Abstract
v-rel is the oncogenic member of the Rel/NF-κB family of transcription factors. The mechanism by which v-Rel induces transformation of avian lymphoid cells and fibroblasts is not precisely known. However, most models propose that v-rel disrupts the normal transcriptional regulatory network. In this study we evaluated the role of AP-1 family members in v-Rel-mediated transformation. The overexpression of v-Rel, c-Rel, and c-RelΔ resulted in a prolonged elevation of c-fos and c-jun expression and in a sustained repression of fra-2 at both the mRNA and protein levels in fibroblasts and lymphoid cells. Moreover, the transforming abilities of these Rel proteins correlated with their ability to alter the expression of these AP-1 factors. v-Rel exhibited the most pronounced effect, whereas c-Rel, with poor transforming ability, elicited only moderate changes in AP-1 levels. Furthermore, c-RelΔ, which exhibits enhanced transforming potential relative to c-Rel, induced intermediate changes in AP-1 expression. To directly evaluate the role of AP-1 family members in the v-Rel transformation process, a supjun-1 transdominant mutant was used. The supjun-1 mutant functions as a general inhibitor of AP-1 activity by inhibiting AP-1-mediated transactivation and by reducing AP-1 DNA-binding activity. Coinfection or sequential infection of fibroblasts or lymphoid cells with viruses carrying rel oncogenes and supjun-1 resulted in a reduction of the transformation efficiency of the Rel proteins. The expression of supjun-1 inhibited the ability of v-Rel transformed lymphoid cells and fibroblasts to form colonies in soft agar by over 70%. Furthermore, the expression of supjun-1 strongly interfered with the ability of v-Rel to morphologically transform avian fibroblasts. This is the first report showing that v-Rel might execute its oncogenic potential through modulating the activity of early response genes.
The v-rel oncogene of the avian reticuloendotheliosis virus (REV-T) encodes a member of the Rel/NF-κB family of transcription factors (reviewed in references 11, 26, and 27). REV-T induces a rapidly fatal lymphoma in young chickens and transforms both immature hematopoietic cells and fibroblasts in culture (7, 9, 21, 32, 48, 52, 56). The product of the v-rel oncogene, p59v-rel (v-Rel), is a nuclear phosphoprotein that is a truncated and mutated version of the avian proto-oncoprotein c-Rel (69, 78). The deletion of the C terminus of c-Rel to produce v-Rel resulted in the removal of a cytoplasmic retention sequence and transactivation sequences (29, 62). Like other members of the Rel/NF-κB family, c-Rel and v-Rel have a conserved N terminus, the Rel homology region. This region contains sequences important for DNA binding, homo- and heterodimerization, and nuclear localization (5, 24, 29, 45). Both v-Rel and c-Rel form homodimers and heterodimers with other family members and bind to κB sites located in the promoter and enhancer elements of various effector genes (42). DNA-binding complex formation and transactivation activity are necessary for the full transforming potential of v-Rel (20, 32, 50, 58, 64). Due to the deletion of sequences involved in transcriptional activation however, v-Rel exhibits a lower transcriptional activity than does c-Rel (5, 40, 54, 62).
The regulation of Rel/NF-κB family members is mediated, in part, by their interaction with IκB proteins. IκB proteins sequester these transcription factors in the cytoplasm and inhibit their DNA binding (reviewed in references 6, 8, and 28). In response to external stimuli that result in the activation of Rel/NF-κB complexes, IκB-α is proteolytically degraded, allowing the nuclear translocation of these transcription complexes (35, 44, 72). The gene encoding IκB-α is then upregulated by the nuclear Rel/NF-κB factors, establishing an autoregulatory loop that results in a transient response to exogenous stimuli (17, 18, 41, 51). We have previously shown that v-Rel activates the transcription of IκB-α far less efficiently than c-Rel and the induction of IκB-α occurs with delayed kinetics (38, 65). Moreover, avian IκB-α transcription is synergistically regulated by Rel and AP-1 factors (49). v-Rel, however, is less effective in the synergistic stimulation of the IκB-α promoter than is c-Rel. Because v-Rel is impaired in the induction of IκB-α, the activity of v-Rel is less sensitive to autoregulation by IκB-α. This is likely to be an important feature in the establishment of transformation by v-Rel. The functional interaction between AP-1 factors and Rel suggested by the IκB-α promoter analysis led us to define whether AP-1 factors play a role in the v-Rel transformation process. While this work was in progress, Fuji et al. demonstrated that the c-jun promoter is activated by v-Rel, providing additional evidence that c-Jun expression may be involved in the v-Rel transformation pathway (22).
Functional AP-1 activity is required for cellular transformation by certain oncogenes (v-src, v-yes, v-fps, c-Ha-ras, and N-terminally truncated c-raf) but is not essential for transformation induced by others (v-ros, v-myc) (70). AP-1 is a dimeric transcription factor composed of proteins belonging to two different families of proto-oncogene products, Jun (c-Jun, JunB, and JunD) and Fos (c-Fos, FosB, Fra-1, and Fra-2) (reviewed in references 4 and 31). AP-1 binds to a specific DNA target sequence, TGA(C/G)TCA, known as the TPA response element, to increase the transcriptional activities of target genes (3). fos and jun are immediate-early response genes (47). This family of genes is highly, rapidly, and transiently activated upon stimulation of quiescent cells by external stimuli, leading to cell proliferation. Moreover, the altered expression of several individual Fos or Jun proteins can result in the transformation of cells in culture, either alone or in cooperation with other activated oncogene products (14, 55, 60). The AP-1 transcription system is subject to complex regulation (4). Therefore, analysis of the individual activities of different members of the AP-1 family in v-Rel-transformed cells is required to understand the general mechanism by which v-Rel may affect AP-1 activity. To test the potential role of AP-1 in v-Rel transformation, the expression of the known chicken AP-1 components (c-jun, c-fos, fra-2, and junD) was analyzed at the mRNA and protein levels. In this study, we demonstrate that c-jun, c-fos, and fra-2 are differentially expressed in v-Rel-transformed cells compared with control cells, while the expression of junD is unaffected. Moreover, the transforming ability of oncogenic Rel proteins (v-Rel, c-RelΔ, and c-Rel) correlates with their ability to alter the expression of c-Jun and other AP-1 factors. These observations suggest an involvement of AP-1 factors in the Rel-induced transformation pathway. To provide direct evidence for a functional role for AP-1 factors in v-Rel transformation, we have used the targeted inhibition of AP-1 activity by a supjun-1 transdominant mutant. The supJun mutant is missing a transactivation domain but retains both dimer-forming and DNA-binding activities (70). The simultaneous or sequential introduction of the supjun-1 transdominant mutant with v-rel or c-relΔ in fibroblasts and lymphoid cells had a significant inhibitory effect on the transformation potential of these oncogenes.
MATERIALS AND METHODS
Cells and growth conditions.
Chicken embryo fibroblast (CEF) cultures were prepared from 10- to 11-day-old embryos (SPAFAS, Norwich, Conn.) and were routinely grown in Dulbecco’s modified Eagle’s medium (JRH Biosciences) supplemented with 5% fetal calf serum (Summit Biotechnology), 3% chicken serum (GIBCO-BRL), and antibiotics as previously described (48). For serum starvation, cultures of 5 × 106 cells/100-mm plate were grown for 18 h in Dulbecco’s modified Eagle’s medium supplemented with 0.2% calf serum. Subsequently, these cells were stimulated by the addition of 10% calf serum for the times indicated. DT95 cells, an avian B-cell line derived from a chicken infected with avian leukosis virus, and the v-rel-transformed lymphoid cell line 160/2 (T-cell line) were maintained as previously described (37). C4-1 cells (RECC-UTC4-1), a non-virus-producing lymphoblastoid pre-B-cell line obtained from reticuloendotheliosis virus transformation of spleen cells (52, 81), were grown under the same conditions as the other lymphoid cell lines.
Plasmid constructs and virus production.
All recombinant techniques were carried out by conventional procedures (63). Oligomer TNFE2 (obtained from J. Palma) containing five AP-1 consensus binding sites (TGACTCA) was cloned into the BglII site of the pGL2-promoter vector (Promega, Madison, Wis.) to create the construct (AP-1)5SV40-luc. The other reporter constructs used in transactivation experiments include −73/+63coll CAT and −60/+63coll CAT, which contain the collagenase promoter with or without the AP-1-binding site, respectively (both kindly provided by P. Vogt) (1). The transdominant mutant supjun-1, as well as v-rel, c-fos, and c-jun, was cloned into the expression vector pRc/RSV (Invitrogen) for transient-transactivation studies. supjun-1 was excised from the pDS3 vector (kindly provided by H. Iba) with BglII, blunt ended, and ligated with a blunt-ended XbaI-digested vector, pRc/RSV. Similarly, a 1.6-kb XbaI-ClaI fragment of v-rel with previously described modifications and a 1.3-kb KpnI fragment of c-fos (obtained from R. Muller) were blunt ended and ligated as described for supjun-1 (57). The c-jun construct was provided by P. Vogt.
To deliver the v-rel and c-relΔ oncogenes or the supjun-1 transdominant mutant into cells to obtain stable cell lines, we used a replication-competent retrovirus vector, composed of pREP and pDS3 SalI fragments containing the 5′ and 3′ halves of the provirus, respectively (61). The supjun-1 expression vector was generated by insertion of a 5′-end-truncated human c-jun sequence into the BglII site of pDS3 prior to ligation with the pREP SalI fragment (70). The v-rel expression vector was constructed by insertion of the 1.6-kb XbaI-ClaI fragment, which was filled in with Klenow and ligated with the blunt-ended, BglII-cut pDS3. The C-terminal deletion construct of c-Rel (c-RelΔ) was generated as described previously, and a EcoRV-EagI fragment was blunted and cloned into a blunt-ended BglII site of pDS3 as described for v-rel (48).
For the production of recombinant viruses, supjun-1, v-rel, and c-relΔ in the pDS3 vector were completely digested with SalI and ligated to the SalI-digested pREP-A or pREP-B to create the replication-competent viruses (70). Ligated DNAs (4 μg) were transfected into CEF cultures by a calcium phosphate precipitate technique, and replication-competent virus stocks were collected from the cultures 6 or 7 days after transfection, aliquoted, and kept frozen at −70°C (16). The infectious titer of virus stocks was measured in CEF cultures by an in situ expression assay as described below.
In situ immunohistochemical detection of virus-infected cells.
To determine the infectious unit titer (IU) of the collected virus stocks, in situ immunohistochemical detection was used for virus-infected cells with an antibody against gag viral proteins. CEF cultures to be tested for viral infection were plated (7 × 105 cells/60-mm petri dish) 24 h before infection. The following day, the growth medium was replaced with 0.5 ml of medium containing various dilutions of the viral stock. After a 2-h incubation, the medium was removed and cells were overlaid with 0.7% agar medium and grown to full confluency for an additional 5 days. The agar was removed, and the cells were washed with phosphate-buffered saline (PBS) and fixed in 2% paraformaldehyde in PBS for 30 min. The fixed cells were permeabilized by incubation in acetone–methanol (1:1) for 2 min. Subsequently, the cells were treated with the primary antibody, i.e., polyclonal rabbit anti-p27gag serum (Life Sciences, St. Petersburg, Fla.) diluted 1:500 in PBS with 2% bovine serum albumin (BSA) for 1 h at room temperature. After being washed in PBS, the cells were incubated under the same conditions with an alkaline phosphatase-conjugated goat anti-rabbit serum (Kirkegaard & Perry Laboratories, Gaithersburg, Md.) for 1 h. The alkaline phosphatase activity was detected in the presence of 1 mg of Fast Red TR (Sigma) per ml and 0.2 mg of Napthol AS-MX phosphate (Sigma) per ml in 0.1 M Tris (pH 8.2)–0.1 M NaCl. The reaction was stopped after 10 to 20 min by extensive washing in PBS, and macroscopically visible red-stained infectious centers were counted.
Virus infections.
Fresh CEF cultures (plated at 7 × 105 cells/60-mm dish) were infected with retroviruses by incubating the virus for 2 h with the cells in the presence of Polybrene (Sigma) at 8 μg/ml. The incubation was then continued overnight after dilution of Polybrene to 2 μg/ml with medium. The medium was replaced the next day, and cells were subcultured as necessary. Usually after two passages, cells exhibited morphological transformation following infection with retroviruses containing v-rel.
To obtain cell populations of doubly infected CEF cultures expressing both v-Rel and the supJun mutant at similar levels, two sets of vectors that differ with respect to envelope subgroup specificity (A and B) were used for each gene. CEF cultures were first infected with the virus from subgroup A first and, 4 days later, superinfected with the virus from subgroup B to deliver the second gene (70). Alternatively, doubly infected cultures were obtained through the application of both types of viruses simultaneously (coinfection). We routinely infect CEF cultures at a multiplicity of infection of 2. For superinfection of lymphoid cell lines with supjun-1, a multiplicity of infection of 5 to 10 was used.
DNA transfections and reporter assays.
Secondary cultures of CEFs were seeded at a density of 7 × 105 cells per 60-mm-diameter dish 1 day before transfection. DNA was transfected into CEF cultures by calcium phosphate precipitation techniques as previously described (49). Briefly, 0.1 μg of chloramphenicol acetyltransferase (CAT) reporter DNA (−73/+63coll CAT, −60/+63coll CAT) was cotransfected with 3 μg of vector DNA (pRc/RSV; Invitrogen) or vectors containing the c-jun or supjun-1 genes (3 and 8 μg, respectively). The total DNA concentration was kept constant by adding an unrelated plasmid DNA (pBluescript). At 24 h after the glycerol shock, the cells were harvested (34). CAT activity in cell extracts containing equal amounts of protein was determined with the CAT enzyme-linked immunosorbent assay kit (Boehringer Mannheim Corp. no. 1363727) as specified by the manufacturer. The absorbance of the samples was measured at 410 nm with a microtiter plate enzyme-linked immunosorbent assay reader (Dynatech no. MR5000). The same procedure with several modifications was applied to deliver luciferase constructs. A 1-μg portion of luciferase construct [(AP-1)5SV40-luc] was cotransfected with vectors expressing c-jun, c-fos, v-rel, c-rel, and supjun-1 as indicated. At 36 h posttransfection, the cells were harvested (65). Luciferase activity for equal amounts of protein was determined by the luciferase assay system (Promega no. E4030) with an MLX microtiter plate luminometer (Dynatech).
In vitro transcription and translation.
Template plasmids containing c-jun, c-fos, or supjun-1 in the pTZ18R vector, cloned in the sense orientation with the T7 promoter, were linearized by restriction enzymes which cut the polylinker 3′ of the inserted gene. For in vitro protein synthesis, the TNT T7 quick coupled transcription/translation system (Promega no. L1170) was used as specified by the manufacturer. To determine the size and quantity of the in vitro-translated proteins, parallel reactions were done in the presence of [35S]methionine. These in vitro-translated proteins were analyzed by electrophoresis through sodium dodecyl sulfate (SDS)–10% polyacrylamide gels and visualized by autoradiography.
EMSA analysis.
Nuclear extracts for gel shift analysis were prepared as described previously (65, 66). For the electrophoretic mobility shift assay (EMSA) reactions, 5 μg of nuclear extracts was incubated for 15 min at room temperature in a total reaction volume of 25 μl containing 20 mM Tris (pH 7.5), 75 mM KCl, 40 μM EDTA, 5% glycerol, 1 mM dithiothreitol, 100 μg of BSA per ml, and 1 μg of poly(dI-dC). Then 0.2 ng of double-stranded AP-1 oligonucleotide (end labeled with 32P) (Santa Cruz Biotechnology, Inc. no. sc-2501) was added to the reaction mixture. The mixture was then incubated for an additional 20 min at room temperature. For competition analysis, a 100-fold molar excess of unlabeled oligonucleotide was added. For supershift analysis, 2 μl of the appropriate antiserum was added, and the mixture was incubated on ice for 45 min before the addition of the labeled oligonucleotide. Samples were then analyzed by electrophoresis in a 5% polyacrylamide gel with 0.25× Tris-borate-EDTA. Following the electrophoresis, the gels were dried and DNA-protein complexes were visualized by autoradiography.
In vitro-translated proteins were mixed proportionally according to the efficiency of the in vitro translations (total volume, 4.5 μl) and incubated at 37°C for 45 min to allow protein-protein association. Then 5 μl of binding buffer (10 mM HEPES [pH 7.9], 50 mM NaCl, 4 mM MgCl2, 0.1 mM EDTA, 4 mM spermidine, 2 mM dithiothreitol, 100 μg of BSA per ml, 15 μg of salmon sperm DNA per ml, 15% glycerol) and 1 μg of poly(dI-dC) (Pharmacia) were added, and the mixture was incubated for a further 15 min at room temperature. Then 1 ng of 32P-labeled, double-stranded DNA probe containing the FSE2–AP-1 sequence was added, and the incubation was continued at 4°C for 15 min (71). The samples were loaded on a 5% polyacrylamide gel and electrophoresed at 4°C for an extended period to provide maximum separation of complexes. This extended electrophoresis resulted in the free probe being electrophoresed from the gel.
The oligonucleotides used for EMSA are listed below. Bold letters represent the 12-O-tetradecanoylphorbol-13-acetate responsive element, while underlined letters indicate the nucleotides that are different between the AP-1 and AP-1m oligonucleotides. AP-1 5′-CGCTTGATGACTCAGCCGGAA 3′ 3′-GCGAACTACTGAGTCGGCCTT 5′ AP-1m 5′-CGCTTGATGACTTGGCCGGAA-3′ 3′-GCGAACTACTGAACCGGCCTT-5′ FSE2 5′-TCGACTATTAAAAACATGACTCAGAGGAAAAC-3′ 3′-GATAATTTTTGTACTGAGTCTCCTTTTGAGCT-5′
Protein analysis.
Antibodies used for protein detection include a polyclonal antiserum made against a bacterially expressed c-Jun (USC30-4), kindly provided by P. Vogt. A c-Jun/AP-1 affinity-purified polyclonal antiserum corresponding to a highly conserved DNA-binding domain (residues 247 to 263) of mouse c-Jun was purchased from Santa Cruz Biotechnology (no. cs-44X). Since this antiserum is broadly cross-reactive with Jun proteins of chicken, mouse, and human origin, it was used to detect the human supJun mutant. To detect chicken Fra-2 protein, we used anti-Fra-2 (Q-20) serum, which is cross-reactive with avian Fra-2 (Santa Cruz Biotechnology no. sc-604X). This antiserum was raised against a peptide corresponding to amino acids 3 to 22 mapping at the N terminus of Fra-2 of human origin (this peptide sequence differs from the corresponding chicken sequence by two amino acids). Two v-Rel-specific antipeptide antisera were generated against the 13 C-terminal residues from the env sequences (anti-E1) or the 10 N-terminal env residues (anti-R5). c-Rel specific antiserum (anti-A5) was raised against C-terminal residues 531 to 541.
Cell lysates of virus infected cells were prepared as previously described (53). A 20-μg portion of total cellular protein or lysate derived from 2 × 105 cells was then resolved by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) followed by electrophoretic transfer to nitrocellulose membrane (Optitran BA-S83; Schleicher & Schuell, Keene, N.H.) (80). For detection of supJun protein, 50 μg of nuclear extract was subjected to SDS-PAGE on a discontinuous 4, 10, and 18% step polyacrylamide gel (0.75 mm thick) by using a Hoefer minislab electrophoretic unit with 0.1 M Tris–0.1 M Tricine–0.1% SDS as the cathode buffer and 0.2 M Tris (pH 8.9) as the anode buffer. Immunoblots were treated with 5% nonfat dried milk and then incubated with specific antisera (polyclonal rabbit antisera diluted 1:1,000 in 5% nonfat dried milk). The secondary antibody was goat anti-rabbit serum conjugated with horseradish peroxidase (Jackson Immunoresearch Lab., Inc. West Grove, Pa, no. 111-035-003) used at a 1:5,000 dilution in 5% nonfat dried milk. The protein bands were visualized by the enhanced chemiluminescence Western blotting detection system (Dupont NEN). The results were quantified by densitometric analysis.
For 35S metabolic labeling, cells were incubated for 3 h in methionine- and cysteine-free medium containing 5% fetal calf serum for exponentially growing cells and 0.2% fetal calf serum for starved cells. The cells were then labeled with 500 μCi of [35S]methionine-[35S]cysteine label mix (NEG-072 express protein labeling mix [NEN Life Science Products]) per ml for 1 h. For growth stimulation, serum-starved cells (starved for 21 h) were exposed to 10% calf serum, which was added to the culture simultaneously with the label mix. Subsequently, the cells were harvested and nuclear extracts were prepared from labeled cells by the same method as that used for gel shifts. Proteins in nuclear extracts were subjected to a first immunoprecipitation with the Fra-2 antiserum, and supernatant fluids from these precipitations were reprecipitated with an anti-c-Jun antiserum.
Northern blot analysis.
Total cellular RNAs were prepared from cells by acidic guanidinium thiocyanate-phenol-chloroform extraction (19). For Northern blot analysis, RNA (15 or 20 μg) was separated on 1% agarose–formaldehyde gels (63). This was followed by capillary transfer to nylon filters (Hybond-N+), which were then stained with methylene blue to confirm equal loading and RNA transfer. The filters were hybridized at 65°C in the presence of 10% dextran sulfate, 5× SSPE (0.9 M NaCl, 0.05 M sodium phosphate, 5 mM EDTA), 5× Denhardt’s solution (0.1% BSA, 0.1% Ficoll, 0.1% polyvinylpyrrolidone), 0.5% SDS, and 50 μg of salmon sperm DNA per ml with one of the following randomly primed cDNA probes: 1-kb XbaI fragment of chicken c-jun, 1.3-kb KpnI fragment containing a full-length cDNA of c-fos, 1.46-kb EcoRI-PstI fragment of chicken junD, or 0.26-kb PvuII-HindIII fragment of chicken fra-2 (23, 33, 59, 60, 70). As a control for RNA loading, the filters were rehybridized with a 1.2-kb EcoRI fragment of human glyceraldehyde phosphate dehydrogenase (GAPDH). The results were quantified by densitometric analyses.
Soft agar colony formation.
CEF cultures (105 cells) sequentially infected or coinfected with two species of viruses were seeded in soft agar (0.37%) on top of a bed of hard agar (0.75%) 4 days after infection as previously described (48). The cells were refed with additional soft agar medium at weekly intervals. The plates were scored for the development of colonies 4 weeks after seeding. Lymphoid cells, C4-1 or 160/2, were superinfected with supjun-1 virus or DS3 virus (carrying no insert) and were seeded into 0.37% soft agar 10 days after infection (104 cells/60-mm plate). Colonies were counted 2 weeks after seeding, and the mean value from three plates was determined.
RESULTS
AP-1 factors are differentially expressed in Rel-transformed cells.
We have previously observed that the steady-state level of c-Jun in v-rel-transformed fibroblasts is elevated relative to that in nontransformed cells, suggesting that early-response genes may play a role in the v-rel-induced transformation process. To test this hypothesis, we have analyzed the effect of v-rel expression on c-jun as well as the other known members of the AP-1 family. CEF cultures were infected with retrovirus expression vectors containing no insert (DS3), v-rel, c-relΔ, or the c-rel proto-oncogene. v-rel is a strongly transforming oncogene, c-rel is a poorly transforming proto-oncogene, and c-relΔ, whose product lacks the 40 C-terminal amino acids of c-Rel, exhibits intermediate transforming activity (48). Ten days after infection, the time when cells exhibit the morphological change associated with the transformed phenotype, cell extracts were prepared, RNA was isolated, and the expression of various AP-1 factors (c-jun, c-fos, fra-2, and junD) was analyzed by Northern analysis.
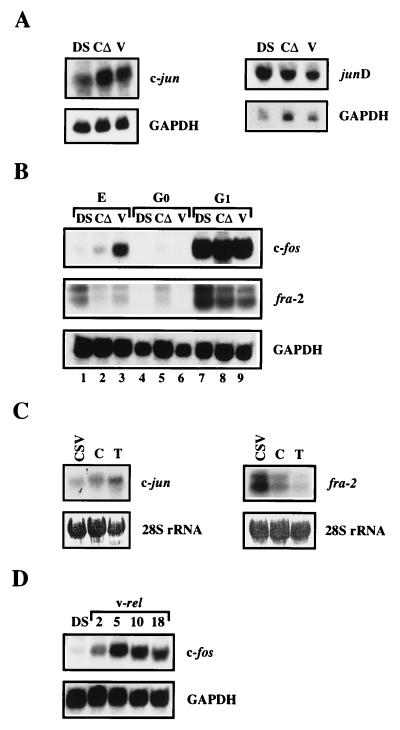
As shown in Fig. 1A, the c-jun mRNA level was elevated twofold in v-rel as well as in c-relΔ-transformed CEF cultures. A twofold increase in the c-jun mRNA level was also observed in the v-rel-overexpressing lymphoid cell line DT95, whereas c-rel overexpression in these cells resulted in only a slight increase (1.3-fold) in the level of c-jun transcripts (Fig. 1C). In contrast to c-jun, junD mRNA levels were the same in control and transformed cells (Fig. 1A). Since c-fos mRNA is not usually detected in exponentially growing CEF cultures, total RNA was also extracted from serum-starved and serum-stimulated cells. As shown in Fig. 1B, c-fos mRNA (lanes 7 to 9) was highly expressed in the control cells (lane 7) as well as in the rel-transformed cells (lanes 8 and 9) after serum stimulation but was undetectable in serum-starved cells (lanes 4 to 6). However, in exponentially growing v-Rel-transformed cells, the level of c-fos mRNA was increased (4.6-fold) compared to that in the vector-infected control cells (lanes 1 and 3), whereas in c-RelΔ-expressing cells, only a slight increase (1.6-fold) was observed (lane 2). To determine if the elevation of the level of c-fos mRNA in v-Rel-transformed cells is a transient or a prolonged feature, RNA was harvested at different time points after v-rel infection and the levels were evaluated by Northern analysis (Fig. 1D). Two days after infection, as soon as v-Rel was expressed in infected CEF cultures (data not shown), the c-fos mRNA level was increased (twofold) and remained high (four- to fivefold increase) relative to control cells at the later time points examined. The upregulation of c-fos mRNA was much more pronounced in v-Rel-transformed cells than in c-RelΔ-transformed cells (Fig. 1B, lanes 2 and 3; Fig. 1C, lanes C and T). Surprisingly, Rel overexpression seems to have a suppressive effect on the mRNA levels of another member of the fos family, fra-2. In both cell types examined, i.e., CEFs (Fig. 1B) and the lymphoid cell line DT95 (Fig. 1C), a lower level of fra-2 was detected following infection with rel expressing viruses in exponentially growing cells (Fig. 1B, lanes 2 and 3; Fig. 1C, lanes C and T). For v-Rel- and c-RelΔ-transformed CEF cultures, about a 2.5-fold reduction compared to control cells was observed, whereas in DT95 cells, a more pronounced suppression following v-Rel overexpression (4.5-fold decrease) was detected. Furthermore, the lower level of expression of the fra-2 mRNA was also maintained in Rel-transformed cells after growth stimulation compared to the expression in control cells (DS3) (Fig. 1B, compare lane 7 with lanes 8 and 9). While the fra-2 mRNA was highly stimulated by serum in control cells (lanes 4 and 7), the same growth stimulus resulted in lower induction and overall expression of the fra-2 mRNA in v-Rel-transformed cells (compare lanes 6 and 9). In conclusion, Rel overexpression seems to have a differential effect on the mRNA levels of the various members of the AP-1 family: it upregulates c-jun and c-fos, downregulates fra-2, and does not appear to influence the expression of junD.
FIG. 1.
Expression levels of mRNAs encoding AP-1 components in Rel-overexpressing cells. (A) Expression of c-jun and junD mRNA in CEF cultures infected with viruses carrying no insert (DS) or the oncogene c-relΔ (CΔ) or v-rel (V). A 15-μg sample of each RNA was used for Northern blot hybridization, first with radiolabeled c-jun cDNA and then with a human GAPDH probe to demonstrate the RNA loading (left two panels). For junD mRNA analysis, 20 μg of total RNA was used and the membrane was hybridized with junD cDNA followed by rehybridization with GAPDH (right two panels). (B) Serum stimulation of c-fos and fra-2 mRNA. Three different cell pools of DS3-, c-relΔ-, and v-rel-infected CEF cultures were grown in serum (exponentially growing cells [E]), or were serum starved for 18 h in the presence of 0.2% calf serum (quiescent cells [G0]) and then stimulated by the addition of 10% calf serum for 90 min prior to RNA extraction (serum-stimulated cells [G1]). Total RNA (20 μg) isolated from these various cultures was sequentially analyzed by Northern blot hybridization with radiolabeled chicken c-fos, fra-2, and human GAPDH DNA probes. (C) Expression levels of endogenous c-jun and fra-2 mRNA in infected DT95 lymphoid cells. DT95 cells were infected with REV-C (C) or REV-T (T) in the presence of the CSV helper virus. Three weeks after infection, the expression of these oncoproteins was confirmed by Western blot analysis (see Fig. 2B) and total RNA (20 μg) was subjected to Northern blot analyses with c-jun and fra-2 DNA probes as described above. (D) Time course of c-fos mRNA induction in CEF cultures following infection with a retrovirus expressing v-rel. Total RNA (20 μg per lane) was extracted at the time points (days) indicated and sequentially analyzed by Northern blot hybridization with radiolabeled c-fos and then GAPDH probes.
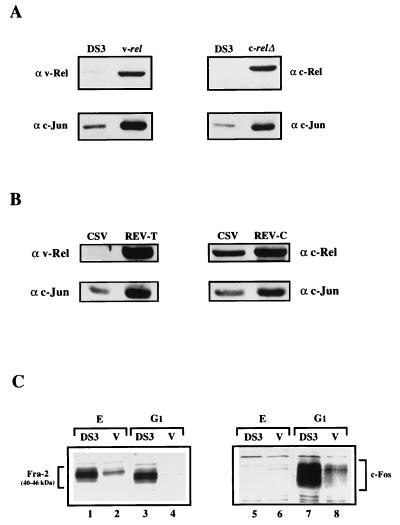
Next, we analyzed the effect of v-Rel on AP-1 expression at the protein level. The steady-state expression of endogenous c-Jun in exponentially growing CEF cultures infected with different viruses was analyzed by Western immunoblotting. After blotting, the membranes were cut into two parts. The upper portion was incubated with v-Rel- or c-Rel-specific antiserum to evaluate the expression of the virus-delivered oncogenes v-rel or c-rel and c-relΔ, respectively. The lower portion was incubated with anti-c-Jun antiserum to detect endogenous c-Jun. As shown in Fig. 2A, v-rel-infected CEF cultures expressed 4-fold more c-Jun and c-relΔ-infected CEF cultures expressed 3.5-fold more c-Jun than did control DS3 cells. In DT95 lymphoid cells infected with REV-T (carrying the v-rel oncogene) or REV-C (carrying full-length c-rel) in the presence of the chicken syncytial virus (CSV) helper virus, a similar 3.7-fold or 2-fold increase, respectively, was observed (Fig. 2B). These results consistently demonstrated that in both cell types examined, there is a correlation between the transformation potential of v-rel, c-rel, or c-relΔ and the expression pattern of c-jun. Generally, the strongly transforming oncoprotein v-Rel had the most pronounced effect on c-Jun expression, while full-length c-Rel had only a moderate effect. c-RelΔ, which exhibits enhanced transforming ability compared to c-Rel, also has a stronger effect on c-Jun levels than did c-Rel (48). Unfortunately, we were not able to make the comparison for JunD, because JunD expression was undetectable with the available antisera (data not shown).
FIG. 2.
Expression of endogenous c-Jun, Fra-2, and c-Fos in cells overexpressing Rel proteins. (A) Proteins in whole-cell lysates from control and c-relΔ- or v-rel-expressing CEF cultures (each analysis contained the equivalent of 2 × 105 cells) were resolved on SDS–10% polyacrylamide gel and then subjected to immunoblotting with the enhanced chemiluminescence Western blotting detection system. Membranes were cut into two parts; the upper portion was incubated with anti-v-Rel (α v-Rel) serum to detect p59v-rel or anti-c-Rel serum (α c-Rel) to detect p64c-relΔ, and the lower portion was incubated with anti-c-Jun (α c-Jun) antiserum to detect c-Jun (40 kDa). The expression of these proteins was monitored at 10 days postinfection. (B) Proteins in whole-cell lysates corresponding to 2 × 105 cells per lane from DT95 infected with REV-T (v-rel) or REV-C (c-rel) in the presence of the CSV helper virus were analyzed by Western blot analysis as described above for CEF cultures. (C) Exponentially growing (E) and serum-stimulated (G1) CEF cultures (expressing v-rel [V] or DS3 [DS]) were 35S labeled for 60 min, and proteins in nuclear extracts were immunoprecipitated with anti-Fra-2 serum (lanes 1 to 4). Then the supernatant fluids from these Fra-2 precipitations were subjected to a second precipitation with anti-c-Jun serum (lanes 5 to 8) to detect coprecipitated c-Fos. Proteins in immunoprecipitates were resolved by SDS-PAGE (10% polyacrylamide) and visualized by autoradiography after exposure of the X-ray film for 3 weeks.
Due to the low level of Fra-2 and c-Fos expression, radioimmunoprecipitations were performed to detect changes in the expression of these proteins. Control and v-Rel-transformed cells which were grown under normal growth conditions—exponentially growing cells (E) or serum-starved cells stimulated with 10% calf serum (G1) were labeled with [35S]Met-[35S]Cys, and nuclear extracts were prepared. Proteins in these extracts were immunoprecipitated with Fra-2 antiserum (Fig. 2C, lanes 1 to 4), and the supernatant fluids from these Fra-2 precipitations were subjected to a second precipitation with c-Jun antiserum to detect associated c-Fos (lanes 5 to 8). By using this approach, it was demonstrated that Fra-2 proteins (the molecular masses of unphosphorylated and phosphorylated forms range from 40 to 46 kDa) were expressed at a higher level in control cells than in v-rel-infected cells and that this pattern of expression did not change substantially after serum stimulation (lanes 1 to 4). c-Fos proteins were detected by secondary coimmunoprecipitations with anti-c-Jun antiserum. In serum-stimulated CEF cultures (infected with DS3 or v-rel), highly phosphorylated c-Fos was observed at the expected molecular mass, whereas in exponentially growing cells, c-Fos was barely detected. These protein analyses correlated well with the mRNA analysis which shows that the Fra-2 was less abundant in v-Rel-transformed cells and that the level was not dramatically increased by serum stimulation compared to that in control cells. For the detection of c-Fos, we relied on coimmunoprecipitation of c-Fos with c-Jun (chicken c-Fos antiserum was not available). These results might not reflect the actual quantitative level of c-Fos in the examined cells. This may explain why the elevated expression of c-Fos was not detected in exponentially growing v-Rel-transformed cells as was observed at the mRNA level.
In conclusion, the altered mRNA and protein expression patterns of various AP-1 members in Rel-transformed cells compared to those in control cells led us to examine the role of AP-1 in the Rel-induced transformation pathway.
Expression of a supjun-1 transdominant mutant downregulates c-jun.
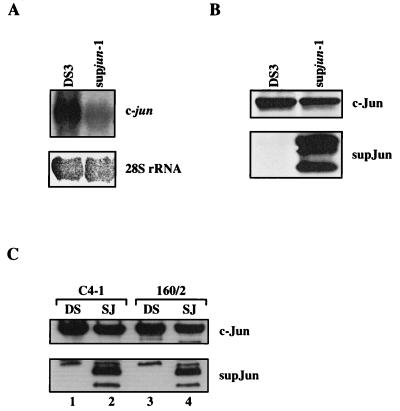
We used the targeted inhibition of AP-1 by a supjun-1 transdominant mutant to define a functional role for AP-1 factors in v-Rel transformation. Since c-jun expression is positively regulated by its own product (2), the presence of supJun is likely to downregulate c-jun expression. To test this hypothesis, the expression of endogenous c-jun in supjun-1-expressing CEF cultures versus control cells (DS3 infected) was assayed by Northern and Western blot analyses. Figure 3A shows that in supjun-1-expressing CEF cultures, the c-jun mRNA level was decreased twofold. Similarly, the expression of the supJun protein reduced the steady-state level of the c-Jun protein approximately twofold as determined by densitometry (Fig. 3B). The membrane was stained with Ponceau S to verify equal protein loading, and then the top half of this Western blot was incubated with c-Jun antiserum to detect endogenous c-Jun (40 kDa) and the bottom half was incubated with a panspecific Jun antiserum to detect the human supJun protein. The multiple proteins detected in the bottom panel most probably represent breakdown products of supJun (expected size, 15 kDa). In addition, a 1.5-fold decrease in the expression of endogenous c-Jun was detected in the REV-T-transformed lymphoid cell lines C4-1 and 160/2 superinfected with the virus expressing supjun-1 (Fig. 3C). These results indicate that one of the inhibitory effects of supjun-1 on AP-1 activity may be the direct downregulation of c-jun.
FIG. 3.
Effect of supjun-1 expression on the level of endogenous c-jun. (A) Level of the c-jun transcript in CEF cultures infected with DS3 and supjun-1 viruses (top panel). CEF cultures were harvested 10 days after infection, and total RNA was extracted and subjected to Northern blot analyses (20 μg per lane) with a radiolabeled c-jun cDNA probe. RNA loading was demonstrated by showing 28S rRNA (bottom panel). (B) Nuclear extracts (50 μg) from infected CEF cultures were analyzed on a discontinuous 4, 10, and 18% step polyacrylamide gel as described in Materials and Methods and then subjected to immunoblotting. After blotting, the membranes were cut into two parts. The top half was incubated with anti-c-Jun serum to detect endogenous c-Jun (40 kDa), and the bottom half was incubated with a panspecific Jun antiserum to detect the human supJun protein. (C) Nuclear extracts (50 μg) from v-Rel-transformed lymphoid cells C4-1 and 160/2 were isolated, and protein expression was analyzed 10 days after superinfection with DS3 or supjun-1 viruses as described for panel B.
SupJun inhibits c-Jun as well as v-Rel-induced transactivation.
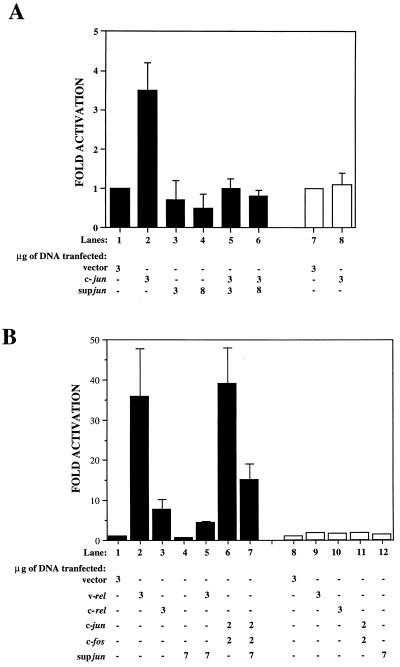
SupJun, which is missing the transactivation domain of c-Jun but retains its specific DNA binding activity and leucine zipper domain, is expected to be a transdominant inhibitor of AP-1-mediated transcriptional regulation (70). To demonstrate that supJun inhibits AP-1-mediated transactivation, we have used a reporter construct from the human collagenase promoter (containing a single AP-1-responsive element) linked to the CAT gene (−73/+63coll CAT). This reporter was cotransfected with expression plasmids (Rc/RSV; Invitrogen) carrying c-jun, supjun-1, or both. Figure 4A, lane 2, demonstrated a 3.5-fold increase over basal-level endogenous activity in CAT when c-jun was transfected into CEF cultures. This activity was mediated by AP-1, since a reporter CAT construct with the AP-1 site deleted (−60/+63coll CAT) showed no increase in CAT activity when cotransfected with c-jun (lane 8). Moreover, cotransfection with increasing amounts of supjun-1 with a constant amount of c-jun resulted in suppression of CAT activity (lanes 5 and 6). In addition, supjun-1 was able to suppress the basal level of endogenous AP-1 activity (lanes 3 and 4).
FIG. 4.
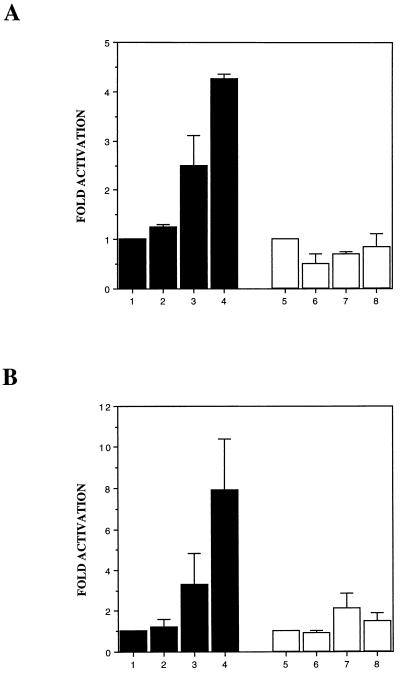
(A) Inhibition of c-Jun-induced transactivation of collagenase promoter by supjun-1. Transactivation was assessed by measuring CAT activity in extracts of CEF cultures cotransfected with or without supjun-1 and the −73/+63coll CAT reporter gene (lanes 1 to 6, solid bars) or −60/+63coll CAT with the AP-1 site deleted (lanes 7 and 8, open bars) 24 h posttransfection (34). (B) Inhibition of v-Rel-induced transactivation by supjun-1. CEF cultures were cotransfected with expression vectors for the genes indicated, along with a luciferase reporter construct with or without multiple AP-1 binding sites cloned in front of the SV40 promoter, respectively (lanes 1 to 7, solid bars; lanes 8 to 12, open bars). Luciferase activity for equal amounts of protein was determined as described in Materials and Methods 36 h after transfection (65). The fold activation was determined by dividing the actual chloramphenicol or luciferase activity obtained in the presence of c-Jun, c-Fos, v-Rel, and supJun expression vectors by the activity of the reporter when the cells were cotransfected with an “empty” expression vector, pRc/RSV (defined as baseline activity 1.0). The mean results from three to five experiments are shown. Standard errors were determined; however, in some cases they are not visible in the given activation scale for the luciferase assays.
It has been shown recently that v-rel can transactivate the c-jun promoter (22). Based on this observation, one might expect that v-rel, by activating c-jun, could also potentially modulate the collagenase reporter construct. We were, however, unable to show that v-rel can mediate the transactivation of the CAT-linked collagenase reporter construct (data not shown). However, using a luciferase construct with multiple AP-1-binding sites cloned upstream of the simian virus 40 (SV40) promoter [(AP-1)5SV40-luc], we observed a strong activation of luciferase activity not only with vectors expressing c-fos and c-jun genes (Fig. 4B, lane 6) but also with those expressing v-rel and to a lesser extent c-rel (lanes 2 and 3). This strong activation by v-rel (a 35.8-fold increase) was inhibited by cotransfection with supjun-1 (lane 5). The activation of the (AP-1)5SV40-luc construct was AP-1 mediated, since the SV40-luc construct lacking AP-1-binding sites was not activated by any of the genes examined (lanes 8 to 12). Figure 4 shows the results obtained from at least three different experiments. The much stronger activation of the (AP-1)5SV40-luc construct than of the collagenase-CAT reporter was probably due to the presence of multiple AP-1-binding sites in the former construct instead of just a single AP-1 site in the latter construct. Furthermore, the luciferase assay system is more sensitive than the CAT assay system. The results obtained from these transactivation experiments confirm that the supjun-1 mutant can inhibit c-jun- and v-rel-induced transactivation of AP-1-controlled promoters in CEFs.
AP-1 reporters are activated in Rel-transformed cells.
In the previous section, it was demonstrated that Rel proteins increase the expression of AP-1 reporter constructs in transient-transfection assays (Fig. 4A and B). Next, we wanted to assay the expression of AP-1 reporter constructs in transformed cells. CEF cultures were infected with control virus carrying no insert, DS3, v-rel, or c-relΔ. At 8 to 14 days later, when rel-infected cells exhibited all features of morphological transformation, the cells were transfected with −73/+63coll CAT or (AP-1)5SV40-luc reporters. The transfection efficiency was monitored by the expression of a cotransfected RSVβ-Gal reporter. The expression of both AP-1 reporter constructs was elevated in Rel-transformed cells compared to that in uninfected CEF cultures or those infected with control virus carrying no insert (DS3) (Fig. 5A and B). A more pronounced activation of the AP-1 reporter constructs was detected in v-Rel-transformed cells than in c-RelΔ-transformed cells. The expression of Rel proteins had no significant impact on the expression of reporter constructs lacking AP-1 sites (−60/+63coll CAT, SV40-luc), indicating that the specific activation of these constructs in the Rel-transformed cells was mediated by AP-1 sites (Fig. 5). The activation of the −73/+63coll CAT reporter construct in these experiments, as opposed to the transient-transfection assays described in the previous section, is probably due to the higher expression of Rel proteins in transformed cells. The results of these experiments confirm that AP-1 activity is significantly elevated in Rel-transformed cells.
FIG. 5.
(A) Activation of the collagenase promoter in Rel-transformed CEF cultures. Transactivation was assayed by measuring CAT activity in extracts of v-Rel- or c-RelΔ-transformed CEF cultures transfected with the −73/+63coll CAT reporter gene (lanes 1 to 4, solid bars) or −60/+63coll CAT with the AP-1 sites deleted (lanes 5 to 8, empty bars) 24 h posttransfection. Lanes: 1 and 5, uninfected CEF cultures; 2 and 6, DS3-infected CEF cultures; 3 and 7, c-RelΔ-transformed CEF cultures; 4 and 8, v-Rel-transformed CEF cultures. (B) Activation of (AP-1)5SV40-luc in Rel-transformed CEF cultures. Lanes: 1 and 5, uninfected CEF cultures; 2 and 6, DS3-infected CEF cultures; 3 and 7, c-RelΔ-transformed CEF cultures; 4 and 8, v-Rel-transformed CEF cultures. Luciferase activity for equal amounts of protein was determined as described in Materials and Methods 36 h after transfection (65). Fold activation was determined by dividing the actual chloramphenicol or luciferase activity obtained from v-Rel- or c-RelΔ-transformed CEF cultures by the activity of the reporter from normal CEF cultures or those infected with an “empty” expression vector, DS3 (defined as baseline activity 1.0). The mean results from two experiments with standard errors are shown.
SupJun interferes with AP-1 DNA binding.
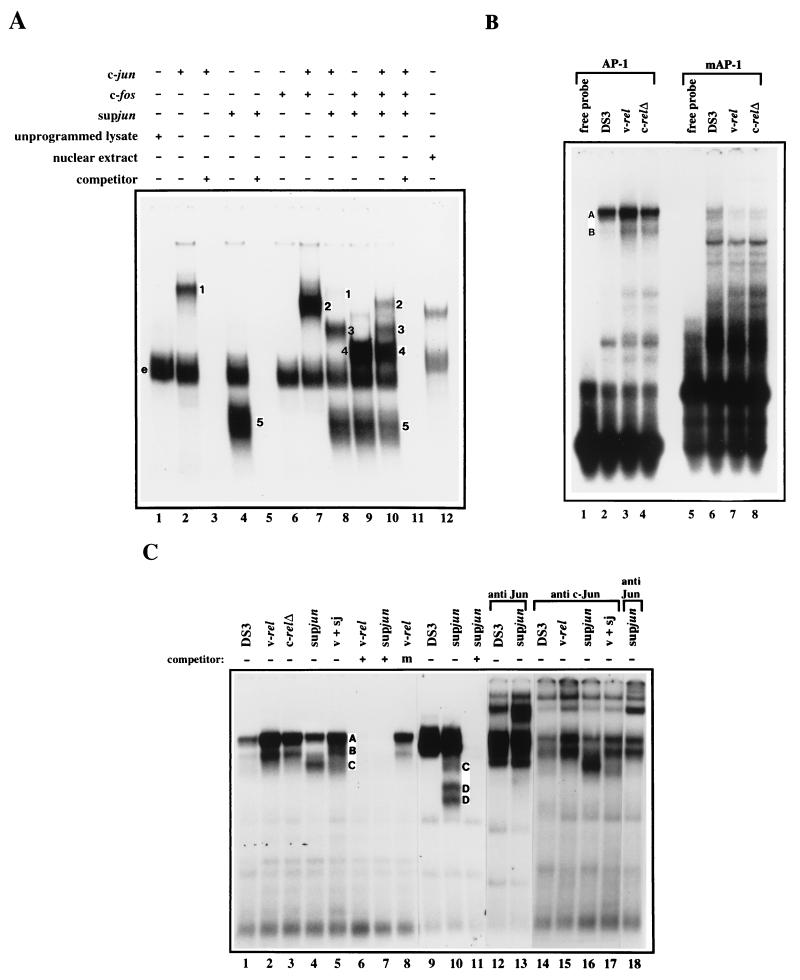
To determine if supJun could affect wild-type c-Jun and c-Fos DNA binding, we tested the ability of c-Jun and c-Fos to bind DNA containing an AP-1 site in the presence and/or absence of the supJun protein. Proteins were synthesized separately by using a reticulocyte lysate-based in vitro transcription-translation system and then mixed and incubated at 37°C for 45 min to allow the formation of dimers. The preincubated protein mixture was then tested for DNA binding by an EMSA. The binding reactions and electrophoresis were done at 4°C to detect the binding of not only heterodimers but also the less stable homodimers. Figure 6A shows the results of one such assay. The addition of c-Jun, c-Fos, or supJun alone to the labeled AP-1 oligonucleotide resulted in the retardation of a specific band corresponding to c-Jun/c-Jun homodimers (lane 2, band 1) and supJun/supJun homodimers (lane 4, band 5) bound to DNA. These complexes are AP-1 specific, since incubation with an unlabeled AP-1 oligonucleotide abolished their binding (lanes 3, 5, and 11). The major complex migrating in the middle of the gel (labeled e) probably represents endogenous AP-1-like activity in the reticulocyte lysate (lane 1), since it was competed with an excess of unlabeled AP-1 oligonucleotide (lanes 3, 5, and 11). c-Fos alone was unable to bind DNA (lane 6). The addition of c-Jun and c-Fos, supJun and c-Jun, or supJun and c-Fos to the labeled AP-1 oligonucleotide resulted in the retardation of specific complexes corresponding to c-Jun/c-Fos heterodimers (lane 7, band 2), supJun/c-Jun heterodimers (lane 8, band 3), and supJun/c-Fos heterodimers (lane 9, band 4) bound to DNA. As seen in lanes 8 and 9, the supJun protein competed with c-Jun and c-Fos for binding to DNA on AP-1 sites either as supJun homodimers or as c-Jun/supJun or c-Fos/supJun heterodimers. In addition, when c-Jun, c-Fos, and supJun were mixed, supJun/c-Fos heterodimers represent the major DNA-binding complex, and this preferred dimerization decreased both c-Jun/c-Fos heterodimer formation and DNA binding (lane 10). Therefore, the supJun protein can form dimers with itself or with c-Jun or c-Fos, and the resulting complexes can competitively inhibit wild-type c-Jun or c-Fos from binding to AP-1 sites.
FIG. 6.
(A) DNA-binding activity of in vitro-translated c-Jun, c-Fos, and supJun. Proteins were synthesized separately by using a reticulocyte lysate-based in vitro transcription-translation system and then mixed and incubated at 37°C for 45 min to allow dimerization. The DNA-binding activity was determined by an EMSA with 1 ng of a 32P-labeled oligonucleotide containing the AP-1-binding site (FSE2) and 4.5 μl of reticulocyte lysate mixture in the presence (+) (lanes 3, 5, and 11) or absence (−) (lanes 2, 4, and 6 to 10) of a 100-fold molar excess of unlabeled oligonucleotide. The positions of the shifted c-Jun-, c-Fos-, and supJun homo- and heterodimer-containing complexes are indicated by the numbers 1 to 5 (1, c-Jun/c-Jun; 2, c-Jun/c-Fos; 3, supJun/c-Jun; 4, supJun/c-Fos; 5, supJun/supJun). The complex labeled e is due to the reticulocyte lysate since it was seen when unprogrammed lysate was mixed with the oligonucleotide (lane 1). (B) Complex formation between oligonucleotides containing consensus AP-1 or mutant AP-1 sequences and nuclear proteins from CEF cultures. Nuclear extracts (5 μg) from CEF cultures infected with viruses carrying no insert (DS3) (lanes 1 and 4), v-rel (lanes 2 and 5), or c-relΔ (lane 3 and 6) were assayed 10 days after infection for AP-1-binding activity with 0.2 ng of a 32P-labeled AP-1 oligonucleotide (lanes 1 to 3) or mutant AP-1 oligonucleotide (lanes 4 to 6) (Santa Cruz). Bands A and B represent AP-1 DNA-binding activity in DS3- and Rel-infected CEF cultures, respectively. (C) Endogenous AP-1 DNA-binding activity in nuclear extracts from infected CEF cultures and lymphoid cells. Nuclear extracts (5 μg) from CEF cultures infected with viruses carrying no insert (DS3) (lanes 1 and 14), v-rel (lanes 2, 6, 8, and 15), c-relΔ (lane 3), or supjun-1 (lanes 4, 7, 16, and 18) or coinfected with v-rel and supjun-1 (v + sj) (lanes 5 and 17) were assayed 10 days after infection for AP-1-binding activity with 0.2 ng of a 32P-labeled AP-1 oligonucleotide (Santa Cruz). Lanes 1 to 8 and 14 to 18 show binding activity in nuclear extracts isolated from CEF cultures, whereas lanes 9 to 13 show activity originating from v-Rel-transformed C4-1 lymphoid cells superinfected with DS3 (lanes 9 and 12) or supjun-1 (lanes 10, 11, and 13) viruses (2.5 μg per lane). The specificity of AP-1 binding was determined by competition assays with a 100-fold molar excess of unlabeled oligonucleotide (+) or mutant oligonucleotide (m). The top bands, A and B (lane 8), are most probably formed by dimers of the Fos/Jun family. The lower bands, C and D, were detected in nuclear extracts from supJun-expressing cells (lanes 4, 5, and 10) and represent supJun heterodimers with Fos/Jun proteins and supJun homodimers, respectively.
To compare protein complexes formed by in vitro-translated c-Fos, c-Jun, and supJun with complexes formed with nuclear extracts from supjun-1-expressing CEF cultures, proteins in nuclear extracts were electrophoresed on the same gel adjacent to the in vitro-translated proteins (Fig. 6A, lane 12). A similar profile of protein complexes was observed between the in vitro-translated and the nuclear extract samples. The slower migration of the in vitro-translated proteins is most probably due to differences in posttranslational modifications.
Next we examined the AP-1 DNA-binding activity in nuclear extracts derived from CEF cultures infected with viruses containing no insert, expressing v-Rel, or expressing c-RelΔ (Fig. 6B). Nuclear extracts prepared from CEF cultures infected with DS3 generated two complexes (A and B), of which the top band (A) represents the major complex. Nuclear extracts from CEF cultures expressing v-Rel or c-RelΔ demonstrated greatly enhanced bandshifts (2.5- to 3-fold increase) with the same mobility observed in DS3-infected CEF cultures (compare lane 2 with lanes 3 and 4). These complexes do not bind to a mutant AP-1 oligonucleotide (lanes 6 to 8). In addition, competition experiments were performed with a consensus AP-1 or mutated AP-1 oligonucleotide as the competitor (Fig. 6C). In the presence of the mutated AP-1 oligonucleotide, the top bands (A and B) were clearly visible, even at a 100-fold molar excess (lane 8). In contrast, a 100-fold excess of the consensus AP-1 oligonucleotide completely abolished all complexes (lanes 6, 7, and 11). The slower-migrating complex (A) appears to contain c-Jun since it was sensitive to treatment with c-Jun antisera (Fig. 6C, lanes 1 and 14). Unfortunately, we were unable to identify any other AP-1 members in these complexes, since antiserum against chicken c-Fos is not available and none of the purchased cross-reactive Fra-2 antisera were able to supershift or inhibit specific protein-DNA complexes (data not shown). The treatment of extracts from v-Rel-transformed cells with anti-c-Jun serum resulted in a significant reduction of AP-1-binding activity and the formation of two supershifted DNA-protein complexes (lane 15). To determine the binding profile of supJun in vivo and whether it functions by interfering with the ability of endogenous AP-1 factors to bind to DNA, nuclear extracts from supjun-1-infected cells were examined. These extracts generated additional complexes, as shown in lanes 4, 5 and 10. Complexes designated C were detected mainly in nuclear extracts derived from CEFs (lanes 4 and 5). They most probably represent supJun heterodimers with Fos/Jun partners. Nuclear extracts from lymphoid cells (C4-1) superinfected with viruses expressing supjun-1 generated two additional fast-migrating bands (designated D [lane 10]) which correspond to supJun homodimers. It is not clear if these two bands represent different modifications of supJun dimers, but both forms supershifted upon addition of a panspecific Jun antiserum (lane 13). The observed differences in the nature of the supJun complexes in CEFs and lymphoid cells may be related to the level of supJun expression as well as to the differences in the level of AP-1 factors expressed in these two cell systems. Extracts from CEF cultures coinfected with v-rel- and supjun-1-expressing viruses consistently generated lower levels of the higher-molecular-weight complexes (designated A and B) when compared to CEF cultures infected with v-rel alone (compare lanes 2 and 5). This indicates that supJun, by forming heterodimers with Fos/Jun family members, competitively inhibits these factors from binding to AP-1 sites. In conclusion, v-Rel-overexpressing cells exhibited elevated levels of c-Jun as well as elevated levels of AP-1-binding activity. Moreover, the expression of the supJun transdominant mutant decreased AP-1-binding activity, probably by modulating c-jun expression (as shown in the previous section) and also by competing with wild-type Fos/Jun family members for binding to AP-1 sites.
SupJun inhibits v-Rel- and c-RelΔ-induced transformation.
Consistent with the results of others, our data demonstrate that supJun can function as a general inhibitor of AP-1 activity, as measured through DNA-binding and transactivation assays (70). Since elevated AP-1 activity in v-Rel-transformed cells indicates a possible involvement of these early response genes in the v-Rel-induced transformation process, the effect of supJun on v-Rel transformation was examined. To perform these studies, CEF cultures were infected with v-rel- and supjun-1-expressing viruses to determine if supJun could block the transformation of this cell type. Since transformed cells are known to display anchorage-independent growth, transformation efficiency was assayed by growth in agar suspension. Secondary cultures of CEFs were coinfected or sequentially infected with viruses carrying the rel oncogenes and supjun-1. Four days after the second infection, the cells were plated in soft agar and the expression of the oncoproteins (v-Rel or c-RelΔ) and supJun was confirmed by Western blot analysis as described in Materials and Methods (data not shown). No difference in the expression of v-Rel or c-RelΔ was observed in cells expressing supJun from that in cells expressing these oncoproteins alone (data not shown). While infection of CEF cultures with the DS3 vector alone failed to transform CEFs, the v-Rel- or c-RelΔ-containing viruses produced multiple colonies in soft agar. On average, 105 v-rel-infected CEFs seeded in soft agar produced 229 colonies and 105 c-relΔ-infected CEFs produced 201 colonies. The total numbers of colonies varied to some extent from experiment to experiment but were not substantially reduced if v-rel infection was followed by superinfection with DS3 of a different subgroup (B) or if sequential infection was done in the reverse order. However, coinfection of v-rel with supjun-1 or infection of v-rel followed by supjun-1 superinfection (or vice versa) demonstrated a substantial inhibitory effect on colony formation in soft agar (Table 1). A similar suppressive effect of supJun was detected in c-RelΔ transformation (Table 1). The suppressive effect of supJun expression on colony formation was measured by determining the relative transformation efficiency, which represents the transformation efficiency of Rel- and DS3-infected CEFs (defined as 100%). The results show that transformation induced by rel was efficiently suppressed by infection with supjun-1 when evaluated by colony-forming activity (Table 1).
TABLE 1.
Suppression of colony formation of Rel-transformed cells by supjun-1 infection
| Cells and viruses | No. of coloniesc | Relative transformation efficiencyd
|
|
|---|---|---|---|
| Mean | Range | ||
| CEFs | |||
| Sequential infection with:a | |||
| DS3 (A) then c-relΔ (B) | 196 | 100 | |
| supjun (A) then c-relΔ (B) | 34 | 17 | 8–27 |
| c-relΔ (A) then DS3 (B) | 207 | 100 | |
| c-relΔ (A) then supjun-1 (B) | 56 | 27 | 17–36 |
| DS3 (A) then v-rel (B) | 290 | 100 | |
| supjun-1 (A) then v-rel (B) | 75 | 25 | 10–40 |
| v-rel (A) then DS3 (B) | 168 | 100 | |
| v-rel (A) then supjun-1 (B) | 49 | 29 | 24–33 |
| Coinfectiona | |||
| DS3 (A) plus c-relΔ (B) | 219 | 100 | |
| supjun-1 (A) plus c-relΔ (B) | 49 | 22 | 15–29 |
| DS3 (B) plus c-relΔ (A) | 209 | 100 | |
| supjun-1 (B) plus c-relΔ (A) | 63 | 30 | 27–42 |
| DS3 (A) plus v-rel (B) | 307 | 100 | |
| supjun-1 (A) plus v-rel (B) | 77 | 25 | 10–40 |
| DS3 (B) plus v-rel (A) | 144 | 100 | |
| supjun-1 (B) plus v-rel (A) | 43 | 29 | 20–40 |
| Lymphoid cell linesb | |||
| C4-1, DS3 (B) | 681 | 100 | |
| C4-1, supjun-1 (B) | 136 | 20 | 7–25 |
| 160/2, DS3 (A) | 314 | 100 | |
| 160/2, supjun-1 (A) | 69 | 22 | 8–29 |
CEFs were infected with the primary virus and 4 days later were subcultured into fresh dishes and infected with the secondary virus. Alternatively, CEFs were coinfected with viruses of two different subgroups simultaneously. Virus subgroups are indicated in parentheses. Then 105 doubly infected CEFs were seeded in soft agar 4 days after the second infection, and colonies were scored 4 weeks later.
104 lymphoid cells (C4-1 or 160/2 cells) were seeded 10 days after superinfection with DS3 or supjun-1 into soft agar, and colonies were scored 2 weeks later.
Each number represents the mean number of colonies per 60-mm dish in two to six experiments.
The transformation efficiency of Rel/supjun-1 relative to Rel/DS3, which represents 100%.
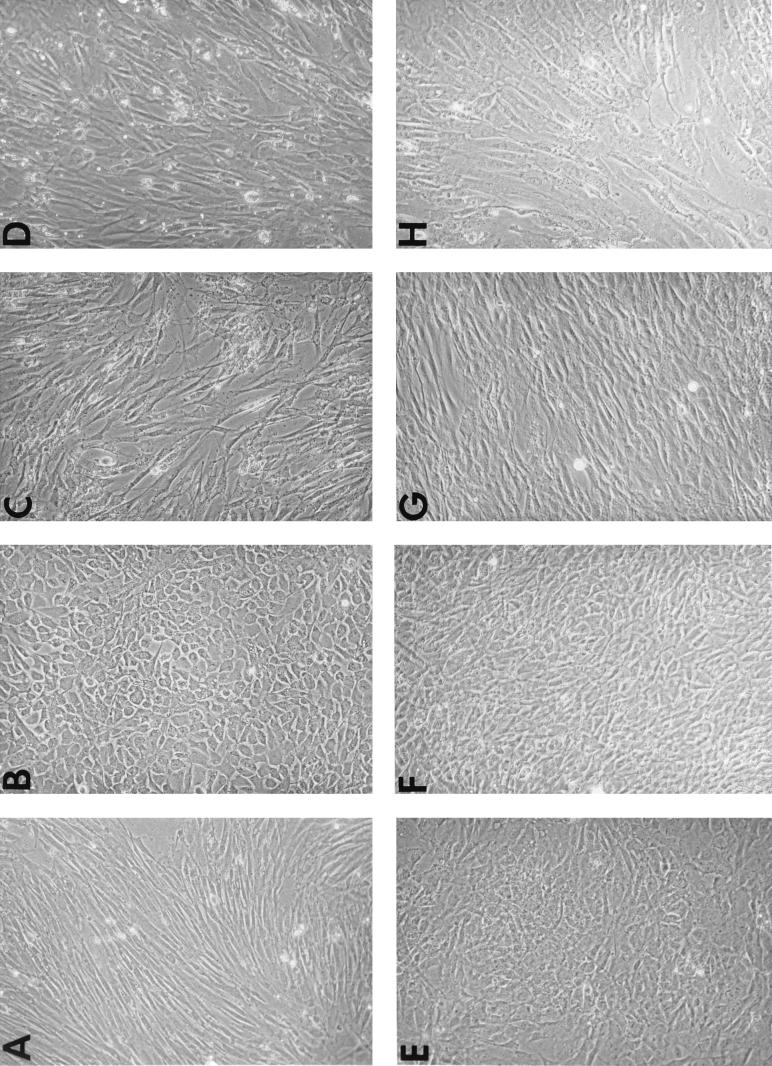
In addition, the suppressive effect of supJun was observed in the cellular morphology (Fig. 7). When DS3-infected CEF cultures were subsequently infected with v-rel or c-relΔ, the cells were clearly morphologically transformed, assuming a polygonal cellular morphology with a high saturation density (compare Fig. 7A with Fig. 7B and E). In contrast, CEF cultures infected with supjun-1 followed by v-rel or c-relΔ retained a morphology similar to that of cells infected with the vector alone (compare Fig. 7A with Fig. 7C and G). When the order of introduction of the rel genes and supjun-1 was reversed, the inhibitory effect of supJun on Rel transformation was not substantially changed (Fig. 7D and H).
FIG. 7.
Cellular morphology of sequentially infected CEF cultures. CEF cultures were infected with the virus from subgroup A first and superinfected 4 days later with the virus from subgroup B to deliver the second gene. (A) CEF cultures infected with the virus carrying no insert, DS3. (B and C) CEF cultures infected first with c-relΔ followed by DS3 (B) or supjun-1 (C). (F and G) CEF cultures infected first with v-rel followed by DS3 (F) or supjun-1 (G). When the order of introduction of the rel genes and supjun-1 or DS3 was reversed, the inhibitory effect of supJun expression on the Rel-induced transformation was not substantially changed: DS3 followed by v-rel resulted in a transformed phenotype (E), whereas infection of CEF cultures with the supjun-1 rendered them resistant to subsequent transformation by v-Rel (H) or c-RelΔ (D). Infected cells were passaged in vitro for 10 days after the second superinfection and then photographed at a magnification of ×200.
Since the natural targets of the v-rel oncogene are lymphoid cells, we also examined whether supJun can inhibit or reduce the colony-forming activity of v-rel-transformed lymphoid cells. Two lymphoid cell lines which were derived from REV-T-infected spleen cells were used in this study (C4-1 and 160/2 represent B and T cells, respectively). All cell lines examined expressed a high level of the v-Rel oncoprotein (data not shown). For the introduction of supjun, a highly concentrated virus stock was used and the efficiency of virus infection was determined by indirect immunofluorescence with a primary antibody to the p19gag protein. Seven days after infection, the majority of the cells (60 to 98%) showed strong expression of the gag protein whereas uninfected cells were completely negative. The expression of supJun was verified by Western blot analysis and is shown in Fig. 3C. The supjun-1 viral infection did not seem to impair the growth potential of the tested lymphoid cells when they were grown in mass culture at high density. However, when cells superinfected with supjun-1 were seeded in soft agar, a dramatic decrease in the number and size of colonies was observed compared to those for cells infected with control virus (DS3) (Table 1). Compared to control cells, the supjun-1-superinfected lymphoid cells fail to grow when plated out in a limiting-dilution assay (data not shown). The in vivo function of supJun in CEFs and lymphoid cells was clearly dependent on its level of expression. When a high level of expression was reached, a more pronounced inhibitory effect on soft agar colony formation was consistently observed. In conclusion, the results obtained from both CEF cultures and v-Rel-transformed lymphoid cells consistently demonstrated that the transdominant mutant supJun, through its inhibition of AP-1 activity, can also inhibit the oncogenic potential of v-Rel-transformed cells. This implies a direct involvement of AP-1 factors in the v-Rel transformation pathway.
DISCUSSION
The exact mechanisms by which oncogenes regulate the activity of their “downstream targets” are still not understood. One of the models developed to explain the mechanism by which v-Rel effects transformation of avian lymphoid cells is that v-Rel binds to DNA and directly alters the expression of cellular genes (30). The role that v-Rel plays in gene expression has been controversial. Some studies suggest that v-Rel may act as a dominant negative repressor of κB-dependent transcription (5, 40, 54, 62). The repressive function, however, is not sufficient for transformation by v-Rel. A C-terminal deletion mutant of v-Rel fully represses κB site-dependent transcription but fails to transform cells (64, 77). v-Rel also functions as a transcriptional activator. A few cellular genes are activated by v-Rel more efficiently than by c-Rel, and these genes might play an important role in v-Rel-induced transformation (10, 22, 37, 68, 76).
In this paper, we provide conclusive evidence that AP-1 factors are directly involved in the v-Rel transformation pathway. supJun functions as a general inhibitor of AP-1 activity in avian fibroblast and lymphoid cells. Coinfection or sequential infection with viruses expressing the v-rel oncogene and supjun-1 significantly reduces the ability of lymphoid cells to form colonies in soft agar and interferes with the ability of v-Rel to morphologically transform avian fibroblasts (Table 1; Fig. 7). Fibroblast cultures transformed by v-Rel revert to a normal phenotype after infection by a retroviral vector encoding supjun-1 (Fig. 7).
The major components of AP-1, c-Fos and c-Jun, belong to the family of the immediate-early genes whose transcription is highly, rapidly, and transiently activated upon stimulation by external stimuli (4, 47). c-Fos and c-Jun have been proposed to act as nuclear third-messenger molecules that function in coupling short-term signals to long-term adaptive changes in cell phenotype by regulating the expression of specific target genes. Moreover, c-Fos and c-Jun are metabolically unstable, being degraded rapidly with half-lives of about 10 and 60 min, respectively (47, 75). The instabilities of these proteins are presumably important for down regulation of the transactivation of target genes by AP-1, which appears to be essential for regulating cellular function or normal cell cycle progression. When continuously expressed at high levels, c-fos and c-jun transform cells (14, 55). Our results demonstrate that v-Rel induces the expression of c-fos and c-jun, suppresses fra-2 expression, and has no effect on junD expression. It is, however, unclear whether the elevated expression is due to increased mRNA stability or increased mRNA transcription. Other Rel proteins (c-Rel and c-RelΔ) in addition to v-Rel were evaluated for their regulatory effect on AP-1 components. The transforming ability of Rel proteins correlated with their ability to alter the expression of AP-1 factors. Moreover, our results indicate that the modulations of the expression of c-fos, c-jun, and fra-2 by v-Rel are not transient. The sustained elevated levels of c-fos and c-jun mRNA and protein as well as suppressed levels of fra-2 were observed at a number of time points in cells expressing v-Rel. It is assumed that the prolonged induction or repression of these AP-1 components results in long-term changes in the composition of AP-1 complexes. Because different members of the fos and jun families have distinct transcriptional activities and possibly sequence recognition properties, changes in the composition of the AP-1 complex are likely to affect the level and spectrum of genes under their control.
A strong differential effect of v-Rel on the expression of specific AP-1 members suggests that v-Rel is highly selective in the activation or repression of genes even within one family of transcription factors. However, the mechanism by which v-Rel modulates the transcription of different AP-1 members is not understood. Recently, it has been shown that v-Rel activates the promoter of c-jun. Two regions in the promoter are required: a κB site located between −87 and −76 and a region between −52 and +148 (22). Whether the expression of c-fos and fra-2 is regulated directly by activating or repressing promoters through specific sequences or by an indirect mechanism remains to be determined. Generally, changes in AP-1 activity in response to extracellular signals are regulated both at the level of transcription of the fos and jun genes and by posttranslational modifications of preexisting AP-1 factors (reviewed in references 31, 39, and 43). The observed elevated levels of c-fos and c-jun mRNA in v-Rel-transformed cells resulted in the increased synthesis of Fos-Jun dimers and increased AP-1 activity (Fig. 1, 2, and 6B). This increased AP-1 activity might further induce c-jun transcription through the AP-1 motif, known as the TPA response element, in its promoter (positive autoregulation). However, the expression of the transdominant mutant supJun in CEF cultures leads to a decreased expression of endogenous c-jun at both mRNA and protein levels (Fig. 3). It is most likely that supJun/c-Jun heterodimers, due to the absence of a transactivation domain in supJun, mediate less efficient autoregulation of the c-jun promoter, leading to a decreased expression of c-jun (quenching mechanism) (12). It would be interesting to determine the effect of supjun-1 on the expression of endogenous c-fos, but such an analysis is not feasible due to the barely detectable c-fos levels in exponentially growing cells. However, for fra-2, whose basal expression may be determined, similar experiments indicate that the level of Fra-2 and the extent of Fra-2 hyperphosphorylation were not affected by the expression of supJun (70).
The induction of transcription from the c-fos promoter is an early and complex event. Several cis elements mediate c-fos induction to a diverse spectrum of extracellular stimuli (15, 25, 36, 43, 73, 74). This results in increased synthesis of c-Fos, which, upon translocation to the nucleus, combines with preexisting Jun proteins to form AP-1 dimers that are more stable than those formed by the Jun protein alone. However, the induction of c-fos transcription is a transient event because of its rapid repression by its own gene product (negative autoregulation) (13, 46, 67, 79, 82). Decreased levels of Fos, in turn, reduce both AP-1 activity and c-jun transcription (31). The complex control of the fos promoter presumably allows tight regulation of fos transcription under different physiological conditions and in different cellular environments (31). In contrast, our results clearly show that v-Rel overexpression is accompanied by sustained elevated levels of c-fos mRNA and probably c-Fos protein (Fig. 1D). v-Rel, by an as yet unknown mechanism, interferes with the c-fos negative autoregulatory circuit leading to long-term c-fos upregulation. Deregulated expression of c-Jun and c-Fos, in turn, might lead to cell transformation. Therefore, it is likely that AP-1 components or at least their subsets are common target molecules which are regulated by the κB pathway and mediate abnormal cell proliferation. Interestingly, other oncogenes have also been shown to modulate AP-1 activity (4, 31, 70). In cells transformed by v-src, c-Ha-ras, and N-terminally truncated c-raf, the elevated levels of c-Jun and Fra-2 were observed while c-Fos was not detectable. Since our results show a different modulation pattern of the AP-1 family members, it is obvious that different oncogenes use different mechanisms to alter AP-1 activity. These may include, besides alterations in the expression of various jun and fos genes, posttranslational modifications of preexisting and newly synthesized AP-1 complexes. The activity of the AP-1 transcription factor is regulated by phosphorylation (reviewed in references 39 and 43). Phosphorylation by protein kinases can affect the interaction of the transcription factor transactivation domain with the transcriptional machinery (39). Further studies are needed to determine if the expression of v-Rel alters the phosphorylation status of different AP-1 factors, which might represent another important mechanism mediating v-Rel-induced transformation.
ACKNOWLEDGMENTS
We thank H. Iba for providing the pREP(A), pREP(B), pDS3, pfraRPA, and psupjun-1 plasmids and P. Vogt for providing avian c-jun cDNA, c-Jun antisera (USC-3, USC-4), and reporter plasmids −73/+63coll CAT and −60/+63coll CAT. In addition we are grateful to R. Hrdlickova for providing the lymphoid cell line DT95 and the v-Rel-transformed lymphoid cell line 160/2.
This work was supported by National Institutes of Health grant CA 33192, the National Cancer Institute, and the Texas Advanced Research Program. Part of this work was supported by a grant from the Academy of Sciences of the Czech Republic (A5052503) to J.K.
REFERENCES
- 1.Angel P, Baumann I, Stein B, Delius H, Rahmsdorf H J, Herrlich P. 12-O-tetradecanoyl-phorbol-13-acetate induction of the human collagenase gene is mediated by an inducible enhancer element located in the 5′-flanking region. Mol Cell Biol. 1987;7:2256–2266. doi: 10.1128/mcb.7.6.2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Angel P, Hattori K, Smeal T, Karin M. The jun proto-oncogene is positively autoregulated by its product, Jun/AP-1. Cell. 1988;55:875–885. doi: 10.1016/0092-8674(88)90143-2. [DOI] [PubMed] [Google Scholar]
- 3.Angel P, Imagawa M, Chiu R, Stein B, Imbra R J, Rahmsdorf H J, Jonat C, Herrlich P, Karin M. Phorbol ester-inducible genes contain a common cis element recognized by a TPA-modulated trans-acting factor. Cell. 1987;49:729–739. doi: 10.1016/0092-8674(87)90611-8. [DOI] [PubMed] [Google Scholar]
- 4.Angel P, Karin M. The role of Jun, Fos and the AP-1 complex in cell-proliferation and transformation. Biochim Biophys Acta. 1991;1072:129–157. doi: 10.1016/0304-419x(91)90011-9. [DOI] [PubMed] [Google Scholar]
- 5.Ballard D W, Walker W H, Doerre S, Sista P, Molitor J A, Dixon E P, Peffer N J, Hannink M, Greene W C. The v-rel oncogene encodes a κB enhancer binding protein that inhibits NF-κB function. Cell. 1990;63:803–814. doi: 10.1016/0092-8674(90)90146-6. [DOI] [PubMed] [Google Scholar]
- 6.Beg A A, Baldwin A S., Jr The IκB proteins: multifunctional regulators of Rel/NF-κB transcription factors. Genes Dev. 1993;7:2064–2070. doi: 10.1101/gad.7.11.2064. [DOI] [PubMed] [Google Scholar]
- 7.Beug H, Müller H, Grieser S, Doederlein G, Graf T. Hematopoietic cells transformed in vitro by REV-T avian reticuloendotheliosis virus express characteristics of very immature lymphoid cells. Virology. 1981;115:295–309. doi: 10.1016/0042-6822(81)90112-4. [DOI] [PubMed] [Google Scholar]
- 8.Blank V, Kourilsky P, Israel A. NF-κB and related proteins: Rel/dorsal homologies meet ankyrin-like repeats. Trends Biochem Sci. 1992;17:135–140. doi: 10.1016/0968-0004(92)90321-y. [DOI] [PubMed] [Google Scholar]
- 9.Boehmelt G, Madruga J, Dörfler P, Briegel K, Schwartz H, Enrietto P J, Zenke M. Dendritic cell progenitor is transformed by a conditional v-Rel estrogen receptor fusion protein v-RelER. Cell. 1995;80:341–352. doi: 10.1016/0092-8674(95)90417-4. [DOI] [PubMed] [Google Scholar]
- 10.Boehmelt G, Walker A, Kabrun N, Mellitzer G, Beug H, Zenke M, Enrietto P J. Hormone-regulated v-rel estrogen receptor fusion protein: reversible induction of cell transformation and cellular gene expression. EMBO J. 1992;11:4641–4652. doi: 10.1002/j.1460-2075.1992.tb05566.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bose H R., Jr The Rel family: models for transcriptional regulation and oncogenic transformation. Biochim Biophys Acta. 1992;114:1–17. doi: 10.1016/0304-419x(92)90002-g. [DOI] [PubMed] [Google Scholar]
- 12.Brown P H, Alani R, Preis L H, Szabo E, Birrer M J. Suppression of oncogene-induced transformation by a deletion mutant of c-jun. Oncogene. 1993;8:877–886. [PubMed] [Google Scholar]
- 13.Cahill M A. c-Fos transrepression revisited. FEBS Lett. 1997;400:9–10. doi: 10.1016/s0014-5793(96)01349-x. [DOI] [PubMed] [Google Scholar]
- 14.Castellazzi M, Dangy J-P, Mechta F, Hirai S-I, Yaniv M, Samarut J, Lassailly A, Brun G. Overexpression of avian or mouse c-jun in primary chick embryo fibroblasts confers a partially transformed phenotype. Oncogene. 1990;5:1541–1547. [PubMed] [Google Scholar]
- 15.Cavigelli M, Dolfi F, Claret F-X, Karin M. Induction of c-fos expression through JNK-mediated TCF/Elk-1 phosphorylation. EMBO J. 1995;14:5957–5964. doi: 10.1002/j.1460-2075.1995.tb00284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen C, Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheng Q, Cant C A, Moll T, Hofer-Warbinek R, Wagner E, Birnstiel M L, Bach F H, de Martin R. NF-κB subunit specific regulation of the IκBα promoter. J Biol Chem. 1994;269:13551–13557. [PubMed] [Google Scholar]
- 18.Chiao P J, Miyamoto S, Verma I M. Autoregulation of IκBα activity. Proc Natl Acad Sci USA. 1994;91:28–32. doi: 10.1073/pnas.91.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 20.Diehl J A, Hannink M. Heterologous C-terminal sequences disrupt transcriptional activation and oncogenesis by p 59v-rel. J Virol. 1993;67:7161–7171. doi: 10.1128/jvi.67.12.7161-7171.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Franklin R B, Kang C-Y, Wan K M-M, Bose H R., Jr Transformation of chick embryo fibroblasts by reticuloendotheliosis virus. Virology. 1977;83:313–321. doi: 10.1016/0042-6822(77)90176-3. [DOI] [PubMed] [Google Scholar]
- 22.Fujii M, Minamino T, Nomura M, Miyamoto K-I, Tanaka J, Seiki M. Selective activation of the proto-oncogene c-jun promoter by the transforming protein v-Rel. Oncogene. 1996;12:2193–2202. [PubMed] [Google Scholar]
- 23.Fujiwara K T, Ashida K, Nishina H, Iba H, Miyajima N, Nishizawa M, Kawai S. The chicken c-fos gene: cloning and nucleotide sequence analysis. J Virol. 1987;61:4012–4018. doi: 10.1128/jvi.61.12.4012-4018.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ghosh S, Gifford A M, Riviere L R, Tempst P, Nolan G P, Baltimore D. Cloning of the p50 DNA binding subunit of NF-κB: homology to rel and dorsal. Cell. 1990;62:1019–1029. doi: 10.1016/0092-8674(90)90276-k. [DOI] [PubMed] [Google Scholar]
- 25.Gille H, Kortenjann M, Strahl T, Shaw P E. Phosphorylation-dependent formation of a quaternary complex at the c-fos SRE. Mol Cell Biol. 1996;16:1094–1102. doi: 10.1128/mcb.16.3.1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gilmore T D. Malignant transformation by mutant Rel proteins. Trends Genet. 1991;7:318–322. doi: 10.1016/0168-9525(91)90421-l. [DOI] [PubMed] [Google Scholar]
- 27.Gilmore T D. Role of rel family genes in normal and malignant lymphoid cell growth. Cancer Surv. 1992;15:69–87. [PubMed] [Google Scholar]
- 28.Gilmore T D, Morin P J. The IκB proteins: members of a multifunctional family. Trends Genet. 1993;9:427–433. doi: 10.1016/0168-9525(93)90106-r. [DOI] [PubMed] [Google Scholar]
- 29.Gilmore T D, Temin H M. v-rel oncoproteins in the nucleus and in the cytoplasm transform chicken spleen cells. J Virol. 1988;62:703–714. doi: 10.1128/jvi.62.3.703-714.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gilmore T D, White D W, Sarkar S, Sif S. Malignant transformation of cells by the v-Rel oncoprotein. In: Cooper G M, Temin R G, Sugden B, editors. The DNA provirus: Howard Temin’s scientific legacy. Washington, D.C: American Society for Microbiology; 1995. pp. 109–128. [Google Scholar]
- 31.Gutman A, Wasylyk B. Nuclear targets for transcription regulation by oncogenes. Trends Genet. 1991;7:49–54. doi: 10.1016/0168-9525(91)90231-E. [DOI] [PubMed] [Google Scholar]
- 32.Hanning M, Temin H M. Transactivation of gene expression by nuclear and cytoplasmic rel proteins. Mol Cell Biol. 1989;9:4323–4336. doi: 10.1128/mcb.9.10.4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hartl M, Hutchins J T, Vogt P K. The chicken junD gene and its product. Oncogene. 1991;6:1623–1631. [PubMed] [Google Scholar]
- 34.Havarstein L S, Morgan I M, Wong W-Y, Vogt P K. Mutations in the Jun Delta region suggest an inverse correlation between transformation and transcription activation. Proc Natl Acad Sci USA. 1992;89:618–622. doi: 10.1073/pnas.89.2.618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Henkel T, Zabel U, van Zee K, Müller J M, Fanning E, Baeuerle P A. Intramolecular masking of the nuclear location signal and dimerization domain of the precursor for the p50 NF-κB subunit. Cell. 1992;68:1121–1133. doi: 10.1016/0092-8674(92)90083-o. [DOI] [PubMed] [Google Scholar]
- 36.Hill C S, Treisman R. Differential activation of c-fos promoter elements by serum, lysophosphatidic acid, G proteins and polypeptide growth factors. EMBO J. 1995;14:5037–5047. doi: 10.1002/j.1460-2075.1995.tb00186.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hrdlickova R, Nehyba J, Humphries E H. v-rel induces expression of three avian immunoregulatory surface receptors more efficiently than c-rel. J Virol. 1994;68:308–319. doi: 10.1128/jvi.68.1.308-319.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hrdlickova R, Nehyba J, Roy A, Humphries E H, Bose H R., Jr The relocalization of v-Rel from the nucleus to the cytoplasm coincides with induction of expression of Ikba and nfkb1 and stabilization of IκB-α. J Virol. 1995;69:403–413. doi: 10.1128/jvi.69.1.403-413.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hunter T, Karin M. The regulation of transcription by phosphorylation. Cell. 1992;70:375–387. doi: 10.1016/0092-8674(92)90162-6. [DOI] [PubMed] [Google Scholar]
- 40.Inoue J-I, Kerr L D, Ransone L J, Bengal E, Hunter T, Verma I M. c-rel activates but v-rel suppresses transcription from κB sites. Proc Natl Acad Sci USA. 1991;88:3715–3719. doi: 10.1073/pnas.88.9.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ito C Y, Kazantsev A G, Baldwin A S., Jr Three NF-κB sites in the IκBα promoter are required for induction of gene expression by TNFα. Nucleic Acids Res. 1994;22:3787–3792. doi: 10.1093/nar/22.18.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kabrun N, Hodgson J W, Doemer M, Mak G, Franza B R, Jr, Enrietto P J. Interaction of the v-rel protein with an NF-κB DNA binding site. Proc Natl Acad Sci USA. 1991;88:1783–1787. doi: 10.1073/pnas.88.5.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Karin M. The regulation of AP-1 activity by mitogen-activated protein kinases. J Biol Chem. 1995;270:16483–16486. doi: 10.1074/jbc.270.28.16483. [DOI] [PubMed] [Google Scholar]
- 44.Kerr L D, Inoue J-I, Davis N, Link E, Baeuerle P A, Bose H R, Jr, Verma I M. The Rel-associated pp40 protein prevents DNA binding of Rel and NF-κB: relationship with IκBβ and regulation by phosphorylation. Genes Dev. 1991;5:1464–1476. doi: 10.1101/gad.5.8.1464. [DOI] [PubMed] [Google Scholar]
- 45.Kieran M, Blank V, Logeat F, Vanderkerckhove J, Lottspeich F, Le Bail O, Urban M B, Kourilsky P, Baeuerle P A, Israel A. The DNA binding subunit of NF-κB is identical to factor KBF1 and homologous to the rel oncogene product. Cell. 1990;62:1007–1018. doi: 10.1016/0092-8674(90)90275-j. [DOI] [PubMed] [Google Scholar]
- 46.König H, Ponta H, Rahmsdorf U, Büscher M, Schönthal A, Rahmsdorf H J, Herrlich P. Autoregulation of fos: the dyad symmetry element as the major target of repression. EMBO J. 1989;8:2559–2566. doi: 10.1002/j.1460-2075.1989.tb08394.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kovary K, Bravo R. Expression of different Jun and Fos proteins during the G0-to-G1 transition in mouse fibroblasts: in vitro and in vivo associations. Mol Cell Biol. 1991;11:2451–2459. doi: 10.1128/mcb.11.5.2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kralova J, Schatzle J D, Bargmann W, Bose H R., Jr Transformation of avian fibroblast overexpression of c-rel proto-oncogene and a variant of c-rel lacking 40 C-terminal amino acids. J Virol. 1994;68:2073–2083. doi: 10.1128/jvi.68.4.2073-2083.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kralova J, Schatzle J D, Liss A S, Bargmann W, Bose H R., Jr Synergistic stimulation of avian IκBα transcription by rel and fos/jun factors. Oncogene. 1996;12:2595–2604. [PubMed] [Google Scholar]
- 50.Kumar S, Rabson A B, Gélinas C. The RxxRxRxxC motif conserved in all Rel/κB proteins is essential for the DNA binding activity and redox regulation of the v-Rel oncoprotein. Mol Cell Biol. 1992;12:3094–3106. doi: 10.1128/mcb.12.7.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Le Bail O, Schmidt-Ullrich R, Israel A. Promoter analysis of the gene encoding the IκBα/ MAD-3 inhibitor of NF-κB: positive regulation by members of the rel/NF-κB family. EMBO J. 1993;12:5043–5049. doi: 10.1002/j.1460-2075.1993.tb06197.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lewis R B, McClure J, Rup B, Niesel D W, Garry R F, Hoelzer J D, Nazerian K, Bose H R., Jr Avian reticuloendotheliosis virus: identification of the hematopoietic target cell for transformation. Cell. 1981;25:421–431. doi: 10.1016/0092-8674(81)90060-x. [DOI] [PubMed] [Google Scholar]
- 53.Lim M Y, Davis N, Zhang J, Bose H R., Jr The v-rel oncogene product is complexed with the cellular proteins including its proto-oncogene product and heat shock protein 70. Virology. 1990;175:149–160. doi: 10.1016/0042-6822(90)90195-w. [DOI] [PubMed] [Google Scholar]
- 54.McDonnel P C, Kumar S, Rabson A B, Gélinas C. Transcriptional activity of rel family proteins. Oncogene. 1992;7:163–170. [PubMed] [Google Scholar]
- 55.Miller A, Curran T, Verma I M. c-fos protein can induce cellular transformation: a novel mechanism of activation of a cellular oncogene. Cell. 1984;36:51–60. doi: 10.1016/0092-8674(84)90073-4. [DOI] [PubMed] [Google Scholar]
- 56.Moore B E, Bose H R., Jr Expression of the v-rel oncogene in reticuloendotheliosis virus-transformed fibroblasts. Virology. 1988;162:377–387. doi: 10.1016/0042-6822(88)90478-3. [DOI] [PubMed] [Google Scholar]
- 57.Morrison L E, Boehmelt G, Beug H, Enrietto P J. Expression of v-rel in a replication competent virus: transformation and biochemical characterization. Oncogene. 1991;6:1657–1666. [PubMed] [Google Scholar]
- 58.Morrison L E, Boehmelt G, Enrietto P J. Mutations in the rel-homology domain alter the biochemical properties of v-rel and render it transformation defective in chicken embryo fibroblasts. Oncogene. 1992;7:1137–1147. [PubMed] [Google Scholar]
- 59.Nishimura T, Vogt P K. The avian cellular homolog of the oncogene jun. Oncogene. 1988;3:659–663. [PubMed] [Google Scholar]
- 60.Nishina H, Sato H, Suzuki T, Sato M, Iba H. Isolation and characterization of fra-2, an additional member of the fos gene family. Proc Natl Acad Sci USA. 1990;87:3619–3623. doi: 10.1073/pnas.87.9.3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Okuno H, Suzuki T, Yoshida T, Hashimoto Y, Curran T, Iba H. Inhibition of jun transformation by a mutated fos gene: design of an anti-oncogene. Oncogene. 1991;6:1491–1497. [PubMed] [Google Scholar]
- 62.Richardson P M, Gilmore T G. v-Rel is an inactive member of the Rel family of transcriptional activating proteins. J Virol. 1991;65:3122–3130. doi: 10.1128/jvi.65.6.3122-3130.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sambrook J, Fritsch F F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 64.Sarkar S, Gilmore T D. Transformation by the v-Rel oncoprotein requires sequences carboxy-terminal to the Rel homology domain. Oncogene. 1993;8:2245–2252. [PubMed] [Google Scholar]
- 65.Schatzle J D, Kralova J, Bose H R., Jr Avian IκBα is transcriptionally induced by c-Rel and v-Rel with different kinetics. J Virol. 1995;69:5383–5390. doi: 10.1128/jvi.69.9.5383-5390.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schreiber S, Matthias P, Müller M M, Schaffner W. Rapid detection of octamer binding proteins with ‘mini-extracts’ prepared from a small number of cells. Nucleic Acids Res. 1989;17:6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shaw P E, Frasch S, Nordheim A. Repression of c-fos transcription is mediated through p 67SRF bound to the SRE. EMBO J. 1989;8:2567–2574. doi: 10.1002/j.1460-2075.1989.tb08395.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sif S, Capobianco A J, Gilmore T D. The v-Rel oncoprotein increases expression from Sp1 site-containing promoters in chicken embryo fibroblasts. Oncogene. 1993;8:2501–2509. [PubMed] [Google Scholar]
- 69.Stephens R M, Rice N R, Hiebsch R R, Bose H R, Jr, Gilden R V. Nucleotide sequence of v-rel: the oncogene of reticuloendotheliosis virus. Proc Natl Acad Sci USA. 1983;80:6229–6233. doi: 10.1073/pnas.80.20.6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Suzuki T, Murakami M, Onai N, Fukuda E, Hashimoto Y, Sonobe M H, Kameda T, Ichinose M, Miki K, Iba H. Analysis of AP-1 function in cellular transformation pathways. J Virol. 1994;68:3527–3535. doi: 10.1128/jvi.68.6.3527-3535.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Suzuki T, Okuno H, Yoshida T, Endo T, Nishina H, Iba H. Difference in transcriptional regulatory function between c-Fos and Fra-2. Nucleic Acids Res. 1991;19:5537–5542. doi: 10.1093/nar/19.20.5537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Traechner EB-M, Wilk S, Baeuerle P A. A proteasome inhibitor prevents activation of NF-κB and stabilizes a newly phosphorylation form of IκBα that is still bound to NF-κB. EMBO J. 1994;13:5433–5441. doi: 10.1002/j.1460-2075.1994.tb06878.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Treisman R. The serum response element. Trends Biochem Sci. 1992;17:423–426. doi: 10.1016/0968-0004(92)90013-y. [DOI] [PubMed] [Google Scholar]
- 74.Treisman R. Ternary complex factors: growth factor regulated transcriptional activators. Curr Opin Genet Dev. 1994;4:96–101. doi: 10.1016/0959-437x(94)90097-3. [DOI] [PubMed] [Google Scholar]
- 75.Tsurumi C, Ishida N, Tamura T, Kakizuka A, Nishida E, Okumura E, Kishimoto T, Inakagi M, Okazaki K, Sagata N, Ichihara A, Tanaka K. Degradation of c-Fos by the 26S proteasome is accelerated by c-Jun and multiple protein kinases. Mol Cell Biol. 1995;15:5682–5687. doi: 10.1128/mcb.15.10.5682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Walker A K, Enrietto P J. Analysis of the role of v-rel in transcriptional regulation of high mobility group protein 14. Oncogene. 1996;12:2515–2525. [PubMed] [Google Scholar]
- 77.Walker W H, Stein B, Ganchi P A, Hoffman J A, Kaufman P A, Ballard D W, Hannink M, Greene W C. The v-rel oncogene: insights into the mechanism of transcriptional activation, repression, and transformation. J Virol. 1992;66:5018–5029. doi: 10.1128/jvi.66.8.5018-5029.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wilhelmsen K C, Eggleton K, Temin H M. Nucleic acid sequence of the oncogene v-rel in reticuloendotheliosis virus strain T and its cellular homolog, the proto-oncogene c-rel. J Virol. 1984;52:172–182. doi: 10.1128/jvi.52.1.172-182.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wilson T, Treisman R. Fos C-terminal mutations block down-regulation of c-fos transcription following serum stimulation. EMBO J. 1988;7:4193–4201. doi: 10.1002/j.1460-2075.1988.tb03316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang J, Olson W, Ewert D, Bargmann W, Bose H R., Jr The v-rel oncogene of avian reticuloendotheliosis virus transforms immature and mature lymphoid cells in B cell lineage in vitro. Virology. 1991;183:457–466. doi: 10.1016/0042-6822(91)90975-h. [DOI] [PubMed] [Google Scholar]
- 81.Zhang J, Bose H R., Jr Acquisition of new proviral copies in avian lymphoid cells transformed by reticuloendotheliosis virus. J Virol. 1989;63:1107–1115. doi: 10.1128/jvi.63.3.1107-1115.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zinck R, Hipskind R A, Pingoud V, Nordheim A. c-fos transcriptional activation and repression correlate temporally with the phosphorylation status of TCF. EMBO J. 1993;12:2377–2387. doi: 10.1002/j.1460-2075.1993.tb05892.x. [DOI] [PMC free article] [PubMed] [Google Scholar]