Abstract
The mammalian mRNA for selenium-dependent glutathione peroxidase 1 (Se-GPx1) contains a UGA codon that is recognized as a codon for the nonstandard amino acid selenocysteine (Sec). Inadequate concentrations of selenium (Se) result in a decrease in Se-GPx1 mRNA abundance by an uncharacterized mechanism that may be dependent on translation, independent of translation, or both. In this study, we have begun to elucidate this mechanism. We demonstrate using hepatocytes from rats fed either a Se-supplemented or Se-deficient diet for 9 to 13 weeks that Se deprivation results in an ∼50-fold reduction in Se-GPx1 activity and an ∼20-fold reduction in Se-GPx1 mRNA abundance. Reverse transcription-PCR analyses of nuclear and cytoplasmic fractions revealed that Se deprivation has no effect on the levels of either nuclear pre-mRNA or nuclear mRNA but reduces the level of cytoplasmic mRNA. The regulation of Se-GPx1 gene expression by Se was recapitulated in transient transfections of NIH 3T3 cells, and experiments were extended to examine the consequences of converting the Sec codon (TGA) to either a termination codon (TAA) or a cysteine codon (TGC). Regardless of the type of codon, an alteration in the Se concentration was of no consequence to the ratio of nuclear Se-GPx1 mRNA to nuclear Se-GPx1 pre-mRNA. The ratio of cytoplasmic Se-GPx1 mRNA to nuclear Se-GPx1 mRNA from the wild-type (TGA-containing) allele was reduced twofold when cells were deprived of Se for 48 h after transfection, which has been shown to be the extent of the reduction for the endogenous Se-GPx1 mRNA of cultured cells incubated as long as 20 days in Se-deficient medium. In contrast to the TGA allele, Se had no effect on expression of either the TAA allele or the TGC allele. Under Se-deficient conditions, the TAA and TGC alleles generated, respectively, 1.7-fold-less and 3-fold-more cytoplasmic Se-GPx1 mRNA relative to the amount of nuclear Se-GPx1 mRNA than the TGA allele. These results indicate that (i) under conditions of Se deprivation, the Sec codon reduces the abundance of cytoplasmic Se-GPx1 mRNA by a translation-dependent mechanism and (ii) there is no additional mechanism by which Se regulates Se-GPx1 mRNA production. These data suggest that the inefficient incorporation of Sec at the UGA codon during mRNA translation augments the nonsense-codon-mediated decay of cytoplasmic Se-GPx1 mRNA.
Many organisms, including bacteria, Saccharomyces cerevisiae, and vertebrates, appear to have established mechanisms that eliminate the production of mRNAs that prematurely terminate translation because of a frameshift or nonsense mutation (reviewed in references 32, 33, 37, and 42). Therefore, the discovery of mRNAs in both bacterial and mammalian cells in which one or more UGA codons are purposefully used as selenocysteine (Sec) codons rather than as nonsense codons (reviewed in references 10 and 30) raises the interesting issue of whether these mRNAs are reduced in abundance when the UGA codon is recognized as nonsense.
Cues from (i) biochemical and genetic studies of Escherichia coli (reviewed in reference 10), (ii) the discovery of mammalian homologs to some of the bacterial factors that mediate the cotranslational incorporation of Sec (27, 29, 31), and (iii) information on mammalian mRNA sequences that mediate the incorporation of Sec (7, 34) have contributed to elucidating the mechanism by which a UGA codon is recognized as a Sec codon. In mammals, recognition requires selenium (Se), a metabolic pathway that converts Se to selenocysteyl-tRNA[Ser]Sec, and at least one cis-acting Sec insertion sequence element that presumably associates with the specialized elongation factor. Studies of the type I iodothyronine deiodinase (5′ DI) gene, in which the single TGA codon was converted to a cysteine (Cys) codon, indicate that cells transiently overexpressing the gene are able to produce 20- to 400-fold-more protein from the Cys-containing allele than from the TGA-containing allele (8). Therefore, at least in transiently transfected cells, Sec is incorporated at the UGA codon of 5′ DI mRNA only inefficiently, as may be the case for other selenoprotein mRNAs.
Consistent with the concept that UGA-containing selenoprotein mRNAs can be reduced in abundance when the UGA codon(s) is recognized as nonsense, Se deprivation reduces the abundance of certain selenoprotein mRNAs, some more effectively than others. For example, rats or mice fed a Se-deficient diet for 42 to 135 days manifest an 80 to 95% drop in the level of Se-dependent glutathione peroxidase 1 (Se-GPx1) mRNA in liver (28, 43, 46, 48) that is not attributable to a decrease in Se-GPx1 gene transcription (6, 13, 14, 18, 46). The level of endogenous Se-GPx1 mRNA is also decreased in Se-deficient cultured cells, although not by more than 70%, even if the cells derive from liver (1, 13, 19, 49). Se deficiency reduces the level of other selenoprotein mRNAs, including those for 5′ DI (23), selenoprotein P (23), and selenoprotein W (47). However, Se deficiency does not reduce the level of all selenoprotein mRNAs, as exemplified by mRNA for phospholipid hydroperoxide glutathione peroxidase (PHGPx) (5, 6, 28). Consistent with Se having different effects on different mRNAs, the results of incubating H4 rat hepatoma cells with actinomycin D indicate that Se deficiency decreases the half-life of total-cell Se-GPx1 mRNA but is of no consequence to the half-life of cytoplasmic PHGPx mRNA (5). It is possible that Se regulates the level of selenoprotein mRNAs not only in translation-dependent mechanisms but also in translation-independent mechanisms, and this possibility has been proposed given that (i) feeding Se to Se-deficient rats results in an increase in the level of Se-GPx1 mRNA before a detectable increase in Se-GPx1 activity (45, 48) and (ii) the level of Se-GPx1 activity in rats fed different amounts of Se does not always parallel the level of Se-GPx1 mRNA (6). The mechanism by which Se deprivation reduces the expression of Se-GPx1 cDNA in Chinese hamster ovary (CHO) cells has been demonstrated to be dependent on sequences within the 3′ untranslated region (49), which may also be consistent with a translation-independent mechanism.
In order to clarify if Se (i) regulates the level of Se-GPx1 mRNA via the Sec codon, (ii) affects Se-GPx1 gene expression independently of the Sec codon, or (iii) does both, Se deficiency was induced in rat liver and in NIH 3T3 cells transiently transfected with one of several Se-GPx1 alleles that harbored different sequences at their Sec codons. Results indicate that Se deficiency reduces the abundance of cytoplasmic Se-GPx1 mRNA without altering the ratio of nuclear Se-GPx1 mRNA to nuclear Se-GPx1 pre-mRNA. Regardless of the Se concentration, changing the Sec codon from TGA to a TAA nonsense codon decreased the ratio of cytoplasmic Se-GPx1 mRNA to nuclear Se-GPx1 mRNA while changing the Sec codon to a TGC cysteine codon increased the ratio of cytoplasmic Se-GPx1 mRNA to nuclear Se-GPx1 mRNA. Neither change altered the ratio of nuclear Se-GPx1 mRNA to nuclear Se-GPx1 pre-mRNA. These data indicate that Se deprivation regulates the abundance of cytoplasmic Se-GPx1 mRNA, presumably by eliciting the nonsense-codon-mediated decay of cytoplasmic mRNA.
MATERIALS AND METHODS
Animals and diets.
Six weaned male Long-Evans hooded rats weighing between 40 and 60 g were randomly assigned to either a Se-supplemented or a Se-deficient dietary group. Rats in the Se-supplemented group were fed a Torula yeast-based diet with 0.5 mg of Se per kg of body weight as sodium selenite (Na2SeO3), whereas rats in the Se-deficient group were fed an unsupplemented diet (40). Animals, housed four per cage and subjected to a cycle of 12 h of light and 12 h of dark, were fed ad libitum and provided with double-distilled drinking water for the 9- to 13-week experimental period.
Hepatocyte isolation.
Hepatocytes were isolated from rat liver according to the procedure outlined by Seglen (44) and Boyer et al. (11), as modified by our laboratory. Animals weighing between 350 and 440 g were anesthetized, their superior venae cavae were ligated just below their hearts, their inferior venae cavae were cannulated just above their renal veins with a 16-gauge needle, and their portal veins were cut to allow the exit of perfusate. Their livers were perfused in situ for 10 min at an approximate flow rate of 40 ml per min with calcium-free and magnesium-free Hanks balanced salt solution (Life Technologies) supplemented with 25 mM sodium bicarbonate and 0.5 mM EGTA. Perfusion was continued at the same flow rate with Hanks balanced salt solution containing 1.2 mM calcium and 0.93 mM magnesium and supplemented with 25 mM sodium bicarbonate and 0.5% type I or type IV collagenase (Sigma). Prior to perfusion, 100% oxygen was bubbled through bottles of both Hanks buffers for 15 min in a 37°C water bath. Following perfusion, the livers were placed in ice-cold Dulbecco’s modified Eagle’s medium (DMEM; Life Technologies) supplemented with 10% heat-inactivated fetal calf serum (Hyclone Laboratories) and teased apart with forceps to create a single cell suspension. Cells were filtered through two layers of gauze and pelleted at 500 rpm for 2 min at 4°C. The cell pellet was washed twice with serum-free DMEM and suspended in DMEM. Cell viability was determined by exclusion of 0.4% trypan blue (Sigma) to be ∼70%. Of the ∼2.8 × 108 cells obtained per liver, evaluation by light microscopy indicated that ∼95% were hepatocytes. Cells were pelleted and either used to isolate nuclear and cytoplasmic RNAs or frozen at −80°C for later measurements of Se-GPx1 activity and purification of total RNA.
Plasmid DNAs.
In order to generate pmCMV-GPx1, a plasmid carrying the rat Se-GPx1 gene driven by the mouse cytomegalovirus (mCMV) promoter, the 1,485-bp XbaI-NspI fragment from pKS-rGPx (25) was inserted into the SacI and XbaI sites of pUC19. Prior to insertion, the SacI and NspI sites were made blunt with Klenow fragment and deoxynucleoside triphosphates. Subsequently, the 538-bp XbaI-EcoRI fragment from pMH4 (a gift from Jack Gauldie, McMaster University) that harbors the mCMV promoter was inserted upstream of the Se-GPx1 gene at the XbaI and MboII sites. Prior to insertion, the EcoRI and MboII sites were made blunt.
Overlap-extension PCR (24) was used to convert the Sec-encoding TGA codon at position 46 within exon 1 to either a TGC codon for cysteine (Cys) or a TAA termination codon. In order to create the TGC Cys codon, two overlapping fragments were generated from pGPx1211 (37). A 214-bp fragment was generated with the flanking sense oligonucleotide 5′ GTCCAATATCTTCAAGCTTATGTCTGCTGCTCGG 3′, which corresponds to 19 bp of 5′ untranslated sequences (which was mutagenized to harbor an HindIII site for purposes unrelated to and not affecting this work) plus codons 0 through 4, and the mutagenic antisense oligonucleotide 5′ CGTGGTGCCGCAGAGGGACGC 3′, which corresponds to codons 43 through 49 and harbors the italicized mutagenic nucleotide. An overlapping 706-bp fragment was generated with the mutagenic sense oligonucleotide 5′ GCGTCCCTCTGCGGCACCACG 3′, which corresponds to codons 43 through 49, and the flanking antisense oligonucleotide 5′ CAAAAACGTGCCCATCTAGACACGGAATTCC 3′, which corresponds to bp 851 through 881 of the 3′ untranslated region. In order to generate full-length cDNA of 854 bp harboring the TGC Cys codon, the two PCR products were mixed and subjected to a second PCR with both flanking oligonucleotides. The 129-bp SacII-XmaI fragment harboring the Cys codon was then inserted into the SacII and XmaI sites of pmCMV-GPx1, creating pmCMV-GPx1-TGC(46). A similar approach was used to create cDNA harboring the TAA codon. A 262-bp fragment was generated with the sense flanking oligonucleotide 5′ GAGCTGCGTTCTACGTGGG 3′, which corresponds to nucleotides within the mCMV promoter, and the antisense mutagenic oligonucleotide 5′ GGGTCGTGGTGCCTTAGAGGG 3′, which corresponds to codons 44 through 51. An overlapping 556-bp fragment was generated with the sense mutagenic oligonucleotide 5′ CCCTCTAAGGCACCACGACCC 3′, which corresponds to codons 44 through 51, and the antisense flanking oligonucleotide 5′ ATGTCGTTGCGGCACACCGGG 3′, which corresponds to codons 150 through 157. A 217-bp KpnI-SmaI fragment containing the TAA mutation was inserted into the KpnI and SmaI sites of pmCMV-GPx1 to create pmCMV-GPx1-TAA(46). The integrity of all three clones [pmCMV-GPx1, pmCMV-GPx1-TAA(46), and pmCMV-GPx1-TGC(46)] was verified by DNA sequencing. pmCMV-GPx1-TAA(46) harbored an additional mutation within codon 15 (TAT→TGT) that likely arose during the PCR.
In order to generate an intronless pmCMV-GPx1, the 457-bp XmaI-DraIII fragment that extends from exon 1 into exon 2 was excised from pmCMV-GPx1 and replaced with the corresponding 251-bp XmaI-DraIII fragment from pGPx1211.
Cell transfections.
Both NIH 3T3 cells and L cells were maintained in minimal essential medium (Life Technologies) containing 5% calf serum and 10% fetal calf serum. NIH 3T3 cells (107 cells/15-cm-diameter dish, 40 to 50% confluency) were transiently transfected with the pmCMV-GPx1 test plasmid (25 μg) and the pmCMV-G1 reference plasmid (25 μg) (34) with calcium phosphate (50). After 12 h, cells were washed twice with phosphate-buffered saline and cultured in either serum-free medium or serum-free medium supplemented with 50 ng of selenous acid per ml (26). Serum-free medium consisted of a 1:1 (vol/vol) mixture of Ham’s F-12 (Life Technologies) and DMEM plus 25 μg of bovine holo-transferrin per ml, 10 μg of bovine insulin per ml, 10 ng of mouse epidermal growth factor per ml, and 25 μg of human high-density lipoprotein per ml (17). Cells were harvested after another 48 h.
Se-GPx1 enzyme activity and protein assay.
Hepatocytes, L cells and NIH 3T3 cells were suspended in glutathione peroxidase homogenization buffer (25 mM sucrose, 10 mM potassium phosphate, 1 mM EDTA, 2 mM glutathione [pH 7.4]) and lysed by sonication (Branson model 450 sonifier) at 4°C with two 30-s pulses at a power setting of 2. Cell lysates were centrifuged at 25,000 × g for 30 min at 4°C, and protein concentration was measured spectrophotometrically by a protein dye-binding assay (Bio-Rad) with bovine serum albumin as a standard. Se-GPx1 activity was measured spectrophotometrically by the procedure as modified by Reddy et al. (41) of coupling the reduction of H2O2 by glutathione to the oxidation of NADPH with glutathione reductase (36). A molar extinction coefficient of 6,200 M−1 cm−1 for NADPH was used in the calculation. One unit of enzyme activity was defined as 1 μM NADPH oxidized min−1 mg of protein−1.
RNA isolation, Northern hybridization, and RT-PCR analyses.
Hepatocytes and NIH 3T3 cells were separated into nuclear and cytoplasmic fractions (method 1 [4]). Cytoplasmic RNA was purified by CsCl gradient centrifugation (4), and nuclear RNA was purified with Trizol reagent (Life Technologies). For reverse transcriptase PCR (RT-PCR), RNA (25 μg) was treated with RQ1 DNase (1 U; Promega Corp.) in DNase buffer (40 mM Tris-HCl [pH 7.9], 10 mM NaCl, 6 mM MgCl2, 10 mM CaCl2) for 1 h at 37°C. Alternatively, total RNA was isolated with Trizol reagent.
For Northern hybridization, RNA (25 μg) was denatured with glyoxal, electrophoresed in a 1.5% agarose gel, and transferred to a nylon membrane (Zetabind). The membrane was hybridized (16) to two fragments: (i) a 311-bp EcoRI-EcoRI fragment that derives from pGPx1211 (38) and that includes 44 bp of the 5′ untranslated region and 267 bp of the translated region of Se-GPx1 cDNA and (ii) a 428-bp fragment of the cytoplasmic β-actin translated region that was generated by RT-PCR with hepatocyte RNA and two primers (5′ CACTGGCATTGTGATGGA 3′ [sense] and 5′ ACGGATGTCAACGTCACA 3′ [antisense] [21]). Prior to hybridization, each fragment was 32P labeled by random priming (Promega). Quantitative analysis was performed with a PhosphorImager and ImageQuant software (Molecular Dynamics).
For all RT-PCRs (16), cDNA was synthesized from 0.16 to 5.0 μg of total, nuclear, or cytoplasmic RNA with RT (Superscript; GIBCO) and random hexamers (Promega). Se-GPx1 and β-actin cDNAs that derived from rat hepatocyte RNA were amplified in separate tubes. Each PCR mixture contained 1/20 of the RT reaction mixture, 0.12 mM each deoxynucleotide (n = 4), 4 μCi of [α-32P]dATP (3,000 Ci/mmol; Amersham), 0.5 μM each primer (n = 2), and 2.5 U of Taq DNA polymerase (Promega). To amplify β-actin RNA, the primers consisted of 5′ CACTGGCATTGTGATGGA 3′ (sense) and 5′ ACGGATGTCAACGTCACA 3′ (antisense) and amplification was for 21 cycles. To amplify Se-GPx1 RNA, the primers consisted of 5′ ATGTCTGCTGCTCGGCTCTCCGCGG 3′ (sense) and 5′ CTTCFTCACCATTCACCTCGCCTT 3′ (antisense) and amplification was for 32 cycles. mCMV-GPx1 and mCMV-G1 cDNAs from transfected NIH 3T3 cells were amplified in 19 cycles with a sense primer, 5′ ACCACCGTAGAACGCAGATCG 3′, that corresponds to the common mCMV promoter region. The antisense primer used to amplify mCMV-GPx1 cDNA, 5′ CTTCTCACCATTCACCTCGCACTT 3′, corresponds to Se-GPx1 exon 2. The antisense primer used to amplify mCMV-G1 cDNA, 5′ CGGGGTGAAGCTCCTTGCCAAG 3′, corresponds to exon 3 of the human β-globin gene (16). Notably, Se-GPx1 cDNA that derived from the endogenous NIH 3T3 cell gene was not amplified. For all PCRs, each cycle consisted of denaturation at 94°C for 75 s, annealing at 55°C for 40 s, and extension at 72°C for 40 s. One-tenth of each PCR mixture was electrophoresed in a 4% polyacrylamide gel, and RT-PCR products were quantitated by PhosphorImaging.
RESULTS
Se deprivation has no effect on the abundance of nuclear Se-GPx1 pre-mRNA or nuclear Se-GPx1 mRNA but reduces the abundance of cytoplasmic Se-GPx1 mRNA in rat liver.
One way to determine if Se regulates the level of rat Se-GPx1 mRNA through the UGA codon or if regulation also takes place independently of Sec incorporation is to analyze the expression of Se-GPx1 alleles harboring a mutated TGA codon. This might be accomplished by transfecting cultured cells with normal and mutated rat Se-GPx1 alleles, provided the exogenous TGA-containing allele is regulated like the endogenous Se-GPx1 gene of the intact animal. As a start, the metabolism of Se-GPx1 RNA in the livers of rats fed either a Se-supplemented or a Se-deficient diet for 10 to 13 weeks was evaluated. Liver was chosen because it has the highest level of Se-GPx1 activity of any tissue that has been analyzed (2) and retains Se only inefficiently during Se deprivation (3, 12). Furthermore, a sufficient number of a single type of cell can be isolated from one animal, since one liver routinely yields ∼2.8 × 108 cells, ∼95% of which are hepatocytes.
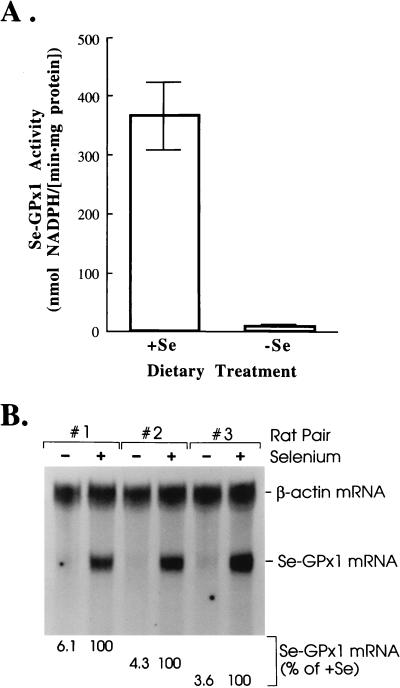
Hepatocytes from three individually analyzed rats fed a Se-deficient diet had an average of only ∼2% of the Se-GPx1 activity and ∼5% of the level of Se-GPx1 mRNA of hepatocytes from three individually analyzed rats fed a Se-supplemented diet (Fig. 1), consistent with Se-GPx1 activity and mRNA measurements of rat liver made by others (18, 23, 28, 43). For RNA quantitation, Northern blot analysis was used to normalize the level of Se-GPx1 mRNA to the level of β-actin mRNA, which is Se unresponsive, to control for variations in levels of RNA loaded between samples.
FIG. 1.
Hepatocytes from rats fed a Se-supplemented diet for 12 weeks have 2% of the Se-GPx1 activity and 5% of the level of Se-GPx1 mRNA of hepatocytes from rats fed a Se-supplemented diet for the same amount of time. Rats were analyzed in pairs in which one was fed a Se-supplemented diet and the other was fed a Se-deficient diet, and the periods of feeding for both members of the pair were the same. Hepatocytes were isolated according to the methods of Seglen (44) and Boyer et al. (11), as modified by us (see Materials and Methods). (A) Se-GPx1 activity was measured according to the method of Paglia and Valentine (36), as modified by Reddy et al. (39). Hepatocytes from three different animals per dietary group were analyzed, and duplicate measurements were taken for each animal. Measurements are presented as means ± standard deviations. (B) Total hepatocyte RNA (25 μg) was electrophoresed in agarose, transferred to a nylon membrane, and hybridized to a 311-bp EcoRI-EcoRI fragment of Se-GPx1 cDNA (38) and a 428-bp fragment, generated by RT-PCR, that consists of the open reading frame of cytoplasmic β-actin cDNA. Lanes marked with a minus sign contain RNAs isolated from three different animals fed a Se-deficient diet. Lanes marked with a plus sign contain RNAs isolated from three different animals fed a Se-supplemented diet. The level of Se-GPx1 mRNA was normalized to the level of β-actin mRNA in order to control for variations in the amounts of RNAs loaded in lanes. The normalized value for each rat fed a Se-supplemented diet (+Se) was considered to be 100%, and the normalized value for the corresponding rat fed a Se-deficient diet was calculated as a percentage of that of the rat fed a Se-supplemented diet.
While Se deficiency has been shown to have no effect on the level of Se-GPx1 gene transcription (6, 13, 14, 18, 46), the consequence of Se deficiency on the processing of nuclear Se-GPx1 pre-mRNA or nuclear mRNA has never been successfully evaluated because of detection difficulties (18). Therefore, RT-PCR, which is a more sensitive assay than Northern hybridization, was used to determine if the reduction in Se-GPx1 mRNA abundance brought about by Se deficiency takes place in the nuclear fraction, the cytoplasmic fraction, or both. cDNA was made from each RNA fraction with random hexamers. Specific pairs of primers were subsequently used to amplify exon 1 through exon 2 of Se-GPx1 cDNA and, as a control, exon 1 through exon 2 of β-actin cDNA, i.e., the first through the last exon of each cDNA. Conditions were empirically established to provide a linear relationship between the amount of each RT-PCR product and the amount of input RNA, as evidenced by an analysis of serial dilutions of cytoplasmic RNA (Fig. 2). In agreement with the results of Northern hybridization (Fig. 1B), Se deficiency in two individually analyzed rats reduced the abundance of cytoplasmic Se-GPx1 mRNA to ∼3% of the level observed for rats fed a Se-supplemented diet (Fig. 2; Table 1). Notably, Se deficiency had no effect on the level of either nuclear Se-GPx1 pre-mRNA or nuclear Se-GPx1 mRNA, which, respectively, comprised averages of 9 and 29% of the level of cytoplasmic Se-GPx1 mRNA in Se-fed rats (Fig. 2; Table 1). By evaluating the data another way, the ratio of nuclear Se-GPx1 mRNA to nuclear Se-GPx1 pre-mRNA was unaffected by Se while the ratio of cytoplasmic Se-GPx1 mRNA to nuclear Se-GPx1 mRNA in Se-deficient rats was 3 to 4% of the ratio in Se-fed rats (Table 1). Together with demonstrations with actinomycin D that Se deficiency decreases the stability of Se-GPx1 mRNA in total RNA (5), these data indicate that Se deficiency reduces the half-life of cytoplasmic Se-GPx1 mRNA without affecting the metabolism of nuclear RNA.
FIG. 2.
The reduced level of Se-GPx1 mRNA in Se-deficient hepatocytes is due to a decrease in the level of cytoplasmic Se-GPx1 mRNA but not nuclear Se-GPx1 mRNA. Nuclear (N) and cytoplasmic (C) RNA was isolated (method 1 [4]) from hepatocytes of each pair of rats fed either a Se-deficient (−) or a Se-supplemented (+) diet. Se-GPx1 and β-actin RNAs in each sample were analyzed by RT-PCR. The left-most four lanes consist of serial dilutions of cytoplasmic RNA (5.0, 1.3, 0.6, and 0.3 μg, from left to right), which were used to establish that there is a linear relationship between the amount of input RNA and the amount of each RT-PCR product. Size standards for RT-PCR products of Se-GPx1 pre-mRNA and Se-GPx1 mRNA were provided by the PCR amplification of, respectively, 25 pg of pmCMV-GPx1 (Se-GPx1 gene) and 25 pg of pGPx1211 (Se-GPx1 cDNA) with the same primers that were used to amplify hepatocyte cDNA.
TABLE 1.
Se-GPx1 RNA in rat liver
| Rat pair | Se | %a of:
|
||||
|---|---|---|---|---|---|---|
| pre- mRNAN | mRNAN | mRNAC | mRNAN/ pre-mRNAN | mRNAC/ mRNAN | ||
| 1 | − | 6.94 | 25.92 | 3.78 | 105 | 3 |
| + | 6.06 | 23.81 | 100 | 100 | 100 | |
| 2 | − | 10.41 | 27.49 | 3.15 | 108 | 4 |
| + | 13.22 | 37.43 | 100 | 100 | 100 | |
Values derive from a quantitative analysis of the experimental results shown in Fig. 2. The levels of nuclear Se-GPx1 pre-mRNA (pre-mRNAN) and nuclear Se-GPx1 mRNA (mRNAN) were normalized to the level of nuclear β-actin mRNA, and the level of cytoplasmic Se-GPx1 mRNA (mRNAC) was normalized to the level of cytoplasmic β-actin mRNA. For each rat pair, normalized values for both Se-deficient (−) and Se-supplemented (+) rats were calculated as percentages of the normalized value for Se-GPx1 cytoplasmic mRNA in the Se-supplemented (+) rat, which was considered to be 100%. The ratios (mRNAN/pre-mRNAN and mRNAC/mRNAN) for each Se-deficient rat were calculated as percentages of the corresponding ratios for the Se-supplemented rat of each pair, which were considered to be 100.
The rat Se-GPx1 gene, when transiently expressed in NIH 3T3 cells, is qualitatively regulated by Se as it is in rat liver.
Having concluded that Se deficiency reduces the abundance of cytoplasmic but not nuclear Se-GPx1 mRNA in intact animals, we wanted to determine if Se deficiency does the same when the Se-GPx1 gene is transiently expressed in cultured cells. If it does, there would be a way of testing the effects of mutations within the TGA codon on Se-GPx1 gene expression. Mouse L and mouse NIH 3T3 cells were chosen for their superior transfection efficiencies, our ability to distinguish rat and mouse Se-GPx1 RNAs by RT-PCR, and there being no appreciable difference between cultured liver cells and cultured fibroblasts (or any other cultured cell type evaluated) in the extents to which Se deficiency reduces Se-GPx1 mRNA abundance (1, 13, 19, 23, 49).
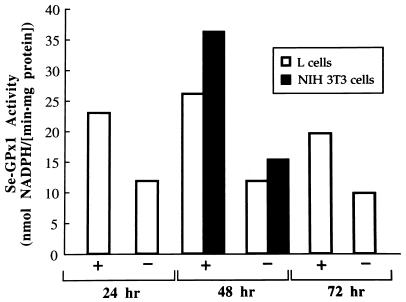
First, in order to determine the rate of decline in Se-GPx1 activity after Se deprivation, cells were mock transfected and transferred 12 h later to Se-deficient medium. After 24 h in Se-deficient medium, Se-GPx1 activity in L cells had decreased to 51% of the activity in L cells grown in Se-supplemented medium (Fig. 3). This decrease was not significantly different from the decreases observed after 48 and 72 h in Se-deficient medium (Fig. 3). Similarly, the activity in NIH 3T3 cells after 48 h in Se-deficient medium was 55% of the activity in NIH 3T3 cells grown in Se-supplemented medium (Fig. 3). These data resemble data from the time-dependent decline in Se-GPx1 activity observed for other cultured cells grown in Se-deficient medium (1, 5, 23).
FIG. 3.
Incubating either L cells or NIH 3T3 cells in Se-deficient medium for 24, 48, or 72 h decreases Se-GPx1 activity to 50 to 55% of the activity of Se-supplemented cells. Activity was measured according to the method of Paglia and Valentine (36), as modified by Reddy et al. (39). Each cell line was mock transfected with either DEAE dextran (L cells) or calcium phosphate (NIH 3T3 cells) and incubated for an additional 12 h before the medium was changed to either Se-deficient (−) or Se-supplemented (+) medium. Se-GPx1 activities are presented as the means of duplicate measurements for each time point.
Northern hybridization with total RNA revealed that the level of endogenous Se-GPx1 mRNA in Se-deficient NIH 3T3 cells was reduced by only 31% compared to the level in Se-supplemented NIH 3T3 cells (data not shown). The level in Se-deficient cells was not expected to be further reduced with a longer period of Se deficiency, since Se deprivation of cultured HepG2 or H4IIIE cells for 4 days yielded only a two- to threefold decrease in the level of endogenous Se-GPx1 mRNA (23). Furthermore, Se deprivation of cultured HL-60 cells for as long as 20 days yielded only a 1.2- to 2.3-fold decrease in the level of endogenous Se-GPx1 mRNA (13), and the analysis of CHO cells that had been stably transfected with Se-GPx1 cDNAs and Se deprived for 7 days yielded less than a 2-fold decrease in the level of exogenous Se-GPx1 mRNA (49). According to these data, stable transfections offer no advantage over transient transfections in achieving a larger effect of Se deprivation on Se-GPx1 mRNA abundance. The greater extent to which Se deprivation mediates a reduction in the abundance of Se-GPx1 mRNA in the liver cells of animals relative to that in cultured cells may reflect a difference in mechanism. However, data suggest that the variation in extents may be limited to a quantitative difference rather than a mechanistic difference since the cellular compartment of the reduction and the molecule targeted for reduction are the same in liver cells and cultured cells (see below). Since our extensive experience with RT-PCR (see references 4, 16, and 51) has proven the technique to be sufficiently sensitive and accurate to measure twofold differences in levels of RNA between samples, we reasoned that the transient transfection of either NIH 3T3 or L cells would provide a suitable means to study changes in Se-GPx1 mRNA concentration in response to Se nutrition.
NIH 3T3 cells were transiently transfected with a test plasmid, pmCMV-GPx1, which harbors a rat Se-GPx1 allele driven by the mCMV promoter (Fig. 4A), and a reference plasmid, pmCMV-G1 (52), which harbors a β-globin allele similarly driven by the mCMV promoter. RNA produced from the reference plasmid was used to control for variations in the efficiencies of cell transfection and RNA recovery. Twelve hours after transfection, cells were placed in either Se-deficient or Se-supplemented medium, and after an additional 48 h, either total or nuclear and cytoplasmic RNA was isolated. RT-PCR was used to quantitate the levels of plasmid-derived Se-GPx1 and β-globin RNAs in a way that did not detect NIH 3T3 cell transcripts.
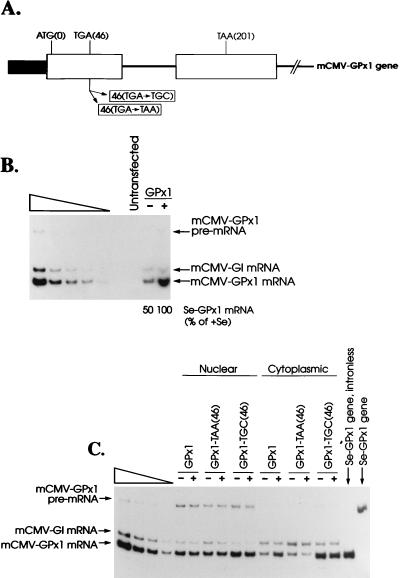
FIG. 4.
(A) Structures of the mCMV-GPx1 gene and derivative alleles. The shaded box represents the 550-bp XbaI-EcoRI fragment that harbors the mCMV promoter. The open boxes represent exons, the intervening line represents the single 216-bp intron, and the right-most line represents 3′ flanking DNA. ATG(0), TGA(46), and TAA(201) represent, respectively, the initiation codon, Sec codon, and termination codon. Mutations that convert the Sec codon to either a Cys codon (TGC) or a premature termination codon (TAA) are indicated below the gene structure. (B) The level of mCMV-GPx1 mRNA in total RNA of NIH 3T3 cells grown in Se-deficient medium (−) is 50% of the level in NIH 3T3 cells grown in Se-supplemented (+) medium. Cells were either untransfected or transiently transfected with pmCMV-GPx1 (25 μg) and the reference plasmid pmCMV-G1 (25 μg). Total RNA was isolated, and RT-PCR was used to quantitate mCMV-GPx1 and mCMV-G1 mRNAs. The left-most five lanes contain twofold serial dilutions of RNA from Se-supplemented cells in order to demonstrate a linear relationship between the amounts of input RNA and RT-PCR products. The level of mCMV-GPx1 mRNA was normalized to the level of mCMV-G1 mRNA. The normalized value for mCMV-GPx1 mRNA in Se-deficient cells was then calculated as a percentage of the normalized value for mCMV-GPx1 mRNA in Se-supplemented cells, which was considered to be 100%. The values from two independently performed experiments did not differ by more than 3%. (C) The ratio of cytoplasmic mCMV-GPx1 mRNA to nuclear mCMV-GPx1 mRNA is decreased in Se-deficient NIH 3T3 cells by a decay pathway that is dependent on recognition of the Sec codon as a termination codon. Transfections and analyses of RNA were as described in the legend to Fig. 4B, except that nuclear and cytoplasmic RNAs were purified and analyzed as described in the legend to Fig. 2. The left-most four lanes contain twofold dilutions of cytoplasmic RNA from Se-supplemented NIH 3T3 cells transiently transfected with pmCMV-GPx1-TGC(46). GPx1, GPx1-TAA(46), and GPx1-TGC(46) signify pmCMV-GPx1 plasmids harboring at position 46 the wild-type sequence (TGA), a nonsense codon (TAA), and a Cys codon (TGC), respectively. The two right-most lanes, which reflect PCR analyses of intronless pmCMV-GPx1 DNA and pmCMV-GPx1 DNA, provide molecular weight standards for pre-mRNA and mRNA, respectively.
As anticipated, Se deficiency reduced the level of Se-GPx1 mRNA in total RNA to 50% of the level observed under Se-supplemented conditions (Fig. 4B). By analyzing nuclear and cytoplasmic RNA, this reduction was found to be characteristic of cytoplasmic but not nuclear Se-GPx1 mRNA: Se deprivation reduced the ratio of cytoplasmic to nuclear Se-GPx1 mRNA to 52% of the ratio observed under conditions of Se supplementation but had no effect on the ratio of nuclear Se-GPx1 mRNA to nuclear Se-GPx1 pre-mRNA (Fig. 4C; Table 2). Therefore, the transient expression of pmCMV-GPx1 in Se-deficient NIH 3T3 cells results in the production of an abnormally low level of cytoplasmic but not nuclear Se-GPx1 mRNA, as does Se deficiency induced in the livers of rats fed a Se-deficient diet. This finding suggests, but does not prove, that the difference in the extent to which Se deficiency affects the level of Se-GPx1 mRNA in the cells of animals and in cultured cells is quantitative rather than mechanistic.
TABLE 2.
Rat Se-GPx1 RNA in transiently transfected mouse NIH 3T3 cells
| Codon 46 | Se | %a of:
|
||||
|---|---|---|---|---|---|---|
| premRNAN | mRNAN | mRNAC | mRNAN/ pre-mRNAN | mRNAC/ mRNAN | ||
| Sec (UGA) | − | 73 | 237 | 52 | 105 | 50 |
| + | 84 | 260 | 100 | 100 | 100 | |
| Ter (UAA) | − | 39 | 138 | 34 | 114 | 30 |
| + | 50 | 194 | 30 | 102 | 29 | |
| Cys (UGC) | − | 145 | 502 | 157 | 110 | 143 |
| + | 157 | 487 | 154 | 101 | 153 | |
Values derive from a quantitative analysis of the experimental results shown in Fig. 4. The levels of nuclear pre-mRNA (pre-mRNAN), nuclear mRNA (mRNAN), and cytoplasmic mRNA (mRNAC) were normalized to the corresponding level of β-globin mRNA and calculated as percentages of the normalized value for the cytoplasmic mRNA Sec (UGA) in Se-supplemented (+) medium, which was given a value of 100%. Each ratio (mRNAN/pre-mRNAN and mRNAC/mRNAN) was calculated as a percentage of the corresponding ratio for mCMV-GPx1-UGA(46)RNA in Se-supplemented (+) medium, which was given a value of 100.
Se regulates Se-GPx1 gene expression through the Sec TGA codon.
Conceivably, Se deficiency reduces the abundance of cytoplasmic Se-GPx1 mRNA by more than one mechanism. For example, some investigators have hypothesized the binding of one or more Se-regulated proteins to a sequence within Se-GPx1 RNA that stabilizes the RNA (48, 49). As another example, our interest in the nonsense-codon-mediated decay of mRNA has led us to propose that Se deficiency destabilizes Se-GPx1 mRNA by evoking the premature termination of translation at the UGA(46) codon. Se deficiency has been shown to reduce the levels of both selenocysteine tRNA[Ser]Sec isoacceptors (20, 22), which is likely to reduce the efficiency of Sec insertion into position 46 of the growing Se-GPx1 polypeptide chain.
In order to determine if Se-GPx1 mRNA is susceptible to nonsense-codon-mediated decay, the TGA(46) codon within pmCMV-GPx1 was changed to either a TAA nonsense codon or a TGC Cys codon (Fig. 4A). Each change would eliminate the sensitivity of Se-GPx1 mRNA to Se if the TGA codon is required for Se sensitivity. NIH 3T3 cells were transiently transfected with each pmCMV-GPx1 plasmid together with the reference pmCMV-G1 plasmid, and gene expression was quantitated by RT-PCR after cell growth in Se-deficient or Se-supplemented medium. Results indicate that neither the TAA-containing allele nor the TGC-containing allele is sensitive to Se: neither the ratio of nuclear Se-GPx1 mRNA to nuclear Se-GPx1 pre-mRNA nor the ratio of cytoplasmic Se-GPx1 mRNA to nuclear Se-GPx1 mRNA was affected by a change in Se concentration (Fig. 4C; Table 2). Therefore, Se regulates Se-GPx1 gene expression in a mechanism that is dependent on the TGA codon. For reasons not understood, the level of Se-GPx1 pre-mRNA was highest for the TGC-containing allele and lowest for the TAA-containing allele (Fig. 4C; Table 2), even though the TGC-containing and TAA-containing alleles were generated from the TGA-containing allele simply by swapping the appropriate 129-bp fragment. This variation, however, is unaffected by Se and not relevant to our studies.
Evidence that the Sec codon reduces the abundance of cytoplasmic Se-GPx1 mRNA by eliciting nonsense-codon-mediated mRNA in the cytoplasm.
Relative to results with the TGA-containing allele expressed under Se-deficient conditions, the TAA-containing allele produced a 1.7-fold-lower ratio of cytoplasmic to nuclear Se-GPx1 mRNA and the TGC-containing allele produced a 3-fold-higher ratio of cytoplasmic to nuclear Se-GPx1 mRNA (Fig. 4C; Table 2). These findings suggest that the premature termination of translation at position 46 mediates the decay of cytoplasmic Se-GPx1 mRNA, whether the codon is a UGA codon in Se-deficient cells or a UAA codon in either Se-deficient or Se-supplemented cells. mRNA from the TAA-containing allele, regardless of the Se concentration, is degraded more efficiently than mRNA from the TGA-containing allele under Se-deficient conditions. This result indicates that the UAA codon mediates translation termination more efficiently than the UGA codon even when cells are incubated in Se-deficient medium, suggesting that the cells are never completely depleted of Se.
DISCUSSION
Our results indicate that the responsiveness of the Se-GPx1 gene to Se deprivation is (i) dependent on the efficiency with which the TGA codon at position 46 mediates the incorporation of Sec and (ii) likely attributable to the nonsense-codon-mediated decay of cytoplasmic Se-GPx1 mRNA. First, liver cells of rats fed a Se-deficient diet, like Se-deficient cultured cells that transiently express the rat Se-GPx1 gene, are characterized by an abnormally low cytoplasmic level of rat Se-GPx1 mRNA that is not attributable to alterations in the metabolism of nuclear RNA (Fig. 1B and 4B; Table 1). Second, changing the TGA codon to either a TAA nonsense codon or a TGC Cys codon makes the Se-GPx1 gene insensitive to Se concentration (Fig. 4; Table 2). The efficiency with which cytoplasmic Se-GPx1 mRNA is degraded in Se-deficient medium is highest for the TAA-containing allele, intermediate for the TGA-containing allele, and lowest for the TGC-containing allele (Fig. 4C; Table 2). These data suggest that, even in Se-deficient medium, there may be some incorporation of Sec at the UGA codon that abrogates nonsense-codon-mediated mRNA decay. Consistent with data indicating that incorporation of Sec at the UGA codons of mammalian selenoprotein mRNAs is less than 100% under Se-supplemented conditions (8, 9), the ratio of cytoplasmic to nuclear Se-GPx1 mRNA is 1.5-fold higher for mRNA harboring the UGC codon than for mRNA harboring the UGA codon under Se-supplemented conditions (Fig. 4C; Table 2). Therefore, it appears that a fraction of cytoplasmic Se-GPx1 mRNA harboring the UGA codon, even under Se-supplemented conditions, is subject to nonsense-codon-mediated decay. It is currently not known how the efficiency of UGA-mediated translation termination correlates with the efficiency of UGA-mediated mRNA decay.
Interestingly, not all selenoprotein mRNAs are reduced in abundance when the Se concentration is reduced. As an example, mRNA for PHGPx essentially escapes reduction (6). It follows that mRNA sequences residing outside of a UGA codon for Sec must determine whether the codon will mediate a reduction in mRNA abundance.
The mechanism by which nonsense codons mediate a reduction in mRNA abundance is a topic of considerable controversy, with some investigators proposing that nonsense codons can influence pre-mRNA splicing (reviewed in references 32 and 33). With regard to instances in which fully spliced mRNA is the target of decay, decay may take place (i) in association with nuclei, presumably, but not certainly, during the process of mRNA export from the nucleus to the cytoplasm, or (ii) in the cytoplasm (reviewed in references 32 and 33). To date, it is not possible to predict where a particular mRNA will be degraded other than by noting that most mammalian cell mRNAs that have been studied are degraded in association with nuclei. Therefore, studies of Se-GPx1 mRNA decay add to our understanding of the under-represented type of mRNA that is degraded in the cytoplasm.
The analyses of insertions and deletions within mRNAs for triosephosphate isomerase and β-globin indicate that nonsense codons located more than ∼50 nucleotides upstream of the final exon-exon junction reduce mRNA abundance but that those residing closer to the junction or downstream of the junction have no effect on mRNA abundance (15, 52, 53). A priori, there is no reason why this criterion could not be used to predict whether a Sec codon mediates Se-GPx1 mRNA decay when it is inefficiently utilized. The TGA codon within the Se-GPx1 gene resides 105 bp upstream of the sole intron (25), consistent with indications from our research that Se-GPx1 mRNA is susceptible to nonsense-codon-mediated decay. Furthermore, deletion of the intron eliminates decay (35), which is also consistent with what we know about nonsense-codon-mediated decay (52).
While the simplest interpretation of our data is that the UGA codon of Se-GPx1 mRNA mediates mRNA decay by the same mechanism as the UAA codon, this, in fact, may not be true. Our data, combined with data of Weiss and Sunde (49), demonstrate that the UGA codon and the 3′ untranslated region are required for the reduction in Se-GPx1 mRNA abundance brought about by Se deprivation. While it is quite possible that the interplay between the codon and the 3′ untranslated region is solely a means for Sec incorporation, our data do not rule out the proposal by Weiss and Sunde (49) that Se-GPx1 mRNA is also directly protected from cytoplasmic decay by the formation of an Se-dependent RNA binding complex that depends on the presence of the UGA codon but involves sequences within the 3′ untranslated region. Future studies will examine the finer points of the mechanism by which the UGA codon mediates a reduction in the abundance of Se-GPx1 mRNA.
ACKNOWLEDGMENTS
We thank Jing Zhang, Yimei Qian, and Gerry Jahries for advice and Donna Ovak for typing the manuscript.
This work was supported by Public Health Service research grant GM52822 to L.E.M. from the National Institutes of Health.
REFERENCES
- 1.Baker R D, Baker S S, LaRosa K, Whitney C, Newburger P E. Selenium regulation of glutathione peroxidase in human hepatoma cell line Hep3B. Arch Biochem Biophys. 1993;304:53–57. doi: 10.1006/abbi.1993.1320. [DOI] [PubMed] [Google Scholar]
- 2.Behne D, Wolters W. Distribution of selenium and glutathione peroxidase in the rat. J Nutr. 1983;113:456–461. doi: 10.1093/jn/113.2.456. [DOI] [PubMed] [Google Scholar]
- 3.Behne D, Hilmert H, Scheid S, Gessner H, Elger W. Evidence for specific selenium target tissues and new biologically important selenoproteins. Biochim Biophys Acta. 1988;966:12–21. doi: 10.1016/0304-4165(88)90123-7. [DOI] [PubMed] [Google Scholar]
- 4.Belgrader P, Cheng J, Zhou X, Stephenson L S, Maquat L E. Mammalian nonsense codons can be cis-effectors of nuclear mRNA half-life. Mol Cell Biol. 1994;14:8219–8228. doi: 10.1128/mcb.14.12.8219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bermano G, Arthur J R, Hesketh J E. Selective control of cytosolic glutathione peroxidase and phospholipid hydroperoxide glutathione peroxidase mRNA stability by selenium supply. FEBS Lett. 1996;387:157–160. doi: 10.1016/0014-5793(96)00493-0. [DOI] [PubMed] [Google Scholar]
- 6.Bermano G, Nicol F, Dyer J A, Sunde R A, Beckett G J, Arthur J R, Hesketh J E. Tissue-specific regulation of selenoenzyme gene expression during selenium deficiency in rats. Biochem J. 1995;311:425–430. doi: 10.1042/bj3110425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berry M J, Banu L, Harney J W, Larsen P R. Functional characterization of the eukaryotic SECIS elements which direct selenocysteine insertion at UGA codons. EMBO J. 1993;12:3315–3322. doi: 10.1002/j.1460-2075.1993.tb06001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berry M J, Maia A L, Kieffer J D, Harney J W, Larsen P R. Selenocysteine insertion or termination: factors affecting UGA codon fate and complementary anticodon: codon mutations. Endrocrinology. 1992;131:1848–1852. doi: 10.1093/nar/22.18.3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berry M J, Harney J W, Ohama T, Hatfield D L. Selenocysteine insertion or termination: factors affecting UGA codon fate and complementary anticodon:codon mutations. Nucleic Acids Res. 1994;22:3753–3759. doi: 10.1093/nar/22.18.3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Böck A, Forchhammer K, Heider J, Baron C. Selenoprotein synthesis: an expansion of the genetic code. Trends Biochem Sci. 1991;16:463–467. doi: 10.1016/0968-0004(91)90180-4. [DOI] [PubMed] [Google Scholar]
- 11.Boyer J L, Phillips J M, Graf J. Preparation and specific applications of isolated hepatocyte couplets. Methods Enzymol. 1990;192:501–509. doi: 10.1016/0076-6879(90)92090-z. [DOI] [PubMed] [Google Scholar]
- 12.Buckman T D, Sutphin M S, Eckhert C D. A comparison of the effects of dietary selenium on selenoprotein expression in rat brain and liver. Biochim Biophys Acta. 1993;1163:176–184. doi: 10.1016/0167-4838(93)90179-u. [DOI] [PubMed] [Google Scholar]
- 13.Chada S, Whitney C, Newburger P E. Post-transcriptional regulation of glutathione peroxidase gene expression by selenium in the HL-60 human myeloid cell line. Blood. 1989;74:2535–2541. [PubMed] [Google Scholar]
- 14.Chang M, Reddy C C. Active transcription of the selenium-dependent glutathione peroxidase gene in selenium-deficient rats. Biochem Biophys Res Commun. 1991;181:1431–1436. doi: 10.1016/0006-291x(91)92099-6. [DOI] [PubMed] [Google Scholar]
- 15.Cheng J, Belgrader P, Zhou X, Maquat L E. Introns are cis effectors of the nonsense-codon-mediated reduction in nuclear mRNA abundance. Mol Cell Biol. 1994;14:6317–6325. doi: 10.1128/mcb.14.9.6317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheng J, Maquat L E. Nonsense codons can reduce the abundance of nuclear mRNA without affecting the abundance of pre-mRNA or half-life of cytoplasmic mRNA. Mol Cell Biol. 1993;13:1892–1902. doi: 10.1128/mcb.13.3.1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chiang L, Silnutzer J, Pipas J, Barnes D W. Selection of transformed cells in serum-free media. In Vitro Cell Dev Biol. 1985;21:707–712. doi: 10.1007/BF02620926. [DOI] [PubMed] [Google Scholar]
- 18.Christensen M J, Burgener K W. Dietary selenium stabilizes glutathione peroxidase mRNA in rat liver. J Nutr. 1992;122:1620–1626. doi: 10.1093/jn/122.8.1620. [DOI] [PubMed] [Google Scholar]
- 19.Chu F-F, Esworthy R S, Akman S, Doroshow J H. Modulation of glutathione peroxidase expression by selenium: effect on human MCF-7 breast cancer cell transfectants expressing a cellular glutathione peroxidase cDNA and doxorubicin-resistant MCF-7 cells. Nucleic Acids Res. 1990;18:1531–1539. doi: 10.1093/nar/18.6.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Diamond A M, Choi I S, Crain P F, Hashizume T, Pomerantz S C, Cruz R, Steer C J, Hill K E, Burk R F, McCloskey J A, Hatfield D L. Dietary selenium affects methylation of the Wobble nucleoside in the anticodon of selenocysteine tRNA[Ser]Sec. J Biol Chem. 1993;19:14215–14223. [PubMed] [Google Scholar]
- 21.Hall J C, Reddy N G. Protein D is differentially expressed and regulated in the rat epididymis. Biochem Biophys Res Commun. 1992;183:1109–1116. doi: 10.1016/s0006-291x(05)80305-5. [DOI] [PubMed] [Google Scholar]
- 22.Hatfield D, Lee B J, Hampton L, Diamond A M. Selenium induces changes in the selenocysteine tRNA[Ser]Sec population in mammalian cells. Nucleic Acids Res. 1991;19:939–943. doi: 10.1093/nar/19.4.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hill K E, Lyons P R, Burk R F. Differential regulation of rat liver selenoprotein mRNAs in selenium deficiency. Biochem Biophys Res Commun. 1992;185:260–263. doi: 10.1016/s0006-291x(05)80984-2. [DOI] [PubMed] [Google Scholar]
- 24.Ho S N, Hunt H D, Horton R M, Pullen J K, Pease L R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 25.Ho Y, Howard A J. Cloning and characterization of the glutathione peroxidase gene. FEBS Lett. 1992;301:5–9. doi: 10.1016/0014-5793(92)80198-p. [DOI] [PubMed] [Google Scholar]
- 26.Kelner M J, Bagnell R D, Uglik S F, Montoya M A, Mullenbach G T. Heterologous expression of selenium-dependent glutathione peroxidase affords cellular resistance to paraquat. Arch Biochem Biophys. 1995;323:40–46. doi: 10.1006/abbi.1995.0007. [DOI] [PubMed] [Google Scholar]
- 27.Kim I Y, Stadtman T C. Selenophosphate synthetase: detection in extracts of rat tissues by immunoblot assay and partial purification of the enzyme from the archaean Methanococcus vannielii. Proc Natl Acad Sci USA. 1995;92:7710–7713. doi: 10.1073/pnas.92.17.7710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lei X G, Evenson J K, Thompson K M, Sunde R A. Glutathione peroxidase and phospholipid hydroperoxide glutathione peroxidase are differentially regulated in rats by dietary selenium. J Nutr. 1995;125:1438–1446. doi: 10.1093/jn/125.6.1438. [DOI] [PubMed] [Google Scholar]
- 29.Lesson A, Mehta A, Singh R, Chisolm G M, Driscoll D M. An RNA-binding protein recognizes a mammalian selenocysteine insertion sequence element required for cotranslational incorporation of selenocysteine. Mol Cell Biol. 1997;17:1977–1985. doi: 10.1128/mcb.17.4.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Low S C, Berry M J. Knowing when not to stop: selenocysteine incorporation in eukaryotes. Trends Biol Sci. 1996;21:203–208. [PubMed] [Google Scholar]
- 31.Low S C, Harney J W, Berry M J. Cloning and functional characterization of human selenophosphate synthetase, an essential component of selenoprotein synthesis. J Biol Chem. 1995;270:21659–21664. doi: 10.1074/jbc.270.37.21659. [DOI] [PubMed] [Google Scholar]
- 32.Maquat L E. When cells stop making sense: effects of nonsense codons on RNA metabolism in vertebrate cells. RNA. 1995;1:453–465. [PMC free article] [PubMed] [Google Scholar]
- 33.Maquat L E. Defects in RNA splicing and the consequence of shortened translational reading frames. Am J Hum Genet. 1996;59:279–286. [PMC free article] [PubMed] [Google Scholar]
- 34.Martin G W, III, Harney J W, Berry M J. Selenocysteine incorporation in eukaryotes: insights into mechanism and efficiency from sequence, structure, and spacing proximity studies of the type 1 deiodinase SECIS element. RNA. 1996;2:171–182. [PMC free article] [PubMed] [Google Scholar]
- 35.Moriarty P M, Reddy C C, Maquat L E. The presence of an intron within the rat gene for selenium-dependent glutathione peroxidase 1 is not required to protect nuclear RNA from UGA-mediated decay. RNA. 1997;3:1369–1373. [PMC free article] [PubMed] [Google Scholar]
- 36.Paglia D E, Valentine W N. Studies in the quantitative and qualitative characterization of glutathione peroxidase. J Lab Clin Med. 1967;70:158–169. [PubMed] [Google Scholar]
- 37.Peltz S W, Feng H, Welch E, Jacobson A. Nonsense-mediated decay in yeast. Prog Nucleic Acid Res Mol Biol. 1994;47:271–298. doi: 10.1016/s0079-6603(08)60254-8. [DOI] [PubMed] [Google Scholar]
- 38.Reddy A P, Hsu B L, Reddy P S, Li N Q, Kedam T, Reddy C C, Tam M F, Tu C P D. Expression of glutathione peroxidase 1 gene in selenium-deficient rats. Nucleic Acids Res. 1988;16:5561–5568. doi: 10.1093/nar/16.12.5557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reddy A P, Hsu B L, Reddy P S, Li N Q, Thyagaraju K, Reddy C C, Tam M F, Tu C P D. Expression of glutathione peroxidase 1 gene in selenium-deficient rats. Nucleic Acids Res. 1988;16:5557–5568. doi: 10.1093/nar/16.12.5557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reddy C C, Thomas C E, Scholz R W. Xenobiotic metabolism: nutritional effects. ACS Symp Ser. 1985;277:253–265. [Google Scholar]
- 41.Reddy C C, Tu C P, Burgess J R, Ho C Y, Scholz R W, Massaro E J. Evidence for the occurrence of selenium-independent glutathione peroxidase activity in rat liver microsomes. Biochem Biophys Res Commun. 1981;101:970–978. doi: 10.1016/0006-291x(81)91844-1. [DOI] [PubMed] [Google Scholar]
- 42.Ruiz-Echevarria M J, Peltz S W. Utilizing the GCN4 leader region to investigate the role of the sequence determinants in nonsense-mediated mRNA decay. EMBO J. 1996;15:2810–2819. [PMC free article] [PubMed] [Google Scholar]
- 43.Saedi M, Smith C G, Frampton J, Chambers I, Harrison P R, Sunde R A. Effect of selenium status on mRNA levels for glutathione peroxidase in rat liver. Biochem Biophys Res Commun. 1988;53:855–861. doi: 10.1016/s0006-291x(88)81174-4. [DOI] [PubMed] [Google Scholar]
- 44.Seglen P O. Preparation of rat liver cells. III. Enzymatic requirements for tissue dispersion. Exp Cell Res. 1973;82:391–398. doi: 10.1016/0014-4827(73)90357-1. [DOI] [PubMed] [Google Scholar]
- 45.Toyoda H, Himeno S-I, Imura N. The regulation of glutathione peroxidase gene expression relevant to species difference and the effects of dietary selenium manipulation. Biochim Biophys Acta. 1989;1008:301–308. doi: 10.1016/0167-4781(89)90020-1. [DOI] [PubMed] [Google Scholar]
- 46.Toyoda H, Himeno S-I, Imura N. Regulation of glutathione peroxidase mRNA level by dietary selenium manipulation. Biochim Biophys Acta. 1990;1049:213–215. doi: 10.1016/0167-4781(90)90042-z. [DOI] [PubMed] [Google Scholar]
- 47.Vendeland S C, Beilstein M A, Yeh J Y, Ream W, Whanger P D. Rat skeletal muscle selenoprotein W: cDNA clone and mRNA modulation by dietary selenium. Proc Natl Acad Sci USA. 1995;92:8749–8753. doi: 10.1073/pnas.92.19.8749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weiss S L, Evenson J K, Thompson K M, Sunde R A. The selenium requirement for glutathione peroxidase mRNA level is half of the selenium requirement for glutathione peroxidase activity in female rats. J Nutr. 1996;126:2260–2267. doi: 10.1093/jn/126.9.2260. [DOI] [PubMed] [Google Scholar]
- 49.Weiss S L, Sunde R A. Selenium regulation of classical glutathione peroxidase expression requires the 3′ untranslated region in Chinese hamster ovary cells. J Nutr. 1997;127:1304–1310. doi: 10.1093/jn/127.7.1304. [DOI] [PubMed] [Google Scholar]
- 50.Wigler M, Sweet R, Sim G K, Wold B, Pellicer A, Lacy E, Maniatis T, Silverstein S, Axel R. Transformation of mammalian cells with genes from procaryotes and eucaryotes. Cell. 1979;16:777–785. doi: 10.1016/0092-8674(79)90093-x. [DOI] [PubMed] [Google Scholar]
- 51.Zhang J, Maquat L E. Evidence that the decay of nucleus-associated nonsense mRNA for human triosephosphate isomerase involves nonsense codon recognition after splicing. RNA. 1996;2:235–243. [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang, J., X. Sun, Y. Qian, and L. E. Maquat. At least one intron is required for nonsense-mediated decay of triosephosphate isomerase mRNA: a possible link between nuclear splicing and cytoplasmic translation. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 53.Zhang, J., X. Sun, Y. Qian, and L. E. Maquat. Intron function in the nonsense-mediated decay of β-globin mRNA: indications that nuclear splicing can influence cytoplasmic translation. Submitted for publication.