Abstract
The rapid emergence of multiple strains of Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2) has sparked profound concerns regarding the ongoing evolution of the virus and its potential impact on global health. Classified by the World Health Organization (WHO) as variants of concern (VOC), these strains exhibit heightened transmissibility and pathogenicity, posing significant challenges to existing vaccine strategies. Despite widespread vaccination efforts, the continual evolution of SARS-CoV-2 variants presents a formidable obstacle to achieving herd immunity. Of particular concern is the coronavirus spike (S) protein, a pivotal viral surface protein crucial for host cell entry and infectivity. Mutations within the S protein have been shown to enhance transmissibility and confer resistance to antibody-mediated neutralization, undermining the efficacy of traditional vaccine platforms. Moreover, the S protein undergoes rapid molecular evolution under selective immune pressure, leading to the emergence of diverse variants with distinct mutation profiles. This review underscores the urgent need for vigilance and adaptation in vaccine development efforts to combat the evolving landscape of SARS-CoV-2 mutations and ensure the long-term effectiveness of global immunization campaigns.
Keywords: SARS-CoV-2 mutations, Spike protein, Variant of concern (VOC), Vaccine efficacy, Immune escape, Evolutionary adaptation
1. Introduction
RNA viruses, such as coronaviruses, influenza viruses, HCV and HIV, are renowned for their elevated mutation rates, attributed to their replication mechanisms and the absence of viral RNA polymerase proofreading activity [1]. Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), the causative agent of Coronavirus Disease 2019 (COVID-19), belongs to the category of positive-sense single-stranded RNA (+ssRNA) viruses. With a diameter of approximately 120 nm, the CoV genome spans about 30,000 bases (27–32 kb) [2]. Mutations act as the fundamental units of evolution, enabling the natural selection of advantageous traits, such as increased virulence, adaptability, and progression [3].
The global response to COVID-19 has prompted extensive investigations into the genetic evolution of the virus and the development of vaccines to mitigate its spread. The genetic diversity of coronaviruses is bolstered by their expansive genome, high mutation rate driven by viral polymerase infidelity, and frequent recombination events (up to 25 % of the total genome in vivo), rendering them formidable pathogens [3].
Central to the infection process of SARS-CoV-2 is its spike (S) protein, crucial for host cell entry. Notably, the S gene, particularly the S1 and receptor-binding domain (RBD) coding regions, exhibits the highest nonsynonymous mutation rates across most coronaviruses [[4], [5], [6]]. This viral surface protein is pivotal for viral adhesion and cell entry and undergoes rapid molecular evolution as the primary target of the host immune system [7,8]. Adaptive mutations within the viral genome have the potential to alter the virus's pathogenicity, with even single amino acid changes profoundly impacting immune evasion and challenging vaccine development [9,10]. The virus has vigorously spread among humans since the pandemic due to continuous recombination and mutational processes in its genome [11,12]. The proliferation of SARS-CoV-2 among human populations during the pandemic is fueled by continuous genome recombination and mutational processes. Variants of concern (VOCs), including Alpha (B.1.1.7), Beta (B.1.351), Gamma (P.1), Delta (B.1.617.2), Omicron (B.1.1.529.1), and IHU (B.1.640.2), as well as variants of interest (Lambda and Mu), and those under surveillance, present heightened contagiousness, reduced antibody neutralization, and evasion of detection or therapeutic efficacy [[13], [14], [15], [16], [17], [18]].
This review provides an overview of SARS-CoV-2 variant emergence, highlighting specific mutations that enhance transmission. As the threat of emerging and re-emerging viruses like SARS-CoV-2 persists, understanding the role of viral and receptor genes and their associated mutations remains paramount for prediction and prevention strategies. It underscores the critical importance of current SARS-CoV-2 mutations in shaping the future success of traditional vaccine platforms. The ongoing evolution of SARS-CoV-2 mutations poses a significant challenge to the efficacy of traditional vaccine platforms, as mutations can diminish vaccine effectiveness and potentially compromise immune responses. Understanding the impact of these mutations on vaccine efficacy is essential for developing strategies to address the evolving landscape of COVID-19 and ensure the continued success of vaccination efforts worldwide.
1.1. Spike glycoprotein
The S protein of SARS-CoV-2 comprises an N-terminal domain (NTD) S1 and a C-terminal membrane-proximal S2 subunit, with the S1 subunit further divided into S1A, S1B, S1C, and S1D domains. Specifically, the S1A domain recognizes carbohydrates like sialic acid, crucial for viral attachment to host cell surfaces and interaction with the human angiotensin-converting enzyme 2 (ACE2) receptor [[19], [20], [21]]. Additionally, the furin cleavage site, harboring the PRRA sequence motif between the S1 and S2 subunits, along with the S2 subunit's second proteolytic cleavage site, S20, upstream of the fusion peptide, play pivotal roles in viral entry into host cells [22,23]. Notably, these cleavage sites correlate with the extreme mutation density observed in the S-protein region [24]. Furthermore, the NTD represents another mutation-prone region within the S protein. Conversely, nonstructural proteins (NSPs), including those encoded by ORF1a (NSP1-11), ORF1b (NSP12-16), ORF3a, ORF6, ORF7a, ORF7b, ORF8, and ORF10, typically function as enzymes or proteins involved in virus replication, methylation, and initiating host immune responses [[24], [25], [26]]. Notably, mutant hotspots within NSP1 ORF1a/ORF1ab and ORF8 have been associated with alterations in virulence and infectivity [[27], [28], [29], [30]]. Particularly, NSP1 is critical for interfering with type I interferon induction in the host and promoting virus replication [31]. Fig. 1 depicts the genome structure of SARS-CoV2, including open reading frames and highlights the locations of mutations across the genome.
Fig. 1.
The genomic structure of SARS-CoV-2, highlighting mutations found in various variants. The SARS-CoV-2 genome spans about 30,000 base pairs (bp) and includes ORFs (open reading frames) and structural elements, each serving specific functions. Notably, the S protein plays a crucial role in attaching to and entering host cells. It is noteworthy that SARS-CoV-2 variants exhibit numerous mutations resulting in changes to amino acids, particularly in the receptor binding domain and the S1/S2 subunit of the S protein. Research has shown that these alterations enhance the virus's transmissibility.
1.2. Tropism
The tropism of SARS-CoV-2 for different cell types is influenced by the expression levels of key host proteins involved in virus binding and entry [32]. Apart from ACE2, other receptors such as CD209L/L-SIGN, CD209/DC-SIGN, CD209L, CLEC4M, CLEC4G, BSG, and ASK (ACE2/ASGR1/KREMEN1) have been implicated in virus entry and fusion, alongside various proteases including furin, transmembrane serine protease 2 (TMPRSS2), cathepsin L (CTSL), cathepsin B (CTSB), elastase, and trypsin [[33], [34], [35]]. Notably, while CTSL is uniformly expressed in all strain assessments, the levels of ACE2 and TMPRSS2 exhibit variation, suggesting a potential correlation between their distribution in the lungs or trachea and SARS-CoV-2 tropism [[35], [36], [37]]. Some COVID-19-related deaths have been attributed to inflammation and organ damage caused by the immune system rather than direct pathogen-induced effects [38]. Additionally, studies have indicated that immunosuppression with corticosteroids may prevent fatalities from severe SARS-CoV-2 infection [39,40].
2. Variants of concern (VOCs)
2.1. Alpha VOC
The Alpha variant (B.1.1.7 lineage) harbors 17 mutations in the virus genome, with nine mutations, including N501Y, in the S protein, notably enhancing viral attachment to host cells via increased affinity for ACE2 receptors [41,42]. Emerging in the United Kingdom at the end of December 2020, the Alpha variant exhibits increased transmissibility and severity compared to ancestral variants, quickly becoming the dominant strain [[43], [44], [45]]. Moreover, a new variant combining B.1.1.7 with the E484K mutation has significantly enhanced serum antibody levels, potentially affecting immunological responses and the longevity of neutralizing antibody responses [46]. Epidemiological studies have indicated higher mortality rates and increased ICU admissions among individuals infected with the Alpha variant, contributing to its rapid spread and heightened disease severity [[47], [48], [49]].
2.2. Beta VOC
First identified in South Africa in October 2020, the Beta variant (B.1.351 lineage) contains ten mutations in the S protein, including K417 N, E484K, and N501Y, which augment ACE2 receptor binding affinity and enhance transmissibility [42,45,50]. Notably, the Beta variant poses challenges for existing therapies and vaccines, showing resistance to monoclonal antibody therapy, convalescent sera, and revaccinated sera, leading to reduced neutralization [51]. Evidence suggests that Alpha and Beta variants increase transmissibility, particularly among younger age groups and children [52].
2.3. Gamma VOC
Originating in Brazil in early January 2021, the Gamma variant (B.1.1.28 lineage) shares 12 mutations in the S protein, including N501Y, K417 N, and E484K, associated with enhanced infectivity and severity compared to earlier strains [47,53,54]. Notably, the Gamma variant exhibits increased virulence and enhanced neutralization resistance against monoclonal antibody therapies, post-vaccination sera, and convalescent sera [51].
2.4. Delta VOC
First reported in India in December 2020, the Delta variant (B.1.617.2 lineage) has seven mutations in the S protein, including L452R and T478, which enhance ACE2 receptor binding affinity and contribute to increased transmissibility and disease severity compared to previous variants [[55], [56], [57], [58]]. The Delta variant represents a significant concern due to its rapid spread and increased mortality rates among infected individuals.
2.5. Epsilon variant of concern (VOC)
Previously classified as a variant of interest (VOI), the Epsilon variant, also known as the B.1.427 lineage, emerged in California in January 2021 and has since been designated as a VOC. This variant harbor two mutations in the spike (S) protein, namely L452R and D614G. Moreover, the B.1.429 lineage, a subtype of the Epsilon variant, bears four mutations in the S protein (S131, W152C, L452R, and D614G), with L452R being situated in the receptor-binding domain (RBD) in both variants [[59], [60], [61]]. The B.1.429 lineage is presumed to exhibit higher transmissibility compared to previous circulating variants, with an estimated 20 % increase in viral transmissibility, significantly impacting neutralization [62,63]. Epsilon variants have shown increased transmissibility by up to 24 %, attributed to elevated viral shedding, largely associated with the L452R mutation, which stabilizes the interaction with the ACE2 receptor [[64], [65], [66]].
2.6. Kappa variant of concern (VOC)
The Kappa variant, initially identified in India in December 2020 and initially categorized as a variant of interest (VOI), became a VOC by the end of March 2021. By that time, the Kappa variant had accounted for over half of the sequences recorded from India [67]. Similar to the Epsilon variant, Kappa bears eight mutations in the S protein (T951, G142D, E154K, L452R, E484Q, D614G, P681R, and Q1071H), with the notable mutations L452R and E484Q located in the RBD [68,69].
2.7. Omicron variant of concern (VOC)
The Omicron variant, also known as lineage B.1.1.529.1 (BA.1), was designated as the fifth VOC by the World Health Organization (WHO) in November 2021, first identified in South Africa and subsequently replaced by the BA.2 sub-lineage in Denmark [70,71]. This variant carries numerous mutations, some of which are of particular concern. Cases of the B.1.1.529 lineage increased across all regions of South Africa, with evidence suggesting an increased risk of reinfection associated with this variant [72]. Omicron harbors approximately 34 mutations in the S protein, including A67V, Δ69–70, T95I, G142D, Δ143-145, Δ211, L212I, ins214EPE, G339D, S371L, S373P, S375F, K417 N, N440K, G446S, S477 N, T478K, E484A, Q493R, G496S, Q498R, N501Y, Y505H, T547K, D614G, H655Y, N679K, P681H, N764K, D796Y, N856K, Q954H, N969K, and L981F [[73], [74], [75]]. Notably, fifteen of these mutations (G339D, S371L, S373P, S375F, K417 N, N440K, G446S, S477 N, T478K, E484A, Q493R, G496S, Q498R, N501Y, and Y505H) are located in the RBD [[73], [74], [75]]. The rapid dissemination and high replication rate of the Omicron variant, estimated to be approximately 70 times faster than the Delta variant and previous strains, can be attributed to the synergistic effects of multiple mutations, enhancing its binding affinity and ability to evade neutralizing antibodies [[76], [77], [78], [79], [80]].
2.8. IHU variant of concern (VOC)
The IHU variant, lineage B.1.640.2, named after the IHU Méditerranée Infection Institute, was first detected in France in November 2021. This variant, likely of Cameroonian origin, was reported to carry 46 mutations and 37 deletions. Among these are 14 amino acid substitutions, including N501Y, E484K, F490S, D1139H, P681H, and D614G, combined with T859 N, along with nine deletions located in the S protein, rendering it more resistant to vaccines [81,82,83]. Table 1 illustrates the Variant of Concerns along with their key mutations.
Table 1.
Some of the important spike Mutations of SARS-CoV-2 variants.
| Type of VOCs | Main circulating region | Emerging time | Key mutations | Influences of mutations | References |
|---|---|---|---|---|---|
| Alpha (B.1.1.7) | UK | December 2020 | N501Y | Increase binding affinity and transmissibility | [41,42] |
| E484K | Increase binding affinity and transmissibility Increase capacity of immune escape | ||||
| 69-70del | |||||
| D614G | Reinfection Increase binding affinity and transmissibility |
||||
| Beta (B.1.351) | South Africa | October 2020 | K417 N | Increase binding affinity and transmissibility Increase capacity of immune escape | [42,45,50] |
| E484K | Increase binding affinity and transmissibility Increase capacity of immune escape | ||||
| N501Y | Increase binding affinity and transmissibility | ||||
| D614G | Reinfection Increase binding affinity and transmissibility |
||||
| Gamma (B.1.1.28) | Brazil | January 2021 | K417 N | Increase binding affinity and transmissibility Increase capacity of immune escape | [47,53,54] |
| E484K | Increase binding affinity and transmissibility Increase capacity of immune escape | ||||
| N501Y | Increase binding affinity and transmissibility | ||||
| H655Y | Increase the capacity of immune escape | ||||
| Delta (B.1.617.2) | India | December 2020 | L452R | Increase binding affinity and transmissibility | [[56], [57], [58]] |
| T478K | Increase binding affinity and transmissibility | ||||
| K417 N | Increase the capacity of immune escape. | ||||
|
Epsilon (B.1.427) Epsilon (B.1.429) |
USA | January 2021 | L452R | Increase binding affinity and transmissibility Immune escape capacity, and SARS-CoV2 infectivity |
[[59], [60], [61]] |
| D614G | Increased binding affinity and transmissibility Increased SARS-CoV2 infectivity | ||||
| S131 | Increase the infectivity of the variant | ||||
| W152C | Increase binding affinity and transmissibility Increase capacity of immune escape |
||||
| L452R | Increase binding affinity and transmissibility, immune escape capacity, and SARS-CoV2 infectivity. | ||||
| D614G | Reinfection Increase binding affinity and transmissibility |
||||
| Kappa (B.1.617.1) | India | December 2020 | L452R | Increased capacity of immune escape Increased SARS-CoV2 infectivity |
[68,69] |
| E484Q | Increased capacity of immune escape Increased SARS-CoV2 infectivity |
||||
| Omicron (B.1.1.529.1) | South Africa | November 2021 | N501Y | Increase binding affinity and transmissibility | [[73], [74], [75]] |
| D614G | Reinfection Increase binding affinity and transmissibility |
||||
| K417 N | Increase the capacity of immune escape Increase binding affinity and transmissibility |
||||
| S477 N | Increase binding affinity and transmissibility | ||||
| G339D | Increase the capacity of immune escape | ||||
| G496S | Increase the capacity of immune escape | ||||
| N440K | Increase the capacity of immune escape | ||||
| E484A | Increase the capacity of immune escape and infectivity Increase binding affinity and transmissibility |
||||
| Q493 R/K | Increase the capacity of immune escape Increase binding affinity and transmissibility |
||||
| Y505H | Increase the capacity of immune escape and infectivity | ||||
| IHU (B.1.640.2) | France | November 2021 | N501Y | Increase binding affinity and transmissibility | [81,82,83] |
| E484K | Increase binding affinity and transmissibility Increase capacity of immune escape |
||||
| D1139H | Increase binding affinity and transmissibility | ||||
| D614G combined with T859 N | Increase binding affinity and transmissibility |
2.9. Temporal divergence of SARS-CoV-2 variants
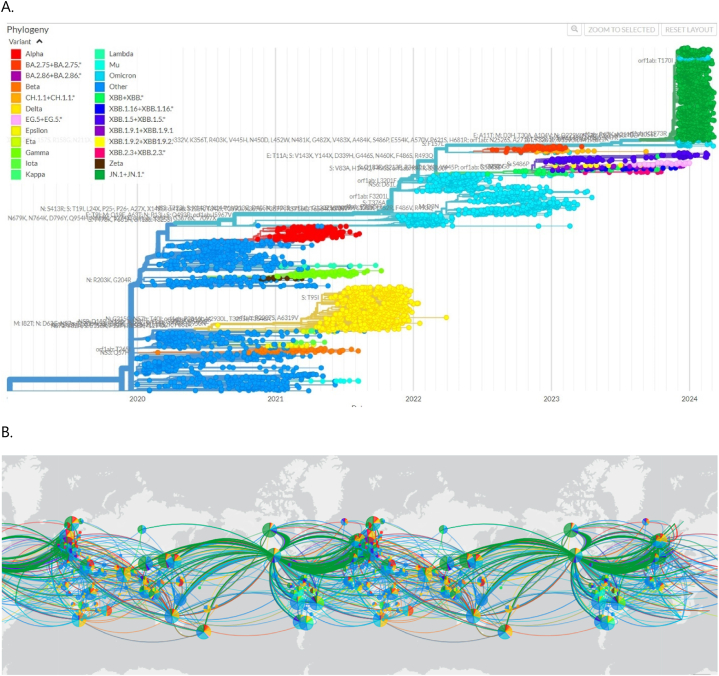
Understanding the temporal evolution of SARS-CoV-2 is paramount for deciphering its genetic landscape and the emergence of new variants over time. The period spanning from December 2019 to March 2024 has been marked by significant shifts in the genomic composition of the virus, reflecting its dynamic nature and adaptive strategies. Fig. 2 provides a visual representation of the phylodynamics of SARS-CoV-2 variants, offering insights into the chronological divergence and global dissemination of different viral lineages. In Fig. 2A, a time-stamped maximum likelihood phylogeny presents a subset of 4255 complete genomes of SARS-CoV-2 obtained from the GISAID database. This phylogeny delineates the evolutionary relationships among various viral strains sampled across the specified timeframe. Variants of concern (VOCs) are highlighted using distinct color schemes, aiding in the identification and tracking of genetically significant viral lineages. The visualization, created by Nextstrain using data sourced from GISAID, serves as a valuable tool for studying the evolutionary dynamics and genetic diversity of SARS-CoV-2. Additionally, Fig. 2B illustrates the distribution frequencies of VOCs across different regions worldwide. By mapping the prevalence of VOCs using distinct color schemes, this visualization offers insights into the global spread and geographic distribution of different SARS-CoV-2 variants. Analyzing the temporal and spatial distribution patterns of VOCs provides crucial information for understanding viral transmission dynamics and informing public health interventions.
Fig. 2.
The phylodynamics of the pandemic coronavirus worldwide: (A) A time-stamped maximum likelihood phylogeny depicting a representative subset of 4255 complete genomes of SARS-CoV-2 sampled from December 2019 to March 2024 from the GISAID database. Variants of concern (VOCs) are highlighted using various color schemes. This visualization was created by Nextstrain using data sourced from GISAID. (B) The distribution frequencies of VOCs across the globe are indicated using distinct color schemes. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
The temporal divergence of SARS-CoV-2 variants, as depicted in Fig. 2, underscores the ongoing evolution and adaptation of the virus in response to selective pressures, including host immunity and environmental factors. This evolutionary perspective is essential for anticipating future trends in viral evolution, guiding surveillance efforts, and informing the development of effective vaccination strategies tailored to the evolving landscape of SARS-CoV-2 variants.
3. Spike protein mutations: implications for SARS-CoV-2 transmissibility and immune evasion
Approximately one-third of the Spike (S) protein sequences exhibit susceptibility to mutations. Among the various mutations observed in the Receptor Binding Domain (RBD) of Variant of Concerns (VOCs), notable ones include N501Y, K417 N, K417T, E484K, L452R, T477K, E484Q, S477 N, Q498R, and E484A [84,85]. Notably, mutations within the Angiotensin-Converting Enzyme 2 (ACE2) receptor (S19P, K26R) and substantial RBD mutations (N501Y, E484K, S477 N) have been found to enhance viral transmissibility and binding affinity [86,87]. It is noteworthy that the interaction between the S-RBD and ACE2 is nuanced, influenced by factors such as temperature and conformational states of the Spike protein [88]. The RBD typically binds to ACE2 in the “up” conformation, rather than the “down” perfusion trimer. Moreover, all reported VOCs contain mutations in both the RBD and N-terminal domain (NTD), with the N501Y mutation being particularly prevalent in Alpha, Beta, Gamma, and Omicron variants, augmenting the affinity of the S protein for ACE2 and thereby facilitating viral entry into host cells. Studies have indicated that individual mutations, such as N501Y, significantly enhance the affinity between the RBD and ACE2, substantially increasing transmissibility [89]. Notably, variants like Beta and Gamma, despite carrying mutations like N417/K848/Y501-RBD, exhibit lower binding affinity to ACE2 compared to variants with N501Y and E484K mutations [90,91].
The B.1.351 variant, in addition to the N501Y mutation, carries K417 N and E484K mutations, potentially contributing to increased transmissibility [84,92]. Similarly, the K417 N mutation, present in the Delta Plus sublineage and the Omicron variant (BA.1), underscores the potential for cross-neutralization by antibodies targeting this site [93,94]. The location of E484 within the RBD's receptor binding motif (RBM) is crucial, as it interacts with the K31 domain on ACE2, influencing binding affinity [95,96]. Furthermore, the E484K mutation confers resistance to antibody-mediated neutralization, posing challenges for immunity [84,97,98]. Notably, variant P.1 carries mutations like K417T, E484K, and N501Y, impacting antibody-mediated neutralization and transduction efficiency akin to the B.1.351 variant [99,100]. Variants like B.1.617.2 and B.1.427/B.1.429 harbor mutations like L452R, which affect neutralization and infectivity [[101], [102], [103]]. Of particular interest is the B.1.617.1 lineage, characterized by the “double mutant” E484Q and L452R mutations, conferring immune evasion capabilities [[104], [105], [106]]. The Omicron variant, with its numerous deletions and mutations, exhibits enhanced transmissibility and antibody escape [[107], [108], [109]].
Mutations such as S477 N and E484K further enhance binding affinity and immune evasion, thereby affecting the virus's infectivity [86,110]. Notably, the rare mutation Q498R, though not currently prevalent, has been observed to increase ACE2 binding, potentially impacting viral transmissibility [87,111,112]. Moreover, mutations like N460K, G339D, and G446S, along with deletions and mutations in Omicron, contribute to resistance against monoclonal antibodies, further complicating therapeutic strategies [113,114]. The D614G substitution, pervasive across VOCs, is associated with increased infectivity, likely due to its effect on furin cleavage and S protein stability [42,[115], [116], [117]]. Additionally, mutations like S477 N and P681R, found in variants like Delta and Omicron, influence viral replication and entry into host cells [118,119].
These mutations underscore the dynamic nature of SARS-CoV-2 evolution, with implications for transmission dynamics, immune evasion, and therapeutic strategies. Understanding the interplay between Spike protein mutations and viral phenotype is crucial for devising effective public health interventions.
4. Host cell receptors in SARS-CoV-2 infection: implications for host susceptibility and viral entry
Variations in the Angiotensin-Converting Enzyme 2 (ACE2) receptor have emerged as potential factors influencing virus-host interactions, thereby modulating host susceptibility and the severity of COVID-19 [120,121]. Notable ACE2 variants such as S19P, I21V, E23K, K26R, T27A, N64K, T92I, Q102P, and H378R have been associated with increased susceptibility to SARS-CoV-2, while variants like K31R, N33I, H34R, E35K, E37K, D38V, N33I, Y50F, N51S, M62V, K68E, F72V, Y83H, G326E, G352V, D355 N, Q388L, and D509Y may confer protective effects by reducing binding to the Spike (S) protein [122]. Biochemical analyses have confirmed altered affinity for the S protein, with variants like K31R and E37K exhibiting reduced binding, while variants such as K26R and T92I demonstrate increased affinity compared to the wild-type ACE2 [122,123]. Additionally, mutations such as S19W, T27W, and N330Y have been found to enhance SARS-CoV-2 S-RBD binding [122,123]. However, certain ACE2 mutations, including Y180, L456, R460, P500, A501, S502, L503, F504, H505, N506, S507, Y207, E208, V209, N210, G211, V212, Y215, D216, Y217, P565, T567, H373, H374, E375, M376, G377, H378, I379, A380, Y381, F315, H401, V404, G405, and M408, have been identified as highly destabilizing ACE2-S interactions, while others like 98, A99, Q102, N103, and G104 correlate with stabilizing ACE2-S complex formation [124].
Moreover, the transmembrane glycoprotein CD147 has emerged as another potential receptor for SARS-CoV-2, particularly in cells with low expression levels of ACE2, providing an alternate mechanism for viral entry [124,125]. Furthermore, interactions with novel SARS-CoV-2 S binding proteins, including NID1, CNTN1, and APOA4, have been identified, with APOA4 exhibiting comparable affinity to ACE2 [126,127]. Additionally, various receptors and proteases, such as ASK (ACE2/ASGR1/KREMEN1), CD209/CD-SIGN, CD209L/L-SIGN, CLEC4G, BSG, and proteases like furin, TMPRSS2, GTSL, CTSB, elastase, and trypsin, have been implicated as important factors in S-host cell binding [[127], [128], [129]].
4.1. Impact of SARS-CoV-2 variants on clinical presentation
The clinical spectrum of SARS-CoV-2 infection ranges from asymptomatic cases to severe illnesses affecting multiple organ systems, including the nervous, digestive, cardiovascular, and respiratory systems [130]. Host factors, including immune status due to vaccination or prior infection, play a significant role in determining disease severity, alongside viral mutations that give rise to distinct strains [129,131,132]. Variants with specific genomic features may exhibit varying degrees of virulence and tissue tropism. For instance, strains with a preference for alveolar replication may lead to pneumonia and acute respiratory distress syndrome, while those favoring replication in the respiratory mucosa, such as Omicron, may manifest with nasal congestion, cough, and sore throat [133,134]. The Gamma variant has been associated with symptoms like anosmia and dysgeusia, while the Delta variant, characterized by higher viral loads, may cause changes in olfactory discrimination and affect the gastrointestinal system, resulting in symptoms like nausea, vomiting, and diarrhea [[135], [136], [137]].
Studies have shown that the Delta variant, in particular, exhibits increased virulence compared to the original Wuhan strain and even the Alpha variant [138,139]. Animal studies suggest that Delta mutations promote cell-to-cell fusion and syncytia formation, contributing to increased pathogenicity [140]. The clinical manifestations and severity of disease can also be influenced by mutations affecting interferon production and immune evasion mechanisms [141]. In contrast, the Omicron variant tends to cause milder symptoms compared to Alpha and Delta variants, with a lower likelihood of severe illness and death [142,143]. However, the Delta variant remains associated with more severe respiratory infections and a higher demand for specialized hospital care [142,143].
In summary, the evolving landscape of SARS-CoV-2 variants underscores the dynamic interplay between viral genetics, host factors, and clinical outcomes, highlighting the importance of continued surveillance and adaptive public health strategies.
4.2. Impact of mutations in variants of concern on COVID-19 vaccine effectiveness
The significance of the S protein lies in its role in membrane fusion and binding to ACE2, which has led to its recognition as a crucial target for vaccine design [144]. The success in vaccine development against SARS-CoV-2 has been remarkable, with various vaccine platforms deployed to combat the pandemic. These include mRNA-based vaccines, inactivated whole virus vaccines, vector vaccines, subunit vaccines, virus-like particles, and live attenuated vaccines. To achieve herd immunity and curb the spread of COVID-19, it is imperative to vaccinate adolescents and children, who contribute significantly to virus transmission. Vaccination efforts for this age group have shown promising results, with both Pfizer and Moderna vaccines demonstrating high efficacy and safety profiles [145,146]. Unvaccinated individuals face a higher risk of infection and severe outcomes from COVID-19 compared to those who are fully vaccinated. However, despite the mass vaccination campaigns, the emergence of SARS-CoV-2 variants poses challenges to vaccine efficacy, as seen with the N439K spike mutation [147]. Meta-analysis studies have indicated that full vaccination, especially with mRNA-based vaccines, maintains considerable efficacy against Alpha, Beta/Gamma, and Delta variants [146]. Furthermore, sera from individuals vaccinated with certain vaccines, such as the Sputnik V vaccine, have demonstrated robust neutralization activity against various VOCs [148]. Initially, widely used COVID-19 vaccines were developed based on the spike antigen of early virus strains, primarily the Wuhan-Hu-1 reference sequence. Despite some minor changes observed in the Alpha variant, laboratory experiments showed moderate evasion from vaccine-induced antibodies for variants like Beta, Gamma, and Delta. The continuous evolution of SARS-CoV-2 underscores the need for ongoing research to develop more effective vaccination strategies against future variants. This includes exploring the possibility of annual updates to COVID-19 vaccines, similar to influenza vaccines, and the development of universal vaccines capable of providing protection against a broad range of variants. Understanding the evolutionary trends of the virus is crucial in devising strategies to combat the potential emergence of more infectious and lethal strains [[149], [150], [151], [152], [153], [154]]. The emergence of the Omicron variant, comprising various sub-lineages like BA.1, BA.2, BA.4, and BA.5, has presented a significant challenge due to its ability to infect vaccinated and previously infected individuals. Omicron variants exhibit extensive mutations in the spike receptor-binding domain (RBD) and the amino-terminal domain (NTD), resulting in poor neutralization by first-generation vaccines and pre-Omicron antibodies. Consequently, booster doses are crucial to maintain vaccine effectiveness against Omicron, although the duration of protection decreases over time. This rapid antigenic shift has prompted the development of second-generation vaccines specifically targeting Omicron variants or adopting universal vaccine approaches. Additionally, the incremental acquisition of mutations during persistent infections contributes to antigenic drift, further complicating efforts to combat SARS-CoV-2 evolution [[155], [156], [157], [158], [159], [160], [161], [162], [163], [164], [165], [166], [167]].
In a phase 2 trial study, Gorilla adenovirus-based (GRAd) vaccines induced strong VOC-cross-reactive T-cell responses, highlighting promising aspects of cellular immunity [168]. Among vaccine platforms, mRNA-based vaccines like BNT162b2 (Pfizer BioNTech COVID-19 vaccine) and mRNA-1273 (Moderna) have shown better resilience to genetic alterations compared to other types of vaccines [169,170]. However, some mutations, such as D614G and P681R, have demonstrated moderate resistance to certain monoclonal antibodies targeting the RBD region, posing challenges for vaccine and monoclonal antibody-based therapies [171,172]. Additionally, variants carrying mutations like E484K have shown enhanced resistance to vaccine-elicited antibodies and monoclonal antibodies, thereby threatening the efficacy of specific vaccines [173,174].
The emergence of variants like B.1.617, carrying mutation E484Q, has shown enhanced entry and pathogenesis, along with resistance to neutralization by antibodies used in COVID-19 treatment and plasma from vaccinated individuals, indicating enhanced evasion of humoral immunity induced by both infection and vaccination [175]. Furthermore, multiple mutations observed in the spike protein of VOCs have been associated with high infectivity and resistance to pre-existing antibodies, convalescent plasma, or vaccine-induced immunity. Studies have shown strong resistance to neutralization by sera from individuals vaccinated with certain vaccines, such as AstraZeneca, against variants like B.1.1.7 and B.1.351 [176]. Additionally, the B.1.351 variant has moderately affected the neutralization ability of sera from individuals vaccinated with inactivated BBIBP-CorV and ZF2001 RBD recombinant vaccines [177].
5. Impact of vaccine escape mutations on traditional vaccine efficacy and optimal vaccine platforms for resistance
The emergence of mutations that confer resistance to traditional COVID-19 vaccines poses significant challenges to global vaccination efforts [178,179]. Mutations like E484K and P681R have demonstrated enhanced resistance to vaccine-induced immunity, jeopardizing the efficacy of vaccines such as Pfizer-BioNTech BNT162b2 and Novavax [[178], [179], [180], [181]]. The impact of these mutations on vaccine efficacy has garnered significant attention as the virus continues to evolve [178,179]. Recent data suggest that mRNA-based vaccines, such as Pfizer-BioNTech and Moderna, have demonstrated robust efficacy against various variants, including Alpha, Beta, Gamma, and Delta [182,183]. These vaccines have shown better resilience to genetic alterations and have maintained high levels of neutralizing antibodies against diverse Variants of Concern (VOCs). Moreover, mRNA vaccines have proven to be adaptable, with the potential for rapid modification to target specific mutations, making them promising candidates for addressing emerging variants [[182], [183], [184], [185]].
Adenoviral vector vaccines, such as Johnson & Johnson's Janssen vaccine and the AstraZeneca vaccine, have also shown efficacy against certain variants [186,187]. However, concerns have been raised about their effectiveness against variants carrying mutations like E484K, which confer resistance to vaccine-induced immunity [[174], [175], [176]]. Nonetheless, adenoviral vector vaccines offer advantages such as ease of storage and distribution, making them valuable tools for global vaccination campaigns, especially in resource-limited settings [[186], [187], [188]]. Whole inactivated vaccines, such as Sinovac's CoronaVac and Sinopharm's BBIBP-CorV, have demonstrated efficacy against specific variants but may exhibit reduced effectiveness against others. Studies have shown varying levels of neutralizing antibody titers against VOCs, highlighting the need for ongoing monitoring and potential booster doses to maintain immunity [[189], [190], [191]]. Recombinant protein vaccines, such as Novavax's NVX-CoV2373, have shown promising results in clinical trials, with efficacy against certain variants comparable to mRNA vaccines. These vaccines utilize protein subunits of the virus to stimulate an immune response, offering potential advantages in terms of safety and scalability [192,193].
Regarding mutations that could potentially hamper the effectiveness of current vaccines, several key variants have been identified. Mutations in the spike protein, such as D614G, P681R, and E484K, have demonstrated resistance to certain monoclonal antibodies and vaccine-induced immunity, posing challenges for vaccination efforts. Variants like B.1.617, carrying mutation E484Q, have shown enhanced entry and pathogenesis, along with resistance to neutralization by antibodies used in COVID-19 treatment and plasma from vaccinated individuals [[194], [195], [196], [197], [198]].
6. Conclusion
In conclusion, the ongoing evolution of SARS-CoV-2 variants presents a formidable challenge to global health and vaccination efforts. The emergence of variants of concern (VOCs) with heightened transmissibility and immune evasion capabilities underscores the urgent need for vigilance and adaptation in vaccine development strategies. The spike protein mutations, particularly within the receptor-binding domain (RBD), play a critical role in enhancing viral transmissibility and immune evasion. Variants like Delta, Beta, Gamma, and Omicron exhibit extensive mutations in the spike protein, rendering them poorly neutralized by first-generation vaccines and pre-existing antibodies. This necessitates the deployment of booster doses and the development of second-generation vaccines targeting specific variants or adopting universal vaccine approaches. While mRNA vaccines have demonstrated robust efficacy against various VOCs, concerns remain regarding the effectiveness of other vaccine platforms, particularly against mutations like E484K and P681R. Adenoviral vector vaccines, whole inactivated vaccines, and recombinant protein vaccines have shown varying levels of efficacy against specific variants, highlighting the importance of ongoing monitoring and potential adaptation of vaccination strategies. In terms of vulnerability to illness based on vaccine type, populations in countries where specific vaccines are predominant may face different levels of risk. For example, populations vaccinated primarily with adenoviral vector vaccines may have slightly reduced protection against certain variants compared to those vaccinated with mRNA vaccines. However, the overall impact may vary depending on factors such as vaccine coverage, population demographics, and public health measures. Furthermore, the impact of mutations on vaccine effectiveness underscores the dynamic nature of the virus and the need for continuous research and development to stay ahead of its evolution. Understanding the interplay between viral genetics, host factors, and clinical outcomes is crucial for devising effective public health interventions and ensuring the long-term success of vaccination campaigns worldwide. As we navigate the evolving landscape of SARS-CoV-2 mutations, collaborative efforts between researchers, public health officials, and vaccine manufacturers are essential to combat the pandemic and protect global health.
Availability of data and materials
The datasets are available from the corresponding author upon request.
CRediT authorship contribution statement
Niloofar Faraji: Writing – review & editing, Writing – original draft, Investigation, Conceptualization. Tahereh Zeinali: Writing – review & editing, Writing – original draft, Conceptualization. Farahnaz Joukar: Supervision, Methodology, Conceptualization. Maryam Sadat Aleali: Investigation. Narges Eslami: Investigation. Mohammad Shenagari: Supervision, Methodology, Conceptualization. Fariborz Mansour-Ghanaei: Supervision, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors gratefully acknowledge the support contributors to this study.
Contributor Information
Mohammad Shenagari, Email: shenagari@gmail.com.
Fariborz Mansour-Ghanaei, Email: fmansourghanaei@gmail.com.
References
- 1.Sharma A., Lal S.K. Is tetherin a true antiviral: the influenza a virus controversy. Rev. Med. Virol. 2019;29 doi: 10.1002/rmv.2036. [DOI] [PubMed] [Google Scholar]
- 2.Wang M.Y., Zhao R., Gao L.J., Gao X.F., Wang D.P., Cao J.M. SARS-CoV-2: structure, Biology, and structure-based therapeutics development. Front. Cell. Infect. Microbiol. 2020;10 doi: 10.3389/fcimb.2020.587269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Singh J., Pandit P., McArthur A.G., Banerjee A., Mossman K. Evolutionary trajectory of SARS-CoV-2 and emerging variants. Virol. J. 2021;18:166. doi: 10.1186/s12985-021-01633-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Woo P.C.Y., Huang Y., Lau S.K.P., Yuen K.Y. Coronavirus genomics and bioinformatics analysis. Viruses. 2010;2:1804–1820. doi: 10.3390/v2081803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vijgen L., Lemey P., Keyaerts E., Van Ranst M. Genetic variability of human respiratory coronavirus OC43. J. Virol. 2005;79:3223–3225. doi: 10.1128/JVI.79.5.3223-3225.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sánchez C.M., Gebauer F., Suñé C., Mendez A., Dopazo J., Enjuanes L. Genetic evolution and tropism of transmissible gastroenteritis coronaviruses. Virology. 1992;190:92–105. doi: 10.1016/0042-6822(92)91195-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Q., Xiang R., Huo S., Zhou Y., Jiang S., Wang Q., et al. Molecular mechanism of interaction between SARS-CoV-2 and host cells and interventional therapy. Signal Transduct. Targeted Ther. 2021;6:233. doi: 10.1038/s41392-021-00653-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gadanec L.K., McSweeney K.R., Qaradakhi T., Ali B., Zulli A., Apostolopoulos V. Can SARS-CoV-2 virus Use multiple receptors to enter host cells? Int. J. Mol. Sci. 2021;22 doi: 10.3390/ijms22030992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saputri D.S., Li S., van Eerden F.J., Rozewicki J., Xu Z., Ismanto H.S., et al. Flexible, functional, and Familiar: characteristics of SARS-CoV-2 spike protein evolution. Front. Microbiol. 2020;11:1–6. doi: 10.3389/fmicb.2020.02112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giovanetti M., Benedetti F., Campisi G., Ciccozzi A., Fabris S., Ceccarelli G., et al. Evolution patterns of SARS-CoV-2: Snapshot on its genome variants. Biochem. Biophys. Res. Commun. 2021;538:88–91. doi: 10.1016/j.bbrc.2020.10.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., et al. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182:812–827.e19. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Şenel E., Türk S., Malkan Ü.Y., Peker M.Ç., Türk C., Güner H.R., et al. Pathobiological alterations affecting the distinct clinical courses of pediatric versus adult COVID-19 syndrome. Turk. J. Med. Sci. 2023;53:1194–1204. doi: 10.55730/1300-0144.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tracking SARS-CoV-2 Variants. World Heal Organ. n.d..
- 14.Aleem A., Akbar Samad A.B., Vaqar S. Treasure Island (FL); 2023. Emerging Variants of SARS-CoV-2 and Novel Therapeutics against Coronavirus (COVID-19) [PubMed] [Google Scholar]
- 15.Malik Y.A. Covid-19 variants: impact on transmissibility and virulence. Malays. J. Pathol. 2022;44:387–396. [PubMed] [Google Scholar]
- 16.Misra G., Hora S., Ginwal S., Singh N., Anvikar A. SARS-CoV-2 variants impact on key signaling Pathways Metamorphoses into severity. Braz. Arch. Biol. Technol. 2023;66:1–12. doi: 10.1590/1678-4324-2023220261. [DOI] [Google Scholar]
- 17.Zhou H., Møhlenberg M., Thakor J.C., Tuli H.S., Wang P., Assaraf Y.G., et al. Sensitivity to vaccines, therapeutic antibodies, and viral entry inhibitors and advances to counter the SARS-CoV-2 omicron variant. Clin. Microbiol. Rev. 2022;35 doi: 10.1128/cmr.00014-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thye A.Y.K., Law J.W.F., Pusparajah P., Letchumanan V., Chan K.G., Lee L.H. Emerging sars-cov-2 variants of concern (Vocs): an impending global crisis. Biomedicines. 2021;9:1–25. doi: 10.3390/biomedicines9101303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paul D., Pyne N., Paul S. Mutation profile of SARS-CoV-2 spike protein and identification of potential multiple epitopes within spike protein for vaccine development against SARS-CoV-2. Virusdisease. 2021;32:703–726. doi: 10.1007/s13337-021-00747-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mohammadi M., Shayestehpour M., Mirzaei H. The impact of spike mutated variants of SARS-CoV2 [Alpha, Beta, Gamma, Delta, and Lambda] on the efficacy of subunit recombinant vaccines. Brazilian J Infect Dis an Off Publ Brazilian Soc Infect Dis. 2021;25 doi: 10.1016/j.bjid.2021.101606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim C.H. Sars-cov-2 evolutionary adaptation toward host entry and recognition of receptor o-acetyl sialylation in virus–host interaction. Int. J. Mol. Sci. 2020;21:1–34. doi: 10.3390/ijms21124549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lavie M., Dubuisson J., Belouzard S. SARS-CoV-2 spike furin cleavage site and S2’ basic residues modulate the entry process in a host cell-dependent manner. J. Virol. 2022;96 doi: 10.1128/jvi.00474-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roy S., Ghosh P., Bandyopadhyay A., Basu S. Capturing a crucial ‘disorder-to-order transition’at the heart of the coronavirus molecular pathology—triggered by highly persistent, interchangeable salt-bridges. Vaccines. 2022 Feb 16;10(2):301. doi: 10.3390/vaccines10020301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bhattacharyya C., Das C., Ghosh A., Singh A.K., Mukherjee S., Majumder P.P., et al. SARS-CoV-2 mutation 614G creates an elastase cleavage site enhancing its spread in high AAT-deficient regions. Infect Genet Evol J Mol Epidemiol Evol Genet Infect Dis. 2021;90 doi: 10.1016/j.meegid.2021.104760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takeda M. Proteolytic activation of SARS-CoV-2 spike protein. Microbiol. Immunol. 2022;66:15–23. doi: 10.1111/1348-0421.12945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gupta S., Gupta D., Bhatnagar S. Analysis of SARS-CoV-2 genome evolutionary patterns. Microbiol. Spectr. 2024;12 doi: 10.1128/spectrum.02654-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hassan S.S., Choudhury P.P., Roy B. Rare mutations in the accessory proteins ORF6, ORF7b, and ORF10 of the SARS-CoV-2 genomes. Meta Gene. 2021;28 doi: 10.1016/j.mgene.2021.100873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hassan S.S., Choudhury P.P., Dayhoff GW 2nd, Aljabali A.A.A., Uhal B.D., Lundstrom K., et al. The importance of accessory protein variants in the pathogenicity of SARS-CoV-2. Arch. Biochem. Biophys. 2022;717 doi: 10.1016/j.abb.2022.109124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Han G.-Z. Pangolins harbor SARS-CoV-2-related coronaviruses. Trends Microbiol. 2020;28:515–517. doi: 10.1016/j.tim.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guruprasad L. Human coronavirus spike protein-host receptor recognition. Prog. Biophys. Mol. Biol. 2021;161:39–53. doi: 10.1016/j.pbiomolbio.2020.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iaconis G., Jackson B., Childs K., Boyce M., Goodbourn S., Blake N., et al. Rotavirus NSP1 inhibits type I and type III interferon induction. Viruses. 2021;13 doi: 10.3390/v13040589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murgolo N., Therien A.G., Howell B., Klein D., Koeplinger K., Lieberman L.A., et al. SARS-CoV-2 tropism, entry, replication, and propagation: considerations for drug discovery and development. PLoS Pathog. 2021;17:1–18. doi: 10.1371/JOURNAL.PPAT.1009225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Amraei R., Rahimi N. COVID-19, renin-angiotensin system and endothelial dysfunction. Cells. 2020;9:1–18. doi: 10.3390/cells9071652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rahimi N. C-type lectin CD209L/L-SIGN and CD209/DC-SIGN: cell adhesion molecules turned to pathogen recognition receptors. Biology. 2021;10:1–15. doi: 10.3390/biology10010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eslami N., Aghbash P.S., Shamekh A., Entezari-Maleki T., Nahand J.S., Sales A.J., et al. SARS-CoV-2: receptor and Co-receptor tropism probability. Curr. Microbiol. 2022;79:133. doi: 10.1007/s00284-022-02807-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zamorano Cuervo N., Grandvaux N. ACE2: evidence of role as entry receptor for SARS-CoV-2 and implications in comorbidities. Elife. 2020;9 doi: 10.7554/eLife.61390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Puray-Chavez M., LaPak K.M., Schrank T.P., Elliott J.L., Bhatt D.P., Agajanian M.J., et al. Systematic analysis of SARS-CoV-2 infection of an ACE2-negative human airway cell. Cell Rep. 2021;36 doi: 10.1016/j.celrep.2021.109364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zeinali T., Faraji N., Joukar F., Khan Mirzaei M., Kafshdar Jalali H., Shenagari M., et al. Gut bacteria, bacteriophages, and probiotics: tripartite mutualism to quench the SARS-CoV2 storm. Microb. Pathog. 2022;105704 doi: 10.1016/j.micpath.2022.105704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cano E.J., Fonseca Fuentes X., Corsini Campioli C., O'Horo J.C., Abu Saleh O., Odeyemi Y., et al. Impact of corticosteroids in coronavirus disease 2019 outcomes: systematic review and meta-analysis. Chest. 2021;159:1019–1040. doi: 10.1016/j.chest.2020.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Crisan Dabija R., Antohe I., Trofor A., Antoniu S.A. Corticosteroids in SARS-COV2 infection: certainties and uncertainties in clinical practice. Expert Rev. Anti Infect. Ther. 2021;19:1553–1562. doi: 10.1080/14787210.2021.1933437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020;11:1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bhattacharya M., Chatterjee S., Sharma A.R., Agoramoorthy G., Chakraborty C. D614G mutation and SARS-CoV-2: impact on S-protein structure, function, infectivity, and immunity. Appl. Microbiol. Biotechnol. 2021;105:9035–9045. doi: 10.1007/s00253-021-11676-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z., et al. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181:894–904.e9. doi: 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xia H., Cao Z., Xie X., Zhang X., Chen J.Y.-C., Wang H., et al. Evasion of type I interferon by SARS-CoV-2. Cell Rep. 2020;33 doi: 10.1016/j.celrep.2020.108234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guruprasad L. Human SARS CoV-2 spike protein mutations. Proteins Struct Funct Bioinforma. 2021;89:569–576. doi: 10.1002/prot.26042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin J.-W., Tang C., Wei H.-C., Du B., Chen C., Wang M., et al. Genomic monitoring of SARS-CoV-2 uncovers an Nsp1 deletion variant that modulates type I interferon response. Cell Host Microbe. 2021;29:489–502.e8. doi: 10.1016/j.chom.2021.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Challen R., Brooks-Pollock E., Read J.M., Dyson L., Tsaneva-Atanasova K., Danon L. Risk of mortality in patients infected with SARS-CoV-2 variant of concern 202012/1: matched cohort study. BMJ. 2021;372:n579. doi: 10.1136/bmj.n579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Davies N.G., Jarvis C.I., van Zandvoort K., Clifford S., Sun F.Y., Funk S., et al. Increased mortality in community-tested cases of SARS-CoV-2 lineage B.1.1.7. Nature. 2021;593:270–274. doi: 10.1038/s41586-021-03426-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martínez-García L., Espinel M.A., Abreu M., González-Alba J.M., Gijón D., McGee A., et al. Emergence and spread of B.1.1.7 lineage in primary care and clinical impact in the morbi-mortality among hospitalized patients in Madrid, Spain. Microorganisms. 2021;9 doi: 10.3390/microorganisms9071517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Khetran S.R., Mustafa R. Mutations of SARS-CoV-2 structural proteins in the Alpha, Beta, Gamma, and delta variants: bioinformatics analysis. JMIR Bioinforma Biotechnol. 2023;4 doi: 10.2196/43906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Walensky R.P., Walke H.T., Fauci A.S. SARS-CoV-2 variants of concern in the United States—challenges and opportunities. JAMA. 2021;325:1037–1038. doi: 10.1001/jama.2021.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Volz E., Mishra S., Chand M., Barrett J.C., Johnson R., Geidelberg L., et al. Assessing transmissibility of SARS-CoV-2 lineage B.1.1.7 in England. Nature. 2021;593:266–269. doi: 10.1038/s41586-021-03470-x. [DOI] [PubMed] [Google Scholar]
- 53.Tian D., Sun Y., Zhou J., Ye Q. The global epidemic of SARS-CoV-2 variants and their mutational immune escape. J. Med. Virol. 2022;94:847–857. doi: 10.1002/jmv.27376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang P., Nair M.S., Liu L., Iketani S., Luo Y., Guo Y., Wang M., Yu J., Zhang B., Kwong P.D., Graham B.S. Antibody resistance of SARS-CoV-2 variants B. 1.351 and B. 1.1. 7. Nature. 2021 May;593(7857):130–135. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- 55.Dhawan M., Sharma A., Priyanka Thakur N., Rajkhowa T.K., Choudhary O.P. Delta variant (B.1.617.2) of SARS-CoV-2: mutations, impact, challenges and possible solutions. Hum. Vaccines Immunother. 2022;18 doi: 10.1080/21645515.2022.2068883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.He X., He C., Hong W., Zhang K., Wei X. The challenges of COVID-19 Delta variant: prevention and vaccine development. MedComm. 2021;2:846–854. doi: 10.1002/mco2.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.de Souza A.S., de Freitas Amorim V.M., Guardia G.D.A., Dos Santos F.F., Ulrich H., Galante P.A.F., et al. Severe acute respiratory syndrome coronavirus 2 variants of concern: a perspective for emerging more transmissible and vaccine-resistant strains. Viruses. 2022;14 doi: 10.3390/v14040827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Barroso-Arévalo S., Sánchez-Morales L., Pérez-Sancho M., Domínguez L., Sánchez-Vizcaíno J.M. First detection of SARS-CoV-2 B.1.617.2 (delta) variant of concern in a symptomatic cat in Spain. Front. Vet. Sci. 2022;9 doi: 10.3389/fvets.2022.841430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Carroll T., Fox D., van Doremalen N., Ball E., Morris M.K., Sotomayor-Gonzalez A., et al. The B.1.427/1.429 (epsilon) SARS-CoV-2 variants are more virulent than ancestral B.1 (614G) in Syrian hamsters. PLoS Pathog. 2022;18 doi: 10.1371/journal.ppat.1009914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Deng X., Garcia-Knight M.A., Khalid M.M., Servellita V., Wang C., Morris M.K., et al. Identification of SARS-CoV-2 spike mutations that attenuate monoclonal and serum antibody neutralization. Cell Host Microbe. 2021;29:477–488. doi: 10.1016/j.chom.2021.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kubik S., Arrigo N., Bonet J., Xu Z. Mutational hotspot in the SARS-CoV-2 Spike protein N-terminal domain conferring immune escape potential. Viruses. 2021;13:2114. doi: 10.3390/v13112114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Deng X., Garcia-Knight M.A., Khalid M.M., Servellita V., Wang C., Morris M.K., et al. Transmission, infectivity, and neutralization of a spike L452R SARS-CoV-2 variant. Cell. 2021;184:3426–3437.e8. doi: 10.1016/j.cell.2021.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tao K., Tzou P.L., Nouhin J., Gupta R.K., de Oliveira T., Kosakovsky Pond S.L., et al. The biological and clinical significance of emerging SARS-CoV-2 variants. Nat. Rev. Genet. 2021;22:757–773. doi: 10.1038/s41576-021-00408-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Khateeb J., Li Y., Zhang H. Emerging SARS-CoV-2 variants of concern and potential intervention approaches. Crit. Care. 2021;25:244. doi: 10.1186/s13054-021-03662-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Devaux C.A., Fantini J. ACE2 receptor polymorphism in humans and animals increases the risk of the emergence of SARS-CoV-2 variants during repeated intra- and inter-species host-switching of the virus. Front. Microbiol. 2023;14 doi: 10.3389/fmicb.2023.1199561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu Z., VanBlargan L.A., Bloyet L.-M., Rothlauf P.W., Chen R.E., Stumpf S., et al. Identification of SARS-CoV-2 spike mutations that attenuate monoclonal and serum antibody neutralization. Cell Host Microbe. 2021;29:477–488. doi: 10.1016/j.chom.2021.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Singh J., Malhotra A.G., Biswas D., Shankar P., Lokhande L., Yadav A.K., et al. Relative consolidation of the Kappa variant pre-dates the massive second wave of COVID-19 in India. Genes. 2021;12 doi: 10.3390/genes12111803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ghosh N., Nandi S., Saha I. A review on evolution of emerging SARS-CoV-2 variants based on spike glycoprotein. Int. Immunopharm. 2022;105 doi: 10.1016/j.intimp.2022.108565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cherian S., Potdar V., Jadhav S., Yadav P., Gupta N., Das M., et al. SARS-CoV-2 spike mutations, L452R, T478K, E484Q and P681R, in the second wave of COVID-19 in Maharashtra, India. Microorganisms. 2021;9 doi: 10.3390/microorganisms9071542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Manjunath R., Gaonkar S.L., Saleh E.A.M., Husain K. A comprehensive review on Covid-19 Omicron (B.1.1.529) variant. Saudi J. Biol. Sci. 2022;29 doi: 10.1016/j.sjbs.2022.103372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shuai H., Chan J.F.W., Hu B., Chai Y., Yoon C., Liu H., et al. The viral fitness and intrinsic pathogenicity of dominant SARS-CoV-2 Omicron sublineages BA.1, BA.2, and BA.5. EBioMedicine. 2023;95 doi: 10.1016/j.ebiom.2023.104753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dhawan M., Saied A.A., Mitra S., Alhumaydhi F.A., Bin Emran T., Wilairatana P. Omicron variant (B.1.1.529) and its sublineages: what do we know so far amid the emergence of recombinant variants of SARS-CoV-2? Biomed. Pharmacother. 2022;154 doi: 10.1016/j.biopha.2022.113522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shanmugaraj B., Malla A., Khorattanakulchai N., Phoolcharoen W. SARS-CoV-2 omicron variant: could it be another threat? J. Med. Virol. 2022;94:1284–1288. doi: 10.1002/jmv.27532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ou J., Lan W., Wu X., Zhao T., Duan B., Yang P., et al. Tracking SARS-CoV-2 Omicron diverse spike gene mutations identifies multiple inter-variant recombination events. Signal Transduct. Targeted Ther. 2022;7:138. doi: 10.1038/s41392-022-00992-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mcgonagle D. the company‘s public news and information; 2020. Since January 2020 Elsevier Has Created a COVID-19 Resource Centre with Free Information in English and Mandarin on the Novel Coronavirus COVID-19. The COVID-19 Resource Centre Is Hosted on Elsevier Connect. [Google Scholar]
- 76.Shrestha L.B., Foster C., Rawlinson W., Tedla N., Bull R.A. Evolution of the SARS-CoV-2 omicron variants BA.1 to BA.5: implications for immune escape and transmission. Rev. Med. Virol. 2022;32 doi: 10.1002/rmv.2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sharma V., Rai H., Gautam D.N.S., Prajapati P.K., Sharma R. Emerging evidence on omicron (B. 1.1. 529) SARS‐CoV‐2 variant. J. Med. Virol. 2022;94:1876–1885. doi: 10.1002/jmv.27626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mohapatra R.K., Kandi V., Sarangi A.K., Verma S., Tuli H.S., Chakraborty S., Chakraborty C., Dhama K. The recently emerged BA.4 and BA.5 lineages of Omicron and their global health concerns amid the ongoing wave of COVID-19 pandemic - correspondence. Int. J. Surg. 2022;103 doi: 10.1016/j.ijsu.2022.106698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cox M., Peacock T.P., Harvey W.T., Hughes J., Wright D.W., Willett B.J., Thomson E., Gupta R.K., Peacock S.J., Robertson D.L., Carabelli A.M. SARS-CoV-2 variant evasion of monoclonal antibodies based on in vitro studies. Nat. Rev. Microbiol. 2023;21:112–124. doi: 10.1038/s41579-022-00809-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.RECOVERY Collaborative Group. Horby P.W., Mafham M., Peto L., Campbell M., Pessoa-Amorim G., Spata E., Staplin N., Emberson J.R., Prudon B., Hine P. Casirivimab and imdevimab in patients admitted to hospital with COVID-19 (RECOVERY): a randomised, controlled, open-label, platform trial. medRxiv. 2021:6. [Google Scholar]
- 81.Colson P., Delerce J., Burel E., Dahan J., Jouffret A., Fenollar F., Yahi N., Fantini J., La Scola B., Raoult D. Emergence in Southern France of a new SARS-CoV-2 variant of probably Cameroonian origin harbouring both substitutions N501Y and E484K in the spike protein. medRxiv. 2021 doi: 10.1101/2021.12.24.21268174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Han J., Liu T., Zhang X., Yang Y., Shi Y., Li J., Ma M., Zhu W., Gong L., Xu Z. D3AI-Spike: a deep learning platform for predicting binding affinity between SARS-CoV-2 spike receptor binding domain with multiple amino acid mutations and human angiotensin-converting enzyme 2. Comput. Biol. Med. 2022;151 doi: 10.1016/j.compbiomed.2022.106212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Beatty J.D., Beatty B.G., Vlahos W.G. Measurement of monoclonal antibody affinity by non-competitive enzyme immunoassay. J. Immunol. Methods. 1987;100:173–179. doi: 10.1016/0022-1759(87)90187-6. [DOI] [PubMed] [Google Scholar]
- 84.Pondé R.A.A. Physicochemical effect of the N501Y, E484K/Q, K417N/T, L452R and T478K mutations on the SARS-CoV-2 spike protein RBD and its influence on agent fitness and on attributes developed by emerging variants of concern. Virology. 2022;572:44–54. doi: 10.1016/j.virol.2022.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Almalki S.S., Izhari M.A., Alyahyawi H.E., Alatawi S.K., Klufah F., Ahmed W.A., Alharbi R. Mutational analysis of circulating omicron SARS-CoV-2 lineages in the Al-baha region of Saudi arabia. J. Multidiscip. Healthc. 2023;16:2117–2136. doi: 10.2147/jmdh.s419859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Barton M.I., MacGowan S.A., Kutuzov M.A., Dushek O., Barton G.J., van der Merwe P.A. Effects of common mutations in the SARS-CoV-2 Spike RBD and its ligand, the human ACE2 receptor on binding affinity and kinetics. Elife. 2021;10 doi: 10.7554/eLife.70658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bate N., Savva C.G., Moody P.C.E., Brown E.A., Evans S.E., Ball J.K., Schwabe J.W.R., Sale J.E., Brindle N.P.J. In vitro evolution predicts emerging SARS-CoV-2 mutations with high affinity for ACE2 and cross-species binding. PLoS Pathog. 2022;18 doi: 10.1371/journal.ppat.1010733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Verkhivker G.M. Molecular simulations and network modeling reveal an allosteric signaling in the SARS-CoV-2 spike proteins. J. Proteome Res. 2020;19:4587–4608. doi: 10.1021/acs.jproteome.0c00654. [DOI] [PubMed] [Google Scholar]
- 89.Watanabe Y.H., Takano M., Yoshida M. ATP binding to nucleotide binding domain (NBD)1 of the ClpB chaperone induces motion of the long coiled-coil, stabilizes the hexamer, and activates NBD2. J. Biol. Chem. 2005;280:24562–24567. doi: 10.1074/jbc.M414623200. [DOI] [PubMed] [Google Scholar]
- 90.He X., He C., Hong W., Yang J., Wei X. Research progress in spike mutations of SARS-CoV-2 variants and vaccine development. Med. Res. Rev. 2023;43:932–971. doi: 10.1002/med.21941. [DOI] [PubMed] [Google Scholar]
- 91.Zinatizadeh M.R., Zarandi P.K., Zinatizadeh M., Yousefi M.H., Amani J., Rezaei N. Efficacy of mRNA, adenoviral vector, and perfusion protein COVID-19 vaccines. Biomed. Pharmacother. 2022;146 doi: 10.1016/j.biopha.2021.112527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tian F., Tong B., Sun L., Shi S., Zheng B., Wang Z., Dong X., Zheng P. N501Y mutation of spike protein in SARS-CoV-2 strengthens its binding to receptor ACE2. Elife. 2021;10 doi: 10.7554/eLife.69091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Moyo-Gwete T., Madzivhandila M., Mkhize N.N., Kgagudi P., Ayres F., Lambson B.E., Manamela N.P., Richardson S.I., Makhado Z., van der Mescht M.A., de Beer Z., de Villiers T.R., Burgers W.A., Ntusi N.A.B., Rossouw T., Ueckermann V., Boswell M.T., Moore P.L. Shared N417-dependent epitope on the SARS-CoV-2 omicron, Beta, and delta Plus variants. J. Virol. 2022;96 doi: 10.1128/jvi.00558-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kumar S., Karuppanan K., Subramaniam G. Omicron (BA.1) and sub-variants (BA.1.1, BA.2, and BA.3) of SARS-CoV-2 spike infectivity and pathogenicity: a comparative sequence and structural-based computational assessment. J. Med. Virol. 2022;94:4780–4791. doi: 10.1002/jmv.27927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang W.B., Liang Y., Jin Y.Q., Zhang J., Su J.G., Li Q.M. E484K mutation in SARS-CoV-2 RBD enhances binding affinity with hACE2 but reduces interactions with neutralizing antibodies and nanobodies: binding free energy calculation studies. J. Mol. Graph. Model. 2021;109 doi: 10.1016/j.jmgm.2021.108035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 97.Yang W.T., Huang W.H., Liao T.L., Hsiao T.H., Chuang H.N., Liu P.Y. SARS-CoV-2 E484K mutation narrative review: epidemiology, immune escape, clinical implications, and future considerations. Infect. Drug Resist. 2022;15:373–385. doi: 10.2147/IDR.S344099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Harvey W.T., Carabelli A.M., Jackson B., Gupta R.K., Thomson E.C., Harrison E.M., Ludden C., Reeve R., Rambaut A., Peacock S.J., Robertson D.L. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021;19:409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hu L., Xu Y., Wu L., Feng J., Zhang L., Tang Y., Zhao X., Mai R., Chen L., Mei L., Tan Y., Du Y., Zhen Y., Su W., Peng T. The E484K substitution in a SARS-CoV-2 spike protein subunit vaccine resulted in limited cross-reactive neutralizing antibody responses in mice. Viruses. 2022 doi: 10.3390/v14050854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Trombetta C.M., Marchi S., Viviani S., Manenti A., Benincasa L., Ruello A., Bombardieri E., Vicenti I., Zazzi M., Montomoli E. Serum neutralizing activity against B.1.1.7, B.1.351, and P.1 SARS-CoV-2 variants of concern in hospitalized COVID-19 patients. Viruses. 2021 doi: 10.3390/v13071347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhang X., Hong B., Wei P., Pei P., Xu H., Chen L., Tong Y., Chen J., Luo S.Z., Fan H., He C. Pathogen-host adhesion between SARS-CoV-2 spike proteins from different variants and human ACE2 studied at single-molecule and single-cell levels. Emerg. Microb. Infect. 2022;11:2658–2669. doi: 10.1080/22221751.2022.2128887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Jiang Y., Wu Q., Song P., You C. The variation of SARS-CoV-2 and advanced research on current vaccines. Front. Med. 2021;8 doi: 10.3389/fmed.2021.806641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Di Giacomo S., Mercatelli D., Rakhimov A., Giorgi F.M. Preliminary report on severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Spike mutation T478K. J. Med. Virol. 2021;93:5638–5643. doi: 10.1002/jmv.27062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gomari M.M., Tarighi P., Choupani E., Abkhiz S., Mohamadzadeh M., Rostami N., Sadroddiny E., Baammi S., Uversky V.N., Dokholyan N.V. Structural evolution of Delta lineage of SARS-CoV-2. Int. J. Biol. Macromol. 2023;226:1116–1140. doi: 10.1016/j.ijbiomac.2022.11.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sanyaolu A., Okorie C., Marinkovic A., Haider N., Abbasi A.F., Jaferi U., Prakash S., Balendra V. The emerging SARS-CoV-2 variants of concern. Ther Adv Infect Dis. 2021;8:9–11. doi: 10.1177/20499361211024372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.McCallum M., Bassi J., De Marco A., Chen A., Walls A.C., Di Iulio J., Tortorici M.A., Navarro M.J., Silacci-Fregni C., Saliba C., Sprouse K.R., Agostini M., Pinto D., Culap K., Bianchi S., Jaconi S., Cameroni E., Bowen J.E., Tilles S.W., Pizzuto M.S., Guastalla S.B., Bona G., Pellanda A.F., Garzoni C., Van Voorhis W.C., Rosen L.E., Snell G., Telenti A., Virgin H.W., Piccoli L., Corti D., Veesler D. SARS-CoV-2 immune evasion by the B.1.427/B.1.429 variant of concern. Science. 2021;373:648–654. doi: 10.1126/science.abi7994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Karim S.S.A., Karim Q.A. Omicron SARS-CoV-2 variant: a new chapter in the COVID-19 pandemic. Lancet (London, England) 2021;398:2126–2128. doi: 10.1016/S0140-6736(21)02758-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Setiabudi D., Sribudiani Y., Hermawan K., Andriyoko B., Nataprawira H.M. The Omicron variant of concern: the genomics, diagnostics, and clinical characteristics in children. Front Pediatr. 2022;10 doi: 10.3389/fped. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Paz M., Aldunate F., Arce R., Ferreiro I., Cristina J. An evolutionary insight into severe acute respiratory syndrome coronavirus 2 omicron variant of concern. Virus Res. 2022 doi: 10.1016/j.virusres.2022.198753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Yao H., Song Y., Chen Y., Wu N., Xu J., Sun C., Zhang J., Weng T., Zhang Z., Wu Z., Cheng L., Shi D., Lu X., Lei J., Crispin M., Shi Y., Li L., Li S. Molecular architecture of the SARS-CoV-2 virus. Cell. 2020;183:730–738.e13. doi: 10.1016/j.cell.2020.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zappa M., Verdecchia P., Angeli F. Severe acute respiratory syndrome coronavirus 2 evolution: how mutations affect XBB.1.5 variant. Eur. J. Intern. Med. 2023;112:128–132. doi: 10.1016/j.ejim.2023.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhang J., Cai Y., Lavine C.L., Peng H., Zhu H., Anand K., Tong P., Gautam A., Mayer M.L., Rits-Volloch S., Wang S., Sliz P., Wesemann D.R., Yang W., Seaman M.S., Lu J., Xiao T., Chen B. Structural and functional impact by SARS-CoV-2 Omicron spike mutations. Cell Rep. 2022;39 doi: 10.1016/j.celrep.2022.110729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zahradník J., Marciano S., Shemesh M., Zoler E., Harari D., Chiaravalli J., Meyer B., Rudich Y., Li C., Marton I., Dym O., Elad N., Lewis M.G., Andersen H., Gagne M., Seder R.A., Douek D.C., Schreiber G. SARS-CoV-2 variant prediction and antiviral drug design are enabled by RBD in vitro evolution. Nat Microbiol. 2021;6:1188–1198. doi: 10.1038/s41564-021-00954-4. [DOI] [PubMed] [Google Scholar]
- 114.Zahradník J., Marciano S., Shemesh M., Zoler E., Chiaravalli J., Meyer B., et al. SARS-CoV-2 RBD in vitro evolution follows contagious mutation spread, yet generates an able infection inhibitor. bioRxiv. 2021:2001–2021. [Google Scholar]
- 115.Gazali F.M., Wijayanti N., Hakim M.S., Supriyati E., Arguni E., Daniwijaya M.E.W., et al. The high mutation rate at the D614G hotspot-furin cleavage site region increases the priming efficiency of the Spike protein by furin protease: analysis of Indonesian SARS-CoV-2 G614 variants obtained during the early COVID-19 pandemic. Virusdisease. 2023;34:1–10. doi: 10.1007/s13337-023-00827-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gobeil S.M.-C., Janowska K., McDowell S., Mansouri K., Parks R., Manne K., et al. D614G mutation alters SARS-CoV-2 spike conformation and enhances protease cleavage at the S1/S2 junction. Cell Rep. 2021;34 doi: 10.1016/j.celrep.2020.108630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Guo C., Tsai S.-J., Ai Y., Li M., Anaya E., Pekosz A., et al. The D614G mutation redirects SARS-CoV-2 spike to lysosomes and suppresses deleterious traits of the furin cleavage site insertion mutation. Sci. Adv. 2022;8 doi: 10.1126/sciadv.ade5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Schrörs B., Riesgo-Ferreiro P., Sorn P., Gudimella R., Bukur T., Rösler T., et al. Large-scale analysis of SARS-CoV-2 spike-glycoprotein mutants demonstrates the need for continuous screening of virus isolates. PLoS One. 2021;16 doi: 10.1371/journal.pone.0249254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lasek-Nesselquist E., Pata J., Schneider E., George K. A tale of three SARS-CoV-2 variants with independently acquired P681H mutations in New York State. medRxiv. 2021;(12) 2021-03. [Google Scholar]
- 120.Bakhshandeh B., Sorboni S.G., Javanmard A.R., Mottaghi S.S., Mehrabi M.R., Sorouri F., et al. Variants in ACE2; potential influences on virus infection and COVID-19 severity. Infect. Genet. Evol. 2021;90 doi: 10.1016/j.meegid.2021.104773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Chaudhary M. COVID-19 susceptibility: potential of ACE2 polymorphisms. Egypt J Med Hum Genet. 2020;21:1–8. doi: 10.1186/s43042-020-00099-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Suryamohan K., Diwanji D., Stawiski E.W., Gupta R., Miersch S., Liu J., et al. Human ACE2 receptor polymorphisms and altered susceptibility to SARS-CoV-2. Commun. Biol. 2021;4:475. doi: 10.1038/s42003-021-02030-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Jami G., Ataee M., Esmaeili V., Chamani S., Rezaei A., Naghizadeh A. Characterization of the angiotensin-converting enzyme 2 (ACE2), the main receptor for the SARS-CoV-2 virus. American Journal of Clinical and Experimental Immunology. 2023;12(3):24. [PMC free article] [PubMed] [Google Scholar]
- 124.Benetti E., Tita R., Spiga O., Ciolfi A., Birolo G., Doddato G., et al. ACE2 gene variants may underlie interindividual variability and susceptibility to COVID-19 in the Italian population. Eur. J. Hum. Genet. 2020;28:1602–1614. doi: 10.1038/s41431-020-0691-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Behl T., Kaur I., Aleya L., Sehgal A., Singh S., Sharma N., et al. CD147-spike protein interaction in COVID-19: get the ball rolling with a novel receptor and therapeutic target. Sci. Total Environ. 2022;808 doi: 10.1016/j.scitotenv.2021.152072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ang M., Moriyama A., Colby K., Sutton G., Liang L., Sharma N., et al. Corneal transplantation in the aftermath of the COVID-19 pandemic: an international perspective. Br. J. Ophthalmol. 2020;104:1477–1481. doi: 10.1136/bjophthalmol-2020-317013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Brockbank S.M.V., Soden J., Faba-Rodriguez R., Ribeiro L.R., Geh C., Thomas H., et al. BioRxiv.; 2021. SARS-CoV-2 Comprehensive Receptor Profiling: Mechanistic Insight to Drive New Therapeutic Strategies. 2021.03.11.434937. [Google Scholar]
- 128.Li Q., Wu J., Nie J., Zhang L., Hao H., Liu S., et al. The impact of mutations in SARS-CoV-2 spike on viral infectivity and antigenicity. Cell. 2020;182:1284–1294.e9. doi: 10.1016/j.cell.2020.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Murugesan M., Mathews P., Paul H., Karthik R., Mammen J.J., Rupali P., et al. Protective effect conferred by prior infection and vaccination on COVID-19 in a healthcare worker cohort in South India. PLoS One. 2022;17 doi: 10.1371/journal.pone.0268797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Halaji M., Heiat M., Faraji N., Ranjbar R. Epidemiology of COVID-19: an updated review. J. Res. Med. Sci. 2021;26:82. doi: 10.4103/jrms.JRMS_506_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Yaghubi T., Shakoori V., Nasiri S., Keivan M., Tavakol C., Ahanjide S., et al. Clinical characteristics and outcomes of COVID-19 patients with a history of cardiovascular disease. J. Curr. Biomed. Rep. 2022;3(1):1–7. [Google Scholar]
- 132.Kalurazi T.Y., Shakoori V., Nasiri S., Foumani A.A., Hesni E., Mahfoozi L., et al. Clinical characteristics and laboratory findings of patients with COVID-19 in Rasht, Iran. J Curr Biomed Reports. 2022;3:91–97. [Google Scholar]
- 133.Khan M.Z.I., Nazli A., Al-Furas H., Asad M.I., Ajmal I., Khan D., et al. An overview of viral mutagenesis and the impact on pathogenesis of SARS-CoV-2 variants. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.1034444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Al-Qahtani A.A. Mutations in the genome of severe acute respiratory syndrome coronavirus 2: implications for COVID-19 severity and progression. J. Int. Med. Res. 2022;50 doi: 10.1177/03000605221086433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Stolp B., Stern M., Ambiel I., Hofmann K., Morath K., Gallucci L., et al. SARS-CoV-2 variants of concern display enhanced intrinsic pathogenic properties and expanded organ tropism in mouse models. Cell Rep. 2022;38 doi: 10.1016/j.celrep.2022.110387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Menni C., Valdes A.M., Polidori L., Antonelli M., Penamakuri S., Nogal A., et al. Symptom prevalence, duration, and risk of hospital admission in individuals infected with SARS-CoV-2 during periods of omicron and delta variant dominance: a prospective observational study from the ZOE COVID Study. Lancet. 2022;399:1618–1624. doi: 10.1016/S0140-6736(22)00327-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Luna-Muschi A., Borges I.C., de Faria E., Barboza A.S., Maia F.L., Leme M.D., et al. Clinical features of COVID-19 by SARS-CoV-2 Gamma variant: a prospective cohort study of vaccinated and unvaccinated healthcare workers. J. Infect. 2022;84:248–288. doi: 10.1016/j.jinf.2021.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Tian D., Sun Y., Zhou J., Ye Q. The global epidemic of the SARS-CoV-2 delta variant, key spike mutations and immune escape. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.751778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Hu Z., Huang X., Zhang J., Fu S., Ding D., Tao Z. Differences in clinical characteristics between delta variant and wild-type SARS-CoV-2 infected patients. Front. Med. 2022;8 doi: 10.3389/fmed.2021.792135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Saito A., Irie T., Suzuki R., Maemura T., Nasser H., Uriu K., et al. Enhanced fusogenicity and pathogenicity of SARS-CoV-2 Delta P681R mutation. Nature. 2022;602:300–306. doi: 10.1038/s41586-021-04266-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Guo K., Barrett B.S., Mickens K.L., Vladar E.K., Morrison J.H., Hasenkrug K.J., et al. Interferon resistance of emerging SARS-CoV-2 variants. BioRxiv Prepr. Serv. Biol. 2021;119(32) doi: 10.1073/pnas.2203760119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Chatterjee S., Bhattacharya M., Nag S., Dhama K., Chakraborty C. A detailed overview of SARS-CoV-2 omicron: its sub-variants, mutations and pathophysiology, clinical characteristics, immunological landscape, immune escape, and therapies. Viruses. 2023;15 doi: 10.3390/v15010167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Alam M.S. Insight into SARS-CoV-2 Omicron variant immune escape possibility and variant independent potential therapeutic opportunities. Heliyon. 2023;9 doi: 10.1016/j.heliyon.2023.e13285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Krammer F. SARS-CoV-2 vaccines in development. Nature. 2020;586:516–527. doi: 10.1038/s41586-020-2798-3. [DOI] [PubMed] [Google Scholar]
- 145.Regan J.J. Use of updated COVID-19 vaccines 2023–2024 formula for persons aged≥ 6 months: recommendations of the Advisory Committee on Immunization Practices—United States, September 2023. MMWR. Morbidity and Mortality Weekly Report. 2023;72 doi: 10.15585/mmwr.mm7242e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Ali K., Berman G., Zhou H., Deng W., Faughnan V., Coronado-Voges M., et al. Evaluation of MRNA-1273 SARS-CoV-2 vaccine in adolescents. N. Engl. J. Med. 2021;385:2241–2251. doi: 10.1056/NEJMoa2109522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Moro P.L., McNeil M.M. Successes of the CDC monitoring systems in evaluating post-authorization safety of COVID-19 vaccines. Expet Rev. Vaccine. 2022 Mar 4;21(3):281–284. doi: 10.1080/14760584.2022.2019020. [DOI] [PubMed] [Google Scholar]
- 148.Radion E.I., Mukhin V.E., Kholodova A.V., Vladimirov I.S., Alsaeva D.Y., Zhdanova A.S., Ulasova N.Y., Bulanova N.V., Makarov V.V., Keskinov A.A. Functional characteristics of serum Anti-SARS-CoV-2 antibodies against Delta and Omicron variants after vaccination with Sputnik V. Viruses. 2023;15:1349. doi: 10.3390/v15061349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Hogan A.B., Doohan P., Wu S.L., Mesa D.O., Toor J., Watson O.J., Winskill P., Charles G., Barnsley G., Riley E.M., Khoury D.S. Estimating long-term vaccine effectiveness against SARS-CoV-2 variants: a model-based approach. Nat. Commun. 2023 Jul 19;14(1):4325. doi: 10.1038/s41467-023-39736-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Waltz E. Omicron-targeted vaccines do No better than original jabs in early tests. Nature. 2022;10 doi: 10.1038/d41586-022-00003-y. [DOI] [PubMed] [Google Scholar]
- 151.Scheaffer S.M., Lee D., Whitener B., Ying B., Wu K., Liang C.Y., Jani H., Martin P., Amato N.J., Avena L.E., et al. Bivalent SARS-CoV-2 mRNA vaccines increase breadth of neutralization and protect against the BA.5 omicron variant in mice. Nat Med. 2023;29:247–257. doi: 10.1038/s41591-022-02092-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Callaway E. The next generation of coronavirus vaccines: a graphical guide. Nature. 2023;614:22–25. doi: 10.1038/d41586-023-00220-z. [DOI] [PubMed] [Google Scholar]
- 153.Kozlov M. Should COVID vaccines Be given yearly? Proposal divides US scientists. Nature. 2023 doi: 10.1038/d41586-023-00234-7. [DOI] [PubMed] [Google Scholar]
- 154.Dolgin E. Pan-coronavirus vaccine pipeline takes form. Nat. Rev. Drug Discov. 2022;21:324–326. doi: 10.1038/d41573-022-00074-6. [DOI] [PubMed] [Google Scholar]
- 155.Tegally H., et al. Emergence of SARS-CoV-2 omicron lineages BA.4 and BA.5 in South Africa. Nat Med. 2022 doi: 10.1038/s41591-022-01911-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Cele S., et al. Omicron extensively but incompletely escapes Pfizer BNT162b2 neutralization. Nature. 2022;602:654–656. doi: 10.1038/s41586-021-04387-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Willett B.J., et al. Distinct antigenic properties of the SARS-CoV-2 Omicron lineages BA.4 and BA.5. Preprint at bioRxiv. 2022. [DOI]
- 158.Viana R., et al. Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in southern Africa. Nature. 2022 doi: 10.1038/s41586-022-04411-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Cao Y., et al. BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection. Nature. 2022 doi: 10.1038/s41586-022-04980-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Meng B., et al. Altered TMPRSS2 usage by SARS-CoV-2 Omicron impacts tropism and fusogenicity. Nature. 2022 doi: 10.1038/s41586-022-04474-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Khan K., et al. Omicron BA.4/BA.5 escape neutralizing immunity elicited by BA.1 infection. Nat. Commun. 2022;13:4686. doi: 10.1038/s41467-022-32396-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Simon-Loriere E., Schwartz O. Towards SARS-CoV-2 serotypes? Nat. Rev. Microbiol. 2022;20:187–188. doi: 10.1038/s41579-022-00708-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.van der Straten K., et al. Antigenic cartography using sera from sequence-confirmed SARS-CoV-2 variants of concern infections reveal antigenic divergence of Omicron. Immunity. 2022 doi: 10.1016/j.immuni.2022.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Fang Z., et al. Omicron-specific mRNA vaccination alone and as a heterologous booster against SARS-CoV-2. Nat. Commun. 2022;13:3250. doi: 10.1038/s41467-022-30878-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Kherabi Y., Launay O., Luong Nguyen L.B. COVID-19 vaccines against Omicron variant: real-world data on effectiveness. Viruses. 2022 Sep 20;14(10):2086. doi: 10.3390/v14102086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Kirsebom F.C.M., et al. COVID-19 vaccine effectiveness against the omicron (BA.2) variant in England. Lancet Infect. Dis. 2022 doi: 10.1016/S1473-3099(22)00309-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Higdon M.M., et al. Duration of effectiveness of vaccination against COVID-19 caused by the omicron variant. Lancet Infect. Dis. 2022;22:1114–1116. doi: 10.1016/S1473-3099(22)00409-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Capone S., Fusco F.M., Milleri S., Borrè S., Carbonara S., Lo Caputo S., Leone S., Gori G., Maggi P., Cascio A., Lichtner M. GRAd-COV2 vaccine provides potent and durable immunity in randomised placebo-controlled phase 2 trial (COVITAR) medRxiv. 2022:10. doi: 10.1016/j.xcrm.2023.101084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Zhang G., Tang T., Chen Y., Huang X., Liang T. mRNA vaccines in disease prevention and treatment. Signal Transduct. Targeted Ther. 2023;8:365. doi: 10.1038/s41392-023-01579-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Echaide M., Chocarro de Erauso L., Bocanegra A., Blanco E., Kochan G., Escors D. mRNA vaccines against SARS-CoV-2: advantages and caveats. Int. J. Mol. Sci. 2023;24:5944. doi: 10.3390/ijms24065944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Yaqinuddin A., Shafqat A., Kashir J., Alkattan K. Effect of SARS-CoV-2 mutations on the efficacy of antibody therapy and response to vaccines. Vaccines. 2021;9:914. doi: 10.3390/vaccines9080914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Saito A., Nasser H., Uriu K., Kosugi Y., Irie T., Shirakawa K., Sadamasu K., Kimura I., Ito J., Wu J., et al. 2021. SARS-CoV-2 Spike P681R Mutation Enhances and Accelerates Viral Fusion. [Google Scholar]
- 173.Jangra S., Ye C., Rathnasinghe R., Stadlbauer D., Krammer F., Simon V., Martinez-Sobrido L., Garcia-Sastre A., Schotsaert M., Group P.V.I.S. The E484K mutation in the SARS-CoV-2 spike protein reduces but does not abolish neutralizing activity of human convalescent and post-vaccination sera. medRxiv. 2021 [Google Scholar]
- 174.Collier D.A., De Marco A., Ferreira I.A.T.M., Meng B., Datir R.P., Walls A.C., Kemp S.A., Bassi J., Pinto D., Silacci-Fregni C., et al. Sensitivity of SARS-CoV-2 B.1.1.7 to mRNA vaccine-elicited antibodies. Nature. 2021;593:136–141. doi: 10.1038/s41586-021-03412-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Hoffmann M., Hofmann-Winkler H., Krüger N., Kempf A., Nehlmeier I., Graichen L., Arora P., Sidarovich A., Moldenhauer A.S., Winkler M.S., Schulz S. SARS-CoV-2 variant B. 1.617 is resistant to bamlanivimab and evades antibodies induced by infection and vaccination. Cell Rep. 2021 Jul 20;36(3) doi: 10.1016/j.celrep.2021.109415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Mengist H.M., Kombe A.J.K., Mekonnen D., Abebaw A., Getachew M., Jin T. Mutations of SARS-CoV-2 spike protein: implications on immune evasion and vaccine-induced immunity. Semin. Immunol. 2021;55 doi: 10.1016/j.smim.2021.101533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Hu J., Peng P., Wang K., Fang L., Luo F., Jin A., Liu B., Tang N., Huang A. Emerging SARS-CoV-2 variants reduce neutralization sensitivity to convalescent sera and monoclonal antibodies. Cell. Mol. Immunol. 2021;18:1061–1063. doi: 10.1038/s41423-021-00648-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Centers for Disease Control and Prevention COVID-19 vaccine breakthrough case investigations team. COVID-19 vaccine breakthrough infections reported to cdc—United States, january 1–april 30, 2021. MMWR Morb. Mortal. Wkly. Rep. 2021;70:792–793. doi: 10.15585/mmwr.mm7021e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Goldberg Y., Mandel M., Bar-On Y.M., et al. Waning immunity after the BNT162b2 vaccine in Israel. N. Engl. J. Med. 2021;385:e85. doi: 10.1056/NEJMoa2114228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Puranik A., Lenehan P.J., Silvert E., et al. Comparison of two highly-effective mRNA vaccines for COVID-19 during periods of Alpha and Delta variant prevalence. medRxiv. 2021 doi: 10.1101/2021.08.06.21261707. [DOI] [Google Scholar]
- 181.Sadoff J., Gray G., Vandebosch A., et al. Safety and efficacy of single-dose Ad26.COV2.S vaccine against COVID-19. N. Engl. J. Med. 2021;384:2187–2201. doi: 10.1056/NEJMoa2101544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Madhi S.A., Baillie V., Cutland C.L., et al. Efficacy of the ChAdOx1 nCoV-19 Covid-19 vaccine against the B.1.351 variant. N. Engl. J. Med. 2021;384:1885–1898. doi: 10.1056/NEJMoa2102214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Ozsahin D.U., Gelisen M.I., Taiwo M., Agachan Y., Rahi D., Uzun B. Decision analysis of the COVID-19 vaccines. The EuroBiotech Journal. 2021 Jun;5(s1):20–25. [Google Scholar]
- 184.Lora A.J., Long J.E., Huang Y., Baden L.R., El Sahly H.M., Follmann D., Goepfert P., Gray G., Grinsztejn B., Kotloff K., Rouphael N. Rapid development of an integrated network infrastructure to conduct phase 3 COVID-19 vaccine trials. JAMA Netw. Open. 2023;6(1) doi: 10.1001/jamanetworkopen.2022.51974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Logunov D.Y., Dolzhikova I.V., Shcheblyakov D.V., et al. Safety and efficacy of an rAd26 and rAd5 vector-based heterologous prime-boost COVID-19 vaccine: an interim analysis of a randomised controlled phase 3 trial in Russia. Lancet. 2021;397:671–681. doi: 10.1016/S0140-6736(21)00234-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Pilkington V., Keestra S.M., Hill A. Global COVID-19 vaccine inequity: failures in the first year of distribution and potential solutions for the future. Front. Public Health. 2022 Mar 7;10 doi: 10.3389/fpubh.2022.821117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Dunkle L.M., Kotloff K.L., Gay C.L., Áñez G., Adelglass J.M., Barrat Hernández A.Q., Harper W.L., Duncanson D.M., McArthur M.A., Florescu D.F., McClelland R.S. Efficacy and safety of NVX-CoV2373 in adults in the United States and Mexico. N. Engl. J. Med. 2022 Feb 10;386(6):531–543. doi: 10.1056/NEJMoa2116185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Travieso T., Li J., Mahesh S., Mello J.D., Blasi M. The use of viral vectors in vaccine development. npj Vaccines. 2022;7:75. doi: 10.1038/s41541-022-00503-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Sharma E., Revinipati S., Bhandari S., Thakur S., Goyal S., Ghose A., Bajpai S., Muhammad W., Boussios S. Efficacy and safety of COVID-19 vaccines—an update. Diseases. 2022;10:112. doi: 10.3390/diseases10040112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Islam M.A. A review of SARS-CoV-2 variants and vaccines: viral properties, mutations, vaccine efficacy, and safety. Infectious Medicine. 2023;12 doi: 10.1016/j.imj.2023.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Hu L., Sun J., Wang Y., Tan D., Cao Z., Gao L., Guan Y., Jia X., Mao J. A review of inactivated COVID-19 vaccine development in China: focusing on safety and efficacy in special populations. Vaccines. 2023;11:1045. doi: 10.3390/vaccines11061045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Vohra-Miller S., Schwartz I.S. NVX-CoV2373, a protein-based vaccine against SARS-CoV-2 infection. CMAJ (Can. Med. Assoc. J.) 2022;194 doi: 10.1503/cmaj.220688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Marchese A.M., Kalkeri R., Vadivale M., Suntronwong N., Toback S., Poovorawan Y. Pivoting to protein: the immunogenicity and safety of protein-based NVX-CoV2373 as a heterologous booster for inactivated and viral vector COVID-19 vaccines. Expet Rev. Vaccine. 2023;22:620–628. doi: 10.1080/14760584.2023.2232020. [DOI] [PubMed] [Google Scholar]
- 194.Mengist H.M., Kombe A.J.K., Mekonnen D., Abebaw A., Getachew M., Jin T. Mutations of SARS-CoV-2 spike protein: implications on immune evasion and vaccine-induced immunity. Semin. Immunol. 2021;55 doi: 10.1016/j.smim.2021.101533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Rayati Damavandi A., Dowran R., Al Sharif S., Kashanchi F., Jafari R. Molecular variants of SARS-CoV-2: antigenic properties and current vaccine efficacy. Med. Microbiol. Immunol. 2022;211:79–103. doi: 10.1007/s00430-022-00729-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Ao D., Lan T., He X., Liu J., Chen L., Baptista‐Hon D.T., Zhang K., Wei X. SARS‐CoV‐2 Omicron variant: immune escape and vaccine development. MedComm. 2022;3:e126. doi: 10.1002/mco2.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Zandi M., Shafaati M., Shenagari M., Naziri H. Targeting CD47 as a therapeutic strategy: a common bridge in the therapy of COVID-19-related cancers. Heliyon. 2023;9 doi: 10.1016/j.heliyon.2023.e17959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Ebrahim-Saraie H.S., Dehghani B., Mojtahedi A., Shenagari M., Hasannejad-Bibalan M. Functional and structural characterization of SARS-Cov-2 spike protein: an in-silico study. Ethiopian Journal of Health Sciences. 2021;31(2) doi: 10.4314/ejhs.v31i2.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets are available from the corresponding author upon request.