in wild populations. eLife 7:e33423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Noble C, Min J, Olejarz J, Buchthal J, Chavez A, et al. 2019. Daisy-chain gene drives for the alteration of local populations. PNAS 116(17):8275–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Noble C, Olejarz J, Esvelt KM, Church GM, Nowak MA. 2017. Evolutionary dynamics of CRISPR gene drives. Sci. Adv 3(4):e1601964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Normandin AM, Fitzgerald LM, Yip J. 2022. Hurdles in responsive community engagement for the development of environmental biotechnologies. Synth. Biol 7(1):ysac022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.North AR, Burt A, Godfray HCJ. 2019. Modelling the potential of genetic control of malaria mosquitoes at national scale. BMC Biol. 17(1):26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.North AR, Godfray HCJ. 2018. Modelling the persistence of mosquito vectors of malaria in Burkina Faso. Malar. J 17(1):140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Nuckolls NL, Bravo Núñez MA, Eickbush MT, Young JM, Lange JJ, et al. 2017. wtf genes are prolific dual poison-antidote meiotic drivers. eLife 6:e26033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Oberhofer G, Ivy T, Hay BA. 2018. Behavior of homing endonuclease gene drives targeting genes required for viability or female fertility with multiplexed guide RNAs. PNAS 115(40):E9343–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Oberhofer G, Ivy T, Hay BA. 2019. Cleave and Rescue, a novel selfish genetic element and general strategy for gene drive. PNAS 116(13):6250–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Oberhofer G, Ivy T, Hay BA. 2020. Gene drive and resilience through renewal with next generation Cleave and Rescue selfish genetic elements. PNAS 117(16):9013–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Oberhofer G, Ivy T, Hay BA. 2020. 2-Locus Cleave and Rescue selfish elements harness a recombination rate-dependent generational clock for self limiting gene drive. bioRxiv 2020.07.09.196253. 10.1101/2020.07.09.196253 [DOI] [Google Scholar]
- 176.Oberhofer G, Ivy T, Hay BA. 2021. Gene drive that results in addiction to a temperature-sensitive version of an essential gene triggers population collapse in Drosophila. PNAS 118(49):e2107413118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Oberhofer G, Ivy T, Hay BA. 2021. Split versions of Cleave and Rescue selfish genetic elements for measured self limiting gene drive. PLOS Genet. 17(2):e1009385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Orgel LE, Crick FH. 1980. Selfish DNA: the ultimate parasite. Nature 284(5757):604–7 [DOI] [PubMed] [Google Scholar]
- 179.Orozeo-Dávila D, Quintero L, Hernández E, Solís E, Artiaga T, et al. 2017. Mass rearing and sterile insect releases for the control of Anastrepha spp. pests in Mexico—a review. Entomol. Exp. Appl 164(3):176–87 [Google Scholar]
- 180.Papathanos PA, Bourtzis K, Tripet F, Bossin H, Virginio JF, et al. 2018. A perspective on the need and current status of efficient sex separation methods for mosquito genetic control. Parasit. Vectors 11(Suppl. 2):654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Pescod P, Bevivino G, Anthousi A, Shelton R, Margiotta M, et al. 2023. Measuring the impact of genetic heterogeneity and chromosomal inversions on the efficacy of CRISPR-Cas9 gene drives in different strains of Anopheles gambiae. bioRxiv 2023.03.31.535088. 10.1101/2023.03.31.535088 [DOI] [PubMed] [Google Scholar]
- 182.Peters LL, Barker JE. 1993. Novel inheritance of the murine severe combined anemia and thrombocytopenia (Scat) phenotype. Cell 74(1):135–42 [DOI] [PubMed] [Google Scholar]
- 183.Pfitzner C, White MA, Piltz SG, Scherer M, Adikusuma F, et al. 2020. Progress toward zygotic and germline gene drives in mice. CRISPR J. 3(5):388–97 [DOI] [PubMed] [Google Scholar]
- 184.Pham TB, Phong CH, Bennett JB, Hwang K, Jasinskiene N, et al. 2019. Experimental population modification of the malaria vector mosquito, Anopheles stephensi. PLOS Genet. 15(12):e1008440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Raban RR, Akbari OS. 2022. An introduction to the molecular genetics of gene drives and thoughts on their gradual transition to field use. In Transgenic Insects: Techniques and Applications, ed. Benedict MQ, Scott MJ, pp. 1–21. Wallingford, UK: CABI [Google Scholar]
- 186.Raban RR, Marshall JM, Akbari OS. 2020. Progress towards engineering gene drives for population control. J. Exp. Biol 223(Suppl. 1):jeb208181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Rašić G, Lobo NF, Jeffrey Gutiérrez EH, Sánchez C HM, Marshall JM. 2021. Monitoring needs for gene drive mosquito projects: lessons from vector control field trials and invasive species. Front. Genet 12:780327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Ravindran S 2012. Barbara McClintock and the discovery of jumping genes. PNAS 109(50):20198–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Reeves RG, Bryk J, Altrock PM, Denton JA, Reed FA. 2014. First steps towards underdominant genetic transformation of insect populations. PLOS ONE 9(5):e97557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Reid W, Williams AE, Sanchez-Vargas I, Lin J, Juncu R, et al. 2022. Assessing single-locus CRISPR/Cas9-based gene drive variants in the mosquito Aedes aegypti via single-generation crosses and modeling. G3 12(12):jkac280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Ribeiro JM, Kidwell MG. 1994. Transposable elements as population drive mechanisms: specification of critical parameter values. J. Med. Entomol 31(1):10–16 [DOI] [PubMed] [Google Scholar]
- 192.Robinson AS. 1976. Progress in the use of chromosomal translocations for the control of insect pests. Biol. Rev. Camb. Philos. Soc 51(1):1–24 [DOI] [PubMed] [Google Scholar]
- 193.Roggenkamp E, Giersch RM, Schrock MN, Turnquist E, Halloran M, Finnigan GC. 2018. Tuning CRISPR-Cas9 gene drives in Saccharomyces cerevisiae. G3 8(3)999–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Roggenkamp E, Giersch RM, Wedeman E, Eaton M, Turnquist E, et al. 2017. CRISPR-UnLOCK: multipurpose Cas9-based strategies for conversion of yeast libraries and strains. Front. Microbiol 8:1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Samuel GH, Pohlenz T, Dong Y, Coskun N, Adelman ZN, et al. 2023. RNA interference is essential to modulating the pathogenesis of mosquito-borne viruses in the yellow fever mosquito. PNAS 120(11):e2213701120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Sánchez C HM, Bennett JB, Wu SL, Rašić G, Akbari OS, Marshall JM. 2020. Modeling confinement and reversibility of threshold-dependent gene drive systems in spatially-explicit Aedes aegypti populations. BMC Biol. 18(1):50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Schairer CE, Taitingfong R, Akbari OS, Bloss CS. 2019. A typology of community and stakeholder engagement based on documented examples in the field of novel vector control. PLOS Negl. Trop. Dis 13(11):e0007863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Schairer CE, Triplett C, Akbari OS, Bloss CS. 2022. California residents’ perceptions of gene drive systems to control mosquito-borne disease. Front. Bioeng. Biotechnol 10:848707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Schairer CE, Triplett C, Buchman A, Akbari OS, Bloss CS. 2020. Interdisciplinary development of a standardized introduction to gene drives for lay audiences. BMC Med. Res. Methodol 20(1):273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Seidel HS, Ailion M, Li J, van Oudenaarden A, Rockman MV, Kruglyak L. 2011. A novel sperm-delivered toxin causes late-stage embryo lethality and transmission ratio distortion in C. elegans. PLOS Biol. 9(7):e1001115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Serebrovskii AS. 1940. On the possibility of a new method for the control of insect pests. Zool. Zhurnal 19:618–30 [Google Scholar]
- 202.Serebrovsky AS. 1969. On the possibility of a new method for the control of insect pests. In Sterile-Male Technique for Eradication or Control of Harmful Insects, pp. 123–237. Vienna: Int. Atom. Energy Ag. [Google Scholar]
- 203.Shapiro RS, Chavez A, Porter CBM, Hamblin M, Kaas CS, et al. 2018. A CRISPR-Cas9-based gene drive platform for genetic interaction analysis in Candida albicans. Nat. Microbiol 3(1):73–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Simoni A, Hammond AM, Beaghton AK, Galizi R, Taxiarchi C, et al. 2020. A male-biased sex-distorter gene drive for the human malaria vector Anopheles gambiae. Nat. Biotechnol. 38(9):1054–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Simoni A, Siniscalchi C, Chan YS, Huen DS, Russell S, et al. 2015. Development of synthetic selfish elements based on modular nucleases in Drosophila melanogaster. Nucleic Acids Res. 42(11):7461–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Smidler AL, Pai JJ, Apte RA, Sánchez CHM, Corder RM, et al. 2023. A confinable female-lethal population suppression system in the malaria vector, Anopheles gambiae. Sci. Adv 9(27):eade8903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Snow JW. 1988. Radiation, insects and eradication in North America: an overview from screwworm to bollworm. In Modern Insect Control: Nuclear Techniques and Biotechnology, pp. 3–13. Vienna: Int. At. Energy Agency [Google Scholar]
- 208.Spinner SAM, Barnes ZH, Puinean AM, Gray P, Dafa’alla T, et al. 2022. New self-sexing Aedes aegypti strain eliminates barriers to scalable and sustainable vector control for governments and communities in dengue-prone environments. Front. Bioeng. Biotechnol 10:975786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Taitingfong RI, Triplett C, Vásquez VN, Rajagopalan RM, Raban R, et al. 2023. Exploring the value of a global gene drive project registry. Nat. Biotechnol 41:9–13 [DOI] [PubMed] [Google Scholar]
- 210.Tanaka H, Stone HA, Nelson DR. 2017. Spatial gene drives and pushed genetic waves. PNAS 114(32):8452–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Taxiarchi C, Beaghton A, Don NI, Kyrou K, Gribble M, et al. 2021. A genetically encoded anti-CRISPR protein constrains gene drive spread and prevents population suppression. Nat. Commun 12(1):3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Terns MP, Terns RM. 2011. CRISPR-based adaptive immune systems. Curr. Opin. Microbiol 14(3):321–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Terradas G, Bennett JB, Li Z, Marshall JM, Bier E. 2023. Genetic conversion of a split-drive into a full-drive element. Nat. Commun 14(1):191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Terradas G, Buchman AB, Bennett JB, Shriner I, Marshall JM, et al. 2021. Inherently confinable split-drive systems in Drosophila. Nat. Commun 12:1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Thomas DD, Donnelly CA, Wood RJ, Alphey LS. 2000. Insect population control using a dominant, repressible, lethal genetic system. Science 287(5462):2474–76 [DOI] [PubMed] [Google Scholar]
- 216.Unckless RL, Clark AG, Messer PW. 2017. Evolution of resistance against CRISPR/Cas9 gene drive. Genetics 205(2):827–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Vella MR, Gunning CE, Lloyd AL, Gould F. 2017. Evaluating strategies for reversing CRISPR-Cas9 gene drives. Sci. Rep 7(1):11038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Verkuijl SAN, Anderson MAE, Alphey L, Bonsall MB. 2022. Daisy-chain gene drives: the role of low cut-rate, resistance mutations, and maternal deposition. PLOS Genet. 18(9):e1010370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Verkuijl SAN, Ang JXD, Alphey L, Bonsall MB, Anderson MAE. 2022. The challenges in developing efficient and robust synthetic homing endonuclease gene drives. Front. Bioeng. Biotechnol 10:856981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Verkuijl SAN, Gonzalez E, Li M, Ang JXD, Kandul NP, et al. 2022. A CRISPR endonuclease gene drive reveals distinct mechanisms of inheritance bias. Nat. Commun 13(1):7145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Wang G-H, Gamez S, Raban RR, Marshall JM, Alphey L, et al. 2021. Combating mosquito-borne diseases using genetic control technologies. Nat. Commun 12(1):4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Ward CM, Su JT, Huang Y, Lloyd AL, Gould F, Hay BA. 2011. Medea selfish genetic elements as tools for altering traits of wild populations: a theoretical analysis. Evolution 65(4):1149–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Warmbrod KL, Kobokovich AL, West R, Gronvall GK, Montague M. 2022. The need for a tiered registry for US gene drive governance. Health Secur. 20(1):43–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Weitzel AJ, Grunwald HA, Weber C, Levina R, Gantz VM, et al. 2021. Meiotic Cas9 expression mediates gene conversion in the male and female mouse germline. PLOS Biol. 19(12):e3001478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Werren JH, Baldo L, Clark ME. 2008. Wolbachia: master manipulators of invertebrate biology. Nat. Rev. Microbiol 6(10):741–51 [DOI] [PubMed] [Google Scholar]
- 226.Werren JH, Nur U, Wu CI. 1988. Selfish genetic elements. Trends Ecol. Evol 3(11):297–302 [DOI] [PubMed] [Google Scholar]
- 227.Whitten MJ. 1971. Insect control by genetic manipulation of natural populations. Science 171(3972):682–84 [DOI] [PubMed] [Google Scholar]
- 228.Wiedenheft B, Sternberg SH, Doudna JA. 2012. RNA-guided genetic silencing systems in bacteria and archaea. Nature 482(7385):331–38 [DOI] [PubMed] [Google Scholar]
- 229.Windbichler N, Menichelli M, Papathanos PA, Thyme SB, Li H, et al. 2011. A synthetic homing endonuclease-based gene drive system in the human malaria mosquito. Nature 473(7346):212–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Windbichler N, Papathanos PA, Catteruccia F, Ranson H, Burt A, Crisanti A. 2007. Homing endonuclease mediated gene targeting in Anopheles gambiae cells and embryos. Nucleic Acids Res. 35(17):5922–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Winegard TC. 2019. The Mosquito: A Human History of Our Deadliest Predator. New York: Penguin [Google Scholar]
- 232.World Health Organ. 2020. World Malaria Report 2020: 20 Years of Global Progress and Challenges. Geneva: World Health Organ. [Google Scholar]
- 233.Xu X-RS, Bulger EA, Gantz VM, Klanseck C, Heimler SR, et al. 2020. Active genetic neutralizing elements for halting or deleting gene drives. Mol. Cell 80(2):246–62.e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Yan Y, Finnigan GC. 2018. Development of a multi-locus CRISPR gene drive system in budding yeast. Sci. Rep 8(1):17277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Yan Y, Finnigan GC. 2019. Analysis of CRISPR gene drive design in budding yeast. Access Microbiol. 1(9):e000059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Yang E, Metzloff M, Langmüller AM, Xu X, Clark AG, et al. 2022. A homing suppression gene drive with multiplexed gRNAs maintains high drive conversion efficiency and avoids functional resistance alleles. G3 12(6):jkac081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Yen P-S, James A, Li J-C, Chen C-H, Failloux A-B. 2018. Synthetic miRNAs induce dual arboviral-resistance phenotypes in the vector mosquito Aedes aegypti. Commun. Biol 1:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Yoshida S, Matsuoka H, Luo E, Iwai K, Arai M, et al. 1999. A single-chain antibody fragment specific for the Plasmodium berghei ookinete protein Pbs21 confers transmission blockade in the mosquito midgut. Mol. Biochem. Parasitol 104(2):195–204 [DOI] [PubMed] [Google Scholar]
- 239.Yoshida S, Shimada Y, Kondoh D, Kouzuma Y, Ghosh AK, et al. 2007. Hemolytic C-type lectin CEL-III from sea cucumber expressed in transgenic mosquitoes impairs malaria parasite development. PLOS Pathog. 3(12):e192. [DOI] [PMC free article] [PubMed] [Google Scholar]