Abstract
The barley powdery mildew fungus, Blumeria hordei (Bh), secretes hundreds of candidate secreted effector proteins (CSEPs) to facilitate pathogen infection and colonization. One of these, CSEP0008, is directly recognized by the barley nucleotide‐binding leucine‐rich‐repeat (NLR) receptor MLA1 and therefore is designated AVRA1. Here, we show that AVRA1 and the sequence‐unrelated Bh effector BEC1016 (CSEP0491) suppress immunity in barley. We used yeast two‐hybrid next‐generation interaction screens (Y2H‐NGIS), followed by binary Y2H and in planta protein–protein interactions studies, and identified a common barley target of AVRA1 and BEC1016, the endoplasmic reticulum (ER)‐localized J‐domain protein HvERdj3B. Silencing of this ER quality control (ERQC) protein increased Bh penetration. HvERdj3B is ER luminal, and we showed using split GFP that AVRA1 and BEC1016 translocate into the ER signal peptide‐independently. Overexpression of the two effectors impeded trafficking of a vacuolar marker through the ER; silencing of HvERdj3B also exhibited this same cellular phenotype, coinciding with the effectors targeting this ERQC component. Together, these results suggest that the barley innate immunity, preventing Bh entry into epidermal cells, requires ERQC. Here, the J‐domain protein HvERdj3B appears to be essential and can be regulated by AVRA1 and BEC1016. Plant disease resistance often occurs upon direct or indirect recognition of pathogen effectors by host NLR receptors. Previous work has shown that AVRA1 is directly recognized in the cytosol by the immune receptor MLA1. We speculate that the AVRA1 J‐domain target being inside the ER, where it is inapproachable by NLRs, has forced the plant to evolve this challenging direct recognition.
Keywords: barley, ER quality control, ERdj3b, immunity, pathogen effectors, powdery mildew, signal peptide‐independent ER‐uptake, yeast two‐hybrid next‐generation interaction screening
HvERdj3B, identified as a host target of AVRA1 and BEC1016, is an immunity‐associated chaperone in the endoplasmic reticulum, into which these effectors enter signal peptide‐independently.
1. INTRODUCTION
Plants have evolved a multilayered and interconnected innate immune system to protect themselves against pathogen attacks (Ngou et al., 2022; Yuan, Jiang, et al., 2021). An initial layer, designated pathogen‐associated molecular pattern‐triggered immunity (PTI) is activated by pattern‐recognition receptors (PRRs) located on the plasma membrane (PM) (Zipfel & Oldroyd, 2017). However, plant pathogens can secrete large numbers of effectors into the host to interfere with many host cellular processes in order to suppress PTI and establish parasitism (Figueroa et al., 2021; Kanja & Hammond‐Kosack, 2020). In response, plants have evolved effector‐triggered immunity (ETI) that is initiated upon direct or indirect recognition of effectors by specific intracellular nucleotide‐binding leucine‐rich repeat (NLR) receptors and often involves a programmed cell death response, termed the hypersensitive reaction (HR) (Jones et al., 2016; Ngou et al., 2022; Thordal‐Christensen, 2020). With some exceptions, NLR receptors represent the most utilized class of resistance proteins in agriculture (Kourelis & van der Hoorn, 2018; Ngou et al., 2022; Sun et al., 2020; van Wersch et al., 2020). These innate immune systems rely on basic cellular processes, such as protein quality control systems. An intact endoplasmic reticulum (ER) quality control (ERQC) is required to transport certain de novo‐synthesized PRRs through the ER for them to be trafficked to their destination at the PM (Tintor & Saijo, 2014). For example, trafficking of the Arabidopsis EF‐Tu receptor, EFR, is particularly dependent on the ERQC, where it requires the chaperones ER DnaJ3B protein (ERdj3B) and binding immunoglobulin protein (BiP), as well as stromal cell‐derived factor 2 (SDF2), for its proper folding and glycosylation (Nekrasov et al., 2009). While ERdj3B is an ER luminal J‐domain protein, homologous proteins elsewhere in the cell have been implicated in plant immunity as well. For instance, in rice infected by the blast fungus, Magnaporthe oryzae, cytosolic and mitochondria‐associated J‐domain proteins are essential for immunity, and interestingly, the latter protein is targeted by the disease‐promoting fungal effector MoCDIP4 (Xu, Zhong, et al., 2020; Zhong et al., 2018). J‐domain proteins are related to the bacterial DnaJ chaperones, also known as heat shock proteins 40 (HSP40). They interact with HSP70 proteins, such as BiP in the ER, to which they deliver misfolded client proteins and stimulate the HSP70 ATPase activity (Fatima et al., 2021; Pobre et al., 2019).
Pathogen effectors manipulate diverse aspects of host biology during infection (Bray Speth et al., 2007). Several recent genomics, transcriptomics and proteomics studies have identified some 350 to 800 candidates for secreted effector proteins (CSEPs) in the closely related grass powdery mildew fungi (Bindschedler et al., 2009; Frantzeskakis et al., 2018; Menardo et al., 2017; Müller et al., 2019; Pedersen et al., 2012; Spanu et al., 2010). However, only a subset of these have been studied in detail. In case of the barley powdery mildew fungus, Blumeria hordei (Bh), host‐induced gene silencing (HIGS) analyses have demonstrated that approximately 20 CSEPs contribute to virulence, which is c. 25% of those studied (Aguilar et al., 2016; Ahmed et al., 2015, 2016; Li et al., 2021; Pliego et al., 2013; Yuan, Jin, et al., 2021; Zhang et al., 2012). Moreover, Li et al. (2021) screened about 100 CSEPs from Bh and found 15 of them to suppress BAX‐induced programmed cell death in Nicotiana benthamiana. Two of those, CSEP0139 and CSEP0182, also suppressed BAX‐induced programmed cell death in barley. To date, only a few plant targets of Bh effectors have been identified. CSEP0055 targets the barley defence proteins PR1 and PR17 (Zhang et al., 2012), while CSEP0105 and CSEP0162 target the barley small heat shock proteins Hsp16.9 and Hsp17.5 (Ahmed et al., 2015). CSEP0064 and CSEP0264 target PR10, while CSEP0264, historically designated Blumeria effector candidate (BEC)1054, also targets eukaryotic elongation factor 1α (Pennington et al., 2019). Likewise, CSEP0027 was recently found to target a barley catalase (Yuan, Jin, et al., 2021). Lately, CSEP0162 was found also to interact with barley MON1 (Liao et al., 2023).
In addition to virulence functions, six Bh CSEPs are directly recognized as avirulence proteins by NLRs encoded by alleles of the Mla powdery mildew resistance locus (Bauer et al., 2021; Lu et al., 2016; Saur et al., 2019; Seeholzer et al., 2010). For instance, CSEP0008 (gene ID BLGH_03023) is recognized by MLA1, and thus named AVRA1 (Lu et al., 2016). Other recognized CSEPs are CSEP0059 (AVRA7), CSEP0174 (AVRA9), CSEP0141 (AVRA10 and AVRA22), and CSEP0372 (AVRA13) (Bauer et al., 2021; Cao et al., 2023; Lu et al., 2016; Saur et al., 2019). AVRA6 is represented by three near‐identical copies in the DH14 genome, BLGH_00709 (CSEP0254), BLGH_00708 and BLGH_07091 (Bauer et al., 2021; Cao et al., 2023), which may be expressed in an isolate‐specific manner (Velásquez‐Zapata, Smith, et al., 2023). In the wheat powdery mildew fungus, Blumeria graminis f. sp. tritici (Bgt), eight effector candidates with avirulence function have been characterized (Bourras et al., 2015, 2019; Hewitt et al., 2021; Kunz et al., 2023; Manser et al., 2021; Müller et al., 2022; Praz et al., 2017) as well as a suppressor of Avr recognition (Bourras et al., 2019). All described Blumeria Avr proteins belong to the superfamily of RNase‐like proteins associated with haustoria (RALPH) effectors with features resembling catalytically inactive RNases (Kusch et al., 2023; Spanu, 2017). While data suggest wheat NLRs also directly recognize Bgt Avr proteins (Kunz et al., 2023), the Pm2a/AvrPm2 recognition involves a wheat zinc finger‐type transcription factor, interacting with both the NLR and the RALPH effector (Manser et al., 2023). However, whether the hitherto described Blumeria Avr proteins have effector functions promoting virulence has received little attention.
Early studies have shown that effectors screened against immune‐associated proteins of Arabidopsis identified several hub proteins that are targeted by multiple effectors (Mukhtar et al., 2011; Weßling et al., 2014). To test this observation in the barley–Bh system, we initiated a screen for protein–protein interactions with several CSEPs, focusing on cloned AVR effectors (Lu et al., 2016), as well as those that displayed unique transcript abundance during infection and/or a significant HIGS phenotype (Pliego et al., 2013). Here, we benefit from recent advances in yeast two‐hybrid (Y2H) analyses, termed next‐generation interaction screening (NGIS) (Suter et al., 2015), which uses deep sequencing to score the output from Y2H screens (Erffelinck et al., 2018; Lewis et al., 2012; Pashkova et al., 2016; Trigg et al., 2017; Weimann et al., 2013; Yachie et al., 2016). These approaches facilitate a quantitative measure of which preys interact with each bait protein (Suter et al., 2015) and identify reproducible protein–protein interactions (PPI) with 70%–90% accuracy (Pashkova et al., 2016; Trigg et al., 2017; Velásquez‐Zapata et al., 2021; Weimann et al., 2013). This then allows functional PPI to be positioned within interactome networks (Mukhtar et al., 2011; Velásquez‐Zapata et al., 2021, 2022; Weßling et al., 2014).
We aimed to use our NGIS pipeline to find barley targets of Bh effectors, and in the present study found that AVRA1 (CSEP0008, gene ID BLGH_03023) and BEC1016 (CSEP0491, gene ID BLGH_07006) both target the barley ER‐luminal J‐domain protein, HvERdj3B. BEC1016 has no documented avirulence function but, unlike AVRA1, it has previously documented virulence contribution (Pliego et al., 2013). Using a split green fluorescent protein (GFP) system, we could show that both effectors translocate into the ER lumen, signal peptide‐independently, allowing them to target HvERdj3B. Silencing HvERdj3B, as well as overexpression of AVRA1 and BEC1016, not only enhanced the formation of fungal haustoria as an immunity‐related phenotype, but it also hampered trafficking of a vacuolar marker protein through the ER as a common cellular phenotype. Together, our results suggest that barley immunity involves ERQC, and that this is sensitive to Bh effectors.
2. RESULTS
2.1. AVRA1 and BEC1016 suppress PTI
The two Bh effector candidates, AVRA1 and BEC1016, target the same barley protein (see below). They are both RALPH effectors, but their amino acid sequences are very different (Figure S1). AVRA1 is a singleton in Bh, Bgt and in other Blumeria lineages. Meanwhile, BEC1016 belongs to a CSEP family (#5 in Bh and #6 in Bgt) with multiple paralogues in the Blumeria lineages (Menardo et al., 2017; Pedersen et al., 2012). AVRA1 and BEC1016 exhibit similar patterns of transcript accumulation in incompatible and compatible interactions up to the time of Bh appressorium formation, when penetration of epidermal cells and initiation of the first haustoria occur. However, they diverge during development of haustoria, when the AVR A1 transcript in particular has a notable increase in the compatible interaction (Figure S2a,b). As an avirulence protein recognized by MLA1, AVRA1 is predicted also to have virulence function. However, silencing of AVR A1 did not result in a significant decrease in haustoria in the original HIGS assay, unlike the case for BEC1016 (Pliego et al., 2013). As an alternative approach to study virulence functions, we transiently overexpressed AVRA1 and BEC1016 (without signal peptide) in leaf epidermal cells of 1‐week‐old Golden Promise (susceptible) barley plants using particle bombardment. Two days later, the leaves were inoculated with Bh isolate C15 (AVR A1 ), and after another 2 days transformed cells were scored for presence of haustoria, as evidence for successful penetration and fungal infection. Our data revealed that overexpression of AVRA1, as well as BEC1016, resulted in higher haustoria numbers (Figure 1a).
FIGURE 1.
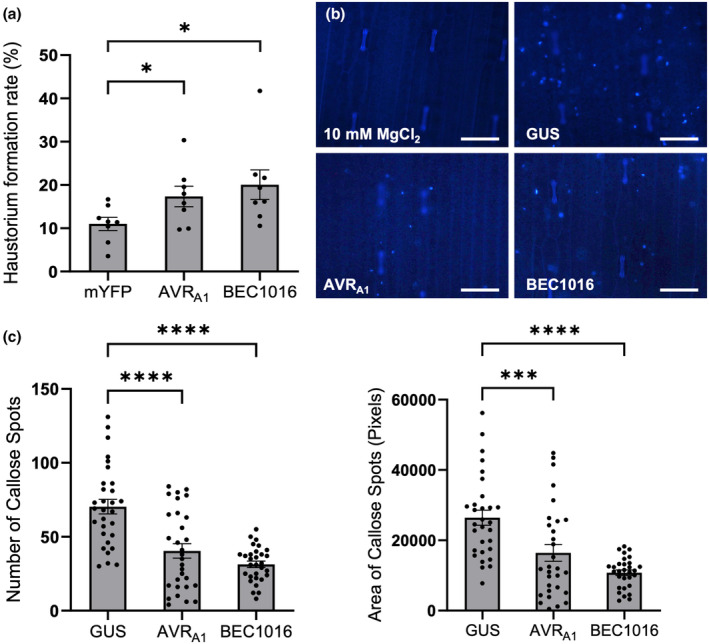
Influence of AVRA1 and BEC1016 on Blumeria hordei (Bh) infection and callose deposition in barley. (a) Effect of transient overexpression of AVRA1 and BEC1016 on Bh invasion. pUbi promoter‐driven overexpression constructs were co‐bombarded with a β‐glucuronidase (GUS) reporter construct into leaf epidermal cells of 1‐week‐old Golden Promise barley and 2 days later inoculated with Bh. After another 2 days, the fungal haustorium formation was scored in GUS‐expressing cells. Data shown are mean values of eight independent experiments. Error bars, SE. *p < 0.05 determined by Student's t test. (b,c) AVRA1 and BEC1016 can reduce bacterium‐triggered callose deposition in barley plants. Leaves of 8‐day‐old Golden Promise barley plants were infiltrated with Pseudomonas fluorescens EtHAn, transformed with pEDV6‐GUS, pEDV6‐AVRA1 or pEDV6‐BEC1016, and 24 h later the callose deposition were assessed by aniline blue staining and UV‐fluorescence microscopy. (b) Representative images. Scale bars, 100 μm. (c) Number (left) and area (right) of callose spots were scored on 10 images (each 1110 μm × 740 μm/5184 × 3456 pixels) recorded from each of three leaves after each treatment. Data shown are from one representative experiment. ***p < 0.001, ****p < 0.0001 determined by one‐way analysis of variance in GraphPad Prism. All raw data for (a) and (c), plus four other experiments like the one in (c) are available in Table S1.
Pathogen‐induced callose depositions function as a chemical and physical reinforcement of the plant cell wall towards invading pathogens (Voigt, 2014). To determine whether AVRA1 and BEC1016 affect callose deposition, they were introduced into barley leaf cells using EtHAn, a strain of Pseudomonas fluorescens modified to express the type III secretion system (T3SS) (Thomas et al., 2009; Upadhyaya et al., 2014). To verify protein transfer to the barley cells, EtHAn transformed with a construct expressing β‐glucuronidase (GUS) fused to a signal peptide for T3SS resulted in clear and uniform blue staining inside the leaf cells after reaction buffer incubation. There was no sign of stained bacteria in the apoplast (Figure S3), indicating GUS to be efficiently transferred into the barley cell. While EtHAn itself triggers callose formation, it can at the same time be studied how this is influenced by effectors (Sohn et al., 2007; Xu, Tang, et al., 2020; Figure 1b). Twenty‐four hours after infiltration of EtHAn expressing GUS (negative control), AVRA1 or BEC1016, both the total number and total area of callose depositions were significantly reduced by the effectors (Figure 1b,c). The number of bacteria after 24 h was the same for the three strains (Figure S4), supporting that the effectors directly influence callose deposition. Taken together, it appears that AVRA1 and BEC1016 target and inhibit PTI, and thereby promote Bh penetration. This result is consistent with previous transcriptome analyses from barley–Bh interactions with non‐corresponding MLA:AVR pairs showing suppression of PTI‐associated transcripts (Caldo et al., 2004, 2006; Moscou et al., 2011; Surana et al., 2017). By applying the same set of EtHAn strains to P‐02, carrying the Mla3 powdery mildew resistance allele, and inoculated with the Bh isolate A6, carrying AVR A3 , we saw that the Mla3‐mediated HR was not influenced by AVRA1 and BEC1016 (Figure S5).
2.2. Both AVRA1 and BEC1016 interact with barley J‐domain protein, HvERdj3B
To identify host–pathogen PPI, three independent replications of batch Y2H‐NGIS were performed according to Elmore et al. (2023), Velásquez‐Zapata et al. (2021), and Velásquez‐Zapata, Elmore, and Wise (2023). The screens involved parallel histidine selection (for enrichment of yeast cells with bait and prey interactions) versus non‐selected controls (selection for bait and prey, but not their interactions). Deep Illumina sequencing was performed across the GAL4 activation domain (AD) and into the prey protein coding sequences for all cultures. Two complementary software packages, designated NGPINT and Y2H‐SCORES, were used to quantify the outcome of Y2H‐NGIS (Banerjee et al., 2021; Velásquez‐Zapata et al., 2021; Velásquez‐Zapata, Elmore, & Wise, 2023). NGPINT facilitates the identification of GAL4 AD–prey in‐frame coding sequence fusion reads. Then, Y2H‐SCORES uses appropriate normalization methods, count data and statistical models to build a set of ranking scores to predict three properties expected from true interactors, that is, enrichment in selection versus non‐selection conditions, specificity to a bait screening and in‐frame selection of the prey (Velásquez‐Zapata et al., 2021). Prey proteins that interact with the selected baits, as indicated by significant enrichment under selection, are reconstituted in silico with their mapped fusion reads to identify the prey sequence containing the interaction domain for further studies. Note that this approach delineates the specific interaction region, and not necessarily the full‐length clone, which is inferred from the reference genome annotation.
Using the AVRA1 and BEC1016 effectors as baits to screen a three‐frame Y2H library made from our infection time course (see Experimental Procedures), we identified HORVU1Hr1G022990, herein designated HvERdj3B (GenBank ID: AK376215.1 and KAE8793706.1) as a candidate interactor for both (Figures 2 and S6). Ranking (Table S2) and visualization of the candidate interactors for each bait identified a prey fragment covering the C‐terminal two‐thirds of HvERdj3B (aa 142–350). Subsequently, binary Y2H according to Dreze et al. (2010) confirmed interaction between this HvERdj3B fragment and AVRA1 as well as BEC1016 (Figure S7).
FIGURE 2.
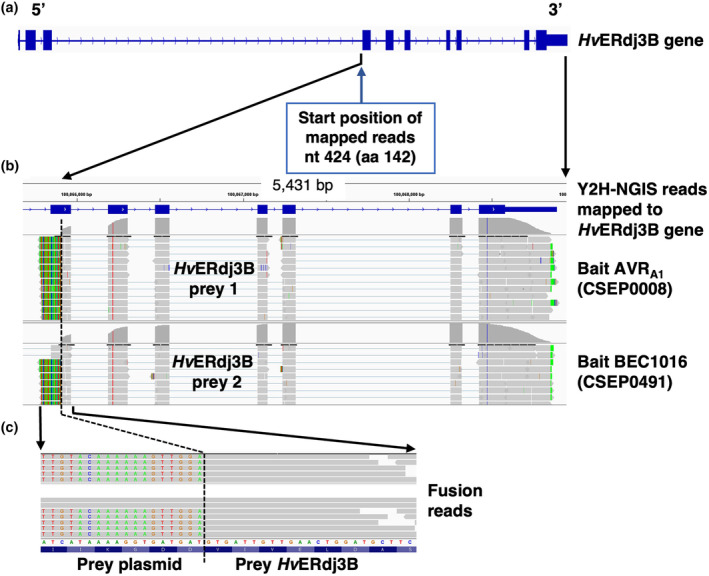
Yeast two‐hybrid next‐generation interaction screen (Y2H‐NGIS) used to identify interactions between AVRA1, BEC1016 and HvERdj3B. (a) HvERdj3B gene model with exons depicted as blue boxes and introns as hash marks. (b) Integrative genomic view (IGV) obtained from the software NGPINT after Y2H‐NGIS analysis of the HvERdj3B prey reads selected by both the AVRA1 and the BEC1016 bait. Prey fragments were reconstructed from the reads mapped to the HvERdj3B gene (shown in grey across the exons) in each Y2H‐NGIS dataset and located towards the 3′ end of the gene in both cases. (c) Expanded view of the 5′ fusion reads that allowed the determination of the frame and nucleotide‐resolved prey fragments. The fusion reads contain a prey plasmid sequence (shown in different colours as mismatches from the reference gene) and the prey sequence. See also Figure S6.
To study the specificity of these interactions further, we performed extensive binary Y2H assays with an independent colourimetric Y2H system (see Experimental Procedures). The two effectors (without signal peptide) were fused to either the Gal4 transcription factor DNA‐binding domain (BD) or transcription‐activation domain (AD) and combined with either the full‐length (without signal peptide), the C‐terminal two‐thirds (amino acids 142–350) or the central part (amino acids 142–231) of HvERdj3B fused to either the AD or the BD. In both orientations of the setup, both effectors were found to interact with the C‐terminal two‐thirds and the central part, but not the full‐length of HvERdj3B (Figure 3). It is common to recover only a fragment and not a full‐length protein interaction in Y2H assays. Several factors like protein size, domain toxicity and accessibility can explain a negative interaction in yeast, which may vary when tested in planta (Galletta & Rusan, 2015). Nevertheless, these results suggest that the N‐terminus of HvERdj3B is not necessary for effector binding and that the interaction occurs only at the central part of HvERdj3B. In turn, this explains why reads did not appear from the 5′ end of the coding sequence in the Y2H‐NGIS sequencing results (Figure 2).
FIGURE 3.
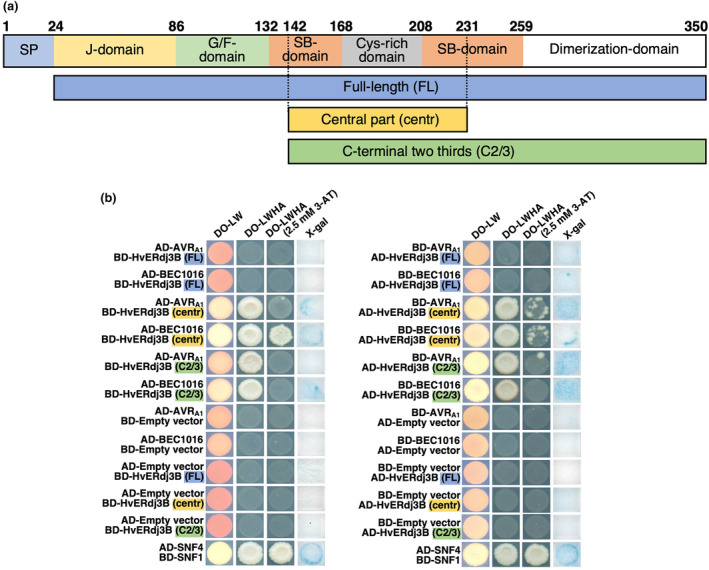
AVRA1 and BEC1016 interact with the central part of barley HvERdj3B in yeast two‐hybrid (Y2H) assay. (a) Domain structure of the 350 amino acid HvERdj3B protein and the Y2H fragments. The domains are predicted according to Chen et al. (2017) (see Figure S6). SP, signal peptide; G/F, glycine/phenylalanine; SB, substrate binding. Below the three Y2H fragments are shown. (b) Yeast transformed with the destination vectors pDEST‐AS2‐1 [GAL4 binding‐domain (BD)] and pDEST‐ACT2‐1 [GAL4 activation‐domain (AD)] (Robertson, 2004) in different combinations of domain fusions to effectors and the full‐length (FL), the central part (centre) and the C‐terminal two‐thirds (C2/3) of HvERdj3B. Growth on dropout (DO) medium lacking leucine (L) and tryptophan (W) indicated presence of both constructs. Growth on DO medium lacking L, W, histidine (H) and adenine (A) ± 2.5 mM 3‐amino‐1,2,4‐triazole (3AT) indicated protein–protein interaction. β‐galactosidase assay (X‐gal), also indicating protein–protein interaction, was performed on filter paper prints of DO‐LW‐grown yeast. SNF1/SNF4 was used as a positive control and empty vectors were used as negative control.
To confirm interactions between AVRA1 or BEC1016 and HvERdj3B in planta, a bimolecular fluorescence complementation (BiFC) assay (Hu & Kerppola, 2003) was carried out in N. benthamiana leaf epidermal cells after Agrobacterium‐mediated transformation. In this study, only the full‐length HvERdj3B (without signal peptide) was used. Fluorescence was observed only when the HvERdj3B‐cGFP fusion was combined with nGFP fused to the N‐terminal or the C‐terminal of the effectors (Figures 4a and S8). The specificity of the interactions was further documented by co‐immunoprecipitation (Co‐IP) (Figure 4b).
FIGURE 4.
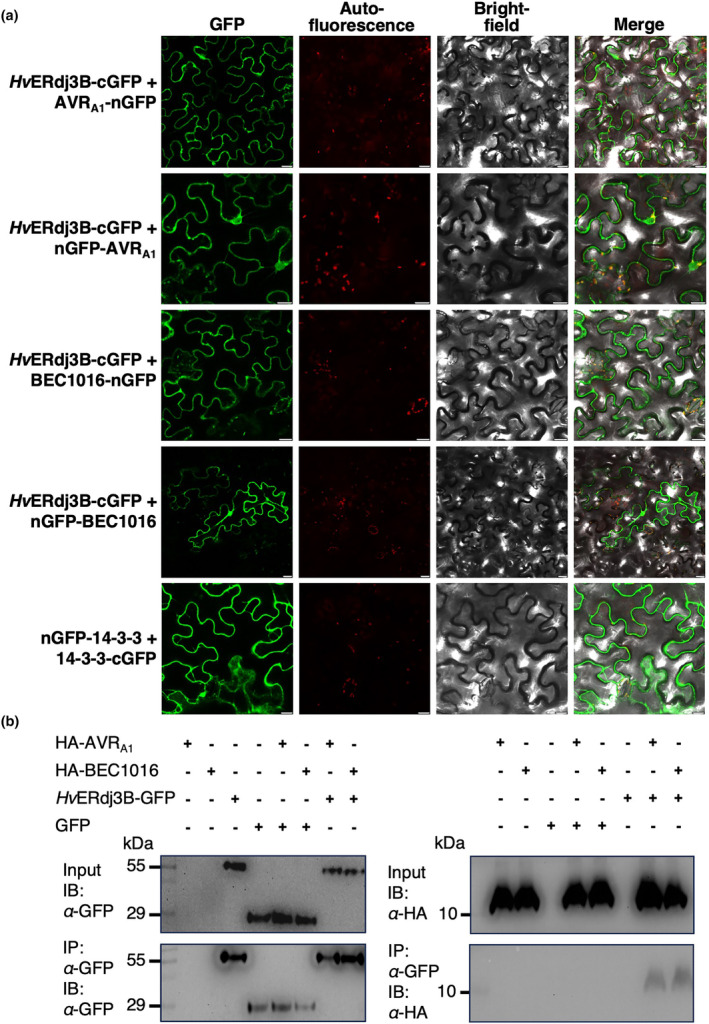
Interactions of HvERdj3B and AVRA1 as well as BEC1016 confirmed in living plant cells. (a) Reconstitution of fluorescent GFP (bimolecular fluorescence complementation) was observed in Nicotiana benthamiana epidermal cells expressing the shown combinations of the C‐ and N‐terminal parts of GFP fused with HvERdj3B (without signal peptide) and AVRA1/BEC1016 (without signal peptide). The remaining combinations did not allow fluorescent GFP to reconstitute (see Figure S8). 14‐3‐3 dimerization was used as positive control. Expression constructs were introduced using Agrobacterium infiltration, and epidermal cells were observed by laser scanning confocal microscopy 48 h later. Size bars, 20 μm. (b) Co‐immunoprecipitation of HvERdj3B and AVRA1/BEC1016. Construct combinations for expression of the indicated proteins (all without signal peptide) were introduced in leaves of N. benthamiana using Agrobacterium infiltration. Three days later the proteins were extracted from the leaves and analysed by SDS‐PAGE/immunoblotting (IB) with anti‐GFP and anti‐HA antibodies before and after immunoprecipitation (IP) using anti‐GFP magnetic beads. Expected protein molecular weight: GFP, 27 kDa, HvERdj3B‐GFP, 65 kDa, HA‐AVRA1, 13 kDa and HA‐BEC1016, 13 kDa.
Taken together, these results confirmed that amino acids 142–231 of the J‐domain protein HvERdj3B interact with the diverged RALPH effectors AVRA1 and BEC1016. According to alignment with the most closely related mammalian J‐domain protein, this segment corresponds with the predicted substrate‐binding and the Cys‐rich domains (Chen et al., 2017; Figure S6). While this domain is inaccessible for binding in the full‐length HvERdj3B in Y2H experiments, it appears accessible in planta, which is a prerequisite for the interactions to be functionally relevant.
2.3. HvERdj3B is an ER luminal J‐domain protein required for immunity during powdery mildew attack
HvERdj3B encodes a J‐domain protein with an N‐terminal signal peptide for ER targeting. The mature protein shares 77% amino acid identity with its closest Arabidopsis homologue, AtERdj3B (Figure S6), which is localized to the ER (Yamamoto et al., 2008). Significant accumulation of the HvERdj3B transcript was observed during Bh penetration, peaking at 20 h after inoculation, which would be consistent with a role for it in immunity (Figure S2c). Indeed, AtERdj3B has previously been implicated in immunity in Arabidopsis, where it is required for triggering of responses to the bacterial elongation factor‐Tu (Nekrasov et al., 2009). To investigate whether HvERdj3B contributes to the immunity to Bh, we silenced the HvERdj3B gene in barley plants using transient‐induced gene silencing (TIGS) (Douchkov et al., 2005). Two RNAi constructs were generated with different HvERdj3B fragments and, together with a GUS reporter construct, bombarded into single epidermal cells of leaves of 1‐week‐old Golden Promise plants. Two days later, the leaves were inoculated with Bh isolate C15 and formation of fungal haustoria was assessed after another 2 days. Our data revealed that silencing of HvERdj3B significantly enhanced the penetration rate, as HvERdj3B‐RNAi‐1 and HvERdj3B‐RNAi‐2 on average tripled the number of haustoria (p < 0.001). The Mlo RNAi‐positive control reduced the number of haustoria by c. 40% (Figure 5a). This increase in the level of infection suggests that HvERdj3B plays an essential role in barley immunity.
FIGURE 5.
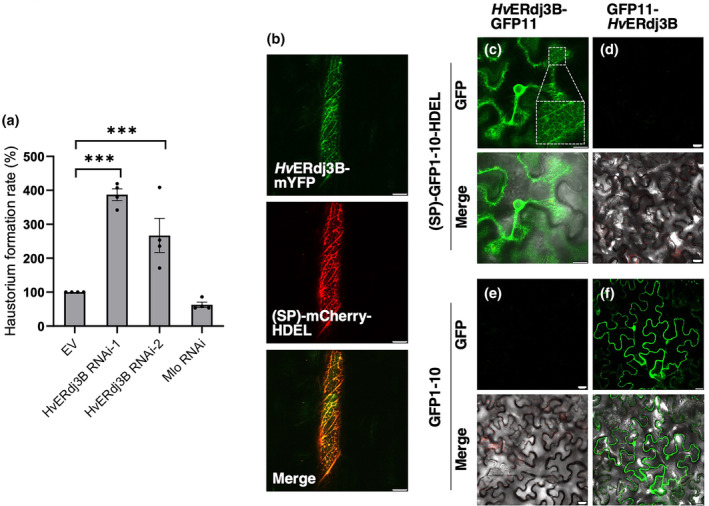
HvERdj3B is an endoplasmic reticulum luminal protein contributing to preinvasive immunity. (a) One‐week‐old barley leaves, bombarded with RNAi and β‐glucuronidase (GUS) reporter constructs, were inoculated with Blumeria hordei and scored for fungal haustorium formation, calculated as the ratio of haustoria‐containing transformed cells (GUS‐expressing cells) divided by the total number of transformed cells. Data shown are mean values of four independent experiments. EV, empty vector. Error bars, SE. ***p < 0.001 calculated using a logistic regression model. Raw data are available in Table S1. (b) Localization of HvERdj3B in barley leaf epidermal cells. A ubiquitin promoter‐driven expression construct encoding HvERdj3B (with signal peptide [SP]) fused to the N‐terminus of mYFP was co‐transformed with an SP‐mCherry‐HDEL construct into barley epidermal cells using particle bombardment. The cells were observed by confocal laser scanning microscopy 48 h later. (c–f) Localization of HvERdj3B in Nicotiana benthamiana leaf epidermal cells using the split GFP system. HvERdj3B (with SP)‐GFP11 and (SP)‐GFP1‐10‐HDEL reconstitute reticulate fluorescent GFP, while GFP11‐HvERdj3B (without SP) and GFP1‐10 reconstitute cytosolic and nuclear fluorescent GFP. Expression constructs were introduced using Agrobacterium infiltration, and epidermal cells were observed by laser scanning confocal microscopy 48 h later. Size bars, 20 μm.
To reveal HvERdj3B's subcellular localization, it was expressed with its signal peptide and a C‐terminal mYFP fusion in barley leaf epidermal cells. Laser scanning confocal microscopy indicated that HvERdj3B is an ER protein as the mYFP signal has a reticulate pattern that overlaps with the (SP)‐mCherry‐HDEL ER marker (Figure 5b). This localization of HvERdj3B found in barley was confirmed in N. benthamiana (Figure S9). Next, we tested if HvERdj3B, as expected, is an ER luminal protein. Here, we made use of a split GFP system different from BiFC. GFP is barrel‐shaped and consists of 11 β‐sheets (GFP1–11), which can be split into two fragments, β‐sheets 1–10 (GFP1–10) and β‐sheet 11 (GFP11). The two parts of GFP have affinity for one another, and when they colocalize in the same compartment, a fluorescent GFP complex will be assembled (Xie et al., 2017). Thus, when a HvERdj3B‐GFP11 fusion protein was co‐expressed with an SP‐GFP1–10‐HDEL construct that delivers and retains GFP1–10 in the ER lumen, perinuclear and reticulate GFP signals appeared (Figure 5c). No visible signal appeared when HvERdj3B‐GFP11 was co‐expressed with cytosolic GFP1–10 (Figure 5e). Furthermore, when GFP11‐HvERdj3B was co‐expressed with either (SP)‐GFP1–10‐HDEL or with cytosolic GFP1–10, GFP signal was absent or present in the cytosol and nucleus, respectively (Figure 5d,f). In summary, these results show that HvERdj3B is important for barley immunity to Bh and that it is located inside the ER lumen. This complements accumulating evidence of the role of ERQC and ERdj3B in plant immunity (Nekrasov et al., 2009; Tintor & Saijo, 2014).
2.4. AVRA1 and BEC1016 translocate from the plant cytosol to the ER lumen
For interactions to occur between either AVRA1 or BEC1016 and HvERdj3B in barley epidermal cells attacked by Bh, the two effectors are required to be secreted from the fungus, when their signal peptides are removed, and to be taken up, either across the plant plasma membrane or the extrahaustorial membrane, into the plant cytosol. Secondly, in the plant cell, the effectors are required to pass the ER membrane into the ER lumen. Indirect evidence that AVRA1 translocates into the plant epidermal cell cytosol comes from the observation that it functions as an avirulence protein that binds to the cytosolic NLR‐protein MLA1 and triggers HR (Lu et al., 2016). The fact that the number of haustoria is increased when BEC1016 is expressed in the epidermal cell cytosol after particle bombardment suggests that BEC1016 also functions inside the plant cell (Figure 1a). To inquire whether AVRA1 and BEC1016 can translocate across the ER membrane, we started by overexpressing AVRA1‐GFP and BEC1016‐GFP (both without signal peptide) in N. benthamiana epidermal cells. Here, they were both localized to the cytosol and nucleus together with free mCherry, and no obvious ER signal could be distinguished as there apparently was a complete overlap between the GFP and mCherry fluorescent signals (Figure 6). Expression in barley epidermal cells also suggested the effectors mainly to be cytosolic (Figure S10).
FIGURE 6.
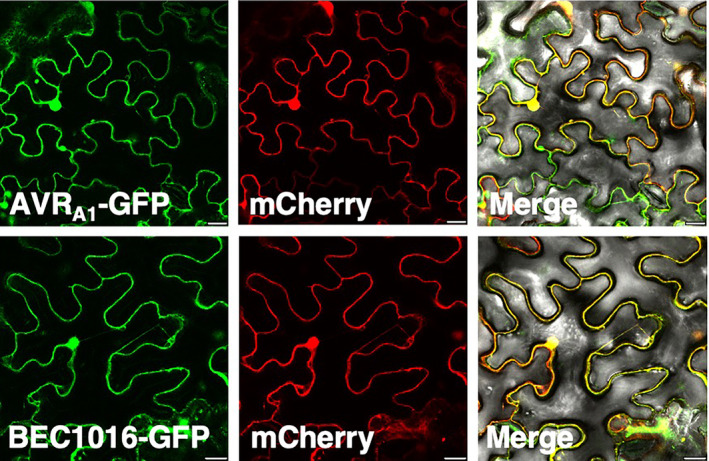
AVRA1 and BEC1016 localized in the cytosol and nucleus of Nicotiana benthamiana epidermal cells. Constructs for expression of effectors (without signal peptide) were introduced using Agrobacterium infiltration, and epidermal cells were observed by laser scanning confocal microscopy 48 h later. Size bars, 20 μm.
However, to interact with HvERdj3B, the two effectors will have to enter the plant ER. To test this more rigorously, we again used our split GFP system (Xie et al., 2017). We generated constructs for fusing GFP11 to both the N‐ and C‐terminus of AVRA1, BEC1016 and CSEP0105, all without signal peptide. CSEP0105, which interacts with cytosolic small heat shock proteins (Ahmed et al., 2015), was used as a negative control. These were then co‐expressed with constructs for cytosolic GFP1–10 and ER‐luminal (SP)‐GFP1–10‐HDEL in all combinations (Figure 7). GFP11 fused to the N‐ and C‐terminal of CNX™, the transmembrane domain of the ER membrane protein calnexin 1 (Xie et al., 2017), was used as reference (Figure 7a,b,i,j). All three GFP11‐effector fusions combined with GFP1–10 resulted in cytosolic and nuclear GFP signal (Figure 7c–h). Yet, when combined with (SP)‐GFP1–10‐HDEL, removal of the cytosolic and nuclear GFP signal visualized distinct reticulate and perinuclear GFP signals in the case of AVRA1 and BEC1016 (Figures 7k–n and S11). Because this was not seen for CSEP0105 (Figure 7o,p) and because the ER‐localized GFP11‐CNX™/GFP1–10‐HDEL combination also gave a reticulate network signal (Figure 7i), the data indicate that a fraction of the cytosolic AVRA1 and BEC1016 specifically translocate into the ER lumen post‐translationally and independently of their signal peptides.
FIGURE 7.
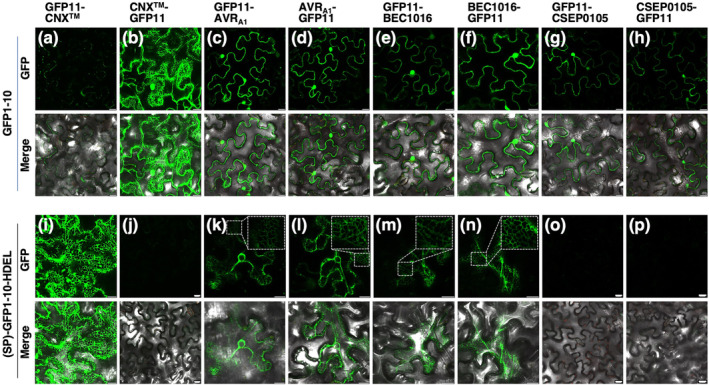
AVRA1 and BEC1016 are partially translocated into the endoplasmic reticulum (ER) lumen. The GFP1–10/GFP11 self‐constituting fluorescence protein demonstrates cytosolic and ER‐luminal protein localizations in Nicotiana benthamiana epidermal cells. (a,b) and (i,j) GFP11‐CNX™ and CNX™‐GFP11 confirm (SP)‐GFP1–10‐HDEL to be ER‐luminal and GFP1–10 to be cytosolic, respectively. (c–h) and (k–p) AVRA1, BEC1016 and CSEP0105 (all without signal peptide) are cytosolic while AVRA1 and BEC1016 (both without signal peptide) are also ER‐luminal, indicated by their reticulate and perinuclear signals (see also Figure S11). Expression constructs were introduced using Agrobacterium infiltration, and epidermal cells were observed by laser scanning confocal microscopy 48 h later. Size bars, 20 μm.
2.5. Silencing of HvERdj3B and expression of AVRA1 and BEC1016 affect ER trafficking
Having observed an importance of AVRA1, BEC1016 and HvERdj3B in immunity and localization of these three proteins in the plant ER lumen, we were prompted to test whether they can affect ER function. For this, we made use of the vacuolar marker RFP‐AFVY, expressed with an N‐terminal signal peptide. The C‐terminal AFVY amino acid sequence, which guides the protein to the vacuole, is derived from the vacuolar storage protein phaseolin (Hunter et al., 2007). When the (SP)‐RFP‐AFVY construct was expressed together with the empty‐vector control, a clear vacuolar localization was observed (Figure 8). However, when co‐expressed with AVRA1 or BEC1016, the RFP signal was also detected around the nucleus and to some extent in reticular structures, which are signs of (SP)‐RFP‐AFVY being retained in the ER. The same signal pattern was observed when HvERdj3B was silenced using the RNAi‐1 hairpin construct, also used in Figure 5a above (Figure 8). When expression of the effectors and silencing of HvERdj3B was combined, a similar pattern of the RFP signal was again seen. This same cellular phenotype, induced by expression of the effectors and silencing of HvERdj3B, coincides with hampering of the ERQC and with this J‐domain protein being targeted by the effectors in the ER lumen.
FIGURE 8.
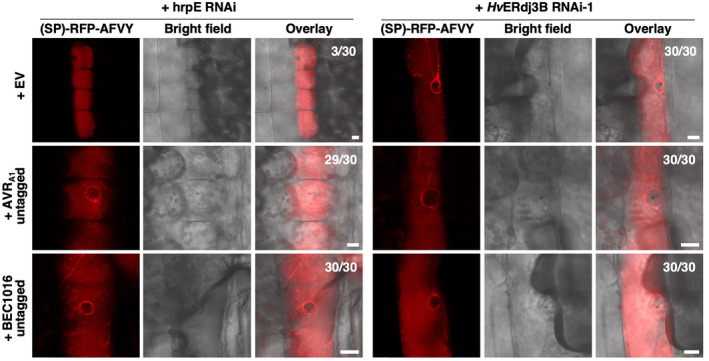
Overexpression of AVR1A and BEC1016 or RNAi of the target HvERdjB3 cause endoplasmic reticulum (ER) retention of a vacuolar marker. The vacuolar marker construct p35S::SP‐RFP‐AFVY was co‐expressed with either the pUbi::GW empty vector (EV), pUbi::AVRA1 or pUbi::BEC1016 (both without signal peptide), along with either the control RNAi construct (hrpE‐RNAi) or an RNAi construct for HvERdj3B, in barley epidermal cells upon particle bombardment. The vacuolar marker (SP)‐RFP‐AFVY was partially mislocalized in an ER‐like structure after overexpression of AVRA1 and BEC1016 or after RNA‐interference of HvERdj3B. This was evident by its presence in a perinuclear ring and occasionally in reticular structures. Counts indicate occurrence of mislocalization of the vacuolar marker in 30 cells. Scale bars, 10 μm.
3. DISCUSSION
Obligate biotrophic fungi feed on living plant tissues for nutrient uptake via haustoria. To suppress host defences and promote colonization, these pathogens deliver a large repertoire of effectors into host cells (Frantzeskakis et al., 2018; Menardo et al., 2017). We focused on Bh AVRA1 and BEC1016, as examples of avirulence proteins and proteins having a documented contribution to fungal virulence, respectively (Lu et al., 2016; Pliego et al., 2013). Here, we found that bacterial T3SS‐based delivery of both these effectors in fact reduced defence‐associated callose deposition and that overexpression of them enhanced the development of Bh haustoria, while they did not affect Mla3‐mediated HR. Thus, our data may illustrate that Blumeria AVR proteins can also suppress PTI. Through Y2H‐NGIS, these effectors were found to target the same host protein, HvERdj3B, which we further substantiated with binary Y2H as well as in planta BiFC and Co‐IP assays. Silencing of the HvERdj3B gene in barley led to increased formation of Bh haustoria, indicating that this ER protein is involved in immunity to fungal penetration. This, and the observation that AVRA1 and BEC1016 can enter the ER, and that expression of them as well as silencing of HvERdj3B cause arrest of a vacuolar marker in the ER, supports the immunity‐suppressing function of these effectors to be mediated through targeting of this J‐domain protein.
ERQC and ER stress responses are well documented to contribute to plant immunity. Our data are consistent with a previous study showing that a T‐DNA line mutated in HvERdj3B's closest homologue in Arabidopsis, AtERdj3B, is more susceptible to bacterial pathogens (Nekrasov et al., 2009). Here, it was reported that AtERdj3B is involved in PTI. In the ER, it forms a complex with SDF2 and the Hsp70 BiP1, which are required for proper trafficking and function of the plasma membrane leucine‐rich‐repeat receptor kinases, EFR. HvERdj3B may have a similar function and may be involved in proper functioning of PRRs in barley. Besides being required for processing of PRRs, ERQC is required for secretion of antimicrobial proteins and linked to pathogen‐induced programmed cell death (Kørner et al., 2015; Li et al., 2009; Moreno et al., 2012; Qiang et al., 2012). In addition, a recent study showed that the ER proteins UBAC2 and PICC interact with each other and are required for proper delivery of callose synthase to the plasma membrane and for flagellin‐triggered callose deposition (Wang et al., 2019). As studies in Arabidopsis have demonstrated that callose is important for immunity towards penetration by powdery mildew fungi (Voigt, 2016), it will be interesting to see if future studies can demonstrate that the ERdj3B/BiP complex is required for UBAC2/PICC‐dependent callose deposition. Therefore, even if ERdj3B is essential for maturation of certain PRRs in the ER, there might be additional ways by which it contributes to plant immunity and thus how Bh uses AVRA1 and BEC1016 to aid the penetration process in barley.
Localization studies initially suggested both effectors to be cytosolic and nuclear, while HvERdj3B was found to be ER‐luminal. Therefore, we used the split GFP system to demonstrate that these effectors are also translocated into the ER post‐translationally. There are other studies showing that pathogen effectors enter the ER to facilitate infection by targeting ER‐localized host proteins. For instance, the RXLR effector PcAvr3a12 from an Arabidopsis oomycete pathogen, Phytophthora capsici, can post‐translationally enter the ER and target the ER‐localized host PPIase FKBP15‐2, which is involved in ER stress‐sensing and required for ER stress‐mediated plant immunity (Fan et al., 2018). Furthermore, it has been reported that an essential effector, PsAvh262, secreted by the soybean pathogen Phytophthora sojae, translocates from the host cytosol into the ER to stabilize BiPs, thereby suppressing ER stress‐triggered cell death and facilitating infection (Jing et al., 2016). In both cases, it was shown by BiFC that effector–target interactions occur inside the ER. Yet another recent study showed that a nematode effector (CLE) is translocated from the plant cell cytosol to the ER, and then secreted as an extracellular receptor ligand (Wang et al., 2021). Here, post‐translational ER uptake was demonstrated using the GFP1–10/11 system, which we also used. Together, these observations suggest that these effectors hijack one or more signal peptide‐independent plant ER uptake mechanisms.
Interactions with HvERdj3B were found for both AVRA1 and BEC1016, and both enter the ER post‐translationally and signal peptide‐independently. They have very diverse amino acid sequences but appear derived from an ancient RNase and may potentially be structurally related (Cao et al., 2023; Pedersen et al., 2012). Therefore, we used AlphaFold2 to predict the structures of AVRA1 and BEC1016, and in an overlay between those we can suggest shared loops on the protein surfaces (Figure S1). Yet, no identical or similar amino acids could be identified in these loops, which otherwise could have been suggested to mediating the interaction with HvERdj3B.
ER post‐translational uptake of proteins through the Sec61 translocon complex is known to engage the ERQC machinery (Hassdenteufel et al., 2019; Zimmermann et al., 2011). Thus, we consider that interactions of AVRA1 and BEC1016 with HvERdj3B may facilitate one‐way movement through this translocon into the ER lumen. Previously, we found that silencing of the barley Sec61 β‐ and γ‐subunits both reduce the plant's susceptibility to Bh (Xu et al., 2015; Zhang et al., 2013). At the time, these findings were difficult to explain as preventing ER uptake of immunity‐associated proteins, such as PR‐proteins and PM‐bound PRR, should increase plant susceptibility. However, our present finding of a susceptibility‐promoting role of AVRA1 and BEC1016 (Figure 1), which both enter the ER, may make the consequences of Sec61 silencing (Xu et al., 2015; Zhang et al., 2013) more meaningful.
NLR‐recognition of effectors often occurs indirectly after they have impacted effector targets, which are guarded and associated with the NLRs (Carter et al., 2019; Ngou et al., 2022; Thordal‐Christensen, 2020). However, an effector target that contributes to immunity, like HvERdj3B, inside the ER may not be guarded by an NLR, as to date, such receptors have not been localized in this compartment. Instead, this favours a requirement for direct effector monitoring by NLRs in the cytosol or the nucleus, which agrees with the observed direct recognition and interaction of AVRA1 and the NLR, MLA1 (Lu et al., 2016). One may speculate whether targeting an ER luminal host protein thus has provided an evolutionary benefit to the pathogen. The effector may escape from indirect recognition and force the plant to make more demanding inventions of specific recognition of each effector, rather than guarding proteins targeted by more effectors, potentially from different pathogens. Whether the recently discovered ER‐localized non‐NLR wheat powdery mildew resistance protein Pm4 (Sánchez‐Martín et al., 2021) guards an ER‐luminal process remains unknown. Pm4 is a protein kinase, and it indeed has an ER‐luminal loop that may have such a guarding function.
4. EXPERIMENTAL PROCEDURES
4.1. Plant and fungal materials
Seedlings of barley (Hordeum vulgare) cv. Golden Promise were grown at 16 h light (150 μmol s−1 m−2, 20°C)/8 h of darkness (20°C) to be used for inoculations, gene amplifications, TIGS, overexpression and callose deposition assays after effector‐delivery using P. fluorescens EtHAn. Bh isolates C15 (AVR A1 ) and A6 (AVR A3 ) were propagated on barley P‐02 (Mla3) and P‐01 (Mla1) plants, respectively, in a cycle of 1 week. Four‐ to six‐week‐old N. benthamiana plants were used for BiFC and subcellular localization studies after Agrobacterium‐mediated leaf cell transformation.
4.2. Gateway plasmid construction
Coding DNA sequences (CDS) for the effectors AVRA1 and BEC1016, without their signal peptides, were amplified using primer pairs listed in Table S3. PCR was performed on cDNA from barley leaves infected with Bh, and the fragments were cloned into pENTR/D‐TOPO vectors (Invitrogen), with or without stop codons. The CDS for the full‐length barley HvERdj3B (HORVU1Hr1G022990; GenBank ID: AK376215.1 and KAE8793706) with stop codon was synthesized by TWIST Bioscience (San Francisco, CA, USA). Fragments of HvERdj3B were amplified from the synthesized clone using primers listed Table S3 and cloned into pENTR/D‐TOPO. Subsequently, the entry clone inserts were transferred to destination vectors using Gateway LR cloning reactions (Invitrogen). For overexpression constructs, the CDSs for HvERdj3B (with signal peptide), AVRA1 and BEC1016 (without signal peptides) were transferred into pUbi‐mYFP‐Gateway‐Nos, pUbi‐Gateway‐mYFP‐Nos, pUbi‐Gateway‐Nos destination vectors (Kwaaitaal et al., 2010) and p2WFHB‐Gateway‐GFP (Karimi et al., 2002). The vacuolar marker SP‐RFP‐AFVY construct, where the AFVY vacuolar targeting signal is derived from phaseolin, was amplified from ‘sp‐RFP‐AFVY’ (Hunter et al., 2007). The SP‐mCherry‐HDEL construct was from Nelson et al. (2007). RNAi constructs were generated in the CaMV 35S promoter‐driven hairpin destination vector pIPKTA30N (Douchkov et al., 2005). The HvERdj3B‐RNAi‐1 and ‐2 constructs contain HvERdj3B CDS fragments from positions 283 to 579 and 532 to 879, respectively. These sequences were predicted using the si‐Fi21 open‐source software (Lück et al., 2019). None of them were found to have off‐targets. For Co‐IP, the CDS for AVRA1 and BEC1016 (without signal peptide) were transferred into destination vector pEarleyGate201, which is a Gateway‐compatible vector encoding an N‐terminal HA tag (Earley et al., 2006). The CDS of HvERdj3B (without signal peptide) was transferred into destination vector pK7FWG2, a Gateway‐compatible vector encoding a C‐terminal GFP tag (Karimi et al., 2002). All the constructs were sequenced for confirmation.
4.3. Barley single cell transient‐induced gene silencing and overexpression
Barley leaf epidermal cell transformation was obtained after particle bombardment as described by Douchkov et al. (2005) and Nowara et al. (2010), using the biolistic PDS‐1000/He Particle Delivery System from Bio‐Rad. For each bombardment, six detached first leaves of 1‐week‐old Golden Promise barley plants were used. The particle coating was performed using 7 μg of DNA for each construct together with 2.4 mg of gold, 1 μg/μL of protamine (Sivamani et al., 2009) and 0.625 M CaCl2 (Rasco‐Gaunt et al., 1999). For the bombardments, a hepta adapter and rupture discs bursting at a helium pressure of 1100 psi were used. After bombardment, the leaves were transferred onto 1% phytoagar Petri dishes containing 40 mg/mL benzimidazole. For effector overexpression and TIGS studies, the constructs were co‐transformed with a GUS reporter gene construct driven by the pUbi promoter into the epidermal cells. Two days later, the leaves were inoculated with Bh, and after another 2 days, they were stained for GUS activity by vacuum‐infiltrating and incubating in 2 mM X‐Gluc, 100 mM sodium phosphate, 100 mM EDTA, 1.4 mM potassium ferricyanide, 1.4 mM potassium ferrocyanide and 0.1% Triton X‐100 at 37°C overnight. Transformed (blue) cells were assessed microscopically for the presence of haustoria as an indication of fungal infection. The rate of haustoria formation was calculated as the number of blue cells with haustoria divided by the total number of blue cells. An mYFP expression construct and the empty vector pIPKTA30N were used as negative controls and the Mlo RNAi (pIPKTA36) (Douchkov et al., 2005) construct was used as positive control. The effect of constructs was analysed by a logistic regression model with random effects, assuming a binominal distribution, with construct as fixed effect and the experiment × construct × leaf interaction as random effect. The analyses were performed in PC‐SAS (release 9.4, SAS Institute).
4.4. A. tumefaciens infiltration‐mediated transformation of N. benthamiana leaf cells
T‐DNA construct were introduced into A. tumefaciens GV3101 by electroporation. The transformed bacterial cells were grown on Luria Bertani agar plates supplemented with rifampicin, spectinomycin and gentamycin antibiotics. For leaf cell transformation, overnight liquid cultures (28°C) of recombinant A. tumefaciens were harvested by centrifugation and resuspended in 10 mM MgCl2, 10 mM MES and 200 μM acetosyringone to OD600 = 0.6. A strain of A. tumefaciens with a construct expressing the p19 silencing suppressor (Brioudes et al., 2022) was managed in the same way. The resuspended A. tumefaciens transformants, including the one with the P19 construct, were mixed in equal ratios and infiltrated into leaves of 4‐ to 6‐week‐old N. benthamiana plants.
4.5. Yeast two‐hybrid next‐generation interaction screen
The Bh effectors AVRA1 and BEC1016 were used as baits in Y2H‐NGIS to mine for novel PPI (Elmore et al., 2023; Velásquez‐Zapata, Elmore, & Wise, 2023). Bait sequences (without signal peptides) were fused with the GAL4 transcription factor binding domain (GAL4‐BD) in the p97‐BD, Leu2p vector and transformed into Saccharomyces cerevisiae Y8930 (Dreze et al., 2010). For the prey library, first seedling leaves from an infection time course of the resistant barley line CI 16151 (Mla6) and four fast‐neutron‐derived immune mutants were sampled from a split‐plot design at 0, 16, 20, 24, 32 and 48 h after inoculation (HAI) with Bh isolate 5874 (AVR a1 , AVR a3 , AVR a6 and AVR a12 ) (Chapman et al., 2021; Surana, 2017; Velásquez‐Zapata et al., 2022). mRNAs isolated from the 90 experimental units (5 genotypes × 6 time points × 3 biological replications) were used for RNA‐Sequencing (data at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE101304), small RNA sequencing (Hunt et al., 2019), and was also pooled to prepare a three‐frame cDNA prey library in the Gateway‐compatible ARS4/CEN6 GAL4 activation domain (AD) vector p86‐AD, Trp1p (Dreze et al., 2010; Velásquez‐Zapata et al., 2021; Yu et al., 2015). After transforming the library into S. cerevisiae Y8800, 1.1 × 107 primary clones were mated with each bait clone and subsequently cultured in three independent replicates under two conditions (selection and non‐selection). At the end of each culture, prey plasmids were isolated, and prey cDNA fragments were amplified by low‐cycle PCR to maintain the bait:prey ratios resulting from the batch culture mating. Amplicon libraries were sequenced using the Illumina HiSeq 2500 platform at the Iowa State University DNA facility, collecting 5–10 million reads per sample. Sequence output from NGIS screens confirmed that the prey library contained 78.4% of annotated genes in the barley Morex V3 assembly (Mascher et al., 2021), and nearly 99% of the expressed genes from our CI 16151 transcriptome (NCBI‐GEO GSE101304). Y2H‐NGIS data from these experiments were processed using the NGPINT and Y2H‐SCORES software (Banerjee et al., 2021; Velásquez‐Zapata et al., 2021) to reconstruct the prey fragments and rank them as interactors for each bait. Outputs from these pipelines allowed us to identify interacting prey fragments, their frame with the Gal4‐AD as well as enrichment and specificity scores that assess their properties as Y2H interactors.
4.6. Binary Y2H assays
Using the output from the Y2H‐NGIS, we designed primers to reclone identified prey fragments into p86‐AD to confirm interactions using binary Y2H (Dreze et al., 2010) under three levels of selective media: Diploid selection (SC − LW) and specific selection (SC − LWH + 0.1 mM 3‐AT) using three dilutions (100, 10−1 and 10−2) as shown in Figure S7. After the first confirmation of HvERdj3B as prey, additional AVRA1, BEC1016 and HvERdj3B constructs were made in the destination vectors pDEST‐AS2‐1 (GAL4‐BD) and pDEST‐ACT2‐1 (GAL4‐AD) (Robertson, 2004). These constructs were transformed into the haploid yeast strains, Y189 and Y190, respectively. Subsequent matings, selections and LacZ reporter assays were made according to the Matchmaker Gold Yeast Two‐Hybrid System user manual (Clontech).
4.7. In planta protein–protein interaction studies
For the BiFC assay, the CDS for full‐length barley HvERdj3B and both effectors, without signal peptides, and with and without stop codons, were transferred to CaMV 35S promoter‐driven BiFC binary destination vectors (Kamigaki et al., 2016) as N‐ and C‐terminal fusions of nGFP (amino acids 1 to 174) and cGFP (amino acids 175 to 239). Two‐by‐two combinations of constructs were transformed into N. benthamiana leaves by A. tumefaciens infiltration. Two days after infiltration, the fluorescence signals of all eight possible AVRA1/HvERdj3B combinations and of all eight possible BEC1016/HvERdj3B combinations were evaluated by a laser scanning confocal microscopy.
For the Co‐IP assay, N. benthamiana was agro‐infiltrated with constructs for expression of HvERdj3B‐GFP in combination with either HA‐AVRA1 or HA‐BEC1016, all without signal peptides. Two days later, Co‐IP was performed according to Gruner et al. (2021) using 15 μL α‐GFP‐magnetic beads (Chromotek). Anti‐GFP antibody (sc9966), anti‐HA antibody (sc7392 HRP) and secondary antibody m‐IgGκ BP‐HRP (sc‐516102) were purchased from Santa Cruz Biotechnology.
4.8. Localization of effectors in the ER
The GFP1–10/GFP11 split GFP system (Xie et al., 2017) was used to document presence of effectors in the ER lumen. CDS for the effectors AVRA1 and BEC1016, without their signal peptides, were transferred to the T‐DNA Gateway destination vectors pGFP11‐GW and pGW‐GFP11 to fuse them to GFP11. T‐DNA constructs for GFP11‐CNX™ and CNX™‐GFP11 were from Xie et al. (2017). These were co‐expressed with constructs for cytosolic GFP1–10 and ER‐luminal SP‐GFP1–10‐HDEL from Xie et al. (2017).
4.9. Confocal microscopy
The microscopy was performed using a Leica SP5‐X laser scanning microscope at the Center for Advanced Biomaging (CAB) at the University of Copenhagen. To improve the subcellular localization, the leaves were mounted with perfluorodecalin (Alfa Aesar A18288) and imaged with a 20× water immersion lens. GFP was excited at 488 nm and the emission from 514 to 540 nm was collected. mYFP was excited at 514 nm and the emission from 527 to 586 nm was collected. mCherry was excited at 587 nm and the emission from 595 to 650 nm was collected. To restrict bleed‐through, all imaging was done using sequential scan mode.
4.10. Callose deposition and HR assays
CDSs for the GUS reporter gene, effectors AVRA1 and BEC1016, without signal peptides and with stop codons were transferred to the AvrRPS4 promoter‐driven destination vector pEDV6, which also fuses 136 amino acids of the AvrRPS4 N‐terminus to the effectors (Fabro et al., 2011), and transformed into P. fluorescens EtHAn (Thomas et al., 2009) by electroporation. Transformed EtHAn strains were grown overnight in King's B medium containing ampicillin, chloramphenicol, tetracycline and gentamicin at 28°C. The bacteria were harvested by centrifugation and resuspended in 10 mM MgCl2 to a final OD600 = 0.3 and infiltrated into leaves of barley. This method for introducing proteins into barley leaf cells was validated by performing a GUS assay as above. Furthermore, growth of EtHAn in barley leaves was quantified by extracting bacteria from homogenized leaves at 0 and 1 days post‐infiltration (dpi). The colony‐forming units assessed from the 1 dpi samples was normalizing to the average of those obtained from the 0 dpi samples.
Callose responses were studied after EtHAn infiltration of first leaves of 8‐day‐old barley. Twenty‐four hours later, the callose response was assayed in the infiltrated leaf areas using a Nikon ECLIPSE Ni‐U fluorescence microscope. For one repeat, ten 1110 × 740 μm (5184 × 3456 pixels) images from random sites at one leaf were used to quantify the number of callose deposits and the accumulated area of callose deposits using the Fiji open‐source platform (Jin & Mackey, 2017; Schindelin et al., 2012). The outcomes of effectors were calculated relative to GUS as control and the total number and total area of the callose deposits were assessed independently.
The HR was studied after EtHAn infiltration and Bh inoculation of 7‐day‐old barley. To ascertain even Bh spore distribution, first leaves, still attached to the plants, were mounted horizontally side‐by‐side on plastic plates and pots were inoculated together in an inoculation tower. Four days later, trypan blue staining was performed on 4‐cm leaf pieces according to Kock and Slusarenko (1990), and quantified by counting in a light microscope.
4.11. Differential transcript accumulation
RNA‐Sequencing data were extracted from an infection time course of barley CI 16151 and the derived fast‐neutron mutant, mla6‐m18982, at 0, 16, 20, 24, 32 and 48 h after inoculation with Bh isolate 5874 (AVR a1 , AVR a6 ; NCBI‐GEO GSE101304) and analysed as described in (Chapman et al., 2021; Velásquez‐Zapata et al., 2022). Genes differentially expressed at an adjusted p of <0.001 for barley and <0.003 for Bh were considered significant.
4.12. Structural prediction of proteins
The Bh AVRA1 and BEC1016 amino acid sequences were analysed in AlphaFold2 (https://colab.research.google.com/github/sokrypton/ColabFold/blob/main/AlphaFold2.ipynb#pdb100) to make the structural predictions presented in Figure S1.
Supporting information
Figure S1. Attempted alignment of the amino acid sequences of AVRA1 (CSEP0008, BLGH_03023) and BEC1016 (CSEP0491, BLGH_07006).
Figure S2. Transcript accumulation on barley inoculated with Blumeria hordei isolate 5874.
Figure S3. Pseudomonas fluorescens EtHAn‐delivery of β‐glucuronidase into barley leaf cells.
Figure S4. Growth of Pseudomonas fluorescens EtHAn in barley leaves is independent on effector construct.
Figure S5. AVRA1 and BEC1016 do not affect MLA3‐mediated hypersensitive response.
Figure S6. Alignment of HvERdj3B with human HsERdj3 (NP_057390.1) and Arabidopsis AtERdj3B (At3g62600).
Figure S7. Binary yeast two‐hybrid assay used to validate interactions between AVRA1, BEC1016 and HvERdj3B.
Figure S8. Bimolecular fluorescence complementation study of the interaction between AVRA1, BEC1016 and HvERdj3B.
Figure S9. Subcellular localization of HvERdj3B in Nicotiana benthamiana epidermal cells.
Figure S10. Localization of AVRA1 and BEC1016 in barley leaf epidermal cells.
Figure S11. BEC1016 can translocate into the endoplasmic reticulum.
Table S1. Raw data from penetration rate, callose deposition and hypersensitive response assessments.
Table S2. Summary Y2H‐SCORES data for the AVRA1 and BEC1016 baits.
Table S3. Primer sequences for plasmid constructions.
ACKNOWLEDGEMENTS
We thank our late and good friend, Dr Patrick Schweizer (Leibniz Institute of Plant Genetics and Crop Plant Research, Gatersleben, Germany) for providing the RNAi vector and the Mlo RNAi construct, Dr Masumi Robertson (Commonwealth Scientific and Individual Research Organization Plant Industry) for the Gateway Y2H vectors (pDEST‐ACT2 and pDEST‐AS2‐1), Greg Fuerst (USDA‐ARS at Iowa State University) for conducting the Bh time‐course infection experiment and expert isolation of RNA for the 3‐frame Y2H library, Associate Professor Shoi Mano for the BiFC‐vectors, and Drs Pietro Spanu (Imperial College, U.K.) and Ralph Panstruga (RWTH Aachen, Germany) for CSEP clones. Research supported in part by China Scholarship Council for PhD‐student scholarship 201708340064 to Z.Z., Novo Nordisk Foundation Challenge grant NNF19OC0056457 to H.T.C., Villum Fonden Experiment Programme grant 00028131 to H.T.C., Fulbright – Minciencias 2015 & Schlumberger Faculty for the Future fellowships to V.V.Z., USDA‐ARS Postdoctoral Research Associateship and USDA‐NIFA‐ELI Postdoctoral Fellowship 2017‐67012‐26086 to J.M.E., Oak Ridge Institute for Science and Education under U.S. Department of Energy contract number DE‐SC0014664 to S.B., and National Science Foundation – Plant Genome Research Program grant 13‐39348, USDA‐National Institute of Food and Agriculture grant 2020‐67013‐31184, USDA‐Agricultural Research Service projects 3625‐21000‐067‐00D and 5030‐21220‐068‐000‐D to R.P.W. and Marie Skłodowska‐Curie Actions Postdoctoral Fellowship project 101104193 to S.D. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the CSC, NNF, VF, USDA, NIFA, ARS, DOE, ORAU/ORISE, NSF or MSCA. USDA is an equal opportunity provider and employer.
Li, Z. , Velásquez‐Zapata, V. , Elmore, J.M. , Li, X. , Xie, W. , Deb, S. et al. (2024) Powdery mildew effectors AVRA1 and BEC1016 target the ER J‐domain protein HvERdj3B required for immunity in barley. Molecular Plant Pathology, 25, e13463. Available from: 10.1111/mpp.13463
Contributor Information
Roger P. Wise, Email: roger.wise@usda.gov.
Hans Thordal‐Christensen, Email: htc@plen.ku.dk.
DATA AVAILABILITY STATEMENT
Infection‐time‐course RNA‐Sequencing data are at NCBI‐GEO under the acc. no. GSE101304 (https://www.ncbi.nlm.nih.gov/geo/query/.acc.cgi?acc=GSE101304). R code and the ReadMe file for the NGPINT and Y2H‐SCORES software used to identify Bh AVRA1 and BEC1016 interactors are provided at the GitHub pages (https://github.com/Wiselab2/NGPINT_V2; https://github.com/Wiselab2/Y2H‐SCORES). Raw Y2H‐Seq reads are at NCBI‐GEO under acc. nos. GSE164954 (AVRA1/CSEP0008) and GSE166108 (BEC1016/CSEP0491). Otherwise, Y2H‐NGIS scores are available in the supporting information. GenBank accession numbers for AVR A1 (CSEP0008, BLGH_03023) and BEC1016 (CSEP0491, BLGH_07006), are CCU81904.1 and CCU83284.1, respectively. HvERdjB3 is represented in GenBank as an mRNA‐derived DNA sequence from Haruna Nijo (AK376215.1) and as a protein sequence from the genome sequence of Tibetan Hulless barley (KAE8793706).
REFERENCES
- Aguilar, G.B. , Pedersen, C. & Thordal‐Christensen, H. (2016) Identification of eight effector candidate genes involved in early aggressiveness of the barley powdery mildew fungus. Plant Pathology, 65, 953–958. [Google Scholar]
- Ahmed, A.A. , Pedersen, C. , Schultz‐Larsen, T. , Kwaaitaal, M. , Jørgensen, H.J. & Thordal‐Christensen, H. (2015) The barley powdery mildew candidate secreted effector protein CSEP0105 inhibits the chaperone activity of a small heat shock protein. Plant Physiology, 168, 321–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed, A.A. , Pedersen, C. & Thordal‐Christensen, H. (2016) The barley powdery mildew effector candidates CSEP0081 and CSEP0254 promote fungal infection success. PLoS One, 11, e0157586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee, S. , Velásquez‐Zapata, V. , Fuerst, G. , Elmore, J.M. & Wise, R.P. (2021) NGPINT: a next‐generation protein–protein interaction software. Briefings in Bioinformatics, 22, bbaa351. [DOI] [PubMed] [Google Scholar]
- Bauer, S. , Yu, D. , Lawson, A.W. , Saur, I.M. , Frantzeskakis, L. , Kracher, B. et al. (2021) The leucine‐rich repeats in allelic barley MLA immune receptors define specificity towards sequence‐unrelated powdery mildew a virulence effectors with a predicted common RNase‐like fold. PLoS Pathogens, 17, e1009223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindschedler, L.V. , Burgis, T.A. , Mills, D.J. , Ho, J.T. , Cramer, R. & Spanu, P.D. (2009) In planta proteomics and proteogenomics of the biotrophic barley fungal pathogen Blumeria graminis f. sp. hordei . Molecular & Cellular Proteomics, 8, 2368–2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourras, S. , Kunz, L. , Xue, M. , Praz, C.R. , Müller, M.C. , Kälin, C. et al. (2019) The AvrPm3–Pm3 effector–NLR interactions control both race‐specific resistance and host‐specificity of cereal mildews on wheat. Nature Communications, 10, 2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourras, S. , McNally, K.E. , Ben‐David, R. , Parlange, F. , Roffler, S. , Praz, C.R. et al. (2015) Multiple avirulence loci and allele‐specific effector recognition control the Pm3 race‐specific resistance of wheat to powdery mildew. The Plant Cell, 27, 2991–3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray Speth, E. , Lee, Y.N. & He, S.Y. (2007) Pathogen virulence factors as molecular probes of basic plant cellular functions. Current Opinion in Plant Biology, 10, 580–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brioudes, F. , Jay, F. & Voinnet, O. (2022) Suppression of both intra‐ and intercellular RNA silencing by the tombusviral P19 protein requires its small RNA binding property. New Phytologist, 235, 824–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldo, R.A. , Nettleton, D. , Peng, J. & Wise, R.P. (2006) Stage‐specific suppression of basal defense discriminates barley plants containing fast‐ and delayed‐acting Mla powdery mildew resistance alleles. Molecular Plant–Microbe Interactions, 19, 939–947. [DOI] [PubMed] [Google Scholar]
- Caldo, R.A. , Nettleton, D. & Wise, R.P. (2004) Interaction‐dependent gene expression in Mla‐specified response to barley powdery mildew. The Plant Cell, 16, 2514–2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, Y. , Kümmel, F. , Logemann, E. , Gebauer, J.M. , Lawson, A.W. , Yu, D. et al. (2023) Structural polymorphisms within a common powdery mildew effector scaffold as a driver of coevolution with cereal immune receptors. Proceedings of the National Academy of Sciences of the United States of America, 120, e2307604120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter, M.E. , Helm, M. , Chapman, A.V.E. , Wan, E. , Restrepo Sierra, A.M. , Innes, R.W. et al. (2019) Convergent evolution of effector protease recognition by Arabidopsis and barley. Molecular Plant–Microbe Interactions, 32, 550–565. [DOI] [PubMed] [Google Scholar]
- Chapman, A.V.E. , Hunt, M. , Surana, P. , Velásquez‐Zapata, V. , Xu, W. , Fuerst, G. et al. (2021) Disruption of barley immunity to powdery mildew by an in‐frame Lys‐Leu deletion in the essential protein SGT1. Genetics, 217, iyaa026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, K.C. , Qu, S. , Chowdhury, S. , Noxon, I.C. , Schonhoft, J.D. , Plate, L. et al. (2017) The endoplasmic reticulum HSP 40 co‐chaperone ERdj3/DNAJB11 assembles and functions as a tetramer. EMBO Journal, 36, 2296–2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douchkov, D. , Nowara, D. , Zierold, U. & Schweizer, P. (2005) A high‐throughput gene‐silencing system for the functional assessment of defense‐related genes in barley epidermal cells. Molecular Plant–Microbe Interactions, 18, 755–761. [DOI] [PubMed] [Google Scholar]
- Dreze, M. , Monachello, D. , Lurin, C. , Cusick, M.E. , Hill, D.E. , Vidal, M. et al. (2010) High‐quality binary interactome mapping. Methods in Enzymology, 470, 281–315. [DOI] [PubMed] [Google Scholar]
- Earley, K.W. , Haag, J.R. , Pontes, O. , Opper, K. , Juehne, T. , Song, K. et al. (2006) Gateway‐compatible vectors for plant functional genomics and proteomics. The Plant Journal, 45, 616–629. [DOI] [PubMed] [Google Scholar]
- Elmore, J.M. , Velásquez‐Zapata, V. & Wise, R.P. (2023) Next‐generation yeast two‐hybrid screening to discover protein–protein interactions. Methods in Molecular Biology, 2690, 205–222. [DOI] [PubMed] [Google Scholar]
- Erffelinck, M.L. , Ribeiro, B. , Perassolo, M. , Pauwels, L. , Pollier, J. , Storme, V. et al. (2018) A user‐friendly platform for yeast two‐hybrid library screening using next generation sequencing. PLoS One, 13, e0201270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabro, G. , Steinbrenner, J. , Coates, M. , Ishaque, N. , Baxter, L. , Studholme, D.J. et al. (2011) Multiple candidate effectors from the oomycete pathogen Hyaloperonospora arabidopsidis suppress host plant immunity. PLoS Pathogens, 7, e1002348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, G. , Yang, Y. , Li, T. , Lu, W. , Du, Y. , Qiang, X. et al. (2018) A Phytophthora capsici RXLR effector targets and inhibits a plant PPIase to suppress endoplasmic reticulum‐mediated immunity. Molecular Plant, 11, 1067–1083. [DOI] [PubMed] [Google Scholar]
- Fatima, K. , Naqvi, F. & Younas, H. (2021) A review: molecular chaperone‐mediated folding, unfolding and disaggregation of expressed recombinant proteins. Cell Biochemistry and Biophysics, 79, 153–174. [DOI] [PubMed] [Google Scholar]
- Figueroa, M. , Ortiz, D. & Henningsen, E.C. (2021) Tactics of host manipulation by intracellular effectors from plant pathogenic fungi. Current Opinion in Plant Biology, 62, 102054. [DOI] [PubMed] [Google Scholar]
- Frantzeskakis, L. , Kracher, B. , Kusch, S. , Yoshikawa‐Maekawa, M. , Bauer, S. , Pedersen, C. et al. (2018) Signatures of host specialization and a recent transposable element burst in the dynamic one‐speed genome of the fungal barley powdery mildew pathogen. BMC Genomics, 19, 381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galletta, B.J. & Rusan, N.M. (2015) A yeast two‐hybrid approach for probing protein–protein interactions at the centrosome. Methods in Cell Biology, 129, 252–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruner, K. , Leissing, F. , Sinitski, D. , Thieron, H. , Axstmann, C. , Baumgarten, K. et al. (2021) Chemokine‐like MDL proteins modulate flowering time and innate immunity in plants. Journal of Biological Chemistry, 296, 100611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassdenteufel, S. , Nguyen, D. , Helms, V. , Lang, S. & Zimmermann, R. (2019) ER import of small human presecretory proteins: components and mechanisms. FEBS Letters, 593, 2506–2524. [DOI] [PubMed] [Google Scholar]
- Hewitt, T. , Müller, M.C. , Molnár, I. , Mascher, M. , Holušová, K. , Šimková, H. et al. (2021) A highly differentiated region of wheat chromosome 7AL encodes a Pm1a immune receptor that recognizes its corresponding AvrPm1a effector from Blumeria graminis . New Phytologist, 229, 2812–2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, C.D. & Kerppola, T.K. (2003) Simultaneous visualization of multiple protein interactions in living cells using multicolor fluorescence complementation analysis. Nature Biotechnology, 21, 539–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt, M. , Banerjee, S. , Surana, P. , Liu, M. , Fuerst, G. , Mathioni, S. et al. (2019) Small RNA discovery in the interaction between barley and the powdery mildew pathogen. BMC Genomics, 20, 610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter, P.R. , Craddock, C.P. , Di Benedetto, S. , Roberts, L.M. & Frigerio, L. (2007) Fluorescent reporter proteins for the tonoplast and the vacuolar lumen identify a single vacuolar compartment in Arabidopsis cells. Plant Physiology, 145, 1371–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, L. & Mackey, D.M. (2017) Measuring callose deposition, an indicator of cell wall reinforcement, during bacterial infection in Arabidopsis . Methods in Molecular Biology, 1578, 195–205. [DOI] [PubMed] [Google Scholar]
- Jing, M. , Guo, B. , Li, H. , Yang, B. , Wang, H. , Kong, G. et al. (2016) A Phytophthora sojae effector suppresses endoplasmic reticulum stress‐mediated immunity by stabilizing plant binding immunoglobulin proteins. Nature Communications, 7, 11685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, J.D. , Vance, R.E. & Dangl, J.L. (2016) Intracellular innate immune surveillance devices in plants and animals. Science, 354, aaf6395. [DOI] [PubMed] [Google Scholar]
- Kamigaki, A. , Nito, K. , Hikino, K. , Goto‐Yamada, S. , Nishimura, M. , Nakagawa, T. et al. (2016) Gateway vectors for simultaneous detection of multiple protein−protein interactions in plant cells using bimolecular fluorescence complementation. PLoS One, 11, e0160717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanja, C. & Hammond‐Kosack, K.E. (2020) Proteinaceous effector discovery and characterization in filamentous plant pathogens. Molecular Plant Pathology, 21, 1353–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi, M. , Inzé, D. & Depicker, A. (2002) GATEWAY™ vectors for Agrobacterium‐mediated plant transformation. Trends in Plant Science, 7, 193–195. [DOI] [PubMed] [Google Scholar]
- Kock, E. & Slusarenko, A. (1990) Arabidopsis is susceptible to infection by a downy mildew fungus. The Plant Cell, 2, 437–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kørner, C.J. , Du, X. , Vollmer, M.E. & Pajerowska‐Mukhtar, K.M. (2015) Endoplasmic reticulum stress signaling in plant immunity‐at the crossroad of life and death. International Journal of Molecular Sciences, 16, 26582–26598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kourelis, J. & Van Der Hoorn, R.A. (2018) Defended to the nines: 25 years of resistance gene cloning identifies nine mechanisms for R protein function. The Plant Cell, 30, 285–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz, L. , Sotiropoulos, A.G. , Graf, J. , Razavi, M. , Keller, B. & Müller, M.C. (2023) The broad use of the Pm8 resistance gene in wheat resulted in hypermutation of the AvrPm8 gene in the powdery mildew pathogen. BMC Biology, 21, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusch, S. , Qian, J. , Loos, A. , Kümmel, F. , Spanu, P.D. & Panstruga, R. (2023) Long‐term and rapid evolution in powdery mildew fungi. Molecular Ecology. Available from: 10.1111/mec.16909 [DOI] [PubMed] [Google Scholar]
- Kwaaitaal, M. , Keinath, N.F. , Pajonk, S. , Biskup, C. & Panstruga, R. (2010) Combined bimolecular fluorescence complementation and Forster resonance energy transfer reveals ternary SNARE complex formation in living plant cells. Plant Physiology, 152, 1135–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, J.D. , Wan, J. , Ford, R. , Gong, Y. , Fung, P. , Nahal, H. et al. (2012) Quantitative interactor screening with next‐generation sequencing (QIS‐Seq) identifies Arabidopsis thaliana MLO2 as a target of the Pseudomonas syringae type III effector HopZ2. BMC Genomics, 13, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J. , Zhao‐Hui, C. , Batoux, M. , Nekrasov, V. , Roux, M. , Chinchilla, D. et al. (2009) Specific ER quality control components required for biogenesis of the plant innate immune receptor EFR. Proceedings of the National Academy of Sciences of the United States of America, 106, 15973–15978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X. , Jin, C. , Yuan, H. , Huang, W. , Liu, F. , Fan, R. et al. (2021) The barley powdery mildew effectors CSEP0139 and CSEP0182 suppress cell death and promote B. graminis fungal virulence in plants. Phytopathology Research, 3, 7. [Google Scholar]
- Liao, W. , Nielsen, M.E. , Pedersen, C. , Xie, W.J. & Thordal‐Christensen, H. (2023) Barley endosomal MONENSIN SENSITIVITY1 is a target of the powdery mildew effector CSEP0162 and plays a role in plant immunity. Journal of Experimental Botany, 74, 118–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, X. , Kracher, B. , Saur, I.M. , Bauer, S. , Ellwood, S.R. , Wise, R. et al. (2016) Allelic barley MLA immune receptors recognize sequence‐unrelated avirulence effectors of the powdery mildew pathogen. Proceedings of the National Academy of Sciences of the United States of America, 113, E6486–E6495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lück, S. , Kreszies, T. , Strickert, M. , Schweizer, P. , Kuhlmann, M. & Douchkov, D. (2019) siRNA‐finder (si‐fi) software for RNAi‐target design and off‐target prediction. Frontiers in Plant Science, 10, 1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manser, B. , Koller, T. , Praz, C.R. , Roulin, A.C. , Zbinden, H. , Arora, S. et al. (2021) Identification of specificity‐defining amino acids of the wheat immune receptor Pm2 and powdery mildew effector AvrPm2. The Plant Journal, 106, 993–1007. [DOI] [PubMed] [Google Scholar]
- Manser, B. , Zbinden, H. , Herren, G. , Steger, J. , Isaksson, J. , Bräunlich, S. et al. (2023) Wheat zinc finger protein TaZF interacts with both the powdery mildew AvrPm2 protein and the corresponding wheat Pm2a immune receptor. Plant Communications, 4, 100769. [DOI] [PubMed] [Google Scholar]
- Mascher, M. , Wicker, T. , Jenkins, J. , Plott, C. , Lux, T. , Koh, C.S. et al. (2021) Long‐read sequence assembly: a technical evaluation in barley. The Plant Cell, 33, 1888–1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menardo, F. , Praz, C.R. , Wicker, T. & Keller, B. (2017) Rapid turnover of effectors in grass powdery mildew (Blumeria graminis). BMC Evolutionary Biology, 17, 223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno, A.A. , Mukhtar, M.S. , Blanco, F. , Boatwright, J.L. , Moreno, I. , Jordan, M.R. et al. (2012) IRE1/bZIP60‐mediated unfolded protein response plays distinct roles in plant immunity and abiotic stress responses. PLoS One, 7, e31944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscou, M.J. , Lauter, N. , Caldo, R.A. , Nettleton, D. & Wise, R.P. (2011) Quantitative and temporal definition of the Mla transcriptional regulon during barley–powdery mildew interactions. Molecular Plant–Microbe Interactions, 24, 694–705. [DOI] [PubMed] [Google Scholar]
- Mukhtar, M.S. , Carvunis, A.R. , Dreze, M. , Epple, P. , Steinbrenner, J. , Moore, J. et al. (2011) Independently evolved virulence effectors converge onto hubs in a plant immune system network. Science, 333, 596–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller, M.C. , Kunz, L. , Schudel, S. , Lawson, A.W. , Kammerecker, S. , Isaksson, J. et al. (2022) Ancient variation of the AvrPm17 gene in powdery mildew limits the effectiveness of the introgressed rye Pm17 resistance gene in wheat. Proceedings of the National Academy of Sciences of the United States of America, 119, e2108808119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller, M.C. , Praz, C.R. , Sotiropoulos, A.G. , Menardo, F. , Kunz, L. , Schudel, S. et al. (2019) A chromosome‐scale genome assembly reveals a highly dynamic effector repertoire of wheat powdery mildew. New Phytologist, 221, 2176–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekrasov, V. , Li, J. , Batoux, M. , Roux, M. , Chu, Z.H. , Lacombe, S. et al. (2009) Control of the pattern‐recognition receptor EFR by an ER protein complex in plant immunity. EMBO Journal, 28, 3428–3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, B.K. , Cai, X. & Nebenführ, A. (2007) A multicolored set of in vivo organelle markers for co‐localization studies in Arabidopsis and other plants. The Plant Journal, 51, 1126–1136. [DOI] [PubMed] [Google Scholar]
- Ngou, B.P.M. , Ding, P. & Jones, J.D. (2022) Thirty years of resistance: zig‐zag through the plant immune system. The Plant Cell, 34, 1447–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowara, D. , Gay, A. , Lacomme, C. , Shaw, J. , Ridout, C. , Douchkov, D. et al. (2010) HIGS: host‐induced gene silencing in the obligate biotrophic fungal pathogen Blumeria graminis . The Plant Cell, 22, 3130–3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pashkova, N. , Peterson, T.A. , Krishnamani, V. , Breheny, P. , Stamnes, M. & Piper, R.C. (2016) DEEPN as an approach for batch processing of yeast 2‐hybrid interactions. Cell Reports, 17, 303–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen, C. , van Themaat, E.V. , McGuffin, L.J. , Abbott, J.C. , Burgis, T.A. , Barton, G. et al. (2012) Structure and evolution of barley powdery mildew effector candidates. BMC Genomics, 13, 694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennington, H.G. , Jones, R. , Kwon, S. , Bonciani, G. , Thieron, H. , Chandler, T. et al. (2019) The fungal ribonuclease‐like effector protein CSEP0064/BEC1054 represses plant immunity and interferes with degradation of host ribosomal RNA. PLoS Pathogens, 15, e1007620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pliego, C. , Nowara, D. , Bonciani, G. , Gheorghe, D.M. , Xu, R. , Surana, P. et al. (2013) Host‐induced gene silencing in barley powdery mildew reveals a class of ribonuclease‐like effectors. Molecular Plant–Microbe Interactions, 26, 633–642. [DOI] [PubMed] [Google Scholar]
- Pobre, K.F. , Poet, G.J. & Hendershot, L.M. (2019) The endoplasmic reticulum (ER) chaperone BiP is a master regulator of ER functions: getting by with a little help from ERdj friends. Journal of Biological Chemistry, 294, 2098–2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praz, C.R. , Bourras, S. , Zeng, F. , Sánchez‐Martín, J. , Menardo, F. , Xue, M. et al. (2017) AvrPm2 encodes an RNase‐like avirulence effector which is conserved in the two different specialized forms of wheat and rye powdery mildew fungus. New Phytologist, 213, 1301–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiang, X. , Zechmann, B. , Reitz, M.U. , Kogel, K.H. & Schäfer, P. (2012) The mutualistic fungus Piriformospora indica colonizes Arabidopsis roots by inducing an endoplasmic reticulum stress–triggered caspase‐dependent cell death. The Plant Cell, 24, 794–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasco‐Gaunt, S. , Riley, A. , Barcelo, P. & Lazzeri, P.A. (1999) Analysis of particle bombardment parameters to optimise DNA delivery into wheat tissues. Plant Cell Reports, 19, 118–127. [DOI] [PubMed] [Google Scholar]
- Robertson, M. (2004) Two transcription factors are negative regulators of gibberellin response in the HvSPY‐signaling pathway in barley aleurone. Plant Physiology, 136, 2747–2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez‐Martín, J. , Widrig, V. , Herren, G. , Wicker, T. , Zbinden, H. , Gronnier, J. et al. (2021) Wheat Pm4 resistance to powdery mildew is controlled by alternative splice variants encoding chimeric proteins. Nature Plants, 7, 327–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saur, I.M. , Bauer, S. , Kracher, B. , Lu, X. , Franzeskakis, L. , Müller, M.C. et al. (2019) Multiple pairs of allelic MLA immune receptor‐powdery mildew AVRA effectors argue for a direct recognition mechanism. eLife, 8, e44471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin, J. , Arganda‐Carreras, I. , Frise, E. , Kaynig, V. , Longair, M. , Pietzsch, T. et al. (2012) Fiji: an open‐source platform for biological‐image analysis. Nature Methods, 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeholzer, S. , Tsuchimatsu, T. , Jordan, T. , Bieri, S. , Pajonk, S. , Yang, W. et al. (2010) Diversity at the Mla powdery mildew resistance locus from cultivated barley reveals sites of positive selection. Molecular Plant–Microbe Interactions, 23, 497–509. [DOI] [PubMed] [Google Scholar]
- Sivamani, E. , DeLong, R.K. & Qu, R. (2009) Protamine‐mediated DNA coating remarkably improves bombardment transformation efficiency in plant cells. Plant Cell Reports, 28, 213–221. [DOI] [PubMed] [Google Scholar]
- Sohn, K.H. , Lei, R. , Nemri, A. & Jones, J.D. (2007) The downy mildew effector proteins ATR1 and ATR13 promote disease susceptibility in Arabidopsis thaliana . The Plant Cell, 19, 4077–4090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanu, P.D. (2017) Cereal immunity against powdery mildews targets RNase‐like proteins associated with Haustoria (RALPH) effectors evolved from a common ancestral gene. New Phytologist, 213, 969–971. [DOI] [PubMed] [Google Scholar]
- Spanu, P.D. , Abbott, J.C. , Amselem, J. , Burgis, T.A. , Soanes, D.M. , Stüber, K. et al. (2010) Genome expansion and gene loss in powdery mildew fungi reveal tradeoffs in extreme parasitism. Science, 330, 1543–1546. [DOI] [PubMed] [Google Scholar]
- Sun, Y. , Zhu, Y.X. , Balint‐Kurti, P.J. & Wang, G.F. (2020) Fine‐tuning immunity: players and regulators for plant NLRs. Trends in Plant Science, 25, 695–713. [DOI] [PubMed] [Google Scholar]
- Surana, P. (2017) Membrane trafficking in resistance gene‐mediated defense against the barley powdery mildew fungus. [PhD thesis]. Ames, IA: Iowa State University. [Google Scholar]
- Surana, P. , Xu, R. , Fuerst, G. , Chapman, A.V.E. , Nettleton, D. & Wise, R.P. (2017) Interchromosomal transfer of immune regulation during infection of barley with the powdery mildew pathogen. G3: Genes, Genomes, Genetics, 7, 3317–3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suter, B. , Zhang, X. , Pesce, C.G. , Mendelsohn, A.R. , Dinesh‐Kumar, S.P. & Mao, J.H. (2015) Next‐generation sequencing for binary protein–protein interactions. Frontiers in Genetics, 6, 346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, W.J. , Thireault, C.A. , Kimbrel, J.A. & Chang, J.H. (2009) Recombineering and stable integration of the Pseudomonas syringae pv. syringae 61 hrp/hrc cluster into the genome of the soil bacterium Pseudomonas fluorescens Pf0‐1. The Plant Journal, 60, 919–928. [DOI] [PubMed] [Google Scholar]
- Thordal‐Christensen, H. (2020) A holistic view on plant effector‐triggered immunity presented as an iceberg model. Cellular and Molecular Life Sciences, 77, 3963–3976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tintor, N. & Saijo, Y. (2014) ER‐mediated control for abundance, quality, and signaling of transmembrane immune receptors in plants. Frontiers in Plant Science, 5, 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trigg, S.A. , Garza, R.M. , MacWilliams, A. , Nery, J.R. , Bartlett, A. , Castanon, R. et al. (2017) CrY2H‐seq: a massively multiplexed assay for deep‐coverage interactome mapping. Nature Methods, 14, 819–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyaya, N.M. , Ellis, J.G. & Dodds, P.N. (2014) A bacterial type III secretion‐based delivery system for functional assays of fungal effectors in cereals. Methods in Molecular Biology, 1127, 277–290. [DOI] [PubMed] [Google Scholar]
- van Wersch, S. , Tian, L. , Hoy, R. & Li, X. (2020) Plant NLRs: the whistleblowers of plant immunity. Plant Communications, 1, 100016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velásquez‐Zapata, V. , Elmore, J.M. , Banerjee, S. , Dorman, K.S. & Wise, R.P. (2021) Next‐generation yeast‐two‐hybrid analysis with Y2H‐SCORES identifies novel interactors of the MLA immune receptor. PLoS Computational Biology, 17, e1008890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velásquez‐Zapata, V. , Elmore, J.M. , Fuerst, G.S. & Wise, R.P. (2022) An interolog‐based barley interactome as an integration framework for immune signaling. Genetics, 221, iyac056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velásquez‐Zapata, V. , Elmore, J.M. & Wise, R.P. (2023) Bioinformatic analysis of yeast two‐hybrid next‐generation interaction screen data. Methods in Molecular Biology, 2690, 223–239. [DOI] [PubMed] [Google Scholar]
- Velásquez‐Zapata, V. , Smith, S. , Surana, P. , Chapman, A.V.E. & Wise, R.P. (2023) Transcriptome‐based host epistasis and pathogen co‐expression in barley‐powdery mildew interactions. bioRxiv 10.1101/2023.09.18.558274 [preprint]. [DOI]
- Voigt, C.A. (2014) Callose‐mediated resistance to pathogenic intruders in plant defense‐related papillae. Frontiers in Plant Science, 5, 168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt, C.A. (2016) Cellulose/callose glucan networks: the key to powdery mildew resistance in plants? New Phytologist, 212, 303–305. [DOI] [PubMed] [Google Scholar]
- Wang, J. , Dhroso, A. , Liu, X. , Baum, T.J. , Hussey, R.S. , Davis, E.L. et al. (2021) Phytonematode peptide effectors exploit a host post‐translational trafficking mechanism to the ER using a novel translocation signal. New Phytologist, 229, 563–574. [DOI] [PubMed] [Google Scholar]
- Wang, Z. , Li, X. , Wang, X. , Liu, N. , Xu, B. , Peng, Q. et al. (2019) Arabidopsis endoplasmic reticulum‐localized UBAC2 proteins interact with PAMP‐INDUCED COILED‐COIL to regulate pathogen‐induced callose deposition and plant immunity. The Plant Cell, 31, 153–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weimann, M. , Grossmann, A. , Woodsmith, J. , Özkan, Z. , Birth, P. , Meierhofer, D. et al. (2013) A Y2H‐seq approach defines the human protein methyltransferase interactome. Nature Methods, 10, 339–342. [DOI] [PubMed] [Google Scholar]
- Weßling, R. , Epple, P. , Altmann, S. , He, Y. , Yang, L. , Henz, S.R. et al. (2014) Convergent targeting of a common host protein‐network by pathogen effectors from three kingdoms of life. Cell, Host & Microbe, 16, 364–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, W. , Nielsen, M.E. , Pedersen, C. & Thordal‐Christensen, H. (2017) A split‐GFP gateway cloning system for topology analyses of membrane proteins in plants. PLoS One, 12, e0170118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, G. , Zhong, X. , Shi, Y. , Liu, Z. , Jiang, N. , Liu, J. et al. (2020) A fungal effector targets a heat shock–dynamin protein complex to modulate mitochondrial dynamics and reduce plant immunity. Science Advances, 6, eabb7719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Q. , Tang, C. , Wang, L. , Zhao, C. , Kang, Z. & Wang, X. (2020) Haustoria – arsenals during the interaction between wheat and Puccinia striiformis f. sp. tritici . Molecular Plant Pathology, 21, 83–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, W. , Meng, Y. , Surana, P. , Fuerst, G. , Nettleton, D. & Wise, R.P. (2015) The knottin‐like Blufensin family regulates genes involved in nuclear import and the secretory pathway in barley‐powdery mildew interactions. Frontiers in Plant Science, 6, 409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yachie, N. , Petsalaki, E. , Mellor, J.C. , Weile, J. , Jacob, Y. , Verby, M. et al. (2016) Pooled‐matrix protein interaction screens using barcode fusion genetics. Molecular Systems Biology, 12, 863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, M. , Maruyama, D. , Endo, T. & Nishikawa, S.I. (2008) Arabidopsis thaliana has a set of J proteins in the endoplasmic reticulum that are conserved from yeast to animals and plants. Plant and Cell Physiology, 49, 1547–1562. [DOI] [PubMed] [Google Scholar]
- Yu, C.P. , Chen, S.C. , Chang, Y.M. , Liu, W.Y. , Lin, H.H. , Lin, J.J. et al. (2015) Transcriptome dynamics of developing maize leaves and genomewide prediction of cis elements and their cognate transcription factors. Proceedings of the National Academy of Sciences of the United States of America, 112, E2477–E2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, H. , Jin, C. , Pei, H. , Zhao, L. , Li, X. , Li, J. et al. (2021) The powdery mildew effector CSEP0027 interacts with barley catalase to regulate host immunity. Frontiers in Plant Science, 12, 733237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, M. , Jiang, Z. , Bi, G. , Nomura, K. , Liu, M. , Wang, Y. et al. (2021) Pattern‐recognition receptors are required for NLR‐mediated plant immunity. Nature, 592, 105–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, W.‐J. , Hanisch, S. , Kwaaitaal, M. , Pedersen, C. & Thordal‐Christensen, H. (2013) A component of the Sec61 ER protein transporting pore is required for plant susceptibility to powdery mildew. Frontiers in Plant Science, 4, 127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, W.J. , Pedersen, C. , Kwaaitaal, M. , Gregersen, P.L. , Mørch, S.M. , Hanisch, S. et al. (2012) Interaction of barley powdery mildew effector candidate CSEP0055 with the defence protein PR17c. Molecular Plant Pathology, 13, 1110–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, X. , Yang, J. , Shi, Y. , Wang, X. & Wang, G.L. (2018) The DnaJ protein OsDjA6 negatively regulates rice innate immunity to the blast fungus Magnaporthe oryzae . Molecular Plant Pathology, 19, 607–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann, R. , Eyrisch, S. , Ahmad, M. & Helms, V. (2011) Protein translocation across the ER membrane. Biochimica et Biophysica Acta, 1808, 912–924. [DOI] [PubMed] [Google Scholar]
- Zipfel, C. & Oldroyd, G.E. (2017) Plant signalling in symbiosis and immunity. Nature, 543, 328–336. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Attempted alignment of the amino acid sequences of AVRA1 (CSEP0008, BLGH_03023) and BEC1016 (CSEP0491, BLGH_07006).
Figure S2. Transcript accumulation on barley inoculated with Blumeria hordei isolate 5874.
Figure S3. Pseudomonas fluorescens EtHAn‐delivery of β‐glucuronidase into barley leaf cells.
Figure S4. Growth of Pseudomonas fluorescens EtHAn in barley leaves is independent on effector construct.
Figure S5. AVRA1 and BEC1016 do not affect MLA3‐mediated hypersensitive response.
Figure S6. Alignment of HvERdj3B with human HsERdj3 (NP_057390.1) and Arabidopsis AtERdj3B (At3g62600).
Figure S7. Binary yeast two‐hybrid assay used to validate interactions between AVRA1, BEC1016 and HvERdj3B.
Figure S8. Bimolecular fluorescence complementation study of the interaction between AVRA1, BEC1016 and HvERdj3B.
Figure S9. Subcellular localization of HvERdj3B in Nicotiana benthamiana epidermal cells.
Figure S10. Localization of AVRA1 and BEC1016 in barley leaf epidermal cells.
Figure S11. BEC1016 can translocate into the endoplasmic reticulum.
Table S1. Raw data from penetration rate, callose deposition and hypersensitive response assessments.
Table S2. Summary Y2H‐SCORES data for the AVRA1 and BEC1016 baits.
Table S3. Primer sequences for plasmid constructions.
Data Availability Statement
Infection‐time‐course RNA‐Sequencing data are at NCBI‐GEO under the acc. no. GSE101304 (https://www.ncbi.nlm.nih.gov/geo/query/.acc.cgi?acc=GSE101304). R code and the ReadMe file for the NGPINT and Y2H‐SCORES software used to identify Bh AVRA1 and BEC1016 interactors are provided at the GitHub pages (https://github.com/Wiselab2/NGPINT_V2; https://github.com/Wiselab2/Y2H‐SCORES). Raw Y2H‐Seq reads are at NCBI‐GEO under acc. nos. GSE164954 (AVRA1/CSEP0008) and GSE166108 (BEC1016/CSEP0491). Otherwise, Y2H‐NGIS scores are available in the supporting information. GenBank accession numbers for AVR A1 (CSEP0008, BLGH_03023) and BEC1016 (CSEP0491, BLGH_07006), are CCU81904.1 and CCU83284.1, respectively. HvERdjB3 is represented in GenBank as an mRNA‐derived DNA sequence from Haruna Nijo (AK376215.1) and as a protein sequence from the genome sequence of Tibetan Hulless barley (KAE8793706).


