Abstract
Pancreatic ductal adenocarcinoma (PDAC) is an aggressive malignancy characterized by an immunosuppressive tumor microenvironment enriched with cancer associated fibroblasts (CAFs). This study utilized a convergence approach to identify tumor cell and CAF interactions through the integration of single-cell data from human tumors with human organoid co-culture experiments. Analysis of a comprehensive atlas of PDAC single-cell RNA sequencing (scRNA-seq) data indicated that CAF density is associated with increased inflammation and epithelial-mesenchymal transition (EMT) in epithelial cells. Transfer learning using transcriptional data from patient-derived organoid and CAF co-cultures provided in silico validation of CAF induction of inflammatory and EMT epithelial cell states. Further experimental validation in co-cultures demonstrated integrin beta 1 (ITGB1) and vascular endothelial factor A (VEGF-A) interactions with neuropilin-1 (NRP1) mediating CAF-epithelial cell crosstalk. Together, this study introduces transfer learning from human single-cell data to organoid co-culture analyses for experimental validation of discoveries of cell-cell crosstalk and identifies fibroblast-mediated regulation of EMT and inflammation.
Keywords: pancreatic ductal adenocarcinoma (PDAC), scRNAseq, organoids, cancer associated fibroblasts (CAFs), atlas
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is challenging to treat and the mortality rate remains high largely due to its detection at advanced stages and heterogeneous tumor microenvironment (TME)(1). Molecular changes in the epithelial cell population during carcinogenesis promote changes in the surrounding non-epithelial cell populations, resulting in a dense and immunosuppressive TME(1). The TME is characterized by its heterogeneity and includes a variety of cell types, including mesenchymal cells, such as cancer associated fibroblasts (CAFs), and populations of myeloid and lymphoid lineages(1). Identifying phenotypic heterogeneity across cell types and understanding their functional diversity have historically been limited by available genomic technologies, representing a major barrier to our understanding of PDAC biology.
Single-cell RNA sequencing (scRNA-seq) has recently enabled a more nuanced study of the PDAC TME. Prior work with PDAC single-cell datasets has provided a roadmap to help identify individual cell populations and associated transcriptional regulation of the TME(2–5). These data identified previously underappreciated cellular heterogeneity in human and mouse models of PDAC and are complemented by studies exploring the signaling pathways driving tumor phenotype(6,7). Single-cell technologies have furthered our capacity to identify discrete subpopulations in both tumor and stromal cells. While the identification and characterization of CAF subpopulations have opened new avenues of research (8,9), the mechanisms of intercellular interaction and the role of CAF heterogeneity on tumor cell phenotypes remain poorly understood.
In this study, we developed a multidisciplinary approach to integrate computational methods for single-cell analysis with new experimental approaches for human organoid co-culture that together can untangle the complexities of intercellular interactions in the PDAC TME. To discover the molecular changes resulting from interactions between tumor cells and CAFs, we collated a comprehensive atlas of six published scRNA-seq datasets generated from small cohorts of PDAC patients(8,10–14). Computational analyses of epithelial cell heterogeneity reveal a transcriptional program associated with malignant epithelial cell phenotypic transitions that includes co-occurrence of inflammatory signaling and epithelial to mesenchymal transition (EMT). While this cell state transition correlates with CAF density in the single-cell cohort, additional cell types in the TME may also contribute to these phenotypes. To isolate the impact of CAFs on epithelial cells, we complement computational analyses of the atlas dataset with a three-dimensional co-culture of human patient-derived organoids (PDOs) and patient-derived CAFs. Our experimental system uniquely enables culturing of patient-matched fibroblasts and tumor cells, isolating CAF-tumor cell interactions from resected human tumors in a manner not feasible in patient biospecimens(15,16). Integration of the reference atlas of scRNA-seq data of PDAC with our organoid co-culture through transfer learning enables comprehensive, in silico and experimental investigation of the inflammatory and EMT program from human tumor cell-CAF crosstalk. This convergence approach associates integrin beta 1 (ITGB1) and vascular endothelial factor A (VEGF-A) interaction with Neuropilin-1 (NRP1) with CAF and epithelial crosstalk that can regulate the malignant epithelial cell state transitions defined from single-cell analysis of human tumors.
Materials and Methods
scRNA-seq dataset integration for the PDAC atlas
The six different datasets provided gene expression data with different versions (GRCh37 or GRCh38) and nomenclatures (Ensembl identifiers vs. HUGO gene nomenclature) of the human reference genome. Available patient metadata are summarized in Table 1. All analyses were performed in R (V 3.6–4.1) or Python (version 3.8). Unified integration of the measured features revealed 15,219 genes that could be matched between all datasets with assured certainty. Next, cells with unfavorable quality, defined as mitochondrial counts >15% and unique features of <50 or >5,000, were removed. Computational pre-processing was performed with the Monocle3(17) R package. Dimensionality reduction into a unified manifold approximation and projection (UMAP) was based on the first 100 principal components and batch correction was applied per manuscript to account for potential dataset-intrinsic biases (technical or biological) using Batchelor as utilized by the Monocle3 pipeline(18). Annotation of cell types is described in detail in the Supplemental Methods and in Supplemental Table 1. Plotting was performed with the ggplot2 R package and Excel (Microsoft, Redmond, WA). For high-performance computing tasks, we leveraged the MARCC (Maryland Advanced Research Computing Center, Baltimore, MD) and AWS (Amazon Web Services, Seattle, WA) servers.
Table 1.
Clinical patient data extracted from the six harmonized published datasets integrated in the PDAC atlas.
| Peng et al | Steele et al | Lin et al | Elyada et al | Moncada et al | Bernard et al | |
|---|---|---|---|---|---|---|
| Year of publication | 2019 | 2020 | 2020 | 2019 | 2020 | 2019 |
| Country of enrollment | China | USA | South Korea, USA | USA | USA | USA |
| PDAC pancreas tissue samples | ||||||
| Treatment-naïve, % | 100% | 100% | 100% | 100% | 100% | 100% |
| Patients, N | 24 | 16 | 10 | 6 | 3 | 2 |
| Sex, N | ||||||
| Female | 13 | 6 | 4 | 2 | Unknown | 0 |
| Male | 11 | 10 | 6 | 4 | Unknown | 2 |
| Age, years median (range) | 59 (36–72) | 66 (42–80) | 66 (41–80) | 73 (64–87) | Unknown | 61 (59–62) |
| Diabetes, N | 10 | 6 | Unknown | Unknown | Unknown | Unknown |
| Staging, N | ||||||
| Localized | 24 | 12 | 10 | 5 | Unknown | 1 |
| Metastatic | 0 | 4 | 0 | 1 | Unknown | 1 |
| Grading, N | ||||||
| Well differentiated | 3 | Unknown | 0 | 0 | Unknown | 0 |
| Moderately differentiated | 8 | ≥6 | 6 | 3 | Unknown | 0 |
| Poorly differentiated | 13 | Unknown | 2 | 3 | Unknown | 2 |
| Anaplastic | 0 | Unknown | 2 | 0 | Unknown | 0 |
| Tumor location, N | ||||||
| Head or uncinate | 15 | Unknown | Unknown | Unknown | Unknown | 1 |
| Neck, body or tail | 9 | Unknown | Unknown | Unknown | Unknown | 1 |
| Specimen, N | ||||||
| Surgery (resected) | 24 | 6 | 10 | 6 | 3 | 2 |
| Biopsy (FNA) | 0 | 10 | 0 | 0 | 0 | 0 |
| Survival | Unknown | Unknown | Unknown | Unknown | Unknown | Unknown |
| Non-malignant pancreas tissue samples | ||||||
| Patients, N | 11 | 3 | 0 | 2 | 0 | 0 |
| Sex, N | ||||||
| Female | 6 | 2 | --- | 1 | --- | --- |
| Male | 5 | 1 | --- | 1 | --- | --- |
| Histology, N | ||||||
| Normal | 10 | 1 | --- | 0 | --- | --- |
| Normal-adjacent to adenocarcinoma | 1 | 2 | --- | 2 | --- | --- |
| Specimen, N | ||||||
| Surgery (resected) | 11 | 3 | --- | 2 | --- | --- |
| Biopsy (EUS-FNA) | 0 | 0 | --- | 0 | --- | --- |
PDAC, pancreatic ductal adenocarcinoma
EUS-FNA, endoscopic ultrasound guided-fine needle aspiration
CoGAPS analysis of expression patterns
Non-negative matrix factorization (NMF) of transcript counts was conducted using CoGAPS (V 3.5.8)(19,20). Given a matrix of single-cell data with normalized expression values, CoGAPS factorizes this matrix into two related matrices of gene weights (amplitude matrix) and sample weights (pattern matrix) for random subsets of the data based on the nsets parameter followed by relearning of the amplitude matrix on the full dataset. CoGAPS was run on log2 transformed counts of 15,176 genes from 25,442 cells in Peng et al and Steele et al annotated as epithelial_normal, epithelial_cancer, or epithelial_unspecified(10,11).
Standard parameters were set to 8 Patterns, 50,000 iterations, seed 367, sparse optimized, and distributed: “Single-Cell”. Sparsity parameters were alpha = 0.01, max Gibb mass 100. Distributed CoGAPS parameters were 15 nSets, cut 10, minNS 8, maxNS 23.
Marker genes for each pattern were identified using the patternMarkers function in CoGAPS (V3.9.5) with the “cut” threshold to provide subsets of the top-ranking genes associated with each pattern(21). Overrepresentation analysis was then conducted using the fora function in the fgsea R package (V1.18.0) to find enrichment of any hallmark gene sets from the Molecular Signatures Database(22,23) among the pattern markers for each CoGAPS pattern. The universe used in the overrepresentation analysis was all human genes with HGNC symbols in the GRCh38.p13 genome assembly (n = 39,535)(24,25).
In vitro assays
Organoid and CAF co-culture and single-cell analysis
Patients with PDAC undergoing endoscopic biopsy or surgical resection were enrolled in IRB-approved tissue acquisition protocols at Johns Hopkins Hospital (NA_00001584). All samples were acquired following written informed consent in accordance with Declaration of Helsinki guidelines. PDOs were generated from patient surgical specimens following a combination of mechanical and enzymatic dissociation as previously described(15,16). CAFs were extracted from surgical resection specimens after straining remnant tissue through a 70μm cell strainer and washed twice with human organoid wash media (Advanced DMEM/F12, 10mM HEPES, 1x GlutaMAX, 100μg/mL Primocin, 0.1% BSA) with centrifugation between washes. Following establishment, cell lines were tested for mycoplasma routinely using a combination of MycoStrips (InvivoGen) carried out in the lab and the EZ-PCR assay run through by the Johns Hopkins Genetic Resources Core Facility (GRCF). Cell line authentication is done by matching established primary lines to source tissue using short tandem repeat profiling at the GRCF. For co-culture, organoids were combined with patient-matched CAFs in Matrigel (Corning, 356234) at a 1:10 ratio of organoids to CAFs. In parallel, CAFs and PDOs were plated separately in Matrigel. Co-cultures and monocultures were plated in 24-well tissue culture dishes and extracted after 12 hours using Cell Recovery Solution (Corning, 354253) and incubated on ice at 4°C for 45 minutes for Matrigel depolymerization. Cells were then pelleted and washed in human organoid wash media prior to pelleting again. Organoids were dissociated to single cells using TrypLE Express (ThermoFisher Scientific, 12604013) following manufacturer instructions. Single-cells were barcoded using the MULTI-seq protocol as previously described(26). Single-cell transcriptomics library prep was completed using the 10x Genomics Chromium Single Cell 3’ Gene Expression Dual Index Library (V3.1) according to manufacturer specifications. Library preparations quality were analyzed using the 2100 Bioanalyzer (Agilent). Sequencing was completed at the Johns Hopkins Genetic Resources Core Facility (GCRF). Cellranger (V6.0.0) was used to generate the feature-barcode matrices, aligned to the hg38 genome. Multiseq10x (V1.0) was used as the preprocessing pipeline companion to split the MULTI-seq FASTQs into cell barcode, unique molecular identifiers (UMI), and sample barcode sequences. Reads that did not align with >1 mismatch to any reference sequence and reads representing duplicated UMIs on a cell-by-cell basis were removed. Demultiplex (V1.0.2) was used for demultiplexing the data. The 3DGE data were log normalized, linear dimension was reduced using principle component analysis, and differentially expressed genes were identified in Seurat by Wilcoxon Rank Sum Test (V4.0.1). Additional annotations of Moffitt classifiers, denoting classical and basal epithelial subtypes, and CAF subtypes were added to the Seurat object metadata based on the clustering and module scores(27). Co-culture cell types were parsed based on these annotations and the barcode distinctions. Projection of the discovered CoGAPS Pattern 7 onto the 12HR MULTI-seq expression data was completed using ProjectR (V1.8.0). The MULTI-seq expression data and CoGAPs feature loadings were run through the projectR function of the package(28). The projection results were combined with the MULTI-seq metadata and plotted using ggplot2 (V3.3.5) and Wilcoxon results added using ggpubr (V0.4.0)(28).
Flow Cytometry and Cell Sorting
Organoids were extracted using Cell Recovery Solution (Corning, 354253) and incubated on ice at 4°C for 45 minutes for Matrigel depolymerization. Cells were pelleted and washed in human organoid wash media. Organoids were dissociated to single cells using TrypLE Express and washed in MACS buffer (PBS + 5 mM EDTA + 1% Fetal bovine serum). Cells were resuspended in PBS + Zombie NIR catalog no. 423106 (dilution 1:1000) + Human TruStain FcX catalog no. 422302 (dilution 1:100) for 10 minutes at room temperature in the dark. Cells were quenched with MACS buffer, spun down, and then resuspended in surface stain for 20 minutes on ice at 4°C in the dark; antibodies purchased from Biolegend: APC EpCAM catalog no. 324208 (dilution 1:200), EpCAM PE/Cy7 catalog no. 324221 (dilution 1:200), PE/Cy7 HLA-A, B, C catalog no. 311429 (dilution 1:200), AF700 HLA-DR catalog no. 307626 (dilution 1:200), PerCP/Cy5.5 PD-L1 catalog no. 329738 (dilution 1:100), APC Neuropilin-1 catalog no. 354506 (dilution 1:100), R&D PE VEGFR1 VEGFR2 catalog no. FABSP3P (dilution 1:100). Cells were washed twice in MACS buffer. Flow cytometry analyses were performed on the Beckman Coulter Cytoflex.
For co-culture cell sorting, 1mL of 1mg/mL Dispase II (Thermofisher, 17105041) in organoid wash media was added to each co-culture and monoculture dome to depolymerize Matrigel for 1 hr at 37°C. Digest was quenched with 1mL of wash media and cells were spun down. Cells resuspended in PBS + Zombie NIR catalog no. 423106 (dilution 1:1000) + Human TruStain FcX catalog no. 422302 (dilution 1:100) for 10 minutes at room temperature in the dark. Cells were quenched with MACS buffer, spun down, and then resuspended in surface stain APC EpCAM catalog no. 324208 (dilution 1:200) and FAP R&D Systems, catalog no FAB3715P-100 (dilution 1:75) on ice at 4°C for 20 minutes in the dark. Cells were washed twice in MACS buffer and filtered through 70um filter. Cell sorting was performed on BD Fusion Sorter.
Stimulation of PDOs with IFNγ
1X105 organoids were plated and treated with 200ng/mL of human recombinant IFNγ (PeproTech, 300–02) for 0hr – 96hr for stimulation. Cells were then harvested to be analyzed by mixed methods. Cells were first extracted, stained, and analyzed by flow cytometry. Flow cytometric analyses were performed on the Beckman Coulter Cytoflex. Intact Matrigel domes with PDO that had been stimulated and left untreated were harvested and formalin fixed and paraffin embedded for downstream analysis. Lastly, cells were harvested from treated and untreated Matrigel domes and lysed for total cellular RNA.
VEGF supplementation
1X105 CAF were plated per well of a 24 well plate and allowed to adhere overnight. Recombinant VEGF (PeproTech, 100–20) was added to cultures at increasing concentrations from 10ng/mL to 50ng/mL for either 4 or 24 hours to identify correct timing and concentration of treatment. Changes in surface proteins were seen at 50ng/mL after 4hr by flow cytometric analysis that was acquired on Beckman Coulter Cytoflex. PDO were treated with 50ng/mL of rVEGF for 4 hr to assess cellular surface marker changes and analyzed by flow cytometry on Beckman Coulter Cytoflex.
Quantitative PCR (qPCR)
To evaluate gene expression of MHC-II genes in patient-derived organoids, total RNA extraction using the RNeasy Mini Kit (Qiagen, Catalog Number: 74104) was completed for each patient-derived organoid line according to manufacturer specifications. cDNA synthesis was performed using Invitrogen TaqMan Reverse Transcription Reagents (Catalog Number: N8080234), following manufacturer’s instructions. Real-time quantitative PCR was completed using the ThermoFisher Taqman Gene Expression Assays according to manufacturer’s protocol in the QuantStudio 6 Flex System (Applied Biosystems) mRNA targets included: ITGB1 (Hs01127536_m1), VEGFA (Hs00900055_m1), NRP-1 (Hs00826128_m1) HLA-DRA (Hs00219575_m1), HLA-DRB1 (Hs04192464), HLA-DQB1 (Hs03054971_m1), and HLA-DPB1 (Hs03045105_m1). Relative gene expression was quantified using the 2−ΔΔCt method as previously described(29), and GAPDH (Hs02786624_g1) was used as the endogenous control. Data were analyzed using Applied Biosystems QuantStudio™ Real Time PCR System Software (V1.7.1).
Inference of transitions in cellular phenotypes and intercellular interactions
Within each cell group, additional analyses were performed to compute heterogeneity of cellular phenotypes, state transitions, and inter-cellular signaling across the atlas datasets. First, cell cycle scores and phases were computed with tricycle (V1.2.0)(30). Further unsupervised exploratory analysis of transitions in epithelial cell states was performed with CoGAPS (V3.5.8)(19) analysis across epithelial populations in tumor and normal samples from Peng et al(10) and Steele et al(11). Single cell CoGAPs was run for 8, 10, and 12 patterns. Eight patterns were selected as the final analysis because 12 patterns returned 10 patterns suggesting an overfitting of the data. Further the 8-pattern run resulted in all 8 patterns that were analogous with the other patterns found in the higher dimensional runs. Finally, the impact of fibroblast cells on epithelial cells was computed by estimating intercellular signaling with Domino (V0.1.1)(31) independently for each of the datasets in the atlas.
For Domino analysis, pyScenic (V0.11.0) for Python was first used to generate the gene regulatory network and co-expression modules, the regulon predictions, and the area under the curve (AUC) matrix of cellular enrichment(32). This was completed by providing the extracted counts matrix, a list of transcription factors, motif annotations, and cisTarget motifs for the hg38 genome(32). With the use of the AUC and regulon predictions, a domino object is created and the signaling network built. This allowed for the visualization of global signaling network, gene networks and incoming signaling heatmaps for each narrow subtype annotation, a heatmap of the correlation between transcription factors and receptors, and lastly, the global transcription factor-ligand-receptor network between all subtype annotations(31).
Availability of data and materials:
The scRNA-seq data analyzed in this study were obtained from the NIH gene Expression Omnibus (GEO Database), the Genome Sequence Archive (GSA), and dbGaP at GSE155698 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE155698), GSE111672 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE111672), GSE154778 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE154778), CRA001160 (https://ngdc.cncb.ac.cn/gsa/browse/CRA001160), and phs001840.v1.p1 (https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs001840.v1.p1). The Bernard et al dataset was kindly provided by the corresponding author. PDAC atlas is available for free use at www.sciserver.com after the creation of an account. Once created, please join the “Genomics” Science Domain [https://apps.sciserver.org/dashboard/science/94704] to be granted access. Next, navigate to the data volumes section of the files tab, where you will see a “STAC” directory [https://apps.sciserver.org/dashboard/files/datavolumes/53]. Here, you will see all of our public datasets. Just select the cellxgene link next to the “PDAC” title for access to our cellxgene host of the PDAC Atlas dataset.
Submission of the MULTIseq and Bulk RNA-seq data to dbGaP is in process. All analysis scripts are available from: https://github.com/fertiglab/PDAC_Atlas
All other raw data generated in this study are available upon request from the corresponding author.
Results
A transcriptional reference of CAF and PDAC tumor cells is generated from an integrative analysis of 6 scRNA-seq datasets
To explore signaling between CAFs and tumor cells in PDAC, we integrated six published human scRNA-seq datasets into a comprehensive atlas. In total, the atlas contains 174,394 total cells from 61 PDAC (142,807 cells) and 16 non-malignant pancreatic tissue samples (31,587 cells) (Figure 1A-B, Supplemental Figure 1A)(8,10–14). All samples were of pancreatic origin and from treatment-naïve patients (Table 1). Of the 61 PDAC samples, 52 samples originated from patients with early-stage localized disease, 6 samples originated from patients with metastatic disease, and 3 samples were from patients whose stage was unknown. Of the 16 non-malignant control samples, 5 were specified by the authors as normal-adjacent to a known adenocarcinoma and 11 were derived from samples described as normal-adjacent to non-malignant pathologies. All control samples and the majority of PDAC samples were obtained from resected surgical specimens. Ten PDAC samples originated from fine-needle biopsies, four of which were from patients presenting with metastatic disease.
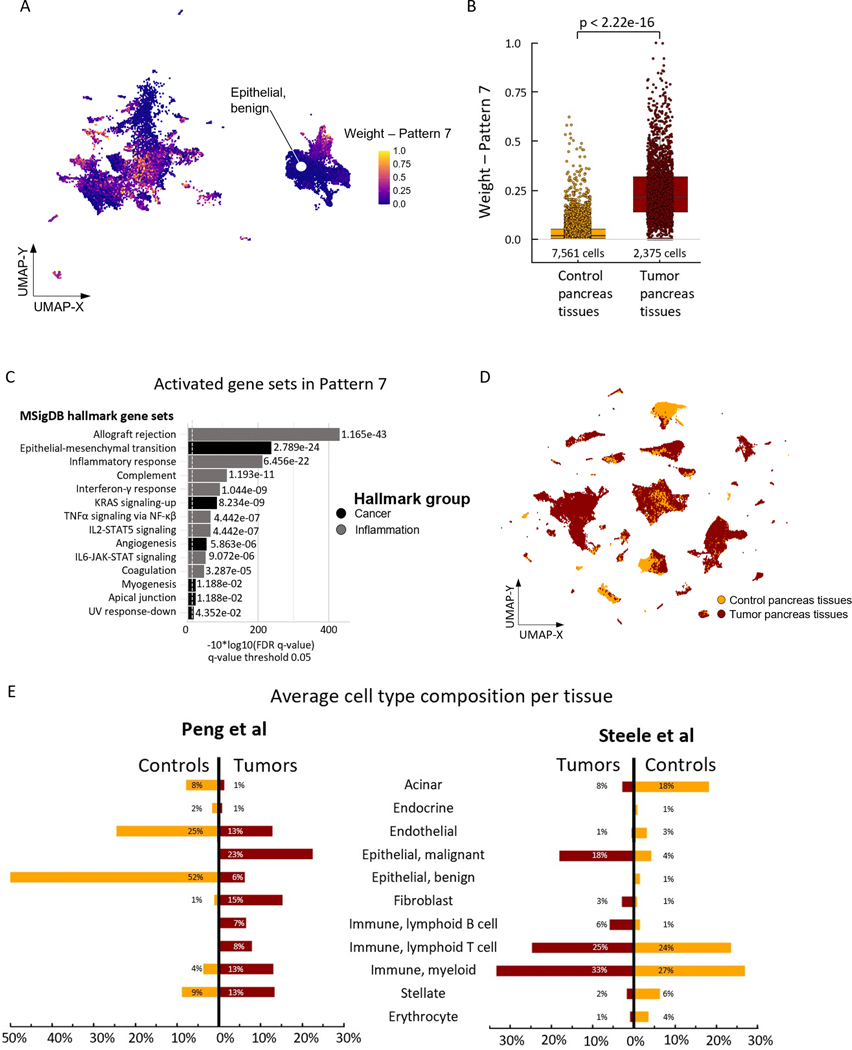
Figure 1.
Summary of atlas composition. (A) Complete atlas with assigned cell types. (B) Heatmap of differentially expressed genes used for cell type annotations. (C) Relative contribution of the 77 different samples with 140,250 cells, separated by tumor (below line) and control tissue (above line). (D) Mean number of cells per tissue by dataset origin. (E) Cell mapping by dataset origin from the six manuscripts in the complete atlas.
After filtering cells based on biological and technical quality metrics, the resulting atlas included 140,250 cells (19.6% dropout). The median and mean cell counts per patient were 1,455 and 1,821, respectively (interquartile range: 828 – 2,200). Following computational pre-processing, we performed a clustering analysis for cell type annotation (supplemental methods). To make inferences on biology from an integrated atlas, an assessment of inter-dataset variation was undertaken to minimize technical artifacts. Most of the contributing patient samples and cells (35 and 54,813, respectively) originated from Peng et al(10) (Figure 1C-E) with a mean of 1,566 cells per tissue. The mean number of cells per sample was highest (4,754 cells) in the dataset by Moncada et al(13), a set that contributed PDAC samples from three patients (Figure 1C-E). However, mapping each dataset on the atlas’ UMAP demonstrated contribution from all six datasets across clusters, visually verifying our integration approach (Figure 1E).
Within PDAC samples, the predominant cell populations were malignant epithelial cells (32,515 or 29.3%), cells of myeloid origin (28,971 cells or 26.1%), and T cells (17,284 cells or 15.6%) (Supplemental Figure 1B). Cell cycle analyses of the PDAC samples revealed high cell cycle activity primarily in the malignant epithelial, acinar, myeloid and CAF populations (Supplemental Figure 1C-D). The mesenchymal cell populations were composed of 8,953 CAFs (8.1%) and 6,049 stellate cells (5.4%). Altogether, the components of the TME (immune and mesenchymal cell populations combined) contributed 67,690 cells or 60% of the total cell count. In the myeloid and lymphoid cell clusters, we annotated subpopulations to include macrophages, mast cells, neutrophils, regulatory T cells and natural killer/cytotoxic T cells (Supplemental Figure 2). Epithelial cell classification as benign or malignant was completed using copy number variation (CNV, Supplemental Figure 3) and differential gene expression as markers of malignant etiology. Malignant epithelial cells were classified using the classical and basal gene markers first reported by Moffitt et al(27) (Supplemental Figure 4). In the CAF cluster, cells were subtyped according to inflammatory CAF (iCAF) and myofibroblastic CAF (myCAF) gene markers, as the antigen-presenting CAF (apCAF) population is rare (Supplemental Figure 5)(8).
CoGAPS identifies phenotypic states of EMT and inflammatory signaling co-occurrence in cancer-associated epithelial cells correlates with CAF density
Determining the intercellular regulatory programs in tumor cells first requires delineating the distinct phenotypic states in epithelial cells during tumor progression. Previously, we have demonstrated that our single-cell Bayesian non-negative matrix factorization algorithm CoGAPS can concurrently define cellular states and dynamics from scRNA-seq data(19). Therefore, we applied CoGAPS to investigate patterns of gene expression within the tumor and normal epithelial clusters in the atlas. Batch effects were mitigated by identifying gene expression patterns maintained across the two largest sample cohorts that contain both PDAC and non-malignant samples: Peng et al(10) (18,261 epithelial cells) and Steele et al(11) (7,181 epithelial cells). These two datasets account for 61.0% of all epithelial cells in the atlas and represent 49.3% of all malignant cells, and 97.1% of all benign cells. The populations of benign cells also have a mixture of those obtained from normal controls and those obtained from tumor-adjacent normal cells, providing the opportunity to fully characterize epithelial cell state transitions associated with malignancy.
We applied CoGAPS to interrogate patterns of gene expression across these epithelial populations, identifying 8 distinct patterns of transcriptional phenotypes (Supplemental Figure 6). The molecular function of the cellular phenotype for each pattern was annotated by an overrepresentation test across genes identified by the CoGAPS pattern marker statistic(33) and Hallmark gene sets from the Molecular Signatures Database(23). The highest ranked pathways associated with Pattern 1 were UV response and TGFβ signaling that did not reach statistical significance (Supplemental Table 2). Pattern 2 identified pathways of estrogen response and KRAS signaling that are relevant to the growth factor receptor signaling that occurs during PDAC development. Metabolic pathways including cell cycle, oxidative phosphorylation, and glycolysis were significant in Patterns 3–5. Pattern 6 identified pathways of apoptosis activity and Pattern 8 was dominated by genes inherent to the response to hypoxia. Notably, genes associated with Pattern 7 were classified by CoGAPS to include increased pathway activities in inflammatory, fibrogenic, and malignant progression-associated gene sets, including interferon gamma (IFNγ) response and epithelial-mesenchymal transition (EMT) (Figure 2A-C, Supplemental Table 3). Plotting these patterns along the tumor cells in the UMAP demonstrates that the cells associated with the Pattern of cellular proliferation (Pattern 2) are distinct from those demonstrating the co-occurrence of inflammatory signaling and EMT in Pattern 7.
Figure 2.
Identification of Pattern 7 and cell type distribution in the atlas. (A) Complete atlas subset of the epithelial cell populations from the Peng et al(11) and Steele et al(10) data with assigned weights of Pattern 7 as resulting from CoGAPS analyses. (B) Boxplot of Pattern 7 weights within the Epithelial, benign cell population demonstrating differences between control and tumor pancreas tissues. p < 2.22 e-16, generated by Wilcoxon test. (C) Overrepresented MSigDB hallmark gene sets in cells expressing Pattern 7 genes. (D) Cell mapping by tumor vs. control pancreas tissue in the complete atlas. (E) Differences in cell type composition for selected tumor vs. control pancreas tissues originating from Peng et al(11) and Steele et al(10).
Tumor-associated epithelial cells enriched for Pattern 7 are predominantly classified as unspecified as opposed to more clearly-defined tumor cells. We also observe that Pattern 7 weights were significantly higher in the benign epithelial cells derived from PDAC samples relative to the epithelial cells in the non-malignant control samples (Figure 2B). The enrichment of Pattern 7 within the epithelial cells from PDAC samples suggests a potentially transformative process that drives a phenotype of inflammation and EMT in the epithelial compartment similar to previous data from non-epithelial cell types in PDAC(34). We hypothesize the pathways included in Pattern 7 contribute to epithelial cell plasticity during tumorigenesis as well as in established PDAC. This hypothesis is further supported in an independent study from our group by Bell et al(35) in which they apply atlas Pattern 7 to spatial transcriptomics data from pancreatic intraepithelial neoplasia and identify a tradeoff of Pattern 7 with patterns associated with cellular proliferation through ProjectR transfer learning(28).
We also hypothesized that factors in the microenvironment regulate the inflammatory and EMT signaling in Pattern 7. To investigate intercellular interactions as drivers for Pattern 7, we first reviewed cell type composition in tumor and adjacent normal samples to identify subpopulations in the TME. Notably, the non-epithelial populations in the atlas largely originated from tumor samples (Figure 2D). When comparing the average cell type composition between the control and tumor samples within the Peng et al(10) dataset, the fractions of fibroblast, myeloid and lymphoid populations were greater in tumor samples, with a proportional decrease in cells of endothelial origin, acinar cells and total epithelial cells (Figure 2E). Within the Steele et al(11) dataset, differences between the fibroblast/CAF, myeloid and lymphoid populations, while present, were less pronounced (Figure 2E). To understand the impact of CAF signaling on epithelial gene expression, we correlated the mean CoGAPS weights from pattern 7 in the epithelial compartment with the presence of CAFs using the datasets from Peng et al(10) and Steele et al(11). We identified a direct association between an increasing fibroblast proportion in the TME and the mean weight of CoGAPS Pattern 7 in the epithelial cells (Supplemental Figure 7). This association was lost when fibroblast populations were further divided into iCAFs and myCAFs (Supplemental Figure 7). This correlation leads to the hypothesis that CAFs promote expression of Pattern 7 in malignant epithelial cells. Yet, while CAF presence enriches the expression of Pattern 7, uncovering the mechanisms that drive CAF-mediated signaling in the epithelial compartment requires further validation using approaches beyond correlative associations in human single-cell data.
Transfer of Pattern 7 from the PDAC atlas to PDAC PDO-CAF co-culture demonstrates that CAFs induce EMT and inflammatory signaling in epithelial cells
To establish an association between PDO-CAF interactions and the inflammatory and EMT signaling observed through the correlative analysis of Pattern 7, we established PDAC PDOs co-cultured with patient-derived, patient-matched CAFs. Each PDO-CAF co-culture leverages our previous experience generating PDOs(15,16) while in parallel extracting CAFs from surgical resection specimens (Figure 3A, Supplemental Figure 8). As a result, our PDO co-culture uniquely enables us to isolate the effects of CAFs on human PDAC tumor cells without the confounding of additional cell types as occurs in human tissue. To compare the signaling processes in our co-culture to Pattern 7 identified in the atlas, we performed scRNA-seq profiling of a 12-hour PDO-CAF co-culture with PDO and CAF monoculture from the same patient as controls. We used multiplex analysis, with an established MULTI-seq protocol(26), to assess transcriptional heterogeneity in the epithelial and CAF compartments. Clustering analysis and UMAP visualization of the scRNA-seq data separated the fibroblasts from epithelial cells. This clustering analysis demonstrated that cells from the co-culture and monoculture conditions retained the gene expression patterns of their cellular identity, without apparent sub-clustering of co-culture from monoculture conditions (Figure 3B-C).
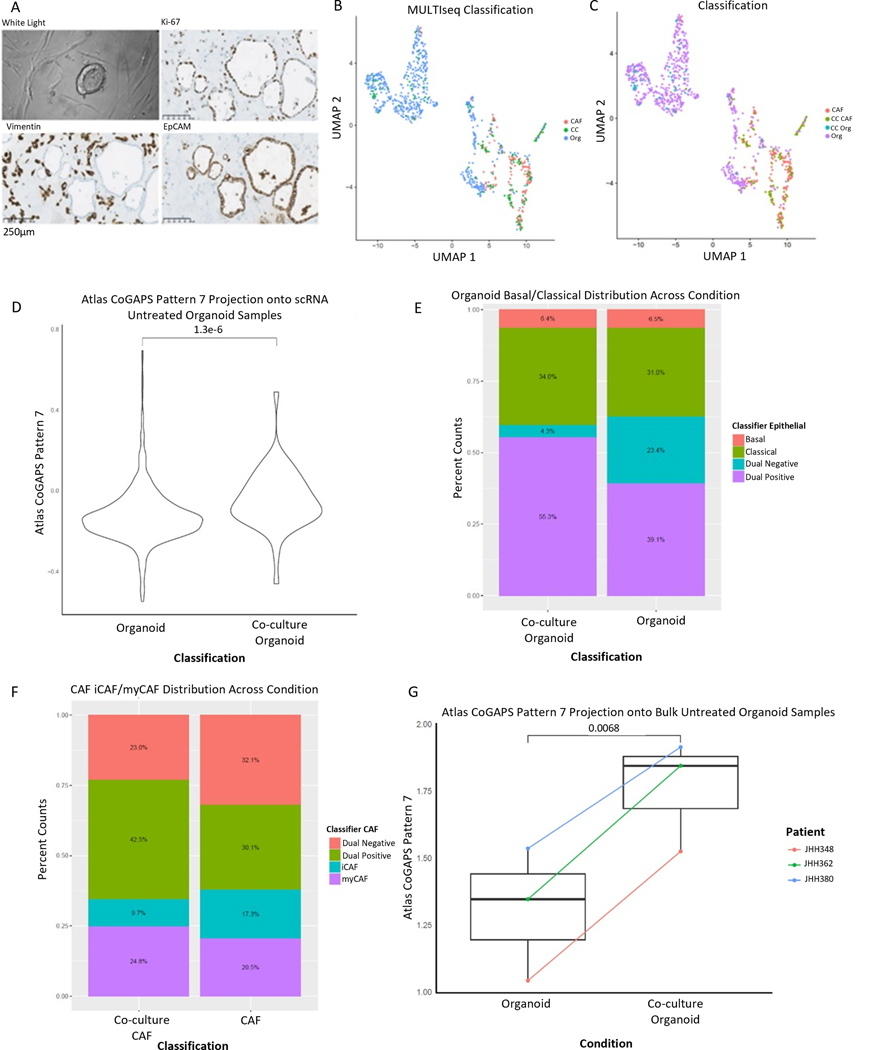
Figure 3.
Patient-derived organoids co-cultured with CAFs recapitulate the Pattern 7 identified in tumor epithelial cells and demonstrate dynamic cellular phenotypes. (A) Representative brightfield image of co-culture. Representative IHC of co-culture demonstrating proliferation by Ki-67 after co-culture (top right), vimentin positive CAFs (bottom left), and EpCAM positive organoids (bottom right). Images obtained at 20x magnification; scale bars represent 250μm. (B) UMAP demonstrating culture conditions: organoid monoculture (Org), CAF monoculture (CAF), co-culture (CC). (C) UMAP demonstrating cell-type calls after co-culture: Organoid monoculture (Org), CAF monoculture (CAF), CAFs from co-culture (CC CAF), organoids from co-culture (CC Org). (D) Pattern 7 was enhanced in organoid cells from co-culture relative to organoid cells from monoculture, p=1.3e-6 by Wilcoxon. (E) Co-culture demonstrates plasticity in epithelial representation in the co-culture condition with a greater percentage of cells representing both basal and classical markers (dual positive) present in co-culture. (F) Co-culture demonstrates plasticity in CAF representation in the co-culture condition with a greater percentage of cells representing both iCAF and myCAF markers (dual positive) present in co-culture. (G) Pattern 7 is enhanced in bulk RNA-seq after co-culture, p=0.0068 by paired T-test.
Though clustering analysis identified few gene expression changes, we hypothesized that the co-culture causes the increase in inflammatory signaling and EMT gene signature observed in Pattern 7 and associates with CAFs through correlation analysis in the atlas dataset. Therefore, we evaluated the occurrence of Pattern 7 in PDO-CAF co-culture compared to PDO monoculture through transfer learning with our ProjectR method(28) between the scRNA-seq data from the PDAC atlas and the scRNA-seq data from our PDAC PDO cultures. We identified increased expression in the Pattern 7 signature in PDAC organoids co-cultured with CAFs, relative to those from monoculture (Figure 3D). We also found that enrichment of Pattern 7 is associated with a shift in epithelial cell subtype, so that fewer cells distinctly classify as exclusively basal or classical after co-culture (Figure 3E). Similarly, the CAF population also displays plasticity with increased proportions of CAFs expressing gene markers for both iCAFs and myCAFs (Figure 3F). Further application of this projectR(28) analysis for transfer learning to bulk RNA-seq data from a broader panel of 3 co-cultured lines also demonstrate an enrichment of Pattern 7 in the co-culture condition with CAFs relative to monoculture (Figure 3G). Technically, this analysis approach extends our previous studies leveraging transfer learning to performing in silico validation of phenotypic states within a cell type to the validation of intercellular regulatory programs. The robustness of the inferred signaling patterns between human tissue and organoid co-culture demonstrate that this dynamic, reductionist in vitro system can provide an experimental model to validate intercellular signaling. In this case, based on the biological findings from our analyses, we sought to further leverage the organoid co-culture to experimentally examine the CAF-induction of inflammatory signaling and EMT in PDAC epithelial cells.
PDOs demonstrate that CAFs in co-culture enrich IFNγ response and induces major histocompatibility complex type II (MHC-II) expression in epithelial cells
We used our PDO culture system to further validate the expression of Pattern 7 as identified in the atlas’ epithelial populations. Multiple MHC-II genes and IFNγ response were included in Pattern 7, which we then examined more deliberately in the epithelial cells from the control and tumor samples (Supplemental Figure 9). To validate that pattern 7 is a true epithelial cell signature and not a result of unidentified doublets, we demonstrated that this CoGAPS pattern also occurred in a restricted set of epithelial cells from the PDAC atlas that do not express canonical immune genes (Supplemental Figure 10). Based on these findings, we hypothesize that there exists a signaling cascade in which CAFs induce IFNγ signaling in malignant epithelial cells and induce MHC-II gene expression.
We then first sought to validate that IFNγ can induce MHC-II expression in PDAC epithelial cells using our PDO model. Prior work has shown that 24 hours of 200ng/mL of IFNγ added into cell media induces MHC-I and PD-L1 upregulation in organoids in colorectal cancer and NSCLC(36). To extend these findings to our PDOs, we screened eleven PDO lines by flow cytometry (gating strategy Supplemental Figure 11) for both constitutive and IFNγ-induced cell surface protein expression of MHC-I and II and PD-L1 (Supplemental Figure 12A). At 24 hours, both MHC-I and PD-L1 demonstrated a robust increase in expression in response to IFNγ stimulation.
We then examined MHC-II surface expression over a longer duration given the representation of MHC-II genes in the Pattern 7 gene set. HLA-DR was used as a representative marker for MHC-II expression. As expected, HLA-DR expression was limited at baseline and following 24 hours of IFNγ stimulation. Longer-term exposure to IFNγ for 96 hours resulted in upregulation of HLA-DR expression, consistent with induced gene expression changes (Supplemental Figure 12B-D). To further evaluate IFNγ-induced changes in MHC-II alleles which lack specific antibodies, we validated HLA-DRB1, HLA-DRA, HLA-DPB1, HLA-DQB1 by qPCR and observed a variable IFNγ-induced response in gene expression (Supplemental Figure 12E). These data were then confirmed in formalin fixed and paraffin-embedded (FFPE) PDAC PDOs using IHC with antibodies against HLA-DR and HLA-DR/DP/DQ (Supplemental Figure 12F-G).
While the PDO experiments rely on direct induction of IFNγ stimulation to induce MHC-II expression, the correlation of CAF density with Pattern 7 and its enrichment in co-culture led us to hypothesize that CAFs can mediate upregulation of MHC-II in malignant epithelial cells. To test this hypothesis in co-culture, we identified the presence of MHC-II expression from the co-culture scRNA-seq data (Supplemental Figure 13A-C). We then expanded beyond 12 hours to examine temporal changes in protein-level gene expression in the epithelial compartment by co-culturing PDOs and CAFs for 24 and 96 hours. At each timepoint, we used Fluorescence Activated Cell Sorting (FACS) of the co-culture to have a pure population of cells to query MHC-II gene expression changes by qPCR. Similar to our findings with IFNγ treatment, HLA-DRA and HLA-DRB expression increased with increasing time in co-culture up to 96 hours (Supplemental Figure 13D-E). This further implicates CAFs as critical mediators of epithelial cell plasticity, which we hypothesize plays a role in antigen presentation in PDAC. Our observation of MHC-II expression in human PDAC tumor cells is further supported recent study of tissue microarrays of patient PDAC samples(37).
Ligand-receptor analysis of the PDAC atlas identifies ITGB1 as a ligand signaling from fibroblasts to epithelial cells and VEGF-A as a ligand signaling from epithelial cells to fibroblasts
Our combined computational and experimental analyses suggest CAF-epithelial cell signaling impacts PDAC behavior but does not provide direct signaling mechanisms responsible for these interactions. Therefore, we examined the signaling networks between CAFs and epithelial cells in silico from the PDAC atlas and validated them experimentally in our co-culture. To infer these pathways, we selected the epithelial tumor and CAF populations originating from PDAC samples in the atlas derived from the Peng et al(10) (12,120 epithelial cells, 63.8%; 5,823 CAFs, 84.0%) and Steele et al(11) (6,883 epithelial cells, 36.2%; 1,110 CAFs, 16%) datasets. We then analyzed these data with Domino, a computational method that infers intercellular interactions in scRNA-seq data by quantifying coordinated gene expression changes between ligands of one cell type and receptors of another and subsequent transcription factor activation(31). The resulting analysis generated a putative global signaling network in PDAC based on population-specific gene expression of ligands and receptors with established signaling relationships, identifying ITGB1 and VEGF-A as important contributors to communication between epithelial tumor cells and CAFs (Figure 4A).
Figure 4.
Domino evaluation of intercellular interactions in the atlas with PDO-CAF co-culture. (A) Signaling network between epithelial and CAF subpopulations from tumor pancreas tissues in the Peng et al(10) dataset as derived from the Domino R package. Nodes of the subpopulations are sized according to the amounts of expressed targeting ligands. The thicknesses of the intercellular connections are scaled based on the strength of signaling with their color indicating the signals’ origin (directionality). (B) Heatmap plotting mean normalized expression of each ligand for each group, demonstrating ITGB1 as a ligand originating in the CAF populations with the epithelial cells receiving this signal from the Peng et al(10) dataset. (C) ITGB1 expression in human PDAC tissue shown first as H&E stain at 5X magnification with focused section at 40X magnification from H&E and ITGB1 IHC with hematoxylin nuclear stain. H&E and IHC completed on sequentially cut slides. (D) Schema demonstrating the workflow for co-culture setup and disassembly with flow sort prior to qPCR. (E) ITGB1 expression in monoculture and co-culture CAFs and epithelial cells after 24 or 96 hours of co-culture. Plotted are the Fold Change values comparing our PDO co-culture to monoculture using GAPDH as an endogenous control. Comparisons of monoculture and co-culture conditions are statistically supported using the two-tailed students t-test with equal variance in PRISM (V9.2.0 [283]). Significance is measured as: ****, p<0.0001; ***, p<0.001; **, p<0.01; *, p<0.05; ns, not significant. Panel D created with BioRender.com
In the atlas, across epithelial subtypes, ITGB1 was identified in both the Peng et al(10) and Steele et al(11) datasets as a ligand from both iCAFs and myCAFs directed at the epithelial subpopulations (Figure 4B, Supplemental Figure 14A). To validate signaling through ITGB1, we first used IHC to confirm ITGB1 is expressed in patient PDAC tumor tissue by examining expression in two patients with PDAC included in our rapid autopsy cohort (Figure 4C). Taking this into our PDO-CAF co-culture, ITGB1 expression was identified both in PDOs and CAFs using qPCR. Supporting our hypothesis, when PDOs and CAFs are co-cultured, ITGB1 gene expression is upregulated in CAFs (Figure 4D-E). This demonstrates that epithelial presence contributes to the CAF-specific expression of ITGB1. Although ITGB1 expression is not canonically thought to be tissue- or cell-type specific, integrins are known to play a key role in the structure and function of the extracellular matrix(38). High levels of expression of ITGB1 are associated with poor outcomes in PDAC, as well as progression and metastasis(39). Further, ITGB1 has been implicated in EMT in PDAC through ERK/MAPK signaling(40).
Across both datasets, VEGF-A was expressed by epithelial populations and predicted to serve as a key mediator of crosstalk with fibroblasts (Figure 5A, Supplemental Figure 14B). To examine the relevance of this inference in human tissue, we first validated VEGF expression in human PDAC tissue by IHC in two patients from our rapid autopsy cohort and then set out to examine this relationship in PDO-CAF co-culture (Figure 5B). Cells were FACS sorted after 24 or 96 hours of co-culture and qPCR was performed to assess VEGF-A expression in the co-culture as compared to the monoculture. At both timepoints, VEGF-A expression increased in cells derived from the co-culture relative to those extracted from monoculture, consistent with the Domino inference (Figure 5C). To assess levels of secreted VEGF-A, we next used an ELISA assay to confirm an increase in secretion following co-culture, further implying enhanced signaling when both cell types are spatially permitted to interact (Figure 5D). VEGF is a key factor driving angiogenesis; therefore, we wanted to verify that the increase in VEGF-A secretion correlated with this core function. To test the effect of CAF and PDO-secreted factors on angiogenesis, we cultured human umbilical vein endothelial (HUVEC) cells in PDO conditioned media, CAF conditioned media, or co-culture conditioned media, to identify the presence and initiation of new vasculature. Compared to PDO conditioned media alone, segment length of the HUVEC cell network was greater when cultured with co-culture conditioned media, suggesting propagation of endothelial cells in co-culture conditioned media and enhanced angiogenesis (Figure 5E). Interestingly, segment length was also enhanced in CAF conditioned media alone. Evaluating nodes in our HUVEC assay, both co-culture conditioned media and CAF conditioned media increased the number of nodes in comparison to PDO conditioned media alone (Figure 5E). This supports the notion of increased angiogenesis signaling, likely mediated by VEGF-A, produced through epithelial and CAF crosstalk. While VEGF-A is most classically associated with angiogenesis, it has also been implicated in EMT and inflammation in nasopharyngeal cancers and inflammatory bowel disease, respectively, reinforcing the complexity of VEGF-A signaling and the utility of both a computational and in vitro approach(41,42).
Figure 5.
Domino evaluation of intercellular interactions through VEGF-A with PDO-CAF co-culture validation. (A) Heatmap plotting mean normalized expression of each ligand for each group, demonstrating VEGF-A as a ligand originating in the epithelial populations with the CAFs receiving this signal from the Peng(10) dataset. (B) VEGF-A expression in human PDAC tissue shown first as H&E stain at 5X magnification with focused section at 40X magnification from H&E and VEGF-A IHC with hematoxylin nuclear stain. H&E and IHC completed on sequentially cut slides. (C) Differential expression by qPCR of VEGF-A in monoculture and co-culture CAFs and epithelial cells after 24 or 96 hours of co-culture. Plotted are the Fold Change values comparing our PDO co-culture to monoculture using GAPDH as an endogenous control. (D) VEGF concentration on ELISA of monoculture and co-culture. N=7 PDO-CAF cultures from 7 distinct patients. (E) Angiogenesis assay in HUVEC cells cultured in organoid, CAF, or co-culture conditioned media (CM). Top: transillumination at 4X magnification after 8 hours in culture. Bottom Left: Quantification of segment length of HUVEC networks following culture for 8 hours with increased segment length after culture in CAF CM with trend towards increased segment length in co-culture CM. Bottom Right: Quantification of Nodes in angiogenesis assay after 8 hours in culture. (F) Representative flow cytometry contour plots examining VEGFR1 and VEGFR2 surface expression in CAF (top) and PDO (bottom) monoculture or co-culture. (G) Quantification of CAF (top) VEGFR1 and VEGF2 percent positive, PDO (middle) percent positive VEGFR1 and VEGFR2 and PDO (bottom) Median Fluorescence Intensity expression in monoculture and co-culture across 5 patients. Colored circles on bar graphs correspond to distinct PDOs across experimental conditions. Comparisons of monoculture and co-culture conditions are statistically supported using the two-tailed students t-test with equal variance in PRISM (V9.2.0 [283]). Significance is measured as: ****, p<0.0001; ***, p<0.001; **, p<0.01; *, p<0.05; ns, not significant.
PDO-CAF co-culture validates neuropilin-1 (NRP1) as a dynamically expressed binding partner for VEGF-A
We next explored the relationship between VEGF-A and its known receptor binding partners to better understand the mechanism of VEGF signaling in co-culture. We first used flow cytometry to screen receptor expression in five patient co-cultures with matched monoculture, demonstrating no appreciable difference in the expression of VEGFR1 or VEGFR2 between co-culture and monoculture (Figure 5F-G). Even following recombinant VEGF treatment, receptor modulation was modest, leading us to consider other binding partners that may play a role in VEGF-A crosstalk between PDOs and CAFs (Supplemental Figure 15).
To identify other binding partners, we returned to the individual network diagrams generated from Domino. In both the iCAF and myCAF gene networks, neuropilin-1 (NRP1) was identified as a binding partner for VEGF-A (Figure 6A). NRP1 is an essential co-receptor for VEGF-A, most notably in endothelial cells, but is less often studied than VEGFR1 or VEGFR2(43). Evaluating the expression of NRP1 by qPCR, we found no significant change in NRP1 gene expression in co-culture when compared to monoculture (Supplemental Figure 16A). However, upon evaluation of surface expression by flow cytometry, we identified decreased expression of NRP1 on PDOs in three of four co-cultures examined compared to monoculture alone (Figure 6B-C, Supplemental Figure 16B). Additionally, three of the four PDOs were treated with recombinant VEGF in monoculture, driving a decrease in NRP1 protein expression in two of the three PDOs (Supplemental Figure 16C). Downregulation of NRP1 was not identified in CAF lines, suggesting this signaling pathway is more active in the epithelial compartment than the CAF compartment (Figure 6B-C). These data support the notion of an inverse relationship between NRP1 surface receptor availability and secreted VEGF-A that is driven by endocytosis and receptor recycling(44). We then confirmed with IHC in three patients with PDAC that NRP1 and VEGF-A are co-expressed, reinforcing NRP1 is a relevant VEGF-A binding partner in PDAC (Figure 6D). Together, these data demonstrate a role for NRP1 in VEGF-A signaling in PDAC independent of changes in surface expression of VEGFR1/2.
Figure 6.
NRP1 is an inferred binding partner for VEGF-A and overexpression has survival implications (A) Ligand-receptor interaction network between epithelial and CAF subpopulations from tumor pancreas tissues in the Peng et al(10) dataset as derived from the Domino R package. (B) Representative flow cytometry contour plots evaluation of NRP1 expression in CAF (top) and PDO (bottom) from co-culture and monoculture. Associated quantification in bar graphs on the right. (C) Left: Quantification of NRP1 expression on CAFs in monoculture and in co-culture with PDO, Right: Quantification of NRP1 expression on PDOs in monoculture and co-culture resulting in significant decrease of NRP1 after co-culture with CAFs (D) Representative H&E of human PDAC tumor at 5X and 40X with IHC with hematoxylin nuclear stain on sequential slides examining VEGF-A and NRP1 expression (N=3 patients). Quantification of NRP1 and VEGFA expression from 11 Regions of Interest (ROI) from JHH394 as determined by pathologist. Correlation statistics by simple linear regression. Comparisons of monoculture and co-culture conditions are statistically supported using the two-tailed students t-test with equal variance in PRISM (V9.2.0 [283]). Significance is measured as: ****, p<0.0001; ***, p<0.001; **, p<0.01; *, p<0.05; ns, not significant. (E) Proposed interaction of tumor epithelial cells and CAFs through VEGF-A and ITGB1 enriching epithelial cell inflammatory signaling and EMT. Panel E created with BioRender.com
Discussion
PDACs are composed of a complex TME that actively restricts therapeutic access and limits cancer cell killing. In addition, studies suggest that cancer cells alter normal stromal cell behavior, inducing fibroblasts to become CAFs that support PDAC development and progression(45). However, the molecular and phenotypic impacts of CAF and PDAC tumor cell interactions remain poorly characterized. In this study, we demonstrate that CAFs are associated with inflammatory signaling and EMT in cancer-associated epithelial cells. This pattern is enriched in epithelial cells from tumor samples, irrespective of whether the epithelial cells are directly in the tumor. This further supports the idea of co-opted epithelial cells surrounding the tumor that contribute to the complex TME. A study from Bell et al(35) suggests that this Pattern 7 may also be enriched during tumorigenesis, with an inverse relationship between inflammation and EMT and proliferation as tumorigenesis progresses. This leads us to hypothesize in future studies that CAFs not only play a role in dictating epithelial cell behavior in PDAC, but they may play a similarly crucial role during tumorigenesis.
Transfer learning between the human atlas scRNA-seq data and PDO-CAF co-culture data allows more direct experimental association of CAFs as contributors to inflammatory and EMT related pathways than exclusively relying on correlations in the human atlas data. Whereas previous studies applying transfer learning to relate phenotypic states in human tumors to biological models have been applied for single cell types(28,46,47), this is the first study, to our knowledge, that extends transfer learning to relate cell-cell interactions between human tumors and an in vitro system. This analysis occurs at a transcriptome-wide level, providing a means for in silico validation of cellular phenotypes in culture. Still, understanding the specific molecular components requires gene-level analysis. Multiple MHC-II genes were included in Pattern 7, and MHC-II expression was induced following treatment with IFN-γ as well as in the presence of CAFs in our PDO-CAF co-culture. Classically expressed on the surface of professional antigen presenting cells, MHC-II is recognized by CD4+ T cells, which are increasingly appreciated for their role in clinical response to immune checkpoint inhibitors(48). Recent work demonstrates MHC-II expression in PDAC tissue using IHC in a cohort of 63 patients, and posit this expression could support neo-antigen-based immunotherapy by leveraging the MHC-II and CD4+ T cell interaction(37). Fully examining these phenomena requires expanding our computational and experimental approaches to also include immune cells of both myeloid and lymphoid lineage, which is an ongoing challenge in co-culture systems. These data further demonstrate the need for immune cell inclusion in future studies, particularly when asking questions related to the tumor immune microenvironment or mechanisms of response or resistance to immunotherapy.
Gaining an understanding of specific signaling mechanisms that drive epithelial-CAF interactions is important to furthering our understanding of PDAC biology and disrupting its progression. Evaluating signaling interactions between epithelial cells and CAFs using Domino identified crosstalk from CAFs to epithelial cells through ITGB1 and from epithelial cells to CAFs through VEGF-A. ITGB1 is part of a larger family of integrins, and high expression is associated with decreased survival in PDAC as well as EMT, inflammation, and angiogenesis(49). ITGB1 is important in epithelium-CAF crosstalk and is historically a contributor to disease progression and an enhanced mesenchymal phenotype(50). Integrins are crucial components of the tumor extracellular matrix, and our recent work suggests the cell crosstalk they mediate may also play a role in clinical response to immune checkpoint inhibitors(51).
The role of VEGF-A in epithelial-CAF crosstalk is of particular interest given the complex role of VEGF-A across malignancies. In addition to its association with angiogenesis, VEGF-A plays a broader role in tumor behavior and has established autocrine and paracrine signaling across diverse cell populations in the TME, contributing to cell phenotypes extending well beyond angiogenesis(52,53). We further identified dynamic surface expression of NRP1 on the surface of PDOs in response to recombinant VEGF-A treatment or co-culture with CAFs that was independent of changes in surface expression of VEGFR1 or VEGFR2. NRP1 is an established binding partner for VEGF-A, but this interaction has not previously been evaluated in PDAC epithelial cells and CAFs(54,55). Moreover, the VEGF-A/NRP1 axis is increasingly appreciated for contributing to EMT in breast cancer and gastric cancer and is a potential target for therapeutic intervention(56,57). We hypothesize that NRP1 overexpression in PDAC is dynamic in response to enhanced VEGF-A presence, and that this interaction not only contributes to angiogenesis but also EMT. NRP1 is also a binding partner for TGFβ, and VEGF-A can promote inflammation through macrophage recruitment, suggesting this relationship may also contribute to inflammatory signaling in epithelial tumors (Figure 6E)(52,58). While VEGF inhibition has demonstrated limited efficacy in PDAC(59–61), we hypothesize that modulation of VEGF-A has the potential to reprogram other cell types in the TME when modulation of this pathway is combined with other TME modulating agents as is an area of active investigation across tumor types (53,62,63).
While our study robustly demonstrates that computationally inferred intercellular interactions in the TME are preserved between human scRNA-seq datasets and PDO co-culture models, there are also shortcomings to this study. Our collated scRNA-seq atlas of PDAC tumors is restricted to treatment naïve biospecimens from 61 patients, with limited representation of some cell types and limited clinical annotations of the samples. This presents a challenge when trying to relate intercellular dynamics and signaling to patient outcomes for target discovery. Nonetheless, adapting our validated suite of computational tools to this atlas and the organoid co-culture provides novel insight into the role of cellular crosstalk. Domino is one method for prioritizing ligand-receptor interactions and was used in this study to highlight the strong association between VEGF-A and NRP1(64). While ligand-receptor network inference tools cannot be used to exhaustively infer mechanisms of cell-cell communication, they can still provide mechanistic insights into mechanisms of cell-cell communication. While additional computational tools will enable more direct inference of molecular changes from cellular interactions(65), the unique application of transfer learning between scRNA-seq data and organoid co-culture enables investigation of intercellular signaling using both computational discovery and experimental validation. Currently, this analysis relies on inferences resulting from a pipeline combining NMF-based pattern detection with CoGAPS(28), transfer learning with ProjectR(31), and additional ligand-receptor networks from Domino(10). While CoGAPS and ProjectR allow for unsupervised discovery and query of novel cell states, Domino is limited to investigation of pre-specified pairs of cell types and catalogued ligand-receptor interactions. Additional methods that enable discovery of multicellular interactions and their impacts on cellular phenotypes are needed to model the complex processes that underlie the PDAC TME. We also recognize that in this study we are not emphasizing the role of specific CAF (i.e. iCAF, myCAF) and epithelial (i.e. basal, classical) populations in inflammation and EMT; these populations should be deliberately interrogated in the future as we aim to better understand the complexities of these cell type interactions.
Considering the totality of these data, we propose that CAF presence enhances epithelial tumor cell inflammation and EMT with concurrent increase in MHC-II expression. Our interrogation of direct interactions provides insight into the role of epithelial and CAF crosstalk in EMT and the immunosuppressive TME. Future work will further delineate these interactions while also examining the impact of the immune compartment in driving cell phenotypes and plasticity.
Supplementary Material
Statement of Significance:
Adaptation of transfer learning to relate human scRNA-seq data to organoid-CAF co-cultures facilitates discovery of human pancreatic cancer intercellular interactions and uncovers crosstalk between CAFs and tumor cells through VEGF-A and ITGB1.
Acknowledgements:
This work was funded by the Hopper-Belmont Foundation (JWZ), The Lustgarten Foundation (EMJ), Johns Hopkins University Discovery Award (EJF, LW), Maryland Cancer Moonshot Research Grant to the Johns Hopkins Medical Institutions (FY24) (EJF), NIH/NCI (U01CA253403 to EJF; P01CA247886 to EMJ; P30CA006973; R50 CA243627 to LD; K08CA248710 to RAB). TT Seppälä was supported by fellowship grants and research funding from Sigrid Juselius Foundation, Instrumentarium Science Foundation, Emil Aaltonen Foundation, Jane and Aatos Erkko Foundation, Relander Foundation, and the iCAN precision medicine flagship of the Finnish Academy. Stand Up To Cancer–Lustgarten Foundation Pancreatic Cancer Interception Translational Cancer Research Grant (SU2C-AACR-DT26-17 to RAB). Stand Up To Cancer (SU2C) is a division of the Entertainment Industry Foundation and funding is administered by the American Association for Cancer Research, the scientific partner of SU2C). Stand Up To Cancer -Lustgarten Foundation (2015-002 to DTT).
The authors would like to thank Dr. Chris McGinnis for his technical expertise in the completion of the MULTI-seq experiment. We appreciate the help of the SciServer and IDIES team and Drs. Loyal Goff and Frederick Tan for their assistance in making the atlas available. We would also like to thank the authors of the 6 datasets used in the generation of the atlas: the laboratories of Dr. Hector Alvarez, Dr. Haiyong Han, Dr. Marina Pasca di Magliano, Dr. David Tuveson, Dr. Wenming Wu, and Dr. Itai Yanai. Sequencing was completed through the Genetic Resources Core Facility, RRID:SCR_018669. We appreciate the Legacy Gift Rapid Autopsy program for the tissue used in immunohistochemistry. Finally, we are grateful to Aviva Fertig for sharing her mommy with us during a pandemic.
Conflict of Interest Statement:
T.T.S. is the CEO and co-owner of Healthfund Finland and reports consultation fees from Boehringer Ingelheim Finland and Amgen. E.M.J is a paid consultant for Adaptive Biotech, Achilles, DragonFly, Candel Therapeutics, Genocea, and Roche. She receives funding from Lustgarten Foundation and Bristol Myer Squibb. She is the Chief Medical Advisor for Lustgarten and SAB advisor to the Parker Institute for Cancer Immunotherapy (PICI) and for the C3 Cancer Institute. She is a founding member of Abmeta. E.J.F is on the SAB for Resistance Biology, Consultant for Mestag Therapeutics and Merck. D.T.T. has received consulting fees from ROME Therapeutics, Tekla Capital, Ikena Oncology, Foundation Medicine, Inc., NanoString Technologies, and Pfizer that are not related to this work. D.T.T. is a founder and has equity in ROME Therapeutics, PanTher Therapeutics and TellBio, Inc., which is not related to this work. D.T.T. receives research support from ACD-Biotechne, PureTech Health LLC, and Ribon Therapeutics, which was not used in this work. D.T.T.’s interests were reviewed and are managed by Massachusetts General Hospital and Mass General Brigham in accordance with their conflict of interest policies. L.Z. reports personal fees from Biosion, Alphamab, NovaRock, Xilio, Ambrx, Novagenesis, and Snow Lake Capitals; and other support from Alphamab and Mingruizhiyao outside the submitted work. A.C.K. reports support from Vescor Therapeutics, Rafael Pharma, and AbbVie outside the submitted work; in addition, A.C.K. has a patent for targeting alanine transport pending, a patent for KRAS-regulated metabolic pathways issued, a patent for targeting GOT1 as a therapeutic approach issued, and a patent for autophagy control of iron metabolism issued. D.P.R. reports personal fees and other support from MPM, other support from Boehringer Ingelheim and Exact Sciences, and personal fees from UpToDate and McGraw Hill outside the submitted work. J.W.Z. reports grant funding from Genentch outside the submitted work.
Footnotes
Competing interests:
All other authors declare no potential conflict of interest.
References
- 1.Ho WJ, Jaffee EM, Zheng L. The tumour microenvironment in pancreatic cancer - clinical challenges and opportunities. Nat Rev Clin Oncol. 2020. Sep;17(9):527–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peran I, Madhavan S, Byers SW, McCoy MD. Curation of the Pancreatic Ductal Adenocarcinoma Subset of the Cancer Genome Atlas Is Essential for Accurate Conclusions about Survival-Related Molecular Mechanisms. Clin Cancer Res. 2018. Aug 15;24(16):3813–9. [DOI] [PubMed] [Google Scholar]
- 3.Chen K, Wang Q, Li M, Guo H, Liu W, Wang F, et al. Single-cell RNA-seq reveals dynamic change in tumor microenvironment during pancreatic ductal adenocarcinoma malignant progression. EBioMedicine. 2021. Apr;66:103315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hosein AN, Huang H, Wang Z, Parmar K, Du W, Huang J, et al. Cellular heterogeneity during mouse pancreatic ductal adenocarcinoma progression at single-cell resolution. JCI Insight [Internet]. 2019. Jul 23;5(16). Available from: 10.1172/jci.insight.129212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carstens JL, Yang S, Correa de Sampaio P, Zheng X, Barua S, McAndrews KM, et al. Stabilized epithelial phenotype of cancer cells in primary tumors leads to increased colonization of liver metastasis in pancreatic cancer. Cell Rep. 2021. Apr 13;35(2):108990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ligorio M, Sil S, Malagon-Lopez J, Nieman LT, Misale S, Di Pilato M, et al. Stromal Microenvironment Shapes the Intratumoral Architecture of Pancreatic Cancer. Cell. 2019. Jun 27;178(1):160–175.e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raghavan S, Winter PS, Navia AW, Williams HL, DenAdel A, Lowder KE, et al. Microenvironment drives cell state, plasticity, and drug response in pancreatic cancer. Cell. 2021. Dec 9;184(25):6119–6137.e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elyada E, Bolisetty M, Laise P, Flynn WF, Courtois ET, Burkhart RA, et al. Cross-Species Single-Cell Analysis of Pancreatic Ductal Adenocarcinoma Reveals Antigen-Presenting Cancer-Associated Fibroblasts. Cancer Discov. 2019. Aug;9(8):1102–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Öhlund D, Handly-Santana A, Biffi G, Elyada E, Almeida AS, Ponz-Sarvise M, et al. Distinct populations of inflammatory fibroblasts and myofibroblasts in pancreatic cancer. J Exp Med. 2017. Mar 6;214(3):579–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peng J, Sun B-F, Chen C-Y, Zhou J-Y, Chen Y-S, Chen H, et al. Single-cell RNA-seq highlights intra-tumoral heterogeneity and malignant progression in pancreatic ductal adenocarcinoma. Cell Res. 2019. Sep;29(9):725–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Steele NG, Carpenter ES, Kemp SB, Sirihorachai V, The S, Delrosario L, et al. Multimodal Mapping of the Tumor and Peripheral Blood Immune Landscape in Human Pancreatic Cancer. Nat Cancer. 2020. Nov;1(11):1097–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin W, Noel P, Borazanci EH, Lee J, Amini A, Han IW, et al. Single-cell transcriptome analysis of tumor and stromal compartments of pancreatic ductal adenocarcinoma primary tumors and metastatic lesions. Genome Med. 2020. Sep 29;12(1):80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moncada R, Barkley D, Wagner F, Chiodin M, Devlin JC, Baron M, et al. Integrating microarray-based spatial transcriptomics and single-cell RNA-seq reveals tissue architecture in pancreatic ductal adenocarcinomas. Nat Biotechnol. 2020. Mar;38(3):333–42. [DOI] [PubMed] [Google Scholar]
- 14.Bernard V, Semaan A, Huang J, San Lucas FA, Mulu FC, Stephens BM, et al. Single-Cell Transcriptomics of Pancreatic Cancer Precursors Demonstrates Epithelial and Microenvironmental Heterogeneity as an Early Event in Neoplastic Progression. Clin Cancer Res. 2019. Apr 1;25(7):2194–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seppälä TT, Zimmerman JW, Sereni E, Plenker D, Suri R, Rozich N, et al. Patient-derived Organoid Pharmacotyping is a Clinically Tractable Strategy for Precision Medicine in Pancreatic Cancer. Ann Surg. 2020. Sep 1;272(3):427–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seppälä TT, Zimmerman JW, Suri R, Zlomke H, Ivey GD, Szabolcs A, et al. Precision medicine in pancreatic cancer: Patient derived organoid pharmacotyping is a predictive biomarker of clinical treatment response. Clin Cancer Res. 2022. Apr 1;OF1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol. 2014. Apr;32(4):381–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haghverdi L, Lun ATL, Morgan MD, Marioni JC. Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nat Biotechnol. 2018. Jun;36(5):421–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sherman TD, Gao T, Fertig EJ. CoGAPS 3: Bayesian non-negative matrix factorization for single-cell analysis with asynchronous updates and sparse data structures. BMC Bioinformatics. 2020. Oct 14;21(1):453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fertig EJ, Ding J, Favorov AV, Parmigiani G, Ochs MF. CoGAPS: an R/C++ package to identify patterns and biological process activity in transcriptomic data. Bioinformatics. 2010. Nov 1;26(21):2792–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stein-O’Brien GL, Carey JL, Lee WS, Considine M, Favorov AV, Flam E, et al. PatternMarkers & GWCoGAPS for novel data-driven biomarkers via whole transcriptome NMF. Bioinformatics. 2017. Jun 15;33(12):1892–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005. Oct 25;102(43):15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015. Dec 23;1(6):417–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Durinck S, Moreau Y, Kasprzyk A, Davis S, De Moor B, Brazma A, et al. BioMart and Bioconductor: a powerful link between biological databases and microarray data analysis. Bioinformatics. 2005. Aug 15;21(16):3439–40. [DOI] [PubMed] [Google Scholar]
- 25.Durinck S, Spellman PT, Birney E, Huber W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat Protoc. 2009. Jul 23;4(8):1184–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McGinnis CS, Patterson DM, Winkler J, Conrad DN, Hein MY, Srivastava V, et al. MULTI-seq: sample multiplexing for single-cell RNA sequencing using lipid-tagged indices. Nat Methods. 2019. Jul;16(7):619–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moffitt RA, Marayati R, Flate EL, Volmar KE, Loeza SGH, Hoadley KA, et al. Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat Genet. 2015. Oct;47(10):1168–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sharma G, Colantuoni C, Goff LA, Fertig EJ, Stein-O’Brien G. projectR: an R/Bioconductor package for transfer learning via PCA, NMF, correlation and clustering. Bioinformatics. 2020. Jun 1;36(11):3592–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001. Dec;25(4):402–8. [DOI] [PubMed] [Google Scholar]
- 30.tricycle [Internet]. Github; [cited 2022. Mar 29]. Available from: https://github.com/hansenlab/tricycle [Google Scholar]
- 31.Cherry C, Maestas DR, Han J, Andorko JI, Cahan P, Fertig EJ, et al. Computational reconstruction of the signalling networks surrounding implanted biomaterials from single-cell transcriptomics. Nat Biomed Eng. 2021. Oct;5(10):1228–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aibar S, González-Blas CB, Moerman T, Huynh-Thu VA, Imrichova H, Hulselmans G, et al. SCENIC: single-cell regulatory network inference and clustering. Nat Methods. 2017. Nov;14(11):1083–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stein -O’, Brien GL. PatternMarkers & GWCoGAPS for novel data-driven biomarkers via whole transcriptome NMF. Bioinformatics. 2017;33:1892–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khalafalla FG, Khan MW. Inflammation and Epithelial-Mesenchymal Transition in Pancreatic Ductal Adenocarcinoma: Fighting Against Multiple Opponents. Cancer Growth Metastasis. 2017. May 15;10:1179064417709287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bell ATF, Mitchell JT, Kiemen AL, Fujikura K, Fedor H, Gambichler B, et al. Spatial transcriptomics of FFPE pancreatic intraepithelial neoplasias reveals cellular and molecular alterations of progression to pancreatic ductal carcinoma [Internet]. bioRxiv. 2022. Available from: 10.1101/2022.07.16.500312 [DOI] [Google Scholar]
- 36.Dijkstra KK, Cattaneo CM, Weeber F, Chalabi M, van de Haar J, Fanchi LF, et al. Generation of Tumor-Reactive T Cells by Co-culture of Peripheral Blood Lymphocytes and Tumor Organoids. Cell. 2018. Sep 6;174(6):1586–1598.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baleeiro RB, Bouwens CJ, Liu P, Di Gioia C, Dunmall LSC, Nagano A, et al. MHC class II molecules on pancreatic cancer cells indicate a potential for neo-antigen-based immunotherapy. Oncoimmunology. 2022. May 27;11(1):2080329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li J, Peng L, Chen Q, Ye Z, Zhao T, Hou S, et al. Integrin β1 in Pancreatic Cancer: Expressions, Functions, and Clinical Implications. Cancers [Internet]. 2022. Jul 11;14(14). Available from: 10.3390/cancers14143377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Iwatate Y, Yokota H, Hoshino I, Ishige F, Kuwayama N, Itami M, et al. Transcriptomic analysis reveals high ITGB1 expression as a predictor for poor prognosis of pancreatic cancer. PLoS One. 2022. Jun 1;17(6):e0268630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sheng W, Chen C, Dong M, Wang G, Zhou J, Song H, et al. Calreticulin promotes EGF-induced EMT in pancreatic cancer cells via Integrin/EGFR-ERK/MAPK signaling pathway. Cell Death Dis. 2017. Oct 26;8(10):e3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen L, Lin G, Chen K, Liang R, Wan F, Zhang C, et al. VEGF promotes migration and invasion by regulating EMT and MMPs in nasopharyngeal carcinoma. J Cancer. 2020. Oct 21;11(24):7291–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scaldaferri F, Vetrano S, Sans M, Arena V, Straface G, Stigliano E, et al. VEGF-A links angiogenesis and inflammation in inflammatory bowel disease pathogenesis. Gastroenterology. 2009. Feb;136(2):585–95.e5. [DOI] [PubMed] [Google Scholar]
- 43.Parker MW, Xu P, Li X, Vander Kooi CW. Structural basis for selective vascular endothelial growth factor-A (VEGF-A) binding to neuropilin-1. J Biol Chem. 2012. Mar 30;287(14):11082–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Salikhova A, Wang L, Lanahan AA, Liu M, Simons M, Leenders WPJ, et al. Vascular endothelial growth factor and semaphorin induce neuropilin-1 endocytosis via separate pathways. Circ Res. 2008. Sep 12;103(6):e71–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bussard KM, Mutkus L, Stumpf K, Gomez-Manzano C, Marini FC. Tumor-associated stromal cells as key contributors to the tumor microenvironment. Breast Cancer Res [Internet]. 2016. Dec;18(1). Available from: 10.1186/s13058-016-0740-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Davis-Marcisak EF, Fitzgerald AA, Kessler MD, Danilova L, Jaffee EM, Zaidi N, et al. Transfer learning between preclinical models and human tumors identifies a conserved NK cell activation signature in anti-CTLA-4 responsive tumors. Genome Med. 2021. Aug 11;13(1):129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stein-O’Brien GL, Clark BS, Sherman T, Zibetti C, Hu Q, Sealfon R, et al. Decomposing cell identity for transfer learning across cellular measurements, platforms, tissues, and species. Cell Syst. 2021. Feb 17;12(2):203. [DOI] [PubMed] [Google Scholar]
- 48.Tay RE, Richardson EK, Toh HC. Revisiting the role of CD4+ T cells in cancer immunotherapy-new insights into old paradigms. Cancer Gene Ther. 2021. Feb;28(1–2):5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Benesch MG, Wu R, Menon G, Takabe K. High beta integrin expression is differentially associated with worsened pancreatic ductal adenocarcinoma outcomes. Am J Cancer Res. 2022. Dec 15;12(12):5403–24. [PMC free article] [PubMed] [Google Scholar]
- 50.Yang Z, Zhou X, Liu Y, Gong C, Wei X, Zhang T, et al. Activation of integrin β1 mediates the increased malignant potential of ovarian cancer cells exerted by inflammatory cytokines. Anticancer Agents Med Chem. 2014;14(7):955–62. [DOI] [PubMed] [Google Scholar]
- 51.Li K, Tandurella JA, Gai J, Zhu Q, Lim SJ, Thomas DL 2nd, et al. Multi-omic analyses of changes in the tumor microenvironment of pancreatic adenocarcinoma following neoadjuvant treatment with anti-PD-1 therapy. Cancer Cell. 2022. Nov 14;40(11):1374–1391.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cursiefen C, Chen L, Borges LP, Jackson D, Cao J, Radziejewski C, et al. VEGF-A stimulates lymphangiogenesis and hemangiogenesis in inflammatory neovascularization via macrophage recruitment. J Clin Invest. 2004. Apr;113(7):1040–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bourhis M, Palle J, Galy-Fauroux I, Terme M. Direct and Indirect Modulation of T Cells by VEGF-A Counteracted by Anti-Angiogenic Treatment. Front Immunol. 2021. Mar 29;12:616837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mehta V, Fields L, Evans IM, Yamaji M, Pellet-Many C, Jones T, et al. VEGF (Vascular Endothelial Growth Factor) Induces NRP1 (Neuropilin-1) Cleavage via ADAMs (a Disintegrin and Metalloproteinase) 9 and 10 to Generate Novel Carboxy-Terminal NRP1 Fragments That Regulate Angiogenic Signaling. Arterioscler Thromb Vasc Biol. 2018. Aug;38(8):1845–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fukahi K, Fukasawa M, Neufeld G, Itakura J, Korc M. Aberrant expression of neuropilin-1 and −2 in human pancreatic cancer cells. Clin Cancer Res. 2004. Jan 15;10(2):581–90. [DOI] [PubMed] [Google Scholar]
- 56.Luo M, Hou L, Li J, Shao S, Huang S, Meng D, et al. VEGF/NRP-1axis promotes progression of breast cancer via enhancement of epithelial-mesenchymal transition and activation of NF-κB and β-catenin. Cancer Lett. 2016. Apr 1;373(1):1–11. [DOI] [PubMed] [Google Scholar]
- 57.Jin Q, Ren Q, Chang X, Yu H, Jin X, Lu X, et al. Neuropilin-1 predicts poor prognosis and promotes tumor metastasis through epithelial-mesenchymal transition in gastric cancer. J Cancer. 2021. Apr 30;12(12):3648–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Glinka Y, Stoilova S, Mohammed N, Prud’homme GJ. Neuropilin-1 exerts co-receptor function for TGF-beta-1 on the membrane of cancer cells and enhances responses to both latent and active TGF-beta. Carcinogenesis. 2011. Apr;32(4):613–21. [DOI] [PubMed] [Google Scholar]
- 59.Van Cutsem E, Vervenne WL, Bennouna J, Humblet Y, Gill S, Van Laethem J-L, et al. Phase III trial of bevacizumab in combination with gemcitabine and erlotinib in patients with metastatic pancreatic cancer. J Clin Oncol. 2009. May 1;27(13):2231–7. [DOI] [PubMed] [Google Scholar]
- 60.Singh RR, O’Reilly EM. New treatment strategies for metastatic pancreatic ductal adenocarcinoma. Drugs. 2020. May;80(7):647–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sahai V, Saif MW, Kalyan A, Philip PA, Rocha-Lima CM, Ocean A, et al. A phase I/II open-label multicenter single-arm study of FABLOx (metronomic 5-fluorouracil plus nab-paclitaxel, bevacizumab, leucovorin, and oxaliplatin) in patients with metastatic pancreatic cancer. J Pancreat Cancer. 2019. Sep 25;5(1):35–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang J, Yan J, Liu B. Targeting VEGF/VEGFR to Modulate Antitumor Immunity. Front Immunol. 2018. May 3;9:978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Voron T, Colussi O, Marcheteau E, Pernot S, Nizard M, Pointet A-L, et al. VEGF-A modulates expression of inhibitory checkpoints on CD8+ T cells in tumors. J Exp Med. 2015. Feb 9;212(2):139–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Armingol E, Officer A, Harismendy O, Lewis NE. Deciphering cell–cell interactions and communication from gene expression. Nat Rev Genet. 2021. Feb;22(2):71–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Deshpande A, Loth M, Sidiropoulos DN, Zhang S, Yuan L, Bell A, et al. Uncovering the spatial landscape of molecular interactions within the tumor microenvironment through latent spaces [Internet]. bioRxiv. 2022. Available from: 10.1101/2022.06.02.490672 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The scRNA-seq data analyzed in this study were obtained from the NIH gene Expression Omnibus (GEO Database), the Genome Sequence Archive (GSA), and dbGaP at GSE155698 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE155698), GSE111672 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE111672), GSE154778 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE154778), CRA001160 (https://ngdc.cncb.ac.cn/gsa/browse/CRA001160), and phs001840.v1.p1 (https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs001840.v1.p1). The Bernard et al dataset was kindly provided by the corresponding author. PDAC atlas is available for free use at www.sciserver.com after the creation of an account. Once created, please join the “Genomics” Science Domain [https://apps.sciserver.org/dashboard/science/94704] to be granted access. Next, navigate to the data volumes section of the files tab, where you will see a “STAC” directory [https://apps.sciserver.org/dashboard/files/datavolumes/53]. Here, you will see all of our public datasets. Just select the cellxgene link next to the “PDAC” title for access to our cellxgene host of the PDAC Atlas dataset.
Submission of the MULTIseq and Bulk RNA-seq data to dbGaP is in process. All analysis scripts are available from: https://github.com/fertiglab/PDAC_Atlas
All other raw data generated in this study are available upon request from the corresponding author.