Abstract
β-Thalassemia is brought about by defective β-globin (HBB [hemoglobin subunit β]) formation and, in severe cases, requires regular blood transfusion and iron chelation for survival. Genome editing of hematopoietic stem cells allows correction of underlying mutations as curative therapy. As potentially safer alternatives to double-strand-break–based editors, base editors (BEs) catalyze base transitions for precision editing of DNA target sites, prompting us to reclone and evaluate two recently published adenine BEs (ABEs; SpRY and SpG) with relaxed protospacer adjacent motif requirements for their ability to correct the common HBBIVSI-110(G>A) splice mutation. Nucleofection of ABE components as RNA into patient-derived CD34+ cells achieved up to 90% editing of upstream sequence elements critical for aberrant splicing, allowing full characterization of the on-target base-editing profile of each ABE and the detection of differences in on-target insertions and deletions. In addition, this study identifies opposing effects on splice correction for two neighboring context bases, establishes the frequency distribution of multiple BE editing events in the editing window, and shows high-efficiency functional correction of HBBIVSI-110(G>A) for our ABEs, including at the levels of RNA, protein, and erythroid differentiation.
Keywords: MT: RNA/DNA editing, CRISPR/Cas9, gene therapy, hemoglobinopathies, β-thalassemia, near-PAMless, base editing, hematopoietic stem and progenitor cell, nucleofection
Graphical abstract
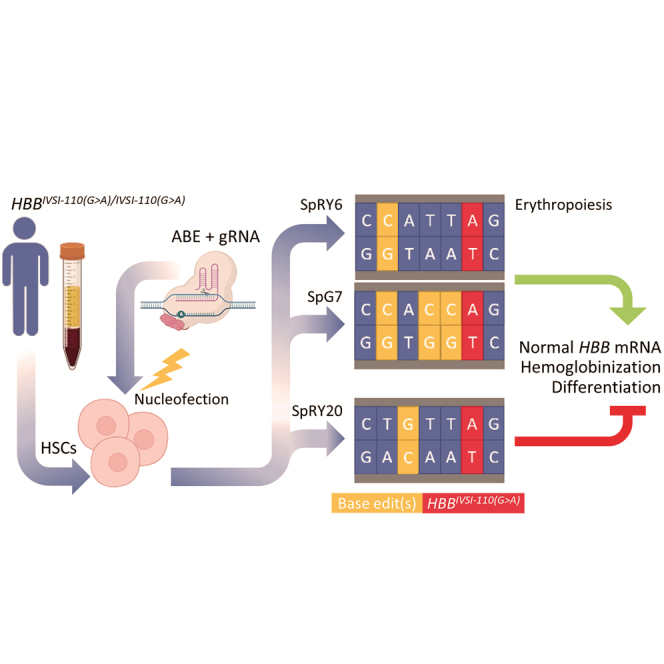
Lederer and colleagues have targeted HBBIVSI-110(G>A) thalassemia with novel base editors and shown that editing of an upstream base results in phenotypic correction. Alternatively, combinatorial base editing of three HBBIVSI-110(G>A)-proximal upstream bases has similarly high correction efficiency. By contrast, editing of an interspersed base is instead detrimental to normal splicing.
Introduction
Hereditary quantitative defects in β-globin chain synthesis cause blood disorders covered by the umbrella term β-thalassemia, characterized by clinical signs that include hepatosplenomegaly, anemia, and bone marrow expansion.1 The highest national carrier rate for β-thalassemia in the EU at 12% is found on the island of Cyprus, where 76% of disease alleles bear the globally common and severe β-globin (HBB)IVSI-110(G>A) mutation (IVSI-110).2 This G>A point mutation in intron 1 creates an aberrant splice acceptor site and results in the retention of a 19-nucleotide stretch of intronic sequence, including a premature termination codon, in a large proportion of the mature HBB mRNA.3 Consequentially reduced quantity of HBB prompts homotetramerization and precipitation of surplus α-globin chains (HBA, encoded by the HBA1 and HBA2 genes) in precursor cells, which in IVSI-110 homozygotes leads to premature death of red blood cells and ineffective erythropoiesis.4 As the only conventional curative treatment and despite considerable improvements in event- and disease-free survival, bone marrow transplantation is unsuitable for most patients owing to the limited availability of matching donors and potentially lethal complications, such as graft-versus-host disease, for suboptimal matches.5 Frequency and severity of IVSI-110 have prompted the development of mutation-specific therapy approaches, which have been pursued by our group6,7,8 and several others.9,10,11,12 We explored efficient double-strand break (DSB)-based disruption of aberrant regulatory elements to restore normal splicing7,8 and found by clonal analyses that deletion of longer upstream sequence stretches, but also of individual bases, may fully restore correct splicing.7 Although designer nucleases have started to accelerate mutation-specific therapy development,13 the proven inherent risks of genotoxicity for DSB-based approaches and the comparative safety of newer, DSB-independent editors may point the way for the next generation of advanced therapies.13,14 Of these, base editors (BEs) combine high-efficiency precision editing with the safety of a DNA nickase and have proven to be effective for use in hematopoietic stem and progenitor cells (HSPCs) in experimental treatment for β-hemoglobinopathies, including the IVSI-110 mutation.10 One class of these editors, adenine BEs (ABEs) prompt A>G transitions restricted to a small editing window close to the Cas-encoded protospacer adjacent motif (PAM). This study utilizes ABEs with relaxed PAM requirements15 with the aim of correcting the IVSI-110 splice defect in patient-derived CD34+ cells by altering an upstream sequence element that we have previously demonstrated to be critical for aberrant splicing and a suitable target for functional splice correction.7,8 Such disruption of aberrant regulatory elements7 allows greater flexibility than base editing directed at the primary mutation and might thus be more widely applicable across different intronic mutations.
Results
Novel ABEs show high DNA-editing efficiency
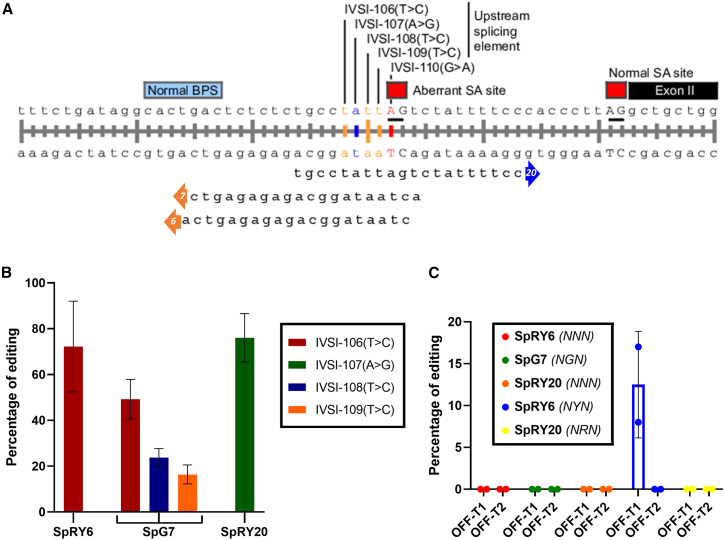
For use with near-PAMless BEs,15 NGN-restricted SpG and NRN>NYN-restricted SpRY, gRNAs were designed to target the base four nucleotides upstream of the IVSI-110 mutation (Figure 1A) by placing it within the 4-nucleotide editing window,16 hereafter referred to as IVSI-106, in a strategy suggested by our published clonal analyses of functional splice correction.7 In order to avoid the suboptimal efficiency associated with plasmid-based expression,17 we performed transient BE delivery by gRNA/mRNA nucleofection.18 After testing different ABE/gRNA combinations for editing efficiency (data not shown), we proceeded with the gRNAs eliciting the highest efficiencies per ABE for analyses in thalassemic, patient-derived CD34+ cells. According to EditR analysis,19 the approach achieved up to 90% editing at the IVSI-106 target base with the more permissive SpRY ABE using gRNA6 and up to 60% on-target editing with the more stringent SpG ABE using gRNA7, albeit with bystander edits (<30%) of both other Ts in the base-editing window (Figure 1B). To explore additional editing of context bases, we employed SpRY ABEs with differently oriented gRNAs targeting the IVSI-107 adenine base adjacent to the IVSI-106 target. By evaluating different gRNA lengths, we observed a positive correlation for gRNA length and efficiency of editing for patient-derived CD34+ cells (Figure S1), with optimized editing conditions and a gRNA length of 20 nucleotides (gRNA20) resulting in IVSI-107 editing of up to 88%. It is noteworthy in this context that SpRY6, while employing the less efficient NYN PAM (i.e., GTG) for its on-target site, shows efficiency similar to that of SpRY20, the activity of which is based on the NRN PAM (i.e., CAC), for which SpRY exerts its highest efficiency.15
Figure 1.
gRNA design strategy for targeting of the IVSI-110 region
(A) The three gRNAs capable of editing the region upstream of the IVSI-110 mutation are shown. Predicted editing windows extended from just upstream of the mutated IVSI-110G>A base (for gRNA20) to the third upstream T base (IVSI-106, for gRNAs 6 and 7). T>C base changes were detected for SpRY6 and SpG7 and are highlighted in orange, whereas an A>G base change was detected for SpRY20 and is highlighted in blue. (B) Bar graph depicting average editing efficiency of ABEs according to EditR analysis, across CD34+ cells derived from different patients across independent experiments (n = 3 for SpRY20, n = 5 for other treatments). (C) Bar graph depicting the percentage of editing at the top two off-target sites (OFF-T1 and OFF-T2, respectively) for SpRY6, SpG7, and SpRY20, as indicated (n = 2). Off-target identities are detailed in Tables S2–S4. For (A) and (B), error bars show the standard deviation of the mean.
In silico specificity predictions of gRNA/ABEs predicts higher safety for SpG7
In order to estimate the safety of our editors, we utilized the CRISPOR20 in silico tool to help ascertain the predicted off-target events of our editors (Table 1; see Tables S2–S4 for full off-target information).21 Based on CRISPOR and in line with its relatively stringent PAM (NGN), SpG7 had the lowest predicted number (67 total and 3 intragenic for up to 3 mismatches) of off-target hits. Off-target predictions for SpRY6 (121 total and 9 intragenic for up to 3 mismatches) and SpRY20 (264 total and 14 intragenic for up to 3 mismatches) give a high number of total potential off-target sites for SpRY20, but a comparably low number of potential intragenic off-target sites, in line with the high U:A content of the gRNA, which would typically favor intergenic sites.22 Though based on the same ABE protein, off-target predictions for SpRY6 are approximately half those for SpRY20. We conducted investigation of the off-targeting activity of our ABEs by sequencing the top two predicted off-target regions, as ranked by the CRISPOR algorithm, for each editor with all its potential PAM(s), i.e., NGN for SpG7, NYN/NNN for SpRY6, and NRN/NNN for SpRY20. There were no off-targets detected for the five loci sequenced for SpG7 and SpRY20 (Figure 1C). Validating our assay, out of the four genes analyzed for SpRY6, the intronic KIAA1328 site, predicted to be the top off-target for SpRY6 (NYN), showed up to 17% editing in ABE-treated patient cells (Figure S2), indicating superior specificity and safety for SpG7 and SpRY20. The off-target locus KIAA1328 encodes the protein hinderin, which competes with the structural maintenance of chromosome (SMC) protein 3 for binding to the hinge domain of SMC1, thereby preventing their heterodimerization and consequent formation of the cohesion complex, necessary for sister chromatid separation and chromosomal stability.23
Table 1.
In silico prediction of gRNA off-target effects
| gRNA identity | gRNA sequence | MIT specificity score24 | CrisprSCAN25 | Off-targetsa |
|---|---|---|---|---|
| gRNA7 with ABE_SpG (NGN) | ACTAATAGGCAGAGAGAGTC | 24 | 42 | 0|1|2|64|8000|1|1|1|13 |
| gRNA6 with ABE_SpRY (NYN) | CTAATAGGCAGAGAGAGTCA | 17 | 43 | 0|1|4|116|14580|1|1|7|27 |
| gRNA20 with ABE_SpRY (NRN) | TGCCTATTAGTCTATTTTCC | 9 | 6 | 0|1|8|255|34890|0|0|14|24 |
Based on analyses using CRISPOR, numbers shown indicate the number of (first row: total, second row: intragenic) off-targets with 0|1|2|3|4 gRNA mismatches.
NGS analysis reveals editing profile of ABEs
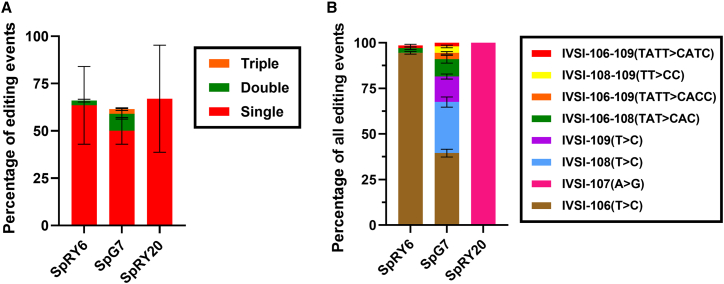
Targeted deep sequencing of HBB accurately discerned that the editing efficiency of SpG7 as the cumulation of all three cis edits in the IVSI-110 upstream region was close to that of SpRY6, leading us to predict a similar effect by both editors on splice correction. This analysis also allowed us to determine the absolute and relative contribution of individual and combined editing events per allele. As shown in Figure 2, the edited bulk population consists of all possible combinations of editing events, including single (81% of total editing events), double (15%), and triple (4%) base edits. Based on our next-generation sequencing (NGS) data, a small but notable increase in insertion-deletion (indel) frequency is seen for SpRY6 and SpG7 compared to the control samples, so that the percentages of reads that contained at least one small insertion overlapping the gRNA binding site were, on average, 0.96% and 0.39% for SpRY6 and SpG7, respectively, while the observed percentage for the control samples was 0.23%. The indel rates normalized for segment length and corrected for baseline indel frequencies showed similar results (Figures S3–S7), with SpRY6 target rates being 3-fold higher at 4.1 × 10−4 insertion frequency per position compared to the control (1.3 × 10−4 insertion frequency per position). SpG7 insertion rates were approximately 1.42-fold higher, while SpRY20 remained equal or lower, compared to the control. Deletion rates were similar across all samples. In agreement with our results, increased indel frequency following base editing in the same HBB target region has also been noted by others.10
Figure 2.
Next-generation DNA sequencing data of edited samples
(A) Graph illustrating frequency of editing events relative to homozygous IVSI-110(G>A) control. (B) Graph illustrating percentage contribution of editing events. For (A) and (B), error bars show the standard deviation of the mean.
Post-editing functional analyses
All edited cells were subjected to induced erythroid differentiation for a period of 18 days post-editing and were functionally analyzed for hallmarks of β-thalassemia at different levels.
ABE editing of different context bases impacts RNA expression differentially
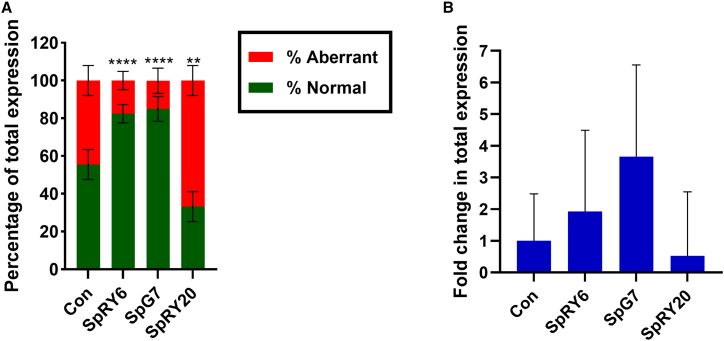
Analysis by RT-qPCR of normal and aberrant splicing at the RNA level, as a critical parameter of correction for IVSI-110,7,8,26 indicates a significantly higher and comparable percentage of normal HBB mRNA for SpG7 (85% ± 6.7%) and SpRY6 (80% ± 4.8%) compared to the untreated control (56% ± 8.0%) (Figure 3A). However, for total HBB transcript levels normalized to HBA (Figure 3B), a quantitative increase of expression was observed by a factor of 1.9 × 0.41±1 ± 2.7 for SpRY6, and a higher factor of 3.7 × 0.64±1 ± 2.1 for SpG7, compared to control (1.0 ± 1.1 × 0.48±1). Inversely, SpRY20-treated samples indicated a significantly lower percentage (30% ± 8.0%) of normal mRNA expression, as well as decreased overall HBB expression (0.5 ± 1.6 × 0.24±1), as compared to untreated control. These results were confirmed via additional analyses based on RT-droplet digital (dd)PCR (Figure S8). Corresponding in silico predictions indicate an effect of the introduced base changes on DNA elements that regulate splicing and may elicit differences in transcript levels and ratios at the intron-exon border of IVSI (Figure S9). For SpRY20, in contrast to normal and other edited sequences, these predictions include the formation of a novel donor site between the aberrant and normal splice acceptors, the loss of a cryptic branchpoint site (BS), and a reduction in the overall ratio of splice enhancer:silencer elements as three in silico results that may guide future experimental substantiation.
Figure 3.
Assessment of splicing at the transcriptional level in patient-derived CD34+ cells on day 11 of erythroid differentiation
(A) Mean proportion of normal and aberrant HBB mRNA. The groupwise comparison to untreated control for aberrant RNA levels was performed using one-way ANOVA with Dunnett’s multiple comparison analysis. ∗∗∗∗p < 0.0001 and ∗∗p = 0.0015. (B) Relative mRNA expression levels of HBB to HBA. The groupwise comparison to untreated control was performed using Kruskal-Wallis analysis with Dunn’s multiple comparisons test, and only the proportional upward variation is indicated. For (A) and (B), n = 3 for SpRY20, n = 5 for other treatments. For (A) and (B), error bars show the standard deviation of the mean.
β-Globin and HbA levels are significantly increased after IVSI-106 targeting and significantly decreased after IVSI-107 targeting by ABEs
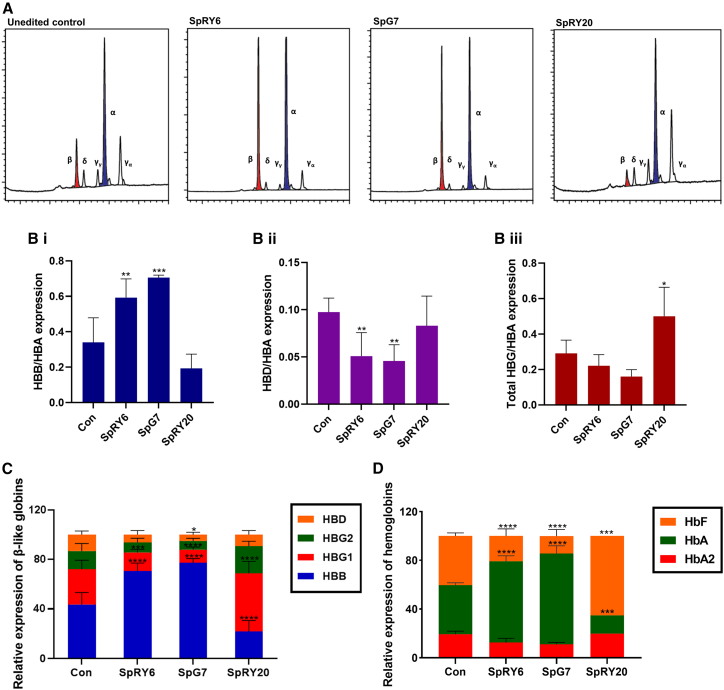
High-performance liquid chromatography (HPLC) analyses revealed a significantly increased HBB protein level relative to HBA for both SpRY6 (0.59 ± 0.10) and SpG7 (0.71 ± 0.01) (by reversed-phase [RP]-HPLC; Figures 4A–4C) compared to control cells (0.34 ± 0.14). A correspondingly significantly decreased HBD level relative to HBA is seen for SpG7 and SpRY6 (0.05 ± 0.02 for both) compared to untreated control (0.10 ± 0.01). Total HBG (as the sum of HBG2 and HBG1) relative to HBA is slightly decreased for SpG7 (0.16 ± 0.04) and SpRY6 (0.22 ± 0.06) compared to untreated control (0.29 ± 0.07). Assessment of proportions of total globin chains for the editors revealed significantly increased HBB levels for SpRY6 (0.71 ± 0.06) and SpG7 (0.77 ± 0.03) compared to untreated control (0.44 ± 0.10). As for HBG1, levels were significantly decreased for SpG7 (0.11 ± 0.02) and SpRY6 (0.15 ± 0.03) compared to untreated control (0.29 ± 0.07). In the case of HBG2, levels were decreased for SpG7 (0.07 ± 0.02) and SpRY6 (0.08 ± 0.03) compared to untreated control (0.15 ± 0.06). Finally, for SpG7, we detected a significantly decreased level of HBD (0.05 ± 0.02) compared to untreated controls (0.13 ± 0.03), with decreased expression also seen for SpRY6 (0.06 ± 0.03). As for the proportion of hemoglobins, HbA (by cation-exchange [CE]-HPLC; Figure 4D) was significantly increased following SpRY6 and SpG7 treatments compared to untreated control (40% ± 2.0%), with expression levels of 67% ± 4.5% and 75% ± 6.3%, respectively. Fetal hemoglobin (HbF) expression was also significantly decreased for both corrective editors at 21% ± 5.9% for SpRY6 and 14% ± 5.3% for SpG7 as compared to untreated control levels (40% ± 2.5%). HbA2 was likewise decreased in these samples, albeit not at a significant level, at 13% ± 3.5% for SpRY6 and 11% ± 1.6% for SpG7 compared to 19% ± 2.5% for the control. Compared to control values and consistent with decreased HBB/HBA transcript expression detected for SpRY20 treatment (Figure 3), protein levels of HBB relative to both HBA (0.19 ± 0.08) and HbA (15%) were decreased, with commensurate increased HbF levels (65%). In fact, compared to untreated control, SpRY20-treated cells expressed significantly higher levels of total HBG (0.50 ± 0.16) and notably lower levels of HBD (0.08 ± 0.03) relative to HBA. Expressed as proportions of β-like globin relative to HBA, SpRY20 showed a significantly decreased HBB (0.22 ± 0.09) and notably decreased HBD (0.09 ± 0.03), but significantly increased HBG1 (0.47 ± 0.10) and a notable increase in HBG2 (0.22 ± 0.04), compared to control values.
Figure 4.
HPLC-based protein measurements of patient-derived CD34+ cells on day 14 of erythroid differentiation
(A) Representative chromatograms of RP-HPLC-based detection of human globin chains in patient cells indicating induction of HBB after SpRY6 or SpG7 treatment and reduction of HBB with SpRY20. (B) Bar graphs depicting proportion of HBB-like globin species relative to HBA expressed post-treatment for all editors and untreated controls. Groupwise comparisons were performed using one-way ANOVA with Dunnett’s multiple comparison test for HBB (navy, i) and HBG (maroon, iii) and with Holm-Sidak’s multiple comparisons test for HBD (purple, ii). For HBB/HBA, ∗∗∗p < 0.0001 and ∗∗p = 0.0031; for HBD/HBA, ∗∗p = 0.0085 for SpRY6 and ∗∗p = 0.0060 for SpG7; for total HBG/HBA, ∗p = 0.0108. (C) Bar graph depicting proportion of each globin species post-treatment. Groupwise comparisons were performed using mixed-effects analysis with Dunnett’s multiple comparison test. ∗∗∗∗p < 0.0001, ∗∗∗p = 0.0004, and ∗p = 0.0454. All as measured by RP-HPLC (n = 3 for SpRY20, n = 5 for other treatments). (D) Bar graph depicting post-editing expression of hemoglobin species after control, SpRY6, and SpG7 treatments (n = 3) as measured by CE-HPLC. SpRY20 data are shown as a single analysis owing to insufficient material for replicate samples. Groupwise comparisons were performed using parametric mixed-effects analysis with Dunnett’s multiple comparison test. ∗∗∗∗p < 0.0001 and ∗∗∗p = 0.0002. For (B) through (D), error bars show the standard deviation of the mean, and only the upward variation is given.
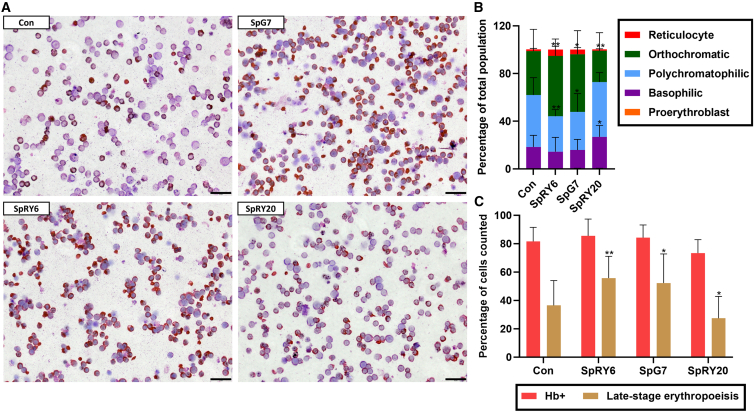
ABE-mediated changes of RNA and protein levels significantly impact erythroid morphology
Microscopy of cytocentrifuged samples during erythroid differentiation and differential scoring of cells in the erythroid lineage is highly informative for the restoration of normal hematopoiesis. Here, SpRY6- and SpG7-treated cells showed comparably greater hemoglobinization of 86% ± 11.9% and 84% ± 8.9%, respectively, compared to untreated control (82% ± 9.9%), and samples treated with these editors showed a significant shift toward late-stage cells (orthochromatic erythroblasts and reticulocytes) of 56% ± 15.4% and 52% ± 20.5%, respectively, compared to untreated control (37% ± 17.5%), indicating more advanced and higher rates of erythropoiesis (Figure 5). The basophilic subpopulation was slightly decreased for SpRY6 (14% ± 11.9%) and SpG7 (16% ± 8.9%) compared to untreated control (18% ± 9.8%). Likewise, the proportions of polychromatophilic cells for SpRY6 (30% ± 5.5%) and SpG7 (32% ± 15.2%) are decreased when compared to untreated controls (44% ± 14.5%). As for the more differentiated orthochromatic cells, a significantly increased percentage is seen for SpRY6 (51% ± 14.2%) and SpG7 (48% ± 19.8%) as compared to untreated controls (37% ± 18.2%). In the case of reticulocytes, the most differentiated erythroid subpopulation quantified, the percentage is increased for SpG7 (4% ± 1.6%) and SpRY6 (5% ± 2.8%) compared to untreated controls (1% ± 0.9%) but not at a statistically significant level. In contrast, SpRY20-treated cells exhibited delayed differentiation and lower hemoglobinization (73% ± 9.6%), and showed a significantly decreased late-stage differentiation (27% ± 15.4%), with a significantly increased basophilic erythroblast cell population (27% ± 9.6%), a slightly increased polychromatophilic subpopulation (46% ± 7.9%), and a decreased orthochromatic subpopulation (26% ± 15.1%) compared to untreated controls, indicating worsening of the β-thalassemia phenotype, in line with protein and RNA parameters.
Figure 5.
Cytocentrifugation and scoring of patient-derived CD34+ cells on day 11 of erythroid differentiation
(A) Representative cytocentrifugation images of stained treated and control cells. Size marker: 20 μm. (B) Bar chart representing differential scoring of the proportion of erythroid subpopulations. Groupwise comparison was made to untreated control using parametric mixed-effects analysis with Dunnett’s multiple comparison test. For basophilic: ∗p = 0.0283. For polychromatophilic: ∗∗p = 0.0030 and ∗p = 0.0151. For orthochromatophilic: ∗∗p = 0.0032 for SpRY6, ∗∗p = 0.0016 for SpRY20, and ∗p = 0.0171. (C) Bar chart indicating the number of cells in late-stage erythropoiesis (equal to the sum of orthochromatophilic cells and reticulocytes from B) and hemoglobin-positive (Hb+) cells, equivalent to late-stage erythropoietic cells plus polychromatophilic (dianisidine-staining cells). Analyses are of patient-derived cells following treatment (n = 3 for SpRY20, n = 5 for other treatments). Groupwise comparison was made to untreated controls and significance obtained using parametric mixed-effects analysis with Dunnett’s multiple comparison test. ∗∗p = 0.0028 and ∗p = 0.0137 for SpG7 and ∗p = 0.0295 for SpRY20. For (B) and (C), error bars show the standard deviation of the mean, and only the upward variation is given.
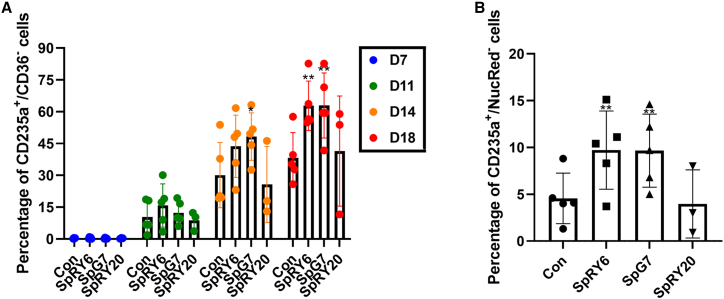
Flow cytometric lineage and enucleation markers confirm the effect of ABEs on erythroid lineage differentiation
Analysis of erythroid differentiation via flow cytometry showed a gradual improvement of late-stage erythroid differentiation (CD235a+/CD36−) starting from day 11 of induced erythroid differentiation in SpRY6- and SpG7-edited samples compared to controls. In particular, we observed significantly higher percentages of CD235a+/CD36− stained cells in SpRY6-edited samples (day 14: 44% ± 14.7%; day 18: 63% ± 11.7%) and SpG7 (day 14: 48% ± 11.2%; day 18: 63% ± 15.3%) compared to controls (day 14: 30% ± 15.4%; day 18: 38% ± 11.9%), indicating greater rates of late-stage differentiation and thus rescue of ineffective erythropoiesis in the SpRY6- and SpG7-treated samples (Figures 6A and S10). Specifically, our data indicate increased differentiation of SpG7-treated cells compared to its counterpart, SpRY6, in agreement with the detection of greater total HBB mRNA levels for the former. Additionally, significantly increased percentages of enucleated cells (NucRed−) were seen for cells edited with SpRY6 (9.7% ± 4.2%) and SpG7 (9.7% ± 3.9%) compared to control cells (4.6% ± 2.7%) (Figures 6B and S11), corroborating the correction of erythrocyte formation. Opposing results were noted for SpRY20, with fewer enucleated cells (4.0% ± 3.6%) and fewer CD235a+/CD36− cells (day 14: 26% ± 17.9%; day 18: 41% ± 26.0%) in comparison to untreated patient cells, albeit not at a statistically significant level.
Figure 6.
Flow cytometry analysis of edited vs. control cells undergoing differentiation
(A) CD235a+/CD36− staining. Cells stained for differentiation markers were scored on four collection timepoints (days 7, 11, 14, and 18). Groupwise comparisons were performed using two-way ANOVA with Dunnett’s multiple comparison test. Day 14: ∗p = 0.0379; day 18: ∗∗p = 0.0032 for SpRY6 and ∗∗p = 0.0030 for SpG7. (B) CD235a+/NucRed− staining. Corresponding representative flow cytometry scatterplots are shown in Figures S10 and S11 and corresponding data for cell proliferation in Figure S12. On the final day of differentiation (day 18), cells were additionally stained with the nucleation marker NucRed; the chart shows the proportion of enucleated cells in the CD235a+ cell population. Significance was obtained using two-way ANOVA with Dunnett’s multiple comparison test. ∗∗p = 0.0036 for SpRY6 and ∗∗p = 0.0039 for SpG7. Statistical analyses were performed in comparison to control for both analyses of live patient cells following treatment (n = 3 for SpRY20, n = 5 for other treatments). For (A) and (B), error bars show the standard deviation of the mean.
Discussion
We have presented here an approach for correction of the IVSI-110(G>A) β-thalassemia splicing mutation using our optimized ABEs, SpRY6 and SpG7, which substantially improve the disease phenotype of edited patient-derived CD34+ cells by modifying a critical upstream splice element, IVSI-106, required for aberrant splicing, with demonstrable correction of disease hallmarks at the RNA, protein, morphology, and cytometry levels. Building on our earlier work,7 these findings confirmed previous observations and thereby establish the identity of the IVSI-106 nucleotide rather than a change of position for neighboring elements as critical for regulation of HBB splicing.
Additionally, we identified an immediately adjacent splice element, IVSI-107, as required for normal instead of aberrant splicing, based on the observation that SpRY20-mediated A>G base transition of IVSI-107 aggravated those same β-thalassemia disease parameters corrected by the IVSI-106(T>C) transition. At a general level, our observation underscores the intricate balance of regulatory splice elements and the substantial and variable consequences different editing events may have in even well-characterized regulatory regions. More specifically, and on a route parallel to that chosen by the Miccio group in their elegant work on BE-based correction of the primary IVSI-110 mutation,10 our study pinpoints and validates an alternative therapeutic target while opening up the path to exploring the interplay of factors affecting the splicing of HBB intron 1.
Interestingly, our NGS data showing the same-molecule distribution of base-editing events for SpG7 suggests that all combinations of three possible edits were represented, resembling chance distribution of edits within the editing window. This effectively resulted in an overall editing efficiency far higher than would be expected from the individual editing efficiencies per base and from the assumption of initial saturation of the most efficient editing event. As for similar analyses of on-target base editing for the IVSI-110(G>A) mutation by Hardouin et al.,10 editing distribution was reproducible between different patients in the present study. However, whereas here over 80% of editing events for SpG7, upstream of the aberrant splice site, were single base edits, in the previous study, 75% of editing events at the IVSI-110 locus were double or triple editing events proximal to the normal splice acceptor. For both studies, highly efficient functional correction renders it unlikely that individual editing events may have detrimental rather than corrective effects, but corresponding functional dissection of individual edits would call for clonal analyses, which are outside the scope of also the current study. Instead, our analyses prioritized evaluation of the therapeutic potential for SpG7, due to its more potent correction, lower indel rates, and lower predicted and observed off-target activity compared to SpRY6, toward its full preclinical evaluation with long-term repopulating cells in NBSGW mice and comparison to equivalent DSB-based editors as the current clinical benchmark.7,27,28 In the same vein and with a focus on on-target events, the present study performed an experimental evaluation of off-target sites for the top two predictions per editor and, in the process, detected a significant off-target site for SpRY6, KIAA1328, and consequently highlighted SpG7 as safer for therapeutic application. While such experimental evaluation of off-target sites, albeit limited, compares favorably to application of only in silico analyses for ex vivo cultures elsewhere,29 more comprehensive genotoxicity studies, as well as upscaling of manipulations and use of good-manufacturing-practice–level reagents and environments will be required to provide further safety and efficiency data toward clinical translation of our approach. Of note, SpRY20, though based on the same enzyme as SpRY6, in contrast to the latter, does not show a marked increase in on-target indel frequency. This may be due to the complementary orientation of gRNA20 compared to gRNA6, which, together with additional sequence context effects, might lead to alternative interactions, editing intermediates, and therefore nickase activity, along with inadvertent DSB behavior.
Intriguingly, the detrimental effect of SpRY20 can be explained by at least two mutually inclusive explanations. BSs are a requirement for splicing catalysis and have been found to be numerous in most human introns.30 In the case of HBB intron 1, the dominant and strongest branchpoint is at position −37, but a cryptic branchpoint at position −24 exists, albeit with a lower binding energy.30 In both cases, lariat formation occurs at an adenine31 that bulges from the U2/BS helix, later taking on the role of reactive nucleophile,32 which in the case of HBB intron 1 is base IVSI-107. SpRY20 action changes this residue and thereby limits the spliceosome to using only the distal BS. In fact, Reed et al. observed that the substitution of a guanine in place of the BS adenine (A>G, akin to action by SpRY20) altered 3′ splice site selection.31 Moreover, the IVSI-107(A>G) base change predictably gives rise to a transcript containing a recognition motif (UGUU) for two members of the CELF family of RNA-binding proteins. CELF1 and -2 can repress or activate splicing33 and can do the latter by displacing silencer elements in the polypyrimidine tract34 or physically occluding trans-acting factors involved in branchpoint recognition.30 Thereby, the creation of this tetranucleotide, which is sufficient for recognition35 by the CELF proteins, might favor aberrant splicing, possibly by displacing an inhibitory element or recruiting enhancing factors exacerbating the IVSI-110 phenotype. Altogether, aggravation specifically of the characteristic hallmarks of HBBIVSI-110(G>A) thalassemia by SpRY20, combined withits off-target activity being being below the detection level for the top two off-target sites, point to SpRY20 on-target action rather than to off-target action as the basis of its effects. In the same vein, HbF induction by SpRY20 may be explained by decreased expression of β-globin, which is commonly associated with detection of increased γ-globin levels, for which a range of complementary mechanisms have been proposed at the transcriptional, translational, and post-translational levels.36,37,38,39 The interconnectedness of, first, the expression of different genes on the β-globin locus and, second, sequence elements and steps of gene expression, from DNA accessibility to post-translational action, for each gene on that locus, poses a persistent challenge to distinguishing cause and effect for corresponding expression events. However, beyond the scope of the current study, surer insights into mechanisms underlying the effects of SpRY20 and of the therapeutic BEs presented here may be gained by further experimentation, such as by the modification of individual bases and protein factors suspected of involvement, in conjunction with monitoring of the resulting transcripts.
Many advanced treatment options are being developed for hemoglobinopathies and have achieved validation of gene addition and genome-editing approaches in clinical trials, where prolonged or persistent transfusion dependence for severe β-thalassemia genotypes is still a frequent occurrence. The ongoing CRISPR-Therapeutics/Vertex ex vivo genome-editing trial utilizes efficient genome disruption to induce HbF with good efficacy data but with the inherent risk of mutagenesis typical of DSB-based DNA-editing approaches. To address the issue of safety, clinical trials aiming for HbF induction now also include BEs with high on-target fidelity.40 However, therapies based on HbF may be more affected by genetic modifiers, such as those affecting γ-globin expression, than therapies based on HbA, which has evolved for optimal expression and function in adults.
The present study addresses many concerns over advanced therapies and their safety in the therapy of β-thalassemia by applying DSB-independent and efficient base editing to restore HbA and normal phenotypes based on methodology of virus- and DNA-free, highly transient delivery of editors for cell manipulation. Our approach is tailored for the HBBIVSI-110(G>A) mutation and, in many affected individuals, may thus prove superior to universal therapeutic approaches. Beyond HBBIVSI-110(G>A), exploitation of context bases for phenotypic correction gives greater flexibility in the choice of efficient and safe DNA editors than does targeting the causative mutation alone. DSB-independent genome editors might therefore be of utility as a therapeutic approach in a range of other disorders caused by abnormal splicing, in particular where the underlying mutation itself is inaccessible to direct editing.
Materials and methods
Isolation and culture of PB-derived hCD34+ HSPCs
Utilization of primary samples was based on informed consent according to the Declaration of Helsinki and was covered by bioethics approval from the Cyprus National Bioethics Committee (license numbers ΕΕΒΚ/ΕΠ/2018/52 and ΕΕΒΚ/ΕΠ/2022/05). Isolation of CD34+ cells from peripheral blood (PB) mononuclear cells was performed using the CD34+ MicroBead Kit, LS MiniMACS Column, by magnetic separation using an autoMACS Separator (Miltenyi Biotec, Bergisch Gladbach, Germany) from five IVSI-110-homozygous patients with fully informed consent. Isolated cells were then expanded and, after editing, induced to differentiate into the erythroid lineage in a three-phase program in which medium contents and concentration levels varied. Briefly, in phase 1 of differentiation (days 1–7), 0.2–1.0 × 104 cells/mL were maintained in Iscove’s modified Dulbecco’s medium (Gibco, Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 5% human serum (Sigma-Aldrich, St. Louis, MO, USA), 2 IU/mL heparin (Sigma-Aldrich), 10 μg/mL insulin (Sigma-Aldrich), 330 μg/mL holo-transferrin (Sigma-Aldrich), 1 μM hydrocortisone (Medochemie, Limassol, Cyprus), 100 ng/mL stem cell factor (SCF; R&D Systems, Minneapolis, MN, USA), 5 ng/mL interleukin-3 (IL-3; R&D Systems), 3 U/mL erythropoietin (Sandoz GmbH, Kundl, Austria), 1× penicillin-streptomycin (Thermo Fisher Scientific), and GlutaMAX (Thermo Fisher Scientific). In phase 2 (days 7–11), IL-3 and hydrocortisone were omitted, and cell concentration was kept at 1–2 × 105 cells per mL. In phase 3 (days 11–18), SCF was omitted compared to phase 2, and the concentration was maintained at 1–2 × 106 cells per mL.
Design and synthesis of gRNAs
gRNAs were manually designed based on the availability of an appropriate PAM adjacent to the target site and on positioning of the editing window for the desired base substitution.16
All gRNAs were purchased from Synthego (Redwood City, CA, US) for use in combination with in-vitro-synthesized Cas mRNA (for BEs).
In vitro transcription
The Cas mRNA for BEs was synthesized with the HiScribe T7 Anti-Reverse Cap Analog mRNA kit with tailing (New England Biolabs [NEB], Ipswich, MA, US) following the manufacturer’s instructions, as described elsewhere.18
Cloning procedure
Original plasmids were received from Addgene (Watertown, MA, USA) and cultured, purified, and verified by analytical restriction digest using standard methods.41 Excision of GFP from the nearly PAM-less pCMV-T7-ABEmax(7.10)-SpG-P2A-EGFP and pCMV-T7-ABEmax(7.10)-SpRY-P2A-EGFP plasmids (Addgene #140002 and #140003; gifts from Benjamin Kleinstiver) involved isolation of the ABEs from their backbone and insertion into a classical, GFP-free BE backbone (pCMV_ABEmax; Addgene #112093; gift from David Liu). Resulting bacterial clones were confirmed by DNA sequencing, deposited to Addgene, and are available under accession numbers #195278 and #195279.
Nucleofection
Nucleofection of in-vitro-transcribed mRNA was performed using the Lonza 4D-Nucleofector with the P3 Primary Cell 4D-Nucleofector X Kit (both Lonza, Basel, Switzerland) with the CA-137 program according to our published procedures18 and manufacturer’s instructions, respectively.
PCR
Amplification was performed using the Q5 High-Fidelity PCR Kit (NEB) and the Veriti thermal cycler (TFS, Waltham, Massachusetts, USA) according to the manufacturer’s instructions. Primer sequences (Metabion International AG, Martinsried, Germany) can be found in Table S1.
gDNA extraction and sequencing
Genomic DNA from eukaryotic cells was extracted using QIAmp DNA Blood Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Cycle sequencing was performed using the BigDye Terminator v.1.1 Cycler sequencing kit (Applied Biosystems, Waltham, Massachusetts, USA) and the Veriti thermal cycler (TFS). Performa DTR Gel Filtration Cartridges (Edge Biosystems, Gaithersburg, Maryland, USA) were used for purification of sequencing products, all in accordance with manufacturers’ instructions, and analyzed on a Hitachi 3031xl Genetic Analyzer with Sequence Detection Software v.5.2 (Applied Biosystems).
Targeted deep sequencing
The Devyser Thalassemia kit was used to analyze HBB and HBA gene clusters and to generate a target-specific library, following the manufacturer’s protocol (Dvysr, Hägersten, Sweden). This was then subjected to NGS using the MiSeq Sequencing platform, employing the MiSeq Reagent MicroKit v.2 (Illumina, San Diego, CA, USA) (300 cycles) with a high coverage of 200×.
HPLC
RP-HPLC of globin chains and CE-HPLC of hemoglobins were performed by modification of our published procedures and manufacturer’s instructions, respectively.26,42
Cell counting
Cell counting was performed manually using a Bright-Line hemocytometer (Sigma-Aldrich, Munich, Germany), and cell viability was measured using trypan blue solution, 0.4% (Thermo Fisher Scientific). Cells were counted under a light microscope (Eclipse TS100, Nikon, Chiyoda, Japan).
Cytocentrifugation and microscopy
For cell preparations on slides, the Tharmac Cellspin II cytocentrifuge with an EASY rotor (Tharmac, Wiesbaden, Germany) was used, followed by dianisidine and standard May-Grünwald/Giemsa (Fluka, Munich, Germany) staining, as described elsewhere.26 The proliferation rate for post-edited cells on the final day of differentiation is shown in Figure S12.
Assessment of RNA expression
RNA was isolated using TRIzol Reagent (Thermo Fisher Scientific) following the associated user manual offered by the company (MAN0001271). cDNA was then created and used as template for qPCR analysis as described elsewhere.4
For ddPCR, experiments were performed using the QX200 AutoDG ddPCR system (Bio-Rad Laboratories, Hercules, CA, USA) following the manufacturer’s protocol. Briefly, each 25 μL reaction included 12.5 μL ddPCR Supermix for probes (no dUTP), 450 nM of each primer, and 2 μL of the sample. Droplet generation was performed on the AutoDG instrument, followed by PCR amplification on a BioRad T100 Thermal Cycler (Bio-Rad Laboratories) using the following thermal protocol: 10 min at 95°C, 40 cycles of 30 s at 95°C, 60 s at 50°C, and final hold at 4°C. Each PCR step included a 2°C/s ramp rate. The plate was read using the QX200 droplet reader and analyzed using the QuantaSoft software (v.1.7.4.0917) (Bio-Rad Laboratories).
Primer and probe sequences (Metabion International AG) can be found in Table S1.
Flow cytometry analysis
Antibodies for early, anti-Hu CD36 APC and late, anti-Hu CD235a PE (both from ExBio, Vestec, Czech Republic) were used for surface marker analysis. NucRed Live 647 ReadyProbe Reagent (Thermo Fisher Scientific) staining was also utilized. Samples were run on the BD FACSAria III (BD Biosciences, Franklin Lakes, NJ, USA), and post-run analysis was performed on the FCS Express 7 software.
Statistical and bioinformatics analyses
Statistical analyses, such as normality tests (Shapiro-Wilks) and correspondingly parametric or non-parametric groupwise comparisons, as specified for each figure panel, were performed using GraphPad Prism 9 software (GraphPad, La Jolla, CA, USA).
For DNA-level evaluation of base editing, the EditR software (https://moriaritylab.shinyapps.io/editr_v10/) was used.19
Splice site analyses were performed using the Human Splicing Finder software (https://www.genomnis.com/access-hsf).43
NGS data processing was performed using third-party tools and custom scripts in R programming language v.4.3.1.44 NGS bioinformatics analysis was performed using a combination of third-party publicly available software and custom-written scripts. Specifically, all FASTQ files were processed using the nf-core/Sarek pipeline written in the nextflow framework.45 Reads underwent quality control, filtering, and trimming of residual adapter sequences following alignment to the GRCh37 genome using BWA-MEMMEM.46 The aligned BAM files were marked for duplicates and processed with samtools v.1.1047 to extract the reads aligned to the region of interest (ROI), which was defined as the 20 bases targeted by the gRNA ± 50 bases upstream and downstream of the target site.
The resulting ROI BAM files were parsed and processed in R using the Rsamtools,48 tidyverse,49 and matrixStats50 packages. Specifically, using the CIGAR notation, all reads were represented in a matrix format of dimensions d ∗ w where d is the depth of sequencing and w is the number of bases at the ROI. Base compositions were calculated as proportions for each position (column):
where Pi,j is the proportion of the base i calculated as the counts of i over the depth dj at the jth position.
Similarly, proportions of 7-nucleotide sequences overlapping the region of the targeted bases at chr11: 5,248,050–5,248,056 were calculated using the table() function in R.
Alignments at the ROI that comprised at least one small indel were extracted and analyzed further to calculate their frequencies at the target site (gRNA 20 bp segments) and flanking regions (sequences in the ROI that do not overlap the gRNA binding site). The rates were calculated as follows:
where Nxr is the number of reads in region r, which contains an indel over the mean depth and length lr (in bases) of the specific region. Fold changes of Rxr were calculated by first correcting for baseline rates by subtracting the respective flanking region rates Rx,Flanks (assumed as baseline) from the Rx,Target of each sample and then dividing by the corresponding corrected mean rate of the controls.
The alignments, base and read proportions, and indel rates were visualized using the ggplot2 package in R. All NGS processing was performed on a Linux (Ubuntu 20.04.6 LTS) server with a 64-core CPU and 1 TB RAM.
Data and code availability
All data are available in the article. Correspondence and requests for materials should be addressed to C.W.L. Reprints and permissions information is available.
Acknowledgments
The project was co-funded by the European Regional Development Fund and the Republic of Cyprus through the Research and Innovation Foundation (EXCELLENCE/1216/0092 and EXCELLENCE/0421/0086), by a Telethon Cyprus PhD studentship, by an A. G. Leventis Foundation educational grant, by COST Action CA21113 (GenE-HumDi), supported by COST (European Cooperation in Science and Technology), and by core funding of the Cyprus Institute of Neurology & Genetics. Reagent and equipment support by the project “New Infrastructure for the Diagnosis and Therapy of Patients” was funded by the Norway Grants 2014–2021. We are grateful to Thessalia Papasavva for advice on NGS analyses and to all of our patients for generously donating their samples and for making our work possible. The graphical abstract was in part created using BioRender.com.
Author contributions
Conceptualization, formal analysis, data curation, and visualization, B.N., P.P., and C.W.L.; investigation and methodology, B.N., P.L.P., N.Y.P., M.T., L.K., X.F., C.G.C., P.P., and C.W.L.; resources, M.S., S.C., M.K., P.P., and C.W.L.; supervision, M.K., P.P., and C.W.L.; writing – original draft preparation, B.N., M.T., P.P., and C.W.L.; writing – review & editing, B.N., P.L.P., M.T., P.P., and C.W.L.; project administration, M.K., P.P., and C.W.L.; funding acquisition: M.K., P.P., and C.W.L. All authors have read and agreed to the published version of the manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2024.102183.
Supplemental information
References
- 1.Origa R. β-Thalassemia. Genet. Med. 2017;19:609–619. doi: 10.1038/gim.2016.173. [DOI] [PubMed] [Google Scholar]
- 2.Kountouris P., Lederer C.W., Fanis P., Feleki X., Old J., Kleanthous M. IthaGenes: An interactive database for haemoglobin variations and epidemiology. PLoS One. 2014;9 doi: 10.1371/journal.pone.0103020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Busslinger M., Moschonas N., Flavell R.A. β+ Thalassemia: Aberrant splicing results from a single point mutation in an intron. Cell. 1981;27:289–298. doi: 10.1016/0092-8674(81)90412-8. [DOI] [PubMed] [Google Scholar]
- 4.Patsali P., Papasavva P., Christou S., Sitarou M., Antoniou M.N., Lederer C.W., Kleanthous M. Relative and absolute quantification of aberrant and normal splice variants in hbbivsi−110 (G > a) β-thalassemia. Int. J. Mol. Sci. 2020;21:1–17. doi: 10.3390/ijms21186671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Angelucci E. Hematopoietic stem cell transplantation in thalassemia. Hematology (United States) 2010;2010:456–462. doi: 10.1182/asheducation-2010.1.456. [DOI] [PubMed] [Google Scholar]
- 6.Patsali P., Papasavva P., Stephanou C., Christou S., Sitarou M., Antoniou M.N., Lederer C.W., Kleanthous M. Short-hairpin RNA against aberrant HBBIVSI-110(G>A) mRNA restores β-globin levels in a novel cell model and acts as mono- and combination therapy for β-thalassemia in primary hematopoietic stem cells. Haematologica. 2018;103:e419–e423. doi: 10.3324/haematol.2018.189357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Patsali P., Mussolino C., Ladas P., Floga A., Kolnagou A., Christou S., Sitarou M., Antoniou M.N., Cathomen T., Lederer C.W., Kleanthous M. The Scope for Thalassemia Gene Therapy by Disruption of Aberrant Regulatory Elements. J. Clin. Med. 2019;8 doi: 10.3390/jcm8111959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patsali P., Turchiano G., Papasavva P., Romito M., Loucari C.C., Stephanou C., Christou S., Sitarou M., Mussolino C., Cornu T.I., et al. Correction of IVS I-110(G>A) β-thalassemia by CRISPR/Cas- And TALEN-mediated disruption of aberrant regulatory elements in human hematopoietic stem and progenitor cells. Haematologica. 2019;104:E497–E501. doi: 10.3324/haematol.2018.215178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.El-Beshlawy A., Mostafa A., Youssry I., Gabr H., Mansour I.M., El-Tablawy M., Aziz M., Hussein I.R. Correction of aberrant pre-mRNA splicing by antisense oligonucleotides in β-thalassemia Egyptian patients with IVSI-110 mutation. J. Pediatr. Hematol. Oncol. 2008;30:281–284. doi: 10.1097/MPH.0b013e3181639afe. [DOI] [PubMed] [Google Scholar]
- 10.Hardouin G., Antoniou P., Martinucci P., Felix T., Manceau S., Joseph L., Masson C., Scaramuzza S., Ferrari G., Cavazzana M., Miccio A. Adenine base editor–mediated correction of the common and severe IVS1-110 (G>A) β-thalassemia mutation. Blood. 2023;141:1169–1179. doi: 10.1182/blood.2022016629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu Y., Zeng J., Roscoe B.P., Liu P., Yao Q., Lazzarotto C.R., Clement K., Cole M.A., Luk K., Baricordi C., et al. Highly efficient therapeutic gene editing of human hematopoietic stem cells. Nat. Med. 2019;25:776–783. doi: 10.1038/s41591-019-0401-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Puthenveetil G., Scholes J., Carbonell D., Qureshi N., Xia P., Zeng L., Li S., Yu Y., Hiti A.L., Yee J.K., Malik P. Successful correction of the human β-thalassemia major phenotype using a lentiviral vector. Blood. 2004;104:3445–3453. doi: 10.1182/blood-2004-04-1427. [DOI] [PubMed] [Google Scholar]
- 13.Papasavva P., Kleanthous M., Lederer C.W. Rare Opportunities: CRISPR/Cas-Based Therapy Development for Rare Genetic Diseases. Mol. Diagn. Ther. 2019;23:201–222. doi: 10.1007/s40291-019-00392-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fu Y., Foden J.A., Khayter C., Maeder M.L., Reyon D., Joung J.K., Sander J.D. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walton R.T., Christie K.A., Whittaker M.N., Kleinstiver B.P. Unconstrained genome targeting with near-PAMless engineered CRISPR-Cas9 variants. Science. 2020;368:290–296. doi: 10.1126/science.aba8853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rees H.A., Liu D.R. Base editing: precision chemistry on the genome and transcriptome of living cells. Nat. Rev. Genet. 2018;19:770–788. doi: 10.1038/s41576-018-0059-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Youn H., Chung J.K. Modified mRNA as an alternative to plasmid DNA (pDNA) for transcript replacement and vaccination therapy. Expet Opin. Biol. Ther. 2015;15:1337–1348. doi: 10.1517/14712598.2015.1057563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Papaioannou N.Y., Patsali P., Naiisseh B., Papasavva P.L., Koniali L., Kurita R., Nakamura Y., Christou S., Sitarou M., Mussolino C., et al. High-efficiency editing in hematopoietic stem cells and the HUDEP-2 cell line based on in vitro mRNA synthesis. Front. Genome Ed. 2023;5 doi: 10.3389/fgeed.2023.1141618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kluesner M.G., Nedveck D.A., Lahr W.S., Garbe J.R., Abrahante J.E., Webber B.R., Moriarity B.S. EditR: A Method to Quantify Base Editing from Sanger Sequencing. CRISPR J. 2018;1:239–250. doi: 10.1089/crispr.2018.0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Concordet J.P., Haeussler M. CRISPOR: Intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Res. 2018;46:W242–W245. doi: 10.1093/nar/gky354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Doench J.G., Fusi N., Sullender M., Hegde M., Vaimberg E.W., Donovan K.F., Smith I., Tothova Z., Wilen C., Orchard R., et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9 Synthesis of an arrayed sgRNA library targeting the human genome. Nat. Biotechnol. 2016;34:184–191. doi: 10.1038/nbt.3437.Optimized. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qi W.-H., Yan C.-C., Li W.-J., Jiang X.-M., Li G.-Z., Zhang X.-Y., Hu T.-Z., Li J., Yue B.-S. Distinct patterns of simple sequence repeats and GC distribution in intragenic and intergenic regions of primate genomes. Aging (Albany NY) 2016;8:2635–2654. doi: 10.18632/aging.101025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Patel C.A., Ghiselli G. Hinderin, a five-domains protein including coiled-coil motifs that binds to SMC3. BMC Cell Biol. 2005;6:3. doi: 10.1186/1471-2121-6-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hsu P.D., Scott D.A., Weinstein J.A., Ran F.A., Konermann S., Agarwala V., Li Y., Fine E.J., Wu X., Shalem O., et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moreno-Mateos M.A., Vejnar C.E., Beaudoin J.D., Fernandez J.P., Mis E.K., Khokha M.K., Giraldez A.J. CRISPRscan: Designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Nat. Methods. 2015;12:982–988. doi: 10.1038/nmeth.3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loucari C.C., Patsali P., Van Dijk T.B., Stephanou C., Papasavva P., Zanti M., Kurita R., Nakamura Y., Christou S., Sitarou M., et al. Rapid and Sensitive Assessment of Globin Chains for Gene and Cell Therapy of Hemoglobinopathies. Hum. Gene Ther. Methods. 2018;29:60–74. doi: 10.1089/hgtb.2017.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Turchiano G., Andrieux G., Klermund J., Blattner G., Pennucci V., el Gaz M., Monaco G., Poddar S., Mussolino C., Cornu T.I., et al. Quantitative evaluation of chromosomal rearrangements in gene-edited human stem cells by CAST-Seq. Cell Stem Cell. 2021;28:1136–1147.e5. doi: 10.1016/j.stem.2021.02.002. [DOI] [PubMed] [Google Scholar]
- 28.Frangoul H., Altshuler D., Cappellini M.D., Chen Y.-S., Domm J., Eustace B.K., Foell J., de la Fuente J., Grupp S., Handgretinger R., et al. CRISPR-Cas9 Gene Editing for Sickle Cell Disease and β-Thalassemia. N. Engl. J. Med. 2021;384:252–260. doi: 10.1056/nejmoa2031054. [DOI] [PubMed] [Google Scholar]
- 29.Xu S., Luk K., Yao Q., Shen A.H., Zeng J., Wu Y., Luo H.Y., Brendel C., Pinello L., Chui D.H.K., et al. Editing aberrant splice sites efficiently restores β-globin expression in b-thalassemia. Blood. 2019;133:2255–2262. doi: 10.1182/blood-2019-01-895094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pineda J.M.B., Bradley R.K. Most human introns are recognized via multiple and tissue-specific branchpoints. Genes Dev. 2018;32:577–591. doi: 10.1101/gad.312058.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reed R., Maniatis T. The role of the mammalian branchpoint sequence in pre-mRNA splicing. Genes Dev. 1988;2:1268–1276. doi: 10.1101/gad.2.10.1268. [DOI] [PubMed] [Google Scholar]
- 32.Cretu C., Gee P., Liu X., Agrawal A., Nguyen T.V., Ghosh A.K., Cook A., Jurica M., Larsen N.A., Pena V. Structural basis of intron selection by U2 snRNP in the presence of covalent inhibitors. Nat. Commun. 2021;12:4491. doi: 10.1038/s41467-021-24741-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goo Y.H., Cooper T.A. CUGBP2 directly interacts with U2 17S snRNP components and promotes U2 snRNA binding to cardiac troponin T pre-mRNA. Nucleic Acids Res. 2009;37:4275–4286. doi: 10.1093/nar/gkp346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gromak N., Matlin A.J., Cooper T.A., Smith C.W.J. Antagonistic regulation of α-actinin alternative splicing by CELF proteins and polypyrimidine tract binding protein. RΝΑ. 2003;9:443–456. doi: 10.1261/rna.2191903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jensen K.B., Musunuru K., Lewis H.A., Burley S.K., Darnell R.B. The tetranucleotide UCAY directs the specific recognition of RNA by the Nova K-homology 3 domain. Proc. Natl. Acad. Sci. USA. 2000;97:5740–5745. doi: 10.1073/pnas.090553997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hardison R.C. Promoter competition in globin gene control. Blood. 2022;139:2089–2091. doi: 10.1182/blood.2022015642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rees D.C., Porter J.B., Clegg J.B., Weatherall D.J. Why are hemoglobin F levels increased in HbE/beta thalassemia? Blood. 1999;94:3199–3204. [PubMed] [Google Scholar]
- 38.Efremov D.G., Dimovski A.J., Sukarova E., Schilirò G., Zisovski N., Efremov G.D., Burrone O.R., Huisman T.H. Gamma-mRNA and Hb F levels in beta-thalassaemia. Br. J. Haematol. 1994;88:311–317. doi: 10.1111/j.1365-2141.1994.tb05024.x. [DOI] [PubMed] [Google Scholar]
- 39.Qiu C.C., Abraham E.C. Evidence for posttranslational control of fetal hemoglobin synthesis. Am. J. Hematol. 1988;29:58–59. doi: 10.1002/ajh.2830290116. [DOI] [PubMed] [Google Scholar]
- 40.Han W., Qiu H.-Y., Sun S., Fu Z.-C., Wang G.-Q., Qian X., Wang L., Zhai X., Wei J., Wang Y., et al. Base editing of the HBG promoter induces potent fetal hemoglobin expression with no detectable off-target mutations in human HSCs. Cell Stem Cell. 2023;30:1624–1639.e8. doi: 10.1016/j.stem.2023.10.007. [DOI] [PubMed] [Google Scholar]
- 41.Green M.R., Sambrook J. Cold Spring Harbor Laboratory Press, Cold Spring Harbor; 2012. Molecular Cloning: A Laboratory Manual Fourth. [Google Scholar]
- 42.Ou C.N., Buffone G.J., Reimer G.L., Alpert A.J. High-performance liquid chromatography of human hemoglobins on a new cation exchanger. J. Chromatogr. 1983;266:197–205. doi: 10.1016/s0021-9673(01)90893-3. [DOI] [PubMed] [Google Scholar]
- 43.Desmet F.-O., Hamroun D., Lalande M., Collod-Béroud G., Claustres M., Béroud C. Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009;37:e67. doi: 10.1093/nar/gkp215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ihaka R., Gentleman R. R: A Language for Data Analysis and Graphics. J. Comput. Graph Stat. 1996;5:299–314. doi: 10.2307/1390807. [DOI] [Google Scholar]
- 45.Garcia M., Juhos S., Larsson M., Olason P.I., Martin M., Eisfeldt J., DiLorenzo S., Sandgren J., Díaz De Ståhl T., Ewels P., et al. Sarek: A portable workflow for whole-genome sequencing analysis of germline and somatic variants. F1000Res. 2020;9:63. doi: 10.12688/f1000research.16665.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv. 2013 doi: 10.48550/arXiv.1303.3997. Preprint at. [DOI] [Google Scholar]
- 47.Danecek P., Bonfield J.K., Liddle J., Marshall J., Ohan V., Pollard M.O., Whitwham A., Keane T., McCarthy S.A., Davies R.M., Li H. Twelve years of SAMtools and BCFtools. GigaScience. 2021;10 doi: 10.1093/gigascience/giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morgan M. An Introduction to Rsamtools. Ind Commer Train. 2011;10:11–18. [Google Scholar]
- 49.Wickham H., Averick M., Bryan J., Chang W., McGowan L., François R., Grolemund G., Hayes A., Henry L., Hester J., et al. Welcome to the Tidyverse. J. Open Source Softw. 2019;4:1686. doi: 10.21105/joss.01686. [DOI] [Google Scholar]
- 50.Bengtsson H., Ahlmann-Eltze C., Bravo H.C., Gentleman R., Gleixner J., Hickey P., Hossjer O., Jaffee H., Jiang D., Langfelder P., et al. matrixStats: Functions that Apply to Rows and Columns of Matrices (and to Vectors) 2023. https://github.com/HenrikBengtsson/matrixStats Version 1.2.0.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available in the article. Correspondence and requests for materials should be addressed to C.W.L. Reprints and permissions information is available.