Abstract
The article divulges the crystal growth, synthesis, and X-ray structure characterization of one centrosymmetric cadmium complex, [Cd{CdL(μ2-1,3-acetate)}2] using Salen ligand (SL). The complex is further characterized using spectroscopic and analytical techniques, including DRS, SEM-EDX, PXRD, and ICP-MS. The crystallographic study showed that the complex has a monoclinic space P21/c. Addison parameters (Ʈ) show the hexagonal geometry of the central Cd(II) metal ion. Hirshfeld surface and 2-D fingerprint confirm supramolecular contacts despite weak C–H⋯O and C–H···π interactions. Energy frameworks, FMOs, global reactivity parameters, MEP, and energy bandgap explain the complex reactivity outlook. The complex inter- and intramolecular bonding interactions were explored through natural bond orbital (NBO), QTAIM, NCI-RDG, Electron Location Function (ELF), and Localized Orbital Locator (LOL) quantization methods. In addition, the complex and its synthetic components in vitro antibacterial efficacy were investigated using Gram-positive and Gram-negative microbial strains. SAR (structure-activity relationship) correlates with biological potency. Molecular docking assessed antimicrobial potency with proteins S. aureus (PDB ID: 1JIJ), C. albicans (PDB ID: 1M7A), E. coli (PDB ID: 1T9U), P. aeruginosa (PDB ID: 2UV0), and A. Niger (PDB ID: 3K4P). The findings are backed by the Protein-Ligand Interaction Profiler (PLIP). The antifungal potency and cell viability test of C. albicans were conducted using photodynamic therapy (APDT).
Keywords: Acetate, Salen ligand, Cd(II), DFT, Antimicrobial, APDT
Graphical abstract
Highlights
-
•
An X-ray characterized Cd-complex, [Cd{CdL(μ2-1,3-acetate)}2], was synthesized.
-
•
HS, Energy frameworks, HOMO-LUMO, and MEP properties were analysed.
-
•
NBO, QTAIM, NCI-RDG, and ELF-LOL explained complex bonding outlooks.
-
•
The antimicrobial activities tie up via MD and PLIP experiments.
-
•
APDT evaluated the antifungal efficacy and cell viability test of C. albicans.
1. Introduction
The astonishing discovery of Schiff bases (SBs) by German chemist Hugo Schiff in 1864 [1] (Scheme S1) revolutionized the coordination chemistry research. From today's research viewpoint, H. Schiff is a cornerstone of each branch of coordination chemistry. Among the SBs, the Salen ligand (SL) is a particular class of N/O-donor ligands (N2O2/N2O4-type) [[2], [3], [4], [5], [6], [7], [8], [9], [10], [11]] coordinated with Mn + ions via the N-atom of azomethine group (-CH N) and phenolic O-atom after de-protonation [12] (Scheme S2). However, synthetic chemists have expanded in contemporary research using SL due to its excellent properties and flexibility of complex crystal structures [[13], [14], [15], [16], [17], [18], [19], [20]]. In modern research, coordination chemists have prepared multidimensional metal complexes using SL or SL-like compartments [3,9,21,22] (Scheme S3). In addition, to enhance the formation of homo/heteronuclear complexes, several co-ions, AcO−, SCN−, NO3−, [N(CN)2]-, cyanate (OCN−), and azido ions (N3−) have been used [3]. There has been a growing interest in researching metal acetate complexes [2,4,12,[23], [24], [25], [26], [27]], particularly concerning their μ-acetate bridging flexibility (Scheme S4), which began after exploring the family of diverse coordination networks of cadmium acetate crystals [28]. The study of acetate complexes is growing due to their wide application in supramolecular chemistry, molecular magnetic materials, metal-organic chemical vapour deposition precursors [28], electronic and optoelectronic equipment [3,29], and luminescence properties. In contrast, Cd-compound structures in the solid state are often unpredictable [28]. Yet, various coordination geometries are explored, including tetrahedral (Td), trigonal bipyramidal (TBP), octahedral (Oct), distorted pentagonal bipyramidal (PBP), and dodecahedral [30]. The sizeable ionic radius of Cd(II) accounts for the structural variations in its coordination geometry. The d10 electronic configuration eliminates CFSE values and permits diverse geometries. Further, supramolecular interactions (SE) are crucial in structural biological chemistry [31] and harmonizing various biological and physicochemical factors sometimes involves utilizing multiple non-covalent supramolecular interactions [32]. In recent decades, photosensitizers (PSs) have been developed to treat infectious diseases through a methodology known as photodynamic inactivation (PDI) of microorganisms [32]. This innovative methodology has opened the door to a new research frontier. PDI involves three stages: PS accumulation into microbial cells and ROS generation when illuminated. Finally, the intermediates react with macromolecules in cells, causing irreversible damage and cell death. Intriguingly, synthetic researchers have recently entered the research field of antifungal photodynamic therapy (APDT) and cell viability testing [[33], [34], [35]]. The standard cell viability test was conducted for the duration of yeast eradication [36,37]. One of the most prevalent microbes linked to fungal infections is C. albicans. Antimicrobial photoinactivation (APDI) can be used as an alternative therapy instead of standard treatment, and it was examined for various photo-oxidizable substrates. It is well-established that Cd(II) compounds coordinated with SB/SL demonstrate effective antibacterial and fungal properties [[38], [39], [40], [41], [42]]. However, recently, antimicrobial research has expanded beyond SB/SL compounds to include 3D Ag-MOCP, Cd-CP, 2D Hg-MOCP, Cu-CP, and Nano-structures of Zn(II) complexes [[43], [44], [45], [46], [47]]. The literature extensively covers antimicrobial research on various transition metal complexes [48,49]. Still, our Cd(II)-Salen complex stands out due to its unique crystal structure by μ2-1,3-acetate bridging evidenced by SCXRD, supramolecular contacts, in-depth biological spectrum (antimicrobial and PDT), MD/PLIP, SAR, and quantum chemical investigations. The article clarifies the uniqueness and novelty, particularly in the section on DFT and PDT investigations against C. albicans, using a Newport Xenon arc lamp of 300W as a UV–visible region light source. Studies on the subject above are scarce in the existing literature, especially on the cadmium acetate complexes. Currently, researchers are exploring research on DFT-quantized steel corrosion [50,51] and sensor activity [52]. Studies on DFT are famous for exploring novel properties of synthesized compounds, including energy frameworks, MEP, global chemical parameters, energy bandgap, FMO, NBO, QTAIM, ELF-LOL, and NCI-RDG [[53], [54], [55], [56], [57], [58], [59]]. Contrarily, MD/PLIP research studies establish a reliable biological correlation between experimental and theoretical data [[60], [61], [62]].
In this article, we endeavour to investigate a Cd(II)-Salen complex encompassing synthesis, characterization, and crystallographic features. DFT explored the complex chemical reactivity and bonding rapport. MD/PLIP and SAR compared the antimicrobial efficacy of complex and SL. The cell viability of C. albicans was systematically achieved by photodynamic therapy (PDT).
2. Experimental
2.1. Materials and instrumentation
The current research chemicals used are reagent grade (RG). PerkinElmer instrument offers a service to measure the CHN. Model number PerkinElmer Spectrum RX 1 instrument analyses IR spectra. Bruker RFS 27 (4000-50 cm−1) is used for Raman spectra as KBr pellets. NMR spectra were collected using a Bruker FT-NMR spectrometer model ranging from 400 MHz to 75.45 MHz The Oxford XMX N model collected the EDX spectrum data. The JEOL (JSM-6390LV) and BRUKER AXS investigated the SEM figures and PXRD. The ICP-MS analysis was used for the instrument Thermo-Scientific, iCAP RQ model, along with Helium KED mode (Kinetic Energy Discrimination). The U-3501 spectrophotometer model is designed to analyse synthesized compounds' UV–visible spectra. We used the PerkinElmer Lambda 365 model (200–1000 nm) for the diffuse reflectance spectroscopic (DRS) study, utilizing a DRS integrating sphere with a 50 mm diameter and an 8° angle of incidence.
2.2. Bacterial/fungal cultures
The microbial strains utilized in this study were received from (MTCC). The MTCC stands for Microbial Type Culture Collection. The bacterial strains were cryopreserved in the MHB under experimental conditions at −80 °C and 30 % glycerol, maintained by vol/vol. Generally, MHB is meant for Mueller-Hinton broth. Before testing, they were subculture on Mueller-Hinton agar (MHA) (30 °C for one day). The examined compounds were dissolved in dimethyl sulfoxide (DMSO) to create stock solutions. This process was done under sterile conditions to ensure purity and accuracy. Overall, solutions were completely preserved in dark environments for 2-weeks. Then, the referenced solutions were diluted in dimethyl sulfoxide using the serial dilution technique when necessary.
2.2.1. Minimum inhibitory concentration (MIC)
The agar dilution method (ADM) determines the investigated compounds' MIC (minimum inhibitory concentration). Two mediums were used: Muller Hinton agar and yeast malt extract. A solution with a 5 mg/mL concentration was added for the intended purposes. The attention of compounds concentration presents in the plates containing medium ranged. It will generally range from 0.005 to 3 mg/mL 0.1 mL of active microbial strains (antibacterial/fungal) were streaked onto plates with a sterile toothpick. The term MIC refers to the concentration of an extract that inhibits microbial growth at the lowest level. Bacteria and yeast samples were incubated at two different temperatures. It covers at 37 °C and 25 °C, respectively. The duration of the experiment is one day. The control group was replaced with DMSO to compare with the test group. The plates were incubated in two different ways for Candida albicans. A plate was exposed to white light for 180 min. After that, it was set at 37.1 °C for one day. The second dish was incubated in darkness at 37 °C for 24 h. The MIC value is the lowest substance concentration that can visibly inhibit fungi growth.
2.2.2. Light source
The photoinactivation experimental studies were conducted for 180 min at a light dose of 360 J/cm2 and 50 mW/cm2 irradiance under white-light LED irradiation (400–800 nm, visible range). According to the standard protocol, the 96-well plates, which were exposed to radiation, remained closed and 10 cm away from the light source. The experiments were performed in triplicate [63].
2.2.3. Cell viability curve test
A cell viability curve experiment determined the duration of C. albicans growth suppression to the treatment. Previously, the complex's MIC against the isolates determines the BMP (Broth Microdilution Process). After preparing two 96-well plates containing the minimum inhibitory concentration (MIC), +ve, and a -ve control. One necessary plate was incubated in dark conditions at 37 °C for 120 min, while the other was irradiated at white light. At minute intervals (0, 10, 15, 30, 60, 120, and 180), each 10 μL aliquot successively was taken from the wells and placed on Savoured dextrose agar plates for seeding. The reference plates were incubated at 37.1 °C for one day. Finally, after completion of the said techniques, the colonies were counted according to the standard protocol. After that, the colony-forming units (CFU) were evaluated and reported in terms of Log10 CFU/mL [64].
2.3. Synthesis of ligand (SL)
The widely used SL was made using the method described in Ref. [65]. Herein we submitted the ligand characterization data. Yield: Satisfactory (93 %), Anal. Calc. for C17H18N2O2: C, 72.32; H, 6.43; N, 9.92 Found: C, 72.28; H, 6.50; N, 9.88 %. IR (KBr cm−1) selected bands: ν(C꞊N), 1632, ν(C-Ophenolic)1278, ν(OH), 3441, 1H NMR (DMSO‑d6, 400 MHz): δ (ppm): 13.53 (s, 1H, OH), 8.59 (s, 1H, N CH), 6.87–7.45 (m, 8H, Ar–H), 3.40–3.69 (t, 2H, N–CH2), 2.01–2.51 (m, 2H, CH2), 13C NMR (DMSO‑d6, 75.45 MHz): δ (ppm): 116.92–132.76 (Arom-C), 161.16 (C–OH), 166.67 (CH]N), UV–Vis λmax (CH3OH): 254, and 331 nm.
2.4. Synthesis and crystal growth of [Cd{CdL(μ2-1,3-acetate)}2]
In a porcelain mortar, solid Cd(OAc)2.4H2O (0.267g, 1 mmol) and Salen ligand (SL) (0.0742g, 1 mmol) were ground and then poured in 40 mL of hot methanol. The solution was agitated with a magnetic stirrer for roughly 30 min. Then, a 5 mL DCM (dichloromethane) solution was added to develop a better diffracting quality crystal. The solution was refluxed for 3 h and filtered through the Whatman 41 filter paper. The overall solution crystallizes slowly at room temperature. Needle-sized, yellow, single crystals suitable for SCXRD were obtained after a few days. After filtration, the crystals were carefully preserved and air-dried in a desiccator. Yield: 72 %, Anal. Calc. for C38H38Cd3N4O8: C, 44.92; H, 3.77; N, 5.51, Cd, 33.19. Found: C, 44.88; H, 3.70; N, 5.48, Cd, 33.13 %. ICP-MS for Cd, analysis 32.98 %. FT-IR (KBr cm−1) selected bands: ν(C꞊N), 1645 m, ν(Cd–N), 554, ν(Cd–O), 460, ν(AcO), 1409, 1611, ν(C–H), 2856–2950, ν(N–H), 3030, ν(Ar–O), 1198, FT-Raman (cm−1) selected bands: ν(C꞊N), 1642, ν(Cd–N), 545, ν(Cd–O), 454, ν(AcO), 1445, 1602, ν(C–H), 2839–2951, ν(N–H), 3065, ν(Ar–O), 1225, 1H NMR (DMSO‑d6, 400 MHz): δ (ppm): 2.14 (s, 3H), 6.85–7.33 (s, Arom-H), 8.31 (w, 1H), 3.73 (s, 2H), 13C NMR (DMSO‑d6, 75.45 MHz): δ (ppm): 117.00–132.30 (Arom-C), 161.12–165.47 (CH]N), UV–Vis λmax (DMF): 279 and 341 nm and DRS: 283, 302, 322 nm and 358 nm.
2.5. X-ray crystallography
The diffraction data was collected at room temperature on a smart Bruker CCD diffractometer employing Mo Kα radiation with a wavelength of 0.71073 Å. We have used several crystallographic programs to solve the acetate complex crystal structure. The crystallographic programs are step-by-step and mentioned in sequential order. First, information frames, index reflections, and scale and lattice parameter collections were performed using SMART and SAINT [66]. 2ndly, the absorption correction was performed by SADAB [67]. SHELXTL has carried out for space group structure determination. F2 does the crystal least-squares refinements. The crystal structure was determined using SHELXL-2014 [68] and Olex-2 software [69] with full-matrix least-squares refinement against F2. The anisotropic shift parameters were used to increase the accuracy of non-H atom refinement. By fixing the positions of the H-atoms, we have successfully achieved a highly sensitive isotropic that meets our desired standards. The crystal structure in the checkcif report has no A-level alert (R = 0.02), indicating high-quality crystal data. The details regarding the crystallographic information and structure refinement parameters are displayed in Table 1.
Table 1.
Crystal data and full structure refinement.
| Empirical formula | C38H38Cd3N4O8 |
|---|---|
| Formula weight | 1015.95 |
| Temperature/K | 293(2) |
| Crystal system | monoclinic |
| Space group | P21/c |
| a/Å | 10.5826(12) |
| b/Å | 20.5198(19) |
| c/Å | 8.4416(8) |
| α/° | 90 |
| β/° | 91.748(4) |
| γ/° | 90 |
| Volume/Å3 | 1832.3(3) |
| Z | 2 |
| ρcalcg/cm3 | 1.842 |
| μ/mm−1 | 1.783 |
| F(000) | 1004.0 |
| Crystal size/mm3 | 0.300 × 0.200 × 0.100 |
| Radiation | MoKα (λ = 0.71073) |
| 2Θ range for data collection/° | 5.532 to 54.316 |
| Index ranges | −13 ≤ h ≤ 13, −26 ≤ k ≤ 26, −10 ≤ l ≤ 10 |
| Reflections collected | 34724 |
| Independent reflections | 4068 [Rint = 0.1175, Rsigma = 0.0481] |
| Data/restraints/parameters | 4068/0/242 |
| Goodness-of-fit on F2 | 1.068 |
| Final R indexes [I ≥ 2σ (I)] | R1 = 0.0298, wR2 = 0.0776 |
| Final R indexes [all data] | R1 = 0.0331, wR2 = 0.0802 |
| Largest diff. peak/hole/e Å−3 | 1.03/-1.17 |
| CCDC | 2314897 |
2.6. DFT methodology
The CIF structure, extracted from X-ray diffraction analyses of the studied compound, was optimized in the gas phase using the DFT/B3LYP-D3/lanL2DZ implemented in the Gaussan09 package [[70], [71], [72], [73], [74]]. Solid and transparent MEP plots were generated using Gauss View 5 [75]. TD-DFT/B3LYP-D3 was employed to calculate the frontier molecular orbitals (FMOs), global reactivity parameters, and the energy bandgap energy of our studied complex [76,77]. Subsequently, our complex's inter- and intramolecular bonding interactions were determined through natural bond orbital (NBO). The QTAIM topological parameters, their extensions, NCI-RDG, the Electron Location Function (ELF), and the Localized Orbital Locator (LOL) were generated using the Multiwfn program [78]. The Crystal Explorer v17.5 software can create HS and 2-D fingerprint plots by processing CIF input files of the cadmium complex. In HS, different colour codes signify that Red is always shorter contact, white is for contacts within the van der Waals separation, and blue means no close connection [79]. The Crystal Explorer v17 software utilized the HF/3-21G energy calculation method to analyse intermolecular interaction energies. The Molecular docking (MD) technique exists to compare the biological effects of ligand (SL) and metal complexes. Here MD simulations were carried out to evaluate its potential antimicrobial activity with proteins that are S. aureus (PDB ID: 1JIJ) [80], C. albicans (PDB ID: 1M7A) [81], E. coli (PDB ID: 1T9U) [82], P. aeruginosa (PDB ID: 2UV0) [83], and A. Niger (PDB ID: 3K4P) [84]. The investigation utilizes software like HEX. As mentioned above, the bacteria interact with the SL and its Cd complex. According to research, biological activities are accelerated [85]. HEX 8.0.0 has successfully investigated the bacteria and the complex molecule files [[86], [87]].. Concerning the docking study, the components are recommended: correlation is of shape only, mode of 3-D FFT, 0.6-D grid, 180° receptor (R), L ranges, 360° twist range (TR), and 40° distance range (DR). The computational high-profile PLIP study established SL and the complex attractions with the bacterial strains [88,89].
3. Results and discussion
3.1. Synthetic perspective
The previous literature method prepared the Salen ligand (SL) [65]. The self-assembly of SL and cadmium acetate tetrahydrate, [Cd(OAc)2.4H2O] in CH3OH solvent (1:1 M ratio), produced an air-stable acetate-bridged cadmium crystal complex with satisfactory yield (71 %). SL and its complex structure have been thoroughly characterized. The compound structure is characterized by IR/Raman/NMR, UV–vis, DRS (diffuse reflectance spectroscopy), SCXRD, powder X-ray diffraction (PXRD), SEM/EDX, combined CHN, and ICP-MS techniques. The PXRD analysis has confirmed the phase's purity and crystallinity. The research on acetate compounds began after discovering the crystal networks of cadmium acetate [28], leading to more complex investigations. However, the literature on Salen ligand (SL) involving cadmium acetate complexes is limited. Apart from that, there has always been little exploration in the literature of the properties of Cd(II)-l-acetate complexes characterized by X-ray [2,4,12,[23], [24], [25], [26], [27], [28]]. Therefore, our research team is searching for new cadmium acetate Salen complexes to discover their novel properties. The current study investigates a novel cadmium complex, [Cd{CdL(μ2-1,3-acetate)}2], which exhibits unique properties through DFT analysis, which includes HS, NBO, QTAIM, ELF-LOL, NCI-RDG, SAR, MD, and PLIP. The complex also shows antibacterial and antifungal efficacy and is effective in antifungal photodynamic activities (APDT) and cell viability curve tests. All are novel works related to the literature published on Cd(II)-l-acetate complexes (L = SB/SL) [2,4,12,[23], [24], [25], [26], [27], [28]]. Scheme 1 highlights the overall synthetic protocol, including the Ligand (SL) and the centrosymmetric Cd(II) complex.
Scheme 1.
The synthetic outline of Salen ligand (SL) and the complex.
3.2. Characterization
3.2.1. FTIR and Raman spectra
We have conducted a thorough study using priority-based IR (Fig. S1-Fig. S3) and Raman spectroscopy (Fig. S4) to characterize the cadmium complex. First, the (-CH N) bonding feature in the ligand and the complex was identified based on IR and Raman's study. The Nitrogen-donor atom in the (-CH N) group coordinates with the cadmium (II). It confirms the presence of the linkage in the complex—formation of the ligand's ν(C N) group results at 1627 cm−1. In the complex, the reference bonding is found to be 1640 cm−1 [65]. In addition, complex IR spectroscopy identified at 1225 cm−1 for ν(Ar−O), 2839–2951, 454, and 545 cm−1, correspondence stretching bands like ν(C–H), ν(Cd–O), and ν(Cd–N), respectively. Further, 1445 and 1602 cm−1 for ν(AcO−) bridging, 3065 for ν(N–H) [2,4,12,[23], [24], [25], [26], [27], [28]]. We have conducted a Raman spectroscopic study to understand the complex structural framework better. The complex Raman spectra have confirmed the existence of the (-HC N) group at a frequency of 1642 cm−1. Raman peaks like 1225 cm−1, 2839–2951, 454, 545 cm−1, 1445 and 1602 cm−1, 3065 cm-1 were also found in the complex. All these values confirm the presence of ν(Ar–O), ν(C–H), ν(Cd–O), ν(Cd–N), ν(Aco−) and ν(N–H). Table 2 has been submitted to aid in understanding the IR/Raman stretching frequencies of the synthesized complex.
Table 2.
FTIR and Raman stretching values (cm−1).
| Complex | ν(C N) | ν(M − N) | ν(M − O) | ν(AcO−) | ν(C–H) | ν(N–H) | ν(Ar–O) |
|---|---|---|---|---|---|---|---|
| IR | 1645 | 554 | 460 | 1409 1611 |
2856-2950 | 3030 | 1198 |
| Raman | 1642 | 545 | 454 | 1445 1602 |
2839–2951 | 3065 | 1225 |
3.2.2. UV–vis and DRS spectrum
The UV–vis absorption spectra in solution and solid-state (DRS) were used to investigate the possible optical transitions in SL and the cadmium(II) complex (Fig. S5-Fig. S6A). The UV spectrum was analysed in methanol (SL) and dimethyl sulfoxide (DMSO) solvents (complex), one after the other. The SL absorption bands exhibit near 260 and 302 nm, respectively. Generally, π→π* and n→π* transitions are associated with these bands. The complex dissolved in DMF shows UV bands at 279 and 341 nm, respectively. Based solely on the ligand's UV–Vis domain, these bands result from the SL to Metal (M) CT, which are likely to be π→π* or n→ π* types. It was seen that the complex causes a shift towards longer wavelengths compared to the free SL. Such an effect is produced due to the coordination of SL (N and O-donor atoms) with Cd(II) ions. We also conducted diffuse reflectance spectroscopy (DRS) for the complex to check if any shoulder peak existed in the DRS spectroscopy. Herein, three slightly shifted bands are observed, along with the shoulder. The two absorption bands at 283, 302 nm, and 322 nm correspond to the intra-ligand π→π* or n→ π* type transitions. A new shoulder peak is identified at 358 nm. The broad DRS nature supports SL's complexation with Cd(II) metal ions. Cd(II) has a fully occupied d10 configuration; no d-d transition is initiated by the complex's Cd(II) ions. The absorption bands match those of X-ray-characterized Cd(II)-acetate complexes [2,90,91].
3.2.3. NMR spectra
An NMR spectroscopic study (1H/13C NMR) was conducted in a complete phase to characterize the structural frameworks of the Salen ligand (SL) (Fig. S7-Fig. S8) and its complex (Fig. S9-Fig. S10). We prioritize performing the 1H NMR characterization before proceeding with the 13C NMR approach. In our analysis of SL, we can't observe a peak between δ 5.0–8.1 ppm, which reflects the lack of a –NH2 group. The O–H peak protons are δ 13.53 ppm, supporting that SL possesses an uncoordinated -OH group. The δ 6.87–7.45 ppm is representative of the Aro-H present in SL. The proximity of –OH and imino N groups causes an δ 8.31 ppm downshift in the complex protons connected to the imino-C [55]. The complex identifies the Aro-H peaks at δ 6.85–7.33 ppm. The methoxy (–OCH3) group is characterized by the well-known proton peak at 3.76 ppm [92]. The O–H proton peak disappeared in the complex's 1H NMR spectra. After deprotonation, it ensures that the O-atom is coordinated with the Cd(II) ions [93,94]. The 13C NMR spectra of SL explored the (CH N)–C at 166.67 ppm, Aro-C at 116.92–132.76 ppm, and C–OH at 161.16 ppm, respectively. Similarly, reference peaks were identified in the complex at 161.12–165.47 ppm and 117.00–132.30 ppm. We can determine the structural frameworks of SL and the cadmium(II) acetate complex by analysing the combined NMR spectra.
3.2.4. EDX-SEM
We use EDX and SEM techniques to analyse the elemental composition, morphology, and complexity of SL and the complex. Fig. S11 shows that the compound contains C, O, and Cd metal ions. According to this profile, the highest peak in the EDX spectrum is reached by C, followed by O and Cd. In addition, the weight % contribution of C, O, and Cd metal reflect the EDX profiles. The EDX study confirms the interaction between Cd(II) metal ions, SL, and acetate co-ligands. SEM is an essential diagnostic process to determine synthesized compounds' morphology [95]. In the current analysis, the SEM image of SL displays an irregular ice-type morphology (Fig. S12). The acetate complex structure also reveals a morphology based on ice in a distributed manner.
3.2.5. Inductively coupled plasma mass spectrometry (ICP-MS)
We characterize the complex based on the Cd metal % utilizing ICP-MS techniques (where *112Cd isotope is used). The cadmium complex was digested using an acid digestion, commonly conc. HNO3. It was then filtered, diluted, and analysed. The process found 32.98 % Cd, ensuring nearly agreement with the calculated cadmium. The detection limit is up to 0.01 ppm.
3.2.6. Powder X-ray diffraction
The powder X-ray diffraction analysis (PXRD) successfully verified the phase purity and crystallinity of the cadmium complex. In the true sense, we used PXRD (Fig. S13) to determine the structure of the acetate complex, ensuring sample consistency. Herein, experimental PXRD (EPXRD) patterns of bulk samples are consistent with those simulated from SCXRD results. CCDC Hg-software was used to generate Complex Simulated Patterns (CSP) from single crystal structural data in CIF (Crystal Information Files). Therefore, according to the PXRD profile, the complex bulk material is a single crystal.
4. X-ray single-crystal structure
4.1. Crystal structure description
The centrosymmetric cadmium acetate compound which was found to be crystallized in the monoclinic space group P21/c, was determined by single-crystal X-ray diffraction studies. The intricate structural framework comprises isolated trinuclear [Cd{LCd(μ2-1,3-OAc)}2] units. Table 1 provides crystallographic data and refinement details. The different cell parameters for the cadmium complex were determined through crystallography as: a = 10.5826 (12) Å, b = 20.5198 (19) Å, c = 8.4416 (8) Å, and α = γ = 90° and β = 91.748° (4), Z = 2, and Volume/Å3 = 1832.3(3). Table S1 contains important crystallographic parameters that have been collected for the complex. The complex comprises two terminals known as 'metalloligands' (CdL). One central cadmium atom is connected to two acetate co-ligands through a bridging of μ2-1,3, as shown in the perspective asymmetric unit view (Fig. 1a–b). The marked Cd(1) and Cd(1)* of Cd(II) centers are bound to two nitrogen atoms from an imine group, [N(1), N(2) for Cd(1) and N(1)*, N(2)* for Cd(1)*] and two O-atoms from phenoxy group [O(1), O(2) for Cd(1) and O(1)*, O(2)* for Cd(1)*] of the [SL]2- [* = 1-x,1-y,1-z]. An O-atom coordinates the 5th site, specifically O(3) for Cd(1) and O(3)* for Cd(1)*, from an AcO− co-ligand. The Addison parameters (Ʈ) were used to calculate the geometrical distortion from SQP (Square pyramid) to TBP (Trigonal bipyramid) around both Cd(II) centers [96]. A zero (0) value indicates an ideal SQP, while one (1) indicates a TBP. The Cd(1) and Cd(1)* centers have Ʈ values of 0.015 each, indicating square pyramidal geometries around Cd(II) centers (Fig. 2b). Fig. 2a shows the hexagonal geometry of the central Cd(II) metal ion according to Addison parameters (Ʈ). The N(1)-Cd(1)-O(2), and N(2)-Cd(1)-O(1) trans angles around Cd(1)] are found to be 140.8(2)°, and 141.71(8) °]. For Cd(1) metal center, the deviations of coordinating atoms from the mean plane are- O(1): 0.131(17) Å, O(2): 0.156(17) Å, N(1): 0.161(3) Å, N(2): 0.141(3) Å. From the same plane, the deviation of Cd(1) is −0.590(17) Å. The chelate rings comprising six members and containing cadmium atoms exhibit a puckered envelope conformation. The following are the chelating parameters: q = 0.639(3) Å, θ = 176.9(3), and φ = 95(4). Here the chelate ring are built by [Cd(1)-N(1)-C(8)-C(9)-C(10)-N(2)]. The overall geometry of the central Cd(II) metal ion is hexa-coordinated. Two [SL]2- phenoxy oxygen atoms and two oxygen atoms from AcO− form a distorted OCT (octahedral geometry) (Fig. 2a). The distances between Cd–O(phenoxy) atoms are larger, ranging from 2.269 Å to 2.295 Å, compared to those between Cd–O(acetate) atoms, which is 2.336 Å. The metal-metal distance of Cd(1)–Cd(2) is 3.263 Å. The crystallographic features are comparable to the X-ray-characterized literature published on cadmium acetate complexes (Fig. S14).
Fig. 1a–b.
The asymmetric unit (a) and perspective view of the complex (b) (selective atom numbering scheme).
Fig. 2a–b.
Perspective view of (a) octahedral geometry of cadmium (II) center and (b) square pyramidal geometry of cadmium(II) center.
4.1.1. Supramolecular interactions
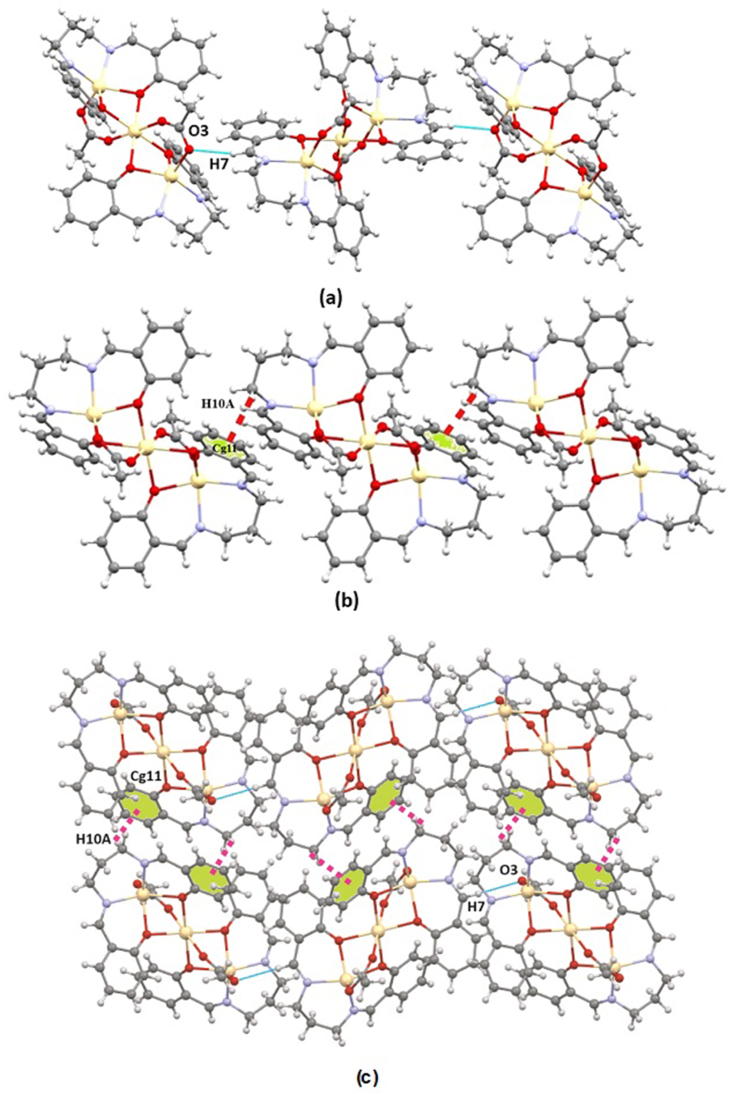
The complex's crystal packing exhibits feeble H-bonding interactions. These interactions are developed by C–H⋯O and C–H···π categories. Fig. 3a shows a symmetry-operated (2-x,1-y,1-z) inter-molecular H-bond between H(7) atoms and O(3) atom, where H(7) is attached to C(7) atom. The significant geometric parameters are D-H = 0.93 Å, H … A = 2.52 Å, D-H … A = 167°, D … A = 3.437(3) Å. A hydrogen atom, H(10A), attached to C-atom, C(10), forms a C–H···π interaction with a symmetry operated (2-x, 1-y,1-z) ring, Cg(11) (Fig. 3b). The different geometric parameters in the complex are H … Cg = 2.75 Å, X–H … Cg = 125°, X … Cg = 3.409(3) Å. Crystal packing obtained from, C–H⋯O and C–H···π interactions resulted in a 2D structure shown in Fig. 3c. The hexagonal shape of 3D crystal packing of the complex is shown in Fig. 4.
Fig. 3a–c.
Perspective view of C–H⋯O (a), C–H···π (b) interactions in the complex, and for (c) Perspective view of supramolecular interactions with selective atom numbering scheme (pink dotted line refers to C–H···π and the blue dotted line refers to C–H⋯O interactions).
Fig. 4.
Perspective view of the hexagonal shape of 3D crystal packing of the complex.
5. DFT quantization's
5.1. Hirshfeld surface
The interactions between molecules in the synthesized compound are measured through Hirshfeld surface analysis (HSA). This method is a visual aid that helps to understand and analyse various intermolecular interactions using graphical representations. Fig. 5a–c (top) explores the HS of the complex. Using dnorm (−0.5 to 1.5 Å), shape index (−1.0 to 1.0 Å), and curvedness (−4.0 to 0.4 Å), Hirshfeld surface has been mapped. According to such analysis, the most significant contacts were identified by H⋯O (18.5 %). Apart from that, minor contacts like H⋯N (2.1 %) and Cd⋯O (3.5 %) are present in the HS profile. Molecular interactions across the molecular region are presented from different colours of fragment patches (Fig. 5e). In Fig. 5d, di denotes distances from the Hirshfeld surface to the nearest nuclei inside the surface. 2-D fingerprint plots visualize such minor contacts (Fig. 5, Bottom). Overall, HS and 2-D fingerprint tools are introduced in the cadmium complex in three categories of (%) contacts: H⋯O, H⋯N, and Cd⋯O.
Fig. 5a–c.
(Top) HS mapped with dnorm (a), shape index (b), and curvedness (c) di (d) fragment patch (e), (Bottom) 2-D fingerprint plots.
5.1.1. Interaction energy and energy frameworks
According to such analysis, a cluster of molecules involves employing crystallographic symmetry operations (CSP) to a central molecule. These operations are based on the symmetrical arrangement of atoms within the molecule, resulting in multiple identical molecules arranged in a symmetrical pattern around the central molecule. The distance between each molecule and the central molecule is carefully controlled, typically at a radius of 3.8 Å (Fig. S15). [97]. The total energy between molecules (Etot) is calculated using four types of standard energy: electrostatic energy (Eele), polarization energy (Epol), dispersion energy (Edis), and exchange-repulsion energy (Erep) (Table 3). The four terms have different weights, with scale factors of 1.019, 0.651, 0.901, and 0.811 [98]. The complex has an interaction energy (IE) of −251.6 kJ/mol, comprising Eele = −81.7, Epol = −36.3, Edis = −301.2, and Erep = 156.4. According to Fig. 6a–c, the dispersion energy (DE) is the most dominant among all the other interaction energies. The reason for this occurrence is the existence of heavy atoms (Z high) that possess more electrons or electron clouds. Classical H-bonds are absent, resulting in a reduction in electrostatic energy (EE). The energy frameworks depict intermolecular interaction energies visually [99]. In a molecular system, the interactions between pairs of molecules can be visualized by drawing cylinders that connect the centroids of the two molecules. Herein, there is a direct relationship between the radius of a cylinder and the corresponding interaction energy (IE) [100]. Preparing three energy frameworks involves using red cylinders to represent Eele, green cylinders to depict Edis, and blue cylinders to symbolize Etot. These frameworks provide a visual representation of the energy distribution and help in understanding the energy components of the system (Fig. 6a–c).
Table 3.
Different interaction energies of the molecular pairs (kJ/mol).
Fig. 6a–c.
Perspective views of electrostatic energy (a), dispersion energy (b), and total energy diagrams (c) constructed from the energy framework for a cluster of molecules in the complex. (The cylindrical radius is proportional to the relative strength of the corresponding energies, Scale factor used is 100 with cut-off values of 10 kJ/mol).
5.1.2. FMO and reactivity parameters
In this study, Fig. 7 shows HOMO-LUMO distributions, associated energy levels, and quantum chemical parameters (Table 4). As depicted in the graph, it is evident that both the HOMO and LUMO distributions extend across the aromatic moieties. This finding indicates the presence of significant delocalized electron concentrations around all the central Cd metals, suggesting a potential electron charge transfer occurring on the surface of our studied compound. Based on the calculated results, the HOMO is −8.98 eV, while the LUMO registers at −5.85 eV, resulting in a corresponding Eg of 3.13 eV. In general, a significant energy gap indicates more excellent stability and lower reactivity of a molecule, whereas a small energy gap suggests higher reactivity and reduced stability. Additional chemical parameters, like ionization potential (IP), electron affinity (EA), chemical potential (μ), global hardness (η), Mulliken electronegativity (χ), and electrophilicity index (ω), were evaluated by employing Koopman's theorem. The obtained low value of hardness (1.57 eV) indicates the ease with which electrons can transition from a ground state (S0) to an excited state (S1), characterizing the highly charged transfer occurring on the surface. Additionally, these parameters suggest that the complex exhibits moderate reactivity and stability, as reflected in the values of μ (−7.42 eV) and η (1.57 eV). The large value of χ indicates the effectiveness of this studied system in quickly forming an electronic charge transfer with the guest. Therefore, this complex may hold promise for biomedical applications. Furthermore, the elevated value of ω (17.55 eV) suggests the complex's readiness for reaction.
Fig. 7.
HOMO-LUMO distribution of the complex.
Table 4.
Calculated HOMO-LUMO energy levels and quantum chemical parameters.
| Quantum chemical parameters | |
|---|---|
| εHOMO (eV) | −8.98 |
| εLUMO (eV) | −5.85 |
| Ega | 3.13 |
| Ionization Potential, IP (eV)b | 8.98 |
| Electron Affinity, EA (eV)c | 5.85 |
| Chemical potential, μ (eV)d | −7.42 |
| Mulliken electronegativity, χ (eV)e | 7.42 |
| Global hardness, η (eV)f | 1.57 |
| Electrophilicity index, ω (eV)g | 17.55 |
| a: , b: , c: , d: , e: , f: , g: | |
5.1.3. MEP surface
Fig. 8 depicts the MEP maps of the studied complex, featuring various colours. The red region indicates areas with a negative electrostatic potential, where an electrophile (e.g., +ve) is likely to attack. The blue region is the local reactive site nucleophiles can strike due to its + ve electrostatic potential. A highly negative electrostatic potential is concentrated around O-atoms, indicating them as local nucleophilic sites in the SL and aromatic ring moieties. Meanwhile, acceptor sites surrounding the electronegative atoms suggest that this material exhibits acceptor characteristics, making it suitable for testing as a sensitive element for new sensor devices. Conversely, the electrophilic sites, marked by positive electrostatic potentials, are predominantly situated on the periphery, precisely above the hydrogen atoms. The analysis affirms the existence of intermolecular interactions between the central and peripheral regions.
Fig. 8.
MEP surface of the cadmium complex.
5.1.4. Natural bond orbitals (NBO)
Natural bond orbital (NBO) analysis is a powerful tool for examining inter- and intramolecular bonding, providing a convenient foundation for exploring charge transfer (CT) and conjugative interactions within molecular systems. NBO analysis often utilizes second-order perturbation energies (E(2)) as a quantitative tool to investigate bonding and antibonding interactions. This energy is determined using the standard expression:
| (1) |
Here, qi represents occupancy of donor orbital, F(i,j)2 denotes the off-diagonal matrix elements, and εi and εj are the donor (D) and acceptor (A) orbital energies (OE), respectively. Herein E(2) numerical data's quantify the interaction strength between occupied Lewis NBOs (OLNBO, donors) and unoccupied Lewis NBOs (UOLNBO, acceptors), providing insight into the degree of delocalization within the molecular species [101]. Table S2 lists the diverse interactions between donor (i) and acceptor (j) orbitals, with their 2nd-order interaction energy (IE) E(2) ≥ 10 kcal/mol corresponding to the analysed complex. Based on the results summarized in Table S2, the stabilization of the analysed complex is effectively influenced by six types of interactions: LP*→LP*, LP→LP*, π→π*, π*→π*, LP→σ*, and LP→π*. The significant transitions between donor-acceptor orbitals involve π*(C11–C22)→π*(C9–C12), π*(C16–C25)→π*(C20–C31), LP(O5)→LP*(C42), LP(O6)→LP*(C42), π*(C56–C67)→π*(C54–C57), π*(C61–C70)→π*(C65–C76), LP(O50)→LP*(C87), LP(O51)→LP*(C87), with the corresponding highest perturbation energies being 168.23, 168.20, 173.65, 150.56, 168.23, 168.21, 173.65, and 150.56 kcal/mol, respectively. In summary, the stability of the analysed complex is attributed to those above hyper-conjugative and non-bonding interactions.
5.1.5. QTAIM approach
QTAIM explores a clear explanation and visualization of interactions among diverse components within a given molecular complex. It offers a straightforward depiction of atoms and bonds, employing topological parameters determined at each bond critical point (BCP) [102,103]. The studied complex's AIM graph (Fig. 9) showcases the essential BCPs. Table 5 represents different AIM parameters. According to Fig. 9 and due to the symmetrical nature of the complex, we consider only BCPs (1-12). Based on the calculated data, the AIM parameters, namely the total electron density , Laplacian electron density , and electron energy density , data employed to evaluate the interactions illustrated at the BCPs using the following criteria [104]: weak interaction (electrostatic) occurs when > 0 and > 0; moderate interaction (H-bonding) is evident when > 0 and < 0; and strong interaction (covalent interactions) takes place when < 0 and < 0. As illustrated in Table 5, the values of and fall within the ranges of 0.004–0.052 a.u. and 0.0016 to 0.59 a.u., respectively. In contrast, the values of encompass both positive and negative values, indicating electrostatic interaction at BCP (4–8) and hydrogen bonding at BCP (1–3, 9–12). Furthermore, the energy of interactions ( is employed to measure the strength of bonds at the BCPs. The results confirm that the interactions at BCPs (1–8) are indeed the strongest, with values falling within the range of −85 to −122 kJ mol−1. These robust interactions involve the bonding between the metal atom (Cd), oxygen (O), and nitrogen (N) atoms. The high interaction energy coexisting in BCPs between Cd and oxygen (O) neighboring atoms exhibits the stability of the complex and the well-fixed metal in the centre of the aromatic ring. This idea confirms the sound, relaxed system of interactions in the crystal backing, as demonstrated in the experimental section. Ellipticity ε(r) serves as an indicator of structural stability. An ellipticity value between 0.5 and 1.0 indicates an unstable bond interaction, while a value of ε below 0.5 signifies a stable bond interaction. Consequently, all the BCPs exhibit low values of ε, suggesting that the studied complex demonstrates high stability.
Fig. 9.
AIM graph of the complex, highlighting the Bond Critical Points (BCPs).
Table 5.
The AIM parameters, computed at specific bond critical points (BCPs).
| BCPs | |||||||
|---|---|---|---|---|---|---|---|
| 1 | 0.042 | 0.33 | 0.073 | −0.073 | 0.0003 | 0.049 | −95.830 |
| 2 | 0.040 | 0.29 | 0.065 | −0.065 | 0.0004 | 0.063 | −85.328 |
| 3 | 0.040 | 0.29 | 0.065 | −0.065 | 0.0004 | 0.063 | −85.328 |
| 4 | 0.046 | 0.35 | 0.079 | −0.081 | −0.001 | 0.068 | −106.332 |
| 5 | 0.050 | 0.32 | 0.075 | −0.080 | −0.005 | 0.068 | −105.020 |
| 6 | 0.050 | 0.32 | 0.075 | −0.080 | −0.005 | 0.068 | −105.020 |
| 7 | 0.052 | 0.39 | 0.089 | −0.093 | −0.004 | 0.074 | −122.085 |
| 8 | 0.046 | 0.35 | 0.079 | −0.081 | −0.001 | 0.068 | −106.332 |
| 9 | 0.004 | 0.02 | 0.003 | −0.002 | 0.001 | 0.412 | −2.625 |
| 10 | 0.004 | 0.02 | 0.003 | −0.002 | 0.001 | 0.412 | −2.625 |
| 11 | 0.006 | 0.03 | 0.006 | −0.005 | 0.001 | 0.056 | −6.563 |
| 12 | 0.004 | 0.016 | 0.003 | −0.002 | 0.0009 | 0.244 | −2.625 |
: electron density, : Laplacian of electron density, : Lagrangian kinetic energy, : potential energy density, : energy density.
: ellipticity of electron density, and :interaction energy (kJ.mol−1).
5.1.6. NCI
To enhance our understanding of potential inter- and intra-molecular interactions within the analysed complex, non-covalent interaction (NCI) analysis was employed. These attractive or repulsive forces between atoms, not involving the sharing or transfer of electrons [59] are visually represented through this analytical approach. It has demonstrated effectiveness in distinguishing various types of interactions, including H-bonds, van der Waals interactions, and repulsive steric interactions. The NCI-graph (Fig. 10) explains all interactions in the studied complex in 3D. The graph shows feeble van der Waals interactions with green disks. In contrast, blue and red disks represent strong, attractive H-bonds and repulsive steric effects. H-bonds occur between Cd and O-atoms of the ligand (SL) moiety. Additionally, a steric effect manifests within the central aromatic rings. These findings highlight various van der Waals interactions in our complex. These results conclude that our complex is well-stabilized through van der Waals and hydrogen bonding interactions, confirming the excellent stability of the studied compound and the good fixation of the central metal Cd in interactions with oxygen (O) atoms in its active area.
Fig. 10.
NCI index of the studied complex.
5.1.7. ELF-LOL profile
The Electron Localization Function (ELF) and the Localized Orbital Locator (LOL) are undoubtedly significant methods for the gain of chemical bonding nature. These approaches help to locate critical areas in molecular space where electrons are concentrated. These areas include bonding, non-bonding, and lone-pair regions [58,105]. ELF and LOL share comparable chemical mappings as they both rely on kinetic energy density. However, ELF is based on the density of electron pairs, while LOL manifests as a gradient of localized orbitals, specifically utilized when localized orbitals overlap [106]. The shaded surface, ELF, and LOL colour-filled maps are presented in Fig. 11. The colour scales for ELF and LOL were plotted within the ranges of 0.0–1.0 and 0.0 to 0.8, respectively. Localized electrons, bonding, and non-bonding are present in regions where the ELF value exceeds 0.5. Conversely, regions with ELF values below 0.5 indicate the presence of delocalized electrons. The interpretation of LOL aligns with ELFs, providing supporting evidence for charge delocalization [107]. In Fig. 11, the ELF and LOL maps display various colours, predominantly shades of blue and red. The presence of red regions around the hydrogen atoms indicates a high localization of bonding and non-bonding electrons. This observation supports the possibility of electronic charge transfer on the surface of the studied complex, reinforcing the existence of a donor-acceptor couple between entities and confirming the system's stability. This claim aligns with the findings of the QTAIM analysis. Notably, blue regions are observed, particularly around cadmium and a few carbon, oxygen, and nitrogen atoms, indicating the presence of a delocalized electron cloud enveloping them. Thus, the ELF and LOL maps illustrate intra- and intermolecular charge delocalization within the studied molecule.
Fig. 11.
Shaded surface and ELF-LOL Color-filled maps. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
5.1.8. MD and PLIP
Molecular dynamics (MD) calculations (using optimized structure, Fig. S16A) can help better understand the biological potencies of molecules exhibited (SL & complex). Thus, molecular dynamics calculations were performed. Thus, molecular dynamics calculations were performed. The function of proteins depends on the chemical interactions (CI) between ligands (SL)/complexes and the proteins themselves. It is commonly accepted that different categories of chemical interactions [[108], [109], [110]] can enhance molecules' activity. After performing MD calculations, a comparison is made based on the Etotal energy value of SL and its metal complex (Table 6) [111]. The most negative Etotal energy value compounds have the highest activity. According to the MD calculations, the cadmium metal complex demonstrated a higher activity level than the SL (Fig. S16B).
Table 6.
Etotal energy value of SL and the complex.
| Etotal of SL | Etotal of Complex | |
|---|---|---|
| S. aureus | −223.51 | −353.85 |
| C. albicans | −230.56 | −351.65 |
| E. coli | −249.82 | −390.00 |
| P. aeruginosa | −244.08 | −388.70 |
| A. Niger | −257.61 | – |
After performing MD simulations, PLIP investigated interactions in detail for SL and the metal complex. Tables S3–S7 comprehensively summarizes the numerical values of interactions between the SL/metal complex and proteins. After conducting the PLIP analysis, we examined the interaction between SL and the complex with various bacteria (Fig. 12, Top-Bottom, L-R)-Fig. 13, Top-Middle-Bottom, L-R). Fig. 12 (Top, L) illustrates that SL interacts hydrophobically and through hydrogen bonds with five points on the S. aureus protein: LEU, VAL, LYS, PHE, and GLY. Fig. 12 (Top, R) examines the metal complex interacting hydrophobically and through salt bridges with the S. aureus protein at eight points, including LYS, ASP, LEU, PHE, HIS, and ALA. Fig. 12 (Bottom, L) displays ten points where the SL interacts with the C. albicans protein. The interactions involve Hydrophobic Interactions and Hydrogen bonds with ALA, ASN, HIS, PHE, SER, ARG, and VAL. Fig. 12, Bottom, R) displays the cadmium complex interacting with the C. albicans protein THR, ARG, ILE, ASN, GLU, and LEU at eight points through Hydrophobic Interactions and Hydrogen bonds. Fig. 13 (Top, L) shows the hydrophobic interactions of SL with three amino acids (ILE, ASP, and ALA) on the E. coli protein. Fig. 13 (Top, R) illustrates ten different points where the complex interacts hydrophobically and via hydrogen bonds with E. coli protein TYR, ASP, THR, PRO, VAL, TRP, LYS, and SER. Fig. 13 (Middle, L) shows that the SL makes hydrophobic interactions and hydrogen with the P. aeruginosa protein, which are ILE, ASP, and ALA, at three different points. Fig. 13 (Middle, R) shows the cadmium complex that makes hydrophobic interactions and hydrogen with the P. aeruginosa protein ASP, PHE, ILE, LYS, and ARG at 10 points. Fig. 13 (Bottom, L) shows that SL makes Hydrophobic Interactions and Hydrogen with the A. Niger protein that are HIS, ILE, TRP, ALA, HIS, PHE, VAL, and ASN at eight different points. Fig. 13 (Bottom, R) explores that complex makes Hydrophobic Interactions and Hydrogen with the A. Niger protein that are PHE, GLU, ASN, LEU, TRP, and ASP at seven different points. After conducting MD calculations to examine the interactions of SL and Cd complex with bacteria, it was observed that the molecule interacts with various proteins. The PLIP analysis supports this result.
Fig. 12.
(Top-Bottom, L-R). Demonstration of the interaction of SL (left) and Complex (right) with the S. aureus protein (Top), and the interaction of SL (left) and Complex (right) with the C. albicans protein (Bottom).
Fig. 13.
(Top-Middle-Bottom, L-R). Demonstration of the interaction of SL (left) and Complex (right) with the E. coli protein (Top), the interaction of SL (left) and Complex (right) with the P. aeruginosa protein (Middle), and the interaction of SL (left) and Cd metal complex (right) with the A. Niger protein (Bottom).
6. Antibacterial and fungal activities
This research article explores the in vitro antibacterial and fungal activity of Ligand (SL), metal salts (M = Cd(OAc)2.4H2O), and the complex (Fig. S17). The study extensively investigated the antimicrobial activity of M and Cd(II)-complex with Salen ligand (SL). Antimicrobial functions favouring Gm-ve (E. coli and P. aeruginosa), Gm + ve (S. aureus, B. cereus), and antifungal potentiality (C. albicans and A. Niger) of SL, M, and complex are systematically displayed in a tabulated manner (Table 7-Table 8). The reference tables demonstrate some antimicrobial and antifungal activity against the specified microorganisms. In antimicrobial evaluation, SL was active against the Gm-ve strain P. aeruginosa. M showed moderate activity, and complex showed higher activity than standard antibiotics. SL, M, and complex showed weak to moderate activities against S. aureus. But, against B. cereus, they are shown to have suitable antibacterial activities. Complex showed very high activity, which is close to standard drug. In the case of the bacterial strain E. coli, all compounds exhibited low to moderate activity. According to the results, the complex showed improved antibacterial function against P. aeruginosa. The results indicate that the complex has a more pronounced antibacterial activity against Gm + ve and Gm-ve bacteria than SL and M. However, the complex is a more potent antibacterial agent against P. aeruginosa. In addition, the tested compounds have variable inhibitory effects against antifungal strains. The complex shows high antifungal activity against the selected stains. These compounds have shown great potential in acting as effective antifungal agents. SL and M offer low to modest function with C. albicans and A. Niger.
Table 7.
Mean zone diameter of SL, M, and the complex.
| Mean zone diameter (mm)a |
||||||
|---|---|---|---|---|---|---|
| S. aureus | P. aeruginosa | B. cereus | E. coli | C. albicans | A. Niger | |
| SL | 14 ± 0.2 | 0.9 ± 0.3 | 16 ± 0.2 | 0.7 ± 0.3 | 0.9 ± 0.4 | 0.7 ± 0.1 |
| M | 26 ± 0.2 | 26 ± 0.1 | 22 ± 0.4 | 10 ± 0.2 | 12 ± 0.2 | 12 ± 0.4 |
| Complex | 23 ± 0.1 | 33 ± 0.2 | 30 ± 0.1 | 14 ± 0.3 | 30 ± 0.4 | 29 ± 0.2 |
| Chloramphenicol | 25.0 ± 0.4 | 30 ± 0.2 | 32 ± 0.1 | 26.0 ± 0.1 | – | – |
| Nystatin | – | – | – | – | 19.0 ± 0.1 | 19.2 ± 0.2 |
| DMSO | – | – | – | – | – | – |
[Each value represents a mean ± standard deviation (SD) of three replications. aThe zone diameters have been calculated (mm). –: not detected inhibition. Chloramphenicol act as positive control against bacteria and Nystatin against fungal stains.].
Table 8.
MIC values of SL, M, and the complex.
| MIC (μg/mL) |
||||||
|---|---|---|---|---|---|---|
| S. aureus | P. aeruginosa | B. cereus | E. coli | C. albicans (light) | A. Niger | |
| SL | 125 | 500 | 125 | 500 | 500 | 500 |
| M | 25 | 125 | 62.5 | 250 | 125 | 62.5 |
| Complex | 50 | 12.5 | 12.5 | 125 | 12.5 | 12.5 |
6.1. Structure-activity relationship
The Cd(II)-Salen complex demonstrates more potent antibacterial activity against both Gm + ve and Gm-ve bacteria than SL and M. The increased activity of the Cd complex, as compared to the SL, has been rationalized by Overtone's principle (OP) [112,113] and Tweedy's chelation theory (TCT) [114]. The question arises: why do the compounds exhibit antibacterial properties? We analyse based on the SAR (structure-activity relationship) after considering significant factors like azomethine (-CH N) structural framework, Cd metal charge, size, and Lipophilicity factors. Already, the SCXRD reveals that Cd(II) metal ions in the complex coordinate with azomethine N-atoms and O-derived from the deprotonated form of [SL]2-. Therefore, the (-CH N) group in the complex's structural frameworks was attributed to the occurrence [115]. Further, the Cd metal ion's positive charge is partially distributed with SL's N/O-donor atoms during the chelation process, reducing the impact of Cd metal ion polarization. Lipophilicity boosts the potency of Cd metal complexes and is vital for higher antimicrobial activity when complexed with SL. Besides, the size of the particles affects the antibacterial activity of SB metal complexes [115]. According to the research, tiny particles are more effective in killing bacteria than large ones. Microbial protoplasts can be damaged by tiny particles that penetrate cell walls and cytoplasmic membranes. This harm includes condensing DNA molecules, preventing replication, halting enzymes, and metabolism, and disrupting membrane permeability. The SAR discussion finally concluded that the Cd(II)-Salen complex has more potent antimicrobial properties than SL or M. Therefore, it can be used as an effective antimicrobial agent shortly.
6.2. Antifungal photodynamic therapy (APDT)
The cadmium complex demonstrated excellent antifungal efficacy against the studied C. albicans strain in vitro. After 2 h of irradiation, PDT studies were conducted on C. albicans using a 300 W Newport Xenon arc lamp for photodynamic therapy. The optimal irradiation time was determined to be 2 h, as no increase in antimicrobial activity was observed beyond this point. The antimicrobial strain's photodynamic antifungal activity is proposed to operate through membrane disruption and cell cycle arrest mechanisms [116,117]. The complex sample after irradiation is designated as C*. Fungal cell death may occur due to reactive oxygen species (ROS) [118]. It may be the primary method of inhibiting fungi in Photodynamic Therapy (PDT). The excited PS can seriously affect microbial cells by generating highly reactive species through type-I or type-II inhibition (Fig. S18). Therefore, the complex is effective against C. albicans in vitro and is an excellent photosensitizer [119].
6.2.1. Cell viability curve test
The cell viability curve test evaluated the cadmium acetate complex's kinetics in vitro fungicidal activity against the fungal strain of C. albicans. The cell count (viable) was compared with the +ve growth control on a logarithmic scale. The process is conducted by exposing the sample to white light and darkness (Fig. S19). The C. albicans strain was photochemically inactivated within 30 min of exposure to white light. The examined microorganism was eradicated. The cell samples remained viable after 3 h (180 min). The tested concentration did not inhibit fungal growth without light (dark conditions). According to the results, the deactivation was influenced by the radiation source type and the use of a Photosensitizer (PS). According to research, two factors can effectively halt fungal growth: light and molecular O2 [120]. The photodynamic activity of the complex is explained using a structure containing imine (-CH N) and phenolic groups (-OH). Amino groups (-NH2) can become protonated in biological systems [121]. It provides an amphiphilic character that increases cell wall interaction, thereby improving the photodynamic activity against C. albicans. Herein, ground-state (GS) molecular structures successfully establish the cadmium complex's photodynamic properties. The properties were interesting because the complex's photochemistry is due to a phenolic –OH group [122]. It causes slight and insignificant changes to the metal centre after coordination with SL. The steric and complex geometry distortion affects the photophysical activities. Phenols containing aromatic ligands with traditional PSs can reinforce PDT's effectiveness by increasing ROS production, thus providing a better therapeutic effect.
7. Conclusion
This article presented synthesizing, spectroscopic findings, and crystallographic characterizing of one μ2-1,3-acetate bridge Cd(II)-Salen complex. The compound is characterized using diffuse reflectance spectroscopy (DRS) and other analytical techniques like PXRD, SEM-EDX, and ICP-MS. SCXRD revealed the complex attained monoclinic space group P21/c and hexagonal geometry around the central Cd(II) metal ion. The intricate crystal packing displays supramolecular interactions C–H⋯O and C–H···π. HS and 2-D fingerprint plots confirm the presence of H⋯O, H⋯N, and Cd⋯O contacts. DFT quantization, like energy frameworks, FMO, MEP, NBO, QTAIM, NCI-RGD, and ELF-LOL studies, was conducted to investigate the cadmium complex reactivity and versatile bonding features. The antibacterial efficacy of the complex and its synthetic components were thoroughly investigated against Gram-positive (S. aureus, B. cereus) and antifungal potentiality (C. albicans and A. Niger) and Gram-negative (E. coli and P. aeruginosa) microbial strains. MD revealed that the cadmium complex exhibited higher antibacterial activity than the SL, which achieved a higher negative Etotal value (−390.00) than the SL (−249.82) against E. coli. The complex, SL, MIC (μg/mL), and mean zone diameter (nm) values have been compared against these selected microbial strains. Herein, the MIC range of values for SL is 125 → 500, whereas for complex, it is 50 → 12.5. Similarly, SL's mean zone diameter range is 14 ± 0.2 → 0.7 ± 0.1. While for complex, this variation is 23 ± 0.1 → 29 ± 0.2. The numeric value implies that the complex has more potent antimicrobial activity than SL and M. Moreover, the complex antimicrobial potency has been demonstrated based on the SAR approach. PLIP experiments rationalize the complex antimicrobial research. Finally, we evaluated the antifungal potency and cell viability of photodynamic therapy (PDT) on C. albicans. The study is presented with an emphasis on mechanisms. The article will benefit the researchers' community solely by focusing on theoretical DFT work and fungal photodynamic therapy (PDT).
Research funding
This research has not received funding from any public, commercial, or non-profit agencies.
Data availability statement
The data that has been used is confidential.
Funding
This research has not received funding from any public, commercial or non-profit agencies.
CRediT authorship contribution statement
Dhrubajyoti Majumdar: Writing – review & editing, Writing – original draft, Visualization, Validation, Supervision, Software, Resources, Project administration, Methodology, Investigation, Formal analysis, Data curation, Conceptualization. Jessica Elizabeth Philip: Visualization, Validation, Software, Formal analysis, Data curation. Bouzid Gassoumi: Visualization, Validation, Software, Methodology, Formal analysis, Data curation, Conceptualization. Sahbi Ayachi: Visualization, Validation, Supervision, Software, Methodology, Formal analysis, Data curation. Balkis Abdelaziz: Visualization, Validation, Methodology, Formal analysis, Data curation. Burak Tüzün: Visualization, Validation, Software, Resources, Formal analysis, Data curation. Sourav Roy: Visualization, Validation, Software, Investigation, Formal analysis, Data curation, Conceptualization.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests:Dr Dhrubajyoti Majumdar reports administrative support, equipment, drugs, or supplies, and statistical analysis were provided by Tamralipta Mahavidyalaya Department of Chemistry. Dr Dhrubajyoti Majumdar reports a relationship with Tamralipta Mahavidyalaya that includes: board membership, employment, and non-financial support. None has patent Not applicable pending to Not applicable. All the authors declared no conflicts of interest or personal relationships that could have influenced the current research work. Further, no author serve as an editorial capacity for the journal like Dr Dhrubajyoti Majumdar and Dr Sourav Roy. If there are other authors, they declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
All authors involved in the current research would like to express their gratitude to the Central Laboratory of Tamralipta Mahavidyalaya, Purba Medinipur, Tamluk-721636, West Bengal, India, for their support, which was funded by the DST-FIST project (Level-0) under the Department of Science and Technology (DST), Govt. of India. We greatly acknowledge the sophisticated Raman spectra, Diffuse reflectance spectroscopy (DRS), ICP-MS, and SEM-EDX analysis conducted by SAIF, IIT Madras in Chennai-600036, and STIC, Cochin in Kerala-682022.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e29856.
Contributor Information
Dhrubajyoti Majumdar, Email: dmajumdar30@gmail.com.
Sourav Roy, Email: souravscott@gmail.com.
Appendix B. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Schiff Hugo. third ed. 1970-1979. The Great Soviet Encyclopaedia. [Google Scholar]
- 2.Roy S., Bauza A., Frontera A., Chattopadhyay S. Inorg. Chim. Acta. 2016;450:321–329. [Google Scholar]
- 3.Roy S., Drew M.G.B., Bauza A., Frontera A., Chattopadhyay S. Dalton Trans. 2017;46:5384–5397. doi: 10.1039/c6dt04906k. [DOI] [PubMed] [Google Scholar]
- 4.Roy S., Bhattacharyya A., Purkait S., Bauza A., Frontera A., Chattopadhyay S. Dalton Trans. 2016;45:15048–15049. doi: 10.1039/c6dt02587k. [DOI] [PubMed] [Google Scholar]
- 5.Majumdar D.J., Frontera A., Gomila R.M., Das S., Bankura K.P. RSC Adv. 2022;12:6352–6363. doi: 10.1039/d1ra09346k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Majumdar D.J., Philip J.E., Tuzun B., Frontera A., Gomila R.M., Roy S., Bankura K.P., Inorg J. Organomet. Polym. 2022;32:4320–4339. [Google Scholar]
- 7.Majumdar D.J., Tuzun B., Pal T.K., Das S., Bankura K.P., Inorg J. Organomet. Polym. 2022;32:1159–1176. [Google Scholar]
- 8.Majumdar D.J., Agrawal Y., Thomas R., Ullah Z., Santra M.K., Das S., Pal T.K., Bankura K.P., Mishra D. Appl. Organomet. Chem. 2019;34 [Google Scholar]
- 9.Roy S., Dey A., Drew M.G.B., Roy P.P., Chattopadhyay S. New J. Chem. 2019;43:5020–5031. [Google Scholar]
- 10.Majumdar D.J., Dey S., Kumari A., Pal T.K., Bankura K.P., Mishra D. Spectrochim. Acta Part A. 2021;254 doi: 10.1016/j.saa.2021.119612. [DOI] [PubMed] [Google Scholar]
- 11.Majumdar D.J., Das D., Sreejith S.S., Das S., Biswas J.K., Mondal M., Ghosh D., Bankura K.P., Mishra D. Inorg. Chim. Acta. 2019;489:244–254. [Google Scholar]
- 12.Majumdar A., Rosair G.M., Mallick A., Chattopadhyay N., Mitra S. Polyhedron. 2006;25:1753–1762. [Google Scholar]
- 13.Gupta K.C., Sutar A.K. Coord. Chem. Rev. 2008;252:1420–1450. [Google Scholar]
- 14.De Clercq B., Lefebvre F., Verpoort F. Appl. Catal. A. 2003;247:345–364. [Google Scholar]
- 15.Papish E.T., Taylor M.T., Jernigan F.E., III, Rodig M.J., Shawhan R.R., Yap G.P.A., Jove F.A. Inorg. Chem. 2006;45:2242–2250. doi: 10.1021/ic051579a. [DOI] [PubMed] [Google Scholar]
- 16.Khalaji A.D., Nikookar M., Fejfarova K., Dusek M. J. Mol. Struct. 2014;1071:6–10. [Google Scholar]
- 17.Nassar M.Y., Mohamed T.Y., Ahmed I.S. J. Mol. Struct. 2013;1050:81–87. [Google Scholar]
- 18.Ma D.-L., He H.-Z., Leung K.-H., Chan D.S.-H., Leung C.-H. Angew. Chem. Int. Ed. 2013;52:7666–7682. doi: 10.1002/anie.201208414. [DOI] [PubMed] [Google Scholar]
- 19.Long J., Rouquette J., Thibaud J.-M., Ferreira R.A.S., Carlos L.D., Donnadieu B., Vieru V., Chibotaru L.F., Konczewicz L., Haines J., Guari Y., Larionova J. Angew. Chem. Int. Ed. 2015;54:2236–2240. doi: 10.1002/anie.201410523. [DOI] [PubMed] [Google Scholar]
- 20.Yang X., Schipper D., Jones R.A., Lytwak L.A., Holliday B.J., Huang S. J. Am. Chem. Soc. 2013;135:8468–8471. doi: 10.1021/ja4031243. [DOI] [PubMed] [Google Scholar]
- 21.Majumdar D.J., Roy S., Frontera A. RSC Adv. 2022;12:35860–35872. doi: 10.1039/d2ra07080d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roy S., Dey A., Gomila R.M., Ortega-Castro J., Frontera A., Ray P.P., Chattopadhyay S. Dalton Trans. 2022;51:5721–5734. doi: 10.1039/d2dt00197g. [DOI] [PubMed] [Google Scholar]
- 23.Filipovic N.R., Bacchi A., Pelizzi G., Markovic R., Mitic D., Andelcovic K.K. J. Coord. Chem. 2005;58:1541–1550. [Google Scholar]
- 24.Gusev A.N., Shul’gin V.F., Kiskin M.A., Eremenko I.L. Russ. J. Coord. Chem. 2011;37:119–123. [Google Scholar]
- 25.Hakimi M., Moeini K., Mardani Z., Khorrami F. J. Kor. Chem. Soc. 2013;57:352–356. [Google Scholar]
- 26.Kruszynski R., Malinowska A., Czakis-Sulikowska D., Lamparska A. J. Coord. Chem. 2009;62:911–922. [Google Scholar]
- 27.Lo W.-K., Wong W.-K., Wong W.-Y., Guo J. Eur. J. Inorg. Chem. 2005:3950–3954. [Google Scholar]
- 28.Vickers S.M., Frischmann P.D., MacLachlan M.J. Inorg. Chem. 2011;50:2957–2965. doi: 10.1021/ic1023828. [DOI] [PubMed] [Google Scholar]
- 29.Roy S., Dey A., Ray P.P., Ortega-Castro J., Frontera A., Chattopadhyay S. Chem. Commun. 2015;51:12974–12976. doi: 10.1039/c5cc04323a. [DOI] [PubMed] [Google Scholar]
- 30.Pons J., García-Anton J., Jimenez R., Solans X., Font-Bardia M., Ros J. Inorg. Chem. Commun. 2007;10:1554–1556. [Google Scholar]
- 31.Biswas S., Gómez-García C.J., Clemente-Juan J.M., Benmansour S., Ghosh A. Inorg. Chem. 2014;53:2441–2449. doi: 10.1021/ic4023536. [DOI] [PubMed] [Google Scholar]
- 32.Pucelik B., Dabrowski J.M. Adv. Inorg. Chem. 2022;79:65–103. doi: 10.1016/bs.adioch.2021.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liang J., Sun D., Yang Y., Li M., Li H., Chen L. Eur. J. Med. Chem. 2021;224 doi: 10.1016/j.ejmech.2021.113696. [DOI] [PubMed] [Google Scholar]
- 34.Sen P., Nyokong T. Polyhedron. 2019;173 [Google Scholar]
- 35.Majumdar D.J., Philip J.E., Tuzun B., Roy S. Inorg. Chem. Commun. 2023;155 [Google Scholar]
- 36.Mesquita M.J.Q., Dias C., S Neves M.G.P.M., Almeida A., Faustino M.A.F. Molecules. 2018;23:2424. [Google Scholar]
- 37.Winckler K.D. J. Photochem. Photobiol. B Biol. 2007;86:43–44. doi: 10.1016/j.jphotobiol.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 38.Majumdar D.J., Das S., Biswas J.K., Mondal M. J. Mol. Struct. 2017;1134:617–624. [Google Scholar]
- 39.Majumdar D.J., Surendra Babu M.S., Das S., Mohapatra C., Biswas J.K., Mondal M. ChemistrySelect. 2017;2:4811–4822. [Google Scholar]
- 40.Majumdar D.J., Biswas J.K., Mondal M., Surendra Babu M.S., Das S., Metre R.K., Sreejith S.S. ChemistrySelect. 2018;3:2912–2925. [Google Scholar]
- 41.Majumdar D.J., Dey S., Sreejith S.S., Biswas J.K., Mondal M., Shukla P., Das S., Pal T., Das D., Bankura K.P., Mishra D. J. Mol. Struct. 2019;1179:694–708. [Google Scholar]
- 42.Cozzi P.G., Dolci L.S., Garelli A., Montalti M., Prodi L., Zaccheroni N. New J. Chem. 2003;27:692–697. [Google Scholar]
- 43.Aghaee M., Mohammadi K., Hayati P., S-Badr P., Yazdian F., Alonso A.G., Rostammia S., Eshghi F. J. Solid State Chem. 2022;310 [Google Scholar]
- 44.Bhattacharjee T., Adhikary S., Dutta A., Daniliuc C.-G., Montazerozohori M., Naghiha R., Hayati P. Polyhedron. 2022;211 [Google Scholar]
- 45.Mehrabadi Z., Ahmadi S., Gutierrez A., Karimi M., Hayati P., S-Badr P., Moaser A.G., Rostamnia S., Hasanzadeh A., Khaksar S., Rouhani S., Msagati T.A.M. J. Mol. Struct. 2021;1226 [Google Scholar]
- 46.Souri B., Hayati P., Rezvani A.R., M-Merono R., Janczak J. Inorg. Nano-Met. Chem. 2020;50:80–93. [Google Scholar]
- 47.Rashidi N., Jaafar M., Fard S., Hayati P., Janczak J., Yazdian F., Rouhani S., Msagati T.A.M. J. Mol. Struct. 2021;1231 [Google Scholar]
- 48.El-Gammal Ola A., El-Bindary A.A., Mohamed F. Sh, Rezk G.N., El-Bindary M.A. J. Mol. Liq. 2022;346 [Google Scholar]
- 49.Abou-Melha K.S., Al-Hazmi G.A., Althagafi I., Alharbi A., Shaaban F., El-Metwaly N.M., El-Bindary A.A., El-Bindary M.A. J. Mol. Struct. 2021;334 [Google Scholar]
- 50.M.A.A.H. Allah. Balakit A.A., Salman H.E., Abdulridha A.A., Sert Y. J. Adhes. Sci. Technol. 2023;37:525–547. [Google Scholar]
- 51.A. A. Abdulridha, M. A. A. H. Allah. Makki S.Q., Sert Y., Salman H.E., Balakit A.A. J. Mol. Liq. 2020;315 [Google Scholar]
- 52.Balakit A.A., Makki S.Q., Sert Y., Ucun F., Alshammari M.B., Thordarson P., Ei-Hiti G.A. J. Adhes. Sci. Technol. 2020;32:519–526. [Google Scholar]
- 53.Gadre S.R., Suresh C.H., Mohan N. Molecules. 2021;26(11):3289. doi: 10.3390/molecules26113289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Emori W., Louis H., Adalikwu S.A., Timothy R.A., Cheng C.R., Gber T.E., Adeyinka A.S. Polycycl. Aromat. Compd. 2022:1–18. [Google Scholar]
- 55.Singh H. Chem. Phys. 2019;524:1–13. [Google Scholar]
- 56.Khemalapure S.S., Hiremath S.M., Hiremath C.S., Katti V.S., Basanagouda M.M., Khanal G.P., Karthick T. J. Mol. Struct. 2020;1220 [Google Scholar]
- 57.Elekofehinti O.O., Iwaloye O., Olawale F., Chukwuemeka P.O., Folorunso I.M. In. Silico. Pharmacol. 2021;9:1–16. doi: 10.1007/s40203-021-00108-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chérif I., Raissi H., Abiedh K., Gassoumi B., Teresa Caccamo M., Magazu S., Haj Said A., Hassen F., Boubaker T., Ayachi S. J. Photochem. Photobiol., A. 2023;443 doi: 10.1016/j.saa.2023.122939. [DOI] [PubMed] [Google Scholar]
- 59.Nemati-Kande E., Karimian R., Goodarzi V., Ghazizadeh E. Appl. Surf. Sci. 2020;510 [Google Scholar]
- 60.Majumdar D.J., Philip J.E., Dubey A., Tufail A., Roy S. Heliyon. 2023 doi: 10.1016/j.heliyon.2023.e16103. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 61.Majumdar D.J., Dubey A., Tufail A., Sutradhar D., Roy S. Heliyon. 2023 doi: 10.1016/j.heliyon.2023.e16057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Majumdar D.J., Tuzun B., Pal T.K., Saini R., Bankura K.P., Mishra D. Polyhedron. 2021;210 [Google Scholar]
- 63.Rossi G.G., Guterres K.B., da Silveira C.H., Moreira K.S., L Burgo T.A., Iglesias B.A., de Campos M.M.A. Microb. Pathog. 2020;148 doi: 10.1016/j.micpath.2020.104455. [DOI] [PubMed] [Google Scholar]
- 64.Schwalbe R., Steele-Moore L., Goodwin A.C. CRC Press; Boca Raton, FL, USA: 2007. Antimicrobial Susceptibility Testing Protocols. [Google Scholar]
- 65.Majumdar D.J., Roy S., Dey A., Sutradhar D. J. Mol. Struct. 2023;1294 [Google Scholar]
- 66.SMART & SAINT Software Reference Manuals Version 6.45; Bruker Analytical X-Ray Systems. Inc.; Madison, WI: 2003. [Google Scholar]
- 67.SHELXTL Reference Manual Ver. 6.1; Bruker Analytical X-Ray Systems. Inc.; Madison, WI: 2000. [Google Scholar]
- 68.Sheldrick G.M. Vol. 6.12. Bruker AXS Inc.; Madison: 2001. (SHELXTL, a Software for Empirical Absorption Correction Ver). [Google Scholar]
- 69.Dolomanov O.V., Bourhis L.J., Gildea R.J., Howard J.A.K., Puschmann aH. J. Appl. Crystallog. 2009;42:339–341. [Google Scholar]
- 70.Lee C., Yang W., Parr R.G. Phys. Rev. B. 1988;37:785–789. doi: 10.1103/physrevb.37.785. [DOI] [PubMed] [Google Scholar]
- 71.Hay P.J., Wadt W.R. J. Chem. Phys. 1985;82:270–283. [Google Scholar]
- 72.G09|Gaussian.com, http://gaussian.com/glossary/g09/.
- 73.Jabli S., Gassoumi B., Nasri S., Ghalla H., Vollbert E.M., Molton F., Loiseau F., Roisnel T., Nasri H. J. Mol. Struct. 2023;1294 [Google Scholar]
- 74.Borah B.P., Majumder S., Nath P.P., Choudhury A.K., Bhuyan J. J. Mol. Struct. 2023;1275 [Google Scholar]
- 75.Roy D.D., Todd A.K., John M.M. Gaussian Inc; Wallingford: 2009. Gauss View 5.0. 8. [Google Scholar]
- 76.Kerraj S., Salah M., Belaaouad S., Mohammed M. Polyhedron. 2023;230 [Google Scholar]
- 77.Washburn S., Kaswan R.R., Shaikh S., Moss A., D'Souza F., Wang H. J. Phys. Chem. A. 2023;127:9040–9051. doi: 10.1021/acs.jpca.3c05261. [DOI] [PubMed] [Google Scholar]
- 78.Lu T., Chen F. J. Comp. Chem. 2012;33:580–592. doi: 10.1002/jcc.22885. [DOI] [PubMed] [Google Scholar]
- 79.Spackman M.A., Jayatilaka D. CrystEngComm. 2009;11:19–32. [Google Scholar]
- 80.Qiu X., Janson C.A., Smith W.W., Green S.M., McDevitt P., Johanson K., Jarvest R.L. Protein Sci. 2001;10(10):2008–2016. doi: 10.1110/ps.18001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Whitlow M., Howard A.J., Stewart D., Hardman K.D., Kuyper L.F., Baccanari D.P., Tansik R.L. J. Bio. Chem. 1997;272(48):30289–30298. doi: 10.1074/jbc.272.48.30289. [DOI] [PubMed] [Google Scholar]
- 82.Yu E.W., Aires J.R., McDermott G., Nikaido H. J. Bacteriol. 2005;187(19):6804–6815. doi: 10.1128/JB.187.19.6804-6815.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bottomley M.J., Muraglia E., Bazzo R., Carfì A. J. Bio. Chem. 2007;282(18):13592–13600. doi: 10.1074/jbc.M700556200. [DOI] [PubMed] [Google Scholar]
- 84.Oakley A.J. Biochem. Biophys. Res. 2010;397(4):745–749. doi: 10.1016/j.bbrc.2010.06.024. [DOI] [PubMed] [Google Scholar]
- 85.Ozmen U.O., Tuzun B., Ayan E.B., Cevrimli B.S. J. Mol. Struct. 2023;1286 [Google Scholar]
- 86.D. Ritchie, T. Orpailleur, Hex 8.0. 0 User Manual. Protein Docking Using Spherical Polar Fourier Correlations Copyright C. (21013). [PubMed]
- 87.Zahirović A., Hadžalić S., Višnjevac A., Fočak M., Tüzün B., Žilić D., Osmanković I. J. Inorg. Biochem. 2023 doi: 10.1016/j.jinorgbio.2023.112232. [DOI] [PubMed] [Google Scholar]
- 88.Salentin S., Schreiber S., Haupt V.J., Adasme M.F., Schroeder M. Nucleic Acids Res. 2015;43(W1):W443–W447. doi: 10.1093/nar/gkv315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Karimi S., Rezaeivala M., Mokhtare M., Tuzun B., Sayin K. J. Taiwan Inst. Chem. Eng. 2023;147 [Google Scholar]
- 90.Lo W.-K., Wong W.-K., Wong W.-Y., Guo J. Eur. J. Inorg. Chem. 2005:3950–3954. [Google Scholar]
- 91.Prakasha A., Singha B.K., Bhojakb N., Adhikari D. Spectrochim. Acta Part A. 2010;76:356–362. doi: 10.1016/j.saa.2010.03.019. [DOI] [PubMed] [Google Scholar]
- 92.Gonul I., Demirbag B., Ocakoglu K., Ayaz F. J. Coord. Chem. 2020;73:2878–2888. [Google Scholar]
- 93.Sadhukhan D., Roy A., Rosair G., Charbonniere L., Mitra S. BCSJ. 2011;84:211–217. [Google Scholar]
- 94.Amirnasr M., Schenk K.J., Salavati M., Dehghanpour S., Taeb A., Tadjarodi A. J. Coord. Chem. 2003;56:231–243. [Google Scholar]
- 95.Dutta B., Paul S., Halder S. Heliyon. 2023;9 doi: 10.1016/j.heliyon.2023.e13504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Addition A.W., Rao T.N., Reedjik J., Van Rinj J., Verscholar C.G. J. Chem. Soc. Dalton Trans. 1984:1349. [Google Scholar]
- 97.Turner M.J., Grabowsky S., Jayatilaka D., Spackman M.A. J. Phys. Chem. Lett. 2014;5:4249–4255. doi: 10.1021/jz502271c. [DOI] [PubMed] [Google Scholar]
- 98.Venkatesan P., Thamotharan S., Ilangovan A., Liang H., Sundius T. Spectrochim. Acta, Part A. 2016;153:625–636. doi: 10.1016/j.saa.2015.09.002. [DOI] [PubMed] [Google Scholar]
- 99.Turner M.J., Thomas S.P., Shi M.W., Jayatilaka D., Spackman M.A. Chem Commun. 2015;51:3735–3738. doi: 10.1039/c4cc09074h. [DOI] [PubMed] [Google Scholar]
- 100.Yuan G., Shao K.Z., Du D.Y., Wang X.L., Su Z.M., Ma J.F. CrystEngComm. 2012;14:1865–1873. [Google Scholar]
- 101.Saji R., Prasana J., Muthu S., George J., Kuruvilla T., Raajaraman B. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2020;226(117614) doi: 10.1016/j.saa.2019.117614. [DOI] [PubMed] [Google Scholar]
- 102.Chérif I., Raissi H., Abiedh K., Gassoumi B., Caccamo M.T., Magazu S., Said A.H., Hassen F., Boubaker T., Ayachi S. Spectrochim. Acta, Part A. 2023;301 doi: 10.1016/j.saa.2023.122939. [DOI] [PubMed] [Google Scholar]
- 103.Bader R.F., Austen M.A. J. Chem. Phys. 1997;107:4271–4285. [Google Scholar]
- 104.Rozas I., Alkorta I., Elguero J. J. Am. Chem. Soc. 2000;122:11154–11161. [Google Scholar]
- 105.Silvi B., Savin A. Nature. 1994;371:683–686. [Google Scholar]
- 106.Rizwana F., Prasana J.C., Muthu S., Abraham C.S. Comput. Biol. Chem. 2019;78:9–17. doi: 10.1016/j.compbiolchem.2018.11.014. [DOI] [PubMed] [Google Scholar]
- 107.Abdelaziz B., Mazouz Z., Gassoumi B., Boukortt N.E.I., Patanè S., Ayachi S. J. Mol. Liq. 2024;395 [Google Scholar]
- 108.Poustforoosh A., Faramarz S., Negahdaripour M., Tüzün B., Hashemipour H. J. Biomol. Struct. Dyn. 2023:1–15. doi: 10.1080/07391102.2023.2213344. [DOI] [PubMed] [Google Scholar]
- 109.Bensalah J., Ouaddari H., Erdoğan S., Tüzün B., Gaafar A.R., Nafidi H.A., Habsaoui A. Inorg. Chem. Commun. 2023 [Google Scholar]
- 110.Hermi S., Mrad M.H., Alotaibi A., Tüzün B., Böhme U., Alotaibi K.M., Nasr C.B. Inorg. Chem. Commun. 2023 [Google Scholar]
- 111.Sayin K. J. Coord. Chem. 2018;71(20):3292–3303. [Google Scholar]
- 112.El-gammal O.A., El-Bindary A.A., Mohamed F.S., Rezk G.N., El-Bindary M.A. J. Mol. Liq. 2021 [Google Scholar]
- 113.El-Bindary M.A., El-Bindary A.A. Appl. Organomet. Chem. 2022;36(4) [Google Scholar]
- 114.Tweedy B.G. Phytopathology. 1964;55:910. [Google Scholar]
- 115.El-Zahed M.M., Diab M.A., El-Sonbati A.Z., Saad M.H., Eldesoky A.M., El-Bindary M.A. Appl. Organomet. Chem. 2023;37(11) [Google Scholar]
- 116.Kumari M., Giri V.P., Pandey S., Kumar M., Katiyar R., Nautiyal C., Mishra A. Pestic. Biochem. Physiol. 2019;157:45–52. doi: 10.1016/j.pestbp.2019.03.005. [DOI] [PubMed] [Google Scholar]
- 117.Kim K., Sung W.S., Suh B.K., Moon S., Choi J., Kim J.G., Lee D.G. Biometals. 2009;22:235–242. doi: 10.1007/s10534-008-9159-2. [DOI] [PubMed] [Google Scholar]
- 118.Das M., Khan R., Jayabalan S.K., Behera S., Yun S., Tripathy S.K., Mishra A. Sci. Rep. 2016;6 doi: 10.1038/srep36403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Guterres K.B., Rossi G.G., de Campos M.M.A., Moreira K.S., Burgo T.A.L., Iglesias B.A. Photodiagnosis Photodyn. Ther. 2022;38 doi: 10.1016/j.pdpdt.2022.102770. [DOI] [PubMed] [Google Scholar]
- 120.Amorim C.F., Iglesias B.A., Pinheiro T.R., Lacerda L.E., Sokolonski A.R., Pedreira B.O., Moreira K.S., Burgo T.A., Meyer R., Azevedo V. Photodiagnosis Photodyn. Ther. 2023;42 doi: 10.1016/j.pdpdt.2023.103343. [DOI] [PubMed] [Google Scholar]
- 121.Liu S., Han J., Wang W., Chang Y., Wang R., Wang Z., Li G., Zhu D., Bryce M.R. Dalton Trans. 2022;51:16119–16125. doi: 10.1039/d2dt02960j. [DOI] [PubMed] [Google Scholar]
- 122.Gonul I., Demirbag B., Ocakoglu K., Ayaz F. J. Coord. Chem. 2020;73:2878–2888. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that has been used is confidential.