Abstract
The Epstein-Barr virus latent membrane protein 1 (LMP1) oncoprotein causes multiple cellular changes, including induction of epidermal growth factor receptor (EGFR) expression and activation of the NF-κB transcription factor. LMP1 and the cellular protein CD40, which also induces EGFR expression, interact with the tumor necrosis factor receptor-associated factor (TRAF) proteins. The LMP1 carboxy-terminal activation region 1 signaling domain interacts specifically with the TRAFs and is essential for EGFR induction through a mechanism independent of NF-κB alone. LMP1 and CD40 share a common TRAF binding motif, PXQXT. In this study, the PXQXT motifs in both LMP1 and CD40 were altered and mutant proteins were analyzed for induction of EGFR expression. Replacement of the T residue with A in CD40 completely blocked induction of the EGFR, while the same mutation in LMP1 did not affect EGFR induction. Replacement of both P and Q residues with A’s in LMP1 reduced EGFR induction by >75%, while deletion of PXQXT blocked EGFR induction. These results genetically link EGFR induction by LMP1 to the TRAF signaling pathway. Overexpression of TRAF2 potently activates NF-κB, although TRAF2 did not induce expression of the EGFR either alone or in combination with TRAF1 and TRAF3. In vivo analyses of the interaction of the TRAFs with LMP1 variants mutated in the PXQXT domain indicate that high-level induction of EGFR expression requires interaction with TRAF1, -2, and -3. However, exogenous expression of TRAF3 decreased EGFR induction mediated by either LMP1 or CD40. These data suggest that TRAF-mediated activation of EGFR expression requires assembly of a complex containing the appropriate stoichiometry of TRAF proteins clustered at the cell membrane with LMP1.
The Epstein-Barr virus (EBV)-encoded latent membrane protein 1 (LMP1) is essential for immortalization of B lymphocytes infected in vitro with EBV and is expressed in lymphomas which develop in immunocompromised individuals (32, 47, 66). LMP1 is also expressed in all cases of premalignant lesions related to the human malignancy nasopharyngeal carcinoma (NPC) and in a majority of NPC biopsies (12, 46, 67). LMP1 is the only EBV protein shown to have transforming properties in rodent fibroblasts, highlighting the importance of this viral oncoprotein in cellular transformation associated with EBV infection (1, 43, 60). The function of LMP1 and the involvement of LMP1 in the activation of cellular signal transduction pathways are just beginning to be understood. Expression of LMP1 in B lymphocytes induces the transcription of many genes, including those encoding activation antigens such as CD23 and CD40, adhesion molecules such as ICAM-1, LFA-1, and LFA-3, and molecules which inhibit programmed cell death such as Bcl-2, Mch-1, and A20 (16, 22, 33, 35, 36, 62–64). Expression of LMP1 in epithelial cells induces expression of the genes encoding the epidermal growth factor receptor (EGFR) and the A20 molecule (38–40). Induction of these genes is likely to play an important role in the transformation of epithelial cells by LMP1. LMP1 activates the NF-κB transcription factor and also activates the N-terminal Jun kinase (JNK1) (19, 21, 25, 34, 42, 45). Activation of these pathways is likely to mediate some but not all of the LMP1-induced changes in gene expression described above.
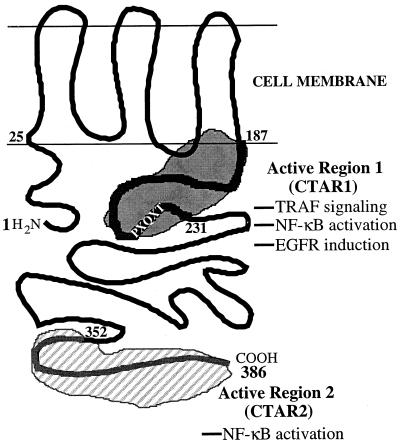
LMP1 consists of a 24-amino-acid cytoplasmic domain located at the amino terminus, a membrane-spanning hydrophobic domain consisting of six transmembrane domains connected by short turns between amino acids 25 and 187, followed by a 200-amino-acid cytoplasmic domain at the carboxy terminus (Fig. 1) (13, 23). The six transmembrane domains of LMP1 are essential for the activation of signaling pathways, indicating that proper aggregation in the plasma membrane is required for LMP1 function (25, 37, 40). Mutational analysis of LMP1 with respect to activation of cellular signaling pathways further divides the carboxy-terminal domain into three regions. Carboxy-terminal activation region 1 (CTAR1), located between amino acids 187 and 231, is essential for EGFR induction and is a relatively weak activator of NF-κB (25, 40, 42). CTAR2, located between amino acids 352 and 386, is responsible for activation of JNK and is the major NF-κB activation domain (6, 25, 34, 40, 42). The amino acids separating these two domains (amino acids 232 to 351) are not associated with any known function.
FIG. 1.
Model depicting the molecular structures and locations of functional domains in LMP1. LMP1 contains a 24-amino-acid cytoplasmic amino terminus, a transmembrane hydrophobic domain, and a 200-amino-acid cytoplasmic carboxy terminus. The carboxy terminus contains the major signaling domains in LMP1. CTAR1 mediates interaction with the TRAFs, induces EGFR expression, and is the minor NF-κB-activating region; CTAR2 is the major NF-κB-activating region. The location of the TRAF-interacting motif, PXQXT, is also indicated.
LMP1 has many similarities to the tumor necrosis factor receptor (TNFR) superfamily. The TNFR superfamily comprises a large group of transmembrane proteins including CD40 and CD30 that transmit extracellular signals but do not contain the intrinsic tyrosine kinase activity often associated with membrane receptors (56, 58, 59). Instead, the TNFR family members initiate signaling events through interaction of their cytoplasmic domains with the recently identified tumor necrosis factor receptor-associated factors (TRAFs) (8, 44, 48, 49, 53). The mechanisms by which the TRAFs activate signaling cascades are just beginning to be understood.
Several lines of evidence indicate that LMP1 functions as a constitutively activated member of the TNFR superfamily. First, expression of LMP1 in B-lymphoid cells induces many of the phenotypic changes characteristic of EBV immortalization (4, 5, 11, 17, 52). Activation of CD40, in conjunction with interleukin-4 stimulation, can mimic EBV-induced immortalization of human lymphocytes in short-term culture, suggesting parallel roles for CD40 and LMP1 in signal transduction pathways (4, 17, 50). CD40, like LMP1, can induce expression of the EGFR in epithelial cells, which also indicates a similarity between LMP1 and CD40 signaling (40). Second, signaling from CTAR1 involves the TRAF signaling pathway, and LMP1 interacts with TRAF1, -2, -3, and -5 (6, 10, 44, 51). The precise TRAF interaction motif in CTAR1 has been identified by homology with the TNFR family members CD40 and CD30 (7, 18, 24). The TRAFs bind to the PXQXT motif located between amino acids 204 and 208 of LMP1 (10, 15, 51). TRAF2 appears to be a positive regulator of TRAF-mediated NF-κB activation; in support of this finding, NF-κB activation by CTAR1 in LMP1 involves interactions with TRAF1 and TRAF2 (10, 31). In contrast, TRAF3 appears to be a negative regulator of the TRAF pathway, and accordingly, overexpression of TRAF3 blocks NF-κB activation from CTAR1 but not CTAR2 (10, 11, 40, 48). The mechanism of NF-κB activation by CTAR2 may involve interaction with the TNFR-associated death domain protein (TRADD) and studies have localized CTAR2 to amino acids 379 to 384 (14, 29). Although the interaction with TRADD may activate NF-κB through the interaction with TRAF2, the details of CTAR2 signaling remain unknown (10, 29, 31).
Recent experiments indicate that the CTAR1 domain in LMP1 is essential for induction of EGFR expression (40). This induction of EGFR expression by LMP1 occurred at the RNA level, indicating that LMP1 either activates the EGFR promoter or stabilizes the EGFR mRNA (39, 40). Expression of an LMP1 mutant deleted for CTAR2, LMP1(1-231), induced high levels of EGFR expression. In contrast, an LMP1 mutant deleted for CTAR1, LMP1(Δ187-351), activated NF-κB to high levels in both transient and stable assays but was completely defective for EGFR induction (40). These results indicate that NF-κB activation alone is unable to induce EGFR expression. However, since the CTAR1 signaling domain is linked to both EGFR induction and NF-κB activation, the contribution of NF-κB to the induction of EGFR expression could not be determined.
The experiments presented in this study were designed to determine the contribution of NF-κB activation from CTAR1 to the induction of EGFR expression, to analyze the roles of the TRAF proteins in these signaling pathways, and to identify residues in the PXQXT motif in CTAR1 which are responsible for EGFR induction and interaction with the TRAFs.
MATERIALS AND METHODS
Cell lines and establishment of derivatives.
C33A epithelial cells, derived from a human cervical carcinoma (human papillomavirus negative), were grown at 37°C in Dulbecco’s modified Eagle’s medium (DMEM-H) supplemented with 10% fetal bovine serum (Gibco-BRL) and antibiotics. Cells were routinely grown in 150-mm-diameter cell culture dishes and subcultured three times weekly. C33A cell lines expressing CD40, wild-type LMP1 [LMP1(WT)], and mutants of LMP1 were obtained by selection in 600 μg of G418 (Geneticin; Gibco-BRL) following transfection with an expression construct containing the appropriate gene. Briefly, 5 × 105 cells were seeded in 60-mm-diameter tissue culture dishes and Lipofectin transfected as specified by the manufacturer (Gibco-BRL) with 5 μg of DNA. Cells were fed with fresh medium 24 h posttransfection, and 48 h posttransfection, the medium was replaced with fresh medium containing G418. Cell lines were created by pooling >50 G418-resistant colonies.
Plasmids and expression vectors.
The expression vector used for most of the constructs in this study is pcDNA3 (Invitrogen), which contains the cytomegalovirus (CMV) immediate-early promoter, polyadenylation sequence from the bovine growth hormone gene, and neomycin resistance cassette. The LMP1 cDNA was subcloned into the EcoRI site of pcDNA3. The construction of pcDNA3 vectors expressing FLAG-tagged LMP1 and LMP1 mutants has been described previously (40). The CD40 cDNA was subcloned into the HindIII and XhoI sites of pcDNA3. pSG5- and pcDNA3-based expression vectors for TRAF1, TRAF2, and TRAF3 were kindly provided by Elliott Kieff. The pSG5 vector contains the simian virus early promoter and intron sequences from the rabbit β-globin gene (Stratagene). TRAF3 was also subcloned into the pMEP4 expression vector (Invitrogen). The pMEP4 vector contains the human metallothionein promoter, herpes simplex virus thymidine kinase polyadenylation sequence, and hygromycin resistance cassette. pMEP4 and pMEP4-TRAF3 were transfected into C33A cells as described above and selected 48 h following transfection in medium containing 100 μg of hygromycin (Boehringer Mannheim) per ml. To coexpress TRAF3 with either LMP1 or CD40, pcDNA3-based vectors for LMP1 or CD40 were transfected into the pMEP4-TRAF3 cell line and selected in G418. Transfection of a fivefold excess of pSG5-TRAF1 with pcDNA3-TRAF2 in the pMEP4-TRAF3 cell lines, followed by selection in G418, created a cell line overexpressing all three TRAFs. The CMV-IκBα(S32/36A) expression vector, which encodes an IκBα molecule in which serines 32 and 36 have been replaced by alanines, was a kind gift of Dean Ballard (55). Cell lines expressing IκBα(S32/36A) were created by transfecting a fivefold excess of CMV-IκBα(S32/36A) with pcDNA3-based plasmids and selected in G418.
Construction of TRAF interaction domain LMP1 and CD40 mutants.
The LMP1 cDNA was subcloned into the EcoRI site of the pGEM3Z vector to facilitate construction of LMP1 mutants. PCR using Vent polymerase (New England Biolabs) was used to produce all mutant sequences. For the LMP1 mutants, following PCR amplification, the PCR products were digested with BsaBI and MscI and ligated into pGEM3Z-LMP1 which was digested with the same enzymes (except for EGRHH221-225AGAAA, which was digested with BsaBI and NaeI). The nucleotides representing the mutated codons are boldfaced in the sequences below. The sense oligonucleotide used in the PCR was 5′-AACAAAACTGGTGGACTCTATTGGTTGATCTCCTTTGGC-3′. For the P204A mutant, the antisense oligonucleotide was 5′-ATTCATGGCCAGAATCATCGGTAGCTTGTTGAGCGTGCG-3′. For the PQ204/206AA mutant, the antisense oligonucleotide was 5′-ATTCATGGCCAGAATCATCGGTAGCTGCTTGAGCGTGCG-3′. For the T208A mutant, the antisense oligonucleotide was 5′-ATTCATGGCCAGAATCATCGGCAGCTTGTTGAGGGTGCG-3′. For the DD209/210AA mutant, the antisense oligonucleotide was 5′-ATTCATGGCCAGAAGCAGCGGTAGCTTGTTGAGGGGCCG-3′. For the Δ204-208 mutant, the antisense oligonucleotide was 5′-TTCATGGCCAGAATCATCGTGCGGGAGGGAGTCATC-3′). For the 204-208AAAAA mutant, the antisense oligonucleotide was 5′-ATTCATGGCCAGAATCATCGGCAGCTGCTGCAGCGTGCG-3′. For the EGRH221-225AGAAA mutant, the antisense oligonucleotide was 5′-GTCGCCGGCTCCACTCACGAGCAGGGCGGCTGCGCCCGCGTTGGA-3′. The mutated LMP1 cDNAs were then subcloned into the EcoRI sites of pcDNA3. To construct FLAG-LMP1(WT) expression constructs containing these point mutants, the SfiI-BstEII fragments from the full-length constructs was subcloned into FLAG-LMP1(WT) cut with the same enzymes. To construct FLAG-LMP1(1-231) expression constructs containing the point mutations, the SfiI-MscI fragment from the full-length constructs was subcloned into the same sites in FLAG-LMP1(1-231).
The T234A mutant of CD40 was constructed in the same way except that the mutant PCR products were digested with BamHI and BstEII and subcloned into pcDNA3-CD40 digested with the same enzymes. The sense oligonucleotide was 5′-TCTGTGGTCCCCAGGATCCGGCTGAGAGCCCTGGTGGTGA-3′; the antisense oligonucleotide was 5′-CTCCTGGGTGACCGGTTGGCATCCATGTAAAGCTTCCTGCAC-3′. All mutations were confirmed by DNA sequencing using a model 377 DNA sequencer (Perkin-Elmer, Applied Biosystems Division) at the UNC-CH Automated DNA Sequencing Facility.
Immunoprecipitations.
C33A cells were transfected in duplicate with each of the mutated FLAG-LMP1 molecules (full length). Briefly, 1.5 × 106 cells were seeded in 100-mm-diameter tissue culture dishes and 24 h later cells were Lipofectin treated as specified by the manufacturer with 15 μg of each plasmid. Six plates were transfected with each of the LMP1 mutants; 24 h after lipofection, DMEM-H containing 10% fetal bovine serum was added, and 48 h after lipofection, cells were harvested. Whole-cell extracts were prepared by washing cells once in cold phosphate-buffered saline and then lysing cells in 900 μl of lysis buffer (20 mM HEPES, 0.5% Nonidet P-40 [NP-40], 250 mM NaCl, 10% glycerol, 2 mM EDTA, 1 mM phenylmethylsulfonyl fluoride [PMSF], 2.5 μg of aprotinin per ml, 25 μg of leupeptin per ml) for 20 min at 4°C. The supernatant was clarified by centrifugation and was stored at −80°C until use. FLAG-LMP1 and associated proteins were immunopurified with 100 μl of anti-FLAG M2 affinity gel (Sigma) for >3 h with constant rocking at 4°C. Immunocomplexes were washed three times in lysis buffer, diluted in 2× sample buffer, boiled for 5 min, and electrophoresed on sodium dodecyl sulfate (SDS)–10% polyacrylamide gels. Bound LMP1 and TRAF proteins were detected by immunoblotting as described below. For experiments with overexpression of TRAF1 or TRAF3, 5 × 105 cells were seeded in 60-mm-diameter tissue culture dishes and 24 h later were subjected to lipofection in duplicate with 2.5 μg of LMP1 construct and 2.5 μg of either TRAF1 or TRAF3 construct. For experiments with TRAF2 overexpression, 1.5 × 106 cells were seeded in 100-mm-diameter tissue culture dishes and transfected with 7.5 μg of mutant LMP1 constructs and 7.5 μg of TRAF2. Duplicate plates were combined, and whole-cell extracts were prepared by lysing cells in 250 μl of lysis buffer. FLAG-LMP1 and associated proteins were immunopurified with 25 μl of anti-FLAG M2 affinity gel. Bound LMP1 and TRAF proteins were detected by immunoblotting as described below.
Nuclear and cytoplasmic extracts.
Cell lines expressing various derivatives of LMP1 were seeded in 100-mm-diameter dishes and were harvested for extract preparation when the cells were ∼75% confluent. Extracts were prepared by a method described previously (9, 38). Briefly, cells were washed once with ice-cold phosphate-buffered saline, scraped from the dishes, collected in microcentrifuge tubes, and lysed for 3 min on ice in 5 pellet volumes of cytoplasmic extraction buffer (10 mM HEPES [pH 7.6], 60 mM KCl, 1 mM EDTA, 0.1% NP-40, 1 mM dithiothreitol, 1 mM PMSF, 2.5 μg each of aprotinin, leupeptin, and pepstatin per ml). Nuclei were collected (1,500 rpm, 4°C, 5 min), and the supernatants containing the cytoplasmic fraction were transferred to fresh tubes. Nuclei were washed with 300 μl of cytoplasmic extraction buffer without NP-40, collected as before, and lysed in 3 pellet volumes of nuclear extraction buffer (20 mM Tris [pH 8.0], 420 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM PMSF, 25% glycerol, 2.5 μg each of aprotinin, leupeptin, and pepstatin per ml). The final salt concentration was adjusted to ∼400 mM with NaCl, and nuclear pellets were vortexed and incubated on ice for 10 min. Cytoplasmic and nuclear extracts were cleared (14,000 rpm, 4°C, 10 min) and transferred to fresh tubes. Glycerol was added to the cytoplasmic extracts to a final concentration of 20%, and protein concentrations were determined with the Bio-Rad protein assay dye reagent.
Immunoblot analysis.
Cytoplasmic or whole-cell extracts (100 μg) were solubilized in 2× sample buffer, boiled for 10 min, separated on SDS–8 or 10% polyacrylamide gels, and transferred to supported nitrocellulose filters (Schleicher & Schuell), using a Bio-Rad Trans-blot wet electrophoretic transfer apparatus. Filters were stained with Ponceau S stain to visualize equal loading of protein in all lanes. Nonspecific reactivity was blocked by incubation overnight in Tris-buffered saline containing 0.1% Tween 20 and 5% nonfat dried milk (BLOTTO). Supernatant from the S12 monoclonal antibody (generous gift of David A. Thorley-Lawson) was used at a 1:10 dilution in BLOTTO for the detection of LMP1. A rabbit antiserum raised against the carboxy-terminal 100 amino acids of the EGFR fused to glutathione S-transferase (GST) (ERCT) was used at a 1:1,500 dilution in BLOTTO for the detection of the EGFR (kind gift of H. Shelton Earp). Rabbit antisera directed against TRAF1 (S-19), TRAF2 (C-20), TRAF3 (C-20), and CD40 (C-20) were used at a 1:200 dilution as specified by the manufacturer (Santa Cruz Biotechnology, Inc.). Antiserum directed against the A20 protein (generous gift of Vishva Dixit) was used at 1:500. Antiserum directed against IκBα (Rockland) was used at 1:2,000. Appropriate secondary anti-mouse or anti-rabbit antibodies (Amersham) were used at a dilution of 1:2,000 in BLOTTO to detect bound primary antibody. Reactive proteins were detected by incubation of washed filters (TBST) in the Amersham enhanced chemiluminescence system followed by exposure to autoradiographic film.
Electrophoretic mobility shift assays (EMSAs).
Nuclear extracts (10 μg) were incubated for 15 min at room temperature with a 32P-labeled probe containing a κB site from the class I major histocompatibility complex promoter (5′-CAGGGCTGGGGATTCCCCATCTCCACAGTTTCACTTC-3′) in binding buffer (10 mM Tris [pH 7.7], 50 mM NaCl, 0.5 mM EDTA, 1 mM dithiothreitol, 10% glycerol) with 2 μg of poly(dI-dC) · poly(dI-dC) (Pharmacia Biotech) (2, 3, 20, 54). Complexes were separated on 5% polyacrylamide gels in high-ionic-strength Tris-glycine-EDTA buffer (25 mM Tris, 190 mM glycine, 1 mM EDTA), dried, and subjected to autoradiography. For supershift experiments, 1- to 5-μl aliquots of rabbit polyclonal antibodies against the NF-κB subunits p50 (sc-114; Santa Cruz Biotechnology), p52 (catalog no. 100-4185; Rockland), and p65 (catalog no. 100-4165; Rockland) were incubated with nuclear extracts for 15 min prior to the addition of poly(dI-dC) · poly(dI-dC) and 32P-labeled probe. Complexes were then analyzed as described above.
RESULTS
LMP1 signaling domains induce different NF-κB complexes.
Previous analysis of the function of LMP1 mutants has indicated that two different signaling domains can activate the NF-κB transcription factor (Fig. 1) (6, 10, 25, 31, 40, 42). However, only the TRAF signaling domain, CTAR1, can induce expression of the EGFR. This finding suggested that a specific component of the TRAF signaling pathway that is not activated by CTAR2 is essential for the induction of EGFR expression.
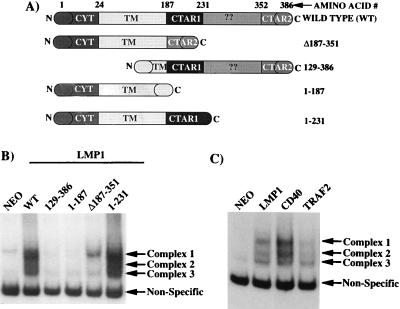
To determine if CTAR1 activated a specific subset of NF-κB components distinct from CTAR2, the LMP1 mutants described in Fig. 2A were stably transfected into C33A epithelial cells. LMP1(Δ187-351) lacks the TRAF interaction domain, CTAR1, but retains the major NF-κB activation domain, CTAR2. LMP1(129-386) is deleted for the first four transmembrane domains and is unable to insert properly into the cell membrane (37, 61). LMP1(1-187) is deleted for the entire carboxy-terminal cytoplasmic domain and therefore does not contain either CTAR1 or CTAR2. LMP1(1-231) retains CTAR1 but is deleted for CTAR2. Nuclear extracts from C33A cell lines expressing individual LMP1 mutants were analyzed by EMSA (Fig. 2B). As expected LMP1(WT), LMP1(Δ187-351), and LMP1(1-231) each induced the translocation of NF-κB components into the nucleus. The LMP1 mutants LMP1(129-386) and LMP1(1-187), which have been described as unable to activate NF-κB in transient reporter gene assays, did not induce the translocation of NF-κB components into the nucleus. Full-length LMP1(WT) induced three different NF-κB complexes (complexes 1, 2, and 3). Interestingly, while LMP1(1-231) induced the same three NF-κB complexes, LMP1(Δ187-351) induced only complex 1 and minimal or undetectable levels of complex 3. Immunoblot analyses of the expression levels of LMP1(1-231) and LMP1(Δ187-351) indicated that these mutants were expressed at equal levels (data not shown) (40). Supershift analyses of these complexes indicated that complex 3 contains p50-p50 homodimers, complex 2 contains p50-p52 heterodimers, and complex 1 contains p52 and p65 as well as additional unknown components as previously described (38, 45). Since CTAR1 and CTAR2 induced different NF-κB complexes, the repertoire of NF-κB complexes induced by LMP1 was compared to those of CD40 and TRAF2, which activate NF-κB through the TRAF pathway. EMSA analysis indicated that LMP1, CD40, and TRAF2 all induced three NF-κB complexes in C33A cells (Fig. 2C). Thus, CTAR1-mediated activation of complexes 1, 2, and 3 parallels the activation of NF-κB mediated by the TRAF signaling pathway. These results further distinguish the unique nature of CTAR2-mediated activation of NF-κB since this signaling domain activates complex 1 only.
FIG. 2.
Mutational analysis of LMP1 mutants with respect to NF-κB activation and EGFR induction. (A) Diagram of LMP1 mutants used in this experiment. The domains of LMP1 retained by the individual mutants are depicted. (B) EMSA analysis of nuclear NF-κB binding activity in extracts from C33A cells stably transfected with the indicated LMP1 constructs. The arrows indicate the specific complexes induced by LMP1. (C) EMSA analysis of NF-κB binding activity of LMP1 compared to CD40 and TRAF2.
Inhibition of NF-κB activation with a constitutively active IκBα reduces EGFR induction by LMP1 and CD40.
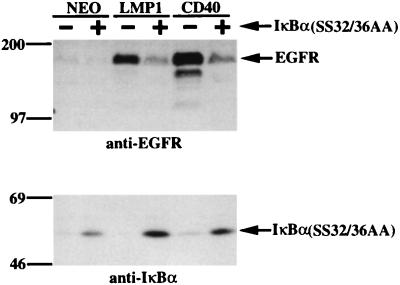
To determine if the specific NF-κB complexes induced by CTAR1 contribute to EGFR induction, C33A cell lines which express LMP1(WT) or CD40 either in the presence or in the absence of a constitutively active IκBα inhibitor of NF-κB were established (55). This IκBα molecule contains serine-to-alanine mutations at amino acids 32 and 36, IκBα(SS32/36AA), and is unable to be phosphorylated and degraded (55). Extracts from these cell lines were analyzed by immunoblotting for EGFR induction (Fig. 3, top). In control cells, LMP1 and CD40 induced high levels of EGFR expression as expected. However, in the presence of IκBα(SS32/36AA), LMP1- and CD40-mediated induction of EGFR expression was reduced by approximately 50 to 70%. IκBα(SS32/36AA) expression in these cell lines was confirmed by immunoblot analysis (Fig. 3, bottom). Since the antibody also reacts with endogenous IκBα, low-level expression of IκBα was also detected and was particularly evident in the CD40-expressing cell line. These results indicate that although NF-κB by itself is insufficient to induce EGFR expression, activation of NF-κB by the TRAF pathway contributes to high levels of EGFR induction.
FIG. 3.
EGFR induction is decreased in the presence of a constitutively active IκBα. LMP1 or CD40 was transfected into C33A cells in both the absence and the presence of a constitutively active IκBα(SS32/36AA). The resulting cell lines were analyzed by immunoblotting for EGFR expression (top) and the constitutively active IκBα(SS32/36AA) (bottom). Migration of molecular mass standards (in kilodaltons) is shown.
Activation of NF-κB and induction of EGFR expression by full-length LMP1 containing mutations in the TRAF binding domain PXQXT.
LMP1 contains a five-amino-acid sequence, PQQAT, which constitutes the minimal TRAF binding core (Fig. 4A). LMP1 shares this motif with the other TRAF-interacting proteins, and the consensus sequence is defined as PXQXT (Fig. 5A). Alanine substitution mutations were made in this motif and in surrounding residues, and the locations of residues mutated in the core PXQXT motif are indicated by asterisks in Fig. 4A. Each of the LMP1 mutant constructs was transfected into C33A cells, and cell lines expressing individual LMP1 constructs were established following selection in G418.
FIG. 4.
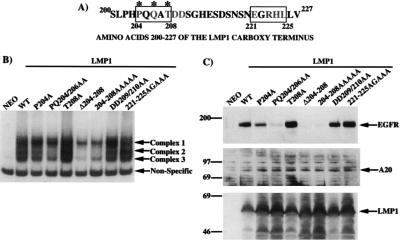
Effects of mutations in the LMP1 TRAF binding motif PXQXT on NF-κB activation and EGFR induction. (A) Amino acids 200 to 227 of the carboxy-terminal domain of LMP1. The TRAF binding PXQXT motif is indicated by a box, and asterisks denote the residues of the core element. Amino acids 221 to 225 show some similarity with a region in the CD40 TRAF interaction domain and are also indicated by a box. The shaded residues indicate those that were mutated and analyzed in the experiments. (B) EMSA of nuclear NF-κB binding activity in extracts from C33A cells stably transfected with the indicated LMP1 constructs. Induced complexes are indicated by arrows. (C) Immunoblot analysis of EGFR and A20 induction by LMP1 mutants. Migration of molecular mass standards (in kilodaltons) is shown.
FIG. 5.
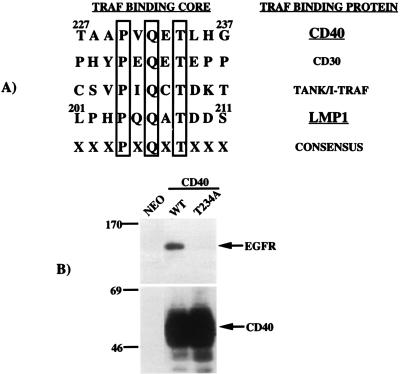
Effects of mutations in the CD40 TRAF binding motif PXQXT on EGFR induction. (A) Alignment of the amino acid sequences of several proteins known to interact with the TRAF molecules reveals a common motif, PXQXT, which is essential for interaction with the TRAFs. (B) The effect of replacement of Thr-234 with Ala in the PXQXT motif in CD40 on the induction of EGFR expression was analyzed by immunoblotting (top). Both wild-type and mutant CD40 proteins were expressed at equal levels as determined by immunoblotting (bottom). Migration of molecular mass standards (in kilodaltons) is shown.
Nuclear extracts from each cell line were analyzed by EMSA for the presence of TRAF-mediated activation of NF-κB (Fig. 4B). Total disruption of the PXQXT domain, in mutants LMP1(Δ204-208) and LMP1(204-208AAAAA), greatly reduced induction of complexes 2 and 3. The complexes induced by these mutants were very similar to those induced by LMP1(Δ187-351), indicating that complete disruption of the TRAF domain in CTAR1 eliminates NF-κB activation from this domain, leaving only activity due to CTAR2 signaling (compare Fig. 4B and 2B). Mutation of proline 204 to alanine, LMP1(P204A), slightly decreased the levels of complexes 2 and 3, while mutation of proline 204 and glutamine 206 to alanines, LMP1(PQ204/206AA), further decreased the levels of complexes 2 and 3. These results indicate that increasingly disruptive mutations of the PXQXT motif (P204A the least and Δ204-208 the strongest) reduces the strength of the TRAF signal which activates NF-κB complexes 2 and 3. Interestingly, mutation of threonine 208 to alanine, LMP1(T208A), did not reduce the TRAF-mediated component of NF-κB activation. This result was somewhat surprising since this position appears to be conserved in the TRAF-interacting proteins and the analogous mutation in CD40 abolishes TRAF binding and NF-κB activation (24, 26–28, 52). Two additional substitution mutants in CTAR1 which are located outside the PXQXT motif were also tested for activation of NF-κB. Replacement of aspartic acid residues 209 and 210 [LMP1(DD209/210AA)] or of residues between amino acids 221 and 225 [LMP1(221-225AGAAA)] has been shown to reduce TRAF binding to GST-LMP1 proteins (10, 51). Mutation of these residues did not reduce the TRAF-mediated component of NF-κB activation, indicating that these mutants retain sufficient TRAF interaction to activate NF-κB (Fig. 4B).
These cell lines were also analyzed by immunoblotting for induction of EGFR expression (Fig. 4C, top). Mutations of amino acids in the PXQXT motif also reduced the ability of LMP1 to induce EGFR expression. Specifically the LMP1(P204A) mutant had slightly reduced EGFR induction, while the double mutant, LMP1(PQ204/206AA), had greatly reduced EGFR induction. Interestingly, the LMP1(T208A) mutant induced levels of EGFR expression comparably to LMP1(WT). The LMP1(DD209/210) and LMP1(221-225AGAAA) mutants also induced EGFR expression comparably to LMP1(WT). As would be predicted, the two mutants with completely mutated PXQXT motifs, LMP1(Δ204-208) and LMP1(204-208AAAAA), were incapable of inducing EGFR expression.
The effect of mutations in the PXQXT motif were also analyzed in the context of an LMP1 mutant consisting of amino acids 1 to 231. FLAG-LMP1(1-231) containing either a single substitution of alanine for proline 204 (P204A) or a double substitution of alanines for proline 204 and glutamine 206 (PQ204/206AA) was defective for induction of EGFR expression, while substitution of alanine for threonine 208 (T208A) did not affect EGFR induction (data not shown). These results reveal that PXQXT mutations in LMP1(1-231) have a more severe effect than mutations in the full-length molecule and indicate that the overall structure of LMP1 contributes to the signaling capacity of LMP1 as well as the stability of LMP1-TRAF interactions.
Since all of these LMP1 mutants retain CTAR2, mutations in the PXQXT motif should disrupt TRAF signaling but should not completely block activation of NF-κB. Immunoblot analysis of the expression of the A20 protein, whose expression is regulated by NF-κB, indicated that CTAR2-mediated activation of NF-κB complex 1 was sufficient to induce expression of A20 despite the presence of disruptive mutations in the CTAR1 PXQXT motif (Fig. 4C, middle). Immunoblot analysis of LMP1 expression indicated that the mutants were all expressed at approximately equal levels (Fig. 4C, bottom). Importantly, these results demonstrate that mutations in the PXQXT motif in CTAR1 can specifically disrupt TRAF-mediated NF-κB activation and TRAF-mediated EGFR induction but do not affect the stability of LMP1 or NF-κB activation mediated by CTAR2.
Induction of EGFR expression by CD40 is blocked by mutation of the PXQXT motif.
Alignment of the TRAF interaction domains of several TRAF binding proteins including LMP1 and CD40 revealed a homology limited to the PXQXT motif which does not extend to amino acids surrounding this core motif (Fig. 5A). Substitution of threonine 234 with alanine (T234A) not only disrupts interaction with TRAF3 but also prevents CD40-mediated signaling (24, 26, 52). Expression constructs for wild-type [CD40(WT)] and mutant [CD40(T234A)] proteins were transfected into C33A cells, and cell lines were established. Previous studies have indicated that overexpression of CD40 can activate signaling pathways in the absence of ligand, which is most likely due to the clustering of CD40 in the cell membrane (48). Therefore the experiments with overexpressed CD40 in C33A cells were analyzed in the absence of ligand. These cell lines were analyzed by immunoblot for EGFR expression (Fig. 5B, top) and CD40 expression (Fig. 5B, bottom). As expected, CD40(WT) was a potent inducer of EGFR expression whereas CD40(T234A) was incapable of inducing EGFR expression. Expression of wild-type and mutant CD40 proteins was confirmed in both cell lines, and the two proteins were expressed at similar levels. These data indicate that the TRAF signaling pathway activated by CD40 is essential for EGFR induction. Interestingly, while the T234A mutation in the TRAF binding motif in CD40 completely blocked EGFR induction (Fig. 5B), the parallel mutation T208A in LMP1 (Fig. 4C) did not block LMP1-mediated induction of EGFR expression. These results suggest that while the core PXQXT motif in both of these proteins is essential for TRAF signaling, other residues surrounding the motif may contribute to TRAF interaction and activation of signaling pathways.
Interaction of LMP1 PXQXT mutants with TRAF1, TRAF2, and TRAF3.
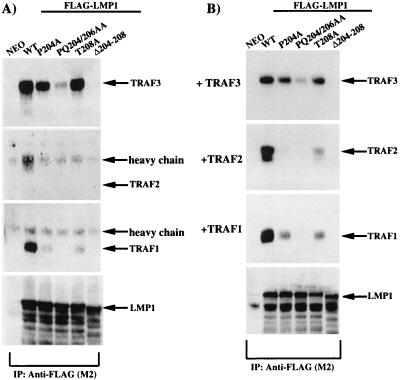
The results presented in Fig. 4C reveal that the PXQXT motif in CTAR1 is essential for LMP1 to induce expression of the EGFR. To further explore this unique TRAF-mediated signal, the ability of each of the full-length LMP1 constructs with point mutations in the TRAF binding PXQXT motif to interact with TRAF1, TRAF2, and TRAF3 was analyzed. Expression constructs for FLAG-LMP1(WT) and FLAG-LMP1 containing the point mutations described above were transfected into C33A cells. Wild-type and mutant FLAG-LMP1 constructs were immunopurified with anti-FLAG affinity gel, immunocomplexes were subjected to SDS-polyacrylamide gel electrophoresis, and bound TRAF proteins were analyzed by immunoblotting (Fig. 6A). TRAF1 and TRAF3 strongly interacted with FLAG-LMP1(WT), but an interaction between FLAG-LMP1(WT) and TRAF2 could not be detected. Compared to FLAG-LMP1(WT), the interaction of FLAG-LMP1(P204A) with TRAF3 was slightly weaker, FLAG-LMP1(PQ204/206AA) interacted very weakly with TRAF3, and FLAG-LMP1(Δ204-208) was unable to interact with TRAF3. Interestingly, the interaction of FLAG-LMP1(T208A) with TRAF3 was similar to that of FLAG-LMP1(WT). FLAG-LMP1(P204A) and FLAG-LMP1(T208A) interacted very weakly with TRAF1, while FLAG-LMP1(PQ204/206AA) and FLAG-LMP1(Δ204-208) were unable to interact with TRAF1. As for FLAG-LMP1(WT), an interaction between the LMP1 mutants and TRAF2 was not detected. Reprobing of the immunoblots with anti-LMP1 antibody indicated that all LMP1 mutants were present at equal levels in the immunoprecipitation reactions (a representative immunoblot is shown in the lowest panel of Fig. 6A). These results indicated that mutations in the LMP1 TRAF binding motif PXQXT distinctly affect interaction with TRAF1 and TRAF3.
FIG. 6.
In vivo analysis of TRAF binding to LMP1 proteins containing mutations in the PXQXT TRAF binding domain. (A) C33A cells were transfected with FLAG-LMP1 expression constructs containing mutations in the PXQXT motif. (B) C33A cells were transfected with FLAG-LMP1 and TRAF expression constructs. LMP1 was immunoprecipitated with anti-FLAG affinity gel, and TRAF binding to each mutant was analyzed by immunoblotting. Immunoblots were also reprobed with the anti-LMP1 monoclonal antibody S12 to ensure that equal amounts of LMP1 were immunoprecipitated in all reactions.
Since the interaction between LMP1 and endogenous TRAF2 was not detectable, the abilities of the various FLAG-LMP1 constructs to interact with overexpressed TRAF2 were analyzed (Fig. 6B). When TRAF2 was overexpressed in C33A cells, the interaction with FLAG-LMP1(WT) was readily detectable. FLAG-LMP1(T208A) retained the ability to interact weakly with TRAF2, while FLAG-LMP1(P204A) interaction with TRAF2 was even weaker and was detectable only on longer exposures of the TRAF2 immunoblot (Fig. 6B and data not shown). FLAG-LMP1(PQ204/206AA) and FLAG-LMP1(Δ204-208) did not interact with TRAF2. As described above, reprobing of the immunoblots with anti-LMP1 antibody indicated that all LMP1 mutants were present at equal levels in the immunoprecipitation reactions (a representative immunoblot is shown in the lowest panel of Fig. 6B). These results indicated that TRAF2 can also interact with the PXQXT motif in CTAR1 and that TRAF1, TRAF2, and TRAF3 each bind to LMP1 in a unique manner. The interaction between FLAG-LMP1 and the various point mutants with overexpressed TRAF1 and TRAF3 was identical to that observed with endogenous TRAF1 and TRAF3 (compare Fig. 6B and A). Analysis of the strength of binding between LMP1 and overexpressed TRAF proteins indicated that only 4% of total TRAF2 interacted with LMP1, while at least 25% of TRAF1 and 50% of TRAF3 were bound to LMP1 (data not shown). The lack of detectable interaction between LMP1 and endogenous TRAF2 could therefore reflect the very weak interaction with TRAF2 or could indicate that TRAF2 expression in C33A cells is very low.
Interestingly, the interaction of TRAF3 with LMP1 specifically correlated with the ability of LMP1 to induce expression of the EGFR (compare the uppermost panels in Fig. 4C and 6A). Mutants P204A, PQ204/206AA, and Δ204-208 were increasingly more defective in EGFR induction (Fig. 4C) and in TRAF3 binding (Fig. 6A), while the T208A mutation decreased binding of TRAF1 and TRAF2 but did not affect TRAF3 binding or EGFR induction. However, LMP1(WT) and LMP1(T208A), which both induced high levels of EGFR expression, retained binding to TRAF1, -2, and -3, suggesting that all three TRAF proteins contribute to EGFR induction.
Effect of overexpression of individual TRAF proteins on EGFR induction.
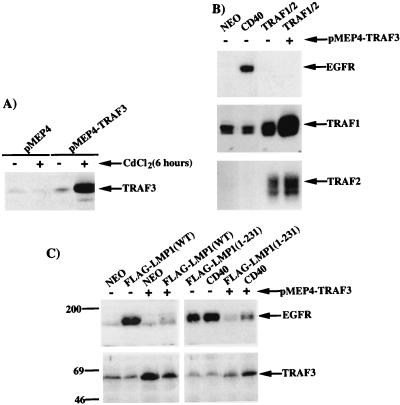
Previous reports have indicated that overexpressed TRAF2 acts as a positive regulator and overexpressed TRAF3 acts as a negative regulator of TRAF-mediated activation of NF-κB (7, 8, 48, 57). To analyze the effects of these molecules on the induction of EGFR expression, cell lines overexpressing these proteins were established. The pMEP4 expression vector with the inducible metallothionein promoter was used to establish a stable cell line overexpressing TRAF3 (pMEP4-TRAF3), in which TRAF3 expression could be further induced with CdCl2 (Fig. 7A). pMEP4 and pMEP4-TRAF3 cell lines were stably transfected with expression vectors for CD40, or a combination of TRAF1 and TRAF2, and the resulting cell lines were analyzed by immunoblotting for induction of the EGFR (Fig. 7B, top). Expression of CD40 in the control pMEP4 cell line clearly resulted in the induction of EGFR expression. Coexpression of TRAF1 and TRAF2 in the control pMEP4 cell line did not induce EGFR expression, although expression of TRAF2 activated the same NF-κB complexes as CD40 (Fig. 2C). Expression of TRAF1 and TRAF2 in these cell lines was confirmed by immunobloting (Fig. 7B, lower two panels). Expression of TRAF1 or TRAF2 individually did not induce EGFR expression (data not shown). Coexpression of TRAF1 and TRAF2 in the TRAF3-overexpressing cell line also did not induce EGFR expression (Fig. 7B, top). Therefore, overexpression of TRAF1, TRAF2, and TRAF3 is unable to induce EGFR expression. These data suggest that a proper complex of multiple TRAF proteins must assemble at the plasma membrane with LMP1 or CD40 in order to mediate EGFR induction.
FIG. 7.
Effects of overexpression of individual TRAF proteins on EGFR induction. (A) Establishment of a TRAF3-expressing cell line. pMEP4 or pMEP4-TRAF3 was transfected into C33A cells, and following selection in hygromycin, the cell lines were analyzed by immunoblotting for TRAF3 expression. The cell lines were either left untreated or treated with 50 μM CdCl2 for 6 h to induce expression of the metallothionein-driven TRAF3 expression construct. (B) TRAF1 and TRAF2 were stably overexpressed in the pMEP4 and pMEP4-TRAF3 cell lines, and extracts were analyzed by immunoblotting. + and − indicate the pMEP4-TRAF3 and pMEP4 cell lines. The upper panel is an immunoblot for EGFR expression, and the lower panels are immunoblots for TRAF1 and TRAF2 expression. (C) The LMP1(WT), LMP1(1-231), or CD40 expression construct was transfected into either the pMEP4 or pMEP4-TRAF3 cell line. Resulting cell lines were analyzed by immunoblotting for EGFR expression (top) or TRAF3 expression (bottom). The immunoblot shown in the upper left panel with FLAG-LMP1(WT) is a longer exposure reflecting the somewhat weaker induction of EGFR expression by FLAG-LMP1(WT) than by FLAG-LMP1(1-231) or CD40. Migration of molecular mass standards (in kilodaltons) is shown.
To analyze the effect of TRAF3 overexpression on LMP1- and CD40-mediated induction of EGFR expression, FLAG-LMP1(WT), FLAG-LMP1(1-231), or CD40 was expressed in either the pMEP4 or pMEP4-TRAF3 cell line (Fig. 7C). As expected, expression of each of these proteins in the control pMEP4 cell line resulted in the induction of EGFR expression (Fig. 7C, top). In contrast, expression of FLAG-LMP1(WT) or FLAG-LMP1(1-231) in the pMEP4-TRAF3 cell line decreased EGFR induction >90% compared to the pMEP4 control, while CD40-mediated EGFR induction was decreased ∼75% (Fig. 7C, top). Analysis of TRAF3 expression in these cell lines confirmed that TRAF3 was overexpressed in the absence of CdCl2 (Fig. 7C, bottom). Increasing TRAF3 expression with CdCl2 did not further reduce EGFR expression (data not shown). Since overexpression of TRAF3 decreased EGFR induction mediated by LMP1, the effect of overexpression of TRAF1 or TRAF2 was also analyzed in cell lines expressing LMP1. In contrast to TRAF3, neither TRAF1 or TRAF2 affected the induction of EGFR expression by LMP1 (data not shown). These results indicate that overexpression of TRAF3 decreases induction of EGFR expression by either FLAG-LMP1(WT), FLAG-LMP1(1-231), or CD40. Since TRAF3 has the strongest binding capacity for LMP1 and because TRAF3 overexpression likely displaces other essential TRAFs, these data suggest that induction of EGFR expression is dependent on the appropriate stoichiometry of TRAF1, -2, and -3 with LMP1 at the plasma membrane.
DISCUSSION
Of the two signaling domains in the LMP1 carboxy-terminal cytoplasmic terminus, only CTAR1 is capable of inducing EGFR expression (40). Both domains activate NF-κB, although CTAR1 is a weaker activator (25% of the wild-type level) in transient reporter gene assays compared to CTAR2 (75% of the wild-type level) (25, 40, 42). Since only CTAR1 is capable of inducing EGFR expression, the specific NF-κB components induced by CTAR1 and CTAR2 were analyzed. The data presented here indicate that in C33A cells, CTAR1 activates a more diverse and robust nuclear mobilization of NF-κB components as determined by EMSA (Fig. 2B). LMP1(WT) induces three specific NF-κB complexes (complexes 1, 2, and 3). LMP1(1-231), which retains CTAR1, induces the same three complexes, while LMP1(Δ187-351), which retains CTAR2, induces only complex 1. The data suggest that in C33A cells, the difference in NF-κB activation potential by CTAR1 and CTAR2 in transient reporter gene assays may be reflected in the different repertoires of NF-κB complexes induced by these domains. The relatively stronger activation of NF-κB by CTAR2 in transient reporter gene assays may result from activation of only complex 1 (p52-p65). Complex 3 (p50-p50), which is induced strongly by CTAR1, may dampen the NF-κB-activating effects of p52-p65 by competing for κB sites in the reporter gene. This could effectively lower the amount of CTAR1-mediated NF-κB activity in reporter gene assays without reflecting the effects of the individual NF-κB components on specific gene activation. It is possible that complex 2 (p50-p52), which is induced by CTAR1, contributes to EGFR induction mediated by the TRAF domain. The NF-κB-regulated A20 gene is induced to equal levels by either LMP1 signaling domain; therefore, activation of complex 1 in C33A cells by mutants deleted in CTAR1, such as like LMP1(Δ204-208) or LMP1(Δ187-351), is sufficient to induce expression of A20. The data presented here indicate that one of the distinct NF-κB complexes induced by CTAR1-mediated TRAF signaling may contribute to high-level induction of EGFR expression.
Mutational analyses of the PXQXT motif in CTAR1 revealed that the presence of these amino acids in LMP1 is essential for induction of EGFR expression. Complete deletion of the motif or substitution with alanines eliminated induction of EGFR expression as well as the CTAR1-specific NF-κB complexes. These experiments reveal that LMP1(Δ204-208) is functionally identical to LMP1(Δ187-351). LMP1(PQ204/206AA) induced low levels of complex 2 and reduced levels of complex 3 compared to LMP1(WT) and also induced low levels of EGFR expression. The induction of EGFR expression and activation of NF-κB complexes 2 and 3 by LMP1 in C33A cells are the first defined biochemical events that are specific for TRAF activation through the TRAF-interacting PXQXT motif.
These studies are also the first to analyze the effects of mutations in the PXQXT motif on the ability of LMP1 to interact with the TRAFs in vivo in comparison with EGFR induction and NF-κB activation. LMP1(P204A) and LMP1(PQ204/206AA) were reduced in the ability to induce EGFR expression and the TRAF-specific NF-κB complexes. In contrast, LMP1(T208A) induced EGFR expression and activated NF-κB comparably to LMP1(WT). Analysis of LMP1 interaction with the TRAFs indicated that there was a close correlation between EGFR induction and TRAF3 association with LMP1. LMP1(P204A) and LMP1(PQ204/206AA) were reduced in the ability to interact with TRAF3 comparably with their ability to induce EGFR expression. LMP1(T208A) associated strongly with TRAF3 and induced EGFR expression comparably to LMP1(WT). While interaction of LMP1 with TRAF3 appears to directly correlate with EGFR induction, TRAF2 and TRAF1 apparently contribute to high-level induction of EGFR expression. LMP1(T208A) still interacted weakly with TRAF1 and TRAF2 and induced high levels of EGFR expression, while LMP1(PQ204/206AA) was unable to interact with TRAF1 and TRAF2 and induced low levels of EGFR expression. These results suggest that interaction of TRAF1, -2, and -3 with LMP1 contributes to induction of EGFR expression. It is also possible that the interaction of TRAF1 and TRAF2 with LMP1 contributes to high-level EGFR induction through the activation of NF-κB. The activation of NF-κB and augmentation of EGFR induction by TRAF1 and TRAF2 is also indicated by the finding that EGFR induction was decreased in the presence of a constitutively activate IκBα(SS32/36AA) molecule which behaves as an NF-κB repressor.
Overexpression of TRAF2 in C33A cells either alone or in combination with TRAF1 or TRAF3 does not induce EGFR expression even though overexpression of TRAF2 resulted in activation of the same three NF-κB complexes induced by LMP1 and CD40. These results suggest that induction of EGFR expression requires clustering of the TRAFs through the action of a membrane-bound receptor such as LMP1 or CD40 and also that the activation of NF-κB complexes 2 and 3 by TRAF2 is insufficient to induce EGFR expression. The interaction of LMP1 with TRAF3 appears to be required for EGFR induction; however, interaction of TRAF3 with LMP1 is not the sole signaling molecule required for LMP1-mediated EGFR induction, since overexpression of TRAF3 decreases LMP1-mediated EGFR induction. Studies have shown that overexpression of TRAF3 could displace more weakly interacting TRAF molecules, which would effectively block the LMP1 signal; thus, it is likely that TRAF3 overexpression displaces a TRAF which is also essential for EGFR induction (10, 51). These results indicate that activation of this signaling pathway requires the correct assembly and stoichiometry of the TRAF molecules with LMP1 at the cell membrane.
The involvement of TRAF3 in LMP1-mediated induction of EGFR expression suggests that although TRAF2 appears to be the major regulator of NF-κB activation, the other TRAFs also mediate distinct TRAF signaling events. Interestingly, recent studies with TRAF3-deficient mice suggest a positive role for TRAF3 in TRAF signaling pathways (65). These mice become increasingly runted with age, correlating with a depletion of peripheral leukocytes and death by day 10. These results suggest that TRAF3 may play a critical role in survival of B precursor cells (65). The importance of TRAF3 in EGFR induction also suggests that TRAF3 is a positive factor during TRAF signaling events. TRAF3 is likely to be particularly important for the growth-promoting effects of LMP1 on epithelial cells, where EGFR expression is necessary for proliferation (39).
Although the PXQXT motif is the sole TRAF binding site in LMP1, surrounding residues in LMP1 contribute to the strength of interaction with the TRAFs (10). Specifically, GST-LMP1(199-214) with the P204A mutation decreased TRAF1, -2, and -3 binding by >99%, while GST-LMP1(187-231) or FLAG-LMP1(1-231) with the P204A mutation demonstrated strong binding to TRAF3, reduced binding to TRAF1, and little or no binding to TRAF2 (10). In the context of full-length LMP1(WT) proteins, the results presented here confirm the previous observations and extend the analysis to include mutations in other residues in the PXQXT motif. Particularly interesting is the strength with which LMP1(T208A) binds to the TRAFs. Full-length LMP1(T208A) retained TRAF3 binding comparably to LMP1(WT) and retained the ability to interact with TRAF1 and TRAF2 (Fig. 6). The same mutation in GST-LMP1(199-214) abrogated binding to TRAF1, -2, and -3 by greater then 95% (10). Comparison of functional LMP1 signaling with regard to EGFR induction also highlights the differences between full-length LMP1 and LMP1(1-231). While LMP1(1-231) mutated in either P204A or PQ204/206AA is completely unable to induce EGFR expression, full-length LMP1 proteins with these same mutations are still able to induce intermediate to low levels of EGFR expression. These results confirm the suggestion that amino acids surrounding the PXQXT motif in LMP1 contribute to the strength of interactions with the TRAFs and also enhance the functional effects of signaling from CTAR1.
LMP1 has many of the same signaling properties as the TNFR family members CD40 and CD30, which share the core TRAF binding motif PXQXT (7, 10, 18, 24). In agreement with this observation is the finding that CD40, like LMP1, can interact with the TRAF molecules and induce expression of the EGFR (8, 24, 40, 53). CD40 has been shown to interact with TRAF2, -3, -5, and -6 (24, 26–28, 52). These studies revealed that the T234A mutation in the PXQXT motif of CD40 prevented interaction with TRAF2, -3, and -5 and prevented CD40 signaling. The data presented in this study indicate that the T234A mutant was completely deficient in EGFR induction, although the analogous mutation in LMP1 did not block EGFR induction. These results and the data for the LMP1(1-231) mutants indicate that each amino acid in the PXQXT motif, as well as surrounding residues, contributes to TRAF interaction and reveals that the TRAFs interact differently with the CD40 and LMP1 TRAF domains. This is an important difference and may facilitate the design of specific inhibitors that could affect LMP1 signaling while leaving TRAF signaling pathways from cellular proteins like CD30 and CD40 largely unaffected.
Activation of TRAF signaling pathways by LMP1 is essential for the immortalization of lymphocytes in vitro by EBV and probably also in the establishment of epithelial malignancies like NPC (30). Activation of NF-κB by LMP1 mediates important aspects of the transformation process; however, pathways other than NF-κB activated by the TRAF signaling pathway are also likely to be important in mediating the transforming effects of LMP1. Deletion of the PXQXT motif in LMP1 abolishes interaction with the TRAF proteins and prevents transformation of lymphocytes in vitro although the mutated LMP1 induces high levels of NF-κB (30). The data presented in this study demonstrate that deletion of the PXQXT motif prevents induction of EGFR expression in epithelial cells. The recruitment of a proper combination of TRAF molecules to CTAR1 in LMP1 results in the activation of signaling pathways, leading to activation of NF-κB and induction of molecules like the EGFR. Recent studies indicate that induction of E-selectin by the TRAF pathway involves activation of both the c-Jun and NF-κB transcription factors (41). However, these two essential pathways were both activated by TRAF2 (41). High-level EGFR induction by the TRAF pathway also requires two signals; one requires activation of NF-κB through LMP1-TRAF2 interaction, and the second is dependent on assembly of an LMP1 complex containing TRAF1, -2, and -3. The study of pathways activated by LMP1-TRAF interactions is essential to further understand LMP1 function and is likely to identify signaling pathways and downstream targets which are critical to lymphoid and epithelial cell transformation.
ACKNOWLEDGMENTS
We thank Katherine Fries and Frank Scholle for critical reading of the manuscript, H. Shelton Earp for the anti-ERCT rabbit antiserum, D. A. Thorley Lawson for the anti-LMP1 monoclonal antibody S12, Vishva Dixit for the anti-A20 monoclonal antibody, Dean Ballard for the IκB(SS32/36AA) construct, George Mosialos for TRAF1 and TRAF3 constructs, Ken Kaye for the TRAF2 construct, and Hitoshi Kikutani for the CD40 cDNA construct. J.L.C. acknowledges the support of A. S. Baldwin, Jr.
This work was supported by Public Health Service grants CA19014, CA32979, and CA52406 from the National Institutes of Health to N.R.-T. and CA72771 to A.S.B. W.E.M. was supported in part by predoctoral NIH National Service Award AI07419-02, and J.L.C. was supported by NIH/NRSA fellowship 1 F32 AG05745-01.
REFERENCES
- 1.Baichwal V R, Sugden B. The multiple membrane-spanning segments of the BNLF-1 oncogene from Epstein-Barr virus are required for transformation. Oncogene. 1989;4:67–74. [PubMed] [Google Scholar]
- 2.Baldwin A S, Jr, Sharp P A. Binding of a nuclear factor to a regulatory sequence in the promoter of the mouse H-2kb class I major histocompatibility gene. Mol Cell Biol. 1987;7:305–313. doi: 10.1128/mcb.7.1.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baldwin A S, Jr, Sharp P A. Two transcription factors, NF-kappa B and H2TF1, interact with a single regulatory sequence in the class I major histocompatibility complex promoter. Proc Natl Acad Sci USA. 1988;85:723–727. doi: 10.1073/pnas.85.3.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Banchereau J, de Paoli P, Valle A, Garcia E, Rousset F. Long-term human B cell lines dependent on interleukin-4 and antibody to CD40. Science. 1991;251:70–72. doi: 10.1126/science.1702555. [DOI] [PubMed] [Google Scholar]
- 5.Berberich I, Shu G, Clark E A. Cross-linking CD40 on B-cells rapidly activates nuclear factor-kB. J Immunol. 1994;153:4357–4366. [PubMed] [Google Scholar]
- 6.Brodeur S R, Cheng G, Baltimore D, Thorley-Lawson D. Localization of the major NF-κB-activating site and the sole TRAF3 binding site site of LMP1 defines two distinct signaling motifs. J Biol Chem. 1997;272:19777–19784. doi: 10.1074/jbc.272.32.19777. [DOI] [PubMed] [Google Scholar]
- 7.Cheng G, Baltimore D. TANK, a co-inducer with TRAF2 of TNF-and CD40L-mediated NF-κB activation. Genes Dev. 1996;10:963–973. doi: 10.1101/gad.10.8.963. [DOI] [PubMed] [Google Scholar]
- 8.Cheng G, Cleary A L, Ye Z-S, Hong D I, Lederman S, Baltimore D. Involvement of CRAF1, a relative of TRAF, in CD40 signaling. Science. 1995;267:1494–1498. doi: 10.1126/science.7533327. [DOI] [PubMed] [Google Scholar]
- 9.Cheshire J L, Baldwin A S., Jr Synergistic activation of NF-κB by tumor necrosis factor alpha and gamma interferon via enhanced IκBα degradation and de novo IκBβ degradation. Mol Cell Biol. 1997;17:6746–6754. doi: 10.1128/mcb.17.11.6746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Devergne O, Hatzivassiliou E, Izumi K, Kaye K, Kleijnen M, Kieff E, Mosialos G. TRAF1, TRAF2, and TRAF3 effect NF-κB activation by an Epstein-Barr virus LMP1 domain important for B-lymphocyte transformation. Mol Cell Biol. 1996;16:7098–7108. doi: 10.1128/mcb.16.12.7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eliopoulus A G, Stack M, Dawson C, Kaye K, Hodgkin L, Sihota S, Rowe M, Young L. Epstein-Barr virus-encoded LMP1 and CD40 mediate IL-6 production in epithelial cells via an NF-κB pathway involving TNF receptor-associated factors. Oncogene. 1997;14:2899–2916. doi: 10.1038/sj.onc.1201258. [DOI] [PubMed] [Google Scholar]
- 12.Fahraeus R, Fu H L, Ernberg I, Finke J, Rowe M, Klein G, Falk K, Nilsson E, Yadav M, Busson P, et al. Expression of Epstein-Barr virus-encoded proteins in nasopharyngeal carcinoma. Int J Cancer. 1988;42:329–338. doi: 10.1002/ijc.2910420305. [DOI] [PubMed] [Google Scholar]
- 13.Fennewald S, van Santen V, Kieff E. The nucleotide sequence of a messenger RNA transcribed in latent growth transforming virus infection indicates that it may encode a membrane protein. J Virol. 1984;51:411–419. doi: 10.1128/jvi.51.2.411-419.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Floettmann J E, Rowe M. Epstein-Barr virus latent membrane protein-1 (LMP1) C-terminus activation region 2 (CTAR2) maps to the far C-terminus and requires oligomerization for NF-κB activation. Oncogene. 1997;15:1851–1858. doi: 10.1038/sj.onc.1201359. [DOI] [PubMed] [Google Scholar]
- 15.Franken M, Devergne O, Rosenzweig M, Annis B, Kieff E, Wang F. Comparative analysis identifies conserved tumor necrosis factor receptor-associated factor 3 binding sites in the human and simian Epstein-Barr virus oncogene LMP1. J Virol. 1996;70:7819–7826. doi: 10.1128/jvi.70.11.7819-7826.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fries K L, Miller W E, Raab-Traub N. Epstein-Barr virus latent membrane protein 1 blocks p53-mediated apoptosis through the induction of the A20 gene. J Virol. 1996;70:8653–8659. doi: 10.1128/jvi.70.12.8653-8659.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galibert L, Durand I, Banchereau J, Rousset F. CD40-activated surface IgD-positive lymphocytes constitute the long term IL-4-dependent proliferating B cell pool. J Immunol. 1994;152:22–29. [PubMed] [Google Scholar]
- 18.Gedrich R W, Gilfillan M C, Duckett C S, Dongen J L, Thompson C B. CD30 contains two binding sites with different specificities for members of the tumor necrosis factor-receptor-associated factor family of signal transducing proteins. J Biol Chem. 1996;271:12852–12858. doi: 10.1074/jbc.271.22.12852. [DOI] [PubMed] [Google Scholar]
- 19.Hammarskjold M L, Simurda M C. Epstein-Barr virus latent membrane protein transactivates the human immunodeficiency virus type 1 long terminal repeat through induction of NF-κB activity. J Virol. 1992;66:6496–6501. doi: 10.1128/jvi.66.11.6496-6501.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haskill S, Beg A A, Tompkins S M, Morris J M, Yurochko A D, Sampson-Johanes A, Mondal K, Ralph P, Baldwin A S., Jr Characterization of an immediate-early gene induced in adherent monocytes that encodes I kappa B-like activity. Cell. 1991;65:1281–1289. doi: 10.1016/0092-8674(91)90022-q. [DOI] [PubMed] [Google Scholar]
- 21.Hatzivassiliou E, Miller W E, Raab-Traub N, Kieff E, Mosialos G. A fusion of the Epstein-Barr virus latent membrane protein 1 (LMP1) transmembrane domains to the CD40 cytoplasmic domain is similar to LMP1 in constitutive activation of epidermal growth factor expression, NF-κB, and stress activated protein kinase. J Immunol. 1998;160:1116–1121. [PubMed] [Google Scholar]
- 22.Henderson S, Rowe M, Gregory C, Croom-Carter D, Wang F, Longnecker R, Kieff E, Rickinson A. Induction of bcl-2 expression by Epstein-Barr virus latent membrane protein 1 protects infected B cells from programmed cell death. Cell. 1991;65:1107–1115. doi: 10.1016/0092-8674(91)90007-l. [DOI] [PubMed] [Google Scholar]
- 23.Hennessy K, Fennewald S, Hummel M, Cole T, Kieff E. A membrane protein encoded by Epstein-Barr virus in latent growth-transforming infection. Proc Natl Acad Sci USA. 1984;81:7201–7211. doi: 10.1073/pnas.81.22.7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu H M, O’Rourke K, Boguski M S, Dixit V M. A novel RING finger protein interacts with the cytoplasmic domain of CD40. J Biol Chem. 1994;269:30069–30072. [PubMed] [Google Scholar]
- 25.Huen D S, Henderson S A, Croom-Carter S, Rowe M. The Epstein-Barr virus latent membrane protein-1 (LMP1) mediates activation of NF-kB and cell surface phenotype via two effector regions in its carboxy-terminal cytoplasmic domain. Oncogene. 1995;10:549–560. [PubMed] [Google Scholar]
- 26.Inui S, Kaisho T, Kikutani H, Stamenkovic I, Seed B, Clark E, Kishimoto T. Identification of the intracytoplasmic region essential for signal transduction through a B-cell activation molecule, CD40. Eur J Immunol. 1990;20:1747–1753. doi: 10.1002/eji.1830200819. [DOI] [PubMed] [Google Scholar]
- 27.Ishida T, Mizushima S, Azuma S, Kobayshi N, Tojo T, Suzuki K, Aizawa S, Watanabe T, Mosialos G, Kieff E, Yamamoto T, Inoue J. Identification of TRAF6, a novel tumor necrosis factor receptor-associated factor protein that mediates signaling from an amino-terminal domain of the CD40 cytoplasmic domain. J Biol Chem. 1996;271:28745–28748. doi: 10.1074/jbc.271.46.28745. [DOI] [PubMed] [Google Scholar]
- 28.Ishida T, Tojo T, Aoki T, Kobayashi N, Ohishi T, Watanabe T, Yamamoto T, Inoue J. TRAF5, a novel tumor necrosis factor receptor-associated factor family protein, mediated CD40 signaling. Proc Natl Acad Sci USA. 1996;93:9437–9442. doi: 10.1073/pnas.93.18.9437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Izumi K, Kieff E. The Epstein-Barr virus oncogene product latent membrane protein 1 engages the tumor necrosis factor receptor-associated death domain protein to mediate B-lymphocyte growth transformation and activate NF-κB. Proc Natl Acad Sci USA. 1997;94:12592–12597. doi: 10.1073/pnas.94.23.12592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Izumi K M, Kaye K M, Kieff E D. The Epstein-Barr virus LMP1 amino acid sequence that engages tumor necrosis factor receptor associated factors is critical for primary B lymphocyts growth transformation. Proc Natl Acad Sci USA. 1997;94:1447–1452. doi: 10.1073/pnas.94.4.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaye K, Devergne O, Harada J, Izumi K, Yalamanchili R, Kieff E, Mosialos G. Tumor necrosis receptor associated factor 2 is a mediator of NF-kB activation by latent infection membrane protein 1, the Epstein-Barr virus transforming protein. Proc Natl Acad Sci USA. 1996;93:11085–11090. doi: 10.1073/pnas.93.20.11085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaye K M, Izumi K M, Kieff E. Epstein-Barr virus latent membrane protein 1 is essential for B-lymphocyte growth transformation. Proc Natl Acad Sci USA. 1993;90:9150–9154. doi: 10.1073/pnas.90.19.9150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kieff E. Epstein-Barr virus and its replication. In: Fields B N, Knipe D M, Howley P M, editors. Virology. 3rd ed. Philadelphia, Pa: Lippinscott-Raven Publishers; 1996. pp. 2343–2396. [Google Scholar]
- 34.Kieser A, Kilger E, Gires O, Ueffing M, Kolch W, Hammerschmidt W. Epstein-Barr virus latent membrane protein-1 triggers AP-1 activity via the c-Jun N-terminal kinase cascade. EMBO J. 1997;16:6478–6485. doi: 10.1093/emboj/16.21.6478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laherty C D, Hu H M, Opipari A W, Wang F, Dixit V M. The Epstein-Barr virus LMP1 gene product induces A20 zinc finger protein expression by activating NF-κB. J Biol Chem. 1992;267:24157–24160. [PubMed] [Google Scholar]
- 36.Liebowitz D, Kieff E. Epstein-Barr virus latent membrane protein: induction of B-cell activation antigens and membrane patch formation does not require vimentin. J Virol. 1989;63:4051–4054. doi: 10.1128/jvi.63.9.4051-4054.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liebowitz D, Mannick J, Takada K, Kieff E. Phenotypes of Epstein-Barr virus LMP1 deletion mutants indicate transmembrane and amino-terminal cytoplasmic domains necessary for effects in B-lymphoma cells. J Virol. 1992;66:4612–4616. doi: 10.1128/jvi.66.7.4612-4616.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miller, W. E., J. L. Cheshire, A. S. Baldwin, Jr., and N. Raab-Traub. The NPC derived C15 LMP1 protein confers enhanced activation of NF-κB and induction of the EGFR in epithelial cells. Oncogene, in press. [DOI] [PubMed]
- 39.Miller W E, Earp H S, Raab-Traub N. The Epstein-Barr virus latent membrane protein1 induces expression of the epidermal growth factor receptor. J Virol. 1995;69:4390–4398. doi: 10.1128/jvi.69.7.4390-4398.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miller W E, Mosialos G, Kieff E, Raab-Traub N. Epstein-Barr virus LMP1 induction of the EGFR is mediated through a TRAF signaling pathway distinct from NF-κB activation. J Virol. 1997;71:586–594. doi: 10.1128/jvi.71.1.586-594.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Min W, Pober J. TNF initiates E-selectin transcription in human endothelial cells through parallel TRAF-NF-kB and TRAF-RAC/CDC42-JNK-c-Jun/ATF-2 pathways. J Immunol. 1997;159:3508–3518. [PubMed] [Google Scholar]
- 42.Mitchell T, Sugden B. Stimulation of NF-κB-mediated transcription by mutant derivatives of the latent membrane protein of Epstein-Barr virus. J Virol. 1995;69:2968–2976. doi: 10.1128/jvi.69.5.2968-2976.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moorthy R K, Thorley-Lawson D A. All three domains of the Epstein-Barr virus-encoded latent membrane protein LMP-1 are required for transformation of rat-1 fibroblasts. J Virol. 1993;67:1638–1646. doi: 10.1128/jvi.67.3.1638-1646.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mosialos G, Birkenbach M, Yalamanchili R, VanArsdale T, Ware C, Kieff E. The Epstein-Barr virus transforming protein LMP1 engages signalling proteins for the tumor necrosis factor receptor family. Cell. 1995;80:389–399. doi: 10.1016/0092-8674(95)90489-1. [DOI] [PubMed] [Google Scholar]
- 45.Paine E, Scheinman R I, Baldwin A S, Jr, Raab-Traub N. Expression of LMP1 in epithelial cells leads to the activation of a select subset of NF-κB/Rel family proteins. J Virol. 1995;69:4572–4576. doi: 10.1128/jvi.69.7.4572-4576.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rajadurai P, Prasad U, Sadler R, Flynn K, Raab-Traub N. Clonal proliferations of cells infected with Epstein-Barr virus in preinvasive lesions related to nasopharyngeal carcinoma. N Engl J Med. 1995;333:693–698. doi: 10.1056/NEJM199509143331103. [DOI] [PubMed] [Google Scholar]
- 47.Rickinson A B, Kieff E. Epstein-Barr virus. In: Fields B N, Knipe D M, Howley P M, editors. Virology. 3rd ed. Philadelphia, Pa: Lippinscott-Raven Publishers; 1996. pp. 2397–2446. [Google Scholar]
- 48.Rothe M, Sarma V, Dixit V M, Goeddel D V. TRAF-2 mediated activation of NF-κB by TNF receptor 2 and CD40. Science. 1995;269:1424–1427. doi: 10.1126/science.7544915. [DOI] [PubMed] [Google Scholar]
- 49.Rothe M, Wong S C, Henzel W J, Goeddel D V. A novel family of putative signal transducers associated with the cytoplasmic domain of the 75 kDa tumor necrosis factor receptor. Cell. 1994;78:681–692. doi: 10.1016/0092-8674(94)90532-0. [DOI] [PubMed] [Google Scholar]
- 50.Saeland S, Duvert V, Moreau I, Banchereau J. Human B cell precursors proliferate and express CD23 after CD40 ligation. J Exp Med. 1993;178:113–120. doi: 10.1084/jem.178.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sandberg M, Hammerschmidt W, Sugden B. Characterization of LMP1’s association with TRAF1, TRAF2, and TRAF3. J Virol. 1997;71:4649–4656. doi: 10.1128/jvi.71.6.4649-4656.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sarma V, Lin Z, Clark L, Rust B M, Tewari M, Noelle R J, Dixit V M. Activation of the B-cell surface receptor CD40 induces A20, a novel zinc finger protein that inhibits apoptosis. J Biol Chem. 1995;270:12343–12346. doi: 10.1074/jbc.270.21.12343. [DOI] [PubMed] [Google Scholar]
- 53.Sato T, Irie S, Reed J C. A novel member of the TRAF family of putative signal transducing proteins binds to the cytosolic domain of CD40. FEBS Lett. 1995;358:113–118. doi: 10.1016/0014-5793(94)01406-q. [DOI] [PubMed] [Google Scholar]
- 54.Scheinman R I, Begg A A, Baldwin A S., Jr NF-κB p100 (Lyt-10) is a component of H2TF1 and can function as an IκB-like molecule. Mol Cell Biol. 1993;13:6089–6101. doi: 10.1128/mcb.13.10.6089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Scherer D C, Brockman J A, Chen Z, Maniatis T, Ballard D. Signal-induced degradation of IκBα requires site-specific ubiquitination. Proc Natl Acad Sci USA. 1995;92:11259–11263. doi: 10.1073/pnas.92.24.11259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Smith C, Farrah T, Goodwin R. The TNF receptor superfamily of cellular and viral proteins: activation, costimulation, and death. Cell. 1994;76:959–962. doi: 10.1016/0092-8674(94)90372-7. [DOI] [PubMed] [Google Scholar]
- 57.Takeuchi M, Rothe M, Goeddel D V. Anatomy of TRAF2. J Biol Chem. 1996;271:19935–19942. doi: 10.1074/jbc.271.33.19935. [DOI] [PubMed] [Google Scholar]
- 58.Tartaglia L V, Goeddel D V. Two TNF receptors. Immunol Today. 1992;13:151–153. doi: 10.1016/0167-5699(92)90116-O. [DOI] [PubMed] [Google Scholar]
- 59.Vandenabeele P, Declercq W, Beyaert R, Fiers W. Two tumour necrosis factor receptors: structure and function. Trends Cell Biol. 1995;5:392–399. doi: 10.1016/s0962-8924(00)89088-1. [DOI] [PubMed] [Google Scholar]
- 60.Wang D, Liebowitz D, Kieff E. An EBV membrane protein expressed in immortalized lymphocytes transforms established rodent cells. Cell. 1985;43:831–840. doi: 10.1016/0092-8674(85)90256-9. [DOI] [PubMed] [Google Scholar]
- 61.Wang D, Liebowitz D, Kieff E. The truncated form of the Epstein-Barr virus latent-infection membrane protein expressed in virus replication does not transform rodent fibroblasts. J Virol. 1988;62:2337–2346. doi: 10.1128/jvi.62.7.2337-2346.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang D, Liebowitz D, Wang F, Gregory C, Rickinson A, Larson R, Springer T, Kieff E. Epstein-Barr virus latent infection membrane protein alters the human B-lymphocyte phenotype: deletion of the amino terminus abolishes activity. J Virol. 1988;62:4173–4184. doi: 10.1128/jvi.62.11.4173-4184.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang F, Gregory C, Sample C, Rowe M, Liebowitz D, Murray R, Rickinson A, Kieff E. Epstein-Barr virus latent membrane protein (LMP1) and nuclear proteins 2 and 3C are effectors of phenotypic changes in B lymphocytes: EBNA-2 and LMP1 cooperatively induce CD23. J Virol. 1990;64:2309–2318. doi: 10.1128/jvi.64.5.2309-2318.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang S, Rowe M, Lundgren E. Expression of the Epstein-Barr virus transforming protein LMP1 causes a rapid and transient stimulation of the bcl-2 homologue Mcl-1 levels in B-cell lines. Cancer Res. 1996;56:4610–4613. [PubMed] [Google Scholar]
- 65.Xu Y, Cheng G, Baltimore D. Targeted disruption of TRAF3 leads to postnatal lethality and defective T-lymphocyte immune responses. Immunity. 1996;5:407–415. doi: 10.1016/s1074-7613(00)80497-5. [DOI] [PubMed] [Google Scholar]
- 66.Young L, Alfieri C, Hennessy K, Evans H, OHara C, Anderson K C, Ritz J, Shapiro R S, Rickinson A, Kieff E, Cohen J I. Expression of Epstein-Barr virus transformation-associated genes in tissues of patients with EBV lymphoproliferative disease. N Engl J Med. 1989;321:1080–1085. doi: 10.1056/NEJM198910193211604. [DOI] [PubMed] [Google Scholar]
- 67.Young L, Dawson C, Clark D, Rupani H, Busson P, Tursz T, Johnson A, Rickinson A. Epstein-Barr virus gene expression in nasopharyngeal carcinoma. J Gen Virol. 1988;69:1051–1065. doi: 10.1099/0022-1317-69-5-1051. [DOI] [PubMed] [Google Scholar]