Abstract
The orphan nuclear receptor steroidogenic factor 1 (SF-1) is a critical developmental regulator in the urogenital ridge, because mice targeted for disruption of the SF-1 gene lack adrenal glands and gonads. SF-1 was recently shown to interact with DAX-1, another orphan receptor whose tissue distribution overlaps that of SF-1. Naturally occurring loss-of-function mutations of the DAX-1 gene cause the human disorder X-linked adrenal hypoplasia congenita (AHC), which resembles the phenotype of SF-1-deficient mice. Paradoxically, however, DAX-1 represses the transcriptional activity of SF-1, and AHC mutants of DAX-1 lose repression function. To further investigate these findings, we characterized the interaction between SF-1 and DAX-1 and found that their interaction indeed occurs through a repressive domain within the carboxy terminus of SF-1. Furthermore, we demonstrate that DAX-1 recruits the nuclear receptor corepressor N-CoR to SF-1, whereas naturally occurring AHC mutations of DAX-1 permit the SF-1–DAX-1 interaction, but markedly diminish corepressor recruitment. Finally, the interaction between DAX-1 and N-CoR shares similarities with that of the nuclear receptor RevErb and N-CoR, because the related corepressor SMRT was not efficiently recruited by DAX-1. Therefore, DAX-1 can serve as an adapter molecule that recruits nuclear receptor corepressors to DNA-bound nuclear receptors like SF-1, thereby extending the range of corepressor action.
Members of the nuclear receptor superfamily of transcription factors are critical players in a myriad of developmental, physiologic, and neoplastic processes (reviewed in references 7, 31, 39, 40, and 50). The modular structural motifs within nuclear receptors provide functional regions responsible for their activities. The domain structure typically consists of an amino-terminal ligand-independent activation domain (activation function 1 [AF-1], or domain A/B) that is usually not conserved among nuclear receptor subfamilies; a conserved DNA binding domain (DBD) (domain C), consisting of two zinc-binding modules; an intervening hinge region (domain D); and a carboxy-terminal ligand binding domain (domain E). Domain E participates in receptor hetero- or homodimerization (8, 46), as well as in transcriptional repression and ligand-induced transcriptional activation (4, 12, 16, 22, 37). In fact, domains D and E serve as the interface for a multitude of cooperating proteins whose purpose is to transduce activating or repressive transcriptional signals (reviewed in reference 23). Corepressor molecules N-CoR (nuclear receptor corepressor) and SMRT (silencing mediator of retinoid and thyroid receptors) usually dissociate from receptor when ligand is bound (12, 22, 33). N-CoR and SMRT repress transcription by recruiting a complex of proteins, including mSin3A and mRPD3 (HDAC1), which in turn deacetylate histones (1, 20, 45). Orphan nuclear receptors, which have no known high-affinity endogenous ligands, also interact with corepressor molecules (19, 66, 67), but the mechanisms which regulate those interactions are not known. While it is possible that the associations are constitutive, it is likely that unknown ligands and/or other interactive proteins influence these interactions.
The nuclear receptor steroidogenic factor 1 (SF-1) is an orphan receptor, and it is able to bind DNA with high affinity as well as activate transcription as a monomer (63). While 25-hydroxycholesterol has been shown to potentiate SF-1 activity (34), no high-affinity-specific ligand has been isolated. The SF-1 gene is expressed constitutively in specific endocrine tissues: the hypothalamus, anterior pituitary, adrenal gland, gonads, and placenta (26, 27, 29, 51). The importance of SF-1 is underscored by its ability to regulate the expression of many genes characteristic of these tissues. For instance, experiments with cultured cell lines have shown that each steroidogenic enzyme gene and the steroidogenic acute regulatory protein are regulatable by SF-1 (3, 11, 13, 21, 25, 27, 36, 41–43, 49, 58, 64, 70). Moreover, SF-1 is a regulator of the genes encoding Müllerian inhibitory substance in vitro and in vivo (17, 55), the α-subunit of the gonadotropins (6), the β-subunit of luteinizing hormone (18, 32, 35), the adrenocorticotropin receptor (10), the prolactin receptor (24), and oxytocin (62). Furthermore, SF-1 is of particular interest biologically because of its integral role in mammalian development: it serves as a critical regulator of the development of the adrenal glands, gonads, and ventromedial hypothalamus; additionally, it is a dominant coordinator of the steroidogenic cell phenotype (14, 26, 38, 51, 57). Much remains unknown about SF-1, however, such as target genes in the developing urogenital ridge, as well as the molecules which modify its activity at specific spatial and temporal locations in development. Such molecules could contribute to the diversity of functions exhibited by this transcription factor.
DAX-1 is an atypical orphan receptor, harboring a conserved domain E, but instead of domains A to D, it possesses an amino-terminal domain that harbors repetitive regions rich in glycine and alanine residues. The gene encoding DAX-1 was positionally cloned as the gene mutated in X-linked adrenal hypoplasia congenital (AHC), a birth defect characterized by adrenal insufficiency and hypogonadotropic hypogonadism at puberty (44, 68). The expression of DAX-1 (for dose-sensitive sex reversal-AHC critical region on the X chromosome, gene 1) in the mouse overlaps that of SF-1, namely, the hypothalamus, pituitary, adrenal gland, and gonads (28, 59). Furthermore, the similarity of the phenotype of AHC patients to that of mice who lack SF-1 suggests that these receptors cooperate in development. Paradoxically, DAX-1 is an inhibitor of steroidogenesis, because it suppresses transcriptional activation by SF-1 through direct physical interaction (30), and DAX-1 serves as a DNA-bound repressor in some promoter contexts (69). Because the developmental and physiologic roles of DAX-1 and SF-1 appear to be interrelated, it is important to determine what mechanism DAX-1 employs to repress transcription.
To dissect the transcriptional activity of the SF-1–DAX-1 complex, we have localized the domain within SF-1 through which DAX-1 interacts. Furthermore, we have found that DAX-1 recruits the nuclear receptor corepressor N-CoR to SF-1, which otherwise do not interact. However, AHC mutations of DAX-1 do not readily recruit N-CoR to SF-1, thus decreasing their ability to repress SF-1 activity. These findings extend the range of corepressor action by demonstrating that a nuclear receptor normally refractory to repression by N-CoR (i.e., SF-1) is susceptible in the presence of DAX-1. DAX-1 is therefore a potentially important variable in corepressor signaling.
MATERIALS AND METHODS
Plasmids.
All cloning that employed PCR was performed with the high-fidelity KlenTaq polymerase enzyme (5). DAX-1 was cloned by reverse transcription-PCR from mouse adrenal gland total RNA into pBSKS (Stratagene). The template was sequenced, compared to the mouse DAX-1 sequence (2, 59), and then used for cloning into subsequent vectors. Murine SF-1 cDNA was obtained and used as previously described (14, 15, 65). Murine RARα cDNA was obtained from S. Adler (Washington University, St. Louis, Mo.). pCMX-N-CoR (amino acids 1 to 2453) was obtained from A. Hörlein (DKFC, Heidelberg, Germany); pCMX-N-CoR (amino acids 1510 to 2453) was obtained from M. Lazar (University of Pennsylvania). pCMX-SMRT was obtained from J. D. Chen (University of Massachusetts). GAL4 DBD (amino acids 1 to 147) fusions were made in the pM vector series (52). Fragments of the SF-1 ligand binding domain (LBD) (amino acids 120 to 462, 120 to 451, 120 to 447, 120 to 437, 120 to 380, 120 to 277, 220 to 462, 226 to 462, 230 to 462, and 245 to 462), DAX-1 LBD (amino acids 256 to 472 and 256 to 369), or RARα (amino acids 162 to 462) were generated by PCR and cloned in frame into the appropriate pM vector. Full-length or mutant cDNAs were expressed from the pCMVneo vector (9). For VP16 fusions, full-length DAX-1 (amino acids 1 to 472), partial DAX-1 (amino acids 1 to 369), or N-CoR (amino acids 1550 to 2453) was amplified by PCR and cloned downstream of and in frame with VP16 (residues 411 to 487), which harbors a synthetic Kozak initiator methionine within pCMVneo. The SMRT gene (encoding amino acids 565 to 1495) was amplified with a primer providing a synthetic initiator methionine and cloned upstream of VP16 in pCMVneo. Point mutagenesis of DAX-1 was performed by inverse PCR, after which template DNA was removed by digestion with DpnI (New England Biolabs, Inc.). Mutants R267P and F449D of DAX-1 correspond to the human DAX-1 numbering; for the mouse sequence used in these studies, the mutations are actually R269P and F451D.
Transfected SF-1 activity was measured from the SF-1 luciferase reporter (SF-1/Luc), which harbors two SF-1 binding sites (5′-TCA AGGTCA) spaced by five nucleotides upstream of the prolactin minimal promoter and the luciferase reporter gene (a derivative of pPrl36 [15]). For GAL4 fusions, activity was measured from the plasmid ΔGKI (obtained from P. Webb, University of California, San Francisco), which harbors five GAL4 binding sites upstream of the thymidine kinase minimal promoter and luciferase (15).
Transfections.
CV-1 monkey kidney cells were grown in Dulbecco’s modified Eagle medium, 10% fetal bovine serum, and antibiotics at 5% CO2. JEG-3 human choriocarcinoma cells were maintained in minimal essential medium with Earle’s salts, 10% fetal bovine serum, and antibiotics at 5% CO2. Transfections were performed in 12-well plates (cells plated at 5 × 104/well 24 h prior to addition of DNA) by calcium phosphate precipitation in CV-1 media, and JEG-3 cells were placed in 10% CO2 for 3 h prior to the addition of DNA. Cells were harvested 48 h after the addition of DNA (1 μg of total DNA per well), and lysates were analyzed for luciferase activity as previously described (15). For each experiment, transfections were performed at least five times in duplicate. All data sets presented yielded ≤10% variation between duplicates. In each case, the results presented are of one representative experiment. All luciferase activities were normalized to β-galactosidase activity, derived from a cotransfected pRSV-β-gal plasmid. For two-hybrid experiments performed with GAL4 DBD fusions, all fold activities were corrected for background reporter activity derived from expression plasmids cotransfected with GAL4 DBD alone (pM2 vector). For experiments performed with native SF-1, fold activation levels were corrected for background activity derived from expression plasmids cotransfected with nonrecombinant pCMVneo.
Physical interaction assay.
All proteins were translated in vitro with the TNT in vitro transcription-translation kit (Promega), in the presence of [35S]Met (DuPont/NEN). Partial N-CoR was translated from pCMX-N-CoR (amino acids 1510 to 2453). GAL4 DBD fusions of DAX-1 were translated from pCITE 3 (Novagen, Inc.). Protein A-agarose beads (Gibco BRL, Inc.) were incubated overnight at 4°C with mouse anti-GAL4 DBD antibody (sc-510 [Santa Cruz]) at a final concentration of 15 μg/ml. After being washed in immunoprecipitation (IP) buffer (20 mM HEPES-KOH [pH 7.9], 100 mM KCl, 1 mM EDTA, 5% glycerol, 0.5% Nonidet P-40, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride), beads were incubated for 20 min with 500 μg of bovine serum albumin per ml at 4°C. Three microliters of each in vitro-translated protein was incubated at 4°C in 200 μl of IP buffer. After 20 min, 30 μl of blocked antibody-protein A-agarose conjugate was added to each reaction mixture, which was allowed to incubate at 4°C rocking for 2 h. Beads were washed four times with IP buffer and incubated for 5 min with 30 μl of 2× sodium dodecylsulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer at 70°C, and eluates were electrophoresed. After signal enhancement, gels were dried down and exposed to film.
Western blotting.
COS-1 cells were seeded in six-well plates in CV-1 media at 1.5 × 105 cells/well. Twenty-four hours later, cells were transfected with expression vectors encoding GAL4 DBD–DAX-1 LBD fusion proteins. Forty-eight hours after the addition of DNA, cells were lysed in radioimmunoprecipitation assay buffer (1× phosphate-buffered saline, 0.1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride). Ten micrograms of total cell lysate was subjected to SDS-PAGE, transferred to nitrocellulose, and blotted with the anti-GAL4 DBD antibody used above, by using a horseradish peroxidase-conjugated goat anti-mouse secondary antibody. The signal was developed by enhanced chemiluminescence (Amersham) and exposed to film for 5 min.
RESULTS
Repression of SF-1 by DAX-1 requires an SF-1-repressive domain between residues 437 and 447.
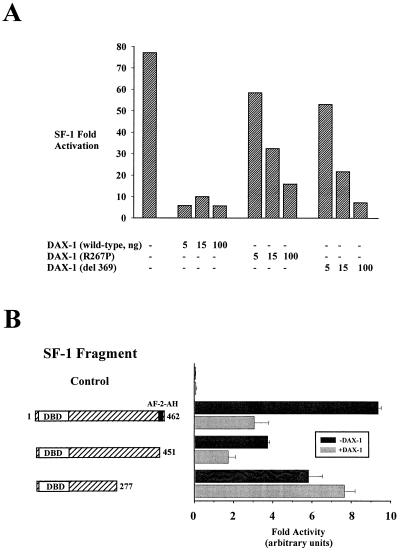
To assess the ability of DAX-1 to repress SF-1 transcriptional activation, we cotransfected JEG-3 human choriocarcinoma cells with wild-type SF-1 and increasing concentrations of mouse DAX-1 (DAX-1 does not bind to an SF-1 response element [data not shown and reference 30]). In addition, we employed two mutated versions of DAX-1 which correspond to naturally occurring human mutations that cause AHC (R267P and del 369 [44]). With an SF-1-responsive reporter, wild-type DAX-1 was a potent repressor of SF-1 activity, whereas the naturally occurring DAX-1 mutants were much less potent SF-1 repressors (Fig. 1A).
FIG. 1.
Repression of SF-1 transactivation by DAX-1 requires the most carboxy-terminal repressive domain within SF-1. (A) JEG-3 cells were cotransfected with full-length SF-1 (50 ng) and increasing amounts of the wild type, mutant R267P, or mutant del 369 DAX-1 (0, 5, 15, or 100 ng), along with 250 ng of the SF-1/Luc reporter, which harbors two SF-1 response elements upstream of the prolactin minimal promoter TATA box. (B) JEG-3 cells were cotransfected with empty expression vector (control), full-length SF-1 (amino acids 1 to 462), SF-1 (amino acids 1 to 451), or SF-1 (amino acids 1 to 277) (30 ng) in the presence or absence of exogenous wild-type DAX-1 (10 ng) and 250 ng of SF-1/Luc reporter. Luciferase activities were measured and standardized as described in Materials and Methods.
Because SF-1 does not possess an AF-1 domain (domain A/B), it relies entirely on the carboxy terminus (domains D and E) to modulate transcription. Having characterized two regions responsible for transcriptional activation (15), we used progressive truncations to unveil regions that mediate trans-repression. These experiments illustrated that the region including SF-1 amino acids 277 to 451 possesses repressive activity (data not shown). To determine if repression of SF-1 by DAX-1 requires this portion of the SF-1 LBD, we transfected full-length SF-1 (amino acids 1 to 462), the AF-2-AH deletion (amino acids 1 to 451), or the truncation (amino acids 1 to 277) into JEG-3 cells and determined their abilities to be repressed by DAX-1 (wild type). Full-length SF-1 (amino acids 1 to 462) or SF-1 (amino acids 1 to 451) is repressed by DAX-1, but SF-1 (amino acids 1 to 277) is not (Fig. 1B). DAX-1 could similarly repress fusions of these SF-1 truncations to the VP16 activation domain (data not shown), a heterologous domain that fully restores transcriptional activation of SF-1 derivatives which lack the carboxy-terminal AF-2-AH (14). Because DAX-1 is unable to repress SF-1 which lacks this carboxy-terminal domain (between residues 277 and 451), DAX-1 most likely mediates repression of SF-1 through this region.
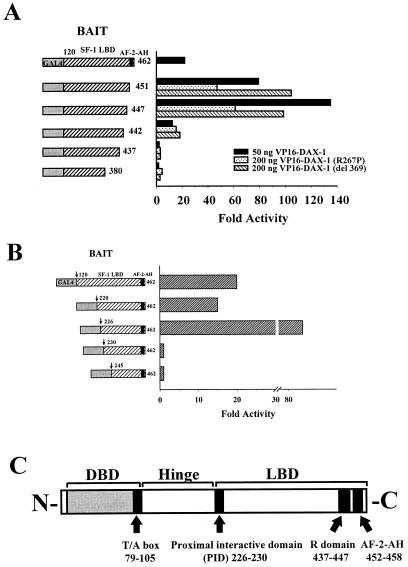
To determine if SF-1 amino acids 277 to 451 are in fact required for a physical interaction between SF-1 and DAX-1, we performed two-hybrid experiments with CV-1 cells by cotransfecting GAL4–SF-1 carboxy-terminal fusions along with the entire DAX-1 cDNA fused to the VP16 activation domain. In addition, we utilized fusions of VP16 and the two DAX-1 AHC mutants used above to confirm that they were in fact capable of interacting with SF-1 (30). As measured by activity from the GAL4 reporter, fusions of GAL4 DBD to SF-1 amino acids 120 to 462, 120 to 451, and 120 to 447 were all capable of strong interactions with all three versions of DAX-1 (only VP16–DAX-1 [wild type] is shown for the GAL4–SF-1 [amino acids 120 to 462] construct) (Fig. 2A). The magnitude of the SF-1–DAX-1 interaction was diminished with the AHC-inducing mutants of DAX-1, but could be partially overcome by increasing the amount of VP16–DAX-1 mutant (only the higher concentration of VP16–DAX-1 mutants is shown). In addition, SF-1 LBD fusions which lack AF-2-AH exhibited enhanced interaction with DAX-1, which is a feature that other nuclear receptor-corepressor interactions have manifested (19). The fusion protein containing SF-1 amino acids 120 to 442 interacted weakly with all three VP16–DAX-1 partners, and those containing SF-1 amino acids 120 to 437 or 120 to 380 did not interact at all with VP16–DAX-1 (wild type or mutant). Thus, the SF-1 region circumscribed by amino acids 437 to 447 (termed the R domain [YLYHKHLGNEM]) is required for interaction with, and therefore repression by, DAX-1. Furthermore, while strong interactions occur between SF-1 and DAX-1 AHC mutants, a component of the diminished repression of SF-1 by AHC mutants of DAX-1 may also be attributable to attenuated interaction with SF-1 (see Discussion).
FIG. 2.
Interaction between SF-1 and DAX-1 requires two domains within SF-1. CV-1 cells were cotransfected with fusions of GAL4 DBD and SF-1 carboxy terminus (20 ng), fusions of VP16 and DAX-1 (wild type [amino acids 1 to 472]), DAX-1 (R267P), or DAX-1 (del 369) (50 or 200 ng), and 250 ng of GAL4 reporter. (A) Fusions of GAL4 DBD with SF-1 amino acids 120 to 462, 120 to 451, 120 to 447, 120 to 442, 120 to 437, or 120 to 380 were tested with a two-hybrid assay against all three forms of VP16–DAX-1. Only activity from VP16–DAX-1 (wild type) is shown for GAL4–SF-1 (amino acids 120 to 462). (B) Fusions of GAL4 DBD with SF-1 amino acids 120 to 462, 220 to 462, 226 to 462, 230 to 462 or 245 to 462 were tested in a two-hybrid assay against VP16–DAX-1 (wild type). Luciferase activities were measured and standardized as described in Materials and Methods. Fold activities were calculated by determining the degree of enhancement by VP16–DAX-1 of the activity of each GAL4–SF-1 construct compared to the degree of enhancement observed with GAL4 DBD (see Materials and Methods). (C) Schematic diagram of SF-1, demonstrating the DBD, T/A box, PID, R domain, and AF-2-AH. See text for details.
Previously, we determined that two distinct domains within the SF-1 carboxy terminus were required for the interaction of SF-1 with and potentiation by the coactivator SRC-1 (15). Therefore, we questioned whether or not the interaction of SF-1 with DAX-1 involved an analogous arrangement of multiple domains. To determine if additional regions from the hinge or LBD of SF-1 were required for interaction with DAX-1 in concert with the R domain, we made progressive amino-terminal truncations from the SF-1 hinge or LBD, maintaining the natural stop codon at the carboxy terminus (after residue 462). Using the two-hybrid assay with CV-1 cells, we demonstrated that fusions of GAL4 DBD to SF-1 residues 120 to 462, 220 to 462, and 226 to 462 were able to interact with VP16–DAX-1 (wild type), as well as with the VP16 fusion to mutants of DAX-1 (Fig. 2B and data not shown). However, fusions of the GAL4 DBD to SF-1 residues 230 to 462 or 245 to 462 were unable to interact with VP16–DAX-1 (Fig. 2B). Therefore, in addition to the R domain of SF-1, residues 226 to 230 (ELILQ) are required for the interaction between SF-1 and DAX-1. Interestingly, we had previously shown that residues 187 to 245 of SF-1 are required for the interaction between SF-1 and the coactivator SRC-1 (15). Further delimitation of this domain has shown that the SF-1–SRC-1 interaction in fact requires the same residues (226 to 230) as those required for the SF-1–DAX-1 interaction (49a). Thus, the region between amino acids 226 and 230 (proximal interactive domain [PID]) mediates interactions with both SRC-1 and DAX-1 (see Discussion). A schematic of the SF-1 functional domains incorporates these regions (Fig. 2C).
DAX-1, but not AHC mutants, recruits N-CoR to SF-1.
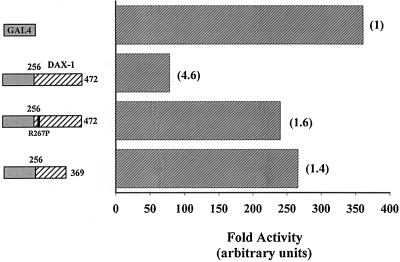
DAX-1 represses SF-1 by interacting with specific domains within SF-1, but the mechanism of repression by DAX-1 is unclear. While the ability of AHC mutants to interact with SF-1 is partially abrogated, the repression of SF-1 by these mutants is more severely affected than this diminished interaction would produce, suggesting that AHC mutations may also attenuate the actual repression function of DAX-1. All AHC mutations discovered to date mutate or eliminate the LBD of DAX-1 (44, 68), indicating that the DAX-1 LBD may transduce a repression function. To determine if DAX-1 LBD acts as a repressor of transcription when bound directly to DNA, we fused DAX-1 LBD (amino acids 256 to 472) to GAL4 DBD (amino acids 1 to 147) and compared its transcriptional activity to that of GAL4 DBD alone on a GAL4 reporter in CV-1 cells. Additionally, we transfected fusions of GAL4 DBD and DAX-1 LBD that correspond to naturally occurring AHC mutations (amino acids 256 to 472 [R267P] and 256 to 369). As shown in Fig. 3, GAL4–DAX-1 (amino acids 256 to 472) represses basal transcriptional activity, but fusions of DAX-1 which harbor AHC LBD mutations do not readily repress. Thus, the DAX-1 LBD harbors a repression function, and mutations of DAX-1 that cause AHC disturb this repressive ability. A Western blot demonstrates the equivalent stability of these transfected GAL4–DAX-1 fusions (see Fig. 6E). A similar blot demonstrated the equivalent stability of the GAL4–SF-1 fusions described above (data not shown).
FIG. 3.
DAX-1 acts as a repressor of transcription when tethered to DNA. Expressors driving GAL4 DBD alone or fusions of GAL4 DBD and DAX-1 (amino acids 256 to 472, 256 to 472 [R267P], and 256 to 369) (20 ng) were transfected with 250 ng of GAL4 reporter as described in Materials and Methods. Fold repression relative to the activity that observed with GAL4 DBD is given in parentheses. Luciferase activities were determined and standardized as described above.
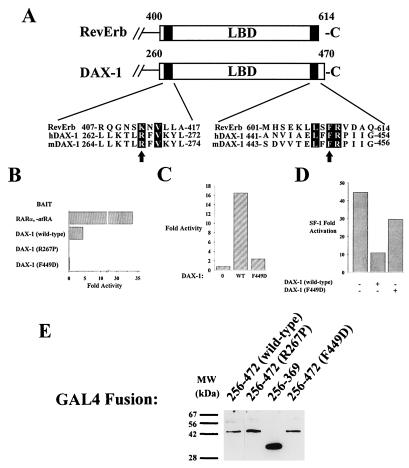
FIG. 6.
Conservation of N-CoR-interactive domains within DAX-1 and RevErb. (A) Two of the three previously delimited domains within RevErb required for interaction with N-CoR (66, 67) share similarities with regions of DAX-1 which are mutated in AHC and which are also required for interaction with N-CoR. Arrows demarcate residues analyzed in this study: R267 of DAX-1 is mutated in some AHC kindreds (R267P), and F449 is one of three shared residues in the carboxy-terminal-interactive domain of RevErb (B, C, and D). (B) Mutation F449D prevents the two-hybrid interaction between DAX-1 and N-CoR. A two-hybrid experiment was performed and results were analyzed as described in the legend to Fig. 5A (500 ng of VP16–N-CoR used in this experiment). (C) Mutation F449D in the context of full-length DAX-1 abrogates the potentiation of the two-hybrid SF-1–N-CoR interaction. Ten nanograms of DAX-1 (wild type [WT]) or F449D was transfected with 20 ng of VP16–N-CoR, 20 ng of GAL4–SF-1 (120 to 462), and 200 ng of GAL4 reporter. (D) Mutant F449D of DAX-1 is not a potent repressor of SF-1 transactivation. Fifty nanograms of SF-1 expressor was cotransfected with 20 ng of DAX-1 (wild-type) or DAX-1 (F449D) mutant, along with 250 ng of SF-1/Luc reporter. The experiment was analyzed as described in the legend to Fig. 2A. (E) Western blotting (performed as described in Materials and Methods with anti-GAL4 DBD antibody) demonstrates expression levels of GAL4 DBD–DAX-1 mutant fusions. Lanes: 1, GAL4–DAX-1 (amino acids 256 to 472); 2, GAL4–DAX-1 (amino acids 256 to 472 [R267P]); 3, GAL4–DAX-1 (amino acids 256 to 369); 4, GAL4–DAX-1 (amino acids 256 to 472 [F449D]). MW, molecular mass.
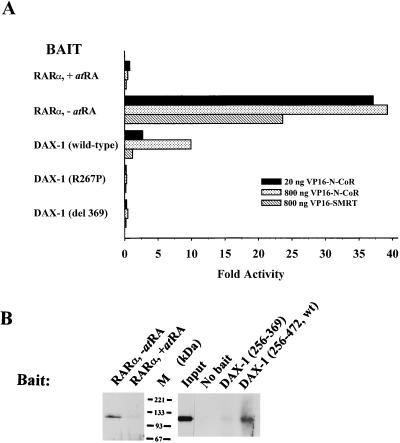
The mechanism of transcriptional repression by DAX-1 is unknown. Several nuclear receptors repress transcription through a mechanism that involves the recruitment of the nuclear receptor corepressors N-CoR and SMRT (see the introduction). To determine if DAX-1 shares this function, we performed two-hybrid experiments with CV-1 cells by using GAL4–DAX-1 LBD (amino acids 256 to 472) and VP16–N-CoR (amino acids 1550 to 2453) or VP16-SMRT (amino acids 565 to 1495) fusion proteins. These fragments of N-CoR or SMRT harbor regions previously shown to be required for interaction with nuclear receptors (54, 56). For a positive control, we utilized a GAL4-RARα fusion, which demonstrates a >20-fold interaction with VP16–N-CoR or VP16-SMRT in the absence of ligand (all-trans-retinoic acid [atRA]), but no interaction in the presence of ligand. While wild-type DAX-1 LBD interacts with N-CoR, two mutants of DAX-1 with mutations which correspond to the AHC-inducing mutations (amino acids 256 to 472 [R267P] and 256 to 369) do not (Fig. 4A). Interestingly, DAX-1 (wild type) and SMRT do not readily interact, which is analogous to the relationship between RevErb (as well as RORα) and these corepressors (references 19 and 67 and as described below). Furthermore, the interaction between DAX-1 and N-CoR is weaker than that between unliganded RARα and N-CoR, requiring a higher concentration of added VP16–N-CoR to obtain an interaction, which could indicate that the complex between DAX-1 and N-CoR is weaker and may require additional stabilizing proteins (see Discussion).
FIG. 4.
DAX-1 LBD interacts with N-CoR. (A) CV-1 cells were cotransfected with fusions of GAL4 DBD and RARα (amino acids 162 to 462) or DAX-1 LBD (amino acids 256 to 472, 256 to 472 [R267P], or 256 to 369 [del 369]) (20 ng), fusions of VP16 and N-CoR (1550 to 2453) or SMRT (565 to 1495) (20 or 800 ng), and 200 ng of GAL4 reporter. Luciferase activities were measured and standardized as described in Materials and Methods. Fold activities were calculated by determining the degree of enhancement by VP16–N-CoR or VP16-SMRT of the activity of each GAL4–DAX-1 (or GAL4-RARα) construct, compared to the degree of enhancement observed with GAL4 DBD (see Materials and Methods). (B) In vitro interaction between N-CoR and nuclear receptors. The experiment was performed as described in Materials and Methods. Precipitation of N-CoR (amino acids 1510 to 2453) by RARα (as well as 1/10 loaded input N-CoR) was exposed to film for 15 min. Precipitation of N-CoR by the DAX-1 wild type (amino acids 256 to 472) or mutant (amino acids 256 to 369) was exposed to film for 2 h. Where indicated, 10−7 M atRA was added to transfection media or to the protein incubation. M, molecular mass.
The two-hybrid interaction in mammalian cells between DAX-1 and N-CoR was confirmed with a physical interaction assay. An in vitro-translated N-CoR fragment (amino acids 1510 to 2453) was tested for binding to GAL4 DBD fusions of DAX-1 in a coimmunoprecipitation assay with an anti-GAL4 DBD antibody. As demonstrated with two-hybrid interaction, the GAL4–DAX-1 wild type (amino acids 256 to 472) precipitates N-CoR more readily than the GAL4–DAX-1 mutant (amino acids 256 to 369), but less readily than RARα (Fig. 4B). The in vitro relationship between wild-type and mutated DAX-1 is similar to that between unliganded and liganded RARα (incubated with 10−7 M atRA). The interaction between N-CoR and RARα in vitro is more robust than that between N-CoR and DAX-1, because the N-CoR and DAX-1 lanes were exposed to film for a longer period.
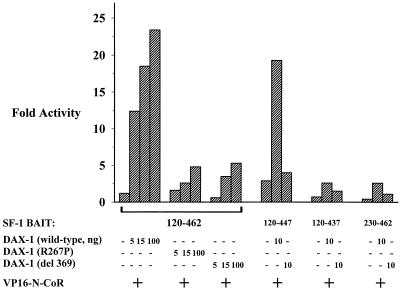
Thus far, we have demonstrated that GAL4 fusions of DAX-1 interact with the corepressor N-CoR. To determine if wild-type DAX-1 is able to recruit N-CoR to SF-1, we performed a modified two-hybrid experiment between SF-1 and N-CoR. We tested several of the previously employed GAL4 DBD fusions of SF-1 (amino acids 120 to 462, 120 to 447, 120 to 437, and 230 to 462) for the ability to interact with fusions of VP16 and N-CoR (amino acids 1550 to 2453) in the presence or absence of exogenous full-length wild-type or mutated DAX-1. The SF-1 LBD (amino acids 120 to 462) was unable to significantly interact with N-CoR, unless wild-type DAX-1 was added, which permitted an ∼20-fold increase (Fig. 5). On the other hand, AHC mutants of DAX-1 were not able to potentiate the SF-1–N-CoR interaction, even at high concentrations of added mutant DAX-1. Fusion of SF-1 amino acids 120 to 447, a sequence which lacks AF-2-AH, permitted very weak interaction with N-CoR in the absence of added DAX-1, but this interaction was enhanced when DAX-1 was added. Finally, the DAX-1-enhanced SF-1–N-CoR interaction explicitly requires the same domains of SF-1 which are required for an SF-1–DAX-1 interaction, because removal of the R domain (achieved with the amino acids 120-to-437 construct) or the PID (achieved with the amino acid 230-to-462 construct) abrogates interaction between SF-1 and N-CoR in the presence or absence of added DAX-1 (Fig. 5). In a control experiment, added full-length N-CoR did not modify the two-hybrid interaction between GAL4–SF-1 and VP16–DAX-1 (data not shown). It is important to note that the potentiation of the SF-1–N-CoR interaction requires low levels of added DAX-1 and less VP16–N-CoR than the direct DAX-1–N-CoR two-hybrid assay.
FIG. 5.
DAX-1 potentiates two-hybrid interaction between SF-1 and N-CoR. CV-1 cells were cotransfected with GAL4 DBD fusions of the SF-1 carboxy terminus (amino acids 120 to 462, 120 to 447, 120 to 437, or 230 to 462) (20 ng), VP16–N-CoR (20 ng [Fig. 5]); increasing amounts of wild-type, mutant R267P, or del 369 DAX-1 (0, 5, 10, 15, or 100 ng); and GAL4 reporter (250 ng). Luciferase activities were measured and standardized as described in Materials and Methods. Fold activities were calculated by determining the degree of enhancement by VP16–N-CoR of the activity of each GAL4–SF-1 construct in the presence or absence of DAX-1 constructs compared to the degree of enhancement observed with GAL4 DBD (see Materials and Methods).
Conservation of interactive motifs confirms DAX-1–N-CoR interaction.
The specific interaction of DAX-1 with N-CoR but not SMRT also occurs with nuclear receptors RevErb and RORα, which were shown to interact with both N-CoR and SMRT in glutathione S-transferase precipitation assays but with only N-CoR in DNA-dependent binding assays (19, 67). To determine if RevErb and DAX-1 employ a similar mechanism of corepressor recruitment, we examined the two N-CoR-interactive domains delimited within RevErb LBD (amino acids 407 to 418 and 602 to 614 [66, 67]) and compared them to regions within DAX-1 that might be involved with a DAX-1–N-CoR interaction (Fig. 6A). The amino-terminal interactive domain of RevErb LBD shares homology with two residues within the DAX-1 LBD that are mutated in some AHC kindreds (44), R267 (conserved within RevErb as K412) and V269 (conserved with RevErb as V414). Furthermore, the R267P DAX-1 mutant is incapable of efficient SF-1 repression as well as N-CoR recruitment (Fig. 2A and 6). The carboxy-terminal N-CoR interactive domain of RevErb shares three conserved residues within the carboxy terminus of DAX-1, which is often deleted in AHC kindreds. Strikingly, mutation of a conserved Phe residue in this domain of DAX-1 (F449D) abrogates the repression function of GAL4–DAX-1 (amino acids 256 to 472) and completely abolishes the two-hybrid interaction between DAX-1 and N-CoR (Fig. 6B and data not shown). Moreover, DAX-1’s ability to potentiate the two-hybrid interaction between GAL4–SF-1 (amino acids 120 to 462) and VP16–N-CoR is diminished when DAX-1 (F449D) is added in lieu of wild-type DAX-1 (Fig. 6C). Finally, DAX-1 (F449D) is a less potent repressor of SF-1-mediated transcriptional activation in JEG-3 cells than wild-type DAX-1, behaving similarly to the AHC DAX-1 mutants (Fig. 6D). All three assays confirm the required integrity of this carboxy-terminal domain for repression by DAX-1. Therefore, RevErb and DAX-1 probably employ similar motifs to interact with N-CoR, and their conservation supports the specificity of corepressor recruitment exhibited by each of them. To confirm that each mutation of DAX-1 does not diminish the stability of the resulting proteins, we performed Western blotting of transfected fusions of GAL4 DBD and wild-type DAX-1, as well as mutants R267P, del 369, and F449D. These of DAX-1 mutants do not diminish protein stability (Fig. 6E). Therefore, the inability of DAX-1 mutants to repress SF-1 and recruit N-CoR reflects an impairment in the execution of normal DAX-1 function.
DISCUSSION
DAX-1 is a structurally unique nuclear receptor whose gene is mutated in the human disorder AHC. The amino-terminal region of DAX-1 does not harbor a classical DBD, but instead possesses a structurally uncharacterized region that binds DNA (69). The conserved LBD (domain E) is usually deleted or mutated in AHC-inducing DAX-1 alleles (44, 68). Because nuclear receptor domains D and E provide surfaces for interactions with a variety of proteins, including coactivators, corepressors, and molecules with unidentified function (23), domain E of DAX-1 may be an important target for molecules which influence fundamental developmental and physiologic processes. The results presented herein demonstrate that DAX-1 is able to repress transcriptional activation by SF-1 and is also able to interact with the nuclear receptor corepressor N-CoR. Collectively, less-repressive AHC mutations in DAX-1 leave intact the ability to bind DNA (69) as well as interact with SF-1 (shown herein and within reference 30) but disrupt the ability of DAX-1 to interact with N-CoR. Therefore, a primary responsibility of DAX-1 in the developing adrenal gland may be to recruit N-CoR, or a related corepressor, to appropriate target promoter contexts. Absence of corepressor recruitment may in turn result in aberrant maturation of the adrenocortical phenotype. While seemingly paradoxical, the requirement for transcriptional repression in development has precedence, particularly in Drosophila melanogaster with proteins such as Krüppel (47, 53). While corepressors may be presumed to play critical roles in cycling physiologic cascades, as well as in genetic mechanisms that protect against neoplastic events, loss-of-function studies of mice targeted for deletion of N-CoR and/or SMRT could also unveil developmental roles in the genesis of many tissue types.
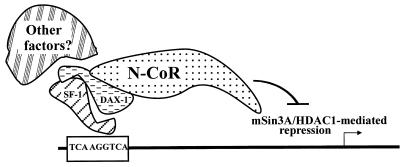
The interaction among SF-1, DAX-1, and N-CoR requires a number of domains within each molecule. To interact with DAX-1, SF-1 requires at least two domains: a carboxy-terminal repressive domain (R domain [amino acids 437 to 447]) and a PID that was previously shown to be required for transcriptional activation by SF-1 through interaction with the SRC-1 family of coactivators (reference 15 and data not shown). Thus, the PID (which lies within the amino terminus of domain E) is required for SF-1 to interact with both the activator and repressor molecules. It is not known whether the interactions between SF-1 and SRC-1 or SF-1 and DAX-1 are mutually exclusive. The PID may fold as part of a tertiary structure that allows other motifs within SF-1 to fold correctly and serve as interfaces with other regulatory molecules. The role of intramolecular contacts in the generation of higher-order structures within nuclear receptor LBDs has strong precedence among the retinoid and thyroid receptors (8, 48, 61). To interact with SF-1, DAX-1 uses its DBD (30), while the repression function of DAX-1 is mediated by at least two regions of the LBD, both of which are required to recruit N-CoR to DNA. Interestingly, both the R domain of SF-1 and the carboxy-terminal N-CoR interaction domain within DAX-1 (Fig. 6A) lie within helix 11 of the LBD, according to alignments and structural analyses of RXRα and RARγ (8, 48). The sequences of these regions within SF-1 and DAX-1 are not identical, however, which is consistent with the distinct function of these two domains. A schematic model for the interaction among SF-1, DAX-1, and N-CoR is shown in Fig. 7. The attenuated ability of DAX-1 AHC mutants to repress SF-1 clearly arises because of a loss of repressive function by the DAX-1 LBD (Fig. 3), which is in turn caused by an inability to recruit N-CoR (Fig. 4 and 5). However, our results also suggest that a component of the weakened repression of SF-1 by AHC mutants of DAX-1 may be due to a diminished ability to interact with SF-1 (Fig. 2B). Indeed, the interaction among SF-1, DAX-1, and N-CoR may be interdependent and could involve other nuclear proteins that govern the stability of the putative complex.
FIG. 7.
Schematic model of mechanism of DAX-1 repression of SF-1. SF-1, bound to its response element on DNA, recruits DAX-1, requiring the PID and the R domain within the carboxy terminus of SF-1. To interact with SF-1, DAX-1 employs its structurally degenerate DBD (30). DAX-1 in turn recruits N-CoR to DNA, requiring at least two conserved regions within DAX-1 LBD, either of which is mutated or eliminated in different AHC kindreds. Other molecules may stabilize or modify the interaction among the three proteins, depending on the cell or promoter context.
Despite the clear ability of wild-type DAX-1 to interact with N-CoR, the magnitude of interaction was nevertheless lower than that of unliganded RARα with N-CoR in two-hybrid as well as physical interaction assays. There are several potential explanations for this finding. First, the interaction may be stabilized in cells by unknown proteins involved in DAX-1 or N-CoR signaling. Second, posttranslational modification, such as phosphorylation, of DAX-1 or N-CoR may modify the interaction. Third, unknown N-CoR and SMRT family members could serve as higher-affinity targets for DAX-1. Irrespective of the precise reason, influences over receptor-corepressor interaction most likely extend beyond the known roles of ligand-induced conformational changes, as well as polarity dependence of DNA response elements (12, 22, 33). For those receptors which may have no high-affinity endogenous ligand, other factors are likely to hold influence. These factors could plausibly play roles in specific cell types under particular physiologic conditions, allowing for greater DAX-1–N-CoR interaction in vivo.
Recent studies have shown that DAX-1 is a transcriptional repressor and inhibitor of steroidogenesis (30, 68, 69). Nevertheless, the AHC phenotype (which is caused by loss-of-function mutations of DAX-1) suggests that the role of DAX-1 in the developing and mature urogenital ridge, as well as the hypothalamus and pituitary, most likely also influences processes outside the regulation of steroidogenesis. Because DAX-1 is able to repress transcription when bound to hairpin structures of DNA (69), or when indirectly bound through another transcription factor, like SF-1, its scope of action is likely to be fairly large. Because DAX-1 has been shown to be an inhibitor of retinoid transcriptional activation as well (68), it will be interesting to determine if the DBD of DAX-1 permits interaction with other receptors, extending its range of action even further. Therefore, the expression of DAX-1 could be a critical determinant of nuclear function, and it is not surprising that DAX-1 expression is dynamically regulated. For example, its expression in the developing gonads appears to diminish at certain developmental periods (28, 59). Furthermore, expression of DAX-1 in the testicular Sertoli cell is cyclical and is inhibited by cyclic AMP (60).
The ability of DAX-1 to repress transcription when bound directly or indirectly to DNA indicates that the mechanism of repression utilized by DAX-1 may be conserved in either case. This is further supported by the fact that all known AHC mutations of DAX-1 mutate or delete the LBD, and as a group, these mutations appear to disrupt the ability to interact with N-CoR. Thus, DAX-1 is likely to play important roles in nuclear receptor corepressor signaling. However, it is conceivable that DAX-1 uses different repressive mechanisms in particular cellular and/or promoter contexts. Alternatively, DAX-1 could under certain conditions activate transcription, because it does harbor a conserved AF-2-AH. Therefore, DAX-1 probably has functions outside the realm of SF-1 regulation, and their delineation will be important for our understanding of the role of DAX-1 in organogenesis, steroidogenesis, and reproductive function.
ACKNOWLEDGMENTS
We thank M. Lazar, A. Hörlein, J. D. Chen, and P. Webb for providing plasmids used in this study. We thank S. Audrain for sequencing constructs. Finally, we thank J. Svaren and T. Wilson for critical review of the manuscript.
This work was supported by a grant from the National Cancer Institute (PO1-49712-07) (J.M.), an MSTP training grant (P.A.C.), and NIH grant HD-34110 (Y.S.).
REFERENCES
- 1.Alland L, Muhle R, Hou J, Potes J J, Chin L, Schrieber-Agus N, DePinho R. Role for N-CoR and histone deacetylase in Sin3-mediated transcriptional repression. Nature. 1997;387:49–55. doi: 10.1038/387049a0. [DOI] [PubMed] [Google Scholar]
- 2.Bae D S, Schaefer M L, Partan B W, Muglia L. Characterization of the mouse DAX-1 gene reveals evolutionary conservation of a unique amino-terminal motif and widespread expression in mouse tissue. Endocrinology. 1996;137:3921–3927. doi: 10.1210/endo.137.9.8756567. [DOI] [PubMed] [Google Scholar]
- 3.Bakke M, Lund J. Mutually exclusive interactions of two nuclear orphan receptors determine activity of a cAMP-responsive sequence in the bovine CYP17 gene. Mol Endocrinol. 1995;9:327–339. doi: 10.1210/mend.9.3.7776979. [DOI] [PubMed] [Google Scholar]
- 4.Baniahmad A, Leng X, Burris T P, Tsai S Y, Tsai M-J, O’Malley B W. The τ4 activation domain of the thyroid hormone receptor is required for release of a putative corepressor(s) necessary for transcriptional silencing. Mol Cell Biol. 1995;15:76–86. doi: 10.1128/mcb.15.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barnes W M. PCR amplification of up to 35-kb DNA with high fidelity and high yield from lambda bacteriophage templates. Proc Natl Acad Sci USA. 1994;91:2216–2220. doi: 10.1073/pnas.91.6.2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barnhart K M, Mellon P L. The orphan nuclear receptor, SF-1, regulates the glycoprotein hormone alpha-subunit gene in pituitary gonadotropes. Mol Endocrinol. 1994;8:878–885. doi: 10.1210/mend.8.7.7527122. [DOI] [PubMed] [Google Scholar]
- 7.Beato M, Herrlich P, Shutz G. Steroid hormone receptors: many actors in search of a plot. Cell. 1995;83:851–857. doi: 10.1016/0092-8674(95)90201-5. [DOI] [PubMed] [Google Scholar]
- 8.Bourguet W, Ruff M, Chambon P, Gronemeyer H, Moras D. Crystal structure of the ligand-binding domain of the human nuclear receptor RXR-α. Nature. 1995;375:377–382. doi: 10.1038/375377a0. [DOI] [PubMed] [Google Scholar]
- 9.Brewer C B. Cytomegalovirus plasmid vectors for permanent lines of polarized epithelial cells. Methods Cell Biol. 1994;43:233–245. doi: 10.1016/s0091-679x(08)60606-8. [DOI] [PubMed] [Google Scholar]
- 10.Cammas F M, Pullinger G D, Barker S, Clark A J L. The mouse adrenocorticotropin receptor gene: cloning and characterization of its promoter and evidence for a role for the orphan nuclear receptor steroidogenic factor 1. Mol Endocrinol. 1997;11:867–876. doi: 10.1210/mend.11.7.9938. [DOI] [PubMed] [Google Scholar]
- 11.Caron K M, Ikeda Y, Soo S-C, Stocco D M, Parker K L, Clark B J. Characterization of the promoter region of the mouse gene encoding the steroidogenic acute regulatory protein. Mol Endocrinol. 1997;11:138–147. doi: 10.1210/mend.11.2.9880. [DOI] [PubMed] [Google Scholar]
- 12.Chen J D, Evans R M. A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature. 1995;377:454–457. doi: 10.1038/377454a0. [DOI] [PubMed] [Google Scholar]
- 13.Clemens J W, Lala D S, Parker K L, Richards J S. Steroidogenic factor-1 binding and transcriptional activity of the cholesterol side-chain cleavage promoter in rat granulosa cells. Endocrinology. 1994;134:1499–1508. doi: 10.1210/endo.134.3.8119192. [DOI] [PubMed] [Google Scholar]
- 14.Crawford P, Sadovsky Y, Milbrandt J. Nuclear receptor steroidogenic factor-1 directs embryonic stem cells toward the steroidogenic lineage. Mol Cell Biol. 1997;17:3997–4006. doi: 10.1128/mcb.17.7.3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crawford P A, Polish J A, Ganpule G, Sadovsky Y. The activation function-2 domain of steroidogenic factor-1 is required, but not sufficient for potentiation by SRC-1. Mol Endocrinol. 1997;11:1626–1635. doi: 10.1210/mend.11.11.9970. [DOI] [PubMed] [Google Scholar]
- 16.Danielian P S, White R, Lees J A, Parker M G. Identification of a conserved region required for hormone dependent transcriptional activation by steroid hormone receptors. EMBO J. 1992;11:1025–1033. doi: 10.1002/j.1460-2075.1992.tb05141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giuili G, Shen W-H, Ingraham H A. The nuclear receptor SF-1 mediates sexually dimorphic expression of Mullerian inhibiting substance, in vivo. Development. 1997;124:1799–1807. doi: 10.1242/dev.124.9.1799. [DOI] [PubMed] [Google Scholar]
- 18.Halvorson L M, Kaiser U B, Chin W W. Stimulation of luteinzing hormone beta gene promoter activity by the orphan receptor, steroidogenic factor-1. J Biol Chem. 1996;271:6645–6650. doi: 10.1074/jbc.271.12.6645. [DOI] [PubMed] [Google Scholar]
- 19.Harding H P, Atkins G B, Jaffe A B, Seo W J, Lazar M A. Transcriptional activation and repression by RORα, and orphan nuclear receptor required for cerebellar development. Mol Endocrinol. 1997;11:1737–1746. doi: 10.1210/mend.11.11.0002. [DOI] [PubMed] [Google Scholar]
- 20.Heizel T, Lavinsky R, Mullen T-M, Soderstrom M, Laherty C, Torchia J, Yang W-M, Brard G, Ngo S, Davie J, Seto E, Eisenmann R, Rose D, Glass C, Rosenfeld M. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature. 1997;387:43–48. doi: 10.1038/387043a0. [DOI] [PubMed] [Google Scholar]
- 21.Honda S-I, Morohashi K-I, Nomura M, Takeya H, Kitajima M, Omura T. Ad4BP regulating steroidogenic P450 gene is a member of steroid hormone receptor superfamily. J Biol Chem. 1993;268:7494–7502. [PubMed] [Google Scholar]
- 22.Horlein A J, Naar A M, Heinzel T, Torchia J, Gloss B, Kurokawa R, Ryan A, Kamei Y, Soderstrom M, Glass C K, Rosenfeld M G. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature. 1995;377:397–404. doi: 10.1038/377397a0. [DOI] [PubMed] [Google Scholar]
- 23.Horwitz K B, Jackson T A, Bain D L, Richer J K, Takimoto G S, Tung L. Nuclear receptor coactivators and corepressors. Mol Endocrinol. 1996;10:1167–1177. doi: 10.1210/mend.10.10.9121485. [DOI] [PubMed] [Google Scholar]
- 24.Hu Z, Zhuang L, Guan X, Meng J, Dufau M L. Steroidogenic factor-1 is an essential transcriptional activator for gonad-specific expression of promoter I of the rat prolactin receptor gene. J Biol Chem. 1997;272:14263–14271. doi: 10.1074/jbc.272.22.14263. [DOI] [PubMed] [Google Scholar]
- 25.Ikeda Y, Lala D S, Luo X, Kim E, Moisan M P, Parker K L. Characterization of the mouse FTZ-F1 gene, which encodes a key regulator of steroid hydroxylase gene expression. Mol Endocrinol. 1993;7:852–860. doi: 10.1210/mend.7.7.8413309. [DOI] [PubMed] [Google Scholar]
- 26.Ikeda Y, Luo X, Abbud R, Nilson J H, Parker K L. The nuclear receptor SF-1 is essential for the formation of the ventromedial hypothalamic nucleus. Mol Endocrinol. 1995;9:478–486. doi: 10.1210/mend.9.4.7659091. [DOI] [PubMed] [Google Scholar]
- 27.Ikeda Y, Shen W-H, Ingraham H A, Parker K L. Developmental expression of mouse steroidogenic factor-1, an essential regulator of the steroid hydroxylases. Mol Endocrinol. 1994;8:654–662. doi: 10.1210/mend.8.5.8058073. [DOI] [PubMed] [Google Scholar]
- 28.Ikeda Y, Swain A, Weber T J, Hentges K E, Zanaria E, Lalli E, Tamai K T, Sassone-Corsi P, Lovell-Badge R, Camerino G, Parker K L. Steroidogenic factor 1 and Dax-1 colocalize in multiple cell lineages: potential links in endocrine development. Mol Endocrinol. 1996;10:1262–1272. doi: 10.1210/mend.10.10.9121493. [DOI] [PubMed] [Google Scholar]
- 29.Ingraham H A, Lala D S, Ikeda Y, Luo X, Shen W-H, Nachtigal M W, Abbud R, Nilson J H, Parker K L. The nuclear receptor SF-1 acts at multiple levels of the reproductive axis. Genes Dev. 1994;8:2302–2312. doi: 10.1101/gad.8.19.2302. [DOI] [PubMed] [Google Scholar]
- 30.Ito M, Yu R, Jameson J L. DAX-1 inhibits SF-1-mediated transactivation via a carboxy-terminal domain that is deleted in adrenal hypoplasia congenita. Mol Cell Biol. 1997;17:1476–1483. doi: 10.1128/mcb.17.3.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kastner P, Mark M, Chambon P. Nonsteroid nuclear receptors: what are genetic studies telling us about their role in real life? Cell. 1995;83:859–869. doi: 10.1016/0092-8674(95)90202-3. [DOI] [PubMed] [Google Scholar]
- 32.Keri R A, Nilson J H. A steroidogenic factor-1 binding site is required for activity of the luteinizing hormone beta subunit promoter in gonadotropes of transgenic mice. J Biol Chem. 1996;271:10782–10785. doi: 10.1074/jbc.271.18.10782. [DOI] [PubMed] [Google Scholar]
- 33.Kurokawa R, Soderstrom M, Horlein A, Halachmi S, Brown M, Rosenfeld M G, Glass C K. Polarity-specific activities of retinoic acid receptors determined by a co-repressor. Nature. 1995;377:451–454. doi: 10.1038/377451a0. [DOI] [PubMed] [Google Scholar]
- 34.Lala D S, Syka P M, Lazarchik S B, Mangelsdorf D J, Parker K L, Heyman R A. Activation of the orphan nuclear receptor steroidogenic factor 1 by oxysterols. Proc Natl Acad Sci USA. 1997;94:4895–4900. doi: 10.1073/pnas.94.10.4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee S L, Sadovsky Y, Swirnoff A H, Polish J A, Goda P, Gavrilina G, Milbrandt J. Luteinizing hormone deficiency and female infertility in mice lacking the transcription factor NGFI-A (Egr-1) Science. 1996;273:1219–1222. doi: 10.1126/science.273.5279.1219. [DOI] [PubMed] [Google Scholar]
- 36.Leers-Sucheta S, Morohashi K, Mason J, Melner M. Synergistic activation of the human type II 3b-HSD/D5-D4 isomerase promoter by the transcription factor SF-1/Ad4BP and phorbol ester. J Biol Chem. 1997;272:7960–7967. doi: 10.1074/jbc.272.12.7960. [DOI] [PubMed] [Google Scholar]
- 37.Leng X, Blanco J, Tsai S Y, Ozato K, O’Malley B W, Tsai M-J. Mouse retinoid X receptor contains a separable ligand-binding and transactivation domain in its E region. Mol Cell Biol. 1995;15:255–263. doi: 10.1128/mcb.15.1.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luo X, Ikeda Y, Parker K L. A cell-specific nuclear receptor is essential for adrenal and gonadal development and sexual differentiation. Cell. 1994;77:481–490. doi: 10.1016/0092-8674(94)90211-9. [DOI] [PubMed] [Google Scholar]
- 39.Mangelsdorf D J, Evans R M. The RXR heterodimers and orphan receptors. Cell. 1995;83:841–850. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- 40.Mangelsdorf D J, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, Evans R M. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morohashi K, Honda S, Inomata Y, Handa H, Omura T. A common trans-acting factor, Ad4-binding protein, to the promoters of steroidogenic P-450s. J Biol Chem. 1992;267:17913–17919. [PubMed] [Google Scholar]
- 42.Morohashi K-I, Iida H, Nomura M, Hatano O, Honda S-I, Tsukiyama T, Niwa O, Hara T, Takakusu A, Shibata Y, Omura T. Functional difference between Ad4BP and ELP, and their distributions in steroidogenic tissues. Mol Endocrinol. 1994;8:643–653. doi: 10.1210/mend.8.5.8058072. [DOI] [PubMed] [Google Scholar]
- 43.Morohashi K-I, Zanger U M, Honda S-I, Hara M, Waterman M R, Omura T. Activation of CYP11A and CYP11B gene promoters by the steroidogenic cell-specific transcription factor, Ad4BP. Mol Endocrinol. 1993;7:1196–1204. doi: 10.1210/mend.7.9.8247022. [DOI] [PubMed] [Google Scholar]
- 44.Muscatelli F, Strom T M, Walker A P, Zanaria E, Recan D, Meindi A, Bardoni B, Guioli S, Zehtner G, Rabl W, Schwarz H P, Kaplan J-C, Camerino G, Meitinger T, Monaco A P. Mutations in the DAX-1 gene give rise to both adrenal hypoplasia congenita and hypogonadotropic hypogonadism. Nature. 1994;372:672–676. doi: 10.1038/372672a0. [DOI] [PubMed] [Google Scholar]
- 45.Nagy L, Kao H-Y, Chakravarti D, Lin R J, Hassig C A, Ayer D E, Scrieber S L, Evans R M. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell. 1997;89:373–380. doi: 10.1016/s0092-8674(00)80218-4. [DOI] [PubMed] [Google Scholar]
- 46.Perlmann T, Umesono K, Rangarajan P N, Forman B M, Evans R M. Two distinct dimerization interfaces differentially modulate target gene specificity of nuclear hormone receptors. Mol Endocrinol. 1996;10:958–966. doi: 10.1210/mend.10.8.8843412. [DOI] [PubMed] [Google Scholar]
- 47.Preiss A, Rosenberg U B, Kienlin A, Seifert E, Jackle H. Molecular genetics of Kruppel, a gene required for segmentation of the Drosophila embryo. Nature. 1985;313:27–32. doi: 10.1038/313027a0. [DOI] [PubMed] [Google Scholar]
- 48.Renaud J-P, Rochel N, Ruff M, Vivat V, Chambon P, Gronemeyer H, Moras D. Crystal structure of the RARγ ligand-binding domain bound to all-trans retinoic acid. Nature. 1995;378:681–689. doi: 10.1038/378681a0. [DOI] [PubMed] [Google Scholar]
- 49.Rice D A, Mouw A R, Parker K L. A shared promoter element regulates the expression of three steroidogenic enzymes. Mol Endocrinol. 1991;5:1552–1561. doi: 10.1210/mend-5-10-1552. [DOI] [PubMed] [Google Scholar]
- 49a.Sadovsky, Y. Unpublished data.
- 50.Sadovsky Y, Crawford P A. Developmental and physiologic roles of the nuclear receptor steroidogenic factor 1 in the reproductive system. J Soc Gynecol Invest. 1998;5:6–12. doi: 10.1016/s1071-5576(97)00096-8. [DOI] [PubMed] [Google Scholar]
- 51.Sadovsky Y, Crawford P A, Woodson K G, Polish J A, Clements M A, Tourtellotte L M, Simburger K, Milbrandt J. Mice deficient in the orphan receptor steroidogenic factor 1 lack adrenal glands and gonads but express P450 side-chain-cleavage enzyme in the placenta, and have normal embryonic serum levels of corticosteroids. Proc Natl Acad Sci USA. 1995;92:10939–10943. doi: 10.1073/pnas.92.24.10939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sadowski I, Ptashne M. A vector for expressing GAL4(1–147) fusions in mammalian cells. Nucleic Acids Res. 1989;17:7539. doi: 10.1093/nar/17.18.7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sauer F, Jackle H. Dimerization and the control of transcription by Kruppel. Nature. 1993;364:454–457. doi: 10.1038/364454a0. [DOI] [PubMed] [Google Scholar]
- 54.Seol W, Mahon M J, Lee Y K, Moore D D. Two receptor interacting domains in the nuclear hormone receptor corepressor RIP13/N-CoR. Mol Endocrinol. 1996;10:1646–1655. doi: 10.1210/mend.10.12.8961273. [DOI] [PubMed] [Google Scholar]
- 55.Shen W-H, Moore C C D, Ikeda Y, Parker K L, Ingraham H A. Nuclear receptor steroidogenic factor 1 regulates the Mullerian inhibiting substance gene: a link to the sex determination cascade. Cell. 1994;77:651–661. doi: 10.1016/0092-8674(94)90050-7. [DOI] [PubMed] [Google Scholar]
- 56.Shibata H, Nawaz Z, Tsai S Y, O’Malley B W, Tsai M J. Gene silencing by chicken ovalbumin upstream promoter-transcription factor I (COUP-TFI) is mediated by transcriptional corepressors, nuclear receptor-corepressor (N-CoR) and silencing mediator for retinoic acid receptor and thyroid hormone receptor (SMRT) Mol Endocrinol. 1997;11:714–724. doi: 10.1210/mend.11.6.0002. [DOI] [PubMed] [Google Scholar]
- 57.Shinoda K, Lei H, Yoshii H, Nomura M, Nagano M, Shiba H, Sasaki H, Osawa Y, Ninomiya Y, Niwa O, Morohashi K-I, Li E. Developmental defects of the ventromedial hypothalamic nucleus and pituitary gonadotroph in the Ftz-F1 disrupted mice. Dev Dyn. 1995;204:22–29. doi: 10.1002/aja.1002040104. [DOI] [PubMed] [Google Scholar]
- 58.Sugawara T, Holt J A, Kiriakidou M, Strauss J F., III Steroidogenic factor 1-dependent promoter activity of the human steroidogenic acute regulatory protein (StAR) gene. Biochemistry. 1996;35:9052–9059. doi: 10.1021/bi960057r. [DOI] [PubMed] [Google Scholar]
- 59.Swain A, Zanaria E, Hacker A, Lovell-Badge R, Camerino G. Mouse Dax1 expression is consistent with a role in sex determination as well as in adrenal and hypothalamus function. Nat Genet. 1996;12:404–409. doi: 10.1038/ng0496-404. [DOI] [PubMed] [Google Scholar]
- 60.Tamai K T, Monaco L, Alastalo T P, Lalli E, Parvinen M, Sassone-Corsi P. Hormonal and developmental regulation of DAX-1 expression in Sertoli cells. Mol Endocrinol. 1996;10:1561–1569. doi: 10.1210/mend.10.12.8961266. [DOI] [PubMed] [Google Scholar]
- 61.Wagner R L, Apriletti J W, McGrath M E, West B L, Baxter J D, Fletterick R J. A structural role for hormone in the thyroid hormone receptor. Nature. 1995;378:690–697. doi: 10.1038/378690a0. [DOI] [PubMed] [Google Scholar]
- 62.Wehrenberg U, Ivell R, Jansen M, von Goedecke S, Walther N. Two orphan receptors binding to a common site are involved in the regulation of the oxytocin gene in the bovine ovary. Proc Natl Acad Sci USA. 1994;91:1440–1444. doi: 10.1073/pnas.91.4.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wilson T E, Fahrner T J, Milbrandt J. The orphan receptors NGFI-B and steroidogenic factor 1 establish monomer binding as a third paradigm of nuclear receptor-DNA interaction. Mol Cell Biol. 1993;13:5794–5804. doi: 10.1128/mcb.13.9.5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wilson T E, Mouw A R, Weaver C A, Milbrandt J, Parker K L. The orphan nuclear receptor NGFI-B regulates expression of the gene encoding steroid 21-hydroxylase. Mol Cell Biol. 1993;13:861–868. doi: 10.1128/mcb.13.2.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Woodson K G, Crawford P A, Sadovsky Y, Milbrandt J. Characterization of the promoter of SF-1, an orphan nuclear receptor required for adrenal and gonadal development. Mol Endocrinol. 1997;11:117–126. doi: 10.1210/mend.11.2.9881. [DOI] [PubMed] [Google Scholar]
- 66.Zamir I, Harding H P, Atkins G B, Hörlein A, Glass C K, Rosenfeld M G, Lazar M A. A nuclear hormone receptor corepressor mediates transcriptional silencing by receptors with distinct repression domains. Mol Cell Biol. 1996;16:5458–5465. doi: 10.1128/mcb.16.10.5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zamir I, Zhang J, Lazar M A. Stoichiometric and steric principles governing repression by nuclear hormone receptors. Genes Dev. 1997;11:835–846. doi: 10.1101/gad.11.7.835. [DOI] [PubMed] [Google Scholar]
- 68.Zanaria E, Muscatelli F, Bardoni B, Strom T M, Guioli S, Guo W, Lalli E, Moser C, Walker A P, McCabe E R B, Meitinger T, Monaco A P, Sassone-Corsi P, Camerino G. An unusual member of the nuclear hormone receptor superfamily responsible for X-linked adrenal hypoplasia congenita. Nature. 1994;372:635–641. doi: 10.1038/372635a0. [DOI] [PubMed] [Google Scholar]
- 69.Zazopoulos E, Lalli E, Stocco D M, Sassone-Corsi P. DNA binding and transcriptional repression by DAX-1 blocks steroidogenesis. Nature. 1997;390:311–315. doi: 10.1038/36899. [DOI] [PubMed] [Google Scholar]
- 70.Zhang P, Mellon S H. The orphan nuclear receptor steroidogenic factor-1 regulates the cAMP-mediated transcriptional activation of rat cytochrome P450c17 (17a-hydroxylase/c17-20 lyase) Mol Endocrinol. 1996;10:147–158. doi: 10.1210/mend.10.2.8825555. [DOI] [PubMed] [Google Scholar]