Abstract
eIF4E plays key roles in protein synthesis and tumorigenesis. It is phosphorylated by the kinases MNK1 and MNK2. Binding of MNKs to eIF4G enhances their ability to phosphorylate eIF4E. Here, we show that mTORC1, a key regulator of mRNA translation and oncogenesis, directly phosphorylates MNK2 on Ser74. This suppresses MNK2 activity and impairs binding of MNK2 to eIF4G. These effects provide a novel mechanism by which mTORC1 signaling impairs the function of MNK2 and thereby decreases eIF4E phosphorylation. MNK2[S74A] knock-in cells show enhanced phosphorylation of eIF4E and S6K1 (i.e., increased mTORC1 signaling), enlarged cell size, and increased invasive and transformative capacities. MNK2[Ser74] phosphorylation was inversely correlated with disease progression in human prostate tumors. MNK inhibition exerted anti-proliferative effects in prostate cancer cells in vitro. These findings define a novel feedback loop whereby mTORC1 represses MNK2 activity and oncogenic signaling through eIF4E phosphorylation, allowing reciprocal regulation of these two oncogenic pathways.
Electronic supplementary material
The online version of this article (10.1007/s00018-020-03491-1) contains supplementary material, which is available to authorized users.
Keywords: Rapamycin, Prostate cancer, eIF4E, Protein synthesis, mRNA translation
Introduction
Eukaryotic initiation factor 4E (eIF4E) interacts with the 5′-terminal cap structure of mRNAs and plays a crucial role in mRNA translation (protein synthesis) by mediating recruitment of ribosomes to mRNAs. eIF4E also binds the scaffold protein, eIF4G, which in turn associates with other proteins including eIF3, which recruits the 40S ribosomal subunit.
Dysregulation of eIF4E, through its increased expression, its phosphorylation or activation of signaling through mTORC1 (the mammalian [or mechanistic] target of rapamycin complex 1), is strongly implicated in oncogenesis [1, 2]. eIF4E has thus emerged as a potential target in cancer therapy, because, for example, phosphorylation of eIF4E is needed for tumorigenesis in prostate cancer [1]. Therefore, understanding the interplay between the pathways that converge on eIF4E is of critical importance. eIF4E is phosphorylated, on Ser209 [3, 4], by protein kinases termed MAP kinase-interacting kinases (MNKs). The MNKs are the only kinases that phosphorylate eIF4E on Ser209 [5] and phosphorylation of eIF4E affects the translation of specific mRNAs [6].
Notably, the MNKs are linked to oncogenesis and tumor progression [7, 8]. MNKs interact with eIF4G, which allows them to efficiently phosphorylate eIF4E [9]. MNKs bind to, and are phosphorylated by, MAP kinases, in particular ERK, leading to their activation [10]. ERK signaling is activated in a significant proportion of human tumors [11].
There are two MNK genes in mammals, MKNK1 and MKNK2. In humans, each gives rise to two mRNAs by alternative splicing to produce four distinct proteins which contain the same N-terminal and catalytic regions but differ at their C-termini [12–14]. The longer ‘a’ forms, which contain MAP kinase-binding sites, are termed MNK1a and MNK2a, while the shorter ‘b’ forms, which lack this feature, are termed MNK1b and MNK2b. MNK1a and MNK2a differ in their intrinsic activity, regulation and subcellular localisation [10]. MNK1a is acutely activated by signaling through ERK and p38 MAP kinase [4, 15]. In contrast, MNK2a has high basal activity that is only enhanced slightly by these pathways [12]. Indeed, it is not clear how the activity of MNK2a or its ability to phosphorylate eIF4E is regulated.
eIF4E binds to small phosphoproteins, called eIF4E-binding proteins (4E-BPs), which compete with eIF4G for binding to eIF4E [16]. 4E-BPs are negatively regulated by phosphorylation by mTORC1, a heteromeric protein kinase [17] which promotes multiple anabolic processes. When bound to a 4E-BP, eIF4E cannot bind eIF4G and promote translation initiation. By catalysing the phosphorylation of 4E-BPs, and bringing about their release from eIF4E, mTORC1 positively regulates translation initiation [18] and allows eIF4E and MNKs to come into closer proximity through their interactions with eIF4G.
mTORC1 is activated by the phosphatidylinositol (PI) 3-kinase/Akt [protein kinase B (PKB/Akt)] and Ras/Raf/MEK/MAP kinase signaling pathways that are frequently constitutively activated in cancers [11, 19]. Rapamycin and its analogues, which interfere with substrate access to the active site of mTORC1, have been tested clinically to treat a number of cancers [20]. So-called ‘second generation’ mTOR inhibitors (mTORis), which act as ATP-competitive inhibitors and thus inhibit both mTORC1 and mTORC2, are also currently being evaluated in clinical trials [20].
Rapamycin increases the phosphorylation of eIF4E in various cancer cell lines [21–26]. Since phosphorylation of eIF4E is associated with cell transformation and migration, this could limit the effectiveness of drugs like rapamycin in cancer therapy. As such, it is critical to understand the links between mTORC1 signaling and eIF4E phosphorylation. Although it is well established that mTORC1 and MNKs are often activated in cancer and may drive oncogenesis, there is surprisingly little information on the links between these pathways.
We have previously shown that rapamycin increases the activity of MNK2a [21]. Here, we show that mTORC1 directly phosphorylates MNK2 on Ser74, which suppresses MNK2 activity and decreases the association of MNK2a with eIF4G, an interaction that is required for efficient phosphorylation of eIF4E [9]. We also demonstrate that complete inhibition of mTORC1 disrupts the interaction between MNK2 and eIF4G, thus rendering MNK2 unable to phosphorylate any remaining eIF4G-bound eIF4E. Phosphorylation of MNK2 by mTORC1 thus provides a feedback mechanism which can restrain MNK signaling. Mutation of Ser74 to alanine in MNK2 results in enhanced eIF4E phosphorylation and increased mTORC1 activity, and is associated with increased cell size, and elevated cellular invasive and enhanced transformation capacities. Finally, MNK2 Ser74 was inversely correlated with disease progression in human prostate tumors. Furthermore, selective inhibition of MNK signalling exhibited strong anti-proliferative effects in prostate cancer cells in vitro.
Taken together, our findings identify a novel regulatory link between the MNK2 and mTORC1 signaling pathways, which has clear implications for the design of future cancer treatment strategies targeting these oncogenic signaling pathways.
Results
mTOR directly phosphorylates MNK2a on Ser74
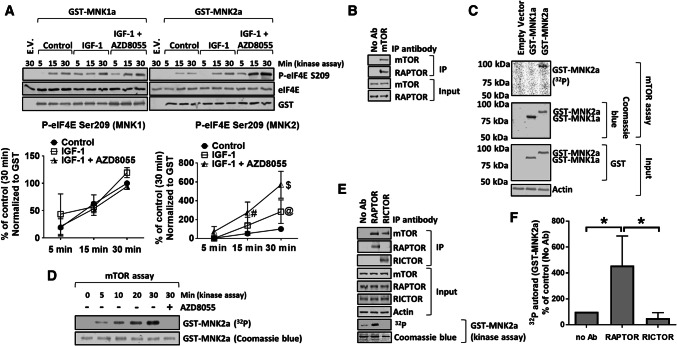
To assess the effects of mTOR signaling on MNK activity, we expressed GST-tagged MNK2a in HEK293 cells, which were subsequently treated with insulin growth factor-1 (IGF-1) with or without AZD8055, a second-generation mTORi [27]. After purification of GST-tagged MNK2a, its activity was measured in vitro using recombinant eIF4E as a substrate (Fig. 1a). IGF-1 had little effect on MNK2a activity, consistent with its limited ability to activate ERK in these cells (Fig. S1; compare with data for TPA (12-O-tetradecanoylphorbol-13-acetate) [28]). Treatment of IGF-1-stimulated cells with AZD8055 markedly enhanced the activity of MNK2a (Fig. 1a). AZD8055 did not affect MNK1a activity (Fig. 1a), implying that the activity MNK1a is not modulated by mTORCs.
Fig. 1.
MNK2a is a direct substrate for mTOR. a HEK293 cells were transfected with vectors encoding WT GST-MNK2a; 32 h later, cells were starved of serum for 16 h and then transferred to KRB for 30 min. Cells were then treated with IGF-1 for 30 min in the presence or absence of 1 µM AZD8055. GST-MNK1a/2a was then isolated from the lysates on glutathione beads and subjected to MNK kinase assay for the indicated times using recombinant eIF4E as substrate. Assay products were separated and analysed by SDS-PAGE/Western blotting (WB). E.V empty vector. b mTOR was immunoprecipitated (IP) from lysates of HEK293 cells cultured in growth medium. The presence of mTOR and RAPTOR in the immunoprecipitates and input lysates was assessed by SDS-PAGE/WB. c HEK293 cells were transfected with vectors for WT GST-MNK1a or GST-MNK2a; cells were starved of serum for 16 h followed by incubation in KRB for 1 h. Cells were then lysed and GST-MNKs were pulled down and then eluted from glutathione beads. The presence of GST-MNK1a or GST-MNK2a was verified by SDS-PAGE/WB. Eluted GST-MNKs were incubated with immunoprecipitates (containing mTOR) from b for 30 min in assays including [γ-32P]ATP. Radioactivity was detected using a phosphorimager. d mTOR kinase assays were carried out as in c for the indicated periods of time. e mTORC1 or 2 was immunoprecipitated with antibodies to RAPTOR or RICTOR, respectively. Kinase assays were then carried out using [γ-32P]ATP and GST-MNK2a as substrate (for 30 min; with detection by phosphorimager). f Quantification of data in e. Quantification of data in panels a, f is presented as means ± SD. n = 3. *0.01 ≤ P < 0.05 (one-way ANOVA). @0.01 ≤ P < 0.05; #0.001 ≤ P < 0.01; $P < 0.001 (two-way ANOVA)
Given that MNK2 binds to the RAPTOR component of mTORC1 [29], a property shared with known mTORC1 substrates [30, 31], we tested whether mTORC1 could phosphorylate MNK2. mTOR was immunoprecipitated from HEK293 cell lysates (Fig. 1b) and its activity was tested against GST-MNK proteins, expressed in and purified from HEK293 cells (Fig. 1c, d). mTOR phosphorylated GST-MNK2a but not GST-MNK1a (human, unless otherwise specified; Fig. 1c, d). To assess whether mTORC1 or mTORC2 phosphorylated MNK2a, we immunoprecipitated these complexes separately using antibodies to RAPTOR or RICTOR, respectively. Anti-RAPTOR immunoprecipitates phosphorylated MNK2a, while anti-RICTOR immunoprecipitates did not (Fig. 1e, f), indicating that MNK2a is a specific substrate for mTORC1, consistent with RAPTOR-binding MNK2 [29].
It would be informative to test the behavior of endogenous MNK2a; however, no suitable antibodies were available. We did test three antibodies marketed as detecting ‘total’ MNK2 but none recognized the endogenous MNK2 protein (Fig. S2).
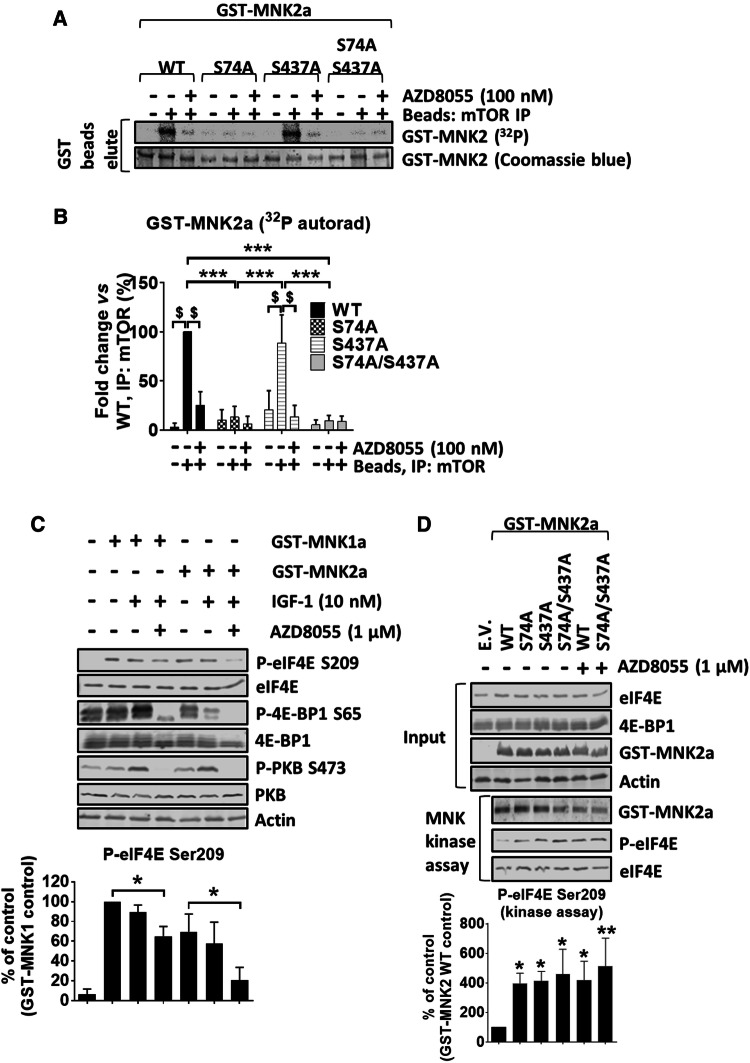
Using the human PC3 prostate cancer cell line, we previously identified two highly conserved residues in MNK2a (Ser74 and Ser437, see Fig. S3) whose phosphorylation was altered in response to rapamycin treatment [21], although the mechanism explaining this effect was unclear. We, therefore, generated mutants of MNK2a in which Ser74 or/and Ser437 were changed to alanine (S74A and S437A, respectively) and tested whether they were still phosphorylated by mTOR (Fig. 2a, b). As expected, immunoprecipitated mTOR phosphorylated recombinant GST-4E-BP1 (a positive control). This was inhibited by AZD8055 (Fig. S4A), as was the phosphorylation of MNK2a (Fig. 2a, b), confirming that mTOR (rather than a contaminating kinase) phosphorylates MNK2a. While the GST-MNK2a[S437A] was phosphorylated to a similar extent as wild-type (WT) MNK2a, the S74A and S74A/S437A mutants of GST-MNK2a were not phosphorylated by mTOR. Similarly, immunoprecipitated mTORC1 (from a RAPTOR pull-down) phosphorylated GST-MNK2a[WT] and GST-MNK2a[S437A], but not GST-MNK2a[S74A] or [S74A/S437A] (Fig. S4B and C). In contrast, neither GST-MNK1a nor GST-MNK2a, could be phosphorylated by S6K1 [32], while a kinase-dead version of eEF2K {GST-eEF2K[K170M] [33]}, an established S6K substrate [32] was (Fig. S4D and E). Taken together, these data demonstrate that mTOR directly phosphorylates MNK2a on Ser74, while Ser437 appears to be the target of a different, yet to be identified, kinase.
Fig. 2.
mTOR phosphorylates MNK2a on Ser74. a mTOR kinase assays were performed with mTOR IPed as in Fig. 1b, and, as substrates, GST-MNK2a (WT or mutants) that had been purified as in Fig. 1c. b Quantitation of a. c HEK293 cells were treated as in Fig. 1a, after SDS-PAGE, phosphorylated (P-) or total proteins were detected as indicated. d HEK293 cells were transfected with indicated GST-MNK2a constructs. Cells were starved of serum for 16 h and then kept in KRB in the presence or absence of AZD8055 for 30 min. GST-MNK2a proteins were then pulled down with glutathione beads and assayed for kinase activity against recombinant eIF4E as substrate. Quantification of data in panel B and D is presented as means ± SD, n = 3. *0.01 ≤ P < 0.05; ***P < 0.001 (one-way ANOVA); $P < 0.001 (two-way ANOVA)
AZD8055 decreased the elevated level of eIF4E phosphorylation seen in HEK293 cells overexpressing MNK2a (presumably reflecting the disruption of eIF4G–MNK2 binding, see Fig. 3b–e below; also, when MNKs are overexpressed, the role of eIF4G as a scaffold is reduced or eliminated), but not in cells overexpressing MNK1a (Figs. 2c, S4F). Overexpression of GST-MNK2a[S74A], [S437A] or [S74A/S437A] further enhanced eIF4E phosphorylation compared to the level seen when WT GST-MNK2a was overexpressed (Fig. 2d). We also generated constructs in which the corresponding serine residues in GST-MNK1a were substituted with alanine (S39A and S396A mutants, respectively). Expressing them exogenously in HEK293 cells led to similar increases in eIF4E phosphorylation as were observed with WT GST-MNK1a (Fig. S4F), indicating these residues do not alter MNK1 function.
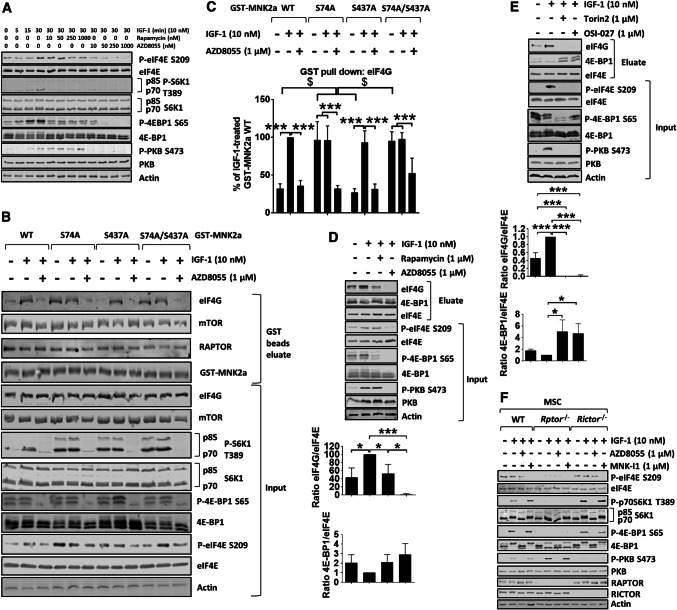
Fig. 3.
mTOR kinase inhibitors, but not rapamycin, reduce eIF4E phosphorylation in vitro and in vivo. a HEK293 cells were starved of serum for 16 h, and then kept in KRB in the presence of the indicated concentrations of rapamycin or AZD8055 for 30 min, before stimulation with IGF-1 (10 nM) for the indicated times. b HEK293 cells were transfected with GST-MNK2a constructs. 32 h later, cells were starved of serum for 16 h and then kept in KRB in the presence of AZD8055 for 30 min, before stimulation with IGF-1 for another 30 min and lysis. GST-MNK2a proteins were then pulled down with glutathione beads. Both eluates and input lysates were subjected to immunoblotting analysis for the indicated proteins. c Quantification of eIF4G binding from b. Results are given as means ± SD. d HEK293 cells were starved of serum for 16 h and then kept in KRB in the absence or presence of rapamycin or AZD8055 for 30 min. eIF4E and associated proteins were then isolated by affinity chromatography on immobilised m7GTP, followed by analysis using SDS-PAGE/WB. e HEK293 cells were starved of serum for 16 h and then kept in KRB in the absence or presence of Torin 2 or OSI-027 for 30 min, followed by stimulation with IGF-1 for another 30 min. eIF4E and associated proteins were isolated by affinity chromatography on immobilised m7GTP, followed by SDS-PAGE/WB. f WT, Rptor−/− and Rictor−/− MSCs were serum starved for 4 h before incubation in KRB in the presence of AZD8055 or MNK-I1 for 30 min, followed by the addition of IGF-1 for another 30 min. Quantification of data in panels c–e is presented as means ± SD, n = 3. *0.01 ≤ P < 0.05; **0.001 ≤ P < 0.01; ***P < 0.001 (one-way ANOVA); $P < 0.001 (two-way ANOVA)
MNK2a associates with mTORC1 but not mTORC2 [29]. To identify the region of MNK2a responsible for binding mTORC1, truncated versions of MNK2a, as short as containing the first 59 amino acids, were immunoprecipitated with mTOR (Fig. S4G). This region of MNK2a is longer than the N-terminal part of MNK1a (Fig. S3A). The mTORC1 component, RAPTOR [30, 34], binds partner proteins (i.e., substrates for mTORC1) via their TOR signaling (TOS) motifs [35, 36]. Known TOS motifs contain a crucial phenylalanine residue [35, 36]. We, therefore, mutated each of the phenylalanine residues within residues 1–59 of MNK2a individually to alanine residues. Each of the resulting GST-MNK2a mutants still bound to RAPTOR (Fig. S4H, I), suggesting that MNK2a may contain multiple TOS motifs and/or non-canonical TOR-binding motif(s). Therefore, the RAPTOR-binding site of MNK2a lies within its N-terminus, but MNK2a does not bind RAPTOR through a single canonical TOS motif.
Inhibition of mTOR kinase activity disrupts the association of MNK2 with eIF4G
The above data indicate that mTORC1-mediated phosphorylation of MNK2a impairs its activity. This is consistent with the ability of the ‘allosteric’ mTORC1 inhibitor, rapamycin, to increase P-eIF4E. However, given that both rapamycin and the ‘second generation’ mTORis, such as AZD8055 [27], completely block the activity of both mTOR complexes, we wanted to compare their effects on P-eIF4E levels. In HEK293 cells (Fig. 3a quantified in Fig. S5A), AZD8055, but not rapamycin, attenuated eIF4E phosphorylation. Indeed, every mTORi tested, including AZD8055 (Fig. 3a), Torin 2 [37] and OSI-027 [38] (Fig. S5B), reduced eIF4E phosphorylation. Thus, consistent with previous studies [1, 23], rapamycin and mTORis exert opposing effects on eIF4E phosphorylation.
MNK2a showed only low binding to eIF4G under serum-starved conditions, while MNK2a/eIF4G binding increased markedly following treatment of cells with IGF-1 (Fig. 3b, c). This provides a mechanism by which IGF-1 can enhance the ability of MNK2a to phosphorylate eIF4E, even though IGF-1 has little effect on the intrinsic activity of MNK2a (Fig. 1a). The IGF-1-induced binding of eIF4G to MNK2a was not affected by rapamycin (Fig. S5C and D). In contrast, it was strongly decreased by AZD8055 (Figs. 3b, c, S5C, D). These findings also help to explain why AZD8055 decreases P-eIF4E levels while rapamycin does not.
To assess whether phosphorylation of the mTORC1 site in MNK2a, Ser74, played a role in modulating the association of eIF4G with MNK2a, we examined the behavior of the MNK2a[S74A] mutant. It showed strikingly higher basal association with eIF4G, which was prevented by AZD8055 (Figs. 3b, c, S5C, D). The S74A/S437A mutant behaved similarly to MNK2a[S74A], while the MNK2a[S437A] variant showed IGF-1-induced binding to eIF4G, similar to WT MNK2a (Fig. 3b, c). Thus, phosphorylation of Ser74 inhibits the basal association of MNK2a with eIF4G. Ser74 lies immediately C-terminal to the eIF4G-binding site on MNK2 (Fig. S3A).
Interestingly, rapamycin did not decrease the association with eIF4G of WT MNK2a or the S74A mutant (Fig. S5C, D). The AZD8055-induced decrease in eIF4G/MNK2a binding cannot be explained by dephosphorylation of Ser74 and must involve additional events. These might include mTOR-dependent alterations in the phosphorylation of eIF4G [39], in which at least seven phosphorylation events are sensitive to rapamycin, and/or its conformation/interaction with other partner proteins, perhaps its binding to eIF4E, which is blocked by mTORis but not by rapamycin (Fig. 3d, e). It was also notable that GST-MNK2a associated with mTOR and/or RAPTOR (i.e., mTORC1) but GST-MNK1a did not (Fig. S6A–E), consistent with the data presented above and the findings of Brown and Gromeier [29].
The effects of AZD8055 might involve the inhibition of mTORC1, mTORC2 or both. To distinguish between these possibilities, we generated cells that lacked RAPTOR or RICTOR. Mesenchymal stem cells (MSCs) were isolated from Rptorfl/fl and Rictorfl/fl mice and tamoxifen used to induce the expression of Cre recombinase, resulting in deletion of Rptor and Rictor (Fig. 3f). GST-MNK2a did not bind to mTOR in tamoxifen-treated Rptorfl/fl MSCs (Fig. S6F), implying that MNK2a interacts with mTOR via RAPTOR. Interestingly, S6K1 phosphorylation was enhanced in cells lacking RICTOR, most likely as a result of the available mTOR protein binding RAPTOR to form the mTORC1 complex, while phosphorylation of 4E-BP1 was not affected (Fig. 3f). This may well reflect the facts that S6Ks and 4E-BP1 are, respectively, ‘weak’ and ‘strong’ substrates for mTORC1 [40]. Conversely, phosphorylation of PKB/Akt at Ser473 was enhanced in Rptor−/− cells, most likely as a result of more mTOR forming mTORC2 and/or release of mTORC1 from upstream negative feedback mechanisms mediated through mTORC2 [17]. As expected, phosphorylation of 4E-BP1 was strongly decreased in Rptor−/− cells (Fig. 3f). Importantly, P-eIF4E was also markedly lower in Rptor−/−cells (Fig. 3f). These data suggest that complete inhibition of mTORC1, rather than suppression of mTORC2 function, impairs the phosphorylation of eIF4E.
The inhibitory effect of AZD8055 on the phosphorylation of eIF4E by endogenous MNKs could also reflect sequestration of eIF4E by dephosphorylated 4E-BPs, causing decreased levels of eIF4G/MNK complexes. Lysates from HEK293 cells which had been pre-treated with rapamycin, or an mTORi, and then stimulated with IGF-1, were subjected to affinity chromatography on beads with immobilised 7-methyl GTP (m7GTP, cap analog, Fig. 3d, e). As expected from its ability to stimulate mTORC1 signaling, IGF-1 treatment reduced the association of eIF4E with 4E-BP1 and enhanced its binding to eIF4G (Fig. 3d, e). Rapamycin prevented the IGF-1-induced binding of eIF4G to eIF4E (Fig. 3d, e), whereas mTORis completely blocked eIF4G:eIF4E binding and increased the association of 4E-BP1 with eIF4E (Fig. 3d, e). Since MNKs interact with eIF4G, which promotes their ability to phosphorylate eIF4E [9], it is likely that mTORis decrease P-eIF4E levels by disrupting eIF4G–eIF4E binding, thereby impeding MNKs’ ability to phosphorylate eIF4E.
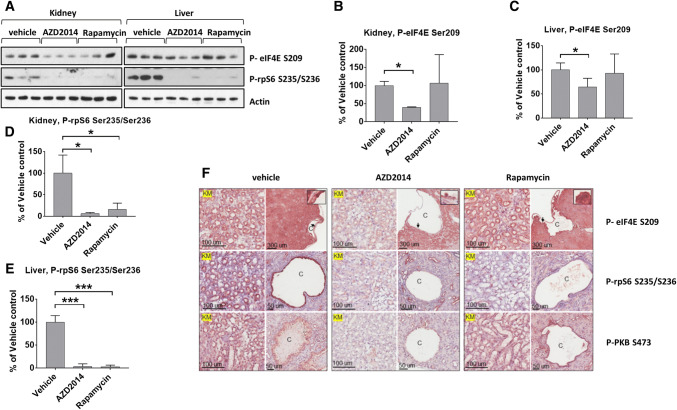
Given the above data, and the roles of mTOR and MNK signaling in cancer, it was important to test the effects of mTOR inhibition on eIF4E phosphorylation in vivo. To do so, we used Tsc2+/− mice which spontaneously develop renal lesions due to aberrant activation of mTOR signaling [41]. AZD2014, an mTORi that is suitable for use in vivo [42], blocked mTORC1 signaling (i.e., P-rpS6) in both liver and kidney as well as in renal lesions (Fig. 4; see IHC data in Fig. 4f). Importantly, AZD2014 also reduced P-eIF4E levels in tissues and renal lesions (Fig. 4). These data suggest that mTORis are likely to exert similar ‘anti-cancer’ effects to those of a combination of rapalogs and MNK inhibitors.
Fig. 4.
AZD2014 attenuate eIF4E phosphorylation in TSC2+/− mice. a Tsc2+/− mice at the age of 12 months were treated for two months with either AZD2014 or rapamycin, or with vehicle control. Lysates prepared from tissues from the mice were analysed by Western blot for phosphorylation of rpS6 at Ser235/Ser236 and eIF4E at Ser209. Actin was used as a loading control. Representative blots are shown. b Quantification of A (kidney, P-eIF4E Ser209). c Quantification of a (liver, P-eIF4E Ser209). d Quantification of a (kidney, P-rpS6 Ser235/Ser236). e Quantification of a (liver, P-rpS6 Ser235/Ser236). f Paraffin sections of kidney prepared from the drug-treated Tsc2+/− mice were analysed by immunohistochemistry. Representative IHC-stained images show phosphorylation of rpS6 at Ser235/Ser236, PKB/Akt at S473 and eIF4E at Ser209. Black arrows point to stained cells as shown in the boxed areas. Black lines are scale bars. c cystic lesions; KM, kidney medulla. For panels b–e, results are presented as means ± SD, n = 3. *0.01 ≤ P < 0.05; **0.001 ≤ P < 0.01; ***P < 0.001 (one-way ANOVA); $P < 0.001 (two-way ANOVA)
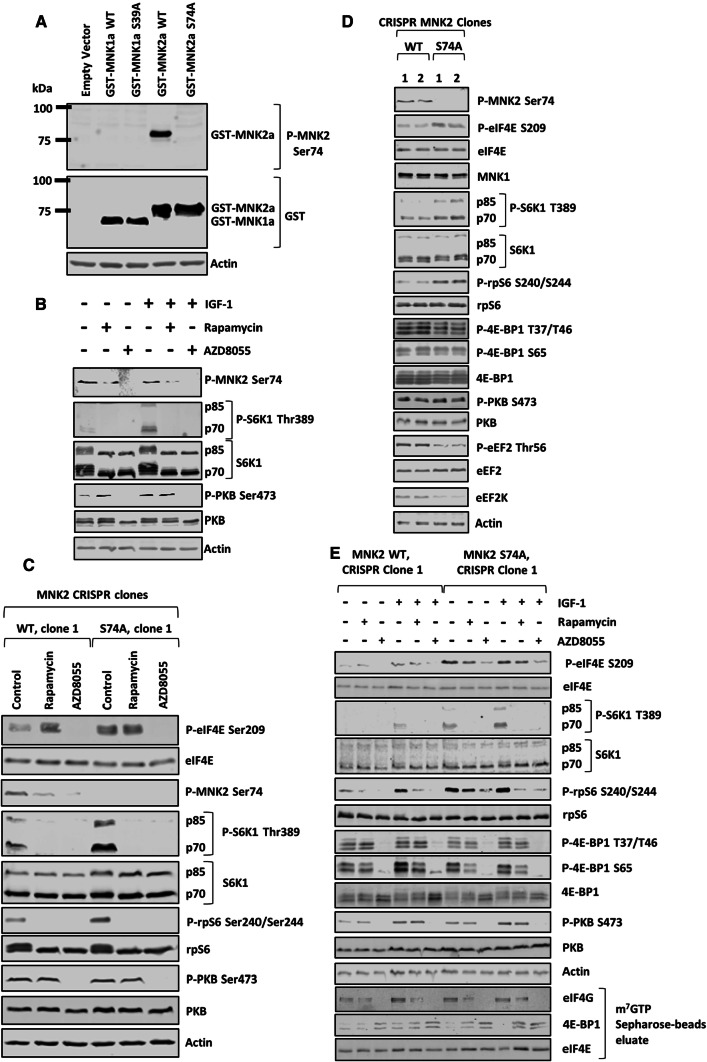
MNK2[S74A] knock-in HEK293 cells show increased size, and enhanced migratory, invasive and transformation capacities
To assess levels of MNK2[Ser74] phosphorylation, we developed a phosphospecific antibody against P-MNK2[Ser74] (Fig. 5a). As expected, this antibody did not recognize MNK1 (Fig. 5a). The phosphospecific antibody revealed that Ser74 is basally phosphorylated and this is not affected by treating cells with IGF-1 (Fig. 5b). Consistent with Ser74 being an mTOR substrate, rapamycin decreased its phosphorylation, and AZD8055 blocked it completely (Fig. 5b, c).
Fig. 5.
Genetic knock-in mutation of Ser74 of MNK2a to alanine in HEK293 cells enhances phosphorylation of eIF4E and mTORC1 downstream targets. a Cells were transfected with empty vector or the indicated GST-MNK constructs for 48 h, before lysates were made and subjected to immunoblot analysis. b Cells were serum starved for 16 h, pre-treated with 1 μM rapamycin or 1 μM AZD8055 before stimulation with 10 nM IGF-1 for 30 min. c MNK2 WT and S74A knock-in cells were treated with 1 μM rapamycin or 1 μM AZD8055 for 24 h before lysis and immunoblotting analysis was performed for the indicated P- or total proteins. d Lysates from MNK2 WT and S74A cells cultured in growth medium for 24 h were subjected to immunoblot analysis for the indicated proteins. e MNK2 WT and S74A cells were serum starved for 16 h, treated with rapamycin (1 μM) or AZD8055 (1 μM) for 30 min, and then stimulated with 10 nM IGF-1 for another 30 min. eIF4G, 4E-BP1 and eIF4E were isolated from lysates by m7GTP affinity chromatography and analysed by SDS-PAGE/WB against the indicated proteins from the eluates as well as the input lysates
Next, we used CRISPR/Cas9 genome editing to generate MNK2[S74A] knock-in HEK293 cells, where this mutation was introduced into both chromosomal copies of the MKNK2 gene (Fig. S7). As negative controls, we used clones obtained from the same process but which retained copies of the WT MKNK2 gene. Our custom-made P-MNK2[Ser74] antibody only detected one band (52 kDa) that corresponds to MNK2a (Fig. S8A), and did not detect any signal in MNK2[S74A] knock-in HEK293 cells (Fig. 5c). Importantly, the levels of eIF4E phosphorylation in MNK2[S74A] cells were higher than in their WT counterparts, and this phosphorylation was no longer sensitive to rapamycin (Fig. 5c). MNK1 levels did not alter between WT and MNK2[S74A] knock-in HEK293 cells (Fig. 5d). This implies that rapamycin induces phosphorylation of eIF4E by inhibiting the phosphorylation by mTORC1 of Ser74 in MNK2. Brown and Gromeier [29] recently reported that active MNK2 displaces DEPTOR (DEP domain-containing mTOR-interacting protein) from mTORC1, while promoting its binding to TELO2 (telomere length regulation protein 2), thereby facilitating the interaction of mTORC1 with its substrates and, in effect, activating it.
We, therefore, also studied the effect of MNK2[S74A] knock-in on mTORC1 downstream targets. We observed increases in P-eIF4E, P-S6K1 and P-rpS6, but not P-4E-BP1, compared to WT cells (Figs. 5c–e, S8, S9). We also observed a reduction in levels of eukaryotic elongation factor 2 (eEF2) kinase (eEF2K), which is negatively regulated by mTORC1 via S6K1 [43]; and the phosphorylation of its substrate eEF2 (Thr56) [44] in MNK2[S74A] cells (Figs. 5d, S8). Levels of association of eIF4E with eIF4G or 4E-BP1 were similar in WT and MNK2[S74A] cells (Figs. 5e, S8, S9). We did not observe any difference in 4E-BP1 phosphorylation in MNK2[S74A] cells as compared to WT cells, likely, because mutation of MNK2, and its consequent increased activity, mainly affects “weak” mTORC1 substrates such as S6Ks rather than strong ones such as 4E-BPs [40]. Nevertheless, the observed high levels of P-S6K1 and P-rpS6 are consistent with Brown and Gromeier’s report [29] that MNK2 promotes mTORC1 activation. In agreement with data showing that mTORC1 phosphorylates MNK2 at Ser74 (Fig. 2), rapamycin and AZD8055 each inhibited MNK2[Ser74] phosphorylation (Fig. 5c).
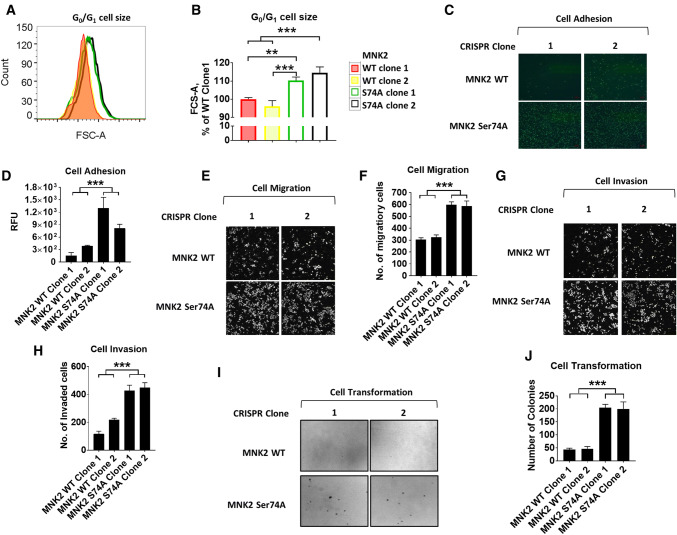
To examine the effect of abrogating MNK2[Ser74] phosphorylation, and thus enhancing MNK2a activity, on the expression levels of specific proteins, we applied stable isotope labelling with amino acids in cell culture (SILAC) methodology coupled with mass spectrometry [45] (Fig. S10; Table S1). Notably, levels of proteins involved in mRNA translation, such as ribosomal proteins, were increased in MNK2[S74A] cells (Fig. S10B, C). Consistent with this and the role of S6K1 in regulating cell size [46], MNK2 [S74A] cells were larger than WT cells (Fig. 6a, b). Consistent with earlier data showing that eIF4E phosphorylation plays roles in the transformation and invasiveness of cancer cells [1, 47, 48], MNK2[S74A] cells displayed enhanced adhesion (Fig. 6c, d), migration (Fig. 6e, f) and invasiveness (Fig. 6g, h). They also exhibited greater transformation ability as shown by their augmented anchorage-independent growth in soft agar assays (Fig. 6i, j).
Fig. 6.
MNK2 S74A HEK293 cells exhibit increased cell size, and enhanced invasive and transformation capacities. a G0/G1 size of CRISPR MNK2a WT and S74A cells was determined by flow cytometric analysis. b Quantification of a. c Cell adhesion assays for CRISPR MNK2a WT and S74A cells. d Quantification of data in c. e Cell migration assays for CRISPR MNK2a WT and S74A cells. f Quantification of data in e. g Cell invasion assays for CRISPR MNK2a WT and S74A cells. h Quantification of g. i Soft agar assays were performed in six-well plates and cultures maintained for 3 weeks before staining with 0.5% crystal violet. The numbers of colonies in each well larger than 100 µM were counted. j Quantification of i. For panels d, f, h and j, results are presented as means ± SD, n = 3. **0.001 ≤ P < 0.01; ***P < 0.001 (one-way ANOVA); @0.01 ≤ P < 0.05 and $P < 0.001 (two-way ANOVA)
These data imply that the phosphorylation of MNK2 on Ser74 by mTORC1 normally suppresses oncogenesis, likely due to its inhibitory effect on MNK2 activity (Fig. S11). Thus, while mTORC1 signaling is strongly implicated in tumorigenesis and a target for cancer therapy [49], its roles are multilayered and some effects of mTOR inhibition may actually aid tumorigenesis or cancer cell survival.
Specific inhibition of MNKs does not affect signaling through mTORC1 or mTORC2
Because mTORC1 phosphorylates MNK2, which inhibits MNK2 activity, we also studied the effect of inhibiting the MNKs on the mTORC1 pathway. To do so, we first used three different small molecule inhibitors of the MNKs: CGP57380 [50], MNK-I1 [47] and MNK-7g [51]. CGP57380 was the first small molecule MNK inhibitor to be reported [50] but suffers from substantial drawbacks, in particular relatively low potency and off-target effects, i.e. ability to inhibit several other protein kinases [52]. MNK-I1 is substantially (at least ten-fold) more potent than CGP57380 against MNK1 and MNK2 and lacks its off-target effects [47]. MNK-7g is another recently discovered specific MNK inhibitor which is more selective against MNK2 [51].
Three different types of human cells [A549 (human lung adenocarcinoma), HEK293T and MDA-MB-231 (human breast adenocarcinomas)] were pre-treated with these MNK inhibitors. Cells were stimulated with IGF-1 for 30 min (to activate mTORC1 signaling) which enhanced eIF4E phosphorylation (Fig. S12). CGP57380 inhibited MNK activity at concentrations > 3 μM, although complete inhibition required higher concentrations in HEK293T cells. MNK-I1 and MNK-7g blocked MNK activity at lower concentrations than CGP57380 (Fig. S12). As expected, IGF-1 increased the phosphorylation of S6K1, rpS6, 4E-BP1 and PKB/Akt (on Ser473, an mTORC2 site) (Fig. S12), indicating that IGF1 activates both mTORCs in all three cell types. Neither MNK-I1 nor MNK-7g significantly affected the IGF-1-induced phosphorylation of 4E-BP1, S6K1 or the S6K1 substrate rpS6 (Fig. S12). CGP57380 did not affect the phosphorylation of 4E-BP1 or S6K, but did impair S6 phosphorylation in all three cell lines, suggesting that it affects the activation or activity of S6Ks, without affecting mTORC1 itself. Moreover, none of the MNK inhibitors affected the phosphorylation of PKB/Akt at Ser473 (i.e., mTORC2 signaling).
Taken together, these data indicate that MNKs do not regulate mTORC1 signaling in response to IGF-1 in any of these human cell lines and also show that CGP57380 is not a reliable probe for the roles of the MNKs, due to its off-target effects, e.g., on S6K signaling.
Knock-out of MNKs does not affect IGF-1-stimulated signaling through mTORC1 or mTORC2
As an adjunct to using small molecule MNK inhibitors, we also studied mTOR signaling in cells lacking MNK1, MNK2 or both isoforms to explore the effects of MNKs on mTOR signaling, i.e., mouse embryonic fibroblasts (MEFs) from WT or MNK knockout (KO) mice. Importantly, IGF1 promoted the phosphorylation of S6K1, 4E-BP1 and PKB/Akt to similar extents in MEFs from MNK1-KO, MNK2-KO, MNK-DKO and WT mice (Fig. S13A, quantified in Fig. S14). The stimulation of mTORC1 and mTORC2 signaling was not affected by CGP57380 or MNK-I1 in WT MEFs, although they did strongly inhibit phosphorylation of eIF4E (Fig. S13A). These findings provide further strong evidence that MNKs do not affect the activity of mTORC1 or mTORC2 or its activation by IGF-1.
As shown (Fig. S13B, C, quantified in Fig. S15), we observed no differences between WT and MNK-DKO MEFs in the enhancement of the phosphorylation of S6K1, rpS6, 4E-BP1 or PKB/Akt by epidermal growth factor (EGF) or FBS. We extended our analysis of the possible role of MNKs in mTOR signaling to mouse cells, using 3T3-L1 cells. They are used widely as a reliable model for adipogenesis, which is induced by a ‘cocktail’ containing insulin, rosiglitazone, dexamethasone and the phosphodiesterase inhibitor, IBMX, over 7–9 days. This cocktail activated signaling through ERK, PKB/Akt and mTORC1 (Fig. S13D, quantified in Fig. S16), including stimulation of MNK activity (enhanced eIF4E phosphorylation). MNK-I1, MNK-7g and CGP57380 each inhibited eIF4E phosphorylation, confirming that they inhibit MNK function, but none of the compounds affected the phosphorylation of PKB/Akt at Ser473, or phosphorylation of 4E-BP1 at Ser65 (Fig. S13D), showing that signaling through mTORC2 or mTORC1 is not affected by MNK inhibition in these murine cells. However, CGP57380, but not MNK-I1 or MNK-7g, impaired the phosphorylation of S6, again suggesting that CGP57380 impairs the activation or activity of S6Ks.
MNKs do assist the activation of mTORC1 signaling by TPA
Since MNKs are activated by signaling through ERK, we tested their role in regulating mTORC1 signaling in response to an alternative stimulus, the phorbol ester TPA, which strongly activates ERK (and thus MNK1) but does not affect PI 3-kinase/PKB (Akt) signaling [28] (NB these pathways can independently regulate mTORC1; Fig. S1). In WT MEFs, the addition of the phorbol ester TPA rapidly increased the phosphorylation of eIF4E (indicating activation of MNK signaling) and of S6K and its substrate rpS6 (Fig. S13E, quantified in Fig. S17). In MNK-DKO cells, the TPA-induced phosphorylation of S6K and rpS6 was delayed and remained markedly blunted after 60 min (Fig. S13E). These data suggest that MNKs do aid the activation of mTORC1 by TPA in MEFs. Also, the IC50 of the MNK inhibitor eFT508 [53] did not differ between WT and MNK2 [S74A] cells (Fig. S18).
The reason that MNKs are not required for activation of mTORC1 signaling by EGF, IGF-1 or FBS likely reflects the fact that these agents, unlike TPA, activate PI 3-kinase/PKB signaling, which provides an alternative mechanism to turn on mTORC1 through the PKB-mediated phosphorylation and inactivation of TSC1/2 [54].
MNK2[Ser74] phosphorylation is inversely associated with high Gleason score in prostate cancer patients and promotes proliferation and survival of tumor cells
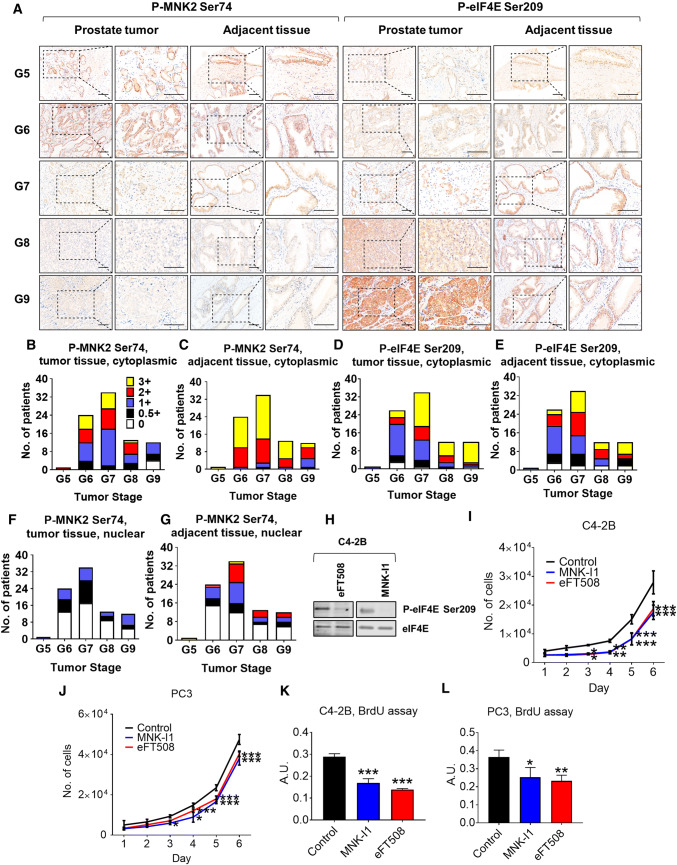
Previous studies, Furic et al. [1] have shown that phosphorylation of eIF4E positively correlates with high Gleason score in prostate cancer patients, suggesting that phosphorylation of eIF4E is a key event in prostate cancer progression. We, therefore, carried out tissue IHC microarray analysis to examine levels of MNK2[Ser74] and eIF4E[Ser209] phosphorylation in 84 individual prostate tumors and their adjacent non-cancer tissue specimens (Fig. 7a–g, Table S2). We noted that in tumors with a high Gleason score, MNK2 was less phosphorylated at Ser74 whereas, as expected [1], eIF4E was more phosphorylated. MNK2[Ser74] was also more phosphorylated in adjacent non-cancer tissue compared to tumor (Fig. 7a–g). We could not determine the relative expression levels of MNK2a and 2b, which could also influence the levels of nuclear P-MNK2[Ser74]. Since low levels of P-MNK2[Ser74] indicate more active MNK2 (which probably explains the observed increase in eIF4E phosphorylation; shown in Figs. 2d, 7a, f, g), these data indicate an inverse correlation between the inhibitory phosphorylation of MNK2 on Ser74 and prostate cancer progression. Two structurally unrelated MNK inhibitors, MNK-I1 and eFT508 [53], also inhibited the proliferation of prostate cancer cells in vitro (Fig. 7h–l). Therefore, inhibition of MNK2 activity, or in combination of mTORis, may be a beneficial approach in tackling prostate cancer.
Fig. 7.
Loss of MNK2 Ser74 phosphorylation correlates with advanced prostate tumors in patients. a Levels of P-MNK2 Ser74 in prostate cancer patient samples were assessed by tissue microarray analysis against 84 paired prostate tumors and adjacent normal tissue. Representative IHC images from Gleason score 5–9 (G5–G9) were shown. Scale bar 50 μm. b–i Average immunoreactivity in a was graded in a blinded manner on a scale of 0 (none), 0.5 + (very weak); 1 + (weak); 2 + (intermediate); and 3 + (strong) in b–e: cytoplasmic; f, g: nuclear; b, c, f, g: P-MNK2 Ser74; d, e: P-eIF4E Ser209. b, d, f: tumor tissues; c, e, g: adjacent normal tissues. h C4-2B cells were treated with vehicle (control), eFT508 (2 μM) or MNK-I1 (2 μM) for 24 h before lysis and immunoblotting analysis for P- (Ser209) or total eIF4E. C4-2B (i) or PC3 (j) cells were treated with vehicle (control), eFT508 (2 μM) or MNK-I1 (2 μM), the number of cells were counted for the indicated periods of time. C4-2B (k) or PC3 (l) cells were treated with vehicle (control), eFT508 (2 μM) or MNK-I1 (2 μM) for 48 h, before subjected to BrdU incorporation assays. In panels h–l data are shown as means ± SD, n = 3. *0.01 ≤ P < 0.05; **0.001 ≤ P < 0.01; ***P < 0.001 (one-way ANOVA)
We also tested the effects of eFT508 in an ex vivo prostate tumor model; in about half the explants, eFT508 inhibited cell proliferation, but did not in others (Table S3). This variability in response clearly requires further investigation, but nevertheless indicates the potential of targeting MNK in prostate cancer.
Discussion
The MNKs are the only kinases that phosphorylate eIF4E. eIF4E and its phosphorylation play key roles in mRNA translation and its control, and in oncogenesis [16]. Here, we show that mTORC1 directly phosphorylates MNK2 on Ser74. Importantly, our data show that mTORC1 signaling modulates the ability of MNK2a to phosphorylate its substrate eIF4E in two ways; by phosphorylating Ser74, mTORC1 signaling represses (1) MNK2a’s high basal activity and (2) MNK2a’s binding to the scaffold protein eIF4G. This provides acute control of MNK2a function, in contrast to its weak regulation by upstream MAP kinase signaling [12].
Conversely, preventing MNK2[Ser74] phosphorylation (by partially inhibiting mTORC1 with rapamycin or mutating Ser74, for example) enhances MNK2 activity and increases the level of phosphorylation of its substrate eIF4E, an event which promotes tumorigenesis [1, 55, 56]. We find that MNK2[Ser74] phosphorylation correlates inversely with prostate cancer progression in patients (an effect which promotes MNK2a activity), while MNK inhibition exhibited anti-tumor activity in prostate cancer cell lines. Previously, it has been reported that ratios MNK2a:MNK2b mRNAs are downregulated in several types of cancer including breast, lung, colon [57] and brain [58] cancers. However, relative MNK2 protein expression levels in tumours remain unknown, and we could not detect total MNK2 levels in our tissue microarray experiment (Fig. 7a). No reliable total MNK2 antibody is currently available (Fig. S2); therefore, we still cannot exclude the possibility that the low P-MNK2[Ser74] levels we saw in advanced prostate tumours is due to low protein expression levels of MNK2.
In summary, while MNK1a activity is tightly and rapidly activated by MAP kinases, MNK2a’s ability to phosphorylate eIF4E is negatively regulated by mTORC1 signaling.
Several groups have reported that the activation of MNKs confers resistance to rapalogs in cancer cells [22–25], and that combinational administration of MNK inhibitors and rapalogs completely prevented tumor growth in lymphoma [25], medulloblastoma [23], glioma [24] and prostate cancer [22], although the mechanisms involved remained unclear. Here, we show that, while inhibiting mTORC1 increases MNK2 activity in vitro, mTORis actually decrease eIF4E phosphorylation in vivo. This reflects the fact that mTORis prevent the association of eIF4G with MNK2a and with eIF4E. Loss of these interactions hinders MNK-mediated phosphorylation of eIF4E, leading to decreased phosphorylation of eIF4E. The ability of mTORis to impair mTOR signaling and eIF4E phosphorylation should contribute them to being more effective anti-tumor agents than rapamycin and rapalogs, which have so far met only with limited success in tumor therapy [49].
mTORC1 signaling is enhanced in MNK2[S74A] cells as shown by the increased phosphorylation and activity of S6K, a positive regulator of cell size [46]. Importantly, and consistent with these changes and the role of mTORC1 in cell size control [34], MNK2[S74A] knock-in cells exhibit larger cell size, enhanced adhesion, migration, invasiveness and anchorage-independent growth. These effects presumably reflect the displacement of MNK2-driven DEPTOR from mTORC1 and enhancing binding of mTORC1 to TELO2 (as described previously [29]).
The activation by TPA of S6Ks (a surrogate readout for mTORC1) is impaired in MNK-DKO cells. However, we observed this MNK-mTORC1 signaling link only under conditions where ERK signaling is the main pathway being activated, and only for S6Ks, an example of a ‘weak’ mTORC1 substrate [40]. Disabling the MNKs did not affect the activation of mTORC1 by other stimuli tested (serum, IGF-1, EGF, which activate PI 3-kinase/Akt signaling) or did it affect signaling through mTORC2, which phosphorylates Ser473 in PKB/Akt. Thus, MNKs can positively regulate mTORC1 activity, but in a more limited way than reported previously [29].
In agreement with that study [29], we find that MNK2a, but not MNK1a, binds to mTORC1 and that this interaction is mediated by the N-terminal region of MNK2, which is substantially different from the N terminus of MNK1, involving either a non-canonical TOS motif or multiple TOS motifs.
mTORC1 plays an essential role in maintaining cell mass and function, yet its over-activation brings detrimental effects impacting on pathogenesis such as the development of tumors. As revealed by previous studies, stimulation of mTORC1 also evokes several negative feedback loops to prevent its over-activation [17]. These include inhibitory phosphorylation of insulin receptor substrate-1 (IRS1) by S6K1 [59] and the phosphorylation and stabilization of growth factor bound-receptor protein 10 (Grb10) by mTORC1, an event which subsequently destabilizes IRS1 [39, 60]. Here, we define an additional, distinct negative feedback loop whereby activation of mTORC1 catalyses inhibitory phosphorylation of MNK2 at Ser74, which prevents ‘hyper-activation’ of eIF4E phosphorylation (Fig. S17). Notably, we demonstrate that MNK2 Ser74 phosphorylation is decreased in prostate cancer patients and is associated with elevated levels of eIF4E phosphorylation, an event which has been shown to contribute to prostate cancer progression [1]. Interestingly, lower levels of mTORC1 signaling (and thus, presumably, higher MNK2a activity) are associated with worse outcomes in prostate cancer patients [61]. However, activation of mTORC1 signaling is a more common feature of solid tumours [62]. We also show that pharmacological inhibition of the MNKs by two potent MNK inhibitors, MNK-I1 and eFT508, the latter of which is in phase 2 trials for lymphoma and solid tumours [53], impairs the proliferation of prostate cancer cells in vitro.
In conclusion, our data provide important new insights into the crosstalk between mTORC1 and the MNKs which are relevant for tumor cell biology, in particular the role of the Ser74 phosphorylation site in MNK2, which is directly phosphorylated by mTORC1. Our data thus identify the molecular events by which rapamycin and rapalogs enhance MNK2 activity. Complete inhibition of mTORC1 activity impairs the association between MNK2a and eIF4G as well as the binding of eIF4G to eIF4E. Both effects operate to decrease the phosphorylation of eIF4E. These new data on the interplay between the oncogenic mTORC1 and MNK2/eIF4E pathways provide new insights into the cell signaling events which integrate the mTORC1 and MAP kinase signaling pathways and impinge on the control of mRNA translation, in particular eIF4E, a crucial regulatory and oncogenic initiation factor. As such, these findings are very relevant to tumor biology and are likely to be valuable in the design and evaluation of novel anti-cancer therapies.
Materials and methods
Chemicals and reagents
All chemicals were from Merck (Frenchs Forest, NSW, Australia) unless otherwise specified. [γ-32P]ATP and [35S]methionine/cysteine were purchased from Perkin Elmer (Gladesville, NSW, Australia). AZD8055, Torin 2 and OSI-027 were purchased from Jomar Life Research (Scoresby, VIC, Australia). MNK-I1 [47] and MNK-7g [51] were previous described.
Cell culture
MEFs from MNK1-KO, MNK2-KO and MNK-DKO mice and matched wild-type counterparts were prepared from embryos at embryonic day 13.5. MEFs, human lung carcinoma A549 cells, human embryonic kidney HEK293 cells, human breast carcinoma MDA-MB-231 cells, mouse 3T3-L1 fibroblasts (a kind gift from Dr Yeesim Khew-Goodall from the Centre for Cancer Biology [SA Pathology], Adelaide) were maintained in Dulbecco’s modified Eagle medium (DMEM) media containing 10% (v/v) foetal bovine serum (FBS) and 1% penicillin/streptomycin (growth medium). Cells were cultured at 37 °C in 5% CO2 and 95% air. Prostate cancer cells (C4-2B and PC3) were maintained in Roswell Park Memorial Institute-1640 (RPMI-1640) media containing 10% (v/v) foetal bovine serum (FBS) and 1% penicillin/streptomycin (growth medium).
Animal study
All experimental procedures involving mice were approved, as appropriate, by the animal ethics committees of Cardiff University or the South Australian Health & Medical Research Institute (SAHMRI), and were conducted in accordance with the regulations of Scientific Procedures both in the UK (Act 1986) and in South Australia (Act 1985).
As described previously [63], Tsc2+/− mice were backcrossed to the Balb/c strain. To determine the effect of AZD2014 and rapamycin on molecular signaling, 9 Tsc2+/− mice at the age of 12 months were randomly allocated into three groups (3/group) and treated with vehicle, 20 mg/kg AZD2014 or 5 mg/kg rapamycin five times a week for 8 weeks. These mice were then humanely killed for tissue collection. AZD2014 (APExBIO, Houston, TX, USA) at 4 mg/ml and rapamycin (LC Laboratories, Woburn, MA, USA) at 1 mg/ml were prepared in vehicle solution (2.5% PEG-400, 2.5% Tween-80 and 2.5% DMSO), respectively.
Treatment and lysis
Unless specified, prior to treatments, cells were serum starved for 16 h, the media were then removed and the cells were washed twice and incubated with modified Krebs–Ringer bicarbonate buffer (KRB) (115 mM NaCl, 5 mM KCl, 10 mM NaHCO3, 2.5 mM MgCl2, 2.5 mM CaCl2, 20 mM HEPES, pH 7.4, 0.5X MEM amino acids, 0.5X MEM non-essential amino acids solution and 0.5X Glutamax) for 30 min, before being treated with indicated inhibitors or the adipogenic cocktail (350 nM insulin, 500 µM IBMX, 0.5 µM dexamethasone and 2 µM rosiglitazone).
After treatments, cells were lysed by scraping into ice-cold lysis buffer containing 1% (v/v) Triton X-100, 20 mM Tris–HCl pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 2.5 mM Na2H2P2O7, 1 mM β-glycerophosphate, 1 mM Na3VO4, 1 mM dithiothreitol and protease inhibitor cocktail, unless otherwise stated. 3T3-L1 cells were lysed in RIPA buffer containing 1% (v/v) Igepal, 50 mM Tris–HCl pH 7.5, 150 mM NaCl, 1 mM EGTA, 50 mM β-glycerophosphate, 0.5 mM Na3VO4, 0.1% (v/v) β-mercaptoethanol and protease inhibitor cocktail. Lysates were spun at 16,000×g for 10 min, the supernatants were kept and total protein concentration was quantified by Bradford assay (Bio-Rad, Gladesville, NSW, Australia) following the manufacturer’s instructions. Normalized lysates were either kept at − 20 °C or subjected to further analysis.
Immunohistochemistry (IHC)
Formalin-fixed and paraffin-embedded sections were prepared for IHC as described previously [41, 64]. SignalStain Boost Rabbit specific IHC Detection Reagent (Cell Signaling Technology, Danvers, MA, USA) and ImmPACT NovaRED Peroxidase Substrate mmPACT (Vector Laboratories, Peterborough, Cambridgeshire, UK) were used to stain antigens. Primary antibodies were used for IHC indicated antibodies. Images were captured with a Nanozoomer scanner. The percentage of Ki-67 and cleaved caspase-3-positive nuclei were determined by blind manual counting of at least 200 malignant cells over 5–10 fields at 40× magnification.
Isolation of MSCs
Murine compact bone MSCs were derived from the long bones of 6–7-week-old Rptorfl/fl and Rictorfl/fl mice [65] as previously described [66, 67]. Cord blood (CB) MSCs (passages 2–3) were then infected with a lentivirus carrying a tamoxifen-inducible self-deleting Cre recombinase (LEGO-CreERT2-iG2) [68] in the presence of 4 μg/ml polybrene and CB MSCs with stable lentiviral integration were selected on the basis of GFP expression. As previously described [66, 67], cells were treated with 0.5 μM 4-hydroxytamoxifen for 8 days to induce Rptor or Rictor deletion, and ethanol (0.05%, v/v) was used as a vehicle control to generate wild-type control MSCs. MSCs were maintained in α-MEM supplemented with 20% FCS, 2 mM l-glutamine, 100 µM l-ascorbate-2-phosphate, 50 IU/ml penicillin, 50 μg/ml streptomycin sulphate, 1 mM sodium pyruvate and 15 mM HEPES, and passaged by detachment with a 0.05% (w/v) trypsin–EDTA solution upon reaching 80–90% confluence.
Vectors, mutagenesis and cell transfection
Vectors for glutathione S-transferase (GST)-MNK1a and GST-MNK2a (both of human and mouse origin) have been described previously [4]. Point mutations were introduced by PCR mutagenesis using the Pfu DNA polymerase (Promega, Alexandria, VIC, Australia) and corresponding primers as listed in Table S4. HEK293 cells were transfected by the calcium phosphate method as previously described [69]. MEFs were transfected using lipofectamine® 3000 (Thermofisher Scientific, Adelaide, SA, Australia) according to manufacturer’s instructions.
SDS-PAGE/WB analysis
SDS-PAGE (sodium dodecyl sulphate–polyacrylamide gel electrophoresis) and western blot (WB) analyses were performed as previously described [69]. See Table S5 for a complete list of primary antibodies used in this study. Data shown are representative of at least three independent experiments. Blots were quantified using the FIJI software [70].
Immunoprecipitation and mTORC1 assays
Cells were harvested in CHAPS lysis buffer [40 mM HEPES pH 7.5, 120 mM NaCl, 1 mM EDTA pH8, 10 mM Na2H2P2O7, 10 mM β-glycerophosphate, 50 mM NaF, 0.5 mM Na3VO4, 0.3% (m/v) CHAPS and protease inhibitor cocktail] and mTOR complexes were immunoprecipitated with protein G beads as previously described [71]. For mTORC1 assays, after washing with lysis buffer, mTOR complex immunoprecipitates were further washed twice in mTOR reaction buffer [25 mM HEPES at pH 6.0, 6.6 or 7.4, 50 mM NaCl, 20% (v/v) glycerol, 10 mM MgCl2, 4 mM MnCl2 and 1 mM dithiothreitol], and assays were performed in 20 μl mTOR reaction buffer containing 1 μg recombinant 4E-BP1 or purified GST-MNK (see below) as (test) substrates, 1 μM MNK-I1, 50 mM unlabelled ATP and 1 μCi [γ-32P]ATP at 30 °C for 30 min. Reactions were stopped by adding 20 μl of 2× Laemmli sample buffer; samples were then heated at 100 °C for 4 min followed by SDS-PAGE. Proteins were stained with Coomassie blue and phosphorylated 4E-BP1 or MNK was detected by Cyclone plus phosphorimager system (Perkin Elmer).
Pull-down and purification of GST-tagged recombinant proteins
GST-MNK constructs were transfected into HEK293 cells, and for detection of mTOR/RAPTOR-binding to GST-MNK2, cells were harvested in CHAPS lysis buffer. Lysates were centrifuged at 16,000×g for 10 min, and the supernatants were then incubated with GST·Bind™ Resin at 4 °C for 2 h. After incubation, resins were washed thrice with CHAPS lysis buffer, proteins were then dissolved in 2× Laemmli sample buffer for SDS-PAGE/WB analysis.
For purification of GST-MNKs or MNK assays (see below), HEK293 cells were lysed in triton lysis buffer, incubated with GST·Bind™ Resin at 4 °C for 1 h. Resins were then washed twice with high salt triton lysis buffer [1% (v/v) Triton X-100, 50 mM Tris–HCl pH 7.5, 350 mM NaCl, 0.2 mM EDTA, 2.5 mM Na2H2P2O7, 50 mM β-glycerophosphate, 1 mM Na3VO4, 1 mM MgCl2, 1 mM dithiothreitol and protease inhibitor cocktail] to remove contaminating kinase activity, and twice with GST-purification washing buffer (20 mM Tris–HCl, pH 8.0, 100 mM KCl, 10% glycerol, 1 mM dithiothreitol and protease inhibitor cocktail). Proteins were eluted from the resins with elution buffer (GST-purification washing buffer plus 40 mM reduced glutathione), dialysed over-night in Slide-A-Lyzer dialysis cassettes (Thermofisher Scientific) in dialysis buffer (20 mM Tris–HCl, pH 8.0, 100 mM KCl, 5% glycerol and 5 mM β-mercaptoethanol). Quantity of purified proteins were estimated by SDS-PAGE analysis followed by Coomassie blue staining, and were either used immediately or stored at − 80 °C.
Purification of GST-eEF2K[K170M] was performed as previously described [72]. Human recombinant pGEX-6P-GST-eEF2K[K170M] [72] were expressed in E. coli BL21 (DE3) cells. After the absorbance at 600 nm reached 0.5, GST-eEF2K[K170M] expression was induced by the addition of 0.5 mM isopropyl β-d-1-thiogalactopyranoside. Cells were grown overnight at 18 °C prior to harvesting. Cells were broken by sonication in lysis buffer comprising 50 mM Tris–HCl pH 8.0, 100 mM KCl, 10% (v/v) glycerol, 1 mM dithiothreitol, 10% triton and protease inhibitor cocktail. Lysates were clarified by centrifugation at 16,000×g for 15 min at 4 °C. GST-eEF2K[K170M] was then purified on glutathione-Sepharose, washed once in lysis buffer and once in wash buffer (lysis buffer without 10% triton), and then eluted with elution buffer (50 mM Tris–HCl pH 8.0, 100 mM KCl, 10 mM reduced glutathione, 14 mM β-mercaptoethanol and protease inhibitor cocktail). Elutes were dialysed overnight at 4 °C in Slide-A-Lyzer cassettes (Thermofisher Scientific) against the dialysis buffer [20 mM Tris–HCl pH 7.5, 100 mM KCl, 5% (v/v) glycerol and 5 mM β-mercaptoethanol]. Recombinant GST-eEF2KK170M was kept at − 80 °C before further experiments.
MNK assays
After pull-down with GST·Bind™ Resin (see above) and washed the resins twice with high salt triton lysis buffer, resins were then washed once with MNK reaction buffer (20 mM HEPES pH 7.5, 50 mM KCl and 2 mM MgCl2). Assays were performed in 20 μl MNK reaction buffer containing 50 mM ATP and 130 ng recombinant eIF4E [69] as the substrate, at 30 °C for 30 min or the indicated periods of time. Reactions were stopped by adding 20 μl of 2× Laemmli sample buffer; samples were then heated at 100 °C for 4 min followed by SDS-PAGE/WB analysis.
S6K1 assays
S6K1 assays were performed as previously described [73]. HEK293 cells were transfected with HA-S6K1 for 48 h, cells were then lysed in ice-cold lysis buffer [1% (v/v) Triton X-100, 20 mM Tris–HCl pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 2.5 mM Na2H2P2O7, 1 mM β-glycerophosphate, 1 mM Na3VO4, 1 mM dithiothreitol and protease inhibitor cocktail]. HA-S6K1 was immunoprecipitated with the anti-HA antibody immobilized on protein G-Sepharose beads. Immune complexes were washed three times with lysis buffer, followed by a single wash with kinase assay buffer (50 mM HEPES, pH 7.5, 10 mM MgCl2, 1 mM dithiothreitol and 10 mM β-glycerophosphate). The kinase reaction was initiated by resuspending the beads in 25 μl of kinase assay buffer supplemented with 50 μM ATP, 1 μCi of [γ-32P]ATP, plus 3 μg of recombinant GST-MNKs or 1 μg of recombinant GST-eEF2K[K170M]. The reaction was carried out at 30 °C for 10 min and terminated by the addition of Laemmli sample buffer. Samples were boiled for 3 min at 100 °C and separated by SDS-PAGE.
m7GTP pull-down
After treatments, cells were lysed in ice-cold triton lysis buffer and precleared by centrifugation for 10 min at 16,000×g, before binding to γ-AH-m7GTP agarose (Jena Bioscience, Jena, Germany) for 1 h at 4 °C. The beads were then washed thrice with triton lysis buffer and resuspended in 2× Laemmli sample buffer before SDS-PAGE/WB analysis.
Subcellular fractionation
An adapted method [74] was applied to separate cytoplasmic and nuclear fractions from cell lysates. Briefly, cells were lysed in hypotonic buffer (HB) [10 mM Tris pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 0.2% (v/v) Triton X-100 plus protease inhibitor cocktail]. Samples were incubated in a thermomixer comfort (Eppendorf, Stevenage, UK) at 500 rpm, 4 °C for 30 min, before centrifuged at 1000×g, The supernatant was centrifuged again at 4500×g to remove debris, a final concentration of 200 mM NaCl was added to the clear supernatant which is kept as the cytoplasmic fraction. Pellets were resuspended in high salt buffer [10 mM Tris pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 400 mM NaCl, 0.4% (v/v) Triton X-100 and protease inhibitors] and incubated in a thermomixer comfort at 500 rpm, 4 °C for 30 min, followed by centrifugation at 16,000×g. Equal volume of HB was added to the supernatant, and this constituted the nuclear fraction.
Generation of CRISPR-directed MNK2a[S74A] knock-in HEK293 cells
The MNK2a[S74A] CRISPR targeting vector is derived from the GeneArt® CD4 CRISPR Nuclease Vector (Thermofisher Scientific); a stuffer fragment was added to the linear vector to regenerate the tandem BaeI cut sites necessary to permit guide RNA sequence insertion, and an AgeI/EcoRI fragment containing the CD4 selection cassette was replaced with an EGFP coding sequence, creating the vector known as pKJ388-EGFP.
To make the final MNK2a[S74A] targeting vector, the ssDNA oligonucleotides 5′-CGGGCCACCGACAGCTTCTCGTTTT-3′ and 5′-GAGAAGCTGTCGGTGGCCCGCGGTG-3′ were annealed and ligated into BaeI-digested pKJ388-EGFP. 2 µg of this vector was combined with 12 µg of the ssDNA repair template 5′-GsGsGCAGCGGGGCGGGCGTGAGAGGGACCCTGGCTTTTCCCCGCTCCCGGCTCCCCCAATGCCCGCCATCCCCGCTCACCTTCAAACCTGCCTGAGAAGGCGTCGGTGGCCCGGCCGCGCTTCTTCTTCTTGCCCsCsT-3′ (where ‘s’ indicates a phosphor-thioate linkage) and 1 × 106 cells in a total volume of 100 µl of SE buffer. This material was placed in a 4D-X cuvette (Lonza, Mt Waverley, VIC, Australia) and run through the CN-114 nucleofection program.
Cells were then plated into a 100 mm dish and grown for 48 h. Single GFP-positive cells were sorted into individual wells of a 96-well plate using a BD FACSFusion flow cytometer (Becton, Dickinson & Company, Adelaide, SA, Australia). After approximately 10 days of further growth, cell colonies were split one fraction for further growth, and one for genomic DNA analysis. In addition to the introduction of the S74A mutation [and alteration of the adjacent protospacer adjacent motif (PAM) site], the ssDNA repair template silently inserted a BsaHI cut site adjacent to the S74A locus; digestion of a PCR product encompassing this region of the MNK2a gene allowed for the rapid selection of correctly edited cells, which were further confirmed by sanger sequencing analysis (performed by the Australian Genome Research Facility, Adelaide, Australia).
Stable isotope-labelling with amino acids in cell culture (SILAC)
As previously described, MNK2 S74A HEK293 cells were passaged six times in DMEM containing 77 mg/l L-[13C]6, [15N]2-Lys and 44.5 mg/l L-[13C]6, [15N]4-Arg (heavy-labelled, in place of normal lysine and arginine) to reach essentially 100% labelling, while wildtype cells were also passaged six times in DMEM containing normal lysine and arginine (light-labelled). 24 h after the last passage, cells were scraped from the culture plates with 1× phosphate buffered saline (PBS, Thermofisher Scientific), before further analysis.
Mass spectrometric analysis
Sample preparation
Three wildtype and three MNK2[S74A] knock-in HEK293 cell lysates (0.5 ml in total) were homogenised using a Precellys bead mill (Bertin technologies, Montigny le Bretonneux, France). Total protein was determined in each sample using bicinchoninic acid (BCA) assay (Thermofisher Scientific) following manufacturer’s instructions. Wildtype and knock-in samples were combined in three pairings containing of 150 µg of protein from each genotype, thus producing three mixed samples containing 300 µg of total protein each. Sample volume was made up to 123 µl through the addition of 25 mM ammonium bicarbonate. Acid degradable surfactant, Rapigest SF (Waters Corporation, Milford, MA, USA) made up to 1% (w/v) in 25 mM ammonium bicarbonate was added to each sample (7.5 µl), and samples were mixed and heated to 80 °C for 10 min. Proteins were reduced by addition of 3.8 µl 100 mM dithiothreitol in 25 mM ammonium bicarbonate followed by mixing and heating to 60 °C for 10 min. Samples were cooled to room temperature and 3.8 µl of iodoacetamide in 25 mM ammonium bicarbonate was added to alkylate cysteine residues. Samples were allowed to incubate in darkness for 30 min. 12 µl of 0.25 mg/ml trypsin (ThermoFisher, Scoresby, VIC, Australia) was then added to the samples in 25 mM ammonium bicarbonate for 16 h at 37 °C. After trypsin digestion, 0.8 µl of trifluoro acetic acid was added to the samples and they were incubated for 40 min at 37 °C to degrade the Rapigest surfactant. Samples were then centrifuged at 16,000×g for 20 min in a Biofuge Pico (Heraeus, Kendro Laboratory Products Pty Ltd, Sydney, NSW, Australia). The supernatant was used without further purification.
Mass spectrometry (MS)
All MS analyses were performed on a Xevo G2-XS QTof mass spectrometer (Waters Corporation, Milford, MA, USA) coupled to an Acquity M-class nano/microflow UPLC system (Waters Corporation). Injection volume used was 3.75 µl, equivalent to 7.5 µg of total protein. Peptides were separated on a reverse phase analytical column (HSS T3, 1.8 µm particle size, 300 µm inner diameter, 150 mm length, Waters Corporation) using a solvent gradient. The mobile phases used were: (A) 0.1% (v/v) aqueous formic acid and (B) 0.1% (v/v) formic acid in acetonitrile, both LC–MS grade from Honeywell, Burdick and Jackson (Chem-Supply, Gillman, SA, Australia). The gradient was held at 97% A for 5 min, then dropped to 60% A over 60 min, followed by a 5 min wash step at 15% A, and finally re-equilibration at 97% A for 7 min. Flow rate was 7 µl/min.
The mass spectrometer was operated in SONAR mode. The m/z range used was 50–2000, scan time was 0.5 s, the quadrupole was scanned from m/z 400 to 900 with a 20 Da window. Collision voltage in low energy scans was set to 5 V and in elevated energy scans it was ramped from 14 V (m/z 400) to 36 V (m/z 900). During acquisition, the instrument switched continuously between low and elevated energy scans.
MS data analysis
MS data were processed and databank searched using Progenesis QI for proteomics v4.1 (Nonlinear Dynamics, Newcastle upon Tyne, UK) using data thresholding settings of LE 350 and HE 25. All other settings were left as default. The protein databank used was a non-redundant FASTA formatted UniProt database (human). SILAC quantification was performed using Proteolabels software (Omic Analytics, Liverpool, UK). Functional protein association analysis was performed using STRING (https://string-db.org).
Flow cytometry analysis
Following treatment, cells were trypsinized and fixed in 90% methanol, pelleted by centrifugation at 200×g for 5 min and resuspended in 1 ml PBS containing 20 μg/ml propidium iodide and 100 μg/ml RNase A. The intensity of signals was recorded using FACSCantoTM II flow cytometry (Becton, Dickinson & Company), data were analysed using FlowJo software version 10.2 (Becton, Dickinson & Company). FCS (forward scatter) values from cells in G0/G1 phase (10,000 gated events per sample) were collected to determine cell size.
Protein synthesis measurements
Cells were incubated in methionine-free DMEM medium for 30 min, pre-treated with 1 μM AZD8055 for 30 min, before the addition of 10 μCi EasyTag™ l-[35S]-Methionine (Perkin Elmer) and 10 nM IGF-1. Cells were further incubated for 1 h before lysed in ice-cold triton lysis buffer. Incorporated radioactivity was determined as previously described [75].
Cell adhesion assays
Cells were trypsinized and counted following overnight serum starvation. 3 × 105 cells per well were seeded into 12 well plates which were coated with 10 μg/ml Type I bovine collagen (catalogue No. 04902, Stemcell Technologies, Tullamarine, VIC, Australia) for 1 h at 37 °C. The collagen was aspirated and the wells allowed to air-dry prior to cell seeding. The cells were allowed to adhere for 30 min, trypsinized and resuspended in serum-free medium with Calcein-AM (catalogue No. C3099, Thermofisher Scientific) at a final concentration of 1 μg/ml. The cells were then seeded into a 96-well plate and left for 30 min at 37 °C, 5% CO2. The fluorescence signals were then monitored using the GloMax Discover with 475 nm excitation and 500–550 nm emission wavelengths. Images were taken using the ZEISS Axio Cert microscope A1 inverted microscope (ZEISS Australia, Lonsdale, SA, Australia) with the 10× objective lens and GFP filter with 470/440 emission and 525/550 excitation.
Transwell migration and invasion assays
The bottom side of the Transwell membranes (6.5 mm Transwell inserts with 8 μm pores, Merck, catalogue No. CLS3422) were coated with 10 μg/ml Type I bovine collagen (Stemcell Technologies) for 1 h at 37 °C. For invasion assays, matrigel (Merck, catalogue No. 356231) was diluted in serum-free medium to a final concentration of 0.5 mg/ml and then layered over the Transwell after drying it in a volume of 100 μl. The gel was then allowed to set for 1 h at 37 °C. The remaining unset liquid was then removed prior to cell seeding. The collagen was then aspirated and 600 μl of medium containing 20% (v/v) FBS was then put into the bottom chamber of the Transwell plate. Following overnight starvation in serum-free medium, cells were trypsinized and seeded at a density of 1 × 105 cells in 100 μl of serum-free medium to the top of the Transwell. The cells were allowed to migrate or invade over 48 h, after which the medium was aspirated and 500 μl of 3.7% formaldehyde was added to the bottom chamber. The cells were fixed for 15 min. The formaldehyde was then aspirated and replaced with 500 μl of 100% methanol for 20 min. The methanol was then replaced with DAPI diluted in PBS to a concentration of 5 μg/ml for 15 min. The membranes were then cut out and mounted on a slide. Six representative images were taken of each membrane with the 10× objective lens using the Nikon Eclipse Ni Fluorescent Microscope (Nikon, Sydney, NSW, Australia) with the DAPI filter (340–380 nm excitation, 435–485 nm emission). The cells were counted using the NIS Elements BR 4.40.00 software (Nikon).
Soft agar assays
HEK293 cells were plated at a density of 5000 cells/well in 1 ml DMEM containing 0.3% (m/v) agarose over a base layer of 1 ml DMEM containing 0.5% (m/v) agarose. 1 ml of DMEM was added to prevent drying. Cultures were maintained for 4 weeks with media being changed twice per week. Cells were stained with 0.5% (m/v) crystal violet in PBS containing 2% (v/v) ethanol for 1 h before colonies larger than 100 µm were counted.
Immunofluorescence and confocal imaging
Cells were rinsed in PBS and fixed in 4% PFA. Cells were then incubated with rabbit anti-GST and mouse anti-mTOR antibodies [Thermofisher Scientific; 1/400 dilution for both, in PBS containing 0.5% (m/v) BSA and 0.08% (m/v) Saponin] and were then stained with donkey anti-rabbit-Cy3 and donkey anti-mouse-Alexa Fluor 488 antibodies [Jackson ImmunoResearch, PA, USA; 1/400 dilution for both, in PBS containing 0.5% (m/v) BSA and 0.08% (m/v) Saponin]. The cells were counter-stained with DAPI (Thermofisher Scientific) and were then mounted for imaging. Images were acquired on the Leica TCS SP8X confocal microscope (Leica Microsystems, Wetzlar, Germany) and were processed using the FIJI software [70].
Tissue microarrays (TMA)
Formalin-fixed and paraffin-embedded human prostate cancer tissue microarrays were performed as previously described [76]. The human prostate cancer microarray (catalogue No. HProA180PG05, Shanghai Biochip, Shanghai, China) contains 180 cylinders from 90 paired prostate cancer and adjacent normal tissues. 6 patient samples did not show significant binding and were, therefore, excluded from this study. Sections (4 μm) were placed on slides coated with 3-aminopropyltriethoxysilane. For IHC analysis, TMA slides were first deparaffinized and then rehydrated. After antigen retrieval, endogenous peroxidase activity was blocked by incubating with 3% (v/v) H2O2 in methanol, nonspecific binding sites were blocked with 1% (m/v) BSA. Samples were incubated with the P-MNK2 Ser74 antibody (1:50 dilution) overnight at 4 °C followed by incubation with SignalStain® Boost IHC Detection Reagent (HRP, Rabbit) (catalogue No. 8114, cell signaling technologies, Danvers, MA, USA), the sections were developed in diaminobenzidine solution and counterstained with hematoxylin. Immunostaining was scored according to the intensity in comparison to negative controls (without primary antibody). The average immunoreactivity was graded on a scale of 0 (none), 0.5 + (very weak); 1 + (weak); 2 + (intermediate); and 3 + (strong). Scoring was undertaken by a pathologist in a blinded manner.
Proliferation assays
Cell proliferation rates were determined by either cell counting using haemocytometer, or BrdU incorporation assays using BrdU Cell Proliferation Assay Kit (Cell Signaling Technology).
Ex vivo human prostate tumour culture
Prostate cancer tissue was obtained with written informed consent through the Australian Prostate Cancer BioResource from men undergoing robotic radical prostatectomy at the St. Andrews Hospital, Adelaide, SA, Australia. An 8 mm core of tissue was dissected into 1–2 mm3 pieces and cultured on pre-cut and pre-soaked Gelfoam sponges (Pfizer, Thebarton, SA, Australia) in 24-well plates containing 500 μl of phenol red-free Roswell Park Memorial Institute-1640 medium (Thermofisher Scientific) with 10% FBS, antibiotic/antimycotic solution, 0.01 mg/ml hydrocortisone and 0.01 mg/ml insulin, and either vehicle (DMSO), 5 or 10 μM eFT508. Tissues were cultured at 37 °C for 48 h, then formalin-fixed and paraffin embedded.
Statistical analysis
Statistical analysis was performed using a one-way or two-way analysis of variance with an unpaired student t test or Dunnett’s post hoc test with the means of three independent experiments unless otherwise specified. GraphPad Prism software package was used to calculate P values. Results are means ± S.D. One-way: *0.01 ≤ P < 0.05; **0.001 ≤ P < 0.01; ***P < 0.001. Two-way: @0.01 ≤ P < 0.05; #0.001 ≤ P < 0.01; $P < 0.001.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We gratefully acknowledge financial support from the South Australian Health & Medical Research Institute (SAHMRI), Australia; the Wales Gene Park, UK; the Tuberous Sclerosis Association, UK; Science Foundation for Distinguished Young Scholar of Shanghai, China (Kaikai Shen, No. 2017067) and Xinglin Scholar Talent Program from Shanghai University of Traditional Chinese Medicine, China (Kaikai Shen, No. A1-U17205010436). JX is supported by SAHMRI E/MCR seed funding Grant. LMB is supported by a Principal Cancer Research Fellowship produced with the financial and other support of Cancer Council SA's Beat Cancer Project on behalf of its donors and the State Government of South Australia through the Department of Health. LMB acknowledges Grant support from The Movember Foundation/Prostate Cancer Foundation of Australia (MRTA3) and Cancer Australia (1138766). We would also like to thank Dr Randall Grose from the Australian Cancer Research Foundation (ACRF) Cellular and Imaging Cytometry Core Facility at SAHMRI for his assistance and expertise in FACS cell sorting.
Author contributions
JX performed most of the experiments. KS, ATJ, JY, ART, MY, MHS, SI, DW, JM, RVL, SDP, PJT, MFS, MK, SKM, ST and XW also provided data and designed the experiments. MHS, KBJ, MFS, SF, LMB, ACWZ and CGP helped designed the experiments and provided supervision. JX and CGP wrote the manuscript with help from KS, ART, MHS, SI, MFS, XW, LMB and ACWZ. All authors commented on the manuscript.
Compliance with ethical standards
Conflict of interests
The authors declare that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Furic L, Rong L, Larsson O, Koumakpayi IH, Yoshida K, Brueschke A, Petroulakis E, Robichaud N, Pollak M, Gaboury LA, et al. eIF4E phosphorylation promotes tumorigenesis and is associated with prostate cancer progression. Proc Natl Acad Sci USA. 2010;107:14134–14139. doi: 10.1073/pnas.1005320107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Biffo S, Manfrini N, Ricciardi S. Crosstalks between translation and metabolism in cancer. Curr Opin Genet Dev. 2017;48:75–81. doi: 10.1016/j.gde.2017.10.011. [DOI] [PubMed] [Google Scholar]
- 3.Flynn A, Proud CG. Serine 209, not serine 53, is the major site of phosphorylation in initiation factor eIF-4E in serum-treated Chinese hamster ovary cells. J Biol Chem. 1995;270:21684–21688. doi: 10.1074/jbc.270.37.21684. [DOI] [PubMed] [Google Scholar]
- 4.Waskiewicz AJ, Flynn A, Proud CG, Cooper JA. Mitogen-activated protein kinases activate the serine/threonine kinases Mnk1 and Mnk2. EMBO J. 1997;16:1909–1920. doi: 10.1093/emboj/16.8.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ueda T, Watanabe-Fukunaga R, Fukuyama H, Nagata S, Fukunaga R. Mnk2 and Mnk1 are essential for constitutive and inducible phosphorylation of eukaryotic initiation factor 4E but not for cell growth or development. Mol Cell Biol. 2004;24:6539–6549. doi: 10.1128/MCB.24.15.6539-6549.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Proud CG. Phosphorylation and signal transduction pathways in translational control. Cold Spring Harb Perspect Biol. 2018;20:20. doi: 10.1101/cshperspect.a033050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Proud CG. Mnks, eIF4E phosphorylation and cancer. Biochim Biophys Acta. 2015;1849:766–773. doi: 10.1016/j.bbagrm.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 8.Bramham CR, Jensen KB, Proud CG. Tuning specific translation in cancer metastasis and synaptic memory: control at the MNK-eIF4E axis. Trends Biochem Sci. 2016;41:847–858. doi: 10.1016/j.tibs.2016.07.008. [DOI] [PubMed] [Google Scholar]
- 9.Pyronnet S, Imataka H, Gingras AC, Fukunaga R, Hunter T, Sonenberg N. Human eukaryotic translation initiation factor 4G (eIF4G) recruits mnk1 to phosphorylate eIF4E. EMBO J. 1999;18:270–279. doi: 10.1093/emboj/18.1.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buxade M, Parra-Palau JL, Proud CG. The Mnks: MAP kinase-interacting kinases (MAP kinase signal-integrating kinases) Front Biosci. 2008;13:5359–5373. doi: 10.2741/3086. [DOI] [PubMed] [Google Scholar]
- 11.Samatar AA, Poulikakos PI. Targeting RAS-ERK signalling in cancer: promises and challenges. Nat Rev Drug Discov. 2014;13:928–942. doi: 10.1038/nrd4281. [DOI] [PubMed] [Google Scholar]
- 12.Scheper GC, Morrice NA, Kleijn M, Proud CG. The mitogen-activated protein kinase signal-integrating kinase Mnk2 is a eukaryotic initiation factor 4E kinase with high levels of basal activity in mammalian cells. Mol Cell Biol. 2001;21:743–754. doi: 10.1128/MCB.21.3.743-754.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scheper GC, Parra JL, Wilson M, Van Kollenburg B, Vertegaal AC, Han ZG, Proud CG. The N and C termini of the splice variants of the human mitogen-activated protein kinase-interacting kinase Mnk2 determine activity and localization. Mol Cell Biol. 2003;23:5692–5705. doi: 10.1128/MCB.23.16.5692-5705.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parra-Palau JL, Scheper GC, Wilson ML, Proud CG. Features in the N and C termini of the MAPK-interacting kinase Mnk1 mediate its nucleocytoplasmic shuttling. J Biol Chem. 2003;278:44197–44204. doi: 10.1074/jbc.M302398200. [DOI] [PubMed] [Google Scholar]
- 15.Fukunaga R, Hunter T. MNK1, a new MAP kinase-activated protein kinase, isolated by a novel expression screening method for identifying protein kinase substrates. EMBO J. 1997;16:1921–1933. doi: 10.1093/emboj/16.8.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gingras AC, Raught B, Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem. 1999;68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- 17.Xie J, Proud CG. Signaling crosstalk between the mTOR complexes. Translation (Austin) 2014;2:e28174. doi: 10.4161/trla.28174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saxton RA, Sabatini DM. mTOR signaling in growth, metabolism, and disease. Cell. 2017;169:361–371. doi: 10.1016/j.cell.2017.03.035. [DOI] [PubMed] [Google Scholar]
- 19.Fruman DA, Rommel C. PI3K and cancer: lessons, challenges and opportunities. Nat Rev Drug Discov. 2014;13:140–156. doi: 10.1038/nrd4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xie J, Wang X, Proud CG. mTOR inhibitors in cancer therapy. F1000Res. 2016;5:F1000. doi: 10.12688/f1000research.9207.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stead RL, Proud CG. Rapamycin enhances eIF4E phosphorylation by activating MAP kinase-interacting kinase 2a (Mnk2a) FEBS Lett. 2013;587:2623–2628. doi: 10.1016/j.febslet.2013.06.045. [DOI] [PubMed] [Google Scholar]
- 22.D'Abronzo LS, Bose S, Crapuchettes ME, Beggs RE, Vinall RL, Tepper CG, Siddiqui S, Mudryj M, Melgoza FU, Durbin-Johnson BP, et al. The androgen receptor is a negative regulator of eIF4E phosphorylation at S209: implications for the use of mTOR inhibitors in advanced prostate cancer. Oncogene. 2017;36:6359–6373. doi: 10.1038/onc.2017.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eckerdt F, Beauchamp E, Bell J, Iqbal A, Su B, Fukunaga R, Lulla RR, Goldman S, Platanias LC. Regulatory effects of a Mnk2-eIF4E feedback loop during mTORC1 targeting of human medulloblastoma cells. Oncotarget. 2014;5:8442–8451. doi: 10.18632/oncotarget.2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grzmil M, Huber RM, Hess D, Frank S, Hynx D, Moncayo G, Klein D, Merlo A, Hemmings BA. MNK1 pathway activity maintains protein synthesis in rapalog-treated gliomas. J Clin Invest. 2014;124:742–754. doi: 10.1172/JCI70198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marzec M, Liu X, Wysocka M, Rook AH, Odum N, Wasik MA. Simultaneous inhibition of mTOR-containing complex 1 (mTORC1) and MNK induces apoptosis of cutaneous T-cell lymphoma (CTCL) cells. PLoS ONE. 2011;6:e24849. doi: 10.1371/journal.pone.0024849. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Wang X, Yue P, Chan CB, Ye K, Ueda T, Watanabe-Fukunaga R, Fukunaga R, Fu H, Khuri FR, Sun SY. Inhibition of mammalian target of rapamycin induces phosphatidylinositol 3-kinase-dependent and Mnk-mediated eukaryotic translation initiation factor 4E phosphorylation. Mol Cell Biol. 2007;27:7405–7413. doi: 10.1128/MCB.00760-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chresta CM, Davies BR, Hickson I, Harding T, Cosulich S, Critchlow SE, Vincent JP, Ellston R, Jones D, Sini P, et al. AZD8055 is a potent, selective, and orally bioavailable ATP-competitive mammalian target of rapamycin kinase inhibitor with in vitro and in vivo antitumor activity. Cancer Res. 2010;70:288–298. doi: 10.1158/0008-5472.CAN-09-1751. [DOI] [PubMed] [Google Scholar]
- 28.Herbert TP, Kilhams GR, Batty IH, Proud CG. Distinct signalling pathways mediate insulin and phorbol ester-stimulated eukaryotic initiation factor 4F assembly and protein synthesis in HEK 293 cells. J Biol Chem. 2000;275:11249–11256. doi: 10.1074/jbc.275.15.11249. [DOI] [PubMed] [Google Scholar]
- 29.Brown MC, Gromeier M. MNK controls mTORC1: substrate association through regulation of TELO2 binding with mTORC1. Cell Rep. 2017;18:1444–1457. doi: 10.1016/j.celrep.2017.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hara K, Maruki Y, Long X, Yoshino K, Oshiro N, Hidayat S, Tokunaga C, Avruch J, Yonezawa K. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell. 2002;110:177–189. doi: 10.1016/s0092-8674(02)00833-4. [DOI] [PubMed] [Google Scholar]
- 31.Wang L, Harris TE, Lawrence JC., Jr Regulation of proline-rich Akt substrate of 40 kDa (PRAS40) function by mammalian target of rapamycin complex 1 (mTORC1)-mediated phosphorylation. J Biol Chem. 2008;283:15619–15627. doi: 10.1074/jbc.M800723200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang X, Li W, Williams M, Terada N, Alessi DR, Proud CG. Regulation of elongation factor 2 kinase by p90(RSK1) and p70 S6 kinase. EMBO J. 2001;20:4370–4379. doi: 10.1093/emboj/20.16.4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pigott CR, Mikolajek H, Moore CE, Finn SJ, Phippen CW, Werner JM, Proud CG. Insights into the regulation of eukaryotic elongation factor 2 kinase and the interplay between its domains. Biochem J. 2012;442:105–118. doi: 10.1042/BJ20111536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim DH, Sarbassov DD, Ali SM, King JE, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110:163–175. doi: 10.1016/s0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- 35.Schalm SS, Blenis J. Identification of a conserved motif required for mTOR signaling. Curr Biol. 2002;12:632–639. doi: 10.1016/s0960-9822(02)00762-5. [DOI] [PubMed] [Google Scholar]
- 36.Schalm SS, Fingar DC, Sabatini DM, Blenis J. TOS motif-mediated raptor binding regulates 4E-BP1 multisite phosphorylation and function. Curr Biol. 2003;13:797–806. doi: 10.1016/s0960-9822(03)00329-4. [DOI] [PubMed] [Google Scholar]
- 37.Liu Q, Wang J, Kang SA, Thoreen CC, Hur W, Ahmed T, Sabatini DM, Gray NS. Discovery of 9-(6-aminopyridin-3-yl)-1-(3-(trifluoromethyl)phenyl)benzo[h][1,6]naphthyridin-2(1H)-one (Torin2) as a potent, selective, and orally available mammalian target of rapamycin (mTOR) inhibitor for treatment of cancer. J Med Chem. 2011;54:1473–1480. doi: 10.1021/jm101520v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carayol N, Vakana E, Sassano A, Kaur S, Goussetis DJ, Glaser H, Druker BJ, Donato NJ, Altman JK, Barr S, et al. Critical roles for mTORC2- and rapamycin-insensitive mTORC1-complexes in growth and survival of BCR-ABL-expressing leukemic cells. Proc Natl Acad Sci USA. 2010;107:12469–12474. doi: 10.1073/pnas.1005114107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu Y, Yoon SO, Poulogiannis G, Yang Q, Ma XM, Villen J, Kubica N, Hoffman GR, Cantley LC, Gygi SP, et al. Phosphoproteomic analysis identifies Grb10 as an mTORC1 substrate that negatively regulates insulin signaling. Science. 2011;332:1322–1326. doi: 10.1126/science.1199484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kang SA, Pacold ME, Cervantes CL, Lim D, Lou HJ, Ottina K, Gray NS, Turk BE, Yaffe MB, Sabatini DM. mTORC1 phosphorylation sites encode their sensitivity to starvation and rapamycin. Science. 2013;341:1236566. doi: 10.1126/science.1236566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang J, Kalogerou M, Samsel PA, Zhang Y, Griffiths DF, Gallacher J, Sampson JR, Shen MH. Renal tumours in a Tsc2(+/−) mouse model do not show feedback inhibition of Akt and are effectively prevented by rapamycin. Oncogene. 2015;34:922–931. doi: 10.1038/onc.2014.17. [DOI] [PubMed] [Google Scholar]
- 42.Pike KG, Malagu K, Hummersone MG, Menear KA, Duggan HM, Gomez S, Martin NM, Ruston L, Pass SL, Pass M. Optimization of potent and selective dual mTORC1 and mTORC2 inhibitors: the discovery of AZD8055 and AZD2014. Bioorg Med Chem Lett. 2013;23:1212–1216. doi: 10.1016/j.bmcl.2013.01.019. [DOI] [PubMed] [Google Scholar]
- 43.Wang X, Regufe da Mota S, Liu R, Moore CE, Xie J, Lanucara F, Agarwala U, Pyr Dit Ruys S, Vertommen D, Rider MH, et al. Eukaryotic elongation factor 2 kinase activity is controlled by multiple inputs from oncogenic signaling. Mol Cell Biol. 2014;34:4088–4103. doi: 10.1128/MCB.01035-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Price NT, Redpath NT, Severinov KV, Campbell DG, Russell JM, Proud CG. Identification of the phosphorylation sites in elongation factor-2 from rabbit reticulocytes. FEBS Lett. 1991;282:253–258. doi: 10.1016/0014-5793(91)80489-p. [DOI] [PubMed] [Google Scholar]
- 45.Huo Y, Iadevaia V, Yao Z, Kelly I, Cosulich S, Guichard S, Foster LJ, Proud CG. Stable isotope-labelling analysis of the impact of inhibition of the mammalian target of rapamycin on protein synthesis. Biochem J. 2012;444:141–151. doi: 10.1042/BJ20112107. [DOI] [PubMed] [Google Scholar]
- 46.Pende M, Kozma SC, Jaquet M, Oorschot V, Burcelin R, Le Marchand-Brustel Y, Klumperman J, Thorens B, Thomas G. Hypoinsulinaemia, glucose intolerance and diminished beta-cell size in S6K1-deficient mice. Nature. 2000;408:994–997. doi: 10.1038/35050135. [DOI] [PubMed] [Google Scholar]
- 47.Beggs JE, Tian S, Jones GG, Xie J, Iadevaia V, Jenei V, Thomas G, Proud CG. The MAP kinase-interacting kinases regulate cell migration, vimentin expression and eIF4E/CYFIP1 binding. Biochem J. 2015;467:63–76. doi: 10.1042/BJ20141066. [DOI] [PubMed] [Google Scholar]
- 48.Robichaud N, del Rincon SV, Huor B, Alain T, Petruccelli LA, Hearnden J, Goncalves C, Grotegut S, Spruck CH, Furic L, et al. Phosphorylation of eIF4E promotes EMT and metastasis via translational control of SNAIL and MMP-3. Oncogene. 2015;34:2032–2042. doi: 10.1038/onc.2014.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim J, Guan KL. mTOR as a central hub of nutrient signalling and cell growth. Nat Cell Biol. 2019;21:63–71. doi: 10.1038/s41556-018-0205-1. [DOI] [PubMed] [Google Scholar]
- 50.Tschopp C, Knauf U, Brauchle M, Zurini M, Ramage P, Glueck D, New L, Han J, Gram H. Phosphorylation of eIF-4E on Ser 209 in response to mitogenic and inflammatory stimuli is faithfully detected by specific antibodies. Mol Cell Biol Res Commun. 2000;3:205–211. doi: 10.1006/mcbr.2000.0217. [DOI] [PubMed] [Google Scholar]
- 51.Jin X, Merrett J, Tong S, Flower B, Xie J, Yu R, Tian S, Gao L, Zhao J, Wang X, et al. Design, synthesis and activity of Mnk1 and Mnk2 selective inhibitors containing thieno[2,3-d]pyrimidine scaffold. Eur J Med Chem. 2018;162:735–751. doi: 10.1016/j.ejmech.2018.10.070. [DOI] [PubMed] [Google Scholar]
- 52.Bain J, Plater L, Elliott M, Shpiro N, Hastie CJ, McLauchlan H, Klevernic I, Arthur JS, Alessi DR, Cohen P. The selectivity of protein kinase inhibitors: a further update. Biochem J. 2007;408:297–315. doi: 10.1042/BJ20070797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reich SH, Sprengeler PA, Chiang GG, Appleman JR, Chen J, Clarine J, Eam B, Ernst JT, Han Q, Goel VK, et al. Structure-based design of pyridone-aminal eFT508 targeting dysregulated translation by selective mitogen-activated protein kinase interacting kinases 1 and 2 (MNK1/2) inhibition. J Med Chem. 2018;61:3516–3540. doi: 10.1021/acs.jmedchem.7b01795. [DOI] [PubMed] [Google Scholar]
- 54.Huang J, Manning BD. The TSC1-TSC2 complex: a molecular switchboard controlling cell growth. Biochem J. 2008;412:179–190. doi: 10.1042/BJ20080281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ruggero D, Montanaro L, Ma L, Xu W, Londei P, Cordon-Cardo C, Pandolfi PP. The translation factor eIF-4E promotes tumor formation and cooperates with c-Myc in lymphomagenesis. Nat Med. 2004;10:484–486. doi: 10.1038/nm1042. [DOI] [PubMed] [Google Scholar]
- 56.Wendel HG, De Stanchina E, Fridman JS, Malina A, Ray S, Kogan S, Cordon-Cardo C, Pelletier J, Lowe SW. Survival signalling by Akt and eIF4E in oncogenesis and cancer therapy. Nature. 2004;428:332–337. doi: 10.1038/nature02369. [DOI] [PubMed] [Google Scholar]
- 57.Maimon A, Mogilevsky M, Shilo A, Golan-Gerstl R, Obiedat A, Ben-Hur V, Lebenthal-Loinger I, Stein I, Reich R, Beenstock J, et al. Mnk2 alternative splicing modulates the p38-MAPK pathway and impacts Ras-induced transformation. Cell Rep. 2014;7:501–513. doi: 10.1016/j.celrep.2014.03.041. [DOI] [PubMed] [Google Scholar]
- 58.Mogilevsky M, Shimshon O, Kumar S, Mogilevsky A, Keshet E, Yavin E, Heyd F, Karni R. Modulation of MKNK2 alternative splicing by splice-switching oligonucleotides as a novel approach for glioblastoma treatment. Nucleic Acids Res. 2018;46:11396–11404. doi: 10.1093/nar/gky921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Um SH, Frigerio F, Watanabe M, Picard F, Joaquin M, Sticker M, Fumagalli S, Allegrini PR, Kozma SC, Auwerx J, et al. Absence of S6K1 protects against age- and diet-induced obesity while enhancing insulin sensitivity. Nature. 2004;431:200–205. doi: 10.1038/nature02866. [DOI] [PubMed] [Google Scholar]
- 60.Hsu PP, Kang SA, Rameseder J, Zhang Y, Ottina KA, Lim D, Peterson TR, Choi Y, Gray NS, Yaffe MB, et al. The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science. 2011;332:1317–1322. doi: 10.1126/science.1199498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stelloo S, Sanders J, Nevedomskaya E, de Jong J, Peters D, van Leenders GJ, Jenster G, Bergman AM, Zwart W. mTOR pathway activation is a favorable prognostic factor in human prostate adenocarcinoma. Oncotarget. 2016;7:32916–32924. doi: 10.18632/oncotarget.8767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Murugan AK. mTOR: Role in cancer, metastasis and drug resistance. Semin Cancer Biol. 2019;20:20. doi: 10.1016/j.semcancer.2019.07.003. [DOI] [PubMed] [Google Scholar]
- 63.Onda H, Lueck A, Marks PW, Warren HB, Kwiatkowski DJ. Tsc2(+/−) mice develop tumors in multiple sites that express gelsolin and are influenced by genetic background. J Clin Invest. 1999;104:687–695. doi: 10.1172/JCI7319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Centenera MM, Gillis JL, Hanson AR, Jindal S, Taylor RA, Risbridger GP, Sutherland PD, Scher HI, Raj GV, Knudsen KE, et al. Evidence for efficacy of new Hsp90 inhibitors revealed by ex vivo culture of human prostate tumors. Clin Cancer Res. 2012;18:3562–3570. doi: 10.1158/1078-0432.CCR-12-0782. [DOI] [PubMed] [Google Scholar]
- 65.Bentzinger CF, Romanino K, Cloetta D, Lin S, Mascarenhas JB, Oliveri F, Xia J, Casanova E, Costa CF, Brink M, et al. Skeletal muscle-specific ablation of raptor, but not of rictor, causes metabolic changes and results in muscle dystrophy. Cell Metab. 2008;8:411–424. doi: 10.1016/j.cmet.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 66.Martin SK, Fitter S, Dutta AK, Matthews MP, Walkley CR, Hall MN, Ruegg MA, Gronthos S, Zannettino AC. Brief report: the differential roles of mTORC1 and mTORC2 in mesenchymal stem cell differentiation. Stem Cells. 2015;33:1359–1365. doi: 10.1002/stem.1931. [DOI] [PubMed] [Google Scholar]
- 67.Fitter S, Matthews MP, Martin SK, Xie J, Ooi SS, Walkley CR, Codrington JD, Ruegg MA, Hall MN, Proud CG, et al. mTORC1 plays an important role in skeletal development by controlling preosteoblast differentiation. Mol Cell Biol. 2017;37:20. doi: 10.1128/MCB.00668-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Weber K, Bartsch U, Stocking C, Fehse B. A multicolor panel of novel lentiviral "gene ontology" (LeGO) vectors for functional gene analysis. Mol Ther. 2008;16:698–706. doi: 10.1038/mt.2008.6. [DOI] [PubMed] [Google Scholar]
- 69.Tian S, Wang X, Proud CG. Oncogenic MNK signalling regulates the metastasis suppressor NDRG1. Oncotarget. 2017;8:46121–46135. doi: 10.18632/oncotarget.17555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xie J, Ponuwei GA, Moore CE, Willars GB, Tee AR, Herbert TP. cAMP inhibits mammalian target of rapamycin complex-1 and -2 (mTORC1 and 2) by promoting complex dissociation and inhibiting mTOR kinase activity. Cell Signal. 2011;23:1927–1935. doi: 10.1016/j.cellsig.2011.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xie J, Mikolajek H, Pigott CR, Hooper KJ, Mellows T, Moore CE, Mohammed H, Werner JM, Thomas GJ, Proud CG. Molecular mechanism for the control of eukaryotic elongation factor 2 kinase by pH: role in cancer cell survival. Mol Cell Biol. 2015;35:1805–1824. doi: 10.1128/MCB.00012-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Moore CE, Xie J, Gomez E, Herbert TP. Identification of cAMP-dependent kinase as a third in vivo ribosomal protein S6 kinase in pancreatic beta-cells. J Mol Biol. 2009;389:480–494. doi: 10.1016/j.jmb.2009.04.020. [DOI] [PubMed] [Google Scholar]
- 74.Natarajan TG, Kallakury BV, Sheehan CE, Bartlett MB, Ganesan N, Preet A, Ross JS, FitzGerald KT. Epigenetic regulator MLL2 shows altered expression in cancer cell lines and tumors from human breast and colon. Cancer Cell Int. 2010;10:13. doi: 10.1186/1475-2867-10-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Xie J, El Sayed NM, Qi C, Zhao X, Moore CE, Herbert TP. Exendin-4 stimulates islet cell replication via the IGF1 receptor activation of mTORC1/S6K1. J Mol Endocrinol. 2014;53:105–115. doi: 10.1530/JME-13-0200. [DOI] [PubMed] [Google Scholar]
- 76.Shen K, Xi Z, Xie J, Wang H, Xie C, Lee CS, Fahey P, Dong Q, Xu H. Guttiferone K suppresses cell motility and metastasis of hepatocellular carcinoma by restoring aberrantly reduced profilin 1. Oncotarget. 2016;7:56650–56663. doi: 10.18632/oncotarget.10992. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.