Extended Data Fig. 6 |. Supplementary validation and characterization of specific base edits in arrayed format.
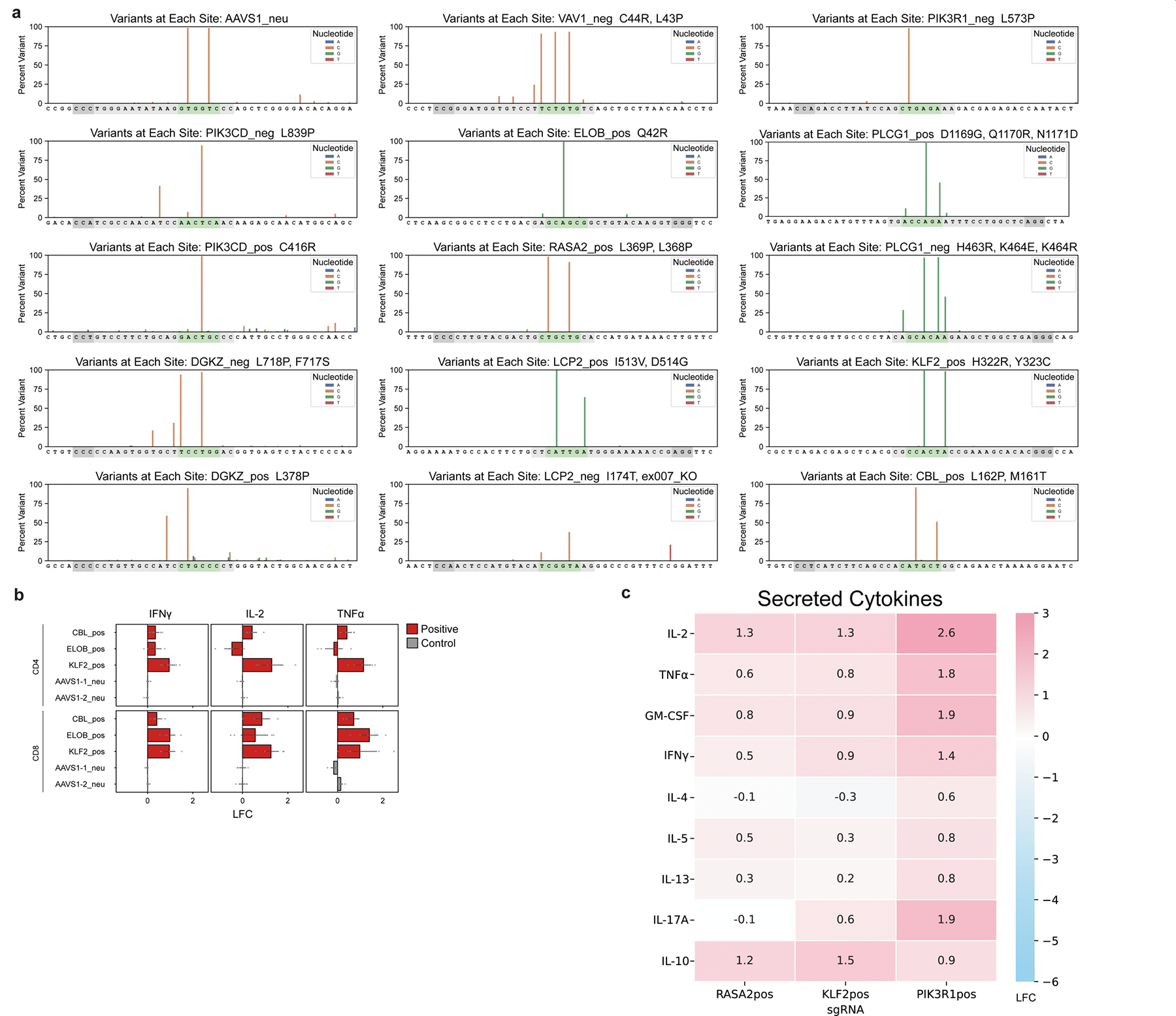
a, Editing by ABE mRNA and synthetic sgRNA co-electroporation for each sgRNA chosen for validation, assessed by deep amplicon sequencing and analyzed with Crispresso254. Guide sequences are in gray, predicted editing window in green, and PAM in dark gray. b, LFC (log2-fold changes) of levels of the indicated cytokines over control (mean of 2 AAVS1 control gRNAs) measured by intracellular staining and flow cytometry are plotted. n = 6 human donors; mean ± SE; *p < 0.05, **p < 0.01, ***p < 0.001. P-values were derived using a two-tailed independent two-sample t-test. c, Cytokine secretion in culture supernatants for T cells with the indicated base edits were measured by Luminex. Heatmap represents LFC (log2-fold changes) over the mean of cells edited with two AAVS1 control sgRNAs. n = 4 human donors.
