Abstract
Mutations in the SEC238 and SRP54 genes of the yeast Yarrowia lipolytica not only cause temperature-sensitive defects in the exit of the precursor form of alkaline extracellular protease and of other secretory proteins from the endoplasmic reticulum and in protein secretion but also lead to temperature-sensitive growth in oleic acid-containing medium, the metabolism of which requires the assembly of functionally intact peroxisomes. The sec238A and srp54KO mutations at the restrictive temperature significantly reduce the size and number of peroxisomes, affect the import of peroxisomal matrix and membrane proteins into the organelle, and significantly delay, but do not prevent, the exit of two peroxisomal membrane proteins, Pex2p and Pex16p, from the endoplasmic reticulum en route to the peroxisomal membrane. Mutations in the PEX1 and PEX6 genes, which encode members of the AAA family of N-ethylmaleimide-sensitive fusion protein-like ATPases, not only affect the exit of precursor forms of secretory proteins from the endoplasmic reticulum but also prevent the exit of the peroxisomal membrane proteins Pex2p and Pex16p from the endoplasmic reticulum and cause the accumulation of an extensive network of endoplasmic reticulum membranes. None of the peroxisomal matrix proteins tested associated with the endoplasmic reticulum in sec238A, srp54KO, pex1-1, and pex6KO mutant cells. Our data provide evidence that the endoplasmic reticulum is required for peroxisome biogenesis and suggest that in Y. lipolytica, the trafficking of some membrane proteins, but not matrix proteins, to the peroxisome occurs via the endoplasmic reticulum, results in their glycosylation within the lumen of the endoplasmic reticulum, does not involve transport through the Golgi, and requires the products encoded by the SEC238, SRP54, PEX1, and PEX6 genes.
One of the hallmarks of eukaryotic cells is the coexistence of functionally distinct subcellular organelles (compartments), with each organelle possessing a specific set of enzymes required for its particular metabolic role. One organelle, the peroxisome, is present in most eukaryotic cells (22). Peroxisomes compartmentalize more than 50 enzymes involved in different metabolic functions, including the β-oxidation of fatty acids and the decomposition of H2O2 by catalase (42, 49). The importance of peroxisomes for normal human development and physiology is demonstrated by the lethality of various peroxisome biogenesis disorders (23).
Changes in the abundance and composition of peroxisomes in response to changes in environmental conditions must be coordinated with the biogenesis and functioning of other organelles in order to achieve an overall balance in cellular function. An example of such interorganellar communication is the tripartite path of communication among mitochondria, the nucleus, and peroxisomes, which regulates the expression of genes encoding peroxisomal proteins (6, 32). Some peroxisomal proteins may also play an important role in the biogenesis of other organelles. The peroxisome-associated protein Car1p has been shown to be essential for karyogamy in the filamentous fungus Podospora anserina (3). A functional relationship between peroxisome biogenesis and a specific process in cell morphogenesis, i.e., the dimorphic transition from the yeast to the mycelial form, has also been demonstrated recently in the yeast Yarrowia lipolytica (46).
While the role of the endoplasmic reticulum (ER) as the entry point for all compartments of the secretory and endocytic pathways is well established (27, 36, 41), the significance of the ER for peroxisome biogenesis remains unclear. Recent data have suggested a dual role for the ER in peroxisome biogenesis in supplying phospholipid for the formation of the peroxisomal membrane (45) and in protein trafficking to peroxisomes (2, 4, 13, 50, 51). We have applied a combined genetic, biochemical, and morphological approach to study the importance of the ER for peroxisome biogenesis in Y. lipolytica. An analysis of Y. lipolytica sec238A, srp54KO, pex1-1, and pex6KO mutants that are deficient in the exit of secretory proteins from the ER, in protein secretion, and in peroxisome biogenesis has provided evidence for an essential role for the ER in the assembly of peroxisomes.
MATERIALS AND METHODS
Yeast strains and microbial techniques.
The Y. lipolytica strains used in this study are listed in Table 1. The new nomenclature for peroxisome assembly genes and proteins has been used (8). Media, growth conditions, and genetic techniques for Y. lipolytica have been described (33, 35, 43). Medium components were as follows. YEPD contains 1% yeast extract, 2% peptone, and 2% glucose. 2×-YEPD contains 2% yeast extract, 4% peptone, and 4% glucose. YPBO contains 0.3% yeast extract, 0.5% peptone, 0.5% K2HPO4, 0.5% KH2PO4, 1% Brij 35, and 1% (wt/vol) oleic acid. 2×-YPBO contains 0.6% yeast extract, 1% peptone, 1% K2HPO4, 1% KH2PO4, 2% Brij 35, and 2% (wt/vol) oleic acid. YND contains 0.67% yeast nitrogen base without amino acids and 2% glucose. YNO contains 0.67% yeast nitrogen base without amino acids, 0.05% (vol/vol) Tween 40, and 0.1% (wt/vol) oleic acid. YND and YNO were supplemented with adenine, leucine, histidine, and lysine, each at 50 μg/ml, as required.
TABLE 1.
Y. lipolytica strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| E122 | MATA ura3-302 leu2-270 lys8-11 | 15 |
| DX547-1A | MATA ade1 leu2-270 | 19 |
| sec238A mutant | MATA ade1 sec238A | 50 |
| srp54KO mutant | MATA ade1 leu2-270 srp54::ADE1 | 26 |
| pex1-1 mutant | MATA ura3-302 leu2-270 lys8-11 pex1-1 | 35 |
| pex2KO mutant | MATA ura3-302 leu2-270 lys8-11 pex2::LEU2 | 13 |
| pex6KO mutant | MATA ura3-302 leu2-270 lys8-11 pex6::LEU2 | 34 |
| pex9KO mutant | MATA ura3-302 leu2-270 lys8-11 pex9::LEU2 | 10 |
Electron and immunofluorescence microscopy.
Electron microscopy (16) and double-labeling, indirect immunofluorescence microscopy (43) were performed as previously described.
Subcellular fractionation.
The initial step in the subcellular fractionation of YPBO-grown cells was performed as described previously (43) and included the differential centrifugation of lysed and homogenized spheroplasts at 1,000 × gmax for 8 min at 4°C in a Beckman JS13.1 rotor to yield a postnuclear supernatant (PNS) fraction. The PNS fraction was further subjected to differential centrifugation at 20,000 × gmax for 30 min at 4°C in a Beckman JS13.1 rotor to yield pellet (20KgP) and supernatant (20KgS) fractions. The 20KgS fraction was further subfractionated by differential centrifugation at 245,000 × gmax for 1 h at 4°C in a Beckman TLA120.2 rotor to yield pellet (245KgP) and supernatant (245KgS) fractions. Protease protection analysis of different subcellular fractions was performed as previously described (45). Separation of the organelles, i.e., peroxisomes, plasma membrane, mitochondria, ER, Golgi, and vacuoles, present in the 20KgP fraction of YPBO-grown cells was performed by isopycnic centrifugation on a discontinuous sucrose (25, 35, 42, and 53%, wt/wt) gradient in a Beckman VTi50 rotor at 100,000 × gmax for 1 h at 4°C.
Purification of peroxisomes (43, 45), ER (46), and plasma membranes (46) and in vitro disassembly of the complex formed by peroxisomes and the ER in mutant strains (45) have already been described. Peroxisomes and the ER were more than 97% pure, as determined by marker protein analyses. Contamination of purified peroxisomes by ER elements and vice versa was less than 0.5%.
Radiolabeling, immunoprecipitation, and endo H digestion.
Yeast cultures were grown in YPBO or YEPD at the temperatures indicated in Results. Aliquots of cells were sedimented in a clinical centrifuge, resuspended at a concentration of 2 U of optical density at 600 nm (OD600)/ml in YNO (YPBO grown) or YND (YEPD grown) medium, and incubated at the temperatures indicated for 30 min. Radiolabeling was performed in the same medium containing l-[35S]methionine (ICN Biomedicals, Mississauga, Ontario, Canada) at a concentration of 40 μCi/OD600 U for 3 min (in YNO) or 1.5 min (in YND) at the temperatures indicated and chased with an equal volume of 2×-YPBO or 2×-YEPD, respectively, supplemented with 10 mM l-methionine. Samples were taken at the postchase times indicated in Results. Reactions were terminated by the addition of an equal volume of ice-cold 20 mM NaN3, and cells were immediately separated from the culture supernatant by centrifugation in a microcentrifuge at 20,000 × gmax for 3 min at 4°C. Immunoprecipitation of pulse-labeled Kar2p and Sec14p from fractions of discontinuous sucrose gradients was performed as previously described (52). Digestion of immunoprecipitated, pulse-labeled Pex2p, Pex16p, thiolase (THI), acyl coenzyme A oxidase (AOX), and isocitrate lyase (ICL) with endoglycosidase H (endo H) was performed as previously described (26). Samples were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Gels were treated with 22.2% 2,5-diphenyloxazole in either dimethyl sulfoxide or glacial acetic acid (7), dried, and exposed to preflashed Kodak X-Omat AR X-ray film at −80°C with intensifying screens.
Antibodies.
Guinea pig polyclonal antibodies to Y. lipolytica ICL, THI, Pex2p, Pex5p, and Pex16p and to Saccharomyces cerevisiae AOX and rabbit polyclonal anti-SKL antibodies have already been described (11, 12, 43). Rabbit polyclonal antibodies to S. cerevisiae malate synthase (MLS) (12) and to Y. lipolytica alkaline extracellular protease (AEP) (26), Sls1p (5), Kar2p (46), and Sec14p (25) were described previously. Anti-MLS antibodies were kindly provided by Andreas Hartig (Institute of Biochemistry and Molecular Cell Biology, Vienna, Austria). Anti-AEP and anti-Kar2p antibodies were generous gifts of David Ogrydziak (University of California, Davis). Anti-Sls1p and anti-Sec14p antibodies were generous gifts of Claude Gaillardin (Institut National Agronomique Paris-Grignon, Thiveral-Grignon, France). Mouse monoclonal antibody SPA-827 specific for grp78 (BiP) was from StressGen Biotechnologies (Victoria, British Columbia, Canada).
Analytical procedures.
Enzymatic activities of catalase, cytochrome c oxidase (43), NADPH:cytochrome c reductase, α-mannosidase, vanadate-sensitive plasma membrane ATPase (39), guanosine diphosphatase (1), and alkaline phosphatase (44) were determined by established methods. Inorganic phosphate liberated in assays for the activities of guanosine diphosphatase and vanadate-sensitive plasma membrane ATPase was measured as previously described (21). SDS-PAGE (20) and immunoblotting using a semidry electrophoretic transfer system (ET-20; Tyler Research Instruments, Edmonton, Alberta, Canada) (19) were performed as previously described. Antigen-antibody complexes were detected by enhanced chemiluminescence (Amersham Life Sciences, Oakville, Ontario, Canada). Quantitation of immunoblots was performed as previously described (43).
RESULTS
The sec238A and srp54KO mutations cause temperature-sensitive growth in oleic acid-containing medium.
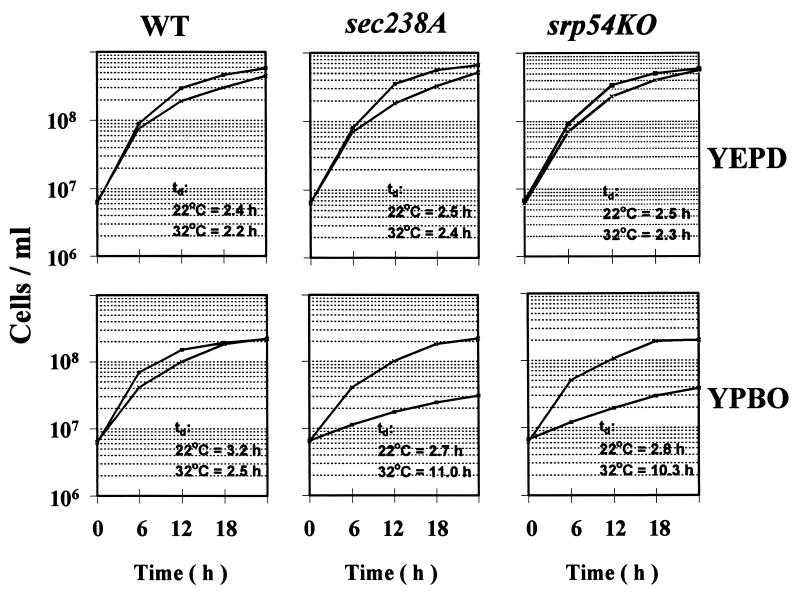
A characteristic feature of Y. lipolytica is its extensive peroxisome proliferation during growth in oleic acid-containing medium. The assembly of functionally intact peroxisomes is absolutely required for growth in media containing oleic acid as the sole carbon source (33). In contrast, growth in glucose-containing medium does not require intact peroxisomes (33). We have previously demonstrated that sec238A and srp54KO mutants grown in glucose-containing YEPD medium show temperature-sensitive defects in protein secretion (46). To study the possible effects of the sec238A and srp54KO mutations on peroxisome biogenesis, we first tested the ability of sec238A and srp54KO mutants to grow in oleic acid-containing YPBO medium at either 22 or 32°C (Fig. 1). Both strains were affected in growth in YPBO only at 32°C, with doubling times of 11.0 and 10.3 h for the sec238A and srp54KO mutants, respectively, compared to 2.5 h for the isogenic wild-type strain. No effect of either the sec238A or the srp54KO mutation on growth in YPBO at 22°C was observed. Neither the sec238A nor the srp54KO mutation affected growth in YEPD medium at either 22 or 32°C.
FIG. 1.
The sec238A and srp54KO mutations cause temperature-sensitive growth in YPBO but do not affect growth in YEPD. Growth curves and doubling times (td) of wild-type (WT) strain DX547-1A and of the sec238A and srp54KO mutants grown in YEPD or YPBO at 22°C (×) or 32°C (▪). Cells were pregrown twice in YEPD at 22 or 32°C until an OD600 of 1.8 to 2.0 (cell titer, ∼1.1 × 108 to 1.3 × 108 cells/ml) and inoculated into YEPD or YPBO at an initial OD600 of 0.1 (cell titer, ∼6.1 × 106 to 6.5 × 106 cells/ml).
The sec238A and srp54KO mutations significantly reduce the size and number of peroxisomes at the restrictive temperature.
We have previously reported (46) that the sec238A and srp54KO mutations affect protein secretion and exit of the precursor form (pAEP) of AEP, a major secretory protein of Y. lipolytica (14, 17, 26), from the ER in YEPD medium at the restrictive temperature of 32°C (46). Similar results were observed for the sec238A and srp54KO mutations on a carbon source, oleic acid, which can be metabolized only by functionally intact peroxisomes (data not shown). These results, combined with the observation that the sec238A and srp54KO mutant strains are retarded in their growth in YPBO medium at 32°C (Fig. 1), suggested to us that these two temperature-sensitive mutations not only compromise the exit of secretory proteins from the ER and their delivery to the extracellular medium but also might affect some aspect(s) of peroxisome biogenesis.
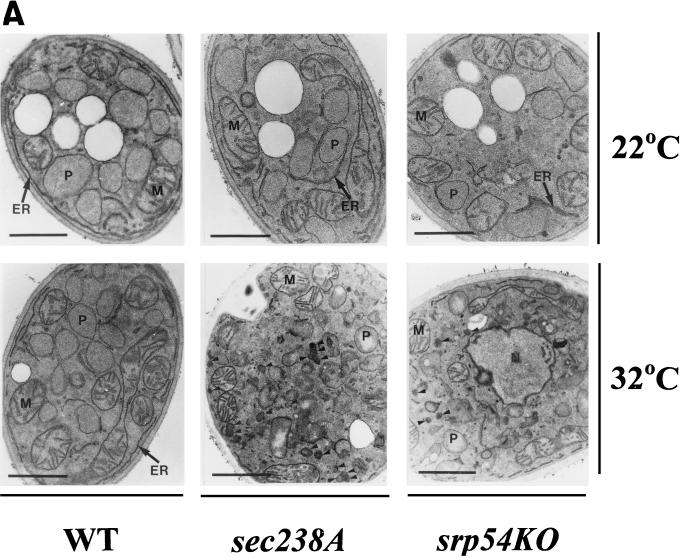
Electron microscopical analysis showed no significant differences in peroxisome size and number between the wild-type and mutant strains grown in YPBO at 22°C, the temperature permissive for growth and protein secretion (Fig. 2A). However, the size and number of peroxisomes in sec238A and srp54KO mutant cells at the restrictive temperature, 32°C, were significantly reduced compared to those of wild-type cell peroxisomes (Fig. 2A). The sec238A and srp54KO mutant cells also accumulated 100- to 120-nm vesicles (Fig. 2A, arrowheads) at the restrictive temperature. These vesicles were rarely observed in wild-type cells (Fig. 2A). Moreover, in contrast to the peroxisomes in wild-type cells, peroxisomes in sec238A and srp54KO mutant cells at either 22 or 32°C were closely apposed to ER membranes (Fig. 2A).
FIG. 2.
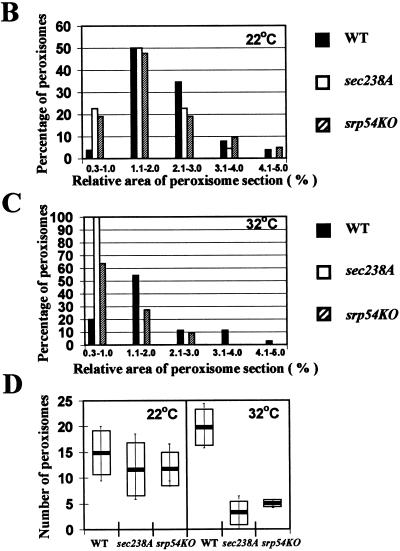
The sec238A and srp54KO mutations significantly reduce the size and number of peroxisomes at the restrictive temperature. (A) Cells were grown for 9 h in YPBO at either 22 or 32°C, fixed in KMnO4, and processed for electron microscopy. Abbreviations: P, peroxisome; M, mitochondrion; N, nucleus; WT, wild type. Arrowheads, 100- to 120-nm vesicles accumulating in sec238A and srp54KO mutant cells at 32°C. Bars, 1 μm. (B to D) Results of morphometric analyses performed on random electron microscopic sections of cells of wild-type and mutant strains grown in YPBO at either 22 or 32°C. (B and C) Percentages of peroxisomes having the indicated relative areas of peroxisome section. The relative area of peroxisome section is calculated as the area of peroxisome section/area of cell section × 100. (D) Numbers of peroxisomes in wild-type and mutant strains. Peroxisomes were counted in electron micrographs, and the data were expressed as the number of peroxisomes per cubic micrometer of cell section volume. The bar in the middle of each box indicates the mean peroxisome number. The limits of each box indicate the sample standard deviation on each side of the mean. The lines extend to indicate the extremes of the distribution.
Morphometric analysis of random cell sections of the wild-type and mutant strains grown in YPBO at either 22 or 32°C showed that the sec238A and srp54KO mutations significantly reduced the size of peroxisomes at the restrictive temperature compared to the wild-type condition. In sec238A and srp54KO cells, there was an accumulation of exclusively (sec238A) or mostly (srp54KO) small peroxisomes at the restrictive temperature of 32°C, with their relative areas of peroxisome section ranging from 0.3 to 1.0% (Fig. 2C). In wild-type cells at 32°C, more than half (54.7%) of all peroxisomes had relative areas of peroxisome section ranging from 1.1 to 2.0%, while 25.7% of all peroxisomes had relative areas of peroxisome section ranging from 2.1 to 5.0% (Fig. 2C). The effect of either mutation on peroxisome size was much less pronounced at the permissive temperature (Fig. 2B).
The sec238A and srp54KO mutations also showed a four- to fivefold decrease in the number of peroxisomes per cell at the restrictive temperature (3.18 ± 2.45 and 4.9 ± 0.83 peroxisomes per μm3 of cell section volume, respectively) compared to the number of peroxisomes in wild-type cells (19.84 ± 3.53 peroxisomes per μm3 of cell section volume) (Fig. 2D). The effect of either mutation on the number of peroxisomes per cell was much less pronounced at the permissive temperature (14.88 ± 4.34, 11.53 ± 5.24, and 11.56 ± 3.36 peroxisomes per μm3 of cell section volume for the wild-type, sec238A, and srp54KO strains, respectively) (Fig. 2D).
The sec238A and srp54KO mutations affect the subcellular localization of peroxisomal matrix and membrane proteins at the restrictive temperature.
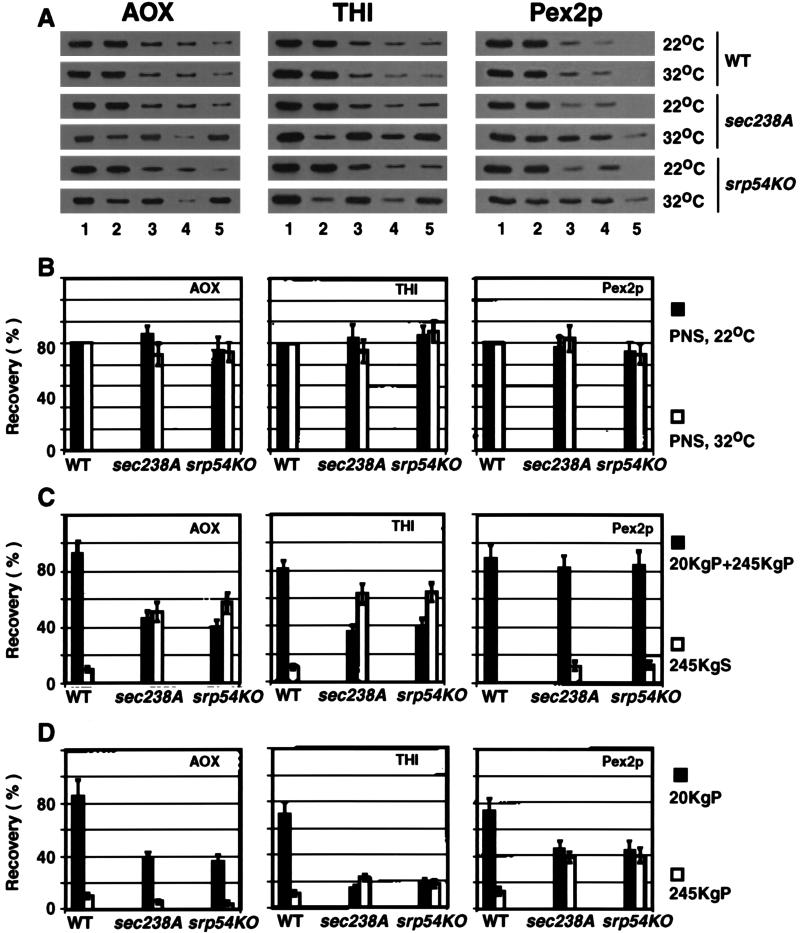
A significant decrease in the size and number of peroxisomes, which was observed in the sec238A and srp54KO mutants grown in YPBO at the restrictive temperature of 32°C, suggested that these mutations either compromise the synthesis of peroxisomal proteins or affect their import into the peroxisome at the restrictive temperature, thereby leading to the mislocalization of peroxisomal matrix and membrane proteins. When the wild-type, sec238A, and srp54KO strains were shifted from YEPD to YPBO and incubated for 8 h at 32°C, the levels of all peroxisomal proteins were greatly induced and reached a steady state in all three strains. No appreciable differences in the rate of induction and in the steady-state levels of all peroxisomal matrix (AOX, MLS, 62- and 64-kDa anti-SKL-reactive polypeptides, ICL, THI, catalase, and multifunctional enzyme) and membrane (Pex2p and Pex16p) proteins were observed between the wild-type strain and both mutants at 32°C (data not shown). Therefore, neither the sec238A nor the srp54KO mutation affected the synthesis of peroxisomal proteins at the temperature restrictive for the exit of secretory proteins from the ER, protein secretion, and growth in oleic acid-containing medium. In contrast, the distribution of peroxisomal matrix and membrane proteins to different subcellular fractions was altered in both mutants compared to the wild-type strain at the restrictive temperature. In wild-type cells, the major fraction (82 to 100%) of peroxisomal matrix and membrane proteins was associated with the low-speed (20KgP) and high-speed (245KgP) pelletable fractions (Fig. 3A and C show the data for AOX, THI, and Pex2p; data for other proteins not shown). These peroxisomal proteins were resistant to exogenously added protease (trypsin) and were therefore in membrane-enclosed structures (data not shown). In sec238A and srp54KO mutant cells at 32°C, a large fraction (57 to 65%) of peroxisomal matrix proteins was mislocalized to the 245KgS (cytosolic) fraction (Fig. 3A and C show the data for AOX and THI; data for other peroxisomal matrix proteins not shown). These proteins were accessible to external protease (data not shown). Some fraction of peroxisomal membrane proteins Pex2p and Pex16p was also mislocalized to the cytosol in sec238A and srp54KO mutant cells at 32°C (Fig. 3C shows the data for Pex2p; data for Pex16p not shown). However, the mislocalization of Pex2p and Pex16p to the cytosol in both mutants was much less pronounced than that of peroxisomal matrix proteins. Up to 15% of Pex2p and up to 9% of Pex16p were mislocalized to the 245KgS fraction in mutant cells (compare data for Pex2p to those for AOX and THI in Fig. 3A and C; data for Pex16p not shown).
FIG. 3.
The sec238A and srp54KO mutations affect the subcellular localization of peroxisomal matrix and membrane proteins at the restrictive temperature. The recoveries of the peroxisomal matrix proteins AOX and THI and of the peroxisomal integral membrane protein Pex2p in different subcellular fractions of the wild-type (WT) and sec238A and srp54KO mutant strains are presented. Strains were grown for 9 h in YPBO at either 22 or 32°C and subjected to subcellular fractionation. (A) Equal fractions (0.15% of the total volume for AOX and THI, 0.9% of the total volume for Pex2p) of the PNS (lanes 1), 20KgP (lanes 2), 20KgS (lanes 3), 245KgP (lanes 4), and 245KgS (lanes 5) were analyzed by immunoblotting with anti-AOX, anti-THI, and anti-Pex2p antibodies. Immunoblots were scanned densitometrically, and the recovery of peroxisomal proteins in different subcellular fractions was quantitated. Values for the PNS (B), 20KgP plus 245KgP and 245KgS (C), and 20KgP and 245KgP (D) signals are presented for the wild-type and mutant strains grown in YPBO at 32°C. Values for signals in different subcellular fractions of mutant strains in panels B, C, and D are relative to the signal for a particular protein in corresponding fractions of the wild-type strain grown at the same temperature. All values for signals are means ± standard deviations for three independent experiments.
The relative distributions of pelletable, protease-protected pools of THI (matrix protein) and of Pex2p and Pex16p (membrane proteins) between the 20KgP and 245KgP fractions were also altered by the sec238A and srp54KO mutations at the restrictive temperature. In wild-type cells, the major fraction (70.6 to 73.8%) of these proteins was found in the 20KgP fraction, while 10.7 to 12.8% was associated with the 245KgP fraction (Fig. 3D shows the data for THI and Pex2p; data for Pex16p not shown). In contrast, in sec238A and srp54KO mutant cells, pelletable pools of THI, Pex2p, and Pex16p were equally distributed between the 20KgP and 245KgP fractions (Fig. 3D shows the data for THI and Pex2p; data for Pex16p not shown). This effect of the sec238A and srp54KO mutations on the distribution of the pelletable pools of peroxisomal proteins between the high- and low-speed fractions was due to the accumulation of a population of high-speed-pelletable vesicles containing THI, Pex2p, and Pex16p but not AOX (Fig. 3D) or any of the other peroxisomal matrix proteins tested (data not shown). These high-speed-pelletable vesicles derive by budding from the ER and initially contain Pex2p and Pex16p. THI is imported into the vesicles soon after they bud from the ER and before they fuse with peroxisomes (48).
Data from immunofluorescence analysis were in agreement with the results of subcellular fractionation and indicated that the sec238A and srp54KO mutations cause the mislocalization of a large fraction of peroxisomal matrix proteins (THI, anti-SKL-reactive proteins, ICL, and MLS) to the cytosol in cells grown for 9 h in YPBO at the restrictive temperature only (data not shown).
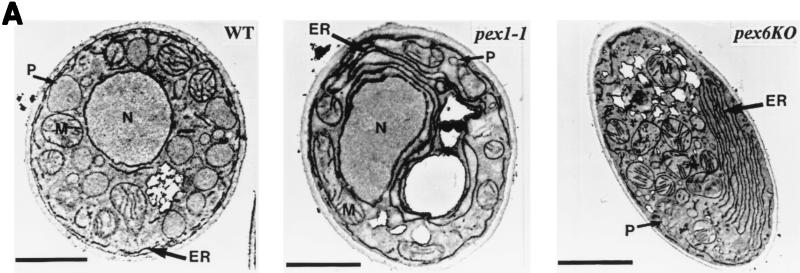
The pex1-1 and pex6KO mutations cause the accumulation of an extensive network of ER membranes and significantly reduce the size and number of peroxisomes.
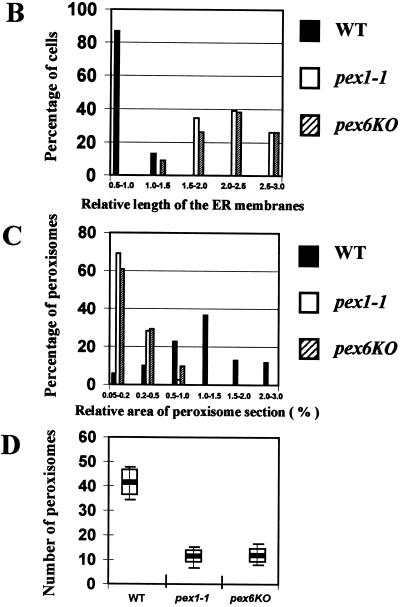
We have previously shown that some, but not all, pex mutations, i.e., the pex1-1, pex2KO, pex6KO, and pex9KO mutations, significantly reduce the rate and efficiency of protein secretion and affect the exit of secretory proteins from the ER during growth in YEPD at both 22 and 32°C (46). Similar results were obtained for these mutations during growth in YPBO at both temperatures (data not shown). We assessed the morphological effects of mutations in the PEX1 and PEX6 genes, which encode members of the AAA family of N-ethylmaleimide-sensitive fusion protein-like ATPases (8), during growth in YPBO. Electron microscopical analysis showed an extensive proliferation of ER membranes in cells of YPBO-grown pex1-1 and pex6KO mutants (Fig. 4A). The extent of ER membrane accumulation was assessed by comparing the lengths of ER membranes to the cell circumference (relative length of ER membranes) in random cross sections. While most (86.8%) of the wild-type cells had relative ER membrane lengths ranging from 0.5 to 1.0, all pex1-1 mutant cells and most (90.9%) pex6KO mutant cells had relative ER membrane lengths ranging from 1.5 to 3.0 (Fig. 4B). Morphometric analysis of random sections of cells of the wild-type and mutant strains also demonstrated that the pex1-1 and pex6KO mutations significantly reduced the size of peroxisomes. pex1-1 and pex6KO mutant cells accumulated mostly small peroxisomes, with relative areas of peroxisome section ranging from 0.05 to 0.2% (Fig. 4C). In contrast, in wild-type cells, more than one-third (36.6%) of all peroxisomes had relative areas of peroxisome section ranging from 1.0 to 1.5%, while 24.7% of all peroxisomes had relative areas of peroxisome section ranging from 1.5 to 3.0% (Fig. 4C). The pex1-1 and pex6KO mutations also caused a three- to fourfold decrease in the number of peroxisomes per cell (10.78 ± 2.23 and 11.27 ± 2.71 peroxisomes per μm3 of cell section volume, respectively) compared to the number of peroxisomes in wild-type cells (41.65 ± 5.2 peroxisomes per μm3 of cell section volume) (Fig. 4D).
FIG. 4.
The pex1-1 and pex6KO mutations cause the accumulation of ER membranes and significantly reduce the size and number of peroxisomes. (A) Cells were grown for 9 h in YPBO at 32°C, fixed in KMnO4, and processed for electron microscopy. Abbreviations are as defined in the legend to Fig. 2. Bars, 1 μm. (B to D) Morphometric analysis was performed on random electron microscopic sections of cells of the wild-type (WT) and mutant strains grown in YPBO at 32°C. (B) The percentage of cells having the indicated relative lengths of ER membranes. The relative length of the ER membranes is calculated as the length of ER membranes relative to cell circumference. (C) Percentage of peroxisomes having the indicated relative area of peroxisome section. The relative area of peroxisome section is calculated as the [(area of peroxisome section/area of cell section) × 100]. (D) Numbers of peroxisomes in the wild-type and mutant strains. Peroxisomes were counted in electron micrographs, and the data were expressed as the number of peroxisomes per cubic micrometer of cell section volume. The bar in the middle of each box indicates the mean peroxisome number. The limits of each box indicate the sample standard deviation on each side of the mean. The lines extend to indicate the extremes of the distribution.
The sec238A, srp54KO, pex1-1, and pex6KO mutations cause accumulation of peroxisomal membrane proteins Pex2p and Pex16p in the ER.
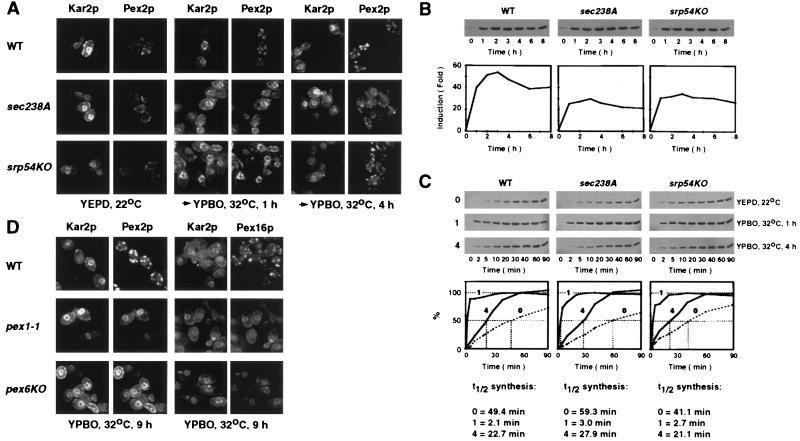
In wild-type Y. lipolytica cells grown in oleic acid-containing medium, Pex2p and Pex16p are, correspondingly, integral (12) and peripheral (11) peroxisomal membrane proteins. Wild-type, sec238A, and srp54KO cells were grown in YEPD at 22°C, shifted to YPBO, and incubated for 1 or 4 h at 32°C. Indirect immunofluorescence microscopy of YEPD- and YPBO-grown wild-type cells with anti-Pex2p (Fig. 5A) and anti-Pex16p (data not shown) antibodies showed a punctate pattern of staining characteristic of peroxisomes. Double-labeling, indirect immunofluorescence with antibodies to Pex2p and to the ER luminal protein Kar2p (Fig. 5A), or to Pex16p and to Kar2p (data not shown), showed that in YEPD- and YPBO-grown wild-type cells, punctate structures decorated by antibodies to peroxisomal proteins failed to colocalize with structures recognized by antibodies to Kar2p. Antibodies to Kar2p showed fluorescence patterns characteristic of the ER, including bright staining of the perinuclear region and cell periphery, and occasionally of filamentous extensions into the cytosol (37, 40, 46). The fluorescence patterns generated by antibodies to Pex2p and to Kar2p (Fig. 5A), or to Pex16p and to Kar2p (data not shown), in sec238A and srp54KO mutant cells in YEPD at 22°C were similar to those seen in wild-type cells and showed no colocalization of Pex2p and Pex16p with ER elements. However, a shift of sec238A and srp54KO mutant cells from YEPD to YPBO and incubation for 1 h at 32°C, the temperature restrictive for the exit of pAEP from the ER and for protein secretion (data not shown) (46), caused the localization of a significant fraction of both Pex2p (Fig. 5A) and Pex16p (data not shown) to the ER. The association of both peroxisomal membrane proteins with the ER was transient in sec238A and srp54KO mutant cells shifted to YPBO at 32°C. At 4 h after the shift, antibodies to both Pex2p (Fig. 5A) and Pex16p (data not shown) yielded only a punctate pattern of staining characteristic of peroxisomes and did not decorate ER elements.
FIG. 5.
Peroxisomal membrane proteins Pex2p and Pex16p transiently or permanently accumulate in the ER of sec238A, srp54KO, pex1-1, and pex6KO mutant cells. (A) Wild-type (WT) strain DX547-1A and sec238A and srp54KO mutant strains were grown in YEPD at 22°C until the cell titer was 2.0 × 108 to 2.3 × 108 cells/ml. Cell cultures were separated into three aliquots. Two aliquots were transferred to YPBO and incubated for 1 or 4 h at 32°C. The third aliquot was processed immediately for double-labeling, indirect immunofluorescence using rabbit anti-Y. lipolytica Kar2p and guinea pig anti-Y. lipolytica Pex2p primary antibodies. Primary antibodies were detected with fluorescein-conjugated goat anti-rabbit immunoglobulin G and rhodamine-conjugated donkey anti-guinea pig immunoglobulin G secondary antibodies. (B) Wild-type strain DX547-1A and sec238A and srp54KO mutant strains were grown in YEPD at 22°C until the cell titer was 2.0 × 108 to 2.3 × 108 cells/ml. Cells were transferred to YPBO and incubated at 32°C. Aliquots of cells were taken at the times indicated. The levels of Pex2p in whole-cell lysates of the wild-type and mutant strains were determined. The lysate from 6 × 108 cells was applied to each lane. Blots were probed with anti-Pex2p antibodies. Immunoblots were scanned densitometrically, and the level of Pex2p in wild-type and mutant cells at the times indicated was quantitated. (C) Wild-type strain DX547-1A and sec238A and srp54KO mutant strains were grown in YEPD at 22°C until the cell titer was 2.0 × 108 to 2.3 × 108 cells/ml. Cultures were separated into three aliquots. Cells from one aliquot were immediately labeled with l-[35S]methionine. The other two aliquots were transferred to YPBO, incubated for 1 or 4 h at 32°C, and then labeled with l-[35S]methionine. Aliquots of cells from each of the three subcultures of the wild-type and mutant strains were taken at the times indicated, and radiolabeling was terminated by the addition of an equal volume of ice-cold 20 mM NaN3 and 20 mM l-methionine. Pex2p was immunoprecipitated from whole-cell lysates derived from 6 × 108 cells. Immunoprecipitates were resolved by SDS-PAGE and visualized by fluorography. Fluorograms were quantitated by densitometry. Values for the level of Pex2p in wild-type and mutant cells at the indicated times of labeling are relative to the maximum level of Pex2p in cells of the corresponding strain preincubated in YPBO for 1 h at 32°C, which was set at 100%. The half-times (t1/2) of synthesis of Pex2p by wild-type and mutant cells were calculated. 0, 1, and 4 represent aliquots of a particular strain either pregrown in YEPD at 22°C (0) or preincubated in YPBO for 1 h (1) or 4 h (4) at 32°C before radiolabeling. (D) Wild-type strain E122 and pex1-1 and pex6KO mutant strains were grown in YEPD at 32°C until the cell titer was 1.8 × 107 to 2.1 × 107 cells/ml. Cells were transferred to YPBO and incubated for 9 h at 32°C. Wild-type and mutant cells were processed for double-labeling, indirect immunofluorescence using rabbit anti-Y. lipolytica Kar2p and guinea pig anti-Y. lipolytica Pex2p or rabbit anti-Y. lipolytica Kar2p and guinea pig anti-Y. lipolytica Pex16p primary antibodies. Primary antibodies were detected with fluorescein-conjugated goat anti-rabbit immunoglobulin G and rhodamine-conjugated donkey anti-guinea pig immunoglobulin G secondary antibodies.
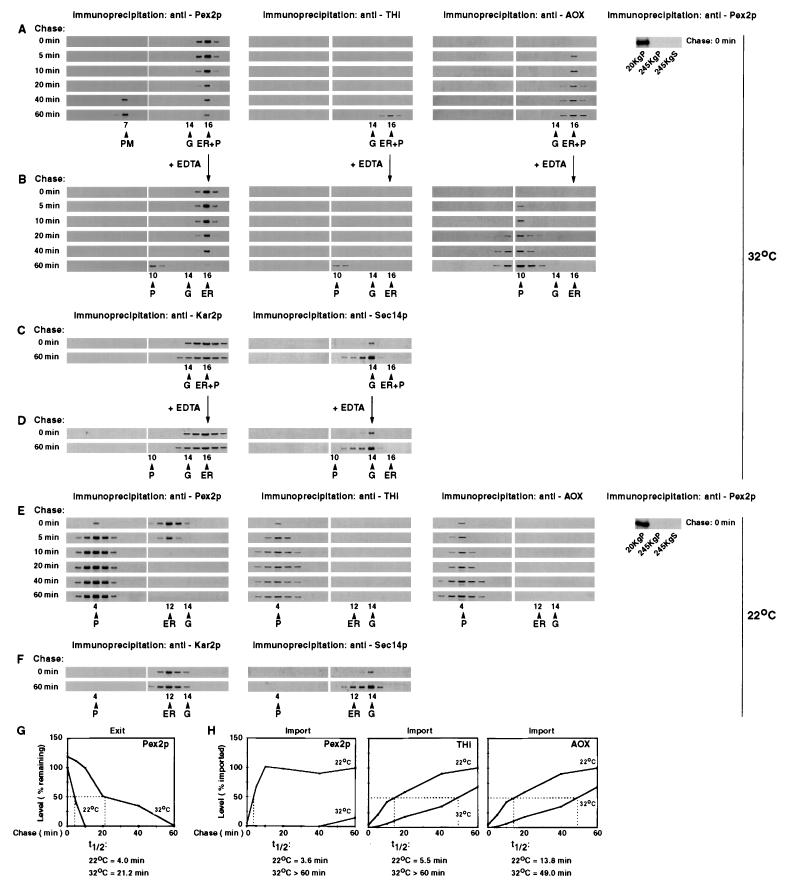
How can the transient association of Pex2p and Pex16p with the ER seen by immunofluorescence of sec238A and srp54KO mutant cells shifted to YPBO at 32°C be explained? An analysis of the kinetics of induction of Pex2p in cells incubated in YPBO indicated that by 1 h after the shift from YEPD to YPBO, the steady-state levels of Pex2p were increased 40-, 25-, and 31-fold in wild-type, sec238A, and srp54KO cells, respectively (Fig. 5B). In contrast, by 4 h after the shift, no significant increase in the steady-state levels of Pex2p was observed in wild-type and mutant cells compared to the levels of Pex2p in cells induced for 1 h (Fig. 5B). Therefore, the fluorescence patterns generated by antibodies to Pex2p in cells shifted from YEPD to YPBO and incubated at 32°C for either 1 or 4 h reflected the intracellular localization of the bulk of Pex2p (69.1 to 97.6% of the total Pex2p, as judged from the data in Fig. 5B), which was synthesized 1 h after the shift. Furthermore, the rates of synthesis of Pex2p after the shift from YEPD to YPBO for 1 h dramatically increased and reached half-times of 2.1, 3.0, and 2.7 min for the wild-type, sec238A, and srp54KO strains, respectively (Fig. 5C). In wild-type cells incubated in YPBO for 1 h, the rate of synthesis of Pex2p was comparable to its rate of exit from the ER, which had a half-time of 3.0 min (see Fig. 6A and F). In contrast, in sec238A and srp54KO mutant cells incubated in YPBO for 1 h, the rates of synthesis of Pex2p were 7.1 to 8.8 times faster than its rates of exit from the ER (half-times of 21.2 and 23.7 min for the sec238A and srp54KO mutant strains, respectively; see Fig. 7A and G for data on the sec238A mutant strain). Therefore, the bulk of Pex2p in sec238A and srp54KO mutant cells shifted to YPBO for 1 h was associated with the ER, as detected by immunofluorescence (Fig. 5A). The Pex2p already present in peroxisomes of sec238A and srp54KO mutant cells pregrown in YEPD at 22°C represents only a minor fraction (3.1 to 3.9%, as judged from the data in Fig. 5B) of the total Pex2p in cells of both mutants incubated in YPBO for 1 h at 32°C. These low levels of Pex2p might not be sufficient to generate the immunofluorescent punctate pattern of staining characteristic of peroxisomes, as the bulk of Pex2p is localized to the ER in sec238A and srp54KO mutant cells incubated in YPBO for 1 h at 32°C. In contrast, in sec238A and srp54KO mutant cells incubated in YPBO for 4 h, the rates of synthesis of Pex2p reached half-times of 27.9 and 21.1 min, respectively (Fig. 5C), and these rates were comparable to the rates of exit of Pex2p from the ER, which had half-times of 21.2 and 23.7 min for the sec238A (see Fig. 7A and G) and srp54KO (data not shown) mutant strains, respectively. Under these conditions, the bulk of Pex2p can exit from the ER of sec238A and srp54KO mutant cells, and therefore, Pex2p was localized by immunofluorescence in punctate structures characteristic of peroxisomes rather than colocalized in elements containing the ER lumenal protein Kar2p (Fig. 5A).
FIG. 6.
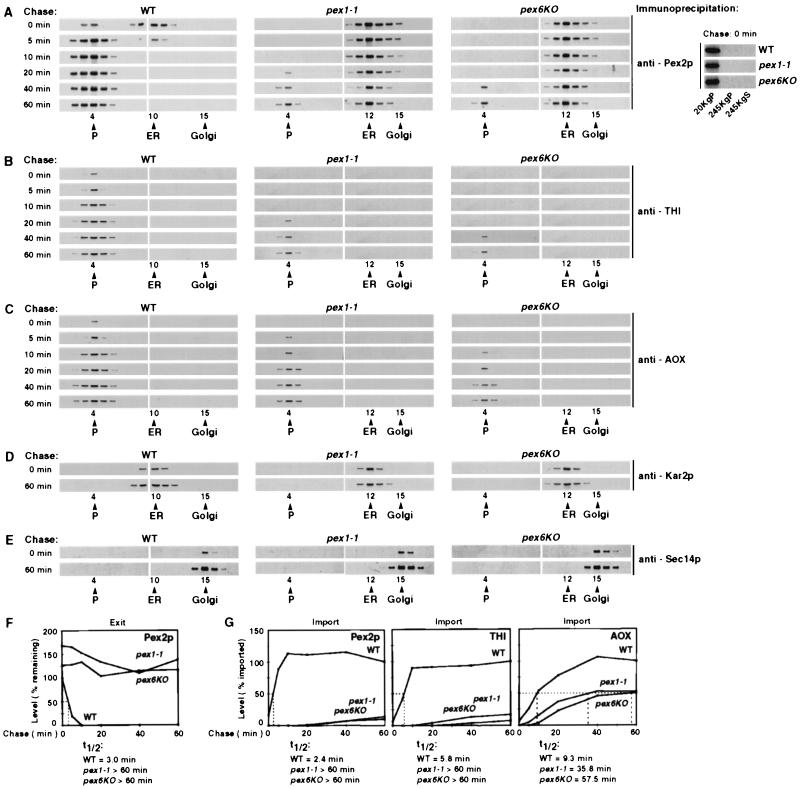
The pex1-1 and pex6KO mutations prevent the exit of Pex2p from the ER and differentially affect the peroxisomal import of Pex2p, THI, and AOX. Wild-type (WT) and mutant strains grown in YPBO at 32°C were pulse-labeled for 3 min with l-[35S]methionine and chased with unlabeled l-methionine. Samples were taken at the indicated times postchase. Cells were subjected to subcellular fractionation, and the 20KgP fractions were further fractionated by isopycnic centrifugation on discontinuous sucrose density gradients. Cells taken at chase time 0 were also subjected to subcellular fractionation to yield 20KgP, 245KgP, and 245KgS (cytosolic) fractions. Pex2p (A), THI (B), AOX (C), Kar2p (D), and Sec14p (E) were immunoprecipitated from gradient fractions. Pex2p (A) was also immunoprecipitated from 20KgP, 245KgP, and 245KgS fractions of wild-type and mutant cells taken at chase time 0. Immunoprecipitates were resolved by SDS-PAGE and visualized by fluorography. Peak fractions in which protein markers of peroxisomes (P), the ER, and the Golgi (G) localized are indicated. (F) The first three panels of the fluorograms in panel A were quantitated by densitometry. The levels of ER-associated Pex2p in wild-type and mutant cells at the indicated times postchase are relative to the level of ER-associated Pex2p in wild-type cells at chase time 0, which was set at 100%. The half-times (t1/2) for the exit of Pex2p from the ER of wild-type and mutant cells were calculated. (G) The first three parts of the fluorograms in panel A and the fluorograms in panels B and C were quantitated by densitometry. The levels of Pex2p, THI, and AOX imported into peroxisomes in wild-type and mutant cells at the indicated chase times are relative to the levels of the respective proteins imported into peroxisomes of wild-type cells after a 60-min chase, which were set at 100%. The half-times for the import of Pex2p, THI, and AOX into peroxisomes of wild-type and mutant cells were calculated.
FIG. 7.
The sec238A mutation significantly delays, but does not prevent, the exit of Pex2p from the ER and differentially affects the peroxisomal import of Pex2p, THI, and AOX. (A to D) The sec238A mutant strain grown in YPBO at 32°C was pulse-labeled for 3 min with l-[35S]methionine and chased with unlabeled l-methionine. Samples were taken at the indicated chase times. Cells were subjected to subcellular fractionation, and the 20KgP fractions were further fractionated by isopycnic centrifugation on discontinuous sucrose density gradients. Pex2p, THI, AOX, Kar2p, and Sec14p were immunoprecipitated from gradient fractions. Immunoprecipitates were resolved by SDS-PAGE and visualized by fluorography. Fractions containing either the complex formed between peroxisomes and the ER or the Golgi alone (as judged from the distributions of protein markers) were combined and treated with 30 mM EDTA under conditions allowing complex disassembly. Treated samples were fractionated by isopycnic centrifugation on discontinuous sucrose density gradients. Pex2p, THI, AOX, Kar2p, and Sec14p were immunoprecipitated from gradient fractions. Immunoprecipitates were resolved by SDS-PAGE and visualized by fluorography. (E and F) The sec238A mutant strain was grown in YPBO at 32°C, shifted to 22°C for 2 h, pulse-labeled at 22°C for 3 min with l-[35S]methionine, and chased with unlabeled l-methionine. Samples were taken at the indicated chase times. Cells were subjected to subcellular fractionation, and the 20KgP fractions were further fractionated by isopycnic centrifugation on discontinuous sucrose density gradients. Pex2p, THI, AOX, Kar2p, and Sec14p were immunoprecipitated from gradient fractions. Immunoprecipitates were resolved by SDS-PAGE and visualized by fluorography. Peak fractions in which markers of peroxisomes (P), the plasma membrane (PM), the ER, and the Golgi (G) localized are indicated. (A and E) sec238A mutant cells taken at chase time 0 at 32°C (A) or 22°C (E) were also subjected to subcellular fractionation to yield 20KgP, 245KgP, and 245KgS (cytosolic) fractions. Pex2p was immunoprecipitated from different subcellular fractions, and immunoprecipitates were resolved by SDS-PAGE and visualized by fluorography. (G) Fluorograms in panels B and E (left side) were quantitated by densitometry. The levels of ER-associated Pex2p in sec238A mutant cells grown at 22 or 32°C at the indicated chase times are relative to the level of ER-associated Pex2p in sec238A mutant cells grown at 22°C at chase time 0, which was set at 100%. The half-times (t1/2) for the exit of Pex2p from the ER for the sec238A mutant strain grown at 22 or 32°C were calculated. (H) Fluorograms in panel B and of the first three parts of panel E were quantitated by densitometry. The levels of Pex2p, THI, and AOX imported into peroxisomes in sec238A mutant cells grown at 22 or 32°C at the indicated times postchase are relative to the levels of the corresponding proteins imported into peroxisomes of the sec238A mutant strain grown at 22°C after a 60-min chase, which were set at 100%. The half-times for the import of Pex2p, THI, and AOX into peroxisomes of the sec238A mutant strain grown at 22 or 32°C were calculated.
While the localization of both Pex2p and Pex16p to ER elements in the sec238A and srp54KO mutants at the restrictive temperature was transient, both proteins were permanently associated with the ER in pex1-1 and pex6KO mutant cells. The fluorescence patterns generated by antibodies to Pex2p and to Kar2p, or to Pex16p and to Kar2p, in pex1-1 and pex6KO mutant cells either grown in YEPD at 22°C (data not shown) or shifted to YPBO and incubated at 32°C for 9 h (Fig. 5D) were superimposable and showed staining characteristic of the ER (cf. Fig. 5A).
Our data are suggestive of trafficking of Pex2p and Pex16p to the peroxisomal membrane via the ER in wild-type cells. The exit of both peroxisomal membrane proteins from the ER at 32°C is significantly delayed, but not prevented, by the sec238A and srp54KO mutations and is completely blocked by the pex1-1 and pex6KO mutations at 22 and 32°C. In contrast, the trafficking of peroxisomal matrix proteins, including THI, MLS, ICL, and anti-SKL-reactive proteins, apparently does not occur via the ER. Double-labeling, indirect immunofluorescence with antibodies to any of these peroxisomal proteins and to Kar2p showed no association of matrix proteins with the ER in any mutant cells under the conditions in which a significant fraction of Pex2p and Pex16p localized to the ER (data not shown).
Membrane proteins Pex2p and Pex16p, but not matrix proteins, traffic to peroxisomes via the ER.
We studied the trafficking of peroxisomal membrane and matrix proteins by immunoprecipitation of pulse-labeled and chased proteins from fractions of discontinuous sucrose density gradients of 20KgP subcellular fractions. In wild-type cells, the majority of pulse-labeled, unchased Pex2p cofractionated with the ER marker (Kar2p; Fig. 6A and D, left) but not with markers of the Golgi (Sec14p; Fig. 6A and E, left; data for guanosine diphosphatase not shown), plasma membrane, mitochondria, or vacuoles (data not shown). By 5 min of chase, most of this ER-associated Pex2p was chased into fractions containing peroxisomal proteins (Fig. 6A to C, left parts). The transit of Pex2p from the ER to the peroxisome was complete by 10 min of chase (Fig. 6A, left). In contrast, the majority of pulse-labeled Pex2p in pex1-1 and pex6KO mutant cells localized to ER elements even by 60 min of chase (Fig. 6A and D, second and third parts). Only a minor fraction of pulse-labeled Pex2p in pex1-1 and pex6KO mutant cells was chased into fractions containing peroxisomal proteins (Fig. 6A to C, second and third parts). No pulse-labeled Pex2p in either wild-type or mutant cells cofractionated with markers of the Golgi (Sec14p; Fig. 6A and E; data for guanosine diphosphatase not shown), plasma membrane, mitochondria, or vacuoles (data not shown). Similar results were obtained for the trafficking of Pex16p (data not shown). Immunoprecipitation of pulse-labeled Pex2p and Pex16p from the 20KgP, 245KgP, and 245KgS (cytosolic) fractions showed no cytosolic pools of pulse-labeled Pex2p or Pex16p prior to the appearance of either protein in peroxisomes, i.e., at the start of the chase in wild-type, pex1-1, or pex6KO cells (Fig. 6A, right, shows the data for Pex2p; data for Pex16p not shown). Therefore, Pex2p and Pex16p found in peroxisomes derive from the ER rather than being imported directly from the cytosol. Accordingly, the targeting of both peroxisomal membrane proteins to the ER in wild-type, pex1-1, and pex6KO cells is a prerequisite for their import into peroxisomes and is not a targeting pathway alternative to a cytosol-to-peroxisome targeting pathway for Pex2p and Pex16p. These data extend the results of double-labeling, indirect immunofluorescence (Fig. 5) and demonstrate that the delivery of the membrane proteins Pex2p and Pex16p to the peroxisome occurs via the ER, does not involve their transport through the Golgi (or any other organelle) as an intermediate step, and is largely prevented by the pex1-1 and pex6KO mutations. In contrast, the trafficking of the peroxisomal matrix proteins THI (Fig. 6B), AOX (Fig. 6C), and ICL (data not shown) does not occur via the ER (or any other organelle). None of these proteins was associated with the ER at any time in either wild-type or mutant cells.
In wild-type cells, Pex2p and Pex16p exited the ER with half-times of 3.0 and 3.4 min, respectively, which were very close to the half-times for their import into peroxisomes (2.4 and 3.2 min, respectively) (Fig. 6F and G show the data for Pex2p; data for Pex16p not shown). These data suggest some coordination between the exit of Pex2p and Pex16p from the ER and their import into the peroxisome. The kinetics of import of Pex2p and Pex16p and of the matrix protein THI into peroxisomes in wild-type cells were different from those of other peroxisomal proteins. While the import of Pex2p (Fig. 6G), Pex16p (data not shown), and THI (Fig. 6G) into peroxisomes in wild-type cells reached a steady state by 10 min of chase, steady-state levels of peroxisomal import for AOX (Fig. 6G) and ICL (data not shown) were observed only after 40 to 60 min of chase. Moreover, while both the pex1-1 and pex6KO mutations severely affected the import of Pex2p (Fig. 6G), Pex16p (data not shown), and THI (Fig. 6G) into the peroxisome, their effects on the peroxisomal import of AOX (Fig. 6G) and ICL (data not shown) were much less pronounced. These data suggest that the import of Pex2p, Pex16p, and THI into the peroxisome could occur by a mechanism distinct from that used for the import of AOX and ICL. Indeed, the import of Pex2p, Pex16p, and THI into peroxisomes involves the budding from the ER of vesicles that initially contain Pex2p and Pex16p and import THI after they bud from the ER and before they fuse with peroxisomes (48).
We also studied the trafficking of peroxisomal membrane and matrix proteins in sec238A mutant cells grown in YPBO at 22 or 32°C. At 22°C, the temperature permissive for protein secretion, the major portion of pulse-labeled, unchased Pex2p and Pex16p cofractionated with the ER marker Kar2p (Fig. 7E and F, left, show the data for Pex2p and Kar2p; data for Pex16p not shown). By 10 min of chase, all of Pex2p and Pex16p was chased into fractions containing peroxisomal proteins (Fig. 7E). Similar to the data presented above for wild-type, pex1-1, and pex6KO cells, no cytosolic pool of either peroxisomal membrane protein was found in sec238A mutant cells at the start of the chase, i.e., prior to the appearance of Pex2p and Pex16p in peroxisomes (Fig. 7E, right, shows the data for Pex2p; data for Pex16p not shown). Therefore, pulse-labeled Pex2p and Pex16p chased into the peroxisomal fractions of sec238A mutant cells grown at 22°C are not imported into peroxisomes directly from the cytosol but rather are delivered to peroxisomes from the ER. The trafficking of Pex2p and Pex16p from the ER to the peroxisome in sec238A mutant cells did not involve their transport through the Golgi or any other organelle, as no Pex2p (Fig. 7E, left) or Pex16p (data not shown) cofractionated with markers of the Golgi (Fig. 7F, right, shows the data for Sec14p; data for guanosine diphosphatase not shown), plasma membrane, mitochondria, or vacuoles (data not shown). None of the peroxisomal matrix proteins tested, including THI (Fig. 7E, second part), AOX (Fig. 7E, third part), and ICL (data not shown), was associated with the ER at any time. These data confirm that the transit of matrix proteins to the peroxisome does not occur via the ER.
In sec238A mutant cells grown at 32°C, the temperature restrictive for protein secretion, the peroxisomes pelletable at low speed (20,000 × gmax) form a complex with the ER in vitro (47). Due to the formation of this complex, peroxisomal matrix proteins (Fig. 7A shows the data for THI and AOX; data for ICL not shown) and ER marker proteins (Fig. 7C, left, shows the data for Kar2p; data for Sls1p and NADPH:cytochrome c reductase not shown) peaked in fraction 16 of a discontinuous sucrose density gradient of the 20KgP subcellular fraction. While the distribution of peroxisomal proteins around fraction 16 coincided with the distribution of ER luminal and membrane proteins, their distribution did not coincide with the distribution of markers for the Golgi apparatus (Fig. 7C, right, shows the data for Sec14p; data for guanosine diphosphatase not shown), mitochondria, vacuoles, or plasma membrane (data not shown). Peroxisomes (Fig. 7B) could be separated from the ER (Fig. 7D) in the complex by treatment with EDTA (45). These peroxisomes were intact and essentially free from contamination by other organelles, as judged by protease protection, flotation on a sucrose gradient, and electron microscopy (data not shown). By using these conditions for disassembly, we demonstrated that in sec238A mutant cells grown at the restrictive temperature, the major portion of pulse-labeled, unchased Pex2p (Fig. 7B, left panel) and Pex16p (data not shown) localized to the ER. The exit of Pex2p and Pex16p from the ER was significantly delayed, but not blocked, by the sec238A mutation at 32°C (Fig. 7B and G show the data for Pex2p; data for Pex16p not shown). The negative effect of the sec238A mutation at 32°C on the exit of Pex2p and Pex16p from the ER was much less pronounced than that of the pex1-1 and pex6KO mutations (see above). However, the rates of import of both Pex2p and Pex16p into peroxisomes were comparable in sec238A mutant cells grown at 32°C and in pex1-1 and pex6KO mutant cells (half-times for the peroxisomal import of both proteins were >60 min in all of these strains grown at the restrictive temperature). Furthermore, while the sec238A mutation at 32°C severely affected the import of Pex2p (Fig. 7H), Pex16p (data not shown), and THI (Fig. 7H) into the peroxisome, its effect on the import of AOX (Fig. 7H) and ICL (data not shown) was much less pronounced. Therefore, these data also suggest that the import of Pex2p, Pex16p, and THI into peroxisomes may occur by a mechanism distinct from that serving for the import of AOX and ICL.
Pex2p and Pex16p are subjected to N-linked core glycosylation in the ER lumen and are delivered from the ER to the peroxisome in glycosylated form.
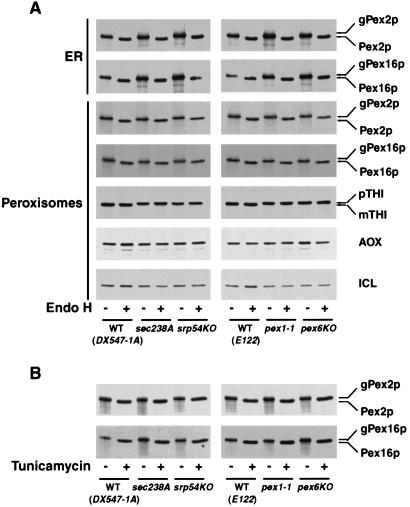
Pex2p and Pex16p were immunoprecipitated from the ER or peroxisomes of cells pulse-labeled with radiolabeled methionine and chased with unlabeled methionine for 5 or 60 min, respectively. Immunoprecipitated Pex2p and Pex16p were treated with endo H, which cleaves the bond between two N-acetyl-d-glucosamine residues in the core of N-linked oligosaccharides attached to the polypeptide backbone (7). Treatment with endo H increased the electrophoretic mobilities of both the ER- and peroxisome-associated forms of Pex2p and Pex16p from wild-type and mutant cells (Fig. 8). The difference in apparent molecular mass between endo H-treated and untreated polypeptides was approximately 2 kDa, which corresponds to the mass of a single core oligosaccharide chain (7). In contrast, endo H treatment of pulse-labeled and chased THI (mature form in wild-type cells and precursor form in mutant cells), AOX, and ICL immunoprecipitated from peroxisomes of wild-type and mutant strains did not change their electrophoretic mobilities (Fig. 8). Therefore, these matrix proteins were not core glycosylated.
FIG. 8.
Pex2p and Pex16p exist as glycosylated forms in the ER and peroxisomes of wild-type (WT) and mutant strains. (A) Wild-type and mutant cells grown in YPBO at 32°C were pulse-labeled for 3 min with l-[35S]methionine and subjected to either a 5- or 60-min chase with unlabeled l-methionine, followed by immunoprecipitation and endo H treatment of ER-associated Pex2p and Pex16p (5-min chase) or of peroxisomal Pex2p, Pex16p, THI, AOX, and ICL (60-min chase). Cells were subjected to subcellular fractionation, and the 20KgP fractions were further fractionated by isopycnic centrifugation on discontinuous sucrose density gradients. Pex2p and Pex16p were immunoprecipitated from peak fractions in which protein markers of the ER or of peroxisomes localized, while THI, AOX, and ICL were subjected to immunoprecipitation from fractions in which peroxisomal matrix proteins peaked (Fig. 6 and 7). Immunoprecipitates were divided into two equal aliquots. One aliquot was digested with endo H (+), while the second was mock digested (−). Endo H-treated and untreated proteins were resolved by SDS-PAGE and visualized by fluorography. (B) Wild-type and mutant cells grown in YPBO at 32°C were separated into two equal aliquots. Tunicamycin (+) (final concentration, 10 μg/ml) was added to one aliquot 5 min before the addition of l-[35S]methionine, while the second aliquot was mock treated (−). Cells from both aliquots were radiolabeled for 20 min with l-[35]methionine. Radiolabeling was terminated by the addition of an equal volume of ice-cold 20 mM NaN3 and 20 mM l-methionine. Pex2p and Pex16p were immunoprecipitated from whole-cell lysates. Immunoprecipitates were resolved by SDS-PAGE and visualized by fluorography. The positions of the glycosylated (gPex2p and gPex16p) and unglycosylated forms of Pex2p and Pex16p and of the precursor (pTHI) and mature (mTHI) forms of THI are indicated at the right.
We also tested the effects of tunicamycin treatment of cells on the glycosylation of Pex2p and Pex16p. Tunicamycin is a specific inhibitor of N-linked protein glycosylation in vivo (7). Wild-type and mutant cells grown in YPBO at 32°C were radiolabeled with l-[35S]methionine in the presence or absence of tunicamycin. Treatment of cells with tunicamycin resulted in increased electrophoretic mobilities of radiolabeled Pex2p and Pex16p from wild-type and mutant cells relative to the mobilities of the two proteins from untreated cells (Fig. 8B). The difference in apparent molecular mass between Pex2p and Pex16p from tunicamycin-treated and untreated cells was approximately 2 kDa and similar to that observed between endo H-treated and untreated forms of both proteins, confirming that both Pex2p and Pex16p are core glycosylated in vivo and contain a single core oligosaccharide chain.
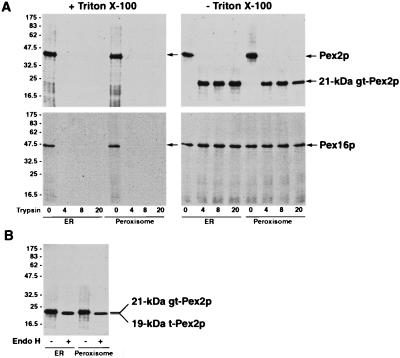
We performed protease protection analysis of Pex2p and Pex16p to analyze the topology of the N-linked core glycosylation of these proteins in both the ER and peroxisomes. In wild-type Y. lipolytica cells, Pex2p is an integral peroxisomal membrane protein that contains one predicted membrane-spanning α-helix and one canonical Asn-Xaa-Thr sequence for N-linked glycosylation carboxyl to this potential transmembrane domain (12). Treatment of the ER and peroxisomes from radiolabeled wild-type cells with trypsin in the absence of detergent, followed by immunoprecipitation with anti-Pex2p antibodies, revealed that a 21-kDa fragment of Pex2p was protected from degradation by trypsin in both the ER and peroxisomes (Fig. 9A, upper right part). This 21-kDa fragment is core glycosylated in both the ER and peroxisomes and contains a single N-linked oligosaccharide chain, as endo H treatment increased the electrophoretic mobilities of both the ER- and peroxisome-associated forms of this fragment, resulting in the formation of a 19-kDa polypeptide (Fig. 9B). An analysis of the primary structure of Pex2p (12) shows that since (i) a trypsin-sensitive cleavage site precedes the potential membrane-spanning α-helix of Pex2p, (ii) the only canonical site for N-linked glycosylation is carboxyl to this transmembrane domain and is located towards the carboxyl terminus of Pex2p, and (iii) the predicted molecular mass (18.3 kDa) of a fragment of Pex2p that includes the membrane-spanning domain and the amino acid residues carboxyl to it is similar to the molecular mass of the identified protease-protected fragment of Pex2p after endo H treatment (19 kDa), we suggest that this carboxyl-terminal part of Pex2p is oriented towards the lumen of the ER and towards the matrix of the peroxisome. Therefore, N-linked core glycosylation of Pex2p occurs in the ER lumen, Pex2p is delivered to the peroxisome in glycosylated form, and the transmembrane topology of Pex2p is the same in the peroxisome as in the ER.
FIG. 9.
Orientations of Pex2p and Pex16p in the ER and in peroxisomes and topology of N-linked core glycosylation of Pex2p. (A) Wild-type cells grown in YPBO at 32°C were pulse-labeled for 3 min with l-[35S]methionine and subjected to either a 5- or 60-min chase with unlabeled l-methionine for immunoprecipitation and endo H treatment of the ER-associated (5-min chase) or peroxisomal (60-min chase) forms of Pex2p and Pex16p. Cells were subjected to subcellular fractionation, and the 20KgP fractions were further fractionated by isopycnic centrifugation on discontinuous sucrose density gradients. ER and peroxisomes purified as described in the legend to Fig. 6 were incubated with 0, 4, 8, and 20 μg of trypsin in the presence (+) or absence (−) of 0.5% (vol/vol) Triton X-100 for 40 min on ice. Reactions were terminated by addition of SDS-PAGE sample buffer and boiling for 5 min. Reaction mixtures were cooled, and Pex2p and Pex16p were immunoprecipitated. Immunoprecipitates were resolved by SDS-PAGE and visualized by fluorography. The positions of Pex2p and Pex16p and of the glycosylated, trypsin-resistant fragment of Pex2p (21-kDa gt-Pex2p) are indicated at the right. (B) Samples of the 21-kDa trypsin-resistant fragment of Pex2p from both the ER and peroxisomes were divided into two equal aliquots. One aliquot was digested with endo H (+), while the second was mock digested (−). Endo H-treated and untreated proteins were resolved by SDS-PAGE and visualized by fluorography. The positions of the glycosylated, trypsin-resistant fragment of Pex2p (21-kDa gt-Pex2p) and the trypsin-resistant fragment of Pex2p deglycosylated after endo H treatment (19-kDa t-Pex2p) are indicated at the right. The values at the left are molecular mass standards (in kilodaltons).
Pex16p is an intraperoxisomal peripheral membrane protein in wild-type Y. lipolytica cells (11). Treatment of the ER and peroxisomes of wild-type cells with trypsin in the absence of detergent, followed by immunoprecipitation of radiolabeled Pex16p, revealed that Pex16p is protected from protease digestion in both the ER and peroxisomes (Fig. 9A, bottom right part). Therefore, N-linked core glycosylation of Pex16p occurs in the ER lumen and Pex16p is targeted from the ER lumen to the peroxisomal matrix in glycosylated form.
DISCUSSION
Here we report that in wild-type Y. lipolytica, trafficking of the peroxisomal membrane proteins Pex2p and Pex16p to the peroxisome occurs via the ER and results in the glycosylation of both proteins in the ER lumen but does not involve their transit through the Golgi as an intermediate step. We also demonstrate that mutations in the SEC238, SRP54, PEX1, and PEX6 genes not only cause defects in the exit of pAEP and other secretory proteins from the ER and in protein secretion but also significantly delay or prevent the exit of Pex2p and Pex16p from the ER and affect the assembly of functionally intact peroxisomes. The SRP54 gene encodes the Y. lipolytica homolog of the Srp54p component of the signal recognition particle involved in protein translocation across the ER membrane (24), while the PEX1 and PEX6 genes encode members of the AAA family of N-ethylmaleimide-sensitive fusion protein-like ATPases that are essential not only for peroxisome biogenesis but also for homo- and heterotypic fusion events required for the assembly of other organelles (46). While previous morphological studies of mammalian and yeast cells have shown a close association between peroxisomes and the ER (for reviews, see references 15 and 22) and recent genetic and biochemical data have suggested a dual role for the ER in supplying phospholipids for the formation of the peroxisomal membrane (45) and in protein trafficking to peroxisomes (2, 4, 13, 46, 50, 51), this study provides the first genetic evidence that the ER is required for the assembly of functionally intact peroxisomes in Y. lipolytica. A requirement for the ER in peroxisome assembly is also supported by our data showing that not only Pex1p and Pex6p, but also other peroxins essential for peroxisome biogenesis in Y. lipolytica, are required for the exit of secretory proteins from the ER and for their delivery to the extracellular medium (46; this study).
The sec238A and srp54KO mutations at the restrictive temperature of 32°C, and the pex1-1 and pex6KO mutations at both 22 and 32°C, affected the export of AEP to the extracellular medium and caused the accumulation of pAEP in the ER. The rate and efficiency of secretion of all other secretory proteins were affected in these mutants to similar extents. These data suggest that in Y. lipolytica, Sec238p, Srp54p, Pex1p, and Pex6p are essential for the exit of secretory proteins from the ER. Unexpectedly, none of the sec238A, srp54KO, pex1-1, or pex6KO mutations affected the growth of Y. lipolytica in glucose-containing YEPD medium, even at temperatures at which protein secretion was impaired. In contrast, previous studies have shown that all temperature-sensitive mutants of the yeast S. cerevisiae affected in protein secretion are unable to grow in glucose-containing media at temperatures restrictive for secretion (29, 30, 52). The observed differences in growth between the secretory mutants of these two yeast species are probably due to essential differences in the relationship between protein secretion and cell surface growth in these yeasts. S. cerevisiae apparently has one major pathway that is common for the secretion of proteins into the cell envelope/extracellular medium and for the export of materials for plasma membrane and cell wall synthesis (18, 28–31, 38). Therefore, any mutational block in protein secretion also affects cell surface growth. In contrast, our recent data have demonstrated that protein secretion and cell surface growth in Y. lipolytica are served by distinct pathways (46). Therefore, the sec238A, srp54KO, pex1-1, and pex6KO mutations affect protein secretion but do not compromise the export of plasma membrane and cell wall-associated proteins during the yeast mode of growth of Y. lipolytica in YEPD.
While neither the sec238A nor the srp54KO mutation affected growth in YEPD, which does not require intact peroxisomes, both mutations caused temperature-sensitive growth in oleic acid-containing YPBO medium, the metabolism of which requires the assembly of functionally intact peroxisomes. Furthermore, the pex1-1 and pex6KO mutations affected growth in YPBO at both 22 and 32°C but did not compromise growth in YEPD at either temperature. Growth defects of these mutants in YPBO were not caused by nonspecific (secondary) effects of the gene mutations on cell viability and/or on the level of synthesis of total phospholipid in YPBO (data not shown). Rather, the combined data of electron and immunofluorescence microscopy and of pulse-chase analysis demonstrated that Sec238p, Srp54p, Pex1p, and Pex6p perform an essential and specific role in the import of matrix and membrane proteins into the peroxisome. None of the sec238A, srp54KO, pex1-1, or pex6KO mutations compromised the synthesis of peroxisomal proteins (data not shown). However, all of these mutations at the restrictive temperature significantly reduced the size and number of peroxisomes, caused the mislocalization of a large fraction of peroxisomal matrix proteins and of some fraction of the peroxisomal membrane proteins Pex2p and Pex16p to the cytosol, and selectively affected the rates and efficiencies of import of individual peroxisomal proteins into the organelle.
The combined data of immunofluorescence microscopy, pulse-chase analysis, subcellular fractionation, protease protection, and endo H digestion showed that in Y. lipolytica wild-type cells, the trafficking of Pex2p and Pex16p to the peroxisome occurs via the ER and results in the core glycosylation of both proteins in the ER but does not involve their transit through the Golgi. N-linked core glycosylation of Pex2p and Pex16p occurs in the ER lumen, and both proteins are delivered from the ER to the peroxisome in glycosylated form. The transmembrane topology of the integral membrane protein Pex2p in the ER and the peroxisomal membrane is the same, with the carboxyl terminus of Pex2p oriented towards the lumen of the ER and towards the matrix of the peroxisome. Glycosylated Pex16p is localized within the ER lumen and the peroxisomal matrix and is associated with the membranes of both organelles. The structural features of Pex2p and Pex16p that mediate their targeting from the cytosol to the ER, and from the ER to the peroxisomal membrane, remain to be determined. Pex2p of Y. lipolytica (11) contains a recently identified peroxisomal targeting signal, termed mPTS, for peroxisomal integral membrane proteins (9). One characteristic feature of peroxisomal membrane proteins containing mPTS motifs, including Pex2p of Y. lipolytica and Pmp47 of S. cerevisiae (9), is the presence of a canonical Asn-Xaa-Thr sequence for N-linked glycosylation in mPTS. Pex16p of Y. lipolytica also contains this canonical sequence (11). Our data demonstrate that both Pex2p and Pex16p in Y. lipolytica are glycosylated in the ER lumen and probably contain a single core N-linked oligosaccharide chain. The importance of this N-glycosylation for the targeting of Pex2p and Pex16p to the ER and/or peroxisome is unknown. Interestingly, the amino-terminal 16 amino acids preceding the mPTS of the peroxisomal integral membrane protein, Pex3p, of the yeast Hansenula polymorpha, while insufficient to target Pex3p to peroxisomes, have been shown to be able to sort catalase, lacking its carboxyl-terminal peroxisomal targeting signal 1, to the ER and nuclear envelope (2). However, fusion of both the amino-terminal 16 amino acids and mPTS of Pex3p to the truncated catalase led to targeting of the fusion protein to the peroxisomal membrane (2). We suspect that the targeting of some peroxisomal membrane proteins, including Pex2p and Pex16p of Y. lipolytica and Pex3p of H. polymorpha, from the cytosol to the ER, and from the ER to the peroxisomal membrane, can be mediated by distinct targeting signals. In contrast to the transit of the peroxisomal membrane proteins Pex2p and Pex16p, the trafficking of the peroxisomal matrix proteins THI, AOX, and ICL in Y. lipolytica cells does not occur via the ER and does not involve the glycosylation of these proteins.
In conclusion, the results described herein provide evidence for the essential role of the ER in the assembly of functionally intact peroxisomes. We show that in Y. lipolytica, the delivery of Pex2p and Pex16p to the peroxisomal membrane occurs via the ER, results in their glycosylation in the ER lumen, does not involve their transit through the Golgi, and requires the products of the SEC238, SRP54, PEX1, and PEX6 genes. The molecular mechanisms by which Sec238p, Srp54p, Pex1p, and Pex6p function in the exit of peroxisomal membrane proteins from the ER and in peroxisomal protein import are currently being investigated.
ACKNOWLEDGMENTS
This work was supported by a grant from the Natural Sciences and Engineering Research Council of Canada to R.A.R. R.A.R. is a Medical Research Council of Canada Senior Scientist and an International Research Scholar of the Howard Hughes Medical Institute.
We thank Honey Chan for help with electron microscopy.
REFERENCES
- 1.Abeijon C, Orlean P, Robins P W, Hirschberg C B. Topography of glycosylation in yeast: characterization of GDP-mannose transport and lumenal guanosine diphosphatase activities in Golgi-like vesicles. Proc Natl Acad Sci USA. 1989;86:6935–6939. doi: 10.1073/pnas.86.18.6935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baerends R J S, Rasmussen S W, Hilbrands R E, van der Heide M, Faber K N, Reuvekamp P T W, Kiel J A K W, Cregg J M, van der Klei I J, Veenhuis M. The Hansenula polymorpha PER9 gene encodes a peroxisomal membrane protein essential for peroxisome assembly and integrity. J Biol Chem. 1996;271:8887–8894. doi: 10.1074/jbc.271.15.8887. [DOI] [PubMed] [Google Scholar]
- 3.Berteaux-Lecellier V, Picard M, Thompson-Coffe C, Zickler D, Panvier-Adoutte A, Simonet J-M. A nonmammalian homolog of the PAF1 gene (Zellweger syndrome) discovered as a gene involved in caryogamy in the fungus Podospora anserina. Cell. 1995;81:1043–1051. doi: 10.1016/s0092-8674(05)80009-1. [DOI] [PubMed] [Google Scholar]
- 4.Bodnar A G, Rachubinski R A. Characterization of the integral membrane polypeptides of rat liver peroxisomes isolated from untreated and clofibrate-treated rats. Biochem Cell Biol. 1991;69:499–508. doi: 10.1139/o91-074. [DOI] [PubMed] [Google Scholar]
- 5.Boisramé A, Beckerich J-M, Gaillardin C. Sls1p, an endoplasmic reticulum component, is involved in the protein translocation process in the yeast Yarrowia lipolytica. J Biol Chem. 1996;271:11668–11675. doi: 10.1074/jbc.271.20.11668. [DOI] [PubMed] [Google Scholar]
- 6.Chelstowska A, Butow R A. RTG genes in yeast that function in communication between mitochondria and the nucleus are also required for expression of genes encoding peroxisomal proteins. J Biol Chem. 1995;270:18141–18146. doi: 10.1074/jbc.270.30.18141. [DOI] [PubMed] [Google Scholar]
- 7.Coligan J E, Dunn B M, Ploegh H L, Speicher D W, Wingfield P T. Current protocols in protein science. New York, N.Y: Wiley Interscience; 1995. [Google Scholar]
- 8.Distel B, Erdmann R, Gould S J, Blobel G, Crane D I, Cregg J M, Dodt G, Fujiki Y, Goodman J M, Just W W, Kiel J A K W, Kunau W-H, Lazarow P B, Mannaerts G P, Moser H, Osumi T, Rachubinski R A, Roscher A, Subramani S, Tabak H F, Valle D, van der Klei I, van Veldhoven P P, Veenhuis M. A unified nomenclature for peroxisome biogenesis. J Cell Biol. 1996;135:1–3. doi: 10.1083/jcb.135.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dyer J M, McNew J A, Goodman J M. The sorting sequence of the peroxisomal integral membrane protein PMP47 is contained within a short hydrophilic loop. J Cell Biol. 1996;133:269–280. doi: 10.1083/jcb.133.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eitzen G A, Aitchison J D, Szilard R K, Veenhuis M, Nuttley W M, Rachubinski R A. The Yarrowia lipolytica gene PAY2 encodes a 42-kDa peroxisomal integral membrane protein essential for matrix protein import and peroxisome enlargement but not for peroxisome membrane proliferation. J Biol Chem. 1995;270:1429–1436. doi: 10.1074/jbc.270.3.1429. [DOI] [PubMed] [Google Scholar]
- 11.Eitzen G A, Szilard R K, Rachubinski R A. Enlarged peroxisomes are present in oleic acid-grown Yarrowia lipolytica overexpressing the PEX16 gene encoding an intraperoxisomal peripheral membrane peroxin. J Cell Biol. 1997;137:1265–1278. doi: 10.1083/jcb.137.6.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eitzen G A, Titorenko V I, Smith J J, Veenhuis M, Szilard R K, Rachubinski R A. The Yarrowia lipolytica gene PAY5 encodes a peroxisomal integral membrane protein homologous to the mammalian peroxisome assembly factor PAF-1. J Biol Chem. 1996;271:20300–20306. doi: 10.1074/jbc.271.34.20300. [DOI] [PubMed] [Google Scholar]
- 13.Elgersma Y. Transport of proteins and metabolites across the peroxisomal membrane in Saccharomyces cerevisiae. Ph.D. thesis. Amsterdam, The Netherlands: University of Amsterdam; 1995. [Google Scholar]
- 14.Fabre E, Tharaud C, Gaillardin C. Intracellular transit of a yeast protease is rescued by trans-complementation with its prodomain. J Biol Chem. 1992;267:15049–15055. [PubMed] [Google Scholar]
- 15.Fahimi H D, Baumgart E, Völkl A. Ultrastructural aspects of the biogenesis of peroxisomes in rat liver. Biochimie. 1993;75:201–208. doi: 10.1016/0300-9084(93)90078-7. [DOI] [PubMed] [Google Scholar]
- 16.Goodman J M, Trapp S B, Hwang H, Veenhuis M. Peroxisomes induced in Candida boidinii by methanol, oleic acid and d-alanine vary in metabolic function but share common integral membrane proteins. J Cell Sci. 1990;97:193–204. doi: 10.1242/jcs.97.1.193. [DOI] [PubMed] [Google Scholar]
- 17.He F, Yaver D, Beckerich J-M, Ogrydziak D, Gaillardin C. The yeast Yarrowia lipolytica has two, functional, signal recognition particle 7S RNA genes. Curr Genet. 1990;17:289–292. doi: 10.1007/BF00314874. [DOI] [PubMed] [Google Scholar]
- 18.Kaiser C A, Schekman R. Distinct sets of SEC genes govern transport vesicle formation and fusion early in the secretory pathway. Cell. 1990;61:723–733. doi: 10.1016/0092-8674(90)90483-u. [DOI] [PubMed] [Google Scholar]
- 19.Kyhse-Andersen J. Electroblotting of multiple gels: a simple apparatus without buffer tank for rapid transfer of proteins from polyacrylamide to nitrocellulose. J Biochem Biophys Methods. 1984;10:203–209. doi: 10.1016/0165-022x(84)90040-x. [DOI] [PubMed] [Google Scholar]
- 20.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 21.Lanzetta P A, Alvarez L J, Reinach P S, Candia O A. An improved assay for nanomole amounts of inorganic phosphate. Anal Biochem. 1979;100:95–97. doi: 10.1016/0003-2697(79)90115-5. [DOI] [PubMed] [Google Scholar]
- 22.Lazarow P B, Fujiki Y. Biogenesis of peroxisomes. Annu Rev Cell Biol. 1985;1:489–530. doi: 10.1146/annurev.cb.01.110185.002421. [DOI] [PubMed] [Google Scholar]
- 23.Lazarow P B, Moser H W. Disorders of peroxisome biogenesis. In: Scriver C R, Beaudet A L, Sly W S, Valle A D, editors. The metabolic basis of inherited disease. 7th ed. New York, N.Y: McGraw-Hill; 1994. pp. 2287–2324. [Google Scholar]
- 24.Lee I H, Ogrydziak D M. Yarrowia lipolytica SRP54 homolog and translocation of Kar2p. Yeast. 1997;13:499–513. doi: 10.1002/(SICI)1097-0061(199705)13:6<499::AID-YEA100>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 25.Lopez M C, Nicaud J-M, Skinner H B, Vergnolle C, Kader J C, Bankaitis V A, Gaillardin C. A phosphatidylinositol/phosphatidylcholine transfer protein is required for differentiation of the dimorphic yeast Yarrowia lipolytica from the yeast to the mycelial form. J Cell Biol. 1994;125:113–127. doi: 10.1083/jcb.125.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matoba S, Fukayama J, Wing R A, Ogrydziak D M. Intracellular precursors and secretion of alkaline extracellular protease of Yarrowia lipolytica. Mol Cell Biol. 1988;8:4904–4916. doi: 10.1128/mcb.8.11.4904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mellman I, Simons K. The Golgi complex: in vitro veritas? Cell. 1992;68:829–840. doi: 10.1016/0092-8674(92)90027-A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Novick P, Ferro S, Schekman R. Order of events in the yeast secretory pathway. Cell. 1981;25:461–469. doi: 10.1016/0092-8674(81)90064-7. [DOI] [PubMed] [Google Scholar]
- 29.Novick P, Field C, Schekman R. Identification of 23 complementation groups required for post-translational events in the yeast secretory pathway. Cell. 1980;21:205–215. doi: 10.1016/0092-8674(80)90128-2. [DOI] [PubMed] [Google Scholar]
- 30.Novick P, Schekman R. Secretion and cell-surface growth are blocked in a temperature-sensitive mutant of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1979;76:1858–1862. doi: 10.1073/pnas.76.4.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Novick P, Schekman R. Export of major cell surface proteins is blocked in yeast secretory mutants. J Cell Biol. 1983;96:541–547. doi: 10.1083/jcb.96.2.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nunnari J, Walter P. Regulation of organelle biogenesis. Cell. 1996;84:389–394. doi: 10.1016/s0092-8674(00)81283-0. [DOI] [PubMed] [Google Scholar]
- 33.Nuttley W M, Brade A M, Gaillardin C, Eitzen G A, Glover J R, Aitchison J D, Rachubinski R A. Rapid identification and characterization of peroxisomal assembly mutants in Yarrowia lipolytica. Yeast. 1993;9:507–517. [Google Scholar]
- 34.Nuttley W M, Brade A M, Eitzen G A, Veenhuis M, Aitchison J D, Szilard R K, Glover J R, Rachubinski R A. PAY4, a gene required for peroxisome assembly in the yeast Yarrowia lipolytica, encodes a novel member of a family of putative ATPases. J Biol Chem. 1994;269:556–566. [PubMed] [Google Scholar]
- 35.Ogrydziak D M, Cheng S-C, Scharf S J. Characterization of Saccharomycopsis lipolytica mutants producing lowered levels of alkaline extracellular protease. J Gen Microbiol. 1982;128:2271–2280. doi: 10.1099/00221287-128-6-1225. [DOI] [PubMed] [Google Scholar]
- 36.Palade G. Intracellular aspects of the process of protein synthesis. Science. 1975;189:347–358. doi: 10.1126/science.1096303. [DOI] [PubMed] [Google Scholar]
- 37.Pidoux A L, Armstrong J. Analysis of the BiP gene and identification of an ER retention signal in Schizosaccharomyces pombe. EMBO J. 1992;11:1583–1591. doi: 10.1002/j.1460-2075.1992.tb05203.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pryer N K, Wuestehube L J, Schekman R. Vesicle-mediated protein sorting. Annu Rev Biochem. 1992;61:471–516. doi: 10.1146/annurev.bi.61.070192.002351. [DOI] [PubMed] [Google Scholar]
- 39.Roberts C J, Raymond C K, Yamashiro C T, Stevens T H. Methods for studying the yeast vacuole. Methods Enzymol. 1991;194:644–661. doi: 10.1016/0076-6879(91)94047-g. [DOI] [PubMed] [Google Scholar]
- 40.Rose M D, Misra L M, Vogel J P. KAR2, a karyogamy gene, is the yeast homolog of the mammalian BiP/GRP78 gene. Cell. 1989;57:1211–1221. doi: 10.1016/0092-8674(89)90058-5. [DOI] [PubMed] [Google Scholar]
- 41.Rothman J E, Orci L. Molecular dissection of the secretory pathway. Nature (London) 1992;355:409–415. doi: 10.1038/355409a0. [DOI] [PubMed] [Google Scholar]
- 42.Subramani S. Protein import into peroxisomes and biogenesis of the organelle. Annu Rev Cell Biol. 1993;9:445–478. doi: 10.1146/annurev.cb.09.110193.002305. [DOI] [PubMed] [Google Scholar]
- 43.Szilard R K, Titorenko V I, Veenhuis M, Rachubinski R A. Pay32p of the yeast Yarrowia lipolytica is an intraperoxisomal component of the matrix protein translocation machinery. J Cell Biol. 1995;131:1453–1469. doi: 10.1083/jcb.131.6.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thieringer R, Shio H, Han Y S, Cohen G, Lazarow P B. Peroxisomes in Saccharomyces cerevisiae: immunofluorescence analysis and import of catalase A into isolated peroxisomes. Mol Cell Biol. 1991;11:510–522. doi: 10.1128/mcb.11.1.510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Titorenko V I, Eitzen G A, Rachubinski R A. Mutations in the PAY5 gene of the yeast Yarrowia lipolytica cause the accumulation of multiple subpopulations of peroxisomes. J Biol Chem. 1996;271:20307–20314. doi: 10.1074/jbc.271.34.20307. [DOI] [PubMed] [Google Scholar]
- 46.Titorenko V I, Ogrydziak D M, Rachubinski R A. Four distinct secretory pathways serve protein secretion, cell surface growth, and peroxisome biogenesis in the yeast Yarrowia lipolytica. Mol Cell Biol. 1997;17:5210–5226. doi: 10.1128/mcb.17.9.5210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Titorenko, V. I., and R. A. Rachubinski. Unpublished data.
- 48.Titorenko, V. I., and R. A. Rachubinski. Unpublished data.
- 49.Van den Bosch H, Schutgens R B H, Wanders R J A, Tager J M. Biochemistry of peroxisomes. Annu Rev Biochem. 1992;61:157–197. doi: 10.1146/annurev.bi.61.070192.001105. [DOI] [PubMed] [Google Scholar]
- 50.Walton P A, Gould S J, Rachubinski R A, Subramani S, Feramisco J R. Transport of microinjected alcohol oxidase from Pichia pastoris into vesicles in mammalian cells: involvement of the peroxisomal targeting signal. J Cell Biol. 1992;118:499–508. doi: 10.1083/jcb.118.3.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Waterham H R, Titorenko V I, Swaving G J, Harder W, Veenhuis M. Peroxisomes in the methylotrophic yeast Hansenula polymorpha do not necessarily derive from pre-existing organelles. EMBO J. 1993;12:4785–4794. doi: 10.1002/j.1460-2075.1993.tb06167.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wuestehube L J, Duden R, Eun A, Hamamoto S, Korn P, Ram R, Schekman R. New mutants of Saccharomyces cerevisiae affected in the transport of proteins from the endoplasmic reticulum to the Golgi complex. Genetics. 1996;142:393–406. doi: 10.1093/genetics/142.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]