Abstract
In mammals, males and females show marked differences in immune responses. Males are globally more sensitive to infectious diseases, while females are more susceptible to systemic autoimmunity. X-chromosome inactivation (XCI), the epigenetic mechanism ensuring the silencing of one X in females, may participate in these sex biases. We perturbed the expression of the trigger of XCI, the noncoding RNA Xist, in female mice. This resulted in reactivation of genes on the inactive X, including members of the Toll-like receptor 7 (TLR7) signaling pathway, in monocyte/macrophages and dendritic and B cells. Consequently, female mice spontaneously developed inflammatory signs typical of lupus, including anti–nucleic acid autoantibodies, increased frequencies of age-associated and germinal center B cells, and expansion of monocyte/macrophages and dendritic cells. Mechanistically, TLR7 signaling is dysregulated in macrophages, leading to sustained expression of target genes upon stimulation. These findings provide a direct link between maintenance of XCI and female-biased autoimmune manifestations and highlight altered XCI as a cause of autoimmunity.
Perturbation of X-chromosome inactivation, a female-specific process, induces signs of lupus, a disease that mostly affects women.
INTRODUCTION
In mammals, there is a marked sexual dimorphism in immune responses and functions. This is partly explained by stronger innate and adaptive immunity in adult females, conferring a better response to various types of pathogens and vaccines compared to males. Such enhanced immunity in females may, however, lead to over-reactivity when not properly controlled, hence resulting in autoimmune manifestations. The determinants of this sexual bias are not fully understood. While these differences are partly attributable to sex hormones, X-linked factors represent other likely contributors (1–6). Men with Klinefelter’s syndrome, who bear an extra X chromosome (47, XXY) in a male hormonal context, have a risk equivalent to women to develop relatively rare immune disorders such as systemic lupus erythematosus (SLE) (7), Sjogren’s syndrome (8), or Systemic Sclerosis (9). Moreover, the female bias in autoimmune diseases such as SLE is observed before puberty (10). Incidentally, the X chromosome has a high density of genes involved in immune functions (11, 12), and some of these, including TLR7, TASL, CXCR3, or CD40LG, tend to be overexpressed in autoimmune conditions, suggesting a causal link between autoimmunity and X-linked gene regulation (2, 5, 13). A key role for Toll-like receptor 7 (TLR7), a single-stranded RNA (ssRNA) sensor essential for the defense against RNA viruses that can also be engaged by endogenous ligands, has been established in SLE pathogenesis (11). Expression of two copies of Tlr7 in male mice is sufficient to induce full-blown autoimmunity (14, 15). Recently, a genetic variant of human TLR7 (TLR7Y264H, gain of function) has been identified in a young girl with juvenile SLE. Mice carrying this Tlr7Y264H mutation spontaneously develop a lupus-like disease due to aberrant TLR7 signaling, which results in the accumulation of pathogenic unconventional T-bet+ CD11c+ memory B cells, also known as age-associated B cells (ABCs) (16). Hence, the link between Tlr7 overexpression and autoimmunity has been firmly established. However, how such Tlr7 up-regulation is initially triggered, whether perturbation of X-chromosome expression may be involved and, more generally, how broad alteration of X-linked gene expression would affect the fitness of the immune system is unknown.
X-linked gene expression is equalized between the sexes through the transcriptional silencing of most genes of one of the two X chromosomes, at random, in females. This X-chromosome inactivation (XCI) is an essential process established during early embryogenesis and maintained afterward in daughter cells throughout in utero and postnatal life. XCI is triggered by the accumulation of the Xist long noncoding RNA (lncRNA) on one of the two X chromosomes [the future inactive X (Xi)]. Xist then recruits a series of factors inducing gene silencing in cis (17). While, in most cell types, the repressed state is thought to be locked by several layers of chromatin modifications, XCI maintenance in immune cells exhibit certain specific features. First, a number of X-linked genes including TLR7, TASL, and CXCR3 tend to escape from XCI and are transcribed from the Xi in a substantial proportion of human immune cells in physiological conditions (3, 11, 18–20). Second, Xi hallmarks including Xist lncRNA accumulation and enrichment in repressive histone modifications (notably histone H3 lysine K27 tri-methylation H3K27me3 and histone H2A lysine K119 ubiquitination H2AK119ub) are almost completely lost during B and T lymphopoiesis and regained upon lymphocyte activation (21). These marks are either reduced or completely absent in natural killer cells, dendritic cells (DC), and macrophages (MΦ) (22). This tendency is exacerbated in B lymphocytes from patients with SLE (21) or in stimulated B cells from the lupus mouse model (NZB/W F1) (23, 24). In addition, in human, distinct sets of proteins interact with the XIST lncRNA in myeloid compared to lymphoid lineages, suggesting that XIST could mediate silencing using different mechanisms depending on the immune cell type (25). Third, knocking out (KO) Xist when hematopoietic cells are specified during development results in differentiation defects during hematopoiesis, up-regulation of natural XCI escapees (26), and aggressive blood cancer in adult mice (27). The same Xist KO has, however, relatively minor effects when induced in other, non-immune, cell types (28, 29). In human, repressing XIST in a B cell line results in increased expression of a subset of X-linked genes that tend to be overexpressed in ABCs of patients with SLE (25). Together, these findings suggest that XCI displays specific regulatory requirements in hematopoietic cells. However, whether such a unique XCI plasticity may lead to over-reactivity of female immune system when not properly controlled remains to be formerly addressed (13).
To characterize the impact of perturbed X-linked gene regulation on immune functions in vivo, we used the Ftx KO background (Ftx−/−) as a mean to mildly impair XCI while bypassing lethality associated with Xist deficiency. This mutation also mimics XCI alterations that are susceptible to occur in vivo. Ftx is a noncoding gene of the X-inactivation center (Fig. 1A), which acts as a cis-activator of Xist during the initiation of XCI (30, 31). Consequently, deletion of the Ftx promoter results in reduced Xist expression, impaired accumulation of Xist lncRNAs on the Xi, and incomplete X-linked silencing during mouse ES cell differentiation (32) as well as during female development (33, 34). Here, we show that, in immune cells of Ftx−/− females, XCI is progressively destabilized, resulting in the erosion of silencing of selected X-linked genes with immune functions. These include genes of the TLR7 pathway for which escape from XCI is enhanced. This occurs coincidently with the development of autoimmune manifestations such as splenomegaly, higher percentages of activated T and B lymphocytes, and higher levels of immunoglobulins and of autoantibodies against nucleic acids (NAs) and ribonucleoprotein (RNP) complexes in the serum. Autoantibody production is furthermore associated with the accumulation of CD11c+ ABCs and germinal center (GC) B cells in these mice. Mechanistically, MΦ of 1-year-old Ftx−/− females exhibit sustained pro-inflammatory cytokine expression upon exogenous activation of the TLR7 pathway, suggesting that Tlr7 enhanced escape from XCI perpetuates an over-reactive immune environment. Together, these observations provide a direct link between XCI deregulation—a female-specific biological process—and changes in immune cell features and point to alteration in XCI maintenance as a potential trigger of various forms of female-biased autoimmune manifestations.
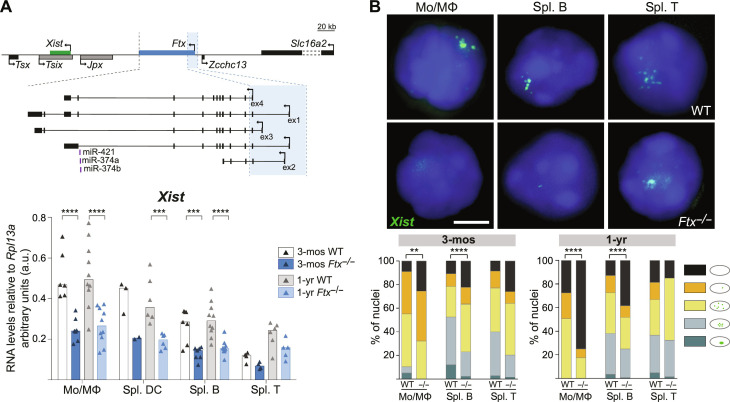
Fig. 1. Perturbation of X-inactivation in immune cells of Ftx−/− females.
(A) Map of the X inactivation center showing the location of Xist (green) and Ftx and the Ftx promoter region that has been deleted (blue shading). Other noncoding regulators of Xist are shown in gray. Underneath, Xist RNA levels measured by reverse transcription quantitative polymerase chain reaction (RT-qPCR) in wild-type (WT) and Ftx−/− females at 3 months (3-mos) and 1-year (1-yr) of age. Monocytes/macrophages (Mo/MΦ) were collected from the bone marrow (BM). Other cell types were collected from the spleen. Each triangle represents RNA levels in a mouse. Bar plots show median values (t test, ***P < 0.005 and ****P < 0.001). (B) Representative images of RNA fluorescence in situ hybridization (RNA-FISH) for Xist (green) on WT and Ftx−/− female cells of the indicated cell type. Note that Xist lncRNAs tend to be delocalized from the Xi even in WT mice as previously described (21). The percentages of cells with different patterns of Xist RNA distribution in the cell populations are shown on the histograms (chi-square test, **P < 0.01 and ****P < 0.001; N > 2 mice; n > 100 nuclei per mice). Scale bar, 5 μm.
RESULTS
Ftx deletion induces aberrant Xist expression profiles in immune cells of adult females
To study the effect of XCI perturbation on immune functions during adult life, we created a mouse line carrying a deletion of Ftx transcription start sites similar to the mutations generated previously (Fig. 1A) (32, 34). Ftx KO animals were born in Mendelian ratios (table S1). Male and females developed normally, appeared healthy overall and fertile with no difference in life span compared to wild-type (WT) animals, as reported in (33, 34). As expected, Ftx expression was completely abolished in immune cells of Ftx−/− females as measured by reverse transcription quantitative polymerase chain reaction (RT-qPCR) (fig. S1A) and by RNA fluorescence in situ hybridization (RNA-FISH) (fig. S1B). Xist expression appeared to be significantly reduced in most Ftx−/− immune cells (half the levels of WT) (Fig. 1A), and Xist lncRNAs hardly clustered on the Xi in immune cell nuclei from 3 months of age onward (Fig. 1B). This indicates that Ftx deletion affects Xist expression in adult immune cells. This perturbation can be considered as mild because it does not lead to a loss of Xist lncRNAs in all the cells (Fig. 1B).
Altered Xist expression results in overexpression and reactivation of genes on the Xi
To determine whether and how X-linked gene silencing is changed upon Xist perturbation, we measured the RNA levels of a battery of X-linked genes. We chose genes with different functions and XCI features including immune-related and unrelated genes, genes known to be associated with autoimmune phenotypes, housekeeping genes, and genes known to escape from XCI (Fig. 2A and fig. S2). We used RT-qPCR on a selection of immune cell types from 3-month- and 1-year-old females to accurately quantify the expression levels (Fig. 2A). At 3 months of age, no significant variation of X-linked RNA levels was observed between WT and mutant cells with the exception of Cxcr3 in T cells (Fig. 2, A and B). In contrast, in 1-year-old females, a number of genes were significantly overexpressed in immune cells of Ftx−/− versus WT mice (Fig. 2A). Many of these genes (Tlr7, Tlr8, Tasl/CXorf21, Cybb, and Cxcr3) naturally escape from XCI in a significant proportion of immune cells (35). Examination of RNA levels in individual mice showed high animal-to-animal variability (Fig. 2B), which may reflect variable frequencies of cells in which XCI was not properly established in the hematopoietic stem population.
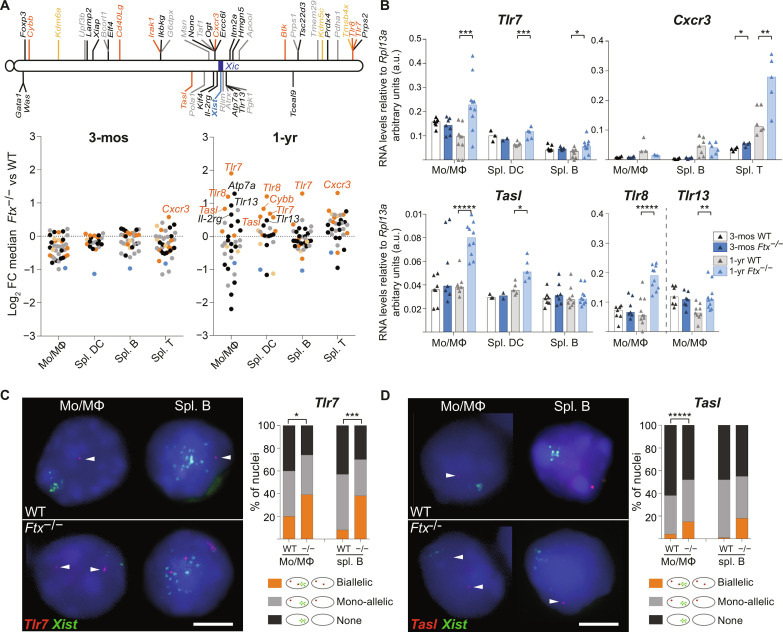
Fig. 2. Aberrant X-inactivation results in overexpression of several X-linked genes associated with autoimmune conditions.
(A) Map of the X chromosome showing genes known to constitutively escape from XCI (yellow), genes with immune function and a tendency to escape from XCI (orange), other genes with immune function (black), and housekeeping genes (gray). Underneath, log2 fold change (FC) between median RNA levels as measured by RT-qPCR in KO versus WT mice for each gene, in the indicated cell type, either in 3-month-old (left) or in 1-year-old mice (right). Each dot represents an X-linked gene. The names of genes showing significantly different RNA levels between KO versus WT are indicated on the graphs (t test, P < 0.05; n ≥ 3 mice per genotype). (B) RNA levels of Tlr7, Cxcr3, Tasl, Tlr8, and Tlr13 as measured by RT-qPCR in cell types showing detectable expression. Each triangle represents RNA levels in a mouse. Bar plots show median values (t test, *P < 0.05, **P < 0.01, ***P < 0.005, and *****P < 0.001). (C) Representative images of RNA-FISH for Tlr7 (red) and Xist (green) on WT and Ftx−/− cells from 1-year-old female mice. The percentages of cells with biallelic, mono-allelic, or no signals are shown on the histogram (chi-square test, *P < 0.05 and ***P < 0.005; N > 2 mice; n > 100 nuclei per mice). Scale bar, 5 μm. (D) Same as (C) for Tasl transcription (chi-square test, *****P < 0.001; N > 2 mice; n > 100 nuclei per mice).
Higher mRNA levels may result from higher expression of alleles on the active X, from increased expression of alleles that already escaped from XCI in these cells, or from reactivation of Xi alleles in additional cells, which means, in this latter configuration, a relaxation of XCI-mediated silencing of these genes. To discriminate between these possibilities, we performed double RNA-FISH for Tlr7 and Xist or for Tasl and Xist on monocyte/macrophages (Mo/MΦ) from the bone marrow (BM) of 1-year-old females, in which the differential of expression in Ftx−/− versus WT by RT-qPCR was the most pronounced (Fig. 2B). We detected significantly higher percentages of nuclei with two pinpoint signals indicating biallelic expression in Ftx−/− compared to WT cells (Fig. 2, C and D). This shows that Tlr7 or Tasl mRNA overexpression in 1-year-old Ftx−/− Mo/MΦ results from an increase of the proportion of cells in which these genes escape from XCI in the population compared to WT Mo/MΦ population. Similar increase in the frequency of biallelically expressing cells was also observed for Tlr7 in B lymphocytes from 1-year-old Ftx−/− mice (Fig. 2C).
Not all known escapees appeared overexpressed (fig. S2A), and some genes supposedly subject to XCI (Tlr13, Il-2rg, and Atp7a) showed higher RNA levels in Ftx−/− compared to WT cells (Fig. 2, A and B, and fig. S2B). This suggests that labile expression from the Xi may facilitate, but is not a prerequisite for, overexpression and that genes may escape from XCI upon Xist perturbation specifically. In contrast, expression of X-linked housekeeping genes was not significantly perturbed in Ftx−/− immune cells (fig. S2C). We also observed some genes with lower expression in Ftx−/− cells compared to WT (Fig. 2A and fig. S2D) that may constitute secondary targets of X-linked gene overexpression.
Together, these results indicate that impaired Xist expression in Ftx−/− immune cells does not lead to global reactivation of the Xi. Rather, it appears to induce or enhance escape from XCI of specific X-linked genes, leading to increased expression levels of those genes as time goes by.
Specific X-linked factors involved in autoimmunity tend to be overexpressed upon XCI perturbation
The most notable feature of X-linked genes affected by Xist perturbation is the high number of genes involved in innate immune response (TLR7, TLR8, TLR13, TASL, and CXCR3), which have been reported to be associated with or to have a causative role in different autoimmune diseases (table S2). They include many endosomal TLRs (TLR7, TLR8, and TLR13) or plasma membrane chemokine receptor (CXCR3). No significant deregulation of two other membrane receptor genes, Il-4ra and Il-6ra, that are located on autosomes could be detected in Ftx−/− immune cells, which confirms a specific effect on X-linked receptor genes (fig. S3A). TLR7, TLR8, and TLR13 trigger, upon activation, the nuclear factor κB pathway (36), but neither Irak1 nor Ikbkg (NEMO), two X-linked core effectors of the TLR-signaling pathway (36), showed significant changes in their expression level in Ftx−/− immune cells (fig. S3B). This suggests either that they are not sensitive to Xist perturbation or that their expression is controlled by additional regulation.
Female mice with impaired XCI develop a splenomegaly during aging
We then characterized changes in the immune system upon XCI perturbation. We could not detect any difference in spleen weight or morphology between Ftx−/− and WT females at 3 months of age. In contrast, spleens from 1-year-old and, more markedly, from 2-year-old females appeared significantly larger in Ftx−/− conditions (Fig. 3A). Histological analyses did not reveal any defects in cell organization and any signs of fibrosis or of cell infiltration in Ftx−/− spleens compared to WT at any age (Fig. 3B and fig. S4A). Both the time-course progression of the splenomegaly and the fact that it never occurred in males (fig. S4B) are consistent with the notion that XCI molecular alteration is the driving force responsible for the phenotype of Ftx−/− mice.
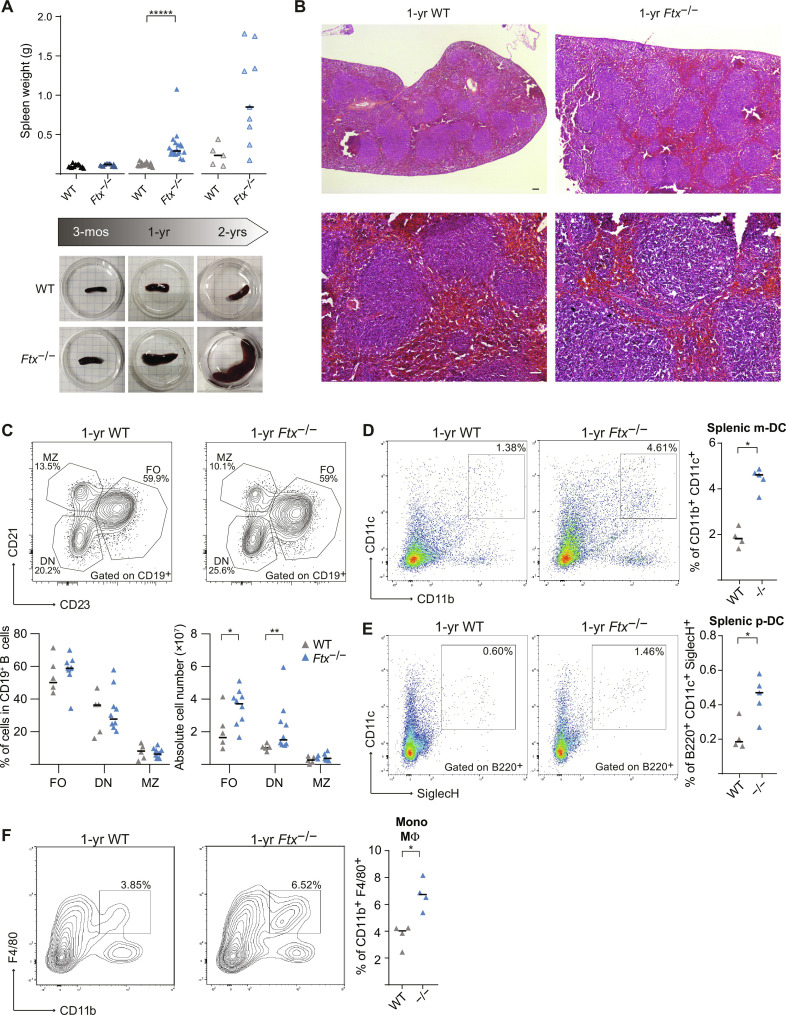
Fig. 3. Ftx−/− females develop a splenomegaly associated with a deregulation of B and myeloid cell populations.
(A) Spleen weight of WT and Ftx−/− females at 3 months, 1 year, and 2 years of age. Median values are shown (t test, *****P < 0.001). Underneath, representative images of WT and Ftx−/− spleens from 3-month-, 1-year-, and 2-year-old females. (B) Representative images of hematoxylin-eosin staining on sections of spleens from 1-year-old WT and Ftx−/− females. Scale bars, 100 μm. (C) Representative flow cytometry analysis of follicular (FO; CD21+CD23+), double-negative (DN; CD21−CD23−), and marginal zone (MZ; CD21+CD23−) B cells among CD19+ B cells in spleen from 1- to 1.5-year-old WT and Ftx−/− females. Percentages and absolute number are shown on the graphs. Median values are shown (Mann-Whitney test, *P < 0.05 and **P < 0.01). (D) Representative flow cytometry analysis of splenic myeloid DCs (m-DC) in WT and Ftx−/− 1-year-old females. On the right, percentages of CD11b+CD11c+ splenic m-DC in leucocytes. Each triangle represents a mouse. Median values are shown (Mann-Whitney test, *P < 0.05). (E) Same as (D) for CD11c+B220+SiglecH+ splenic plasmacytoid DCs (p-DC) (Mann-Whitney test, *P < 0.05). (F) Same as (D) for CD11b+F4/80+ monocyte/macrophages (Mann-Whitney test, *P < 0.05).
The splenomegaly in 1-year-old Ftx−/− females resulted from a multilineage cell expansion preferentially targeting myeloid and DCs (table S3). Follicular and CD21−CD23− double-negative B cells appeared increased in absolute number but not in percentages, while marginal zone B cell counts remained unchanged upon Ftx deficiency (Fig. 3C). In contrast, percentages of both myeloid DCs (Fig. 3D) and plasmacytoid DCs (Fig. 3E) and of Mo/MΦ were significantly higher in Ftx−/− compared to WT animals (Fig. 3F). No differences were observed between aged-matched Ftx−/− and WT males (table S4).
Perturbation of XCI leads to autoimmune manifestations
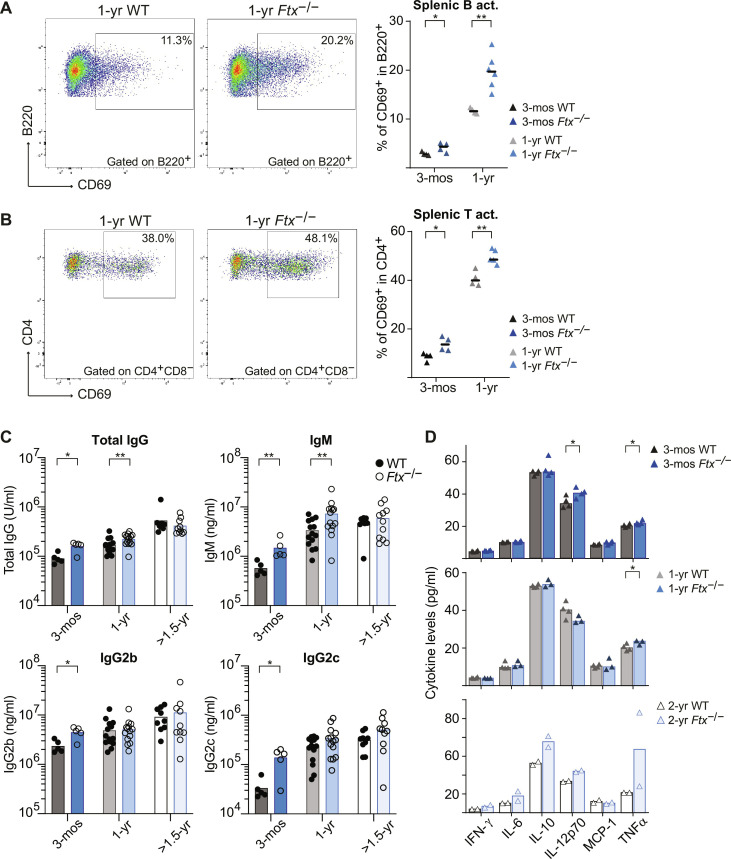
Mo/MΦ and DC expansion are typical of inflammation reported in mouse models of SLE, including NZB/W F1 and Yaa mice in which Tlr7 is overexpressed (15, 24). Accordingly, we observed higher frequencies of spontaneously activated CD69+ B and T cells in the spleen of Ftx−/− females compared to WT from 3 months of age onward (Fig. 4, A and B). This is associated with higher levels of immunoglobulin M (IgM), total IgG, IgG2b, and IgG2c immunoglobulins in the serum of Ftx−/− versus WT females (Fig. 4C). These changes in immune regulation were, however, not associated with any marked increase in cytokine levels in the serum of 3-month- or 1-year-old Ftx−/− females (Fig. 4D). Only a mild increase of interleukin-12p70 (IL-12p70) and of tumor necrosis factor–α (TNFα) levels, one of the cytokines induced by TLR7 signaling pathway (35), was detected at 3 months and/or 1 year of age, suggesting that cytokine levels are efficiently controlled despite inflammation signs.
Fig. 4. Ftx deficiency in female mice promotes signs of inflammation.
(A) Representative flow cytometry analysis of spontaneously activated B220+CD69+ B cells in spleen from 1-year-old WT and Ftx−/− females. Percentages in leucocytes are shown on the graphs beneath. Each triangle represents a mouse. Median values are shown (Mann-Whitney test, *P < 0.05 and **P < 0.01). (B) Same as (A) for spontaneously activated CD4+CD69+ T cells (Mann-Whitney test, *P < 0.05 and **P < 0.01). (C) Total IgG, IgM, IgG2b, and IgG2c natural antibody levels in sera of 3-month-, 1-year, and >1.5-year-old WT or Ftx−/− females measured by ELISA. Each circle represents a mouse. Mean values are shown (Mann-Whitney test, *P < 0.05 and **P < 0.01). (D) Cytokines levels in the blood analyzed with cytometric bead array assays on sera from 3-month-, 1-year-, or 2-year-old WT and Ftx−/− females. Each triangle represents a mouse. Median values are shown (t test, *P < 0.05).
XCI alteration induces a lupus-like syndrome in female mice
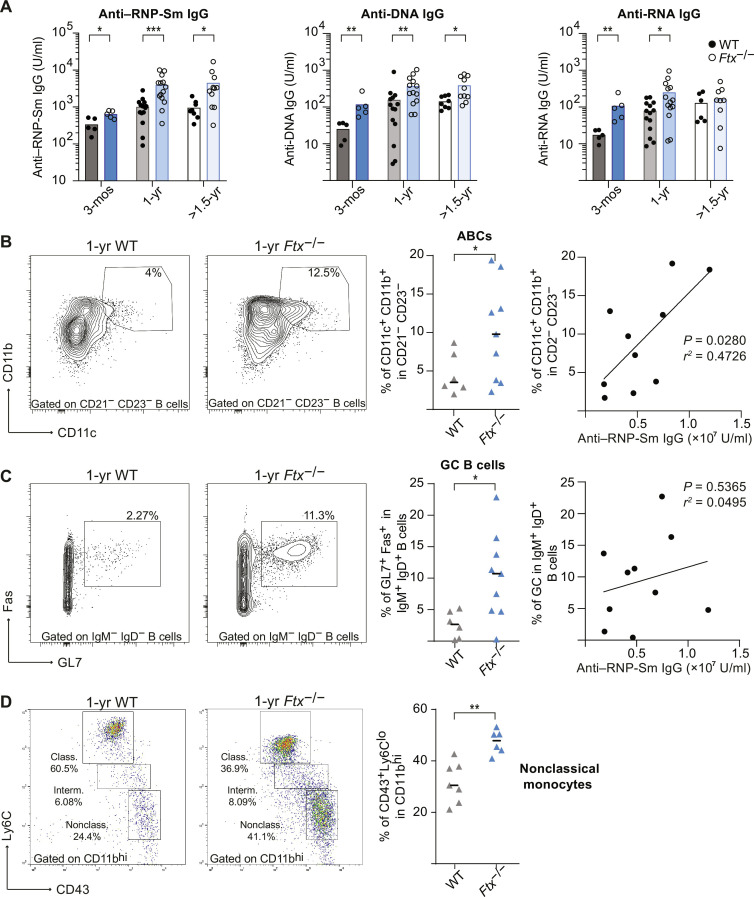
Lupus-like syndromes are specifically defined by high quantities of circulating autoantibodies against RNP complex (RNP-Sm) or against NAs including anti-ssRNA and anti-DNA antibodies. Enzyme-linked immunosorbent assay (ELISA) quantifications of anti-RNP-Sm, anti-RNA, and anti-DNA IgG in sera of 3-month-, 1-year-, and >1.5-year-old Ftx−/− versus WT females showed significantly higher levels in Ftx−/− animals (Fig. 5A). Coincidentally with autoantibodies overproduction, we detected higher frequencies of ABCs and GC B cells in Ftx−/− compared to WT spleens (Fig. 5, B and C). In particular, anti-RNP-Sm levels significantly correlated with the percentages of ABCs (Fig. 5B), which are consistently found overrepresented in SLE and other autoimmune disorders (37). In contrast, GC B cell percentages did not correlate with anti-RNP-Sm antibody levels (Fig. 5C).
Fig. 5. Ftx deficiency in female mice induces anti-NA and anti-RNP-Sm autoantibody production and the development of ABC-like cells.
(A) Anti–RNP-Sm IgG, anti-DNA IgG, and anti-RNA IgG autoantibody levels in sera of 3-month-, 1-year-, and >1.5-year-old WT or Ftx−/− females measured by ELISA. Each circle represents a mouse. Mean values are shown (Mann-Whitney test, *P < 0.05, **P < 0.01, and ***P < 0.005). (B) Representative flow cytometry analysis of ABC-like cells (CD11c+CD11b+) among DN (CD21−CD23−) in spleen from 1-year-old to 1.5-year-old WT and Ftx−/− females. Percentages are shown on the graphs. Each triangle represents a mouse. Median values are indicated. (Welch’s t test, *P < 0.05). The right panel shows the correlation between the frequency of ABCs (CD11c+CD11b+) relative to anti–RNP-Sm IgG levels in each Ftx−/− female (Pearson correlation). (C) Representative flow cytometry analysis of GC cells (Fas+GL7+) among switch memory B cells (IgM−IgD−) in spleen from 1-year-old to 1.5-year-old WT and Ftx−/− females. Percentages are shown on the graphs. Each triangle represents a mouse. Median values are indicated (Welch’s t test, *P < 0.05). The right panel shows the correlation between the frequency of GC cells (Fas+GL7+) and anti–RNP-Sm IgG Ab levels in each Ftx−/− females (Pearson correlation). (D) Representative flow cytometry analysis of monocyte populations including nonclassical (CD11bhiCD43+Ly6Clo) scavenger monocytes in the blood of 1-year-old WT and Ftx−/− females. Percentages in leucocytes are shown on the graphs beneath. Each triangle represents a mouse. Median values are shown (Mann-Whitney test, **P < 0.01).
This strongly suggests that ABCs constitute the major producer of this class of autoantibodies as previously reported in human SLE (38, 39). In SLE, ABCs may originate from the extra-follicular pathway and develop into autoreactive plasma cells upon TLR7 signal (38). In agreement, the numbers of long-lived plasma cells were increased in the spleen of >1-year-old KO animals (fig. S5, A and B).
Given that the effect of Xist perturbation on X-linked gene expression is especially pronounced in Mo/MΦ, we examined the populations of circulating Mo/MΦ in 1-year-old females. We detected an overrepresentation of Ly6Clo nonclassical monocytes in the blood of Ftx−/− females (Fig. 5D), a monocyte population recruited at inflammatory tissues in lupus-like contexts (40–42), but we did not observe signs of inflammation in peripheral tissues like kidneys, a clinical manifestation that is observed at late stages of SLE.
Thus, Ftx−/− females progressively develop a splenomegaly during aging, which is accompanied by multiple markers of SLE, including high levels of spontaneous lymphocyte activation, increased percentages of ABC-like and GC B cells, overproduction of immunoglobulins (IgM and IgG), including IgG autoantibodies to NA and RNP-Sm, and a predominance of atypical Ly6Clo monocytes in the circulation.
XCI impairment triggers an overexpression of target cytokines in TLR7-stimulated macrophages
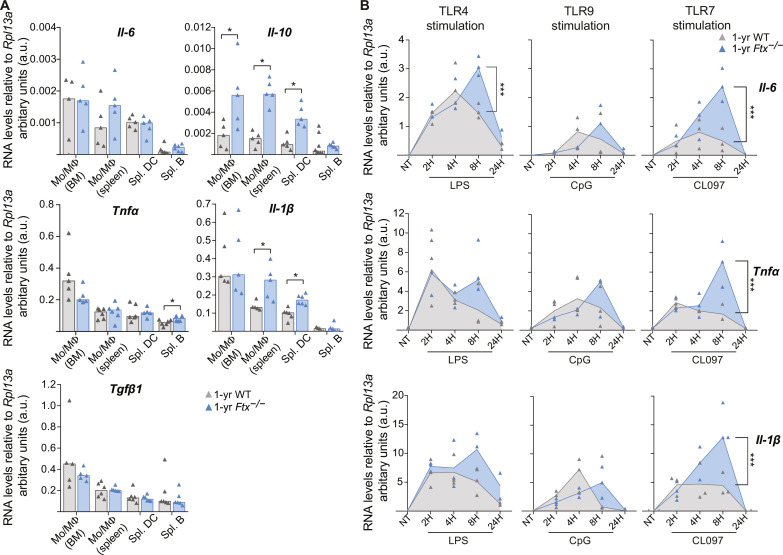
To test whether TLR7 pathway hyperactivity could contribute to Ftx−/− lupus-like phenotype and to identify the targeted cell populations, we first measured the basal expression of cytokine genes normally induced upon activation of TLR7 pathway (i.e., Tnfα, Il-1β, Il-6, and Il-10) by RT-qPCR in BM or splenic Mo/MΦ, in splenic DC, and in splenic B cells of 1-year-old females (Fig. 6A).
Fig. 6. Hyperactive TLR7 pathway in Ftx−/− female macrophages.
(A) Analysis of cytokine RNA levels by RT-qPCR in the indicated cell population collected from WT or Ftx−/− 1-year-old females. Each triangle represents a mouse. Bar plots show median values (t test, *P < 0.05). (B) RT-qPCR analysis of cytokine RNA levels in BM-derived macrophages (granulocyte-macrophage colony-stimulating factor differentiation of total BM cells) activated with either lipopolysaccharide (LPS; TLR4 pathway), CpG (TLR9 pathway), or the TLR7 agonist CL097. Cells were either not treated (NT) or treated for 2, 4, 8, or 24 hours (2H, 4H, 8H, and 24H, respectively). Each triangle represents a mouse. Median values are shown [analysis of variance (ANOVA), ***P < 0.001].
Basal RNA levels of Il-10, Il-1β, and Tnfα appeared significantly higher in Mo/MΦ and/or in splenic B cells of Ftx−/− mice. Il-6 also displayed similar tendencies. In contrast, Tgfβ1, a gene that is not a direct target of the TLR7 pathway, was expressed at the same levels in WT and Ftx KO contexts (Fig. 6A). Such changes in the basal production of cytokines may result from secondary effect of the inflammatory/autoimmune phenotype in Ftx-deficient mice or from cell-intrinsic mechanisms, and we therefore analyzed the kinetics of TLR-signaling in ex vivo differentiated BM-derived MΦ (fig. S6). These MΦ were then stimulated with TLR ligands, specific for TLR4 [lipopolysaccharide (LPS)], TLR9 (CpG), or TLR7 (CL097), and mRNA expression kinetics of key pro-inflammatory cytokines TNFα, IL-1β, and IL-6 were measured by RT-qPCR. Although all three cytokine transcripts were up-regulated in response to TLR agonist ligands, a higher and sustained expression of these target cytokines was specifically observed in response to TLR7 activation in Ftx-deficient MΦ and not in WT cells (Fig. 6B). A significantly higher up-regulation of Il-6 was also detected upon TLR4 stimulation in Ftx−/− compared to WT MΦ (Fig. 6B).
We conclude that Ftx−/− MΦ intrinsically exhibit a hyperfunctional secretory phenotype, characterized by sustained transcriptional activation of the TLR7/TLR8 signaling pathway upon activation. This is likely to induce an overproduction of target cytokines driving adaptive immunity and the development of lupus-like autoimmunity in Ftx−/− females.
DISCUSSION
With recent worldwide waves of viral infections and increased realization that women are more resistant to these infections than men, understanding the basis of sexual dimorphism in immune system competence has emerged as critical to the design of innovative therapeutic strategies (43). In this framework, we show that perturbing XCI, a female-specific epigenetic process that is established early during development, directly affects female immune response in mouse adult life. More specifically, certain X-linked genes with immune functions that naturally show a tendency to escape from XCI are expressed from the inactive X in higher percentages of immune cells upon XCI perturbation, resulting in overall higher levels of mRNAs. This deregulation is associated with the progressive development of autoimmune manifestations indicative of an over-reactive immune system. This suggests that, under normal conditions, a low level of escape from XCI of specific X-linked genes leads to a slightly higher dosage of the corresponding immune factors compared to males, which may endow females with enhanced plasticity in immune responses. This hypothesis is corroborated by former observations that correlate higher expression of some X-linked genes (Tlr7, Cxcr3, and Tlr8) in females compared to males with better protection against viral or parasite infections in both humans and mice (44–46). Moreover, recent studies have identified loss-of-function mutations of TLR7 associated with severe forms of COVID-19 in young men, demonstrating the essential role of TLR7 pathway in the protective response against severe acute respiratory syndrome coronavirus 2 (47–49).
Different X-linked genes appear overexpressed upon XCI alteration depending on immune cell types (i.e., Cxcr3 in T cells; Tlr7 in B cells; Tlr7, Cybb, and Tlr8 in splenic DCs; and Tlr7, Tlr8, Tlr13, Tasl, Il-2rg, and Atp7a in Mo/MΦ), suggesting that various molecular pathways could be affected in immune cells. The number of affected genes might be underestimated because we only tested a subset of immune-related X-linked genes. The phenotype that we observe probably results from the combination of effects ensuing from distinct reactivation events on the Xi in different cell types and the more global changes in gene expression that these initial events triggered. Additional regulatory loops may also be at play. Some overexpressed X-linked genes such as Tlr7 and Tasl are type I interferon (IFN-I)–stimulated genes (50). Hence, overexpression of such genes at the mRNA levels could result from a combination of XCI escape and enhanced IFN-I signaling associated with lupus pathogenesis (51). Accordingly, we observe higher numbers of pDCs in Ftx-deficient mice compared to WT. IFN-I produced by pDCs is known to propagate anti-NA antibody response and SLE-like disease and could further activate of X-linked IFN-stimulated gene transcription (51).
Several lines of evidence support the conclusion that Tlr7 reactivation is a major contributor to the lupus-like syndrome of Ftx−/− females. First, we observe increased monocytosis characterized by the selective expansion in the blood of the Ly6Clo nonclassical monocyte subset, which have been commonly reported in TLR7-driven lupus mouse models (40–42). Moreover, in the NZM/NZW lupus model, Ly6Clo monocytes, which spontaneously accumulate with age, express high levels of TLR7 protein, and administration of TLR7 agonist ligands accelerates Ly6Clo monocyte augmentation in the blood and promotes nephritis (40). Second, GC B cells accumulate in the spleen of Ftx−/− mice. Spontaneous GC formation is well described in lupus mouse models and has been shown to be either strictly dependent on B cell–intrinsic TLR7 expression (52) or promoted upon enhanced TLR7-signaling (16). Last, the formation of pathogenic CD11c+ ABCs is significantly enhanced in Ftx−/− females, as observed in many lupus-associated conditions in mice (16, 53, 54) and humans (38, 39). Anti-RNP-Sm autoantibody production strongly correlates with ABC development in Ftx−/− females, suggesting a major role of ABC in B cell systemic autoimmunity. Extrafollicular ABCs and their plasma cell products, rather than GC-derived B cells, have been shown to drive the development of pathogenic B cell subsets in spontaneous TLR7-driven lupus models (16, 51).
What is the molecular mechanism underlying reactivation of specific X-linked genes in Ftx−/− females? Intriguingly, we noticed that X-linked genes sensitive to Xist perturbation tended either to cluster (Tlr7/Tlr8 and Atp7a/Tlr13) or to be located in gene-poor regions (Cxcr3 and Cybb) (Fig. 2A), suggesting a mechanism that operates regionally over several kilobases rather than an action restricted to promoters. In this regard, silencing of TLR7 and, more generally, of X-linked genes lacking promoter DNA methylation in human B cells is thought to depend on continuous association with XIST RNA and XIST-dependent H3K27 deacetylation of distal enhancers and not promoters (25, 55). In female mouse Mo/MΦ, the methylation status of the promoter of genes targeted by reactivation is unknown, but it is conceivable that, in the Ftx−/− context, reduced deacetylation of local enhancers following exacerbated Xist RNA delocalization from the Xi contributes to enhance reactivation of neighboring genes or leads to a spreading of escape to genes subject to XCI in WT cells. Another intriguing observation is the sustained cytokine gene expression upon TLR7 activation of Ftx−/− macrophages. Overexpression of Tlr7 probably leads to TLR7 accumulation at the membrane of late endosome or lysosomes, resulting in prolonged ligand-receptor contact and continuous production of pro-inflammatory cytokines. Deficiency in components of the SMRC8-WDR4-C9ORF72 complex, a regulator of autophagy and lysosomal function, causes inflammation due to excessive endosomal TLR signaling in MΦ (56). Alternatively, impaired re-localization of Xist RNA to the Xi upon stimulation, a phenomenon that has recently been reported in activated T cells from the MRL/Lpr lupus model (57) and in activated B and T cells of Ciz1−/− mice suffering from lymphoproliferative disorder (58), may lead to sub-efficient secondary repression of Tlr7 Xi copy. Last, it is tempting to postulate the existence of an active process that would have been evolutionary selected to maintain low levels of XCI escape of specific X-linked genes because this may both confer a greater resistance to infection to female individuals.
To our knowledge, none of the known XIST regulators, including XIST itself, has been identified as a SLE susceptible locus in genome-wide association studies. Alterations of XCI as a cause of different autoimmune manifestations have not been reported so far but not thoroughly investigated either. Two independent studies have recently reported that (i) XIST levels were elevated in blood leucocytes from women with SLE (59) and that (ii) transgenic expression of Xist lncRNAs in male mice can promote autoantibodies directed against Xist RNP in the context of pristane-induced lupus in a permissive genetic background (SJL/J) (60). This result seems in apparent contradiction with observations of the present study in which reduced levels of Xist in female immune cells, associated with altered XCI, trigger spontaneous systemic autoimmunity. The study by Dou et al. (60) indicates that Xist RNP itself harbors immunogenic properties upon release from dying male cells that are not naturally expressing Xist, whereas, in the present study, altered XCI is constitutive and triggers spontaneous autoimmunity in aged female mice of the non-autoimmune prone background (C57/BL6). These two findings are, however, not mutually exclusive and may reflect the wide variety of causes, manifestations, and onset of autoimmunity. Of note, in the spontaneous model of lupus in genetically predisposed mice, disease can develop in the absence of Xist. This has been well established in male mice carrying the Y-linked genomic modifier Yaa, in which a portion of the X chromosome has translocated onto the Y chromosome, resulting in a twofold increase in Tlr7 expression (14, 54). In this model, spontaneous systemic autoimmunity is more severe in males than in females, suggesting that it is the translocated Yaa locus rather than Xist expression, which is the critical factor controlling spontaneous systemic B cell autoimmunity. Further studies using uniformized genetic backgrounds associated with tailored ways to either increase or reduce Xist expression will be required to establish the effect of Xist RNP and of XCI alteration on the kinetics and types of autoimmune manifestations that are promoted. Together, it is possible that autoreactivity to Xist RNP and escape from XCI may both synergize to promote female-biased autoimmunity in the context of tissue damage (60).
Molecular and cellular phenotypes resulting from impaired Xist expression progress gradually with age. Although we cannot exclude that Xist expression depends on Ftx in immune cells specifically, it is more likely that XCI perturbation in Ftx−/− females that initiates in the embryo (33) is transmitted, as such, to the hematopoietic lineage. This perturbation has, however, few effects in immune cells from 3-month-old females—the most marked phenotypic changes are detected around 1-year of age—and does not really affect life expectancy or animal fitness. Whether this evolutive phenotype relates to changes in heterochromatin features that are known to occur during aging (61, 62) and/or whether it is accelerated by the gradual loss of immune cell differentiation potential (63) remains to be determined. In this regard, it is tempting to speculate that deregulation of XCI may also contribute to autoimmune conditions specific to postmenopausal women [rheumatoid arthritis (64), some forms of Sjögren’s syndrome (65), atherosclerosis or of ischemic heart diseases (43, 66), and inflammaging (67)] when estrogen levels are low and cannot account for the sexual dimorphism observed in these diseases. In conclusion, we have established a direct link between XCI maintenance and function of the immune system. This paves the way for exploring further the role of XCI regulators in female-biased unexplained forms of autoimmune conditions and opens up alternative potentials for therapeutic strategies.
MATERIALS AND METHODS
Generation of Ftx-deficient mice
Ftx-deficient mice were generated in the Institut Clinique de la Souris (Illkirch, France). The strategy used to create Ftx−/− mice on a C57BL/6N background is depicted in fig. S7. Ftx−/− females were generated either by mating Ftx−/Y males with Ftx+/− females or by mating Ftx−/Y males with Ftx−/− females. Similar results were obtained on Ftx−/− females from either cross. Mice were maintained under specific pathogen–free conditions in the animal facility of the Jacques Monod Institute (Paris, France) and handled following the European Community guidelines (project authorization no. 05353.02 approved by the ethical comity of the French Ministry for Scientific Research). Virgin females with no signs of inflammation from self-inflicted or other injuries have been used exclusively.
Serological analyses
For anti-DNA and anti-RNA IgG ELISA, 96-well ELISA plates were first coated overnight with poly-l-lysine (Sigma-Aldrich) and then overnight with DNA from calf thymus (Sigma-Aldrich) or yeast RNA (Sigma-Aldrich), respectively. For anti–RNP-Sm IgG ELISA, Nunc Maxisorp plates were coated overnight with RNP-Sm antigen (Native Calf Thymus, Arotec). Plates were blocked with phosphate-buffered saline (PBS) with 1% bovine serum albumin (BSA), and sera were then titrated and compared to a standard serum from a pool of SLE1, SLE2, and SLE3 mice. Anti-DNA, anti-RNA, and anti–RNP-Sm IgG are expressed in arbitrary units per milliliter. For each isotype, 1 U/ml corresponds to the standard serum concentration resulting in 50% of the maximum optical density (OD) read at 405 nm. IgGs were revealed with goat anti-mouse biotinylated IgG (SouthernBiotech) followed by alkaline phosphatase–conjugated streptavidin (Jackson ImmunoResearch Laboratories), and absorbance at 405 to 650 nm was read.
For total IgG, IgM, IgG2b, and IgG2c ELISA, 96-well ELISA plates were coated with goat anti-mouse IgG (1 μg/ml; H+L) (Jackson ImmunoResearch Laboratories) in PBS for 2 hours at 37°C and then overnight at 4°C. Serially diluted sera were applied. Specific antibodies were detected with biotinylated goat anti-mouse total IgG, IgM, IgG2b, or IgG2c, respectively (SouthernBiotech), followed by incubation with streptavidin coupled with alkaline phosphatase (Jackson ImmunoResearch Laboratories). Plates were read at 405 to 650 nm with an ELISA reader (Varioskan Flash, Thermo Fisher Scientific). IgM (clone MADNP5), IgG2b (clone MADNP3) from PARIS-anticorps (Cergy-Pontoise, France), and IgG2c (Invitrogen) were used as standards. Results are expressed in nanograms per milliliter. For total IgG, a standard serum from a pool of mice was used. Total IgG levels were expressed in arbitrary units per milliliter (1 U/ml corresponds to the standard serum concentration resulting in 50% of the maximum OD read at 405 nm). Levels of inflammatory cytokines in sera were measured using a cytometric bead array mouse inflammation kit (552364, BD Biosciences) according to the manufacturer’s instructions.
Flow cytometry analyses
BM, spleen, blood, and peritoneal cavity cells were stained using the following antibodies: CD3 PerCP-Vio770 (130-119-656, Miltenyi Biotec), CD4-allophycocyanin (APC) (130-123-207, Miltenyi Biotec), CD5-APC-Vio770 (130-120-165, Miltenyi Biotec), CD8-fluorescein isothiocyanate (FITC) (130-118-468, Miltenyi Biotec), CD11b APC (553312, BD Pharmingen), CD11c phycoerythrin (PE)–Vio770 (130-110-840, Miltenyi Biotec), CD19-FITC (557398, BD Pharmingen), CD21-APC-Vio770 (130-111-733, Miltenyi Biotec), CD23-PE-Vio770 (130-118-764, Miltenyi Biotec), CD38-PE (130-123-571, Miltenyi Biotec), CD43-PE (130-112-887, Miltenyi Biotec), CD69-PE (130-115-575, Miltenyi Biotec), CD138 PE-Vio615 (130-108-989, Miltenyi Biotec), F4/80 FITC (130-117-509, Miltenyi Biotec), Ter119 PE (130-112-909, Miltenyi Biotec), SiglecH APC-Vio770 (130-112-299, Miltenyi Biotec), B220-APC (130-110-847, Miltenyi Biotec), B220 VioBlue (130-110-851, Miltenyi Biotec), IgM-VioBlue (130-116-318, Miltenyi Biotec), IgD-PE (130-111-496, Miltenyi Biotec), GL7-PE-Cy7 (144619, BioLegend), Ly6C-FITC (130111-915, Miltenyi Biotec), streptavidin FITC (554060, BD Biosciences), CD138 BV605 (563147, BD-Horizon), CD23 BV605 (101637, BioLegend), I-A/I-E BV711 (107643, BioLegend), CD19 BV786 (563333, BD Horizon), T and B cell activation antigen (GL7) PE (561530, BD Pharmingen), CD95 PE-Cy7 (557653, BD Pharmingen), IgM APC-eFluor 780 (47-5790-82, Invitrogen), CD45R/B220 APC/cyanine7 (103224, BioLegend), CD11b eFluor 450 (48-0112-82, eBioscience), CD267 (TACI) BV421 (742840, BD Biosciences), IgD BUV395 (564274, BD Horizon), streptavidin APC (4317-82, eBioscience), CD3 Biotin (100304, BioLegend), Biotin CD11c (568970, BD Biosciences), CD21 PercP Cy5.5 (562797, BD Biosciences), and Fixable Viability Dye eFluor 506 (65-0866-18, Invitrogen) following the recommendations of the manufacturers. Flow cytometry was conducted on a BD FACSAria Fusion (BD Biosciences) at the ImagoSeine platform of the Jacques Monod Institute (Paris, France). The instrument is equipped with 18 detectors and five lasers (ultraviolet, 355 nm, 20 mW; violet, 405 nm, 10 mW; blue, 488 nm, 13 mW; yellow-green, 561 nm, 50 mW; and red, 633 nm, 11 mW). The Cytometer Setup and Tracking RUO beads (lot ID 82169) were used to establish reference fluorescence intensities for measuring instrument sensitivity over time. At least 50,000 single cells were acquired. Data files were exported to Flow Cytometry Standard (FCS) file 3.1. All manual analysis was performed using FlowJo 10.8.1 (BD Biosciences). Signal stability over time was checked using flowClean plugin. Forward scatter (FSC) height and area are plotted against each other and used to eliminate doublets and aggregates.
Cell sorting and activation
Magnetic sorting
After isolation, spleen or BM cells were stained for 30 min on ice with Anti-CD23 Magnetic Microbeads (130-098-784, Miltenyi Biotec), Anti-F4/80 Magnetic Microbeads (130-110-443, Miltenyi Biotec), Anti-CD11b Magnetic Microbeads (130-126-725, Miltenyi Biotec), CD11c Magnetic Microbeads (130-108-338, Miltenyi Biotec), Anti-CD90.2 Magnetic Microbeads (130-121-278, Miltenyi Biotec), or Anti-CD19 Magnetic Microbeads (130-121-301, Miltenyi Biotec); washed with 1× PBS with 3% fetal bovine serum (FBS); and centrifuged at 300g for 10 min. Cells suspension was purified on LS column (130–042-401, Miltenyi Biotec). Purity of sorted cells was controlled by cytometry using corresponding antibodies.
For differentiation of BM-derived macrophages (BMDMs), total BM cells were cultured with granulocyte-macrophage colony-stimulating factor (100 ng/ml; 130-095-742, Miltenyi Biotec) in complete RPMI 1640 (RPMI 1640 and GlutaMAX, 10% FBS, 1 mM sodium pyurvate, 10 mM Hepes, 0.1 mM nonessential amino acids (NEAA), and 50 μM β-mercaptoethanol). Differentiation efficiency was checked by cytometry using a CD11c, CD11b, F4/80, CD3, and B220 antibody panel.
Cell activation
BMDMs were stimulated with LPS (1 ng/ml; Sigma-Aldrich) for TLR4 activation, CpG ODN 2395 (100 nM; tlrl-2395, InvivoGen) for TLR9 activation, or CL097 (50 nM; tlrl-c97, InvivoGen) for TLR 7 activation. Matched negative controls ODN 5328 (InvivoGen) or CL075 (InvivoGen) were used for ODN 2395 and CL097, respectively.
RNA fluorescence in situ hybridization
Cell preparation
Magnetic sorted cells or fluorescence-activated cell sorting–sorted cells were allowed to sediment on poly-lysine–coated slides (Thermo Fisher Scientific) in a 50-μl drop of 1× PBS for 15 min at room temperature, fixed for 10 min in an ice-cold 3% paraformaldehyde/1× PBS solution (Electron Microscopy Sciences), and permeabilized for 5 to 10 min in ice-cold cytoskeletal (CSK) buffer [10 mM 30 Pipes, 300 mM sucrose, 100 mM NaCl, and 3 mM MgCl2 (pH6.8)] supplemented with 0.5% Triton X-100 (Sigma-Aldrich) and 2 mM vanadyl-ribonucleoside complex (VRC; New England Biolabs).
Probe preparation
One microgram of purified fosmid/bacterial artificial chromosome DNA purified using standard alkaline lysis protocol was labeled with fluorescent deoxyuridine triphosphates (SpectrumOrange and SpectrumGreen from Abott Molecular and Cy5-UTPs from GE HealthCare Life Sciences) in a 50-μl nick-translation reaction for 3 hours at 15°C. Xist, p510 plasmid (68); Ftx, fosmid probe (WI1-1177B13, BACPAC); Tlr7, fosmid probes (WI1-1548 K9, WI1-977A7, BACPAC); and Tasl, fosmid probes (WI1-1708P12, WI1-841O8, BACPAC).
Hybridization
One hundred nanograms of probe was co-precipitated with 3 μg of Cot-I DNA (Invitrogen) and 10 μg of Sheared Salmon Sperm DNA (Invitrogen) using 1/10 3 M NaOAc and 2.5 volumes of ethanol for 3 hours at −20°C. Precipitated probes were washed with 70% ethanol and resuspended in formamide (deionized formamide > 99.5%; Sigma-Aldrich) and then denatured for 7 min at 75°C. After probe denaturation, an equivalent volume of 2× hybridization buffer [4× saline sodium citrate (SSC; Ambion), 20% dextran sulfate, BSA (2 mg/ml; New England Biolabs), and 2 mM VRC (New England Biolabs)] was added, and slides were hybridized overnight at 37°C in a humid chamber. The slides were subsequently washed three times in 50% formamide/2× SSC (pH 7.2) at 42°C for 5 min each and three times in 2× SSC at 42°C for 5 min each. Slides were mounted in Vectashield containing 4′,6-diamidino-2-phenylindole (DAPI) (Vector Laboratories).
Microscopy and image analysis
Images were taken on an Axioplan 2 Imaging fluorescence microscope (Zeiss) with a cooled Coolsnap camera (Roper Scientifics) or a DMI-6000 inverted fluorescence microscope with a motorized stage (Leica) and a charge-coupled device camera HQ2 (Roper Scientific), both controlled by the MetaMorph 7.04 software (Roper Scientifics), using a Plan-NEOFLUAR 63×/1.25 oil objective (Zeiss), a Plan-NEOFLUAR 100×/1.30 oil objective (Zeiss), or a HCX PL APO 63×/1.4 oil objective (Leica). Optical sections were collected at 0.2-mm steps through each nucleus at different wavelengths (in nanometers) {Zeiss: DAPI (345, 455), FITC (488, 507), and CY3 (625, 660); Leica: DAPI (360, 470), FITC (470, 525), and CY3 (550, 570)}. Approximately 40 optical sections per nucleus were collected. Stacks were processed using Icy (http://icy.bioimageanalysis.org), and the images are represented as two-dimensional projections of the stacks (maximum projection).
Acknowledgments
We thank the Microscopy Platform-UMR7216 Epigenetic and Cell Fate center for access to instruments and technical advice. We acknowledge the ImagoSeine core facility of the Institut Jacques Monod, member of the France BioImaging infrastructure and GIS-IBiSA; in particular, we thank the ImagoSeine cytometry platform and S. Many and D. Korenkov for technical assistance. We thank the Cytometry core facility at INFINITY INSERM U1291 (Toulouse). We thank the cytometry platform of St Louis Hospital–INSERM UMRS 976. We thank the animal facility of the Jacques Monod Institute and particularly L. Pontoizeau for taking good care of the Ftx KO colony for such a long time. We thank C. Chureau for help in the generation of Ftx KO mice. We thank S. Polo, P.-A. Defossez, S. Ait-Si-Ali, and J.-F. Ouimette for critical reading of the manuscript.
Funding: This work was supported by the Agence Nationale de la Recherche (ANR) (ANR-10-INBS-04) and La Ligue contre le cancer (R03/75-79) to the ImagoSeine core facility of the Institut Jacques Monod. This study was supported by the ATIP/Avenir program (to C.R.) from the Centre National de la Recherche Scientifique (CNRS) and from the Institut National de la Santé et de la Recherche Medicale (INSERM), by ERC starting grant no. 206875 (to C.R.), by the ANR-nonCodiX-14-CE10-0017-01 (to C.R.), and by La Ligue contre le cancer (to C.R.). Works in J.-C.G.’s laboratory were supported by the FOREUM Foundation for Research in Rheumatology (to J.-C.G.) and by the ANR-23-CE15–0002-01 (to J.-C.G.). L.F. was supported by a fellowship from the Région Occitanie/Pyrénées-Méditerranée (no. 1901175) and FOREUM.
Author contributions: Conceptualization: C.H., M.G., J.-C.G., C.R., and C.M. Methodology: C.H., J.-C.G., C.R., and C.M. Investigation: C.H., L.F., A.D., M.M., N.V., F.C., M.S., M.G., and C.M. Visualization: C.H., L.F., F.C., M.S., J.-C.G., and C.M. Supervision: M.G., C.R., and C.M. Writing—original draft: C.H. and C.M. Writing—review and editing: C.H., L.S., M.S., M.G., J.-C.G., C.R., and C.M.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S7
References
Other Supplementary Material for this manuscript includes the following:
Tables S1 to S5
REFERENCES AND NOTES
- 1.Schurz H., Salie M., Tromp G., Hoal E. G., Kinnear C. J., Möller M., The X chromosome and sex-specific effects in infectious disease susceptibility. Hum. Genomics 13, 2 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sauteraud R., Stahl J. M., James J., Englebright M., Chen F., Zhan X., Carrel L., Liu D. J., Inferring genes that escape X-Chromosome inactivation reveals important contribution of variable escape genes to sex-biased diseases. Genome Res. 31, 1629–1637 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sierra I., Anguera M. C., Enjoy the silence: X-chromosome inactivation diversity in somatic cells. Curr. Opin. Genet. Dev. 55, 26–31 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Credendino S. C., Neumayer C., Cantone I., Genetics and epigenetics of sex bias: Insights from human cancer and autoimmunity. Trends Genet. 36, 650–663 (2020). [DOI] [PubMed] [Google Scholar]
- 5.Yuen G. J., Autoimmunity in women: An eXamination of eXisting models. Clin. Immunol. 210, 108270 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klein S. L., Flanagan K. L., Sex differences in immune responses. Nat. Rev. Immunol. 16, 626–638 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Scofield R. H., Bruner G. R., Namjou B., Kimberly R. P., Ramsey-Goldman R., Petri M., Reveille J. D., Alarcón G. S., Vilá L. M., Reid J., Harris B., Li S., Kelly J. A., Harley J. B., Klinefelter’s syndrome (47,XXY) in male systemic lupus erythematosus patients: Support for the notion of a gene-dose effect from the X chromosome. Arthritis Rheum. 58, 2511–2517 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harris V. M., Sharma R., Cavett J., Kurien B. T., Liu K., Koelsch K. A., Rasmussen A., Radfar L., Lewis D., Stone D. U., Kaufman C. E., Li S., Segal B., Wallace D. J., Weisman M. H., Venuturupalli S., Kelly J. A., Alarcon-Riquelme M. E., Pons-Estel B., Jonsson R., Lu X., Gottenberg J.-E., Anaya J.-M., Cunninghame-Graham D. S., Huang A. J. W., Brennan M. T., Hughes P., Alevizos I., Miceli-Richard C., Keystone E. C., Bykerk V. P., Hirschfield G., Xie G., Siminovitch K. A., Ng W.-F., Nordmark G., Bucher S. M., Eriksson P., Omdal R., Rhodus N. L., Rischmueller M., Rohrer M., Wahren-Herlenius M., Witte T., Mariette X., Lessard C. J., Harley J. B., Sivils K. L., Scofield R. H., Klinefelter’s syndrome (47,XXY) is in excess among men with Sjögren’s syndrome. Clin. Immunol. 168, 25–29 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scofield R. H., Lewis V. M., Cavitt J., Kurien B. T., Assassi S., Martin J., Gorlova O., Gregersen P., Lee A., Rider L. G., O’Hanlon T., Rothwell S., Lilleker J., Consortium M. G., Liu X., Kochi Y., Terao C., Igoe A., Stevens W., Sahhar J., Roddy J., Rischmueller M., Lester S., Proudman S., Chen S., Brown M. A., Mayes M. D., Lamb J. A., Miller F. W., 47XXY and 47XXX in Scleroderma and Myositis. ACR Open Rheumatol. 4, 528–533 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cattalini M., Soliani M., Caparello M. C., Cimaz R., Sex differences in pediatric rheumatology. Clin. Rev. Allergy Immunol. 56, 293–307 (2019). [DOI] [PubMed] [Google Scholar]
- 11.Souyris M., Mejía J. E., Chaumeil J., Guéry J.-C., Female predisposition to TLR7-driven autoimmunity: Gene dosage and the escape from X chromosome inactivation. Semin. Immunopathol. 41, 153–164 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Bost C., Arleevskaya M. I., Brooks W. H., Plaza S., Guery J.-C., Renaudineau Y., Long non-coding RNA Xist contribution in systemic lupus erythematosus and rheumatoid arthritis. Clin. Immunol. 236, 108937 (2022). [DOI] [PubMed] [Google Scholar]
- 13.Youness A., Miquel C.-H., Guéry J.-C., Escape from X chromosome inactivation and the female predominance in autoimmune diseases. Int. J. Mol. Sci. 22, 1114 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pisitkun P., Deane J. A., Difilippantonio M. J., Tarasenko T., Satterthwaite A. B., Bolland S., Autoreactive B cell responses to RNA-related antigens due to TLR7 gene duplication. Science 312, 1669–1672 (2006). [DOI] [PubMed] [Google Scholar]
- 15.Deane J. A., Pisitkun P., Barrett R. S., Feigenbaum L., Town T., Ward J. M., Flavell R. A., Bolland S., Control of toll-like receptor 7 expression is essential to restrict autoimmunity and dendritic cell proliferation. Immunity 27, 801–810 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brown G. J., Cañete P. F., Wang H., Medhavy A., Bones J., Roco J. A., He Y., Qin Y., Cappello J., Ellyard J. I., Bassett K., Shen Q., Burgio G., Zhang Y., Turnbull C., Meng X., Wu P., Cho E., Miosge L. A., Andrews T. D., Field M. A., Tvorogov D., Lopez A. F., Babon J. J., López C. A., Gónzalez-Murillo Á., Garulo D. C., Pascual V., Levy T., Mallack E. J., Calame D. G., Lotze T., Lupski J. R., Ding H., Ullah T. R., Walters G. D., Koina M. E., Cook M. C., Shen N., de Lucas Collantes C., Corry B., Gantier M. P., Athanasopoulos V., C. G., TLR7 gain-of-function genetic variation causes human lupus. Nature 605, 349–356 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Loda A., Collombet S., Heard E., Gene regulation in time and space during X-chromosome inactivation. Nat. Rev. Mol. Cell Biol. 23, 231–249 (2022). [DOI] [PubMed] [Google Scholar]
- 18.Souyris M., Cenac C., Azar P., Daviaud D., Canivet A., Grunenwald S., Pienkowski C., Chaumeil J., Mejía J. E., Guéry J.-C., TLR7 escapes X chromosome inactivation in immune cells. Sci. Immunol. 3, eaap8855 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Tukiainen T., Villani A.-C., Yen A., Rivas M. A., Marshall J. L., Satija R., Aguirre M., Gauthier L., Fleharty M., Kirby A., Cummings B. B., Castel S. E., Karczewski K. J., Aguet F., Byrnes A., GTEx Consortium, Lappalainen T., Regev A., Ardlie K. G., Hacohen N., MacArthur D. G., Landscape of X chromosome inactivation across human tissues. Nature 550, 244–248 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berletch J. B., Ma W., Yang F., Shendure J., Noble W. S., Disteche C. M., Deng X., Escape from X inactivation varies in mouse tissues. PLOS Genet. 11, e1005079 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang J., Syrett C. M., Kramer M. C., Basu A., Atchison M. L., Anguera M. C., Unusual maintenance of X chromosome inactivation predisposes female lymphocytes for increased expression from the inactive X. Proc. Natl. Acad. Sci. U.S.A. 113, E2029–E2038 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Syrett C. M., Sindhava V., Sierra I., Dubin A. H., Atchison M., Anguera M. C., Diversity of epigenetic features of the inactive X-chromosome in NK cells, dendritic cells, and macrophages. Front. Immunol. 9, 3087 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Syrett C. M., Sierra I., Beethem Z. T., Dubin A. H., Anguera M. C., Loss of epigenetic modifications on the inactive X chromosome and sex-biased gene expression profiles in B cells from NZB/W F1 mice with lupus-like disease. J. Autoimmun. 107, 102357 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Helyer B. J., Howie J. B., Renal disease associated with positive lupus erythematosus tests in a crossbred strain of mice. Nature 197, 197–197 (1963). [DOI] [PubMed] [Google Scholar]
- 25.Yu B., Qi Y., Li R., Shi Q., Satpathy A. T., Chang H. Y., B cell-specific XIST complex enforces X-inactivation and restrains atypical B cells. Cell 184, 1790–1803.e17 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang T., Ou J., Yildirim E., Xist exerts gene-specific silencing during XCI maintenance and impacts lineage-specific cell differentiation and proliferation during hematopoiesis. Nat. Commun. 13, 4464 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yildirim E., Kirby J. E., Brown D. E., Mercier F. E., Sadreyev R. I., Scadden D. T., Lee J. T., Xist RNA is a potent suppressor of hematologic cancer in mice. Cell 152, 727–742 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang L., Kirby J. E., Sunwoo H., Lee J. T., Female mice lacking Xist RNA show partial dosage compensation and survive to term. Genes Dev. 30, 1747–1760 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang L., Yildirim E., Kirby J. E., Press W., Lee J. T., Widespread organ tolerance to Xist loss and X reactivation except under chronic stress in the gut. Proc. Natl. Acad. Sci. U.S.A. 117, 4262–4272 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chureau C., Chantalat S., Romito A., Galvani A., Duret L., Avner P., Rougeulle C., Ftx is a non-coding RNA which affects Xist expression and chromatin structure within the X-inactivation center region. Hum. Mol. Genet. 20, 705–718 (2011). [DOI] [PubMed] [Google Scholar]
- 31.Chureau C., Prissette M., Bourdet A., Barbe V., Cattolico L., Jones L., Eggen A., Avner P., Duret L., Comparative sequence analysis of the X-inactivation center region in mouse, human, and bovine. Genome Res. 12, 894–908 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Furlan G., Gutierrez Hernandez N., Huret C., Galupa R., van Bemmel J. G., Romito A., Heard E., Morey C., Rougeulle C., The Ftx noncoding locus controls X chromosome inactivation independently of its RNA products. Mol. Cell 70, 462–472.e8 (2018). [DOI] [PubMed] [Google Scholar]
- 33.Hosoi Y., Soma M., Shiura H., Sado T., Hasuwa H., Abe K., Kohda T., Ishino F., Kobayashi S., Female mice lacking Ftx lncRNA exhibit impaired X-chromosome inactivation and a microphthalmia-like phenotype. Nat. Commun. 9, 3829 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soma M., Fujihara Y., Okabe M., Ishino F., Kobayashi S., Ftx is dispensable for imprinted X-chromosome inactivation in preimplantation mouse embryos. Sci. Rep. 4, 5181 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiwrajka N., Anguera M. C., The X in seX-biased immunity and autoimmune rheumatic disease. J. Exp. Med. 219, e20211487 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Duan T., Du Y., Xing C., Wang H. Y., Wang R.-F., Toll-like receptor signaling and its role in cell-mediated immunity. Front. Immunol. 13, 812774 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jenks S. A., Cashman K. S., Woodruff M. C., Lee F. E.-H., Sanz I., Extrafollicular responses in humans and SLE. Immunol. Rev. 288, 136–148 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jenks S. A., Cashman K. S., Zumaquero E., Marigorta U. M., Patel A. V., Wang X., Tomar D., Woodruff M. C., Simon Z., Bugrovsky R., Blalock E. L., Scharer C. D., Tipton C. M., Wei C., Lim S. S., Petri M., Niewold T. B., Anolik J. H., Gibson G., Lee F. E.-H., Boss J. M., Lund F. E., Sanz I., Distinct effector B cells induced by unregulated toll-like receptor 7 contribute to pathogenic responses in systemic lupus erythematosus. Immunity 49, 725–739.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang S., Wang J., Kumar V., Karnell J. L., Naiman B., Gross P. S., Rahman S., Zerrouki K., Hanna R., Morehouse C., Holoweckyj N., Liu H., Autoimmunity Molecular Medicine Team, Manna Z., Goldbach-Mansky R., Hasni S., Siegel R., Sanjuan M., Streicher K., Cancro M. P., Kolbeck R., Ettinger R., IL-21 drives expansion and plasma cell differentiation of autoreactive CD11chiT-bet+ B cells in SLE. Nat. Commun. 9, 1758 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nomura A., Mizuno M., Noto D., Aoyama A., Kuga T., Murayama G., Chiba A., Miyake S., Different spatial and temporal roles of monocytes and monocyte-derived cells in the pathogenesis of an imiquimod induced lupus model. Front. Immunol. 13, 764557 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Santiago-Raber M.-L., Amano H., Amano E., Baudino L., Otani M., Lin Q., Nimmerjahn F., Verbeek J. S., Ravetch J. V., Takasaki Y., Hirose S., Izui S., Fcgamma receptor-dependent expansion of a hyperactive monocyte subset in lupus-prone mice. Arthritis Rheum. 60, 2408–2417 (2009). [DOI] [PubMed] [Google Scholar]
- 42.Kikuchi S., Santiago-Raber M.-L., Amano H., Amano E., Fossati-Jimack L., Moll T., Kotzin B. L., Izui S., Contribution of NZB autoimmunity 2 to Y-linked autoimmune acceleration-induced monocytosis in association with murine systemic lupus. J. Immunol. 176, 3240–3247 (2006). [DOI] [PubMed] [Google Scholar]
- 43.Mauvais-Jarvis F., Bairey Merz N., Barnes P. J., Brinton R. D., Carrero J.-J., DeMeo D. L., De Vries G. J., Epperson C. N., Govindan R., Klein S. L., Lonardo A., Maki P. M., McCullough L. D., Regitz-Zagrosek V., Regensteiner J. G., Rubin J. B., Sandberg K., Suzuki A., Sex and gender: Modifiers of health, disease, and medicine. Lancet 396, 565–582 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fink A. L., Engle K., Ursin R. L., Tang W.-Y., Klein S. L., Biological sex affects vaccine efficacy and protection against influenza in mice. Proc. Natl. Acad. Sci. U.S.A. 115, 12477–12482 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oghumu S., Varikuti S., Stock J. C., Volpedo G., Saljoughian N., Terrazas C. A., Satoskar A. R., Cutting edge: CXCR3 escapes X chromosome inactivation in T cells during infection: Potential implications for sex differences in immune responses. J. Immunol. 203, 789–794 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Youness A., Cenac C., Faz-López B., Grunenwald S., Barrat F. J., Chaumeil J., Mejía J. E., Guéry J.-C., TLR8 escapes X chromosome inactivation in human monocytes and CD4+ T cells. Biol. Sex Differ. 14, 60 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Asano T., Boisson B., Onodi F., Matuozzo D., Moncada-Velez M., Renkilaraj M. R. L. M., Zhang P., Meertens L., Bolze A., Materna M., Korniotis S., Gervais A., Talouarn E., Bigio B., Seeleuthner Y., Bilguvar K., Zhang Y., Neehus A.-L., Ogishi M., Pelham S. J., Le Voyer T., Rosain J., Philippot Q., Soler-Palacín P., Colobran R., Martin-Nalda A., Rivière J. G., Tandjaoui-Lambiotte Y., Chaïbi K., Shahrooei M., Darazam I. A., Olyaei N. A., Mansouri D., Hatipoğlu N., Palabiyik F., Ozcelik T., Novelli G., Novelli A., Casari G., Aiuti A., Carrera P., Bondesan S., Barzaghi F., Rovere-Querini P., Tresoldi C., Franco J. L., Rojas J., Reyes L. F., Bustos I. G., Arias A. A., Morelle G., Kyheng C., Troya J., Planas-Serra L., Schlüter A., Gut M., Pujol A., Allende L. M., Rodriguez-Gallego C., Flores C., Cabrera-Marante O., Pleguezuelo D. E., de Diego R. P., Keles S., Aytekin G., Akcan O. M., Bryceson Y. T., Bergman P., Brodin P., Smole D., Smith C. I. E., Norlin A.-C., Campbell T. M., Covill L. E., Hammarström L., Pan-Hammarström Q., Abolhassani H., Mane S., Marr N., Ata M., Al Ali F., Khan T., Spaan A. N., Dalgard C. L., Bonfanti P., Biondi A., Tubiana S., Burdet C., Nussbaum R., Kahn-Kirby A., Snow A. L., COVID Human Genetic Effort, COVID-STORM Clinicians, COVID Clinicians, Imagine COVID Group, French COVID Cohort Study Group, CoV-Contact Cohort, Amsterdam UMC Covid-19 Biobank, NIAID-USUHS COVID Study Group, Bustamante J., Puel A., Boisson-Dupuis S., Zhang S.-Y., Béziat V., Lifton R. P., Bastard P., Notarangelo L. D., Abel L., Su H. C., Jouanguy E., Amara A., Soumelis V., Cobat A., Zhang Q., Casanova J.-L., X-linked recessive TLR7 deficiency in ~1% of men under 60 years old with life-threatening COVID-19. Sci. Immunol. 6, eabl4348 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van der Made C. I., Simons A., Schuurs-Hoeijmakers J., van den Heuvel G., Mantere T., Kersten S., van Deuren R. C., Steehouwer M., van Reijmersdal S. V., Jaeger M., Hofste T., Astuti G., Corominas Galbany J., van der Schoot V., van der Hoeven H., Wanda Hagmolen Of Ten Have, Klijn E., van den Meer C., Fiddelaers J., de Mast Q., Bleeker-Rovers C. P., Joosten L. A. B., Yntema H. G., Gilissen C., Nelen M., van der Meer J. W. M., Brunner H. G., Netea M. G., van de Veerdonk F. L., Hoischen A., Presence of genetic variants among young men with severe COVID-19. JAMA 324, 663–673 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fallerini C., Daga S., Mantovani S., Benetti E., Picchiotti N., Francisci D., Paciosi F., Schiaroli E., Baldassarri M., Fava F., Palmieri M., Ludovisi S., Castelli F., Quiros-Roldan E., Vaghi M., Rusconi S., Siano M., Bandini M., Spiga O., Capitani K., Furini S., Mari F., GEN-COVID Multicenter Study, Renieri A., Mondelli M. U., Frullanti E., Association of toll-like receptor 7 variants with life-threatening COVID-19 disease in males: Findings from a nested case-control study. eLife 10, e67569 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Odhams C. A., Roberts A. L., Vester S. K., Duarte C. S. T., Beales C. T., Clarke A. J., Lindinger S., Daffern S. J., Zito A., Chen L., Jones L. L., Boteva L., Morris D. L., Small K. S., Fernando M. M. A., Cunninghame Graham D. S., Vyse T. J., Interferon inducible X-linked gene CXorf21 may contribute to sexual dimorphism in systemic lupus erythematosus. Nat. Commun. 10, 2164 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Soni C., Perez O. A., Voss W. N., Pucella J. N., Serpas L., Mehl J., Ching K. L., Goike J., Georgiou G., Ippolito G. C., Sisirak V., Reizis B., Plasmacytoid dendritic cells and type I interferon promote extrafollicular B cell responses to extracellular self-DNA. Immunity 52, 1022–1038.e7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Soni C., Wong E. B., Domeier P. P., Khan T. N., Satoh T., Akira S., Rahman Z. S. M., B cell-intrinsic TLR7 signaling is essential for the development of spontaneous germinal centers. J. Immunol. 193, 4400–4414 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Manni M., Gupta S., Ricker E., Chinenov Y., Park S. H., Shi M., Pannellini T., Jessberger R., Ivashkiv L. B., Pernis A. B., Regulation of age-associated B cells by IRF5 in systemic autoimmunity. Nat. Immunol. 19, 407–419 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ricker E., Manni M., Flores-Castro D., Jenkins D., Gupta S., Rivera-Correa J., Meng W., Rosenfeld A. M., Pannellini T., Bachu M., Chinenov Y., Sculco P. K., Jessberger R., Prak E. T. L., Pernis A. B., Altered function and differentiation of age-associated B cells contribute to the female bias in lupus mice. Nat. Commun. 12, 4813 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dossin F., Pinheiro I., Żylicz J. J., Roensch J., Collombet S., Le Saux A., Chelmicki T., Attia M., Kapoor V., Zhan Y., Dingli F., Loew D., Mercher T., Dekker J., Heard E., SPEN integrates transcriptional and epigenetic control of X-inactivation. Nature 578, 455–460 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McAlpine W., Sun L., Wang K.-W., Liu A., Jain R., San Miguel M., Wang J., Zhang Z., Hayse B., McAlpine S. G., Choi J. H., Zhong X., Ludwig S., Russell J., Zhan X., Choi M., Li X., Tang M., Moresco E. M. Y., Beutler B., Turer E., Excessive endosomal TLR signaling causes inflammatory disease in mice with defective SMCR8-WDR41-C9ORF72 complex function. Proc. Natl. Acad. Sci. U.S.A. 115, E11523–E11531 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jiwrajka N., Toothacre N. E., Beethem Z. T., Sting S., Forsyth K. S., Dubin A. H., Driscoll A., Stohl W., Anguera M. C., Impaired dynamic X-chromosome inactivation maintenance in T cells is a feature of spontaneous murine SLE that is exacerbated in female-biased models. J. Autoimmun. 139, 103084 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ridings-Figueroa R., Stewart E. R., Nesterova T. B., Coker H., Pintacuda G., Godwin J., Wilson R., Haslam A., Lilley F., Ruigrok R., Bageghni S. A., Albadrani G., Mansfield W., Roulson J.-A., Brockdorff N., Ainscough J. F. X., Coverley D., The nuclear matrix protein CIZ1 facilitates localization of Xist RNA to the inactive X-chromosome territory. Genes Dev. 31, 876–888 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Crawford J. D., Wang H., Trejo-Zambrano D., Cimbro R., Talbot C. C., Thomas M. A., Curran A. M., Girgis A. A., Schroeder J. T., Fava A., Goldman D. W., Petri M., Rosen A., Antiochos B., Darrah E., The XIST lncRNA is a sex-specific reservoir of TLR7 ligands in SLE. JCI Insight 8, e169344 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dou D. R., Zhao Y., Belk J. A., Zhao Y., Casey K. M., Chen D. C., Li R., Yu B., Srinivasan S., Abe B. T., Kraft K., Hellström C., Sjöberg R., Chang S., Feng A., Goldman D. W., Shah A. A., Petri M., Chung L. S., Fiorentino D. F., Lundberg E. K., Wutz A., Utz P. J., Chang H. Y., Xist ribonucleoproteins promote female sex-biased autoimmunity. Cell 187, 733–749.e16 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Djeghloul D., Kuranda K., Kuzniak I., Barbieri D., Naguibneva I., Choisy C., Bories J.-C., Dosquet C., Pla M., Vanneaux V., Socié G., Porteu F., Garrick D., Goodhardt M., Age-associated decrease of the histone methyltransferase SUV39H1 in HSC perturbs heterochromatin and B lymphoid differentiation. Stem Cell Rep. 6, 970–984 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu Y., Sinke L., Jonkman T. H., Slieker R. C., BIOS Consortium, van Zwet E. W., Daxinger L., Heijmans B. T., The inactive X chromosome accumulates widespread epigenetic variability with age. Clin. Epigenetics 15, 135 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Denkinger M. D., Leins H., Schirmbeck R., Florian M. C., Geiger H., HSC aging and senescent immune remodeling. Trends Immunol. 36, 815–824 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Krasselt M., Baerwald C., Sex, symptom severity, and quality of life in rheumatology. Clin Rev Allergy Immunol 56, 346–361 (2019). [DOI] [PubMed] [Google Scholar]
- 65.Wang Y., Roussel-Queval A., Chasson L., Hanna Kazazian N., Marcadet L., Nezos A., Sieweke M. H., Mavragani C., Alexopoulou L., TLR7 signaling drives the development of Sjögren’s syndrome. Front. Immunol. 12, 676010 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Björkegren J. L. M., Lusis A. J., Atherosclerosis: Recent developments. Cell 185, 1630–1645 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Márquez E. J., Chung C., Marches R., Rossi R. J., Nehar-Belaid D., Eroglu A., Mellert D. J., Kuchel G. A., Banchereau J., Ucar D., Sexual-dimorphism in human immune system aging. Nat. Commun. 11, 751 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Debrand E., Chureau C., Arnaud D., Avner P., Heard E., Functional analysis of the DXPas34 locus, a 3′ regulator of Xist expression. Mol. Cell. Biol. 19, 8513–8525 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Itoh Y., Golden L. C., Itoh N., Matsukawa M. A., Ren E., Tse V., Arnold A. P., Voskuhl R. R., The X-linked histone demethylase Kdm6a in CD4+ T lymphocytes modulates autoimmunity. J. Clin. Invest. 129, 3852–3863 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Syrett C. M., Paneru B., Sandoval-Heglund D., Wang J., Banerjee S., Sindhava V., Behrens E. M., Atchison M., Anguera M. C., Altered X-chromosome inactivation in T cells may promote sex-biased autoimmune diseases. JCI Insight 4, 126751 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Demaria O., Pagni P. P., Traub S., de Gassart A., Branzk N., Murphy A. J., Valenzuela D. M., Yancopoulos G. D., Flavell R. A., Alexopoulou L., TLR8 deficiency leads to autoimmunity in mice. J. Clin. Invest. 120, 3651–3662 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Libert C., Dejager L., Pinheiro I., The X chromosome in immune functions: When a chromosome makes the difference. Nat. Rev. Immunol. 10, 594–604 (2010). [DOI] [PubMed] [Google Scholar]
- 73.Becker-Herman S., Meyer-Bahlburg A., Schwartz M. A., Jackson S. W., Hudkins K. L., Liu C., Sather B. D., Khim S., Liggitt D., Song W., Silverman G. J., Alpers C. E., Rawlings D. J., WASp-deficient B cells play a critical, cell-intrinsic role in triggering autoimmunity. J. Exp. Med. 208, 2033–2042 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mohr A., Atif M., Balderas R., Gorochov G., Miyara M., The role of FOXP3+ regulatory T cells in human autoimmune and inflammatory diseases. Clin. Exp. Immunol. 197, 24–35 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yu H.-H., Yang Y.-H., Chiang B.-L., Chronic granulomatous disease: A comprehensive review. Clin Rev Allergy Immunol 61, 101–113 (2021). [DOI] [PubMed] [Google Scholar]
- 76.Borziak K., Finkelstein J., X-linked genetic risk factors that promote autoimmunity and dampen remyelination are associated with multiple sclerosis susceptibility. Mult. Scler. Relat. Disord. 66, 104065 (2022). [DOI] [PubMed] [Google Scholar]
- 77.Lian X., Xiao R., Hu X., Kanekura T., Jiang H., Li Y., Wang Y., Yang Y., Zhao M., Lu Q., DNA demethylation of CD40l in CD4+ T cells from women with systemic sclerosis: A possible explanation for female susceptibility. Arthritis Rheum. 64, 2338–2345 (2012). [DOI] [PubMed] [Google Scholar]
- 78.Liu Y., Liao J., Zhao M., Wu H., Yung S., Chan T. M., Yoshimura A., Lu Q., Increased expression of TLR2 in CD4+ T cells from SLE patients enhances immune reactivity and promotes IL-17 expression through histone modifications. Eur. J. Immunol. 45, 2683–2693 (2015). [DOI] [PubMed] [Google Scholar]
- 79.Su L.-C., Xu W.-D., Huang A.-F., IRAK family in inflammatory autoimmune diseases. Autoimmun. Rev. 19, 102461 (2020). [DOI] [PubMed] [Google Scholar]
- 80.de Jesus A. A., Hou Y., Brooks S., Malle L., Biancotto A., Huang Y., Calvo K. R., Marrero B., Moir S., Oler A. J., Deng Z., Montealegre Sanchez G. A., Ahmed A., Allenspach E., Arabshahi B., Behrens E., Benseler S., Bezrodnik L., Bout-Tabaku S., Brescia A. C., Brown D., Burnham J. M., Caldirola M. S., Carrasco R., Chan A. Y., Cimaz R., Dancey P., Dare J., DeGuzman M., Dimitriades V., Ferguson I., Ferguson P., Finn L., Gattorno M., Grom A. A., Hanson E. P., Hashkes P. J., Hedrich C. M., Herzog R., Horneff G., Jerath R., Kessler E., Kim H., Kingsbury D. J., Laxer R. M., Lee P. Y., Lee-Kirsch M. A., Lewandowski L., Li S., Lilleby V., Mammadova V., Moorthy L. N., Nasrullayeva G., O’Neil K. M., Onel K., Ozen S., Pan N., Pillet P., Piotto D. G., Punaro M. G., Reiff A., Reinhardt A., Rider L. G., Rivas-Chacon R., Ronis T., Rösen-Wolff A., Roth J., Ruth N. M., Rygg M., Schmeling H., Schulert G., Scott C., Seminario G., Shulman A., Sivaraman V., Son M. B., Stepanovskiy Y., Stringer E., Taber S., Terreri M. T., Tifft C., Torgerson T., Tosi L., Van Royen-Kerkhof A., Wampler Muskardin T., Canna S. W., Goldbach-Mansky R., Distinct interferon signatures and cytokine patterns define additional systemic autoinflammatory diseases. J. Clin. Invest. 130, 1669–1682 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lee Y., Wessel A. W., Xu J., Reinke J. G., Lee E., Kim S. M., Hsu A. P., Zilberman-Rudenko J., Cao S., Enos C., Brooks S. R., Deng Z., Lin B., de Jesus A. A., Hupalo D. N., Piotto D. G., Terreri M. T., Dimitriades V. R., Dalgard C. L., Holland S. M., Goldbach-Mansky R., Siegel R. M., Hanson E. P., Genetically programmed alternative splicing of NEMO mediates an autoinflammatory disease phenotype. J. Clin. Invest. 132, e128808 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ye X.-P., Yuan F.-F., Zhang L.-L., Ma Y.-R., Zhang M.-M., Liu W., Sun F., Wu J., Lu M., Xue L.-Q., Shi J.-Y., Zhao S.-X., Song H.-D., Liang J., Zheng C.-X., ITM2A expands evidence for genetic and environmental interaction in graves disease pathogenesis. J. Clin. Endocrinol. Metab. 102, 652–660 (2017). [DOI] [PubMed] [Google Scholar]
- 83.McDonald C., Xanthopoulos C., Kostareli E., The role of Bruton’s tyrosine kinase in the immune system and disease. Immunology 164, 722–736 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang J., Zhang Y., Yang J., Zhang L., Sun L., Pan H.-F., Hirankarn N., Ying D., Zeng S., Lee T. L., Lau C. S., Chan T. M., Leung A. M. H., Mok C. C., Wong S. N., Lee K. W., Ho M. H. K., Lee P. P. W., Chung B. H.-Y., Chong C. Y., Wong R. W. S., Mok M. Y., Wong W. H. S., Tong K. L., Tse N. K. C., Li X.-P., Avihingsanon Y., Rianthavorn P., Deekajorndej T., Suphapeetiporn K., Shotelersuk V., Ying S. K. Y., Fung S. K. S., Lai W. M., Garcia-Barceló M.-M., Cherny S. S., Tam P. K.-H., Cui Y., Sham P. C., Yang S., Ye D. Q., Zhang X.-J., Lau Y. L., Yang W., Three SNPs in chromosome 11q23.3 are independently associated with systemic lupus erythematosus in Asians. Hum. Mol. Genet. 23, 524–533 (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S7
References
Tables S1 to S5