Abstract
NAD(P)-dependent steroid dehydrogenase-like (NSDHL), an essential enzyme in human cholesterol synthesis and a regulator of epidermal growth factor receptor (EGFR) trafficking pathways, has attracted interest as a therapeutic target due to its crucial relevance to cholesterol-related diseases and carcinomas. However, the development of pharmacological agents for targeting NSDHL has been hindered by the absence of the atomic details of NSDHL. In this study, we reported two X-ray crystal structures of human NSDHL, which revealed a detailed description of the coenzyme-binding site and the unique conformational change upon the binding of a coenzyme. A structure-based virtual screening and biochemical evaluation were performed and identified a novel inhibitor for NSDHL harboring suppressive activity towards EGFR. In EGFR-driven human cancer cells, treatment with the potent NSDHL inhibitor enhanced the antitumor effect of an EGFR kinase inhibitor. Overall, these findings could serve as good platforms for the development of therapeutic agents against NSDHL-related diseases.
Electronic supplementary material
The online version of this article (10.1007/s00018-020-03490-2) contains supplementary material, which is available to authorized users.
Keywords: NSDHL, Cholesterol synthesis pathway, Membrane-anchored protein, Structure-based drug design, EGFR
Introduction
The cholesterol synthesis pathway is ubiquitous in animal cells and plays a pivotal role in producing cholesterol, which is not only a major component of all plasma membranes but also a precursor of steroid hormones [1]. The biosynthesis of cholesterol begins with acetylated coenzyme A, and cholesterol molecules are finally synthesized in more than 20 reactions, which are complicated and strictly regulated [2]. Dysregulation and imbalances in the cholesterol synthesis pathway have been considered major causes of human diseases, such as hypercholesterolemia, cardiovascular disease, cancer, and neuropathy, which lead to mortality and morbidity worldwide [3–5].
Several molecules that target enzymes involved in sterol biosynthesis have been developed. However, a majority of the agents are antifungal and target specific stages in the ergosterol synthesis pathway of fungi [6–9]. In addition, these agents are not considered as good medications due to their resistance and detrimental physiological side effects. Statins that inhibit 3-hydroxy-3-methylglutaryl-coenzyme A reductase (HMGCR) are the most successful drugs for treating hypercholesterolemia and cardiovascular diseases [2, 10]. Nevertheless, it has been reported that the prolonged use of statins is strongly associated with side effects [11]. Thus, the discovery of novel drugs that regulate the cholesterol synthesis pathway is needed to counter cholesterol-related diseases in a specific manner.
Enzymes, including HMGCR, that act upstream of the cholesterol synthesis pathway have been widely investigated, whereas many enzymes that participate in the downstream pathway from lanosterol to cholesterol have not been thoroughly investigated. NAD(P)-dependent steroid dehydrogenase-like (NSDHL, sterol-4-α-carboxylate 3-dehydrogenase, decarboxylating), which is an essential human enzyme in the downstream cholesterol synthesis pathway, catalyzes NAD+-dependent oxidative decarboxylation of the C4 methyl groups of 4α-carboxysterols [12, 13]. NSDHL is localized in the endoplasmic reticulum (ER) membrane, on which the enzymatic reaction occurs, and in lipid droplets, which are ER-derived cytoplasmic structures for storing lipids and cholesterols [14]. In addition to its association with cholesterol biosynthesis, this enzyme is also strongly related to cancer growth and the signaling of proto-oncogenes [4, 15, 16]. In particular, it has been reported that NSDHL regulates the expression and signaling of epidermal growth factor receptor (EGFR), and the loss of Nsdhl gene expression sensitizes cancer cells to EGFR-targeting inhibitors [17]. Moreover, the accumulation of sterol metabolites as a result of NSDHL deficiency has been shown to induce the suppression of tumor growth [4]. Despite the multiple important roles of NSDHL in cholesterol-associated diseases, its specific inhibitors have not yet been developed because of a lack of structural information on human NSDHL protein.
This study aimed to elucidate the crystal structures of NSDHL and identify a structure-based inhibitor of NSDHL for novel and effective treatment against cholesterol imbalance and cancers. We described two crystal structures of NSDHL on the atomic level and identified key information regarding the active site of the enzyme, which was used to guide the discovery of the inhibitors. The inhibitory profiles of 12 analogs were determined through a competitive inhibitor assay, and compound 9, which exhibited a half-maximal inhibitory concentration (IC50) of approximately 8 μM, was found to be the most effective inhibitor. Furthermore, we verified that compound 9 altered EGFR protein turnover and suppressed EGFR signaling cascades, thus leading to sensitization to erlotinib, an EGFR kinase inhibitor, in both erlotinib-sensitive and erlotinib-resistant cancer cell lines. Taken together, these findings reveal considerable insights into the structure of human NSDHL that can lead to develop potent and novel inhibitors reducing EGFR activity and thus provide novel therapeutic strategies against EGFR-driven cancers.
Materials and methods
Gene cloning, protein expression, and purification
The gene-encoding NSDHL from Homo sapiens was amplified by polymerase chain reaction (PCR) using a clone provided by Korea Human Gene Bank (Medical Genomics Research Center, KRIBB, Korea) (Clone ID: hMU006554) as a template. The PCR primers used for the NSDHL crystallization and mutation studies are shown in Supplementary Table S2. For the experiments, a truncated construct (31–267) of the full sequence (1–373) was employed. For the wild-type and two mutants (G205S and K232Δ) of NSDHL sequences (31–267), the applied procedures, from cloning to purification, were nearly identical. The PCR product and pET21a vector (Merck Millipore, Germany) were digested with both NdeI and XhoI (NEB, UK) and ligated. After verifying the DNA sequences, the recombinant protein was overexpressed in E. coli C41(DE3) (Sigma Aldrich, USA) and grown at 37 °C in Luria–Bertani (LB) broth. When the cells reached an OD600 of 0.5, 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) was added to induce protein expression. Then, the cells were transferred to a 15 °C incubator and grown for an additional 20 h. The cells were harvested by centrifugation at 4300×g for 10 min. The cell pellet was resuspended in lysis buffer (50 mM Tris, 500 mM NaCl, (pH 7.8), 5 mM imidazole, 0.5 mM TCEP, 10% (v/v) glycerol) and then sonicated at 4 °C. The lysate was centrifuged at 18,000×g for 60 min at 4 °C, and the supernatant was loaded into either an open Ni–NTA column (Qiagen, USA) or a HiTrap Chelating HP column (GE Healthcare) that was pre-equilibrated with lysis buffer. The column was washed with a 30-fold excess volume of buffer containing 30 mM imidazole. The protein was eluted at an imidazole concentration range of 100–250 mM. After concentrating the collected fractions, the buffer was changed to buffer A [40 mM HEPES, 150 mM NaCl, (pH 8.0), 0.5 mM TCEP, 5% (v/v) glycerol] by dialysis using an Amicon Ultra-15 Centrifugal Filter Unit with a 10 k molecular weight cut-off (Millipore, USA). The protein was further purified by gel filtration on a HiLoad 16/600 Superdex 200 prep‐grade column (GE Healthcare). Crystallization of coenzyme-free NSDHL (NSDHLapo) was carried out with a protein solution of 0.26 mM. To obtain the crystals of NAD(H)-bound NSDHL, 1 mM NAD+ or NADH was added to the final protein solution prior to crystallization. The NAD+-bound NSDHL crystal (NSDHLholo) was solely obtained in this study.
Crystallization and X-ray data collection
Crystallization was performed with manufactured screening kits using the sitting-drop vapor diffusion method. The best NAD+-bound NSDHL crystals were obtained in the presence of 1 mM NAD at 20 °C by mixing equal volumes (0.3 μl each) of the protein solution (at a concentration of 10 mg/ml in buffer A) and reservoir solution, which consisted of 0.1 M Tris–HCl (pH 8.0), 0.2 M calcium chloride, and 44% (v/v) PEG400. Cocrystallization with NADH did not generate crystals, whereas many crystal hits were observed when NAD+ was used for cocrystallization. The crystallization of NSDHLapo (NAD+-free NSDHL) was carried out in the absence of NAD+ using the sitting-drop vapor diffusion method at 20 °C by mixing equal volumes (0.5 μl each) of the protein solution (at a concentration of 10 mg/ml in buffer A) and reservoir solution, which consisted of 0.02 M glycine (pH 10.0), 0.5 M magnesium chloride, 0.02 M lithium chloride, and 33% (w/v) PEG1000. Prior to data collection, 20% (v/v) glycerol was added to each crystallization solution to protect the crystals from flash cooling by liquid nitrogen. Diffraction datasets for the NSDHLholo crystals were collected using an MAR300HE CCD detector at beamline BL44XU of SPring-8 (Japan). For the NSDHLapo crystals, the data were collected using a Pilatus 6 M detector at beamline 11C of Pohang Light Source (Republic of Korea). Both datasets were processed and scaled with the HKL-2000 software package [18].
Structure determination, refinement, and analysis
The crystal structure of NSDHLholo was determined using the molecular replacement method, which used a monomer model of GOX2253 from Gluconobacter oxydans as a search model (PDB code: 3WJ7) [19], in the Phaser-MR program within the Phenix software suite [20]. The structure of NSDHLapo was also determined by the molecular replacement method using a monomer of the NSDHLholo model as a template. In both models, the initial structures were further refined using Refmac [21] and Phenix.refine [20] software. The models were manually constructed, and ligands were added using the Coot software [22]. Five percent of the data were randomly set aside as the test data for the calculation of Rfree [23]. The stereochemistry of the final structures was evaluated using MolProbity software [24]. The structural deviations were calculated using the secondary structure-matching (SSM) superpose option in Coot software [22]. The solvent-accessible surface areas were calculated using PISA [25], and the protein–ligand interactions were calculated using LigPlot [26].
Isothermal titration calorimetry (ITC)
ITC analysis was carried out using a MicroCal iTC200 instrument (GE Healthcare). All experiments were performed at 25 °C under stirring at 1000 rpm in buffer A [40 mM HEPES, 150 mM NaCl, (pH 8.0), 0.5 mM TCEP, 5% (v/v) glycerol]. All titrations of NSDHL into coenzymes [NAD+, NADH, NADP+, and NADPH (Santa Cruz Biotechnology, USA)] were performed using an initial injection volume of 0.4 μl followed by 20 identical 2 μl injections with a 5 s delay time per injection and intervals of 150 s between the injections. The stock solutions of the coenzyme and NSDHL were separately diluted in buffer A with a compound concentration of 1 mM and a protein concentration of 0.1 mM prior to the titrations. The NSDHL proteins were titrated into buffer A to estimate the heat background generated by the buffer, and all data were calculated by subtracting the background. To ensure reproducibility, all titrations were performed in triplicate. A one-site fitting model was applied, and a nonlinear least squares algorithm was used to obtain the binding constant (Kd), change in enthalpy (ΔH), and stoichiometry (N). Thermodynamic parameters were subsequently calculated with the following formula: ΔG = ΔH − TΔS = − RT ln K, where ΔG, ΔH, ΔS, T, and R are the changes in free energy, enthalpy, entropy of binding, experimental temperature, and gas constant, respectively. All heat generation data were collected and processed using the MicroCal Origin 7.0 software package.
Size-exclusion chromatography with multiangle light scattering (SEC-MALS)
SEC-MALS experiments for NSDHLapo and NSDHLholo were performed using a fast protein liquid chromatography (FPLC) system (GE Healthcare) connected to a Wyatt MiniDAWN TREOS MALS instrument and a Wyatt Optilab rEX differential refractometer (Santa Barbara, CA, USA). A Superdex 200 10/300 GL (GE Healthcare) gel filtration column pre-equilibrated with a buffer (50 mM HEPES, 500 mM NaCl, 500 mM imidazole, pH 8.0) was normalized using the ovalbumin protein. Proteins (5 mg/ml) were injected at a flow rate of 0.4 ml/min. The data were analyzed using the Zimm model for fitting static light scattering data and were graphed using EASI graph with an ultraviolet (UV) peak in ASTRA 6 software (Wyatt).
Thermal shift assay (TSA)
Protein solutions of wild-type NSDHL (31–267) and its mutants (G205S and K232Δ) diluted in buffer A were added to each well of a MicroAmp 96-well reaction plate (Applied Biosystems) at a 5 µM concentration. The assay was initiated with a final volume of 25 μl after a 5 × solution of SYPRO Orange (Sigma Aldrich) was added to each well. The temperature was ramped from 25 to 95 °C at a rate of 1 °C/min using an Applied Biosystems 7500 Fast Real-Time PCR Instrument System (Thermo Fisher Scientific). Fluorescence data were plotted as a sigmoidal curve. Midpoint melting temperature (Tm) values were calculated using Boltzmann sigmoidal fitting in GraphPad Prism five (GraphPad software). The data for each curve were normalized to the maximum and minimum fluorescence signals.
High-throughput virtual screening
Docking-based virtual screening was performed using Schrödinger Suite 2017–4 with the X-ray crystal structure of NAD+-bound NSDHL (PDB code: 6JKH). Protein preparation was revised using Protein Preparation Wizard in Maestro v11.4 software (Schrödinger Release 2017–4: Maestro 11.4, Schrödinger, LLC, New York, NY, USA, 2017), and the receptor grid box was generated with a 30 Å × 30 Å × 30 Å cube centered on complexed NAD+. The 388,852 molecule-containing KCB library was browsed for ligand preparation using LigPrep v4.4 (Schrödinger Release 2017–4: LigPrep 4.4, Schrödinger, LLC, New York, NY, USA, 2017) and the OPLS_2005 force field [27], and during the process, tautomer and ionization states at pH 7.0 ± 2.0 were generated using the Epik v4.2 module (Schrödinger Release 2017–4: Epik 4.2, Schrödinger, LLC, New York, NY, USA, 2017). Ligand docking was performed using Glide v7.7 (Schrödinger Release 2017–4: Glide 7.7, Schrödinger, LLC, New York, NY, USA, 2017) and the standard precision (SP) method. The top 5000 compounds were selected according to the Glide score. Then, rescoring of the top-ranking SP pose of each compound was conducted with the extra precision (XP) Glide scoring function. Based on the top 2000 compounds combined with visual inspection, 495 compounds were selected for biochemical testing. Among the 495 compounds that underwent in vitro biochemical screening, 2 compounds were identified as new chemical inhibitors. To find more analogs of active compounds, we performed a second round of virtual screening based on the fingerprint similarity method. The similarity between single pairs of compounds was calculated using Pipeline Pilot 2017 (Pipeline Pilot 2017: Dassault Systèmes, San Diego, CA, USA, 2017) for comparisons of the degree of similarity with two molecules. The database of KCB library compounds was screened to find analogs with a Tanimoto coefficient > 0.8 for comparisons between 2 active compounds, and 162 compounds were selected by visual inspection. Among the 162 compounds, 11 compounds were finally selected as potent inhibitors by in vitro biochemical screening.
NADH-based competitive binding assay for identifying inhibitors
To identify active hits from the extensive chemical pool, we conducted a high-throughput assay. The fluorescence signal of NADH increases when it binds to proteins, such as dehydrogenases [28], therefore, this property was used for the assay.
The assay was performed in a total volume of 100 µl in 96-well flat-bottom black microplates (SPL Life Sciences). Two microliters of 100 µM library compounds were added to each well in duplicate. Forty-eight microliters of 32 µM NSDHL in assay buffer [50 mM HEPES (pH 8.0), 20% (v/v) glycerol] was added to each well. The plates were then incubated for 30 min at room temperature. The fluorescence intensity was measured using a SpectraMax M5 microplate reader (Molecular Devices) at Ex/Em = 340/460 nm at room temperature after adding 50 µl of 80 µM NADH solution in assay buffer. The inhibition (%) was calculated using the following equation: inhibition (%) = [1 − (F[chemical] /F[control])] × 100, where F[chemical] and F[control] are the fluorescence signals of the wells treated with the chemical and the dimethyl sulfoxide (DMSO) control, respectively. IC50 curves were fitted using the nonlinear regression function in GraphPad Prism5 (GraphPad software). The Ki values were converted from the IC50 and Kd values of NADH obtained from the ITC data. The data were generated using the Cheng-Prusoff equation for competitive inhibition.
Molecular docking
The docking of compound 9 was performed using Schrödinger Suite 2017–4 with the X-ray crystal structure of NAD+-bound NSDHL. Protein preparation was revised using Protein Preparation Wizard in Maestro v11.4 (Schrödinger Release 2017–4: Maestro 11.4, Schrödinger, LLC, New York, NY, USA, 2017), and the receptor grid box was generated with a 30 Å × 30 Å × 30 Å cube centered on complexed NAD+. The ligands were minimized using a Merck molecular force field (MMFF) with a dielectric constant of 80.0 in MacroModel v11.8 (Schrödinger Release 2017–4: MacroModel 11.8, Schrödinger, LLC, New York, NY, USA, 2017). Ligand docking was performed using Glide v7.7 software (Schrödinger Release 2017–4: Glide 7.7, Schrödinger, LLC, New York, NY, USA, 2017) with the SP method. The proposed binding models of the compounds were visualized using PyMOL v1.5 (Schrödinger, LLC, 2012) [29].
Surface plasmon resonance (SPR)
The direct binding affinity of NSDHL with compound 9 was investigated by SPR analysis. SPR measurements were performed at 25 °C using the Biacore T200 apparatus (GE Healthcare). For immobilization, the Amine Coupling Kit containing 0.1 M N-hydroxysuccinimide and 0.4 M 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride was applied to a CM5 sensor chip (GE Healthcare) with a buffer containing 10 mM HEPES, 200 mM NaCl, pH 8.0, and 0.005% (v/v) Tween20 according to the manufacturer’s protocol. Subsequently, 50 μg/mL NSDHL dissolved in 10 mM sodium phosphate at pH 7.4 was injected at a rate of 5 μl/min at regular intervals until the immobilization level reached approximately 8000 response units (RU). To deactivate the remaining activated carboxyl groups, 1.0 M ethanolamine at pH 8.5 was applied to the surface of the sensor chip for 400 s. The control experiment was treated as described above without protein, and the response of the control was subtracted from each sample dataset. Compound 9 at twofold diluted concentrations of 0.625–20.0 μM in PBS buffer containing 5% DMSO and 0.005% Tween 20 was injected over the NSDHL-immobilized chip at a rate of 30 μl/min for 120 s, followed by dissociation for 300 s in multicycle reactions. The sensor chip surface was regenerated for 10 s with 10 mM NaOH between cycles. The SPR response data were fitted to the simple bimolecular 1:1 Langmuir isotherm binding model to determine the equilibrium dissociation constant (KD) using Biacore T200 evaluation software 3.0 (GE Healthcare).
Cellular viability assay
A431 (epidermoid carcinoma cell line), A549 and H1299 (non-small-cell lung cancer cell lines), and K562 (myelogenous leukemia cell line) cells were obtained from the Korean Cell Line Bank. The A431 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM), and A549, H1299, and K562 cells were cultured in Roswell Park Memorial Institute (RPMI)-1640 medium, which were both supplemented with 10% (v/v) fetal bovine serum (FBS), l-glutamine, and antibiotics (penicillin and streptomycin). The cells were cultured at 37 °C in a humidified incubator with 5% CO2. To determine the cytotoxicity of the inhibitors, A431, A549, and H1299, and K562 cells were plated in 96-well flat-bottom plates at a concentration of 5 × 103 and 1 × 105 cells per well, respectively. After 24 h, the medium was removed, and compound 9 (0.16–500 μM) or the vehicle (DMSO) was added to each well. After an additional incubation for 72 h, the plates were treated with a stock solution of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazolium bromide (MTT) reagent, followed by additional incubation for 2 h. After the removal of the MTT solution, DMSO was added to the plates to dissolve the purple crystals. The absorbance was measured using a SpectraMax M5 multi-detection microplate reader (Molecular Devices, USA) at a wavelength of 570 nm.
The cytotoxicity assay of the NSDHL inhibitor with the EGFR-targeting drug erlotinib was performed in an erlotinib-sensitive cell line (A431) and erlotinib-resistant cell line (A549 and H1299). Then, each cell line was plated in 96-well cell culture plates at a concentration of 5 × 103 cells per well. After 24 h, A431 cells were treated with either DMSO (0.2%), erlotinib (0.5 μM), compound 9 (200 or 500 μM), or compound 9 and erlotinib (either 200 or 500 μM, and 0.5 μM, respectively). A549 and H1299 cells were subjected to the same treatment conditions except for 5 μM of erlotinib. All plates were incubated for 48 h after the treatment. Cell viability was measured using MTT reagent as described above, and the absorbance of the DMSO-treated groups was normalized to the average viability. We used the sensitization index (SI) [17] to determine whether our inhibitor enhances the cytotoxicity of the EGFR-targeting drug. The SI was calculated for each individual well on a 96-well plate as follows: SI = (XE/DE)/(XD/DD), where X and E are the viability of each well treated with compound 9 and erlotinib, respectively. D is the viability of cells treated with DMSO as a negative control to compound 9 or erlotinib. SI = 1 indicates no interaction, and SI < 1 indicates the enhanced cytotoxic effect. We verified that DMSO had no effect on the cell viability assay.
Flow cytometry
A431 cells were plated in 6-well plates at 50 ~ 60% confluency. Each well was treated with either compound 9 (100 μM) or the vehicle (the corresponding volume of DMSO) at 24 h after cell plating and was incubated for 72 h. After washing twice with phosphate-buffered saline (PBS), the cells were starved for an additional 24 h in serum-free DMEM at 37 °C. At the end of the incubation, the medium was changed to serum-free DMEM containing 500 ng/ml EGF, and the plates were then incubated for 8, 24 and 48 h. After washing the cells with PBS twice, the cells for all conditions were harvested simultaneously and then washed with fluorescence-activated cell sorting (FACS) buffer (1% FBS and 0.01% NaN3 in PBS) and stained with either phycoerythrin (PE)-conjugated anti-EGFR (Abcam, Cambridge, UK, ab130738) or its isotype control at 4 °C for 20 min in the dark. Fluorescence was measured using a FACSAria III flow cytometer (BD Biosciences), and the data were analyzed by FlowJo software (Tree Star). The EGFR level of each dataset was determined using geometric mean fluorescence intensity (gMFI), and the RFI was calculated using the following formula: gMFI of PE-EGFR/gMFI of the isotype control antibody. Three independent experiments were carried out to ensure reproducibility.
Immunoblotting
A431 cells were lysed with radioimmunoprecipitation assay buffer [50 mM Tris (pH 7.4), 150 mM NaCl, 1 mM EDTA, 1% NP-40, 0.25% sodium deoxycholate] supplemented with protease (Roche) and phosphatase inhibitors (Sigma). For immunoblotting, cell lysates were separated using 7% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose membranes. The membranes were blocked with 5% nonfat dry milk, incubated with primary antibodies overnight at 4 °C and subsequently reacted with horseradish peroxidase-conjugated secondary antibodies (GTX213110-01 and GTX213111-01, GeneTex) for 1 h at room temperature. Bands were visualized using enhanced chemiluminescence with a ChemiDoc™ MP imager (Bio-Rad).
Antibodies specific for the following factors were used for immunoblotting at the indicated dilutions: phospho-EGFR (Tyr1068) (1: 5000, #2234, Cell Signaling Technology), EGFR (1:5000, SC-03, Santa Cruz), phospho-AKT (Ser473) (1:2000, #4060, Cell Signaling), AKT (1:1000, #9272, Cell Signaling Technology), phospho-ERK1/2 (Thr202/Tyr204) (1:2000, #9101, Cell Signaling Technology), ERK1/2 (1:1000, #9102, Cell Signaling Technology) and β-actin (1:10,000, MAB1501R, Millipore). Original images of all blots are shown in Supplementary Fig. S12.
Public data analysis
Genomic alterations of Nsdhl in EGFR-driven human cancers in the TCGA PanCancer Atlas datasets were analyzed using cBioportal (https://www.cbioportal.org/) [30]. The analysis of overall survival (OS) of patients with lung cancer were performed using the Kaplan-Meier (KM) plotter online database (https://www.kmplot.com) [31]. Three public datasets of human lung cancers (GSE31210 [32], GSE30219 [33], and caArray [34]) were selected in the KM plotter, and the mean expression of two Nsdhl probes (209 279_s_at and 215093_at) was used in the analyses. To stratify the patients into two groups with high or low expression levels of Nsdhl, the cut-off value of Nsdhl expression was determined based on the computed best-performing thresholds.
Statistical analysis
Statistical analysis was performed using GraphPad Prism5 (GraphPad software). Statistically significant values were compared using unpaired two-tailed Student’s t test or one-way analysis of variance (ANOVA) with Bonferroni’s post hoc test. P < 0.05 was used to indicate a statistically significant difference, and the data are presented as the mean ± standard error of the mean (SEM).
Data availability
The coordinates and structural factors were deposited in the Research Collaboratory for Structural Bioinformatics (RCSB) PDB under the accession codes 6JKG for human NSDHLapo (NAD+-free form) and 6JKH for human NSDHLholo (NAD+-bound form).
Results
Overall structures of human NSDHL
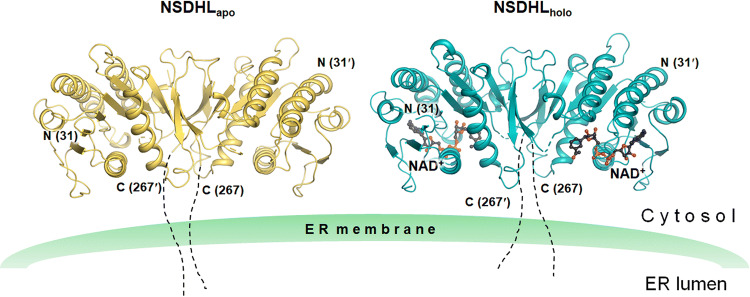
In this study, we obtained two crystal structures, including the single domain, from an extensive number of trials aimed at crystallizing various NSDHL constructs as well as the full-length construct including the transmembrane region. The two crystal structures consisted of a ligand-free form (NSDHLapo) and an NAD+-bound form (NSDHLholo) from the truncated construct (31–267) (Fig. 1).
Fig. 1.
Overall structures of the apo and holo forms of NSDHL. Two structures of NSDHL (31–267) are anchored in the ER membrane. NSDHLapo and NSDHLholo are shown as ribbon diagrams and colored in yellow and cyan, respectively. NAD+ of NSDHLholo (black carbon) is shown as a ball and stick, and disordered loops are shown as dashed lines. Truncated C-terminal residues are shown with dotted lines. The structures were constructed using PyMOL [29]
NSDHLapo and NSDHLholo were refined to Rwork/Rfree = 22.8%/29.1% at a 2.9 Å resolution and Rwork/Rfree = 17.3%/23.8% at a 3.0 Å resolution, respectively. NSDHLapo and NSDHLholo consist of 432 and 429 amino acid residues in 2 monomers (2 NAD+ molecules), respectively, and 3 and 18 water molecules in the asymmetric unit, respectively (Supplementary Table S1). The monomers of NSDHLapo and NSDHLholo comprise five α-helices and nine β-strands (Supplementary Fig. S1). In both models, we observed disordered regions, including (i) N-terminal residues [Lys31–Gln34 (chain A of NSDHLapo) and Lys31–Asn35 (chain B of NSDHLapo and chain A and B of NSDHLholo)], (ii) a loop between β4 and α3 [Pro109–Asn112 (chain B of NSDHLapo)], (iii) loops between β5 and α4 [Gly152–Lys167 (chain A and B of NSDHLapo), Val153–Met166 (chain A of NSDHLholo), and Asp154–Pro168 (chain B of NSDHLholo)], (iv) loops between β6 and β7 [Phe204–Asp208 (chain A and B of NSDHLapo) and Gly202–Pro209 (chain A and B of NSDHLholo)], and (v) a loop between β8 and α5 [Gly229–Lys232 (chain B of NSDHLapo)].
Two NSDHLapo (and NSDHLholo) monomers in the asymmetric unit were found to be highly similar to each other, with the root mean square (r.m.s.) deviation of 0.78 Å (0.67 Å for NSDHLholo) for 210 equivalent Cα (207 equivalent Cα for NSDHLholo) atom pairs. Two monomer structures of NSDHLapo were nearly identical to those of NSDHLholo, with r.m.s. deviations of 0.61–0.83 Å for 203–208 equivalent Cα pairs. Despite the structural similarities between NSDHLapo and NSDHLholo, greater r.m.s. deviations were observed in the following several regions: the loops between β2 and β3 and between β7 and β8. The loop between β2 and β3 showed a maximum deviation of 2.56 Å when two Cα in Asp74 were compared with each other, whereas the loop between β7 and β8 deviated with a maximum deviation of 2.79 Å when two Cα in Gly222 were compared with each other. A structural similarity search using the Dali server [35] revealed other structures that are similar to the monomer structures of NSDHLapo and NSDHLholo (Supplementary Fig. S2).
Diverse features of human NSDHL structures
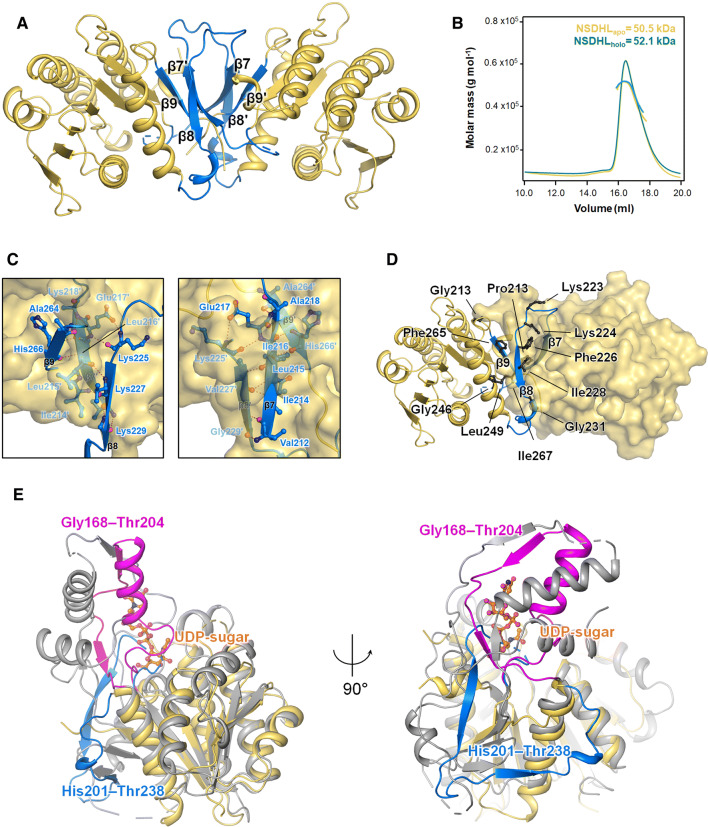
Although the full-length structure of NSDHL has two domains (a substrate-binding domain and a coenzyme-binding domain) along with a short transmembrane domain, the two structures of NSDHL (31–267) in this study had a single domain architecture consisting of an α/β doubly wound scaffold of the short-chain dehydrogenase/reductase (SDR) family fold [36]. The structures of NSDHL consisted of a seven-stranded parallel β-sheet, which was located between two arrays of two or three α-helices (α1 and α6 on one side and α2, α3, and α4 on the other side). The central β-sheet and the two groups of helices were mainly held together through hydrophobic interactions. The coenzyme-binding site was located within two βαβαβ Rossmann folds [37] of β1-α1-β2-(loop)-β3 and β4-α3-β5-α4-β6 (Supplementary Figure S1). The dimeric interfaces in NSDHLapo and NSDHLholo were composed of a pair of three-stranded antiparallel β-sheets (β8-β7ʹ-β9 and β8ʹ-β7-β9ʹ) (Fig. 2a). The interface areas of NSDHLapo and NSDHLholo per dimer were 4450 and 4050 Å2, respectively. The dimeric structures of the NSDHL crystals were consistence with the size-exclusion chromatography with multiangle light scattering (SEC-MALS) results, which showed the dimeric forms in solution (Fig. 2b). The residues involved in the hydrophilic and hydrophobic interactions at the interfaces of the NSDHL structures were nearly identical. The central β7 (and β7ʹ) had hydrophilic interactions with β8ʹ and β9ʹ (β8 and β9 for β7ʹ) of the other monomer (Fig. 2c), and the residues involved in the hydrophobic interactions of the dimeric interface are shown in Fig. 2d.
Fig. 2.
Distinct dimeric interface of the NSDHL structure. a Representation (blue) of the dimeric interface consisting of a pair of three-stranded antiparallel β-sheets (β8-β7ʹ-β9 and β8ʹ-β7-β9ʹ). b NSDHLapo (yellow line) and NSDHLholo (cyan line) were analyzed by SEC-MALS. The thick line indicates the measured molecular mass. c Detailed hydrophilic interactions are shown as red dotted lines, and the interacting residues are shown as a ball and stick with the corresponding β-strands of chain B. d Residues contributing to the hydrophobic interactions at the interface are shown as black balls and sticks. All residues of chain A and chain B are shown as a ribbon diagram and a surface view, respectively. e Superimposed views of NSDHL and B. thuringiensis Pen (PDB code 6BWC). The region (His201–Thr238, covering β7–β8) of NSDHL and the corresponding region (Gly168–Thr204) of Pen are shown in blue and magenta, respectively. The sugar substrate of Pen (orange carbon) is shown as a ball and stick. All figures are shown using NSDHLapo due to no major difference between NSDHLapo and NSDHLholo in this description
Despite substantial structural similarities between NSDHL and other homologs (Supplementary Fig. S2), strikingly different structural features of NSDHL were observed. Compared with the Gly168−Thr204 region of Bacillus thuringiensis Pen (Protein Data Bank (PDB) code: 6BWC) [38], which is composed of one α-helix and two short β-strands, the corresponding region (His201−Thr238) of NSDHL was found to have two long β-strands (β7 and β8) (Fig. 2e). In addition, while the Gly168–Thr204 region of Pen forms the binding pocket for its sugar substrate, the His201–Thr238 region of NSDHL is positioned within the coenzyme-binding region and forms a dimeric interface with C-terminal β9 (Fig. 2d).
NAD+-binding site in NSDHL
In the NAD+-binding site, 11 hydrogen bonds were found between the surface residues of NSDHL and NAD+, in addition to many other kinds of hydrophobic interactions (Fig. 3a, b). In the adenosine ribose moiety of NAD+, Asp84 and the backbone amides of Leu85 and Ile69 showed hydrogen bonds with the adenine ring, and the ring was stabilized by the hydrophobic interaction with Ile69 and Gly83 (Fig. 3a, b). A backbone amide of Ser106 formed a hydrogen bond with the oxygen of the adenine-ribose ring. Furthermore, the proximal residues Cys104 and Ala105 had hydrophobic interactions with the ribose ring (Fig. 3a, b). The bridging of the acidic Asp68 with the two hydroxyl groups of adenine-ribose implies that NSDHL may prefer NAD+ to NADP+ as a coenzyme (Fig. 3a). This finding is consistent with the general concept that a single coenzyme is chosen among more than two coenzymes involved in enzyme-catalyzed reactions. The glycine-rich motif GGxGxxG (Gly44–Gly50), which is the characteristic sequence of the complex SDR family [39], was observed in the NAD+-binding site (Fig. 3b). In addition, this motif was also found to accommodate the adenine-ribose and pyrophosphate moieties by hydrophilic (backbone amides of Phe48 and Leu49) and hydrophobic interactions (Gly44, Ser46, and Gly47) (Fig. 3a, b). In the nicotinamide-ribose moiety, the catalytic residues Tyr172 and Lys176, which are conserved as the YxxxK motif in the SDR family [39], formed hydrogen bonds with two hydroxyl groups of the ribose moiety (Fig. 3a). An additional hydrogen bond between a water molecule and a hydroxyl group was also observed in chain A of NSDHLholo (Fig. 3a). All moieties of NAD+ were clearly defined, except for the nicotinamide rings. The absence of a sterol substrate may have resulted in the poor observation of the nicotinamide rings in both monomers of NSDHLholo. This structural information on the interactions between NSDHL and its coenzyme served as the basis for the design of the inhibitors.
Fig. 3.
NAD+-binding site and the unique conformational change in NSDHL induced by the binding of NAD+. a, b Detailed views of the NAD+-binding site shown with hydrophilic and hydrophobic interactions. The hydrophilic interactions are shown as red dotted lines. The interacting residues and a water molecule are labeled and shown as orange sticks and a red sphere, respectively (a). The residues contributing to hydrophobic interactions are labeled and shown as pink sticks with the electrostatic potential surface view (b). NAD+ is shown with a 2mFo−DFc electron density map, which is contoured at 1.0 σ in blue mashes. c, d Unique open and closed forms observed in NSDHLholo and NSDHLapo, respectively. Both structures are shown as surface views, and two regions relevant to the conformational change are highlighted in magenta and blue, respectively, in the black rectangular boxes. In the detailed views (right) enlarged from the black rectangular boxes, the key residues are also shown as sticks in the transparent surface views. The sterol precursor in the expected substrate-binding site is shown as a yellow dashed model. The red-colored sterol precursor model indicates possible blockage of a substrate in the substrate-binding site. The key residues observed in NSDHLapo are shown with a 2mFo−DFc map of 1.0 σ in blue mashes (d)
Conformational change induced by the binding of NAD+ allows the binding of a sterol precursor to NSDHL
The structural analysis of NSDHLapo and NSDHLholo showed that the B-factors of the key residues in the NAD+-binding site compared with the overall B-factors were lower for NSDHLholo (36/47 Å2) than for NSDHLapo (85/85 Å2) (Supplementary Fig. S3). Similarly, the key residues and the overall structure of NSDHLholo showed a clearer electron density map than those of NSDHLapo, suggesting that the binding of NAD+ induces the rigid conformation of NSDHL. The structural comparison of NSDHLapo and NSDHLholo also revealed strikingly different conformations. In the structure of NSDHLholo, the region surrounded by the loops (His201–Leu211, where Gly202–Pro209 were unclear, and Pro109–Asn112), which is likely to constitute the substrate-binding site and the active site, had an open conformation (Fig. 3c). On the other hand, the loop (His201–Leu211) of NSDHLapo, where three additional residues (Gly202, Ile203, and Pro209) were newly observed, occupied the position of the nicotinamide ring located in the active site and obstructed the expected substrate-binding region, implying a closed conformation in the absence of NAD+ (Fig. 3d).
The structural importance of the loop (His201–Leu211) was also confirmed in TSA of two NSDHL mutants (G205S and K232Δ), which are the most common causes of congenital hemidysplasia with ichthyosiform erythroderma and limb defects (CHILD) and CK syndromes [40], respectively. G205S and K232Δ had a midpoint melting temperature (Tm) of 39.1 and 37.8 °C, respectively, whereas wild-type NSDHL had a Tm of 49.6 °C, indicating that NSDHL mutants have unstable folding and are degraded near the physiological temperature (Supplementary Fig. S4). Interestingly, Gly205, which constitutes the loop (His201–Leu211), and Lys232′, which is present in the other monomer of the dimeric interface, were observed near the loop (Supplementary Fig. S5). Taken together, the conformational change between two NSDHL structures may facilitate the uptake of a sterol substrate and may suggest that the catalytic reaction of NSDHL is the result of a precisely ordered and sequential mechanism [41]. Moreover, we identified that the loop (His201–Leu211) plays a pivotal role in the conformational change and stability for binding its substrate and coenzyme.
Human NSDHL favorably employs NAD(H) as its coenzyme in the cholesterol synthesis pathway
An in silico prediction of the coenzyme specificity of NSDHL has been reported [42]. As mentioned above, the major factor influencing coenzyme specificity is the presence of the acidic residue Asp68, which is conserved in the NAD(H)-dependent SDR family [39]. To better understand the coenzyme-binding mechanism of NSDHL, we explored the preference of coenzymes by analyzing their binding affinity to NSDHL.
In ITC experiment for the quantification of the binding affinity, the titration curves of NAD+ or NADH to NSDHL were fitted well to the single-site binding model. Interestingly, NADH (Kd = 21.4 ± 1.1 μM) showed a sevenfold greater binding affinity to NSDHL than NAD+ (Kd = 151.5 ± 70.2 μM) (Supplementary Fig. S6a, b). However, the NADP+ and NADPH titrations did not show any significant differences (Supplementary Fig. S6c, d), indicating weak binding affinity beyond the limit (high concentration). We concluded that NSDHL employs NAD+ as an electron acceptor for decarboxylation of the C4-methyl sterol in the cholesterol synthesis pathway.
Development of NSDHL inhibitors based on the structural information
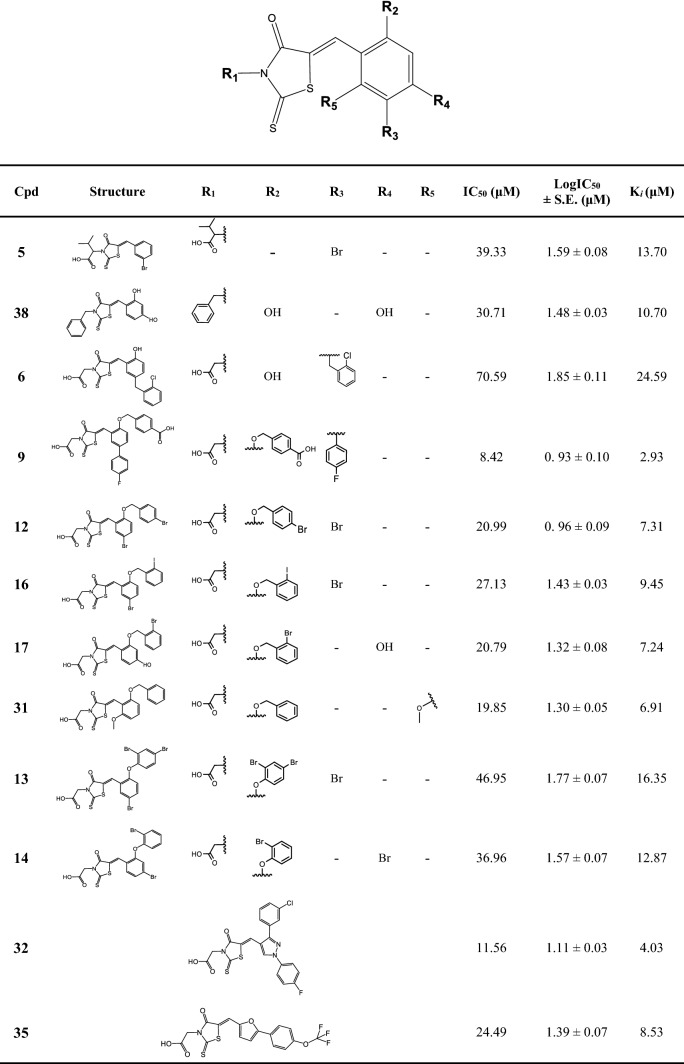
Detailed information on the overall structures and their coenzyme-binding site provided the basis for the design of the NSDHL inhibitors. We selected 495 putative compounds from chemical libraries of the Korea Chemical Bank (KCB), which consists of 388,852 molecules, by a structure-based virtual screening approach. Two compounds (1 and 5) (Supplementary Fig. S7) in the initial set of 495 compounds were identified to moderately inhibit NSDHL by the NADH-based competitive binding assay. Between these two compounds, compound 5 showed more potent inhibitory activity, with IC50 and Ki values of 39.33 μM and 13.70 μM, respectively (Supplementary Fig. S7 and Table 1).
To obtain a potent NSDHL inhibitor, we further conducted a fingerprint similarity search and selected 162 compounds. The NADH-based competitive binding assay indicated that 11 analogs (compounds 6 − 38), which were derived from compound 5, showed various inhibitory profiles (Table 1 and Supplementary Fig. S8). To comprehend the chemical structure of each biochemical property, structure–activity relationship (SAR) analysis was performed. The main scaffold [(Z)-5-benzylidene-2-thioxothiazolidin-4-one], and structures and inhibitory potentials of all the 12 obtained analogs are shown in Table 1. In this study, compound 9 was identified as the most potent analog, with an IC50 of 8.42 μM and a Ki of 2.93 μM, out of all the selected derivatives (Table 1 and Fig. 4a). The inhibitory activity of compound 9 was fivefold greater than that of the parent compound 5.
Table 1.
Chemical structures and inhibitory potentials of NSDHL inhibitors.
*Data represent the mean of two or three independent experiments. S.E. indicates standard error of LogIC50. Cpd 32 and 35 belong to the other chemical scaffold although they possess the identical 2-thioxothiazolidin-4-one moiety.
Fig. 4.
The most potent inhibitor of NSDHL and the proposed docking model. a Catalytic mechanism of NSDHL employing NAD+ as a coenzyme in the cholesterol synthesis pathway, and the chemical structure and inhibition curve of compound 9. The inhibition of NSDHL by compound 9 is shown as a T line. The data points in the curve are presented as the average value of three independent experiments and the error bars indicate the SEM. b The overall view of a docking model of compound 9 based on the NSDHLholo structure. NSDHL and compound 9 are shown as a surface view (white) and a ball and stick (pale-yellow carbon), respectively. c A detailed view showing the proposed binding mode of compound 9. The hydrophilic interactions are shown as red dotted lines, and the key residues are shown as blue sticks. The residues contributing to hydrophobic interactions are shown as salmon-colored sticks. R1, R2, and R3, indicating key substitution sites, are shown as dashed line circles (yellow, green, and red, respectively). d SPR analyses for measurement of the binding affinity between NSDHL and compound 9. An SPR sensorgram shows the direct binding of with increasing concentrations of compound 9 (0.625, 1.25, 2.50, 5.00, 10.0, and 20.0 µM) to immobilized NSDHL. The response data and the fitted data are presented as lines and dotted lines, respectively. The calculated Kon, Koff, and KD values are also shown on the right side
Except for compounds 5 and 38, all analogs have -CH2COOH groups on R1. More hydrophobic substitutions on R1, such as -C(CH3)2COOH of compound 5 and the benzyl group of compound 38, may have resulted in no significant change in the activity. In most analogs, the substitutions on R2, R3, R4, and R5 with small groups, such as -OH, -Br, and -OCH3, rarely influenced inhibitory activity. However, the positioning of a phenylmethanol moiety on R2 showed increased inhibitory activity. In particular, the presence of a carboxyl group on the phenylmethanol moiety (compound 9) showed the strongest inhibitory potential. Furthermore, despite the structural similarity with the phenylmethanol moiety, a phenol moiety on R2 (compounds 13 and 14) showed a moderately lower inhibitory activity than the phenylmethanol moiety. In addition, when R3 was substituted with either 4-fluorobenzene (compound 9) or 1-chloro-2-methylbenzene (compound 6), a marked difference in the inhibitory potentials was observed. Compound 9, which was the most potent inhibitory analog in this study, showed eightfold stronger inhibitory activity than compound 6, which was the poorest inhibitory analog among the derivatives. However, substituting a benzene ring of the main scaffold with either a pyrazole (compound 32) or furan ring (compound 35) still showed a moderate effect on the inhibitory activity.
Proposed binding mode between compound 9 and NSDHL
To better understand the correlation between the structures and activities of the compounds, molecular docking of the most potent inhibitor, compound 9, was performed using Schrödinger Suite with the X-ray crystal structure of NSDHLholo. The docking model of compound 9 bound to NSDHL showed the predictive binding mode of compound 9 (Fig. 4b). Two polar interactions were predicted in binding between NSDHL and compound 9 as follows: (i) the backbone amide of Leu49 forming a hydrogen bond with the carboxyl oxygen of the -CH2COOH group positioned on R1 and (ii) the backbone amide of Leu84 forming a hydrogen bond with the carboxyl oxygen of phenylmethanol positioned on R2 (Fig. 4c). In addition, residues Gly46–Phe48 of the glycine-rich motif GGxGxxG, which is involved in the binding of nucleotides in the SDR family, were found to constitute the binding site cavity for the main core body of compound 9 with Cys104–Ser106 residues. The residues Phe67–Ile69, Gly83, and Asp84 were found to form the hydrophobic pocket for the phenylmethanol group on R2. Furthermore, Gln70 and Gly72 stabilized the hydrophobic fluorobenzene group on R3 of compound 9 (Fig. 4c).
We further validated that compound 9 directly binds to NSDHL. The sensorgram of SPR showed the binding affinity of compound 9 to NSDHL to be KD = 2.351 μM (Fig. 4d), verifying the direct binding of compound 9 to NSDHL. In addition, we confirmed that the binding of compound 9 to wild-type or mutant NSDHL was also associated with a significant increase in Tm, as determined by fluorescent thermal shift analysis (Supplementary Fig. S9). The temperature-induced unfolding of wild-type NSDHL was largely shifted by a mean of 4.7 ºC upon ligand binding. However, the G47S and C104T mutants that were designed to hinder the binding of compound 9 on the basis of the docking prediction (Fig. 4c) showed mean ∆Tm values of 0.6 and 2.0 ºC, respectively, much lower than that of the wild type (Supplementary Fig. S9). These results reveal that Gly47 and Cys104 affect the binding of compound 9 to NSDHL, which is consistent with the proposed binding mode of compound 9.
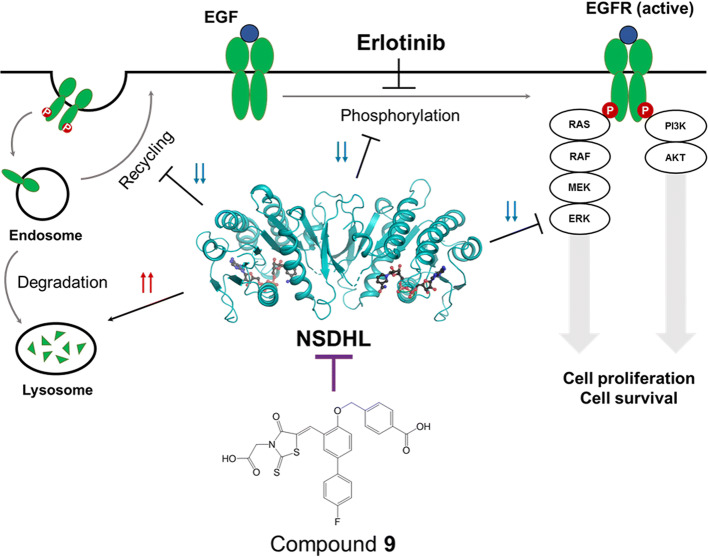
Therapeutic potential of NSDHL inhibition in EGFR-driven cancer
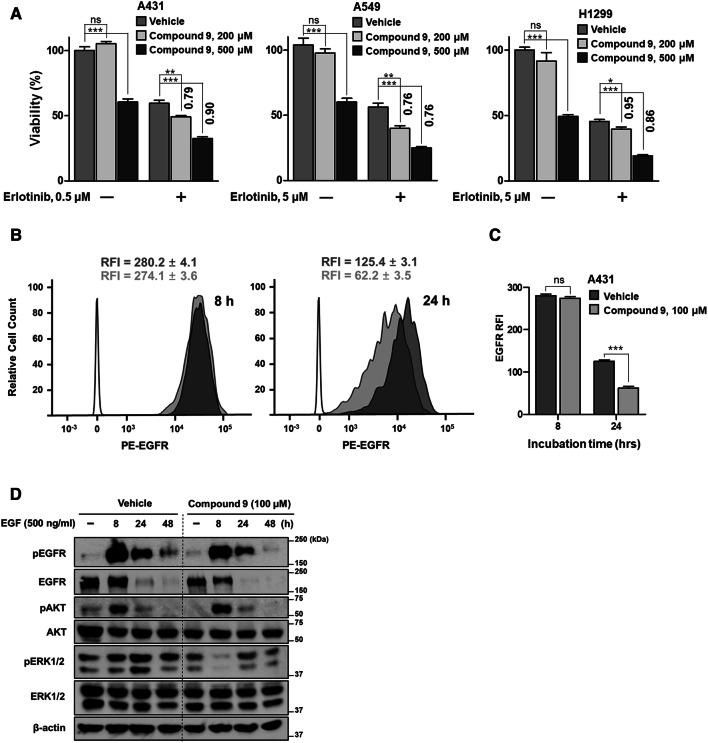
Drugs targeting the EGFR signaling pathway are strongly linked to the internalization and degradation of EGFR [43]. Considering that the downregulation of EGFR expression followed by depletion of Nsdhl leads to the sensitization of cancer cells to EGFR-targeting drugs [17], we postulated that our compound can inhibit EGFR activity and increase the antitumor efficacy of EGFR kinase inhibitors. By measuring the cytotoxicity of compound 9 in various epithelial cancer cells expressing EGFR with different response rates to erlotinib, which is an EGFR kinase inhibitor, we confirmed that compound 9 alone did not affect the viability of erlotinib-sensitive and erlotinib-resistant cell lines (Supplementary Fig. S10a) except at the highest concentration (500 μM). The compound also did not have a cytotoxic effect on three other mammalian cell lines, K562, HepG2, and Vero, except at very high concentrations (500 µM) (Supplementary Fig. S10b). Considering the weak interaction of nonoptimized compound 9, we used a high concentration (200 and 500 μM) of compound 9 to identify the biological effect despite the cytotoxic concentration of compound 9. Notably, compared to treatment with compound 9 alone, the combined treatment with erlotinib and compound 9 augmented the growth-inhibitory effect of erlotinib in both erlotinib-sensitive and erlotinib-resistant cell lines (sensitization index (SI) < 1) (Fig. 5a), indicating that despite the cytotoxic effect of the high concentration of compound 9, it could induce sensitization toward erlotinib in cancer cells harboring EGFR. To further elucidate the potential utility of NSDHL inhibitors in EGFR-driven human cancers, we additionally investigated the clinical relevance of NSDHL in these types of cancers. The Cancer Genome Atlas (TCGA) datasets showed a high frequency of genomic alterations in Nsdhl in various types of EGFR mutation- or amplification-harboring human cancers, including lung adenocarcinoma (n = 503), lung squamous cell carcinoma (n = 466), and head and neck squamous cell carcinoma (n = 488) (Supplementary Fig. S11a). In addition, high Nsdhl expression levels caused by its gene amplification are associated with poor overall survival in lung cancer patients (Supplementary Fig. S11b). No mutations occurred at the key residues of the NAD+-binding site in NSDHL in these cancers. Overall, this evidence indicates that the specific inhibition of NSDHL may represent a valuable approach for the treatment of EGFR-driven carcinomas.
Fig. 5.
Enhanced antitumor effect of erlotinib with the NSDHL inhibitor and suppression of EGFR activity by compound 9. a Enhanced growth inhibition of erlotinib with compound 9 on erlotinib-sensitive cell line (A431) and erlotinib-resistant cell lines (A549 and H1299) at 48 h. Numbers above the columns indicate the SI that shows relative increase in erlotinib cytotoxicity in SI < 1 (the equation is described in the Methods section). b Flow cytometry analysis of cell-surface EGFR expression in response to NSDHL inhibition. A431 cells pretreated with either the vehicle or compound 9 (100 µM) for 72 h were serum starved for 24 h and then stimulated with EGF (500 ng/ml) for the indicated times. The histograms show the amount of phycoerythrin (PE)-conjugated EGFR in the indicated group of cell lines. The empty peaks indicate the negative control (isotype control antibody), and the filled peaks indicate PE-conjugated EGFR in the vehicle-treated (dark gray) and compound 9-treated groups (light gray). c Comparison of the RFI between the vehicle-treated and compound 9-treated groups. d Lysates from the indicated cells that were prepared as mentioned above (b) were subjected to immunoblotting for the analysis of the NSDHL inhibition effect on the EGFR-dependent signaling pathway. All columns represent the average value of three or four independent experiments, and the error bars indicate the SEM (ns, not significant. *P < 0.05. **P < 0.01. ***P < 0.001; ANOVA with Bonferroni’s post hoc test and unpaired two-tailed Student’s t test)
The NSDHL inhibitor accelerates EGFR degradation to suppress EGFR-dependent signaling
The binding of a ligand [i.e., epidermal growth factor (EGF) or TGFα] to the cell-surface EGFR naturally induces its internalization, recycling, and degradation [44]. In this endocytosis pathway, NSDHL has been known to play a crucial role in the regulation of the endocytic trafficking of EGFR [17]. In addition, loss of NSDHL activity induces the acceleration of EGFR degradation pathway to lysosomes and rapidly terminates EGFR-dependent signaling [17]. Thus, we hypothesized that our inhibitor may suppress EGFR expression and its signaling pathway, which would subsequently enhance the antitumor effect of erlotinib. To investigate the effect of compound 9 on EGFR endocytic trafficking, we performed a flow cytometry analysis to evaluate the expression levels of cell-surface EGFR in response to treatment with compound 9 in A431 cells, which show an abnormally high-level expression of EGFR. No major shift was observed in the cytometric peak between compound 9 and the vehicle groups at 8 h after EGF treatment of A431 cells (Fig. 5b). At 24 h, a left shift of the peaks (Fig. 5b) and a reduction in the relative fluorescence intensity (RFI) (Fig. 5c) were observed in both groups, indicating a reduction in the EGFR expression level. Interestingly, compared with the vehicle-treated group, the compound 9-treated group showed substantially decreased fluorescence values (p = 0.0002) (Fig. 5c), suggesting that the rate of EGFR degradation in lysosomes was increased, while the rate of EGFR recycling to the cell surface was decreased by compound 9.
To further clarify the inhibitory effect of compound 9 on the EGFR signaling pathway, we performed immunoblotting for analysis of the activities of EGFR and its key downstream signaling effectors, such as protein kinase B (PKB, also known as AKT) and extracellular regulated kinases (ERKs). The EGF-induced phosphorylation levels of EGFR were more rapidly decreased by treatment with compound 9 than in the control. Furthermore, the total EGFR expression began to decrease after 8 h of EGF treatment in the compound 9-treated group, while prolonged EGFR expression was observed at that time of EGF stimulation in the control group (Fig. 5d). In the absence of EGF, the compound 9-treated group showed the suppressed phosphorylation of AKT, but comparable phosphorylation levels of AKT in the control group were observed at 8, 24, and 48 h of EGF stimulation (Fig. 5d). Interestingly, compound 9 markedly reduced the phosphorylation of ERK1/2 at 8 h of EGF stimulation. Then, ERK signaling recovered, but still showed a weaker signal in the compound 9-treated group than in the control group (Fig. 5d). These data indicated that compound 9 induces EGFR degradation and subsequently suppresses the EGFR/ERK signaling pathway. Based on these findings, we concluded that compound 9 may be useful for inhibiting EGFR signaling and improving the antitumor effect of EGFR-targeting drugs on cancer cells (Fig. 6).
Fig. 6.
Proposed mode of action of compound 9 in the regulation of EGFR signaling
Discussion
Contrary to the research interest in the cholesterol synthesis pathway, investigations of either the NSDHL structure or NSDHL inhibitors were limited. This study provides a foundation for the investigation of NSDHL inhibitors by clearly identifying the structures of NSDHL on the atomic level in both apo and holo forms. This study also identified a novel inhibitor, compound 9, which was the most potent inhibitor among the 12 analogs included in this study. The biological potential of compound 9 was investigated, and the results showed that it not only induces the suppression of EGFR expression and signaling, but also has the enhanced antitumor effect when it is combined with an EGFR kinase inhibitor.
The structural analysis of the two forms of NSDHL identified an early part of the ordered sequential mechanism of NSDHL. The transition from the closed form to the open form, which is induced by the binding of NAD+, suggests that NSDHL allows for the uptake of a sterol substrate via a unique structural intermediate. The loop (His201–Leu211), which plays an important role in this conformational change, may contribute to the formation of the active site cavity and regulate specific substrate recognition, which is similar with other SDR family [45]. The disorder of the loop region in our structures may be attributed to the absence of the substrate-binding domain, which was consistently reported in other studies [46]. When the thermal stability of the two NSDHL mutants (G205S and K232Δ), which cause CHILD and CK syndromes, respectively [40], was investigated, the Tm values of G205S and K232Δ were similar to physiological temperature and corresponded to previously reported Tm values in a study that demonstrated protein degradation of NSDHL-K232Δ at 37 °C [47]. These two mutation sites were closely related to the loop (His201–Leu211) in our structures, suggesting that a single mutation of Gly205 or Lys232 may influence the loop orientation (His201–Leu211), which may cause a substantial instability of the NSDHL structure.
Our study revealed that NSDHL strongly prefers NAD(H) over NADP(H). Although NSDHL is expected to employ NAD+ as a coenzyme for its catalytic reaction, NADH showed the strongest binding affinity to NSDHL in this study. We suggest that this unexpected affinity may have been attributed to the strict regulation of the cholesterol synthesis pathway to maintain a steady flow of sterol intermediates. After the catalytic reaction, NADH, which is a reactant of NAD+, may serve as a product inhibitor with its strong binding affinity and prevent the unnecessary depletion of a sterol precursor, such as isocitrate dehydrogenase [48, 49]. By revealing the unique conformation and reaction mechanism of NSDHL, this study provides a foundation for the development of inhibitors. Furthermore, the good fit of NAD+ within the coenzyme-binding pocket of NSDHL serves as a rationale for developing small-molecule inhibitors and as an attractive starting platform.
The inhibition of NSDHL and the consequent accumulation of C4-methyl sterol can cause severe X-linked genetic disorders, such as CHILD and CK syndromes [40]. Concerns have been raised on the therapeutic potential of targeting NSDHL for these phenotypes that are bound to be generated in the process of inhibiting NSDHL. Nevertheless, a previous study has demonstrated that a deficiency of Nsdhl is tolerated in adult animals, suggesting that the therapeutic exploitation of NSDHL could be a viable strategy in adults with minimal toxicity [4]. To the best of our knowledge, the natural compound FR171456 [50], which has been identified as an inhibitor of ERG26/NSDHL, is the only molecule that has been reported for the inhibition of NSDHL. In this study, a combined analysis that included NSDHL-NAD+ complex structure-based virtual screening and in vitro biochemical screening identified compound 9 as the most potent inhibitor among the 12 selected analogs of a novel scaffold that is different with the previous natural compound. The docking model based on the NSDHL-NAD+ complex structure suggested that a specific substitution on R1, R2, and R3 of the main scaffold leads to the potent inhibitory activity of compound 9. Considering that the -CH2COOH group on R1 was found to be located near the spacious hydrophilic pocket in the docking model, future studies may identify more potent inhibitors by optimizing and modifying R1. Identification of the direct binding affinity between NSDHL and compound 9 verified that compound 9 was rationally designed as a competitive inhibitor for the coenzyme-binding sites of NSDHL. Furthermore, despite solubility and stability problems of mutants selected near the R2 or R3 region, the thermal stability deviation of two mutants selected near the R1 region with compound 9 indicated that the proposed docking model of compound 9 is reliable.
One of the significant challenges in this study was the therapeutic availability of the NSDHL inhibitor as a novel anti-cancer drug in EGFR-driven human cancer. Consistent with the previous report showing the close connection between NSDHL and EGFR signaling [17], our findings demonstrated that NSDHL inhibition alone did not overcome EGFR-induced cancer cell growth while the combined inhibition with NSDHL and EGFR could promote the sensitivity of cancer cells to an EGFR-targeted drug. This sensitization effect induced by the high concentration of compound 9 in EGFR-driven cancer cells might be considered an off-target effect. However, the validation of the direct binding of compound 9 to NSDHL and the consistent tendency of the biological data induced by compound 9 comparable to the results by gene depletion of NSDHL [17] suggest that compound 9 can be a potential lead compound targeting EGFR-driven cancer for future study. Considering the clinical evidence on the frequent genomic alteration of Nsdhl in EGFR-driven cancers and a previous finding that EGFR/KRAS-dependent tumor growth was suppressed by the accumulation of a sterol intermediate [4], a level of NSDHL activity high enough to cause an imbalance in cholesterol synthesis might contribute to the development of EGFR-driven cancer by altering the activity of EGFR. Although further evaluations are required to support the link between NSDHL-mediated cholesterol synthesis and EGFR signaling in the progression of cancer, our findings indicated that NSDHL inhibition has an inhibitory effect against EGFR expression, activity, and downstream signaling, particularly the RAS/ERK pathway, and these results were consistent with observations in previous studies [4, 17]. These previous results and our data indicate that NSDHL inhibitors can be considered a reasonable effector when used with other anti-EGFR drugs that target a broad class of tumors with activated EGFR and RAS oncogenes.
In conclusion, this study unraveled the structures of NSDHL for the first time with the description of the unique conformational transition upon binding of a coenzyme and identified a novel inhibitor based on the elucidated structures. Furthermore, this study verified that the inhibition of NSDHL by compound 9 can suppress EGFR expression and EGFR-dependent signaling and induce sensitization to erlotinib. Therefore, our findings provide valuable insights into the therapeutic potential of targeting NSDHL and serves as a basis for developing therapeutic agents against NSDHL-mediated diseases.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We thank the beamline (BL) staff members at the Pohang Light Source, Korea (BL-5C and BL-11C), and SPring-8, Japan [beamline BL44XU under the approval of the Japan Synchrotron Radiation Research Institute (proposal No. 2017B6773)], for assistance with the X-ray diffraction experiments. All chemical libraries used in this study were kindly provided by the Korean Chemical Bank at the Korean Research Institute of Chemical Technology. This work was funded by Korea Ministry of Science, Information, Communication, Technology, and Future Planning and the National Research Foundation (NRF) of Korea Grants (NRF-2018R1A2A1A19018526 and NRF-2018R1A5A2024425 to B.-J.L.; NRF-2016R1C1B2014609, NRF-2018R1A6B4023605 and NRF-2019R1H1A1102102 to S.J.L.; and NRF-2017R1C1B2012225 to H.S.K.). This work was also supported by the 2018 BK21 Plus Project for Medicine, Dentistry, and Pharmacy and the National Cancer Center Grant of Korea (NCC-1811040; NCC-1910032; and NCC-1910023).
Abbreviations
- NAD
Nicotinamide adenine dinucleotide
- NADP
Nicotinamide adenine dinucleotide phosphate
- r.m.s.
Root mean square
- SEM
Standard error of mean
Author contributions
D-GK designed and performed most experiments, analyzed data, and wrote the paper. SC initiated the project and performed crystallization of NSDHLholo and mutation studies. K-YL and S-HC helped to perform in vitro competition assay and structural analysis. H-JY collected X-ray data of crystals and solved the structure of NSDHLholo. J-YL selected compounds for in vitro competition assay from chemical library and carried out molecular docking. DK and Y-KO offered facilities and reagents for cell experiments. K-SS and C-HK guided D-GK to perform and analyze the flow cytometry experiments. YC and HHL performed SEC-MALS. Y-SJ and S-JC performed and analyzed data on the kinetic solubility of compound 9. MB and K-YJ synthesize compound 9. HSK and HJL performed and analyzed TSA experiments. SJP and J-YL offered valuable scientific feedback in the paper. J-YL also supervised experiments in her fields. SJL and B-JL oversaw all aspects of this project.
Compliance with ethical standards
Conflict of interest
The authors declare no competing financial interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Sang Jae Lee, Email: sangjaelee@postech.ac.kr.
Bong-Jin Lee, Email: lbj@nmr.snu.ac.kr.
References
- 1.Hanukoglu I. Steroidogenic enzymes: structure, function, and role in regulation of steroid hormone biosynthesis. J Steroid Biochem Mol Biol. 1992;43(8):779–804. doi: 10.1016/0960-0760(92)90307-5. [DOI] [PubMed] [Google Scholar]
- 2.Acimovic J, Rozman D. Steroidal triterpenes of cholesterol synthesis. Molecules. 2013;18(4):4002–4017. doi: 10.3390/molecules18044002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sharpe LJ, Brown AJ. Controlling cholesterol synthesis beyond 3-hydroxy-3-methylglutaryl-CoA reductase (HMGCR) J Biol Chem. 2013;288(26):18707–18715. doi: 10.1074/jbc.R113.479808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gabitova L, Restifo D, Gorin A, Manocha K, Handorf E, Yang DH, Cai KQ, Klein-Szanto AJ, Cunningham D, Kratz LE, Herman GE, Golemis EA, Astsaturov I. Endogenous sterol metabolites regulate growth of EGFR/KRAS-dependent Tumors via LXR. Cell Rep. 2015;12(11):1927–1938. doi: 10.1016/j.celrep.2015.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ghosh S, Mukherjee S, Basu A. Chandipura virus perturbs cholesterol homeostasis leading to neuronal apoptosis. J Neurochem. 2015;135(2):368–380. doi: 10.1111/jnc.13208. [DOI] [PubMed] [Google Scholar]
- 6.Onyewu C, Blankenship JR, Del Poeta M, Heitman J. Ergosterol biosynthesis inhibitors become fungicidal when combined with calcineurin inhibitors against Candida albicans, Candida glabrata, and Candida krusei. Antimicrob Agents Chemother. 2003;47(3):956–964. doi: 10.1128/AAC.47.3.956-964.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barrett-Bee K, Dixon G. Ergosterol biosynthesis inhibition: a target for antifungal agents. Acta Biochim Pol. 1995;42(4):465–479. doi: 10.18388/abp.1995_4900. [DOI] [PubMed] [Google Scholar]
- 8.de Souza W, Rodrigues JC. Sterol biosynthesis pathway as target for anti-trypanosomatid drugs. Interdiscip Perspect Infect Dis. 2009;2009:642502. doi: 10.1155/2009/642502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roemer T, Boone C. Systems-level antimicrobial drug and drug synergy discovery. Nat Chem Biol. 2013;9(4):222–231. doi: 10.1038/nchembio.1205. [DOI] [PubMed] [Google Scholar]
- 10.Lim S, Oh PC, Sakuma I, Koh KK. How to balance cardiorenometabolic benefits and risks of statins. Atherosclerosis. 2014;235(2):644–648. doi: 10.1016/j.atherosclerosis.2014.06.001. [DOI] [PubMed] [Google Scholar]
- 11.Ramkumar S, Raghunath A, Raghunath S. Statin therapy: review of safety and potential side effects. Acta Cardiol Sin. 2016;32(6):631–639. doi: 10.6515/ACS20160611A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baudry K, Swain E, Rahier A, Germann M, Batta A, Rondet S, Mandala S, Henry K, Tint GS, Edlind T, Kurtz M, Nickels JT., Jr The effect of the erg26-1 mutation on the regulation of lipid metabolism in Saccharomyces cerevisiae. J Biol Chem. 2001;276(16):12702–12711. doi: 10.1074/jbc.M100274200. [DOI] [PubMed] [Google Scholar]
- 13.Mo C, Valachovic M, Randall SK, Nickels JT, Bard M. Protein-protein interactions among C-4 demethylation enzymes involved in yeast sterol biosynthesis. Proc Natl Acad Sci USA. 2002;99(15):9739–9744. doi: 10.1073/pnas.112202799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caldas H, Herman GE. NSDHL, an enzyme involved in cholesterol biosynthesis, traffics through the Golgi and accumulates on ER membranes and on the surface of lipid droplets. Hum Mol Genet. 2003;12(22):2981–2991. doi: 10.1093/hmg/ddg321. [DOI] [PubMed] [Google Scholar]
- 15.He M, Kratz LE, Michel JJ, Vallejo AN, Ferris L, Kelley RI, Hoover JJ, Jukic D, Gibson KM, Wolfe LA, Ramachandran D, Zwick ME, Vockley J. Mutations in the human SC4MOL gene encoding a methyl sterol oxidase cause psoriasiform dermatitis, microcephaly, and developmental delay. J Clin Invest. 2011;121(3):976–984. doi: 10.1172/JCI42650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu XY, Dangel AW, Kelley RI, Zhao W, Denny P, Botcherby M, Cattanach B, Peters J, Hunsicker PR, Mallon AM, Strivens MA, Bate R, Miller W, Rhodes M, Brown SD, Herman GE. The gene mutated in bare patches and striated mice encodes a novel 3beta-hydroxysteroid dehydrogenase. Nat Genet. 1999;22(2):182–187. doi: 10.1038/9700. [DOI] [PubMed] [Google Scholar]
- 17.Sukhanova A, Gorin A, Serebriiskii IG, Gabitova L, Zheng H, Restifo D, Egleston BL, Cunningham D, Bagnyukova T, Liu H, Nikonova A, Adams GP, Zhou Y, Yang DH, Mehra R, Burtness B, Cai KQ, Klein-Szanto A, Kratz LE, Kelley RI, Weiner LM, Herman GE, Golemis EA, Astsaturov I. Targeting C4-demethylating genes in the cholesterol pathway sensitizes cancer cells to EGF receptor inhibitors via increased EGF receptor degradation. Cancer Discov. 2013;3(1):96–111. doi: 10.1158/2159-8290.CD-12-0031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 19.Yin B, Cui DB, Zhang LJ, Jiang SQ, Machida S, Yuan YA, Wei DZ. Structural insights into substrate and coenzyme preference by SDR family protein Gox2253 from Gluconobater oxydans. Proteins. 2014;82(11):2925–2935. doi: 10.1002/prot.24603. [DOI] [PubMed] [Google Scholar]
- 20.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53(Pt 3):240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 22.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brunger AT. Free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature. 1992;355(6359):472–475. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]
- 24.Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 1):12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372(3):774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 26.Wallace AC, Laskowski RA, Thornton JM. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 1995;8(2):127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
- 27.Jorgensen WL, Maxwell DS, Tirado-Rives J. Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J Am Chem Soc. 1996;118(45):11225–11236. doi: 10.1021/ja9621760. [DOI] [Google Scholar]
- 28.Lakowicz JR, Szmacinski H, Nowaczyk K, Johnson ML. Fluorescence lifetime imaging of free and protein-bound NADH. Proc Natl Acad Sci USA. 1992;89(4):1271–1275. doi: 10.1073/pnas.89.4.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schrodinger LLC. The PyMOL molecular graphics system. Version. 2015;1:8. [Google Scholar]
- 30.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gyorffy B, Surowiak P, Budczies J, Lanczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS ONE. 2013;8(12):e82241. doi: 10.1371/journal.pone.0082241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Okayama H, Kohno T, Ishii Y, Shimada Y, Shiraishi K, Iwakawa R, Furuta K, Tsuta K, Shibata T, Yamamoto S, Watanabe S, Sakamoto H, Kumamoto K, Takenoshita S, Gotoh N, Mizuno H, Sarai A, Kawano S, Yamaguchi R, Miyano S, Yokota J. Identification of genes upregulated in ALK-positive and EGFR/KRAS/ALK-negative lung adenocarcinomas. Cancer Res. 2012;72(1):100–111. doi: 10.1158/0008-5472.CAN-11-1403. [DOI] [PubMed] [Google Scholar]
- 33.S Rousseaux A Debernardi B Jacquiau AL Vitte A Vesin H Nagy-Mignotte D Moro-Sibilot PY Brichon S Lantuejoul P Hainaut J Laffaire A Reynies de DG Beer JF Timsit C Brambilla E Brambilla S Khochbin 2013 Ectopic activation of germline and placental genes identifies aggressive metastasis-prone lung cancers Sci Transl Med 5(186):186ra166 Doi: 10.1126/scitranslmed.3005723 [DOI] [PMC free article] [PubMed]
- 34.Shedden K, Taylor JM, Enkemann SA, Tsao MS, Yeatman TJ, Gerald WL, Eschrich S, Jurisica I, Giordano TJ, Misek DE, Chang AC, Zhu CQ, Strumpf D, Hanash S, Shepherd FA, Ding K, Seymour L, Naoki K, Pennell N, Weir B, Verhaak R, Ladd-Acosta C, Golub T, Gruidl M, Sharma A, Szoke J, Zakowski M, Rusch V, Kris M, Viale A, Motoi N, Travis W, Conley B, Seshan VE, Meyerson M, Kuick R, Dobbin KK, Lively T, Jacobson JW, Beer DG, Director's Challenge Consortium for the Molecular Classification of Lung A Gene expression-based survival prediction in lung adenocarcinoma: a multi-site, blinded validation study. Nat Med. 2008;14(8):822–827. doi: 10.1038/nm.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Holm L, Rosenstrom P (2010) Dali server: conservation mapping in 3D. Nucleic Acids Res 38 (Web Server issue): W545–549. doi: 10.1093/nar/gkq366 [DOI] [PMC free article] [PubMed]
- 36.Orengo CA, Jones DT, Thornton JM. Protein superfamilies and domain superfolds. Nature. 1994;372(6507):631–634. doi: 10.1038/372631a0. [DOI] [PubMed] [Google Scholar]
- 37.Rao ST, Rossmann MG. Comparison of super-secondary structures in proteins. J Mol Biol. 1973;76(2):241–256. doi: 10.1016/0022-2836(73)90388-4. [DOI] [PubMed] [Google Scholar]
- 38.Delvaux NA, Thoden JB, Holden HM. Molecular architectures of Pen and Pal: Key enzymes required for CMP-pseudaminic acid biosynthesis in Bacillus thuringiensis. Protein Sci. 2018;27(3):738–749. doi: 10.1002/pro.3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kavanagh KL, Jornvall H, Persson B, Oppermann U. Medium- and short-chain dehydrogenase/reductase gene and protein families : the SDR superfamily: functional and structural diversity within a family of metabolic and regulatory enzymes. Cell Mol Life Sci. 2008;65(24):3895–3906. doi: 10.1007/s00018-008-8588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morimoto M, Souich C, Trinh J, McLarren KW, Boerkoel CF, Hendson G. Expression profile of NSDHL in human peripheral tissues. J Mol Histol. 2012;43(1):95–106. doi: 10.1007/s10735-011-9375-x. [DOI] [PubMed] [Google Scholar]
- 41.Cook PF, Cleland WW. Enzyme kinetics and mechanism. London: Garland Science; 2007. [Google Scholar]
- 42.Bhatia C, Oerum S, Bray J, Kavanagh KL, Shafqat N, Yue W, Oppermann U. Towards a systematic analysis of human short-chain dehydrogenases/reductases (SDR): Ligand identification and structure-activity relationships. Chem Biol Interact. 2015;234:114–125. doi: 10.1016/j.cbi.2014.12.013. [DOI] [PubMed] [Google Scholar]
- 43.Okada Y, Kimura T, Nakagawa T, Okamoto K, Fukuya A, Goji T, Fujimoto S, Sogabe M, Miyamoto H, Muguruma N, Tsuji Y, Okahisa T, Takayama T. EGFR downregulation after anti-EGFR therapy predicts the antitumor effect in colorectal cancer. Mol Cancer Res. 2017;15(10):1445–1454. doi: 10.1158/1541-7786.MCR-16-0383. [DOI] [PubMed] [Google Scholar]
- 44.Tomas A, Futter CE, Eden ER. EGF receptor trafficking: consequences for signaling and cancer. Trends Cell Biol. 2014;24(1):26–34. doi: 10.1016/j.tcb.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Perinbam K, Balaram H, Guru Row TN, Gopal B. Probing the influence of non-covalent contact networks identified by charge density analysis on the oxidoreductase BacC. Protein Eng Des Sel. 2017;30(3):265–272. doi: 10.1093/protein/gzx006. [DOI] [PubMed] [Google Scholar]
- 46.Yamamoto K, Kusunoki M, Urabe I, Tabata S, Osaki S. Crystallization and preliminary X-ray analysis of glucose dehydrogenase from Bacillus megaterium IWG3. Acta Crystallogr D Biol Crystallogr. 2000;56(Pt 11):1443–1445. doi: 10.1107/S090744490000994X. [DOI] [PubMed] [Google Scholar]
- 47.McLarren KW, Severson TM, du Souich C, Stockton DW, Kratz LE, Cunningham D, Hendson G, Morin RD, Wu D, Paul JE, An J, Nelson TN, Chou A, DeBarber AE, Merkens LS, Michaud JL, Waters PJ, Yin J, McGillivray B, Demos M, Rouleau GA, Grzeschik KH, Smith R, Tarpey PS, Shears D, Schwartz CE, Gecz J, Stratton MR, Arbour L, Hurlburt J, Van Allen MI, Herman GE, Zhao Y, Moore R, Kelley RI, Jones SJ, Steiner RD, Raymond FL, Marra MA, Boerkoel CF. Hypomorphic temperature-sensitive alleles of NSDHL cause CK syndrome. Am J Hum Genet. 2010;87(6):905–914. doi: 10.1016/j.ajhg.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cooper BF, Rudolph FB. Product inhibition applications. Methods Enzymol. 1995;249:188–211. doi: 10.1016/0076-6879(95)49036-1. [DOI] [PubMed] [Google Scholar]
- 49.Maeting I, Schmidt G, Sahm H, Stahmann KP. Role of a peroxisomal NADP-specific isocitrate dehydrogenase in the metabolism of the riboflavin overproducer Ashbya gossypii. J Mol Catalysis B. 2000;10(1):335–343. doi: 10.1016/S1381-1177(00)00135-1. [DOI] [Google Scholar]
- 50.Helliwell SB, Karkare S, Bergdoll M, Rahier A, Leighton-Davis JR, Fioretto C, Aust T, Filipuzzi I, Frederiksen M, Gounarides J, Hoepfner D, Hofmann A, Imbert PE, Jeker R, Knochenmuss R, Krastel P, Margerit A, Memmert K, Miault CV, Movva NR, Muller A, Naegeli HU, Oberer L, Prindle V, Riedl R, Schuierer S, Sexton JA, Tao J, Wagner T, Yin H, Zhang J, Roggo S, Reinker S, Parker CN. FR171456 is a specific inhibitor of mammalian NSDHL and yeast Erg26p. Nat Commun. 2015;6:8613. doi: 10.1038/ncomms9613. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The coordinates and structural factors were deposited in the Research Collaboratory for Structural Bioinformatics (RCSB) PDB under the accession codes 6JKG for human NSDHLapo (NAD+-free form) and 6JKH for human NSDHLholo (NAD+-bound form).