Abstract
Tumor necrosis factor alpha (TNFα) is a key regulatory cytokine whose expression is controlled by a complex set of stimuli in a variety of cell types. Previously, we found that the monocyte/macrophage-enriched nuclear transcription factor C/EBPβ played an important role in the regulation of the TNFα gene in myelomonocytic cells. Abundant evidence suggests that other transcription factors participate as well. Here we have analyzed interactions between C/EBPβ and c-Jun, a component of the ubiquitously expressed AP-1 complex. In phorbol myristate acetate (PMA)-treated Jurkat T cells, which did not possess endogenous C/EBPβ, expression of c-Jun by itself had relatively little effect on TNFα promoter activity. However, the combination of C/EBPβ and c-Jun was synergistic, resulting in greater than 130-fold activation. This effect required both the leucine zipper and DNA binding domains, but not the transactivation domain, of c-Jun, plus the AP-1 binding site centered 102/103 bp upstream of the transcription start site in the TNFα promoter. To determine if C/EBPβ and c-Jun might cooperate to regulate the cellular TNFα gene in myelomonocytic cells, U937 cells that possess endogenous C/EBPβ and were stably transfected with either wild-type c-Jun or the transactivation domain deletion mutant (TAM-67) were examined. U937 cells expressing ectopic wild-type c-Jun or TAM-67 secreted over threefold more TNFα than the control line in response to PMA plus lipopolysaccharide. Transient transfection of the U937 cells expressing TAM-67 suggested that TAM-67 binding to the −106/−99-bp AP-1 binding site cooperated with endogenous C/EBPβ in the activation of the −120 TNFα promoter-reporter. DNA binding assays using oligonucleotides derived from the TNFα promoter suggested that C/EBPβ and c-Jun interact in vitro and that the interaction may be DNA dependent. Our data demonstrate that the TNFα gene is regulated by the interaction of the ubiquitous AP-1 complex protein c-Jun and the monocyte/macrophage-enriched transcription factor C/EBPβ and that this interaction contributes to the expression of the cellular TNFα gene in myelomonocytic cells. This interaction was unique in that it did not require the c-Jun transactivation domain, providing new insight into the cell-type-specific regulation of the TNFα gene.
Tumor necrosis factor alpha (TNFα) contributes to the pathogenesis of many chronic inflammatory diseases, including rheumatoid arthritis, diabetes, hepatitis, and some causes of pulmonary inflammation and fibrosis (14, 16, 27, 29, 49, 53, 54). The controlled expression of TNFα is critical during sepsis and adipocyte differentiation and in obesity (9, 19, 20, 22, 49). Although macrophages are the principal source of TNFα secretion in conditions such as rheumatoid arthritis and sepsis (27, 49), the cytokine is produced by a wide variety of cells, including lymphocytes, adipocytes, mast cells, keratinocytes, and astrocytes.
A number of transcription factors contribute to the complex regulation of the TNFα gene, and interactions between factors may vary depending on the cell type and the particular extracellular stimuli. The transcription factor C/EBPβ (also called NF-IL6, NF-M, LAP, IL6-DBP, AGP/EBP, and CRP2) (1) has been shown to be important in TNFα gene activation in myelomonocytic cells (38). It binds to the TNFα promoter at a site between 74 and 100 bp upstream of the transcription start site (38) and may also be important for the expression of TNFα in other cell types, such as hepatocytes and adipocytes, which also express C/EBPβ.
Several types of evidence suggest that C/EBPβ works in concert with other transcription factors to regulate the TNFα promoter in a cell-type-specific fashion. For example, the TNFα promoter contains potential binding sites for several additional transcription factors, including AP-1, AP-2, NF-κB, NFAT, Ets, SP-1, and cyclic AMP response element (CRE) (see Fig. 1) (13, 15, 25, 33, 38, 39, 48). The transcription factors AP-1, Ets, and NFAT have been shown to play important roles in the activation of the TNFα gene (13, 15, 23, 33, 39, 48). In addition, C/EBPβ synergizes with a variety of transcription factors, including c-Jun, NF-κB, Myb, and the glucocorticoid receptor, to regulate other genes (7, 17, 21, 23, 26, 32, 35, 42, 43). However, very little is known about interactions between C/EBPβ and other transcription factors in the cell-type-specific regulation of the TNFα gene.
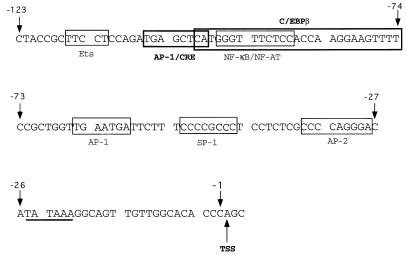
FIG. 1.
Sequence of the human TNFα promoter region −123 bp upstream from the transcription start site (TSS). Previously described binding sites for C/EBPβ (−100 to −74 bp), AP-1 (−106 to −99 and −65 to −59 bp), AP-2 (−36 to −28 bp), SP-1 (−52 to −45 bp), Ets (−116 to −112 bp), NFAT and NF-κB (−97 to −88 bp), and CRE (−106 to −99) are indicated on the sequence representing the first 123 bp of the TNFα promoter (13, 15, 25, 33, 38, 39, 48). The C/EBPβ binding site was defined by using the 32P-oligonucleotide employed in this study (38). The sites highlighted by the bold boxes are the focus of this study.
Here we describe interactions between C/EBPβ and the transcription factor AP-1 which affect the regulation of the TNFα gene. We showed that c-Jun binds adjacent to C/EBPβ on the TNFα promoter and synergistically activates the C/EBPβ-dependent expression of the TNFα gene in phorbol myristate acetate (PMA)-treated Jurkat T cells. This cooperation required the DNA binding domain of c-Jun and an intact AP-1 binding site on the TNFα promoter, suggesting that both C/EBPβ and c-Jun must bind in order to coactivate the gene. U937 cells stably overexpressing wild-type c-Jun secreted increased TNFα. Interestingly, C/EBPβ also cooperated with the mutant form of c-Jun lacking the transcriptional transactivation domain, in both Jurkat T cells and U937 cells, suggesting that the cooperation between c-Jun and C/EBPβ is not just the additive effect of independent transcriptional activation domains.
MATERIALS AND METHODS
Plasmid vector constructs.
The TNFα promoter reporter constructs containing 615, 120, or 95 bp 5′ of the transcription start site, linked to a luciferase gene, have been described elsewhere (13, 38), as have the expression vectors (cytomegalovirus encoding wild-type [CMV]-C/EBPβ) and dominant negative (CMV-5D229) versions of C/EBPβ (38). c-jun and c-jun mutant human cDNAs were cloned into the CMV vector plasmid and have been previously characterized (2, 6, 18, 36). Mutations had been generated by deletion of the leucine zipper (c-Jun-LZ), the DNA binding (c-Jun-DBD), and the transactivation (transactivation domain mutant TAM-67) domains (2, 6, 18, 36). Human c-fos was cloned into a pSV vector (2, 6). The pCDM8 (Invitrogen, San Diego, Calif.), pSV, and CMV vectors were used as controls. A promoter-reporter construct containing dual c-Jun binding sites from the interleukin-2 (IL-2) promoter [TRE(IL-2)-Luc] was used as a control (36). The p300- and CREB-binding protein (CBP)-expressing plasmids have been previously described (10, 12). Luciferase-expressing plasmids possessing the AP-1 and C/EBPβ binding sites of the TNFα gene were constructed by using pT81-Luc, which possessed a weak tk promoter. Two plasmids, constructed by using oligonucleotides representing −115 to −74 bp of the TNFα promoter, possess 0 (pT81TNF0bp) or 10 (pT81TNF10bp) irrelevant bp inserted between −98 and −97 bp. Since the two sites overlapped, −99 and −100 were included on both sides of the inserted base pairs. Plasmids were screened by restriction digestion, and the sequences were confirmed by DNA sequencing employing the dideoxynucleotide method.
Transfection and luciferase assay.
Jurkat T cells were maintained in RPMI 1640 medium supplemented with 10% heat-inactivated fetal calf serum, l-glutamine, and penicillin-streptomycin (complete medium). Transfection of Jurkat T cells was performed by the DEAE-dextran method as previously described (38), keeping the total plasmid concentration constant (8 μg). Transfection of U937 cells was performed by electroporation (38), keeping the total DNA concentration at 16 μg/transfection. After transfection, cells were placed in complete medium for 4 to 6 h, and then PMA (10 ng/ml), alone or with lipopolysaccharide (LPS; 1 μg/ml), or ionomycin (0.5 μM) or control buffer, as indicated in the individual experiments, was added for an additional 12 h. Cells were harvested, washed, and lysed by freeze-thawing three times, and luciferase activities were determined on cell lysates as previously described (38), using a Monolight 2010 luminometer (Analytical Luminescence Laboratory, San Diego, Calif.). Promoter activities were expressed as relative light units (RLU), normalized for the total protein in each extract.
Oligonucleotides and electrophoretic mobility shift assay.
Synthetic oligonucleotide probes were prepared which spanned the following regions of the TNFα promoter: −100 to −74 (−100/−74) (29), −115 to −74 (−115/−74), and −115 to −98 (−115/−98) (Fig. 1). Oligonucleotides representing the C/EBPβ binding site of the IL-6 promoter and an AP-1 binding site from the collagenase promoter have been previously described (1, 3). Recombinant C/EBPβ was expressed in insect cells, using a baculovirus vector, as previously described (38). Recombinant c-Jun was obtained commercially (Promega Corp., Madison, Wis.), as were antibodies specific for C/EBPβ and c-Jun (Santa Cruz Inc., Santa Cruz, Calif.). Electrophoretic mobility shift assays were performed as described previously (38), using 1 ng of 32P-labeled probe per reaction. DNA-binding complexes were analyzed by electrophoresis on a 4% nondenaturing polyacrylamide gel in buffer containing 67 mM Tris-HCl (pH 8.0), 10 mM EDTA, and 33 mM sodium acetate. Gels were dried and exposed to radiographic film overnight at −80°C.
Cell lines.
Jurkat and U937 cell lines were obtained from the American Type Culture Collection (Rockville, Md.) and maintained in complete medium as described above (38). U937 cell lines stably transfected with plasmids expressing a neomycin resistance gene alone or together with plasmids expressing the wild-type c-Jun or TAM-67 have been previously described and characterized (18, 45). These cell lines were maintained in medium containing 400 μg of G418 per ml.
TNFα secretion and quantitation.
U937 cell lines expressing the neomycin resistance gene alone or together with wild-type c-Jun or TAM-67 were plated at 0.5 × 106/ml in complete medium. PMA (10 ng/ml), or PMA plus LPS (1 μg/ml), or control medium was added, and cells were incubated for 18 h. Supernatants were harvested and frozen at −20°C. TNFα was measured by enzyme-linked immunosorbent assay using commercially available reagents as described previously (8).
RESULTS
C/EBPβ and c-Jun proteins synergistically activate the TNFα gene.
Our prior data (38) showed that C/EBPβ was important in activating a promoter reporter gene construct possessing as little as 120 bp of the TNFα promoter (−120 TNFα-luciferase). However, this promoter reporter also possesses two functional AP-1 binding sites centered at 102/103 and 62 bp upstream from the transcription start site (Fig. 1). We used cotransfection assays in Jurkat T cells, which do not express endogenous C/EBPβ, under conditions in which c-Jun was not significantly activated (38, 44), in order to determine if there was an interaction between C/EBPβ and AP-1 complex transcription factors in regulation of the TNFα promoter.
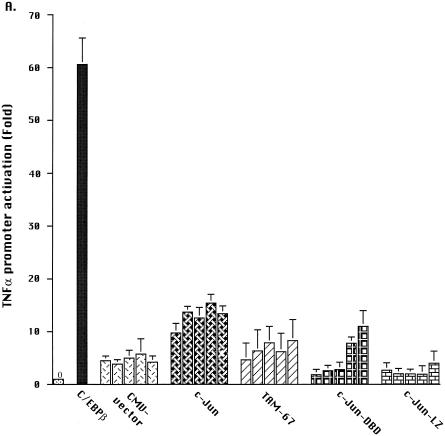
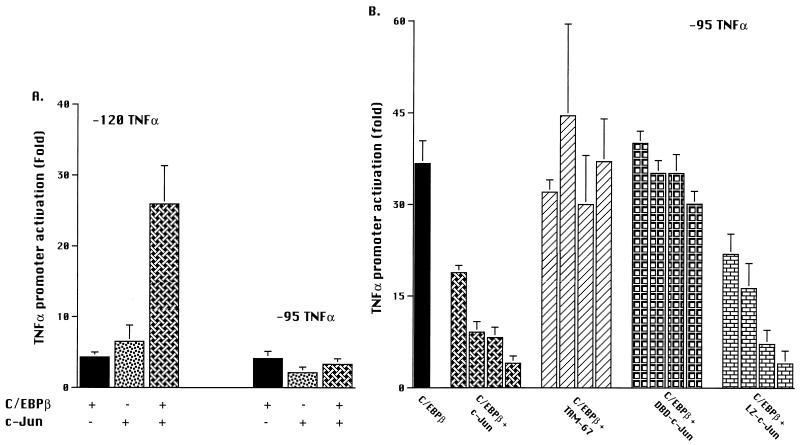
First, the ability of individual proteins to activate the −120 TNFα promoter reporter construct was assessed. Consistent with our earlier observations (38), cotransfection of 2 μg of the C/EBPβ expression vector with the −120 TNFα promoter reporter construct resulted in a 46 ± 7 (standard error [SE])-fold enhancement of PMA-induced activation in Jurkat T cells (Fig. 2A). Increasing the concentration of CMV-C/EBPβ up to 4 μg/transfection resulted in greater activation (>70-fold [data not shown]). Ectopic expression of wild-type c-Jun resulted in up to 14 ± 3 (SE)-fold stimulation of TNFα promoter reporter activity (Fig. 2A). Wild-type c-Jun was also able to activate the TNFα promoter reporter to a limited extent in cells that were not treated with PMA (data not shown), consistent with earlier reports (25). However, no other expression vector, including CMV-C/EBPβ, resulted in any activation of the TNFα promoter, either alone or in combination, in the absence of PMA treatment. This finding suggests that in Jurkat T cells, PMA enhanced the activity of c-Jun but was essential for the transcriptional activation mediated by C/EBPβ.
FIG. 2.
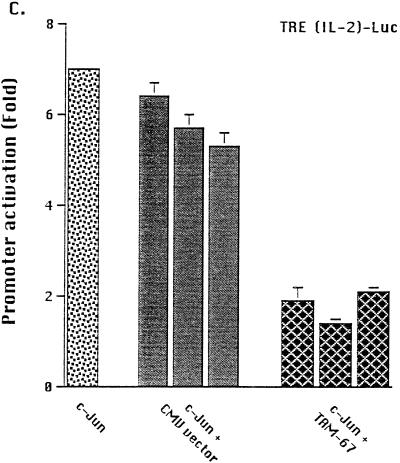
Both c-Jun and TAM-67 synergize with C/EBPβ to activate the TNFα promoter. (A) Effects of C/EBPβ, c-Jun, and c-Jun mutants on PMA-induced TNFα promoter activation. Jurkat T lymphocytes were transfected by the DEAE-dextran method with plasmid vectors expressing C/EBPβ, c-Jun, the c-Jun mutants c-Jun-LZ, c-Jun-DBD, and TAM-67, and the TNFα promoter reporter construct (3 μg/transfection) containing 120 bp 5′ of the transcription start site. The C/EBPβ plasmid was transfected at a suboptimal concentration (2 μg), while the plasmids expressing wild-type and mutant c-Jun and the CMV vector control were added at increasing concentrations (0.25, 0.5, 1, 2, and 3 μg, represented by the columns from left to right), with the total DNA added kept constant (8 μg). Luciferase activity was reported as RLU, corrected for the total protein in each lysate. Fold activation is expressed as a mean ± 1 SE for three or more experiments. (B) Effects of cotransfection of C/EBPβ with c-Jun or its mutants on PMA-induced activation of the TNFα promoter. Jurkat T lymphocytes were transfected with plasmid vectors expressing C/EBPβ plus c-Jun or its mutants and the −120 TNFα promoter reporter construct (3 μg). Various concentrations of control, c-Jun, or mutant c-Jun (0.25 to 3 μg) were added to C/EBPβ (2 μg). In addition, to more readily compare the results of different experiments, prior to analyses, the results of each experiment were normalized to the values obtained for C/EBPβ, which was defined as 100%. Data for wild-type c-Jun are the means ± 1 SE of six experiments, while those involving the mutants are the means of four experiments. Differences between C/EBPβ alone and with mutant or wild-type Jun were determined by t test for matched pairs. (C) Effects of TAM-67 and CMV vector control on c-Jun-induced activation of TRE(IL-2)-Luc promoter-reporter in PMA-stimulated Jurkat T cells. The TRE(IL-2)-Luc promoter reporter contains two copies of the c-Jun binding site of the IL-2 promoter. TRE(IL-2)-Luc (3 μg) was transfected with an optimal concentration of c-Jun (0.25 μg) and increasing concentrations of TAM-67 (0.25, 2, and 5 μg) or control CMV vector (0.25, 2, and 5 μg). The results presented are the means ± 1 SE of a single experiment that was representative of four experiments.
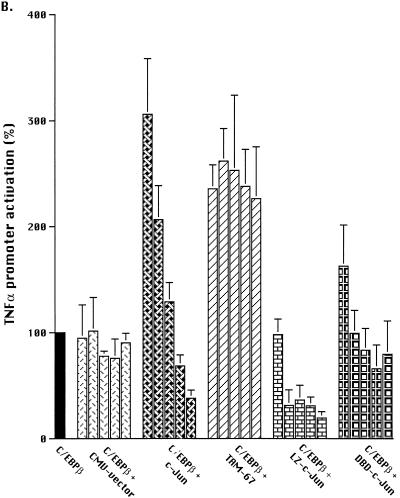
Next, we examined cooperation between C/EBPβ and c-Jun by cotransfecting plasmids expressing both proteins (Fig. 2B). The data for each experiment, prior to statistical analysis, were normalized to the activation induced by the CMV-C/EBPβ, which was defined as 100%. As shown in Fig. 2B, there was no change in TNFα promoter activity when additional CMV vector was cotransfected with the plasmid expressing C/EBPβ. However, cotransfection of C/EBPβ plus low concentrations of the c-Jun-expressing plasmid (0.25 or 0.5 μg) resulted in a significant (P < 0.05) 3-fold synergistic activation of the TNFα promoter (Fig. 2B), representing >130-fold activation above the PMA-treated baseline. Higher concentrations of the plasmid expressing c-Jun (2.0 and 3.0 mg) resulted in significant (P < 0.02) inhibition of C/EBPβ-induced TNFα promoter activity. No such suppression was observed with comparable concentrations of CMV-C/EBPβ vector (data not shown) or the control CMV vector (Fig. 2B), excluding squelching by the CMV promoter as the cause of this suppression and suggesting that the inhibition was the effect of c-Jun overexpression.
To determine if the effects of c-Jun were specific, another component of the AP-1 complex, c-Fos, was also examined. The overexpression of c-Fos resulted in little or no activation of the TNFα promoter reporter in PMA-treated Jurkat T cells (data not shown). Similarly, expression of c-Fos or c-Fos plus c-Jun with C/EBPβ did not result in enhanced activation of the TNFα promoter at any concentration (data not shown). Thus, expression of c-Jun appeared to specifically enhance (at low concentrations) or inhibit (at high concentrations) the ability of C/EBPβ to activate the TNFα promoter.
The c-Jun transactivation domain is not required for enhancement of C/EBPβ-induced activation of the TNFα gene.
Deletion mutants were used to identify the regions of c-Jun responsible for the synergistic activation of the TNFα promoter. The TAM-67, c-Jun-DBD, and c-Jun-LZ domain deletion mutants of c-Jun alone resulted in little or no activation of the TNFα promoter reporter in PMA-treated Jurkat T cells (Fig. 2A). In addition, no synergy was observed when c-Jun-LZ was coexpressed with C/EBPβ (Fig. 2B). These observations suggest that protein-protein interactions, most likely involving the c-Jun leucine zipper, were necessary for the synergistic activation of the TNFα gene by C/EBPβ and c-Jun. To determine if c-Jun DNA binding was required, the c-Jun-DBD mutant, which is capable of dimerization but not DNA binding, was also used. No synergy with C/EBPβ was observed (Fig. 2B), suggesting that binding of c-Jun homodimers or formation of a complex of c-Jun and C/EBPβ was necessary for the observed synergistic activation. Unexpectedly, the ectopic expression of TAM-67, which has the c-Jun DNA binding and dimerization domains intact but lacks the transactivation domain, enhanced the activation of the TNFα promoter by C/EBPβ (P < 0.02 at 0.25 and 0.5 μg) (Fig. 2B). The TAM-67 results suggest that the observed synergism of c-Jun with C/EBPβ was not due to additive effects of heterologous transactivation domains but more likely involved direct protein-protein interactions or cooperative DNA binding to the TNFα promoter.
The ability of the TAM-67 transactivation domain deletion mutant of c-Jun to activate gene expression on its own and to inhibit wild-type c-Jun was examined. TAM-67 had no effect on the TNFα promoter reporter (Fig. 2A), indicating that it was not sufficient to activate this promoter on its own. To determine if TAM-67 inhibited c-Jun activity in our system, TAM-67- and c-Jun-expressing plasmids were cotransfected along with TRE(IL-2)-Luc, a c-Jun-responsive reporter plasmid derived from the IL-2 gene promoter. When transfected alone, no activation of the TRE(IL-2)-Luc promoter reporter was noted in PMA-activated Jurkat cells (data not shown), consistent with lack of substantial activation of endogenous c-Jun under these conditions (44). However, cotransfection of TAM-67, but not the empty CMV vector, effectively inhibited the activation of this promoter by ectopically expressed c-Jun (Fig. 2C). These observations confirm that the TAM-67 deletion mutant can act in a dominant-negative fashion and that alone, it is incapable of activating TNFα gene expression. However, TAM-67 was able to synergize with C/EBPβ to activate the TNFα promoter (Fig. 2B), suggesting that it did so via protein-protein interactions which were independent of the c-Jun transactivation domain.
c-Jun must bind the TNFα promoter to synergize with C/EBPβ.
Prior studies have indicated that AP-1 binding sites centered 62 and 102/103 bp upstream of the transcription start site were involved in TNFα gene activation (25, 39). The contribution of these sites for the enhancement of C/EBPβ-induced activation by c-Jun was examined. Since synergistic interactions between c-Jun and C/EBPβ were evident in assays using the −615 AP-1 mutant TNFα promoter reporter, which possesses a 2-base substitution at −65 and −66 bp of the TNFα promoter (data not presented), we reasoned that the AP-1 binding site at that position was not required for synergism with C/EBPβ. Therefore, we compared the −95 TNFα-luciferase promoter reporter construct, which possesses the C/EBPβ binding site (38, 50) but lacks the AP-1 site centered at −103/−102 bp (25, 33), to the −120 TNFα promoter reporter, in which the AP-1 site is present. The initial cotransfection assays were performed at a limiting concentration of CMV-C/EBPβ (1 μg), which we had determined in preliminary studies more sensitively detected the potential synergistic effects of added c-Jun. The two promoter reporter constructs were comparably activated by cotransfection of 1 μg of C/EBPβ expression vector in PMA-treated Jurkat T cells (Fig. 3A). However, unlike the results observed with the −120 construct, no synergy between C/EBPβ and c-Jun was observed when the −95 TNFα promoter was used. Additional experiments were performed with the −95 promoter reporter construct, using 2 μg of CMV-C/EBPβ and each of the c-Jun constructs (Fig. 3B). Again in contrast to the results observed with the −120 TNFα promoter (Fig. 2B), cotransfection of TAM-67 with the −95 TNFα promoter reporter had no effect on C/EBPβ-induced activation (Fig. 3B). Since the −95 TNFα promoter reporter plasmid still possessed the AP-1 binding site centered 62 bp upstream of the transcription start site, the data confirm that this AP-1 site was not responsible for the synergistic enhancement of C/EBPβ-induced activation of the TNFα promoter by c-Jun in PMA-treated Jurkat T cells. Thus, synergism between c-Jun and C/EBPβ required a c-Jun binding site in the promoter as well as expression of c-Jun proteins containing both the DNA binding and dimerization domains, strongly suggesting that DNA binding by c-Jun was essential.
FIG. 3.
The AP-1 binding site is necessary for synergistic activation of the TNFα promoter. (A) Comparison of −120 and −95 TNFα promoter reporter constructs on C/EBPβ-plus-c-Jun activation in PMA-induced Jurkat T cells. Jurkat T lymphocytes were transfected with plasmid vectors expressing C/EBPβ (1 μg) and c-Jun (0.25 μg), separately or together, plus the −120 TNFα or the −95 TNFα promoter reporter (3 μg/transfection). The experiments were performed and analyzed as described for Fig. 2A. The results presented are the means ± 1 SE of three experiments. (B) Effects of cotransfection of c-Jun and c-Jun mutants on activation of the −95 TNFα promoter reporter construct. Jurkat T cells were transfected with the −95 TNFα-luciferase promoter reporter (3 μg), the CMV-C/EBPβ expression vector (2 μg), and various concentrations of wild-type or mutant c-Jun expression vectors (each at 0.25, 1, 3, and 5 μg/transfection). The experiments were performed, and cells were harvested and analyzed, as described for Fig. 2A. The results presented are the means ± 1 SE of two experiments.
The transactivation domain of c-Jun mediates suppression of C/EBPβ-induced activation in Jurkat T cells.
Cotransfection of either wild-type c-Jun or the c-Jun-LZ mutant resulted in significant (P < 0.05 to 0.005 at 0.5 to 3 μg) suppression of the C/EBPβ-induced activation of both the −120 and the −95 TNFα promoter reporters (Fig. 2B and 3B), suggesting that dimerization or protein-protein interactions mediated by the leucine zipper were not necessary for suppression. In contrast to the results with wild-type c-Jun, suppression was not observed at any concentration of TAM-67 with either the −120 or −95 TNFα promoter reporter construct (Fig. 2B and 3B). These observations suggest that the c-Jun transactivation domain contributed to the inhibition of C/EBPβ-induced activation of the TNFα promoter and that this repression was independent of the AP-1 binding site. The explanation for the lack of suppression observed with the c-Jun DNA binding mutant (Fig. 2B and 3B) remains unclear. The suppressive effect was not specific for c-Jun since c-Fos, at similar concentrations, also suppressed C/EBPβ-induced activation (data not shown).
Potential contribution of other transcription factors or coactivators to C/EBPβ–c-Jun-mediated TNFα activation.
Earlier studies demonstrated that NFAT was important in the activation of the TNFα gene in T cells under certain conditions (15, 48). Jurkat T cells possess NFAT, which requires two signals, such as PMA plus ionomycin, for optimal activation. Since activation of the TNFα gene by NFAT was inhibited by cyclosporin A (48), this inhibitor was used to determine if the TNFα promoter expression induced in response to C/EBPβ plus PMA in Jurkat T cells was mediated by the activation of NFAT. Following transfection with CMV-C/EBPβ and stimulation with PMA, no inhibition of TNFα promoter activation was observed by adding cyclosporin A (data not shown). In contrast, activation of the TNFα promoter induced by PMA plus ionomycin, in the absence of C/EBPβ, was inhibited >90% by cyclosporin A, suggesting that cyclosporin A inhibited NFAT-induced TNFα promoter activation, as previously described (15, 48).
CBP and p300 have been identified as nuclear phosphoproteins capable of interacting with a variety of transcription factors, including c-Jun and C/EBPβ (4, 5, 28a). Vectors expressing either p300 or CBP were cotransfected into Jurkat T cells together with c-Jun, C/EBPβ, and the −120 TNFα promoter reporter. No enhancement of activation of the TNFα promoter by either CBP or p300 was observed in Jurkat T cells either unstimulated or treated with PMA (data not shown). Additionally, neither CBP nor p300 expression reversed the c-Jun-induced suppression of TNFα promoter reporter activation by C/EBPβ (data not shown). These data suggest that neither NFAT, CBP, nor p300 contributed to the enhanced activation of the TNFα promoter by C/EBPβ plus c-Jun.
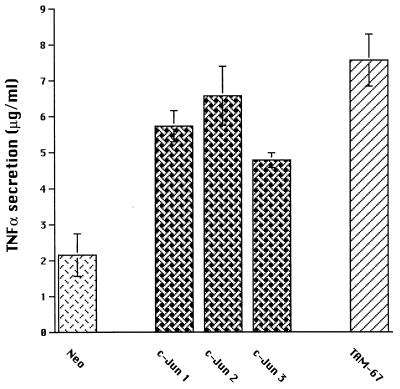
Overexpression of c-Jun and TAM-67 was associated with increased TNFα secretion in U937 cell lines.
The results described above implicate c-Jun in the regulation of the TNFα promoter but do not address whether similar mechanisms also regulate the chromosomal TNFα gene. Since monocytes/macrophages are the principal source of TNFα, we used the myelomonocytic cell line U937 to test whether ectopic expression of wild-type c-Jun or TAM-67 would affect the synthesis and secretion of TNFα. U937 cells constitutively express C/EBPβ, and the concentration was increased by differentiation in response to PMA (31). Therefore, we examined TNFα secretion in three independently derived stably transfected U937 cell lines overexpressing c-Jun and one line expressing TAM-67, each of which have been previously characterized (18, 45), as well as a similar line transfected only with the neomycin resistance gene. Western blots confirmed the increased wild-type c-Jun, both constitutively and following PMA-LPS treatment, in the c-jun-transfected lines compared to the control (data not shown). The TAM-67-transfected line expressed abundant TAM-67, and the wild-type c-Jun level was comparable to that in the neomycin-transfected control cell line (data not shown). Interestingly, no TNFα was secreted by these lines constitutively or when the cells were differentiated with PMA alone (data not shown). However, following differentiation plus activation by LPS, TNFα was secreted by each of the lines. TNFα secretion was significantly increased (P < 0.02) in the lines overexpressing either wild-type c-Jun or TAM-67 compared to the control (Fig. 4), suggesting that wild-type and TAM-67 c-Jun proteins can enhance TNFα protein production by myelomonocytic cells.
FIG. 4.
Overexpression of wild-type c-Jun or TAM-67 in U937 cells resulted in increased TNFα secretion. U937 cell lines expressing the neomycin resistance gene alone (Neo) or together with wild-type c-Jun or with TAM-67 were cultured in complete medium at 0.5 × 106/ml. Three independent lines overexpressing c-Jun (c-Jun 1, 2, and 3) and one expressing TAM-67 (TAM-67) were used. Cells were incubated in medium alone or in medium containing PMA or PMA plus LPS. Little or no TNFα was secreted when cells were cultured in medium alone or with PMA (data not shown). The data presented are the results obtained following incubation with PMA plus LPS. Supernatants were harvested at 18 h, and TNFα was quantitated. The results presented are the means ± 1 SE of three independent experiments.
TAM-67–C/EBPβ interaction in U937 cells was associated with enhanced activation of the TNFα promoter reporter.
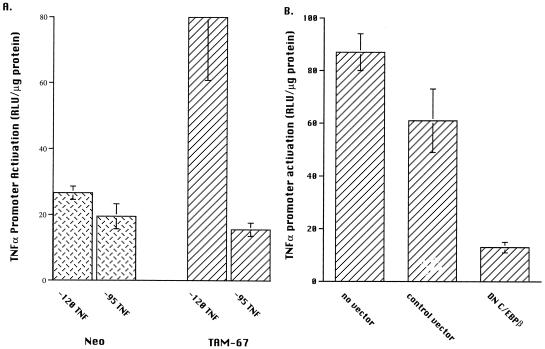
To determine if the AP-1 site centered 102/103 bp 5′ of the transcription start site was involved in the enhanced activation observed in the U937 line expressing TAM-67, this line and the control U937 line were transfected with either the −120 or the −95 TNFα-luciferase promoter reporters. The TAM-67 line, rather than a c-Jun-overexpressing line, was used to avoid potential activation by the wild-type c-Jun through the AP-1 site centered at −62 bp of the promoter. The −95 TNFα construct does not possess the AP-1 site centered at −102/−103 bp (Fig. 1) but does retain the ability to become activated by C/EBPβ (Fig. 3B). In the control neomycin-transfected U937 line, the −120 TNFα promoter-reporter demonstrated no increased activation compared to the −95 TNFα promoter reporter (Fig. 5A). In contrast, the −120 TNFα construct demonstrated a fourfold activation compared to the −95 TNFα construct in the U937 line stably expressing TAM-67 (Fig. 5A). This observation suggests that the enhanced activation of the TNFα promoter in U937 cells overexpressing TAM-67 was mediated through the AP-1 site centered at −102/−103 bp. This observation further documents that the −62 bp AP-1 site was not involved in the enhanced activation of the −120 TNFα promoter reporter observed in the TAM-67 cells, similar to the results observed in the Jurkat T cells.
FIG. 5.
Effect of TAM-67 on the activation of the TNFα promoter reporter in U937 cells. (A) The AP-1 binding site centered 102/103 bp 5′ of the transcription start site contributes to the activation observed in TAM-67-expressing U937 cells. U937 cells stably transfected with TAM-67 were transiently transfected with either the −120 or the −95 TNFα promoter reporter construct (3 μg). Sixteen hours later, the cells were harvested and RLU was determined and corrected for total protein in each lysate. The results presented are the means ± 1 SE of a transfection performed in duplicate and are representative of two experiments. (B) DN C/EBPβ inhibits activation of the −120 TNFα promoter reporter in U937 cells stably expressing TAM-67. U937 cells stably transfected with TAM-67 were cotransfected with the −120 TNFα promoter-reporter (3 μg) plus DN C/EBPβ (6 μg) or the CMV control vector (6 μg). Cells were harvested and analyzed as described above. The results presented represent the means ± 1 SE of an experiment performed in replicate and are representative of four experiments.
The U937 cell line stably transfected with TAM-67 was next used to determine if C/EBPβ was cooperating with TAM-67 to activate the TNFα promoter. A dominant negative version of C/EBPβ, with the transactivation domain deleted, was cotransfected together with the −120 TNFα promoter-reporter. Cotransfection of this construct, DN C/EBPβ, resulted in significant (P < 0.05) suppression of TNFα activation in the U937 cell line expressing TAM-67, while the control vector had no significant effect (Fig. 5B). The suppression may be even more impressive than is apparent since TAM-67 and wild-type C/EBPβ were constitutively present at the time of the transfection, while DN C/EBPβ required additional time for expression. When the TAM-67-transfected U937 cells were treated with PMA plus LPS, conditions required for TNFα secretion, cotransfection of DN C/EBPβ with the TNFα promoter-reporter also resulted in significant (P < 0.05) suppression (data not shown). Together, these observations suggest that C/EBPβ interacted with TAM-67 to effect the enhanced activation of the −120 TNFα promoter observed in these cells.
c-Jun affects the DNA binding of C/EBPβ to the TNFα promoter.
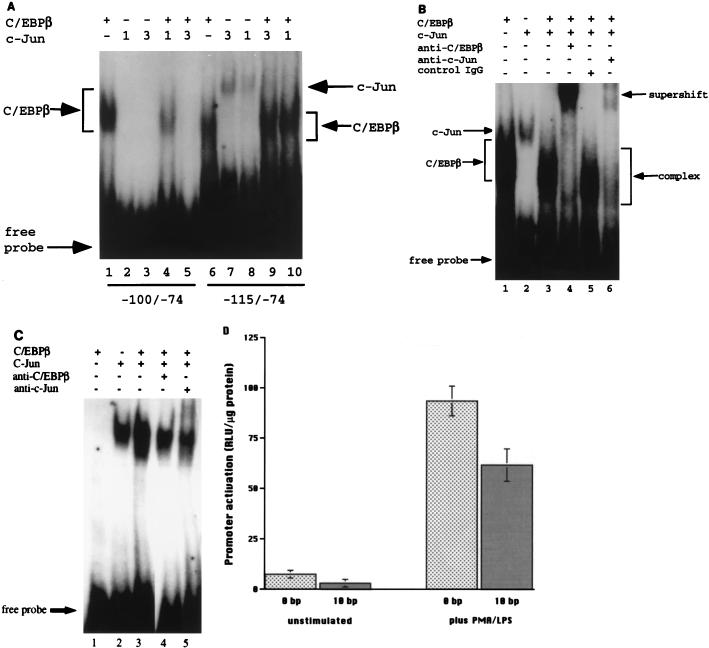
The transfection assays described above suggested that c-Jun must bind the TNFα promoter to enhance C/EBPβ activity. To further characterize the interactions between c-Jun and C/EBPβ, in vitro DNA binding assays were performed with three different 32P-labeled oligonucleotides derived from the TNFα promoter. The −115/−98 oligonucleotide contained only the crucial c-Jun binding site described above. The −100/−74 oligonucleotide contained the C/EBPβ binding site but no AP-1 site, and the −115/−74 oligonucleotide spanned both sites. C/EBPβ bound readily to the 32P-labeled −100/−74 bp oligonucleotide (Fig. 6A, lane 1). This binding has been shown to be specific since it was efficiently competed by adding an excess of either unlabeled −100/−74 oligonucleotide or another C/EBPβ-binding oligonucleotide derived from the IL-6 promoter, and it was supershifted by monospecific antibody to C/EBPβ (38). Others have further narrowed the C/EBPβ binding site to −95 to −87 bp of the TNFα promoter (50). The C/EBPβ binds as a broad band because it is synthesized as a mixture of three translation products of 45, 38, and 20 kDa, all of which bind DNA (1, 41). In contrast, c-Jun did not bind to this oligonucleotide (Fig. 6A, lanes 2 and 3). Addition of increasing concentrations of c-Jun resulted in reduced binding of C/EBPβ to the −100/−74 oligonucleotide (Fig. 6A, lanes 4 and 5), similar to results reported for assay using a C/EBPβ-binding oligonucleotide from the IL-6 promoter (21). This inhibition was often not as complete as is apparent in Fig. 6A, although inhibition was reproducible at higher concentrations of c-Jun.
FIG. 6.
The effect of C/EBPβ and c-Jun interaction is DNA dependent. (A) Transcription factor c-Jun inhibits binding of C/EBPβ to the −100/−74 but not the −115/−74 oligonucleotide. TNFα promoter oligonucleotides −100/−74 (lanes 1 to 5) and −115/−74 (lanes 6 to 10) were 32P labeled and used as probes in the gel shift assay. c-Jun (1 and 3 μg; lanes 2, 3, 7, and 8) and C/EBPβ (1.0 μl of baculovirus nuclear extract; lanes 1 and 7), alone or combined (lanes 4, 5, 9, and 10), were loaded on a 4% polyacrylamide gel and run under nonreducing conditions. (B) c-Jun and C/EBPβ form a complex binding to the −115/−74 TNFα promoter oligonucleotide. C/EBPβ (1 μl) and c-Jun (1 μg) were added alone (lanes 1 and 2, respectively) or together (lanes 3 to 6) to the 32P-labeled −115/−74 oligonucleotide. Antibody to C/EBPβ (lane 4), control antibody (lane 5), or anti-c-Jun (lane 6) was added to the reaction mixture prior to loading on the gel. IgG, immunoglobulin G. (C) C/EBPβ alters the mobility of c-Jun bound to the −115/−98 TNFα promoter oligonucleotide. C/EBPβ (1 μl) and c-Jun (1 μg), alone (lanes 1 and 2) or together (lanes 3 to 5), were incubated with the 32P-labeled −115/−98 oligonucleotide for 20 min at room temperature prior to running the gel. Antibodies to C/EBPβ and c-Jun (lanes 4 and 5) were incubated with the transcription factors for 20 min at room temperature prior to adding the radiolabeled oligonucleotide. (D) The proximity of AP-1 and C/EBPβ binding sites affects activation. A single copy of each of the AP-1 and C/EBPβ binding sites from the TNFα promoter was inserted into pT81-Luc either unchanged (0 bp) or with 10 irrelevant oligonucleotides (10 bp) separating the two sites. The plasmids (5 μg) were transfected by electroporation into wild-type U937 cells, keeping the total concentration of plasmid constant (16 μg). After transfection the cells were treated with PMA and LPS as described in the text. Cells were harvested and analyzed as described in Materials and Methods. The parent plasmid was transfected in each experiment, and the results were subtracted prior to analysis. The results presented as the means ± 1 SE are representative of four independent experiments.
Both c-Jun and C/EBPβ bound to the −115/−74 oligonucleotide (Fig. 6A, lanes 6 to 8). However, in contrast to the results observed with the shorter oligonucleotide lacking the c-Jun binding site, c-Jun did not inhibit C/EBPβ binding to the −115/−74 oligonucleotide but resulted in the formation of a complex (Fig. 6A, lanes 9 and 10; Fig. 6B, lane 3 versus lanes 1 and 2) that contained both c-Jun and C/EBPβ. Monospecific antibodies to each transcription factor (Fig. 6B, lanes 4 and 6), but not a control antibody (Fig. 6B, lane 5), resulted in a supershift of a majority of the complex. As the ratio of C/EBPβ to c-Jun was increased, greater residual C/EBPβ binding was observed (data not shown). When limiting concentrations of c-Jun and or C/EBPβ were used, no synergistic or cooperative binding to the −115/−74 TNFα promoter oligonucleotide was observed (data not shown).
Others have observed that C/EBPβ may bind to AP-1 sites, thereby affecting gene expression (21, 23). Therefore, we used the −115/−98 oligonucleotide to determine if C/EBPβ was capable of binding to it or interfering with c-Jun binding. c-Jun but not C/EBPβ bound to this site (Fig. 6C, lanes 2 and 1, respectively). Binding by c-Jun was inhibited by excess unlabeled −115/−98 oligonucleotide and a consensus AP-1-binding oligonucleotide but not an unrelated oligonucleotide (data not shown), and the c-Jun–DNA complex reacted with antibodies specific for c-Jun (Fig. 6C, lane 5). Although there was no specific binding of C/EBPβ to this oligonucleotide (Fig. 6C, lane 1), addition of C/EBPβ caused a portion of the c-Jun–DNA complex to migrate more rapidly (Fig. 6C, lane 3), and this effect was reversed when antibodies specific for C/EBPβ were added (Fig. 6C, lane 4). This finding rules out the possibility that the changes in migration were due to nonspecific increases in protein concentration and suggests that C/EBPβ and c-Jun interact in vitro, in a DNA binding-dependent fashion.
Next, the functional effect of the proximity of the AP-1 and C/EBPβ binding sites of the TNFα promoter was examined. We constructed luciferase expression vectors that possessed a single copy of these two binding sites (−106 to −74 bp) from the TNFα promoter and one that possessed an irrelevant 10-bp segment between the two binding sites. When transfected into U937 cells differentiated with PMA and treated with LPS, a significant (P < 0.05) reduction of activation was observed when the 10 bp was inserted (Fig. 6D). No change in DNA binding efficiency by C/EBPβ and c-Jun was observed following the insertion of 10 bp between the C/EBPβ and AP-1 binding sites in the −115/−74 oligonucleotide (data not shown). These observations support the importance of the proximity of these two binding sites in the synergistic activation of the TNFα promoter.
DISCUSSION
Our prior data indicated that C/EBPβ was capable of binding to the −100/−74-bp region of the TNFα promoter and of activating the TNFα gene (38). Expression of DN C/EBPβ inhibited endogenous C/EBPβ in myelomonocytic cells, suppressing the activation of the TNFα promoter (38). Since C/EBPβ is readily detected in the nuclei of monocytes not actively synthesizing TNFα, activation of C/EBPβ and/or interaction with an additional transcription factor may be necessary for expression of the TNFα gene. Our data demonstrate that c-Jun is capable of interacting with C/EBPβ to activate the TNFα promoter in a unique manner that does not require the c-Jun transactivation domain.
Transient transfection of Jurkat T cells demonstrated that the macrophage-enriched nuclear transcription factor C/EBPβ and the ubiquitous c-Jun interacted, resulting in synergistic activation of the TNFα promoter. These interactions involved the AP-1 and C/EBPβ binding sites, located −106/−99 and −100/−74 bp, respectively, 5′ from the transcription start site of the TNFα gene (Fig. 1). The data indicate that the enhanced activation was independent of the c-Jun transactivation domain. Both wild-type and TAM-67 c-Jun were capable of amplifying the activation observed with C/EBPβ, indicating that the enhancement by c-Jun did not result from the additive or synergistic effects of heterologous transactivation domains.
Studies were performed with U937 cell lines that constitutively express endogenous C/EBPβ to determine the potential relevance of the observations obtained with the Jurkat T cells. The overexpression of c-Jun and the expression of TAM-67 were both associated with enhanced secretion of TNFα in differentiated cells stimulated with LPS, suggesting the possibility that the enhanced expression of the TNFα gene in U937 cells was dependent on an interaction between c-Jun/TAM-67 and C/EBPβ. The results of the transient transfections in U937 cells expressing TAM-67 support a direct effect of TAM-67 and C/EBPβ in the activation of the TNFα promoter in these cells. Alternatively, it is possible that c-Jun or TAM-67 affected differentiation or another mediator or transcription factor. Since c-Jun and TAM-67 affect differentiation differently, an effect on this process seems an unlikely explanation (18, 45). Although we cannot exclude the possibility that the effect on the TNFα gene was not secondary to that of another mediator or transcription factor, this is this first characterization, to our knowledge, in which the wild-type and transactivation domain mutant c-Jun interacted similarly in the activation of a gene.
In Jurkat T cells, the synergistic effect required that c-Jun and TAM-67 bind to the AP-1 site centered 102/103 bp upstream of the transcription start site. Truncations that abolished the AP-1 binding site or mutations of c-Jun that did not bind DNA did not manifest the synergistic effect. Similarly, truncation of this AP-1 site resulted in diminished activation of the TNFα promoter reporter in U937 cells expressing TAM-67. Our data indicate that the C/EBPβ transactivation domain was required for the enhanced activation of the TNFα gene observed by the interaction between C/EBPβ and c-Jun/TAM-67, since DN C/EBPβ inhibited the activation of the TNFα promoter reporter in TAM-67-expressing U937 cells. These observations, together with the proximity of the two binding sites, suggest that physical interactions between C/EBPβ and c-Jun/TAM-67, mediated by their leucine zippers (21), may contribute to the enhanced TNFα activation.
The importance of the close proximity of these two binding sites is supported by the gel shift experiments. With the −115/−74 oligonucleotide, both c-Jun and C/EBPβ bound preferentially to the same oligonucleotide and were supershifted by monospecific antibody against either protein. Previous studies documented the inhibition of binding by C/EBPβ to an IL-6 promoter oligonucleotide by c-Jun (21). Although similar inhibition of binding was noted with the shorter −100/−74 TNFα oligonucleotide that did not bind c-Jun, suppression of binding of C/EBPβ to the −115/−74 oligonucleotide, capable of binding both C/EBPβ and c-Jun, was not observed. C/EBPβ binds weakly to the TNFα promoter (38). Binding of c-Jun/TAM-67 at the neighboring cis site may facilitate the interaction of C/EBPβ and c-Jun that is mediated through their leucine zipper domains (21), stabilizing the C/EBPβ-DNA interaction and resulting in enhanced activation. Similar observations have been described for NF-κB p65 interaction with c-Fos or c-Jun and C/EBPβ interaction with NF-κB, despite lack of enhanced or cooperative binding to DNA in gel shift experiments (37, 42). Consistent with this possibility, insertion of 10 bp between the AP-1 and C/EBPβ binding sites of the TNFα promoter resulted in diminished activation in PMA-LPS-treated U937 cells.
We did not find that interaction with NFAT, CBP, or p300 was responsible for the enhanced activation observed in this study. Consistent with the results observed with CBP and p300, E1A 12S demonstrated minimal or no effect on TNFα promoter activity in PMA-treated Jurkat T cells and in LPS stimulated THP-1 myelomonocytic cells (28). If CBP or p300 were involved, inhibition by E1A might have been expected (4, 5, 28). Consistent with our data concerning NFAT, cyclosporin A did not inhibit TNFα transcription in macrophages (34). Additionally, in studies not presented, we have observed that c-Jun also interacted with NF-κB proteins to enhance TNFα promoter activation. However, the transactivation domain of c-Jun was required, suggesting that the enhanced expression of TNFα observed in U937 cells in response to TAM-67 was not due to interactions with NF-κB. Additionally, the NF-κB binding site (−97 to −88 bp of the TNFα gene) is partially removed in the −95 TNFα promoter reporter construct, and NF-κB fails to activate this promoter (data not shown).
The importance of the interactions between proteins binding to the −106/−99 and −97/−88 regions of the TNFα promoter has been identified by others (48). ATF-2/Jun and NFATp, respectively, were identified as binding to these sites (48). Independent and noncooperative binding at these two sites was responsible for optimal activation of the TNFα gene by anti-CD3 or ionomycin in Ar-5 T cells (48). Our observations concerning the interaction between c-Jun and C/EBPβ binding to these two regions were similar. We did not find evidence for cooperativity of binding of c-Jun and C/EBPβ to the −115/−74 oligonucleotide or for the binding of heterodimers when the −115/−98 and the −100/−74 TNFα oligonucleotides were independently used.
Our data suggest that the suppression observed with c-Jun in our cotransfection assays using the −120 TNFα promoter reporter was due to squelching by the transactivation domain of c-Jun and not inhibition of binding to the promoter or the formation of heterodimers. The TAM-67 mutant, which lacks a transactivation domain, failed to suppress the C/EBPβ-induced activation of either the −120 or the −95 TNFα promoter reporter in PMA-activated Jurkat T cells. This result suggests that the inhibition of C/EBPβ binding, as seen in the gel shift experiments using the −100/−74 oligonucleotide (Fig. 6) and proposed as a mechanism of inhibition of activation of a C/EBPβ-driven promoter reporter (21), was not responsible for the inhibition of TNFα promoter activation observed with wild-type c-Jun in Jurkat T cells in assays employing the −120 TNFα promoter reporter. We cannot exclude the possibility that overexpression of c-Jun inhibited C/EBPβ binding to the −95 TNFα promoter, since the effect of TAM-67 on C/EBPβ binding to the −100/−74 oligonucleotide was not examined. Supporting the interpretation that interaction through the leucine zipper was not required for suppression, cotransfection of c-Jun-LZ also resulted in suppression of C/EBPβ-induced activation. The explanation for the lack of inhibition by the c-Jun-DBD is not clear, since wild-type c-Jun was suppressive of C/EBPβ-induced activation with the −95 TNFα promoter-reporter, which lacked the AP-1 binding site centered at −102/−103 bp. Our data do not support the consumption of CBP or p300 by c-Jun as a cause for the suppression, since their overexpression failed to reverse the inhibition. Suppression was not specific for c-Jun, however, since coexpression of c-Fos, which failed to enhance C/EBPβ-induced activation, also was suppressive. Since the CMV vector control did not suppress, inhibition was not due simply to promoter squelching. Suppression of TNFα secretion was not observed in the U937 cell lines stably transfected with wild-type c-Jun. It is possible that this phenomenon does not occur in these cells or that the concentration of c-Jun was not high enough to allow this effect to be observed.
Although PMA was not required to activate the TNFα promoter reporter constructs in U937 cells, the requirement for PMA in the Jurkat T cells deserves comment. PMA may be necessary to phosphorylate and activate C/EBPβ (24, 30, 46, 47, 51, 52). It is possible that the PMA induces an additional endogenous factor required for TNFα activation. We did not find evidence to suggest that NFAT, CBP, or p300 was involved. Binding to the −115/−74 TNFα oligonucleotide was observed constitutively with nuclear extracts from Jurkat cells not treated with PMA (data not presented). This binding was not due to C/EBPβ and, although not further characterized, was greatly reduced following the addition of PMA, suggesting that PMA may inhibit a suppressive factor. For example, in pro-B-cell lines, CHOP was shown to bind C/EBPβ, inhibiting its ability to activate the Idl gene (40). As an additional mechanism, PMA may also increase the concentration of C/EBPβ, which was under the control of a CMV promoter. PMA resulted in a two- to fivefold increase in the expression of a CMV-luciferase promoter reporter in Jurkat T cells (data not presented). Activation of the TNFα promoter was very sensitive to the concentration of C/EBPβ-expressing plasmid transfected, less than 5-fold at 1 μg and >40-fold at 2 μg. The ratio of the 20-kDa inhibitory and the 38- and 45-kDa stimulatory versions of C/EBPβ are important in the overall ability of C/EBPβ to become transcriptionally active within a cell. PMA may effect this ratio in favor of activation. This process is thought to be regulated at the level of mRNA translation. We have not been able to definitively ascribe the effects of PMA to any one of these mechanisms.
AP-1 and C/EBPβ are also known to cooperate in the regulation of other genes, including the chicken myelomonocytic growth factor (cGMF) gene and the human TSF-6 and collagenase-1 genes (11, 23, 32). An AP-1 binding site in the proximal promoter and a C/EBPβ site approximately 2,000 bp upstream appeared to regulate the human collagenase-1 gene independent of one another (11). For the cGMF and the TSF-6 genes, mutation of the AP-1 binding sites in the promoter reporters reduced, and mutation of the C/EBPβ sites essentially abolished, activation of the genes. Similarly, deletion of the AP-1 site centered at −102/−103 bp of the TNFα promoter abolished the synergistic effect of the coexpression of c-Jun on C/EBPβ-induced activation. Although we have not developed point mutations in the TNFα promoter reporter constructs to precisely define the location important for C/EBPβ-induced activation, we previously demonstrated that the −36 TNFα promoter reporter, lacking the C/EBPβ binding site, was not active in myelomonocytic U937 cells which constitutively express high concentrations of C/EBPβ or in Jurkat T cells transfected with C/EBPβ (38). Further studies will be required to determine the importance of the interaction between c-Jun and C/EBPβ in the activation of the cellular TNFα gene in monocytes/macrophages and other cell types expressing C/EBPβ.
ACKNOWLEDGMENTS
This work was supported in part from grants provided by NIH (RO1-AR-43642 and PO60-AR-30692), the Veterans Administration Research Service, and the Greater Chicagoland Chapter of the Arthritis Foundation (to R.P.) and by NIH (R01-CA-58443) and The Council for Tobacco Research, Inc. (3849) (to S.N.).
We thank Guobin Song for helpful discussions and thank Guobin Song and Craig Wlodarek for assistance with some of the transient transfection assays.
REFERENCES
- 1.Akira S, Isshiki H, Sugita T, Tanabe O, Kinoshita S, Nishio Y, Nakajima T, Hirano T, Kishimoto T. A nuclear factor for IL-6 expression (NF-IL6) is a member of a C/EBPβ family. EMBO J. 1990;9:1897–1906. doi: 10.1002/j.1460-2075.1990.tb08316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alani R, Brown P, Binétruy B, Dosaka H, Rosenberg R, Angel P, Karin M, Birrer M J. The transactivating domain of the c-Jun proto-oncoprotein is required for cotransformation of rat embryo cells. Mol Cell Biol. 1991;11:6286–6295. doi: 10.1128/mcb.11.12.6286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Angel P, Imagawa M, Chiu R, Stein B, Imbra R J, Rahmsdorf H J, Jonat C, Herrlich P, Karin M. Phorbol ester-inducible genes contain a common cis element recognized by a TPA-modulated trans-acting factor. Cell. 1987;49:729–739. doi: 10.1016/0092-8674(87)90611-8. [DOI] [PubMed] [Google Scholar]
- 4.Arias J, Alperts A S, Brindle P, Claret F X, Smeal T, Karin M, Feramisco J, Montminy M. Activation of cAMP and mitogen responsive genes relies on a common nuclear factor. Nature. 1994;370:226–229. doi: 10.1038/370226a0. [DOI] [PubMed] [Google Scholar]
- 5.Bannister A J, Oehler T, Whilhelm D, Angel P, Kouzarides T. Stimulation of c-Jun activity by CBP:c-Jun residues Ser 63/73 are required for CBP induced stimulation in vivo and CBP binding in vitro. Oncogene. 1995;11:2509–2514. [PubMed] [Google Scholar]
- 6.Brown P H, Alani R, Preis L H, Szabo E, Birrer M J. Suppression of oncogene-induced transformation by a deletion mutant of c-jun. Oncogene. 1992;8:877–886. [PubMed] [Google Scholar]
- 7.Burk O, Mink S, Ringwald M, Klempnauer K H. Synergistic activation of the chicken mim-1 gene by v-myb and C/EBPβ transcription factors. EMBO J. 1993;12:2027–2038. doi: 10.1002/j.1460-2075.1993.tb05852.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Callahan M, Lovis R M, Rammohan C, Lu Y, Pope R M. Autocrine regulation of collagenase gene expression by TNF-α in U937 cells. J Leukocyte Biol. 1996;59:125–132. doi: 10.1002/jlb.59.1.125. [DOI] [PubMed] [Google Scholar]
- 9.Cao Z, Umek R M, McNight S L. Regulated expression of three C/EBPβ isoforms during adipose conversion of 3T3-L1 cells. Genes Dev. 1991;5:1538–1552. doi: 10.1101/gad.5.9.1538. [DOI] [PubMed] [Google Scholar]
- 10.Chrivia J C, Kwok R P S, Lamb N, Hagiwara M, Montminy M R, Goodman R H. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 1993;365:855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- 11.Doyle G A R, Pierce R A, Parks W C. Transcriptional induction of collagenase-1 in differentiated monocyte-like (U937) cells is regulated by AP-1 and an upstream C/EBP-β site. J Biol Chem. 1997;272:11840–11649. doi: 10.1074/jbc.272.18.11840. [DOI] [PubMed] [Google Scholar]
- 12.Eckner R, Ewen M E, Newsome D, Gerdes M, DeCaprio J A, Lawrence J B, Livingston D M. Molecular cloning and functional analysis of the adenovirus E1A-associated 300-kD protein (p300) reveals a protein with properties of a transcriptional adaptor. Genes Dev. 1994;8:869–884. doi: 10.1101/gad.8.8.869. [DOI] [PubMed] [Google Scholar]
- 13.Economou J S, Rhoades K, Essner R, McBride W H, Gassonand J C, Morton D L. Genetic analysis of the human tumor necrosis factor α/cachectin promoter region in a macrophage cell line. J Exp Med. 1989;170:321–326. doi: 10.1084/jem.170.1.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elliot M J, Maini R N, Feldman M, Kalden J R, Antoni C, Smolen J S, Leeb B, Breedveld F C, Macfarlane J D, Bijl H, Woody J N. Randomized double-blind comparison of chimeric monoclonal antibody to tumor necrosis factor a (cA2) versus placebo in rheumatoid arthritis. Lancet. 1994;344:1105–1110. doi: 10.1016/s0140-6736(94)90628-9. [DOI] [PubMed] [Google Scholar]
- 15.Goldfeld A E, McCaffrey P G, Strominger J L, Rao A. Identification of a novel cyclosporin-sensitive element in the human tumor necrosis factor α gene promoter. J Exp Med. 1993;178:1365–1379. doi: 10.1084/jem.178.4.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gonzalez-Amaro R, Garcia-Monzon C, Garcia-Buey L, Moreno-Otero R, Alonso J L, Yague E, Pivel J P, Lopez-Cabrera M, Fernandez-Ruiz E, Sanchez-Madrid F. Induction of tumor necrosis factor alpha production by human hepatocytes in chronic viral hepatitis. J Exp Med. 1994;179:841–848. doi: 10.1084/jem.179.3.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grove M, Plumb M. C/EBPβ, NF-κB, and c-Ets family members and transcriptional regulation of the cell-specific and inducible macrophage inflammatory protein 1α immediate-early gene. Mol Cell Biol. 1993;13:5276–5289. doi: 10.1128/mcb.13.9.5276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grant S, Freemerman A J, Birrer M J, Martin H A, Turner A J, Szabo E, Chelliah J, Jarvis W D. Effect of 1-β-d-arabinofuranosylcytosine on apoptosis and differentiation in human monocytic leukemia cells (U937) expressing c-Jun dominant-negative mutant protein (TAM67) Cell Growth Differ. 1996;7:603–610. [PubMed] [Google Scholar]
- 19.Hotamisligil G S, Arner P, Caro J F, Atkinson R L, Spiegelman B M. Increased adipose tissue expression of tumor necrosis factor-alpha in human obesity and insulin resistance. J Clin Invest. 1995;95:2409–2415. doi: 10.1172/JCI117936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hotamisligil G S, Shargill N S, Spiegelman B M. Adipose expression of tumor necrosis factor-alpha: direct role in obesity-linked insulin resistance. Science. 1993;259:87–91. doi: 10.1126/science.7678183. [DOI] [PubMed] [Google Scholar]
- 21.Hsu W, Kerpolla T K, Chen P-L, Curran T, Chen-Kiang S. Fos and Jun repress transcription activation by NF-IL6 through association at the basic zipper region. Mol Cell Biol. 1994;14:268–276. doi: 10.1128/mcb.14.1.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kern P A, Saghizadeh M, Ong J M, Bosch R J, Deem R. The expression of tumor necrosis factor in human adipose tissue. Regulation by obesity, weight loss, and relationship to lipoprotein lipase. J Clin Invest. 1995;95:2111–2119. doi: 10.1172/JCI117899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klampfer L, Lee T H, Hsu W, Vilcek J, Chen-Kiang S. NF-IL6 and AP-1 cooperatively modulate the activation of the TSG-6 gene by tumor necrosis factor alpha and interleukin-1. Mol Cell Biol. 1994;14:6561–6569. doi: 10.1128/mcb.14.10.6561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kowenz-Leutz E, Twamley G, Ansieau S, Leutz A. Novel mechanism of C/EBPβ (NF-M) transcriptional control: activation through derepression. Genes Dev. 1994;8:2781–2791. doi: 10.1101/gad.8.22.2781. [DOI] [PubMed] [Google Scholar]
- 25.Kramer B, Wiegmann K, Kronke M. Regulation of the human TNFα promoter by the transcription factor Ets. J Biol Chem. 1995;270:6577–6583. doi: 10.1074/jbc.270.12.6577. [DOI] [PubMed] [Google Scholar]
- 26.LeClair K P, Blanar M A, Sharp P A. The p50 subunit of NF-κB associates with the NF-IL6 transcription factor. Proc Natl Acad Sci USA. 1992;89:8145–8149. doi: 10.1073/pnas.89.17.8145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.MacNaul K L, Hutchinson N I, Parson J N, Bayne E K, Tocci M J. Analysis of IL-1 and TNFα-gene expression in human rheumatoid synoviocytes and normal monocytes by in situ hybridization. J Immunol. 1990;145:4154–4166. [PubMed] [Google Scholar]
- 28.Metcalf J P. Adenovirus E1A 13S gene product upregulates tumor necrosis factor gene. Am J Physiol. 1996;270:L535–L540. doi: 10.1152/ajplung.1996.270.4.L535. [DOI] [PubMed] [Google Scholar]
- 28a.Mink S, Haenig B, Klempnauer K H. Interaction and functional collaboration of p300 and C/EBPβ. Mol Cell Biol. 1997;17:6609–6617. doi: 10.1128/mcb.17.11.6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miyazaki Y, Araki K, Vesin C, Garcia I, Kapanci Y, Whitsett J A, Piguet P F, Vassalli P. Expression of a tumor necrosis factor-alpha transgene in murine lung causes lymphocytic and fibrosing alveolitis. A mouse model of progressive pulmonary fibrosis. J Clin Invest. 1995;96:250–259. doi: 10.1172/JCI118029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakajima T, Kinoshita S, Sasagawa T, Sasaki K, Naruto M, Kishimoto T, Akira S. Phosphorylation at threonine-235 by a ras-dependent mitogen-activated protein kinase cascade is essential for transcription factor NF-IL6. Proc Natl Acad Sci USA. 1993;90:2207–2211. doi: 10.1073/pnas.90.6.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Natsuka S, Akira S, Nishio Y, Hashimoto S, Sugita T, Isshiki H, Kishimoto T. Macrophage differentiation-specific expression of NF-IL6, a transcription factor for interleukin-6. Blood. 1991;79:460–466. [PubMed] [Google Scholar]
- 32.Ness S A, Kowenz-Leutz E, Casini T, Graf T, Leutz A. Myb and MF-M: combinatorial activators of myeloid genes in heterologous cell types. Genes Dev. 1993;7:749–759. doi: 10.1101/gad.7.5.749. [DOI] [PubMed] [Google Scholar]
- 33.Newell C L, Deisseroth A B, Lopez-Berestein G. Interaction of nuclear proteins with an AP-1/CRE-like promoter sequence in the human TNFα gene. J Leukocyte Biol. 1994;56:27–35. doi: 10.1002/jlb.56.1.27. [DOI] [PubMed] [Google Scholar]
- 34.Nguyen D T, Eskandari M K, DeFore L E, Raiford C L, Strieter R M, Kunkel S L, Remick D G. Cyclosporin A modulation of tumor necrosis factor gene expression and effects in vitro and in vivo. J Immunol. 1990;144:3822–3828. [PubMed] [Google Scholar]
- 35.Nishio Y, Isshiki H, Kishimoto T, Akira S. A nuclear factor for interleukin-6 expression (NF-IL6) and the glucocorticoid receptor synergistically activate transcription of the rat 1-acid glycoprotein gene via direct protein-protein interaction. Mol Cell Biol. 1993;13:1854–1862. doi: 10.1128/mcb.13.3.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Petrak D, Memon S A, Birrer M J, Ashwell J D, Zacharchuk C M. Dominant negative mutant of c-jun inhibits NF-AT transcriptional activity and prevents IL-2 gene transcription. J Immunol. 1994;153:2046–2051. [PubMed] [Google Scholar]
- 37.Plevy S E, Gemberling J H, Hsu S, Dorner A J, Smale S T. Multiple control elements mediate activation of the murine and human interleukin 12 p40 promoters: evidence of functional synergy between C/EBP and Rel proteins. Mol Cell Biol. 1997;17:4572–4588. doi: 10.1128/mcb.17.8.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pope R M, Leutz A, Ness S. C/EBPβ regulation of the tumor necrosis factor alpha gene. J Clin Invest. 1994;94:1449–1455. doi: 10.1172/JCI117482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rhoads K L, Golub S H, Economou J S. The regulation of the human tumor necrosis factor a promoter region in macrophage, T cell and B cell lines. J Biol Chem. 1992;267:22102–22107. [PubMed] [Google Scholar]
- 40.Saisanit S, Sun X-H. Regulation of the pro-B-cell-specific enhancer of the Idl gene involves the C/EBP family of proteins. Mol Cell Biol. 1997;17:844–850. doi: 10.1128/mcb.17.2.844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sears R C, Sealy L. Multiple forms of C/EBPβ bind the EFII enhancer sequence in the Rous sarcoma virus long terminal repeat. Mol Cell Biol. 1994;14:4855–4871. doi: 10.1128/mcb.14.7.4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stein B, Cogswell P C, Baldwin A S., Jr Functional and physical associations between NF-κB and C/EBPβ family members: a Rel domain-bZip interaction. Mol Cell Biol. 1993;13:3964–3974. doi: 10.1128/mcb.13.7.3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sterneck E, Christine M, Katz S, Leutz A. Autocrine growth induced by kinase type oncogenes in myeloid cells requires AP-1 and NF-M, a myeloid specific, C/EBPβ-like factor. EMBO J. 1992;11:115–126. doi: 10.1002/j.1460-2075.1992.tb05034.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Su B, Jacinto E, Hibi M, Kallunki T, Karin M, Ben-Neriah Y. JNK is involved in signal integration during costimulation of T lymphocytes. Cell. 1994;77:727–736. doi: 10.1016/0092-8674(94)90056-6. [DOI] [PubMed] [Google Scholar]
- 45.Szabo E, Preis L H, Birrer M J. Constitutive cJun expression induces partial macrophage differentiation in U-937 cells. Cell Growth Differ. 1994;5:439–446. [PubMed] [Google Scholar]
- 46.Trautwein C, Caelles C, van der Geer P, Hunter T, Karin M, Chojkier M. Transactivation by NF-IL6/LAP is enhanced by phosphorylation of its activation domain. Nature. 1993;364:544–547. doi: 10.1038/364544a0. [DOI] [PubMed] [Google Scholar]
- 47.Trautwein C, van der Geer P, Karin M, Hunter T, Chojkier M. Protein kinase A and C site-specific phosphorylations of LAP (NF-IL6) modulate its binding affinity to DNA recognition elements. J Clin Invest. 1994;93:2554–2561. doi: 10.1172/JCI117266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsai E Y, Jain J, Pesavento P A, Rao A, Goldfeld A F. Tumor necrosis factor alpha gene regulation in activated T cells involves ATF-2/Jun and NFATp. Mol Cell Biol. 1996;16:459–467. doi: 10.1128/mcb.16.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vassalli P. The pathophysiology of tumor necrosis factors. Annu Rev Immunol. 1992;10:411–452. doi: 10.1146/annurev.iy.10.040192.002211. [DOI] [PubMed] [Google Scholar]
- 50.Wedel A, Sulski G, Ziegler-Heitbrock H W L. CCAAT/enhanced binding protein is involved in the expression of the tumor necrosis factor gene in human monocytes. Cytokine. 1996;8:335–341. doi: 10.1006/cyto.1996.0046. [DOI] [PubMed] [Google Scholar]
- 51.Wegner M, Cao Z, Rosenfeld M G. Calcium-regulated phosphorylation within the leucine zipper of C/EBPβ. Science. 1992;256:370–373. doi: 10.1126/science.256.5055.370. [DOI] [PubMed] [Google Scholar]
- 52.Williams S C, Baer M, Dillner A J, Johnson P F. CRP2 (C/EBPβ) contains a bipartite regulatory domain that controls transcriptional activation, DNA binding and cell specificity. EMBO J. 1995;14:3170–3183. doi: 10.1002/j.1460-2075.1995.tb07319.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang X D, Tisch R, Singer S M, Cao Z A, Liblau R S, Schreiber R D, McDevitt H O. Effect of tumor necrosis factor alpha on insulin-dependent diabetes mellitus in NOD mice. I. The early development of autoimmunity and the diabetogenic process. J Exp Med. 1994;180:995–1004. doi: 10.1084/jem.180.3.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang Y, Lee T C, Guillemin B, Yu M C, Rom W N. Enhanced IL-1 beta and tumor necrosis factor-alpha release and messenger RNA expression in macrophages from idiopathic pulmonary fibrosis or after asbestos exposure. J Immunol. 1993;150:4188–4196. [PubMed] [Google Scholar]