Abstract
Phocaeicola dorei and Phocaeicola vulgatus are very common and abundant members of the human gut microbiome and play an important role in the infant gut microbiome. These species are closely related and often confused for one another; yet, their genome comparison, interspecific diversity, and evolutionary relationships have not been studied in detail so far. Here, we perform phylogenetic analysis and comparative genomic analyses of these two Phocaeicola species. We report that P. dorei has a larger genome yet a smaller pan-genome than P. vulgatus. We found that this is likely because P. vulgatus is more plastic than P. dorei, with a larger repertoire of genetic mobile elements and fewer anti-phage defense systems. We also found that P. dorei directly descends from a clade of P. vulgatus¸ and experienced genome expansion through genetic drift and horizontal gene transfer. Overall, P. dorei and P. vulgatus have very different functional and carbohydrate utilisation profiles, hinting at different ecological strategies, yet they present similar antimicrobial resistance profiles.
Keywords: Phocaeicola, Gut microbiome, Comparative genomics, Pangenome, Horizontal gene transfer
Subject terms: Microbiology, Microbiome
Introduction
Phocaeicola (Bacteroides) dorei and Phocaeicola (Bacteroides) vulgatus are gram-negative, non-spore-forming, non-motile anaerobic rods commonly present in the human gut microbiota1. The genus Bacteroides (first described in2) was recently re-structured after a careful phylogenetic analysis that concluded that a number of Bacteroides species, including P. vulgatus and P. dorei, are phylogenetically closer to the genus Phocaeicola than to Bacteroides fragilis, the type strain of the genus Bacteroides3. P. vulgatus was first identified in 1933 as a common microbe in the faeces of adults, hence the name ‘vulgatus’ meaning common or ordinary. P. dorei was first isolated and characterised in 2006 from adult faeces4. P. vulgatus and P. dorei are indeed widely abundant and ubiquitous bacteria in the human gut5. They colonise the gut soon after birth in vaginally delivered infants, increasing in abundance after the introduction of solid food1. In caesarean section-born infants, the establishment of Phocaeicola species in the gut is delayed and it can take up to 18 months to match the relative abundance of Phocaeicola (Bacteroides) present in the gut of vaginally delivered infants6–8.
P. dorei and P.vulgatus have a wide range of carbohydrate utilisation mechanisms, as well as vitamin and hormone production genes. Different strains of P. vulgatus have varying effects on inflammatory diseases, including alleviating inflammation, reducing atherosclerosis, modulating the gut microbiota and regulating the levels of cytokines9–11. The anti-inflammatory effect of P. vulgatus has been associated with the production of short chain fatty acids (SCFAs) and capsular polysaccharides9,10. Strains of P. dorei have been associated with reduction of cholesterol12, improvement of influenza symptoms13, and improving atherosclerosis by reducing inflammation and lipopolysaccharide production11. The beneficial effects of P. dorei and P. vulgatus reported in the literature are associated with specific strains, not with the whole species, and, in some cases, the use of alternative strains could cause the opposite effect10. Another important consideration is that there are several studies associating P. dorei with metabolic and immunological conditions, such as type one diabetes14.
Initial 16S rRNA sequences and matrix-assisted laser desorption ionisation time-of-flight mass spectrometry (MALDI-TOF) based on β-glucosidase pointed towards the fact that P. dorei and P. vulgatus are two different species15,16. However, P. dorei and P. vulgatus are very similar, and there were cases of misidentification between the two species because of their mass spectra similarity17. Some other studies also identified strong population structure in P. vulgatus, with the presence of subspecies co-existing within the same subjects18,19. While P. vulgatus is closely related to P. dorei, a comparative genomics analysis has not yet been performed.
In the present study, we carried out a comparative analysis of P. dorei and P. vulgatus to investigate the genomic differences between these two species, their role in the human gut and their phylogenetic relationship. We included genomes obtained from whole-genome sequencing from isolates as well as metagenome-assembled genomes (MAGs) from the Unified Human Gastrointestinal Genome (UHGG)20 and from the Early-Life Gut Genomes (ELGG)21 databases. We also included genomes of isolates publicly available on NCBI22. We obtained a total of 3951 genomes, of which 1086 were P. dorei and 2865 were P. vulgatus genomes. We investigated genomic and functional differences, pan-genomes, phylogeny, as well as Carbohydrate-Active enZymes (CAZymes) content, antimicrobial resistance (AMR) genes, mobile genetic elements and horizontal gene transfer (HGT) events.
Methods
Data and metadata collection
Assemblies (MAGs from metagenomes and whole genome sequences from isolates) of the two species studied here were downloaded from the UHGG20 and from the ELGG21 databases. Genomes of isolates publicly available on NCBI (November 2022) were also included. Redundant isolates from different databases were systematically searched and removed. Metadata were retrieved from the UHGG and the ELGG databases. For NCBI isolates, metadata were retrieved using both NCBI-Datasets and Entrez v10.223, searching for ‘biosample’, ‘isolation_source’, ‘host’, ‘disease’, ‘host_disease’, ‘sample_type’, ‘env_broad_scale’, ‘geo_loc_name’. We also used ffq24 for ENA-related metadata25. The metadata obtained was manually curated.
Species identification and filtering criteria
Assemblies that were below 90% completeness and 5% contamination were filtered out using checkM2 v0.1.326 in line with27,28. The UHGG does not differentiate between P. dorei and P. vulgatus. To assign each assembly to one of the two species, the average nucleotide identity (ANI) was calculated with the NCBI reference genomes of the two species using fastANI v1.32 and we considered a threshold higher or equal to 97.5% as the species identity threshold. Results were validated using GGDC 3.0, an in silico DNA-DNA hydridization method. All of the assemblies were assigned to a species. Additionally, the taxonomy of each assembly was checked using GTDB-tk v1.5.027. Assemblies that did not correspond to their assigned species were removed. After these filtration steps, a total of 1086 P. dorei and 2865 P. vulgatus genomes were obtained.
Genome annotation and pan-genome reconstruction
Each assembly was annotated using Prokka v1.1429. The annotated ‘.gff’ files obtained from Prokka were then used for the pan-genome analysis while the protein files ‘.faa’ were used for annotation with eggNOG-mapper30. Pan-genomes analysis were carried out using Roary v3.1331 with the following options: ‘-e -n -g 1000000 -v -cd 90’, in line with recommendations of28 for pan-genomes analysis including MAGs. Rarefaction curves for the pangenomes (total and conserved genes) were performed using the R package micropan with ‘n.perm = 100’ (100 random combinations). The pan-genomes obtained from Roary were annotated for carbohydrate enzymes using dbCAN v3.032 on the CAzy database. Glycoside-hydrolase (GH) family numbers were compiled from dbCAN results, including HMM33 and DIAMOND34 using custom scripts. Later, the results were filtered to only show the GH families related to human milk oligosaccharide (HMO) genes, using both the list produced by35 and36. Heatmaps were produced using the R package ComplexHeatmap v2.10.0. Differential abundance analysis was conducted using Maaslin2 with parameters ‘transform = LOG, max_significance = 0.05, fixed_effects = species’ and default parameters otherwise37. For AMR genes, Resistance Gene Identifier v6.0.0 and the Comprehensive Antibiotic Resistance Database (CARD)38 were used on the two pan-genomes and subsequently an AMR profile for each isolate was assigned.
Mobile genetic elements and horizontal gene transfer analysis
To look for mobile genetic elements, the ‘mobileOG-pl’ pipeline on the mobile-OG database v1.639 was used, with the parameters ‘-k 15 -e 1e−20 -p 90 -q 90’. We screened for insertion sequences (IS), integrative and conjugative elements (ICEs), bacteriophages, and plasmids. For detection of putative HGTs, HGTECTOR v2.0b340 was used with the option ‘-m diamond’. This tool was used on P. dorei and P. vulgatus separately, specifying their taxonomic identification number from NCBI. To identify prophages, virsorter2 v2.2.341 was used, with parameters ‘-min-length 1500’. Clustered Regularly Interspaced Short Palindromic Repeats (CRISPRs) were checked using MinCED v0.4.2 with default parameters42.
Phylogenetic tree
The phylogenetic tree of P. dorei and P. vulgatus was reconstructed using a multi-step approach. First, the tool PopPUNK (population partitioning using nucleotide K-mers)43 was used to reconstruct a draft phylogenetic tree. Briefly, PopPUNK is an annotation and alignment-free method to cluster genomes based on k-mers of variable lengths. It first estimates core and accessory genome distances between genomes by performing pairwise comparisons through k-mer matching between two sequences at multiple-k lengths to distinguish divergence in shared sequences. Then, it creates clusters using core and accessory divergences using a specified clustering model. PopPUNK v2.6.0 was used with all P. dorei and P. vulgatus filtered genomes as well as the NCBI reference genome ‘GCF_013358205.1’ for Phocaeicola sartorii (i.e., closest Phocaeicola species related to P. dorei and P. vulgatus) as an extra-group. The parameters ‘-fit-model dbscan’, choosing an HDBSCAN (density-based clustering based on hierarchical density estimates) model was used to find clusters in the core and accessory distances. The model was refined using ‘-fit-model refine’ and generated the draft phylogenetic tree with the function ‘poppunk_visualise’ and parameter ‘-microreact’, which creates a neighbour-joining tree from the core-distances.
GToTtree was used to obtain a coarse-grained tree to validate our approach. Briefly, eZtree finds single copy makers genes for a set of genomes and aligns them for phylogenetic reconstruction. The phylogenetic tree was obtained with Fasttree v2.1.10. The core alignment file ‘core_gene_alignment.aln’ with FastTree v2.1.10 was used to produce a phylogenetic tree (Price, Dehal, and Arkin 2010). The tree and associated metadata were plotted using R packages ‘ggtree’ v3.1.5, ‘phytools’ v0.7-90, ‘tidyverse’ v1.3.1.
Statistical analysis
Statistical significance was calculated using Wilcoxon rank-sum test (using the function ‘wilcox.test’ as implemented in R 4.0.2, hereto referred as Wilcoxon test), with paired option. The p values were adjusted for false discovery using Benjamini–Hochberg procedure.
Results
P. vulgatus has smaller genomes but a bigger pan-genome than P. dorei
Assemblies and annotated genomes of both species were downloaded from the UHGG20, the ELGG21 and from NCBI22 (November 2022). Genomes with less than 90% completeness and more than 5% contamination were filtered out (Fig. 1D,E). ANI of more than 97.5% with respective reference genomes was used to separate P. dorei from P. vulgatus, since these species were not separated in the UHGG and ELGG databases.
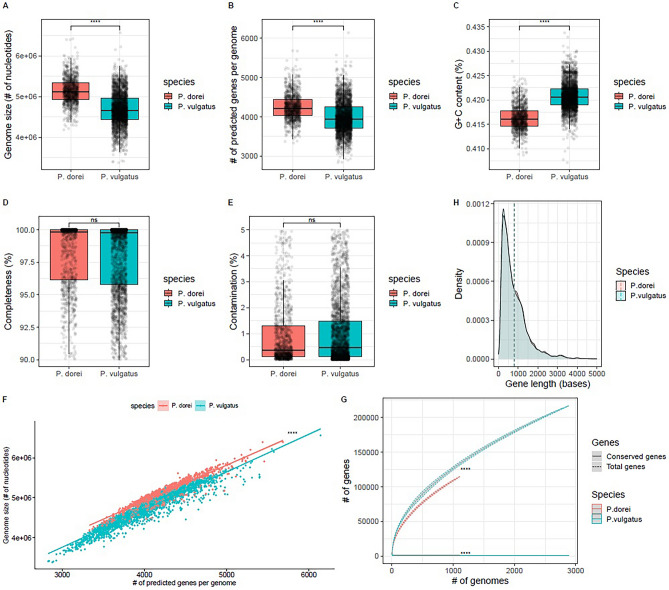
Figure 1.
Overview of the characteristics of the genomes of P. dorei and P. vulgatus. (Wilcoxon tests, ****: adj. p value < 0.0001, ns: non-significant). (A) Genome size comparison (total number of nucleotides). (B) Comparison between the number of predicted genes per genome. (C) Comparison between the GC content (%). (D, E) Completeness and Contamination level of the assemblies used in this study (%). (F) Genome size as a function of the number of predicted genes per genome. The lines correspond to linear regression for each species with 95% confidence intervals (R2 = 0.901 and R2 = 0.920 for P. dorei and P. vulgatus, respectively, t-test adj. p values < 0.0001). (G) Average cumulative number of conserved genes (plain lines) and total genes (dashed lines) as a function of added genomes in the pan-genome for P. dorei and P. vulgatus (averaged over 100 random combinations). Grey area correspond to standard deviation. (H) Distribution of gene lengths (in bases) for P. dorei and P. vulgatus.
The characteristics of the genomes of the two species are shown in Fig. 1A–D and Supplementary Table 1. The average genome size of P. dorei was 5.14 × 106 nucleotides and it was significantly larger than P. vulgatus at 4.69 × 106 nucleotides (Wilcoxon test, adj. p value < 0.0001) (Fig. 1A). On average, the number of predicted genes per genome was significantly higher in P. dorei (Wilcoxon test, adj. p value < 0.0001) (Fig. 1B). As expected, the number of predicted genes increased linearly with genome size for both species (R2 = 0.901 and R2 = 0.920 for P. dorei and P. vulgatus, respectively, t-test adj. p values < 0.0001) (Fig. 1F), but when normalizing by genome size (i.e., looking at the number of genes per unit of genome length) P. vulgatus had more genes per unit of genome length than P. dorei (this corresponds to the y-intercept of the lines in Fig. 1F).
This was surprising and might indicate that P. dorei had more non-coding regions per genome than P. vulgatus and/or larger genes. Nonetheless, no statistical differences were observed in the gene-length distribution between the two species (Fig. 1H), with an average gene-length of 826.84 bases and 795.03 bases for P. dorei and P. vulgatus, respectively. The average GC content was also different between the two species, with P. vulgatus having a higher GC content (41.16% and 42.10% on average for P. dorei and P. vulgatus respectively Fig. 1C). Coding regions usually have a higher GC content, compatible with the hypothesis that P. dorei has more non-coding regions per genome.
In contrast to what was observed for the genome size, P. vulgatus had a larger pan-genome (i.e., total number of genes for the same number of genomes) than P. dorei (Wilcoxon test, adj. p value < 0.0001), with a significantly larger gene repertoire overall (114,820 genes and 217,018 genes for P. dorei and P. vulgatus, respectively) (Fig. 1G). Both pan-genomes were open according to a power-law regression (Heaps' law, Bdorei = 0.486, Bvulgatus = 0.494)44, reflecting the plastic nature of the two species genomes. The number of core genes was higher in P. dorei than in P. vulgatus (1968 and 951 genes, respectively), but this might be prone to artefacts as MAGs were included in this study and they are known to artificially decrease the number of core genes28. The larger pan-genome of P. vulgatus could indicate that P. vulgatus had more time to accumulate genes at the species level, or it could be more prone to acquiring new genes.
P. dorei directly descends from P. vulgatus according to phylogeny
To investigate the evolution and phylogeny of the two species, a phylogenetic tree including all the assemblies was generated (Fig. 2A,B). A phylogenetic tree was built using the tool PopPUNK, including the species Phocaeicola sartorii as an extra-group. There was a marked difference between P. dorei and P. vulgatus assemblies. P. dorei appeared to directly descend from a clade of P. vulgatus. We thus propose that P. dorei and P. vulgatus could form a unique species where P. dorei is a sub-species of P. vulgatus. This is coherent with, for example, the strong sub-species structure found for P. vulgatus in metagenomics data by18 that could be showing both P. dorei and P. vulgatus. As illustrated previously, the genome size of P. dorei assemblies were larger than P. vulgatus. Nonetheless, some clades of P. vulgatus appeared to have larger than average (i.e., similar to P. dorei) genome sizes (Fig. 2A). Whether the mechanisms for genome size increase are the same as for P. dorei is unclear. A subset of strains that were at the transition between P. dorei and P. vulgatus (Fig. 2B) appeared to increase in genome size across the branch of the tree leading to P. dorei.
Figure 2.
Phylogeny and genomic comparison of P. dorei and P. vulgatus. (A) Phylogenetic tree (natural logarithmic scale) including metadata regarding host, country of origin, isolation from disease state, isolation from infants, and including all P. dorei and P. vulgatus assemblies used in this study. The blue and red bars indicate genome size relative to the average genome size of all assemblies. (B) Phylogenetic tree at the normal scale displaying only the average genome size. (C) Comparison of synteny between P. dorei and P. vulgatus reference genomes.
Genome synteny based on the reference genomes seems to be well conserved between P. dorei and P. vulgatus (Fig. 2C). There was a large inversion/complex modification from 1.3 to 1.95 Mb, and few other minor inversions. Additional regions in P. dorei that were missing in P. vulgatus and likely responsible for genome expansion are homogeneously spread across the genomes, hinting at potential gene acquisition by HGT during evolution.
P. vulgatus strains have larger genome plasticity but have fewer horizontal gene transfer events
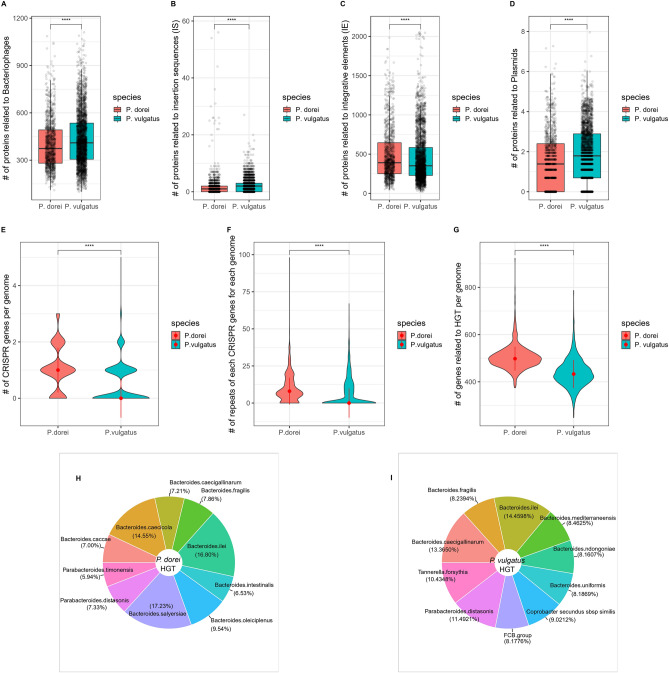
To investigate the plasticity of the two species, we looked at their mobile genetic element content (i.e., mobilome) and the presence of HGT events. In line with39, we considered different categories of mobile genetic elements: IS, ICEs, bacteriophages, and plasmids. On average, P. dorei had significantly less proteins associated with IS, bacteriophages and plasmids than P. vulgatus while it had more proteins associated with ICEs (Wilcoxon test, adj. p value < 0.0001, Fig. 3A–D). P. dorei had subsequently more genes related to HGT than P. vulgatus (503 and 438 genes per genome on average, respectively) (Wilcoxon test, adj. p value < 0.0001, Fig. 3G). For both of them, these genes were inherited from species belonging to the Bacteroidales order, and the majority from the Bacteroides genus (Fig. 3H,I). Most of the donor species were from the human gut. Of note, P. dorei and P. vulgatus had HGT coming from Bacteroides caecicola (5 genes per genome on average) and Bacteroides caecigallinarum (2.5 genes per genome on average), respectively. These species live in the caecum of the domestic chicken (Gallus domesticus), indicating that it could serve as a reservoir of P. dorei and P. vulgatus species where HGT can occur.
Figure 3.
Mobile genetic elements and HGTs. P. dorei and P. vulgatus mobilome. (A) Number of proteins related to bacteriophage per genome. (B) Number of proteins related to IS per genome. (C) Number of proteins related to ICEs. (D) Number of proteins related to plasmids. (E) Number of CRISPR genes per genome. (F) Number of repeats per CRISPR gene per genome. (G) Number of genes associated with HGT. (H) Top 10 species associated with HGT in P. dorei. (I) Top 10 species associated with HGT in P. vulgatus.
To take a closer look at potential anti-phage defense systems, CRISPRs were identified using MinCED. P. dorei had significantly more CRISPR systems per genome than P. vulgatus, with averages of 1 and 0.47, respectively (Wilcoxon test, adj. p value < 0.0001) (Fig. 3E). P. dorei also had significantly more repeats per CRISPR system per genome than P. vulgatus, indicating a larger use of this anti-phage defense system (9.77 and 6.96 repeats, respectively; Wilcoxon test, adj. p value < 0.0001) (Fig. 3F). This could also be indicative of a stronger innate immune response (e.g., prophage integrated signals) mediated by CRISPR systems.
P. dorei and P. vulgatus are functionally different
To gain more insights into the functional differences between P. dorei and P. vulgatus, we annotated the genomes using eggNOG-mapper and extracted the Clusters of Orthologous Groups (COGs) categories for each gene. We then compared P. dorei and P. vulgatus for each category. Out of the 25 COG categories identified, 19 were differentially more present in P. dorei than P. vulgatus (Wilcoxon test, adj. p value < 0.0001) (Fig. 4). This difference remained true when normalising by the genome size. In particular, COG categories C (energy production and conversion), E (amino-acids transport and metabolism), G (carbohydrate transport and metabolism), P (inorganic ion transport and metabolism), T (signal transduction mechanisms) and M (cell wall/membrane/envelope biogenesis) were more present in P. dorei than in P. vulgatus. However, all these differences are related to central metabolism and could be caused by genome expansion.
Figure 4.
COG categories comparison for P. dorei and P. vulgatus (****, Wilcoxon test, adj. p value < 0.0001. (***, Wilcoxon test, adj. p value < 0.001).
P. dorei and P. vulgatus strains have distinct carbohydrate enzyme profiles, hinting at different yet overlapping ecological niches and strategies
There is a high diversity of carbohydrate molecules in the human gut. Bacteria use carbohydrates as a carbon source, for attachment to the host, or other functions during infection (e.g., immunomodulation). CAZymes such as GH, glycosyl transferases, polysaccharide lyases and carbohydrate esterases, are enzymes involved in the assembly, modification and breakdown of carbohydrates45. The amount and variety of CAZymes present in a given organism can be used as an indicator of adaptation to and fitness in a certain environment. Carbohydrate utilisation is an important factor driving bacterial evolution, as it is associated with the niche each organism occupies and how well it can adapt to environmental changes. To investigate the CAZymes present in P. dorei and P. vulgatus, all the genomes collected using dbCAN v3.032 were annotated on the CAZy database.
When looking at the CAZymes profile of each assembly, a clustered heatmap (hierarchical Ward-linkage clustering based on the Pearson correlation coefficients) showed that they all cluster according to the species they belong to, with only 2 exceptions, thus indicating a clear separation in terms of GH profile between P. dorei and P. vulgatus (Fig. 5A). In particular, the GH families GH144 (specific to β-1,2-glucan), GH88 (unsaturated glucuronyl hydrolases), GH146 (β-arabinofuranosidase) were more associated with P. dorei while the GH families GH101 (specific to glycoproteins, especially mucins), GH130 (acting on β-mannosides), GH110 (active on blood group B oligosaccharide) were more associated with P. vulgatus (differential abundance analysis using Maaslin2, adj. p value < 0.05). This marked CAZyme signature difference in P. dorei and P. vulgatus suggests that both species occupy a different yet overlapping ecological niche for carbohydrate utilisation and/or ecological strategies. P. dorei had a higher alpha-diversity (Shannon index) of GH families per genome than P. vulgatus (Fig. 5B, Wilcoxon test, adj. p value < 0.0001), possibly indicating a higher degree of adaptability to complex carbohydrates-rich environments. Additionnally, strains isolated from or present in disease states presented a lower alpha-diversity of GH families compared to strains not isolated in disease states in P. vulgatus, but not in P. dorei (Supplementary Fig. 1, Wilcoxon test, adj. p value < 0.05).
Figure 5.
Carbohydrate enzyme profile of P. dorei and P. vulgatus. (A) Heat map showing the different CAZymes present in each assembly, with P. dorei and P. vulgatus genomes grouping separately (only GH families present in more than 4 genomes are shown here). (B) Heat map showing only GH families associated with HMO utilization genes according to36,46. (C) Alpha diversity (Shannon index) of P. dorei and P. vulgatus GH families profile. (D) Number of genes per genome belonging to the GH family GH95 (1,2-α-L-fucosidase) (****, Wilcoxon test, adj. p value < 0.0001).
P. dorei and P. vulgatus are among the species in the Phocaeicola genus that colonise the human gut soon after birth in vaginally delivered infants1. The ability to digest HMOs is an important advantage to colonise the gut of breastfed infants since HMOs are the third most abundant component in breast milk and they cannot be digested by the host36,46. Recently, specific GH families associated with HMO utilisation were identified in P. dorei using transcriptomics36. Using this list of GH families, we investigated the most abundant GH families related to HMO utilisation present in both species (Fig. 5C). P. dorei had a higher number of these GH families associated with HMO utilisation. The GH families GH2 (beta-galactosidase), GH28 (polygalacturonases), GH29 (1,3/1,4-alpha-fucosidase), GH92 (exo-acting α-mannosidases), GH97 (α-glucosidase and α-galactosidase), GH95 (1,2-alpha-L-fucosidase), GH3 (exo-acting β-D-glucosidases and α-L-arabinofuranosidases), GH51 (L-arabinfuranosidases), GH36 (α-galactosidase and α-N-acetylgalactosaminidase), and GH35 (β-galactosidases) were more abundant per genome in P. dorei (Wilcoxon test, adj. p value < 0.05). Conversely, the GH families GH20 (lacto-N-biosidase), GH109 (α-N-acetylgalactosaminidase), GH33 (sialidase) and GH43 (α-L-arabinofuranosidases) were more abundant per genome in P. vulgatus (Wilcoxon test, adj. p value < 0.05). It seems that P. dorei is more adapted to HMO utilisation in the infant gut microbiome, with for example more genes associated with alpha-fucosidase enzymes which are widely used by bifidobacteria to feed on the widely available 2′-fucosyllactose47 (Fig. 5D). However, the HMO utilisation mechanisms in Bacteroides and Phocaeicola are different than the well-characterized mechanisms in Bifidobacterium and not necessarily associated with specific GH families, therefore more studies are needed to fully understand Bacteroides HMO utilisation genes. For example, it has recently been shown that GH33 is used by P. dorei for utilisation of sialylated HMOs48. As there are more GH33 genes in P. vulgatus, P. dorei and P. vulgatus could have different HMO utilisation strategies. The difference could also be due to genome expansion in P. dorei.
P. dorei and P. vulgatus strains have close but different antimicrobial resistance genes (AMR) profiles
To assess the AMR genes of both species, Resistance Gene Identifier (RGI) and the CARD database were used. A total of 23 AMR families were identified (Fig. 6A), with a round average of 4 AMR genes per genome in both species (Fig. 6B). The most abundant resistance genes were fluoroquinolone and/or tetracycline, glycopeptide, macrolide. P. vulgatus and P. dorei assemblies do not cluster separately according to AMR genes present on their genomes when looking at a clustered heatmap (hierarchical Ward-linkage clustering based on the Pearson correlation coefficients) (Fig. 6A). Overall, the AMR profiles of P. dorei and P. vulgatus were very similar, with 17 of the 23 AMR families present in both species without significant statistical differences. Six AMR families were only found in P. vulgatus assemblies. The only AMR family present in higher abundance per genome in P. dorei was tetracycline antibiotic resistance, while fluoroquinolone antibiotic/tetracycline antibiotic resistance and aminoglycoside antibiotic resistance were more abundant per genome in P. vulgatus (Wilcoxon test, adj. p values < 0.05) (Fig. 6C–E). P. vulgatus thus had a more diverse yet similar AMR profile than P. dorei. Additionally, there were differences in AMR content for strains isolated or present in disease for P. dorei and P. vulgatus (Supplementary Fig. 2). Of note, fluoroquinolone/tetracycline and glycopeptide antibiotic resistance gene families were higher in strains isolated or present in disease for both species. For P. vulgatus, macrolide antibiotic resistance was more abundant in strains isolated or present in disease, while tetracycline antibiotic resistance alone was more abundant in non-disease.
Figure 6.
Antibiotic resistance profile of P. dorei and P. vulgatus. (A) Heat map of P. dorei and P. vulgatus assemblies showing the different antibiotic resistance genes present in each strain. (B) Comparison of the number of AMR genes present in the genomes of P. dorei and P. vulgatus. (C–E) AMR families with different abundances per genome in both species (Wilcoxon test, ****, adj. p value < 0.0001, ***, adj. p value < 0.001, **, adj. p value < 0.01, ns non-significant).
Discussion
P. vulgatus and P. dorei are common, abundant and important commensals of the human gut1,19. Both species are among the depleted bacteria in CS-born infants and the specific roles and differences between these two species in the human gut and their contribution to health and disease have not yet been explored. In the current study, we investigated, for the first time, the genomic details of 3951 assemblies of P. vulgatus and P. dorei and performed genomic comparison and phylogenetic analysis to gain insight into the ecology and evolution of these bacteria with high genome plasticity (a summary of key differences can be found in Supplementary Table 2).
We showed that P. vulgatus has a bigger pan-genome but smaller genomes overall when compared to P. dorei. In other words, P. vulgatus, as a species, had a larger collection of genes, but individually, P. dorei isolates had a larger collection of genes. Both species had an open pan-genome, indicating a high degree of genome plasticity and adaptability in the context of the human gut. To illustrate this diversity/adaptability, the core genes only represent 1.71% and 0.44% of the pan-genome of P. dorei and P. vulgatus, respectively, with only a small portion of their genome shared between all the assemblies. Because P. vulgatus pan-genome is larger, we thus assume that P. vulgatus strains have higher genetic plasticity, in line with49. This is confirmed by the higher proportion of ISs, bacteriophages and plasmids in P. vulgatus, with concurrently less CRISPR-Cas systems within the genomes, all of which play a role in the bacterial genome instability and driving genome diversification5051. P. vulgatus pan-genome could for example be bigger thanks to the higher number of bacteriophages potentially carrying cargo. Nonetheless, P. dorei appears to have had more HGTs, which constitutes a paradox. Looking at the phylogeny and synteny, we hypothesise that P. dorei experienced genome expansion directly from a clade of P. vulgatus, probably driven by HGT from Bacteroides species. Even though P. dorei has a higher degree of genome conservation/stability compared to P. vulgatus, it could have experienced HGT thanks to ICEs carrying cargo. Since cells need to be in close proximity for ICEs to be transferred, the variety of genes that can be transferred using this mechanism is limited to the genes available in the surrounding environment. Also, although P. dorei had more CRISPR-Cas systems, there are other anti-phage defense systems that have not been investigated here and could be prevalent in P. vulgatus52.
There is further evidence that P. dorei evolved directly from a clade of P. vulgatus. P. dorei is more recent than P. vulgatus, as indicated by the fact that P. vulgatus was discovered 73 years before P. dorei, and it might have undergone recent population bottleneck. P. dorei had less genes per unit of genome (Fig. 1F) as well as lower GC content (Fig. 1C), and both could be related to genetic drift53,54. In this case, accumulations of pseudo-genes and mobile genetic elements could be the reasons for less genes per unit of genome. Other explanations could be gene-duplication events, more non-coding regions and genome re-arrangement.
AMR and carbohydrate utilisation are major forces driving bacterial evolution. We showed that there was no significant difference in the AMR profile of P. dorei and P. vulgatus for most AMR families. Also, P. dorei and P. vulgatus assemblies did not group separately according to AMR families present in their genome (Fig. 6A). On the other hand, both species grouped separately according to their GH family’s profile, with only a few exceptions (Fig. 5A). These data showed the importance of carbohydrate utilisation and CAZyme profile on species differentiation and evolution. The phylogenetic tree (Fig. 2) showed that a large proportion of the genome assemblies publicly available came from infants’ gut, demonstrating the importance of these two species in early life. We analysed the GH families associated with HMO utilization present in each genome (Fig. 5B). Most P. dorei species grouped together, with a few exceptions. P. dorei had a higher number of genes associated with HMO utilization present on individual genomes, possibly indicating a better fitness for the infant gut environment than P. vulgatus.
Supplementary Information
Acknowledgements
We are grateful to the Stanton lab for their helpful comments and discussions. We thank the reviewers for their helpful comments. We would also like to acknowledge funding from Science Foundation Ireland/APC Microbiome Ireland and European Union (ERC, BACtheWINNER, Project No. 101054719). Views and opinions expressed are however those of the author(s) only and do not necessarily reflect those of the European Union or the European Research Council Executive Agency. Neither the European Union nor the granting authority can be held responsible for them.
Author contributions
E.S.M and G.G prepared and wrote the manuscript, prepared the figures, and performed the bioinformatics analysis. All authors reviewed the manuscript.
Data availability
All the data used in this manuscript are freely available online. We used the Unified Human Gastrointestinal Genome (UHGG) catalog, deposited in the European Nucleotide Archive under study accession ERP116715 and available from the MGnify FTP site (http://ftp.ebi.ac.uk/pub/databases/metagenomics/mgnify_genomes/). We also used the Early-Life Gut Genomes (ELGG) catalog, deposited in the Zenodo repository under https://doi.org/10.5281/zenodo.6969520. Note that there is no accession number for this catalog, as it was built from previously deposited data with accession numbers listed here: https://static-content.springer.com/esm/art%3A10.1038%2Fs41467-022-32805-z/MediaObjects/41467_2022_32805_MOESM12_ESM.xlsx. Finally, we downloaded genomes of isolates publicly available on the National Center for Biotechnology Information (NCBI) (November 2022): https://www.ncbi.nlm.nih.gov/. All corresponding genomes accession numbers and links are available in Supplementary Table 1.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Emilene Da Silva Morais and Ghjuvan Micaelu Grimaud.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-59148-7.
References
- 1.Wang S, Zeng S, Egan M, Cherry P, Strain C, Morais E, Boyaval P, Stanton C, Ross P. Metagenomic analysis of mother-infant gut microbiome reveals global distinct and shared microbial signatures. Gut Microbes. 2021;13(1):1911571. doi: 10.1080/19490976.2021.1911571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Castellani A, Chalmers A. Manual of Tropical Medicine. Williams Wood and Co.; 1919. [Google Scholar]
- 3.García-López M, Meier-Kolthoff JP, Tindall BJ, Gronow S, Woyke T, Kyrpides NC, Hahnke RL, Göker M. Analysis of 1,000 type-strain genomes improves taxonomic classification of Bacteroidetes. Front. Microbiol. 2019;10:2083. doi: 10.3389/fmicb.2019.02083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bakir MA, et al. Bacteroides dorei sp. Nov., isolated from human faeces. Int. J. Syst. Evol. Microbiol. 2006;56(7):1639–1643. doi: 10.1099/ijs.0.64257-0. [DOI] [PubMed] [Google Scholar]
- 5.Rigottier-Gois L, Rochet V, Garrec N, Suau A, Doré J. Enumeration of Bacteroides species in human faeces by fluorescent in situ hybridisation combined with flow cytometry using 16S rRNA probes. Syst. Appl. Microbiol. 2003;26(1):110–118. doi: 10.1078/072320203322337399. [DOI] [PubMed] [Google Scholar]
- 6.Mitchell CM, Mazzoni C, Hogstrom L, Bryant A, Bergerat A, Cher A, Pochan S. Delivery mode affects stability of early infant gut microbiota. Cell Rep. Med. 2020;1(9):100156. doi: 10.1016/j.xcrm.2020.100156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shao Y, Forster SC, Tsaliki E, Vervier K, Strang A, Simpson N, Kumar N, et al. Stunted microbiota and opportunistic pathogen colonization in caesarean-section birth. Nature. 2019;574(7776):117–121. doi: 10.1038/s41586-019-1560-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yassour M, Vatanen T, Siljander H, Hämäläinen A-M, Härkönen T, Ryhänen SJ, Franzosa EA. Natural history of the infant gut microbiome and impact of antibiotic treatment on bacterial strain diversity and stability. Sci. Transl. Med. 2016;8(43):343ra81. doi: 10.1126/scitranslmed.aad0917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang C, Xiao Y, Yu L, Tian F, Zhao J, Zhang H, Chen W, Zhai Q. Protective effects of different Bacteroides vulgatus strains against lipopolysaccharide-induced acute intestinal injury, and their underlying functional genes. J. Adv. Res. 2022;36:27–37. doi: 10.1016/j.jare.2021.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li S, Wang C, Zhang C, Luo Y, Cheng Q, Yu L, Sun Z. Evaluation of the effects of different Bacteroides vulgatus strains against DSS-induced colitis. J. Immunol. Res. 2021 doi: 10.1155/2021/9117805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yoshida N, Emoto T, Yamashita T, Watanabe H, Hayashi T, Tabata T, Hoshi N, Hatano N, Ozawa G, Sasaki N, Mizoguchi T. Bacteroides vulgatus and Bacteroides dorei reduce gut microbial lipopolysaccharide production and inhibit atherosclerosis. Circulation. 2018;22:2486–2498. doi: 10.1161/CIRCULATIONAHA.118.033714. [DOI] [PubMed] [Google Scholar]
- 12.Gérard P, Lepercq P, Leclerc M, Gavini F, Raibaud P, Juste C. Bacteroides sp. strain D8, the first cholesterol-reducing bacterium isolated from human feces. Appl. Environ. Microbiol. 2007;73(18):5742–5749. doi: 10.1128/AEM.02806-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Song L, Huang Y, Liu G, Li X, Xiao Y, Liu C, Zhang Y, Li J, Xu J, Lu S, Ren Z. A novel immunobiotics bacteroides dorei ameliorates influenza virus infection in mice. Front. Immunol. 2022;12:6000. doi: 10.3389/fimmu.2021.828887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davis-Richardson A, Ardissone A, Dias R, Simell V, Leonard M, Kemppainen K, Drew J, Schatz D, Atkinson M, Kolaczkowski B, Ilonen J. Bacteroides dorei dominates gut microbiome prior to autoimmunity in finnish children at high risk. Front. Microbiol. 2014;5:678. doi: 10.3389/fmicb.2014.00678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bakir M, Sakamoto M, Kitahara M, Matsumoto M, Benno Y. Bacteroides dorei sp. nov., isolated from human faeces. Int. J. Syst. Evol. Microbiol. 2006;56(7):1639–1643. doi: 10.1099/ijs.0.64257-0. [DOI] [PubMed] [Google Scholar]
- 16.Pedersen RM, Marmolin ES, Justesen US. Species differentiation of Bacteroides dorei from Bacteroides vulgatus and Bacteroides ovatus from Bacteroides xylanisolvens–back to basics. Anaerobe. 2013;24:1–3. doi: 10.1016/j.anaerobe.2013.08.004. [DOI] [PubMed] [Google Scholar]
- 17.Cobo F, Pérez-Carrasco V, Rodríguez-Guerrero E, Sampedro A, Rodríguez-Granger J, García-Salcedo JA, Navarro-Marí JM. Misidentification of Phocaeicola (Bacteroides) i in two patients with bacteremia. Anaerobe. 2022;75:102544. doi: 10.1016/j.anaerobe.2022.102544. [DOI] [PubMed] [Google Scholar]
- 18.Costea PI, Coelho LP, Sunagawa S, Munch R, Huerta-Cepas J, Forslund K, Hildebrand F, Kushugulova A, Zeller G, Bork P. Subspecies in the global human gut microbiome. Mol. Syst. Biol. 2017;13:960. doi: 10.15252/msb.20177589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garud NR, Good BH, Hallatschek O, Pollard KS. Evolutionary dynamics of bacteria in the gut microbiome within and across hosts. PLoS Biol. 2019;17(1):e3000102. doi: 10.1371/journal.pbio.3000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Almeida A, Nayfach S, Boland M, Strozzi F, Beracochea M, Shi Z, Pollard K, Sakharova E, Parks D, Hugenholtz P, Segata N. A unified catalog of 204,938 reference genomes from the human gut microbiome. Nat. Biotechnol. 2021;29(1):105–114. doi: 10.1038/s41587-020-0603-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zeng S, Patangia D, Almeida A, Zhou Z, Mu D, Paul Ross R, Stanton C, Wang S. A compendium of 32,277 metagenome-assembled genomes and over 80 million genes from the early-life human gut microbiome. Nat. Commun. 2022;13(1):5. doi: 10.1038/s41467-022-32805-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.National Library of Medicine (US), National Center for Biotechnology Information. National Center for Biotechnology Information (NCBI)[Internet]. Available from: https://www.ncbi.nlm.nih.gov/ (1988).
- 23.Entrez Programming Utilities (E-Utilities). in Encyclopedia of Genetics, Genomics, Proteomics and Informatics (2008).
- 24.Gálvez-Merchán Á, Min KH, Pachter L, Booeshaghi AS. Metadata retrieval from sequence databases with ffq. Bioinformatics. 2023;39(1):667. doi: 10.1093/bioinformatics/btac667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cummins C, Ahamed A, Aslam R, Burgin J, Devraj R, Edbali O, Gupta D, Harrison P, Haseeb M, Holt S, Ibrahim T. The european nucleotide archive in 2021. Nucleic Acids Res. 2022;50:D106–D110. doi: 10.1093/nar/gkab1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chklovski, A. et al. CheckM2: A rapid, scalable and accurate tool for assessing microbial genome quality using machine learning. bioRxiv (2022). [DOI] [PubMed]
- 27.Chaumeil, P. -A. et al. GTDB-Tk: A toolkit to classify genomes with the genome taxonomy database. Bioinformatics (2019). [DOI] [PMC free article] [PubMed]
- 28.Li, T., Yin, Y. Critical assessment of pan-genomics of metagenome-assembled genomes. bioRxiv (2022). [DOI] [PMC free article] [PubMed]
- 29.Seemann T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics. 2014;30(14):2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 30.Cantalapiedra CP, Hernández-Plaza A, Letunic I, Bork P, Huerta-Cepas J. eggNOG-mapper v2: Functional annotation, orthology assignments, and domain prediction at the metagenomic scale. Mol. Biol. Evol. 2021;38(12):5825–5829. doi: 10.1093/molbev/msab293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MT, Parkhill J. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31(22):3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zheng J, Ge Q, Yan Y, Zhang X, Huang L, Yin Y. dbCAN3: Automated carbohydrate-active enzyme and substrate annotation. Nucleic Acids Res. 2023;51:W115–W121. doi: 10.1093/nar/gkad328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Finn RD, Clements J, Eddy SR. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011;39(2):W29–W37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Buchfink B, et al. Fast and sensitive protein alignment using DIAMOND. Nat. Methods. 2015;12(1):59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 35.Ioannou A, Knol J, Belzer C. Microbial glycoside hydrolases in the first year of life: An analysis review on their presence and importance in infant gut. Front. Microbiol. 2021;12:631282. doi: 10.3389/fmicb.2021.631282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kijner S, Cher A, Yassour M. The infant gut commensal Bacteroides dorei presents a generalized transcriptional response to various human milk oligosaccharides. Front. Cell. Infect. Microbiol. 2022;12:854122. doi: 10.3389/fcimb.2022.854122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mallick H, et al. Multivariable association discovery in population-scale meta-omics studies. PLoS Comput. Biol. 2021;17(11):e1009442. doi: 10.1371/journal.pcbi.1009442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alcock B, Huynh W, Chalil R, Smith K, Raphenya A, Wlodarski M, Edalatmand A, Petkau A, Syed S, Tsang K, Baker S. CARD 2023: Expanded curation, support for machine learning, and resistome prediction at the comprehensive antibiotic resistance database. Nucleic Acids Res. 2023;51(D1):D690–D699. doi: 10.1093/nar/gkac920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brown CL, Mullet J, Hindi F, Stoll JE, Gupta S, Choi M, Keenum I, Vikesland P, Pruden A, Zhang L. MobileOG-db: A manually curated database of protein families mediating the life cycle of bacterial mobile genetic elements. Appl. Environ. Microbiol. 2022;88(18):e00991–e1022. doi: 10.1128/aem.00991-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu Q, et al. HGTector: An automated method facilitating genome-wide discovery of putative horizontal gene transfers. BMC Genom. 2014;15(717):1–18. doi: 10.1186/1471-2164-15-717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guo JBBZA, Varsani A, Dominguez-Huerta G, Delmont T, Pratama A, Gazitúa M, Vik D, Sullivan M, Roux S. VirSorter2: A multi-classifier, expert-guided approach to detect diverse DNA and RNA viruses. Microbiome. 2021;9:1–13. doi: 10.1186/s40168-020-00990-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Skennerton, C. et al. Minced—Mining CRISPRs in Environmental Datasets. Available at https://github.com/ctSkennerton/minced (2016).
- 43.Lees J, Harris S, Tonkin-Hill G, Gladstone R, Lo S, Weiser J, Corander J, Bentley S, Croucher N. Fast and flexible bacterial genomic epidemiology with PopPUNK. Genome Res. 2019;29:304–316. doi: 10.1101/gr.241455.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Costa SS, Guimarães LC, Silva A, Soares SC, Baraúna RA. First steps in the analysis of prokaryotic pan-genomes. Bioinf. Biol. Insights. 2020;14:1177932220938064. doi: 10.1177/1177932220938064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Drula E, Garron M-L, Dogan S, Lombard V, Henrissat B, Terrapon N. The carbohydrate-active enzyme database: Functions and literature. Nucleic Acids Res. 2022;50(1):D571–D577. doi: 10.1093/nar/gkab1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Salli K, Hirvonen J, Siitonen J, Ahonen I, Anglenius H, Maukonen J. Selective utilization of the human milk oligosaccharides 2′-fucosyllactose, 3-fucosyllactose, and difucosyllactose by various probiotic and pathogenic bacteria. J. Agric. Food Chem. 2020;69(1):170–182. doi: 10.1021/acs.jafc.0c06041. [DOI] [PubMed] [Google Scholar]
- 47.Sela D, Garrido D, Lerno L, Wu S, Tan K, Eom H, Joachimiak A, Lebrilla C, Mills D. Bifidobacterium longum subsp. infantis ATCC 15697 alpha-fucosidases are active on fucosylated human milk oligosaccharides. Appl. Environ. Microbiol. 2012;78:795–803. doi: 10.1128/AEM.06762-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yassour, M. et al., Identification of a novel human milk oligosaccharides utilization cluster in the infant gut commensal Bacteroides dorei, 27 April 2023, PREPRINT (Version 1) available at research square (2023). [DOI] [PMC free article] [PubMed]
- 49.Lange A, Beier S, Steimle A, Autenrieth IB, Huson DH, Frick JS. Extensive mobilome-driven genome diversification in mouse gut-associated Bacteroides vulgatus mpk. Genome Biol. Evol. 2016;8(4):1197–1207. doi: 10.1093/gbe/evw070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Darmon E, Leach DR. Bacterial genome instability. Microbiol. Mol. Biol. Rev. 2014;78(1):1–39. doi: 10.1128/MMBR.00035-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li Y, Wang Y, Liu J. Genomic insights into the interspecific diversity and evolution of mobiluncus, a pathogen associated with bacterial vaginosis. Front. Microbiol. 2022;13:939406. doi: 10.3389/fmicb.2022.939406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Johnson MC, Laderman E, Huiting E, Zhang C, Davidson A, Bondy-Denomy J. Core defense hotspots within Pseudomonas aeruginosa are a consistent and rich source of anti-phage defense systems. Nucleic Acids Res. 2023;51(10):4995–5005. doi: 10.1093/nar/gkad317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bert E. Genomic GC content drifts downward in most bacterial genomes. Plos One. 2021;16(5):e0244163. doi: 10.1371/journal.pone.0244163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bobay L-M, Ochman H. The evolution of bacterial genome architecture. Front. Genet. 2017;8:72. doi: 10.3389/fgene.2017.00072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Croucher NJ, Page AJ, Connor TR, Delaney AJ, Keane JA, Bentley SD, Harris SR. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 2015;43(3):e15–e15. doi: 10.1093/nar/gku1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Seemann T. Snippy: Fast Bacterial Variant Calling from NGS Reads. https://github.com/tseemann/snippy (2018).
- 57.Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30(9):1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Campbell, D. E. et al. Interrogation of the integrated mobile genetic elements in gut-associated Bacteroidaceae with a consensus prediction approach. bioRxiv 2021-09 (2021).
- 59.Liu, Z. Dynamics of bacterial recombination in the human gut microbiome. bioRxiv 2022-08 (2022). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data used in this manuscript are freely available online. We used the Unified Human Gastrointestinal Genome (UHGG) catalog, deposited in the European Nucleotide Archive under study accession ERP116715 and available from the MGnify FTP site (http://ftp.ebi.ac.uk/pub/databases/metagenomics/mgnify_genomes/). We also used the Early-Life Gut Genomes (ELGG) catalog, deposited in the Zenodo repository under https://doi.org/10.5281/zenodo.6969520. Note that there is no accession number for this catalog, as it was built from previously deposited data with accession numbers listed here: https://static-content.springer.com/esm/art%3A10.1038%2Fs41467-022-32805-z/MediaObjects/41467_2022_32805_MOESM12_ESM.xlsx. Finally, we downloaded genomes of isolates publicly available on the National Center for Biotechnology Information (NCBI) (November 2022): https://www.ncbi.nlm.nih.gov/. All corresponding genomes accession numbers and links are available in Supplementary Table 1.