Abstract
Unraveling the transport of drugs and nanocarriers in cerebrovascular networks is important for pharmacokinetic and hemodynamic studies but is challenging due to the complexity of sensing individual particles within the circulatory system of a live animal. Here, we demonstrate that a DNA-stabilized silver nanocluster (DNA-Ag16NC) that emits in the first near-infrared window (NIR-I) upon two-photon excitation in the second NIR window (NIR-II) can be used for multi-photon in vivo fluorescence correlation spectroscopy (FCS) for the measurement of cerebral blood flow rates in live mice with high spatial and temporal resolution. To ensure bright and stable emission during in vivo experiments, we loaded DNA-Ag16NCs into liposomes which served the dual purposes of concentrating the fluorescent label and protecting it from degradation. DNA-Ag16NC-loaded liposomes enabled the quantification of cerebral blood flow velocity within individual vessels of a living mouse.
Keywords: DNA-AgNCs, liposomes, cerebral blood flow (CBF), FCS, in vivo
Graphical Abstract
Cerebrovascular networks, and their coupling with the central nervous system, regulate ion exchange, signaling molecule transport, nutrient delivery, oxygen consumption, and waste removal in the mammalian brain, which ensures the brain’s normal function.1, 2 Cerebrovascular diseases, caused by hemorrhage and ischemia, are often related to cerebrovascular malformation and hemodynamic interruptions.3, 4 A variety of imaging techniques have been developed to characterize cerebrovascular structures, including magnetic resonance imaging,5 computed tomography,6 and fluorescence imaging.7–10 However, studying in vivo hemodynamic processes inside cerebrovascular systems, especially in capillaries (<10 μm), remains challenging and is limited by the need for spatial resolution beyond the capability of existing techniques.11–14 The cerebrovascular network also serves as a critical route for drug delivery.15 Thus, monitoring nanocarriers in real-time would provide essential information for pharmacokinetic and hemodynamic studies. While several techniques enable the static visualization of nanocarriers after delivery using optical imaging of regions of interest such as tumors, organs, or injury sites,16–21 the capability to track cargo in real-time, while in the circulatory system, would provide additional insight into drug delivery and blood flow dynamics.
We recently demonstrated the ability to achieve the necessary spatial and temporal resolution using in vivo multi-photon fluorescence correlation spectroscopy (FCS) for measuring flow rates within individual capillaries in the central nervous system (CNS) of live mice.22 In FCS, intensity fluctuations of single molecules are recorded as they move in and out of the confocal volume, which gives rise to temporal correlations which can reveal characteristics such as the rates of diffusion or flow rates. By measuring the intensity fluctuations of intravenously injected nanoparticles, we were able to reach sub-capillary resolution and produce 2D cross-sectional flow velocity profiles. In our previous implementation, we used CF488-Dextran with multi-photon excitation at 920 nm and emission detected at 515 nm. However, as scattering and autofluorescence from tissue limits the penetration depth and accuracy of the FCS results,23–25 there is a need to extend both excitation and emission wavelengths further into the near-infrared (NIR), where these effects are diminished.26 DNA-stabilized silver nanoclusters (DNA-AgNCs) are a class of emitters that have been reported to have NIR emission and high two-photon absorption cross-sections, though there are only relatively few published studies concerning their multi-photon cross-sections.27, 28
DNA-AgNCs are composed of a limited number of silver atoms and cations wrapped in one or more DNA strands, and their optical properties can be tuned by varying the DNA sequence.29, 30 In addition to the tunability of DNA-AgNCs, many species possess high photostability, high quantum yield,31 and low toxicity.32 While many DNA-AgNCs are reported to be bright and have high quantum yields, it is challenging to achieve single-molecule sensitivity and chemical stability in dense and biologically complex environments within the vasculature.33 An approach for both enhancing the overall emission intensity and protecting fluorophores from degradation is to up-concentrate them in nanoparticles. Liposomes have emerged as promising candidates for the encapsulation of drugs and fluorescent indicators which has laid the foundation for several classes of drug delivery systems.34, 35 The loading of DNA-AgNCs into nanocarriers could serve as a platform for integrating these emitters into biological applications. Liposome self-assembly in aqueous solutions combined with their structure comprised of an internal aqueous phase surrounded by a lipid bilayer make liposomes well suited to host DNA-AgNCs.36 Since DNA-AgNCs have a negatively charged phosphate backbone, the use of cationic liposomes should enhance the encapsulation efficiency due to electrostatic interactions.37 Encapsulating DNA-AgNCs into cationic liposomes could simultaneously protect them from degradation, while in the circulatory system, and make them brighter compared to individual DNA-AgNCs. Combining NIR-emitting DNA-AgNC-loaded38–40 liposomes and multiphoton FCS for monitoring nanocarriers in the cerebral vasculature in vivo has the potential to promote applications for improved pharmacokinetic and hemodynamic studies.
In this study, structurally stable DNA-Ag16NCs,41 with emission in the first NIR region (NIR-I) and a two-photon absorption band in the second NIR window (NIR-II), were loaded into liposomes for the quantification of cerebral blood flow rates in live mice (Scheme 1). The loading of DNA-Ag16NCs (1~2 nm) into liposomes (~100 nm) did not affect their photophysical properties and provided bright nanoparticles that could be detected by FCS. As both excitation and emission are in the near-infrared (NIR-II excitation and NIR-I emission), there is less scattering and autofluorescence due to light-tissue interactions during in vivo experiments as compared to fluorophores that emit in the visible region. In order to optimize DNA-AgNC-loaded liposomes for in vivo studies, we performed a series of calibration measurements with known volumetric flow rates in a microfluidic device. As a second green emissive dye is used for imaging the cerebrovascular network, we optimized the excitation conditions to limit crosstalk between the spectral channels. This allowed us to simultaneously visualize the cerebrovascular network and correlate the emission of the spectrally distinct DNA-Ag16NC-loaded liposomes for mapping blood flow velocities by two-photon imaging and FCS, respectively.
Scheme 1.
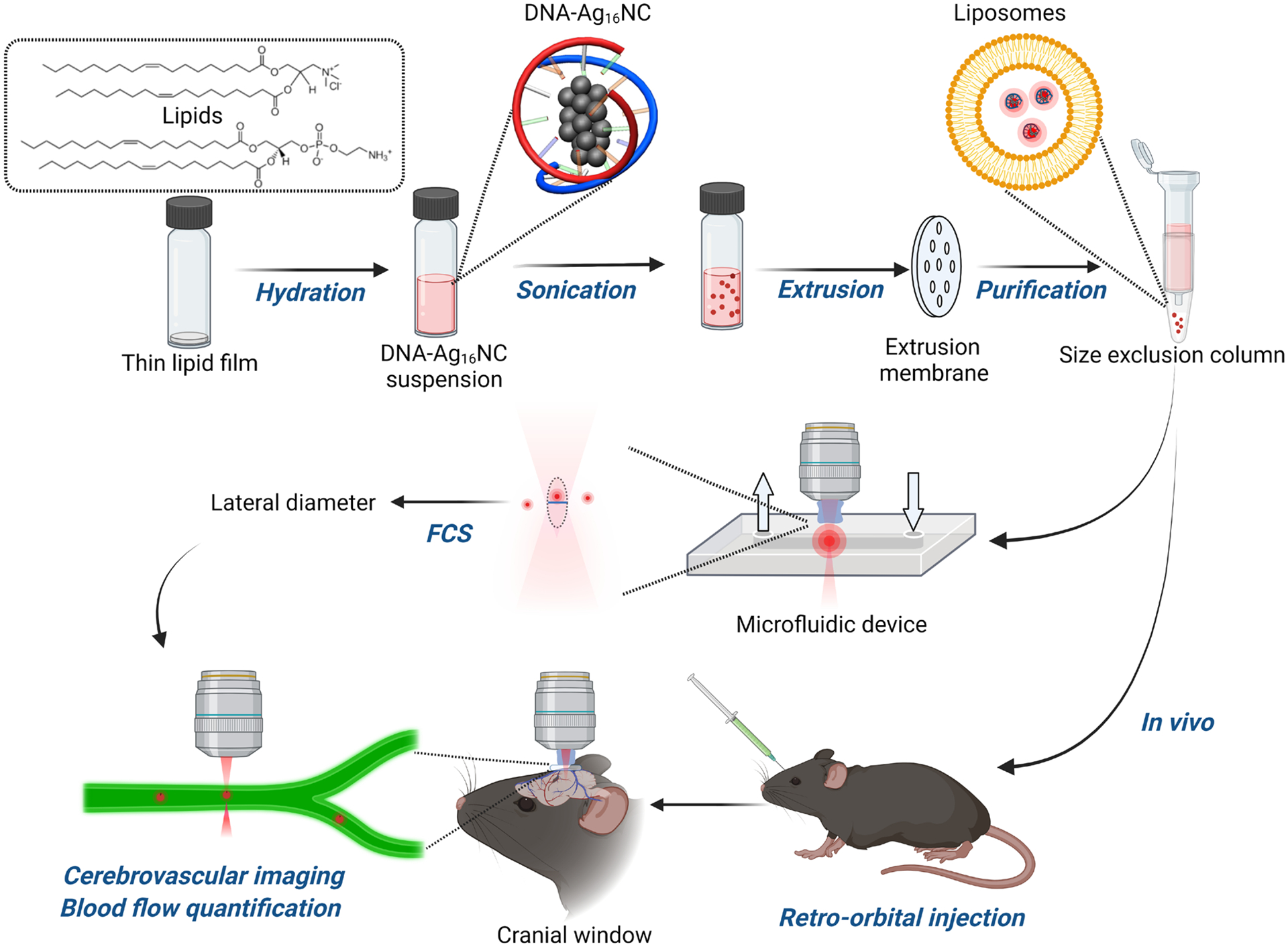
Illustration of the approach presented in this article for loading DNA-Ag16NCs into liposomes and measuring them in a flow chamber and the cerebrovascular network of a living mouse.
RESULTS AND DISCUSSION
Optical properties of DNA-Ag16NCs.
The one- and two-photon excitation properties of DNA-Ag16NCs were characterized with a spectrophotometer, fluorometer, and two-photon confocal laser scanning microscope. The synthesized DNA-Ag16NCs have an absorption peak at 525 nm and a NIR-I fluorescence maximum at around 735 nm (Figure 1a).42 The UV absorption peak around 260 nm is attributed to the DNA bases rather than AgNCs.43 Some DNA-AgNCs have been reported to show anti-Stokes upconversion fluorescence (UCF), which has been used for background-free imaging.44 The DNA-Ag16NC used in this article has been reported to show negligible UCF.45 Nevertheless, we investigated the response of DNA-Ag16NCs in 10 mM ammonium acetate (NH4OAc) when excited by an ultrafast laser in the NIR wavelength range from 760 nm to 1100 nm (Figure 1b). Two excitation bands can be observed, one in the NIR-II window at 1045 nm, corresponding to the main absorption, and the edge of an additional band at around 760 nm. The ammonium acetate solution by itself showed no features under similar excitation conditions (Figure S1). For the subsequent in vivo studies, dextran-conjugated FITC (FITC-Dextran 70 kDa) was chosen for visualizing the cerebrovascular network. To ensure a proper choice of excitation and emission wavelengths, FITC-Dextran was characterized under ultrafast laser excitation. FITC-Dextran also showed two excitation bands, which were both confined to the NIR-I region. In a previous study, we used CF488-Dextran for in vivo FCS measurements, and an equivalent characterization yielded very similar results as to FITC-Dextran (Figure S2).
Figure 1.
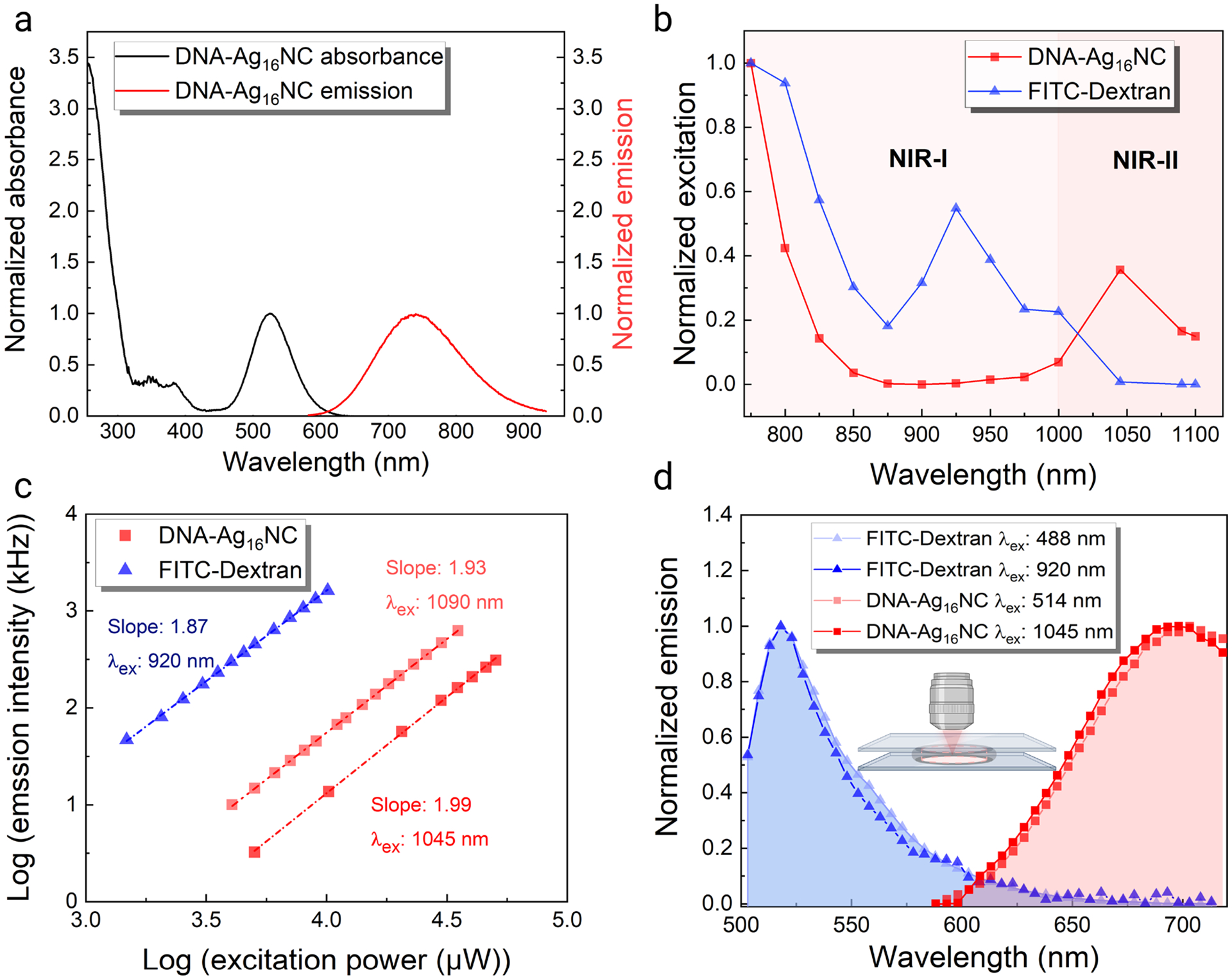
One- and two-photon optical properties of DNA-Ag16NCs and FITC-Dextran 70 kDa. (a) Normalized UV-Vis absorption and fluorescence spectra (λex = 520 nm) of DNA-Ag16NCs. Spectra have been adapted from Cerretani et al.42 Adapted with permission from (Cerretani et al., 2021). Copyright 2021 Tom Vosch. (b) Normalized two-photon excitation spectra of 12.5 μM DNA-Ag16NC and 6.3 μM FITC-Dextran with emission collected in the red/NIR I channel (590–740 nm) and green channel (525–560 nm), respectively. The power was kept constant (3 mW for DNA-Ag16NC and 5 mW for FITC-Dextran) at every wavelength. Note that intensities can differ due to the wavelength-dependent diffraction limited spot size. (c) Excitation intensity dependence of 12.5 μM DNA-Ag16NC and 6.3 μM FITC-Dextran. Note that every DNA-Ag16NC contains two DNA strands, and the concentration here refers to the concentrations of DNA-Ag16NC particles and dextran conjugates (FITC:Glucose = 1:250). (d) Normalized emission spectra of 12.5 μM DNA-Ag16NCs and 6.3 μM FITC-Dextran under one- and two-photon excitation. Note that the minor discrepancy between the emission spectra of DNA-Ag16NCs in (a) and (d) can be attributed to limited sensitivity of the two-photon confocal setup detectors at longer wavelengths.
To determine if the multiphoton excitation bands of DNA-Ag16NCs and FITC-Dextran arise from UCF or 2-photon excitation, we measured the excitation intensity dependence for each fluorophore. For a two-photon process, the fluorescence intensity is expected to increase quadratically with excitation intensity, which yields a slope of two when presented in a log-log plot.28, 46 The slope of FITC-Dextran when excited at 920 nm is 1.87 (Figure 1c), which slightly deviates from the theoretical value of 2.00 but still indicates an overall two-photon absorption process. The DNA-Ag16NCs had a slope of 1.99 at 1045 nm excitation and a 1.93 at 1090 nm (Figure 1c). At 800 nm, the slope dropped to 1.76 (Figure S3), which likely indicates the combination of a two-photon and a more linear dependent UCF process.47 Figure 1d shows that the emission spectra under one- and two-photon excitation overlap, indicating that the same emissive state is populated in both cases. For our chosen pair of DNA-Ag16NCs and FITC-Dextran, minimal spectral crosstalk is ensured by the proper choice of excitation and emission wavelengths (1045/1090 nm excitation with detection in the red/NIR I channel for DNA-Ag16NCs and 920 nm excitation with detection in the green channel for FITC-Dextran).
Characterization of DNA-Ag16NC-loaded liposomes.
To increase the brightness and chemical stability of DNA-Ag16NCs in the circulatory system, we loaded them into cationic liposomes (see Experimental Section). We prepared DNA-Ag16NCs-loaded liposomes by extrusion through a polycarbonate membrane with a 100 nm pore size. Successful encapsulation was visually confirmed by imaging the prepared liposomes using widefield fluorescence microscopy (Figure 2a). Additionally, TIRF experiments were performed, where a lipophilic dye, DiD, was used to label the membrane of the liposomes loaded with DNA-Ag16NCs. By exciting at 561 nm (specific to DNA-Ag16NCs) and 640 nm (specific to DiD), significant colocalization was observed, indicating efficient loading of the liposomes with DNA-Ag16NCs (Figure S4).
Figure 2.
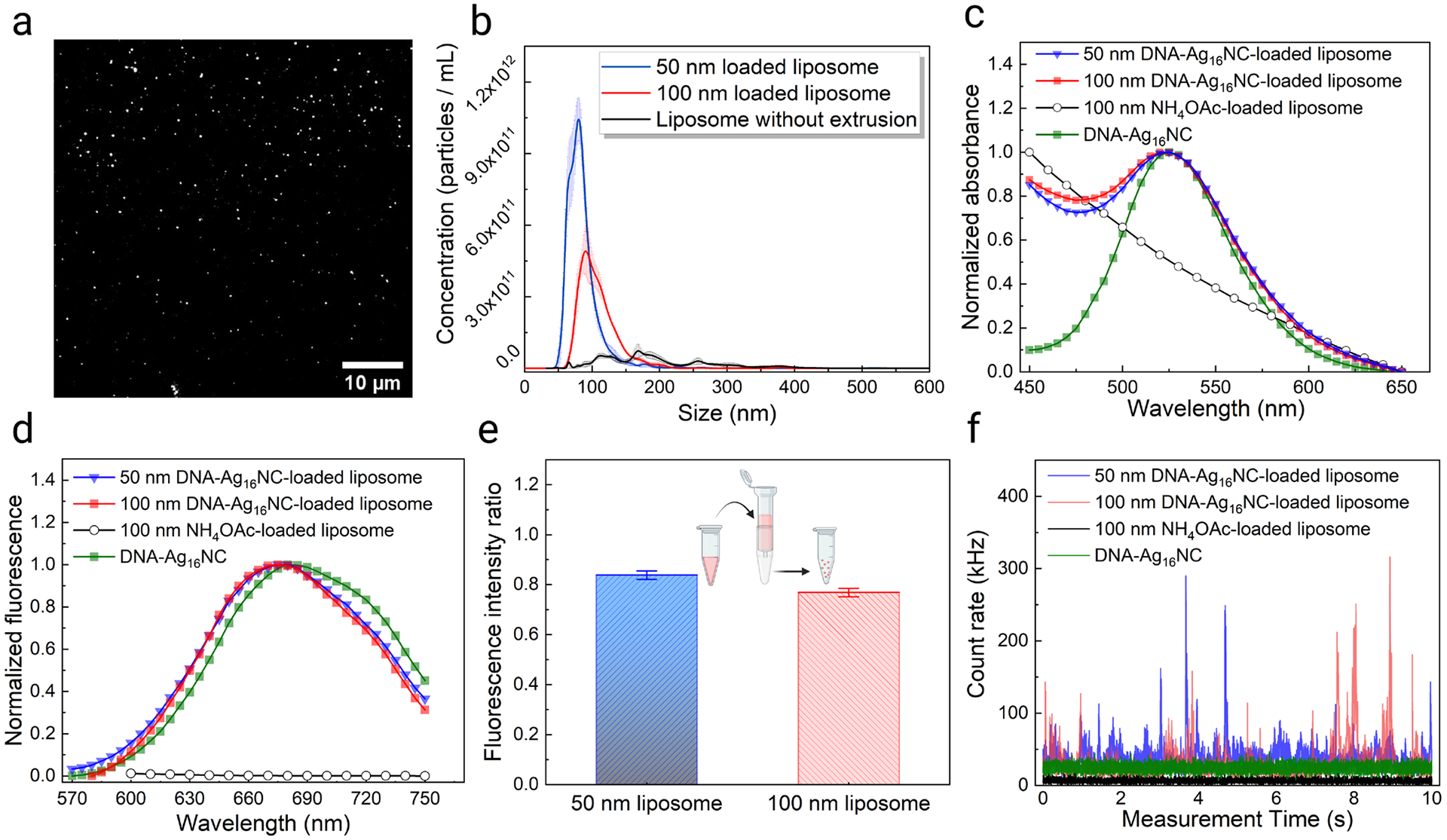
Characterization of liposomes loaded with DNA-Ag16NCs. (a) Fluorescence image of 100 nm liposomes loaded with 12.5 μM DNA-Ag16NCs. (b) Size distribution and concentration of synthesized liposomes. The DNA-Ag16NC concentration was kept at 12.5 μM in all three cases. (c) Normalized absorption and (d) fluorescence spectra of DNA-Ag16NC-loaded liposomes of different sizes. (e) Fluorescence intensity ratio before and after the size exclusion column purification of 50 nm and 100 nm loaded liposomes from free DNA-Ag16NCs. The fluorescence intensity ratios were calculated using emission at 680 nm, before and after purification. The error bar represents the standard deviation of measured ratios at 680±10 nm. (f) Emission intensity fluctuations of a 50 nM DNA-Ag16NCs solution, NH4OAc-loaded liposomes, and DNA-Ag16NC-loaded liposomes under 1045 nm two-photon excitation. Emission was collected in the red/NIR I channel (590–740 nm).
Nanoparticle tracking analysis (NTA) was used to evaluate the size distribution and concentration of the prepared liposomes extruded through different size membranes (Figure 2b). The accuracy of the NTA system was first confirmed using 100 nm standard beads, which gave a mean size of 97.4 nm ± 0.6 nm (Figure S5). Then, the size of the liposomes with and without extrusion was evaluated. The mean size of liposomes without extrusion was 194.2 nm with a standard deviation of 81.7 nm (Table S1), indicating a non-uniform distribution. Extrusion resulted in a decrease of liposome size, a more uniform distribution, and an increase in liposome concentration (Figure 2b and Table S1).
The absorption and fluorescence spectra of the encapsulated liposomes were measured to exclude the effect of close packing within the lipid carriers on the optical properties of DNA-Ag16NCs. We found that the primary absorption peak for DNA-Ag16NCs was still present near 520 nm independent of the vesicle size (Figure 2c). We loaded liposomes at higher concentrations and observed no change in the location of the absorption peak (Figure S6a). However, due to light scattering by the liposomes,48 this absorption feature was present on top of a steep slope. The origin of the steep slope was confirmed by measuring the absorption spectrum of 100 nm liposomes loaded only buffer (10 mM NH4OAc). DNA-Ag16NCs encapsulated in liposomes show similar fluorescence properties under 520 nm excitation as free DNA-Ag16NCs in solution (Figure 2d and Figure 1a). The emission spectrum of 10 mM NH4OAc-loaded liposomes showed no significant emission between 600 nm and 750 nm. Liposomes were purified by a size exclusion column to remove free DNA-Ag16NCs prior to characterization. To obtain an estimate of how well DNA-Ag16NCs are encapsulated in liposomes, the fluorescence intensity ratios before and after the size exclusion column purification were compared for 100 nm and 50 nm liposomes (Figure 2e). The average fluorescence intensity ratios (Post column/pre column) calculated at 680 nm were 0.77 and 0.84 for 100 nm and 50 nm DNA-Ag16NC-loaded liposomes, respectively. A higher loading concentration of DNA-Ag16NCs did not result in a significant change in this ratio for the same liposome size (Figure S6b).
DNA-Ag16NC-loaded liposomes were then characterized under two-photon 1045 nm excitation to evaluate their performance in FCS measurements. While free DNA-Ag16NCs at FCS level concentrations (50 nM) were too dim to be observed at the single molecule level49 (as characterized by the low intensity trace in Figure 2f that lacks any observable spikes), large intensity fluctuations (Figure 2f) could be seen for the DNA-Ag16NC-loaded liposomes indicating loaded liposomes diffusing in and out of the focal volume. Larger liposomes and higher DNA-Ag16NCs loading concentrations led to stronger fluorescence intensities (Figure 2f and Figure S6c). The corresponding autocorrelation functions reveal slow diffusion (Figure S7) around 100 ms and 54 ms for the 100 nm and 50 nm liposomes, respectively (Table S2). The average number of liposomes in the confocal volume, obtained by fitting the autocorrelation function, was consistent with that measured with NTA where the 50 nm liposome had a ~1.6 fold higher concentration. Thus, by loading DNA-Ag16NCs into liposomes, we have shown that they retain their optical properties while becoming sufficiently bright to be detected with FCS.
Characterization of the loaded liposomes in the flow chamber.
Before we utilized DNA-Ag16NC-loaded liposomes in vivo, we performed additional characterization in a microfluidic flow chamber with known flow rates to optimize the experimental parameters and demonstrate the feasibility of the approach. The channel dimensions were 50 mm×5 mm×0.2 mm, and flow rates inside were controlled via a programmable syringe pump (Figure 3a). First, the 10 mM ammonium acetate solution was tested inside the chamber under 1045 nm excitation and a 0.1 mL/min flow rate. Low counts, equivalent to background levels, were observed for this control solution (Figure 3b). Liposomes with different sizes (50 nm and 100 nm) and DNA-Ag16NC concentrations (12.5 μM and 37.5 μM) were prepared and introduced into the flow chamber under the same conditions. The concentrations are estimated based on DNA absorption, thus the reported concentrations reflect the concentration of DNA-Ag16NCs at the liposome synthesis stage.41 Based on the stronger fluorescence intensity spikes, it appears that larger liposomes and higher DNA-Ag16NC concentrations allowed more DNA-AgNCs to be encapsulated into single liposomes (Figure 3c). By fitting the corresponding autocorrelation functions (Figure 3d), we obtained the residence time, particle number, and intensity in counts per particle of each synthesized liposome. We found that 100 nm liposomes, encapsulating 37.5 μM DNA-Ag16NCs, had the highest fluorescence intensity per particle of 172,000 counts per second, while the 50 nm liposome with 12.5 μM DNA-Ag16NCs had the lowest intensity of 45,000 counts per second. It should be noted that the residence time of liposomes in the laser focal volume, regardless of diameter, was similar during solution flow (between 467 μs and 485 μs) (Table S3), as opposed to the differences seen without solution flow when the molecules were simply diffusing (Table S2). This indicates that flow dominated the movement of these liposomes, and that liposome size and free diffusion became irrelevant in calculating the waist size of the excitation volume and flow rates. To verify that the concentration of liposomes does not alter the observed flow properties during FCS measurements, we performed a set of studies at different liposome concentrations. The observed flow rate and residence time remained consistent regardless of the vesicle concentration (i.e., average number of vesicles in the focal volume) (Figure S8).
Figure 3.
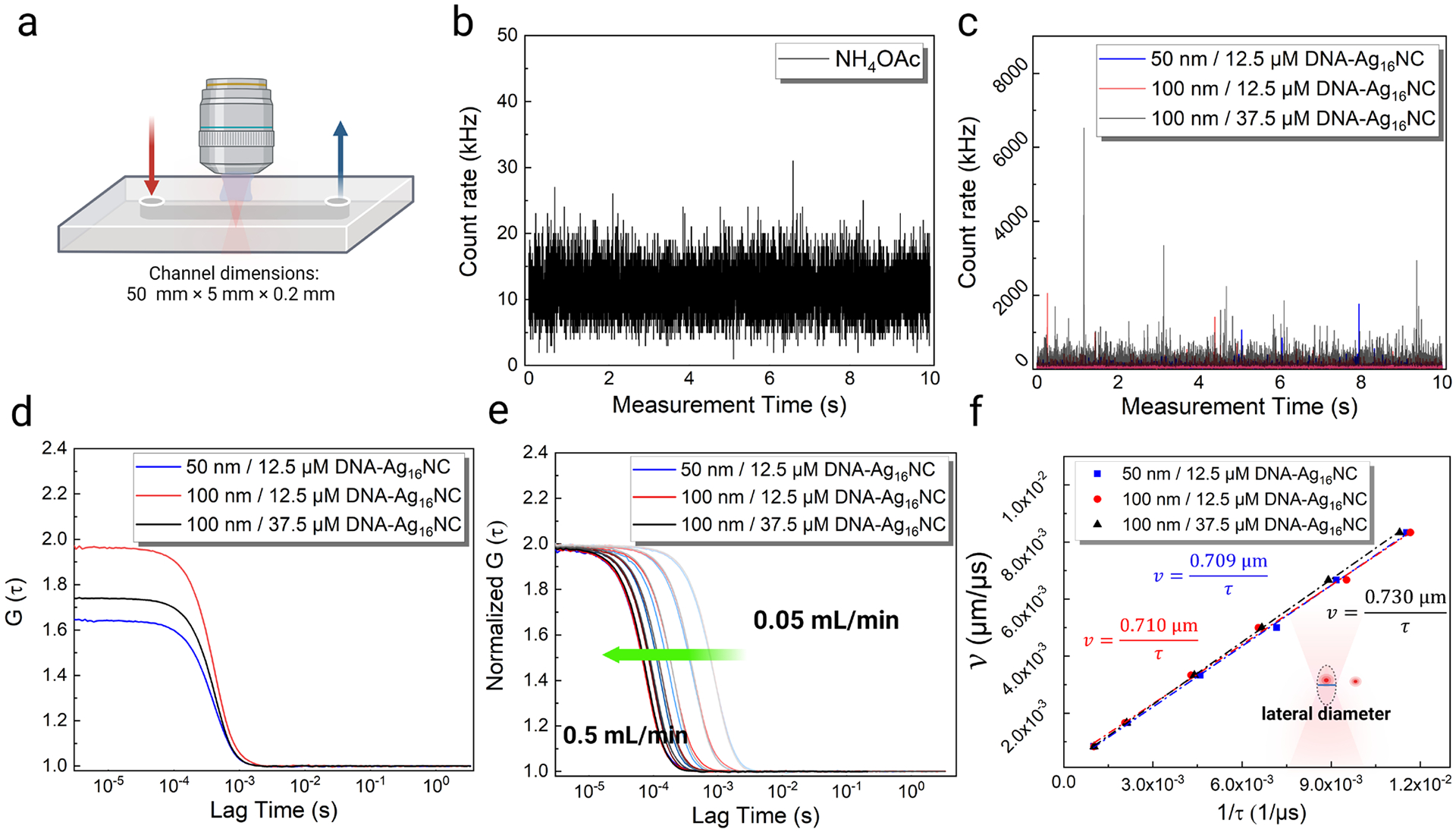
Characterization of the liposomes loaded with DNA-Ag16NCs in the microfluidic flow chamber. (a) Schematic showing the flow chamber dimensions and flow direction. Emission intensity fluctuations of (b) 10 mM NH4OAc and (c) liposomes loaded with DNA-Ag16NCs in the flow chamber under 1045 nm excitation and 0.1 mL/min flow rate. Note that only a 10 s section of a 100 s long time trace is shown here. (d) The corresponding autocorrelation functions of liposomes loaded with DNA-Ag16NCs. (e) Normalized autocorrelation functions of the DNA-Ag16NC-loaded liposomes under different flow rates (0.05 mL/min to 0.5 mL/min from right to left, as the green arrow shows). (f) The proportional fitting curves of the liposome flow velocity as a function of 1/residence time.
Interestingly, we note a significant increase in the residence time for these liposomes when the excitation power was increased from 5 mW to 25 mW under the same flow rate of 0.2 mL/min, regardless of the liposome size and DNA-Ag16NC concentration (Figure S9, Table S4). This is consistent with previous reports50 of optical trapping, where higher excitation intensities lead to more retardation.51, 52 Moreover, our three types of liposomes were subjected to flow rates in the range of 0.05–0.50 mL/min, and intensity traces under 1045 nm (11.7 mW) excitation were recorded (Figure S10–S12). The corresponding autocorrelation functions show a gradual shortening of the residence time, as the flow rate is increased (Figure 3e). For all the investigated flow rates, the residence time between the three liposome types is similar, as seen from the fitted values and from the overlap of the autocorrelation functions (Table S5 and Figure 3e). By fitting the flow velocity (v) as a function of 1/residence time (1/τf), a nearly constant lateral diameter (d) was obtained for the three types of liposomes (Figure 3f), which eliminated the influence of liposome size and DNA-Ag16NCs loading concentration on the acquired FCS residence time.
Excitation wavelength optimization and lateral diameter calibration in the flow chamber.
For in vivo experiments, it was essential to ensure minimal interactions between DNA-Ag16NC-loaded liposomes and FITC-Dextran (used for visualizing the blood vessels) as well as to ensure a proper choice of excitation wavelengths for minimizing spectral crosstalk between the two emitters. We utilized 70 kDa molecular weight FITC-labeled Dextran, which was large enough to constrain it from escaping through the blood brain barrier. Before conducting in vivo experiments, we optimized the excitation and emission conditions in the microfluidic flow chamber with both DNA-Ag16NC-loaded liposomes and FITC-Dextran (Figure 4a). As shown in Figures 1b and 1d, it is possible to selectively excite and detect the emission of DNA-Ag16NCs or FITC-Dextran, by a proper choice of wavelengths (Figure S13). Accordingly, exciting FITC-Dextran or 100 nm liposomes loaded with 10 mM NH4OAc at 1045 nm (11.7 mW and 0.1 mL/min flow) does not yield any significant intensity fluctuations in our red/NIR I emission channel (590–740nm; Figure 4b). But the mixture of FITC-Dextran and NH4OAc-loaded liposomes led to a small amount of undesired intensity spikes, which could affect subsequent FCS measurements with DNA-Ag16NCs. The origin of the spikes upon mixing FITC-Dextran and 10 mM NH4OAc-loaded 100 nm liposomes (i.e., not loaded with DNA-Ag16NCs) is not known and could be due to FITC-Dextran aggregation. However, as shown below, 1045 nm excitation led to non-specific emission signals during in vivo imaging as well. To overcome these problems, we used an excitation wavelength of 1090 nm which eliminated the non-specific intensity fluctuations. Despite the lower excitation efficiency of DNA-Ag16NCs at 1090 nm (Figure 1b), the choice of this longer wavelength was preferable, as it remedied the issues experienced with FITC-Dextran and liposomes (Figure 4c) and performed better during in vivo experiments. As determined from the power dependence studies, excitation at 1090 nm still proceeds as a two-photon absorption process (Figure 1c). Since high concentrations of FITC-Dextran 70 kDa (~1 μM) will be used for visualizing the vessels, we introduced a mixture of 100 nm liposomes loaded with 37.5 μM DNA-Ag16NCs and 1 μM FITC-Dextran into the flow chamber (set to a flow rate of 0.1 mL/min). As shown in Figure 4d, the mixture gave large emission intensity fluctuations in the red/NIR I channel and negligible intensity spikes in the green emission channel (525–560 nm) under 1090 nm excitation (12 mW). This enabled us to generate autocorrelation functions for DNA-Ag16NC-loaded liposomes in the flow chamber without interference from background fluctuations that were observed at 1045 nm excitation (Figure 4e). Furthermore, upon 1090 nm excitation, the derived residence time and number of particles from the mixture of FITC-Dextran and DNA-Ag16NC-loaded liposomes (446 μs and 1.01) were nearly the same as DNA-Ag16NC-loaded liposomes alone (453 μs and 1.07) under the same conditions (Table S6). Meanwhile, the measured residence time and particle numbers of the mixture when using 1045 nm excitation (523 μs and 1.99) deviated significantly from liposomes measured in the absence of FITC-Dextran (472 μs and 1.34) under 1045 nm excitation (Figure 3d, Table S3 and Table S6). Hence, 1090 nm excitation was used for all subsequent experiments.
Figure 4.
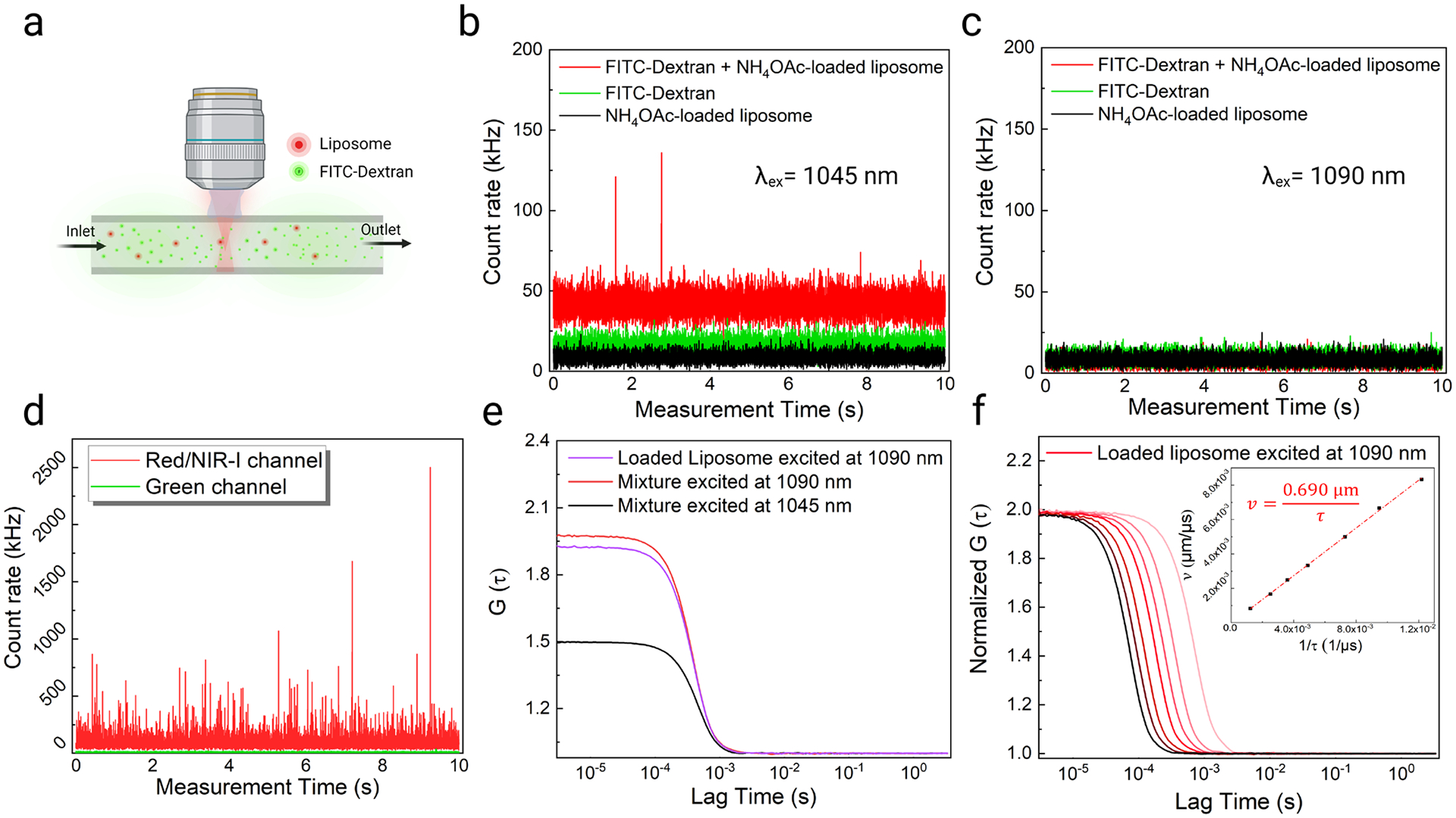
Excitation wavelength optimization and lateral diameter calibration in the flow chamber. (a) Schematic showing the mixture of liposomes and FITC-Dextran in the flow chamber. Emission intensity fluctuations of 1 μM FITC-Dextran, 10 mM NH4OAc-loaded liposomes, and their mixture in the flow chamber under (b) 1045 nm and (c) 1090 nm excitation at a flow rate of 0.1 mL/min. Emission was collected from 590 nm to 740 nm (red/NIR I channel). (d) Emission intensity fluctuations (in the red/NIR I and green channels) of a mixture of 1 μM FITC-Dextran and 37.5 μM DNA-Ag16NC-loaded liposomes in the flow chamber under 1090 nm excitation at a flow rate of 0.1 mL/min. (e) Autocorrelation functions of DNA-Ag16NC-loaded liposomes and their mixture with FITC-Dextran under 0.1 mL/min flow at different excitation wavelengths. (f) Normalized autocorrelation functions of the DNA-Ag16NC-loaded liposome under 1090 nm excitation at different flow rates. The inset figure shows the proportional fitting curve of liposome flow velocity and 1/residence time.
As liposome size did not alter residence time during flow, 100 nm liposomes were selected due to their larger encapsulation capability and higher emission intensity (Figure 3c). 100 nm liposomes encapsulated with 12.5 μM DNA-Ag16NCs showed clear single-particle emission fluctuations under 1090 nm excitation (Figure S14). Subsequently, 100 nm liposomes were used to encapsulate DNA-Ag16NCs (12.5 μM) for in vivo blood flow measurements. As demonstrated in Figure 3f, it is necessary to calibrate the confocal lateral diameter for a given excitation wavelength and emitter. We show in Figure 4f a similar calibration procedure for 1090 nm excitation, which yields a lateral diameter (d) of 0.690 μm. This derived lateral diameter is then used to calculate flow velocities from extracted residence time (τf) during in vivo FCS measurements within the cerebrovascular network (Equation 5 in Experimental Section).
In vivo monitoring of liposomes and cerebrovascular blood flow velocity measurements.
Prior to conducting FCS measurements in vivo, we performed a series of control experiments without FITC-Dextran or DNA-Ag16NC-loaded liposomes to assess the background levels expected under NIR-II excitation for unlabeled brains. For this, we prepared a skull-opened cranial window in a live mouse (Figure S15, see Experimental Section for details).22 In animals that had not been injected with a fluorescent label, we visualized blood vessels through the cranial window using 920 nm excitation, where the vessel wall appeared as a shadow within the brain’s autofluorescence in the green emission channel (Figure 5a). The emission intensity in the red/NIR channel was monitored inside the vasculature during 1090 nm (Figure 5b) or 1045 nm excitation (Figure S16a, b) to determine the level of interference from autofluorescence. Excitation at 1090 nm resulted in much less background than excitation at 1045 nm within individual vessels. As mentioned previously, this further supported our choice of 1090 nm excitation for DNA-Ag16NC-loaded liposomes for in vivo measurements.
Figure 5.
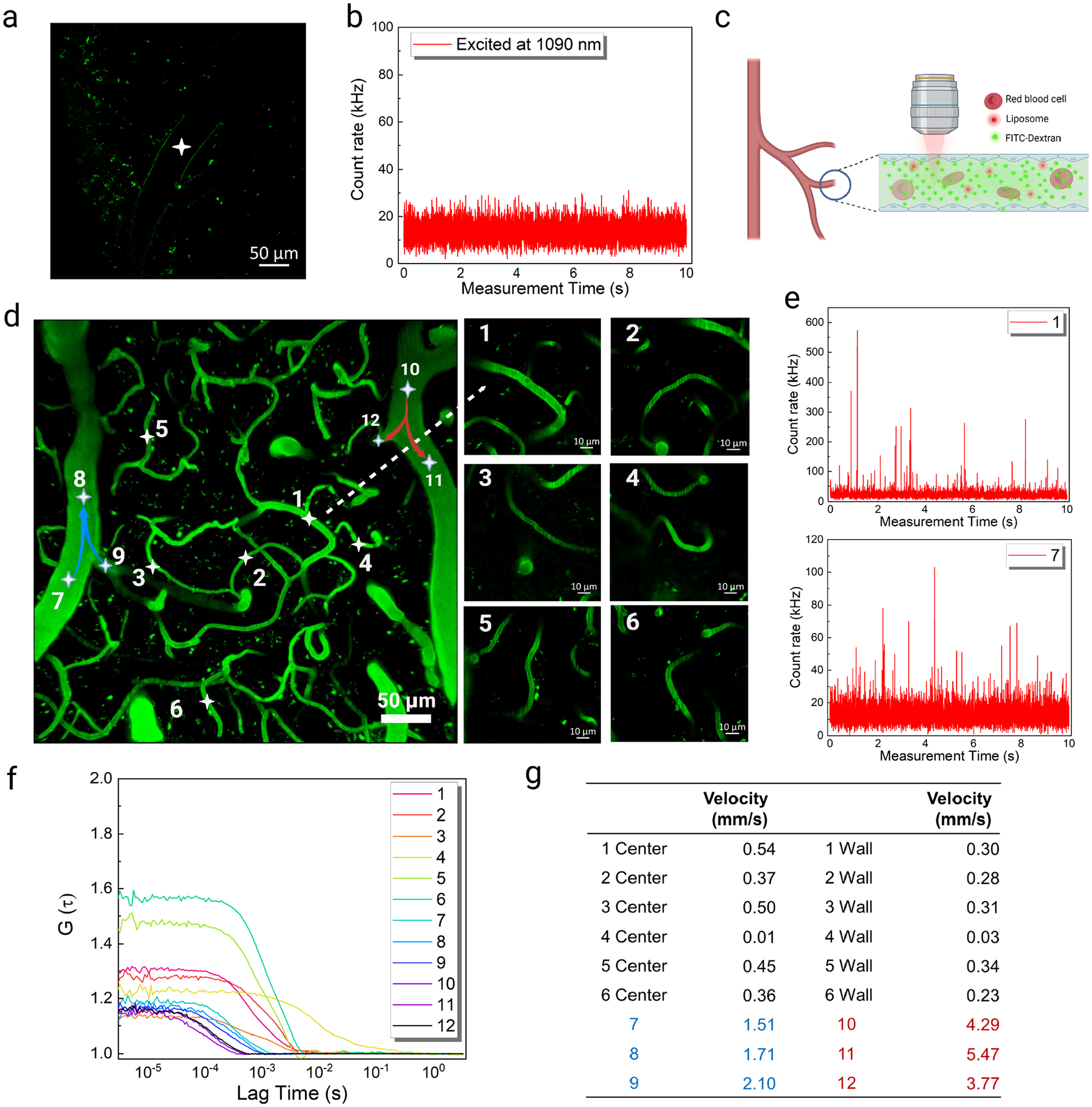
In vivo monitoring of liposomes loaded with DNA-Ag16NCs and cerebrovascular blood flow velocity measurements. (a) Cerebral vascular image obtained in the green channel under 920 nm (25 mW) excitation in a mouse with nothing injected; the white asterisk indicates the FCS position. (b) Emission intensity fluctuations in the red/NIR I channel under 1090 nm (12 mW) excitation with nothing injected. (c) Schematic showing the cerebrovascular blood flow after FITC-Dextran and DNA-Ag16NC-loaded liposome injection. (d) In vivo images of the mouse cerebral vasculature after FITC-Dextran and DNA-Ag16NC-loaded liposome delivery. Left: maximum intensity projection of the 3D image with 35 μm thickness. Right: enlarged capillary images. (e) Emission intensity fluctuations at the center of positions 1 and 7 under 1090 nm excitation. (f) The corresponding autocorrelation functions at the positions under 1090 nm excitation. For positions 1–6, the autocorrelation functions at the center were represented. (g) Measured blood flow velocities at the center and wall of the cerebrovascular. Data from arterioles locations were highlighted in red and venules in blue.
Next, we delivered a 100 μL mixture of FITC-Dextran and DNA-Ag16NC-loaded liposomes into the bloodstream using retro-orbital injection (Figure 5c), yielding a final concentration in the animal that was similar to that in our flow chamber measurements (1 μM of FITC-Dextran and single to a few liposomes in the confocal volume). The vasculature images were acquired from the green emission channel by exciting FITC-Dextran at 920 nm (25 mW). The dynamic characterization of DNA-Ag16NC-loaded liposomes in the vasculature was monitored by two-photon FCS at 1090 nm. A 35 μm thick vasculature area 100 μm beneath the surface of the brain was selected for analysis, which included arterioles (~28 μm in diameter), venules (~32 μm in diameter), and capillaries (<10 μm in diameter). It should be noted that not all of the selected vessels were in the same focal plane (Figure S17). All focal planes were merged into one image with a maximum intensity projection to visualize all FCS points in a single image (Figure 5d). The locations of interest inside the vessels were selected during live imaging, and the DNA-Ag16NC-loaded liposomes were then monitored with FCS in the red/NIR I channel under 1090 nm (12 mW) excitation. Intensity fluctuations from within vessels clearly show distinguishable intensity spikes (Figure 5e). The flow velocities of 12 vessels were mapped by recording similar intensity time traces, as in Figure 5e, and analyzing the corresponding autocorrelation functions (Figure 5f). Using the extracted residence time and calibrated confocal lateral diameter (0.690 μm), we then calculated the flow velocity in each vessel (Figure 5g). The flow velocities in capillaries (positions 1–6) were all slower than 1 mm/s, while the flow velocities in arterioles and venules (positions 7–12), which have a larger diameter, were all faster than 1 mm/s. However, the flow velocities in the arterioles (positions 10–12) were significantly faster than in venules (positions 7–9), even though their diameters were smaller. The blood flow velocities measured here are similar to those previously observed for similarly sized vessels.10, 13, 53, 54 The high spatial resolution of this technique even allowed us to map the flow velocity at multiple points within a single capillary. The blood flow velocities at the center were usually faster than those at the vessel wall apart from position 4, where a sharp turn in the vessel obstructed the flow and gave rise to slower velocities. Overall, encapsulating DNA-Ag16NCs into liposomes made them detectable by two-photon FCS in vivo and provided high spatial-temporal resolution for flow velocity measurements. Previous studies have shown that liposomes are metabolized through phagocytotic uptake in the liver and spleen.55 Cationic liposomes, similar to those used for this study have been widely used for in vivo studies in mouse models and therapeutic delivery in humans.56 These liposomes have been found to accumulate at low levels in a variety of organs including the lungs, kidneys, and liver and have shown no indication of toxicity.57 Previous studies have also shown that DNA-AgNCs are non-toxic to mammalian cells.58–61 To further investigate the biosafety of DNA-Ag16NC-loaded liposomes, the behavior and body weights of mice were monitored after delivery of a mixture of loaded liposomes and FITC-Dextran. These mice were monitored to determine any changes in movement, grooming, nest building, and interactions with cage mates. The animals appeared alert and active when the lid was removed (Figure S18a) and no abnormities were observed during daily hands-on evaluations. Animals were weighed daily for 2 weeks and no abnormal change in weight was observed (Figure S18b, c). We then extracted and sectioned several organs (brain, heart, liver, spleen, and kidney) and stained them with hematoxylin and eosin (H&E) for pathological analysis. No abnormal tissue, organ damage, or inflammatory lesions were observed (Figure S19).
CONCLUSIONS
We have demonstrated that we can load DNA-Ag16NC into liposomes and measure cerebral blood flow rates within individual capillaries in the complex vessel architecture of a live mouse using two-photon in vivo FCS. Encapsulating DNA-Ag16NCs into liposomes did not affect their optical properties but yielded nanoparticles that were sufficiently bright to be detected with two-photon FCS. DNA-Ag16NC exhibited NIR-I emission and two-photon excitation in the NIR-II window, which are very desirable wavelengths for in vivo measurements because of the decreased autofluorescence from tissue compared to the visible region where commonly used two-photon fluorophores emit. From control experiments in a microfluidic device and live mice, we found that exciting at 1090 nm, rather than at 1045 nm, yielded the most reliable two-photon FCS performance. With our optimized excitation conditions and calibrated confocal lateral diameter, we monitored the emission of DNA-Ag16NC-loaded liposomes inside cerebral capillaries in vivo and mapped flow velocities with high spatial resolution. With recent demonstrations of DNA-AgNCs exhibiting upconversion fluorescence, it would be interesting in future studies to explore the use of their anti-Stokes emission for in vivo FCS, allowing for increased brightness and/or reduced excitation intensities on top of a lower background.
EXPERIMENTAL SECTION
Synthesis and purification of DNA-Ag16NC.
DNA-Ag16NCs were synthesized following a previously published procedure.31 Briefly, silver nitrate (Sigma Aldrich, USA) and hydrated DNA with sequence 5’-CACCTAGCGA-3’ (IDT DNA Technologies, USA) were mixed in 10 mM ammonium acetate (NH4OAc) solution (pH 7), and the silver cations were then reduced by freshly prepared NaBH4 (Sigma Aldrich, USA) after 15 minutes. Three days later, the final mixture was purified by high-performance liquid chromatography (HPLC), and the DNA-Ag16NCs fraction was collected after 17 minutes of elution. Finally, the solvent was exchanged to 10 mM NH4OAc by spin-filtration (3 kDa cut-off membrane, Sigma Aldrich, USA), which further concentrated the DNA-Ag16NCs. The concentration of the DNA-Ag16NCs reported in this paper was estimated based on the DNA absorbance.
Optical characterization of DNA-Ag16NCs.
The two-photon optical properties of DNA-Ag16NCs were characterized with an upright confocal laser scanning microscope (LSM 880, Carl Zeiss, Germany). For the multiphoton excitation and emission, at least 300 μL DNA-Ag16NCs were trapped in an O-ring sandwiched by two #1.5 coverslips. The multiphoton excitation spectra were acquired with the LSM 880 microscope equipped with a tunable ultrafast laser (InSight X3, Spectra-Physics, USA). A 760 nm long-pass dichroic mirror (BS-MP 760, Carl Zeiss, Germany) was used to transmit the NIR laser (775 nm – 1100 nm), excite the DNA-Ag16NCs and reflect the emission to the GaAsP detector (BiG-2, Carl Zeiss, Germany). Also, a 10x/0.5 NA objective (Carl Zeiss, Germany) with a long working distance ensured the focus inside the solution under the top coverslip. The excitation power was fixed at 3 mW, and emission in the red emission channel (FF01–665/150, Semrock, USA) was collected with the FCS module in the Zen Black 3.0 software. The excitation spectra of FITC-Dextran 70 kDa (Sigma Aldrich, USA) and CF488-Dextran (Biotium, USA) were measured in the green emission channel (BP 525–560, Carl Zeiss, Germany). Excitation power dependence studies were conducted using the same settings. As for the emission spectra, both one-photon and two-photon excitation were implemented for DNA-Ag16NCs and dextran conjugated dyes, and the emission was collected with the Lambda mode in the same software.
Preparation of DNA-Ag16NC-loaded liposomes.
The encapsulation of DNA-Ag16NCs was conducted using a thin film hydration method normally used for siRNA or mRNA delivery.62–64 First, cationic lipid 1,2-Dioleoyl-3-trimethylammonium-propane (DOTAP) and neutral lipid 1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE) powders (Avanti, USA) were dissolved in chloroform separately to make a 25 mg/mL stock solution. Then, 6 μmol DOTAP and 6 μmol DOPE were mixed uniformly in a 4 mL amber vial. After that, the chloroform in the lipid mixture was evaporated under nitrogen flow, leaving the dried lipids film at the bottom of the vial. To encapsulate the DNA-Ag16NCs into liposomes, 500 μL of 12.5 μM, 22.5 μM or 37.5 μM DNA-Ag16NCs in 10 mM NH4OAc were added to the premade lipids film. Next, the mixture was incubated at room temperature for 30 minutes and followed by 15 minutes of water-bath sonication and gentle vortex mixing. The synthesized multilayer giant liposomes were extruded through a membrane (Whatman, USA) with a 50 nm or 100 nm pore size with a Mini-Extruder (Avanti, USA). After 21 back-and-forth extrusions, the unilamellar liposomes, with the corresponding size, were generated. Finally, the synthesized liposomes were purified by a size exclusion column with a 6,000 MW limit (BIO-RAD, USA) and collected after the centrifuge.
Characterization of DNA-Ag16NC-loaded liposomes.
A fluorescence microscope (BS-7000B, Bestscope, China) was adopted to visualize the DNA-Ag16NC-loaded liposomes. The DNA-Ag16NC-loaded liposomes were diluted 100 times, and a 100 μL sample was dropped on a glass bottom dish. The liposomes settled down at room temperature for 10 minutes, and the suspension was removed from the dish. The fluorescence images were acquired by a 60x water immersion objective under green light excitation. The size distribution of the liposomes was determined by NTA (Malvern Panalytical, Netherlands). The synthesized and purified liposomes were diluted 50,000 times so that there were around 40 liposomes/frame for accurate size measurements. Besides the size characterization, the optical properties of the DNA-Ag16NC-loaded liposomes were also explored with the FlexStation 3 Microplate Reader (Molecular Devices, USA). A 50 μL solution of synthesized liposomes or DNA-Ag16NCs was filled in the wells of a clear Greiner 96-well microplate. The absorption spectra were measured from 450 nm to 650 nm with a 5 nm interval, and fluorescence spectra were measured from 570 nm to 750 nm under 520 nm excitation.
Evaluation of DNA-Ag16NCs encapsulation.
Two methods were used to evaluate the encapsulation of DNA-Ag16NCs into the liposomes. First, the fluorescence intensity ratio65 was measured with the FlexStation 3 Microplate Reader for the liposomes before and after the size exclusion column purification, which indicated the loading ability of synthesized liposomes. The fluorescence intensities at 690 nm were collected for 100 nm and 50 nm liposomes, and the fluorescence intensity ratios were calculated. The self-built total internal reflection fluorescence (TIRF) microscope was then used to evaluate DNA-Ag16NCs encapsulation. DNA-Ag16NC-loaded liposomes were further labeled by a lipophilic carbocyanine dye (DiD) by incubating the liposomes with 5 μM DiD at room temperature for 30 minutes. The labeled liposomes were then purified with a spin column (G-25, cytiva, USA) to remove the free DiD molecules. A 561 nm or 640 nm laser was used to excite, respectively, the DNA-Ag16NCs or DiD, and the emission was collected through a 673/44 nm band-pass filter (ET673/44, Chroma, USA). The encapsulation could be qualitatively evaluated by assessing the degree of colocalization from the TIRF images under 561 nm and 640 nm excitation.
Two-photon FCS of DNA-Ag16NC-loaded liposomes in the flow chamber.
The two-photon FCS analysis was conducted using the same experimental equipment (LSM 880, Carl Zeiss, Germany) used for the optical characterization of the DNA-Ag16NCs. The difference was that a water immersion objective (W Plan-Apochromat 20x/1.0, Carl Zeiss, Germany) with a larger numerical aperture (NA) replaced the one used for measuring excitation and emission spectra. As for FCS analysis of static DNA-Ag16NC and loaded liposomes, the sample was held by an O-ring sandwiched by two #1.5 coverslips, and the emission intensity fluctuations inside the solution were recorded with ten seconds acquisition and ten repetitions with the FCS module of Zen Black software. DNA-Ag16NCs and liposomes were diluted to a single-particle level for FCS analysis. Then, the autocorrelation functions of these particles were calculated and fitted with a 3D diffusion model66 in the same software:
| (1) |
Where 〈N〉 is the number of molecules contained in the focal volume, and τD is the lateral diffusion time. r0 and z0 are the lateral and axial diameters of the confocal volume, respectively. In contrast to the freely diffusing liposome measurements, FCS analysis of liposomes under flow was performed inside a microfluidic flow chamber (stick-Slide/0.2 luer, ibidi, Germany; shown in Figure 3a). This chamber was connected to a syringe with a known diameter by a tube, and the flow rates were controlled by a syringe pump (NE-1000, New Era, USA). Similarly, the autocorrelation functions of DNA-Ag16NC-loaded liposomes under active flow can be described with a flow model considering the diffusion66:
| (2) |
Here, τf is the average residence time of the molecule crossing the confocal volume under the flow, which is inversely proportional to the flow velocity (vf) if the lateral diameter (d) is constant. Furthermore, the FCS fitting model could be simplified by ignoring the free diffusion if the flow dominates the dynamic changes of the liposomes in the flow chamber:
| (3) |
The lateral diameter (d) of the two-photon confocal volume could be calibrated and extracted by varying the flow rates (Q; 0.05, 0.1, 0.2, 0.3, 0.4, and 0.5 mL/min) of liposomes under the same conditions.
| (4) |
Where, the A, w and h are cross-sectional area, width, and height of the flow chamber and Q is the set flow rate.
Animal preparation.
All animal experiments were conducted following the approved protocols and in conformity with the guidelines of the Institutional Animal Care and Use Committee (IACUC) at the University of Kentucky. All the mice were normally housed and fed for 9–12 months. The health of the mice was assessed by their behaviors before the surgery, and these mice were euthanized by cervical dislocation under anesthesia after the experiments.
Cranial window surgery.
A skull-opened cranial window was created for vasculature imaging and FCS measurements in vivo. Several published protocols67–70 were referenced and modified based on the available equipment in our lab. First, the mouse was anesthetized with 3% isoflurane (Henry Schein, USA) for induction in a plastic chamber connected to the low-flow Anesthesia System (Kent Scientific, USA), and its breathing pattern was monitored until loss of response to toe pinch. Then, the mouse was transferred to a warming pad with its head fixed on a stereotactic apparatus by a pair of ear bars (Figure S15a). Next, eye ointment was applied to its eyes to prevent drying, and 70% alcohol wipes were used to sterilize the operating area. The isoflurane concentration and the mouse body temperature were maintained at 2.0–2.5% and 36.5–37.5 °C during surgery, respectively. The skin and film over the skull were removed by sterilized scissors and a blunt scalpel. A circle of 3–4 mm in diameter was drawn using a dental drill on the parietal bone of the mouse and stopped until the thin bone was left. After that, saline was dropped on the drilling area to lift the skull from the dura, and the skull could be removed using forceps with an ultra-fine tip. A gelatin sponge presoaked with saline could be used to stop minor bleeding from the edge of the cranial window when removing the skull. The exposed brain was expeditiously covered by a 5 mm round coverslip and sealed by cyanoacrylate glue (Loctite 416) at the edge. A 3D-printed head holder (Figure S15a) was embedded on the mouse skull with dental cement to immobilize its head on an apparatus during the imaging. Finally, the mouse was transferred to the heating pad under the LSM880 microscope for imaging and FCS measurements and supplied with 1.7–2% isoflurane.
Cerebrovascular visualizing dye and liposome delivery.
In total, a 100 μL mixture of FITC-Dextran 70 kDa (50 μM) and DNA-Ag16NC-loaded liposomes (30 μL) was delivered to the mouse by retro-orbital injection,71 yielding concentrations similar to that in the flow chamber (1 μM of FITC-Dextran and single to few liposomes in the confocal volume). The injection was implemented before the imaging as the mouse was anesthetized. The FITC-Dextran 70 kDa was used to visualize the cerebrovascular by two-photon imaging, and liposomes were monitored by in vivo FCS. In vivo toxicities of DNA-Ag16NCs and DNA-Ag16NC-loaded liposomes were assessed by monitoring the body weights of control mice after administration for ten days. For this, separate mice were used that did not undergo skull-opened imaging.
In Vivo two-photon imaging of mouse cerebrovascular.
The cerebrovascular was imaged by the same LSM 880 microscope with a long working distance water immersion objective (W Plan-Apochromat 20x/1.0, Carl Zeiss, Germany). The cranial window of the mouse was adjusted to be parallel to the focal plane and be centered from the eyepiece view. Then, the 920 nm laser (25 mW) was used to excite the FITC-Dextran 70 kDa for vascular visualization, and the image was acquired in the green emission channel (525–560 nm). A 3D image, 100 μm beneath the brain surface, was created by scanning the selected area every 0.625 μm in the z-direction, yielding a total depth of 35 μm with 56 layers. The maximum intensity projection (Figure S17) was adopted from the 3D image to visualize all the capillaries that were not in the same focal plane. The obtained images were further analyzed with Zen Blue 3.0 (Carl Zeiss, Germany) and Image J software (NIH, USA).
In Vivo liposome monitoring and blood flow velocity quantification by FCS.
Based on the cerebrovascular images, DNA-Ag16NC-loaded liposomes were monitored with the FCS module in these vasculatures. Any point of interest in the image could be selected and analyzed with two-photon FCS. These liposomes were excited at 1045 nm or 1090 nm, and the fluorescence intensity fluctuations in the vasculatures were recorded in the red emission channel (590–740 nm). A short acquisition time (10 s) and repeated measurements helped minimize the artifacts introduced by mouse breathing. The concentration and residence time of the liposomes were obtained by fitting the autocorrelation functions. Then, the blood flow velocity (vf) was calculated based on the calibrated lateral diameter and liposome residence time:
| (5) |
The DNA-Ag16NC-loaded liposomes inside vasculatures, including the arteriole, venules, and capillaries, were monitored, and the blood flow velocities were mapped with two-photon FCS.
In Vivo biosafety evaluation of DNA-Ag16NC-loaded liposomes.
The same amount (as used for in vivo FCS experiments) of DNA-Ag16NC-loaded liposomes and FITC-Dextran was delivered by retro-orbital injection for three mice of the same type. The behavior and body weights of these mice were monitored for 14 days. After 14 days, the mice were perfused with 30 mL PBS and 50 mL 4% paraformaldehyde (PFA, thermos scientific, USA) successively. Then, the main organs (brain, heart, liver, spleen, and kidney) were dissected and stored in 4% PFA. The tissues were fixed in 4% PFA for 24 hours prior to transfer to a 30% sucrose solution for 48 hours. The tissues were sectioned in a cryostat (−18°C) at 30 μm thickness and stained by hematoxylin and eosin (H&E). At last, the tissues were dried completely and mounted to slides by mounting media. The tissue slides were imaged by a 20x objective of a slide scanner (Axioscan, Zeiss, Germany).
Supplementary Material
ACKNOWLEDGMENTS
T.V. and C.I.R. acknowledge financial support from the Novo Nordisk Foundation (NNF20OC0064207). C.I.R. acknowledges support from the NIGMS/NIH (R01GM138837 and R01GM138882)
Footnotes
Supporting Information
The Supporting Information is available free of charge.
Additional experiment details and supplementary Figures S1 – S19. NIR excitation spectra of 10mM NH4OAc, two-photon characterization of CF488-Dextran, excitation intensity dependence of DNA-Ag16NCs under 800 nm excitation, TIRF images of liposomes, optical properties of liposomes loaded with 22.5 μM DNA-Ag16NCs, diffusion autocorrelation functions of liposomes, emission intensity fluctuations and autocorrelation functions of different concentration of liposomes loaded with DNA-Ag16NCs, emission intensity fluctuations of liposomes and FITC-Dextran 70 kDa, a mouse with a cranial window, in vivo detection of liposomes under 1045 nm excitation, in vivo imaging of cerebral vasculatures, mice behaviors and body weights monitoring, and H&E staining images of tissues from mice.
The authors declare no competing financial interest.
REFERENCES
- (1).Attwell D; Buchan AM; Charpak S; Lauritzen M; Macvicar BA; Newman EA Glial and neuronal control of brain blood flow. Nature 2010, 468, 232–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).McConnell HL; Kersch CN; Woltjer RL; Neuwelt EA The Translational Significance of the Neurovascular Unit. J. Biol. Chem 2017, 292, 762–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Kisler K; Lazic D; Sweeney MD; Plunkett S; El Khatib M; Vinogradov SA; Boas DA; Sakadzi S; Zlokovic BV In vivo imaging and analysis of cerebrovascular hemodynamic responses and tissue oxygenation in the mouse brain. Nat. Protoc 2018, 13, 1377–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Luo Y; Wang Y; Fu J Nanomaterials in Cerebrovascular Disease Diagnose and Treatment. Part. Part. Syst. Charact 2021, 38, 2000311. [Google Scholar]
- (5).Pathak AP; Kim E; Zhang J; Jones MV Three-dimensional imaging of the mouse neurovasculature with magnetic resonance microscopy. PLoS One 2011, 6, 22643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Starosolski Z; Villamizar CA; Rendon D; Paldino MJ; Milewicz DM; Ghaghada KB; Annapragada AV Ultra High-Resolution In vivo Computed Tomography Imaging of Mouse Cerebrovasculature Using a Long Circulating Blood Pool Contrast Agent. Sci. Rep 2015, 5, 10178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Coelho-Santos V; Berthiaume AA; Ornelas S; Stuhlmann H; Shih AY Imaging the construction of capillary networks in the neonatal mouse brain. Proc. Natl. Acad. Sci. U. S. A 2021, 118, 2100866118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Heo C; Park H; Kim YT; Baeg E; Kim YH; Kim SG; Suh M A soft, transparent, freely accessible cranial window for chronic imaging and electrophysiology. Sci. Rep 2016, 6, 27818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Koletar MM; Dorr A; Brown ME; McLaurin J; Stefanovic B Refinement of a chronic cranial window implant in the rat for longitudinal in vivo two-photon fluorescence microscopy of neurovascular function. Sci. Rep 2019, 9, 5499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Xu Z; Zhang Z; Deng X; Li J; Jiang Y; Law WC; Yang C; Zhang W; Chen X; Wang K; et al. Deep-Brain Three-Photon Imaging Enabled by Aggregation-Induced Emission Luminogens with Near-Infrared-III Excitation. ACS Nano 2022, 16, 6712–6724. [DOI] [PubMed] [Google Scholar]
- (11).Joris PJ; Mensink RP; Adam TC; Liu TT Cerebral Blood Flow Measurements in Adults: A Review on the Effects of Dietary Factors and Exercise. Nutrients 2018, 10, 530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Saha M; Dremin V; Rafailov I; Dunaev A; Sokolovski S; Rafailov E Wearable Laser Doppler Flowmetry Sensor: A Feasibility Study with Smoker and Non-Smoker Volunteers. Biosensors (Basel) 2020, 10, 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Qureshi MM; Liu Y; Mac KD; Kim M; Safi AM; Chung E Quantitative blood flow estimation in vivo by optical speckle image velocimetry. Optica 2021, 8, 1092–1101. [Google Scholar]
- (14).Kim TN; Goodwill PW; Chen Y; Conolly SM; Schaffer CB; Liepmann D; Wang RA Line-scanning particle image velocimetry: an optical approach for quantifying a wide range of blood flow speeds in live animals. PLoS One 2012, 7, 38590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Pardridge WM A Historical Review of Brain Drug Delivery. Pharmaceutics 2022, 14, 1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Cash KJ; Li C; Xia J; Wang LV; Clark HA Optical drug monitoring: photoacoustic imaging of nanosensors to monitor therapeutic lithium in vivo. ACS nano 2015, 9, 1692–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Cai Y; Qi J; Lu Y; He H; Wu W The in vivo fate of polymeric micelles. Adv. Drug Deliv. Rev 2022, 188, 114463. [DOI] [PubMed] [Google Scholar]
- (18).Wang J; Tang W; Yang M; Yin Y; Li H; Hu F; Tang L; Ma X; Zhang Y; Wang Y Inflammatory tumor microenvironment responsive neutrophil exosomes-based drug delivery system for targeted glioma therapy. Biomaterials 2021, 273, 120784. [DOI] [PubMed] [Google Scholar]
- (19).Rosigkeit S; Meng M; Grunwitz C; Gomes P; Kreft A; Hayduk N; Heck R; Pickert G; Ziegler K; Abassi Y; et al. Monitoring Translation Activity of mRNA-Loaded Nanoparticles in Mice. Mol. Pharm 2018, 15, 3909–3919. [DOI] [PubMed] [Google Scholar]
- (20).Palanikumar L; Choi ES; Oh JY; Park SA; Choi H; Kim K; Kim C; Ryu JH Importance of Encapsulation Stability of Nanocarriers with High Drug Loading Capacity for Increasing in Vivo Therapeutic Efficacy. Biomacromolecules 2018, 19, 3030–3039. [DOI] [PubMed] [Google Scholar]
- (21).Pierrat P; Wang R; Kereselidze D; Lux M; Didier P; Kichler A; Pons F; Lebeau L Efficient in vitro and in vivo pulmonary delivery of nucleic acid by carbon dot-based nanocarriers. Biomaterials 2015, 51, 290–302. [DOI] [PubMed] [Google Scholar]
- (22).Fu X; Sompol P; Brandon JA; Norris CM; Wilkop T; Johnson LA; Richards CI In Vivo Single-Molecule Detection of Nanoparticles for Multiphoton Fluorescence Correlation Spectroscopy to Quantify Cerebral Blood Flow. Nano Lett. 2020, 20, 6135–6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Yang Y; Fan X; Li L; Yang Y; Nuernisha A; Xue D; He C; Qian J; Hu Q; Chen H; et al. Semiconducting Polymer Nanoparticles as Theranostic System for Near-Infrared-II Fluorescence Imaging and Photothermal Therapy under Safe Laser Fluence. ACS Nano 2020, 14, 2509–2521. [DOI] [PubMed] [Google Scholar]
- (24).Qi J; Sun C; Li D; Zhang H; Yu W; Zebibula A; Lam JWY; Xi W; Zhu L; Cai F; et al. Aggregation-Induced Emission Luminogen with Near-Infrared-II Excitation and Near-Infrared-I Emission for Ultradeep Intravital Two-Photon Microscopy. ACS Nano 2018, 12, 7936–7945. [DOI] [PubMed] [Google Scholar]
- (25).Samanta S; Huang M; Li S; Yang Z; He Y; Gu Z; Zhang J; Zhang D; Liu L; Qu J AIE-active two-photon fluorescent nanoprobe with NIR-II light excitability for highly efficient deep brain vasculature imaging. Theranostics 2021, 11, 2137–2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Hong G; Antaris AL; Dai H Near-infrared fluorophores for biomedical imaging. Nat. Biomed.l Eng 2017, 1, 0010. [Google Scholar]
- (27).Patel SA; Richards CI; Hsiang JC; Dickson RM Water-soluble Ag nanoclusters exhibit strong two-photon-induced fluorescence. J. Am. Chem. Soc 2008, 130, 11602–11603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Yau SH; Abeyasinghe N; Orr M; Upton L; Varnavski O; Werner JH; Yeh HC; Sharma J; Shreve AP; Martinez JS; et al. Bright two-photon emission and ultra-fast relaxation dynamics in a DNA-templated nanocluster investigated by ultra-fast spectroscopy. Nanoscale 2012, 4, 4247–4254. [DOI] [PubMed] [Google Scholar]
- (29).Gonzalez-Rosell A; Cerretani C; Mastracco P; Vosch T; Copp SM Structure and luminescence of DNA-templated silver clusters. Nanoscale Adv. 2021, 3, 1230–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Copp SM; Gorovits A; Swasey SM; Gudibandi S; Bogdanov P; Gwinn EG Fluorescence Color by Data-Driven Design of Genomic Silver Clusters. ACS Nano 2018, 12, 8240–8247. [DOI] [PubMed] [Google Scholar]
- (31).Bogh SA; Carro-Temboury MR; Cerretani C; Swasey SM; Copp SM; Gwinn EG; Vosch T Unusually large Stokes shift for a near-infrared emitting DNA-stabilized silver nanocluster. Methods Appl. Fluoresc 2018, 6, 024004. [DOI] [PubMed] [Google Scholar]
- (32).Yang M; Chen X; Su Y; Liu H; Zhang H; Li X; Xu W The Fluorescent Palette of DNA-Templated Silver Nanoclusters for Biological Applications. Front. Chem 2020, 8, 601621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Schmitt S; Huppertsberg A; Klefenz A; Kaps L; Mailander V; Schuppan D; Butt HJ; Nuhn L; Koynov K Fluorescence Correlation Spectroscopy Monitors the Fate of Degradable Nanocarriers in the Blood Stream. Biomacromolecules 2022, 23, 1065–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Tenchov R; Bird R; Curtze AE; Zhou Q Lipid Nanoparticles horizontal line From Liposomes to mRNA Vaccine Delivery, a Landscape of Research Diversity and Advancement. ACS Nano 2021, 15, 16982–17015. [DOI] [PubMed] [Google Scholar]
- (35).Goh WJ; Zou S; Ong WY; Torta F; Alexandra AF; Schiffelers RM; Storm G; Wang JW; Czarny B; Pastorin G Bioinspired Cell-Derived Nanovesicles versus Exosomes as Drug Delivery Systems: a Cost-Effective Alternative. Sci. Rep 2017, 7, 14322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Kim M; Jeong M; Hur S; Cho Y; Park J; Jung H; Seo Y; Woo HA; Nam KT; Lee K; Lee H Engineered ionizable lipid nanoparticles for targeted delivery of RNA therapeutics into different types of cells in the liver. Sci. Adv 2021, 7, 4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Remaut K; Sanders NN; De Geest BG; Braeckmans K; Demeester J; De Smedt SC Nucleic acid delivery: Where material sciences and bio-sciences meet. Mater. Sci. Eng. R Rep 2007, 58, 117–161. [Google Scholar]
- (38).Ruck V; Cerretani C; Neacsu VA; Liisberg MB; Vosch T Observation of microsecond luminescence while studying two DNA-stabilized silver nanoclusters emitting in the 800–900 nm range. Phys. Chem. Chem. Phys 2021, 23, 13483–13489. [DOI] [PubMed] [Google Scholar]
- (39).Liisberg MB; Shakeri Kardar Z; Copp SM; Cerretani C; Vosch T Single-Molecule Detection of DNA-Stabilized Silver Nanoclusters Emitting at the NIR I/II Border. J. Phys. Chem. Lett 2021, 12, 1150–1154. [DOI] [PubMed] [Google Scholar]
- (40).Neacsu VA; Cerretani C; Liisberg MB; Swasey SM; Gwinn EG; Copp SM; Vosch T Unusually large fluorescence quantum yield for a near-infrared emitting DNA-stabilized silver nanocluster. Chem. Commun 2020, 56, 6384–6387. [DOI] [PubMed] [Google Scholar]
- (41).Cerretani C; Kanazawa H; Vosch T; Kondo J Crystal structure of a NIR-Emitting DNA-Stabilized Ag(16) Nanocluster. Angew. Chem. Int. Ed. Engl 2019, 58, 17153–17157. [DOI] [PubMed] [Google Scholar]
- (42).Cerretani C; Palm-Henriksen G; Liisberg MB; Vosch T The effect of deuterium on the photophysical properties of DNA-stabilized silver nanoclusters. Chem. Sci 2021, 12, 16100–16105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).O’Neill PR; Gwinn EG; Fygenson DK UV Excitation of DNA Stabilized Ag Cluster Fluorescence via the DNA Bases. J. Phys. Chem. C 2011, 115, 24061–24066. [Google Scholar]
- (44).Krause S; Carro-Temboury MR; Cerretani C; Vosch T Anti-Stokes fluorescence microscopy using direct and indirect dark state formation. Chem. Commun 2018, 54, 4569–4572. [DOI] [PubMed] [Google Scholar]
- (45).Liisberg MB; Krause S; Cerretani C; Vosch T Probing emission of a DNA-stabilized silver nanocluster from the sub-nanosecond to millisecond timescale in a single measurement. Chem. Sci 2022, 13, 5582–5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Xu C; Webb WW Measurement of two-photon excitation cross sections of molecular fluorophores with data from 690 to 1050 nm. J. Opt. Soc. Am. B 1996, 13, 481–491. [Google Scholar]
- (47).Krause S; Cerretani C; Vosch T Disentangling optically activated delayed fluorescence and upconversion fluorescence in DNA stabilized silver nanoclusters. Chem. Sci 2019, 10, 5326–5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Bruun K; Hille C Study on intracellular delivery of liposome encapsulated quantum dots using advanced fluorescence microscopy. Sci. Rep 2019, 9, 10504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Chen J; Kumar A; Cerretani C; Vosch T; Zigmantas D; Thyrhaug E Excited-State Dynamics in a DNA-Stabilized Ag(16) Cluster with Near-Infrared Emission. J. Phys. Chem. Lett 2023, 14, 4078–4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Kunst BH; Schots A; Visser AJ Detection of flowing fluorescent particles in a microcapillary using fluorescence correlation spectroscopy. J. Anal. Chem 2002, 74, 5350–5357. [DOI] [PubMed] [Google Scholar]
- (51).Park S; Choi SQ; Song C; Kim MW; Choi MC Surface charge effects on optical trapping of nanometer-sized lipid vesicles. Soft Matter 2014, 10, 8406–8412. [DOI] [PubMed] [Google Scholar]
- (52).Jauffred L; Oddershede LB Two-photon quantum dot excitation during optical trapping. Nano Lett. 2010, 10, 1927–1930. [DOI] [PubMed] [Google Scholar]
- (53).Unekawa M; Tomita M; Tomita Y; Toriumi H; Miyaki K; Suzuki N RBC velocities in single capillaries of mouse and rat brains are the same, despite 10-fold difference in body size. Brain Res. 2010, 1320, 69–73. [DOI] [PubMed] [Google Scholar]
- (54).Meng G; Zhong J; Zhang Q; Wong JSJ; Wu J; Tsia KK; Ji N Ultrafast two-photon fluorescence imaging of cerebral blood circulation in the mouse brain in vivo. Proc. Natl. Acad. Sci. U. S. A 2022, 119, 2117346119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Sercombe L; Veerati T; Moheimani F; Wu SY; Sood AK; Hua S Advances and Challenges of Liposome Assisted Drug Delivery. Front. Pharmacol 2015, 6, 286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Saraf S; Jain A; Tiwari A; Verma A; Panda PK; Jain SK Advances in liposomal drug delivery to cancer: An overview. J. Drug Deliv. Sci. Technol 2020, 56, 101549. [Google Scholar]
- (57).Hattori Y; Tang M; Torii S; Tomita K; Sagawa A; Inoue N; Yamagishi R; Ozaki KI Optimal combination of cationic lipid and phospholipid in cationic liposomes for gene knockdown in breast cancer cells and mouse lung using siRNA lipoplexes. Mol. Med. Rep 2022, 26, 253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Rolband L; Yourston L; Chandler M; Beasock D; Danai L; Kozlov S; Marshall N; Shevchenko O; Krasnoslobodtsev AV; Afonin KA DNA-Templated Fluorescent Silver Nanoclusters Inhibit Bacterial Growth While Being Non-Toxic to Mammalian Cells. Molecules 2021, 26, 4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Lyu D; Li J; Wang X; Guo W; Wang E Cationic-Polyelectrolyte-Modified Fluorescent DNA-Silver Nanoclusters with Enhanced Emission and Higher Stability for Rapid Bioimaging. Anal. Chem 2019, 91, 2050–2057. [DOI] [PubMed] [Google Scholar]
- (60).Liu S; Yan Q; Cao S; Wang L; Luo SH; Lv M Inhibition of Bacteria In Vitro and In Vivo by Self-Assembled DNA-Silver Nanocluster Structures. ACS Appl. Mater. Interfaces 2022, 14, 41809–41818. [DOI] [PubMed] [Google Scholar]
- (61).Xie X; Sun T; Xue J; Miao Z; Yan X; Fang W; Li Q; Tang R; Lu Y; Tang L; et al. Ag Nanoparticles Cluster with pH-Triggered Reassembly in Targeting Antimicrobial Applications. Adv. Funct. Mater 2020, 30, 2000511. [Google Scholar]
- (62).Hattori Y; Tamaki K; Ozaki K.-i.; Kawano K; Onishi H Optimized combination of cationic lipids and neutral helper lipids in cationic liposomes for siRNA delivery into the lung by intravenous injection of siRNA lipoplexes. J. Drug Deliv. Sci. Technol 2019, 52, 1042–1050. [Google Scholar]
- (63).Even-Chen S; Cohen R; Barenholz Y Factors affecting DNA binding and stability of association to cationic liposomes. Chem. Phys. Lipids 2012, 165, 414–423. [DOI] [PubMed] [Google Scholar]
- (64).Spencer AC; Torre P; Mansy SS The encapsulation of cell-free transcription and translation machinery in vesicles for the construction of cellular mimics. J. Vis. Exp 2013, 80, 51304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Ullmann K; Leneweit G; Nirschl H How to Achieve High Encapsulation Efficiencies for Macromolecular and Sensitive APIs in Liposomes. Pharmaceutics 2021, 13, 691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Schwille P; Haustein E Fluorescence correlation spectroscopy. A tutorial for the Biophysics Textbook Online (BTOL). Biophysical Society, Rockville, MD, 2002; pp 10–25. [Google Scholar]
- (67).Holtmaat A; Bonhoeffer T; Chow DK; Chuckowree J; De Paola V; Hofer SB; Hubener M; Keck T; Knott G; Lee WC; et al. Long-term, high-resolution imaging in the mouse neocortex through a chronic cranial window. Nat. Protoc 2009, 4, 1128–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Mostany R; Portera-Cailliau C A craniotomy surgery procedure for chronic brain imaging. J. Vis. Exp 2008, 12, 1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Lin X; Zhao T; Xiong W; Wen S; Jin X; Xu XM Imaging Neural Activity in the Primary Somatosensory Cortex Using Thy1-GCaMP6s Transgenic Mice. J. Vis. Exp 2019, 143, 56297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Goldey GJ; Roumis DK; Glickfeld LL; Kerlin AM; Reid RC; Bonin V; Schafer DP; Andermann ML Removable cranial windows for long-term imaging in awake mice. Nat. Protoc 2014, 9, 2515–2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Yardeni T; Eckhaus M; Morris HD; Huizing M; Hoogstraten-Miller S Retro-orbital injections in mice. Lab Anim. 2011, 40, 155–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


