Abstract
In the ciliated protozoan Tetrahymena thermophila the ribosomal DNA (rDNA) minichromosome replicates partially under cell cycle control and is also subject to a copy number control mechanism. The relationship between rDNA replication and rRNA gene transcription was investigated by the analysis of replication, transcription, and DNA-protein interactions in a mutant rDNA, the rmm3 rDNA. The rmm3 (for rDNA maturation or maintenance mutant 3) rDNA contains a single-base deletion in the rRNA promoter region, in a phylogenetically conserved sequence element that is repeated in the replication origin region of the rDNA minichromosome. The multicopy rmm3 rDNA minichromosome has a maintenance defect in the presence of a competing rDNA allele in heterozygous cells. No difference in the level of rRNA transcription was found between wild-type and rmm3 strains. However, rmm3 rDNA replicating intermediates exhibited an enhanced pause in the region of the replication origin, roughly 750 bp upstream from the rmm3 mutation. In footprinting of isolated nuclei, the rmm3 rDNA lacked the wild-type dimethyl sulfate (DMS) footprint in the promoter region adjacent to the base change. In addition, a DMS footprint in the origin region was lost in the rmm3 rDNA minichromosome. This is the first reported correlation in this system between an rDNA minichromosome maintenance defect and an altered footprint in the origin region. Our results suggest that a promoter region mutation can affect replication without detectably affecting transcription. We propose a model in which interactions between promoter and origin region complexes facilitate replication and maintenance of the Tetrahymena rDNA minichromosome.
Vegetatively dividing cells of the single-celled eukaryote Tetrahymena thermophila contain two nuclei: the transcriptionally silent germ line micronucleus and the transcriptionally active macronucleus. These nuclei have distinct physical and functional characteristics. Unlike the diploid micronucleus, the macronucleus contains approximately 200 chromosomes as a result of genome fragmentation, is polygenomic, and does not form a conventional mitotic spindle during nuclear division. Development of the macronucleus entails DNA elimination and rearrangement, telomere addition, and DNA amplification (41). The macronuclear rRNA genes are located on a high-copy-number ribosomal DNA (rDNA) minichromosome (14, 18, 21, 41).
In the micronucleus of Tetrahymena, the ribosomal RNA gene (rDNA) is present in a single copy per haploid genome (50). This single-copy rDNA is amenable to genetic analysis (20, 22, 27, 48). During macronuclear development, each rDNA allele, along with 5′ and 3′ flanking DNA sequences, is excised from the micronuclear chromosome and formed into a 21-kb palindromic linear molecule containing two divergently transcribed rRNA genes, in a head-to-head configuration, and newly added telomeric DNA (Fig. 1A). This molecule is amplified to approximately 104 copies in the developing macronucleus and is maintained at this copy number during subsequent cell divisions (18, 20, 21, 28).
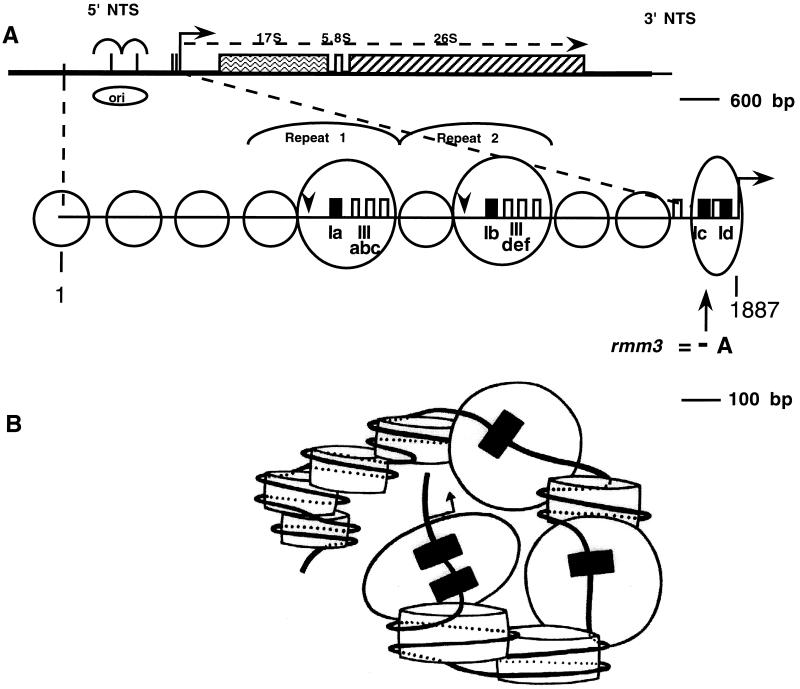
FIG. 1.
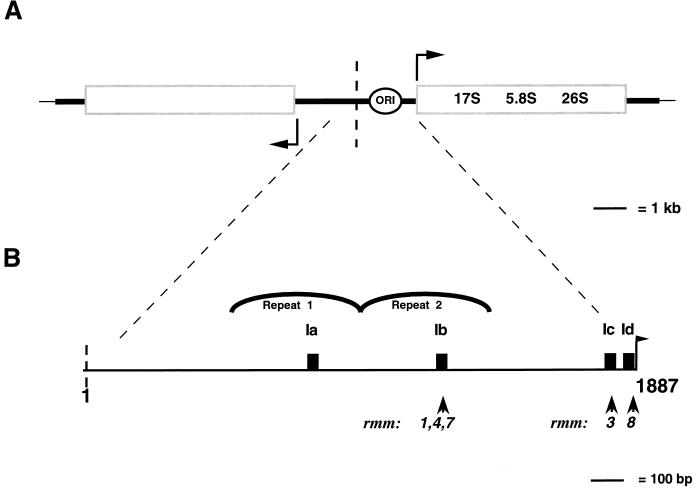
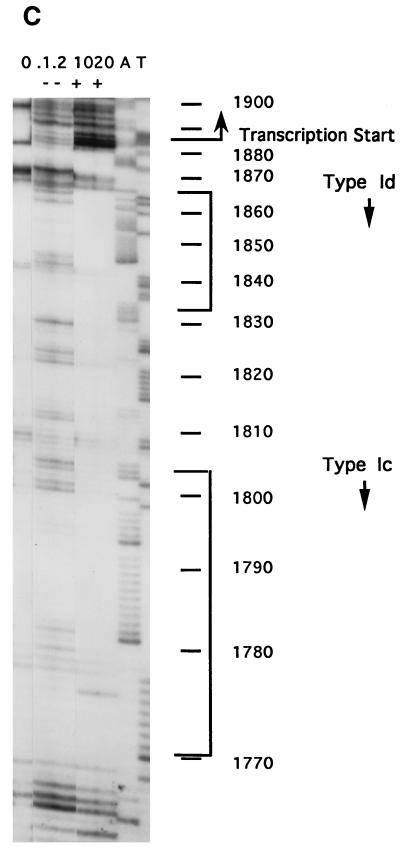
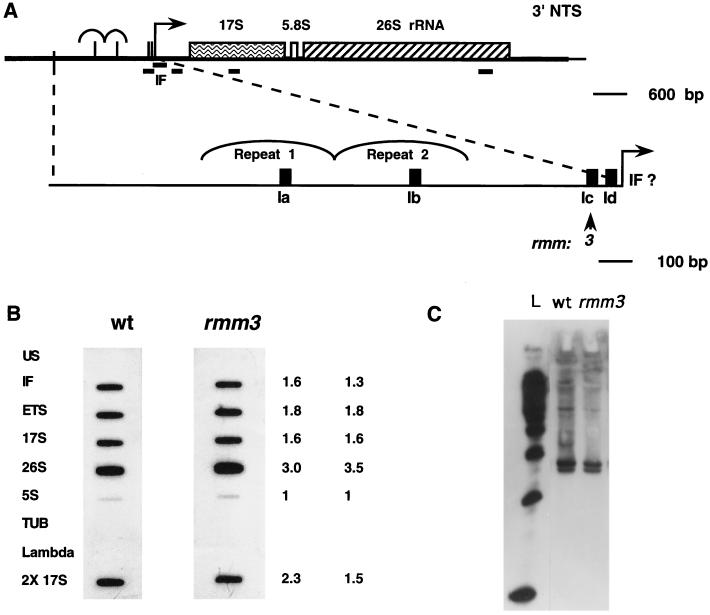
Organization of regions of the Tetrahymena macronuclear rDNA minichromosome. (A) Structure of the Tetrahymena macronuclear rDNA minichromosome. One half of the palindromic 21-kb rDNA minichromosome is shown. Each half carries a single 35S rRNA transcription unit (open rectangle) containing the 17S, 5.8S, and 26S rRNA coding sequences. Each half also contains an origin of DNA replication (ORI) within the 5′ NTS (open oval). Heavy black lines, 5′ and 3′ NTSs of the rRNA genes; vertical dashed line, center of the molecule; thin black line: telomeric DNA; bent arrow, start site of rRNA transcription. (B) rDNA maintenance mutant base changes within the 1.9-kb 5′ NTS. The ∼400-bp repeats 1 and 2 are indicated. Filled rectangles, type I elements (Ia to Id). Positions of base changes are indicated both for the previously identified mutations rmm1, -3, and -4 and for the mutations identified in this work, rmm7 and -8. Sequences of type Ia, Ib, and Ic repeats: TTTTTTTGGCAAAAAAAAAAACAAAAATAGTAA; sequence of type Id repeat: ATTCTTTGGCAAAAAAAATAAAAATAATATCAG/GG (the slash in Id indicates the 3′ boundary of this repeat). Mutated residues and repeats are in boldface and are as follows: rmm1 and -4, −A in type Ib (−720); rmm3, −A in type Ic (−100); rmm7, +A in type Ib (−720); rmm8, G to A 2 nt downstream of type Id (−19) (numbers in parentheses show the distance of the mutation from the start site of transcription [approximate for rmm1, -3, -4, and -7]). (C) DNase I footprint near the start site of rRNA transcription in wild-type C3 cells. Lanes: +, footprinting of rDNA in isolated nuclei; −, control footprinting performed in parallel with purified, deproteinized T. thermophila DNA; A and T, DNA sequencing lanes. Final DNase I concentrations (units per milliliter) are shown above the lanes; nucleotide positions in the rDNA (numbered outward from center of the molecule, starting at 1 as indicated) are shown at right. Bent arrow, transcription start site at position 1887.
A genetic screen to identify Tetrahymena mutants with altered macronuclear rDNA maturation or maintenance (rmm mutants) has been reported previously (22, 27, 48). Maintenance mutant minichromosomes are processed and amplified in the developing macronucleus but are lost from the macronucleus when in competition with a wild-type rDNA allele (27, 48). These mutants may be defective in DNA replication, minichromosome segregation, and/or copy number control. The genetic screen to identify rmm mutants was based on an in vivo competition between two naturally occurring rDNA alleles, C3 and B. Soon after mating between C3 and B strains, progeny cells, heterozygous for the two alleles, contain roughly equal amounts of each allele in the macronucleus (27, 35). However, within 70 generations the B rDNA allele is lost from the cell, even in the presence of selective pressure for this allele (27). Hence, the B rDNA allele is a naturally occurring minichromosome maintenance mutation (27, 37, 48). rDNA minichromosome maintenance mutants have been generated by mutagenizing C3 strain cells, crossing them with B strain rDNA marked with a drug-resistant 17S rRNA gene (27), and screening for drug-resistant (B allele-expressing) progeny in a C3 × B (C3/B) cross. Further analysis identified them as true rDNA maintenance mutants (19, 27), and linkage analysis indicated that most of the mutations were cis acting (19).
The 21-kb linear palindromic rDNA minichromosome has both halves derived from the same micronuclear allele. The minichromosome contains an example of a well-mapped eukaryotic cellular DNA replication origin (18, 21). In vegetatively growing cells, the physical origin of replication is within the 1.9-kb 5′ nontranscribed spacer (NTS) of the rRNA gene and localizes to roughly the repeat 1-repeat 2 region (Fig. 1B) (6, 31). Repeats 1 and 2 consist of two >90% homologous, ∼400-bp tandem direct repeats. The 5′ NTS also functions as an autonomously replicating sequence in Tetrahymena macronuclei (38, 51). The 5′ NTS contains the rDNA promoter region (39), deletion of which reduces the transformation efficiency of 5′-NTS constructs (13).
Several conserved sequence elements within the 5′ NTS were identified by sequence comparison of the 5′ NTSs of several tetrahymenid ciliates (7). Three type III sequence elements, sites of action of topoisomerase I (2), are present in each repeat, and two are located near the promoter (7). Four type I elements (Ia, Ib, Ic, and Id) are found within the 5′ NTS, one each in repeats 1 and 2 (Ia and Ib, respectively) and two (Ic and Id) near the start site of rRNA transcription (Fig. 1B) (7). Factors which bind the 33-nucleotide (nt) type I elements in vitro have been identified (17, 46), although their function is unknown.
Previously, base changes within type I elements which correlated with the presence of minichromosome maintenance defects were reported. In one mutant (rmm3), the base change was located in the type Ic element in the rRNA promoter region (1), and in two other mutants (rmm1 and rmm4), they were located in the type Ic element approximately 700 bp upstream, within the repeat 2 region of the 5′ NTS (Fig. 1B) (27, 48). Strikingly, the base change in each of these rDNA minichromosome maintenance mutants is a deletion of an A residue in the central run of 11 A’s in a type I sequence element. While minichromosome maintenance must entail replication, segregation, and copy number control (27, 28), the proximity of the upstream maintenance mutant base changes (rmm1 and rmm4) to the mapped replication origin suggested that these base changes affect rDNA minichromosome replication (21, 27).
Here we have analyzed replication, transcription, and DNA-protein interactions in the rmm3 rDNA minichromosome maintenance mutant. In addition, as further evidence of the importance of both the promoter and the repeat 2 region in rDNA minichromosome maintenance, we report that two other independently generated rDNA maintenance mutants, the rmm7 and rmm8 mutants (18, 22), have base changes in the repeat 2 and the rDNA promoter regions, respectively.
We show that the rmm3 rDNA base change causes a replication phenotype but has no gross effect on rRNA transcription. Within the rmm3 rDNA minichromosome, dimethyl sulfate (DMS) footprints are altered both at the site of the rmm3 base change in the promoter region and at two additional, newly identified footprinted sites, approximately 700 and 1,100 bp upstream, located in corresponding positions in repeats 1 and 2. These findings imply that physical or functional interactions between the promoter region and the repeat 1-2 region are important for wild-type chromosome maintenance. Our results also provide the first correlation between the loss of footprints in the 5′ NTS of the rDNA and a minichromosome maintenance phenotype.
MATERIALS AND METHODS
Cell strains and culture.
The wild-type cells used were C3 491-1a, a single progeny cell clone from a cross between C3 368-5, isolated from the wild, and an A* strain (generously provided by Eduardo Orias, University of California, Santa Barbara). The B strain used in mating experiments was SB1915 [ChxA2/ChxA2 Pmr/Pmr (cycl-S pm-S) II]. The rmm3 strain was a heterokaryon (obtained from Eduardo Orias) which was mated with an A* strain and then with itself in order to produce homozygous progeny. A slow-growth phenotype of the original rmm3 strain cells was backcrossed out (27, 34). Cells were grown as previously described (27). Cells were starved by being washed twice in 1× Dryl’s medium plus calcium and were then resuspended in this medium for between 16 and 24 h. Cells were refed by the addition of 5% proteose peptone-yeast extract-Sequestrene medium (PPYS) (5) to a final concentration of 2%.
Confirmation of the rmm3 phenotype.
Starved C3 491-1a or C3-rmm3 cells were paired with starved SB1915 (B strain rDNA) cells in six-well dishes for 24 h and then refed as described above. C3/B progeny were selected by the addition of cycloheximide to a final concentration of 15 μg/ml at 24 h after refeeding. Cells were replica plated to fresh medium every 24 h in order to maintain log-phase growth. DNA was prepared every 24 h (every six to seven generations) according to a standard protocol (20) except that 25 μl of NDS (0.5 M EDTA [pH 9.5], 10 mM Tris [pH 9.5], 2% sodium dodecyl sulfate) at 55°C and 25 μl of pronase (2 mg/ml) was added to 50 μl of pelleted cells, and 2 volumes of double-distilled H2O was added to samples prior to phenol-chloroform and chloroform extraction. DNA was resuspended in 25 μl of TE (10 mM Tris [pH 7.5], 1 mM EDTA) with 10 μg of RNase per ml, and 1 μg was cut with SphI. B rDNA contains an SphI site (at nt 1018) that is absent in C3. The Southern-blotted DNA was probed with a 1.9-kb 5′-NTS probe according to standard protocols (4). Relative amounts of B and C3 chromosomes were determined with a PhosphorImager by comparison of the 14-kb C3-specific band to the 6-kb B-specific band (two 6-kb bands are produced per B molecule). Results are graphed as the percentage of C3 chromosome present at each time point.
Sequencing of recombinant molecules.
The B strain rDNA has previously been sequenced in its entirety (12). The C3-specific primer, 36 r.c., and primer 12 (see below) were used to amplify DNAs from B/C3 and B/C3-rmm3 progeny cells. In control reactions with B rDNA alone, no product was obtained. Reactions with C3 rDNA or B/C3 rDNA gave a single product of the expected size. These PCR products were sequenced directly with primer 11 by using the fmol sequencing system (Promega). GATC reactions were done for the last time point of both crosses (generation 116); for the other time points, C-only reactions were done.
Sequencing of the rmm3 minichromosome.
The 5′ NTS of rmm3 rDNA was PCR amplified from the rmm3 homozygotes described above and sequenced directly. C3 rDNA contains an additional 42 bases at nt 1226 (27) which is not accounted for in the nucleotide numbering of the rDNA. The following primer combinations were used (nucleotide numbers, in parentheses, refer to the published sequence of B rDNA [12]): 19 and 5 r.c. (264 to 594); 20 and 60 (474 to 819), 60 r.c. and 36 (798 to 1226 + 30), 5 and 6 (615 to 989), Ib and 63 r.c. (1041 to 1454), 9 and 10 (1321 to 1699), 11 and 240 r.c. (1665 to 2128), and 63 and 12 (1494 to 1948). The sequences of these primers (5′→3′) are as follows (the first nucleotide is given in parentheses): 19, TAC AAA TTT ACA AAT TTT CAA GC (264); 5 r.c., CTA TTT ACT CAT ATT CCT AAA AC (594); 20, AAA GCA TCT AAA AAT GGA CA (474); 60, TTC AAC TCT CAA AAA AAG TG (819); 60 r.c., ATC ACT TTT TTT GAG AGT TG (798); 36, CAC GAA GTC TCA AAA GTT G (1226 + 34); 5, TTA GGA ATA TGA GTA AAT AG (575); 6, AAT GAT ATA CGC ATG CTG TTA (1029); Ib, AAC AAT TTT AAC AAC ATG CGT ATA TC (1001); 63 r.c., CTC CGC TGA ATA TTA AGC GAG (1513); 9, TGA TTT AGG AGA AAT TTT GAG (1321); 10, CGC TAT TTT TCA CTA AGT CTA (1699); 11, GCT CTA AAT TAA ATT AGA CTT AGT G (1665); 12, TCT TAC TGA AGC TCA AAT CGA GCT G (1948); 240 r.c., GCA TTG AAT TTA CAG CCT TCA TG (2128); and 63, CTC GCT TAA TAT TCA GCG GAG (1494). Also used in this work was primer 36 r.c. (CTT TTG CAA CTT TTG AGA CTT CG).
2D gel electrophoresis.
Wild type C3 491-1a and C3-rmm3 cells were grown in 200 ml of 2% PPYS to a density of 2 × 105 to 3 × 105 cells/ml for log-phase cell samples. In addition, cells were starved for 20 h and refed to synchronize them and to enrich for rDNA replicating intermediates (6, 11, 31). Aliquots (100 ml) were processed for DNA at 70, 80, and 90 min after refeeding. DNA was prepared as described previously (31) except that proteinase K was added to a final concentration of 8 mg/ml. Forty micrograms of DNA in a 1-ml volume was digested with a 10-fold excess of HindIII (20 U/liter) for 3 h at 37°C. The 4.2-kb central HindIII fragment spans both 5′ NTSs. DNA electrophoresis was performed in two-dimensional (2D) neutral-neutral gels essentially as reported by Brewer and Fangman (3). The first-dimension gel was 0.4% agarose containing 0.1 μg of ethidium bromide per ml and was run at 1.4 V/cm for 24 h in 1× Tris-borate-EDTA at room temperature. The second-dimension gel was 1% agarose–1× Tris-borate-EDTA containing 0.5 μg of ethidium bromide per ml and was run at 3 to 6 V/cm for 15 to 20 h until 1n and 2n sizes were separated by about 2 cm. DNA in the gel was depurinated in 0.25 M HCl for 10 to 15 min, denatured in 0.5 M NaOH, transferred to Nytran by capillary blotting in 10× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) for 12 to 24 h, and UV cross-linked. The hybridization probe was 32P-end-labeled oligonucleotide 60 r.c. (see above). Blots were washed at 45°C for 30 minutes and autoradiographed for 2 to 9 days.
Nuclear run-on assays.
For nuclear run-on assays, cell ghosts were prepared from 50 ml of cells according to the procedure of Love et al. (29) except that aurinotricarboxylic acid was omitted. Two hundred microliters of the cell ghost suspension was incubated with 2.5 μl (each) of 100 mM ribonucleoside triphosphates (except U) and [32P]UTP (120 μCi) for 2 min at 30°C as described previously (29). The labeled RNA was partially hydrolyzed with NaOH, hydrolysis was quenched with 125 μl of 1 M HEPES, and RNA was ethanol precipitated and resuspended in 100 μl of double-distilled water. Filters loaded with DNA were prepared as described previously (4) by using PCR-generated DNA fragments for the 35S rRNA and 5S RNA gene probes (between 250 ng and 1 μg of DNA loaded in each slot) and 5 μg of lambda DNA (nonspecific DNA control) per slot. The pTub9 plasmid, generously supplied by M. Gorovsky (University of Rochester), contains 283 nt of the Tetrahymena tubulin gene sequence (PolII transcribed). The plasmid was linearized with HindIII, and 2.5 μg of DNA was loaded per slot. In order to ensure that DNA was not limiting on the blot, twice as much DNA from one sample was hybridized to one slot in each experiment (2 × 17S). For each sample, the equivalent number of 32P RNA counts was added to 1.4 ml of hybridization solution (50% formamide, 5× Denhardt’s solution, 0.25% sodium dodecyl sulfate, 5× SSPE [1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA {pH 7.7}], and 150 μg of denatured salmon sperm DNA per ml) and hybridized at 42°C for 36 h. Blots were washed at room temperature for 15 min in 2× SSC, for 60 min at 65°C in 2× SSC, and for 30 min at 37°C in 2× SSC with 10 μg of RNase per ml. Autoradiography was done at room temperature for 10 min. Filters were analyzed with a PhosphorImager for quantitation. The raw data was normalized twice, first to the number of U’s in the transcribed region and then to the 5S gene hybridization of that blot.
Northern analysis.
RNA was prepared from 10 ml of log-phase, starved, or starved and refed cells with Tri-Reagent (Molecular Research Center, Inc.). After extraction with chloroform, RNA was precipitated with isopropanol, washed in 70% ethanol, and resuspended in 20 μl of double-distilled water. Samples were run on a 6% acrylamide (19:1)–0.6× Tris-borate-EDTA–7.5 M urea minigel, electroblotted to Nytran, and hybridized to various 5′-32P-labeled oligonucleotides at 45°C, stripping the blot between probes by a mock overnight hybridization. Sizes of the initiation fragment (IF) RNAs were estimated by running them on a DNA sequencing gel and assuming a migration difference of 10% between DNA and RNA.
Genomic footprinting.
Genomic footprinting was performed essentially as described previously (39). Nuclei were prepared as described previously (36). Two hundred milliliters of cells was grown to a density of 2 × 105 to 3 × 105 cells/ml, collected, and processed. For DMS footprinting, nuclei were resuspended in 800 μl of buffer A (60 mM KCl, 15 mM NaCl, 0.5 mM spermidine, 0.15 mM spermine, 15 mM Tris-HCl [pH 7.4], 2 mM CaCl2, 15 mM β-mercaptoethanol [BME]) and divided into 100-μl aliquots. Tubes were preincubated at 25°C for 2 min, and 10 μl of DMS (1:100 in double-distilled H2O; final concentration, 10 mM) was added, mixed for 10 s, and incubated for various periods of time. Reactions were stopped with the addition of 900 μl of 0.3 M BME in buffer A at 4°C. For the 0− time point, no DMS was added, for the 0+ time point, 0.3 M BME in buffer A was added prior to the addition of DMS. Nuclei were pelleted in an Eppendorf microcentrifuge at 6,500 rpm for 6 min at 4°C. Nuclei were resuspended in 200 μl of proteinase K solution (20 mM Tris-HCl [pH 8.0], 5 mM EDTA, 1% sodium dodecyl sulfate, and 0.5 mg of proteinase K [added fresh] per ml) and 0.3 M BME in buffer A in a 1:1 mix and incubated overnight at 37°C. Samples were extracted twice with phenol-chloroform and then chloroform, and DNA was ethanol precipitated. DNA methylated as chromatin was resuspended in 20 μl of double-distilled H2O. Naked DNA was prepared from nuclei by addition of 100 μl of proteinase K solution to a 100-μl nuclear suspension, incubation at 37°C overnight, and extraction and precipitation as described above. Unmethylated naked DNA was resuspended in 100 μl of buffer A and treated with DMS as described above for nuclei. These reactions were stopped by the addition of 35 μl of 3× stop mix (7.5 M NH4 acetate, 1.0 M BME, 0.2 mg of tRNA per ml) at 4°C, and DNA was ethanol precipitated. Methylated DNA was resuspended in 20 μl of double-distilled H2O and either heat cleaved (95°C for 5 min) or cleaved with pyrollidine (1:10 in double-distilled H2O) at 90°C for 15 min (the same results were obtained with either cleavage method), recovered by ethanol precipitation, resuspended in double-distilled H2O, lyophilized twice, and resuspended in 20 μl of TE. Primer extension was done as previously described (39) with primer 11 for the promoter region and primer 36 for the origin region (see above). The AT ladder was obtained by primer extending plasmid DNA in the presence of ddATP or ddTTP.
Sequencing of the 5′ NTSs of new maintenance mutants.
A TaqI fragment containing the 5′ NTS of the rDNA of each of the rmm5, -7, -8, and -9 maintenance mutants was cloned into Bluescript directly from the hemizygous strains SF104, SF108, SF112, and SF116, respectively (22). The four mutants were sequenced in parallel along with a wild-type 5′ NTS cloned from the hemizygous strain SF137 (22). By using either single-stranded or double-stranded DNA sequencing, for each 5′ NTS one strand was entirely sequenced and 75% of the other strand was sequenced.
Relative to C3 wild-type rDNA, B rDNA has a 42-bp deletion at nt 1226, which has been shown to be responsible for its defective maintenance phenotype (27, 48). Engberg and Nielsen (12) reported several unconfirmed C3 polymorphisms. We confirmed those at nt 1417 and 1442, a C-to-CA change at nt 1507, and a T-to-TTT change at nt 8560 in C3 wild-type rDNA, but nt 8567 and 8573 were the same in both strains, not different as reported previously (12).
RESULTS
The rmm3 rRNA promoter region mutant is defective in minichromosome maintenance.
The rRNA promoter in T. thermophila has not been defined functionally in vitro. However, DNase I footprinting of wild-type C3 nuclei reveals a series of enhancements and protections of DNA in chromatin that extend from the start site of transcription through the type Ic repeat (39) (Fig. 1C). Determination of the exact role that these sequences play in rRNA transcription will depend on functional analysis of mutants with mutations in this region. In the absence of functional data, but with the evidence of the DNase I footprint, we refer to the sequences from the start site of transcription through the type Ic repeat as the promoter region.
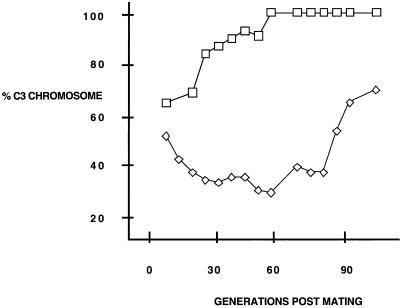
In order to dissect the role of the type I elements and the promoter region in minichromosome maintenance, we analyzed transcription, rDNA replicating intermediates, and protein-DNA interactions in C3-rmm3 (rmm3) rDNA, which contains a deletion of an A residue in the promoter-distal type Ic element (1) (Fig. 1B). We confirmed the effect of the rmm3 mutation on rDNA maintenance by crossing B strain cells with either C3-rmm3 or wild-type control C3 cells. Progeny were maintained in log-phase growth, and total DNA was isolated every six to seven generations. The ratio of the C3 rDNA allele (either wild type or mutant) to the B rDNA allele over time was determined. This ratio reflects the outcome of an in vivo competition, during the course of vegetative cell divisions, between the two rDNA alleles present in the macronucleus at the end of macronuclear development. The maintenance defect of the C3-rmm3 chromosome is illustrated in Fig. 2; compare the percent C3 (wild type or mutant) rDNA in progeny cells of the two crosses over time. In B/C3 cells the B rDNA minichromosome, a naturally occurring maintenance mutant, was completely lost within 60 generations, as seen previously (27). In contrast, in B/C3-rmm3 cells the B allele was never lost completely. Instead, the C3-rmm3 rDNA allele was preferentially lost until recombination (see below) between the two maintenance mutant alleles generated a C3 chromosome that had lost its maintenance defect, after approximately 90 generations (Fig. 2).
FIG. 2.
An in vivo competition assay demonstrates the maintenance defect of rmm3 rDNA. Wild-type C3 (□) or C3-rmm3 (◊) cells were crossed with B strain cells. Progeny were maintained in log-phase growth, and total DNA was isolated every six to seven generations, cut with SphI, and Southern blotted. The graph shows the percentage of C3 rDNA as a function of increasing number of generations.
The rmm3 base change is restored to wild-type in recombinant minichromosomes that have lost their maintenance defect.
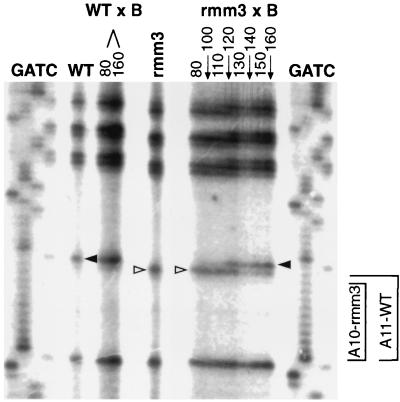
In order to determine whether the single A residue deletion within the type Ic repeat was associated with the maintenance defect of the C3-rmm3 chromosome, we sequenced the C3 promoter region in the rDNA populations of the B/C3-rmm3 cells described above. It was shown previously that in B/C3-rmm4 crosses, recombination between macronuclear rDNA minichromosome alleles eventually restored maintenance properties of the recombinant molecules, also confirming that the rmm4 base change was responsible for the maintenance defect (48). Similarly, the type Ic repeat of the C3 chromosome of progeny from the B/C3-rmm3 cross was restored to wild type in parallel with the reversion of the maintenance defect of the chromosome (Fig. 3; compare with Fig. 2). Furthermore, the sequence of the recombinant rDNA from generation 116 was C3 at each of six upstream C3/B polymorphisms (nt 1277, 1308, 1366, 1417, 1442, and 1507) (data not shown), indicating the specific recombination of the region of the type Ic repeat between the C3-rmm3 and the B chromosome. The only base change from the wild type in rmm3 rDNA in this region is the A deletion within the type Ic element; this strongly suggests that this base change is responsible for the maintenance phenotype of rmm3 rDNA. Similar kinetics were found previously for progeny of a B/C3-rmm4 cross (48). It was estimated that the frequency of recombination was between 10−3 and 10−7 per rDNA per generation, assuming a 5% growth advantage of recombinant cells and depending on the timing of the recombination event (48).
FIG. 3.
Loss of the rmm3 −A base change in the population of recombinant rDNA minichromosomes that have lost their maintenance defect. The rRNA promoter region of the rDNA minichromosome was amplified from total cell DNA isolated from each strain or progeny cell population. The C nucleotide sequencing reaction at the type Ic repeat region is shown for each sample or time point. Lanes: WT and rmm3, C sequencing reactions of wild-type C3 and rmm3 homozygotes respectively; rmm3 × B, progeny of the same B/C3-rmm3 cross shown in Fig. 2, from generations 60 to 116, as indicated above the lanes; WT × B, DNA from progeny of the B/C3 wild-type cross shown in Fig. 2 (generations 60 and 116 only); GATC, sequencing ladders of rDNA plasmid DNA. C3 rDNA has a run of 11 A residues between two C residues in the type Ic repeat; the C3-rmm3 mutant rDNA has only 10 A residues (side brackets). Hence, the next C residue in rmm3 rDNA (open arrowheads) is displaced downward by 1 nt relative to that in wild type rDNA (filled arrowheads).
rmm3 rDNA has altered DNA replication intermediates.
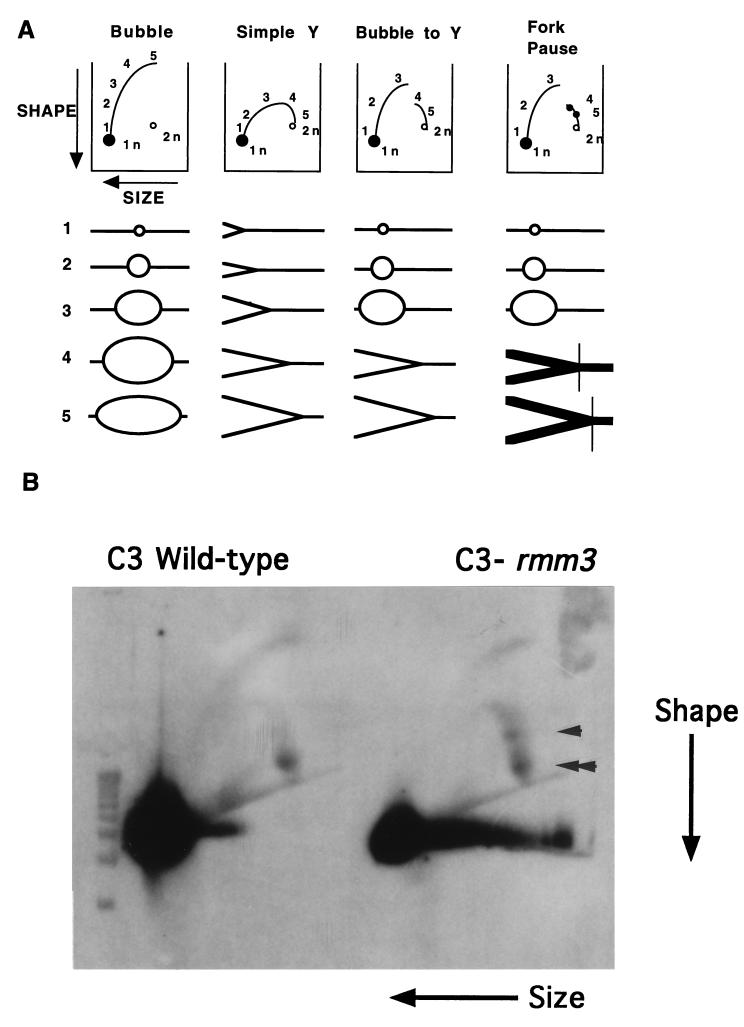
rDNA minichromosome replicating intermediates from C3 wild-type and C3-rmm3 homozygous cells were compared by using neutral-neutral 2D gel electrophoresis (3). Gels are run in the first dimension to separate DNA by size and in the second dimension to separate DNA molecules by shape. DNA molecules containing replication forks or bubbles are preferentially retarded in the second dimension, producing characteristic patterns (Fig. 4A) (3). DNAs from C3 wild-type and C3-rmm3 homozygous cells were prepared and restricted with HindIII. This produces a 4.2-kb central restriction fragment in the rDNA minichromosome. The 2D gel pattern of this central fragment indicates that it contains a replication bubble located asymmetrically within the fragment (Fig. 4A, Bubble to Y) (31), as predicted from earlier results with electron microscopy and density labeling (6, 21). Three replication fork pauses within the 5′ NTS of the Tetrahymena rDNA, which map to repeat 1, repeat 2, and the promoter, respectively, have been described. The repeat 2 and promoter pauses are shown schematically in Fig. 4A (Fork Pause). The role and cause of these pauses have not yet been determined (31).
FIG. 4.
2D gel electrophoretic analysis of rDNA replicating intermediates. (A) Neutral-neutral 2D gel patterns of representative restriction fragments containing replicating DNA intermediates (3). Bubble, origin located in the center of the fragment; Simple Y, fragment passively replicated by a fork originating outside; Bubble to Y, replication bubble located asymmetrically within the fragment. Arrows indicate the directions of DNA migration. 1n, unreplicated bulk DNA fragment; 2n, almost fully replicated DNA fragment. Replicating molecules depicted below each panel run above the arc of linear DNA (connects 1n to 2n) (not illustrated here). Each number along an arc identifies the replicating molecule that runs at that position. Fork Pause, a fork pause leads to the accumulation and hence overrepresentation of a particular replicating intermediate, resulting in increased hybridization at a spot on the arc of replicating molecules (black dots 4 and 5) (see panel B). Vertical line across dark replicating intermediates, location of the fork pause. (B) Enhanced accumulation of rDNA replicating intermediates at a specific pause site in the 5′ NTS of C3-rmm3 cells. DNA from log-phase C3 wild-type or C3-rmm3 cells was restricted with HindIII. Neutral-neutral 2D gels were run as described in Materials and Methods. Double arrowhead, promoter pause (dot 5 in panel A); arrowhead, repeat 2 pause (dot 4 in panel A).
Figure 4B shows the 2D gel patterns obtained from homozygous C3 and homozygous C3-rmm3 cells. Under the conditions used here, only the repeat 2 pause and the promoter pause were seen. Compared to wild-type rDNA, rmm3 rDNA replicating intermediates exhibited an enhancement of the repeat 2 pause relative to the promoter pause, indicating that in rmm3 rDNA elongation of replication is slowed or blocked preferentially at repeat 2. This enhancement of the repeat 2 pause in the rmm3 mutant was also seen both in cells synchronized by starvation and refeeding (Fig. 4B and data not shown) and in log-phase cells (data not shown). The repeat 2 pause site has been mapped to nt 1075 (±35 nt) (31). This site is roughly 750 nt upstream of the rmm3 base change.
The rmm3 rRNA promoter region mutation does not detectably affect the level of rRNA transcription.
We analyzed rRNA transcription in homozygous C3-rmm3 cells in two ways: by comparing polymerase densities along the rRNA gene by nuclear run-on assays and by determining steady-state levels of a small rRNA gene transcript by Northern analysis. The 35S RNA PolI rRNA transcript is processed to produce the 17S, 5.8S, and 26S rRNAs (Fig. 5A) (24). A short transcript which maps to the 5′ external transcribed spacer (ETS) just upstream of the 17S gene has been described previously and was termed the IF (10, 23, 24). It was proposed that the IF is a product of premature transcription termination at the PolI promoter, because it does not correspond to any known processing product of the 35S rRNA (23).
FIG. 5.
No effects on rRNA transcription are detected in rmm3 cells. (A) Map of rDNA minichromosome showing locations of the PCR-generated DNA probes used to analyze run-on transcription in wild-type and C3-rmm3 maintenance mutant strains; one half of the palindromic minichromosome is shown. Thin line at the end of the 3′ NTS, telomere. The expanded view of the 5′ NTS (symbols are as in Fig. 1) shows the location of the rmm3 mutation in the promoter-distal type Ic repeat. Probes: US (upstream probe), IF, ETS, 17S rRNA, and 26S rRNA. Bent arrowhead, start site of rRNA transcription; IF?, see text and panel C. (B) rmm3 homozygotes are not defective in the initiation of transcription. Run-on assays were performed on cell ghosts prepared from log-phase cells. DNA probes were those in panel A and the 5S rRNA gene, gamma tubulin gene (TUB), and lambda phage DNA. wt, wild type. (C) “IrFrt” transcripts are not altered in C3-rmm3 cells. A Northern blot of total cell RNA prepared from log-phase wild-type C3 (wt) and C3-rmm3 cells, probed with primer 12, is shown L, 100-bp ladder.
Run-on assays were performed with log-phase cell ghosts (29), as described in Materials and Methods. Figure 5A indicates the locations of the DNA probes along the 35S transcript. The hybridization intensity along the 35S rRNA gene was normalized first to the number of 32P-U residues in each RNA segment, as shown in Fig. 5B (left column), and then (right column) to the intensity of the 5S rRNA gene, transcribed by RNA polymerase III (25). RNA polymerase I density throughout the 35S pre-rRNA transcription unit was equivalent in log-phase C3 wild-type and C3-rmm3 mutant cells (Fig. 5B). In addition, there was no evidence that the initiator fragment is a premature termination product, since the intensities of hybridization of labeled RNA to the IF and ETS probes were similar in both wild-type and rmm3 mutant cells.
The initiator fragment was first reported as an approximately 230-nt RNA which hybridized to a probe derived from ETS sequences upstream of the 17S rRNA gene (10, 24). We prepared RNA from log-phase C3 wild-type and C3-rmm3 homozygous cells and ran these on an acrylamide gel. Northern analysis revealed two transcripts of approximately 225 and 240 nt that hybridized to probes containing sequences from the first 200 nt of the 35S transcript (Fig. 5C). Neither RNA hybridized to an oligonucleotide probe that extended from nt −21 to −4 from the start site of transcription, while both hybridized to an oligonucleotide probe that extended from nt +38 to +62. Only the longer RNA hybridized to a probe extending from nt +219 to +241 (data not shown). We conclude that these two initiator fragment RNAs initiate at or near nt +1 and extend for approximately 225 and 240 nt, respectively. The rmm3 base change had no effect on the sizes and no consistent effect on the abundances of these RNAs in either log-phase (Fig. 5C) or starved (data not shown) cells. Thus, we detected no effect of the rmm3 base change on rRNA gene transcription by Northern analysis or by run-on analysis.
A DMS promoter footprint is lost in isolated nuclei from rmm3 cells.
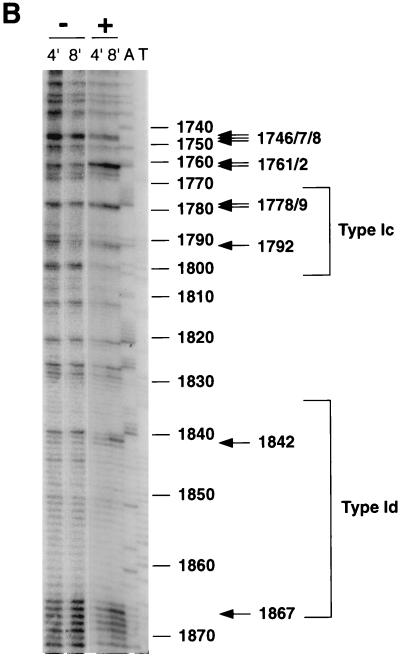
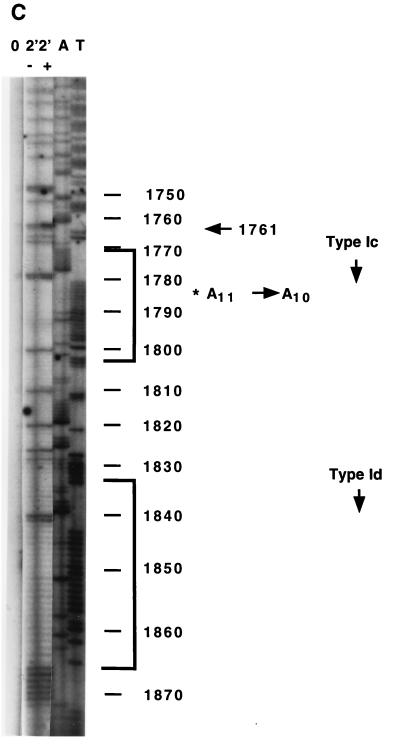
Footprinting of the promoter region in wild-type C3 nuclei revealed enhancements of DMS methylation at residues both within and upstream of the type Ic and type Id elements. Comparison of DNA methylated as purified DNA with that methylated as chromatin in wild-type nuclei revealed residues with enhanced methylation in chromatin, as summarized schematically in the map shown in Fig. 6A. Figure 6B shows the DMS footprinting data for the promoter region of wild-type C3 rDNA. The enhanced methylation at nt 1792 was at a C residue, indicating that this nucleotide is single stranded in chromatin, since C’s are inaccessible to DMS when base paired in double-stranded DNA. The other sites with enhanced methylation were G residues, indicating an alteration of the major groove of double-stranded DNA at these positions (32). Since rmm3 rDNA has a base change in the promoter-distal type Ic element, we investigated the effect of this mutation on the promoter region footprints. Surprisingly, the single-base deletion in rmm3 rDNA significantly altered the DMS footprint throughout the promoter region, leaving chromatin with an accessibility to DMS which was nearly identical to that of purified DNA, with the exception of the enhancements at nt 1761 and 1762 (Fig. 6C). Thus, despite the observation that the level of rRNA transcription is not altered in these cells, rmm3 rDNA in isolated nuclei had lost the wild-type DMS footprint at the promoter region, with the exception of the enhancements upstream of the type Ic repeat. Similarly, the wild-type DMS footprint on the opposite strand in the promoter region was also lost in rmm3 cells (data not shown).
FIG. 6.
Summary of DMS footprinting of rDNA in isolated nuclei. (A) Map of the 5′-NTS regions in the rDNA shown footprinted in panels B and C and Fig. 7 and schematic summary of the footprinting results. Smaller bracket, region of the promoter footprint; larger bracket, region of the origin region footprint; circles, positioned nucleosomes (Nuc 1 to 7) of the 5′ NTS of the rDNA (15, 36); filled rectangles, type I elements (a, b, c, and d); open rectangles, type III elements (a to f), the sites of action of topoisomerase I (2); bent arrow, start site of rRNA transcription; arrowheads, positions of the footprinted residues shown in panels B and C and in Fig. 7 and 8 (topoisomerase I [Topo I], Nuc 5, and nt 701 and 1132). (B) DMS promoter footprint in wild-type C3 rDNA. Naked DNA, or DNA in chromatin of isolated nuclei, from wild-type C3 cells was treated with 10 mM DMS for 8 min. Treated DNA was extended with primer 12. Lanes: −, DMS-treated naked DNA; +, DMS-treated chromatin. rDNA nucleotide numbers are indicated on the side; type Ic and Id elements are bracketed. Nucleotides with enhanced DMS reactivity in chromatin are indicated by arrows. (C) C3-rmm3 lacks the wild-type DMS promoter footprint. Naked DNA, or DNA in chromatin of isolated nuclei, from C3-rmm3 maintenance mutant cells was treated with 10 mM DMS for 2 min (the same patterns were obtained by treatment for 8 min [data not shown]). All else was as in panel B above.
Loss of the only identifiable footprints in the nonnucleosomal portions of the origin region in rmm3 rDNA.
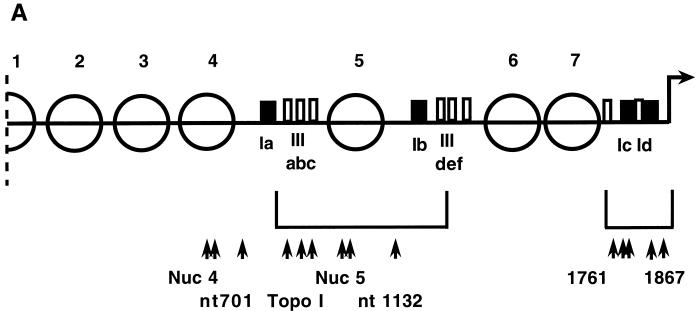
The chromatin structure of the rDNA 5′ NTS is extremely ordered, with two-thirds of it being packaged in highly positioned nucleosomes (15, 36). There are three nonnucleosomal regions: one in the promoter region described above and one within each of repeats 1 and 2. The latter approximately 270-nt nonnucleosomal regions, termed domains 1 and 2, were originally defined by their hypersensitivity to DNase I (36). They are highly homologous to one another in sequence (12).
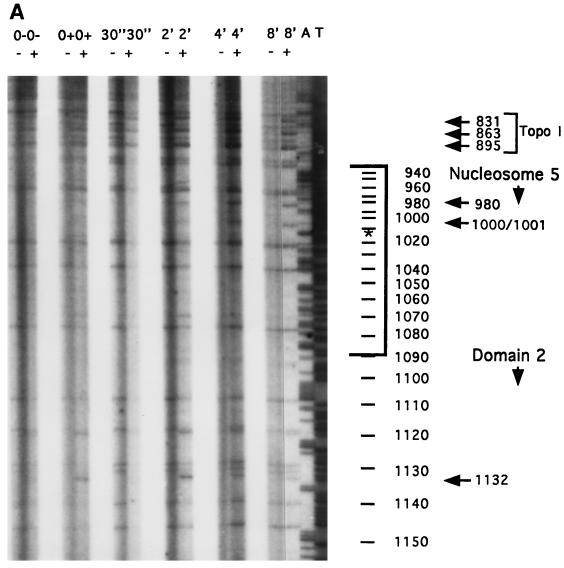
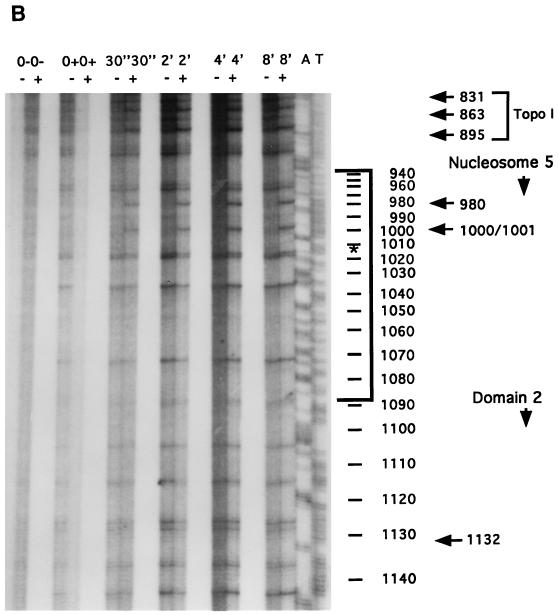
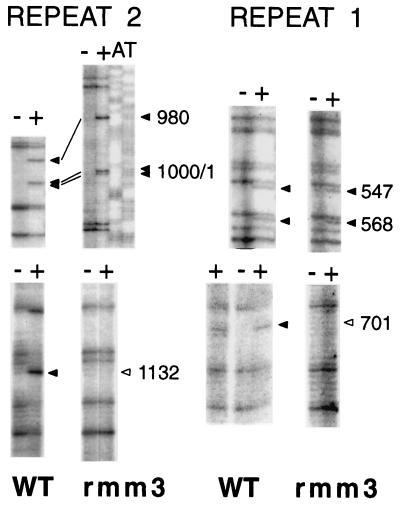
We used high-resolution footprinting to assess the reactivity of chromatin within domains 1 and 2 to DNase I, DMS, and KMnO4. With each reagent the patterns of the two domains were virtually identical (15). No DNase I-footprinted residues were found in either domain (data not shown). There were both DMS- and KMnO4-footprinted residues at the sites of action of topoisomerase I, that is, within the type III sequence elements (2, 7) (Fig. 7 and data not shown). There were no other KMnO4 footprints in the domains. However, there was an additional DMS footprint within each domain: a DMS-reactive A residue 61 nt upstream of the 5′ boundary of each type I element, at nt 701 and 1132 in repeats 1 and 2, respectively (Fig. 7A and 8 and data not shown).
FIG. 7.
DMS-reactive A at residue position 1132 in domain 2 (in repeat 2) of C3 wild-type rDNA but not in rmm3 mutant rDNA. (A) DMS footprinting of wild-type C3 rDNA. Chromatin in nuclei isolated from wild-type C3 strain cells was footprinted with DMS. Samples were treated for 0 or 30 s or 2, 4, or 8 min, as indicated. Lanes: −, naked DNA; +, chromatin. The 30-s (+) timepoint was underloaded. The footprinted region shown extends from just upstream of the type Ib repeat towards the center of the molecule, through a highly positioned nucleosome, labeled Nucleosome 5 (see Fig. 6A). The position of nucleosome 5 is bracketed. The asterisk marks its center. Nucleotide numbers are on the side. (B) C3-rmm3 rDNA has no DMS-reactive A at nucleotide 1132 in domain 2. Chromatin from C3-rmm3 maintenance mutant cells was footprinted with DMS as described for panel A. The footprinted region is as described for panel A.
FIG. 8.
DMS reactivity of A residues in repeats 1 and 2 in wild-type (WT) and rmm3 nuclei. (Top panels) DMS-reactive A residues (filled arrowheads) in nucleosome 5 (repeat 2) and nucleosome 4 (repeat 1) have similar reactivities in wild-type and rmm3 nuclei. (Bottom panels) The A residues at corresponding positions (nt 1132 and 701) in repeats 2 and 1, respectively, are DMS reactive in wild-type nuclei (filled arrowheads) but not in rmm3 nuclei (open arrowheads). For all reactions, marker A and T sequencing reactions were run next to the lanes shown (shown only for rmm3 repeat 2 nucleosome 5 region). Footprinting was performed as described for Fig. 7, in separate experiments, and with DNA and nuclear preparations different from those shown in Fig. 7.
Footprinting was focused on domain 2 in repeat 2, the site of the rmm1, rmm4, and rmm7 base changes. The same DMS-reactive A residue at position 1132 in domain 2 was observed in wild-type C3 rDNA, C3-rmm8 rDNA, C3-rmm7 rDNA, C3-rmm1 rDNA, B strain rDNA, and rDNA of cells transformed with the rDNA vector prD4-1 (Fig. 7A and data not shown). However, this DMS-reactive A residue was specifically absent in rmm3 rDNA (Fig. 7B) of both starved and log-phase cells. DMS footprints of three other A residues, located at positions 980, 1000, and 1001, were present in both wild-type C3 and C3-rmm3 rDNAs (Fig. 7B and 8, top panels) and in the other strains tested (data not shown), as were the footprints at the three topoisomerase I sites, at G residues. These provided internal controls for the reactivities of the samples with DMS. The corresponding A residue at position 701 in repeat 1 was also reactive to DMS in chromatin of wild-type C3, but not C3-rmm3, nuclei (Fig. 8, bottom panels). In both the wild-type C3 and C3-rmm3 rDNA chromatin, the control A residues in nucleosome 4 in repeat 1 were also DMS reactive (Fig. 8, top panels).
The specific loss of the DMS-reactive A residues in domains 1 and 2 of rmm3 maintenance mutant rDNA strongly suggests that these sites play a role in wild-type chromosome maintenance; these nonnucleosomal portions of the origin region are likely to be sites at which origin recognition and/or auxiliary factors bind (21, 31, 36).
Two new minichromosome maintenance mutants have novel base changes in the promoter region and the origin region, respectively.
Four rDNA maintenance mutants (rmm5, -7, -8, and -9 mutants) were generated by the genetic screen described previously (22). The entire 5′ NTS of each mutant rDNA was subcloned and sequenced. No base changes from the wild-type C3 rDNA sequence were found within the 5′ NTS of rmm5 or rmm9 rDNA, indicating that sequences outside the 5′ NTS play a role in rDNA minichromosome maintenance. In rmm7 and rmm8 rDNA, a base change within the 5′ NTS was identified: in rmm7 rDNA an insertion of an A residue in the origin region type Ib repeat and in rmm8 rDNA a G-to-A transition at position −19 from the start site of transcription, within a run of 6 G’s in the promoter (Fig. 1B). This mutation is in a strongly conserved, apparently critical region of the rRNA promoter, because an addition of a G residue to the same run of 6 G’s inactivates the promoter (39, 51). However, the rmm8 promoter is functional, since rmm8 homozygotes are viable. Analysis of recombinant molecules generated in a B/C3-rmm8 cross demonstrated that, like for the B/C3-rmm3 cross described above, in recombinant rDNA molecules the C3 rDNA maintenance mutant phenotype was lost in parallel with the restoration of the rmm8 G-to-A mutation to wild type (data not shown).
The locations of the base changes in the rmm7 and rmm8 maintenance mutants, within the repeat 2 region and the rRNA gene promoter region, respectively (Fig. 1B), further underscore the importance of these two regions in wild-type rDNA minichromosome maintenance. That the rmm7 base change is an addition of an A residue, and not a deletion of an A residue as for rmm1, -3, and -4, suggests that the type I elements may directly bind a control factor(s) or may be located between bound factors whose interaction is important for wild-type minichromosome maintenance.
DISCUSSION
The T. thermophila rDNA minichromosome, a naturally occurring eukaryotic cellular chromosome, is the smallest and most abundant of the macronuclear chromosomes with a well-defined chromatin structure (5, 15, 36), making it an appealing substrate for studies of chromosome structure and maintenance. We have used this macronuclear minichromosome as a model system for studies of chromosome maintenance. A link between transcription and replication in this molecule was suggested by previous findings: first, conserved type I elements are located at both the promoter region and the repeat 1-2 region, which contains the physical origin of replication of the minichromosome; second, base changes at both regions result in a defective minichromosome maintenance phenotype. Here we have shown that the single-base deletion in the promoter region type Ic element of the rmm3 rDNA maintenance mutant, while not grossly altering in vivo rRNA transcription, alters rDNA replication: it enhances a replication fork pause ∼750 bp upstream of the rmm3 base change itself, indicative of an effect on the elongation phase of rDNA replication. Finally, footprinting of wild-type and rmm3 minichromosomes in isolated nuclei demonstrated that the mutant rDNA had lost wild-type DMS footprints not only in the promoter region, which is close to the rmm3 base change, but also at distant upstream sites, in corresponding nucleosome-free positions in repeats 1 and 2. The DMS-reactive A residues at nt 701 and 1132 are 61 nt upstream of the type Ia and Ib elements, respectively (7, 31), and the footprinted site at nt 1132 is about 60 nt upstream of the pause site at nt 1075.
Our results suggest that factors that bind at the promoter function both in transcription and in chromosome maintenance and replication. The loss of the promoter footprint in rmm3 rDNA suggests that the promoter region complex is less stable in vitro in isolated rmm3 mutant nuclei than in wild-type cells; in vivo the result is a detectable effect on chromosome maintenance and replication with no apparent effect on transcription.
Links between DNA replication and transcription have been demonstrated in many systems (16), and transcription factors have been shown to play various roles in viral and cellular DNA replication origin function (9, 16). We propose that within the Tetrahymena macronuclear rDNA minichromosome, interactions between a complex at the promoter region and complexes at the repeat 1 and repeat 2 regions promote wild-type rDNA minichromosome maintenance. This is modeled in Fig. 9. We suggest that the interaction between factors at the promoter and the repeat 1-2 region may be facilitated by the highly positioned nucleosomes that package the majority of the 5′ NTS (Fig. 9A) (15, 36). In other systems, positioned nucleosomes located between transcription factors facilitate interactions between these factors (30, 42). The identities of such interacting factors in the Tetrahymena rDNA minichromosome have yet to be determined. In other organisms the transcription factors assembled at the rRNA promoter consist of TATA-binding protein and several transcription-associated factors at the core promoter element, with an additional factor at an upstream element (16, 45). The Tetrahymena TATA-binding protein has been identified (44), but other rDNA transcription factors have not yet been identified. No factors which bind to the origin region in vivo have been identified, with the exception of histones and topoisomerase I (2, 15, 36).
FIG. 9.
Proposed protein-DNA interactions in the rDNA 5′ NTS and promoter. (A) Map of the rDNA 5′ NTS and promoter region. Numbering of nucleotides is indicated at the rDNA center and transcription start site (1 and 1887). Nucleosomal protections (small circles) and the promoter region footprint (oval), described in this and other work (15, 36, 39) are indicated. The large circles indicate putative protein complexes at the nonnucleosomal domains 1 and 2 of repeats 1 and 2, respectively. Arrowheads demonstrate the positions of the DMS-reactive A residues identified in this work in domains 1 and 2 (positions 701 and 1132), whose reactivity is lost in rmm3 rDNA. The position of the rmm3 mutation (−A) is indicated. (B) Model of potential interactions between complexes at the promoter and upstream regions. Cylinders, highly positioned nucleosomes that package most of the DNA of the 5′ NTS (15); circles, DNA replication origin recognition factors bound to the DNA; oval, factors bound at the rRNA promoter.
DNA replication and maintenance of bovine papillomavirus (BPV) resemble those of the Tetrahymena rDNA minichromosome in that the DNA remains episomal, is amplified and subsequently maintained at a stable copy number, and is replicated primarily once per cell cycle, with some abrogation of cell cycle control (8, 21, 26). While the rRNA promoter is not absolutely required for transformation of Tetrahymena rDNA plasmids, its presence increases the transformation efficiency of rDNA origin constructs (13). Hence, the complex at the rDNA promoter of the Tetrahymena rDNA minichromosome may be functionally analogous to that involving the BPV transcriptional activator BPV E2, which complexes with the DNA replication initiation protein E1 at the BPV replication origin (40, 43, 47, 49). An rDNA complex might similarly facilitate binding of E1-analogous factors at the repeat 1-2 origin region.
The relationship between the enhanced replication fork pause, the loss of the origin region and promoter footprints, and the rmm3 maintenance phenotype has yet to be determined. The increased accumulation of replicating intermediates in the 5′ NTS of rmm3 cells indicates that at least one defect in rmm3 rDNA maintenance is at the level of replication elongation. This defect alone would affect rDNA minichromosome maintenance as a result of a combination of factors: copy number control of the rDNA minichromosome, the apparent abrogation of cell cycle control in the macronucleus, and the absence of a mechanism to ensure faithful segregation of replicated alleles (28, 41). Thus, a slowly replicating rDNA allele could be gradually displaced in the macronucleus by one which replicated more quickly, with eventual loss of the mutant allele.
Merchant et al. (33) have reported the characterization of Mcm10, a trans-acting factor important for minichromosome maintenance in Saccharomyces cerevisiae. MCM10 is identical to DNA43, a gene implicated in the initiation of DNA replication. Interestingly, mcm10-1 mutant cells display a replication elongation defect similar to that of C3-rmm3 rDNA: a replication fork pause is induced in the region of the replication origins affected in the mutant. This similarity between these minichromosome maintenance mutations, one cis acting (rmm3) and the other trans acting (mcm10-1), suggests that they may have similar mechanistic bases.
The subtle effects of the rmm3 promoter region mutation on rDNA minichromosome maintenance in competition with another rDNA allele were detectable because macronuclear division is amitotic and chromosomes do not segregate faithfully and because copy number control requires some abrogation of cell cycle control in the macronucleus (28, 41). Thus, the unusual features of the Tetrahymena macronucleus have allowed us to uncover an interplay between the promoter region and upstream nonnucleosomal regions of the rDNA minichromosome.
ACKNOWLEDGMENTS
We thank Alan Wolffe and David Allis for advice and encouragement during the course of this work and Jagoree Roy and John Prescott for critical readings of the manuscript. We thank Jeff Kapler for sharing results prior to publication, Drena Larson and Eduardo Orias for sharing unpublished results, and Eduardo Orias and members of his laboratory for the rmm3 strain. We thank Dudi Tzfati for help with the figures, Ann Froderberg for artwork and other support, and Tom Porter for help with manuscript preparation.
This work was supported by NIH grant GM32565 to E.H.B. R.C.G. was supported in part by the MSTP program at the University of California, San Francisco.
REFERENCES
- 1.Blackburn, E., D. Larson, and E. Orias. Unpublished data.
- 2.Bonven B J, Gocke E, Westergaard O. A high affinity topoisomerase I binding sequence is clustered at DNAase I hypersensitive sites in Tetrahymena R-chromatin. Cell. 1985;41:541–551. doi: 10.1016/s0092-8674(85)80027-1. [DOI] [PubMed] [Google Scholar]
- 3.Brewer B J, Fangman W L. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell. 1987;51:463–471. doi: 10.1016/0092-8674(87)90642-8. [DOI] [PubMed] [Google Scholar]
- 4.Brown T. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1993. pp. 2.9.1–2.9.6. [Google Scholar]
- 5.Budarf M L, Blackburn E H. Chromatin structure of the telomeric region and 3′-nontranscribed spacer of Tetrahymena ribosomal RNA genes. J Biol Chem. 1986;261:363–369. [PubMed] [Google Scholar]
- 6.Cech T R, Brehm S L. Replication of the extrachromosomal ribosomal RNA genes of Tetrahymena thermophilia. Nucleic Acids Res. 1981;9:3531–3543. doi: 10.1093/nar/9.14.3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Challoner P B, Amin A A, Pearlman R E, Blackburn E H. Conserved arrangements of repeated DNA sequences in nontranscribed spacers of ciliate ribosomal RNA genes: evidence for molecular coevolution. Nucleic Acids Res. 1985;13:2661–2680. doi: 10.1093/nar/13.7.2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DePamphilis M L. Eukaryotic DNA replication: anatomy of an origin. Annu Rev Biochem. 1993;62:29–63. doi: 10.1146/annurev.bi.62.070193.000333. [DOI] [PubMed] [Google Scholar]
- 9.Dutta A. Trans-plication factors? Curr Biol. 1993;3:709–712. doi: 10.1016/0960-9822(93)90076-z. [DOI] [PubMed] [Google Scholar]
- 10.Engberg J, Din N, Saiga H, Higashinakagawa T. Nucleotide sequence of the 5′-terminal coding region for pre-rRNA and mature 17S rRNA in Tetrahymena thermophila rDNA. Nucleic Acids Res. 1984;12:959–972. doi: 10.1093/nar/12.2.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Engberg J, Mowat D, Pearlman R E. Preferential replication of the ribosomal RNA genes during a nutritional shift-up in Tetrahymena pyriformis. Biochim Biophys Acta. 1972;272:312–320. [Google Scholar]
- 12.Engberg J, Nielsen H. Complete sequence of the extrachromosomal rDNA molecule from the ciliate Tetrahymena thermophila strain B1868VII. Nucleic Acids Res. 1990;18:6915–6919. doi: 10.1093/nar/18.23.6915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaertig, J. Personal communication.
- 14.Gall J G. The molecular biology of ciliated protozoa. Orlando, Fla: Academic Press, Inc.; 1986. [Google Scholar]
- 15.Gallagher, R. C., and E. H. Blackburn. Unpublished data.
- 16.Heintz N H. Transcription factors and the control of DNA replication. Curr Opin Cell Biol. 1992;4:459–467. doi: 10.1016/0955-0674(92)90012-2. [DOI] [PubMed] [Google Scholar]
- 17.Hou Z, Umthun A R, Dobbs D L. A single-stranded DNA binding protein that specifically recognizes cis-acting sequences in the replication origin and transcriptional promoter region of Tetrahymena rDNA. Biochemistry. 1995;34:4583–4592. doi: 10.1021/bi00014a011. [DOI] [PubMed] [Google Scholar]
- 18.Kapler G M. Developmentally regulated processing and replication of the Tetrahymena rDNA minichromosome. Curr Opin Gen Dev. 1993;3:730–735. doi: 10.1016/s0959-437x(05)80091-7. [DOI] [PubMed] [Google Scholar]
- 19.Kapler, G. M., and E. H. Blackburn. Unpublished data.
- 20.Kapler G M, Blackburn E H. A weak germ-line excision mutation blocks developmentally controlled amplification of the rDNA minichromosome of Tetrahymena thermophila. Genes Dev. 1994;8:84–95. doi: 10.1101/gad.8.1.84. [DOI] [PubMed] [Google Scholar]
- 21.Kapler G M, Dobbs D L, Blackburn E H. DNA replication in Tetrahymena. In: DePamphilis M L, editor. DNA replication in eukaryotic cells. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 915–932. [Google Scholar]
- 22.Kapler G M, Orias E, Blackburn E H. Tetrahymena thermophila mutants defective in the developmentally programmed maturation and maintenance of the rDNA minichromosome. Genetics. 1994;137:455–466. doi: 10.1093/genetics/137.2.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kister K P, Kaffenberger W, Eckert W A. In vitro synthesis and processing of pre-rRNA in isolated macronuclei from Tetrahymena. Eur J Cell Biol. 1988;46:233–243. [PubMed] [Google Scholar]
- 24.Kister K P, Muller B, Eckert W A. Complex endonucleolytic cleavage pattern during early events in the processing of pre-rRNA in the lower eukaryote, Tetrahymena thermophila. Nucleic Acids Res. 1983;11:3487–3502. doi: 10.1093/nar/11.11.3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumazaki I, Hori H, Osawa S, Mita T, Higashinakagawa T. The nucleotide sequences of 5S rRNAs from three ciliated protozoa. Nucleic Acids Res. 1982;10:4409–4413. doi: 10.1093/nar/10.14.4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lambert P F. Papillomavirus DNA replication. J Virol. 1991;65:3417–3420. doi: 10.1128/jvi.65.7.3417-3420.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Larson D D, Blackburn E H, Yaeger P C, Orias E. Control of rDNA replication in Tetrahymena involves a cis-acting upstream repeat of a promoter element. Cell. 1986;47:229–240. doi: 10.1016/0092-8674(86)90445-9. [DOI] [PubMed] [Google Scholar]
- 28.Larson D D, Umthun A R, Shaiu W-L. Copy number control in the Tetrahymena macronuclear genome. J Protozool. 1991;38:258–263. doi: 10.1111/j.1550-7408.1991.tb04439.x. [DOI] [PubMed] [Google Scholar]
- 29.Love H J, Allen N A, Zhao Q A, Bannon G A. mRNA stability plays a major role in regulating the temperature-specific expression of a Tetrahymena thermophila surface protein. Mol Cell Biol. 1988;8:427–432. doi: 10.1128/mcb.8.1.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu Q, Wallrath L, Elgin S. The role of a positioned nucleosome at the Drosophila melanogaster hsp26 promoter. EMBO J. 1995;14:4738–4746. doi: 10.1002/j.1460-2075.1995.tb00155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.MacAlpine D M, Zhang Z, Kapler G M. Type I elements mediate replication fork pausing at conserved upstream sites in the Tetrahymena thermophila ribosomal DNA minichromosome. Mol Cell Biol. 1997;17:4517–4525. doi: 10.1128/mcb.17.8.4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maxam A M, Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci USA. 1977;74:560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Merchant A M, Kawasaki Y, Chen Y, Lei M, Tye B K. A lesion in the DNA replication initiation factor Mcm10 induces pausing of elongation forks through chromosomal replication origins in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:3261–3271. doi: 10.1128/mcb.17.6.3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Orias, E. Personal communication.
- 35.Orias E, Bradshaw A D. Stochastic developmental variation in the ratio of allelic rDNAs among newly differentiated, heterozygous macronuclei of Tetrahymena thermophila. Dev Genet. 1992;13:87–93. doi: 10.1002/dvg.1020130114. [DOI] [PubMed] [Google Scholar]
- 36.Palen T E, Cech T R. Chromatin structure at the replication origins and transcription-initiation regions of the ribosomal RNA genes of Tetrahymena. Cell. 1984;36:933–942. doi: 10.1016/0092-8674(84)90043-6. [DOI] [PubMed] [Google Scholar]
- 37.Pan W-C, Orias E, Flacks M, Blackburn E H. Allele-specific, selective amplification of a ribosomal RNA gene in Tetrahymena thermophila. Cell. 1982;28:596–604. doi: 10.1016/0092-8674(82)90214-8. [DOI] [PubMed] [Google Scholar]
- 38.Pan W J, Blackburn E H. Tandem repeats of the 5′ non-transcribed spacer of Tetrahymena rDNA function as high copy number autonomous replicons in the macronucleus but do not prevent rRNA gene dosage regulation. Nucleic Acids Res. 1995;23:1561–1569. doi: 10.1093/nar/23.9.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pan W J, Gallagher R C, Blackburn E H. Replication of an rRNA gene origin plasmid in the Tetrahymena thermophila macronucleus is prevented by transcription through the origin from an RNA polymerase I promoter. Mol Cell Biol. 1995;15:3372–3381. doi: 10.1128/mcb.15.6.3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Piirsoo M, Ustav E, Mandel T, Stenlund A, Ustav M. cis and trans requirements for stable episomal maintenance of the BPV-1 replicator. EMBO J. 1996;15:1–11. [PMC free article] [PubMed] [Google Scholar]
- 41.Prescott D M. The DNA of ciliated protozoa. Microbiol Rev. 1994;58:233–267. doi: 10.1128/mr.58.2.233-267.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schild C, Claret F X, Wahli W, Wolffe A P. A nucleosome-dependent static loop potentiates estrogen-regulated transcription from the Xenopus vitellogenin B1 promoter in vitro. EMBO J. 1993;12:423–433. doi: 10.1002/j.1460-2075.1993.tb05674.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sedman J, Stenlund A. Co-operative interaction between the initiator E1 and the transcriptional activator E2 is required for replicator specific DNA replication of bovine papillomavirus in vivo and in vitro. EMBO J. 1995;14:6218–6228. doi: 10.1002/j.1460-2075.1995.tb00312.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stargell L A, Gorovsky M A. TATA-binding protein and nuclear differentiation in Tetrahymena thermophila. Mol Cell Biol. 1994;14:723–734. doi: 10.1128/mcb.14.1.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Steffan J S, Keys D A, Dodd J A, Nomura M. The role of TBP in rDNA transcription by RNA polymerase I in Saccharomyces cerevisiae: TBP is required for upstream activation factor-dependent recruitment of core factor. Genes Dev. 1996;10:2551–2563. doi: 10.1101/gad.10.20.2551. [DOI] [PubMed] [Google Scholar]
- 46.Umthun A R, Hou Z, Sibenaller Z A, Shaiu W L, Dobbs D L. Identification of DNA-binding proteins that recognize a conserved type I repeat sequence in the replication origin region of Tetrahymena rDNA. Nucleic Acids Res. 1994;22:4432–4440. doi: 10.1093/nar/22.21.4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Winokur P L, McBride A A. Separation of the transcriptional activation and replication functions of the bovine papillomavirus-1 E2 protein. EMBO J. 1992;11:4111–4118. doi: 10.1002/j.1460-2075.1992.tb05504.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yaeger P C, Orias E, Shaiu W L, Larson D D, Blackburn E H. The replication advantage of a free linear rRNA gene is restored by somatic recombination in Tetrahymena thermophila. Mol Cell Biol. 1989;9:452–460. doi: 10.1128/mcb.9.2.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang L, Li R, Mohr I J, Clark R, Botchan M R. Activation of BPV-1 replication in vitro by the transcription factor E2. Nature. 1991;353:628–632. doi: 10.1038/353628a0. [DOI] [PubMed] [Google Scholar]
- 50.Yao M-C. Amplification of ribosomal RNA genes. In: Gall J D, editor. The molecular biology of ciliated protozoa. New York, N.Y: Academic Press, Inc.; 1986. pp. 179–201. [Google Scholar]
- 51.Yu G-L, Blackburn E H. Amplification of tandemly repeated origin control sequences confers a replication advantage on rDNA replicons in Tetrahymena thermophila. Mol Cell Biol. 1990;10:2070–2080. doi: 10.1128/mcb.10.5.2070. [DOI] [PMC free article] [PubMed] [Google Scholar]