Abstract
Identifying genes with prognostic significance that can act as biomarkers in solid tumors can help stratify patients and uncover novel therapy targets. Here, our goal was to expand our previous ranking analysis of survival-associated genes in various solid tumors to include colon cancer specimens with available transcriptomic and clinical data. A Gene Expression Omnibus search was performed to identify available datasets with clinical data and raw gene expression measurements. A combined database was set up and integrated into our Kaplan-Meier plotter, making it possible to identify genes with expression changes linked to altered survival. As a demonstration of the utility of the platform, the most powerful genes linked to overall survival in colon cancer were identified using uni- and multivariate Cox regression analysis. The combined colon cancer database includes 2,137 tumor samples from 17 independent cohorts. The most significant genes associated with relapse-free survival with a false discovery rate below 1% in colon cancer carcinoma were RBPMS (hazard rate [HR] = 2.52), TIMP1 (HR = 2.44), and COL4A2 (HR = 2.36). The three strongest genes associated with shorter survival in stage II colon cancer include CSF1R (HR = 2.86), FLNA (HR = 2.88), and TPBG (HR = 2.65). In summary, a new integrated database for colon cancer is presented. A colon cancer analysis subsystem was integrated into our Kaplan-Meier plotter that can be used to mine the entire database (https://www.kmplot.com). The portal has the potential to be employed for the identification and prioritization of promising biomarkers and therapeutic target candidates in multiple solid tumors including, among others, breast, lung, ovarian, gastric, pancreatic, and colon cancers.
Graphical abstract
Public summary
-
•
We established a large integrated transcriptomic database of colorectal cancer suitable for biomarker discovery.
-
•
The cohort was used to uncover genes with the highest correlation to relapse-free survival in colon cancer.
-
•
The top genes were filtered to include hits with higher expression and potential druggability.
-
•
This pipeline can be used to prioritize clinically useful biomarkers of solid tumors by excluding likely failures.
Introduction
Transcriptomics involves the analysis of the transcriptome, which encompasses all RNA transcripts synthesized within a specific cell. Analyzing transcriptome-level data can offer more accurate prognostic insights than examining the mutation status, as we have previously demonstrated for lung cancer.1 Another example is the expression of TSPAN6 as a new predictive marker for Epidermal Growth Factor Receptor (EGFR)-targeted therapies beyond RAS pathway mutations in colorectal cancer (CRC).2 However, precision medicine still encounters a substantial challenge in translating the findings of transcriptomic investigations, as it necessitates the consolidation of numerous molecular changes within a tumor to discern which ones are causative and clinically significant.3
The integration of multiple available cohorts into a unified database has the potential to facilitate the discovery and validation of the most robust prognostic biomarkers. For this purpose, we have previously established the Kaplan-Meier (KM) plotter, a platform for performing survival analysis in real time using transcriptomic data of large patient cohorts.4 In this platform, the patient samples are divided into two groups conferring to various quantile expressions of the investigated gene, and the two cohorts are compared by a Cox regression and a KM survival plot.5 However, until now, it was not possible to perform a survival analysis of CRC using an integrated database of publicly available cohorts.
CRC is the second most common sex-independent cancer. In the United States, there have been a total of 153,000 new cases, with 107,000 occurring in the colon and 46,000 in the rectum.6 Mortality from CRC is on a slightly decreasing trend with about a 2% reduction per year.7 Survival rates vary by stage, with over 89%–90% survival of patients with a localized stage and a mere 14%–15% survival of those in whom the tumor has spread to distant metastases.8
Survival can be improved by the utilization of biomarkers that can stratify patients into effective treatment regimens.9 For example, the response rate to the anti-EGFR cetuximab10 and the vascular endothelial growth factor receptor inhibitor bevacizumab11 is higher in tumors with Kirsten rat sarcoma viral oncogene (KRAS) wild-type status compared to those with KRAS mutations. 5-fluorouracil (5-FU) has been linked to increased toxicity and diminished clinical response in patients with microsatellite instability (MSI) status, as well as in those who have dihydropyrimidine dehydrogenase deficiency.12 Immunotherapy using the PD-1 inhibitor dostarlimab was approved for patients with locally advanced rectal cancer whose tumors show mismatch repair deficiency (dMMR) as determined by measuring the loss of expression of four genes, MLH1, MSH1, MSH6, and PMS2.13 Notably, over 200 genes had higher predictive power than MLH1 when analyzing PD-1 resistance-associated genes in our recent study.14 The urgency to identify additional biomarkers is underscored by the fact that, as per national guidelines, only KRAS, NRAS, BRAF, MSI status, and dMMR are currently recommended for assessing treatment response and forecasting outcomes in CRC.
The goal of this study was to rank gene expression-based biomarkers associated with prognosis in diverse cohorts of patients with colon cancer. Our research had two specific objectives: first, we aimed to establish a comprehensive transcriptomic database of colon cancer cases, incorporating pathological and follow-up data, to serve as the basis for biomarker discovery. Secondly, we employed our previously established online platform for mining the CRC database and conducted an in-depth analysis to identify genes strongly correlated with survival in three cohorts: all patients with CRC, stage II patients, and advanced-stage patients. The results of our project have the potential to assist researchers in prioritizing genes in future investigations aimed at pinpointing clinically relevant biomarkers and therapeutic targets in various cohorts of CRC.
Results
Combined database
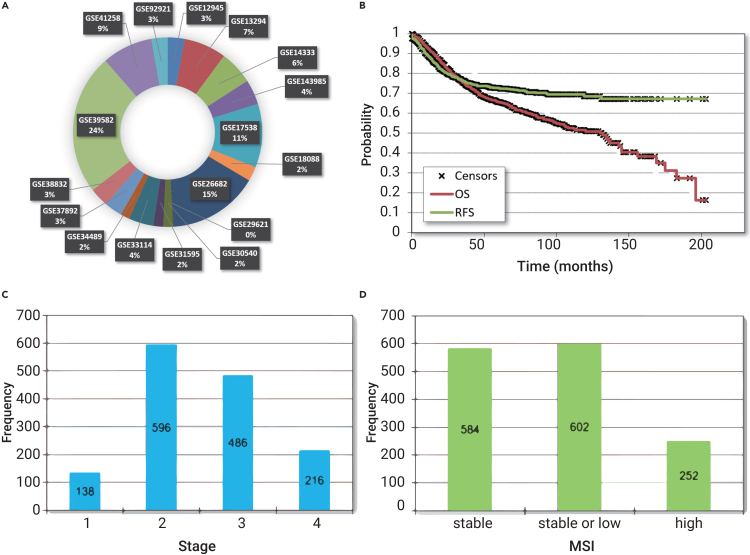
Altogether, 2,885 colon cancer samples were identified. Duplicate samples were identified by searching for identical expression values. After the removal of duplicate samples, 2,137 samples from 17 datasets were integrated into the final database. All in all, 53% of the samples were male, and the average age at diagnosis was 67.4 ± 12.9 years. Of all patients, 1,338 had relapse-free (RFS) survival data, and 1,061 samples had overall survival (OS) time. The mean follow-up for RFS was 49.8 ± 37.1 months and for time to death was 54.6 ± 39.3 months. Of all patients, 1,436 had stage data, and these can be split into stage I (n = 138, 9.6%), stage II (n = 596, 41.5%), stage III (n = 486, 33.8%), and stage IV (n = 216, 15%). Microsatellite status was available for 1,438 tumors, of which 584 (40.6%) were classified as stable, 602 (41.9%) were stable or low, and 252 (17.5%) were high. The tumor was localized proximal in 439 cases (40.8%) and distal in 638 cases (59.2%). Utilizing the JetSet best probes, we obtained gene expression data for a total of 10,090 distinct genes. Among these, 2,750 genes exhibit potential druggability, as indicated by the DGIdb database. You can find an overview of the clinical attributes of the entire study cohort in Figure 1, and for a comprehensive breakdown of each dataset included, please refer to Table 1.
Figure 1.
Clinicopathological characteristics of all tumor samples contained within the integrated database
The list of all datasets with proportional sample numbers (A), relapse-free and overall survival across all available patients (B), distribution of stage (C), and microsatellite instability status (D).
Table 1.
Aggregate characteristics of the colon cancer cohorts included in the analysis
| Dataset | Sample number, n (%) | Age, years, mean ± SD | Relapse-free survival |
Overall survival |
Male, n (%) | Stage, n (%) |
MSI, n (%) |
Location, n (%) |
Mutations, n (%) |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Months, mean ± SD | Events, n (%) | Months, mean ± SD | Events, n (%) | I | II | III | IV | Stable | Stable/low | High | Proximal | Distal | BRAF | KRAS | TP53 | ||||
| GSE12945 | 62 (2.9) | 64.5 ± 11.8 | 45.2 ± 14.1 | 12 (19.4) | 42.2 ± 16.0 | 12 (19.4) | 28 (45.2) | 13 (21.0) | 23 (37.1) | 21 (33.9) | 5 (8.1) | N/A | N/A | N/A | 0 (0) | 33 (100) | N/A | N/A | N/A |
| GSE13294 | 155 (7.3) | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | 77 (49.7) | 0 (0) | 78 (50.3) | N/A | N/A | N/A | N/A | N/A |
| GSE14333 | 123 (5.8) | 67.0 ± 11.5 | 39.7 ± 23.8 | 22 (22.2) | N/A | N/A | 72 (58.5) | 22 (17.9) | 43 (35.0) | 37 (30.1) | 21 (17.1) | N/A | N/A | N/A | 47 (39.2) | 73 (60.8) | N/A | N/A | N/A |
| GSE143985 | 91 (4.3) | N/A | 70.1 ± 37.6 | 15 (16.5) | N/A | N/A | N/A | 0 (0) | 55 (60.4) | 36 (39.6) | 0 (0) | 85 (94.4) | 0 (0) | 5 (5.6) | N/A | N/A | 2 (2.2) | 35 (38.5) | 53 (58.2) |
| GSE17538 | 232 (10.9) | 64.7 ± 13.4 | 47.2 ± 29.5 | 55 (31.1) | 50.6 ± 33.7 | 92 (39.7) | 122 (52.6) | 28 (12.1) | 72 (31.0) | 76 (32.8) | 56 (24.1) | N/A | N/A | N/A | 78 (47.0) | 88 (53.0) | N/A | N/A | N/A |
| GSE18088 | 53 (2.5) | 65.4 ± 12.2 | N/A | 13 (24.5) | N/A | N/A | 26 (49.1) | N/A | N/A | N/A | N/A | 34 (64.2) | 0 (0) | 19 (35.8) | 28 (52.8) | 25 (47.2) | N/A | N/A | N/A |
| GSE26682 | 331 (15.5) | 72.2 ± 10.8 | N/A | N/A | N/A | N/A | 179 (54.1) | N/A | N/A | N/A | N/A | 218 (65.9) | 78 (23.6) | 35 (10.6) | N/A | N/A | N/A | N/A | N/A |
| GSE29621 | 2 (0.1) | N/A | 70.5 ± 12.7 | 1 (50.0) | 73.8 ± 17.2 | 1 (50.0) | 1 (50.0) | 0 (0) | 1 (50.0) | 1 (50.0) | 0 (0) | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
| GSE30540 | 35 (1.6) | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
| GSE31595 | 37 (1.7) | 74.1 ± 10.1 | 47.7 ± 30.2 | 8 (21.6) | N/A | 11 (29.7) | 15 (40.5) | 0 (0) | 20 (54.1) | 17 (45.9) | 0 (0) | N/A | N/A | N/A | 23 (62.2) | 14 (37.8) | N/A | N/A | N/A |
| GSE33114 | 90 (4.2) | 70.6 ± 12.8 | 40.4 ± 26.5 | 20 (22.2) | N/A | N/A | 42 (46.7) | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
| GSE34489 | 33 (1.5) | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
| GSE37892 | 65 (3.0) | 68.6 ± 11.8 | 41.7 ± 21.0 | 13 (20.0) | N/A | N/A | 34 (52.3) | 0 (0) | 55 (84.6) | 10 (15.4) | 0 (0) | 0 (0) | 52 (100) | 0 (0) | 29 (45.3) | 35 (54.7) | 0 (0) | 22 (33.8) | 8 (12.3) |
| GSE38832 | 70 (3.3) | N/A | 27.8 ± 19.8 | 6 (11.1) | 26.2 ± 19.2 | 11 (15.7) | N/A | 14 (20.0) | 21 (30.0) | 19 (27.1) | 16 (22.9) | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
| GSE39582 | 514 (24.1) | 66.6 ± 13.3 | 48.2 ± 41.5 | 170 (33.6) | 56.8 ± 39.6 | 177 (34.7) | 281 (54.7) | 33 (6.5) | 213 (41.8) | 204 (40.0) | 60 (11.8) | 0 (0) | 439 (85.4) | 75 (14.6) | 202 (39.3) | 312 (60.7) | 51 (9.9) | 195 (37.9) | 182 (35.4) |
| GSE41258 | 185 (8.7) | 63.5 ± 13.9 | 66.4 ± 47.2 | 36 (30.5) | 68.4 ± 48.3 | 93 (50.3) | 98 (53.0) | 28 (15.1) | 50 (27.0) | 49 (26.5) | 58 (31.4) | 117 (63.2) | 33 (17.8) | 35 (18.9) | 32 (35.6) | 58 (64.4) | N/A | N/A | N/A |
| GSE92921 | 59 (2.8) | N/A | 72.1 ± 36.1 | 6 (10.2) | N/A | N/A | N/A | 0 (0) | 43 (72.9) | 16 (27.1) | 0 (0) | 53 (91.4) | 0 (0) | 5 (8.6) | N/A | N/A | 2 (3.4) | 23 (39.0) | 35 (59.3) |
| All samples | 2,137 (100) | 67.5 ± 12.9 | 49.8 ± 37.1 | 377 (26.7) | 54.6 ± 39.3 | 397 (36.2) | 898 (53.0) | 138 (10) | 596 (42) | 486 (34) | 216 (15) | 584 (41) | 602 (42) | 252 (18) | 439 (41) | 638 (59) | 55 (8) | 275 (40) | 278 (55) |
SD, standard deviation; N/A, not available.
Survival analysis across all genes in all colon cancer samples
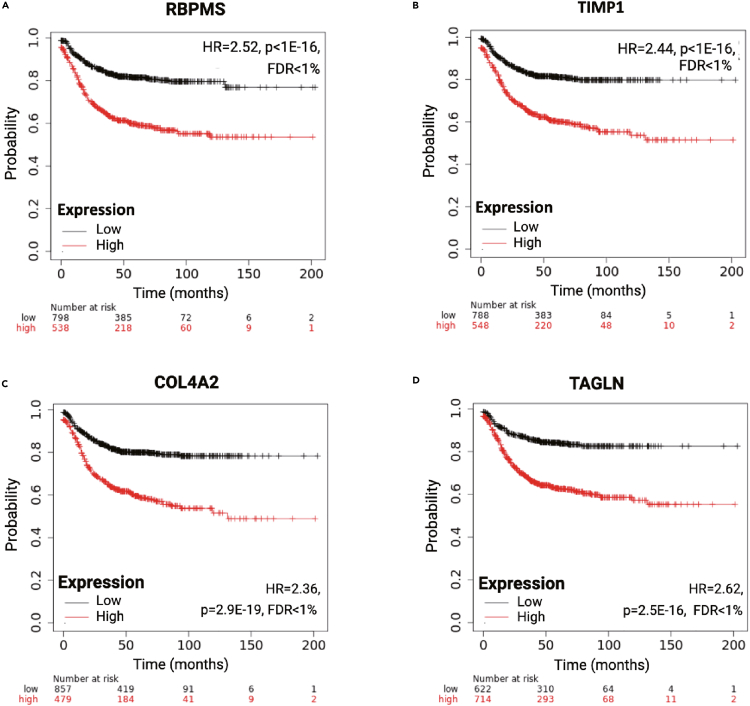
The first analyzed cohort included all CRC samples with available RFS follow-up. The four most significant genes, where higher expression was linked to shorter RFS in CRC, including RNA-binding protein mRNA processing factor (RBPMS), TIMP metallopeptidase inhibitor 1 (TIMP1), COL4A2, and transgelin (TAGLN), are displayed in Figure 2, and the ten most significant genes are listed in Table 2. The aforementioned key genes maintained their significance in a multivariate analysis that took into account gender, stage, MSI, and location (the remaining clinical variables lacked sufficient data). The Cox regression results for all investigated genes in all available colon cancer specimens with RFS data are provided in Table S1, and the results for samples with OS data are in Table S2.
Figure 2.
Most robust genes associated with relapse-free survival in colon carcinoma
The four strongest genes with higher expression correlated to shorter survival are depicted, including RBPMS (A), TIMP1 (B), COL4A2 (C), and TAGLN (D). HR = Hazard Rate, FDR = False Discovery Rate.
Table 2.
The ten strongest genes with higher gene expression correlated to shorter relapse-free survival length in three colon cancer cohorts including all patients, stage II patients, and stage III and IV tumors
| Gene symbol | p value | FDR | HR | Cutoff value |
|---|---|---|---|---|
| All patients | ||||
| RBPMS | 1.1E−17 | 5.7E−16 | 2.49 | 812 |
| TIMP1 | 6.5E−17 | 3.3E−15 | 2.44 | 9,073.5 |
| COL4A2 | 4.0E−16 | 2.1E−14 | 2.35 | 4,413 |
| TAGLN | 4.4E−16 | 2.2E−14 | 2.58 | 2,601.4 |
| LOXL2 | 2.0E−15 | 6.9E−14 | 2.33 | 1,333 |
| S100A11 | 1.0E−14 | 5.1E−13 | 2.31 | 8,250.3 |
| NOTCH3 | 1.0E−14 | 5.9E−13 | 2.26 | 544 |
| FAM127A | 2.1E−14 | 1.1E−12 | 2.24 | 1,523.6 |
| LMO2 | 2.1E−13 | 7.9E−12 | 2.17 | 471 |
| SERPINE1 | 2.8E−13 | 1.8E−11 | 2.18 | 537 |
| Stage II | ||||
| CSF1R | 1.1E−06 | 1.3E−05 | 2.86 | 323.5 |
| FLNA | 1.1E−06 | 2.9E−05 | 2.88 | 764.4 |
| TPBG | 1.5E−06 | 2.6E−05 | 2.65 | 1,199.4 |
| B2M | 2.0E−06 | 1.0E−04 | 2.62 | 30,461.9 |
| LOXL2 | 2.1E−06 | 4.9E−05 | 2.63 | 1,333.0 |
| TAGLN | 4.4E−06 | 7.5E−05 | 2.75 | 2,568.7 |
| LMO2 | 7.9E−06 | 8.1E−05 | 2.48 | 403.4 |
| YWHAE | 1.1E−05 | 2.8E−04 | 2.62 | 3,600.2 |
| PLOD1 | 1.7E−05 | 1.5E−04 | 2.43 | 689.0 |
| ARRB2 | 2.4E−05 | 3.7E−04 | 2.63 | 207.3 |
| Stage III/IV | ||||
| BHLHE40 | 3.1E−08 | 6.7E−07 | 2.06 | 3,756.5 |
| COL4A2 | 2.1E−07 | 2.7E−06 | 2.02 | 4,054.0 |
| TSC22D3 | 3.7E−07 | 9.3E−06 | 1.95 | 1,522.2 |
| NPR3 | 1.9E−06 | 6.8E−05 | 1.91 | 230.8 |
| A2M | 2.6E−06 | 2.7E−05 | 1.86 | 4,767.2 |
| NR1D1 | 2.7E−06 | 3.7E−05 | 2.32 | 347.2 |
| NOTCH3 | 2.9E−06 | 1.8E−05 | 2.27 | 236.1 |
| LOXL2 | 3.7E−06 | 6.2E−05 | 1.88 | 1,166.3 |
| TAGLN | 5.7E−06 | 5.6E−05 | 2 | 2,413.7 |
| KLK6 | 6.0E−06 | 3.1E−04 | 1.85 | 203.3 |
The stage II and stage III/IV gene lists were restricted to druggable genes with at least one possible inhibitor. HR, hazard rate; FDR, false discovery rate.
We used the muTarget platform to identify genes correlated to the mutation status of the most commonly altered genes including APC (Adenomatous polyposis coli), KRAS, TP53, and BRAF. APC gene mutations were found in 73.5% of colon cancer cases in TCGA cohort and are thought to be an early event in the development of the disease. KRAS mutations were present in 40.6% of cases, TP53 mutations were found in 53.5% of cases, BRAF mutations were detected in 15.9% of cases, and all three were associated with a more aggressive form of the disease. We have identified altogether 124, 169, 447, and 1,576 differentially expressed genes for APC, KRAS, TP53, and BRAF mutations, respectively. When restricting to genes up-regulated in tumors with a mutation and simultaneously linked to shorter RFS at a false discovery rate (FDR) <5%, one gene reached significance for APC mutations, four genes for KRAS mutations, three genes for TP53 mutations, and 106 genes for BRAF mutations. Differential expression among APC, KRAS, and TP53 mutated and wild-type tumors as well as correlation to RFS are presented in Table 3, and the complete list of BRAF-mutation-linked and survival-associated genes is provided in Table S3.
Table 3.
Genes whose expression is higher in APC-/KRAS-/TP53-/BRAF-mutated CRC tumors and are correlated to relapse-free survival. These genes might represent optimal targets for future pharmacological developments
| Mutation | Gene symbol | Mean mutant (n = 291) | Mean wild (n = 105) | FC (mutant/wild) | MW p value | Direction | RFS: Cox p value | FDR | RFS: HR | Cutoff |
|---|---|---|---|---|---|---|---|---|---|---|
| APC | MPP1 | 1,006.61 | 640.55 | 1.57 | 1.44E−06 | up | 8.13E−03 | 2.74E−02 | 1.37 | 452 |
| KRAS | S100A2 | 469.6 | 218.91 | 2.15 | 1.30E−05 | up | 6.15E−03 | 1.71E−02 | 1.36 | 292 |
| KRAS | TGFBI | 35,480.06 | 21,977.36 | 1.61 | 1.52E−08 | up | 2.46E−03 | 1.10E−02 | 1.41 | 9,067.7 |
| KRAS | KCNN4 | 3,761.12 | 2,382.31 | 1.58 | 2.82E−10 | up | 5.87E−03 | 1.85E−02 | 1.44 | 527 |
| KRAS | TBXAS1 | 1,163.81 | 773.79 | 1.5 | 1.08E−07 | up | 1.10E−03 | 5.73E−03 | 1.43 | 284 |
| TP53 | GDPD5 | 1,316.4 | 764.68 | 1.72 | 7.44E−09 | up | 3.00E−03 | 1.07E−02 | 1.51 | 256 |
| TP53 | MPP1 | 1,092.34 | 698.95 | 1.56 | 3.14E−08 | up | 8.13E−03 | 2.74E−02 | 1.37 | 452 |
| TP53 | ZSCAN18 | 258.77 | 177.81 | 1.46 | 2.97E−04 | up | 2.71E−09 | 1.30E−08 | 1.97 | 300 |
| BRAF | RAMP1 | 1,330.98 | 394.09 | 3.38 | 1.87E−11 | up | 1.24E−06 | 2.36E−05 | 1.71 | 276 |
| BRAF | SERPINB5 | 4,249.44 | 1,652.96 | 2.57 | 8.96E−08 | up | 2.09E−03 | 4.48E−03 | 1.52 | 382 |
| BRAF | SLC43A3 | 934.53 | 383.75 | 2.44 | 4.38E−08 | up | 4.60E−03 | 1.01E−02 | 1.43 | 295 |
| BRAF | TRIB2 | 1,371.95 | 569.11 | 2.41 | 4.32E−15 | up | 7.73E−03 | 5.76E−02 | 1.34 | 564 |
For BRAF, only the top ten genes (based of FC mutant/wild) are shown. FC, fold change; MW, Mann-Whitney; RFS, relapse-free survival; FDR, false discovery rate; HR, hazard rate; NS, not significant.
Druggable genes in stage II
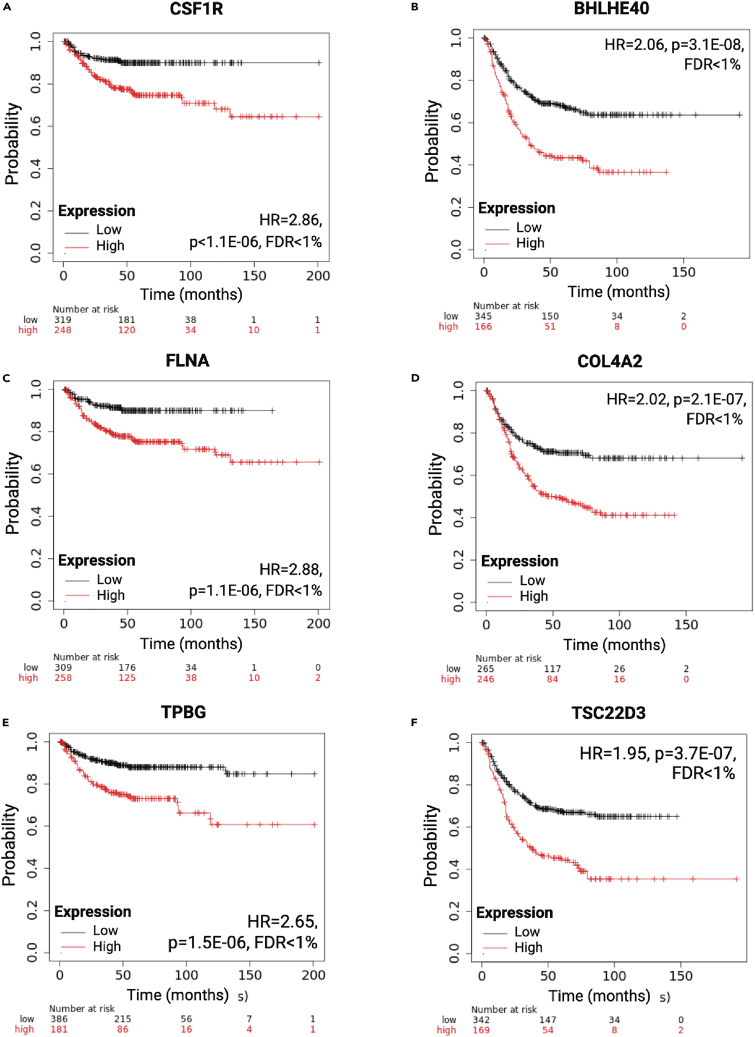
When restricting the analysis to stage II tumors, the correlation between survival and gene expression with an FDR below 1% and a hazard rate (HR) over one was significant for 72 druggable genes. This analysis was restricted to druggable genes to focus our search on potential therapy targets. When ranked by the achieved p value, the five most significant genes include colony-stimulating factor 1 receptor (CSF1R; HR = 2.86), filamin A (FLNA; HR = 2.88), trophoblast glycoprotein (TPBG; HR = 2.65), beta-2-microglobulin (B2M; HR = 2.62), and lysyl oxidase-like 2 (LOXL2; HR = 2.63). The KM plots for the top three genes are displayed in Figure 3, and the top ten genes are listed in Table 2. The complete list of all stage II-linked druggable genes is available in Table S4.
Figure 3.
Most significant druggable genes associated with shorter relapse-free survival
The top three genes in stage II patients include CSF1R (A), FLNA (C), and TPBG (E), and the three strongest genes in patients with stage III/IV colorectal cancer include BHLHE40 (B), COL4A2 (D), and TSC22D3 (F). HR = Hazard Rate, FDR = False Discovery Rate.
Druggable genes in stage III and IV tumors
In the third setting, we included a combined cohort of all stage III and all stage IV patients. The selection of this cohort is based on the fact that these patients routinely receive chemotherapy. The analysis results were filtered for druggable genes. When ordered by the Cox regression p value, the five strongest genes were basic-helix-loop-helix family member E40 (BHLHE40; HR = 2.6), COL4A2 (HR = 2.02), TSC22 domain family member 3 (TSC22D3; HR = 1.95), natriuretic peptide receptor 3 (NPR3; HR = 1.91), and A2M (HR = 1.86). Survival plots for the three top genes are available in Figure 3, and the strongest genes are listed in Table 2. The complete list of all genes significant in stage III and IV patients is accessible in Table S5.
GO analysis results
To reveal the underlying biological mechanisms, we thoroughly examined the comprehensive list of significant genes for overrepresented Gene Ontology (GO) categories using the TNM plotter’s functional analysis tools. The list included genes associated with RFS and OS in all patients with CRC with an FDR below 1%, a cutoff exceeding 200, and an HR greater than one. The most significant GO categories linked to RFS include extracellular matrix organization (GO: 0030198, p = 9.7E−38), cell-substrate adhesion (GO: 0031589, p = 2.4E−18), and cell-matrix adhesion (GO: 0007160, p = 1.2E−13). Interestingly, the same categories reached the highest significance for the OS-linked gene list as well—for a detailed account of all biological processes in each of the two settings, please refer to Table S6.
Discussion
Biomarkers linked to survival, with heightened expression correlating to unfavorable outcomes, present promising targets for therapeutic interventions in the field of oncology. An illustrative instance is HER2, initially identified as an indicator of poor prognosis in breast cancer. The availability of trastuzumab for patients with HER2-positive breast tumors enhanced OS and disease-free survival by almost 40%.20 Presently, the prognosis for early-stage HER2-positive breast cancer is notably positive, with the majority of patients potentially remaining free from the disease. In our present investigation, we leveraged a substantial cohort of patients with colon cancer with comprehensive clinical follow-up and transcriptome-level gene expression data. Using this dataset, we aimed to pinpoint new biomarker candidates and the most noteworthy druggable targets in three cohorts of patients with colon cancer.
The analysis performed using the integrated database was preceded by multiple levels of quality control. These include the removal of redundant samples, the utilization of the best available probe set for each gene, and the automated analysis of each available cutoff value with simultaneous calculation of the FDR. In addition, a filter was added to include only genes where the cutoff value was at least twice the background intensity. While the online available KM plotter allows the reproduction of the results, we suggest using similar quality control steps when validating the role of a selected gene. These quality control steps enable avoiding false discoveries and the identification of clinically useful biomarker candidates and therapy targets.
Among the genes most strongly linked to shorter RFS in all available patients with colon cancer was RBPMS, a gene that regulates the cytokinesis of embryonic cardiomyocytes, which has not been linked to CRC previously. Another top gene was TIMP1, which blocks peptidases involved in the degradation of the extracellular matrix. TIMP1 promotes cellular proliferation and invasion,21 and higher expression of the gene was already described previously as a prognostic marker in 190 patients with CRC.22 Similarly, the high expression of TAGLN was also identified previously in colon cancer in connection with worse survival.23 These results show that our analysis was robust and that we were able to validate a set of top genes associated with survival in colon cancer.
Adjuvant chemotherapy is designed to eliminate micrometastatic disease detected during surgery, avert the emergence of distant metastatic conditions, and ultimately achieve a cure for these patients. Chemotherapy is recommended for stage III and IV patients and not suggested for stage I tumors. However, in stage II colon cancer, adjuvant treatment remains a subject of debate. Both national and international guidelines for adjuvant treatment in stage II colon cancer propose a spectrum of therapeutic choices, spanning from close observation to the administration of chemotherapy through single-agent or combination regimens.24 In this field, novel prognostic biomarkers could help to stratify patients into clinically useful cohorts. Interestingly, two of the five most significant genes uncovered in our analysis have links to the immune system, including CSF1R and B2M. Inhibition of CSF1R has been recently identified as a factor enhancing immunotherapy,25 and an orally active CSF1R inhibitor has already been tested in a murine model of colon cancer.26 Three of the top genes were linked to extra- and intracellular structures, including FLNA, an actin-binding protein crosslinking actin filaments; TPBG, a gene involved in cell adhesion; and LOXL2, which catalyzes the formation of crosslinks in collagens and elastin. Collectively, these results suggest that immunotherapy should be further evaluated in stage II tumors and that the genes linked to extracellular structures might serve as potential biomarkers of worse outcomes.
Patients with stage III and IV tumors have a tumor that already spread to nearby lymph nodes (stage III) or distant organs (stage IV). These tumors need chemotherapy to improve survival, with more regimes available for stage IV tumors. The genes with the strongest prognostic power include BHLHE40, a transcription factor involved in cellular differentiation that also promotes colon cancer proliferation27; TSC22D3, a gene linked to the anti-inflammatory and immunosuppressive effects of interleukin-10 with no implications in colon cancer; and NPR3. Higher expression of NPR3 has been recently uncovered as a marker of worse outcomes in CRC.28 The list of genes with higher expression linked to worse prognosis in stage III and IV tumors could represent potential new therapy targets.
Two notable constraints within our study warrant mention. Firstly, our data exclusively encompass an examination of patient cohorts from prior publications, which precludes the possibility of an independent prospective validation of the outcomes. The second constraint is rooted in our reliance on gene expression data. This stems primarily from the unavailability of protein expression data for a sufficiently expansive cohort of proteins across a comprehensive set of patients, complete with clinical profiles and follow-up information. To address these limitations, a prospective large-scale investigation that concurrently assesses a multitude of genes and proteins may be essential to secure the requisite validation for the identified target genes. Note that the performed multiple testing correction is limited to the genes analyzed in the current manuscript. We suggest using our multiple testing correction tool available at www.multipletesting.com when analyzing other sets of biomarker candidates.29
In brief, here, we established a comprehensive transcriptomic database for colon cancer cases, which includes pathological details and follow-up information. Utilizing this dataset, we pinpointed the key genes that exhibited a strong correlation with RFS across all stages of colon cancer, as well as specifically in stage II and stage III/IV tumors. Our analysis focused on druggable genes that showed elevated expression in patients with a poorer prognosis, aiming to identify the most reliable genes with potential therapeutic value. The presented analysis can also be used as a step-by-step guide for utilizing the online available KM plotter for the identification and validation of new gene expression-based prognostic biomarkers in various other types of solid tumors including, among others, breast, gastric, kidney, liver, lung, ovarian, pancreatic, and thyroid cancers.
Materials and methods
Identification of colon cancer cohorts
Our search for colon cancer cohorts was conducted through the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) and the Genomic Data Commons Data Portal (https://portal.gdc.cancer.gov/). We specifically included samples that had available transcriptome-level data and restricted our initial selection to those comprising a minimum of 30 patients. The final combined database has fewer patients in some cohorts because duplicate gene arrays have been removed. To ensure the consistency of our data and eliminate potential discrepancies arising from variations in sensitivity, specificity, and dynamic range across different gene expression detection technologies, our search focused on tumor samples analyzed using the following in situ oligonucleotide array platforms: GPL96 (Affymetrix Human Genome U133A Array), GPL571 (GeneChip Human Genome U133A 2.0 Array), and GPL570 (Affymetrix Human Genome U133 Plus 2.0 Array). These platforms offer the advantage of employing identical probe sequences to assess the expression levels of individual genes.
Normalization and gene annotation
Gene expression data obtained from the gene arrays underwent a two-step normalization process. Initially, MAS5 normalization was applied, followed by a subsequent scaling normalization to standardize the mean expression to 1,000 within each array. Only probes present in the GPL96 platform were utilized to ensure consistency and mitigate potential platform-specific variations, particularly considering the substantial number of additional probes in the GPL570 arrays. To determine the most reliable probe set for each gene, the JetSet algorithm was employed.15 Quality control measures were implemented to assess background intensity, noise levels, the percentage of present calls, the presence of bioBCD spikes, and the 3′/5′ ratios of GAPDH and ACTB.
Assembly of clinical and pathological data
Clinical information was sourced from the supplementary materials provided in the original publications or extracted from the series matrix files accessible through GEO. We gathered pertinent data, including RFS time and event details, OS time and event, MSI status, tumor localization, stage, grade, TNM (Tumour, Node, and Metastasis) status, gender, and KRAS, BRAF, and TP53 mutation status, as well as treatment information for each sample whenever it was available.
Survival analysis
We used Cox proportional hazards regression analysis to calculate differential survival rates. Initially, a univariate analysis was conducted for each gene independently. In order to ensure that we did not miss any correlations due to the use of a specific cutoff value, we considered all available cutoff values between the lower and upper quartiles of expression for each gene. Additionally, we computed the FDR using the Benjamini-Hochberg method to correct for multiple hypothesis testing.16 Then, we selected the cutoff value with the highest level of significance, also marked by the lowest level of FDR. In cases where multiple cutoff values had identical significance, we chose the cutoff with the highest HR for the final analysis. To reduce noise when identifying the top genes with the most robust correlation with survival, we only considered genes with a cutoff value exceeding 200, which is twice the background intensity of approximately 100. Furthermore, we filtered for genes associated with a worse prognosis at higher expression levels (HR > 1), as these genes hold potential as future therapeutic targets. For the selected top genes, we conducted multivariate Cox regression analysis to validate the correlation between clinical and pathological variables as well as gene expression on survival. To visualize differences in survival, KM plots were generated using the cutoff values identified in the univariate analysis.
Integration of the colon cancer cohort into the KM plotter
We integrated the colon cancer cohort into our previously established KM plotter, which can be accessed at https://www.kmplot.com.17 The plotter enables the analysis of the correlation between survival outcome and any gene or combination of genes in various tumor types including, among others, breast, esophageal, gastric, kidney, liver, lung, ovarian, pancreatic, thyroid, and uterine cancers. Filtering can be done for all available pathological (stage, TNM, mutation status, MSI, grade, localization) and clinical (gender, treatment) parameters. In addition, the analysis can be executed by setting the targeted outcome to RFS, OS, or post-progression survival. Post-progression survival only includes those tumors that had a relapse (an event for RFS) and the OS is known, regardless of the OS event status. The KM-plotter platform provides a valuable opportunity for the future analysis and validation of recently discovered gene expression-based biomarkers and signatures in different types of solid tumors and across various patient subgroups, including those that have not been explored to date. Note that in the KM plotter, the analysis results can be exported as a table, thereby enabling the visualization of the results in other software as well (e.g., Microsoft Excel).
Filtering for the most robust potentially druggable genes
By analyzing the similarities in both sequence and structure to established drug targets, a set of proteins that could be influenced by small drug-like molecules has been identified. This set of genes is commonly referred to as the “druggable genome.” To aid researchers in interpreting genome-wide studies within the context of druggable genomes, the DGIdb database was created.18 DGIdb serves as a comprehensive platform that consolidates information from various sources, including human genes associated with diseases, drugs, drug-gene interactions, and potential druggability. In our demonstration analysis, we used DGIdb as a filter to further narrow down our analysis results to genes that exhibit potential druggability.
GO analysis
To better understand the broader functions linked to alterations in RFS, we performed a GO analysis through the GO functional analysis tool provided by the TNM plotter.19 We conducted three distinct analyses, including of those genes that were identified as significant with an FDR below 0.01. Specifically, we focused on genes demonstrating higher expression associated with poorer survival for each of the three CRC cohorts. The goal of each analysis was to pinpoint significant biological processes with an FDR threshold of less than 1%.
Acknowledgments
This project was supported by the National Research, Development and Innovation Office (PharmaLab, RRF-2.3.1-21-2022-00015). The author thanks András Lánczky for his help in updating the online platform. The support of ELIXIR Hungary (www.bioinformatics.hu) is acknowledged.
Author contributions
B.G. conceived the study, conducted the research, analyzed the data, and wrote the manuscript.
Declaration of interests
The author declares no conflict of interest.
Declaration of generative AI and AI-assisted technologies in the writing process
A GPT-based AI grammar checker was used to improve the English of the manuscript.
Published Online: April 9, 2024
Footnotes
It can be found online at https://doi.org/10.1016/j.xinn.2024.100625.
Lead contact website
Supplemental information
References
- 1.Nagy Á., Pongor L.S., Szabó A., et al. KRAS driven expression signature has prognostic power superior to mutation status in non-small cell lung cancer. Int. J. Cancer. 2017;140(4):930–937. doi: 10.1002/ijc.30509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrijes R., Hejmadi R.K., Pugh M., et al. Tetraspanin 6 is a regulator of carcinogenesis in colorectal cancer. Proc. Natl. Acad. Sci. USA. 2021;118(39) doi: 10.1073/pnas.2011411118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Van Allen E.M., Wagle N., Levy M.A. Clinical analysis and interpretation of cancer genome data. J. Clin. Oncol. 2013;31(15):1825–1833. doi: 10.1200/JCO.2013.48.7215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Györffy B., Lanczky A., Eklund A.C., et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res. Treat. 2010;123(3):725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- 5.Lánczky A., Győrffy B. Web-based survival analysis tool tailored for medical research (KMplot): Development and implementation. J. Med. Internet Res. 2021;23(7) doi: 10.2196/27633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Siegel R.L., Miller K.D., Wagle N.S., et al. Cancer statistics, 2023. CA A. CA A Cancer J. Clin. 2023;73(1):17–48. doi: 10.3322/caac.21763. [DOI] [PubMed] [Google Scholar]
- 7.Cronin K.A., Lake A.J., Scott S., et al. Annual report to the nation on the status of cancer, part I: National cancer statistics. Cancer. 2018;124(13):2785–2800. doi: 10.1002/cncr.31551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mattiuzzi C., Sanchis-Gomar F., Lippi G. Concise update on colorectal cancer epidemiology. Ann. Transl. Med. 2019;7(21):609. doi: 10.21037/atm.2019.07.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patel J.N., Fong M.K., Jagosky M. Colorectal cancer biomarkers in the era of personalized medicine. J. Phys. Math. 2019;9(1):3. doi: 10.3390/jpm9010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lièvre A., Bachet J.-B., Le Corre D., et al. KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Res. 2006;66(8):3992–3995. doi: 10.1158/0008-5472.CAN-06-0191. [DOI] [PubMed] [Google Scholar]
- 11.Hurwitz H.I., Yi J., Ince W., et al. The clinical benefit of bevacizumab in metastatic colorectal cancer is independent of K-RAS mutation status: analysis of a phase III study of bevacizumab with chemotherapy in previously untreated metastatic colorectal cancer. Oncol. 2009;14(1):22–28. doi: 10.1634/theoncologist.2008-0213. [DOI] [PubMed] [Google Scholar]
- 12.Bertagnolli M.M., Niedzwiecki D., Compton C.C., et al. Microsatellite instability predicts improved response to adjuvant therapy with irinotecan, fluorouracil, and leucovorin in stage III colon cancer: cancer and leukemia group B protocol 89803. J. Clin. Orthod. 2009;27(11):1814–1821. doi: 10.1200/JCO.2008.18.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cercek A., Lumish M., Sinopoli J., et al. PD-1 blockade in mismatch repair–deficient, locally advanced rectal cancer. N. Engl. J. Med. 2022;386(25):2363–2376. doi: 10.1056/NEJMoa2201445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kovács S.A., Fekete J.T., Győrffy B. Predictive biomarkers of immunotherapy response with pharmacological applications in solid tumors. Acta Pharmacol. Sin. 2023;44(9):1879–1889. doi: 10.1038/s41401-023-01079-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Q., Birkbak N.J., Gyorffy B., et al. Jetset: Selecting the optimal microarray probe set to represent a gene. BMC Bioinf. 2011;12:474. doi: 10.1186/1471-2105-12-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Benjamini Y., Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. Roy. Stat. Soc. B. 1995;57(1):289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- 17.Győrffy B. Discovery and ranking of the most robust prognostic biomarkers in serous ovarian cancer. Geroscience. 2023;45(3):1889–1898. doi: 10.1007/s11357-023-00742-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Freshour S.L., Kiwala S., Cotto K.C., et al. Integration of the drug–gene interaction database (DGIdb 4.0) with open crowdsource efforts. Nucleic Acids Res. 2021;49(D1):D1144–D1151. doi: 10.1093/nar/gkaa1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bartha Á., Győrffy B. TNMplot.com: a web tool for the comparison of gene expression in normal, tumor and metastatic tissues. Indian J. Manag. Sci. 2021;22(5):2622. doi: 10.3390/ijms22052622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ross J.S., Fletcher J.A. The HER-2/neu oncogene in breast cancer: Prognostic factor, predictive factor, and target for therapy. Stem Cell. 1998;16(6):413–428. doi: 10.1002/stem.160413. [DOI] [PubMed] [Google Scholar]
- 21.Ma B., Ueda H., Okamoto K., et al. TIMP1 promotes cell proliferation and invasion capability of right-sided colon cancers via the FAK/Akt signaling pathway. Cancer Sci. 2022;113(12):4244–4257. doi: 10.1111/cas.15567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Macedo F.C., Cunha N., Pereira T.C., et al. A prospective cohort study of TIMP1 as prognostic biomarker in gastric and colon cancer. Chin. Clin. Oncol. 2022;11(6):43. doi: 10.21037/cco-22-69. [DOI] [PubMed] [Google Scholar]
- 23.Yokota M., Kojima M., Higuchi Y., et al. Gene expression profile in the activation of subperitoneal fibroblasts reflects prognosis of patients with colon cancer. Int. J. Cancer. 2016;138(6):1422–1431. doi: 10.1002/ijc.29851. [DOI] [PubMed] [Google Scholar]
- 24.Varghese A. Chemotherapy for stage II colon cancer. Clin. Colon Rectal Surg. 2015;28(4):256–261. doi: 10.1055/s-0035-1564430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi G., Yang Q., Zhang Y., et al. Modulating the tumor microenvironment via oncolytic viruses and CSF-1R inhibition synergistically enhances anti-PD-1 immunotherapy. Mol. Ther. 2019;27(1):244–260. doi: 10.1016/j.ymthe.2018.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee K.-H., Yen W.-C., Lin W.-H., et al. Discovery of BPR1R024, an orally active and selective CSF1R inhibitor that exhibits antitumor and immunomodulatory activity in a murine colon tumor model. J. Med. Chem. 2021;64(19):14477–14497. doi: 10.1021/acs.jmedchem.1c01006. [DOI] [PubMed] [Google Scholar]
- 27.Wang J., Li B., Yang S., et al. Upregulation of INHBA mediated by the transcription factor BHLHE40 promotes colon cancer cell proliferation and migration. J. Clin. Lab. Anal. 2022;36(7) doi: 10.1002/jcla.24539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martinez-Romero J., Bueno-Fortes S., Martín-Merino M., et al. Survival marker genes of colorectal cancer derived from consistent transcriptomic profiling. BMC Genom. 2018;19(Suppl 8):857. doi: 10.1186/s12864-018-5193-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Menyhart O., Weltz B., Győrffy B. MultipleTesting.com: a tool for life science researchers for multiple hypothesis testing correction. PLoS One. 2021;16(6) doi: 10.1371/journal.pone.0245824. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.