Abstract
Ash1 (for asymmetric synthesis of HO) was first uncovered in genetic screens that revealed its role in mating-type switching. Ash1 prevents HO expression in daughter cells. Because Ash1 has a zinc finger-like domain related to that of the GATA family of transcription factors, it presumably acts by repressing HO transcription. Nonswitching diploid cells also express Ash1, suggesting it could have functions in addition to regulation of HO expression. We show here that Ash1 has an essential function for pseudohyphal growth. Our epistasis analyses are consistent with the deduction that Ash1 acts separately from the mitogen-activated protein kinase cascade and Ste12. Similarly to the case in yeast form cells, Ash1 is asymmetrically localized to the nuclei of daughter cells during pseudohyphal growth. This asymmetric localization reveals that there is a previously unsuspected daughter cell-specific function necessary for pseudohyphal growth.
The developmental process requires that parent cells assign distinctive fates to progeny. This capacity to differentiate often hinges on the action of a few specific molecules. Mating-type switching of haploid Saccharomyces cerevisiae is an example of such a phenomenon. While mother cells switch mating type, daughter cells are unable to switch because transcription of the HO endonuclease is repressed (23). Unlike a mother cell, a daughter cell expresses Ash1 (for asymmetric synthesis of HO), which, by an unknown mechanism, blocks HO expression (4, 33). This asymmetric distribution of Ash1, therefore, is critical to the different fates of mother and daughter cells. Interestingly, Ash1 asymmetry is preserved in nonswitching a/α diploids, leading to speculation that there may be other developmental processes under Ash1 control (33).
One developmental process that exists in both the haploid and diploid cell types of S. cerevisiae is filamentous growth. Haploid yeast cells respond to long incubations on rich medium by growing into the agar substrate in a process termed invasive growth (27). Such growth is characterized by a slight elongation of cells, along with a switch from an axial to a bipolar budding pattern. In the diploid pseudohyphal response, nitrogen starvation cues a more dramatic transition resulting in changes in cell shape, cell separation, agar invasion, and cell cycle (14). Perhaps the most striking change is in morphogenesis, as cells become highly elongated (12, 15). Unlike yeast form cells, these elongated cells remain attached after the cell cycle is complete, showing incomplete cell separation (12). The chains of elongated cells are competent to grow invasively into the agar surface, like their haploid counterparts (12). Finally, pseudohyphal cells exhibit a unique cell cycle in which the G1 delay before Start is largely eliminated and the G2 phase is significantly lengthened (15). The macroscopic result of these changes is a colony of cells with multiple projections radiating away from the bulk of cells (12).
Identification of the molecular components of filamentous growth is currently under way. Indeed, several components have been implicated in mediating the pseudohyphal response to nutritional deprivation. These include Ste20 (a PAK family member), the enzymes of the mitogen-activated protein kinase (MAPK) activation cascade (Ste11 and Ste7), and the Ste12 transcription factor and its negative regulators, Rst1/Dig1 and Rst2/Dig2 (6, 18, 27, 34). While deletion mutants lacking Ste12, Ste11, or Ste7 still form pseudohyphal filaments if they express activated variants of Ras2 (Ras2-V19) or Cdc42 (Cdc42-V12), a sterile 20 deletion mutant expressing Ras2-V19 or Cdc42-V12 does not (21, 22, 28). These results have led to the postulation of a branch in the pathway emerging at the level of Ste20 or possibly a parallel pathway to which Ste20 contributes (28).
This report demonstrates an essential function for Ash1 in the pseudohyphal-growth response. Epistasis experiments suggest that Ash1 does not operate directly upon the MAPK activation cascade or the transcriptional regulators that are downstream of the cascade. Interestingly, deletion of both Ste12 and Ash1 is required to block pseudohyphal-filament formation stimulated by a constitutively activated Ras2 variant. Therefore, it appears that both Ash1 and Ste12 function after Ras2 but on separate arms of a branched pathway. Further, we show that Ash1 maintains its asymmetric localization to daughters as cells undergo pseudohyphal growth. A mechanistic implication of this behavior is that the pseudohyphal-growth process requires a key daughter cell-specific function.
MATERIALS AND METHODS
Plasmids.
Several deletion alleles were used to construct various strains for the studies described here. pNC409 carries an allele that is a deletion of the entire coding region of ASH1. The ash1-Δ1 allele was constructed by using pNC543, a clone that we isolated from the pYES-R genomic overexpression library. This plasmid contains the complete ASH1 coding region under control of the GAL promoter in the pYES-R (CEN4 URA3) vector (9). A 100-bp fragment from the ASH1 5′ untranslated region was amplified by PCR with pNC543 as the template and synthetic oligonucleotides 196 (5′-GCGCACATGCATGCCAAAATTCTATCTT) and 197 (5′-GCTCTAGATTTTCCTTTTCCCGT) as primers. A 500-bp fragment of the ASH1 3′ untranslated region was similarly amplified by using oligonucleotides 204 (5′-CGCGCGGATCCAATTGTACATT) and 205 (5′-CGCCGGAATTCGTAGAATTAGA) as primers. The two PCR-amplified fragments were inserted at the respective XbaI-SphI and EcoRI-BamHI cloning sites in pNC343 (5). These two cloning sites in pNC343 flank the hisG-URA3-hisG cassette. This cassette allows for Ura selection of the deletion allele in gene replacement manipulations and subsequent deletion of the URA3 marker by selecting for recombination events that occur between the repeated hisG sequences (1). pNC527 contains the leu2-Δ1 fragment from p307 inserted as a SalI fragment into the SalI site of the integrating vector pRS306 (URA3 selectable marker) (32). The ste12Δ::LEU2 allele from pSUL16 has been described previously (8).
A fusion of Ash1 to green fluorescence protein (GFP) was constructed to monitor localization of Ash1. pKS+GFP contains a 700-bp cassette encoding GFP in the vector pBluescript KS+ (7). This GFP sequence was fused to ASH1 via the annealed oligonucleotides 271 (5′-GATCCTGGGGACTTCCCAGGAGATTTACCGCGGCCGCCG) and 272 (5′-GATCCGGCGGCCGCGGTATAATCTCCTGGGAAGTCCCCAG), which encode the polypeptide linker GGRGKSPGKSP. The oligonucleotide linker first was inserted into the BamHI site that is immediately after the ATG of ASH1 in pAS163. pAS163 is the 2μm URA3 vector, pRS426, with an epitope-tagged allele of ASH1 (32, 33). This allele has three tandem copies of the Myc epitope-coding sequence inserted immediately after the ATG initiation codon of ASH1. The linker insertion that we made at the BamHI site of pAS163 replaces the Myc epitope sequences. The NotI fragment of pKS+GFP was then inserted between the ATG codon and the segment encoding the polypeptide linker. The resulting plasmid, pNC513, expresses an in-frame GFP-linker-ASH1 fusion from the 2μm URA3 vector, pRS426 (32). The integrating plasmid pNC514 was constructed by cloning an XhoI-SacII fragment containing the GFP-linker-ASH1 fusion from pNC513 into the XhoI-SacII sites of pRS306 (32). Standard molecular biological techniques were employed in plasmid constructions (30).
pNC248 is a 2μm URA3 vector (YEp52) carrying the STE12M-668 allele from pNC247 (2). pCG37 is the 2μm URA3 vector, pRS202, containing a 2.6-kb PHD1 allele fused to a FLU1 epitope tag (11). pIL30-URA3 is a URA3 CEN plasmid with the FG[Ty]-lacZ reporter gene (17). pMW2 is a URA3 CEN plasmid for expressing Ras2-V19 (35). We isolated pNC544 from the genomic overexpression pYES-R library (9). This isolate expresses the Ste11296–717 variant from the GAL promoter.
Yeast genetic procedures, media, and strain constructions.
Standard yeast genetic procedures and various media were as described previously (12, 31). Yeast transformations were done by the lithium acetate method or electroporation (3, 10). Gene replacements were done according to the method of Rothstein (29).
The strains used in this study and their genotypes are listed in Table 1. Diploid strain SC110, which is heterozygous for the ash1-Δ1::hisG-URA3-hisG allele, was made by gene replacement in strain L5783 with the EcoRI-SphI fragment of pNC409. Haploid strains SC112, SC113, and SC114 are ash1-Δ1::hisG-URA3-hisG or ASH1 meiotic segregants from strain SC110. Strains SC121 and SC122 are ash1-Δ1::hisG derivatives of SC112 and SC113, respectively, that were isolated by the 5-fluoro-orotic acid method (1). Diploid strain SC125 was constructed by mating haploid strains SC121 and SC122. Diploid strain SC126 was constructed by transforming strain SC125 with the GFP-linker-ASH1 allele contained on the NheI fragment of pNC514. The transformation resulted in the integration of GFP-ASH1 5′ to the ash1-Δ1::hisG sequence on one chromosome. Strains SC127 and SC128, which have the leu2-Δ1 allele, were constructed by targeting HpaI-linearized pNC527 to LEU2 and then identifying LEU2 “popouts” among 5-fluoro-orotic acid-resistant isolates that lost the adjacent URA3 sequence. Strains SC135 and SC136 are ste12Δ::LEU2 derivatives of SC127 and SC128, respectively. They were constructed by gene replacement with the SacI-SphI fragment of pSUL16. Diploid strain SC137, which is homozygous for ste12Δ::LEU2, was constructed by first transforming SC135 and SC136 with the STE12 replicating plasmid pNC247, mating the resulting transformants, and then curing the diploid strain of pNC247.
TABLE 1.
S. cerevisiae strainsa
| Strain | Genotype | Reference or source |
|---|---|---|
| L5783 | MATa ura3-52 | 18 |
| MATα ura3-52 | ||
| HLY351 | MATa ura3-52 ste7Δ::LEU2 | 18 |
| MATα ura3-52 ste7Δ::LEU2 | ||
| HLY352 | MATa ura3-52 ste12Δ::LEU2 | 18 |
| MATα ura3-52 ste12Δ::LEU2 | ||
| HLY492 | MATa ura3-52 ste20Δ::LEU2 | 18 |
| MATα ura3-52 ste20Δ::LEU2 | ||
| HLY506 | MATa ura3-52 ste11Δ::LEU2 | 18 |
| MATα ura3-52 ste11Δ::LEU2 | ||
| SC110 | MATa ura3-52 ash1-Δ1::hisG-URA3-hisG | This work |
| MATα ura3-52 ASH1 | ||
| SC112 | MATa ura3-52 ash1-Δ1::hisG-URA3-hisG | This work |
| SC113 | MATα ura3-52 ash1-Δ1::hisG-URA3-hisG | This work |
| SC114 | MATa ura3-52 ASH1 | This work |
| SC121 | MATa ura3-52 ash1-Δ1::hisG | This work |
| SC122 | MATα ura3-52 ash1-Δ1::hisG | This work |
| SC125 | MATa ura3-52 ash1-Δ1::hisG | This work |
| MATα ura3-52 ash1-Δ1::hisG | ||
| SC126 | MATa ura3-52 GFP-ASH1::URA3::ash1-Δ1::hisG | This work |
| MATα ura3-52 ash1-Δ1::hisG | ||
| SC127 | MATa ura3-52 ash1-Δ1::hisG leu2-Δ1 | This work |
| SC128 | MATα ura3-52 ash1-Δ1::hisG leu2-Δ1 | This work |
| SC135 | MATa ura3-52 ash1-Δ1::hisG leu2-Δ1 ste12Δ::LEU2 | This work |
| SC136 | MATα ura3-52 ash1-Δ1::hisG leu2-Δ1 ste12Δ::LEU2 | This work |
| SC137 | MATa ura3-52 ash1-Δ1::hisG leu2-Δ1 ste12Δ::LEU2 | This work |
| MATα ura3-52 ash1-Δ1::hisG leu2-Δ1 ste12Δ::LEU2 |
All strains are isogenic with strain Σ1278b, which is ho− and unable to switch mating types.
Assays for the yeast pseudohyphal form.
The qualitative growth assay for filament formation in colonies of diploid cells was performed as described previously (11, 12). Essentially, cells were streaked onto solid synthetic low-ammonia dextrose medium (SLAD) and incubated at 30°C for the specified periods of time, and representative colonies were photographed. A filament is defined as a chain of attached cells that projects away from the colony.
Invasive-growth assays were done as described by Roberts and Fink (27). Essentially, cells were streaked onto plates of complete medium (yeast extract-peptone-dextrose [YPD]), with care taken to avoid scratching the agar surface. After incubation of plates at 30°C for 4 days and at room temperature for 1 day, the culture plates were washed with a gentle stream of deionized water. This treatment washes noninvasive cells off of the plate but leaves the invasive cells behind as a visible residue in the agar surface.
Diploid pseudohyphal-form cells were distinguished from yeast form cells based on cell shape and budding pattern. After 3 days of growth on SLAD at 30°C, colonies were scraped from plates and suspended in water. Analysis of cell elongation was performed by microscopic examination and visual estimation of the length-to-width ratios (l/w) of at least 100 cells. This inspection easily distinguished three categories of cells: round yeast form cells (l/w, ∼1), oval cells (l/w, 1 to 2), and elongated pseudohyphal-form cells (l/w, >2). Bud scar staining of the suspended cells was done with calcofluor white (Fluorescent Brightener no. 28 F6259; Sigma) as described by Mosch and Fink (21). A unipolar budding pattern was assigned to cells with two or more bud scars at the same pole. A bipolar budding pattern was assigned to cells with one or more scars at opposite poles.
The filamentous growth reporter gene, FG[Ty]-lacZ (pIL30-URA3), was used to monitor transcriptional activation during pseudohyphal growth that is mediated by the MAPK activation cascade and Ste12. For these experiments, activities of the lacZ reporter gene product, β-galactosidase, were compared by using whole-cell protein extracts prepared from yeast form and pseudohyphal-form cultures. Cultures of yeast form cells were grown in YPD to a density of 1 × 107 to 2 × 107 cells/ml at 30°C. Cells were harvested by centrifugation, washed, and suspended to a density of 4 × 108 to 5 × 108 cells/ml for cell lysis. Pseudohyphal-form cells were grown on solid SLAD for 3 days at 30°C. Cells were scraped from the surface of the plates, washed, and suspended to a density of 4 × 108 to 5 × 108 cells/ml for cell lysis. Cell lysis by the glass bead method and quantitative β-galactosidase assays were done as described previously except that the optical density (OD) at 420 nm was determined by using a fixed end point after addition of Na2CO3 (26).
Microscopy.
Imaging of colonies was routinely done with a Zeiss Axiophot microscope with a 10× objective (Plan-NEOFLUAR 10/0.30). Images were recorded either by video with a Sony DXC-760MD camera or by photography on TMAX 400 film. Fluorescence imaging of GFP-Ash1 was done on a Microphot FXA with a 60× objective (Plan-APO 60/1.4) and collected with a cooled charge-coupled device camera (C4880-Hamamatsu) as described previously (36, 37). To obtain suitable colonies, thin slabs of SLAD medium were made directly on microscope slides. One hundred to 200 cells were dispersed on these slides, covered with a coverslip, and visualized after 12 h at 30°C.
RESULTS
Phenotypes of strains lacking or overexpressing Ash1.
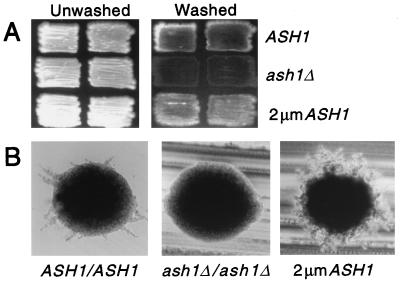
Ash1 was first uncovered in genetic screens that revealed its role in mating-type switching. Because nonswitching diploid cells also express Ash1, we anticipated that it would have additional functions (13, 33). To test whether Ash1 might be needed for diploid functions such as sporulation or pseudohyphal differentiation, we constructed a strain that is homozygous for a complete deletion of the gene. Diploid cells completely lacking Ash1 grow normally. Therefore, Ash1 is nonessential for the viability of either diploid or haploid cell types (13, 33). Because strains lacking or expressing Ash1 showed the same sporulation efficiency and spore viability, Ash1 does not appear to have an essential role in sporulation (data not shown). However, Ash1 is essential for pseudohyphal growth. Hypha-like projections, or filaments, radiating from colonies on solid SLAD are the consequence of the life cycle transition of diploid cells to a pseudohyphal form. Colonies of wild-type cells made such filaments after 2 days of growth on SLAD (Fig. 1B, ASH1/ASH1). Colonies lacking Ash1 did not form pseudohyphal filaments, while the strain overexpressing Ash1 made more filaments than did the wild-type reference strain (Fig. 1B, ash1Δ/ash1Δ and 2μm ASH1).
FIG. 1.
Role of Ash1 in filamentous growth. (A) Invasive growth. Haploid ASH1 [SC114(pRS426)], ash1Δ [SC112(pRS426)], and 2μm ASH1 [SC112(pAS163)] strains were grown on a YPD plate for 5 days. Photographs show patches before and after the plate was washed with water. (B) Pseudohyphal-colony formation. Diploid ASH1/ASH1 [L5783(pRS426)], ash1Δ/ash1Δ [SC125(pRS426)], and 2μm ASH1 [SC125(pAS163)] strains were streaked out on nitrogen starvation medium (SLAD) and grown for 2.5 days at 30°C. Photographs show representative colonies of each strain.
Cells comprising the filaments of diploid pseudohyphal colonies differ in morphology and budding pattern from yeast form cells. Whereas yeast form cells are round or ellipsoidal and have a bipolar budding pattern, pseudohyphal-form cells are elongated and have a unipolar budding pattern. Therefore, to characterize Ash1 effects at the cellular level, we compared the cell morphologies and budding patterns of ASH1, 2μm ASH1, and ash1Δ diploid strains (Table 2). To score these parameters on individual cells, colonies were washed off SLAD plates after 2 days growth at 30°C and suspended in water for microscopic examination. The ratio of elongated to round and oval cells found in the ASH1 colonies is 0.31 and presumably reflects the mixture of yeast and pseudohyphal-form cells comprising the colonies. The unipolar-to-bipolar budding pattern ratio (0.3) also reflects a mixture of yeast and pseudohyphal-form cells. Notably, the fractions of elongated cells (0.57) and cells with a unipolar budding pattern (0.70) are greater in colonies overexpressing Ash1 than in colonies of wild-type cells. This outcome is anticipated, because the 2μm ASH1 strain has a larger number of filaments emanating from the core of yeast form cells (Fig. 1B). By contrast, colonies of cells lacking Ash1 have an insignificant fraction of elongated cells (0.05) and also a lower fraction of cells with a unipolar budding pattern (0.2). These defects of the ash1Δ/ash1Δ mutant are consistent with the absence of colony filaments that are characteristic of pseudohyphal-form cells (Fig. 1B).
TABLE 2.
Shapes and budding patterns of ASH1, 2μm ASH1, and ash1Δ strainsa
| Characteristic | No. of cells of genotype:
|
||
|---|---|---|---|
| ASH1 | 2μm ASH1 | ash1Δ | |
| Cell shapeb | |||
| Round | 44 | 55 | 124 |
| Oval | 109 | 72 | 67 |
| Long | 47 | 72 | 9 |
| Long/(round + oval) | 0.31 | 0.57 | 0.05 |
| Budding patternc | |||
| Bipolar | 71 | 62 | 59 |
| Unipolar | 22 | 41 | 12 |
| Unipolar/bipolar | 0.3 | 0.7 | 0.2 |
Comparisons were made with cells from colonies (on SLAD) of diploid strain L5783 (ASH1/ASH1) transformed with the 2μm vector (pRS426) and diploid strain SC125 (ash1Δ/ash1Δ) transformed with the 2μm vector (pRS426) or the 2μm ASH1 plasmid (pAS163).
Cell shape was assessed by visual estimation of the length-to-width ratio (l/w) for individual cells: round, l/w ∼ 1; oval, l/w = 1 to 2; long, l/w > 2.
Budding pattern was assessed by the pattern of bud scars observed after staining with calcofluor: unipolar, two or more bud scars at the same pole; bipolar, one or more scars at opposite poles.
Haploid cells undergo a related invasive-growth response which causes cells to grow into the agar surface. When patches of wild-type cells on plates are washed off with a stream of water, a residue of embedded cells typical of invasive growth is left behind (Fig. 1A, ASH1). By contrast, patches of the strain lacking Ash1 left little or no residue (Fig. 1A, ash1Δ). Furthermore, overexpression of Ash1 apparently enhanced invasive growth, because there was more of a residue than for the reference wild-type strain (Fig. 1A, 2μm ASH1). These findings establish that Ash1 also has an essential role in invasive growth.
Relationship of Ash1 to other components of the pseudohyphal-response pathway.
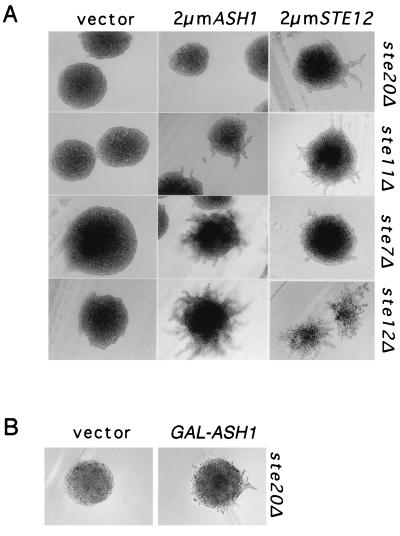
Single deletions that eliminate the sequentially acting protein kinases Ste20, Ste11, and Ste7 or the transcription factor Ste12 block the transition to a pseudohyphal form (Fig. 2A, 2μm vector) (18, 27). It is well established that Ste12 acts downstream of the kinase cascade and that its overproduction suppresses the pseudohyphal defect in different steΔ mutant strains (Fig. 2A, 2μm STE12) (18, 27). To test whether Ash1 has a similar relationship to the MAPK cascade, we tested whether Ash1 overexpression would allow filament formation in the same steΔ mutant strains. Overexpression of Ash1 (2μm ASH1) restored pseudohyphal growth to the strains that lack Ste7 or Ste12 (Fig. 2A, ste7Δ or ste12Δ). Colonies of cells lacking Ste11 but overexpressing Ash1 also made hypha-like projections after 1 day of growth on SLAD (Fig. 2A, 2μm ASH, ste11Δ). However, the response was transient, because after 2 days of growth, the colony was overtaken by yeast form cells (data not shown).
FIG. 2.
Effects of Ash1 overexpression on pseudohyphal-colony formation in strains with deletions of MAPK activation pathway components. (A) ste20Δ/ste20Δ (HLY492), ste11Δ/ste11Δ (HLY506), ste7Δ/ste7Δ (HLY351), and ste12Δ/ste12Δ (HLY352) diploid strains containing either vector (pRS426), 2μm ASH1 (pAS163), or 2μm STE12 (pNC248) were grown on SLAD. (B) The ste20Δ/ste20Δ strain (HLY492) containing either vector (pRS426) or GAL-ASH1 (pNC543) was streaked out on nitrogen starvation medium containing galactose. Photographs show representative colonies of each strain after 2 (A) or 3 (B) days of growth at 30°C.
By contrast, overexpression of Ash1 from a high-copy-number plasmid was insufficient to promote pseudohyphal growth at any time after incubation on SLAD in the strain lacking Ste20 (Fig. 2A, 2μm ASH1, ste20Δ). In this regard, Ash1 acts differently from Ste12, because overexpression of Ste12 from a high-copy-number plasmid does allow pseudohyphal growth in the strain lacking Ste20 (Fig. 2A, 2μm STE12, ste20Δ). Interestingly, ectopic expression of Ash1 from the GAL1 promoter bypassed the need for Ste20 (Fig. 2B). This condition may be permissive simply because there is a larger amount of Ash1 produced from the GAL1 promoter. Alternatively, the heterologous promoter may allow a broader spatial and temporal pattern of Ash1 expression than can be achieved from its own promoter. (See “Localization and expression of Ash1” below). Nevertheless, these analyses show that Ash1 function can bypass the need for the kinase cascade and Ste12. This outcome suggests that Ash1 acts downstream or independently of the MAPK cascade.
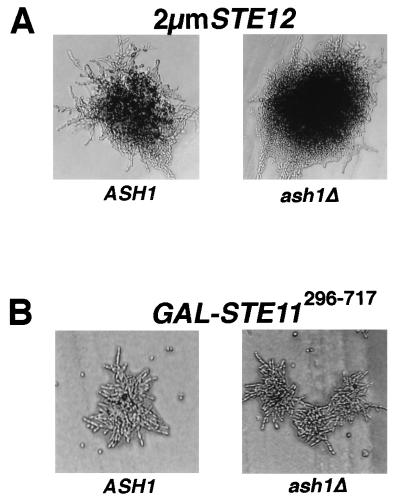
Constitutive activation of the MAPK cascade by expression of a gain-of-function STE11 allele (STE11-4) or bypassing the cascade with GAL-STE12 has been reported to enhance pseudohyphal growth (18). Therefore, such alleles can be used for epistasis tests that are the reciprocal of those done with the deletion strains. We made use of the constitutive Ste11 variant Ste11296–717 and 2μm Ste12 overexpression to artificially induce pseudohyphal growth and then test whether the response would be blocked in a strain devoid of Ash1 (ash1Δ/ash1Δ). Colonies of diploid strains overexpressing Ste12 (2μm STE12) and either lacking or expressing Ash1 made similar filaments on SLAD (Fig. 3A). Ste11296–717-promoted pseudohyphal growth was also the same for strains lacking or expressing Ash1 (Fig. 3B). These results support the deduction that Ash1 functions separately from Ste12 and the MAPK activation cascade.
FIG. 3.
Comparison of ASH1 and ash1Δ mutant strains for pseudohyphal-colony formation promoted by hyperactivation of the pathway. (A) ASH1/ASH1 (L5783) and ash1Δ/ash1Δ (SC125) diploid strains containing 2μm STE12 (pNC248) were grown on SLAD for 2 days at 30°C. (B) ASH1/ASH1 (L5783) and ash1Δ/ash1Δ (SC125) diploid strains containing GAL-STE11296–717 (pNC544) were grown on nitrogen starvation medium containing galactose for 1 day at 30°C. Photographs are of representative colonies.
The Ste12 transcription factor is thought to regulate the expression of genes that are critical for pseudohyphal growth. Although no pseudohyphal gene that is under Ste12 control has been identified, the Ste12-dependent response element from the yeast transposon Ty1 is responsive to conditions that promote pseudohyphal growth (22). If Ash1 functions separately from Ste12, transcriptional activation of the Ty1 UAS by nitrogen starvation should be unaffected in cells lacking Ash1. To test this prediction, we measured expression of a pseudohyphal reporter gene (FG[Ty]-lacZ) in homozygous ASH1 and ash1Δ strains (22). Log-phase cultures (YPD) of the strains expressing or lacking Ash1 produced the same background amount of reporter gene product (β-galactosidase activity, 12 ± 6 and 15 ± 3 units of β-galactosidase activity [milli-OD/min/mg], respectively). Nitrogen-deprived cultures (SLAD) of cells expressing Ash1 produced ∼15-fold-larger amounts of reporter gene product (175 ± 18 milli-OD/min/mg). Under these conditions (SLAD), cells lacking Ash1 similarly induced reporter gene expression (140 ± 3 milli-OD/min/mg). These results show that Ash1 is not required for Ste12 function.
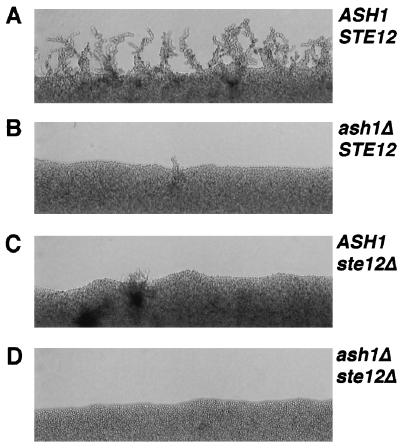
Because Ash1 and Ste12 appear to function separately in the pseudohyphal response, we expected that the phenotype of the double mutant would be more severe than that of either single mutant. Some pseudohyphae emanate from patches of homozygous ste12Δ or ash1Δ strains after 4 days of growth on SLAD, showing that neither single mutation completely blocks filament formation (Fig. 4B and C). By contrast, the double homozygous mutant (ste12Δ ash1Δ) is devoid of any filaments after the same incubation period (Fig. 4D). No filaments were apparent in the double-mutant strain even after 10 days on SLAD, which is longest incubation time that we have monitored (data not shown). The additive effect of Ash1 and Ste12 supports a model that assigns Ash1 a role in the pseudohyphal response separate from that of Ste12 and the MAPK activation cascade.
FIG. 4.
Synthetic pseudohyphal-colony phenotype of ash1Δ and ste12Δ mutants. ASH1/ASH1 STE12/STE12 [L5783(pRS426)], ash1Δ/ash1Δ STE12/STE12 [SC125(pRS426)], ASH1/ASH1 ste12Δ/ste12Δ [HLY352(pRS426)], and ash1Δ/ash1Δ ste12Δ/ste12Δ [SC137(pRS426)] diploid strains were patched onto SLAD and grown at 30°C for 4 days. Photographs show a representative region of an edge from each patch.
Relationship of Ste12 and Ash1 to other regulators of pseudohyphal growth.
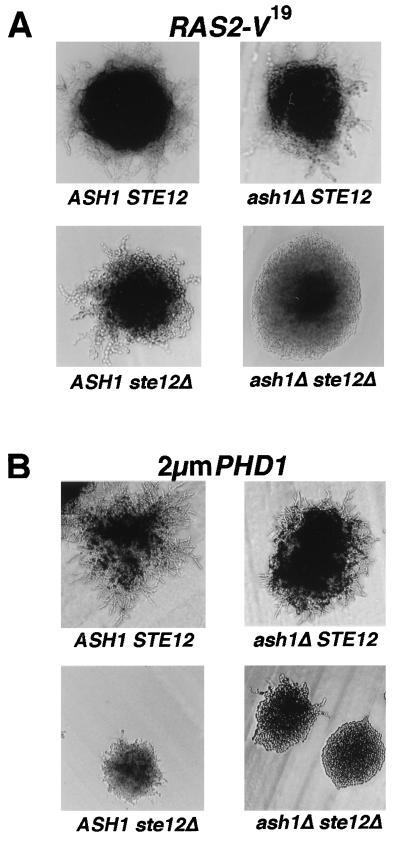
Expression of an activated variant of Ras2, Ras2-V19, has been reported to induce pseudohyphal growth (12). Additionally, strains that express Ras2-V19 show an eightfold increase in FG[Ty]-lacZ reporter gene expression compared with a reference Ras2 strain. Because this effect on FG[Ty]-lacZ expression is blocked in mutants that lack Ste20, Ste11, Ste7, or Ste12, it has been proposed that Ras2 functions upstream of the MAPK activation cascade and Ste12 (22). If Ras2 function is mediated solely by Ste12, this model predicts that an absence of Ste12 should also block pseudohyphal-filament formation promoted by Ras2-V19. However, we find that cells expressing Ras2-V19 but lacking Ste12 still form pseudohyphal filaments (Fig. 5A, ASH1 ste12Δ). This result opens the possibility that components separate from the MAPK cascade might also mediate Ras2-V19 effects on pseudohyphal growth. The results of our epistasis analyses suggested that Ash1 functions separately from Ste12 and hypothetically could fulfill such a role. To test this possibility, we compared pseudohyphal-filament formation promoted by Ras2-V19 in homozygous ash1Δ single-mutant and ash1Δ ste12Δ double-mutant diploid strains. While the absence of Ash1 alone was insufficient to block Ras2-V19-promoted pseudohypha formation, the absence of both Ash1 and Ste12 did block the effect (Fig. 5A, ash1Δ STE12 and ste12Δ ash1Δ). These results are consistent with a model in which Ste12 an Ash1 have a compensatory role in mediating the Ras2-dependent signal(s) for pseudohyphal growth.
FIG. 5.
Comparison of ash1Δ and ste12Δ single- or double-mutant strains for pseudohyphal-colony formation promoted by hyperactivation of the pathway. ASH1/ASH1 STE12/STE12 (L5783), ash1Δ/ash1Δ STE12/STE12 (SC125), ASH1/ASH1 ste12Δ/ste12Δ (HLY352), and ash1Δ/ash1Δ ste12Δ/ste12Δ (SC137) diploid strains containing either RAS2-V19 (pMW2) (A) or 2μm PHD1 (pCG37) (B) were grown on SLAD for 2 days at 30°C. Photographs are of representative colonies.
Phd1 is a presumed transcription factor that is also implicated in the regulation of pseudohyphal growth. While the absence of Phd1 is insufficient to prevent pseudohyphal growth, its overexpression enhances pseudohyphal growth (11). Additionally, overexpression of Phd1 can suppress the pseudohyphal-growth defect in a strain that lacks Ste12 (Fig. 5B, ASH1 ste12Δ) (21). We were curious to learn how Phd1 overexpression would affect pseudohyphal growth of homozygous ash1Δ single- and ash1Δ ste12Δ double-mutant strains. Phd1 overexpression promoted vigorous pseudohyphal growth in the strain lacking Ash1 and allowed some pseudohyphal growth even in the strain lacking both Ash1 and Ste12 (Fig. 5B, ash1Δ STE12 and ash1Δ ste12Δ). Unlike Ras2-V19, overproduction of Phd1 can act independently of both Ste12 and Ash1 to promote pseudohyphal growth.
Localization and expression of Ash1.
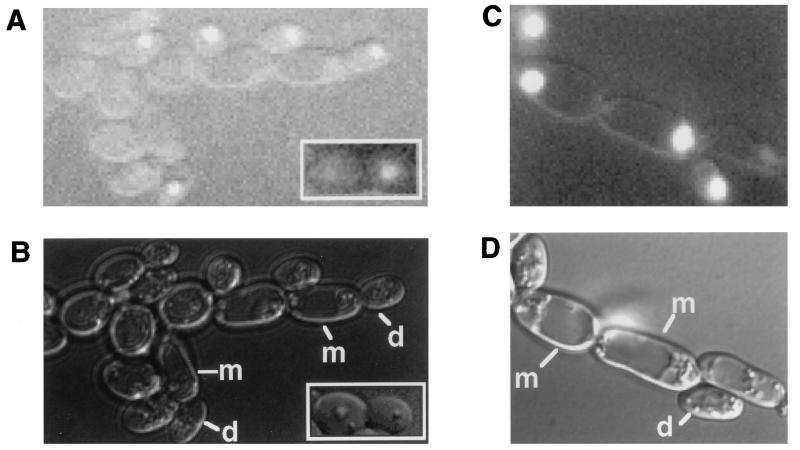
Ash1 localizes to the nuclei of daughter cells in both haploid and diploid yeast form cells (13, 33). To learn if Ash1 also localizes to the nuclei of pseudohyphal-form daughter cells, we constructed a GFP-tagged version of Ash1. The allele encoding the fusion protein was integrated at the ASH1 locus of the homozygous ash1Δ diploid strain. The GFP-Ash1 fusion fully complemented the pseudohyphal-growth defect of the ash1Δ/ash1Δ strain. Fluorescent and differential interference contrast photographs of over 20 pseudohyphal mother-daughter pairs were analyzed. Similarly to the case in yeast form cells, GFP-Ash1 localized exclusively to the nuclei of pseudohyphal daughter cells (Fig. 6A).
FIG. 6.
Localization of GFP-Ash1 during pseudohyphal growth. Fluorescent (A and C) and differential interference contrast (B and D) views of cells are shown. (A and B) Diploid strain SC126, which is heterozygous for GFP-ASH1, grown on SLAD for 12 h. Insets show a mother-daughter pair of yeast form cells of the same diploid strain grown on SD-Ura. (C and D) Diploid ash1Δ/ash1Δ strain SC125 with pNC513, which expresses GFP-ASH1 from the 2μm vector pRS426, grown on SLAD for 12 h. m, mother cell; d, daughter cell.
The intensity of the signal in pseudohyphal cells on SLAD differed little from that observed with yeast form cells grown on nitrogen-rich medium (Fig. 6A, inset). This result is consistent with our finding that steady-state amounts of Ash1 mRNA were not greater in cultures grown on SLAD than in those grown on liquid nitrogen-rich medium (data not shown). Based on the size and position of nuclei in the cells expressing Ash1, it appears that Ash1 becomes expressed late in M phase, similar to what has been observed for yeast form haploid cells (13, 33).
Also similar to what has been reported for yeast form cells, overexpression of Ash1 in pseudohyphal cells leads to a more symmetric localization pattern (Fig. 6C). Because overexpression of Ash1 is not inhibitory to pseudohyphal growth, the presence of Ash1 in mother cells is not disruptive (Fig. 1B and 6D). This result provides additional support for the idea that Ash1 is formally a positive regulator of pseudohyphal growth. Because of its positive regulatory role and the finding that under normal conditions Ash1 is detectable only in daughter cells, we conclude that there must be a daughter cell-specific function needed for the transition from yeast to pseudohyphal-form cells.
DISCUSSION
Pseudohyphal differentiation of S. cerevisiae requires Ash1.
Ash1 deletion mutants are defective for pseudohyphal and invasive growth. These defects establish that Ash1 is formally a positive regulator of filamentous growth. In this regard, the role of Ash1 in pseudohyphal-form growth is different from its negative regulatory role in mating-type switching (13, 33). Ash1 has a zinc finger-like domain related to that of the GATA family of transcription factors (24, 33). This relationship and the nuclear location of Ash1 suggested that it is most likely a transcriptional regulator. This view encouraged speculation that Ash1 might negatively regulate mating-type switching by binding directly to sequences in the HO promoter and repressing its transcription or by binding to and interfering with the Swi5 transcriptional activator of HO (13, 33). The contrasting role for Ash1 in pseudohyphal differentiation suggests that similar to other members of GATA family, Ash1 might function as both an activator and a repressor of transcription (24). On the other hand, if Ash1 functions only as a transcriptional repressor, its role in pseudohyphal differentiation would involve repression of a negative regulator of the process.
Relationship of Ash1 to other known regulators of the pseudohyphal process.
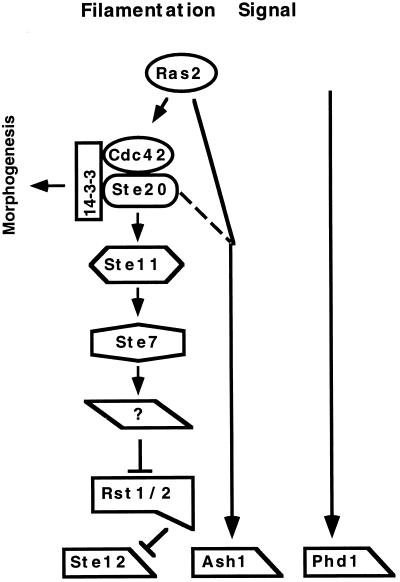
Nitrogen starvation stimulates the transition of S. cerevisiae to a pseudohyphal form. This signal is mediated, at least in part, by a MAPK activation cascade and its downstream transcription factor, Ste12 (18) (Fig. 7). Signaling through this branch of the pathway involves the monomeric G protein Cdc42, as well as the 14-3-3 homologs Bmh1 and Bmh2, which associate with Ste20 (22, 28). More recently, Gpa2-Gα has been shown to be essential for pseudohyphal growth. Gpa2 and also Ras2 appear to regulate a separate branch of the nitrogen-sensing pathway that leads to increases in cyclic AMP levels (16, 20). Ras2 has also been implicated as an upstream regulator of the MAPK cascade and Ste12. This suggestion is based on the finding that an activated Ras2 variant (Ras2-V19) stimulates expression of a pseudohyphal reporter gene (FG[Ty]-lacZ) and that this effect depends on components of the MAPK cascade (22). As such, Ras2 could coordinate activities of the MAPK and cyclic AMP branches of the pseudohyphal signaling network.
FIG. 7.
Model for the pseudohyphal-response pathway. The diagram shows the postulated relationship of signal transmission components that function after the starvation signal(s) for induction of pseudohyphal growth. Arrows indicate activation or stimulation. Lines with bars indicate repression or inhibition. See the text for discussion of the evidence suggesting these relationships.
The results of our epistasis analyses are consistent with the deduction that Ash1 acts separately from the MAPK activation cascade and Ste12 but is still downstream from Ras2 (Fig. 7). Furthermore, Ste12 and Ash1 have additive effects in the response, because a deletion of both is needed to completely block normal or Ras2-V19-promoted filamentation. Additional support for the view that Ash1 acts separately from Ste12 comes from the finding that the presence or absence of Ash1 has no effect on expression of a Ste12-dependent pseudohyphal reporter gene (FG[Ty]-lacZ).
Our epistasis analyses also suggest that Ste20 may have roles in the pseudohyphal response in addition to activation of the MAPK cascade. This deduction stems from the observation that moderate overexpression of Ash1 can bypass deletion mutants of the MAPK activation cascade but not of Ste20. One possibility is that Ste20 regulates both the Ash1 and Ste12 branches of the pathway (22, 28) (Fig. 7). Alternatively, Ste20 could control morphogenetic alterations that while not essential, nevertheless facilitate pseudohyphal growth. This postulated dual role would be analogous to the dual role that has recently been uncovered for Ste20 in mating differentiation (25).
Although Ste12 and Ash1 appear to have separate and additive functions, we nevertheless found that hyperactivation of Ste12 or Ash1, either by their overproduction or by Ras2-V19, can bypass the need of one for the other. This interesting compensatory relationship is not unique to Ash1 and Ste12. Phd1, another presumed transcription factor, also appears to have functions that are additive with Ste12. Similar to our results with Ash1, filament formation was blocked completely only in strains that lacked both Ste12 and Phd1 (19). Also similar to our findings, overexpression of Phd1 compensates for the absence of Ste12 (19). Because overexpression of Phd1 also promotes filamentation in strains lacking both Ash1 and Ste12, Phd1 may be on yet another arm of the pseudohyphal signaling network.
The genes required for pseudohyphal growth that are regulated by these presumed or actual transcription factors have yet to be identified. Nevertheless, the ability of Ash1, Ste12, and Phd1 to compensate for one another suggests some interesting possibilities for regulation of pseudohyphal-gene expression. One possibility is that the three transcription factors regulate the same subset of genes that are essential for filamentous growth. According to this model, the critical genes are expressed optimally only when the three transcription factors and the stimuli to which they respond are present. Residual expression of these genes when any two of the transcription factors are present is sufficient to support the amount of filamentation seen with the single-mutant strains. Conversely, hyperactivation of any one transcription factor could increase expression of the critical genes to levels sufficient for filamentation. It is equally feasible that Ste12, Ash1, and Phd1 control separate sets of genes whose products have partially overlapping functions. In this case, hyperactivation of any of the transcription factors could lead to overexpression of one or the other set and again allow adequate activity to support filamentous growth.
Daughter cell-specific functions in pseudohyphal growth.
Similarly to the case in yeast form cells, Ash1 localizes to daughter cells of pseudohyphal-form yeast, revealing a difference between daughter and mother cells. Because Ash1 mutants do not form pseudohyphae, the implicit daughter-specific functions are essential for filamentous growth. In yeast form cells Ash1 expression is restricted to daughter cells in late anaphase and G1 (13, 33). Based on the size and position of the nuclei in cells expressing Ash1, it appears that Ash1 has a similar cell cycle restriction in its expression during pseudohyphal growth. Because of these spatial and temporal restrictions on Ash1 expression, the gene products subject to its regulatory control must be critical for pseudohyphal development only in the apical cells of the colony during this window of the cell cycle. An important implication of these restrictions is that the pseudohyphal fate is established solely by daughter cells. Once the transition from yeast to pseudohyphal form is made, cells appear to maintain the pseudohyphal form independently of Ash1 and perhaps other regulators of this growth mode. Full comprehension of what role Ash1 might have in establishing the pseudohyphal fate awaits identification of its targets.
ACKNOWLEDGMENTS
This research was supported by Public Health grant GM-39852 from the National Institutes of Health.
We thank H. Liu and G. Fink for providing yeast strains and pIL30 and pCG37 plasmids, A. Sil and I. Herskowitz for providing pAS163, J. Pringle for providing pKS+GFP, M. Ward and S. Garrett for providing pMW2, and S. Ramer and S. Elledge for providing the pYES-R yeast genomic DNA expression library. We are also grateful to E. D. Salmon and E. Yeh for use of the microscope facility and for sharing their expertise with us and to R. Duronio, J. Heitman, and S. Kron for critical reading of the manuscript and helpful discussions.
REFERENCES
- 1.Alani E, Cao L, Kleckner N. A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics. 1987;116:541–545. doi: 10.1534/genetics.112.541.test. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baur M, Esch R K, Errede B. Cooperative binding interactions required for function of the Ty1 sterile responsive element. Mol Cell Biol. 1997;17:4330–4337. doi: 10.1128/mcb.17.8.4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Becker D M, Guarente L. High efficiency transformation of yeast by electroporation. Methods Enzymol. 1990;194:182–187. doi: 10.1016/0076-6879(91)94015-5. [DOI] [PubMed] [Google Scholar]
- 4.Bobola N, Jansen R-P, Shin T H, Nasmyth K. Asymmetric accumulation of Ash1p in postanaphase nuclei depends on a myosin and restricts yeast mating-type switching to mother cells. Cell. 1996;84:699–709. doi: 10.1016/s0092-8674(00)81048-x. [DOI] [PubMed] [Google Scholar]
- 5.Cade R, Errede B. MOT2 encodes a negative regulator of gene expression that affects basal expression of pheromone-responsive genes in Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:3139–3149. doi: 10.1128/mcb.14.5.3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cook J G, Bardwell L, Kron S J, Thorner J. Two novel targets of the MAP kinase Kss1 are negative regulators of invasive growth in Saccharomyces cerevisiae. Genes Dev. 1996;10:2831–2848. doi: 10.1101/gad.10.22.2831. [DOI] [PubMed] [Google Scholar]
- 7.DeVirgilio C, DeMarini D J, Pringle J R. SPR28, a sixth member of the septin gene family in Saccharomyces cerevisiae that is expressed specifically in sporulating cells. Microbiology. 1996;142:2897–2905. doi: 10.1099/13500872-142-10-2897. [DOI] [PubMed] [Google Scholar]
- 8.Dolan J W, Kirkman C, Fields S. The yeast Ste12 protein binds to the DNA sequence mediating pheromone induction. Proc Natl Acad Sci USA. 1989;86:5703–5707. doi: 10.1073/pnas.86.15.5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elledge S J, Mulligan J T, Ramer S W, Spottswood M, Davis R. λYES: a multifunctional cDNA expression vector for the isolation of genes by complementation of yeast and Escherichia coli mutations. Proc Natl Acad Sci USA. 1991;88:1731–1735. doi: 10.1073/pnas.88.5.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gietz D, St. Jean J A, Woods R A, Schiestl R H. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gimeno C J, Fink G R. Induction of pseudohyphal growth by overexpression of PHD1, a Saccharomyces cerevisiae gene related to transcriptional regulators of fungal development. Mol Cell Biol. 1994;14:2100–2112. doi: 10.1128/mcb.14.3.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gimeno C J, Ljungdahl P O, Styles C A, Fink G R. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell. 1992;68:1077–1090. doi: 10.1016/0092-8674(92)90079-r. [DOI] [PubMed] [Google Scholar]
- 13.Jansen R-P, Dowzer C, Michaelis C, Galova M, Nasmyth K. Mother cell-specific HO expression in budding yeast depends on the unconventional myosin Myo4p and other cytoplasmic proteins. Cell. 1996;84:687–697. doi: 10.1016/s0092-8674(00)81047-8. [DOI] [PubMed] [Google Scholar]
- 14.Kron S J, Gow N A. Budding yeast morphogenesis: signalling, cytoskeleton, and cell cycle. Curr Opin Cell Biol. 1995;7:845–855. doi: 10.1016/0955-0674(95)80069-7. [DOI] [PubMed] [Google Scholar]
- 15.Kron S J, Styles C A, Fink G R. Symmetric cell division in pseudohyphae of the yeast Saccharomyces cerevisiae. Mol Biol Cell. 1994;5:1003–1022. doi: 10.1091/mbc.5.9.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kubler E, Mosch H-U, Rupp S, Lisanti M P. Gpa2p, a G-protein alpha-subunit, regulates growth and development in Saccharomyces cerevisiae via a cAMP-dependent mechanism. J Biol Chem. 1997;272:20321–20323. doi: 10.1074/jbc.272.33.20321. [DOI] [PubMed] [Google Scholar]
- 17.Laloux I, Jacobs E, Dubois E. Involvement of SRE element of Ty1 transposon in TEC1-dependent transcriptional activation. Nucleic Acids Res. 1994;22:999–1005. doi: 10.1093/nar/22.6.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu H, Styles C A, Fink G R. Elements of the yeast pheromone response pathway required for filamentous growth of diploids. Science. 1993;262:1741–1744. doi: 10.1126/science.8259520. [DOI] [PubMed] [Google Scholar]
- 19.Lo H-J, Kohler J R, DiDomenico B, Loebenberg D, Cacciapuoti A, Fink G R. Nonfilamentous C. albicans mutants are avirulent. Cell. 1997;90:939–949. doi: 10.1016/s0092-8674(00)80358-x. [DOI] [PubMed] [Google Scholar]
- 20.Lorenz M C, Heitman J. Yeast pseudohyphal growth is regulated by GPA2, a G protein alpha homolog. EMBO J. 1997;16:7008–7018. doi: 10.1093/emboj/16.23.7008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mosch H-U, Fink G R. Dissection of filamentous growth by transposon mutagenesis in Saccharomyces cerevisiae. Genetics. 1997;145:671–684. doi: 10.1093/genetics/145.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mosch H-U, Roberts R L, Fink G R. Ras2 signals via the Cdc42/Ste20/mitogen-activated protein kinase module to induce filamentous growth in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1996;93:5352–5356. doi: 10.1073/pnas.93.11.5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nasmyth K. Regulating the HO endonuclease in yeast. Curr Opin Gen Dev. 1993;3:286–294. doi: 10.1016/0959-437x(93)90036-o. [DOI] [PubMed] [Google Scholar]
- 24.Orkin S H. GATA-binding transcription factors in hematopoietic cells. Blood. 1992;80:575–581. [PubMed] [Google Scholar]
- 25.Peter M, Neiman A M, Park H-O, van Lohuizen M, Herskowitz I. Functional analysis of the interaction between the small GTP binding protein Cdc42 and the Ste20 protein kinase in yeast. EMBO J. 1996;15:7046–7059. [PMC free article] [PubMed] [Google Scholar]
- 26.Rhodes N, Connell L, Errede B. STE11 is a protein kinase required for cell type specific transcription and signal transduction in yeast. Genes Dev. 1990;4:1862–1874. doi: 10.1101/gad.4.11.1862. [DOI] [PubMed] [Google Scholar]
- 27.Roberts R L, Fink G R. Elements of a single MAP kinase cascade in Saccharomyces cerevisiae mediate two developmental programs in the same cell type: mating and invasive growth. Genes Dev. 1994;8:2974–2985. doi: 10.1101/gad.8.24.2974. [DOI] [PubMed] [Google Scholar]
- 28.Roberts R L, Mosch H-U, Fink G R. 14-3-3 proteins are essential for RAS/MAPK cascade signaling during pseudohyphal development in S. cerevisiae. Cell. 1997;89:1055–1065. doi: 10.1016/s0092-8674(00)80293-7. [DOI] [PubMed] [Google Scholar]
- 29.Rothstein R J. Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast. Methods Enzymol. 1991;101:202. doi: 10.1016/0076-6879(91)94022-5. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 31.Sherman F, Fink G R, Hicks J. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1986. [Google Scholar]
- 32.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sil A, Herskowitz I. Identification of an asymmetrically localized determinant, Ash1p, required for lineage-specific transcription of the yeast HO gene. Cell. 1996;84:711–722. doi: 10.1016/s0092-8674(00)81049-1. [DOI] [PubMed] [Google Scholar]
- 34.Tedford K, Kim S, Sa D, Stevens K, Tyers M. Regulation of the mating pheromone and invasive growth responses in yeast by two MAP kinase substrates. Curr Biol. 1997;7:228–238. doi: 10.1016/s0960-9822(06)00118-7. [DOI] [PubMed] [Google Scholar]
- 35.Ward M P, Gimeno C J, Fink G R, Garrett S. SOK2 may regulate cyclic AMP-dependent protein kinase-stimulated growth and pseudohyphal development by repressing transcription. Mol Cell Biol. 1995;15:6854–6863. doi: 10.1128/mcb.15.12.6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang S S, Yeh E, Salmon E D, Bloom K. Identification of a mid-anaphase checkpoint in budding yeast. J Cell Biol. 1997;136:345–354. doi: 10.1083/jcb.136.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yeh E, Skibbens R V, Cheng J W, Salmon E D, Bloom K. Spindle dynamics and cell cycle regulation of Dyenin in the budding yeast, Saccharomyces cerevisiae. J Cell Biol. 1995;130:687–700. doi: 10.1083/jcb.130.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]