Abstract
Infections caused by antibiotic-resistant bacteria represent a serious threat to global public health. Recently, due to its increased resistance to carbapenems and β-lactams, Klebsiella pneumoniae has become one of the main causes of septicemia, pneumonia, and urinary tract infections. It is crucial to take immediate action and implement effective measures to prevent further spread of this issue. This study aims to report the prevalence and antibiotic resistance rates of K. pneumoniae strains isolated from clinical specimens from 2015 to 2020 at the University Hospital of Salerno, Italy. More than 3,800 isolates were collected from urine cultures, blood cultures, respiratory samples, and others. K. pneumoniae isolates showed broad resistance to penicillin and cephalosporins, and increased susceptibility to fosfomycin and gentamicin. Extended spectrum beta-lactamase (ESBL) isolates accounted for 20–22%. A high percentage of strains tested were resistant to carbapenems, with an average of 40% to meropenem and 44% to ertapenem. The production of ESBLs and resistance to carbapenems is one of the major public health problems. Constant monitoring of drug-resistant isolates is crucial for developing practical approaches in implementing antimicrobial therapy and reducing the spread of K. pneumoniae in nosocomial environments.
1. Introduction
Globally, antimicrobial resistance (AMR) is one of the most serious threats to public health. In 2019, over one million deaths were attributed to multidrug-resistant strains (MDRs) [1]. Furthermore, owing to the ease of transmission of MDRs, particularly in healthcare environment, it is expected that more than 5 million deaths will occur by 2030 [2, 3].
Currently, among Enterobacteriaceae family, Klebsiella pneumoniae (K. pneumoniae) and Escherichia coli are regarded as the primary opportunistic pathogens species, causing various infections such as pneumonia, urinary tract infections (UTIs), bloodstream infections (BSIs), and surgical site infection [4, 5]. Moreover, the use of medical devices, such as urinary catheters, venous catheters, and respiratory support equipment represent important risk factors in nosocomial infections promoted by opportunistic pathogen bacteria [6]. Infections produced by K. pneumoniae have grown over the past decade and constitute a particular concern to immunocompromised individuals [7]. Increased healthcare costs, prolonged hospitalization, untreated infections, and mortality rates are the consequences of this situation [8, 9]. Treatment failure is attributed to the ability of these species to produce extended-spectrum beta-lactamases (ESBLs) capable of neutralizing cephalosporins, used to treat Gram-negative infections, including pneumonia caused by K. pneumoniae. ESBLs can hydrolyze and inactivate β-lactam antibiotics, including penicillin, cephalosporins (first, second, and third generation), and aztreonam, but not carbapenems [10]. Recently, it has been noted that ESBL-producing bacteria show a high rate of resistance to other classes of antibiotics, such as fluoroquinolones, sulfonamides, and aminoglycosides [11]. K. pneumoniae has developed resistance to carbapenems through the production of enzymes (carbapenemases) that represent the first-line therapy for severe infection with ESBL-producing K. pneumoniae. Resistance to carbapenems is attributable to one or more carbapenemase genes (blaKPC, blaNDM, blaVIM, blaOXA−48, and blaIMP−Like), whose expression usually leads to carbapenem resistance [12]. However, since the first carbapenem-resistant strain of K. pneumoniae (KPC) was isolated and characterized in 1996 by Yigit et al., carbapenem-resistant strains have increased rapidly and have received considerable public interest due to the scarcity of antibiotic treatment choices [13, 14]. The limited therapy choices resulting from the inefficacy of novel beta-lactam/beta-lactamase inhibitor combos, such as ceftazidime-avibactam and meropenem/vaborbactam against NDM-producing pathogens (New Delhi metallo beta-lactamase) constituted an additional barrier [15]. K. pneumoniae is the most common bacterial species among the carbapenem-resistant Enterobacteriaceae strains, recognized by WHO as one of the most important priority pathogens (included in ESKAPE pathogens) due to reduced treatment options and potential for community spread [16]. In previous studies, the mortality associated with KPC infections was expected to range between 33% and 42% in developed countries [17]. The hospital surveillance programs aim to screen patients colonized by KPC and provide isolation of patients in the ward to limit the contagion. Recent studies reported an increase in carbapenem resistance in K. pneumoniae in China, from 3% in 2005 to 21% in 2017 [18]. In Europe, the KPC represents the fastest growing antibiotic resistance with a six-fold increase in the number of deaths between 2007 and 2015 [19]. Other reports from the US, England, and Argentina highlighted the rapid increase of carbapenem-resistant Klebsiella infections [20–22]. Surveillance programs are particularly needed for carbapenemase-producing K. pneumoniae (KPC-Kp) and E. coli (CP-Ec), which are often responsible for outbreaks, as described in the latest report of the European Antibiotic Surveillance Network (EARS-Net). Due to this increase in antibiotic resistance, several studies have been conducted around the world to monitor the antibiotic resistance of K. pneumoniae [23]. Furthermore, it is desirable to support genotypic tests, such as DNA microarray and PCR, for the study of resistance mechanisms, mostly present in isolates at a local level, which can improve the understanding of the molecular mechanisms underlying antibiotic resistance [24]. In Pakistan, Uddin et al. reported a high prevalence of K. pneumoniae MDR, of which 98% were ESBL producers; furthermore, NDM-positive isolates were resistant to newer formulations, including meropenem/vaborbactam (MEV), ceftazidime/avibactam (CZA), and ceftolozane/tazobactam (C/T) [25, 26]. In Saudi Arabia, surveillance of K. pneumoniae antibiotic resistance revealed an increased rate of resistance among different classes of antibiotics [27]. For example, in Aseer, several studies conducted between 2015 and 2019 showed an increased rate of resistance against penicillin and cephalosporin. The results of these investigations are useful to the clinician to choose an effective and correct treatment and to contrast the phenomenon of antibiotic resistance. The aim of this study was to evaluate the antimicrobial resistance rates of K. pneumoniae isolated from clinical specimens from 2015 to 2020 in our University Hospital to improve clinical practice and therapeutic treatment.
2. Materials and Methods
2.1. Clinical Isolate Collection
Out of 3,841 K. pneumoniae strains were isolated from patients admitted to the University Hospital “San Giovanni di Dio e Ruggi d'Aragona” in Salerno, from January 2015 to December 2020. We collected the demographic data, including age and gender, department unit, and the site of infections for the enrolled patients. The samples included blood, urine, bronchial-aspirate, sputum, swabs (vaginal swab, sternal swab, and wound swab), and other samples (catheters, sperm culture, ascitic fluid, abscess, and liquor). Throughout the entire study period, we made sure to exclude any cultures that contained the same pathogen isolated from a single subject in case of multiple episodes of infection. To avoid duplicity in our data, we only included nonduplicated clinical isolates that were collected during the first episode of infection in the study period. We defined duplicate isolates as isolates from the same patient with an indistinguishable pattern of susceptibilities and excluded them accordingly.
2.2. Species Identification and Antimicrobial Susceptibility Test
All samples were collected in sterile containers and were analyzed in the microbiology laboratory, following the European Committee on Antimicrobial Susceptibility Testing (EUCAST) guidelines. Species were grown on blood agar, chocolate agar, and MacConkey agar (bioMérieux, Marcy-l'Étoile, France) and incubated at 37°C for 18–24 hours. All the biological samples indicated were processed following the official lines for processing biological clinical samples for microbiological investigations and diagnosis of infectious diseases. Identification and antibiotic resistance profile were determined by the Vitek2 system using an identification card (ID-GN) and susceptibility cards (AST-397 and AST-379 only for urine cultures). The Vitek-2 automated system involves the preparation of the inoculum, a 0.45% saline solution in which the bacterial isolates are suspended with a standard turbidity of 0.5 McFarland used for the identification and determination of the MIC at different antibiotics (AST). The ID-GN panel contained 46 fluorometric tests that included pH change and enzymatic reaction tests subjected to a kinetic fluorescence measurement. According to the manufacturer's instructions, quality control test was performed routinely once a week, and the reference Gram-negative strains were Klebsiella oxytoca ATCC 700,324 and Enterobacter ATCC 700,323. For each antibiotic, interpretation of results was done according to EUCAST clinical breakpoints. The following antibiotics were tested: ampicillin (AMP), amoxicillin/clavulanic acid (AUG), piperacillin/tazobactam (SXT), ceftazidime (CAZ), cefotaxime (CTX), cefepime (FEP), ertapenem (ETP), meropenem (MRP), imipenem (IMP), ciprofloxacin (CIP), gentamicin (CN), tigecycline (TGC), fosfomycin (FOS), and trimethoprim/sulfamethoxazole (S/T).
2.3. Ethical Consideration Statement
The study was conducted according to the guidelines of the Declaration of Helsinki. Approval for this study was obtained from the authority of San Giovanni di Dio e Ruggi d'Aragona Hospital. The data retrieved were anonymous and not linked to any patient. This is a retrospective study, which involves the collection of secondary data regarding bacterial isolates and their respective antimicrobial susceptibility data. The study did not have access to any sensitive patient information, only age and gender. All analyzed data contained exam codes that replaced and anonymized sensitive patient data.
2.4. Statistical Analysis
The demographic data of patients, including age, gender, isolated strain(s), and drug sensitivity results, were used for the analysis. The age- and sex-standardized incidences were calculated. The chi-framework test was used to compare the differences among antibiotic sensitivities over the range of years considered in the study. A chi-square test was used to verify the possible associations between the categorical variables, while the Cochran–Armitage trend test was used to verify the existence of a trend. The existence of a trend was checked only for antibiotics that showed statistically significant differences in the distribution of resistance during the years under consideration. Considering a significance level alpha = 0.05 for both tests, therefore those associations with a p value <0.05 were considered statistically significant. The IBM Statistical Package for Social Sciences Version 22.00 (SPSS Inc., Chicago, IL, USA) was used for data analysis.
3. Results
A total of 3,841 K. pneumoniae strains were isolated from January 2015 to December 2020 at the University Hospital “San Giovanni di Dio e Ruggi d'Aragona,” Salerno, Italy. Among the patients, 50.2% were females. The analysis of incidence by age group showed that patients between 61 and 75 years were 31%, followed by the age group 40–60 and 76–95 age group (21%), while for younger patients, the incidence was lower (8–10%) (Supplementary Table 1).
Additionally, there is a higher incidence of cases in the Intensive Care Unit (19.2%), General Surgery (12.9%), Nephrology (12.2%), and Internal Medicine (9.2%). This is shown by the analysis of the distribution of diagnostic materials across hospital departments from 2015 to 2020 (refer to Supplementary Table 2).
The highest percentage of K. pneumoniae strains were isolated from urine samples (45.3%), followed by vaginal and wound swabs (19.9%), sputum and bronchial-aspirate samples (15.4%), blood cultures (12.5%), and medical device samples (6.9%) (Figure 1).
Figure 1.
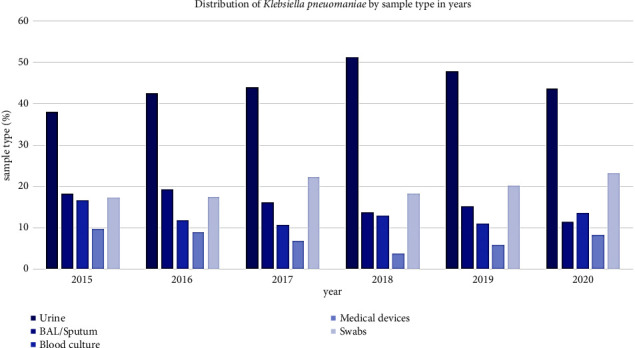
Prevalence of Klebsiella pneumoniae isolated from various clinical specimens. Clinical samples' detail: bronchoalveolar lavage (BAL), vaginal swab, sternal swab, wound swab (swabs), and urinary and vascular catheters (catheters).
These data showed an increase by year of species isolated from urine samples, from 38% in 2015 to 48%–43% in 2019 and 2020, respectively.
In our study, K. pneumoniae showed a high rate of resistance to ampicillin (98.5%), amoxicillin/clavulanic acid (63.7%), piperacillin/tazobactam (55%), ceftazidime (61.9%), cefotaxime (64.7%), and cefepime (50.2%) (Table 1). Also, the rates of resistance to ciprofloxacin and trimethoprim/sulfamethoxazole were higher than 50%, with 62.4% and 54.3%, respectively. Moreover, a low rate of resistance was shown for the carbapenems class, with imipenem at 28%, a slightly higher rate for meropenem (39.3%) and ertapenem (42.5%).
Table 1.
Resistance rates of the clinical isolates of Klebsiella pneumoniae to antimicrobial agents.
| Klebsiella pneumoniae | 2015 | 2016 | 2017 | 2018 | 2019 | 2020 | ∗ | ∗∗ | |
|---|---|---|---|---|---|---|---|---|---|
| Class | Antibiotic | R % (n.) |
R % (n.) |
R % (n.) |
R % (n.) |
R % (n.) |
R % (n.) |
||
| β-lactam/β-lactamase inhibitor combinations | Amoxicillin/clavulanic acid | 65.7 (463) | 66.1 (498) | 58.7 (494) | 66.6 (572) | 64.6 (814) | 60.3 (610) | 0.026 | 0.206 |
| Piperacillin/tazobactam | 64.1 (465) | 63.3 (550) | 53.6 (584) | 47.2 (707) | 56.9 (850) | 48.8 (637) | <0.0001 | <0.0001 | |
|
| |||||||||
| Cephalosporins | Ceftazidime | 61.8 (466) | 65.7 (551) | 60.0 (587) | 64.6 (704) | 64.7 (853) | 54.3 (668) | 0.0001 | 0.0319 |
| Cefotaxime | 67.0 (466) | 69.1 (551) | 62.9 (587) | 66.9 (707) | 66.8 (840) | 56.0 (680) | <0.0001 | 0.000452 | |
| Cefepime | 56.0 (466) | 59.2 (551) | 46.5 (585) | 39.6 (652) | 51.9 (476) | 51.6 (556) | <0.0001 | 0.0048 | |
|
| |||||||||
| Carbapenems | Ertapenem | 44.3 (411) | 51.2 (551) | 43.9 (586) | 33.1 (706) | 45.4 (852) | 37.4 (431) | <0.0001 | 0.0016 |
| Meropenem | 49.0 (465) | 48.6 (551) | 39.7 (585) | 30.2 (706) | 40.8 (850) | 32.1 (663) | <0.0001 | <0.0001 | |
| Imipenem | 29.2 (465) | 33.3 (540) | 33.5 (585) | 20.9 (651) | 23.0 (478) | 28.1 (570) | <0.0001 | 0.0021 | |
|
| |||||||||
| Fluoroquinolones | Ciprofloxacin | 63.7 (466) | 62.3 (551) | 61.2 (587) | 64.8 (707) | 66.4 (853) | 55.5 (676) | 0.0005 | 0.16 |
|
| |||||||||
| Aminoglycosides | Gentamicin | 31.5 (466) | 31.6 (551) | 43.1 (587) | 40.5 (707) | 36.9 (852) | 29.4 (660) | <0.0001 | 0.796 |
|
| |||||||||
| Tetracyclines | Tigecycline | 45.5 (279) | 52.1 (363) | 37.1 (456) | 30.8 (448) | 28.5 (228) | NA | <0.0001 | <0.0001 |
|
| |||||||||
| Others | Fosfomycin | 22.7 (463) | 27.9 (498) | 22.9 (493) | 23.2 (573) | 37.0 (662) | 40.8 (441) | <0.0001 | <0.0001 |
| Trimethoprim/sulfamethoxazole | 62.2 (466) | 63.7 (551) | 63.2 (587) | 65.2 (707) | 45.4 (853) | 33.5 (681) | <0.0001 | <0.0001 | |
∗ p value with chi-square, ∗∗p value with Cochran–Armitage trend test.
Finally, the antibiotics that showed a resistance rate of less than 50% were tigecycline (38.8%), gentamicin (35.8%), and fosfomycin (29.2%) (Table 1).
Furthermore, some antibiotics showed a reduction of resistance rate over the years. Among them, trimethoprim/sulfamethoxazole in 2015 showed a resistance rate of 62,2% while in 2020 it was 33.5%, the same trend for tigecycline (from 45.5% in 2015 to 28.5% in 2019) and meropenem (from 49.0% in 2015 to 32.1% in 2020). On the contrary, fosfomycin showed an increase in the resistance rate from 22.7% in 2015 to 40.8% in 2020. Lastly, the ESBL isolates were between 20 and 25% each year.
4. Discussion
In recent decades, nosocomial infections have posed a major threat to healthcare systems worldwide, mainly in developing countries [28]. K. pneumoniae, being one of the most common pathogens, plays a significant role as the causative agent of severe infections [29]. As the main nosocomial opportunistic pathogen, it is continuously exposed to various antibiotics, leading to the development of resistance mechanisms and the spread of MDR strains. However, the antibiotic resistance rate of K. pneumoniae may vary depending on geographic location, population, and antibiotic management [30]. This study investigated the prevalence and antibiotic resistance profiles of K. pneumoniae isolates from different specimen sources in inpatients from 2015 to 2020. The analysis of incidence by age group revealed a low rate in pediatric patients (0–18 years), with the highest incidence in adult patients aged between 61 and 75 years (31%) and 76– and 95 years (21%). These findings align with Cassini et al., who reported the highest infection rate in Italian individuals aged 65 years or older [19]. K. pneumoniae was mainly isolated from urine cultures (45.3%) followed from vaginal and wound swabs (19.9%), sputum and bronchial-aspirate samples (15.4%), blood cultures (12.5%), and medical device samples (6.9%). In contrast, the study conducted by Romanus et al. showed a following distribution of 30.5% in the urinary tract, 23.6% in respiratory samples and 40% in blood cultures [31]. Another study reported a major portion of bacteria isolated from sputum and bronchoalveolar specimens (49.5%), with a higher rate than in other samples, while the isolates from urine were the second population (16.3%), followed by blood source (8.7%) [32].
In our study, we found that between 20 and 25% of ESBL K. pneumoniae isolates consistent with a previous study in Nigeria where the frequency of this phenotype was 17% [31]. It is essential to note that the prevalence of the ESBL phenotype could vary according to geographical location; for instance, this type of resistance ranges from 14% to 16% for France and England, respectively, to 5% in the United States [33]. Moreover, the Annual Epidemiological Report for 2020 in the European Union showed 33.9% of K. pneumoniae isolated were ESBL, and in Italy, this rate was over 50%. On the other hand, in Iran, studies by Kashefieh et al. and Kiaei et al. reported a percentage of ESBL isolates of 65% and 41.4%, respectively [34, 35]. Finally, in China, Xu et al. reported 33.7% of K. pneumoniae isolates were ELBS-producing [35].
The high incidence of ESBL-producing K. pneumoniae is likely due to the common consumption of third generation cephalosporins in society [36]. According to our study, the highest incidence of resistance was to ampicillin (98.5%). This result aligns with studies conducted in Iran and Russia, where the resistance rate was 97% [37, 38].
In this study, the most effective antibiotics against K. pneumoniae were fosfomycin, imipenem, gentamicin, and tigecycline, with susceptibility of 71%, 72%, 64%, and 61%, respectively.
Fosfomycin is a broad-spectrum antibiotic against Gram-negative and Gram-positive bacteria and has played a major role in treating MDR of Enterobacteriaceae in recent years [39]. Our results regarding fosfomycin for the treatment of K. pneumoniae align with other studies where the resistance rate was 16% and 10.9%. Multiple mechanisms of fosfomycin resistance may be attributed to antimicrobial modifying enzymes, target site modification, or reduced permeability [40].
Due to the limited availability of new antimicrobial agents, the evaluation of older antimicrobial drugs, including fosfomycin, polymyxins, and aminoglycosides, appears as a possible strategy for managing infections induced by multidrug-resistant (MDR) bacteria. Fosfomycin, discovered in 1969, has a bactericidal action against Gram-positive and Gram-negative bacterial species [41]. In recent years, fosfomycin has been increasingly used in Europe and Asia to treat infections caused by carbapenem-resistant Enterobacteriaceae isolates [39, 42].
However, the increased use of fosfomycin favors the spread of resistant isolates, as reported by recent studies evaluating the antimicrobial activity of fosfomycin against carbapenem-resistant K. pneumoniae [43, 44]. These studies report different resistance rates of KPC isolates to fosfomycin in China (61.9%), Taiwan (36.4%), Germany (32%), and the United States (15%).
However, there are few studies on the correlation between fosfomycin resistance and carbapenem resistance in K. pneumoniae isolates, reporting the prevalence and mechanisms of fosfomycin resistance in KPC clinical isolates.
Another effective class of antibiotics for the treatment of this opportunistic pathogen is carbapenems. With the advent of the ESBL-positive phenotype and increasing resistance to these antibiotics, carbapenems have emerged as the drugs of choice for the treatment of serious infections caused by K. pneumoniae ESBL [45]. In our study, the resistance rate to imipenem was 28%, consistent with other studies conducted previously in which the resistance rates were 25.7% [34, 46]. These data are concerning because it is much higher than the rate reported in Europe (10%).
Finally, another interesting class of antibiotics is represented by aminoglycosides which is currently the most widely used drugs for the treatment of multiresistant K. pneumoniae. In our study, the gentamicin sensitivity rate was 64%, similar to the study by Hesam et al., which showed a gentamicin resistance rate of 34.6% [47]. In contrast, in the study by Kashelfieh et al., the resistance rate was 51% [34].
5. Conclusions
This study, through a comprehensive statistical analysis, provided an evaluation of the antibiotic susceptibility trends of K. pneumoniae isolates, revealing increased sensitivity to piperacillin/tazobactam, cefepime, meropenem, imipenem and, conversely, increased resistance to fosfomycin. In conclusion, this study aims to underline the importance of monitoring the evolution of antimicrobial susceptibility models in our province, implementing surveillance systems for multiresistant microorganisms and verifying the effectiveness of the empirical therapies adopted, minimizing therapeutic failures and spread of the phenomenon of antibiotic resistance. Innovative and continuous surveillance strategies are an indispensable resource to preserve antibiotics' effectiveness and patients' health. Finally, improving hygienic conditions and prevention strategies are necessary to slow the spread of multiresistant microorganisms and infections acquired in nosocomial environments.
Acknowledgments
The authors would like to thank the staff of University Hospital “San Giovanni di Dio e Ruggi d'Aragona” for their contributions.
Contributor Information
Giovanni Boccia, Email: gboccia@unisa.it.
Gianluigi Franci, Email: gfranci@unisa.it.
Data Availability
Epidemiological data used to support the results of this study are included within the article.
Consent
Patient consent in this study was not necessary because our study used laboratory management data and clinical information on patients collected from databases. Moreover, it maintained the patient incognito.
Disclosure
The resignation was given, as our study used laboratory management data and clinical information on patients collected from databases. This is a retrospective study and not directly associated with patients. This study was consistent with the principles of the Helsinki Declaration.
Conflicts of Interest
The authors declare that there are no conflicts of interest regarding the publication of this paper.
Supplementary Materials
Supplementary Tables include Supplementary Table 1: distribution of cases for gender and age class and Supplementary Table 2: distribution of diagnostic materials by hospital department from 2015 to 2020.
References
- 1.Murray C. J. L., Ikuta K. S., Sharara F., et al. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. The Lancet . 2022;399(10325):629–655. doi: 10.1016/s0140-6736(21)02724-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chia P. Y., Sengupta S., Kukreja A; S. L., Sl Ponnampalavanar S., Ng O. T., Marimuthu K. The role of hospital environment in transmissions of multidrug-resistant gram-negative organisms. Antimicrobial Resistance and Infection Control . 2020;9(1):p. 29. doi: 10.1186/s13756-020-0685-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.WHO. Antimicrobial Resistance Expected to Cause 5.2 Million Deaths in the Western Pacific by 2030. https://www.who.int/westernpacific/news/item/13-06-2023-antimicrobial-resistance-expected-to-cause-5.2-million-deaths-in-the-western-pacific-by-2030 .
- 4.Navon-Venezia S., Kondratyeva K., Carattoli A. Klebsiella pneumoniae: a major worldwide source and shuttle for antibiotic resistance. FEMS Microbiology Reviews . 2017;41(3):252–275. doi: 10.1093/femsre/fux013. [DOI] [PubMed] [Google Scholar]
- 5.Paczosa M. K., Mecsas J. Klebsiella pneumoniae: going on the offense with a strong defense. Microbiology and Molecular Biology Reviews: Microbiology and Molecular Biology Reviews . 2016;80(3):629–661. doi: 10.1128/mmbr.00078-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu X., Sai F., Li L., Zhu C., Huang H. Clinical characteristics and risk factors of catheter-associated urinary tract infections caused by Klebsiella Pneumoniae. Annals of Palliative Medicine . 2020;9(5):2668–2677. doi: 10.21037/apm-20-1052. [DOI] [PubMed] [Google Scholar]
- 7.Choby J. E., Howard-Anderson J., Weiss D. S. Hypervirulent Klebsiella pneumoniae – clinical and molecular perspectives. Hypervirulent Klebsiella pneumoniae clinical and molecular perspectives Journal of internal medicine . 2020;287(3):283–300. doi: 10.1111/joim.13007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang D., Sharma L., Dela Cruz C. S., Zhang D. Clinical epidemiology, risk factors, and control strategies of Klebsiella pneumoniae infection. Frontiers in Microbiology . 2021;12 doi: 10.3389/fmicb.2021.750662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adar A., Zayyad H., Azrad M., et al. Clinical and demographic characteristics of patients with a new diagnosis of carriage or clinical infection with carbapenemase-producing enterobacterales: a retrospective study. Frontiers in Public Health . 2021;9 doi: 10.3389/fpubh.2021.616793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rawat D., Nair D. Extended-spectrum ß-lactamases in gram negative bacteria. Journal of Global Infectious Diseases . 2010;2(3):263–274. doi: 10.4103/0974-777x.68531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tekele S. G., Teklu D. S., Tullu K. D., Birru S. K., Legese M. H. Extended-spectrum Beta-lactamase and AmpC beta-lactamases producing gram negative bacilli isolated from clinical specimens at International Clinical Laboratories, Addis Ababa, Ethiopia. PLoS One . 2020;15(11) doi: 10.1371/journal.pone.0241984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nordmann P., Dortet L., Poirel L. Carbapenem resistance in Enterobacteriaceae: here is the storm. Trends in Molecular Medicine . 2012;18(5):263–272. doi: 10.1016/j.molmed.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 13.Chattopadhya D., Devi L., Broor S., Rautela R., Grover S., Chakravarti A. Increasing prevalence of Escherichia coli and Klebsiella pneumoniae producing CTX-M-type extended-spectrum beta-lactamase, carbapenemase, and NDM-1 in patients from a rural community with community acquired infections: a 3-year study. International Journal of Applied and Basic Medical Research . 2020;10(3):156–163. doi: 10.4103/ijabmr.IJABMR_360_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yigit H., Queenan A. M., Anderson G. J., et al. Novel carbapenem-hydrolyzing β-lactamase, KPC-1, from a carbapenem-resistant strain ofKlebsiella pneumoniae. Antimicrobial Agents and Chemotherapy . 2001;45(4):1151–1161. doi: 10.1128/aac.45.4.1151-1161.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Assessment E. R. R. Regional Outbreak of New Delhi Metallo-Betalactamase-Producing Carbapenem-Resistant Enterobacteriaceae . Solna, Sweden: European Centre for Disease Prevention and Control; 2019. [Google Scholar]
- 16.WHO. WHO publishes list of bacteria for which new antibiotics are urgently needed. https://www.who.int/news/item/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed .
- 17.Xu L., Sun X., Ma X. Systematic review and meta-analysis of mortality of patients infected with carbapenem-resistant Klebsiella pneumoniae. Annals of Clinical Microbiology and Antimicrobials . 2017;16(1):p. 18. doi: 10.1186/s12941-017-0191-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liao W., Liu Y., Zhang W. Virulence evolution, molecular mechanisms of resistance and prevalence of ST11 carbapenem-resistant Klebsiella pneumoniae in China: a review over the last 10 years. Journal of Global Antimicrobial Resistance . 2020;23:174–180. doi: 10.1016/j.jgar.2020.09.004. [DOI] [PubMed] [Google Scholar]
- 19.Cassini A., Högberg L. D., Plachouras D., et al. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: a population-level modelling analysis. The Lancet Infectious Diseases . 2019;19(1):56–66. doi: 10.1016/s1473-3099(18)30605-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Solomon S. L., Oliver K. B. Antibiotic resistance threats in the United States: stepping back from the brink. American Family Physician . 2014;89(12):938–941. [PubMed] [Google Scholar]
- 21.Gu D., Dong N., Zheng Z., et al. A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. The Lancet Infectious Diseases . 2018;18(1):37–46. doi: 10.1016/s1473-3099(17)30489-9. [DOI] [PubMed] [Google Scholar]
- 22.Gomez-Simmonds A., Nelson B., Eiras D. P., et al. Combination regimens for treatment of carbapenem-resistant Klebsiella pneumoniae bloodstream infections. Antimicrobial Agents and Chemotherapy . 2016;60(6):3601–3607. doi: 10.1128/aac.03007-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Santella B., Serretiello E., De Filippis A., et al. Lower respiratory tract pathogens and their antimicrobial susceptibility pattern: a 5-year study. Antibiotics . 2021;10(7):p. 851. doi: 10.3390/antibiotics10070851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Uddin F., Sohail M., Shaikh Q. H., et al. PCR and microarray analysis of AmpC and ESBLs producing Pseudomonas aeruginosa isolates from intensive care units. Gene Reports . 2021;23 doi: 10.1016/j.genrep.2021.101178. [DOI] [Google Scholar]
- 25.Uddin F., Imam S. H., Khan S., et al. NDM production as a dominant feature in carbapenem-resistant Enterobacteriaceae isolates from a tertiary care hospital. Antibiotics . 2021;11(1):p. 48. doi: 10.3390/antibiotics11010048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Razaq L., Uddin F., Ali S., et al. Vitro activity of new β-lactamase inhibitor combinations against blaNDM, blaKPC, and ESBL-producing enterobacteriales uropathogens. In Vitro Activity of New β-Lactamase Inhibitor Combinations against blaNDM, blaKPC, and ESBL-Producing Enterobacteriales Uropathogens, Antibiotics (Basel, Switzerland) . 2023;12(10):p. 1481. doi: 10.3390/antibiotics12101481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jalal N. A., Al-Ghamdi A. M., Momenah A. M., et al. Prevalence and antibiogram pattern of Klebsiella pneumoniae in a tertiary care hospital in makkah, Saudi Arabia: an 11-year experience. Antibiotics . 2023;12(1):p. 164. doi: 10.3390/antibiotics12010164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nimer N. A. Nosocomial infection and antibiotic-resistant threat in the Middle East. Infection and Drug Resistance . 2022;15:631–639. doi: 10.2147/idr.S351755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Effah C. Y., Sun T., Liu S., Wu Y. Klebsiella pneumoniae: an increasing threat to public health. Annals of Clinical Microbiology and Antimicrobials . 2020;19:p. 1. doi: 10.1186/s12941-019-0343-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Asbell P. A., Pandit R. T., Sanfilippo C. M. Antibiotic resistance rates by geographic region among ocular pathogens collected during the ARMOR surveillance study. Ophthalmol Ther . 2018;7(2):417–429. doi: 10.1007/s40123-018-0141-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Romanus I. I., Egwu O. A., Ngozi A. T., Chidiebube N. A., Chika E. P. Extended spectrum beta-lactamase (ESBL) mediated resistance to antibiotics among Klebsiella pneumoniae in Enugu Metropolis. Macedonian Journal of Medical Sciences . 2009;2:196–199. [Google Scholar]
- 32.Zhang J., Li D., Huang X., Long S., Yu H. The distribution of K. pneumoniae in different specimen sources and its antibiotic resistance trends in sichuan, China from 2017 to 2020. Frontiers of Medicine . 2022;9 doi: 10.3389/fmed.2022.759214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Castanheira M., Simner P. J., Bradford P. A. Extended-spectrum β-lactamases: an update on their characteristics, epidemiology and detection. JAC-Antimicrobial Resistance . 2021;3 doi: 10.1093/jacamr/dlab092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kashefieh M., Hosainzadegan H., Baghbanijavid S., Ghotaslou R. The molecular epidemiology of resistance to antibiotics among Klebsiella pneumoniae isolates in Azerbaijan, Iran. Journal of Tropical Medicine . 2021;2021:9. doi: 10.1155/2021/9195184.9195184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kiaei S., Moradi M., Hosseini-Nave H., Ziasistani M., Kalantar-Neyestanaki D. Endemic dissemination of different sequence types of carbapenem-resistant Klebsiella pneumoniae strains harboring bla (NDM) and 16S rRNA methylase genes in Kerman hospitals, Iran, from 2015 to 2017. Infection and Drug Resistance . 2018;12:45–54. doi: 10.2147/idr.S186994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paterson D. L., Bonomo R. A. Extended-spectrum β-lactamases: a clinical update. Clinical Microbiology Reviews . 2005;18(4):657–686. doi: 10.1128/cmr.18.4.657-686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sheykhsaran E., Bannazadeh Baghi H., Soroush Barhaghi M. H., et al. The rate of resistance to tetracyclines and distribution of tetA, tetB, tetC, tetD, tetE, tetG, tetJ and tetY genes in Enterobacteriaceae isolated from Azerbaijan, Iran during 2017. Physiology and Pharmacology . 2018;22:205–212. [Google Scholar]
- 38.Khaertynov K. S., Anokhin V. A., Rizvanov A. A., et al. Virulence factors and antibiotic resistance of Klebsiella pneumoniae strains isolated from neonates with sepsis. Frontiers of Medicine . 2018;5:p. 225. doi: 10.3389/fmed.2018.00225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Falagas M. E., Kastoris A. C., Kapaskelis A. M., Karageorgopoulos D. E. Fosfomycin for the treatment of multidrug-resistant, including extended-spectrum β-lactamase producing, Enterobacteriaceae infections: a systematic review. The Lancet Infectious Diseases . 2010;10(1):43–50. doi: 10.1016/S1473-3099(09)70325-1. [DOI] [PubMed] [Google Scholar]
- 40.Castaneda-Garcia A., Blazquez J., Rodriguez-Rojas A. Molecular mechanisms and clinical impact of acquired and intrinsic fosfomycin resistance. Antibiotics . 2013;2:217–236. doi: 10.3390/antibiotics2020217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Falagas M. E., Vouloumanou E. K., Samonis G., Vardakas K. Z. Fosfomycin. Clinical Microbiology Reviews . 2016;29(2):321–347. doi: 10.1128/CMR.00068-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ruiz Ramos J., Salavert Lleti M. Fosfomycin in infections caused by multidrug-resistant Gram-negative pathogens. Revista Española de Quimioterapia: publicacion oficial de la Sociedad Espanola de Quimioterapia . 2019;32(Suppl 1):45–54. [PMC free article] [PubMed] [Google Scholar]
- 43.Cho Y. H., Jung S. I., Chung H. S., et al. Antimicrobial susceptibilities of extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae in health care-associated urinary tract infection: focus on susceptibility to fosfomycin. International Urology and Nephrology . 2015;47(7):1059–1066. doi: 10.1007/s11255-015-1018-9. [DOI] [PubMed] [Google Scholar]
- 44.Liu H. Y., Lin H. C., Lin Y. C., Yu S. H., Wu W. H., Lee Y. J. Antimicrobial susceptibilities of urinary extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae to fosfomycin and nitrofurantoin in a teaching hospital in Taiwan. Journal of Microbiology, Immunology, and Infection . 2011;44(5):364–368. doi: 10.1016/j.jmii.2010.08.012. [DOI] [PubMed] [Google Scholar]
- 45.Subha A., Ananthan S. Extended spectrum beta lactamase (ESBL) mediated resistance to third generation cephalosporins among Klebsiella pneumoniae in Chennai. Indian Journal of Medical Microbiology . 2002;20(2):92–95. [PubMed] [Google Scholar]
- 46.Tavakol M., Momtaz H. Determination of antibiotic resistance profile in Klebsiella pneumonia strains isolated from urinary tract infections of patients hospitalized in Peyambaran hospital (Tehran-Iran) KAUMS Journal (FEYZ) . 2017;21:74–82. [Google Scholar]
- 47.Mokhtari H., Eslami G., Zandi H., Dehghan-Banadkouki A., Vakili M. Evaluating the frequency of aac(6′)-IIa, ant(2″)-I, intl1, and intl2 genes in aminoglycosides resistant Klebsiella pneumoniae isolates obtained from hospitalized patients in yazd, Iran. Avicenna Journal of Medical Biotechnology . 2018;10(2):115–119. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Tables include Supplementary Table 1: distribution of cases for gender and age class and Supplementary Table 2: distribution of diagnostic materials by hospital department from 2015 to 2020.
Data Availability Statement
Epidemiological data used to support the results of this study are included within the article.


