Abstract
Active transcriptional repression has been characterized as a function of many regulatory factors. It facilitates combinatorial regulation of gene expression by allowing repressors to be dominant over activators under certain conditions. Here, we show that the Engrailed protein uses two distinct mechanisms to repress transcription. One activity is predominant under normal transient transfection assay conditions in cultured cells. A second activity is predominant in an in vivo active repression assay. The domain mediating the in vivo activity (eh1) is highly conserved throughout several classes of homeoproteins and interacts specifically with the Groucho corepressor. While eh1 shows only weak activity in transient transfections, much stronger activity is seen in culture when an integrated target gene is used. In this assay, the relative activities of different repression domains closely parallel those seen in vivo, with eh1 showing the predominant activity. Reducing the amounts of repressor and target gene in a transient transfection assay also increases the sensitivity of the assay to the Groucho interaction domain, albeit to a lesser extent. This suggests that it utilizes rate-limiting components that are relatively low in abundance. Since Groucho itself is abundant in these cells, the results suggest that a limiting component is recruited effectively by the repressor-corepressor complex only on integrated target genes.
Transcriptional repressors that can function at a distance, analogously to transcriptional activators, with separable DNA binding and effector domains, have been termed active repressors (18). Many higher eukaryotic transcription factors have been found to possess such activities (reviewed in references 13 and 23). One such protein that has been well-characterized both in cultured cells and in vivo is the product of the engrailed locus of Drosophila. The Engrailed protein (EN) contains a homeodomain (HD) related in DNA binding specificity to that of members of the Antennapedia class (3) but representing a separate, conserved class with two known members in both insects and mammals. Several members of the Antennapedia class have been shown to be transcriptional activators, including the fushi tarazu protein FTZ. FTZ is a strong, context-independent activator in cultured cells (16, 37) and participates in a direct positive feedback on its own gene in Drosophila embryos (10, 31, 39). By swapping HDs between FTZ and EN, it was shown that EN domains can confer a dominant negative activity on the FTZ HD, counteracting endogenous FTZ protein to generate a ftz mutant phenotype in embryos (21). Indications that this repression is active, rather than simply a disruption of binding by factors that normally interact with ftz, include the dominant repression of the endogenous en gene, another FTZ target in vivo, even in regions in which FTZ is not expressed, and the loss of repression of the endogenous ftz gene upon deletion of a portion of EN from the chimeric repressor that is also required for active repression in culture. This deleted protein, even though it is unable to repress endogenous ftz, still interacts with FTZ target sites in the ftz upstream enhancer, since it is still capable of repressing a transgene driven by this enhancer by competing for binding sites with the endogenous FTZ protein (21). Using a novel assay, we have confirmed this active repression by EN in vivo and have compared the domains required for repression in vivo with those required for active repression in culture. We find that the EN repression function is contributed by multiple domains in both assays but that different domains have different potencies in the two systems. One conserved region (eh1 [25]) is particularly important in vivo (32) but shows very little activity in standard active repression assays involving transient transfection of cultured cells (see reference 11; confirmed in this report). This region mediates interaction with the Groucho (GRO) corepressor. GRO is related to the yeast corepressor TUP1, which mediates active repression by the HD protein α2 (22), as well as to mammalian homologs of the transducin-like Enhancer of Split (TLE) family (35). GRO has been shown to be recruited to DNA by members of other DNA binding protein families, including the Hairy-related basic-helix-loop-helix (HLH) proteins (29) and Runt domain proteins (1). Two other repression domains (one immediately flanking the EN HD) are more potent in transient transfections of cultured cells than in vivo. The differences between their functional characteristics and those of eh1, which mediates the interaction with GRO, suggest that they utilize a distinct mechanism. This distinction appears to hinge on the integrated state of the target gene in vivo, since on integrated target genes in the same cultured cells, the relative potencies of different repression domains closely parallel those seen in vivo.
MATERIALS AND METHODS
Embryo preparation and staining.
P-element transformations (33), cuticle preparations (36), and in situ hybridization to fixed embryos (7) were performed essentially as described previously. Antibody staining was performed essentially as described elsewhere (28) with a polyclonal α-EN antiserum (a kind gift of Charles Girdham and Patrick O’Farrell) that had been prepared against full-length, partially purified, glutathione S-transferase (GST)-tagged EN and affinity purified against a His-tagged peptide with the N-terminal 150 amino acids of EN. Either alkaline phosphatase (AP) or peroxidase-coupled secondary antibodies (Vector Laboratories) were used both for microscopic examination of fixed embryos, for which either 5-bromo-4-chloro-3-indolylphosphate toluidinium (BCIP) and nitroblue tetrazolium (for AP) or 3,3′-diamino-benzidine (DAB) substrates were used for staining (Boehringer Mannheim), and for quantitation of antibody signals, for which the AP substrate p-nitrophenyl phosphate (Sigma) was used as described before (28). Incubation times were determined to be in the linear range of the assay by incubating sets of embryos with different signal intensities for various times.
Heat shocks were administered to embryos on 35-mm collection plates by floating the plates on 37°C water inside a sealed container in order to minimize evaporative cooling. Standard heat shock conditions employed a 15-min incubation followed by return to a 25°C humidified environment.
Transfections and Western blots.
Cell culture assays for passive and active repression were performed with Drosophila S2 cells as described before (18), with 2 μg of one of two target genes (T3N6D-33CatA and N6T3D-33CatB [18]) per 60-mm culture dish. Active repression assays with each of these target genes gave qualitatively similar results. The values shown in Fig. 2, 3, 5, and 6 were from transfections with the former plasmid. For active repression assays, 0.04 μg of pPAc-GR (38) was used to express the rat glucocorticoid receptor (GR). For passive repression assays, 0.3 μg of FTZ expression plasmid pPAc-ftz (16, 37) was used. Chloramphenicol acetyltransferase (CAT) assays, as well as β-galactosidase assays for expression of the cotransfected reference gene pLac82SU (5), were performed as described elsewhere (18). Cotransfected plasmids used to express EFE and its derivatives were the same as those used for P-element transformation (see below). See figure legends for additional details.
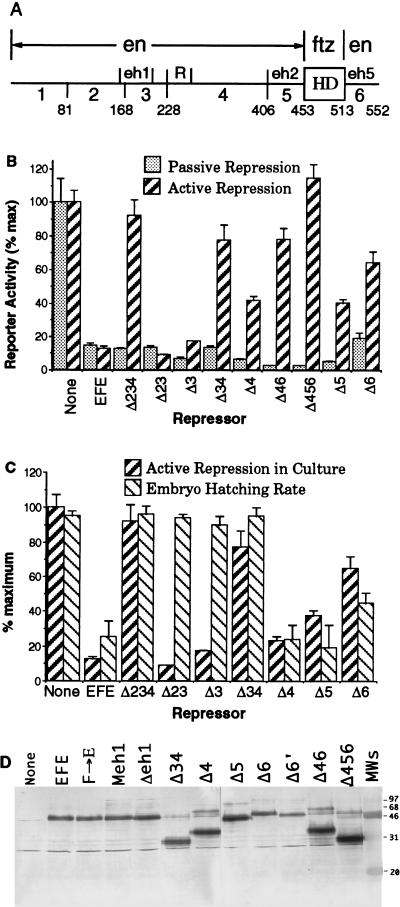
FIG. 2.
(A) Features of the EFE chimeric protein. The diagram indicates which portions of the coding sequence derive from EN and which derive from FTZ, our numerical designations of regions of EN (1 to 6 [not including the FTZ HD]), and the locations of known features within those regions (eh1, eh2, eh5, and R). eh1, eh2, and eh5 are peptide sequences found in all known EN homologs (25) from widely divergent species, including insects and mammals; eh1 is also similar to regions of other classes of HD proteins (32), and R is an autonomous active RD identified in cell culture studies (11). Homologies eh2 and eh5 are part of the conserved regions flanking the EN HD, which also include a sequence termed eh3 (immediately flanking the N terminus of the EN HD) that has been implicated in nuclear localization (16a) and thus was left intact in our analyses. Locations of region boundaries in the amino acid sequence are indicated at the bottom. Deletions and other alterations of these regions are described in detail in subsequent figures or in the text. (B) Repression by EFE and derivatives in cultured cells. Drosophila S2 cells were cotransfected with a CAT reporter plasmid, which contains binding sites for both the GR and the FTZ HD, separated by 40 bp, upstream of a basal promoter, and a plasmid that expresses either FTZ or GR (see Materials and Methods for details). Each of the latter two activate reporter expression by 50- to 100-fold above the basal level (shown as 100%). The ability of either EFE or the indicated derivatives to repress this activated transcription was determined by cotransfection of an appropriate expression plasmid. The same amount of a given expression plasmid was used in both repression assays, but the amounts were adjusted among the derivatives to give approximately equal levels of passive repression to allow a more accurate assessment of the potency for active repression. Thus, 4 μg of expression plasmid was used for Δ234, Δ3, and Δ6; 3 μg was used for EFE, Δ23, and Δ34; 1 μg was used for Δ46, Δ456, and Δ5; and 0.5 μg was used for Δ4. The nonrepressed level was determined by cotransfection of 3 μg of empty parental expression plasmid, which is a P-element transformation vector (see Materials and Methods). CAT activities were determined and normalized to the activities of a cotransfected reference gene (see Materials and Methods for details). The graph represents the averages and ranges for at least two independent transfections in at least two separate experiments. (C) Comparison between active repression in culture and hatching rates in vivo in response to EFE derivatives. Active repression was determined as described above, except that the amounts of expression plasmid for Δ4 and Δ5 were the same as that for EFE (3 μg). Hatching rates were determined for the wild-type recipient strain (none) and for transgenic lines expressing the indicated EFE derivatives following induction of expression by a 15-min heat pulse at 37°C between 2.5 and 3 h after egg deposition. Both hatched and unhatched egg casings were counted 28 h after egg deposition (hatching normally occurs at 24 h). Error bars indicate the ranges of values obtained with at least four collection plates (with at least 100 eggs per plate) in at least two separate experiments. Similar results were obtained with at least two independent homozygous insert lines for each derivative. Hatching rates in the absence of induction were higher than 95% for each line. (D) Western blot analysis of proteins from transfected cultures. Nuclear extracts of S2 cell cultures were transiently transfected with expression plasmid for the indicated EFE derivatives followed by PAGE, electroblotting, and immunodetection with polyclonal antiserum to the N-terminal region of EN (antiserum affinity-purified by using regions 1 and 2, which were contained within each of these derivatives). Cultures in 60-mm dishes that were 20% confluent were each transfected with 20 μg of expression plasmid and harvested 60 h later, and nuclear extracts were prepared as previously described (11). See Table 1 footnotes for a description of Δ6′.
FIG. 3.
Mutations in eh1 more strongly affect activity in vivo, while mutating eh5 (in Δ6) has a stronger effect in culture. Passive and active repression by EFE and derivatives and hatching rates of transgenic lines were determined as described in the legend to Fig. 2. In each case, hatching rate is an accurate reflection of the ability to repress endogenous ftz and generate ftz mutant cuticle patterns (see text). Δ6 is a 9-aa deletion, aa 523 to 531, within the conserved region flanking the EN HD (eh5 [Fig. 2A]). Δ4 removes the RD identified by Han and Manley (11), while Δ5 removes the conserved region N terminal to the HD. Note that mutating either regions 4 and 6 together or the three regions that contribute strongly to repression in cultured cells (Δ456) abolishes activity in culture, but not in vivo, whereas mutating eh1 has the converse effect.
FIG. 5.
Repression of integrated target genes in cultured Drosophila cells. A pool of S2 cells stably transfected with the same CAT-expressing reporter used in Fig. 2 and 3 (selected on 200 μg of hygromycin B per ml after cotransfection of reporter with the hygromycin-resistant gene expression plasmid pCop-hygro) were transiently transfected with the activator expression plasmid encoding GR (see Materials and Methods), either alone or with the indicated repressor expression plasmids. Parallel transfections with empty expression vector were used to determine the background of expression without activation, which was subtracted from the results shown. This background (B.G.) amounted to 50 to 80% of the maximum activity, which is the activity with GR alone. (A) EFE and derivatives (with the FTZ HD) were transfected in parallel cultures. Each received 0.1 μg of GR plasmid, 5 μg of the indicated repressor expression plasmid, and 0.5 ng of the reference gene. Values given were normalized to the amount of CAT activity (divided by reference gene activity) with activator, but with empty repressor expression vector (pCaSpeR-hs), which is shown as 100%. The averages and ranges of two independent transfections are shown. Similar results were obtained for four additional independent transfections in two separate experiments. (B) EN and derivatives (with the EN HD) were transfected in parallel cultures as described for panel A, and expression levels were normalized to the level with activator alone, as in panel A. Similar results were obtained in four additional independent transfections in two separate experiments, each with a different pool of stably transfected S2 cells.
FIG. 6.
Transient transfections with low amounts of reporter and repressor plasmids. S2 cells were transfected as described in the legend to Fig. 2, except that reporter plasmid was reduced by 4-fold to 0.5 μg per 60-mm culture dish, activator plasmid was reduced by 5-fold to 8 ng, and repressor expression plasmids (for EN and EN derivatives) were 12-fold lower (0.4 μg). Total DNA was reduced by 2-fold to 5 μg per dish. The averages and ranges of two independent transfections for each plasmid, normalized to the activity of a cotransfected reference gene and to the activated level without repressor (shown as 100%, corresponding to 18-fold activation above the nonactivated level), are shown. Similar results were obtained in four independent transfections in two additional experiments, one using 0.2 μg of each repressor expression plasmid.
Western blots were performed on nuclear extracts of transiently transfected S2 cells, as previously described (11), except that 60-mm culture dishes were transfected with 20 μg of each expression plasmid, and the polyclonal α-EN antibody preparation described above was used.
Plasmid constructions and Drosophila strains.
Expression plasmids for EFE derivatives were modifications of a P-element transformation vector capable of providing inducible expression of EFE in transformed Drosophila from a heat shock promoter, as described by John et al. (21). Modifications were made using either PCR-based methods (for Δ5 and Δ6), synthetic DNA adaptors to create deletions adjacent to unique restriction sites (for Δ234, Δ23, Δ34, Δ3, and Δ4), or a combination of the two (Δeh1, F→E, and Meh1). Resulting deletion end points and amino acid substitutions are described in figure legends and the text. All regions containing synthetic or PCR-synthesized DNA were subsequently sequenced (automated) to confirm the expected structure. Appropriate restriction fragments were combined to generate the combined deletion plasmids Δ46 and Δ456. Details are available on request. These plasmids were introduced into flies using standard methodologies (33). Homozygous viable insertions on either the second or third chromosome were used in all analyses of repression activity. Additional details are either contained in figure legends or text or are available on request.
Yeast two-hybrid system and in vitro interaction assays.
A Drosophila embryonic library (39) in pACT (6) was screened with an EN clone in pAS2 (14) encoding amino acids (aa) 1 to 349 in frame with the Gal4 DNA binding domain as bait. After transformation of Saccharomyces cerevisiae Y190 (14) with bait and library plasmids, 2 × 106 cells viable on synthetic medium lacking Leu and Trp (DOBA -Leu -Trp; Clontech [with both plasmids]) were plated at a density of 300/cm2 onto DOBA medium (-Leu -Trp -His) with 30 mM 3-aminotriazole, grown at 30°C until single colonies were visible, replica plated onto DOBA medium (-Leu -Trp), and grown overnight, and replicas were transferred to filters. Cells on the filters were permeabilized by freeze-thaw and were stained for β-galactosidase activity. Positive colonies were restreaked and tested for expression of β-galactosidase. Plasmids were isolated from positive colonies and tested by cotransformation against several negative control bait plasmids, and the original interaction was verified. Clones surviving all tests were grouped by partial sequencing and restriction mapping.
GST fusion proteins were expressed in Escherichia coli DH5α with pGEX-5x-1 (Pharmacia) and were purified over glutathione-agarose columns. Equal amounts of each (based on Coomassie staining of sodium dodecyl sulfate-polyacrylamide gel electrophoresis [SDS-PAGE] gels) were mixed with 35S-labeled GRO synthesized with pET15-b (Novagen) and the TNT-coupled rabbit reticulocyte lysate system (Promega) in binding buffer (20 mM HEPES [pH 7.9], 50 mM KCl, 2.5 mM MgCl, 10% glycerol, 1 mM dithiothreitol, 0.2% Nonidet P-40 [NP-40], 2.5 mM phenylmethylsulfonyl fluoride); the mixture was rolled overnight at 4°C, centrifuged, and washed four times with 1 ml of modified radioimmunoprecipitation assay (RIPA) buffer (10 mM Tris · HCl [pH 7.5], 250 mM NaCl, 1 mM EDTA, 0.2% NP-40); and the retained material was analyzed by SDS-PAGE and autoradiography. Parallel incubations of GRO protein in binding buffer and an aliquot of this in modified RIPA buffer showed no indication of degradation.
RESULTS
An in vivo assay for active repression.
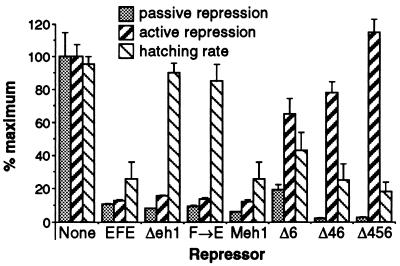
We sought to determine whether the requirements for active repression by EN, as defined in transient transfection assays with cultured cells, were substantially the same as or different from the activities required for repression of endogenous genes in vivo. Previous studies suggested that repression of the endogenous ftz gene by a chimeric repressor termed EFE (EN with its HD replaced by that of FTZ) was analogous to active repression in culture, principally because the activity required a region of the protein well separated from the HD in the primary sequence (32). To validate the use of this assay in a detailed comparison of repression activities, we wanted to test definitively whether this in vivo assay involved active repression, rather than simply a competition for binding sites with an activator. The assay that we developed does not depend on the modularity of repression and targeting domains. Previous studies showed that the EFE derivative F→E (which carries a single amino acid substitution in the conserved eh1 repression domain) had lost most of its ability to repress the endogenous ftz gene. With the new assay, we asked whether F→E can act as an activator when it is competing with the fully active EFE for sites in vivo. This assay can distinguish whether F→E has lost active repression function per se or simply the ability to compete for binding sites. If it had lost only DNA binding ability, then producing F→E in combination with EFE, even if it were still able to partially displace EFE, would not prevent repression but instead would have no effect or might augment repression, if the total occupancy of the site increased.
We expressed EFE from a transgene by heat induction, and in a parallel line expressed both EFE and F→E. The effects on ftz repression and on the developmental consequences of ftz repression were assessed. If F→E could actively repress the ftz gene when bound, but bound poorly, we would expect to see an increase in ftz repression. However, if it were able to displace EFE from target sites but failed to repress, we might expect to see a reduction in repression. Indeed, we saw a significant decrease in ftz repression on a population average basis. However, the range of phenotypes obtained did overlap (Fig. 1 and data not shown). Therefore, we assessed the degree of relief of repression by quantifying the consequences for pattern formation. We categorized pattern defects in the larval cuticle at the end of embryogenesis as either less severe than, equally severe as, or more severe than the ftz pair-rule mutant phenotype. Coexpression of F→E with EFE significantly reduced the percentage of embryos showing severe pattern defects (either pair-rule or stronger), relative to that produced by EFE alone (Fig. 1). This was verified by analyzing two different fly lines containing the EFE and F→E transgenes (Fig. 1d). To determine whether the two transgenes were expressed independently, we performed Western blot analysis on nuclear extracts from embryos. Using an antiserum that recognizes the N-terminal region of EN (which is shared by the two proteins), we observed a twofold increase in staining intensity in the doubly transgenic lines relative to the single transgenic lines (not shown). Thus, coexpression of F→E with EFE can abrogate the effects of EFE, even though both can compete for FTZ binding to the endogenous ftz gene (21). This shows that EFE requires a strong active repression function to repress ftz in this assay.
FIG. 1.
Passive activation by F→E in vivo. F→E is a derivative of the EN-FTZ chimera EFE, which carries a single amino acid change in the conserved eh1 repression domain (see text). Passive activation refers to the relief of repression by F→E when it competes with the active repressor EFE for target sites. Transgenic lines were heat pulsed for 6 min at 37°C, between 2 h and 40 min and 2 h and 46 min after the end of a 15-min collection. (a) Recipient strain showing the normal pattern of endogenous ftz gene expression; (b) transgenic embryo carrying a heat-inducible EFE transgene; (c) transgenic embryo carrying both the same EFE transgene and, on a separate chromosome, an inducible transgene encoding the point-mutated derivative EFE-F→E. (a to c) Embryos from each line were heat shocked and stained in parallel for endogenous ftz RNA by in situ hybridization as previously described (32). The probe does not detect the ftz HD sequence contained in the EFE transgenes. Representative embryos from each strain are shown (see text). (d) Hatching rates were determined, cuticles were prepared 28 h later, and the severity of pattern defects was assessed for lines carrying an EFE transgene insert on chromosome III (EFE3), either without or with an EFE-F→E insert on chromosome II (F→E2), or carrying an EFE transgene on the second chromosome (EFE2), either without or with EFE-F→E on the third chromosome (F→E3). Embryos showing a pair-rule pattern of defects in the ventral denticle bands, those showing more severe defects than the pair-rule pattern, and those showing less severe defects were each counted. Very few embryos showed ambiguities between different regions, consistent with pre- vious studies (21) which showed that ftz-dependent pattern elements are deleted preferentially in response to EFE induction, resulting in mostly pair-rule deletions. The percentage of cuticles showing severe (pair-rule or more) defects was multiplied by the fraction that had failed to hatch, and the results are shown as percentages of severe pattern defects. This assumes that all hatched embryos had less severe defects, as previously determined by analyzing hatched larval cuticles (data not shown). Values shown are the averages and ranges from at least two separate experiments with at least 120 embryos per experiment.
Repression activity in cultured cells is determined by multiple EN domains.
Previous results showed that EFE, like EN, was capable of repressing transcription in cultured cells independent of the context of its binding sites. Specifically, both basal-level transcription of various promoters and transcription activated by a variety of activators are effectively repressed, even when binding sites for the activator and repressor are separated by more than 400 bp (11, 17, 18). Repression occurring over a distance, which depends on specific binding sites in the target gene, as well as on an activity of the repressor functionally separable from DNA binding activity, was termed active repression. In contrast, passive repression, wherein the repressor directly competes with activators for binding sites, requires only a DNA binding domain, such as the HD (18).
We compared EFE derivatives for their abilities to both passively and actively repress transcription in cultured cells (Fig. 2B). These transient transfections utilize a reporter gene previously described (18), which can be activated either from consensus HD binding sites, to which both the FTZ and EN HDs bind effectively in vitro, or from separate sites, by the GR. When the reporter gene is activated by FTZ, repression can occur by a purely passive mechanism, but when the activator is the GR, active repression domains (RDs) are absolutely required for repression (18). Thus, passive repression in culture is a measure of total DNA binding activity and, indirectly, of protein levels (see below) and serves as an internal control for comparing the intrinsic active repression activities of different derivatives.
Deletion of various domains of EFE, either alone or in combination, resulted in proteins that can passively repress to different degrees (Fig. 2B). This reflects their ability to compete for FTZ binding sites in the cells. In fact, several derivatives, i.e., Δ4, Δ46, Δ456, and Δ5, passively repressed this FTZ-activated expression better than EFE. Western blots of nuclear extracts from transiently transfected cultures showed protein levels that closely paralleled passive repression activity (Fig. 2D; Table 1). Thus, the differences in passive repression activity can be accounted for by changes in protein stability. Δ4, Δ46, Δ456, and Δ5 all showed increased expression levels relative to EFE, while Δ3 and Δ34 showed similar levels (Δ34 showed a slight increase) (Fig. 2D and Table 1 footnotes), and Δ6 gave a somewhat reduced level. This comparison supports the idea that deletions within EN-derived regions of EFE do not significantly affect the binding activity of the FTZ HD in the cultured cells and that all of these derivatives bind to the consensus sites in cultured cells with equal affinity. All derivatives shown retain the FTZ HD and a nuclear localization signal from EN (see the legend to Fig. 2A). Previous results showed that an HD capable of binding to the consensus HD binding sites in the reporter gene was required for activity in this assay and that a deletion derivative in which part of region 1 was removed failed to repress, probably due to its being a highly unstable protein (16a, 18).
TABLE 1.
Summary of results with a series of deletion derivatives of EFEa
| EFE derivative | % Lethality (with 15-min heat shock)b | Ability to produce ftz mutant cuticlec | Repression of endogenous ftz gened | Level of protein 10 min after heat shocke | Apparent protein stability in vivof | Passive repression (in cell culture)g | Protein levels in cell cultureh | Active repression (in cell culture)i
|
|
|---|---|---|---|---|---|---|---|---|---|
| With same amt of expression plasmid | With equal levels of passive repression | ||||||||
| EFE | >70 | +++ | +++ | +++ | + | ++ | ++ | +++ | +++ |
| Δ234 | <10 | − | − | +++m | ++ | ++ | ND | +/− | +/− |
| Δ23 | 10–20 | +/−j | + | ++m | ++ | ++ | ND | +++ | +++ |
| Δ3 | 5–15 | +/− | +/− | +++ | + | ++ | ++ | +++ | +++ |
| Δ34 | <10 | − | − | +++ | +++ | ++ | +++ | +/− | +/− |
| Δ4 | >70 | ++ | +++l | +++ | ++ | ++++ | ++++ | +++ | + |
| Δ46 | >60 | + | ++ | +++ | ++ | +++ | ++++ | ++ | +/− |
| Δ5 | >70 | ++ | +++l | +++ | + | +++ | +++ | +++ | + |
| Δ6 | 50–70 | ++ | ++ | +++ | + | ++ | ++ | + | ++ |
| Δ6′ | 10–30 | +/−k | +/− | +++ | + | ++ | ++ | +/− | + |
Deletion end points are given in Fig. 2A, except for Δ46, which combines the deletions of Δ4 and Δ6, and Δ6′, which is an almost complete deletion of region 6 (aa 517 to 552 are replaced by the six foreign C-terminal aa IRWHCS). Repression levels were confirmed in two parallel transfections in at least two independent experiments with at least two different plasmid preparations for each derivative. For embryo assays (the first five columns), consistent results were obtained with at least two independent transformant lines for each derivative, except Δ6′. Since similar results were obtained with Δ6 and Δ6′, we did not analyze Δ6′ further. See Fig. 3.
Lethality (embryos not hatching) was determined as described in the legend for Fig. 2C. Control wild-type embryos heat shocked in parallel showed less than 10% lethality, which non-heat-shocked controls indicated was mostly due to unfertilized eggs. Non-heat-shocked embryos from each derivative also showed hatching rates higher than 90%.
Summary of data from cuticle preparations of similarly heat-shocked embryos prepared 20 h after egg laying. Note that this is a stronger heat shock than that shown in Fig. 1. +++ indicates more than 70% of embryos having preferential deletions of ftz-dependent parts of the ventral denticle bands, including the following categories: pair-rule deletions of ftz-dependent bands, 20 to 30%; deletions of a subset of the ftz-dependent bands, 20 to 30%; and deletions of all of the ftz-dependent bands plus some additional bands, 15 to 30%. Totals of 10 to 20% of these cuticles showed normal patterns, most likely due to embryos that were partially developed at the time of egg laying, and therefore escaped the effects of EFE (see reference 21 and below). ++ indicates preferential deletion of ftz-dependent bands in about 60% of cuticles (with the appearance of 10 to 20% of cuticles with defects in non-ftz-dependent bands without complete deletion of ftz-dependent bands), indicating a loss of specificity (see text). + indicates preferential deletions of ftz-dependent bands in 10 to 50% of cuticles. Both heat-shocked, wild-type controls and each of the transformant lines showed deletion of either A2 or A4 in about 3% of cuticles, in addition to the defects described above.
Comparison of the abilities of these derivatives to repress endogenous ftz expression, analyzed in preparations similar to those shown in Fig. 1. +++, strong repression in more than 50% of embryos and clear repression in more than 70%; ++, clear repression in 40 to 60% of embryos; +, clear repression in 10 to 30% of embryos; +/−, 10% or fewer embryos apparently repressed; −, no apparent activity, except a weak, transient reduction in ftz RNA levels.
Apparent levels of protein produced following a 15-min heat induction, as indicated by quantitation of the signal following α-EN staining, as described by Smith and Jaynes (32). +++, an initial signal 10 min after heat shock of between 70 and 120% of that of EFE, which parallel staining of wild-type embryos showed to be two- to threefold higher than that due to endogenous EN expression (in older embryos in which EN is fully induced) when normalized to the fraction of expressing cells; ++, a signal estimated to be about 50% of that due to EFE.
Summary of data derived from α-EN staining at three time points after induction, i.e., the initial one summarized above as well as 30 and 60 min later. These data indicate apparent protein half-lives of 20 to 30 min (+), 30 to 50 min (++), and >60 min (+++). Since each transgene-derived RNA decays rapidly following induction (within about 15 min), these estimates should closely parallel the actual protein half-lives.
Each derivative is capable of passively repressing FTZ-activated reporter gene expression (assayed as described in the legend to Fig. 2, with 3 μg of each producer plasmid). ++, ++, +++, and ++++, repression to levels between 15 and 30%, 5 and 15%, 5 and 2%, and <2%, respectively, of the FTZ-induced level in the absence of repressor. Control experiments with increasing amounts of producer plasmid have shown that in this range of repression, the percentage of repression is approximately linear with regard to the amount of repressor present (16a).
Summary of the results of Western blot analysis following transient transfection of S2 cells with equal amounts of each producer plasmid (as described in the legend to Fig. 3, except with 20 μg of each plasmid). ++++, a 2.5- to 4-fold increase over EFE; +++, a 1.5- to 2.5-fold increase; ++, an increase of between 1.0 and 1.5 times the EFE level; ++, a 1.5-fold reduction relative to EFE; ND, no determination. The small discrepancies seen with Δ34 and Δ46 between relative protein levels and passive repression in culture suggest that the ability to repress FTZ-activated expression has a small but measurable dependence on active repression function, in addition to its strong dependence on the occupancy of FTZ binding sites in the reporter.
Active repression with either equal amounts of producer or amounts adjusted to give equal levels of passive repression (as described in the legend to Fig. 2). +++, repression to less than 15% of the unrepressed level of GR-activated expression; ++, repression to 15 to 40% of this level; ++, repression to 40 to 50% of this level; +, repression to 50 to 70% of this level; +/−, more than 70% reporter activity.
30-min heat shock severely reduces hatching rate relative to that of wild type but still produces no ftz mutant cuticles.
30-min heat shock reduces hatching rate relative to that of wild type and produces some ftz mutant cuticles.
Pattern of ftz repression altered to preferentially affect stripes most affected by ablating ftz RNA.
Levels estimated based on antigenicity of that portion of EN used to affinity purify antiserum still present.
In active repression assays, potency was reduced by deletions in either region 4, 5, or 6. These assays utilized the same reporter gene, in this case activated by a heterologous activator (the rat GR) through separate binding sites. Previous work had shown that activation by GR depends on the GR binding sites and that repression in this assay satisfies the above criteria for active repression (18). For Δ4 and Δ5, which showed stronger passive repression than EFE when equal amounts of expression plasmid were used for transfection, the levels of active repression were about equal to that of EFE (using equal amounts of expression plasmid). In order to test their potency for active repression, therefore, we reduced the amount of expression plasmid (Fig. 2B) to compensate for the apparent increase in binding site occupancy. Under the conditions used, the degree of passive repression was still greater than that with EFE, but the degree of active repression was significantly less (Fig. 2B). The levels of expression plasmid used for the other derivatives that gave stronger passive repression, Δ46 and Δ456, were also reduced for comparison (Fig. 2B), although their loss of potency was seen even without reducing their levels (data not shown). Region 3 appears to contribute slightly to active repression, since a small but reproducible reduction in activity was seen for Δ3, and since Δ34 had lost more activity than Δ4 alone. The loss of activity of derivatives that remove region 4 is consistent with previous results that localized an active RD to the N-terminal portion of that region (11). However, Δ4 retains considerable active repression activity. This additional activity can be attributed to three other regions, mostly to the conserved sequences that normally flank the EN HD, i.e., those deleted in Δ5 and Δ6 (Fig. 2B), and a barely detectable activity can be attributed to region 3.
Multiple domains also contribute to active repression in vivo but have different relative potencies.
We assessed the activity of EFE derivatives in vivo by multiple criteria. A set of transgenic flies were utilized (32), each expressing a deletion derivative of EFE from a heat-inducible promoter. A brief heat pulse induces ubiquitous expression from the transgene. Such expression of EFE causes rapid and persistent loss of ftz expression in the trunk region (Fig. 1) (32). Repression of ftz and other FTZ target genes results in the generation of pair-rule deletions in the cuticle pattern at the end of embryogenesis that mimic those seen in ftz mutants (21). Such heat treatment had no effect on endogenous ftz expression in wild-type embryos (Fig. 1A) (21). In testing derivatives from which EN-derived portions of EFE were deleted, we discovered that multiple regions contribute to activity. In addition to examining their ability to generate a ftz mutant phenotype at the end of embryogenesis (Table 1), we looked at their general ability to disrupt development of embryos when ectopically expressed. For this, we tested the ability of transgenic embryos to hatch following induction. For each derivative, we found a close correlation between its ability to repress the endogenous ftz gene and its ability to prevent hatching (Table 1; Fig. 2C). We also found that each derivative that prevented hatching caused preferential deletion of ftz-dependent pattern elements (Table 1) (see reference 21 for details of the developmental effects of EFE). However, Δ4 and Δ5, which repress ftz more strongly than EFE, appear to have lost some specificity in vivo, since they also caused a higher incidence of other defects (results summarized in Table 1).
The ability of EFE to cause an ftz mutant phenotype and to prevent hatching, like active repression in culture, depends not only on the HD but also on domains of EN outside the HD. In order to directly compare the in vivo and cell culture activities of EFE derivatives, we used hatching rates as a quantitative measure of in vivo activity. As stated above, this provides an accurate representation of the relative ability to repress endogenous ftz. When this is compared side by side with the ability to actively repress in culture (Fig. 2C), we see a generally good correlation, with one notable exception. Removing region 3 (or regions 2 and 3 together) has a much greater impact on activity in vivo than in culture. (If region 4 is additionally removed, in Δ234 and Δ34, activity is lost in both assays.) Region 3 contains a well-conserved motif previously noted in all known EN class homeoproteins (25) and more recently found to be shared with several other classes of homeoproteins (32). The active repression values used in this comparison differ from those of Fig. 2B in that the amounts of expression plasmid were not reduced for Δ4 and Δ5, but were equal to those used for EFE. Since the induction protocol in vivo was the same for all derivatives, this provides a more direct comparison of protein efficacies between the two assays. Although the correspondence in activity between the cell culture and in vivo assays breaks down for Δ3 and Δ23, the overall correlation between active repression in culture and ftz repression in vivo is much closer than that between passive repression in culture and ftz repression in vivo (compare Fig. 2C with Fig. 2B). This confirms our conclusion (Fig. 1) that EFE activity in vivo is dependent on its active repression function.
Each of the deletions that remove region 3 caused substantial loss of activity (Fig. 2C). However, the overall level of ftz expression was still noticeably repressed by Δ3, and the remaining stripes were often discontinuous either laterally or dorsally (Table 1; data not shown). The additional deletion of region 2 resulted in no additional repression of ftz but caused an increase in nonspecific defects (Table 1), suggesting that it may have acquired neomorphic activity. In contrast, additional deletion of region 4 caused additional loss of activity, to the point that Δ34 produced no ftz mutant cuticles (Table 1) (see reference 21 for a description of a mild, transient effect of Δ34). Nonetheless, Δ34, as described above, is still able to reduce the activity of the ftz upstream enhancer, indicating that it retains targeting activity in vivo. In contrast, deletion of either region 4 or 5 alone resulted in an increase in repression activity (Table 1) (note that Δ5 is a partial deletion of region 5 [aa 407 to 440]). In the case of Δ4, this is attributable to increased protein stability, which apparently masks a loss of potency, since deletion of region 4 in addition to region 3, or in addition to regions 2 and 3, causes a clear loss of activity. In fact, deleting region 4 alone causes nonspecific defects (Table 1), suggesting that the protein level is high enough to cause interaction with target genes other than ftz. In addition, when the strength of transgene induction was reduced to yield a level of ftz repression similar to that caused by EFE, nonspecific defects persisted, indicating that higher levels of Δ4 are required to give the same amount of repression, relative to EFE, again suggesting a loss of potency in active repression (Table 1; data not shown). (In the case of Δ5, the situation is more complex; see below and Discussion.) In region 6, a deletion of the most conserved 9 aa within the EN C-terminal tail caused a partial loss of repression activity (Fig. 2; Table 1 [for simplicity, we refer to this directed deletion as Δ6]). Thus, three regions can be seen to contribute specifically to repression activity in embryos, i.e., regions 3, 4, and 6, with region 3 being the most essential for strong activity. In contrast, only two of these, regions 4 and 6, contribute strongly to activity in transient assays in culture.
The eh1 homology mediates repression in vivo but not in transient transfections of cultured cells.
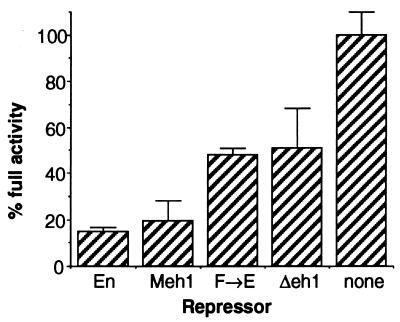
The clear difference in potency of Δ3 between the in vivo and cell culture assays is striking. The ftz repression activity of region 3 was previously attributed (32) to the engrailed homology region eh1 (25). In order to test whether eh1 is required for repression by EFE in cultured cells, we tested both a small deletion within eh1, and a single point mutant at the most conserved position (see reference 32 for a description of the conservation). Both a 15-aa deletion removing the most conserved portion of eh1 and a change of the invariant Phe to Glu (F→E [used in the experiments depicted in Fig. 1]) resulted in derivatives of EFE with strongly reduced abilities to generate the ftz mutant phenotype (Table 1) and to prevent hatching (Fig. 3). The levels of ftz RNA are reduced only slightly relative to that of the wild type following induction of each of these derivatives (32). Thus, each of these changes in eh1 had an effect on EFE activity indistinguishable from that of removing region 3 entirely. To determine whether the conservation of this region from flies to mammals had preserved function, we tested a substitution of the 15-aa region of the Drosophila protein with the corresponding region from the mouse EN1 protein. This resulted in four nonconservative and three neutral substitutions and one conservative substitution within the region. This replacement fully restored the ability of EFE to prevent hatching (Fig. 3 [Meh1]) and to produce an ftz mutant cuticle pattern in Drosophila embryos (data not shown), indicating that the function required for this activity, presumably active repression, is conserved. In contrast to the drastic effect of mutating region 3, combining two deletions that each reduce active repression in culture, Δ4 and Δ6, resulted in a protein (Δ46) with strong repression activity in vivo (Fig. 3 and data not shown) (see also reference 32). Previously, examination of protein levels produced in embryos showed that for those mutated in region 3, the less active proteins were produced at slightly higher levels than were the more active ones, while all were about equally stable (32). For Δ46, the levels were slightly higher initially, and the protein was considerably more stable than EFE, perhaps contributing significantly to its activity. However, Δ46 is less stable than Δ34 (32), but nonetheless has much greater activity in vivo (compare Fig. 2C and Fig. 3), consistent with the strong in vivo activity of region 3. Western blot analysis of extracts from transfected cultures (Fig. 2D) showed that, as found for Δ46 and other EFE derivatives (described above), passive repression activities parallel the levels of protein for the derivatives shown in Fig. 3. Thus, Δeh1, F→E, and Meh1 showed levels of protein indistinguishable from that of EFE, while Δ46 and Δ456 showed increased levels (two- to threefold) and Δ6 showed slightly decreased levels (about twofold [data not shown]). Hatching rate was found to accurately parallel the ability of each derivative to generate an ftz mutant phenotype. In addition, all derivatives were localized to nuclei (data not shown). Thus, eh1 mediates the in vivo activity of EFE, while removal of other regions that effect active repression in culture have a less dramatic impact on activity in vivo. Strikingly, as with deletion of region 3, these mutations fail to strongly affect the repression activity of EFE in transient transfections of cultured cells. This series of comparisons clearly shows that while the active repression function of EFE is required for its function in both assays, different domains of EN are responsible for the predominant repression activity in each case. Thus, these different domains, exemplified by eh1 on the one hand and regions 4 and 6 on the other, are likely to function by distinct mechanisms.
eh1 is required for interaction with the corepressor GRO.
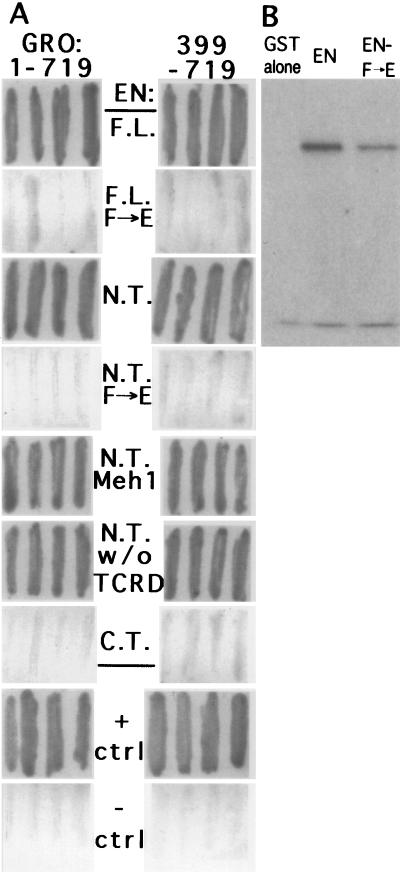
Using as bait an N-terminal fragment of EN (aa 1 to 350) that contains both eh1 and the cell culture RD of region 4, we screened a yeast expression library for interacting proteins using a two-hybrid system (6). After carrying out several tests for specificity and grouping the clones by partial sequencing and restriction mapping, we obtained (from 2 × 106 initial colonies) clones representing 38 distinct cDNAs. We specifically looked for candidate eh1 region interactors by rescreening each group of clones with the same N-terminal EN region but containing the F→E mutation. Only one showed a significant reduction in interaction intensity with the point mutant, and, in this case, the interaction was essentially abolished (Fig. 4A). This group, represented by four identical isolates, encoded the C-terminal conserved (WD40 repeat) region of GRO. To further test the specificity of interaction between EN and GRO, we removed the region 4 RD from the N-terminal clone of EN. This resulted in no apparent reduction in the interaction (Fig. 4A). Full-length EN also interacted strongly with the C terminus of GRO, and, conversely, full-length GRO interacted strongly with both the N-terminal region of EN and full-length EN. In each case, the interaction was virtually abolished by the point mutant F→E (Fig. 4A). Strong interaction was restored (Fig. 4A) by substituting the eh1 region from the mouse EN1 protein (25). Thus, the requirements for interaction with GRO in this system are the same as the requirements in vivo for the repression activity of the eh1 region.
FIG. 4.
(A) Interaction of EN and GRO in yeast. Using a two-hybrid system, we tested the abilities of several EN regions to interact with either full-length GRO (aa 1 to 719) or the GRO WD40 repeat region (aa 399 to 719). The EN derivatives used as bait (fused to the GAL4 DBD) are indicated between the panels. F.L., full length (aa 1 to 552); N.T., N-terminal region (aa 1 to 348); F→E, point-mutated derivatives in which the invariant Phe (aa 175) in eh1 was changed to Glu; Meh1, the 15-aa core of the eh1 homology (aa 172 to 186) in the Drosophila protein replaced by the homologous region from the mouse EN1 protein; w/o TCRD (aa 1 to 227), the cell culture RD removed from the N-terminal region; C.T., the C-terminal region of EN (aa 348 to 552); + ctrl, positive control, i.e., mouse p53 as bait interacting with simian virus 40 large T antigen (both sides); − ctrl, negative control, i.e., mouse p53 and GRO (aa 1 to 719 on the left or 399 to 719 on the right). (B) GST-EN interacts with the GRO WD40 repeats in vitro. The EN N-terminal region (aa 1 to 348, without and with the F→E point mutation) fused in frame with GST was produced in E. coli, purified via the GST tag, and mixed with in vitro-translated GRO (aa 399 to 719). Following incubation with glutathione-agarose beads, centrifugation, washing, elution (see Materials and Methods), and SDS-PAGE, interacting proteins were visualized by autoradiography. The lower band present also in the GST alone lane is seen even without programming the system with GRO-encoding DNA (not shown).
To test whether this interaction is the result of a direct EN-GRO dimerization, we fused the EN N-terminal region with GST. GST-EN, or GST-EN(F→E), was mixed with in vitro-translated GRO (aa 399 to 719) labeled with [35S]methionine. Following pulldown of GST with glutathione-agarose beads, elution, and SDS-PAGE, labeled peptides were visualized by autoradiography. A highly specific interaction was seen between EN and GRO, since no detectable GRO was captured by GST alone, while the F→E point mutation in the eh1 region strongly reduced the interaction (Fig. 4B). The residual interaction that remains with F→E suggests the possibility that other sequences in the N-terminal region of EN contribute to the interaction with GRO. However, the other repression domains do not appear to contribute, since the strength of the interaction in yeast cells is not reduced when they are removed (Fig. 4A).
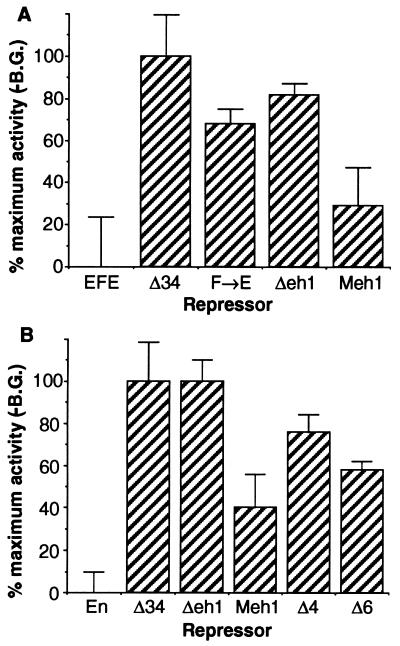
Stably integrated target genes respond to eh1 in cultured cells.
A number of possibilities are suggested by the differences in relative potencies of different RDs when the in vivo and transient transfection assays are compared. For example, a corepressor required for eh1 function in vivo might be missing in S2 cells. However, GRO is present in abundance in these cells (8). Alternatively, if the function of eh1 in vivo requires a normal chromatin environment, the chromatin state of the target gene might be sufficiently different in transient transfections to preclude its function. To attempt to distinguish between these possibilities, we tested whether we could see a more stringent requirement for the eh1 region if the target gene in the cell culture assay were integrated stably into the genome. To this end, we established stably transformed populations of S2 cells containing the same target gene used in the previous transient assays (Fig. 5). We then transfected these cells with activator plasmid encoding GR, along with each of the EFE derivatives shown in Fig. 5A. In sharp contrast to the results with transiently transfected target genes, the repression activity now showed a strong dependence on the eh1 homology region. Rather than causing a reduction of 10% or less in active repression activity (Fig. 3), the point mutation Phe-to-Glu in this region (F→E in Fig. 5A) caused a 70% loss of activity. In addition, replacing Drosophila eh1 with the corresponding mouse EN1 region clearly restored activity (Meh1 in Fig. 5A), rather than having an unmeasurable effect, as it did in transient transfection assays in the cells (Fig. 3). As in the transient transfections, Δ34 had little or no activity in this active repression assay. To confirm and extend these results to the normal EN protein (with its native HD), we transfected the stably transformed cells with the EN derivatives shown in Fig. 5B. Here again, removing eh1 caused a precipitous loss of activity, in contrast to the standard transient transfection assay, in which its removal had no discernible effect (data not shown). Consistently, replacing the Drosophila sequence with the mouse homologous region again restored much of the activity (Fig. 5B [Meh1]). Direct comparison with the Δ4 and Δ6 EN derivatives showed that removing eh1 caused more of a loss of activity than removing these other RDs, in sharp contrast to the results with the transiently transfected target gene (compare Fig. 3 and 2). Thus, when an integrated target gene is assayed, the relative potency of EN domains closely parallels that seen in vivo. The eh1 region apparently has an activity that is invisible in the normal transient transfection analysis; this is due not to a difference in cellular environment relative to the in vivo situation but rather to some difference in the assay itself. Perhaps the activity of eh1 requires a more normal chromatin environment than that occurring on transiently transfected DNA.
Based on the relative expression levels of transiently transfected target genes and stably integrated ones, about the same total number of target genes are being expressed per cell in each case. Both basal expression levels and activated levels are consistent with this estimate. Independent estimates of the percentage of expressing cells following transient transfection are about 2%, while all of the cells express in the stably transformed cultures (data not shown). Thus, we estimate that the average number of expressing copies of target gene per cell is about 50-fold higher in the transient transfection assay. There is the possibility that factors required for repression by the eh1 domain were titrated out by the larger number of target genes per cell in the transient transfections. To address this possibility, we examined the effect of reducing the number of target genes and lowering the levels of activator and repressor in transient assays. As shown in Fig. 6, the dependence of repression activity on the eh1 region is increased under these conditions. Rather than an approximately 1.5-fold decrease in repression activity in standard transient transfection assays (Fig. 3), we saw an approximately 3-fold decrease, and the activity was restored by replacing the Drosophila eh1 region with the mouse version. This reasonably clear-cut difference from the standard assay required reducing both target gene and repressor levels, suggesting that factors important for repression by eh1 can be titrated out by either excess target genes or excess repressor (data not shown). The involvement of titratable factors in repression by eh1 is not inconsistent with the requirement for a normal chromatin environment for its activity, since repressive chromatin components are known to be in limiting supply in vivo (see Discussion).
DISCUSSION
Multiple EN domains contribute to active repression.
Analysis of EN repression function in two assays has shown that multiple domains contribute to activity. In the first assay, EN was retargeted in vivo to the endogenous ftz gene (by replacing the EN HD with that of FTZ), resulting in repression of the ftz gene. In the second assay, this chimeric repressor, EFE, actively repressed artificial target genes in cultured cells. Strikingly, one region predominantly affects repression activity in vivo. This region (region 3 [Fig. 2A]) contains the single conserved domain (eh1) not closely associated with the HD in the primary sequence. Deleting the core of this homology region, which is found in all EN class homeoproteins, or mutating the most conserved amino acid, Phe 175 (F→E), strongly reduces repression activity in vivo, to a degree equivalent to deleting all of region 3 (Fig. 2 and 3). In contrast, none of these mutations strongly affects repression in cultured cells (Fig. 3), although the effect of deleting regions 3 and 4 appears to be significantly greater than that of deleting region 4 alone (Fig. 2B and C). We established that this region contributes to active repression per se and is not simply defective in binding to endogenous sites by showing that the point-mutated protein F→E can actually reduce repression when it is coexpressed with the unmutated EFE (Fig. 1). We interpret this to mean that F→E displaces EFE from sites in the ftz gene but is defective in active repression.
One region (region 4) that contributes to repression activity contains a previously defined active RD (Fig. 2A [R]) from studies using cell culture assays similar to those used here (11, 18). Removing this region results in a more stable protein both in vivo and in culture (32) (Fig. 2D; Table 1), allowing the deleted protein to repress effectively in both assays. However, the potency of repression appears to be reduced in both cases (Fig. 2; Table 1). When both regions 4 and 6 are deleted, very little activity remains in culture, while repression in vivo is still strong (Fig. 3). Thus, in vivo, region 3 contributes the predominant repression activity, while in transient transfection assays in culture, regions 4 and 6 contribute the predominant activity.
A conserved region that normally flanks the C terminus of the EN HD (and thus flanks the FTZ HD in EFE) contributes to the potency of repression in both assays (specifically deleted in Δ6 [Fig. 2 and 3]). This is interesting in light of the involvement of conserved regions flanking the HDs of HOX gene products in determining their functional specificities in vivo (24, 26, 40). Such regions may contribute to functional specificity in more than one way. They may cause differences in transcriptional activities among these proteins that lead to different activities on common target genes, in addition to providing selective targeting to different target genes.
The conserved region that flanks the N terminus of the HD also contributes to potency in culture (and removing it increases the apparent stability of the protein [Fig. 2D; Table 1]). However, removing this region has a complicated effect in vivo. Without increasing the stability of the protein in vivo (32), this deletion (Δ5) actually increases activity (Fig. 2). This might indicate an effect on targeting in vivo that is not reflected in the transfection assays. Perhaps targeting in vivo by the FTZ HD involves both protein-protein and protein-DNA interactions, while targeting in the cell culture assays (i.e., binding to the target sites in the reporter genes) involves only protein-DNA interaction. If region 5 interacts with other proteins in vivo, which interferes with the protein-protein interactions of the FTZ HD necessary for targeting to the ftz gene, then removing it would lead to increased ftz repression by EFE. This suggests that the conserved region N terminal to the EN HD normally participates in targeting in vivo by the EN HD to sites not recognized by the FTZ HD. Since this region has been shown to be required in vitro for interaction with the Extradenticle protein (30), a homeoprotein cofactor implicated in targeting by HOX proteins (reviewed in 27), perhaps such an interaction can occur in vivo even in the context of the FTZ HD.
The general correlation of activity in the two repression assays, the complexities noted above notwithstanding, suggests that the two repression assays measure a similar function of EN-derived domains, that is, active repression. This correlation confirms the previous conclusions (21) that repression of the endogenous ftz gene by EFE requires the active repression function contributed by the EN portion of the molecule. In addition, it shows that multiple EN domains, including each of the conserved blocks outside the HD, which are found in all known EN homologs, contribute to this activity, suggesting that active repression is a primary function of both EN and its homologs.
The eh1 region interacts with the GRO corepressor.
We identified an EN corepressor in a yeast two-hybrid screen, using as bait an N-terminal region of EN that contained both eh1 and the region 4 RD. The interacting clone that we obtained encodes the C-terminal region of GRO, which consists of a tandem array of WD40 repeats highly homologous to the C terminus of both the yeast corepressor TUP1 and mammalian TLE proteins (35). This region of TUP1 mediates its interaction with the α2 protein (22), which, like EN, is a homeodomain-containing repressor. GRO is also recruited to DNA by both Hairy-related bHLH repressors (29) and Runt domain proteins (1). We found that the F→E mutation in eh1, which abolishes the repression activity of eh1 in embryos, virtually eliminates interaction with GRO (both full length and the WD40 repeat region) in the yeast assay, in the context of both full-length EN and the N-terminal region (Fig. 4A). This was confirmed in vitro by GST pulldown assays with the N-terminal region of EN (Fig. 4B). Furthermore, just as substituting the mouse eh1 region for that of Drosophila restores repression activity in Drosophila embryos, the same substitution restores interaction with GRO in the yeast assay (Fig. 4A). Similarly, Jiménez et al. (20) recently showed that the eh1 region of EN is required for GRO-dependent repression by a Hairy-EN fusion protein in Drosophila embryos. The GRO-EN interaction appears to be completely independent of the cell culture RD of region 4, since removing it entirely has no apparent effect on the strength of the interaction in yeast (Fig. 4A). Thus, the requirements for EN-GRO interaction correlate well with the requirements for repression by the eh1 region, while the apparent lack of involvement of the region 4 RD in interaction with GRO is consistent with its distinct functional characteristics, as discussed below.
Distinct mechanisms of active repression in vivo and in culture.
A detailed comparison of the relative potencies of different RDs in the in vivo and cell culture assays leads to the conclusion that multiple mechanisms of active repression are likely to be encoded by EN. The most striking example is highlighted by the comparisons of Fig. 3, in which it is shown that alterations of region 3 and the eh1 homology that it contains clearly have distinct effects from alterations in regions 4 and 6. Region 3, which interacts with GRO, primarily affects activity in vivo, while regions 4 and 6 have much stronger effects in transient transfection assays in culture. This difference is not due simply to one assay being more stringent than the other; rather, the eh1 domain is dispensable for repression in transient transfections, but not in vivo, while the R domain and region 6 are dispensable in vivo, but not in transient assays. This distinction suggests that these two types of RD confer mechanistically different activities on EFE that are each preferentially active in different contexts. Three possibilities for the critical difference in context are (i) the cell type in which the assay is done (cultured cells versus embryonic tissues), (ii) the target gene assayed (reporter genes in culture versus the endogenous ftz gene), and (iii) the integration state of the target gene (transiently transfected DNA versus a normal chromatin environment). The first two of these possibilities are ruled out by our assays of stably integrated target genes in cultured cells (discussed below).
The fact that multiple domains contribute to repression activity in the two assays and the likelihood that they utilize distinct mechanisms suggest that the evolution of EN has involved strong selection for repression function. This possibility is reinforced by the observation that none of our deletion derivatives showed significant activation function, either alone or in combination with other activators, on appropriate reporter genes in culture, even when all identified RDs were removed (Fig. 2) (our unpublished observations). Indeed, preliminary data suggest that even the EN HD contributes to repression activity in the normal EN molecule, since single domain deletions that significantly affect repression activity in the context of the FTZ HD (i.e., in EFE) do not affect the repression activity of EN itself to the same degree (our unpublished observation). The idea that EN might be primarily a repressor in vivo conflicts, on the surface, with results from ectopic expression assays in embryos, in which EN has been shown to induce expression of its own gene (15), as well as with the positive regulatory action of EN on hedgehog (34). That these interactions might be indirect, through repression of a repressor, is suggested by our results. However, it remains possible that protein-protein interactions allow EN to have a net positive regulatory effect on some direct target genes. It is worthy of note in this context that a similar positive autoregulatory effect of Even-skipped (19), a strong repressor in both cell culture assays (12, 16), and in vitro (2), has been attributed to indirect effects in vivo, involving repression of other repressors (9).
Stable integration of target genes reveals a GRO-dependent repression activity invisible in transient transfections.
One difference between the in vivo assay for active repression by EFE and the standard transient assay in cultured cells is the state of the target gene. We tested whether the activity of the eh1 region might be sensitive to this difference by testing stable transformants. Cultured cells stably transformed with the same target gene that showed very little sensitivity to mutation of eh1 in transient assays were transfected either with EFE or with derivatives mutated in the eh1 region that no longer interact strongly with GRO. This transient-on-top-of-stable assay allowed us to directly compare the activities of different repressors in the same population of cells containing the stably integrated target gene. When eh1 was mutated in the context of either EFE or normal EN, the ability to repress the integrated target gene was severely compromised. This is in striking contrast to the effect in the standard transient transfection assay, in which removing eh1 had very little effect. In addition, replacing Drosophila eh1 with mouse eh1 restored activity. Thus, when the target gene is integrated into a chromosome, its repression by the different EN domains closely parallels that of a natural target gene in vivo. This suggests that the state of the target gene is important for repression by the conserved eh1 domain and, by inference, GRO, but not by the other class of EN domains that are more active in transient transfections. One plausible explanation for this difference is that one RD class, exemplified by eh1, represses by stabilizing or inducing a repressive chromatin structure, while the other, exemplified by regions 4 and 6, acts on another target, perhaps the basal transcriptional machinery.
We also tested whether simply reducing the levels of target gene and repressor might allow us to see the activity of eh1 in a transient assay. Such a possibility was suggested by the fact that components involved in chromatin-based repression in vivo, such as those involved in the phenomenon of position effect variegation, appear to be in limiting supply, since they are apparently titrated out by adding abnormal amounts of heterochromatin to the genome (4). Our estimates of the number of copies of our target gene present in the average transfected cell showed that there were about 50-fold more copies under our standard transient assay conditions than in the stably transformed cells, raising the possibility that a repression mechanism involving low-abundance endogenous factors might be less effective in the transient assay. When we reduced the levels of target gene and repressor expression plasmid (as well as activator levels), we were indeed able to see an increased effect of removing eh1 (Fig. 6), suggesting that it requires endogenous factors to function that are in limiting supply. This factor is unlikely to be GRO itself, since it is present in abundance in these cells (8). Rather, it is likely that factors recruited by the EN-GRO complex are limiting. These results suggest that the EN-GRO complex can function to some degree on transiently transfected templates. Perhaps repressive chromatin can be built on a limited number of these templates, until an essential component is used up. However, it should be noted that even using the lowest levels of plasmids that allowed us to reliably quantify our results, we were unable to reproduce the strong dependence on eh1 that occurred with the stably integrated target gene (compare Fig. 5 and 6). This suggests that only when the target gene is integrated into a normal chromatin environment is the GRO interaction domain fully functional in repression.
ACKNOWLEDGMENTS
Thanks go to Tadaatsu Goto (deceased), Alexander Mazo, Michael Caudy, Al Fisher, and Claude Desplan for helpful advice and discussions; to Charles Girdham and Pat O’Farrell for polyclonal anti-EN antiserum; to Al Fisher and M. Caudy for plasmids and reagents; to Aleyamma John and Sheryl T. Smith for excellent technical assistance; and to A. Mazo for comments on the manuscript.
This work was supported by NIH award R01-GM50231.
REFERENCES
- 1.Aronson B D, Fisher A L, Blechman K, Caudy M, Gergen J P. Groucho-dependent and -independent repression activities of Runt domain proteins. Mol Cell Biol. 1997;17:5581–5587. doi: 10.1128/mcb.17.9.5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Biggin M D, Tjian R. A purified Drosophila homeodomain protein represses transcription in vitro. Cell. 1989;58:433–440. doi: 10.1016/0092-8674(89)90424-8. [DOI] [PubMed] [Google Scholar]
- 3.Desplan C, Theis J, O’Farrell P H. The sequence specificity of homeodomain-DNA interaction. Cell. 1988;54:1081–90. doi: 10.1016/0092-8674(88)90123-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dimitri P, Pisano C. Position effect variegation in Drosophila melanogaster: Relationship between suppression effect and the amount of Y chromosome. Genetics. 1989;122:793–800. doi: 10.1093/genetics/122.4.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dorsett D, Viglianti G A, Rutledge B J, Meselson M. Alteration of hsp82 gene expression by the gypsy transposon and suppressor genes in Drosophila melanogaster. Genes Dev. 1989;3:454–468. doi: 10.1101/gad.3.4.454. [DOI] [PubMed] [Google Scholar]
- 6.Durfee T, Becherer K, Chen P, Yeh S, Yang Y, Kilburn A E, Lee W, Elledge S J. The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- 7.Edgar B, O’Farrell P H. The three postblastoderm cell cycles of Drosophila embryogenesis are regulated in G2 by string. Cell. 1990;62:469–80. doi: 10.1016/0092-8674(90)90012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fisher A L, Ohsako S, Caudy M. The WRPW motif of the hairy-related basic helix-loop-helix repressor proteins acts as a 4-amino-acid transcription repression and protein-protein interaction domain. Mol Cell Biol. 1996;16:2670–77. doi: 10.1128/mcb.16.6.2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fujioka M, Jaynes J B, Goto T. Early even-skipped stripes act as morphogenetic gradients at the single cell level to establish engrailed expression. Development. 1995;121:4371–4382. doi: 10.1242/dev.121.12.4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guichet A, Copeland J W R, Erdélyi M, Hlousek D, Závorszky P, Ho J, Brown S, Percival-Smith A, Krause H M, Ephrussi A. The nuclear receptor homologue Ftz-F1 and the homeodomain protein Ftz are mutually dependent cofactors. Nature. 1997;385:548–552. doi: 10.1038/385548a0. [DOI] [PubMed] [Google Scholar]
- 11.Han K, Manley J L. Functional domains of the Drosophila Engrailed protein. EMBO J. 1993;12:2723–2733. doi: 10.1002/j.1460-2075.1993.tb05934.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Han K, Manley J L. Transcriptional repression by the Drosophila Even-skipped protein, definition of a minimal repression domain. Genes Dev. 1993;7:491–503. doi: 10.1101/gad.7.3.491. [DOI] [PubMed] [Google Scholar]
- 13.Hanna-Rose W, Hansen U. Active repression mechanisms of eukaryotic transcription repressors. Trends Genet. 1996;12:229–234. doi: 10.1016/0168-9525(96)10022-6. [DOI] [PubMed] [Google Scholar]
- 14.Harper J W, Adami G R, Wei N, Keyomarsi K, Elledge S J. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 15.Heemskerk J, DiNardo S, Kostriken R, O’Farrell P H. Multiple modes of engrailed regulation in the progression towards cell fate determination. Nature. 1991;352:404–410. doi: 10.1038/352404a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jaynes J B, O’Farrell P H. Activation and repression of transcription by homoeodomain-containing proteins that bind a common site. Nature. 1988;336:744–749. doi: 10.1038/336744a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16a.Jaynes, J. B., and P. H. O’Farrell. Unpublished observations.
- 17.Jaynes J B, Vincent J, O’Farrell P H. Drosophila homeodomain-containing proteins regulate transcription. In: Mahowald A P, editor. Genetics of pattern formation and growth control. New York, N.Y: Wiley-Liss Inc.; 1990. pp. 47–64. [Google Scholar]
- 18.Jaynes J B, O’Farrell P H. Active repression of transcription by the engrailed homeodomain protein. EMBO J. 1991;10:1427–1433. doi: 10.1002/j.1460-2075.1991.tb07663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang J, Hoey T, Levine M. Autoregulation of a segmentation gene in Drosophila: combinatorial interaction of the even-skipped homeo box protein with a distal enhancer element. Genes Dev. 1991;5:265–277. doi: 10.1101/gad.5.2.265. [DOI] [PubMed] [Google Scholar]
- 20.Jiménez G, Paroush Z, Ish-Horowicz D. Groucho acts as a corepressor for a subset of negative regulators, including Hairy and Engrailed. Genes Dev. 1997;11:3072–3082. doi: 10.1101/gad.11.22.3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.John A, Smith S T, Jaynes J B. Inserting the Ftz homeodomain into Engrailed creates a dominant transcriptional repressor that specifically turns off Ftz target genes in vivo. Development. 1995;121:1801–1813. doi: 10.1242/dev.121.6.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keleher C A, Redd M J, Schultz J, Carlson M, Johnson A D. Ssn6-Tup1 is a general repressor of transcription in yeast. Cell. 1992;68:709–719. doi: 10.1016/0092-8674(92)90146-4. [DOI] [PubMed] [Google Scholar]
- 23.Latchman D S. Inhibitory transcription factors. Int J Biochem Cell Biol. 1996;28:965–974. doi: 10.1016/1357-2725(96)00039-8. [DOI] [PubMed] [Google Scholar]
- 24.Lin L, McGinnis W. Mapping functional specificity in the Dfd and Ubx homeo domains. Genes Dev. 1992;6:1071–1081. doi: 10.1101/gad.6.6.1071. [DOI] [PubMed] [Google Scholar]
- 25.Logan C, Hanks M C, Noble-Topham S, Nallainathan D, Provart N J, Joyner A L. Cloning and sequence comparison of the mouse, human, and chicken engrailed genes reveal potential functional domains and regulatory regions. Dev Genet. 1992;13:345–358. doi: 10.1002/dvg.1020130505. [DOI] [PubMed] [Google Scholar]
- 26.Mann R S, Hogness D S. Functional dissection of Ultrabithorax proteins in D. melanogaster. Cell. 1990;60:597–610. doi: 10.1016/0092-8674(90)90663-y. [DOI] [PubMed] [Google Scholar]
- 27.Mann R S, Chan S K. Extra specificity from extradendicle: the partnership between HOX and PBX/EXD homeodomain proteins. Trends Genet. 1996;12:258–262. doi: 10.1016/0168-9525(96)10026-3. [DOI] [PubMed] [Google Scholar]
- 28.Manoukian A S, Krause H M. Concentration-dependent activities of the even-skipped protein in Drosophila embryos. Genes Dev. 1992;6:1740–1751. doi: 10.1101/gad.6.9.1740. [DOI] [PubMed] [Google Scholar]
- 29.Paroush Z, Finley R, Kidd T, Wainwright S M, Ingham P, Brent R, Ish-Horowicz D. Groucho is required for Drosophila neurogenesis, segmentation, and sex determination and interacts directly with hairy-related bHLH proteins. Cell. 1994;79:805–815. doi: 10.1016/0092-8674(94)90070-1. [DOI] [PubMed] [Google Scholar]
- 30.Peltenburg L T, Murre C. Engrailed and Hox homeodomain proteins contain a related Pbx interaction motif that recognizes a common structure present in Pbx. EMBO J. 1996;15:3385–3393. [PMC free article] [PubMed] [Google Scholar]
- 31.Schier A F, Gehring W J. Direct homeodomain-DNA interaction in the autoregulation of the fushi tarazu gene. Nature. 1992;356:804–807. doi: 10.1038/356804a0. [DOI] [PubMed] [Google Scholar]
- 32.Smith S T, Jaynes J B. A conserved region of engrailed, shared among all en-, gsc-, Nk1-, Nk2-, and msh-class homeoproteins, mediates active transcriptional repression in vivo. Development. 1996;122:3141–3150. doi: 10.1242/dev.122.10.3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spradling A C, Rubin G M. Genetic transformation of Drosophila with transposable element vectors. Science. 1982;218:341–347. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- 34.Tabata T, Eaton S, Kornberg T B. The Drosophila hedgehog gene is expressed specifically in posterior compartment cells and is a target of engrailed regulation. Genes Dev. 1992;6:2635–2645. doi: 10.1101/gad.6.12b.2635. [DOI] [PubMed] [Google Scholar]
- 35.Tata F, Hartley D A. The role of the Enhancer of Split complex during cell fate determination in Drosophila. In: Ingham P, Brown A, Arias A M, editors. Signals, polarity, and adhesion in development. Development 1993 supplement. Cambridge, United Kingdom: The Company of Biologists Ltd.; 1993. pp. 139–148. [PubMed] [Google Scholar]
- 36.Wieschaus E, Nüsslein-Volhard C. Looking at embryos. In: Roberts D B, editor. Drosophila: a practical approach. Oxford, England: IRL Press; 1986. pp. 199–228. [Google Scholar]
- 37.Winslow G M, Hayashi S, Krasnow M, Hogness D S, Scott M P. Transcriptional activation by the Antennapedia and fushi tarazu proteins in cultured Drosophila cells. Cell. 1989;57:1017–1030. doi: 10.1016/0092-8674(89)90340-1. [DOI] [PubMed] [Google Scholar]
- 38.Yoshinaga S K, Peterson C L, Herskowitz I, Yamamoto K R. Roles of SWI1, SWI2, and SWI3 proteins for transcriptional enhancement by steroid receptors. Science. 1993;258:1598–1604. doi: 10.1126/science.1360703. [DOI] [PubMed] [Google Scholar]
- 39.Yu Y, Li W, Su K, Yussa M, Han W, Perrimon N, Pick L. The nuclear hormone receptor Ftz-F1 is a cofactor for the Drosophila homeodomain protein Ftz. Nature. 1997;385:552–554. doi: 10.1038/385552a0. [DOI] [PubMed] [Google Scholar]
- 40.Zeng W, Andrew D J, Mathies L D, Horner M A, Scott M P. Ectopic expression and function of the Antp and Scr homeotic genes: the N terminus of the homeodomain is critical to functional specificity. Development. 1993;118:339–352. doi: 10.1242/dev.118.2.339. [DOI] [PubMed] [Google Scholar]