Abstract
This HPLC method is suitable for chitin quantitation (reported as glucosamine) in food raw materials like insects (mealworm larvae, crickets), shrimps, mushrooms and fungi in a research (non-routine) laboratory using a C18 column with HPLC system <600 bar with UV detection capability (at 265 nm). To remove interferences, the sample is defatted (Soxhlet) and deproteinized (by alkali) prior to acid hydrolysis in 6 M HCl. A five-point linear calibration (5–100 µg/mL) is used. The use of fluorescence detection (λex = 260 nm, λem = 350 nm) is also possible with this method [1].
-
•
18 min HPLC run time
-
•
LOD = 0.05 µg/mL and LOQ = 5 µg/mL
Keywords: Liquid chromatography, PDA, DAD, Diode-array detector, Fluorescence detector, FLD, Cricket, Shrimp, Lobster, Crab, Crustaceans, Insects, Mealworm, Mushroom, Fungi, Tenebrio molitor, Chitosan, Reverse phase
Method name: Chitin quantitation in food raw materials by HPLC—C18-DAD with off-line derivatization
Graphical abstract
Specifications table
| Subject area: | Food Science |
| More specific subject area: | Analytical chemistry, food chemistry |
| Name of your method: | Chitin quantitation in food raw materials by HPLC—C18-DAD with off-line derivatization |
| Name and reference of original method: | J.Z.Q. Zhou, T. Waszkuc, F. Mohammed, Determination of glucosamine in raw materials and dietary supplements containing glucosamine sulfate and/or glucosamine hydrochloride by high-performance liquid chromatography with FMOC-Su derivatization: Collaborative study, J AOAC Int. 88 (2005) 1048–1058 [2] |
| Resource availability: | n/a |
Chemicals
-
1.
Petroleum ether – 40–65 °C, >90 %, ACS grade, CAS 64742-49-0 (e.g. Lachner 20045-CT0-M1000-1)
-
2.
Hydrochloric acid – 35 %, ACS grade, CAS 7647-01-0 (e.g. Lachner 10033-A35-M1000-1)
-
3.
Sodium hydroxide – pellets, ACS grade, CAS 1310-73-2 (e.g. Lachner 10006-AP2-G1000-1)
-
4.
Acetonitrile – for chromatography/HPLC grade, CAS 75-05-8 (e.g. Fisher A998-212)
-
5.
Trifluoroacetic acid – for chromatography/HPLC grade, CAS 76-05-1 (e.g. Merck 80457–10 ML by Supelco)
-
6.
Triethylamine – 99.5 %, CAS 121-44-8 (e.g. Merck 471283 by Sigma-Aldrich)
-
7.
N-(9-Fluorenylmethoxycarbonyloxy)succinimide (FMOC N-hydroxysuccinimide ester or FMOC—OSu) – a derivatization agent for amino acids, >98 %, for chromatography, CAS 82911-69-1 (e.g. Merck 46920-5G-F)
-
8.
D-(+)-glucosamine hydrochloride - >99 %, analytical standard, (e.g. Merck 346299 by Millipore)
-
9.
Chitin from shrimp shells – practical grade, powder, used as in-house reference material (e.g. Merck, Sigma-Aldrich, C7170)
-
10.
Water – HPLC grade, >18.2 MΩ, prepared on site by Milli-Q water purification system, filtered through 0.2 µm membrane
-
11.
pH calibration solutions – pH 7.00, 4.00, 2.00
-
12.
Compressed nitrogen – cylinder, 20 L, 200 bar, ≥99.99 % (e.g. Linde)
-
13.
Pure cotton fiber from a drug store
Store all chemicals as required by the supplier.
Materials
-
1.
Analytical balance – readability 0.1 mg and 0.01 mg
-
2.
N2 evaporator – for 1.5 mL HPLC vials, dry (stainless steel block), 60 °C, with timer, hooked up to N2 cylinder
-
3.
Ultrasonic bath – 5 L volume, 60 kHz
-
4.
Volumetric flasks – borosilicate glass, grade A, 25, 100, 1000 mL
-
5.
Glass pipettes – 5 mL
-
6.
Soxhlet extractors – glass, with condenser (hooked up to cool tap water or cooling fluid), with round bottom flasks (500 mL)
-
7.
Soxhlet extractor heater – for up to 100 °C
-
8.
Thimbles for Soxhlet extraction – able to fit Soxhlet extractor above, cellulose, 30 × 80 mm (or bigger), e.g. Ahlstrom Munksjo
-
9.
Magnetic stirrer and PTFE stir bars
-
10.
Flasks for mobile phase – 100, 250, 500, 1000 and 2500 mL with GL45 screw (e.g. Schott Duran)
-
11.
Pipettors – variable volume up to 100, 1000, 5000 and 10000 µL
-
12.
Pipette tips – for pipettors listed above
-
13.
Vortex – up to 3000 vibrations/min, e.g. IKA
-
14.
Volumetric flasks – glass, 100 and 1000 mL
-
15.
Volumetric cylinder – borosilicate glass, 50, 250, 500 and 1000 mL
-
16.
Glass beads (3–4 mm) or boiling stones
-
17.
Syringes – 1 or 2 mL, polypropylene, non-sterile, luer lock or luer tip
-
18.
Syringe filters (HPLC) – with nylon membrane, 0.22 µm, 13 mm in diameter
-
19.
Blender
-
20.
Round or folded filter paper – cellulose, fast filtering, 15 cm in diameter, suitable to accommodate 50 mL of liquid when folded
-
21.
Filtration funnel – glass or plastic, with stem
-
22.
Beakers – glass, 500-mL
-
23.
Petri dishes – glass, about 15 cm in diameter
-
24.
Sieve – screen 1 mm and 3 mm, approx. 20 cm in diameter
-
25.
Ceramic mortar with pestle – 10 cm in diameter
-
26.
(Micro-)centrifuge tubes – conical, 1.5 or 2 mL, polypropylene, with attached cap
-
27.
15-mL glass Hungate type anaerobic culture tubes (125 × 16 mm) with butyl rubber stopper and screw cap (e.g. VWR #BELC2047-16125)
-
28.
Centrifuge tubes – conical, 50 mL, polypropylene with a screw cap
-
29.
Centrifuge – with rotor for 50-mL conical centrifuge tubes (above) and able to achieve 10,000 rpm
-
30.
Laboratory oven – with variable temperature setup up to 110 °C
-
31.
pH meter – readability 0.01 pH unit, glass gel pH electrode with temperature compensation
-
32.
pH indicator strips – 7-12 pH scale
-
33.
Mobile phase filtration apparatus – borosilicate glass, 1 L reservoir, nylon membrane 47 mm (diameter) and 0.2 µm porosity connected to a vacuum pump
-
34.
HPLC vials – screw top, 1.5 ml, clear, with graduation and caps with silicone/PTFE membrane
-
35.
HPLC vials – screw top, 1.5 ml, amber, with graduation and caps with silicone/PTFE membrane
-
36.
HPLC column and guard column – Agilent Poroshell 120 EC—C18, 150 × 3 mm, 2.7 µm with UHPLC guard Poroshell 120 EC—C18, 5 × 3 mm, 2.7 µm (Agilent # 693975-302 and #823750-911)
-
37.
HPLC system – at least binary pump capable of handling pressures up to 600 bar, flow up to 1 mL/min, sampler cooling at 5 °C, 5 µL injection and column thermostat 30 °C (e.g. Agilent 1260 Infinity II consisting of a degasser, quaternary pump, autosampler, DAD detector)
-
38.
water purification system – Milli-Q water purification system for HPLC grade water, >18.2 MΩ, filtered through 0.2 µm membrane
Solutions
Adjust the amounts of all solutions below according to the number of samples analyzed.
-
1.
Mobile phase A is HPLC grade acetonitrile
Note: One sample run requires 8 mL of this mobile phase (for further details see section Gradients)
-
2.
Mobile phase B (water with TFA, pH 2.4)
Note: One sample run requires 8 mL of this mobile phase (for further details see section Gradients)
Work in a fume hood. Into a 1 L mobile phase flask, measure 1000 mL MQ H2O and place a magnetic stir bar into the flask. Place the bottle on a magnetic stirrer and let stir. Adjust the pH with trifluoroacetic acid (TFA) to pH 2.40 ± 0.05. For final small pH adjustments, use diluted trifluoroacetic acid (e.g. water:TFA, 2:1, V/V). The solution is stable for two weeks if kept refrigerated (4 °C). Do not filter this mobile phase since the vacuum filtration would cause the TFA concentration to decrease significantly.
-
3.
Sodium hydroxide solution (1mol/L)
Note: For each sample 80 mL of this solution is needed.
Into 1 L volumetric flask weigh 40 g sodium hydroxide pellets, dissolve in about 700 mL MQ H2O, cool down under tap water while dissolving, equilibrate the solution to room temperature, then make up with MQ H2O up to the mark, stopper and mix. The solution is stable for 3 months at room temperature. There is no need to check the concentration by titration since this solution is used for sample deproteination and not for analytical purposes.
-
4.
Hydrochloric acid solution (6mol/L)
Note: For each sample 10 mL of this solution is needed.
Work in the fume hood. Into 1000 mL volumetric flask transfer 530 mL HCl (35 %) with a volumetric cylinder, add slowly about 200 mL MQ H2O, mix and cool the flask under running tap water, add another 200 mL MQ H2O, mix and keep cooling under running tap water. Let the flask equilibrate to room temperature and fill up to the mark with MQ H2O, stopper and mix well. There is no need to check the concentration by titration since this solution is used for sample hydrolysis and not for analytical purposes.
-
5.
Glusosamine standard stock solution (1mg/mL)
Note: For each sample set 100 mL of this solution is needed.
Into a 100-mL volumetric flask, weigh 120.3 mg glucosamine hydrochloride, dissolve in 50 mL MQ H2O using ultrasonic bath for 2 min, then make up to the mark with MQ H2O, stopper, mix. Prepare fresh solution each day of analysis. Calculate the exact concentration of glucosamine using the purity as given on the certificate of analysis supplied by the vendor and keep in mind that a hydrochloride salt has been used for making this solution (don't forget to correct for that).
-
6.
Glucosamine calibration solutions (5, 35, 70 and 100µg GLCN/mL)
Into four 1.5-mL clear HPLC screw top vials, pipette 5, 35, 70 and 100 µL glucosamine standard stock solution (1 mg/mL). Place the HPLC vials into the N2 evaporator under a gentle stream of nitrogen, into the block heated to 60 °C for 15 min and evaporate the solution to dryness. After the evaporation, visually check that the vial is completely dry. Then, calibration solutions undergo the steps as described in subsection D (see section Sample preparation). Prepare fresh calibration solutions each day of analysis.
-
7.
Redissolution solution (0.76% TEA, V/V)
Note: For each HPLC vial 100 µL of this solution is needed.
Work in a fume hood. Into 25-mL volumetric flask pipette 190 µL triethylamine, fill up with MQ H2O up to the mark, stopper and mix. Prepare fresh solution each day of analysis.
-
8.
Quencher (ACN:MP-A, 1:1, V/V)
Note: For each HPLC vial 100 µL of this solution is needed.
In a 100-mL glass bottle mix 50 mL acetonitrile and 50 mL mobile phase A (water with TFA, pH 2.4), mix. The solution is stable for four weeks if kept refrigerated (4 °C).
-
9.
Derivatization solution (5mg FMOC—OSu/mL)
Note: For each HPLC vial 100 µL of this solution is needed. Prepare not earlier than before working on section D.
Into a 2-mL amber HPLC screw cap vial weigh 5.00 mg FMOC—OSu and add 1.000 mL acetonitrile, cap and mix. Store in a dark place until further use for no more than 4 h. Prepare fresh solution each day of analysis.
-
10
Needle wash solution (50% ACN)
Note: For each injection about 1 mL of this solution is needed
In a 100-mL glass bottle, mix 50 mL acetonitrile with 50 mL MQ H2O, mix
Sample preparation
The sample (at least 30 g) undergoes subsequently defatting, deproteination and hydrolysis. Make sure the sample is homogeneous and the sample has been taken (sampled) properly. If needed, grind the sample to create particles 1–3 mm in size (with particles <1 mm it might be difficult to work with during sample preparation procedure). Do not forget to prepare a blank sample and an in-house reference sample (chitin from shrimp shells) for method monitoring purposes. Samples are measured in triplicates.
-
A)
Defatting
This part takes about 24 h.
-
1.
With a pencil, label an empty cellulose Soxhlet extraction thimble. Using an analytical balance, record the weight of the empty labelled thimble (mthimble) and a cotton plug (mplug) with 0.1 mg precision.
-
2.
Into the labeled empty thimble (from step 1), put 10–15 g of sample (pack loosely to allow the petroleum ether to penetrate the sample entirely), record the net sample weight (msample) with 0.1 mg precision and use the pre-weighted cotton plug (step 1) to gently seal the top of the thimble. Do not overfill the thimble (between the cotton plug and the thimble top rim should be at least 1.5 cm free space). Place the thimble with the sample into the Soxhlet extractor.
-
3.
Work in a fume hood. Into a 500-mL round bottom extraction flask which fits the Soxhlet extractor, place a few boiling stones (or glass beads) and 300 mL of petroleum ether.
-
4.
Assemble the whole Soxhlet apparatus placing the round bottom flask onto the Soxhlet heater, hook it up to the pre-cooled condenser (8–15 °C) and heat the flask containing petroleum ether to 70 °C for 4 h. The extraction takes place in the fume hood.
-
5.
After 4 h, turn off the apparatus and let it cool down. Safely dispose the petroleum ether with the fats extracted (they are not needed for the chitin analysis). Then, take out the thimble containing the defatted sample and let it dry standing upright (in a stand) overnight (for at least 16 h) in a fume hood. Keep the fume hood turned on during the drying phase.
-
6.
Next day, using an analytical balance, weigh the thimble containing the defatted sample and cotton plug (mafterextr) with 0.1 mg precision.
-
7.
Gently remove the cotton plug, make sure the cotton fibers have not contaminated the sample and place the sample into a suitable properly labeled container (polypropylene or glass). Visually, by touch check that the sample is not wet and is powdery and loose. Store at room temperature until further analysis (no longer than two weeks).
-
8.
Calculate the content of the fat-free residue in percent rounding the result to one decimal place:
For mass descriptions see the text in section A. Use masses in grams recorded with 0.1 mg precision. mafterextr – sum of the weight of the dry sample residue after the fat has been removed + thimble + cotton plug [g]; mthimble – weight of the cellulose (Soxhlet) thimble [g]; mplug – weight of the cotton plug [g]; msample – net weight of the sample before fat extraction [g].
-
B)
Deproteination
Since this method uses FMOC—OSu derivatization of glucosamine which is also commonly used for amino acid derivatization, it is important to remove all proteins from the sample to get a clean chromatogram and good separation without any interferences. This part also deacetylates the chitin and takes about 52 h to complete.
-
9.
Preheat the laboratory oven to 80 °C.
-
10.
Fold a fast filtering filter paper and label it with a pencil. Use analytical balance to weigh the labeled filter paper (mfilter) with 0.1 mg precision. Place the filter paper into a filtration funnel mounted to a stand and underneath it a 500-mL beaker.
Note: Filter paper attracts static charge, use anti-static ionizer (discharger) with your analytical balance to get the exact weight of the filter paper. Alternatively, take ten measurements and use the average weight of the filter paper (less precise, will slightly affect your results).
-
11.
Into a 50-mL conical centrifuge polypropylene tube weigh 1.0 g of the defatted sample (as prepared in section A). Record the weight to 0.1 mg (mdefat).
Note: Typically, 200–600 mg of chitin is obtained by using 1 g of the defatted sample which is sufficient for further processing.
-
12.
Add 40 mL 1 M sodium hydroxide solution, cap and immediately shake vigorously by hand. Check visually that all sample is dispersed properly in the solution and that there are no lumps.
-
13.
If there are lumps, use vortex, ultrasonic bath or very vigorous shaking to disintegrate them.
-
14.
Place the 50-mL tubes into an ultrasonic bath for 10 min, inverting them every 2 min to prevent ultrasonic clumping.
-
15.
Place the tubes (in a rack) into the pre-heated oven for 5 h at 80 °C. Shake the tubes by hand every 60 min to mix the contents. Contents are hot, use heat resistant gloves!
-
16.
After 5 h, take the tubes out of the oven and centrifuge them at 10,000 rpm (10,730 rcf) at laboratory temperature for 10 min. Use medium acceleration and medium deceleration. Contents are hot, use heat resistant gloves!
Note: Make sure all the tubes (with their contents and lids) have similar weight ± 0.5 g to protect the centrifuge from damage: find the heaviest plastic tube in the set and adjust the weights of all the other plastic tubes by adding a few drops of 1 M sodium hydroxide solution to match their weight of the heaviest tube (± 0.5 g) in the set.
-
17.
After centrifugation, carefully decant the liquid over the respective (pre-weighted) filter paper. Leave almost no solution in the centrifuge tube.
-
18.
Again, add 40 mL 1 M sodium hydroxide solution to the plastic tube containing the sample, cap and immediately shake vigorously by hand to disintegrate the sediment (after centrifugation, the sediment tends to clump). Check visually that there are no lumps and the sediment is well dispersed in the solution.
-
19.
Place the plastic tubes (in a rack) back into the oven for additional 19 h at 80 °C. Invert the tubes every 60 min to mix the contents (except at night). Contents are hot, use heat resistant gloves!
-
20.
After 19 h, take the tubes out of the oven, let them cool down to room temperature, shake each tube by hand and filter the contents over the respective filter paper. Use wash bottle filled with MQ H2O to remove all the remaining residues from the tube and the cap onto the filter paper.
-
21.
Wash the residues on the filter paper with about 350 mL MQ H2O to get rid of all the sodium hydroxide solution. Using a pH strip indicator, check the pH of the wash liquid coming out of the funnel's stem. The pH should be <8, use more MQ H2O to wash the residues on the filter paper until the liquid pH is below 8.
Note: This is important, since sodium hydroxide residues on the sample and filter paper may slightly increase the weight of the sample and affect chitin in the sample. In some cases, the use of vacuum filtration system is necessary.
-
22.
Using an analytical balance weigh (an empty and labelled) Petri dish (mpetri). Record the weight with 0.1 mg precision.
-
23.
Take out the filter paper with the washed residue and unfold it. With a spatula, gently and carefully transfer the sample solids left on the filter paper onto the pre-weight Petri dish.
Note: Place the residue only in one small spot (about 1 cm) on the Petri dish. Fold the filter paper and place it on the Petri dish next to the deproteinized wet sample. Do not scratch the filter paper with the spatula to remove all sample solids! The paper fibers would increase the weight of the deproteinized sample. It is ok to leave some sample solids on the filter paper.
-
24.
Place the Petri dish with filter paper into the oven overnight (at least 16 h) at 80 °C to dry.
-
25.
Then, take out the Petri dish (containing the dried filter paper and dried sample residue) out of the oven, place it into a desiccator to cool down and record its weight with 0.1 mg precision (mafterdepr).
-
26.
Into a 2-mL micro centrifuge tube, scrape the dry deproteinized sample from the Petri dish glass. Discard the dried filter paper.
Note: Do not use any sample residue left on the filter paper. The deproteinized sample can be stored for two weeks at room temperature.
-
27.
Calculate the protein-free residue in percent rounding the result to one decimal place:
For mass descriptions see text in section B. Use masses in grams recorded with 0.1 mg precision. mafterdepr – weight sum after deproteination and drying i.e. Petri dish + dried filter paper + dried deproteinized sample residue [g]; mfilter – weight of the filter paper [g]; mpetri – weight of Petri dish [g]; mdefat – weight of the fat-free sample (as prepared in section A) [g]
-
C)
Hydrolysis
In this part of sample preparation, do not forget to include a blank sample and an in-house reference sample (chitin from shrimp shells). This part also demineralizes (decalcifies) the sample. This part takes about 28 h.
-
28.
Preheat the laboratory oven to 110 °C. In the oven, preheat one or two 1 L glass bottles with a cap, depending on the number of samples to be analyzed. One bottle fits about 15 samples.
-
29.
Work in a fume hood. Into an appropriately sized volumetric cylinder put sufficient amount of 6 M HCl needed for all samples and bubble with stream of N2 gas for at least 30 min.
Note: The hydrolysis is in done in oxygen-free environment to prevent possible decompositions (our laboratory uses the same setup as for amino acid hydrolysis).
-
30.
Pulverize/grind the defatted and deproteinized sample (as prepared in section A and B) in a ceramic mortar with pestle or by any other grinding technique suitable for very small sample sizes. The particles created should be less than 1 mm to ensure good hydrolysis. Use 1 mm screen sieve if necessary.
-
31.
Into a 15-mL glass Hungate anaerobic culture tube weigh (this time with 0.01 mg precision!) 10 mg of defatted and deproteinized sample (mdeprotdefat). To monitor method performance and hydrolysis process, include 3 other tubes into the sample set. Use nothing for the blank (I), 10 mg chitin from shrimp shells (supplied by Merck) as an in-house reference sample (II) and 10 mg of glucosamine standard (III).
-
32.
in a fume hood, add 10.00 mL of nitrogen flushed 6 M HCl (with a glass pipette, step 29), flush the headspace above the liquid with nitrogen (about 3 s) and close tightly.
Note: Work quickly (yet safely) after flushing the headspace and allow no air into the Hungate tube. Shake by hand to disperse the sample. Check visually that there are no lumps.
-
33.
Place the Hungate tubes into a rack and put them into the ultrasonic bath for 15 min, mixing every 3 min by hand to avoid ultrasonic clumping.
-
34.
Place the Hungate tubes into the preheated 1-L glass bottles with a cap, close tightly and put into the oven for 24 h at 110 °C. Contents are hot, use heat resistant gloves!
Note: Placing the Hungate tubes into the 1-L glass bottles with a cap serves as oven corrosion protection in case of tube breakage/leakage.
-
35.
Very carefully mix the insides of the Hungate tubes every 60 min by very carefully inverting the 1-L bottles ten times. Skip the mixing process during the night. Check the tubes for damages or breakage. Contents are hot, use heat resistant gloves!
-
36.
After 24 h, take the Hungate tubes out of the oven and let cool down to room temperature. Visually check that the liquid level is the same in all tubes (If not, then the tube was not closed properly and some hydrolysis solution has evaporated. In this case, do not further process the sample).
Note: Open the 1-L bottles containing the Hungate tubes in the fume hood in case some of the 6 M HCl has escaped during the hydrolysis or in case of a broken tube.
-
37.
Mix the tubes using vortex for 10 s and let stand for 30 min to sediment any unhydrolyzed residue (mostly mineral ashes).
-
38.
Into a labelled 1.5-mL HPLC vial (clear one) pipette 100 µL of the respective sample hydrolyzate.
-
39.
Work in a fume hood. Evaporate the hydrolyzate in the 1.5-mL HPLC vial under stream of nitrogen at 60 °C for 15 min. After evaporation, visually check each vial that it is completely dry.
Note: Since 6 M HCl is being evaporated, use preferably plastic needles for N2 evaporation. If this is not possible, wipe the N2 evaporator after use well with distilled water to prevent corrosion on metallic parts of the equipment.
-
D)
Joint sample preparation for calibration standards, samples, blanks and in-house reference sample
Do this part immediately after completing section C. This part takes about 3 h. The use of homoserine as an internal standard is possible.
-
40.
Preheat the laboratory oven to 50 °C.
-
41.
To the evaporated calibration standards (described in section Solutions), samples (as prepared in sections A-C), blank and an in-house reference sample (as prepared in section C) in the 1.5-mL HPLC vials add 100 µL redissolution solution (0.76 % TEA) and vortex for 5 s.
Note: Choose such vortex speed that the liquid washes the sides of the vial at least up to 2/3 of its total volume.
-
42.
Let stand on the lab bench for 10 min (to re-dissolve the dry residue properly).
-
43.
Again vortex the HPLC vials for another 5 s.
-
44.
Add 100 µL derivatization solution (5 mg FMOC—OSu/mL), cap the HPLC vials tightly and vortex for 5 s.
-
45.
Put all the vials with calibration standards, samples, blank and an in-house reference sample into the oven for 30 min at 50 °C.
Note: All the vials (the entire sample set) must go into the oven at once! Use the same solutions for all vials (check beforehand that you have sufficient volumes). Do not prepare the sample set in batches!
-
46.
After 30 min, take the samples out of the oven and refrigerate them at 4 °C for 30 min.
-
47.
Vortex the cooled HPLC vials for 5 s.
-
48.
Uncap the HPLC vials and add 800 µL quencher (ACN:MP-A, 1:1, V/V) and vortex uncapped for 5 s.
-
49.
Filter the contents through a syringe filter with 0.2 µm nylon membrane into a separate clean 1.5-mL HPLC vial, cap and analyze by HPLC.
HPLC parameters for sample analysis
Elution type: gradient I (see Table 1)
Table 1.
Solvent lines for a quarternary HPLC system: A: ACN, B: water + TFA, pH = 2.4, C: ACN, D: MQ H2O; a at 111 min, the flow is decreased to 0 mL/min.
| I: Sample analysis |
II: Column pre-conditioning (before sample analysis) |
III: Column flush and storage (end of sample analysis) |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Time [min] | % A | %B | Time [min] | % A | %B | %C | %D | Time [min] | % A | %B | %C | %D |
| 0 | 30 | 70 | 0 | 0 | 0 | 90 | 10 | 0 | 30 | 70 | 0 | 0 |
| 6 | 30 | 70 | 20 | 0 | 0 | 90 | 10 | 0.1 | 0 | 0 | 30 | 70 |
| 11 | 100 | 0 | 25 | 0 | 0 | 30 | 70 | 30 | 0 | 0 | 30 | 70 |
| 12 | 100 | 0 | 35 | 0 | 0 | 30 | 70 | 50 | 0 | 0 | 90 | 10 |
| 15 | 30 | 70 | 35.1 | 30 | 70 | 0 | 0 | 110 | 0 | 0 | 90 | 10 |
| 18 | 30 | 70 | 55 | 30 | 70 | 0 | 0 | 111a | 0 | 0 | 90 | 10 |
| mobile phase consumption mL/run | 8 | 8 | 6 | 13 | 22 | 10 | 0 | 0 | 70 | 177 | ||
Mobile phase A: acetonitrile
Mobile phase B: water with TFA, pH 2.4
Total run time: 18 min
Column temperature: 30 ± 0.8 °C
Autosampler temperature: 5 ± 1 °C
Needle wash time: 3 s
Needle wash solution: 50 % (aqueous) acetonitrile (v/v)
Flow rate: 0.8 mL/min
Injection volume: 5 µL
HPLC column and guard column: Agilent Poroshell 120 EC—C18, 150 × 3 mm, 2.7 µm with UHPLC guard Poroshell 120 EC—C18, 5 × 3 mm, 2.7 µm
Detector parameters: DAD, UV lamp on, 265 nm (bandwidth 4 nm, sampling rate 3 Hz, slit 4 nm, no reference wavelength)
Column storage: 90 % (aqueous) acetonitrile (v/v)
Typical GLCN peak parameters
Retention time (tR): 3.33 min (GLCN peak 1) and 3.99 min (GLCN peak 2)
Column void time (t0): 0.87 min
Peak symmetry: 0.813 (GLCN peak 1) and 0.858 (GLCN peak 2)
Plate number (N): 1193 (GLCN peak 1) and 1375 (GLCN peak 2)
Selectivity (α): 1.27 between GLCN peak 1 and GLCN peak 2
Retention factor (k): 2.82 (GLCN peak 1) and 3.59 (GLCN peak 2)
Resolution (Rs): 1.64 GLCN peak 1 vs. GLCN peak 2
Gradients
Prerun system check
-
•
Make sure the baseline is stable in the second water injection in the sample sequence.
Typical sample sequence (injections)
-
1.
One injection of water using gradient II (to put the column into the working mode)
-
2.
Two injections of water using gradient I (sample analysis) to sufficiently equilibrate the column
-
3.
One injection of blank using gradient I (sample analysis)
-
4.
Four injections of calibration solutions (from low to high) using gradient I (sample analysis)
-
5.
One injection of water using gradient I (sample analysis) to check for carry-over after the injection of highest concentration standard
-
6.
One injection of in-house reference material using gradient I (sample analysis)
-
7.
Inject samples* 1 to 15 using gradient I (sample analysis)
-
8.
Inject one calibration solution 35 µg GLCN/mL using gradient I (sample analysis)
-
9.
Inject samples* 16 to 30 using gradient I (sample analysis)
-
10.
Inject one calibration solution 35 µg GLCN/mL using gradient I (sample analysis)
-
11.
Repeat steps 6 and 7 if you have more samples
-
12.
One injection of water using gradient I (sample analysis) to check for any carry-over after sample set
-
13.
One injection of water using gradient III (column flush and storage) to flush the column after sample analysis and to put it into the storage solution.
Note: * each sample is prepared in triplicates. Repeat full calibration after every 45 samples. Insert water injections if you want to check for carry-over.
Integration
Since glucosamine is a carbohydrate, it's cyclic form comes in two stereoisomeric (anomeric) structures (alpha and beta). Therefore, glucosamine-FMOC derivative has two peaks. The two peaks show area ratio of 1.79 ± 2 % (peak 2/peak 1), thus for quantification, use only the second (higher) peak. Place the integration baseline as depicted on Fig. 6.
Fig. 6.
Integration of chitin.
Note: In samples you might want to calculate the GLCN peak2/peak1 area ratio to check for possible interferences.
Calibration
Calculate the actual concentration of calibration standards used in µg/mL. Don't forget to correct for standard purity and the fact that hydrochloride salt has been used. Use linear calibration (including the origin and no weighing) based on peak area. R2 should be greater than 0.995. An example of a calibration line is given in Fig. 7. Monitor the injections of the 35 µg GLCN/mL calibration solutions during the entire chromatographic sequence. The average area of (second) glucosamine peak should not differ by more than 3 % from the average across the sequence.
Fig. 7.
An example of a linear calibration curve at 5, 35, 70 and 100 µg GLCN/mL including the origin.
Results
Using the chromatography software, integrate all chromatograms (see Fig. 6). Use current calibration curve (see section Calibration) to report results in µg GLCN/mL in the chromatography software. Then, recalculate the results in Microsoft excel sheet using the fat-free and protein-free mass percent (see formulas in section A and B). Round the final result of CLCN content (mg/g) to the whole number (do not use decimal places).
GLCN software result – content of GLCN [µg/mL]; fatfree residue – result as given by the formula in section A [in%, rounded to one decimal place]; proteinfree residue – result as given by the formula in section B [in%, rounded to one decimal place]; mdeprotdefat – sample weight used for hydrolysis [mg, recorded with 0.01 mg precision]
Method monitoring
In a Microsoft excel sheet monitor and evaluate the performance of the method over time e.g. the in-house reference sample, average column pressure and average retention time of glucosamine peaks per sample set analyzed. You might include also other peak or column parameters.
Method validation
To obtain method performance data has been a rather challenging task for several reasons. To our knowledge, no certified reference material for chitin is available to date and second, chitin is also not available as an analytical standard. The only chitin available is technical grade, coming from shrimp shells and is lacking purity assessment. Chitin is a polymer substance that is insoluble in water. This property is utilized in the first two steps in sample preparation (defatting and deproteination) to obtain chitin-containing residue that is then hydrolyzed into glucosamine. To validate the performance of an analytical method without a certified reference material is possible if spiking experiments are performed. However, without the ability to use chitin as a standard substance with a known purity, the method validation becomes very difficult. Spiking experiments with glucosamine are not feasible since glucosamine is water soluble and would get lost during deproteination step. Alternatively, the use of chitosan is questionable because it is a deacetylated form of chitin meaning it is a different substance. Also, spiking any food matrix other than shrimp shells with technical grade shrimp chitin is raising questions too. For these reasons, the validation parameters given in Table 2 have to be considered as estimates.
Table 2.
Validation performance data.
| Glucosamine | Chitin from shrimp shellsa | Forest mushrooms | Mealworm larvae | |
|---|---|---|---|---|
| Chitin content [mg GLCN/g] | n/a | 780.7 | 52.9 | 105.8 |
| Repeatability CV(r) [%] | 3.1 | 6.2 | 7.5 | 8.9 |
| Repeatability r [%] | 5.8 | 9.6 | 10.4 | 14.5 |
| intermediate reproducibility CV(iR) [%] | 7.3 | 13.1 | 19.3 | 16.7 |
| Intermediate reproducibility iR [%] | 8.8 | 17.9 | 26.7 | 21.9 |
used as an in-house reference sample (purchased from Merck, article C7170).
Validation data have been gathered in a single-laboratory validation settings using two analysts on 6 different dates during three-month period in 2023 and two 300 g matrices; the results were measured in triplicates for each matrix. HPLC system injection repeatability was <1 % GLCN peak area based on six consecutive injections (5 µg GLCN/mL). Linearity of the calibration curve has been proven up to 250 µg GLCN/mL. Due to the lack of blank matrices, it has not been possible to reliably determine the limit of detection (LOD1) by spiking experiments, yet it is estimated to be 0.05 µg GLCN/mL. The limit of quantitation (LOQa) has been defined as the lowest concentration of the linear working range (5 µg GLCN/mL). Due to the lack of certified reference material the trueness and recovery of the method cannot be accurately reported. If chitin from shrimp shells could be considered as pure (=100 %), then the mean recovery would be 79.5 %. In case of GLCN (omitting defatting and deproteination) the mean recovery would be 86.4 %.
Fluorescence detection (alternative detection)
Excitation wavelength: 260 nm.
Emission wavelength: 350 nm.
Sampling rate: 3 Hz.
Signal polarity: positive.
LOD for fluorescence detection is 0.065 µg/mL.
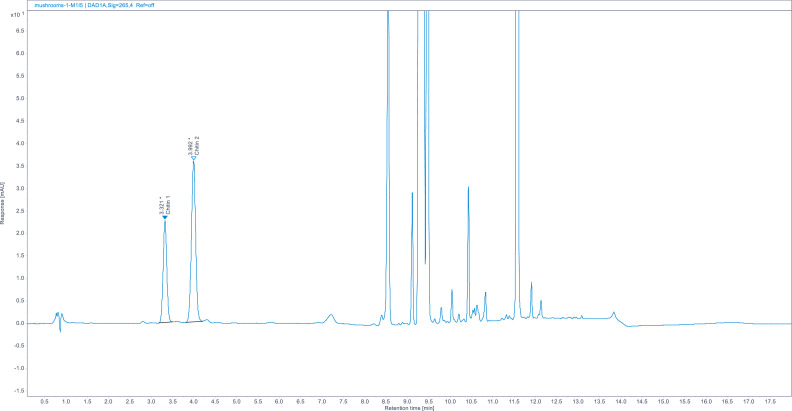
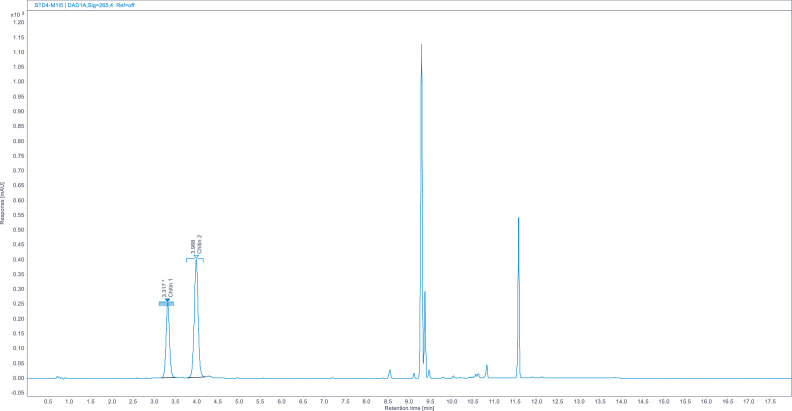
Chromatograms. (Fig. 1, Fig. 2, Fig. 3, Fig. 5, Fig. 8).
Fig. 4.
Zoomed chromatogram of forest mushrooms.
Fig. 1.
Full chromatogram of GLCN standard (70 µg/mL).
Fig. 2.
Full chromatogram of mixed forest mushrooms. Chromatograms of other food matrices (shrimp, mealworm larvae, crickets, fungi) look very similar showing no additional peaks. For zoomed-in chromatogram see Fig. 4.
Fig. 3.
A close up of overlaid GLCN standards (5, 30, 70, 100 µg/mL).
Fig. 5.
An example of column pressure during sample analysis.
Fig. 8.
Full chromatogram of mushroom sample in fluorescence detection (λex = 260 nm, λem = 350 nm).
Additional information
Chitin is a water insoluble nitrogen-containing polysaccharide made from N-acetyl-D-glucosamine containing β-(1→4)-linkages. In food, chitin is considered as a source of (animal derived) fiber with prebiotic properties to gut microflora. Chitin content varies widely in nature from 1 % (yeasts) up to 64 % (butterfly cuticles) and is mostly found in filamentous or mushroom forming fungi, insects and crustaceans. In marine animals it is often penetrated with calcium carbonate [3]. Plants can sense chitin in pathogens and thus activate their defense system. Chitosan, a very attractive biopolymer, is a partially deacetylated chitin. Since glucosamine itself lacks a chromophore, it is detected at 265 nm after an off-line FMOC derivatization procedure (commonly used in amino acid chromatography) as glucosamine-FMOC adduct.
Important notes to the method:
-
•
no certified reference material with chitin reference value is commercially available
-
•
homocysteine can be used as an internal standard (final concentration 25 µg/mL) [4]
-
•
samples with <5 % fat don't have to be defatted as described in step A
-
•
the use of well calibrated pipettors is mandatory
Ethics statements
Not applicable, this is a chemical method.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by the Ministry of Agriculture of the Czech Republic, institutional support MZE-RO0323 and by the Ministry of Education, Youth and Sport of the Czech Republic in METROFOOD-CZ project MEYS Grant No: LM2023064.
Footnotes
The LOD and LOQ values presented were estimated based on signal-to-noise ratio of 3:1 or 10:1 resp. by successive dilution of GLCN standard solution to obtain S/N≈3 or S/N≈10.
Data availability
Data will be made available on request.
References
- 1.Han X., Heinonen M. Development of ultra-high performance liquid chromatographic and fluorescent method for the analysis of insect chitin. Food Chem. 2021;334 doi: 10.1016/j.foodchem.2020.127577. [DOI] [PubMed] [Google Scholar]
- 2.Zhou J.Z.Q., Waszkuc T., Mohammed F. Determination of glucosamine in raw materials and dietary supplements containing glucosamine sulfate and/or glucosamine hydrochloride by high-performance liquid chromatography with FMOC-Su derivatization: collaborative study. J. AOAC Int. 2005;88:1048–1058. [PMC free article] [PubMed] [Google Scholar]
- 3.Abidin N.A.Z., Kormin F., Abidin N.A.Z., Anuar N.A.F.M., Abu Bakar M.F. The potential of insects as alternative sources of chitin: an overview on the chemical method of extraction from various sources. Int. J. Mol. Sci. 2020;21:4978. doi: 10.3390/ijms21144978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ekblad A., Nasholm T. Determination of chitin in fungi and mycorrhizal roots by an improved HPLC analysis of glucosamine. Plant Soil. 1996;178:29–35. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.