Abstract
Aberrant cholesterol metabolism causes neurological disease and neurodegeneration, and mitochondria have been linked to perturbed cholesterol homeostasis via the study of pathological mutations in the ATAD3 gene cluster. However, whether the cholesterol changes were compensatory or contributory to the disorder was unclear, and the effects on cell membranes and the wider cell were also unknown.
Using patient-derived cells, we show that cholesterol perturbation is a conserved feature of pathological ATAD3 variants that is accompanied by an expanded lysosome population containing membrane whorls characteristic of lysosomal storage diseases. Lysosomes are also more numerous in Drosophila neural progenitor cells expressing mutant Atad3, which exhibit abundant membrane-bound cholesterol aggregates, many of which co-localize with lysosomes. By subjecting the Drosophila Atad3 mutant to nutrient restriction and cholesterol supplementation, we show that the mutant displays heightened cholesterol dependence.
Collectively, these findings suggest that elevated cholesterol enhances tolerance to pathological ATAD3 variants; however, this comes at the cost of inducing cholesterol aggregation in membranes, which lysosomal clearance only partly mitigates.
Keywords: mitochondrial disease, cholesterol disorders, lysosomes, ATAD3, AAA+ ATPase, lysosomal storage disorders
Muñoz-Oreja et al. show that pathological mutations in the mitochondrial membrane protein ATAD3A cause cells to increase their cholesterol levels in a compensatory response, leading to cholesterol aggregation in cell membranes. Cells struggle to recycle these difficult-to-digest membranes, giving rise to neurological disease.
Introduction
The brain contains an order of magnitude more cholesterol than other tissues and defective cholesterol metabolism causes neurological disorders, such as Niemann Pick type C disease (NPC),1 and it is implicated in Alzheimer’s disease.2 Notwithstanding its special importance in the brain, cholesterol is essential in every cell and tissue as it determines the properties of biological membranes that are central to organelle biology and function. Specifically, as cholesterol increases membrane rigidity, excess cholesterol reduces membrane flexibility, which can interfere in the myriad activities of membrane-associated proteins.3-5
ATAD3A (ATPase family AAA domain-containing protein 3A) encodes a mitochondrial transmembrane protein whose dysfunction can perturb cellular cholesterol metabolism.6,7 ATAD3A is a member of the AAA+ family (ATPases associated with various cellular activities8) that has been linked to multiple activities and processes in mitochondria, which complicates the interpretation of the effects of pathological ATAD3 mutations. For example, oxidative phosphorylation dysfunction owing to ATAD3 deficiency displays considerable variability among tissues and individuals,6,9,10 and may not be evident in fibroblasts.7,10 Likewise, while there is ample evidence that a fraction of the human ATAD3 protein is tightly associated with mitochondrial DNA (mtDNA) and ATAD3 deficiency and dysfunction impact mtDNA topology and distribution from humans to plants,6,7,11-14 mtDNA depletion is rare in ATAD3 disorders.6,7,9,10,15 Furthermore, a substantial amount of ATAD3 is distributed throughout the mitochondrial network, leading to the suggestion that it forms filaments or scaffolds.12,16 Cholesterol may unify these apparently disparate features of ATAD3, as its association with cholesterol microdomains may be critical to its functional and structural roles.12,16,17
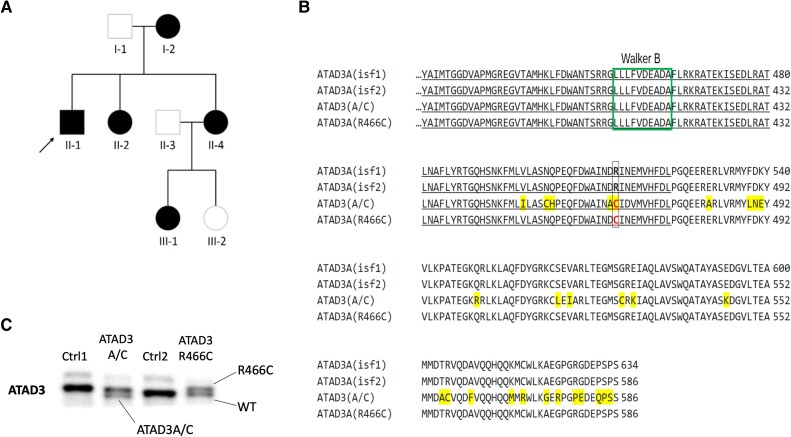
While most species have a single ATAD3 gene, humans possess three paralogues located in tandem on chromosome 1p36.33, ATAD3A, ATAD3B and ATAD3C, which evolved recently by segmental duplication of a single ancestral gene.18 To date, the allelic spectrum of ATAD3A-associated diseases encompasses null, hypomorphic and anti-morphic alleles (Supplementary Fig. 1),6,9,15,19-23 including the dominant de novo variants p.R528W and p.G355D, as well as the recessive changes p.Thr53Ile and p.Thr84Met.15,22,24 These variants are associated with diverse neurological and syndromic manifestations, ranging from cerebellar atrophy, axonal neuropathy and optic neuropathy to cardiomyopathy and seizures. The more severe manifestations are associated with large-scale genomic alterations of the ATAD3 gene cluster to which the region is predisposed by the genomic architecture of the three highly homologous ATAD3 genes.6,7,15,25 Biallelic deletions of ATAD3 genes via non-allelic homologous recombination (NAHR) cause a fatal infantile syndrome, characterized by pontocerebellar hypoplasia, seizure and respiratory insufficiency.6,15 Reciprocal NAHR-mediated de novo duplication, between ATAD3C exon 7 and the homologous ATAD3A exon 11 results in an additional copy of ATAD3B and the creation of an ATAD3A/C fusion gene, which is also lethal owing to a severe cardiomyopathy accompanied by encephalopathy, hypotonia and seizures, with corneal opacities.7,10 The ATAD3A/C protein is predicted to have 29 amino acid substitutions in the C-terminal region compared to ATAD3A.7 We speculated that the loss of the arginine finger, p.R466C, was one of the most pathologically important substitutions of the ATAD3A/C fusion protein, as it is predicted to play a critical role in ATP hydrolysis through interacting with the γ-phosphate of ATP in the neighbouring subunit of the hexamer.7 However, the specific contribution of p.R466C could not be distinguished from those of the other 28 substitutions of the ATAD3A/C fusion protein. There are also many outstanding questions as to the role of perturbed cholesterol metabolism in ATAD3 disease, which in turn raise important questions about cholesterol’s role in the mitochondria, and the extent to which disturbances of cholesterol and mitochondrial metabolism intersect in brain disorders. Hence, further study of ATAD3 mutants and new models could clarify the molecular disease mechanism, as well as shed light on ATAD3 function.
Here, we identified multiple individuals in a family carrying a heterozygous ATAD3A variant, c.1396C>T, p.R466C that ablates an arginine finger implicated in ATP hydrolysis, and who manifest a syndromic dominant optic atrophy with neurological involvement. Human cells carrying this or another pathological ATAD3 variant (the ATAD3A/C fusion gene)7 display elevated free cholesterol and a greatly expanded lysosome population, many packed with membrane whorls that are characteristic of lysosomal storage diseases. In flies, the corresponding arginine finger mutation, p.R472C, increases the animal’s dependence on cholesterol, but causes cholesterol aggregation in membranes and increases lysosomal numbers, most of which contain cholesterol aggregates. These findings suggest that excess membrane-embedded cholesterol is a cellular and pathological abnormality in ATAD3 disease that can cascade to lysosomal insufficiency; however, extrapolating from the fly model, maintaining cholesterol at normal levels would be still more deleterious.
Materials and methods
Subjects
Affected family members were examined after informed written consent was obtained. All the procedures and examinations, including the skin biopsy for Subject II-1, were performed under project number 2014/1435 and approved by the Regional Committee for Medical and Health Research Ethics, Western Norway (IRB no. 000018729).
Whole exome sequencing
Whole exome sequencing (WES) was performed using genomic DNA. DNA samples were prepared using the SeqCap EZ MedExome target enrichment kit (Roche NimbleGen) and then underwent paired-end 150 nucleotide sequencing on the Illumina NextSeq500 (Illumina). Alignment and variant calling were as previously described26,27 Data annotation and interpretation were performed using the Cartagenia Bench Lab, NGS module (Cartagenia).
Cell culture and treatments
Primary human fibroblasts and SHSY-5Y neuroblastoma cells were routinely maintained in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco) containing 25 mM glucose, 1 mM pyruvate supplemented with 10% fetal bovine serum (FBS, Gibco), 5% penicillin and streptomycin (P/S, Gibco), 2 mM GlutaMAX (Gibco), 5% CO2, at 37°C. All the cell lines were regularly confirmed free of mycoplasma, using the Venor Gem Classic Mycoplasma PCR Detection Kit (Minerva Biolabs).
For microscopy experiments, ∼4700 fibroblasts/well were seeded on 96-well black microplates (Ibidi). After at least 24 h, cells were treated with the various reagents as follows: 2.5 µM U18666A (U18) 48 h (Abcam), 50 µM chloroquine diphosphate salt (CLQ) 6 h (Sigma) and 100 nM rapamycin/sirolimus (Rapa) for 24 h (Acofarma), as described in the main text and figure legends.
For immunoblotting experiments, cells were grown in 25 cm2 flasks until high confluency was obtained and—where indicated—treated with 50 µM CLQ for 6 h (Sigma) to block autophagic flux.
Immunocytochemistry
Cells were fixed with 4% paraformaldehyde (PFA) (Electron Microscopy Sciences) for 20 min at room temperature. After washing with Dulbecco's PBS (DPBS, Gibco), the cells were permeabilized and blocked for 1 h at room temperature with 10% donkey (GeneTex) or goat serum (Sigma) in 0.1% Triton X-100, PBS (PBST 0.1%). Cells were then incubated with primary antibodies (Supplementary Table 2) in PBST overnight at 4°C. After three washes for 5 min with PBST 0.1%, cells were incubated with the appropriate secondary antibodies (Supplementary Table 2) 1:450 dilution in 10% goat/donkey serum in PBST 0.1% for 1–2 h at room temperature. Cells were then washed 3× with PBST 0.1% and 1× with PBS, and Mounting Medium with DAPI (Ibidi) was added.
For free cholesterol and neutral lipid labelling, after PFA fixation and DPBS washing, cells protected from light were incubated with 50 µg/ml Filipin III (Sigma) in PBS for 1 h and with 1 µg/ml BODIPYTM 493/503 (Invitrogen) in PBS for 15 min, respectively. These treatments were followed by two washes of 5 min with PBS, and a 5 min incubation with 0.5 µM SYTOXTM Deep Red (Thermo Scientific) in PBS to stain the nuclei. After a final PBS wash, cells were preserved in Mounting Medium (Ibidi).
PFO-GST labelling of membrane-bound cholesterol. Cells were blocked and permeabilized with 10% goat serum in 0.1% PBST for 1 h and incubated with 15 µg/ml recombinant PFO-GST for 3 h. After washing the cells three times with 0.1% PBST, anti-GST (Supplementary Table 2) was applied in 10% goat serum 0.1% PBST, overnight at 4°C. After three washes with 0.1% PBST, secondary antibody cocktail was applied (Supplementary Table 2) for 1 h at room temperature. Finally, cells were washed three times in PBS and mounting medium with DAPI (Ibidi) was added to the wells.
Amplex Red assay of cholesterol from enriched plasma membrane preparations. Plasma membranes were enriched from equal numbers of control and mutant fibroblasts following essentially the protocol of Bezrukov et al.28 Briefly, cells were incubated twice for 5 min with ice-cold purified water and the cellular debris produced by the osmotic shock removed by washing three times with PBS. The plasma membranes were detached with trypsin, pelleted by centrifugation and resuspended in PBS. Total lipids were extracted with a 2:1 mixture of chloroform and methanol28 and the cholesterol content measured with the Amplex Red Cholesterol Assay Kit (A12216, Thermo Scientific) in an Haloled 96 fluorometer (Dynamica), following the manufacturer’s instructions.
To determine lysosome distribution and activity, cells were treated with 10 nM LysoTrackerTM Red DND-99 (Invitrogen) in DMEM for 45 min followed by DMEM FluoroBriteTM (Gibco) to reduce auto-fluorescence.
Image capture and analysis
Fluorescent images were acquired with a LSM 900 Zeiss confocal microscope. Laser power, gain and offset parameters were kept constant for each experiment, any subsequent adjustments to contrast and brilliance were applied equally to all images. The image analysis was performed using Fiji ImageJ software and GraphPad (Prism) was used to quantify and represent the data as charts.
Immunodetection of proteins
Adherent cells were detached with trypsin (0.4% w/v solution, Gibco) and lysed on ice with RIPA buffer (150 mM NaCl, 1.0% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris, pH 8.0), 1× Halt™ Protease and Phosphatase Inhibitor Cocktail (Thermo Scientific). After incubating on ice for 40 min, the samples were centrifuged for 20 min at 15 000g, 4°C. Protein concentration of the supernatants was determined using the DC protein assay kit (Bio-Rad). Protein samples were prepared in 1× Laemmli loading buffer (Bio-Rad) with DTT and resolved on 7%, 8% or 12% Mini-PROTEAN TGX™ Precast Gels (Bio-Rad), using Tris/Glycine/SDS running buffer (Bio-Rad). After electrophoresis, proteins were transferred to low-fluorescence PVDF transfer membranes (Thermo Scientific) and blocked with 3% bovine serum albumin (BSA) in PBS, 0.1% Tween for 1 h at room temperature. Membranes were incubated overnight with primary antibodies at 4°C (Supplementary Table 2) in PBS with 3% BSA and 0.1% Tween, at 4°C and, after washing, incubated with the appropriate secondary antibody (Supplementary Table 2) for 1 h at room temperature. Proteins were detected using SuperSignalTM West Pico PLUS chemiluminiscent substrate (Thermo Scientific) and immunoblots were acquired via an iBright FL1500 Imaging System and quantified with iBright Analysis Software.
Transmission electron microscopy
Cells in an eight-well chamber slide were washed with phosphate buffer 0.1 M (NaH2PO4·H2O and Na2HPO4) and fixed with 3% glutaraldehyde for 10 min at 37°C and 2 h at room temperature. After five phosphate buffer washes of 5 min and air drying, the samples were cut and processed (Centro de Investigación Principe Felipe, Valencia) and images were taken with a 200 kV high-resolution TECNAI G2 20 TWIN transmission electron microscope (University of the Basque Country).
Cloning, transgenesis and Drosophila maintenance
The pUASTattB-dAtad3R472C-V5 construct was generated by performing site-directed mutagenesis PCR using pUASTattB-dAtad3a WT-V521 as a template and the following primers: (R472C)F: 5′-tttgattatgccatcaacgatTGCctggatgaaatggtggagttc-3′, (R472C) R: 5′-gaactccaccatttcatccagGCA atcgttgatggcataatcaaa-3′. For the construction of pUASTattB-mKate-ThetaTox-D4, we amplified mKate-ThetaTox-D4 coding sequence from pAAV-GFAP-mKate-FL-ThetaTox, using the following primers: mKate-D4_F: 5′-attttgAGATCTcaaa ATG GAGCTGATTAAGGAGAACATG-3′; and mKate-D4_R: 5′-aactaaGCGGCCGC TTAGTTGTAGGTGATGCTGCT-3′.
The amplified PCR product was cloned into the BglII and NotI sites of pUASTattB vector.29 The DNA clones were amplified and purified by the PureLink® HiPure Plasmid Midiprep Kits (Invitrogen). The sequences of mid-prep DNA clones were verified by Sanger sequencing and injected into the following embryos: pUASTattB-mKate-ThetaTox-D4 (y1 w1118; PBac{y+-attP-3B}VK00033); and pUASTattB-dAtad3R472C-V5 (y1 w1118; PBac{y+-attP-3B}VK00037).30 Transgenic flies were selected with a W+ marker and balanced.
The insc-Gal4 driver (w*; P{GawB}inscMz1407; # 8751) and UAS-GFP-LAMP (on the second chromosome; 42714) were obtained from the Bloomington Drosophila Stock Center at Indiana University (BDSC). dAtad3 RNAi line (v22445) was obtained from VDRC stock centre. To generate flies carrying both insc-Gal4 and UAS-GFP-LAMP, we performed recombination of these two genetic components. The flies carrying insc-Gal4, UAS-GFP-LAMP were verified by genomic PCR using the following primers. pUAST-F: 5′-AGTGCAAGTTAAAGTGAATC-3′ EGFP-R: 5′-CGCCTTCTTGACGAGTTCTTC-3′.
To determine the effects of dAtad3R472C expression on the levels of cholesterol-containing membranes and lysosomes in the Drosophila neuroblasts, we crossed the w*; insc-Gal4, UAS-GFP-Lamp flies with the flies carrying y, w; UAS-dAtad3R472C-V5; UAS-mKate-ThetaTox-D4. For control, we used files carrying UAS-empty31 and UAS-mKate-ThetaTox-D4.
All flies were maintained at room temperature (21°C). All crosses were maintained at 25°C. One litre of the standard diet comprised 45.45 g cornmeal, 9.1 g soy flour, 15.4 g yeast, 100 ml syrup, 6.8 g agar, 4.27 ml propionic acid; while the modified diets were as follows: modified diet (MD)—22.72 g cornmeal, 4.55 g soy flour, 7.7 g yeast, 135 ml syrup, 6.8 g agar, 4.27 ml propionic acid; MD2—11.36 g cornmeal, 2.27 g soy flour, 3.85 g yeast, 152.5 ml syrup, 6.8 g agar, 4.27 ml propionic acid, with or without 0.1 or 1 g/l cholesterol (0433 VWR).
Drosophila larval brain dissection and immunohistochemistry
For Drosophila brain staining, dissection of third instar larvae was performed as previously described.32 Briefly, brains attached to the cuticle from the third instar larvae were fixed in 500 µl of 4% formaldehyde for 30 min at room temperature. The fixation solution was discarded and the samples were washed in PBS containing 0.3% Triton X-100. Incubations with primary (overnight at 4°C) and secondary (1 h at room temperature) antibodies were as detailed in Supplementary Table 2. Samples were mounted in Vectashield (Vector Laboratories). Imaging was performed by LSM710 confocal microscope (Zeiss). Images were processed with the Zeiss LSM Image Browser and Adobe Photoshop.
Statistical analysis
Normal distribution was determined by the Shapiro-Wilk test. For normally distributed data, single comparisons were tested using the independent Student’s t-test. Non-parametric data were analysed by the Mann-Whitney U-test. Statistical significance was set at *P ≤ 0.05, **P ≤ 0.01 and ***P ≤ 0.001. Precise P-values are indicated in the figure legends until P < 10−7. Unless otherwise indicated, data are presented as mean ± standard error of the mean (SEM). Statistical analyses were performed using GraphPad Prism 8.0.
Results
A monoallelic ATAD3A mutant associated with optic atrophy and peripheral neuropathy
We investigated a three-generation family with five affected members presenting with slowly progressive optic atrophy and signs of peripheral neuropathy (Fig. 1A and Supplementary material). Subject II-1 is the index case (Fig. 1A), from whom primary fibroblasts were derived. Exome sequencing revealed a missense monoallelic variant in ATAD3A c.1396C>T, p.R466C (NM_001170535.2) (Fig. 1B). The variant was present in all five affected family members, indicating a dominant inheritance, and is not found in the gnomAD database33 or in our in-house database. p.R466C is strongly predicted to be a pathogenic substitution (PolyPhen-2, MutationTaster, SIFT and Align),34 all the more because it affects a conserved arginine finger that mediates ATP hydrolysis in other members of the AAA+ protein family (Supplementary Fig. 2A).35 Thus, the clinical and genetic profiles suggest that the ATAD3A c.1396C>T variant causes dominant optic atrophy ‘plus’ (DOA+).
Figure 1.
Identification of a family with DOA+ associated with a dominant point mutation in ATAD3A. (A) Pedigree of the family in which affected members carry ATAD3A c.1396C>T, p.R466C (NM_001170535.2) on one allele. (B) Amino acid sequence alignment between parts of ATAD3A isoforms 1 and 2 affected by the ATAD3A/C gene fusion7 and the arginine finger point mutation. The A/C fusion protein differs at 29 amino acid positions (highlighted in yellow).6 The individuals with the R466C point mutation have an ATAD3 sequence identical in length to ATAD3A isoform 2 and it only differs in the ATP-binding residue in position 466, which it shares with the fusion protein A-C (in red font). Underlined are the residues of the conserved protein kinase domain that form the ATPase region [p.Ile348–p.Asp474; PFam PF00004]. Marked in a green box is the Walker B ATP binding motif. Residue numbering from [Q9NVI7-2/NM_001170535.2]. (C) SDS-PAGE (8% gel) of whole cell protein from controls (Ctrl), ATAD3 R466C and ATAD3A/C mutant cell lines, immunolabelled with an ATAD3 N-terminal antibody. DOA+ = dominant optic atrophy ‘plus’.
Although PAGE separates proteins chiefly according to molecular mass, in some cases single amino acid substitutions are detectable by SDS-PAGE, such as a cysteine for an arginine substitution in a bacterial histidine-transport protein.36 Separation of ATAD3A on a 6% or 8% denaturing PAGE also produced a small but discernible mobility shift in the R466C variant, compared to the wild-type protein, irrespective of strong reducing conditions (Fig. 1C and Supplementary Fig. 2B). Under the same gel conditions, we discovered retrospectively that the ATAD3A/C fusion protein arising from the gene cluster duplication7 has increased mobility compared to wild-type ATAD3A (Fig. 1C and Supplementary Fig. 2B). Thus, immunoblotting can distinguish several ATAD3 mutants, based either on protein abundance or mobility (this report).6,7
ATAD3A R466C is associated with perturbed cholesterol and lipid metabolism and mtDNA aggregation
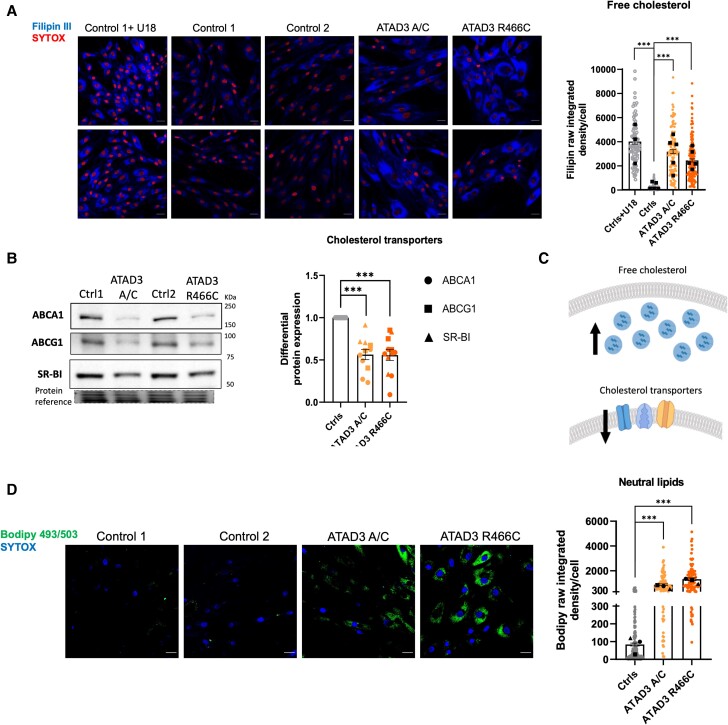
Next, we analysed ATAD3A.R466C fibroblasts to determine whether they shared molecular phenotypes previously associated with ATAD3 deficiency and disease. Analysis of the mitochondrial network revealed mitochondrial clumping and fragmentation, similar to cells with ATAD3A/C (Supplementary Fig. 3A).7 Furthermore, immunostaining of cellular DNA, either via incorporated nucleoside analogue, 5-bromo-2'-deoxyuridine (BrdU) (Supplementary Fig. 3B) or anti-DNA antibody (Supplementary Fig. 3C), revealed significantly more mtDNA clustering in ATAD3A.R466C fibroblasts compared to controls, as per other ATAD3 mutants.6,7 A third previously described feature of ATAD3 mutants evident in ATAD3A.R466C fibroblasts was perturbed cholesterol metabolism, as Filipin-labelled free cholesterol was almost 10 times higher compared to control cells, and similar to that of cells carrying the ATAD3A/C duplication, or control cells treated with the intracellular cholesterol trafficking inhibitor U18666A (U18) (Fig. 2A). We previously hypothesized that the increase in free cholesterol in ATAD3 mutant cells could mitigate the problem of impaired cholesterol delivery to mitochondria through the law of mass action.6 An adaptive response of this type could include reducing cholesterol export via the plasma membrane localized transporters ABCA1, ABCG1 and SR-BI; indeed we found their expression in the ATAD3 mutant cells was half that of the controls (Fig. 2B and Supplementary Fig. 3D).
Figure 2.
ATAD3 mutant fibroblasts display increased unesterified cholesterol, decreased cholesterol export capacity and elevated neutral lipids. (A) Representative images of Filipin III stained ATAD3 mutants and control (Ctrl) fibroblasts, treated with and without the cholesterol trafficking inhibitor U18666A (U18). Quantification of Filipin signal (measured as raw integrated density per total area of the cell), where each point represents a cell and each colour a different cell line/condition [>100 cells per line, independent experiments (represented by black squares) are n = 5, except Ctrls+U18 where n = 3]. Scale bar = 30 µm. Differences between groups were analysed by unpaired, two-tailed Mann-Whitney U-test (Ctrls versus ATAD3 A/C, ***P < 10−7; Ctrls versus ATAD3.R466C, ***P < 10−7; Ctrls versus Ctrls+U18, ***P = 10−10). (B) Abundance of cholesterol efflux proteins (ABCA1, ABCG1 and SR-BI) in ATAD3.R466C and ATAD3 A/C mutants versus controls estimated via immunoblotting of proteins separated by SDS-PAGE. Quantification of the three factors (circles = ABCA1; squares = ABCG1; triangles = SR-BI) relative to controls. Each dot represents an independent experiment and different colours denote each of the cell lines (n = 3–5 independent experiments). Differences between groups were analysed by unpaired, two-tailed Student’s t-test (Ctrls versus ATAD3 A/C, ***P = 2.34 × 10−7; Ctrls versus ATAD3 R466C, ***P = 6.15 × 10−7). (C) Interpretation of A and B to illustrate the increase in free cholesterol and decrease in the abundance of cholesterol efflux plasma membrane proteins. (D) Representative images of control and ATAD3 mutant fibroblasts grown in standard medium (without oleate) and incubated with BODIPYTM 493/503 to stain accumulated neutral lipids (green) and SYTOXTM Deep Red to mark the nuclei (converted to blue). Chart of BodipyTM 493/503 signal (measured as raw integrated density per total area of the cell) from controls and ATAD3 mutant fibroblasts (>100 cells per line, n = 3 independent experiments). As in A, each point represents a cell and each colour a different cell line/condition. Differences between groups were analysed by unpaired, two-tailed Student’s t-test (Ctrls versus ATAD3 A/C, ***P < 10−7; Ctrls versus ATAD3 R466C, ***P < 10−7).
Cholesterol and lipid metabolism are closely intertwined, and in an Atad3 conditional knockout mouse cortical and hippocampal neurons accumulated lipid droplets.16 Here we employed BodipyTM 493/503 to stain lipid droplets and other non-polar lipids, as it does so with greater accuracy and sensitivity than the alternatives, such as Nile red.37 While neutral lipids and lipid droplets were barely detectable by BodipyTM staining in control human fibroblasts, unless grown with oleic acid,38 BodipyTM-stained lipids were present at high abundance in ATAD3.R466C and ATAD3A/C fibroblasts cultured in the absence of oleate (Fig. 2C). Together, the data indicate that perturbed lipid metabolism is a general feature of ATAD3 deficiency, and that the patient-derived fibroblasts recapitulate the features of Atad3 loss in murine neurons.
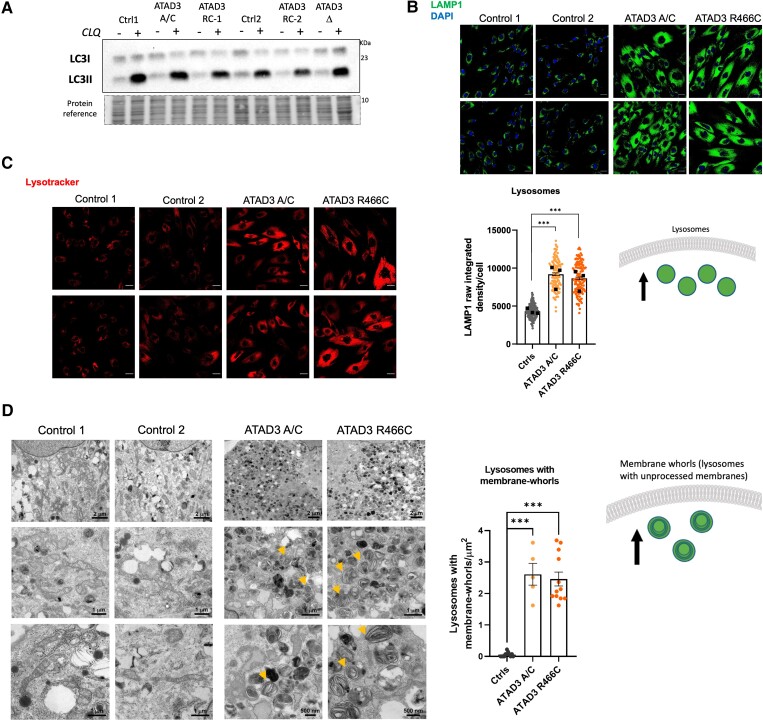
ATAD3 mutant cells display increased lysosome numbers, many with membrane whorls, without altered autophagic flux
In an earlier study, elevated mitophagy and reduced mitochondrial numbers were associated with the transgenic expression of dAtad3R534W,15 and altered autophagic flux was reported for the p.G533D variant in fibroblasts of a patient with hereditary spastic paraplegia.22 The latter study also reported that ATAD3A.G355D fibroblasts and cultured neurons had increased lysosomal mass. Analysis of the autophagic flux, by comparing cells treated with and without the lysosomal inhibitor CLQ,39 revealed no appreciable difference between ATAD3 mutant lines and controls (Supplementary Fig. 4A). That is, lipidated LC3A/B and phosphorylated SQSTM/p62 accumulated to similar extents in all the cell lines, both in standard and nutrient-restricted growth conditions (Fig. 3A and Supplementary Fig. 4A). Notwithstanding that autophagic flux and capacity were unimpaired in the ATAD3 mutants, when we assessed the lysosomal content of the cell, by immunostaining the lysosomal marker LAMP1, we found that ATAD3A.R466C and ATAD3A/C cells had approximately twice the lysosomal content of controls (Fig. 3B). The lysosomes in the ATDA3 mutant cells stained strongly with Lysotracker that is an indicator of their acidity, and thus activity (Fig. 3C), which suggested the lysosomes are functional, as well as more numerous.
Figure 3.
ATAD3 dysfunction increases lysosome numbers maintaining autophagic flux. (A) ATAD3 A/C, deletion (Δ) and R466C cells were treated with and without chloroquine (CLQ), to block autophagy, in parallel with control cells (Ctrl). Lipidated LC3 (LC3II) was detected by immunoblotting after fractionating whole cell lysates via SDS-PAGE. (B) Top: ATAD3A mutant and control fibroblasts stained with an antibody against lysosome-associated membrane protein 1 (LAMP1, green) with DAPI (blue) stained nuclei. Bottom: Chart of LAMP1 signal (measured as raw integrated density per cell) from controls and ATAD3 mutant fibroblasts (>100 cells per line, n = 3 independent experiments). Scale bar = 30 µm. Differences between groups were analysed by unpaired, two-tailed Student’s t-test (Controls versus ATAD3 A/C, ***P < 10−7) and unpaired, two-tailed Mann-Whitney U-test (Controls versus ATAD3 R466C, ***P < 10−7). Cartoon illustrates the increase in the lysosomal pool. (C) ATAD3 mutant and control fibroblasts stained with an indicator of acidified lysosomes, Lysotracker Red. (D) Left: Transmission electron micrographs of ATAD3 mutant and control fibroblasts showing cytoplasmic content at different magnifications. Note the many structures with membrane whorls that are characteristic of lysosomal storage diseases, some of which are arrowed. Right: The chart indicates as dots the number of lysosomes with membrane -whorls in an area of 13.554 µm2 in micrographs of ×2500 magnification. Differences between groups were analysed by unpaired, two-tailed Student’s t-test (Controls versus ATAD3 A/C, ***P = 5 × 10−9) and unpaired, two-tailed Mann-Whitney U-test (Controls versus ATAD3 R466C, ***P = 7.4 × 10−7). The accompanying cartoon illustrates the accumulation of lysosomes with membrane-whorls in the ATAD3 mutant cells.
Repression of mTOR (mammalian target of rapamycin) stimulates lysosomal production and activity, which occurs naturally in response to nutrient restriction (Supplementary Fig. 4A), or chemical inhibition with rapamycin (Supplementary Fig. 4B).39 However, the lysosome changes in the ATAD3 mutant cells (Fig. 3B and C) are not the result of mTOR inhibition, as mTOR is—if anything—more active in ATAD3 mutant than control cells, evidenced by the phosphorylated state of serine235/236 of the ribosomal protein S6 (Supplementary Fig. 4A).40
Although the lysosomes in the ATAD3 mutant cells are functional, evidenced by lysotracker signal and autophagic flux measurements, transmission electron microscopy analysis of ATAD3 mutant fibroblasts revealed many lysosomes containing membrane whorls (Fig. 3D); such unprocessed membranes are characteristic of lysosomal storage diseases.41,42 Collectively, the results for lysosome number, content and autophagic flux (Fig. 3) suggest that more lysosomes are needed to maintain autophagic flux in ATAD3 mutant cells, which implies the degradation process is functional but slower than normal.
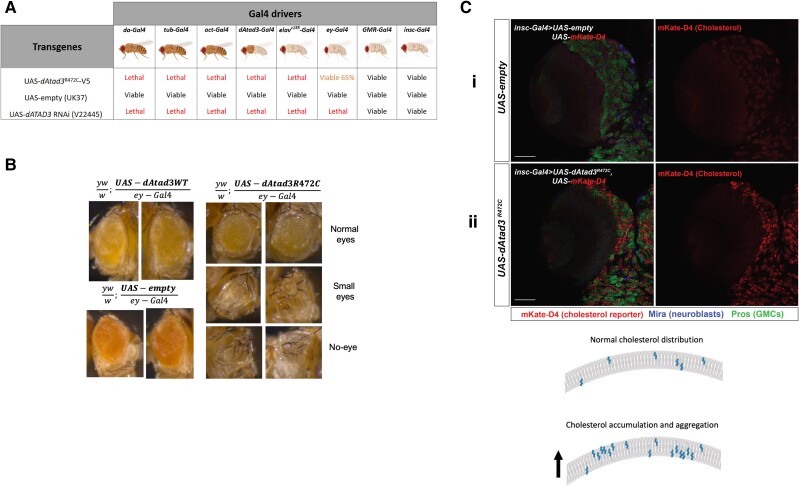
The Atad3 arginine finger mutant results in cholesterol aggregation and lethality in flies
To further study the consequences of the loss of ATAD3’s arginine finger implicated in ATP hydrolysis and explore the pathogenic mechanism in vivo, we created a transgenic Drosophila harbouring the orthologous mutation (UAS-dAtad3R472C). This fly allowed us to express dAtad3R472C in a tissue-specific manner with a range of Gal4 drivers. When ubiquitously expressed (da-Gal4, tub-Gal4 and Act-Gal4), dAtad3R472C caused lethality, similar to dAtad3 gene silencing (Fig. 4A). Even when expression was restricted to the nervous and muscular systems (dAtad3-Gal4),21 or neurons alone (elavC155-Gal4), Atad3R472C expression led to lethality (Fig. 4A). Using an eyeless-Gal4 driver (ey-Gal4) that limits expression to the eye and part of the brain, dAtad3R472C caused partial lethality (65% viable) (Fig. 4A), and among the viable flies, one-third had an abnormal or missing eye (Fig. 4B and Supplementary Table 1). When expression was restricted exclusively to the neuroblasts (insc-Gal4), or delayed, with the late-onset eye and neuronal driver (GMR-Gal4), we obtained viable progeny in numbers equal to controls (Fig. 4A). These findings indicate that the mutation in the conserved arginine finger of ATAD3A associated with ATP hydrolysis is highly deleterious in flies, as well as humans, as it impairs Drosophila development, unless its expression is heavily restricted.
Figure 4.
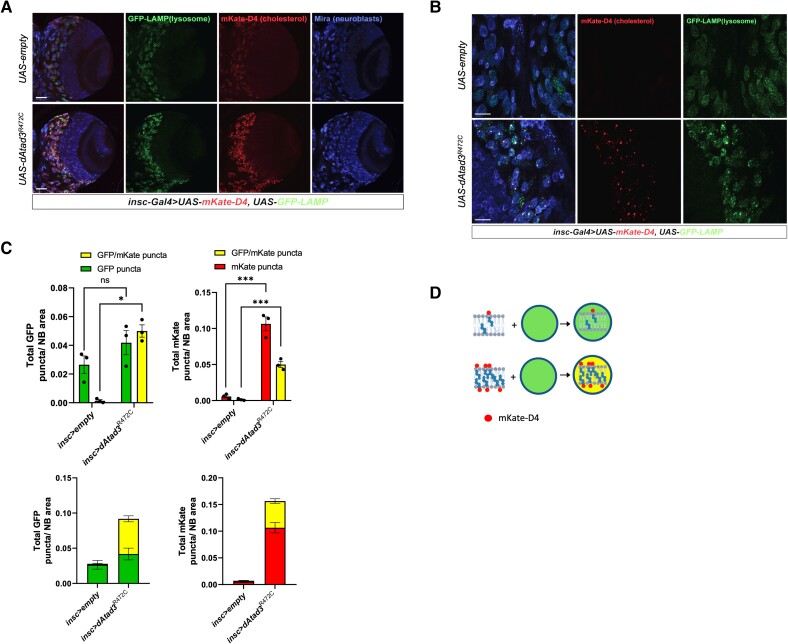
Drosophila, dAtad3R472C is highly deleterious and increases membrane-bound cholesterol. dAtad3R472C was expressed in Drosophila using the UAS-Gal4 system. (A) UAS-dAtad3R472C expressed under different Gal4 drivers led to lethality, except with GMR-Gal4 (late onset eye and neuronal driver), insc-Gal4 (neuroblast driver) and ey-Gal4 (eye discs)56 drivers, similar to that produced when expressing dAtad3 RNAi (UAS-dAtad3RNAi). (B) Light microscope images of the abnormal eye phenotypes of flies expressing dAtad3R472C under the ey-Gal4 driver seen in ∼33% of the viable progeny, compared to the normal eyes of flies expressing no transgene (UAS-empty), or wild-type transgenic Atad3 (UAS-dAtad3WT). (C) Confocal micrographs of Drosophila larvae brain lobes carrying insc-Gal4 and UAS-mKate-D4 together with UAS-empty [control (i) or UAS-dAtad3R472C (ii)]. Neuroblasts are labelled Miranda (blue) and ganglion mother cells (GMCs) by Prospero (green). mKate-D4 (red) is a membrane-bound cholesterol reporter. Scale bar = 50 µm. The cartoon represents the cholesterol distribution in membranes and its predicted accumulation and aggregation based on C(ii).
To determine whether ATAD3 deficiency causes increased cholesterol in vivo, as observed in human cells, we evaluated the effect of dAtad3R472C on cholesterol levels in Drosophila neuroblasts. As we were unable to stain free cholesterol in larval brains, owing to the high background of Filipin III in histological samples, we used an alternative method to label cholesterol in the fly, which had the advantage of labelling membrane-bound, rather than free cholesterol. Perfringolysin O (PFO) is a pore-forming bacterial toxin that contains a cholesterol-binding domain (D4), whose mechanism of membrane-anchoring is known.43 Hence, PFO and D4 can serve as reporters of membrane-bound cholesterol in cultured cells when conjugated to a fluorescent reporter.44,45 Here, we created an in vivo construct to express D4 conjugated to the fluorescent tag mKate in flies: UAS-mKate-D4. Expression of UAS-mKate-D4 with UAS-empty (control) produced a barely discernible red (mKate) signal in neuroblasts of control larvae [Fig. 4C(i)]. In contrast, the expression of UAS-mKate-D4 with UAS-dAtad3R472C yielded numerous red foci in the neuroblasts [Fig. 4C(ii)], indicating that dAtad3R472C produces membrane-bound cholesterol aggregates in Drosophila neuroblasts.
The detection of PFO/D4-based reporters varies according to the proportion of cholesterol that is accessible to D4, as well as the local concentration of cholesterol in membranes or liposomes, thus the reporter does not label the plasma membrane in all cell types46 and was barely detectable in control neuroblasts (Fig. 4C-i). In human fibroblasts, recombinant PFO47 labelled the cell interior and, in the ATAD3 mutant cells, the PFO signal was 7-fold higher than controls (Supplementary Fig. 5A) and partially coincided with lysosomes (Supplementary Fig. 5B), but not with mitochondria or the endoplasmic reticulum (Supplementary Fig. 5C). Although PFO did not detect plasma membrane cholesterol, Amplex red assays (ThermoTM) of cholesterol using enriched plasma membrane fractions28 indicated that those of the ATAD3.R466C mutant cells have a cholesterol content double that of control fibroblasts (Supplementary Fig. 5D). Collectively, the human cell and Drosophila neural stem cell data demonstrate that ATAD3 mutants have abnormally high membrane-bound cholesterol levels, which is the expected consequence of high levels of free cholesterol.
Atad3 mutant neuroblasts display elevated lysosomes with membrane-bound cholesterol aggregates
To determine whether dAtad3R472C expression affected lysosomal content in Drosophila, like the human ATAD3 mutant cells, we used an established lysosomal reporter, UAS-GFP-LAMP.48 Flies carrying insc-Gal4 and UAS-GFP-LAMP were crossed with flies carrying UAS-empty (control) or dAtad3R472C and the UAS-mKate-D4 cholesterol reporter. In neuroblasts expressing the two markers, the dAtad3R472C expressing cells with high levels of cholesterol in membranes also had higher numbers of lysosomes compared to control cells (Fig. 5A and B). The two markers were frequently overlapping or juxtaposed, and quantification revealed that more than a quarter of the D4-mKate puncta coincided with the LAMP-GFP signal, suggesting that many of the cholesterol aggregates had been ingested by lysosomes (Fig. 5C and D). Moreover, a substantial majority of the excess lysosomes coincided with D4-mKate signal, further suggesting that the increase in lysosome numbers is a direct response to the membrane-bound cholesterol aggregates. Hence, these results suggest a model in which lysosomes respond to the excessive cholesterol in membranes to clear it from the cells (refer to the ‘Discussion’ section). This model assumes that the elevated cholesterol in the ATAD3/Atad3 mutant lines is responsible for the lysosomal changes. To determine whether elevated free cholesterol is sufficient to produce lysosomal changes, human SHSY-5Y neuroblastoma cells49 and control fibroblasts were treated with 35 µM cholesterol or intracellular free-cholesterol was increased by U18666A exposure, as before. Both treatments increased lysosomal mass and activity (Supplementary Fig. 6). These findings demonstrate that elevated free cholesterol cascades to lysosomes and support the idea that the lysosomal changes in the ATAD3 mutants are a direct consequence of the perturbed cholesterol metabolism.
Figure 5.
ATAD3A mutant neuroblasts show increased membrane-associated cholesterol together with increased lysosome content. (A and B) Confocal micrographs of Drosophila larval brain lobes carrying insc-Gal4 (neuroblast driver), UAS-GFP-LAMP and UAS-mKate-D4 together with UAS-empty (control), or UAS-dAtad3R472C at magnifications of ×20 (scale bar = 50 µm) and ×63 (scale bar = 20 µm), respectively. Neuroblasts are labelled Miranda (blue), GFP-LAMP (green) marks lysosomes, while mKate-D4 (red) marks cholesterol-containing membranes. (C) Top left: Quantification of GFP-LAMP puncta without (green) and with mKate-D4 (yellow) per neuroblast (NB) area, for UAS-empty (control) and dAtad3R472C expressed under neuroblast driver insc-Gal4. Differences between groups were analysed by unpaired, two-tailed Student’s t-test. Green fluorescent protein (GFP) dAtad3 versus GFP empty non-significant, P = 0.2149; GFP/mKate empty versus GFP/mKate dAtad3, *P = 0.0353. Below the total number of lysosomes in a single column (green + yellow) for UAS-empty (control) and dAtad3R472C. Top right: Quantification of mKate-D4 puncta without (red) and with GFP-LAMP (yellow) per neuroblast (NB) area, for UAS-empty (control) and dAtad3R472C expressed under neuroblast driver insc-Gal4. Differences between groups were analysed by unpaired, two-tailed Student’s t-test: mKate empty versus mKate dAtad3, ***P = 0.0005; GFP/mKate empty versus GFP/mKate dAtad3, ***P = 0.0004. (D) Schematic representation of the data: cholesterol in membranes is labelled red by mKate-D4, while GFP-LAMP stains lysosomes green; hence, mKate-D4 foci and lysosomes that co-localize appear yellow.
dAtad3R472C increases cholesterol dependence
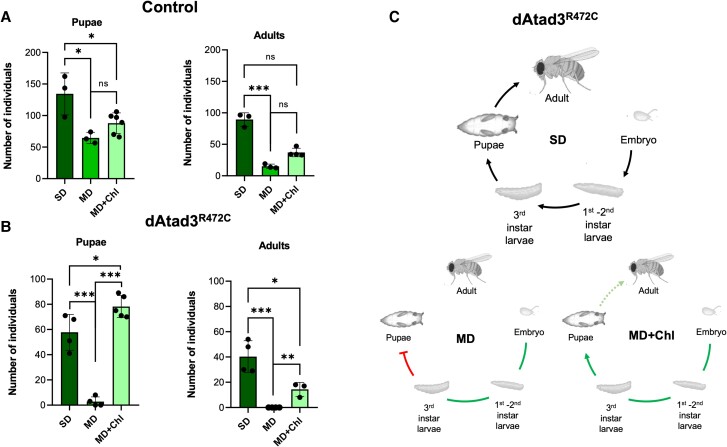
A priori it appeared unlikely that a mitochondrial inner membrane protein, such as ATAD3, would directly regulate plasma membrane cholesterol transporters (Fig. 2B) or enzymes of cholesterol biogenesis.6 Moreover, as ATAD3 had been linked to mitochondrial cholesterol uptake and cholesterol microdomains associated with mtDNA,17,50 we inferred that the increase in the free cholesterol pool was a compensatory mechanism to mitigate a shortage of cholesterol in mitochondrial membranes. None of our attempts to lower cholesterol levels in human fibroblasts was effective, as inhibition of cholesterol biosynthetic enzymes has the opposite effect, as does the inhibition of cholesterol trafficking (Supplementary Fig. 7). However, the lack of cholesterol biosynthesis in flies afforded us the opportunity to manipulate cholesterol availability through the diet. First, we employed two modified diets (MD, MD2), high in sugar but low in the other components of the standard fly laboratory diet (SD). Although there were countless larvae in all cases, the two modified diets both substantially reduced the number of pupae and adult flies (Supplementary Fig. 8A). As the dAtad3 mutant (ey-Gal4 > UAS-dAtad3R472C) had lower viability than the control flies (ey-Gal4 > UAS-empty) on the standard diet, it was not surprising that there were no adult mutants and very few mutant pupae when raised on the modified diets (Supplementary Fig. 8B). Supplementing the less stringent diet (MD) of the control flies with cholesterol yielded two to three times more adults (P = 0.057 or P = 0.049 dependent on the method of analysis) and a third more pupae, although the latter was not statistically significant (Fig. 6A). In contrast, the effect of cholesterol was much more marked in the Atad3R472C fly: it increased the number of pupae an order of magnitude, to levels slightly higher than the mutants raised on the standard diet and allowed some to reach adulthood (Fig. 6B). Thus, the Atad3 mutant fly displays increased dependence on cholesterol (Fig. 6C), which suggests that the elevated cholesterol represents a reprogramming of cellular metabolism designed to mitigate the effects of mutant ATAD3/Atad3.
Figure 6.
Cholesterol supplementation disproportionally benefits Atad3 mutant flies. Flies were maintained on a standard diet (SD) or a modified diet (MD) with or without cholesterol (Chl). (A) Each point indicates the number of control pupae and adults from crosses of flies that carry the UAS construct without a transgene (UAS-empty) and the ey-Gal4 driver. Pupae: differences between groups were analysed by unpaired, two-tailed Student’s t-test (SD versus MD, *P = 0.025; SD versus MD+Chl, *P = 0.022; MD versus MD+Chl, non-significant P = 0.058). Adults: Differences between groups were analysed by unpaired, two-tailed Student’s t-test (SD versus MD, ***P = 0.0003) and unpaired, two-tailed Mann-Whitney U-test (SD versus MD+Chl, non-significant P = 0.057; MD versus MD+Chl, non-significant P = 0.0571). (B) Number of pupae and adults from crosses of flies that carry the UAS-dAtad3R472C transgene (dAtad3R472C) and the ey-Gal4 driver. Pupae: Differences between groups were analysed by unpaired, two-tailed Student’s t-test (SD versus MD, ***P = 0.0003; SD versus MD+Chl, *P = 0.032; MD versus MD+Chl, ***P = 9.38 × 10−7). Adults: differences between groups were analysed by unpaired, two-tailed Student’s t-test (SD versus MD, ***P = 0.0007; SD versus MD+Chl, *P = 0.022; MD versus MD+Chl, **P = 0.003). n = 3 or 4 independent experiments. Data are presented as the mean ± standard deviation. (C) Top: The complete life cycle of the fly. Bottom: The major developmental stages that are impeded by Atad3R472C raised on the modified diet (MD) and that are mitigated by cholesterol supplementation.
Discussion
This study indicates that the expanded pool of free cholesterol associated with pathological ATAD3 variants causes cholesterol accumulation and aggregation in membranes. This is expected to contribute to ATAD3 disease as excess cholesterol disrupts membrane architecture4,5 and is pathological in other contexts.3 Nevertheless, the altered cholesterol metabolism is not a maladaptation to ATAD3 dysfunction; rather, it is a compensatory response, as cholesterol supplementation markedly enhances the viability of the Atad3 mutant fly, particularly at the pupal stage of development (Fig. 6). Thus, the beneficial effects of cholesterol for a fly with a defective mitochondrial membrane protein (Atad3) suggest that the sterol is an essential component of mitochondria. Considering that the bulk of the cholesterol in human mitochondria is associated with the mtDNA,50 and the equivalent sterol in yeasts supports mtDNA maintenance,51,52 the elevated cholesterol might prevent mtDNA abnormalities and loss. Furthermore, cholesterol microdomains, in conjunction with ATAD3, may provide critical structural support to cristae junctions of the mitochondrial inner membrane (see later).
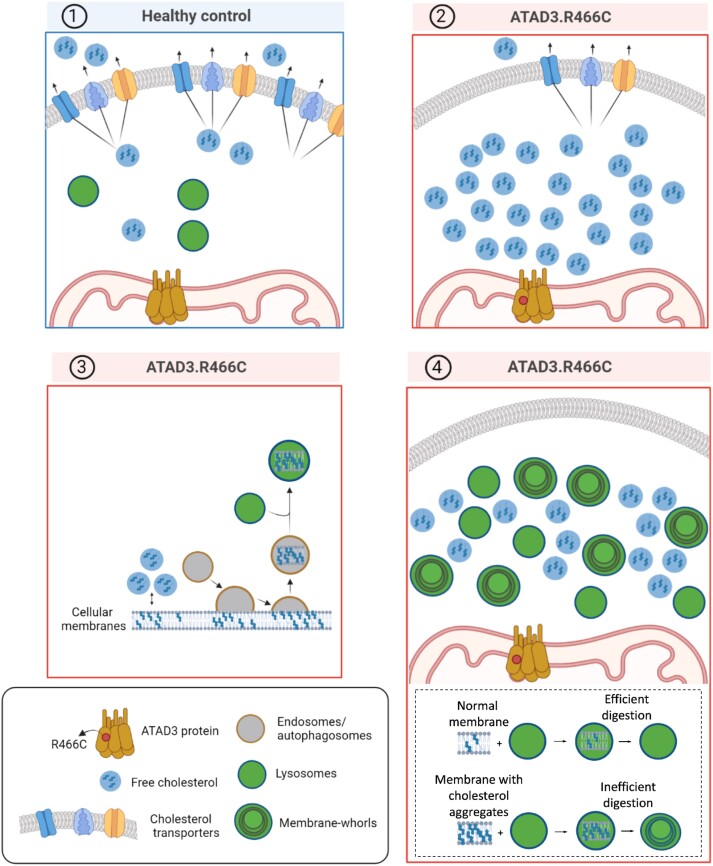
Whatever the precise role of cholesterol in mitochondrial membranes, the elevated free cholesterol comes at the cost of increased cholesterol incorporation and aggregation in cell membranes (Fig. 4C). One way to mitigate the problem of an over-abundance of membrane-embedded cholesterol would be to remove the affected regions via autophagy. Concordantly, lysosome numbers were greatly increased, both in cultured human fibroblasts and Drosophila neuroblasts with mutant Atad3 (Figs 3B and 5). In the dAtad3 mutant neuroblasts, the lysosomes contain a substantial amount of the membrane-bound cholesterol aggregates, based on the cholesterol reporter signal, mKate-D4 coinciding with the lysosomal marker LAMP-GFP (Fig. 5), and this is concordant with the accumulation of membrane whorls in ATAD3 mutant fibroblasts (Fig. 3D). Collectively, the data from primary human fibroblasts and flies suggest a model (Fig. 7), in which defective ATAD3 results in a mitochondrial cholesterol deficit that is attenuated by increasing intracellular cholesterol levels (Fig. 2A). However, the superabundance of cholesterol results in its unregulated insertion in membranes creating areas with excess cholesterol (Fig. 4C), which are targeted for removal by the endolysosomal pathway (Figs 3 and 5). As cholesterol-engorged membranes are difficult to digest, they become the major lysosomal cargo in the Drosophila neuroblasts and the human fibroblasts with mutant ATAD3 (Figs 3D and 5); and although the substantial expansion of the lysosome population is effective at maintaining autophagic flux (Fig. 3A), it is unable to prevent membrane-bound cholesterol reaching levels much higher than controls (Figs 4C and 5). Although autophagic flux keeps the cholesterol aggregates in membranes within tolerable limits, lysosomal dysfunction or exhaustion is clearly a threat, given that membranes with high cholesterol content become the major lysosomal cargo early in fly development (Fig. 5), and the striking similarity with established lysosomal disorders in the hsATAD3 mutant fibroblasts (Fig. 3D).41 On the other hand, the benefits of cholesterol supplementation in the fly (Fig. 6) suggest that lowering cholesterol to normal levels would worsen the situation in flies, and given the similar phenotypes in the fibroblasts, quite possibly in human subjects.
Figure 7.
Consequence of elevated cholesterol in ATAD3 disease. (1) Healthy control cell with normal cholesterol and lysosome levels (blue panel). In contrast, ATAD3 mutant cells (red panels) require more cholesterol than normal and the free cholesterol pool can be increased by reducing the number of cholesterol transporters (2), or remodelling cholesterol biosynthesis.6 The abundant free cholesterol leads to cholesterol aggregation in membranes and activation of the endolysosomal pathway to remove the aberrant membranes (3); however, cholesterol-engorged membranes are difficult to digest leading to many lysosomes with membrane whorls that are characteristic of lysosomal storage diseases (4).
The changes in cholesterol and lysosomes in the ATAD3 mutants coincide closely with NPC disease, as it too features mtDNA aggregation,6 as well as elevated intracellular cholesterol and lysosomes with membrane whorls.53 Some investigators have proposed lowering intracellular cholesterol as a therapeutic strategy for NPC, and positive results were reported for a phase 1b trial.54 Instead, extrapolating from the fly model of ATAD3 deficiency, we speculate that the elevated cholesterol mitigates the lack of the cholesterol trafficking protein. If true, there will be at best a short-term benefit from cholesterol sequestering drugs, or a narrow therapeutic window in NPC disease, as lowering intracellular cholesterol to normal levels will precipitate the progressive depletion of critically important cholesterol microdomains.
The new pathogenic variant, ATAD3A c.1396C>T, p.R466C, offers a simplified genetic picture of ATAD3 dysfunction compared to the ATAD3A/C gene fusion where it occurs as one of 29 substitutions (Fig. 1B).7,10 The evidence for this single amino acid substitution being deleterious is compelling: the mutant allele follows a dominant pattern of inheritance with full penetrance—all five family members with p.R466C display symptoms, with optic atrophy and peripheral neuropathy as the core features. Moreover, this variant has not been reported in any healthy subject. The mutation ablates the arginine finger involved in ATP hydrolysis in AAA+ family proteins, and the equivalent mutation in SPASTIN is dominant and renders the hexameric protein inactive.35,55 The dominancy of the p.R466C ATAD3A variant is supported by the data from Drosophila. Ubiquitous, pan-neuronal and neuromuscular system expression of the orthologous Drosophila mutant Atad3R472C caused lethality, strongly suggesting that Atad3R472C is a dominant-negative or a gain-of-function mutation (Fig. 4A). Eye-specific expression of dAtad3R472C resulted in partial lethality (Fig. 4A) and escapers frequently exhibited a marked developmental defect in the eye (Fig. 4B). Neuroblast-specific expression of dAtad3R472C also indicated that the loss of the arginine finger produces molecular changes in cholesterol and lysosomes in Drosophila (Figs 4C and 5) that parallel those of human fibroblasts with ATAD3A.R466C and ATAD3A/C (Figs 2 and 3). Hence, cellular cholesterol homeostasis appears to depend on the ATPase activity of ATAD3. While ATAD3 has also been proposed to serve as a scaffold for cristae maintenance,16 the structural and enzymatic roles of ATAD3 need not be independent, as altered mitochondrial cholesterol uptake due to ATPase-deficient ATAD3 has the potential to disrupt cristae organization via altered membrane architecture. In any case, the behaviour of the ATAD3 mutants suggests they are unable to recruit or interact with cholesterol (microdomains), which in turn implies that ATAD3 regulates cholesterol in mitochondrial membranes.
The ATAD3 duplication causes a fatal infantile disorder, whereas the new point mutant permits largely normal development and many decades of life, yet, both cellular disease models produce marked cholesterol and lysosomal abnormalities. This could mean that the cholesterol perturbations are only one element of ATAD3 disease, while the severe cardiac dysfunction associated with the duplication syndrome7,10 and the brain developmental abnormality of the deletion syndrome6 might reflect another function of ATAD3. Alternatively, as increased cholesterol appears to be a ubiquitous response to ATAD3 dysfunction, it may be triggered even in the milder forms of the disease owing to mitochondrial cholesterol deficiency. In the latter scenario, the capacity of elevated cholesterol to mitigate ATAD3 disease, and any adverse secondary consequences, are factors superimposed on the severity of the individual mutations. Moreover, abrogation of the same arginine finger in the fly and human produced marked differences in severity, despite the striking similarities in cholesterol and lysosomal changes in the two models. Thus, there remains much to learn about species- and tissue-specific effects of ATAD3 dysfunction, which may depend on a number of genetic modifiers. One species difference that might reduce the severity of the point mutant in humans is the fine control of cholesterol homeostasis via sterol biosynthesis that is unavailable to the fly. Although the two paralogues of ATAD3A in humans, ATAD3B and ATAD3C, which the fly also lacks, might attenuate the loss-of-function of ATAD3A, their impact in humans is limited in the case of the biallelic deletions and monoallelic duplications, as both these mutants cause fatal infantile syndromes despite retaining one or both of ATAD3B and ATAD3C.6,7,10
Finally, to our knowledge, this is the first application of the D4-cholesterol reporter in vivo. The reporter’s ability to reveal changes in cholesterol in biological membranes has considerable potential to answer many questions related to perturbed cholesterol homeostasis in a wide variety of animal models of human diseases, from neurodegeneration to neurological and cardiac disease.
Supplementary Material
Acknowledgements
We would like to thank the patients and their families for support, encouragement and motivation. We thank Mario Soriano-Navarro (Centro de Investigación Principe Felipe) for processing the TEM samples and Dr Ana Martinez-Amesti (Sgiker University of the Basque Country) for help in acquiring the TEM images. We thank Jon Ondaro for technical advice and reagents. We would also like to express our gratitude to Alejandro Carretero for the mKate-D4 insert.
Contributor Information
Mikel Muñoz-Oreja, Department of Neurosciences, Biogipuzkoa Health Research Institute, 20014 San Sebastian, Spain; University of the Basque Country—Bizkaia Campus, 48940 Bilbao, Spain; CIBERNED (Center for Networked Biomedical Research on Neurodegenerative Diseases, Ministry of Economy and Competitiveness, Institute Carlos III), 28031 Madrid, Spain.
Abigail Sandoval, Aging and Metabolism Research Program, Oklahoma Medical Research Foundation, Oklahoma City, OK 73104, USA.
Ove Bruland, Department of Medical Genetics, Haukeland University Hospital, Bergen 5021, Norway.
Diego Perez-Rodriguez, Department of Clinical and Movement Neurosciences, UCL Queen Square Institute of Neurology, Royal Free Campus, London NW3 2PF, UK.
Uxoa Fernandez-Pelayo, Department of Neurosciences, Biogipuzkoa Health Research Institute, 20014 San Sebastian, Spain.
Amaia Lopez de Arbina, Department of Neurosciences, Biogipuzkoa Health Research Institute, 20014 San Sebastian, Spain.
Marina Villar-Fernandez, Department of Neurosciences, Biogipuzkoa Health Research Institute, 20014 San Sebastian, Spain.
Haizea Hernández-Eguiazu, Department of Neurosciences, Biogipuzkoa Health Research Institute, 20014 San Sebastian, Spain.
Ixiar Hernández, University of the Basque Country—Bizkaia Campus, 48940 Bilbao, Spain.
Yohan Park, Aging and Metabolism Research Program, Oklahoma Medical Research Foundation, Oklahoma City, OK 73104, USA.
Leire Goicoechea, Department of Cell Death and Proliferation, Institute of Biomedical Research of Barcelona (IIBB), CSIC, 08036 Barcelona, Spain; Liver Unit, Hospital Clinic i Provincial de Barcelona, Institut d'Investigacions Biomèdiques August Pi i Sunyer (IDIBAPS), 08036 Barcelona, Spain; Centro de Investigación Biomédica en Red (CIBEREHD), 08036 Barcelona, Spain.
Nerea Pascual-Frías, Department of Neurosciences, Biogipuzkoa Health Research Institute, 20014 San Sebastian, Spain; Center for Cooperative Research in Biomaterials (CIC BiomaGUNE), Basque Research and Technology Alliance (BRTA), 20014 San Sebastian, Spain.
Carmen Garcia-Ruiz, Department of Cell Death and Proliferation, Institute of Biomedical Research of Barcelona (IIBB), CSIC, 08036 Barcelona, Spain; Liver Unit, Hospital Clinic i Provincial de Barcelona, Institut d'Investigacions Biomèdiques August Pi i Sunyer (IDIBAPS), 08036 Barcelona, Spain; Centro de Investigación Biomédica en Red (CIBEREHD), 08036 Barcelona, Spain.
Jose Fernandez-Checa, Department of Cell Death and Proliferation, Institute of Biomedical Research of Barcelona (IIBB), CSIC, 08036 Barcelona, Spain; Liver Unit, Hospital Clinic i Provincial de Barcelona, Institut d'Investigacions Biomèdiques August Pi i Sunyer (IDIBAPS), 08036 Barcelona, Spain; Centro de Investigación Biomédica en Red (CIBEREHD), 08036 Barcelona, Spain; Research Center for ALPD, Keck School of Medicine, University of Southern California, Los Angeles, CA 90033, USA.
Itxaso Martí-Carrera, Department of Neurosciences, Biogipuzkoa Health Research Institute, 20014 San Sebastian, Spain; University of the Basque Country—Bizkaia Campus, 48940 Bilbao, Spain; CIBERNED (Center for Networked Biomedical Research on Neurodegenerative Diseases, Ministry of Economy and Competitiveness, Institute Carlos III), 28031 Madrid, Spain; Pediatric Neurology, Hospital Universitario Donostia, 20014 San Sebastián, Spain.
Francisco Javier Gil-Bea, Department of Neurosciences, Biogipuzkoa Health Research Institute, 20014 San Sebastian, Spain.
Mazahir T Hasan, Laboratory of Brain Circuits Therapeutics, Achucarro Basque Center for Neuroscience, Barrio Sarriena, s/n, E-48940 Leioa, Spain; IKERBASQUE, Basque Foundation for Science, 48013 Bilbao, Spain.
Matthew E Gegg, Department of Clinical and Movement Neurosciences, UCL Queen Square Institute of Neurology, Royal Free Campus, London NW3 2PF, UK.
Cecilie Bredrup, Department of Ophthalmology, Haukeland University Hospital, Bergen 5021, Norway; Department of Clinical Medicine (K1), University of Bergen, Bergen 5020, Norway.
Per-Morten Knappskog, Department of Clinical Science (K2), University of Bergen, Bergen 5020, Norway.
Gorka Gereñu-Lopetegui, Department of Neurosciences, Biogipuzkoa Health Research Institute, 20014 San Sebastian, Spain; University of the Basque Country—Bizkaia Campus, 48940 Bilbao, Spain; CIBERNED (Center for Networked Biomedical Research on Neurodegenerative Diseases, Ministry of Economy and Competitiveness, Institute Carlos III), 28031 Madrid, Spain; IKERBASQUE, Basque Foundation for Science, 48013 Bilbao, Spain.
Kristin N Varhaug, Department of Clinical Medicine (K1), University of Bergen, Bergen 5020, Norway; Department of Neurology, Haukeland University Hospital, Bergen 5021, Norway.
Laurence A Bindoff, Department of Ophthalmology, Haukeland University Hospital, Bergen 5021, Norway; Department of Clinical Medicine (K1), University of Bergen, Bergen 5020, Norway; Department of Neurology, Haukeland University Hospital, Bergen 5021, Norway.
Antonella Spinazzola, Department of Clinical and Movement Neurosciences, UCL Queen Square Institute of Neurology, Royal Free Campus, London NW3 2PF, UK.
Wan Hee Yoon, Aging and Metabolism Research Program, Oklahoma Medical Research Foundation, Oklahoma City, OK 73104, USA.
Ian J Holt, Department of Neurosciences, Biogipuzkoa Health Research Institute, 20014 San Sebastian, Spain; University of the Basque Country—Bizkaia Campus, 48940 Bilbao, Spain; CIBERNED (Center for Networked Biomedical Research on Neurodegenerative Diseases, Ministry of Economy and Competitiveness, Institute Carlos III), 28031 Madrid, Spain; Department of Clinical and Movement Neurosciences, UCL Queen Square Institute of Neurology, Royal Free Campus, London NW3 2PF, UK; IKERBASQUE, Basque Foundation for Science, 48013 Bilbao, Spain.
Data availability
The data that support the findings of this study are available from the corresponding author, upon reasonable request.
Funding
M.M.O. was supported by a predoctoral fellowship from the University of the Basque Country (PIF18/317) and later partially supported by the Ikerbasque, Basque Foundation for Science IKUR strategy Neurodegenprot project. A.L. and U.F.P. were recipients of pre-doctoral fellowships from the Basque Government (PRE_2019_1_0184 and PRE_2018_1_0253). The study was supported by funding to I.J.H. from the Instituto de Salud Carlos III (PI17-00380; PI20/00096) and the Basque Government Department of Health (Osasun Saila, Eusko Jaurlaritzako) (grants 2021111070; 2022333050; 2018111043; 2018222031). A.Sp. receives support from Miriam Marks Senior Fellowship, Brain Research UK (202021-26), the Research Councils UK (MR/X002365/1) and the Lily Foundation. W.H.Y. is supported by the National Institute of Neurological Disorders and Stroke (5R01 NS121298-03) of the National Institutes of Health, Oklahoma Center for Adult Stem Cell Research (OCASCR) (221009 and 241006) and Presbyterian Health Foundation (4411-09-10-0).
Competing interests
The authors report no competing interests.
Supplementary material
Supplementary material is available at Brain online.
References
- 1. Pfrieger FW. The Niemann-Pick type diseases—A synopsis of inborn errors in sphingolipid and cholesterol metabolism. Prog Lipid Res. 2023;90:101225. [DOI] [PubMed] [Google Scholar]
- 2. Jeong W, Lee H, Cho S, Seo J. ApoE4-Induced cholesterol dysregulation and its brain cell type-specific implications in the pathogenesis of Alzheimer’s disease. Mol Cells. 2019;42:739–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Luo J, Yang H, Song BL. Mechanisms and regulation of cholesterol homeostasis. Nat Rev Mol Cell Biol. 2020;21:225–245. [DOI] [PubMed] [Google Scholar]
- 4. Shamitko-Klingensmith N, Molchanoff KM, Burke KA, Magnone GJ, Legleiter J. Mapping the mechanical properties of cholesterol-containing supported lipid bilayers with nanoscale spatial resolution. Langmuir. 2012;28:13411–13422. [DOI] [PubMed] [Google Scholar]
- 5. Borochov H, Abbott RE, Schachter D, Shinitzky M. Modulation of erythrocyte membrane proteins by membrane cholesterol and lipid fluidity. Biochemistry. 1979;18:251–255. [DOI] [PubMed] [Google Scholar]
- 6. Desai R, Frazier AE, Durigon R, et al. . ATAD3 gene cluster deletions cause cerebellar dysfunction associated with altered mitochondrial DNA and cholesterol metabolism. Brain. 2017;140:1595–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gunning AC, Strucinska K, Muñoz Oreja M, et al. . Recurrent De Novo NAHR reciprocal duplications in the ATAD3 gene cluster cause a neurogenetic trait with perturbed cholesterol and mitochondrial metabolism. Am J Hum Genet. 2020;106:272–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hanson PI, Whiteheart SW. AAA+ proteins: Have engine, will work. Nat Rev Mol Cell Biol. 2005;6:519–529. [DOI] [PubMed] [Google Scholar]
- 9. Peeters-Scholte CMPCD, Adama van Scheltema PN, Klumper FJCM, et al. . Genotype-phenotype correlation in ATAD3A deletions: Not just of scientific relevance. Brain. 2017;140:e66. [DOI] [PubMed] [Google Scholar]
- 10. Frazier AE, Compton AG, Kishita Y, et al. . Fatal perinatal mitochondrial cardiac failure caused by recurrent de novo duplications in the ATAD3 locus. Med 2021;2:49–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. He J, Mao C-C, Reyes A, et al. . The AAA+ protein ATAD3 has displacement loop binding properties and is involved in mitochondrial nucleoid organization. J Cell Biol. 2007;176:141–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. He J, Cooper HM, Reyes A, et al. . Mitochondrial nucleoid interacting proteins support mitochondrial protein synthesis. Nucleic Acids Res. 2012;40:6109–6121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kim M, Schulz V, Brings L, Schoeller T, Kühn K, Vierling E. mTERF18 and ATAD3 are required for mitochondrial nucleoid structure and their disruption confers heat tolerance in Arabidopsis thaliana. New Phytol. 2021;232:2026–2042. [DOI] [PubMed] [Google Scholar]
- 14. Peralta S, Goffart S, Williams SL, et al. . ATAD3 controls mitochondrial cristae structure in mouse muscle, influencing mtDNA replication and cholesterol levels. J. Cell Sci. 2018;131:jcs217075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Harel T, Yoon WH, Garone C, et al. . Recurrent De Novo and biallelic variation of ATAD3A, encoding a mitochondrial membrane protein, results in distinct neurological syndromes. Am J Hum Genet. 2016;99:831–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Arguello T, Peralta S, Antonicka H, et al. . ATAD3A has a scaffolding role regulating mitochondria inner membrane structure and protein assembly. Cell Rep. 2021;37:110139 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Issop L, Fan J, Lee S, et al. . Mitochondria-associated membrane formation in hormone-stimulated Leydig cell steroidogenesis: Role of ATAD3. Endocrinology. 2015;156:334–345. [DOI] [PubMed] [Google Scholar]
- 18. Li S, Lamarche F, Charton R, et al. . Expression analysis of ATAD3 isoforms in rodent and human cell lines and tissues. Gene. 2014;535:60–69. [DOI] [PubMed] [Google Scholar]
- 19. Frazier AE, Holt IJ, Spinazzola A, Thorburn DR. Reply: Genotype-phenotype correlation in ATAD3A deletions: Not just of scientific relevance. Brain. 2017;140:e67. [DOI] [PubMed] [Google Scholar]
- 20. Dorison N, Dorison N, Gaignard P, et al. . Mitochondrial dysfunction caused by novel ATAD3A mutations. Mol Genet Metab. 2020;131:107–113. [DOI] [PubMed] [Google Scholar]
- 21. Yap ZY, Park YH, Wortmann SB, et al. . Functional interpretation of ATAD3A variants in neuro-mitochondrial phenotypes. Genome Med. 2021;13:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cooper HM, Yang Y, Ylikallio E, et al. . ATPase-deficient mitochondrial inner membrane protein ATAD3A disturbs mitochondrial dynamics in dominant hereditary spastic paraplegia. Hum Mol Genet. 2017;26:1432–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Peralta S, González-Quintana A, Ybarra M, et al. . Novel ATAD3A recessive mutation associated to fatal cerebellar hypoplasia with multiorgan involvement and mitochondrial structural abnormalities. Mol Genet Metab. 2019;128:452–462. [DOI] [PubMed] [Google Scholar]
- 24. Al Madhoun, A, Alnaser F, Melhem M, et al. . Ketogenic diet attenuates cerebellar atrophy progression in a subject with a biallelic variant at the ATAD3A locus. Appl Clin Genet. 2019;12:79–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Carvalho CMB, Lupski JR. Mechanisms underlying structural variant formation in genomic disorders. Nat Rev Genet. 2016;17:224–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Holtan JP, Aukrust I, Jansson RW, et al. . Clinical features and molecular genetics of patients with ABCA4-retinal dystrophies. Acta Ophthalmol. 2021;99:e733–e746. [DOI] [PubMed] [Google Scholar]
- 27. Bredrup C, Johansson S, Bindoff LA, et al. . High myopia-excavated optic disc anomaly associated with a frameshift mutation in the MYC-binding protein 2 gene (MYCBP2). Am J Ophthalmol. 2015;159:973–979.e2. [DOI] [PubMed] [Google Scholar]
- 28. Bezrukov L, Blank PS, Polozov IV, Zimmerberg J. An adhesion-based method for plasma membrane isolation: Evaluating cholesterol extraction from cells and their membranes. Anal Biochem. 2009;394:171–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bischof J, Maeda RK, Hediger M, Karch F, Basler K. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc Natl Acad Sci U S A. 2007;104:3312–3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Venken KJT, He Y, Hoskins RA, Bellen HJ. P[acman]: A BAC transgenic platform for targeted insertion of large DNA fragments in D. Melanogaster. Science. 2006;314:1747–1751. [DOI] [PubMed] [Google Scholar]
- 31. Yoon WH, et al. . Loss of nardilysin, a mitochondrial co-chaperone for α-ketoglutarate dehydrogenase, promotes mTORC1 activation and neurodegeneration. Neuron. 2017;93:115–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hafer N, Schedl P. Dissection of larval CNS in Drosophila melanogaster. J Vis Exp. 2006;1:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Collins RL, Brand H, Karczewski KJ, et al. . A structural variation reference for medical and population genetics. Nature. 2020;581:444–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Evans KJ, Gomes ER, Reisenweber SM, Gundersen GG, Lauring BP. Linking axonal degeneration to microtubule remodeling by spastin-mediated microtubule severing. J. Cell Biol. 2005;168:599–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Noel D, Nikaido K, Ames GFL. A single amino acid substitution in a histidine-transport protein drastically alters its mobility in sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Biochemistry. 1979;18:4159–4165. [DOI] [PubMed] [Google Scholar]
- 37. Gocze PM, Freeman DA. Factors underlying the variability of lipid droplet fluorescence in MA-10 Leydig tumor cells. Cytometry. 1994;17:151–158. [DOI] [PubMed] [Google Scholar]
- 38. Rohwedder A, Zhang Q, Rudge SA, Wakelam MJO. Lipid droplet formation in response to oleic acid in Huh-7 cells is mediated by the fatty acid receptor FFAR4. J. Cell Sci. 2014;127:3104–3115. [DOI] [PubMed] [Google Scholar]
- 39. Agholme L, Abdalla FC, Abeliovich H, et al. . Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kimball SR, Shantz LM, Horetsky RL, Jefferson LS. Leucine regulates translation of specific mRNAs in L6 myoblasts through mTOR-mediated changes in availability of eIF4E and phosphorylation of ribosomal protein S6. J Biol Chem. 1999;274:11647–11652. [DOI] [PubMed] [Google Scholar]
- 41. Parkinson-Lawrence EJ, Shandala T, Prodoehl M, et al. . Lysosomal storage disease: Revealing lysosomal function and physiology. Physiology. 2010;25:102–115. [DOI] [PubMed] [Google Scholar]
- 42. Martinet W, Timmermans JP, De Meyer GRY. Methods to assess autophagy in situ–transmission electron microscopy versus immunohistochemistry. Methods Enzymol. 2014;543:89–114. [DOI] [PubMed] [Google Scholar]
- 43. Ramachandran R, Heuck AP, Tweten RK, Johnson AE. Structural insights into the membrane-anchoring mechanism of a cholesterol-dependent cytolysin. Nat Struct Biol. 2002;9:823–827. [DOI] [PubMed] [Google Scholar]
- 44. Waheed AA, Shimada Yukiko, Heijnen HFG, et al. . Selective binding of perfringolysin O derivative to cholesterol-rich membrane microdomains (rafts). Proc Natl Acad Sci U S A. 2001;98:4926–4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wilhelm LP, Voilquin L, Kobayashi T, Tomasetto C, Alpy F. Intracellular and plasma membrane cholesterol labeling and quantification using filipin and GFP-D4. Methods Mol. Biol. 2019;1949:137–152. [DOI] [PubMed] [Google Scholar]
- 46. Maekawa M. Domain 4 (D4) of perfringolysin O to visualize cholesterol in cellular membranes-the update. Sensors (Basel). 2017;17:504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Goicoechea L, Arenas F, Castro F, et al. . GST-Perfringolysin O production for the localization and quantification of membrane cholesterol in human and mouse brain and liver. STAR Protoc. 2022;3:101068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pulipparacharuvil S, Akbar MA, Ray S, et al. . Drosophila vps16a is required for trafficking to lysosomes and biogenesis of pigment granules. J Cell Sci. 2005;118:3663–3673. [DOI] [PubMed] [Google Scholar]
- 49. Biedler JL, Roffler-Tarlov S, Schachner M FL. Multiple neurotransmitter synthesis by human neuroblastoma cell lines and clones—PubMed. Cancer Res. 1978;38(11 Pt 1):3751–3757. [PubMed] [Google Scholar]
- 50. Gerhold JM, Cansiz-Arda Ş, Lõhmus M, et al. . Human mitochondrial DNA-protein complexes attach to a cholesterol-rich membrane structure. Sci Rep. 2015;5:15292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Cirigliano A, et al. . Ergosterol reduction impairs mitochondrial DNA maintenance in S. cerevisiae. Biochim Biophys Acta Mol cell Biol Lipids. 2019;1864:290–303. [DOI] [PubMed] [Google Scholar]
- 52. Westermeyer C, Macreadie IG. Simvastatin reduces ergosterol levels, inhibits growth and causes loss of mtDNA in Candida glabrata. FEMS Yeast Res. 2007;7:436–441. [DOI] [PubMed] [Google Scholar]
- 53. Blanchette-Mackie EJ. Intracellular cholesterol trafficking: Role of the NPC1 protein. Biochim Biophys Acta. 2000;1486:171–183. [DOI] [PubMed] [Google Scholar]
- 54. Ory DS, Ottinger EA, Farhat NY, et al. . Intrathecal 2-hydroxypropyl-β-cyclodextrin decreases neurological disease progression in Niemann-Pick disease, type C1: A non-randomised, open-label, phase 1–2 trial. Lancet. 2017;390:1758–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hazan J, Fonknechten N, Mavel D, et al. . Spastin, a new AAA protein, is altered in the most frequent form of autosomal dominant spastic paraplegia. Nat Genet. 1999;23:296–303. [DOI] [PubMed] [Google Scholar]
- 56. Hazelett DJ, Bourouis M, Walldorf U, Treisman J. E. Decapentaplegic and wingless are regulated by eyes absent and eyegone and interact to direct the pattern of retinal differentiation in the eye disc. Development. 1998;125:3741–3751. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author, upon reasonable request.