Abstract
The human BLM gene is a member of the Escherichia coli recQ helicase family, which includes the Saccharomyces cerevisiae SGS1 and human WRN genes. Defects in BLM are responsible for the human disease Bloom’s syndrome, which is characterized in part by genomic instability and a high incidence of cancer. Here we describe the cloning of rad12+, which is the fission yeast homolog of BLM and is identical to the recently reported rhq1+ gene. We showed that rad12 null cells are sensitive to DNA damage induced by UV light and γ radiation, as well as to the DNA synthesis inhibitor hydroxyurea. Overexpression of the wild-type rad12+ gene also leads to sensitivity to these agents and to defects associated with the loss of the S-phase and G2-phase checkpoint control. We showed genetically and biochemically that rad12+ acts upstream from rad9+, one of the fission yeast G2 checkpoint control genes, in regulating exit from the S-phase checkpoint. The physical chromosome segregation defects seen in rad12 null cells combined with the checkpoint regulation defect seen in the rad12+ overproducer implicate rad12+ as a key coupler of chromosomal integrity with cell cycle progression.
The fission yeast Schizosaccharomyces pombe undergoes a dose-dependent G2 delay in response to DNA damage caused by radiation (1, 27). Cells remain arrested at this G2 checkpoint while DNA damage is repaired, then enter mitosis and resume progression through the cell cycle. Six checkpoint rad genes have been identified in S. pombe: rad1+, rad3+, rad9+, rad17+, rad26+, and hus1+ (1, 2, 9, 27). Mutations in any of these genes result in almost identical phenotypes. The mutant strains are all sensitive to radiation and to agents which transiently inhibit DNA replication, and all lack the G2 checkpoint, preventing mitotic entry in the presence of such damage (1, 2, 9, 27). While little is known about G2 checkpoint genes in humans, homologs of S. pombe rad3+ (4, 5) and rad9+ (19) have recently been isolated. Recent studies have implicated cell cycle gene mutations in causing cancer. It is likely that mutations in genes regulating the G1 and G2 checkpoints also have the potential to increase cancer susceptibility.
The phenomenon of genomic instability has been firmly established as a crucial step in the genesis of cancer (14, 31, 33). While genomic instability is seen as a step in the progression of cancer cells, there are also several genetic diseases that lead to genomic instability and, ultimately, to cancer. Among these diseases is the rare autosomal recessive disorder Bloom’s syndrome (12). The defective gene in Bloom’s syndrome, BLM, encodes a putative DNA helicase that is homologous to the Escherichia coli RecQ and Saccharomyces cerevisiae Sgs1 helicases (11, 34). While the BLM and SGS1 proteins contain centrally located helicase domains that are homologous to RecQ, they are much larger than RecQ and show homology, both 5′ and 3′, to the helicase core (26, 35, 37).
We describe here the cloning of the fission yeast rad12+ gene, which encodes a helicase similar to that encoded by BLM. We show that rad12+ is a negative regulator of the S-phase checkpoint and that rad12+ functions through the rad9+ checkpoint control gene. The regulation of rad9+ by rad12+ has implications for the regulation of the S-phase checkpoint by BLM in human cells.
MATERIALS AND METHODS
S. pombe genetic manipulations.
S. pombe was cultured by standard techniques (17). The complete genotypes of the strains used in this study are summarized in Table 1. The construction of novel strains for use in this study is described below. Unless otherwise noted, random spore analysis was used to identify the appropriate progeny.
TABLE 1.
Strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| 972 | h−S | 17 |
| Sp8 | h−S ade6-210 leu1-32 | 3 |
| Sp11 | h+N ade6-216 leu1-32 | 3 |
| Sp12 | h+N/h−S ade6-210/ade6-216 | 3 |
| Sp30 | h−S ade6-210 leu1-32 ura4-D18 | This study |
| Sp215 | h+N ade6-704 | 15 |
| Sp263 | h+N leu1-32 | 10 |
| Sp272 | h−S rad12-502 | 7 |
| Sp275 | h+N leu1-32 rad12-502 | 10 |
| Sp301 | h−S ade6-210 leu1-32 rad12-502 | This study |
| Sp302 | h+N ade6-210 leu1-32 rad12-502 | This study |
| Sp303 | h−S/h+N ade6-210/ade6-216 leu1-32/leu1-32 rad12-502/rad12+ | This study |
| Sp304 | h+N/h−S ade6-210/ade6-216 leu1-32/leu1+ rad12-502/rad12-502 | This study |
| Sp325 | h−S rad9::ura4+ | This study |
| Sp326 | h+N rad9::ura4+ | 18 |
| Sp340 | h−S ade6-704 rad12-502 | This study |
| Sp345 | h+N rad12-502 rad9::ura4+ | This study |
| Sp358 | h−S ade6-210 leu1-32 ura4-D18 rad12::ura4+ | This study |
| Sp359 | h+N leu1-32 pgR12OP | This study |
Sp301 (h−S ade6-210 leu1-32 rad12-502) and Sp302 were constructed by crossing Sp8 (h−S ade6-210 leu1-32) with Sp275 (h+N rad12-502 leu1-32). Sp303 (h−S/h+N ade6-210/ade6-216 leu1-32/leu1-32 rad12-502/rad12+) was constructed by crossing Sp301 with Sp11, for 2 days at 25°C, on sporulation medium. Diploids were rescued on minimal medium, with selection for adenine prototrophy, and were further propagated on YE medium (5 g of yeast extract/liter, 30 g of glucose/liter, 20 g of agar/liter) with selection of white (adenine-prototrophic) diploid colonies. In a similar way, Sp304 (h+N/h−S ade6-210/ade6-216 leu1-32/leu1+ rad12-502/rad12-502) was constructed by crossing Sp301 with Sp302. Sp325 (h−S rad9::ura4+) was constructed by outcrossing Sp326 (h+N rad9::ura4+) five times with 972 and once with Sp263 (h+N leu1-32). Sp340 was constructed by crossing Sp215 (h+N ade6-704) with Sp272. Sp345 (h+N rad12-502 rad9::ura4+) was constructed by crossing Sp325 with Sp275. The products of this cross were subjected to tetrad analysis, and the identity of the double mutant was confirmed by outcrossing. The construction of Sp358 and Sp359 are described in detail below.
Positional cloning of the rad12+ gene.
Cosmids spanning the rad15+ region (no. 227 through 139 [22]) were partially digested with Sau3A1, and DNA fragments of between 1 and 4 kb were gel purified. This DNA was shotgun cloned into pYC12, which bears the sup3-5 opal suppressor. The constructs were sequenced at either end, and these sequences were compared to the known genomic DNA sequences available from the S. pombe genome sequencing project (Sanger Centre, Cambridge, United Kingdom). Seven clones spanning the rad15+-containing region were chosen; they were transformed into a strain bearing the ade6-704 mutation (Sp215), and stable ade+ transformants were identified. The integrants were backcrossed to rad12-502 ade6-704 double mutants (Sp340), and the relative distances between the integration sites and rad12+ were determined by tetrad analysis. This analysis predicted that rad12+ was most likely contained in a region corresponding to the end of SPAC2G11 (EMBL accession no. Z54354 [16]).
To clone the candidate gene SPAC2G11.12, cosmids mapping to this region (22) were screened by Southern blotting. Primer sets BLS1-BLS2 and BLS3-BLS4 (Table 2), corresponding to the 5′ and 3′ regions of SPAC2G11.12, respectively, were synthesized and used to produce radioactive probes by PCR amplification carried out with [α-32P]dCTP and [α-32P]dGTP in the reaction mixture. Cosmids 514 and 908 were positive in this assay. The rad12+ gene was isolated as a 5,127-bp NheI-SalI restriction fragment and subcloned into XbaI- and SalI-digested pBluescript SK(−), creating pgR12.
TABLE 2.
Oligonucleotides used in this study
| Oligonucleotide name | Sequence | Locationa |
|---|---|---|
| BLS1 | CGA AAA TGT CAC TAG CCC CA | 68–87 |
| BLS2 | TTC GCT GAC GAT TGG GTT AG | 587–568 |
| BLS3 | TGG CCA TAG ATG ACA GCA GA | 3397–3416 |
| BLS4 | TTC CGT TGA CCA TCC ACT TC | 3757–3738 |
| BLS5 | GAC ATC TGC AGC GGC TGT TGG AAT T | (−20)–(−6)b |
| BLS6 | ACT CAT CAT TAA TCG GAT CC | 1290–1271 |
| UVDE1 | ATG CTT AGG CTA TTG A | 121–136 |
| UVDE2 | CAA CAG ACT CAT CAA T | 508–493 |
| UVDE7 | TCA AAA AGT ATG ACG | 1940–1926 |
| UVDE10 | CAA GCT GGC AAA TAA | 1143–1157 |
| LEU1 | GAG GAA TAT CCT CAC C | 631–647 |
| LEU2 | TAT CAG CGG TAG AAG C | 1074–1058 |
Locations of the primers are given with respect to the initiating ATG of the respective genes. Numbering in the location column is from the 11th nucleotide to the end.
The 10 nucleotides at the 5′ end of this oligonucleotide do not correspond to the rad12+ sequence but rather generate a Pst1 restriction site.
Generation of the rad12 null strain Sp358.
The rad12::ura4+ disruption plasmid was constructed by digesting pgR12 DNA with HindIII, thereby removing the 2,102 bp from nucleotide no. 780 through 2882 (counting the start of translation as nucleotide no. 1). The ura4+ gene was isolated from plasmid pART1 as a 1,900-nucleotide HindIII fragment and ligated into HindIII-digested pgR12, creating pgR12::ura4+. S. pombe Sp30 (h−S ade6-210 leu1-32 ura4-D18) cells were transformed with SalI-linearized pgR12::ura4+. Transformants were selected on minimal plates containing 150 μg each of leucine and adenine per ml. The identities of cell lines in which rad12::ura4+ had replaced the wild-type rad12+ gene were confirmed by Southern blotting.
Preparation of S. pombe whole-cell extracts and performance of UVDE assays.
Whole-cell extracts were prepared from 109 S. pombe cells as described previously (7). The protein concentrations of extracts prepared this way were approximately 20 to 30 mg/ml. The cells used to produce UV-induced extracts were grown in YEA (5 g of yeast extract, 30 g of glucose, and 75 mg of adenine per liter) to late log phase, collected by centrifugation, washed with water, and resuspended in 1 volume of water. The cells were placed in a large petri dish or glass tray and irradiated with constant mixing. Inductions were performed with 254-nm UV light, using an effective dose of 50 J/m2 (as assayed by viability) and a dose rate of 2.68 J/m2/s (7). The cells were transferred to fresh YEA and incubated with shaking at 30°C for the appropriate time periods. The cells were then collected by centrifugation and mixed with an equal volume of extraction buffer prior to being frozen at −70°C. Extracts were prepared as described above.
UV damage endonuclease (UVDE) assays were carried out essentially as described previously (10). Briefly, whole-cell extract (100 μg) was incubated at 37°C for 5 to 15 min with 0.02 pmol of 3′-end-labelled 6-4 photoproduct 51-mer in 45 mM HEPES-KOH (pH 7.8)–70 mM KCl-7 mM MgCl2 in a 20-μl reaction volume. The samples were treated with proteinase K and extracted with phenol-chloroform, and the DNA was analyzed on denaturing 15% polyacrylamide–urea gels. The gels were dried and exposed to X-ray film, and the results were quantitated on an image analysis system (Fuji).
Northern hybridization.
Total RNA was isolated from cells grown in YEA to late log phase (2 × 107/ml). Approximately 0.3 ml of packed cells was resuspended in 4 ml of TRIzol (Gibco BRL), and then 0.5-mm-diameter glass beads were added up to the meniscus. The cells were lysed by three rounds of high-speed vortexing, each for 30 s, with 2 to 3 min of cooling on ice in between rounds. Then 4 ml of TRIzol and 1.6 ml of CHCl3 were added, the solution was mixed, and the aqueous phase was separated by centrifugation. The aqueous layer was extracted with phenol-chloroform, and the RNA was precipitated with an equal volume of isopropanol. Poly(A)+ mRNA was isolated on Oligotex columns (Qiagen) according to the manufacturer’s specifications. Northern blot analysis was carried out as described elsewhere (25), using 3 μg of each mRNA sample. The mRNA was transferred to a Zetablot membrane (Bio-Rad), and 32P-labelled probes were synthesized by PCR amplification of two regions of the uve1+ gene with primer pairs UVDE1-UVDE2 and UVDE7-UVDE10 (Table 2), using the process described above. uve1+ mRNA amounts were normalized to leu1+ mRNA levels by probing with a 32P-labelled PCR probe complementary to leu1+, which was synthesized with primers LEU1 and LEU2 (Table 2). Radioactive imaging and quantitation were carried out with a PhosphorImager (Molecular Dynamics).
HU treatment, fluorescent microscopy, and FACS analysis.
S. pombe cells were grown in minimal medium plus the necessary supplements to a density of approximately 5 × 106/ml, and hydroxyurea (HU) was added to a concentration of 12 mM. Samples were taken at various time points and either diluted and plated or fixed with 70% ethanol. Plated cells were incubated at 30°C for 3 days and then colonies were counted. After 24 h, the fixed cells were treated in one of two ways. Cells were washed once with 50 mM sodium citrate, resuspended in 0.5 ml of 50 mM sodium citrate, and then treated with 250 μg of DNase-free RNase at 37°C for 1 h. DNA was stained by the addition of 0.5 ml of a 2.5-μg/ml solution of propidium iodide in 50 mM sodium citrate. Cells prepared in this way were used for both fluorescence microscopy and flow cytometry. Fluorescence-activated cell sorter (FACS) analysis was performed on a Coulter EPICS Elite flow cytometer. Alternatively, cells were recovered from fixation, resuspended in 50 mM sodium citrate (pH 7.0), and stored at 4°C until they were stained. To stain the DNA, cells were collected and stained by resuspension in a 0.5-μg/ml solution of 4,6-diamidino-2-phenylindole (DAPI) in 50 mM sodium citrate. In both cases, the cells were viewed by fluorescence microscopy.
UV and gamma irradiation experiments.
S. pombe UV irradiations were performed with a 254-nm germicidal lamp at a dose rate of 2.68 J/m2/s. Cells were grown to mid-log phase (2 × 106 to 1 × 107/ml) and plated at various concentrations depending on the dose of UV used. For each irradiation, approximately 1,000 cells were plated on minimal selective medium. Following irradiation, the plates were incubated at 30°C for 3 to 4 days and then the numbers of viable colonies were determined. The results presented are based on three independent experiments.
S. pombe gamma irradiations were carried out on cells grown to densities of 5 × 106 to 5 × 107 in YEA. Cells were plated onto YEA plates and irradiated in a Gamma Cell 220 irradiator (Nordion International Inc., Kanata, Ontario, Canada) at a dose rate of approximately 1 Gy/s. The plates were incubated at 30°C for 3 days, and the colonies were then counted.
Generation and analysis of the rad12+-overproducing (OP) strain Sp359.
To create a cell line that produces high levels of wild-type rad12+, the rad12+ sequence was subcloned into pART1, behind the adh promoter. This plasmid was constructed in two steps. First, primers BLS5 and BLS6 were synthesized. BLS5 introduced a PstI restriction site just upstream of the translation-initiating ATG sequence of rad12. BLS6 primers inside the gene, downstream of a unique BamHI site. Two micrograms of pgR12, linearized with SalI, was used as a template for 10 rounds of PCR amplification with 100 pM each BLS5 and BLS6 and the thermostable DNA polymerase PfuI (Stratagene). The PCR product was digested with PstI and BamHI, gel purified, and ligated into PstI- and BamHI-digested pARTI, creating pPBr12. The 3′ portion of the rad12+ gene was then isolated. Five micrograms of pgR12 DNA was digested with SalI and then incubated with 200 μM each deoxynucleoside triphosphate plus 5 U of T4 DNA polymerase for 15 min at 14°C to create a blunt end. The DNA was then digested with BamHI and ligated into SmaI- and BamHI-digested pPBr12. The resulting plasmid, pgR12OP, places the rad12 sequence just in front of the adh promoter. One microgram of pgR12OP was transformed into Sp263 (h−S leu1-32), and selection for transformants was performed on minimal-medium plates.
To determine the effect of overexpression of rad12+ on cell survival following UV irradiation, Sp263 cells transformed with pgR12OP and 972 cells were grown in minimum medium to mid-log phase and plated onto minimal-medium plates at 200 to 300 cells per plate. The cells were then irradiated with 60 J of 254-nm UV light/m2. Colonies were counted after 3 days. Sp263 cells transformed with pgR12OP and 972 cells were assayed for their checkpoint responses following UV radiation. Both cell types were grown to mid-log phase, irradiated at 60 J/m2, allowed to recover in liquid minimal medium for 40 min, fixed with 70% ethanol, and stained with a 50-μg/ml solution of propidium iodide. Cells were examined by fluorescence microscopy.
RESULTS
rad12+ encodes a DNA helicase homologous to the Bloom’s syndrome disease gene.
Initial attempts to clone the rad12+ gene by complementation of the radiation sensitivity phenotype were unsuccessful. One possible explanation for this was that the rad12-502 mutation was dominant. Diploid strains that were wild type (Sp12) or either homozygous (Sp304) or heterozygous (Sp303) for rad12-502 were constructed and tested for sensitivity to UV light. The rad12-502 heterozygote was as resistant to UV light as the wild-type homozygote, indicating that the rad12-502 allele is recessive.
Our prior genetic analysis indicated that rad12+ is closely linked to rad15+, on chromosome 1, so we adopted a positional cloning strategy to identify the rad12+ gene. Subclones of cosmids spanning this region (22) were used to generate targeted homologous-recombination constructs bearing the sup3-5 marker, as described in Materials and Methods. Preliminary analysis indicated that rad12+ was located in a region that included four genes, one of which was SPAC2G11.12, the S. pombe homolog of the Bloom’s syndrome susceptibility gene, BLM (Fig. 1a). Because of the phenotypes associated with Bloom’s syndrome, this helicase was considered a strong candidate for the rad12+ gene, and we tested this hypothesis directly.
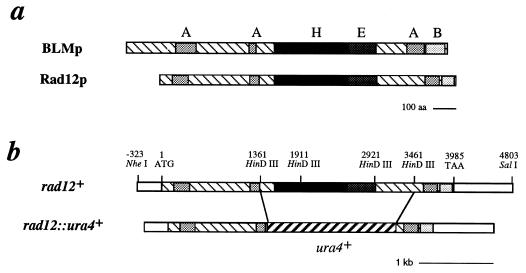
FIG. 1.
Schematic representation of rad12+. (a) Alignment of Rad12p and BLMp sequences, showing the conserved acidic (A), basic (B), RecQp-like helicase (H), and extended high-level-homology domains. The acidic and basic regions exhibit greater than 30% acidic or basic residues over a span of at least 20 amino acids. The highly conserved helicase domain has been previously defined (13). The extended region of high-level homology is a region of 37% identity (57% similarity) that extends for an additional 199 amino acid residues past the C terminus of the helicase domain. (b) Corresponding schematic representation of the rad12+ gene and the construct used to generate the gene disruption, rad12::ura4+ (Sp359).
An NheI-SalI fragment containing the entire SPAC2G11.12 gene was subcloned into pBluescript SK, generating the plasmid pgR12. A gene disruption construct was generated by replacement of the central 2.1 kb, including the helicase domain, with the ura4+ gene (Fig. 1b). The construct was used to generate the rad12::ura4+ null strain Sp358. This null strain was crossed with a rad12-502 mutant, and the resultant progeny were analyzed by random spore analysis. Of 2,600 colonies tested, no wild-type (radiation-resistant) recombinants were identified, indicating that rad12+ encodes the Bloom’s syndrome-like helicase SPAC2G11.12. In addition, the rad12-502 mutation has been sequenced and shown to result in an altered ATP binding site (T to I at amino acid 543 [MPTGGGK] [23]). This is the same gene that was reported by Stewart et al. (32) to be the rqh1+ gene, which was originally identified as hus2+ in a screen of HU-sensitive S. pombe mutants (9). We continue with the use of the rad12+ designation, because this was the first mutation isolated and published for this gene (24).
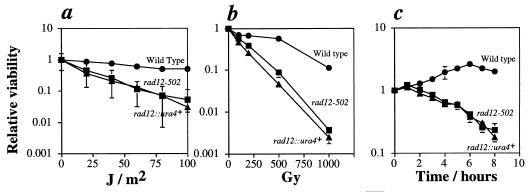
Survival curves demonstrated that rad12-502 and rad12::ura4+ have comparable sensitivities to UV light and to transient DNA inhibition by HU (Fig. 2a and c). Our studies further showed that the loss of rad12+ leads to γ-ray sensitivity (Fig. 2b). The sensitivity of rad12-502 to γ radiation contradicts an earlier report (24). In addition, the rad12::ura4+ cells have the unusual morphology exhibited by rad12-502 cells (see below). With both UV and γ irradiation, rad12-502 cells were slightly less sensitive than rad12::ura4+, indicating that the rad12-502 mutation is not a null allele.
FIG. 2.
Sensitivity of the rad12::ura4+ mutant to DNA damage and transient inhibition of replication. Shown are UV light (a), γ radiation (b), and HU (c) survival curves of the wild-type (Sp30) (•), rad12-502 (Sp272) (▪), and rad12::ura4+ (Sp358) (▴) strains. (a and b) Strains were grown to mid-log phase, plated at appropriate dilutions, and treated with the indicated doses of radiation. Colonies were allowed to grow 4 days at 32°C, at which time the numbers of colonies were determined. Colony numbers were normalized to those of the nonirradiated samples. (c) Strains were grown to mid-log phase; then HU was added to 12 mM. Samples were removed from the HU at the indicated times and transferred to plates for viability testing. Colonies were allowed to grow 4 days at 32°C, at which time the numbers of colonies were determined. Colony numbers were normalized to those of the samples harvested at the time of addition of HU (time = 0). In all cases, error bars represent the standard deviations of the data. Where error bars are not visible, they are smaller than the symbols representing the data points.
rad12+ acts upstream of rad9+ in release from the S-phase checkpoint and UVDE regulation.
Our initial interest in the rad12+ gene grew from our studies of the UVDE. In an early screen of the S. pombe rad1 through rad23 mutants, only rad12-502 had reduced UVDE activity (10). By contrast, we found that extracts prepared from a rad9-192 mutant strain showed consistently high levels of UVDE activity. rad9+ is one of the six checkpoint rad genes, responsible for G2- and S-phase checkpoint control, that have been identified in S. pombe. UVDE activity is approximately fivefold higher in whole-cell extracts prepared from rad9-192 or rad9::ura4+ cells than in extracts prepared from wild-type cells (Fig. 3a). This level of UVDE activity is comparable to that seen in wild-type cells in which UVDE levels have been induced by irradiating with UV light prior to assaying for activity (7) (Fig. 3a). In addition, when rad9::ura4+ cells were assayed for UVDE activity after exposure to UV light, UVDE levels did not increase significantly. Figure 3b shows that basal uve1+ mRNA levels were identical in wild-type and rad9 null cells, when normalized to control (leu1+) mRNA levels. Further, the time courses of uve1+ mRNA induction following irradiation were identical in wild-type and rad9 mutants (Fig. 3c). In both wild-type and rad9 mutant cells, maximal uve1+ mRNA levels were observed 10 min after treatment with UV. Also, the extents of induction were comparable; when normalized to control leu1+ mRNA, and compared to uninduced mRNA levels, the maximal observed uve1+ mRNA inductions were 3.4-fold in wild type and 3.7-fold in rad9 mutant cells. This indicates that the elevation of UVDE activity in rad9 mutants is the result of a posttranscriptional regulation which functions in addition to the previously reported transcriptional regulation of uve1+ following UV treatment (7).
FIG. 3.
rad9+ acts downstream of rad12+ in the repression of basal levels of UVDE activity. (a) Cells were grown to late log phase and then treated with UV where indicated. Cells were allowed to recover for the indicated time periods and then harvested, and whole-cell extracts were prepared. The extracts were assayed for UVDE activity, as measured by cleavage of an oligonucleotide substrate with a single internal UV photoproduct from a 51mer to a 31mer. WT, wild type. (b) Basal levels of uve1+ mRNA were determined by Northern analysis in wild-type and rad9 null cells. Samples were subjected to electrophoresis, transfer, and hybridization against uve1+ and leu1+ probes simultaneously. (c) Time course of uve1+ mRNA induction in wild-type (972) and rad9 null (Sp325) cells after treatment with 50 J of UV light/m2. Inductions were performed as described in Materials and Methods. Cells were harvested at the indicated times (in minutes) after induction. Poly(A)+ RNA was subjected to Northern analysis with probes directed against uve1+ and leu1+. (d) UVDE activity was assayed as for panel a, using strains rad12-502 (Sp275) (lane 3), rad9::ura4+ (Sp325) (lane 2), and rad12-502 rad9::ura4+ (Sp345) (lane 1) mutants.
While UVDE activity is limiting in whole-cell extracts prepared from rad12-502 mutants (10), when rad12-502 cells were UV irradiated prior to extract preparation, UVDE activity increased to the levels seen in UV-induced wild-type extracts (7). This suggests that rad12+ is not required for the induction of UVDE activity in response to UV damage but rather, like rad9+, is involved in the regulation of basal UVDE levels. To establish the epistasis of rad9+ and rad12+, a rad9::ura4+ rad12-502 double mutant was constructed and the genotype was confirmed by outcrossing. UVDE activity in the double mutant was elevated to levels similar to those observed in rad9::ura4+ single mutants (Fig. 3d). This indicates that rad9+ acts downstream of rad12+ in the regulation of UVDE activity.
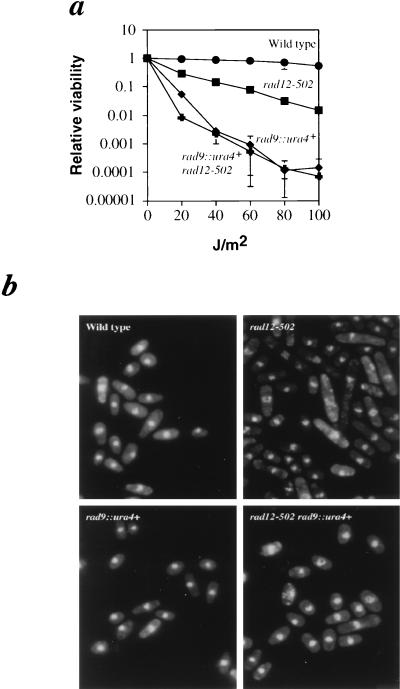
We next tested for epistasis between rad12+ and rad9+. UV survival curves for the double mutant are presented in Fig. 4a. At low UV doses (∼20 J/m2), the rad9::ura4+ rad12-502 double mutant was slightly more sensitive to UV than the single rad9::ura4+ mutant. However, at all other doses, the sensitivity of the double mutant was indistinguishable from that of the rad9::ura4+ single mutant. This epistasis indicates that the pathways for rad9+ and rad12+ are partially overlapping, though not identical. The increased sensitivity at low UV doses is indicative of a second function for either Rad9p or Rad12p under these conditions.
FIG. 4.
rad12+ and rad9+ are epistatic; the aberrant morphology of rad12 mutants is suppressed in a rad9::ura4+ background. (a) UV survival curves were constructed as described in Materials and Methods. Wild-type (972) (•) cells are highly resistant to UV light. rad12-502 cells (Sp275) (▪) are moderately sensitive to UV light. rad12-502 rad9::ura4+ double-mutant cells (Sp345) (✚) are no more sensitive to UV light than rad9::ura4+ single-mutant cells (Sp325) (⧫). (b) Late-log-phase cells which were either wild type (972), rad12-502 (Sp275), rad9::ura4+ (Sp325), or rad12-502 rad9::ura4+ (Sp345) were fixed with ethanol, stained with propidium iodide, and examined by fluorescence microscopy. rad12-502 cell populations exhibit many elongated cells, which are not present in rad12-502 rad9::ura4+ double mutants.
Additional evidence that Rad12p functions upstream of Rad9p was shown by another criterion. As mentioned above, rad12-502 and rad12::ura4+ cells have an abnormal morphology; approximately 20% of the cells are elongated to two to three times the wild-type length (Fig. 4b). This morphological abnormality is absent in rad9::ura4+ rad12-502 double mutants. The absence of this phenotype in a rad9::ura4+ background indicates that the elongation is rad9+ dependent. This suggests that rad9+ acts downstream of rad12+ in maintaining normal cell morphology and that rad12+ influences the cell cycle through rad9+.
Overexpression of Rad12 leads to a checkpoint defect.
The results described above indirectly show that Rad12p acts as a negative regulator of Rad9p. This predicts that overexpression of Rad12p should repress Rad9p levels, resulting in cells which are checkpoint defective. To test this directly, we produced a Rad12p-overexpressing strain by cloning rad12+ into the vector pART1, placing rad12+ under the control of the strong adh promoter. This construct was transformed into the wild-type S. pombe strain Sp263, creating the rad12+ OP strain Sp359. We examined this strain, as well as rad9 and rad12 mutant strains, for the ability to enter and exit the S-phase checkpoint, which responds to transient inhibition of DNA replication. Log-phase cultures of wild-type, rad9::ura4+, rad12::ura4+, and rad12+ OP were prepared and treated with 12 mM HU. At hourly intervals after addition of HU, cells were fixed in ethanol. In all cases, after 5 h, >80% of cells exhibited G1/S DNA content, as assayed by flow cytometry (Fig. 5a). At this time, the cells were collected by centrifugation and fresh medium lacking HU was added. The cells were incubated for an additional 140 min, with aliquots being collected and fixed with ethanol every 20 min. FACS analysis showed that all but the rad9::ura4+ mutant resumed DNA synthesis with similar kinetics, with the peak of replication occurring 60 to 80 min after release from the block (Fig. 5a). Even though the viability of the rad9::ura4+ culture was about 1% after the 5-h HU block (see below), about 50% of these cells resumed DNA synthesis to an extent that was detectable by FACS (Fig. 5a).
FIG. 5.
rad12+ OP cells are defective in the S-phase checkpoint. Wild-type (Sp263), rad9::ura4+ (Sp325), rad12::ura4+ (Sp358), and rad12+ OP (Sp359) strains were grown to mid-log phase at 25°C, arrested with HU, and then released 5 h later. (a) Aliquots were fixed with ethanol each hour during the HU block and every 20 min after HU block release. Samples were stained with propidium iodide and analyzed by flow cytometry. (b) The indicated strains were prepared identically to those in panel a. The 0-h (log phase), 5-h post-HU addition (HU blocked), and 140-min post-HU removal (HU released) samples were stained with propidium iodide and examined by fluorescence microscopy.
Log-phase, HU-blocked (5 h), and HU-released (140 min) cells from the above-described time course were stained with propidium iodide and examined microscopically (Fig. 5b). Wild-type cells entered the S-phase checkpoint normally and had intact interphase nuclei during the HU block. These cells resumed normal division after release from the HU block. By contrast, rad9::ura4+ cells were defective in the S-phase checkpoint and segregated chromosomes prematurely during the HU block. By the time of release from the HU block, most of the rad9::ura4+ cells displayed the typical cut phenotype associated with premature lethal mitosis. The rad12::ura4+ mutants entered the S-phase checkpoint normally and exhibited interphase nuclei similar to those of wild-type cells. However, on release from S phase, many of these cells failed to segregate their chromosomes normally. This result confirms previously published data (32) and indicates that rad12+ function is required for a proper exit from S phase. The rad12+ OP strain, like the rad9::ura4+ mutant, exhibited premature chromosomal segregation in the presence of HU. This was a clear indication that the S-phase checkpoint is deficient in these cells. After release from the HU block, the rad12+ OP strains exhibited many aberrantly segregated nuclei, although other cells appeared normal.
Viability studies of these strains were also performed. Figure 6 shows that while wild-type cells roughly doubled in relative viability after addition of HU, the rad9::ura4+, rad12::ura4+, and rad12+ OP strains lost viability to various degrees. The viability loss of the rad12+ OP was the least severe, but this strain still exhibited only about 25% of the viability of the wild type at later time points. This confirms the microscopic examination of these cells, which indicated that while the rad12+ OP is S-phase checkpoint deficient, the deficiency is not as severe as that of a true checkpoint null strain (rad9::ura4+).
FIG. 6.
Sensitivity of the rad12+ OP to HU treatment. HU survival curves of the wild type (972) (•), rad12::ura4+ (Sp358) (▪), rad9::ura4+ (Sp325) (⧫), and rad12+ OP (Sp359) (▴). Strains were grown to mid-log phase; then 12 mM HU was added. Samples were removed from HU at the indicated times and plated. Colonies were allowed to grow for 4 days at 32°C. Colony numbers were normalized to the samples harvested at time zero, the time of addition of HU. In all cases, error bars represent the standard deviations of the data. Where error bars are not visible, they are smaller than the symbols representing the data points.
DISCUSSION
The studies reported here implicate the rad12+ gene product as a negative regulator of the S-phase checkpoint acting via the rad9+ gene. The cloning of the rad12+ gene revealed that it is a member of a family of genes related to the recQ gene of E. coli, which includes the human BLM and WRN genes, and the SGS1 gene of Saccharomyces cerevisiae (8, 11, 37). Defects in BLM and WRN lead to the human diseases Bloom’s syndrome and Werner’s syndrome, respectively. rad12+ is allelic to the recently reported rqh1+ gene of S. pombe (32). This family of genes is characterized by a central RecQ-like helicase domain and various other regions of structural similarity extending both N and C terminally. However, the role of these genes in normal cell growth is unknown. Mutations in the different genes lead to somewhat different phenotypes. Bloom’s syndrome is characterized by high rates of sister chromatid exchanges (SCE) and an extremely high incidence of cancer with early onset (12). Werner’s syndrome is a disease of premature aging, with high cancer rates but no increase in SCE (6, 28). Loss of SGS1 function in Saccharomyces cerevisiae suppresses the slow-growth phenotype seen in the topoisomerase mutant top3, and two-hybrid studies have shown that Sgs1 associates with both Top2p and Top3p (11, 36). The mutant rad12-502 allele was first identified as a UV-sensitive mutant, while the allelic rqh1-h2 (hus2-22) was identified in a screen for HU sensitivity (9, 24). In addition, we have demonstrated here that rad12 mutant cells are sensitive to gamma rays. rad12+ is the only gene in this family that has been shown conclusively to be required for protection from UV light, γ radiation, or HU. Most recently, two papers have been published which implicate SGS1 in cell aging (29, 30). Morphologically, this is manifested as premature disintegration of the nucleolus. This implies that SGS1 may function in a manner more analogous to WRN than to BLM. It will be of great interest to determine whether this is the case for rad12+.
Our interest in rad12+ began with a screen of radiation-sensitive mutants of S. pombe that were defective in UVDE activity. In this screen, rad12-502 cells had low UVDE activity (10). We had also observed that UVDE activity is inducible by UV radiation (7). This was based on measurements of UVDE activity in extracts prepared from wild-type cells which were irradiated with 254-nm light. Interestingly, UVDE activity is also inducible in rad12-502, with UVDE activity increasing to levels comparable to those seen in induced wild-type extracts (7). This observation demonstrated that rad12+ is a positive regulator of basal UVDE activity levels. In contrast, rad9 mutant cells (either rad9-192 or rad9::ura4+) were shown here to have elevated levels of UVDE when assayed in extracts prepared from untreated cells. Furthermore, UVDE activity is not induced to significantly higher levels when rad9 mutant cells are UV irradiated. This indicates that rad9+ is a negative regulator of basal UVDE activity levels and that the loss of rad9+ leads to derepression of UVDE activity. These data led us to speculate that rad9+ and rad12+ are members of the same pathway, and the series of studies reported here is consistent with this conclusion.
UV survival data of the rad12-502 rad9::ura4+ double mutant demonstrate that these two genes share a common overlapping, but not identical, pathway (Fig. 4). An increased sensitivity to UV light was observed in the double mutant at very low doses (<40 J/m2), indicative of a second function for either Rad9p or Rad12p under these conditions. To determine the order of these genes in the common pathway, we measured UVDE activity in our double mutant rad12-502 rad9::ura4+. Extracts prepared from this double mutant have elevated levels of UVDE, comparable to those of rad9 mutant cells. This result is consistent with rad12+ acting upstream of rad9+. While this is the simplest model to explain the regulation of UVDE by rad9+ and rad12+, it is not the only possible one. We cannot exclude the possibility that rad9+ and rad12+ act in different pathways with respect to UVDE regulation, with rad9+ exerting a stronger effect and rad12+ exerting a weaker effect.
A second, independent experiment supporting the ordering of rad12+ upstream of rad9+ came from studies on cell morphology in rad12 mutants. As seen in Fig. 4b, cycling rad12-502 cells have a subpopulation (about 20%) which are elongated, a phenotype suggesting that these cells are undergoing a cell cycle delay. There are two explanations for this result. One is that rad12 mutant cells undergo increased spontaneous DNA damage and are G2 delayed in response to the damage. The alternative explanation is that the levels of checkpoint gene products are higher in rad12 mutant cells and that this leads to spontaneous G2 arrest. This phenotype is suppressed in rad12-502 rad9::ura4+ double mutants, demonstrating that this effect is rad9+ dependent. Again, this places rad12+ upstream of rad9+ and leads us to favor the hypothesis that the elongated phenotype of rad12 mutants is due to upregulation of the S-phase checkpoint.
If rad12+ negatively regulates rad9+, then its overproduction in cells should lead to a checkpoint-deficient phenotype. To test this theory, rad12+ was cloned behind the strong adh promoter, creating rad12+ OP. Cells overproducing rad12+ were found to be sensitive to transient exposure to HU treatment (Fig. 6), which is consistent with an S-phase checkpoint defect. We went on to carefully study this potential S-phase checkpoint defect in the rad12+ OP strain by FACS and fluorescence microscopy analyses. HU treatment blocks DNA replication in all cells tested, leading to arrest at the S-phase checkpoint. Despite this block in replication, checkpoint-deficient cells continue to divide and cells are seen to have a characteristic cut phenotype, in which their DNA prematurely segregates and nuclei become fragmented. As seen in Fig. 5b, rad9::ura4+ cells show a typical checkpoint defect following treatment with HU. At 5 h after addition of HU, rad9::ura4+ cells have not arrested cell division, despite the fact that DNA synthesis has halted, as determined by FACS analysis (Fig. 5a). Very similar defects are seen in the rad12+ OP strain, although the frequency of nuclear fragmentation here is lower than that observed in rad9 null mutants (20% versus >50%). This is not surprising and suggests that rad12+ is not the sole regulator of rad9+.
By contrast, rad12::ura4+ cells demonstrate quite a different checkpoint deficiency. At 5 h post-HU treatment, rad12::ura4+ cells are not dividing but continue to elongate, just as wild-type cells behave. Thus, rad12+ is not required for entry into the S-phase checkpoint. However, when rad12 null cells are now released from the HU block and replication is completed (as assayed by FACS analysis), these cells do not properly segregate their chromosomes, indicating that exiting from S phase is defective (Fig. 5b).
These results are consistent with the observations of Stewart et al. on rqh1 mutant cells (32). They reported that an rqh1 null mutant had high recombination frequencies and increased chromosome loss, a phenotype similar to the high SCE frequencies seen in human BLM mutant cells. In addition, they showed that while rqh1 mutant cells arrested following HU treatment, they were defective in exiting S phase. They speculated that this defect could be attributed to the loss of its function in suppressing recombination. In their model, they envisioned that the loss of rqh1+ leads to increased recombination and to intermediates that cannot be resolved at mitosis, leading to the cut phenotype seen in HU-treated rqh1 mutant cells. This could be tied to its potential relationship with topoisomerases as seen in SGS1 in Saccharomyces cerevisiae (11, 36). If this is in fact the case, Rad12p would be physically required for completion of S phase. Taken together with the work presented here, which showed that rad12+ regulates the S-phase checkpoint, this would implicate Rad12p as a protein which couples the physical completion of replication with the regulatory release from the S-phase checkpoint.
There are a number of issues still to be resolved with respect to comparisons between different members of this gene family. First, in S. pombe, no connection between rad12+ and any topoisomerase has been established. Further, it has been reported that when the helicase activity of Saccharomyces cerevisiae SGS1 was specifically altered it did not function like the original sgs1 mutant in restoring normal growth in top3 mutant cells or causing slow growth in a top1 mutant background (20). This indicates that the helicase activity of Sgs1 is not essential for its interaction with the topoisomerase. Interestingly, we showed here that the rad12-502 mutant, which is defective in the helicase’s ATP binding domain, is still sensitive to HU, characteristic of the observed defect in existing from the S-phase checkpoint.
The comparisons between rad12+ and BLM are also unclear. rad12 mutants are clearly defective in existing from the S-phase checkpoint. However, it is unclear whether rad12+ is involved in regulation of the G2 checkpoint responding to DNA damage. However, this seems likely, since rad12 mutants are sensitive to UV light and γ radiation. By contrast, BLM mutant cells are not sensitive to DNA damage, and it is not known whether they are sensitive to inhibition of DNA synthesis. One potential downstream target of BLM, the human homolog of rad9+, HRAD9, complements the HU sensitivity of S. pombe rad9::ura4 cells efficiently, but the radiation sensitivity is complemented poorly or not at all (19). This may suggest that BLM is involved in regulating the S-phase checkpoint pathway only and not the G2 checkpoint pathway monitoring DNA damage.
One conclusion that can be made from this work is that a helicase is acting in an inhibitory manner toward checkpoint control. If single-stranded DNA intermediates of repair initiate the signal which leads to checkpoint arrest (21), one would expect helicases to act in general as positive regulators of checkpoint control. The loss of such a helicase might be expected to result in a checkpoint-defective phenotype, which is not the case for rad12-502 (1). Our data are more consistent with a model in which the DNA-protein complexes formed during replication or repair initiate the signal that leads to checkpoint arrest.
ACKNOWLEDGMENTS
S.D. is a Career Scientist of the Cancer Care Ontario (CCO), and this work was supported in part by CCO startup funds and MRCC grant MT-14352 to S.D. G.A.F. is supported by NIH grant CA72647. H.B.L. is supported in part by an NIH Research Career Development Award (CA68446) and by NIH grants GM52493 and CA73043 and American Cancer Society grant CN-106.
REFERENCES
- 1.al-Khodairy F, Carr A M. DNA repair mutants defining G2 checkpoint pathways in Schizosaccharomyces pombe. EMBO J. 1992;11:1343–1350. doi: 10.1002/j.1460-2075.1992.tb05179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.al-Khodairy F, Fotou E, Sheldrick K S, Griffiths D J, Lehmann A R, Carr A M. Identification and characterization of new elements involved in checkpoint and feedback controls in fission yeast. Mol Biol Cell. 1994;5:147–160. doi: 10.1091/mbc.5.2.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beach D, Rodgers L, Gould J. ran1+ controls the transition from mitotic division to meiosis in fission yeast. Curr Genet. 1985;10:297–311. doi: 10.1007/BF00365626. [DOI] [PubMed] [Google Scholar]
- 4.Bentley N J, Holtzman D A, Flaggs G, Keegan K S, DeMaggio A, Ford J C, Hoekstra M, Carr A M. The Schizosaccharomyces pombe rad3 checkpoint gene. EMBO J. 1996;15:6641–6651. [PMC free article] [PubMed] [Google Scholar]
- 5.Cimprich K A, Shin R B, Keith C T, Schreiber S L. cDNA cloning and gene mapping of a candidate human cell cycle checkpoint protein. Proc Natl Acad Sci USA. 1996;93:2850–2855. doi: 10.1073/pnas.93.7.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Darlington G J, Dutkowski R, Brown W T. Sister chromatid exchange frequencies in progeria and Werner syndrome patients. Am J Hum Genet. 1981;33:762–766. [PMC free article] [PubMed] [Google Scholar]
- 7.Davey S, Nass M L, Ferrer J V, Sidik K, Eisenberger A, Mitchell D L, Freyer G A. The fission yeast UVDR DNA repair pathway is inducible. Nucleic Acids Res. 1997;25:1002–1008. doi: 10.1093/nar/25.5.1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ellis N A, Groden J, Ye T Z, Straughen J, Lennon D J, Ciocci S, Proytcheva M, German J. The Bloom’s syndrome gene product is homologous to RecQ helicases. Cell. 1995;83:655–666. doi: 10.1016/0092-8674(95)90105-1. [DOI] [PubMed] [Google Scholar]
- 9.Enoch T, Carr A M, Nurse P. Fission yeast genes involved in coupling mitosis to completion of DNA replication. Genes Dev. 1992;6:2035–2046. doi: 10.1101/gad.6.11.2035. [DOI] [PubMed] [Google Scholar]
- 10.Freyer G A, Davey S, Ferrer J V, Martin A M, Beach D, Doetsch P W. An alternative eukaryotic DNA excision repair pathway. Mol Cell Biol. 1995;15:4572–4577. doi: 10.1128/mcb.15.8.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gangloff S, McDonald J P, Bendixen C, Arthur L, Rothstein R. The yeast type I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolog: a potential eukaryotic reverse gyrase. Mol Cell Biol. 1994;14:8391–8398. doi: 10.1128/mcb.14.12.8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.German J. Bloom’s syndrome. Dermatol Clin. 1995;13:7–18. [PubMed] [Google Scholar]
- 13.Gorbalenya A E, Koonin E V, Donchenko A P, Blinov V M. Two related superfamilies of putative helicases involved in replication, recombination, repair and expression of DNA and RNA genomes. Nucleic Acids Res. 1989;17:4713–4730. doi: 10.1093/nar/17.12.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hartwell L. Defects in a cell cycle checkpoint may be responsible for the genomic instability of cancer cells. Cell. 1992;71:543–546. doi: 10.1016/0092-8674(92)90586-2. [DOI] [PubMed] [Google Scholar]
- 15.Hawthorne D C, Leupold U. Suppressor mutations in yeast. Curr Top Microbiol Immunol. 1974;64:1–47. doi: 10.1007/978-3-642-65848-8_1. [DOI] [PubMed] [Google Scholar]
- 16.Hoheisel J D, Maier E, Mott R, McCarthy L, Grigoriev A V, Schalkwyk L C, Nizetic D, Francis F, Lehrach H. High resolution cosmid and P1 maps spanning the 14 Mb genome of the fission yeast S. pombe. Cell. 1993;73:109–120. doi: 10.1016/0092-8674(93)90164-l. [DOI] [PubMed] [Google Scholar]
- 17.Leupold U. Genetical methods for Schizosaccharomyces pombe. Methods Cell Physiol. 1970;4:169–177. [Google Scholar]
- 18.Lieberman H B, Hopkins K M, Laverty M, Chu H M. Molecular cloning and analysis of Schizosaccharomyces pombe rad9, a gene involved in DNA repair and mutagenesis. Mol Gen Genet. 1992;232:367–376. doi: 10.1007/BF00266239. [DOI] [PubMed] [Google Scholar]
- 19.Lieberman H B, Hopkins K M, Nass M, Demetrick D, Davey S. A human homolog of the Schizosaccharomyces pombe rad9+ checkpoint control gene. Proc Natl Acad Sci USA. 1996;93:13890–13895. doi: 10.1073/pnas.93.24.13890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu J, Mullen J R, Brill S J, Kleff S, Romeo A M, Sternglanz R. Human homologues of yeast helicase. Nature. 1996;383:678–679. doi: 10.1038/383678a0. [DOI] [PubMed] [Google Scholar]
- 21.Lydall D, Weinert T. Yeast checkpoint genes in DNA damage processing: implications for repair and arrest. Science. 1995;270:1488–1491. doi: 10.1126/science.270.5241.1488. [DOI] [PubMed] [Google Scholar]
- 22.Mizukami T, Chang W I, Garkavtsev I, Kaplan N, Lombardi D, Matsumoto T, Niwa O, Kounosu A, Yanagida M, Marr T G, Beach D. A 13 kb resolution cosmid map of the 14 Mb fission yeast genome by nonrandom sequence-tagged site mapping. Cell. 1993;73:121–132. doi: 10.1016/0092-8674(93)90165-m. [DOI] [PubMed] [Google Scholar]
- 23.Murray J M, Lindsay H D, Munday C A, Carr A M. Role of Schizosaccharomyces pombe RecQ homolog, recombination, and checkpoint genes in UV damage tolerance. Mol Cell Biol. 1997;17:6868–6875. doi: 10.1128/mcb.17.12.6868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nasim A, Smith B P. Genetic control of radiation sensitivity in Schizosaccharomyces pombe. Genetics. 1975;79:573–582. doi: 10.1093/genetics/79.4.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rosen K M, Lamperti E D, Villa-Komaroff L. Optimizing the Northern blot procedure. BioTechniques. 1990;8:398–402. [PubMed] [Google Scholar]
- 26.Rothstein R, Gangloff S. Hyper-recombination and Bloom’s syndrome: microbes again provide clues about cancer. Genome Res. 1996;5:258–262. doi: 10.1101/gr.5.5.421. [DOI] [PubMed] [Google Scholar]
- 27.Rowley R, Subramani S, Young P G. Checkpoint controls in Schizosaccharomyces pombe: rad1. EMBO J. 1992;11:1335–1342. doi: 10.1002/j.1460-2075.1992.tb05178.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salk D, Bryant E, Au K, Hoehn H, Martin G M. Systematic growth studies, cocultivation, and cell hybridization studies of Werner syndrome cultured skin fibroblasts. Hum Genet. 1981;58:310–316. doi: 10.1007/BF00294930. [DOI] [PubMed] [Google Scholar]
- 29.Sinclair D A, Guarente L. Extrachromosomal rDNA circles—a cause of aging in yeast. Cell. 1997;91:1033–1042. doi: 10.1016/s0092-8674(00)80493-6. [DOI] [PubMed] [Google Scholar]
- 30.Sinclair D A, Mills K, Guarente L. Accelerated aging and nucleolar fragmentation in yeast sgs1 mutants. Science. 1997;277:1313–1316. doi: 10.1126/science.277.5330.1313. [DOI] [PubMed] [Google Scholar]
- 31.Smith M L, Fornace A J., Jr Genomic instability and the role of p53 mutations in cancer cells. Curr Opin Oncol. 1995;7:69–75. [PubMed] [Google Scholar]
- 32.Stewart E, Chapman C R, al-Khodairy F, Carr A M, Enoch T. rqh1+, a fission yeast gene related to the Bloom’s and Werner’s syndrome genes, is required for reversible S phase arrest. EMBO J. 1997;16:2682–2692. doi: 10.1093/emboj/16.10.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tlsty T D, Briot A, Gualberto A, Hall I, Hess S, Hixon M, Kuppuswamy D, Romanov S, Sage M, White A. Genomic instability and cancer. Mutat Res. 1995;337:1–7. doi: 10.1016/0921-8777(95)00016-d. [DOI] [PubMed] [Google Scholar]
- 34.Umezu K, Nakayama K, Nakayama H. Escherichia coli RecQ protein is a DNA helicase. Proc Natl Acad Sci USA. 1990;87:5363–5367. doi: 10.1073/pnas.87.14.5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Watt P M, Hickson I D, Borts R H, Louis E J. SGS1, a homologue of the Bloom’s and Werner’s syndrome genes, is required for maintenance of genome stability in Saccharomyces cerevisiae. Genetics. 1996;144:935–945. doi: 10.1093/genetics/144.3.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Watt P M, Louis E J, Borts R H, Hickson I D. Sgs1: a eukaryotic homolog of E. coli RecQ that interacts with topoisomerase II in vivo and is required for faithful chromosome segregation. Cell. 1995;81:253–260. doi: 10.1016/0092-8674(95)90335-6. [DOI] [PubMed] [Google Scholar]
- 37.Yu C E, Oshima J, Fu Y H, Wijsman E M, Hisama F, Alisch R, Matthews S, Nakura J, Miki T, Ouais S, Martin G M, Mulligan J, Schellenberg G D. Positional cloning of the Werner’s syndrome gene. Science. 1996;272:258–262. doi: 10.1126/science.272.5259.258. [DOI] [PubMed] [Google Scholar]