SUMMARY
It is thought that mRNA-based vaccine-induced immunity to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) wanes quickly, based mostly on short-term studies. Here, we analyzed the kinetics and durability of the humoral responses to SARS-CoV-2 infection and vaccination using >8,000 longitudinal samples collected over a 3-year period in New York City. Upon primary immunization, participants with pre-existing immunity mounted higher antibody responses faster and achieved higher steady-state antibody titers than naive individuals. Antibody kinetics were characterized by two phases: an initial rapid decay, followed by a stabilization phase with very slow decay. Booster vaccination equalized the differences in antibody concentration between participants with and without hybrid immunity, but the peak antibody titers decreased with each successive antigen exposure. Breakthrough infections increased antibodies to similar titers as an additional vaccine dose in naive individuals. Our study provides strong evidence that SARS-CoV-2 antibody responses are long lasting, with initial waning followed by stabilization.
In brief
SARS-CoV-2 mRNA-based vaccine-induced immunity is thought to wane quickly based on short-term studies. Using longitudinal data, Srivastava et al. find that participants with hybrid immunity show faster, higher antibody responses after initial vaccination, but boosters evened out differences. Modeling of antibody kinetics revealed an initial rapid decay followed by a stabilization phase, challenging the idea that vaccine immunity fades quickly.
Graphical Abstract
INTRODUCTION
The emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in late 2019 sparked the global coronavirus disease 2019 (COVID-19) pandemic that is now in its 4th year. Vaccines to mitigate the impact of the pandemic were developed at record speed and have saved millions of lives. However, the emergence of SARS-CoV-2 variants1 and waning immunity2 have decreased the effectiveness of the vaccines against symptomatic disease.3 These two issues, the emergence of antigenically distinct SARS-CoV-2 variants and waning immunity, are often conflated and used interchangeably but represent two different phenomena.4 Most vaccines used in North America and Europe are based on lipid nanoparticles (LNPs) containing messenger RNA (mRNA) produced by Pfizer/BioNTech (BNT162b2) or Moderna (mRNA-1273), and the common perception now is that mRNA-based vaccine-induced immunity wanes quickly.5 However, this assumption is mostly based on data from short-term studies that include a very limited number of data points following peak responses.2,5
In March of 2020, the densely populated New York metropolitan area was hit with an exponential increase of severe SARS-CoV-2 infections, resulting in a staggering number of fatalities and a severely overburdened healthcare system.6–8 Due to short-ages of personal protective equipment, essential workers in the health care system were at high risk for infection. In response to this crisis, we established (1) a specific and sensitive SARS-CoV-2 binding enzyme-linked immunosorbent assay (ELISA) to measure humoral immune responses,9 and (2) an observational longitudinal cohort of health care workers of the Mount Sinai Health System to determine the kinetics of these humoral responses. This study, named Protection Associated with Rapid Immunity to SARS-CoV-2 (PARIS),10 aims to capture the dynamics of SARS-CoV-2 antibody responses to infection as well as vaccinations, to determine re-infection rates, and to assess correlates of protection in the context of individual immune histories.
With over 8,000 longitudinal study visits across a single cohort during the first 3 years of the pandemic, our investigation represents one of the most extensive and in-depth assessments of the longevity of SARS-CoV-2 immune responses to date. Using this longitudinal cohort, we determined the kinetics of antibody responses to spike protein after infections, during the primary immunization series, during monovalent and bivalent booster vaccination, as well as during breakthrough infections. Our findings indicate that, in contrast to common perception, COVID-19 mRNA vaccination induces long-lasting antibody responses in humans. The PARIS Study also provides insights into the effect of booster vaccination and breakthrough infections on the stability of antibody responses.
RESULTS
The longitudinal observational study design informs on individual immune histories
PARIS is an observational longitudinal study that enrolled 501 adults, mostly healthcare workers (Table 1) with or without pre-existing SARS-CoV-2 immunity. The first participants were enrolled in April 2020, when New York City emerged as one of the very early epicenters of the pandemic in the United States. We have conducted over 8,000 study visits with data and biospecimen collection spanning a 3-year period (April 2020 to March 2023).
Table 1.
Demographics and immune histories of the PARIS study participants
| Seropositivity pre-vaccine | Yes | No | Total |
|---|---|---|---|
|
| |||
| N = 183 | N = 313 | N = 496 | |
|
| |||
| Age | |||
|
| |||
| Mean – Avg (Std) | 42 (11.7) | 40 (11.6) | 41 (11.6) |
| Range | 21–77 | 22–74 | 21–77 |
|
| |||
| Gender - N (%) | |||
|
| |||
| Female | 118 (64%) | 216 (69%) | 334 (67%) |
| Male | 61 (33%) | 88 (28%) | 149 (30%) |
| Missing/other | 4 (2%) | 9 (3%) | 13 (3%) |
|
| |||
| Ancestry - N (%) | |||
|
| |||
| African American | 15 (8%) | 16 (5%) | 31 (6%) |
| Asian | 21 (11%) | 50 (16%) | 71 (14%) |
| White | 104 (57%) | 175 (56%) | 279 (56%) |
| Missing/other | 43 (23%) | 72 (23%) | 115 (23%) |
|
| |||
| Ethnicity - N (%) | |||
|
| |||
| Hispanic | 31 (17%) | 36 (12%) | 67 (14%) |
| Non-Hispanic | 133 (73%) | 230 (73%) | 363 (7%) |
| Missing/other | 19 (10%) | 47 (1%) | 66 (13%) |
|
| |||
| Primary vaccine series | |||
|
| |||
| Moderna | 35 (19%) | 76 (24%) | 111 (22%) |
| Pfizer | 127 (69%) | 215 (69%) | 342 (69%) |
| Not received | 15 (8%) | 20 (6%) | 35 (7%) |
| Other | 5 (3%) | 0 (0%) | 5 (1%) |
| Unknown | 1 (1%) | 2 (1%) | 3 (1%) |
|
| |||
| Vaccine dose 3 (%) | |||
|
| |||
| Moderna | 29 (16%) | 67 (21%) | 96 (19%) |
| Pfizer | 89 (49%) | 174 (56%) | 263 (53%) |
| Not received | 64 (35%) | 69 (22%) | 133 (27%) |
| Other | 0 (0%) | 1 (<1%) | 1 (<1%) |
| Unknown | 1 (1%) | 2 (1%) | 3 (1%) |
|
| |||
| Vaccine dose 4 (%) | |||
|
| |||
| Moderna | 1 (1%) | 3 (1%) | 4 (1%) |
| Moderna bivalent | 3 (2%) | 9 (3%) | 12 (2%) |
| Pfizer | 5 (3%) | 16 (5%) | 21 (4%) |
| Pfizer bivalent | 16 (9%) | 38 (12%) | 54 (11%) |
| Bivalent unspecified | 1 (1%) | 3 (1%) | 4 (1%) |
| Unknown | 1 (1%) | 1 (<1%) | 2 (<1%) |
| Not received | 156 (85%) | 243 (78%) | 399 (80%) |
|
| |||
| Vaccine dose 5 | |||
|
| |||
| Moderna bivalent | 0 (0%) | 1 (<1%) | 1 (<1%) |
| Pfizer bivalent | 5 (3%) | 9 (3%) | 14 (3%) |
| Not received | 178 (97%) | 303 (97%) | 481 (97%) |
|
| |||
| SARS-CoV-2 infections - N (%) | |||
|
| |||
| Never | 0 (0%) | 155 (50%) | 155(31%) |
| Once | 136 (74%) | 137 (44%) | 273 (55%) |
| Twice | 45 (25%) | 20 (6%) | 65 (13%) |
| Thrice | 2 (1%) | 1 (<1%) | 3 (1%) |
|
| |||
| Job description - N (%) | |||
|
| |||
| Clinician | 52 (28%) | 99 (32%) | 151 (30%) |
| PA/Nursing | 37 (20%) | 84 (27%) | 121 (24%) |
| PhD/Trainee | 19 (10%) | 47 (15%) | 66 (13%) |
| Social worker | 6 (3%) | 2 (1%) | 8 (2%) |
| Family member | 11 (6%) | 3 (1%) | 14 (3%) |
| Midwife | 2 (1%) | 3 (1%) | 5 (1%) |
| Other | 52 (28%) | 65 (21%) | 117 (24%) |
| Missing | 4 (2%) | 10 (3%) | 14 (3%) |
Of the participants, 67% were female, and 56% self-identified as white. The mean age at study enrollment was 41 years (Table 1). At the first study visit, 62% of the participants had no measurable SARS-CoV-2 spike-binding antibodies (naive, seronegative). At each study visit, we collected data and biospecimen (e.g., blood and saliva). Study visits were scheduled at shorter intervals (2–4 weeks) from study entry through week 8 after enrollment, but the intervals between visits were extended (to approximately 4–8 weeks) for follow-up visits after week 8. Ad hoc study visits were included at short intervals (e.g., weekly) after immune events such as vaccination or infection. Serum samples were used to measure antibodies binding to the ancestral spike protein using an established in-house method that was developed early in the pandemic.9 Of note, as with all antibody binding assays, binding antibody values are influenced by both the abundance of antibodies as well as their affinity/avidity. This needs to be taken into consideration when interpreting study results since changes in titer may be influenced by increases in antibodies but also by increases in antibody affinity.
465 of 501 PARIS participants (93%) were vaccinated against SARS-CoV-2 using mRNA vaccines: 342/465 received two doses of Pfizer BNT162b2, and 111/465 received two doses of Moderna mRNA-1273. A small proportion of participants received one dose of the Johnson & Johnson vaccine (Ad26.COV2.S) or two doses of the CoviShield™ (AZD1222) vaccine as their primary SARS-CoV-2 immunization regimen (Table 1). The number of vaccine doses administered to a given PARIS participant ranged from two to six doses. Briefly, 366/465 (79%) of PARIS participants who completed their primary immunizations subsequently elected to receive a 3rd vaccine dose (“booster”). Of these 366 participants, 97 (27%) received a 2nd booster (4th vaccine dose overall; 27/97 monovalent ancestral booster vaccine versus 70/97 bivalent ancestral/BA.5 vaccine). Of these 97 participants, 15 opted for a 3rd booster (5th vaccine dose overall). Of these 15 participants, one opted for a 4th booster (6th vaccine dose overall). Lastly, 14/27 participants with two monovalent boosters elected to get the bivalent booster.
Infection prior to immunization shapes antibody responses to primary immunization resulting in higher antibody levels compared with participants with vaccine-only immunity
38% of the study participants entered the PARIS study having detectable spike-binding immunoglobulin G (IgG) antibodies, albeit at highly variable titers (Figure 1, area under the curve [AUC] range at baseline: 12.5– 4,189). Infection-induced anti-spike antibodies showed relatively high stability over approximately 10 months of pre-vaccine follow-up. A detailed analysis of this phase of the study has been reported previously.11
Figure 1. Longitudinal SARS-CoV-2 spike-binding antibody titers in 496 PARIS participants over 3 years provide personalized immune histories.
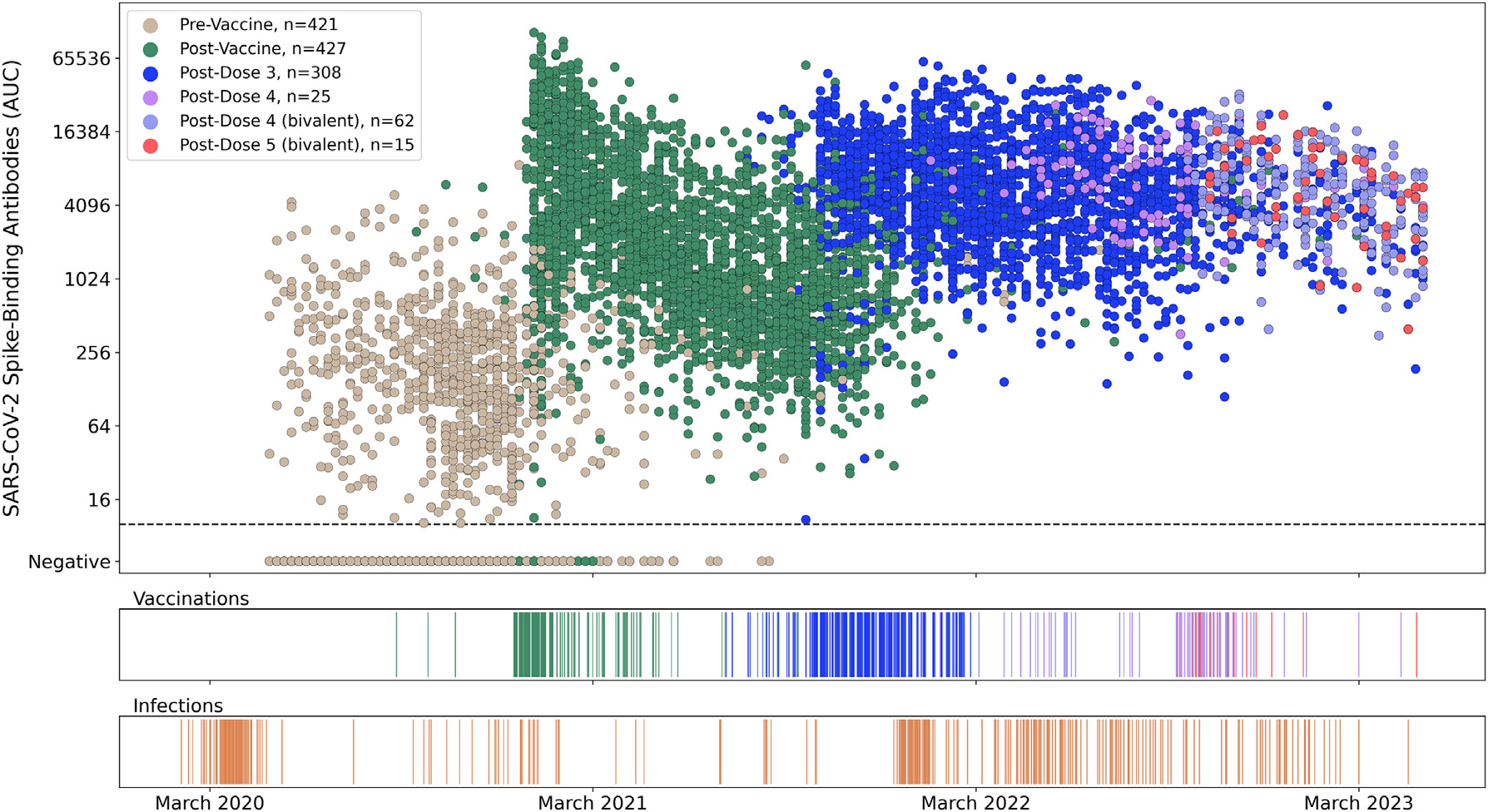
(A) Each dot represents a distinct study visit at which spike-binding antibody titers were measured. Samples are colored by prior vaccination status (2,091 samples pre-vaccination, 3,180 samples post-primary immunization [dose 1 and dose 2], 2,364 samples post-dose 3, 110 samples post-monovalent dose 4, 240 samples post-bivalent dose 4, and 56 samples post-bivalent dose 5). Each SARS-CoV-2 antibody titer measurement (AUC, area under the curve) is anchored on the Monday of the corresponding week. A small amount of normally distributed noise has been added to the log2-transformed data (σ = 0.1), with >95% of transformed values within 15% of the original value to preserve participants’ confidentiality.
(B and C) SARS-CoV-2 infections (B) and vaccinations (C) events are depicted on the same timeline as the antibody values. Three participants received their primary immunization as part of the Pfizer vaccine trials. The vaccination event colors in the ribbon graph correspond to colors of points after the respective event in the antibody scatterplot shown in (A).
See also Figure S1.
Vaccinations with mRNA vaccines became first available to healthcare workers in mid-December 2020 (December 15th, 2020, for Pfizer BNT162b2; December 28th, 2020, for Moderna mRNA-1273). In naive individuals, the first dose of the mRNA vaccine induced no to relatively low antibody titers (Figure S2A), but the 2nd vaccine dose, administered 16–31 days after the first (Pfizer BNT162b2: median 21 days [range: 16–30]; Moderna mRNA-1273: median 27 days [21–31]), increased antibody titers to 5,936 (AUC, peak at 35 days post first dose, Figure 2A; Figure S2). By contrast, individuals with pre-existing immunity induced by infection reached peak titers faster, approximately 10 days after the first dose, and achieved considerably higher antibody titers (AUC 19,594, Figure 2A; Figure S2). After completing the primary immunization regimen, participants with pre-existing immunity had >3-fold higher peak responses compared with participants without pre-existing immunity (p < 0.001).
Figure 2. Immunogenicity of the different SARS-CoV-2 vaccine doses is dependent upon infection history prior to primary immunization.
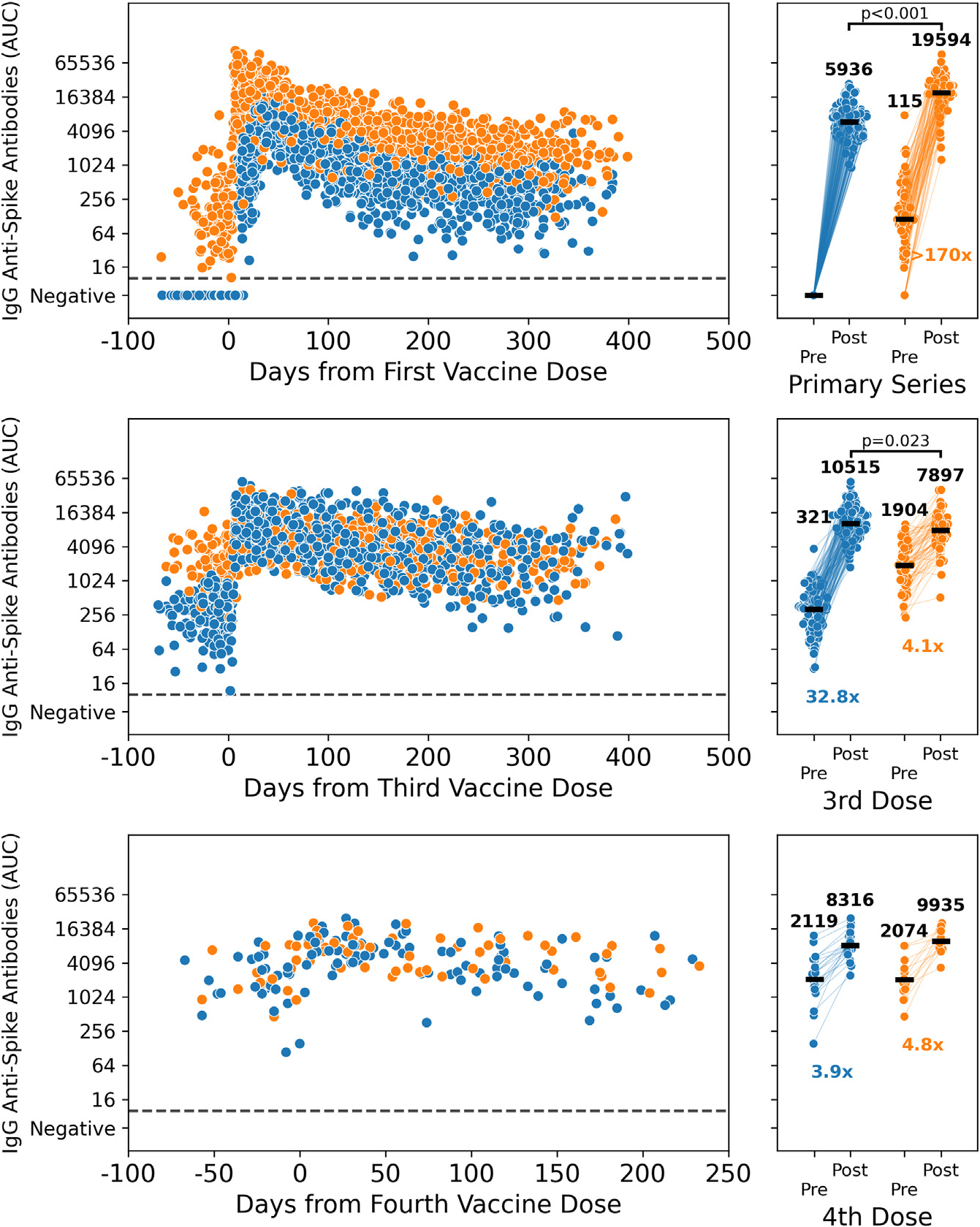
Longitudinal spike-binding antibodies measurements (n = 4,620) for PARIS participants with (orange) or without (blue) pre-existing SARS-CoV-2 immunity.
(A) Longitudinal antibody follow-up post-vaccination for 179 participants with no prior SARS-CoV-2 infection (blue, 1,671 samples) and 111 participants with a pre-vaccine SARS-CoV-2 infection (orange, 1,083 samples). The right panel has matched pre- and post-vaccine time points for 150 previously naive participants and 92 participants with hybrid immunity.
(B) Longitudinal antibody follow-up pre- and post-3rd dose for participants with (83 participants, 585 samples) and without (160 participants, 1,106 samples) prior SARS-CoV-2 infection. The right panel has matched pre- and post-3rd dose time points for 64 participants with prior infection and 126 participants without hybrid immunity.
(C) Longitudinal antibody follow-up before and after the 4th vaccine dose is shown for 15 participants with SARS-CoV-2 infection prior to 4th dose (67 samples) and 26 participants without hybrid immunity (108 samples). The right panel has matched pre- and post-4th dose time points for 13 participants with prior infection and 21 participants without hybrid immunity.
Time points post-breakthrough infection were excluded from the analysis. Pre-vaccination time points were collected within 10 weeks prior to vaccination, whereas peak post-vaccine time points were 1–5 weeks after administration of the vaccine dose (the 2nd dose, in the case of the primary series). The increase in spike-binding antibodies post-vaccination was statistically significant for all recipients (p < 0.0001, Wilcoxon signed rank). Peak antibody titers post-primary vaccination were higher in the hybrid immunity group (p < 0.0001, Mann-Whitney U) compared with the vaccine-only immunity group. After 3rd dose, peak antibody titers were modestly elevated in the vaccination-only immunity group compared with those in the hybrid immunity group (p = 0.023, Mann-Whitney U). Peak antibody titers post-dose 4 were comparable between groups). See also Figure S2.
During the approximately 400-day-long follow-up period, we found an initial steep 5-fold drop of antibody titers in vaccinated individuals with and without pre-existing immunity followed by a stabilization phase (Figure 2A). Based on a simple rolling geometric mean, post-vaccine data were observed to have two rough “phases” with different rates of decay (Figure 3A). This observation resembled a biphasic decay that is well approximated by a two-component, three-parameter, one-phase exponential decay model framework, prompting us to explore the kinetics of the antibody response in further detail. We fitted a nonlinear mixed-effects (NLME) model to describe the antibody dynamics from 2 weeks up to 1 year after the completion of the primary vaccination series. The same model was also fit to antibody dynamics after the 3rd vaccine dose. Specifically, our model had two components: a rapid-decay component with a half-life measured in weeks to months (e.g., antibodies produced by the plasmablast response) and a steady-state component to capture convergence to stable titers during prolonged follow-up (e.g., antibodies produced by long-lived plasma cells) (Figure S3A).
Figure 3. Modeling antibody kinetics after primary and booster vaccinations show an initial decay followed by a stabilization phase.
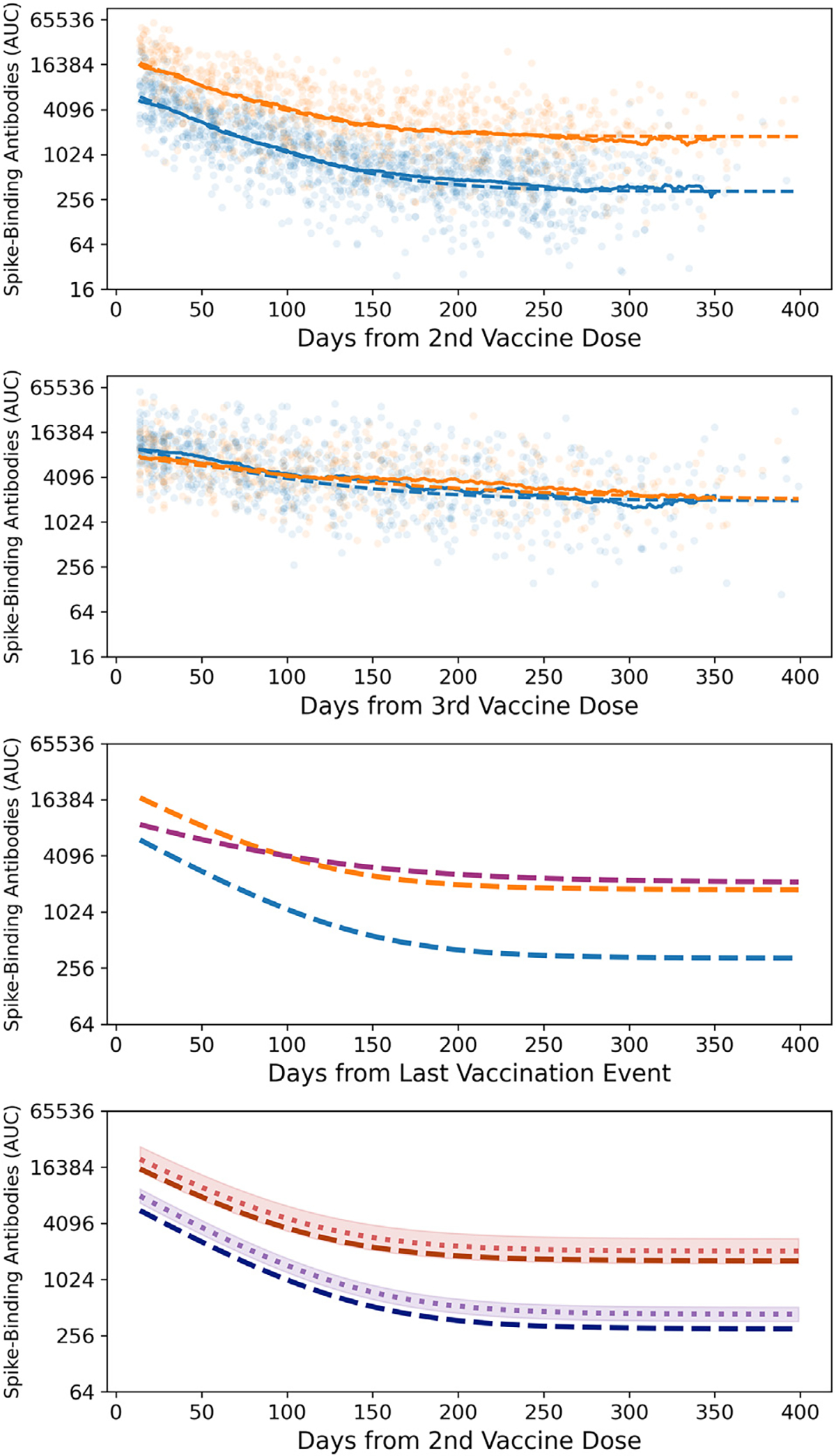
(A) We generated independent model predictions for the longitudinal post-vaccination antibody titers in 359 participants with (orange dashed line, 126 participants, 850 samples) and without (blue dashed line, 233 participants, 1,443 samples) SARS-CoV-2 infection prior to primary immunization. The rolling 49-day geometric means for each group are shown as solid lines.
(B) We fitted the same model to the longitudinal post-boost antibody titers of 223 participants (80 participants with hybrid immunity, 482 samples; 143 vaccine-only participants, 844 samples). The rolling 49-day geometric means for each group are shown as solid lines.
(C) The model estimates for the dynamics predicted by the post-vaccine model stratified by infection status shown in (A) to a combined post-boost model in dark pink (223 participants, 1,326 samples).
(D) The impact of vaccine type on antibody titers, with the fixed effect due to vaccine type separating model predictions for Pfizer recipients (dark blue and dark red dashed lines) and those for Moderna recipients (purple and red dotted lines). 95% confidence intervals of the fixed effect size are represented by the shaded area. Time points after breakthrough infections were excluded from the analysis. 95% confidence intervals are based on a T distribution with standard deviations calculated as part of the model fitting procedure. The vaccine type had a statistically significant effect in participants with vaccine-only immunity (67% increase, p < 0.001).
See also Figures S3 and S4 and Tables S1 and S2.
NLME models were fit using the NLMixed procedure in statistical analysis system (SAS) to account for variable longitudinal sampling between participants. A per-participant random effect for each component accounted for the significant variability in both antibody response magnitude and kinetics. Fixed effects due to demographic parameters and vaccine type were fit as multiplicative shifts in the expected geometric mean AUC across the entire time course. Datasets were stratified by immune status prior to vaccination, with models fit independently to each group. Results for the final models including demographic factors are reported, with independent models fitted for post-vaccine and post-boost antibody dynamics as well as for participants with (“hybrid immunity”) and without (“vaccine-only”) pre-vaccine SARS-CoV-2 infection (Table S1).
The PARIS NLME model fits closely the observed antibody dynamics post-vaccination, irrespective of prior infection status (Figures 3A and 3B). It also provides the means for a personalized two-component immunity score accounting for an individual’s SARS-CoV-2 immune history reflecting both the magnitude of the peak response and the stability of that response (Figure S3). These two components with distinct half-lives distinguish the PARIS model from other available models that consider exponential decay in binding antibodies after vaccination. Previous work comparing the fit of a simple exponential decay and a two-component model with post-vaccine data12 showed similar dynamics. The larger dataset of the PARIS cohort, particularly the longer time course (e.g., >1,000 days) and the denser longitudinal sampling (e.g., monthly), allowed us to select the two-component model. Specifically, we found that the two-component model has the smallest Akaike Information Criterion (AIC) value and the smallest mean squared error when compared with both a simple exponential decay model and a power law model (Figure S4). In all cases except the post-boost model for participants with hybrid immunity, the AIC for the alternative models was, at least, 30 above the AIC for the two-component model (Table S2). The simple exponential model was a less likely but plausible alternative for the post-boost data in the hybrid immunity group (AIC difference of 3.8), but the broader context provided strong support for our model over these two alternatives. When comparing our two-phase decay model with the simple exponential decay model post-vaccination, we noted that the latter overestimates the rate of decay in months 6 to 12 in our cohort (Figure S3B).
For the post-vaccine model (Figure 3A), the initial antibody titers measured at 14 days after the 2nd dose were 6,100 AUC in the naive group (blue) and 17,000 AUC in the hybrid immunity group (2.8-fold higher, in orange). This “peak” response was composed of two components in each case. In the naive group, the short-lived component had a magnitude of 5,700 AUC (95% confidence interval [CI]: 5,200–6,400) with a half-life of 29.6 days (CI: 28.1–31.2), and the stable component had a magnitude of 330 AUC (CI: 290–370). In the hybrid immunity group, the short-lived component had a magnitude of 15,000 AUC (CI: 13,000–18,000) with a half-life of 30.7 days (CI: 28.4–33.5), and the stable component had a magnitude of 1,800 AUC (CI: 1,500–2,100). Because of the larger difference in the stable component, the gap between the hybrid immunity group and the vaccine-only group increased to 4.8-fold at 180 days and 5.4-fold at 360 days after completion of the primary immunization regimen, respectively. 6 months after vaccination, the predicted geometric mean antibody titer (GMT) decreased by 0.95-fold to 445 AUC in the vaccine-only immunity group. In the hybrid immunity group, the predicted GMT was 2,142 corresponding to a decrease of 0.91-fold. At 12 months post-vaccination, the predicted GMT was 329 AUC in the vaccine-only immunity group, corresponding to a decrease of 0.26-fold over the previous 6 months. In the hybrid immunity group, the predicted GMT was 1,783 AUC, corresponding to a decrease of 0.17-fold over the same time period. Thus, although the kinetics of decay in the stable plateau phase are comparable between the two groups, the predicted antibody titers for each group differed by more than 4-fold (e.g., AUC 329 versus AUC 1,783).
In addition to examining broad patterns in antibody kinetics, we specifically quantified the impact of vaccine type as well as demographic factors, including gender, age, race, and ethnicity. Effects were modeled as a constant multiplicative increase or decrease in GMT over the entire time course. Of these, only age and vaccine type were statistically significant (p < 0.05 in any group). The effect of vaccine type is illustrated in Figure 3D for the post-vaccine model. There was a statistically significant effect in the vaccine-only group, with a 1.43-fold increase in antibody titers (CI: 1.20–1.70) in the Moderna mRNA-1273 vaccine recipients compared with the Pfizer BNT162b2 recipients. There was no statistically significant impact of vaccine type noted in the hybrid immunity group. The effect of age was modest but nonetheless statistically significant in the naive group: each decade of additional age lived corresponded to a 0.1-fold decrease in antibody titers (CI: 0.04–0.16) (Table S1). In the hybrid immunity group, there was no statistically significant effect.
In summary, based on our data and modeling, antibody titers achieved a steady state 7–9 months after the primary vaccination series. Stable antibody titers were higher for individuals with hybrid immunity as compared with naive vaccinees suggesting induction of long-lived serum antibody titers even after the primary vaccination series. In addition, vaccine type and age had a modest but measurable influence on antibody titers in participants without hybrid immunity.
Booster immunizations raise the antibody set point in participants with vaccine-only immunity
Third dose booster vaccination was authorized for healthcare workers in September 2021 (September 22, 2021, for Pfizer BNT162b2 and October 20, 2021, for Moderna mRNA-1273). Booster vaccination resulted in 27-fold increases in peak spike antibody responses in previously naive individuals (Figure 2B). By contrast, hybrid immune individuals displayed only a 4-fold increase due to their higher baseline. 4 weeks after booster vaccination, the peak titers were similar between previously naive participants (mean AUC: 10,162) and those with hybrid immunity (mean AUC: 8,001, Figure 2B). This stands in stark contrast to the humoral immune responses mounted upon the primary series (Figures 1 and 2A; Figure S2B). The 3rd vaccine dose seemed to act as an “equalizer” lifting the antibody titers of participants with vaccine-only immunity for the first time in our longitudinal study to the magnitude of those with infection- and vaccine-induced immunity.
Antibody kinetics post-boost were different compared with kinetics after the primary series (Figure 3B). Peak antibody titers post-boost were moderately but statistically significantly lower than in the hybrid immune group post-primary vaccination series. The initial waning of antibodies was slower, and titers stabilized at similar levels to those of the hybrid immune group post-primary vaccination. Importantly, titers settled at comparable setpoints for both previously naive and participants with hybrid immunity and stabilized again after approximately 7 to 9 months.
The 4th vaccine dose did boost serum antibody titers, albeit to a much lower fold increase compared with prior vaccine doses (vaccine-only 3.9-fold, hybrid immune 4.8-fold) (Figure 2C). The peak antibody titers after the 2nd booster vaccine were comparable to the peak titers achieved after the first booster vaccination. Antibody waning kinetics were also very similar to the kinetics observed after administration of the 3rd vaccine dose (Figures 1 and 2B; Figure S2). For individuals receiving a 3rd/4th booster dose (5th or 6th vaccine dose overall), the data were too sparse for an in-depth analysis, but antibody titers were generally within the same range observed post-3rd and post-4th vaccine doses Figure 1).
We next used our model for the analysis of post-boost antibody kinetics. For the post-boost model, antibody titers at 14 days after 3rd dose were 9,500 AUC in the naive group and 7,400 AUC in the hybrid immunity group (Figure 3B). In the naive group, the short-lived component had a magnitude of 7,500 AUC (CI: 6,500–8,700) with a half-life of 44 days (CI: 39–50), and the stable component had a magnitude of 2,000 AUC (CI: 1,500–2,500). In the hybrid immunity group, the short-lived component had a magnitude of 5,400 AUC (CI: 4,300–6,800) with a half-life of 73 days (CI: 60–93), and the stable component had a magnitude of 2,000 AUC (CI: 1,400–2,700). The predicted GMTs decreased by 0.78-fold in the vaccine-only immunity group and 0.62-fold in the hybrid immunity group at 180 days after the peak response (14 days post-boost). The rate of decay slowed over the next 6 months, with the predicted antibody titers at day 360 of the modeled period decreasing by 0.21-fold in the naive group and 0.30-fold in the hybrid immunity group. Despite this difference in shape, the geometric mean magnitude of the antibody response was similar across both groups throughout the entire time course studied, with fold differences between groups with and without hybrid immunity ranging from 0.72 to 1.22 and no detectable difference in the magnitudes of the stable component of the model. Because of this, antibody kinetics after boost were well approximated by a single model trained on a combined dataset disregarding prior infection status (Figure 3B).
We next compared antibody kinetics after the first booster immunization to those mounted after the primary vaccination (Figure 3C). In the model, peak antibody titers after boost were between those for the naive and hybrid immunity groups post-primary vaccination, and the stable component of the response post-boost was comparable to that of the hybrid immunity group post-vaccine. Of note, in the post-boost dataset, the effect due to primary vaccine type followed a pattern like that observed after the primary immunization. Participants without hybrid immunity who received the Moderna mRNA-1273 vaccine for their primary series showed a 1.41-fold increase relative to Pfizer recipients (CI: 1.10–1.82). The hybrid immunity group displayed no statistically significant difference nor trend due to primary vaccine type. Taken together, our data indicated that booster vaccine doses increased the set point at which long-term serum antibody responses stabilized.
Hybrid immunity provided protection against pre-Omicron variants, shaping responses to subsequent vaccination and breakthrough infections with Omicron variants
Although during the early phase of the pandemic, re-infections were rare, breakthrough infections increased with the emergence of variants of concern, especially the Omicron variant. Viral variants of concern started to circulate in the New York City metropolitan area in early 2021 (Figure 4A). In our study, we identified breakthrough infections based on participant self-reporting (rapid antigen tests or PCR testing at external facilities), nucleic acid amplification tests, and/or an increase in antibody titers (e.g., >4-fold).
Figure 4. Hybrid immunity provides protection from infection with pre-Omicron variants and determines responses to subsequent vaccine doses and/or breakthrough infections.
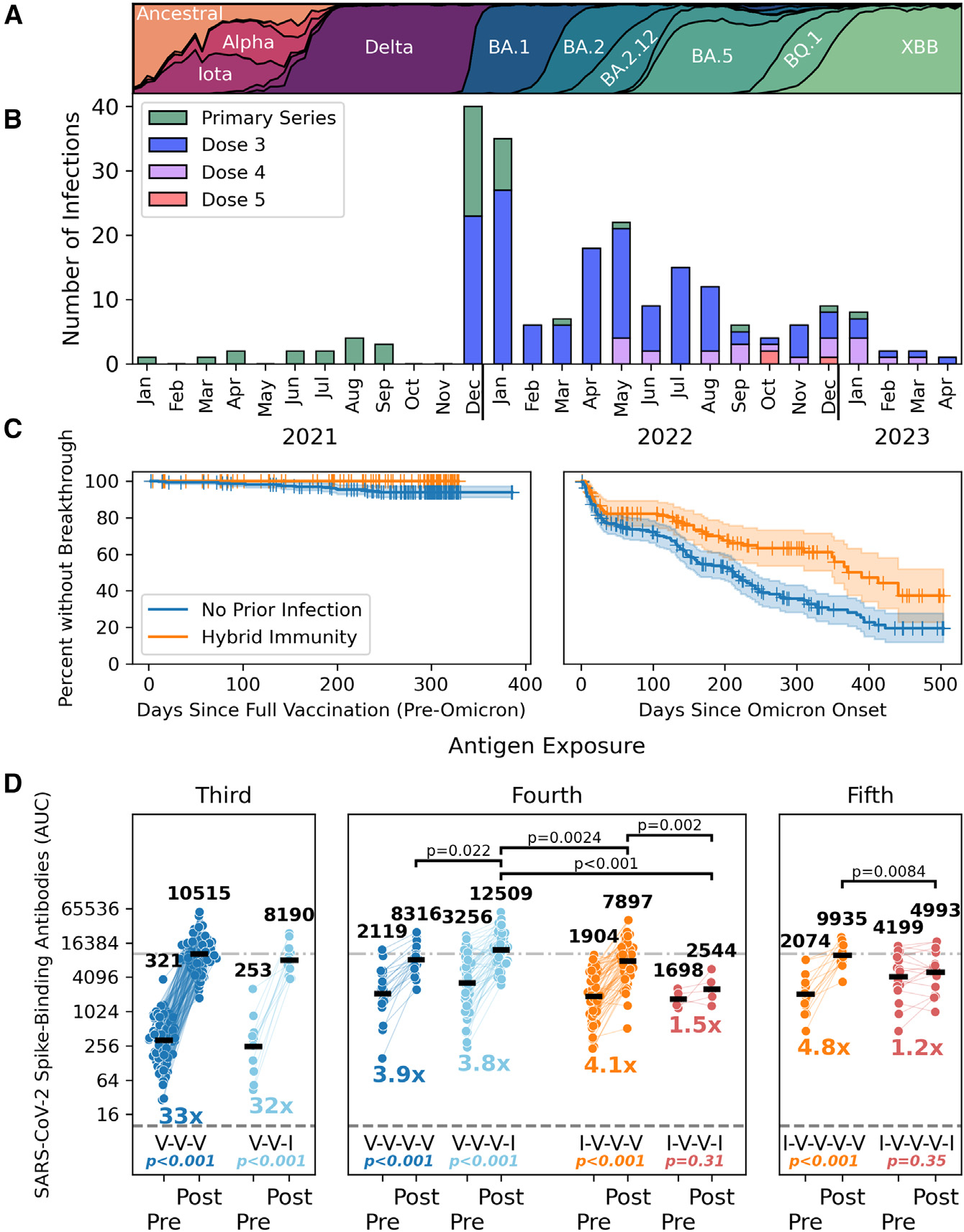
(A) The changing SARS-CoV-2 variant landscape in New York City is depicted for the period spanning January 2021 to April 2023.
(B) The number of breakthrough infections in vaccinated PARIS participants is shown by calendar month (2021–2023). The number of vaccine doses that participants with breakthrough infections received prior to infection is identified by the different colors.
(C) The frequency of breakthrough infections changed after the emergence of Omicron variants in mid-December 2021. Participants with hybrid immunity were only experiencing breakthrough infections with antigenically diverse Omicron variants (right panel). The differences between vaccinated participants with and without hybrid immunity before (left) and after (right) the appearance of Omicron variants is captured by Kaplan-Meier plots.
(D) The impact of infection and vaccination on spike-binding antibody titers is shown. Vaccination events are shown in blue and orange, with infection events shown in light blue and red. Participants are categorized by number of prior exposures and SARS-CoV-2 infection status prior to initial immunization. Second breakthrough infections are excluded. 3rd dose (dark blue, n = 126) is compared with post-vaccine break-through (light blue, n= 12). 4th dose (dark blue, n = 21) is paired with the breakthrough post-3rd dose (light blue, n = 54). 3rd dose in previously infected participants (orange, n = 64) is paired with breakthrough re-infection post-vaccine (red, n = 5). Finally, antibody titers mounted in response to the 4th dose in previously infected participants (orange, n = 13) are compared those mounted after breakthrough re-infections after the 3rd dose (red, n = 17).
Statistical comparisons within groups (Wilcoxon signed rank test) and fold change in geometric means are reported below each group. Statistically significant differences in post-exposure antibodies at the p < 0.05 level (Mann-Whitney U test) are reported.
A total of 225 SARS-CoV-2 infections were recorded among participants over the duration of the PARIS study, with most new infections (214/225) occurring after immunization (at least one vaccine dose administered). We documented three infections after the first vaccine dose and 37 infections after the 2nd vaccine dose but before the booster vaccination (Figure 4B). There were 174 breakthrough infections identified in participants who had received at least one booster vaccine dose. The bulk of breakthrough infections were observed after December 20, 2021, when Omicron BA.1 started to spread widely in our community (192/214 new infections). At that point in time, 299 PARIS participants had received a vaccine dose within the prior 6 months (10/289 2nd dose of the primary immunization series, 288/289 a booster vaccine dose). 55/299 participants who received a vaccine dose within 6 months prior to the Omicron wave experienced a breakthrough infection between December 2021 and February 2022. Nine infections occurred at least 14 days after the participant received a bivalent booster. Of note, one person had three consecutive infections with Omicron variants. Before highly antigenically distinct Omicron variants started to circulate in the New York City metropolitan area beginning mid-December 2021 (Figure 4A), breakthrough infections occurred exclusively in participants with vaccine-only immunity (Figure 4C, left panel). After the emergence of the different Omicron lineages, this picture changed, and breakthrough infections also became more common in hybrid immune individuals (Figure 4C, right panel), but hybrid immunity continued to have a protective effect as compared with vaccine-only immunity (p = 0.00029, log rank test).
We next explored whether breakthrough infections differed from booster vaccination with respect to the increased antibody magnitude. We compared the data for 176 breakthrough infections (at least 14 days after primary vaccination) to the effect observed after receiving an additional vaccine dose (Figure 4D). To start, we compared the effect of a booster dose after the primary immunization series (VVV) in participants who had no recorded infection to the change in antibodies in participants who experienced a breakthrough infection after the primary immunization series (VVI). Breakthrough infection in participants who had only had vaccine-induced antibodies was the 3rd antigen encounter and increased antibody titers by more than 30-fold, in a manner comparable to an mRNA vaccine booster dose. If the breakthrough infection was the 4th antigen encounter, the fold induction was lower (due to a higher baseline), resulting in an approximately from 4- to 5-fold increase, which was comparable to a 4th vaccine dose. For the 4th antigen encounter, in the case of a participant whose breakthrough infection was not their first infection and who initially had hybrid immunity (IVVI), however, saw a relatively lower fold change in antibody responses as opposed to participants with hybrid immunity who received a booster dose (IVVV). This difference may be due to the hybrid immunity limiting virus replication during breakthrough infections, leading to less immune stimulation. A similar observation was made when the breakthrough infection as opposed to booster dose was the 5th exposure in participants with hybrid immunity. Especially in participants with vaccine-only immunity, one could argue that breakthrough infections, representing the very first encounter with the virus, are equivalent to vaccine booster doses in terms of antibody response, whereas in individuals with hybrid immunity, the vaccine has a better effect on boosting systemic antibodies when compared with a second infection.
Hybrid immunity influenced the reactogenicity of consecutive SARS-CoV-2 vaccinations
In addition to immunogenicity, we analyzed the reactogenicity profiles associated with the different vaccine doses in study participants with and without immunity prior to the primary immunizations. After each vaccine dose, participants were provided a survey to provide self-reported side effects experienced. This analysis builds on preliminary data published in early 2021,12 which reported that participants with pre-existing immunity experience more systemic side effects after the first vaccine dose compared with naive participants. We now have data not only on the reactogenicity experienced after the 2nd and 3rd vaccine doses but also longitudinal data on how vaccine associated side effects changed for a given participant.
We collected information regarding local (injection site: pain, erythema, induration, and lymphadenopathy) and systemic (e.g., chills, fatigue, fever, headache, arthralgia, myalgia nausea/emesis, and pharyngitis) side effects using a survey that allowed participants to self-report the signs and symptoms experienced after each of the first three vaccine doses. 391 PARIS participants provided survey responses regarding symptoms experienced after the 1st vaccine dose, 333 participants provided survey responses to the 2nd dose, and 254 participants responded to the side effects survey after a booster dose. From all survey responses received, we selected the subset of 228 participants (69% female, 70% naive at primary vaccination) who submitted surveys after each of the 1st, 2nd, and 3rd vaccine doses. Of note, 69% of these respondents received three doses of Pfizer BNT162b2, 21% three doses of Moderna mRNA-1273, and 11% had a mix of vaccine types. Participants without three mRNA vaccine doses were excluded. The distribution of vaccine types was broadly similar across participants with differing immune histories (Table S3).
Overall, the vaccines were well tolerated with most participants experiencing mild to moderate side effects. Of the participants who reported severe side effects, none required medical attention. 60%–72% of the participants who completed all three surveys reported at least one symptom (dose 1: 60%, dose 2: 72%, dose 3: 67%). Across all vaccine doses and independent of infection history, injection site pain was the most frequently reported local side effect, whereas fatigue was the most frequently reported systemic side effect. 11/160 participants with vaccine only immunity (naive) and 5/68 participants with hybrid immunity reported a breakthrough infection prior to the booster dose, which may have influenced the reactogenicity experienced to this vaccine dose. Participants with hybrid immunity experienced overall more side effects (Figure 5) although the difference in frequency relative to naive participants became smaller with each additional vaccination. After the first dose, both local and systemic side effects were more common in participants with pre-existing immunity compared with naive participants (Figure 5). After the 2nd dose, naive participants reported a higher incidence of systemic side effects, whereas the frequency of local injection site symptoms remained relatively unchanged. The booster vaccination resulted in less painful injection site reactions in the participants with hybrid immunity (dose 1/2: 63% versus dose 3: 49%), whereas there was no such difference in the vaccine-only immunity group (dose 2: 52% versus dose 3: 53%). Both groups also reported lymphadenopathy (e.g., axillary) to occur more frequently (7%–11%), whereas fatigue remained unchanged (43%–44%). The incidence of chills jumped from 6% to 28% in the naive group, whereas participants with hybrid immunity only experienced a 2-fold increase (19% to 32%) upon booster vaccination. Table S4 provides a summary of the frequency of side effects, and Figure S5 shows the frequency of mild, moderate, and severe side effects across the three vaccine doses. When analyzing the data, age had no observable trend for the incidence or severity of reported vaccine reactogenicity. Female participants reported more side effects in most categories when compared with male participants, but this could also be a bias from the side effects respondents’ group as most participants are female. In the naive participant subgroup, Moderna mRNA-1273 vaccinees experienced more chills, nausea, arthralgia, fever, headache, erythema, and induration at the injection site after the 2nd and 3rd dose of the vaccine compared with Pfizer BNT162b2 vaccinees. The median duration between 2nd and 3rd vaccine doses was 272 days (range: 155–400).
Figure 5. Hybrid immunity modulates the reactogenicity of sequential SARS-CoV-2 vaccination (doses 1, 2, and 3).
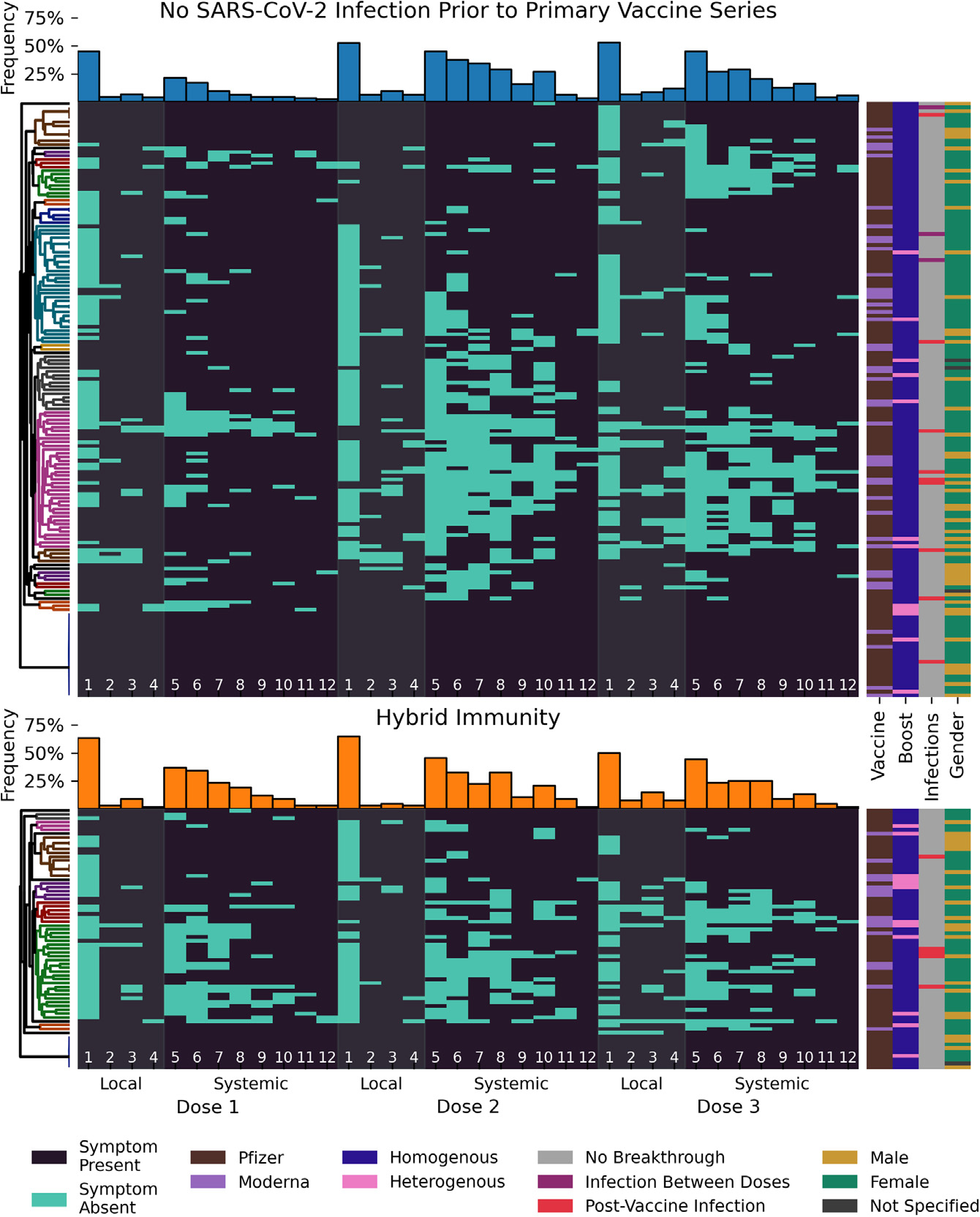
(A and B) Each line represents the post vaccine side effects reported by the same participant. Data shown are based on 684 surveys completed by 228 participants (160 initially naive [70%] in A and 68 with hybrid immunity [30%] in B) after the 1st, 2nd, and 3rd vaccine dose. The order of the symptoms depicted is divided into side effects at the local injection site (pain [1], erythema [2], induration [3], lymphadenopathy [4], gray bands) and systemic in nature (fatigue [5], headache [6], myalgia [7], chills [8], arthralgia [9], fever [10], nausea/emesis [11], pharyngitis [12], dark gray background).
Participants are split into groups based on pre-vaccine infection status and ordered by UPGMA clustering based on the Jaccard metric with optimal leaf ordering (trees shown on the left, colored clusters contain all nodes at distance less than 0.7). The presence of a symptom is indicated by a light blue bar. Bar plots for the overall frequency of each symptom are shown above the longitudinal data for each group. Vaccine type, boost type, infection status, and gender for each participant are annotated on the right. See also Figure S5 and Tables S3 and S4.
To better capture how vaccine reactogenicity changed between the three vaccine doses and to visualize putative symptom clusters, we performed unweighted pair group method with arithmetic mean (UPGMA) clustering based on Jaccard distance with optimal leaf ordering (Figure 5). This ordering showed that although overall frequencies of side effects appeared comparable between doses 2 and 3, this did not hold true at the individual participant level. Indeed, some participants with vaccine-only immunity experienced little or no symptoms after the 2nd dose but reported several systemic side effects after the 3rd dose, whereas others had far fewer side effects after the booster vaccination compared with the 2nd dose.
Taken together, these real-world data indicate that pre-existing immunity modulated the reactogenicity with additional SARS-CoV-2 vaccine doses resulting in more pronounced side effects. Booster vaccine doses produced slightly fewer systemic side effects relative to the 2nd vaccine dose in naive participants, whereas the opposite is true for those with hybrid immunity.
DISCUSSION
The PARIS study10 aimed to investigate the durability of immune responses mounted to SARS-CoV-2 infection and vaccination. Indeed, the SARS-CoV-2 pandemic provided unprecedented challenges but also opportunities to address fundamental questions in human immunology. A large proportion of the human population was exposed—for the first time and within a very limited time window of the pandemic—to a new respiratory viral pathogen and to a series of sequential immunizations that included an antigen to which the whole population was naive.
We used our extensive longitudinal antibody data with frequent sampling to address several important open questions about SARS-CoV-2 antibody-based immunity. It is often assumed that immunity after vaccination with mRNA vaccines wanes quickly. In some cases, this is based on the observation that vaccine effectiveness decreases over time.2 However, declining vaccination effectiveness is impacted not just by waning immune responses, but also by virus evolution.13 In other cases, the observation window is limited to the peak response and a brief period, thereafter, producing a linear decay model that does not accurately reflect B cell biology. Finally, many studies analyze receptor-binding-domain-focused neutralizing antibodies only but ignore the large number of other epitopes present on the spike protein. Our study design allows us to follow individuals longitudinally from the onset of the pandemic, through primary immunization series and now after booster immunizations providing an invaluable opportunity to comprehensively analyze long-term kinetics of antibody-based immunity. We found that anti-spike antibody kinetics after the primary immunization series follow a pattern that would be expected from basic B cell biology. After primary vaccination, serum antibodies produced by plasmablasts reach a high peak.14 Plasmablast responses are the first line of defense when it comes to B cells.14 These cells proliferate quickly, circulate in the periphery, and produce large amounts of antibodies—but typically disappear within 2 weeks.15,16 The antibodies they produce have a longer half-life (e.g., IgG1 has a half-life of approximately 4 weeks17) but eventually also wane over the period of weeks to months. This is consistent with the 28-to-34-day half-life of the short-lived component of our PARIS model following the primary vaccination series. However, serum antibody responses then stabilized after the initial few months. The more stable long-term serum antibody response is likely produced by long-lived plasma cells, which have in the meantime migrated from lymph nodes into the bone marrow.18 Again, the observations in our cohort are in excellent agreement with what we know about B cell biology. Waning kinetics were slower after the booster doses, and the setpoint at which titers stabilized became higher after each booster dose. We noted a clear “ceiling” effect with respect to the overall peak antibody titers reached. Although initial titers after the primary vaccination series in previously infected individuals could reach almost 20,000 AUC, overall, the titers in general seemed to peak at approximately 10,000 AUC. This ceiling seemed to slowly become lower with repeated exposures and booster doses.
A key contribution of our study is the development of a robust model suitable for dissecting the biological factors contributing to the durability of SARS-CoV-2 immune responses. Leveraging our extensive longitudinal antibody data, we could identify patterns and associations that shed light on the mechanisms underpinning long-term antibody protection. Our model allows us to interrogate the determinants of antibody dynamics, including prior SARS-CoV-2 infection and vaccine type as well as age. We found that individuals with initial infections followed by vaccination had antibody decay kinetics similar to individuals who were naive before the primary vaccination series; however, their peak titers were higher,12 and so was the titer at which their long-term serum antibodies eventually settled. The booster dose acted as an equalizer, bringing the serum antibody to similar levels in the group that was initially infected and the group that was initially naive. Other factors that influenced antibody titers, including long-term stable titers, were age and vaccine type. As observed in other studies that stated superiority of Moderna mRNA-1273 over Pfizer BNT162b2 in terms of induced immune responses or protection,19,20 we saw that the Moderna mRNA-1273 vaccine induced approximately 1.3-fold higher titers than the Pfizer BNT162b2 vaccine did, although differences were only significant in initially naive individuals.
Our study also allowed us to determine infection rates in our cohort. We found that during the pre-Omicron era, very few breakthrough infections occurred, and only in the previously naive group. During this time not a single breakthrough infection was detected in the hybrid immune group over the course of 11 months. This changed considerably during the Omicron era when most of our participants experienced breakthrough infections over the course of 18 months. This observation makes sense since Omicron has a strong escape phenotype, and its spike mutations undermine neutralizing antibodies induced by the ancestral or earlier variant spikes.1 However, a notable protective effect remained in the hybrid immune group as compared with initially naive individuals. We also assessed the immune responses to breakthrough infections. When comparing the number of antigen exposures, we found that a breakthrough infection after the primary vaccination series in previously naive individuals induced antibody titers comparable to a 3rd dose of vaccine (booster dose). We noted a similar pattern for the 4th exposure (4th vaccine dose versus breakthrough infection after the 3rd vaccine dose). This suggests that these initial breakthrough infections in vaccinated but previously naive individuals do, in fact, act similarly to a booster dose in terms of inducing antibody responses. The picture changed in individuals who had an initial infection followed by two or three vaccinations. When comparing breakthrough infection versus one additional vaccination in those highly exposed individuals, we saw that vaccination still robustly boosted serum antibody titers, but breakthrough infection did not, at least not as robustly as vaccination. We speculate that virus replication in these individuals was highly restricted due to their strong pre-existing immunity and that therefore the presence of less antigen leads to a lower induction of immunity.
Our longitudinal study setting also allowed us to investigate vaccine reactogenicity as it was experienced after the 1st, 2nd, and 3rd vaccine dose in a longitudinal manner at individual participant level. These data are of great relevance since reactogenicity can increase vaccine hesitancy and reduce uptake. In PARIS, SARS-CoV-2 mRNA vaccines were generally well tolerated with mild to modest side effects occurring in two-thirds of the vaccinees. As reported by us and others,12,21 both local and systemic side effects were more common in individuals with hybrid immunity. Our participant-level analysis of vaccine reactogenicity reveals the landscape of reactogenicity with several symptom clusters being apparent in the participant group with no infection prior to vaccination. In future studies, we plan on analyzing whether certain pattern of vaccine side effects predict the degree of immunogenicity. These real-world data on side effects associated with sequential vaccine doses are important to provide guidance for individuals concerned regarding future immunizations.
In summary, we show that antibody responses to SARS-CoV-2 mRNA vaccination follow a classical biphasic decay with a rapid waning phase initially followed by a transition into a stabilization phase after 7 to 9 months. We also show that hybrid immunity showed better protection against breakthrough infection in both the pre-Omicron but also the post-Omicron era, and we show that breakthrough infections in vaccinated, previously naive individuals have an effect like a booster dose. These data suggest that COVID-19 mRNA vaccination does induce long-lasting spike-specific antibodies consistent with B cell biology.
Limitations of the study
We measured the SARS-CoV-2 spike-binding activity of polyclonal sera throughout this longitudinal study. The affinity of individual antibodies could change over time in response to antigen exposure, which, in turn, may impact antibody binding. However, the impact of antibody affinity on the sensitivity of our serological assay is likely negligible since even low-affinity antibodies can be detected with our experimental setting as we have shown, for example, for influenza virus H7 monoclonal antibodies (mAbs).22 We have also investigated the relationship between binding, neutralization, and polyclonal avidity for SARS-CoV-2 responses using polyclonal sera from our study participants23 and found a modest correlation between binding antibody AUC and an increase in avidity. We plan to assess, therefore, antibody avidity in our longitudinal cohort using different methods as previously described.23
We were unable to analyze mucosal immune responses. We and others have previously shown that secretory immunoglobulin A (IgA) antibodies are not induced or boosted by mRNA vaccination in individuals who were previously naive but that titers increase in hybrid immune individuals after their first vaccination.24,25 We plan, therefore, to conduct similar studies at additional time points in our cohort in the future. Because our purpose was to analyze serum antibody kinetics, this study did not measure neutralizing antibody responses since binding antibodies comprise all antibodies to the spike and not just antibodies to certain neutralizing epitopes. Similarly, this study focused on antibodies to the full-length spike rather than receptor-binding domain (RBD). However, since we have observed differential antibody kinetics for spike and RBD26 (which is part of the spike), we plan to measure RBD antibodies in the future. Of note, binding antibodies have been tied to protection from severe COVID-19 outcomes in the absence of neutralizing antibodies in a recent study.27 Finally, this study examines reactivity to ancestral SARS-CoV-2 spike and not to later variant spikes or nucleoprotein. Assays utilizing the ancestral spike also detect antibodies induced by variant breakthrough infections and existing bivalent vaccines. Indeed, it has been shown that no-to-little specific antibodies are generated to spikes of emerging variants with most of the responses being cross-reactive28,29 in a manner consistent with backboosting.30
Consortia
The members of PARIS Study Group are Hala Alshammary, Angela A. Amoako Dalles Andre, Mahmoud Awawda, Maria C. Bermüdez-González, Katherine F. Beach, Dominika Bielak, Gianna Y. Cai, Rachel L. Chernet, Christian Cognigni, Yuexing Chen, Lily Q. Eaker, Emily D. Ferreri, Daniel L. Floda, Miriam Fried, Joshua Z. Hamburger, Denise Jurczyszak, Hyun Min Kang, Neko Lyttle, Julia C. Matthews, Jacob Mauldin, Wanni A. Mendez, Jacob Mischka, Sara Morris, Lubbertus C.F. Mulder, Ismail Nabeel, Jessica R. Nardulli, Jose Polanco, Annika Oostenink, Aria Rooker, Kayla T. Russo, Ashley-Beathrese Salimbangon, Miti S. Saksena, Amber A. Shin, Levy A. Sominsky, Daniel Stadlbauer, Leeba Sullivan, Morgan van Kesteren, Temima Yellin, Ania Wajnberg.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Requests for information, resources and reagents should be directed to and will be fulfilled by the lead contact, Viviana Simon, (viviana.simon@mssm.edu).
Materials availability
Recombinant Spike and RBD proteins used in this study will be shared with other research groups upon completion of a Material Transfer Agreement (MTA).
Data and code availability
All SARS-CoV-2 IgG binding antibody titer and demographic data are available from ImmPort via the following identifier SDY2468.
SAS code for the model fitting has been deposited in GitHub and is publicly available from the date of publication. DOI is listed in the key resources table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| Anti-human IgG (Fab-specific) horseradish peroxidase antibody | Sigma-Aldrich | Cat# A0293; RRID: AB_257875 |
| Primary Antibody – Anti RBD mAb generated in house (CR3022) | Tian et al.31 | N/A |
|
| ||
| Biological samples | ||
|
| ||
| Sera from PARIS study participants (adults) | This Paper, Icahn School of Medicine Personalized Virology Initiative PARIS Study | IRB-20–03374; STUDY-20–00442 |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| Recombinant SARS-CoV-2 spike receptor binding domain (RBD) | Krammer laboratory at the Icahn School of Medicine at Mount Sinai This protein has been used in other studies and was produced as previously described at Amanat et al.26 |
https://labs.icahn.mssm.edu/krammerlab/reagents/ |
| Recombinant SARS-CoV-2 spike (S) | Krammer laboratory at the Icahn School of Medicine at Mount Sinai This protein has been used in other studies and was produced as previously described at Amanat et al.26 |
https://labs.icahn.mssm.edu/krammerlab/reagents/ |
| SIGMAFAST™ OPD (o-Phenylenediamine dihydrochloride) | Sigma-Aldrich | Cat# P9187 |
| 3-molar hydrochloric acid | Thermo Fisher Scientific | Cat# S25856 |
| Non-Fat Dry Milk Omniblok | American Bio | Cat# AB10109–01000 |
|
| ||
| Deposited data | ||
|
| ||
| Figure Source Data (F1-F4) Antibody titers (AUC) measured by ancestral full-length spike-binding ELISA Participant demographic information |
This Paper | ImmPort SDY2468 |
|
| ||
| Software and algorithms | ||
|
| ||
| GraphPad Prism v10 | GraphPad Software Inc. | RRID:SCR_002798 |
| Python Programming Language v3.9.17 | Python Software Foundation | RRID:SCR_008394 |
| MatPlotLib v3.7.1 | Hunter32 | RRID:SCR_008624 |
| seaborn v0.12.2 | Waskom33 | RRID:SCR_018132 |
| NumPy v1.24.3 | Harris et al.34 | RRID:SCR_008633 |
| SciPy v1.11.1 | Virtanen et al.35 | RRID:SCR_008058 |
| Statistical Analysis System v9.4 (SAS) | SAS Institute Inc. | RRID:SCR_008567 |
| Non-linear mixed effects model (SAS code) | This Paper | https://doi.org/10.5281/zenodo.10467603 |
| REDCap 13.1.37 - © 2024 | Vanderbilt University | RRID:SCR_003445 |
| Canva® Canva Pro | https://www.canva.com/ | |
EXPERIMENTAL MODEL AND STUDY PARTICIPANT DETAILS
Human samples
In Spring 2020, most of New York City was shut down except for essential personnel needed to fight the SARS-CoV-2 pandemic. Personal Protective Equipment was in high demand and needed to be distributed to essential workers providing high risk services. Health care workers performing a wide array of different jobs (e.g., from housekeeping, students, nurses to medical doctors) fell into this category.
The observational longitudinal PARIS Study (Protection Associated with Rapid Immune response to SARS-CoV-2) enrolled adult individuals who worked at the Mount Sinai Health System (MSHS) in New York City in April 2020. Adult family members of MSHS employees were also eligible for participation. The study protocol IRB-20–03374 / STUDY-20–00442 was approved by the Institutional Review Board of the Mount Sinai Hospital. All study participants signed written informed consent forms prior to providing data or biospecimen.
In the initial two months of the study, participants were asked to provide saliva samples weekly and to provide a blood sample every two weeks. After the two-month study visit, participants returned monthly for a period of one year, thereafter study visit frequency varied between one and two months. Nasopharyngeal or ante-near swabs were collected when participants reported symptoms suggestive of a respiratory infection. Ad hoc study visits were performed in these situations as well as after vaccination. If participants reported testing positive for a viral respiratory infection, details about the diagnostic test performed, symptom onset and disease severity were collected.
A total of 501 health care workers were enrolled in PARIS between April 2020 and August 2021. Prior to the SARS-CoV-2 vaccine rollout in December of 2020 412/501 participants had been enrolled. Based on serological status at baseline prior to vaccination, 38% of participants had SARS-CoV-2-spike-binding IgG antibodies above the limit of detection of an in-house ELISA test. One single participant reported a SARS-CoV-2 infection prior to enrollment into the study but was seronegative at baseline.
Of the 501 participants enrolled, one (0.2%) has received four mRNA booster doses (total vaccine doses: 6), 13 (2.6%) have received three mRNA booster doses (total vaccine doses: 5), 83 (16.6%) have received two mRNA booster doses (total vaccine doses: 4) and 269 (53.7%) received one booster vaccine dose. 465 participants (93% of the total cohort) have been vaccinated with the remainder of the participants having withdrawn or being lost to follow up since immunization became mandatory for health care workers. Indeed, 36 participants withdrew from the study and their vaccination status is unknown. Of the 465 participants who were vaccinated, 99 (21.3%) elected not to receive any booster vaccine dose. Over the three years of the PARIS study, we have documented over 8,000 study visits collecting blood, saliva and/or nasal swabs.
METHOD DETAILS
Vaccine reactogenicity
Qualitative survey data capturing general health questions, exposures, and changes to perceived risk to SARS-CoV-2 infection were captured monthly using a custom build REDCap database. When participants received a SARS-CoV-2 vaccination, a survey to collected information about side effects was sent out after the 1st and 2nd dose in the primary immunization series as well as after the 1st booster vaccine dose.
After vaccination, surveys were sent to participants to provide data on side effects experienced after dose 1, 2 and 3. The questions for the survey were designed to capture diversity of possible effects broken into two categories: systemic effects and local injection site effects. The symptoms recorded (Table S3) included side effects reported in publications from Pfizer and Moderna about their respective vaccines.36,37
Dates by which vaccines became available to health care workers were as follows: Pfizer BioNTech: 12/15/2020 (initial) and 09/22/2021 (booster); Moderna: 12/28/2022 (initial), 10/20/21 (booster). Bivalent boosters (both Pfizer/BioNTech, Moderna) were distributed starting September 2022. Three participants received vaccine in the Pfizer Phase 3 vaccine trial.
The surveys for the first two doses were released early in 2021 and the booster vaccine survey was launched in the fall of 2021. 245 participants completed all three surveys. Participants who did not receive mRNA vaccination as their primary immunization or booster dose were excluded from the analysis as well as participants with known immunomodulatory comorbidities leaving a total complete survey response from 228 participants. For each indicated side effect, branching logic questions were designed to ask about the duration of the side effect and perceived severity. For fever and nausea/vomiting, quantitative ranges were asked. For fever, we asked about the highest temperature known. For nausea/vomiting we asked about the number of times a day the participant felt ill and vomited. The other side effects, if indicated as severe, had follow-up questions to ask if medical care was required. None of the participants who reported a severe side effect required hospitalization for treatment. All survey data was captured using REDCap database system. Severity scoring was tabulated in a REDCap report output (not reported = 0, mild = 1, moderate = 2, severe = 3) per effect per vaccine dose. The dataset for all three doses was combined and subgroups based on infection history pre-vaccine, vaccine type, and gender were created. Percent totals of reported incidence and severity were calculated per subgroup and then compared to understand differences within the PARIS cohorts and observe trends between groups across doses.
Enzyme-linked immunosorbent assay (ELISA)
Antibody titers were measured using a two-step well-established ELISA method9,38 in which serum samples are screened at a single dilution (1:50) for IgG against the recombinant receptor binding domain (RBD) of the spike protein from SARS-CoV-2 (Wuhan-Hu-1), followed by detection of antibodies against the full-length spike protein (also Wuhan-Hu-1). All serum samples were initially screened in a high-throughput assay using the recombinant RBD protein. In brief, 96-well microtitre plates (Thermo Fisher) were coated with 50 μl recombinant RBD protein at a concentration of 2 μg/ml overnight at 4 °C. The next day, the plates were washed three times with phosphate-buffered saline (PBS; Gibco) supplemented with 0.1% Tween-20 (PBS-T; Fisher Scientific) using an automatic plate washer (BioTek). The plates were blocked with 200 μl blocking solution consisting of PBS-T with 3% (w/v) milk powder (American Bio) and incubated for 1 h at room temperature. Serum samples were heated at 56 °C for 1 h before use to reduce the risk from any potential residual virus in the serum. The blocking solution was taken off the plates and 100 μl of the serum samples diluted 1:50 in PBS-T containing 1% (w/v) milk powder was added to the respective wells of the microtitre plates. After 2 h, the plates were washed three times with PBS-T and 50 μl anti-human IgG (Fab-specific) horseradish peroxidase antibody (produced in goat; Sigma, A0293) diluted 1:3,000 in PBS-T containing 1% milk powder was added to all wells and incubated for 1 h at room temperature. The microtitre plates were washed three times with PBS-T and 100 μl SigmaFast o-phenylenediamine dihydrochloride (Sigma) was added to all wells. The reaction was stopped after 10 min with 50 μl per well 3 M hydrochloric acid (Thermo Fisher) and the plates were read at a wavelength of 490 nm with a plate reader (BioTek). Serum samples that exceeded an optical density cut-off value of 0.15 at 490 nm (OD490) were categorized as presumptive positive and were tested in a second step in confirmatory ELISAs using the full-length, recombinant spike protein. To perform the confirmatory ELISAs, the plates were coated and blocked as described above except full-length spike protein at a concentration of 2 μg/ml was added to the plates. After one hour, the blocking solution was removed, presumptive positive serum samples that were serially diluted (from 1:80/1:100 to 204,800) in 1% milk prepared in PBS-T were added and the plates were incubated for two hours at room temperature. The remainder of the assay was performed as described above. The cut-off value was set to an OD490 value of 0.15 and true-positive samples were defined as samples that exceeded an OD490 value of 0.15 at a 1:80/1:100 serum dilution. An OD of 0.15 was determined as negative based on data from pre-pandemic/COVID-19 negative samples as described earlier.8,9,38 Trending and quality controls were included in every run to allow for comparison across the entire dataset.
The end-point titer was calculated and defined as the last dilution before the signal dropped below an OD490 of 0.15. The area under the curve (AUC) values were calculated and plotted using Prism 9 software (GraphPad). AUC values >10 were plotted as above the limited of detection in the figures included in this manuscript.
SARS-CoV-2 variants
Genomic data on the viral variants circulating within the New York City metropolitan area were obtained through the New York City Department of Health and Mental Hygiene (DOHMH) web portal. The DOHMH maintains a publicly available database updated regularly since the beginning of the pandemic. SARS-CoV-2 variant tracking within the database began on January 1st, 2021, and was updated weekly until May 18th, 2023 when the COVID-19 emergency was declared over in the US. Data on specific viral variants was only available for cases sequenced by and/or shared with the New York City DOHMH. Sequences were not available for all the SARS-CoV-2 positive test cases reported to DOHMH. Certain variants within the New York City data were collapsed into single categories due to the limited size of variant. The following variants were combined: XBB and XBB 1.5; BQ1 and BQ 1.1, Omicron BF.7, BA.4 and BA.4.6; B.1.427 and B.1.429 (Epsilon), and finally the B.1.526 lineages with and without E484K (Iota).
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical Analysis
Statistical testing for paired samples was done using the Wilcoxon signed rank test, with unpaired samples compared using the Mann-Whitney U test. The protectiveness of hybrid immunity was evaluated using a log rank test. UPGMA clustering for Figure 5 and all statistical tests were run using Scipy (version 1.11.1) in Python 3.9.15. Curve fitting for Figure S3 was done using the stats-models package (version 0.13.2). All figures were rendered using Matplotlib (3.7.1) and Seaborn (0.12.2). The antibody kinetic model was fit using the NLMixed procedure in SAS9.4, using the trust region optimization technique and the adaptive gaussian quadrature integration method, assuming a normal distribution for the random effects and the dependent variable.
Non-Linear Mixed models for spike binding antibodies
Data were selected for the post-vaccine model under the following criteria: Timepoints with a discrepancy between the results generated by the research grade SARS-CoV-2 spike-binding ELISA and the Kantaro spike-binding assay performed in the CLIA certified Pathology laboratories of the Mount Sinai Hospital in the top or bottom percentile over the entire dataset were excluded. Participants who did not receive two doses of an mRNA vaccine as well as participants with unknown SARS-CoV-2 infection status prior to immunization were also excluded. Similarly, data from participants without at least two valid timepoints 14 days post-dose 2 were excluded. Samples obtained after subsequent vaccine doses or breakthrough infections were censored.
Data were selected for the post-boost model using the following criteria: Participants excluded from the post-vaccine model dataset were also excluded from the post-boost dataset. Participants with a breakthrough infection pre-3rd dose were excluded. Samples after 4th doses of the vaccine or breakthrough infections were censored. Participants without a minimum of two valid time points at least 14 days post-dose 3 were excluded.
| (Equation 1) |
Antibody decay has been observed in interventional settings where external antibody treatments were administered. Unlike clinical treatments, natural immunity has multiple sources of antibody production. Since there are multiple sources of antibody production naturally within the body, single component exponential decay models are insufficient as they will over-predict the change in decay rates. The model used here (Equation 1) has two components which are assumed to have distinct decay rates. When reporting aggregate antibody data, the geometric mean is generally used due to a high variability within and among participants (across multiple orders of magnitudes in the datasets used here). By fitting the model directly within the log-transformed space, the output curve directly estimates the geometric mean rather than the arithmetic mean.
All models were generated using the NLMIXED procedure in SAS9.4. This procedure is capable of fitting arbitrary nonlinear functions including both fixed and random effects. The specific model structure used in this case is shown in Figure S3B. A two-phase fitting procedure was used. First, a simplified model without random effects was run from a naìve starting point, and the fitted parameter values were used as the initial values for the mixed effects model. In order to account for underlying variability across participants that was not captured by the fixed effects, a random effect was added to each component for the final mixed effect model. In addition to allowing for variability in the overall magnitude of the antibody response, this approach captures observed variability in the kinetics of the response (fold-reduction relative to peak varies among participants). The model was fit independently to datasets for participants with and without pre-vaccine SARS-CoV-2 infection (hybrid immunity) using a feature in SAS to build stratified models. The same model framework was used for both post-vaccine and post-boost models. Data from a post-boost model fit to the combined dataset without accounting for pre-vaccine immune status is also presented, reflecting a close correspondence in post-boost kinetics in both stratified groups.
| (Equation 3) |
| (Equation 2) |
The impact of demographic factors was assessed in the stratified post-vaccine and post-boost models. In all cases, these factors were assessed as a multiplicative factor across the entire time course studied (manifesting on graphs with log-transformed y axes as a y shift). All demographically informed models were constructed iteratively, beginning with a model including all factors considered (age, gender, race, ethnicity, and primary vaccine type). After each model fitting run, the factor with the highest assessed p value was removed if that p value did not meet a relaxed threshold for statistical significance (p < 0.1). The model was then re-run with the reduced set of fixed effects and finalized when all factors hit this relaxed threshold. This stepwise subtractive modeling practice helps to account for confounding factors and avoids double controlling for effects that alter the result through the same mechanism. Results from the model fitting in SAS were exported to csv files for visualization in Python.
The simple exponential decay model (Equation 2) and the power law model (Equation 3) used in model validation were fit with the NLMixed procedure on both stratified datasets, with a per-participant random effect on each of the two fitted parameters. AIC was calculated for each of the 3 models to compare fit quality as part of the procedure output.
Supplementary Material
Highlights.
COVID-19-vaccine-induced immunity wanes but stabilizes at an individual setpoint
Pre-existing immunity results in rapid antibody responses upon vaccination
Boosters equalize antibody titers between individuals with and without hybrid immunity
Antibody kinetics show two phases: an initial rapid decay followed by a steady state
ACKNOWLEDGMENTS
We thank all the participants of our longitudinal PARIS study for their generous and continued support of our COVID-19 research programs. This work was funded in part by the Centers of Excellence for Influenza Research and Response (CEIRR, contract # 75N93021C00014), the Collaborative Influenza Vaccine Innovation Centers (CIVICs contract # 75N93019C0005), National Institute of Allergy and Infectious Diseases (NIAID) U19 AI168631, NIAID P01 AI172531, philanthropic donations, and institutional funds provided to the Mount Sinai Center for Vaccine Research and Pandemic Preparedness. This work was supported in part through the computational and data resources and staff expertise provided by Scientific Computing and Data at the Icahn School of Medicine at Mount Sinai and supported by the Clinical and Translational Science Award (CTSA) grant UL1TR004419 from the National Center for Advancing Translational Sciences. This effort was also in part supported by the Serological Sciences Network (SeroNet) in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract No. 75N91021F00001 via 21X092F1 Mod 01. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government.
Footnotes
DECLARATION OF INTERESTS
The Icahn School of Medicine at Mount Sinai has filed patent applications relating to SARS-CoV-2 serological assays and NDV-based SARS-CoV-2 vaccines that list F.K. as co-inventor. V.S. is also listed on the SARS-CoV-2 serological assays patent. Mount Sinai has spun out a company, Kantaro, to market serological tests for SARS-CoV-2. F.K. has consulted for CureVac, Merck, and Pfizer, and is currently consulting for 3rd Rock Ventures, GSK, Gritstone, and Avimex, and he is a co-founder and scientific advisory board member of CastleVax. The Krammer laboratory is also collaborating with Pfizer on animal models of SARS-CoV-2.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.immuni.2024.01.017.
REFERENCES
- 1.Carreño JM, Alshammary H, Tcheou J, Singh G, Raskin AJ, Kawabata H, Sominsky LA, Clark JJ, Adelsberg DC, Bielak DA, et al. (2022). Activity of convalescent and vaccine serum against SARS-CoV-2 Omicron. Nature 602, 682–688. [DOI] [PubMed] [Google Scholar]
- 2.Menegale F, Manica M, Zardini A, Guzzetta G, Marziano V, d’Andrea V, Trentini F, Ajelli M, Poletti P, and Merler S (2023). Evaluation of waning of SARS-CoV-2 vaccine-induced immunity: A systematic review and meta-analysis. JAMA Netw. Open 6, e2310650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zaeck LM, GeurtsvanKessel CH, and de Vries RD (2023). COVID-19 vaccine effectiveness and evolving variants: understanding the immunological footprint. Lancet Respir. Med. 11, 395–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DeGrace MM, Ghedin E, Frieman MB, Krammer F, Grifoni A, Alisoltani A, Alter G, Amara RR, Baric RS, Barouch DH, et al. (2022). Defining the risk of SARS-CoV-2 variants on immune protection. Nature 605, 640–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feikin DR, Higdon MM, Abu-Raddad LJ, Andrews N, Araos R, Goldberg Y, Groome MJ, Huppert A, O’Brien KL, Smith PG, et al. (2022). Duration of effectiveness of vaccines against SARS-CoV-2 infection and COVID-19 disease: results of a systematic review and meta-regression. Lancet 399, 924–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gonzalez-Reiche AS, Hernandez MM, Sullivan MJ, Ciferri B, Alshammary H, Obla A, Fabre S, Kleiner G, Polanco J, Khan Z, et al. (2020). Introductions and early spread of SARS-CoV-2 in the New York City area. Science 369, 297–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wajnberg A, Mansour M, Leven E, Bouvier NM, Patel G, Firpo-Betancourt A, Mendu R, Jhang J, Arinsburg S, Gitman M, et al. (2020). Humoral response and PCR positivity in patients with COVID-19 in the New York City region, USA: an observational study. Lancet Microbe 1, e283–e289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stadlbauer D, Tan J, Jiang K, Hernandez MM, Fabre S, Amanat F, Teo C, Arunkumar GA, McMahon M, Capuano C, et al. (2021). Repeated cross-sectional sero-monitoring of SARS-CoV-2 in New York City. Nature 590, 146–150. [DOI] [PubMed] [Google Scholar]
- 9.Amanat F, Stadlbauer D, Strohmeier S, Nguyen THO, Chromikova V, McMahon M, Jiang K, Arunkumar GA, Jurczyszak D, Polanco J, et al. (2020). A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat. Med. 26, 1033–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Simon V, Kota V, Bloomquist RF, Hanley HB, Forgacs D, Pahwa S, Pallikkuth S, Miller LG, Schaenman J, Yeaman MR, et al. (2022). Paris and SPARTA: finding the Achilles’ heel of SARS-CoV-2. mSphere 7, e0017922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kubale J, Gleason C, Carreño JM, Srivastava K, Singh G, PARIS Study Team, Gordon A, Krammer F, and Simon V. (2022). SARS-CoV-2 spike-binding antibody longevity and protection from reinfection with antigenically similar SARS-CoV-2 variants. mBio 13, e0178422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krammer F, Srivastava K, Alshammary H, Amoako AA, Awawda MH, Beach KF, Bermúdez-González MC, Bielak DA, Carreño JM, Chernet RL, et al. (2021). Antibody responses in seropositive persons after a single dose of SARS-CoV-2 mRNA vaccine. N. Engl. J. Med. 384, 1372–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krammer F (2023). The role of vaccines in the COVID-19 pandemic: what have we learned? Semin. Immunopathol. 10.1007/s00281-023-00996-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Akkaya M, Kwak K, and Pierce SK (2020). B cell memory: building two walls of protection against pathogens. Nat. Rev. Immunol. 20, 229–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wrammert J, Smith K, Miller J, Langley WA, Kokko K, Larsen C, Zheng NY, Mays I, Garman L, Helms C, et al. (2008). Rapid cloning of high-affinity human monoclonal antibodies against influenza virus. Nature 453, 667–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ellebedy AH, Jackson KJ, Kissick HT, Nakaya HI, Davis CW, Roskin KM, McElroy AK, Oshansky CM, Elbein R, Thomas S, et al. (2016). Defining antigen-specific plasmablast and memory B cell subsets in human blood after viral infection or vaccination. Nat. Immunol. 17, 1226–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mankarious S, Lee M, Fischer S, Pyun KH, Ochs HD, Oxelius VA, and Wedgwood RJ (1988). The half-lives of IgG subclasses and specific antibodies in patients with primary immunodeficiency who are receiving intravenously administered immunoglobulin. J. Lab. Clin. Med. 112, 634–640. [PubMed] [Google Scholar]
- 18.Turner JS, Kim W, Kalaidina E, Goss CW, Rauseo AM, Schmitz AJ, Hansen L, Haile A, Klebert MK, Pusic I, et al. (2021). SARS-CoV-2 infection induces long-lived bone marrow plasma cells in humans. Nature 595, 421–425. [DOI] [PubMed] [Google Scholar]
- 19.Ioannou GN, Locke ER, Green PK, and Berry K (2022). Comparison of Moderna versus Pfizer-BioNTech COVID-19 vaccine outcomes: A target trial emulation study in the U.S. Veterans Affairs healthcare system. EClinicalMedicine 45, 101326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bajema KL, Dahl RM, Evener SL, Prill MM, Rodriguez-Barradas MC, Marconi VC, Beenhouwer DO, Holodniy M, Lucero-Obusan C, Brown ST, et al. (2021). Comparative effectiveness and antibody responses to moderna and Pfizer-BioNTech COVID-19 vaccines among hospitalized veterans - five Veterans Affairs Medical Centers, United States, February 1–September 30, 2021. MMWR Morb. Mortal. Wkly. Rep. 70, 1700–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Atmar RL, Lyke KE, Deming ME, Jackson LA, Branche AR, El Sahly HM, Rostad CA, Martin JM, Johnston C, Rupp RE, et al. (2022). Homologous and heterologous Covid-19 booster vaccinations. N. Engl. J. Med. 386, 1046–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Henry Dunand CJ, Leon PE, Huang M, Choi A, Chromikova V, Ho IY, Tan GS, Cruz J, Hirsh A, Zheng NY, et al. (2016). Both neutralizing and non-neutralizing human H7N9 influenza vaccine-induced monoclonal antibodies confer protection. Cell Host Microbe 19, 800–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Singh G, Abbad A, Tcheou J, Mendu DR, Firpo-Betancourt A, Gleason C, Srivastava K, Cordon-Cardo C, Simon V, Krammer F, and Carreno JM (2023). Binding and avidity signatures of polyclonal sera from individuals with different exposure histories to severe acute respiratory syndrome coronavirus 2 infection, vaccination, and Omicron breakthrough infections. J. Infect. Dis. 228, 564–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sano K, Bhavsar D, Singh G, Floda D, Srivastava K, Gleason C, PARIS Study Group, Carreño JM, Simon V, and Krammer F. (2022). SARS-CoV-2 vaccination induces mucosal antibody responses in previously infected individuals. Nat. Commun. 13, 5135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sheikh-Mohamed S, Isho B, Chao GYC, Zuo M, Cohen C, Lustig Y, Nahass GR, Salomon-Shulman RE, Blacker G, Fazel-Zarandi M, et al. (2022). Systemic and mucosal IgA responses are variably induced in response to SARS-CoV-2 mRNA vaccination and are associated with protection against subsequent infection. Mucosal Immunol. 15, 799–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Amanat F, Thapa M, Lei T, Ahmed SMS, Adelsberg DC, Carreño JM, Strohmeier S, Schmitz AJ, Zafar S, Zhou JQ, et al. (2021). SARS-CoV-2 mRNA vaccination induces functionally diverse antibodies to NTD, RBD, and S2. Cell 184, 3936–3948.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vikström L, Fjällström P, Gwon YD, Sheward DJ, Wigren-Byström J, Evander M, Bladh O, Widerström M, Molnar C, Rasmussen G, et al. (2023). Vaccine-induced correlate of protection against fatal COVID-19 in older and frail adults during waves of neutralization-resistant variants of concern: an observational study. Lancet Reg Health Eur. 30, 100646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alsoussi WB, Malladi SK, Zhou JQ, Liu Z, Ying B, Kim W, Schmitz AJ, Lei T, Horvath SC, Sturtz AJ, et al. (2023). SARS-CoV-2 Omicron boosting induces de novo B cell response in humans. Nature 617, 592–598. [DOI] [PubMed] [Google Scholar]
- 29.Carreño JM, Singh G, Simon V, and Krammer F; PVI study group (2023). Bivalent COVID-19 booster vaccines and the absence of BA.5-specific antibodies. Lancet Microbe 4, e569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fonville JM, Wilks SH, James SL, Fox A, Ventresca M, Aban M, Xue L, Jones TC, Le NMH, Pham QT, et al. (2014). Antibody land-scapes after influenza virus infection or vaccination. Science 346, 996–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tian X, Li C, Huang A, Xia S, Lu S, Shi Z, Lu L, Jiang S, Yang Z, Wu Y, et al. (2020). Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg Microbes Infect 9, 382–385. 10.1080/22221751.2020.1729069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hunter JD (2007). Matplotlib: A 2D Graphics Environment. Computing in Science & Engineering 9, 90–95. 10.1109/MCSE.2007.55. [DOI] [Google Scholar]
- 33.Waskom ML (2021). Seaborn: statistical data visualization. Journal of Open Source Software 6, 3021. 10.21105/joss.03021. [DOI] [Google Scholar]
- 34.Harris CR, Millman KJ, van der Walt SJ, Gommers R, Virtanen P, Cournapeau D, Wieser E, Taylor J, Berg S, Smith NJ, et al. (2020). Array programming with NumPy. Nature 585, 357–362. 10.1038/s41586-020-2649-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Virtanen P, Gommers R, Oliphant TE, Haberland M, Reddy T, Cournapeau D, Burovski E, Peterson P, Weckesser W, Bright J, et al. (2020). SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat Methods 17, 261–272. 10.1038/s41592-019-0686-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Polack FP, Thomas SJ, Kitchin N, Absalon J, Gurtman A, Lockhart S, Perez JL, Pérez Marc G, Moreira ED, Zerbini C, et al. (2020). Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N. Engl. J. Med. 383, 2603–2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baden LR, El Sahly HM, Essink B, Kotloff K, Frey S, Novak R, Diemert D, Spector SA, Rouphael N, Creech CB, et al. (2021). Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N. Engl. J. Med. 384, 403–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stadlbauer D, Amanat F, Chromikova V, Jiang K, Strohmeier S, Arunkumar GA, Tan J, Bhavsar D, Capuano C, Kirkpatrick E, et al. (2020). SARS-CoV-2 seroconversion in humans: A detailed protocol for a serological assay, antigen production, and test setup. Curr. Protoc. Microbiol. 57, e100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All SARS-CoV-2 IgG binding antibody titer and demographic data are available from ImmPort via the following identifier SDY2468.
SAS code for the model fitting has been deposited in GitHub and is publicly available from the date of publication. DOI is listed in the key resources table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| Anti-human IgG (Fab-specific) horseradish peroxidase antibody | Sigma-Aldrich | Cat# A0293; RRID: AB_257875 |
| Primary Antibody – Anti RBD mAb generated in house (CR3022) | Tian et al.31 | N/A |
|
| ||
| Biological samples | ||
|
| ||
| Sera from PARIS study participants (adults) | This Paper, Icahn School of Medicine Personalized Virology Initiative PARIS Study | IRB-20–03374; STUDY-20–00442 |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| Recombinant SARS-CoV-2 spike receptor binding domain (RBD) | Krammer laboratory at the Icahn School of Medicine at Mount Sinai This protein has been used in other studies and was produced as previously described at Amanat et al.26 |
https://labs.icahn.mssm.edu/krammerlab/reagents/ |
| Recombinant SARS-CoV-2 spike (S) | Krammer laboratory at the Icahn School of Medicine at Mount Sinai This protein has been used in other studies and was produced as previously described at Amanat et al.26 |
https://labs.icahn.mssm.edu/krammerlab/reagents/ |
| SIGMAFAST™ OPD (o-Phenylenediamine dihydrochloride) | Sigma-Aldrich | Cat# P9187 |
| 3-molar hydrochloric acid | Thermo Fisher Scientific | Cat# S25856 |
| Non-Fat Dry Milk Omniblok | American Bio | Cat# AB10109–01000 |
|
| ||
| Deposited data | ||
|
| ||
| Figure Source Data (F1-F4) Antibody titers (AUC) measured by ancestral full-length spike-binding ELISA Participant demographic information |
This Paper | ImmPort SDY2468 |
|
| ||
| Software and algorithms | ||
|
| ||
| GraphPad Prism v10 | GraphPad Software Inc. | RRID:SCR_002798 |
| Python Programming Language v3.9.17 | Python Software Foundation | RRID:SCR_008394 |
| MatPlotLib v3.7.1 | Hunter32 | RRID:SCR_008624 |
| seaborn v0.12.2 | Waskom33 | RRID:SCR_018132 |
| NumPy v1.24.3 | Harris et al.34 | RRID:SCR_008633 |
| SciPy v1.11.1 | Virtanen et al.35 | RRID:SCR_008058 |
| Statistical Analysis System v9.4 (SAS) | SAS Institute Inc. | RRID:SCR_008567 |
| Non-linear mixed effects model (SAS code) | This Paper | https://doi.org/10.5281/zenodo.10467603 |
| REDCap 13.1.37 - © 2024 | Vanderbilt University | RRID:SCR_003445 |
| Canva® Canva Pro | https://www.canva.com/ | |


