Abstract
Chitin deacetylase (CDA) modifies chitin into chitosan by removing acetyl groups, but its inherent instability poses a challenge for successful crystallisation. Despite limited successes in crystallizing CDAs, prior attempts with recombinant chitin deacetylase (BaCDA) failed due to poor stability. To address this, we propose an enzyme buffer formulation as a cost-effective strategy to enhance stability, prolong shelf life, and increase the likelihood of crystallisation. Utilizing the high-throughput screening technique FTSA, we developed a screening method correlating BaCDA stability with its activity. The optimised formulation comprises 50 mM Tris-HCl buffer pH 7, 1 M NaCl, 20 % glycerol, and 1 mM Mg2+ as excipients. This formulation significantly improves BaCDA's thermostability (140.47 % increase) and enzyme activity (2.9-fold enhancement). BaCDA remains stable in the formulated buffer at −20 °C and −80 °C for 30 days and at 4 °C for 15 days. The current study has designed a high-throughput screening method approach to assess the stability of CDA enzyme formulations. The results of this study could contribute to the exploration of formulation elements that enhance the structural stability of CDA, thereby facilitating investigations into the enzyme's structure-function relationships.
Keywords: Fluorescence thermal shift assay (FTSA), Formulation, Shelf-life, Chitin deacetylase, SYPRO orange
Graphical abstract
Highlights
-
•
Limited Crystallisation Success: Only a few CDAs have been successfully crystallised to date, highlighting the difficulty in stabilizing this enzyme.
-
•
Developing enzyme buffer: Simplify, cut costs, boost stability, and extend shelf life for improved crystallization success.
-
•
FTSA Screening Technique: The Fluorescence-Thermal-Shift-Assay (FTSA) is utilized as a high-throughput screening technique to select suitable buffer excipients.
-
•
The presence of the formulated buffer boosted BaCDA's thermostability by 140.47% and enzyme activity by 2.9 fold. In this formulation, BaCDA remains stable at -20°C and -80°C for 30 days and at 4°C for 15 days.
-
•
Implications for Structural Stability: The results contribute to exploring formulation elements that enhance the structural stability of CDA, facilitating investigations into the enzyme's structure-function relationships.
1. Introduction
Chitosan, derived from chitin, finds extensive utility, especially in the biomedical sector, owing to its biological attributes as a safe, antimicrobial, anti-tumour, and environmentally degradable polymer. At the industrial scale, this conversion is accomplished via thermochemical processes, leading to environmental concerns and a lack of product reproducibility. Hence, our research groups have been using chitin deacetylase on the green route [[1], [2], [3], [4], [5], [6]]. Despite nearly four decades since the initial CDA discovery, only a limited number of CDAs have successfully undergone crystallisation. The main challenge in crystallising CDAs lies in their inherent instability [7]. Enhancing the stability during storage and prolonging the shelf-life of CDAs can significantly improve cost-effectiveness in downstream processes. This can be accomplished through the formulation process, which uses various excipients such as buffers, pH modifiers, salts, metal ions, preservatives, amino acids, polymers, sugars, and polyols [8,9].
In the present study, we employed the fluorescence thermal shift assay (FTSA) to assess the thermal stability of the BaCDA enzyme formulation and its shelf life using the SYPRO orange dye. The detection is based on the dye's interaction with the protein's hydrophobic regions, which become exposed when the protein undergoes denaturation, which is read as increased fluorescence [10]. This modified formulation can help in using BaCDA for structural studies.
2. Materials and methods
2.1. BaCDA enzyme preparation
In the previous work, the BaCDA gene was cloned into a pET-22 (b) vector and expressed in E. coli Rosetta pLysS cells. The over-expressed BaCDA was purified homogeneously using Ni-NTA affinity chromatography. After purification, the BaCDA was dialysed to remove imidazole and concentrated to 1 mg/mL using a 10 kDa Amicon Ultra-15 centrifugal filter. This purified BaCDA was then ready for further experiments [6].
2.2. Enzyme activity assay
Enzyme activity was measured using an acetate assay kit. To carry out the assay, 100 μL of a reaction containing 40 μL of ethylene glycol chitin (EGC) (1 mg/mL), 20 μL of BaCDA (1 mg/mL), and 40 μL of 50 mM Tris-HCl (pH 7) buffer was incubated for 1 h at 30 °C at a speed of 800 rpm. BaCDA was removed by passing the mixture through a 3 kDa spin column to stop the reaction. The manufacturer's protocol determined acetate concentration in the 10 μL of flow-through. Accordingly, the enzyme activity was calculated. Specifically, one unit enzyme was defined as the enzyme required to release one μmol of acetate from the reaction per mg of the enzyme. We conducted the enzymatic assays in triplicate for each experiment.
2.3. Fluorescence-based thermal shift assay
2.3.1. Enzyme and dye ratio optimisation
The thermal shift assay used the Bio-Rad CFX96 Touch Deep Well Real-Time PCR System's melting curve program. The temperature was increased by 1 °C within a range of 10 °C–90 °C, with a 30-s hold time, and the final reaction volume for all experiments was 25 μL. In a preliminary investigation, SYPRO Orange and BaCDA concentrations were titrated to determine an appropriate melt curve. A 50X stock solution of SYPRO Orange dye was prepared, and concentrations ranging from 2.5X to 20X were used in the assay. For BaCDA, concentrations within the 2.5–20 μg range were utilized in the assay. Optimal BaCDA and dye concentrations were determined and used in subsequent assays.
2.3.2. Effect of pH on stability of BaCDA
Buffers with a pH range of 4–10 were selected for the screening process. The buffers included in the study were 50 mM citrate buffer with a pH of 4–6, 50 mM Bis-tris buffer with a pH of 6–7, 50 mM phosphate buffer with a pH of 6–8, 50 mM Tris-HCl with a pH of 7–8, 50 mM boric acid buffer with a pH of 8–9, and 50 mM carbonate buffer with a pH of 9–10. Apart from assessing thermal stability, the enzyme activity was also evaluated under these pH conditions [7].
2.3.3. Effect of salt concentration on the stability of BaCDA
The enzyme activity of BaCDA was determined in the presence of NaCl concentrations ranging from 50 mM to 2 M for thermal stability screening.
2.3.4. Effect of additives on the stability of BaCDA
The study involved testing different additives, such as glycerol and metal ions, to determine their impact on the thermostability of BaCDA. The glycerol concentration was tested at different levels ranging from 5 % to 30 %. Additionally, the study examined the effects of metal ions such as K+, Ca2+, Co2+, Mn2+, Mg2+, Ni2+, and Zn2+ at a concentration of 1 mM. All additives were also tested to determine any potential impact on enzyme activity.
2.4. Effect of storage on stability
The BaCDA enzyme was purified using the previously described method and then dialysed against a storage buffer that contained Tris-HCl pH 7, 1 M NaCl, 20 % glycerol and 1 mM Mg2+. The enzyme was then stored at three different temperatures, 4 °C, −20 °C, and −80 °C, to determine its stability. After 7, 15, and 30 days, samples were taken and analysed using SDS-PAGE and acetate assay to assess the enzyme's stability and activity.
3. Results
3.1. Stability of BaCDA
By prior research, BaCDA was eluted using 100 mM imidazole, and the collected fractions were subsequently subjected to dialysis to eliminate imidazole. The resulting purified BaCDA exhibited a molecular weight of 29 kDa, as verified through SDS-PAGE (Supplementary Fig. 1)
3.1.1. Dye and enzyme ratio
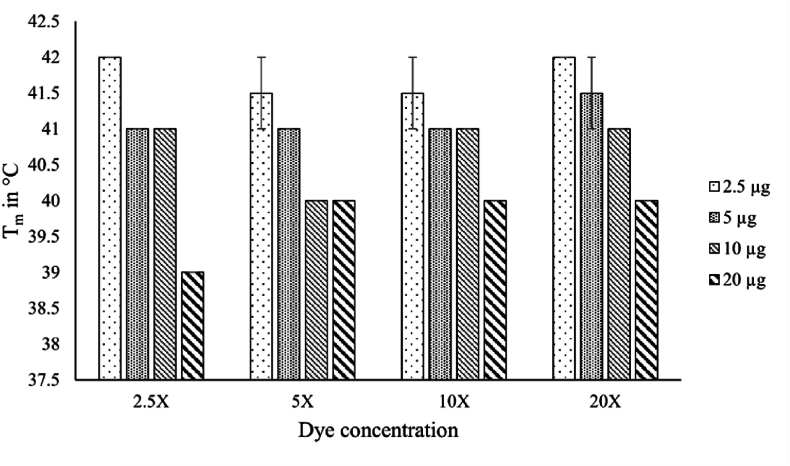
To increase the sensitivity of the method, different concentrations of dye (ranging from 2.5X to 20X) and enzyme (ranging from 2.5 μg to 20 μg) were used (Fig. 1). A common trend observed in all combinations was that increasing the enzyme concentration resulted in a decrease in relative fluorescence units (RFU). The highest melting temperature (Tm) was obtained using 2.5X dye concentration and 2.5 μg of enzyme. Thus, these conditions were maintained for further optimisation steps. The highest melting temperature obtained was considered 100 %.
Fig. 1.
Dye and enzyme ratio optimisation. The Tm of BaCDA was found to be 42 °C in water with 20X dye and 2.5 μg enzyme.
3.1.2. Buffer and pH screening
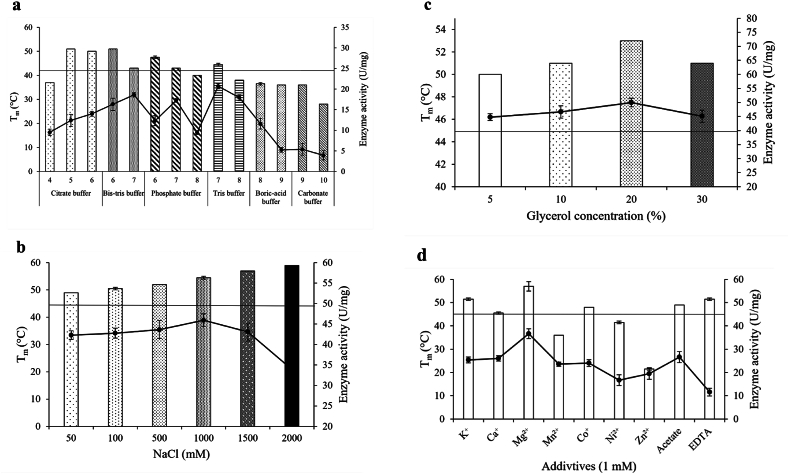
Different buffers with a pH range from 4 to 10 were tested to determine the effect on the stability of BaCDA. The results showed that the Tm of BaCDA increased to 119 % in citrate buffer at pH 5 and Bis-Tris buffer at pH 6, but this did not match with the activity profile of BaCDA. The maximum activity was observed in the Tris-HCl buffer at pH 7. However, the stability of the enzyme improved by 5.95 % in the presence of Tris-HCl buffer at pH 7 compared to the results from the initial experiment. Therefore, all further experiments were conducted with Tris-HCl buffer at pH 7.
3.1.3. Salt concentration screening
The study aimed to investigate how different NaCl concentrations affect the BaCDA's Tm. The experiment tested NaCl concentrations ranging from 50 mM to 2 M, and it was discovered that the Tm of BaCDA increased to 140 % in the presence of 2 M NaCl (Supplementary Fig. 2b). However, using 2 M NaCl resulted in decreased enzyme activity. Further testing revealed that BaCDA activity was highest in the presence of 1 M NaCl, with the Tm at 130.95 % (Fig. 2b). Consequently, the ideal concentration of NaCl to be included in the storage buffer cocktail is 1 M NaCl.
Fig. 2.
Investigation of Tm of BaCDA in different conditions. (a) The Tm and enzyme activity of BaCDA at different buffer and pH conditions. The bar graphs represent the Tm (°C), and the line graph represents the enzyme activity (U/mg). (b) The Tm and enzyme activity of BaCDA at different concentrations of NaCl (mM). The bar graphs represent the Tm (°C), and the line graph represents the enzyme activity (U/mg). (c) The Tm and enzyme activity of BaCDA at different glycerol concentrations (%). The bar graphs represent the Tm (°C), and the line graph represents the enzyme activity (U/mg). (d) The Tm and enzyme activity of BaCDA in the presence of different metal ions (1 mM). The bar graphs represent the Tm (°C), and the line graph represents the enzyme activity (U/mg).
3.1.4. Additive screening
An experiment was conducted to extend the shelf life of BaCDA by adding glycerol to its formulation. Different concentrations of glycerol, ranging from 5 % to 30 %, were tested to determine the optimal concentration that would provide thermal stability to BaCDA. Results showed that the maximum enzyme activity and Tm of BaCDA were observed at a glycerol concentration of 20 %. The Tm of BaCDA with 20 % glycerol was 126.19 %, confirming that a 20 % glycerol concentration is ideal for the storage buffer cocktail.
BaCDA is a type of metalloenzyme. To determine its properties, we tested its thermal stability in the presence of various metal ions. Our results showed that the presence of Mg2+ increased the Tm of BaCDA by 140.47 % (Supplementary Fig. 2d). Moreover, we observed that the enzyme activity was also improved in the presence of Mg2+ in the buffer (Fig. 2d). Based on these findings, we included 2 mM Mg2+ as the metal ion additive in the storage buffer cocktail.
3.2. Effect of storage temperature on the shelf life of BaCDA
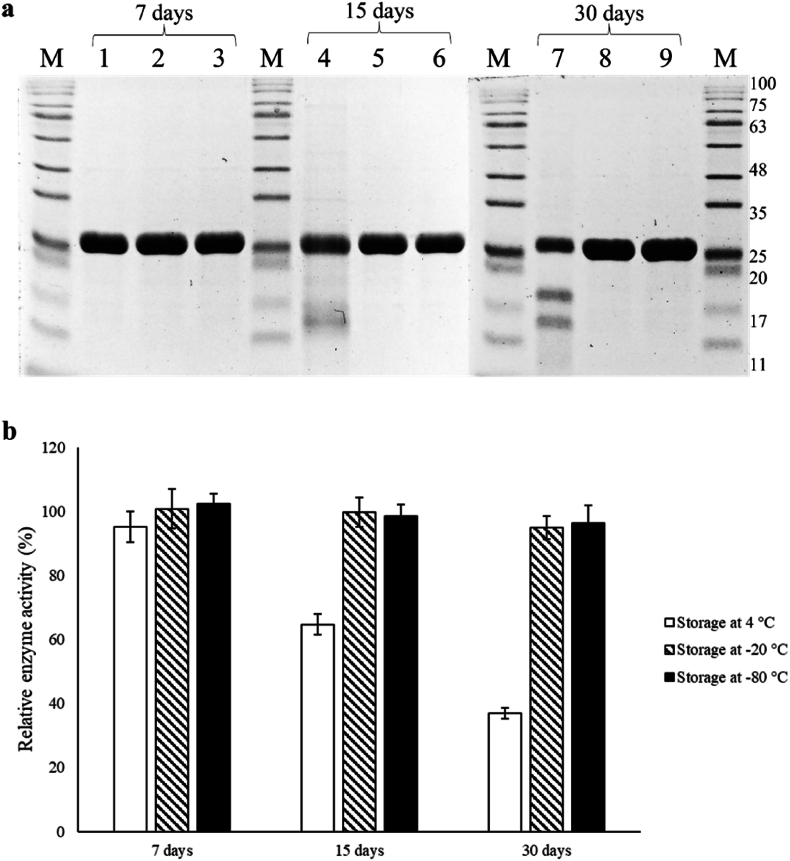
The storage stability study results showed that when the BaCDA enzyme was stored at 4 °C, degradation occurred as observed in the SDS-PAGE (Fig. 3a). There was a decrease in enzyme activity to 95.23 ± 4.87 %, 64.68 ± 3.23 %, and 36.90 ± 1.64 % on days 7, 15, and 30, respectively (Fig. 3b). In contrast, no degradation was observed when the enzyme was stored at −20 and −80 °C, as seen on SDS-PAGE. When held at −20 °C, the BaCDA maintained its activity at 99.76 ± 4.62 % and 95.08 ± 3.61 % on the 15th and 30th day, respectively. Similarly, when stored at −80 °C, the BaCDA retained its activity at 98.56 ± 3.72 % and 96.34 ± 5.63 % on the 15th and 30th day, respectively. These findings suggest that the optimised storage buffer cocktail is suitable for long-term storage of the BaCDA enzyme with only a marginal loss in enzyme activity.
Fig. 3.
Investigation of storage stability of BaCDA. (a) SDS-PAGE for the stability of BaCDA stored at 4 °C, −20 °C and −80 °C was analysed at 7 days, 15 days and 30 days. (b) Relative enzyme activity of BaCDA stored at 4 °C, −20 °C, and −80 °C was analysed at 7 days, 15 days and 30 days.
4. Discussion
The fluorescence-based thermal shift assay (FTSA) is a high-throughput screening technique to investigate the thermal stability of proteins [11]. FTSA has been employed to study the stability of various proteins [[12], [13], [14]]. CDAs are enzymes that are known for their instability. Hence, only a limited number of CDAs from organisms such as Encephalitozoon cuniculi (ECU11_0510), C. lindemuthianum (ClCDA), Bombyx mori (BmCDA), Aspergillus niger (AngCDA), and (SmPgdA) have been successfully crystallised and documented in the Protein Data Bank (PDB) [[15], [16], [17], [18], [19]]. BmCDA, for instance, exhibited instability and underwent autocleavage when exposed to a crystallisation reagent; however, truncation of the enzyme enhanced its stability, leading to its successful crystallisation. As part of a previous study, attempts were made to crystallise Bacillus aryabhattai CDA, but the efforts were unsuccessful due to the enzyme's instability.
The enzyme excipient formulation was tested and assessed using an FTSA-based approach to improve stability. Researcher Kathy Huynh and colleagues introduced the initial protocol for the FTSA study utilizing SYPRO dye. Subsequently, the SYPRO dye to BaCDA ratio was improvised on this [20]. Additionally, Ruth Kellner and her team have developed 158 conditions for studying protein stability. The results of the FTSA were evaluated based on DSF and enzyme activity under these 158 conditions. They also recommended using 5X dye at the final concentration in general. However, our study observed that the best melting curve was obtained with 2.5 μL enzyme and 2.5X dye in a 25 μL reaction [21]. BaCDA is highly unstable without excipients, and its Tm was found to be 42 °C. The stability of BaCDA was found to be highest at pH 6 when Bis-Tris and phosphate buffer were present. This might be because the pH was far from the pKa of BaCDA, which was found to be 9.2. On the other hand, the enzyme activity was at its maximum at pH 7 in the Tris-HCl buffer, and the Tm was 44.5 °C. The results indicate that the Tris-HCl buffer condition suits BaCDA activity and stability. The optimum pH for CDA reported in Aspergillus nidulans, Colletotrichum lindemuthianum, Flammulina velutipes and Rhodococcus sp. is 7 for their activity [22]. The Tris-HCl buffer helped to crystallise ECU11_0510, ClCDA, and BmCDA. Moreover, Tris-HCl buffer is commonly used for enzyme stability. It interacts with the peptide backbone and holds good with BaCDA stability [23]. The presence of salt provides ionic strength to the enzyme's physiological structure. Therefore, we tested different concentrations of NaCl. BaCDA showed stability even at a high concentration of NaCl up to 2 M, but the activity was inhibited. The enzyme activity was maximum at 1 M NaCl with a Tm of 55 °C, which agrees with our previous work. Several enzymes are reported for their halophilic nature, and marine-based enzymes tend to be halophilic; BaCDA is one of them [6,24]. AsnCDA and BmCDA were crystallised in 100 mM and 20 mM ionic strength, respectively. Glycerol is a commonly used preservative for stabilizing proteins [25]. It can be used at concentrations ranging from 10 to 50 %, but its impact on protein activity varies depending on its structural conformation [26]. For instance, the activity and melting temperature of BaCDA increased in the presence of 20 % glycerol but decreased in the presence of 30 %. On the other hand, cryoprotectant concentrations of 10 %, 15 %, and 25 % (v/v) glycerol were used for AsnCDA, ClCDA, and BmCDA, respectively, to preserve their crystalline structure. CDAs are generally classified as metalloenzymes, and many exhibit activity in the presence of metal ions [22]. As such, the activity and Tm of BaCDA were also investigated in the presence of metal ions. The Tm was increased in the presence of K+, Ca2+, Co2+, Mg2+, acetate and EDTA. The Tm was decreased in the presence of Mn2+, Ni2+ and Zn2+. The activity of BaCDA was enhanced to the maximum in the presence of Mg2+. Its melting temperature (Tm) was found to be 59 °C. Other CDAs from various organisms, including B. amyloliquefaciens, Lichtheimia corymbifera, Penicillium oxalicum, and Rhizopus circinans, also displayed enhanced activity in the presence of Mg2+ [27]. Similarly, Zn2+ promoted the crystallisation of ClCDA, SmPgdA, and ECU11_0510. After screening, a buffer cocktail consisting of 1 M NaCl, 20 % glycerol, and 1 mM Mg2+ in the presence of 50 mM Tris-HCl pH 7 buffer was prepared.
The stability of BaCDA was examined at 4 °C, −20 °C, and −80 °C for 7, 15, and 30 days. It was observed that the BaCDA degraded in the presence of elution buffer when stored at 4 °C, −20 °C, and −80 °C for 7 days. However, in the optimised buffer cocktail, the BaCDA was stable at −20 °C and −80 °C for up to 30 days without significant loss of enzyme activity. The BaCDA stored at 4 °C was stable for up to 7 days, and after 15 days of storage, the enzyme degraded and lost its activity. Different CDAs are crystallised in 5 min–10 days under different conditions. Therefore, by optimising the buffer cocktail for BaCDA, suitable storage and temperature conditions were determined. Correlating the stability and activity of the enzyme helped design a single buffer cocktail, which reduces the possible stress on the enzyme and increases the possibility of crystallisation.
5. Conclusion
A sensitive method for screening excipients using FTSA was developed for highly unstable CDA enzymes. The thermal stability of BaCDA activity was found to be correlated. A buffer cocktail that was well-suited for BaCDA activity and storage was formulated. Overall, the prepared buffer cocktail increased the thermostability and enzyme activity of BaCDA by 140 % and ∼2.9-fold, respectively. Therefore, the FTSA-based screening method effectively designed a suitable buffer cocktail for the most fragile CDA enzyme. The storage stability study also demonstrated that the enzyme can be stored at −20 and −80 °C for up to 30 days without any significant loss of activity. These results could assist in crystallisation studies as well.
CRediT authorship contribution statement
Goutam Mohan Pawaskar: Investigation, Methodology, Validation, Visualization, Writing – original draft. Ritu Raval: Conceptualization, Formal analysis, Investigation, Project administration, Supervision, Validation, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors thank the Council of Scientific and Industrial Research (CSIR), India, for providing a Senior Research Fellowship to Mr. Goutam Mohan Pawaskar with file no. 09/1165(0007)/2019-EMR-I dated on March 31, 2019.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbrep.2024.101718.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
Data availability
Data will be made available on request.
References
- 1.Mathew G.M., Huang C.C., Sindhu R., Binod P., Sirohi R., Awsathi M.K., Pillai S., Pandey A. Enzymatic approaches in the bioprocessing of shellfish wastes. 3 Biotech. 2021;11:367. doi: 10.1007/s13205-021-02912-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Raval R., Simsa R., Raval K. Expression studies of Bacillus licheniformis chitin deacetylase in E. coli Rosetta cells. Int. J. Biol. Macromol. 2017;104:1692–1696. doi: 10.1016/j.ijbiomac.2017.01.151. [DOI] [PubMed] [Google Scholar]
- 3.Bhat P., Pawaskar G.M., Raval R., Cord-Landwehr S., Moerschbacher B., Raval K. Expression of Bacillus licheniformis chitin deacetylase in E. coli pLysS: sustainable production, purification and characterisation. Int. J. Biol. Macromol. 2019;131:1008–1013. doi: 10.1016/j.ijbiomac.2019.03.144. [DOI] [PubMed] [Google Scholar]
- 4.Raval R., Raval K., Moerschbacher B.M. Enzymatic modification of chitosan using chitin deacetylase isolated from Bacillus cereus. Open Access Sci. Reports. 2013;2:2–5. doi: 10.4172/scientificreports.617. [DOI] [Google Scholar]
- 5.Pawaskar G.M., Pangannaya S., Raval K., Trivedi D.R., Raval R. Screening of chitin deacetylase producing microbes from marine source using a novel receptor on agar plate. Int. J. Biol. Macromol. 2019;131:716–720. doi: 10.1016/j.ijbiomac.2019.03.118. [DOI] [PubMed] [Google Scholar]
- 6.Pawaskar G.M., Raval K., Rohit P., Shenoy R.P., Raval R. Cloning, expression, purification and characterization of chitin deacetylase extremozyme from halophilic Bacillus aryabhattai B8W22. 3 Biotech. 2021;11:1–13. doi: 10.1007/s13205-021-03073-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Simpson R.J. Stabilization of proteins for storage. Cold Spring Harb. Protoc. 2010;5 doi: 10.1101/pdb.top79. [DOI] [PubMed] [Google Scholar]
- 8.Kamerzell T.J., Esfandiary R., Joshi S.B., Middaugh C.R., Volkin D.B. Protein-excipient interactions: mechanisms and biophysical characterization applied to protein formulation development. Adv. Drug Deliv. Rev. 2011;63:1118–1159. doi: 10.1016/j.addr.2011.07.006. [DOI] [PubMed] [Google Scholar]
- 9.Wang W. Advanced protein formulations. Protein Sci. 2015;24:1031–1039. doi: 10.1002/pro.2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boivin S., Kozak S., Meijers R. Optimization of protein purification and characterization using Thermofluor screens. Protein Expr. Purif. 2013;91:192–206. doi: 10.1016/j.pep.2013.08.002. [DOI] [PubMed] [Google Scholar]
- 11.Reinhard L., Mayerhofer H., Geerlof A., Mueller-Dieckmann J., Weiss M.S. Optimization of protein buffer cocktails using Thermofluor. Acta Crystallogr., Sect. F: Struct. Biol. Cryst. Commun. 2013;69:209–214. doi: 10.1107/S1744309112051858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mezzasalma T.M., Kranz J.K., Chan W., Struble G.T., Schalk-Hihi C., Deckman I.C., Springer B.A., Todd M.J. Enhancing recombinant protein quality and yield by protein stability profiling. J. Biomol. Screen. 2007;12:418–428. doi: 10.1177/1087057106297984. [DOI] [PubMed] [Google Scholar]
- 13.Jaito N., Eurwilaichitr L., Nimchua T. Rapid screening of additive formulations for enhancing xylanase stability in pulp bleaching and storage conditions. Chem. Eng. Trans. 2020;79:307–312. doi: 10.3303/CET2079052. [DOI] [Google Scholar]
- 14.Crossen J., Diamond S.L. Thermal shift assay to probe melting of thrombin, fibrinogen, fibrin monomer, and fibrin: gly-Pro-Arg-Pro induces a fibrin monomer-like state in fibrinogen. Biochim. Biophys. Acta - Gen. Subj. 2021:73–85. doi: 10.1016/j.bbagen.2020.129805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blair D.E., Hekmat O., Schüttelkopf A.W., Shrestha B., Tokuyasu K., Withers S.G., Van Aalten D.M.F. Structure and mechanism of chitin deacetylase from the fungal pathogen Colletotrichum lindemuthianum. Biochemistry. 2006;45:9416–9426. doi: 10.1021/bi0606694. [DOI] [PubMed] [Google Scholar]
- 16.Urch J.E., Hurtado-Guerrero R., Brosson D., Liu Z., Eijsink V.G.H., Texier C., Van Aalten D.M.F. Structural and functional characterization of a putative polysaccharide deacetylase of the human parasite Encephalitozoon cuniculi. Protein Sci. 2009;18:1197–1209. doi: 10.1002/pro.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu L., Zhou Y., Qu M., Qiu Y., Guo X., Zhang Y., Liu T., Yang J., Yang Q. Structural and biochemical insights into the catalytic mechanisms of two insect chitin deacetylases of the carbohydrate esterase 4 family. J. Biol. Chem. 2019;294:5774–5783. doi: 10.1074/jbc.RA119.007597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bonin M., Hameleers L., Hembach L., Roret T., Cord-Landwehr S., Michel G., Moerschbacher B.M. In silico and in vitro analysis of an Aspergillus Niger chitin deacetylase to decipher its subsite sugar preferences. J. Biol. Chem. 2021;279 doi: 10.1016/j.jbc.2021.101129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dong M.D., Urch J.E., Ten Cate J.M., Rao V.A., Van Aalten D.M.F., Crielaard W. Streptococcus mutans SMU.623c codes for a functional, metal-dependent polysaccharide deacetylase that modulates interactions with salivary agglutinin. J. Bacteriol. 2009;91:394–402. doi: 10.1128/JB.00838-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huynh K., Partch C.L. Analysis of protein stability and ligand interactions by thermal shift assay. Curr. Protoc. Protein Sci. 2015;79:28.9.1–28.9.14. doi: 10.1002/0471140864.ps2809s79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kellner R., Malempré R., Vandenameele J., Brans A., Hennen A.F., Rochus N., Di Paolo A., Vandevenne M., Matagne A. Protein formulation through automated screening of pH and buffer conditions, using the Robotein® high throughput facility. Eur. Biophys. J. 2021;50:473–490. doi: 10.1007/s00249-021-01510-y. [DOI] [PubMed] [Google Scholar]
- 22.Zhao Y., Park R.D., Muzzarelli R.A.A. Chitin deacetylases: properties and applications. Mar. Drugs. 2010;8:24–46. doi: 10.3390/md8010024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taha M., Lee M.J. Interactions of TRIS [tris(hydroxymethyl)aminomethane] and related buffers with peptide backbone: thermodynamic characterization. Phys. Chem. Chem. Phys. 2010;12:12840–12850. doi: 10.1039/c0cp00253d. [DOI] [PubMed] [Google Scholar]
- 24.Moreno M. de L., Pérez D., García M.T., Mellado E. Halophilic bacteria as a source of novel hydrolytic enzymes. Life. 2013;3:38–51. doi: 10.3390/life3010038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vagenende V., Yap M.G.S., Trout B.L. Mechanisms of protein stabilization and prevention of protein aggregation by glycerol. Biochemistry. 2009;48:11084–11096. doi: 10.1021/bi900649t. [DOI] [PubMed] [Google Scholar]
- 26.Strategy G. 2010. Protein Expression and Purification Core Facility Protein Expression; pp. 5–8. [Google Scholar]
- 27.Grifoll-Romero L., Pascual S., Aragunde H., Biarnés X., Planas A. Chitin deacetylases: structures, specificities, and biotech applications. Polymers. 2018;10:1–29. doi: 10.3390/polym10040352. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.