Abstract
We examined the effects of mutations in the Saccharomyces cerevisiae RAD27 (encoding a nuclease involved in the processing of Okazaki fragments) and POL3 (encoding DNA polymerase δ) genes on the stability of a minisatellite sequence (20-bp repeats) and microsatellites (1- to 8-bp repeat units). Both the rad27 and pol3-t mutations destabilized both classes of repeats, although the types of tract alterations observed in the two mutant strains were different. The tract alterations observed in rad27 strains were primarily additions, and those observed in pol3-t strains were primarily deletions. Measurements of the rates of repetitive tract alterations in strains with both rad27 and pol3-t indicated that the stimulation of microsatellite instability by rad27 was reduced by the effects of the pol3-t mutation. We also found that rad27 and pol3-01 (an allele carrying a mutation in the “proofreading” exonuclease domain of DNA polymerase δ) mutations were synthetically lethal.
All eukaryotic genomes thus far examined contain many simple repetitive DNA sequences, tracts of DNA with one or a small number of bases repeated multiple times (48). These repetitive regions can be classified as microsatellites (small repeat units in tandem arrays 10 to 60 bp in length) and minisatellites (larger repeat units in tandem arrays several hundred base pairs to several kilobase pairs in length). In this paper, arrays with repeat units 14 bp or less will be considered microsatellites and arrays with longer repeat units will be considered minisatellites.
Previous studies show that simple repetitive sequences are unstable relative to “normal” DNA sequences, frequently undergoing additions or deletions of repeat units, in Escherichia coli (24), Saccharomyces cerevisiae (12), and mammals (59). This mutability has two important consequences. First, it results in polymorphic loci that are useful in genetic mapping and forensic studies (15, 59). Second, although these repetitive tracts are usually located outside of coding sequences, alterations in the lengths of microsatellites or minisatellites located within coding sequences can produce frameshift mutations or novel protein variants (20, 22, 26).
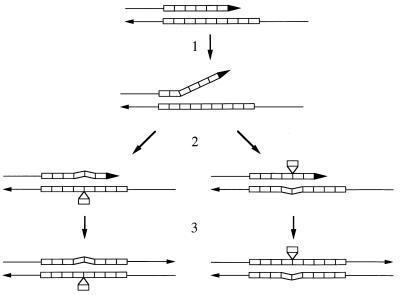
From studies of the effects of various mutations on microsatellite stability in yeast and E. coli (40) and the analysis of mutational changes caused by DNA polymerase in vitro (21), it is likely that most alterations reflect DNA polymerase slippage events (47). These events involve the transient dissociation of the primer and template strands during the replication of a microsatellite (Fig. 1). If the strands reassociate to yield an unpaired repeat on the primer strand, the net result is an addition of repeats (following a second round of DNA replication). Unpaired repeats on the template strand would result in a deletion by the same mechanism.
FIG. 1.
“Classical” model for the generation of microsatellite alterations by DNA polymerase slippage. Two single strands of a replicating DNA molecule are shown, with each repeat unit indicated by a rectangle. Arrows indicate the 3′ ends of the strand, and the top and bottom strands represent the elongating primer strand and the template strand, respectively. Step 1, the primer and template strand dissociate; step 2, the primer and template strands reassociate in a misaligned configuration, resulting in an unpaired repeat on either the template strand (left side) or primer strand (right side); step 3, DNA synthesis is completed. If the unpaired repeats are not excised by the DNA mismatch repair system, after the next round of DNA synthesis one DNA molecule will be shortened by one repeat (left side) or lengthened by one repeat (right side).
A number of mutations have been shown to elevate microsatellite instability. In E. coli (24, 46), yeast (44, 45), and mammalian cells (27), mutations in genes affecting DNA mismatch repair dramatically elevate the instability of a dinucleotide microsatellite. The most likely explanation of this result is that the DNA mismatches (unpaired repeats) resulting from DNA polymerase slippage events are efficiently removed from the newly synthesized strand by the DNA mismatch repair system. Thus, in the absence of mismatch repair, tract instability is elevated. From genetic studies, it has been found that mismatch repair in yeast efficiently corrects DNA mismatches involving 1- to 14-base loops (the size of the repeat units in microsatellites) but fails to correct mismatches involving loops larger than 16 bases (the size of the repeat units in minisatellites) (3, 41, 53). An inefficient mechanism, not involving the classical DNA mismatch repair system, is capable of correcting large DNA loops formed during meiotic recombination (19).
In addition to mutations affecting DNA mismatch repair, some mutations affecting DNA replication in yeast destabilize microsatellites. Yeast strains bearing a null mutation in the RAD27 (RTH1) gene have high levels of instability of the dinucleotide poly(GT) and the trinucleotide CAG, specifically elevating single-repeat insertions (18, 39). RAD27 encodes the homolog of the mammalian FEN-1 protein, a 5′-to-3′ exonuclease (10, 11, 33). This nuclease activity is required for removing the terminal ribonucleotide residue from the 5′ end of the Okazaki fragment (9, 14, 35, 54, 55, 57); this step is necessary for the two adjoining fragments to be ligated together. FEN-1 appears to be active as either an exonuclease in the presence of a single-stranded gap upstream of the 5′ terminus or an endonuclease on a 5′ flap structure (13, 34). Since yeast strains that contain a null mutation in RAD27 grow poorly but are viable (38, 43), it is likely that less efficient nuclease activities that are also capable of 5′ Okazaki fragment processing are present in yeast. In addition to destabilizing dinucleotide microsatellites, rad27 strains have high levels of spontaneous mitotic recombination, elevated rates of forward mutation, and increased sensitivity to the alkylating agent methyl methanesulfonate (MMS) (18, 38, 43). In contrast to the mutations normally seen in mismatch repair mutants, i.e., point mutations or small frameshifts, the types of mutations observed in the absence of Rad27p are duplications of sequences flanked by short direct repeats (4 to 7 bp in length) (49). These duplications were not affected by the DNA mismatch repair system.
The same class of sequences that are duplicated in the rad27 strains show an elevated rate (up to 1,000-fold) of deletion in strains containing a temperature-sensitive allele (pol3-t) of the yeast gene encoding DNA polymerase δ (52, 53). This mutant (initially named tex1) was isolated in a strain that exhibited an increased excision rate of a bacterial transposon with long terminal repeats inserted within a yeast gene (7). The pol3-t allele, which encodes a mutation (Gly641 to Ala641) (51) located near the putative nucleotide binding and active-site domains of the enzyme (58), is thought to diminish the rate of lagging-strand synthesis resulting in long stretches of single-stranded DNA on the lagging-strand template (8). This single-stranded DNA may have the potential to form intrastrand base-paired structures, creating interactions between short direct repeats. These interactions would result in an increased frequency of deletions caused by DNA polymerase slippage.
Since rad27 and pol3-t mutations elevate the rates of duplications and deletions associated with short separated repeats in nonrepetitive DNA sequences, Kunkel et al. (22) suggested that these mutations could also destabilize minisatellites. In this paper, we examine the effects of rad27 and pol3-t mutations on the stability of simple repeats in which the repeat unit length varies between 1 and 20 bp. Our results show that both mutations destabilize both microsatellites and minisatellites, but that the mechanisms involved in the destabilization are different for the two mutations.
MATERIALS AND METHODS
Yeast strains and plasmids.
All yeast strains used in this study were derived from S. cerevisiae AMY125 (α ade5-1 leu2-3 trp1-289 ura3-52 his7-2; obtained from A. Morrison and A. Sugino, Osaka University, Osaka, Japan). MS71 (a LEU2 derivative of AMY125, other markers identical) has been described previously (44). YRTH29 (MS71 rad27Δ [rho−]) was provided by S. Prakash (University of Texas Medical Branch, Galveston, Tex.) (18). RJK56, a [rho+] derivative of YRTH29, was isolated by dissection of a diploid obtained by mating YRTH29 with EAS18 (an a mating type derivative of MS71). Spores were screened for growth on medium containing glycerol, and the presence of rad27Δ was confirmed by Southern hybridization. RJK88 is isogenic with RJK56, except that it has the opposite mating type. MS72 (MS71 pol2-4) and MS73 (MS71 pol3-01) have been described previously (45). In spores derived from diploids obtained by crossing rad27Δ strain RJK88 to pol2-4 (MS72) or pol3-01 (MS73) strains, the presence of pol2-4 and pol3-01 was diagnosed in the spore colonies by PCR and/or restriction analysis (30, 32). rad27Δ was detected in the spore colonies by their failure to grow on plates containing 0.025% MMS (43).
As described in detail in the Results section, when diploid strain RJK54 (heterozygous for rad27 and pol3-01) was sporulated and the resulting spores were analyzed, we found no viable spores with the double-mutant phenotype. To determine whether this property was haploid specific, we attempted to construct a diploid strain homozygous for rad27 and pol3-01. The RJK54 strain was transformed with plasmid pBL304 (YCp50 POL3; provided by P. Burgers, Washington University School of Medicine), and the resulting transformant was sporulated. Two haploid segregants of opposite mating type (RJK138 and RJK139 with a and α mating types, respectively) and the rad27 pol3-01 genotype containing the plasmid were identified. These strains were mated to form a diploid (RJK140-2) that also contained pBL304. Another diploid (RJK153) heterozygous for the rad27 and pol3-01 mutations (otherwise isogenic to RJK140-2) and containing the pBL304 plasmid was also constructed.
Strains with the pol3-t mutation were constructed by using plasmid p171. Plasmid p171 was derived from plasmid p170. The p170 plasmid was constructed by the insertion of a 2.2-kb EcoRV-HindIII fragment (derived from YIpAM26) (30) into PvuII- and HindIII-treated pFL34 (5); this 2.2-kb fragment contains a portion of the wild-type POL3 gene that includes the region of the pol3-t mutation. The region of POL3 with the mutant pol3-t substitution was obtained by PCR amplification from a yeast strain (pol3-t DM) (8) that contained the mutant allele. Amplification was performed with primers from positions 1757 to 1774 (5′ CTAATGGCGTTAGTTAAC 3′) and 2314 to 2297 (5′ ACCCACCGTCGCTCCTGT 3′) in POL3. An HpaI-BamHI fragment of this PCR product was used to replace the HpaI-BamHI fragment of p170 to create plasmid p171. The HpaI-BamHI region of p171 was sequenced to verify that there were no additional mutations. MS71 and RJK56 were transformed with HpaI-linearized p171. Temperature-sensitive Ura+ transformants contained the full length pol3-t allele and a truncated POL3 flanking the URA3 gene. Ura− temperature-sensitive clones were selected on medium containing 5-fluoro-orotate (5-FOA). The presence of the pol3-t mutation was verified by PCR of genomic DNA with subsequent digestion with MboII, since the pol3-t mutation results in the loss of an MboII site. The resulting strains with the pol3-t substitution were MS71-pol3-t and RJK56-pol3-t.
To assess the effect of rad27 and pol3-t on simple-repeat instability, we transformed strains RJK56, MS71-pol3-t, and RJK56-pol3-t with plasmids pMD28, pSH44, pBK10, and pEAS20 (12, 41). In addition, strain RJK56 was transformed with plasmids pBK1 and pBK3 (41). These plasmids contain repetitive DNA tracts inserted in frame within the URA3 gene. The sequences of the repetitive tracts contained within each of these plasmids are as follows: pMD28, (G)18; pSH44, (GT)16; pBK1, (CAGT)16; pBK3, (CAACG)15; pBK10, (CAATCG GT)10; pEAS20, (CAACGCAATGCGTTGGATCT)3.
To examine the effects of rad27 and pol3-t on deletion formation, we used two mutant lys2 genes with insertions of different sizes: lys2-InsLD (31-bp insert flanked by 7-bp direct repeats) and lys2-InsLE (61-bp insert flanked by 6-bp repeats). These alleles are the same as the lys2::InsD and lys2::InsE alleles described previously (52, 53) except that the sequences of the insertions were modified to contain stop codons in all three reading frames. As a result, strains carrying these inserts can revert to Lys+ only by precise deletions between the flanking short direct repeats. Plasmids pLD-Int and pLE-Int were used to insert the lys2-InsLD and lys2-InsLE sequences into strains MS71, RJK56, MS71-pol3-t, and RJK56-pol3-t by the two-step transplacement procedure. These plasmids were provided by N. Degtyareva (St. Petersburg State University, St. Petersburg, Russia) and are identical to p92(InsD) and p93(InsE) described previously (52) except for the nonsense codons in the insertions.
Analysis of repeat instability in plasmids.
The mutant strains transformed with plasmids pMD28, pSH44, pBK10, pEAS20, pBK1, and pBK3 were phenotypically Ura+ because the simple repetitive DNA sequences within the coding sequence of the plasmid-borne URA3 gene are in frame. Alterations in tract length that result in an out-of-frame insertion can be selected on medium containing 5-FOA (12). Instability rates for each strain were determined by measuring the frequency at which 5-FOA-resistant (5-FOAR) colonies appeared in multiple independent cultures and converting these frequencies into a rate estimate by the method of the median (23), as described previously (12). Ninety-five percent confidence intervals for the rate measurements were calculated as described previously (61).
Since strains carrying the rad27Δ and pol3-t mutations are temperature sensitive, cells were preincubated on synthetic dextrose (SD) medium at 30°C, a temperature that is permissive for growth for all the mutant strains tested. Cells plated on medium containing 5-FOA were then incubated at 22°C for 4 days (pol3-t and rad27 pol3-t strains) or at 30°C for 3 days (rad27 and wild-type strains); the lower temperature was used for the pol3-t strains because of their more temperature-sensitive phenotype. In an experiment in which we compared the rates of minisatellite instability of two sets of cultures of a rad27 strain (containing pEAS20) incubated on 5-FOA plates, one set at 22°C for 4 days and one set at 30°C for 3 days, no significant difference was found (data not shown). A total of 12 to 20 cultures were examined for each of at least two independent transformants of each strain. The rates for independent transformants were averaged. For all of the transformants tested, the measured rates from the independent transformants of the same genotype were within 2.5-fold of each other. The types of alterations generated within independent 5-FOAR colonies were determined by PCR amplification of the region containing the repeat tract and analysis by gel electrophoresis or by direct sequencing of this region as described previously (41).
Analysis of rates of reversion of insertion mutations in the LYS2 locus.
Reversion rates and 95% confidence intervals for the reversion rates of the lys2-InsLD and lys2-InsLE alleles were determined as described previously (52). Rate determinations were based on 12 independent cultures for each genotype. For all strains tested, cells were pregrown on enriched media at 30°C. For measurement of the Lys+ reversion rate, cultures were then plated on synthetic media lacking lysine at 22°C. Deletion of the inserts in Lys+ revertants was verified by PCR and sequence analysis.
RESULTS
Minisatellite and microsatellite instability in rad27 and pol3-t strains.
It has been previously demonstrated that a null mutation of the yeast RAD27 (RTH1) gene results in an ∼100-fold increase in the instability of poly(GT) tracts (18). It was suggested that Rad27p might have a role in DNA mismatch repair, although this interpretation was subsequently questioned (49). In this study, we examine the effects of a null mutation of RAD27 (rad27Δ mutation) on the instability of a variety of microsatellites with repeat units varying between 1 and 8 bp and a minisatellite sequence with a 20-bp repeat length. To measure the stability of these repetitive tracts, we employed a plasmid-based frameshift assay described previously (12). In this assay, each of the repetitive tracts is inserted in frame within the coding sequence of a plasmid-borne URA3 gene. Cells transformed with these plasmids are Ura+. Deletions or additions of one or more repeat units in the repeat tract resulting in an out-of-frame insertion are detected by plating the cells on medium containing 5-FOA, which selects for ura3 mutant cells (4). The reporter plasmids used for this study were previously employed to measure the effects of DNA mismatch repair mutations on simple-repeat instability (41, 44, 45).
The rates of alterations for tracts with different repeat units in the rad27Δ strains are shown in Table 1. All the repetitive tracts were destabilized by the rad27 mutation. The effect was strongest on mononucleotide or dinucleotide repeats, i.e., 100- to 200-fold destabilization. Our measurement of the effects of rad27 on dinucleotide microsatellites agrees well with that reported previously (18). In general, the smaller the repeat unit length, the greater the effect of a loss of Rad27p activity. A similar trend was also observed for strains bearing mutations in msh2 (41). The minisatellite containing three 20-bp tandem repeats was destabilized about 10-fold by the rad27 mutation. No other yeast mutation has been reported to affect minisatellite stability. Null mutations in a number of yeast genes affecting DNA mismatch repair (msh2, msh3, msh6, mlh1, and pms1) do not destabilize this minisatellite (41).
TABLE 1.
Rates of alterations of repetitive DNA sequences in wild-type yeast strains and strains with mutations in genes encoding components of the DNA replication system
| Size of repeat unit (bp)a | Relevant genotype | Rate of 5-FOAR cells in independent expts (10−5)b | Avg rate of tract altera- tions (10−5)c | Fold increase vs wild type |
|---|---|---|---|---|
| 1 (18) | Wild type | 0.59 (0.36–1.3) | 0.67 | 1 |
| 0.80 (0.70–1.1) | ||||
| 0.61 (0.40–0.68) | ||||
| rad27 | 89 (53–140) | 92 | 140 | |
| 120 (89–150) | ||||
| 68 (52–99) | ||||
| pol3-t | 5.4 (3.4–6.5) | 4.0 | 6 | |
| 7.6 (3.7–21) | ||||
| rad27 pol3-t | 9.0 (6.9–12) | 10 | 15 | |
| 32 (14–46) | ||||
| 11 (8.8–21) | ||||
| 2 (16.5) | Wild type | 0.49 (0.37–0.97) | 0.48 | 1 |
| 0.51 (0.36–0.76) | ||||
| rad27 | 99 (79–110) | 78 | 165 | |
| 58 (36–80) | ||||
| pol3-t | 1.4 (1.1–2.2) | 1.7 | 4 | |
| 3.0 (2.2–4.6) | ||||
| rad27 pol3-t | 28 (13–89) | 40 | 80 | |
| 53 (28–92) | ||||
| 4 (16) | Wild type | 0.87 (0.63–1.4) | 0.84 | 1 |
| 0.90 (0.80–1.5) | ||||
| rad27 | 24 (14–35) | 20 | 24 | |
| 17 (13–18) | ||||
| 5 (15) | Wild type | 3.4 (2.9–7.7) | 3.0 | 1 |
| 2.8 (2.4–5.5) | ||||
| rad27 | 52 (45–72) | 46 | 15 | |
| 41 (31–61) | ||||
| 8 (10) | Wild type | 0.96 (0.72–1.2) | 0.94 | 1 |
| 1.1 (0.74–2.0) | ||||
| rad27 | 20 (14–30) | 19 | 20 | |
| 22 (20–29) | ||||
| pol3-t | 11 (8.6–14) | 9.0 | 10 | |
| 8.0 (5.2–8.7) | ||||
| rad27 pol3-t | 7.9 (3.5–11) | 5.3 | 6 | |
| 4.0 (2.8–5.1) | ||||
| 20 (3) | Wild type | 6.9 (5.4–11) | 7.4 | 1 |
| 7.8 (6.5–10) | ||||
| rad27 | 96 (61–120) | 83 | 11 | |
| 70 (64–120) | ||||
| pol3-t | 95 (58–140) | 96 | 13 | |
| 98 (75–120) | ||||
| rad27 pol3-t | 130 (110–140) | 150 | 20 | |
| 180 (140–270) |
The number in parentheses is the number of tandem repeats in the repetitive tract.
In each experiment, the rate of formation of 5-FOAR derivatives was determined by measuring the frequency of 5-FOAR cells in about 20 independent cultures and converting these frequency measurements into a rate by the method of the median (23). Numbers in parentheses indicate 95% confidence intervals calculated as described previously (61). Each independent rate measurement was obtained from an independent transformant. Rates for wild-type strains were derived from values reported previously (41).
The rate of tract alteration from each frequency measurement was obtained by multiplying the rate of formation of 5-FOAR derivatives by the fraction of the 5-FOAR cells that had tract alterations. Rates for wild-type strains were derived from values reported previously (41).
Since Rad27p is a component of the DNA replication system, it is possible that other mutations affecting DNA replication would lead to minisatellite and/or microsatellite instability. Consequently, we examined the effects of the pol3-t mutation, a mutation in the catalytic subunit of DNA polymerase δ (described in the Introduction). As shown in Table 1, pol3-t, like rad27, destabilizes both micro- and minisatellites, but the effects of pol3-t on the mononucleotide and dinucleotide repeats are much smaller than those observed with rad27 strains. The effects of pol3-t on the 8- and 20-bp repeats are roughly the same as those observed for rad27. Overall, these data suggest that the larger the size of the repeat unit, the greater the effect of the pol3-t mutation on repeat instability. The different relationships between repeat unit length and the magnitude of destabilization observed for rad27 and pol3-t argue that these effects involve different mechanisms.
Types of tract alterations accumulated in rad27Δ and pol3-t strains.
The types of changes observed for each simple repeat in the two mutant strains are presented in Table 2. It has been previously shown that a strain containing the rad27 mutation accumulates almost exclusively 2-bp insertions within the poly(GT) tract (18). Our results indicate that all of the alterations examined in the rad27 strain for both the mononucleotide and dinucleotide tracts were insertions and, with only one exception, were single-unit insertions. Compared to the mononucleotide and dinucleotide microsatellites, the repeat tracts containing 4- or 5-bp repeat units exhibited a significantly greater number of deletions in the rad27 strain (by the Fisher exact test; P < 0.01 for all comparisons). A 2- to 3-fold bias and a 10-fold bias for insertions over deletions were observed for the tetranucleotide and the octanucleotide tracts, respectively. The minisatellite containing 20-bp repeats also exhibited a twofold bias for insertions in the rad27 mutant. Although nineteen deletions were observed in rad27 strains with the microsatellite assay plasmids, it is interesting that no single-repeat deletions were seen. In contrast, about one-half of the deletions within the minisatellite were 20-bp single-unit deletions.
TABLE 2.
Types of alterations within repetitive tracts in wild-type and mutant strains
| Relevant genotype | No. of bases in repeat unit | No. of tracts containing indicated no. of additions (+) or deletions (−) of repeat units
|
||||||
|---|---|---|---|---|---|---|---|---|
| −2 | −1 | 0 | +1 | +2 | Othera | Total | ||
| Wild typeb | 1 | 2 | 3 | 0 | 24 | 1 | 30 | |
| rad27 | 1 | 0 | 0 | 0 | 20 | 0 | 20 | |
| pol3-t | 1 | 0 | 2 | 8 | 10 | 0 | 20 | |
| rad27 pol3-t | 1 | 0 | 0 | 9 | 14 | 0 | 1 (−5) | 24 |
| Wild typeb | 2 | 0 | 3 | 1 | 12 | 0 | 2 (−7), 1 (+4) | 19 |
| rad27 | 2 | 0 | 0 | 0 | 21 | 0 | 1 (+7) | 22 |
| pol3-t | 2 | 3 | 5 | 5d | 3 | 0 | 1 (−7), 5 (−8), 2 (−10), 1 (−11) | 25 |
| rad27 pol3-t | 2 | 0 | 0 | 0 | 16 | 3 | 1 (+4) | 20 |
| Wild typeb | 4 | 2 | 2 | 1 | 5 | 1 | 1 (−5), 4 (−7), 3 (−8) | 19 |
| rad27 | 4 | 2 | 0 | 0 | 9 | 2 | 3 (+4), 2 (+8), 1 (+10), 1 (−4), 2 (−5), 1 (−7), 1 (−8) | 24 |
| Wild typeb | 5 | 14 | 0 | 1 | 4 | 1 | 5 (−4), 2 (−7) | 27 |
| rad27 | 5 | 7 | 0 | 0 | 1 | 8 | 3 (−4), 2 (+4) | 21 |
| Wild typeb | 8 | 1 | 1 | 2 | 6 | 5 | 7 (−4), 2 (−5) | 24 |
| rad27 | 8 | 1 | 0 | 2 | 14 | 6 | 1 (−4) | 24 |
| pol3-t | 8 | 2 | 6 | 1 | 3 | 2 | 7 (−4), 2 (−5) | 23 |
| rad27 pol3-t | 8 | 4 | 1 | 2 | 2 | 0 | 8 (−4), 2 (−5) | 19 |
| Wild typec | 20 | 12 | 24 | 0 | 5 | 1 | 42 | |
| rad27 | 20 | 6 | 7 | 0 | 17 | 10 | 40 | |
| pol3-t | 20 | 11 | 9 | 0 | 1 | 0 | 21 | |
| rad27 pol3-t | 20 | 6 | 14 | 0 | 3 | 0 | 23 | |
The numbers of independent isolates with tracts of a particular size; the numbers of additions (+) or deletions (−) of repeat units are shown in parentheses.
Data were reported previously (41).
Data were combined from this work and research reported previously (41).
Two of the five isolates with no alterations in their poly(GT) tracts had 19-bp deletions (flanked by 6-bp direct repeats) of URA3 sequences near the tracts.
Although the bias for insertions over deletions in the rad27 strains varies considerably for the different repeat units, we observed that the destabilizing effect of rad27 on insertions is at least 4- to 10-fold greater than the enhancement of deletions for all repeat unit sizes tested (Table 3). The rates for insertions in rad27 strains were 30- to 200-fold greater than those observed in wild-type strains, while the deletion rates were never more than 10-fold greater than those observed in the wild type. Therefore, while the absence of Rad27p can lead to an elevated rate of deletions within repetitive DNA, the predominant effect of this loss is an accumulation of insertions.
TABLE 3.
Comparison of rates of insertions and deletions in wild-type and mutant yeast strains
| Size of repeat unit (bp) | Relevant genotype | Rate ofa:
|
|
|---|---|---|---|
| Insertions/division (10−5) | Deletions/division (10−5) | ||
| 1 | Wild type | 0.56 (1) | 0.11 (1) |
| rad27 | 92 (165) | <4.6 (<42) | |
| pol3-t | 3.3 (6) | 0.67 (6) | |
| rad27 pol3-t | 9.3 (17) | 0.67 (6) | |
| 2 | Wild type | 0.35 (1) | 0.13 (1) |
| rad27 | 78 (220) | <3.5 (<27) | |
| pol3-t | 0.26 (0.7) | 1.4 (11) | |
| rad27 pol3-t | 40 (110) | <2.0 (<15) | |
| 4 | Wild type | 0.28 (1) | 0.56 (1) |
| rad27 | 14 (50) | 5.8 (10) | |
| 5 | Wild type | 0.58 (1) | 2.4 (1) |
| rad27 | 24 (41) | 22 (9) | |
| 8 | Wild type | 0.47 (1) | 0.47 (1) |
| rad27 | 17 (37) | 1.7 (4) | |
| pol3-t | 2.0 (4) | 7.0 (15) | |
| rad27 pol3-t | 0.62 (1.3) | 4.7 (10) | |
| 20 | Wild type | 1.1 (1) | 6.3 (1) |
| rad27 | 56 (51) | 27 (4) | |
| pol3-t | 4.6 (4) | 91 (14) | |
| rad27 pol3-t | 20 (18) | 130 (21) | |
Rates of insertion and deletion were calculated by multiplying the rate of tract alteration of all types (data from Table 1) by the fractions of alterations observed as insertions or deletions (data from Table 2). Numbers in parentheses indicate fold increases in the rates for the mutant strains relative to the rates determined for the wild-type strains for each repeat size. For the rad27 strains with the mono- and dinucleotide tracts and for the rad27 pol3-t strain with the dinucleotide tract, no deletions were observed. The minimal values shown were calculated by multiplying the total rate of tract alterations by the fractions obtained by dividing 1 by the total numbers of tracts analyzed.
In contrast to the bias in favor of insertion formation observed in rad27 strains, microsatellites (with the exception of the mononucleotide tract) and the minisatellite sequences usually underwent deletions in pol3-t strains (Tables 2 and 3). Many of these deletions were large, over 14 bp. The rates with which deletions occurred in pol3-t strains for the repetitive tracts containing 2-, 8-, and 20-bp repeat units were 11- to 15-fold greater than the rates observed for a wild-type strain, while the enhancement of insertions was much smaller (Table 3). For the mononucleotide microsatellite, the enhancement for insertions due to the pol3-t mutation was about the same as the enhancement for deletions (Table 3). In summary, the different types of tract alterations observed in rad27 and pol3-t strains confirm our conclusion that the associated mutations destabilize repetitive DNA sequences by different mechanisms.
Micro- and minisatellite instability in rad27 pol3-t strains.
One method of determining whether two gene products are involved in the same pathway is to determine whether the phenotype of the double-mutant strain is the same as observed for either of the single-mutant strains. Consequently, we measured the stability of the repetitive tracts in double-mutant rad27 pol3-t strains. For the mono- and dinucleotide repeats, the rates observed in the double mutant were intermediate between those observed in the two single-mutant strains (Table 1). The rate of instability for the mononucleotide tract in the double mutant was an order of magnitude lower than the rate seen in the rad27 single mutant and only about two times greater than the rate observed for the pol3-t mutant. The rate of dinucleotide instability in the strain bearing both mutations was approximately half the rate observed in the rad27 strain.
The types of alterations observed in the rad27 pol3-t strain were also examined (Table 2). For the mononucleotide repeat, as observed for the wild-type, rad27, and pol3-t strains, almost all tract changes involved the addition of a single repeat. In the rad27 pol3-t strain, as found for the pol3-t strain, a substantial fraction of the plasmids analyzed did not have an alteration in the tract. The types of alterations of the dinucleotide tracts in the double mutant were similar to those observed in the rad27 mutant, almost exclusively small insertions. From the types of alterations and the rate measurements, we conclude that wild-type DNA polymerase δ is required for most of the destabilizing effects of the rad27 mutation on mononucleotides but is not required for most of the destabilization observed with the dinucleotide tracts. This conclusion will be discussed further below.
The octameric-repeat tract, which was destabilized 10- to 20-fold by the rad27 and pol3-t single mutants, was destabilized only 6-fold in the double mutant (Table 1). In contrast to what was seen for the microsatellites containing shorter repeat unit lengths, the changes seen within the octanucleotide repeat were primarily deletions (Table 2). The alterations resembled those seen in the pol3-t mutant rather than those found for the rad27 mutant. Therefore, the pol3-t mutation eliminates the moderate effect of rad27 on octanucleotide repeat instability.
The rate of minisatellite instability (20-bp repeats) in the double mutant was somewhat greater than for either single mutant, suggesting the possibility of an additive interaction (Table 1). Since the confidence limits for the rate estimates for all three strains overlap, however, an epistatic interaction is also possible. In support of this possibility, the alterations found in the double mutant were very similar to those observed in the pol3-t strain. In summary, for all classes of repetitive DNA sequences except the dinucleotide repeats, the rates of alterations and the types of alterations in the double-mutant strain resemble those found in the pol3-t strain rather than those found in the rad27 strain.
Reversion rates of insertion mutations in the LYS2 gene in rad27, pol3-t, and rad27 pol3-t strains.
It has been previously demonstrated that the pol3-t mutation increased the deletion rate of large insertions flanked by 4- to 7-bp direct repeats (8, 52, 53). We examined the reversion rates of two insertions closely related to those used in previous studies: lys2-InsLD, a 31-bp sequence flanked by 7-bp direct repeats, and lys2-InsLE, a 61-bp sequence flanked by 6-bp repeats. Precise deletion of the insertion and one of the direct repeats restores the original reading frame, resulting in a Lys+ phenotype.
A comparison of the effects of rad27 and pol3-t on the reversion rates for lys2-InsLD and lys2-InsLE is shown in Table 4. While the pol3-t mutation increased the reversion rates of both mutations more than 100-fold, the effect of rad27 on the reversion rate of lys2-InsLD was more modest (4-fold). The tendency for the rad27 mutation to cause additions rather than deletions, which was observed with the micro- and minisatellites (Table 2), is consistent with the small effect of the mutation on the lys2 insertions. Based on the observation of a single deletion at the CAN1 locus in a rad27 strain, it was proposed that rad27 enhances the rate of deletions of unique sequences flanked by direct repeats, but to a lesser extent than it enhances duplications (49). The data presented in Table 4 support this proposal.
TABLE 4.
Reversion rates in wild-type and mutant strains with lys2 mutations caused by 31- (lys2-InsLD) or 61-bp (lys2-InsLE) insertions
| Relevant genotype | Rate of reversion (10−8)a for insertion:
|
|
|---|---|---|
| lys2-InsLD | lys2-InsLE | |
| Wild type | 0.32 (1.0) [0.17–0.55] | <0.02 (1.0) |
| rad27 | 1.2 (4) [0.5–3.3] | 0.13 (>7.0) [0.05–0.3] |
| pol3-t | 49 (150) [32–177] | 15 (>750) [12–27] |
| rad27 pol3-t | 48 (150) [39–66] | 8.4 (>400) [2.6–17] |
Numbers in parentheses indicate the fold increases in the rates for the mutant strains relative to the rates determined for the wild-type strains. Numbers in brackets represent 95% confidence intervals on the unnormalized rates.
The reversion rate in the rad27 pol3-t double mutant was approximately the same as that observed in the pol3-t single mutant. Since the rad27 effect is much less than that observed for pol3-t, however, we cannot rule out an additive effect of the two mutations.
Synthetic lethality of rad27Δ and pol3-01 (an allele carrying a mutation in the proofreading exonuclease domain of DNA polymerase δ) mutations.
Since the rad27 pol3-t mutant had an unexpected effect (a reduction in the rate of microsatellite instability relative to the rad27 single mutant), we decided to examine microsatellite instability in a strain with rad27 and pol3-01 mutations. The pol3-01 mutation results in a loss in the proofreading exonuclease activity of DNA polymerase δ (42) without affecting DNA elongation (29). Strains with this mutation exhibit about a 10-fold destabilization of a dinucleotide microsatellite (45). We attempted the construction of a rad27 pol3-01 double mutant by sporulation of a diploid that was heterozygous for both mutations. We found that spore viability was low. Of 40 tetrads, the numbers of tetrads with two, three, and four viable spores were 11, 22, and 7, respectively; in other sporulated diploids involving the same genetic background without these mutations, about two-thirds of the tetrads have four viable spores. We found that none of the spore colonies derived from 17 tetrads containing three or four viable spores had the double-mutant combination; the presence of the mutations in spores was scored by sensitivity to MMS (rad27 mutation) or by PCR analysis (pol3-01 mutation), as described in Materials and Methods. For 10 tetrads with three viable spores, the ratio of spores scored as wild type/rad27/pol3-01/rad27 pol3-01 was 9:10:11:0. For seven tetrads containing four viable spores, two of the spores were pol3-01 RAD27 and two of the spores were POL3 rad27 in all tetrads. The synthetic lethality of these two mutations suggests that the proofreading activity of DNA polymerase δ may be involved, directly or indirectly, in the processing of DNA lesions resulting from a loss of Rad27p activity.
Previously, it was shown that haploids with the genotype pms1 pol3-01 were inviable, although diploids homozygous for these mutations were viable (31). It was suggested that the mutation rate in the double-mutant strains was too high to be tolerated in a strain with only one copy of essential genes but low enough to allow a diploid strain to survive. Consequently, we attempted to construct a diploid with the rad27 pol3-01 genotype. As outlined in Materials and Methods, we generated a diploid (RJK140-2) that was homozygous for the rad27 and pol3-01 mutations and that contained a plasmid-borne copy of the POL3 gene; the plasmid in this strain (pBL304) had URA3 as the selectable marker. In addition, we constructed an isogenic diploid (RJK153) with the same plasmid but heterozygous for the rad27 and pol3-01 mutations. When these diploids were plated on medium containing 5-FOA to select for the loss of pBL304, although the RJK153 strain produced 5-FOAR derivatives at a very high rate (>10−3/division), the RJK140-2 strain had a very low rate of 5-FOAR colonies (<10−6/division). When 10 independent cultures of RJK140-2 were examined on medium containing 5-FOA, only one culture yielded any 5-FOAR colonies. When one of these 5-FOAR colonies was examined by PCR analysis, we found that it had retained both POL3 and pol3-01 alleles, indicating that the 5FOAR strain had not lost the complementing POL3 gene. In summary, these results indicate that diploid cells homozygous for the rad27 and pol3-01 mutations are inviable.
We also examined the viability of spores derived from a diploid heterozygous for rad27 and pol2-4. The pol2-4 mutation eliminates the proofreading exonuclease activity of DNA polymerase ɛ (28), an essential enzyme required for the replication of chromosomal DNA. This mutant allele has no effect on the stability of a dinucleotide microsatellite (45). Of nine tetrads dissected, eight contained four viable spores, while the remaining tetrad had three viable spores. From four of these tetrads with four viable spores, the observed ratio of wild type/pol2-4/rad27/pol2-4 rad27 spores was 5:3:3:5. Thus, the requirement for the proofreading exonuclease for the viability of rad27 strains is specific to DNA polymerase δ.
DISCUSSION
Our results can be summarized by the following statements: (i) the rad27 mutation increases the frequency of additions more than that of deletions for all simple repetitive DNA sequences examined; (ii) the pol3-t mutation increases the frequency of deletions more than that of insertions for all repetitive DNA sequences examined except the mononucleotide microsatellite; (iii) for all simple repetitive DNA sequences examined, except the dinucleotide repeat, the rad27 pol3-t double mutant had approximately the same rate of instability as the pol3-t single mutant; (iv) the combination of rad27 and pol3-01 was synthetically lethal, although the rad27 pol2-4 combination was not; and (v) both rad27 and pol3-t destabilize a minisatellite as well as the microsatellites. Each of these conclusions will be discussed further below.
Microsatellite instability in rad27 strains.
Two previous studies reported that rad27 stimulates insertion mutations. Johnson et al. (18) showed that rad27 destabilized poly(GT) microsatellites about 100-fold and that the altered tracts had one-repeat unit insertions. Since strains with both the rad27 mutation and mutations affecting DNA mismatch repair had approximately the same microsatellite instability as the single-mutant strains, Johnson et al. suggested that the Rad27p nuclease had a role in DNA mismatch repair. Tishkoff et al. (49) found that forward mutations at the CAN1 locus in rad27 strains almost always represented duplications. The mutational spectrum observed for rad27 strains was different from that observed for msh2 strains (strains defective in DNA mismatch repair), indicating that Rad27p and Msh2p function in different pathways. In addition, they demonstrated that rad27 strains had elevated levels of mitotic recombination and that the rad27 mutation was synthetically lethal in combination with mutations affecting double-strand break repair. They suggested that replication errors in rad27 strains are repaired primarily by a nonmutagenic double-strand break repair pathway. The duplication mutations represented a fraction of replication errors corrected by a mutagenic recombination pathway or by DNA polymerase slippage events.
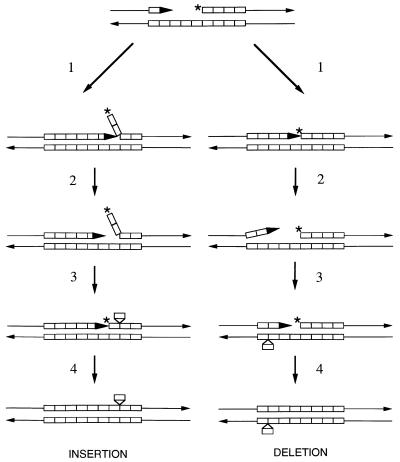
We suggest that the microsatellite instability observed in our experiments is likely to reflect DNA polymerase slippage errors rather than a mutagenic recombination pathway. As shown on the left side of Fig. 2, failure to remove the terminal ribonucleotide from the Okazaki fragment in a rad27 strain would be expected to cause a delay in the ligation of adjacent Okazaki fragments. Displacement of the end of the Okazaki fragment by continued synthesis from an adjacent primer would result in the formation of a DNA flap. We suggest that the displacement synthesis associated with flap formation activates the 3′-to-5′ exonuclease of DNA polymerase, producing a single-stranded gap adjacent to the flap. Reassociation of the flap DNA with this region could produce a DNA molecule with one or more unpaired repeats. Such displaced repeats on the primer strand, if unrepaired, would lead to additions. As shown on the right side of Fig. 2, the terminal nucleotide on the Okazaki fragment could also lead to a block of DNA synthesis and activation of the proofreading exonuclease without flap formation. If a secondary structure is formed on the resulting single-stranded region, subsequent synthesis across the gap would lead to a mismatched repeat on the template strand. This mismatch, if uncorrected, would result in a deletion. Thus, the model shown in Fig. 2 explains the increased frequencies of microsatellite additions and deletions observed in rad27 strains. This model is also consistent with the relatively small effect of rad27 on the rate of reversion of the lys2-InsLD and lys2-InsLE insertions (Table 4). Since additions are more frequent than deletions, we argue that the pathway indicated on the left side of the figure is preferred.
FIG. 2.
Generation of microsatellite insertions and deletions in rad27 strains. In this diagram, as in Fig. 1, individual lines depict single strands and rectangles represent single repeat units. The large arrow indicates the 3′ end of an elongating Okazaki fragment primer, and small arrows represent the 3′ termini of the DNA molecule. The asterisk shows an unexcised ribonucleotide at the 5′ end of an Okazaki fragment. The left and right sides of the figure show insertion and deletion pathways, respectively. From our analysis of microsatellite alterations in rad27 strains, we suggest that the left pathway is more frequently used than the right pathway. Insertion pathway: step 1, the elongating primer displaces the 5′ end of the adjacent Okazaki fragment; step 2, the 3′-to-5′ proofreading exonuclease of DNA polymerase δ is activated, resulting in a small gap; step 3, the displaced flap of the Okazaki fragment reanneals, leaving one displaced repeat; step 4, the ribonucleotide at the end of the Okazaki fragment is removed by an inefficient Rad27p-independent mechanism, and DNA synthesis is completed. Deletion pathway: step 1, Ligation of one Okazaki fragment to an adjacent fragment is blocked by the unprocessed ribonucleotide; step 2, the 3′-to-5′ exonuclease is activated, leading to a gap, and the 3′ end of the primer unpairs one or two repeats from the template strand; step 3, The primer strand reanneals to the template strand displaced by a single repeat unit; step 4, as in the insertion pathway, the ribonucleotide at the end of the Okazaki fragment is removed by an inefficient Rad27p-independent mechanism and DNA synthesis is completed.
There are a number of additional features that must be added to the model to explain all of the results. First, rad27 has its strongest destabilizing effects on repetitive DNA sequences that have small repeat units. This result could indicate that flap formation only involves a small number of base pairs or that the reassociation following dissociation of primer and template strands involved unpaired repeats more often when the repeat units were small than when they were large. Second, microsatellites are greatly destabilized by the msh2 mutation and other mutations affecting DNA mismatch repair (41, 45). If rad27 results in an increased frequency of displaced repeats, one might expect that rad27 msh2 double-mutant strains would have a greatly elevated level of microsatellite instability analogous to the synergistic effect of the pol3-t and mismatch repair mutations on the rate of frameshifts in short homonucleotide tracts (53). Instead, the double-mutant strains have a level of instability that is only slightly greater than those of the single mutants (18). We suggest that the displaced repeats formed in the rad27 strains are not efficiently recognized by the DNA mismatch repair system. One possibility is that the DNA mismatch repair system only functions efficiently during elongation of its associated primer. If the displaced repeat shown in Fig. 2 results from a postreplication slippage event, it could not be repaired. Third, we assume that, in the absence of Rad27p, the terminal ribonucleotide is removed by an inefficient processing system.
Although alterations in the lengths of microsatellites in rad27 strains are likely to reflect DNA slippage events, rad27 mutants also have elevated levels of mitotic recombination (49, 56). This elevation could reflect the processing of the DNA flap structure or gap structure shown in Fig. 2 into a recombinogenic double-strand break (as suggested in reference 49) or the formation of a double-strand break by replication of DNA with an unligated nick. Many yeast mutants that produce elevated levels of DNA lesions, such as cdc9 strains (deficient in DNA ligase) have high levels of mitotic recombination (37). The large insertions detected at the CAN1 locus by Tishkoff et al. (49) could represent either mutagenic processing of double-strand DNA breaks during recombination or long-range DNA polymerase slippage events. Recombination is unlikely to explain the microsatellite alterations observed in rad27 strains for two reasons. First, the rates of alterations for the mono- and dinucleotide microsatellites are about 50-fold higher than those observed for the large duplications seen at the CAN1 locus. Second, if the alterations were a consequence of recombination, one would expect a broad range of types of additions. For the mono- and dinucleotide repeats, we observe almost exclusively single-repeat additions.
Microsatellite instability in pol3-t strains.
In contrast to the microsatellite insertions observed in rad27 strains, the microsatellite alterations observed in pol3-t strains were usually deletions rather than additions (Table 3). An exception to this generalization is that approximately equal numbers of additions and deletions were found in the pol3-t strain with the mononucleotide assay plasmid. Although the different behavior of the mononucleotide may be significant, it should be pointed out that a substantial fraction (40%) of the 5-FOAR derivatives in the pol3-t strain with the mononucleotide assay plasmid did not have a sequence alteration in the tract. Thus, the estimate of the rate of tract instability for this strain would be expected to be less accurate than for strains in which almost all 5-FOAR derivatives have altered tracts.
Many of the deletions observed in pol3-t strains were greater than 14 bp. In addition, as observed in this study (Table 4) and other studies (8, 52, 53), the pol3-t mutation elevates the frequency of reversion of insertion mutations at the LYS2 locus. If the mutant DNA polymerase encoded by pol3-t substantially slowed the rate of DNA elongation on the lagging strand, then pol3-t strains would be expected to have longer single-stranded gaps (8). Such gaps would result in an increased probability of the types of DNA polymerase slippage events that lead to deletions caused by a mechanism related to that shown in Fig. 2 (right side of diagram).
One other feature of the data deserves further comment. The pol3-t mutation had a small (3- to 13-fold) destabilizing effect on the microsatellites and minisatellites compared to its effect on the reversion of the lys2-InsLD and lys2-InsLE insertions (150- to more than 750-fold). For microsatellites with small repeat units, we suggest that even the relatively small single-stranded regions on the template strand between neighboring Okazaki fragments are sufficient to generate deletions by DNA polymerase slippage. Thus, the difference in microsatellite instability between wild-type and pol3-t strains is expected to be small. In contrast, with the lys2 insertion mutations, large single-stranded regions (at least 38 bp for lys2-InsLD and 67 bp for lys2-InsLE) are necessary to allow the pairing of direct repeats necessary for the slippage event. Since these large single-stranded regions will be very rare in wild-type cells, but perhaps common in pol3-t strains, one will observe a very large effect of pol3-t. In addition, DNA mismatch repair is expected to have a different influence on the observed frequencies of addition and deletion for different types of repeats. Small unpaired loops resulting from DNA polymerase slippage in microsatellites are efficiently corrected by the DNA mismatch repair system (45), whereas large loops (expected in slippage events involving lys2-InsLD and lys2-InsLE) are not (53).
Microsatellite and minisatellite instability in rad27 pol3-t strains.
As discussed above, although rad27 and pol3-t mutations both destabilize simple repetitive DNA sequences, the rad27 mutation primarily increases insertion frequency and the pol3-t mutation primarily increases deletion frequency. If these two effects represent independent mechanisms, one would expect strains with both mutations to show additive effects on the rates of mutations and the types of changes. For the strains with 1-, 8-, or 20-bp repeats, the rates of instability and the types of changes approximate those found in pol3-t strains rather than those seen in rad27 strains (Tables 1 to 3). Thus, for repetitive DNA sequences with 1-, 8-, or 20-bp repeats, the increase in the rate of insertions caused by the rad27 mutation requires a wild-type DNA polymerase δ. In the context of the model shown in Fig. 2 (left side), one interpretation of this result is that the DNA polymerase slippage events leading to additions require the displacement of one Okazaki fragment by DNA synthesis from a neighboring DNA fragment. If DNA elongation is slowed as a consequence of the pol3-t mutation, one should see a reduced rate of rad27-induced insertion mutations.
Unlike the other repeats examined, the dinucleotide repeat in the double-mutant strain had about the same rate and types of alterations as those observed in the rad27 strain. To ensure that this result was not an artifact due to an additional mutation in the rad27 pol3-t strain, we retransformed an independently-derived rad27 pol3-t strain with the dinucleotide assay plasmid. No significant differences from our original results were found. Although the explanation of the different behavior of the dinucleotide repeat is not clear, one possibility is that the unusual Z-DNA structure of poly(GT) tracts (62) allows slippage of the ends of the Okazaki fragment without displacement by DNA polymerase. Alternatively, the reduction in the rate of DNA elongation by the pol3-t mutation could be sequence specific. Thus, if the rate of replication of poly(GT) tracts was only slightly reduced by this mutation, pol3-t would have little effect on dinucleotide stability (as observed; Table 1) and on the rate of insertions caused by rad27.
In summary, our results are best explained by the model in which the rad27 mutation increases the rate of insertions by slippage events that produce unpaired repeats on the primer strand and that the pol3-t mutation increases the rate of deletions by slippage events that produce unpaired repeats on the template strand. These same processes may be responsible for the microsatellite insertions and deletions that occur in wild-type cells. One difference between the models shown in Fig. 1 and 2 is that microsatellite additions and deletions occur by quite different mechanisms in the Fig. 2 model.
We previously reported that the instability of poly(GT) tracts increases exponentially with tract length and that the ratio of additions to deletions in microsatellites increases as a function of tract length (60). When poly(GT) tracts are 51 bp or smaller, both additions and deletions of single repeats are common; when tracts are 99 bp or larger, only additions of single repeats are observed. This altered ratio of additions to deletions is difficult to explain by the model shown in Fig. 1. If the probability of ending or starting an Okazaki fragment within a poly(GT) tract is much greater for long tracts than for short tracts, then one might observe enhanced levels of microsatellite additions relative to deletions by the model shown in Fig. 2. Finally, it should be pointed out that although some observations are most easily interpretable as reflecting slippage events as diagrammed in Fig. 2, slippage events as shown in Fig. 1 may also occur.
Synthetic lethality of rad27 and pol3-01 mutations.
The rad27 mutation was lethal in combination with a proofreading exonuclease mutant of DNA polymerase δ (pol3-01 mutant) but not in combination with a proofreading exonuclease mutant of DNA polymerase ɛ (pol2-4 mutant). We suggest three possible explanations of this result. First, the mutational load in the rad27 pol3-01 strain may be too high for viability. This possibility is unlikely because the homozygous diploid strain, which would be expected to be much more resistant to the effects of a high mutation rate, was also inviable. A second possibility is that DNA polymerase δ proofreading activity is required for the processing of Okazaki fragments in the absence of Rad27p. The most direct involvement (although perhaps the least likely) is that the proofreading exonuclease activity (a 3′-to-5′ DNA exonuclease) can remove the 5′ ribonucleotide from the Okazaki fragment. Alternatively, the involvement may be less direct. For example, as shown in Fig. 2 (right side), the failure to remove the 5′ ribonucleotide would result in a block to further DNA synthesis. This block could activate the 3′-to-5′ exonuclease, generating a short single-stranded gap. This structure could then act as a substrate for nucleases to excise the remaining 5′ ribonucleotide. It is interesting to note that a null mutation of the EXO1 gene, which encodes a double-strand-specific 5′-to-3′ exonuclease, is also synthetically lethal with rad27 (50), as is a temperature-sensitive allele of DNA2, which encodes a helicase (6). These proteins could be involved in the Rad27p-independent excision of the terminal ribonucleotide.
A final possibility to explain the rad27 pol3-01 synthetic lethality is that death results from aberrant processing of the rad27-caused DNA lesion. For example, in rad27 POL3 cells, activation of the proofreading exonuclease may prevent extensive flap formation. Extensive flaps formed in the double-mutant strain could be detected by checkpoint proteins, resulting in cell cycle arrest.
Strains with both rad27 and pol2-4, i.e., with the mutation eliminating the exonuclease domain of DNA polymerase ɛ, are viable. If DNA polymerase ɛ primarily functions on the leading strand during DNA replication (28), this result is consistent with the conclusion that POL3 may be involved with the processing of Okazaki fragments in rad27 strains, since Okazaki fragments are much more common on the lagging strand than on the leading strand.
Minisatellite instability in rad27 and pol3-t strains.
Minisatellite sequences in mammalian cells often represent tandem arrays between 0.5 and 30 kb in length in which each repeat unit is greater than 20 bp; sequence variation between repeat units within a single array is common (1). In general, these arrays exhibit high levels of meiotic instability and low levels of somatic instability. The patterns of alterations detected in the germ cells suggest that meiotic instability is primarily a consequence of meiotic recombination (17), whereas mitotic instability is likely to reflect either DNA replication slippage or unequal intragenic sister strand crossover (16). Although there are no previous studies linking minisatellite instability to specific mutations, enhanced somatic instability of minisatellites is found in some human tumor cell lines (2, 36). Our findings suggest the possibility that these lines might have mutations in genes encoding DNA polymerase or polymerase cofactors. Since our assay plasmid for minisatellite instability in yeast has only three repeats, it should be pointed out that the effects of rad27- or pol3-t-like mutations could be qualitatively or quantitatively different for longer minisatellites such as those commonly found in the human genome.
Since the types and rate of alterations observed for the minisatellite sequence are similar to those observed for the microsatellite, we suggest that the length alterations of the minisatellite also reflect DNA polymerase slippage events. We cannot rule out, however, the possibility that the length alterations reflect aberrant end joining of a double-strand DNA break (48) and/or unequal recombination events.
Patterns of microsatellite and minisatellite changes in mutant yeast strains.
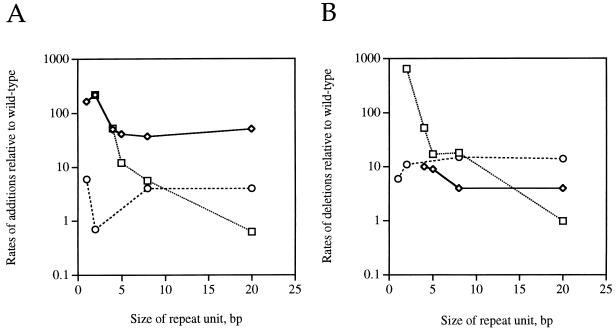
Yeast strains with mutations in genes affecting DNA mismatch repair (MSH2, MLH1, PMS1, MSH3, and MSH6) and DNA replication (RAD27, POL30, and POL3) have altered microsatellite stability. For many of these genes, the effects on the stability of repetitive DNA tracts with different repeat unit lengths have been examined. A summary of some of these data is shown in Fig. 3. The effects of the mutations on the rates of additions (Fig. 3A) and deletions (Fig. 3B) are given. It is clear that different mutants generate different spectra of changes. The only exception to this generalization is that the effects of msh2, mlh1, and pms1 on the rates of instability of all repeats were the same (41).
FIG. 3.
Instability of repetitive DNA sequences as a function of repeat unit length in msh2, rad27, and pol3-t strains. In this figure, we show the destabilizing effects of various mutations (normalized to a wild-type rate of one for each class of repeat units) as a function of the size of the repeat unit. The data in this figure are derived from Tables 1 and 3 for rad27 (diamonds) and pol3-t (circles) strains and from reference 41 for the msh2 (squares) strains. (A) Rates of insertions as a function of repeat unit length; (B) rates of deletions as a function of repeat unit length.
A substantial fraction of familial colorectal cancers are associated with microsatellite instability and mutations in human homologs of yeast mismatch repair genes MSH2, MLH1, and PMS1 (20). In contrast, about half of the sporadic cancers exhibiting microsatellite instability do not have a mutation in a known DNA mismatch repair gene (25). It would be useful to examine the stabilities of microsatellites of different repeat lengths in individual sporadic tumors in order to look for patterns matching those observed in yeast mutant strains. A match of patterns could help guide the search for the human genes mutated in the sporadic tumor. For example, if one found a cancer cell line that had a pattern of instability that mimicked the pol3-t pattern shown in Fig. 3, one could examine the cancer cell line for mutations in the human DNA polymerase δ gene.
ACKNOWLEDGMENTS
We thank E. Sia and D. Kirkpatrick for helpful discussions and comments, R. Pukkila-Worley for technical assistance, Natlalya Degtyareva for plasmids pLD-Int and pLE-Int, and P. Burgers for plasmid pBL304.
Our research was supported by NIH grants GM52319 (T.D.P.) and GM17879 (R.J.K.) and DOE Interagency Agreement DE-A105-94ER61940 (H.T.T., M.A.R., and D.A.G.).
REFERENCES
- 1.Armour J A L, Jeffreys A J. Biology and applications of human minisatellite loci. Curr Opin Genet Dev. 1992;2:850–857. doi: 10.1016/s0959-437x(05)80106-6. [DOI] [PubMed] [Google Scholar]
- 2.Armour J A L, Patel I, Thein S L, Fey M F, Jeffreys A J. Analysis of somatic mutations at human minisatellite loci in tumours and cell lines. Genomics. 1989;4:328–334. doi: 10.1016/0888-7543(89)90338-8. [DOI] [PubMed] [Google Scholar]
- 3.Bishop D K, Williamson M S, Fogel S, Kolodner R D. The role of heteroduplex correction in gene conversion in Saccharomyces cerevisiae. Nature. 1987;328:362–364. doi: 10.1038/328362a0. [DOI] [PubMed] [Google Scholar]
- 4.Boeke J D, Lacroute F, Fink G R. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoroorotic acid resistance. Mol Gen Genet. 1984;167:345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- 5.Bonneaud N, Ozier-Kalogeropoulos O, Li G Y, Labouesse M, Minvielle-Sebastia L, Lacroute F. A family of low and high copy replicative, integrative and single-stranded S. cerevisiae/E. coli shuttle vectors. Yeast. 1991;7:609–615. doi: 10.1002/yea.320070609. [DOI] [PubMed] [Google Scholar]
- 6.Budd M E, Campbell J L. A yeast replicative helicase, Dna2 helicase, interacts with yeast FEN-1 nuclease in carrying out its essential function. Mol Cell Biol. 1997;17:2136–2142. doi: 10.1128/mcb.17.4.2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gordenin D A, Proscyavichus Y Y, Malkova A L, Trofimova M V, Peterzen A. Yeast mutants with increased bacterial transposon Tn5 excision. Yeast. 1991;7:37–50. doi: 10.1002/yea.320070105. [DOI] [PubMed] [Google Scholar]
- 8.Gordenin D A, Malkova A L, Peterzen A, Kulikov V N, Pavlov Y I, Perkins E, Resnick M A. Transposon Tn5 excision in yeast: influence of DNA polymerases α, δ, and ɛ and repair genes. Proc Natl Acad Sci USA. 1992;89:3785–3789. doi: 10.1073/pnas.89.9.3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goulian M, Richards S H, Heard C J, Bigsby B M. Discontinuous DNA synthesis by purified mammalian proteins. J Biol Chem. 1990;265:18461–18471. [PubMed] [Google Scholar]
- 10.Harrington J J, Lieber M R. The characterization of a mammalian DNA structure-specific endonuclease. EMBO J. 1994;13:1235–1246. doi: 10.1002/j.1460-2075.1994.tb06373.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harrington J J, Lieber M R. Functional domains within FEN-1 and RAD2 define a family of structure-specific endonucleases: implications for nucleotide excision repair. Genes Dev. 1994;8:1344–1355. doi: 10.1101/gad.8.11.1344. [DOI] [PubMed] [Google Scholar]
- 12.Henderson S T, Petes T D. Instability of simple sequence DNA in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:2749–2757. doi: 10.1128/mcb.12.6.2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang L, Rumbaugh J A, Murante R S, Lin R J R, Rust L, Bambara R A. Role of calf RTH-1 nuclease in removal of 5′-ribonucleotides during Okazaki fragment processing. Biochemistry. 1996;35:9266–9277. doi: 10.1021/bi9603074. [DOI] [PubMed] [Google Scholar]
- 14.Ishimi Y, Claude A, Bullock P, Hurwitz J. Complete enzymatic synthesis of DNA containing the SV40 origin of replication. J Biol Chem. 1988;263:19723–19733. [PubMed] [Google Scholar]
- 15.Jeffreys A J, Allen M J, Armour J A, Collick A, Dubrova Y, Fretwell N, Guram T, Jobling M, May C A, Neil D L, Neumann R. Mutation processes at human minisatellites. Electrophoresis. 1995;16:1577–1585. doi: 10.1002/elps.11501601261. [DOI] [PubMed] [Google Scholar]
- 16.Jeffreys A J, Neumann R. Somatic mutation processes at a human minisatellite. Hum Mol Genet. 1997;6:129–136. doi: 10.1093/hmg/6.1.129. [DOI] [PubMed] [Google Scholar]
- 17.Jeffreys A J, Tamaki K, MacLeod A, Monckton D G, Neil D L, Armour J A L. Complex gene conversion events in germline mutation at human minisatellites. Nat Genet. 1994;6:136–145. doi: 10.1038/ng0294-136. [DOI] [PubMed] [Google Scholar]
- 18.Johnson R E, Kovvali G K, Prakash L, Prakash S. Requirement of the yeast RTH1 5′ to 3′ exonuclease for the stability of simple repetitive DNA. Science. 1995;269:238–240. doi: 10.1126/science.7618086. [DOI] [PubMed] [Google Scholar]
- 19.Kirkpatrick D T, Petes T D. Repair of DNA loops involves DNA-mismatch and nucleotide-excision repair proteins. Nature. 1997;387:929–931. doi: 10.1038/43225. [DOI] [PubMed] [Google Scholar]
- 20.Kolodner R D. Mismatch repair: mechanisms and relationship to cancer susceptibility. Trends Biochem Sci. 1995;20:397–402. doi: 10.1016/s0968-0004(00)89087-8. [DOI] [PubMed] [Google Scholar]
- 21.Kunkel T A. Biological asymmetries and the fidelity of eukaryotic DNA replication. Bioessays. 1992;14:303–308. doi: 10.1002/bies.950140503. [DOI] [PubMed] [Google Scholar]
- 22.Kunkel T A, Resnick M A, Gordenin D A. Mutator specificity and disease: looking over the FENce. Cell. 1997;88:155–158. doi: 10.1016/s0092-8674(00)81832-2. [DOI] [PubMed] [Google Scholar]
- 23.Lea D E, Coulson C A. The distribution of the number of mutants in bacterial populations. J Genet. 1949;49:264–285. doi: 10.1007/BF02986080. [DOI] [PubMed] [Google Scholar]
- 24.Levinson G, Gutman G A. Slipped-strand mispairing: a major mechanism for DNA sequence evolution. Mol Biol Evol. 1987;4:203–221. doi: 10.1093/oxfordjournals.molbev.a040442. [DOI] [PubMed] [Google Scholar]
- 25.Liu B, Nicolaides M C, Markowitz S, Wilson J K V, Parsons J R, Jen J, Papadopolous N, Peltomaki P, de la Chapelle A, Hamilton S R, et al. Mismatch repair gene defects in sporadic colorectal cancers with microsatellite instability. Nat Genet. 1995;9:48–55. doi: 10.1038/ng0195-48. [DOI] [PubMed] [Google Scholar]
- 26.Markowitz S, Wang J, Myeroff L, Parsons R, Sun L, Lutterbaugh J, Fan R S, Zborowska E, Kinzler K W, Vogelstein B, Brattain M, Wilson J K V. Inactivation of the type II TGF-β receptor in colon cancer cells with microsatellite instability. Science. 1995;268:1336–1338. doi: 10.1126/science.7761852. [DOI] [PubMed] [Google Scholar]
- 27.Modrich P, Lahue R. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu Rev Biochem. 1996;65:101–133. doi: 10.1146/annurev.bi.65.070196.000533. [DOI] [PubMed] [Google Scholar]
- 28.Morrison A, Araki H, Clark A B, Hamatake R K, Sugino A. A third essential DNA polymerase in S. cerevisiae. Cell. 1990;62:1143–1151. doi: 10.1016/0092-8674(90)90391-q. [DOI] [PubMed] [Google Scholar]
- 29.Morrison A, Bell J B, Kunkel T A, Sugino A. Eukaryotic DNA polymerase amino acid sequence required for 3′—5′ exonuclease activity. Proc Natl Acad Sci USA. 1991;88:9473–9477. doi: 10.1073/pnas.88.21.9473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morrison A, Johnson A L, Johnston L H, Sugino A. Pathway correcting DNA replication errors in Saccharomyces cerevisiae. EMBO J. 1993;12:1467–1473. doi: 10.1002/j.1460-2075.1993.tb05790.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morrison A, Sugino A. Roles of POL3, POL2 and PMS1 in maintaining accurate DNA replication. Chromosoma. 1992;102:S147–S149. doi: 10.1007/BF02451799. [DOI] [PubMed] [Google Scholar]
- 32.Morrison A, Sugino A. The 3′ to 5′ exonucleases of both DNA polymerases δ and ɛ participate in correcting errors of DNA replication in Saccharomyces cerevisiae. Mol Gen Genet. 1994;242:289–296. doi: 10.1007/BF00280418. [DOI] [PubMed] [Google Scholar]
- 33.Murante R S, Huang L, Turchi J J, Bambara R A. The calf 5′- to 3′-exonuclease is also an endonuclease with both activities dependent on primers annealed upstream of the point of cleavage. J Biol Chem. 1994;269:1191–1196. [PubMed] [Google Scholar]
- 34.Murante R S, Rust L, Bambara R A. Calf 5′ to 3′ exo/endonuclease must slide from a 5′ end of the substrate to perform structure-specific cleavage. J Biol Chem. 1995;270:30377–30383. doi: 10.1074/jbc.270.51.30377. [DOI] [PubMed] [Google Scholar]
- 35.Murante R S, Rumbaugh J A, Barnes C J, Norton J R, Bambara R A. Calf RTH-1 nuclease can remove the initiator RNAs of Okazaki fragments by endonuclease activity. J Biol Chem. 1996;271:25888–25897. doi: 10.1074/jbc.271.42.25888. [DOI] [PubMed] [Google Scholar]
- 36.Nagel S, Borisch B, Thein S L, Oestreicher M, Nothiger F, Birrer S, Tobler A, Fey M F. Somatic mutations detected by minisatellite and microsatellite DNA markers reveal clonal intratumor heterogeneity in gastrointestinal cancers. Cancer Res. 1995;55:2866–2870. [PubMed] [Google Scholar]
- 37.Petes T D, Malone R E, Symington L S. Recombination in yeast. In: Broach J, Jones E, Pringle J, editors. The molecular and cellular biology of the yeast Saccharomyces: genome dynamics, protein synthesis, and energetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1991. pp. 407–521. [Google Scholar]
- 38.Reagan M S, Pittenger C, Siede W, Friedberg E C. Characterization of a mutant strain of Saccharomyces cerevisiae with a deletion of the RAD27 gene, a structural homolog of the RAD2 nucleotide excision repair gene. J Bacteriol. 1995;177:364–371. doi: 10.1128/jb.177.2.364-371.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schweitzer J K, Livingston D M. Expansions of CAG repeat tracts are frequent in a yeast mutant defective in Okazaki fragment maturation. Hum Mol Genet. 1998;7:69–74. doi: 10.1093/hmg/7.1.69. [DOI] [PubMed] [Google Scholar]
- 40.Sia E A, Jinks-Robertson S, Petes T D. Genetic control of microsatellite stability. Mutat Res. 1997;383:61–70. doi: 10.1016/s0921-8777(96)00046-8. [DOI] [PubMed] [Google Scholar]
- 41.Sia E A, Kokoska R J, Dominska M, Greenwell P, Petes T D. Microsatellite instability in yeast: dependence on repeat unit size and DNA mismatch repair genes. Mol Cell Biol. 1997;17:2851–2858. doi: 10.1128/mcb.17.5.2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Simon M, Giot L, Faye G. The 3′ to 5′ exonuclease activity located in the DNA polymerase δ subunit of Saccharomyces cerevisiae is required for accurate replication. EMBO J. 1991;10:2165–2170. doi: 10.1002/j.1460-2075.1991.tb07751.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sommers C H, Miller E J, Dujon B, Prakash S, Prakash L. Conditional lethality of null mutations in RTH1 that encodes the yeast counterpart of a mammalian 5′- to 3′-exonuclease required for lagging strand DNA synthesis in reconstituted systems. J Biol Chem. 1995;270:4193–4196. doi: 10.1074/jbc.270.9.4193. [DOI] [PubMed] [Google Scholar]
- 44.Strand M, Earley M C, Crouse G F, Petes T D. Mutations in the MSH3 gene preferentially lead to deletions within tracts of simple repetitive DNA in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1995;92:10418–10421. doi: 10.1073/pnas.92.22.10418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Strand M, Prolla T A, Liskay R M, Petes T D. Destabilization of tracts of simple repetitive DNA in yeast by mutations affecting DNA mismatch repair. Nature. 1993;365:274–276. doi: 10.1038/365274a0. [DOI] [PubMed] [Google Scholar]
- 46.Strauss B S, Sagher D, Acharya S. Role of proofreading and mismatch repair in maintaining the stability of nucleotide repeats in DNA. Nucleic Acids Res. 1997;25:806–813. doi: 10.1093/nar/25.4.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Streisinger G, Okada Y, Emrich J, Newton J, Tsugita A, Terzaghi E, Inouye M. Frameshift mutations and the genetic code. Cold Spring Harbor Symp Quant Biol. 1966;31:77–84. doi: 10.1101/sqb.1966.031.01.014. [DOI] [PubMed] [Google Scholar]
- 48.Tautz D, Schlotterer C. Simple sequences. Curr Opin Genet Dev. 1994;4:832–837. doi: 10.1016/0959-437x(94)90067-1. [DOI] [PubMed] [Google Scholar]
- 49.Tishkoff D X, Filosi N, Gaida G M, Kolodner R D. A novel mutation avoidance mechanism dependent on S. cerevisiae RAD27 is distinct from DNA mismatch repair. Cell. 1997;88:253–263. doi: 10.1016/s0092-8674(00)81846-2. [DOI] [PubMed] [Google Scholar]
- 50.Tishkoff D X, Boerger A L, Bertrand P, Filosi N, Gaida G M, Kane M F, Kolodner R D. Identification and characterization of Saccharomyces cerevisiae EXO1, a gene encoding an exonuclease that interacts with MSH2. Proc Natl Acad Sci USA. 1997;94:7487–7492. doi: 10.1073/pnas.94.14.7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tran H, Degtyareva N, Gordenin D, Resnick M A. Altered replication and inverted repeats induce mismatch repair-independent recombination between highly diverged DNAs in yeast. Mol Cell Biol. 1997;17:1027–1036. doi: 10.1128/mcb.17.2.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tran H T, Degtyareva N P, Koloteva N N, Sugino A, Masumoto H, Gordenin D A, Resnick M A. Replication slippage between distant short repeats in Saccharomyces cerevisiae depends on the direction of replication and the RAD50 and RAD52 genes. Mol Cell Biol. 1995;15:5607–5617. doi: 10.1128/mcb.15.10.5607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tran H T, Gordenin D A, Resnick M A. The prevention of repeat-associated deletions in Saccharomyces cerevisiae by mismatch repair depends on size and origin of deletions. Genetics. 1996;143:1579–1587. doi: 10.1093/genetics/143.4.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Turchi J J, Bambara R A. Completion of mammalian lagging strand DNA replication using purified proteins. J Biol Chem. 1993;268:15136–15141. [PubMed] [Google Scholar]
- 55.Turchi J J, Huang L, Murante R S, Kim Y, Bambara R A. Enzymatic completion of mammalian lagging-strand DNA replication. Proc Natl Acad Sci USA. 1994;91:9803–9807. doi: 10.1073/pnas.91.21.9803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vallen E A, Cross F R. Mutations in RAD27 define a potential link between G1 cyclins and DNA replication. Mol Cell Biol. 1995;15:4291–4302. doi: 10.1128/mcb.15.8.4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Waga S, Bauer G, Stillman B. Reconstitution of complete SV40 DNA replication with purified replication factors. J Biol Chem. 1994;269:10923–10934. [PubMed] [Google Scholar]
- 58.Wang T S-F. Cellular DNA polymerases. In: DePamphilis M L, editor. DNA replication in eukaryotic cells. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 461–493. [Google Scholar]
- 59.Weber J L, Wong C. Mutation of human short tandem repeats. Hum Mol Genet. 1993;2:1123–1128. doi: 10.1093/hmg/2.8.1123. [DOI] [PubMed] [Google Scholar]
- 60.Wierdl M, Dominska M, Petes T D. Microsatellite instability in yeast: dependence on the length of the microsatellite. Genetics. 1997;146:769–779. doi: 10.1093/genetics/146.3.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wierdl M, Greene C N, Datta A, Jinks-Robertson S, Petes T D. Destabilization of simple repetitive DNA sequences by transcription in yeast. Genetics. 1996;143:713–721. doi: 10.1093/genetics/143.2.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zimmerman S B A. The three-dimensional structure of DNA. Annu Rev Biochem. 1982;51:395–427. doi: 10.1146/annurev.bi.51.070182.002143. [DOI] [PubMed] [Google Scholar]