Abstract
Mitochondrial DNA (mtDNA) of Saccharomyces cerevisiae contains highly conserved sequences, called rep/ori, that are associated with several aspects of its metabolism. These rep/ori sequences confer the transmission advantage exhibited by a class of deletion mutants called hypersuppressive petite mutants. In addition, because they share features with the mitochondrial leading-strand DNA replication origin of mammals, rep/ori sequences have also been proposed to participate in mtDNA replication initiation. Like the mammalian origins, where transcription is used as a priming mechanism for DNA synthesis, yeast rep/ori sequences contain an active promoter. Although transcription is required for maintenance of wild-type mtDNA in yeast, the role of the rep/ori promoter as a cis-acting element involved in the replication of wild-type mtDNA is unclear, since mitochondrial deletion mutants need neither transcription nor a rep/ori sequence to maintain their genome. Similarly, transcription from the rep/ori promoter does not seem to be necessary for biased inheritance of mtDNA. As a step to elucidate the function of the rep/ori promoter, we have attempted to detect transcription-dependent DNA transactions in the mtDNA of a hypersuppressive petite mutant. We have examined the mtDNA of the well-characterized petite mutant a-1/1R/Z1, whose repeat unit shelters the rep/ori sequence ori1, in strains carrying either wild-type or null alleles of the nuclear genes encoding the mitochondrial transcription apparatus. Complex DNA transactions were detected that take place around GC-cluster C, an evolutionarily conserved GC-rich sequence block immediately downstream from the rep/ori promoter. These transactions are strictly dependent upon mitochondrial transcription.
The mitochondrial DNA (mtDNA) genome of Saccharomyces cerevisiae has been the subject of extensive genetic analysis over the years (reviewed in reference 16). Respiratory-deficient deletion mutants of mtDNA (called petite or [rho−] mutants) arise spontaneously at a high frequency. Only a small fragment of the ∼80-kb wild-type mtDNA genome ([rho+]) is retained in a petite genome, where it is amplified so that the mass of mtDNA in the petite genome is equivalent to that in a wild-type cell. The retained fragment is referred to as the repeat unit of the petite genome. Some petite mutants give rise to diploid progeny composed almost exclusively of petite mutants when crossed with cells harboring [rho+] mtDNA. These mutants, termed hypersuppressive (HS) petite mutants, are thought to present an mtDNA genome with a replicative and/or segregation advantage over [rho+] DNA. While the basis for this biased inheritance is still unknown, molecular characterization of HS genomes has provided important insights (3, 11, 12). All HS genomes consist of a tandem repetition of a short DNA fragment containing one of four highly conserved sequences that are all present in the [rho+] genome. These sequences, called rep or ori, comprise a 300-bp segment sharing about 80% identity. Embedded within this segment are three perfectly conserved GC-rich blocks (GC clusters A, B, and C), as well as a functional promoter for transcription, upstream from GC-cluster C. The integrity of GC clusters A and B is required for rep/ori activity; similarly, deletion of GC cluster C and its surrounding region inactivates the rep/ori sequence (25; reference 1 and references therein). The preferential transmission of HS genomes in crosses with [rho+] genomes has been postulated to reflect an advantage conferred by the increased density of rep/ori sequences in the amplified HS genomes.
The rep/ori sequences have been posited as active origins of mtDNA replication, and hybridization studies have provided evidence that they function as bidirectional origins of replication (reference 2 and references therein). In addition, because their structural organization is reminiscent of that at the origin for leading-strand synthesis in the D-loop region of vertebrate mtDNA, yeast mtRNA polymerase has been proposed to play a role in producing primers for DNA synthesis at the rep/ori sequences (2; reviewed in references 7 and 38). In this light, GC cluster C, also found in the D-loop region of vertebrates (where it is called conserved sequence block II), promotes the polymerase-dependent formation of a persistent RNA-DNA hybrid upon transcription of origin-containing templates (44, 45). Formation of such a hybrid is thought to represent an important step in the sequence of events that lead to initiation of mtDNA synthesis (44, 45).
A relatively simple machinery ensures mtDNA transcription in yeast cells (reviewed in references 26 and 40). The mtRNA polymerase is composed of a core catalytic subunit and a promoter recognition factor, sc-mtTFB. The nuclear genes encoding these components, RPO41 and MTF1, respectively, are essential for maintenance of [rho+] mtDNA, which is consistent with the role postulated for mtRNA polymerase in replication initiation. However, cells lacking a functional mitochondrial transcription apparatus are still able to propagate [rho−] genomes, and a rep/ori sequence need not be present in these deletion mutants (19–21). Thus, either alternative modes of replication exist that use other sequences and/or different mechanisms or transcription plays no role in [rho−] mtDNA replication. In principle this can be postulated for wild-type as well as [rho−] DNA, and therefore the priming of DNA replication in yeast mitochondria remains an open question.
Transcription of HS genomes is initiated at the promoter present within rep/ori sequences in vivo (1), and the presence of an active promoter seems to be an important property of these genomes. In support of this view is the structure of three additional sequences closely related to the rep/ori sequences that are detected in wild-type mtDNA. However, these have never been recovered alone in screens for spontaneous HS petites (11). While these sequences also harbor GC clusters A, B, and C, their promoter has been inactivated by the insertion of a mobile GC-rich element called cluster gamma (11, 43). Why these sequences, which have been termed inactive origins, do not give rise to HS petite mutants is unknown, but at least two hypotheses can be put forth. First, activity of the promoter is required for the formation of an HS genome from a wild-type mtDNA (for instance, the excision and/or amplification of the repeat unit). Second, lack of a functional promoter causes a rep/ori sequence to lose its replication-segregation advantage so that, once formed, the deletion genome is rapidly competed by wild-type mtDNA molecules. The requirement of mtDNA transcription for preferential inheritance of HS mtDNA has recently been tested with cells carrying a deletion allele of the RPO41 gene (30). Since the mitochondrial transcription machinery is essential for maintenance of [rho+] mtDNA, such a requirement was measured in crosses between HS [rho−] and neutral [rho−] mutants (that is, petite mutants whose mtDNA does not harbor a rep/ori sequence and is not preferentially transmitted to the progeny of crosses with [rho+] cells). HS genomes were found to remain preferentially inherited despite the inactivation of RPO41, and it was therefore concluded that transcription from the rep/ori promoter is not necessary for the biased inheritance of HS genomes (30). However, the issue of whether inheritance of mtDNA obeys the same molecular rules in [rho−] × [rho−] and [rho+] × [rho−] crosses remains unsolved. Therefore, the possibility still exists that mtDNA transcription is required for preferential inheritance of HS genomes in crosses between HS petite mutants and wild-type cells.
As a first step toward addressing the role played by mtRNA polymerase in the metabolism of rep/ori sequences, we have attempted to detect transcription-dependent DNA transactions in mtDNA of an HS petite mutant. In this study, we have investigated the mtDNA from the well-characterized petite mutant a-1/1R/Z1, whose 416-bp repeat unit harbors the rep/ori sequence ori1 (1, 2). We report the detection by direct labeling of free 5′ DNA ends of DNA strands that have the same polarity as transcripts derived from the promoter. The 5′ ends of these DNAs map downstream from the promoter, at the 5′ boundary of GC cluster C, a site where transitions from RNA to DNA have previously been proposed to occur (2). Gene disruption experiments were performed to demonstrate that production of these DNA strands is strictly dependent upon the presence of functional alleles of the RPO41 and MTF1 genes. In addition, restriction enzyme analyses and 3′-end-labeling experiments have revealed the presence of double-stranded breaks in the mtDNA of petite a-1/1R/Z1. These complex DNA transactions also depend upon the organelle transcription machinery, and their role in the metabolism of HS genomes will be discussed. We have tested the requirement of the DNA-binding protein ABF2, the yeast homolog of mammalian mitochondrial transcription factor A (mtTFA), which has been shown to be involved in wild-type mtDNA stability (14), for the maintenance of HS mtDNA. In contrast to what is observed with wild-type mtDNA under nonselective conditions (14), ABF2 is not required for the maintenance of HS mtDNA. Moreover, rpo41 abf2 double mutants are still able to propagate the ori1 genome of petite mutant a-1/1R/Z1, indicating that ABF2 is not required for mtDNA maintenance in the absence of mtRNA polymerase. Finally, the transcription-dependent mtDNA transactions reported in this study do not depend on ABF2, further indicating that unlike its mammalian homolog (reviewed in reference 40), the ABF2 protein plays a limited role in mtDNA transcription.
MATERIALS AND METHODS
Strains.
The HS S. cerevisiae strain containing the ori1 mitochondrial genome used in this study is a-1/1R/Z1 (MATa ade1 [ori1]) (reference 2 and references therein). All other ori1 strains are derivatives of a-1/1R/Z1. These strains are listed in Table 1. In order to construct host strains suitable for gene disruption experiments, Ura− derivatives of strains a-1/1R/Z1 and aori1Δrpo41 were selected on 5′-fluoro-orotic acid-containing medium as described previously (4). Induction of [rho0] variants of αRPO41 and αrpo41::URA3 by treatment with ethidium bromide was performed according to the method of Fox et al. (24).
TABLE 1.
Yeast strains used in this study
| Strain | Relevant genotype | Source |
|---|---|---|
| a-1/1R/Z1 | MATa ade1 [ori1] | G. Bernardi |
| aori1FOA | MATa ade1 ura3 [ori1] | This study |
| aori1rpo41Δ | aori1FOA with rpo41::URA3 allele | This study |
| aori1mtf1Δ | aori1FOA with mtf1::URA3 allele | This study |
| aori1abf2Δ | aori1FOA with abf2::URA3 allele | This study |
| aori1nuc1Δ | aori1FOA with nuc1::URA3 allele | This study |
| aori1rpo41ΔFOA | ura3 derivative of aori1rpo41D | This study |
| aori1rpo41Δabf2Δ | aori1rpo41ΔFOA with abf2::URA3 allele | This study |
| aori1rpo41Δnuc1Δ | aori1rpo41ΔFOA with nuc1::URA3 allele | This study |
| αRPO41 | MATα ade2 ura3 leu2 trp1 his3 | E. van Dyck |
| αrpo41::URA3 | αRPO41 with rpo41::URA3 allele | This study |
Cloning of yeast ori1 origin.
mtDNA from petite a-1/1R/Z1 was digested with the single cutter DraI, and a 416-bp fragment corresponding to the repeat unit was cloned into pBluescript II KS− previously cut with SmaI to create plasmid pEKSori1-2.
Plasmid constructions and gene disruptions.
Plasmid pUC3′5′-B1 containing an rpo41::URA3 deletion allele was obtained from Walton Fangman’s laboratory and is identical to prpo41Δ.n described by Lorimer et al. (30). This plasmid was digested with SacI and SalI and used to transform yeast cells and to construct a chromosomal rpo41 null mutant. Plasmid pEl-I consists of a 2.9-kb XhoI fragment containing the MTF1 gene inserted into λYES-R (18). A 1.1-kb BamHI fragment containing the URA3 marker was inserted at a unique BamHI site within the MTF1 open reading frame (ORF) of this plasmid to create pmtf1::URA3. After digestion of pmtf1::URA3 with XhoI, a 4-kb fragment containing the disrupted gene was gel purified and used to transform yeast and construct an mtf1 null mutant. Plasmid pAMIA consists of a 1.6-kb EcoRI fragment containing the ABF2 gene inserted into pUC118 (14). A 1.1-kb BamHI fragment containing URA3 was inserted at a unique BglII site within the ABF2 ORF of this plasmid to create pabf2::URA3. EcoRI digests of pabf2::URA3 were used to transform yeast cells and construct an abf2 null mutant. Plasmid pUZ5 is a 2.1-kb fragment containing the NUC1 gene inserted into pBR322 and was a gift from H. P. Zassenhaus. A 1.1-kb BamHI fragment containing URA3 was inserted at a unique BamHI site within the NUC1 ORF of this plasmid to create pnuc1::URA3. To construct a nuc1 null mutant, plasmid pnuc1::URA3 was digested with BglII and used to transform yeast cells. Gene disruptions were confirmed by Southern blot analysis.
mtDNA preparation and analysis.
mtDNA was prepared from whole cells and purified by two successive centrifugations on CsCl gradients in the presence of bisbenzimide (Hoechst 33258) according to the method of Rickwood et al. (37). Next, 5′ end labeling of mtDNA was performed as follows: 840 ng of mtDNA was treated with 2 U of calf intestine alkaline phosphatase (Boehringer Mannheim) at 37°C for 1 h in the manufacturer’s buffer. The reaction products were extracted with phenol-chloroform (1:1) and precipitated with ethanol in the presence of 0.3 M sodium acetate. Pellets were resuspended in water and the 5′ termini were labeled with 10 U of T4 polynucleotide kinase (Boehringer Mannheim) and 50 μCi of [γ-32P]ATP (6,000 Ci/mmol; Dupont) in a 20-μl reaction volume containing the manufacturer’s buffer. After incubation at 37°C for 30 min, the reaction products were extracted and precipitated as before. The DNA pellets were resuspended in water, and aliquots were subjected to restriction enzyme digestions in a 20-μl reaction volume. After digestion, 50 μl of 0.5 M sodium acetate (pH 5.2) was added, and the reaction products were extracted with phenol-chloroform (1:1). One microliter of carrier tRNA (10 mg/ml) was added to the aqueous phase, and the DNA was precipitated with ethanol. Pellets were resuspended in formamide and denatured prior to electrophoresis on a polyacrylamide–7 M urea gel. Alternatively, loading buffer was added directly to the restriction reactions and samples were run on native polyacrylamide gels. Then 3′ end labeling was carried out with terminal deoxynucleotidyltransferase and cordycepin 5′-triphosphate (CoTP) as follows: 640 ng of mtDNA was incubated for 30 min at 37°C in a 20-μl reaction volume containing 15 U of enzyme (Gibco BRL) and 50 μCi of [α-32P]CoTP (5,000 Ci/mmol; Dupont) in the manufacturer’s buffer. The reaction products were then extracted with phenol-chloroform (1:1) prior to precipitation. The DNA was resuspended in water and subjected to restriction enzyme digestion and gel electrophoresis as described above. RNase treatment of the mtDNA was carried out in a 50-μl reaction volume containing 10 mM Tris-HCl (pH 8.0), 100 mM MgCl2, 10 mM dithiothreitol, 50 mM KCl, 1 μg of RNase A (Boehringer Mannheim), 0.5 U of RNase T1 (Pharmacia), and 0.87 U of RNase H (Pharmacia). After incubation at 37°C for 15 min, the reaction products were extracted with phenol-chloroform (1:1) and precipitated with ethanol. Alkaline treatment of the mtDNA was performed as follows: the DNA was incubated in a 40-μl reaction volume containing 50 mM KOH at 65°C for 30 min. Then 4 μl of 3 M sodium acetate (pH 5.2) was added, and the DNA was precipitated with ethanol.
Total DNA preparation.
Total DNA was prepared by the minilysate methods of Rickwood et al. (37).
mtRNA analysis.
Total RNA was isolated by the method of Schmitt et al. (39). Samples were treated with RNase-free DNase I (Boehringer Mannheim) before being subjected to Northern blot analysis. mtRNA was probed with the BamHI-EcoRI fragment containing ori1 from plasmid pEKSori1-2. Hybridization probes were made with the Prime-It Kit (Stratagene, Inc.).
DNA sequencing.
The DNA sequencing ladder used to map the free 5′ DNA ends detected in the mtDNA of petite a-1/1R/Z1 was obtained by dideoxy sequencing of plasmid pEHSori1-2 with the 15-mer oligonucleotide ori1-Z1-4 (5′-ATCTTCTTGTTTATC-3′) as a primer. The sequence of this primer corresponds to nucleotides 141 to 155 in the repeat unit of petite a-1/1R/Z1 as presented by Baldacci et al. (2). Partial chemical sequencing of the DNA strands detected after 5′ end labeling and restriction of mtDNA from a-1/1R/Z1 was performed on gel-purified DNA as described by Maxam and Gilbert (32).
RESULTS
Free 5′ DNA ends in the mtDNA of a hypersuppressive petite mutant.
The HS petite mutant a-1/1R/Z1 has been well characterized, both at the genetic and molecular level (1, 2; reference 12 and references therein). The 416-bp repeat unit of this petite mutant contains the rep/ori sequence ori1 (Fig. 1A), and its small size implies a high degree of amplification of the genome. We therefore reasoned that DNA intermediates normally associated with rep/ori activity would have a greater chance of being detected. This should permit the investigation of the possible role of the promoter present in ori1 in the metabolism of a-1/1R/Z1 mtDNA and any requirement for mitochondrial transcription proteins.
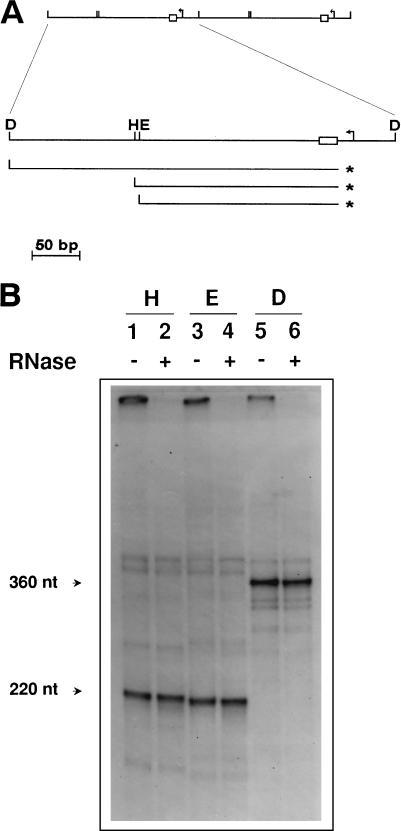
FIG. 1.
Detection and mapping of free 5′ ends in the mtDNA of strain a-1/1R/Z1. (A) Schematic diagram of the repeat units of the ori1 petite mutant showing the results of the 5′ mapping experiment illustrated in panel B. The upper part shows two repeat units of the mitochondrial genome of a-1/1R/Z1. Below is an enlargement of one such repeat showing the promoter (bent arrow) and the GC cluster C element (box) present in the ori1 sequence, as well as the positions of the DraI (D), HpaII (H), and EcoRV (E) restriction sites used in the analysis. Also indicated are the position of the predominant 5′ end detected (asterisks) and the strands containing this terminus upon restriction enzyme digestion and denaturation. (B) Detection of free 5′ ends by direct labeling with [γ-32P]ATP. mtDNA from a-1/1R/Z1 was dephosphorylated and labeled with [γ-32P]ATP prior to restriction enzyme digestion and electrophoresis on a denaturing polyacrylamide gel. Lanes were pretreated with RNases as indicated (+). The restriction enzymes used were HpaII (lanes 1 and 2), EcoRV (lanes 3 and 4), and DraI (lanes 5 and 6).
We first assayed for 5′ mtDNA ends that could correspond to start sites for DNA synthesis within the ori1 sequence. After dephosphorylation, the free 5′ DNA ends present in isolated mtDNA were radiolabeled with [γ-32P]ATP, and aliquots of the mtDNA were subjected to restriction digestion and electrophoresis under denaturing conditions. Each of the restriction enzymes used in this analysis, HpaII, EcoRV, and DraI, cleaves the repeat unit only once (Fig. 1A). A predominant species of constant intensity was obtained by this procedure, the size of which varied with the restriction enzyme used (Fig. 1B, lanes 1, 3, and 5). From the restriction map of a-1/1R/Z1 presented in Fig. 1A, it was possible to determine the polarity of the strand whose 5′ end had been labeled, as well as to map this end. Thus, the product obtained after HpaII digestion was slightly larger than that obtained with EcoRV, indicating that the polarity of the labeled strand was the same as that of the transcripts derived from the promoter. In addition, the labeled 5′ end mapped at the vicinity of GC cluster C. Both the nature of this DNA strand and its polarity were confirmed by partial chemical sequencing (data not shown). Additional species could be seen upon long exposure, but their overall pattern was complex and could not be correlated with the restriction map of a-1/1R/Z1. Assignment of their exact nature was therefore not possible.
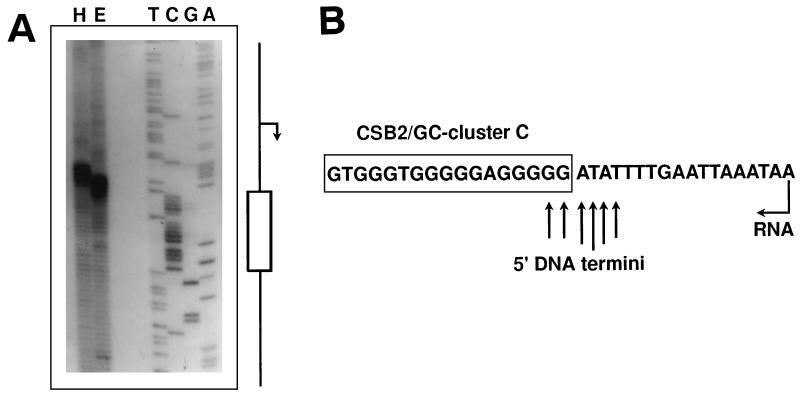
To determine precisely the position of the 5′ end mapped by restriction analysis, a DNA sequence ladder of the ori1 sequence was prepared by using as the primer an oligonucleotide starting at the EcoRV cleavage site and extending toward GC cluster C (see Materials and Methods; Fig. 1A). This ladder was then run next to the products obtained after EcoRV digestion of the labeled mtDNA (Fig. 2A). The labeled DNA strand appeared as a population of molecules whose 5′ termini mapped at the 5′ boundary of GC cluster C (Fig. 2B), a site where transitions from RNA to DNA have previously been proposed to occur (2).
FIG. 2.
Mapping of the predominant 5′ end detected in the mtDNA of a-1/1R/Z1. (A) mtDNA samples labeled as described in Materials and Methods were digested with HpaII (H) or EcoRV (E) and displayed on a sequencing gel. Sequencing reactions generated with a primer starting at the EcoRV cleavage site and extending toward GC cluster C on pEKSori1-2 DNA were used as the markers. The positions of the promoter (bent arrow) and GC cluster C (box) on the sequencing ladder are indicated. (B) Schematic diagram showing the position of the 5′ ends in the ori1 sequence. A portion of the ori1 sequence encompassing the GC cluster C and the start site of transcription (bent arrow) is shown. 5′ DNA termini are indicated by arrows whose lengths reflect their relative intensities on the sequencing gel.
In vertebrates, primers for mtDNA synthesis at the origin for leading-strand DNA replication are provided by transcripts derived from an upstream promoter present at this origin (reviewed in reference 7). A unique demonstration of this process is offered in mouse mtDNA, where mtDNA species were recovered that still possessed ribonucleotides covalently bound to their 5′ ends (5). We tested whether ribonucleotide residues were present at the 5′ ends of the strands detected by direct labeling. We therefore treated the mtDNA with a mixture of RNase A, RNase T1, and RNase H prior to dephosphorylation and labeling. As illustrated in Fig. 1B (lanes 2, 4, and 6), the pattern obtained after restriction enzyme digestion of the RNase-treated mtDNA was identical to that of the untreated mtDNA, suggesting the absence of ribonucleotides. To rule out that this resulted from the inaccessibility of ribonucleotides to RNases, labeled mtDNA restricted with EcoRV was analyzed on a denaturing gel, and the strands of interest were recovered and subjected to alkaline treatment prior to analysis on a denaturing polyacrylamide gel. Since these strands remained unaffected by this treatment (data not shown), it was concluded that no detectable ribonucleotides are covalently joined to their 5′ ends.
Production of free 5′ DNA ends at ori1 is dependent upon mitochondrial transcription.
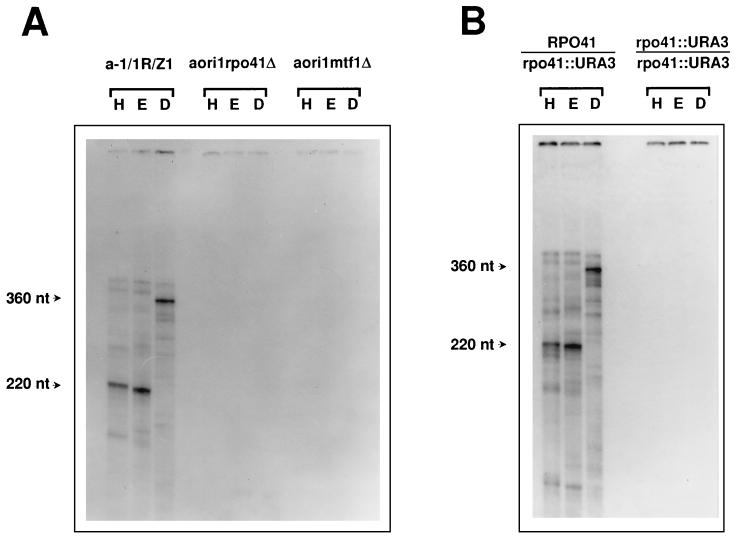
Since the free 5′ DNA ends we detected at ori1 map to a proposed site of transition from RNA to DNA (2) and to assess the role of the promoter in this transaction, we examined whether transcription was required for production of these mtDNA ends. For this purpose, derivatives of a-1/1R/Z1 were constructed that contained disrupted alleles of the RPO41 or MTF1 gene. These null strains, aori1rpo41Δ and aori1mtf1Δ, were found to maintain the ori1 genome stably, and Northern blot analysis confirmed the absence of mitochondrial transcription in these mutants (Fig. 3). mtDNA was then prepared from aori1rpo41Δ and aori1mtf1Δ, and the pattern was obtained after 5′ labeling and restriction digestion of this DNA was compared to that of strain a-1/1R/Z1. As illustrated in Fig. 4A, inactivation of either RPO41 or MTF1 resulted in the complete disappearance of the free 5′ DNA ends detected in the mtDNA from the wild-type strain, suggesting that mtDNA transcription is required for production of these ends. To confirm this result, aori1rpo41Δ was crossed with the [rho0] strains αRPO41 and αrpo41::URA3. The resulting diploids contained the mtDNA genome of a-1/1R/Z1 and were isogenic except for the RPO41 locus. mtDNA was then prepared from these strains and analyzed as described above (Fig. 4B). Restoration of RPO41 function in the RPO41/rpo41::URA3 diploid resulted in the reappearance of the free 5′ DNA ends detected previously. As expected, no such ends were observed in the mtDNA from the homozygous rpo41 null diploid. It was therefore concluded that the major mitochondrial transcription proteins must be present for the production of free 5′ DNA ends at the boundary of GC cluster C in ori1.
FIG. 3.
Northern blot analysis of mitochondrial transcripts from various strains carrying the ori1 genome. Portions (10 μg) of total cellular RNA from various derivatives of a-1/1R/Z1 were displayed on a 2% agarose gel and transferred to a membrane prior to hybridization with the cloned ori1 sequence from plasmid pEKSori1-2. The sources of RNA were a-1/1R/Z1 (lane 1), aori1rpo41Δ (lane 2), aori1mtf1Δ (lane 3), and aori1abf2Δ (lane 4), all of which carried the mtDNA genome of a-1/1R/Z1. Two [rho0] controls were also included: αRPO41 (lane 5) and αrpo41::URA3 (lane 6). Transcription of the mitochondrial genome of a-1/1R/Z1 leads to products of small molecular weight, a finding that is in agreement with published results for this and other petite mutants (1).
FIG. 4.
Production of free 5′ DNA ends in the ori1 genome is dependent upon mitochondrial transcription. (A) Detection of 5′ ends in the mtDNA from a-1/1R/Z1, aori1rpo41Δ, and aori1mtf1Δ (the sources of DNA are indicated above the autoradiograms). (B) The same experiment carried out with mtDNA from diploid strains carrying the RPO41/rpo41::URA3 and rpo41::URA3/rpo41::URA3 alleles, respectively. All experimental details are identical to those described for Fig. 1B. Abbreviations: H, HpaII; E, EcoRV; D, Dral.
Mitochondrial transcription-dependent DNA transactions in the mtDNA of a-1/1R/Z1.
To characterize further the DNA transactions taking place at ori1, we labeled the free 3′ DNA ends present in the mtDNA of a-1/1R/Z1 with radiolabeled cordycepin triphosphate and terminal deoxynucleotidyltransferase (see Materials and Methods). After appropriate restriction enzyme digestion, we analyzed mtDNA by electrophoresis on a polyacrylamide-urea gel. In contrast to what had been observed with 5′-labeled mtDNA, several species of relatively identical intensities were produced, whose sizes were consistent with the restriction map of a-1/1R/Z1 mtDNA (Fig. 5A). In particular, five of these bands identified DNA strands whose 3′ ends all mapped in a ∼40-bp interval centered around GC cluster C. Four of these strands had the same polarity as transcripts derived from the promoter (strands numbered 2, 3, 4, and 5 in Fig. 5C), and their 3′ ends were located, respectively, 10 and 3 bp downstream from the GC-cluster C, at the 5′ boundary of GC cluster C, and 15 bp upstream from this cluster. The remaining strand had opposite polarity and its end mapped at the 3′ end of GC cluster C. As was the case in the 5′-labeling experiments, additional species could be seen, but no simple correlation was established between their pattern and the restriction map of a-1/1R/Z1 mtDNA. Interestingly, the position of one of these 3′ ends at the 5′ boundary of GC cluster C matches exactly that of the predominant end detected by 5′ labeling (strand number 4 in Fig. 5C). Finally, none of these bands could be observed when the mtDNA was prepared from aori1rpo41Δ, confirming that these transactions were dependent upon mtRNA polymerase (Fig. 5A).
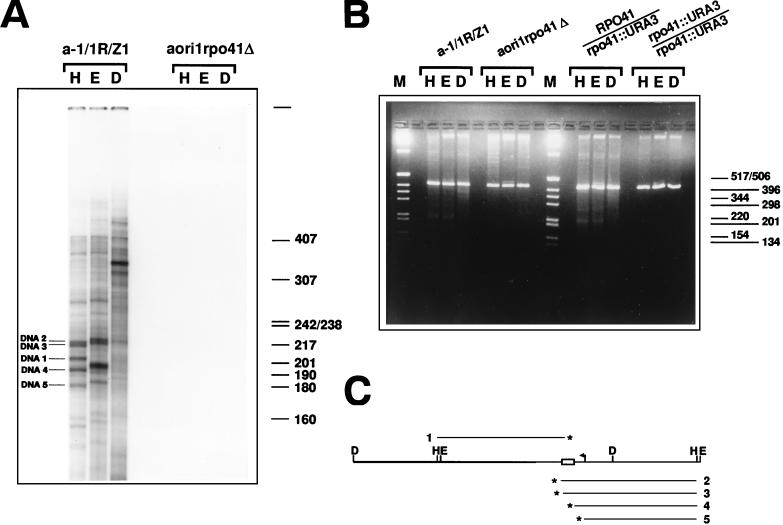
FIG. 5.
Detection of transcription-dependent 3′ DNA ends and DSBs in the mtDNA of strain a-1/1R/Z1. (A) Detection of 3′ ends in the mtDNA from a-1/1R/Z1 and aori1rpo41Δ. The 3′-end-labeled samples were restricted with HpaII (H), EcoRV (E), and DraI (D) prior to denaturation and electrophoresis on a denaturing polyacrylamide gel. The five DNA strands whose polarities were determined and whose 3′ ends were mapped are indicated (DNA1 to DNA5). The indicated size markers are those of 32P-labeled HpaII fragments of pBR322 DNA. (B). Detection of DSBs in the mtDNA of various derivatives of a-1/1R/Z1. Portions (480 ng) of mtDNA from strains a-1/1R/Z1 and aori1rpo41Δ, as well as the diploids 2n RPO41/rpo41::URA3 and 2n rpo41::URA3/rpo41::URA3 were digested with HpaII (H), EcoRV (E), or DraI (D) and displayed on a 2% agarose gel under nondenaturing conditions. In addition to the 416-bp repeat unit generated upon digestion with these single cutters, smaller DNA fragments can also be seen, whose presence depends upon a functional RPO41. M, molecular weight markers (1-kb DNA ladder; Gibco BRL). (C) Schematic diagram showing the map position and polarity for five DNA strands identified by 3′ end labeling. The polarity of each strand was determined from the size of the species obtained after digestion with HpaII and EcoRV, and the restriction map of the repeat unit. A fragment covering a little more than one repeat unit of the tandemly repeated ori1 genome was drawn so that the map positions of strands of both polarities could be shown relative to the HpaII and EcoRV sites. The asterisks indicate the 3′ end of each DNA (numbered 1 to 5). Details of the repeat unit are the same as in Fig. 1A.
The discovery of several DNA strands with free 3′ ends prompted us to examine mtDNA prepared from various ori1 strains for the presence of double-stranded DNA breaks (DSBs). DNA from strains a-1/1R/Z1 and aori1rpo41Δ, as well as the diploids 2n RPO41/rpo41::URA3 and 2n rpo41::URA3/rpo41::URA3, was restricted with HpaII, EcoRV, and DraI, and displayed on a neutral agarose gel (Fig. 5B). In addition to the 416-bp band corresponding to the repeat unit, several smaller DNA fragments of minor intensity were visible in the mtDNA from strains carrying a functional RPO41 gene. In contrast, these bands were absent in samples from the rpo41 null strains (Fig. 5B), as well as strain aori1mtf1Δ (data not shown). Thus, these additional bands were dependent upon a functional transcription apparatus. The mobilities of these bands varied with the restriction enzyme used, and the most prominent fragments had sizes of approximately 200 to 220 bp with HpaII and EcoRV, and approximately 340 to 350 bp with DraI. The lengths of these fragments are consistent with the presence of multiple DSBs at the vicinity of GC cluster C and suggest that the free 3′ ends detected around GC cluster C result from DSBs occurring in this region.
Since there are 3′ ends that match the predominant 5′ DNA ends detected at GC cluster C, we wanted to know if these 5′ ends could in fact belong to a double-stranded DNA. For this purpose, we used a native polyacrylamide gel to examine mtDNA from strain a-1/1R/Z1 after 5′ labeling and restriction digestion. A pattern nearly identical to that obtained with denaturing gels was observed. In particular, the apparent size of the most prominent product of the labeling reaction was the same as that determined under denaturing conditions (data not shown); thus, this labeled DNA is probably double stranded over its entire length.
The transcription-dependent DNA transactions detected at ori1 are not affected by inactivation of the mitochondrial nuclease gene NUC1.
The NUC1 gene encodes the major endo- or exonuclease in yeast mitochondria (46). To determine whether the NUC1 gene product plays a role in the transcription-dependent DNA transactions, we constructed a strain carrying a null allele of the NUC1 gene to compare the mtDNA from this strain with that of a wild-type strain.
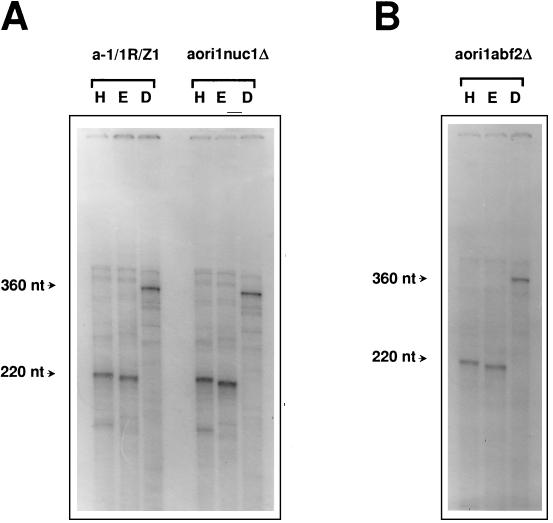
Maintenance of the ori1 HS mitochondrial genome remained unaffected upon disruption of the NUC1 gene, as anticipated from the phenotype of a nuc1 null strain carrying a wild-type mitochondrial genome (46). No difference was observed between the pattern obtained after 5′ labeling and the pattern obtained after restriction enzyme digestion of the mtDNA from the wild-type strain and the nuc1 mutant (Fig. 6A), indicating that NUC1 is not essential for metabolism of the DNA intermediates studied here. Finally, the ori1 HS genome was stably maintained in an rpo41 nuc1 double mutant (data not shown).
FIG. 6.
Detection of 5′ ends in the mtDNA of nuc1 and abf2 null strains. (A) 5′-End-labeled mtDNA samples from a-1/1R/Z1 and aori1nuc1Δ were treated and displayed as described for Fig. 1B. (B) Results of an identical experiment performed on mtDNA from strain aori1abf2Δ. H, HpaII; E, EcoRV; D, DraI.
ABF2 is not required for promoter activity at the ori1 sequence, and it is not essential for maintenance of the ori1 HS genome, even in the absence of a functional RPO41 gene.
The high-mobility-group (HMG) protein encoded by the ABF2 gene is the most abundant DNA binding protein in yeast mitochondria. Two regions of significant ABF2 binding have been detected in vitro in the rep/ori sequences, one of which is a site directly adjacent to the promoter, and the hypothesis has been raised that ABF2 might function at these origins as a trans-acting factor involved in DNA replication (14, 15). We thus tested whether ABF2 played a role in the activity of the promoter present in the rep/ori sequences and whether the ABF2 gene product would be required for maintenance of an HS genome in the presence or absence of RPO41.
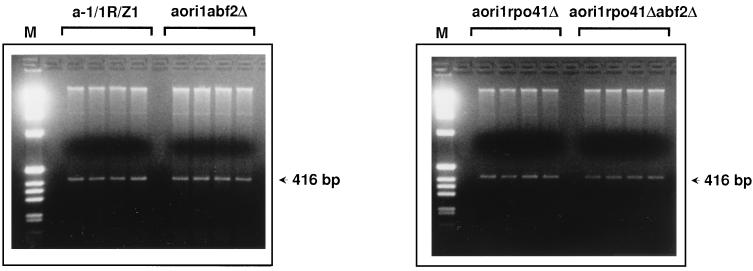
Derivatives of strain a-1/1R/Z1 were constructed that carried either a single abf2 null allele or both rpo41 and abf2 null alleles, and these strains were first tested for maintenance of the ori1 HS genome. Isolated colonies from either RPO41 ABF2 cells, RPO41 abf2 cells, rpo41 ABF2 cells, or rpo41 abf2 cells were inoculated in liquid medium containing glucose and grown for about 20 generations. Four cultures were assayed for each strain. Cells were then harvested, and total DNA was prepared and subjected to restriction enzyme analysis with HpaII, which cleaves the repeat unit of the mtDNA only once (Fig. 7). Due to its amplification in the HS genome, the 416-bp fragment diagnostic of the repeat unit of the ori1 mtDNA is clearly visible against the background of nuclear DNA. As illustrated in Fig. 7, no mtDNA loss is observed upon disruption of the ABF2 gene, regardless of the integrity of the RPO41 gene.
FIG. 7.
Maintenance of mtDNA in abf2 null derivatives of strains a-1/1R/Z1 and aori1rpo41Δ. Colonies isolated from strains a-1/1R/Z1 (RPO41 ABF2), aori1abf2Δ (RPO41 abf2::URA3), aori1rpo41Δ (rpo41::URA3 ABF2), and aori1rpo41Δabf2Δ (rpo41::URA3 abf2::URA3) were grown in liquid medium for about 20 generations. Four independent colonies were cultivated for each strain. Total cellular DNA was prepared from the cultures, restricted with HpaII, and displayed on a 2% agarose gel. The sources of DNA are indicated above the gels. The 416-bp fragment corresponding to the repeat unit of the ori1 genome is indicated by arrows. M, molecular weight markers (1-kb DNA ladder; Gibco BRL).
To assess the importance of ABF2 for promoter activity at the ori1 sequence, total RNA was prepared from an abf2 null strain carrying the ori1 HS genome and examined by Northern blot analysis with the cloned ori1 sequence as a probe (Fig. 3). The pattern of transcription, as well as the number of transcripts obtained for this strain, was found to be comparable to that of a wild-type strain, indicating that ABF2 is not required for activity of the promoter at a rep/ori sequence.
Finally, mtDNA was purified from an abf2 null strain and the pattern obtained after 5′ labeling, and restriction enzyme digestion of this DNA was compared with that of a wild-type strain. No differences were observed between these patterns (Fig. 6B). Thus, ABF2 is not required in the transcription-dependent DNA transactions detected in the mtDNA of the HS petite mutant a-1/1R/Z1.
DISCUSSION
A subset of mtDNA promoters of S. cerevisiae does not function in the transcription of genes. These are located within highly conserved sequences, called rep/ori sequences, that were originally discovered in the mitochondrial genome of HS petite mutants and which are thought to shelter the cis-acting elements conferring the transmission advantage of these mutated genomes. Some structural elements of the rep/ori sequences, notably an active promoter and a GC-rich region, are also present at the origin for leading-strand DNA replication in the mtDNA of vertebrates. These sequences have been suggested as candidates for active origins of replication that also function in wild-type mtDNA, with the promoter being required for priming of DNA synthesis by transcripts generated by mtRNA polymerase. However, a direct test of this hypothesis has not been achieved. Indeed, while transcription is required for maintenance and expression of wild-type mtDNA, the available evidence suggests that, at least for the replication of petite genomes, yeast cells have evolved mechanisms that show no stringent need for specific initiation sequences. For example, electron microscopic evidence has been taken to indicate that yeast mtDNA replicates via a rolling-circle mechanism (31). Moreover, recombination and, in particular, the transformation of recombination intermediates into replication forks have been proposed for the replication of yeast mtDNA. Recently, important connections between recombination and segregation of mtDNA have also been established (28, 29, 32). However, the molecular mechanisms of replication and segregation of yeast mtDNA remain elusive, and the role played by the promoter in the maintenance of rep/ori sequences is unknown.
As a strategy to gain insight into the function of the rep/ori promoter, we sought to identify transcription-dependent mtDNA transactions in the genome of the HS petite mutant a-1/1R/Z1, whose 416-bp repeat unit shelters the rep/ori sequence ori1. Upon direct 5′ labeling and restriction analysis of mtDNA purified from this HS petite, we have detected free 5′ ends of DNA strands that have the same polarity as transcripts derived from the promoter present in ori1 and whose termini map to the 5′ boundary of GC cluster C. This position is consistent with mapped positions of transition between RNA and DNA determined for this petite mutant by Baldacci et al. (2). By use of strains carrying null alleles of the RPO41 or MTF1 gene, the production of these ends was shown to be dependent on the presence of mtRNA polymerase. Taken together, our results are consistent with the notion that these free 5′ DNA ends reflect start sites for transcription-mediated DNA synthesis of one strand of the ori1 sequence. However, this conclusion cannot be reached unambiguously because no detectable 5′ ribonucleotides were found at these termini. Aside from the inherent susceptibility of ribonucleotides to degradation, this observation could be explained by a rapid turnover of the ribonucleotides used as primers within the mitochondria.
Some of the DNA ends detected in this study are indicative of DSBs in the region of GC cluster C, and these may account, at least in part, for the results of Baldacci et al. (2). The role of DSBs in recombination initiation is well documented for both nuclear and mtDNA in yeast cells (17, 35, 41, 49). It is tempting to speculate that the breaks detected in ori1 represent early intermediates of recombination taking place in the HS mtDNA genome. Our observation that these DSBs are dependent upon mtRNA polymerase suggests a role for transcription in the recombination of HS genomes. Transcripts from the ori1 sequence may also participate directly in DNA recombination. This possibility is suggested by the recent observation that in vitro transcription of templates containing rep/ori sequences induces the formation of a persistent RNA-DNA hybrid (44). Indeed, in the case of the immunoglobulin α switch region, where a similar, stable RNA-DNA hybrid is formed upon transcription, such a hybrid has been proposed to become an integral part of the recombination intermediate structure (36).
That the DSBs detected in this study occur around GC cluster C may not be surprising, since GC clusters of several sequence types in yeast mtDNA have been found to be recombinogenic (6, 13, 49). Further work is required to determine whether the transcription-dependent DSBs we have observed are correlated with recombination structures. Similarly, it will be interesting to learn to what extent inactivation of RPO41 affects recombination in HS mtDNA genomes.
In this study we carried out the disruption of the NUC1 and ABF2 genes in various strains harboring the ori1 mtDNA genome. Our objective was twofold. First, we wanted to learn the effect of inactivating these genes on the transcription-dependent DNA transactions that take place at ori1. Second, we wanted to know whether rpo41 abf2 and rpo41 nuc1 double mutants were capable of maintaining mtDNA. This latter question bears directly on our attempt to establish genetic evidence that RPO41 plays a role in the maintenance of HS genomes. Indeed, the loss of mtDNA in rpo41 abf2 and/or rpo41 nuc1 double mutants would be indicative of alternative mechanisms for mtDNA maintenance, one being dependent upon RPO41.
The NUC1 gene encodes the major endo- or exonuclease in yeast mitochondria. Many activities (RNase activity on single-stranded RNA, endonuclease activity on single-stranded and double-stranded DNA, 5′-exonuclease activity on double-stranded DNA) have been associated in vitro with the NUC1 protein (10). A homolog of NUC1 present in mammalian mitochondria, called Endo G, has been proposed to play a role in mtDNA primer formation (9; but see also discussion in references 8, 42, and 44). However, genetic evidence suggests that in yeast cells NUC1 functions mainly in mtDNA recombination, with its 5′-exonuclease activity involved in the production of heteroduplex DNA (47). Compared to wild-type cells, nuc1 disruptants produce only slightly more petite mutants (46), the production of which is increased when cells are disrupted at both the NUC1 gene and the CCE1/MGT1 gene encoding the mitochondrial cruciform structure resolvase (27). The multiple activities ascribed to NUC1 prompted us to test its role in the mtDNA transactions observed in a-1/1R/Z1. Our results have shown that these transactions are not affected by inactivation of NUC1 and that NUC1 is not required for HS mtDNA maintenance in the absence of RPO41. What activity is responsible for formation of the DSBs upon transcription of the ori1 sequence is therefore currently unknown. Of course, nucleases other than NUC1 may play a role in these transactions. Genetic evidence that such nucleases are present in mitochondria has been reported (34, 47) (although the petite strain in the present study would not have a nuclease with an mtDNA-encoded subunit [34]).
The HMG protein encoded by the ABF2 gene is the most abundant DNA-binding protein in yeast mitochondria. A homolog of yeast ABF2, termed mtTFA, is present in vertebrate mitochondria, and both proteins show the general ability to compact, unwind, and bend DNA (reference 22 and references therein). However, while vertebrate mtTFA has evolved a crucial role in mitochondrial transcriptional activation (reviewed in reference 40), the function of ABF2 in yeast seems limited to mtDNA segregation and maintenance. abf2 null mutants lose their wild-type mtDNA on glucose medium (14), a phenotype that can be rescued by the E. coli histone-like protein HU (33). Yet, these mutants are able to maintain and express mtDNA under selective pressure (i.e., on nonfermentable substrates such as glycerol) (14) or even on dextrose minimal medium under specific growth conditions (48), indicating that ABF2 plays no essential role in the transcription of mtDNA. That E. coli histone-like protein HU, which does not belong to the HMG family, is able to functionally complement the loss-of-function mtDNA phenotype of abf2 null mutants, suggests that ABF2 function in DNA maintenance does not involve specific protein-protein interactions. Aside from a role in mtDNA packaging and given the documented role of DNA-binding proteins, including HU, in DNA replication initiation, it is plausible that ABF2 directly participates in the formation of an open complex to allow replication to initiate at a rep/ori sequence.
For these reasons it was relevant to test the function of ABF2 in the transcription and maintenance of HS genomes, as well as its requirement in the DNA transactions that take place at a rep/ori sequence. We found that ABF2 is not required for transcription of ori1 or for the transcription-dependent DNA transactions reported in this study. These findings strengthen the notion that, unlike its vertebrate homologs, yeast ABF2 plays little, if any, direct role in mtDNA transcription. Furthermore, abf2 null mutants were found to stably maintain the HS genome of a-1/1R/Z1, even in the absence of RPO41. Thus, unlike wild-type mtDNA, the maintenance of HS genomes is not affected by ABF2 gene disruption, further supporting the idea that segregation of wild-type and HS mtDNA genomes obeys different rules.
ACKNOWLEDGMENTS
We are grateful to H. P. Zassenhaus for the gift of plasmid pUZ5. We thank the members of the Clayton lab for stimulating discussions and Michel Ghislain, Denise Thinès, Françoise Foury, and André Goffeau (Louvain-La-Neuve) for their continuous support during the redaction of the manuscript.
E.V.D. was supported by Fogarty International Center Postdoctoral Fellowship 1FO5TWO4940. This work was supported by National Institute of General Medical Sciences grant R37-GM33088-27.
ADDENDUM IN PROOF
A very recent and precise mapping of RNA primers of yeast ori5 petite mutation mtDNAs shows that the ori5 promoter is the start site for replication (Nucleic Acids Res. 26:1309–1316, 1998), consistent with the model for mammalian mtDNA replication.
REFERENCES
- 1.Baldacci G, Bernardi G. Replication origins are associated with transcription initiation sequences in the mitochondrial genome of yeast. EMBO J. 1982;1:987–994. doi: 10.1002/j.1460-2075.1982.tb01282.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baldacci G, Chérif-Zahar B, Bernardi G. The initiation of DNA replication in the mitochondrial genome of yeast. EMBO J. 1984;3:2115–2120. doi: 10.1002/j.1460-2075.1984.tb02099.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blanc H, Dujon B. Replicator regions of the yeast mitochondrial DNA responsible for suppressiveness. Proc Natl Acad Sci USA. 1980;77:3942–3946. doi: 10.1073/pnas.77.7.3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boeke J D, LaCroute F, Fink G R. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet. 1984;197:345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- 5.Chang D D, Hauswirth W W, Clayton D A. Replication priming and transcription initiate from precisely the same site in mouse mtDNA. EMBO J. 1985;4:1559–1567. doi: 10.1002/j.1460-2075.1985.tb03817.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clark-Walker G D. In vivo rearrangement of mitochondrial DNA in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1989;86:8847–8851. doi: 10.1073/pnas.86.22.8847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clayton D A. Replication and transcription of vertebrate mitochondrial DNA. Annu Rev Cell Biol. 1991;7:453–478. doi: 10.1146/annurev.cb.07.110191.002321. [DOI] [PubMed] [Google Scholar]
- 8.Clayton D A. A nuclear function for RNase MRP. Proc Natl Acad Sci USA. 1994;91:4615–4617. doi: 10.1073/pnas.91.11.4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Côté J, Ruiz-Carillo A. Primers for mitochondrial DNA replication generated by endonuclease G. Science. 1993;261:765–769. doi: 10.1126/science.7688144. [DOI] [PubMed] [Google Scholar]
- 10.Dake E, Hoffman T J, McIntire S, Hudson A, Zassenhaus H P. Purification and properties of the major nuclease from mitochondria of Saccharomyces cerevisiae. J Biol Chem. 1988;263:7691–7702. [PubMed] [Google Scholar]
- 11.de Zamaroczy M, Marotta R, Faugeron-Fonty G, Goursot R, Mangin M, Baldacci G, Bernardi G. The origins of replication of the yeast mitochondrial genome and the phenomenon of suppressivity. Nature (London) 1981;292:75–78. doi: 10.1038/292075a0. [DOI] [PubMed] [Google Scholar]
- 12.de Zamaroczy M, Faugeron-Fonty G, Baldacci G, Goursot R, Bernardi G. The ori sequences of the mitochondrial genome of a wild-type yeast strain: number, location, orientation and structure. Gene. 1984;32:439–457. doi: 10.1016/0378-1119(84)90019-2. [DOI] [PubMed] [Google Scholar]
- 13.Dieckmann C L, Gandy B. Preferential recombination between GC clusters in yeast mitochondrial DNA. EMBO J. 1987;6:4197–4203. doi: 10.1002/j.1460-2075.1987.tb02767.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diffley J F X, Stillman B. A close relative of the nuclear, chromosomal high-mobility group protein HMG1 in yeast mitochondria. Proc Natl Acad Sci USA. 1991;88:7864–7868. doi: 10.1073/pnas.88.17.7864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Diffley J F X, Stillman B. DNA binding properties of an HMG1-related protein from yeast mitochondria. J Biol Chem. 1992;267:3368–3374. [PubMed] [Google Scholar]
- 16.Dujon B. Mitochondrial genes and function. In: Broach J, Jones E, Strathern J, editors. The molecular biology of the yeast Saccharomyces cerevisiae. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1981. pp. 505–635. [Google Scholar]
- 17.Dujon B. Group I introns as mobile genetics elements: facts and mechanistic speculations—a review. Gene. 1989;82:91–114. doi: 10.1016/0378-1119(89)90034-6. [DOI] [PubMed] [Google Scholar]
- 18.Elledge S J, Mulligan J T, Ramer S W, Spottswood M, Davis R. lambda YES: a multifunctional cDNA expression vector for the isolation of genes by complementation of yeast and Escherichia coli mutations. Proc Natl Acad Sci USA. 1991;88:1731–1735. doi: 10.1073/pnas.88.5.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fangman W L, Dujon B. Yeast mitochondrial genomes consisting of only A/T base pairs replicate and exhibit suppressiveness. Proc Natl Acad Sci USA. 1984;81:7156–7160. doi: 10.1073/pnas.81.22.7156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fangman W L, Henly J W, Churchill G, Brewer B J. Stable maintenance of a 35-base-pair yeast mitochondrial genome. Mol Cell Biol. 1989;9:1917–1921. doi: 10.1128/mcb.9.5.1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fangman W L, Henly J W, Brewer B J. RPO41-independent maintenance of [rho−] mitochondrial DNA in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:10–15. doi: 10.1128/mcb.10.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fisher R P, Lisowsky T, Parisi M A, Clayton D A. DNA wrapping and bending by a mitochondrial high mobility group-like transcriptional activator protein. J Biol Chem. 1992;267:3358–3367. [PubMed] [Google Scholar]
- 23.Foury F, Van Dyck E. A PIF-dependent recombinogenic signal in the mitochondrial DNA of yeast. EMBO J. 1985;4:3525–3530. doi: 10.1002/j.1460-2075.1985.tb04112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fox T D, Folley L S, Mulero J J, McMullin T W, Thorness P E, Hedin L O, Costanzo M C. Analysis and manipulation of yeast mitochondrial genes. Methods Enzymol. 1991;194:149–165. doi: 10.1016/0076-6879(91)94013-3. [DOI] [PubMed] [Google Scholar]
- 25.Goursot R, Goursot R, Bernardi G. Temperature can reversibly modify the structure and the functional efficiency of ori sequences of the yeast mitochondrial genome. Gene. 1988;69:141–145. doi: 10.1016/0378-1119(88)90387-3. [DOI] [PubMed] [Google Scholar]
- 26.Jaehning J A. Mitochondrial transcription: is a pattern emerging? Mol Microbiol. 1993;8:1–4. doi: 10.1111/j.1365-2958.1993.tb01197.x. [DOI] [PubMed] [Google Scholar]
- 27.Kleff S, Kemper B, Sternglanz R. Identification and characterization of yeast mutants and the gene for a cruciform cutting endonuclease. EMBO J. 1992;11:699–704. doi: 10.1002/j.1460-2075.1992.tb05102.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ling F, Makishima F, Morishima N, Shibata T. A nuclear mutation defective in mitochondrial recombination in yeast. EMBO J. 1995;14:4090–4101. doi: 10.1002/j.1460-2075.1995.tb00081.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lockshon D, Zweifel S G, Freeman-Cook L L, Lorimer H E, Brewer B J, Fangman W L. A role for recombination in the segregation of mitochondrial DNA in yeast. Cell. 1995;81:947–955. doi: 10.1016/0092-8674(95)90014-4. [DOI] [PubMed] [Google Scholar]
- 30.Lorimer H E, Brewer B J, Fangman W L. A test of the transcription model for biased inheritance of yeast mitochondrial DNA. Mol Cell Biol. 1995;15:4803–4809. doi: 10.1128/mcb.15.9.4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maleszka R, Skelly P J, Clark-Walker G D. Rolling circle replication of DNA in yeast mitochondria. EMBO J. 1991;10:3923–3929. doi: 10.1002/j.1460-2075.1991.tb04962.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maxam A, Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65:499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- 33.Megraw T L, Chae C-B. Functional complementarity between the HMG1-like yeast mitochondrial histone HM and the bacterial histone-like protein HU. Proc Natl Acad Sci USA. 1993;268:12758–12763. [PubMed] [Google Scholar]
- 34.Morishima N, Nakagawa K, Shibata T. A sequence-specific endonuclease, Endo.Scel, can efficiently induce gene conversion in yeast mitochondria lacking a major exonuclease. Curr Genet. 1993;23:537–541. doi: 10.1007/BF00312648. [DOI] [PubMed] [Google Scholar]
- 35.Nakagawa K, Morishima N, Shibata T. An endonuclease with multiple cutting sites, Endo.Scel, initiates genetic recombination at its cutting site in yeast mitochondria. EMBO J. 1992;11:2707–2715. doi: 10.1002/j.1460-2075.1992.tb05336.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reaban M E, Lebowitz J, Griffin J A. Transcription induces the formation of a stable RNA-DNA hybrid in the immunoglobulin α switch region. J Biol Chem. 1994;269:21850–21857. [PubMed] [Google Scholar]
- 37.Rickwood D, Dujon B, Darley-Usmar V M. Yeast mitochondria. In: Campbell I, Duffus J H, editors. Yeast: a practical approach. Oxford, England: IRL Press; 1988. pp. 185–254. [Google Scholar]
- 38.Schinkel A H, Tabak H F. Mitochondrial RNA polymerase: dual role in transcription and replication. Trends Genet. 1989;5:149–154. doi: 10.1016/0168-9525(89)90056-5. [DOI] [PubMed] [Google Scholar]
- 39.Schmitt M E, Brown T A, Trumpower B L. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 1990;18:3091–3092. doi: 10.1093/nar/18.10.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shadel G S, Clayton D A. Mitochondrial transcription initiation: variation and conservation. J Biol Chem. 1993;268:16083–16086. [PubMed] [Google Scholar]
- 41.Shinohara A, Ogawa T. Homologous recombination and the roles of double-strand breaks. Trends Biochem Sci. 1995;20:387–391. doi: 10.1016/s0968-0004(00)89085-4. [DOI] [PubMed] [Google Scholar]
- 42.Stohl L L, Clayton D A. Saccharomyces cerevisiae contains an RNase MRP that cleaves at a conserved mitochondrial RNA sequence implicated in replication priming. Mol Cell Biol. 1992;12:2561–2569. doi: 10.1128/mcb.12.6.2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weiller G, Schueller C M E, Schweyen R J. Putative target sites for mobile G+C-rich clusters in yeast mitochondrial DNA: single elements and tandem arrays. Mol Gen Genet. 1989;218:272–283. doi: 10.1007/BF00331278. [DOI] [PubMed] [Google Scholar]
- 44.Xu B, Clayton D A. A persistent RNA-DNA hybrid is formed during transcription at a phylogenetically conserved mitochondrial DNA sequence. Mol Cell Biol. 1995;15:580–589. doi: 10.1128/mcb.15.1.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu B, Clayton D A. RNA-DNA hybrid formation at the human mitochondrial heavy-strand origin ceases at replication start sites: an implication for RNA-DNA hybrids serving as primers. EMBO J. 1996;15:3135–3143. [PMC free article] [PubMed] [Google Scholar]
- 46.Zassenhaus H P, Hofmann T J, Uthayashanker R, Vincent R D, Zona M. Construction of a yeast mutant lacking the mitochondrial nuclease. Nucleic Acids Res. 1988;16:3282–3296. doi: 10.1093/nar/16.8.3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zassenhaus H P, Denniger G. Analysis of the role of the NUC1 endo/exonuclease in yeast mitochondrial DNA recombination. Curr Genet. 1994;25:142–149. doi: 10.1007/BF00309540. [DOI] [PubMed] [Google Scholar]
- 48.Zelenaya-Troitskaya O, Perlman P S, Butow R A. An enzyme in yeast mitochondria that catalyzes a step in branched-chain amino acid biosynthesis also functions in mitochondrial DNA stability. EMBO J. 1995;14:3268–3276. doi: 10.1002/j.1460-2075.1995.tb07330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zinn A R, Pohlman J K, Perlman P S, Butow R A. In vivo double-strand breaks occur at recombinogenic G+C-rich sequences in the yeast mitochondrial genome. Proc Natl Acad Sci USA. 1988;85:2686–2690. doi: 10.1073/pnas.85.8.2686. [DOI] [PMC free article] [PubMed] [Google Scholar]