Abstract
Quail, as an advantageous avian model organism due to its compact size and short reproductive cycle, holds substantial potential for enhancing our understanding of skeletal muscle development. The quantity of skeletal muscle represents a vital economic trait in poultry production. Unraveling the molecular mechanisms governing quail skeletal muscle development is of paramount importance for optimizing meat and egg yield through selective breeding programs. However, a comprehensive characterization of the regulatory dynamics and molecular control underpinning quail skeletal muscle development remains elusive. In this study, through the application of HE staining on quail leg muscle sections, coupled with preceding fluorescence quantification PCR of markers indicative of skeletal muscle differentiation, we have delineated embryonic day 9 (E9) and embryonic day 14 (E14) as the start and ending points, respectively, of quail skeletal muscle differentiation. Then, we employed whole transcriptome sequencing to investigate the temporal expression profiles of leg muscles in quail embryos at the initiation of differentiation (E9) and upon completion of differentiation (E14). Our analysis revealed the expression patterns of 12,012 genes, 625 lncRNAs, 14,457 circRNAs, and 969 miRNAs in quail skeletal muscle samples. Differential expression analysis between the E14 and E9 groups uncovered 3,479 differentially expressed mRNAs, 124 lncRNAs, 292 circRNAs, and 154 miRNAs. Furthermore, enrichment analysis highlighted the heightened activity of signaling pathways related to skeletal muscle metabolism and intermuscular fat formation, such as the ECM-receptor interaction, focal adhesion, and PPAR signaling pathway during E14 skeletal muscle development. Conversely, the E9 stage exhibited a prevalence of pathways associated with myoblast proliferation, exemplified by cell cycle processes. Additionally, we constructed regulatory networks encompassing lncRNA‒mRNA, miRNA‒mRNA, lncRNA‒miRNA-mRNA, and circRNA-miRNA‒mRNA interactions, thus shedding light on their putative roles within quail skeletal muscle. Collectively, our findings illuminate the gene and non-coding RNA expression characteristics during quail skeletal muscle development, serving as a foundation for future investigations into the regulatory mechanisms governing non-coding RNA and quail skeletal muscle development in poultry production.
Key words: quail, skeletal muscle, whole transcriptome sequencing, differentially expressed RNAs, RNA regulatory network
INTRODUCTION
Poultry skeletal muscle is valued for its high-quality protein content, contributing to the diverse array of poultry-based food products available for human consumption (Petracci et al., 2013; Liu et al., 2019b). In the wake of the 2019 COVID-19 pandemic, quails were officially reclassified as poultry rather than being categorized solely as specialty economic poultry in the Chinese National Livestock and Poultry Genetic Resources Catalog (http://www.moa.gov.cn/xw/zwdt/202005/t20200529_6345528.htm), highlighting the increasing importance of the quail industry. Quails, beyond their role in egg production, hold significant value in meat production (Quaresma et al., 2022). Understanding the developmental intricacies of quail skeletal muscle can greatly enhance meat quality and assist in selecting quails with desirable traits for egg production. However, a comprehensive understanding of the developmental characteristics specific to quail skeletal muscle has yet to be fully elucidated.
The development of animal skeletal muscle primarily involves a series of well-coordinated steps, including the gradual differentiation of myoblasts into myotubes, followed by myotube fusion and the thickening of muscle fibers (Braun and Gautel, 2011; Abmayr and Pavlath, 2012). Additionally, the activation and differentiation of previously dormant satellite cells contribute to the maturation of skeletal muscle fibers (Sousa-Victor et al., 2022). This intricate process of transitioning myotubes into mature skeletal muscle is orchestrated by the interplay of multiple genes and epigenetic factors (Buckingham and Rigby, 2014; Dong et al., 2020; Rugowska et al., 2021). Key regulators, such as Myogenic Differentiation 1 (MyoD) and Myogenin (MyoG), govern the differentiation of myoblasts (Buckingham and Rigby, 2014). Apart from the influence of functional genes, epigenetic regulation plays a crucial role in the growth and development of skeletal muscle. Mechanisms such as DNA methylation, histone modifications, RNA modifications, and noncoding RNAs (ncRNAs, including microRNAs, long noncoding RNAs, and circular RNAs) collectively contribute to the complex orchestration of muscle development processes (Bianconi and Mozzetta, 2022). NcRNAs predominantly regulate specific biological processes by precisely controlling the expression of coding genes (Dong et al., 2020; Cai et al., 2022b). Nevertheless, a comprehensive understanding of the gene regulatory network governing quail skeletal muscle development is still under investigation, and the distinct characteristics of ncRNA expression in skeletal muscle remain to be elucidated. Furthermore, the involvement of epigenetic regulation in skeletal muscle development has not been extensively reported. The utilization of whole transcriptome sequencing provides a powerful tool for identifying unique expression patterns of genes and ncRNAs within tissues and cells, facilitating the prediction of intricate gene-ncRNA interactions and their regulatory networks (Guo et al., 2023).
In this study, we collected quail leg muscle tissues spanning from the seventh day of incubation (E7) until E15 and the first day posthatching (P1). The purpose was to explore the developmental patterns exhibited by quail embryo leg muscles. Through the preparation of tissue sections, we aimed to discern the distinguishing characteristics associated with quail leg muscle development during the embryonic stage. Combining the results of the previous qPCR of markers indicative of skeletal muscle differentiation (Liu et al., 2023c), we selected leg muscle tissues at 2 critical stages: the initial phase of differentiation (embryonic day 9, E9) and the subsequent completion of differentiation (embryonic day 14, E14), which were subjected to comprehensive whole transcriptome sequencing. By employing this approach, we successfully delineated the distinct expression patterns of genes and ncRNAs within quail embryonic leg muscles. Moreover, we ventured into predicting the regulatory roles that long noncoding RNAs (lncRNAs), microRNAs (miRNAs), and circular RNAs (circRNAs) potentially exert on genes. Furthermore, we conducted predictive analyses on competing endogenous RNA (ceRNA) regulatory networks, encompassing miRNA-centered interactions among lncRNAs-mRNAs and circRNAs-mRNAs. The outcomes derived from our experimental endeavors will undoubtedly furnish a solid foundation for comprehending the developmental traits inherent in quail skeletal muscles while simultaneously unraveling the intricate molecular regulatory networks governing quail leg muscles.
MATERIAL AND METHODS
Ethical Statement
The animal experiments described in this paper were conducted in accordance with relevant national regulations on animal ethics and welfare, as well as the ethical guidelines and animal experimental safety review system of Jiangxi Agricultural University ([2018]30). All animals were treated with utmost care and humanely.
Animal and Tissue Collection
The quail eggs used in this study belonged to the Longcheng line of Korean quail (purchased from the Hengyan Poultry Industry in Jiangxi Province, China). The incubation, sampling, and gender identification of the eggs were performed as previously described (Liu et al., 2023c). A total of 220 quail eggs were incubated at a temperature of 38°C and a relative humidity of 60%. Upon hatching, the temperature was adjusted to 37°C, and the humidity was adjusted to 70%. Quail embryo leg muscle tissues at developmental stages ranging from E7 to E15 of incubation, as well as leg muscle tissues from P1, were collected. Over a specified timeframe, calf samples from 2 quails were harvested and preserved in 4% paraformaldehyde, intended for future histological analysis. Concurrently, a combination of thigh and calf samples from an additional twelve quails were gathered and rapidly frozen in liquid nitrogen, earmarked for upcoming RNA-based investigations. In the context of histological sample preparation, we independently procured musculature from the calves of the quail. Subsequently, we fabricated paraffin sections, and conducted Hematoxylin and Eosin (HE) staining. With respect to the RNA-seq samples, considering the diminutive size of the quail's legs and the necessity to excise the skin and bones to procure skeletal muscle tissue, we elected to amalgamate the thigh and calf muscles. This composite sample was then utilized for RNA extraction, RNA sequencing, and the ensuing validation of RNA-seq. After gender identification using the chromodomain helicase DNA binding protein 1 (CHD1) gene primer, female individuals were selected for subsequent RNA sequencing and quantitative PCR (qPCR) validation experiments.
Morphologic Analysis of Quail Leg Muscles
Two leg muscle samples from individual quails were collected at each stage and fixed in 4% paraformaldehyde. Two slices were prepared from each sample, followed by HE staining. The simplified steps for HE staining were as follows: staining with hematoxylin for 2 min, rinsing twice in water, immersion in 60% ethanol for 2 min, 70% ethanol for 2 min, 80% ethanol for 2 min, 90% ethanol for 2 min, eosin for 2 s, 95% ethanol for 1 min, dehydration in absolute ethanol for 6 min, clearing in xylene for 15 min, and finally mounting with neutral resin. The prepared slices were scanned using a microscope to obtain images for subsequent analysis.
RNA Extraction and Sequencing Library Preparation
TRIzol reagent (Invitrogen, Carlsbad, CA) was employed to extract total RNA from quail leg muscle tissues. The integrity of the RNA was evaluated using agarose gel electrophoresis and an Agilent 2100 Bioanalyzer (Agilent Technologies, Waldbronn, Germany). Only RNA samples that exhibited no degradation or contamination were utilized for subsequent RNA library preparation.
For the construction of mRNA, lncRNA, and circRNA libraries, we adopted a ribosomal RNA (rRNA) depletion method. In essence, rRNAs were selectively removed to enrich the presence of mRNAs and ncRNAs. The enriched RNAs were fragmented using fragmentation buffer and then reverse transcribed into cDNA utilizing random primers. Subsequently, second-strand cDNA synthesis was accomplished through the use of DNA polymerase I, RNase H, dNTPs, and buffer. The resulting cDNA fragments were purified utilizing the QiaQuick PCR extraction kit (Qiagen, Venlo, Netherlands). They were further subjected to end repair, poly(A) tail addition, and ligation with Illumina sequencing adapters. To digest the second-strand cDNA, uracil-N-glycosylase (UNG) was employed. The digested products were then subjected to size selection via agarose gel electrophoresis, followed by PCR amplification and sequencing using the Illumina HiSeq 4000 platform (Genedenovo, Guangzhou, China).
For the miRNA libraries, RNA molecules within the size range of 18-30 nt were isolated using polyacrylamide gel electrophoresis (PAGE). Following this step, 3′ adapters were added, enabling further enrichment of RNAs ranging from 36 to 48 nt in size. Ligation of the 5′ adapters was subsequently performed, facilitating reverse transcription through PCR amplification. PCR products within the size range of 140 to 160 bp were enriched to generate a cDNA library, which was then sequenced using the Illumina HiSeq Xten platform (Genedenovo).
mRNA and LncRNA Sequencing Data Analysis
To ensure the generation of high-quality clean reads, we applied fastp (version 0.18.0) (Chen et al., 2018) to further filter the reads. After excluding rRNA sequences using Bowtie2 (version 2.2.8) (Langmead and Salzberg, 2012), the remaining clean reads underwent mapping against the reference genome of quail (Coturnix japonica 2.1, accessible at https://www.ncbi.nlm.nih.gov/assembly/GCF_001577835.2) via HISAT2 (version 2.1.0) (Kim et al., 2015). Subsequently, transcript reconstruction was performed with Stringtie (version 1.3.4) (Pertea et al., 2015). To identify known mRNAs, lncRNAs, and new transcripts, we aligned all reconstructed transcripts to the reference genome and categorized them into twelve distinct groups using Cuffcompare (Trapnell et al., 2012). To classify reliable novel genes, we established specific criteria, including a transcript length exceeding 200 bp and an exon count greater than 2. Furthermore, we employed CNCI (version 2) (Sun et al., 2013), CPC (version 0.9-r2) (http://cpc.cbi.pku.edu.cn/) (Kong et al., 2007), and FEELNC (version v0.2) (https://github.com/tderrien/FEELnc) (Wucher et al., 2017) to evaluate the protein-coding potential of these novel transcripts. By considering the intersection of nonprotein-coding potential results from both tools, we confidently identified lncRNAs. To gauge the expression abundance and variations of each transcription region, we calculated the fragments per kilobase of transcript per million mapped reads (FPKM) value using RSEM (Li and Dewey, 2011). To gain further insights, we performed principal component analysis (PCA) by utilizing the gmodels package in R (http://www.r-project.org/).
For subsequent analyses, we performed differential expression analysis separately for coding RNAs and lncRNAs. DESeq2 (Love et al., 2014) was employed to assess differential expression between E14 and E9 for both genes and lncRNAs. Genes/lncRNAs displaying a false discovery rate (FDR) below 0.05 and an absolute fold change exceeding and equal to 2 were considered significantly differentially expressed. Moving forward, we subjected the differentially expressed genes (DEGs) to enrichment analysis encompassing Gene Ontology (GO) functions, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways, and gene set enrichment analysis (GSEA). This analysis was conducted using the omicsmart online tool (https://www.omicsmart.com/, Genedenovo), offering a comprehensive understanding of the functional implications and pathways associated with the identified DEGs.
Prediction of LncRNA's Regulatory Role on mRNA
To predict the potential binding between antisense lncRNAs and mRNAs, we utilized RNAplex (version 0.2) (http://www.tbi.univie.ac.at/RNA/RNA plex.1.html) (Tafer and Hofacker, 2008). This program incorporates the ViennaRNA package and calculates the minimum free energy based on thermodynamic structure analysis to determine the optimal base pairing relationships. For the prediction of cis-regulatory target genes, we annotated the genomic positions of lncRNAs. If a lncRNA was located within 10 kb upstream or downstream of a gene, it was considered to potentially intersect with cis-regulatory elements in that region. For trans-regulatory effects, we employed the Pearson correlation coefficient method to assess the expression correlation between lncRNAs and genes across samples. LncRNA-mRNA pairs with an absolute correlation value greater than 0.95 were selected as trans-regulatory interactions. Subsequently, we conducted GO and KEGG enrichment analyses specifically for the identified mRNAs involved in these trans-regulatory interactions.
CircRNA Sequencing Data Analysis
In the analysis of circRNA sequencing data, we followed a similar approach to the analysis of lncRNAs for data filtering and genome alignment. Once the reads were aligned with the reference genome, those that could be mapped to the genomes were discarded, while the unmapped reads were collected for circRNA identification. Specifically, we focused on the unmapped reads where 20 nucleotides from both ends aligned to the reference genome. To identify circRNAs, we utilized the find_circ tool (Memczak et al., 2013). A candidate circRNA was considered if it had support from at least 2 unique back-spliced reads observed in one or more samples. Following this step, we conducted statistical analysis to determine the type, chromosome distribution, and length distribution of the identified circRNAs.
To quantify circRNAs, we normalized the back-spliced junction reads to reads per million mapped reads (RPM). For the detection of differentially expressed circRNAs (DECs) across samples or groups, we employed the edgeR package (version 3.12.1) (http://www.r-project.org/) (Robinson et al., 2010). We deemed circRNAs as DECs when they displayed a fold change of at least 2 and a p value less than 0.05 in E14 vs. E9. Subsequently, we used the parental genes of the DECs to perform GO and KEGG enrichment analysis using omicsmart (https://www.omicsmart.com/).
miRNA Sequencing Data Analysis
In the analysis of miRNA sequencing data, we performed a stringent filtering process to ensure the integrity and reliability of the dataset. Reads containing more than one low-quality base (Q-value ≤ 20), reads lacking 3′ adapters, reads containing 5′ adapters, reads with both 3′ and 5′ adapters but without a small RNA fragment between them, reads containing polyA in the small RNA fragment, and reads shorter than 18 nt (excluding adapters) were removed. Once the dataset was cleaned, all the remaining clean tags were aligned against the small RNAs in the GenBank database (Release 209.0) as well as the Rfam database (Release 11.0) to exclude sequences corresponding to rRNA, scRNA, snoRNA, snRNA, and tRNA. Additionally, alignment with the quail reference genome was performed, and any tags mapping to exons, introns, or repeat sequences were eliminated. The clean tags that remained were further screened against the miRBase database (Release 22) to identify known miRNAs. The known miRNAs are typically named by appending x, y, or z to the miRNA family name. An “x” suffix indicates alignment with the 5′ arm of the precursor of that miRNA family in other species in miRBase, “y” denotes alignment with the 3′ arm, and “z” signifies alignment with a miRNA whose 3′ or 5′ arm origin is unspecified in the database. Any unannotated tags were aligned with the reference genome, and potential novel miRNA candidates were determined based on their genomic positions and the hairpin structures predicted using miRDeep2 (Friedländer et al., 2012).
To quantify miRNA expression levels accurately, we calculated and normalized them to transcripts per million (TPM). Differential expression analysis of miRNAs was conducted using edgeR, comparing E14 and E9. We identified miRNAs with a fold change ≥ 2 and a P value < 0.05 as significant differentially expressed miRNAs (DEMs). In predicting miRNA target genes, we set strict parameters for Miranda software, requiring binding free energy values below −10 kcal/mol and binding scores above 50. We also used TargetScan to ensure the seed sequence (2–8 nt) at the miRNA's 5′ end aligns with the RNA transcript's 3′UTR. The intersecting results from both tools were then displayed for a complete prediction overview. Subsequently, the target genes of the DEMs were subjected to GO and KEGG enrichment analysis using omicsmart, providing valuable insights into the functional implications and pathways associated with these miRNAs.
Prediction of CeRNA Regulatory Network
To identify potential interacting ceRNAs among differentially expressed RNAs, we implemented a comprehensive filtering approach encompassing 3 key aspects. First, we examined the targeting relationship and inversely correlated expression levels between miRNAs and candidate ceRNAs. At this stage, we set more rigorous standards. Alongside the initial TargetScan prediction, we mandated that Miranda software predictions have binding free energy values below −15 kcal/mol and binding scores exceeding 70. Additionally, we considered the positively correlated expression patterns exhibited by candidate ceRNAs, as well as the enrichment level of shared miRNA binding sites. First, we focused on differentially expressed RNAs (mRNAs, lncRNAs, circRNAs) that displayed regulatory relationships with DEMs, employing a stringent filtering criterion based on the Spearman rank correlation coefficient. Specifically, we selected target gene pairs with coefficients equal to or less than –0.7. Encompassing the ceRNA hypothesis, whereby ceRNAs exhibit positive correlations in their expression levels, we further refined our analysis by identifying ceRNA pairs with a Pearson correlation coefficient of 0.9 or higher. Subsequently, utilizing the hypergeometric cumulative distribution function test, we identified ceRNA pairs with a P value below 0.05 as the final set of ceRNA pairs. Upon completion of the prediction process, GO and KEGG enrichment analyses were performed individually for lncRNAs, circRNAs, and the shared ceRNAs (mRNAs). Finally, the ceRNA regulatory network was visualized using Cytoscape (version 3.7.1), effectively portraying the intricate interplay among ceRNAs.
Validation of Sequencing Data
The sequencing data were validated using a qPCR approach. To quantify mRNA, lncRNA, and circRNA, we designed specific primers, with GAPDH chosen as the internal reference. The quantification reagents and procedures employed were in accordance with the methodology described in our previously published paper (Chen et al., 2022a). For miRNA quantification, we synthesized specific primers (RiboBio, Guangzhou, China), with U6 selected as the internal reference. The quantification reagents and procedures utilized were consistent with those outlined in our previously published paper (Li et al., 2020). All the primers utilized in this study can be found in Supplementary Table 1. Values are reported as the mean ± SEM. A two-tailed T test was employed to assess the differences between the 2 groups. Significance levels are denoted as follows: * for P < 0.05, ⁎⁎ for P < 0.01, and ⁎⁎⁎ for P < 0.001.
RESULTS
Developmental Patterns of Leg Muscles in Quail Embryos
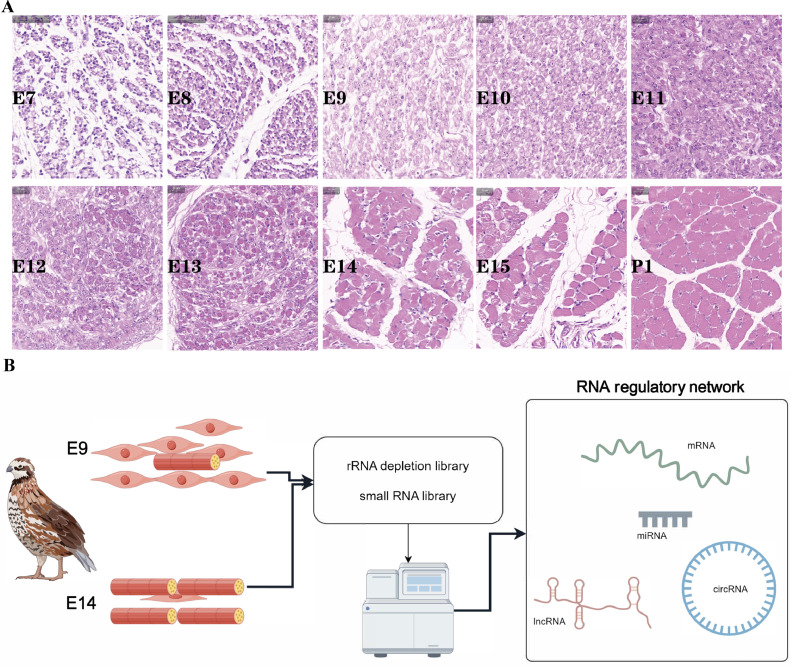
The aim of this study was to identify the developmental patterns of skeletal muscles in quail. Specifically, we focused on analyzing leg muscle tissues from E7 to P1. Through histological analysis of paraffin sections, we observed interesting changes during different time periods. Between E7 and E9, there was a gradual increase in the number of multinucleated myoblasts, indicating the main proliferation phase of myoblasts (Figure 1A). From E10 to E13, myoblasts primarily underwent differentiation, gradually transforming into multinucleated myotubes (Figure 1A). At E14, the myotubes further fused together to form muscle fibers. By P1, most of the muscle fibers in quail legs had formed, resembling those found in adult animals (Figure 1A). Our previous quantitative analysis of the differentiation marker genes MYOG, MYOD, and MSTN in quail leg muscles supported these findings and allowed us to identify E9 and E14 as distinct developmental stages for quail leg muscle (Liu et al., 2023c). However, it is important to note that collecting adequate samples during E7 and E8 proved challenging due to the relatively small size of quail leg muscle tissues, making sample collection and impurity removal more difficult. Therefore, for subsequent whole transcriptome experiments, we selected leg muscle tissues from E9 and E14 as our experimental samples (Figure 1B).
Figure 1.
The developmental patterns of leg muscles in quail embryos. (A) HE staining of embryonic leg muscle. E7, E8, E9, E10, E11, E12, E13, E14, E15, and P1 represent embryonic day 7 to day 15 and posthatching day 1, respectively. (B) Schematic illustration of the experimental strategy.
Expression of Leg Muscle Genes in the Quail Embryonic Stage
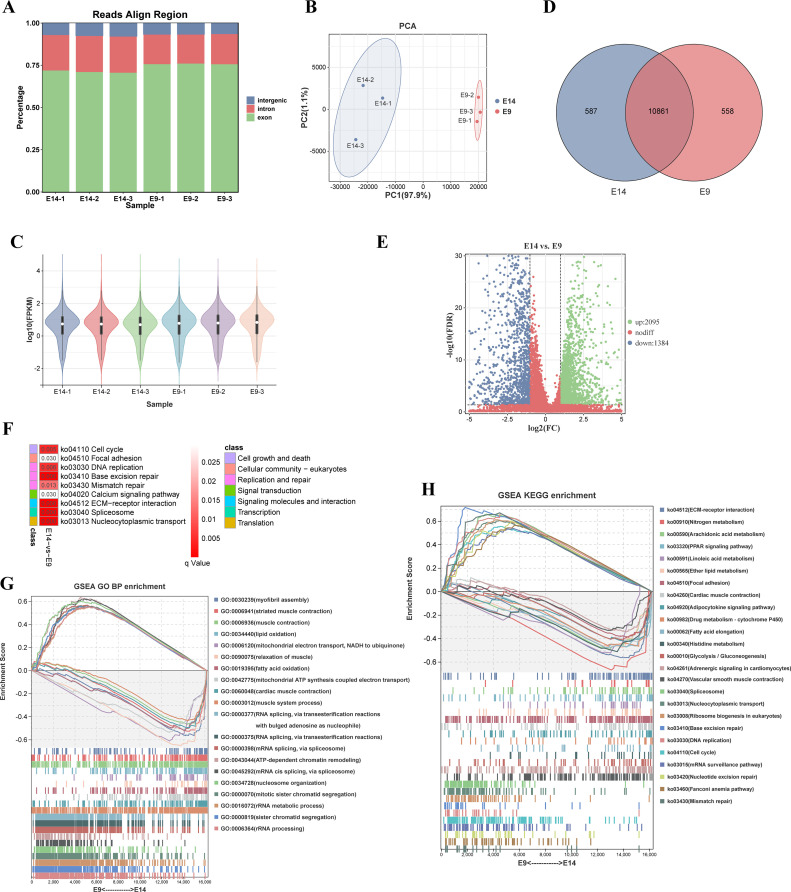
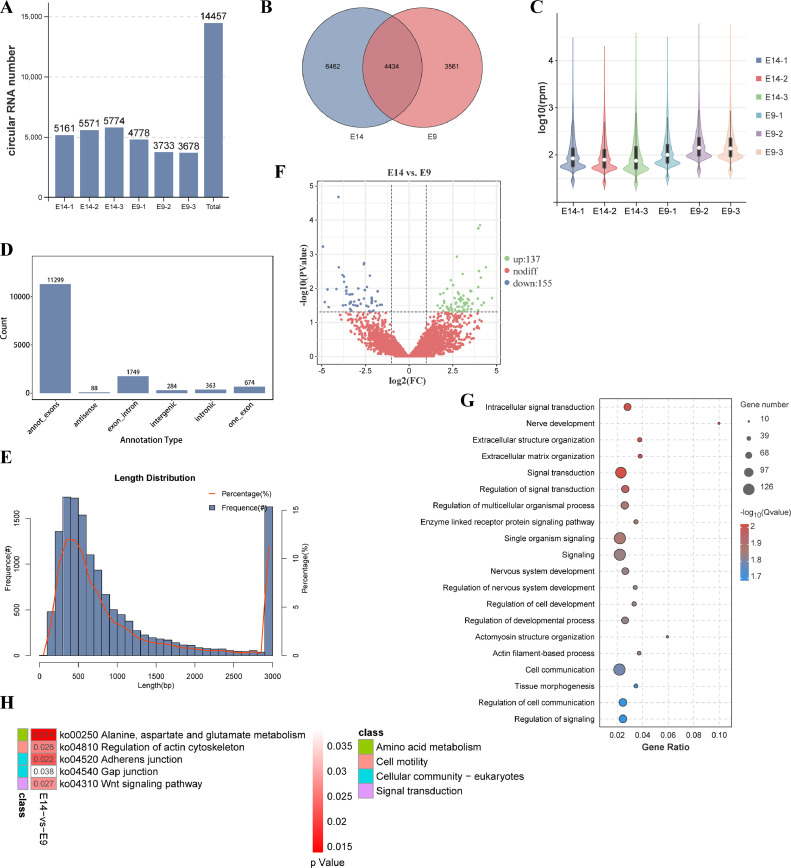
After performing ribosomal RNA depletion and constructing libraries, an average of 83,418,700.33 clean reads were obtained per sample (Table 1). These reads aligned to the quail genome with approximately 70% mapping successfully, while the remaining 30% were classified as unaligned reads (Table 1). Analysis of the alignment positions on the quail genome revealed that within the aligned reads, approximately 73% mapped to exonic regions, approximately 20% mapped to intronic regions, and approximately 7% mapped to intergenic regions (Figure 2A). Principal component analysis showed well-defined clustering of replicates for both groups, with particular emphasis on the striking similarity among the 3 replicates at E9 (Figure 2B). Upon annotating the expressed genes in both groups, it was discovered that 10,861 genes were commonly expressed, whereas 587 and 558 genes exhibited specific expression in the E14 and E9 groups, respectively (Figure 2C). Furthermore, gene expression level calculations demonstrated comparable distribution patterns across all 6 samples (Figure 2D).
Table 1.
Overview of the rRNA depletion sequencing data.
| Sample | raw data | cleandata(%) | Q20(%) | Q30(%) |
|---|---|---|---|---|
| E14-1 | 76,584,182 | 75,889,356 (99.09%) | 10,779,011,924 (95.71%) | 10,099,810,949 (89.68%) |
| E14-2 | 90,952,768 | 90,020,540 (98.98%) | 12,775,891,976 (95.86%) | 12,023,041,711 (90.22%) |
| E14-3 | 85,740,366 | 84,869,396 (98.98%) | 12,007,289,026 (95.28%) | 11,220,165,389 (89.03%) |
| E9-1 | 102,461,308 | 101,509,706 (99.07%) | 14,494,543,971 (96.15%) | 13,654,072,458 (90.58%) |
| E9-2 | 75,306,064 | 74,435,608 (98.84%) | 10,563,042,987 (95.59%) | 9,895,863,730 (89.55%) |
| E9-3 | 77,268,204 | 76,420,174 (98.90%) | 10,892,968,194 (95.96%) | 10,249,273,660 (90.29%) |
Figure 2.
mRNA dynamics in quail leg muscle development. (A) Chart illustrating the mapping ratio of clean reads produced from the rRNA depletion library. (B) PCA plot of mRNA expression across all samples, with the E14 group represented in blue and the E9 group in red. (C) Bar graph showcasing the expression levels of all discovered genes across 6 individual samples. (D) Venn diagram demonstrating the shared and unique genes detected in both the E14 and E9 stages. (E) Volcano plot visualizing the differential mRNA expression between the E14 and E9 groups. (F) Graphical representation of KEGG enrichment analysis for DEGs found in E14 vs. E9, plotted based on the respective q value of each KEGG pathway. (G) GSEA GO enrichment analysis of DEGs between E14 and E9, highlighting the top 10 biological process terms with maximum NES adjacent to their smallest q values and the top 10 terms with minimum NES next to their smallest q values. (H) Illustration of the GSEA KEGG enrichment analysis for DEGs in E14 and E9, featuring the top 20 KEGG signaling pathways sorted by smallest q value.
Subsequently, we focused on identifying and functionally analyzing differentially expressed genes. By applying the screening criteria of FDR < 0.05 and |log2FC| ≥ 1 in the E14 vs. E9 comparison, we identified a total of 2,095 upregulated genes and 1,384 downregulated genes (Figure 2E, Supplementary Table 2). Notably, the top 5 genes with the smallest FDR values were LOC107322047 (myosin heavy chain), sarcolipin (SLN), myelin basic protein (MBP), synaptophysin like 2 (SYPL2), and perilipin 1 (PLIN1). GO enrichment analysis of the differentially expressed genes (DEGs) highlighted their significant association with biological processes related to the mitotic cell cycle, chromosome organization, muscle development, and muscle contraction (Supplementary Table 3).
Furthermore, KEGG enrichment analysis demonstrated significant enrichment of the DEGs in signaling pathways such as the cell cycle, focal adhesion, DNA replication, calcium signaling pathway, and ECM-receptor interaction (Figure 2F, Supplementary Table 4). To evaluate the overall gene expression changes between the 2 stages, we performed GSEA for GO and KEGG analyses. The results revealed that during the E14 embryonic stage, alterations were primarily observed in GO biological process (BP) terms and KEGG pathways related to contractile myofibril assembly, muscle contraction, muscle system process, ECM-receptor interaction, PPAR signaling pathway and so on (Figure 2G, 2H, Supplementary Table 5 and 6). Conversely, changes in gene expression during the E9 embryonic stage were associated with GO BP terms including RNA splicing, mitotic sister chromatid segregation, and KEGG pathways such as cell cycle, spliceosome, and DNA replication (Figure 2G, 2H, Supplementary Table 5 and 6). These enrichments align with our tissue section results and suggest that at E9, quail embryonic leg muscles are in a state of proliferation and the beginning of differentiation, while at E14, there is a substantial presence of muscle fibers.
Characteristics and Expression of lncRNAs in Quail Embryonic Leg Muscles
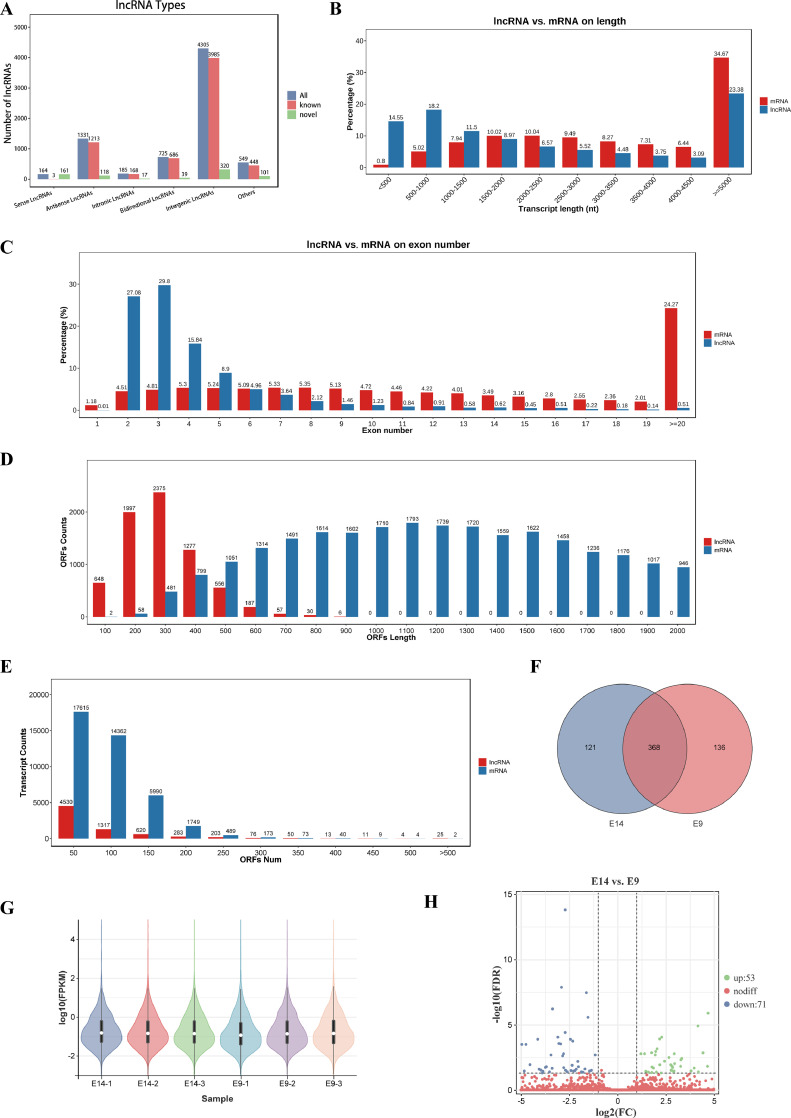
First, we annotated the predicted lncRNAs for their genomic positions and categorized them into 6 main types: intergenic lncRNAs, bidirectional lncRNAs, intronic lncRNAs, antisense lncRNAs, sense lncRNAs, and other types. Among these, the most abundant category was intronic lncRNAs (Figure 3A). Analyzing their length distribution, we found that mRNAs and lncRNAs with lengths equal to or greater than 5,000 nt accounted for approximately 34.67 and 23.38%, respectively. Interestingly, approximately 32.76% of lncRNAs had lengths equal to or less than 1000 nt, whereas only a mere 5.82% of mRNAs fell within this range (Figure 3B). Delving deeper into their exon content, we observed that the majority of lncRNAs contained 2, 3, 4, or 5 exons (81.62%), while mRNA exons displayed a wider distribution without any significant clustering (Figure 3C). Turning our attention to the open reading frame (ORF) lengths and counts, we discovered that lncRNAs had ORFs that were all below 900 nt (300 aa), whereas mRNAs exhibited a broader range of ORF lengths (Figure 3D). Furthermore, when examining the number of lncRNAs and mRNAs with different ORF lengths, we noticed that most lncRNAs and mRNAs had ORF numbers below 300, while the number of mRNA transcripts with different numbers of ORFs was far greater than the number of lncRNAs (Figure 3E).
Figure 3.
Identification and characterization of lncRNAs in quail leg muscle. (A) The number of different lncRNA types. (B) The length distribution of lncRNAs and mRNAs in quail leg muscle. (C) The distribution of the exon number of lncRNAs and mRNAs contained. (D) The distribution of the ORF length (nt) in mRNAs and lncRNAs. (E) The number of lncRNAs and mRNAs containing different ORF numbers. Every 50 ORFs count together. (F) Venn diagram of lncRNAs identified in E14 and E9. (G) The expression levels of all lncRNAs in all 6 samples. (H) Volcano plot of lncRNA expression in E14 vs. E9.
A total of 625 lncRNAs were identified across all samples, 121 of which exhibited specific expression in the E14 group, 136 in the E9 group, and 368 in both groups (Figure 3F, Supplementary Table 7). Evaluating the expression levels of each lncRNA in every sample, we observed similar distribution patterns among the 6 samples (Figure 3G). Applying the criteria of FDR < 0.05 and |log2FC| ≥ 1 for differential analysis between the E14 and E9 comparisons, we identified 53 upregulated and 71 downregulated lncRNAs (Figure 3H). These findings underscore the distinct characteristics of lncRNAs compared to mRNAs and highlight their differential expression during the E14 and E9 stages.
Regulation of mRNA by LncRNA
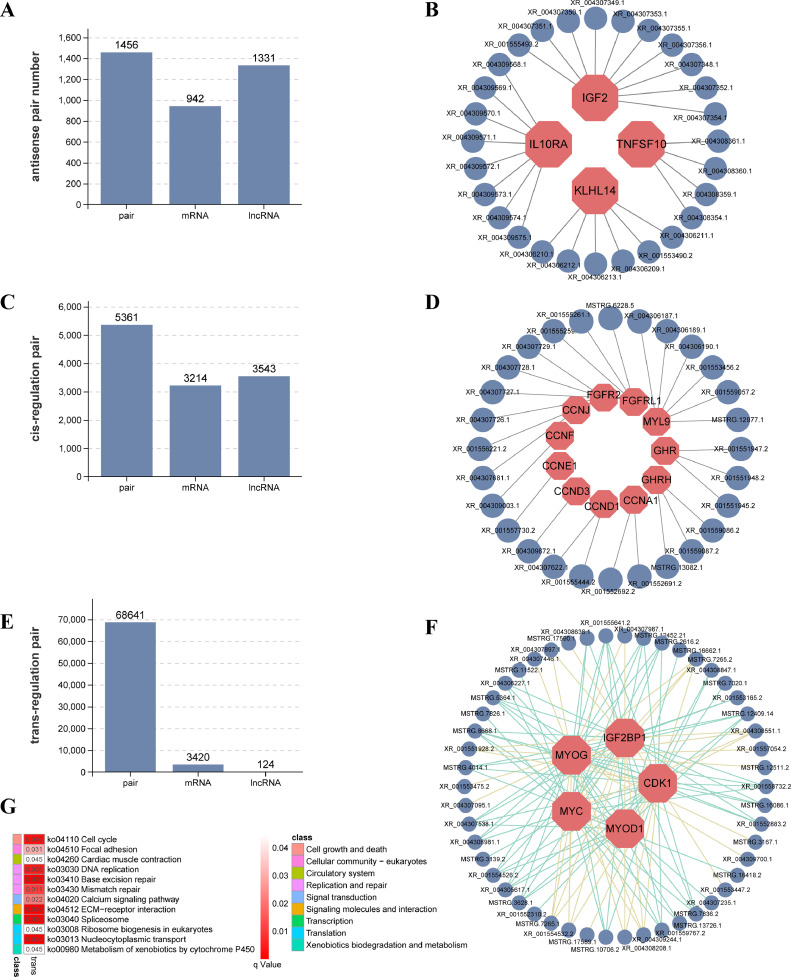
To unravel the functionality of lncRNAs, we delved into the prediction of lncRNA‒mRNA pairs involving antisense-RNA, cis-regulation, and trans-regulation. For antisense-RNA, a total of 1,456 pairs were uncovered (Figure 4A, Supplementary Table 8), with differentially expressed lncRNAs (DELs) such as MSTRG.14280.2, MSTRG.3139.2, MSTRG.3199.1, MSTRG.7020.1, and XR_001559540.2 exhibiting regulation over DEGs such as LOC107324334, GLI3, CAVIN4, ESRRB, and PIK3C2B, respectively (Supplementary Table 9). Additionally, muscle development-related genes such as IGF2, IL10RA, KLHL14, and TNFSF10 were also found to be under the influence of several antisense lncRNAs (Figure 4B). Our predictions illuminated a total of 3,543 lncRNAs, 3,214 mRNAs, and 5,361 cis-regulated lncRNA‒mRNA pairs, with the majority of the lncRNAs displaying their cis-regulatory properties on individual mRNAs (Figure 4C, Supplementary Table 10). Crucial genes associated with muscle growth, such as MYL9 and GHR, and those linked to cell proliferation, such as the CCN family, were all subject to the guidance of lncRNAs in a cis-regulatory manner (Figure 4D). Moving on to trans-regulation, due to the larger number of mRNAs affected by lncRNAs, we limited our focus to predicting downstream genes influenced by DELs. A substantial count of 3,420 mRNAs emerged as being trans-regulated by lncRNAs (Figure 4E, Supplementary Table 11). We also visualized some key myoblast regulators, MYOG, MYOD1, MYC, CDK1 and IGFBP1, and their trans-regulatory lncRNAs (Figure 4F). Strikingly, mirroring the enrichment analysis results of DEGs, the trans-regulated mRNAs predominantly exhibited enrichment in vital signaling pathways encompassing the cell cycle, ECM-receptor interaction, calcium signaling pathway, spliceosome, and beyond (Figure 4G). These findings support the extensive involvement of lncRNAs through diverse mechanisms, including antisense-RNA, cis-regulation, and trans-regulation, in the development of quail leg muscles.
Figure 4.
The regulation of lncRNAs on the genes in quail leg muscle. (A) The number of antisense lncRNAs and lncRNA‒mRNA pairs. (B) The antisense lncRNAs IGF2, TNFSF10, KLHL14 and IL10RA. (C) The number of cis-regulation lncRNA‒mRNA pairs. (D) Cis-acting lncRNAs on CCN family genes and muscle growth-related genes. (E) The number of trans-regulation lncRNA‒mRNA pairs in quail leg muscle. (F) The trans-acting lncRNAs on MYOG, MYOD1, MYC, IGF2BP1 and CDK1. (G) KEGG enrichment analysis of the genes trans-regulated by lncRNAs.
Characteristics and Expression of CircRNAs in Quail Embryonic Leg Muscles
We investigated the unique features and expression patterns of circRNAs in quail embryonic leg muscles. Employing back-splicing and expression quantification, we identified a total of 14,457 circRNAs across the samples analyzed (Figure 5A, Supplementary Table 12). Among these, 4,434 circRNAs were found to be expressed in both groups, while 6,462 and 3,561 exhibited exclusive expression in the E14 and E9 groups, respectively (Figure 5B). Comparison of circRNA expression levels revealed relatively higher expression in samples E9-2 and E9-3 (Figure 5C). When categorizing the circRNAs, it was observed that exonic circRNAs were the most abundant in quail leg muscles, with a total of 11,299 identified (Figure 5D). Interestingly, 674 circRNAs were formed from a single exon (Figure 5D). Additionally, we found 1,749 circRNAs derived from exon‒intron regions, while intronic circRNAs, intergenic circRNAs, and antisense strand circRNAs were less prevalent, consisting of 363, 284, and 88 circRNAs, respectively (Figure 5D). Analyzing the length distribution of circRNAs, we noted that the majority fell below 1,000 bp, with the 400 to 600 bp range accounting for over 45% of all circRNAs (Figure 5E).
Figure 5.
Characterization and expression of circular RNA in quail leg muscle. (A) The circular RNA detected in each sample and the total number of all samples. (B) Venn diagram of the circular RNA detected in E14 and E9. (C) All circular RNA expression levels in all 6 samples. (D) The number of all circular RNA types detected in quail muscle. (E) The length distribution of all circular RNAs detected in quail leg muscle. (F) Volcano plot of all circular RNAs in E14 vs. E9. (G) The GO enrichment analysis with the parental genes of the DECs. The top 20 biological process terms are presented. (H) KEGG enrichment analysis of the parental genes of the DECs. All significant KEGG signaling pathways (P < 0.05) are shown.
Following the characterization of circRNAs in quail leg muscles, we performed a selection process based on a P value < 0.05 and |log2FC| ≥ 1 to identify DECs between the E14 and E9 stages. Among these, 137 circRNAs displayed upregulated expression, while 155 circRNAs exhibited downregulated expression (Figure 5F, Supplementary Table 13). Notably, several circRNAs associated with skeletal muscle development, such as GAS2 circRNA, MEF2A circRNA, and SVIL circRNA, showed differential expression during quail skeletal muscle development (Supplementary Table 13). Enrichment analysis was conducted for both the parental genes of all circRNAs and the DECs. The results indicated significant enrichment of the parental genes of DECs in biological process terms including intracellular signal transduction, extracellular matrix organization, signal transduction, actomyosin structure organization, and more (Figure 5G, Supplementary Table 14). Furthermore, KEGG pathway analysis revealed notable enrichment of the parental genes of DECs in signaling pathways such as alanine, aspartate, and glutamate metabolism, regulation of actin cytoskeleton, adherens junction, Wnt signaling pathway, and gap junction signaling pathway (Figure 5H, Supplementary Table 15). Additionally, all circRNA parental genes exhibited significant enrichment in cell proliferation- and muscle growth-related signaling pathways, including the cell cycle, GnRH signaling pathway, insulin signaling pathway, and ECM-receptor interaction (Supplementary Table 16). These findings underscore the widespread presence of circRNAs in quail leg muscle tissue and suggest their potential involvement through their parental genes in the regulation of skeletal muscle development.
Identification and Expression of miRNAs during Quail Embryonic Leg Muscle Development
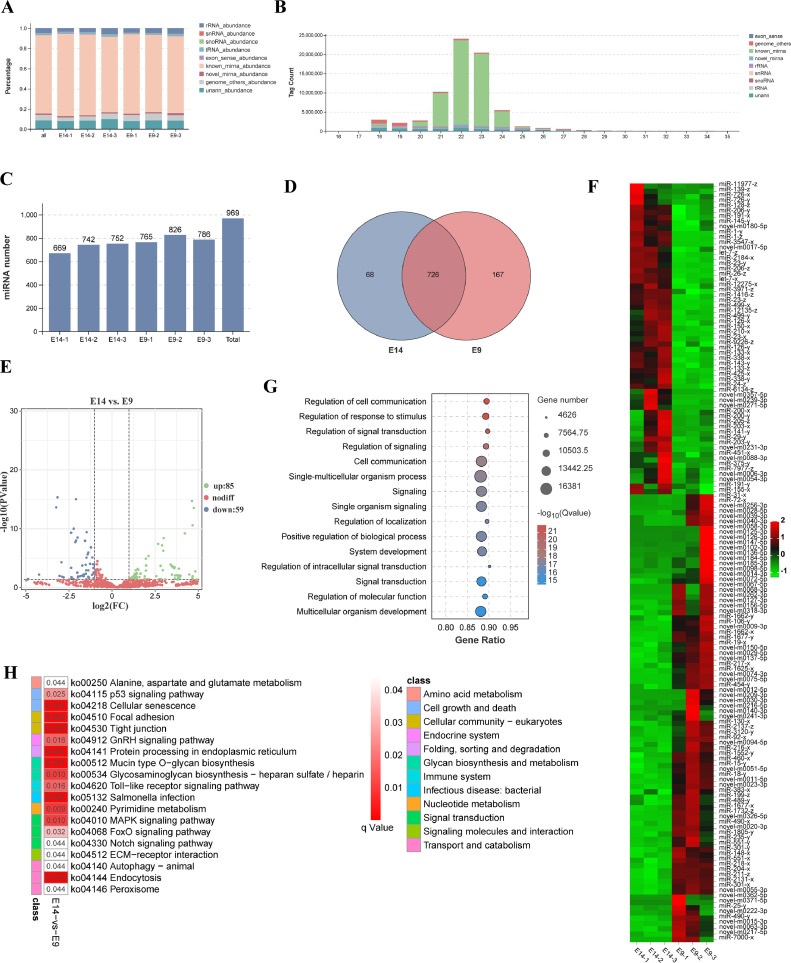
To investigate the role of miRNAs in quail embryonic leg muscles, we conducted short RNA sequencing on 6 muscle samples and compared the data with existing databases. Our analysis revealed that approximately 78% of the total tags corresponded to known miRNA tags, while novel miRNA tags accounted for approximately 1.5% (Figure 6A). Length distribution analysis showed that most of the tags fell within the 18 to 30 nt range, and the identified miRNAs were primarily distributed in the 20 to 24 nt range (Figure 6B). We successfully identified a total of 969 miRNAs, including 556 known miRNAs and 408 novel miRNAs (Figure 6C, Supplementary Table 17). Among them, 726 miRNAs were expressed in both groups, while 68 and 167 miRNAs were specifically expressed in the E14 and E9 groups, respectively (Figure 6D). Differential expression analysis revealed 85 significantly upregulated and 59 significantly downregulated miRNAs (Figure 6E). The top 5 DEMs were miR-1-y, miR-1-z, miR-203-y, novel-m0020-3p, and miR-499-x (Supplementary Table 18). Interestingly, several previously identified miRNAs associated with skeletal muscle development, such as the miR-1 family, miR-133 family, let-7 family, miR-206 family, and miR-203-y, were among the DEMs (Figure 6F, Supplementary Table 18). Notably, most of the differentially expressed novel miRNAs exhibited higher expression in the E9 group than in the E14 group (Figure 6F). To further explore their potential functions, we performed target gene prediction for all identified miRNAs. Enrichment analysis indicated that the target genes of DEMs were significantly enriched in BP terms such as regulation of cell communication, signal transduction, and system development (Figure 6G, Supplementary Table 19). Moreover, KEGG enrichment analysis showed significant enrichment of the target genes in signaling pathways including focal adhesion, tight junction, GnRH signaling pathway, and ECM-receptor interaction (Figure 6H, Supplementary Table 20). These findings highlight the involvement of multiple miRNAs, particularly myomiRNAs, in regulating quail leg muscle development by influencing muscle-related pathways through their target genes.
Figure 6.
The detection and expression of miRNAs in quail leg muscle. (A) The percentage of all kinds of small RNAs detected in all 6 samples. (B) The length distribution of all small RNAs detected in the current study. (C) The miRNAs detected in all 6 samples. (D) Venn diagram of the miRNAs detected in E14 and E9. The miRNAs with transcripts per million (TPM) higher than 0.1 were submitted to Venn diagram analysis. (E) Volcano plot of the miRNAs in E14 vs. E9. (F) Heatmap of all DEMs in E14 vs. E9. (G) The GO enrichment analysis with all target genes of the DEMs. The top 15 biological process terms are shown. (H) KEGG enrichment analysis with all target genes of DEMs. The significantly enriched KEGG signaling pathways (q < 0.05) are presented.
Construction of the ceRNA Regulatory Network in Quail Leg Muscles
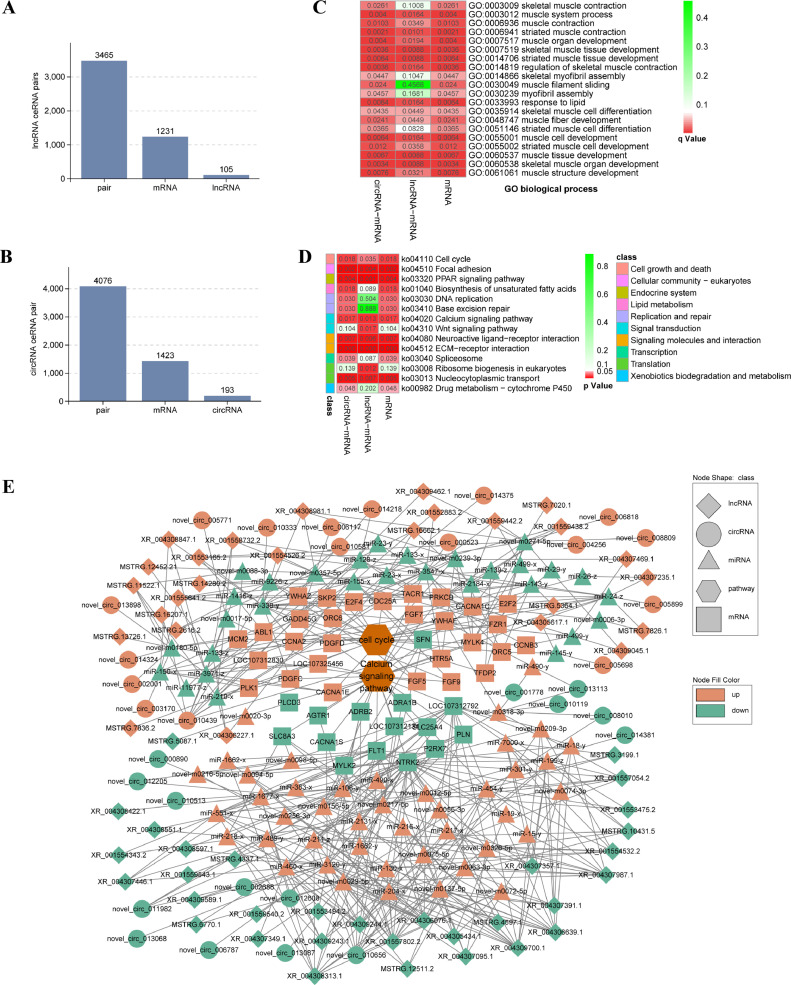
Building a ceRNA regulatory network allows us to gain insights into how ncRNAs regulate specific biological processes through their interactions with mRNAs. In this study, we utilized all differentially expressed RNA molecules from the E14 vs. E9 comparison to construct the ceRNA network (Table 2). After meticulous filtering, we identified 105 lncRNAs, 1,231 mRNAs, and 3,465 lncRNA‒mRNA pairs (Figure 7A, Supplementary Table 21). Additionally, we discovered 193 circRNAs, 1,423 mRNAs, and 4,076 circRNA-mRNA pairs that exhibited ceRNA regulatory relationships (Figure 7B, Supplementary Table 22). Enrichment analysis was then performed on the mRNA targets within the sets of lncRNA‒miRNA-mRNA-circRNA, lncRNA‒miRNA-mRNA, and circRNA-miRNA‒mRNA relationships. The results revealed that potential ceRNA-regulated mRNAs were significantly enriched in muscle development-related BP terms, including skeletal muscle tissue development, positive regulation of cell proliferation, muscle cell differentiation, and muscle tissue development (Figure 7C, Supplementary Table 23). Furthermore, these mRNAs were found to be associated with signaling pathways such as the cell cycle, focal adhesion, ECM−receptor interaction, calcium signaling pathway, and PPAR signaling pathway (Figure 7D, Supplementary Table 24). Moreover, based on the KEGG enrichment analysis of DEGs, DEL trans-regulated mRNAs, and mRNAs within the ceRNA network, we observed significant enrichment in signaling pathways such as the cell cycle, focal adhesion, ECM-receptor interaction, and calcium signaling pathway. Of particular interest were the ceRNA regulatory networks associated with these signaling pathways. Within the cell cycle signaling pathway and calcium signaling pathway, numerous miRNAs related to muscle development mediated the ceRNA regulatory effects exerted by lncRNAs and circRNAs on the genes involved in this pathway (Figure 7E). In the ceRNA regulatory network associated with the cell cycle and calcium signaling pathway, 2 miRNAs, namely novel-m0318-3p, and miR-338-y, each have the ability to target a multitude of genes within these 2 pathways. These 2 miRNAs, in conjunction with the relevant lncRNAs and circRNAs, are considered significant candidate RNAs for future investigations into the development of skeletal muscle in quails. These intricate ceRNA regulatory networks lay the foundation for further investigations into the roles of these genes and ncRNAs in quail skeletal muscle development.
Table 2.
All differentially expressed RNA numbers.
| Comparison | miRNA |
mRNA |
lncRNA |
circRNA |
||||
|---|---|---|---|---|---|---|---|---|
| up | down | up | down | up | down | up | down | |
| E14 vs. E9 | 85 | 59 | 2095 | 1384 | 53 | 71 | 137 | 155 |
| total | 144 | 3479 | 124 | 292 | ||||
Figure 7.
CeRNA prediction of all differentially expressed RNAs. (A) The number of ceRNA pairs of lncRNA‒mRNA. (B) The number of ceRNA pairs of circRNA-mRNA. (C) The GO enrichment analysis of the mRNAs in circRNA-mRNA pairs, mRNAs in lncRNA‒mRNA pairs and mRNAs in both kinds of pairs. The myogenesis- and muscle growth-related biological process terms are shown. (D) KEGG enrichment analysis of the mRNAs in circRNA-mRNA pairs, mRNAs in lncRNA‒mRNA pairs and mRNAs in both kinds of pairs. The KEGG signaling pathways with P values less than 1 are presented. (E) All RNA molecules and the ceRNA regulatory network of the cell cycle pathway and the calcium signaling pathway in quail leg muscle. In the network visualization, nodes of various shapes represent different entities: hexagons for signaling pathways, squares for genes, triangles for miRNAs, diamonds for lncRNAs, and circles for circRNAs. The color of the nodes indicates the expression level of the RNAs: orange for upregulated RNAs, green for downregulated RNAs. Additionally, the 2 nodes in deep orange specifically denote signaling pathways.
Sequencing Data Validation
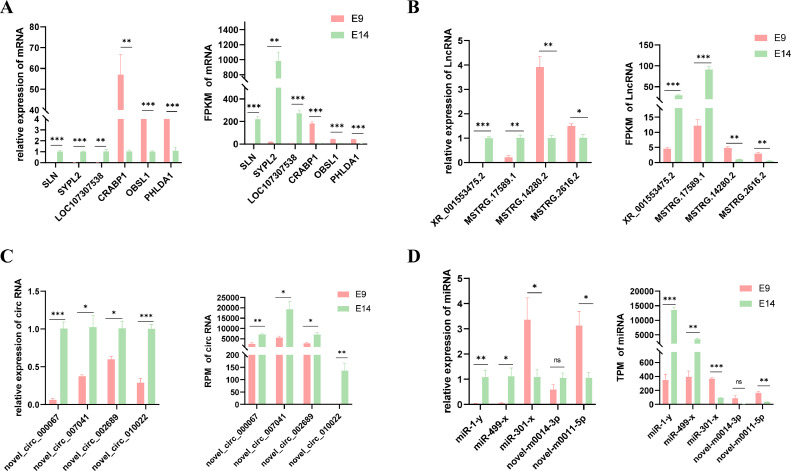
To verify the accuracy of our sequencing data, we performed qPCR on a selection of 6 genes (SLN, SYPL2, LOC107307538, CRABP1, OBSL1, and PHLDA1), as shown in Figure 8A. Similarly, we conducted qPCR analysis on 4 lncRNAs (XR_001553475.2, MSTRG.17589.1, MSTRG.14280.2, and MSTRG.2616.2), as depicted in Figure 8B, and 4 circular RNAs (novel_circ_000067, novel_circ_007041, novel_circ_002689, and novel_circ_010022), as presented in Figure 8C. Additionally, we examined the expression levels of 5 miRNAs (miR-1-y, miR-499-x, miR-301-x, novel-m0014-3p, novel-m0011-5p) using qPCR, as displayed in Figure 8D. The results obtained from the qPCR were consistent with the trends observed in the sequencing data, providing evidence for the accuracy of our sequencing analysis.
Figure 8.
qPCR validation of the sequencing data. Values are presented as the mean ± SEM. * indicates P < 0.05, ** indicates P < 0.01, *** indicates P < 0.001.
DISCUSSION
Quail, being a small-sized animal with a short reproductive cycle, is an ideal avian model for research (Serralbo et al., 2020). However, the specific characteristics of quail skeletal muscle development and the expression patterns of mRNAs, lncRNAs, circRNAs, and miRNAs in this context remain largely unexplored. In this study, we aimed to shed light on these aspects by examining quail leg muscle tissues collected at different time points, ranging from E7 to E15 and P1. To understand the developmental patterns of quail leg muscles, we employed paraffin sections. Subsequently, we conducted comprehensive transcriptome sequencing on leg muscle tissues representing the early differentiation stage (E9) as well as the stage of completed differentiation (E14). This allowed us to uncover the distinct expression profiles of mRNAs, lncRNAs, circRNAs, and miRNAs in quail leg muscles and detect any differential expression across various stages. Additionally, we constructed a molecular regulatory network that elucidated the interactions between ncRNAs and mRNAs.
During the embryonic period and the first week after hatching, the determination of muscle fiber quantity in poultry is crucial (Li et al., 2015). The proliferation and differentiation processes of myoblasts during embryonic development heavily influence the number of muscle fibers and overall muscle content after hatching (Bi et al., 2017). Hence, our study initially focused on identifying the developmental patterns of quail leg muscles during the embryonic stage. Similar to mammals and other avian species, quail leg muscle development involves essential steps such as myogenic cell differentiation, fusion of muscle fibers, and thickening of muscle fibers. Encouragingly, our findings align with previous observations made in Chinese yellow-broiler chickens, substantiating the consistency in muscle development patterns (Liu et al., 2023b). Specifically, we examined leg muscle tissues from E9 embryos, which mark the early appearance of multinucleated muscle fibers and the initiation of myogenic cell differentiation. Conversely, by E14, muscle fibers and fascia are well formed, and intermuscular fat starts to emerge. Prior studies have highlighted the highest expression levels of the differentiation marker genes MYOG and MYOD1 at E9, which are substantially decreased by E14 (Liu et al., 2023c; b). Remarkably, our mRNA sequencing analysis echoes these reports, providing further support for these temporal dynamics (Supplementary Table 2). We also conducted differential expression analysis of mRNAs, revealing elevated expression of genes associated with skeletal muscle fiber development and fast-to-slow muscle fiber transition (including the MYOZ family, MYOM family, MyHC, TNNC family, and TNNT family, among others) at E14. In contrast, E9 exhibited heightened expression of cell cycle-related regulatory factors (such as the CDK family and CCN family). We performed enrichment analyses encompassing GO, KEGG, and GSEA, which consistently corroborated the results we obtained from paraffin sections. Notably, biological processes linked to muscle fiber development and lipid metabolism showed significant upregulation at E14, while E9 was enriched in processes related to the cell cycle, mitosis, and nucleotide metabolism. These enrichments were further confirmed across different stages of skeletal muscle development through RNA-seq.
In recent years, increasing research has provided ample evidence demonstrating the crucial role of ncRNAs in the process of skeletal muscle development (Liu et al., 2022; Cai et al., 2022b). LncRNAs, a specific type of RNA molecule exceeding 200 nt in length that typically lacks coding capacity, have been extensively reported to be highly expressed in the skeletal muscle tissues of various avian species, including chickens, geese, and pigeons (Li et al., 2019; Chen et al., 2022b; Luo et al., 2022). In this study, we focused on identifying the characteristics of lncRNAs in quail leg muscles, discovering that the length distribution of lncRNAs in quail leg muscles is similar to that found in chicken leg muscles (Li et al., 2017). Furthermore, our statistical analysis revealed that approximately 75% of quail leg muscle lncRNAs contain 2-4 exons, which aligns with findings from pigeon pectoral muscle lncRNAs (Luo et al., 2022). The regulation of skeletal muscle development by lncRNAs encompasses diverse mechanisms, such as acting as miRNA sponges for ceRNA regulation (Li et al., 2019), modulating functional genes through cis or trans regulatory actions (Dong et al., 2020), functioning as antisense transcripts to regulate gene function (Cai et al., 2022a), serving as protein binders to modulate protein activity (Liu et al., 2019a), encoding micropeptides (Cai et al., 2017), and participating in the regulation of genomic DNA methylation (Huang et al., 2022). Our investigation successfully identified 625 lncRNAs and their unique characteristics in quail leg muscles. Notably, a comparison between the E14 and E9 stages resulted in the identification of 1,124 DELs, suggesting their potential importance in the developmental processes of quail skeletal muscles.
MiRNAs, another widely distributed class of single-stranded RNA molecules measuring approximately 18 to 26 nt in length, play a vital role in posttranscriptional gene regulation (Oikawa et al., 2023). MiRNAs achieve their main function by binding to mRNAs and inhibiting their translation (Du and Zamore, 2007). The ability of miRNAs to interact with other long RNAs has positioned them as key players in the ceRNA hypothesis (Tay et al., 2014). Notably, mRNAs, lncRNAs, circRNAs, and pseudogenes can competitively bind to miRNAs with similar harbor sites, thereby exerting mutual regulatory effects on each other (Tay et al., 2014). Apart from their intracellular functions, miRNAs are also present in extracellular vesicles and the bloodstream, where they regulate the functions of other tissues and cells through systemic interactions (Banack et al., 2020). The regulation exerted by miRNAs on skeletal muscle development occurs during various stages, including myoblast proliferation, differentiation, myotube fusion, and fiber hypertrophy or atrophy (Li et al., 2019). By binding to specific target genes, miRNAs modulate the expression of genes associated with myogenesis. Some miRNAs are highly expressed in muscle cells and possess the ability to regulate muscle-specific genes (Horak et al., 2016). For instance, miR-1 and miR-133 are widely recognized as muscle-specific miRNAs responsible for regulating myoblast differentiation and determining muscle fiber types (Chen et al., 2006). In our study, we identified a total of 969 miRNAs present in quail leg muscles at E9 and E14, among which 144 displayed differential expression between these 2 stages. These DEMs included crucial muscle-related miRNAs such as miR-1, miR-133, and let-7, alongside others such as miR-206, miR-203, and miR-15, which are known to be involved in poultry myoblast proliferation, differentiation, and myotube fusion (Luo et al., 2014; Ma et al., 2015; Li et al., 2019). These findings indicate that the role of miRNAs in quail skeletal muscle development is similar to that observed in other avian species. Importantly, the miR-15 family was found to mediate the regulation of lncIRS1 through a ceRNA mechanism, ultimately influencing the expression of the parental gene IRS1 and subsequently modulating the IGF1-PI3K/AKT pathway that governs chicken skeletal muscle development (Li et al., 2019). Moreover, the transcription factor c-Myc was found to inhibit the expression of gga-let-7a at the transcriptional level, while let-7a, in turn, negatively regulated c-Myc expression through its target genes, thereby impacting downstream genes and non-coding RNA expression, thus playing a role in skeletal muscle development Luo et al., 2019). Intriguingly, among the DEMs, let-7-x displayed sequence identity to gga-let-7a and exhibited high expression levels during both stages of quail skeletal muscle development, suggesting its potential significance in this process.
CircRNAs are a unique class of ncRNAs characterized by covalently linked 5′ and 3′ ends, which impart increased stability (Patop et al., 2019). These circular RNA molecules can originate from different sources, including exons, introns, both exons and introns, intergenic regions, and the antisense strand of genes (Ouyang et al., 2018). In our study, we identified all 5 types of circRNAs in quail skeletal muscle, with a notable abundance of circRNAs originating from exons (Figure 5D). Extensive research has demonstrated that circRNAs have a widespread presence in the cells of eukaryotic organisms and play significant roles in various biological processes (Xu et al., 2020; Shi et al., 2020; Cai et al., 2022b). They exert their functional roles through multiple mechanisms, such as acting as miRNA sponges to competitively bind and regulate miRNAs (Cai et al., 2022b), encoding peptides (Yin et al., 2020), and interacting with RNA-binding proteins (Du et al., 2016). While previous studies have reported the existence of circRNAs in the tissues of birds such as chickens, ducks, geese, and pigeons (Ouyang et al., 2018; Wu et al., 2022; Ma et al., 2022; Liu et al., 2023a), there is currently limited information available regarding circRNAs in quails. During poultry muscle development, circRNAs primarily function by binding to miRNAs. A previous study showed significant differential expression of circRBFOX2 at different stages of chicken embryo development (Ouyang et al., 2018). Interestingly, this circRNA contains binding sites for miR-1a and miR-206, and it acts as a competing endogenous RNA (ceRNA) to promote myoblast proliferation (Ouyang et al., 2018). Additionally, through predictive analysis, circ-FAM188B has the potential to encode Circ-FAM188B-103aa. Experimental validation confirmed that Circ-FAM188B-103aa shares the same functionality as its host gene transcript (FAM188B) and promotes skeletal muscle satellite cell proliferation but inhibits their differentiation (Yin et al., 2020). In our study, we identified a total of 14,457 circRNAs in quail skeletal muscle across 2 developmental stages. Among these circRNAs, 292 exhibited significant differential expression during leg muscle development, indicating their substantial presence and potential importance in the regulation of quail skeletal muscle development.
Building a regulatory network that encompasses the interplay between genes and ncRNAs allows for a comprehensive understanding of gene expression patterns and regulatory mechanisms. It also aids in the identification of differentially expressed RNAs, leading to a deeper exploration of cellular functions and the discovery of novel signaling pathways (Fu et al., 2019). After conducting data analysis on various RNA types, we proceeded to construct a regulatory network for the differentially expressed RNAs. Specifically, we built networks involving miRNAs and their target genes, lncRNAs and their target genes, as well as a ceRNA network encompassing 4 RNA types. The enrichment analysis of these 3 RNA regulatory networks revealed their association with biological processes related to muscle development. In the ceRNA network, we observed a significant enrichment of DEGs in biological processes such as muscle organ development, skeletal muscle tissue development, skeletal muscle contraction, myofibril assembly, and response to lipid stimuli (Figure 7C). Muscle organ development, skeletal muscle tissue development, and skeletal muscle contraction are closely tied to the morphological changes occurring in skeletal muscle through the action of actin/myosin complexes (https://www.informatics.jax.org/vocab/gene_ontology). Myofibril assembly represents the orderly arrangement of thick and thin filaments, forming muscle fibers (https://www.ebi.ac.uk/QuickGO/term/GO:0045214). These enriched processes were consistently identified across different stages of skeletal muscle (Qu et al., 2021; Zhu et al., 2023). Additionally, the DEMs within the ceRNA network showed notable enrichment in focal adhesion, ECM-receptor interaction, calcium signaling pathway, and cell cycle pathways. Focal adhesion consists of an intricate interplay between extracellular matrix molecules and intracellular signaling molecules, playing pivotal roles in cellular migration, proliferation, and signal transduction (Jung et al., 2022). ECM-receptor interactions involve the binding between cell surface receptors and components of the extracellular matrix, contributing significantly to processes such as cell adhesion, migration, proliferation, and differentiation, which are crucial for cell development, tissue formation, and maintenance (Rahbari et al., 2016; San et al., 2021). The calcium signaling pathway is involved in the release, transmission, and response to calcium ions (Clapham, 2007). By modulating calcium ion concentrations, cells regulate a series of calcium-dependent proteases, ion channels, and transcription factors, ultimately governing cellular functions and physiological responses. The cell cycle, a highly intricate signaling pathway comprising multiple stages and regulatory factors, ensures accurate replication and division of cells in a timely manner (Helmbrecht et al., 2000). Hence, the differentially expressed lncRNAs, circRNAs, and miRNAs identified in our study are likely to play vital roles in regulating quail skeletal muscle development through the aforementioned biological processes or signaling pathways.
Drawing from a range of KEGG enrichment analyses (Figures 2F, 3E, 7D), we opted to visualize the genes and associated miRNAs, lncRNAs, and circRNAs of ceRNA within the cell cycle and calcium signaling pathway, all of which were enriched across multiple analyses. Upon visualization of the ceRNA regulatory network, both novel-m0318-3p and miR-338-y were found to concurrently target genes within the 2 aforementioned pathways. Specifically, novel-m0318-3p targets the tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein epsilon (YWHAE), neurotrophic receptor tyrosine kinase 2 (NTRK2), novel_circ_001778, novel_circ_008010, novel_circ_010119, novel_circ_013113, and XR_001557054.2. Meanwhile, miR-338-y targets E2F transcription factor 4 (E2F4), cyclin A2 (CCNA2), origin recognition complex subunit 6 (ORC6), platelet derived growth factor D (PDGFD), protein kinase C beta (PRKCB), tachykinin receptor 1 (TACR1), calcium voltage-gated channel subunit alpha1 C (CACNA1C), fibroblast growth factor 9 (FGF9), platelet derived growth factor C (PDGFC), XR_001553165.2, XR_001554526.2, XR_001555641.2, XR_004308847.1, XR_004308981.1, MSTRG.16662.1, XR_001559438.2, novel_circ_010333, and novel_circ_010581 (Figure 7E). Previous studies have shown that miR-338 has been associated with the regulation of muscle growth and myogenesis (Duran et al., 2022). Furthermore, miR-338 has been found to modulate the expression of genes involved in muscle fiber type and metabolic enzyme activities (Liu et al., 2016). The sequence of miR-338-y identified in this study bears similarity to the animal miR-338-3p sequence (https://mirbase.org/). MiR-338-3p has been implicated in various cellular processes, including tumorigenesis, metastasis, and apoptosis across different types of cancer (Zhang et al., 2017; Luan and Wang, 2018; Lu et al., 2019). However, the specific role of miR-338-3p in skeletal muscle development remains unexplored. As such, further research is warranted to clarify the specific contributions of miR-338-3p and novel-m0318-3p to skeletal muscle development. The results and discussions suggest that novel-m0318-3p, miR-338-y, and associated genes and RNAs may regulate quail skeletal muscle development via the cell cycle and calcium signaling pathway.
CONCLUSION
In this study, we identified the developmental patterns of leg muscles in quail embryos and extensively characterized the distinctive features of mRNA and ncRNAs present in quails. Through a comprehensive analysis, we investigated the dynamic expression profiles of mRNAs, lncRNAs, circRNAs, and miRNAs during the initial stages of differentiation and completion of differentiation in quail leg muscles. Leveraging enrichment analysis, we predicted the potential functions of these differentially expressed RNAs in the intricate process of skeletal muscle development. Furthermore, we constructed regulatory networks encompassing lncRNA‒mRNA, miRNA‒mRNA, lncRNA‒miRNA-mRNA, and circRNA-miRNA‒mRNA interactions in quail skeletal muscle, shedding light on the underlying pathways that may play significant roles. Our findings also suggest that novel-m0318-3p, miR-338-y, and their respective target RNAs could potentially exert a substantial regulatory influence on the development of skeletal muscle in quails. These noteworthy findings serve as a solid foundation for further exploration into the regulatory mechanisms governing quail skeletal muscle development and provide essential data support for studying non-coding RNAs in the context of quail species.
Acknowledgments
ACKNOWLEDGMENTS
This research was supported by grants from the Jiangxi Natural Science Foundation of China (20212BAB215016, 20232BAB205061), the Modern Poultry Industry Technology System in Jiangxi Province (JXARS-09), the Jiangxi Joint Key Project of Quail Improvement (2022JXCQZY02), and the Graduate Innovation Special Fund Projects in Jiangxi Province (YC2022-s410, YC2023-S392).
Data Availability Statement: We have deposited the raw sequence data from the rRNA depletion libraries and small RNA libraries mentioned in this paper into the Genome Sequence Archive (GSA). The accession numbers assigned to these datasets are GSA: CRA012844 and CRA012844. These datasets can be publicly accessed at https://ngdc.cncb.ac.cn/gsa.
DISCLOSURES
All authors declare that there are no conflicts of interest.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.psj.2024.103603.
Appendix. Supplementary materials
REFERENCES
- Abmayr S.M., Pavlath G.K. Myoblast fusion: lessons from flies and mice. Development. 2012;139:641–656. doi: 10.1242/dev.068353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banack S.A., Dunlop R.A., Cox P.A. An miRNA fingerprint using neural-enriched extracellular vesicles from blood plasma: towards a biomarker for amyotrophic lateral sclerosis/motor neuron disease. Open. Biol. 2020;10 doi: 10.1098/rsob.200116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi P., Ramirez-Martinez A., Li H., Cannavino J., McAnally J.R., Shelton J.M., Sánchez-Ortiz E., Bassel-Duby R., Olson E.N. Control of muscle formation by the fusogenic micropeptide myomixer. Science (1979) 2017;356:323–327. doi: 10.1126/science.aam9361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianconi V., Mozzetta C. Epigenetic control of muscle stem cells: time for a new dimension. Trends Genet. 2022;38:501–513. doi: 10.1016/j.tig.2022.01.001. [DOI] [PubMed] [Google Scholar]
- Braun T., Gautel M. Transcriptional mechanisms regulating skeletal muscle differentiation, growth and homeostasis. Nat. Rev. Mol. Cell Biol. 2011;12:349–361. doi: 10.1038/nrm3118. [DOI] [PubMed] [Google Scholar]
- Buckingham M., Rigby P.W.J. Gene regulatory networks and transcriptional mechanisms that control myogenesis. Dev. Cell. 2014;28:225–238. doi: 10.1016/j.devcel.2013.12.020. [DOI] [PubMed] [Google Scholar]
- Cai B., Li Z., Ma M., Wang Z., Han P., Abdalla B.A., Nie Q., Zhang X. LncRNA-Six1 encodes a micropeptide to activate Six1 in Cis and is Involved in cell proliferation and muscle growth. Front. Physiol. 2017;8:230. doi: 10.3389/fphys.2017.00230. Accessed December 24, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai B., Ma M., Zhang J., Kong S., Zhou Z., Li Z., Abdalla B.A., Xu H., Zhang X., Lawal R.A., Nie Q. Long noncoding RNA ZFP36L2-AS functions as a metabolic modulator to regulate muscle development. Cell Death. Dis. 2022;13:1–12. doi: 10.1038/s41419-022-04772-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai B., Ma M., Zhou Z., Kong S., Zhang J., Zhang X., Nie Q. circPTPN4 regulates myogenesis via the miR-499-3p/NAMPT axis. J. Anim. Sci. Biotechnol. 2022;13:2. doi: 10.1186/s40104-021-00664-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B., Liu S., Zhang W., Xiong T., Zhou M., Hu X., Mao H., Liu S. Profiling analysis of N6-methyladenosine mRNA methylation reveals differential m6A patterns during the embryonic skeletal muscle development of ducks. Animals. 2022;12:2593. doi: 10.3390/ani12192593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.-F., Mandel E.M., Thomson J.M., Wu Q., Callis T.E., Hammond S.M., Conlon F.L., Wang D.-Z. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet. 2006;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Zhang S., Chen G., Deng X., Zhang D., Wen H., Yin Y., Lin Z., Zhang X., Luo W. Transcriptome sequencing reveals pathways related to proliferation and differentiation of Shitou goose myoblasts. Animals. 2022;12:2956. doi: 10.3390/ani12212956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Zhou Y., Chen Y., Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapham D.E. Calcium Signaling. Cell. 2007;131:1047–1058. doi: 10.1016/j.cell.2007.11.028. [DOI] [PubMed] [Google Scholar]
- Dong A., Preusch C.B., So W.-K., Lin K., Luan S., Yi R., Wong J.W., Wu Z., Cheung T.H. A long noncoding RNA, LncMyoD, modulates chromatin accessibility to regulate muscle stem cell myogenic lineage progression. Proc. Natl. Acad. Sci. U.S.A. 2020;117:32464–32475. doi: 10.1073/pnas.2005868117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du W.W., Yang W., Liu E., Yang Z., Dhaliwal P., Yang B.B. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic. Acids. Res. 2016;44:2846–2858. doi: 10.1093/nar/gkw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du T., Zamore P.D. Beginning to understand microRNA function. Cell Res. 2007;17:661–663. doi: 10.1038/cr.2007.67. [DOI] [PubMed] [Google Scholar]
- Duran B.O.S., Zanella B.T.T., Perez E.S., Mareco E.A., Blasco J., Dal-Pai-Silva M., Garcia de la serrana D. Amino acids and IGF1 regulation of fish muscle growth revealed by transcriptome and microRNAome integrative analyses of Pacu (Piaractus mesopotamicus) myotubes. Int. J. Mol. Sci. 2022;23:1180. doi: 10.3390/ijms23031180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedländer M.R., Mackowiak S.D., Li N., Chen W., Rajewsky N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic. Acids. Res. 2012;40:37–52. doi: 10.1093/nar/gkr688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X.-Z., Zhang X.-Y., Qiu J.-Y., Zhou X., Yuan M., He Y.-Z., Chun C.-P., Cao L., Ling L.-L., Peng L.-Z. Whole-transcriptome RNA sequencing reveals the global molecular responses and ceRNA regulatory network of mRNAs, lncRNAs, miRNAs and circRNAs in response to copper toxicity in Ziyang Xiangcheng (Citrus junos Sieb. Ex Tanaka) BMC. Plant Biol. 2019;19:509. doi: 10.1186/s12870-019-2087-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S., Liu Y., Xu Y., Gai K., Cong B., Xing K., Qi X., Wang X., Xiao L., Long C., Guo Y., Chen L., Sheng X. Identification of key genes affecting sperm motility in chicken based on whole-transcriptome sequencing. Poult. Sci. 2023;102 doi: 10.1016/j.psj.2023.103135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmbrecht K., Zeise E., Rensing L. Chaperones in cell cycle regulation and mitogenic signal transduction: a review. Cell Prolif. 2000;33:341–365. doi: 10.1046/j.1365-2184.2000.00189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horak M., Novak J., Bienertova-Vasku J. Muscle-specific microRNAs in skeletal muscle development. Dev. Biol. 2016;410:1–13. doi: 10.1016/j.ydbio.2015.12.013. [DOI] [PubMed] [Google Scholar]
- Huang W., Li H., Yu Q., Xiao W., Wang D.O. LncRNA-mediated DNA methylation: an emerging mechanism in cancer and beyond. J. Exp. Clin. Cancer Res. 2022;41:100. doi: 10.1186/s13046-022-02319-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung J., Khan M.M., Landry J., Halavatyi A., Machado P., Reiss M., Pepperkok R. Regulation of the COPII secretory machinery via focal adhesions and extracellular matrix signaling. J. Cell Biol. 2022;221 doi: 10.1083/jcb.202110081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Langmead B., Salzberg S.L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L., Zhang Y., Ye Z.-Q., Liu X.-Q., Zhao S.-Q., Wei L., Gao G. CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic. Acids. Res. 2007;35:W345–W349. doi: 10.1093/nar/gkm391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Dewey C.N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC. Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Xiong T., Zhou M., Wan L., Xi S., Liu Q., Chen Y., Mao H., Liu S., Chen B. Characterization of microRNAs during embryonic skeletal muscle development in the Shan Ma Duck. Animals. 2020;10:1417. doi: 10.3390/ani10081417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Cai B., Abdalla B.A., Zhu X., Zheng M., Han P., Nie Q., Zhang X. LncIRS1 controls muscle atrophy via sponging miR-15 family to activate IGF1-PI3K/AKT pathway. J. Cachexia Sarcopenia Muscle. 2019;10:391–410. doi: 10.1002/jcsm.12374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Ouyang H., Zheng M., Cai B., Han P., Abdalla B.A., Nie Q., Zhang X. Integrated analysis of long non-coding RNAs (LncRNAs) and mRNA expression profiles reveals the potential role of LncRNAs in skeletal muscle development of the chicken. Front Physiol. 2017;7:687. doi: 10.3389/fphys.2016.00687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Wang Z., Zhang D., Shen Q., Pan T., Hui T., Ma J. Characterization of key aroma compounds in Beijing roasted duck by gas chromatography–olfactometry–mass spectrometry, odor-activity values, and aroma-recombination experiments. J. Agric. Food Chem. 2019;67:5847–5856. doi: 10.1021/acs.jafc.9b01564. [DOI] [PubMed] [Google Scholar]
- Liu J., Liu S., Zhang W., Hu X., Mao H., Liu S., Chen B. Transcriptome RNA sequencing reveals that circular RNAs are abundantly expressed in embryonic breast muscle of duck. Vet. Sci. 2023;10:75. doi: 10.3390/vetsci10020075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Zhang W., Luo W., Liu S., Jiang H., Liu S., Xu J., Chen B. Cloning of the RNA m6A Methyltransferase 3 and its impact on the proliferation and differentiation of quail myoblasts. Vet. Sci. 2023;10:300. doi: 10.3390/vetsci10040300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Zhou X., Chen Y., Nie Y., Huang H., Chen H., Mo D. Not all the number of skeletal muscle fibers is determined prenatally. BMC Dev Biol. 2015;15:42. doi: 10.1186/s12861-015-0091-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Li T., Song G., He Q., Yin Y., Lu J.Y., Bi X., Wang K., Luo S., Chen Y.-S., Yang Y., Sun B.-F., Yang Y.-G., Wu J., Zhu H., Shen X. Insight into novel RNA-binding activities via large-scale analysis of lncRNA-bound proteome and IDH1-bound transcriptome. Nucleic. Acids. Res. 2019;47:2244–2262. doi: 10.1093/nar/gkz032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Ren L., Liu A., Wang J., Wang J., Wang Q. Genome-wide identification and characterization of long non-coding RNAs in embryo muscle of chicken. Animals. 2022;12:1274. doi: 10.3390/ani12101274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Yin L., Yuan Y., Tang Y., Lin Z., Liu Y., Yang J. Developmental characteristics of skeletal muscle during the embryonic stage in Chinese yellow quail (Coturnix japonica) Animals. 2023;13:2317. doi: 10.3390/ani13142317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Trakooljul N., Hadlich F., Muráni E., Wimmers K., Ponsuksili S. MicroRNA-mRNA regulatory networking fine-tunes the porcine muscle fiber type, muscular mitochondrial respiratory and metabolic enzyme activities. BMC. Genomics. 2016;17:531. doi: 10.1186/s12864-016-2850-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M., Huang H., Yang J., Li J., Zhao G., Li W., Li X., Liu G., Wei L., Shi B., Zhao C., Fu Y. miR-338-3p regulates the proliferation, apoptosis and migration of SW480 cells by targeting MACC1. Exp. Ther. Med. 2019;17:2807–2814. doi: 10.3892/etm.2019.7260. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Luan X., Wang Y. LncRNA XLOC_006390 facilitates cervical cancer tumorigenesis and metastasis as a ceRNA against miR-331-3p and miR-338-3p. J. Gynecol. Oncol. 2018;29:e95. doi: 10.3802/jgo.2018.29.e95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W., Chen J., Li L., Ren X., Cheng T., Lu S., Lawal R.A., Nie Q., Zhang X., Hanotte O. c-Myc inhibits myoblast differentiation and promotes myoblast proliferation and muscle fibre hypertrophy by regulating the expression of its target genes, miRNAs and lincRNAs. Cell Death. Differ. 2019;26:426–442. doi: 10.1038/s41418-018-0129-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W., Wu H., Ye Y., Li Z., Hao S., Kong L., Zheng X., Lin S., Nie Q., Zhang X. The transient expression of miR-203 and its inhibiting effects on skeletal muscle cell proliferation and differentiation. Cell Death. Dis. 2014;5:e1347. doi: 10.1038/cddis.2014.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y., Hu S., Yan P., Wu J., Guo H., Zhao L., Tang Q., Ma J., Long K., Jin L., Jiang A., Li M., Li X., Wang X. Analysis of mRNA and lncRNA expression profiles of breast muscle during Pigeon (Columba livia) development. Genes. (Basel) 2022;13:2314. doi: 10.3390/genes13122314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma G., Wang Y., Li Y., Cui L., Zhao Y., Zhao B., Li K. MiR-206, a key modulator of skeletal muscle development and disease. Int. J. Biol. Sci. 2015;11:345–352. doi: 10.7150/ijbs.10921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H., Bian S., Li Y., Ni A., Zhang R., Ge P., Han P., Wang Y., Zhao J., Zong Y., Yuan J., Sun Y., Chen J. Analyses of circRNAs profiles of the lactating and nonlactating crops in pigeon (Columba livia) Poult. Sci. 2022;102 doi: 10.1016/j.psj.2022.102464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., Maier L., Mackowiak S.D., Gregersen L.H., Munschauer M., Loewer A., Ziebold U., Landthaler M., Kocks C., le Noble F., Rajewsky N. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- Oikawa S., Yuan S., Kato Y., Akimoto T. Skeletal muscle-enriched miRNAs are highly unstable in vivo and may be regulated in a Dicer-independent manner. FEBS. J. 2023;290:5692–5703. doi: 10.1111/febs.16917. [DOI] [PubMed] [Google Scholar]
- Ouyang H., Chen X., Wang Z., Yu J., Jia X., Li Z., Luo W., Abdalla B.A., Jebessa E., Nie Q., Zhang X. Circular RNAs are abundant and dynamically expressed during embryonic muscle development in chickens. DNA Research. 2018;25:71–86. doi: 10.1093/dnares/dsx039. Accessed March 13, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patop I.L., Wüst S., Kadener S. Past, present, and future of circRNAs. EMBO J. 2019;38 doi: 10.15252/embj.2018100836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M., Pertea G.M., Antonescu C.M., Chang T.-C., Mendell J.T., Salzberg S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015;33:290–295. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petracci M., Bianchi M., Mudalal S., Cavani C. Functional ingredients for poultry meat products. Trends. Food Sci. Technol. 2013;33:27–39. [Google Scholar]
- Qu Z., Liu A., Liu C., Tang Q., Zhan L., Xiao W., Huang J., Liu Z., Zhang S. Theaflavin Promotes Mitochondrial Abundance and Glucose Absorption in Myotubes by Activating the CaMKK2-AMPK Signal Axis via Calcium-Ion Influx. J. Agric. Food Chem. 2021;69:8144–8159. doi: 10.1021/acs.jafc.1c02892. [DOI] [PubMed] [Google Scholar]
- Quaresma M.A.G., Antunes I.C., Ferreira B.G., Parada A., Elias A., Barros M., Santos C., Partidário A., Mourato M., Roseiro L.C. The composition of the lipid, protein and mineral fractions of quail breast meat obtained from wild and farmed specimens of Common quail (Coturnix coturnix) and farmed Japanese quail (Coturnix japonica domestica) Poult. Sci. 2022;101 doi: 10.1016/j.psj.2021.101505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahbari N.N., Kedrin D., Incio J., Liu H., Ho W.W., Nia H.T., Edrich C.M., Jung K., Daubriac J., Chen I., Heishi T., Martin J.D., Huang Y., Maimon N., Reissfelder C., Weitz J., Boucher Y., Clark J.W., Grodzinsky A.J., Duda D.G., Jain R.K., Fukumura D. Anti-VEGF therapy induces ECM remodeling and mechanical barriers to therapy in colorectal cancer liver metastases. Sci. Transl. Med. 2016;8 doi: 10.1126/scitranslmed.aaf5219. 360ra135-360ra135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rugowska A., Starosta A., Konieczny P. Epigenetic modifications in muscle regeneration and progression of Duchenne muscular dystrophy. Clin Epigenet. 2021;13:13. doi: 10.1186/s13148-021-01001-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San J., Du Y., Wu G., Xu R., Yang J., Hu J. Transcriptome analysis identifies signaling pathways related to meat quality in broiler chickens – the extracellular matrix (ECM) receptor interaction signaling pathway. Poult. Sci. 2021;100 doi: 10.1016/j.psj.2021.101135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serralbo O., Salgado D., Véron N., Cooper C., Dejardin M.-J., Doran T., Gros J., Marcelle C. Transgenesis and web resources in quail. Elife. 2020;9:e56312. doi: 10.7554/eLife.56312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Jia X., Xu J. The new function of circRNA: translation. Clin. Transl. Oncol. 2020;22:2162–2169. doi: 10.1007/s12094-020-02371-1. [DOI] [PubMed] [Google Scholar]
- Sousa-Victor P., García-Prat L., Muñoz-Cánoves P. Control of satellite cell function in muscle regeneration and its disruption in ageing. Nat. Rev. Mol. Cell Biol. 2022;23:204–226. doi: 10.1038/s41580-021-00421-2. [DOI] [PubMed] [Google Scholar]
- Sun L., Luo H., Bu D., Zhao G., Yu K., Zhang C., Liu Y., Chen R., Zhao Y. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic. Acids. Res. 2013;41:e166. doi: 10.1093/nar/gkt646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tafer H., Hofacker I.L. RNAplex: a fast tool for RNA–RNA interaction search. Bioinformatics. 2008;24:2657–2663. doi: 10.1093/bioinformatics/btn193. [DOI] [PubMed] [Google Scholar]
- Tay Y., Rinn J., Pandolfi P.P. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–352. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Li H., Zhao X., Baki G., Ma C., Yao Y., Li J., Yao Y., Wang L. Differential expression of circRNAs of testes with high and low sperm motility in Yili geese. Front. Genet. 2022;13:970097. doi: 10.3389/fgene.2022.970097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wucher V., Legeai F., Hédan B., Rizk G., Lagoutte L., Leeb T., Jagannathan V., Cadieu E., David A., Lohi H., Cirera S., Fredholm M., Botherel N., Leegwater P.A.J., Béguec C.L.e, Fieten H., Johnson J., Alföldi J., André C., Lindblad-Toh K., Hitte C., Derrien T. FEELnc: a tool for long non-coding RNA annotation and its application to the dog transcriptome. Nucleic. Acids. Res. 2017;45:e57. doi: 10.1093/nar/gkw1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Sun Y., You B., Huang C.-P., Ye D., Chang C. Androgen receptor reverses the oncometabolite R-2-hydroxyglutarate-induced prostate cancer cell invasion via suppressing the circRNA-51217/miRNA-646/TGFβ1/p-Smad2/3 signaling. Cancer Lett. 2020;472:151–164. doi: 10.1016/j.canlet.2019.12.014. [DOI] [PubMed] [Google Scholar]
- Yin H., Shen X., Zhao J., Cao X., He H., Han S., Chen Y., Cui C., Wei Y., Wang Y., Li D., Zhu Q. Circular RNA CircFAM188B encodes a protein that regulates proliferation and differentiation of chicken skeletal muscle satellite cells. Front. Cell Dev. Biol. 2020;8:522588. doi: 10.3389/fcell.2020.522588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G., Zheng H., Zhang G., Cheng R., Lu C., Guo Y., Zhao G. MicroRNA-338-3p suppresses cell proliferation and induces apoptosis of non-small-cell lung cancer by targeting sphingosine kinase 2. Cancer Cell Int. 2017;17:46. doi: 10.1186/s12935-017-0415-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Hamill R.M., Mullen A.M., Kelly A.L., Gagaoua M. Molecular mechanisms contributing to the development of beef sensory texture and flavour traits and related biomarkers: Insights from early post-mortem muscle using label-free proteomics. J. Proteomics. 2023;286 doi: 10.1016/j.jprot.2023.104953. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.