Abstract
Hyperspectral imaging analysis combined with machine learning was applied to identify eight edible vegetable oils, and its classification performance was compared with the chemical method based on fatty acid compositions. Furthermore, the degree of adulteration in vegetable oils was quantitatively investigated using machine learning-enabled hyperspectral approaches. The hyperspectral absorbance spectra of palm oil with a high degree of saturation were distinctly different from those of the other liquid oils. The flaxseed and olive oils exhibited the dominant hyperspectral intensities at 1170/1671 and 1212/1415 nm, respectively. Linear discriminant analysis demonstrated that two linear discriminants could explain a significant portion of the total variability, accounting for 96.0% (fatty acid compositions) and 98.9% (hyperspectral images). When the hyperspectral results were used as datasets for three machine learning models (decision tree, random forest, and k-nearest neighbor), several instances to incorrectly classify grapeseed and sunflower oils were detected, while olive, palm, and flaxseed oils were successfully identified. The machine learning models showed a great classification performance that exceeded 98.9% from the hyperspectral images of the vegetable oils, which was comparable to the fatty acid composition-based chemical method in identifying edible vegetable oils. In addition, the random forest model was the most effective in ascertaining adulteration levels in binary oil blends (R2 > 0.992 and RMSE < 2.75).
Keywords: Artificial intelligence, AI, Classification, Gas chromatography, Food
Graphical abstract
Highlights
-
•
Hyperspectral imaging (HSI) was coupled to machine learning (ML) for identifying oils.
-
•
Two linear discriminants explained 98.9% of the total variation in the HSI datasets.
-
•
Random forest ML model offered performance advantages in the oil classification tasks.
-
•
HSI-based ML showed the classification performance comparable to the chemical method.
1. Introduction
Edible vegetable oils from various botanical sources have been extensively used in a variety of food products, depending on their unique physicochemical features such as the fatty acid composition and the degree of saturation. For example, palm oil has been commonly utilized as a frying medium for fried foods such as snacks and ramen, since it has oxidative stability derived from its high saturated fatty acid content. On the other hand, olive oil with a high level of unsaturated fatty acids, is a primary ingredient in salad dressings (Kim et al., 2010). It is therefore crucial to identify vegetable oils in the food industry for determining their appropriate applications in addition to preventing adulteration. A chemical method with gas chromatography (GC), has been dominantly used to identify vegetable oils by analyzing their fatty acid compositions. However, this GC method may be time-consuming, labor-intensive, and environmentally unsustainable, so a new methodology is needed to analyze vegetable oils in more efficient and non-destructive ways.
Vegetable oils consist of organic molecules containing carbon, hydrogen, oxygen, and other elements whose vibrational frequencies are located in the near-infrared region of the electromagnetic spectrum (Wu et al., 2009; Yang et al., 2005). As a result, a chemometric method with near-infrared spectroscopy has been recently used to characterize edible oils by investigating the characteristic absorption and intensity of near-infrared radiation. Moh et al. (1999) developed a near-infrared spectroscopic technique to measure the peroxide values of refined palm oils, showing a high correlation coefficient between the spectral values and the peroxide values (R2 = 0.994). He et al. (2020) and Jiang et al. (2021) successfully verified the feasibility of near-infrared spectroscopy to detect the storage periods of 4 different edible oils and their acid values during storage, respectively. Near-infrared spectroscopic techniques were also used to detect the adulteration of edible oils such as extra-virgin olive (Vanstone et al., 2018), coriander (Kaufmann et al., 2022), and Sacha inchi (Plukenetia volubilis) (Cruz-Tirado et al., 2023) oils. Furthermore, near-infrared spectroscopic analysis has been combined with other techniques such as hyperspectral imaging in order to enhance the capabilities and versatility of analytical techniques (Li et al., 2020b). As a cutting-edge technology, hyperspectral imaging allows for the non-destructive and rapid analysis of the chemical properties of materials and their spatial distribution (Aviara et al., 2022). This technique has been employed in the analysis of diverse agricultural and food commodities, encompassing oils. There are several preceding studies on the use of hyperspectral imaging analysis in characterizing the qualities of oil seeds. Using hyperspectral imaging, Jin et al. (2016) determined the oil content in peanuts and Da Silva Medeiros et al. (2022) quantified the level of oil and fatty acids in Brassicas seeds. As the demand for premium vegetable oils rises, hyperspectral imaging analysis started to gain scientific attention as an analytical tool to detect the adulteration of vegetable oils. Thus, different varieties of sesame oil (Kim et al., 2010; Xie et al., 2014) and extra virgin olive oil with cheaper edible oils (Malavi et al., 2023) were identified from their hyperspectral images. The primary emphasis in hyperspectral analysis, nonetheless, has centered on the detection of adulteration in high-value vegetable oils, despite the widespread utilization of various vegetable oils within the food industry.
Machine learning, which is an area of artificial intelligence, focuses on creating algorithms that enable computers to analyze data, identify patterns, and make predictions (Deng et al., 2021). It is recognized that there are three main types of machine learning algorithms: supervised learning, unsupervised learning, and reinforcement learning (Jin, 2020). Supervised learning uses labeled input data to learn from and make predictions. On the other hand, unsupervised learning discovers patterns and structures with unlabeled input data mainly for clustering and dimensionality reduction. In the case of reinforcement learning, it involves an agent interacting with an environment to learn through rewards and punishments. More recently, machine learning started to be combined with the hyperspectral imaging analysis. Lu et al. (2017) have successfully applied machine learning techniques to hyperspectral images for predicting rice starch content with an accuracy of over 86.9%. A hyperspectral technique coupled with machine learning was also applied to rapidly discriminate the adulteration of sesame and rapeseed oils, showing an accuracy of 100% with random forest (Weng et al., 2019). Also, Mishra et al. (2022) associated the content of aflatoxin B1 in single-kernel almonds with the hyperspectral data using partial least squares regression. More recently, two machine learning algorithms including support vector machine and linear discriminant analysis were employed to qualitative discriminate the authenticity of the camellia seed oil, reaching 100% accuracy (Rady and Adedeji, 2020). However, research that integrates hyperspectral imaging with a wider variety of machine learning models for categorizing edible vegetable oils from diverse botanical sources is still limited. Moreover, from a machine learning perspective, hyperspectral imaging techniques have not yet been systematically compared with chemical methods such as gas chromatography which have been still dominantly used in the food industry, to our best knowledge.
In this study, eight edible vegetable oils were subjected to hyperspectral imaging analysis, which was coupled to machine learning algorithms for identifying the edible vegetable oils. The classification performance of the three machine learning models (decision tree, random forest, and k-nearest neighbor) was compared with that of the fatty acid composition-based chemical methods, based on accuracy and f1-score values. Furthermore, the degree of adulteration in vegetable oil blends was investigated using hyperspectral imaging analysis combined with machine learning.
2. Materials and methods
2.1. Materials
Eight edible vegetable oils – canola (Sajo Haepyo Co., Ltd., Seoul, Korea), corn (Sajo Haepyo Co., Ltd., Seoul, Korea), grapeseed (Sajo Haepyo Co., Ltd., Seoul, Korea), olive (Sajo Haepyo Co., Ltd., Seoul, Korea), soybean (CJ Beksul Co., Ltd, Seoul, Korea), palm (Lotte Foods Co., Ltd., Seoul, Korea), flaxseed (Goccia d'oro, Baldissero d’Alba, Italy), and sunflower (Sajo Haepyo Co., Ltd., Seoul, Korea) oils were purchased from a commercial source. All chemicals used in this study were of analytical grade.
2.2. Determination of fatty acid composition
The fatty acid compositions of the vegetable oils were analyzed using a gas chromatograph coupled with an Agilent 5975 series mass selective detector (Santa Clara, CA, USA). For saponification and derivatization of fatty acids, oil (4 mg) was dissolved in 2 mL of 0.5 M KOH in methanol in a 20 mL glass vial and incubated at 80 °C for 60 min, followed by a reaction with 2 mL of 10% BF3-methanol solution (Sigma-Aldrich, St. Louis, MO, USA) at 100 °C for 20 min. The reaction was terminated by adding deionized water (4 mL) and hexane (2 mL). The reaction mixture was then vigorously vortexed and centrifuged. The organic phase was separated, and the residual aqueous phase was further washed with fresh hexane (2 mL) twice. The collected organic phases were combined and evaporated under a vacuum. The fatty acid methyl esters (FAMEs) were dissolved in 1 mL of hexane and filtered using a 0.45 μm PVDF syringe filter. The filtrate was suitably diluted with hexane and subjected to GC/MS (6890N, Agilent Technologies, Santa Clara, CA, USA). An HP-INOWAX capillary column (30 m × 0.32 mm × 0.25 μm, Agilent Technologies) was used to separate FAMEs, and purified helium was used as a carrier gas at a flow rate of 2 mL/min. The injector was operated in a split mode at 250 °C. The GC oven temperature was programmed to initially hold at 100 °C for 5 min, rise to 250 °C at a rate of 3 °C/min, and finally hold for 5 min. The mass detector was operated in a scan mode in the range of 50–700 m/z with an ionization energy of 70 eV. The peak deconvolution and identification of the FAMEs were performed using AMDIS software with NIST 11 mass spectral and retention index libraries (Ver 2.71, National Institute of Standards and Technology). The concentrations of FAMEs were estimated using their corresponding FAME calibration curves constructed using FAME Mix C8 – C24 reference standards (Supelco Analytical, Bellefonte, PA, USA) in hexane.
2.3. Hyperspectral measurement
The hyperspectral images of the oil samples were acquired with a Specim FX17 camera (Spectral Imaging Ltd, Oulu, Finland), which provided 224 spectral bands over a wavelength range of 900–1700 nm. Each oil sample (45 g) in a Petri dish was presented on a mobile platform illuminated using two 170 W halogen lamps. The moving speed of the mobile platform was 40 mm/s, the exposure time was 4 ms, and the objective distance was 315 cm. The hyperspectral system was located in a dark room to prevent interference from external light sources. The corrected images (R) were obtained by calibrating the original images (OI) of the samples, the black image (B) was acquired by covering the camera lens with its cap, and the white image (W) was obtained with a standard white bar.
The imaging processing of the hyperspectral images was conducted by ENVI software (L3Harris Geospatial Solutions, Broomfield, CO, USA) and Python interface (3.8.13 version) with the spectral module (Spy, 0.21 version). A region of interest (ROI) with a size of 25 × 25 pixels was obtained from the center of the calibrated hyperspectral images and smoothed by employing the Savitzky-Golay filter (Yang et al., 2015).
The hyperspectral signals were then transformed into an absorbance profile as follows.
| A (absorbance) = -log10R |
2.4. Machine learning analysis
Machine learning models for oil classification were constructed in a Python programming environment with a Jupyter Notebook. The fatty acid compositions and hyperspectral images obtained were used as the machine learning datasets. In the fatty acid composition dataset (117 × 9 matrix), the fatty acid compositions and the oil types were assigned to X and Y vectors, respectively. In addition, 224 hyperspectral bands were assigned to X vectors in the hyperspectral dataset (75,000 × 225 matrix). The min-max normalization library was applied to convert the X data into a fixed range of 0–1. The fatty acid composition and hyperspectral results were subjected to linear discriminant analysis (LDA) which is a linear model for classification and dimensionality reduction (Bandos et al., 2009; Giansante et al., 2003). The LDA was directly applied to the pixels from the hyperspectral cube (25 × 25 x 224) where the number of samples was greater than the number of features in the hyperspectral datasets (Setser and Smith, 2018). The Python scikit-learn and matplot libraries were utilized for conducting LDA and visualizing the corresponding results.
Three different machine learning classification models (decision tree, random forest, and k-nearest neighbor) were furthermore utilized to classify the oil samples based on their fatty acid composition and hyperspectral imaging results. The models of the decision tree, random forest, and k-nearest neighbor were trained with the optimal hyperparameters selected using a bayesian optimization process - (max_depth = None, min_samples_leaf = 1, and min_samples_split = 2), (max_depth = None, min_samples_leaf = 1, and min_samples_split = 2), and (the number of neighbors = 10, metric = ‘minkowski’, leaf_size = 100, p = 1, and weights = ‘distance’), respectively. The classification performance of the proposed models was assessed in terms of accuracy and F1-score, which were determined from the confusion matrix as follows (Tharwat, 2021).
where TP is true positive, FP is false positive, TN is true negative, and FN is false negative.
The three machine learning algorithms were furthermore applied in order to predict the adulteration levels in two paired edible oils (olive/canola oils and sunflower/grapeseed oils). For each pair, 11 mixtures were prepared by blending the two oil samples at a mass ratio of 0:100, 10:90, 20:80, 30:70, 40:60, 50:50, 60:40, 70:30, 80:20, 90:10, and 0:100 (w/w), which were expressed as concentration of olive oil in canola oil and sunflower oil in grapeseed oil (0%, 10%, 20%, …, 90%, and 100%). The model hyperparameters were optimally determined using a bayesian algorithm (decision tree: max_depth = 96, min_samples_leaf = 5, and min_samples_split = 2), (random forest: max_depth = 5, min_samples_leaf = 1, and min_samples_split = 2), and (k-nearest neighbor: the number of neighbors = 5, metric = ‘minkowski’, leaf_size = 30, p = 2, and weights = ‘uniform’). The coefficient of determination (R2) and the root mean squared error (RMSE) were applied to evaluate the prediction performance of the models.
2.5. Statistical analysis
The fatty acid composition and hyperspectral experiments of the oil samples were carried out twelve and fifteen times, respectively, and the values were reported as mean ± standard deviation.
3. Results and discussion
The fatty acid compositions of eight vegetable oils were analyzed as shown in Table 1. Oleic acid was the primary fatty acid in the canola, olive, and palm oils, whereas the grapeseed, sunflowerseed, soybean, and corn oils had the highest level of linoleic acid. Also, linolenic acid was predominantly detected in the flaxseed oil. The palm oil contained a high level of palmitic acid. These results were in a great agreement with several preceding studies that analyzed the fatty acid compositions of various vegetable oils (Giakoumis, 2018; Kim et al., 2010; Mancini et al., 2015). All vegetable oils except for palm oil mostly consisted of unsaturated fatty acids. Especially, the canola, flaxseed, and grapeseed oils showed the lowest ratio of saturated to unsaturated fatty acids, whereas the highest ratio was observed in the palm oil, which behaves like a semi-solid material at room temperature (Norhaizan et al., 2013).
Table 1.
Fatty acid compositions of various vegetable oils.
| Fatty acid | Grapeseed | Canola | Olive | Sunflowerseed | Soybean | Palm | Corn | Flaxseed |
|---|---|---|---|---|---|---|---|---|
| Myristic acid (14:0) | – | – | – | – | – | 1.49 ± 0.14 | – | – |
| Palmitic acid (16:0) | 5.12 ± 0.47 | 3.81 ± 0.11 | 9.11 ± 1.55 | 5.59 ± 0.99 | 8.08 ± 0.69 | 39.14 ± 3.28 | 7.41 ± 1.09 | 3.97 ± 0.69 |
| Palmitoleic acid (16:1) | – | 0.22 ± 0.14 | 0.95 ± 0.19 | – | – | – | – | – |
| Stearic acid (18:0) | 3.28 ± 0.29 | 2.21 ± 0.52 | 4.09 ± 0.22 | 3.97 ± 0.52 | 3.88 ± 0.85 | 5.80 ± 0.39 | 1.93 ± 0.12 | 3.87 ± 0.64 |
| Oleic acid (18:1) | 22.46 ± 0.96 | 52.69 ± 0.48 | 74.32 ± 1.42 | 31.37 ± 2.31 | 21.41 ± 1.52 | 42.10 ± 3.34 | 28.02 ± 0.63 | 13.82 ± 0.44 |
| Linoleic acid (18:2) | 69.13 ± 1.04 | 27.75 ± 0.34 | 8.41 ± 1.94 | 59.07 ± 2.29 | 59.17 ± 2.04 | 11.48 ± 0.49 | 60.64 ± 0.46 | 21.85 ± 0.35 |
| Linolenic acid (18:3) | – | 13.06 ± 0.22 | 1.76 ± 0.75 | – | 7.46 ± 0.74 | – | 1.99 ± 0.23 | 56.48 ± 0.74 |
| Arachidic acid (20:0) | – | 0.26 ± 0.20 | 1.37 ± 0.60 | – | – | – | – | – |
| Saturated fatty acid | 8.40 | 6.28 | 14.57 | 9.56 | 11.96 | 46.43 | 9.34 | 7.84 |
| Unsaturated fatty acid | 91.59 | 93.72 | 85.44 | 90.44 | 88.04 | 53.58 | 90.65 | 92.15 |
| Saturated/Unsaturated fatty acid | 0.09 | 0.07 | 0.17 | 0.11 | 0.13 | 0.87 | 0.10 | 0.09 |
The quality attributes of various edible vegetable oils have been widely characterized using near-infrared spectroscopy. A hyperspectral imaging technique that operates in the near-infrared region, was thus utilized to identify the oil samples that show distinct absorption peaks derived from specific molecular vibrations such as C–H functional groups (Li et al., 2020a). Fig. 1 exhibits the average hyperspectral absorbance spectra of the vegetable oils. The liquid oils seemed to have similar absorbance spectra, while those of the palm oil were distinctly different. The spectra of the vegetable oils were mainly characterized by three absorption bands at 1212, 1415, and 1671 nm (Dong et al., 2022). These spectral patterns were similar to those reported by Chu et al. (2018) and Troshchynska et al. (2019) who determined the near-infrared spectra of camellia and flaxseed oils, respectively. Similarly, significant peaks at 1200 and 1450 nm were found in palm and sea buckthorn seed oils whose near-infrared spectra were investigated in the wavelength of 950–1650 nm (Basri et al., 2017) and 1000–2500 nm (Li et al., 2016), respectively. In addition, Borghi et al. (2020) reported that vegetable oils high in unsaturated fatty acids showed an intense peak at around 1170 nm. Therefore, small peaks that were observed at nearly 1170 nm might be associated with –HC CH- and their intensity seemed to be more dominant in the vegetable oils with a high degree of unsaturation, compared to the palm oil. (Caporaso et al., 2018). Among the edible vegetable oils tested, the signal intensities at 1170 and 1671 nm were the highest in the flaxseed oil, while those at 1212 and 1415 nm were the most dominant in the olive oil.
Fig. 1.
Hyperspectral absorbance curves of various vegetable oils.
Fig. 2 demonstrates the 2-dimensional plots from the linear discriminant analysis (LDA) of the fatty acid composition and the hyperspectral imaging datasets. In Fig. 2, the x- and y-axis display the total variance explained by the first and second linear discriminants, respectively. Thus, 96.0% and 98.9% of the total variation in the fatty acid composition and hyperspectral datasets, respectively, could be explained by the two linear discriminants. When the oil samples were described by different symbols in Fig. 2, the clusters of the samples were visually observed based on their similarity. Fig. 2(a) shows that the two linear discriminants from the fatty acid compositions clearly separated the flaxseed oil high in linolenic acid and low in oleic acid, compared to the other oil samples. In addition, the canola, olive, palm, and soybean oils seemed to be separable while there seemed to be overlapping among the grapeseed, corn, and sunflowerseed oil samples with similar fatty acid compositions (Table 1). Fig. 2(b) exhibits the LDA score plot from the hyperspectral imaging dataset. It was very interesting to note that the flaxseed, olive, and palm oils were distinctly separable in the hyperspectral LDA plot, like the fatty acid composition dataset (Fig. 2(a)). About 98.9 % of the total variance of the data was displayed in this LDA plot, showing that the first and second linear discriminant accounted for 90.7% and 8.2% of the variance, respectively. Unlike the fatty acid composition dataset, the palm oils in the LDA score plot were positively positioned on the first linear discriminant, while the other oil samples were negatively positioned. A fairly distinct separation appeared to be achieved in the palm, flaxseed, and olive oils. Especially, the flaxseed and olive oils were located in the positive and negative region of y-axis, respectively, whereas the others were positioned in the middle. This distinct separation of the flaxseed and olive oils might be related to the signal intensities differences in their hyperspectral spectra as mentioned in Fig. 1. However, it appeared that the oil samples other than palm, flaxseed, and olive oils were not visually separable. As a supervised method, LDA seemed to be effective in visualizing the clusters of the oil samples, since it maximizes the variability between the classes and reduces the variability within the classes (Xing et al., 2019).
Fig. 2.
Linear discriminant analysis of oil features ((a) fatty acid compositions and (b) hyperspectral images).
Fig. 3 exhibits the experimental procedure to utilize machine learning analysis in classifying edible vegetable oils. As shown in Fig. 3, the fatty acid composition and the hyperspectral results were divided into training and testing datasets, which were used for training the models and for evaluating the model performance, respectively. The datasets were then subjected to three different machine learning algorithms (decision tree, random forest, and k-nearest neighbor). The classification performances of the models were evaluated in terms of confusion matrix, accuracy, and F1-score.
Fig. 3.
Schematic diagram of machine learning classification of vegetable oils.
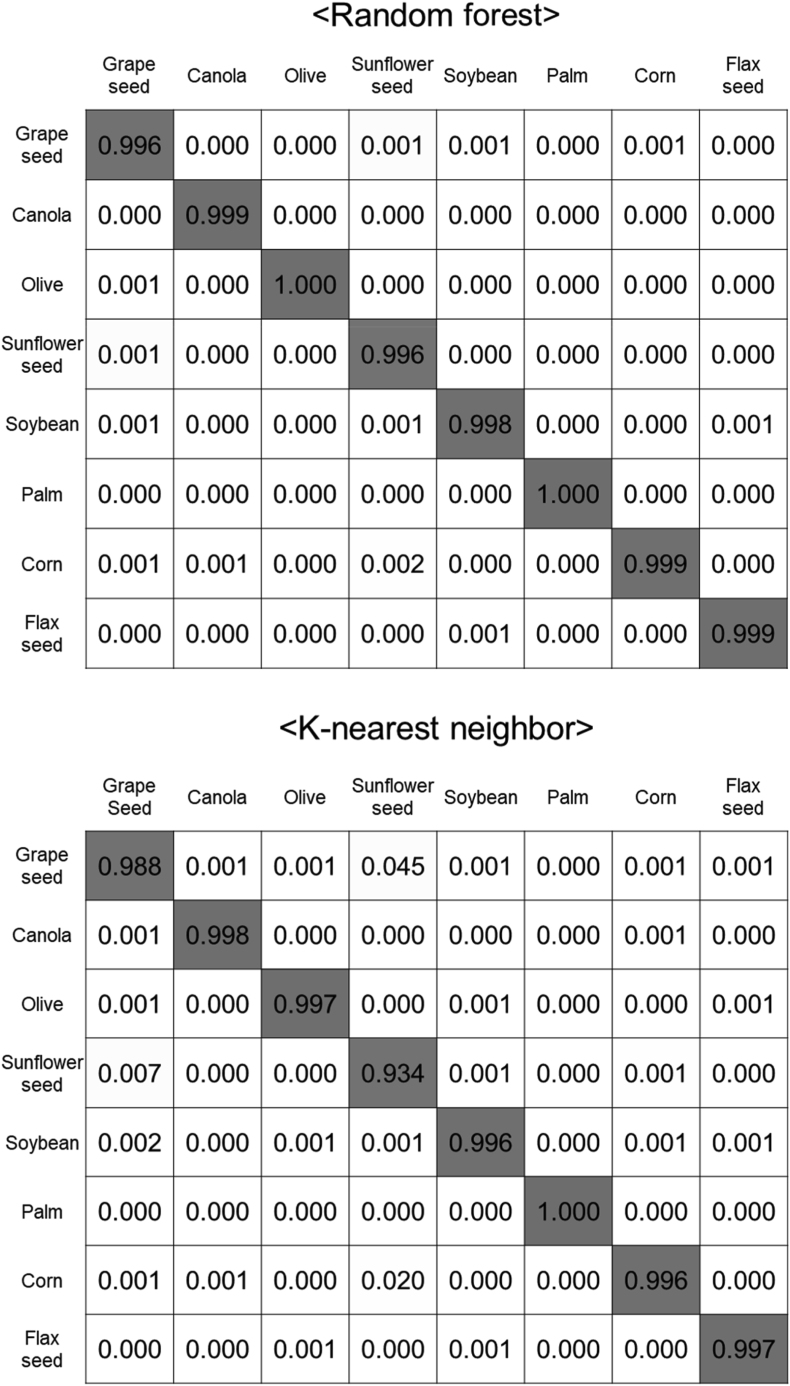
A confusion matrix is a tabular way of visualizing the performance of a classification model (Krstinić et al., 2020; Ortega et al., 2020; Valero-Carreras et al., 2023). Fig. 4 presents the confusion matrices of the three different models developed, based on the fatty acid composition and hyperspectral results. The rows in the confusion matrix represent the true values, the columns represent the predicted values, and the diagonals indicate the correct predictions. As can be seen in Fig. 4(a), the classification performance of the decision tree became slightly lower for sunflower, soybean, and flaxseed oils. In the case of random forest and k-nearest neighbor algorithms, several instances to incorrectly classify sunflower oils were detected. However, all three classifiers showed a fairly decent classification ratio higher than 0.92, demonstrating that they were effective in correctly identifying the oil samples based on their fatty acid compositions. Fig. 4(b) presents the confusion matrices of the machine learning models based on the hyperspectral images. More instances of incorrectly classifying the oil samples were detected in the decision tree and k-nearest neighbor models, while most of the oil samples were successfully classified by the random forest. In the decision tree and k-nearest neighbor models, the grapeseed and sunflower oils were frequently confused with each other. While the soybean oils were misclassified as grapeseed and sunflower oils, the corn oils were mispredicted as sunflower oils. However, there were few cases to misclassify olive, palm, and flaxseed oils, regardless of the classification models. These trends were in a good agreement with the score plot of the LDA (Fig. 2) where these three oil samples (olive, palm, and flaxseed oils) were distinctly isolated from the other oil samples.
Fig. 4.
Confusion matrix of machine learning classifiers based on (a) fatty acid composition dataset and (b) hyperspectral image dataset.
The classification performances of the machine learning algorithms were furthermore compared in terms of their accuracy and F1-score (Table 2). When the fatty acid compositions were used as a dataset, the accuracies were found to be more than 97.5% regardless of the machine learning models. The use of the hyperspectral imaging results as a machine learning dataset seemed to slightly improve the performance of the classification models by showing great classification success of more than 98.9%. The decision tree algorithm showed relatively low accuracies for the two experimental datasets, whereas both the random forest and k-nearest neighbor exhibited a higher classification performance. As also presented in Table 2, f1-scores were used as a measure to evaluate the performance of the classification models. In the case of the fatty acid composition dataset, the f1-score values of the decision tree, random forest, and k-nearest neighborhood were determined to be 0.98, 0.99, and 0.99, respectively. Like the fatty acid composition dataset, the f1-score values were high in the random forest and k-nearest neighbor, followed by decision tree, however the f1-score values were almost close to 1.0 for all the classification models with the hyperspectral datasets. A decision tree algorithm has a flowchart-like tree structure for both classification and regression problems (Patel and Prajapati, 2018). Random forest is an ensemble classifier that is composed of multiple decision trees that are independently trained on a random subset of data, making predictions based on majority voting (Schonlau and Zou, 2020). K-nearest neighbor is a non-parametric supervised learning classifier for estimating the likelihood that a data point belongs to one group or another based on how its neighbors are classified (Cunningham and Delany, 2021). Compared to the decision tree, the superb classification of random forest and k-nearest neighbor could be attributed to their ability to classify the samples by taking a majority of votes in the form of ensemble learning (Sinta et al., 2014; Zhang and Suganthan, 2014). Several preceding studies reported the better classification performance of k-nearest neighbor and random forest in classifying breast cancer and in identifying associated risk factors for type 2 diabetes, respectively (Esmaily et al., 2018; Rajaguru and SR, 2019). As a result, the machine learning models based on the hyperspectral results showed the performance of oil classification comparable to the fatty acid composition-based chemical method.
Table 2.
Classification performance of machine learning classifiers.
| Accuracy (%) | Decision tree | Random forest | K-nearest neighbor |
|---|---|---|---|
| Fatty acid composition | 0.975 ± 0.023 | 0.992 ± 0.019 | 0.992 ± 0.019 |
| Hyperspectral images | 0.989 ± 0.006 | 0.999 ± 0.000 | 0.990 ± 0.001 |
|
F1-score |
Decision tree |
Random forest |
K-nearest neighbor |
| Fatty acid composition | 0.979 ± 0.022 | 0.992 ± 0.017 | 0.992 ± 0.017 |
| Hyperspectral images | 0.989 ± 0.006 | 0.999 ± 0.000 | 0.990 ± 0.001 |
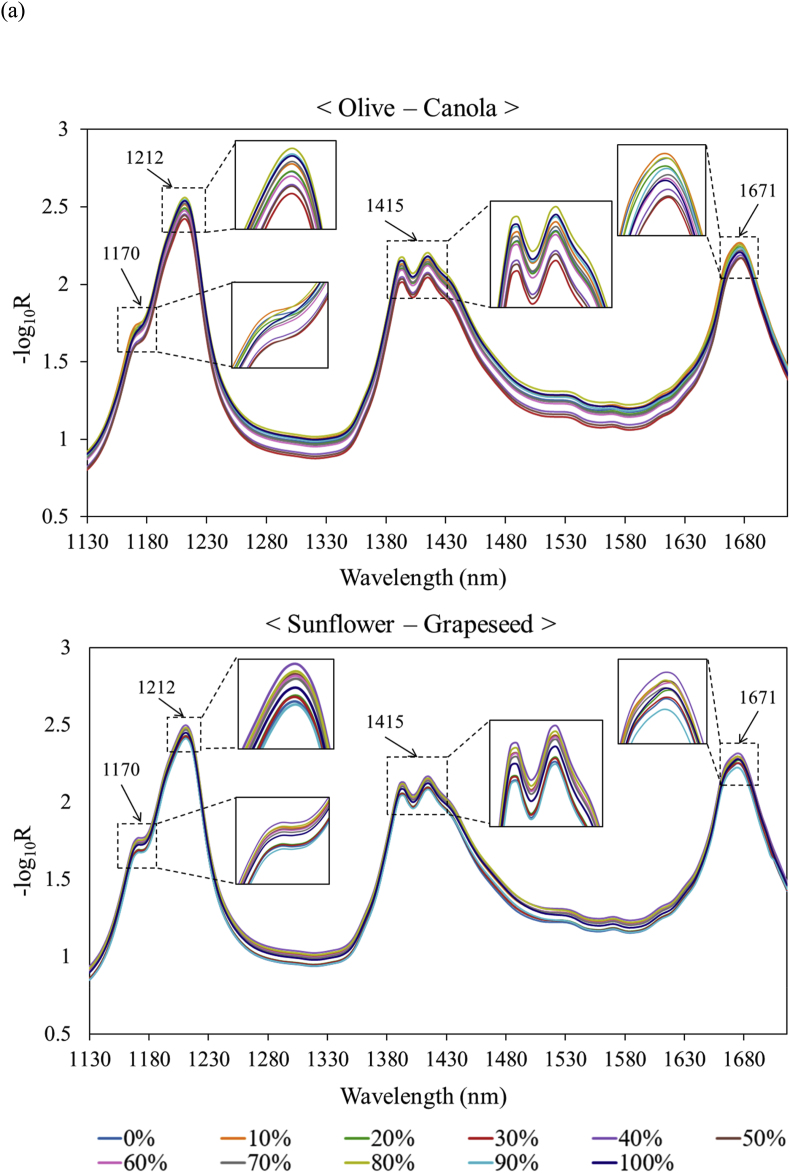
The hyperspectral images of two binary oil blends (olive/canola oils and sunflower/grapeseed oils) were measured to ascertain adulteration levels by quantifying the concentration of an oil sample within the oil blends. Olive and canola oils were selected based on their fatty acid compositions (distinct saturated/unsaturated fatty acid ratios), as evidenced in Table 1. Also, given the propensity for confusion between sunflower and grapeseed oils by machine learning models, as illustrated in Fig. 4, both oils also underwent hyperspectral imaging analysis for the detection of adulteration. Fig. 5(a) shows the average hyperspectral absorbance spectra of the two oil blends. When their hyperspectral results were subjected to LDA (Fig. 5(b)), 97.4% and 98.4% of the total variability was explained by the first and second components for olive/canola oils and sunflower/grapeseed oils, respectively. The first linear discriminant appeared to separate the oil samples according to adulteration levels. Thus, the samples with a higher proportion of canola and grapeseed oils were located in the positive zone of the first linear discriminants, while those containing a higher proportion of olive and sunflower oils were located in the negative region.
Fig. 5.
Hyperspectral absorbance curves of vegetable oil blends (a) and their linear discriminant analysis plots (b).
Hyperspectral imaging technology has been applied to determine the adulteration levels of food ingredients. Malavi et al. (2023) successfully utilized hyperspectral imaging for quantifying adulteration in extra virgin olive oils blended with various oils at different concentrations up to 20% (R2 = 0.97), and Zhao et al. (2018) quantitatively detected peanut and walnut powders in whole wheat flour using hyperspectral imaging (with a determination coefficient of prediction (R2 = 0.987). Thus, the hyperspectral results experimentally measured were applied to the three machine learning models that provide both regression and classification tasks as also shown in Fig. 3. Fig. 6 exhibits the scatter plots comparing the actual concentrations of olive/canola oils and sunflower/grapeseed oils with the predicted ones. Overall, it seemed that they provided decent prediction performance, as evidenced by higher R2 (0.974–0.997) and lower RMSE (1.81–4.90) values. In particular, the random forest model gave rise to high prediction performance for both oil blends, compared to decision tree and k-nearest neighbor models.
Fig. 6.
Actual versus predicted concentration scatter plots of vegetable oils.
Although hyperspectral imaging is particularly useful for heterogeneous samples with distinct regions, its application to homogeneous samples such as edible oils can still be justified for quality control, adulteration detection, and scientific research. Its ability to reveal hidden variations and provide valuable spatial information in a non-destructive and rapid way, makes it a versatile analytical tool, even in cases where samples appear uniform.
4. Conclusion
Hyperspectral analysis coupled with machine learning was applied as a non-destructive method for classifying eight edible vegetable oils from different plant sources, and its classification performance was compared with that of the fatty acid composition-based chemical results. The linear discriminant analysis showed that two linear discriminants were appropriate to explain 96.0% and 98.9% of the total variation in the fatty acid composition and hyperspectral imaging datasets, respectively. The random forest offered performance advantages in the classification tasks of vegetable oils with the best accuracy and F1-score values, compared to the decision tree and k-nearest neighbor models. In addition, the quantitative investigation of adulteration levels in vegetable oils was successfully conducted utilizing hyperspectral methodologies facilitated by machine learning techniques. Thus, this study showed promising results of utilizing hyperspectral imaging combined with machine learning as an alternative to the conventional chemical method for oil classification and adulteration detection. This can provide a highly accurate method for predicting the type and level of unknown oil samples, consequently helping food manufacturers to identify and control the potential quality issues of vegetable oils.
CRediT authorship contribution statement
Jeongin Hwang: Conceptualization, Methodology, Formal analysis, Investigation, Writing – original draft, Project administration. Kyeong-Ok Choi: Formal analysis, Investigation, Writing – review & editing. Sungmin Jeong: Conceptualization, Methodology, Formal analysis, Investigation, Writing – original draft, Writing – review & editing, Supervision, Validation. Suyong Lee: Conceptualization, Methodology, Writing – original draft, Supervision, Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Ministry of Science and ICT (2022R1A2C1002813) and the Ministry of Education (2022R1A6A1A03055869).
Handling Editor: Aiqian Ye
Contributor Information
Sungmin Jeong, Email: sungmin@sejong.ac.kr.
Suyong Lee, Email: suyonglee@sejong.ac.kr.
Data availability
Data will be made available on request.
References
- Aviara N.A., Liberty J.T., Olatunbosun O.S., Shoyombo H.A., Oyeniyi S.K. Potential application of hyperspectral imaging in food grain quality inspection, evaluation and control during bulk storage. J Agric Food Res. 2022;8 doi: 10.1016/j.jafr.2022.100288. [DOI] [Google Scholar]
- Bandos T.V., Bruzzone L., Camps-Valls G. Classification of hyperspectral images with regularized linear discriminant analysis. IEEE Geosci Remote Sens. 2009;47(3):862–873. doi: 10.1109/TGRS.2008.2005729. [DOI] [Google Scholar]
- Basri K.N., Hussain M.N., Bakar J., Sharif Z., Khir M.F.A., Zoolfakar A.S. Classification and quantification of palm oil adulteration via portable NIR spectroscopy. Spectrochim. Acta Mol. Biomol. Spectrosc. 2017;173:335–342. doi: 10.1016/j.saa.2016.09.028. [DOI] [PubMed] [Google Scholar]
- Borghi F.T., Santos P.C., Santos F.D., Nascimento M.H., Correa T., Cesconetto M., Pires A.A., Ribeiro A.V., Lacerda Jr V., Romao W. Quantification and classification of vegetable oils in extra virgin olive oil samples using a portable near-infrared spectrometer associated with chemometrics. Microchem. J. 2020;159 doi: 10.1016/j.microc.2020.105544. [DOI] [Google Scholar]
- Caporaso N., Whitworth M.B., Grebby S., Fisk I.D. Rapid prediction of single green coffee bean moisture and lipid content by hyperspectral imaging. J. Food Eng. 2018;227:18–29. doi: 10.1016/j.jfoodeng.2018.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu X., Wang W., Li C., Zhao X., Jiang H. Identifying camellia oil adulteration with selected vegetable oils by characteristic near-infrared spectral regions. J Innov Opt Health Sci. 2018;11(2) doi: 10.1142/S1793545818500062. [DOI] [Google Scholar]
- Cruz-Tirado J., Muñoz-Pastor D., de Moraes I.A., Lima A.F., Godoy H.T., Barbin D.F., Siche R. Comparing data driven soft independent class analogy (DD-SIMCA) and one class partial least square (OC-PLS) to authenticate sacha inchi (Plukenetia volubilis L.) oil using portable NIR spectrometer. Chemometr. Intell. Lab. Syst. 2023;242 doi: 10.1016/j.chemolab.2023.105004. [DOI] [Google Scholar]
- Cunningham P., Delany S.J. K-nearest neighbour classifiers-A tutorial. ACM Comput. Surv. 2021;54(6):1–25. doi: 10.1145/3459665. [DOI] [Google Scholar]
- Da Silva Medeiros M.L., Cruz-Tirado J., Lima A.F., de Souza Netto J.M., Ribeiro A.P.B., Bassegio D., Godoy H.T., Barbin D.F. Assessment oil composition and species discrimination of Brassicas seeds based on hyperspectral imaging and portable near infrared (NIR) spectroscopy tools and chemometrics. J. Food Compos. Anal. 2022;107 doi: 10.1016/j.jfca.2022.104403. [DOI] [Google Scholar]
- Deng X., Cao S., Horn A.L. Emerging applications of machine learning in food safety. Annu. Rev. Food Sci. Technol. 2021;12:513–538. doi: 10.1146/annurev-food-071720-024112. [DOI] [PubMed] [Google Scholar]
- Dong F., Bi Y., Hao J., Liu S., Lv Y., Cui J., Wang S., Han Y., Rodas-González A. A combination of near-infrared hyperspectral imaging with two-dimensional correlation analysis for monitoring the content of alanine in beef. Biosensors. 2022;12(11):1043. doi: 10.3390/bios12111043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esmaily H., Tayefi M., Doosti H., Ghayour-Mobarhan M., Nezami H., Amirabadizadeh A. A comparison between decision tree and random forest in determining the risk factors associated with type 2 diabetes. J Health Sci Res. 2018;18(2):412. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7204421/ [PubMed] [Google Scholar]
- Giakoumis E.G. Analysis of 22 vegetable oils' physico-chemical properties and fatty acid composition on a statistical basis, and correlation with the degree of unsaturation. Renew. Energy. 2018;126:403–419. doi: 10.1016/j.renene.2018.03.057. [DOI] [Google Scholar]
- Giansante L., Di Vincenzo D., Bianchi G. Classification of monovarietal Italian olive oils by unsupervised (PCA) and supervised (LDA) chemometrics. J. Sci. Food Agric. 2003;83(9):905–911. doi: 10.1002/jsfa.1426. [DOI] [Google Scholar]
- He Y., Jiang H., Chen Q. High-precision identification of the actual storage periods of edible oil by FT-NIR spectroscopy combined with chemometric methods. Anal. Methods. 2020;12(29):3722–3728. doi: 10.1039/D0AY00779J. [DOI] [PubMed] [Google Scholar]
- Jiang H., He Y., Chen Q. Determination of acid value during edible oil storage using a portable NIR spectroscopy system combined with variable selection algorithms based on an MPA‐based strategy. J. Sci. Food Agric. 2021;101(8):3328–3335. doi: 10.1002/jsfa.10962. [DOI] [PubMed] [Google Scholar]
- Jin H., Ma Y., Li L., Cheng J.-H. Rapid and non-destructive determination of oil content of peanut (Arachis hypogaea L.) using hyperspectral imaging analysis. Food Anal. Methods. 2016;9:2060–2067. doi: 10.1007/s12161-015-0384-3. [DOI] [Google Scholar]
- Jin W. Research on machine learning and its algorithms and development. J Phys: Conf Ser. 2020;1544 doi: 10.1088/1742-6596/1544/1/012003. [DOI] [Google Scholar]
- Kaufmann K.C., Sampaio K.A., García-Martín J.F., Barbin D.F. Identification of coriander oil adulteration using a portable NIR spectrometer. Food Control. 2022;132 doi: 10.1016/j.foodcont.2021.108536. [DOI] [Google Scholar]
- Kim J., Kim D.N., Lee S.H., Yoo S.-H., Lee S. Correlation of fatty acid composition of vegetable oils with rheological behaviour and oil uptake. Food Chem. 2010;118(2):398–402. doi: 10.1016/j.foodchem.2009.05.011. [DOI] [Google Scholar]
- Krstinić D., Braović M., Šerić L., Božić-Štulić D. International Conference on Soft Computing, Artificial Intelligence and Machine Learning. 2020. Multi-label classifier performance evaluation with confusion matrix. Copenhagen, Denmark. [DOI] [Google Scholar]
- Li X., Chen K., He Y. In situ and non-destructive detection of the lipid concentration of scenedesmus obliquus using hyperspectral imaging technique. Algal Res. 2020;45 doi: 10.1016/j.algal.2019.101680. [DOI] [Google Scholar]
- Li X., Zhang L., Zhang Y., Wang D., Wang X., Yu L., Zhang W., Li P. Review of NIR spectroscopy methods for nondestructive quality analysis of oilseeds and edible oils. Trends Food Sci. Technol. 2020;101:172–181. doi: 10.1016/j.tifs.2020.05.002. [DOI] [Google Scholar]
- Li Z., Wang J., Xiong Y., Li Z., Feng S. The determination of the fatty acid content of sea buckthorn seed oil using near infrared spectroscopy and variable selection methods for multivariate calibration. Vib. Spectrosc. 2016;84:24–29. doi: 10.1016/j.vibspec.2016.02.008. [DOI] [Google Scholar]
- Lu X., Sun J., Mao H., Wu X., Gao H. Quantitative determination of rice starch based on hyperspectral imaging technology. Int. J. Food Prop. 2017;20(Suppl. 1):S1037–S1044. doi: 10.1080/10942912.2017.1326058. [DOI] [Google Scholar]
- Malavi D., Nikkhah A., Raes K., Van Haute S. Hyperspectral imaging and chemometrics for authentication of extra virgin olive oil: a comparative approach with FTIR, UV-VIS, Raman, and GC-MS. Foods. 2023;12(3):429. doi: 10.3390/foods12030429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancini A., Imperlini E., Nigro E., Montagnese C., Daniele A., Orrù S., Buono P. Biological and nutritional properties of palm oil and palmitic acid: effects on health. Molecules. 2015;20(9):17339–17361. doi: 10.3390/molecules200917339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra G., Panda B.K., Ramirez W.A., Jung H., Singh C.B., Lee S.-H., Lee I. Application of SWIR hyperspectral imaging coupled with chemometrics for rapid and non-destructive prediction of aflatoxin B1 in single kernel almonds. LWT. 2022;155 doi: 10.1016/j.lwt.2021.112954. [DOI] [Google Scholar]
- Moh M., Che Man Y., Van de Voort F., Abdullah W. Determination of peroxide value in thermally oxidized crude palm oil by near infrared spectroscopy. J. Am. Oil Chem. Soc. 1999;76(1):19–23. doi: 10.1007/s11746-999-0042-2. [DOI] [Google Scholar]
- Norhaizan M.E., Hosseini S., Gangadaran S., Lee S.T., Kapourchali F.R., Moghadasian M.H. Palm oil: features and applications. Lipid Technol. 2013;25(2):39–42. doi: 10.1002/lite.201300254. [DOI] [Google Scholar]
- Ortega J., Lagman A.C., Natividad L.R.Q., Bantug E.T., Resureccion M.R., Manalo L. Analysis of performance of classification algorithms in mushroom poisonous detection using confusion matrix analysis. Int. J. Adv. Trends Comput. Sci. Eng. 2020;9(1.3):451–456. doi: 10.30534/ijatcse/2020/7191.32020. [DOI] [Google Scholar]
- Patel H.H., Prajapati P. Study and analysis of decision tree based classification algorithms. Int. J. Comput. Sci. Eng. 2018;6(10):74–78. doi: 10.26438/ijcse/v6i10.7478. [DOI] [Google Scholar]
- Rady A., Adedeji A.A. Application of hyperspectral imaging and machine learning methods to detect and quantify adulterants in minced meats. Food Anal. Methods. 2020;13:970–981. doi: 10.1007/s12161-020-01719-1. [DOI] [Google Scholar]
- Rajaguru H., SR S.C. Analysis of decision tree and k-nearest neighbor algorithm in the classification of breast cancer. Asian Pac J Cancer Prev. 2019;20(12):3777. doi: 10.31557/APJCP.2019.20.12.3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schonlau M., Zou R.Y. The random forest algorithm for statistical learning. STATA J. 2020;20(1):3–29. doi: 10.1177/1536867X20909688. [DOI] [Google Scholar]
- Setser A.L., Smith R.W. Comparison of variable selection methods prior to linear discriminant analysis classification of synthetic phenethylamines and tryptamines. Forensic Chem. 2018;11:77–86. doi: 10.1016/j.forc.2018.10.002. [DOI] [Google Scholar]
- Sinta D., Wijayanto H., Sartono B. Ensemble k-nearest neighbors method to predict rice price in Indonesia. Appl. Math. Sci. 2014;8(160):7993–8005. doi: 10.12988/ams.2014.49721. [DOI] [Google Scholar]
- Tharwat A. Classification assessment methods. Appl. Comput. Inform. 2021;17(1):168–192. doi: 10.1016/j.aci.2018.08.003. [DOI] [Google Scholar]
- Troshchynska Y., Bleha R., Kumbarová L., Sluková M., Sinica A., Štětina J. Characterisation of flaxseed cultivars based on NIR diffusion reflectance spectra of whole seeds and derived samples. Czech J. Food Sci. 2019;37(5) doi: 10.17221/270/2018-CJFS. [DOI] [Google Scholar]
- Valero-Carreras D., Alcaraz J., Landete M. Comparing two SVM models through different metrics based on the confusion matrix. Comput. Oper. Res. 2023;152 doi: 10.1016/j.cor.2022.106131. [DOI] [Google Scholar]
- Vanstone N., Moore A., Martos P., Neethirajan S. Detection of the adulteration of extra virgin olive oil by near-infrared spectroscopy and chemometric techniques. Food Qual Saf. 2018;2(4):189–198. doi: 10.1093/fqsafe/fyy018. [DOI] [Google Scholar]
- Weng S., Pan F., Yu S., Guo B. 2019 International Conference on Electronic Engineering and Informatics (EEI), Nanjing, China. 2019. Rapid distinguish of edible oil adulteration using a hyperspectral spectroradiometer. [DOI] [Google Scholar]
- Wu D., Chen X., Shi P., Wang S., Feng F., He Y. Determination of α-linolenic acid and linoleic acid in edible oils using near-infrared spectroscopy improved by wavelet transform and uninformative variable elimination. Anal. Chim. Acta. 2009;634(2):166–171. doi: 10.1016/j.aca.2008.12.024. [DOI] [PubMed] [Google Scholar]
- Xie C., Wang Q., He Y. Identification of different varieties of sesame oil using near-infrared hyperspectral imaging and chemometrics algorithms. PLoS One. 2014;9(5) doi: 10.1371/journal.pone.0098522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing C., Yuan X., Wu X., Shao X., Yuan J., Yan W. Chemometric classification and quantification of sesame oil adulterated with other vegetable oils based on fatty acids composition by gas chromatography. LWT. 2019;108:437–445. doi: 10.1016/j.lwt.2019.03.085. [DOI] [Google Scholar]
- Yang H., Irudayaraj J., Paradkar M.M. Discriminant analysis of edible oils and fats by FTIR, FT-NIR and FT-Raman spectroscopy. Food Chem. 2005;93(1):25–32. doi: 10.1016/j.foodchem.2004.08.039. [DOI] [Google Scholar]
- Yang X., Hong H., You Z., Cheng F. Spectral and image integrated analysis of hyperspectral data for waxy corn seed variety classification. Sensors. 2015;15(7):15578–15594. doi: 10.3390/s150715578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Suganthan P.N. Random forests with ensemble of feature spaces. Pattern Recogn. 2014;47(10):3429–3437. doi: 10.1016/j.patcog.2014.04.001. [DOI] [Google Scholar]
- Zhao X., Wang W., Ni X., Chu X., Li Y.-F., Sun C. Evaluation of near-infrared hyperspectral imaging for detection of peanut and walnut powders in whole wheat flour. Appl. Sci. 2018;8(7):1076. doi: 10.3390/app8071076. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.