Abstract
In a two-hybrid screen for proteins that interact with human PCNA, we identified and cloned a human protein (hCdc18) homologous to yeast CDC6/Cdc18 and human Orc1. Unlike yeast, in which the rapid and total destruction of CDC6/Cdc18 protein in S phase is a central feature of DNA replication, the total level of the human protein is unchanged throughout the cell cycle. Epitope-tagged protein is nuclear in G1 and cytoplasmic in S-phase cells, suggesting that DNA replication may be regulated by either the translocation of this protein between the nucleus and the cytoplasm or the selective degradation of the protein in the nucleus. Mutation of the only nuclear localization signal of this protein does not alter its nuclear localization, implying that the protein is translocated to the nucleus through its association with other nuclear proteins. Rapid elimination of the nuclear pool of this protein after the onset of DNA replication and its association with human Orc1 protein and cyclin-cdks supports its identification as human CDC6/Cdc18 protein.
The recent identification of multiple eukaryotic proteins that bind directly or indirectly to origins of DNA replication has set the stage for a thorough exploration of how DNA replication is initiated and regulated in eukaryotes. Central to the process is the origin recognition complex (ORC), a tight complex of six polypeptides identified initially in Saccharomyces cerevisiae because it binds in a sequence-specific manner to origins of DNA replication and is involved in the initiation of chromosomal DNA replication (1, 3, 27). Homologs of three of the subunits of the ORC (Orc1, Orc2, and Orc4) have been identified in humans (16, 32) and in other eukaryotes (14, 17, 22, 28).
An additional molecule, CDC6 in S. cerevisiae and Cdc18 in Schizosaccharomyces pombe, is essential for the onset of DNA replication and known to associate physically with the ORC and cdc2 kinase (2, 12, 15, 18, 22, 24, 37). The yeast CDC6/Cdc18 proteins decrease in concentration as cells proceed through S phase, with overexpression of Cdc18 in S. pombe resulting in the rereplication of DNA in G2 (29). The Xenopus CDC6/Cdc18 and ORC are required for DNA replication and for the loading of the minichromosome maintenance (MCM) proteins onto the chromatin (6, 11, 13, 19, 34, 35). Collectively, a model has emerged that emphasizes the central role of CDC6/Cdc18 in cooperating with ORC and MCM proteins to form a prereplication complex at origins of DNA replication in G1. Once replication begins, the concordant removal of CDC6/Cdc18 prevents the loading of MCM proteins onto the origin-bound ORC in G2, thereby preventing rereplication of DNA. After mitosis, synthesis of new CDC6/Cdc18 protein allows the loading of MCM proteins on the chromatin, perhaps by forming a bridge between the chromatin-bound ORC and the MCM proteins (4). The prereplication initiation complex thus formed in G1 is ready to initiate DNA replication upon the activation of S-phase-promoting factors like cyclin-cdk’s and CDC7 kinase.
We identified and cloned a human protein that is structurally homologous to yeast CDC6/Cdc18 and Orc1 proteins by virtue of its interaction with human PCNA in a two-hybrid assay. A similar human protein was independently identified by Williams and coworkers because of its sequence homology with Orc1 and CDC6/Cdc18 and was named human CDC6 protein (36). Biochemical experiments with this protein reported here confirm that it is the putative human CDC6/Cdc18 but also indicate that, although the overall model of replication regulation is conserved between humans and yeasts, some aspects are different in detail.
MATERIALS AND METHODS
Yeast two-hybrid screen.
For the yeast two-hybrid screen, the open reading frame encoding human PCNA was cloned by PCR and inserted into the pAS2 vector to create a fusion protein with the GAL4 DNA-binding domain (7). The interacting plasmid from a B-cell cDNA library (human PCNA interacting protein 4; HPB4) had a 1.2-kb cDNA insertion encoding a protein fragment whose sequence was closely related to Cdc18 from S. pombe and to human Orc1 (hOrc1).
Cloning of hCdc18 gene.
Using HPB4 as a probe, we isolated a 1.6-kb cDNA from a human fetal brain cDNA library. This 1.6-kb cDNA overlapped with the HPB4 gene and extended the 5′ terminus by 600 bp. The extreme 5′ terminus of the human Cdc18 (hCdc18) gene was found by employing 5′ rapid amplification of cDNA ends (RACE) with human HeLa Marathon-Ready cDNA (Clontech), which extended the 5′ end of the Cdc18 clone by 300 bases. There is an in-frame stop codon 150 bp 5′ of the first methionine. The 3′ end of the hCdc18 gene cDNA came from the expressed sequence tag (EST) clone T85849 (GenBank), which had a 150-base overlap with the 3′ end of the HPB4 gene sequence, and extends the nucleotide sequence by 1,000 bases and the protein sequence by five amino acids. The hCdc18 gene cDNA was constructed by combining the RACE PCR product, HPB4 gene insert, and T85849 EST clone and contains an open reading frame flanked at both ends by stop codons. The predicted protein consists of 560 amino acids with a calculated molecular mass of 62.7 kDa.
RNA analysis.
Total RNA was extracted from HeLa cells as described previously (10), and 10 μg was subjected to Northern blot analysis at 42°C. The probes used were a 1.2-kb cDNA fragment from the HPB4 gene (nucleotides 520 to 1750 of the hCdc18 gene), a 2.4-kb cDNA fragment from pKG28 (the entire open reading frame encoding hOrc1 [16]), a 1.3-kb HindIII-PstI cDNA fragment (encoding glyceraldehyde-3-phosphate dehydrogenase), and cDNA fragments containing the entire open reading frames encoding cyclins E, A, B, and hOrc2 (16).
Plasmid construction and protein purification.
Full-length hCdc18 (amino acids 1 to 560) and its fragments (amino acids 1 to 179, 179 to 407, and 407 to 560) were generated by PCR with appropriate pairs of oligonucleotide primers and Pfu polymerase (Stratagene) and cloned into BamHI and SalI sites of pGEX vector (Pharmacia) to generate glutathione S-transferase (GST) fusion proteins (9).
The full-length hCdc18 was cloned into mammalian expression vectors pA3M and pAHP to generate plasmids expressing hCdc18 with a myc (pA3M) epitope tag at the C terminus or hemagglutinin (pAHP) epitope tag at the N terminus. Plasmid pEBG expresses GST in mammalian cells (26). The coding region of the Cdc18 gene was cloned into the BamHI site of pEBG to generate pEBG-Cdc18 (sense orientation) and pEBG-Cdc18rev (antisense orientation). The former expresses GST-Cdc18 (90 kDa), and the latter expresses GST fused to a short irrelevant protein expressed from the antisense strand of the Cdc18 gene (30 kDa).
Cell synchronization.
Exponentially growing HeLa or U2OS cells were arrested for 24 h with 10 mM hydroxyurea at early S phase, with 40 ng of nocodazole/ml at M phase, or with 5 μg of aphidicholin/ml at the G1-S transition point. The cell populations were checked by propidium iodide staining of the DNA and by flow cytometry. For serum starvation and release experiments with NIH 3T3, CV1, and WI38 cells, 60%-confluent cultures were placed in Dulbecco’s modified Eagle’s medium (DMEM) containing 0.5% serum for 48 h and split 1:2 into DMEM with 10% calf serum.
Pull-down assay.
Affinity chromatography on glutathione beads coated with various GST fusion proteins (pull-down assay) was done as described previously (8, 9). Incubation of MCM proteins with GST- or GST-Cdc18-coated beads was carried out at 37°C for 10 min followed by 4°C for 1 h. The unbound proteins were washed off with A7.4 buffer containing 100 mM NaCl.
Antibodies and immunoprecipitations.
Antibody against hCdc18 was raised against a GST protein fused to a fragment of hCdc18 protein from amino acids 145 to 326. The antibody was affinity purified on the antigen to confirm the immunoblot experiments. Antibody against hOrc1 was raised against a recombinant His6-tagged fragment of Orc1 from amino acids 647 to 861 created by cloning the 1.1-kb EcoRI-HindIII fragment of EST clone 121313 (GenBank) into pRSETB (Invitrogen). Antibody against hOrc2 was raised against a recombinant His6-tagged fragment of Orc2 from amino acids 27 to 577 created by cloning the XbaI-SacI Orc2 fragment into the PvuII site of pRSETC. Immunoblotting of cell lysates (20 μg of protein) was carried out with the antibodies at the following dilutions: anti-hCdc18, 1:5,000; anti-hOrc1, 1:15,000; anti-hOrc2, 1:2,000; and 9E10 anti-Myc monoclonal antibody ascites, 1:2,000.
Immunofluorescence.
U2OS cells were transfected with pAHP-Cdc18 or with pA3M-Cdc18. The cells were fixed in 3% formaldehyde containing 2% sucrose at room temperature for 10 min, washed twice with phosphate-buffered saline (PBS), and permeabilized in Triton solution (3% bovine serum albumin, 0.5% Triton X-100 in PBS) for 5 min. After being washed with PBS, the cells were incubated for 20 min at 37°C with anti-hemagglutinin (HA) antibody (12CA5) in hybridoma culture supernatant diluted 1:100, anti-Myc monoclonal antibody in mouse ascites diluted 1:100, anti-p21 rabbit polyclonal antibody (Santa Cruz) diluted 1:100, or human autoimmune antibody to PCNA. Cells were then washed three times with PBS and incubated with the secondary antibody (fluorescein isothiocyanate [FITC]-conjugated goat anti-mouse immunoglobulin G [IgG], rhodamine-conjugated goat anti-rabbit IgG, or anti-human IgG [Jackson Laboratory]). After being washed three times with PBS, the cells were stained briefly with DAPI (4′,6-diamidino-2-phenylindole) and photographed under a fluorescent microscope.
Preparation of HeLa whole-cell extract and column chromatography.
HeLa whole-cell extract was prepared according to the procedure of Manley et al. (25). All steps were performed at 4°C. Briefly, a HeLa cell pellet was suspended and incubated on ice for 20 min in 4 volumes of buffer WCE I (10 mM Tris-acetate [pH 7.9], 1 mM EDTA, 5 mM dithiothreitol [DTT], and 1 mM phenylmethylsulfonyl fluoride [PMSF]). The cells were lysed in a Dounce homogenizer with eight strokes and then mixed with another 4 volumes of buffer WCE II (50 mM Tris acetate [pH 7.9], 10 mM Mg acetate, 25% sucrose, 50% glycerol, 2 mM DTT, and 1 mM PMSF). Ammonium sulfate was added to a final concentration of 0.4 M, and after stirring for 30 min, the extract was centrifuged at 40,000 rpm in a Ti45 rotor (Beckman) for 3 h. The supernatant was subjected to 65% ammonium sulfate precipitation, after which the pellet was suspended in 20 mM Tris acetate (pH 7.9) containing 1 mM EDTA, 0.15 M potassium acetate, 20% glycerol, 2 mM DTT, and 1 mM PMSF (buffer H), and the suspension was dialyzed against the same buffer. Starting from 3 × 1010 cells, 3.0 g of protein was recovered. The protein extract was applied to a Bio-Rex 70 (Bio-Rad) column (bed volume, 65 ml) equilibrated in buffer H, and the bound proteins were eluted with 0.3, 0.6, and 1.5 M potassium acetate in buffer H. The 0.3 M fraction containing Orc1 and Cdc18 (total protein, 280 mg) was dialyzed and further fractionated over a DEAE-cellulose column (bed volume, 25 ml) equilibrated in buffer H containing 0.2 M potassium acetate. The proteins were eluted with 0.35 and 1.0 M potassium acetate in buffer H. A portion of 0.2 M flowthrough (total protein recovery, 88 mg) containing 3.5 mg of protein was subjected to Mono-S HR 5/5 fast protein liquid chromatography (Pharmacia) column in 0.2 M KCl containing buffer and eluted with a gradient of 0.2 to 0.6 M KCl containing buffer. The flowthrough containing Orc1 and Cdc18 was further characterized by Superose 12 gel filtration chromatography.
RESULTS
Cloning and sequence analysis of hCdc18.
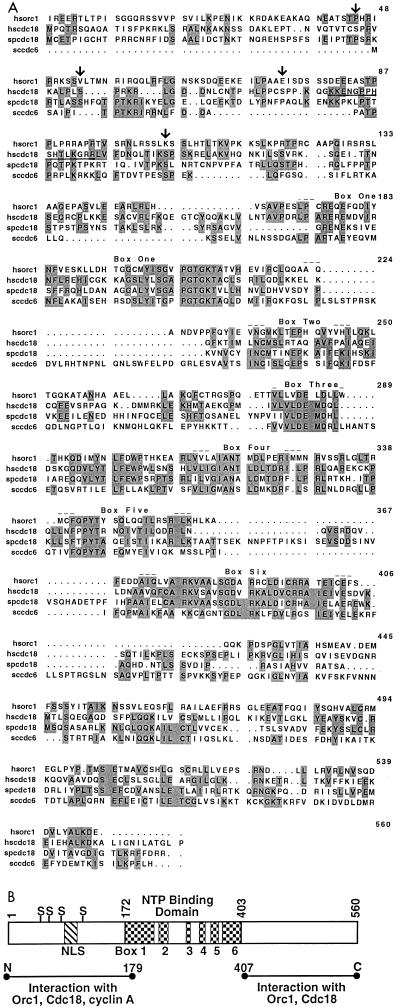
While screening for proteins which interact with the human DNA replication factor PCNA in a two-hybrid assay, we discovered an interacting molecule whose sequence was related to S. pombe Cdc18 (33% identical, 53% similar), S. cerevisiae CDC6 (32% identical, 54% similar), and hOrc1 (28% identical and 52% similar) (Fig. 1). The theoretical mass of the protein is similar to that of yeast CDC6/Cdc18 (60 kDa) and different from that of the Orc1 proteins (100 to 120 kDa). The homology with the Orc1-Cdc18 family is most marked in the middle one-third of the protein, which contains a nucleotide binding motif with the putative P and A loops (20) together with four other conserved boxes unique to this family of proteins (2, 16). The N-terminal one-third has four potential sites of phosphorylation by cyclin-cdk’s (serines at positions 45, 54, 74, and 106) and a putative bipartite nuclear localization sequence (NLS) at positions 80 to 95 (33) (Fig. 1B).
FIG. 1.
(A) Alignment of protein sequences of hCdc18 (hscdc18) with those of hOrc1 (hsorc1), S. pombe Cdc18 (spcdc18), and S. cerevisiae CDC6 (sccdc6). The numbers refer to the sequence of hCdc18. The alignment was done with the PILEUP program of the Genetics Computer Group package. Identical amino acids are shaded. Arrows mark putative substrate sites for cyclin-cdk’s (on the hscdc18 protein), and the putative bipartite NLS is underlined. Conserved boxes 1 to 6 are indicated; boxes 1 and 3 contain the P and A loops, respectively. (B) Schematic representation of the hCdc18 protein divided into three domains. Boxes 1 to 6 in the middle one-third of the protein are the same as in panel A. S, a serine in a putative cyclin-cdk phosphorylation site; NLS, putative bipartite nuclear localization sequence; NTP, nucleoside triphosphate. The horizontal lines represent recombinant fragments of Cdc18 which are sufficient to mediate the interactions with Orc1, Cdc18 (Fig. 4A), and cyclin A (data not shown).
As shown in Fig. 1, the sequence homology of the newly identified protein with hOrc1 leaves open the possibility that it is a member of the Orc1-Cdc18 family and is not necessarily the human CDC6/Cdc18. We have independently identified a hOrc4 which, surprisingly, has sequence similarity to the Orc1-Cdc18 family of proteins, implying that there may be an entire family of human proteins with sequence homology to yeast Orc1 or Cdc18 (32). The protein-protein association and the cell cycle regulation of the new protein reported below, however, provides strong evidence that it is indeed human CDC6/Cdc18.
Although hCdc18 transcripts are increased at the G1-S transition, total Cdc18 protein levels are uniform throughout the cell cycle.
One of the characteristic properties of yeast CDC6/Cdc18 is that the highest levels of the mRNA and protein are obtained in the G1 phase of the cell cycle, with almost all RNA and protein disappearing in the S and G2 phases. Northern blot analysis of total RNA from HeLa cells at various stages in the cell cycle indicates that with hCdc18, the mRNA level peaks at the onset of S phase (simultaneous with the expression of cyclin E) and diminishes at the onset of mitosis (when cyclin B is expressed) (Fig. 2A). This pattern is repeated for hOrc1 but not hOrc2. Consistent with this observation, cells arrested with hydroxyurea (in S phase) have significantly more Cdc18, Orc1, and cyclin E mRNA than cells arrested with nocodazole (in M phase) (Fig. 2B). Such variation of mammalian Orc1 and Cdc18 mRNA during the cell cycle agrees with observations in the yeasts. The cell cycle stage-specific regulation of mammalian Orc1 and mammalian CDC6/Cdc18 mRNA was reported by others to be due to the activity of E2F transcription factor and was postulated to be important for linking the initiation of S phase with activation of E2F (30, 36).
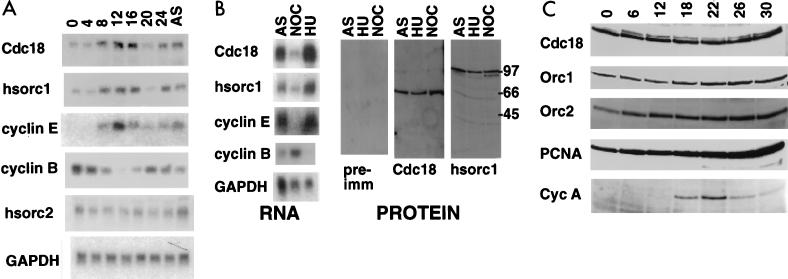
FIG. 2.
Levels of Cdc18 and Orc RNAs and proteins in different stages of the cell cycle. (A) HeLa cells were synchronized in mitosis by nocodazole (50 ng/ml), and after shake off, the cells were replated in nocodazole-free medium. A Northern blot of RNA extracted from the cells at the indicated number of hours after release is shown. AS, asynchronous cells; hsorc1 and -2, hOrc1 and -2; GAPDH, glyceraldehyde-3-phosphate dehydrogenase. (B) HeLa cells were either asynchronous (AS), blocked in S phase with hydroxyurea (HU), or blocked in mitosis with nocodazole (NOC). RNA, Northern blot of total RNA with the indicated probes; PROTEIN, immunoblots of protein extracts with preimmune serum (pre-imm) (for anti-Cdc18 antibody; the same result was obtained with the preimmune serum for anti-Orc1 antibody or rabbit serum against bacterially produced GST-Cdc18 protein (Cdc18) or the His6-tagged C-terminal one-third of hOrc1 protein (hsorc1). (C) NIH 3T3 cells were released from serum starvation and harvested at indicated time-points (h) as the cells moved from G0 through G1 to S. Extracts were immunoblotted for the indicated proteins. Cyc A, cyclin A.
Since cell cycle stage-specific regulation of mRNA does not always produce an equivalent cell cycle stage-specific regulation of the protein, we examined whether the abundance of the mammalian Orc1 and Cdc18 proteins changed during the cell cycle. An antibody raised against recombinant Cdc18 and immunoaffinity purified recognized a protein of 66 kDa in HeLa cell extracts, while one raised against Orc1 recognized a protein of 100 kDa (Fig. 2B). The anti-Cdc18 antibody also recognized a myc epitope-tagged Cdc18 expressed by transient transfection in human cells (data not shown). Although the mRNA levels of Cdc18 and Orc1 were much higher in S-phase (hydroxyurea-blocked) than in M-phase (nocodazole-blocked) cells, the protein levels were virtually unchanged between the two phases of the cell cycle. To ensure that the constant levels of total cellular Cdc18 protein was not an anomaly of transformed HeLa cells, we serum starved untransformed mouse NIH 3T3 cells and followed CDC6/Cdc18 protein levels after serum stimulation (Fig. 2C). The serum-starved cells had a significant amount of Cdc18 protein, with little change in the level of the protein as the cells moved from G0 through G1 into the S phase of the cell cycle upon release from serum starvation (Fig. 2C). The band of slightly reduced mobility seen in G1 is weaker but present in G0 cells and could be due to posttranslational modification of Cdc18 protein as cells enter the cell cycle. Both bands are recognized by affinity-purified anti-Cdc18 antibody. The levels of Orc1 and Orc2 proteins are similarly unchanged in this experiment. The constant levels of Orc2 and PCNA proteins demonstrate that all lanes contain protein from equivalent numbers of cells. The transient appearance of cyclin A shows that the proteins which are expected to cycle as cells passage through G1 into S do so normally in this experiment. Similar results were obtained with untransformed monkey CV-1 and primary human WI38 cells (data not shown). Therefore, the G1-S peak in the expression of Cdc18 or Orc1 mRNAs is not paralleled by changes in the total protein concentration.
Subcellular localization of hCdc18 protein changes during cell cycle.
The previous report on human CDC6/Cdc18 (36) noted that the protein associated with the nuclear fraction was transiently increased in G1 and early S and decreased thereafter. The authors concluded that the human CDC6/Cdc18 protein is regulated by cell cycle stage-specific degradation similar to that noted in the yeasts. Since we did not observe a change in the abundance of total cellular protein throughout the cell cycle, one explanation could be that the subcellular localization of CDC6/Cdc18 protein changed during the cell cycle, with increased nuclear protein seen in G1 and early S. To test this possibility, the subcellular localization of Cdc18 protein was determined by immunofluorescence.
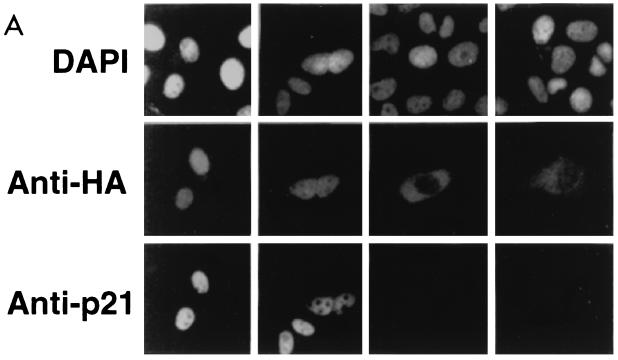
Because the anti-Cdc18 antibody did not immunostain cells, we expressed Cdc18 with an N-terminal HA epitope tag by transient transfection of human U2OS osteosarcoma cells and immunostained the cells with anti-HA antibody (12CA5). The results were confirmed with Cdc18 fused to a C-terminal myc epitope. Of 100 asynchronous cells expressing epitope-tagged Cdc18, 20 to 40% expressed the protein in the cytoplasm and 60 to 80% expressed it in the nucleus in different experiments (Fig. 3A and B). Thirty percent of the cells were in G1, as judged by positive staining for nuclear p21 (CIP1) (23). In all of these G1 cells, HA-hCdc18 protein was detected in the nucleus (Fig. 3A). All cells where HA-hCdc18 protein was found in the cytoplasm were negative for p21 (Fig. 3A), implying that HA-hCdc18 protein was seen in the cytoplasm in some phase other than G1.
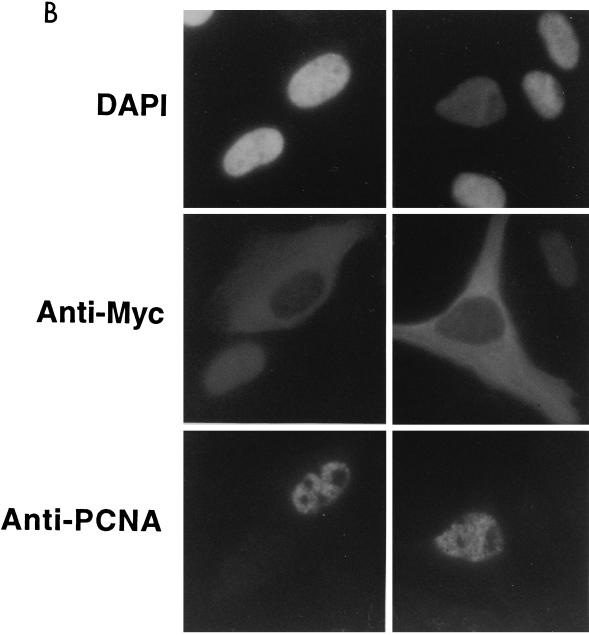
FIG. 3.
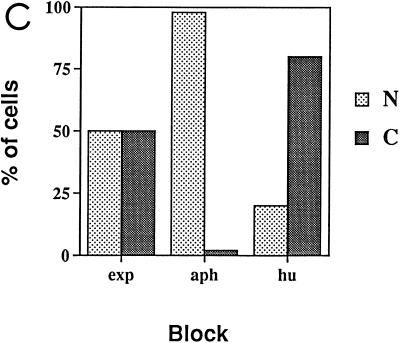
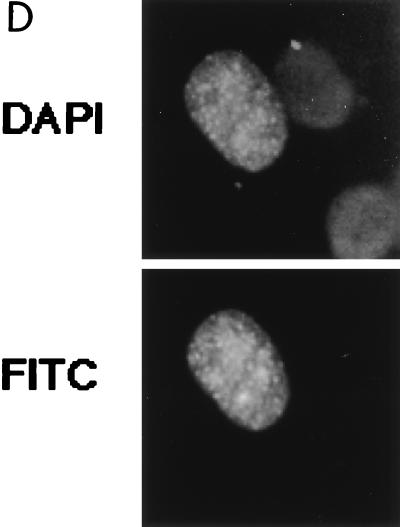
Subcellular localization of epitope-tagged hCdc18 protein. (A) Nuclear hCdc18 protein is seen in G1 cells that are positive for nuclear p21. Asynchronously growing human osteosarcoma U2OS cells were transfected with plasmid expressing HA-Cdc18 protein, and 2 days after transfection the cells were stained with DAPI (top row), anti-HA monoclonal antibody (12CA5) and FITC-conjugated goat anti-mouse secondary antibody (middle row), and anti-p21 (CIP1) rabbit polyclonal antibody and rhodamine-conjugated goat anti-rabbit secondary antibody (bottom row). The two columns on the left show positive nuclear staining for p21 (G1 cells); the two columns on the right show negative staining for p21 (non-G1 cells). Similar results were obtained when the cells were transfected with the plasmid expressing myc-Cdc18 protein. (B) S-phase cells marked by nuclear PCNA have Cdc18 protein in the cytoplasm. Asynchronously growing U2OS cells were transfected with plasmid expressing myc-Cdc18 protein, and the cells were stained with DAPI (top row), anti-myc monoclonal antibody and FITC-conjugated secondary antibody (middle row), and autoimmune human serum containing anti-PCNA antibody and rhodamine-conjugated secondary antibody (bottom row). (C) Subcellular localization of epitope-tagged hCdc18 protein in synchronized cells. U2OS cells were transfected with plasmid expressing HA-Cdc18 protein and then either grown exponentially (exp) or arrested 24 h posttransfection with aphidicholin (aph) or hydroxyurea (hu). HA-Cdc18 protein was detected by immunofluorescence, and the cells containing HA-hCdc18 in the cytoplasm (C) or the nucleus (N) were counted. The results represent the averages of four different experiments with independent transfections. (D) HA-Cdc18 protein with the NLS mutated is still localized to the nucleus. Details are the same as for the top and middle rows of panel A.
To further clarify when during the cell cycle hCdc18 protein appears in the cytoplasm, we expressed myc epitope-tagged hCdc18 protein in U2OS cells and stained the cells with anti-myc and anti-PCNA antibodies (Fig. 3B). Among the 120 cells expressing Myc-hCdc18 protein, 30 to 40% were in S phase, as judged by nuclear staining for PCNA (23). Myc-hCdc18 protein was detected in the cytoplasm of all these cells (Fig. 3B). Whenever myc-Cdc18 protein was detected in the nucleus, the cells were negative for PCNA and were therefore not in S phase. Therefore, hCdc18 protein is present in the nucleus during G1 phase (p21 positive) in time to form prereplication initiation complexes. After origins have fired in S phase (PCNA positive), hCdc18 protein is seen only in the cytoplasm, reappearing in the nucleus some time before the next G1.
The above observations were confirmed by immunofluorescence of HA-hCdc18 protein after transient transfection and cell cycle synchronization of U2OS cells (Fig. 3C). In exponentially growing culture, HA-hCdc18 protein was detected in the nuclei of 50% of the cells expressing the protein. When the cells were arrested at G1-S transition with aphidicholin, HA-hCdc18 protein was detected in the nuclei of 90% of the transfected cells, whereas the protein was detected in the cytoplasm of 80% of the transfected cells arrested at S phase with hydroxyurea.
Thus, although the total amount of hCdc18 protein is unchanged during the cell cycle, the activity is likely to be regulated by changes in its subcellular localization. The increase in nuclear protein in G1 makes this protein similar to yeast CDC6/Cdc18 rather than yeast Orc1, whose levels are unchanged throughout the cell cycle.
The putative NLS of CDC6/Cdc18 protein is not necessary for nuclear localization.
KK(X)10KGRR matches a consensus bipartite (NLS) near the N terminus of CDC6/Cdc18 protein and is conserved in Xenopus CDC6 protein (Fig. 1) (13, 33). Because epitope-tagged hCdc18 protein was easily detected in the nucleus, we tested whether its localization was dependent on this putative NLS by mutating the sequence to KK(X)10KGGG and expressing the protein with an N-terminal HA epitope tag (Fig. 3D). The mutant protein was still detected in the nucleus, implying that the bipartite NLS of CDC6/Cdc18 is not required for nuclear localization of Cdc18 protein. This result also suggests that the nuclear localization of Cdc18 protein could be mediated by its association with other cellular proteins that have their own NLSs.
hCdc18 protein associates with free Orc1 protein.
Further similarity with yeast CDC6/Cdc18 was noted upon examination of the association of human Cdc18 with other components in the replication initiation complex. Yeast CDC6/Cdc18 interacts with the yeast ORC. GST-hCdc18 protein associated well with hOrc1 protein produced by in vitro transcription-translation (Fig. 4A). Studies with the deletion mutants of hCdc18 revealed that either the N- or C-terminal one-third of the protein could independently associate with Orc1 protein. The middle one-third of the protein, containing the domain with maximum homology to Orc1, was incapable of mediating any of these associations. Although homo-oligomerization of CDC6/Cdc18 protein has not been reported in any species, we observed very robust association between GST-Cdc18 protein and S35-labeled Cdc18 protein produced by in vitro transcription-translation. Consistent with the sequence similarities between Cdc18 protein and Orc1 protein, the same regions of Cdc18 protein were sufficient to interact with Cdc18 protein as with Orc1 protein. Despite the discovery of human CDC6/Cdc18 by a two-hybrid screen with PCNA, neither radiolabeled PCNA produced by in vitro transcription-translation nor recombinant purified PCNA produced in bacteria associated with GST-Cdc18, suggesting that the association in the two-hybrid assay may be mediated by unidentified yeast proteins.
FIG. 4.
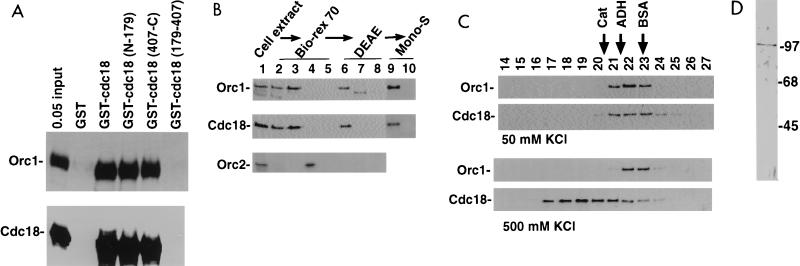
Association of Cdc18 protein with Orc1 protein. (A) Glutathione-agarose beads coated with GST, GST-Cdc18 protein, or indicated deletion derivatives prepared in Escherichia coli were incubated with Orc1 protein or Cdc18 protein produced by in vitro transcription-translation in rabbit reticulocyte lysates. Bound proteins were visualized by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and fluorography. (B) Immunoblots with anti-Orc1 protein, anti-Cdc18 protein, and anti-Orc2 protein antibodies. Lane 1, HeLa cell extract was fractionated over a Bio-Rex 70 column; lane 2, flowthrough of 0.15 M K acetate; lane 3, 0.3 M K acetate eluate containing 7% of input protein; lane 4, 0.6 M K acetate eluate; lane 5, 1.5 M K acetate eluate. The 0.3 M eluate was fractionated on DEAE-Sepharose. Lane 6, flowthrough at 0.2 M K acetate containing 57% of the input protein; lane 7, 0.35 M K acetate eluate; lane 8, 1 M K acetate eluate wash. The flowthrough in lane 6 was passed over a Mono-S column. Lane 9, flowthrough at 0.2 M KCl containing 25% of the input protein; lane 10, eluate at 0.6 M KCl. (C) Orc1 and Cdc18 proteins coelute as a heterodimer of 150 kDa upon gel filtration at 50 mM KCl (top) but are separated at 500 mM KCl (bottom). The flowthrough from the Mono-S column was loaded on a Superose 12 column. Fractions were immunoblotted with anti-Orc1 protein and anti-Cdc18 protein antibodies. The positions of elution of the molecular mass markers catalase (Cat; 300 kDa), alcohol dehydrogenase (ADH; 150 kDa), and bovine serum albumin (BSA; 68 kDa) are indicated. (D) Silver stain of the fraction containing Orc1 protein purified to homogeneity. The numbers indicate molecular masses in kilodaltons.
Although Cdc18 and Orc1 proteins associated with each other in vitro, we did not know what fractions of the cellular Cdc18 and Orc1 proteins were physically present in the same complex. To determine this, HeLa whole-cell extract (made with 0.4 M ammonium sulfate) (25) was fractionated over Bio-Rex-70, DEAE-Sepharose, and Mono-S columns. The bulk of Cdc18 protein in the cell extracts cofractionated with Orc1 protein over the three columns (Fig. 4B). Only 15% of the input protein flows through both DEAE-dextran (binds to negative charge) and Mono-S (binds to positive charge) columns as Cdc18 and Orc1 proteins do. The purification of the Cdc18-Orc1 proteins on column 1 is 14-fold, on column 2 it is 1.75-fold, and on column 3 it is 3.7-fold, for a total purification of 90-fold.
To examine whether the coelution of Cdc18 and Orc1 proteins over three columns was due to their presence in the same physical complex, the fraction from the Mono-S column was subjected to gel filtration. Cdc18 and Orc1 proteins coeluted on a Superose 12 (Pharmacia) column in 50 mM KCl, with Orc1 protein eluting as an apparent monomer of about 100 kDa (about one-half to one fraction lighter than where it coelutes with Cdc18 protein in 50 mM KCl). Cdc18 protein elutes as a multimer of 300 kDa when separated from Orc1 protein, consistent with the same region(s) of the protein being utilized for association with itself as with Orc1 protein.
Another component of the human ORC, the 72-kDa hOrc2 protein, separated from the Orc1-Cdc18 complex in the first column, implying that most of the hOrc1 protein and hCdc18 protein was readily separated from other components of the putative six-subunit human ORC (Fig. 4B). The small size of the Cdc18-Orc1 protein complex (120 to 150 kDa) on the gel filtration column also suggests that Cdc18 protein (66 kDa) is associated only with free Orc1 protein (100 kDa) rather than with the entire six-subunit ORC (expected to be greater than 300 kDa). To confirm this, the fractions from the Mono-S column were purified further by sequential chromatography over Mono-Q, Affigel Blue, and Superose 12 columns to obtain Orc1 protein purified to homogeneity (Fig. 4D). This form of Orc1 protein was still capable of associating with GST-Cdc18 protein (data not shown), indicating that Cdc18 and Orc1 proteins associate directly with each other.
Thus, contrary to the situation in the yeasts or Xenopus egg extract, the human ORC appears to dissociate under the conditions of cell lysis and chromatography used here. It is interesting that the Cdc18-Orc1 protein association appears to be more stable than the association of Orc1 protein with other ORC subunits.
Association of hCdc18 protein with cyclin-cdk2 and its inhibition by a factor present in mitotic cell extracts.
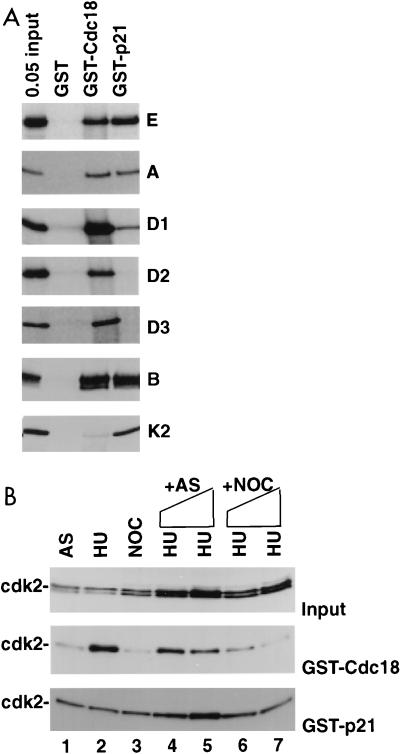
Since Cdc18 protein and cdc2 associate with the ORC in S. pombe cells (22) and a direct interaction has also been reported between S. cerevisiae CDC6 and CDC28 (15, 31), we tested whether human Cdc18 protein associated with any of the cyclin-cdk’s. Cyclins D1, D2, D3, E, A, and B produced by in vitro transcription-translation in a rabbit reticulocyte lysate associated well with GST-Cdc18 protein produced in bacteria (Fig. 5A; compare interaction of cyclins with the positive control, p21). All interactions were stable in the presence of 50 μg of ethidium bromide/ml, showing that the associations are not mediated through nucleic acids (21). cdk2 produced by in vitro transcription-translation did not form a complex with rabbit cyclins (9) and did not associate with bacterially produced GST-Cdc18 protein (Fig. 5A). cdk2 from S-phase extracts of HeLa cells associated with GST-Cdc18 protein (Fig. 5B, lane 2), presumably because of the associated cellular cyclins. The cyclin-cdk’s phosphorylated the N-terminal one-third of Cdc18 protein in vitro (data not shown).
FIG. 5.
Cdc18 protein associates with cyclin-cdk’s through the cyclin, and the association is regulated by a mitotic inhibitor. (A) Indicated S35-labeled proteins were produced by in vitro transcription-translation and tested for association with bacterially produced GST, GST-Cdc18, and GST-p21. 0.05 input, 5% of the input protein incubated with indicated glutathione-agarose beads. Proteins tested were cyclins E, A, D1, D2, D3, and B and cdk2 (K2). (B) Association of cdk2 with GST-Cdc18 is inhibited by a mitotic factor. The immunoblots show input cdk2 (top), cdk2 bound by GST-Cdc18 (middle), and cdk2 bound by GST-p21 (bottom). The extracts are from asynchronous (AS), hydroxyurea-blocked (HU), and nocodazole-blocked (NOC) HeLa cells. Lanes 1 to 3, 0.5-mg extracts directly incubated with glutathione-agarose beads coated with GST fusion proteins produced in bacteria; lanes 4 to 7, the same amount of HU extract used in lane 2 was incubated with increasing amounts of asynchronous cell extracts (lane 4, 0.5 mg; lane 5, 1 mg) or nocodazole-blocked mitotic cell extracts (lane 6, 0.5 mg; lane 7, 1 mg) before incubation with glutathione-agarose beads.
Since a mitotic inhibitor of the association of CDC6 protein with Clb-CDC28 kinases has been reported in S. cerevisiae (15), we tested whether mitotic human cells contained a factor that inhibited the association of cyclin-cdk2 with GST-Cdc18 protein. The association of cyclin-cdk2 with Cdc18 protein was indeed specifically inhibited by the addition of a mitotic extract to an S-phase extract (Fig. 5B, middle blot; compare lanes 6 and 7 to lane 2). The inhibitor was specific for Cdc18-cdk2 association, because p21-cdk2 association was not inhibited (Fig. 5B, bottom blot). When the beads with GST-Cdc18 protein were preincubated with mitotic extracts, washed, and incubated with S-phase extracts, association with cdk2 was unaffected (data not shown), suggesting that the mitotic inhibitor did not stably associate with or modify Cdc18 protein. Thus, the association of the S-phase activator cdk2 with Cdc18 is not only regulated by a requirement for cyclins but also inhibited by a factor present in mitotic cells, similar to the situation in S. cerevisiae.
In vivo association of hCdc18 with Orc1 and cyclin-cdk2.
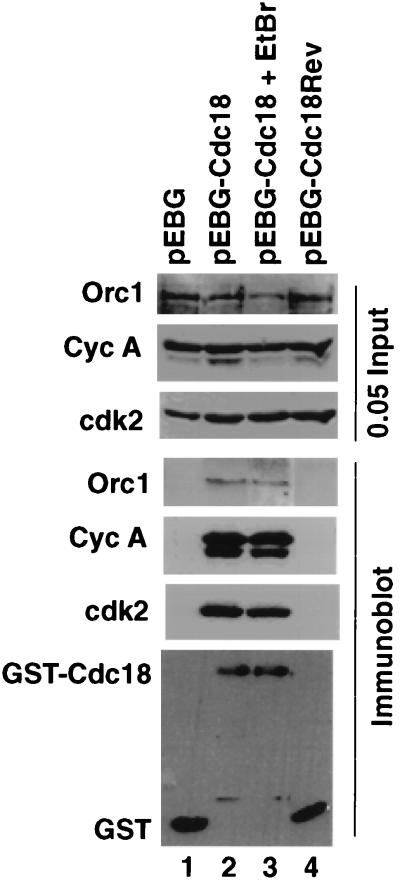
To demonstrate the association of Cdc18 protein with cyclin-cdk2 and with Orc1 protein in vivo, a plasmid expressing GST-Cdc18 protein was transiently transfected into human kidney 293T cells (Fig. 6). Negative controls included cells transfected with plasmids expressing GST or GST fused to a short protein expressed from the antisense strand of the Cdc18 gene (GST-Cdc18rev). Selective precipitation of GST-Cdc18 protein from the cell extracts showed coprecipitation of cyclin A, cdk2, and Orc1 protein, confirming that the cyclin-cdk2-Cdc18 protein and Orc1-Cdc18 protein associations are observed in cells. The addition of ethidium bromide (which intercalates into DNA) (21) did not affect the interactions (Fig. 6, lane 3), implying that the interactions are not mediated through DNA. Orc2 protein was not present in these precipitates (data not shown), confirming that Orc1-Cdc18 protein heterodimers are more stable in extracts of asynchronous cells than an ORC containing both Orc1 and Orc2 proteins.
FIG. 6.
Association of Cdc18 protein with Orc1 protein, cyclin A, and cdk2 in vivo. 293T cells were transfected with pEBG (lane 1), pEBG-Cdc18 (lanes 2 and 3), or pEBG-Cdc18rev (lane 4). Cellular proteins associated with glutathione-agarose beads were detected by immunoblot analyses. In the top three panels, lysates input into the reactions were detected by anti-Orc1 protein (Orc1), anti-cyclin A (Cyc A), and anti-cdk2 (cdk2) antibodies. In the bottom four panels, proteins associated with glutathione-agarose beads were detected by immunoblot analyses. To see the effect of ethidium bromide (EtBr) on the interaction, 50 μg/ml was added to the extract, incubated for 30 min on ice, and centrifuged to remove any particulate matter; the resulting extract was incubated with glutathione-agarose beads (lane 3). 0.05 Input, 5% of the input protein incubated with the indicated glutathione-agarose beads.
DISCUSSION
We have identified a human CDC6/Cdc18 protein and shown that it associates directly with Orc1 protein and cyclin-cdk’s. The protein is homologous to S. pombe Cdc18, S. cerevisiae CDC6, and human Orc1 proteins. The similarity in size of the protein to yeast CDC6/Cdc18 protein and the direct physical association with Orc1 protein and with cyclin-cdk’s in vitro and in vivo support the hypothesis that this protein is the human equivalent of CDC6/Cdc18 protein. In the S. cerevisiae genome there are only two genes that belong to the Orc1-Cdc18 gene family. However, human cells may contain a larger family of Orc1-Cdc18 genes, and a better candidate for hCdc18 may emerge in the future. This caveat aside, we call the protein described in this paper hCdc18 protein.
Although in the yeasts the CDC6/Cdc18 protein is destroyed as cells enter S phase, the situation appears slightly different in humans. Williams and coworkers noted an absence of nuclear hCdc18 protein in serum-starved human WI38 fibroblasts and an increase followed by a decrease of nuclear hCdc18 protein as the cells passed through S phase. This result was postulated to be consistent with the rapid degradation of hCdc18 protein in G2-M (36). However, they also noted the absence of any changes in nuclear Cdc18 protein levels as transformed HeLa cells passed through the cell cycle. In our experiments, which included several controls and were done in both transformed and untransformed cells (including WI38 fibroblasts), the level of total hCdc18 protein does not change significantly during the cell cycle. The protein is also abundantly present in G0 (serum-starved) fibroblasts. Therefore, we postulate that the regulation of hCdc18 protein during the cell cycle is not achieved via changes in total protein level. The constant amount of Cdc18 protein despite the periodic expression of the RNA could be due either to the protein being long-lived or to an increase in the degradation of the protein to compensate for increased synthesis in S phase. An hCdc18 protein isoform of slightly reduced mobility was present in the cell extracts, and although considerably less abundant than the major protein band, increased in G1 relative to G0. Therefore, another possibility is that Williams and coworkers detected only this rare slower-moving isoform with their antibody (which was raised against an N-terminal portion of the protein) while we detected this and the more abundant hCdc18 protein with our antibody.
Changes in the subcellular localization of hCdc18 protein throughout the cell cycle provide a better explanation of the apparent discrepancy between the two reports. Recombinant epitope-tagged hCdc18 protein was seen in G1 nuclei along with a dramatic change in its localization to the cytoplasm in S phase. This could be either because hCdc18 protein is exported out of the nucleus into the cytoplasm in S phase or because the protein is selectively degraded in the nucleus while newly synthesized protein is denied entry into the nucleus and so accumulates in the cytoplasm. There is a parallel between our observations and that with Xenopus Cdc18 protein, which was displaced from the chromatin to perinuclear membrane vesicles during DNA replication (13). Human cells may regulate the functional pool of hCdc18 protein by selectively limiting its concentration in the nucleus once replication is initiated. The next round of replication then becomes dependent on the reentry of hCdc18 protein into the nucleus sometime before the next S phase.
We have also shown that the putative nuclear localization sequence in the N-terminal one-third of hCdc18 protein is not necessary for its nuclear localization. There are no other candidates for an NLS in the sequence of Cdc18 protein, making it likely that the protein is imported into the nucleus by its association with an unknown cellular factor which has its own NLS. Endogenous Cdc18 or Orc1 protein could be a candidate for the unknown cellular factor that chaperones the ectopic Cdc18 protein into the nucleus. Regulation of this association through the stages of the cell cycle may regulate the subcellular localization of Cdc18 protein.
Consistent with the role of Cdc18 protein as an adapter that recruits other DNA replication proteins to the chromatin-bound ORC (4, 13), we observed that the protein associates in vivo and in vitro with one of the components of the ORC, hOrc1 protein. In budding yeast, six polypeptides form a stable ORC which is resistant to high salt levels (2). Similar complexes were identified in other eukaryotes (14, 17, 22, 28). Transfection of epitope-tagged derivatives of hOrc1 and hOrc2 suggested that at least some hOrc1 and hOrc2 proteins are associated with each other (16). However, a human equivalent of the ORC has not yet been identified and our study suggests that endogenous hOrc1 protein is more stably associated with hCdc18 protein than with hOrc2 protein in human cell extracts. We cannot rule out the possibility that the lysis conditions disrupted a preexisting hOrc1-hOrc2 protein complex, that a small fraction of hOrc1 and hOrc2 proteins are associated with each other, or that the hOrc1-hOrc2 protein association is strictly cell cycle regulated. However, under similar lysis conditions a detectable fraction of hOrc2 protein forms a stable complex with the third identified subunit of the human ORC, hOrc4 protein (32). Overall, the association of hCdc18 protein with hOrc1 protein appears more stable than that of hOrc2 protein with hOrc1 protein. It is possible that the interaction of the Cdc18-Orc1 protein heterodimer with hOrc2 protein (and the rest of the ORC) can be a regulated step during the human cell cycle.
The association of hCdc18 protein with cyclin-cdk2 is also consistent with the role of the former as an adaptor protein. Cyclin-cdk2, an activator of S phase, has been localized to replication foci (5), but how it is recruited there is unclear. The association of the kinase with hCdc18 protein provides an attractive mechanism by which cyclin-cdk’s can be recruited to prereplication complexes in G1. The mitotic inhibitor of this association could be important for preventing the inappropriate recruitment of the kinase in M phase. Elsasser and coworkers reported a similar mitotic inhibitor of the association of CDC6 with Clb-CDC28 kinases in S. cerevisiae and provided evidence that this inhibitor is Sic1, a protein that associates with the Clb-CDC28 kinase (15). At this moment we do not know the identity of the mitotic inhibitor of hCdc18-cyclin-cdk association.
In summary, the interprotein associations reported here strongly suggest that the protein we have identified is hCdc18 protein. The constant level of the protein throughout the cell cycle is difficult to reconcile with the existing model of how global degradation of human CDC6/Cdc18 protein regulates its activity. Instead, the results point to two other mechanisms by which hCdc18 protein activity could be regulated to limit the formation of prereplication initiation complexes to once per cell cycle in G1. First, Cdc18 protein is exported from the nucleus (or selectively degraded in the nucleus while new protein accumulates in the cytoplasm) after origins have fired in S phase, necessitating a reentry step later in the cell cycle before the nucleus can replicate again. Second, if the interaction between Cdc18 protein and the S phase-promoting factor cyclin-cdk is essential for the initiation of DNA replication, the mitotic inhibitor of this interaction could prevent a premature interaction until the appropriate time in G1. Future work will be directed towards testing these hypotheses.
ACKNOWLEDGMENTS
We thank B. Stillman for the hOrc1 and hOrc2 clones and B. J. Mayer for the EBG plasmid.
This work was supported by grant CA60499 from the National Cancer Institute and fellowship (J.C.) and career development (A.D.) awards from the U.S. Armed Forces Medical Research Command. P.S. was supported by a grant from the Massachusetts Department of Public Health.
P.S. and J.C. contributed equally to the work.
REFERENCES
- 1.Bell S P, Kobayashi R, Stillman B. Yeast origin recognition complex functions in transcription silencing and DNA replication. Science. 1993;262:1844–1849. doi: 10.1126/science.8266072. [DOI] [PubMed] [Google Scholar]
- 2.Bell S P, Mitchell J, Leber J, Kobayashi R, Stillman B. The multidomain structure of Orc1p reveals similarity to regulators of DNA replication and transcriptional silencing. Cell. 1995;83:563–568. doi: 10.1016/0092-8674(95)90096-9. [DOI] [PubMed] [Google Scholar]
- 3.Bell S P, Stillman B. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature. 1992;357:128–134. doi: 10.1038/357128a0. [DOI] [PubMed] [Google Scholar]
- 4.Botchan M. Coordinating DNA replication with cell division: current status of the licensing concept. Proc Natl Acad Sci USA. 1996;93:9997–10000. doi: 10.1073/pnas.93.19.9997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cardoso M C, Leonhardt H, Nadal G B. Reversal of terminal differentiation and control of DNA replication: cyclin A and Cdk2 specifically localize at subnuclear sites of DNA replication. Cell. 1993;74:979–992. doi: 10.1016/0092-8674(93)90721-2. [DOI] [PubMed] [Google Scholar]
- 6.Carpenter P B, Mueller P R, Dunphy W G. Role for a Xenopus Orc2-related protein in controlling DNA replication. Nature. 1996;379:357–363. doi: 10.1038/379357a0. [DOI] [PubMed] [Google Scholar]
- 7.Chen J, Chen S, Saha P, Dutta A. p21Cip1/Waf1 disrupts the recruitment of human Fen1 by PCNA into the DNA replication complex. Proc Natl Acad Sci USA. 1996;93:11597–11602. doi: 10.1073/pnas.93.21.11597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen J, Jackson P K, Kirschner M W, Dutta A. Separate domains of p21 involved in the inhibition of cdk kinase and PCNA. Nature. 1995;374:386–388. doi: 10.1038/374386a0. [DOI] [PubMed] [Google Scholar]
- 9.Chen J, Saha P, Kornbluth S, Dynlacht B D, Dutta A. Cyclin-binding motifs are essential for the function of p21CIP1. Mol Cell Biol. 1996;16:4673–4682. doi: 10.1128/mcb.16.9.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 11.Chong J, Thommes P, Blow J J. The role of MCM/p1 proteins in the licensing of DNA replication. Trends Biochem Sci. 1996;21:102–106. . (Review.) [PubMed] [Google Scholar]
- 12.Cocker J H, Piatti S, Santocanale C, Nasmyth K, Diffley J F. An essential role for the CDC6 protein in forming the pre-replicative complexes of budding yeast. Nature. 1996;379:180–182. doi: 10.1038/379180a0. [DOI] [PubMed] [Google Scholar]
- 13.Coleman T R, Carpenter P B, Dunphy W G. The Xenopus CDC6 protein is essential for the initiation of a single round of DNA replication in cell free extracts. Cell. 1996;87:53–63. doi: 10.1016/s0092-8674(00)81322-7. [DOI] [PubMed] [Google Scholar]
- 14.Ehrenhofer-Murray A E, Gossen M, Pak D T S, Botchan M R, Rine J. Separation of origin recognition complex functions by cross-species complementation. Science. 1995;270:1671–1674. doi: 10.1126/science.270.5242.1671. [DOI] [PubMed] [Google Scholar]
- 15.Elsasser S, Lou F, Wang B, Campbell J L, Jong A. Interaction between yeast CDC6 protein and B-type cyclin/CDC28 kinases. Mol Biol Cell. 1996;7:1723–1735. doi: 10.1091/mbc.7.11.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gavin K A, Hidaka M, Stillman B. Conserved initiator proteins in eukaryotes. Science. 1995;270:1667–1671. doi: 10.1126/science.270.5242.1667. [DOI] [PubMed] [Google Scholar]
- 17.Gossen M, Pak D T S, Hansen S K, Acharya J K, Botchan M R. A Drosophila homolog of the yeast origin recognition complex. Science. 1995;270:1674–1677. doi: 10.1126/science.270.5242.1674. [DOI] [PubMed] [Google Scholar]
- 18.Kelly T J, Martin G S, Forsburg S L, Stephen R J, Russo A, Nurse P. The fission yeast cdc18+ gene product couples S phase to START and mitosis. Cell. 1993;74:371–382. doi: 10.1016/0092-8674(93)90427-r. [DOI] [PubMed] [Google Scholar]
- 19.Kimura H, Takizawa N, Nozaki N, Sugimoto K. Molecular cloning of cDNA encoding mouse Cdc21 and CDC46 homologs and characterization of the products: physical interaction between P1(MCM3) and CDC46 proteins. Nucleic Acids Res. 1995;23:2097–2104. doi: 10.1093/nar/23.12.2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koonin E V. A common set of conserved motifs in a vast variety of putative nucleic acid-dependent ATPases including MCM proteins involved in the initiation of eukaryotic DNA replication. Nucleic Acids Res. 1993;21:2541–2547. doi: 10.1093/nar/21.11.2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lai J S, Herr W. Ethidium bromide provides a simple tool for identifying genuine DNA-independent protein associations. J Biol Chem. 1992;89:6958–6962. doi: 10.1073/pnas.89.15.6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leatherwood J, Lopezgirona A, Russell P. Interaction of cdc2 and cdc18 with a fission yeast orc2-like protein. Nature. 1996;379:360–363. doi: 10.1038/379360a0. [DOI] [PubMed] [Google Scholar]
- 23.Li R, Hannon G J, Beach D, Stillman B. Subcellular distribution of p21 and PCNA in normal and repair deficient cells following DNA damage. Curr Biol. 1996;6:189–199. doi: 10.1016/s0960-9822(02)00452-9. [DOI] [PubMed] [Google Scholar]
- 24.Liang C, Weinreich M, Stillman B. ORC and CDC6p interact and determine the frequency of initiation of DNA replication in the genome. Cell. 1995;81:667–676. doi: 10.1016/0092-8674(95)90528-6. [DOI] [PubMed] [Google Scholar]
- 25.Manley J L, Fire A, Samuel M, Sharp P. In vitro transcription: whole cell extract. Methods Enzymol. 1983;101:568–582. doi: 10.1016/0076-6879(83)01038-1. [DOI] [PubMed] [Google Scholar]
- 26.Mayer B J, Hirai H, Sakai R. Evidence that SH2 domains promote processive phosphorylation by protein kinase. Curr Biol. 1995;5:296–305. doi: 10.1016/s0960-9822(95)00060-1. [DOI] [PubMed] [Google Scholar]
- 27.Micklem G, Rowley A, Harwood J, Nasmyth K, Diffley J F. Yeast origin recognition complex is involved in DNA replication and transcriptional silencing. Nature. 1993;366:87–89. doi: 10.1038/366087a0. [DOI] [PubMed] [Google Scholar]
- 28.Muzi F M, Kelly T J. Orp1, a member of the Cdc18/CDC6 family of S-phase regulators, is homologous to a component of the origin recognition complex. Proc Natl Acad Sci USA. 1995;92:12475–12479. doi: 10.1073/pnas.92.26.12475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nishitani H, Nurse P. p65cdc18 plays a major role controlling the initiation of DNA replication in fission yeast. Cell. 1995;83:397–405. doi: 10.1016/0092-8674(95)90117-5. [DOI] [PubMed] [Google Scholar]
- 30.Ohtani K, DeGregori J, Leone G, Herendeen D R, Kelly T J, Nevins J R. Expression of HsOrc1 gene, a human ORC1 homolog, is regulated by cell proliferation via the E2F transcription factor. Mol Cell Biol. 1996;16:6977–6984. doi: 10.1128/mcb.16.12.6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Piatti S, Bohm T, Cocker J H, Diffley J F, Nasmyth K. Activation of S-phase-promoting Cdks in late G1 defines a “point of no return” after which CDC6 synthesis cannot promote DNA replication in yeast. Genes Dev. 1996;10:1516–1531. doi: 10.1101/gad.10.12.1516. [DOI] [PubMed] [Google Scholar]
- 32.Quintana D G, Hou Z H, Thome K C, Hendricks M, Saha P, Dutta A. Identification of a novel subunit of the human origin recognition complex with homology to yeast Orc4. J Biol Chem. 1997;272:28247–28251. doi: 10.1074/jbc.272.45.28247. [DOI] [PubMed] [Google Scholar]
- 33.Robbins J, Dilworth S M, Laskey R A, Dingwall C. Two interdependent basic domains in nucleoplasmin nuclear targeting sequence: identification of a class of bipartite nuclear targeting sequence. Cell. 1991;64:615–623. doi: 10.1016/0092-8674(91)90245-t. [DOI] [PubMed] [Google Scholar]
- 34.Rowles A, Chong J P J, Brown L, Howell M, Evan G I, Blow J J. Interaction between the origin recognition complex and the replication licensing system in Xenopus. Cell. 1996;87:287–296. doi: 10.1016/s0092-8674(00)81346-x. [DOI] [PubMed] [Google Scholar]
- 35.Todorov I T, Attaran A, Kearsey S E. BM28, a human member of the MCM2-3-5 family, is displaced from chromatin during DNA replication. J Cell Biol. 1995;129:1433–1445. doi: 10.1083/jcb.129.6.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Williams R S, Shohet R V, Stillman B. A human protein related to yeast CDC6p. Proc Natl Acad Sci USA. 1997;94:142–147. doi: 10.1073/pnas.94.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou C, Huang S H, Jong A Y. Molecular cloning of Saccharomyces cerevisiae CDC6 gene. Isolation, identification, and sequence analysis. J Biol Chem. 1989;264:9022–9029. [PubMed] [Google Scholar]