Abstract
The Hippo signaling is instrumental in regulating organ size, regeneration, and carcinogenesis. The cytoskeleton emerges as a primary Hippo signaling modulator. Its structural alterations in response to environmental and intrinsic stimuli control Hippo signaling pathway activity. However, the precise mechanisms underlying the cytoskeleton regulation of Hippo signaling are not fully understood. RAP2 GTPase is known to mediate the mechanoresponses of Hippo signaling via activating the core Hippo kinases LATS1/2 through MAP4Ks and MST1/2. Here we show the pivotal role of the reciprocal regulation between RAP2 GTPase and the cytoskeleton in Hippo signaling. RAP2 deletion undermines the responses of the Hippo pathway to external cues tied to RhoA GTPase inhibition and actin cytoskeleton remodeling, such as energy stress and serum deprivation. Notably, RhoA inhibitors and actin disruptors fail to activate LATS1/2 effectively in RAP2-deficient cells. RNA sequencing highlighted differential regulation of both actin and microtubule networks by RAP2 gene deletion. Consistently, Taxol, a microtubule-stabilizing agent, was less effective in activating LATS1/2 and inhibiting cell growth in RAP2 and MAP4K4/6/7 knockout cells. In summary, our findings position RAP2 as a central integrator of cytoskeletal signals for Hippo signaling, which offers new avenues for understanding Hippo regulation and therapeutic interventions in Hippo-impaired cancers.
Keywords: Hippo signaling, RAP2 GTPase, cytoskeleton, MAP4K, mechanotransduction, energy stress
The Hippo signaling pathway has consistently emerged as a central player in the orchestration of organ size, tissue regeneration, and carcinogenesis (1, 2). Its core comprises a kinase signaling pathway including MST1/2-MAP4K and LATS1/2, which have the critical function of phosphorylating and thereby inactivating YAP and TAZ, two transcriptional co-factors vital for transcribing genes for cell proliferation, morphogenesis, and mobility (3). These phosphorylation events play an instrumental role in maintaining cellular homeostasis and ensuring regulated tissue growth and development.
Recent advancements in the field have highlighted the significance of the cellular cytoskeleton as a modulator of the Hippo pathway (4, 5, 6, 7, 8, 9). Structural and mechanical alterations in the cytoskeleton, triggered by a range of environmental and intrinsic stimuli, have been proposed to directly and profoundly tune the core kinases of the Hippo signaling pathway, specifically the activity of LATS1/2 (10). Such changes can ripple through downstream processes, culminating in modifications to cell proliferation, apoptosis, and other vital cellular functions. However, despite these insights, the detailed mechanisms through which the actin and tubulin cytoskeletons regulate the Hippo pathway remain to be fully elucidated.
RAP2 GTPase, as recent investigations have established, is intricately tied to the mechanoresponses of Hippo signaling (11, 12). It exerts influence by activating the MAP4Ks and MST1/2 kinases while concurrently inhibiting the RhoA GTPase. These findings have spurred further interest in the precise role and regulation of RAP2 GTPase, especially considering its documented bidirectional relationship with the cytoskeleton (13, 14, 15, 16, 17) and its potential implications on Hippo pathway regulation. Several studies have documented the physical binding of the actin cytoskeleton to the RAP2 GTPase (13, 17), underscoring the importance of this interplay. Moreover, there is growing evidence suggesting that the actin cytoskeleton might wield influence over RAP2 activity, and conversely, RAP2 GTPase could modulate actin dynamics. This reciprocal regulation has far-reaching implications, especially in the context of how stress and biochemical cues regulate the Hippo pathway.
Our current study probes the intricate dynamics between RAP2 GTPase and the cytoskeleton in the contexts of stress and biochemical cues, seeking to shed light on the complex interplay between Hippo signaling and the cytoskeleton. We aim to provide a comprehensive understanding that could pave the way for therapeutic interventions in cancer malignancies and other diseases characterized by Hippo pathway disruptions.
Results
RAP2 mediates Hippo regulation by environmental signals and stresses
We previously reported that RAP2 GTPase mediates the Hippo pathway regulation by matrix stiffness by two downstream effectors, MAP4Ks and ARHGAP29 (11). While the former is a direct kinase of LATS1/2, the latter can activate the signaling pathway of MAP4K-MST1/2 and LATS1/2 through RhoA inhibition and actin depolymerization. Therefore, we speculated that RAP2 GTPases, including RAP2A, RAP2B, and RAP2C, may be also involved in the Hippo pathway regulation by other environmental biochemical signals and stresses beyond mechanical cues. Therefore, we challenged RAP2A/B/C triple knockout (RAP2A/B/C-tKO) cells and parental cells with various signaling activators or stress inducers.
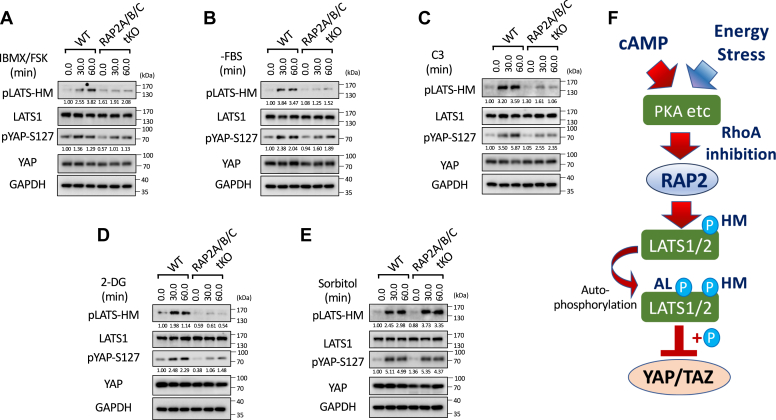
G protein-coupled receptors (GPCRs) modulate the Hippo signaling pathway through the interactions of their downstream Gα proteins with the actin cytoskeleton and Rho GTPases (7, 18). Upon activation by specific ligands, GPCRs, depending on the Gα protein they couple to, promote polymerization or depolymerization of F-actin and subsequently tune down or up the activity of LATS1/2, respectively. Gαs-coupled GPCRs activate LATS1/2 kinases through the actin depolymerization by the cyclic AMP (cAMP)-PKA signaling, which was also observed in this study where the co-application of forskolin (FSK, an adenylyl cyclase activator elevating cAMP) and 3-Isobutyl-l-methylxanthine (IBMX, a phosphodiesterase inhibitor preventing cAMP breakdown) quickly increase phosphorylation of LATS at its hydrophobic motif (HM) by about three folds and subsequent phosphorylation of YAP at Serine 127 by ∼30%, a classical residue for LATS phosphorylation of YAP (Figs. 1A and S1A). In contrast, the increases in the phosphorylation of LATS and YAP were substantially reduced in the RAP2A/B/C-tKO cells.
Figure 1.
RAP2 mediates Hippo regulation by environmental signals and stresses.A, Wild-type (WT) and RAP2A/B/C triple knockout (tKO) cells were treated with cAMP inducers IBMX and FSK for 30 and 60 min. Then the cell lysates were collected for analyses of LATS phosphorylation at hydrophobic motif (HM) and YAP phosphorylation at Serine 127. The results of the Western blot analysis are quantified and displayed beneath each corresponding band. B, WT and RAP2A/B/C tKO cells were serum starved, by replacing 10% FBS DMEM with serum-free DMEM (-FBS), for 30 and 60 min. C, WT and RAP2A/B/C tKO cells were treated with the Rho inhibitor C3 for 30 and 60 min for analyses of the role of RAP2 in Hippo regulation by Rho GTPase. D, WT and RAP2A/B/C tKO cells were treated with an energy stress-inducing agent 2-deoxy-D-glucose (2-DG) for 30 and 60 min. E, WT and RAP2A/B/C tKO cells were treated with an energy stress-inducing agent sorbitol for 30 and 60 min. F, a diagram of a working model illustrating the role of RAP2 GTPase in the Hippo signaling regulation by environmental signals and stress.
The LATS phosphorylation and activation caused by increased cAMP is mainly attributed to RhoA inhibition and actin depolymerization mediated by PKA. On the other hand, the function of RAP2 GTPase is well-known to be regulated by direct actin binding. Considering the critical role of RhoA in Hippo signaling regulation documented in the previous literature, we first asked whether the RhoA inhibition acts through RAP2 GTPase to trigger LATS activation and YAP phosphorylation. Serum starvation has been reported to activate LATS by inactivating RhoA GTPase. Phosphorylation of YAP at Serine 127 induced by serum starvation was substantially compromised in the RAP2A/B/C tKO cells (∼2 folds in the wild-type cells versus 1.1–1.5 folds in the RAP2A/B/C-tKO cells). Consistently, the phosphorylation of the LATS hydrophobic motif by serum starvation was strongly reduced by RAP2 deletion (∼3.5 folds in the wild-type cells versus 1.2–1.5 folds in the RAP2A/B/C-tKO cells) (Figs. 1B and S1B). The phosphorylation of LATS and YAP triggered by the treatment of the RhoA inhibitor Exoenzyme C3 was also compromised in the RAP2A/B/C tKO cells (as quantified in Figs. 1C and S1C), indicating that RAP2 GTPase could affect the phosphorylation of LATS and YAP induced by RhoA inhibition.
We further examined the role of RAP2 GTPase in Hippo signaling regulation by energy stress (Figs. 1D and S1D) and osmotic stress (Figs. 1E and S1E), respectively (19, 20, 21), to the RAP2A/B/C tKO and parental cells. The phosphorylation of LATS and YAP resulting from 2-deoxy-D-Glucose (2-DG) was greatly diminished in the RAP2A/B/C tKO cells, suggesting a prominent role of RAP2 GTPase in the regulation of Hippo signaling by energy stress (Figs. 1D and S1D). This result is consistent with a previous study showing that PKA knockout cells display weaker Hippo activation upon 2-DG-induced energy stress. In contrast, RAP2A/B/C tKO and parental cells responded similar to hyperosmotic stress caused by sorbitol treatment (Figs. 1E and S1E). It should be noted that the role of RhoA in the Hippo signaling regulation by hyperosmotic stress has not been directly defined. These results altogether indicated that RAP2 GTPase is selectively engaged in the Hippo pathway regulation by certain extracellular signals.
In summary, the abovementioned results clearly showed that RAP2 GTPase can mediate the Hippo signaling regulation by environmental signals (i.e., Gαs-coupled GPCR ligands and energy stress) other than matrix stiffness and other environmental cues (Fig. 1F). An interplay between RAP2 GTPase and RhoA GTPase was also implicated in the Hippo regulation in the contexts.
RAP2 is a required component of the MAP4K-mediated non-canonical Hippo signaling (Hpo2) to respond to environmental signals
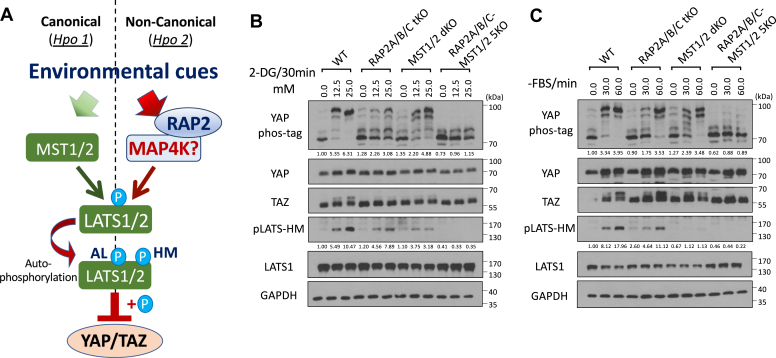
The MAP4K-mediated non-canonical Hippo signaling pathway is a molecular axis that diverges from the classical Hippo pathway mediated by MST1/2. The biological roles of this MAP4K-Hippo signaling pathway have recently been implicated in the mouse models (22, 23, 24), and key Hippo components (SAV1, NF2, and KIBRA) have thus been re-stratified (Hpo1 versus Hpo2) (22).
Our previous study demonstrated that RAP2 acts through MAP4Ks to activate LATS1/2 during mechanosensing (11), positioning RAP2 as a component of the non-canonical MAP4K-Hippo signaling pathway (Hpo2) (22) (Fig. 2A). To test this notion, we applied different doses of 2-DG to induce differential levels of energy stress in MST1/2 dKO and RAP2A/B/C-MST1/2 5KO cells (11) in addition to RAP2A/B/C tKO cells. Though RAP2A/B/C deletion alone could already block a large portion of YAP hyperphosphorylation (seen by YAP band shift in phos-tag gel analysis: ∼6-folds in the wild-type cells versus 2- to 3-folds in the RAP2A/B/C-tKO cells), TAZ hyperphosphorylation (seen by TAZ band shift in regular SDS-PAGE) and LATS hydrophobic motif phosphorylation, combined deletion of RAP2A/B/C and MST1/2 blocked the majority of YAP and LATS phosphorylation to energy stress (Figs. 2B and S2A), suggesting that RAP2 and MST1/2 work in parallel to activate LATS kinases in this context. The band shift of TAZ proteins, which indicates its hyperphosphorylation, also displayed a similar pattern.
Figure 2.
RAP2 is a required component of the MAP4K-mediated non-canonical Hippo signaling (Hpo2) to respond to environmental signals.A, a diagram illustrating the role of RAP2 in the canonical MST1/2-mediated (Hpo1) and the non-canonical MAP4K-mediated (Hpo2) signaling pathways. B, WT, RAP2A/B/C tKO, MST1/2 dKO, and RAP2A/B/C-MST1/2 5KO cells were treated with low dose (12.5 mM) and high dose (25.0 mM) of 2-DG. C, WT, RAP2A/B/C tKO, MST1/2 dKO, and RAP2A/B/C-MST1/2 5KO cells were serum-starved for 30 and 60 min.
Furthermore, we applied serum starvation to deplete RhoA activity in the same set of knockout cells and discovered that the phosphorylation of LATS, YAP, and TAZ (shown by its band shift) could be blocked by deletion of both RAP2A/B/C and MST1/2, but not either alone (Figs. 2C and S2B). This result demonstrated that RAP2A/B/C and MST1/2 function as parallel components in mediating LATS1/2 activation by serum starvation.
Altogether, environmental signals can act through both RAP2A/B/C and MST1/2, likely in parallel, to activate LATS1/2 and inactivate YAP/TAZ, though the results did not rule out the crosstalk between RAP2A/B/C and MST1/2 as previously reported in mechanotransduction (11).
The Hippo signaling regulation by actin cytoskeleton requires RAP2 GTPase
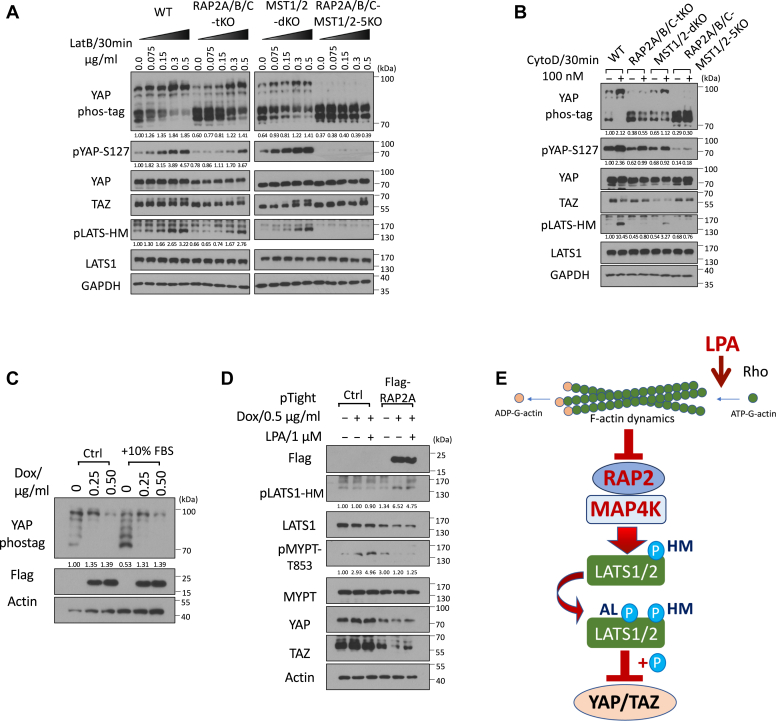
RhoA GTPase controls Hippo pathway activity through modulating actin polymerization and stress fiber formation (3). To understand whether RAP2 GTPase is required by Hippo activation by actin depolymerization resulting from RhoA inhibition, we applied Latrunculin B (LatB), which binds to actin monomers near the nucleotide-binding cleft and thus prevents actin polymerization and thus actomyosin contractile ability (25), to the set of RAP2A/B/C tKO, MST1/2 dKO, and RAP2A/B/C-MST1/2 5KO cells (Figs. 3A and S3A).
Figure 3.
The Hippo signaling regulation by the actin cytoskeleton requires RAP2 GTPase.A, WT, RAP2A/B/C tKO, MST1/2 dKO, and RAP2A/B/C-MST1/2 5KO cells were treated with a range of doses of Latrunculin B (LatB) for 30 min. B, WT, RAP2A/B/C tKO, MST1/2 dKO, and RAP2A/B/C-MST1/2 5KO cells were treated with 100 nM Cytochalasin D (CytoD) for 30 min. C, HEK293A cells transduced with pRetroX-Tet-on-advanced-neo viruses and selected by G418, and then were transduced with pRetroX-Tight-puro-Flag-RAP2A vectors and then selected by Puromycin. The subsequent stable cells were treated with 0.25 or 0.50 μg/ml Doxycycline (Dox) for 24 h in 10% FBS and then another 10% FBS was added to the cells. D, cells transduced with the inducible Flag-RAP2A viral vector and the control vector were treated with 0.50 μg/ml Dox for 24 h and then were treated with 1 μM Lysophosphatidic Acid (LPA) for 30 min. E, a working model of how actin cytoskeleton dynamics acts through RAP2 GTPase to control the Hippo pathway activity.
We observed dose-dependent effects of LatB on YAP phosphorylation in the WT, RAP2A/B/C tKO, and MST1/2 dKO cells, but the YAP phosphorylation of RAP2A/B/C tKO cells, detected by Phos-tag analysis and pYAP-S127 antibody, was reduced when LatB was used at a low concentration. More importantly, the deletion of both RAP2A/B/C and MST1/2 completely blocked the effect of LatB on YAP, TAZ, and LATS (Figs. 3A and S3A). We further confirmed the role of RAP2A/B/C in actin-mediated Hippo regulation by treating cells with Cytochalasin D (CytoD), which binds to the (+) end of F-actin to block the addition of new monomer actin subunits. We observed similar responses of the knockout cells to CytoD (Figs. 3B and S3B), though LatB and CytoD disrupt actin dynamics in different manners, further supporting the general role of RAP2 GTPase in mediating the Hippo pathway regulation by actin cytoskeleton-related effectors (26).
To determine whether RAP2 acts downstream of RhoA, we generated a RAP2A-inducible expression cell line (Fig. 3C). Induction of RAP2 expression by doxycycline (Dox) triggered YAP phosphorylation. An additional 10% FBS, which can activate RhoA GTPase, can decrease YAP phosphorylation in the cells only when RAP2 expression is not induced (as shown in the first versus fourth lane of the Phostag analysis) (Fig. 3C). We also applied a high dose of LPA (1 μM) to the inducible cell line, which showed that LPA failed to block LATS phosphorylation caused by RAP2A expression (Fig. 3D). Furthermore, phosphorylation of MYPT, which indicates actin stress fiber formation, was increased by LPA but strongly blocked by RAP2A induction. A similar expression change of TAZ, in response to LPA and RAP2 induction, was also observed.
Overall, these results established a signaling axis of external stimuli => Rho GTPase => actin remodeling => RAP2 GTPase => Hippo signaling pathway (Fig. 3E).
RAP2 and SAV1 work in parallel to promote YAP phosphorylation by LATS via MAP4K and MST1/2, respectively
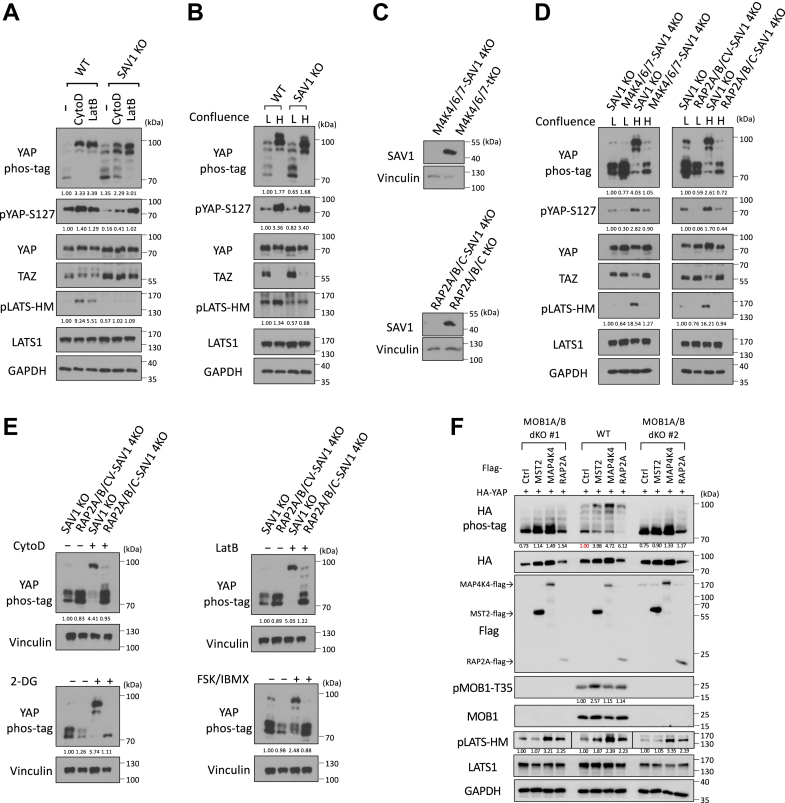
Salvador homolog 1 (SAV1) is an essential component of the canonical Hippo signaling pathway (Hpo1) mediated by MST1/2. SAV1 forms a complex with MST1/2 and thus promotes autophosphorylation of MST1/2. Recent advances have also shown that SAV1 functions as a scaffold protein that anchors MST1/2 at the plasma membrane for the subsequent phosphorylation and activation of LATS1/2, as well as attenuates MST1/2 inactivation by the STRIPAK-PP2A complex. Furthermore, we have found that the LATS phosphorylation by CytoD and LatB is blunted in the SAV1 KO cells (27), indicating that SAV1 is involved in cytoskeleton regulation of LATS1/2 (Fig. 4A). Consistently, SAV1 KO also displayed reduced LATS phosphorylation caused by high confluence (Fig. 4B), though YAP phosphorylation was less affected by SAV1 deletion.
Figure 4.
RAP2 and SAV1 work in parallel to promote YAP phosphorylation by LATS via MAP4K and MST1/2, respectively.A, WT and SAV1 KO cells were treated with 100 nM CytoD and 0.2 μg/ml LatB for 30 min. B, WT and SAV1 KO cells were grown at low and high (4.5× dense) confluence for 24 h and then collected for immunoblot analysis. C, Validating the deletion of SAV1 in MAP4K4/6/7 tKO and RAP2A/B/C tKO cells by CRISPR/Cas9. D, SAV1 KO, MAP4K4/6/7-SAV1 4KO, and RAP2A/B/C-SAV1 4KO cells were grown at low and high confluence. E, SAV1 KO and RAP2A/B/C-SAV1 4KO cells were treated with 100 nM CytoD, 0.2 μg/ml LatB, 2-DG or FSK/IBMX for 30 min. F, WT and MOB1A/B dKO cells were transfected with HA-tagged YAP and Flag-tagged pcDNA3.1 control/MST2/MAP4K4/RAP2A vectors for 24 h and then collected for immunoblot analysis. Note that HA phos-tag quantification is normalized to WT cells transfected with a control vector (red font), and pLATS-HM is from different blots (separated by lines).
To understand the potential interaction and cooperation of SAV1 and the RAP2-MAP4K non-canonical Hippo signaling, we deleted SAV1 in MAP4K4/6/7 tKO and RAP2A/B/C tKO cells to generate MAP4K4/6/7-SAV1 4KO and RAP2A/B/C-SAV1 4KO cells (Fig. 4C). These two 4KO cell lines showed significantly compromised phosphorylation of YAP and LATS caused by high (H) confluence when compared with low (L) confluence (Figs. 4D and S4A), mimicking the responses of MAP4K4/6/7-MST1/2 5KO and RAP2A/B/C-MST1/2 5KO that were generated and characterized previously.
Similarly, phosphorylation of YAP in MAP4K4/6/7-SAV1 4KO and RAP2A/B/C-SAV1 4KO cells cannot be stimulated efficiently by actin disassembly-inducers CytoD and LatB, energy stress-inducer 2-DG, or PKA-activators FSK/IBMX (Figs. 4E and S4B). These results suggested the non-redundant and parallel functions of SAV1 and RAP2A/B/C or MAP4K4/6/7 in mediating Hippo pathway regulation by external signals, while RAP2A/B/C requires MAP4K4/6/7, but not the MST1/2 and SAV1 complex, to activate LATS1/2.
MOB1 is another known key player in the Hippo pathway. Therefore, we determined whether RAP2, MAP4K, or MST1/2 depend on MOB1 to activate LATS1/2 and induce YAP/TAZ phosphorylation (Fig. 4F). We overexpressed RAP2, MAP4K, and MST2 in two independent MOB1A/B double knock-out (dKO) single clones (27) and then determined the phosphorylation status of MOB1, LATS1/2, and YAP/TAZ. Our results indicate that: (1) the canonical Hpo1 arm molecule MST2 phosphorylates MOB1 and requires MOB1 for the phosphorylation of LATS1/2, which is consistent with the consensus understanding; (2) the non-canonical Hpo2 arm molecules (RAP2 and MAP4K4) cannot phosphorylate MOB1 and do not require MOB1 for the phosphorylation of LATS1/2; and (3) MOB1 is essential for YAP regulation, as the knock-out of MOB1A and MOB1B genes nearly blocks YAP phosphorylation, as indicated by the Phos-tag gel analysis. This holds regardless of the ectopic overexpression of Hpo1 arm molecule MST2 or Hpo2 molecules RAP2 and MAP4K4.
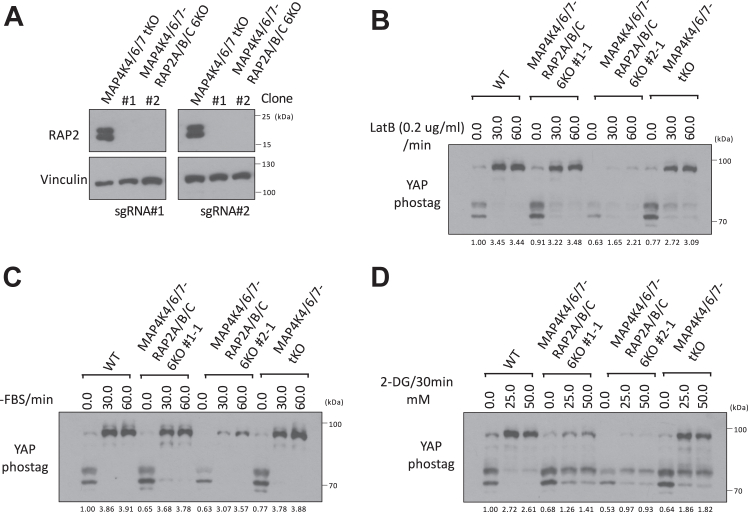
To further test whether RAP2 can play a role in the canonical MST1/2 Hippo pathway, we deleted RAP2A/B/C gene in MAP4K4/6/7 tKO to examine whether it can potentiate Hippo deficiency in MAP4K4/6/7 tKO cells (Fig. 5A). In fact, RAP2A/B/C-MAP4K4/6/7 6KO cells and MAP4K4/6/7 6KO cells showed similar YAP hyperphosphorylation upon the treatment of LatB and serum starvation (Fig. 5, B and C), though there was a different response to 2-DG-induced energy stress (Fig. 5D). Still, YAP hyperphosphorylation by 2-DG in RAP2A/B/C-MAP4K4/6/7 6KO cells cannot be blocked as much in RAP2A/B/C-MST1/2 5KO cells. These results altogether supported a working model where RAP2 functions upstream of MAP4K in the non-canonical Hippo (Hpo2) signaling (22) to mediate the phosphorylation of LATS and YAP triggered by actin disassembly and stress. The role of RAP2 in Non-Canonical (Hpo2) signaling is as crucial as the well-characterized role of SAV1 in Canonical (Hpo1) signaling.
Figure 5.
Combined deletion of RAP2 GTPase and MAP4Ks does not further potentiate Hippo deficiency compared with deletion of RAP2 GTPase or MAP4Ks alone.A, two different sets of sgRNAs targeting RAP2A, RAP2B, and RAP2C genes, as previously described in Ref. (11), were used to delete these three genes in MAP4K4/6/7 tKO cells. Two clones were expanded for further analysis. B–D, WT, MAP4K4/6/7 tKO, and RAP2A/B/C-MAP4K4/6/7 6KO cells were treated with LatB (B), serum starvation (C), and 2-DG (D) for 30 min.
Interplay between RAP2 GTPase and cytoskeleton dynamics
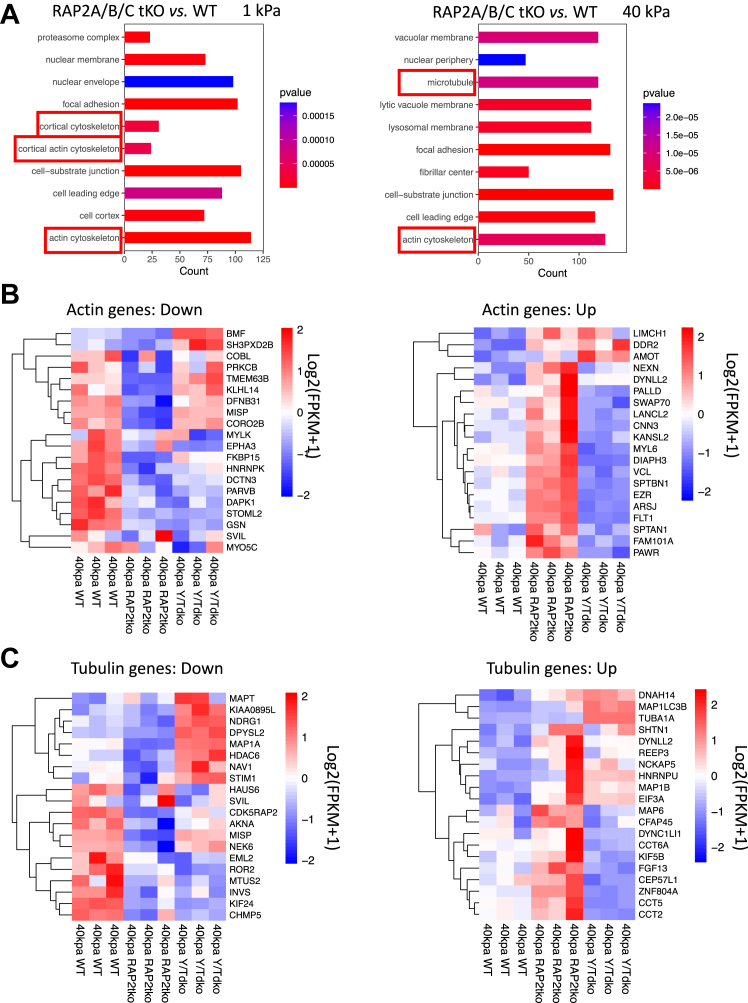
Our previous work proves that RAP2 GTPase, as an intracellular mechanotransducer, conveys extracellular matrix stiffness signals to gene transcription regulation through the Hippo pathway (11). We then re-analyzed our RNA sequencing of RAP2A/B/C tKO and WT cells cultured at low stiffness (1 kPa) and high stiffness (40 kPa) (GSE98547). Gene ontology analyses revealed the expression of many genes involved in the actin cytoskeleton had been altered by RAP2A/B/C gene deletion, particularly at low stiffness where RAP2 GTPase was more active. In addition, at high stiffness, microtubule genes were also strongly differentially expressed in RAP2A/B/C tKO cells (Fig. 6A).
Figure 6.
Differentially expressed actin and tubulin genes in RAP2A/B/C tKO cells.A, gene ontology for cellular components with the RNAseq analysis of RAP2A/B/C tKO cells versus WT cells on soft (1 kPa) and stiff (40 kPa) substrates. Cytoskeleton terms were enriched. B, down- and upregulated actin-related genes, in WT, RAP2A/B/C tKO, and YAP/TAZ dKO (Y/T dKO) were plotted with heat-map. C, down- and upregulated tubulin-related genes, in WT, RAP2A/B/C tKO, and Y/T dKO were plotted with heat-map.
Many altered actin genes in RAP2/B/C tKO cells were potentially YAP/TAZ-targeting genes, as YAP/TAZ dKO (Y/TdKO) cells showed oppositely regulated patterns when compared with RAP2/B/C tKO cells (Fig. 6B), especially those upregulated by RAP2A/B/C deletion at high stiffness. Many tubulin genes are also differentially regulated by RAP2 and YAP/TAZ (Fig. 6C), including microtubule-associated proteins (e.g., MAP1A and MAP6) (28) and chaperonins for tubulin and actin protein folding (e.g., CCT2, CCT5, and CCT6A) (29).
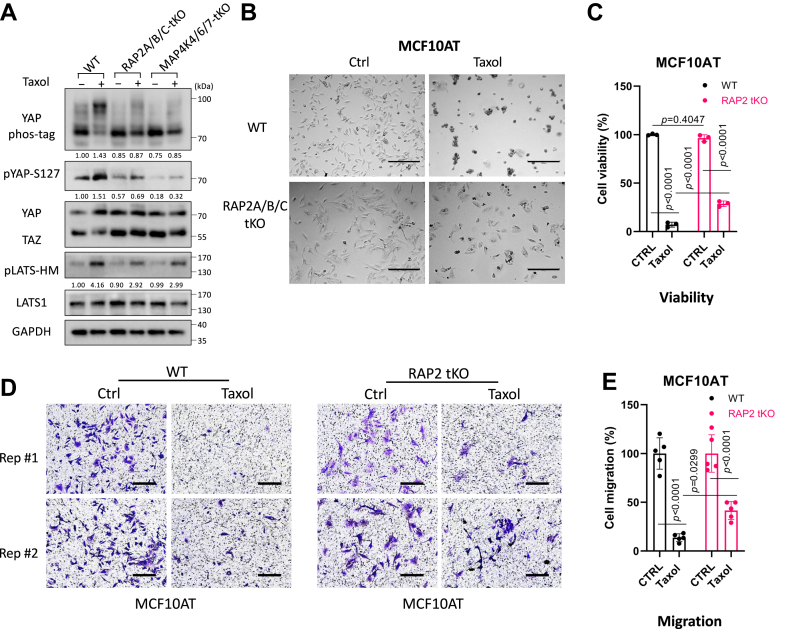
As RAP2 appears to regulate tubulin dynamics in the cells, we asked whether RAP2 is involved in the Hippo pathway regulation by tubulin dynamics. Stabilization of tubulin by Taxol increases phosphorylation of YAP and LATS in the wild-type cells by ∼50% (Figs. 7A and S4C), which is consistent with a previous observation that nocodazole, a tubulin-disrupting agent, decreases phosphorylation of YAP and LATS (8). Interestingly, the effect of Taxol on YAP and LATS is reduced to a low level (2–10% based on the Phos-tag analysis) in RAP2A/B/C tKO and MAP4K4/6/7 tKO cells, suggesting that the RAP2-MAP4K signaling also plays a role in Hippo pathway regulation by tubulin dynamics. As Taxol has been often used as a first-line chemotherapy drug (30) and YAP is known to confer chemoresistance, we compared the responses of RAP2A/B/C tKO MCF10A cells that were transformed by the H-Ras V12 proto-oncogene (MCF10AT) (11) (Fig. 7B). The deletion of RAP2A/B/C substantially reduced the growth inhibition of Taxol on MCF10AT cells (Fig. 7C). Furthermore, although taxol strongly inhibits the migration of MCF10AT cells, this inhibition was attenuated in RAP2A/B/C tKO MCF10AT cells (11) (Fig. 7, D and E). While a previous study implicated that MAP4K mediates the crosstalk of F-actin and microtubule (28), our study hereby suggested that the RAP2-MAP4K non-canonical Hippo signaling and the tubulin/actin cytoskeleton mutually regulate each other to control cell growth and mobility.
Figure 7.
RAP2 GTPase is involved in Hippo signaling regulation by tubulin cytoskeleton dynamics.A, WT, RAP2A/B/C tKO, and MAP4K4/6/7 tKO HEK293A cells were treated with Taxol (3 μM) for 120 min. B and C, Representative bright filed images (B) and cell viability MTS test (C) showed cell survival of WT and RAP2A/B/C tKO MCF10AT cells treated with 50 nM of Taxol for 24 h. For the MTS assay, a Two-way ANOVA and Tukey's multiple comparison test was applied. Scale bar, 300 μm. The data are reported as mean ± SD. D, transwell migration assay on WT and RAP2A/B/C tKO MCF10AT cells treated with 50 nM of Taxol for 24 h. Scale bar, 100 μm. E, quantification of cell migration assay. The data are reported as mean ± SD. Two-way ANOVA and Tukey's multiple comparison tests were used.
Discussion
RAP2 is an essential component of the non-canonical Hippo signaling pathway for sensing environmental and intrinsic cues
Understanding the mechanism by which organs perceive their size and regulate growth has been a major challenge in cellular biology. Central to this conundrum is the Hippo signaling pathway, an evolutionarily conserved network governing organ size, tissue regeneration, and tumorigenesis. In our study, we elucidate a novel aspect of this regulation. This study, together with previous studies (11, 12), has implicated a crucial role of RAP2 GTPases in the Hippo pathway regulation by environmental signals, particularly those cytoskeleton-remodeling signals.
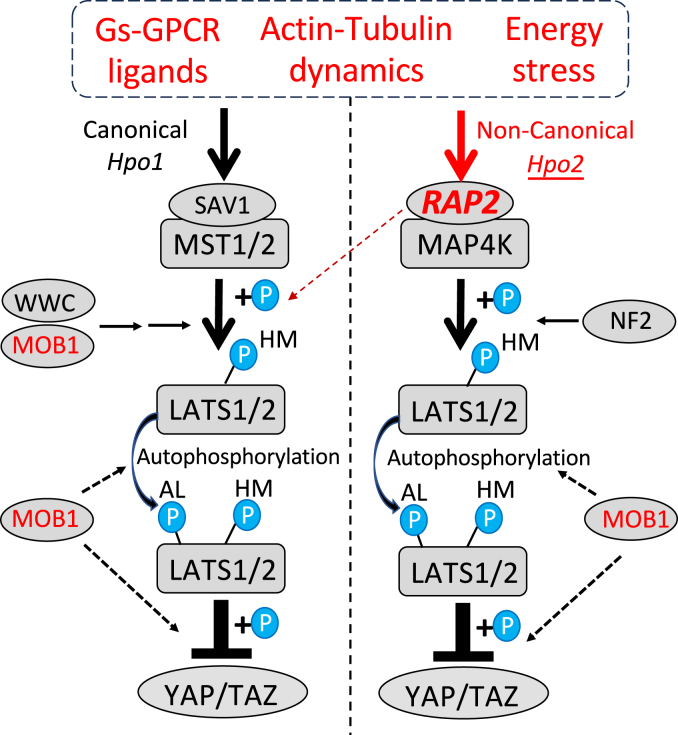
RAP2, much like NF2 and WWC1-3 (22), appears pivotal in facilitating and modulating kinase interactions within the Hippo pathway (Fig. 8). Specifically, our results showcase the capability of RAP2 to interface with LATS1/2 and upstream kinases such as MST1/2 and MAP4K1-7, providing a functional nexus for signal transduction.
Figure 8.
A diagram summarizing new findings in the RAP2-mediated Hippo signaling. RAP2 acts upstream of MAP4K in the Non-Canonical Hippo pathway (Hpo2). The dotted line indicates RAP2 may activate MST1/2 independent of MAP4K in a context-dependent manner. MST1/2 requires MOB1 to phosphorylate LATS1/2 at their hydrophobic motif (HM) but MAP4Ks do not require MOB1 to phosphorylate LATS1/2-HM. However, MOB1 is required for YAP/TAZ phosphorylation by LATS1/2 in both Hpo1 and Hpo2 signaling.
Interestingly, our data seem to place RAP2 in an intricate regulatory network with WWC1-3 and NF2. The latter two proteins have recently been posited as “loading docks” for LATS1/2, integrating upstream signals (22). Drawing from our results, RAP2 might also function as a dynamic regulator, and RAP2 and SAV1 work synergistically to enhance the activation of Hpo2 (non-canonical, MAP4K-mediated) and Hpo1 (canonical, MST1/2-mediated) signaling, respectively. In addition, as RAP2 also regulates actin dynamics, our findings have not fully excluded the role of RAP2 in Hpo1 activation. For instance, in the energy stress context, the Hippo regulation by RAP2 likely engaged RAP2 downstream regulators other than MAP4Ks (Figs. 5D and 8). This observation is consistent with the previous report that RAP2 potentially activates MST1/2 through activating ARHGAP29 and subsequent actin depolymerization (11).
The roles of classical Hippo components SAV1 and MOB1 in the RAP2-mediated LATS activation
In the classical model of the Hippo signaling, SAV1 and MOB1 are considered core components of the Hippo signaling pathway (2, 10), where SAV1 and MOB1 serve as scaffold proteins that assist MST1/2 in the recruitment of LATS1/2 and the subsequent phosphorylation of LATS1/2 at their hydrophobic motif, thus activating LATS1/2.
SAV1 is involved in multiple steps of Hpo1 activation. For instance, SAV1 blocks the repression of MST1/2 by STRIPAK (31, 32), promotes dimerization of MST1/2 to initiate MST1/2 autophosphorylation activation (33, 34), and incorporates multiple upstream signals (e.g., cell-cell contact) and interacts with regulators (e.g., STK25, PP2A with B’ specificity subunits, APC) enhance MST1/2 activities (32, 35, 36, 37, 38, 39). In addition, SAV1 is also a substrate of MST1/2 and its phosphorylation by MST1/2 can be essential for MST1/2 to subsequently phosphorylate LATS1/2. However, it is relatively clear that SAV1 has a little-to-no role in the RAP2-mediated Hpo2 signaling, though SAV1 may be required for the MST1/2-LATS1/2 kinase cascade activation by RAP2/ARHGAP39-mediated actin remodeling (11) (Fig. 8). RAP2 functions towards MAP4K rather differently from SAV1 toward MST1/2. RAP2 mainly regulates MAP4K activity in response to environmental cues at the initiation stage of Hpo2 activation (11).
In contrast, our study suggested more complicated roles for MOB1 across Hpo1 and Hpo2 signaling. MOB1 was initially recognized to bind to LATS1/2 and target LATS1/2 to the plasma membrane, where MST1/2 phosphorylates the hydrophobic motif of LATS1/2 (40). The following studies suggested multiple roles for MOB1 in LATS1/2 phosphorylation and activation in the canonical MST1/2-mediated Hippo signaling (Hpo1) (41, 42, 43, 44). As discussed previously (3, 44), MOB1 binds to autophosphorylated (activated) MST1/2. This binding relieves MOB1 from its autoinhibitory conformation and thus allows MOB1 to bind to LATS1/2. The subsequently formed MST1/2-MOB1-LATS1/2 ternary complex enhances the phosphorylation of the LATS1/2 hydrophobic motif by MST1/2. Furthermore, the binding of MOB1 to LATS1/2 allosterically promotes autophosphorylation at the LATS activation loop, which is required for the full activation of LATS after its hydrophobic motif phosphorylation (3, 41, 44).
In our study with MOB1A/1B dKO cells, we observed that MOB1 is required for MST2-mediated but not for RAP2/MAP4K4-mediated phosphorylation of LATS-HM (Fig. 4F), which is consistent with previous findings (41, 42, 43, 44). In contrast, despite the increased LATS-HM phosphorylation by RAP2/MAP4K4 overexpression in the MOB1A/1B dKO cells, we observed limited YAP phosphorylation (less than two folds as quantified by the Phostag analysis) when Hpo2 is activated in these KO cells. Two potential mechanisms can account for this observation. One is that MOB1 is also required for full LATS activation after MAP4K4-mediated LATS-HM phosphorylation. The other is that MOB1 may still be required for YAP phosphorylation by a fully activated LATS. Future biochemical characterizations will be needed to fully define the role(s) of MOB1 in Hpo2 signaling. Given that the regulation of Hpo1 signaling is highly dependent on its spatial context, conducting imaging analyses of Hpo2 signaling can also be essential for elucidating additional mechanisms of its activation. For example, it is critical to observe where in the cell MAP4K4 is activated by RAP2, as well as where MAP4K4 phosphorylates LATS hydrophobic motif to initiate the downstream signaling cascades.
Interplay of RAP2 GTPase and the cytoskeleton in Hippo pathway regulation
A major revelation from our study lies in understanding the intricate relationship between RAP2 GTPase and the cytoskeleton. It is well established that the cytoskeleton, with its vast network of filaments and tubules, orchestrates a myriad of cellular processes, including signaling pathways such as the Hippo pathway (3, 10). Our data suggest that RAP2, beyond its role in the Hippo pathway, might be integral in shaping cytoskeletal dynamics.
Previous studies have implicated the physical regulation of RAP2 GTPase by actin cytoskeleton (13, 14, 15, 16, 17). Given the pivotal role of the cytoskeleton in mechanotransduction, the potential for RAP2 to act as a mechanosensor becomes an exciting avenue for future exploration.
Our findings highlight instances where RAP2, likely in its active form based on the previous study, modulates Hippo pathways in response to actin and tubulin dynamics. In the future, more characterization will be needed to determine the impact of RAP2 GTPase in the formation of stress fibers formation and alteration in cellular stiffness. Moreover, the migration patterns of these cells were notably different, suggesting a role for RAP2 in cell motility, possibly through cytoskeletal rearrangements. Given its newfound association with the cytoskeleton and the previous finding of RAP2’s role in sensing matrix stiffness, RAP2 can play a dual role: as a modulator of the Hippo pathway and a mediator of cellular responses to mechanical stimuli.
Furthermore, we noted potential functional interplays between RAP2 and known cytoskeleton-associated proteins from the RNA sequencing (Fig. 6). Such interplays might be pivotal for cellular processes like adhesion, spreading, and even differentiation. It also raises the possibility of a feedback loop where RAP2-induced changes in the cytoskeleton could further influence Hippo pathway dynamics independent of its downstream effector MAP4K. To date, the detailed molecular mechanisms by which the Hippo pathway is regulated by the cytoskeleton still need to be defined. Identification of cytoskeleton-associated proteins connected to RAP2-mediated Hippo pathway regulation, which are missing in our current working models (Fig. 8), will help fill this knowledge gap in the molecular mechanisms.
In summary, the interplay of Hippo signaling, RAP2 GTPase, and the cytoskeleton presents potential implications in diseases, especially those characterized by cytoskeletal abnormalities or altered mechanotransduction. Unraveling the exact mechanisms and interactions would not only provide a clearer picture of cellular signaling but also pave the way for therapeutic interventions targeting this interface.
Experimental procedures
Cell culture and treatment
HEK293A cells, a gift from Dr Kun-Liang Guan’s group at UC San Diego, were cultured in DMEM supplemented with 10% fetal bovine serum. MCF10A cells were originally obtained from ATCC and cultured in DMEM/F12 supplemented with 5% horse serum, 20 ng/ml of human EGF, 0.5 mg/ml of hydrocortisone, 100 ng/ml of cholera toxin and 10 μg/ml of Insulin. The H-Ras V12 proto-oncogene transformed MCF10A cell line MCF10AT, a gift from Dr Kun-Liang Guan, was also maintained in the above medium. For conditions of low cell density, we seeded 1.5 × 105 cells in each well of six-well plates. For a denser cell population, we either seeded 6.0 × 105 or 8 × 105 cells in each well. For other treatments, cells at low density were exposed to 2-deoxy-D-Glucose (2-DG) at a concentration of 12.5 or 25 mM, Latrunculin B (LatB) at 0.075 to 0.5 μg/ml, Exoenzyme C3 at 1 μg/ml, Cytochalasin D (CytoD) at 100 nM, Lysophosphatidic acid (LPA) at 1 μM, Sorbitol at 200 mM, Isobutylmethylxanthine, 1-Methyl-3-Isobutylxanthine/Forskolin (IBMX/FSK) at 100 and 10 μM, respectively, and Paclitaxel (Taxol) at 3 μM.
For transient gene overexpression, 1.0 × 105 HEK293A cells were seeded to in each well of 6-well plate. 10 ng of HA-tagged YAP and 500 of Flag-tagged MST2/MAP4K4/RAP2A plasmids were co-transfected into cells using polyJet transfection reagent, following the manufacturer’s protocol. After 24 h of transfection, cells were harvested for further analysis.
To induce gene expression via doxycycline, we used the pRetroX-Tet-on-advanced and pRetrox-Tight-Puro plasmids from Clontech Inc. Stable HEK293A cell lines were developed following the guidelines provided by the manufacturer. To trigger gene expression of RAP2A, we administered 500 ng/ml of Doxycycline for at least 24 h. MCF10AT cells were exposed to 50 nM of Taxol for cell viability and migration assays.
Generation of the Hippo component knockout cell lines
The CRISPR gene-editing method was employed to remove targeted genes. We cloned the guide RNA sequences into the px459 plasmids (Addgene 48319), kindly provided by Dr Feng Zhang. These modified plasmids were then introduced into either HEK293A or MCF10AT cells. A day post-transfection, cells that had been transfected were enriched using 1 μg/ml puromycin for 3 days. Following this, the cells were distributed into 96-well plates, ensuring just a single cell per well. The resulting clones were assessed via Western blot using gene-specific antibodies. We ensured that at least two distinct clones were employed for each gene deletion in the subsequent experiments. The guide RNA sequences for SAV1 and RAP2 from the previous studies were used (11, 27). All the information on single clones is listed as below:
RAP2 A/B/C tKO MCF10AT single clones, RAP2 A/B/C tKO HEK293A single clones, and RAP2 A/B/C MST1/2 5KO HEK293A single clones were previously established (11). Two individual knockout clones, generated with two different sgRNAs for the same target gene, were used where it was applicable to rule out potential “off-target” effects. MST1/2 KO, SAV1 KO, and MAP4K4/6/7 tKO HEK293A single clones were previously established, and the rescue of the MAP4K-KO cells with a MAP4K4 plasmid was also previously performed and described (11, 27). RAP2 A/B/C SAV1 4KO single clone was established from RAP2 A/B/C tKO HEK293A single clone #1 described in (11). MAP4K4/6/7 SAV1 4KO single clone was established from MAP4K4/6/7 tKO single clone. MAP4K4/6/7 RAP2 A/B/C 6KO was established from MAP4K4/6/7 tKO single clone. The KO efficiency, including SAV1 and RAP2, of these single clones was confirmed by Western blot as shown in corresponding figures.
Biochemical assays
Western blot was performed following standard methods, mostly with 9 to 12% SDS PAGE gels. We applied the Phos-tag SDS-PAGE to detect YAP protein phosphorylation. This technique is based on the fact that Mn2+–Phos-tag molecules slows down the migration of proteins (reflected by multiple bands in the same lane) in a phosphorylation-dependent manner (45). In the present work, 7.5% phos-tag gel was used to resolve the phospho-YAP proteins, and the resolved proteins were then transferred to PVDF membranes and probed with anti-YAP antibodies. The information on the antibodies is provided in Table S1. ImageJ software was used to quantify the regular and Phos-tag immunoblots. For the Phos-tag analysis, a weighted-average mean value was calculated for each lane. Specifically, 7.5% Phos-tag SDS-PAGE results in five distinct bands for YAP proteins, with the band distribution from 70 kDa (non-phosphorylated) to 100 kDa (fully phosphorylated). The weights of the five bands from 70 kDa to 100 kDa are 0%, 25%, 50%, 75%, and 100%, respectively. The total amount of phosphorylation is determined by multiplying the grayscale values of the bands by their corresponding weights. The final value of the YAP phosphorylation is then obtained by dividing the total weighted amount of phosphorylated bands by the arithmetic sum of the grayscale values of all five bands. The results of the Western blot analysis are quantified and displayed beneath each corresponding band. Antibodies were validated by comparing wild-type cells and corresponding gene-specific knockout cells.
Cell viability and migration assay
To test the impact of taxol on the cell viability of MCF10AT cells, cells were seeded in 96-well plates at a density of 2.0 × 103 per well. After 24 h, cells were treated with 50 nM of taxol for another 24 h and tested. Cell viability was assessed by MTS using CellTiter 96 Aqueous One Solution Cell Proliferation Assay (Promega). For the transwell migration study, 5.0 × 104 cells were seeded in 12-well transwell inserts with 8 μm pore size. After 24 h, the medium in the upper chamber was changed to 500 μl of no serum DMEM/F12, and 1 ml of complete MCF10AT culture medium was added into the lower chamber. Taxol was only added in the upper chamber. After 24 h, cells were fixed with 10% formalin and visualized with 0.1% crystal violet.
Bioinformatic analysis
The raw data from GSE98547 (11) was downloaded from the GEO database. Differential gene expression analysis was conducted using DESeq2 (46), with the criteria of log2FoldChange >0 and a p-value < 0.05 to identify genes that exhibited differential expression. Gene Ontology (GO) Cellular Component analysis (47) was carried out in R (version 4.3.1), and the top 10 enriched terms were visualized in a plot.
Statistical analysis
All data analyses were performed using GraphPad Prism 9.1.2. The data were analyzed by two-way ANOVA and Tukey's multiple comparison test. All experiments were repeated independently at least twice.
Data availability
All data are contained in the manuscript.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare no conflict of interest with the contents of this article.
Acknowledgments
Author contributions
C. W. and Z. M. conceptualization; C. W., X. C., Y. W., C. D. R., G. Z., L. H., C.-Y. H., X. W., and Z. M. investigation; X. C. formal analysis; C. W., R. O. L., and Z. M. visualization; V. R. S. and R. O. L. methodology; V. R. S. and Z. M., resources; C. W. and Z. M. writing-original draft; X. C., C. D. R., G. Z., C.-Y. H., X. W., V. R. S., R. O. L., and Z. M. writing-review & editing; Z. M. project administration; Z. M. supervision.
Funding and additional information
Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health (NIH) under award number R35GM142504 to Z. M. A NIH/NINDS award to R. O. L. (NINDS R01NS126501) exclusively contributed to the Methodology, Visualization, and Writing work by R. O. L.; R01NS126501 did not support Z. M. for his research reported in this paper. L. H. was supported in part by an NIH/NCI R25 CREATE Summer Undergraduate Research Fellowship (R25CA261632). V. R. S. is the recipient of the Department of Defense Concept Award (W81XWH2010549), Department of Defense Career Development Award (W81XWH2110377), and American Cancer Society’s Research Scholar Grant (RSG-22-071-01-TBE). C. W. was supported by an internal award for postdoctoral trainees from the Sylvester Comprehensive Cancer Center.
Reviewed by members of the JBC Editorial Board. Edited by Enrique De La Cruz
Supporting information
References
- 1.Franklin J.M., Wu Z., Guan K.L. Insights into recent findings and clinical application of YAP and TAZ in cancer. Nat. Rev. Cancer. 2023;23:512–525. doi: 10.1038/s41568-023-00579-1. [DOI] [PubMed] [Google Scholar]
- 2.Zheng Y., Pan D. The Hippo signaling pathway in development and disease. Dev. Cell. 2019;50:264–282. doi: 10.1016/j.devcel.2019.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meng Z., Moroishi T., Guan K.L. Mechanisms of Hippo pathway regulation. Genes Dev. 2016;30:1–17. doi: 10.1101/gad.274027.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rauskolb C., Sun S., Sun G., Pan Y., Irvine K.D. Cytoskeletal tension inhibits Hippo signaling through an Ajuba-Warts complex. Cell. 2014;158:143–156. doi: 10.1016/j.cell.2014.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aragona M., Panciera T., Manfrin A., Giulitti S., Michielin F., Elvassore N., et al. A mechanical checkpoint controls multicellular growth through YAP/TAZ regulation by actin-processing factors. Cell. 2013;154:1047–1059. doi: 10.1016/j.cell.2013.07.042. [DOI] [PubMed] [Google Scholar]
- 6.Dupont S., Morsut L., Aragona M., Enzo E., Giulitti S., Cordenonsi M., et al. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474:179–183. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- 7.Yu F.X., Zhao B., Panupinthu N., Jewell J.L., Lian I., Wang L.H., et al. Regulation of the Hippo-YAP pathway by G-protein-coupled receptor signaling. Cell. 2012;150:780–791. doi: 10.1016/j.cell.2012.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao B., Li L., Wang L., Wang C.Y., Yu J., Guan K.L. Cell detachment activates the Hippo pathway via cytoskeleton reorganization to induce anoikis. Genes Dev. 2012;26:54–68. doi: 10.1101/gad.173435.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deng H., Wang W., Yu J., Zheng Y., Qing Y., Pan D. Spectrin regulates Hippo signaling by modulating cortical actomyosin activity. Elife. 2015;4 doi: 10.7554/eLife.06567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu F.X., Zhao B., Guan K.L. Hippo pathway in organ size control, tissue homeostasis, and cancer. Cell. 2015;163:811–828. doi: 10.1016/j.cell.2015.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meng Z., Qiu Y., Lin K.C., Kumar A., Placone J.K., Fang C., et al. RAP2 mediates mechanoresponses of the Hippo pathway. Nature. 2018;560:655–660. doi: 10.1038/s41586-018-0444-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamashiro Y., Thang B.Q., Ramirez K., Shin S.J., Kohata T., Ohata S., et al. Matrix mechanotransduction mediated by thrombospondin-1/integrin/YAP in the vascular remodeling. Proc. Natl. Acad. Sci. U. S. A. 2020;117:9896–9905. doi: 10.1073/pnas.1919702117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duncan E.D., Han K.J., Trout M.A., Prekeris R. Ubiquitylation by Rab40b/Cul5 regulates Rap2 localization and activity during cell migration. J. Cell Biol. 2022;221 doi: 10.1083/jcb.202107114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Machida N., Umikawa M., Takei K., Sakima N., Myagmar B.E., Taira K., et al. Mitogen-activated protein kinase kinase kinase kinase 4 as a putative effector of Rap2 to activate the c-Jun N-terminal kinase. J. Biol. Chem. 2004;279:15711–15714. doi: 10.1074/jbc.C300542200. [DOI] [PubMed] [Google Scholar]
- 15.McLeod S.J., Shum A.J., Lee R.L., Takei F., Gold M.R. The Rap GTPases regulate integrin-mediated adhesion, cell spreading, actin polymerization, and Pyk2 tyrosine phosphorylation in B lymphocytes. J. Biol. Chem. 2004;279:12009–12019. doi: 10.1074/jbc.M313098200. [DOI] [PubMed] [Google Scholar]
- 16.Taira K., Umikawa M., Takei K., Myagmar B.E., Shinzato M., Machida N., et al. The Traf2- and Nck-interacting kinase as a putative effector of Rap2 to regulate actin cytoskeleton. J. Biol. Chem. 2004;279:49488–49496. doi: 10.1074/jbc.M406370200. [DOI] [PubMed] [Google Scholar]
- 17.Torti M., Bertoni A., Canobbio I., Sinigaglia F., Lapetina E.G., Balduini C. Interaction of the low-molecular-weight GTP-binding protein rap2 with the platelet cytoskeleton is mediated by direct binding to the actin filaments. J. Cell Biochem. 1999;75:675–685. doi: 10.1002/(sici)1097-4644(19991215)75:4<675::aid-jcb13>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 18.Yu F.X., Zhang Y., Park H.W., Jewell J.L., Chen Q., Deng Y., et al. Protein kinase A activates the Hippo pathway to modulate cell proliferation and differentiation. Genes Dev. 2013;27:1223–1232. doi: 10.1101/gad.219402.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mo J.S., Meng Z., Kim Y.C., Park H.W., Hansen C.G., Kim S., et al. Cellular energy stress induces AMPK-mediated regulation of YAP and the Hippo pathway. Nat. Cell Biol. 2015;17:500–510. doi: 10.1038/ncb3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang W., Xiao Z.D., Li X., Aziz K.E., Gan B., Johnson R.L., et al. AMPK modulates Hippo pathway activity to regulate energy homeostasis. Nat. Cell Biol. 2015;17:490–499. doi: 10.1038/ncb3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hong A.W., Meng Z., Yuan H.X., Plouffe S.W., Moon S., Kim W., et al. Osmotic stress-induced phosphorylation by NLK at Ser128 activates YAP. EMBO Rep. 2017;18:72–86. doi: 10.15252/embr.201642681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qi S., Zhong Z., Zhu Y., Wang Y., Ma M., Wang Y., et al. Two Hippo signaling modules orchestrate liver size and tumorigenesis. EMBO J. 2023;42 doi: 10.15252/embj.2023115749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Q., Nirala N.K., Chen H.J., Nie Y., Wang W., Zhang B., et al. The Misshapen subfamily of Ste20 kinases regulate proliferation in the aging mammalian intestinal epithelium. J. Cell Physiol. 2019;234:21925–21936. doi: 10.1002/jcp.28756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma M., Zhong Z., Zhu Y., Gu Y., Jin R., Meng Z., et al. Phosphorylation of NF2 at Serine-13 by MAP4K family kinases mediates pathological angiogenesis. Protein Cell. 2023;14:137–142. doi: 10.1093/procel/pwac005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wakatsuki T., Schwab B., Thompson N.C., Elson E.L. Effects of cytochalasin D and latrunculin B on mechanical properties of cells. J. Cell Sci. 2001;114:1025–1036. doi: 10.1242/jcs.114.5.1025. [DOI] [PubMed] [Google Scholar]
- 26.Song S., Cong W., Zhou S., Shi Y., Dai W., Zhang H., et al. Small GTPases: structure, biological function and its interaction with nanoparticles. Asian J. Pharm. Sci. 2019;14:30–39. doi: 10.1016/j.ajps.2018.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Plouffe S.W., Meng Z., Lin K.C., Lin B., Hong A.W., Chun J.V., et al. Characterization of Hippo pathway components by gene inactivation. Mol. Cell. 2016;64:993–1008. doi: 10.1016/j.molcel.2016.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goodson H.V., Jonasson E.M. Microtubules and microtubule-associated proteins. Cold Spring Harb. Perspect. Biol. 2018;10 doi: 10.1101/cshperspect.a022608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grantham J. The molecular chaperone CCT/TRiC: an essential component of proteostasis and a potential modulator of protein aggregation. Front Genet. 2020;11:172. doi: 10.3389/fgene.2020.00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weaver B.A. How Taxol/paclitaxel kills cancer cells. Mol. Biol. Cell. 2014;25:2677–2681. doi: 10.1091/mbc.E14-04-0916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bae S.J., Ni L., Osinski A., Tomchick D.R., Brautigam C.A., Luo X. SAV1 promotes Hippo kinase activation through antagonizing the PP2A phosphatase STRIPAK. Elife. 2017;6 doi: 10.7554/eLife.30278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bae S.J., Ni L., Luo X. STK25 suppresses Hippo signaling by regulating SAV1-STRIPAK antagonism. Elife. 2020;9 doi: 10.7554/eLife.54863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tran T., Mitra J., Ha T., Kavran J.M. Increasing kinase domain proximity promotes MST2 autophosphorylation during Hippo signaling. J. Biol. Chem. 2020;295:16166–16179. doi: 10.1074/jbc.RA120.015723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karchugina S., Benton D., Chernoff J. Regulation of MST complexes and activity via SARAH domain modifications. Biochem. Soc. Trans. 2021;49:675–683. doi: 10.1042/BST20200559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brandt Z.J., Echert A.E., Bostrom J.R., North P.N., Link B.A. Core Hippo pathway components act as a brake on Yap and Taz in the development and maintenance of the biliary network. Development. 2020;147 doi: 10.1242/dev.184242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin Z., Xie R., Guan K., Zhang M. A WW tandem-mediated dimerization mode of SAV1 essential for Hippo signaling. Cell Rep. 2020;32 doi: 10.1016/j.celrep.2020.108118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neal S.J., Zhou Q., Pignoni F. Protein Phosphatase 2A with B' specificity subunits regulates the Hippo-Yorkie signaling axis in the Drosophila eye disc. J. Cell Sci. 2022;135 doi: 10.1242/jcs.259558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paul A., Annunziato S., Lu B., Sun T., Evrova O., Planas-Paz L., et al. Cell adhesion molecule KIRREL1 is a feedback regulator of Hippo signaling recruiting SAV1 to cell-cell contact sites. Nat. Commun. 2022;13:930. doi: 10.1038/s41467-022-28567-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qi S., Zhu Y., Liu X., Li P., Wang Y., Zeng Y., et al. WWC proteins mediate LATS1/2 activation by Hippo kinases and imply a tumor suppression strategy. Mol. Cell. 2022;82:1850–1864.e7. doi: 10.1016/j.molcel.2022.03.027. [DOI] [PubMed] [Google Scholar]
- 40.Hergovich A., Schmitz D., Hemmings B.A. The human tumour suppressor LATS1 is activated by human MOB1 at the membrane. Biochem. Biophys. Res. Commun. 2006;345:50–58. doi: 10.1016/j.bbrc.2006.03.244. [DOI] [PubMed] [Google Scholar]
- 41.Chan E.H., Nousiainen M., Chalamalasetty R.B., Schäfer A., Nigg E.A., Silljé H.H. The Ste20-like kinase Mst2 activates the human large tumor suppressor kinase Lats1. Oncogene. 2005;24:2076–2086. doi: 10.1038/sj.onc.1208445. [DOI] [PubMed] [Google Scholar]
- 42.Xiong S., Lorenzen K., Couzens A.L., Templeton C.M., Rajendran D., Mao D.Y.L., et al. Structural basis for auto-inhibition of the Ndr1zent. Structure. 2018;26:1101–1115.e6. doi: 10.1016/j.str.2018.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Praskova M., Xia F., Avruch J. MOBKL1A/MOBKL1B phosphorylation by MST1 and MST2 inhibits cell proliferation. Curr. Biol. 2008;18:311–321. doi: 10.1016/j.cub.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ni L., Zheng Y., Hara M., Pan D., Luo X. Structural basis for Mob1-dependent activation of the core Mst-Lats kinase cascade in Hippo signaling. Genes Dev. 2015;29:1416–1431. doi: 10.1101/gad.264929.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen R., Plouffe S.W., Guan K.L. Determining the phosphorylation status of Hippo components YAP and TAZ using Phos-tag. Methods Mol. Biol. 2019;1893:281–287. doi: 10.1007/978-1-4939-8910-2_21. [DOI] [PubMed] [Google Scholar]
- 46.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained in the manuscript.