Abstract
Background
Glioma, a prevalent malignancy of the brain and spinal cord, poses a considerable threat to human health. The association between aberrant sialic acid modification and glioma progression has been suggested, but the precise mechanism is still elusive. ST3GAL4, a sialoglycosyltransferase, is implicated in increased metastatic potential and poor prognosis in various cancers; however, its specific role in glioma requires further elucidation.
Methods
We evaluated ST3GAL4 expression levels and their clinical relevance using the TCGA database, and we assessed immune infiltration via the Tumor Immune Evaluation Resource (TIMER) database. In vitro experiments were performed to determine the effects of ST3GAL4 knockdown on glioma cell malignancy, with additional co-culture assays to assess its impact on macrophage phenotype.
Results
ST3GAL4 expression was markedly elevated in glioma tissues compared to normal brain tissues, with a strong correlation to glioma patient clinical characteristics. Survival analyses and receiver operating characteristic (ROC) curves suggested that ST3GAL4 is a feasible diagnostic and prognostic biomarker for glioma. Knockdown studies revealed that ST3GAL4 inhibition reduces glioma cell line proliferation, migration, and invasion, while causing G1 phase cell cycle arrest. ST3GAL4 appears to mediate glioma progression through extracellular matrix reorganization and EMT signaling pathway activation, further contributing to M2 macrophage polarization and infiltration within the tumor microenvironment.
Conclusion
Our research highlights the critical role of ST3GAL4 in glioma development, positioning it as a promising candidate for diagnostic and therapeutic interventions.
Keywords: ST3GAL4, Glioma, Prognostic biomarker, Extracellular matrix regulation, Macrophage infiltration
1. Introduction
Gliomas, comprising 81 % of malignant brain tumors, are the predominant primary tumors affecting the brain and spinal cord, posing a significant challenge to healthcare systems worldwide [1,2]. Despite advances in treatment modalities, the prognosis for patients with gliomas remains poor [3,4], highlighting the urgent need for a deeper understanding of the molecular mechanisms underlying their pathogenesis and progression.
Sialic acids (Sias) are a group of negatively charged α-keto acid monosaccharide derivatives with nine carbon atoms. These biochemical compounds exert their biological effects through sialylation, a process in which they bind to proteins and lipids [5]. Sialylation typically occurs at the terminal positions of sugar chains and is vital for transmitting biological information in various physiological and pathological processes by interacting with cell surface proteins. In recent research, sialic acids have been implicated in the pathogenesis of several diseases, such as inflammation, microbial infections, and cancer [6]. Elevated serum sialic acid levels are associated with cardiovascular and respiratory diseases. Remarkably, sialic acid is an important tumor marker, with elevated detection in cancers such as those of the lung, liver, and colorectum [[7], [8], [9]].
ST3 beta-galactoside alpha-2,3-sialyltransferase 4 (ST3GAL4) is a pivotal enzyme in glycosylation, facilitating the attachment of sugar molecules to proteins and lipids. Dysregulation of ST3GAL4 has been implicated in the onset of multiple diseases, notably tumors. Overexpression of ST3GAL4 correlates with enhanced tumor cell invasiveness and migration, as well as resistance to apoptosis [10]. Research indicates that elevated levels of ST3GAL4 may promote tumor advancement and metastasis, while aberrant expression is associated with poorer prognoses in cancer patients, including more advanced tumor stages, lymph node metastasis, and lower overall survival rates [11]. However, the specific function of ST3GAL4 in gliomas and its potential as a therapeutic target have not been fully elucidated.
This study aims to investigate the expression patterns of ST3GAL4 in gliomas and explore its clinical relevance. We hypothesize that ST3GAL4 plays a role in glioma malignancy and macrophage infiltration, which may have significant implications for patient prognosis. Through the integration of bioinformatics analysis, in vitro experiments, and clinical data, our research aims to provide a comprehensive understanding of the role of ST3GAL4 in glioma development and assess its potential as a biomarker for diagnosis and prognosis. The findings of this study not only expand our knowledge of glioma pathogenesis but also open up possibilities for the development of novel therapeutic strategies targeting the sialylation pathway.
2. Materials and methods
2.1. Patient sample data acquiring
Acquiring patient sample data involved retrieving messenger RNA (mRNA) expression data and corresponding clinical information from the TCGA database (https://cancergenome.nih.gov). This dataset included 169 glioblastoma multiforme (GBM) samples, 532 lower-grade glioma (LGG) samples, and 5 neighboring brain tissue samples. Data regarding the expression of normal brain tissue were obtained from the GTEx database (https://www.gtexportal.org). These data were downloaded to further examine the differential distribution of ST3GAL4 in primary glioma or normal tissues and explore the potential correlation between ST3GAL4 expression and clinical characteristics of glioma, such as WHO grade, IDH status, 1p/19q codeletion, and histological subtypes.
2.2. Bioinformatics analysis
Bioinformatics analysis on the TCGA GBM datasets was conducted to compare the transcriptomes of the ST3GAL4 high- and low-expression groups. Differentially expressed genes (DEGs) of these groups were identified using functional and signaling enrichment analysis. Additionally, statistical calculations were performed using the R software package (Version 4.2.1). Furthermore, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses were conducted to identify the cellular component (CC), biological process (BP), molecular function (MF), and signaling pathways enriched in the target gene cluster. Gene set enrichment analysis (GSEA) was used to confirm the results obtained from the GO and KEGG analyses by evaluating enrichments in various biological functions and signaling pathways. The cutoff criteria used were P < 0.05, |NES| > 1, and FDR <0.25. Moreover, immune infiltration was analyzed using gene markers specific to 24 immune cell types. The level of tumor immune infiltration was determined using the single-sample GSEA (ssGSEA) method and the GSVA R package, based on TCGA-COADREAD datasets. The correlation between ST3GAL4 expression and the expression levels of these 24 immune cell types was calculated using the Spearman correlation test. Graphs and figures were generated using the ggplot2 R package.
2.3. Immunohistochemistry on human Glioma specimens
Specimens were acquired from the Pathology Department of Xijing Hospital, affiliated with the Fourth Military Medical University in Xi'an, Shaanxi, China. This study incorporated tissue microarrays from 63 glioma patients, categorized by genetic and histological profiles: 11 with IDH-mutant astrocytomas, 11 with IDH-mutant anaplastic astrocytomas, 13 with IDH-wildtype glioblastomas, 11 with IDH-mutant and 1p/19q-co-deleted oligodendrogliomas, 10 with IDH-mutant and 1p/19q-co-deleted anaplastic oligodendrogliomas, along with 7 paracancerous and 7 normal brain tissues. Patient enrollment spanned from 2010 to 2015, encompassing those who had primary or metastatic tumors resected. Ethical clearance was granted by the ethics committee of the university, with informed consent from all participants. Microdissection was meticulously performed on the collected tissues.
For IHC staining, glioma sections were deparaffinized in xylene and rehydrated through graded ethanol washes. Antigen retrieval was achieved by boiling the sections, and endogenous peroxidase was quenched with 3 % H2O2 in methanol at room temperature. Primary antibodies, diluted as per Beyotime Biotech Inc. instructions, were applied to sections and incubated overnight at 4 °C in a moisture-controlled environment. Following incubation, sections were treated with corresponding secondary antibodies for 30 min and with streptavidin-HRP conjugate (Maxim Biomedical, Inc.) for 10 min at room temperature. Detection was performed using MaximBio's DAB substrate for 1–2 min. Sections were counterstained with hematoxylin, dehydrated, and mounted with Eukitt mounting medium.
2.4. Cell culture
U251, HEK293T, and THP-1 cell lines were obtained from Cell Bank of the Chinese Academy of Sciences. U251 and HEK293T cells were cultured in 90 % DMEM with 10 % FBS and 1 % penicillin-streptomycin (Gibco), while THP-1 cells were cultured in 90 % 1640 with 10 % FBS and 1 % penicillin-streptomycin. Cell cultures were incubated at 37 °C with 5 % CO2.
2.5. Vector and lentivirus packaging
Three shRNAs targeting ST3GAL4 were designed, synthesized, and inserted into a shRNA-expressing lentiviral vector containing GFP and puromycin. HEK293T cells were transfected with the plasmid, along with the PMD2G and psPAX2 vectors, using a three-plasmid system. After 48 h of culture, the viral supernatant was collected. U251 cells were infected with the viral supernatant for 12 h and then cultured in a medium supplemented with 1 ng/mL puromycin (ST551, Beyotime) for one week to select stable transfected cell lines. The sequences of the three shRNA constructs targeting ST3GAL4 were as follows: sh-ST3GAL4-1: 5′-GGTCGTCATGGTGTGGTATTC-3'; sh-ST3GAL4-2: 5′-GCAAGGCCTCTAAGCTCTTTG-3'; sh-ST3GAL4-3: 5′-GCGAAAGGGTTTCTGGAAACA-3'. The negative control shRNA sequence was 5′-ACTACCGTTGTTATAGGTG-3'.
2.6. Quantitative real-time PCR (qRT-PCR) assay
RNA was extracted using the Takara miniBEST kit (9767, Takara). cDNA synthesis was performed using the PrimerScript RT kit (RR037Q, Takara). Real-time quantitative polymerase chain reaction (qPCR) analysis was performed using TB Green Premix Ex Taq TM II (RR420Q, Takara). All reagents used in the qRT-PCR assay were purchased from Takara (Kumastu, Shiga, Japan). Calculation was performed using the 2^-ΔΔCT method. The primer sequences used were: ST3GAL4: Forward-GACAGCTCTCCCCAGGAATC, Reverse-TGTCTTCCCGGGAGATGGAA; β-actin: Forward-GACAGCTCTCCCAGGAAT, Reverse-CTCCTTAATGTCACGCACGAT-3; CDH1: Forward- TGCCCAGAAAATGAAAAAGG, Reverse-GTGTATGTGGCAATGCGTTC; Vimentin: Forward-GAGAACTTTGCCGTTGAAGC, Reverse- GCTTCCTGTAGGTGGCAATC; FN1: Forward- CTGGCCGAAAATACATTGTAAA, Reverse-CCACAGTCGGGTCAGGAG; EST1: Forward-ACAGGGTAAGTGAAGGTTAATTCCA, Reverse-AGAAAGATGACTACCTTGCTTGACT; SNAI1: Forward-ACAAGCACCAAGAGTCCG, Reverse-ATGGCAGTGAGAAGGATGTG; ZEB1: Forward-GACAGTGTTACCAGGGAGGAGCA, Reverse-TTCAGGTGCCTCAGGAAAAATGA; N-Cadherin: Forward-ATGGGAAATGGAAACTTGATGGC, Reverse-CAGTTGCTAAACTTCACTGAAAGG; Claudin: Forward-CAGTCAATGCCAGGTACGAA, Reverse-TCACACGTAGTCTTTCCCGC.
2.7. Western blot assay
Total proteins were extracted from whole cell lysates using RIPA lysis buffer (HY–K1001, Sangon), which was supplemented with a protease inhibitor mixture (B11-01, Genstar). The concentration of total protein was determined using the bicinchoninic acid (BCA) assay kit (BL521A, Biosharp). Each sample contained 15 μg of protein and was then separated on a 10 % SDS-PAGE gel at a constant voltage of 120 V. The isolated proteins were transferred to a polyvinylidene fluoride (PVDF) membrane (IPVH00010, Millipore) at a constant current of 360 mA for 1 h. To block non-specific binding, the PVDF membrane was sealed with 5 % skim milk powder that was diluted in Tris buffered saline with 0.05 % Tween 20 (TBST) and incubated at room temperature for 1 h. Next, the PVDF membrane was incubated overnight at 4 °C with the appropriate primary antibody. On the following day, the PVDF membrane was washed three times with TBST and then incubated with the specified secondary antibody at room temperature for 1 h. Finally, the signal band of the target protein was captured using a chromograph.
2.8. Cell proliferation assay
The cell proliferation activity was evaluated using the Cell Counting Kit-8 (CCK-8, GK10001, Glpbio), following the provided instructions. U251 cells were seeded at a density of 4 × 103 cells per well in a 96-well plate. The absorbance at a wavelength of 450 nm was measured using a microplate reader at 24, 48, 72, and 96 h after cell attachment to assess cell proliferation.
2.9. Wound healing assay
U251 cells were seeded at a density of 3 × 105 cells per well in a 12-well plate. After the cells had fully adhered, scratches were created using a sterile pipette tip. Subsequently, the cells were triple-washed with PBS to remove cellular debris and cultured in a medium supplemented with 3 % serum under 37 °C and 5 % CO2 conditions. Images of migration were captured at different time points, including 0 and 24 h. Finally, the migration distance was measured using ImageJ software.
2.10. Transwell assay
The U251 stable cell line underwent a 24-h starvation treatment. Cell counting was performed, and the cell density was adjusted to 1 × 104 cells/ml. In both the control and experimental groups, a 200 μL cell suspension was added to the upper chamber of the Transwell(MCEP24H48, Millipore) without matrix gel. The lower chamber was filled with 600 μL of culture medium containing 10 % FBS. Following a 24-h incubation, the upper chamber was removed and washed twice with PBS. Residual cells in the upper chamber were wiped away using a cotton swab, while the cells that had invaded the bottom surface were fixed with 75 % ethanol for 5 min. Finally, the cells were stained with a 0.4 % crystal violet solution for 20 min and imaged using inverted microscopy for cell counting.
2.11. Clonal formation assay
The stable U251 or shST3GAL4 cell lines were seeded in a 6-well plate at a density of 200 cells per well in culture medium. The experiment was concluded when clonal colonies became visible without the aid of a microscope. Subsequently, the cells were washed twice with PBS, fixed with 75 % ethanol for 30 min, and stained with 0.4 % crystal violet for 15 min. Finally, cell counting was performed by capturing images using a camera.
2.12. Detection of cell cycle and apoptosis
Harvest U251 cells in the logarithmic growth phase from a culture dish and rinse them with PBS. Fix the cells overnight with pre-chilled 70 % ethanol at 4 °C and then rinse them twice with PBS. Subsequently, stain the cells with propidium iodide (PI)/RNase staining solution (550825, BD Pharmingen) in the dark for 30 min. Utilize a flow cytometer to analyze the distribution of cell cycle phases. To detect apoptosis, collect 5 × 105 cells in the logarithmic growth phase and stain them with the PE Annexin V Apoptosis Detection Kit I (559763, BD Pharmingen) in the dark for 15 min. Finally, utilize a flow cytometer to assess cell apoptosis.
2.13. Co-culture experiment with THP-1 and U251 cells
1 × 106 THP-1 cells in the logarithmic growth phase were collected and cultured in a medium supplemented with 20 ng/mL PMA (P8139, Sigma) for 48 h. Subsequently, the THP-1 cells underwent a 48-h period of rest in normal medium before the addition of 1 × 106 U251 tumor cells for co-culture. Following 48 h of co-culture, flow cytometry was employed to assess the expression levels of the M2 macrophage marker CD163 and PD-L1.
2.14. Statistical analysis
Our experimental data were statistically analyzed using SPSS software (Version 25.0), presented as mean values plus-minus the standard deviations (SDs) based on a minimum of three independent assays for robustness. Bioinformatics datasets were curated with R software (Version 4.2.1) from TCGA databases. Statistical significance for two-group comparisons was calculated using a two-tailed Student's t-test, under the assumption of normality and equal variance across groups. For comparisons involving more than two groups, one-way ANOVA was applied, followed by Bonferroni's correction to mitigate the risk associated with multiple comparisons. Survival differences were analyzed through Kaplan-Meier curves, with the log-rank test assessing significance. This method compares the expected and observed survival times to determine if there's a statistically significant difference between groups. The Spearman rank correlation test was employed to evaluate the monotonic relationship between two variables, which does not require the assumption of normal distribution inherent to the Pearson correlation. To investigate multiple variables influencing survival time, we used Cox regression modeling. The diagnostic accuracy of biomarkers was gauged through ROC curve analysis, with the AUC assessing the test's ability to differentiate between conditions. Statistical significance was set at *,P < 0.05; **,P < 0.01, and ***,P < 0.001.
3. Results
3.1. Upregulation of ST3GAL4 in gliomas: association with adverse clinical and pathological features
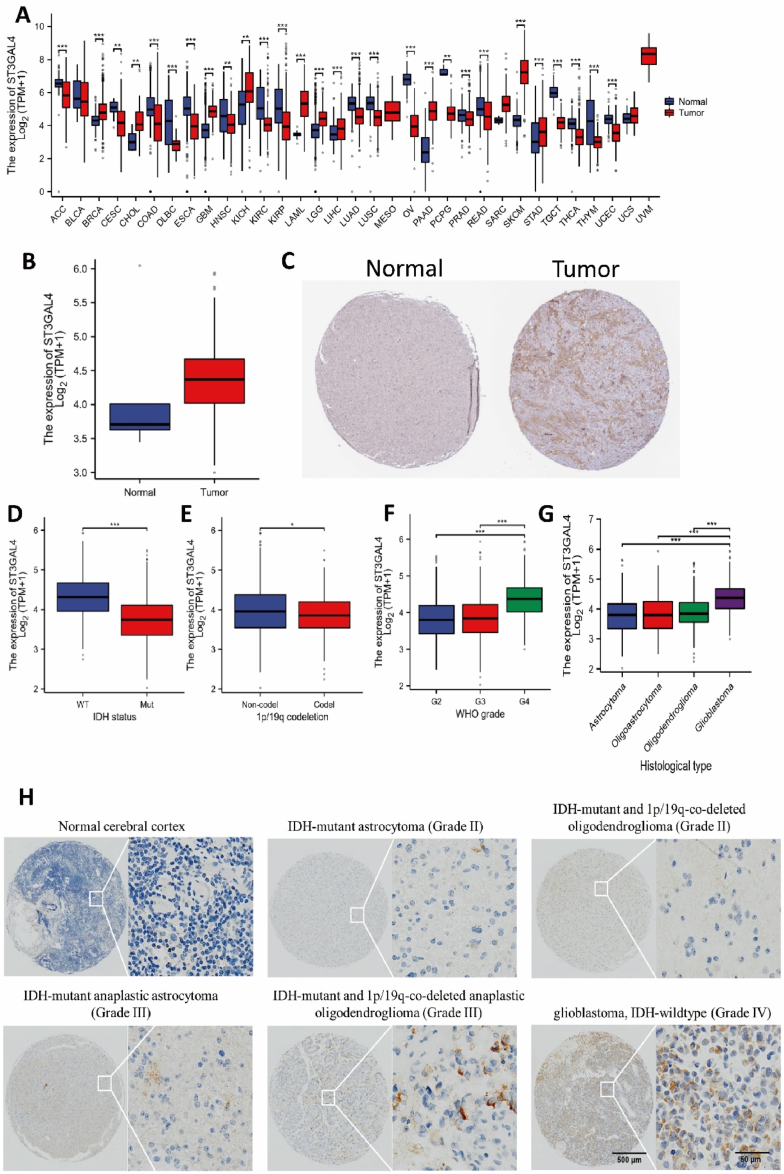
We obtained gene expression data from the TCGA database to assess the expression levels of ST3GAL4 in various tumors and their corresponding normal tissues. Significant differences were observed in the expression levels of ST3GAL4 across different tumor types. In particular, ST3GAL4 expression levels were markedly higher in tumors such as breast invasive carcinoma (BRCA), cholangiocarcinoma (CHOL), and acute myeloid leukemia (LAML) compared to normal tissues. Conversely, tumors such as adrenocortical carcinoma (ACC), kidney renal clear cell carcinoma (KIRC), and pancreatic adenocarcinoma (PAAD) exhibited significantly lower levels of ST3GAL4 expression compared to control tissues (Fig. 1A). These findings suggest that ST3GAL4 exerts diverse functions in different types of tumor cells. Importantly, ST3GAL4 expression was higher in both glioblastoma multiforme (GBM) and low-grade glioma (LGG) tumor tissues (Fig. 1A). An especially pronounced increase in ST3GAL4 expression characterized glioblastoma, IDH-wildtype (Fig. 1B). Elevated levels of ST3GAL4 protein in tumor tissues were further validated by immunohistochemical staining in patients with glioblastoma, IDH-wildtype (Fig. 1C). Notably, lower levels of ST3GAL4 expression were found in patients with IDH-mutant gliomas and those with 1p/19q co-deletion, both of which are typically linked to a more favorable prognosis (Fig. 1D and E). Conversely, higher expression levels of ST3GAL4 were observed in patients with more advanced grades of glioma and more adverse histopathological subtypes (Fig. 1F and G).
Fig. 1.
ST3GAL4 expression and its correlation with clinicopathological characteristics in gliomas. A: Expression levels of ST3GAL4 in different cancer types. ACC, adrenocortical carcinoma; BLCA, bladder urothelial carcinoma; BRCA, breast invasive carcinoma; CESC, cervical squamous cell carcinoma and endocervical adenocarcinoma; CHOL, cholangiocarcinoma; COAD, colon adenocarcinoma; DLBC, lymphoid neoplasm diffuse large B-cell lymphoma; ESCA, esophageal carcinoma; GBM, glioblastoma multiforme; HNSC, head and neck squamous cell carcinoma; KICH, kidney chromophobe; KIRC, kidney renal clear cell carcinoma; KIRP, kidney renal papillary cell carcinoma; LAML, acute myeloid leukemia; LGG, low grade glioma; LIHC, liver hepatocellular carcinoma; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; MESO, mesothelioma; OV, ovarian serous cystadenocarcinoma; PAAD, pancreatic adenocarcinoma; PCPG, pheochromocytoma and paraganglioma; PRAD, prostate adenocarcinoma; READ, rectum adenocarcinoma; SARC, sarcoma; SKCM, skin cutaneous melanoma; STAD, stomach adenocarcinoma; TGCT, testicular germ cell tumors; THCA, thyroid carcinoma; THYM, thymoma; UCEC, uterine corpus endometrial carcinoma; UCS, uterine carcinosarcoma; UVM, uveal melanoma. B: ST3GAL4 is highly expressed in glioblastoma multiforme. C: Immunohistochemical staining of ST3GAL4 in different grades glioma tissues. D–F: Expression levels of ST3GAL4 in various clinical features of gliomas. D: ST3GAL4 expression in gliomas with IDH wild type and mutant type. E: ST3GAL4 expression in gliomas with or without 1p/19q co-deletion. F: Expression level of ST3GAL4 in gliomas with different WHO grades. G: Expression level of ST3GAL4 in gliomas with different pathological types. H: Immunohistochemical staining of ST3GAL4 in normal tissue and glioma tissues with varying pathological types. Scale: 50 μm. P < 0.05, P < 0.01, **P < 0.001.
To delve deeper into ST3GAL4 expression across the spectrum of glioma subtypes, we conducted immunohistochemical analyses on tissue samples from patients with a range of tumor grades, which included IDH-mutant astrocytoma (WHO grade II), IDH-mutant anaplastic astrocytoma (WHO grade III), IDH-mutant and 1p/19q-co-deleted oligodendroglioma (WHO grade II), IDH-mutant and 1p/19q-co-deleted anaplastic oligodendroglioma (WHO grade III), glioblastoma, IDH-wildtype (WHO grade IV), and samples of normal brain tissue. A clear trend emerged showing a stepwise augmentation in ST3GAL4 expression from normal cerebral tissue, through lower-grade diffuse gliomas, up to the highest levels in glioblastoma, IDH-wildtype (Fig. 1H). These findings suggest a positive correlation between ST3GAL4 expression and the clinical advancement of the disease, implying a potential role for ST3GAL4 as an oncogenic factor in glioma.
3.2. Prognostic implications of ST3GAL4 overexpression in Glioma cases
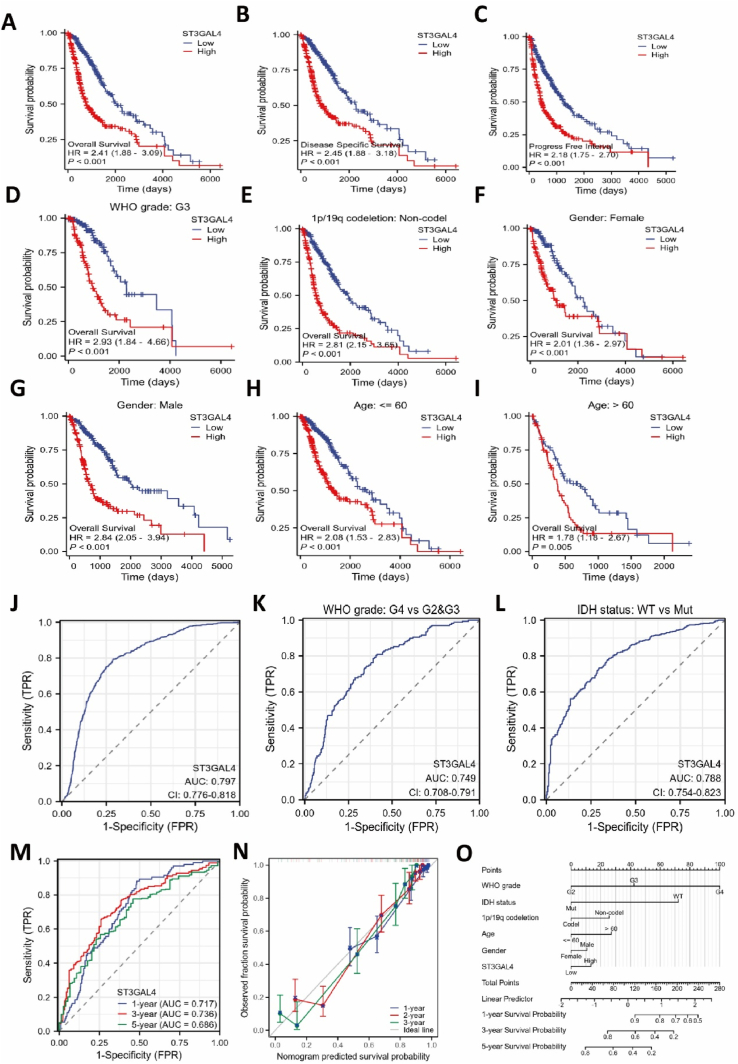
To investigate the correlation between the expression levels of ST3GAL4 and the survival rates of glioma patients, we conducted a thorough analysis of patient prognosis. The Kaplan-Meier curve analysis demonstrated that patients with significantly elevated levels of ST3GAL4 expression had significantly poorer overall survival (OS), and disease-specific survival (DSS) and progression-free interval (PFI) rates compared to those with lower expression levels (Fig. 2A–C). Subgroup analysis yielded notable results, demonstrating that high expression of ST3GAL4 was significantly associated with an unfavorable prognosis for patients diagnosed with WHO grade III glioblastoma (HR = 2.90, P < 0.001). Furthermore, an intriguing finding was the positive correlation between low expression of ST3GAL4 and the survival rate of patients with co-deletion of 1p/19q. Moreover, results from the analysis of age and gender subgroups also indicated a link between high ST3GAL4 expression and a poorer prognosis (Fig. 2D–I). Thus, these findings strongly suggest that significantly elevated expression of ST3GAL4 acts as an independent prognostic indicator for the survival duration of glioma patients.
Fig. 2.
Significant association between ST3GAL4 expression and poor prognosis in gliomas. A–C: Kaplan-Meier curve analysis using the TCGA database was conducted to assess overall survival (A), disease-specific survival (B), and progression-free interval (C) in patients with gliomas. D–I: ST3GAL4 expression within various subgroups: WHO grade (D), 1p/19q codeletion (E), gender (F, G), and age (H, I), correlated with glioma prognosis (P < 0.005). J: Cox analysis was used to evaluate the diagnostic value of ST3GAL4 expression level in gliomas. K–M: Cox analysis to predict the relationship between ST3GAL4 and glioma prognosis, considering various clinical feature states, including WHO grade (K), IDH status (L), and time points (M). N: Calibration plot of the nomogram for OS prediction. O: Nomogram survival prediction chart for predicting the 1-, 3-, and 5-year overall survival rates.
3.3. Diagnostic potential of ST3GAL4 expression levels in Glioma
An ROC curve was generated to assess the diagnostic capability of ST3GAL4 expression, resulting in an area under the curve (AUC) of 0.797 (95 % confidence interval: 0.776–0.818), with a sensitivity of 79.4 %, specificity of 71.3 %, positive predictive value of 0.622, and negative predictive value of 0.857 (Fig. 2J). Additionally, ST3GAL4 had important diagnostic value in distinguishing glioma based on different WHO grades and wild or mutant IDH types (Fig. 2K, L). Moreover, the diagnostic value of ST3GAL4 in glioma was demonstrated at 1 year, 3 years, and 5 years (Fig. 2M). For predicting patient survival at 1, 3, and 5 years, a composite nomogram was constructed that incorporated ST3GAL4 expression data and clinical factors (Fig. 2N). Furthermore, the calibration graph showed strong agreement between observed and projected values, providing further evidence of the positive prognostic impact of ST3GAL4 expression on glioma (Fig. 2O). In conclusion, the analysis results suggest that ST3GAL4 may serve as a potential treatment target for glioma patients.
3.4. Anti-tumor effects of ST3GAL4 knockdown: impaired proliferation and cell cycle arrest
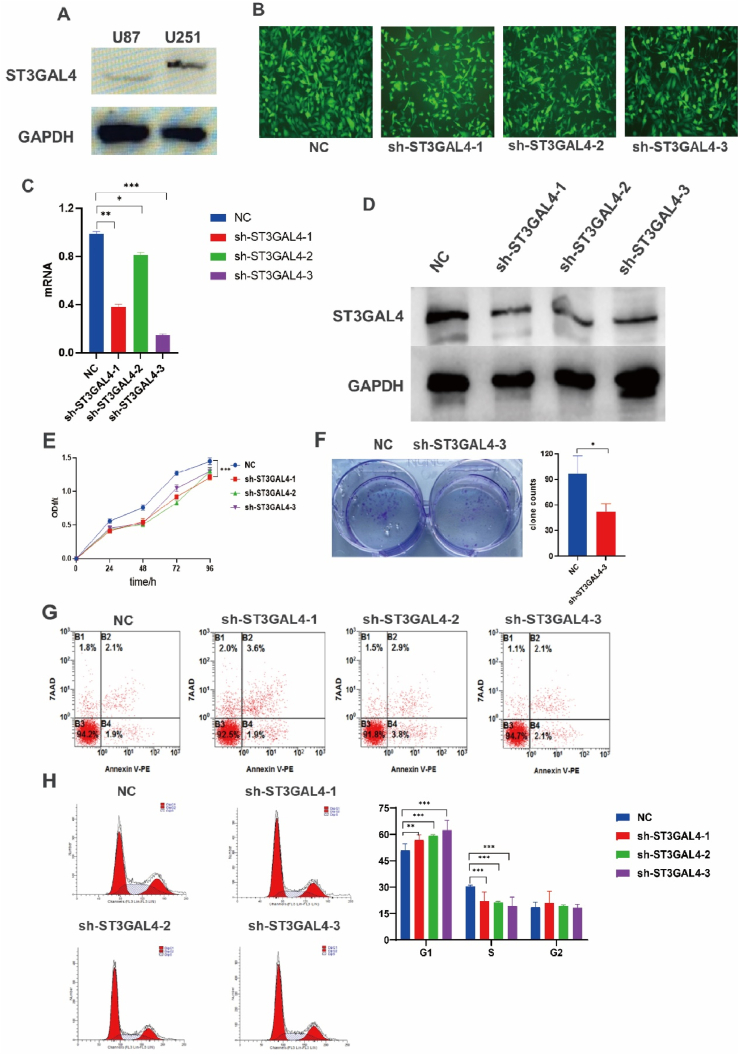
We conducted a comprehensive analysis to validate the functional role of ST3GAL4 in glioblastoma tumor cells, examining its impact on proliferation, migration, colony formation, and cell cycle regulation. We examined the U251 and U87 glioma cell lines for ST3GAL4 protein expression and proceeded with the U251 cells for the subsequent experiments due to their high expression of ST3GAL4 (Fig. 3A). Stable U251 cell lines expressing shRNA were established through lentivirus infection (Fig. 3B). The efficiency of ST3GAL4 knockdown was validated using qPCR and Western blotting, and the sh-ST3GAL4-3 U251 cell line exhibited the most significant knockdown effect (Fig. 3C and D). The results of the CCK-8 cell proliferation experiment revealed a significant inhibition in cell proliferation upon knocking down ST3GAL4 (Fig. 3E). Moreover, inhibiting ST3GAL4 also impaired the ability of tumor cells to form colonies (Fig. 3F). Further analysis using flow cytometry revealed no significant change in cell apoptosis (Fig. 3G). However, the cell cycle assay clearly demonstrated a G1 phase arrest in cells with ST3GAL4 knockdown (Fig. 3H). In conclusion, knocking down ST3GAL4 led to significant inhibition of proliferation and cell cycle arrest.
Fig. 3.
Inhibition of ST3GAL4 hinderd tumor cell growth and cell cycle progression. A: Protein immunoblotting was used to detect the expression levels of ST3GAL4 in U87 and U251 glioma cells. B: Stable U251 cell lines expressing control or ST3GAL4-targeting shRNAs. C: The efficiency of ST3GAL4 knockdown at the mRNA level was assessed using qPCR. D: Protein immunoblotting was used to determine the efficiency of ST3GAL4 knockdown. E, F: The proliferation of stable U251 cells was measured using CCK-8 assay (E) and clonal formation assay (F). G: Apoptosis assay by flow cytometry with U251 cells knocked down for ST3GAL4. H: Cell cycle assay by flow cytometry with U251 cells knocked down for ST3GAL4. *P < 0.05, **P < 0.01, ***P < 0.001.
3.5. Anti-migratory and anti-sialylation effects of ST3GAL4 inhibition in Glioma cells
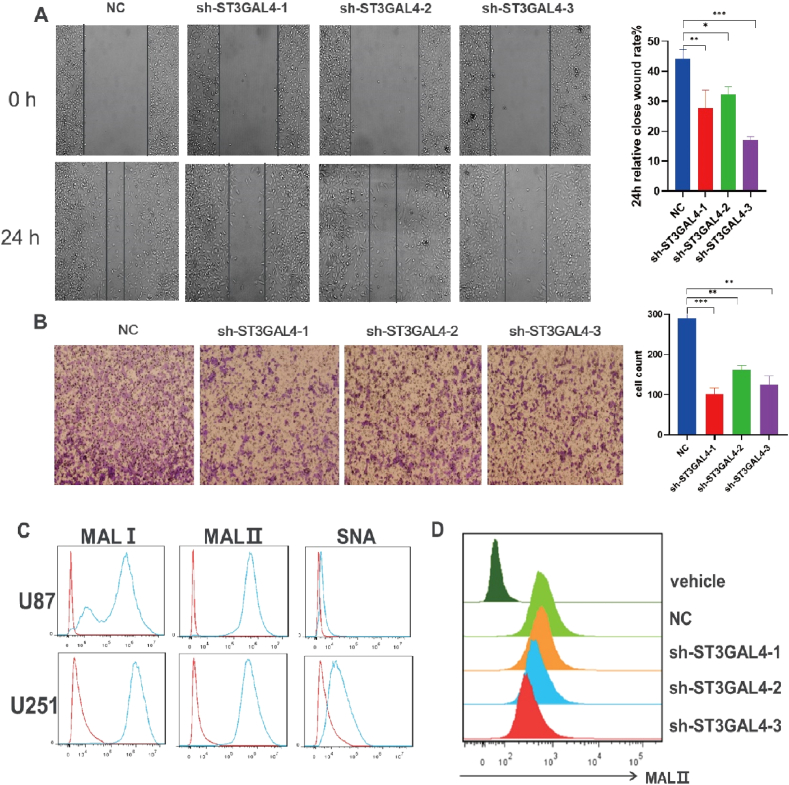
The migration of glioma cells was investigated using the wound healing assay and Transwell. In the wound healing assay, both the width and number of migrating cells were observed 24 h post-scratching in U251 cells. Significantly reduced values were observed compared to the control condition (Fig. 4A). Similarly, the Transwell assay showed a significant decrease in migratory ability in glioma cells with ST3GAL4 knockdown compared to the control condition (Fig. 4B). To assess sialic acid expression in glioma cells, flow cytometry was performed on U251 cells using biotinylated plant lectins derived from Maackia amurensis (MALI/II) and Sambucus nigra (SNA). These lectins acted as probes for identifying sialylated structures with α2,3 or α2,6 configuration, respectively. The results showed high expression levels of MALⅠ and MALⅡ in both U251 and U87 glioma cells, with a much lower level of SNA in U251 cells, in contrast to U87 cells (Fig. 4C). This indicated that α2,3-linked sialic acids were highly expressed in the glioma cell lines. Interestingly, there was a decrease in MALⅡ levels in the ST3GAL4 knockdown condition, indicating a downregulation of α2,3-linked sialic acids in the sh-ST3GAL4 U251 cells (Fig. 4D).
Fig. 4.
Knockdown of ST3GAL4 impeded cell migration and α2,3-linked sialic acid expression in U251 cells. A, B: Cell migration ability was assessed in U251 cells with and without ST3GAL4 knockdown using the wound healing assay (A) and Transwell (B). *P < 0.05, **P < 0.01, ***P < 0.001. C: Flow cytometry analysis of α2,3- and α2,6-linked sialic acid expression in U87 and U251 cells. D: Flow cytometry analysis of α2,3-linked sialic acid expression in control or ST3GAL4 knockdown U251 cells.
3.6. ST3GAL4's role in modulating extracellular matrix and potential to facilitate EMT
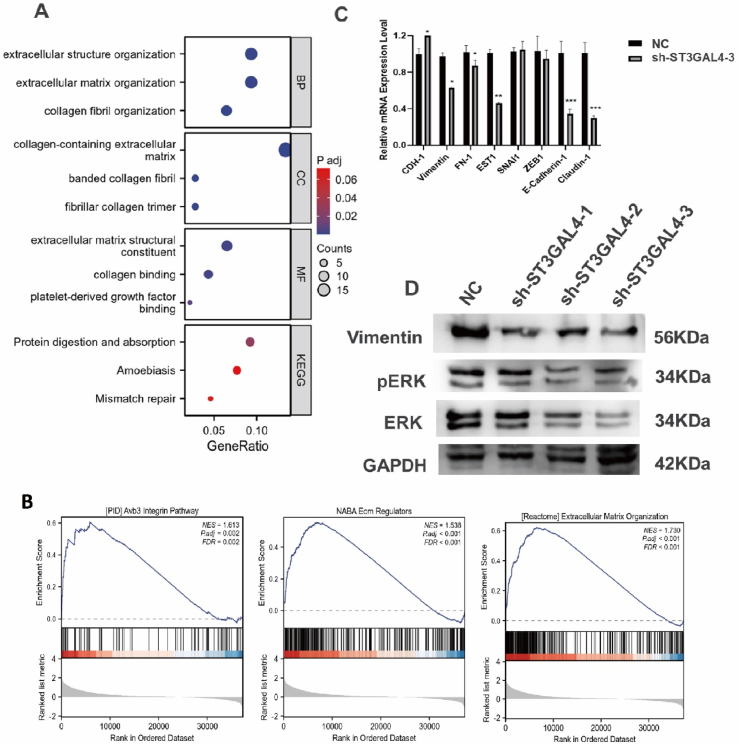
To analyze the mechanism underlying the promotion of glioma cell proliferation and metastasis by ST3GAL4, we conducted GO, KEGG and GSEA using TCGA data. The top 200 differentially expressed genes (DEGs) with high expression in the ST3GAL4 high-expression group were subjected to these analyses. The results demonstrate that these genes are involved in various functions, including extracellular structure organization, extracellular matrix organization, collagen-containing extracellular matrix, and collagen binding (Fig. 5A). Additionally, the GSEA results indicate significant associations between ST3GAL4 and extracellular matrix organization, the integrin pathway, and ECM regulation (Fig. 5B). Therefore, these findings strongly suggest that ST3GAL4 may play a role in modulating the composition and organization of the extracellular matrix. Moreover, previous studies have reported that sialylation of TGF-β may regulate EMT [12]. Considering this, we investigated whether ST3GAL4 exerts its effects via the EMT-related signaling pathway. We performed qPCR to examine the expression of EMT-related genes in ST3GAL4 knockdown U251 cells. As anticipated, the results revealed a significant reduction in mRNA levels of EMT-related genes in the sh-ST3GAL4 cells (Fig. 5C). Furthermore, Western blotting analysis showed a decrease in the expression of the mesenchymal marker vimentin in ST3GAL4 knockdown U251 cells. In addition, a decrease in the expression levels of both total ERK and phosphorylated ERK was observed (Fig. 5D). These findings suggest a potential role of ST3GAL4 in glioma progression via the EMT signaling pathway.
Fig. 5.
Functional enrichment analysis demonstrated the involvement of ST3GAL4 in extracellular matrix organization. A: Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis were performed on the top 200 upregulated differentially expressed genes (DEGs) in glioma patients with high ST3GAL4 expression. B: The gene set enrichment analysis (GSEA) identified pathways related to the extracellular matrix that were significantly enriched. C: qRT-PCR analysis to evaluate the expression of eight representative epithelial-mesenchymal transition (EMT) biomarkers in U251 cells with or without ST3GAL4 knockdown. *P < 0.05, **P < 0.01, ***P < 0.001. D: Western blot analysis to examine the protein expression levels of vimentin, ERK1/2, pERK1/2, and GAPDH in U251 cells.
3.7. Correlation between ST3GAL4 overexpression and macrophage infiltration in the Glioma tumor microenvironment
Within the glioma TME, non-tumor cells, including infiltrating immune cells, promote tumor progression and contribute to treatment resistance and immune evasion through their interactions with glioma cells [13,14]. Existing evidence suggests that the interaction between glioma cells and the immune microenvironment stimulates the production of immune suppressive factors in glioma [[15], [16], [17]]. Tumor-associated macrophages (TAMs) and microglia cells make up the primary immune cell populations in the immune microenvironment of glioma [18,19]. TAMs play a crucial role in promoting tumor progression and inducing immune suppression within the glioma microenvironment [20].
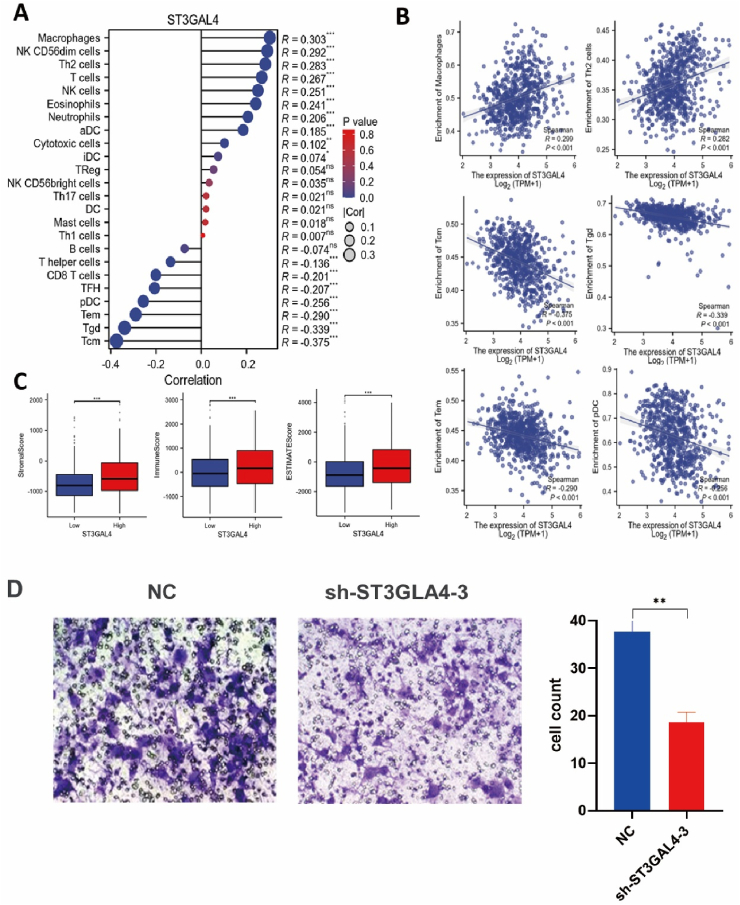
We predicted the infiltration levels of 24 immune cell types in glioma using the single-sample gene set enrichment analysis (ssGSEA) algorithm. The results indicated a positive correlation between ST3GAL4 expression and macrophages, Th2 cells, NK CD56dim cells, and T cells, while a negative correlation was observed with Tcm, Tgd, and CD8 T cells (Fig. 6A B). Furthermore, increased expression of ST3GAL4 was associated with higher stromal and immune infiltration scores in the tumor microenvironment (Fig. 6C). According to existing literature, in the presence of glioma in the brain, macrophages from the bloodstream migrate to the surrounding microenvironment, promoting the development of M2 polarized TAMs that contribute to immune suppression and facilitate tumor growth [21,22]. The analysis above revealed that macrophages showed the most significant relationship with ST3GAL4 expression. To delve deeper into this, we performed a Transwell chamber assay, with the lower chamber containing tumor cells and the upper chamber containing THP-1-differentiated macrophages for a duration of 24 h. The results exhibited that inhibiting ST3GAL4 significantly diminishes the recruitment capability of macrophages in glioma cells (Fig. 6D).
Fig. 6.
ST3GAL4 facilitated macrophage infiltration in the glioma TME. A: Correlation between ST3GAL4 expression and 24 immune cell types. B: Correlation of ST3GAL4 expression with immune infiltration levels of macrophages, Th2 cells, Tcm cells, Tgd cells, Tem cells, and pDCs. C: Correlation between ST3GAL4 expression level and immune matrix scores. ***P < 0.001. D: Transwell assay assessing macrophage recruitment by control and sh-ST3GAL4 U251 cells. *P < 0.05, **P < 0.01, ***P < 0.001.
3.8. Immune checkpoint Co-expression and immunomodulation by ST3GAL4 in Glioma
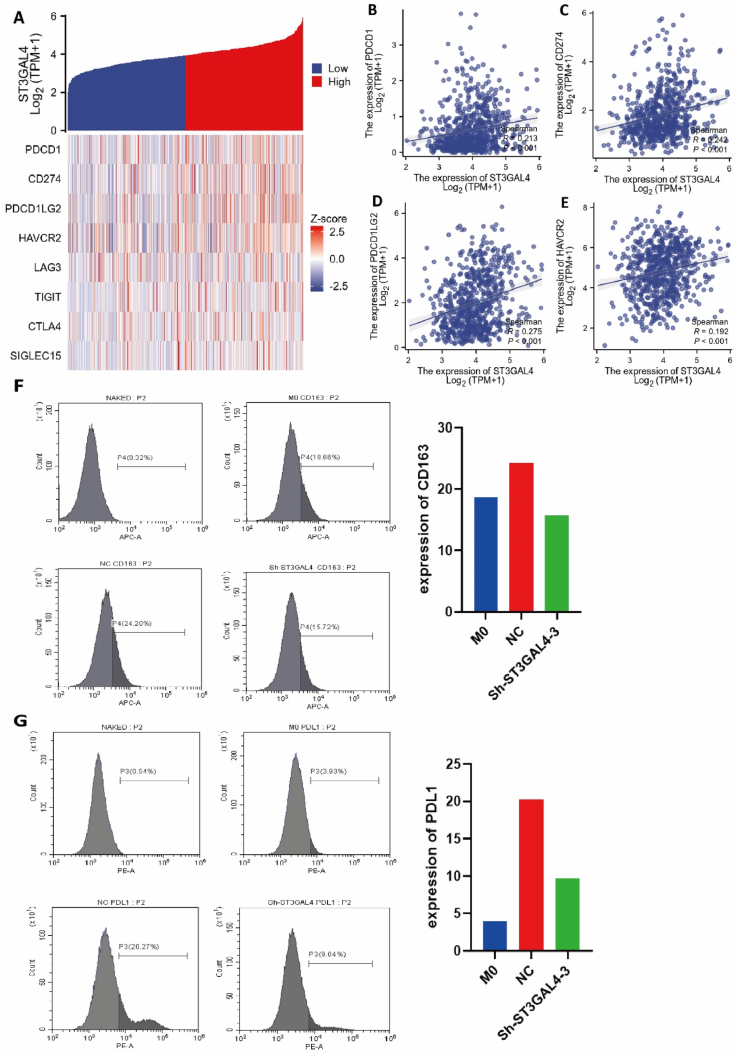
The expression levels of eight immune checkpoint genes (PDCD1, SIGLEC15, CD274, HAVCR2, PDCD1LG2, LAG3, CTLA4, and PDCD1) were compared between the high expression group and the low expression group of ST3GAL4. An increased expression of these classic immune checkpoints was observed in the high expression group of ST3GAL4 (p < 0.001) (Fig. 7A). Subsequently, the relationship between ST3GAL4 and these eight immune checkpoint genes in gliomas was assessed through Spearman correlation analysis. The findings indicated a positive correlation between the expression levels of ST3GAL4 and PDCD1 (p < 0.001, R = 0.213, Fig. 7B), CD274 (p < 0.001, R = 0.242, Fig. 7C), PDCD1LG2 (p < 0.001, R = 0.275, Fig. 7D), and HAVCR2 (p < 0.001, R = 0.192, Fig. 7E), suggesting co-expression of ST3GAL4 with these classic immune checkpoints in gliomas.
Fig. 7.
Correlation between ST3GAL4 and immune checkpoint genes as well as macrophage immunosuppressive markers. A: Heatmap displaying the expression of immune checkpoint genes in groups with high and low ST3GAL4 expression. B–E: Correlation analysis of ST3GAL4 with immune checkpoint gene PDCD1(B), CD274 (C), PDCD1LG2 (D), and HAVCR2 (E). F, G: Expression of CD163 (F) and CD274 (G) on macrophages cocultured with or without U251 cells. Flow cytometry was used to detect CD163 and CD274 in unstained control samples as well as in macrophages alone, macrophages cocultured with control U251 cells, or macrophages cocultured with sh-ST3GAL4 U251 cells for 24 h.
The influence of ST3GAL4-suppressed tumor cells on macrophages was investigated by co-culturing U251 cells (control or sh-ST3GAL4) with THP-1-differentiated macrophages for 24 h. Compared to macrophages cultured alone with a CD163 expression ratio of 19.66 %, macrophages co-cultured with control U251 cells showed an increased CD163 expression ratio of 24.20 %, whereas only 15.72 % of macrophages were CD163 positive when co-cultured with sh-ST3GAL4 U251 cells (Fig. 7F). Similarly, the expression of the immune checkpoint CD274 significantly decreased on macrophages when co-cultured with sh-ST3GAL4 U251 cells compared to co-culturing with control U251 cells (Fig. 7G). These findings suggest that the expression of ST3GAL4 in glioma cells can upregulate the expression of immunosuppressive markers, including CD163 and CD274, on macrophages, potentially resulting in tumor immune evasion.
4. Discussion
Sialylation, a critical post-translational modification, adds sialic acid to glycan chains on glycoproteins and glycolipids, thereby influencing cellular functions including recognition, adhesion, and signal transduction. Notably, the presence of excess sialic acid, or hypersialylation, has been identified as a cancer hallmark, implicated in promoting malignancy by affecting tumor cell behavior, angiogenesis, immune evasion, and apoptosis resistance, thus potentially serving as a cancer prognostic marker.
Sialyltransferases, the enzymes orchestrating sialylation, are categorized into four groups—ST3GAL, ST6GAL, ST6GALNAC, and ST8SIA—based on the glycosidic bonds they form [23,24]. These enzymes' altered expression in cancer can lead to modifications in glycoprotein sialylation patterns, affecting tumor growth and immune response. For example, the ST3GAL family has been linked with cancer progression; specifically, ST3GAL1 is known for its role in synthesizing cancer-associated sialic acids and promoting tumor growth, as well as enabling immune evasion by modifying the complement system. ST3GAL3, similarly, is associated with chemoresistance [25]. ST3GAL4, the focus of this analysis, has been shown to support metastasis by enhancing adhesion and migration of cancer cells, as evidenced by reduced tumor cell motility upon its downregulation [26,27]. While studies have shown ST3GAL4's overexpression increases invasive properties in gastric cancer via sLex antigen production and c-Met signaling, its relationship with glioma pathogenesis remains unexplored.
Our study here leveraged the TCGA database to evaluate ST3GAL4 expression across diverse cancer types, revealing a notable pattern of overexpression in gliomas. This pattern correlates significantly with adverse prognostic indicators, confirming the contributions of aberrant sialylation to cancer progression described in preceding research. Functional assays demonstrated that inhibiting ST3GAL4 diminishes α2,3-sialic acid on glioma cells, attenuating tumorigenesis, cell motility, and cell cycle advancement. Further analysis employing KEGG pathway enrichment disclosed a substantial link between ST3GAL4 and the regulation of the extracellular matrix, a crucial aspect of tumor infiltration and dissemination. GSEA corroborated the association of ST3GAL4 with the extracellular matrix and its probable impact on EMT signaling pathways, which confer metastatic and resistance traits to cancer cells.
Examination of the tumor's immune milieu revealed that ST3GAL4 showed positive associations with a range of immune cells, while inversely correlated with certain T cell subsets. This points to a complex role for ST3GAL4 in manipulating the immune landscape, possibly steering macrophages towards an immunosuppressive state that favors tumor growth. Our analysis also uncovered significant ties between ST3GAL4 and the immune checkpoint CD274 (PD-L1), indicating its potential involvement in immune escape mechanisms.
Our experimental design and integration of bioinformatics with clinical data yield an extensive understanding of ST3GAL4's role in gliomas. Nevertheless, the research has limitations: dependency on public databases may introduce biases, and in vitro loss-of-function experiments lack the intricacies of in vivo human gliomas, warranting further investigation through animal models.
In summary, the findings of our study have significant implications for the diagnosis and prognosis of glioma. ST3GAL4's elevated expression in glioma tissues, particularly in high-grade tumors, suggests its potential as a diagnostic marker. Furthermore, the correlation between ST3GAL4 levels and poor patient outcomes underscores its prognostic value. These findings align with previous research on the role of sialylation in cancer progression, reinforcing the importance of ST3GAL4 as a target for therapeutic intervention.
Ethics approval and consent to participate
The research was approved by the Medical Ethics Committee of the First Affiliated Hospital of the Fourth Military Medical University (Approval number: KY20224001-21).
Funding
This research was funded by grants from the National Natural Science Foundation of China, grant number 82073361; the State Key Laboratory of Cancer Biology Project, grant number CBSKL2022ZZ21; the Xi'an Municipal Health Commission grant, grant number 2022ms06; the Key R&D Plan of Shaanxi Province, grant number 2023-YBSF-667.
Data availability statement
The datasets associated with this paper are available with corresponding authors on request.
CRediT authorship contribution statement
Wenjing Zheng: Writing – original draft, Validation, Methodology, Data curation. Han Zhang: Methodology, Data curation. Yi Huo: Validation. Lingling Zhang: Formal analysis. Longqi Sa: Resources. Lequn Shan: Investigation, Funding acquisition, Conceptualization. Tao Wang: Writing – review & editing, Supervision, Project administration, Investigation, Funding acquisition, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We gratefully acknowledge the Medical Innovation Center of the Fourth Military Medical University for their provision of experimental equipment and services. Additionally, we are indebted to the American Journal Experts (www.aje.cn) for their linguistic assistance during the preparation of this manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e29829.
Contributor Information
Lequn Shan, Email: drshanlq@fmmu.edu.cn.
Tao Wang, Email: wangt@fmmu.edu.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Louis D.N., et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114(2):97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang K., et al. Glioma targeted therapy: insight into future of molecular approaches. Mol. Cancer. 2022;21(1):39. doi: 10.1186/s12943-022-01513-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang G., et al. Tumor-associated microglia and macrophages in glioblastoma: from basic insights to therapeutic opportunities. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.964898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen H., et al. Activated TRPA1 plays a therapeutic role in TMZ resistance in glioblastoma by altering mitochondrial dynamics. BMC Mol Cell Biol. 2022;23(1):38. doi: 10.1186/s12860-022-00438-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burzynska P., et al. Sialic acids as receptors for pathogens. Biomolecules. 2021;11(6) doi: 10.3390/biom11060831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Silva M.L.S. Capitalizing glycomic changes for improved biomarker-based cancer diagnostics. Explor Target Antitumor Ther. 2023;4(3):366–395. doi: 10.37349/etat.2023.00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Munkley J. Aberrant sialylation in cancer: therapeutic opportunities. Cancers. 2022;14(17) doi: 10.3390/cancers14174248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alvarez M.R.S., et al. N-glycan and glycopeptide serum biomarkers in philippine lung cancer patients identified using liquid chromatography-tandem mass spectrometry. ACS Omega. 2022;7(44):40230–40240. doi: 10.1021/acsomega.2c05111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doostkam A., et al. Sialic acid: an attractive biomarker with promising biomedical applications. Asian Biomed. 2022;16(4):153–167. doi: 10.2478/abm-2022-0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guerrero P.E., et al. Knockdown of alpha2,3-sialyltransferases impairs pancreatic cancer cell migration, invasion and E-selectin-dependent adhesion. Int. J. Mol. Sci. 2020;21(17) doi: 10.3390/ijms21176239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Costa A.F., et al. ST3GalIV drives SLeX biosynthesis in gastrointestinal cancer cells and associates with cancer cell motility. Glycoconj. J. 2023;40(4):421–433. doi: 10.1007/s10719-023-10113-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Du J., et al. Dynamic sialylation in transforming growth factor-beta (TGF-beta)-induced epithelial to mesenchymal transition. J. Biol. Chem. 2015;290(19):12000–12013. doi: 10.1074/jbc.M115.636969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Quail D.F., Joyce J.A. The microenvironmental landscape of brain tumors. Cancer Cell. 2017;31(3):326–341. doi: 10.1016/j.ccell.2017.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou X., Yang G., Guan F. Biological functions and analytical strategies of sialic acids in tumor. Cells. 2020;9(2) doi: 10.3390/cells9020273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pang L., et al. Mechanism and therapeutic potential of tumor-immune symbiosis in glioblastoma. Trends Cancer. 2022;8(10):839–854. doi: 10.1016/j.trecan.2022.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pang L., et al. Pharmacological targeting of the tumor-immune symbiosis in glioblastoma. Trends Pharmacol. Sci. 2022;43(8):686–700. doi: 10.1016/j.tips.2022.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klemm F., et al. Interrogation of the microenvironmental landscape in brain tumors reveals disease-specific alterations of immune cells. Cell. 2020;181(7):1643–1660 e17. doi: 10.1016/j.cell.2020.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xuan W., et al. Context-dependent glioblastoma-macrophage/microglia symbiosis and associated mechanisms. Trends Immunol. 2021;42(4):280–292. doi: 10.1016/j.it.2021.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu C., et al. Origin, activation, and targeted therapy of glioma-associated macrophages. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.974996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simonds E.F., et al. Deep immune profiling reveals targetable mechanisms of immune evasion in immune checkpoint inhibitor-refractory glioblastoma. J Immunother Cancer. 2021;9(6) doi: 10.1136/jitc-2020-002181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang C., et al. Tumor purity as an underlying Key factor in glioma. Clin. Cancer Res. 2017;23(20):6279–6291. doi: 10.1158/1078-0432.CCR-16-2598. [DOI] [PubMed] [Google Scholar]
- 22.Khan F., et al. Macrophages and microglia in glioblastoma: heterogeneity, plasticity, and therapy. J. Clin. Invest. 2023;133(1) doi: 10.1172/JCI163446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Y., Chen X. Sialic acid metabolism and sialyltransferases: natural functions and applications. Appl. Microbiol. Biotechnol. 2012;94(4):887–905. doi: 10.1007/s00253-012-4040-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nag S., et al. Sialyltransferases and neuraminidases: potential targets for cancer treatment. Diseases. 2022;10(4) doi: 10.3390/diseases10040114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yeo H.L., et al. Sialylation of vasorin by ST3Gal1 facilitates TGF-beta1-mediated tumor angiogenesis and progression. Int. J. Cancer. 2019;144(8):1996–2007. doi: 10.1002/ijc.31891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perez-Garay M., et al. alpha2,3-Sialyltransferase ST3Gal IV promotes migration and metastasis in pancreatic adenocarcinoma cells and tends to be highly expressed in pancreatic adenocarcinoma tissues. Int. J. Biochem. Cell Biol. 2013;45(8):1748–1757. doi: 10.1016/j.biocel.2013.05.015. [DOI] [PubMed] [Google Scholar]
- 27.Liu W., et al. The role of paxillin aberrant expression in cancer and its potential as a target for cancer therapy. Int. J. Mol. Sci. 2023;24(9) doi: 10.3390/ijms24098245. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets associated with this paper are available with corresponding authors on request.