Abstract
CRISPR-enabled screening is a powerful tool for the discovery of genes that control T cell function and has nominated candidate targets for immunotherapies1–6. However, new approaches are required to probe specific nucleotide sequences within key genes. Systematic mutagenesis in primary human T cells could reveal alleles that tune specific phenotypes. DNA base editors are powerful tools for introducing targeted mutations with high efficiency7,8. Here we develop a large-scale base-editing mutagenesis platform with the goal of pinpointing nucleotides that encode amino acid residues that tune primary human T cell activation responses. We generated a library of around 117,000 single guide RNA molecules targeting base editors to protein-coding sites across 385 genes implicated in T cell function and systematically identified protein domains and specific amino acid residues that regulate T cell activation and cytokine production. We found a broad spectrum of alleles with variants encoding critical residues in proteins including PIK3CD, VAV1, LCP2, PLCG1 and DGKZ, including both gain-of-function and loss-of-function mutations. We validated the functional effects of many alleles and further demonstrated that base-editing hits could positively and negatively tune T cell cytotoxic function. Finally, higher-resolution screening using a base editor with relaxed protospacer-adjacent motif requirements9 (NG versus NGG) revealed specific structural domains and protein–protein interaction sites that can be targeted to tune T cell functions. Base-editing screens in primary immune cells thus provide biochemical insights with the potential to accelerate immunotherapy design.
Complex cellular signalling pathways integrate positive and negative signals to drive T cell activation and effector functions that are critical for productive immune responses against infections and cancer. Dysregulation of T cell activation can lead to autoimmunity, inflammation or immune deficiency. Immune-based therapies, including checkpoint inhibitors and adoptive cellular therapies such as chimeric antigen receptor (CAR) T cells, have transformed cancer treatment, and small-molecule pharmaceuticals and biologic drugs that interfere with T cell activation and cytokine signalling have been approved for use as anti-inflammatory agents and immunosuppressants for autoimmunity and organ transplantation10. However, immunotherapies have important limitations and many patients do not respond to these treatments. Improving the success rates across immunotherapies will require a deeper understanding of T cell regulation and further discovery of new targets for therapeutic manipulation.
CRISPR screens in primary human cells can now identify genes that control specific cellular phenotypes and nominate gene targets for cancer therapies1–3,11. Although CRISPR knockout, activation and interference screens can discover genes that are necessary or sufficient to influence immune cell functions, higher-resolution mutagenesis is required to dissect the functions of critical nucleotides within single genes. The discovery of distinct alleles for the same gene could provide unique insights into protein structure and mechanism and enable fine-tuning of protein function. Base editing is a CRISPR-based technology that introduces specific mutations at sites determined by single guide RNA (sgRNA) molecules without double-strand breaks or the need for homology-directed repair (HDR), rendering it suitable for large-scale screening to interrogate genetic variation at near-single-nucleotide resolution7,8,12,13. The base editors used in this study use a catalytically impaired CRISPR–Cas nuclease (nickase, nCas9) fused to a DNA deaminase, which deaminates cytosines (via its cytosine base editor (CBE) function) or adenines (via the adenine base editor (ABE) function) proximal to the targeted protospacer-adjacent motif (PAM) site within a defined editing window, causing GC-to-TA (CBE) or AT-to-GC (ABE) mutations. Streptococcus pyogenes Cas9-based base editors rely on the presence of an NGG PAM sequence for guide targeting; however, Cas9 variants with modified PAM requirements have been developed and incorporated into base editing enzymes7–9. In human T cells, base editing has been used for highly efficient multiplexed knockout with reduced risk of chromosomal translocations relative to multiplexed double-stranded breaks, and has advanced to applications in clinical-stage CAR T cell products14,15. The ability to introduce targeted genetic variants systematically and at large scale affords a powerful, underutilized discovery tool. An experimental platform for high-throughput pooled mutational screening could uncover an allelic spectrum—natural and synthetic—of genes involved in T cell activity and nominate candidates for manipulation in T cell and other immunotherapies.
Here we used an optimized lentiviral delivery approach3 to deploy base editing (ABE and CBE) for large-scale screens in primary human T cells. A pooled library of around 117,000 guide RNAs targeting the coding regions of 385 genes involved in T cell function was generated to identify base edits that tune activation state by fluorescence-activated cell sorting (FACS) of stimulated T cells into TNF, IFNγ, CD25 or PD-1 low and high bins and quantifying sgRNA abundance in each population. Enriched guides pinpointed protein hotspots that tune T cell activation responses when mutated, revealing a spectrum of alleles for multiple key genes ranging from protein loss to partial function and gain of function. These variant alleles could modulate ex vivo cytotoxic function in addition to cytokine production and secretion. Finally, adopting an evolved version of Cas9 with an NG PAM (SpG Cas9) increased the resolution of mutational mapping, providing structural and functional insights into protein domains and interaction sites. Systematic base-editing mutagenesis can now serve as a powerful platform to perform high-throughput biochemical perturbation studies with cellular readouts in primary human immune cells that will accelerate immunotherapy development for a broad range of indications. This platform provides a step towards a systematic reference atlas for functional alleles controlling immune cell function.
Base-editing screens in primary T cells
To enable large-scale mutagenesis experiments with base editors in primary human T cells, we optimized lentiviral delivery of the adenine base editor16 ABE8e(V106W) (hereafter referred to as ABE) and the cytosine base editor evoCDA1-BE4max17 (hereafter referred to as CBE). Lentiviral base editing in primary human T cells was confirmed for genes encoding the well-characterized T cell transmembrane proteins CD3, CD5 and CD7 (Extended Data Fig. 1). Targeting a splice site (in CD7 using ABE and CBE) or introducing a stop codon (in CD5 and CD7 using CBE) efficiently reduced surface protein expression in both CD4+ and CD8+ T cells concordant with genomic-level editing. By contrast, sgRNAs targeting CD3 that were designed to introduce a distal or synonymous mutation left surface protein expression intact despite high-efficiency DNA editing at the genomic target site. These experiments confirmed that base editors could be transduced stably into primary human T cells to mediate efficient mutagenesis at sites dictated by transduced sgRNAs.
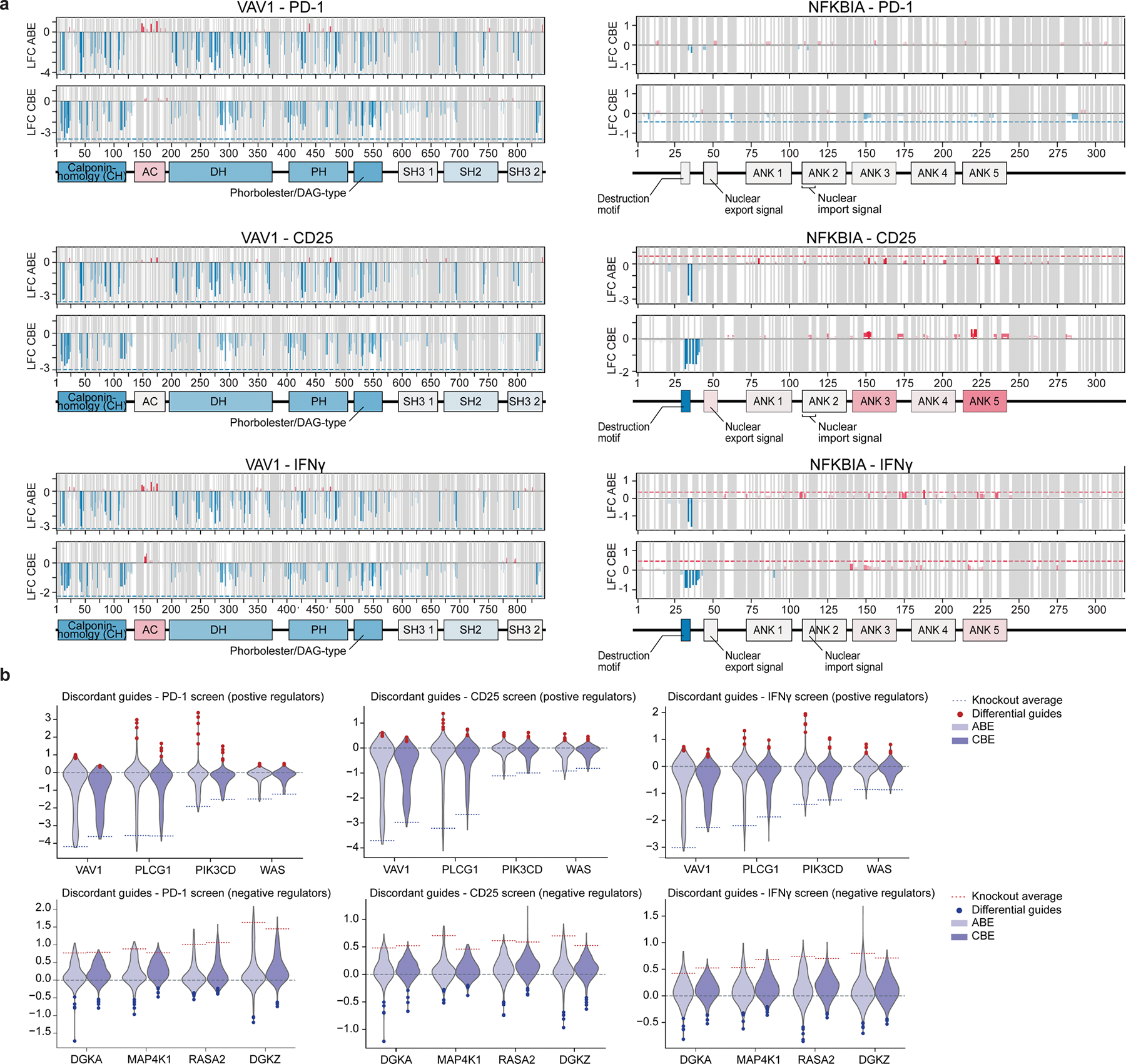
We then designed a library with around 117,000 unique sgRNAs tiling across the coding regions of 385 genes implicated in T cell activity for systematic discovery of mutations that tune T cell activation (Supplementary Tables 1 and 2). The library includes guides targeting every PAM site across the coding regions of the selected genes. The targeted genes encode canonical T cell receptor (TCR) signalling pathway components, T cell transcription factors, co-stimulatory and co-inhibitory proteins, and genes that have been nominated by previous CRISPR-based (CRISPR-knockout, CRISPR interference (CRISPRi) and CRISPR activation (CRISPRa)) screens to have key roles in T cell function1–3,5,18. Primary human CD4+ T cells were isolated, stimulated, transduced with the sgRNA library and with ABE or CBE, and then expanded ex vivo. The pool of base-edited cells was restimulated and then sorted by FACS into bins based on intracellular cytokine production or surface markers of T cell activation (Fig. 1a). Specifically, we performed screens for regulators of IL-2 production, TNF production, CD25 (also known as IL2RA) surface expression, and PD-1 surface expression, based on guide enrichment in high and low FACS bins. To confirm the robustness of the screens, we analysed guides predicted to terminate each gene prematurely (functional knockouts). These base-editing knockouts result from ablation of start codons or splice sites (ABE and CBE) or by the creation of a stop codon (CBE only). We quantified the effects in each screen of the average of the top three sgRNAs (by effect size) predicted to introduce a knockout mutation in the first half of each protein. Notably, knockout of well-known genes involved in TCR signalling—including multiple members of the CD3 family, ZAP70, LCP2 (encoding SLP-76) or LAT—caused decreased TNF production, with high correlations between results from three independent human donors (Fig. 1b and Extended Data Fig. 2a). Conversely, knockout of known negative regulators of T cell activation (such as RASA2, DGKZ and CD5) reproducibly increased TNF production. Furthermore, sgRNAs targeting CD25 and PD-1 were depleted in the CD25 and PD-1 high bins, respectively (Fig. 1c), and functional effects of individual sgRNAs were highly reproducible across different donors for all ABE and CBE screens (Extended Data Fig. 2a). Additionally, we observed substantial overlap of sgRNA hits across screens, representing a core set of regulators related to general T cell activity (Extended Data Fig. 2b). Together, these data confirmed that pooled base edits could be introduced into human T cells for massively high-throughput screening and that their functional effects on activation responses could be assessed reproducibly with a forward-genetic approach.
Fig. 1 |. Base editing screens in primary human T cells identify mutations with effects on activation and cytokine production.
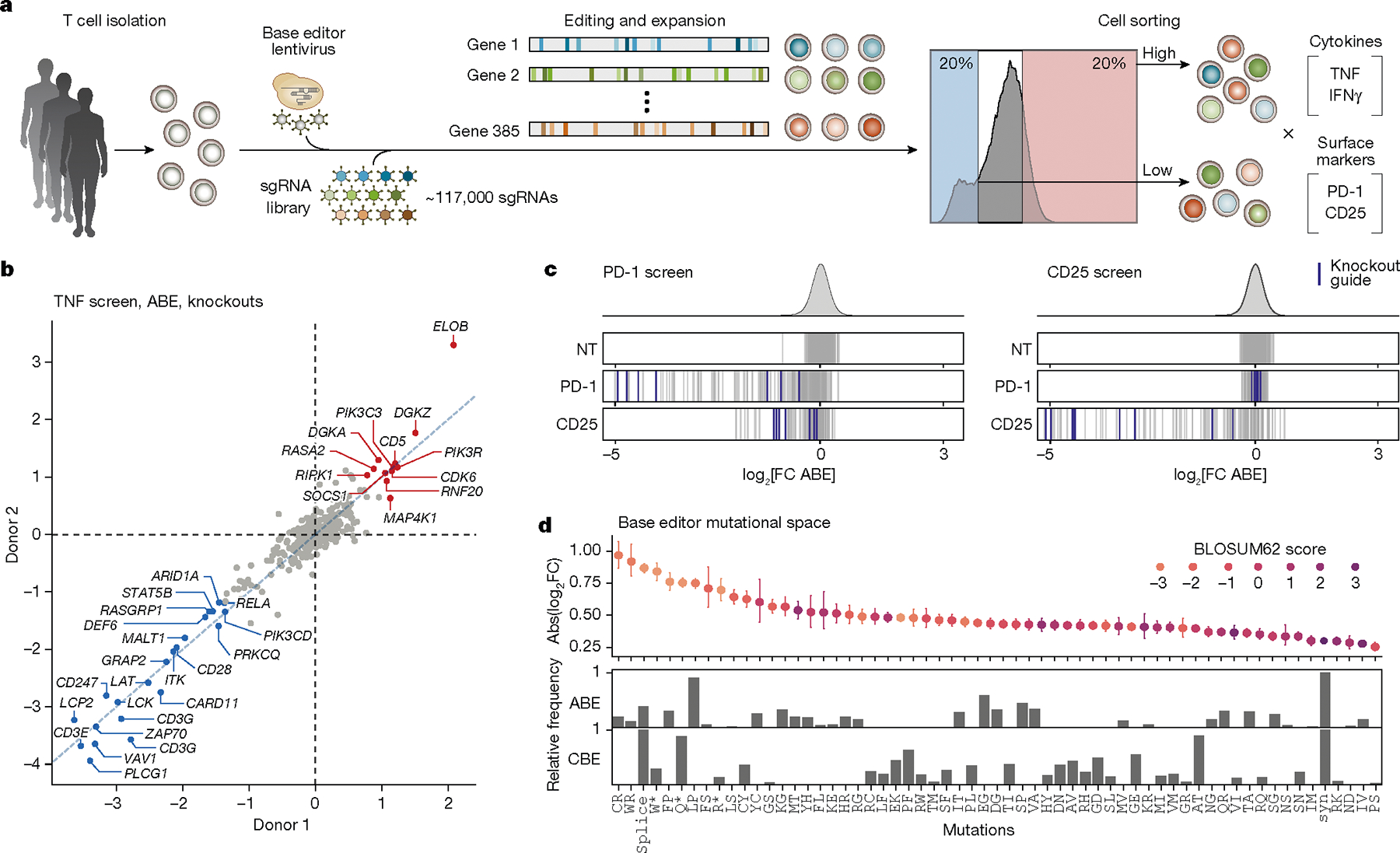
a, Schematic of the ABE and CBE base editor screens performed in primary T cells from three human donors. b, Scatter plot of average mean log2-transformed fold change (log2FC) of top three knockout-inducing sgRNAs (guides that ablate a start codon or edit a splice site within the first half of the coding sequence) for each gene, comparing ABE screens in two donors. Negative regulators of TNF production are shown in red, positive regulators are shown in blue. c, log2FC (high/low sorting bins for PD-1 or CD25 cell surface expression) for individual guide RNAs (vertical lines) directly targeting PD-1 (encoded by PDCD1) or CD25 (encoded by IL2RA) or non-targeting controls. Vertical purple lines indicate guides that are predicted to cause functional knockout (premature gene termination). d, Top, the spectrum of predicted ABE- and CBE-induced mutations for guides targeting genes with demonstrated effects on TNF production (knockout effect size > 1.0 log2FC) is plotted against effect size (mean absolute log2FC ± 95% confidence interval in all screens, from n = 3 donors) and coloured by biological substitution likelihood (BLOSUM62 score19). Frequency of each mutation type in ABE (middle) and CBE (bottom) screens.
We next compared all guides targeting the same gene to evaluate the spectrum of functional effects caused by distinct base edits. Guides predicted to induce nonsense or non-conservative missense mutations (based on BLOSUM62 score19) tended to have larger effect sizes than those predicted to introduce synonymous or more conservative missense mutations, and base edits with the largest effect sizes were enriched in structured regions of proteins as predicted by AlphaFold20,21(Extended Data Fig. 3). These results demonstrate that base-editing screens can recapitulate the expected biochemical effects of the mutational spectrum (Fig. 1d).
Tiling guides across protein sequences using both ABE and CBE enabled us to map domains on the basis of hotspots of editing effects. We plotted the effects of base edits, excluding guides that were predicted to terminate proteins or introduce synonymous mutations, across gene sequences for selected genes in the TNF screen (Fig. 2) and across all screens. We used two techniques to deconvolute the effects of multiple guides targeting a single site, as well as a single guide spanning multiple sites. First, we calculated a simple guide level average across all amino acid residues (Fig. 2a), and second, we adopted a multiple linear regression approach to estimate the effect sizes and significance of each edited base (Extended Data Fig. 5a,b). The residue-level and base-level analyses proved highly similar (Extended Data Fig. 5c). Additionally, we used these methods to assess whether our hits arose from predicted off-target effects of guide RNAs or potential editing-window mis-annotation and saw essentially no difference with the use of off-target filtering or an expanded window for ABE (Extended Data Fig. 5b).
Fig. 2 |. Tiling base edits reveal functional protein domains across gene coding sequences.
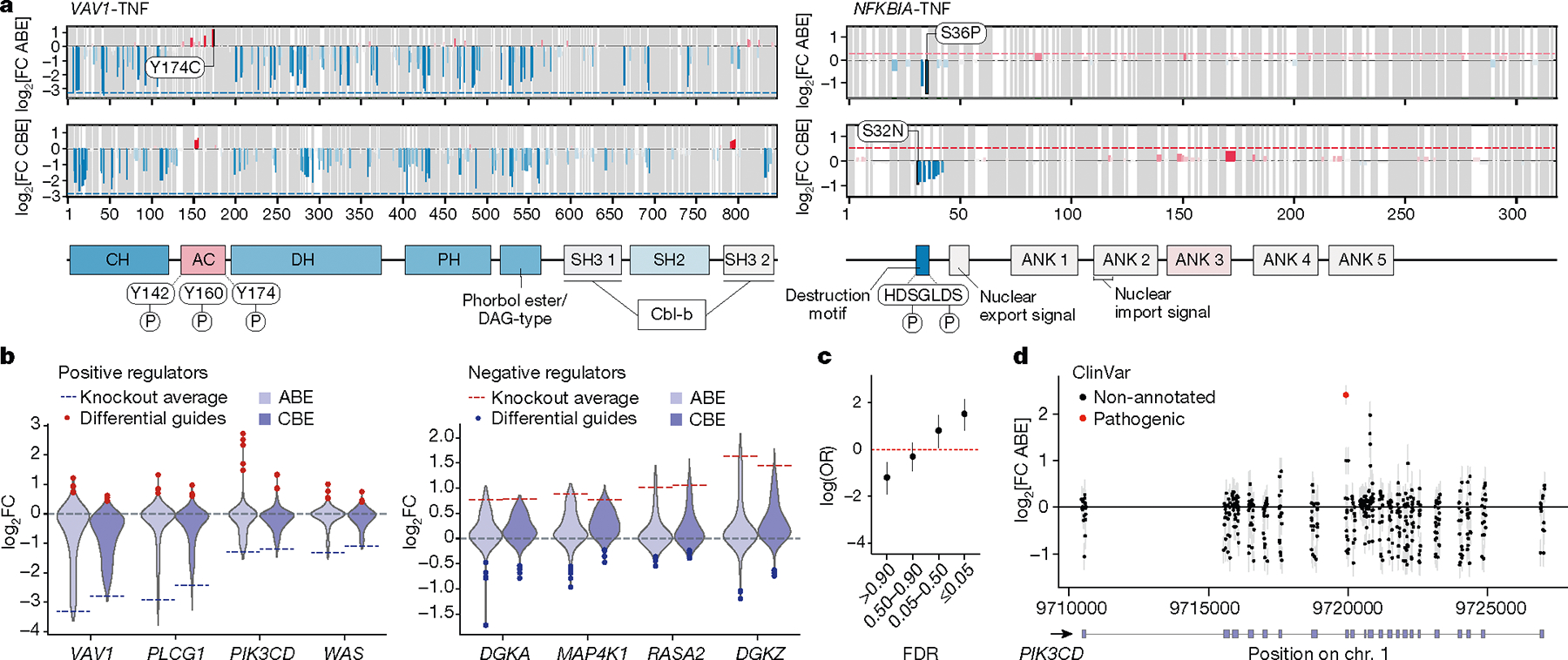
a, Average effects log2FC of non-synonymous, non-terminating base edits at each residue (top, ABE; bottom, CBE) on TNF production are plotted across open reading frames encoding VAV1 (left) and NFKBIA (right). The dotted line indicates the average log2FC of the top three terminating (knockout) guides in the first half of the coding sequence, with blue for negative effects and red for positive effects on TNF levels. Annotated domains from UniProt are shown below and are coloured by the average effect of guides targeting those regions. Known phosphorylated residues within domains with gain-of-function mutations (discordant with knockout effect) are mapped. AC, acidic region; ANK, ankyrin repeats; CH, calponin homology; DAG, diacylgycerol; DH, Dbl homology domain; PH, pleckstrin homology domain; SH2, Src homology 2 domain; SH3, Src homology 3. b, Violin plots show distribution of log2FC of CBE and ABE guides tiling positive (left) and negative (right) regulator genes of TNF. Dotted lines indicate the average log2FC of the top three terminating (knockout) guides in the first half of the coding sequence. Blue and red dots indicate guides with strongly opposing effects with respect to the knockout guides for the same gene (discordant guides). c, Base-editing variants in the TNF ABE screen (from n = 3 donors) binned by MAGeCK guide-level false discovery rate (FDR) are plotted with the log odds ratio of overlap with ClinVar ‘pathogenic’ variants. d, Base-editing variants from the PD-1 screen (from n = 3 donors) across the PIK3CD locus. Those with an exact base-level match in ClinVar are coloured red. c,d, Error bars represent 95% confidence intervals.
The distribution of base-edit effects clearly identified known and putative protein domains in which mutations have strong effects on T cell function. Of note, we found localized domains where base edit effects were discordant with the effect of protein knockout, for example in VAV1 and NFKBIA (Fig. 2a). The cluster of negative edits near the N terminus of NFKBIA pinpoints its destruction motif22, where mutations are predicted to impair degradation of this negative regulator and thus reduce TNF production in edited cells. Similarly, VAV1 mutations largely cause negative effects on T cell function, as expected for a well-known positive regulator of T cell activation, but edits to the acidic domain that autoregulates protein function enhanced TNF production in our screen. Across these genes, the majority of functional sgRNAs disrupted protein function to varying degrees, and the direction of their effects was concordant with those of knockout-inducing sgRNAs. However, the screens indicated a range of quantitative effects of different mutations, suggesting generation of an allelic series with multiple hypomorphic alleles with different effect sizes. Among the genes in our screen, we discovered notably broad spectra of allelic effects for variants of DGKA, MAP4K1, RASA2, DGKZ, VAV1, PLCG1, PIK3CD and WAS, with a subset of mutations having strong effects discordant with those resulting from protein disruption, indicating discovery of specific gain-of-function variants (Fig. 2b and Extended Data Fig. 4b). Cross-referencing our base edit sites with the ClinVar database showed enrichment for pathogenic and probably pathogenic nonsynonymous variants at sites where base editing had functional effects on cytokine production or activation marker expression, including a PIK3CD gain-of-function mutation (Fig. 2c,d and Extended Data Fig. 5d). Thus, base editing screens uniquely enable systematic mapping of alleles—both loss of function and gain of function—that tune human T cell activation responses. Functional base-edited alleles can pinpoint key protein domains and help to assess sites of natural genetic variants for pathogenicity.
Functional testing of selected alleles
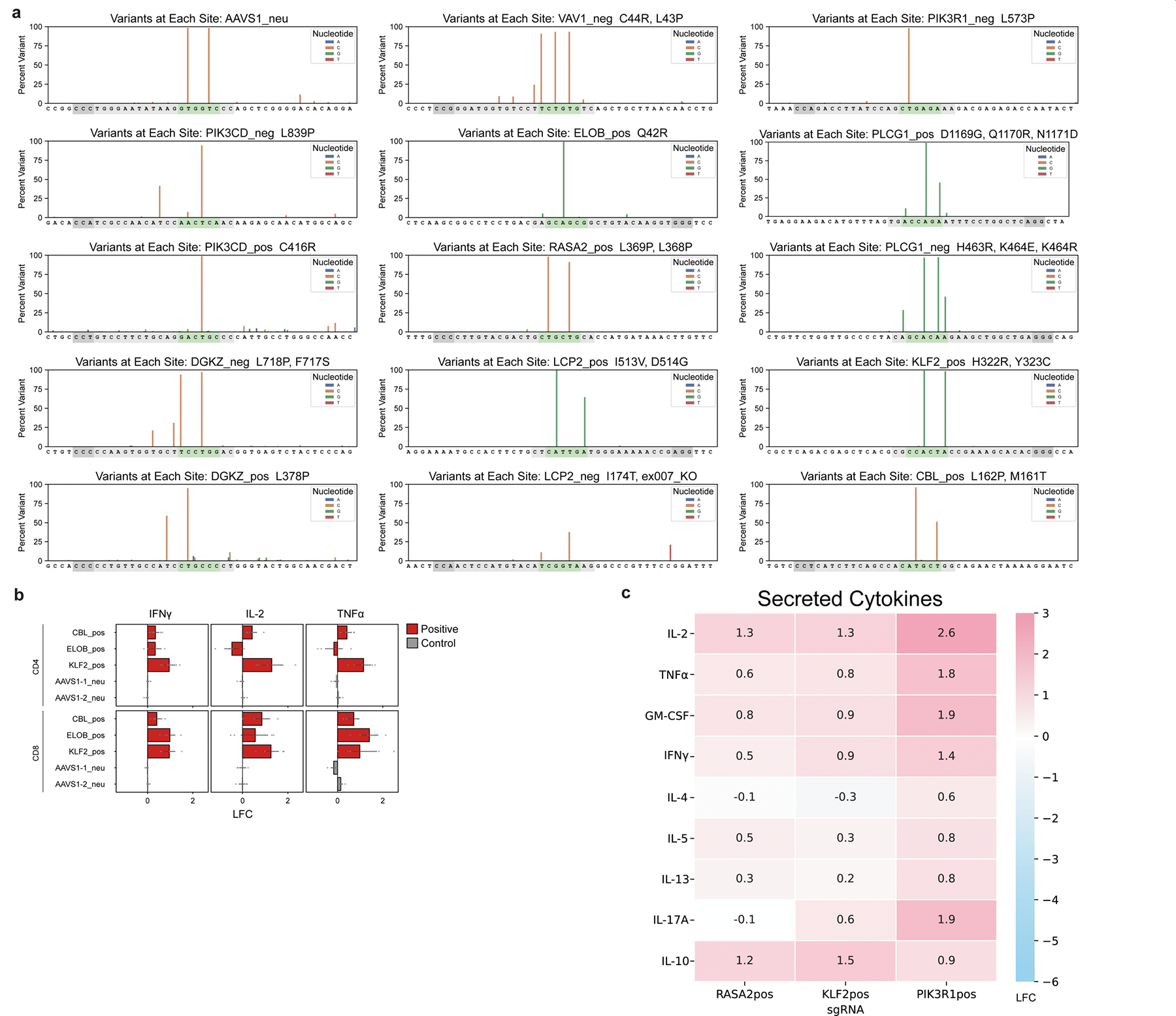
We selected a subset of functional sgRNA hits from our pooled screens to assess more deeply as individual perturbations in arrayed experiments. We base-edited T cells by co-electroporation of ABE8e (V106W) mRNA and synthetic sgRNA15,23–25, providing orthogonal validation for our initial experiments with lentiviral transduction (Fig. 3a). We term guide RNAs enhancing T cell activation (including cytokine production) ‘positive’ sgRNAs, and refer to those dampening activation responses as ‘negative’ sgRNAs. Notably, we assessed both positive and negative sgRNAs nominated by the pooled screens for DGKZ, LCP2, PIK3CD, PLCG1 and VAV1 to confirm discordant functional effects of distinct alleles.
Fig. 3 |. Validation and functional characterization of allelic spectra in key genes.
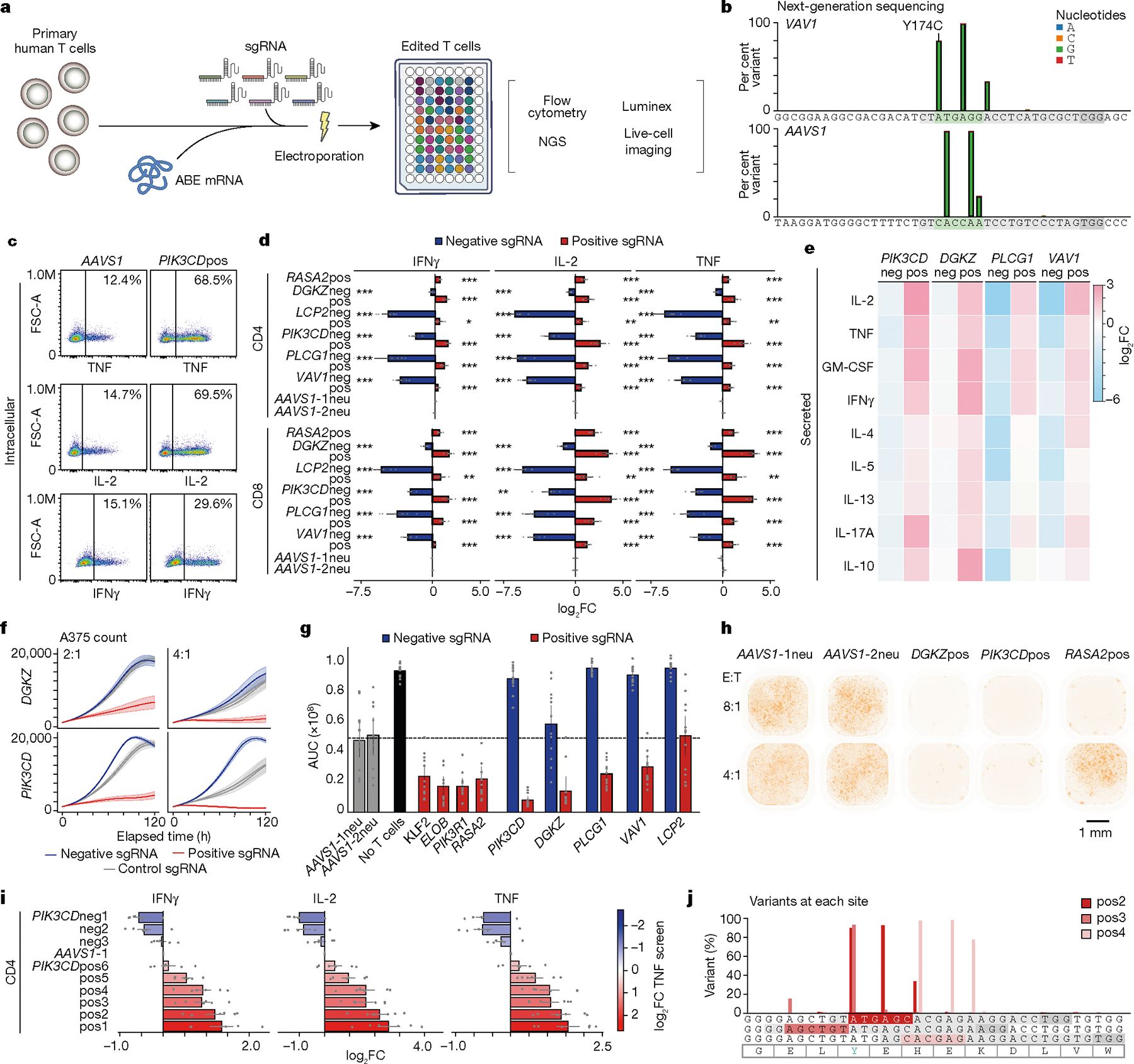
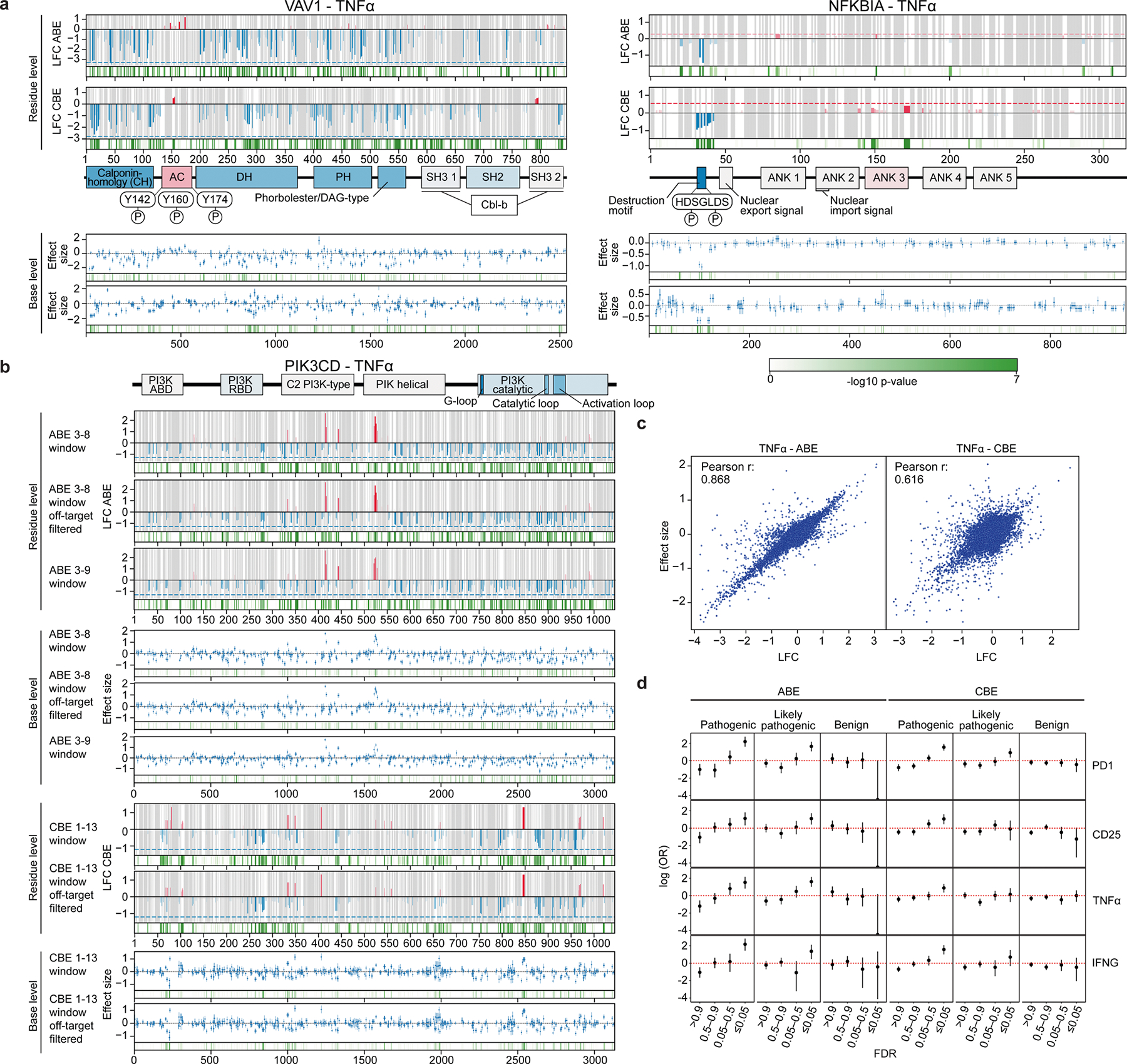
a, Schematic of arrayed validation by co-electroporation of ABE mRNA and synthetic sgRNA. b, ABE base editing at a positive VAV1 (VAV1pos sgRNA) site and AAVS1 control site verified by deep amplicon sequencing and analysed with Crispresso254. Predicted editing window in green, guide sequence in grey, and NGG PAM in dark grey. c, Representative flow cytometry plots for indicated cytokines in control (AAVS1) or PIK3CDpos sgRNA-edited T cells, gated on CD4+ T cells. d, log2FC of intracellular expression of indicated cytokines over control (mean of two AAVS1 guide RNAs) measured by flow cytometry. Negative sgRNAs in the original screen are in blue, positive sgRNAs are in red. n = 6; *P < 0.05, **P < 0.01, ***P < 0.001. pos and neg indicate sgRNAs with positive and negative effects on T cell activation responses, respectively. e, Cytokine secretion in culture supernatants for base-edited T cells measured by Luminex. The heat map represents log2FC over mean of cells edited with two AAVS1 controls. n = 4. f, Cytotoxicity (A375 cell killing) of antigen-specific T cells base edited with control, positive or negative guide RNAs targeting DGKZ or PIK3CD measured by Incucyte imaging over time. n = 6. g, Area under the curve (AUC) of A375 cell counts over time when co-cultured with base edited T cells (x axis) at an effector:target (E:T) ratio of 4. n = 6 donors in technical duplicates. h, Imaging of A375 cells co-cultured with base-edited, antigen-specific T cells using indicated guides and E:T ratios after 120 h. i, Change in production of indicated cytokines in arrayed validation for nine PIK3CD guides relative to AAVS1 control in CD4+ T cells; colour indicates the guide log2FC value in the original ABE TNF screen. n = 6. j, Sequencing of base edits clustering around Y524 (green) show distinct mutations. All n values refer to the number of human donors. Data are mean ± s.e.m. Two-tailed independent two-sample t-test.
First, we assessed the editing efficiency of selected sgRNAs and confirmed very high editing efficiency within defined editing windows as well as occasional off-window editing in positions adjacent to the predicted window (Fig. 3b and Extended Data Figs. 6a and 8b). Next, we assessed production of the cytokines TNF, IL-2 and IFNγ under different strengths of T cell stimulation. We observed high correlations between the effects of individual base edits and the pooled screen results (Fig. 3c,d and Extended Data Figs. 6b and 7). The positive and negative sgRNAs for DGKZ, LCP2, PIK3CD, PLCG1 and VAV1 had discordant effects, tuning cytokine production in opposite directions upon T cell stimulation. We were able to assess more deeply the functional effects of each base edit on tuning stimulation-dependent T cell responses including extracellular secretion of a broad array of cytokines (with Luminex beads; Fig. 3e and Extended Data Fig. 6c) and cell surface expression of multiple markers of activation (by flow cytometry; Extended Data Fig. 7). Together, these results demonstrate the broad effects of targeted base edits in shaping T cell activity and confirm that the base-editing mutagenesis screens identify novel alleles that modulate T cell function bidirectionally.
Given the potent effects of base edits in tuning T cell responses to stimulation, we next explored whether the positive and negative base edits are also sufficient to modulate T cell cytotoxic function. We transduced human T cells with the 1G4 TCR recognizing the NY-ESO-1 cancer-associated antigen presented by HLA-A226 and electroporated cells with ABE8e mRNA and synthetic sgRNAs in arrayed format. The cytotoxic function of base-edited, antigen-specific cells was assessed by co-culturing them with nuclear RFP-labelled A375 melanoma cells that naturally express NY-ESO1 in an HLA-A2 context and performing live cell imaging. Remarkably, negative and positive sgRNAs targeting PIK3CD, DGKZ, PLCG1, VAV1 and LCP2 could dampen or enhance ex vivo cytotoxic function, respectively, relative to control sgRNAs (Fig. 3f,g and Extended Data Fig. 9). A gain-of-function, positive base edit in PIK3CD enhanced T cell cytotoxic function even more potently than a protein-ablating base edit in RASA2—a gene encoding an intracellular T cell checkpoint, identified in our previous CRISPR knockout screens2 (Fig. 3g,h). These co-culture cytotoxicity assays coupled with live-cell imaging further underscore the capacity of specific base edits identified through high-throughput mutagenesis to tune complex primary immune cell functions both negatively and positively.
Forward genetic studies have the potential to uncover allelic series with a spectrum of different quantitative effects on phenotype. We next asked whether we could use the predicted log2FC value from the screen to create an allelic series with a spectrum of gain- or loss-of-function potency. We chose nine sgRNAs targeting PIK3CD on the basis of results of the pooled TNF screen with observed log2FC values (TNF high/low) ranging from −2 to 2.7 and individually measured their effects on T cell cytokine production by flow cytometry. As predicted, we observed a gradient of effect sizes on IFNγ, IL-2 and TNF production that were concordant with screen results (Fig. 3i and Extended Data Fig. 8c). This functional allelic spectrum was not explained by effects on transcript abundance or differential editing efficiency of the guide RNAs. Real-time quantitative PCR and deep sequencing of genomic DNA confirmed stable transcript expression and high on-target editing efficiencies as well as generation of distinct mutations for each of the PIK3CD sgRNAs that cluster around Y524 (Fig. 3j and Extended Data Fig. 8b,d). Despite the substantially lower efficiency (and higher resulting genetic heterogeneity) of knock-in approaches compared to base edits (Extended Data Fig. 8g), we used orthogonal CRISPR-based HDR to introduce defined amino acid changes as further validation of our base-editing results. These experiments were generally consistent with the findings from base editing and demonstrated that nonsynonymous substitutions introduced by HDR at the identified positive sites in PIK3CD can induce the expected gain-of-function phenotypes in PIK3CD, evidenced by trends toward increased cytokine expression and cell surface activation marker expression (Extended Data Fig. 8e,f). Together, these findings demonstrated that base-editing screens can define allelic series of different mutations to precisely fine-tune T cell activity.
Of note, this allelic series nominated by systematic mutagenesis included two sites of naturally occurring PIK3CD mutations that have been identified in patients with primary immune dysregulation. First, the gain-of-function C416R substitution in PIK3CD (introduced by the pos1 sgRNA) was described in patients exhibiting susceptibility to infection, cancer and elevated IgM levels27–30. Furthermore, the sgRNAs pos2, pos3 and pos4 centre around a distinct PIK3CD(Y524S) gain-of-function variant reported to cause activated phosphoinositide 3-kinase δ syndrome30 (APDS). This further confirms that base-editing mutagenesis in primary immune cells can detect pathogenic mutations that cause immune dysregulation in patients and thus also has the potential to clarify the function of variants of unknown significance (Fig. 2d).
High-resolution NG base-editor screen
Our base-editor screens with wild-type-derived nCas9 revealed functional protein domains and hotspots in key T cell genes; however, their resolution is limited by the availability of guide RNAs across the gene. To increase amino acid coverage from around 30% to more than 60% (Extended Data Fig. 10a), we established an ABE base-editor screening platform with NG PAM dependency31,32 (SpG ABE). We demonstrated the ability to use NGG or NG PAM sgRNAs targeting CD3, CD5 and CD7 in conjunction with ABE8e(V106W) fused to SpG Cas9 (Extended Data Fig. 10b). SpG ABE was able to introduce base edits at high efficiency at eight out of nine loci with on-target editing, except for one sgRNA with an NGG PAM that had worked efficiently with Cas9 NGG (Extended Data Fig. 1). Next, we selected 58 genes largely from the NGG base-editor screen for TNF production and designed an sgRNA library tiling those genes with NG PAM restriction, resulting in on average around four times more guides per gene compared with the original screen (Supplementary Tables 4 and 5). Following a similar editing protocol as for the NGG screen, we then sorted either CD4+ or CD8+ cells for high and low TNF production and assessed guide enrichment. Functional sgRNAs were highly reproducible across two donors (Extended Data Fig. 10c), and we found high correlation between NGG and NG screens for guides present in both (Extended Data Fig. 10d).
The screens with SpG ABE enabled us to obtain functional data on a highly expanded set of alleles compared with what was possible for the same genes with the NGG PAM screen (Fig. 4). Although NGG PAM screens confirmed established functional domains, SpG mutagenesis successfully identified functional domains that were less easily appreciated in the lower-resolution data. For example, SpG mutagenesis revealed functional mutations across the WH1 domain of the Wiskott–Aldrich syndrome protein (WAS), where the majority of mutations linked to Wiskott–Aldrich syndrome (characterized by immune dysregulation and thrombocytopenia) are located33. SpG mutagenesis also revealed functional mutations across the RING domain of CBL E3 ubiquitin ligase. In addition, the screens highlighted other protein domains that have not been well-annotated, including a hotspot of base edits in DGKZ that negatively tune TNF production and a hotspot of base edits in RHOH that increase TNF production (Fig. 4).
Fig. 4 |. NG PAM Cas9 base editors enable high-resolution screens.
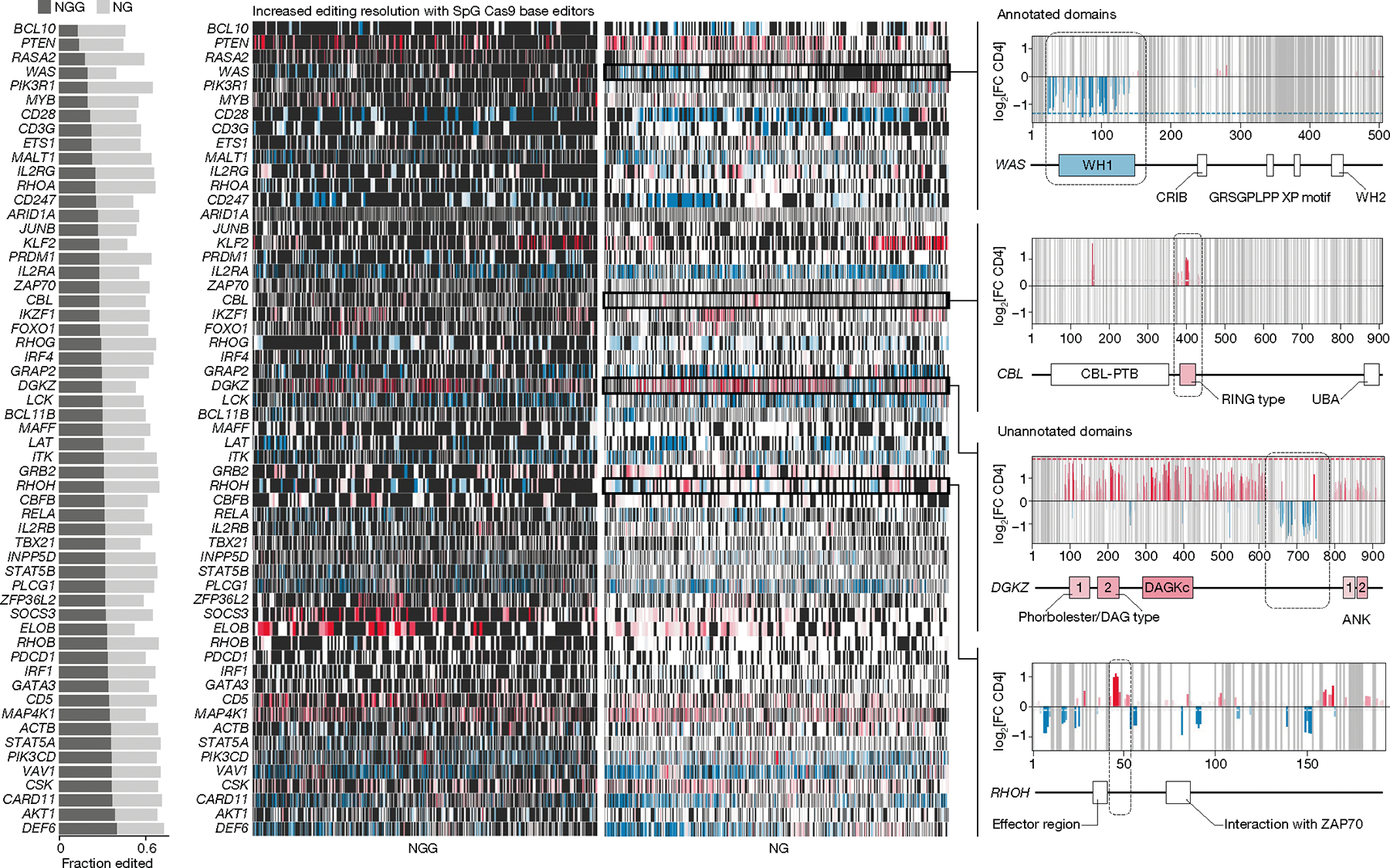
Comparison of base-editing resolution between NGG PAM- and NG PAM-dependent Cas9 for 57 genes overlapping between libraries. Left, the fraction of editable residues with NGG PAM compared with NG PAM ABE base editors. Centre, heat maps of effects of base editing across coding sequences for each gene (rows); black indicates an absence of data (uneditable residue), white is an editable residue with no observed effect, red indicates a positive effect, and blue indicates a negative effect. Right, detailed results for base edits across WAS, CBL, DGKZ and RHOH, with domains confirmed by base editing (WAS and CBL) or suggested as novel functional domains (DGKZ and RHOH) boxed and indicated on the gene plots. X-axis labels on far right plots show amino acid positions. CRIB, Cdc42- and Rac-interactive binding motif; DAGKc, diacylglycerol kinase catalytic domain; PTB, phosphotyrosine-binding domain; WH, WASp homology domains.
We next assessed the relationship between our functional data and known protein structure. We superimposed functional data based on amino acid perturbations across PIK3CD on a known crystal structure of the protein34, revealing three-dimensional regions with hotspots of critical residues where mutations negatively tune TNF production (concordant with the effect of PIK3CD knockout) (Fig. 5a, catalytic loop). SpG mutagenesis also provided a high-resolution functional picture of a 3D pocket where gain-of-function PIK3CD mutations are clustered (with effects on TNF production that are discordant with PIK3CD knockout). PIK3CD was co-crystallized with the PIK3R1 regulatory subunit of PI3K, which was also mutagenized in our screens. The SpG base editor pinpointed specific residues in PIK3R1 that interact with PIK3CD that could also be mutated to enhance T cell activation. Indeed, the effect of base edits targeting these sites in PIK3R1 could increase production of TNF, IL2 and IFNγ (Fig. 5b,c). Base-editing mutagenesis can therefore provide functional insights into critical residues and domains—including protein–protein interaction sites—within 3D protein complex structures in primary human T cells.
Fig. 5 |. High-resolution map of functional residues in a 3D protein complex structure.
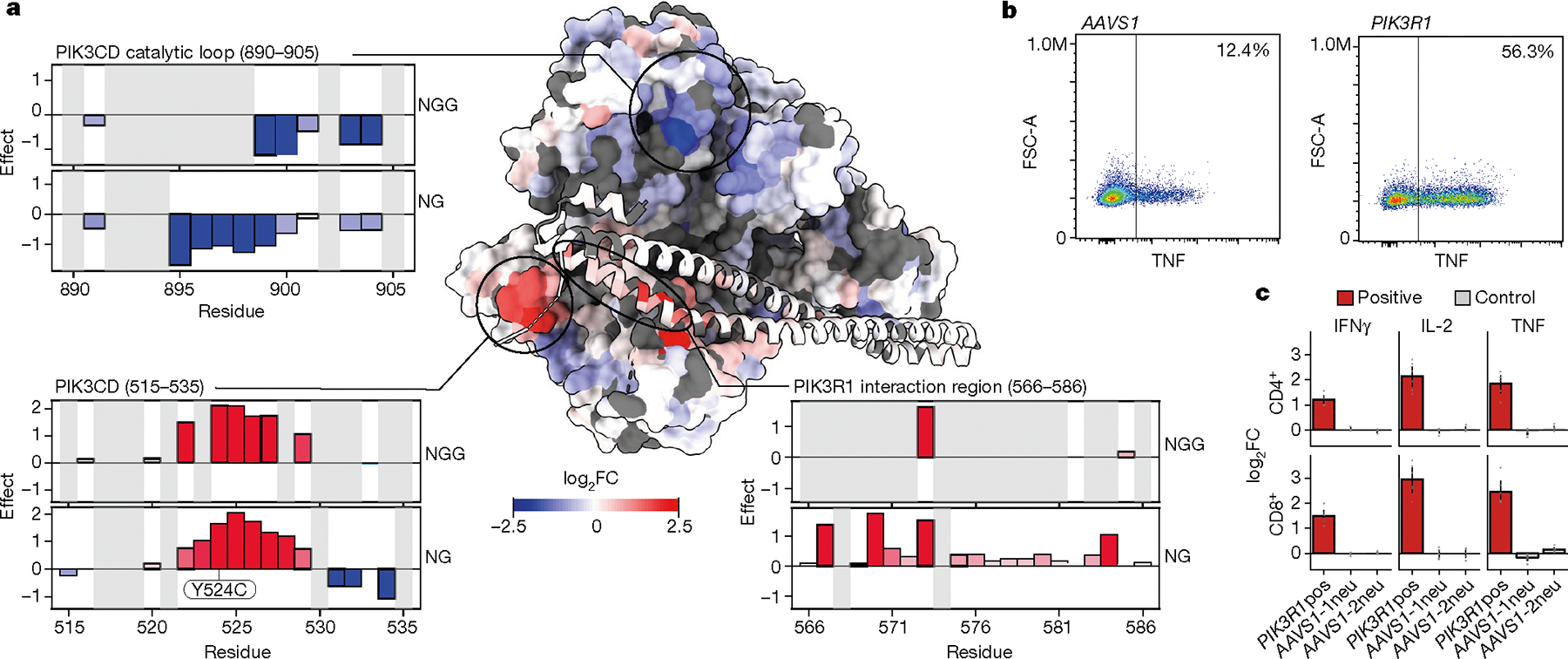
a, Structural model of the PIK3CD–PIK3R1 complex (Protein Data Bank: 7JIS), with residues coloured by log2FC of TNF production from the NG PAM screen. The catalytic loop (residues 890–905) is marked by negative base-editing effects (top left); two other domains—the PIK3R1-interaction region (residues 566–586) (bottom right) and residues 515–535 (bottom left) are marked by positive base-editing effects. b, Flow cytometry plots of CD4+ T cells edited with ABE mRNA and synthetic sgRNA targeting PIK3R1 or AAVS1 (control). c, log2FC in cytokine production for CD4+ or CD8+ T cells edited with positive PIK3R1 sgRNA or AAVS1 controls in arrayed format. n = 6 human donors. Data are normalized to controls and show mean ± s.e.m.
Discussion
Forward genetic studies in model organisms have uncovered a wide range of genetic alleles that tune critical cellular phenotypes, providing transformative insights into biological mechanisms and nominating targets for drug development35. Here we develop a forward genetic platform for massively high-throughput (more than 100,000 sgRNAs) base-editing mutagenesis coupled with complex cellular selections in primary human T cells to dissect the consequences of genetic variants with nucleotide resolution—both naturally occurring and synthetic—that modulate immune cell responses to stimulation and tune their effector functions. We deployed our base-editing mutagenesis platform in primary human T cells, enabling functional identification directly in cell types that control physiological and pathological immune responses. We focused here on screens to identify alleles that tune T cell activation and cytokine production. The platform can be applied to a diverse set of selections with various cellular conditions and is adaptable for studies in additional cell types in the immune system and beyond36. Recently, single-domain, dual-base editors have been developed that possess similar efficiency to the ABE and CBE editors used in this work and could streamline future screens by introducing more missense mutations per guide than ABE or CBE alone37. Although we focus here on mutagenesis of around 400 protein-coding genes, we also foresee opportunities to dissect the cellular functions of noncoding sequences36,38,39.
Base-editing screens offer new opportunities to decode the genetic basis of human diseases. Our systematic forward genetic approach to identify alleles that modulate T cell function nominated variants in PIK3CD that occur naturally in patients and cause the primary immune dysregulation syndrome APDS, which is associated with T cell effector differentiation and senescence, potentially owing to increased susceptibility of PI3K and MTOR signalling pathways to T cell stimulation27,40–42. Furthermore, clinical subtypes of APDS encompass both APDS1—caused by a hyperactivating mutation in PIK3CD—and APDS2—which results from a loss-of-function splice mutation in PIK3R142. These complementary mutations mimic the findings of our study, showing that mutation of specific domains of PIK3CD, including the PIK3R1 interaction domain, are phenocopied by mutations in PIK3R1, which based on the crystal structure engage in the protein–protein interaction. Thus, base editing can functionally dissect protein domains and define mutation sites liable to cause concordant or discordant effects on T cell function, including in physiological settings such as immune dysregulation diseases. Additionally, we highlighted a functional hotspot in WAS, which overlaps with the region where mutations commonly occur in individuals with Wiskott–Aldrich syndrome. Conducting these functional studies directly in the cell types relevant to disease pathogenesis—in this case human T cells—is crucial to accurately defining variant effects on the biological processes active in disease. Systematic mutagenesis studies have the potential to provide mechanistic insights into rare and common disease-associated variants and could help to prioritize likely pathogenic variants among variants of unknown significance in sequencing data from patients, as well as somatic cancer mutations such as those found in VAV1 and PLCG143,44. Indeed, large-scale analysis of the ClinVar variants present among our base-edit sites demonstrated significant enrichment for pathogenic and probably pathogenic variants among mutation sites with effects on T cell functions. Thus, cross-referencing base-resolution datasets such as those in this study and future base-editing level screens will be a valuable tool in prioritizing and interpreting natural variants associated with Mendelian and complex genetic diseases.
Our screens nominated a spectrum of alleles that quantitatively tuned T cell responses. In some cases, these alleles overlapped with naturally occurring patient mutations, but we also identified synthetic alleles that tune T cell activity. Notably, we identified nine PIK3CD alleles—including both loss-of-function and gain-of-function variants—that could tune T cell cytokine production, upon single or double nucleotide changes, over a dynamic range of more than 3 log2FC. Such precise control of T cell activation is a key step in the development of next-generation immunotherapies, including CAR T cells and other adoptive cellular therapies, where safety and efficacy can be limited by both excessive and insufficient activity45–48.
Base-editing mutagenesis screens complement an existing pipeline of functional genetic technologies in human immune cells. In recent years, we and others have optimized genome-wide screens with CRISPR nuclease, CRISPRi and CRISPRa to identify genes that influence human T cell functions when they are ablated, repressed or overexpressed1–6,49–51. We are now able to use the power of pooled sgRNA perturbation libraries coupled with base editors to introduce targeted mutations throughout these key genes and identify functional allelic variants. Looking forward, complementary technologies can be deployed for deeper mutagenesis at sites nominated by base-editing screens, including pooled knockins or EvolvR to test the full complement of possible synthetic mutations to identify optimal variants to engineer into T cell products52,53. Together, these studies will nominate alleles that can be engineered into immune cell therapies, facilitate design of engineered protein and RNA therapies, and accelerate small molecule discovery campaigns by prioritizing promising functional pockets in target complexes. Base-editing mutagenesis will deepen our understanding of molecular mechanisms in human immunology and provide a transformative tool for immunotherapy development.
Online content
Any methods, additional references, Nature Portfolio reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at https://doi.org/10.1038/s41586-023-06835-6.
Methods
Lentiviral plasmids and cloning for ABE and CBE base editors
The lentiviral transfer plasmids containing base editors were generated using pZR071 (Addgene 180264) as a backbone. The pZR071 plasmid was digested with EcoRI and KpnI, followed by ligation of a cassette composed of the EF1a promoter, base editor coding sequence, and a P2A-Blasticidin resistance element. For the plenti-CR033 transfer plasmid, the ABE8e(V106W) coding sequence was obtained from ABE8e(TadA-8e-V106W) (Addgene 138495). For the plenti-CR029 transfer plasmid, the evoCDA1-BE4max sequence was derived from pBT277_evoCDA1-BE4max-in-mammalian-cells (Addgene 122608). For the plenti-CR102 transfer plasmid, the nCas9 from plenti-CR033 was replaced with SpG, which was sourced from pCAG-CBE-4max-SpG-P2A-EGFP (RTW4552) (Addgene 139998).
Library cloning
A combination of literature review, gene ontology annotations and hits from previous CRISPR screens in T cells1–3,5,53 led to a list of 385 genes implicated in regulation of T cell activation, differentiation and function (Supplementary Table 1). For each gene we choose the longest annotated isoform and selected all NGG PAM guides which: (1) overlapped with the coding sequence plus 2 bp on the side of each exon to include splice sites; and (2) contained an editable base (A or C) in the expected editing window (position 3–8 for ABE and 1–13 for CBE). The addition of 1,000 non-targeting sgRNAs derived from the Brunello library55 resulted in a list of 117,249 unique sgRNAs. (Supplementary Table 2) We added type II restriction sites and sequences for amplification, as previously described55. The oligonucleotide pool was obtained from Agilent and amplified with KAPA HiFi HotStart (Roche 07958935001). The PCR product was purified and ligated into LentiGuide-Puro plasmid (Addgene 52963) backbone with the New England Biolabs NEB Golden Gate Assembly Kit (BsmBI-HF v2) (New England Biolabs, E1602L) per the manufacturer’s instructions. Ligation product was transfected into Lucigen Endura electrocompetent cells (LGC, 60242–2) per the manufacturer’s instructions. Cells were expanded at 30 C for 18 h and plasmid was extracted using the QIAGEN Plasmid Plus Mega Kit (Qiagen, 12981). Total amount of transfected cells was calculated by counting colonies of separate ampicillin plates at 1/10,000,000 and 1/100,000,000 dilution. We proceeded with next generation sequencing of the plasmid pool at coverage of >1,000×, confirming representation and distribution of sgRNAs. The plasmid pool was then used for lentivirus production as described under ‘Lentivirus production’. For screens using SpG nCas9, 57 gene hits from the NGG screen and CBLB were selected based on the effect of knockout guides (Supplementary Table 4). Guides were selected for the coding regions of these genes in an identical manner to the NGG library, only for NG PAM guides, an oligonucleotide pool containing 45,941 oligonucleotides was ordered similarly to the first screen using NGG nCas9 (Supplementary Table 5). Library preparation and quality control was performed as described for the NGG library; read counts for quality control sequencing of the libraries used are provided in Supplementary Table 10.
RNA production
For experiments with base editor mRNA, an in vitro transcription (IVT) plasmid containing ABE8e(V106W) with a mutated T7 promoter was designed and cloned as previously described23,56. The IVT templates were produced by PCR of ABE8e(V106W) with the forward primer correcting the T7 mutation and reverse primer appending the polyA tail, thus the final PCR product contained WT T7 promoter, 5′ untranslated region including Kozak sequence, the codon-optimized ABE8e(V106W) coding sequence, 3′ untranslated region, and a 145-bp polyA tail. The PCR product was purified and stored at −20 °C until further use. IVT reactions were performed with the HiScribe T7 High Yield RNA Synthesis Kit (New England Biolabs, E2040S) under full substitution of UTP and in presence of 4 mM CleanCap (TriLink Biotechnologies, N-7113–5), as described23,57. Transcribed mRNA was purified with lithium chloride and eluted in RNA storage solution (Fisher Scientific, AM7000). After quantification on a Nanodrop spectrophotometer and normalization to 1 μg μl−1, mRNA product was assessed on an Agilent 4200 Tapestation system and subsequently stored at −80 °C.
Lentivirus production
Lentiviral particles were produced as previously described3. In brief, for a T75 flask, 12 × 106 Lenti-X HEK 293T cells were seeded in 15 ml complete Opti-MEM. The next morning, cells were transfected with 12 μg transfer plasmid, 12 μg psPAX2 (Addgene 12260) and 5 μg pMD2.G (Addgene 12259) with 56 μl p3000 and 65 μl Lipofectamine 3000 (Fisher Scientific, L3000075). Six hours after transfection, medium was replaced with fresh cOpti-MEM supplemented with 1× viral boost (Alstem, VB100). Another 18 h later, virus containing supernatant was collected and spun down at 500g for 5 min at 4 °C to pellet cell debris. Lentivirus-containing medium was moved to a new vessel and subsequently concentrated 100-fold using Lenti-X Concentrator (Takara Bio, 631232) per the manufacturer’s instructions. Viral particles were stored at −80 °C until further use.
T cell isolation and culture
Human peripheral blood Leukopaks enriched for PBMCs were sourced from Stemcell Technologies (200–0092). The donors were chosen without regard to sex, gender, ethnicity or race. Donors for the screens were <60 years old and non-smokers. T cells were isolated with the EasySep Human T cell isolation kit (100–069) or EasySep Human CD4+ T Cell Isolation Kit (Stemcell Technologies, 100–0696) per the manufacturer’s instructions. Immediately after isolation, T cells were either frozen or used directly for in vitro experiments. Fresh T cells were seeded at 1 × 106 cells per ml and subsequently used for experiments. When frozen, T cells were counted, spun down, and resuspended in Bambanker freezing medium at a concentration of 50 × 106 cells per ml. T cells were stored at −80 °C for up to 2 months or in liquid nitrogen for longer periods. When needed, T cells were thawed and seeded at 1 × 106 cells per ml, similar to fresh T cells. Unless otherwise indicated, T cells were cultured in complete X-VIVO 15 (cX-VIVO) consisting of X-VIVO 15 (Lonza Bioscience, 04–418Q) supplemented with 5% FCS (R&D systems, lot M19187), 4 mM N-acetyl-cysteine (VWR, VWRV0108–25G) and 55 μM 2-mercaptoethanol (Fisher Scientific, 21985023).
Cell lines
Lenti-X HEK 293T cells (Takara Bio, 632180) were cultured in complete DMEM consisting of high glucose DMEM (Fisher Scientific, 10566024) supplemented with 10% FCS (R&D systems, LOT M19187), 1× MEM Non-Essential Amino Acids Solution (Fisher Scientific, 11140050), 1 mM sodium pyruvate (Fisher Scientific, 11360070), 10 mM HEPES (Sigma Aldrich, H0887–100ML), 1,000 U ml−1 penicillin-streptomycin (Fisher Scientific, 15140122). Lenti-X HEK 293T cells were subcultured every 2–3 days and maintained at a confluence of <70% for a maximum of 15 passages. A375-nRFP (a gift from A. Ashworth) cells were cultured in complete RPMI (cRPMI) consisting of RPMI (Fisher Scientific, 21870092) with 10% FCS, 55 μM 2-mercaptoethanol, 2 mM l-Glutamine (Fisher Scientific, 25030081), 1,000 U ml−1 penicillin-streptomycin, 1× MEM non-essential amino acids solution, 1 mM sodium pyruvate and 10 mM HEPES (Sigma Aldrich, H0887–100ML) and subcultured every 2–3 days to keep them at a confluency of <70%. Cell lines were tested negative for mycoplasma contamination.
Arrayed base editing with lentivirus
Frozen human Pan CD3+ T cells were thawed and activated with anti-CD3/anti-CD28 Dynabeads (Life Technologies, 40203D) in presence of 200 IU ml−1 IL-2. The next morning, plenti-CR029 or plenti-CR033 lentivirus was added to the cells at 2% v/v. The following day cells were subcultured 1/1 and IL-2 was replenished. Additionally, lentiguide-Puro with CD3-, CD5- and CD7-targeting sgRNAs were added. One day later, cells were split again 1:1 with half of the cells receiving blasticidin treatment at 10 μg ml−1 and all cells receiving puromycin treatment at 2 μg ml−1. Cells were subcultured and IL-2 was refreshed every two days and assessed by flow cytometry (Supplementary Fig. 2) and Sanger sequencing seven days after initial T cell activation.
For testing our ABE SpG lentiviral base editing approach, we followed a similar strategy with viral particles produced from plenti-CR102.
NGG screen
CD4+ T cells from 3 human donors were isolated as described under ‘T cell isolation and culture’. For ABE screens, 150 × 106 cells per donor were used for subsequent activation in cX-VIVO 15 at a concentration of 1 × 106 cells per ml. For CBE screens, 225 × 106 cells per donor were seeded under similar culture conditions. CD4+ T cell purity was confirmed by flow cytometry (>95%). Cells were subsequently activated with anti-CD3/anti-CD28 Dynabeads (Life Technologies, 40203D) at a 1:1 bead-to-cell ratio and with 200 IU ml−1 IL-2 (R&D systems, 202-GMP-01M). Next morning, 2% v/v plenti-CR033 or 2.2% plenti-CR029 lentivirus was added for ABE and CBE screens, respectively. Two days later, 50% of the original culture volume cX-VIVO was added and IL-2 was supplemented for a final concentration of 200 IU ml−1. lCR005 library lentivirus was added at 1.3% v/v (corresponding to a multiplicity of infection (MOI) ~ 0.3) and cells were mixed. Next day, cells were counted and fresh medium was added to keep the culture at 1 × 106 cells per ml. IL-2 was supplemented to a final concentration of 200 IU ml−1, puromycin and blasticidin were added to 2.5 μg ml−1 and 20 μg ml−1 final concentrations, respectively. Cells were subcultured and expanded every 2 days and kept at ~1 × 106 per ml and 200 IU ml−1 IL-2. Five days after addition of blasticidin and puromycin, cells from each screen were collected, pooled, and counted. Fresh medium without supplements was added to bring T cells to 2 × 106 cells per ml. Next morning, T cells were restimulated with 6.25 μl ml−1 anti-CD3/anti-CD28/anti-CD2 Immunocult (Stemcell Technologies, 10990). For cytokine screens, Protein Transport Inhibitor (Becton Dickinson, 555029) was added at a 1/1,000 dilution 1 h after stimulation, followed by 9 h incubation. Cytokine screen cells were collected 10 h after stimulation and fixed and stained for FACS using the Cytofix/Cytoperm Fixation/Permeabilization Solution Kit (Becton Dickinson, 554714). For PD-1 and CD25 screens, cells were collected one day after activation, stained for Live/Dead and surface protein expression facs sorting and fixed in 4% paraformaldehyde for 30 min at 4 C. T cells were subsequently sorted into TNF, IFN-γ, CD25 or PD-1 high and low bins (Supplementary Fig. 1a). Over the whole course of the screen from initial activation and expansion to sorting, we maintained a coverage of >1,000× (cells/sgRNA) to ensure consistent representation of the library. After staining and fixation, T cells for all screens were washed twice in EasySep (1× PBS with 2% FCS and 1 mM EDTA) and sorted into the respective bin. Sorted T cells were spun down and cell pellets were stored at −80 °C until further use.
NG screen
Fifty million human Pan CD3+ T cells from each two human donors were isolated as described under ‘T cell isolation and culture’. Subsequently, T cells were stimulated with anti-CD3/anti-CD28 Dynabeads at a 1:1 bead-to-cell ratio and 200 IU ml−1 IL-2. Next day, plenti-CR102 ABE8e(V106W)-SpG base editor lentivirus was added at 2% v/v. One day later, T cells were infected with 1.5% v/v (corresponding to an MOI of ~0.6) of the NG base editor library lentivirus followed by selection with blasticidin and puromycin as described before. T cells were subcultured and expanded with fresh medium and IL-2 every 2–3 days and further processed as described under ‘NGG screen’. Over the whole course of the screen from initial T cell activation to sorting, a coverage of >1,000× (cells per sgRNA) was maintained to ensure representation of the library. Finally, human Pan CD3+ T cells were sub-sorted into CD4+ TNF-low and TNF-high as well as CD8+ TNF-low and TNF-high T cells (Supplementary Fig. 1b). Extraction of genomic DNA and quantification of sgRNAs in each population was performed as described under ‘Genomic DNA extraction, library preparation and NGS for base editing screens’.
Genomic DNA extraction, library preparation and NGS for base editing screens
Genomic DNA of sorted T cell pellets was performed as described11. For 5 × 106 T cells, pellets were thawed and incubated overnight at 65 °C in 400 μl lysis buffer (Chip composition) and 16 μl of 5 M NaCl. Next day, 32 μl RNase A (10 mg ml−1 stock concentration) was added and incubated at 37 °C for 2 h followed by a 2 h 55 °C incubation after adding 16 μl (20 mg ml−1) proteinase K. Genomic DNA was then separated with phenol:chloroform:isoamyl alcohol and precipitated with sodium acetate and washed with 70% ethanol. DNA was eluted in water and quantified on a Nanodrop Spectrophotometer. PCR for NGS was performed with KAPA HiFi HotStart polymerase, using previously established primers and PCR protocol55 with annealing temperature at 63 °C. PCR products were purified by solid-phase reversible immobilization and further gel purified, diluted to 10 nM and submitted for NGS on a NovaSeq Instrument (Illumina) at the UCSF Center for Advanced Technologies. Targeted sequencing depth was 1,000× of sgRNA counts per sample.
Real-time qPCR
T cells from four independent human donors were treated as described under ‘Arrayed base editing with mRNA’. One day after restimulation, RNA for expression analyses with qPCR was isolated with the Quick-RNA MicroPrep and RNA Clean and Concentrator-5 Kits (Zymo Research, R1051 and R1016) using TURBO DNase and 10× TURBO DNase Buffer (Invitrogen, AM2239). Real-time qPCR was performed using the Prime-Time One-Step RT-qPCR Master Mix (10007065) with pre-designed primer–probe pairs for PIK3CD, IL2RA and B2M (housekeeping) following the manufacturer’s instructions. Expression relative to B2M was calculated with the 2−ΔΔCT method.
Arrayed base editing with mRNA
For arrayed base editing experiments with ABE mRNA, fresh or previously frozen human Pan T cells were activated with a 1:1 bead-to-cell ratio with anti-CD3/CD28 Dynabeads (Thermo Fisher, 40203D) in the presence of 500 IU ml−1 IL-2 at 1 × 106 cells per ml. Two days after stimulation, T cells were magnetically de-beaded and taken up in P3 buffer with supplement (Lonza Bioscience, V4SP-3096) at 37.5 × 106 cells per ml. Two micrograms of ABE mRNA mixed with 1.5 μg synthetic modified sgRNA (Synthego) was added per 20 μl cells, not exceeding 25 μl total per reaction. Cells were subsequently electroporated on a Lonza 4D Nucleofector using the electroporation code DS137. Immediately after electroporation, 100 μl warm complete X-VIVO15 was added to each electroporation well and cells were incubated for 15 min in a CO2 incubator at 37 °C followed by distribution of each electroporation reaction into three wells of a 96-well flat bottom plate. Each 96 flat bottom well was brought to 200 μl cXVIVO15 and 100 IU ml−1 IL-2. Cells were subcultured and expanded by addition of fresh medium and IL-2 every 2–3 days. Ten days after electroporation, cells were counted and normalized to 2 × 106 live cells per ml through complete medium change and without the addition of IL-2. The next morning, cells were re-stimulated with indicated doses of anti-CD3/anti-CD28/anti-CD2 Immunocult (Stemcell Technologies, 10990). Cell pellets saved for genomic DNA analysis were spun down at 300g for 5 min and resuspended in QuickExtract (VWR, 76081–766) following the manufacturer’s recommended protocol. Next, cells were treated as described under ‘NGG screen’ for cytokine and surface marker flow cytometry and gated for CD4+ and CD8+ cells (Supplementary Figs. 3 and 4). sgRNA sequences used in arrayed experiments are provided in Supplementary Table 11.
Cytotoxicity assays
For in-vitro cytotoxicity assays, fresh or frozen T cells were stimulated as described under ‘NGG screen’. One day after stimulation, cells were infected with 1% v/v 100× concentrated lentivirus carrying an open reading frame element encoding the 1G4 T cell receptor that can recognize the NYESO1 cancer antigen in an HLA-A0201 context. One day later, cells were electroporated and cultured as described under ‘Arrayed base editing with mRNA’. Ten days after electroporation, 300 RFP-expressing A375 melanoma cells were seeded in 50 μl cRPMI per well in a 384 well plate. The outer two positions of the 384 well plate were filled with water and not used for analyses. The next morning, T cells were counted and 1G4 T cell receptor expression was assessed by flow cytometry. TCR positive cells were used for normalization and added to A375 cells according to the indicated E:T ratios. The final volume for one well of a 384-well plate was 90 μl. After adding the T cells on top of A375 cells, the plate was moved to an Incucyte live-cell imaging system (Sartorius) with assessment cycles every 6 h. A375 cells were automatically counted by the Incucyte instrument based on RFP expression and cell counts were exported for plotting.
HDR-mediated knockins
Fresh pan T cells were isolated and activated as described above. Two days after activation, cells were electroporated in presence of Cas9 RNPs and HDR templates consisting of 100-bp single stranded oligonucleotides synthesized by IDT, as described58. Next, cells were expanded and restimulated for cytokine staining as described above and subsequently assessed for cytokine production and activation marker expression, similar to the description in ‘Arrayed base editing with mRNA’. Genomic DNA was isolated from cells and prepared for NGS as described above.
Sorting and flow cytometry
FACS was performed on BD FACSAria Fusion cell sorters equipped with 70 μm nozzles at the Gladstone Flow Core. For each sorting bin, a cell-to-guide RNA ratio of >500 was targeted. Flow cytometry analyses were done using an Attune Nxt machine with plate reader function. Antibodies used for flow cytometry are listed in Supplementary Table 9.
Secreted cytokine analyses
For cytokine analyses in culture medium, cell culture plates were spun down at 500g for 5 min and half of the supernatant was removed and stored at −80 °C. All samples were analysed by Eve Technologies using Luminex xMAP technology. First, a titration series was run to determine optimal dilution factors and to avoid assay saturation. IL-6, IL-8 and IL17A were run at a twofold dilution. All other cytokines were analysed at a 200-fold dilution.
Base-editing screen analyses
After sequencing, fastq files were analysed with MAGeCK59. Samples with poor genomic DNA recovery after sorting (<100× coverage) were excluded from further analysis. First, the command MAGeCK count was used to assess fastq files, representing each sorting bin, for the presence of guideRNA sequences and count them. Next, MAGeCK test was run to assess the distribution of guideRNAs and comparisons across different sorting bins relative to distribution of non-targeting controls. The low bins were used as MAGeCK control input and high bins as treatment input for the NGG screen. The low bins were used as MAGeCK treatment input and high bins as control input for the NG screen. For plotting of the NG screen, log2FC values were inverted. Therefore, a negative log2FC or negative hit indicates enrichment in the low bins while positive hits are enriched in the high bins. For downstream analysis the bottom 2.5 percentile of guides were discarded. As coding regions were selected inclusively based on longest isoforms, for ease of interpretation and analysis, predicted amino-acid-level edit locations were mapped by pairwise alignment to the canonical sequence in UniProt for each protein (Supplementary Tables 1 and 2, NGG screens; Supplementary Tables 4 and 5, NG screens) and the canonical sequence positions used in all further analysis. For correlation of editing effects with BLOSUM62 substitution penalty, the highest penalty mutation was used for guides with multiple predicted mutations and the relative frequencies in Fig. 1d calculated based on the single highest penalty mutation as well.
sgRNA off-target analyses
To assess off target potential, each guide was searched against the human genome for off target sites with up to five mismatches using Cas-OFFinder60. Afterwards, scoring of target sites was performed with the CFD algorithm61,62. Guides were filtered out in off target analysis if there existed >5 off-target sites with a CFD score of 1.012.
Evaluation of base-level effects
To estimate the base-resolution effects in the screen analyses, we first listed all editable bases in the expected editing windows for each guide RNA (A in position 3–8 for ABE, or 3–9 for expanded window analysis and C in position 1–13 for CBE). Next, the effect of each guide RNA in every sample was quantified by dividing the normalized count in the high bin by the normalized count in the low bin. Normalized counts were obtained from the output file of the MAGeCK analysis, as described above. Following this, we log-normalized the effects of all guide RNAs pertaining to each gene and performed a regression analysis based on the presence of editable bases, using a multiple linear regression model. This allowed us to estimate base-level effect sizes as well as statistical significance (two-sided Wald test) for each base in each screen. (Supplementary Tables 6–8) In instances where two bases were covered by the same combination of guide RNAs and thus could not be distinguished, they were assigned identical effect sizes. For residue-level statistics, we averaged log2FC (obtained from MAGeCK) from all non-terminating guides spanning a residue that induces a non-synonymous change, and combined the P values (also from MAGeCK) using Fisher’s method.
To test whether disease-causing human genetic variants were enriched among functional bases in our screening results, we compared our results with the variants in ClinVar63. We downloaded the VCF file for variant functions from ClinVar on May 8th, 2023. Our focus was on variants categorized as ‘pathogenic’, ‘likely-pathogenic’ or ‘benign’ in ClinVar. We restricted our comparison to non-synonymous variants that overlapped with the editing windows of our guide RNAs and had reference/alternative alleles matching our editing (A to G or T to C for ABE, and C to T or G to A for CBE). The overlap between ClinVar variants and variants in our screen filtered to a defined significance threshold was compared with the overlap between ClinVar variants and the remaining variants in the screening using Fisher’s two-sided exact tests.
Statistical and reproducibility
Screen analysis was done with MAGeCK59 as described in the respective screen section. All other statistical analyses (correlation and significance testing) were done with the scipy.stats package.
In Fig. 3h, one representative donor out of six, measured in duplicates, is shown (all data points for an E:T ratio of 4 can be seen in Fig. 3g).
Extended Data
Extended Data Fig. 1 |. Optimization and assessment of base editing efficacy in primary human T cells.
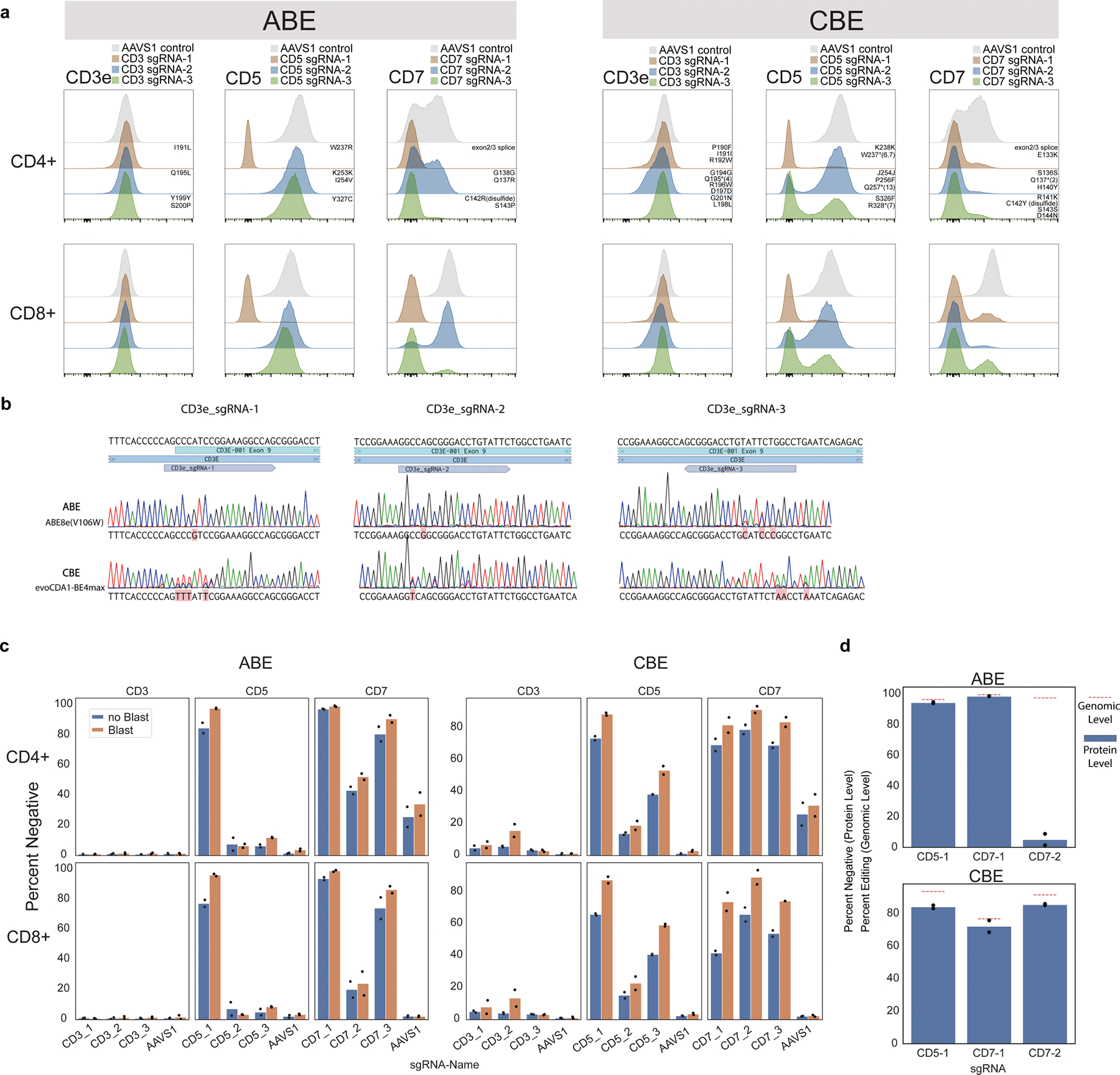
a, Distribution of protein surface expression levels for CD3e, CD5, or CD7 in CD4+ or CD8+ T cells base edited with three sgRNAs targeting each gene or AAVS1 control with ABE (left) and CBE (right). Predicted mutations for each guide are annotated above each distribution in the CD4+ plots. n = 2 independent donors. b, Sanger sequencing traces for ABE and CBE editing using the three CD3e sgRNAs for both ABE and CBE. Consensus sequences are in black above and detected BE mutations are highlighted in red under traces. c, Summary boxplots of T cell editing outcomes expressed as % cells negative for CD3, CD5, or CD7 protein expression using the guides in (a), with (Blast) or without (noBlast) blasticidin selection. n = 2 independent donors. d, Protein level and genomic level editing for efficient sgRNAs targeting CD5 and CD7, shown as percent negative (flow cytometry, protein level) and percent edited (NGS, genomic level). Genomic level editing was analyzed with CRISPResso2. n = 2 independent donors for protein-level assessment and 1 matching donor for genomic-level assessments.
Extended Data Fig. 2 |. ABE and CBE screens are reproducible across human donors.
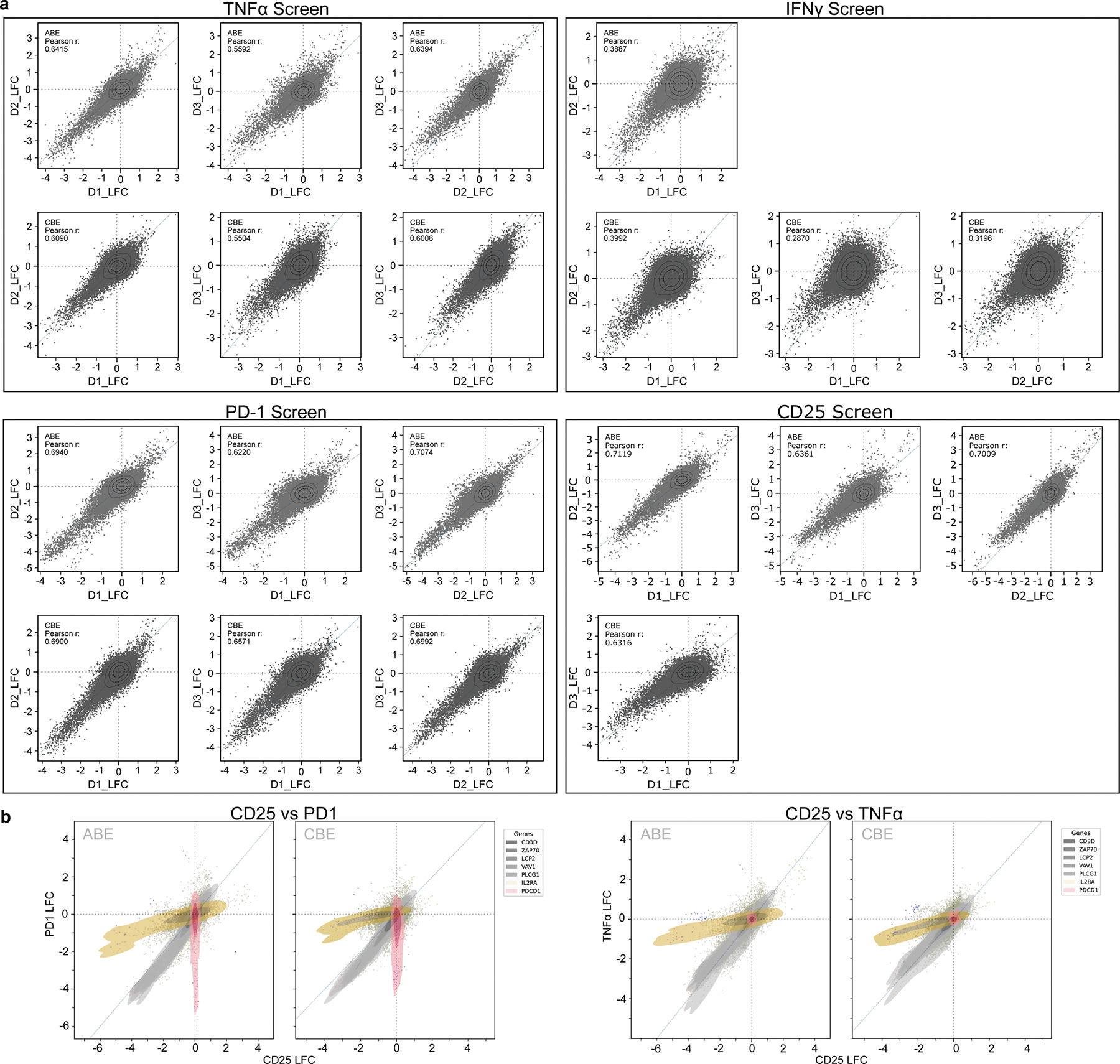
a, Scatter plots showing LFC (log2-fold changes) of pairwise donor-to-donor correlations (high/low bins) for each screen. Three human donors were used for all screens except the IFNγ-ABE and CD25-CBE screens, where two were used for analyses. Pearson correlation coefficient is given for each comparison. b, Comparison of sgRNA level effect sizes between CD25 and PD1 screens (left) or CD25 and TNFα screens (right), shown as LFC (log2-fold changes, high/low bins).
Extended Data Fig. 3 |. Base edits with strong functional effects are enriched in structured regions of proteins.
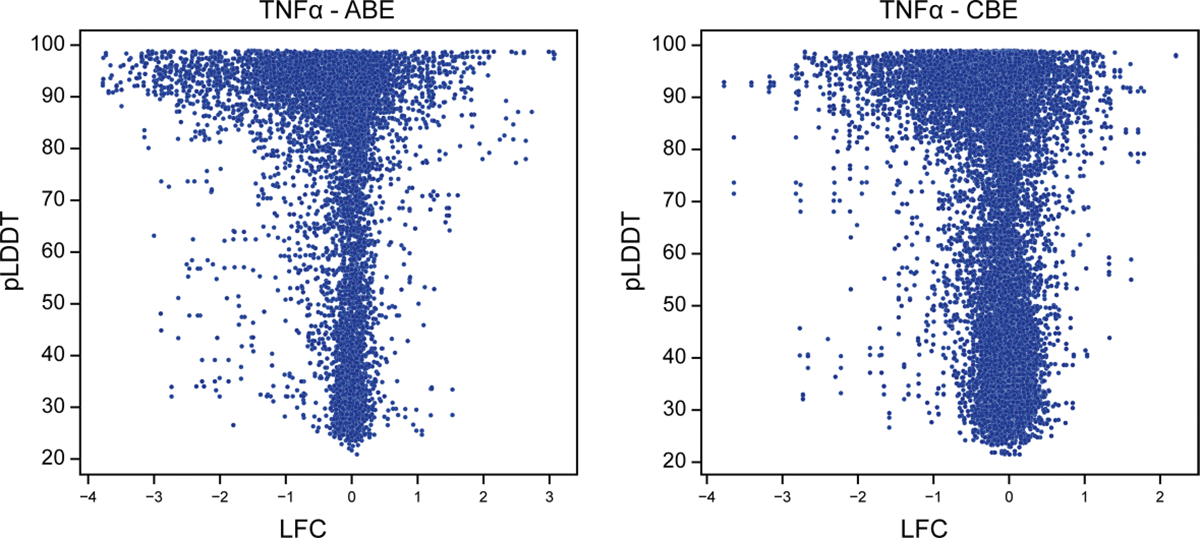
Scatter plot showing the correlation between AlphaFold predicted local Distance Difference Test (pLDDT) scores and screen LFC (log2-fold changes) for residues in proteins whose genes had |LFC | > 1.0 in the TNFα screen. Lower (<50) AlphaFold pLDDTs scores are predictive of structurally disordered regions of proteins. Predictions were obtained from pre-computed AlphaFold structures for canonical UniProt accessions for each screen gene.
Extended Data Fig. 4 |. Tiling base edits reveal mutations with discordant effects across screens.
a, Average effects (LFC, log2-fold changes) of non-synonymous, non-terminating base edits at each residue (ABE, top; CBE, bottom) on PD-1, CD25 expression, and IFNγ production are plotted across open reading frames encoding VAV1 (left) and NFKBIA (right). The dotted line indicates the average log2-fold change of the top 3 terminating (knockout) guides in the first half of the coding sequence, with blue for negative effect and red for positive effect on TNFα levels. Annotated domains from UniProt are shown below and are colored by the average effect of guides targeting those regions. b, Violin plots show distribution of log2-fold changes of CBE (dark purple) and ABE (light purple) guides tiling positive (top) and negative (bottom) regulator genes of PD-1, CD25 expression, and IFNγ production. Dotted lines indicate the average log2-fold change of the top 3 terminating (knockout) guides in the first half of the coding sequence. Blue and red dots indicate guides with strongly opposing effects with respect to the knockout guides for the same gene.
Extended Data Fig. 5 |. Comparative analysis of residue vs. base-level analysis and correlation to clinical variants.
a, Comparison between residue-level analysis derived from average LFC (log2-fold change) and base-level analysis derived from a multiple regression model for VAV1 and NFKBIA. For residue-level: p values were derived with Fisher’s method from MAGeCK results for individual guides; for base-level: p values were derived from a two-sided Wald test. b, Residue-level and base-level analysis for PIK3CD showing comparisons when guides with high off-target scores are filtered from analysis as well as when the predicted editing window for ABE is expanded to include the 9th position. c, Effect size for base-level analysis vs. LFC (log2-fold change) for residue-level analysis is plotted for the TNFα ABE (left) and CBE (right) screens. d, Variants are binned based on MAGeCK guide-level FDR. Enrichments of base edited variants in Clinvar “pathogenic,” “likely pathogenic,” and “benign” categories are plotted for each group for the PD1, CD25, TNFα, and IFNγ screens (from n = 3 donors); error bars represent 95% confidence intervals.
Extended Data Fig. 6 |. Supplementary validation and characterization of specific base edits in arrayed format.
a, Editing by ABE mRNA and synthetic sgRNA co-electroporation for each sgRNA chosen for validation, assessed by deep amplicon sequencing and analyzed with Crispresso254. Guide sequences are in gray, predicted editing window in green, and PAM in dark gray. b, LFC (log2-fold changes) of levels of the indicated cytokines over control (mean of 2 AAVS1 control gRNAs) measured by intracellular staining and flow cytometry are plotted. n = 6 human donors; mean ± SE; *p < 0.05, **p < 0.01, ***p < 0.001. P-values were derived using a two-tailed independent two-sample t-test. c, Cytokine secretion in culture supernatants for T cells with the indicated base edits were measured by Luminex. Heatmap represents LFC (log2-fold changes) over the mean of cells edited with two AAVS1 control sgRNAs. n = 4 human donors.
Extended Data Fig. 7 |. Stimulation responses of base edited T cells in arrayed validation.
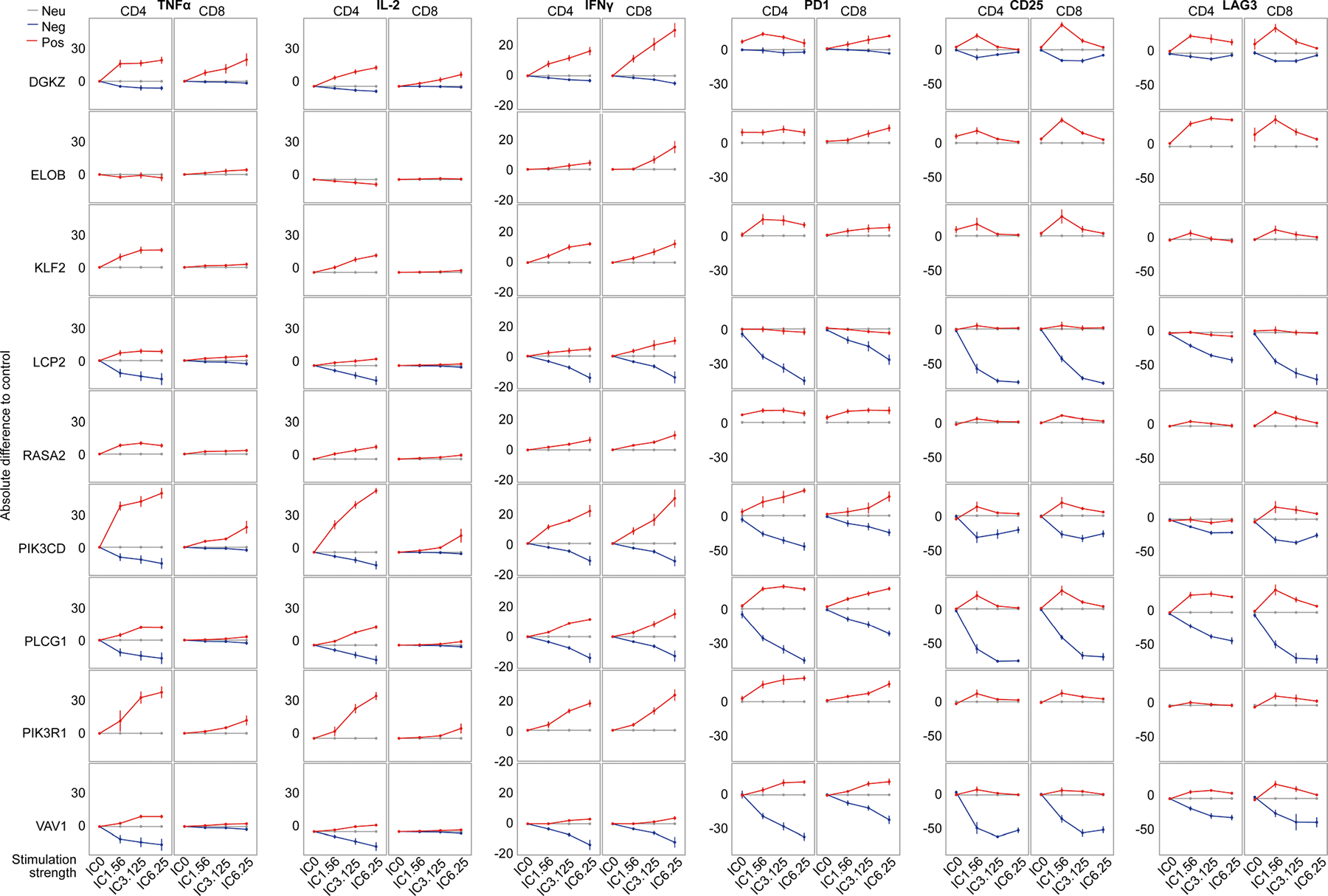
Individual plots of cytokine and cell surface protein expression for CD4+ or CD8+ T cells base edited with the indicated guides (red, positive in TNFα screen; blue, negative in TNFα screen; gray, AAVS1 control), measured by flow cytometry and normalized to AAVS1 control, over a range of anti-CD3/28/2 immunocult (IC) doses ranging from 0 (IC0) to 6.25ul/ml (IC6.25). n = 6 human donors, shown as mean ± SE.
Extended Data Fig. 8 |. Characterization of a PIK3CD allelic series.
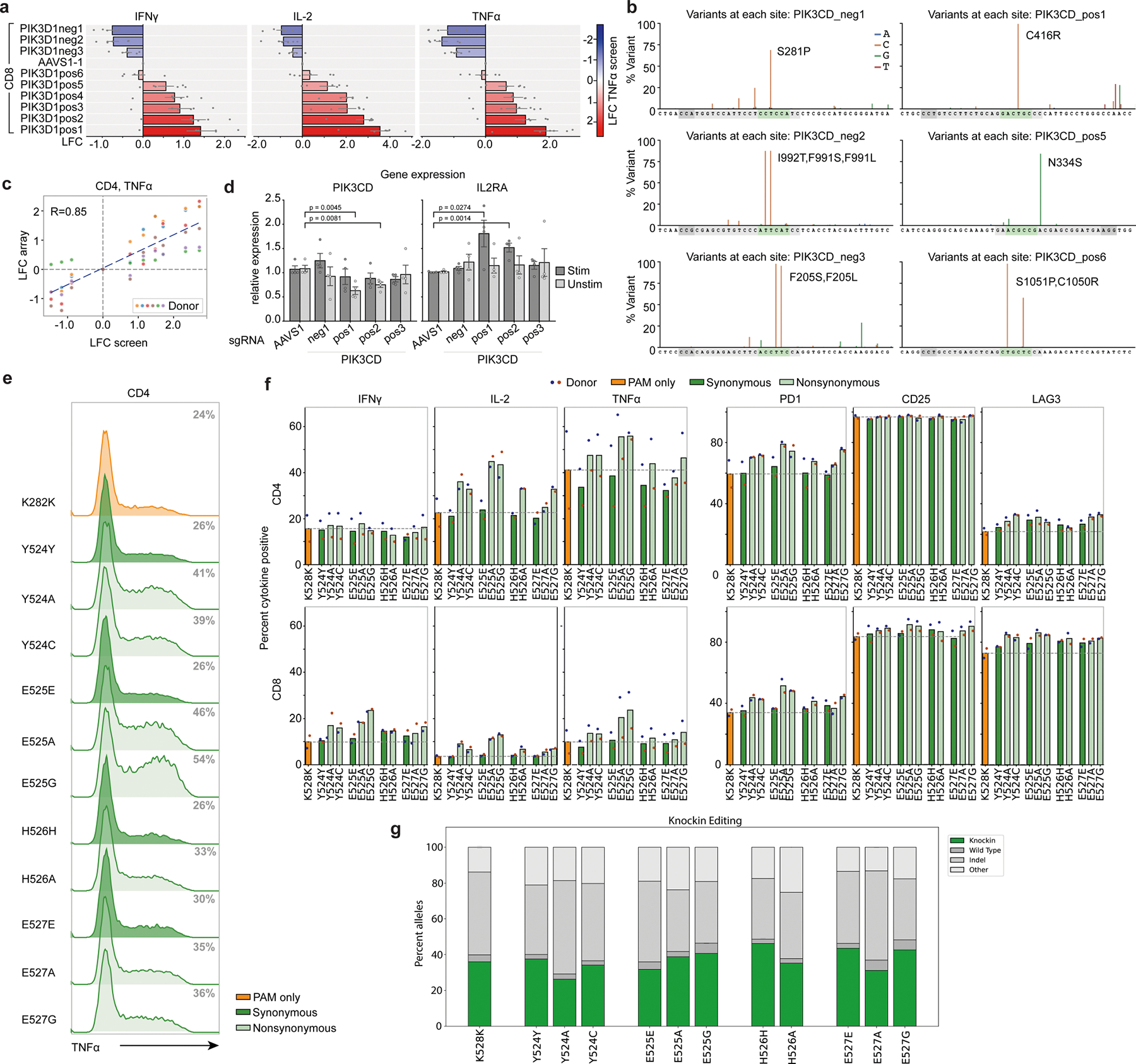
a, Bar graphs corresponding to Fig. 3i show LFC (log2-fold change) in expression of the indicated cytokines in arrayed validation for a series of 9 PIK3CD guides relative to AAVS1 control in CD8+ T cells; color indicates LFC (log2-fold change) value of each guide in the original ABE TNFα screen. n = 6 human donors, shown as mean ± SE. b, A-T to G-C editing by co-electroporation of ABE mRNA and synthetic PIK3CD sgRNAs, assessed by deep amplicon sequencing and analyzed with Crispresso254. Guide sequences are in gray, predicted editing window in green, and PAM in dark gray. c, Correlation between PIK3CD LFC (log2-fold change) values from the TNFα screen and validation experiment (Fig. 3i) in CD4+ T cells. d, mRNA expression of PIK3CD and IL2RA in PIK3CD edited T cells. Mean ± SD, n = 4 independent donors. e-g, Single amino acid substitution by CRISPR/Cas9 knockin. All edits include a synonymous K282K mutation deleting the PAM site. e, Flow cytometry histograms showing TNFα expression in CD4+ gated T cells. Percent positive cells in gray. f, Percent cytokine positive CD4+ and CD8+ gated T cells with the indicated knockins. n = 2 human donors (red and blue dots) shown as mean. g, Corresponding gene editing outcomes for knockin experiments as reported by CRISPResso2. n = 2 human donors shown as mean.
Extended Data Fig. 9 |. Cytotoxic function of base edited T cells in arrayed validation.
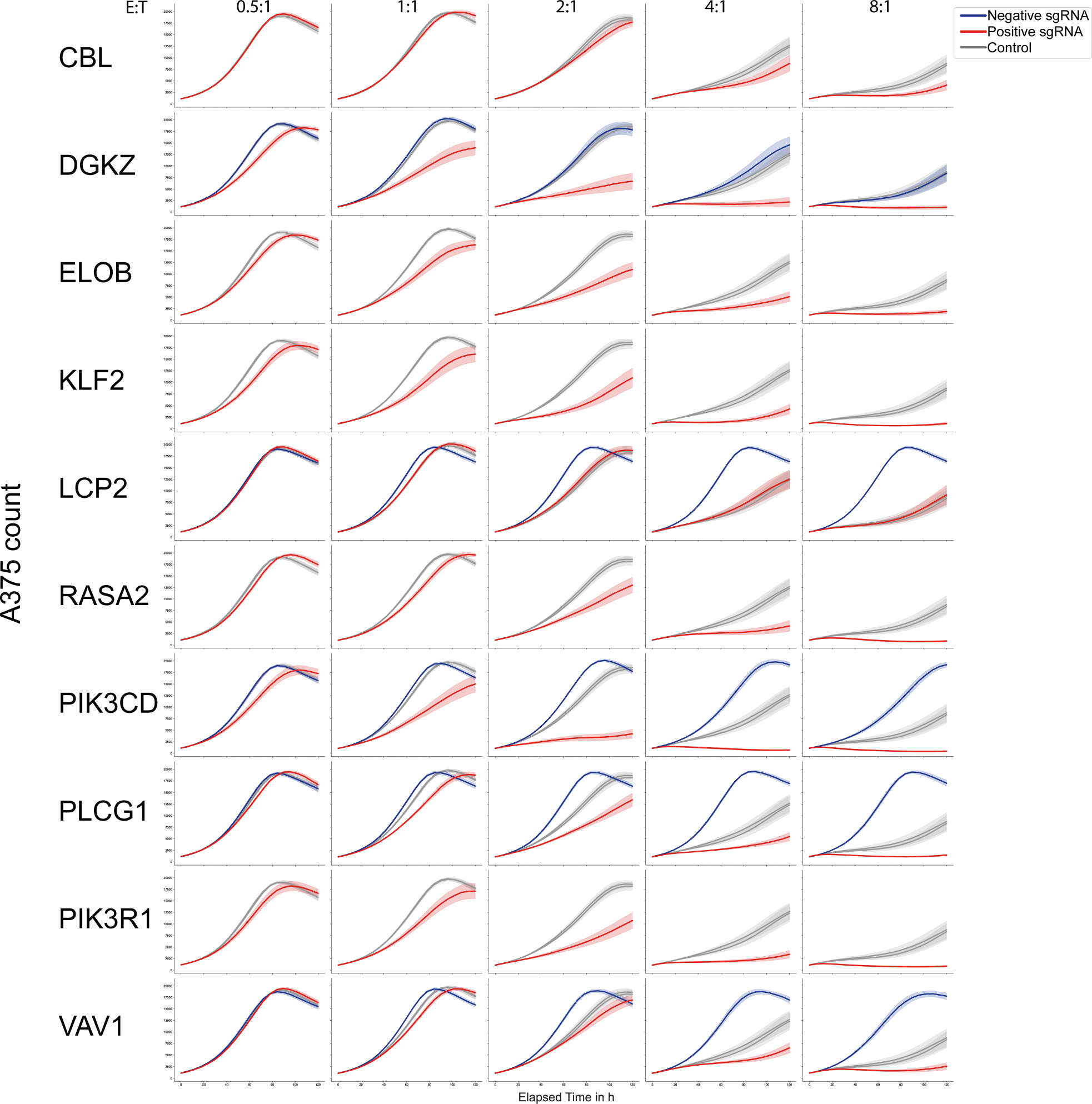
Cytotoxicity (measured by A375 target cell killing) of antigen-specific T cells base edited with control, positive (increased T cell activation, red), or negative (decreased T cell activation, blue) guides targeting selected genes measured with Incucyte live-cell imaging over time. n = 6 human donors, shown as mean (line) ± SE (shaded area).
Extended Data Fig. 10 |. Establishment and performance of NG-PAM dependent base editing with SpG Cas9.
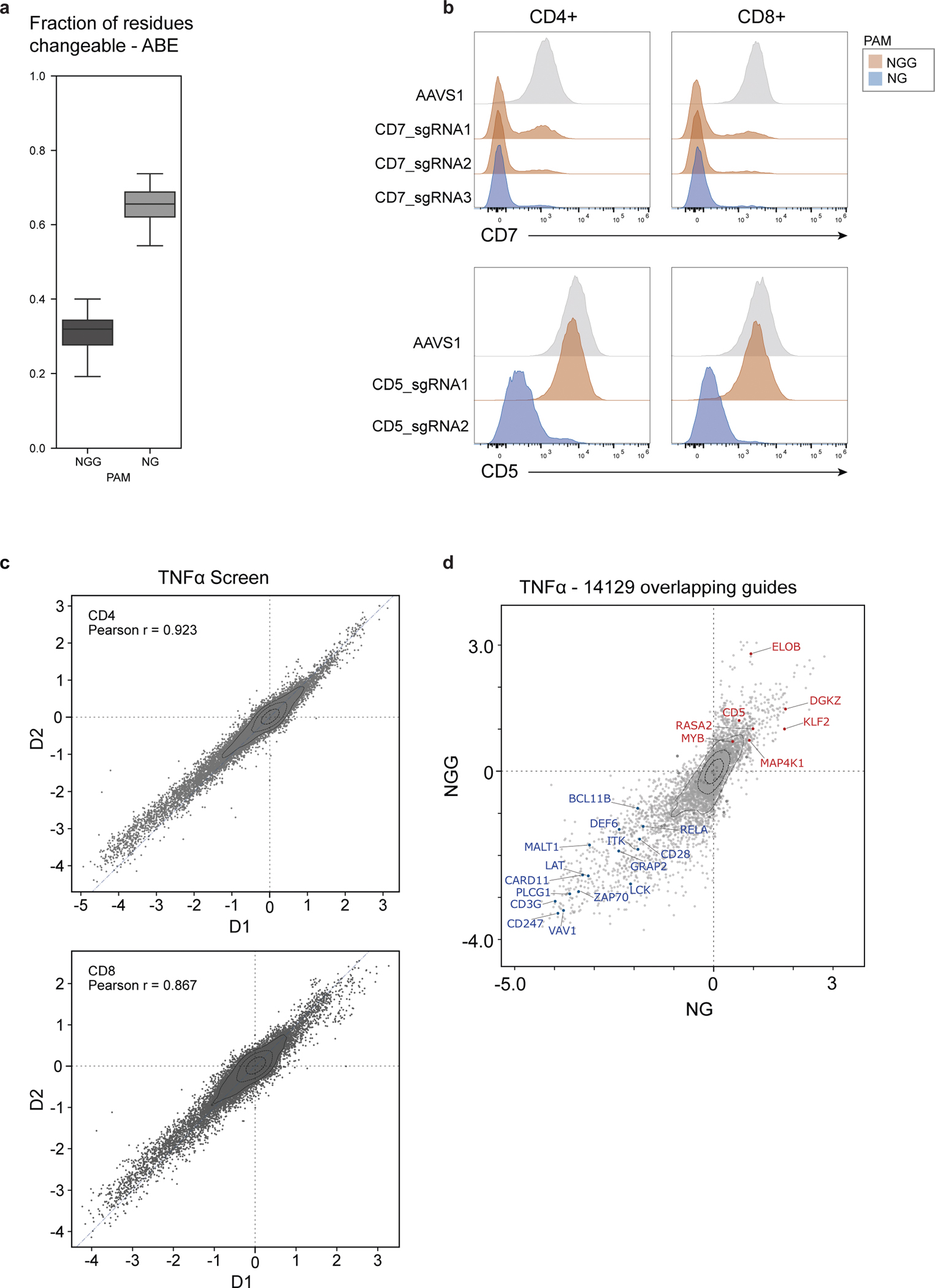
a, Predicted distribution of fraction of editable residues, within the original 385 genes in the NGG-PAM Cas9 screen, using either WT nCas9 (NGG PAM) or SpG Cas9 (NG PAM) for ABE. Box plots show median, center quartiles, and extremes within 1.5 * IQR. b, Distribution of surface protein expression levels for CD5 or CD7 in pan T cells gated for CD4+ (right) or CD8+ (left) base edited with one (CD5) or two (CD7) ABE NGG sgRNAs targeting each gene and one ABE NG sgRNA targeting each gene plus AAVS1 control. c, LFC (log2 fold changes) in the TNFα screens using WT nCas9 (NGG, y-axis) vs. using SpG Cas9 (NG, x-axis) for 13,334 overlapping guides between the two screens. The average knockout effect of specific genes present in both screens is shown with overlaid blue/red dots and labels. Knockout effects were calculated using the top 3 predicted knockout guides. d, Scatter plots showing LFC (log2-fold changes) of pairwise donor-to-donor correlations for each NG screen. Two human donors were used for all NG screens.
Supplementary Material
Acknowledgements
We thank Z. Steinhart, F. Blaeschke and all other members of the Eyquem, Ye and Marson laboratories for helpful discussions and feedback on the manuscript; J. Woo, R. Manlapaz and J. Sawin for organization and management; T. Tolpa and S. Pyle for help with illustrations and figure design; J. Srivastava and the Gladstone Institute’s flow cytometry core for assistance with cell sorting; and R. Innerhofer, M. Hollenstein, T. Koller and K. Schmetterer for covering clinical shifts and for scientific input during the revision process. Some elements in the schematics of Figs. 1 and 3 were sourced from BioRender.com. This work was supported by the NIAID (P01AI138962, 1P01AI55393), NIDDK (R01DK129364), the Larry L. Hillblom Foundation (grant no. 2020-D-002-NET), Northern California JDRF Center of Excellence, a gift from the Byers Family and the James B. Pendleton Charitable Trust. The Marson lab has received funds from the Parker Institute for Cancer Immunotherapy (PICI), the Lloyd J. Old STAR award from the Cancer Research Institute (CRI), the Simons Foundation, the CRISPR Cures for Cancer Initiative and K. Jordan. R.S. was supported by the Max Kade Foundation and the Austrian Society for Laboratory Medicine. C.W. is supported by an NCI F99/K00 Fellowship (K00CA245718). R.D. is supported by an NIAID research supplement to promote diversity in health-related research (P01AI138962). M.O. is supported by Astellas Foundation for Research on Metabolic Disorder and Chugai Foundation for Innovative Drug Discovery Science (C-FINDs). L.A.G. is supported by a NIH New Innovator Award (DP2 CA239597), a Pew-Stewart Scholars for Cancer Research award, and the Goldberg-Benioff Endowed Professorship in Prostate Cancer Translational Biology and the Arc Institute. J.K.P., M.O., and A.M. are supported by the NHGRI (2R01HG008140). Sequencing performed at the UCSF CAT is supported by UCSF PBBR, RRP IMIA, and NIH 1S10OD028511–01 grants. Sorting at the Gladstone flow core is supported by NIH S10 RR028962, James B. Pendleton Charitable Trust, and DARPA. B.R.S. was supported by NIH grants K08CA273529 and L30TR002983. D.D. was supported by the UCSF Scholars At Risk programme.
Footnotes
Competing interests A.M. is a co-founder of Function Bio, Arsenal Biosciences, Spotlight Therapeutics and Survey Genomics, serves on the boards of directors at Function Bio, Spotlight Therapeutics and Survey Genomics, was a board observer at Arsenal Biosciences, is a member of the scientific advisory boards of Function Bio, Arsenal Biosciences, Spotlight Therapeutics, Survey Genomics, NewLimit, Amgen, Tenaya, and Lightcast owns stock in Arsenal Biosciences, Spotlight Therapeutics, NewLimit, Survey Genomics, PACT Pharma, Tenaya, and Lightcast, and has received fees from Arsenal Biosciences, Spotlight Therapeutics, NewLimit, 23andMe, PACT Pharma, Juno Therapeutics, Tenaya, Lightcast, Trizell, Vertex, Merck, Amgen, Genentech, AlphaSights, Clearview Healthcare, Rupert Case Management, Bernstein, GLG, Survey Genomics, and ALDA. A.M. is an investor in and informal advisor to Offline Ventures and a client of EPIQ. C.W., S.E.D., R.S. and A.M. are shareholders of Function Bio. The Marson laboratory has received research support from Juno Therapeutics, Epinomics, Sanofi, GlaxoSmithKline, Gilead, and Anthem. J.E. is a compensated co-founder at Mnemo Therapeutics. J.E. owns stocks in Mnemo Therapeutics and Cytovia Therapeutics. J.E. is a compensated scientific advisor for Enterome, Treefrog Therapeutics and Resolution Therapeutics. The Eyquem lab has received research support from Cytovia Therapeutic, Mnemo Therapeutics and Takeda. J.E. is a holder of patents pertaining to but not resulting from this work. L.A.G. has filed patents on CRISPR approaches and is a co-founder of Chroma Medicine. R.S., C.W. and A.M. are listed as inventors on patent applications related to this work. The other authors declare no competing interests.
Ethics statement
Primary human T cells were sourced from human peripheral blood Leukopaks (Stemcell Technologies, 200–0092) with approved Stemcell IRB.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Code availability
Analysis and visualization were done with matplotlib and seaborn in Python or with ggplot2 in R, with editing for clarity before publication. Code and data used to create key figures have been deposited at Zenodo (https://doi.org/10.5281/zenodo.8415038).
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41586-023-06835-6.
Peer review information Nature thanks Sidi Chen, Johan Henriksson and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Reprints and permissions information is available at http://www.nature.com/reprints.
Data availability
All guide library information from this manuscript is provided in Supplementary Tables 1 and 2 (for the NGG screen) and Supplementary Tables 4 and 5 (for the NG screen). MAGeCK test output for all screens is provided in Supplementary Table 3. Raw sequencing data for all base editing screens have been deposited on GEO (GSE244774). Source data are provided with this paper.
References
- 1.Shifrut E et al. Genome-wide CRISPR screens in primary human T cells reveal key regulators of immune function. Cell 175, 1958–1971.e15 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carnevale J et al. RASA2 ablation in T cells boosts antigen sensitivity and long-term function. Nature 609, 174–182 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schmidt R et al. CRISPR activation and interference screens decode stimulation responses in primary human T cells. Science 375, eabj4008 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belk JA et al. Genome-wide CRISPR screens of T cell exhaustion identify chromatin remodeling factors that limit T cell persistence. Cancer Cell 40, 768–786.e7 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Henriksson J et al. Genome-wide CRISPR screens in T helper cells reveal pervasive crosstalk between activation and differentiation. Cell 176, 882–896.e18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ye L et al. A genome-scale gain-of-function CRISPR screen in CD8 T cells identifies proline metabolism as a means to enhance CAR-T therapy. Cell Metab. 34, 595–614.e14 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Komor AC, Kim YB, Packer MS, Zuris JA & Liu DR Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gaudelli NM et al. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature 551, 464–471 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sangree AK et al. Benchmarking of SpCas9 variants enables deeper base editor screens of BRCA1 and BCL2. Nat. Commun. 13, 1318 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parlakpinar H & Gunata M Transplantation and immunosuppression: a review of novel transplant-related immunosuppressant drugs. Immunopharmacol. Immunotoxicol. 43, 651–665 (2021). [DOI] [PubMed] [Google Scholar]
- 11.Freimer JW et al. Systematic discovery and perturbation of regulatory genes in human T cells reveals the architecture of immune networks. Nat. Genet. 54, 1133–1144 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanna RE et al. Massively parallel assessment of human variants with base editor screens. Cell 184, 1064–1080.e20 (2021). [DOI] [PubMed] [Google Scholar]
- 13.Coelho MA et al. Base editing screens map mutations affecting interferon-γ signaling in cancer. Cancer Cell 41, 288–303.e6 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Georgiadis C et al. Base-edited CAR T cells for combinational therapy against T cell malignancies. Leukemia 35, 3466–3481 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Diorio C et al. Cytosine base editing enables quadruple-edited allogeneic CART cells for T-ALL. Blood 140, 619–629 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Richter MF et al. Phage-assisted evolution of an adenine base editor with improved Cas domain compatibility and activity. Nat. Biotechnol. 38, 883–891 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thuronyi BW et al. Continuous evolution of base editors with expanded target compatibility and improved activity. Nat. Biotechnol. 37, 1070–1079 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith-Garvin JE, Koretzky GA & Jordan MS T cell activation. Annu. Rev. Immunol. 27, 591–619 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henikoff S & Henikoff JG Amino acid substitution matrices from protein blocks. Proc. Natl Acad. Sci. USA 89, 10915–10919 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jumper J et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Varadi M et al. AlphaFold protein structure database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 50, D439–D444 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Winston JT et al. The SCFβ-TRCP–ubiquitin ligase complex associates specifically with phosphorylated destruction motifs in IκBα and β-catenin and stimulates IκBα ubiquitination in vitro. Genes Dev. 13, 270–283 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gaudelli NM et al. Directed evolution of adenine base editors with increased activity and therapeutic application. Nat. Biotechnol. 38, 892–900 (2020). [DOI] [PubMed] [Google Scholar]
- 24.Glaser V et al. Combining different CRISPR nucleases for simultaneous knock-in and base editing prevents translocations in multiplex-edited CAR T cells. Genome Biol. 24, 89 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Webber BR et al. Highly efficient multiplex human T cell engineering without double-strand breaks using Cas9 base editors. Nat. Commun. 10, 5222 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robbins PF et al. Single and dual amino acid substitutions in TCR CDRs can enhance antigen-specific T cell functions. J. Immunol. 180, 6116–6131 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Singh A, Joshi V, Jindal AK, Mathew B & Rawat A An updated review on activated PI3 kinase delta syndrome (APDS). Genes Dis 7, 67–74 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Singh MD, Ni M, Sullivan JM, Hamerman JA & Campbell DJ B cell adaptor for PI3-kinase (BCAP) modulates CD8+ effector and memory T cell differentiation. J. Exp. Med. 215, 2429–2443 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crank MC et al. Mutations in PIK3CD can cause hyper IgM syndrome (HIGM) associated with increased cancer susceptibility. J. Clin. Immunol. 34, 272–276 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thauland TJ, Pellerin L, Ohgami RS, Bacchetta R & Butte MJ Case study: mechanism for increased follicular helper T cell development in activated PI3K delta syndrome. Front. Immunol. 10, 753 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu L et al. Efficient precise in vivo base editing in adult dystrophic mice. Nat. Commun. 12, 3719 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walton RT, Christie KA, Whittaker MN & Kleinstiver BP Unconstrained genome targeting with near-PAMless engineered CRISPR–Cas9 variants. Science 368, 290–296 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Massaad MJ, Ramesh N & Geha RS Wiskott–Aldrich syndrome: a comprehensive review. Ann. N. Y. Acad. Sci. 1285, 26–43 (2013). [DOI] [PubMed] [Google Scholar]
- 34.Methot JL et al. Optimization of Versatile Oxindoles as Selective PI3Kδ Inhibitors. ACS Med. Chem. Lett. 11, 2461–2469 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Appleby MW & Ramsdell F A forward-genetic approach for analysis of the immune system. Nat. Rev. Immunol. 3, 463–471 (2003). [DOI] [PubMed] [Google Scholar]
- 36.Martin-Rufino JD et al. Massively parallel base editing to map variant effects in human hematopoiesis. Cell 186, 2456–2474.e24 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neugebauer ME et al. Evolution of an adenine base editor into a small, efficient cytosine base editor with low off-target activity. Nat. Biotechnol. 41, 673–685 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen Z et al. Integrative dissection of gene regulatory elements at base resolution. Cell Genomics 3, 100318 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zeng J et al. Therapeutic base editing of human hematopoietic stem cells. Nat. Med. 26, 535–541 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jia Y et al. Hyperactive PI3Kδ predisposes naive T cells to activation via aerobic glycolysis programs. Cell. Mol. Immunol. 18, 1783–1797 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lucas CL et al. Dominant-activating germline mutations in the gene encoding the PI(3) K catalytic subunit p110δ result in T cell senescence and human immunodeficiency. Nat. Immunol. 15, 88–97 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lucas CL, Chandra A, Nejentsev S, Condliffe AM & Okkenhaug K PI3Kδ and primary immunodeficiencies. Nat. Rev. Immunol. 16, 702–714 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Robles-Valero J et al. Cancer-associated mutations in VAV1 trigger variegated signaling outputs and T-cell lymphomagenesis. EMBO J. 40, e108125 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Prawiro C et al. A frequent PLCγ1 mutation in adult T-cell leukemia/lymphoma determines functional properties of the malignant cells. Biochim. Biophys. Acta. 1869, 166601 (2023). [DOI] [PubMed] [Google Scholar]
- 45.Fraietta JA et al. Determinants of response and resistance to CD19 chimeric antigen receptor (CAR) T cell therapy of chronic lymphocytic leukemia. Nat. Med. 24, 563–571 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Long AH et al. 4–1BB costimulation ameliorates T cell exhaustion induced by tonic signaling of chimeric antigen receptors. Nat. Med. 21, 581–590 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Feucht J et al. Calibration of CAR activation potential directs alternative T cell fates and therapeutic potency. Nat. Med. 25, 82–88 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guedan S et al. Single residue in CD28-costimulated CAR-T cells limits long-term persistence and antitumor durability. J. Clin. Invest. 130, 3087–3097 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Parnas O et al. A genome-wide CRISPR screen in primary immune cells to dissect regulatory networks. Cell 162, 675–686 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shang W et al. Genome-wide CRISPR screen identifies FAM49B as a key regulator of actin dynamics and T cell activation. Proc. Natl Acad. Sci. USA 115, E4051–E4060 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dong MB et al. Systematic immunotherapy target discovery using genome-scale in vivo CRISPR screens in CD8 T cells. Cell 178, 1189–1204.e23 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Halperin SO et al. CRISPR-guided DNA polymerases enable diversification of all nucleotides in a tunable window. Nature 560, 248–252 (2018). [DOI] [PubMed] [Google Scholar]
- 53.Roth TL et al. Pooled knockin targeting for genome engineering of cellular immunotherapies. Cell 181, 728–744.e21 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Clement K et al. CRISPResso2 provides accurate and rapid genome editing sequence analysis. Nat. Biotechnol. 37, 224–226 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sanson KR et al. Optimized libraries for CRISPR–Cas9 genetic screens with multiple modalities. Nat. Commun. 9, 5416 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen PJ et al. Enhanced prime editing systems by manipulating cellular determinants of editing outcomes. Cell 184, 5635–5652.e29 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jiang T et al. Chemical modifications of adenine base editor mRNA and guide RNA expand its application scope. Nat. Commun. 11, 1979 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shy BR et al. High-yield genome engineering in primary cells using a hybrid ssDNA repair template and small-molecule cocktails. Nat. Biotechnol. 41, 521–531 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li W et al. MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol. 15, 554 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bae S, Park J & Kim J-S Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics 30, 1473–1475 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Doench JG et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR–Cas9. Nat. Biotechnol. 34, 184–191 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Concordet J-P & Haeussler M CRISPOR: intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Res. 46, W242–W245 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Landrum MJ et al. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 46, D1062–D1067 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All guide library information from this manuscript is provided in Supplementary Tables 1 and 2 (for the NGG screen) and Supplementary Tables 4 and 5 (for the NG screen). MAGeCK test output for all screens is provided in Supplementary Table 3. Raw sequencing data for all base editing screens have been deposited on GEO (GSE244774). Source data are provided with this paper.


