SUMMARY
Vitamin B12 (B12) deficiency causes neurological manifestations resembling multiple sclerosis (MS); however, a molecular explanation for the similarity is unknown. FTY720 (fingolimod) is a sphingosine 1-phosphate (S1P) receptor modulator and sphingosine analog approved for MS therapy that can functionally antagonize S1P1. Here, we report that FTY720 suppresses neuroinflammation by functionally and physically regulating the B12 pathways. Genetic and pharmacological S1P1 inhibition upregulates a transcobalamin 2 (TCN2)-B12 receptor, CD320, in immediate-early astrocytes (ieAstrocytes; a c-Fos-activated astrocyte subset that tracks with experimental autoimmune encephalomyelitis [EAE] severity). CD320 is also reduced in MS plaques. Deficiency of CD320 or dietary B12 restriction worsens EAE and eliminates FTY720’s efficacy while concomitantly downregulating type I interferon signaling. TCN2 functions as a chaperone for FTY720 and sphingosine, whose complex induces astrocytic CD320 internalization, suggesting a delivery mechanism of FTY720/sphingosine via the TCN2-CD320 pathway. Taken together, the B12-TCN2-CD320 pathway is essential for the mechanism of action of FTY720.
Graphical Abstract
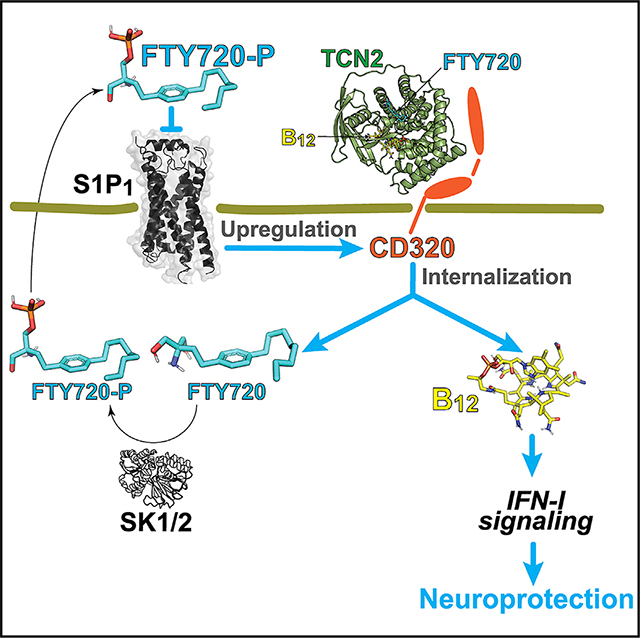
In brief
Jonnalagadda et al. find fingolimod-mediated maintenance of vitamin B12 homeostasis in the CNS. Fingolimod binds directly to TCN2 to form a complex that promotes vitamin B12 availability in astrocytes through internalization of CD320, followed by phosphorylation of fingolimod, which functionally antagonizes S1P1, resulting in upregulation of CD320 in astrocytes.
INTRODUCTION
Multiple sclerosis (MS) is a prototypical neuroinflammatory disease that produces demyelination and neurodegeneration in the CNS.1 An animal model of MS—experimental autoimmune encephalomyelitis (EAE)—involves myelin antigen-primed helper T cells (Th1/Th17) and B cells2,3 that attack the CNS to mimic MS. However, molecular mechanisms involving affected CNS cell types that can be therapeutically accessed to ameliorate disease in MS models and human MS remain incompletely understood. A long-recognized similar spectrum of neurological sequelae observed in MS also occurs with deficiency of vitamin B12 (B12; also called “cobalamin”) such as muscle weakness and cognitive dysfunction,4 suggesting a possibly overlapping disease mechanism; however, such linkage has been considered equivocal in the absence of molecularly defined pathways linking the two.5 The B12 pathway involves formation of a complex between extracellular B12 and transcobalamin 2 (TCN2), which binds to multiple members of the low-density lipoprotein receptor (LDLR) family–megalin/LRP2, cubilin, and CD320/TCblR6–enabling B12 to enter cells. In particular, CD320 mediates CNS B12 access.7
Therapeutic efficacy in MS (and EAE) affecting immune and CNS cell types has been realized through sphingosine 1-phosphate (S1P) receptor modulators.8–10 S1P is a lysophospholipid whose effects are meditated by cognate G protein-coupled receptors (GPCRs).11 FTY720 (known by its generic clinical name fingolimod, which is a structural analog of sphingosine) was the first FDA-approved orally available disease-modifying therapy (DMT) for treating relapsing-remitting MS (RRMS).9,10,12,13 FTY720 is phosphorylated by sphingosine kinases (SK1/2; gene name, SPHK1/2) to produce the active S1P analog FTY720P (fingolimod-P), which binds to four of the five known S1P GPCRs: S1P1,3,4,5.11,14–17 S1P receptor modulators functionally antagonize S1P1 expressed on lymphocytes, resulting in the accumulation of pathogenic lymphocytes in the secondary lymphoid organs.17–19 This effect of reducing the infiltration of pathogenic lymphocytes into the CNS was proposed as a mechanism of action (MOA) of fingolimod.9
However, direct CNS activities of S1P receptor modulators have also been proposed to occur in parallel with immunological activities,10,12,13,20–22 in view of the expression of S1P receptors in the brain23–27 and fingolimod’s preferential accumulation within the CNS.28 EAE studies using astrocyte-specific S1P1 conditional knockout (S1P1-AsCKO) mice implicated S1P1 expressed on astrocytes for FTY720 efficacy29,30 since S1P1-AsCKO mice showed a reduction in circulating lymphocytes in response to FTY720 equivalent to control mice.29 Clinical data also support CNS mechanisms of fingolimod that have been reported to reduce brain volume loss in MS,13,31 contrasting with the increased brain volume loss produced by some anti-inflammatory MS drugs.32
To better understand the CNS cellular targets involved in EAE and MS, we previously used an unbiased in vivo screen to identify the earliest and most affected CNS cell types,30 as detected by immediate-early gene c-fos expression, using a tetracycline transactivator (tTA)-controlled genetic tagging of c-Fos-activated cells that historically marked cellular activation with nuclear green fluorescence protein (GFP).33 This unbiased screen identified immediate-early astrocytes (ieAstrocytes), accounting for over 95% of c-Fos-activated cells during EAE, whose prevalence linearly increased with EAE disease severity30 consistent with ieAstrocytes contributing to the pathogenesis and progression of EAE. Genetic removal or pharmacological inhibition of S1P1 suppressed ieAstrocyte formation,30 indicating that S1P-S1P1 signaling in ieAstrocytes is important in FTY720 efficacy.
Here, the CNS molecular pathways accessed by FTY720 through astrocytes were investigated by analyzing gene expression profiles of isolated ieAstrocytes, under conditions of S1P1 inhibition, using fluorescence-activated nuclear sorting (FANS) combined with RNA sequencing (RNA-seq).34 This strategy identified a B12 pathway, including CD320 and TCN2, that was functionally validated using EAE and human MS brain samples and further supported by the human brain MS resource dataset.35 It also identified a well-known type I interferon (IFN-I) pathway relevant to the target of MS drugs. Notably, physical binding of FTY720 to TCN2 and CNS B12 signaling provide molecular metabolic links to the long-recognized similarities between MS and B12 deficiency.
RESULTS
CD320 was identified by nuclear RNA-seq of ieAstrocytes and was upregulated by S1P1 inhibition
Because we previously identified ieAstrocytes as the key murine astrocyte subset responding to FTY720,30 a nuclear RNA-seq method developed for neurons34 was adapted to assess ieAstrocyte nuclei isolated by FANS from EAE-induced wild-type (WT)fos, S1P1-AsCKOfos (TetTag:S1P1flox/flox:hGFAP-Cre), and WTfos+FTY720 mice (Figures 1A–1C). FANS enabled a more accurate assessment of activated astrocyte transcriptomes by focusing on GFP+ ieAstrocytic nuclei (Figure 1B). Both intronic and exonic reads were assessed from nuclear RNA, which covered ~80% of transcripts in all three groups (Figure S1). Control assessments of S1pr1 (the mouse gene name for S1P1) expression revealed significant loss in the S1P1-AsCKOfos, but not the WTfos or WTfos+FTY720, nuclei (Figure S1), supporting the expected gene deletion in astrocytes by the hGFAP-promoter-driven Cre recombinase.36 In accordance with our previous findings and the definition of ieAstrocytes,30 Fos mRNA expression was decreased in both S1P1-AsCKOfos and WTfos+ FTY720 (Figure S1). These nuclear RNA-seq datasets showed ~92% overlap with a reported gene set for mouse neurotoxic/neuroprotective (A1/A2) reactive astrocytes, but which lacked skewing toward either subtype37 (Figure S1), supporting a reactive yet distinct phenotype from A1/A2 for ieAstrocytes.
Figure 1. FTY720 restores CD320 expression that is downregulated in EAE and MS lesions and is essential for protecting against neuroinflammation.
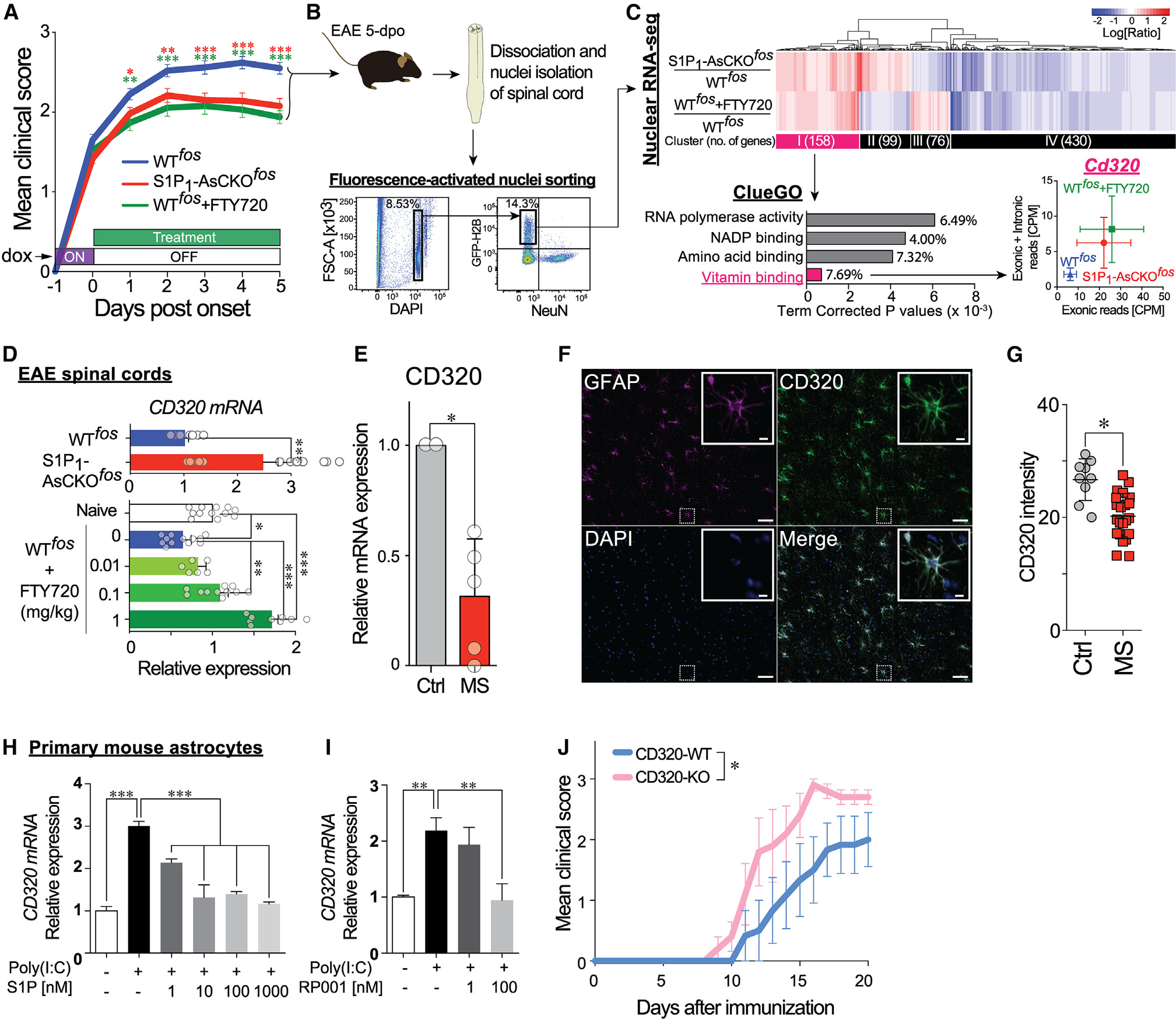
(A) Disease course of EAE-induced WTfos, S1P1-AsCKOfos, and FTY720-treated WTfos (WTfos+FTY720) mice (n = 50, 43, and 32 animals, respectively). Day 0 was designated as the day of clinical sign onset (mean ± SEM, *p < 0.05, **p < 0.01, and ***p < 0.001 by two-way ANOVA with Bonferroni’s multiple comparisons test). The x axis represents “days post-onset,” which is different from “days after immunization.”
(B) Fluorescence-activated nuclei sorting (FANS) to isolate NeuN−GFP+ singlet nuclei from EAE SCs at 5 days post-onset, which is equivalent to 5 days off doxycycline.
(C) Nuclear RNA-seq. Heatmap displays 2-fold up- and downregulated genes in S1P1-AsCKOfos and WTfos+FTY720 vs. WTfos. Top 4 significantly enriched gene ontologies in cluster I are identified by ClueGO38 of Cytoscape.39 Expression levels of Cd320 (count per million [CPM]) are plotted (mean ± SEM).
(D) Expression of Cd320 mRNA in EAE SCs determined by qPCR (mean ± SEM, *p < 0.05, **p < 0.01, and ***p < 0.001 by t test [top] and by one-way ANOVA with Tukey’s multiple comparisons test [bottom]).
(E) Cd320 mRNA expression in human MS normalized to control brains (mean ± SEM, n = 5 and 2 brains, respectively; *p < 0.01 by one-sample t test).
(F) Immunohistochemistry (IHC) for CD320 and GFAP in human control brain. Scale bars, 100 μm and 5 μm (inset).
(G) CD320 immunolabeling is significantly reduced in human MS plaques as compared to control brain. Each symbol represents an assessed region of interest in the sections. *p < 0.001 by Mann-Whitney U test.
(H and I) Cd320 mRNA expression in primary astrocytes stimulated with 10 μg/mL poly(I:C) for 24 h in the presence of S1P (H) or the S1P1-specific agonist RP001
(I). Data are from three independent experiments of three technical replicates (mean ± SEM, **p < 0.01 and ***p < 0.001 by one-way ANOVA with Tukey’s multiple comparisons test).
(J) EAE disease course of CD320-WT and CD320-KO mice (n = 6 and 5 animals, respectively). Data are from three independent experiments (mean ± SEM, *, statistical significance was analyzed by two-way ANOVA; interaction, p = 0.90; time, p < 0.0001; genotype, p < 0.001).
Hierarchical clustering of 2-fold up- or downregulated genes in S1P1-AsCKOfos and WTfos+FTY720 vs. WTfos nuclei revealed four distinct clusters (I–IV) (Figure 1C; Table S1). Cluster I consisted of 158 upregulated genes that were of primary interest in identifying anti-neuroinflammatory genes produced by S1P1 inhibition. The ClueGO plugin38 of Cytoscape39 identified a vitamin binding pathway as the most significantly enriched Gene Ontology (GO) molecular function term (Figure 1C). Reactome pathway analysis40 identified 27 pathways (p < 0.05; Table S2) composed of 8 genes (Cd320, Dhfr, Tlr3, Eif2ak2, Ppp2cb, Hist1h2bf, Cdc7, and Psmb2). Further analyses by quantitative PCR (qPCR) of these 8 genes in the spinal cords (SCs) of chronic EAE mice revealed that Cd320 was significantly downregulated during EAE (Figure S1), which was supported by previously reported RNA-seq data that compared naive vs. EAE astrocytes (~20%).41 Critically, Cd320 expression was increased during amelioration of EAE through S1P1 genetic deletion or pharmacological inhibition by FTY720 exposure in animals (Figure 1D).
CD320 expression was downregulated in human MS brains
CD320 is a type I membrane protein that belongs to the LDLR family that transports the TCN2-B12 complex into CNS cells.42 It has a primary role in maintaining brain B12 homeostasis, as demonstrated by greater than 95% reduction of B12 in the CNS of CD320-KO mice.43 To assess the human relevance of CD320 downregulation in MS, CD320 expression was assessed in human brains by qPCR, which identified a significant reduction (30% controls) in MS plaques (Figure 1E). These results are consistent with publicly available datasets derived from chronic MS plaques.44 CD320 immunoreactivity was examined in human brain sections containing MS plaques as compared to non-diseased controls (Figures 1F–1I). GFAP+ astrocytes expressed CD320 (Figure 1H), the expression of which was significantly downregulated in MS plaques (Figure 1I). These results support the relevance of CD320 and its related pathways to the human MS brain.
Analyses in vitro and in vivo supported worsening of disease with CD320 loss
The restoration of Cd320 expression during EAE by genetic or pharmacological S1P1 inhibition (Figure 1D) indicated that S1P1 activation negatively regulates CD320 expression. Primary mouse astrocytes were examined in culture by first upregulating (2- to 3-fold) Cd320 expression via exposure to polyinosinic-polycytidylic acid (poly(I:C)), a Toll-like receptor 3 (TLR3) agonist45 that ameliorated EAE.46 As expected from the co-elevation of Tlr3 and Cd320 in ieAstrocytes (Figure S1J), poly(I:C) increased Cd320 expression (Figures 1J and 1K). Both S1P and a short-acting S1P1-specific agonist, RP001,47 inhibited poly(I:C)-induced Cd320 expression in a dose-dependent manner (Figures 1J and 1K) consistent with increased S1P levels reported in human MS brain lesions48 and the reduced CD320 observed here (Figures 1E–1I). Linkage between S1P1 and CD320 (CD320 downregulation by S1P1 inhibition) predicted that genetic removal of CD320 should exacerbate EAE. To test this hypothesis, CD320-WT and CD320-KO mice were challenged with EAE, which resulted in a more severe disease course with earlier onset in the CD320-KO compared to WT controls (Figures 1L and S1) that was independent of peripheral blood lymphocyte counts (Figure S1). These results showed that CD320 loss exacerbates EAE.
B12 restriction also worsened EAE, with parallel activation of inflammatory responses and suppression of IFN-I signaling
Downregulation of CD320 in both EAE and MS may result in B12 deficiency within the CNS, although the effects of B12 deficiency on EAE severity have not been previously reported. B12 is an essential vitamin obtained through diet.49 Mice deficient in B12 (B12def) were produced through ~8–9-week dietary restriction50 followed by EAE challenge. B12def mice exhibited severe EAE (Figures 2A and S2), significant B12 reduction in SCs (Figure 2B), and increased histological damage (Figure 2C) but equivalent T cell proliferative responses against MOG35–55 peptide as compared to controls (Figure S2). In addition, CD320 protein levels were also downregulated (by ~40%) in B12def-EAE SCs compared to controls (Figure S2).
Figure 2. Vitamin B12 restriction worsens EAE, activates astrocytes, and reduces IFN-I signaling.
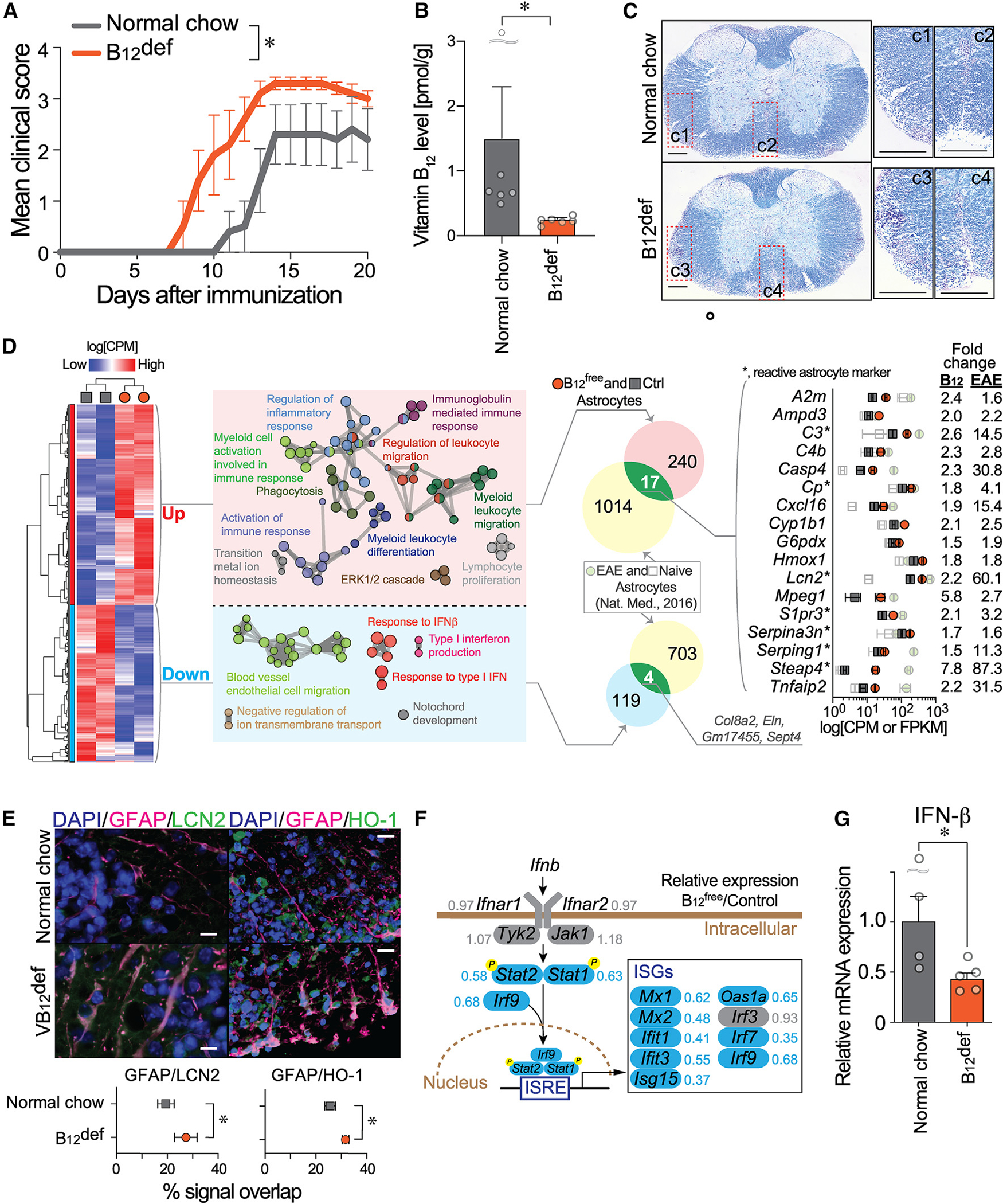
(A) EAE disease course of control (normal chow) and B12-deficient (B12def) diet-fed mice (n = 5 animals). Data are from two independent experiments (mean ± SEM; *, statistical significance was analyzed by two-way ANOVA; interaction, p = 0.32; time, p < 0.0001; diet, p < 0.001).
(B) Vitamin B12 levels in the EAE SCs (n = 6; mean ± SEM; *p < 0.01 by Mann Whitney U test.)
(C) Luxol fast blue staining of EAE SCs at 15 days post-immunization. Regions of interest (C1–C4) are magnified. Scale bar, 200 μm.
(D) Heatmap for up- and downregulated differentially expressed genes (DEGs) produced from RNA-seq data of cultured astrocytes in serum-free B12free and control media. DEGs were analyzed using ClueGO plugin38 of Cytoscape,39 and the functional groups are shown as networks with different colors. DEGs were compared with EAE astrocytes,41 which identified 17 and 4 commonly up- and downregulated genes, respectively. Expression levels of commonly upregulated genes in B free/control (Ctrl) (CPM) and EAE/Ctrl (FPKM [fragment per kilobase of transcripts per million]) are plotted.
(E) IHC validation for upregulated genes; LCN2 or HO-1 (green) and GFAP (red) in the EAE SC at 21 days post-immunization. Scale bar, 10 μm. Overlapping signals of LCN2 or HO-1 in GFAP+ astrocytes are plotted (mean ± SEM, n = 5 animals, *p < 0.05 by unpaired t test).
(F) Detailed mapping of DEGs within the astrocyte signaling cascade for the IFN-I system. The numbers next to the gene names are the expression ratio of B12free to control. ISRE, IFN-stimulated response element; ISGs, IFN-stimulated genes.
(G) IFN-β mRNA expression in the EAE SCs was determined by qPCR (*p < 0.05 by unpaired t test).
To explore the mechanisms underlying increased EAE severity in B12-restricted animals, the transcriptomic consequences of B12 loss in astrocytes were examined. Primary mouse astrocyte cultures were maintained for 2 weeks in B12-free (B12free) vs. control media, both of which supported growth and GFAP positivity. RNA-seq analysis identified 257 upregulated and 123 downregulated differentially expressed genes (DEGs; showing > 1.5-fold, p < 0.05, or < 0.6-fold, p < 0.05, respectively: Figure 2D; Table S3). GO analyses of upregulated DEGs revealed enrichment of multiple pro-inflammatory cascades (Figure 2D). Although no obvious astrocyte class A1/A2 skewing37 was observed (Figure S2), B12free astrocytes shared 17 DEGs with EAE astrocytes41 (Figure 2D), in which 7 genes were shared with A1/A2 reactive astrocyte genes.37 Protein expression of upregulated genes in astrocytes was supported by co-immunolabeling for GFAP and proteins for two identified genes, lipocalin-type 2 (LCN2/Lcn2) and heme oxygenase-1 (HO-1/Hmox1), in B12def EAE SCs (Figure 2E). Interestingly, despite no exposure to inflammatory stimuli, B12free astrocytes displayed a reactive astrocyte phenotype, mplicating B12-CD320 in maintaining astrocytes in quiescent or non-reactive states.
IFN-I signaling pathways, whose genes were markedly downregulated in B12free astrocytes as compared to controls, were identified by GO analysis (Figure 2F). The mRNA expression of an endogenous ligand for IFN-I receptors—IFN-β—showed significant downregulation in B12def EAE SCs over controls (Figure 2G), which appeared to be produced by microglia in the EAE-affected CNS.46,51 Taken together, loss of B12 promotes neuroinflammation by inducing astrocyte pro-inflammatory responses, reducing IFN-I sensitivity in astrocytes, and suppressing IFN-β production in microglia and possibly other immune cell types.
Loss of CD320 or B12 eliminated efficacy of FTY720 in EAE
S1P1 inhibition restored CD320 expression in the CNS of EAE mice (Figure 1D), raising the possibility that FTY720’s beneficial effects in EAE could be reduced or diminished in CD320-KO mice or dietarily B12-restricted mice. EAE-induced WT mice, which served as positive controls, exhibited the expected ameliorating effects of FTY720 treatment on EAE scores (Figure 3A), recovery rate (Figure 3B), reductions in peripheral blood lymphocyte counts (Figure S3), and reduced IFN-β mRNA expression (Figure 3C). In marked contrast, FTY720 did not ameliorate EAE in CD320-KO mice (Figures 3A and 3B) yet still reduced peripheral blood lymphocyte numbers (Figure S3). Moreover, FTY720 did not significantly restore IFN-β mRNA expression in CD320-KO mice while showing a slight increase as compared to vehicle controls (Figure 3C).
Figure 3. Loss of FTY720 efficacy during EAE in CD320-deficient and dietary-B12-restricted mice.
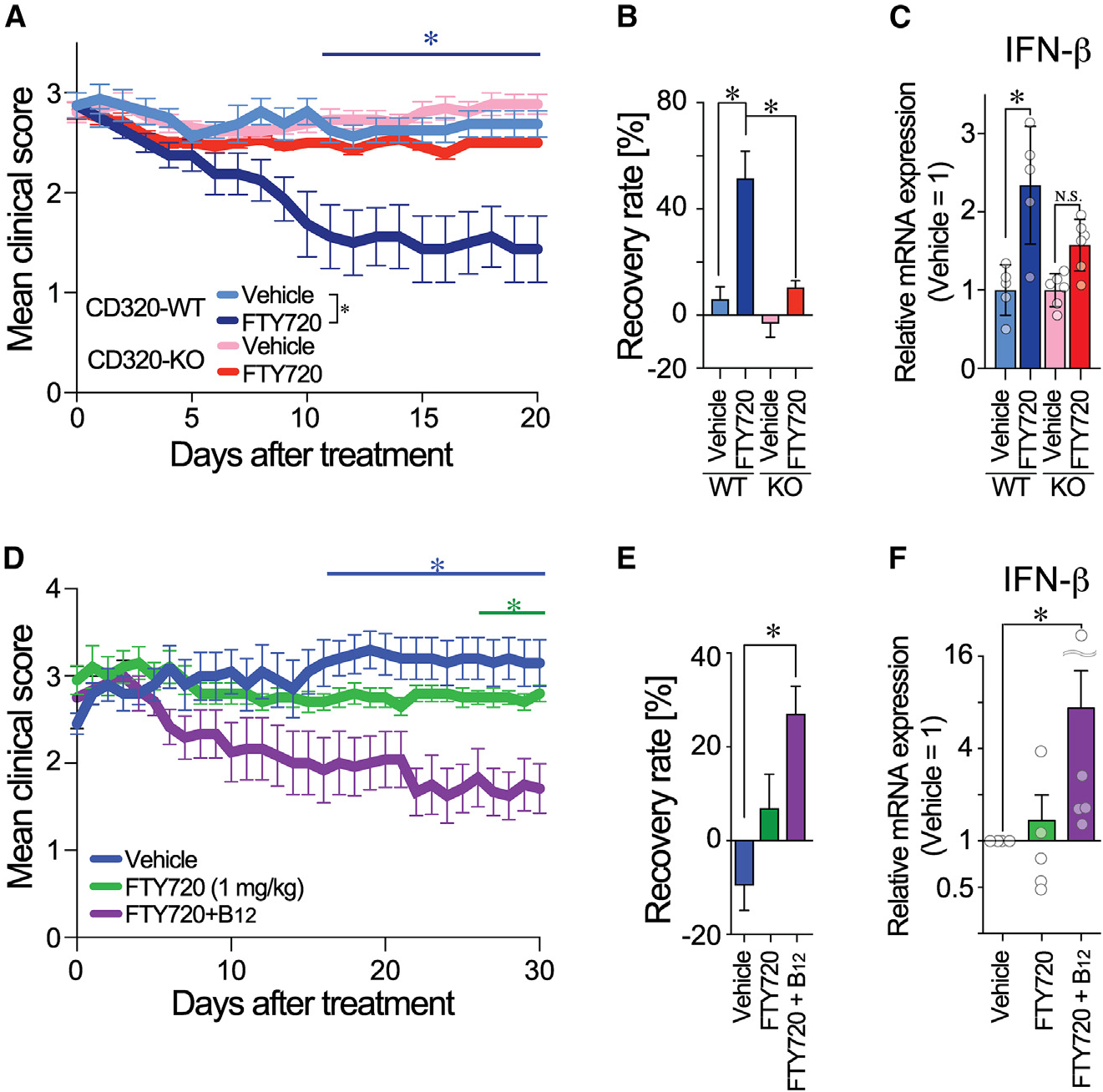
(A) FTY720 treatment on EAE-induced CD320-WT (n = 8) and CD320-KO mice (n = 14). Data are combined from two independent experiments (mean ± SEM; *, statistical significance was analyzed by two-way ANOVA between days 9 and 20; interaction, p = 0.16; time, p < 0.01; genotype, p < 0.01).
(B) Recovery rate with FTY720 treatment (*p < 0.05 by one-way ANOVA with Bonferroni’s multiple comparisons test).
(C) IFN-β mRNA expression in EAE SCs (*p < 0.05 by one-way ANOVA with Bonferroni’s multiple comparisons test; N.S., non-significant, n = 5–6).
(D) Therapeutic treatment of EAE-induced B12def mice with vehicle, FTY720, and FTY720 + B12 (n = 5, 5, and 6 animals, respectively). Data are representative of 2 independent experiments (mean ± SEM; *p < 0.05 vs. vehicle by two-way ANOVA; interaction, p = 0.27, time, p = 0.74; diet, p < 0.0001 with Bonferroni’s multiple comparisons test).
(E) EAE recovery rate with FTY720 treatment (*p < 0.05 by one-way ANOVA with Bonferroni’s multiple comparisons test).
(F) IFN-β mRNA expression in EAE SCs (*p < 0.05 by one-way ANOVA with Bonferroni’s multiple comparisons test, n = 5–6).
Since B12 is essential for protein synthesis, its deficiency suppressed CD320 expression (Figure S2). B12def-EAE mice might also lose responsivity to FTY720. As was the case in CD320-KO mice, FTY720 lost efficacy in B12def-EAE mice (Figures 3D and 3E), even though FTY720 reduced peripheral blood lymphocyte counts (Figure S3). Importantly, B12def-EAE mice that were administered combination therapy (FTY720 + B12) exhibited the greatest response in reducing disease score (Figure 3D), an increased recovery rate (Figure 3E), and elevated IFN-β mRNA expression in SCs (Figure 3F). Therapeutic effects and IFN-β expression correlated with the recovery of CD320 expression observed in the FTY720 + B12 group (Figure S3). Collectively, FTY720 requires either CD320 or B12 to ameliorate EAE and has synergistic therapeutic effects with B12.
FTY720-TCN2 binding showed subnanomolar affinity and induced CD320 internalization
The loss of FTY720 efficacy in CD320-KO mice (Figure 3A) generated a hypothesis that CD320 might be a carrier for FTY720 via direct binding. This was tested using a compensated interferometric reader (CIR) that is a label-free, optical technique for measuring molecular interactions in free solution.52,53 A positive control included CD320 binding to TCN2 (Figure 4A), whose KD value (1.86 nM; 95% confidence interval [CI], 0.13–17.05; R2 = 0.91) was comparable to previously published values.7 However, no binding signals were observed between CD320 and FTY720 or FTY720P (Figure 4B). By comparison, sphingosine (KD = 0.36 nM; 95% CI, −0.01~73.2; R2 = 0.81) and S1P (KD = 11.8 nM; 95% CI, 0.56–infinity; R2 = 0.91) did show binding to CD320 (Figure 4C). Because the hypothesis of FTY720-CD320 interactions was rejected, an alternative hypothesis was that FTY720 instead directly binds to TCN2. As expected, TCN2 bound B12 (KD = 0.19 nM; 95% CI, −0.19 to 11.2; R2 = 0.83) (Figure 4D). TCN2 also bound both FTY720 (KD = 0.24 nM; 95% CI, 0.03–1.79; R2 = 0.96; Figure 4E) and sphingosine (KD = 0.14 nM; 95% CI, −0.02 to 49.6; R2 = 0.88; Figure 4F), whereas little or no binding was identified for FTY720P (Figure 4E) and S1P (Figure 4F). A next-generation S1PR modulator, BAF312 (siponimod), which is not a sphingosine analog and does not require in vivo phosphorylation, did not show binding to TCN2 (Figure S4).
Figure 4. Direct binding of FTY720 with TCN2 induces astrocyte CD320 internalization.
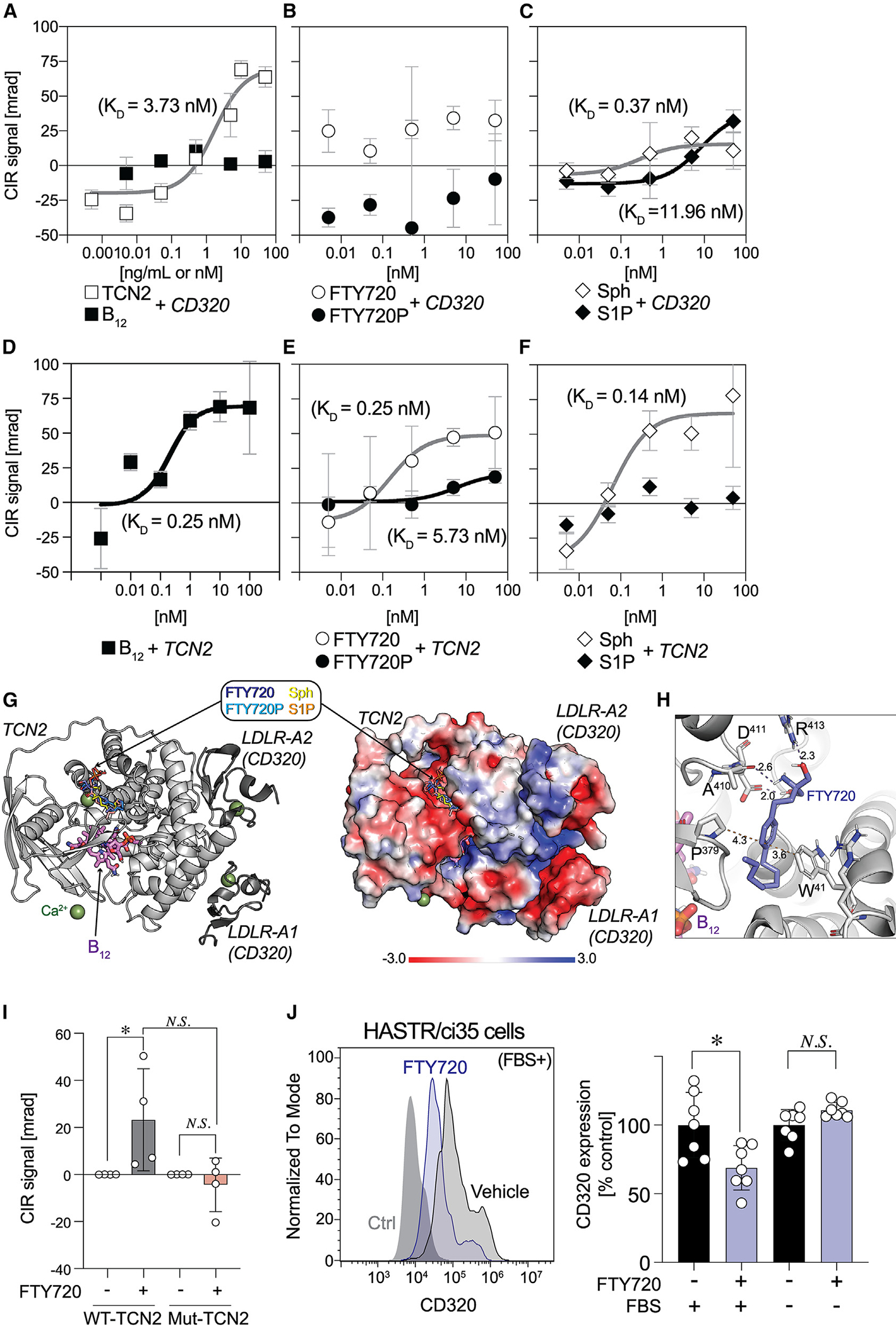
(A‒C) Specific binding curves between CD320 vs. TCN2 and B12 (A), FTY720 and FTY720P (B), and sphingosine (Sph) and S1P (C).
(D‒F) Specific binding curves between TCN2 vs. B12 (D), FTY720 and FTY720P (E), and Sph and S1P (F). Compensated interferometric reader (CIR) signals are plotted against concentrations of binding partners and fitted by non-linear regression using the Michaelis-Menten equation (mean ± SEM, n = 6–7).
(G) Computational modeling of FTY720 binding site on TCN2 (PDB: 4ZRP). Cartoon representation of TCN2 (light gray) and LDLR domains of CD320 (dark gray) (left) and electrostatic surface of these complex (right).
(H) Magnified view of a potential FTY720 binding site in TCN2. Interactions are represented by dashed lines with distances between atoms (Å).
(I) CIR binding signals of WT-TCN2 and mutant TCN2 (Mut-TCN2) to 100 nM FTY720 (mean ± SEM, n = 16 and 8 for WT-TCN2 and Mut-TCN2, respectively, pooled from at least 2 independent experiments; *p < 0.05 by one-way ANOVA with Dunn’s multiple comparisons test; N.S., non-significant).
(J) A representative flow cytometry (FCM) histogram of CD320 expression on HASTR/ci35 cells stimulated with or without FTY720 (1 μM) in the presence or absence of FBS (left). Normalized CD320 expression levels (mean ± SEM, n = 6–7, pooled from 2 independent experiments; *p < 0.05 by Kruskal-Wallis test with Dunn’s multiple comparisons test; N.S., non-significant).
Neither FTY720 nor sphingosine altered the affinity of TCN2 binding to B12 (Figure S4), indicating distinct FTY720 binding sites compared to B12. Molecular modeling identified a potential binding site of FTY720 and sphingosine on the surface of TCN2, which was distinct from a B12 binding pocket (Figure 4G). The model identified interactions between the benzene ring of FTY720 and W41/P379 of TCN2 and between a polar group of FTY720 and D411/R413 of TCN2 (Figure 4H). Reduced binding of FTY720P or S1P to TCN2 might be explained by electrostatic repulsion between the phosphate groups of the ligands vs. the negatively charged surface of the binding site (Figure 4G). Mutagenesis of four putative interacting TCN2 residues (W41/P379/D411/R413) by alanine replacement generated a mutant (Mut-TCN2) used to examine FTY720 binding (Figure 4I). Positive controls using WT-TCN2 protein showed robust CIR signals in the presence of FTY720, while no binding signals were detected when using the Mut-TCN2 (Figure 4I). Overall, these data supported TCN2 as a carrier and possible chaperone for both FTY720 and sphingosine that also binds and transports B12 into astrocytes in conjunction with CD320.
To assess whether FTY720 induces CD320 internalization, CD320 cell surface expression was evaluated by flow cytometry using an immortalized human astrocyte cell line, HASTR/ci35 cells.54 FTY720 exposure reduced CD320 expression in the presence of bovine serum but not in the absence of serum (Figure 4I). It is important to note that bovine serum contains copious amounts of TCN2 that are secreted into the plasma by the vascular endothelium and carry the newly absorbed B12 to all organs.42 Moreover, bovine TCN2 has conserved amino acid residues with human TCN255 that were determined to be essential residues for FTY720/sphingosine binding. Sphingosine also induced CD320 internalization (Figure S4), indicating that FTY720- or sphingosine-bound TCN2 increases astrocytic B12 availability by driving internalization of the FTY720/TCN2-CD320 complex.
DISCUSSION
A molecular link between B12 and MS was identified through a novel CNS mechanism involving astrocytes that potentially explains the controversial relationship between B12 deficiency and MS that has spanned decades without resolution.4 The linkage consisted of (1) Cd320 downregulation in ieAstrocytes and reduced CD320 in human MS lesions; (2) worsening of EAE by both CD320 genetic deletion and B12 deficiency; (3) detection of specific, physical binding between FTY720 and the B12 carrier protein TCN2; and (4) CD320 internalization in astrocytes enhanced by FTY720. This direct CNS effect of fingolimod (FTY720) implicates neuroprotective functions involving the B12-TCN2-CD320 pathway and IFN-I signaling (Figure 5).
Figure 5. Schematic diagram of the FTY720 (fingolimod) CNS mechanism of action (MOA) through B12 and IFN-β signaling.
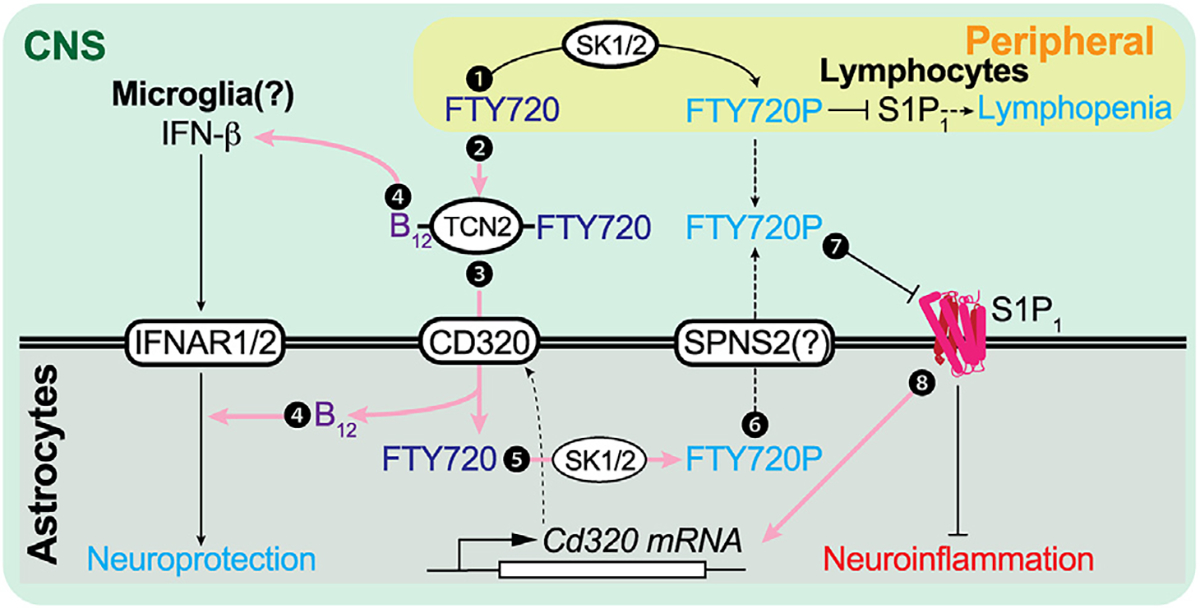
Pink arrows indicate newly identified pathways that contribute to the direct CNS effects of FTY720 beyond functional antagonism of astrocyte S1P1. ① S1P1 inhibition in peripheral lymphocytes is the originally proposed CNS MOA. ② FTY720 is complexed with B12-TCN2. ③ This FTY720-TCN2-B12 complex is taken up by astrocytes via CD320, followed by dissociation of the complex. ④ B12 is essential for astrocyte IFN-I sensitivity and microglial IFN-β production. ⑤ Astrocyte SK1/2 phosphorylates FTY720 to form FTY720P.35 ⑥ The resulting FTY720P may be transported via SPNS256 and functionally antagonizes S1P1 in an autocrine/paracrine manner. ⑦ FTY720P functionally antagonizes astrocyte S1P1 and downregulates cell surface expression of S1P1. Ⓑ Astrocytic S1P1 inhibition increases CD320 expression, enabling increased engagement of and astrocyte internalization of the B12-TCN2-FTY720 complex. SK1/2, sphingosine kinase 1/2; SPNS2, SPNS lysolipid transporter 2; TCN2, transcobalamin; IFNAR, interferon α/β receptor.
CD320 is downregulated in ieAstrocytes during neuroinflammation
Astrocytes have long been considered bystanders in neuroinflammatory diseases.57 However, ieAstrocytes are actively involved in neuroinflammation that tracks with EAE disease severity.30 Molecular neuroinflammatory responses in ieAstrocytes are partly initiated by S1P-S1P1 signaling,30 resulting in c-Fos-dependent CD320 downregulation.58 Astrocytic CD320 downregulation in both EAE SCs (Figures 1A–1D) and human MS plaques (Figures 1E–1I) is consistent with reported increases in S1P levels and S1P1 expression in both EAE SCs and human MS lesions.59,60 CD320 downregulation in MS/EAE lesions could reduce astrocyte B12 availability (i.e., uptake of B12), resulting in a negative feedback loop to downregulate CD320 production (Figure S3). CD320 expression in chronic EAE astrocytes maintained persistent downregulation,41 implicating ieAstrocytes as a potential precursor of reactive astrocytes. Another astrocyte classification nomenclature defined originally in mouse proposed “harmful” A1 and “helpful” A2 reactive astrocytes. An additional analysis of human MS astrocytes retained this nomenclature but utilized non-overlapping gene sets to define the A1 subtype (the A2 subtype was not reported in human brains).37 ieAstrocytes and B12free astrocytes expressed most of the proposed mouse A1/A2/Pan genes, but they could not be specifically categorized as the A1 or A2 subtype (Figures S1 and S2). It is possible that ieAstrocytes and related subtypes temporally precede A1/A2 subtypes. Developing an understanding of MS astrocyte subtypes warrants more investigation; however, our findings implicate ieAstrocytes defined by c-Fos expression as a key element of EAE-/MS-specific reactive astrocytes.
TCN2 functions as a sphingosine and FTY720 chaperone promoting CD320 signaling
In blood, S1P exists as a complex with its protein carrier chaperones that include albumin, apolipoprotein M (ApoM), and ApoA4, which presents S1P to its cognate receptors.61 S1P concentrations are higher in blood than lymph, while sphingosine levels show an inverse trend.62 TCN2 was identified here as a putative chaperone for sphingosine and FTY720. The Human Protein Atlas database reported the second highest expression level for TCN2 within lymphoid organs compared to all other organ systems,63 which is consistent with higher sphingosine levels in the lymph.62 Although neither FTY720 nor sphingosine altered the binding affinity of B12 to TCN2, both induced CD320 internalization (Figures 4 and S4). Sphingosine and its chemical analog FTY720 may thus act as TCN2 co-factors for the bound B12, forming a complex with CD320 and thus enabling B12 to enter astrocytes. Increased sphingosine levels in MS brains and EAE SCs48 may reflect a compensatory mechanism to increase B12 in the brain (Figure 1). The FTY720-TCN2 complex may also affect methylation processes of myelin proteins in oligodendrocytes via B12 signaling, which could be linked to the well-documented reversible demyelination in B12 deficiency.4 It is notable that B12 availability in the CNS through TCN2-CD320 may not be accurately reported by peripherally measured B12 levels, which highlights a need to assess B12 brain levels.
A direct CNS MOA for FTY720 is supported by astrocyte B12 and S1P1 signaling
Direct CNS activities of FTY720 beyond immunological mechanisms have received previous support from studies on S1P1-AsCKO mice challenged with EAE.29,30 These mice showed a loss of FTY720 efficacy as compared to control mice along with a baseline reduction in clinical score despite maintaining FTY720’s immune cell trafficking effects.29 Complementary results were obtained here in EAE mice lacking either CD320 or B12 that were also refractory to FTY720 therapy (Figures 1L and 2A) but showed a baseline increase in clinical disease score consistent with reduced CD320 gene expression from ieAstrocytes (Figure 1D). FTY720 treatment that produces functional S1P1 antagonism also restored CD320 expression (Figure 1D). This process requires SK (SK1 and/or SK2) activity that phosphorylates FTY720 to the active compound, FTY720P. Single-nucleus RNA-seq identified loss of SPHK1/2-expressing cells in secondary progressive MS (SPMS) brains as compared to RRMS brains. Further analyses using SK1/2-AsCKO mice showed a loss of FTY720 efficacy.35 In this light, a failed phase 3 clinical trial investigating FTY720’s efficacy in patients with primary progressive MS (PPMS)64 may be explained by the loss of SKs from the PPMS brain.
These results, along with the expression of brain S1P receptors and accumulation of FTY720 in the CNS,23–28 implicate a direct CNS MOA for FTY720 via astrocyte S1P1 that is amplified by B12 signaling (Figure 5). These direct CNS effects of fingolimod may also explain clinical data on reduced atrophy31,65,66 that contrasts with increased brain atrophy—”pseudoatrophy”—observed with the anti-inflammatory immunosuppressive agent natalizumab,32 as increased brain atrophy has been reported under conditions of reduced B12, including that reported in CD320-KO mice67 and human B12-deficient patients.68
Astrocyte IFN-I components are accessed by FTY720
IFN-I members include IFN-β, which was the first FDA-approved and is the current therapy for patients with MS (IFN-β1b, Beta-seron).69 It was developed as a peripheral immunomodulatory agent, while its CNS effects emerged later.70 More severe EAE in IFN-β-KO mice71 likely reflects the normally protective effects of CNS IFN-β,46,51 a mechanism further supported by silencing an astrocytic IFN-I receptor (IFNAR1/Ifnar1), which aggravates EAE.41 A new relationship between B12-TCN2-CD320-FTY720 and IFN-I signaling was identified here based on B12-deficient conditions that removed FTY720 efficacy, reduced astrocytic IFN-I sensitivity, and reduced endogenous IFN-β production (Figure 2). This relationship may be involved in IFN-β resistance reported in EAE27 and nonresponders to IFN therapy in MS.72–74 Moreover, a prior study reporting the positive effects of B12 on IFN-β therapy in EAE may reflect these CNS mechanisms via not only astrocyte modulation but also through promoting oligodendrocyte maturation by reducing Notch1 and increasing sonic hedgehog signaling.75
An additional relationship between S1P1 signaling and IFN-I was previously reported where S1P1 activation inhibited IFN-I autoamplification via c-Fos activation in astrocytes76 and via IFNAR1 degradation in myeloid cells.77 S1P1 functional antagonism would counter these effects, and it is consistent with IFN-β induction by FTY720 in EAE mice (Figure 3). Notably, reduced IFN-β expression was not rescued by FTY720 alone in EAE mice generated with CD320 loss or B def (Figure 3), underscoring the connected relationships among these elements (Figure 5).
In conclusion, a tangible molecular link between B12 deficiency and MS was identified through the effects on EAE of loss of B12 or CD320 in astrocytes, along with identification of a novel physical binding of B12-TCN2 with FTY720. B12 signaling appears to be capable of impacting fingolimod efficacy as well as disease severity mediated by sphingolipids in MS. The newly identified TCN2 binding of FTY720 promotes B12 and FTY720 availability within astrocytes. These data support the use of CNS-penetrant B12 supplementation during fingolimod clinical treatment and suggest a need for assessing B12 signaling in the CNS beyond peripheral B12 measurements. Other S1P receptor modulators8,9 may access one or more FTY720-B12 pathways, particularly restoring CD320 expression levels in astrocytes, without a requirement for phosphorylation by SK activity. Beyond MS,1 neuroinflammation in the CNS has been associated with other brain diseases including, for example, Alzheimer’s disease78 and neuropsychiatric disorders,79 extending the potential biological and therapeutic relevance of astrocyte-B12 signaling.
Limitations of the study
While we have clearly demonstrated the functional and physical interactions between the sphingolipid and B12 pathways, there are limitations related to the validation of some of the studies. First, TCN2-KO mice have not been available to date, limiting the validation of the in vivo efficacy of FTY720 in EAE-induced TCN2-KO mice. Second, FTY720-induced CD320 internalization in the presence of serum may need to be directly validated using recombinant TCN2, which requires large amounts of recombinant protein and a sophisticated experimental strategy. Finally, B12 supplementation is clinically recommended for patients with MS. However, for patients treated with MS drugs other than fingolimod, B12 delivery to the CNS is more important than simple supplementation. This study supports the endogenous lipid sphingosine as an alternative to fingolimod, but poor absorption of sphingosine limits its use as a supplement. These limitations need to be addressed in the future.
STAR★METHODS
Detailed methods are provided in the online version of this paper and include the following:
RESOURCE AVAILABILITY
Lead contact
Further information and requests for the resources and reagents should be directed and will be fulfilled by the lead contact, Yasuyuki Kihara (kihara-yasuyuki@umin.net).
Materials availability
Mouse lines and HASTR/ci35 cells used in this study are available upon request. This study did not generate new reagents.
Data and code availability
RNA-seq data that supports the findings of this study are provided as Supplemental Tables (Tables S1 and S3). Fastaq files have been deposited into Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/) Accession number is listed in the key resources table.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| anti-CD320 | Proteintech | RRID:AB_2074223 |
| anti-Lcn2 | R&D Systems | RRID:AB_355022 |
| anti-Hmox1 | ThermoFisher Scientific | RRID:AB_2544814 |
| anti-GFAP | Neuromics | RRID:AB_10014322 |
| Alexa Fluor 488 | ThermoFisher Scientific | RRID:AB_143165 |
| Alexa Fluor 568 | ThermoFisher Scientific | RRID:AB_2534098 |
| anti-NeuN | MilliporeSigma | RRID:AB_2571567 |
| anti-rabbit APC | ThermoFisher Scientific | RRID:AB_429727 |
| HRP conjugated anti-rabbit or anti-mouse IgG(H + L) | Cell Signaling Technologies | RRID:AB_2099233 |
|
| ||
| Biological samples | ||
|
| ||
| Human MS and control brain | Rocky Mountain MS Center Tissue Bank | N/A |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| MOG35–55 | EZBiolab | cat # cp7203–5 |
| Pertussis toxin | List Biological Laboratories | cat # 180 |
| Vitamin B12 | Sigma | cat #V2876 |
| Recombinant human CD320 | R&D Systems | cat # 1557-CD-050 |
| Recombinant human TCN2 | R&D Systems | cat # 7895-TC-050 |
| FTY720 | Novartis | N/A |
| FTY720-P | Novartis | N/A |
| Sphingosine | Avanti Polar Lipids | cat # 860490P |
| S1P | Avanti Polar Lipids | cat # 860492P |
|
| ||
| Critical commercial assays | ||
|
| ||
| Picopure™ RNA isolation kit | ThermoFisher Scientific | cat # KIT0204 |
| RNeasy minikit | Qiagen | cat # 74104 |
| SMART-seq v4 Ultra Low Input RNA Kit for Sequencing | Takara Bio | cat # 634888 |
| Nextera XT Library Preparation Kit | Illumina | cat # FC-131-1024 |
| NEB Next Ultra Pure RNA library kit for Illumina | New England Biolabs | cat #E7770S |
|
| ||
| Deposited data | ||
|
| ||
| GSE99115 | Gene Expression Omnibus | https://www.ncbi.nlm.nih.gov/geo/ |
|
| ||
| Experimental models: Cell lines | ||
|
| ||
| HASTR/ci35 | Furihata, T. et al.54 | N/A |
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| Tg(tetO-HIST1H2BJ/GFP)47Efu/J × Tg(Fos105 tTA)1Mmay | Matsuo, N. et al.33 | RRID:IMSR_JAX:005104 |
| S1P1 flox/flox:hGFAP-cre mice | Choi, J.W. et al.29 | N/A |
| CD320-KO | Lai, S.C. et al.43 | N/A |
|
| ||
| Oligonucleotides | ||
|
| ||
| PCR primer sets | This paper | Table S4 |
|
| ||
| Software and algorithms | ||
|
| ||
| FlowJo (10.0.8r1) | BD | N/A |
| Prism | GraphPad | N/A |
| R | r-project.org | RRID:SCR_001905 |
|
| ||
| Other | ||
|
| ||
| Vitamin B12-deficient diet | Research Diets, Inc | N/A |
EXPERIMENTAL MODEL AND STUDY PARTICIPANT DETAILS
Animals
All animal protocols were approved by the Institutional Animal Care and Use Committee (IACUC) of the Sanford Burnham Prebys Medical Discovery Institute and conform to National Institutes of Health guidelines and public law. The TetTag c-Fos reporter mice were generated by crossing Tg(Fos-tTA)1Mmay and Tg(tetO-HIST1H2BJ/GFP)47Efu/J mice, which expressed a green fluorescent protein-histone H2B fusion protein (GFP-H2B) under a tetO promoter controlled by a c-Fos inducible tetracycline transactivator (tTA) protein as described earlier.80 S1P1flox/flox:hGFAP-cre mice29 were crossed with TetTag mice for multiple generations to generate astrocyte specific S1P1 deletion (S1P1-AsCKOfos, TetTag:S1P1flox/flox:hGFAP-cre) and their littermate controls (WTfos, TetTag:S1P1flox/flox). TetTag mice were maintained on doxycycline chow (DOX, 40 mg/kg; Bio-Serv) throughout breeding, birth, and development to prevent GFP-H2B expression until experimental examination. CD320-KO mice were generated as described previously.43 C57BLJ/6 mice were maintained on a vitamin B12 deficient diet (removal of vitamin B12 from #D07012902, Research Diets, Inc.) as needed.
METHOD DETAILS
EAE
EAE was induced in 7- to 13-wk old female mice as described previously.81 Briefly, mice were immunized subcutaneously with 150 μg MOG35–55 (MEVGWYRSPFSRVVHLYRNGK, EZBiolab) in PBS and complete Freund’s adjuvant (BD Biosciences, cat # 263910) containing 4 mg/mL M. Tuberculosis H37Ra (BD Biosciences, cat # 231141) with or without intraperitoneal injection of 250 ng pertussis toxin (List Biological Laboratories, cat # 180) on day 0 and day 2. Daily clinical scores corresponding to the most severe sign observed were given as follows: 0, no sign; 0.5, mild loss of tail tone; 1.0, complete loss of tail tone; 1.5, mildly impaired righting reflex; 2.0, abnormal gait and/or impaired righting reflex; 2.5, hindlimb paresis; 3.0, hindlimb paralysis; 3.5, hindlimb paralysis with hind body paresis; 4.0, hind and fore limb paralysis; and 4.5, death or severity necessitating euthanasia. Treatment of mice was performed by gavaging FTY720 (1 mg/kg; Novartis) and/or vitamin B12 (15 mg/kg; Sigma), unless otherwise noted.
Histological analysis
Formalin-fixed paraffin-embedded (FFPE) sectioning
SCs were incubated overnight in 10% neutral buffered formalin (PROTOCOL, Thermo Fisher Scientific) at room temperature. They were then cut into 3 sections and embedded in warm 2% agar (BD) dissolved in water and were allowed to solidify on crushed ice. The solid block was stored in 70% EtOH (PHARMCO, cat # 241000140CSGL), washed with 95% absolute EtOH, 100% absolute EtOH:Xylene (1:1), Xylene, molten warm Paraffin (Tissue-Tek, cat # 4005), and embedded into paraffin blocks using a manual paraffin embedder (Sakura Tissue-Tek). Sections (10 mm) were cut using a microtome (Leica RM2155) and used for Kluver Berrara staining and immunostaining. Histological scoring was performed as previously described.82 Antigen retrieval with Diva Decloaker (Biocare) was performed followed by blocking with species-appropriate serum. Sections were incubated with primary antibodies including rabbit anti-CD320 (1:25 dilution, Proteintech, cat # 10343-1-AP), goat anti-Lcn2 (1:500 dilution, R&D systems, cat # AF1857), rabbit anti-Hmox1 (1:500 dilution, ThermoFisher Scientific, cat # PA5-27338), and chicken anti-GFAP(1:1000 dilution, Neuromics, cat # CH22102), followed by incubation with secondary antibodies conjugated with Alexa Fluor 488 or Alexa Fluor 568 (1:2000 dilution, ThermoFisher, cat # A11008 or cat # A11041, respectively) and counterstaining with DAPI (1:10,000 dilution, Sigma, cat #D8417). Sections were visualized and images acquired on a Zeiss Apotome.2 (Zen 2 Blue Edition).
RNA-seq
Nuclear isolation
SCs were rapidly dissected and frozen in liquid nitrogen. Samples were equilibrated to 4°C and dounce homogenized in a nuclei extraction buffer made with 0.32 M sucrose/5 mM CaCl2/3 mM Mg(CH3COO)2/0.1 mM EDTA/20 mM Tris-HCl pH 8.0/0.1% Triton X-100 in DEPC-treated H2O. Homogenized samples were filtered (50μm) (Celltrics, Sysmex) and washed in DEPC-treated PBS containing 2 mM EDTA (PBSE-d). Nuclei were purified by centrifugation at 3250g for 12 min in an iso-osmotic iodixanol gradient made of a 35%, 10%, and 5% OptiPrep (Millipore Sigma, cat #D1556) in DEPC-treated H2O containing 20 mM tricine-KOH (pH 7.8), 25 mM KCl, and 30 mM MgCl2.83 Nuclei were recovered in the 35%–10% interface, washed in PBSE-d, and immunolabeled with rabbit anti-NeuN antibody (1:400 dilution, MilliporeSigma, cat # MABN140) in 1% bovine serum albumin (BSA)/PBSE-d for 20 min. Samples were washed in PBSE-d and labeled with a goat anti-rabbit APC conjugated secondary antibody (1:500 dilution, ThermoFisher Scientific, cat # 31984) and DAPI (1:5000 dilution, Sigma, cat #D8417) in 1%BSA/PBSE-d for 10 min. Samples were washed and suspended in PBSE-d. Nuclear populations were analyzed and sorted on a BD FACSAria Fusion. Gating was performed as follows: (i) DAPI positive, (ii) size and granularity consistent with nuclei by forward-scatter area (FSC-A) and side-scatter area (SSC-A), and (iii) single nuclei selected by both FSC-A and forward scatter height (FSC-H) and SSC-A and side-scatter height (SSC-H). Analysis was performed on FlowJo (10.0.8r1).
RNA isolation
Populations of 15,000 DAPI+NeuN−GFP-H2B+ nuclei were directly sorted into an extraction buffer for RNA isolation of Picopure RNA isolation kit (ThermoFisher Scientific, cat # KIT0204). RNase free Deoxyribonuclease I (DNase I) (Qiagen, cat # 1080901) was used to remove genomic DNA. For astrocyte RNA-seq, RNA was extracted using an RNeasy mini kit (Qiagen, cat # 74104) (with DNase I treatment) as per manufacturer’s instructions. RNA concentration and quality was assessed using a 2100 Bioanalyzer (Agilent Technologies).
Library preparation
Between 1 and 3 ng of the nuclear RNA was used in the SMART-seq v4 Ultra Low Input RNA Kit for Sequencing (Takara, cat # 634888) according to the manufacturer’s protocols. Briefly, first-strand synthesis by poly(A)-priming is followed by template switching and extension by reverse transcription, allowing high fidelity amplification of full-length cDNA transcripts by long distance PCR. A sequencing library was produced using 0.75 ng of the amplified cDNA in the Nextera XT Library Preparation Kit (Illumina, cat # FC-131-1024). For astrocyte RNA-seq, 150 ng of RNA per sample was employed in the generation of libraries using the NEB Next Ultra Pure RNA library kit for Illumina (NEB, cat #E7770S) per manufacturer’s instructions.
Nuclear RNA-seq analysis
Libraries were sequenced using the Illumina NextSeq 500 sequencer with 100 base pair single-end reads. Spliced Transcripts Alignment to a Reference (STAR) was used to align to the Mus musculus genome (mm10). All reads that mapped to exons and introns were used in this analysis, as nuclear RNA was previously noted to contain higher levels of intronic mapping (~27%) and intron expression is highly correlated with transcriptional activity.84,85 Partek software (v6.6) was used for nuclear RNA-seq analysis. The CPM values for each group were averaged to analyze fold differences between S1P1-AsCKOfos and WTfos+FTY720 vs. WTfos. RNA-seq and data processing were performed at the Sequencing Core of The Scripps Research Institute.
Astrocyte RNA-seq analysis
Libraries were sequenced using the Illumina NextSeq 500 sequencer with 100 base pair single-end reads. Salmon86 was used for transcript quantification while Counts Per Million (CPM) values were calculated using tximport.87 RNA-seq and data processing were performed at the Sequencing Core of The Scripps Research Institute.
Astrocyte culture
Cerebral hemispheres of postnatal day 0 C57BL/6J pups were harvested and the meninges were removed. They were placed in DMEM/F12 (ThermoFisher Scientific, cat # 11320033), penicillin streptomycin (ThermoFisher Scientific, cat # 10378016), 1 mg/mL DNase I (Millipore Sigma, cat #D5025), and 0.05% trypsin (Millipore Sigma, cat #T1005). After gentle dissociation the tissue was incubated for 30 min at 37°C. Trypsin in the medium was inactivated by increasing the volume with DMEM/F12 containing 10% FBS (Gemini, cat # 100-500), centrifuged at 700 rpm for 5 min, filtered through a 40 μm cell strainer (Millipore Sigma, cat # CLS431750). The cells were cultured in DMEM/F12 containing 10% FBS and grown until 80% confluency before passage. The purity of astrocytes with this method was estimated to be more than 95% by immunostaining for glial fibrillary acidic protein (GFAP). For RNA-seq, after first passage, primary astrocytes were allowed to grow for 2 weeks either in normal media (DMEM/F12) or custom synthesized B12free DMEM/F12 media (AthenaES) containing PS and 0.5% fatty acid free BSA.
Human astrocytes, HASTR/ci35,54 were cultured in Astrocyte Medium (ThermoFisher Scientific, cat # A1261301) at 33°C, 5% CO2 incubator. Cells were plated onto 6 well plates at 5 × 105 cells/well one day before the experiment. Cells were washed with DMEM and stimulated with FTY720 and sphingosine in the presence or absence of serum for 1 h. Cells were detached and sequentially stained with anti-CD320 antibody (1:100 dilution: Proteintech, cat # 10343-1-AP) as the primary antibody and Alexa Fluor 488-conjugated Goat Anti-Rabbit IgG (1:1000 dilution: ThermoFishre Scientific, cat #A-11008) as a secondary antibody. FCM analyses were conducted with Novocyte (Agilent).
Quantitative PCR
The Superscript II first strand cDNA synthesis kit (ThermoFisher Scientific, cat # 18064022) was used to synthesize cDNA per manufacturer’s instructions for RT-PCR. This cDNA was used for qPCR analysis using GoTaq qPCR master mix (Promega, cat # A6001) using a Biorad CFX 384. Gene expression was normalized to b-actin. The primer sets for the target genes used in this study are provided in Table S4.
SDS page and western blotting
SC samples were minced for a few seconds intermittently in homogenization buffer (50 mM Tris-HCl pH 8.0, 150 mM NaCl, 0.5% Triton X-100, 2 mM EDTA, 1x protease inhibitor cocktail, and 250 mM sucrose). The mixture was incubated on ice for 1 h and centrifuged at 8000g for 10 min at 4°C. The supernatant was collected for measuring protein concentration using a Bradford reagent (Bio-rad, cat # 5000001). Samples were denatured in NuPAGE LDS sample buffer (ThermoFisher Scientific, cat # NP0007) and 250 mM β-mercaptoethanol (Millipore Sigma, cat #M6250) at 80°C for 15 min, and run on 4–12% SDS Nu-PAGE gradient gel (ThermoFisher Scientific, cat # NP0323BOX). The proteins were transferred (NuPAGE transfer buffer; ThrermoFisher Scientific, cat # NP00061) onto a PVDF membrane, blocked in 5% non-fat milk followed by incubation with an anti-CD320 Ab (1:100 dilution, Protein Tech, cat # 10343-1-AP). After several washes, the membrane was incubated in secondary antibody (HRP conjugated anti-rabbit or anti-mouse IgG(H + L) (1:10,000 dilution, Cell Signaling, cat # 7074P2 or 32230, respectively). Subsequently the signal was developed using Super Signal West Femto substrate (ThermoFisher Scientific, cat # 34094) and visualized using the molecular imager ChemiDoc XRS+ imaging system (Bio-Rad).
Binding assay
The Compensated Interferometric Reader (CIR) was developed from the backscattering interferometry (BSI) instrument that was described in detail previously.52,53 Recombinant human CD320 (R&D systems, cat # 1557-CD-050) and human TCN2 (R&D systems, cat # 7895-TC-050) were mixed with FTY720 (Novartis), FTY720P (Novartis), sphingosine (Avanti Polar Lipids, cat # 860490P), and S1P (Avanti Polar Lipids, cat # 860492P) and applied to the CIR. Mutant TCN2 protein (W41→A, P379→A, D411→A, R413→A) was obtained from supernatant of Baculovirus expression system (GenScript). The specific binding signal was calculated by subtracting the control signal followed by normalization to the signal without ligand. The obtained signal displayed as milliradian were fitted by non-linear regression using the Michaelis-Menten equation using Prism software (GraphPad).
B12 measurement
Spinal cord tissues were crushed with an SK mill (SK-100; Tokken, Chiba, Japan). B12 was extracted with methanol by probe sonication. Cyanocobalamin-[13C7] (IsoSciences, King of Prussia, PA) was added as an internal standard. The tissue debris was removed by centrifugation at 10,000 × g for 5 min and filtration with YMC Duo-filter (YMC Co., Ltd., Kyoto, Japan) with a pore size of 0.2 μm, and a portion of the methanol extract was injected onto a Xevo TQ-S micro triple quadrupole mass spectrometry system (Waters) equipped with an electrospray ionization (ESI) source. Liquid chromatography (LC) separation was performed on a ZIC-pHILIC column (5 μm, 2.1 mm × 100 mm; Millipore) coupled to a ZIC-pHILIC guard column (2.1 mm × 20 mm; Millipore). Mobile phase A was acetonitrile containing 0.1% formic acid, and mobile phase B was water containing 0.1% formic acid. The LC method consisted of a linear gradient from 95% A to 50% B over 12 min at 0.2 mL/min, a linear gradient to 80% B over 16 min at 0.1 mL/min, 80% B for 20 min at 0.1 mL/min, 95% A for 7 min at 0.1 mL/min, and 95% A for 5 min at 0.2 mL/min (60 min total run time). The column temperature was set at room temperature. The detection was performed in multiple reaction monitoring (MRM) mode. MRM transitions from [M + H]2+ to the fragment ions at m/z 147.1 and 154.1 were used for B12 and the internal standard, respectively. The collision energy was set to 40 eV. The ESI capillary voltage was set at 1.0 kV, and the sampling cone was set at 30 V. The source temperature was set at 150°C, desolvation temperature was set at 500°C, and desolvation gas flow was 1000 L/h. The cone gas flow was set at 50 L/h.
Docking model
AutoDock is an automated docking method to identify binding modes of ligands inside the binding pocket of a biomolecular receptor.88–90 The computational protocol of AutoDock has been reported multiple times. In consistent with “united atom model”, only heavy atoms and polar hydrogens are used while docking simulations. To represent the target receptor, AutoDock uses a regularly spaced 3D grid which is defined by the user. A simplified representation of the receptor is then used during docking simulation. AutoDock family offers several tools, however AutoDock Vina is the latest method from that family.91 AutoDock Vina uses a unique approach to identify ligand binding modes implementing automatic calculation of the grid maps. Simultaneously, it also achieved approximately two orders of magnitude speed-up compared to its earlier version (AutoDock 4) while significantly improving the accuracy of the binding mode predictions based on an extensive training set. In the current report, we used Autodock (Version 4.2.3) and Vina (Version 1.1.2). The 3D coordinate of the protein bound to B12 was obtained from RCSB (http://www.rcsb.org/pdb/). Refining the system structure was achieved through using AutoDockTools. The 3D coordinate of the protein kept rigid while performing the docking. A graphical-user-interface for AutoDock, AutoDockTools (ADT, version 1.5.6 rc3), was used for adding hydrogens and Gasteiger charges and to generate the molecules in AutoDock suitable formats (pdbqt). A default grid spacing parameter (0.375Å) was used. The size of the grid box was chosen to be 100 × 100 × 100 which is large enough to cover the entire receptor and explore all possible binding pockets. For generating the docking parameter file (DPF) and the grid parameter file (GPF), ADT was used. AutoDock (4.2 and up) offers four different search algorithms based on speed and accuracy: 1) Simulated Annealing 2) Genetic Algorithm 3) Local Search and 4) Lamarkian. It has been reported multiples times that Genetic Algorithm (GA) often achieves most reliable results and highly appropriate for systems with many degrees of freedom. Thus, GA was used in the current report.
QUANTIFICATION AND STATISTICAL ANALYSIS
Results are expressed as mean ± SEM. Data were analyzed statistically by means of unpaired t test, and ANOVA with indicated post hoc tests as appropriate, using GraphPad Prism software. Values of p < 0.05 were considered to be statistically significant.
Supplementary Material
Highlights.
CD320 expression is suppressed in MS and EAE astrocytes
S1P1 inhibition upregulates CD320 expression in astrocytes
Fingolimod and sphingosine directly bind to TCN2
Fingolimod-bound TCN2 promotes CD320 internalization in astrocytes
ACKNOWLEDGMENTS
We thank V. Brinkmann and Novartis for gifts of FTY720 and discussions; J. Wong, H. Mirendil, V.A. Blaho, M.H. Lee, M. Kachi, and all the lab members for discussions and support; and D. Jones for editorial assistance. This work was supported by the National Institute of Neurological Disorders and Stroke of the National Institutes of Health under award number R01NS103940 (Y.K.); Novartis (J.C.); and MEXT/JSPS KAKENHI 18H02627, 19KK0199, and 21H04798 (T.Y.) and 18K16246 and 21K08565 (H.-C.L.-O.). Y.K. was supported by fellowships from the Uehara Memorial Foundation, the Kanae Foundation for the Promotion of Medical Science, the Mochida Memorial Foundation for Medical and Pharmaceutical Research, and the Human Frontier Science Program. A.G. was supported by the Medical Scientist Training Program and Pharmacology Training Grant at the University of California San Diego (T32GM007752). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
DECLARATION OF INTERESTS
J.C. has received honoraria, consulting fees, and funding support from Novartis, Bristol-Myers Squibb (Celgene), Biogen, and Janssen Pharmaceuticals. J.C. has an employment relationship with Neurocrine Biosciences, Inc., a company that may potentially benefit from the research results. Dr. Chun’s relationship with Neurocrine Biosciences, Inc. has been reviewed and approved by Sanford Burnham Prebys Medical Discovery Institute in accordance with its conflict of interest policies. D.J. is currently employed by Radionetics Oncology. A.G. is currently affiliated with University of California, Los Angeles. R.M. is currently affiliated with Loyola University Chicago. A.S. is currently affiliated with University of Wisconsin-Milwaukee. R.R. is currently employed by 8five8 Therapeutics.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2023.113545.
REFERENCES
- 1.Noseworthy JH, Lucchinetti C, Rodriguez M, and Weinshenker BG (2000). Multiple sclerosis. N. Engl. J. Med. 343, 938–952. [DOI] [PubMed] [Google Scholar]
- 2.González H, and Pacheco R (2014). T-cell-mediated regulation of neuroinflammation involved in neurodegenerative diseases. J. Neuroinflammation 11, 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hauser SL, Bar-Or A, Comi G, Giovannoni G, Hartung HP, Hemmer B, Lublin F, Montalban X, Rammohan KW, Selmaj K, et al. (2017). Ocrelizumab versus Interferon Beta-1a in Relapsing Multiple Sclerosis. N. Engl. J. Med. 376, 221–234. [DOI] [PubMed] [Google Scholar]
- 4.Miller A, Korem M, Almog R, and Galboiz Y (2005). Vitamin B12, demyelination, remyelination and repair in multiple sclerosis. J. Neurol. Sci. 233, 93–97. [DOI] [PubMed] [Google Scholar]
- 5.Najafi MR, Shaygannajad V, Mirpourian M, and Gholamrezaei A (2012). Vitamin B(12) Deficiency and Multiple Sclerosis; Is there Any Association? Int. J. Prev. Med. 3, 286–289. [PMC free article] [PubMed] [Google Scholar]
- 6.Kozyraki R, and Cases O (2013). Vitamin B12 absorption: mammalian physiology and acquired and inherited disorders. Biochimie 95, 1002–1007. [DOI] [PubMed] [Google Scholar]
- 7.Alam A, Woo J-S, Schmitz J, Prinz B, Root K, Chen F, Bloch JS, Zenobi R, and Locher KP (2016). Structural basis of transcobalamin recognition by human CD320 receptor. Nat. Commun. 7, 12100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chun J, Giovannoni G, and Hunter SF (2021). Sphingosine 1-phosphate Receptor Modulator Therapy for Multiple Sclerosis: Differential Downstream Receptor Signalling and Clinical Profile Effects. Drugs 81, 207–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chun J, Kihara Y, Jonnalagadda D, and Blaho VA (2019). Fingolimod: Lessons Learned and New Opportunities for Treating Multiple Sclerosis and Other Disorders. Annu. Rev. Pharmacol. Toxicol. 59, 149–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kihara Y, and Chun J (2023). Molecular and neuroimmune pharmacology of S1P receptor modulators and other disease-modifying therapies for multiple sclerosis. Pharmacol. Ther. 246, 108432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mizuno H, and Kihara Y (2020). Druggable Lipid GPCRs: Past, Present, and Prospects. Adv. Exp. Med. Biol. 1274, 223–258. [DOI] [PubMed] [Google Scholar]
- 12.Chun J, and Hartung HP (2010). Mechanism of action of oral fingolimod (FTY720) in multiple sclerosis. Clin. Neuropharmacol. 33, 91–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cohen JA, and Chun J (2011). Mechanisms of fingolimod’s efficacy and adverse effects in multiple sclerosis. Ann. Neurol. 69, 759–777. [DOI] [PubMed] [Google Scholar]
- 14.Brinkmann V, Davis MD, Heise CE, Albert R, Cottens S, Hof R, Bruns C, Prieschl E, Baumruker T, Hiestand P, et al. (2002). The immune modulator FTY720 targets sphingosine 1-phosphate receptors. J. Biol. Chem. 277, 21453–21457. [DOI] [PubMed] [Google Scholar]
- 15.Kihara Y, Maceyka M, Spiegel S, and Chun J (2014). Lysophospholipid receptor nomenclature review: IUPHAR Review 8. Br. J. Pharmacol. 171, 3575–3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kihara Y, Mizuno H, and Chun J (2015). Lysophospholipid receptors in drug discovery. Exp. Cell Res. 333, 171–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mandala S, Hajdu R, Bergstrom J, Quackenbush E, Xie J, Milligan J, Thornton R, Shei GJ, Card D, Keohane C, et al. (2002). Alteration of lymphocyte trafficking by sphingosine-1-phosphate receptor agonists. Science 296, 346–349. [DOI] [PubMed] [Google Scholar]
- 18.Arnon TI, Xu Y, Lo C, Pham T, An J, Coughlin S, Dorn GW, and Cyster JG (2011). GRK2-dependent S1PR1 desensitization is required for lymphocytes to overcome their attraction to blood. Science 333, 1898–1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matloubian M, Lo CG, Cinamon G, Lesneski MJ, Xu Y, Brinkmann V, Allende ML, Proia RL, and Cyster JG (2004). Lymphocyte egress from thymus and peripheral lymphoid organs is dependent on S1P receptor 1. Nature 427, 355–360. [DOI] [PubMed] [Google Scholar]
- 20.Chun J, and Brinkmann V (2011). A mechanistically novel, first oral therapy for multiple sclerosis: the development of fingolimod (FTY720, Gilenya). Discov. Med. 12, 213–228. [PMC free article] [PubMed] [Google Scholar]
- 21.Groves A, Kihara Y, and Chun J (2013). Fingolimod: direct CNS effects of sphingosine 1-phosphate (S1P) receptor modulation and implications in multiple sclerosis therapy. J. Neurol. Sci. 328, 9–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Soliven B, Miron V, and Chun J (2011). The neurobiology of sphingosine 1-phosphate signaling and sphingosine 1-phosphate receptor modulators. Neurology 76, S9–S14. [DOI] [PubMed] [Google Scholar]
- 23.McGiffert C, Contos JJA, Friedman B, and Chun J (2002). Embryonic brain expression analysis of lysophospholipid receptor genes suggests roles for s1p(1) in neurogenesis and s1p(1–3) in angiogenesis. FEBS Lett. 531, 103–108. [DOI] [PubMed] [Google Scholar]
- 24.Zhang G, Contos JJ, Weiner JA, Fukushima N, and Chun J (1999). Comparative analysis of three murine G-protein coupled receptors activated by sphingosine-1-phosphate. Gene 227, 89–99. [DOI] [PubMed] [Google Scholar]
- 25.Chun J (1999). Lysophospholipid receptors: implications for neural signaling. Crit. Rev. Neurobiol. 13, 151–168. [DOI] [PubMed] [Google Scholar]
- 26.Chun J, Weiner JA, Fukushima N, Contos JJ, Zhang G, Kimura Y, Dubin A, Ishii I, Hecht JH, Akita C, and Kaushal D (2000). Neurobiology of receptor-mediated lysophospholipid signaling. From the first lysophospholipid receptor to roles in nervous system function and development. Ann. N. Y. Acad. Sci. 905, 110–117. [DOI] [PubMed] [Google Scholar]
- 27.Ishii I, Friedman B, Ye X, Kawamura S, McGiffert C, Contos JJ, Kingsbury MA, Zhang G, Brown JH, and Chun J (2001). Selective loss of sphingosine 1-phosphate signaling with no obvious phenotypic abnormality in mice lacking its G protein-coupled receptor, LP(B3)/EDG-3. J. Biol. Chem. 276, 33697–33704. [DOI] [PubMed] [Google Scholar]
- 28.Foster CA, Howard LM, Schweitzer A, Persohn E, Hiestand PC, Balatoni B, Reuschel R, Beerli C, Schwartz M, and Billich A (2007). Brain penetration of the oral immunomodulatory drug FTY720 and its phosphorylation in the central nervous system during experimental autoimmune encephalomyelitis: consequences for mode of action in multiple sclerosis. J. Pharmacol. Exp. Ther. 323, 469–475. [DOI] [PubMed] [Google Scholar]
- 29.Choi JW, Gardell SE, Herr DR, Rivera R, Lee CW, Noguchi K, Teo ST, Yung YC, Lu M, Kennedy G, and Chun J (2011). FTY720 (fingolimod) efficacy in an animal model of multiple sclerosis requires astrocyte sphingosine 1-phosphate receptor 1 (S1P1) modulation. Proc. Natl. Acad. Sci. USA. 108, 751–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Groves A, Kihara Y, Jonnalagadda D, Rivera R, Kennedy G, Mayford M, and Chun J (2018). A Functionally Defined In Vivo Astrocyte Population Identified by c-Fos Activation in a Mouse Model of Multiple Sclerosis Modulated by S1P Signaling: Immediate-Early Astrocytes (ieAstrocytes). eNeuro 5. ENEURO.0239, 18.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kappos L, Antel J, Comi G, Montalban X, O’Connor P, Polman CH, Haas T, Korn AA, Karlsson G, and Radue EW; FTY720 D2201 Study Group (2006). Oral fingolimod (FTY720) for relapsing multiple sclerosis. N. Engl. J. Med. 355, 1124–1140. [DOI] [PubMed] [Google Scholar]
- 32.Vidal-Jordana A, Sastre-Garriga J, Pérez-Miralles F, Tur C, Tintoré M, Horga A, Auger C, Rió J, Nos C, Edo MC, et al. (2013). Early brain pseudoatrophy while on natalizumab therapy is due to white matter volume changes. Mult. Scler. 19, 1175–1181. [DOI] [PubMed] [Google Scholar]
- 33.Matsuo N, Reijmers L, and Mayford M (2008). Spine-type-specific recruitment of newly synthesized AMPA receptors with learning. Science 319, 1104–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lake BB, Ai R, Kaeser GE, Salathia NS, Yung YC, Liu R, Wildberg A, Gao D, Fung H-L, Chen S, et al. (2016). Neuronal subtypes and diversity revealed by single-nucleus RNA sequencing of the human brain. Science 352, 1586–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kihara Y, Zhu Y, Jonnalagadda D, Romanow W, Palmer C, Siddoway B, Rivera R, Dutta R, Trapp BD, and Chun J (2022). Single-Nucleus RNA-seq of Normal-Appearing Brain Regions in Relapsing-Remitting vs. Secondary Progressive Multiple Sclerosis: Implications for the Efficacy of Fingolimod. Front. Cell. Neurosci. 16, 918041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tien AC, Tsai HH, Molofsky AV, McMahon M, Foo LC, Kaul A, Dougherty JD, Heintz N, Gutmann DH, Barres BA, and Rowitch DH (2012). Regulated temporal-spatial astrocyte precursor cell proliferation involves BRAF signalling in mammalian spinal cord. Development 139, 2477–2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liddelow SA, Guttenplan KA, Clarke LE, Bennett FC, Bohlen CJ, Schirmer L, Bennett ML, Münch AE, Chung WS, Peterson TC, et al. (2017). Neurotoxic reactive astrocytes are induced by activated microglia. Nature 541, 481–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, Fridman WH, Pagè s F, Trajanoski Z, and Galon J (2009). ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 25, 1091–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cline MS, Smoot M, Cerami E, Kuchinsky A, Landys N, Workman C, Christmas R, Avila-Campilo I, Creech M, Gross B, et al. (2007). Integration of biological networks and gene expression data using Cytoscape. Nat. Protoc. 2, 2366–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fabregat A, Sidiropoulos K, Viteri G, Forner O, Marin-Garcia P, Arnau V, D’Eustachio P, Stein L, and Hermjakob H (2017). Reactome pathway analysis: a high-performance in-memory approach. BMC Bioinf. 18, 142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rothhammer V, Mascanfroni ID, Bunse L, Takenaka MC, Kenison JE, Mayo L, Chao CC, Patel B, Yan R, Blain M, et al. (2016). Type I interferons and microbial metabolites of tryptophan modulate astrocyte activity and central nervous system inflammation via the aryl hydrocarbon receptor. Nat. Med. 22, 586–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Quadros EV, and Sequeira JM (2013). Cellular uptake of cobalamin: transcobalamin and the TCblR/CD320 receptor. Biochimie 95, 1008–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lai S-C, Nakayama Y, Sequeira JM, Wlodarczyk BJ, Cabrera RM, Finnell RH, Bottiglieri T, and Quadros EV (2013). The transcobalamin receptor knockout mouse: a model for vitamin B12 deficiency in the central nervous system. Faseb. J. 27, 2468–2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Han MH, Lundgren DH, Jaiswal S, Chao M, Graham KL, Garris CS, Axtell RC, Ho PP, Lock CB, Woodard JI, et al. (2012). Janus-like opposing roles of CD47 in autoimmune brain inflammation in humans and mice. J. Exp. Med. 209, 1325–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alexopoulou L, Holt AC, Medzhitov R, and Flavell RA (2001). Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature 413, 732–738. [DOI] [PubMed] [Google Scholar]
- 46.Khorooshi R, Mørch MT, Holm TH, Berg CT, Dieu RT, Dræby D, Issazadeh-Navikas S, Weiss S, Lienenklaus S, and Owens T (2015). Induction of endogenous Type I interferon within the central nervous system plays a protective role in experimental autoimmune encephalomyelitis. Acta Neuropathol. 130, 107–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cahalan SM, Gonzalez-Cabrera PJ, Sarkisyan G, Nguyen N, Schaeffer MT, Huang L, Yeager A, Clemons B, Scott F, and Rosen H (2011). Actions of a picomolar short-acting S1P(1) agonist in S1P(1)-eGFP knock-in mice. Nat. Chem. Biol. 7, 254–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miller LG Jr., Young JA, Ray SK, Wang G, Purohit S, Banik NL, and Dasgupta S (2017). Sphingosine Toxicity in EAE and MS: Evidence for Ceramide Generation via Serine-Palmitoyltransferase Activation. Neurochem. Res. 42, 2755–2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nielsen MJ, Rasmussen MR, Andersen CBF, Nexø E, and Moestrup SK (2012). Vitamin B12 transport from food to the body’s cells[mdash]a sophisticated, multistep pathway. Nat. Rev. Gastroenterol. Hepatol. 9, 345–354. [DOI] [PubMed] [Google Scholar]
- 50.Ghosh S, Sinha JK, Putcha UK, and Raghunath M (2016). Severe but Not Moderate Vitamin B12 Deficiency Impairs Lipid Profile, Induces Adiposity, and Leads to Adverse Gestational Outcome in Female C57BL/6 Mice. Front. Nutr. 3, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kocur M, Schneider R, Pulm AK, Bauer J, Kropp S, Gliem M, Ingwersen J, Goebels N, Alferink J, Prozorovski T, et al. (2015). IFNbeta secreted by microglia mediates clearance of myelin debris in CNS autoimmunity. Acta Neuropathol. Commun. 3, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mizuno H, Kihara Y, Kussrow A, Chen A, Ray M, Rivera R, Bornhop DJ, and Chun J (2019). Lysophospholipid G protein-coupled receptor binding parameters as determined by backscattering interferometry. J. Lipid Res. 60, 212–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ray M, Nagai K, Kihara Y, Kussrow A, Kammer MN, Frantz A, Bornhop DJ, and Chun J (2020). Unlabeled lysophosphatidic acid receptor binding in free solution as determined by a compensated interferometric reader. J. Lipid Res. 61, 1244–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Furihata T, Ito R, Kamiichi A, Saito K, and Chiba K (2016). Establishment and characterization of a new conditionally immortalized human astrocyte cell line. J. Neurochem. 136, 92–105. [DOI] [PubMed] [Google Scholar]
- 55.Polak DM, Elliot JM, and Haluska M (1979). Vitamin B12 binding proteins in bovine serum. J. Dairy Sci. 62, 697–701. [DOI] [PubMed] [Google Scholar]
- 56.Hisano Y, Kobayashi N, Kawahara A, Yamaguchi A, and Nishi T (2011). The sphingosine 1-phosphate transporter, SPNS2, functions as a transporter of the phosphorylated form of the immunomodulating agent FTY720. J. Biol. Chem. 286, 1758–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rossi DJ, Brady JD, and Mohr C (2007). Astrocyte metabolism and signaling during brain ischemia. Nat. Neurosci. 10, 1377–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jiang W, Sequeira JM, Nakayama Y, Lai SC, and Quadros EV (2010). Characterization of the promoter region of TCblR/CD320 gene, the receptor for cellular uptake of transcobalamin-bound cobalamin. Gene 466, 49–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Van Doorn R, Van Horssen J, Verzijl D, Witte M, Ronken E, Van Het Hof B, Lakeman K, Dijkstra CD, Van Der Valk P, Reijerkerk A, et al. (2010). Sphingosine 1-phosphate receptor 1 and 3 are upregulated in multiple sclerosis lesions. Glia 58, 1465–1476. [DOI] [PubMed] [Google Scholar]
- 60.Kułakowska A, Żendzian-Piotrowska M, Baranowski M, Konończuk T, Drozdowski W, Górski J, and Bucki R (2010). Intrathecal increase of sphingosine 1-phosphate at early stage multiple sclerosis. Neurosci. Lett. 477, 149–152. [DOI] [PubMed] [Google Scholar]
- 61.Blaho VA (2020). Druggable Sphingolipid Pathways: Experimental Models and Clinical Opportunities. Adv. Exp. Med. Biol. 1274, 101–135. [DOI] [PubMed] [Google Scholar]
- 62.Nagahashi M, Yamada A, Aoyagi T, Allegood J, Wakai T, Spiegel S, and Takabe K (2016). Sphingosine-1-phosphate in the lymphatic fluid determined by novel methods. Heliyon 2, e00219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Colwill K, and Renewable Protein Binder Working Group; and Gräslund S (2011). A roadmap to generate renewable protein binders to the human proteome. Nat. Methods 8, 551–558. [DOI] [PubMed] [Google Scholar]
- 64.Lublin F, Miller DH, Freedman MS, Cree BAC, Wolinsky JS, Weiner H, Lubetzki C, Hartung HP, Montalban X, Uitdehaag BMJ, et al. (2016). Oral fingolimod in primary progressive multiple sclerosis (INFORMS): a phase 3, randomised, double-blind, placebo-controlled trial. Lancet 387, 1075–1084. [DOI] [PubMed] [Google Scholar]
- 65.De Stefano N, Silva DG, and Barnett MH (2017). Effect of Fingolimod on Brain Volume Loss in Patients with Multiple Sclerosis. CNS Drugs 31, 289–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yousuf F, Dupuy SL, Tauhid S, Chu R, Kim G, Tummala S, Khalid F, Weiner HL, Chitnis T, Healy BC, and Bakshi R (2017). A two-year study using cerebral gray matter volume to assess the response to fingolimod therapy in multiple sclerosis. J. Neurol. Sci. 383, 221–229. [DOI] [PubMed] [Google Scholar]
- 67.Arora K, Sequeira JM, Hernández AI, Alarcon JM, and Quadros EV (2017). Behavioral alterations are associated with vitamin B12 deficiency in the transcobalamin receptor/CD320 KO mouse. PLoS One 12, e0177156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Deng Y, Wang D, Wang K, and Kwok T (2017). High Serum Folate Is Associated with Brain Atrophy in Older Diabetic People with Vitamin B12 Deficiency. J. Nutr. Health Aging 21, 1065–1071. [DOI] [PubMed] [Google Scholar]
- 69.Oh J, and O’Connor PW (2015). Established disease-modifying treatments in relapsing-remitting multiple sclerosis. Curr. Opin. Neurol. 28, 220–229. [DOI] [PubMed] [Google Scholar]
- 70.Lublin F (2005). History of modern multiple sclerosis therapy. J. Neurol. 252, iii3–iii9. [DOI] [PubMed] [Google Scholar]
- 71.Teige I, Treschow A, Teige A, Mattsson R, Navikas V, Leanderson T, Holmdahl R, and Issazadeh-Navikas S (2003). IFN-beta gene deletion leads to augmented and chronic demyelinating experimental autoimmune encephalomyelitis. J. Immunol. 170, 4776–4784. [DOI] [PubMed] [Google Scholar]
- 72.Feng X, Petraglia AL, Chen M, Byskosh PV, Boos MD, and Reder AT (2002). Low expression of interferon-stimulated genes in active multiple sclerosis is linked to subnormal phosphorylation of STAT1. J. Neuroimmunol. 129, 205–215. [DOI] [PubMed] [Google Scholar]
- 73.Feng X, Wang Z, Howlett-Prieto Q, Einhorn N, Causevic S, and Reder AT (2019). Vitamin D enhances responses to interferon-beta in MS. Neurol. Neuroimmunol. Neuroinflamm. 6, e622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Prosperini L, Capobianco M, and Gianni C (2014). Identifying responders and nonresponders to interferon therapy in multiple sclerosis. Degener. Neurol. Neuromuscul. Dis. 4, 75–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mastronardi FG, Min W, Wang H, Winer S, Dosch M, Boggs JM, and Moscarello MA (2004). Attenuation of experimental autoimmune encephalomyelitis and nonimmune demyelination by IFN-beta plus vitamin B12: treatment to modify notch-1/sonic hedgehog balance. J. Immunol. 172, 6418–6426. [DOI] [PubMed] [Google Scholar]
- 76.Yester JW, Bryan L, Waters MR, Mierzenski B, Biswas DD, Gupta AS, Bhardwaj R, Surace MJ, Eltit JM, Milstien S, et al. (2015). Sphingosine-1-phosphate inhibits IL-1-induced expression of C-C motif ligand 5 via c-Fos-dependent suppression of IFN-beta amplification loop. FASEB J 29, 4853–4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Teijaro JR, Studer S, Leaf N, Kiosses WB, Nguyen N, Matsuki K, Negishi H, Taniguchi T, Oldstone MBA, and Rosen H (2016). S1PR1-mediated IFNAR1 degradation modulates plasmacytoid dendritic cell interferon-alpha autoamplification. Proc. Natl. Acad. Sci. USA. 113, 1351–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Acosta C, Anderson HD, and Anderson CM (2017). Astrocyte dysfunction in Alzheimer disease. J. Neurosci. Res. 95, 2430–2447. [DOI] [PubMed] [Google Scholar]
- 79.Tré panier MO, Hopperton KE, Mizrahi R, Mechawar N, and Bazinet RP (2016). Postmortem evidence of cerebral inflammation in schizophrenia: a systematic review. Mol. Psychiatry 21, 1009–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tayler KK, Lowry E, Tanaka K, Levy B, Reijmers L, Mayford M, and Wiltgen BJ (2011). Characterization of NMDAR-Independent Learning in the Hippocampus. Front. Behav. Neurosci. 5, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kihara Y, Matsushita T, Kita Y, Uematsu S, Akira S, Kira J. i., Ishii S, and Shimizu T (2009). Targeted lipidomics reveals mPGES-1-PGE2 as a therapeutic target for multiple sclerosis. Proc. Natl. Acad. Sci. USA. 106, 21807–21812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kihara Y, Ishii S, Kita Y, Toda A, Shimada A, and Shimizu T (2005). Dual phase regulation of experimental allergic encephalomyelitis by platelet-activating factor. J. Exp. Med. 202, 853–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Graham JM (2001). Isolation of Nuclei and Nuclear Membranes From Animal Tissues. In Current Protocols in Cell Biology (John Wiley & Sons, Inc.). [DOI] [PubMed] [Google Scholar]
- 84.Gaidatzis D, Burger L, Florescu M, and Stadler MB (2015). Analysis of intronic and exonic reads in RNA-seq data characterizes transcriptional and post-transcriptional regulation. Nat Biotech 33, 722–729. http://www.nature.com/nbt/journal/v33/n7/abs/nbt.3269.html#supplementary-information. [DOI] [PubMed] [Google Scholar]
- 85.Grindberg RV, Yee-Greenbaum JL, McConnell MJ, Novotny M, O’Shaughnessy AL, Lambert GM, Araúzo-Bravo MJ, Lee J, Fishman M, Robbins GE, et al. (2013). RNA-sequencing from single nuclei. Proc. Natl. Acad. Sci. USA. 110, 19802–19807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Patro R, Duggal G, Love MI, Irizarry RA, and Kingsford C (2017). Salmon provides fast and bias-aware quantification of transcript expression. Nat Meth 14, 417–419. http://www.nature.com/nmeth/journal/v14/n4/abs/nmeth.4197.html#supplementary-information. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Soneson C, Love MI, and Robinson MD (2015). Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res. 4, 1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, and Olson AJ (2009). AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 30, 2785–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Goodsell DS, Morris GM, and Olson AJ (1996). Automated docking of flexible ligands: applications of AutoDock. J. Mol. Recognit. 9, 1–5. [DOI] [PubMed] [Google Scholar]
- 90.Morris GM, Goodsell DS, Huey R, and Olson AJ (1996). Distributed automated docking of flexible ligands to proteins: parallel applications of AutoDock 2.4. J. Comput. Aided Mol. Des. 10, 293–304. [DOI] [PubMed] [Google Scholar]
- 91.Trott O, and Olson AJ (2010). AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31, 455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data that supports the findings of this study are provided as Supplemental Tables (Tables S1 and S3). Fastaq files have been deposited into Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/) Accession number is listed in the key resources table.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| anti-CD320 | Proteintech | RRID:AB_2074223 |
| anti-Lcn2 | R&D Systems | RRID:AB_355022 |
| anti-Hmox1 | ThermoFisher Scientific | RRID:AB_2544814 |
| anti-GFAP | Neuromics | RRID:AB_10014322 |
| Alexa Fluor 488 | ThermoFisher Scientific | RRID:AB_143165 |
| Alexa Fluor 568 | ThermoFisher Scientific | RRID:AB_2534098 |
| anti-NeuN | MilliporeSigma | RRID:AB_2571567 |
| anti-rabbit APC | ThermoFisher Scientific | RRID:AB_429727 |
| HRP conjugated anti-rabbit or anti-mouse IgG(H + L) | Cell Signaling Technologies | RRID:AB_2099233 |
|
| ||
| Biological samples | ||
|
| ||
| Human MS and control brain | Rocky Mountain MS Center Tissue Bank | N/A |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| MOG35–55 | EZBiolab | cat # cp7203–5 |
| Pertussis toxin | List Biological Laboratories | cat # 180 |
| Vitamin B12 | Sigma | cat #V2876 |
| Recombinant human CD320 | R&D Systems | cat # 1557-CD-050 |
| Recombinant human TCN2 | R&D Systems | cat # 7895-TC-050 |
| FTY720 | Novartis | N/A |
| FTY720-P | Novartis | N/A |
| Sphingosine | Avanti Polar Lipids | cat # 860490P |
| S1P | Avanti Polar Lipids | cat # 860492P |
|
| ||
| Critical commercial assays | ||
|
| ||
| Picopure™ RNA isolation kit | ThermoFisher Scientific | cat # KIT0204 |
| RNeasy minikit | Qiagen | cat # 74104 |
| SMART-seq v4 Ultra Low Input RNA Kit for Sequencing | Takara Bio | cat # 634888 |
| Nextera XT Library Preparation Kit | Illumina | cat # FC-131-1024 |
| NEB Next Ultra Pure RNA library kit for Illumina | New England Biolabs | cat #E7770S |
|
| ||
| Deposited data | ||
|
| ||
| GSE99115 | Gene Expression Omnibus | https://www.ncbi.nlm.nih.gov/geo/ |
|
| ||
| Experimental models: Cell lines | ||
|
| ||
| HASTR/ci35 | Furihata, T. et al.54 | N/A |
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| Tg(tetO-HIST1H2BJ/GFP)47Efu/J × Tg(Fos105 tTA)1Mmay | Matsuo, N. et al.33 | RRID:IMSR_JAX:005104 |
| S1P1 flox/flox:hGFAP-cre mice | Choi, J.W. et al.29 | N/A |
| CD320-KO | Lai, S.C. et al.43 | N/A |
|
| ||
| Oligonucleotides | ||
|
| ||
| PCR primer sets | This paper | Table S4 |
|
| ||
| Software and algorithms | ||
|
| ||
| FlowJo (10.0.8r1) | BD | N/A |
| Prism | GraphPad | N/A |
| R | r-project.org | RRID:SCR_001905 |
|
| ||
| Other | ||
|
| ||
| Vitamin B12-deficient diet | Research Diets, Inc | N/A |


