Abstract
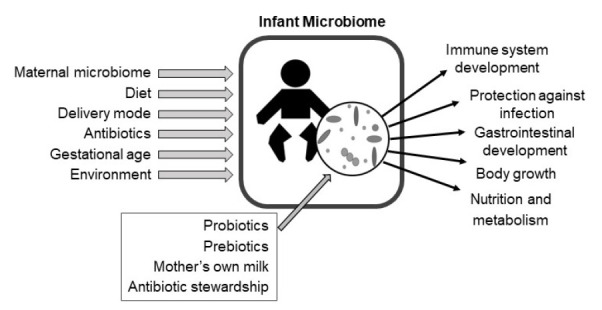
The microbiome is a complex ecosystem comprising microbes, their genomes, and the surrounding environment. The microbiome plays a critical role in early human development, including maturation of the host immune system and gastrointestinal tract. Multiple factors, including diet, anti-biotic use, and other environmental exposures, influence the establishment of the microbiome during infancy. Numerous studies have identified associations between the microbiome and neonatal diseases, including necrotizing enterocolitis, sepsis, and malnutrition. Furthermore, there is compelling evidence that perturbation of the microbiome in early life can have lasting developmental effects that increase an individual’s risk for immune and metabolic diseases in later life. Supplementation of the microbiome with probiotics reduces the risk of necrotizing enterocolitis and sepsis in at-risk infants. This review focuses on the structure and function of the infant microbiome, the environmental and clinical factors that influence its assembly, and its impact on infant health and development.
Keywords: Microbiome, Neonate infant, Host-microbiome interaction
Graphical abstract
Introduction
After birth,the infant’s skin and mucosal surfaces become rapidly colonized by complex microbial communities of bacteria, fungi, and viruses [1-3]. In early infancy, the composition and diversity of these microbial communities diverge by body site, driven by differences in nutrient availability, the chemical environment, and the host’s immune system across sites [3,4]. Multiple environmental factors influence the microbiome in infancy, including the maternal microbiome, the infant’s diet, delivery mode, and antibiotic exposure [1,5-8]. The microbiome plays a critical role in shaping multiple aspects of infant development, including the structure and function of the immune system and intestinal tract [9]. Alterations to the composition and diversity of the infant microbiome are associated with multiple morbidities including necrotizing enterocolitis (NEC), sepsis, growth failure, malnutrition, and others [10-14]. Furthermore, compelling evidence supports that alterations to the microbiome in early life have lasting effects on child development that may increase the risk of immune and metabolic diseases in later life [15-19]. This review will focus on the assembly of microbiome in infancy, its influence on infant health and development, and potential microbiome-targeted therapies to improve health outcomes.
Sources and dynamics of microbiome assembly in infancy
The sources of microbes that seed the infant’s gut microbiome include strains present in the microbiomes of the mother, other caregivers, and the infant’s environment [5,20]. A diversity of organisms from the mother’s oral, vaginal, and skin microbiomes are transiently present in the early neonatal intestinal microbiome, but many are soon replaced by species better adapted to the intestinal environment (Table 1) [3,20]. Of all maternal body sites, the fecal microbiome contributes the highest number of strains that stably colonize the infant gut [5,20]. Approximately half of the species present in the infant’s gut microbiome are shared with the maternal microbiome [20]. Many of these shared species are present at a low relative abundance in the maternal fecal microbiome, as differences in diet and the biochemical environment in the infant gut exert unique selective pressures compared to the mother [20]. The specific bacterial features that confer a selective advantage for some strains to colonize and persist in the infant gut over others are only beginning to be elucidated, but they include genes involved in polysaccharide utilization, surface adhesion, and iron acquisition [21,22]. Bacterial strains within the Bacteroides, Bifidobacterium, and Escherichia genera are commonly shared between mothers and their infants. Infants born by cesarean delivery and infants of mothers who were administered peripartum antibiotics have a lower proportion of shared bacterial strains from the mother’s microbiome than those born by vaginal delivery without maternal antibiotic exposure [5,6,23]. In addition to bacteria, other elements of the microbiome, including bacterial viruses (bacteriophages) and fungi, may be vertically transmitted from mothers to their infants [24,25].
Table 1.
Vertical transmission of bacteria from the maternal microbiome to the infant gut microbiome
| Maternal body site | Contribution to the infant’s fecal microbiome |
|---|---|
| Fecal [1,3,5,20,22,23] | The infant fecal microbiome shares more species and strains in common with the maternal fecal microbiome than other maternal body sites. |
| Strains commonly shared between maternal and infant fecal microbiomes include strains of Bacteroides vulgatus, Bacteroides uniformis, Parabacteroides distasonis, Bifidobacterium adolescentis, Bifidobacterium longum, and Escherichia coli. | |
| In some cases, nondominant strains within a bacterial species in the maternal fecal microbiome are transmitted to the infant instead of the dominant strain. | |
| Vagina [3,5,20] | Shared species are present in the maternal vaginal microbiome and infant fecal microbiome after birth, but typically do not persist in the infant’s fecal microbiome. |
| Species shared between mothers and infants include Lactobacillus spp, Gardnerella vaginalis, and Atopobium vaginae. | |
| Skin [3,20] | A minor proportion of the infant’s fecal microbiome is shared with mother’s skin microbiome at birth, but these species typically do not persist in infant’s fecal microbiome. |
| Oral [3,20] | Shared species are transiently present in the infant fecal microbiome after birth, but typically do not persist |
| Milk [20,27] | Shared Bifidobacterium strains and other taxa are present in mother’s milk and infant fecal microbiome. |
| Retrograde transfer of bacteria from the infant’s oral cavity during breastfeeding may contribute to shared taxa in maternal and infant microbiomes. |
Human milk is another source of bacteria that may colonize the infant’s intestinal tract [20,26,27]. Breast milk contains viable microbial communities often dominated by Staphylococcus, Streptococcus, Acinetobacter, and Pseudomonas [26-28]. While the microbial profiles of human milk and infant stool are generally distinct, there are shared bacterial groups between mother’s milk and her infant’s stool [26,27]. Elucidating the contribution of the breast milk microbiome to the infant gut microbiome has been challenging due to several factors including contamination of milk samples with maternal skin flora, retrograde transfer of the infant’s oral bacteria to the mammary gland and milk during breastfeeding, and technical difficulties extracting and amplifying bacterial DNA from milk [27,29]. Milk from mothers who feed expressed breast milk has fewer bacterial types in common with the infant gut microbiome than that from mothers who directly breastfeed their infants, suggesting that the transfer of bacteria from the infant mouth could be a source of bacteria in mother’s milk [27].
Other sources of bacteria that seed the infant gut include microbes transmitted from other family members and microbes present in the physical environment [23]. The hospital environment may serve as a reservoir of strains that colonize the infant, particularly among infants born preterm [30-33]. In fact, preterm infants often share identical bacterial strains with other infants in the neonatal intensive care unit [21,34].
The assembly of the microbiome in infancy is a highly dynamic process [1,7,35]. Meconium samples collected from infants typically contain low or undetectable amounts of bacterial DNA, but a rapid rise in bacterial density occurs within the first postnatal days [2]. Many of the early colonizers of the infant’s intestinal tract are facultative anaerobes (e.g., Enterococcus, Escherichia, Staphylococcus) [1,2,36]. Over time, an increase is observed in the relative abundance of strictly anaerobic organisms. This is thought to reflect a shift from an aerobic to anerobic environment in the intestine. However, in silico modeling of the meconium microbiome suggests that bacterial growth occurs under anaerobic conditions within hours of birth [2]. After this transition, Bifidobacterium and Bacteroides spp. commonly dominate the infant gut microbiome [1,7,37,38]. In breastfeeding infants, these bacteria outcompete others secondary to their ability to digest human milk oligosaccharides (HMOs) [37,38]. In later infancy, the relative abundance of anaerobic taxa characteristic of the adult fecal microbiome increases (e.g., Ruminococcus, Roseburia) [1]. While the microbiome of young breastfeeding infants is enriched in genes involved in HMO degradation, the microbiome of older infants is enriched with genes required for the digestion of dietary fibers present in solid foods [1,37].
Environmental factors that influence infant microbiome
The infant microbiome is highly individualized. Variation among individual infants often exceeds the variation attributable to environmental exposures and treatments [6,39]. Environmental factors that have been consistently associated with microbiome composition and function include delivery mode, gestational age, diet, and antibiotics.
1. Delivery mode
The early microbiome of infants born by cesarean delivery differs from that of infants born by vaginal delivery [1,6,23,40]. These differences are most apparent in the early neonatal period and diminish over time [1,3,6]. The gut microbiome of infants born vaginally is characterized by a higher relative abundance of Bacteroides spp., Bifidobacterium spp., and Escherichia coli, while the microbiome of infants born by cesarean delivery has higher relative abundance of Klebsiella, Enterococcus, Streptococcus, and Enterobacter spp [1,5-7,39,41]. Many of the taxa that are overrepresented in the gut microbiome of infants born by cesarean delivery are hospital-associated opportunistic pathogens. Cesarean delivery is also associated with a higher abundance of antibiotic resistance genes in the neonatal microbiome [1]. Some studies demonstrated that the differences in the composition of the microbiome of infants born by cesarean versus vaginal delivery are associated with functional changes that may impact immune system development [40,42,43]. For example, one study reported an enrichment of lipopolysaccharide (LPS) biosynthesis pathways in the microbiomes of infants born vaginally [42]. LPS extracts from stool samples of vaginally delivered infants induced stronger proinflammatory cytokine responses in vitro compared to samples from infants born by cesarean delivery. Studies support that maternal-to-infant transmission of bacterial strains continues to occur throughout infancy and the composition of the microbiome of infants born by cesarean delivery becomes increasingly similar to that of infants born by vaginal delivery over time [3,21].
2. Gestational age
Gestational age strongly influences the development of the gut microbiome in infancy. Compared to the microbiome of term infants, the microbiome of preterm infants is characterized by low bacterial diversity,increased relative abundance of Enterobacteriaceae (e.g., Klebsiella, Enterobacter), a paucity of Bifidobacterium, and enrichment of antibiotic resistance genes [4,39,44-46]. The gut microbiome of preterm infants is often dominated by only a few bacterial species [47]. The altered development of the microbiome of preterm versus full-term infants is likely due to a number of factors including frequent antibiotic exposure, the neonatal intensive care unit environment, delays and interruptions in the establishment of enteral feedings, and immaturity of the gut and immune system. In contrast to full-term infants, birth mode does not appear to have a strong influence on the microbiome of preterm infants [36,44,45].
The composition of the preterm infant microbiome also varies by postmenstrual age (PMA; birth gestational age plus chronologic age). At an early PMA, the microbiome of preterm infants is dominated by Staphylococcus and Enterococcus, followed by a transition to dominance by Enterobactericeae (e.g., Klebsiella, Escherichia), followed by an increase in the abundance of Clostridium, Veillonella, and other anaerobes and a late emergence of Bifidobacterium [12,13,35,45]. This predictable progression suggests that host developmental processes influence the assembly of gut microbial communities. For example, postnatal maturation of bile acid synthesis and the enterohepatic cycle contributes to the development of the intestinal microbiome in neonatal mice [48]. Interactions between microbes also contribute to the microbiome development in preterm infants. For example, inhibitory interactions between Klebsiella, Enterococcus, Staphylococcus, and Candida may drive the ordered succession of these organisms in the preterm infant gut [36].
3. Antibiotics
Antibiotics disrupt the richness, diversity, and stability of the infant microbiome [37,44,49,50]. In preterm infants, antibiotic use is associated with enrichment of antibiotic resistance genes in the microbiome and disruption of multiple functional metabolic pathways including short-chain fatty acid production [44,51]. Maternal antibiotic exposure may also influence the infant’s microbiome. For example, intrapartum antibiotic administration is associated with lower bacterial diversity, a lower relative abundance of Bifidobacteriaceae, and an increased relative abundance of Proteobacteria in the exposed versus unexposed infant’s intestinal microbiome [52].
4. Diet
Diet has strong influence on the infant microbiome [1,7]. Human milk contains multiple bioactive factors that influence the microbiome, including immunoglobulins and HMOs [53,54]. HMOs are abundant structurally diverse sugar chains in human milk. HMOs are indigestible by the infant and reach the lower intestinal tract intact. Bifidobacterium and Bacteroides encode the enzymes necessary to break down HMOs and generally become the most abundant bacterial genera in the infant microbiome. While exclusively breastfed infants have lower diversity and higher relative abundances of certain Bifidobacterium and Lactobacillus spp., nonexclusively breastfed infants have higher relative abundances of other taxa including Bacteroides and Eubacterium [1,7,55]. In one study, even brief formula supplementation among otherwise exclusively breastfed infants during the postnatal hospitalization was associated with lower relative abundance of Bifidobacteriaceae at 3–4 months of age compared to exclusively breastfed infants not exposed to supplemental formula [55].
Formula type may also influence the microbiome. For example, the microbiomes of infants fed soy formula or lactosereduced formula supplemented with corn syrup solids have increased relative abundance of Lachnospiraceae [8,56]. Preterm infants who are fed pasteurized donor human milk have altered microbiome profiles compared to those fed their mother’s own milk, including a lower relative abundance of Bifidobacteriaceae [57,58]. Diet also affects the metabolic capacity of the gut microbiome. For example, the microbiomes of breastfed versus formula-fed infants are enriched in multiple amino acid synthesis pathways [8]. In later infancy, the cessation of breastfeeding is associated with a shift toward an adult-like composition including increased abundance of Roseburia, Bilophila, Clostridium, Bacteroides, and Anaerostipes as well as an enrichment of the genes required for fiber degradation [1].
Beneficial effects of microbiome on infant development
The microbiome has a broad range of effects on infant development, including roles in immunity, growth, metabolism, and neurodevelopment [9,59-63]. These effects occur through direct interactions of the microbiome with the host’s immune system at the skin and mucosal interfaces as well as through the production of bioactive metabolites by the microbiome that are absorbed into the circulation [19,61,63,64]. The infant’s immune system and gastrointestinal tract develop upon close interaction with the microbiome. Evidence from studies of germ-free animals reared in a sterile environment demonstrated the importance of the microbiome for normal intestinal and immune development. Germ-free rodents have enlarged ceca, reduced gastrointestinal motility, altered intestinal epithelial cell morphology including longer villi and shorter crypts, and structural and functional deficits in immune development [9,65-67]. Multiple studies reported a sensitive period in early life during which certain aspects of immune development are uniquely influenced by the microbiome, and perturbations of the microbiome during this period may have lasting health consequences [16,18,19,68,69]. Similarly, metabolic programming is uniquely influenced by the microbiome during early life, and disruption of the maternal and infant microbiomes may increase an individual’s susceptibility to later metabolic diseases including obesity [15,17,70,71].
The codevelopment of the microbiome, gastrointestinal tract, and immune system also influence physical growth in infancy. Young germ-free mice demonstrate slower weight gain, length gain, organ and bone growth as well as greater weight loss during periods of malnutrition than mice raised with a conventional microbiome, demonstrating the importance of the microbiome for supporting early life growth [60]. Germ-free mice have lower circulating levels of insulin-like growth factor 1 (IGF-1) and growth hormone sensitivity in the peripheral tissues than those raised with a microbiome [60]. In a mouse model of malnutrition, a select Lactobacillus plantarum strain was sufficient to increase IGF-1 production and sustain postnatal growth through a mechanism involving the interaction between its cell wall components and the pattern recognition receptor nucleotide-binding oligomerization domain–containing-2 in the intestinal epithelium [61]. In human infants and children, malnutrition is associated with persistent immaturity in the intestinal microbiome [14]. Transferring fecal bacteria from children with malnutrition to germ-free mice recapitulates phenotypic features of malnutrition in the animals, supporting a causal role of the microbiome in malnutrition [59]. A dietary intervention designed to modulate the microbiome of children with malnutrition to a composition similar to that of healthy children improves growth and nutritional biomarkers, demonstrating the potential ability of microbiome-targeted therapies to improve growth and nutrition in early life [72].
The commensal gut microbiome excludes pathogens from the gut and protects the host from infection through a phenomenon known as colonization resistance. There are multiple mechanisms of colonization resistance, including outcompeting pathogens for space and nutrients, bacterial secretion of antimicrobial peptides, and stimulation of the host immune system [73]. The relatively low bacterial diversity and increased luminal oxygen availability in the neonatal gut may impair the colonization resistance of the neonatal microbiome and increase susceptibility to pathogen invasion and sepsis [74-76]. Colonization resistance may be further impaired following disruption of the microbiome by antibiotic use [74].
Microbiome-targeted therapies to improve infant health
Given the importance of the microbiome to infant health, there is high interest in understanding the best practices to support healthy microbiome development. Antimicrobial stewardship and breastfeeding support are sample practices that optimize the microbiome and improve infant health. Other microbiome-targeted interventions may include the administration of live bacteria (probiotics) or select nutrients that promote the growth of beneficial bacteria (prebiotics). Microbiome-targeted therapies have proven effective at reducing the incidence of NEC and sepsis, major causes of morbidity and mortality in infants (Table 2).
Table 2.
Studies of probiotics for neonatal diseasesa)
| Study | Description | No. of studies and infants | Results for effect of probiotic and/or synbiotic treatment vs. control |
|---|---|---|---|
| Systematic reviews and meta-analyses | |||
| Sharif et al. [82] 2020 | Included RCTs and quasi-RCTs of probiotics for infants born <32 weeks’ gestation and/ or <1,500 g | 56 Trials with 10,812 infants | NEC: RR, 0.54; 95% CI, 0.45–0.65 |
| Mortality: RR, 0.76; 95% CI, 0.65–0.89 | |||
| Late-onset invasive infection: RR, 0.89; 95% CI, 0.82–0.97 | |||
| Dermyshi et al. [80] 2017 | Included RCTs and observational studies of probiotics for infants born <34 weeks’ gestation and <1,500 g | 30 Trials with 8,622 infants and 14 observational studies with 13,779 infants | Severe NEC: RR, 0.57; 95% CI, 0.47–0.70 in RCTs, RR, 0.51; 95% CI, 0.37–0.70 in observational studies |
| All-cause mortality: RR, 0.77; 95% CI, 0.65–0.92 in RCTs, RR, 0.71; 95% CI, 0.62–0.81 in observational studies | |||
| Late-onset sepsis: RR, 0.88; 95% CI, 0.80–0.97 in RCTs, RR, 0.81; 95% CI, 0.69–0.96 in observational studies | |||
| Sawh et al. [81] 2016 | Included RCTs of probiotics for infants born <37 weeks’ gestation and/or <2,500 g | 42 Trials with 10,520 infants | Severe NEC: RR, 0.53; 95% CI, 0.42–0.66 |
| All-cause mortality: RR, 0.79; 95% CI, 0.68–0.93 | |||
| Culture-proven sepsis RR, 0.88; 95% CI, 0.77–1.00 | |||
| Rao et al. [86] 2016 | Included RCTs of probiotics for infants born <37 weeks’ gestation and/or <2,500 g | 37 Trials with 9,416 infants | Late-onset sepsis: RR, 0.86; 95% CI, 0.78–0.94 |
| Large randomized, controlled trials | |||
| Costeloe et al. [87] 2016 | RCT of probiotics (Bifidobacterium breve) for infants born 23–30 weeks’ gestation | 1,315 Infants | Late-onset sepsis: RR, 0.97; 95% CI, 0.73–1.29 |
| NEC ≥stage 2: RR, 0.93; 95% CI, 0.68–1.27 | |||
| Mortality: RR, 0.93; 95% CI, 0.67–1.30 | |||
| Jacobs et al. [88] 2013 | RCT of probiotics (Bifidobacterium infantis, Streptococcus thermophilus, and Bifidobacterium lactis) for infants born <32 weeks’ gestation and <1,500 g | 1,099 Infants | Late-onset sepsis: RR, 0.81, 95% CI, 0.61–1.08 |
| NEC: RR, 0.46, 95% CI, 0.23–0.93 | |||
| Mortality: RR, 0.97, 95% CI, 0.58–1.62 | |||
| Panigrahi et al. [85] 2017 | RCT of synbiotic (Lactobacillus plantarum plus fructooligosaccharide) for infants born ≥35 weeks and ≥2,000 g | 4,556 Infants | Sepsis and death: RR, 0.60; 95% CI, 0.48–0.74 |
| Culture-positive sepsis: RR, 0.22; 95% CI, 0.09–0.53 | |||
| Culture-negative sepsis: RR, 0.53; 95% CI, 0.30–0.92 | |||
| Lower-respiratory tract infection: RR, 0.66; 95% CI, 0.51–0.88 | |||
RCT, randomized controlled trial; NEC, necrotizing enterocolitis; RR, relative risk; CI, confidence interval.
Table includes a selection of relevant studies; not all published studies are included.
While the pathogenesis of NEC is not fully understood, interactions between an abnormal microbiome with the immature gut and immune system of preterm infants are central to its pathogenesis. Prolonged antibiotic use is associated with an increased risk of NEC, while breastfeeding is protective [77,78]. Breastmilk contains multiple bioactive components including HMOs and immunoglobulin A that likely contribute to its protective effects against NEC [53,79]. NEC is commonly preceded by a bloom in the relative abundance of Proteobacteria in the fecal microbiome [10]. Meta-analyses of randomized trials support that probiotics significantly reduce the risk of NEC and mortality in preterm infants (Table 2) [80-82]. Further, observational studies support the effectiveness of probiotics in clinical practice [83,84].
Sepsis is a major cause of morbidity and mortality in both full-term and preterm infants. A randomized controlled trial of an oral probiotic and prebiotic preparation (Lactobacillus plantarum plus fructooligosaccharide) in 4,556 term infants in rural India showed a striking reduction in the primary outcome of sepsis and death in the treatment arm [85]. The risk of lower-respiratory tract infections was also reduced among infants in the treatment group. Probiotics have been shown to reduce the risk of late-onset sepsis in preterm infants [86]. These studies demonstrate the promise of microbiome-targeted therapies to reduce infant morbidity and mortality rates.
Conclusion
The microbiome has a key role in health and development during infancy.Its composition and function are influenced by environmental exposures throughout infancy. Interventions to promote healthy microbiome development have already proven effective for multiple diseases that affect infants, but the discovery of other means to optimize microbiome development in early life has great potential to improve health outcomes across the lifespan.
Key message
· The infant gut microbiome is highly dynamic and indivi-dualized.
· Microbes are vertically transmitted from mother to infant during delivery and throughout infancy.
· Delivery mode, gestational age, diet, and antibiotic use influ-ence infant microbiome composition and function.
· In animal studies, the microbiome played critical roles in the structural and functional development of the infant gastro-intestinal and immune systems.
· Microbiome-targeted therapies have great potential to reduce infant morbidity and mortality.
Footnotes
Conflicts of interest
No potential conflict of interest relevant to this article was reported.
Funding
Dr. Younge is supported by the National Institutes of Health (K23DK120960, R03DK134684) and the Zeist Foundation.
References
- 1.Backhed F, Roswall J, Peng Y, Feng Q, Jia H, Kovatcheva-Datchary P, et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe. 2015;17:852. doi: 10.1016/j.chom.2015.05.012. [DOI] [PubMed] [Google Scholar]
- 2.Bittinger K, Zhao C, Li Y, Ford E, Friedman ES, Ni J, et al. Bacterial colonization reprograms the neonatal gut metabolome. Nat Microbiol. 2020;5:838–47. doi: 10.1038/s41564-020-0694-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chu DM, Ma J, Prince AL, Antony KM, Seferovic MD, Aagaard KM. Maturation of the infant microbiome community structure and function across multiple body sites and in relation to mode of delivery. Nat Med. 2017;23:314–26. doi: 10.1038/nm.4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Younge N, Araujo-Perez F, Brandon D, Seed PC. Early life skin microbiota of hospitalized preterm and full-term infants. Microbiome. 2018;6:98. doi: 10.1186/s40168-018-0486-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Podlesny D, Fricke WF. Strain inheritance and neonatal gut microbiota development: a meta-analysis. Int J Med Microbiol. 2021;311:151483. doi: 10.1016/j.ijmm.2021.151483. [DOI] [PubMed] [Google Scholar]
- 6.Shao Y, Forster SC, Tsaliki E, Vervier K, Strang A, Simpson N, et al. Stunted microbiota and opportunistic pathogen colonization in caesarean-section birth. Nature. 2019;574:117–21. doi: 10.1038/s41586-019-1560-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stewart CJ, Ajami NJ, O'Brien JL, Hutchinson DS, Smith DP, Wong MC, et al. Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature. 2018;562:583–8. doi: 10.1038/s41586-018-0617-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baumann-Dudenhoeffer AM, D'Souza AW, Tarr PI, Warner BB, Dantas G. Infant diet and maternal gestational weight gain predict early metabolic maturation of gut microbiomes. Nat Med. 2018;24:1822–9. doi: 10.1038/s41591-018-0216-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gensollen T, Iyer SS, Kasper DL, Blumberg RS. How colonization by microbiota in early life shapes the immune system. Science. 2016;352:539–44. doi: 10.1126/science.aad9378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pammi M, Cope J, Tarr PI, Warner BB, Morrow AL, Mai V, et al. Intestinal dysbiosis in preterm infants preceding necrotizing enterocolitis: a systematic review and meta-analysis. Microbiome. 2017;5:31. doi: 10.1186/s40168-017-0248-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stewart CJ, Embleton ND, Marrs ECL, Smith DP, Fofanova T, Nelson A, et al. Longitudinal development of the gut microbiome and metabolome in preterm neonates with late onset sepsis and healthy controls. Microbiome. 2017;5:75. doi: 10.1186/s40168-017-0295-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Younge NE, Newgard CB, Cotten CM, Goldberg RN, Muehlbauer MJ, Bain JR, et al. Disrupted maturation of the microbiota and metabolome among extremely preterm infants with postnatal growth failure. Sci Rep. 2019;9:8167. doi: 10.1038/s41598-019-44547-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grier A, Qiu X, Bandyopadhyay S, Holden-Wiltse J, Kessler HA, Gill AL, et al. Impact of prematurity and nutrition on the developing gut microbiome and preterm infant growth. Microbiome. 2017;5:158. doi: 10.1186/s40168-017-0377-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Subramanian S, Huq S, Yatsunenko T, Haque R, Mahfuz M, Alam MA, et al. Persistent gut microbiota immaturity in malnourished Bangladeshi children. Nature. 2014;510:417–21. doi: 10.1038/nature13421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cox LM, Yamanishi S, Sohn J, Alekseyenko AV, Leung JM, Cho I, et al. Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences. Cell. 2014;158:705–21. doi: 10.1016/j.cell.2014.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Olszak T, An D, Zeissig S, Vera MP, Richter J, Franke A, et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science. 2012;336:489–93. doi: 10.1126/science.1219328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hild B, Dreier MS, Oh JH, McCulloch JA, Badger JH, Guo J, et al. Neonatal exposure to a wild-derived microbiome protects mice against diet-induced obesity. Nat Metab. 2021;3:1042–57. doi: 10.1038/s42255-021-00439-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cahenzli J, Koller Y, Wyss M, Geuking MB, McCoy KD. Intestinal microbial diversity during early-life colonization shapes long-term IgE levels. Cell Host Microbe. 2013;14:559–70. doi: 10.1016/j.chom.2013.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scharschmidt TC, Vasquez KS, Truong HA, Gearty SV, Pauli ML, Nosbaum A, et al. A wave of regulatory T cells into neonatal skin mediates tolerance to commensal microbes. Immunity. 2015;43:1011–21. doi: 10.1016/j.immuni.2015.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ferretti P, Pasolli E, Tett A, Asnicar F, Gorfer V, Fedi S, et al. Mother-to-infant microbial transmission from different body sites shapes the developing infant gut microbiome. Cell Host Microbe. 2018;24:133–45.e5. doi: 10.1016/j.chom.2018.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lou YC, Olm MR, Diamond S, Crits-Christoph A, Firek BA, Baker R, et al. Infant gut strain persistence is associated with maternal origin, phylogeny, and traits including surface adhesion and iron acquisition. Cell Rep Med. 2021;2:100393. doi: 10.1016/j.xcrm.2021.100393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yassour M, Jason E, Hogstrom LJ, Arthur TD, Tripathi S, Siljander H, et al. Strain-level analysis of mother-to-child bacterial transmission during the first few months of life. Cell Host Microbe. 2018;24:146–54.e4. doi: 10.1016/j.chom.2018.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Korpela K, Costea P, Coelho LP, Kandels-Lewis S, Willemsen G, Boomsma DI, et al. Selective maternal seeding and environment shape the human gut microbiome. Genome Res. 2018;28:561–8. doi: 10.1101/gr.233940.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Duranti S, Lugli GA, Mancabelli L, Armanini F, Turroni F, James K, et al. Maternal inheritance of bifidobacterial communities and bifidophages in infants through vertical transmission. Microbiome. 2017;5:66. doi: 10.1186/s40168-017-0282-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schei K, Avershina E, Oien T, Rudi K, Follestad T, Salamati S, et al. Early gut mycobiota and mother-offspring transfer. Microbiome. 2017;5:107. doi: 10.1186/s40168-017-0319-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pannaraj PS, Li F, Cerini C, Bender JM, Yang S, Rollie A, et al. Association between breast milk bacterial communities and establishment and development of the infant gut microbiome. JAMA Pediatr. 2017;171:647–54. doi: 10.1001/jamapediatrics.2017.0378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fehr K, Moossavi S, Sbihi H, Boutin RCT, Bode L, Robertson B, et al. Breastmilk feeding practices are associated with the co-occurrence of bacteria in mothers' milk and the infant gut: the CHILD cohort study. Cell Host Microbe. 2020;28:285–97.e4. doi: 10.1016/j.chom.2020.06.009. [DOI] [PubMed] [Google Scholar]
- 28.Asbury MR, Butcher J, Copeland JK, Unger S, Bando N, Comelli EM, et al. Mothers of preterm infants have individualized breast milk microbiota that changes temporally based on maternal characteristics. Cell Host Microbe. 2020;28:669–82.e4. doi: 10.1016/j.chom.2020.08.001. [DOI] [PubMed] [Google Scholar]
- 29.Asnicar F, Manara S, Zolfo M, Truong DT, Scholz M, Armanini F, et al. Studying vertical microbiome transmission from mothers to infants by strain-level metagenomic profiling. mSystems. 2017;2:e00164–16. doi: 10.1128/mSystems.00164-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Younge NE, Araujo-Perez F, Brandon D, Seed PC. Early-life skin microbiota in hospitalized preterm and full-term infants. Microbiome. 2018;6:98. doi: 10.1186/s40168-018-0486-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brooks B, Firek BA, Miller CS, Sharon I, Thomas BC, Baker R, et al. Microbes in the neonatal intensive care unit resemble those found in the gut of premature infants. Microbiome. 2014;2:1. doi: 10.1186/2049-2618-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brooks B, Olm MR, Firek BA, Baker R, Thomas BC, Morowitz MJ, et al. Strain-resolved analysis of hospital rooms and infants reveals overlap between the human and room microbiome. Nat Commun. 2017;8:1814. doi: 10.1038/s41467-017-02018-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brooks B, Olm MR, Firek BA, Baker R, Geller-McGrath D, Reimer SR, et al. The developing premature infant gut microbiome is a major factor shaping the microbiome of neonatal intensive care unit rooms. Microbiome. 2018;6:112. doi: 10.1186/s40168-018-0493-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raveh-Sadka T, Thomas BC, Singh A, Firek B, Brooks B, Castelle CJ, et al. Gut bacteria are rarely shared by co-hospitalized premature infants, regardless of necrotizing enterocolitis development. Elife. 2015;4:e05477. doi: 10.7554/eLife.05477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.La Rosa PS, Warner BB, Zhou Y, Weinstock GM, Sodergren E, Hall-Moore CM, et al. Patterned progression of bacterial populations in the premature infant gut. Proc Natl Acad Sci U S A. 2014;111:12522–7. doi: 10.1073/pnas.1409497111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rao C, Coyte KZ, Bainter W, Geha RS, Martin CR, RakoffNahoum S. Multi-kingdom ecological drivers of microbiota assembly in preterm infants. Nature. 2021;591:633–8. doi: 10.1038/s41586-021-03241-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vatanen T, Franzosa EA, Schwager R, Tripathi S, Arthur TD, Vehik K, et al. The human gut microbiome in early-onset type 1 diabetes from the TEDDY study. Nature. 2018;562:589–94. doi: 10.1038/s41586-018-0620-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vatanen T, Plichta DR, Somani J, Munch PC, Arthur TD, Hall AB, et al. Genomic variation and strain-specific functional adaptation in the human gut microbiome during early life. Nat Microbiol. 2019;4:470–9. doi: 10.1038/s41564-018-0321-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hill CJ, Lynch DB, Murphy K, Ulaszewska M, Jeffery IB, O'Shea CA, et al. Evolution of gut microbiota composition from birth to 24 weeks in the INFANTMET Cohort. Microbiome. 2017;5:4. doi: 10.1186/s40168-016-0213-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jakobsson HE, Abrahamsson TR, Jenmalm MC, Harris K, Quince C, Jernberg C, et al. Decreased gut microbiota diversity, delayed Bacteroidetes colonisation and reduced Th1 responses in infants delivered by caesarean section. Gut. 2014;63:559–66. doi: 10.1136/gutjnl-2012-303249. [DOI] [PubMed] [Google Scholar]
- 41.Mitchell CM, Mazzoni C, Hogstrom L, Bryant A, Bergerat A, Cher A, et al. Delivery mode affects stability of early infant gut microbiota. Cell Rep Med. 2020;1:100156. doi: 10.1016/j.xcrm.2020.100156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wampach L, Heintz-Buschart A, Fritz JV, Ramiro-Garcia J, Habier J, Herold M, et al. Birth mode is associated with earliest strain-conferred gut microbiome functions and immunostimulatory potential. Nat Commun. 2018;9:5091. doi: 10.1038/s41467-018-07631-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McCauley KE, Rackaityte E, LaMere B, Fadrosh DW, Fujimura KE, Panzer AR, et al. Heritable vaginal bacteria influence immune tolerance and relate to early-life markers of allergic sensitization in infancy. Cell Rep Med. 2022;3:100713. doi: 10.1016/j.xcrm.2022.100713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gibson MK, Wang B, Ahmadi S, Burnham CA, Tarr PI, Warner BB, et al. Developmental dynamics of the preterm infant gut microbiota and antibiotic resistome. Nat Microbiol. 2016;1:16024. doi: 10.1038/nmicrobiol.2016.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Korpela K, Blakstad EW, Moltu SJ, Strommen K, Nakstad B, Ronnestad AE, et al. Intestinal microbiota development and gestational age in preterm neonates. Sci Rep. 2018;8:2453. doi: 10.1038/s41598-018-20827-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen Y, Brook TC, Soe CZ, O'Neill I, Alcon-Giner C, Leelastwattanagul O, et al. Preterm infants harbour diverse Klebsiella populations, including atypical species that encode and produce an array of antimicrobial resistance- and virulence-associated factors. Microb Genom. 2020;6:e000377. doi: 10.1099/mgen.0.000377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Raveh-Sadka T, Firek B, Sharon I, Baker R, Brown CT, Thomas BC, et al. Evidence for persistent and shared bacterial strains against a background of largely unique gut colonization in hospitalized premature infants. ISME J. 2016;10:2817–30. doi: 10.1038/ismej.2016.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Best N, Rolle-Kampczyk U, Schaap FG, Basic M, Olde Damink SWM, Bleich A, et al. Bile acids drive the newborn's gut microbiota maturation. Nat Commun. 2020;11:3692. doi: 10.1038/s41467-020-17183-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bokulich NA, Chung J, Battaglia T, Henderson N, Jay M, Li H, et al. Antibiotics, birth mode, and diet shape microbiome maturation during early life. Sci Transl Med. 2016;8:343ra82. doi: 10.1126/scitranslmed.aad7121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yassour M, Vatanen T, Siljander H, Hamalainen AM, Harkonen T, Ryhanen SJ, et al. Natural history of the infant gut microbiome and impact of antibiotic treatment on bacterial strain diversity and stability. Sci Transl Med. 2016;8:343ra81. doi: 10.1126/scitranslmed.aad0917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu Y, Milburn O, Beiersdorfer T, Du L, Akinbi H, Haslam DB. Antibiotic exposure prevents acquisition of beneficial metabolic functions in the preterm infant gut microbiome. Microbiome. 2022;10:103. doi: 10.1186/s40168-022-01300-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zimmermann P, Curtis N. Effect of intrapartum antibiotics on the intestinal microbiota of infants: a systematic review. Arch Dis Child Fetal Neonatal Ed. 2020;105:201–8. doi: 10.1136/archdischild-2018-316659. [DOI] [PubMed] [Google Scholar]
- 53.Gopalakrishna KP, Macadangdang BR, Rogers MB, Tometich JT, Firek BA, Baker R, et al. Maternal IgA protects against the development of necrotizing enterocolitis in preterm infants. Nat Med. 2019;25:1110–5. doi: 10.1038/s41591-019-0480-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bode L. Human milk oligosaccharides: every baby needs a sugar mama. Glycobiology. 2012;22:1147–62. doi: 10.1093/glycob/cws074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Forbes JD, Azad MB, Vehling L, Tun HM, Konya TB, Guttman DS, et al. Canadian Healthy Infant Longitudinal Development Study I. Association of exposure to formula in the hospital and subsequent infant feeding practices with gut microbiota and risk of overweight in the first year of life. JAMA Pediatr. 2018;172:e181161. doi: 10.1001/jamapediatrics.2018.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jones RB, Berger PK, Plows JF, Alderete TL, Millstein J, Fogel J, et al. Lactose-reduced infant formula with added corn syrup solids is associated with a distinct gut microbiota in Hispanic infants. Gut Microbes. 2020;12:1813534. doi: 10.1080/19490976.2020.1813534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kumbhare SV, Jones WD, Fast S, Bonner C, Jong G, Van Domselaar G, et al. Source of human milk (mother or donor) is more important than fortifier type (human or bovine) in shaping the preterm infant microbiome. Cell Rep Med. 2022;3:100712. doi: 10.1016/j.xcrm.2022.100712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ford SL, Lohmann P, Preidis GA, Gordon PS, O'Donnell A, Hagan J, et al. Improved feeding tolerance and growth are linked to increased gut microbial community diversity in very-low-birth-weight infants fed mother's own milk compared with donor breast milk. Am J Clin Nutr. 2019;109:1088–97. doi: 10.1093/ajcn/nqz006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Blanton LV, Charbonneau MR, Salih T, Barratt MJ, Venkatesh S, Ilkaveya O, et al. Gut bacteria that prevent growth impairments transmitted by microbiota from malnourished children. Science. 2016;351 doi: 10.1126/science.aad3311. 10.1126/science.aad3311 aad3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schwarzer M, Makki K, Storelli G, Machuca-Gayet I, Srutkova D, Hermanova P, et al. Lactobacillus plantarum strain maintains growth of infant mice during chronic undernutrition. Science. 2016;351:854–7. doi: 10.1126/science.aad8588. [DOI] [PubMed] [Google Scholar]
- 61.Schwarzer M, Gautam UK, Makki K, Lambert A, Brabec T, Joly A, et al. Microbe-mediated intestinal NOD2 stimulation improves linear growth of undernourished infant mice. Science. 2023;379:826–33. doi: 10.1126/science.ade9767. [DOI] [PubMed] [Google Scholar]
- 62.Buffington SA, Di Prisco GV, Auchtung TA, Ajami NJ, Petrosino JF, Costa-Mattioli M. Microbial reconstitution reverses maternal diet-induced social and synaptic deficits in offspring. Cell. 2016;165:1762–75. doi: 10.1016/j.cell.2016.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vuong HE, Pronovost GN, Williams DW, Coley EJL, Siegler EL, Qiu A, et al. The maternal microbiome modulates fetal neurodevelopment in mice. Nature. 2020;586:281–6. doi: 10.1038/s41586-020-2745-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107–18. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 65.Gustafsson BE, Midtvedt T, Strandberg K. Effects of microbial contamination on the cecum enlargement of germfree rats. Scand J Gastroenterol. 1970;5:309–14. [PubMed] [Google Scholar]
- 66.Abrams GD, Bishop JE. Effect of the normal microbial flora on gastrointestinal motility. Proc Soc Exp Biol Med. 1967;126:301–4. doi: 10.3181/00379727-126-32430. [DOI] [PubMed] [Google Scholar]
- 67.Abrams GD, Bauer H, Sprinz H. Influence of the normal flora on mucosal morphology and cellular renewal in the ileum. A comparison of germ-free and conventional mice. Lab Invest. 1963;12:355–64. [PubMed] [Google Scholar]
- 68.An D, Oh SF, Olszak T, Neves JF, Avci FY, Erturk-Hasdemir D, et al. Sphingolipids from a symbiotic microbe regulate homeostasis of host intestinal natural killer T cells. Cell. 2014;156:123–33. doi: 10.1016/j.cell.2013.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gollwitzer ES, Saglani S, Trompette A, Yadava K, Sherburn R, McCoy KD, et al. Lung microbiota promotes tolerance to allergens in neonates via PD-L1. Nat Med. 2014;20:642–7. doi: 10.1038/nm.3568. [DOI] [PubMed] [Google Scholar]
- 70.Cho I, Yamanishi S, Cox L, Methe BA, Zavadil J, Li K, et al. Antibiotics in early life alter the murine colonic microbiome and adiposity. Nature. 2012;488:621–6. doi: 10.1038/nature11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Soderborg TK, Clark SE, Mulligan CE, Janssen RC, Babcock L, Ir D, et al. The gut microbiota in infants of obese mothers increases inflammation and susceptibility to NAFLD. Nat Commun. 2018;9:4462. doi: 10.1038/s41467-018-06929-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen RY, Mostafa I, Hibberd MC, Das S, Mahfuz M, Naila NN, et al. A microbiota-directed food intervention for undernourished children. N Engl J Med. 2021;384:1517–28. doi: 10.1056/NEJMoa2023294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ducarmon QR, Zwittink RD, Hornung BVH, van Schaik W, Young VB, Kuijper EJ. Gut microbiota and colonization resistance against bacterial enteric infection. Microbiol Mol Biol Rev. 2019;83:e00007–19. doi: 10.1128/MMBR.00007-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Singer JR, Blosser EG, Zindl CL, Silberger DJ, Conlan S, Laufer VA, et al. Preventing dysbiosis of the neonatal mouse intestinal microbiome protects against late-onset sepsis. Nat Med. 2019;25:1772–82. doi: 10.1038/s41591-019-0640-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mai V, Torrazza RM, Ukhanova M, Wang X, Sun Y, Li N, et al. Distortions in development of intestinal microbiota associated with late onset sepsis in preterm infants. PLoS One. 2013;8:e52876. doi: 10.1371/journal.pone.0052876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kim YG, Sakamoto K, Seo SU, Pickard JM, Gillilland MG 3rd, Pudlo NA, et al. Neonatal acquisition of Clostridia species protects against colonization by bacterial pathogens. Science. 2017;356:315–9. doi: 10.1126/science.aag2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cotten CM, Taylor S, Stoll B, Goldberg RN, Hansen NI, Sanchez PJ, et al. Prolonged duration of initial empirical antibiotic treatment is associated with increased rates of necrotizing enterocolitis and death for extremely low birth weight infants. Pediatrics. 2009;123:58–66. doi: 10.1542/peds.2007-3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lucas A, Cole TJ. Breast milk and neonatal necrotising enterocolitis. Lancet. 1990;336:1519–23. doi: 10.1016/0140-6736(90)93304-8. [DOI] [PubMed] [Google Scholar]
- 79.Masi AC, Embleton ND, Lamb CA, Young G, Granger CL, Najera J, et al. Human milk oligosaccharide DSLNT and gut microbiome in preterm infants predicts necrotising enterocolitis. Gut. 2021;70:2273–82. doi: 10.1136/gutjnl-2020-322771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dermyshi E, Wang Y, Yan C, Hong W, Qiu G, Gong X, et al. The "golden age" of probiotics: a systematic review and meta-analysis of randomized and observational studies in preterm infants. Neonatology. 2017;112:9–23. doi: 10.1159/000454668. [DOI] [PubMed] [Google Scholar]
- 81.Sawh SC, Deshpande S, Jansen S, Reynaert CJ, Jones PM. Prevention of necrotizing enterocolitis with probiotics: a systematic review and meta-analysis. PeerJ. 2016;4:e2429. doi: 10.7717/peerj.2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sharif S, Meader N, Oddie SJ, Rojas-Reyes MX, McGuire W. Probiotics to prevent necrotising enterocolitis in very preterm or very low birth weight infants. Cochrane Database Syst Rev. 2020;10:CD005496. doi: 10.1002/14651858.CD005496.pub5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hartel C, Pagel J, Rupp J, Bendiks M, Guthmann F, Rieger-Fackeldey E, et al. Prophylactic use of Lactobacillus acidophilus/Bifidobacterium infantis probiotics and outcome in very low birth weight infants. J Pediatr. 2014;165:285–9.e1. doi: 10.1016/j.jpeds.2014.04.029. [DOI] [PubMed] [Google Scholar]
- 84.Gray KD, Messina JA, Cortina C, Owens T, Fowler M, Foster M, et al. Probiotic use and safety in the neonatal intensive care unit: a matched cohort study. J Pediatr. 2020;222:59–64.e1. doi: 10.1016/j.jpeds.2020.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Panigrahi P, Parida S, Nanda NC, Satpathy R, Pradhan L, Chandel DS, et al. A randomized synbiotic trial to prevent sepsis among infants in rural India. Nature. 2017;548:407–12. doi: 10.1038/nature23480. [DOI] [PubMed] [Google Scholar]
- 86.Rao SC, Athalye-Jape GK, Deshpande GC, Simmer KN, Patole SK. Probiotic supplementation and late-onset sepsis in preterm infants: a meta-analysis. Pediatrics. 2016;137:e20153684. doi: 10.1542/peds.2015-3684. [DOI] [PubMed] [Google Scholar]
- 87.Costeloe K, Hardy P, Juszczak E, Wilks M, Millar MR; Probiotics in Preterm Infants Study Collaborative Group. Bifidobacterium breve BBG-001 in very preterm infants: a randomised controlled phase 3 trial. Lancet. 2016;387:649–60. doi: 10.1016/S0140-6736(15)01027-2. [DOI] [PubMed] [Google Scholar]
- 88.Jacobs SE, Tobin JM, Opie GF, Donath S, Tabrizi SN, Pirotta M, et al. Probiotic effects on late-onset sepsis in very preterm infants: a randomized controlled trial. Pediatrics. 2013;132:1055–62. doi: 10.1542/peds.2013-1339. [DOI] [PubMed] [Google Scholar]


