Abstract
Breast cancer remains a significant global health concern, emphasizing the critical need for effective treatment strategies, especially targeted therapies. This systematic review summarizes the findings from in vitro and in vivo studies regarding the therapeutic potential of exosomes as drug delivery platforms in the field of breast cancer treatment. A comprehensive search was conducted across bibliographic datasets, including Web of Science, PubMed, and Scopus, using relevant queries from several related published articles and the Medical Subject Headings Database. Then, all morphological, biomechanical, histopathological, and cellular-molecular outcomes were systematically collected. A total of 30 studies were identified based on the Preferred Reporting Items for Systematic Reviews and Meta-Analysis guidelines. These studies underwent assessment using the Systematic Review Centre for Laboratory Animal Experimentation risk of bias assessment tool. The results indicate that exosomes exhibit promise as effective drug delivery platforms, capable of hindering cancer cell viability, proliferation, migration, and angiogenesis. However, a comprehensive assessment is challenging due to some studies deviating from guidelines and having incomplete methodology. Addressing these, future studies should detail methodologies, optimize dosing, and enhance exosome production. Standardization in reporting, consistent protocols, and exploration of alternative sources are crucial.
Keywords: breast cancer, drug delivery, exosome, small extracellular vesicles, systematic review
Graphical abstract
Parang and colleagues offer a comprehensive overview of exosome use in treating breast cancer, the leading malignancy in women. Exosomes, nanoscale vesicles with cargo capacity, present a promising strategy in cancer therapy, potentially mitigating treatment side effects by serving as carriers.
Introduction
Breast cancer
Breast cancer is a complex and heterogeneous disease, standing as the most prevalent malignant tumor among women globally, constituting around 36% of all cancer cases. It poses a significant health concern among women due to its high mortality rate, as indicated by the 5-year survival rate. In 2020, approximately 685,000 women globally succumbed to breast cancer, constituting 16% of all cancer-related fatalities among women, equating to 1 in every 6 such deaths.1
Breast cancers are classified based on their histopathological characteristics and, more recently, molecular features. The gene expression profiling of the hormone receptors (HRs) (estrogen receptor [ER] or progesterone receptor [PR]), human epidermal growth factor 2 (HER-2), and the nuclear protein Ki-67 has led to the identification of five breast cancer subtypes: luminal A (HR+ (ER+ and/or PR+)/HER-2−/Ki-67−), luminal B (HR+/(ER+ and/or PR+)/HER-2−/+, Ki-67+), HER-2 positive (HR− (ER−, PR−)/HER-2+), normal-like (HR+ (ER+ and/or PR+)/HER-2−/Ki-67−), and triple-negative breast cancer (TNBC) or basal-like (HR− (ER−, PR−)/HER-2−/basal marker +).2,3,4,5 Although normal-like and luminal A breast tumors exhibit different expression patterns, they share similar features, pathological markers, and prognosis.6 Among these subtypes, TNBC is of particular interest due to its invasive clinical characteristics and the absence of standard targeted systemic therapy.7
It is worth mentioning that treatment options for breast cancer patients typically include surgery (such as radical mastectomy and breast-conserving therapy), radiotherapy, chemotherapy, hormone therapy, and immune therapy.4,8 However, in the past 25 years, targeted therapy has emerged as a significant advancement in breast cancer treatment. In this context, precision medicine plays a crucial role in providing highly personalized therapies based on the genetic profiling of the patient’s cancer.2,9 By employing targeted agents specific to the genetic characteristics of cancer, targeted therapy reduces systemic toxicity and addresses the lack of specificity associated with conventional chemotherapy as the first-line approach in cancer treatment.10,11
Nanotechnology in cancer treatment
Recently, nanotechnology has emerged as a promising approach for cancer therapy by providing suitable carriers.12 Indeed, these carriers known as nanocarriers, have the capability to transport various types of agents, including chemical drugs, nucleic acids (DNA and RNAs), proteins, and radioactive elements, to the recipient cells. This platform offers enhanced drug solubility and prolongs the half-life of agents. One of the most intriguing aspects of nanocarriers is that targeted nanocarriers can specifically deliver their cargo to the targeted cells. This targeted delivery reduces systemic toxicity and minimizes off-target effects while simultaneously improving treatment efficiency.13,14 Thus, by harnessing the potential of nanocarriers, we have witnessed significant advancements in the delivery of anticancer agents to the desired cells.
Nanocarriers can be classified into two primary categories: synthetic nanoparticles and biological/bio-inspired nanovectors. The first category, synthetic nanoparticles can be further divided into two subgroups: inorganic and organic nanoparticles. Inorganic nanoparticles include metallic nanoparticles, silica and polystyrene nanoparticles, carbon-based nanoparticles, quantum dots, and hybrid inorganic nanoparticles. Organic nanoparticles encompass lipid-based nanoparticles like micelles, macromolecular nanoassemblies such as nanodendrimers, ferritin-based nanoparticles, protein-based nanoparticles, nanogels, and hybrid organic nanoparticles. The second category, biological and bio-inspired nanovectors can be classified into three main subgroups. The first subgroup comprises virus-like particles, including plant and animal viruses, as well as bacteriophages. The second subgroup consists of oncolytic viruses, which are specifically designed to target and destroy cancer cells. The third subgroup includes cellular vesicles, which contain Gram(+) or Gram(−) bacterial minicells, bacterial outer membrane vesicles, and extracellular vesicles (EVs).12,15,16,17,18
EVs are a general term used to describe lipid bilayer membrane vesicles derived from cells possessing a spherical shape.19,20 EVs play a critical role in various pathophysiological processes and hold tremendous potential for clinical applications.21 According to the guidelines provided by the International Society of Extracellular Vesicles, EVs are classified into four major families: oncosomes, apoptotic bodies, plasma membrane-derived vesicles called ectosomes (also known as microvesicles/microparticles), and exosomes, which are referred as small EVs.22,23
Exosomes
Exosomes are nanoscale vesicles with a size ranging from 30 to 200 nm, originating from late endosome/multivesicular bodies. They are characterized by the presence of protein markers such as CD9, CD63, CD81, and TSG 101. Exosomes possess the remarkable ability to transport various cargoes, including proteins, lipids, and nucleic acids.19,22,24,25,26
These nanoscaled vesicles play a decisive role in intercellular communication, as well as in tumorigenesis and cancer development.27,28 Moreover, exosomes have wide-ranging applications, serving as biomarkers for tumor diagnosis and prognosis, as well as drug delivery systems for cancer treatment,29,30 in particular, for breast cancer.31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60 Exosomes are manipulated using various approaches to make them more suitable for breast cancer-targeted therapy.61
In this regard, exosomes can be manipulated using different approaches to enhance their suitability for targeted therapy in breast cancer. Two main strategies are commonly employed for exosome modification. The first approach involves direct modification of exosomes, wherein therapeutic cargoes such as genetic elements, biomolecules, and drugs are directly loaded into exosomes. This approach involves several steps, including incubation, freeze-thaw cycles, electroporation, sonication, extrusion, and membrane permeabilization.62
The second strategy is termed indirect exosome modification, wherein parental cells are modified using physical or genetic techniques, allowing them to serve as sources of the engineered exosomes.61,63 Besides, certain molecules such as antibodies, receptors, or desired ligands can be attached on the surface of the exosome to target cancerous cells.64,65 As a consequence, by utilizing targeted exosomes loaded with therapeutic biodegradable agents, the delivery efficiency can be significantly improved.66
Given the significant potential of exosomes as promising drug delivery carriers for cancer treatment and considering the growing importance of exosome research, this systematic review aims to widely summarize the in vitro and in vivo findings regarding the therapeutic capabilities of exosomes, with a specific focus on breast cancer treatment with more inclusion criteria in comparison with previous reviews written about the application of exosomes as drug delivery agents. This review specifically focuses on the following aspects: quality assessment in in vivo experiments considering factors such as study design methodology, a statistical analysis, reporting quality, adherence to Minimal Information for Studies of Extracellular Vesicles (MISEV2018) guidelines, stability in the context of factors such as storage conditions and freeze-thaw cycles, animal models and their relevance to breast cancer, source of exosomes, isolation and characterization techniques, evaluating their reliability and standardization, as well as evaluating the therapeutic potential of the exosomes contents in breast cancer. By addressing these specific aspects, this systematic review provides an updated overview of the therapeutic potential of utilizing exosomes in breast cancer treatment.
Systematic review and criteria for exclusion and inclusion
This systematic review was performed in accordance with the Preferred Reporting Items for Systematic Reviews and Meta-Analysis (PRISMA) guidelines. On October 6, 2022, a survey was conducted using the Web of Science, PubMed, and Scopus databases as bibliographic datasets. The search queries used were “exosomes OR small extracellular vesicles” AND “drug delivery systems” AND “breast cancer”, which were derived from several relevant published studies and the Medical Subject Headings (MeSH) database.
The articles retrieved from the three databases were pooled and imported to EndNote, where duplicates were removed. Initially, abstracts were retrieved, and then a refined list was generated based on predefined inclusion or exclusion criteria. Next, the related full texts were downloaded and assessed for eligibility. Criteria were utilized in a manner that included only original research publications written in English, with the search keywords in both the title and the abstract.
In the first stage, the entire existing reports on the application of exosomes as a drug delivery system for breast cancer therapy were included. At this stage, reports that were not original such as reviews, letters, commentaries, and conference proceedings, were excluded. In addition, non-peer-reviewed articles, those written in languages other than English, and articles not specifically related to exosome application in breast cancer and drug delivery were also excluded.
In the second stage, both in vitro and in vivo experiments were included if they reported at least one exosomal protein marker and provided information on the exosome size. The summary of this approach is reported as a flowchart in (Figure 1).
Figure 1.
PRISMA flow diagram systematic review preparation
PRISMA, Preferred Reporting Items for Systematic Reviews and Meta-Analysis.
Quality assessment of the studies
The collected studies were assessed for study design, with a focus on assessing the quality of in vivo experiments. Due to a lack of comprehensive guidelines to ensure the quality of in vitro experiments for systematic reviews, the assessment was limited to the quality of in vivo experiments.67 For this purpose, the Systematic Review Centre for Laboratory Animal Experimentation (SYRCLE) risk of bias assessment tool was utilized to evaluate the risk of bias in these articles.68 In addition, the quality of reporting and adherence to exosome characterization methods and purity criteria outlined in the MISEV2018 guidelines were investigated.
Data collection
The study design and results of the study have been extracted individually as data for this systematic review. Texts, tables, figures, supplemental materials, and references were utilized to gather the necessary data. The study design information encompassed details about the animal models employed, such as sex and species, age, disease model, sample size, and the year of study. In addition, details regarding the exosomes, including the extraction and characterization methods, origins, size distribution, storage conditions, and exosomal markers, were extracted.
Details on treatment parameters, including groups of studies, type of therapeutic agents, concentration/quantity and volume used, delivery route of administration, and treatment frequency were also collected. Furthermore, data on the timing of the euthanasia process for the animal models were accumulated. The outcomes extracted from the in vivo and in vitro studies were of qualitative and quantitative nature wherever available. Moreover, general study features, including authors and publication year were extracted. After analyzing the study outcomes, they were qualitatively represented in the tables.
Selection and analysis
All 364 retrieved articles were pooled into EndNote X9.3.3 software. Following the exclusion of 78 duplicate articles, the titles and abstracts of the remaining 286 articles were analyzed. Of these, we excluded 223 articles due to their lack of original data, including books, reviews, and editorial articles (n = 181), articles unrelated to exosomes (n = 19), cancer treatment (n = 2), breast cancer (n = 15), or drug delivery systems (n = 5), and articles not written in English (n = 1).
The full text of the remaining 63 articles was assessed for eligibility criteria, resulting in the exclusion of 33 articles that lacked either in vivo or in vitro results. Specifically, 26 articles lacked in vivo results, two articles only presented in vivo outcomes, three articles did not report at least one exosomal protein marker, and two articles did not determine the exosome size. Finally, a total of 30 reports were included in the present systematic review31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60 (Figure 2).
Figure 2.
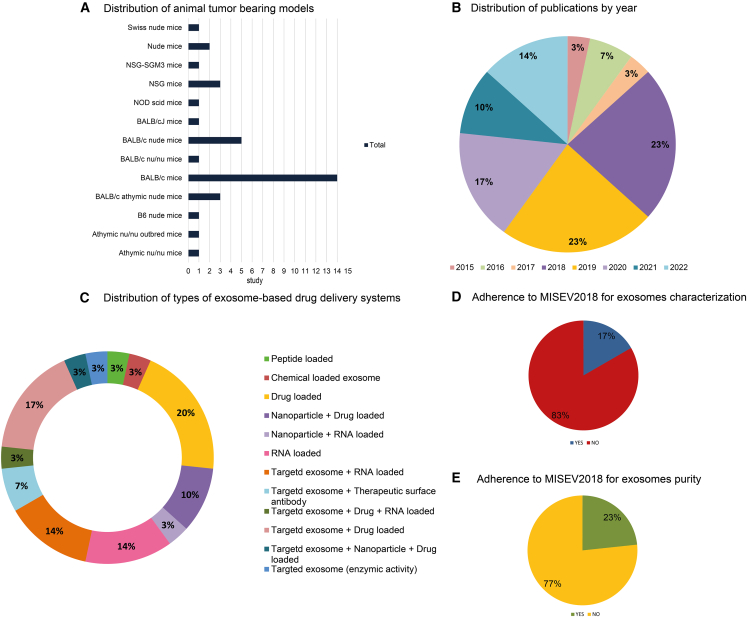
An overview of the study characteristics, including (A) animal models, (B) year of publication, (C) various types of exosome-based drug delivery systems, (D) adherence of articles to MISEV2018 exosome characterization criteria, and (E) adherence of articles to MISEV2018 exosome purity criteria.
In Figure 2A, different animal models used in 30 reports are depicted. These reports collectively employed 814 mice to establish breast tumor-bearing animal models. All the studies included in this review were published from 2015 to 2022 (Figure 2B). The selected 30 studies used a wide range of therapeutic agents to develop exosome-based drug delivery systems that could be effective in breast cancer treatment (Figure 2C). The adherence to MISEV2018 guidelines for exosome characterization and purity is illustrated in Figures 2D and 2E.31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60 In addition, the total sample size of the experiments was not explicitly stated in four studies.32,42,44,46
We summarized the details of the studies in Tables 1, 2, 3, and 4. In Table 1, descriptions of the animal models, including gender, sample size, and the methods used to establish tumor animal models are reported. The source of exosomes, isolation and characterization methods, storage conditions, expression markers of the exosomes, and adherence to relevant guidelines, including MISEV2018 criteria, are distinctly defined in Table 2. The approaches for generating exosome-based drug delivery systems, quantities and concentrations of exosome-based drugs, the routes of administration, injection frequencies, and animal groups for in vivo studies are detailed in Table 3. The key in vitro and in vivo outcomes are demonstrated in Table 4, providing insight into cellular uptake, anticancer evaluation, anti-metastatic effects as well as biodistribution assessment, in vivo antitumor effects, and histopathological and functional evaluations. The collective findings contribute to a comprehensive understanding of exosome-based drug delivery systems, highlighting their potential in advancing breast cancer therapeutics.
Table 1.
Features of breast tumor animal models in summary
| First author | Year | Animal | Gender | Sample size | Established methods | BC type |
|---|---|---|---|---|---|---|
| Wang et al.31 | 2018 | BALB/c athymic nude mice | female | 30 | BT474 (107) cells inoculated into mammary fat pad | human HER-2+ |
| Zhou et al.32 | 2021 | BALB/c athymic nude mice | female | NR | MDA-MB-231-D3H2LN cells (2 × 106) and 1 × 106 of stimulated PBMCs injected subcutaneously into mammary fat pads | human triple negative |
| Zhao et al.33 | 2020 | BALB/c mice | female | 24 | 4T1 cells (1.0 × 106 cells) injected into the mammary fat pad | murine triple negative |
| Xie et al.34 | 2021 | BALB/c nude mice | female | 20 | MDA-MB-231 cells (2 × 106) were mammary implanted in the mice | human triple negative |
| Wang et al.35 | 2019 | BALB/c mice | female | 40 | mice were injected with 1 × 106 of 4T1 cells into their flanks | murine triple negative |
| Tian et al.36 | 2020 | BALB/c mice | female | 25 | 1 × 105 of 4T1 cells were implanted in mice | murine triple negative |
| Si et al.37 | 2022 | BALB/cJ mice | female | 40 | 1 × 106 of 4T1-FLuc cells were injected into the mammary fat pad | murine triple negative |
| NSG mice | female | NR | NSG mice were injected subcutaneously into their right flanks by fresh harvested tumor or fresh frozen tumor tissues minced into small fragments | murine triple negative | ||
| Pi et al.38 | 2018 | athymic nu/nu outbred mice | female | 15 | 2 × 106 of MDA-MB-468 cells for orthotopic mammary fat-pad xenograft tumor | human triple negative |
| Peng et al.39 | 2022 | BALB/c mice | female | 18 | mice were injected with 1.25 × 105 of 4T1 cells in the mammary fat pad | murine triple negative |
| NSG-SGM3 mice | female | 18 | mice were injected with 1 × 106 of Ca1 cells into the mammary fat pad | human triple negative | ||
| BALB/c nude mice | female | 12 | the nude mice were injected with 5 × 106 of MDA-MB-468 cells into the mammary fat pad | human triple negative | ||
| BALB/c mice | female | 20 | mice were injected intravenously with 2.5 × 105 of 4T1-hEGFR cells | murine triple negative | ||
| O’Brien et al.40 | 2018 | BALB/c athymic nude mice | female | 32 | mice received an injection of 1 × 107 HCC-luc cells into the inguinal mammary fat pad | human HER-2+ |
| Naseri et al.41 | 2018 | BALB/c mice | female | 40 | 1 × 106 cells of 4T1 or TUBO tumor cells were inoculated subcutaneously in the flank region of mice | murine triple negative and HER-2+ |
| Martins-Marques et al.42 | 2016 | Swiss nude mice | female | NR | 0.5 × 106 of 4T1 cells in opposite flanks of female mice were injected subcutaneously | murine triple negative |
| Li et al.43 | 2020 | BALB/c nude mice | female | 20 | 1 × 106 of MDA-MB-231 cells per pad were injected in right mammary pad for orthotopic tumor model | human triple negative |
| Jung et al.44 | 2018 | BALB/c nu/nu mice | female | NR | 1 × 106 of MDA-MB-231 were injected subcutaneously into the right flaks of mice | human triple negative |
| Haney et al.45 | 2020 | BALB/c mice | female | 42 | 0.8 × 106 of 8FlmC-FLuc-T11 cells/mouse for orthotopic tumor model | murine triple negative |
| athymic nu/nu mice | female | 42 | 0.1 × 106 of MDA-MB-231 cells were injected to left side mammary fat pad mice | human triple negative | ||
| Hadla et al.46 | 2016 | nude mice | female | NR | 3 × 106 of MDA-MB-231 cells were injected subcutaneously into mice | human triple negative |
| Gong et al.47 | 2019 | BALB/c nude mice | male | 40 | 1 × 107 cells of MDA-MB-231 cell suspension was injected into right flank of male mice | human triple negative |
| Gomari et al.48 | 2019 | B6 nude mice | female | 16 | mouse was injected subcutaneously in flank with 1 × 106 of TUBO cells | murine HER-2+ |
| Feng et al.49 | 2021 | BALB/c mice | female | 16 | 1 × 106 of 4T1-luc cells were injected into the mammary gland of mice | murine triple negative BC |
| Cheng et al.50 | 2022 | NSG mice | female | 20 | mice have a subcutaneous injection in the right hind limbs with 5 × 106 of BT-20 cells and they received intraperitoneal injection of human PBMCs (20 × 106 cells per mouse) | human triple negative |
| Nguyen Cao et al.51 | 2022 | BALB/c nude mice | female | 20 | mice have a subcutaneous injection with 1 × 106 of MCF-7 cells into the right back region | human luminal-A |
| Ahmed et al.52 | 2015 | BALB/c mice | female | 24 | mice have a subcutaneous injection with 1 × 107 4T1 cells into their mammary fat pads | murine triple negative |
| Hong et al.53 | 2019 | BALB/c mice | female | 18 | mice were orthotopically inoculated with 1 × 106 of 4T1 cells into the mammary fat pad | murine triple negative |
| Liu et al.54 | 2019 | BALB/c mice | female | 60 | mice were subcutaneously injected at the right flanks with 1 × 106 of 4T1 cells | murine triple negative |
| Melzer et al.55 | 2019 | NOD SCID mice | female | 12 | 2 × 106 of MDA-hyb1 cells were injected subcutaneously into female NOD SCID mice | human triple negative |
| Shi et al.56 | 2020 | NSG mice | female | 10 | 1.5 × 106 of HCC 1954 cells were subcutaneously implanted into the right flank of NSG mice then 2 i.p. injections of 20 × 106 activated human PBMCs) with a 9-day interval | human HER-2+ |
| Usman et al.57 | 2018 | nude mice | female | 32 | 5 × 106 of CA1a cells were injected subcutaneously in the left and right flanks of female nude mice | human triple negative |
| Wan et al.58 | 2018 | BALB/c mice | female | 36 | 2 × 106 of MDA-MB-231 were inoculated subcutaneously to the flanks of BALB/c mice | human triple negative |
| Wang et al.59 | 2017 | BALB/c mice | female | 42 | 3.0 × 106 of MDA-MB-231 cells were injected subcutaneously into the right flanks of the mice | human triple negative |
| Xiong et al.60 | 2019 | BALB/c mice | female | 30 | 1 × 106 of 4T1 cells were injected into the mammary fat pad of mice | murine triple negative |
Table 2.
Summary of isolation and characterization methods, storage conditions, and marker expression of exosomes
| First author | Year | Origin | Isolation method | Characterization method | Storage condition |
Size distribution (nm) | Exosomal markers | Adherence to MISEV2018 exosome characterization criteria | Adherence to MISEV2018 exosome purity criteria |
|---|---|---|---|---|---|---|---|---|---|
| Wang et al.31 | 2018 | HEK239 and 293FT | ultracentrifugation | NTA, TEM, WB | fresh | 30–100 | CD63 CD81 MFGE8 | no | no |
| Zhou et al.32 | 2021 | human AT-MCS | ultracentrifugation | NTA, TEM, WB | NR | 80–100 | CD9 CD63 CD81 | no | no |
| Zhao et al.33 | 2020 | autologous breast cancer cells | gradient centrifugation | DLS, TEM, WB | −80°C | ∼124 | CD9 TSG101 | yes | yes |
| Xie et al.34 | 2021 | MDA-MB-231 | ultracentrifugation | NTA, TEM, AFM, flow cytometry | −80°C | 80–200 | CD9 CD63 | no | no |
| Wang et al.35 | 2019 | RAW 264.7 | ultracentrifugation | NTA, DLS, TEM, WB | NR | 75.3 | CD9 TSG101 Alix | no | no |
| Tian et al.36 | 2020 | 4T1 | ultracentrifugation | NTA, TEM, WB | NR | 50–100 | CD63 CD81 | no | no |
| Si et al.37 | 2022 | HEK293F | ultrafiltration | NTA, WB | −80°C | 78.2–151.1 | CD63 Hsp70 GAPDH | no | no |
| Pi et al.38 | 2018 | HEK293T | modified differential ultracentrifugation | NTA, DLS, TEM, WB | NR | 96–179 | TSG101 | no | no |
| Peng et al.39 | 2022 | human RBCs | modified differential ultracentrifugation | NTA, TEM, WB, flow cytometry | −80°C | 120–200 | TSG101 Alix GAPDHGPA | yes | yes |
| O’Brien et al.40 | 2018 | human MSCs | ultrafiltration | NTA,TEM, WB | NR | 30–150 | CD63 | no | no |
| Naseri et al.41 | 2018 | mice BMDMSCs | exosome isolation kit (Exoquick) | DLS, TEM, WB | NR | 107.3 | CD63 CD81 | no | no |
| Martins-Marques et al.42 | 2016 | HEK293 CX43+ or CX43− | ultracentrifugation | TEM, WB | fresh | <100 | CD63 CD81 | no | yes |
| Li et al.43 | 2020 | RAW 264.7 | ultracentrifugation with hypotonic treatment | DLS, TEM, WB | NR | ∼100 | CD63 CD81 | no | no |
| Jung et al.44 | 2018 | MDA-MB-231 | exosome isolation kit (Exoquick) | NTA, TEM, WB | NR | 30–200 | CD9 CD63 Hsp70 | no | no |
| Haney et al.45 | 2020 | RAW 264.7 | ultracentrifugation | NTA, DLS TEM,WB | −80°C | ∼110.8 | CD63 TSG101Hsp90 | no | no |
| Hadla et al.46 | 2016 | MDA-MB-231 | AB cell culture-nanovesicles solution | NTA, SEM, WB, fluorescence microscopy | NR | 101 | CD63 Lamp1 FLOT1 TUBA1A | no | no |
| Gong et al.47 | 2019 | THP-1 | ultracentrifugation | NTA, TEM, WB | −80°C | 179.4 | CD63 CD81 ACTIN | no | no |
| Gomari et al.48 | 2019 | MSCs | exosome isolation kit | DLS, TEM, WB | NR | 120 | CD9 CD63 CD81 | no | no |
| Feng et al.49 | 2021 | PH20-expressing HEK293T | multi-step gradient force | NTA, TEM, WB | −80°C | 100 | CD9 CD63 | no | no |
| Cheng et al.50 | 2022 | Expi293F | ultracentrifugation | NTA, TEM, WB | NR | 105–115 | CD9 CD63 CD81 | no | no |
| Nguyen Cao et al.51 | 2022 | HEK293T | exosome isolation kit (Exoquick) | NTA, TEM, WB | −80°C | ∼117 | CD63 CD81 syntenin | yes | yes |
| Ahmed et al.52 | 2015 | HEK293 and 4T1 | ultracentrifugation | AFM, WB | NR | ∼50 | TSG101 Alix | no | no |
| Hong et al.53 | 2019 | HEK293T | ultracentrifugation | DLS, TEM, WB | 4°C | ∼100 | TSG101 CD81 Alix | yes | yes |
| Liu et al.54 | 2019 | 4T1 | ultracentrifugation | NTA, DLS, TEM, WB | −80°C | ∼126.71 | CD9 CD63 | no | no |
| Melzer et al.55 | 2019 | human MSCs | ultracentrifugation | NTA, TEM, WB | −80°C | ∼171.4 | CD63 | no | no |
| Shi et al.56 | 2020 | Expi293 | ultracentrifugation | NTA, TEM, WB | NR | ∼199 | CD9 CD63 CD81 | no | yes |
| Usman et al.57 | 2018 | human RBCs | ultracentrifugation | NTA, TEM, WB | −80°C | 140 | Alix TSG101 stomatin HBA GAPDH |
yes | yes |
| Wan et al.58 | 2018 | mice DCs | ultracentrifugation | TEM, cryo-SEM, cryo-TEM, NTA, WB | −80°C | ∼100 | Annexin II, TSG101 HSC70 CD9 CD59 CD55 |
no | no |
| Wang et al.59 | 2017 | mice primary DCs | total exosome isolation kit | NTA,TEM, fluorescent microscopy | −80°C | ∼77 | CD63 | no | no |
| Xiong et al.60 | 2019 | RAW 264.7 | ultracentrifugation | NTA,TEM, flow cytometry | NR | ∼106 | Alix TSG101 FLOT1 CD9 Hsp70 LFA-1 |
no | no |
BC, breast cancer; BMDMSC, bone marrow-derived mesenchymal stem cell; NR, not reported; MFGE8, milk fat globule-EGF factor 8 protein; SEM, scanning electron microscopy; WB, western blotting.
Table 3.
Summary of exosome-based therapeutic agents, animal studies, and treatment parameters
| First author | Year | Exosome-based therapeutic agent | Methods for generating and loading | Treated groups | Quantity | Volume of injection | Route of administration | Frequency of administration | Euthanasia | |
|---|---|---|---|---|---|---|---|---|---|---|
| Wang et al.31 | 2018 | EXO-DEPT | cell engineering and incubation | 1. untreated (PBS or saline) | NR | 100 μL | i.p. | days 1, 2, 4, 6, 8, 9, 11, 14 | NR | |
| 2. EVs only | 2 × 109 EV | 100 μL | i.p. | days 1, 8 | NR | |||||
| 3. CNOB (6-chloro-9-nitro-5-oxo-5H-benzo(a)phenoxazine) only | 3 mg/kg | 100 μL | i.p. | days 2, 4, 6, 9, 11, 14 | NR | |||||
| 4. undirected loaded EVs + CNOB (3 mg/kg) | 2 × 109 EV | 100 μL | i.p. + i.v. | days 1, 2, 4, 6, 8, 9, 11, 14 | NR | |||||
| 5. Exo-DEPTs + CNOB (3 mg/kg) | 2 × 109 EV | 100 μL | i.p. + i.v. | days 1, 2, 4, 6, 8, 9, 11, 14 | NR | |||||
| Zhou et al.32 | 2021 | EVs-424 | cell transfection with miR-424 mimics | 1. untreated (PBS) | NR | 100 μL | i.t. | days 0, 3, 6, 9, 12 | 45 days | |
| 2. EV-unloaded miR-424 | 30 μg | 100 μL | i.t. | days 0, 3, 6, 9, 12 | 45 days | |||||
| 3. EV-424 | 30 μg | 100 μL | i.t. | days 0, 3, 6, 9, 12 | 45 days | |||||
| Zhao et al.33 | 2020 | CBSA/siS100A4@Exosome | incubation | 1. untreated (saline) | NR | NR | i.v. | days 14, 16, 18, 20 | 30 days | |
| 2. free siS100A4 | 1 mg/kg siRNA | NR | i.v. | days 14, 16, 18 ,20 | 30 days | |||||
| 3. CBSA/siS100A4 (1 mg/kg siRNA) | NR | NR | i.v. | days 14, 16, 18, 20 | 30 days | |||||
| 4. CBSA/siS100A4@Liposome (1 mg/kg siRNA) | NR | NR | i.v. | days 14, 16, 18, 20 | 30 days | |||||
| 5. CBSA/siS100A4@Exosome (1 mg/kg siRNA) | NR | NR | i.v. | days 14, 16, 18, 20 | 30 days | |||||
| 6. CBSA/siNC@Exosome (1 mg/kg siRNA) | NR | NR | i.v. | days 14, 16, 18, 20 | 30 days | |||||
| Xie et al.34 | 2021 | Exo-DOX | sonication | 1. untreated (saline) | NR | NR | i.v. | every 3 days (8 doses) | 7 weeks | |
| 2. exosome | 150 μg/kg | NR | i.v. | every 3 days (8 doses) | 7 weeks | |||||
| 3. free DOX | 300 μg/kg | NR | i.v. | every 3 days (8 doses) | 7 weeks | |||||
| 4. Exo-DOX(300 μg/kg DOX) | NR | NR | i.v. | every 3 days (8 doses) | 7 weeks | |||||
| Wang et al.35 | 2019 | PTX- M1-Exos | sonication | 1. untreated (PBS) | NR | NR | i.v. | every 3 days | 27 days | |
| 2. PTX | 5 mg/kg | NR | i.v. | every 3 days | 27 days | |||||
| 3. M1-Exos | NR | NR | i.v. | every 3 days | 27 days | |||||
| 4. PTX-M1-Exos (5 mg/kg PTX) | NR | NR | i.v. | every 3 days | 27 days | |||||
| Tian et al.36 | 2020 | ID@E-MSNs | incubation for ID@MSNs and sonication for ID@E-MSNs | 1. untreated (PBS) | NR | NR | i.v. | days 0, 3 | 16 days | |
| 2. free ICG | 2 mg/kg | NR | i.v. | days 0, 3 | 16 days | |||||
| 3 free DOX | 0.5 mg/kg | NR | i.v. | days 0, 3 | 16 days | |||||
| 4. ID@MSNs (2 mg/kg ICG + 0.5 mg/kg DOX) | NR | NR | i.v. | days 0, 3 | 16 days | |||||
| 5. ID@E-MSNs (2 mg/kg ICG + 0.5 mg/kg DOX) | NR | NR | i.v. | days 0, 3 | 16 days | |||||
| Si et al.37 | 2022 | mAb-EV-Ver-A | synthesis for Ab attachment via linker and incubation for Ver-A | primary TNBC xenograft | 1. untreated (PBS) | NR | NR | i.v. | every 3 days (4 doses) | 20 days |
| 2. EGFR/CD47 mAb-EV | NR | NR | i.v. | every 3 days (4 doses) | 20 days | |||||
| 3. EGFR/CD47 mAb-EV-Ver-A (0.5 mg/kg Ver-A) | NR | NR | i.v. | every 3 days (4 doses) | 20 days | |||||
| 4. EGFR/CD47 mAb-EV-Ver-A (1.5 mg/kg Ver-A) | NR | NR | i.v. | every 3 days (4 doses) | 20 days | |||||
| 5. EGFR/CD47 mAb-EV-Ver-A (2 mg/kg Ver-A) | NR | NR | i.v. | every 3 days (4 doses) | 20 days | |||||
| 6. EGFR/CD47 mAb-EV-Ver-A (2.5 mg/kg Ver-A) | NR | NR | i.v. | every 3 days (4 doses) | 20 days | |||||
| 7. EGFR mAb-EV-Ver-A (0.5 mg/kg Ver-A) | NR | NR | i.v. | every 3 days (4 doses) | 20 days | |||||
| 8. CD47mAb-EV-Ver-A (0.5 mg/kg Ver-A) | NR | NR | i.v. | every 3 days (4 doses) | 20 days | |||||
| PDX | 1. untreated (PBS) | NR | NR | i.v. | days 0, 4, 9, 15, 21, 26 | NR | ||||
| 2. EV | NR | NR | i.v. | days 0, 4, 9, 15, 21, 26 | NR | |||||
| 3. mAb-EV-Ver-A (0.5 mg/kg Ver-A) | NR | NR | i.v. | days 0, 4, 9, 15, 21, 26 | NR | |||||
| Pi et al.38 | 2018 | EGFRaptamer/EV/siSurvivin | transfection for siRNA and incubation for aptamer display | 1. untreated (PBS) | NR | NR | i.v. | once a week | 9 weeks | |
| 2. EGFRaptamer/EV/siScramble | 0.5 mg siRNA/5 mg EV/kg mice | NR | i.v. | once a week | 9 weeks | |||||
| 3. EGFRaptamer/EV/siSurvivin | 0.5 mg siRNA/5 mg EV/kg mice | NR | i.v. | once a week | 9 weeks | |||||
| Peng et al.39 | 2022 | immRNA and 3p-125b-ASO-loaded RBCEV with EGFR nanobody | transfection for RNA conjugation with tetrameric streptavidin and biotinylated anti-EGFR nanobody | intratumorally NSG-SGM3 models | 1. untreated | NR | NR | i.t. | days 3, 6, 9,1 2, 15 | 18 days |
| 2. NC RNA-EVs | 2.5 mg/kg | NR | i.t. | days 3, 6, 9, 12, 15 | 18 days | |||||
| 3. immRNA-EVs | 2.5 mg/kg | NR | i.t. | days 3, 6, 9, 12, 15 | 18 days | |||||
| intratumorally BALB/c models | 1. untreated | NR | NR | i.t. | days 3, 6, 9, 12, 15 | 18 days | ||||
| 2 .NC RNA-EVs | 5 mg/kg | NR | i.t. | days 3, 6, 9, 12, 15 | 18 days | |||||
| 3. immRNA-EVs | 2.5 mg/kg | NR | i.t. | days 3, 6, 9, 12, 15 | 18 days | |||||
| 4. 3p-125b-ASO-EVs | 5 mg/kg | NR | i.t. | days 3, 6, 9, 12, 15 | 18 days | |||||
| intrapulmonary models | 1. untreated | NR | NR | i.v. | days 2, 4, 6, 8, 10 | 11 days | ||||
| 2. NC RNA-EVs | 25 mg/kg | NR | i.v. | days 2, 4, 6, 8, 10 | 11 days | |||||
| 3. immRNA-EVs | 25 mg/kg | NR | i.v. | days 2, 4, 6, 8, 10 | 11 days | |||||
| 4. Ctrl-VHH-immRNA-EVs | 25 mg/kg | NR | i.v. | days 2, 4, 6, 8, 10 | 11 days | |||||
| 5. EGFR-VHH-. immRNA-EVs | 25 mg/kg | NR | i.v. | days 2, 4, 6, 8, 10 | 11 days | |||||
| O’Brien et al.40 | 2018 | EV-miR379 enriched | cell engineering | 1. MSC-379 | 1 × 106 cell | NR | i.v. | once a week | 6 weeks | |
| 2. 1 × 106 MSC-NTC | 1 × 106 cell | NR | i.v. | once a week | 6 weeks | |||||
| 3. 2.6 × 107 MSC-NTC EVs | 2.6 × 107 particles | NR | i.v. | once a week | 6 weeks | |||||
| 4. MSC-379 EVs | 2.6 × 107 particles | NR | i.v. | once a week | 6 weeks | |||||
| Naseri et al.41 | 2018 | MSCs-Exo-loaded LNA-anti-miR-142-3p | sonication | 4T1 tumor | 1. untreated (PBS) | NR | NR | i.v. | every 48 h | 36 days |
| 2. unloaded MSCs-Exo | 30 μg | NR | i.v. | every 48 h | 36 days | |||||
| 3. MSCs-Exo loaded with LNA-anti-miR negative control | 30 μg | NR | i.v. | every 48 h | 36 days | |||||
| 4. MSCs-Exo loaded with LNA-anti-miR-142-3p | 30 μg | NR | i.v. | every 48 h | 36 days | |||||
| TUBO tumor | 1. untreated (PBS) | NR | NR | i.v. | every 48 h | 40 days | ||||
| 2. unloaded MSCs-Exo | 30 μg | NR | i.v. | every 48 h | 40 days | |||||
| 3. MSCs-Exo loaded with LNA-anti-miR negative control | 30 μg | NR | i.v. | every 48 h | 40 days | |||||
| 4. MSCs-Exo loaded with LNA-anti-miR-142-3p | 30 μg | NR | i.v. | every 48 h | 40 days | |||||
| Martins-Marques et al.42 | 2016 | EV (Cx43+) DOX | electroporation | 1. untreated (PBS) | NR | NR | i.t. | days 5, 8, 11 | day 11 | |
| 2. EV CX43− | NR | NR | i.t. | days 5, 8, 11 | day 11 | |||||
| 3. EV CX43+ | NR | NR | i.t. | days 5, 8, 11 | day 11 | |||||
| 4. DOX | 2 mg/kg | NR | i.t. | days 5, 8, 11 | day 11 | |||||
| 5. EV (CX43−) DOX (2 mg/kg) | NR | NR | i.t. | days 5, 8, 11 | day 11 | |||||
| 6. EV (CX43+) DOX (2 mg/kg) | NR | NR | i.t. | days 5, 8, 11 | day 11 | |||||
| Li et al.43 | 2020 | MEP-D | incubation for DOX loading in PLGA co extrusion for exosome-loading ligation binding peptide |
1. untreated (PBS) | NR | 100 μL | i.v. | every 3 days (6 doses) | 18 days | |
| 2. DOX | 5 mg/kg | 100 μL | i.v. | every 3 days (6 doses) | 18 days | |||||
| 3. PL-D (5 mg/kg DOX) | NR | 100 μL | i.v. | every 3 days (6 doses) | 18 days | |||||
| 4. EP-D (5 mg/kg DOX) | NR | 100 μL | i.v. | every 3 days (6 doses) | 18 days | |||||
| 5. MEP-D (5 mg/kg DOX) | NR | 100 μL | i.v. | every 3 days (6 doses) | 18 days | |||||
| Jung et al.44 | 2018 | SPIO-labeled Olaparib-loaded exosome | incubation for SPIO and electroporation for Olaparib | 1. untreated (PBS) | NR | NR | i.t. | every 2 days | 3 weeks | |
| 2. Olaparib-loaded exosome | 100 μg/mL | NR | i.t. | every 2 days | 3 weeks | |||||
| 3. Olaparib | 100 μM | NR | i.t. | every 2 days | 3 weeks | |||||
| Haney et al.45 | 2020 | EV-PTX and EV-DOX | sonication | T11 models | 1. EV-DOX (2.5 mg/kg DOX) | 1 × 109 particles/100 μL/mouse | 10 mL/kg | i.v. | days 1, 4, 7, 10, 14 | 19 days |
| 2. EV-PTX (0.5 mg/kg PTX) | 1 × 109 particles/100 μL/mouse | 10 mL/kg | i.v. | days 1, 4, 7, 10, 14 | 19 days | |||||
| 3. EV | 1 × 109 particles/100 μL/mouse | 10 mL/kg | i.v. | days 1, 4, 7, 10, 14 | 19 days | |||||
| 4. Doxil (2.5 mg/kg DOX | NR | 10 mL/kg | i.v. | days 1, 4, 7, 10, 14 | 19 days | |||||
| 5. Taxol (0.5 mg/kg PTX) | NR | 10 mL/kg | i.v. | days 1, 4, 7, 10, 14 | 19 days | |||||
| 6. untreated (saline) | NR | 10 mL/kg | i.v. | days 1, 4, 7, 10, 14 | 19 days | |||||
| MDA-MB-231 models | 1. EV-DOX (2.5 mg/kg DOX) | 1 × 109 particles/100 μL/mouse | 10 mL/kg | i.v. | days 1, 4, 7, 10, 14 | 65 days | ||||
| 2. EV-PTX (0.5 mg/kg PTX) | 1 × 109 particles/100 μL/mouse | 10 mL/kg | i.v. | days 1, 4, 7, 10, 14 | 65 days | |||||
| 3. EV (exosome only) | 1 × 109 particles/100 μL/mouse | 10 mL/kg | i.v. | days 1, 4, 7, 10, 14 | 65 days | |||||
| 4. Doxil | 2.5 mg/kg | 10 mL/kg | i.v. | days 1, 4, 7, 10, 14 | 65 days | |||||
| 5. Taxol | 0.5 mg/kg | 10 mL/kg | i.v. | days 1, 4, 7, 10, 14 | 65 days | |||||
| 6. untreated (saline) | NR | 10 mL/kg | i.v. | days 1, 4, 7, 10, 14 | 65 days | |||||
| Hadla et al.46 | 2016 | Exo-DOX | electroporation | 1. untreated | NR | NR | i.p. | twice in week (5 doses) | 16 days | |
| 2. DOX | 3 mg/kg | NR | i.p. | twice in week (5 doses) | 16 days | |||||
| 3. Doxil (liposomal DOX, 6 mg/kg DOX) | NR | NR | i.p. | twice in week (5 doses) | 16 days | |||||
| 4. ExoDOX (6 mg/kg DOX) | NR | NR | i.p. | twice in week (5 doses) | 16 days | |||||
| Gong et al.47 | 2019 | Co-A15-Exo | incubation | 1. untreated (PBS) | NR | NR | i.v. | weekly | 5 weeks | |
| 2. Exo | NR | NR | i.v. | weekly | 5 weeks | |||||
| 3. cho-miR159 | 0.1 nmoL/kg | NR | i.v. | weekly | 5 weeks | |||||
| 4. DOX | 5 mg/kg | NR | i.v. | weekly | 5 weeks | |||||
| 5. A.15 Exo | NR | NR | i.v. | weekly | 5 weeks | |||||
| 6. A.15 Exo/cho-miR159 (0.1 nmoL/kg miR) | NR | NR | i.v. | weekly | 5 weeks | |||||
| 7. A.15 Exo/DOX (5 mg/kg DOX) | NR | NR | i.v. | weekly | 5 weeks | |||||
| 8. Co A15-Exo (0.1 nmoL/kg miR) and (5 mg/kg DOX) | NR | NR | i.v. | weekly | 5 weeks | |||||
| Gomari et al.48 | 2019 | targeted exo-DOX | cell engineering for targeted exosomes electroporation for DOX loading |
1. untreated (PBS) | NR | NR | i.v. | twice in week (6 doses) | NR | |
| 2. untargeted Exo-DOX | 1.5 mg/kg | NR | i.v. | twice in week (6 doses) | NR | |||||
| 3. targeted Exo-DOX | 1.5 mg/kg | NR | i.v. | twice in week (6 doses) | NR | |||||
| 4. free DOX | 1.5 mg/kg | NR | i.v. | twice in week (6 doses) | NR | |||||
| Feng et al.49 | 2021 | DOX@Exo-PH20-FA | cell engineering for Exox-PH20 incubation for FA coating electroporation for DOX loading |
1. untreated (saline) | NR | NR | i.v. | days 7, 10, 13, 16, 19 | 22 days | |
| 2. free DOX | NR | NR | i.v. | days 7, 10, 13, 16, 19 | 22 days | |||||
| 3. DOX@Exos-Con (control) | 100 μg DOX/200 μg total exosome protein | NR | i.v. | days 7, 10, 13, 16, 19 | 22 days | |||||
| 4. DOX@Exos-PH20 | 100 μg DOX/200 μg total exosome protein | NR | i.v. | days 7, 10, 13, 16, 19 | 22 days | |||||
| 5. DOX@Exo-PH20-FA | 100 μg DOX/200 μg total exosome protein | NR | i.v. | days 7, 10, 13, 16, 19 | 22 days | |||||
| Cheng et al.50 | 2022 | GEMINI-Exos | cell engineering | 1. untreated (PBS) | NR | NR | i.v. | every other day (6 doses) | 50 days | |
| 2. exosomes | 10 mg/kg | NR | i.v. | every other day (6 doses) | 50 days | |||||
| 3. PD-1-OX40LExos | 10 mg/kg | NR | i.v. | every other day (6 doses) | 50 days | |||||
| 4. αCD3-αEGFR-Exos | 10 mg/kg | NR | i.v. | every other day (6 doses) | 50 days | |||||
| 5. PD-1-OX40LExos + αCD3-αEGFR-Exos | 10 mg/kg for each |
NR | i.v. | every other day (6 doses) | 50 days | |||||
| 6. αCD3-αEGFR-PD-1-OX40L GEMINI-Exos | 10 mg/kg | NR | i.v. | every other day (6 doses) | 50 days | |||||
| Nguyen Cao et al.51 | 2022 | SBC-EV(ICG/PTX) | incubation | 1. untreated (PBS) | NR | NR | i.v. | day 0 | 14 days | |
| 2. PBS + US (3 min) | NR | NR | i.v. | day 0 | 14 days | |||||
| 3. ICG + PTX (10 mg/kg ICG + 0.4 mg/kg PTX) + US (3 min) | 10 mg/kg ICG + 0.4 mg/kg PTX | NR | i.v. | day 0 | 14 days | |||||
| 4. EV(ICG-PTX) (10 mg/kg ICG + 0.4 mg/kg PTX) + US (3 min) | NR | NR | i.v. | day 0 | 14 days | |||||
| 5. SBC-EV(ICG-PTX) (10 mg/kg ICG + 0.4 mg/kg PTX) + US (3 min) | NR | NR | i.v. | day 0 | 14 days | |||||
| Ahmed et al.52 | 2015 | PTEN-CT-loaded exosome | cell engineering electroporation |
into tail vein | 1. exosome | 150 μg | NR | i.v. | NR | 4 weeks |
| 2. PTEN-CT-loaded exosome | 150 μg | NR | i.v. | NR | 4 weeks | |||||
| into tumor | 1. exosome | 150 μg | NR | i.t. | NR | 4 weeks | ||||
| 2. PTEN-CT-loaded exosome | 150 μg | NR | i.t. | NR | 4 weeks | |||||
| Hong et al.53 | 2019 | Exo-PH20 | cell engineering | 1. PBS + 2 mg/kg anti-PD-L1 | NR | NR | i.t. + i.p. | every 3 days (3 doses) | 18 days | |
| 2. Exo(control) + 2 mg/kg anti-PD-L1 | 50 μg | NR | i.t. + i.p. | every 3 days (3 doses) | 18 days | |||||
| 3. Exo-PH20 + 2 mg/kg anti-PD-L1 | 50 μg | NR | i.t. + i.p. | every 3 days (3 doses) | 18 days | |||||
| Liu et al.54 | 2019 | Exo-DVDMS-sonodynamic therapy | incubation | 1. control | NR | NR | i.v. | NR | 12 days | |
| 2. US1 (2 W, 3 min) + US2 (3 W, 3 min) | – | – | – | NR | 12 days | |||||
| 3. DVDMS | 2 mg/kg | NR | i.v. | NR | 12 days | |||||
| 4. DVDMS + US1 | 2 mg/kg | NR | i.v. | NR | 12 days | |||||
| 5. DVDMS + US2 | 2 mg/kg | NR | i.v. | NR | 12 days | |||||
| 6. DVDMS + US1 + US2 | 2 mg/kg | NR | i.v. | NR | 12 days | |||||
| 7. Exo-DVDMS | 2 mg/kg | NR | i.v. | NR | 12 days | |||||
| 8. Exo-DVDMS + US1 | 2 mg/kg | NR | i.v. | NR | 12 days | |||||
| 9. Exo-DVDMS + US2 | 2 mg/kg | NR | i.v. | NR | 12 days | |||||
| 10. Exo-DVDMS + US1 + US2 | 2 mg/kg | NR | i.v. | NR | 12 days | |||||
| Melzer et al.55 | 2019 | Taxol-loaded exosomes | incubation | 1. control exosomes | NR | 100 μL | i.v. | twice in week (6 doses) | 21 days | |
| 2. Taxol exosomes | NR | 100 μL | i.v. | twice in week (6 doses) | 21 days | |||||
| 3. Taxol | 5 mg/kg | 100 μL | i.v. | twice in week (6 doses) | 21 days | |||||
| Shi et al.56 | 2020 | SMART-Exo | cell engineering | 1. PBS (control) | NR | NR | i.v. | every other days (6 doses) | 33 days | |
| 2. αCD3-αHER2 SMART-Exos | 1.5 × 1010 particles | NR | i.v. | every other days (6 doses) | 33 days | |||||
| Usman et al.57 | 2018 | 125b-ASO-loaded RBCEVs | electroporation | 1. untreated | – | – | i.t. | every 3 days | 44 days | |
| 2. UE-EVs | 8.25 × 1011 particles | NR | i.t. | every 3 days | 44 days | |||||
| 3. NC-ASO E-EVs | 8.25 × 1011 particles | NR | i.t. | every 3 days | 44 days | |||||
| 4. 125b-ASO E-EVs | 8.25 × 1011 particles | NR | i.t. | every 3 days | 44 days | |||||
| Wan et al.58 | 2018 | AS1411-ENV-PTX | ligation for AS141 sonication for PTX |
1. control | NR | NR | i.v. | every 2 days | 21 days | |
| 2. ENV | NR | NR | i.v. | every 2 days | 21 days | |||||
| 3. AS1411-ENV | NR | NR | i.v. | every 2 days | 21 days | |||||
| 4. PTX | 7.5 mg/kg | NR | i.v. | every 2 days | 21 days | |||||
| 5. ENV-PTX (7.5 mg/kg) | NR | NR | i.v. | every 2 days | 21 days | |||||
| 6. AS1411-ENV-PTX (7.5 mg/kg) | NR | NR | i.v. | every 2 days | 21 days | |||||
| Wang et al.59 | 2017 | AS1411-EV-let 7 miRNA | ligation for AS141 electroporation for miRNA |
1. PBS (control) | NR | NR | i.v. | every other day | 25 days | |
| 2. let-7 | NR | NR | i.v. | every other day | 25 days | |||||
| 3. EVs | 150 μg | NR | i.v. | every other day | 25 days | |||||
| 4. T-AS1411 | NR | NR | i.v. | every other day | 25 days | |||||
| 5. AS1411-EVS | 150 μg | NR | i.v. | every other day | 25 days | |||||
| 6. EVs-let-7 | 150 μg | NR | i.v. | every other day | 25 days | |||||
| 7. AS1411-EVs-let-7 | 150 μg | NR | i.v. | every other day | 25 days | |||||
| Xiong et al.60 | 2019 | NPs/Rex | nanoprecipitation process for NPs sonication for loading into Rex |
1. PBS (control) | NR | NR | i.v. | once a week | 28 days | |
| 2. Rex | NR | NR | i.v. | once a week | 28 days | |||||
| 3. Pt(lau)HSA NPs (4.55 mg/kg Pt) | NR | NR | i.v. | once a week | 28 days | |||||
| 4. NPs/Rex (4.55 mg/kg Pt) | NR | NR | i.v. | once a week | 28 days | |||||
| 5. HSA | NR | NR | i.v. | once a week | 28 days | |||||
| 6. cisplatin | 4.55 mg/kg | NR | i.v. | once a week | 28 days | |||||
Ctrl-VHH-immRNA, control exosome containing VHH antibody and immRNA; ENV, extracellular nanovesicle; Exo-DEPT, exosome-delivered enzyme prodrug therapy; DOX@Exo-PH20-FA, exosome express PH20 and modified by FA containing doxorubicin; EP-D, exosome-loaded PLGA and DOX; Exo-DVDMS, exosomes loaded by sinoporphyrin sodium; MSC-NTC, non-transfected mesenchymal stem cells; MEP-D, membrane-coated exosome-loaded PLGA and DOX; NPs/Rex, nanoparticle loaded into RAW-derived exosomes; NC-ASO E-EVs, negative control anti-sense oligonucleotide loaded into electroporated EVs; UE-EVs, un-electroporated EVs; US, ultrasound.
Table 4.
Some central in vitro and in vivo outputs
| First author | Year | In vitro | In vivo |
|---|---|---|---|
| Wang et al.31 | 2018 | Exo-DEPT induced cell death in HER-2-overexpressing breast cancer cells by converting the CNOB into 9-p amino-6-chloro-5H-benzo[a]phenoxazine-5-one (MCHB) | Exo-DEPTs combined with CNOB suppressed orthotopic BT474 xenografts tumor growth |
| Zhou et al.32 | 2021 | miR-424-5p delivered via exosomes promoted pro-inflammation and enhanced antitumor cytotoxicity. Exosomes increased secretion of pro-inflammatory cytokines, decreased production of anti-inflammatory cytokines and promoted the apoptosis in tumor cells | the intratumoral administration of miR-424-5p encapsulated in exosomes significantly repressed tumor growth |
| Zhao et al.33 | 2020 | encapsulating of CBSA and siS100A4 into exosomes protected siRNA from degradation with excellent biocompatibility | CBSA/siS100A4@Exosome had a higher affinity toward lungs in comparison with the CBSA/siS100A4@Liposome, also by acting through gene silencing, it inhibited the growth of malignant breast cancer cells |
| Xie et al.34 | 2021 | Exo-DOX inhibited proliferation of cancer cells | Exo-DOX hindered angiogenesis and breast cancer metastasis to the lungs. CD47 expressed on surface of Exo-DOX prevented the clearance of Exo-DOX from the blood, resulting in the increased antitumor effect of Exo-DOX |
| Wang et al.35 | 2019 | M1-Exos increased the expression of caspase-3 in breast cancer cells and provided a pro-inflammatory environment which enhances the antitumor activity via caspase-3-mediated pathway | administration of PTX-M1-Exos resulted in the higher antitumor effects than that of M1-Exos or PTX groups alone |
| Tian et al.36 | 2020 | ID@E-MSNs were effectively taken up by the tumor cell and accumulated in tumor with the help of the exosome membrane. ID@E-MSNs also improved the photothermal effect of ICG and cytotoxicity of DOX | ID@E-MSNs were accumulated in tumor tissue and suppressed the growth and metastasis of tumor |
| Si et al.37 | 2022 | Ver-A encapsulated into exosomes, had high cytotoxicity against TNBC | animal study demonstrated that targeted exosomes by displaying specific Ab against CD47 and EGFR containing Ver-A could effectively target TNBC and subsequently block tumor growth in both animal groups |
| Pi et al.38 | 2018 | exosomes were able to targetedly deliver siRNA to the cancer cells | targeted siRNA-loaded exosomes that displaying epidermal growth factor receptor aptamer on the surface blocked tumor growth in orthotopic breast cancer models |
| Peng et al.39 | 2022 | RBCEV-delivered RNAs induced cell death in both mouse and human breast cancer cells | EGFR-binding nanobody administered via intrapulmonary delivery facilitated the accumulation of RBCEVs in metastatic cancer cells. Moreover after intratumoral injection of RBCEVs loaded with immRNA or 3p-125bASO, it caused prominent suppression of breast cancer metastasis to the lung |
| O’Brien et al.40 | 2018 | miR-379 encapsulated in the exosomes caused a significant reduction in mRNA and protein expression of cyclooxygenase 2 | administration of cell-free Evs enriched with miR-379 led to an antitumor effect |
| Naseri et al.41 | 2018 | MSCs-Exo could efficiently deliver anti-miR-142-3p to reduce the expression of miR-142-3p and increased the transcription of the regulatory target genes leading to breast cancer cell death | MSCs-Exo with potential to penetrate into the tumor site were suitable nanovehicles to deliver the inhibitory oligonucleotides into the tumor tissues to downregulate the expression levels of miR-142-3p |
| Martins-Marques et al.42 | 2016 | presence of Cx43 in exosomes increased releasing of luciferin from exosomes into tumor cells | presence of Cx43 on the surface of the DOX-loaded exosomes reduced the cardiotoxicity of DOX |
| Li et al.43 | 2020 | targeted exosomes significantly improved the cellular uptake efficiency and the antitumor efficacy of DOX | targeted exosomes loading DOX repressed tumor growth and induced tumor apoptosis |
| Jung et al.44 | 2018 | flow cytometry and fluorescence microscopy showed that exosomes were completely taken up by the cells | distribution of Olaparib/SPIO-labeled exosomes was successively imaged using MPI showing induction apoptosis and arrested tumor growth |
| Haney et al.45 | 2020 | exosomes with high drug loading rate, efficiently accumulated in TNBC cells and showed anti-proliferation effect | drug-loaded exosomes targeted TNBC in vivo and abolished tumor growth |
| Hadla et al.46 | 2016 | DOX-loaded exosomes limited myocardial-endothelial crossing of DOX | DOX encapsulated in exosomes had a low toxicity, which allowed to treat mice at a higher concentration of DOX leading to the reduced volume of breast tumors |
| Gong et al.47 | 2019 | A15 expressed on A15-Exo facilitated co-delivery of DOX and Cho-miR159 to TNBC cells | Cho-miR159 and DOX delivery by vesicular system effectively improved anticancer effects, without adverse impacts |
| Gomari et al.48 | 2019 | flow cytometry results revealed significant differences in binding of targeted exosomes, which carries Dox to HER-2+ breast cancer cells greater than free DOX | imaging outcomes showed the selective distribution of targeted DOX-loaded exosomes in the target tissues of the murine breast cancer model. Doxorubicin-loaded exosomes diminished the tumor growth rate |
| Feng et al.49 | 2021 | in vitro outcomes demonstrated that Exos-PH20-FA targetedly delivered DOX and induced cell death as well as reduced hyaluronidase-induced metastasis of tumor cells | Exos-PH20-FA allowed an enhanced delivery of chemotherapy by tumor-targeting using FA modification. Exos-PH20-FA improved tumor treatment efficiency and reduced the side effects of cancer treatment |
| Cheng et al.50 | 2022 | GEMINI-Exos redirected and activated T cells toward killing EGFR- TNBC cells | activating of anticancer immunity by GEMINI-Exos resulted in highly potent inhibition against established TNBC tumors in mice |
| Nguyen Cao et al.51 | 2022 | SBC-EV (ICG/PTX) efficiently demonstrated anticancer activity against breast tumor cells by using the chemo-sonodynamic therapeutic strategy | high-resolution PA imaging visualized the preferential tumor accumulation of SBC-EV (ICG/PTX) in tumor-bearing mice. A single intravenous injection of the therapeutic exosomes along with US irradiation significantly hindered tumor growth in mice, without systemic toxicity |
| Ahmed et al.52 | 2015 | exosome-mediated delivery of intrinsic PTEN domain reduced proliferation, migration and colony forming | PTEC-CT delivered by exosomes reduced tumorigenesis in breast tumor models |
| Hong et al.53 | 2019 | low-molecular-weight oligo-HA had potential to activate DCs | Exo-PH20 penetrated into tumor tissues via HA degradation. Also, Exo-PH20 treatment successfully activated the maturation and migration of DCs |
| Liu et al.54 | 2019 | endocytosis of Exo-DVDMS by lysosomes. This system induced multiple cell death-signaling pathways | tumor-derived exosomes exhibited high stability and specificity toward the tumors. Furthermore, exosomes served as a functionalized nanostructure inhibited tumor metastasis higher than that of free form |
| Melzer et al.55 | 2019 | human-derived exosomes loaded with Taxol inhibited cancer cell growth and showed cytotoxic effects | Taxol-loaded exosomes revealed a more than 60% reduction in subcutaneous primary tumors. Distant organ metastasis also reduced |
| Shi et al.56 | 2020 | SMART-Exos redirected and activated cytotoxic T cells toward HER-2-expressing breast cancer cells | SMART-Exos redirected and activated cytotoxic T cells toward HER-2− tumor with specific antitumor activity |
| Usman et al.57 | 2018 | RBC-EVs were used for delivery of therapeutic RNA with no observable cytotoxicity in human cells | RBC-EVs delivered therapeutic RNA to xenograft mouse models |
| Wan et al.58 | 2018 | a new and rapid method for production of drug-targeting nanovesicles developed for cancer treatment | targeted exosomes by AS1411peptide and encapsulated with PTX efficiently inhibited tumor growth in mice models |
| Wang et al.59 | 2017 | AS1411-EVs loaded with miRNA let-7 targetedly delivered miRNA to MDA-MB-231 cells | AS1411-EVs loaded with labeled miRNA let-7 selectively targeted tumor tissues in tumor-bearing mice and inhibited tumor growth. Importantly, this drug delivery system had no side effects or immune responses |
| Xiong et al.60 | 2019 | NPs/Rex enhanced colloidal stability of therapeutic agents and reduced cell proliferation as well as arrested cell cycle of breast cancer cells | NPs/Rex prolonged blood circulation of therapeutic agent, increased smart organ tropism, enhanced biocompatibility, and reduced metastatic nodules of lungs |
Biases analysis
In biomedical research, the reliability and validity of study outcomes are crucial for advancing scientific knowledge and informing medical practices. Methodological biases and poor reporting can lead to skewed outcomes, misleading estimates, and hinder the reproducibility of studies. Thus, we assessed both outcomes and methodologies for in vivo sections as shown in Table 5.
Table 5.
SYRCLE risk of bias assessment tool was employed to assess risk of bias
| First author | Year | Selection bias | Performance bias | Detection bias | Attrition bias | Reporting bias | Other | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Sequence generation | Baseline generation | Allocation concealment | Random housing | Blinding | Random outcome assessment | Blinding | Incomplete outcome data | Selective outcome reporting | Other sources of bias | ||
| Wang et al.31 | 2018 | low risk | low risk | unclear risk | unclear risk | unclear risk | low risk | unclear risk | low risk | low risk | unclear risk |
| Zhou et al.32 | 2021 | unclear risk | low risk | unclear risk | unclear risk | unclear risk | low risk | unclear risk | low risk | low risk | unclear risk |
| Zhao et al.33 | 2020 | low risk | low risk | unclear risk | unclear risk | unclear risk | low risk | unclear risk | low risk | low risk | unclear risk |
| Xie et al.34 | 2021 | unclear risk | low risk | unclear risk | unclear risk | unclear risk | low risk | unclear risk | low risk | low risk | unclear risk |
| Wang et al.35 | 2019 | low risk | low risk | unclear risk | unclear risk | unclear risk | low risk | unclear risk | low risk | low risk | unclear risk |
| Tian et al.36 | 2020 | low risk | low risk | unclear risk | unclear risk | unclear risk | low risk | unclear risk | low risk | low risk | unclear risk |
| Si et al.37 | 2022 | low risk | low risk | unclear risk | unclear risk | unclear risk | low risk | unclear risk | low risk | low risk | unclear risk |
| Pi et al.38 | 2018 | low risk | low risk | unclear risk | unclear risk | high risk | low risk | high risk | low risk | low risk | unclear risk |
| Peng et al.39 | 2022 | low risk | low risk | unclear risk | unclear risk | low risk | low risk | low risk | low risk | low risk | unclear risk |
| O’Brien et al.40 | 2018 | unclear risk | low risk | unclear risk | unclear risk | high risk | low risk | low risk | low risk | low risk | unclear risk |
| Naseri et al.41 | 2018 | low risk | low risk | unclear risk | unclear risk | unclear risk | low risk | unclear risk | low risk | low risk | unclear risk |
| Martins-Marques et al.42 | 2016 | unclear risk | low risk | unclear risk | unclear risk | unclear risk | low risk | unclear risk | low risk | low risk | unclear risk |
| Li et al.43 | 2020 | low risk | low risk | unclear risk | unclear risk | unclear risk | low risk | unclear risk | low risk | low risk | unclear risk |
| Jung et al.44 | 2018 | unclear risk | low risk | unclear risk | unclear risk | unclear risk | low risk | unclear risk | low risk | low risk | unclear risk |
| Haney et al.45 | 2020 | low risk | low risk | unclear risk | unclear risk | unclear risk | low risk | unclear risk | low risk | low risk | unclear risk |
| Hadla et al.46 | 2016 | unclear risk | low risk | unclear risk | unclear risk | unclear risk | low risk | unclear risk | low risk | low risk | unclear risk |
| Gong et al.47 | 2019 | low risk | low risk | unclear risk | unclear risk | unclear risk | low risk | unclear risk | low risk | low risk | unclear risk |
| Gomari et al.48 | 2019 | low risk | low risk | unclear risk | unclear risk | unclear risk | low risk | unclear risk | low risk | low risk | unclear risk |
| Feng et al.49 | 2021 | low risk | low risk | unclear risk | unclear risk | unclear risk | low risk | unclear risk | low risk | low risk | unclear risk |
| Cheng et al.50 | 2022 | unclear risk | low risk | unclear risk | unclear risk | unclear risk | low risk | unclear risk | low risk | low risk | unclear risk |
| Nguyen Cao et al.51 | 2022 | low risk | low risk | unclear risk | unclear risk | unclear risk | low risk | unclear risk | low risk | low risk | unclear risk |
| Ahmed et al.52 | 2015 | unclear risk | low risk | unclear risk | unclear risk | unclear risk | low risk | unclear risk | low risk | low risk | unclear risk |
| Hong et al.53 | 2019 | unclear risk | low risk | unclear risk | unclear risk | unclear risk | low risk | unclear risk | low risk | low risk | unclear risk |
| Liu et al.54 | 2019 | low risk | low risk | unclear risk | unclear risk | unclear risk | low risk | unclear risk | low risk | low risk | unclear risk |
| Melzer et al.55 | 2019 | low risk | low risk | unclear risk | unclear risk | unclear risk | low risk | unclear risk | low risk | low risk | unclear risk |
| Shi et al.56 | 2020 | unclear risk | low risk | unclear risk | unclear risk | unclear risk | low risk | unclear risk | low risk | low risk | unclear risk |
| Usman et al.57 | 2018 | low risk | low risk | unclear risk | unclear risk | unclear risk | low risk | low risk | low risk | low risk | unclear risk |
| Wan et al.58 | 2018 | low risk | low risk | unclear risk | unclear risk | unclear risk | low risk | unclear risk | low risk | low risk | unclear risk |
| Wang et al.59 | 2017 | unclear risk | low risk | unclear risk | unclear risk | unclear risk | low risk | unclear risk | low risk | low risk | unclear risk |
| Xiong et al.60 | 2019 | low risk | low risk | unclear risk | unclear risk | unclear risk | low risk | unclear risk | low risk | low risk | unclear risk |
All 30 studies provided essential details regarding animal models, such as age, gender, and species, indicating a low risk for baseline generation bias.31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60 However, the focus shifted to the explicit mention of random allocation of animal groups.
Random allocation and sequence generation bias
Among the 30 studies, 11 did not explicitly mention the random allocation of animal groups, resulting in an unclear risk of sequence generation bias.32,34,40,42,44,46,50,50,52,53,56,59 On the other hand, the remaining 19 studies were classified as low risk for the sequence generation bias, as they adequately addressed the random allocation of animal groups.31,33,35,36,37,38,39,41,43,45,47,48,49,51,54,55,57,58,60 This highlights the importance of transparently reporting randomization procedures to minimize bias.
Outcome assessment and reporting bias
Regarding random outcome assessment and reporting bias, all studies were assigned a low risk, as they reported some quantitative results for in vivo experiments and used statistical methods for data analysis.31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60 This reflects a positive trend in ensuring transparency and reliability in reporting study outcomes.
Blinding and bias in performance and detection
With respect to blinding in performance bias, two studies explicitly stated that blinding was not performed during the experiments, resulting in a high-risk classification.38,40 One study was assigned as low risk for performance bias,39 and the remaining studies had unclear risk for the bias.31,32,33,34,35,36,37,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60 It is worth to mentioning that one study acquired a high risk for blinding in detection bias due to the absence of blinded assessors for the results.38 Three studies stated that they employed the blinded reviewers to assess their outcomes; hence, they were classified as low risk for blinding in detection bias.39,40,57 The remaining 26 studies did not provide information on the blinding of their assessors. Hence, they were designated as an unclear risk for blinding in detection bias.31,32,33,34,35,36,37,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,58,59,60 These findings underscore the importance of transparently reporting blinding procedures to minimize bias in experimental outcomes.
Attrition bias
All studies reported complete data, even in cases where cancer animal models died before the end of the experiment. Consequently, these studies were allocated as low risk for attrition bias,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60 demonstrating a positive aspect of methodological rigor in handling and reporting data.
Allocation concealment and random housing
None of the studies mentioned allocation concealment or random housing; hence, an unclear risk was specified for these two biases.31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60 This indicates a potential area for improvement in reporting methodologies to enhance the overall transparency and reproducibility of in vivo studies.
All the studies had unclear risks in terms of other biases,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60 emphasizing the need for comprehensive reporting to assess potential sources of bias. Addressing biases such as performance and detection bias is crucial for interpreting study results accurately. This assessment highlights the critical importance of transparent and comprehensive reporting in in vivo studies to ensure methodological rigor and minimize biases. While the majority of studies demonstrated low risk in certain aspects, areas such as random allocation, blinding, and reporting of potential biases remain as opportunities for improvement. Enhancing reporting standards can contribute to the credibility and reproducibility of biomedical research, ultimately advancing our understanding of complex biological systems.
Reporting quality
Ensuring the transparency and reproducibility of scientific research relies heavily on the quality of reporting. The reporting quality across the reviewed studies31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60 was inconsistent which may impact the interpretation, reproducibility, and overall reliability of the study outcomes.
In vitro experiments
A notable finding in the examined studies is the lack of reporting on key parameters in in vitro experiments. In terms of in vitro experiments, none of the studies reported either seeding density, passage number of exosome-secreting cells, or cell viability at the time of harvest. Although exosome depletion protocols were reported for all studies that depleted exosomes from medium or blood samples, many did not provide details of centrifugation parameters, such as rotor type, adjusted K factor, and the volume centrifuged for exosome isolation. These details are crucial for understanding the experimental conditions and ensuring the reliability of in vitro results.
In vivo experiments
Among the in vivo experiments, only five studies highlighted how the sample size had been calculated.31,38,39,40,57 Notably, among the 30 studies investigated in this systematic review, two studies reported the outcomes without corresponding details in the methodology section, posing challenges to reproducibility.44,53 Remarkably, among the selected studies, five separate studies reported pharmacokinetics (PK) experiments.45,54,56,58,60 In terms of outcome reporting, almost all of the studies reported actual numerical data,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60 contributing to transparency in outcome reporting. This is essential for the scientific community to critically evaluate study findings and draw meaningful conclusions.
Inconsistencies in reporting practices, especially regarding crucial details in in vitro experiments and sample size calculation in in vivo experiments, highlight the importance of standardizing reporting guidelines. Addressing these gaps can enhance the overall transparency, reproducibility, and reliability of research outcomes.
Animal models
The selection of appropriate animal models is a crucial aspect of breast cancer research, impacting the translatability and relevance of findings to human biology. In all 30 studies, diverse mouse models were utilized to establish tumor-bearing models,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60 showcasing the importance of considering different strains for experimental purposes. Among them, 11 studies used BALB/c33,35,36,41,49,52,53,54,58,59,60, and five selected BALB/c nude mice as tumor-bearing models.34,43,44,47,51 Three studies established tumor-bearing animal models by using BALB/c athymic nude mice.31,32,40 Si et al. utilized BALB/cJ mice for 4T1 firefly luciferase gene (4T1-FLuc) tumor model and NOD SCID gamma (NSG) mice for a TNBC patient cell model.37 Peng et al. applied three types of mice, including BALB/c for 4T1 and high epithelial growth factor receptor (EGFR) tumor models, NSG-SGM3 mice for a cA1a model, and BALB/c nude mice for an MDA-MB-468 model.39 Haney et al. conducted a study utilizing BALB/c mice for an 8FlmC-FLuc-T11 model and athymic nu/nu mice for a MDA-MB-231 model.45
Five studies applied other types of mice as tumor-bearing models, with Pi et al., Cheng et al., and Hadla et al. using athymic nu/nu outbred mice, NSG mice, and nude mice, respectively, and the remaining by Gomari et al. and Martin-Marques et al., utilizing B6 nude and Swiss nude mice as breast cancer mice models.38,42,46,48,50 Furthermore, Shi et al., Melzer et al., and Usman et al. selected NSG mice, NOD SCID mice and nude mice as tumor-bearing mice models.55,56,57
Cell line diversity
In 29 studies, various types of breast cancer cell lines were inoculated into mice,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60 reflecting the common practice of using established cell lines for modeling breast cancer. However, Si et al. implanted tumor cells isolated from TNBC patients,37 offering a more clinically relevant perspective. Interestingly, a single study employed male mice as a breast tumor model,47 introducing an intriguing dimension to the understanding of breast cancer in non-traditional contexts. A summary of this section is shown in Table 1.
The comprehensive overview of animal models and cell lines in breast cancer research highlights the diversity of approaches employed by researchers. The choices made in model selection can significantly impact the relevance and translatability of study outcomes. Understanding the rationale behind these choices provides valuable insights for future research, emphasizing the need for thoughtful consideration of model characteristics in breast cancer studies.
Source of exosomes
We explored the diverse range of sources for exosomes identified across 30 studies, shedding light on the implications of these choices for in vitro and in vivo experiments. Among the 30 studies, 5 used mesenchymal stem cells (MSCs) as their source of exosomes.32,40,41,48,55 Notably Zhou et al., O’Brien et al., and Melzer et al. utilized human MSCs,32,40,55 while Naseri et al. used murine MSCs,41 and Gomari et al. did not specify the origin of their MSCs.48 Four studies considered RAW 264.7 cells as a source for exosome extraction,35,45,60 while the MDA-MB-231 cell line was used in three studies for exosome isolation.34,44,46 In another five studies, exosomes were separated from human embryonic kidney 239 (HEK239) T cells.38,42,49,51,53
Ahmed et al. mentioned HEK239 and 4T1 cell lines as two sources for exosome fabrication for in vitro and in vivo experiments.52 Liu et al. and Tian et al. applied 4T1-derived exosomes as delivery agents.36,54 Wang et al. generated therapeutic exosomes using both HEK239 and 293FT cells, mixing them for targeted breast cancer therapy,31 and showcasing innovative approaches to therapeutic development. Si et al. isolated exosomes from HEK293F cells.37
Furthermore, in two studies performed by Gong et al. and Cheng et al., exosomes were extracted from THP1 (human leukemia monocytic) and Expi293 F (highly transfectable 293 cells) cells and used as a delivery system to transfer therapeutic agents to cancerous cells.47,50 Shi et al. used Expi 293 cells as origins of therapeutic exosomes.56
Wan et al. and Wang et al. departed from traditional cell sources by utilizing dendritic cells (DCs) for exosome harvesting, introducing a novel approach to exosome isolation.58,59 Interestingly, Peng et al. and Usman et al. used red blood cells (RBCs) as sources for exosome isolation.39,57 Moreover, Zhao et al. noted that exosomes isolated from autologous breast cancer cells showed - therapeutic efficacy as a drug carrier.33
The wide array of exosome sources in breast cancer research reflects the innovative and diverse approaches adopted by researchers. The choice of cell lines, stem cells, and unconventional sources has implications for the development of therapeutic exosomes. Understanding the strengths and limitations of each source is crucial for advancing our knowledge and harnessing the full potential of exosomes in breast cancer therapeutics.
Isolation, characterization, and storage conditions of exosomes
The isolation techniques varied across the 30 studies. In the realm of exosome isolation, 19 out of the 30 studies made use of the usual ultracentrifugation method for exosome isolation.31,32,34,35,36,42,43,45,47,48,50,52,53,54,55,56,57,58,60 Two studies introduced a modified ultracentrifugation technique, one using 60% iodixanol at 100,000 × g for 70 min,38 and the other employing 60% sucrose at 50,000 × g for 16 h to extract exosomes.39 Zhao et al. and Feng et al. also employed gradient centrifugation as a modified approach to isolate exosomes.33,49 In one study, exosomes were collected by ultrafiltration,40 whereas five others used solution-based kits for isolation of exosomes.41,44,46,51,59
Across all 30 studies, a size distribution ranging from 30 to 200 nm was reported for exosomes, assessed through various methods such as nanoparticle tracking analysis (NTA), dynamic light scattering (DLS), or atomic force microscopy (AFM). Furthermore, the presence of exosomal markers, including cluster of differentiation markers (CD9, CD63, CD81, CD59, CD55), tumor susceptibility gene 101 (TSG101), ALG-2-interacting protein X (ALIX), heat shock proteins (Hsc70, Hsp70, and 90), lysosomal-associated membrane protein 1 (Lamp1), Flotillin 1(FLOT1), tubulin alpha-1A chain (TUBA1A), syntenin, ACTIN, stomatin, leukocyte-associated antigen-1 (LFA-1), Annexin II, and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was confirmed by western blotting, flow cytometry, and fluorescence microscopy. The integrity of the exosome membrane and morphology was examined by transmission electron microscopy (TEM), scanning electron microscopy, and AFM.31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60
Another parameter evaluated in this section was the storage condition of exosomes. Only two articles reported freshly prepared exosomes.31,42 Hong et al. reported that their exosomes were stored at 4°C,53 whereas 13 others stored exosomes at −80°C before use.33,34,37,39,45,47,49,51,54,55,57,58,59 The remaining 14 studies provided no details on the storage conditions of the exosomes.32,35,36,38,40,41,43,44,46,48,50,52,56,60
These findings highlight the diversity in methodologies employed in exosome research, emphasizing the need for standardized protocols. The varied storage conditions observed may impact the stability and functionality of exosomes, making it imperative for researchers to carefully consider and report such details.
Adherence to MISEV2018 for exosome characterization and purity
To align with MISEV2018 guidelines, vesicles must undergo characterization through the quantitation of exosomes using either protein concentration or particle count. In addition, identification of at least two positive EV protein markers (one transmembrane and one cytosolic marker) is required, along with one source-appropriate negative (non-EV protein marker). Furthermore, two complementary single-vesicle analysis approaches should be employed to investigate morphology and biophysical properties, including count and size distribution.
Among the 30 studies, only 5 fulfilled these criteria.33,39,51,53,57 This indicates a notable gap in adherence to standardized characterization protocols within the broader research landscape on exosomes. Moreover, seven studies reported the purity of exosomes by examining the presence of negative/depleted markers, specifically non-EV contaminants.33,39,42,51,53,56,57 This emphasizes a subset of studies that went beyond mere quantitation and positive marker identification, taking into account the potential presence of contaminants.
The limited number of studies meeting the MISEV2018 criteria suggests the need for increased awareness and adherence to standardized guidelines in the field of exosome research. Consistent and rigorous adherence to these guidelines will enhance the reproducibility and comparability of results, fostering a more robust and reliable foundation for advancements in exosome-related studies.
The therapeutics used as exosomal cargoes
The findings offer a comprehensive overview of the diverse range of therapeutics employed as exosomal cargoes in the context of exosome-based drug delivery systems for breast cancer treatment. The 30 studies included in the analysis showcased a variety of therapeutic agents utilized in these systems (Figure 3).31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60
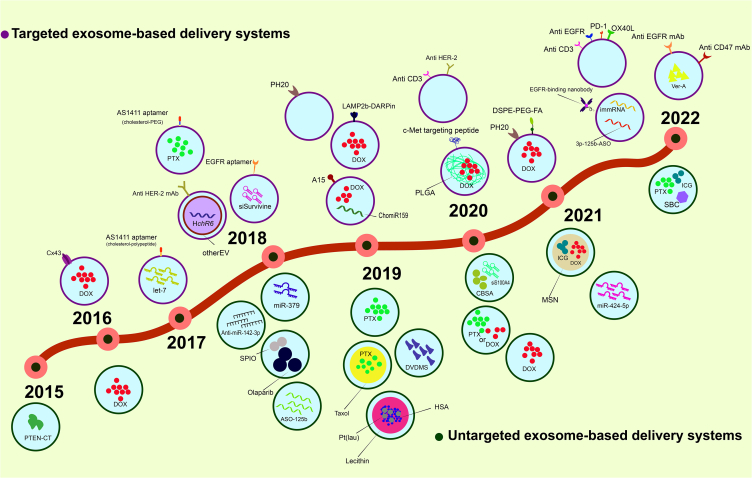
Figure 3.
Time line
A schematic figure shows the timeline of the studies which were assessed in this systematic review.
Chemotherapy drugs, such as paclitaxel (PTX) and DOX, were prominently featured in four studies, emphasizing their relevance in the development of exosome-mediated drug delivery strategies for breast cancer.34,35,45,46 In addition, four studies encapsulated exosomes with antitumor RNAs, including microRNAs miR-424-5p and miR-379, anti-microRNAs such as LNA (locked nucleic acid)-antimiR-142-3p, and antisense oligonucleotide (ASO-125b).32,40,41,57
Six studies utilized a combination of several types of nanoparticles and therapeutic materials.33,36,44,51,55,60 For example, Zhao et al. used cationic bovine serum albumin (CBSA) and small interfering RNA (siRNA) against S100A4 (siS100A4) as exosome cargoes.33 Another study carried out by Tian et al., developed a drug delivery system through loading indocyanine green (ICG) and DOX into the porous silicon nanoparticles (MSNs), designated as ID@E-MSNs, which were then encapsulated with the 4T1-derived exosomes.36 Jung et al. applied exosomes loaded with supermagnetic iron oxide (SPIO) nanoparticles and Olaparib to develop a magnetic particle imaging (MPI) system based on theranostic exosomes.44
Cao et al. also generated a drug delivery system by adding ICG, sodium bicarbonate (SBC), photoacoustic imaging agent (PA), and PTX into exosomes to be used as breast cancer therapy or imaging system.51 Melzer et al. developed a drug delivery system by loading liposomal PTX into human MSC-derived exosomes.55 In addition, Xiong et al. designed an exosome-based delivery system using the nanoparticles that included laurate functionalized Pt(IV) pro-drug (Pt(lau)) and human serum albumin (HSA), which were then stabilized by lecithin and loaded into RAW 246.7-derived exosomes (Rex).60
Furthermore, nine studies focused on targeted exosomes with surface molecules for specific therapeutic agent delivery to cancer cells.31,37,38,39,42,43,47,48,49 Among these, Wang et al. generated exosomes containing the humanized E. coli nitroreductase (HChrR6) mRNA, and incubated them with EVs displaying anti-HER2 scFv (high-affinity single-chain fragment variable antibody against human epidermal growth factor receptor 2) on the surface, enabling them to target HER-2+ human breast tumors.31 Li et al. used exosomes containing DOX-loaded PLGA (polylactic-co-glycolic acid) and surface modified them by adding a peptide targeting mesenchymal-epithelial transition factor (c-Met) with the ability to selectively treat TNBC.43 Hong et al. developed a delivery system utilizing the genetically modified exosomes capable of overexpressing human hyaluronidase (PH20) combined with anti-programmed death ligand 1 (PD-L1) to trigger immune response in breast cancer.53 Similarly, Feng et al. fabricated a DOX@Exos-PH20-FA delivery system using exosomes overexpressing PH20 on the surface and encapsulating DOX. This drug delivery system could target hyaluronan (HA), a hallmark of a wide range of solid tumors, with the help of folic acid (FA) displayed on the surface.49
Gong et al. loaded DOX as well as cholesterol-modified miR-159 (Cho-miR-159) into the THP-1 cell-derived exosomes induced by phorbol 12-myristate 13-acetate to overexpress exosomal metalloproteinase 15 (A15-Exo) for targeted delivery to TNBC cells.47 Pi et al. packaged survivin siRNA within the modified exosomes displaying epidermal growth factor receptor aptamer on the surface to specifically deliver the cargo to TNBC cells.38 Martin-Marques et al. produced exosomes containing DOX and expressing gap junction protein connexin43 (Cx43) on the surface.42
Interestingly, Peng et al. loaded immunomodulatory RNA (immRNA) and 5′ triphosphorylated antisense oligonucleotide (3p-125b-ASO) into exosomes derived from RBCs displaying an epidermal growth factor receptor EGFR-binding nanobody on the surface for targeted immune therapy against breast cancer and its metastatic form.39 Similarly, Gomari et al. used genetically engineered exosomes displaying lysosome-associated membrane glycoprotein 2b and designed ankyrin repeat protein (LAMP2b DARPin) on the surface and loaded them with DOX to selectively target HER-2+ breast cancer cells.48 Wan et al. and Wang et al. employed DC-derived exosomes displaying the polyethylene glycol or polypeptide-cholestrol-AS1411 aptamer, which binds to nucleolin, a protein overexpressed in breast cancer cell membranes, and loaded them with PTX or lethal-7 (let-7) miRNAs.58,59 A drug delivery system developed by Si et al. to deliver verrucarin A (Ver-A) used exosomes expressing EGFR and CD47 on the outer face.37
In addition to the aforementioned studies, two reports were found on genetically manipulated exosomes capable of activating anticancer immunity and inducing cancer cell death through surface-displayed antibodies against CD3+ human T cells, EGFR, programmed death 1 (PD-1) and OX40 ligand (OX40L), or anti-HER-2 antibodies.50,56 In another study, Liu et al. used tumor-derived exosomes to deliver sinoporphyrin sodium (DVDMS) toward breast tumors for sonodynamic therapy.54 Finally, Ahmed et al. developed a novel exosome-mediated system for delivering the C terminus of phosphatase and tensin homolog (PTEN-CT) into various cancer cells.52
The diverse range of therapeutics used as exosomal cargoes in the explored studies showcases the richness of innovation in the field of exosome-based drug delivery for breast cancer treatment. These findings hold promise for the development of more effective and targeted therapeutic strategies in the ongoing efforts to combat breast cancer.
Approaches for generating exosome-based drug delivery systems
The results presented shed light on the diverse approaches employed for generating exosome-based drug delivery systems, reflecting the dynamic landscape of innovation in this field. As stated earlier in the introduction, several techniques have been devised for loading cargo into exosomes. The findings from the 30 studies explored in this section underscore the multiplicity of strategies adopted by researchers.
Six distinct studies investigated the incubation method for establishing an exosomal drug delivery system.33,45,47,51,54,55 This approach involves the co-incubation of therapeutic agents with exosomes, allowing for the passive loading of cargoes into the vesicles. In addition, three studies used sonication to load therapeutic agents into the exosomes,34,35,41 demonstrating the versatility of physical methods in cargo loading. An additional approach involved electroporation, with the goal of developing an exosome-based drug delivery system.42,46,57 Electroporation involves the application of electric pulses to create transient pores in the exosome membrane, facilitating the loading of therapeutic agents.
In addition, five studies utilized an indirect durg-loading approach by employing engineered cells.32,40,50,53,56 This innovative strategy involves modifying parent cells to produce exosomes loaded with therapeutic agents, providing a unique and controlled approach to cargo incorporation. Furthermore, a combined strategy, including the genetically cell engineering method to produce targeted exosomes and electroporation/incubation technique to load druggable molecules into the exosomes, was explored by four different groups.31,48,49,52 This hybrid approach leverages the strengths of both methods to enhance the specificity and efficiency of exosome-based drug delivery.
Finally, nine different studies evaluated a combination of direct methods, including electroporation, EV transfection, and sonication to load therapeutic agents into exosomes. Subsequently, the surface of the exosomes was modified using a ligation method.36,37,38,39,43,44,58,59,60 This comprehensive approach integrates multiple loading techniques with surface modification, offering a multifaceted strategy for tailoring exosomes for specific therapeutic applications.
The diversity in the approaches for generating exosome-based drug delivery systems reflects the complexity of the field and the continuous exploration of novel methods. Each approach has its unique advantages and challenges, and the selection of a specific method may depend on the nature of the therapeutic cargo, the desired targeting specificity, and the intended application.
Quantities and concentrations of exosome-based drugs
The studies varied significantly in the quantities of exosomes used as therapeutic drug carriers. The range of quantities across the 30 studies indicates a lack of standardized dosing, reflecting the experimental nature of exosome-based drug delivery.
All 30 studies utilized a wide range of quantities of exosomes as therapeutic drug carriers.31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60 Among these, five studies specifically reported the number of exosomes as nanocarriers. In this regard, Wang et al. used 2 × 109 particles in 100 μL of phosphate buffered saline (PBS). O’Brien et al. and Haney et al. applied 2.6 × 107 and 1 × 109 of exosomes, respectively. Shi et al. and Usman et al. used 1.5 × 1010 and 8.25 × 1011 exosomes, respectively.31,40,45,56,57 This specificity in reporting nanocarrier quantities suggests a growing awareness of the importance of precision in exosome dosage, a critical factor in optimizing therapeutic outcomes. The variability may stem from different therapeutic goals, targeted diseases, or experimental conditions.
Ten studies only reported information on the concentration of therapeutic agents loaded into the exosomes without specifying the dose of the exosomes used.33,35,36,37,42,46,47,51,58,60 Interestingly, Li et al. and Melzer et al. reported both the volume of the injected exosomes and the concentration of the loaded therapeutic agents.43,55 Two studies addressed the concentration of the exosomes used, with Zhou et al. employing 30 μg/100 μL and Jung et al. using 100 μg/mL of exosomes.32,44 Four studies stated the amount of the administered exosome in the animal models based on the weight of the mice models.38,39,50,54 This approach adds a layer of complexity to dosing considerations, acknowledging the importance of adapting dosage to the characteristics of the recipients. Furthermore, five studies documented the final amount of exosomes in terms of micrograms, with Naseri et al. applying 30 μg, Ahmed et al. and Wang et al. using 150 μg, Hong et al. employing 50 μg, and Feng et al. utilizing 200 μg of exosomes per 100 μg of prescriptive DOX.41,49,52,53,59 This divergence in concentration suggests ongoing exploration of optimal conditions for drug loading and delivery efficiency.
While the variability in quantities and concentrations reflects the early stages of research, it also underscores the importance of refining dosing strategies for optimal therapeutic efficacy. The results highlight the need for further standardization in the field of exosome-based drug delivery.
Assessment of drug loading
The results highlight the diversity of methodologies used to assess drug loading, emphasizing the complexity of characterizing the encapsulation process. The loading and encapsulation efficiency of drugs have been presented in 21 studies, either qualitatively or quantitatively.33,34,36,38,41,42,43,44,45,46,47,48,49,51,52,54,55,57,58,59,60 Among these studies, 5 pointed out the absorbance-based method, where the absorbance of drugs such as DOX was compared before and after loading into the exosomes to determine the encapsulation yield.34,42,46,48,49,54 Similarly, Naseri et al. reported the yield of LNA-anti-miR-142-3p encapsulation into the delivering exosomes by measuring the absorbance at 260 nm before and after the ultracentrifugation process.41
Two studies used gel shift assay to show the encapsulation process of RNAs into the therapeutic exosomes.33,38 Another two studies carried out by Usman et al. and Wang et al. used quantitative reverse-transcription PCR (qRT-PCR)-based techniques to quantify the loading of RNAs into exosomes,57,59 providing a molecular-level assessment of the encapsulation process. These methods contribute to a more nuanced understanding of RNA encapsulation efficiency.
Jung et al. utilized TEM and MPI techniques to demonstrate the encapsulation of SPIO in the theranostic exosomes.44 Likewise, Li et al. used TEM and DLS analysis to report the encapsulation of PLGA and DOX into exosomes.43 Gong et al. reported the encapsulation of DOX and cho-miR159 into the exosome-based delivery system by fluorescence microscopy images.47 Haney et al. also used a similar method based on fluorescence microscopy images to reflect the encapsulation of PTX or doxorubicin into the macrophage-derived exosomes as a drug delivery system.45 These imaging-based methods offer direct visualization of the encapsulation process, providing valuable insights into the distribution of loaded agents within exosomes.
Ahmed et al. employed trypsin digestion and western blotting to verify the encapsulation of PTEN-CT into exosomes.52 Cao et al. and Melzer et al. applied liquid chromatography-tandem mass spectrometry (LC-MS/MS) to quantify the amount of ICG and PTX loaded into the therapeutic exosomes.51,55 This high-precision method ensures accurate measurement of drug concentrations, contributing to a quantitative understanding of drug loading.
Tian et al. reported the encapsulation of MSN, ICG, and DOX into exosomes using the Avanti mini extruder.36 Furthermore, Xiong et al. determined the encapsulation efficiency of HSA/Pt(IV)/lecithin into the therapeutic exosomes using graphite furnace atomic absorption spectroscopy (GFAAS),60 demonstrating the versatility of techniques used in drug quantification. Finally, Wan et al. measured the amount of the loaded PTX employing high-performance liquid chromatography (HPLC).58
The diverse array of techniques employed in assessing drug loading in exosome-based drug delivery systems reflects the multidisciplinary nature of this field. Each method brings its unique advantages, contributing to a comprehensive understanding of encapsulation efficiency. As this area of research continues to evolve, the combination of multiple assessment techniques and the standardization of methodologies will be crucial for establishing robust protocols in the development of effective exosome-based drug delivery systems.
Stability of the exosome-based drug delivery systems
In addition to the importance of in vitro stability of exosomes, the stability of exosomes in blood/serum is crucial due to their interaction with blood proteins, which is a prerequisite for in vivo experiments.
Overall, 17 studies reported on the stability of exosome-based drug delivery systems.33,34,38,43,44,46,47,48,51,52,54,55,56,57,58,59,60 Among these, two studies used a gel shift assay to assess the stability of the therapeutic exosomes containing RNAs.33,38 This method allows for the visual confirmation of stability by examining the interactions between exosomes and loaded RNAs. Six studies determined the size of the exosomes encapsulating the therapeutic agents by NTA and DLS to reveal exosome stability in serum, PBS, or a cell culture medium under various storage conditions.43,44,51,54,58,60 However, Melzer et al. used an NTA-based technique to measure particle mobility to confirm the stability of the liposomal PTX (Taxol)-loaded exosomes.55 This method contributes to a nuanced understanding of exosome stability by examining particle dynamics.
Xie et al. demonstrated the stability of exosomes containing DOX for at least 4 h in mouse serum using a flow cytometry-based approach.34 Likewise Usman et al. assessed the stability of PKH26-labeled EVs encapsulating ASO-125b in mice blood using a flow cytometry-based approach,57 providing real-time insights into the stability of the delivery system in a physiological environment. Shi et al. reported that synthetic multivalent antibody retargeted exosomes (SMART-Exos) had stability in mice blood for at least 6 h using an enzyme-linked immunosorbent assay (ELISA)-based method,56 showcasing the versatility of techniques employed to assess stability.
Hadla et al. performed an experiment in which stability of the exosomes containing DOX was investigated by measuring the DOX concentration in a cell culture medium.46 This approach provides a direct assessment of the stability of the therapeutic cargo in a relevant environment.
On the other hand, a few studies were focused on the stability of the cargoes encapsulated into the exosomes. For instance, Ahmed et al. addressed the stability of PTEN-CT loaded into exosomes using trypsin and western blotting,52 offering a specific analysis of the stability of the therapeutic cargo. Usman et al. measured the fluorescence of labeled RNAs electroporated into exosomes using a microplate reader,57 providing a quantitative measure of cargo stability. Finally, two studies simply stated that the exosomes used as a delivery system were stable without presenting any supporting evidence.47,48
The results underscore the multi-faceted nature of stability assessment in exosome-based drug delivery systems. The diverse array of techniques used in these studies reflects the complexity of evaluating stability in different contexts. As this field advances, standardization of stability assessment protocols and a more detailed presentation of evidence will be crucial for establishing the reliability and reproducibility of exosome-based drug delivery systems in both in vitro and in vivo settings.
Routes of administration and injection frequency of exosome-based drug delivery systems
The choice of administration routes and injection frequencies plays a pivotal role in determining the efficacy and targeted delivery of exosome-based drug delivery systems. Three routes of administration employed in all 30 studies included intratumoral (i.t.), intravenous (i.v.), and intraperitoneal (i.p.) injections.31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60 This variety reflects the versatility of exosome-based drug delivery systems, allowing researchers to tailor their approaches based on specific therapeutic goals, targeted tissues, and the nature of the cargo.
Tail vein injections were utilized in 21 studies to deliver therapeutic exosomes to mouse models.33,34,35,36,37,38,40,41,43,45,47,48,49,50,51,54,55,56,58,59,60 This route is advantageous for systemic delivery, allowing for the distribution of exosomes throughout the body and facilitating their interaction with target tissues. The widespread use of tail vein injections indicates the interest in systemic delivery and the potential for targeting multiple organs or tissues simultaneously.
Four groups delivered cargoes entrapped in exosomes intratumorally to animal models,32,42,44,57 highlighting the interest in localized delivery of therapeutic cargo. Two studies compared i.t. and i.v. routes of injection in different groups and reported their respective findings.39,52 Comparing i.t. and i.v. routes of injection provide valuable insights into the advantages and limitations of these two delivery methods, contributing to the optimization of targeted therapies.
Hadla et al. administered exosomes containing DOX intraperitoneally to mouse models.46 Wang et al. administered therapeutic exosomes loaded with HChrR6 mRNA intraperitoneally, while 6-chloro-9-nitro-5-oxo-5H-benzo(a)phenoxazine, the substrate of HChrR6 enzyme, was intravenously injected into the mice to examine the therapeutic effects of the exosome-based drug delivery system.31 Furthermore, Hong et al. administered exosomes displaying PH20 intratumorally, while anti-PD-L1 was administered intraperitoneally.53
The variety in routes of administration across these studies emphasizes the need for tailored approaches in exosome-based drug delivery. The selection of a specific route depends on factors such as the therapeutic goal, targeted tissues, and the nature of the cargo. As this field advances, a deeper understanding of the advantages and limitations of each administration route will contribute to the development of more effective and targeted exosome-based drug delivery systems. In addition, the consideration of injection frequency is crucial for determining the sustained therapeutic effects and optimizing the overall treatment regimen.
In vivo techniques and the related outcomes associated with the exosome-based drug delivery system
An overview of the biodistribution assessment and in vivo antitumor effects of exosome-based drug delivery systems across 30 studies highlights the significance of understanding the distribution patterns of therapeutic agents and the subsequent impact on tumor reduction and overall antitumor efficacy.
Biodistribution assessment
Biodistribution serves as a crucial method to validate the tumor targeting of exosome-based drug delivery systems in vivo. Among the 13 studies focusing on biodistribution, various strategies, such as in vivo or ex vivo fluorescence/bioluminescence imaging, were utilized to assess the biodistribution of the delivery systems in animals or organs such as the heart, kidneys, liver, lungs, spleen, brain, and tumor. These 13 studies generally suggest that exosomes can enhance the tropism of the therapeutic agents toward specific regions.33,36,38,41,43,47,48,49,51,54,57,59,60 Out of these 13 studies, five employed a targeted exosome-based drug delivery system displaying specific molecules such as peptides and other ligands on the exosome surface to bind to target cells. They demonstrated that targeted exosomes exhibited efficient uptake by specific types of breast tumors with minimal off-target biodistribution.43,47,48,49,59
The diverse techniques used for biodistribution assessment, such as whole-body photoacoustic imaging, ex vivo imaging, and fluorescence imaging, contribute valuable insights. In one study, Cao et al. used whole-body PA imaging to assess the accumulation of drug-loaded exosomes in the mouse model.51 Furthermore, Xie et al. isolated major organ tissues from mouse models to determine the biodistribution of DOX-loaded exosomes using ex vivo imaging. They reported that DOX loaded into the exosomes had been accumulated in the lungs more than the liver, suggesting the potential of exosomes in reducing lung metastatic nudes.34 Likewise, Ahmed et al. utilized fluorescence imaging to assess the biodistribution of PTEN-CT exosomes in cryo-sections of the heart, lungs, liver, kidneys, and spleen.52
In another study performed by Haney et al., mouse model tissues were used to compare the difference between the biodistribution of the drug-loaded exosomes in solid tumor tissues using different routes of injection such as i.p., i.t., and i.v. The results indicated the i.v. injections as the most effective route.45 Peng et al. employed a flow cytometry-based approach to track the intrapulmonary distribution of the EGFR-targeted RBC-derived exosomes.39 Jung et al. utilized microcomputed tomography and MPI methods to assess the biodistribution of SPIO- and Olaparib-loaded exosomes in the whole body of the tumor-bearing animal model.44
Interesting, we found only one study in which the biodistribution of DOX-loaded exosomes was evaluated in tumor tissues of the mouse model using LC-MS/MS analysis.46 In addition, Wan et al. employed an HPLC technique to investigate the biodistribution of the targeted exosome-based PTX delivery system that displayed AS1411 aptamer on surface.58
The findings emphasize the importance of biodistribution assessment in validating the effectiveness of exosome-based drug delivery systems. The diverse range of techniques employed across these studies provides valuable insights into the potential of exosomes for targeted and efficient drug delivery in various in vivo settings.
In vivo antitumor effect of the exosome-based drug delivery system
The findings span various aspects, including reductions in tumor size, volume, and weight, anti-metastatic effects, prevention of body weight loss, increased survival rates, and additional insights gained through histopathology and functional evaluations.
Antitumor efficacy
All 30 studies investigated the in vivo antitumor effects of the exosome-based drug delivery system.31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60 Among these, several studies indicated that the administration of the therapeutic exosomes in mouse tumor-bearing models led to a reduction in tumor size, tumor volume, and tumor weight, thereby demonstrating the in vivo antitumor efficacy of the exosome-based drug delivery system.31,32,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57 This uniformity across a diverse range of studies underscores the reliability and repeatability of the observed antitumor effects.
In addition to the general antitumor impacts of the therapeutic exosomes, five studies emphasized that the therapeutic-loaded exosomes exhibited in vivo anti-metastatic effects by reducing the metastatic nodes in the lungs of the mouse models.33,34,49,54,60 This multifaceted impact on both primary and metastatic tumors enhances the therapeutic value of exosome-based drug delivery systems.
In addition, 12 studies revealed that the use of the encapsulated anticancer therapeutics prevented the body weight loss in mouse models, which is a common side effect of the cancer therapy.35,36,37,43,46,49,50,51,53,56,58,60 This suggests that exosome-based therapies may not only be effective in reducing tumors but also in minimizing adverse effects commonly associated with cancer treatments.
Across all the analyzed 30 studies, four specifically emphasized that the exosome-based cancer therapy resulted in increased survival rates among the mouse models, further highlighting the efficacy of the cancer therapy using therapeutic exosomes.35,41,47,53 This is a key metric that directly translates to the potential clinical benefits of such therapeutic interventions.
Validation through histopathology
A proper method for validating the findings of in vivo antitumor effects of exosome-based drug delivery systems is histopathology, which was employed by 26 studies.33,34,35,36,37,39,40,41,42,43,44,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60 Histopathology serves as a powerful tool, offering both qualitative and quantitative insights into the structural and molecular alterations within tumor tissues, thus providing a comprehensive validation of the therapeutic efficacy.
Among these studies, three specifically applied hematoxylin and eosin (H&E) staining to visualize metastatic nudes in the mice lung tissues treated with the exosome-based drug delivery systems.33,34,49 This technique provides a direct and microscopic assessment of the structural changes in tumor tissues, offering a qualitative validation of the therapeutic effects. The visualization of metastatic nodes in the lungs of mouse models treated with exosome-based drug delivery systems establishes a direct correlation between treatment and structural changes, confirming the in vivo antitumor efficacy.
The application of the terminal deoxynucleotidyl transferase dUTP nick labeling (TUNEL) assay adds a quantitative dimension to the validation process, specifically highlighting the increased apoptotic rates following treatment. This not only supports the antitumor efficacy but also provides insights into the mechanisms underlying the therapeutic impact of exosome-based drug delivery systems. Feng et al. revealed that the administration of DOX@Exos-PH20-FA increased apoptotic rate in all the treated animal groups using the TUNEL assay.49 Peng et al. observed the apoptotic process in tumor tissues of the mouse model by employing TUNEL staining, and they also visualized the metastatic zones in the lung tissues using H&E staining following treatment with exosomes loaded with immRNAs.39 Similarly, Wang et al. utilized the TUNEL assay to demonstrate the apoptosis mechanism in mice tumor tissues. They also utilized H&E staining to assess the side effects of PTX loaded in M1 macrophage-derived exosomes in various organs such as the heart, liver, spleen, lungs, and kidneys.35 Li et al. reported apoptotic induction and antitumor effects of exosomes encapsulating PLGA and DOX, and displaying a c-Met binding peptide on the surface targeted toward TNBC mice models.43
Immunohistochemistry (IHC) antibody staining techniques used by several studies enable the visualization and quantification of specific proteins associated with antitumor effects. This adds specificity to the validation process, linking observed outcomes to molecular changes in tumor tissues. In the study conducted by Tian et al., the anticancer effects of the ID@E-MSNs drug delivery system using H&E and Ki67 IHC antibody in both non-treated and treated tumor tissues of mouse models were unveiled.36 In a separate study, Si et al. prepared targeted exosomes enclosing Ver-A and displaying antibodies against EGFR and CD47 on their surface to evaluate both its antitumor properties and the related side effects in a mice model. They demonstrated that the exosomes specifically targeted the tumor tissues expressing EGFR and CD47 using the IHC approach, while other tissues were not influenced by the targeted exosomes verifying by H&E staining.37 O’Brian et al. stained tumor tissues of the mice models by H&E to reveal the in vivo antitumor effects of miR-379-enriched exosomes. In addition, they used IHC to demonstrate the in vivo anti-angiogenic activity of the exosomes encapsulated with miR379. The rate of tumor growth in both treated and untreated groups was compared using ultrasound and oxygenated/deoxygenated photoacoustic imaging to provide a standard approach for assessment.40
In alignment with this study, Gomari et al. conducted an H&E experiment on tumor tissues of mouse models to verify the significant antitumor activity of DOX loaded in the targeted exosomes. They also utilized an IHC test to confirm the presence of HER-2 ligands for targeted therapy.48 Two other studies used H&E staining of the tumor tissues as well as various animal organs such as the heart, kidneys, liver, spleen, and lungs to assess the in vivo antitumor activity and potential side effects of the therapeutics loaded into exosomes.41,51 H&E and Sirius Red staining were used by Martin-Marques et al. to perform histological analysis on the heart tissues and immunofluorescent staining to assess the presence of Cx43 in doxorubicin-loaded exosomes. Their findings demonstrated a reduction in the doxorubicin-induced cardiotoxicity due to the targeted uptake of the exosome-based delivery system by tumor cells. The study also evaluated apoptosis through analysis of B cell lymphoma 2 protein (Bcl-2) and Bcl-2-associated X protein (Bax) expression levels in tumor sections following treatment.42 Consistently, Hadla et al. also reported lower cytotoxicity as the side effect of DOX for the exosomes loaded with DOX, examined by H&E staining of the heart tissues.46
The combination of H&E staining with other techniques, such as immunofluorescent staining, RT-PCR, and western blot analysis, as demonstrated by various studies, enables a comprehensive assessment of both antitumor activity and potential side effects. This multidimensional approach ensures a thorough understanding of the treatment’s impact on tumor cells, surrounding tissues, and major organs.
Jung et al. employed three methods, including H&E staining, fluorescent imaging of labeled exosomes, and IHC staining against cleaved caspase-3 to explore the antitumor activity of Olaparib/SPIO-loaded exosomes.44 Gong et al. used H&E staining for tumor and heart tissues in mouse models and employed IHC staining with anti-Ki-67, CD31, transcription factor 7 (TCF7), and MYC antibodies in the tumor tissues to confirm the antitumor and the related side effects of the targeted exosomes expressing metalloproteinase 15 (A15) on the surface and loaded with DOX.47
Furthermore, Cheng et al. reported on the anticancer effect of the immune-modulating exosomes (GEMINI-Exos) through modulating the immune system. To this end, they expressed the antibodies specific for human T cell CD3, EGFR, immune checkpoint modulators, PD-1, and OX40L on the surface. Immunohistofluorescence imaging was used to observe the infiltration of T cells into breast tumors implanted in the mouse models, reflecting the anticancer activity of GEMINI-Exos.50 Ahmed et al. utilized the IHC method to examine the expression of PTEN, Ki-67, and caspase-3 in tumor tissues. Their study verified the antitumor activity of the PTEN-CT-loaded exosomes in the animal groups.52 In addition, Liu et al. employed H&E and IHC methods, using antibodies against proliferating cell nuclear antigen (PCNA) and matrix metalloproteinase 9 (MMP9), as well as the TUNEL assay on tumor or lung sections in tumor-bearing mice models. Their findings demonstrated the antitumor and anti-metastatic activity of EXO-DVDMS as a sonodynamic therapy strategy.54
Melzer et al. reported that Taxol-loaded exosomes can reduce metastasis of breast tumor cells. They used fluorescent microscopy and RT-PCR on tissue sections of the mouse models to validate their findings.55 Based on the H&E staining of tumor and lung sections, Usman et al. demonstrated that ASO-125b encapsulated into the RBC-EVs reduced breast-tumor cell proliferation and metastasis to the lungs.57 Xiong et al. utilized the TUNEL assay and the IHC technique with antibodies against Bax, Bcl2, PCNA, and p53 on tumor sections collected from the mouse models. Their study reported that nanoparticles (NPs)/Rex had antitumor effects and highlighted that exosomes reduced the side effects of Pt(IV) using the H&E staining for the major organ tissues of animal models.60
Furthermore, Wan et al. revealed an antitumor property for the targeted DC-derived exosomes encapsulating PTX and displaying AS1411 aptamer on the surface. They used H&E and Ki-67 IHC staining techniques to support their findings.58 In contrast, Wang et al. loaded let-7 miRNA into the exosomes displaying AS1411 aptamer to induce tumor cell death. They utilized western blot analysis to show the expression of cMyc and Kirsten rat sarcoma virus (KRAS) proteins. The study also confirmed the reduction of off-target effects of this delivery system through H&E staining for the major organ tissues in mice.59
The incorporation of immunofluorescence microscopy and flow cytometry in studies reveals the immune-related aspects of exosome-based therapies. Hong et al. used immunofluorescence microscopy to observe tumor sections of mouse models and assess the degradation of HA caused by MSC-derived exosomes displaying PH20. They also demonstrated a number of tumor-infiltrating CD8+ T cells in tumor-draining lymph nodes through flow cytometry.53 Similarly, Shi et al. disclosed the infiltration of CD3+ T cells following the administration of αCD3-αHER2 SMART-Exos.56 They used immunohistofluorescence images and flow cytometry to confirm their findings.
The collective use of histopathological techniques in the evaluated studies establishes a robust foundation for validating the in vivo antitumor effects of exosome-based drug delivery systems. From structural assessments to quantitative apoptosis measurements and molecular correlations, these studies showcase the versatility and depth of histopathological validation.
The comprehensive evaluation of exosome-based drug delivery systems extends beyond traditional histopathological analysis, encompassing a spectrum of functional assessments to provide a holistic understanding of their impact on various physiological and immunological parameters. The studies conducted collectively contribute valuable insights into the immunomodulatory effects, systemic responses, and safety profiles of exosome-based therapies.
Tian et al. conducted functional evaluations of the liver and kidney in the mice treated with ID@E-MSN. They measured the level of alanine aminotransferase (ALT), aspartate aminotransferase (AST), creatinine (CRE), and blood urea nitrogen in the blood samples to assess liver and kidney function.36 This analysis ensures that the therapeutic interventions do not compromise the normal functioning of these vital organs, reinforcing the safety profile of the exosome-based drug delivery system. The titer of different immunological factors was also checked in the mice by Peng et al., who explored the immunomodulatory effect of engineered exosomes.39 The measurement of various immunological factors unveils the potential of exosomes to modulate immune responses, shedding light on their broader impact beyond direct antitumor effects.
Feng et al. determined the hyaluronidase activity of modified exosomes displaying FA and PH20 on the surface, signifying a nuanced exploration of the enzymatic functions associated with exosome interactions. They quantified the levels of interferon-gamma and tumor necrosis factor alpha (TNF-α) in the serum using the ELISA and assessed the percentage of the CD3+CD8+ T cells by flow cytometry,49 providing a detailed understanding of the immunological responses triggered by these modified exosomes.
Similar to the latter study, Cheng et al. investigated CD8+ and T regulatory cells and determined ALT and CRE levels in the blood samples of the mouse models,50 delving into the immune cell dynamics influenced by exosome-based therapies. Xiong et al. reported that exosomes encapsulated with human serum albumin (HSA) and Pt(IV) induced an immune response and reduced the side effects of Pt(IV) underscoring the intricate balance between therapeutic efficacy and safety. They quantified the levels of TNF-α, IL-6, and IL-12, as well as hemolysis, alkaline phosphatase (ALP), AST, and ALT in the serum of the mouse models,60 unveiling the systemic impacts of exosome treatments, guiding the assessment of their overall safety and potential immune-stimulating effects.
Wan et al. conducted a comprehensive range of blood tests to examine the levels of white blood cells, RBCs, ALT, ALP, AST, urea, and CRE in tumor-bearing mice with exosomal PTX. The exosomal PTX had lower side effects than the naked PTX.58 This thorough examination aids in delineating the hematological and biochemical consequences, crucial for understanding the overall impact on the physiological well-being of the treated animals.
The inclusion of these diverse functional assessments enriches the understanding of exosome-based drug delivery systems. Beyond their antitumor efficacy, these studies illuminate the intricate interactions with the immune system, validate the safety of the therapeutic interventions through organ function evaluations, and provide a foundation for considering exosomes not only as drug carriers but also as immunomodulatory agents. The assessment of immunomodulatory effects, hyaluronidase activity, and cytokine levels further broadens the scope of the evaluation, considering the complex interactions between the immune system and the administered exosomes.
The comprehensive exploration of biodistribution patterns and in vivo antitumor effects of exosome-based drug delivery systems demonstrates the versatility and effectiveness of these delivery platforms. The integration of advanced imaging techniques, quantitative analyses, and histopathological assessments contributes to a nuanced understanding of the therapeutic potential and safety profile of exosome-based drug delivery systems in vivo.
In vitro techniques and outcomes
The collective findings from the in vitro assessments underscore the remarkable potential of exosome-based drug delivery systems in targeting breast cancer cells. The integration of various techniques, from traditional 3-(4,5-dimethylthiazol-2-yl)-2.5-diphenyl tetrazolium bromide (MTT) assays to advanced molecular and imaging methods, provides a robust foundation for validating the therapeutic efficacy. Moreover, the identification of anti-metastatic effects broadens the scope of these systems, positioning them as promising candidates for comprehensive cancer treatment strategies.
Cellular uptake and cellular binding assessment
The extensive evaluation of cellular uptake and binding mechanisms is a cornerstone in understanding the effectiveness of exosome-based drug delivery systems. This comprehensive assessment, spanning 30 studies, delves into the intricacies of how these systems interact with tumor cells, utilizing a myriad of techniques to provide a nuanced understanding of their behavior.
To evaluate the uptake or binding of exosome-based drug delivery systems, a comprehensive assessment was conducted across 30 studies.31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60 Among these, 25 studies used fluorescent agents such as PKH-26, PKH-67, 3,3-dioctadecyloxacarbocyanine, cyanine-3, carboxyfluorescein succinimidyl ester, CD63 fusion green fluorescent protein, or ICG. Confocal or inverted fluorescent microscopes were used to track the uptake by tumor cells.31,32,33,34,35,36,37,39,41,43,44,45,46,47,48,51,52,53,54,55,56,57,58,59,60 This traditional yet robust approach allows for real-time visualization, offering insights into the initial stages of cellular interaction.
Out of the 25 studies, 7 incorporated alternative methods in addition to fluorescent imaging.31,37,39,48,54,55,56 Techniques such as quantitative PCR (qPCR), FD500-uptake assay, flow cytometry, and ELISA, showcase the versatility in confirming and quantifying cellular uptake and binding. For instance, Peng et al. employed qPCR to confirm the uptake of the 3p-125b-ASO- and immRNA-loaded exosomes by 4T1 cells.39 Liu et al. employed an FD500-uptake assay to approve the uptake of EXO-DVDMS by breast cancer cells.54 Furthermore, Gomari et al., Si et al., Wang et al., Melzer et al., and Shi et al. used a flow cytometry approach to demonstrate the binding ability of the selective drug-loaded exosomes to targeted breast cancer cells.31,37,48,55,56 Wang et al. and Usman et al. used qPCR to verify the uptake of therapeutic exosomes encapsulating let-7 miR or ASO-125b by target cells.57,59 Pi et al. measured the fluorescent intensity of the Alexa Flour-tagged siRNA to verify the uptake of the siRNA-loaded exosomes by cancer cells.38 Interestingly, Feng et al. utilized a flow cytometry-based assay to explore the selective entry of drug-loaded exosomes via determining the intracellular concentration of DOX.49 By determining the intracellular concentration of DOX, they added a quantitative dimension, shedding light on the efficiency of drug delivery at the cellular level. Finally, Cheng et al. established an ELISA assay to follow the binding ability of the engineered exosomes to breast cancer cells.50 This quantitative approach contributes to a more precise assessment of the binding dynamics.
To further evaluate the subcellular distributions, Li et al., Xie et al., and Liu et al. employed fluorescent microscopy imaging to display lysosomal endocytosis of exosomes,34,43,54 while Xiong et al. utilized a GFAAS technique to demonstrate the subcellular localization of NPs/Rex in 4T1 cells through energy-expenditure endocytosis.60 This detailed examination contributes to understanding the fate of exosomes within cells, providing clues about potential pathways and mechanisms involved in their cellular processing.
These studies illuminate the complex interplay between exosome-based drug delivery systems and tumor cells. By employing a diverse array of techniques, researchers have not only visualized cellular interactions but also validated and quantified these processes, enhancing the robustness of their observations.
In vitro anticancer evaluation of the exosome-based drug delivery systems
The studies presented employ a variety of experimental approaches, ranging from conventional assays like MTT to more specialized techniques such as flow cytometry, luminescence assays, and enzymatic activity assays.
Across all 30 reported studies,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60 in vitro outcomes from diverse analytical approaches, which were considered complementary data, consistently confirmed the anticancer properties of the exosome-based delivery systems. One study reported that the targeted exosome-based delivery of HChrR6 mRNA resulted in cancer cell death, as assessed through MTT assay.31 Another study investigated the apoptotic effect of miR-424-5p-loaded exosomes as a delivery system on tumor cells. This drug delivery system was assessed by two different approaches: CaspaseGlo 3/7 and lactate dehydrogenase release assays. This study identified miR-424-5p as a key regulatory factor that modulates pro-inflammatory cytokines, leading to anticancer activity. In addition, qRT-PCR was employed to assess the expression of PD-L1 by miR-424-5p, leading to an enhanced efficacy of the system in inducing anticancer effects.34 The integration of qRT-PCR to assess PD-L1 expression further enhanced the understanding of the system’s anticancer effects.
Various studies utilized cytotoxicity assays such as MTT, cell counting kit-8 (CCK-8), and CellTiter-Glo luminescence assays to evaluate the impact of exosome-loaded drugs on cancer cells. Xie et al. utilized an MTT assay to assess either cytotoxicity effect of DOX-loaded exosomes or compare the cytotoxic effect between breast cancer cells (MDA-MB-231) and normal human umbilical vein endothelial cells (HUVECs).34 Similarly, Wang et al. proved the anticancer activity of PTX-loaded exosomes using MTT assay. They also utilized Annexin V/PI and caspase-3 activity assays to both measure caspase-3 activity and unveil the apoptotic effects induced by the modified exosomes. In addition, they detected the influence of exosomes on the release of pro-inflammatory cytokines, potentially beneficial in cancer therapy.35 Tian et al. employed the approach of CCK-8 to reveal the anticancer activity of ID@E-MSNs. They also performed Annexin V/PI assay and western blotting for the cleaved caspase-3/9 to validate apoptosis induction in the cancer cells treated with ID@E-MSNs.36 The use of Annexin V/PI and caspase-3 activity assays added depth by unveiling apoptotic effects induced by modified exosomes.
A single report among the 22 studies demonstrated the anticancer efficacy of the targeted exosomes displaying anti-EGFR and loaded with Ver-A, using a wide range of cytotoxicity assays on TNBC cells.37 Pi et al. also used qRT-PCR analysis and MTT assay to demonstrate the efficient delivery of survivin siRNA to cancer cells, and the corresponding in vitro anticancer activity.38 Consistent with other studies, Naseri et al. generated the exosomes capable of delivering miR-142-3p to breast cancer cells, which consequently caused cytotoxic and apoptotic effects.41 Martin-Marques et al. employed a variety of in vitro experiments, including the MTT assay, cell-cycle progression assay, 5-bromo-2′-deoxyuridine incorporation analysis, staining of Ki-67-positive cells, and Trypan blue exclusion to demonstrate the therapeutic efficacy of DOX loaded into the targeted exosomes.42 Li et al. validated the anticancer activity of the targeted exosomes displaying a peptide against c-Met and loaded with PLGA and DOX, using the MTT assay, TUNEL staining, and Annexin V/PI methods.43
On the other hand, Jung et al. applied CCK-8 instead of MTT assay, along with an apoptosis detection kit to evaluate the anticancer properties of Olaparib/SPIO-loaded exosomes.44 Hadla et al. and Haney et al. revealed the cytotoxic effects of the DOX/PTX-loaded exosomes via MTT and CellTiter-Glo luminescence assays.45,46 In addition, they verified that the designed exosome-based drug delivery system caused less cardiotoxicity than DOX alone.46
Gong et al. designed a drug delivery system based on exosomes exhibiting A15 on the surface, carrying DOX and Cho-miR159. Their findings showed an in vitro anticancer efficacy against breast cells, evaluated using the CCK-8 and Annexin V/PI assays. By employing qRT-PCR, they also identified miR159’s potential to reduce TCF7 expression level, enhancing the efficacy of the drug delivery system.47
Both Gomari et al. and Feng et al. developed targeted drug delivery systems by loading DOX into engineered exosomes. The first group employed LAMP2b DARPin and the second used PH20 and FA to specifically target breast cancer cells. They both utilized MTT assay to validate the cytotoxicity effects of the drug-loaded exosomes.48,49 Moreover, Hong et al. demonstrated exosomes overexpressing PH20 on the surface could stimulate cancer immunity responses by activating DCs through the degradation of HA, as measured by in vitro enzymatic activity and DC activation assays.53
Nguyen Cao et al. developed SBC-, ICG-, and PTX-loaded exosomes and used MTT and flow cytometry-based techniques to measure the intracellular reactive oxygen species (ROS) levels to show the cytotoxicity of the therapeutic exosomes. They also reported an apoptotic effect on breast cancer cells for this exosome-based drug delivery system using Annexin V/PI assay.51 Xiong et al. employed Annexin V/PI and caspase-3/7 activity assays to demonstrate the apoptotic effects of Rex delivering HSA/Pt(IV) on breast cancer cells.60
Wan et al. used PI staining and cell-cycle arrest techniques to uncover that DC-derived exosomes displaying AS1411 and encapsulated PTX reduced the proliferation and viability of breast cancer cells.58 Likewise, Wang et al. stated that DC-derived exosomes displaying AS1411 and encapsulated let-7 reduced breast cancer cell viability, as determined by 2-(2-methoxy-4-nitrophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)-2H-tetrazolium, monosodium salt (WST-8) reduction assay.59 In line with the aforementioned studies, one study in which exosomes were encapsulated with PTEN-CT displayed an anti-breast cancer effect using both MTT and Annexin V/PI methods.52
In addition, Liu et al. used both MTT and calcein AM/PI double staining assays to report the cytotoxic effects of Exo-DVDMS-mediated sonodynamic therapy. They also stated that this delivery system elevates the level of intracellular ROS using flow cytometry.54 Melzer et al. uncovered the cytotoxic property of Taxol-loaded exosomes via fluorescent imaging and cell-cycle arrest approaches.55 Peng et al. uncovered the immunomodulatory impact of RNA loaded into the targeted exosomes with antitumor immunity activity through triggering the retinoic acid-inducible gene I pathway which in turn induced apoptosis in breast cancer cells, as measured by an Annexin V/PI assay kit.39
Cheng et al. and Shi et al. reported that GEMINI-Exos and SMART-Exos, respectively, induced antitumor CD3+CD8+ T cell responses, leading to tumor cell death,50,56 showcasing the potential of these systems in harnessing immune responses against cancer. Usman et al. demonstrated that RBC-EVs loaded with ASO-125b diminished cell viability, as revealed by crystal violet staining.57 Finally, Zhao et al. introduced a safe drug delivery system using exosome-mediated siRNA, which showed no detrimental effect on healthy HUVECs.33
The comprehensive nature of these studies not only validates the anticancer potential of exosome-based drug delivery systems but also lays the foundation for future research avenues. Further investigations into the specific mechanisms underlying the observed effects, long-term safety profiles, and potential clinical applications will be crucial for translating these promising findings into practical therapeutic solutions.
Anti-metastatic effects of the exosome-based drug delivery systems
In addition to the previously mentioned cytotoxic and apoptotic effects observed in exosome-based therapeutic delivery systems, selected studies have highlighted other properties, including anti-metastatic, anti-angiogenic, and anti-migration effects, enriching our comprehension of the intricate interactions between exosomes and cancer cells. In one study, O’Brien et al. revealed that microRNA-379 loaded into exosomes causes a reduction in the proangiogenic marker cyclooxygenase 2 mRNA expression.40 This observation underscores the potential of exosome-based systems in modulating angiogenesis, a process tightly linked to metastasis. Similarly, Xie et al. applied the vessel formation assay to unveil the in vitro anti-angiogenic effects of DOX-loaded exosomes, which were found to inhibit angiogenesis and suppress metastasis.34
To further investigate the anti-migration effects of the exosome-based drug delivery systems, eight studies were identified, each employing different methods.33,34,36,42,47,49,52,59 Among these studies, Zhao et al., Feng et al., and Gong et al. reported on the anti-migration efficacy of the exosomes containing siRNA or DOX utilizing transwell and scratch assays. In this context, Zhao et al. demonstrated that siRNA-loaded exosomes decreased the migration capability of breast cancer cells. Consistently, Feng et al. reported that DOX@Exos-PH20-FA inhibited cancer cell migration due to a hyaluronidase activity of PH20 in the drug delivery system. This is while Gong et al. showed that therapeutic exosomes displaying A15 on the surface could inhibit MDA-MB-231 cell migration.33,47,49
Furthermore, Xie et al. reported an anti-migration effect for DOX-loaded exosomes using a transwell experiment.34 Wang et al. used the transwell assay to uncover the anti-migration effect of nucleolin-targeted exosomes encapsulated with let-7 miR.59 Martin-Marques et al. used the colony formation assay to show an anti-migration activity for the targeted exosomes loaded with DOX. Both free doxorubicin and exosomal DOX exhibited similar anti-migration effects. In addition, they assessed cell motility using time-lapse video microscopy and reported that DOX-loaded targeted exosomes displaying Cx43 on the surface significantly reduced cancer cell motility.42
In a separate study, Ahmed et al. utilized soft agar and scratch assays to demonstrate that PTEN-CT loaded into exosomes inhibited the migration of breast cancer cells.52 Notably, Tian et al. evaluated the levels of MMP2/9 via western blotting and stated that ID@E-MSNs reduced the level of MMP2/9, suggesting an anti-metastatic role for the exosome-based drug delivery system.36
The observed anti-angiogenic and anti-migration effects underscore the potential of exosome-based drug delivery systems in addressing critical aspects of cancer progression. These insights pave the way for the development of more effective and targeted therapies, opening new avenues for advancing cancer therapeutics.
The studies collectively contribute to our understanding of the intricate interactions between exosomes and cancer cells, paving the way for the development of more effective and targeted therapies. The diverse approaches in cellular uptake, anticancer evaluation, and anti-metastatic effects collectively establish exosome-based drug delivery systems as a compelling avenue for advancing cancer therapeutics.
Efficacy of exosomes versus other nanocarriers as a drug delivery system
In the realm of cancer therapeutics, the quest for an optimal drug delivery system has fueled extensive research, with exosomes emerging as a promising contender. To underscore the superiority of exosomes as a drug delivery system for cancer cells compared with other nano-sized carriers, all 30 studies were examined in detail.31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60 Among these studies, only seven studies investigated the difference between exosomes and other nanoparticles in terms of the efficiency in delivering therapeutic agents.33,43,45,46,54,55,60 These investigations provide valuable insights into the unique advantages conferred by exosomes in the context of drug delivery for cancer treatment.
One study carried out by Zhao et al. showed that CBSA and siS100A4 loaded into exosomes accumulated to a greater extent in tumor cells and exhibited less cytotoxicity against normal cells when compared with liposomes loaded with the drugs.33 Likewise, Haney et al. and Hadla et al. loaded DOX or PTX into both exosomes and liposomes as nanocarriers, and observed that exosomes encapsulated with the drugs acted more efficiently in terms of having anticancer activity, both in vitro and in vivo and had less cytotoxicity toward the control cells than those encapsulated into liposomes.45,46
Similarly, Melzer et al. conducted a comparison of the therapeutic efficacy between PTX-loaded exosomes and PTX alone, revealing a similar anticancer efficacy. However, the concentration of Taxol was about 1,000-fold lower in exosomes, indicating the enhanced potency of exosomal PTX. They used HUVEC-derived exosomes instead of MSC-derived exosomes and confirmed that PTX-loaded MSC-derived exosomes exhibited greater cytotoxic effects than the PTX-loaded HUVEC-derived exosomes.55 Likewise, Liu et al. reported that 4T1-derived exosomes had more efficacy in delivering the DVDMS toward tumor cells compared with RBC-EVs.54 In this regard, Xiong et al. demonstrated that exosomes strengthen the therapeutic index of HSA/Pt(IV) compared with the naked HSA/Pt(IV).60
In another study, Li et al. used PLGA as a DOX carrier and compared it with the targeted exosomes loaded with both DOX and PLGA. They showed that the antitumor activity of doxorubicin and PLGA within targeted exosomes was more effective than the naked PLGA and DOX. Besides, the exosomes could increase the drug accumulation in the tumor tissue and reduced biodistribution, due to their targeted generation.43
These comparative studies collectively underscore the superiority of exosomes over other nanocarriers in terms of enhanced drug delivery, reduced cytotoxicity, and targeted therapeutic effects. The findings pave the way for further investigations into the underlying mechanisms that contribute to the unique advantages of exosomes.
Future perspectives
Breast cancer remains a significant health concern for women worldwide, highlighting the critical need for effective treatment strategies.69 Recent research, as mentioned earlier in this systematic review, underscores the pivotal role of selective drug delivery systems, also known as targeted therapies, in breast cancer treatment.70 Among these targeted delivery systems, exosomes, naturally occurring vesicles secreted by all types of eukaryotic cells, emerge as promising platforms for drug delivery.71 This systematic review gathers cellular, biochemical, histopathological, and immunological data from various articles to explore the role of exosomes as a therapeutic delivery platform for breast cancer treatment. By analyzing the outcomes and considering previous reviews, coupled with the inherent versatility of exosomes that can be manipulated using different strategies, this review presents exosomes as novel platforms for delivering therapeutic agents to target sites. In this systematic review, we conducted a comprehensive assessment of scientific works utilizing small EVs or exosomes as drug delivery systems, employing stringent inclusion and exclusion criteria to achieve a comprehensive and systematic evaluation of the potential of exosomes as drug delivery agents in breast cancer.
This systematic review concludes that exosomes, as a drug delivery system, exhibit strong attachment to the cells and facile uptake by tumor cells, aligning with the findings of earlier systematic reviews that have examined the binding efficacy of the targeted exosomes to breast cancer cells. In addition, we have observed satisfactory uptake of exosomes loaded with various cargos by different cells.31,37,38,39,42,43,47,48,49,50,53,56,58,59 According to our analysis, therapeutic exosomes serve as safe drug delivery systems, introducing cell death in breast cancer cells through multiple mechanisms with minimal side effects. To our knowledge, no previous study has systematically addressed the mechanism of action of therapeutic exosomes in breast cancer.
Apart from different cell death mechanisms induced by the therapeutic exosomes, this review also assesses the role of the exosomes loaded with various anticancer agents in cellular migration and organ metastasis. Exosomes encapsulated with a wide range of anticancer agents could offer a novel approach to anti-metastatic therapies. Furthermore, we highlight the advantages of exosomes as nanocarriers for breast cancer treatment compared with the synthetic counterparts such as liposome or PLGA. Limited studies have compared the efficacy of exosomes with synthetic nanocarriers for delivering anticancer molecules.33,43,45,46,54,55,60
Moreover, this systematic review emphasizes that the flexibility of exosomes extends not only to the diverse range of therapeutics that can be encapsulated within them but also to the materials employed in cancer imaging. Similar advantages have been addressed in other reviews or systematic reviews10,70,72
Prior to advancing the therapeutic exosomes into clinical trials, it is crucial to conduct in vivo trials to evaluate their effectiveness. Hence, we carefully analyzed all 30 studies with regard to their utilization of tumor mouse models. These studies employed various in vivo models of breast cancer, using tumor-bearing mice to corroborate the potency of the exosome-based drug delivery systems. In this regard, a number of breast cancer cell lines including the TNBC cell lines, MDA-MB-231, MDA-MB-468 and 4T1, HER-2 + TUBO breast cancer cell line, and MCF-7 (hormone-positive breast cancer cell line) were utilized to establish the mice models. Diverse routes of injection, such as i.p., i.t., and i.v. routes were employed to deliver exosomes containing therapeutics into mouse models.31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60 To the best of our knowledge, this is the first systematic review that categorizes the breast tumor-bearing animal models used to assess the therapeutic potential of exosome-based drug delivery systems. Based on the in vivo and in vitro findings, we propose that the therapeutic exosomes hold great promise for efficiently treating breast cancer.31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,70,72
Despite the numerous reported benefits of exosome-based drug delivery systems in the 30 studies reviewed, and their potential use in the treatment of breast cancer, there still remain several issues that need to be addressed before drawing reliable conclusions. Inconsistency in the outcomes and methodologies relating to in vivo biodistribution of the exosome-based drug delivery systems can be regarded as another drawback in this systematic review. These studies are also limited by factors such as lack of stability evaluation in biological fluids, inadequate assessment of storage conditions, insufficient evaluation of encapsulation efficiency in exosomes, and a lack of survival rate in the tumor-bearing mice, all of which can influence our conclusions on the application of exosomes as therapeutic carriers. Furthermore, challenges arise from the absence of appropriate methodologies for randomization processes in animal housing and blinding of performance according to SYRCLE tool bias assessment guidelines, resulting in most articles having an unclear risk of bias.31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60
The field of exosome research has been hindered by several other limitations, including the lack of standardization, limited in vivo studies, insufficient characterization, limited understanding of mechanisms, and challenges associated with large-scale production and cost-effectiveness. These factors have impeded the progress and translation of exosome-based therapies for breast cancer treatment. It is noteworthy that the majority of studies focused on evaluating exosome-based drug delivery systems in tumor-bearing mice, which may be attributed to the ease of establishing the cancer animal models and the lower quantity of exosomes required compared with other animal models. Nonetheless, in vivo studies typically require a larger quantity of exosomes than in vitro experiments.
To address these limitations in future studies, it is crucial to provide more detailed methodologies regarding the doses used, animals studied, and the maintenance conditions of the exosomes to ensure more reliable results. In looking toward future perspectives for exosome-based drug delivery systems in breast cancer treatment, several key considerations and potential directions emerge.
Enhanced reporting standards
Addressing the inconsistency in reporting quality is crucial. Future studies should adhere to standardized reporting guidelines, such as MISEV2018, ensuring transparency and reproducibility. Clear documentation of in vitro experiment details, including seeding density, cell viability, and exosome depletion protocols, will enhance the reliability of study outcomes.
Optimization of exosome sources
Understanding the implications of different exosome sources is essential. Future research could delve into comparative studies, exploring the advantages and disadvantages of using various cell types for exosome isolation. Moreover, investigating the potential of engineered exosomes with enhanced targeting capabilities may offer new avenues for personalized and precise drug delivery.
Standard reporting
In our opinion, for a more informative conclusion, it would be essential to strive for standardized reporting of methodologies and outcomes in future studies. It is crucial to establish standardized methods for the isolation, purification, characterization, and quantification of exosomes. Having consistent protocols will enable better comparison across studies and enhance the reproducibility of data.
Advanced isolation techniques
While current studies employ diverse exosome isolation techniques, future research could focus on developing novel, efficient, and scalable methods. Also, there is a lack of accurate and precise evaluation of the purity of the isolated exosomes. Exploring alternative approaches beyond ultracentrifugation, such as microfluidics or affinity-based methods, may improve the scalability and purity of exosome isolation.
Standardized dosage and drug loading protocols
One issue is the lack of data on doses of the administered exosomes in vivo, and a lack of optimal procedure for achieving the desired dose. More research is required to determine the optimal dosing and delivery route of exosomes for drug delivery in breast cancer. Thus, the PK and biodistribution studies in vivo will assist in designing more effective treatment strategies using exosomes. Furthermore, the 30 selected studies lack sufficient biochemical and immunological analysis to further demonstrate the efficiency of the drug delivery system in cancer therapy and explore the off-targets.31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60 Establishing standardized dosing protocols for exosome-based drug delivery systems is also crucial for comparing outcomes across studies. Future research should aim to define optimal quantities and concentrations of exosomes, considering different therapeutic goals and targeted diseases. In addition, developing universal methodologies for assessing drug loading efficiency will enhance the comparability of results.
Innovative cargo loading strategies
The exploration of novel cargo loading strategies is a promising avenue. Future studies could focus on refining existing methods such as electroporation and sonication or introducing innovative techniques for loading therapeutic agents into exosomes. Achieving efficient and controlled cargo loading is vital for maximizing the therapeutic potential of exosome-based drug delivery systems.
In vivo imaging techniques
Advancements in in vivo imaging techniques can provide deeper insights into the biodistribution and antitumor effects of exosome-based drug delivery systems. Integrating advanced imaging modalities, such as intravital microscopy or molecular imaging, can enhance our understanding of real-time interactions between therapeutic exosomes and target tissues.
Immunomodulatory effects and safety profiling
Future research should delve into the immunomodulatory effects and safety profiles of exosome-based therapies. Comprehensive evaluations beyond traditional histopathological analyses, including assessments of systemic responses and immunological parameters, will contribute to a more nuanced understanding of the overall impact of these systems.
Enhancing the therapeutic efficacy
Another aspect of enhancing the therapeutic efficacy of exosomes is improving tumor cell recognition by incorporating targeting ligands or engineering exosomes with higher specificity toward tumor cells. Future research should also explore more the potential of combining different therapeutics within exosomes as versatile carriers to generate synergistic effects and overcome drug resistance. These cargo molecules can include nucleic acids and proteins, in addition to small-molecule chemotherapeutic drugs.
High cost of production
Indeed, using exosomes as a drug delivery system, particularly in breast cancer, poses challenges due to the high cost of large-scale production and the issue of low yield.73 To overcome this issue, employing three-dimensional cultures in huge bioreactors, which can yield up to 40-fold more exosomes, is highly suggested.73,74 In addition, other non-human sources of exosomes, such as bovine milk, are recommended in this context.75 Thus, developing cost-effective strategies is crucial for scalable exosome production for clinical studies.
Clinical translation
Finally, bridging the gap between preclinical research and clinical applications is paramount. Future studies should prioritize translational research, conducting robust preclinical trials that pave the way for clinical trials. Collaboration between researchers, clinicians, and industry partners is key to advancing exosome-based drug delivery systems from bench to bedside.
Conclusions
The extensive research conducted on exosome applications in drug delivery systems will undoubtedly provide new insights into dealing with different diseases and disorders. Hence, it is of vital importance to understand the progress made and the challenges that currently exist in this field. The studies investigated in this systematic review focused on the potential of exosomes as a delivery platform for breast cancer treatment, both in in vitro and in vivo. The findings support the notion that exosomes can be manipulated using different methods and utilized for theranostic approaches such as targeted therapy in breast cancer.
However, several improvements are still needed to overcome the remaining issues, such as the limited long-term stability of exosomes, inefficient loading of the therapeutics, clearance of exosomes from circulation, and the transition from the bench-scale to clinical production. Moving forward, it is necessary to translate some of the promising preclinical studies into well-designed clinical studies to evaluate the safety, efficacy, and long-term use of exosomes in breast cancer patients. It should be noted that only a few research findings propose exosomes as FDA-approved nanomedicines. In other words, formulations containing exosomes must stick to GMP regulations for quality control. This highlights the challenges that lie ahead in successfully translating exosome-based therapies, particularly for breast cancer therapy, into clinical applications.
Acknowledgments
We thank the Chapman University School of Pharmacy for funding support of K.P. Ethics approval was not required for this systematic review.
Author contributions
All authors contributed to the study conception and design. Material preparation, data collection, and analysis were performed by A.T.G., M.S., and K.P. The first draft of the manuscript was written by A.T.G., with all other authors actively participating in writing additional materials and offering feedback. All authors read and approved the final manuscript.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Keykavous Parang, Email: parang@chapman.edu.
Mona Salimi, Email: salimimona@pasteur.ac.ir.
References
- 1.Arnold M., Morgan E., Rumgay H., Mafra A., Singh D., Laversanne M., Vignat J., Gralow J.R., Cardoso F., Siesling S., Soerjomataram I. Current and future burden of breast cancer: Global statistics for 2020 and 2040. The Breast. 2022;66:15–23. doi: 10.1016/j.breast.2022.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sarhangi N., Hajjari S., Heydari S.F., Ganjizadeh M., Rouhollah F., Hasanzad M. Breast cancer in the era of precision medicine. Mol Biol Rep. 2022;49:10023–10037. doi: 10.1007/s11033-022-07571-2. [DOI] [PubMed] [Google Scholar]
- 3.Grogan Fleege N.M., Cobain E.F. Breast cancer management in 2021: A primer for the obstetrics and gynecology. Best Pract Res Clin Obstet Gynaecol. 2022;82:30–45. doi: 10.1016/j.bpobgyn.2022.02.004. [DOI] [PubMed] [Google Scholar]
- 4.Smolarz B., Nowak A.Z., Romanowicz H. Breast Cancer—Epidemiology, Classification, Pathogenesis and Treatment (Review of Literature) Cancers. 2022;14 doi: 10.3390/cancers14102569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nascimento C., Ferreira F. Tumor microenvironment of human breast cancer, and feline mammary carcinoma as a potential study model. Biochim. Biophys. Acta Rev. Cancer. 2021;1876:188587. doi: 10.1016/j.bbcan.2021.188587. [DOI] [PubMed] [Google Scholar]
- 6.Dai X., Li T., Bai Z., Yang Y., Liu X., Zhan J., Shi B. Breast cancer intrinsic subtype classification, clinical use and future trends. Am J Cancer Res. 2015;5:2929–2943. [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu S.Y., Yu K.D. Breast Cancer Vaccines: Disappointing or Promising? Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.828386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shien T., Iwata H. Adjuvant and neoadjuvant therapy for breast cancer. Jpn. J. Clin. Oncol. 2020;50:225–229. doi: 10.1093/jjco/hyz213. [DOI] [PubMed] [Google Scholar]
- 9.Jacobs A.T., Martinez Castaneda-Cruz D., Rose M.M., Connelly L. Targeted therapy for breast cancer: An overview of drug classes and outcomes. Biochem. Pharmacol. 2022;204 doi: 10.1016/j.bcp.2022.115209. [DOI] [PubMed] [Google Scholar]
- 10.Kumar D.N., Chaudhuri A., Aqil F., Dehari D., Munagala R., Singh S., Gupta R.C., Agrawal A.K. Exosomes as Emerging Drug Delivery and Diagnostic Modality for Breast Cancer: Recent Advances in Isolation and Application. Cancers. 2022;14:1435. doi: 10.3390/cancers14061435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kerr A.J., Dodwell D., McGale P., Holt F., Duane F., Mannu G., Darby S.C., Taylor C.W. Adjuvant and neoadjuvant breast cancer treatments: A systematic review of their effects on mortality. Cancer Treat. Rev. 2022;105 doi: 10.1016/j.ctrv.2022.102375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li A., Zhao Y., Li Y., Jiang L., Gu Y., Liu J. Cell-derived biomimetic nanocarriers for targeted cancer therapy: cell membranes and extracellular vesicles. Drug Deliv. 2021;28:1237–1255. doi: 10.1080/10717544.2021.1938757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martinelli C. Smart Nanocarriers for Targeted Cancer Therapy. Anticancer. Agents Med. Chem. 2021;21:546–557. doi: 10.2174/1871520620666200619181425. [DOI] [PubMed] [Google Scholar]
- 14.Alqosaibi A.I. Nanocarriers for anticancer drugs: Challenges and perspectives. Saudi J. Biol. Sci. 2022;29 doi: 10.1016/j.sjbs.2022.103298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Briolay T., Petithomme T., Fouet M., Nguyen-Pham N., Blanquart C., Boisgerault N. Delivery of cancer therapies by synthetic and bio-inspired nanovectors. Mol. Cancer. 2021;20:55. doi: 10.1186/s12943-021-01346-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sepahdar Z., Miroliaei M., Bouzari S., Khalaj V., Salimi M. Surface Engineering of Escherichia coli-Derived OMVs as Promising Nano-Carriers to Target EGFR-Overexpressing Breast Cancer Cells. Front. Pharmacol. 2021;12 doi: 10.3389/fphar.2021.719289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gujrati V., Prakash J., Malekzadeh-Najafabadi J., Stiel A., Klemm U., Mettenleiter G., Aichler M., Walch A., Ntziachristos V. Bioengineered bacterial vesicles as biological nano-heaters for optoacoustic imaging. Nat. Commun. 2019;10:1114. doi: 10.1038/s41467-019-09034-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sepahdar Z., Saghiri R., Miroliaei M., Salimi M. In silico approach to probe the binding affinity between OMVs harboring the Z(EGFR) affibody and the EGF receptor. J. Mol. Model. 2022;28:113. doi: 10.1007/s00894-022-05043-9. [DOI] [PubMed] [Google Scholar]
- 19.Sun H., Burrola S., Wu J., Ding W.Q. Extracellular Vesicles in the Development of Cancer Therapeutics. Int. J. Mol. Sci. 2020;21 doi: 10.3390/ijms21176097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huyan T., Li H., Peng H., Chen J., Yang R., Zhang W., Li Q. Extracellular Vesicles - Advanced Nanocarriers in Cancer Therapy: Progress and Achievements. Int. J. Nanomedicine. 2020;15:6485–6502. doi: 10.2147/ijn.S238099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu M., Wang M., Jia H., Wu P. Extracellular vesicles: emerging anti-cancer drugs and advanced functionalization platforms for cancer therapy. Drug Deliv. 2022;29:2513–2538. doi: 10.1080/10717544.2022.2104404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Théry C., Witwer K.W., Aikawa E., Alcaraz M.J., Anderson J.D., Andriantsitohaina R., Antoniou A., Arab T., Archer F., Atkin-Smith G.K., et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles. 2018;7 doi: 10.1080/20013078.2018.1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He X., Zhong X., Hu Z., Zhao S., Wei P., Li D. An insight into small extracellular vesicles: Their roles in colorectal cancer progression and potential clinical applications. Clin. Transl. Med. 2020;10 doi: 10.1002/ctm2.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Massaro C., Sgueglia G., Frattolillo V., Baglio S.R., Altucci L., Dell'Aversana C. Extracellular Vesicle-Based Nucleic Acid Delivery: Current Advances and Future Perspectives in Cancer Therapeutic Strategies. Pharmaceutics. 2020;12 doi: 10.3390/pharmaceutics12100980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Al-Masawa M.E., Alshawsh M.A., Ng C.Y., Ng A.M.H., Foo J.B., Vijakumaran U., Subramaniam R., Ghani N.A.A., Witwer K.W., Law J.X. Efficacy and safety of small extracellular vesicle interventions in wound healing and skin regeneration: A systematic review and meta-analysis of animal studies. Theranostics. 2022;12:6455–6508. doi: 10.7150/thno.73436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tominaga N. Anti-Cancer Role and Therapeutic Potential of Extracellular Vesicles. Cancers (Basel) 2021;13 doi: 10.3390/cancers13246303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dai J., Su Y., Zhong S., Cong L., Liu B., Yang J., Tao Y., He Z., Chen C., Jiang Y. Exosomes: key players in cancer and potential therapeutic strategy. Signal Transduct. Target. Ther. 2020;5:145. doi: 10.1038/s41392-020-00261-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gharavi A.T., Hanjani N.A., Movahed E., Doroudian M. The role of macrophage subtypes and exosomes in immunomodulation. Cell. Mol. Biol. Lett. 2022;27:83. doi: 10.1186/s11658-022-00384-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shao J., Zaro J., Shen Y. Advances in Exosome-Based Drug Delivery and Tumor Targeting: From Tissue Distribution to Intracellular Fate. Int. J. Nanomedicine. 2020;15:9355–9371. doi: 10.2147/ijn.S281890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shi Z.Y., Yang X.X., Malichewe C., Li Y.S., Guo X.L. Exosomal microRNAs-mediated intercellular communication and exosome-based cancer treatment. Int. J. Biol. Macromol. 2020;158:530–541. doi: 10.1016/j.ijbiomac.2020.04.228. [DOI] [PubMed] [Google Scholar]
- 31.Wang J.H., Forterre A.V., Zhao J., Frimannsson D.O., Delcayre A., Antes T.J., Efron B., Jeffrey S.S., Pegram M.D., Matin A.C. Anti-HER2 scFv-Directed Extracellular Vesicle-Mediated mRNA-Based Gene Delivery Inhibits Growth of HER2-Positive Human Breast Tumor Xenografts by Prodrug Activation. Mol. Cancer Ther. 2018;17:1133–1142. doi: 10.1158/1535-7163.Mct-17-0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou Y., Yamamoto Y., Takeshita F., Yamamoto T., Xiao Z., Ochiya T. Delivery of miR-424-5p via extracellular vesicles promotes the apoptosis of MDA-MB-231 TNBC cells in the tumor microenvironment. International Journal of Molecular Sciences. 2021;22:1–17. doi: 10.3390/ijms22020844. 844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao L., Gu C., Gan Y., Shao L., Chen H., Zhu H. Exosome-mediated siRNA delivery to suppress postoperative breast cancer metastasis. J. Control. Release. 2020;318:1–15. doi: 10.1016/j.jconrel.2019.12.005. [DOI] [PubMed] [Google Scholar]
- 34.Xie X., Lian S., Zhou Y., Li B., Lu Y., Yeung I., Jia L. Tumor-derived exosomes can specifically prevent cancer metastatic organotropism. J. Control. Release. 2021;331:404–415. doi: 10.1016/j.jconrel.2021.01.030. [DOI] [PubMed] [Google Scholar]
- 35.Wang P., Wang H., Huang Q., Peng C., Yao L., Chen H., Qiu Z., Wu Y., Wang L., Chen W. Exosomes from M1-polarized macrophages enhance paclitaxel antitumor activity by activating macrophages-mediated inflammation. Theranostics. 2019;9:1714–1727. doi: 10.7150/thno.30716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tian R., Wang Z., Niu R., Wang H., Guan W., Chang J. Tumor Exosome Mimicking Nanoparticles for Tumor Combinatorial Chemo-Photothermal Therapy. Front. Bioeng. Biotechnol. 2020;8:1010. doi: 10.3389/fbioe.2020.01010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Si Y., Chen K., Ngo H.G., Guan J.S., Totoro A., Zhou Z., Kim S., Kim T., Zhou L., Liu X. Targeted EV to Deliver Chemotherapy to Treat Triple-Negative Breast Cancers. Pharmaceutics. 2022;14:146. doi: 10.3390/pharmaceutics14010146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pi F., Binzel D.W., Lee T.J., Li Z., Sun M., Rychahou P., Li H., Haque F., Wang S., Croce C.M., et al. Nanoparticle orientation to control RNA loading and ligand display on extracellular vesicles for cancer regression. Nat. Nanotechnol. 2018;13:82–89. doi: 10.1038/s41565-017-0012-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peng B., Nguyen T.M., Jayasinghe M.K., Gao C., Pham T.T., Vu L.T., Yeo E.Y.M., Yap G., Wang L., Goh B.C., et al. Robust delivery of RIG-I agonists using extracellular vesicles for anti-cancer immunotherapy. J. Extracell. Vesicles. 2022;11 doi: 10.1002/jev2.12187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O'Brien K.P., Khan S., Gilligan K.E., Zafar H., Lalor P., Glynn C., O'Flatharta C., Ingoldsby H., Dockery P., De Bhulbh A., et al. Employing mesenchymal stem cells to support tumor-targeted delivery of extracellular vesicle (EV)-encapsulated microRNA-379. Oncogene. 2018;37:2137–2149. doi: 10.1038/s41388-017-0116-9. [DOI] [PubMed] [Google Scholar]
- 41.Naseri Z., Oskuee R.K., Jaafari M.R., Forouzandeh Moghadam M. Exosome-mediated delivery of functionally active miRNA-142-3p inhibitor reduces tumorigenicity of breast cancer in vitro and in vivo. Int. J. Nanomedicine. 2018;13:7727–7747. doi: 10.2147/IJN.S182384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martins-Marques T., Pinho M.J., Zuzarte M., Oliveira C., Pereira P., Sluijter J.P.G., Gomes C., Girao H. Presence of Cx43 in extracellular vesicles reduces the cardiotoxicity of the anti-tumour therapeutic approach with doxorubicin. J. Extracell. Vesicles. 2016;5 doi: 10.3402/jev.v5.32538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li S., Wu Y.J., Ding F., Yang J.P., Li J., Gao X.H., Zhang C., Feng J. Engineering macrophage-derived exosomes for targeted chemotherapy of triple-negative breast cancer. Nanoscale. 2020;12:10854–10862. doi: 10.1039/d0nr00523a. [DOI] [PubMed] [Google Scholar]
- 44.Jung K.O., Jo H., Yu J.H., Gambhir S.S., Pratx G. Development and MPI tracking of novel hypoxia-targeted theranostic exosomes. Biomaterials. 2018;177:139–148. doi: 10.1016/j.biomaterials.2018.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Haney M.J., Zhao Y., Jin Y.S., Li S.M., Bago J.R., Klyachko N.L., Kabanov A.V., Batrakova E.V. Macrophage-Derived Extracellular Vesicles as Drug Delivery Systems for Triple Negative Breast Cancer (TNBC) Therapy. J. Neuroimmune Pharmacol. 2020;15:487–500. doi: 10.1007/s11481-019-09884-9. [DOI] [PubMed] [Google Scholar]
- 46.Hadla M., Palazzolo S., Corona G., Caligiuri I., Canzonieri V., Toffoli G., Rizzolio F. Exosomes increase the therapeutic index of doxorubicin in breast and ovarian cancer mouse models. Nanomedicine. 2016;11:2431–2441. doi: 10.2217/nnm-2016-0154. [DOI] [PubMed] [Google Scholar]
- 47.Gong C., Tian J., Wang Z., Gao Y., Wu X., Ding X., Qiang L., Li G., Han Z., Yuan Y., Gao S. Functional exosome-mediated co-delivery of doxorubicin and hydrophobically modified microRNA 159 for triple-negative breast cancer therapy. J. Nanobiotechnology. 2019;17:93. doi: 10.1186/s12951-019-0526-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gomari H., Forouzandeh Moghadam M., Soleimani M., Ghavami M., Khodashenas S. Targeted delivery of doxorubicin to HER2 positive tumor models. Int. J. Nanomedicine. 2019;14:5679–5690. doi: 10.2147/ijn.S210731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Feng C., Xiong Z., Wang C., Xiao W., Xiao H., Xie K., Chen K., Liang H., Zhang X., Yang H. Folic acid-modified Exosome-PH20 enhances the efficiency of therapy via modulation of the tumor microenvironment and directly inhibits tumor cell metastasis. Bioact. Mater. 2021;6:963–974. doi: 10.1016/j.bioactmat.2020.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cheng Q., Dai Z., Smbatyan G., Epstein A.L., Lenz H.J., Zhang Y. Eliciting anti-cancer immunity by genetically engineered multifunctional exosomes. Mol. Ther. 2022;30:3066–3077. doi: 10.1016/j.ymthe.2022.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nguyen Cao T.G., Kang J.H., Kim W., Lim J., Kang S.J., You J.Y., Truong Hoang Q., Kim W.J., Rhee W.J., Kim C., et al. Engineered extracellular vesicle-based sonotheranostics for dual stimuli-sensitive drug release and photoacoustic imaging-guided chemo-sonodynamic cancer therapy. Theranostics. 2022;12:1247–1266. doi: 10.7150/thno.65516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ahmed S.F., Das N., Sarkar M., Chatterjee U., Chatterjee S., Ghosh M.K. Exosome-mediated delivery of the intrinsic C-terminus domain of PTEN protects it from proteasomal degradation and ablates tumorigenesis. Mol. Ther. 2015;23:255–269. doi: 10.1038/mt.2014.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hong Y., Kim Y.K., Kim G.B., Nam G.H., Kim S.A., Park Y., Yang Y., Kim I.S. Degradation of tumour stromal hyaluronan by small extracellular vesicle-PH20 stimulates CD103(+) dendritic cells and in combination with PD-L1 blockade boosts anti-tumour immunity. J. Extracell. Vesicles. 2019;8 doi: 10.1080/20013078.2019.1670893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu Y., Bai L., Guo K., Jia Y., Zhang K., Liu Q., Wang P., Wang X. Focused ultrasound-augmented targeting delivery of nanosonosensitizers from homogenous exosomes for enhanced sonodynamic cancer therapy. Theranostics. 2019;9:5261–5281. doi: 10.7150/thno.33183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Melzer C., Rehn V., Yang Y., Bähre H., von der Ohe J., Hass R. Taxol-Loaded MSC-Derived Exosomes Provide a Therapeutic Vehicle to Target Metastatic Breast Cancer and Other Carcinoma Cells. Cancers. 2019;11:798. doi: 10.3390/cancers11060798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shi X., Cheng Q., Hou T., Han M., Smbatyan G., Lang J.E., Epstein A.L., Lenz H.J., Zhang Y. Genetically Engineered Cell-Derived Nanoparticles for Targeted Breast Cancer Immunotherapy. Mol. Ther. 2020;28:536–547. doi: 10.1016/j.ymthe.2019.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Usman W.M., Pham T.C., Kwok Y.Y., Vu L.T., Ma V., Peng B., Chan Y.S., Wei L., Chin S.M., Azad A., et al. Efficient RNA drug delivery using red blood cell extracellular vesicles. Nat. Commun. 2018;9:2359. doi: 10.1038/s41467-018-04791-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wan Y., Wang L., Zhu C., Zheng Q., Wang G., Tong J., Fang Y., Xia Y., Cheng G., He X., Zheng S.-Y. Aptamer-Conjugated Extracellular Nanovesicles for Targeted Drug Delivery. Cancer Res. 2018;78:798–808. doi: 10.1158/0008-5472.Can-17-2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang Y., Chen X., Tian B., Liu J., Yang L., Zeng L., Chen T., Hong A., Wang X. Nucleolin-targeted Extracellular Vesicles as a Versatile Platform for Biologics Delivery to Breast Cancer. Theranostics. 2017;7:1360–1372. doi: 10.7150/thno.16532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xiong F., Ling X., Chen X., Chen J., Tan J., Cao W., Ge L., Ma M., Wu J. Pursuing Specific Chemotherapy of Orthotopic Breast Cancer with Lung Metastasis from Docking Nanoparticles Driven by Bioinspired Exosomes. Nano Lett. 2019;19:3256–3266. doi: 10.1021/acs.nanolett.9b00824. [DOI] [PubMed] [Google Scholar]
- 61.Nan W., Zhang C., Wang H., Chen H., Ji S. Direct Modification of Extracellular Vesicles and Its Applications for Cancer Therapy: A Mini-Review. Front. Chem. 2022;10 doi: 10.3389/fchem.2022.910341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sadeghi S., Tehrani F.R., Tahmasebi S., Shafiee A., Hashemi S.M. Exosome engineering in cell therapy and drug delivery. Inflammopharmacology. 2023;31:145–169. doi: 10.1007/s10787-022-01115-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ma Y., Dong S., Li X., Kim B.Y.S., Yang Z., Jiang W. Extracellular Vesicles: An Emerging Nanoplatform for Cancer Therapy. Front. Oncol. 2020;10 doi: 10.3389/fonc.2020.606906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Si Y., Kim S., Zhang E., Tang Y., Jaskula-Sztul R., Markert J.M., Chen H., Zhou L., Liu X.M. Targeted Exosomes for Drug Delivery: Biomanufacturing, Surface Tagging, and Validation. Biotechnol. J. 2020;15 doi: 10.1002/biot.201900163. [DOI] [PubMed] [Google Scholar]
- 65.Estes S., Konstantinov K., Young J.D. Manufactured extracellular vesicles as human therapeutics: challenges, advances, and opportunities. Curr. Opin. Biotechnol. 2022;77 doi: 10.1016/j.copbio.2022.102776. [DOI] [PubMed] [Google Scholar]
- 66.Chen H., Wang L., Zeng X., Schwarz H., Nanda H.S., Peng X., Zhou Y. Exosomes, a New Star for Targeted Delivery. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.751079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tran L., Tam D.N.H., Elshafay A., Dang T., Hirayama K., Huy N.T. Quality assessment tools used in systematic reviews of in vitro studies: A systematic review. BMC Med. Res. Methodol. 2021;21:101. doi: 10.1186/s12874-021-01295-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hooijmans C.R., Rovers M.M., de Vries R.B.M., Leenaars M., Ritskes-Hoitinga M., Langendam M.W. SYRCLE's risk of bias tool for animal studies. BMC Med. Res. Methodol. 2014;14:43. doi: 10.1186/1471-2288-14-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wilkinson L., Gathani T. Understanding breast cancer as a global health concern. Br. J. Radiol. 2022;95 doi: 10.1259/bjr.20211033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang F., Wang M., Guan X. Exosomes and mimics as novel delivery platform for cancer therapy. Front. Pharmacol. 2022;13 doi: 10.3389/fphar.2022.1001417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bunggulawa E.J., Wang W., Yin T., Wang N., Durkan C., Wang Y., Wang G. Recent advancements in the use of exosomes as drug delivery systems. J. Nanobiotechnology. 2018;16:81. doi: 10.1186/s12951-018-0403-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Elsharkasy O.M., Nordin J.Z., Hagey D.W., de Jong O.G., Schiffelers R.M., Andaloussi S.E., Vader P. Extracellular vesicles as drug delivery systems: Why and how? Adv. Drug Deliv. Rev. 2020;159:332–343. doi: 10.1016/j.addr.2020.04.004. [DOI] [PubMed] [Google Scholar]
- 73.Syromiatnikova V., Prokopeva A., Gomzikova M. Methods of the Large-Scale Production of Extracellular Vesicles. Int. J. Mol. Sci. 2022;23 doi: 10.3390/ijms231810522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Haraszti R.A., Miller R., Stoppato M., Sere Y.Y., Coles A., Didiot M.C., Wollacott R., Sapp E., Dubuke M.L., Li X., et al. Exosomes Produced from 3D Cultures of MSCs by Tangential Flow Filtration Show Higher Yield and Improved Activity. Mol. Ther. 2018;26:2838–2847. doi: 10.1016/j.ymthe.2018.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Paolini L., Monguió-Tortajada M., Costa M., Antenucci F., Barilani M., Clos-Sansalvador M., Andrade A.C., Driedonks T.A.P., Giancaterino S., Kronstadt S.M., et al. Large-scale production of extracellular vesicles: Report on the “massivEVs” ISEV workshop. J. Extracell. Biol. 2022;1:e63. doi: 10.1002/jex2.63. [DOI] [PMC free article] [PubMed] [Google Scholar]