Abstract
Mice deficient in hepatocyte nuclear factor 1 alpha (HNF-1α) were produced by use of the Cre-loxP recombination system. HNF-1α-null mice are viable but sterile and exhibit a phenotype reminiscent of both Laron-type dwarfism and non-insulin-dependent diabetes mellitus (NIDDM). In contrast to an earlier HNF-1α-null mouse line that had been produced by use of standard gene disruption methodology (M. Pontoglio, J. Barra, M. Hadchouel, A. Doyen, C. Kress, J. P. Bach, C. Babinet, and M. Yaniv, Cell 84:575–585, 1996), these mice exhibited no increased mortality and only minimal renal dysfunction during the first 6 months of development. Both dwarfism and NIDDM are most likely due to the loss of expression of insulin-like growth factor I (IGF-I) and lower levels of insulin, resulting in stunted growth and elevated serum glucose levels, respectively. These results confirm the functional significance of the HNF-1α regulatory elements that had previously been shown to reside in the promoter regions of both the IGF-I and the insulin genes.
Hepatocyte nuclear factor 1 alpha (HNF-1α) is a homeodomain-containing transcription factor (4, 7). It is enriched in liver but is also expressed in the kidneys, intestine, stomach, and pancreas (4, 5, 8, 31). Many genes that are preferentially expressed in the liver, including those encoding cytochrome P-450 2E1, albumin, phosphoenolpyruvate carboxykinase, UDP-glucuronosyltransferase, and phenylalanine hydroxylase (PAH), contain a functional HNF-1α-binding sequence in their upstream regulatory regions, suggesting that HNF-1α plays an important role in regulating liver-specific expression of these genes (13, 25, 32, 38, 43). The human insulin-like growth factor I (IGF-I) and the rat insulin I genes also contain binding sites for HNF-1α in their promoter regions and are trans-activated by HNF-1α in reporter gene cotransfection assays (10, 28).
A role for HNF-1α in controlling development and metabolism was suggested by analysis of HNF-1α-null mice (31). These mice only survive to about 1 month after birth, possibly due to severe renal dysfunction that resembles Fanconi syndrome in humans (31). This severe renal dysfunction phenotype precludes further study of the role of HNF-1α in physiological homeostasis, especially its role in maturity-onset diabetes of the young (MODY). Recently, HNF-1α was clearly linked to the occurrence of MODY in humans (41). Patients with MODY carry in one allele of their HNF-1α gene sequence mutations that result in inactivation of HNF-1α derived from the mutated allele (40, 41). However, the mechanism by which HNF-1α causes MODY has not yet been elucidated.
Here, we report the generation of an Hnf-1α−/− mouse strain by use of Cre-loxP-mediated deletion to remove both the first exon of the Hnf-1α gene and the gene encoding a selectable marker, PGK.neo, that had been introduced into the mouse genome during the embryonic stem (ES) cell targeting step. The resulting Hnf-1α−/− mice are viable but develop Laron-type dwarfism (19). The Hnf-1α−/− mice also develop non-insulin-dependent diabetes mellitus (NIDDM) 2 weeks after birth. Surprisingly, in contrast to earlier studies (31), no obvious mortality and only minimal renal dysfunction were found in these mice during the first 6 months of postnatal development.
MATERIALS AND METHODS
Targeting vector and generation of Hnf-1α−/− mice.
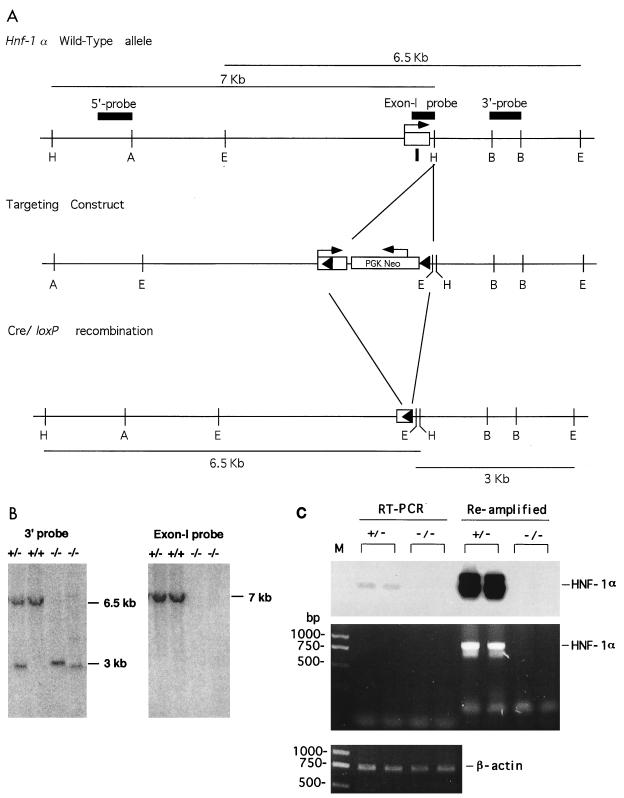
An Hnf-1α genomic clone was isolated from a mouse 129SVJ lambda Dash genomic library (Stratagene), and the 8-kb AvrII-EcoRI fragment containing the Hnf-1α first exon and promoter region was isolated and used for preparing the targeting vector. A 34-bp loxP sequence (15, 36) was inserted into the 5′-untranslated region 40 bp downstream of the transcriptional start site of the Hnf-1α gene (Fig. 1A). The PGK.neo cassette, along with loxP and EcoRI sites, was inserted into the first intron at the HindIII site located 80 bp downstream of the first exon. The orientation of the PGK.neo cassette and loxP sites is shown in Fig. 1. The Hnf-1α targeting vector contained 4.5 kb of homologous DNA upstream of the first loxP site and 3 kb of homologous DNA downstream of the loxP-PGK.neo cassette.
FIG. 1.
Targeted modification of the Hnf-1α gene locus. (A) Hnf-1α gene (top), targeted allele (middle), and schematic of the expected Cre-loxP-mediated deletion of the Hnf-1α gene (bottom). Filled triangles are loxP sites, and arrows show the direction of transcription. The restriction sites are as follows: A, AvrII; B, BamHI; E, EcoRI; H, HindIII; S, SalI. (B) Results of Southern blot analysis of representative F3 mouse tail biopsies. Tail DNA was digested with EcoRI and probed with the 1.0-kb 3′ probe or the first exon probe indicated in panel A. The sizes of the expected bands are shown for DNA from a wild-type allele (+) or a Cre-mediated deleted allele (−). (C) Results of RT-PCR analysis of HNF-1α mRNA in the kidneys of HNF-1α-null mice. The middle and bottom panels show the RT-PCR products for HNF-1α and β-actin mRNAs, respectively, in ethidium bromide-stained agarose gels. The top panel shows the result of probing DNA in the middle panel with a 32P-labeled oligonucleotide derived from the HNF-1α coding region. M, DNA molecular size marker.
ES cells (RW4; Genome Systems) were electroporated with the linearized targeting vector DNA, and G418-resistant clones were selected, expanded, and analyzed by Southern blotting with both a 5′ probe and a 3′ probe to identify specific homologous recombinants (Fig. 1A). The correctly targeted ES cells were injected into C57BL/6J blastocysts to generate chimeric founder mice as described previously (16, 23, 34). Chimeric male founders with close to 100% agouti coat color were bred with C57BL/6J females. Offspring mice having germ line transmission of the loxP-targeted Hnf-1α allele were bred with homozygous EIIa-cre mice (18) as described previously (23). F1 mice carrying the Cre-recombined Hnf-1α deletion were bred to generate the F2 HNF-1α-null mice.
Isolated mouse tail DNA was digested with EcoRI, electrophoresed in a 0.5% agarose gel, transferred to a nylon membrane (GeneScreen; Dupont), and hybridized with a 3′ probe derived from the Hnf-1α gene as indicated in Fig. 1B.
Animal studies.
All mice used in this study were F3 siblings derived from breeding the F2 Hnf-1α+/− mice described earlier. F3 mice were kept in a sterile microisolator and were observed closely throughout the experiment. For measuring the growth rate, three or four mice of each sex and of either genotype were monitored for their body weight once every week, starting 1 week after birth. For measuring the urine chemistry, a group of three 6-week-old mice of each genotype was placed in a mini-metabowl, and urine was collected for 2 h and stored at 4°C before analysis. Urine samples from three groups of each genotype were analyzed. For measuring serum chemistry, both 5-week-old and 3-month-old mice were euthanatized by CO2 asphyxiation, serum was collected, and the selected tissues were snap frozen in liquid nitrogen or fixed in 10% buffered formalin phosphate. Urine and blood chemistry analyses were carried out by Anilytic Inc. (Gaithersburg, Md.).
RNA extraction, RT-PCR, and Northern blot analysis.
Frozen mouse tissues were homogenized in Ultraspec RNA reagent (Biotecx, Houston, Tex.), and total RNA was isolated according to the manufacturer’s protocol. For reverse transcription (RT)-PCR analysis of the HNF-1α and β-actin mRNAs, 2 μg of total RNA was reversed transcribed at 42°C in 20 μl of reaction mixture containing 0.2 μg of oligo-dT primer and 5 U of avian myeloblastosis virus reverse transcriptase. A total of 3 μl of the cDNA product was used in the subsequent PCR amplification (50 μl) with primer sets designed to amplify the HNF-1α and β-actin coding regions. A total of 5 μl of the PCR product was used in a second PCR for reamplification with the same set of primers. The primer sets used to amplify HNF-1α and β-actin were as follows: HNF-1α forward primer, 5′-GATGGTCAAGTCGTACTTGC; HNF-1α backward primer, 5′-GATGTCTGCTCCAAGCTG; β-actin forward primer, 5′-GAACATGGCATTGTTACCAACTG; and β-actin backward primer, 5′-CTGCTTGCTGATCCACATCTGCTG. The oligonucleotide 5′-GATACTTGGTGTAAGGCCGCAGACACT, derived from the HNF-1α coding region (exon 5), was end labeled with [32P]ATP and used to probe the PCR product amplified with the primer set for HNF-1α.
For Northern blot analysis, RNA (15 μg) was denatured, electrophoresed, transferred to a nylon membrane, and probed with either DNA or oligonucleotide probes as previously described (22, 23). Mouse growth hormone (GH) receptor (GHR) cDNA (44) and HNF-1α cDNA (17) were kindly provided by J. J. Kopchick and J. Crabtree, respectively. The oligonucleotides used to probe RNA in this study were as follows: albumin, 5′-CACTACAGCACTTGGTAACATGCTCACTC; IGF-I, 5′-CATCCACAATGCCTGTCTGAGGTGCC; IGF-II, 5′-GATGGTTGCTGGACATCTCCGAAGAGGCTC; and PAH, 5′-CTTGCTACGCTTATCCAGATAGGTG.
Immunohistochemistry.
Formalin-fixed pancreas from 10-day-old mice was used for histological sections (American HistoLabs, Inc., Gaithersburg, Md.) and for immunohistochemical staining with antibodies specific for mouse insulin and glucagon (Histological Consultants, Inc., Alabaster, Ala.).
RESULTS
Generation of Hnf-1α−/− mice.
As shown in Fig. 1A, the Hnf-1α gene targeting vector was constructed to contain a 34-bp loxP sequence in the 5′-untranslated region of the first exon of the Hnf-1α gene locus just 40 bp downstream of the transcription start site, and a loxP-PGK.neo cassette carrying the second loxP site was inserted into the first intron 80 bp downstream of the first exon. No homologous DNA was deleted in the final targeting construct. Cre-mediated recombination at the two loxP sites would thus result in deletion of the translated region of the first exon, which encodes 108 amino acids of the HNF-1α N-terminal region, and of the 5′ region of the first intron, including the exon-intron junction as well as the integrated PGK.neo cassette (Fig. 1A). After transfection of the linearized targeting vector DNA into ES cells, 13% of the G418-resistant ES clones were found to carry a mutant allele in which both loxP sites were present.
Mice exhibiting germ line transmission of the loxP-targeted allele were produced and then bred with a cre transgenic mouse line, EIIa-cre (18). As described previously (23), a cross with EIIa-cre mice generated F1 mice heterozygous for the Cre-mediated HNF-1α knockout. F1 heterozygotes were backcrossed with C57BL/6J mice to yield F2 mice which were heterozygous for the HNF-1α knockout and which had segregated the cre transgene. F2 heterozygotes lacking the cre transgene were selected for interbreeding to generate F3 homozygous null mice, which were used for further analysis. The wild-type and loxP-targeted Hnf-1α alleles can be identified by use of a 3′ probe located in the first intron and a diagnostic EcoRI site introduced into the second loxP sequence (Fig. 1A and B). Deletion of the Hnf-1α first exon region was confirmed by Southern analysis with a probe specific for the Hnf-1α first exon (Fig. 1B). Abolition of HNF-1α transcription in the HNF-1α-null homozygotes was confirmed by probing liver mRNA with a full-length mouse HNF-1α cDNA probe (17) and also by use of an oligonucleotide probe specific for liver PAH mRNA, whose expression is dependent on the presence of HNF-1α (31). In addition, a portion of the coding region of HNF-1α mRNA derived from a region encompassing 800 bp between exons 2 and 5 of the gene was not detected in the kidneys of the HNF-1α-null mice by a highly sensitive RT-PCR method (Fig. 1C). Reamplification of the RT-PCR product confirmed the absence of HNF-1α mRNA in the kidneys of the HNF-1α-null mice.
Growth retardation, infertility, and hyperglycemia in Hnf-1α−/− mice.
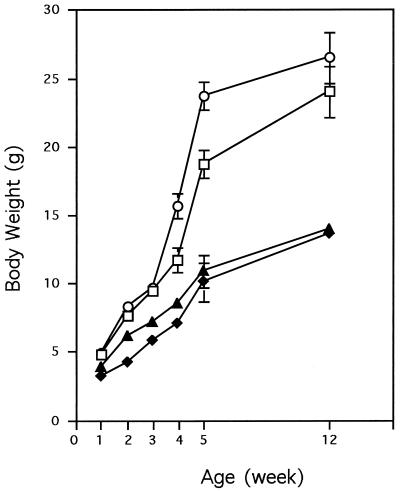
The Hnf-1α−/− mice were born at less than half of the expected frequency, although the litter size was not reduced in the interbreeding of Hnf-1α+/− mice when compared to that observed with the breeding of other lines of mice housed in our animal facility. The neonates were slightly smaller than those of the wild-type and Hnf-1α+/− mice. Unlike that of the previously reported Hnf-1α−/− mice that had been made in a slightly different manner (31), the mortality of the neo-deleted Hnf-1α−/− mice in this study did not differ significantly from that of wild-type or heterozygous mice during the first 6 months of postnatal development. However, the rate of growth of the Hnf-1α−/− mice was significantly lower than that of the heterozygous or wild-type control mice. The body weight of the Hnf-1α−/− mice was only 50 to 60% that of the heterozygous littermates 5 weeks after birth (Fig. 2). At the age of 12 weeks, the Hnf-1α−/− mice were still viable but did not differ significantly in body weight and size from 5-week-old null mice (Fig. 2).
FIG. 2.
Growth curves for HNF-1α-null mice. The value for each point is the average body weight of three to five mice of the same genotype. The standard deviation for each group is shown as a vertical line. Symbols: ○, +/−, male; □, +/−, female; ▴, −/−, male; ⧫, −/−, female.
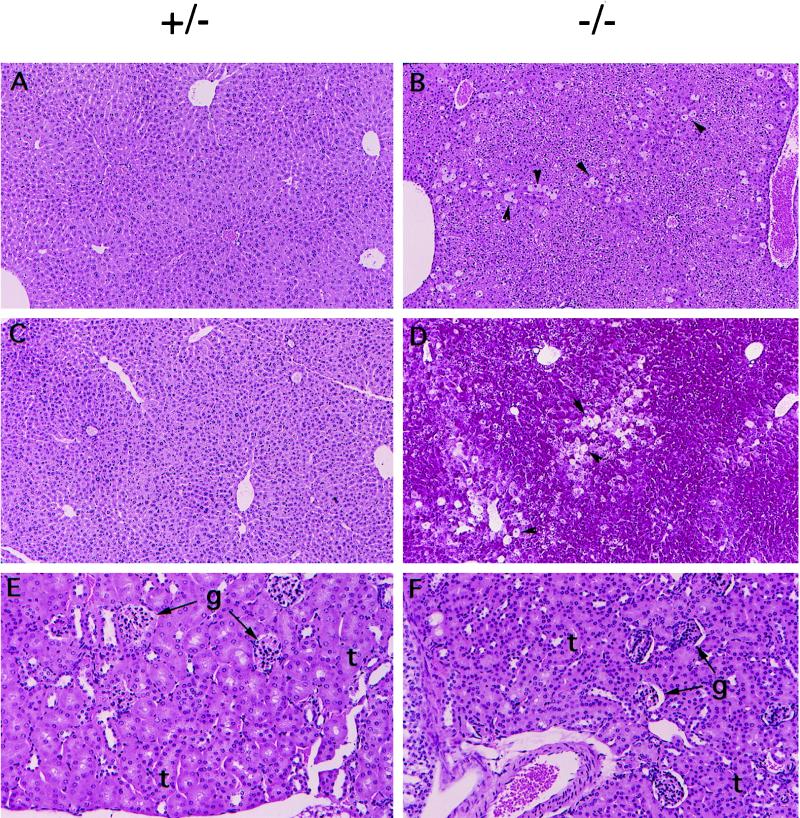
The liver of 5-week-old Hnf-1α−/− mice was about 50% larger than that of normal littermates, while other organs appeared to be allometric to body size, although the mice showed stunted growth (data not shown). The enlarged liver suggests that a dysfunctional liver may be the result of HNF-1α deficiency. Indeed, the levels of the liver enzymes aspartate aminotransferase (AST) and alanine aminotransferase (ALT) in serum were elevated in the Hnf-1α−/− mice, and a progressive severity of liver damage with age was observed (Table 1). In agreement with the elevated activities of AST and ALT, the liver of the Hnf-1α−/− mice developed central lobular hypertrophy with degeneration of individual hepatocytes. The degeneration of hepatocytes did not occur in the liver of young Hnf-1α−/− mice (5 weeks old; data not shown) but was obvious in that of 12-week-old mice (Fig. 3A to D).
TABLE 1.
Serum chemistry of HNF-1α knockout micea
| Age (wk) and type | AST (U/liter) | ALT (U/liter) | Glucose (mg/dl) | Cholesterol (mg/dl) | Albumin (g/dl) | Total protein (g/dl) |
|---|---|---|---|---|---|---|
| 1 | ||||||
| +/− | 300 ± 16 | 40 ± 7 | 103 ± 7 | 99 ± 2 | 1.9 ± 0.1 | 3.1 ± 0.1 |
| −/− | 194 ± 24 | 24 ± 5 | 100 ± 6 | 89 ± 4 | 1.9 ± 0.1 | 3.1 ± 0.1 |
| 5 | ||||||
| +/− | 95 ± 16 | 28 ± 2 | 142 ± 29 | 110 ± 20 | 3.5 ± 0.2 | 4.7 ± 0.1 |
| −/− | 121 ± 57 | 87 ± 35 | 287 ± 79 | 286 ± 52 | 2.8 ± 0.2 | 4.3 ± 0.3 |
| 12 | ||||||
| +/− | 89 ± 20 | 25 ± 4 | 129 ± 30 | 121 ± 19 | 3.4 ± 0.2 | 5.0 ± 0.2 |
| −/− | 243 ± 101 | 242 ± 117 | 332 ± 15 | 307 ± 54 | 2.9 ± 0.2 | 4.5 ± 0.2 |
Serum was collected from groups of mice of either genotype at three different ages and subjected to analysis for levels of AST, ALT, glucose, cholesterol, and albumin. Values presented are the means ± standard errors for at least four serum samples.
FIG. 3.
Histological sections of liver and kidneys from 3-month-old HNF-1α-null mice. (A and B) Hematoxylin-eosin-stained liver sections. The arrowheads show representative degenerating hepatocytes. Original magnification, ×100. (C and D) Periodic acid-Schiff-stained liver sections. The arrows show representative degenerating hepatocytes. Original magnification, ×100. (E and F) Hematoxylin-eosin-stained kidney sections. Original magnification, ×200. g, glomeruli. t, tubules.
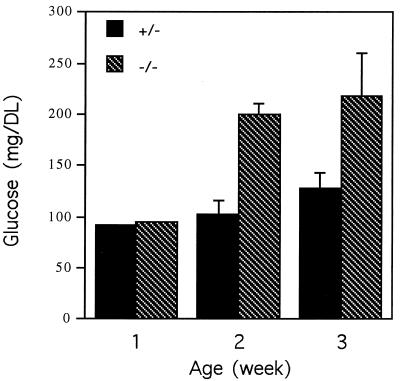
The Hnf-1α−/− mice developed hyperglycemia 2 weeks after birth (Fig. 4 and Table 1), and blood glucose levels remained high afterward. Hepatocytes from the Hnf-1α−/− mice were highly vacuolate and stained positively with periodic acid-Schiff stain, indicating an abnormal accumulation of complex carbohydrate, possibly glycogen, in liver cells (Fig. 3C and D). In addition, the urine glucose level of the 6-week-old mice was 330 times higher than that of the heterozygous littermates (Table 2). The Hnf-1α−/− mice also had significantly higher cholesterol levels than did the heterozygous littermates (Table 1).
FIG. 4.
Serum glucose levels in HNF-1α-null mice during development. The value for each group is the average for at least three serum samples. The standard deviation for each group is shown as a vertical line.
TABLE 2.
Urine chemistry of 6-week-old HNF-1α knockout micea
| Component | Level in Hnf-1α mice
|
|
|---|---|---|
| +/− | −/−b | |
| Glucose (mg/dl) | 17 ± 2 | 5,600 ± 2,510 (330) |
| ALP (U/liter) | 44 ± 20 | 29 ± 6 (0.7) |
| GGT (U/liter) | 40 ± 11 | 11 ± 3 (0.3) |
| Creatinine (mg/dl) | 16 ± 1 | 8 ± 1 (0.5) |
| Protein (mg/dl) | 167 ± 17 | 209 ± 42 (1.3) |
| Sodium (meq/liter) | 55 ± 12 | 49 ± 6 (0.9) |
| Potassium (meq/liter) | 78 ± 3 | 52 ± 9 (0.7) |
| Phosphorus (mg/dl) | 133 ± 8 | 80 ± 2 (0.6) |
Urine was collected from groups of mice of either genotype and subjected to analysis for levels of glucose, alkaline phosphatase (ALP), gamma glutamyl transpeptidase (GGT), creatinine, protein sodium, potassium, and phosphorus. Values presented are the means ± standard errors for three serum samples.
Values in parentheses are the fold increase or decrease for knockout versus control mice.
In contrast to the previously reported Hnf-1α−/− mice (31), the Hnf-1α−/− mice generated in this study showed no signs of renal dysfunction, based on results from the urine chemistry analysis and the kidney histological study (Table 2 and Fig. 3E and F). Except for the elevated glucose level, levels of most urine components analyzed were lower than the normal levels (Table 2). This is probably due to the facts that the Hnf-1α−/− mice urinated more frequently than normal mice and that their diuresis was about two times control levels. Histological analysis of kidneys from the Hnf-1α−/− mice did not reveal any abnormalities in glomeruli or in proximal tubules when compared to kidneys from normal mice (Fig. 3E and F).
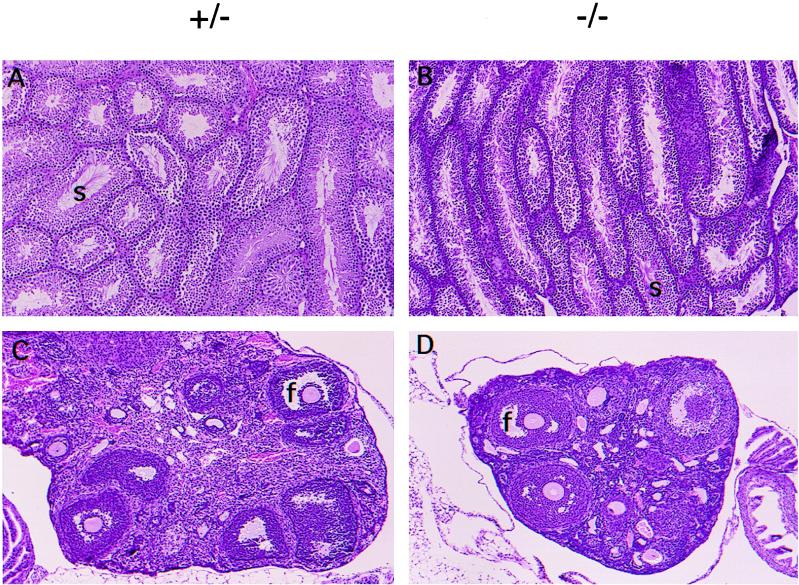
The retarded growth of the Hnf-1α−/− mice resembles that of GH-deficient Little (lit) mice (9). Male and female lit mice are fertile, but all reproductive organs are reduced in size (about 50% normal). In contrast, both female and male Hnf-1α−/− mice are infertile. This was partly due to the lack of sex drive in males and the underdeveloped reproductive organs in both sexes. The Hnf-1α−/− males showed no mating behavior whether paired with wild-type or Hnf-1α−/− females. Conversely, normal males failed to impregnate Hnf-1α−/− females despite repeated attempts at mating. The Hnf-1α−/− females possess an infantile uterus that is thin and flaccid and that exhibits dramatic hypoplasia, especially in the myometrium (data not shown). In Hnf-1α−/− males, the vas deferens as well as other accessory reproductive organs, such as the seminal vesicles and prostate, are vestigial (data not shown). Nonetheless, the histological sections revealed that folliculogenesis was ongoing in the ovaries of Hnf-1α−/− mice and was indistinguishable from that of control females (Fig. 5C and D). In contrast, spermatogenesis was significantly reduced in male Hnf-1α−/− mice (Fig. 5A and B).
FIG. 5.
Histological sections of ovaries and testes from 3-month old HNF-1α-null mice. (A and B) Hematoxylin-eosin-stained testis sections. s, sperm. Original magnification, ×100. (C and D) Hematoxylin-eosin-stained ovary sections. f, follicle. Original magnification, ×100.
HNF-1α deficiency results in reduced expression of IGF-I and IGF-II mRNAs and increased GH resistance.
Dwarfism in lit mice is due to a GH deficiency caused by a mutation in the gene encoding a receptor for the GH-releasing factor (24). Although the dwarfism in the Hnf-1α−/− mice appeared to be similar to that of the lit mice, the Hnf-1α−/− mice were not GH deficient. In particular, at the age of 5 weeks, the Hnf-1α−/− mice had GH levels 100 times and 45 times higher than those of control females and males, respectively (Table 3). However, the blood IGF-I level in the Hnf-1α−/− mice was only 20 to 30% the control level 5 weeks after birth. The findings of dwarfism in the Hnf-1α−/− mice but of elevated blood GH levels and reduced blood IGF-I levels suggest that the mice are resistant to the action of GH and therefore that their dwarfism more closely resembles Laron dwarfism, a human hereditary GH resistance disease that is due to defects in the GHR or in postreceptor mechanisms, leading to an inability of the liver to produce IGF-I (19, 20, 39).
TABLE 3.
Levels of serum growth factors in HNF-1α knockout micea
| Age (wk) and type | GH (ng/ml) in mice
|
IGF-I (ng/ml) in mice
|
Insulin (ng/ml) | ||
|---|---|---|---|---|---|
| Male | Female | Male | Female | ||
| 5 | |||||
| +/+ | 2.1 ± 0.5 | 1.7 ± 1.3 | 426 ± 29 | 430 ± 56 | ND |
| +/− | 4.1 ± 3.5 | 3.0 ± 1.9 | 423 ± 50 | 495 ± 25 | 0.53 ± 0.05 |
| −/− | 91.5 ± 13.0 | 118.0 ± 28.0 | 147 ± 47 | 115 ± 23 | 0.29 ± 0.06 (P < 0.01) |
| 12 | |||||
| +/+ | 6.1 ± 2.8 | 4.5 ± 2.1 | 248 ± 35 | 434 ± 52 | ND |
| +/− | 7.5 ± 4.1 | 2.8 ± 0.4 | 236 ± 42 | 317 ± 43 | 0.54 ± 0.16 |
| −/− | 25.4 ± 9.3 | 31.8 ± 14.7 | 129 ± 8 | 103 ± 3 | 0.34 ± 0.13 (P < 0.10) |
Serum was collected from groups of mice of either genotype at two different ages and subjected to analysis for levels of GH, IGF-I, and insulin. Values presented are the means ± standard errors for at least three serum samples. P values indicate significance for knockout mice with respect to control mice at the same age. ND, not determined.
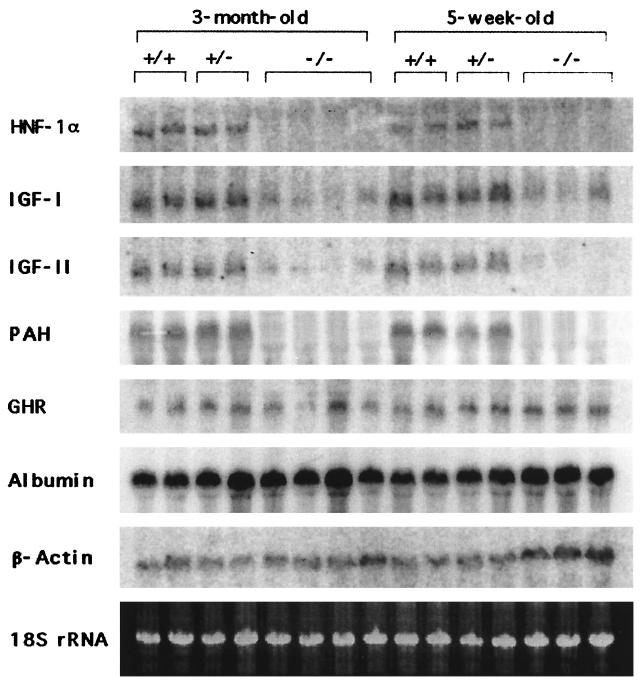
To understand the molecular mechanism of GH resistance in the HNF-1α-null dwarf mice, hepatic IGF mRNA levels were analyzed, since it is well established that many of the growth-promoting effects of GH are mediated by circulating IGFs produced primarily in the liver. As shown in Fig. 6, the deficiency of HNF-1α significantly reduced the IGF-I and IGF-II mRNA levels, suggesting that the reduced circulating IGF-I level in the Hnf-1α−/− mice resulted in large part, if not completely, from the reduced expression of the IGF-I gene in the liver. However, the low-level expression of liver IGFs was not due to a defect in GHR gene expression. The GHR mRNA level in the Hnf-1α−/− mice did not differ from that in the control mice (Fig. 6), indicating that resistance to GH stimulation in Hnf-1α−/− mice is due to a postreceptor mechanism, possibly involving a direct trans-activation effect of HNF-1α on the IGF-I gene promoter, as has been previously demonstrated (27, 28).
FIG. 6.
Inactivation of HNF-1α abolishes liver IGF-I and IGF-II mRNA levels. Northern blotting analysis of representative liver biopsies of Hnf-1α−/− mice is shown. Liver total RNAs (15 μg) from 5-week-old and 3-month-old Hnf-1α−/− mice were denatured and electrophoresed on a formaldehyde-containing 1% agarose gel, blotted to nylon membranes, and probed with the indicated cDNA and oligonucleotide probes. Each lane contains RNA from an individual animal.
HNF-1α deficiency reduces expression of insulin.
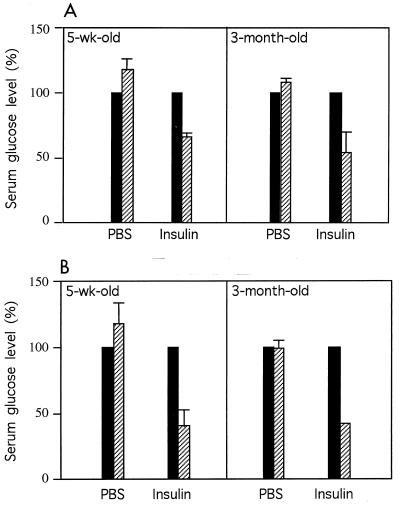
The Hnf-1α−/− mice developed diabetes 2 weeks after birth. The circulating insulin level in the Hnf-1α−/− mice was about 60% the level found in heterozygous mice at the two ages examined (Table 3), indicating that the Hnf-1α−/− mice were not completely deficient in insulin production. Therefore, the diabetes in the Hnf-1α−/− mice is similar to NIDDM, characterized by high blood glucose levels and a relative deficiency of insulin (30). In addition, the Hnf-1α−/− mice were not resistant to the insulin treatment at the two ages examined (Fig. 7). At 30 min after insulin administration, blood glucose levels in the Hnf-1α−/− mice were reduced to below 50% the level before insulin administration (Fig. 7). This response to insulin treatment was greater than that found in the control mice receiving the same amount of insulin (Fig. 7).
FIG. 7.
HNF-1α-null mice are not resistant to insulin treatment. Hnf-1α+/− mice (A) and Hnf-1α−/− mice (B) were administered phosphate-buffered saline (PBS) or insulin solution intraperitoneally at a dosage of 2 mU/mg of body weight/10 μl. Before (▪) and 30 min after (▨) insulin administration, 3 μl of blood was taken from the tail vein of each animal and monitored for glucose concentration. The pretreatment glucose level was designated 100%. After treatment, the glucose level was calculated as a percentage of the pretreatment level. The value for each group is the average for the three serum samples. The standard deviation for each group is shown as a vertical line.
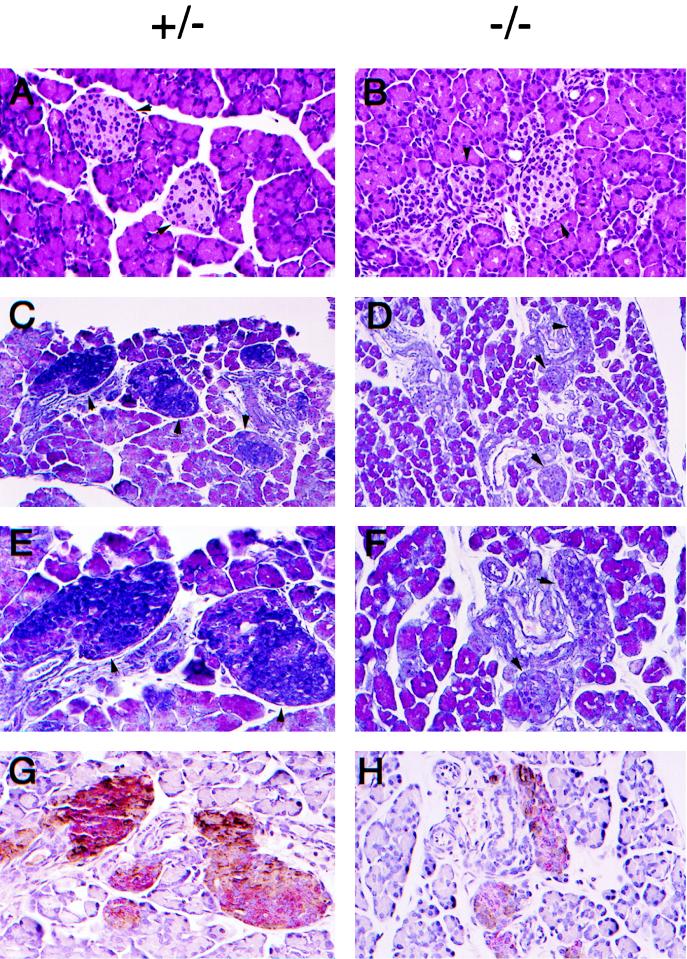
It has been well documented that long-term high blood glucose levels damage the insulin-secreting pancreatic β cells in NIDDM (35). HNF-1α is also expressed in the pancreas and has been shown to trans-activate the rat insulin I gene promoter (8, 10). We therefore examined the histological structure of the pancreas taken from the Hnf-1α−/− mice 10 days after birth, before hyperglycemia fully developed. The pancreas was smaller in the dwarf Hnf-1α−/− mice but was structurally indistinguishable between the genotypes, based on the haematoxylin-eosin staining technique (Fig. 8A and B). The islet of Langerhans, the endocrine tissue in the pancreas that secrets insulin and glucagon, was also histologically indistinguishable from that in the pancreas of control mice (Fig. 8A and B). However, when the Gomori chrome alum-hematoxylin-phloxine staining technique was used to examine the components of tissue in the pancreas, the islet of Langerhans in the Hnf-1α−/− pancreas differed significantly from that in the control pancreas (Fig. 8C to F). The Gomori chrome alum histochemical staining method can be used to distinguish between glucagon- and insulin-secreting cells, which stain pink and blue, respectively (6). No cells in the islet of Langerhans in the Hnf-1α−/− pancreas were stained blue, while the majority of cells (60 to 80%) in the islet of Langerhans in the control pancreas were strongly stained blue (Fig. 8E and F). Nevertheless, the existence of insulin-secreting β cells in the pancreas of Hnf-1α−/− mice was confirmed by immunohistochemical staining with an antibody specific for mouse insulin (Fig. 8G and H). The percentages of glucagon-secreting α cells and insulin-secreting β cells in the pancreas of Hnf-1α−/− mice appeared to be normal, i.e., 20% for α cells and 60 to 70% for β cells. However, the amount of the stained insulin peptide was significantly reduced in the Hnf-1α−/− pancreas (Fig. 8H).
FIG. 8.
Reduced expression of insulin peptide in the pancreatic β cells of HNF-1α-null mice. Histological sections of pancreas from 10-day-old HNF-1α-null mice are shown. (A and B) Hematoxylin-eosin-stained pancreas sections. The arrowheads show representative islet of Langerhans cells. Original magnification, ×200. (C to F) Gomeri chrome alum-hematoxylin-phloxine-stained pancreas sections. Original magnifications, ×200 for C and D and ×400 for E and F. (G and H) Double immunostaining for insulin and glucagon. Formalin-fixed paraffin-embedded pancreas sections were stained with monoclonal antibody to insulin (alkaline phosphatase method labeled and red) and monoclonal antibody to glucagon (peroxidase-diaminobenzidine method labeled and brown). Original magnification, ×400. Different sections of the same islet of Langerhans cells of each genotype are shown in panels E and G and panels F and H.
DISCUSSION
We report here the facilitated generation of adult HNF-1α-null mice with the Cre-loxP recombination system (15, 36). Conditional HNF-1α mice were constructed and mated with EIIA-cre mice to delete, at the F1 stage, the first exon and 5′ sequences of the first intron of the Hnf-1α gene as well as the selection marker, the PGK.neo gene, that had been introduced into the genome during the ES cell targeting step. HNF-1α-null mice for this study were then generated by appropriate crosses of heterozygous HNF-1α-null mice that had already segregated the cre transgene. Deletion of the first exon of Hnf-1α removes the region encoding the first 108 amino acids of HNF-1α, including the entire dimerization domain (4, 7). The removal of the dimerization domain would result in the functional inactivation of HNF-1α by abolishing both its dimerization and DNA-binding capabilities (7). The deletion was also designed to block HNF-1α mRNA maturation, as it removes the splicing donor site for the first (>3-kb) intron (Fig. 1A); as expected, no truncated mRNA was detected by either Northern or RT-PCR analysis.
The role of HNF-1α in postnatal development in mice was established in this study. We show here that the loss of HNF-1α reduced, directly or indirectly, IGF mRNA levels in the liver, leading to low circulating IGF-I levels. This finding may be responsible for the resistance to GH action and for the development of Laron-type dwarfism in the Hnf-1α−/− mice. In mammals, GH plays an important role in postnatal development and growth by maintaining levels of circulating IGF produced mainly in the liver. The circulating IGFs mediate many of the GH functions in growth (26). The present study established that HNF-1α has an essential role in postnatal growth and development, although it is dispensable in early embryonic development (31; this study).
The dwarfism in the HNF-1α-null mice resembles Laron-type dwarfism, which is due to a defect in the GHR or in a postreceptor mechanism that results in the development of resistance to GH stimulation characterized by a significantly higher circulating GH level, a lower IGF-I level, and a phenotype similar to that of mice deficient in GH production (19, 20). The effect of the HNF-1α deficiency on growth may be due to a postreceptor mechanism, since GHR mRNA levels were not affected in the HNF-1α-null mice. Interestingly, the infertility in both sexes of the HNF-1α-null mice did not resemble that caused by GH deficiency, such as that described for lit mice (9). Rather, the infertility in the HNF-1α-null mice was similar to that described for the IGF-I-null mice (3). In addition to being regulated by GH in the liver, the IGF-I mRNA level is also regulated by other hormonal stimuli in a tissue-specific manner and functions in an autocrine-paracrine fashion; an example is ovarian IGF-I regulated by estrogen (14). The similarity between the HNF-1α-null and the IGF-I-null mice suggests that HNF-1α plays an important role in regulating Igf1 gene expression, possibly by a direct interaction with the Igf1 gene promoter. Indeed, the HNF-1α regulatory elements were found in the promoter region of the human IGF-I gene (27, 28). HNF-1α appears to be the major trans-activator in regulating human IGF-I gene expression in Hep3B cells, a human liver carcinoma-derived cell line (1, 28).
Other liver-enriched transcription factors, such as the C/EBP proteins and HNF-3β, were also found to trans-activate the human IGF-I gene in Hep3B cells (27, 29). However, in mice, the expression of the liver IGF-I gene was not affected by diminished C/EBPα protein expression in adults, as demonstrated with a conditional gene knockout system (23). Similar to that of C/EBPα, a deficiency of the C/EBPβ protein did not affect the expression of the mouse Igf1 gene in the liver (21a). On the other hand, HNF-1α appears to be a major transcription factor responsible for Igf1 gene expression in mouse liver, as demonstrated in this study. Nonetheless, whether there is a direct interaction between HNF-1α and the IGF-I gene in vivo awaits confirmation.
The disease MODY is a genetic disorder characterized by autosomal dominant inheritance and an age of onset of 25 years or younger and is responsible for 2 to 5% of NIDDM (12, 21). MODY genes in humans have been mapped to chromosomes 20 (MODY1), 7 (MODY2), and 12 (MODY3) (12, 40). It is now known that the MODY1 and MODY2 genes encode HNF-4α and glucokinase, respectively (12, 41). On the other hand, the MODY3 gene encodes HNF-1α (42). Recently, insulin promoter factor-1 (IPF1) was shown to represent a new genetic locus of MODY, MODY4 (37). The HNF-1α-null mice from this study developed diabetes at the age of 2 weeks. The relative deficiency of blood insulin levels and the normal response to insulin treatment in diabetic Hnf-1α−/− mice were markedly similar to those in the MODY disorder in humans. However, unlike humans, in which the disorder is dominant, the mice did not develop diabetes when only one allele of the gene was inactivated (31; this study). This result may be due to species differences in the level of HNF-1α expression or in target gene responses. Nevertheless, the Hnf-1α−/− mice reported here provide an opportunity to further examine the cause, at the molecular level, of diabetes due to MODY3 in humans. In addition to the liver and kidneys, HNF-1α is expressed in the pancreas and has been shown to trans-activate the rat insulin I gene promoter (8, 10). We demonstrate here that in mice, the expression of insulin in pancreatic β cells is affected by the inactivation of the Hnf-1α−/− gene. However, demonstration that HNF-1α is a major trans-activator for the insulin gene promoter remains to be done.
In spite of their dwarfism and early-onset diabetes, the HNF-1α mice did not show mortality significantly different from that of control mice during the first 6 months of postnatal development. Surprisingly, the mice generated in this study showed no sign of renal dysfunction in histology or urine chemistry, compared to an HNF-1α-null mouse line reported earlier (31). Strain differences between the homozygotes generated here (129 Svj × C57BL/6J × FVB/N [EIIa-cre background]) and the previously reported knockout mice (129 Svj × C57BL/6J) may explain the difference in renal phenotypes for the two animal models. However, in this study, we also chose to remove the selection marker, the PGK.neo gene, from the genome of the targeted mouse ES cells by using Cre-mediated DNA recombination because of concerns that the selection marker gene might have adverse effects on the expression of neighboring genes surrounding the Hnf-1α gene locus and that effects on neighboring genes might then result in sickness or phenotypes unrelated to those caused by the inactivation of HNF-1α itself. The adverse effect caused by the selection marker gene used in gene knockout experiments has been reported (2, 11, 33). Although it is unclear at present what is responsible for the different phenotypes between the previously reported HNF-1α-null mice (31) and the HNF-1α-null mice generated in this study, the new line of Hnf-1α−/− mice should prove useful for studying the role of HNF-1α in liver function and in MODY of NIDDM.
ACKNOWLEDGMENTS
We thank Derek LeRoith for advice on our experiments and Shioko Kimura for assistance with the ES cell culture.
REFERENCES
- 1.Aden D P, Fogel A, Plotkin S, Damjanov I, Knowles B B. Controlled synthesis of HBsAg in a differentiated human liver carcinoma-derived cell line. Nature. 1979;282:615–616. doi: 10.1038/282615a0. [DOI] [PubMed] [Google Scholar]
- 2.Artelt P, Grannemann R, Stocking C, Friel J, Bartsch J, Hauser H. The prokaryotic neomycin-resistance-encoding gene acts as a transcriptional silencer in eukaryotic cells. Gene. 1991;99:249–254. doi: 10.1016/0378-1119(91)90134-w. [DOI] [PubMed] [Google Scholar]
- 3.Baker J, Hardy M P, Zhou J, Bondy C, Lupu F, Bellve A R, Efstratiadis A. Effects of an Igf1 gene null mutation on mouse reproduction. Mol Endocrinol. 1996;10:903–918. doi: 10.1210/mend.10.7.8813730. [DOI] [PubMed] [Google Scholar]
- 4.Baumhueter S, Mendel D B, Conley P B, Kuo C J, Turk C, Graves M K, Edwards C A, Courtois G, Crabtree G R. HNF-1 shares three sequence motifs with the POU domain proteins and is identical to LF-B1 and APF. Genes Dev. 1990;4:372–379. doi: 10.1101/gad.4.3.372. [DOI] [PubMed] [Google Scholar]
- 5.Blumenfeld M, Maury M, Chouard T, Yaniv M, Condamine H. Hepatic nuclear factor 1 (HNF 1) shows a wider distribution than products of its known target genes in developing mouse. Development. 1991;113:589–599. doi: 10.1242/dev.113.2.589. [DOI] [PubMed] [Google Scholar]
- 6.Burkitt H G, Youn B, Heath J W. Wheater’s functional histology: a text and colour atlas. 3rd ed. Edinburgh, Scotland: Churchill Livingstone, Ltd.; 1993. [Google Scholar]
- 7.Chouard T, Blumenfeld M, Bach I, Vandekerckhove J, Cereghini S, Yaniv M. A distal dimerization domain is essential for DNA-binding by the atypical HNF1 homeodomain. Nucleic Acids Res. 1990;18:5853–5863. doi: 10.1093/nar/18.19.5853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Simone W, De Magistris L, Lazzaro D, Gerstner J, Monaci P, Nicosia A, Cortese R. LFB3, a heterodimer-forming homeoprotein of the LEB1 family, is expressed in specialized epithelia. EMBO J. 1991;10:1435–1443. doi: 10.1002/j.1460-2075.1991.tb07664.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eicher E M, Beamer W G. Inherited ateliotic dwarfism in mice: characteristics of the mutation little (lit) J Hered. 1976;67:87–91. doi: 10.1093/oxfordjournals.jhered.a108682. [DOI] [PubMed] [Google Scholar]
- 10.Emens L A, Landers D W, Moss L G. Hepatocyte nuclear factor 1 alpha is expressed in a hamster insulinoma line and transactivates the rat insulin I gene. Proc Natl Acad Sci USA. 1992;89:7300–7304. doi: 10.1073/pnas.89.16.7300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fiering S, Kim C G, Epner E M, Groudine M. An “in-out” strategy using gene targeting and FLP recombinase for the functional dissection of complex DNA regulatory elements: analysis of the beta-globin locus control region. Proc Natl Acad Sci USA. 1993;90:8469–8473. doi: 10.1073/pnas.90.18.8469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Froguel P, Zouali H, Vionnet N, Velho G, Vaxillaire M, Sun F, Lesage S, Stoffel M, Takeda J, Passa P, et al. Familial hyperglycemia due to mutations in glucokinase. Definition of a subtype of diabetes mellitus. N Engl J Med. 1993;328:697–702. doi: 10.1056/NEJM199303113281005. [DOI] [PubMed] [Google Scholar]
- 13.Hansen A J, Lee Y H, Gonzalez F J, Mackenzie P I. HNF 1 alpha activates the rat UDP glucuronosyltransferase UGT2B1 gene promoter. DNA Cell Biol. 1997;16:207–214. doi: 10.1089/dna.1997.16.207. [DOI] [PubMed] [Google Scholar]
- 14.Hernandez E R, Roberts C T, LeRoith D, Adashi E Y. Rat ovarian insulin-like growth factor I (IGF-I) gene expression is granulosa cell-selective: 5′-untranslated mRNA variant representation and hormonal regulation. Endocrinology. 1989;125:572–574. doi: 10.1210/endo-125-1-572. [DOI] [PubMed] [Google Scholar]
- 15.Hoess R H, Ziese M, Sternbeng N. P1 site-specific recombination: nucleotide sequence of the recombining sites. Proc Natl Acad Sci USA. 1982;79:3398–3402. doi: 10.1073/pnas.79.11.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hogan B, Beddington R, Costantini F, Lacy E. Manipulating the mouse embryo: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- 17.Kuo C J, Conley P B, Hsieh C L, Francke U, Crabtree G R. Molecular cloning, functional expression, and chromosomal localization of mouse hepatocyte nuclear factor I. Proc Natl Acad Sci USA. 1990;87:9838–9842. doi: 10.1073/pnas.87.24.9838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lakso M, Pichel J G, Gorman J R, Sauer B, Okamoto Y, Lee E, Alt F W, Westphal H. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc Natl Acad Sci USA. 1996;93:5860–5865. doi: 10.1073/pnas.93.12.5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laron Z. Disorders of growth hormone resistance in childhood. Curr Opin Pediatr. 1993;5:474–480. doi: 10.1097/00008480-199308000-00018. [DOI] [PubMed] [Google Scholar]
- 20.Laron Z, Klinger B. Laron syndrome: clinical features, molecular pathology and treatment. Horm Res. 1994;42:198–202. doi: 10.1159/000184193. [DOI] [PubMed] [Google Scholar]
- 21.Ledermann H M. Is maturity onset diabetes at young age (MODY) more common in Europe than previously assumed? Lancet. 1995;345:648. doi: 10.1016/s0140-6736(95)90548-0. [DOI] [PubMed] [Google Scholar]
- 21a.Lee, Y.-H., and P. F. Johnson. Unpublished observations.
- 22.Lee Y H, Williams S C, Baer M, Sterneck E, Gonzalez F J, Johnson P F. Ability of C/EBPβ but not C/EBPα to synergize with an Sp1 protein is specified by the leucine zipper and activation domain. Mol Cell Biol. 1997;17:2038–2047. doi: 10.1128/mcb.17.4.2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee Y H, Sauer B, Johnson P F, Gonzalez F J. Disruptiona of the c/ebpα gene in adult mouse liver. Mol Cell Biol. 1997;17:6014–6022. doi: 10.1128/mcb.17.10.6014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin S-C, Lin C R, Gukovsky I, Lusis A J, Sawchenko P E, Rosenfeld M G. Molecular basis of the little mouse phenotype and implications for cell type-specific growth. Nature. 1993;364:208–213. doi: 10.1038/364208a0. [DOI] [PubMed] [Google Scholar]
- 25.Liu S Y, Gonzalez F J. Role of the liver-enriched transcription factor HNF-1a in expression of the CYP2E1 gene. DNA Cell Biol. 1995;14:285–293. doi: 10.1089/dna.1995.14.285. [DOI] [PubMed] [Google Scholar]
- 26.Lowe W L., Jr . Biological actions of the insulin-like growth factors. In: LeRoith M D, editor. Insulin-like growth factors: molecular and cellular aspects. Boca Raton, Fla: CRC Press, Inc.; 1991. pp. 49–85. [Google Scholar]
- 27.Nolten L A, Van Schaik F M A, Steenbergh P H, Sussenbach J S. Expression of the insulin-like growth factor I gene is stimulated by the liver-enriched transcription factors C/EBPα and LAP. Mol Endocrinol. 1994;8:1636–1645. doi: 10.1210/mend.8.12.7708053. [DOI] [PubMed] [Google Scholar]
- 28.Nolten L A, Steenbergh P H, Sussenbach J S. Hepatocyte nuclear factor 1α activates promoter 1 of the human insulin-like growth factor I gene via two distinct binding sites. Mol Endocrinol. 1995;9:1488–1499. doi: 10.1210/mend.9.11.8584026. [DOI] [PubMed] [Google Scholar]
- 29.Nolten L A, Steenbergh P H, Sussenbach J S. The hepatocyte nuclear factor 3β stimulates the transcription of the human insulin-like growth factor I gene in a direct and indirect manner. J Biol Chem. 1996;271:31846–31854. doi: 10.1074/jbc.271.50.31846. [DOI] [PubMed] [Google Scholar]
- 30.Polonsky K S. Lilly Lecture 1994. The beta-cell in diabetes: from molecular genetics to clinical research. Diabetes. 1995;44:705–717. doi: 10.2337/diab.44.6.705. [DOI] [PubMed] [Google Scholar]
- 31.Pontoglio M, Barra J, Hadchouel M, Doyen A, Kress C, Bach J P, Babinet C, Yaniv M. Hepatocyte nuclear factor 1 inactivation results in hepatic dysfunction, phenylketonuria, and renal Fanconi syndrome. Cell. 1996;84:575–585. doi: 10.1016/s0092-8674(00)81033-8. [DOI] [PubMed] [Google Scholar]
- 32.Pontoglio M, Faust D M, Doyen A, Yaniv M, Weiss M C. Hepatocyte nuclear factor 1 alpha gene inactivation impairs chromatin remodeling and demethylation of the phenylalanine hydroxylase gene. Mol Cell Biol. 1997;17:4948–4956. doi: 10.1128/mcb.17.9.4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rijli F M, Dolle P, Fraulob V, LeMeur M, Chambon P. Insertion of a targeting construct in a Hoxd-10 allele can influence the control of Hoxd-9 expression. Dev Dyn. 1994;201:366–377. doi: 10.1002/aja.1002010408. [DOI] [PubMed] [Google Scholar]
- 34.Robertson E J, editor. Teratocarcinomas and embryonic stem cells: a practical approach. Washington, D.C: IRL Press; 1987. [Google Scholar]
- 35.Robertson R P, Olson L K, Redmon J B, Zhang H-J, Towle H C. Glucose toxicity and the insulin gene. In: Draznin B, LeRoith D, editors. Molecular biology of diabetes. I. Autoimmunity and genetics; insulin synthesis and secretion. Towota, N.J: Humana Press; 1994. pp. 195–212. [Google Scholar]
- 36.Sauer B. Manipulation of transgenes by site-specific recombination: use of Cre recombinase. Methods Enzymol. 1993;225:890–901. doi: 10.1016/0076-6879(93)25056-8. [DOI] [PubMed] [Google Scholar]
- 37.Stoffers D A, Ferrer F, Clarke W L, Habener J F. Early-onset type-II diabetes mellitus (MODY4) linked to IPF1. Nat Genet. 1997;17:138–139. doi: 10.1038/ng1097-138. [DOI] [PubMed] [Google Scholar]
- 38.Tronche F, Roller A, Bach I, Weiss M C, Yaniv M. Rat albumin promoter cooperation with upstream elements is required when binding of APF/HNF1 to the proximal element is partially impaired by mutation or bacterial methylation. Mol Cell Biol. 1989;9:4759–4766. doi: 10.1128/mcb.9.11.4759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Udy G B, Towers R P, Snell R G, Wilkins R J, Park S-H, Ram P A, Waxman D J, Davey H W. Requirement of STAT5b for sexual dimorphism of body growth rates and liver gene expression. Proc Natl Acad Sci USA. 1997;94:7239–7244. doi: 10.1073/pnas.94.14.7239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vaxillaire M, Rouard M, Yamagata K, Oda N, Kaisaki P J, Borirai V V, Chevre J-C, Boccio V, Cox R D, Lathrop G M, Dussoix P, Philippe J, Timsit J, Charpentier G, Velho G, Bell G I, Froguel P. Identification of nine novel mutations in the hepatocyte nuclear factor alpha gene associated with maturity-onset diabetes of the young (MODY3) Hum Mol Genet. 1997;6:583–586. doi: 10.1093/hmg/6.4.583. [DOI] [PubMed] [Google Scholar]
- 41.Yamagata K, Oda N, Kaisaki P J, Menzel S, Furuta H, Vaxillaire M, Southam L, Cox R D, Lathrop G M, Borirai V V, Chen X, Cox N J, Oda Y, Yano H, Le Beau M M, Yamada S, Nishigori H, Takeda J, Fajans S S, Hattersley A T, Iwasaki N, Hansen T, Pedersen O, Polonsky K S, Turner R C, Velho G, Chevre J-C, Froguel P, Bell G I. Mutations in the hepatocyte nuclear factor-1α gene in maturity-onset diabetes of the young (MODY3) Nature. 1996;384:455–458. doi: 10.1038/384455a0. [DOI] [PubMed] [Google Scholar]
- 42.Yamagata K, Furuta H, Oda N, Kaisaki P J, Menzel S, Cox N J, Fajans S S, Signorini S, Stoffel M, Bell G I. Mutations in the hepatocyte nuclear factor-4α gene in maturity-onset diabetes of the young (MODY1) Nature. 1996;384:458–460. doi: 10.1038/384458a0. [DOI] [PubMed] [Google Scholar]
- 43.Yanuka-Kashles O, Cohen H, Trus M, Aran A, Benvenisty N, Reshef L. Transcriptional regulation of the phosphoenolpyruvate carboxykinase gene by cooperation between hepatic nuclear factors. Mol Cell Biol. 1994;14:7124–7133. doi: 10.1128/mcb.14.11.7124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou Y, Xu B, Wang X, Chen W Y, Kopchick J J. Functional expression of a mouse growth hormone receptor cDNA in transfected mouse L cells. Receptor. 1994;4:143–155. [PubMed] [Google Scholar]