Abstract
Objective
In this study, we aimed to utilize computed tomography (CT)-derived radiomics and various machine learning approaches to differentiate between invasive mucinous adenocarcinoma (IMA) and invasive non-mucinous adenocarcinoma (INMA) preoperatively in solitary pulmonary nodules (SPN) ≤3 cm.
Methods
A total of 538 patients with SPNs measuring ≤3 cm were enrolled, categorized into either the IMA group (n = 50) or INMA group (n = 488) based on postoperative pathology. Radiomic features were extracted from non-contrast-enhanced CT scans and identified using the least absolute shrinkage and selection operator (LASSO) algorithm. In constructing radiomics-based models, logistic regression, support vector machines, classification and regression trees, and k-nearest neighbors were employed. Additionally, a clinical model was developed, focusing on CT radiological features. Subsequently, this clinical model was integrated with the most effective radiomic model to create a combined model. Performance assessments of these models were conducted, utilizing metrics such as the area under the receiver operating characteristic curve (AUC), DeLong's test, net reclassification index (NRI), and integrated discrimination improvement (IDI).
Results
The support vector machine approach showed superior predictive efficiency, with AUCs of 0.829 and 0.846 in the training and test cohorts, respectively. The clinical model had AUCs of 0.760 and 0.777 in the corresponding cohorts. The combined model had AUCs of 0.847 and 0.857 in the corresponding cohorts. Furthermore, compared to the radiomic model, the combined model significantly improved performance in both the training (DeLong test P = 0.045, NRI 0.206, IDI 0.024) and test cohorts (P = 0.029, NRI 0.125, IDI 0.032), as well as compared to the clinical model in both the training (P = 0.01, NRI 0.310, IDI 0.09) and test cohorts (P = 0.047, NRI 0.382, IDI 0.085).
Conclusion
the combined model exhibited excellent performance in distinguishing between IMA and INMA in SPNs ≤3 cm.
Keywords: Invasive mucinous adenocarcinoma, Solitary pulmonary nodules, Radiomics, Machine learning
Highlights
-
•
Support vector machine showed superior performance as a radiomics model.
-
•
The combined model showed excellent distinction between IMA and INMA in SPNs ≤3 cm.
-
•
Use of the combined approach may improve clinical decisions and prognosis.
1. Introduction
Within the spectrum of lung adenocarcinoma (LADC), invasive mucinous adenocarcinoma (IMA) is recognized as a distinct histological subtype distinguished by its unique clinical, radiological, histopathological, and molecular features [1]. Compared with invasive non-mucinous adenocarcinoma (INMA), IMA is less prevalent, constituting approximately 3–10 % of all LADC cases [2,3]. Despite its rarity, IMA presents with significant prognostic heterogeneity. Certain investigations have hinted at a reduced overall survival (OS) in IMA patients in contrast to those with INMA [4,5]. However, contradictory evidence has surfaced, proposing a more promising prognosis for IMA patients when juxtaposed with their INMA counterparts [6,7]. Subgroup analysis further substantiated that patients with IMA have an elevated risk of intrapulmonary recurrence, exacerbating the prognosis relative to INMA [8]. Additionally, patients with IMA display a disease-free survival (DFS) rate between that of low- (lepidic predominance) and intermediate-grade (acinar/papillary predominance) INMA. Notably, their OS aligns with that of patients with intermediate-grade INMA [9].
Extensive inquiry has shed light on the distinguishing characteristics of IMA, elucidating its unique genomic, pathological, and histological attributes relative to INMA. Histologically, IMA showcases reduced occurrences of lymphatic permeation, pleural involvement, and vascular invasion, while displaying an increased spread through air spaces [10,11]. Pathologically, IMA is characterized by goblet-shaped or columnar cells and a profusion of mucin in the cytoplasm [2,12]. Patients diagnosed with IMA exhibit mutation patterns tailored to their specific cases, including alterations in EGFR, ALK, and KRAS [[13], [14], [15], [16]]. Consequently, targeted therapies hold substantial promise for the treatment of patients with IMA. Recognizing the unique characteristics, the classification of IMA as a distinct subtype of LADC was officially established by the World Health Organization (WHO) in 2015 [17].
Previous research endeavors have extensively explored the imaging characteristics of IMA, categorizing them into two primary types discerned from computed tomography (CT) morphological features: solitary pulmonary nodules (SPN) and pneumonia-type presentations [[18], [19], [20], [21]]. Notably, pneumonia-type IMA exhibited significantly inferior OS and DFS compared to that seen with SPN-type IMA [[18], [19], [20], [21]]. In the past, mucinous bronchioloalveolar carcinoma was often linked with multifocal disease or appeared as a pneumonia-like pattern on CT scans [12]. Nevertheless, recent findings indicate that SPN-type IMA is more commonly encountered than pneumonia-type IMA [9,[19], [20], [21]]. Prior investigations have proposed certain CT characteristics, such as air bronchograms, elevated standardized uptake values, and abnormal vascular alterations, as potential predictive indicators for SPN-type IMA [22]. Notably, we must acknowledge that these CT radiological features are highly subjective and lack representation as they rely heavily on the experience of radiologists. Specifically, for SPNs ≤3 cm, conventional CT radiological features exhibit greater inconsistency and atypicality, rendering the diagnosis of IMA in such nodules challenging. Consequently, achieving an accurate differentiation between early-stage (≤3 cm) SPN-type IMA and INMA based solely on preoperative CT images poses a formidable challenge.
Radiomics, a burgeoning domain within medical imaging, involves extracting and analyzing quantitative characteristics from radiographic images. Its distinctive strength lies in its ability to transform medical images into mineable data, enabling a more thorough understanding of tumor phenotypes. This approach contributes significantly to building predictive models that are crucial for personalized diagnostics, prognosis, and treatment planning in lung cancer [23]. Yu et al. [24] devised a nomogram to distinguish pneumonia-type IMA from pneumonia by amalgamating clinical characteristics and CT-based radiomic features. Similarly, Zhang et al. [25] exhibited effective diagnostic accuracy in discerning SPN-type IMA from pulmonary tuberculoma using a radiomics-based nomogram. While these studies provide groundwork for the preoperative diagnosis of IMA through radiomics, there is still a predominant emphasis on diagnosing pneumonia-type IMA, resulting in a noticeable gap in the application to SPN-type IMA. In addition, there is a lack of radiomic models exploring the differentiation between IMA and INMA in SPNs ≤3 cm. Hence, the objective of this investigation was to employ machine learning-boosted radiomics for establishing reliable, highly precise, and efficient predictive models [26] to facilitate accurate discrimination between IMA and INMA in SPNs ≤3 cm. This methodology has the potential to significantly augment clinical decision-making procedures.
2. Materials and methods
2.1. Patient selection
This research was conducted in accordance with the guidelines set forth in the Declaration of Helsinki and was approved by the Ethics Committees at the participating medical institutions (reference numbers: KY2020147 from The Affiliated Hospital of Southwest Medical University and 2021-07-009 from Xiangtan Central Hospital). Given the retrospective design of the study, informed consent was not required. Between May 1, 2019 and September 1, 2023, a total of 1092 SPN-type LADCs subjected to surgical resection were identified at The Affiliated Hospital of Southwest Medical University and Xiangtan Central Hospital. In the most recent categorization provided by the WHO, IMA is delineated by two cytological attributes: the existence of tumor cells, comprising both the columnar cell-type and foveolar-type (or goblet cell-type), and abundant cytoplasmic mucin. Mucin is usually present within and outside the cytoplasm of these cells, typically with their nuclei positioned at the base, and the alveoli surrounding the tumor frequently become filled with mucus [3].
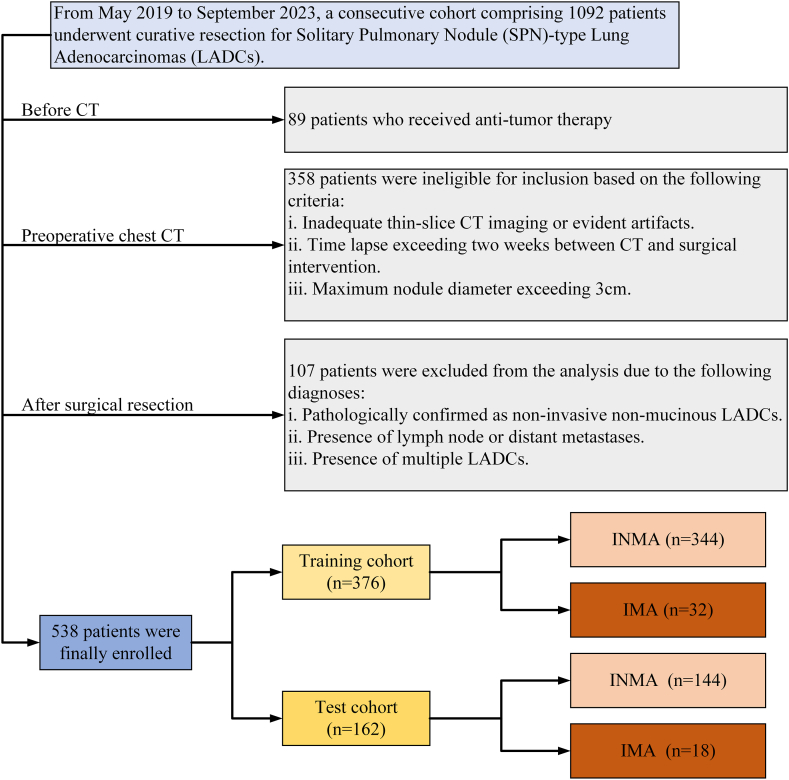
The criteria for inclusion were established as follows: (1) lesions with diameters ranging from 0.5 to 3.0 cm; (2) confirmation of IMA or INMA via postoperative pathology; (3) SPN lacking calcification, cavities, or vacuoles, and without ground-glass density; and (4) availability of complete thin-slice CT image data within 14 days before pathological diagnosis. The exclusion criteria comprised (1) presence of multiple LADCs in postoperative pathology; (2) prior chemoradiation before CT examination or pathological diagnosis; (3) pathological diagnosis of minimally invasive LADC or LADC in situ; or (4) evidence of lymph node or distant metastases, occurrences less common in IMA than in INMA [27]. Ultimately, 538 patients with SPNs ≤3 cm underwent postoperative pathological examinations, resulting in diagnosis of IMA (n = 50) or INMA (n = 488).These cases were then classified as the training (n = 376) and test (n = 162) cohorts, following a 70 %:30 % ratio. The detailed patient inclusion process is illustrated in Fig. 1.
Fig. 1.
Flowchart illustrating the patient enrollment process for the study.
2.2. CT image acquisition
Preoperative chest CT examinations were conducted using multidetector-row CT systems at the respective hospitals. Xiangtan Central Hospital employed either the Revolution CT (GE Healthcare, Chicago, IL) or MX16 CT (Philips Healthcare, Best, The Netherlands), while The Affiliated Hospital of Southwest Medical University utilized the uCT550 or uCT760 (Shanghai United Imaging Healthcare, Shanghai, China). The imaging protocol included obtaining images in the supine position, covering the area from the lung apex to below the costophrenic angle, with breath-hold helical scanning. Volumetric CT parameters consisted of a tube voltage of 120 kV, a tube current–time product ranging from 180 to 280 mA, a matrix size of 512 × 512, and image preprocessing. CT image reconstruction was conducted using a standard algorithm, with a layer thickness of 0.55–0.625 mm and layer interval of 0.55–0.625 mm.
2.3. CT radiological feature estimation
The acquired CT image data were transferred to a dedicated workstation for subsequent processing. Thereafter, multiplanar reconstruction was executed to establish lung and mediastinal windows, facilitating further scrutiny. The lung window parameters were set within a width ranging from 1500 to 2000 HU and a level of −450 to −600 HU, whereas the mediastinal window spanned a width of 250–350 HU and a level of 30–50 HU. Various parameters encompassing size and morphological characteristics were documented and appraised during CT image analysis. Pursuant to the 8th edition of tumor, node, and metastasis staging, the lesion's size on the CT image was delineated as the maximum diameter in the axial plane [27]. Morphological attributes such as shape, boundary, spiculation, lobulation, vascular convergence signs, and vacuole signs were assessed.
Two seasoned thoracic radiologists, both accredited and specialized in chest CT imaging with 8 and 12 years of practice (Y.Z. and H.L.), independently scrutinized the CT morphological features, oblivious to clinical and histological findings. Should any disparities arise in qualitative indicators, consensus was attained through deliberation.
2.4. Tumor segmentation
CT images were imported into ITK-SNAP (version 4.0) for tumor segmentation and meticulously delineated by a radiologist with 8 years of expertise (Y.Z.). To achieve ideal image contrast through curve-based contrast adjustment, we opted for the lung window (width: 1500 HU, level: 500 HU) and the mediastinum window (width: 350 HU, level: 40 HU) for observation purposes. The tumor boundary underwent meticulous delineation, slice by slice, culminating in the establishment of a precise region of interest (ROI) on the CT image in alignment with the mask. Following this, another radiologist (H.L.), with12 years of expertise in the domain, conducted a comprehensive review of the lesion delineation, implementing requisite refinements as needed.
2.5. Radiomic feature extraction
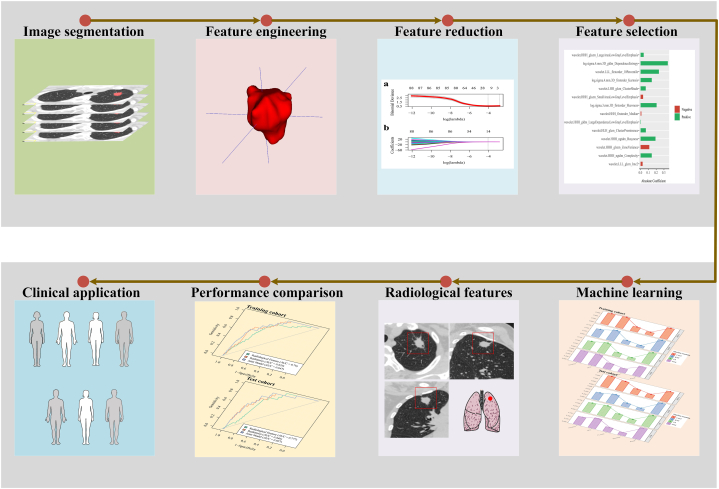
The original CT image and segmented mask were imported into Spyder (Python version 3.8) for further processing, with the pixel values of the images scaled to a range of 0–1. Subsequently, the volumes underwent resampled to a voxel size of 1 × 1 × 1 mm3 using a bicubic interpolation algorithm implemented with the “Numpy” package. Following this step, radiomics features were extracted from the ROI on the CT image corresponding to the mask utilizing the “PyRadiomics” package. This comprehensive tool facilitated the extraction of 1239 distinct radiomic features, covering first-order statistics, shape-based metrics, gray level co-occurrence matrix, the gray level dependence matrix, gray level run length matrix, gray level size zone matrix, and neighboring gray tone difference matrix. Detailed feature classification with specific explanations is presented in Fig. 2.
Fig. 2.
Detailed classification of features accompanied by specific explanations.
To accommodate the discrepancies inherent in CT images procured from various medical facilities and employing diverse protocols, normalization of all radiomic feature intensities was efficiently executed via z-score transformation, as delineated by equation (1):
| z = (x-μ)/σ | (1) |
2.6. Radiomic feature dimensionality reduction and selection
Z-score transformation was followed by the implementation of the minimum redundancy-maximum correlation (mRMR) method. This method aims to reduce feature dimensionality by selecting a subset of features that demonstrate low redundancy and high correlation with the target variable. By iteratively evaluating each feature's pertinence to the target variable and simultaneously considering the redundancy among features, this process determines which features are best suited for inclusion in the reduced feature set. Priority is given to features displaying notable relevance to the target variable and minimal redundancy with other selected features. This iterative process ensures the retention of the most informative and discriminative features while minimizing redundancy, ultimately resulting in a more efficient and effective representation of the top 100 radiomic features. Then, the least absolute shrinkage and selection operator (LASSO) method was then applied to identify features strongly associated with the desired result. The selected features with coefficients >0 were identified using a 10-fold cross-validation strategy. These procedures were implemented within the training cohort and then extended to the test cohort. Fig. 3 provides a visual representation of the radiomics process, illustrating ROI delineation, feature extraction, dimensionality reduction, feature selection, and utilization of machine-learning techniques.
Fig. 3.
Framework outlining the essential steps of computed tomography image acquisition, tumor segmentation, feature extraction, dimensionality reduction, and subsequent machine-learning analysis that is crucial for the development of a radiomic model.
2.7. Machine learning-derived radiomic models
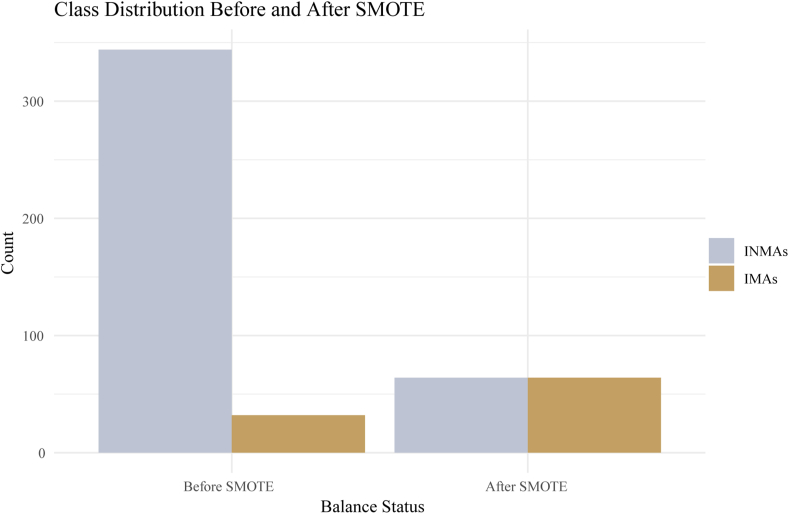
Given the disproportionate distribution between INMA and IMA cases (344:32 in the training cohort), the models may demonstrate a tendency to favor the majority class during the training process. To address this concern, we implemented resampling methodologies, particularly employing the synthetic minority oversampling technique (SMOTE), to rectify the imbalance in distribution within the training cohort during model development. This approach ensures the prevention of data leakage and maintains the representation of real clinical scenarios [28]. Detailed steps regarding the implementation are provided as comments within the provided R code:
Library (DMwR)
| smote_data < - SMOTE (label ∼., data = training cohort, perc.over = 100, k = 5) |
In this study, the “DMwR” package was used to perform resampling of both minority (32) and majority classes (344) in the training cohort (Fig. 1). The oversampling ratio was set to 100 % of the minority class, resulting in the oversampled samples being twice the original minority class size. The label represents IMA and INMA, with the parameter k for selecting neighbors set to 5. Subsequently, this algorithm was applied to both the minority and majority classes, which entailed oversampling the minority class and undersampling the majority class. The distribution after rebalancing was 64:64, as illustrated in Fig. 4.
Fig. 4.
Distribution post rebalancing through the synthetic minority oversampling technique.
After rebalancing the distribution, the features identified through LASSO regression analysis, which exhibited non-zero coefficients, were further refined within the radiomics model conducting machine learning techniques. These refined features were then leveraged to construct robust radiomics models to identify exceptional classifiers within the database. The evaluation in this study encompassed four prominent training models: logistic regression (LR), classification and regression trees (CARTs), k-nearest neighbors (KNN), and support vector machines (SVMs). A comparison of these models involved the utilization of a confusion matrix and the calculation of performance metrics such as the area under the curve (AUC), accuracy, precision, and F1 score. The calculation formula for the F1 score is as follows, as delineated by equation (2):
| (2) |
Subsequently, a meticulous screening process was conducted, prioritizing the comparison of AUCs among diverse machine-learning approaches. In cases where the AUCs were similar, further comparison based on F1 scores was carried out to determine the optimal radiomic model.
2.8. Construction of the clinical and combined models
We initially conducted an LR analysis of the training cohort, considering both clinical and CT radiological features as predictor variables. Univariate logistic analysis was performed, followed by multivariate LR with a significance threshold of P < 0.1 to identify the optimal predictor factors for subsequent model development. Odds ratios (ORs) and their respective 95 % confidence intervals (CIs) were calculated for each factor. Following this, chosen predictors were employed to formulate a clinical model. A consolidated model was created by integrating a clinical model with an optimized radiomic model. The efficacy of these models was scrutinized and validated through receiver operating characteristic (ROC) curve analysis in both the training and test cohorts. Assessment metrics including the AUC, accuracy, sensitivity, and specificity were examined.
2.9. Statistical analysis
Statistical analyses were executed using R version 4.3.1 (R Core Team, Vienna, Austria). Regarding continuous variables, normally distributed data were assessed via Student's t-test, while skewed distributed data were assessed using the Mann–Whitney U test. The chi-squared test was used to evaluate categorical variables. Comparisons among models were performed utilizing the DeLong test alongside reclassification metrics (the net reclassification index [NRI] and integrated discrimination improvement [IDI]). Statistical significance was determined by a P value of <0.05.
3. Results
3.1. Baseline characteristics
This study included a total of 538 patients with SPN-type LADC with a tumor size ≤3 cm, among whom 50 patients (9.3 %) were diagnosed with IMA. Among these patients, 376 were assigned to the training cohort and 162 to the test cohort. Importantly, there were no notable differences in baseline characteristics between the two cohorts, as shown in Table 1.
Table 1.
Comparative analysis of baseline characteristics between the training and test cohorts.
| Variables | Total (n = 538) | Training cohort (n = 376) | Test cohort (n = 162) | p-value |
|---|---|---|---|---|
| Location, N (%) | 0.34 | |||
| RUL | 155 (28.8) | 109 (29) | 46 (28.4) | |
| RLL | 126 (23.4) | 81 (21.5) | 45 (27.8) | |
| RML | 43 (8) | 33 (8.8) | 10 (6.2) | |
| LUL | 126 (23.4) | 94 (25) | 32 (19.8) | |
| LLL | 88 (16.4) | 59 (15.7) | 29 (17.9) | |
| Boundary, N (%) | 0.988 | |||
| Ill-Defined | 121 (22.5) | 84 (22.3) | 37 (22.8) | |
| Well-Defined | 417 (77.5) | 292 (77.7) | 125 (77.2) | |
| Shape, N (%) | 0.832 | |||
| Irregular | 291 (54.1) | 205 (54.5) | 86 (53.1) | |
| Others | 247 (45.9) | 171 (45.5) | 76 (46.9) | |
| Lobulation, N (%) | 0.864 | |||
| Absence | 122 (22.7) | 84 (22.3) | 38 (23.5) | |
| Presence | 416 (77.3) | 292 (77.7) | 124 (76.5) | |
| Spiculation, N (%) | 0.697 | |||
| Absence | 154 (28.6) | 110 (29.3) | 44 (27.2) | |
| Presence | 384 (71.4) | 266 (70.7) | 118 (72.8) | |
| Vascular Convergence Sign, N (%) | 0.431 | |||
| Absence | 280 (52) | 191 (50.8) | 89 (54.9) | |
| Presence | 258 (48) | 185 (49.2) | 73 (45.1) | |
| Vacuole Sign, N (%) | 0.139 | |||
| Absence | 431 (80.1) | 308 (81.9) | 123 (75.9) | |
| Presence | 107 (19.9) | 68 (18.1) | 39 (24.1) | |
| Pleural Indentation, N (%) | 0.559 | |||
| Absence | 129 (24) | 87 (23.1) | 42 (25.9) | |
| Presence | 409 (76) | 289 (76.9) | 120 (74.1) | |
| Sex, N (%) | 0.558 | |||
| Male | 257 (47.8) | 176 (46.8) | 81 (50) | |
| Female | 281 (52.2) | 200 (53.2) | 81 (50) | |
| Age, Median (Q1, Q3) | 61 (54, 67) | 61 (54, 67) | 61 (54, 67.8) | 0.845 |
| Size, Median (Q1, Q3) | 20.6 (15, 26.6) | 20.8 (15, 26.2) | 20.4 (15.2, 27.8) | 0.5 |
| Group, n (%) | 0.429 | |||
| INMA | 488 (90.7) | 344 (91.5) | 144 (88.9) | |
| IMA | 50 (9.3) | 32 (8.5) | 18 (11.1) | |
Abbreviation: LUL Left Upper Lobe, LLL Left Lower Lobe, RUL Right Upper Lobe, RML Right Middle Lobe, RLL Right Lower Lobe, IMA Invasive Mucinous Adenocarcinoma, LADC Lung Adenocarcinoma, INMA invasive non-mucinous adenocarcinoma.
Table 2 provides a comprehensive overview of IMA and INMA characteristics in SPNs ≤3 cm. Across both the training and test cohorts, no significant demographic differences were noted concerning age or sex (all P > 0.05). Nevertheless, distinct disparities were evident in the CT radiological features (all P > 0.05). Notably, SPN-type IMAs exhibited a heightened prevalence of pleural indentation (P = 0.032), vascular convergence signs (P < 0.001), and larger tumor size (P < 0.001) compared to INMAs.
Table 2.
Comparative analysis of radiological features of IMA and INMA within the training and test cohorts.
| Variables | Training cohort |
Test cohort |
||||||
|---|---|---|---|---|---|---|---|---|
| Total (n = 376) | INMA (n = 344) | IMA (n = 32) | p-value | Total (n = 162) | INMA (n = 144) | IMA (n = 18) | p-value | |
| Location, N (%) | 0.878 | 0.504 | ||||||
| RUL | 109 (29) | 102 (29.7) | 7 (21.9) | 46 (28.4) | 39 (27.1) | 7 (38.9) | ||
| RLL | 81 (21.5) | 73 (21.2) | 8 (25) | 45 (27.8) | 39 (27.1) | 6 (33.3) | ||
| RML | 33 (8.8) | 30 (8.7) | 3 (9.4) | 10 (6.2) | 9 (6.2) | 1 (5.6) | ||
| LUL | 94 (25) | 86 (25) | 8 (25) | 32 (19.8) | 31 (21.5) | 1 (5.6) | ||
| LLL | 59 (15.7) | 53 (15.4) | 6 (18.8) | 29 (17.9) | 26 (18.1) | 3 (16.7) | ||
| Boundary, N (%) | 0.297 | 0.401 | ||||||
| Ill-defined | 84 (22.3) | 74 (21.5) | 10 (31.2) | 37 (22.8) | 27 (18.8) | 10 (55.6) | ||
| Well-defined | 292 (77.7) | 270 (78.5) | 22 (68.8) | 125 (77.2) | 117 (81.2) | 8 (44.4) | ||
| Shape, N (%) | 0.984 | 0.14 | ||||||
| Irregular | 205 (54.5) | 187 (54.4) | 18 (56.2) | 86 (53.1) | 73 (50.7) | 13 (72.2) | ||
| Others | 171 (45.5) | 157 (45.6) | 14 (43.8) | 76 (46.9) | 71 (49.3) | 5 (27.8) | ||
| Lobulation, N (%) | 0.24 | 0.075 | ||||||
| Absence | 84 (22.3) | 80 (23.3) | 4 (12.5) | 38 (23.5) | 37 (25.7) | 1 (5.6) | ||
| Presence | 292 (77.7) | 264 (76.7) | 28 (87.5) | 124 (76.5) | 107 (74.3) | 17 (94.4) | ||
| Spiculation, N (%) | 0.117 | 0.782 | ||||||
| Absence | 110 (29.3) | 105 (30.5) | 5 (15.6) | 44 (27.2) | 40 (27.8) | 4 (22.2) | ||
| Presence | 266 (70.7) | 239 (69.5) | 27 (84.4) | 118 (72.8) | 104 (72.2) | 14 (77.8) | ||
| Vascular Convergence Sign, N (%) | 0.001 | 0.019 | ||||||
| Absence | 191 (50.8) | 184 (53.5) | 7 (21.9) | 89 (54.9) | 83 (57.6) | 6 (33.3) | ||
| Presence | 185 (49.2) | 160 (46.5) | 25 (78.1) | 73 (45.1) | 61 (42.4) | 12 (66.7) | ||
| Vacuole Sign, N (%) | 0.89 | 0.144 | ||||||
| Absence | 308 (81.9) | 281 (81.7) | 27 (84.4) | 123 (75.9) | 112 (77.8) | 11 (61.1) | ||
| Presence | 68 (18.1) | 63 (18.3) | 5 (15.6) | 39 (24.1) | 32 (22.2) | 7 (38.9) | ||
| Pleural Indentation, N (%) | 0.032 | 0.048 | ||||||
| Absence | 87 (23.1) | 85 (24.7) | 2 (6.2) | 42 (25.9) | 39 (27.1) | 3 (16.7) | ||
| Presence | 289 (76.9) | 259 (75.3) | 30 (93.8) | 120 (74.1) | 105 (72.9) | 15 (83.3) | ||
| Sex, N (%) | 1 | 0.803 | ||||||
| Absence | 176 (46.8) | 161 (46.8) | 15 (46.9) | 81 (50) | 71 (49.3) | 10 (55.6) | ||
| Presence | 200 (53.2) | 183 (53.2) | 17 (53.1) | 81 (50) | 73 (50.7) | 8 (44.4) | ||
| Age, Median (Q1, Q3) | 61 (54, 67) | 61 (54, 67) | 59 (52.5, 65.2) | 0.285 | 61 (54, 67.8) | 60 (53.8, 67) | 64 (56.8, 71) | 0.191 |
| Size, Median (Q1, Q3) | 20.8 (15, 26.2) | 20.2 (14.6, 25.6) | 23.5 (21.1, 30) | <0.001 | 20.4 (15.2, 27.8) | 19.5 (15, 25.9) | 29 (22.9, 30) | <0.001 |
Abbreviation: LUL Left Upper Lobe, LLL Left Lower Lobe, RUL Right Upper Lobe, RML Right Middle Lobe, RLL Right Lower Lobe, IMA Invasive Mucinous Adenocarcinoma, LADC Lung Adenocarcinoma, INMA invasive non-mucinous adenocarcinoma.
3.2. Clinical model
Univariate and multivariate LR analyses identified CT radiological features as independent risk factors, including the vascular convergence sign (OR, 1.097; 95 % CI, 1.038–1.159; P = 0.001) and size (OR, 1.006; 95 % CI, 1.002–1.011; P = 0.003). These factors were used to develop the clinical model (Table 3).
Table 3.
Univariate and Multivariate Logistic Analysis Differentiating IMA from INMA in SPNs ≤3 cm.
| Variables | Odd Ratio (95%CI) |
p-value |
Odd Ratio (95%CI) |
p-value |
|---|---|---|---|---|
| Univariate analysis | Multivariate analysis | |||
| Location | 1.6 (0.55–4.6) | 0.39 | ||
| Boundary | 0.6 (0.27–1.3) | 0.21 | ||
| Shape | 0.93 (0.45–1.9) | 0.84 | ||
| Lobulation | 2.1 (0.72–6.2) | 0.17 | ||
| Spiculation | 2.4 (0.89–6.3) | 0.084 | ||
| Vascular convergence sign | 4.1 (1.7–9.7) | 0.0014 | 1.097(1.038–1.159) | 0.001 |
| Vacuole sign | 0.83 (0.31–2.2) | 0.71 | ||
| Pleural indentation | 4.9 (1.2–21) | 0.031 | 1.054(0.986–1.127) | 0.121 |
| Sex | 1 (0.48–2.1) | 0.99 | ||
| Age | 0.98 (0.94–1) | 0.18 | ||
| Size | 1.1 (1–1.2) | 0.001 | 1.006(1.002–1.011) | 0.003 |
Abbreviation: IMA invasive mucinous adenocarcinoma, LADC lung adenocarcinoma, SPN solitary pulmonary nodules. INMA invasive non-mucinous adenocarcinoma.
The clinical model integrating the key risk factors attained an AUC of 0.760 (95 % CI: 0.677–0.843) and an F1 score of 0.256 in the training cohort, and an AUC of 0.777 (95 % CI: 0.695–0.860) and an F1 score of 0.4 in the test cohort.
3.3. Radiomic model
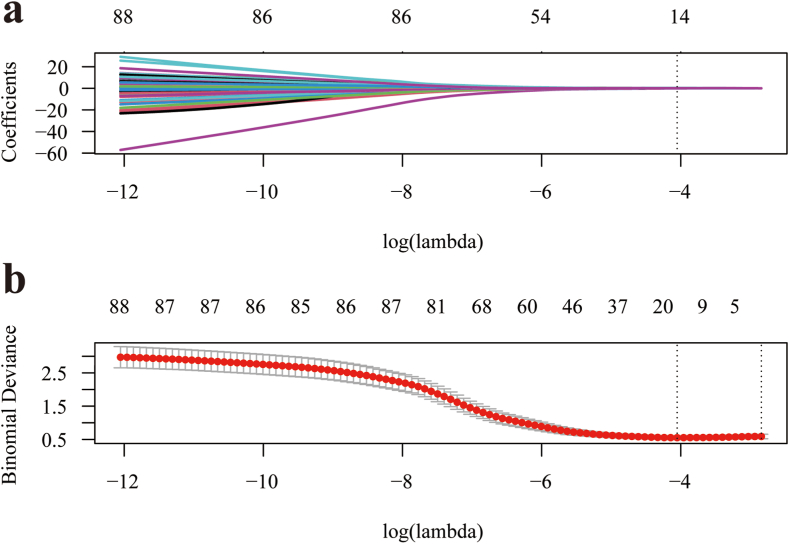
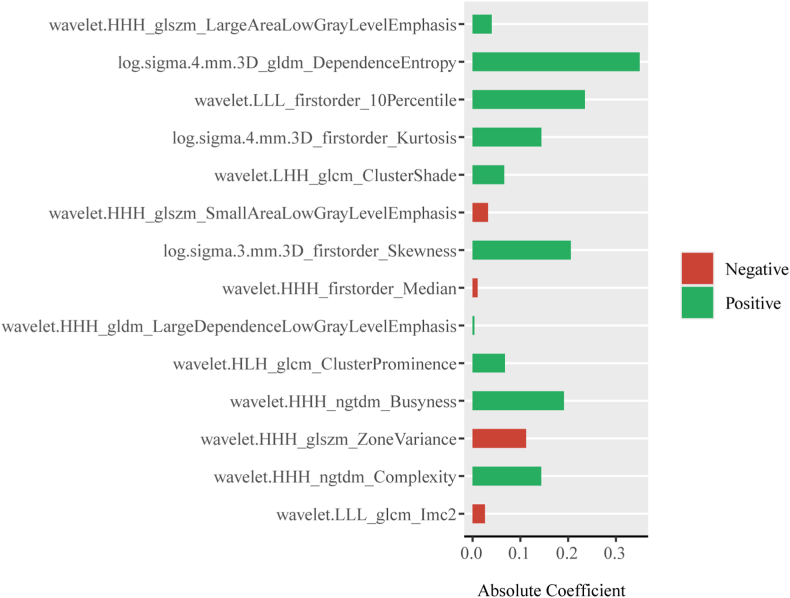
To construct the final model, an optimized subset of features was selected using LASSO regression using the mRMR method to eliminate redundant and irrelevant features. To determine the optimal hyperparameter λ, a 10-fold cross-validation process was conducted, as shown in Fig. 5(a b). The optimal value of λ was found to be 0.017, resulting in the selection of 14 features for further analysis, as illustrated in Fig. 6.
Fig. 5.
Utilization of LASSO regression (a) and 10-fold cross-validation (b) to facilitate the selection of radiomic features. LASSO, least absolute shrinkage and selection operator.
Fig. 6.
Radiomic features selected based on the outcomes of LASSO regression. LASSO, least absolute shrinkage and selection operator.
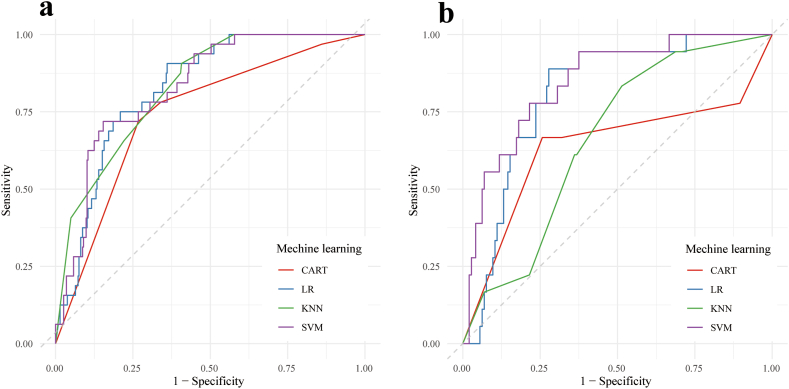
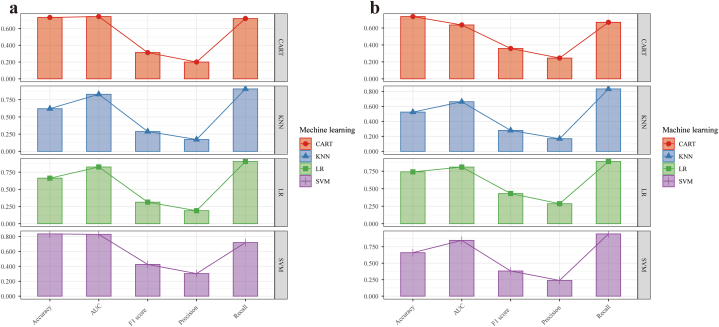
Fig. 7, Fig. 8 show the ROC curves and visualization plots of the AUC, accuracy, precision, and F1 score of the four radiomics models (SVM, KNN, CART, and LR) in both the training and test cohorts. From this analysis, it was evident that the SVM model showcased superior overall performance, boasting an AUC of 0.829 (95 % CI: 0.766–0.891) and an F1 score of 0.426 in the training cohort, as well as an AUC of 0.846 (95 % CI: 0.759–0.934) and an F1 score of 0.382 in the test cohort (Table 4).
Fig. 7.
ROC curve analysis performed on the radiomic model derived from various machine-learning models to differentiate between IMA and INMA in SPNs ≤3 cm within both the training cohort (a) and the test cohort (b). ROC, receiver operating characteristic; IMA, invasive mucinous adenocarcinoma; INMA, invasive non-mucinous adenocarcinoma; SPN, solitary pulmonary nodule.
Fig. 8.
Visual representations generated to demonstrate the accuracy, AUC, F1 score, precision, and recall values corresponding to various machine learning models within both the training cohort (a) and the test cohort (b). AUC, area under the receiver operating characteristic curve.
Table 4.
Diagnostic Performance of Various Machine Learning Models in Distinguishing IMA from INMA in SPNs ≤3 cm.
| Model | Cohorts | AUC | Accuracy | Precision | Recall | F1 score |
|---|---|---|---|---|---|---|
| SVM | Training | 0.829 | 0.835 | 0.303 | 0.719 | 0.426 |
| Test | 0.846 | 0.660 | 0.239 | 0.944 | 0.382 | |
| CART | Training | 0.743 | 0.732 | 0.200 | 0.719 | 0.313 |
| Test | 0.636 | 0.735 | 0.245 | 0.667 | 0.358 | |
| KNN | Training | 0.829 | 0.620 | 0.172 | 0.906 | 0.288 |
| Test | 0.662 | 0.525 | 0.168 | 0.833 | 0.280 | |
| LR | Training | 0.826 | 0.663 | 0.190 | 0.906 | 0.314 |
| Test | 0.809 | 0.741 | 0.286 | 0.889 | 0.432 |
Abbreviation: IMA invasive mucinous adenocarcinoma, LADC lung adenocarcinoma, AUC area under the curve, INMA invasive non-mucinous adenocarcinoma, SVM support vector machine, CART classification and regression trees, KNN k-nearest neighbors, LR logistic regression, SPN solitary pulmonary nodule.
3.4. Combined model construction and validation
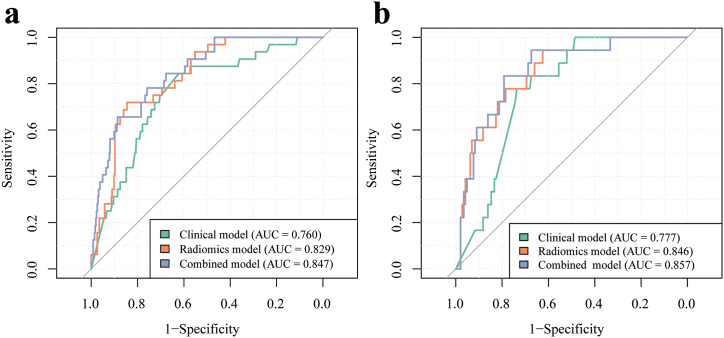
The integration of clinical and SVM-derived radiomic models in the combined model enhanced the comprehensiveness of diagnostic assessment and improved the overall efficacy. Fig. 9(a b) displays the ROC curves of the three models (clinical, radiomic, and combined) for both the training and test cohorts. The combined model demonstrated a significantly improved discriminative capacity, providing a more comprehensive evaluation compared to both the clinical and radiomic models. This enhancement was supported by an AUC of 0.847 (95 % CI: 0.785–0.909) and an F1 score of 0.457 for the training cohort, and an AUC of 0.857 (95 % CI: 0.773–0.940) and an F1 score of 0.476 for the test cohort, as outlined in Table 5. In comparison to the radiomic model, the combined model notably enhanced the holistic assessment in both the training cohort (Delong test, P = 0.045; NRI of 0.206, IDI of 0.024) and the test cohort (Delong test, P = 0.029; NRI of 0.125, IDI of 0.032). Similarly, it outperformed the performance of the clinical model in the training cohort (Delong test, P = 0.01; NRI of 0.310, IDI of 0.09) and the test cohort (Delong test, P = 0.047; NRI of 0.382, IDI of 0.085).
Fig. 9.
ROC curve analysis conducted on radiomic, clinical, and combined models for discriminating between IMA and INMA in SPNs ≤3 cm within both the training cohort (a) and the testing cohort (b). ROC, receiver operating characteristic; IMA, invasive mucinous adenocarcinoma; INMA, invasive non-mucinous adenocarcinoma; SPN, solitary pulmonary nodule.
Table 5.
Diagnostic Performance of Diverse Models in Discriminating Between IMA and INMA in SPNs ≤3 cm.
| Model | Cohorts | AUC | Accuracy | Sensitivity | Specificity | Precision | Recall | F1 |
|---|---|---|---|---|---|---|---|---|
| Clinical | Training | 0.760 | 0.641 | 0.844 | 0.622 | 0.172 | 0.844 | 0.256 |
| Test | 0.777 | 0.741 | 0.778 | 0.736 | 0.269 | 0.778 | 0.4 | |
| Radiomicsa | Training | 0.829 | 0.835 | 0.719 | 0.846 | 0.303 | 0.719 | 0.426 |
| Test | 0.846 | 0.660 | 0.944 | 0.625 | 0.239 | 0.944 | 0.382 | |
| Combined | Training | 0.847 | 0.867 | 0.656 | 0.887 | 0.351 | 0.656 | 0.457 |
| Test | 0.857 | 0.796 | 0.833 | 0.792 | 0.334 | 0.833 | 0.476 |
Abbreviation: SPN solitary pulmonary nodules, IMA invasive mucinous adenocarcinoma, LADC lung adenocarcinoma, AUC area under the curve, INMA invasive non-mucinous adenocarcinomas.
Represents models were constructed using support vector machine (SVM).
4. Discussion
In this study, the SVM-derived radiomic approach demonstrated superior performance compared with that of other machine-learning models when classifying SPN-type IMA and INMA among SPNs ≤3 cm. To further enhance the overall diagnostic efficacy, we developed a combined model, integrating the clinical model with the SVM-derived radiomic model. This combined model successfully achieved satisfactory classification outcomes for SPN-type IMA and INMA, presenting a viable, reliable, and non-invasive approach to evaluating management strategies and treatment options in patients with SPN-type LADC tumors ≤3 cm in size.
The predominant presentation of invasive mucinous adenocarcinoma on computed tomography imaging is commonly identified as a SPN, with ground-glass opacity nodules being less frequently encountered [3]. The distinction in presentation is likely linked to histological characteristics, which include the presence of tumor cells rich in mucin, fibrosis, and central fibrosis with mucin-filled alveolar spaces, resulting in increased CT density [3]. Recent research has reported a higher prevalence of SPN-type IMA compared to pneumonia-type IMA [9,[19], [20], [21]]. However, a comprehensive investigation into the ability of preoperative CT radiological features to differentiate between IMA and INMA in SPNs ≤3 cm is lacking. In our study, multivariate logistic analysis identified independent risk factors for distinguishing between IMA and INMA, encompassing CT radiological features such as the vascular convergence sign and tumor size. These discoveries align with earlier studies [22,29,30], indicating that the seepage of mucus from the nodule's edge and the movement of macrophages along alveolar walls contribute to vascular convergence. However, the clinical model based on CT radiological features exhibited only moderate diagnostic effectiveness, yielding an AUC of 0.760 for the training cohort and 0.770 for the test cohort. This constraint could be attributed to the subjective interpretation of radiological features, lacking consistency, typicality, and generalizability, especially in early-stage SPNs (≤3 cm), where atypical presentations are more common.
Radiomics is an advanced non-invasive approach that uses intelligent algorithms to build models from original medical images, capturing tumor heterogeneity to reveal additional insights [23]. To enhance the stability and generalization of the radiomic model, we incorporated four machine-learning models: KNN, LR, CART, and SVM. Our findings underscored the superior performance of the SVM-derived radiomic model compared with that of other machine-learning approaches. For these machine-learning approaches, LR is utilized in binary classification tasks by estimating the probability of a binary outcome based on predictor variables. CART, a decision tree type, partitions data into subsets recursively considering predictor variables’ values, forming a tree-like structure suited for predictions, particularly in handling non-linear relationships. KNN, recognized for its simplicity, predicts datapoint classification by majority voting of the K-nearest neighbors and is especially effective in non-linear separable data scenarios. SVM, a robust algorithm for both linear and non-linear tasks through dimensional mapping, excels in maximizing class margins, notably in high-dimensional spaces, making it a powerful supervised learning model for classification and regression tasks. Employing the kernel trick, SVM efficiently captures and optimizes complex relationships owing to its strong mathematical foundation. The resilience of SVM against overfitting demonstrates comparable performance to models trained on larger datasets, particularly in high-dimensional and complex data distributions of radiomic features [31,32]. In this study, utilizing SMOTE for resampling addressed the disproportionate distribution between INMA and IMA cases, highlighting the superiority of SVM due to its ability to address overfitting issues effectively even with a constrained sample size.
Ultimately, a combined model was devised by amalgamating vital data from both clinical and radiomic models, showcasing superior efficacy in contrast to employing solely clinical or radiomic models. Our premise rested on the notion that the fusion of diverse and plentiful information within the composite model would heighten diagnostic proficiency in distinguishing between IMA and INMAs. This composite approach leverages integrated insights from various modalities, presenting a holistic perspective for improved diagnostic accuracy.
This study has several limitations. Primarily, we must acknowledge the retrospective nature of our investigation, constrained by the relative scarcity of IMA cases, thus leading to a low incidence rate. There may exist potential selection biases due to the exclusive inclusion of patients with postoperative pathological results. Furthermore, our focus solely on IMA and INMA in SPNs ≤3 cm might restrict the generalizability of our conclusions to different disease stages. The integration of data from two centers introduced variability in acquisition parameters, image quality, and possible co-registration errors, potentially confounding the analysis. Additionally, artifacts and technical limitations might have influenced the reliability and reproducibility of radiomic features. Lastly, the relatively short postoperative follow-up period precluded the development of predictive models for disease outcomes.
In conclusion, our investigation showcased the enhanced diagnostic accuracy of SVM within a radiomic framework, surpassing alternative machine-learning techniques. The amalgamated model, merging clinical insights with radiomic features, offers a holistic strategy for distinguishing between IMA and INMA within SPNs of ≤3 cm. Such an approach holds promise in refining clinical management, predicting patient outcomes, and guiding treatment strategies, thereby aligning with the ethos of precision medicine to optimize personalized and effective healthcare interventions.
Ethics approval and consent to participate
This research upheld the guidelines set forth in the Declaration of Helsinki and obtained authorization from the Ethics Committees of the involved hospitals (with reference numbers: KY2020147 from The Affiliated Hospital of Southwest Medical University and 2021-07-009 from Xiangtan Central Hospital). Due to the retrospective nature of this investigation, the necessity for obtaining informed consent was waived.
Data availability statement
The processed data necessary for replicating these discoveries can be accessed via the following link: https://github.com/online-calculator/Pulmonary-Solid-Nodules-3-cm.
Funding
This research was supported by the Southwest Medical University Project (No. 2021ZKQN102).
CRediT authorship contribution statement
Zhengyuan Xiao: Conceptualization. Jing Chen: Data curation. Xiaolan Feng: Conceptualization. Yinjun Zhou: Conceptualization. Haibo Liu: Conceptualization. Guidong Dai: Supervision, Conceptualization. Wanyin Qi: Writing – review & editing, Writing – original draft, Supervision, Software, Formal analysis, Data curation, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We wish to express our sincere thanks to the research participants for their dedication, which laid the foundation for this study. We also appreciate Editage for their expert editing, which helped to enhance the quality of the manuscript. We are grateful for the combined support that contributed to the success of this paper. Thank you to all involved.
Contributor Information
Guidong Dai, Email: daiguidong2002@163.com.
Wanyin Qi, Email: qiwanyin0508@163.com.
References
- 1.Marx A., Chan J.K.C., Chalabreysse L., Dacic S., Detterbeck F., French C.A., Hornick J.L., Inagaki H., Jain D., Lazar A.J., Marino M., Marom E.M., Moreira A.L., Nicholson A.G., Noguchi M., Nonaka D., Papotti M.G., Porubsky S., Sholl L.M., Tateyama H., Thomas de Montpréville V., Travis W.D., Rajan A., Roden A.C., Ströbel P. The 2021 WHO classification of tumors of the thymus and mediastinum: what is new in thymic epithelial, germ cell, and mesenchymal tumors? J. Thorac. Oncol. 2022;17:200–213. doi: 10.1016/j.jtho.2021.10.010. [DOI] [PubMed] [Google Scholar]
- 2.Travis W.D., Brambilla E., Noguchi M., Nicholson A.G., Geisinger K.R., Yatabe Y., Beer D.G., Powell C.A., Riely G.J., Van Schil P.E., Garg K., Austin J.H., Asamura H., Rusch V.W., Hirsch F.R., Scagliotti G., Mitsudomi T., Huber R.M., Ishikawa Y., Jett J., Sanchez-Cespedes M., Sculier J.P., Takahashi T., Tsuboi M., Vansteenkiste J., Wistuba I., Yang P.C., Aberle D., Brambilla C., Flieder D., Franklin W., Gazdar A., Gould M., Hasleton P., Henderson D., Johnson B., Johnson D., Kerr K., Kuriyama K., Lee J.S., Miller V.A., Petersen I., Roggli V., Rosell R., Saijo N., Thunnissen E., Tsao M., Yankelewitz D. International association for the study of lung cancer/american thoracic society/european respiratory society international multidisciplinary classification of lung adenocarcinoma. J. Thorac. Oncol. 2011;6:244–285. doi: 10.1097/JTO.0b013e318206a221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang W.C., Zhang Y.Z., Nicholson A.G. Pulmonary invasive mucinous adenocarcinoma. Histopathology. 2024;84:18–31. doi: 10.1111/his.15064. [DOI] [PubMed] [Google Scholar]
- 4.Yoshizawa A., Motoi N., Riely G.J., Sima C.S., Gerald W.L., Kris M.G., Park B.J., Rusch V.W., Travis W.D. Impact of proposed IASLC/ATS/ERS classification of lung adenocarcinoma: prognostic subgroups and implications for further revision of staging based on analysis of 514 stage I cases. Mod. Pathol. 2011;24:653–664. doi: 10.1038/modpathol.2010.232. [DOI] [PubMed] [Google Scholar]
- 5.Russell P.A., Wainer Z., Wright G.M., Daniels M., Conron M., Williams R.A. Does lung adenocarcinoma subtype predict patient survival?: a clinicopathologic study based on the new International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society international multidisciplinary lung adenocarcinoma classification. J. Thorac. Oncol. 2011;6:1496–1504. doi: 10.1097/JTO.0b013e318221f701. [DOI] [PubMed] [Google Scholar]
- 6.Warth A., Muley T., Meister M., Stenzinger A., Thomas M., Schirmacher P., Schnabel P.A., Budczies J., Hoffmann H., Weichert W. The novel histologic International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society classification system of lung adenocarcinoma is a stage-independent predictor of survival. J. Clin. Oncol. 2012;30:1438–1446. doi: 10.1200/jco.2011.37.2185. [DOI] [PubMed] [Google Scholar]
- 7.Xu X., Shen W., Wang D., Li N., Huang Z., Sheng J., Rucker A.J., Mao W., Xu H., Cheng G. Clinical features and prognosis of resectable pulmonary primary invasive mucinous adenocarcinoma. Transl. Lung Cancer Res. 2022;11:420–431. doi: 10.21037/tlcr-22-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matsui T., Sakakura N., Koyama S., Nakanishi K., Sasaki E., Kato S., Hosoda W., Murakami Y., Kuroda H., Yatabe Y. Comparison of surgical outcomes between invasive mucinous and non-mucinous lung adenocarcinoma. Ann. Thorac. Surg. 2021;112:1118–1126. doi: 10.1016/j.athoracsur.2020.09.042. [DOI] [PubMed] [Google Scholar]
- 9.Lee H.Y., Cha M.J., Lee K.S., Lee H.Y., Kwon O.J., Choi J.Y., Kim H.K., Choi Y.S., Kim J., Shim Y.M. Prognosis in resected invasive mucinous adenocarcinomas of the lung: related factors and comparison with resected nonmucinous adenocarcinomas. J. Thorac. Oncol. 2016;11:1064–1073. doi: 10.1016/j.jtho.2016.03.011. [DOI] [PubMed] [Google Scholar]
- 10.Duruisseaux M., Antoine M., Rabbe N., Poulot V., Fleury-Feith J., Vieira T., Lavolé A., Cadranel J., Wislez M. The impact of intracytoplasmic mucin in lung adenocarcinoma with pneumonic radiological presentation. Lung Cancer. 2014;83:334–340. doi: 10.1016/j.lungcan.2013.12.016. [DOI] [PubMed] [Google Scholar]
- 11.Ichinokawa H., Ishii G., Nagai K., Yoshida J., Nishimura M., Hishida T., Suzuki K., Ochiai A. Clinicopathological characteristics of primary lung adenocarcinoma predominantly composed of goblet cells in surgically resected cases. Pathol. Int. 2011;61:423–429. doi: 10.1111/j.1440-1827.2011.02677.x. [DOI] [PubMed] [Google Scholar]
- 12.Casali C., Rossi G., Marchioni A., Sartori G., Maselli F., Longo L., Tallarico E., Morandi U. A single institution-based retrospective study of surgically treated bronchioloalveolar adenocarcinoma of the lung: clinicopathologic analysis, molecular features, and possible pitfalls in routine practice. J. Thorac. Oncol. 2010;5:830–836. doi: 10.1097/jto.0b013e3181d60ff5. [DOI] [PubMed] [Google Scholar]
- 13.Shim H.S., Kenudson M., Zheng Z., Liebers M., Cha Y.J., Hoang Ho Q., Onozato M., Phi Le L., Heist R.S., Iafrate A.J. Unique genetic and survival characteristics of invasive mucinous adenocarcinoma of the lung. J. Thorac. Oncol. 2015;10:1156–1162. doi: 10.1097/jto.0000000000000579. [DOI] [PubMed] [Google Scholar]
- 14.Kadota K., Yeh Y.C., D'Angelo S.P., Moreira A.L., Kuk D., Sima C.S., Riely G.J., Arcila M.E., Kris M.G., Rusch V.W., Adusumilli P.S., Travis W.D. Associations between mutations and histologic patterns of mucin in lung adenocarcinoma: invasive mucinous pattern and extracellular mucin are associated with KRAS mutation. Am. J. Surg. Pathol. 2014;38:1118–1127. doi: 10.1097/pas.0000000000000246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo M., Tomoshige K., Meister M., Muley T., Fukazawa T., Tsuchiya T., Karns R., Warth A., Fink-Baldauf I.M., Nagayasu T., Naomoto Y., Xu Y., Mall M.A., Maeda Y. Gene signature driving invasive mucinous adenocarcinoma of the lung. EMBO Mol. Med. 2017;9:462–481. doi: 10.15252/emmm.201606711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shang G., Jin Y., Zheng Q., Shen X., Yang M., Li Y., Zhang L. Histology and oncogenic driver alterations of lung adenocarcinoma in Chinese. Am. J. Cancer Res. 2019;9:1212–1223. [PMC free article] [PubMed] [Google Scholar]
- 17.Travis W.D., Brambilla E., Nicholson A.G., Yatabe Y., Austin J.H.M., Beasley M.B., Chirieac L.R., Dacic S., Duhig E., Flieder D.B., Geisinger K., Hirsch F.R., Ishikawa Y., Kerr K.M., Noguchi M., Pelosi G., Powell C.A., Tsao M.S., Wistuba I. The 2015 World Health organization classification of lung tumors: impact of genetic, clinical and radiologic advances since the 2004 classification. J. Thorac. Oncol. 2015;10:1243–1260. doi: 10.1097/jto.0000000000000630. [DOI] [PubMed] [Google Scholar]
- 18.Beck K.S., Sung Y.E., Lee K.Y., Han D.H. Invasive mucinous adenocarcinoma of the lung: serial CT findings, clinical features, and treatment and survival outcomes. Thorac Cancer. 2020;11:3463–3472. doi: 10.1111/1759-7714.13674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nie K., Nie W., Zhang Y.X., Yu H. Comparing clinicopathological features and prognosis of primary pulmonary invasive mucinous adenocarcinoma based on computed tomography findings. Cancer Imag. 2019;19:47. doi: 10.1186/s40644-019-0236-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang T., Yang Y., Liu X., Deng J., Wu J., Hou L., Wu C., She Y., Sun X., Xie D., Chen C. Primary invasive mucinous adenocarcinoma of the lung: prognostic value of CT imaging features combined with clinical factors. Korean J. Radiol. 2021;22:652–662. doi: 10.3348/kjr.2020.0454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Watanabe H., Saito H., Yokose T., Sakuma Y., Murakami S., Kondo T., Oshita F., Ito H., Nakayama H., Yamada K., Iwazaki M. Relation between thin-section computed tomography and clinical findings of mucinous adenocarcinoma. Ann. Thorac. Surg. 2015;99:975–981. doi: 10.1016/j.athoracsur.2014.10.065. [DOI] [PubMed] [Google Scholar]
- 22.Cha M.J., Lee K.S., Kim T.J., Kim H.S., Kim T.S., Chung M.J., Kim B.T., Kim Y.S. Solitary nodular invasive mucinous adenocarcinoma of the lung: imaging diagnosis using the morphologic-metabolic dissociation sign. Korean J. Radiol. 2019;20:513–521. doi: 10.3348/kjr.2018.0409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tunali I., Gillies R.J., Schabath M.B. Application of radiomics and artificial intelligence for lung cancer precision medicine. Cold Spring Harb Perspect Med. 2021;11 doi: 10.1101/cshperspect.a039537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu X., Zhang S., Xu J., Huang Y., Luo H., Huang C., Nie P., Deng Y., Mao N., Zhang R., Gao L., Li S., Kang B., Wang X. Nomogram using CT radiomics features for differentiation of pneumonia-type invasive mucinous adenocarcinoma and pneumonia: multicenter development and external validation study. AJR Am. J. Roentgenol. 2023;220:224–234. doi: 10.2214/ajr.22.28139. [DOI] [PubMed] [Google Scholar]
- 25.Zhang J., Hao L., Qi M., Xu Q., Zhang N., Feng H., Shi G. Radiomics nomogram for preoperative differentiation of pulmonary mucinous adenocarcinoma from tuberculoma in solitary pulmonary solid nodules. BMC Cancer. 2023;23:261. doi: 10.1186/s12885-023-10734-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parmar C., Grossmann P., Bussink J., Lambin P., Aerts H. Machine learning methods for quantitative radiomic biomarkers. Sci. Rep. 2015;5 doi: 10.1038/srep13087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.MacMahon H., Naidich D.P., Goo J.M., Lee K.S., Leung A.N.C., Mayo J.R., Mehta A.C., Ohno Y., Powell C.A., Prokop M., Rubin G.D., Schaefer-Prokop C.M., Travis W.D., Van Schil P.E., Bankier A.A. Guidelines for management of incidental pulmonary nodules detected on CT images: from the fleischner society 2017. Radiology. 2017;284:228–243. doi: 10.1148/radiol.2017161659. [DOI] [PubMed] [Google Scholar]
- 28.Lv J., Chen X., Liu X., Du D., Lv W., Lu L., Wu H. Imbalanced data correction based PET/CT radiomics model for predicting lymph node metastasis in clinical stage T1 lung adenocarcinoma. Front. Oncol. 2022;12 doi: 10.3389/fonc.2022.788968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu J., Zhang T., Pan J., Zhang Q., Lin X., Chang L., Chen Y.C., Xue X. Characteristics of the computed tomography imaging findings in 72 patients with airway-invasive pulmonary aspergillosis. Med Sci Monit. 2021;27 doi: 10.12659/msm.931162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang X., Qiao W., Kang Z., Pan C., Chen Y., Li K., Shen W., Zhang L. CT features of stage IA invasive mucinous adenocarcinoma of the lung and establishment of a prediction model. Int. J. Gen. Med. 2022;15:5455–5463. doi: 10.2147/ijgm.S368344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sanz H., Valim C., Vegas E., Oller J.M., Reverter F. SVM-RFE: selection and visualization of the most relevant features through non-linear kernels. BMC Bioinf. 2018;19:432. doi: 10.1186/s12859-018-2451-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma X., Qian X., Wang Q., Zhang Y., Zong R., Zhang J., Qian B., Yang C., Lu X., Shi Y. Radiomics nomogram based on optimal VOI of multi-sequence MRI for predicting microvascular invasion in intrahepatic cholangiocarcinoma. Radiol. Med. 2023;128:1296–1309. doi: 10.1007/s11547-023-01704-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The processed data necessary for replicating these discoveries can be accessed via the following link: https://github.com/online-calculator/Pulmonary-Solid-Nodules-3-cm.