Abstract
During the transition from the maternal to the zygotic developmental program, the expression of genes important for pattern formation or cell cycle regulation changes dramatically. Rapid changes in gene expression are achieved in part through the control of mRNA stability. This report focuses on bicoid, a gene essential for formation of anterior embryonic structures in Drosophila melanogaster. bicoid mRNA is synthesized exclusively during oogenesis. Here, we show that bicoid mRNA stability is regulated. While bicoid mRNA is stable in retained oocytes, in unfertilized eggs, and during the first 2 h of embryogenesis, specific degradation is activated at cellularization of the blastoderm. To identify cis-acting sequences required for bicoid mRNA’s regulated stability, fusions between bicoid and genes producing stable mRNAs were introduced into the Drosophila germ line by P-element-mediated transformation. The analysis of the fusion mRNAs identified a bicoid instability element (BIE) contained within a 43-nucleotide sequence immediately following the stop codon. The BIE is sufficient to destabilize the otherwise-stable ribosomal protein A1 mRNA and is separable from the previously identified bicoid mRNA localization signals and from the “nanos response element.” Similar mechanisms may regulate a class of developmentally important maternal genes whose mRNA has a temporal profile similar to that of bicoid.
The degradation of maternally encoded mRNAs at the transition from maternal to zygotic expression is a common occurrence in animal development (4, 13, 14, 29, 36). In Drosophila melanogaster, the degradation of two maternal mRNAs, string and twine, which encode Cdc25 phosphatases, is essential for embryonic development (13). Many other genes important for embryonic development are similarly regulated (33). One of them is bicoid (16), a gene essential for the morphogenesis of the anterior half of the embryo.
Transcription of bicoid is strictly maternal; the mRNA is synthesized in the ovary by the nurse cells and then transported to the oocyte (6). bicoid mRNA localizes to the anterior ends of oocytes and early embryos by a mechanism involving multiple steps (31). Anterior localization is mediated by a 625-nucleotide sequence in the bicoid untranslated region (UTR) (18). After egg deposition, bicoid mRNA is translated (10) and the unstable Bicoid protein forms a concentration gradient decreasing from the anterior tip of the embryo. Different protein concentration thresholds activate different gap genes along the anterior half of the embryo (11, 12). By the beginning of gastrulation, bicoid mRNA has been completely degraded. Thus, bicoid function depends on at least five posttranscriptional processes: (i) mRNA localization at the anterior pole of the oocyte, (ii) translational repression during oogenesis, (iii) diffusion of the protein from a localized source, (iv) instability of the protein, and (v) destabilization of bicoid mRNA. This report focuses on the regulation of bicoid mRNA stability.
Previously, Berleth et al. (6) had determined that bicoid mRNA is absent from embryos over 4 h old. However, it is unknown how degradation of bicoid mRNA is controlled. It is possible that fertilization triggers the destabilization and gradual degradation of bicoid mRNA. Alternatively, the message may be initially stable and then rapidly degraded before the onset of gastrulation. Moreover, the sequences that regulate bicoid mRNA stability are unknown. In the present report we consider the following questions. Is bicoid mRNA constitutively unstable, or is its stability regulated? If regulated, is bicoid mRNA degradation activated at fertilization or later in embryogenesis? What cis-acting elements mediate the selective destabilization of bicoid mRNA? Surprisingly, we found that bicoid mRNA is stable even after egg activation and fertilization and that its degradation is activated during embryogenesis, at cellularization of the blastoderm. Moreover, by assaying different hybrid genes in embryos from transgenic flies, we defined a cis-acting bicoid instability element (BIE) that is both necessary and sufficient for regulated message decay.
MATERIALS AND METHODS
Construction of plasmids.
The genomic bicoid 8.7-kb EcoRI fragment was obtained from the plasmid p902 (provided by P. Macdonald). The relevant parts of the bicoid sequence used in this study are numbered as in reference 6. The ribosomal protein A1 gene (rpA1) 2.4-kb BamHI fragment and the α1-tubulin gene 4.5-kb HindIII fragment were obtained from the plasmids p5D (24) and pDmt-1 (17), respectively.
Construction of the bicoid–rpA1 and bicoid–α1-tubulin hybrid genes. (i) 5B (5′bicoid/3′rpA1).
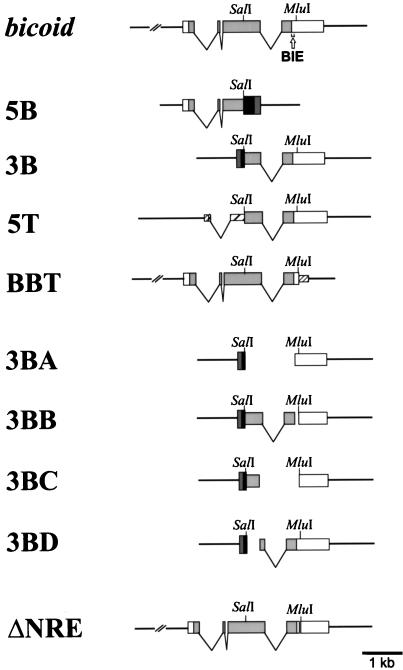
To construct the chimeric gene 5B (5′bicoid/3′rpA1) the vector pGEM3 (Promega) was modified as follows. A linker containing the NotI site was inserted to replace the HindIII site (modified vector kindly provided by A. Riedl). The SalI site of pGEM3 was removed by linearization of the plasmid with SalI and filling in of the restriction site with T4 DNA polymerase, followed by ligation. The 2.4-kb BamHI fragment from the p5D plasmid was subcloned into the modified pGEM3 vector with the NotI site in the 5′ direction from the rpA1 gene to yield pGEM3rpA1. A bicoid 1.5-kb PstI-SalI fragment obtained from p902 by partial digestion was ligated to pGEM3rpA1, which had been digested with PstI-SalI and dephosphorylated. The resulting 2.8-kb insert contained 1,519 bp of the bicoid gene, from the cap site to the SalI site (bicoid positions 1245 to 2764), and a 1.3-kb rpA1 sequence from the unique SalI site within the protein-coding region to the end of the rpA1 mRNA plus 3′ flanking sequences. The HA1 DNA cassette encoding the HA1 epitope YPYDVPDYA from the influenza virus hemagglutinin HA1 protein (32) was inserted at the SalI junction in the aforementioned plasmid by digesting the plasmid with SalI, filling in the site with T4 DNA polymerase, and ligating the ends to a 55-bp StuI-ScaI DNA fragment encoding the HA1 epitope. This plasmid was designated pGEM-5B (Fig. 1). The insertion of the DNA cassette fuses in frame the protein-coding sequences of bicoid and the rpA1 gene. The NotI-BamHI fragment containing the 5′bicoid/3′rpA1 hybrid was subcloned into the transformation vector CaSpeR4/GERM4 to yield CaSpeR-5B. CaSpeR4/GERM4 was constructed as follows: the fragment NotI-EcoRI, from the plasmid pGERM4 (kindly provided by K. Cheung and R. Cohen), which contained a nurse cell-specific enhancer from the heat shock gene hsp26 (388 bp) and the sgs3 promoter from bp −127 to +33, was subcloned into CaSpeR4.
FIG. 1.
Constructs used to localize the BIE. The structure of the bicoid gene is drawn from the work of Berleth et al. (6). 5B contains a hybrid between bicoid 5′ sequences and rpA1 gene 3′ sequences. The glue gene sgs3 promoter containing the nurse cell-specific hsp26 enhancer was used to drive the expression of this construct. 3B contains a hybrid between rpA1 gene 5′ sequences and bicoid 3′ sequences. This hybrid gene is under the control of the rpA1 promoter. 5T contains a hybrid between α1-tubulin gene 5′ sequences and bicoid 3′ sequences. The α1-tubulin promoter was used for this construct. In the BBT construct, the bicoid 3′ UTR sequence downstream of the MluI site was replaced by that of the tubulin gene 3′ UTR. The bicoid promoter was used for this construct (a kind gift from P. Macdonald [18]). 3BA, 3BB, 3BC, and 3BD are four overlapping deletion constructs of the 3B construct. The ΔNRE construct contains a bicoid gene with a deletion of 45 nucleotides in the 3′ UTR between the HpaI and EcoRV restriction sites, downstream of the stop codon (a kind gift from R. Wharton [34]). Horizontal lines, nontranscribed flanking regions; ∨s, introns, ░⃞, bicoid protein-coding region; □, bicoid UTRs; ▪, rpA1 protein-coding region; ▩, rpA1 gene UTRs; , tubulin protein-coding region; ▨, tubulin gene UTRs. A DNA cassette encoding the influenza virus HA1 hemagglutinin epitope was inserted (not shown [32]) to maintain the protein reading frame in constructs 5B, 3B, 5T, 3BA, 3BB, 3BC, and 3BD.
(ii) 3B (5′rpA1/3′bicoid).
The pGEM3rpA1 plasmid was digested with SalI and EcoRI to remove the 3′ end of the gene. The resulting DNA was dephosphorylated and ligated to a bicoid 3.0-kb SalI-EcoRI fragment from p902, yielding pGEM-3B (Fig. 1). The 4.1-kb insert from pGEM-3B contains 1.1 kb of 5′ rpA1 gene sequences, including the promoter and coding region up to the SalI site. The remaining 3.0 kb contain the 3′-most 2,044 bp of the bicoid gene, from the SalI site (bicoid position 2765) to the polyadenylation site plus 3′ flanking sequences extending to the 3′ EcoRI site from the bicoid 8.7-kb EcoRI genomic fragment. The HA1 DNA cassette was inserted at the SalI junction of the aforementioned plasmid by digesting the plasmid with SalI, filling in the site with T4 DNA polymerase, and ligating the ends to a 69-bp NaeI-SmaI fragment containing the sequence encoding the HA1 epitope (32). The insertion of the HA1 DNA cassette fuses in frame the protein-coding sequences of the rpA1 gene and bicoid. The NotI-EcoRI fragment containing the 5′rpA1/3′bicoid hybrid was subcloned into CaSpeR4 to yield CaSpeR-3B.
(iii) 5T (5′α1-tub/3′bicoid).
To construct the chimeric gene 5T (5′α/1-tub/3′bicoid), the vector pBluescript II KS+ (pBSIIKS+; Stratagene) was modified as follows. The BamHI-PstI portion in the polylinker of pBSIIKS+ was removed by digestion with PstI, treatment with nuclease S1, digestion with BamHI, filling with T4 DNA polymerase, and ligation. The 4.5-kb HindIII fragment from pDm αt-1 was subcloned into modified pBSIIKS+ to yield pBSIIKS+α1-tub. The bicoid 3.0-kb SalI-EcoRI fragment obtained from p902 and a 125-bp BamHI-SalI fragment containing the sequence encoding the HA1 epitope (32) were subcloned by a three-fragment ligation into pBSIIKS+α1-tub digested with BamHI and EcoRI and dephosphorylated, yielding pBSII-5T (Fig. 1). The resulting 5.6-kb insert in pBSIIKS+ contains 5′ sequences of the tubulin gene, including the promoter and the first 750 nucleotides of the tubulin mRNA sequence, joined with the 3′-most 2,044 bp of the bicoid gene, from the SalI site to the polyadenylation site plus the 3′ flanking sequences (bicoid position 2765 up to the EcoRI site). The insertion of the HA1 DNA cassette fuses in frame the protein coding sequences of α1-tub and bicoid. The XhoI-NotI fragment containing the 5′α1-tub/3′bicoid hybrid was subcloned into CaSpeR4 to yield CaSpeR4-5T.
The DNA sequences of the blunt-end junctions and the insertion sites of the HA1 DNA cassettes were all checked by DNA sequencing with nearby primers.
Construction of the 5′ rpA1/3′ bicoid deletion derivatives.
Four DNA fragments were prepared by PCR with pGEM-3B as a template and the primers listed in Table 1. These PCR products were digested with NheI and MluI, and the resulting fragments were inserted into the plasmid pGEM-3B, which had been digested with NheI and MluI and dephosphorylated, to yield four pGEM-5′rpA1/Δ3′bicoid plasmids with various deletions in the bicoid sequence: pGEM-3BA (deletion at positions 2765 to 4006), -3BB (deletion at positions 4007 to 4104), -3BC (deletion at positions 3137 to 4104), and -3BD (deletion at positions 2765 to 3136) (Fig. 1).
TABLE 1.
Oligonucleotides used in 3B deletion derivative construction, RNase H treatment, and BIE constructsa
| Oligonucleotide | Sequencea | Restriction enzyme | Gene (positions) |
|---|---|---|---|
| 3BA upper strand | 5′ GGCGCTAGCAATTAGCCTGGACGAGAGGCGT 3′ | NheI | bicoid (4007–4029) |
| 3BA or 3BD lower strand | 5′ GATTACGCCCAAGAGAAACATT 3′ | bicoid (4473–4452) | |
| 3BB or 3BC upper strand | 5′ CGACTACGCCGCTAGCAGTTCG 3′ | HA1 DNA cassette (47–65), bicoid (2765–2767) | |
| 3BB lower strand | 5′ GGGACGCGTCTAATTGAAGCAGTAGGCAAAC 3′ | MluI | bicoid (4012–3991) |
| 3BC lower strand | 5′ GGGACGCGTCTATCCGCCGATGCCGATGCCACAC 3′ | MluI | Stop codon; bicoid (3136–3115) |
| 3BD upper strand | 5′ GCCGCTAGCCCTTGCGCCATCGCCGTTGGCG 3′ | NheI | bicoid (3137–3158) |
| ΔNRE | 5′ GTACAATCAGGAACAACAGTGGT 3′ | bicoid (4079–4057) | |
| BIES | 5′ CCGGCCCTGGACGAGAGGCGTGTTAGAGAGTTTCATTAGCTTTAGGTTG 3′ | NgoMI | bicoid (4013–4055) |
| BIEA | 5′ CCGGCAACCTAAAGCTAATGAAACTCTCTAACACGCCTCTCGTCCAGGG 3′ | NgoMI | bicoid (4055–4013) |
Underlining indicates added nucleotides. Boldface type indicates endonuclease restriction sites.
The parts inserted by PCR in pGEM-5′rpA1/Δ3′bicoid plasmids were verified by DNA sequencing. The NotI-EcoRI fragments containing the 5′rpA1/Δ3′bicoid hybrids were subcloned into CaSpeR4 to yield CaSpeR-3BA, -3BB, -3BC, and -3BD.
rpA1-BIEsense and rpA1-BIEantisense.
To distinguish the BIE-containing rpA1 gene from the endogenous rpA1 gene, a fragment of 500 nucleotides from the human transferrin receptor 3′ UTR, located upstream of the iron regulatory sequence (28), was made by PCR and inserted at the NgoMI site of the 3′ UTR of the rpA1 gene, which was cloned in pGEM3. Then, the 5′ NgoMI site was destroyed by partial digestion and filling in. Two complementary oligonucleotides, the sense BIE (BIES) and the antisense BIE (BIEA) (Table 1), were annealed, and the resulting fragment was inserted at the remaining NgoMI site.
The rpA1 gene (a 2.4-kb BamHI fragment described in reference 24), containing the 0.5-kb transferrin fragment and the 43-nucleotide BIE in either orientation, was subcloned into CaSpeR4 to yield CaSpeR-BIES and CaSpeR-BIEA.
P-element-mediated transformation and establishment of transformed lines.
CaSpeR-5B, -3B, -5T, -3BA, -3BB, -3BC, -3BD, -BIES, and -BIEA constructs (500 μg/ml) were coinjected with phs-π helper plasmid DNA (100 μg/ml) (30) into yellow white mutant embryos (23, 26). Transformants were selected by mating G0 adults with yellow white flies and screening for flies having orange eyes. Homozygous stocks were generated for all the transformant lines used in this work. Transformant lines with BBT and with the nanos response element deleted (ΔNRE transformants) were kind gifts from P. Macdonald and R. Wharton, respectively (18, 34).
RNA analysis.
Ovaries were taken from virgin females deprived of yeast for 4 to 12 days (to prevent egg laying) and from well-fed, mated females. All dissections were done in Ringer’s solution (3). Embryos were collected for 1 h from well-fed females on agar-molasses plates and left to age at 25°C for different lengths of time. The synchrony of the collections was checked by staging an aliquot of dechorionated embryos. The percentage of unfertilized eggs was determined, and only collections containing less than 5% unfertilized eggs were analyzed. A low proportion of unfertilized eggs is important for quantitative analysis of bicoid mRNA decay because bicoid mRNA is stable in laid, unfertilized eggs (see Results). Ovaries and embryos were kept at −80°C after being frozen in liquid nitrogen.
Each sample of ovaries and embryos was homogenized in 1× binding buffer (0.5 M NaCl, 10 mM Tris-HCl [pH 8.0], 1 mM EDTA, 1% sodium dodecylsulfate) containing proteinase K (100 mg/ml) and incubated at 55°C for 2 h. After centrifugation for 10 min at 16,000 × g, the homogenates were passed three times on oligo(dT) columns to recover the poly(A)+ RNA. The columns were washed with 1X binding buffer without sodium dodecyl sulfate and the poly(A)+ RNA was recovered by elution with Tris-EDTA (10 mM Tris [pH 8.0], 1 mM EDTA) and precipitated with ethanol. For the experiments involving BIES and BIEA constructs, total RNA instead of poly(A)+ RNA was analyzed. Oligonucleotide-mediated RNase cleavage of bicoid mRNA was carried out according to the method of Brown and Harland (7). The RNA samples were fractionated by electrophoresis in denaturing 1 or 1.5% agarose-formaldehyde gels (27) and transferred to GeneScreen nylon membranes (NEN Research Products) by using the manufacturer’s recommendations. The membranes were UV irradiated, baked, prehybridized in Church’s buffer (8), and hybridized in the same buffer with α-32P-labeled probes synthesized by random primer labeling (15). Northern blots were quantified with a PhosphorImager (Molecular Dynamics). The following plasmids were used to prepare the probes: bicoid cDNA p1122, pGEM3rpA1, and pBSIIKS+α1-tub.
RESULTS
bicoid mRNA stability is developmentally regulated.
To determine whether bicoid mRNA is constitutively unstable or whether its stability is regulated in development, we measured mRNA decay in retained oocytes, in unfertilized eggs, and in early embryos.
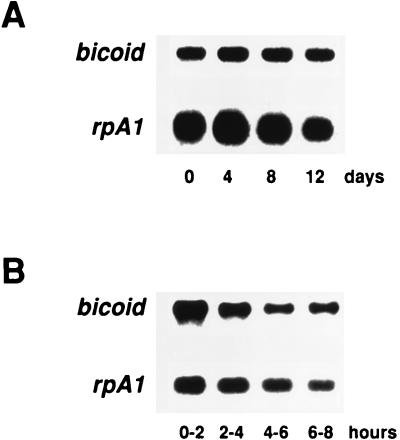
To measure mRNA stability in retained oocytes, we took advantage of the fact that females deprived of yeast retain their oocytes for extended periods of time (35). These females accumulate mature oocytes in which transcription and translation are arrested. Since no transcription takes place in these mature oocytes, determination of bicoid mRNA abundance as a function of time provides a direct measure of turnover rate. Polyadenylated RNAs from ovaries of rapidly laying females and from ovaries of females that retained their oocytes for 4, 8, and 12 days were analyzed on Northern blots. Like that of the stable rpA1 mRNA, the intensity of the bicoid signal remained constant over 12 days (Fig. 2A). Thus, bicoid mRNA is completely stable for up to 12 days in retained oocytes.
FIG. 2.
bicoid mRNA is stable for at least 12 days in retained oocytes and for at least 8 h in activated, unfertilized eggs. Polyadenylated mRNAs were prepared from ovaries of 3BA transgenic females that retained their oocytes for 0, 4, 8, or 12 days (A) and from laid (activated), unfertilized eggs from 3BA transgenic females, 0 to 2, 2 to 4, 4 to 6, or 6 to 8 h after oviposition (B), and analyzed on Northern blots with bicoid and rpA1 mRNA probes. Similar results were obtained in at least two independent experiments. Identical results were obtained for the 3BA hybrid mRNA (data not shown). rpA1 mRNA served as a loading control. mRNA sizes: bicoid mRNA, 2.6 kb; rpA1 mRNA, 0.6 kb.
In many instances, ongoing translation is required for mRNA degradation (5, 33). The absence of translation could explain why bicoid mRNA is completely stable in retained oocytes. However, bicoid mRNA translation is activated in laid eggs, even if they are unfertilized (11). We measured the stability of bicoid mRNA in laid, unfertilized eggs of different ages. As shown in Fig. 2B, the intensity of the bicoid signal relative to the rpA1 signal was constant for up to 8 h. Thus, bicoid mRNA in eggs is stable even if it is translated.
The disappearance of bicoid mRNA during early embryogenesis could be due either to the activation of a degradation pathway soon after fertilization or to the onset of degradation at a later time. Since bicoid is strictly maternal, changes in bicoid mRNA abundance in embryos provides a direct measure of decay. Figure 3 shows a detailed analysis of bicoid mRNA abundance during the first 4 h of embryogenesis. Precisely staged embryos were analyzed on Northern blots (Fig. 3A), and the signals were quantified with a PhosphorImager (Fig. 3B). The results showed that bicoid mRNA is stable during the first 2 h of embryogenesis and is then rapidly degraded between 2 and 3 h after fertilization. After 3 h of embryogenesis, bicoid mRNA was barely detectable. From these results, we estimate the bicoid mRNA half-life to be less than 30 min between 2 and 3 h of development. Thus, bicoid mRNA decay is activated at the end of the syncytial blastoderm stage of embryogenesis.
FIG. 3.
bicoid mRNA is stable during the first 2 h of embryogenesis and is rapidly degraded after this time. (A) Polyadenylated RNAs prepared from 0- to 1-, 1- to 2-, 2- to 3-, and 3- to 4-h-old embryos were analyzed on a Northern blot probed with bicoid and with rpA1 mRNA as a loading control. mRNA sizes are indicated in the legend to Fig. 2. (B) This histogram represents a quantitative analysis of data from seven independent experiments similar to the experiment presented in panel A. The intensity of the bicoid (bcd) mRNA signal (relative to the 0- to 1-h signal, given in arbitrary units) was plotted versus the time of development, in hours. The bicoid mRNA signals were normalized to those of the rpA1 loading control. Error bars represent standard deviations.
A bicoid mRNA destabilizing sequence is contained in the 3′ half of the message.
The initial stability of bicoid mRNA and its subsequent rapid degradation suggest two possible regulatory mechanisms: (i) protection from degradation until 2 h of development and (ii) activation of degradation after 2 h of development. The first mechanism predicts that deletion of the regulatory element would destabilize bicoid mRNA. The second mechanism predicts that deletion of the regulatory element would prevent the destabilization of bicoid mRNA. To distinguish between these two possibilities and to localize the putative regulatory determinant in bicoid mRNA, we constructed fusion genes containing sequences from bicoid and from stable mRNAs. These genes were transformed into flies and mRNA stability was measured in transgenic embryos.
Three constructs were initially tested: 5B, 3B, and 5T (Fig. 1). Construct 5B contains the entire 5′ half of bicoid mRNA, while the reciprocal constructs 3B and 5T contain the 3′ half of bicoid mRNA (Fig. 1). All constructs are driven by promoters that are active during oogenesis and inactive during the first 3 h of embryogenesis (α1-tubulin, rpA1, and hsp26/sgs3 promoters). Thus, the decrease in mRNA abundance during early embryogenesis serves as a measure of their degradation rates. Polyadenylated RNAs were extracted from staged embryos of transgenic lines. The abundance of bicoid, hybrid, and reference (rpA1 or tubulin) mRNAs was measured by Northern blot analysis (Fig. 4). In each lane, the signal from the reference mRNA provided a measure of the amount of mRNA analyzed.
FIG. 4.
5B mRNA is significantly more stable than bicoid mRNA, while 3B and BBT mRNAs are unstable. Polyadenylated RNAs prepared from 0- to 1-, 1- to 2-, 2- to 3-, and 3- to 4-h-old embryos derived from 5B, 3B 5T, and BBT transgenic flies were analyzed on Northern blots hybridized with bicoid and rpA1 or tubulin mRNA probes. The rpA1 and tubulin signals served to quantify the amount of RNA analyzed in each lane. mRNA sizes: bicoid mRNA, 2.6 kb; 5B mRNA, 1.5 kb; 3B mRNA, 1.9 kb; BBT mRNA, 2.2 kb; and rpA1 mRNA, 0.6 kb.
Figures 4 and 5 present the results. While none of the hybrid mRNAs was completely stable, the mRNAs containing 3′ bicoid sequences (3B and 5T) were degraded more efficiently than the mRNAs containing 5′ bicoid sequences (5B). Quantitative analysis showed that 5B mRNA is threefold more abundant than bicoid mRNA at 2 to 3 h of development and eightfold more abundant at 3 to 4 h (Fig. 5). In contrast, the stability of the 3B and 5T mRNAs was regulated similarly to that of bicoid mRNA (Fig. 4B and 5). These results suggest that a major destabilizing sequence resides in the 3′ half of bicoid mRNA, while a weaker destabilizing sequence resides in the 5′ half.
FIG. 5.
The 3′ half of bicoid mRNA contains the main destabilizing element. Each histogram represents the quantitative analysis of experiments such as those shown in Fig. 4 and combines data from at least three independent experiments. The intensities of bicoid (bcd) mRNA signals (relative to the 0- to 1-h signal, given in arbitrary units) were plotted versus the time of development, in hours. The abundance of bicoid mRNA was normalized to rpA1 or tubulin loading controls. Error bars represent standard deviations. No standard deviation is shown for 3B, since the histogram shows the quantification of one experiment. However, the same result as for 3B was obtained for 5T, a fusion with the tubulin gene containing exactly the same bicoid sequence.
bicoid mRNA is localized at the anterior tip of the embryo through sequences contained within 625 nucleotides of its 3′ UTR (18). The selective degradation of bicoid mRNA at cellularization could be dependent on its localization. To address this hypothesis, we measured the stability of a bicoid mRNA in which most of the 3′ UTR was replaced by the 3′ UTR of the tubulin gene (Fig. 1, BBT). Figures 4C and 5 show that BBT mRNA is initially stable and is later degraded to become barely detectable at 3 to 4 h of development, as is the case for endogenous bicoid.
The instability of BBT mRNA suggests that localization of bicoid mRNA is not necessary for its specific degradation in the early embryo and that the main bicoid destabilization sequence is distinct from the localization sequence. This assumption is supported by experiments presented later in this paper.
The main BIE is contained within the 92 nucleotides immediately following the translation termination codon.
The results above suggested that the main instability element is located within the 3′ half of bicoid mRNA, between the SalI and MluI sites (Fig. 1). To determine the location of the instability element more precisely, a series of deletion derivatives (3BA, 3BB, 3BC, and 3BD) from the 3B construct were generated (Fig. 1). All constructs retained the terminal 733-nucleotide segment which is deleted in BBT. In 3BA the complete bicoid 3′ UTR remained, in 3BB the 92 nucleotides after the stop codon were deleted, in 3BC the last quarter of the bicoid translated region plus the 92 nucleotides after the stop codon were deleted, and in 3BD, the third quarter of the bicoid translated region was deleted. All these constructs were under the control of the rpA1 promoter. The constructs were transformed into the germ line and the abundance of the corresponding mRNAs was analyzed as before. As shown in Fig. 6 and 7, the 3BA and 3BD mRNAs were unstable, while the 3BB and 3BC mRNAs were stable. Thus, the 92-nucleotide sequence that was deleted in 3BB is required for mRNA destabilization at the time of cellularization of the blastoderm.
FIG. 6.
3BA and 3BD mRNAs are unstable, while 3BB and 3BC mRNAs are stable. Polyadenylated RNAs prepared from 0- to 1-, 1- to 2-, 2- to 3-, and 3- to 4-h-old embryos from 3BA, 3BB, 3BC, and 3BD transgenic flies were analyzed on Northern blots hybridized with bicoid and rpA1 mRNA probes. rpA1 mRNA served as a loading control. mRNA sizes: bicoid mRNA, 2.6 kb; 3BA mRNA, 1.2 kb; 3BB mRNA, 1.8 kb; 3BC mRNA, 1.4 kb; 3BD mRNA, 1.6 kb; and rpA1 mRNA, 0.6 kb.
FIG. 7.
The main bicoid mRNA destabilizing element is located within the first 43 nucleotides after the stop codon. Each histogram represents the quantitative analysis of experiments such as those shown in Fig. 8 and combines data from at least three independent experiments. The intensities of the bicoid (bcd) mRNA signals (relative to the 0- to 1-h signal, given in arbitrary units) were plotted versus the time of development, in hours. The abundance of bicoid mRNA was normalized to the rpA1 loading control. Error bars represent standard deviations.
The main instability element is distinct from the NRE.
The 92 nucleotides after the stop codon of the bicoid mRNA contain an NRE sequence. This element is also present in hunchback mRNA, where it is necessary for translational repression in the posterior part of the embryo. It is not clear why an NRE is present in bicoid mRNA because Nanos protein and bicoid mRNA are not normally colocalized. However, translation of bicoid mRNA is repressed via the NRE when Nanos is ectopically localized in the anterior part of the embryo in BicaudalD mutants (34). To assess the role of the NRE in bicoid mRNA destabilization, we analyzed ΔNRE transgenic flies carrying a variant of the bicoid gene with a deletion of the 45 3′-most nucleotides of the 92-nucleotide element (Fig. 1). The size difference between the messages derived from wild-type and ΔNRE bicoid genes was too small to separate them on Northern blots. Instead, an RNase H assay was used. Poly(A)+ RNA from different stages was hybridized with an oligonucleotide complementary to a sequence within the NRE and digested with RNase H. This treatment was expected to cleave bicoid mRNA into two fragments (1.6 and 1.0 kb) and leave the ΔNRE mRNA intact. The Northern blot in Fig. 8 and quantitative analysis in Fig. 7 show that the ΔNRE mRNA (2.6-kb fragment) is as unstable as the wild-type bicoid mRNA (1.6- and 1.0-kb fragments). Consequently, the 45 nucleotides containing the NRE are not necessary for the degradation of bicoid mRNA in early embryos. Moreover, these results show that the BIE is contained within the first 43 nucleotides after the stop codon.
FIG. 8.
Deletion of the NRE does not affect bicoid mRNA stability. The hollow arrow in the diagram at the top indicates the position of the NRE sequence that was deleted from the bicoid gene. Polyadenylated RNAs were prepared from control (0- to 1-h-old untransformed embryos) and 0- to 1-, 1- to 2-, 2- to 3-, and 3- to 4-h-old embryos from ΔNRE transgenic flies. The RNAs were hybridized with an oligonucleotide (Table 1) complementary to a sequence missing in the ΔNRE construct and digested with RNase H. The RNAs were analyzed on a Northern blot hybridized with bicoid (bcd) and rpA1 mRNA probes. The bicoid mRNA is cleaved into two fragments of 1.6 and 1.0 kb, while the ΔNRE mRNA is unaffected by the treatment. rpA1 mRNA served as a loading control. The control lane shows that wild-type bicoid mRNA is cleaved to completion by the RNase H treatment. mRNA sizes: ΔNRE mRNA, 2.6 kb, and rpA1 mRNA, 0.6 kb.
The BIE is sufficient to destabilize an otherwise-stable mRNA.
To determine whether the 43-nucleotide BIE is sufficient to confer proper regulation on a heterologous mRNA, the BIE was inserted in the 3′ UTR of the otherwise-stable rpA1 mRNA to yield rpA1-BIE. A 0.5-kb fragment of an unrelated sequence (human transferrin receptor 3′ UTR) was inserted into the rpA1 gene 3′ UTR to distinguish the rpA1-BIE mRNA from the endogenous rpA1 mRNA. The stability of rpA1 mRNA was not affected by the insertion of this sequence. Figure 9 shows that when the BIE is inserted in the sense orientation, rpA1 mRNA is destabilized with the same kinetics as bicoid mRNA (Fig. 9, left; compare with Fig. 3). In contrast, when the BIE is inserted in the antisense orientation, rpA1 is completely stable (Fig. 9, right). Therefore, the BIE is sufficient to destabilize a stable mRNA.
FIG. 9.
The BIE is sufficient to confer regulated instability on a heterologous mRNA. (Top) Schematic diagram of the constructs used to obtain transgenic flies. Stippled rectangles, rpA1 gene UTRs; black rectangles, rpA1-coding regions; white rectangles, 500-nucleotide-long fragments from the transferrin gene (3′ UTR TfR) inserted to tag the recombinant gene; arrows, the 43-nucleotide BIE (BIES, right-pointing arrow; BIEA, left-pointing arrow). (Middle) Representative Northern blots of total RNA extracted from synchronized embryos (ages are at the bottom of each lane) from transgenic flies carrying either the BIES or the BIEA construct. (Bottom) Graphs showing quantification of three independent experiments similar to those shown in the middle part. Error bars represent standard deviations.
DISCUSSION
Little was known about bicoid mRNA metabolism beyond the fact that by 4 h of embryogenesis, the message is completely degraded (6). Among possible models to explain the observed pattern—constitutive instability, destabilization at fertilization, and activation of degradation in developing embryos—we found that the latter model applies. bicoid mRNA is destabilized at the time of cellularization of the blastoderm, which is the time of major activation of the zygotic genome (2, 21), raising the possibility that bicoid degradation requires de novo transcription.
We found that bicoid mRNA is stable for over 1 week in retained oocytes. Perhaps this is not surprising, since these oocytes are metabolically inactive. However, oviposited, unfertilized eggs become completely activated (19). For instance, meiosis is completed and translation is activated, including that of bicoid mRNA (11). Surprisingly, bicoid mRNA was completely stable for at least 8 h in oviposited, unfertilized eggs. This observation makes a “clock” mechanism less likely and supports the concept that zygotic transcription is required for bicoid destabilization. Moreover, the stability of bicoid mRNA in unfertilized eggs may explain the larger-than-normal Bicoid protein amounts in such eggs (11). In contrast to bicoid, an rpA1/fushi tarazu hybrid mRNA is rapidly degraded in activated, unfertilized eggs (25), suggesting that the activities that degrade bicoid and fushi tarazu mRNAs are distinct.
All mRNAs carrying the 43-nucleotide BIE (3B, 5T, BBT, 3BA, 3BD, ΔNRE, and BIES) are unstable after 2 h of development, while all mRNAs lacking the BIE (3BB and 3BC) are stabilized. Therefore, the BIE is necessary for regulation of bicoid mRNA stability. Moreover, since mRNAs lacking the BIE (5B, 3BB, and 3BC) are much more stable in early embryos, it can be argued that the BIE acts to destabilize bicoid mRNA after 2 h of development as opposed to being necessary to protect bicoid mRNA from degradation until 2 h of development. This is in complete agreement with the finding that the BIE is sufficient to destabilize the heterologous rpA1 mRNA. The position of the BIE does not seem to be important because in the BIES construct, the BIE is 600 nucleotides from the stop codon instead of next to it. The BIE appears to act as an independent, developmentally regulated target of mRNA degradation because the sequence context in which it is placed is irrelevant. The BIE is likely to provide a binding site for a factor or complex involved in mRNA degradation. Binding sites could be recognized as secondary structures (5). However, no stable secondary structure was predicted when the computer program of Zuker (38) was used to fold the BIE sequence.
The 5B transcript is much more stable than endogenous bicoid mRNA but is not as stable as rpA1 mRNA. The 5′ half of bicoid mRNA has much weaker destabilizing activity than the BIE, since 5B mRNA abundance is reduced 2-fold from the first hour to the fourth hour, while 3B mRNA abundance is reduced 20-fold. Possibly, another destabilization element much weaker than the BIE is present in the 5′ half of bicoid mRNA. Alternative explanations, such as the creation of a fortuitous destabilizing element at the junction of the chimeric construct, cannot be ruled out.
A connection between mRNA translation and stability was suggested for a number of mRNA degradation pathways (for reviews, see references 5 and 33). However, in the case of bicoid, the translation status seems not to be critical for control of mRNA decay. First, while bicoid mRNA is continuously translated from the time of fertilization, its degradation is activated only at cellularization of the blastoderm. Thus, bicoid mRNA is stable both when translated (in activated, unfertilized eggs and in early embryos) and when not translated (during oogenesis and in retained eggs). Second, when a premature stop codon was introduced by site-directed mutagenesis into codon 218 of the bicoid cDNA and the construct was analyzed in transgenic flies, the transgenic mRNA was degraded with a temporal profile indistinguishable from that of wild-type bicoid mRNA (21a). Third, the rate of translation appears to have no effect on the regulation of bicoid mRNA stability. The 3B mRNA contains the rpA1 gene 5′ UTR, which is poorly translated during early embryogenesis (22). Despite the presence of the rpA1 gene 5′ UTR, 3B mRNA is as unstable as the endogenous bicoid mRNA. Moreover, 5T contains the 5′ half of the actively translated tubulin mRNA linked to the same 3′ half of the bicoid mRNA as in 3B. 5T is degraded with the same kinetics as 3B.
Transcript localization could be a mechanism for targeting specific mRNAs for degradation (20, 37). However, BBT mRNA, which lacks localization sequences but contains the BIE, is unstable. Moreover, bicoid mRNA and fushi tarazu mRNA are apically localized in early embryos (9) and yet, at the same stage, the former is stable while the latter is unstable. Most importantly, BIES mRNA is presumed not to be localized and is degraded with the same kinetics as bicoid mRNA. These observations suggest that bicoid mRNA localization and mRNA degradation are two independent processes.
While no similarities were found by comparing the entire BIE sequence with sequences in databases (using the BLAST program [1]), the short sequence UUUCAUU present in the BIE was found to appear in a subset of the genes listed in the EMBL UTR database. Of about 1,800 UTRs searched, 112 (∼6%) contained the UUUCAUU motif. Additional genes not listed in the UTR database were also found to have this motif (data not shown). Interestingly, a significant proportion of the genes retrieved from the UTR database (∼35%) could be classified among four classes of regulatory genes whose mRNA and protein concentrations are expected to change rapidly as a function of time. In the majority of the genes (∼82%), the motif was present in the 3′ UTR. The consensus for the 21 nucleotides containing the UUUCAUU motif (from a total of 137 motifs; the Genetics Computer Group package, University of Wisconsin, was used) is DDDDDHDUUUACUUDDDHWHH (D = not C, H = not G, and W = A or T), indicating that the UUUCAUU motif is embedded in a region from which the C residue is usually excluded. The presence of the UUUCAUU motif in several classes of genes suggests the existence of a common pathway for mRNA destabilization. Further research is needed to verify the validity of this hypothesis.
ACKNOWLEDGMENTS
We thank Robert Cohen and Kam Cheung (Columbia University) for providing a plasmid containing the sgs3 promoter and the nurse cell-specific enhancer from hsp26. We thank Paul Macdonald (Stanford University) for flies transformed with the BBT construct and for the plasmid p1122, containing the bicoid cDNA. We thank Robin Wharton (Duke University) for flies transformed with the ΔNRE construct. P.S. is very grateful to Lukas Kuehn (ISREC, Epalinges, Switzerland) for giving him the opportunity to finish this study in his laboratory and for his continuous support. We thank Claude Bonnard for his help in use of the Genetics Computer Group package. We thank Marilyn Doman, Ann Riedl, and Maria Goreti Freitas-Sibajev for their comments on the manuscript.
This work was supported by a grant from the National Institutes of Health.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Anderson K W, Lengyel J A. Rates of synthesis of major classes of RNA in Drosophilaembryos. Dev Biol. 1979;70:217–231. doi: 10.1016/0012-1606(79)90018-6. [DOI] [PubMed] [Google Scholar]
- 3.Ashburner M. Drosophila: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 4.Audic Y, Omilli F, Osborne H B. Postfertilization deadenylation of mRNAs in Xenopus laevisembryos is sufficient to cause their degradation at the blastula stage. Mol Cell Biol. 1997;17:209–218. doi: 10.1128/mcb.17.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belasco J, Brawerman G. Control of messenger RNA stability. San Diego, Calif: Academic Press; 1993. [Google Scholar]
- 6.Berleth T, Burri M, Thoma G, Bopp D, Richstein S, Frigerio G, Noll M, Nüsslein-Volhard C. The role of localization of bicoid RNA in organizing the anterior pattern of the Drosophilaembryo. EMBO J. 1988;7:1749–1756. doi: 10.1002/j.1460-2075.1988.tb03004.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown B D, Harland R M. Endonucleolytic cleavage of a maternal homeobox mRNA in Xenopusoocyte. Genes Dev. 1990;4:1925–1935. doi: 10.1101/gad.4.11.1925. [DOI] [PubMed] [Google Scholar]
- 8.Church G M, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davis I, Ish-Horowicz D. Apical localization of pair-rule transcripts requires 3′ sequences and limits protein diffusion in the Drosophilablastoderm embryo. Cell. 1991;67:927–940. doi: 10.1016/0092-8674(91)90366-7. [DOI] [PubMed] [Google Scholar]
- 10.Driever W, Nüsslein-Volhard C. A gradient of bicoid protein in Drosophilaembryos. Cell. 1988;54:83–93. doi: 10.1016/0092-8674(88)90182-1. [DOI] [PubMed] [Google Scholar]
- 11.Driever W, Nüsslein-Volhard C. The bicoid protein is a positive regulator of hunchback transcription in the early Drosophilaembryo. Nature. 1989;337:138–143. doi: 10.1038/337138a0. [DOI] [PubMed] [Google Scholar]
- 12.Driever W, Thoma G, Nüsslein-Volhard C. Determination of spatial domains of zygotic gene expression in the Drosophila embryo by affinity of binding sites for the bicoidmorphogen. Nature. 1989;340:363–367. doi: 10.1038/340363a0. [DOI] [PubMed] [Google Scholar]
- 13.Edgar B A, Datar S A. Zygotic degradation of two maternal Cdc25 mRNAs terminates Drosophila’s early cell cycle program. Genes Dev. 1996;10:1966–1977. doi: 10.1101/gad.10.15.1966. [DOI] [PubMed] [Google Scholar]
- 14.Edgar L G, Wolf N, Wood W B. Early transcription in Caenorhabditis elegansembryos. Development. 1994;120:443–451. doi: 10.1242/dev.120.2.443. [DOI] [PubMed] [Google Scholar]
- 15.Feinberg A P, Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1984;137:266–267. doi: 10.1016/0003-2697(84)90381-6. [DOI] [PubMed] [Google Scholar]
- 16.Frohnhöfer H G, Nüsslein-Volhard C. Organization of anterior pattern in the Drosophila embryo by the maternal gene bicoid. Nature. 1986;324:120–125. [Google Scholar]
- 17.Kalfayan L, Wensink P C. α-Tubulin genes of Drosophila. Cell. 1981;24:97–106. doi: 10.1016/0092-8674(81)90505-5. [DOI] [PubMed] [Google Scholar]
- 18.Macdonald P M, Struhl G. Cis-acting sequences responsible for anterior localization of bicoid mRNA in Drosophilaembryos. Nature. 1988;336:595–598. doi: 10.1038/336595a0. [DOI] [PubMed] [Google Scholar]
- 19.Mahowald A P, Goralski T J, Caulton J H. In vitro activation of Drosophilaeggs. Dev Biol. 1983;98:437–445. doi: 10.1016/0012-1606(83)90373-1. [DOI] [PubMed] [Google Scholar]
- 20.Mason J O, Williams G T, Neuberger M S. The half-life of immunoglobulin mRNA increases during B-cell differentiation: a possible role for targeting to membrane-bound polysomes. Genes Dev. 1988;2:1003–1011. doi: 10.1101/gad.2.8.1003. [DOI] [PubMed] [Google Scholar]
- 21.McKnight S L, Miller O L., Jr Ultrastructural patterns of RNA synthesis during early embryogenesis of Drosophila melanogaster. Cell. 1976;8:305–319. doi: 10.1016/0092-8674(76)90014-3. [DOI] [PubMed] [Google Scholar]
- 21a.Mouland, A., and M. Jacobs-Lorena. Unpublished observations.
- 22.Patel R, Jacobs-Lorena M. Cis-acting sequences in the 5′-untranslated region of the ribosomal protein A1 mRNA mediates its translational regulation during early embryogenesis of Drosophila. J Biol Chem. 1992;267:1159–1164. [PubMed] [Google Scholar]
- 23.Pirrotta V. Vectors for P-mediated transformation in Drosophila. Biotechnol Ser. 1988;10:437–456. doi: 10.1016/b978-0-409-90042-2.50028-3. [DOI] [PubMed] [Google Scholar]
- 24.Qian S, Zhang J-Y, Kay M A, Jacobs-Lorena M. Structural analysis of the DrosophilarpA1 gene, a member of the eukaryotic ’A’ type ribosomal protein family. Nucleic Acids Res. 1987;15:987–1003. doi: 10.1093/nar/15.3.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Riedl, A., and M. Jacobs-Lorena. Unpublished data.
- 26.Rubin G M, Spradling A C. Genetic transformation of Drosophilawith transposable element vectors. Science. 1982;218:348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- 27.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 28.Schneider C, Owen M J, Banville D, Williams J G. Primary structure of human transferrin receptor deduced from the mRNA sequences. Nature. 1984;311:675–678. doi: 10.1038/311675b0. [DOI] [PubMed] [Google Scholar]
- 29.Schultz R M. Regulation of zygotic gene activation in the mouse. Bioessays. 1993;15:531–538. doi: 10.1002/bies.950150806. [DOI] [PubMed] [Google Scholar]
- 30.Steller H, Pirrotta V. P transposons controlled by the heat shock promoter. Mol Cell Biol. 1986;6:1640–1649. doi: 10.1128/mcb.6.5.1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.St. Johnston, D., W. Driever, T. Berleth, S. Richstein, and C. Nüsslein-Volhard. 1989. Multiple steps in the localization of bicoid RNA to the anterior pole of the Drosophila oocyte. Development 107(Suppl.):13–19. [DOI] [PubMed]
- 32.Surdej P, Jacobs-Lorena M. Strategy for epitope tagging the protein coding-region of any gene. BioTechniques. 1994;17:560–565. [PubMed] [Google Scholar]
- 33.Surdej P, Riedl A, Jacobs-Lorena M. Regulation of mRNA stability in development. Annu Rev Genet. 1994;28:263–282. doi: 10.1146/annurev.ge.28.120194.001403. [DOI] [PubMed] [Google Scholar]
- 34.Wharton R P, Struhl G. RNA regulatory elements mediate control of the Drosophila body pattern by the posterior morphogen nanos. Cell. 1991;67:955–967. doi: 10.1016/0092-8674(91)90368-9. [DOI] [PubMed] [Google Scholar]
- 35.Wyman R. The temporal stability of the Drosophilaoocyte. J Embryol Exp Morphol. 1979;50:137–144. [PubMed] [Google Scholar]
- 36.Yasuda G K, Schubiger G. Temporal regulation in the early embryo: is MBT too good to be true? Trends Genet. 1992;8:124–127. doi: 10.1016/0168-9525(92)90369-F. [DOI] [PubMed] [Google Scholar]
- 37.Zambetti G, Stein J, Stein G. Targeting of a chimeric human histone fusion mRNA to membrane-bound polysomes in HeLa cells. Proc Natl Acad Sci USA. 1987;84:2683–2687. doi: 10.1073/pnas.84.9.2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zuker M. On finding all suboptimal foldings of an RNA molecule. Science. 1989;244:48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]